- 1 Department of Chemistry, Bielefeld University, Bielefeld, Germany
- 2 Department of Microbiology, Otto-von-Guericke University, Magdeburg, Germany
- 3 School of Biomolecular and Biomedical Sciences, University College Dublin, Dublin, Ireland
- 4 Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN, USA
- 5 Department of Pathology, Microbiology and Immunology, Vanderbilt University School of Medicine, Nashville, TN, USA
- 6 Veterans Affairs Tennessee Valley Healthcare System, Nashville, TN, USA
- 7 Department of Chemical Biology, Helmholtz Center for Infection Research, Braunschweig, Germany
Helicobacter pylori is a specific gastric pathogen that colonizes the stomach in more than 50% of the world’s human population. Infection with this bacterium can induce several types of gastric pathology, ranging from chronic gastritis to peptic ulcers and even adenocarcinoma. Virulent H. pylori isolates encode components of a type IV secretion system (T4SS), which form a pilus for the injection of virulence proteins such as CagA into host target cells. This is accomplished by a specialized adhesin on the pilus surface, the protein CagL, a putative VirB5 ortholog, which binds to host cell β1 integrin, triggering subsequent delivery of CagA across the host cell membrane. Like the human extracellular matrix protein fibronectin, CagL contains an RGD (Arg-Gly-Asp) motif and is able to trigger intracellular signaling pathways by RGD-dependent binding to integrins. While CagL binding to host cells is mediated primarily by the RGD motif, we identified an auxiliary binding motif for CagL–integrin interaction. Here, we report on a surface exposed FEANE (Phe-Glu-Ala-Asn-Glu) interaction motif in spatial proximity to the RGD sequence, which enhances the interactions of CagL with integrins. It will be referred to as RGD helper sequence (RHS). Competitive cell adhesion assays with recombinant wild type CagL and point mutants, competition experiments with synthetic cyclic and linear peptides, and peptide array experiments revealed amino acids essential for the interaction of the RHS motif with integrins. Infection experiments indicate that the RHS motif plays a role in the early interaction of H. pylori T4SS with integrin, to trigger signaling and to inject CagA into host cells. We thus postulate that CagL is a versatile T4SS surface protein equipped with at least two motifs to promote binding to integrins, thereby causing aberrant signaling within host cells and facilitating translocation of CagA into host cells, thus contributing directly to H. pylori pathogenesis.
Introduction
About 50% of the world’s human population is infected by Helicobacter pylori, a gastric pathogen causing gastritis in all infected individuals and more severe gastric disease in 10–15% of cases (Amieva and El-Omar, 2008; Atherton and Blaser, 2009; Polk and Peek, 2010). H. pylori can infect humans lifelong as the consequence of a highly complex host–pathogen crosstalk, and is an excellent model system to study bacterially induced epithelial cell signaling cascades which are of relevance to neoplasia. H. pylori strains are surprisingly diverse in both their genome sequences and resulting virulence. Multiple bacterial virulence factors such as the cag pathogenicity island (PAI), the protein CagA, and the vacuolating toxin VacA have been identified. Considerable research interest worldwide is currently focused on the effector protein CagA because CagA-positive but not CagA-negative H. pylori strains are associated with the development of severe gastric diseases. A direct causal link between CagA and carcinogenesis in vivo was achieved by the generation of transgenic mice expressing CagA (Ohnishi et al., 2008). The cagPAI has been shown to encode a type IV secretion system (T4SS) for the delivery of CagA into the cytoplasm of host target cells, where CagA is phosphorylated by host tyrosine kinases (Backert and Meyer, 2006; Mueller et al., 2012). T4SSs are typically composed of 11 VirB proteins (encoded by virB1–11) and the so-called coupling protein (VirD4, an NTPase). The H. pylori cagPAI contains up to 32 genes encoding orthologs of all 11 VirB proteins and VirD4 as well as various associated factors (Backert et al., 2002; Backert and Selbach, 2008; Fischer, 2011; Tegtmeyer et al., 2011a). Scanning electron microscopy (SEM) studies showed that the H. pylori T4SS is induced upon host cell contact and forms pilus-like structures protruding from the bacterial membrane (Rohde et al., 2003; Kwok et al., 2007).
Various cell surface molecules are required for T4SS function, suggesting a sophisticated control mechanism by which H. pylori injects CagA (Wessler and Backert, 2008). The first identified host receptor for the T4SS was integrin α5β1, according to a series of experiments including respective knockout cell lines, gene silencing RNAs, function-blocking antibodies, and competition studies (Kwok et al., 2007). The bacterial interaction partner was identified as CagL, a VirB5 ortholog, and specialized adhesin that is targeted to the pilus surface, where it binds to integrin α5β1 and mediates receptor-dependent delivery of CagA into gastric epithelial cells (Backert et al., 2008, 2011). Like fibronectin, an extracellular matrix protein and natural ligand of integrin α5β1, CagL contains an arginine–glycine–aspartate (RGD) motif that was shown to be important for interaction with integrin α5β1 on host cells (Kwok et al., 2007). Binding of CagL during infection or by incubation with recombinant protein in vitro elicits downstream signaling like tyrosine kinase activation of several proteins including EGF-receptor, FAK, and Src (Kwok et al., 2007; Saha et al., 2010; Tegtmeyer et al., 2010) as well as stimulation of ERK1/2 MAP kinase (Wiedemann et al., 2012). In line with these observations, various other structural T4SS proteins have subsequently also been demonstrated to bind to β1 integrins in vitro, including CagA, CagI, and CagY (Jiménez-Soto et al., 2009). In the same study, mutation of the RGD motif in CagL revealed no reduction of injected CagA during infection (Jiménez-Soto et al., 2009). Using mass spectrometry it was further reported that surface exposed CagL associates with two other cagPAI proteins, CagI, and CagH (Shaffer et al., 2011). All three factors are required for CagA translocation into host cells. SEM analysis revealed that these proteins are involved in the formation of T4SS pili. The H. pylori mutant strains ΔcagI and ΔcagL failed to form T4SS pili, while the ΔcagH mutant revealed a hyperpiliated phenotype and produced pili that are elongated and thickened as compared to those of the wild type (WT) bacteria (Shaffer et al., 2011). This suggests that T4SS pilus dimensions may be regulated by CagH. Taken together, the above results indicate that CagH, CagI, and CagL are components of a T4SS subassembly complex involved in the biogenesis of pili that interact with integrin α5β1 (Kwok et al., 2007; Shaffer et al., 2011). However, the exact co-operation of the various integrin-targeting cagPAI proteins is not yet clear and needs to be investigated.
We recently reported on cell-based in vitro binding studies with WM-115 and AGS cells revealing that CagL not only interacts with integrin α5β1, but also with αVβ3 and αVβ5 integrins (Conradi et al., 2011; Wiedemann et al., 2012). Infection and in vitro binding studies showed that CagL induces gastrin expression via a novel integrin αVβ5-integrin linked kinase signaling complex independent of CagA injection (Wiedemann et al., 2012). CagL–integrin αVβ5 interactions were demonstrated by immunoprecipitation and Biacore binding studies. In addition, the adhesion of WM-115 cells to immobilized CagL was inhibited by cyclic RGD peptides with pre-defined conformation, where the sequence was based on the CagL sequence Ala-Leu-Arg-Gly-Asp-Leu-Ala (ALRGDLA; Conradi et al., 2011). The application of the so-called spatial screening approach had previously been applied in the screening of cyclic RGD peptides and resulted in highly active peptides that efficiently and selectively inhibit the interaction of extracellular matrix proteins like vitronectin, fibronectin, and fibrinogen with the integrins αVβ3, α5β1, and αIIbβ3 in the nanomolar IC50 range (Gurrath et al., 1992; Pfaff et al., 1994; Haubner et al., 1996, 1997; Weide et al., 2007). The spatial screening approach makes use of incorporation of single D-amino acids into cyclic peptides in order to lock the conformation and to predictably present the amino acid side chain functional groups in a well-defined manner. D-Amino acids are known to induce and stabilize discrete turn structures in linear and, especially, cyclic peptides. Different cyclic RGD peptides with the CagL basic sequence ALRGDLA and a D-amino acid in different positions were synthesized. The variation of the amino acid stereochemistry contributes to different backbone structures, increases the spatial diversity, and provides constrained peptides with different β- and γ-turns. The structures were evaluated using NMR and molecular dynamics calculations, and activities of the different peptides were determined in cell adhesion assays. These structure–activity relationship studies revealed a β-turn around Asp-D-Leu-Ala-Arg to be favorable for integrin interactions (Conradi et al., 2011). The corresponding cyclopeptides were shown to inhibit integrin-mediated interactions of WM-115 cells with CagL. In particular, the cyclopeptide-7 [c-(-RGDlA-); Table 3, D-amino acids are written in small letters] displayed high activity (Conradi et al., 2011).
In comparison with purified CagLWT, purified CagL mutants (CagLRAD and CagLRGA) exhibited decreased, but not completely abolished affinity to integrin α5β1, αVβ3, and αVβ5 in vitro (Kwok et al., 2007; Conradi et al., 2011; Wiedemann et al., 2012). Other recent studies indicated that the RGD motif of CagL is certainly important but not sufficient for triggering cell signaling (Tegtmeyer et al., 2010; Wiedemann et al., 2012). Taken together, these data strongly underscore the importance of the RGD motif for CagL binding (Conradi et al., 2011), but also suggested the existence of one or more yet unidentified auxiliary motifs that is/are involved in CagL–integrin interactions. Here we report that another CagL motif, a FEANE-containing sequence, in proximity to the RGD motif, participates in binding of CagL to integrins (thus referred to as RHS) and is able to induce cell binding and signaling both in vitro and during H. pylori infection of gastric epithelial cells.
Materials and Methods
Human Cell Culture and H. pylori Strains
The human epithelial melanoma cell line WM-115 (ATCC, Wesel, Germany; CRL-1675™) is an adherent growing cell line with continuous integrin expression (mainly integrin α5β1, αVβ3, and αVβ5) and was cultivated in DMEM medium (PAA, Pasching, Austria) supplemented with 10% fetal bovine serum and 50 μg/ml gentamicin (PAA, Pasching, Austria). The human gastric adenocarcinoma cell line AGS (ATCC CRL-1739™) was cultivated in RPMI 1640 medium, which was supplemented with 10% fetal calf serum (Gibco, Paisley, UK). Cells were cultivated at 37°C and 5.3% (v/v) CO2 and subcultivated in a ratio of 1:3–1:5 every 2–3 days at a confluence of 70% to 90%. The H. pylori strains P12 WT and P12ΔcagL were generated and grown as described (Kwok et al., 2007). To complement the P12ΔcagL mutant strain, genes encoding CagLWT or CagL mutant proteins were introduced into the chromosomal ureA locus, using a pAD1-derived plasmid. CagL proteins expressed from the ureA promoter contain a hemagglutinin (HA) tag introduced following the signal sequence at amino acid position 22 (Shaffer et al., 2011). For infection, H. pylori were grown for 2 days in thin layers and added at a multiplicity of infection (MOI) of 50 or 100 (Selbach et al., 2002; Moese et al., 2004). The number of elongated AGS cells in each experiment was quantified in 10 different 0.25 mm2 fields (Tegtmeyer et al., 2011b). All experiments were done in triplicate.
CagL Protein Expression and Purification
CagLWT and mutants were expressed as C-terminal His-tag fusions in vector pET28a (Novagen®, Merck, Darmstadt, Germany) and purified as described earlier (Kwok et al., 2007). Briefly, E. coli ER2566 (NEB, Ipswich, USA) transformed with the plasmids were grown in 10 ml LB medium at 37°C. After overnight incubation, 500 ml of fresh LB medium were added and shaken for another 2.5–3 h until reaching OD600 = 1. Then 300 μl of a 1-mM IPTG stock solution was added and the bacteria were grown for 1.5 h to induce CagL expression. Bacterial pellets were collected by centrifugation and then resuspended in ice-cold buffer CW (50 mM KH2PO4–K2HPO4, pH 7.5, 200 mM NaCl) supplemented with 20 μM PMSF. After lysis using a French Press, the overexpressed CagL present in the inclusion bodies was solubilized in buffer LW (50 mM KH2PO4–K2HPO4, pH 7.5, 200 mM NaCl, 6 M guanidine hydrochloride) and refolded by a rapid dilution approach in ice-cold refolding buffer (50 mM Tris–HCl, pH 8.2, 20 mM NaCl, 0.1 mM KCl, 1 mM EDTA, 2 mM reduced glutathione, 0.2 mM oxidized glutathione) using a dual-channel syringe pump (KD Scientific Inc., Holliston, USA) with a flow of 0.1 ml/min. After refolding, CagL was further purified by metal-chelate affinity chromatography through Talon® resin (BD Biosciences). Protein concentrations of the resultant samples were determined by NanoDrop 1000 (Thermo Scientific, Waltman, USA) and typically yielded a total amount of about 1.5 mg CagL in 10 ml buffer. Purification of CagL was judged to be of >95% homogeneity by SDS-PAGE/Coomassie Blue staining. The folded conformation of the purified CagL proteins was subsequently confirmed by circular dichroism (CD) as described below.
Site-Directed Mutagenesis
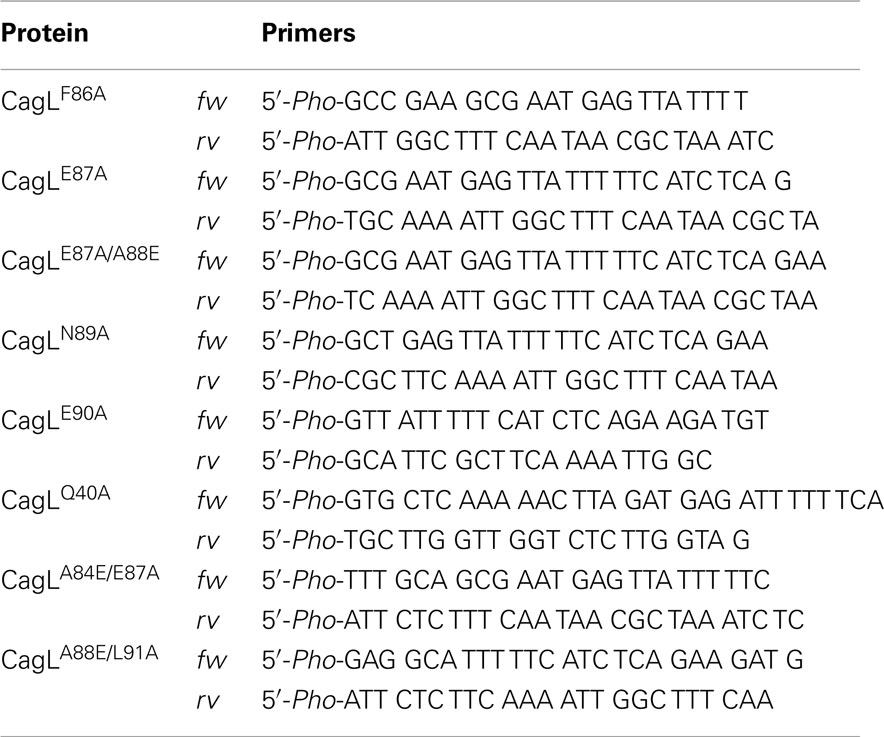
Site-directed mutagenesis of CagL was performed using the corresponding pET28a or pAD1 vectors as DNA template (Table 1). For amplification, Phusion® High-Fidelity DNA Polymerase (NEB, Ipswich, USA) was used, followed by PCR purification (MinElute PCR Purification Kit, Qiagen, Hilden, Germany), digestion with DpnI (Promega, Madison, USA), and ligation using T4 DNA Ligase (Promega). Re-sequencing and Western blotting of E. coli or H. pylori strains, respectively, verified the appropriate expression of CagL in the resulting plasmids.
Cell Adhesion Assays with WM-115 Cells
The cell adhesion assays to immobilized CagL proteins were performed as described previously (Conradi et al., 2011). A 96 well microtiter plate (Nunc Maxisorp™, Thermo Fisher Scientific Inc., Waltham, USA) was coated with 100 μl of protein solution (CagLWT or mutant; 10–20 μg/ml) per well, and immobilized for 18 h at 37°C. The solution was aspirated and free binding sites were blocked with 100 μl of a solution, consisting of 2% (w/v) fatty acid-free BSA (PAA, Pasching, Austria) in PBS for 1 h at 37°C. The adhesion assays were performed with WM-115 human epithelial cancer cells. This cell line was chosen for the cell adhesion assays due to a constitutive expression of integrin α5β1, αVβ3, and αVβ5, and was shown to display reproducible adherence to CagL and fibronectin (Conradi et al., 2011). The WM-115 cells were cultivated to a confluence of 70%, and detached with Trypsin-EDTA (0.05/0.02% in D-PBS; PAA, Pasching, Austria). After washing with DMEM medium, the cells were resuspended in DMEM medium with 1 mg/ml fluorescein diacetate (Sigma-Aldrich, St. Louis, USA), adjusted to a cell density of 1 × 105 cells/ml and incubated at 37°C for 30 min under steady shaking. Two DMEM washing steps were performed to remove excess fluorescein diacetate. The cells were resuspended in DMEM medium containing divalent cations Ca2+ and Mg2+ (2 nM) to a cell density of 1 × 105 cells/ml and incubated in the dark on ice for 30 min. Subsequently, the cell solution was distributed into solutions of peptides ranging in concentrations of millimolar to nanomolar, and the resulting mixtures were incubated at 37°C for 30 min. The solutions were then dispensed on the coated microtiter plate (5 × 103 cells/well) and incubated for 1 h at 37°C. After several washing steps the fluorescence of cells, adherent to the immobilized CagL, was measured (λex 485 nm; λem 514 nm) using an Infinite 200 Microplate Reader (Tecan, Männedorf, Switzerland). The inhibition concentration IC50, confidence interval CI95%, and the square of the correlation coefficient R2 values were evaluated. To test the accuracy of the fit model for the non-linear regression a “Runs test” was performed and high P-values were obtained for all measurements (data not shown), which support the chosen regression model. All evaluation was performed using the GraphPad Prism 4.03 Software (GraphPad, San Diego, USA).
Helicobacter pylori Binding to AGS Cells
Infection of AGS at a density of 3.2 × 105 cells/well was performed using a MOI of 50 or 100 per H. pylori strain (Kwok et al., 2002). After infection for 4 h, the AGS-bacterium co-cultures were washed three times with 1 ml of pre-warmed RPMI medium per well to remove non-adherent bacteria. To determine the total number of colony forming units (CFU) corresponding to host-bound H. pylori, the infected monolayers were incubated with 1 ml of 0.1% saponin in PBS at 37°C for 15 min. The resulting suspensions were diluted and incubated on GC agar plates as described (Kwok et al., 2002). The total CFU of cell-bound H. pylori are given as CFU per well of AGS cells.
CagL Peptide Synthesis
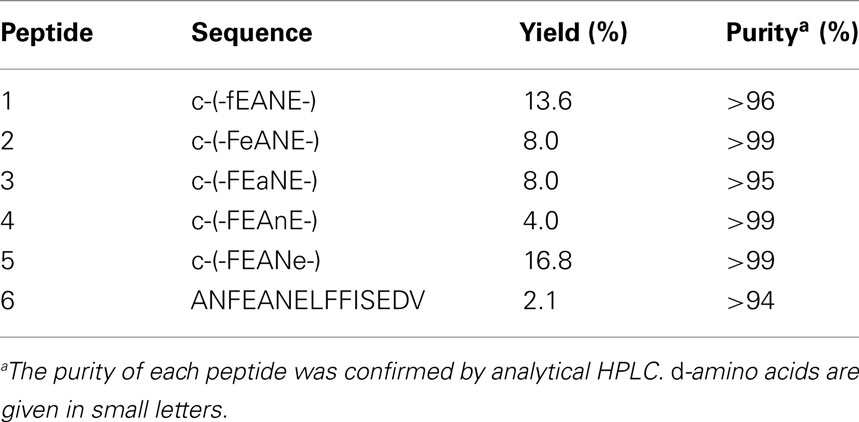
All Fmoc-α-amino acids (9-fluorenylmethylcarbonyl-protected α-amino acids) were purchased from IRIS Biotech (Marktredwitz, Germany) and Advanced ChemTech (Louisville, USA). MALDI-ToF MS analyses were performed on a Voyager-DE (Perseptive Biosystems, Foster City, USA) using 2,5-dihydrobenzoic acid as the matrix. The analytical RP-HPLC was performed with UV detection at 220 nm and the following elution gradients: eluent A: 95% water, 5% acetonitrile, 0.1% TFA; Eluent B: 95% acetonitrile, 5% water, 0.1% TFA; 0.7 ml min−1, 0–5 min 100% A → 100% B; 5–6 min 100% B → 100% A; 6–6.5 min 100% A (Thermo Separation Products apparatus equipped with a Hypersil Gold (3 μm, 150 mm × 2.1 mm) column (Thermo Fisher Scientific, Waltham, USA). Preparative RP-HPLC was performed on a Thermo Separation Products apparatus equipped with a Jupiter C18 (350 Å, 10 μm, 250 mm × 21.2 mm) efficiency column (Phenomenex, Torrance, USA) with water/acetonitrile gradients as the eluent and UV detection at 220 nm. Linear peptides were synthesized by solid phase peptide synthesis on a Liberty 12 channel microwave-assisted automated peptide synthesizer (CEM, Matthews, USA) according to a Fmoc-protocol with 2-chlorotrityl resin (IRIS Biotech) as solid support. The resin loading was 0.8 mmol/g. The C-terminal resin bound amino acids were Phe for cyclopeptides-2 to -5, D-Phe for cyclopeptide-1, and Val for the linear peptide 6. Peptide coupling was performed with three equivalents of Fmoc-amino acid (0.5 M in DMF), three equivalents TBTU (3-[bis-(dimethylamino)methyliumyl]-3H-benzotriazol-1-oxide; 0.5 M in DMF; dimethylformamide), and six equivalents DIPEA [diisopropylethylamine; 2 M in NMP (1-methyl-2-pyrrolidone)]. After washing with DMF the Fmoc group was cleaved with a solution of 20% piperidine in DMF. After synthesis of the bound linear peptides, resin cleavage was performed with 1% TFA (trifluoroacetic acid) in dichloromethane (10 times for 5 min each). The peptides one to five were cyclized under pseudo-high dilution conditions (Malesevic et al., 2004). For slow reagent addition a dual-channel syringe pump (KD Scientific Inc., Holliston, USA) was used. 200 μmol of linear precursor were dissolved in 20 ml DMF, and 600 μmol HATU (1-[bis-(dimethylamino) methyliumyl]-1H-1,2,3-triazolo[4,5-b]pyridine-3-oxide) were dissolved in the same volume of DMF. Each solution was transferred into a syringe, and both solutions simultaneously were added to a stirred solution of 1200 μmol DIPEA and 20 μmol HATU in 10 ml DMF at a rate of 1.00 ml/h (Malesevic et al., 2004). Finally, the mixture was stirred for further 15 min, and the solvent was evaporated under reduced pressure at a temperature below 30°C. Peptide purification was carried out using preparative RP-HPLC. Deprotection of the cyclic peptides took place in a mixture of TFA (95%), TIS (triisopropylsilane; 2.5%), and water (2.5%), with shaking at room temperature for 2 h. The solvent was evaporated and cold diethyl ether (30 ml) was added to the residue. After centrifugation for 1 h at 0°C and 4000 × g, diethyl ether was decanted and the residue was dissolved in water, lyophilized, and purified by preparative RP-HPLC. The yield and purity of the synthesized peptides are given in Table 2.
CagL Peptide SPOT Arrays
The CagL peptide arrays were generated by the SPOT-synthesis technique as described earlier (Beutling et al., 2008). Briefly, the indicated peptides in Figure 5 were synthesized on an amino-functionalized cellulose membrane using Fmoc/tert-butyl chemistry. The spots consist of ∼5 nmol of each peptide (Dikmans et al., 2006). For the binding assays, the peptide arrays were blocked overnight at room temperature with blocking buffer consisting of 2× blocking buffer concentrate (Sigma-Aldrich, St. Louis, MO/USA) and 5% (w/v) sucrose in TBS-T (0.02 M sodium phosphate buffer with 0.15M sodium chloride (pH 7), 0.05% Tween 20). Approximately 1 μg of purified integrins α5β1, αVβ3, and αVβ5 (Chemicon-Millipore, Billerica, MA, USA) in blocking buffer was added to CagL peptide arrays for 4 h at room temperature. Next, the arrays were washed with a 10-fold volume of TBS-T three times and then incubated with α-integrin-β1, α-integrin-αv, or α-integrin-β5 antibodies (Santa Cruz, Santa Cruz, USA). Finally, a chemiluminescence reaction using the ECL Plus kit (Amersham Pharmacia Biotech, Buckinghamshire, UK) was performed as described below for Western blotting.
Circular Dichroism Spectroscopy
Circular dichroism spectroscopy was performed on a Jasco J-810 spectrometer (Jasco, Groß-Umstadt, Germany). For the CD measurements of the CagL proteins a buffer containing 5 mM NaCl and 10 mM Na2HPO4 (pH 7.4) was used. The CagLWT and all mutants were measured in a 1-mm quartz cuvette, adjusted to a concentration of 3.9 nM. The secondary structure was evaluated using the deconvolution function of the Spectra Manager II Software (Jasco) based on CDPro structure analysis methods using Yang’s references (Yang et al., 1986; Sreerama and Woody, 2004).
Antibodies and Western Blotting
Treated/infected cells were harvested in ice-cold PBS containing 1 mM Na3VO4 (Sigma-Aldrich). Western blotting was done as previously described (Tegtmeyer et al., 2009). Rabbit α-CagL antiserum was raised against the C-terminal peptide (C-RSLEQSKRQYLQER) of the protein and was prepared by Biogenes (Berlin, Germany). The α-HA-tag antibody (Invitrogen, Darmstadt, Germany) was also used to detect tagged CagL. The pan-α-phospho-tyrosine antibody PY-99 (Santa Cruz) and α-CagA (Austral Biologicals, San Ramon, CA, USA) were used to investigate the phosphorylation of CagA. The polyclonal α-phospho-ERK1/2-PT202/PY-204 antibody was purchased from NEB (Frankfurt, Germany). The polyclonal α-phospho-Cortactin-PS-405 antibody was described recently (Tegtmeyer et al., 2011b). The α-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Santa Cruz) served as loading control in each Western blot. As secondary antibodies, horseradish peroxidase conjugated α-mouse, α-rabbit, or α-goat polyvalent sheep immunoglobulin were used and antibody detection was performed with the ECL Plus chemiluminescence kit (Amersham Pharmacia Biotech). Band intensities and corresponding kinase activities were quantitated with the Lumi-Imager F1 (Roche Diagnostics, Mannheim, Germany).
Statistical Analysis
All data were evaluated using Student t-test with SigmaStat statistical software (version 2.0). Statistical significance was defined by P ≤ 0.05 (*) and P ≤ 0.005 (**). All error bars shown in figures and those quoted following the ± signs represent standard deviation.
Results
Synthetic RHS-Cyclopeptides Influence WM-115 Cell Adhesion to Immobilized CagL Mutants
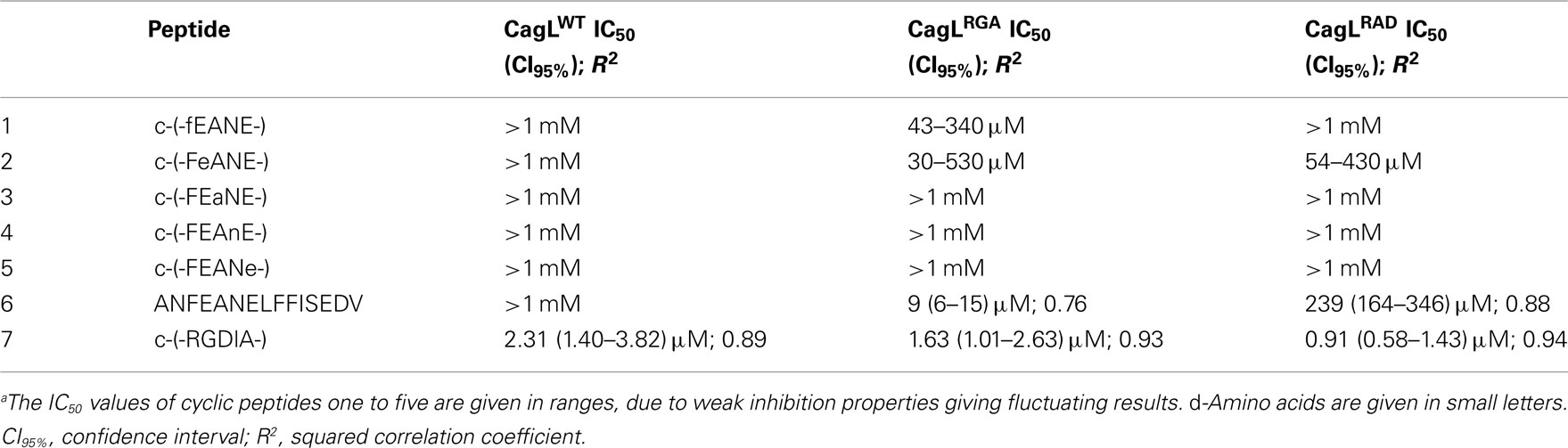
Previous studies indicated that the RGD sequence in CagL is important but not sufficient to trigger host cell receptor binding and signaling (Kwok et al., 2007; Tegtmeyer et al., 2010; Conradi et al., 2011; Wiedemann et al., 2012). The recently published 3D homology model of CagL (Backert et al., 2008) was screened for other surface exposed amino acid sequences to further explore the hypothesis that other CagL features besides the RGD sequence might be important for integrin interactions. Among other possible motifs that are surface exposed and with an orientation that is directed to the plausible CagL–integrin interaction site, the so-called RHS motif in proximity to the RGD motif (Figure 1) was identified as a candidate for host interaction. A series of five cyclopeptides comprising the RHS sequence was designed and synthesized to mimic the exposed motif in CagL (Figure 1). According to the spatial screening approach each of the five amino acids was mutually replaced by its D-configured counterpart to stabilize the overall conformation and increase the spatial peptide backbone diversity (Tables 2 and 3). None of the cyclopeptides-1 to -5 inhibited WM-115 cell adhesion to immobilized CagLWT (IC50 > 1 mM; Table 3). However, WM-115 cell adhesion to the mutant CagLRAD was inhibited by cyclopeptide-2 [c-(-FeANE-)] with an IC50 value in the range of 54–430 μM (Table 3). In addition, cyclopeptides-1 and -2 displayed weak inhibition of WM-115 cell adhesion to the mutant CagLRGA with IC50 values of 43–340 and 30–530 μM, respectively. The other cyclopeptides-3 to -5 did not display any significant effect on the WM-115–CagL interaction (IC50 > 1 mM; Table 3).
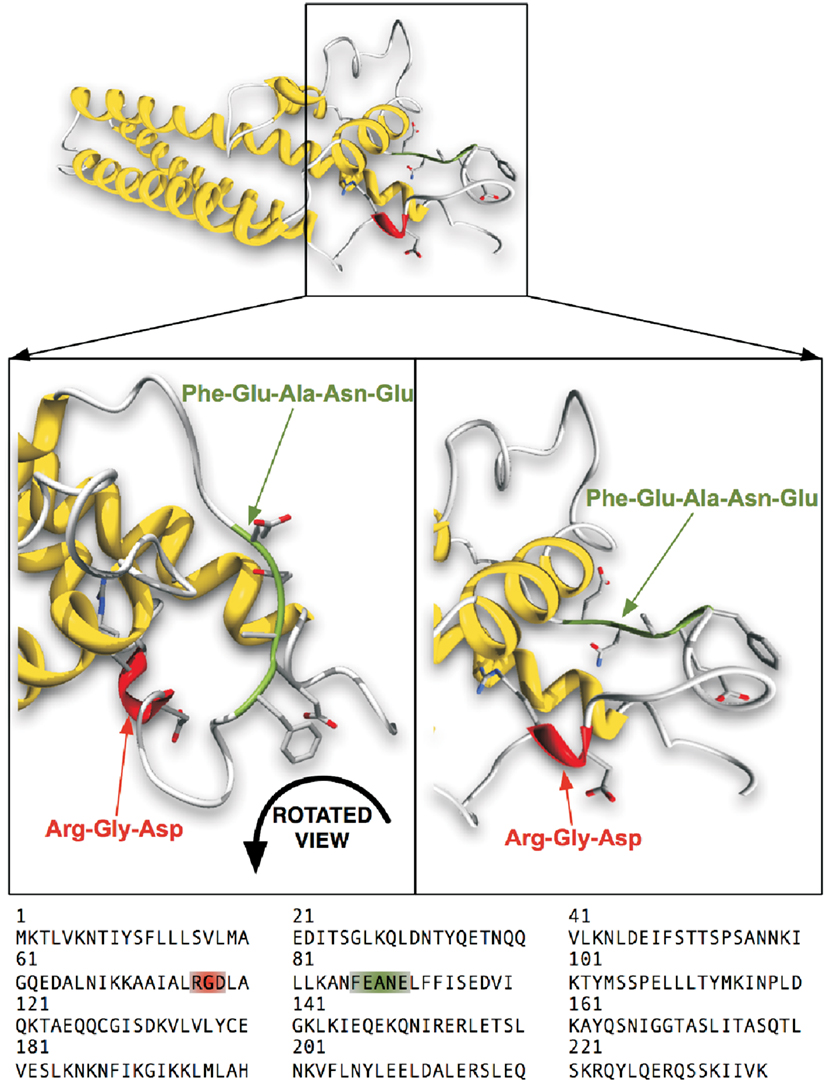
Figure 1. Computational 3D-structure model of CagLWT with highlighted motifs of interest. The ribbon diagram of CagL shown in this figure consists of three major helices and a globular domain with the exposed RGD motif highlighted in the front. The second highlighted FEANE sequence (here called RHS motif) is shown in a flexible loop, also exposed at the surface. The CagL homology model is derived from the VirB5 ortholog TraC protein (PDB: 1R8I) encoded in plasmid pKM101 (Yeo et al., 2003; Backert et al., 2008).
A Linear RHS 15-mer Peptide Interferes with WM-115 Adhesion to the CagLRAD and CagLRGA Mutants
In addition, cell adhesion assays were performed to investigate whether the linear peptide 6 (ANFEANELFFISEDV) has effects on the adhesion of WM-115 cells to immobilized CagLWT and its RGD mutants CagLRGA and CagLRAD. Linear peptide 6 mimics the RHS motif with the Phe-Glu-Ala-Asn-Glu sequence and its adjacent amino acids within the CagL protein sequence (Figure 1). According to its CD spectrum, it does not adopt a discrete conformation in solution (data not shown). While linear peptide 6 did not significantly interfere with CagLWT–WM-115 interaction in the cell adhesion assay (IC50: >1 mM), it exhibited a slight inhibitory activity on the CagLRAD–WM-115 interaction (IC50: 239 μM; CI95%: 164–346 μM; R2: 0.88) and the CagLRGA–WM-115 interaction with an IC50 of 9 μM (CI95%: 6–15 μM; R2: 0.76; Figure 2; Table 3). In comparison to the properties of the earlier mentioned c-(-RGDlA-) peptide to inhibit the WM-115 cell adhesion to CagLWT (IC50: 2.31 μM; CI95%: 1.40–3.82 μM; R2: 0.89) and its RGD mutants CagLRGA (IC50: 1.63 μM; CI95%: 1.01–2.63 μM; R2: 0.93) and CagLRAD (IC50: 0.91 μM; CI95%: 0.58–1.43 μM; R2: 0.94), the inhibition properties of the linear peptide 6 are obviously reduced (Table 3). Nonetheless the results support involvement of the RHS motif in CagL–WM-115 interaction.
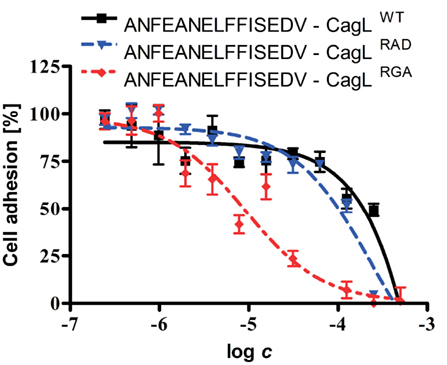
Figure 2. Dose–response curve of a linear RHS motif-containing peptide, inhibiting binding of WM-115 cells to immobilized CagLWT and mutants. The WM-115 cells were pre-incubated in different concentrations of the linear 15-mer peptide ANFEANELFFISEDV, which mimics the exposed RHS motif of CagL. Purified CagLWT and the CagLRAD and CagLRGA mutants were immobilized on plastic surface followed by addition of the WM-115 cells. After 1 h of co-incubation, the amounts of attached cells were determined. The data were evaluated using non-linear fit for the evaluation of the potency of peptide ANFEANELFFISEDV to inhibit the binding of WM-115 cells to immobilized CagL.
The Recombinant CagL Mutants Are Correctly Folded
For further investigation of the RHS motif and to reveal the relevance of specific amino acids in this sequence, CagL point mutants were recombinantly produced in E. coli, where individual amino acids were replaced by alanine residues (Table 4). CD spectroscopy was used to verify the correct folding of the mutants CagLF86A, CagLE87A, CagLE87A/A88E, and CagLE90A in comparison to CagLWT. The secondary structure composition calculated for CagLWT is in good accordance with the proposed CagL homology model (Backert et al., 2008) and displays a high content of α-helices (∼35%), accompanied by β-strands (∼29%, Figure 3A). The mutants CagLF86A, CagLE87A, CagLE87A/A88E, and CagLE90A CD spectra display comparable curve shapes like the CagLWT (Figure 3D). Unfortunately, the amount of CagLN89A protein necessary to estimate the secondary structure and to validate the correct protein folding with CD spectroscopy could not be obtained and, therefore, could not be included in the study. However, to obtain information on the structural stability and to further compare the CagLE87A and CagLE90A mutants with CagLWT, temperature-interval dependent CD measurements were performed ranging from 0 to 60°C (Figures 3A–C). In general, all tested proteins behaved in similar manner, exhibiting temperature-dependent denaturation above 40°C. Below 40°C, the secondary structures are comparable, with only minor structural differences. In comparison, the CagLE90A CD spectrum shows maximum deviations at temperatures below 20°C (Figure 3C). In addition, we performed a pH screening by measuring the CD spectra of CagLWT over a pH range from four to nine in phosphate buffer and could show that the protein is very stable under the tested conditions (Figure 3E).
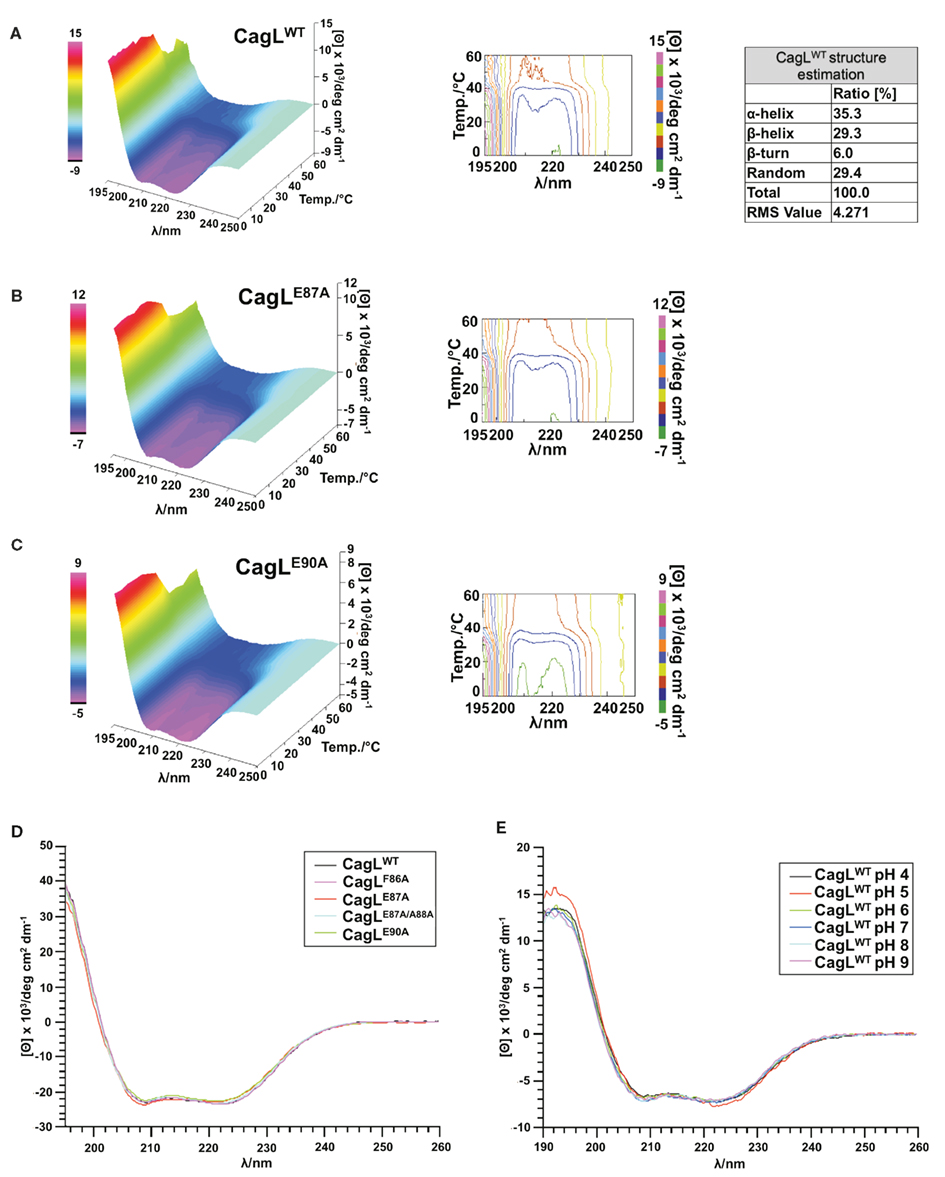
Figure 3. Structural evaluation of purified CagL proteins with CD spectroscopy. (A) The CagLWT secondary structure was evaluated as described previously (Yang et al., 1986). (A–C) The temperature-dependent structure stability of the mutants CagLE87A, CagLE90A, and the CagLWT protein was evaluated in CD measurements. (D) Secondary structure of CagLWT and mutants CagLF86A, CagLE87A, CagLE87A/A88E, and CagLE90A were evaluated to control correct folding of the proteins after expression and purification. (E) pH stability of CagLWT in the pH range four to nine.
WM-115 Cell Adhesion Studies with CagL RHS Mutants
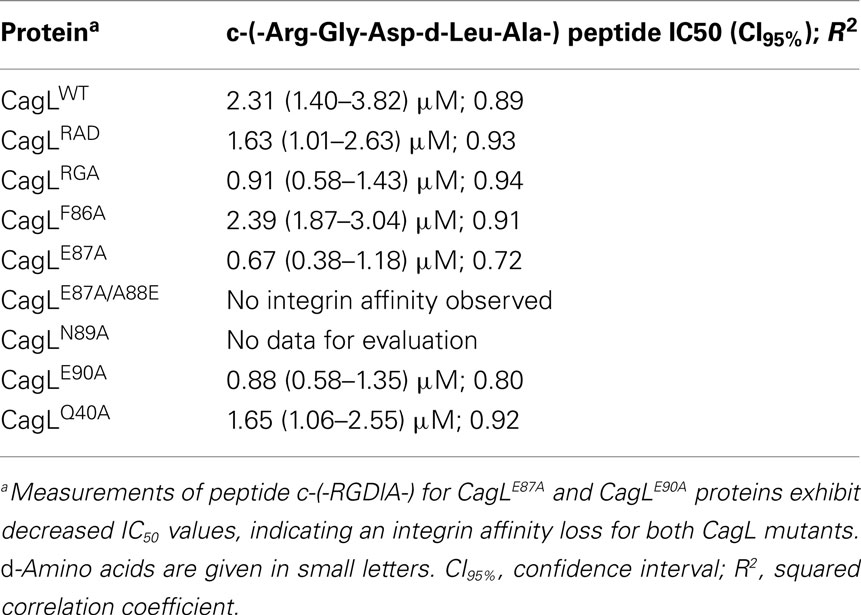
Different CagL mutants with amino acid variations in the RHS motif (CagLF86A, CagLE87A, and CagLE90A) were immobilized and investigated in cell adhesion assays. The WM-115 cells were pre-incubated with the previously described cyclopeptide-7 [c-(-Arg-Gly-Asp-D-Leu-Ala-)] in different concentrations from nano- to millimolar ranges to block the RGD-binding site of the integrins. The dose–response curves obtained in these assays are shown in Figure 4. For comparison, the cell adhesion results of the recently published CagLWT, CagLRGA, and CagLRAD mutants are also given in Table 4 (Conradi et al., 2011). Additionally, the CagLQ40A mutant was included as a negative control to exclude the possibility that genetic mutations in the CagL sequence may generally lead to loss of function. Cyclopeptide-7 inhibited WM-115 cell adhesion to CagLQ40A with an IC50 value of 1.65 μM (CI95% = 1.06–2.55; R2 = 0.92), while the cell adhesion of WM-115 cells to immobilized CagLF86A was inhibited with an IC50 value of 2.39 μM (CI95% = 1.87–3.04 μM; R2 = 0.91), a value that is very similar to that of CagLWT (IC50 = 2.31 μM; CI95% = 1.87–3.04 μM; R2 = 0.91). Interesting results were observed in the inhibition assays for the mutants CagLE87A and CagLE90A. WM-115 cell adhesion to immobilized CagLE87A was inhibited by cyclopeptide-7 with an IC50 of 0.67 μM (CI95% = 0.38–1.18 μM; R2 = 0.72), the lowest value observed for all CagL mutants investigated (Figure 4; Table 4). The IC50 value of 0.88 μM (CI95% = 0.58–1.35 μM; R2 = 0.80) for the inhibition of WM-115 cell adhesion to immobilized CagLE90A by cyclopeptide-7 is similar, and is also comparable to the values observed for CagLRGA (0.91 μM; Figure 4; Table 4). In an additional experiment we tested the double mutant CagLE87A/A88E to reveal more details on the involvement of CagL amino acid Glu87 in the interaction with WM-115 cells. The double mutant CagLE87A/A88E formally is characterized by the shift of an acidic amino acid side chain from position 87 to position 88. The immobilized CagLE87A/A88E protein displayed no binding to the WM-115 cells, which may indicate a loss of integrin affinity. In conclusion, the results show remarkably reduced adhesion of WM-115 cells to immobilized CagLE87A and CagLE90A compared to the CagLWT, which implies a participation of the glutamates in the CagL–WM-115 interaction.
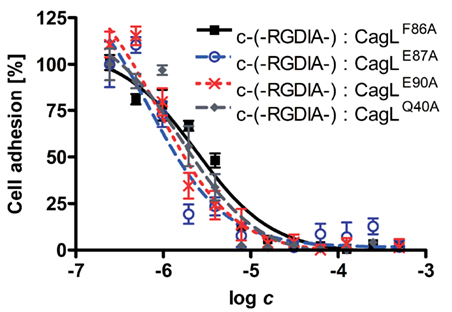
Figure 4. Dose–response curves showing the inhibitory effect of cyclic RGD peptides on WM-115 cell binding to the different immobilized CagL proteins in competitive cell adhesion assays. The purified CagLQ40A, CagLF86A, CagLE87A, and CagLE90A mutant proteins were immobilized on plastic surface. WM-115 cells were pre-incubated with varying concentrations of cyclic peptide c-(-RGDlA-; D-amino acids in small letter), which is known to inhibit WM-115 cell binding to immobilized CagL in the low μM range (Conradi et al., 2011). Subsequently, the WM-115 cells were incubated with the immobilized CagL mutants. The amount of WM-115 cells bound to immobilized CagL was quantified, and the resulting dose–response curves were evaluated with a non-linear fit to determine the binding inhibition potency of peptide c-(-RGDlA-) for the interaction of WM-115 cells with immobilized CagL.
Peptide Array Mapping of the CagL RHS Motif for Integrin Binding
The previously described SPOT technique (Frank, 2002) was applied to identify the CagL amino acid sequence responsible for binding to integrin α5β1. Overlapping linear 15-mer peptides derived from the CagL sequence from amino acid position 60–104 were chemically synthesized on a cellulose membrane by the SPOT method (Frank, 1992). As shown in Figure 5A, adjacent peptides share the same sequence of 12 amino acids but differ by three amino acids at the C- or N-terminal ends, respectively. Purified integrin α5β1 was incubated with these membranes and the binding was assayed as described in the Section “Materials and Methods.” Figure 5B shows that integrin α5β1 binds to the RGD motif-containing arraypeptides-3 and -4. In addition, a very strong signal for arraypeptide-9 covering the RHS motif was recorded (Figure 5B, top and Figure 5C). Interestingly, two other FEANE sequence-containing array peptides (-7 and -8) did not bind integrin α5β1, suggesting that the flanking C-terminal sequences are also important for this interaction. As a control, incubation of the membranes with two other integrins, αVβ3 and αVβ5 also revealed signals for the two RGD containing CagL peptides and some weak signals for arraypeptides-10 or -11, respectively (Figure 5B, middle and Figure 5C). The mock control blot revealed no signals as expected (Figure 5B, bottom).
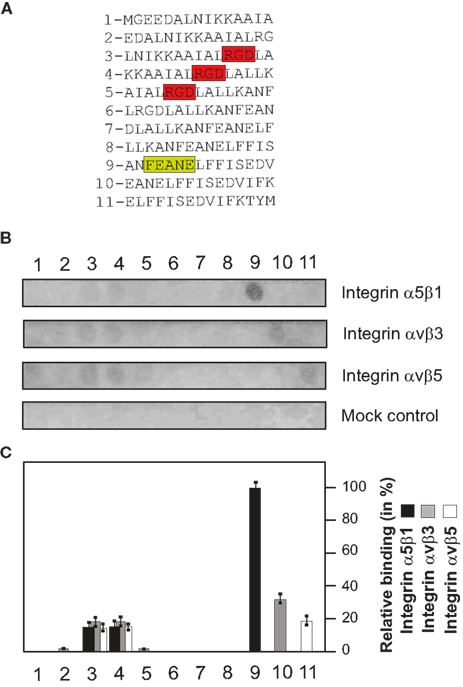
Figure 5. Mapping of the RHS motif and flanking CagL sequences in binding to integrins using a peptide array. (A) The design of the array of 15-mer peptides and corresponding CagL amino acid sequences from position 60–104 is shown. The RGD and RHS motifs are highlighted with color. (B) Purified integrins α5β1, αVβ3, and αVβ5 were incubated with these membranes and bound proteins were identified using antibodies as described in Materials and Methods. As a mock control, buffer-treated membranes followed by incubation with an integrin-β1 antibody revealed no signals, as expected. (C) Densitometric measurement of individual spot intensities using the Lumi-Imager F1 (Roche) revealed the relative amount of bound binding partner per given peptide in%. The strongest spot intensity seen was set as 100%.
Generation of RHS Point Mutations of CagL in H. pylori
To investigate the importance of the RHS motif in CagL directly during infection with H. pylori, two CagL mutants were generated carrying the A84E/E87A and A88E/L91A point mutations. Based on the above described peptide array the two point mutants were constructed to contain one mutation in the RHS sequence and one either N- or C-terminal, to cover additional amino acids that may be relevant for host cell interactions. CagLWT and both mutants were chromosomally integrated into a P12ΔcagL deletion mutant and expressed as HA-tag fusions in the ureA locus as described (Shaffer et al., 2011). The correct expression of each of these CagL variants was verified by Western blotting using an α-HA antibody (Figure 6A). The AGS cells were infected with each of these H. pylori strains for 4 h, followed by analysis of viable bacterial binding to cells, as described in Materials and methods. The results show that each of these strains bound to AGS cells with high efficiency. Some minor differences in binding were seen among the different strains but were not statistically significant (Figure 6B). This suggests that mutation of the RHS motif in CagL has no significant inhibitory impact on the overall capacity of the bacteria to bind to AGS host cells.
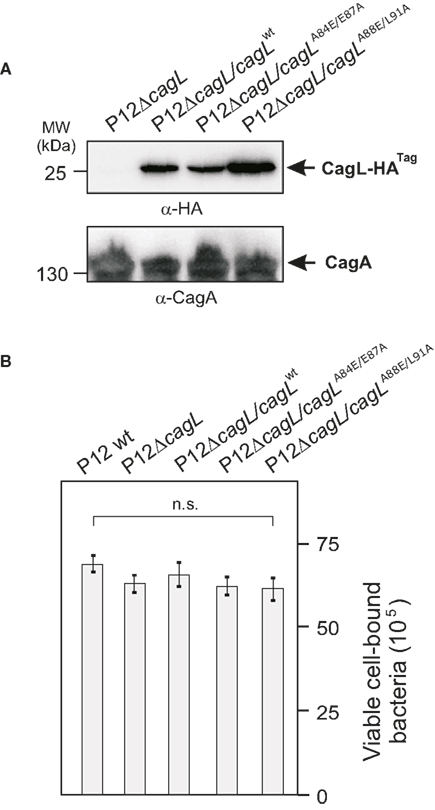
Figure 6. Effect of CagL deletion or point mutations in the CagL RHS motif on binding of H. pylori to AGS cells. (A) A ΔcagL deletion mutant in strain P12 was generated by replacing the cagL gene with an aphA3 cassette. CagLWT and two CagL mutants carrying either the A84E/E87A or A88E/L91A point mutations were expressed as HA-tag fusion proteins from genes integrated into the ureA locus of the P12ΔcagL strain. The correct expression of each of these CagL variants was verified by Western blotting using an α-HA antibody. The α-CagA Western blot was performed as loading control. (B) AGS cells were infected with each of these strains for 4 h using MOI = 100, followed by determination of the amount of viable bound H. pylori per input strain. The quantification data indicate that each of these strains bound to AGS cells equally well with high efficiency. No significant differences in results were seen when MOI = 50 was used (data not shown).
Mutation of the RHS Motif Inhibits CagA Phosphorylation at Early Times of Infection
AGS cells were infected with the different complemented H. pylori strains during a time course of 2 and 4 h, respectively, to investigate whether strains expressing CagL mutant proteins can trigger the injection and phosphorylation of CagA. The results of the α-phospho-tyrosine and α-CagA specific Western blots show that P12ΔcagL re-expressing CagLWT can efficiently inject and phosphorylate CagA in a time-dependent fashion (Figures 7A,B). Infection of AGS with P12ΔcagL expressing the CagLA84E/E87A mutant revealed a ∼46% reduced phospho-CagA signal at 2 h, while very strong phospho-CagA signals were produced at the 4-h time point with almost no difference as compared to the complemented CagLWT control (Figures 7A,B). This suggests that mutation of A84E/E87A in CagL affects the injection and phosphorylation of CagA at very early times of infection. In contrast, infection with H. pylori expressing the CagLA88E/L91A mutant revealed a stronger reduction of phospho-CagA signals (∼69%) at 2 h as compared to the complemented CagLWT control, while this low level of phospho-CagA did only slightly increase after 4 h of infection (Figures 7A,B). This result shows that a strain expressing the CagLA88E/L91A mutant protein has a significant defect in injection and phosphorylation of CagA at both time points of infection.
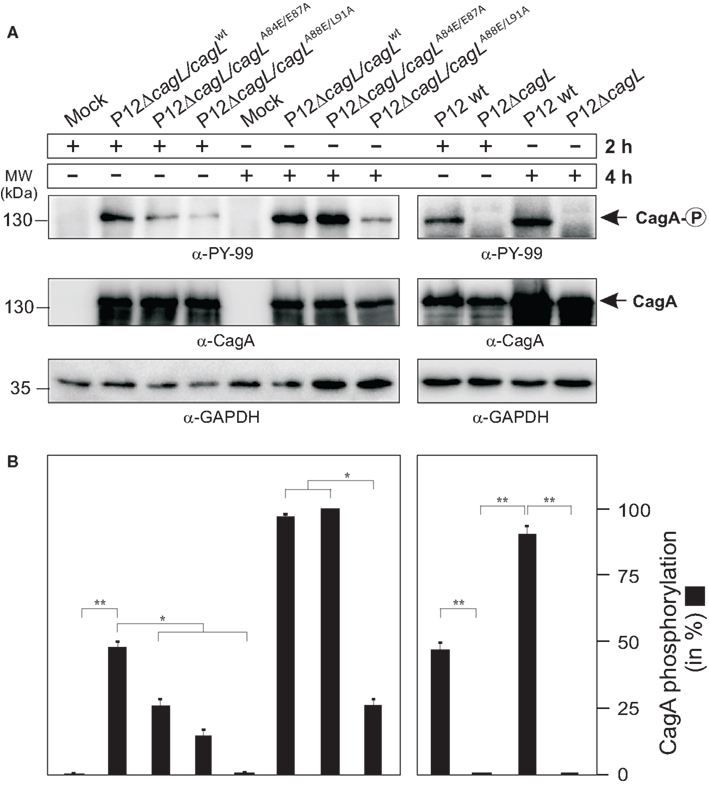
Figure 7. Effects of CagL point mutations in the RHS motif on CagA injection and phosphorylation during H. pylori infection of AGS cells. (A) The P12ΔcagL deletion mutant was complemented with CagLWT or various CagL point mutants. AGS cells were infected with the different indicated H. pylori strains (MOI = 100) during a time course of 2 or 4 h, respectively. The resulting protein lysates were subjected to Western blotting using α-phospho-tyrosine (PY-99) and α-CagA antibodies. The α-GAPDH blot served as loading control in each sample. (B) Quantification of CagA phosphorylation. Densitometric measurement of individual band intensities of the α-CagA and α-phospho-tyrosine (PY-99) blots in (A) was performed with the Lumi-Imager F1 (Roche). Quantification revealed the relative amount of phospho-CagA per sample in %. Similar inhibitory activities of all mutants were seen when MOI = 50 was used (not shown).
Mutation of the RHS Motif Exhibits Significant Signaling Defects in H. pylori-Induced ERK1/2 Activation
Finally the various RHS mutations in CagL were inspected in relation to impacts on cellular downstream signaling involved in the H. pylori-induced AGS cell elongation phenotype. It was recently shown that besides CagA phosphorylation, the activation of the ERK1/2 MAP kinase pathway either by H. pylori infection (Mimuro et al., 2002; Tegtmeyer et al., 2009) or by transfection CagA in the absence of H. pylori (Higashi et al., 2004) is also crucial for phenotypical outcome. Therefore, we tested whether the different CagL-expressing H. pylori strains induced the activation of ERK1/2, using the same conditions as described for Figure 7. The results of the α-phospho-ERK1/2 Western blots show that P12 WT or P12ΔcagL re-expressing CagLWT (but not P12ΔcagL) can efficiently activate this MAP kinase in a time-dependent fashion (Figures 8A,B). Infection of AGS with H. pylori expressing the complemented CagLA84E/E87A mutant revealed a significantly (46%) reduced phospho-ERK1/2 signal at 2 h, and a similarly reduced (38%) phospho-ERK1/2 signal at 4 h time point as compared to the complemented CagLWT control (Figures 8A,B). This suggests that mutation of A84E/E87A in CagL downregulates not only the injection and phosphorylation of CagA, but also reduced the activation of ERK1/2. In addition, infection with H. pylori expressing the CagLA88E/L91A mutant revealed an even stronger reduction of phospho-ERK1/2 signals by about 63 or 70% at the 2- or 4-h time points, respectively. This result indicates that the CagLA88E/L91A mutant has a significantly pronounced defect in activating ERK1/2 during infection.
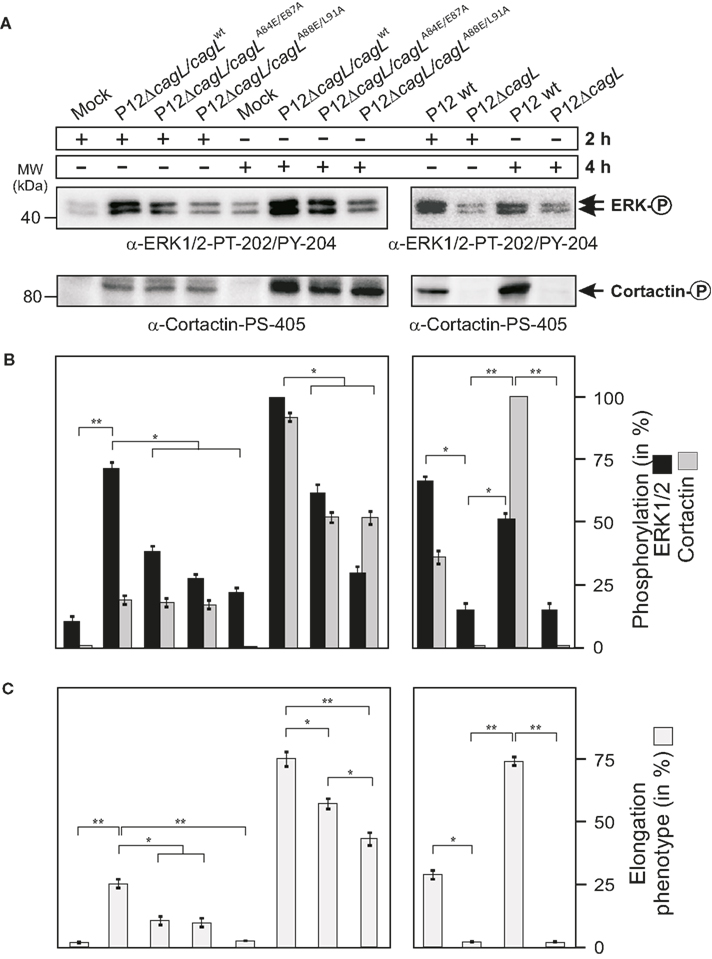
Figure 8. Mutation of CagL in specific amino acids within the RHS motif results in defects in H. pylori-triggered ERK1/2 activation, cortactin phosphorylation, and AGS cell elongation. (A) AGS cells were infected with the different complemented H. pylori strains (MOI = 100) during a time course of 2 or 4 h, respectively. The same protein lysates as shown in Figure 7 were subjected to Western blotting using the indicated α-phospho-ERK1/2 and α-phospho-cortactin antibodies. (B) Densitometric measurement of individual band intensities of the blots in (A) and comparison to total α-ERK1/2 and α-cortactin blots (not shown) was performed with the Lumi-Imager F1 (Roche). The relative amounts of phospho-ERK1/2 and phospho-cortactin per sample were quantified and given in%. (C) Quantification of the AGS cell elongation phenotype in the same set of experiments. Similar inhibitory activities of all mutants were seen when MOI = 50 was used (not shown).
Mutation of the RHS Motif Results in Reduced Cortactin Serine Phosphorylation and AGS Cell Elongation
Very recently, we demonstrated that one important downstream target of activated ERK1/2 during infection is the actin-binding protein cortactin, phosphorylated at serine residue 405 (Tegtmeyer et al., 2011b). Hence it was tested whether the various CagL-expressing strains induce the phosphorylation of cortactin at S-405 during a time course of 2 and 4 h infection. The results of the α-phospho-cortactin Western blots show that P12ΔcagL re-expressing CagLWT but not P12ΔcagL can efficiently phosphorylate cortactin in a time-dependent manner (Figures 8A,B). Infection of AGS cells with H. pylori expressing either CagLA84E/E87A or CagLA88E/L917A mutants revealed the induction of phospho-cortactin signals similar to that of CagLWT with no significant difference at 2 h, but a strongly reduced (about 45–46%) phospho-cortactin signal at the 4-h time point (Figures 8A,B). This suggests that mutation of CagL in the RHS motif (either A84E/E87A or A88E/L91A mutations) downregulates cortactin’s serine phosphorylation to a similar high extent at the 4-h time point, indicating that the CagLA84E/E87A and CagLA88E/L91A mutants share a significantly pronounced defect in activating cortactin during infection. Finally, the elongation phenotype was monitored in the same set of experiments. As shown in Figure 8C, mutation of the RHS motif at A84E/E87A or A88E/L91A inhibited the elongation phenotype by more than 50% at the 2-h time point. At the 4-h time point, the CagLA84E/E87A mutant exhibited a ∼20% reduction and the CagLA88E/L91A mutant revealed a ∼43% reduction as compared to the complemented CagLWT control. This set of experiments reveals that mutation of the RHS motif in H. pylori results in significant defects in signaling to cortactin and cell elongation, especially at very early times of infection.
Discussion
Our previous studies have suggested that the RGD sequence is an important structural motif in the H. pylori CagL pilus protein, able to trigger T4SS-mediated host cell binding and signaling, but this sequence alone is not sufficient (Kwok et al., 2007; Tegtmeyer et al., 2010; Conradi et al., 2011; Wiedemann et al., 2012). Thus, we examined the 3D structural CagL model (Backert et al., 2008) to identify other potential surface exposed CagL–integrin interaction motifs. In this study, we report on the discovery of a novel motif in CagL, called RHS consisting of a Phe-Glu-Ala-Asn-Glu core sequence, which also plays a role in the interaction with host cell integrins. A peptide array of overlapping 15-mer peptides derived from the CagL primary structure showed that peptides containing the RHS motif of CagL can bind particularly to integrin α5β1, whereas they bind only weakly to other integrins, such as αVβ3 and αVβ5. Further studies, including the use of CagL-derived peptides in competitive WM-115 cell binding assays, and the analysis of recombinant CagL point mutants in cell binding in vitro and during infection of AGS cells with H. pylori, collectively indicated that the RHS sequence plays an important role in the interaction of CagL with host cells.
The spatial screening approach (Haubner et al., 1996; Weide et al., 2007) was employed with five conformationally designed synthesized cyclopeptides-1 to -5, wherein each of the five amino acids was mutually replaced by its D-configured correlate to mimic the RHS sequence of CagL. Receptor binding of such cyclopeptides is regarded to be entropically favored, if the receptor-bound conformation is still accessible. However, neither the linear RHS-peptide 6 (ANFEANELFFISEDV) nor the RHS-cyclopeptides-1 to -5 showed a significant effect on WM-115 cell adhesion to immobilized CagLWT. In contrast, RGD-dependent WM-115 cell adhesion could be efficiently influenced by the RGD-cyclopeptide-7. Mutations within the RGD sequence in extracellular matrix proteins such as fibronectin are usually accompanied by loss of integrin affinity (Giancotti and Ruoslahti, 1999; Takahashi et al., 2007; Leiss et al., 2008). This seems not to be fully applicable to CagL, as integrin-expressing WM-115 cells still adhere to the mutated proteins CagLRAD and CagLRGA but with reduced affinity (Conradi et al., 2011). Interestingly, both the linear RHS-peptide ANFEANELFFISEDV and the RHS-cyclopeptide-2 were able to interfere with WM-115 cell adhesion to immobilized CagLRGA and CagLRAD proteins with IC50 values in the micromolar range, indicating significant binding. The studies with the CagLRGA and CagLRAD mutants allowed us to distinguish RGD-dependent interaction from other types of binding. Not all RHS-cyclopeptides displayed such inhibitory biological activity, which clearly proves the influence of peptide conformation and three-dimensional structure on the anti-adhesive properties. We thus conclude that the WM-115 cell adhesion to recombinant CagL is mainly driven by the RGD recognition sequence, but is significantly assisted by the RHS sequence as a proposed auxiliary motif. There is an interesting analogy to fibronectin. Besides the RGD motif, fibronectin harbors another sequence required for maximum binding affinity to integrin α5β1, called the synergy region, located in the ninth type III module adjacent to the RGD site (Nagai et al., 1991; Aota et al., 1994; Bowditch et al., 1994; Chada et al., 2006). Subsequent studies showed that the synergy region is not directly in contact with the integrin α5β1 bound to fibronectin, and proposed an indirect function of the synergy region for the high affinity binding by optimally exposing the flexible RGD motif or by inducing long range electrostatic steering (Baron et al., 1992; Takagi et al., 2003). In the case of CagL, however, a FEANE-containing peptide alone can bind integrin α5β1 as shown in peptide arrays, and the synergy region of fibronectin shares no sequence homology to CagL or the RHS domain.
The role of the RHS motif and neighboring amino acids was further investigated on the protein level with recombinant CagL proteins carrying corresponding point mutants. The secondary structure and temperature stability of all generated point mutants corresponds with that observed for CagLWT according to CD spectra. Given the above assumptions, pre-incubation of the WM-115 cells with the RGD-cyclopeptide-7 should provide information on the contribution of the RHS sequence to CagL binding phenomena. Strikingly, there is an effect of mutations in the RHS motif of CagL on WM-115 cell adhesion. RGD-cyclopeptide-7 was able to inhibit cell adhesion to CagL with mutations in the RHS sequence (CagLF86A, CagLE87A, CagLE90A) even at lower concentrations than for CagLWT. Granted that cell adhesion is governed by both RGD and RHS sequences, a mutation in the CagL RGD sequence (CagLRGA, CagLRAD) would lead to reduced cell adhesion and, hence, require a smaller concentration (reduced IC50) of the RGD-peptide that competes for binding (Table 4; Conradi et al., 2011). Conversely, WM-115 cell adhesion to immobilized CagL with mutations in the RHS motif conferring decreased affinity could be inhibited by the RGD-peptide at a smaller concentration (reduced IC50). This was indeed observed for CagLE87A and CagLE90A. In conclusion, the point mutant CagLF86A and a control mutant CagLQ40A show no significant affinity loss in their binding to WM-115 cells as compared to CagLWT, while the point mutants CagLE87A and CagLE90A display remarkably reduced affinity to WM-115 cells. The replacement of the negatively charged glutamates E87 and E90 by the hydrophobic amino acid alanine resulted in a decreased affinity toward WM-115 cells. In contrast, the exchange of the uncharged amino acids phenylalanine F86 and glutamine Q40 by alanine did not have any consequence in the same assays. These data imply that especially glutamate E87 is important for the interaction with integrin on the surface of host cells.
At first sight, the peptide array experiments in which we investigated the binding of immobilized CagL 15-mer peptides with the different purified integrins seem to implicate a stronger signal of a RHS containing peptide for bound integrin α5β1, but much weaker signals compared to RGD containing peptides on the same membrane. Regarding the conformation of RGD containing peptides, cyclic RGD peptides with a β-turn around Asp-D-Leu-Ala-Arg display a much higher affinity to some integrins than do linear RGD peptides. The linear RGD containing peptides do not prefer this structural β-turn conformation, which could explain the fact that linear RGD peptides on the peptide array display very little affinity to integrins as compared to their cyclopeptide counterparts. While the arraypeptides-7 to -9 contain the whole FEANE amino acids of the RHS motif, only arraypeptide-9 reveals integrin β1 interaction (Figure 5). In arraypeptide-10 and -11 the 15-mer peptide sequence is shifted C-terminally for three amino acids. Arraypeptide-10 therefore is lacking the phenylalanine of the RHS motif and displays a different interaction with integrin subunit β3, while the arraypeptide-11 includes only the final glutamate of the RHS motif and displays weak interaction with the integrin β5 subunit. In cell binding studies with the 15-mer peptide of arraypeptide-9 corresponding to linear peptide 6, a binding inhibition of WM-115 cells to immobilized CagLRAD and CagLRGA could be shown (Tables 2 and 3). Overall the binding potency of this linear peptide was surprisingly better than that observed for the cyclic RHS peptides. These results emphasize the possible involvement of adjacent amino acids next to the FEANE sequence for integrin interactions that probably are located in the C-terminal flanking site. The presented results allow no complete elucidation of amino acids important for integrin interaction, but combining the results of cell binding studies and the peptide array, the CagL F86 amino acid is not involved in the interaction with WM-115 cells (Table 3) and the interaction with purified integrins (Figure 5), while both glutamates E87 and E90 have an impact and are probably exposed on the CagL surface. The observed binding of three different integrins to a relatively small CagL sequence region of about 18 amino acids on peptide arrays is a very interesting finding, and is relevant to recent reports which showed that CagL interacts not only with integrin α5β1 on cells, but also αVβ3 and αVβ5 (Kwok et al., 2007; Conradi et al., 2011; Wiedemann et al., 2012).
Finally, it was investigated if the newly discovered RHS motif in CagL might also play a role during infections with H. pylori. As discussed above, an explicit amino acid region for the so-called RHS motif could not be clearly defined. Therefore two double point mutants CagLA84E/E87A and CagLA88E/L91A were chosen, with one mutation in the RHS region and another located at the N- or C-terminus, respectively. Based on infection experiments, we could show that H. pylori P12 WT, deletion mutant P12ΔcagL, and P12ΔcagL re-expressing CagLWT or CagL point mutants (A84E/E87A and A88E/L91A) bound equally well to cultured AGS cells. This suggests that CagL is not involved in general host cell binding during infection, in agreement with previous reports showing the overall importance of the major canonical adhesins BabA/B, SabA, AlpA/B, or OipA (for review, see Tegtmeyer et al., 2011a). However, this illustrates clearly that the observed signaling defects discussed below are not due to a more general deficiency of the bacteria to adhere to AGS cells. First, H. pylori expressing the generated CagL point mutants exhibited significant defects in injection and phosphorylation of CagA between 2 and 4 h of infection. Second, each of the CagL mutants in H. pylori had significantly pronounced defects in activating the MAP kinases ERK1/2 and serine phosphorylation of cortactin as well as the induction of the AGS cell elongation phenotype during infection. This suggests that mutation of the RHS motif in H. pylori results in various defects in the interaction of CagL with integrin α5β1 with resulting inhibitory consequences for (i) the injection of CagA and (ii) for the downstream signaling to ERK1/2 and cortactin triggering cell elongation, especially at early times of infection.
Taken together, our presented data provide evidence for a novel domain in CagL playing a significant role in binding to integrin α5β1 in vitro and for initiating signaling pathways through certain motifs encoded in the so-called RHS motif comprising the FEANE sequence and its flanking amino acids. We propose that the RGD motif is important for the CagL binding to integrins, while subsequent binding of other integrin-specific CagL motifs may assist in certain intracellular signaling events. Although we could demonstrate that the RHS motif and its adjacent amino acids are relevant in CagL–integrin interactions and have an impact on the resulting signaling, more studies are necessary to further define the specific amino acids relevant for integrin interaction and downstream signaling. In addition, it has been shown that besides CagL, multiple other H. pylori factors are also involved in integrin β1 binding, including CagA, CagI, CagY, and possibly CagH (Kwok et al., 2007; Jiménez-Soto et al., 2009; Shaffer et al., 2011). Their individual role for integrin-targeting/binding is not yet fully clear. Future studies should investigate whether these proteins act together or at different stages of the infection process.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work of Steffen Backert was supported through Priority Program SPP1150 of the Deutsche Forschungsgemeinschaft DFG (Ba1671/3) and another DFG grant (Ba1671/8-1). Timothy L. Cover was supported by the National Institutes of Health (R01 AI068009 and P01 CA116087) and the Department of Veterans Affairs. We thank Sabine Brandt (University Magdeburg) for support in the peptide array hybridizations, and Carrie Shaffer (Vanderbilt University Nashville) for help with the CagL complementation plasmid.
References
Amieva, M. R., and El-Omar, E. M. (2008). Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134, 306–323.
Aota, S., Nomizu, M., and Yamada, K. M. (1994). The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell adhesive function. J. Biol. Chem. 269, 24756–24761.
Atherton, J. C., and Blaser, M. J. (2009). Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119, 2475–2487.
Backert, S., Churin, Y., and Meyer, T. F. (2002). Helicobacter pylori type IV secretion, host cell signalling and vaccine development. Keio J. Med. 51(Suppl. 2), 6–14.
Backert, S., Clyne, M., and Tegtmeyer, N. (2011). Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori. Cell Commun. Signal. 9, 28.
Backert, S., Fronzes, R., and Waksman, G. (2008). VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 16, 409–413.
Backert, S., and Meyer, T. F. (2006). Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9, 207–217.
Backert, S., and Selbach, M. (2008). Role of type IV secretion in Helicobacter pylori pathogenesis. Cell. Microbiol. 10, 1573–1581.
Baron, M., Main, A. L., Driscoll, P. C., Mardon, H. J., Boyd, J., and Campbell, I. D. (1992). 1H NMR assignment and secondary structure of the cell adhesion type III module of fibronectin. Biochemistry 31, 2068–2073.
Beutling, U., Stading, K., Stradal, T., and Frank, R. (2008). Large-scale analysis of protein-protein interactions using cellulose-bound peptide arrays. Adv. Biochem. Eng. Biotechnol. 110, 115–152.
Bowditch, R. D., Hariharan, M., Tominna, E. F., Smith, J. W., Yamada, K. M., Getzoff, E. D., and Ginsberg, M. H. (1994). Identification of a novel integrin binding site in fibronectin. Differential utilization by b3 integrins. J. Biol. Chem. 269, 10856–10863.
Chada, D., Mather, T., and Nollert, M. U. (2006). The synergy site of fibronectin is required for strong interaction with the platelet integrin aIIbb3. Ann. Biomed. Eng. 34, 1542–1552.
Conradi, J., Huber, S., Gaus, K., Mertink, F., Royo Gracia, S., Strijowski, U., Backert, S., and Sewald, N. (2011). Cyclic RGD peptides interfere with binding of the Helicobacter pylori protein CagL to integrins αVβ3 and α5β1. Amino Acids. doi:10.1007/s00726-011-1066-0
Dikmans, A., Beutling, U., Schmeisser, E., Thiele, S., and Frank, R. (2006). SC2: a novel process for manufacturing multipurpose high-density chemical microarrays. QSAR Comb. Sci. 25, 1069–1080.
Fischer, W. (2011). Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 278, 1203–1212.
Frank, R. (1992). Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217–9232.
Frank, R. (2002). The SPOT-synthesis technique: synthetic peptide arrays on membrane supports principles and applications. J. Immunol. Methods 267, 13–26.
Gurrath, M., Müller, G., Kessler, H., Aumailley, M., and Timpl, R. (1992). Conformation/activity studies of rationally designed potent anti-adhesive RGD peptides. Eur. J. Biochem. 210, 911–921.
Haubner, R., Finsinger, D., and Kessler, H. (1997). Stereoisomeric peptide libraries and peptidomimetics for designing selective inhibitors of the αVβ3 integrin for a new cancer therapy. Angew. Chem. Int. Ed. Engl. 36, 1375–1389.
Haubner, R., Gratias, R., Diefenbach, B., Goodman, S. L., Jonczyk, A., and Kessler, H. (1996). Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin αVβ3 antagonists. J. Am. Chem. Soc. 118, 7461–7472.
Higashi, H., Nakaya, A., Tsutsumi, R., Yokoyama, K., Fujii, Y., Ishikawa, S., Higuchi, M., Takahashi, A., Kurashima, Y., Teishikata, Y., Tanaka, S., Azuma, T., and Hatakeyama, M. (2004). Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J. Biol. Chem. 279, 17205–17216.
Jiménez-Soto, L. F., Kutter, S., Sewald, X., Ertl, C., Weiss, E., Kapp, U., Rohde, M., Pirch, T., Jung, K., Retta, S. F., Terradot, L., Fischer, W., and Haas, R. (2009). Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 5, e1000684. doi:10.1371/journal.ppat.1000684
Kwok, T., Backert, S., Schwarz, H., Berger, J., and Meyer, T. F. (2002). Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70, 2108–2120.
Kwok, T., Zabler, D., Urman, S., Rohde, M., Hartig, R., Wessler, S., Misselwitz, R., Berger, J., Sewald, N., König, W., and Backert, S. (2007). Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449, 862–866.
Leiss, M., Beckmann, K., Girós, A., Costell, M., and Fässler, R. (2008). The role of integrin binding sites in fibronectin matrix assembly in vivo. Curr. Opin. Cell Biol. 20, 502–507.
Malesevic, M., Strijowski, U., Bächle, D., and Sewald, N. (2004). An improved method for the solution cyclization of peptides under pseudo-high dilution conditions. J. Biotechnol. 112, 73–77.
Mimuro, H., Suzuki, T., Tanaka, J., Asahi, M., Haas, R., and Sasakawa, C. (2002). Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell 10, 745–755.
Moese, S., Selbach, M., Kwok, T., Brinkmann, V., König, W., Meyer, T. F., and Backert, S. (2004). Helicobacter pylori induces AGS cell motility and elongation via independent signaling pathways. Infect. Immun. 72, 3646–3649.
Mueller, D., Tegtmeyer, N., Brandt, S., Yamaoka, Y., De Poire, E., Sgouras, D., Wessler, S., Torres, J., Smolka, A., and Backert, S. (2012). c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J. Clin. Invest. 122, 1553–1566.
Nagai, T., Yamakawa, N., Aota, S., Yamada, S. S., Akiyama, S. K., Olden, K., and Yamada, K. M. (1991). Monoclonal antibody characterization of two distant sites required for function of the central cell-binding domain of fibronectin in cell adhesion, cell migration, and matrix assembly. J. Cell Biol. 114, 1295–1305.
Ohnishi, N., Yuasa, H., Tanaka, S., Sawa, H., Miura, M., Matsui, A., Higashi, H., Musashi, M., Iwabuchi, K., Suzuki, M., Yamada, G., Azuma, T., and Hatakeyama, M. (2008). Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc. Natl. Acad. Sci. U.S.A. 105, 1003–1008.
Pfaff, M., Tangemann, K., Müller, B., Gurrath, M., Müller, G., Kessler, H., Timpl, R., and Engel, J. (1994). Selective recognition of cyclic RGD peptides of NMR defined conformation by αIIbβ3, αVβ3, and α5β1 integrins. J. Biol. Chem. 269, 20233–20238.
Polk, D. B., and Peek, R. M. (2010). Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer 10, 403–414.
Rohde, M., Püls, J., Buhrdorf, R., Fischer, W., and Haas, R. (2003). A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49, 219–234.
Saha, A., Backert, S., Hammond, C. E., Gooz, M., and Smolka, A. J. (2010). Helicobacter pylori CagL activates ADAM17 to induce repression of the gastric H, K-ATPase alpha subunit. Gastroenterology 139, 239–248.
Selbach, M., Moese, S., Meyer, T. F., and Backert, S. (2002). Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect. Immun. 70, 665–671.
Shaffer, C. L., Gaddy, J. A., Loh, J. T., Johnson, E. M., Hill, S., Hennig, E. E., Mcclain, M. S., McDonald, W. H., and Cover, T. L. (2011). Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 7, e1002237. doi:10.1371/journal.ppat.1002237
Sreerama, N., and Woody, R. W. (2004). Computation and analysis of protein circular dichroism spectra. Meth. Enzymol. 383, 318–351.
Takagi, J., Strokovich, K., Springer, T. A., and Walz, T. (2003). Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 22, 4607–4615.
Takahashi, S., Leiss, M., Moser, M., Ohashi, T., Kitao, T., Heckmann, D., Pfeifer, A., Kessler, H., Takagi, J., Erickson, H. P., and Fässler, R. (2007). The RGD motif in fibronectin is essential for development but dispensable for fibril assembly. J. Cell Biol. 178, 167–178.
Tegtmeyer, N., Hartig, R., Delahay, R. M., Rohde, M., Brandt, S., Conradi, J., Takahashi, S., Smolka, A. J., Sewald, N., and Backert, S. (2010). A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. J. Biol. Chem. 285, 23515–23526.
Tegtmeyer, N., Wessler, S., and Backert, S. (2011a). Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 278, 1190–1202.
Tegtmeyer, N., Wittelsberger, R., Hartig, R., Wessler, S., Martinez-Quiles, N., and Backert, S. (2011b). Serine phosphorylation of cortactin controls focal adhesion kinase activity and cell scattering induced by Helicobacter pylori. Cell Host Microbe 9, 520–531.
Tegtmeyer, N., Zabler, D., Schmidt, D., Hartig, R., Brandt, S., and Backert, S. (2009). Importance of EGF receptor, HER2/Neu and Erk1/2 kinase signalling for host cell elongation and scattering induced by the Helicobacter pylori CagA protein: antagonistic effects of the vacuolating cytotoxin VacA. Cell. Microbiol. 11, 488–505.
Weide, T., Modlinger, A., and Kessler, H. (2007). Spatial screening for the identification of the bioactive conformation of integrin ligands. Top. Curr. Chem. 272, 1–50.
Wessler, S., and Backert, S. (2008). Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 16, 397–405.
Wiedemann, T., Hofbaur, S., Tegtmeyer, N., Huber, S., Sewald, N., Wessler, S., Backert, S., and Rieder, G. (2012). Helicobacter pylori CagL dependent induction of gastrin expression via a novel αVβ5-integrin-integrin linked kinase signalling complex. Gut. [Epub ahead of print].
Yang, J. T., Wu, C. S., and Martinez, H. M. (1986). Calculation of protein conformation from circular dichroism. Meth. Enzymol. 130, 208–269.
Keywords: CagL, binding motifs, cortactin, ERK kinase, integrin interaction, α5β1
Citation: Conradi J, Tegtmeyer N, Woźna M, Wissbrock M, Michalek C, Gagell C, Cover TL, Frank R, Sewald N and Backert S (2012) An RGD helper sequence in CagL of Helicobacter pylori assists in interactions with integrins and injection of CagA. Front. Cell. Inf. Microbio. 2:70. doi: 10.3389/fcimb.2012.00070
Received: 17 November 2011; Accepted: 02 May 2012;
Published online: 23 May 2012.
Edited by:
D. Scott Merrell, Uniformed Services University, USAReviewed by:
Jeong-Heon Cha, Yonsei University, South KoreaNina Salama, Fred Hutchinson Cancer Research Center, USA
Jay V. Solnick, University of California Davis, USA
Copyright: © 2012 Conradi, Tegtmeyer, Woźna, Wissbrock, Michalek, Gagell, Cover, Frank, Sewald and Backert. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Norbert Sewald, Department of Chemistry, Bielefeld University, PO Box 10 01 31, D-33501 Bielefeld, Germany. e-mail:bm9yYmVydC5zZXdhbGRAdW5pLWJpZWxlZmVsZC5kZQ==; Steffen Backert, School of Biomolecular and Biomedical Sciences, Science Center West, University College Dublin, Belfield Campus, Dublin-4, Ireland. e-mail:c3RlZmZlbi5iYWNrZXJ0QHVjZC5pZQ==
†Jens Conradi and Nicole Tegtmeyer have contributed equally to this work.
 Jens Conradi1†
Jens Conradi1†