
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 30 November 2012
Sec. Molecular Bacterial Pathogenesis
Volume 2 - 2012 | https://doi.org/10.3389/fcimb.2012.00151
This article is part of the Research Topic The pathogenic Yersiniae – advances in the understanding of physiology and virulence View all 17 articles
Low molecular weight siderophores are used by many living organisms to scavenge scarcely available ferric iron. Presence of at least a single siderophore-based iron acquisition system is usually acknowledged as a virulence-associated trait and a pre-requisite to become an efficient and successful pathogen. Currently, it is assumed that yersiniabactin (Ybt) is the solely functional endogenous siderophore iron uptake system in highly virulent Yersinia (Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica biotype 1B). Genes responsible for biosynthesis, transport, and regulation of the yersiniabactin (ybt) production are clustered on a mobile genetic element, the High-Pathogenicity Island (HPI) that is responsible for broad dissemination of the ybt genes in Enterobacteriaceae. However, the ybt gene cluster is absent from nearly half of Y. pseudotuberculosis O3 isolates and epidemic Y. pseudotuberculosis O1 isolates responsible for the Far East Scarlet-like Fever. Several potential siderophore-mediated iron uptake gene clusters are documented in Yersinia genomes, however, neither of them have been proven to be functional. It has been suggested that at least two siderophores alternative to Ybt may operate in the highly virulent Yersinia pestis/Y. pseudotuberculosis group, and are referred to as pseudochelin (Pch) and yersiniachelin (Ych). Furthermore, most sporadic Y. pseudotuberculosis O1 strains possess gene clusters encoding all three iron scavenging systems. Thus, the Ybt system appears not to be the sole endogenous siderophore iron uptake system in the highly virulent yersiniae and may be efficiently substituted and/or supplemented by alternative iron siderophore scavenging systems.
Iron is one of the abundant elements on Earth, but it is hardly available to bacteria under aerobic conditions (10−17 M solubility limit for Fe(III) at pH ~7) and/or in mammalian hosts (10−26 M) (Otto et al., 1991). To obtain iron, bacteria have developed different strategies including one that utilizes low molecular weight siderophore molecules with high affinity to Fe3+ ions.
The human pathogenic Yersinia (Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica) are ideal model organisms to study the impact of siderophore-mediated iron uptake systems on virulence as well as on the overall bacterial fitness and physiology. The pathogenic Yersinia can be divided into three main pathogroups, with Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica biogroup 1B making up the highly virulent group (strains able to kill mice at low infection doses); low pathogenic Y. enterocolitica biogroups 2–5, and normally apathogenic Y. enterocolitica biogroup 1A (Carter, 1975; Heesemann, 1987). This grouping is based on the ability of bacteria of the high-pathogenic group to express an efficient siderophore yersiniabactin (Ybt) that plays a significant role in iron acquisition and murine pathogenicity (Carniel et al., 1987; Heesemann, 1987; Rakin et al., 1994). In contrast, two other pathogroups (low and apathogenic) do not produce Ybt.
Several other gene clusters with similarities to iron acquisition genes were uncovered by whole genome sequencing in Y. pestis/Y. pseudotuberculosis genomes, but their implication in iron uptake remains enigmatic (Forman et al., 2010). Therefore, Ybt is currently believed to be the exclusive endogenous siderophore of the highly-pathogenic Yersinia.
The Ybt-siderophore system of Yersinia has been extensively studied during the last 20 years (for a recent review, see Perry and Fetherston, 2011). The structure of Ybt was determined and shown to contain phenolate, thiazoline, and thiazolidine rings and depicts high similarity to pyochelin produced by Pseudomonas aeruginosa and anquibactin of Vibrio anguillarum. The Ybt molecule is assembled by a typical strategy in which non-ribosomal peptide synthetase (NRPS) and polyketide synthetase (PKS) mediate biosynthesis. The strategy follows modular assembly of the siderophore from salicylate, a group from malonyl coenzyme A, three cysteine molecules and three methyl groups (Gehring et al., 1998). Six genes are involved in the Ybt synthesis and are designated irp1—irp5, irp9 in Y. enterocolitica, and irp1–2, ybtU, T, E, S—in Y. pestis and Y. pseudotuberculosis (Figure 1). Irp9 (YbtS) directly converts chorismate into salicylate, the precursor of Ybt (Pelludat et al., 2003). Irp5 (YbtE) salicyl-AMP ligase transfers the activated salicylate to HMWP2 (encoded by irp2). HMWP2 possesses six predicted NRPS domains involved in initial cyclization and condensation reactions. Irp3 (YbtU) reduces the internal thiazoline ring to a thiozolidine structure while the first five domains of HMWP1 (encoded by irp1) switch from NRPS-type assembly line molecules to a PKS-strategy. Irp4 (YbtT) contains a thioesterase domain to remove aberrant structures from the enzymatic complex and displays an editing function together with terminal HMWP1 domains. The mechanism of Ybt secretion into the environment is currently unknown. Irp8 (YbtX in Y. pestis) was supposed to be involved in Ybt export (Fetherston et al., 1999), however, YbtX− mutants are still able to secrete Ybt, so the function of this protein is still enigmatic. Fe-Ybt assimilation occurs through the outer membrane receptor FyuA (Psn in Y. pestis) and the inner cell membrane proteins Irp6 and Irp7 (YbtQ and YbtP in Y. pestis) (Fetherston et al., 1999; Brem et al., 2001; Perry and Fetherston, 2011). A periplasmic ferric Ybt transport protein has not been identified till now.
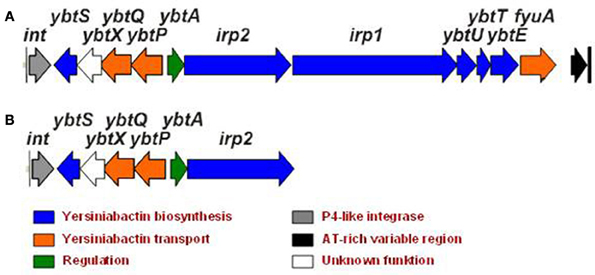
Figure 1. Yersiniabactin gene cluster in Y. pseudotuberculosis O1 (A) and its truncated form in Y. pseudotuberculosis O3 (B).
The biosynthetic and transport genes of Ybt are located on the High-Pathogenicity Island (HPI), a genomic mobile element that exhibits site-specific recombination activity of the P4-like integrase (Rakin et al., 1999; Carniel, 2001). The location of the Ybt-cluster on the mobile element accounts for its wide distribution in Enterobacteriaceae (Schubert et al., 2004; Antonenka et al., 2005).
Expression of the HPI genes is tightly regulated by host- and HPI-encoded factors. The Ybt biosynthetic and transport genes are repressed by iron loaded ferric uptake regulator (Fur)-protein and activated by the AraC-type YbtA transcriptional activator that also represses its own transcription (Fetherston et al., 1999; Anisimov et al., 2005). Besides, the functionality of the Ybt system is activated at the post-transcriptional level by P-pant transferase (YbtD, located outside the HPI) necessary for phosphopantetheinylation of NRP/PK- synthetases (Bobrov et al., 2002).
Inactivation of the Ybt biosynthetic and/or transport genes results in significant virulence attenuation of Yersinia (de Almeida et al., 1993; Heesemann et al., 1993; Rakin et al., 1994; Bearden et al., 1997; Pelludat et al., 1998; Brem et al., 2001). Ybt is the only known siderophore in Y. enterocolitica1B and its inactivation results in loss of the siderophore activity on chrome azurol S indicator plates (CAS-agar, used to detect a siderophore activity) (Schwyn and Neilands, 1987) and virulence attenuation. Also, the absence of Ybt in Y. pestis strains leads to the inability of bacteria to cause lethal infection by peripheral routes (Fetherston et al., 2010). The high significance of Ybt for establishing infection is supported by the comparison of the ybt genes expression in LB media and in the in vivo growth conditions. The ybt genes were up-regulated in the rat bubo after the subcutaneous Y. pestis inoculation (Sebbane et al., 2006) and in the lungs of mice after intranasal inoculation (Lathem et al., 2005; Liu et al., 2009). Interestingly, the ybt genes were not up-regulated in the mice spleen and liver in the pneumonic model of infection (Liu et al., 2009) and were down-regulated in the flea (Vadyvaloo et al., 2007, 2010). The increased expression of the ybt genes in mammalian hosts supports the importance of the Ybt siderophore for the Yersinia pathogenesis. Moreover, Ybt demonstrates a multifunctional character beyond iron binding and acquisition by its ability to protect bacteria from copper toxicity in uropathogenic Escherichia coli (Chaturvedi et al., 2012).
However, Y. pseudotuberculosis YPIII O3 type strain, widely used in animal experiments to address yersiniae virulence (Rosqvist and Wolf-Watz, 1986), completely lacks the Ybt iron acquisition system but demonstrates efficient iron binding on the CAS agar. In contrast, another group of the Y. pseudotuberculosis O3 strains possesses only a truncated form of the ybt gene cluster (Figure 1B) and shows no activity on the CAS agar. These strains also have lower virulence in animal models (Fukushima, 2003).
Furthermore, complete genome examination of epidemic Y. pseudotuberculosis IP31758 O1, an isolate that belongs to the group of strains responsible for the Far East Scarlet-like Fever (FESLF) in the Far East Russia and Japan (Gurleva et al., 1973; Eppinger et al., 2007), reveals the absence of the ybt gene cluster. Nevertheless, these strains demonstrate CAS activity and high virulence in humans and animals. Also the comparison of the genomes of IP32953 sporadic and IP31758 epidemic FESLF Y. pseudotuberculosis O1 strains favors the absence of the ybt genes from the epidemic strain. While the sporadic Y. pseudotuberculosis O1 strains contain a cluster identical to the Y. pestis ybt, the nosocomial “outbreak” FESLF strain, is ybt-deficient.
These results allude to the possibility that siderophore(s) alternative to Ybt are active in Y. pestis/Y. pseudotuberculosis highly virulent group.
A potential siderophore producing locus with a high similarity of gene sequences and organization to the ybt gene cluster was discovered in all sequenced strains of Y. pestis and Y. pseudotuberculosis but not in Y. enterocolitica and has been designated the Yersinia non-ribosomal peptide (ynp) (Perry and Fetherston, 2004; Forman et al., 2010). Because of these similarities, this cluster was initially annotated as a second HPI in Y. pestis CO92 (Parkhill et al., 2001). However, it lacks any elements of a mobile pathogenicity island such as the presence of an integrase encoding gene or recombination sites.
In Y. pseudotuberculosis IP32953 the ynp locus (YPTB3290-3298) (Figure 2) contains putative NRPS and PKS siderophore assembly genes (YPTB3296-3297), ferri-siderophore transport genes (YPTB3290-3291), and a gene coding for the TonB-dependent outer membrane ferri-siderophore receptor (YPTB3298). Although the ynp locus shares many similarities with the ybt locus, the ynp locus appears to lack a salicylate synthase (ybtS) equivalent, suggesting that the putative siderophore may not use salicylate as a biosynthetic precursor. Based on the enzymatic domains encoded within the YPTB3296 and YPTB3297 genes, the structure of the putative Ynp siderophore was predicted to contain three thiazoline rings and have a molecular mass of 404.4/460.37 m/z with and without Fe3+, respectively (Forman et al., 2010).
Differential distribution of the ybt and ynp clusters in Y. pseudotuberculosis O1 is evidenced by the whole genome comparison of the epidemic Y. pseudotuberculosis IP31758 responsible for manifestation of FESLF and the typical “sporadic” Y. pseudotuberculosis IP32953 strains. While the latter has both clusters in its genome, the Far East strain lacks the ybt cluster and contains a frameshift mutation in the N-terminal region of the biosynthetic gene (YPTB3297). Nevertheless, it still demonstrates the CAS-positive phenotype suggesting that either some additional siderophore(s) mediate this activity or the truncated protein retains its function.
In contrast to the “sporadic” Y. pseudotuberculosis strains, the epidemic Y. pestis strains contain at least two mutations in the ynp locus. The NRPS (YPTB3297) is split by an IS100 mobile element into two separate parts and contains a frameshift mutation in the C-terminal region (Forman et al., 2010). In the Microtus endemic Y. pestis 91001 strain, the YPTB3297 gene does not carry the IS100 insertion, but still contains the frameshift mutation, as noted in the epidemic Y. pestis strains. In Y. pestis CO92 the ynp cluster is present in two separate chromosomal loci where YPO1011-1012 codes for the receptor (YPTB3298) and N-terminal part of YPTB3297 while YPO0770-0778 contains the remaining genes of the cluster (YPTB3290-3296). These observations have put the functionality of the ynp locus in Y.pestis into question (Forman et al., 2010). Thus, the ynp locus is likely specific to the sporadic Y. pseudotuberculosis strains and we propose that the encoded siderophore be called pseudochelin (Pch).
Classical models use iron limiting conditions in vitro to induce the expression of iron uptake systems and to identify iron regulated genes. The expression of the ynp genes has been supported by both in vivo and in vitro studies using microarray and quantitative real time PCR. Since a Fur-like binding region is located 184 bp upstream of the putative ynp receptor transcriptional start site in Y. pestis, expression of this locus is likely Fur-dependent and activated under iron limited conditions (Han et al., 2007; Gao et al., 2008). Several studies indicate that simulation of low iron concentrations in vitro results in up-regulation of the putative ynp receptor gene (YPTB3298) in Y. pestis and Y. pseudotuberculosis (Han et al., 2007; Gao et al., 2008; Rosso et al., 2008). When Y. pestis was grown in human plasma the putative ynp receptor and the biosynthetic gene (YPTB3297) were up-regulated relative to growth in LB media (Chauvaux et al., 2007). The Y. pestis murine pneumonic infection model showed nearly equal expression levels for the fyuA and ynp putative siderophore receptor genes 48 h post-intranasal infection providing evidence that pseudochelin is an early infection virulence factor (Lathem et al., 2005). Expression analysis in the rat bubonic infection model complements these findings by confirming up-regulation of the receptor and biosynthetic genes (YPTB3298 and YPTB3296) in vivo as compared to the expression levels found in LB media (Sebbane et al., 2006; Vadyvaloo et al., 2010). A proteomic analysis revealed that the putative Ynp receptor (homolog of YPTB3298) was present at higher concentrations in the outer membrane extracts of Y. pestis KIM6 at 26°C vs. 37°C when cells were grown in complete PMH2 media (Pieper et al., 2009).
Due to their many similarities, co-operation and crosstalk between the ynp and ybt gene clusters has been hypothesized. Since, the ynp operon lacks P-pant, a necessary transferase component used to commence the NRPS/PKS system, it has been suggested that the Ybt siderophore assembly machinery, namely YbtD, could compensate by performing this function (Forman et al., 2010). It has also been proposed that the Y. pestis KIM6 Ybt-negative strain produces Ybt-like molecules that are present in culture supernatants and are capable of ybt transcriptional activation (Miller et al., 2010). Since this data suggest that compounds produced outside of the ybt gene cluster can activate the ybt genes, yersiniabactin-like molecules are present in the media and the predicted structure of the Pch siderophore shares similarities with Ybt, it is likely that the products of the ynp locus participate in this crosstalk. Whether the aberrant Ybt-like molecules are produced by the ynp operon, has not yet been experimentally established, however, this evidence provides insight into their ability to influence other siderophore producing systems (Miller et al., 2010).
Another Yersinia siderophore was found as a component of the autoagglutination factor (AF) purified from the surface of the Ybt-negative strain Y. pestis EV76 (Podladchikova and Rykova, 2006). AF appeared to be a complex antigen composed of the 17,485-kDa protein and an iron-loaded low molecular weight component that demonstrated siderophore activity. The AF protein was identified as Ypo0502 (Podladchikova et al., 2011), the Hcp (hemolysin coregulated protein)-like component of an extracellular apparatus of the type six secretion system (T6SS) recently characterized in gram-negative bacteria (for review, see Records, 2011). Hcp monomers are known to self-assemble on the cell surface into pilus-like filaments through which T6SS-associated proteins and effectors are transported. The physiological consequence of the siderophore interaction with the Hcp-like protein on the Y. pestis cell surface awaits further investigation.
The AF-derived siderophore was considered to be a novel Yersinia siderophore (Podladchikova et al., 2012), as it was purified from the strain (Y.pestis EV76), which has no genes coding for Ybt synthesis. Chemical analysis of the novel siderophore, designated yersiniachelin (Ych), revealed that it contained hydroxamate groups. In available genome sequences of Y. pestis, two gene clusters are able to code for hydroxamate siderophores. The first, homologous to the aerobactin siderophore cluster, carries several mutations in biosynthetic genes and was shown to be non-functional in Y. pestis (Forman et al., 2007). The other cluster (YPO1528-1538 in Y. pestis CO92) designated ysu (Yersinia siderophore uptake), which contains hydroxamate siderophore synthesis and transport genes, has no visible mutations and thus is predicted to encode for the biosynthesis of a functional siderophore (Perry and Fetherston, 2004; Forman et al., 2010). According to bioinformatic data the ysu-cluster is present in all sequenced Y. pestis and Y. pseudotuberculosis strains but not in Y. enterocolitica.
The involvement of the ysu locus in Ych production was confirmed by the analysis of a Y. pestis EV76 knock-out mutant in which three biosynthetic genes of the putative hydroxamate siderophore (ysu IHG) were deleted (Podladchikova et al., 2012). In contrast to the parent strain, the mutant did not produce Ych. These results provide evidence that Ych is encoded by the ysu locus, so we propose that ysu may stand for yersiniachelin synthesis and utilization. The ysu-cluster is similar to the alc-cluster (Figure 3) coding for the hydroxamate siderophore alcaligin (Alc) produced by Bordetella spp and important for the multiplication in vivo and virulence of bacteria (Brickman et al., 2008, 2011; Brickman and Armstrong, 2009). The Alc siderophore is well-studied and known to be assembled from diamine precursors by a process-independent of the NRPS-pathway and performed by a number of enzymes encoded by the alc-cluster on the Bordetella chromosome. The Alc biosynthesis begins with ornithine decarboxylation by Odc (encoded outside the alc-cluster) followed by a N-hydroxylation by AlcA and N-acetylation of the hydroxylamine group by AlcB. Then the product is C-hydroxylated by AlcE and undergoes dimerization and macrocyclization performed by AlcC.
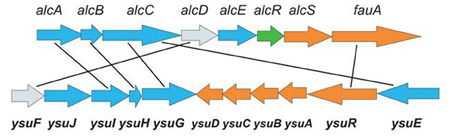
Figure 3. Comparison of the alc-cluster encoding the production of the alcaligin siderophore in Bordetella spp with the ysu-cluster encoding the production of the yersiniachelin siderophore in Y. pestis. Siderophore biosynthetic genes are shown in blue, transport genes—in orange, regulatory gene—in green, and ferric reductase genes with unknown function—in grey.
Comparison of the ysu and alc clusters shows similarities in DNA and protein sequences of a number of genes and differences in genetic organization and content. The homology is obvious in the four siderophore biosynthetic genes (ysuGHIJE and odc, alcABC), the ferri-siderophore outer membrane receptors (ysuR and fauA), as well as the ferric iron reductase genes (ysuF and alcD) whose role in Alc siderophore function is currently unknown. Though homologous in the siderophore biosynthetic and receptor genes, the two clusters have several apparent differences. First, a homolog of the alcE biosynthetic gene coding for an iron-sulfur protein involved in C-hydroxylation of precursor molecules during Alc biosynthesis, is absent from the ysu-cluster and Y. pestis genome. This suggests that the structure of Ych may be slightly different from Alc. Nevertheless, due to the obvious homology of the four siderophore biosynthetic genes several possible structures of the ysu-encoded putative macrocyclic hydroxamate siderophore have recently been proposed (Forman et al., 2010). Second, in the ysu-cluster there is no homolog of the alcS gene which codes for the major facilitator superfamily (MFS)-like protein shown to be involved in the Alc siderophore efflux by Bordetella cells (Brickman and Armstrong, 2005). Currently the mechanism of Ych export from bacteria is obscure. Third, the ysu-cluster carries genes (ysuABCD) coding for ferri-siderophore transporters through the periplasm and cytoplasmic membrane which are absent from the alc cluster. This step of the siderophore-based iron acquisition in Bordetella has been suggested to be performed by an alternative mechanism (Brickman et al., 2011). The FbpA ABC-transporter, (a periplasmic iron-binding protein encoded outside the alc cluster) was shown to transport iron liberated in the periplasm from multiple structurally distinct siderophores through the cytoplasmic membrane. Fourth, the ysu-cluster lacks the alcR homolog coding for the AraC-type transcriptional regulator AlcR which, in complex with ferri-Alc, activates the expression of the siderophore biosynthesis and transport genes by Bordetella (Brickman and Armstrong, 2009).
The expression of the ysu genes by Y. pestis is supported by multiple microarray studies which have been applied to various in vitro growth conditions, ex vivo growth in human plasma and macrophages, as well as in vivo animal infection models. The transcriptomic data on different Y. pestis strains grown in vitro indicate that the ysu genes are transcribed in culture broth independent of temperature (Han et al., 2004; Motin et al., 2004) and are repressed in the presence of iron by the Fur protein and up-regulated in iron-deficient conditions (Zhou et al., 2006; Gao et al., 2008). Comparative transcriptome analysis of Y. pestis in different in vitro conditions (Han et al., 2005, 2007) revealed additional signals sensed by the ysu-locus genes which were upregulated during hyperosmotic and high-salinity stress in OmpR-dependent manner. This data infers that OmpR might play the role of a transcriptional activator for ysu gene expression, the function which is performed by AlcR for the alc genes expression.
The ex vivo transcriptome analysis of Y. pestis and Y. pseudotuberculosis grown in human plasma (Chauvaux et al., 2007; Rosso et al., 2008) showed that the ysu genes were up-regulated in plasma in both species. In contrast, no difference in expression was observed in Ybt-negative Y. pestis KIM5Δpgm inside J774.1 macrophage-like cells (Fukuto et al., 2010). During intracellular multiplication a number of Y. pestis genes involved in iron acquisition (hmu, yfe, and yiu) were down-regulated suggesting that in macrophages Y. pestis is not starved for iron.
The ysu genes were not expressed in vivo in the flea vector (Vadyvaloo et al., 2007, 2010) and the level of their expression in mice lungs, spleen and liver during primary pneumonic plague did not differ from the level detected in LB medium (Lathem et al., 2005; Liu et al., 2009). However, the ysu, ybt and yfe genes were upregulated in the bubo of s.c. infected rats (Sebbane et al., 2006). This result is consistent with the observation that the ysu genes are induced not only by the iron starvation but also by the high osmolarity signal and point to a possible involvement of Ych in the initial stages of infection after s.c. challenge.
The production of the ysu encoded proteins by Y. pestis was confirmed by proteomic analysis (Pieper et al., 2008, 2009). Two ysu encoded proteins, the biosynthetic protein YsuI (AlcA homolog) and siderophore receptor YsuR (FauA homolog), were identified in the membrane proteome of Y. pestis KIM6 grown to stationary phase at 26°C in chemically defined PMH2 medium. The YsuR protein associated with the outer membrane was increased in abundance in iron-deficient conditions (Pieper et al., 2010). Moreover, in iron-starved cells the amount of the siderophore biosynthetic protein YsuG (AlcC homolog) was also increased in the periplasmic cell fraction. Thus, the last steps of the Ych synthesis might take place in the periplasm after the transport of the siderophore precursor through the cytoplasmic membrane.
Thus, in highly virulent Yersinia the expression of the third siderophore system encoded by the ysu locus is substantiated by the mutagenesis of the ysu genes, transcriptomic and proteomic data, as well as by the purification of the Ych siderophore from the ybt-negative Y. pestis strain. The up-regulation of the ysu genes in iron-deplete conditions in vitro point to the possible involvement of Ych in iron acquisition. However, the exposure of Ych associated with the Hcp-like protein YPO502 on the bacterial surface in iron-replete conditions and the up-regulation of its biosynthetic genes by high osmotic and salt stress suggest an additional role for Ych in Y. pestis physiology.
Like most pathogenic bacteria, highly-virulent Yersinia pestis /Y. pseudotuberculosis possess several alternative endogenous siderophore-mediated iron acquisition systems. A complete functional understanding of these novel siderophore systems is currently lacking in the literature and future studies should focus on parsing out the impact these systems have on bacterial physiology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anisimov, R., Brem, D., Heesemann, J., and Rakin, A. (2005). Transcriptional regulation of high pathogenicity island iron uptake genes by YbtA. Int. J. Med. Microbiol. 295, 19–28.
Antonenka, U., Nölting, C., Heesemann, J., and Rakin, A. (2005). Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4attB target presenting shuttle plasmid. Mol. Microbiol. 57, 727–734.
Bearden, S. W., Fetherston, J. D., and Perry, R. D. (1997). Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65, 1659–1668.
Bobrov, A. G., Geoffroy, V. A., and Perry, R. D. (2002). Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect. Immun. 70, 4204–4214.
Brem, D., Pelludat, C., Rakin, A., Jacobi, C. A., and Heesemann, J. (2001). Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147, 1115–1127.
Brickman, T. J., and Armstrong, S. K. (2005). Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187, 3650–3661.
Brickman, T. J., and Armstrong, S. K. (2009). Temporal signaling and differential expression of Bordetella iron transport systems: the role of ferrimones and positive regulators. Biometals 22, 33–41.
Brickman, T. J., Cummings, C. A., Liew, S. Y., Relman, D. A., and Armstrong, S. K. (2011). Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J. Bacteriol. 193, 4798–4812.
Brickman, T. J., Hanawa, T., Anderson, M. T., Suhadolc, R. J., and Armstrong, S. K. (2008). Differential expression of Bordetella pertussis iron transport system genes during infection. Mol. Microbiol. 70, 3–14.
Carniel, E. (2001). The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3, 561–569.
Carniel, E., Mazigh, D., and Mollaret, H. H. (1987). Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect. Immun. 55, 277–280.
Carter, P. B. (1975). Pathogenecity of Yersinia enterocolitica for mice. Infect. Immun. 11, 164–170.
Chaturvedi, K. S., Hung, C. S., Crowley, J. R., Stapleton, A. E., and Henderson, J. P. (2012). The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736.
Chauvaux, S., Rosso, M. L., Frangeul, L., Lacroix, C., Labarre, L., Schiavo, A., et al. (2007). Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology 153, 3112–3123.
de Almeida, A. M., Guiyoule, A., Guilvout, I., Iteman, I., Baranton, G., and Carniel, E. (1993). Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb. Pathog. 14, 9–21.
Eppinger, M., Rosovitz, M. J., Fricke, W. F., Rasko, D. A., Kokorina, G., Fayolle, C., et al. (2007). The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 3:e142. doi: 10.1371/journal.pgen.0030142
Fetherston, J. D., Bertolino, V. J., and Perry, R. D. (1999). YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32, 289–299.
Fetherston, J. D., Kirillina, O., Bobrov, A. G., Paulley, J. T., and Perry, R. D. (2010). The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun. 78, 2045–2052.
Forman, S., Nagiec, M. J., Abney, J., Perry, R. D., and Fetherston, J. D. (2007). Analysis of the aerobactin and ferric hydroxamate uptake systems of Yersinia pestis. Microbiology 153, 2332–2341.
Forman, S., Paulley, J. T., Fetherston, J. D., Cheng, Y. Q., and Perry, R. D. (2010). Yersinia ironomics: comparison of iron transporters among Yersinia pestis biotypes and its nearest neighbor, Yersinia pseudotuberculosis. Biometals 2, 275–294.
Fukushima, H. (2003). Molecular epidemiology of Yersinia pseudotuberculosis. Adv. Exp. Med. Biol. 529, 357–358.
Fukuto, H. S., Svetlanov, A., Palmer, L. E., Karzai, A. W., and Bliska, J. B. (2010). Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect. Immun. 78, 3700–3715.
Gao, H., Zhou, D., Li, Y., Guo, Z., Han, Y., Song, Y., et al. (2008). The iron-responsive fur regulon in Yersinia pestis. J. Bacteriol. 190, 3063–3075.
Gehring, A. M., DeMoll, E., Fetherston, J. D., Mori, I., Mayhew, G. F., Blattner, F. R., et al. (1998). Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5, 573–586.
Gurleva, G. G., Domaradskii, I. V., Smolikova, L. M., Khaliapina, E. E., and Grigor'ian, E. G. (1973). [Biological properties of the causative agent of pseudotuberculosis isolated from scarlatina-like fever patients]. Zh. Mikrobiol. Epidemiol. Immunobiol. 50, 125–129.
Han, Y., Qiu, J., Guo, Z., Gao, H., Song, Y., Zhou, D., et al. (2007). Comparative transcriptomics in Yersinia pestis: a global view of environmental modulation of gene expression. BMC Microbiol. 7:96. doi: 10.1186/1471-2180-7-96
Han, Y., Zhou, D., Pang, X., Song, Y., Zhang, L., Bao, J., et al. (2004). Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol. 48, 791–805.
Han, Y., Zhou, D., Pang, X., Zhang, L., Song, Y., Tong, Z., et al. (2005). Comparative transcriptome analysis of Yersinia pestis in response to hyperosmotic and high-salinity stress. Res. Microbiol. 156, 403–415.
Heesemann, J. (1987). Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol. Lett. 48, 229–233.
Heesemann, J., Hantke, K., Vocke, T., Saken, E., Rakin, A., Stojiljkovic, I., et al. (1993). Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol. Microbiol. 8, 397–408.
Lathem, W. W., Crosby, S. D., Miller, V. L., and Goldman, W. E. (2005). Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 102, 17786–17791.
Liu, H., Wang, H., Qiu, J., Wang, X., Guo, Z., Qiu, Y., et al. (2009). Transcriptional profiling of a mice plague model: insights into interaction between Yersinia pestis and its host. J. Basic Microbiol. 49, 92–99.
Miller, M. C., Fetherston, J. D., Pickett, C. L., Bobrov, A. G., Weaver, R. H., DeMoll, E., et al. (2010). Reduced synthesis of the Ybt siderophore or production of aberrant YBT-like molecules activates transcription of yersiniabactin genes in Yersinia pestis. Microbiology 156, 2226–2238.
Motin, V. L., Georgescu, A. M., Fitch, J. P., Gu, P. P., Nelson, D. O., Mabery, S. L., et al. (2004). Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186, 6298–6305.
Otto, B. R., Verweij, W. R., Sparrius, M., Verweij-van Vught, A. M., Nord, C. E., and MacLaren, D. M. (1991). Human immune response to an iron-repressible outer membrane protein of Bacteroides fragilis. Infect. Immun. 59, 2999–3003.
Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., et al. (2001). Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413, 523–527.
Pelludat, C., Brem, D., and Heesemann, J. (2003). Irp9, encoded by the high-pathogenicity island of Yersinia enterocolitica, is able to convert chorismate into salicylate, the precursor of the siderophore yersiniabactin. J. Bacteriol. 185, 5648–5653.
Pelludat, C., Rakin, A., Jacobi, C. A., Schubert, S., and Heesemann, J. (1998). The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180, 538–546.
Perry, R. D., and Fetherston, J. (2004). “Iron and heme uptake systems,” in Yersinia Molecular and Cellular Biology, eds E. Carniel and B. J. Hinnebusch (Norfolk: Horizon Bioscience), 257–283.
Perry, R. D., and Fetherston, J. D. (2011). Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 13, 808–817.
Pieper, R., Huang, S. T., Clark, D. J., Robinson, J. M., Alami, H., Parmar, P. P., et al. (2009). Integral and peripheral association of proteins and protein complexes with Yersinia pestis inner and outer membranes. Proteome Sci. 7, 5.
Pieper, R., Huang, S. T., Clark, D. J., Robinson, J. M., Parmar, P. P., Alami, H., et al. (2008). Characterizing the dynamic nature of the Yersinia pestis periplasmic proteome in response to nutrient exhaustion and temperature change. Proteomics 8, 1442–1458.
Pieper, R., Huang, S. T., Parmar, P. P., Clark, D. J., Alami, H., Fleischmann, R. D., et al. (2010). Proteomic analysis of iron acquisition, metabolic and regulatory responses of Yersinia pestis to iron starvation. BMC Microbiol. 10:30. doi: 10.1186/1471-2180-10-30
Podladchikova, O., Antonenka, U., Heesemann, J., and Rakin, A. (2011). Yersinia pestis autoagglutination factor is a component of the type six secretion system. Int. J. Med. Microbiol. 301, 562–569.
Podladchikova, O., Rykova, V., Antonenka, U., and Rakin, A. (2012). Yersinia pestis autoagglutination is mediated by HCP-like protein and siderophore Yersiniachelin (Ych). Adv. Exp. Med. Biol. 954, 289–292.
Podladchikova, O. N., and Rykova, V. A. (2006). Isolation and characterization of autoagglutination factor of Yersinia pestis Hms− cells. Biochemistry (Mosc.) 71, 1192–1199.
Rakin, A., Saken, E., Harmsen, D., and Heesemann, J. (1994). The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13, 253–263.
Rakin, A., Schubert, S., Pelludat, C., Brem, D., and Heesemann, J. (1999). “The high-pathogenicity island of yersiniae,” in Pathogenicity Islands and Other Mobile Virulence Elements, eds J. B. Kaper and J. Hacker (Washington, DC: ASM Press), 77–90.
Records, A. R. (2011). The type VI secretion system: a multi-purpose delivery system with a phage-like machinery. Mol. Plant-Microbe Interact. 24, 751–757.
Rosqvist, R., and Wolf-Watz, H. (1986). Virulence plasmid-associated HeLa cell induced cytotoxicity of Yersinia pseudotuberculosis. Microb. Pathog. 1, 229–240.
Rosso, M.-L., Chauvaux, S., Dessein, R., Laurans, C., Frangeul, L., Lacroix, C., et al. (2008). Growth of Yersinia pseudotuberculosis in human plasma: impacts on virulence and metabolic gene expression. BMC Microbiol. 8:211. doi: 10.1186/1471-2180-8-211
Schubert, S., Rakin, A., and Heesemann, J. (2004). The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int. J. Med. Microbiol. 294, 83–94.
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56.
Sebbane, F., Lemaitre, N., Sturdevant, D. E., Rebeil, R., Virtaneva, K., Porcella, S. F., et al. (2006). Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 11766–11771.
Vadyvaloo, V., Jarrett, C., Sturdevant, D., Sebbane, F., and Hinnebusch, B. J. (2007). Analysis of Yersinia pestis gene expression in the flea vector. Adv. Exp. Med. Biol. 603, 192–200.
Vadyvaloo, V., Jarrett, C., Sturdevant, D. E., Sebbane, F., and Hinnebusch, B. J. (2010). Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog. 6:e1000783. doi: 10.1371/journal.ppat.1000783
Keywords: highly virulent Yersinia, iron acquisition, siderophores, pathogenomics, epidemic Yersinia
Citation: Rakin A, Schneider L and Podladchikova O (2012) Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Inf. Microbio. 2:151. doi: 10.3389/fcimb.2012.00151
Received: 29 September 2012; Accepted: 13 November 2012;
Published online: 30 November 2012.
Edited by:
Matthew Francis, Umeå University, SwedenReviewed by:
Michael E. Murphy, University of British Columbia, CanadaCopyright © 2012 Rakin, Schneider and Podladchikova. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Alexander Rakin, Phylogenomics of the Enteropathogenic Yersinia, Max von Pettenkofer-Institute, LMU, Pettenkofer str. 9a, 80336 Munich, Germany. e-mail:cmFraW5AbXZwLnVuaS1tdWVuY2hlbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.