
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 March 2012
Sec. Molecular Bacterial Pathogenesis
Volume 2 - 2012 | https://doi.org/10.3389/fcimb.2012.00032
This article is part of the Research Topic Research Advances in the Study of Campylobacter, Helicobacter & Related Organisms View all 29 articles
Antimicrobial peptides (AMPs) are critical components of host defense limiting bacterial infections at the gastrointestinal mucosal surface. Bacterial pathogens have co-evolved with host innate immunity and developed means to counteract the effect of endogenous AMPs. However, molecular mechanisms of AMP resistance in Campylobacter, an important human food-borne pathogen with poultry as a major reservoir, are still largely unknown. In this study, random transposon mutagenesis and targeted site-directed mutagenesis approaches were used to identify genetic loci contributing Campylobacter resistance to fowlicidin-1, a chicken AMP belonging to cathelicidin family. An efficient transposon mutagenesis approach (EZ::TN™ <KAN-2> Transposome) in conjunction with a microtiter plate screening identified three mutants whose susceptibilities to fowlicidin-1 were significantly increased. Backcrossing of the transposon mutations into parent strain confirmed that the AMP-sensitive phenotype in each mutant was linked to the specific transposon insertion. Direct sequencing showed that these mutants have transposon inserted in the genes encoding two-component regulator CbrR, transporter CjaB, and putative trigger factor Tig. Genomic analysis also revealed an operon (Cj1580c-1584c) that is homologous to sapABCDF, an operon conferring resistance to AMP in other pathogens. Insertional inactivation of Cj1583c (sapB) significantly increased susceptibility of Campylobacter to fowlicidin-1. The sapB as well as tig and cjaB mutants were significantly impaired in their ability to compete with their wild-type strain 81–176 to colonize the chicken cecum. Together, this study identified four genetic loci in Campylobacter that will be useful for characterizing molecular basis of Campylobacter resistance to AMPs, a significant knowledge gap in Campylobacter pathogenesis.
Campylobacter species including C. jejuni and C. coli are the most common bacterial causes of human gastroenteritis in the United States and other developed countries (Allos, 2001). Human Campylobacter illnesses are caused primarily by C. jejuni (∼90%) and secondarily by C. coli (∼10%). Poultry are the major reservoir of Campylobacter and thus the main source for human campylobacteriosis (Friedman et al., 2000). As a comensal, Campylobacter could establish persistent, benign infections in chickens (up to 1010 CFU per gram of feces). To successfully colonize and persist in intestinal tracts of animals and humans, Campylobacter must have evolved a variety of mechanisms to counteract hash in vivo conditions as well as host innate immunity. However, we know less about Campylobacter pathogenicity than we do about other prevalent pathogens, which impeded the development of efficient intervention strategies to prevent and control Campylobacter infections in humans and animal reservoirs.
Endogenous antimicrobial peptides (AMPs) belong to the most ancient and efficient components of host defense. Defensins and cathelicidins are two major families of AMPs in humans and animals. Both defensins and cathelicidines are important defense AMPs expressed by epithelium in small intestine, consequently limiting bacterial infections at the gastrointestinal mucosal surface (Wehkamp et al., 2007). Despite the existence of a broad diversity in AMP sequences and structures, the vast majority of AMPs share a common theme in the mechanism of killing action by disruption of membrane integrity (Yeaman and Yount, 2003; Peschel and Sahl, 2006). Generally, AMPs directly interact with target cells via initial electrostatic and hydrogen bond attraction, then disrupt the structure or function of bacterial membrane by permeating lipid bilayers, forming transmembrane pore, and ultimately lead to cell death. However, transmembrane pore forming is not the only mechanism of bacterial killing by AMP. AMPs may also have intracellular targets (e.g., DNA gyrase, ribosome) and AMP-mediated bacterial death may occur as a result of several independent or cooperative mechanisms of action (Yeaman and Yount, 2003). Expression of defensins and cathelicidins is either constitutive or inducible in gastrointestinal tract (Wehkamp et al., 2007). Epithelial AMPs have been observed to be induced in human intestinal epithelia upon infection by C. jejuni (Zilbauer et al., 2005). Recently, a full panel of chicken defensins and cathelicidins (designated as “fowlicidins”) has been identified and characterized (Lynn et al., 2004; Xiao et al., 2004, 2006; Bommineni et al., 2007; van Dijk et al., 2007). These chicken AMPs displayed potent and broad spectrum of antibacterial activity, highlighting their role in host innate defense and in the development of novel antimicrobial (peptide antibiotic) (Hancock and Sahl, 2006).
Bacterial pathogens have co-evolved with host innate defense and developed means to curtail the effect of endogenous AMPs (Yeaman and Yount, 2003; Peschel and Sahl, 2006). Different AMP resistance mechanisms have been well characterized in various enteric pathogens such as Salmonella enterica serovar Typhimurium and Escherichia coli (Yeaman and Yount, 2003; Peschel and Sahl, 2006). The best studied AMP resistance mechanisms in Gram-negative bacteria involve (1) electrostatic repulsion of AMPs by modification of cell surface, (2) proteolytic cleavage of AMPs, and (3) active extrusion of AMPs by drug efflux pumps. Other mechanisms, such as capsule production, changes in the composition of lipid fatty acids in membrane and maintenance of appropriate membrane potential, have been also implicated in AMP resistance. Covalent modification of LPS in outer membrane via two-component regulatory systems (e.g., PhoP/PhoQ), which consequently reduces the negative charge and/or membrane fluidity of LPS, has been identified as a major mechanism in these Gram-negative pathogens (Ernst et al., 2001; Yeaman and Yount, 2003; Peschel and Sahl, 2006). However, as an important strategy to evade killing by innate immunity and by potential peptide antibiotics, AMP resistance mechanisms are still largely unknown in Campylobacter. Recently, it has been observed that lipid A modification (Cullen and Trent, 2010; van Mourik et al., 2010) and lipooligosaccharide (LOS) production (Lin et al., 2009; Naito et al., 2010) could confer Campylobacter resistance to polymyxin B, an AMP produced by bacterial species Paenibacillus polymyxa, that has been successfully used as a model peptide to study AMP resistance in many Gram-negative pathogens. However, the polymyxin B resistance gene identified from our recent study (Lin et al., 2009) did not contribute C. jejuni resistance to endogenous AMPs (e.g., fowlicidins and bacteriocins), strongly suggesting that polymyxin B may not be a good surrogate for studying Campylobacter resistance to structurally unrelated but physiologically relevant AMPs.
In this study, we chose fowlicidin-1 (F1), a cathelicidin expressed in chicken intestine, as a model peptide to examine mechanisms of AMP resistance and identified three genetic loci (cbrR, cjaB, and tig) involved in F1 resistance in C. jejuni using an efficient in vivo transposon mutagenesis system. In addition, genomic analysis revealed an operon cj1580c-1584c that shared homology to the sap (sensitivity to antimicrobial peptide) operon that conferred resistance to AMP-mediated killing of different pathogens such as Salmonella (Parra-Lopez et al., 1993). Site-directed mutagenesis of the sap operon showed that sapB (cj1583c) is involved in F1 resistance in C. jejuni. Chicken experiments further demonstrated that these genes play an important role in the colonization of Campylobacter in the intestine.
The major bacterial strains and plasmids used in this study are listed in Table 1. Among the 174 Campylobacter isolates used for susceptibility test to chicken AMP F1, 154 isolates were C. jejuni and 20 isolates were C. coli. These Campylobacter strains were isolated from different hosts and geographically diverse locations described our previous study (Hoang et al., 2011a). Campylobacter strains were routinely grown in Mueller–Hinton (MH) broth (Difco) or MH agar at 42°C under microaerophilic conditions generated by using CampyGen Plus gas pack (Oxoid) in an enclosed jar. E. coli strains were grown in Luria–Bertani (LB) broth with shaking (250 rpm) or on agar at 37°C overnight. Antibiotics kanamycin (Kan) or chloramphenicol (Cm) was added in medium at a desired concentration when needed.
The susceptibilities of C. jejuni and C. coli isolates to F1 were determined by a standard microtiter broth dilution method with an inocula of 106 bacterial cells/ml as described previously (Lin et al., 2002). Minimum inhibitory concentrations (MICs) were determined by the lowest concentration of specific antimicrobial showing complete inhibition of bacterial growth after 24 h of incubation at 42°C. The chicken cathelicidin F1 (>95% purity) was synthesized by Bio-Synthesis (Lewisville, TX).
F1 was used as the selective agent to obtain spontaneous F1 resistant mutants in vitro. Briefly, C. jejuni 81–176 was grown on F1-free MH agar plates overnight. The cells were harvested and suspended in MH to final OD600 nm about 1.2. The cell suspensions were plated on MH agar plates containing 16, 32, or 64 μg/ml of F1. Following four days of incubation under microaerophilic conditions at 42°C, the plates were checked for the emergence of F1 resistant mutants. The F1 pre-adapted cells were also used for in vitro selection of F1 resistant mutants. Briefly, C. jejuni 81–176 F1 were grown in MH broth containing sub-lethal concentration of F1 (4 μg/ml) for five consecutive passages (2 days per passage). Following the fifth passages, cultures were centrifuged, and the pellets were suspended in MH broth to final OD600 nm about 1.2. The bacterial cells suspensions were also plated MH agar plates with increasing concentrations of F1 for selecting F1 resistant mutant as described above. Mutants were randomly selected, grown in MH broth, and subjected to MIC test together with the parent strain C. jejuni 81–176.
Chromosomal DNA was isolated from Campylobacter using the Wizard Genomic Purification Kit (Promega) according to the manufacturer's instructions. Natural transformation (biphasic method) was performed following standard procedure (Wang and Taylor, 1990).
C. jejuni 81–176 (JL28 in Table 1, F1 MIC = 8 μg/ml) was subjected to in vivo transposon mutagenesis using EZ::Tn5™ <KAN-2> Transposome (Epicentre) as detailed in our previous publication (Lin et al., 2009). Briefly, one microliter of EZ-Tn5 <KAN-2> TnP transposome complex containing 25 ng transposon was used to electroporate C. jejuni JL28 competent cells. The Kanr transformants were individually picked and inoculated in 96-well microplates. Following 24 h of incubation, cultures of mutants were replicated into microtiter plates containing 4 μg/ml of F1. Those mutants that could not grow in F1-containing media were selected from the initial plates and subjected to a second screening to confirm increased sensitivities to F1. To confirm specific genetic linkage between the transposon insertion and the increased F1 susceptibility of each mutant, backcrossing of the transposon mutations into wild-type C. jejuni 81–176 was performed using natural transformation. The MICs of the backcrossed mutants for F1 were determined together with parent strain. The specific transposon insertion site of each mutant was determined by directly sequencing the genomic DNA (Lin et al., 2009). Sequence analysis was performed using DNAStar software package.
PCR was performed in a volume of 50 μl containing 1 mM each deoxynucleoside triphosphate, 200 nM primers, 2.0 mM MgCl2, 100 ng of Campylobacter genomic DNA, and 2.5 U of Taq DNA polymerase (Promega) or Pfu Turbo DNA polymerase (Stratagene). Cycling conditions varied according to the estimated annealing temperature of the primers and the expected sizes of the products (available upon request). RNA was extracted from C. jejuni strains using RNeasy Mini Kit (Qiagen) followed by treatment of RNA samples with RNase-free DNase (Qiagen). RT-PCR was conducted using the MasterAmp kit (Epicentre). Cycling conditions for RT-PCR included an initial incubation at 55°C for 30 min and 94°C for 2 min followed by 40 cycles of 94°C for 15s, 52°C for 30s, and 68°C for 1 min, and a final extension at 68°C for 5 min. A RT-PCR mixture lacking the RT was included as a negative control.
To complement cjaB mutation in isogenic CjaB mutant JL657, a 2.7-kb complete cjaAB operon including its 174-bp upstream and 193-bp downstream regions, was PCR amplified from NCTC 11168 using primer pairs of CjaAB-F and CjaAB-R (Table 2 and Figure 1). The PCR was performed using pfu DNA polymerase (Stratagene) and the blunt-ended PCR product was purified and ligated to shuttle vector pRY111 (Yao et al., 1993), which was digested with SmaI prior to ligation. The ligation mix was introduced into DH5α by transformation. One transformant (JL690) with a plasmid bearing intact cjaAB operon (pCjaAB) was created. The pCjaAB from JL690 was then transferred to JL657 by tri-parental conjugation using JL48 as a helper strain (Lin et al., 2005). Similar approach was used to complement tig and cbrR mutations using primer pairs of Td-F/Td-R and Cj0643-F/Cj0643-R, respectively (Table 2 and Figure 1). The complemented strains, together with other related strains, were subjected to AMP killing assay as described below.
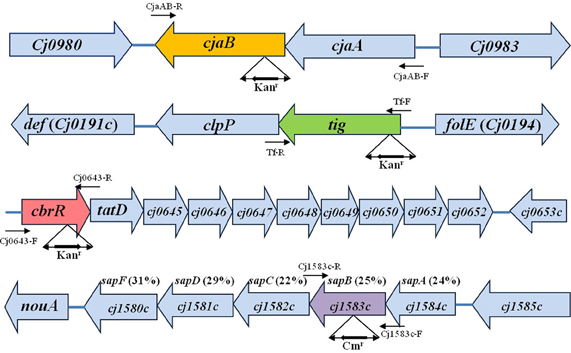
Figure 1. Genomic organization and features of the cjaAB, tig, cbrR, and sap operons in C. jejuni 81–176. The identified ORFs are indicated by boxed arrows. The corresponding gene loci in C. jejuni NCTC 11168 are described in boxed arrows. The locations of major primers used in this study are indicated by arrows. The location and orientation of antibiotic resistance cassette are indicated below each inactivated gene. For sap operon, the aa identity of the putative sap gene to its homolog in S. enreica serovar Typhimurium LT2 is listed in parentheses.
The putative sap genes (Cj1580c to cj1584c, Figure 1) were inactivated by insertional mutagenesis according to our previous publication (Lin et al., 2005). Taking sapB (Cj1583c) as an example, an approximately 2 kb fragment was PCR amplified from genomic DNA of 11168 (JL241) by using primer pair Cj1583c-F and Cj1583c-R (Table 2). The PCR product was cloned into pGEM-T Easy vector (Promega) to generate pSapB (Table 1). The chloramphenicol resistance gene cassette (cat) was PCR amplified from plasmid pRY111 (Yao et al., 1993) by using PfuUltra® High-Fidelity DNA polymerase (Stratagene) and primers of CmF and CmR (Table 2). The resulting blunt-ended PCR product was purified and ligated into pSapB vector, which was digested with SwaI prior to ligation, to generate mutant construct pcmSapB (Table 1). The construct pcmSapB, which serves as suicide vector, was then introduced into 81–176 (JL242) by natural transformation. One transformant, designated JL697, was selected on MH agar containing 5 μg/ml of Cm. The inactivation of putative sapB (Cj1583c) in JL697 was confirmed by PCR (data not shown). A similar site-directed mutagenesis approach was used to create isogenic Cj1584c (sapA), Cj1582c (sapC), Cj1581c (sapD), and Cj1580c (sapF) mutants of 81–176 by using primers pairs Cj1584c-F/R Cj1582c-F/R, Cj1581c-F/R, and Cj1580c-F/R, respectively (Table 2). Notably, due to the absence of a transcriptional terminator downstream of the cat gene and the same orientation of the cat gene with respect to the inactivated gene, inactivation of specific gene likely has no polar effect on the downstream genes; thus, construction of the individual isogenic sap mutants may help us to evaluate their relative contribution to fowlicidin resistance.
The C. jejuni mutants together with their parent strains were subjected to AMP killing assay using 96-well plate as described previously (Mount et al., 2010) with minor modifications. The AMPs used in killing assay include chicken cathelicidin F1, chicken defensin AvBD9 (kindly provided by Dr. Guolong Zhang, Oklahoma State University), bacteriocin OR-7 and E-760 (Lin et al., 2009), and those purchased from Sigma (polymyxin B, colistin, cecropin A, gramicidin, and magananin). Briefly, Campylobacter strains were grown in MH broth to mid-log phase, and the cells were washed with MH broth and diluted to approximately 106 CFU/ml in MH broth. A volume of 180 μl of the diluted cells was mixed with 20 μl MH broth (control) or AMP stock solution (concentration of 10-fold MIC of specific AMP) (treatment). The plates were incubated under microaerophilic conditions at 42°C for 2 h. After 2 h incubation, 20 μl of bacterial culture were taken and serially diluted in MH broth and plated onto MH agar plates. The number of CFU was enumerated after two days of incubation under microaerophilic conditions at 42°C. Percentage survival was calculated by dividing the CFU number of bacteria incubated with AMP relative to those incubated in the presence of MH broth and then multiplied by 100. All assays were carried out in triplicate and two independent experiments were performed. The significance of differences in susceptibility was determined using the Student's t-test.
Because cbrR mutant has been evaluated for its colonization ability in a previous study (Raphael et al., 2005), only sapB as well as tig and cjaB mutants were evaluated together with their parent strain 81–187 in this study. The chicken study was approved by the Institutional Animal Care and Use Committee at The University of Tennessee. Briefly, one-day-old broiler chickens were obtained from a commercial hatchery (Hubbard Hatchery, Pikesville, TN). The chickens were negative for Campylobacter as determined by culturing cloacal swabs prior to use in this study. These chickens were randomly assigned into three treatment group (10 or 11 chicks/group). At four days of age, each chicken was orally inoculated with a 1:1 mixture of wild type 81–176 (JL242) and its isogenic cjaB mutant (group I), tig mutant (group II), or sapB mutant (group III), with a dose of approximately 107CFU of bacteria per chick. For each group, five birds were euthanized and cecal contents were collected at 3, and 10 days post-inoculation (DPI). The cecal contents from each bird were weighed and diluted in MH broth. The cecal suspensions were duplicate plated onto MH agar plates with Campylobacter-specific selective supplements (Oxoid, UK) for total Campylobacter enumeration and onto selective plates supplemented with appropriate antibiotics (30 μg/ml of Kan or 6 μg/ml of Cm) for the specific mutant numbers in each sample. The plating media were tested prior to use to ensure that they supported the growth of the mutant strains. Notably, before inoculation, the motility of the wild-type and its isogenic mutants were confirmed to be at a comparable level.
The number of CFU per gram of cecal contents was calculated for each chicken and was used as an indicator of the colonization level. The detection limit of the plating methods was 100 CFU/g of cecal contents. The bird from which no Campylobacter colonies were detected was assigned a conservative value of 99 CFU/g of cecal contents for the purpose of calculating means and for statistical analysis. Student's t-test was used to examine the significance of differences in Campylobacter colonization levels (log transformed CFU). A P-value of <0.01 was considered significant.
Emergence of acquired AMP resistance is an important issue related to bacterial pathogenesis and development of sustainable peptide antibiotics (Yeaman and Yount, 2003; Hancock and Sahl, 2006; Peschel and Sahl, 2006). Previous studies suggested that high-level resistance to host endogenous AMP is difficult to develop (Peschel and Sahl, 2006), which is also supported by our recent Campylobacter work on the development of acquired resistance to polymyxin B (a widely used model peptide) and bacteriocins (the AMPs produced by bacteria) (Lin et al., 2009; Hoang et al., 2011a,b). Thus, in this study we first performed a screening to determine if high-level F1 resistance is observed in various Campylobacter isolates. Standard MIC test using MH broth showed that majority of C. jejuni and C. coli isolates exhibited low MIC to F1, ranging from 4 μg/ml to 8 μg/ml. Among the 154 tested C. jejuni isolates, 58 and 85 showed MICs of 4 μg/ml and 8 μg/ml, respectively. Only one C. jejuni strain showed slightly higher MIC of 16 μg/ml and only 6.5% of C. jejuni isolates (10 out of 154) showed MICs ≤ 2 μg/ml (eight isolates with MIC of 2 μg/ml and 2 with MIC of 1 μg/ml). C. coli showed similar pattern as C. jejuni and displayed MICs of 0.5 μg/ml (1 isolate), 1 μg/ml (three isolates), 2 μg/ml (three isolates), 4 μg/ml (eight isolates), and 8 μg/ml (five isolates). Together, the MIC survey here showed that none of the strains displayed high-level acquired resistance to F1.
Our recent Campylobacter studies (Lin et al., 2009; Hoang et al., 2011a,b) and work in other bacteria have demonstrated that availability of AMP resistant mutant was very helpful for examination of acquired AMP resistance using functional genomics approaches, such as random transposon mutagenesis and microarray. Thus, in this study we also intended to obtain an in vitro selected F1 resistant mutant. Different in vitro selection methods consistently led to the emergence of F1 resistant colonies on the selective plates containing 32, or 64 μg/ml of F1. Total 24 colonies were randomly selected for MIC test together with their parent strain 81–176. However, after being cultured in F1 free MH broth, none of these isolates displayed higher MIC than 81–176 for F1, indicating all these mutants were false-positive. Despite extensive efforts, no mutant with stable acquired F1 resistance was selected in vitro. In summary, these findings strongly suggest that endogenous AMPs are not favorable for the development of high-level acquired AMP resistance in Campylobacter.
Although we failed to obtain C. jejuni mutants with acquired F1 resistance, examination of intrinsic AMP resistance in wild-type C. jejuni strains would fill a significant knowledge gap and also likely shed light on the mechanisms of acquired AMP resistance in Campylobacter. Thus, in this study a library containing 4800 Kanr mutants were generated for screening the mutants with an increased susceptibility to F1. Three mutants displaying increased sensitivity to F1 were identified. Backcrossing of the transposon mutations into C. jejuni 81–176 further confirmed that the F1 sensitive phenotype in each mutant was linked to the gene with a specific transposon insertion. Direct sequencing of the mutant genomic DNA using transposon-specific primers showed the EZ::TN <KAN-2> transposon inserted into Cj0981c, Cj0193c, and Cj0643 (Figure 1). All the transposon insertions occurred in the coding regions of corresponding genes. The orientations of Kanr cassette within transposon in each mutant were the same as the corresponding disrupted gene (Figure 1). The Cj0981c, Cj0193c, and Cj0643 encode transporter CjaB, putative trigger factor Tig, and the response regulator CbrR, respectively. F1 killing assay further confirmed that the insertional inactivation of cbrR, tig, or cjaB led to significantly increased susceptibility of C. jejuni 81–176 to F1 (Figure 2A). This phenotype was also observed in other strain background (C. jejuni NCTC 11168 and S3B) (Figure 2B). However, mutation of these genes did not confer significantly enhanced susceptibility of these mutants to other tested AMPs (data not shown).
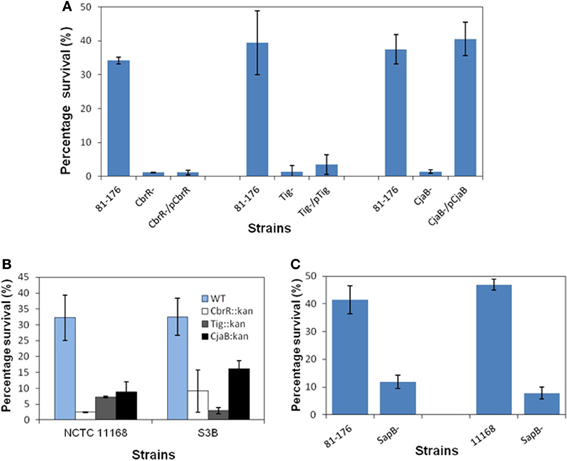
Figure 2. Role of cbrR, tig, cjaB and sapB in F-1 resistance in different C. jejuni strains.(A) Susceptibilities of wild-type 81–176 and its cbrR, tig, and cjaB mutant constructs to F1. (B) Effects of mutations in cbrR, tig, and cjaB on the susceptibilities of C. jejuni NCTC 11168 and S3B to F1. (C) Inactivation of sapB increased susceptibilities of 81–176 and NCTC 11168 to F1. Log-phase cells in MH broth were supplemented with F1 at final concentration of 8 μg/ml and incubated for 2 h at 42°C under microaerophic condition. Samples were diluted and plated on MH agar plates to determine bacterial viability. Percentage survival was calculated by dividing the CFU number of bacteria incubated with AMP relative to those incubated in the presence of MH broth and then multiplied by 100. Each data point represents the mean value obtained from two independent experiments with triplicate measurements in each independent experiment.
Complementation of cjaB mutation in JL657 completely restored F1 resistance level back to the level of parent strain 81–176 (Figure 2A), which is consistent with the report that the cjaB and its upstream gene cjaA formed an operon (Wyszynska et al., 2006; see Figure 1). Complementation of CbrR and Tig mutant had little effect on sensitivity of the mutants to F1 (Figure 2A), likely due to the polar effect of the transposon mutation on the downstream genes in the same operon. The cbrR, a gene encoding a two-component response regulator required for bile salt resistance of Campylobacter (Raphael et al., 2005), appears to form an operon together with its nine downstream genes (Figure 1). For example, the stop codon of cbrR and start codon of tatD overlap by 10 nucleotides (Figure 1). There is no intergenic space between the stop codon of tig and the start codon of downstream clpP, suggesting these two genes are organized into an operon. The clpP is separated from its downstream gene def by a 28 bp intergenic region which is predicted to contain promoter sequence. Thus, tig operon likely contains two genes (tig and clpP) (Figure 1). RT-PCR using specific primers (Table 2) showed that clp and tatD/Cj0645 were barely transcribed in tig and cbrR mutants, respectively, while all the genes were expressed normally in wild-type 81–176 (data not shown), indicating that the insertional mutation in tig and cbrR caused a polar effect on the downstream genes.
The sap (sensitivity to antimicrobial peptide) operon confers resistance to AMP-mediated killing of different pathogens such as Salmonella (Parra-Lopez et al., 1993) and Haemophilus influenza (Mason et al., 2005) although the exact mechanisms of how Sap transporters protect the cells from AMP attack remain unclear. Analysis of NCTC 11168 genome revealed that the operon Cj1580c-Cj1584c shared homology to the identified sap operon in S. enreica serovar Typhimurium LT2 with identical genetic components and organization, which include (Figure 1), in which sapA (Cj1584c) encodes a putative periplasmic peptide binding protein and sapB (Cj1583c) encodes a membrane permease. Comparative genomic analysis of published C. jejuni genomes showed that the putative sapABCDF operon is highly conserved in C. jejuni with nucleotide sequence identity ranging from 98 to 100%. This observation was further confirmed by our PCR survey using sapB specific primer pair (Sap4-F/R, Table 2) and sapA specific primer pair (Sap5-F/R, Table 2) for 27 diverse Campylobacter strains (21 C. jejuni strains and 6 C. coli isolates), in which majority of strains contain sap operon as reflected by the positive PCR results (23 out of 27).
Isogenic SapA, SapB, SapD, and SapF mutants of 81–176 were successfully created in C. jejuni 81–176 (Table 1). Mutation in SapC could not be generated after repeated attempts. RT-PCR analysis using specific primers (Table 2) indicated that insertional inactivation using Cm resistance gene marker did not cause polar effect on the transcription of the genes (Cj1582c, Cj1581c, and Cj1580c) downstream of specific mutated gene (data not shown). As shown in Figure 2C, inactivation of SapB caused significantly in increased sensitivity to F1 in both 81–176 and 11168 strain background. However, mutation of other individual sap genes including sapA (Cj1584c), sapD (Cj1581c), and sapF (Cj1580c) (Figure 1) did not alter sensitivity of the mutants to F1 (data not shown). In addition, isogenic sapB mutant did not display significantly increased susceptibilities to other tested AMPs with diverse sequence and structure (data not shown).
It has been demonstrated that cbrR is required for optimal colonization of Campylobacter in chickens (Raphael et al., 2005). In this study, we compared colonization ability of the isogenic sapB, cjaB, and tig mutants to their parent strain 81–176 using mixed infection in chicken. As shown in Figure 3, inactivation of the sapB, cjaB, or tig gene greatly impaired the colonization ability of C. jejuni 81–176 in chickens. Specifically, when 81–176 and its specific isogenic mutant were co-inoculated into a group of chickens, the colonization level of the specific mutant was significantly lower than that of 81–176 at 3 and 10 days post-inoculation (DPI). In particular, by 10 DPI, the specific mutant of C. jejuni 81–176 (tig−, cjaB−, or sapB− mutant) was no longer detected in any of the cecal samples collected from the chickens inoculated with mixture of 81–176 and its isogenic mutant. It is important to mention that both 81–176 and the isogenic mutants showed similar growth patterns in MH broth (data not shown). We also performed an identical in vitro competition assay in MH broth; all the mutants were not outcompeted by 81–176 and the isogenic mutant displayed an in vitro competitive index (ratio of the mutant to wild-type strain at early stationary phase) of about 1, indicating that growth rate of these mutants is similar to its wild-type 81–176.
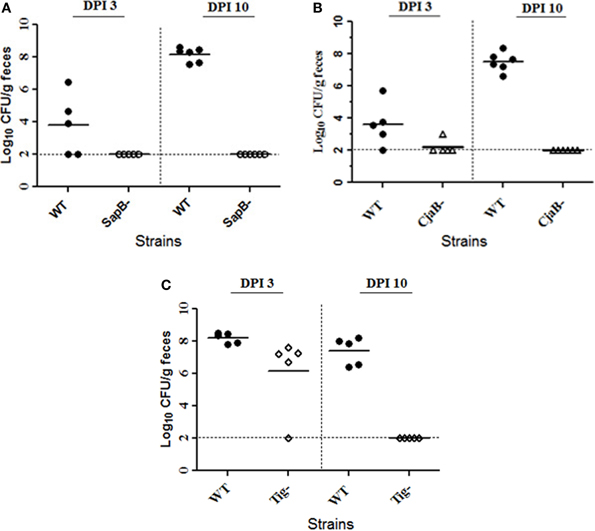
Figure 3. Inactivation of SapB, CjaB, or Tig impaired C. jejuni 81–176 colonization of chickens. Three chicken groups were inoculated with a 1:1 mixture of wild type 81–176 and its isogenic SapB mutant (A), CjaB mutant (B), or Tig mutant (C). For each group, five to six birds were euthanized at the indicated days post-inoculation (DPI) and the cecal contents were collected and used for CFU enumeration. Each symbol indicates the log10 number of CFU/g of cecal contents for a single chicken. The horizontal bars indicate the means of groups at the indicated times. The horizontal dash line indicates the limit of detection.
Development of AMP resistance is an important strategy utilized by many bacterial pathogens including various enteric bacteria to evade host innate immunity and to colonize in various host niches successfully (Ernst et al., 2001; Yeaman and Yount, 2003; Peschel and Sahl, 2006). However, it is still largely unknown how important AMP resistance is for Campylobacter infection and what is the molecular basis of AMP resistance in Campylobacter. To date, there are no any animal knockout experiments demonstrating the critical role of specific host AMP in defense against Campylobacter. However, it has been well established that AMPs (primarily defensins and cathelicidins) are a major component of host innate immunity systems against enteric pathogens. For example, mice deficient in cathelicidin or defensins production are more susceptible to intestinal infections by enteric pathogens (Wilson et al., 1999; Iimura et al., 2005). Thus, both defensins and cathelicidins are important AMPs expressed in the small intestine to protect host against pathogens (Wehkamp et al., 2007). Zilbauer et al. (2005) also observed that epithelial AMPs were induced in human intestinal epithelia upon infection by C. jejuni. In chickens, although different AMPs including the F1 used in this study are expressed in the intestine, the concentrations of specific AMP in any of intestinal niches are still unknown. Specific knock out model should be developed to determine if chicken deficient in F1 production is more susceptible for Campylobacter infection. Notably, colonization of Campylobacter in chicken and other hosts is determined by delicate interactions between Campylobacter and host in which many determinants in addition to AMP are involved. However, given the intimate interaction of Campylobacter with intestinal mucosa and the intracellular stage during systemic Campylobacter infection in humans (invasion of epithelial cells and engulfment by phagocytes), Campylobacter should have co-evolved with host innate defense and developed means to curtail the effect of endogenous AMPs for successful colonization as observed in other enteric pathogens (Ernst et al., 2001; Yeaman and Yount, 2003; Peschel and Sahl, 2006). However, mechanisms of AMP resistance in Campylobacter are still largely unknown.
We have initiated determination of AMP resistance mechanisms in Campylobacter using polymyxin B as a model peptide and have defined seven genetic loci (e.g., galU required for LOS production) contributing to C. jejuni resistance to polymyxin B (Lin et al., 2009). Recent work also showed that production of specific LOS (Naito et al., 2010) and lipidA modification (Cullen and Trent, 2010; van Mourik et al., 2010) were required for polymyxin B resistance in C. jejuni, likely due to their role in electrostatic shielding against polymyxin B. Polymyxin B has been successfully used as a model peptide to study AMP resistance in many Gram-negative pathogens although polymyxin B bears little structural resemblance to the host defense AMPs (Ernst et al., 2001; Yeaman and Yount, 2003; Peschel and Sahl, 2006). However, our previous study (Lin et al., 2009) strongly suggested that polymyxin B may not be a good surrogate for studying AMP resistance in Campylobater. In addition, examination of Campylobacter resistance to bacteriocins revealed a different set of genes which are not involved in resistance to polymyxin B and other endogenous AMPs (Hoang et al., 2011a). Therefore, direct usage of natural host AMP as a model peptide may generate physiologically relevant information for Campylobacter pathogenesis. We chose to use F1 as a surrogate of host defense peptides because of following reasons. First, unlike defensins, the short α-helical fowlicidin-1 (F1) can be easily synthesized at reasonable cost for large scale screening. Second, cathelicidins including F1 are important AMPs expressed in intestine and displayed potent bactericidal effect (Lynn et al., 2004; Xiao et al., 2004, 2006; Peschel and Sahl, 2006; Bommineni et al., 2007). In addition, some genes (e.g., phoPQ) required for cathelicidin resistance could also involve resistance to structurally unrelated AMPs such as defensins (Ernst et al., 2001; Winfield and Groisman, 2004). Thus, genes contributing to F1 resistance may be important for in vivo colonization of C. jejuni in the host. Finally, F1 is produced by chicken, an animal model used in this study, which would make us well correlate the findings obtained from in vitro and in vivo studies.
In this study, inactivation of SapB, CjaB, Tig, and CbrR led to increased susceptibilities to F1 in different C. jejuni strain background. In addition, chicken experiment also showed impaired colonization ability of these mutants in the intestine. The Sap operon encode ABC-type transporter system and have been demonstrated to involve AMP resistance in various Gram-negative bacteria; mutation of the sap operon also resulted in impaired colonization in animal models (Parra-Lopez et al., 1993; Lopez-Solanilla et al., 1998; Chen et al., 2000; McCoy et al., 2001; Lupp et al., 2002; Mason et al., 2005, 2006; Mount et al., 2010). The exact mechanisms of how Sap transporters protect the cells from AMP attack remain unclear. In this study, we demonstrated the role of sapB in Campylobacter resistance to F1; however, mutation of sapB in C. jejuni did not lead to significantly increased susceptibilities to other natural AMPs. We should interpret this finding cautiously. The C. jejunisap may still interact to other structurally unrelated AMPs with lower efficiency than F1; however, limited expression level of Sap in our in vitro killing assay system may diminish the susceptibility difference between Sap mutant and wild-type strain. Mason et al. (2003, 2005) reported that sap expression of H. influenza is greatly up-regulated in vivo. Therefore, the sap operon of Campylobacter is possible up-regulated during in vivo infection and such enhanced expression of sap may ultimately confer C. jejuni resistance to various endogenous AMPs, leading to greatly impaired intestinal colonization observed in this study (Figure 3A). This hypothesis needs to be examined in the future. In terms of CjaAB operon, our chicken study has clearly showed its role in C. jejuni colonization in the intestine, likely due to its contribution to AMP resistance. The CjaB was predicted as an inner membrane transporter with unknown function in Campylobacter (Wyszynska et al., 2006). The underlying mechanism for CjaAB conferring F1 resistance is not clear. It has been observed that efflux pumps are involved in AMP resistance in various bacteria (Yeaman and Yount, 2003; Peschel and Sahl, 2006). Recently, we also demonstrated that CmeABC efflux pump is required for Campylobacter resistance to bacteriocins (Hoang et al., 2011a). Therefore, it is likely that CjaAB function as an efflux pump to extrude F1 out of cells for resistance. However, it is equally possible that CjaAB may function as an influx transporter and facilitate transportation of F1 to Campylobacter cytoplasm where F1 is targeted for degradation.
Given the failure of complementation and lack of expression of the genes downstream of the inactivated tig and cbrR, the phenotypes of isogenic tig and cbrR mutants observed in this study are likely caused by other gene(s) in the same operon or the coordination of all genes within the operon (Figure 1). Trigger factor Tig was found highly conserved in eubacteria, functioning as a chaperon to interact with newly synthesized polypeptides, assist protein folding, and plays a vital role in bacterial virulence (Rassow and Pfanner, 1996; Martinez-Hackert and Hendrickson, 2009). The gene immediately downstream of tig in Campylobacter was annotated as clpP that function as a protease to degrade misfolding peptides in other bacteria (Frees et al., 2007). Notably, the ClpP homolog of Bacillus anthracis has been demonstrated to be involved in resistance to AMPs human cathelicidin LL-3 (McGillivray et al., 2009). Therefore, since degradation of AMPs by protease is one of mechanisms used by bacteria to resist endogenous AMPs (Yeaman and Yount, 2003), the C. jejuni ClpP may involve proteolytic cleavage and play a role in virulence by mediating the role in AMP resistance. This hypothesis needs to be examined in the future. Recently, Cohn et al. (2007) has characterized the C. jejuni ClpP and observed that ClpP influenced heat tolerance of C. jejuni. Therefore, the impaired colonization of tig mutant observed in this study also could be attributed to the temperature sensitivity due to inactivation of ClpP. Similarly, cbrR and nine of its downstream genes appear to organize into an operon (Figure 1) and some downstream genes may be involved in AMP resistance in C. jejuni. For example, Cj0649 encodes a beta-barrel LptD-like protein and the LptD in Pseudomonas aeruginosa appeared to act as a dominant resistance marker against cathelicidins likely due to its function in the assembly of LPS in the outer leaflet of the outer membrane (Srinivas et al., 2010). However, the two component regulators identified in this study (CbrR) and in our recent work (RacR) (Lin et al., 2009) may still involve Campylobacter resistance to AMPs. In response to environmental cues via two-component regulatory systems (e.g., PhoP/PhoQ), Gram-negative bacteria can add covalent modification to LPS and consequently reduce the negative charge and/or membrane fluidity of LPS and protect themselves from attack by AMPs, which has been a major AMP resistant mechanism in enteric bacteria (Ernst et al., 2001; Yeaman and Yount, 2003; Peschel and Sahl, 2006). Mutations occurred in two-component regulators could result in sustained, acquired AMP-resistance. To hunt potential two-component regulators required for AMP resistance, we have performed sequencing of nine regulators, five histidine kinases, and six signal transduction systems in polymyxin B resistant mutant; but no sequence difference was observed in all selected regulators between wild-type 81–176 and its polymyxin B resistant derivatives (Hoang, 2010). Despite accumulating studies on the functional characterization of two-component regulators in C. jejuni, it is still unknown which environmental signals to which these regulators respond. We speculate that some two-component regulatory systems in C. jejuni can sense and integrate multiple environmental cues in the host into a coordinated cellular response and promote Campylobacter resistance to AMPs in different host niches. Testing this hypothesis relies on the identification of specific cues activating two-component regulatory systems, a challenge issue to elucidate physiology and pathogenesis role of two-component regulatory systems in bacterial pathogens.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by grant 1 R21 AI069133–01A2 from NIH.
AMP, antimicrobial peptide; F1, fowlicidin-1.
Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206.
Black, R. E., Levine, M. M., Clements, M. L., Hughes, T. P., and Blaser, M. J. (1988). Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157, 472–479.
Bommineni, Y. R., Dai, H. E., Gong, Y. X., Soulages, J. L., Fernando, S. C., Desilva, U., Prakash, O., and Zhang, G. L. (2007). Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. FEBS J. 274, 418–428.
Chen, H. Y., Weng, S. F., and Lin, J. W. (2000). Identification and analysis of the sap genes from Vibrio fischeri belonging to the atp-binding cassette gene family required for peptide transport and resistance to antimicrobial peptides. Biochem. Biophys. Res. Commun. 269, 743–748.
Cohn, M. T., Ingmer, H., Mulholland, F., Jorgensen, K., Wells, J. M., and Brondsted, L. (2007). Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 73, 7803–7813.
Cullen, T. W., and Trent, M. S. (2010). A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 107, 5160–5165.
Ernst, R. K., Guina, T., and Miller, S. M. (2001). Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3, 1327–1334.
Frees, D., Savijoki, K., Varmanen, P., and Ingmer, H. (2007). Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63, 1285–1295.
Friedman, C. R., Neimann, J., Wegener, H. C., and Tauxe, R. V. (2000). “Epidemiology of Campylobacter jejuni infections in the United States and other Industrialized Nations,” in Campylobacter, 2nd ed., eds I. Nachamkin and M. J. Blaser (Washington, DC: American Society for Microbiology), 121–138.
Hancock, R. E. W., and Sahl, H. G. (2006). Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557.
Hoang, K. V. (2010). Mechanisms of Antimicrobial Peptide Resistance in Campylobacter. Ph.D. dissertation, University of Tennessee. http://trace.tennessee.edu/utk_gra-ddiss/887
Hoang, K. V., Stern, N. J., Saxton, A. M., Xu, F., Zeng, X., and Lin, J. (2011a). Prevalence, development, and molecular mechanisms of bacteriocin resistance in Campylobacter. Appl. Environ. Microbiol. 77, 2309–2316.
Hoang, K. V., Stern, N. J., and Lin, J. (2011b). Development and stability of bacteriocin resistance in Campylobacter spp. J. Appl. Microbiol. 111, 1544–1550.
Iimura, M., Gallo, R. L., Hase, K., Miyamoto, Y., Eckmann, L., and Kagnoff, M. F. (2005). Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J. Immunol. 174, 4901–4907.
Lin, J., Akiba, M., Sahin, O., and Zhang, Q. J. (2005). CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 49, 1067–1075.
Lin, J., Michel, L. O., and Zhang, Q. J. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131.
Lin, J., Wang, Y., and Hoang, K. V. (2009). Systematic identification of genetic loci required for polymyxin resistance in Campylobacter jejuni using an efficient in vivo transposon mutagenesis system. Foodborne Pathog. Dis. 6, 173–185.
Lopez-Solanilla, E., Garcia-Olmedo, F., and Rodriguez-Palenzuela, P. (1998). Inactivation of the sapa to sapf locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell 10, 917–924.
Lupp, C., Hancock, R. E., and Ruby, E. G. (2002). The Vibrio fischeri sapABCEF locus is required for normal growth, both in culture and in symbiosis. Arch. Microbiol. 179, 57–65.
Lynn, D. J., Higgs, R., Gaines, S., Tierney, J., James, T., Lloyd, A. T., Fares, M. A., Mulcahy, G., and O'Farrelly, C. (2004). Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 56, 170–177.
Martinez-Hackert, E., and Hendrickson, W. A. (2009). Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138, 923–934.
Mason, K. M., Bruggeman, M. E., Munson, R. S., and Bakaletz, L. O. (2006). The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and sapd-dependent potassium acquisition. Mol. Microbiol. 62, 1357–1372.
Mason, K. M., Munson, R. S., and Bakaletz, L. O. (2003). Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71, 3454–3462.
Mason, K. M., Munson, R. S., and Bakaletz, L. O. (2005). A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect. Immun. 73, 599–608.
McCoy, A. J., Liu, H., Falla, T. J., and Gunn, J. S. (2001). Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45, 2030–2037.
McGillivray, S. M., Ebrahimi, C. M., Fisher, N., Sabet, M., Zhang, D. X., Chen, Y., Haste, N. M., Aroian, R. V., Gallo, R. L., Guiney, D. G., Friedlander, A. M., Koehler, T. M., and Nizet, V. (2009). ClpX contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. J. Innate Immun. 1, 494–506.
Mount, K. L. B., Townsend, C. A., Rinker, S. D., Gu, X. P., Fortney, K. R., Zwickl, B. W., Janowicz, D. M., Spinola, S. M., Katz, B. P., and Bauer, M. E. (2010). Haemophilus ducreyi sapA contributes to cathelicidin resistance and virulence in humans. Infect. Immun. 78, 1176–1184.
Naito, M., Frirdich, E., Fields, J. A., Pryjma, M., Li, J., Cameron, A., Gilbert, M., Thompson, S. A., and Gaynor, E. C. (2010). Effects of sequential Campylobacter jejuni 81–176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192, 2182–2192.
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T., Holroyd, S., Jagels, K., Karlyshev, A. V., Moule, S., Pallen, M. J., Penn, C. W., Quail, M. A., Rajandream, M. A., Rutherford, K. M., van Vliet, A. H., Whitehead, S., and Barrell, B. G. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668.
Parra-Lopez, C., Baer, M. T., and Groisman, E. A. (1993). Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12, 4053–4062.
Peschel, A., and Sahl, H. G. (2006). The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4, 529–536.
Raphael, B. H., Pereira, S., Flom, G. A., Zhang, Q., Ketley, J. M., and Konkel, M. E. (2005). The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J. Bacteriol. 187, 3662–3670.
Rassow, J., and Pfanner, N. (1996). Protein biogenesis: chaperones for nascent polypeptides. Curr. Biol. 6, 115–118.
Srinivas, N., Jetter, P., Ueberbacher, B. J., Werneburg, M., Zerbe, K., Steinmann, J., van der Meijden, B., Bernardini, F., Lederer, A., Dias, R. L., Misson, P. E., Henze, H., Zumbrunn, J., Gombert, F. O., Obrecht, D., Hunziker, P., Schauer, S., Ziegler, U., Kach, A., Eberl, L., Riedel, K., Demarco, S. J., and Robinson, J. A. (2010). Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013.
van Dijk, A., Veldhuizen, E. J., Kalkhove, S. I., Tjeerdsma-van Bokhoven, J. L., Romijn, R. A., and Haagsman, H. P. (2007). The beta-defensin gallinacin-6 is expressed in the chicken digestive tract and has antimicrobial activity against food-borne pathogens. Antimicrob. Agents Chemother. 51, 912–922.
van Mourik, A., Steeghs, L., van Laar, J., Meiring, H. D., Hamstra, H. J., van Putten, J. P. M., and Wosten, M. M. S. M. (2010). Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J. Biol. Chem. 285, 15828–15836.
Wang, Y., and Taylor, D. E. (1990). Natural transformation in Campylobacter species. J. Bacteriol. 172, 949–955.
Wehkamp, J., Schauber, J., and Stange, E. F. (2007). Defensins and cathelicidins in gastrointestinal infections. Curr. Opin. Gastroenterol. 23, 32–38.
Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M., and Parks, W. C. (1999) Regulation of intestinal alphadefensin activation by the metalloproteinase matrilysin in innate host defense. Science 286, 113–117.
Winfield, M. D., and Groisman, E. A. (2004). Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc. Natl. Acad. Sci. U.S.A. 101, 17162–17167.
Wyszynska, A., Pawlowski, M., Bujnicki, J., Pawelec, D., van Putten, J. P., Brzuszkiewicz, E., and Jagusztyn-Krynicka, E. K. (2006). Genetic characterisation of the cjaAB operon of Campylobacter coli. Pol. J. Microbiol. 55, 85–94.
Xiao, Y., Hughes, A. L., Ando, J., Matsuda, Y., Cheng, J. F., Skinner-Noble, D., and Zhang, G. (2004). A genome-wide screen identifies a single beta-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics 5, 56.
Xiao, Y., Cai, Y., Bommineni, Y. R., Fernando, S. C., Prakash, O., Gilliland, S. E., and Zhang, G. (2006). Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 281, 2858–2867.
Yao, R., Alm, R. A., Trust, T. J., and Guerry, P. (1993). Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130, 127–130.
Yeaman, M. R., and Yount, N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55.
Keywords: Campylobacter, fowlicidin, antimicrobial peptide, resistance
Citation: Hoang KV, Wang Y and Lin J (2012) Identification of genetic loci that contribute to Campylobacter resistance to fowlicidin-1, a chicken host defense peptide. Front. Cell. Inf. Microbio. 21:32. doi: 10.3389/fcimb.2012.00032
Received: 23 November 2011; Accepted: 29 February 2012;
Published online: 16 March 2012.
Edited by:
Alain Stintzi, Ottawa Institute of Systems Biology, CanadaReviewed by:
Eric Déziel, Institut National de la Recherche Scientifique, CanadaCopyright: © 2012 Hoang, Wang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jun Lin, Department of Animal Science, The University of Tennessee, 2640 Morgan Circle Drive, Knoxville, TN 37996-4574, USA. e-mail:amxpbjZAdXRrLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.