- School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, NSW, Australia
Over the last decade Campylobacter concisus, a highly fastidious member of the Campylobacter genus has been described as an emergent pathogen of the human intestinal tract. Historically, C. concisus was associated with the human oral cavity and has been linked with periodontal lesions, including gingivitis and periodontitis, although currently its role as an oral pathogen remains contentious. Evidence to support the role of C. concisus in acute intestinal disease has come from studies that have detected or isolated C. concisus as sole pathogen in fecal samples from diarrheic patients. C. concisus has also been associated with chronic intestinal disease, its prevalence being significantly higher in children with newly diagnosed Crohn’s disease (CD) and adults with ulcerative colitis than in controls. Further C. concisus has been isolated from biopsy specimens of patients with CD. While such studies support the role of C. concisus as an intestinal pathogen, its isolation from healthy individuals, and failure of some studies to show a significant difference in C. concisus prevalence in subjects with diarrhea and healthy controls has raised contention as to its role in intestinal disease. Such findings could argue against the role of C. concisus in intestinal disease, however, the fact that C. concisus strains are genetically diverse raises the possibility that differences exist in their pathogenic potential. Evidence to support this view comes from studies showing strain specific differences in the ability of C. concisus to attach to and invade cells and produce virulence factors, including toxins and hemolytic phospholipase A. Further, sequencing of the genome of a C. concisus strain isolated from a child with CD (UNSWCD) and comparison of this with the only other fully sequenced strain (BAA-1457) would suggest that major differences exist in the genetic make-up of this species which could explain different outcomes of C. concisus infection.
Introduction
Members of the Campylobacter genus are fastidious Gram-negative spiral or curved shaped rods, which grow under microaerobic or anaerobic conditions. The majority of species are motile, having a single polar flagellum at one or both ends of the cell (Vandamme et al., 2005; Man, 2011). The most well-studied species of the Campylobacter genus is the intestinal pathogen Campylobacter jejuni, a bacterium currently recognized as the leading cause of acute diarrhea and gastroenteritis worldwide, accounting for 400 million cases of gastroenteritis in adults and children each year (Blaser et al., 1983; Tauxe et al., 1987; Allos, 2001). Additionally in a small number of those acutely infected with C. jejuni a number of serious sequelae can occur including Guillain–Barré syndrome and reactive arthritis (Altekruse et al., 1999). In addition to C. jejuni, C. coli has also been recognized as an important cause of gastroenteritis accounting for 5–18.6% of cases of gastroenteritis caused by Campylobacter spp. (Friedman et al., 2004; Valenza et al., 2010). While from a clinical perspective C. jejuni and C. coli are major foci of attention worldwide, over the last decade mounting evidence has accrued that would suggest that other members of the Campylobacter genus including C. concisus, C. gracilis, C. upsaliensis, and C. lari may also play a role in intestinal disease. While evidence to support the role of each of these four non-jejuni/coli Campylobacter species in intestinal disease is increasing (a recent review by Man, 2011 provides a comprehensive reference source for the clinical importance of emerging Campylobacter species), evidence for the role of C. concisus is perhaps the most contentious.
Campylobacter concisus was first isolated and named by Tanner et al. (1981). In a study, which compared 46 Gram-negative, asaccharolytic, rod shaped bacteria isolated from humans with periodontal disease with 10 reference strains including Eikenella corrodens, Vibrio succinogenes (now known as Wolinella succinogenes), Bacteroides ureolyticus (now known as C. ureolyticus), and Campylobacter species, Tanner et al. (1981) identified six isolates from patients with gingivitis and periodontitis that were described as non-corroding, microaerophilic, Gram-negative, rods predominantly curved in shape, with deoxyribonucleic acid guanine-plus-cytosine contents of 34–38%, which they proposed should be named “Campylobacter concisus sp. nov.” (type strain, ATCC 33237). Although it is now 30 years since Tanner and colleagues first named C. concisus, advances in our understanding of the required growth conditions, phenotypic characteristics (Table 1; Tanner et al., 1981; Vandamme et al., 2005), genetic make-up, and pathogenic potential of C. concisus have been relatively slow, and somewhat sporadic, with only approximately 80 papers relating to C. concisus being published over the last 30 years.
While initially studies focused upon the role of C. concisus in periodontal disease, over the last 20 years the major research focus has predominantly been investigation of the role of C. concisus in intestinal disease, including enteritis and more recently inflammatory bowel diseases (IBD). Although a relatively large number of studies have examined the relationship between C. concisus and intestinal disease there still remains no consensus as to its role. By enlarge, this relates to the fact that in many studies no significant difference was detected between the prevalence of C. concisus in those with intestinal disease and healthy controls (Van Etterijck et al., 1996; Lawson et al., 1998; Inglis et al., 2011). While these findings per se would argue against such a link, the fact that there is an extremely high degree of genetic heterogeneity among C. concisus strains raises the possibility that only some C. concisus strains have the ability to cause intestinal disease. Given this, a number of studies have attempted to link specific genomospecies of C. concisus with disease outcome, however, although some progress has been made in this area no specific C. concisus genomospecies has been associated with disease outcome. This review aims to provide an overview of our current understanding of the association between C. concisus and oral and intestinal disease, the importance of genetic diversity and its association with disease outcome, the pathogenic potential of C. concisus, as well as potential reservoirs of infection and susceptibility to antibiotics.
Association with Disease
Oral Diseases
As described above C. concisus was first recognized and named as a member of the microflora of the oral cavity by Tanner et al. (1981). The isolation of six C. concisus strains from three subjects with gingivitis and three with periodontitis raised the possibility that an association may exist between C. concisus and oral diseases (Tanner et al., 1981, 1987). Following Tanner’s paper further reports of the isolation of C. concisus from the oral cavity of small numbers of subjects were published. For example, Tyrrell et al. (2003) isolated C. concisus from a patient with oral malodor, while Haffajee et al. (1984) reported the level of C. concisus to be associated with a gain in tooth attachment in a subject with periodontitis.
In one of the few studies to examine the immune response to C. concisus in periodontal disease and controls, Ebersole et al. investigated the longitudinal effects on systemic antibody levels to a range of oral microorganisms known to exist in subjects with different periodontal diseases. This showed that in adult patients with periodontitis undergoing subgingival scaling, antibodies to C. concisus were elevated as compared with that in the normal population (Ebersole et al., 1985). This association was supported by a study by Taubman et al. (1992) who also found higher antibody levels to C. concisus in periodontally diseased subjects when compared to healthy subjects.
Moore et al. (1987) compared the subgingival bacterial flora cultured from children and adults with naturally occurring gingivitis with that in healthy controls and subjects with moderate, severe, and juvenile periodontitis. This showed that in both children and adults with naturally occurring gingivitis, C. concisus was more numerous than in healthy controls or those with periodontitis. Based on their findings Moore et al. suggested that the composition of the subgingival microflora represented a transition between that associated with health and periodontitis. In a similar study Macuch and Tanner (2000) compared the presence of Campylobacter spp. in healthy subjects with that in subjects with gingivitis, initial periodontitis, and established periodontitis. While C. concisus was more frequently isolated from subjects with initial periodontitis (approximately 68%) than healthy individuals (35%), the isolation rates from those with gingivitis (32%), or established periodontitis (32%) did not differ from that in controls (Macuch and Tanner, 2000). Interestingly, the proportion of C. concisus as a percentage of all Campylobacter species was higher in shallow pocket sites (19.7%) as compared with deep pocket sites (6.6%) and healthy controls.
In a further study that examined the composition of the subgingival microbiota of children, Kamma et al. isolated 45 microbial species from subgingival plaque of both permanent and deciduous teeth of systemically healthy children. While C. concisus was not one of the most frequently detected species, its detection rate in permanent teeth was significantly higher than that in deciduous teeth (P < 0.001). Furthermore the presence of C. concisus was significantly associated with the bleeding index in permanent teeth (Kamma et al., 2000a,b), a finding that was in line with a previous study by Kamma et al. that showed C. concisus to be associated with bleeding in young adults with rapidly progressive periodontitis (Kamma et al., 1994; Nakou et al., 1998). Furthermore, Kamma et al. found C. concisus to be present in significantly higher numbers, and more frequently, in smokers as compared with non-smokers (Kamma and Nakou, 1997; Kamma et al., 1999).
Given reports that the enzyme aspartate aminotransferase (AST) is markedly elevated in gingival crevicular fluid (GCF) from sites with severe gingival inflammation and progressive attachment loss, Kamma et al. (2001) determined the association between AST activity in GCF and the subgingival microbiota in periodontal sites of individuals with early onset periodontitis. This showed the isolation frequency of seven bacterial species, including C. concisus, to be significantly higher at AST positive sites than at AST negative sites. Furthermore, C. concisus had the fourth strongest positive association with AST activity (Odds Ratio 3.9; 95% CI: 1.91–22.36; Kamma et al., 2001).
While there have been further sporadic reports of the detection and isolation of C. concisus from subjects with periodontal disease over the last 10 years, there is currently limited evidence to support a role for C. concisus in oral disease. Given that many of the studies reported only the isolation of C. concisus from sites of oral disease, rather than comparing this with that in healthy controls, assessment of the role of C. concisus in oral disease is problematic. Thus, currently it remains unclear whether C. concisus is an oral pathogen, an opportunistic pathogen in inflamed areas or is simply a commensal of the oral cavity.
Enteritis
Johnson and Finegold (1987) published the first evidence that C. concisus did not exclusively colonize the oral cavity. While this finding was of significant interest, it was a study by Vandamme et al. (1989) 2 years later that provided the first convincing evidence that C. concisus was present in the gastrointestinal tract (GIT) of humans. In their study Vandamme et al. (1989) used protein analysis, immunotyping, DNA hybridization, and DNA base analysis to determine the species identity of 14 Campylobacter species previously isolated from a blood culture, two antral biopsies, a duodenal aspirate, two esophageal biopsies and the feces of eight patients with persistent diarrhea, which had previously, based on a small number of phenotypic tests and immune-typing, been provisionally grouped and named EF-group 22. Based on their testing of these isolates as well as a range of Campylobacter reference strains, all 14 were shown to be C. concisus a finding that led Vandamme et al. (1989) to conclude that fecal carriage of C. concisus may be common, and might be associated with gastrointestinal disease.
Further evidence that C. concisus could be isolated from fecal samples of subjects with diarrhea came from a prospective study of Belgian children and adults conducted by Lauwers et al. In this study, C. concisus was isolated from the stools of 2.4% of children (n = 3165) and 1.5% of adults (n = 1265) with diarrhea (Lauwers et al., 1991). Of those positive for C. concisus, 20% of children, and 10.5% of adults harbored another enteropathogen. Interestingly, 54% of the C. concisus-positive children were <1 year of age while only 9% were positive in the age group >5 years. Of the 78 children in whom only C. concisus was isolated, 72% had enteritis symptoms (62% diarrhea and 22% vomiting) while 89.4% of adults had diarrhea. Lauwers et al. (1991) concluded that although C. concisus may be commonly found in the feces of children and adults, further studies were necessary to determine its pathogenic role in the human intestinal tract. In contrast, in a study comparing the isolation rates of Campylobacter species from stool samples of 867 adults with diarrhea and 1077 children (<10 years; 90% with diarrhea), Lindblom et al. (1995) failed to isolate C. concisus from any adult stool sample, although stool samples from four children with diarrhea were positive, which represented 4% of all Campylobacter species isolated. Moreover, following the detection of C. concisus from fecal samples of healthy individuals (3/20; 15%), Lawson et al. (1998) also suggested that C. concisus might only be transiently present within the GIT.
Figura et al. (1993) reported the isolation of C. mucosalis from the feces of two children with enteritis (NCTC 12408 and NCTC 12407) from Sienna, Italy. The classification of these isolates as C. mucosalis was however later challenged by Lastovica et al. (1993) due to the limited number of phenotypic tests used by Figura et al. to classify them. Subsequently, based on rigorous phenotypic analysis, consisting of 64 individual phenotypic tests, On (1994) confirmed NCTC 12408 (NCTC 12407 was no longer extant) to be C. concisus. In a large study also conducted in the Sienna/Tuscany region of Italy, Musmanno et al. investigated the presence of unusual Campylobacter species in fecal samples of 288 Italian children with enteritis. Based on a filtration technique C. jejuni subsp. jejuni was isolated from 6.9% of children, C. coli from 2%, and each of C. jejuni subsp. doylei, C. upsaliensis, and C. concisus from 0.7% of children (Musmanno et al., 1998). While the prevalence of C. concisus in these Italian children was low (0.7%), in the same year a significantly higher isolation rate of C. concisus was reported in South African children (Lastovica, 2006). In his study, which used the Cape Town Protocol to determine the distribution of Campylobacter and related species in diarrheic stools collected from children attending the Red Cross Children’s Hospital in Cape Town, Lastovica reported C. concisus to represent the third most common of the Campylobacter and related species isolated (399/2351; 17.0%), the most common being C. jejuni (33.4%) followed by C. upsaliensis (23.7%). Unfortunately, as this study did not include a healthy control group, the role of C. concisus in diarrheal disease could not be assessed.
The importance of Campylobacter species other than C. jejuni and C. coli in diarrheal disease was also investigated by Aabenhus et al. (2002b) who determined the prevalence of C. concisus infection in 11,550 stool samples obtained from patients with enteric disease, using a filter isolation method. Of the 224 Campylobacter isolates cultured, 110 (49%) were shown to be C. concisus, with concomitant infection, with an established gastrointestinal pathogen being found in 27% of the 103 patients from whom these isolates were cultured. No conclusive association was observed between any specific age group and C. concisus infection. Interestingly, of the infected patients 73 (70.9%) were immunocompromised the majority of whom were adults (84%). Based on these findings, Aabenhus et al. (2002b) concluded that the prevalence of C. concisus was likely to be significantly underestimated, and that although C. concisus could be isolated from both immunocompetent and immunocompromised adult patients, immunocompromised patients appeared to be at a higher risk of infection.
Maher et al. (2003) employed both culture and a 16S/23S PCR/DNA probe assay to detect Campylobacter species in 320 consecutive liquid and semi-solid fecal samples from subjects with acute infectious gastroenteritis, and found Campylobacter DNA, other than C. jejuni or C. coli, to be present in 30% of fecal specimens (including C. curvus, C. concisus, and C. gracilis). Furthermore, Kulkarni et al. (2002) reported the isolation of C. concisus using membrane filtration and selective culture from only 1 of 343 stool samples collected from patients with gastroenteritis.
In a recent epidemiological study conducted over a 22-month period commencing January 2009, Nielsen et al. (2011a) which investigated the prevalence of Campylobacter species and other enteric bacterial pathogens in 10,388 stool samples using a filter isolation technique, found the most prevalent enteric pathogen isolated from these stools was C. jejuni (456 patients) followed by C. concisus (378 patients). Interestingly, C. concisus was found to be most frequently isolated in young children and elderly patients, a finding consistent with that of Engberg et al. (2000). Moreover, in the Nielsen study elderly patients with C. concisus tended to present with a more long-lasting diarrhea.
In the first of a limited number of studies to investigate the pathogenic role of C. concisus in intestinal disease, Van Etterijck et al. (1996) used a filter isolation technique to isolate C. concisus from children with enteritis (n = 174) and a control population (n = 958) with no diarrhea. This study showed that while the isolation rate of C. concisus in children with diarrhea (13%) was higher than that in the control children (9%) this did not reach significance (P = 0.15), which led Van Etterijck et al. to conclude that C. concisus should not be considered a primary pathogen associated with gastrointestinal disease. In a further study which used a range of culture techniques to isolate Campylobacter species from the feces of Danish children and adults with diarrhea, Engberg et al. (2000) reported the isolation of C. concisus from a large number of diarrheal cases, particularly from those at the extremes of age (0–9 and >60 years). However, similar to the findings of Van Etterijck, when the isolation rates of C. concisus in patients with diarrhea were compared with healthy controls, isolation rates in those with diarrhea (5.6%) were not significantly different to that in controls (2.8%).
More recently, Inglis et al. (2011) investigated the presence of Campylobacter species in stools from diarrheic (n = 442) and healthy (n = 58) humans living in southwestern Alberta using PCR. In contrast to other studies, in this population the prevalence of C. concisus DNA as detected by two primer sets was significantly higher (P < 0.001) in healthy subjects (52%) as compared with subjects with diarrhea (34.8%).
Although overall these findings support the presence of C. concisus in the intestinal tract of healthy humans and those with diarrheal disease, evidence to support a role for C. concisus in acute enteritis, although increasing, remains tenuous.
Inflammatory Bowel Diseases
Inflammatory bowel diseases are chronic inflammatory conditions of the GIT, with currently unknown etiology and rising global incidence (Economou and Pappas, 2008). The two major forms of IBD are Crohn’s disease (CD) and ulcerative colitis (UC). Both forms have distinct phenotypical characteristics: CD inflammation may occur in any location along the GIT, with transmural lesions that may extend through the mucosal surface and into the underlying serosa, whilst UC is characterized by continuous sub-mucosal inflammation occurring only in the colon (Colletti, 2004). A histopathological hallmark of CD is the presence of compact granulomas, composed of macrophages, giant cells, and epithelioid cells. The prominent histological features of UC include the presence of an elevated number of polymorphonuclear cells within the lamina propria as well as in the intestinal crypts, in which micro-abscesses form (Colletti, 2004). Research indicates that the etio-pathogenesis of CD and UC are to some extent different; yet, the general hypothesis regarding their etiology is that gastrointestinal microbes or their by-products, in association with a disrupted gastrointestinal epithelium and/or environmental trigger, propagate a dysregulated immune response leading to chronic inflammation in genetically predisposed individuals (Man et al., 2011).
The possible association between Campylobacter species and IBD was first investigated in 1984. While initial studies examining a possible link between C. jejuni and IBD failed to provide evidence for an association (Blaser et al., 1984; Weber et al., 1992; Boyanova et al., 2004), more recent studies would suggest that C. jejuni infection may be associated with increased IBD risk and flare ups of IBD (Gradel et al., 2009).
Interestingly, Aabenhus et al. (2002a) reported the isolation of C. concisus from fecal samples of IBD patients. However, it was not until 2009 that Zhang et al. (2009) compared the prevalence of C. concisus in patients with CD and controls. In this study which investigated a possible association between C. concisus and newly diagnosed pediatric CD, a significantly higher prevalence of C. concisus DNA (P < 0.0001) as well as C. concisus specific IgG antibody levels (P < 0.001) were detected in children with newly diagnosed CD (51%; 0.991 ± 0.447) as compared with controls (2%; 0.329 ± 0.303; Zhang et al., 2009). Additionally, C. concisus UNSWCD was successfully isolated from an intestinal biopsy of a child with CD (Zhang et al., 2009), indicating that viable bacteria were present within the patient’s intestine. These findings were later confirmed in a larger cohort of children where detection of C. concisus DNA from fecal samples showed that 65% (35/54) of CD patients were positive, which was significantly higher than that in healthy (33%, 11/33) and non-IBD controls (37%, 10/27; Man et al., 2010b). Recently, Hansen et al. (2011) also reported the detection rate of C. concisus DNA in intestinal biopsies of pediatric patients with IBD (14/24; 58.3%), specifically CD (8/12; 66.7%), was higher than that in controls (11/26; 42.3%), however in their study the difference was not significant, which most likely relates to the relatively small number of subjects in their study.
These findings in children have recently been supported by studies in adult CD patients, where a significantly higher prevalence of C. concisus DNA was detected in colonic biopsies of CD patients (53%) as compared with controls (18%, P < 0.05; Mahendran et al., 2011). Further indirect evidence of a possible association between C. concisus and CD comes from the finding that rifaximin, an antibiotic with good activity against Campylobacter species, is effective for adults with active CD, and when combined with long-term administration of probiotics can induce and maintain remission of CD (Guslandi et al., 2009).
In a recent study, Kovach et al. (2011) investigated C. concisus immunoreactive proteins in the sera of C. concisus-positive children with CD, and identified 37 proteins of which flagellin B, ATP synthase F1 alpha subunit and outer membrane protein 18 were predominant antigens recognized by all CD patients. At least six of the identified proteins functioned in adhesion to the host cell, suggesting that this bacterium can cross the mucus layer and attach to the intestinal epithelium (Kovach et al., 2011).
Of significant interest is the recent finding by Mukhopadhya et al. (2011) that C. concisus may also be associated with UC. In their study the prevalence of C. concisus DNA in biopsy specimens from adults with UC was shown to be significantly increased (P = 0.0019) in UC patients (33.3%; 23/69) as compared with controls (10.8%; 7/65). This finding is supported by the study of Mahendran et al. (2011) who not only isolated C. concisus from patient biopsies, but also determined that C. concisus positivity in UC patients (77%) was significantly higher than that in controls (36%, P < 0.05). In a study to investigate the presence of Campylobacter species in IBD, Tankovic et al. (2009) employed real-time PCR to detect Campylobacter species in the stools of 8 CD patients, 11 UC patients and 11 symptomatic controls. In addition stools were cultured for 10 days on Skirrow-supplemented blood agar plates in a microaerobic atmosphere. While detection of Campylobacter DNA was higher in IBD patients (21%) than control patients (9%), this did not reach significance (P = 0.46). Further analysis of the Campylobacter PCR positive isolates using species-specific primers showed four of these to be C. concisus. Of these four PCR positive patients, C. concisus was isolated from the stools of two patients. Based on the finding that C. concisus was only present in the stools of IBD patients, the authors suggested that C. concisus may be implicated in IBD (Tankovic et al., 2009).
Finally, attempts to associate oral C. concisus with IBD showed no significant difference to be present in the detection of C. concisus DNA in saliva samples of IBD patients (100%; CD, n = 13, UC, n = 5) and healthy controls (97%, 57/59; Zhang et al., 2010).
Overall, these findings provide considerable evidence that some C. concisus strains may be involved in IBD (Table 2), either as a direct trigger or as an opportunistic pathogen.
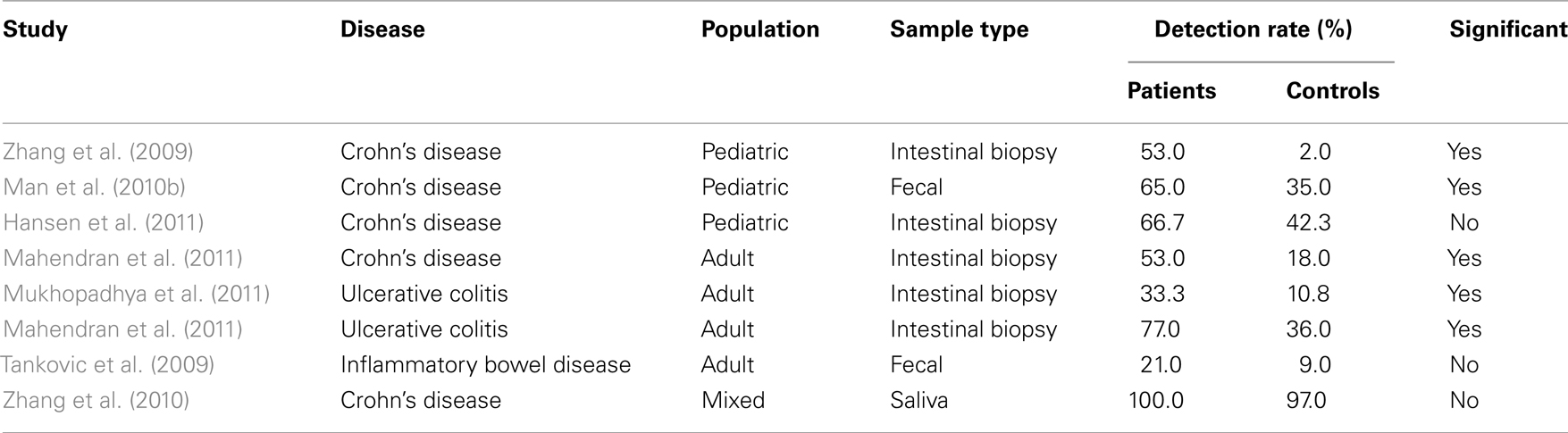
Table 2. Detection rates of Campylobacter concisu s in inflammatory bowel disease patients and controls.
Other Diseases
The majority of research associating C. concisus with human disease has focused on periodontal disease, enteritis, and IBD, however, several studies have associated C. concisus with other human diseases. The most significant of these is the association of this bacterium with Barrett’s esophagus (BE; Macfarlane et al., 2007), a condition in which gastroduodenal reflux leads to squamous epithelial cells lining the esophagus being replaced with columnar epithelial cells. In this study by Macfarlane et al. (2007), high levels of C. concisus and C. rectus were found in four of seven patients (57%) with BE compared with none of the controls. C. concisus was isolated from both the aspirate and mucosal samples, and was the most prevalent bacterium. As a result of their study Macfarlane et al. hypothesized that pathogenic and putative toxin producing Campylobacter species could be involved in the initiation, maintenance, or exacerbation of esophageal disease.
In addition to the above study there have been a number of sporadic reports of the isolation of C. concisus from a range of body sites. For example, Johnson and Finegold (1987) reported the isolation of C. concisus from a diabetic foot ulcer of a patient with underlying osteomyelitis. Further, in a report on Campylobacter species isolated from extra-oro-intestinal abscesses, C. concisus was one member of a polymicrobial flora isolated from a brain abscess of a patient who had previously undergone craniotomy for maxillary sinus carcinoma (de Vries et al., 2008). Despite antibiotic therapy the patient later died from an intracerebral infection due to chronic sinusitis (de Vries et al., 2008). In this case it is likely that C. concisus was an opportunistic pathogen, an interpretation that may also explain the isolation of C. concisus from human blood samples (Lastovica, 2009). Further, following a survey of pediatric records Lastovica reported the isolation of C. concisus from stool specimens of nine liver transplant patients, six patients with biliary atresia and three with renal transplants or renal failure (Lastovica, 2009). While a role for C. concisus in these diseases cannot be confirmed, these results in combination with the finding of micro-abscesses in the livers of two mice infected with C. concisus (Aabenhus et al., 2008), raises the possibility of an association between this bacterium and hepatobiliary disease.
Genetic Typing and Diversity
While the above studies support the role of C. concisus as a human pathogen, its isolation from healthy individuals, and the failure of some studies to show a significant difference in C. concisus prevalence in subjects with disease and healthy controls has raised contention as to its role in disease. The existence of high genetic diversity among C. concisus strains has been proposed as a possible justification for this inconsistency.
An early study conducted by Vandamme et al. (1989) that used DNA–DNA hybridization to investigate diarrheal isolates that fitted the phenotypic description of C. concisus, first raised the possibility that C. concisus was genetically diverse. In this study phenotypically confirmed C. concisus strains were shown to exhibit only 42–50% DNA–DNA hybridization values with the type and reference strains of oral origin, leading Vandamme et al. to conclude that C. concisus was likely to be genetically diverse. Given this, On and Harrington (2000) suggested that, based on current taxonomic guidelines, diarrheal isolates and oral strains should actually be distinct genomic species, however, as they could not be differentiated phenotypically they should be regarded as a “complex species comprising at least two genomospecies.”
Further evidence that considerable genetic diversity existed within C. concisus isolates of fecal origin came from a study by Van Etterijck et al. (1996) who used randomly amplified polymorphic DNA (RAPD) analysis to compare C. concisus strains isolated from fecal samples of 37 children with diarrhea attending the same Belgian day care center. This showed that C. concisus strains isolated from 35 of the 37 children (94.6%) had distinct RAPD profiles, further confirming the genetic heterogeneity of C. concisus strains.
The considerable genetic diversity between oral and fecal isolates of C. concisus was further confirmed by Aabenhus et al. (2002b) who examined the protein profiles of C. concisus strains isolated from Danish subjects with a range of symptoms including diarrhea and dyspepsia using SDS-PAGE. This showed, based on major protein band composition, that 85% of C. concisus fecal strains differed from the oral reference type strain C. concisus ATCC 33237. Given this, Aabenhus et al. assigned the C. concisus isolates into two broad groups, Group 1 which included C. concisus strains resembling the oral C. concisus ATCC 33237 type strain, and Group 2 which comprised C. concisus strains whose protein profiles differed from that of the C. concisus type strain. Interestingly Group 2, which included 83 of the 98 (85%) C. concisus isolates tested, predominantly comprised C. concisus strains isolated from patients with diarrhea, as well as all C. concisus strains isolated from children and immunocompetent patients. In the same year, Matsheka et al. (2002) used pulsed field gel electrophoresis (PFGE), with the restriction enzyme NotI to determine the genetic diversity of 53 C. concisus strains isolated from fecal samples of children with diarrhea. This showed that of the 53 isolates examined, 51 had distinct NotI macrorestriction fragments, while 2 strains were resistant to NotI digestion. The patterns comprised between 1 and 14 restriction fragments, with type and reference strains of two well-defined genomospecies of oral and fecal origin containing 6 and 12 fragments, respectively. Based on these results Matsheka et al. (2002) concluded that C. concisus was genetically diverse and that the species was likely to be “a taxonomic continuum comprised of several genomospecies.” In a later study, Matsheka et al. (2006) typed 100 C. concisus isolates obtained from 98 children with diarrhea and 2 dental isolates from adult patients by DNA fingerprinting. RAPD analysis of the 100 isolates showed 86% of isolates to be genotypically diverse. Of these heterogeneous isolates, 25 had previously been shown by the same group to have unique profiles using PFGE (Matsheka et al., 2002). The remaining 14 strains were shown to have 5 RAPD profiles. The high level of heterogeneity observed in these C. concisus strains was in line with this group’s previous study.
This high level of diversity is consistent with a study by Pruckler et al. (2002) who examined the genetic diversity of 73 phenotypically and genotypically proven C. concisus strains including six ATCC/NCTC reference strains (two of which represented two established genomospecies; Aabenhus et al., 2002b) using PFGE and the restriction enzyme NotI. This showed all strains to have an unique NotI PFGE pattern. Further, composite analysis revealed five clusters, with the reference strains representing the two established genomospecies being clearly separated into different clusters. Moreover, in the same year, Aabenhus et al. (2002a) employed a lectin typing system, which used a panel of 4 lectins, to type 44 clinical isolates of C. concisus obtained from patients presenting with malignancies, HIV, IBD, and other conditions, who had suffered diarrhea and upper gastrointestinal dyspepsia, as well as the type strain ATCC 33237 of oral origin. This typing system grouped the 45 strains into 13 lectin reaction patterns, however, when the authors attempted to correlate the reaction patterns with the clinical category of the patients, no association was observed.
To determine the clinical relevance of C. concisus in gastrointestinal disease, Engberg et al. (2005) investigated the genotypic characteristics of 39 C. concisus isolates from Danish patients with diarrhea, three from healthy individuals and the type strain CCUG 13144. Protein profiling and PCR amplification of the 23S ribosomal RNA (rRNA) gene showed the isolates to cluster into two distinct, but discordant groups. Furthermore, automated ribotyping showed 34 of the 43 isolates to have distinct patterns. Additional analysis of 37 isolates using RAPD showed these to have 37 unique profiles. Interestingly, two strains from the feces of healthy human carriers were distinct from the majority of strains of diarrheal origin. Based on these studies Engberg et al. concluded that C. concisus consists of at least two genomospecies with extensive genetic diversity, however, no clear genotypic differences were observed between isolates from patients with diarrhea and isolates from healthy carriers. In a further study published in the same year, Aabenhus et al. (2005a) investigated the genotype of 62 C. concisus clinical isolates (56 diarrheal strains, 4 oral strains, and the oral CCUG 13114 and intestinal CCUG 19995 type strains) using amplified length fragment polymorphism (AFLP) analysis and correlated the results to clinical data. All strains examined were shown to have unique profiles, however, following numerical analysis of the AFLP profiles, relationships at the taxonomic level were revealed that allowed the strains to be grouped into four distinct clusters. AFLP cluster 1 contained the type strain of oral origin (CCUG 13144) as well as 22 other clinical isolates, 27% of which were isolated from immunocompetent (IC) patients. Cluster 2 contained the reference strain CCUG 19995 originally isolated from a human diarrhea sample, as well as 32 other clinical isolates, 59% of which were isolated from IC patients. Interestingly, five of the seven C. concisus strains isolated from IBD patients were present in cluster 2. AFLP cluster 3 consisted of a single diarrheal isolate from an IC patient, whereas cluster 4 contained five strains all of which were isolated from severely immunodeficient patients, a finding that led the authors to suggest that the strains in AFLP cluster 4 may be less invasive. As result of their study, Aabenhus et al. (2005a) concluded that C. concisus contains at least four distinct genomospecies that may vary in their pathogenic potential.
In a recent study, Kalischuk and Inglis (2011) compared the genotypes of C. concisus fecal isolates from diarrheic and asymptomatic healthy individuals using AFLP analysis and a genomospecies-specific 23S rRNA gene PCR, that grouped isolates into genomospecies A or B. Of the 22 isolates examined, six were assigned to genomospecies A and 12 to genomospecies B, while three isolates generated PCR products with both the genomospecies A and B primer sets, and were designated genomospecies A/B. One isolate failed to amplify with either primer set. Consistent with previous observations the type strain, LMG7788, was assigned to genomospecies A. Based on AFLP analysis the C. concisus isolates were shown to cluster into two phylotypes, distinguished from each other at a 34% similarity level. All isolates assigned to AFLP cluster 1 belonged to genomospecies A and included the type strain LMG7788, four isolates from healthy controls and one isolate from a patient with diarrhea. Of the 17 C. concisus isolates assigned to AFLP cluster 2, 12 (70.6%) belonged to genomospecies B, 3 to genomospecies A/B (17.6%), 1 to genomospecies A (5.9%), and the final untypable isolate (5.9%). While these results are in contrast to the findings of Aabenhus et al. (2005a) who based on AFLP identified four clusters among 62 C. concisus isolates, it should be noted that the majority of isolates (90.3%) in that study formed two main clusters.
In studies conducted in our own laboratory we recently sequenced the genome of C. concisus strain UNSWCD, isolated from a patient with CD and found it to have a 1.8-Mb genome that is substantially smaller than the available 2.1 Mb C. concisus BAA-1457 genome (Deshpande et al., 2011). While 1593 genes were conserved across UNSWCD and BAA-1457, 138 genes (7.8%) from UNSWCD and 281 (13.98%) from BAA-1457 were unique when compared against the other, and substantial functional differences were observed between the two strains. When cutoff values of 70% identity plus at least 85% gene length coverage were employed, searches between C. concisus UNSWCD and BAA-1457 showed that 76% of genes were homologs, whereas those between C. jejuni strains showed 90–91% to be homologs, indicating substantial variation exists within these two C. concisus genomes (Kaakoush et al., 2011a). Furthermore, average percentage identities for all homologs revealed that C. concisus strains had higher variation when compared to the phylogenetically related species C. jejuni (96 vs. 98%, respectively). We investigated the protein profiles of these two strains and a further six C. concisus strains isolated from different disease states, and found BAA-1457 was highly divergent from the other C. concisus strains (Kaakoush et al., 2011a). Interestingly, the healthy control strain ATCC 51561 was divergent from the other strains isolated from acute and chronic gastroenteritis and CD. Attempts to discover markers for the detection of C. concisus through the identification of proteins found in all our strains but not in another bacterial species, found two candidates that may be good taxon-specific markers for this species.
Pathogenesis
Until recently, the pathogenic potential of C. concisus has remained relatively unexplored with comparatively few studies dedicated to elucidating the pathogenic mechanisms within this bacterium. However, following the association of C. concisus with enteritis and IBD, studies have increased significantly.
Motility and Chemotaxis
Owing to its single polar flagellum (Man et al., 2010a), C. concisus has the ability to swim through protective host barriers such as saliva or the intestinal mucus layer, allowing it to achieve close contact with epithelial layers, and thus, a greater chance of triggering disease. One of the first studies to investigate C. concisus pathogenesis was that of Paster and Gibbons (1986) who in an attempt to understand the role of this bacterium in oral disease studied its chemotactic response to a range of compounds. While this showed that C. concisus exhibited a chemotactic response to formate, it was not chemoattracted to many sugars, inorganic salts, amino acids and their derivatives, purines and pyrimidines, fatty acids, or natural mixtures such as saliva, serum, and crevicular fluid (Paster and Gibbons, 1986). Intriguingly, unlike C. jejuni strains, C. concisus strain 288 was not chemoattracted to mucin (Paster and Gibbons, 1986), a finding that may relate to the oral origin of this strain. In contrast, the aggregation and possible attraction of C. concisus strains isolated from human intestinal biopsies to mucin has been observed through scanning electron microscopy (SEM; Kaakoush et al., 2011b).
Production of Toxins and Other Virulence Factors
Campylobacter concisus strains have been shown to produce several toxins and virulence factors. Istivan et al. (1998) reported 20 C. concisus strains isolated from children with enteritis, and one from a healthy control to have hemolytic activity associated with lysis of human and animal erythrocytes. The same group later showed that C. concisus strains had membrane-bound hemolytic phospholipase A2 activity that caused stable vacuolating and cytolytic effects on Chinese hamster ovary (CHO) cells (Istivan et al., 2004), and that this activity was encoded by the pldA gene (Istivan et al., 2008). Additionally, Istivan et al. (2008) detected secreted hemolytic activities in C. concisus strains isolated from children with enteritis. These findings are in accordance with Musmanno et al. (1998) who showed that a C. concisus strain isolated from a child with enteritis produced a cytotoxic-like effect on CHO cells and induced intracytoplasmic vacuole formation, and Kalischuk and Inglis (2011) who also showed that C. concisus isolates had hemolytic activity against sheep red blood cells. Moreover, Engberg et al. (2005) reported strain specific differences in the ability of C. concisus strains to induce cytolethal distending toxin-like effects on monkey kidney (Vero) cells, however, no association with disease outcome was found.
Further evidence of the cytotoxicity of C. concisus isolates was provided more recently by Nielsen et al. (2011b) who observed an elevated level of lactate dehydrogenase release in cells exposed to this bacterium. In contrast, Kalischuk and Inglis (2011) reported that none of their C. concisus isolates caused significant epithelial cytotoxicity (also measured through lactate dehydrogenase release). However, these authors also reported that 64.3% (9 out of 14) of their C. concisus isolates induced epithelial DNA fragmentation and this was correlated with an increase in host cell metabolic activity (Kalischuk and Inglis, 2011).
In studies conducted in our own laboratory we recently identified an invasin InvA, a hemolysin TlyA and zonula occludens toxin Zot within the genome of C. concisus BAA-1457, isolated from a patient with acute enteritis (Kaakoush et al., 2010). A complete list of virulence factors and their putative functions is provided in Kaakoush et al. (2010). Zot, a protein used by virulent pathogens to increase tissue permeability was not a common feature among our C. concisus strains (one out of eight; Kaakoush et al., 2011a). In contrast, Kalischuk and Inglis (2011) detected zot in 42.8% (6 out of 14) of their C. concisus isolates, which may suggest that these six isolates were genetically related to the BAA-1457 strain. We also analyzed the secretome of C. concisus strain UNSWCD isolated from an intestinal biopsy of a patient with CD, and identified a S-layer RTX protein and an outer membrane fibronectin-binding protein CadF (Kaakoush et al., 2010). RTX proteins are pore-forming toxins synthesized by a diverse group of Gram-negative pathogens, whereas CadF is known to be involved in adhesion to the host cell. Analysis of the genome of C. concisus UNSWCD revealed an O-antigen ligase that may play a role in the aggregation and adherence onto host cells, and an Acr protein (α-crystallin) that may contribute to persistent infection within the host and may influence the host response to infection (Kaakoush et al., 2011a), as has been observed in Mycobacterium tuberculosis (Stewart et al., 2005). Furthermore, we identified variations within the flagellin glycosylation pathways of C. concisus strains (Kaakoush et al., 2011a), a factor that may contribute to the differences observed in their pathogenic potential (Man et al., 2010a; Kaakoush et al., 2011b).
Adherence and Invasion into Host Cells
Russell and Ward (1998) investigated the ability of C. concisus strains isolated from children with diarrhea to adhere to and invade HEp-2 cells, and found rates 4- to 100-fold higher than the pathogenic and invasive Campylobacter species C. jejuni and C. coli. In contrast, in a recent study Kalischuk and Inglis (2011) found that C. concisus isolates exhibited comparable epithelial adherence, invasion, and translocation abilities to that of C. jejuni 81–176. Moreover, the authors did not observe any differences in mean adherence, invasion, or translocation between isolates from diarrheic and healthy humans (Kalischuk and Inglis, 2011). In a recent study, Nielsen et al. (2011b) examined six oral and eight fecal C. concisus isolates and found that all strains invaded confluent host cells and impaired epithelial barrier function. The authors concluded that epithelial barrier dysfunction by C. concisus strains could mainly be assigned to apoptotic leaks together with moderate tight junction changes (Nielsen et al., 2011b).
In recent studies we have investigated the invasive phenotype of C. concisus in more detail through the analysis of isolates from different disease states (Man et al., 2010a). These studies showed that the percentage invasion of C. concisus strain UNSWCD, isolated from a child with newly diagnosed CD, was significantly higher than that of C. concisus strains isolated from patients with acute enteritis and a healthy control. Interestingly, C. concisus ATCC 51561 isolated from a healthy subject, showed no evidence of invasion. These findings were confirmed with SEM (Figure 1; Man et al., 2010a). To investigate the possible involvement of host microtubules and microfilaments on the C. concisus UNSWCD invasion process, we pre-treated Caco-2 cells with colchicine or cytochalasin D, and this showed the level of invasion by C. concisus UNSWCD to be significantly attenuated in the presence of both colchicine and cytochalasin D (Man et al., 2010a). We also showed that C. concisus UNSWCD preferentially attached to intercellular junctional spaces (Figure 2) and that this spatial distribution was concomitantly associated with a loss of membrane-associated ZO-1 and occludin. Given that C. concisus UNSWCD was isolated from a patient with CD, we also investigated the potential effect of pre-existing inflammation (driven by TNF-α and IFN-γ) on C. concisus UNSWCD invasion. This showed that the ability of C. concisus UNSWCD to invade Caco-2 cells treated with TNF-α or IFN-γ was significantly increased, suggesting that the presence of inflammation increases the invasive ability of C. concisus (Man et al., 2010a).
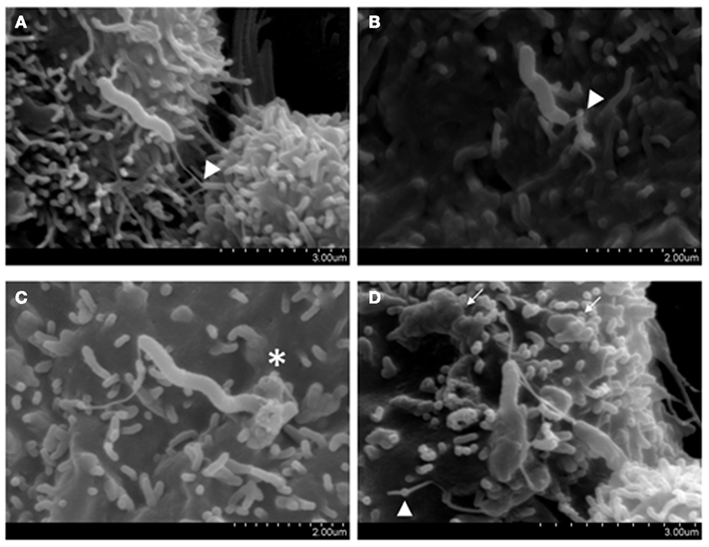
Figure 1. Host attachment and invasion by Campylobacter concisus UNSWCD (adapted from Man et al., 2010a). Caco-2 cells were infected with bacteria at a MOI of 200 for 6 h. (A) The polar flagellum of C. concisus UNSWCD mediated attachment to the microvillus tip (triangles). (B) The flagellum appears to fold around the microvillus (triangle). (C) C. concisus induced a membrane ruffling-like effect (*). (D) C. concisus is observed half internalized in the host cell, resulting in a surface protrusion on the host cell membrane, and the flagellated half remains externally exposed. A host cell infected with multiple bacteria displays cell membrane irregularities and uneven texture because of bacteria-induced protrusions (arrows).
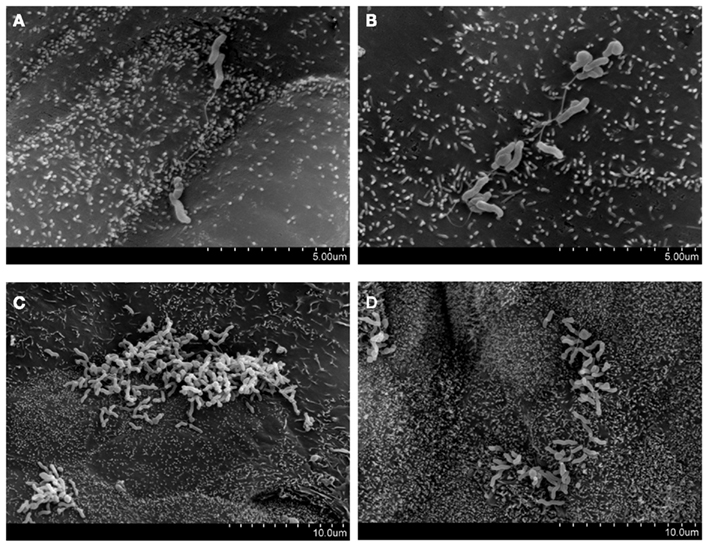
Figure 2. Preferential attachment of Campylobacter concisus UNSWCD to intercellular junctional spaces of host cells (adapted from Man et al., 2010a). HT-29 cells were infected with bacteria at a MOI of 200 for 6 h. (A–D) C. concisus UNSWCD preferentially attached to areas resembling the intercellular junctional space.
In a more recent study, we investigated the invasive potential of eight C. concisus strains isolated from different disease states (three CD, one chronic enteritis, three acute enteritis, and one healthy subject), and found that the percentage invasion of C. concisus strains isolated from chronic intestinal diseases were more than 500-fold higher than that of C. concisus strains isolated from acute intestinal diseases and a healthy control (Kaakoush et al., 2011b). As was previously reported by Russell and Ward (1998), the levels of invasion observed for C. concisus strains in our study were higher than that reported for C. jejuni (Kaakoush et al., 2011b). Importantly, we detected a plasmid harboring virulence determinants from several pathogenic organisms only in the highly invasive strains isolated from chronic intestinal diseases, thus, elucidating the feature that may be responsible for the heterogeneity in the invasive potential of C. concisus (Kaakoush et al., 2011b).
Host Immune Response to Campylobacter concisus
Several studies have attempted to elucidate the host immune response to C. concisus infection. In our own studies we have shown that following infection of intestinal cells, C. concisus UNSWCD stimulated significantly higher levels of IL-8 as compared with Escherichia coli K-12 (Man et al., 2010a), a finding that is supported by Kalischuk and Inglis (2011) who also reported the infection of intestinal cells with C. concisus strains to result in increased expression of IL-8 by more than twofold. Interestingly, in our study the levels of IL-8 stimulated with heat-killed C. concisus were found to be comparable to that of their viable counterparts. Furthermore, in an examination of the THP-1 monocytic cell response to bacterial stimulation we observed that C. concisus UNSWCD induced significantly elevated levels of IL-8 and TNF-α, but not IL-1β when compared with unstimulated controls (Man et al., 2010a). When primary human macrophages were infected with C. concisus UNSWCD a significantly higher level of TNF-α was observed as compared with the unstimulated control, while relatively low levels of IL-1β were produced. Whilst C. concisus UNSWCD induced increased levels of IL-8 in primary human macrophages, the levels observed were not significantly higher than that in the unstimulated control (Man et al., 2010a). Comparison of the results of C. concisus UNSWCD to that of other C. concisus strains, showed strain dependency in their ability to stimulate IL-8 and TNF-α in the different types of host cells examined (Man et al., 2010a).
More recently, we showed that host cells infected with any of our C. concisus strains (CD, chronic enteritis, acute enteritis, and a healthy subject) produced high amounts of IL-12, however, only the C. concisus strains capable of internalizing into host cells induced a significantly increased quantity of IFN-γ with respect to controls (Kaakoush et al., 2011b). These findings, along with the regulation of proteasomal subunits, ubiquitination pathways, and NF-κB inhibitors, led us to hypothesize that invasive C. concisus strains can activate the NF-κB pathway (Kaakoush et al., 2011b).
Pathogenesis in Animal Models
Only one study has observed the pathogenic potential of C. concisus in vivo (Aabenhus et al., 2008). In this study, five clinical C. concisus isolates of intestinal origin and the ATCC 33237 type strain of oral origin were inoculated into immunocompetent BALB/cA mice treated with cyclophosphamide to suppress their immune functions. While C. concisus was isolated from the liver, ileum, and jejunum of some of these mice, this was not consistent across experiments. Further histological examination did not consistently find signs of inflammation in the gut, but occasionally micro-abscesses were found in the liver of infected animals (Aabenhus et al., 2008). Compared to controls, mice infected with some C. concisus strains showed a significant weight loss in the first couple of days, but this effect wore off by the fifth day of infection. Finally, a total of three C. concisus-infected mice died during the course of the experiments.
The findings in relation to animal models of C. concisus infection are of no surprise given the difficulty faced with developing a good animal model of C. jejuni infection. For example, many murine models of C. jejuni infection have resulted in sporadic colonization and/or absence of clinical disease signs (Bereswill et al., 2011), an outcome that is suggested to relate to colonization resistance caused by the murine intestinal flora. The recent development of novel C. jejuni infection models by Bereswill et al. (2011) may however now overcome previous problems relating to both poor colonization and the absence of the classical campylobacteriosis picture observed in humans. To overcome colonization resistance due to the murine flora, Bereswill et al. eradicated the intestinal flora of gnotobiotic mice using antibiotic treatment and then investigated the colonization potential of these mice with C. jejuni. This showed that C. jejuni could colonize all regions of the GIT of these mice. Further, a group of gnotobiotic mice treated with antibiotics were recolonized with human intestinal microflora and following recolonization, were infected with C. jejuni and the colonization and inflammatory response investigated. This showed that C. jejuni could not only stably colonize the gnotobiotic “humanized” intestinal tract but also exhibited the typical pro-inflammatory features of human campylobacteriosis. Given this, the gnotobiotic and “humanized” mouse models may turn out to be an excellent model of C. concisus infection (Bereswill et al., 2011).
Reservoirs of Infection
While the source and routes of transmission for the closely related species C. jejuni and C. coli are well established (Vandamme et al., 2005), little is known about the source and route of transmission of C. concisus. Given the importance of birds and animals as a source of C. jejuni and C. coli in human infection, current studies have focused on detection of C. concisus in domestic animals, sheep, cattle, pigs, and chickens. For example, in a study investigating the presence of C. concisus DNA in the saliva of domestic pets, Petersen et al. (2007) identified this bacterium in 1 of 8 cats but in none of 12 dogs. In another study, which screened fecal samples from healthy dogs for different Campylobacter species, although several Campylobacter species were identified, C. concisus was not detected (Chaban et al., 2009). Interestingly, in a follow up study by the same group, which screened fecal samples from both healthy (n = 70) and diarrheic dogs (n = 65), C. concisus DNA was detected in six diarrheic dogs but not in healthy dogs (P < 0.05; Chaban et al., 2010). In an attempt to determine if farm animals were a reservoir for emerging Campylobacter species, Oporto and Hurtado (2011) detected two isolates from healthy sheep feces that were shown to cluster with C. concisus strains, despite sharing higher similarity with C. mucosalis, however, no C. concisus DNA was detected in swine, beef cattle, or dairy cattle. Interestingly in a study which investigated the presence of C. concisus in chicken (n = 185), pork (n = 179), and beef (n = 186) samples collected throughout the Republic of Ireland, Lynch et al. (2011) found C. concisus to be present in 10 and 3% of chicken and beef samples, respectively. Notably, the detection level of C. concisus in chicken meat was similar to that observed for C. jejuni (13% of samples; Lynch et al., 2011). While C. concisus may be associated with enteritis in dogs (Chaban et al., 2010), collectively current studies would suggest that chickens, and not domestic pets, are a more likely reservoir for C. concisus, thus suggesting the possibility of zoonotic transmission. Furthermore, the presence of viable C. concisus in the saliva of most humans (described above) may support a person to person route of transmission.
Antibiotic Susceptibility
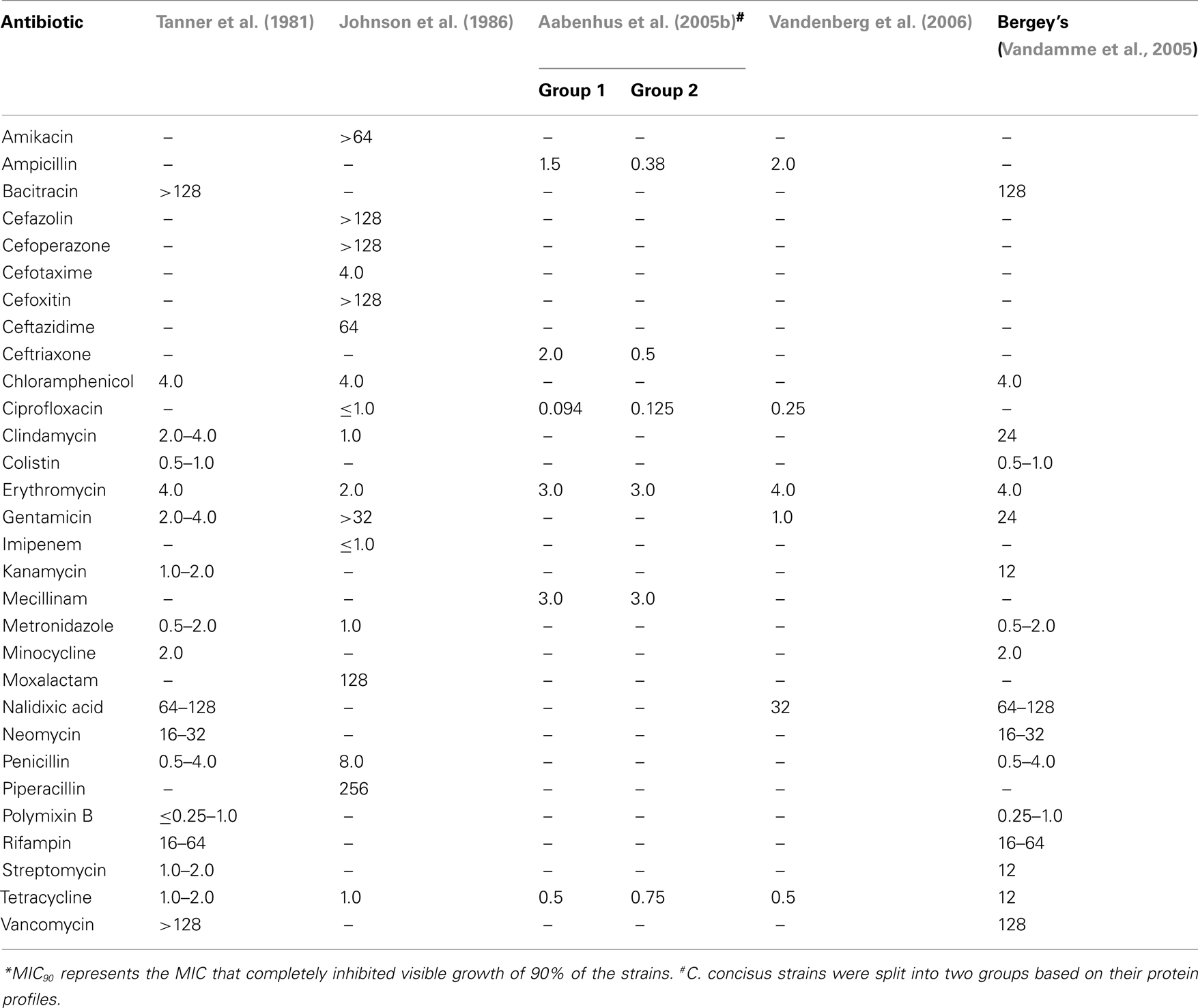
The susceptibility of C. concisus to a variety of antimicrobial agents has been examined in several studies and these are summarized in Table 3 (Tanner et al., 1981; Johnson et al., 1986; Aabenhus et al., 2005b; Vandenberg et al., 2006). In 1981, upon designation of six isolates from the oral cavity of humans into the new species C. concisus, Tanner et al. (1981) measured the effects of 17 antimicrobial agents against these isolates and found them to be resistant to bacitracin, nalidixic acid, rifampin, and vancomycin (Table 3). Five years later, Johnson et al. (1986) observed the effects of 17 antimicrobial agents, 7 of which had been previously investigated by Tanner et al. against a group of five isolates designated “Wolinella spp./C. concisus” (Table 3) and showed that as reported by Tanner et al. isolates were susceptible to chloramphenicol, metronidazole, clindamycin, erythromycin, tetracycline, and penicillin. In contrast to Tanner et al., Johnson et al. (1986) found two of five isolates to be resistant to gentamicin. O’Connor et al. (1990) studied the effect of minocycline on C. concisus and as was observed by Tanner et al. (1981), they found the drug to be effective against the bacterium. However, they also observed an increase in the MIC of minocycline for C. concisus after the bacterium was exposed to sub-lethal concentrations of the antibiotic for 6–7 weeks, a finding that was not observed in most of the bacteria they tested (O’Connor et al., 1990).
In a more recent study Engberg et al. (2005) reported that all 43 C. concisus isolates tested were susceptible to erythromycin, tetracycline, nalidixic acid, streptomycin, ciprofloxacin, gentamicin, colistin, chloramphenicol, sulfamethizole, neomycin, and ampicillin, however, their data was not shown. In a study which characterized and sub-grouped 109 C. concisus strains isolated from 98 patients with gastrointestinal disease into two major groups (Group 1, n = 15; Group 2, n = 94) using protein profiling, Aabenhus et al. (2005b) tested the effects of eight antimicrobial agents against the isolates in each group using a range of techniques (Table 3). While resistance to nalidixic acid was detected in 87–90% of strains (API Campy system and disks from Biodisc AB), all strains were resistant to cefazolin (API Campy system). Based on two techniques (Neosensitabs and E-tests), C. concisus strains were found to be generally susceptible to ampicillin (93–100%), tetracycline (97–100%), ceftriaxone (89–98%), mecillinam (92–100%), erythromycin (93–97%), and ciprofloxacin (87–95%). In contrast, the API Campy system found all isolates to be resistant to erythromycin (Aabenhus et al., 2005b). The authors concluded that the API Campy system was not a valid tool in the identification of C. concisus as they considered the reaction patterns to be too heterogenous (Aabenhus et al., 2005b). In agreement with Aabenhus et al., Vandenberg et al. (2006) reported all 20 C. concisus isolates tested to be susceptible to ampicillin and tetracycline, 5% to be resistant to gentamicin, ciprofloxacin, and erythromycin, and 80% to be resistant to nalidixic acid. Strikingly, Aabenhus et al. (2005b) found that the majority of their strains (94%) were susceptible to cephalothin, a finding that contradicts Roop et al. (1985) who stated that C. mucosalis strains can be differentiated from C. concisus through the latter’s resistance to cephalothin. In agreement with Aabenhus et al., Pruckler et al. (2002) reported that only 5% of their 73 C. concisus isolates to be resistant to cephalothin. In addition, the authors reported that the overall resistance rates for their 73 C. concisus isolates were 93.75% for nalidixic acid, 30% for clindamycin, 7.5% for ciprofloxacin, 3.75% for tetracycline, 2.5% for erythromycin, 1.25% for azithromycin, and 0% for both chloramphenicol and gentamicin (Pruckler et al., 2002).
Notably, Moore et al. (2006) upon examining 457 C. concisus isolates collected between 1998 and 2006, reported that resistance to nalidixic acid increased from 40 to 62%, resistance to ciprofloxacin increased from 6.9 to 18%, resistance to erythromycin increased from 4.8 to 21.7%, whereas ceftriaxone resistance remained stable at 2% over the time frame. Moreover, some isolates were resistant to several classes of antibiotic (Moore et al., 2006).
Other studies reporting the effect of antimicrobial agents against C. concisus include the finding that this bacterium expresses pyrazinamidase activity (Shingaki et al., 1999), a reaction that converts the pro-drug pyrazinamide to its active form pyrazinoic acid.
Interestingly, a number of studies investigating the effect of antibiotic therapy in patients with CD have reported nitroimidazoles and ciprofloxacin, antibiotics to which C. concisus remains sensitive, to have a beneficial effect (Feller et al., 2010). For example, in a recent systematic review and meta-analysis of randomized clinical trials, which investigated the effect of antibiotic therapy on remission in patients with active disease and relapse in patients with inactive disease, three studies using nitroimidazoles (two, metronidazole and one, omidazole), showed a beneficial effect (OR: 3.54; 95% CI: 1.94–6.47). Further, a trial examining treatment with ciprofloxacin for 6 months also showed benefit (OR: 11.3; 95% CI: 2.60–48.8). In contrast, the combined OR (OR: 0.58, 95% CI: 0.29–1.18) obtained from three trials involving treatment with anti-tuberculosis drugs (including rifamycins) showed no beneficial effect (Feller et al., 2010).
Collectively, these studies show that although several classes of antibiotics that are generally effective against anaerobes remain effective against C. concisus, resistance to antimicrobial agents within C. concisus isolates is on the rise.
Conclusion
Significant advances in our understanding of C. concisus have occurred over the last 5 years with over a third of studies on C. concisus published within this time period. Current evidence would suggest that likely reservoirs of this bacterium include humans and chickens. In relation to disease, C. concisus has been associated with periodontal diseases, acute enteritis, and IBDs, with the strongest evidence relating to acute and chronic intestinal diseases. The finding that C. concisus strains can adhere to and invade host cells, translocate across cell monolayers and induce inflammatory immune responses supports the possible role of this bacterium in intestinal diseases. Further studies that compare the prevalence and pathogenic potential of C. concisus strains isolated from different disease outcomes with those from healthy controls are required to reinforce the current evidence suggesting an association between C. concisus and intestinal diseases. A further important step will be the identification of a suitable animal model in which to investigate the pathogenesis of C. concisus infection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Goldstar Award from The University of New South Wales. Nadeem Omar Kaakoush is a recipient of an Early Career Fellowship from the National Health and Medical Research Council, Australia.
References
Aabenhus, R., Hynes, S. O., Permin, H., Moran, A. P., and Andersen, L. P. (2002a). Lectin typing of Campylobacter concisus. J. Clin. Microbiol. 40, 715–717.
Aabenhus, R., Permin, H., On, S. L., and Andersen, L. P. (2002b). Prevalence of Campylobacter concisus in diarrhoea of immunocompromised patients. Scand. J. Infect. Dis. 34, 248–252.
Aabenhus, R., On, S. L., Siemer, B. L., Permin, H., and Andersen, L. P. (2005a). Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. J. Clin. Microbiol. 43, 5091–5096.
Aabenhus, R., Permin, H., and Andersen, L. P. (2005b). Characterization and subgrouping of Campylobacter concisus strains using protein profiles, conventional biochemical testing and antibiotic susceptibility. Eur. J. Gastroenterol. Hepatol. 17, 1019–1024.
Aabenhus, R., Stenram, U., Andersen, L. P., Permin, H., and Ljungh, A. (2008). First attempt to produce experimental Campylobacter concisus infection in mice. World J. Gastroenterol. 14, 6954–6959.
Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206.
Altekruse, S. F., Stern, N. J., Fields, P. I., and Swerdlow, D. L. (1999). Campylobacter jejuni – an emerging foodborne pathogen. Emerging Infect. Dis. 5, 28–35.
Bereswill, S., Fischer, A., Plickert, R., Haag, L. M., Otto, B., Kühl, A. A., Dasti, J. I., Zautner, A. E., Muñoz, M., Loddenkemper, C., Gross, U., Göbel, U. B., and Heimesaat, M. M. (2011). Novel murine infection models provide deep insights into the “ménage à trios” of Campylobacter jejuni, microbiota and host innate immunity. PLoS ONE 6, e20953. doi:10.1371/journal.pone.0020953
Blaser, M. J., Hoverson, D., Ely, I. G., Duncan, D. J., Wang, W. L., and Brown, W. R. (1984). Studies of Campylobacter jejuni in patients with inflammatory bowel disease. Gastroenterology 86, 33–38.
Blaser, M. J., Taylor, D. N., and Feldman, R. A. (1983). Epidemiology of Campylobacter jejuni infections. Epidemiol. Rev. 5, 157–176.
Boyanova, L., Gergova, G., Spassova, Z., Koumanova, R., Yaneva, P., Mitov, I., Derejian, S., and Krastev, Z. (2004). Campylobacter infection in 682 Bulgarian patients with acute enterocolitis, inflammatory bowel disease, and other chronic intestinal diseases. Diagn. Microbiol. Infect. Dis. 49, 71–74.
Chaban, B., Musil, K. M., Himsworth, C. G., and Hill, J. E. (2009). Development of cpn60-based real-time quantitative PCR assays for the detection of 14 Campylobacter species and application to screening of canine fecal samples. Appl. Environ. Microbiol. 75, 3055–3061.
Chaban, B., Ngeleka, M., and Hill, J. E. (2010). Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol. 10, 73. doi:10.1186/1471-2180-10-73
de Vries, J. J., Arents, N. L., and Manson, W. L. (2008). Campylobacter species isolated from extra-oro-intestinal abscesses: a report of four cases and literature review. Eur. J. Clin. Microbiol. Infect. Dis. 27, 1119–1123.
Deshpande, N. P., Kaakoush, N. O., Mitchell, H., Janitz, K., Raftery, M. J., Li, S. S., and Wilkins, M. R. (2011). Sequencing and validation of the genome of a Campylobacter concisus reveals intra-species diversity. PLoS ONE 6, e22170. doi:10.1371/journal.pone.0022170
Ebersole, J. L., Taubman, M. A., Smith, D. J., and Haffajee, A. D. (1985). Effect of subgingival scaling on systemic antibody responses to oral microorganisms. Infect. Immun. 48, 534–539.
Economou, M., and Pappas, G. (2008). New global map of Crohn’s disease: genetic, environmental, and socioeconomic correlations. Inflamm. Bowel Dis. 14, 709–720.
Engberg, J., Bang, D. D., Aabenhus, R., Aarestrup, F. M., Fussing, V., and Gerner-Smidt, P. (2005). Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. Clin. Microbiol. Infect. 11, 288–295.
Engberg, J., On, S. L., Harrington, C. S., and Gerner-Smidt, P. (2000). Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38, 286–291.
Feller, M., Huwiler, K., Schoepfer, A., Shang, A., Furrer, H., and Egger, M. (2010). Long-term antibiotic treatment for Crohn’s disease: systematic review and meta-analysis of placebo-controlled trials. Clin. Infect. Dis. 50, 473–480.
Figura, N., Guglielmetti, P., Zanchi, A., Partini, N., Armellini, D., Bayeli, P. F., Bugnoli, M., and Verdiani, S. (1993). Two cases of Campylobacter mucosalis enteritis in children. J. Clin. Microbiol. 31, 727–728.
Friedman, C. R., Hoekstra, R. M., Samuel, M., Marcus, R., Bender, J., Shiferaw, B., Reddy, S., Ahuja, S. D., Helfrick, D. L., Hardnett, F., Carter, M., Anderson, B., and Tauxe, R. V. (2004). Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3), S285–S296.
Gradel, K. O., Nielsen, H. L., Schønheyder, H. C., Ejlertsen, T., Kristensen, B., and Nielsen, H. (2009). Increased short- and long-term risk of inflammatory bowel disease after Salmonella or Campylobacter gastroenteritis. Gastroenterology 137, 495–501.
Guslandi, M., Zhang, L., Man, S. M., Day, A. S., Leach, S. T., Lemberg, D. A., Dutt, S., Stormon, M., Otley, A., O’Loughlin, E. V., Magoffin, A., Ng, P. H., and Mitchell, H. (2009). Campylobacter concisus: a new character in the Crohn’s disease story? J. Clin. Microbiol. 47, 1614–1615.
Haffajee, A. D., Socransky, S. S., Ebersole, J. L., and Smith, D. J. (1984). Clinical, microbiological and immunological features associated with the treatment of active periodontosis lesions. J. Clin. Periodontol. 11, 600–618.
Hansen, R., Mukhopadhya, I., Russell, R. K., Bisset, W. M., Berry, S. H., Thomson, J. M., El-Omar, E. M., and Hold, G. L. (2011). The role of microaerophilic colonic mucosal bacteria in de novo paediatric inflammatory bowel disease. Gut 60(Suppl. 1), A147.
Inglis, G. D., Boras, V. F., and Houde, A. (2011). Enteric campylobacteria and RNA viruses associated with healthy and diarrheic humans in the Chinook health region of southwestern Alberta, Canada. J. Clin. Microbiol. 49, 209–219.
Istivan, T. S., Coloe, P. J., Fry, B. N., Ward, P., and Smith, S. C. (2004). Characterization of a haemolytic phospholipase A(2) activity in clinical isolates of Campylobacter concisus. J. Med. Microbiol. 53, 483–493.
Istivan, T. S., Smith, S. C., Fry, B. N., and Coloe, P. J. (2008). Characterization of Campylobacter concisus hemolysins. FEMS Immunol. Med. Microbiol. 54, 224–235.
Istivan, T. S., Ward, P. W., Lee, A., Coloe, P. J., and Smith, S. C. (1998). “Hemolysins of Campylobacter concisus,” in Campylobacter, Helicobacter and Related Organisms, eds A. Lastovica, D. G. Newell, and E. Lastovica (Cape Town: University of Cape Town Press), P440–P444.
Johnson, C. C., and Finegold, S. M. (1987). Uncommonly encountered, motile, anaerobic gram-negative bacilli associated with infection. Rev. Infect. Dis. 9, 1150–1162.
Johnson, C. C., Reinhardt, J. F., Mulligan, M. E., George, W. L., and Finegold, S. M. (1986). In vitro activities of 17 antimicrobial agents against the formate/fumarate-requiring, anaerobic gram-negative bacilli. Diagn. Microbiol. Infect. Dis. 5, 269–272.
Kaakoush, N. O., Deshpande, N. P., Wilkins, M. R., Raftery, M. J., Janitz, K., and Mitchell, H. (2011a). Comparative analyses of Campylobacter concisus strains reveal the genome of the reference strain BAA-1457 is not representative of the species. Gut Pathog. 3, 15.
Kaakoush, N. O., Deshpande, N. P., Wilkins, M. R., Tan, C. G., Burgos-Portugal, J. A., Raftery, M. J., Day, A. S., Lemberg, D. A., and Mitchell, H. (2011b). The pathogenic potential of Campylobacter concisus strains associated with chronic intestinal diseases. PLoS ONE 6, e29045. doi:10.1371/journal.pone.0029045
Kaakoush, N. O., Man, S. M., Lamb, S., Raftery, M. J., Wilkins, M. R., Kovach, Z., and Mitchell, H. (2010). The secretome of Campylobacter concisus. FEBS J. 277, 1606–1617.
Kalischuk, L. D., and Inglis, G. D. (2011). Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. BMC Microbiol. 11, 53. doi:10.1186/1471-2180-11-53
Kamma, J. J., Diamanti-Kipioti, A., Nakou, M., and Mitsis, F. J. (2000a). Profile of subgingival microbiota in children with mixed dentition. Oral Microbiol. Immunol. 15, 103–111.
Kamma, J. J., Diamanti-Kipioti, A., Nakou, M., and Mitsis, F. J. (2000b). Profile of subgingival microbiota in children with primary dentition. J. Periodont. Res. 35, 33–41.
Kamma, J. J., and Nakou, M. (1997). Subgingival microflora in smokers with early onset periodontitis. Anaerobe 3, 153–157.
Kamma, J. J., Nakou, M., and Baehni, P. C. (1999). Clinical and microbiological characteristics of smokers with early onset periodontitis. J. Periodont. Res. 34, 25–33.
Kamma, J. J., Nakou, M., and Manti, F. A. (1994). Microbiota of rapidly progressive periodontitis lesions in association with clinical parameters. J. Periodontol. 65, 1073–1078.
Kamma, J. J., Nakou, M., and Persson, R. G. (2001). Association of early onset periodontitis microbiota with aspartate aminotransferase activity in gingival crevicular fluid. J. Clin. Periodontol. 28, 1096–1105.
Kovach, Z., Kaakoush, N. O., Lamb, S., Zhang, L., Raftery, M. J., and Mitchell, H. (2011). Immunoreactive proteins of Campylobacter concisus, an emergent intestinal pathogen. FEMS Immunol. Med. Microbiol. 63, 387–396.
Kulkarni, S. P., Lever, S., Logan, J. M., Lawson, A. J., Stanley, J., and Shafi, M. S. (2002). Detection of Campylobacter species: a comparison of culture and polymerase chain reaction based methods. J. Clin. Pathol. 55, 749–753.
Lastovica, A., Le Roux, E., Warren, R., and Klump, H. (1993). Clinical isolates of Campylobacter mucosalis. J. Clin. Microbiol. 31, 2835–2836.
Lastovica, A. J. (2006). Emerging Campylobacter spp.: the tip of the iceberg. Clin. Microbiol. Newsl. 28, 49–55.
Lastovica, A. J. (2009). Clinical relevance of Campylobacter concisus isolated from pediatric patients. J. Clin. Microbiol. 47, 2360.
Lauwers, S., Devreker, T., and Etterijck, R. V. (1991). Isolation of Campylobacter concisus from human faeces. Microb. Ecol. Health Dis. 4(Suppl.), :S91.
Lawson, A. J., Linton, D., and Stanley, J. (1998). 16S rRNA gene sequences of “Candidatus Campylobacter hominis,” a novel uncultivated species, are found in the gastrointestinal tract of healthy humans. Microbiology 144(Pt 8), 2063–2071.
Lindblom, G. B., Sjogren, E., Hansson-Westerberg, J., and Kaijser, B. (1995). Campylobacter upsaliensis, C. sputorum sputorum and C. concisus as common causes of diarrhoea in Swedish children. Scand. J. Infect. Dis. 27, 187–188.
Lynch, O. A., Cagney, C., Mcdowell, D. A., and Duffy, G. (2011). Occurrence of fastidious Campylobacter spp. in fresh meat and poultry using an adapted cultural protocol. Int. J. Food Microbiol. 150, 171–177.
Macfarlane, S., Furrie, E., Macfarlane, G. T., and Dillon, J. F. (2007). Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin. Infect. Dis. 45, 29–38.
Macuch, P. J., and Tanner, A. C. (2000). Campylobacter species in health, gingivitis, and periodontitis. J. Dent. Res. 79, 785–792.
Mahendran, V., Riordan, S. M., Grimm, M. C., Tran, T. A., Major, J., Kaakoush, N. O., Mitchell, H., and Zhang, L. (2011). Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS ONE 6, e25417. doi:10.1371/journal.pone.0025417
Maher, M., Finnegan, C., Collins, E., Ward, B., Carroll, C., and Cormican, M. (2003). Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter species in clinical specimens of feces. J. Clin. Microbiol. 41, 2980–2986.
Man, S. M. (2011). The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 8, 669–685.
Man, S. M., Kaakoush, N. O., Leach, S. T., Nahidi, L., Lu, H. K., Norman, J., Day, A. S., Zhang, L., and Mitchell, H. M. (2010a). Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J. Infect. Dis. 202, 1855–1865.
Man, S. M., Zhang, L., Day, A. S., Leach, S. T., Lemberg, D. A., and Mitchell, H. (2010b). Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn’s disease. Inflamm. Bowel Dis. 16, 1008–1016.
Man, S. M., Kaakoush, N. O., and Mitchell, H. M. (2011). The role of bacteria and pattern-recognition receptors in Crohn’s disease. Nat. Rev. Gastroenterol. Hepatol. 8, 152–168.
Matsheka, M. I., Elisha, B. G., Lastovica, A. L., and On, S. L. (2002). Genetic heterogeneity of Campylobacter concisus determined by pulsed field gel electrophoresis-based macrorestriction profiling. FEMS Microbiol. Lett. 211, 17–22.
Matsheka, M. I., Lastovica, A. J., Zappe, H., and Elisha, B. G. (2006). The use of (GTG)5 oligonucleotide as an RAPD primer to type Campylobacter concisus. Lett. Appl. Microbiol. 42, 600–605.
Moore, J. E., Barton, M. D., Blair, I. S., Corcoran, D., Dooley, J. S., Fanning, S., Kempf, I., Lastovica, A. J., Lowery, C. J., Matsuda, M., Mcdowell, D. A., Mcmahon, A., Millar, B. C., Rao, J. R., Rooney, P. J., Seal, B. S., Snelling, W. J., and Tolba, O. (2006). The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 8, 1955–1966.
Moore, L. V., Moore, W. E., Cato, E. P., Smibert, R. M., Burmeister, J. A., Best, A. M., and Ranney, R. R. (1987). Bacteriology of human gingivitis. J. Dent. Res. 66, 989–995.
Mukhopadhya, I., Thomson, J. M., Hansen, R., Berry, S. H., El-Omar, E. M., and Hold, G. L. (2011). Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE 6, e21490. doi:10.1371/journal.pone.0021490
Musmanno, R. A., Russi, M., Figura, N., Guglielmetti, P., Zanchi, A., Signori, R., and Rossolini, A. (1998). Unusual species of campylobacters isolated in the Siena Tuscany area, Italy. New Microbiol. 21, 15–22.
Nakou, M., Kamma, J. J., Andronikaki, A., and Mitsis, F. (1998). Subgingival microflora associated with nifedipine-induced gingival overgrowth. J. Periodontol. 69, 664–669.
Nielsen, H. L., Ejlertsen, T., Engberg, J., and Nielsen, H. (2011a). “Clinical epidemiology and manifestations of Campylobacter concisus,” in European Society of Clinical Microbiology and Infectious Diseases, Milan, P990.
Nielsen, H. L., Nielsen, H., Ejlertsen, T., Engberg, J., Gunzel, D., Zeitz, M., Hering, N. A., Fromm, M., Schulzke, J. D., and Bucker, R. (2011b). Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. PLoS ONE 6, e23858. doi:10.1371/journal.pone.0023858
O’Connor, B. C., Newman, H. N., and Wilson, M. (1990). Susceptibility and resistance of plaque bacteria to minocycline. J. Periodontol. 61, 228–233.
On, S. L. (1994). Confirmation of human Campylobacter concisus isolates misidentified as Campylobacter mucosalis and suggestions for improved differentiation between the two species. J. Clin. Microbiol. 32, 2305–2306.
On, S. L., and Harrington, C. S. (2000). Identification of taxonomic and epidemiological relationships among Campylobacter species by numerical analysis of AFLP profiles. FEMS Microbiol. Lett. 193, 161–169.
Oporto, B., and Hurtado, A. (2011). Emerging thermotolerant Campylobacter species in healthy ruminants and swine. Foodborne Pathog. Dis. 8, 807–813.
Paster, B. J., and Gibbons, R. J. (1986). Chemotactic response to formate by Campylobacter concisus and its potential role in gingival colonization. Infect. Immun. 52, 378–383.
Petersen, R. F., Harrington, C. S., Kortegaard, H. E., and On, S. L. (2007). A PCR-DGGE method for detection and identification of Campylobacter, Helicobacter, Arcobacter and related Epsilobacteria and its application to saliva samples from humans and domestic pets. J. Appl. Microbiol. 103, 2601–2615.
Pruckler, J. M., Fields, P. I., and Fitzgerald, C. (2002). Composite analysis of the phenotypic and genotypic characteristics of C. concisus strains. J. Clin. Microbiol. 2001, P230.
Roop, R. M. II, Smibert, R. M., Johnson, J. L., and Krieg, N. R. (1985). DNA homology studies of the catalase-negative campylobacters and “Campylobacter fecalis,” an emended description of Campylobacter sputorum, and proposal of the neotype strain of Campylobacter sputorum. Can. J. Microbiol. 31, 823–831.
Russell, J., and Ward, P. (1998). “Adhesion and invasion of HEp2 cells by Campylobacter concisus from children with diarrhoea,” in Campylobacter, Helicobacter and Related Organisms, eds A. Lastovica, D. G. Newell, and E. Lastovica (Cape Town: University of Cape Town Press), P327–P330.
Shingaki, M., Yokoyama, K., Takahashi, M., Saitoh, K., and Itoh, T. (1999). Detection of pyrazinamidase activity for differentiation of Campylobacter, Arcobacter, and Helicobacter spp. by using a high-performance liquid chromatography method. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 10, 111–116.
Stewart, G. R., Newton, S. M., Wilkinson, K. A., Humphreys, I. R., Murphy, H. N., Robertson, B. D., Wilkinson, R. J., and Young, D. B. (2005). The stress-responsive chaperone alpha-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 55, 1127–1137.
Tankovic, J., Burghoffer, B., and Petit, J. C. (2009). Frequent detection by real-time PCR of bacteria from the Helicobacter and Campylobacter genera in stool samples from inflammatory bowel disease patients,” in 19th European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, R2084.
Tanner, A. C., Dzink, J. L., Ebersole, J. L., and Socransky, S. S. (1987). Wolinella recta, Campylobacter concisus, Bacteroides gracilis, and Eikenella corrodens from periodontal lesions. J. Periodont. Res. 22, 327–330.
Tanner, A. C. R., Badger, S., Lai, C.-H., Listgarten, M. A., Visconti, R. A., and Socransky, S. S. (1981). Wolinella gen. nov., Wolinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and description of Bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. Int. J. Syst. Bacteriol. 31, 432–445.
Taubman, M. A., Haffajee, A. D., Socransky, S. S., Smith, D. J., and Ebersole, J. L. (1992). Longitudinal monitoring of humoral antibody in subjects with destructive periodontal diseases. J. Periodont. Res. 27, 511–521.
Tauxe, R. V., Pegues, D. A., and Hargrett-Bean, N. (1987). Campylobacter infections: the emerging national pattern. Am. J. Public Health 77, 1219–1221.
Tyrrell, K. L., Citron, D. M., Warren, Y. A., Nachnani, S., and Goldstein, E. J. (2003). Anaerobic bacteria cultured from the tongue dorsum of subjects with oral malodor. Anaerobe 9, 243–246.
Valenza, G., Frosch, M., and Abele-Horn, M. (2010). Antimicrobial susceptibility of clinical Campylobacter isolates collected at a German university hospital during the period 2006–2008. Scand. J. Infect. Dis. 42, 57–60.
Van Etterijck, R., Breynaert, J., Revets, H., Devreker, T., Vandenplas, Y., Vandamme, P., and Lauwers, S. (1996). Isolation of Campylobacter concisus from feces of children with and without diarrhea. J. Clin. Microbiol. 34, 2304–2306.
Vandamme, P., Dewhirst, F. E., Paster, B. J., and On, S. L. W. (2005). “Campylobacter,” in Bergey’s Manual of Systematic Bacteriology, eds D. J. Brenner, N. R. Krieg, and J. T. Staley (New York: Springer), 1147–1160.
Vandamme, P., Falsen, E., Pot, B., Hoste, B., Kersters, K., and De Ley, J. (1989). Identification of EF group 22 campylobacters from gastroenteritis cases as Campylobacter concisus. J. Clin. Microbiol. 27, 1775–1781.
Vandenberg, O., Houf, K., Douat, N., Vlaes, L., Retore, P., Butzler, J. P., and Dediste, A. (2006). Antimicrobial susceptibility of clinical isolates of non-jejuni/coli campylobacters and arcobacters from Belgium. J. Antimicrob. Chemother. 57, 908–913.
Weber, P., Koch, M., Heizmann, W. R., Scheurlen, M., Jenss, H., and Hartmann, F. (1992). Microbic superinfection in relapse of inflammatory bowel disease. J. Clin. Gastroenterol. 14, 302–308.
Zhang, L., Budiman, V., Day, A. S., Mitchell, H., Lemberg, D. A., Riordan, S. M., Grimm, M., Leach, S. T., and Ismail, Y. (2010). Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J. Clin. Microbiol. 48, 2965–2967.
Zhang, L., Man, S. M., Day, A. S., Leach, S. T., Lemberg, D. A., Dutt, S., Stormon, M., Otley, A., O’Loughlin, E. V., Magoffin, A., Ng, P. H., and Mitchell, H. (2009). Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J. Clin. Microbiol. 47, 453–455.
Keywords: Campylobacter concisus, oral, intestine, enteritis, Crohn’s disease, pathogenesis, reservoirs of infection, antibiotic susceptibility
Citation: Kaakoush NO and Mitchell HM (2012) Campylobacter concisus – a new player in intestinal disease. Front. Cell. Inf. Microbio. 2:4. doi: 10.3389/fcimb.2012.00004
Received: 29 November 2011;
Accepted: 17 January 2012;
Published online: 03 February 2012.
Edited by:
D. Scott Merrell, Uniformed Services University, USAReviewed by:
Terry Kwok, Monash University, AustraliaGeorgina Hold, University of Aberdeen, UK
Albert Joseph Lastovica, University of the Western Cape, South Africa
Copyright: © 2012 Kaakoush and Mitchell. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hazel Marjory Mitchell, School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, NSW 2052, Australia. e-mail:aC5taXRjaGVsbEB1bnN3LmVkdS5hdQ==
 Nadeem Omar Kaakoush
Nadeem Omar Kaakoush