
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 07 December 2011
Sec. Molecular Bacterial Pathogenesis
Volume 1 - 2011 | https://doi.org/10.3389/fcimb.2011.00013
This article is part of the Research Topic Burkholderia cepacia complex: an embarrassment of niches View all 11 articles
Trimeric autotransporter adhesins (TAAs) are multimeric surface proteins exclusively found in bacteria. They are involved in various biological traits of pathogenic Gram-negative bacteria including adherence, biofilm formation, invasion, survival within eukaryotic cells, serum resistance, and cytotoxicity. TAAs have a modular architecture composed by a conserved membrane-anchored C-terminal domain and a variable number of stalk and head domains. In this study, a bioinformatic approach has been used to analyze the distribution and architecture of TAAs among Burkholderia cepacia complex (Bcc) genomes. Fifteen genomes were probed revealing a total of 74 encoding sequences. Compared with other bacterial species, the Bcc genomes contain a large number of TAAs (two genes to up to eight genes, such as in B. cenocepacia). Phylogenetic analysis showed that the TAAs grouped into at least eight distinct clusters. TAAs with serine-rich repeats are clearly well separated from others, thereby representing a different evolutionary lineage. Comparative gene mapping across Bcc genomes reveals that TAA genes are inserted within conserved synteny blocks. We further focused our analysis on the epidemic strain B. cenocepacia J2315 in which seven TAAs were annotated. Among these, three TAA-encoding genes (BCAM019, BCAM0223, and BCAM0224) are organized into a cluster and are candidates for multifunctional virulence factors. Here we review the current insights into the functional role of BCAM0224 as a model locus.
Bacteria belonging to the Burkholderia cepacia complex (Bcc) have emerged as highly problematic opportunistic human pathogens in immunocompromised individuals and in patients with the genetic disease cystic fibrosis (CF). The virulence of the Bcc members is variable and B. cenocepacia and B. multivorans are the most common species isolated from the respiratory tract of CF patients. Bcc strains posses a wide range of virulence factors that are critical for colonization and disease. Despite the identification and characterization of some, many details of Burkholderia virulence still remains to be clarified (reviewed in Drevinek and Mahenthiralingam, 2010).
To initiate infection in CF patients, Bcc strains must be able to colonize the respiratory epithelium by binding to a diverse group of host cell surface molecules including proteins, glycolipid receptors, and secretory mucins (McClean and Callaghan, 2009). This essential step, although not fully characterized, is mediated by a variety of proteins collectively termed adhesins, which are surface-exposed proteins (Kline et al., 2009). Thus far, only the cable pili-associated adhesin (cbl, 22 kDa) of B. cenocepacia, has been identified to interacts with cytokeratin 13, a 55-kDa protein which is enriched in CF epithelial cells differentiated into squamous phenotype (Sajjan et al., 2000). However, cbl gene is absent in many Bcc isolates (Sajjan et al., 2002), suggesting that other adhesins may play a relevant role in epithelial adhesion and colonization. Among these, the family of the designated trimeric autotransporter adhesins (TAAs) represents a class of proteins found in Gram-negative pathogens that are known to mediate adherence of the bacteria to host tissues and thereby may be relevant for the overall pathogenic potential of Bcc strains.
Trimeric autotransporter adhesins belong to a subtype of an outer membrane family of proteins termed autotransporters, that have been studied and emerging as important virulence factors in a range of pathogenic alpha-, beta-, and gamma-proteobacteria (Linke et al., 2006). Adhesion to extracellular matrix (ECM) proteins and host cells seems to be the major role played by these proteins (Linke et al., 2006). Müller et al. (2011b) have used both static and dynamic adhesion assays, aiming to prove the involvement of three distinct TAAs in bacterial adherence (Müller et al., 2011b). These authors were further able to show that these three TAAs exhibit promiscuous binding to ECM proteins and endothelial cells, albeit with differences in the results obtained in the static and dynamic conditions (Müller et al., 2011b). Despite the importance of TAAs in cell adhesion, these proteins are multifunctional virulence factors involved in several other biological traits of pathogenic Gram-negative bacteria including biofilm formation, cell-to-cell aggregation, protecting the bacterium from host immune responses (serum resistance), and promoting the invasion of host cells (Heise and Dersch, 2006; Serruto et al., 2009).
Trimeric autotransporter adhesins are multi-domain proteins organized in a modular fashion i.e., an integral membrane-anchored C-terminal domain that forms a trimeric 12-stranded beta-barrel pore and permits, through the type V protein secretion pathway (T5SS), the translocation of a passenger domain (divided in two regions, the stalk and an N terminal head) into the extracellular space (Cotter et al., 2005). Among the various TAAs described, YadA from enteropathogenic Yersinia species (Yersinia enterocolitica and Yersinia pseudotuberculosis) represent the structural prototype for this family of proteins (Koretke et al., 2006; Figure 1). TAAs trimerization is essential for their translocation and function, providing stability and potential for multivalent interactions. Although it is poorly understood and controversial, TAAs biogenesis seems to occur dependently of other protein partners (Lehr et al., 2010). The C-terminal translocator domain is highly conserved among TAAs (generally consists of 70–100 amino acid residues) and therefore used as the defining element of the family (Cotter et al., 2005). In contrast, the passenger domains are fibrous, more or less repetitive, varying in length, and sequence motifs (Linke et al., 2006). Further, these tandemly repeated sequences may undergo contraction or expansion, thereby defining their specific activities (Linke et al., 2006; Sheets and St Geme III, 2011). Often, the passenger domains contain immunogenic Hep_Hag (Pfam PF05658) and HIM (Pfam PF05662) domains that thereby make them good candidates for vaccine development (Tiyawisutsri et al., 2007). Hep_Hag and HIM are short repeat motifs found in bacterial hemagglutinins and invasins. Although it has been shown that TAAs were found only in prokaryotes, Müller et al. (2011a) demonstrated that the expression in yeast of the β-barrel domain of the Y. enterocolitica yadA resulted in the synthesis of a trimeric 12-stranded beta-barrel, exclusively targeted to the mitochondrial outer membrane (Müller et al., 2011a).
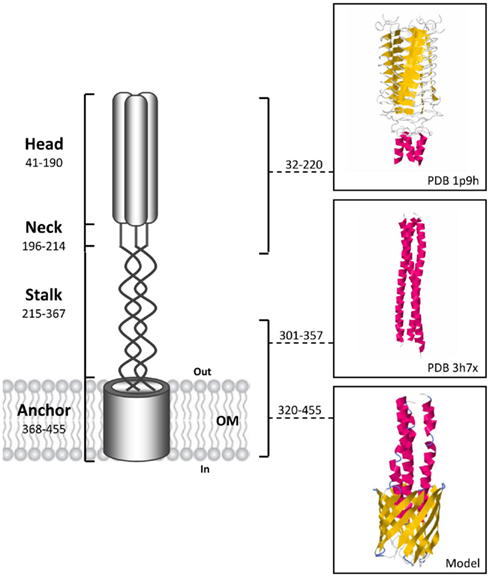
Figure 1. Schematic representation of a full-length trimeric autotransporter adhesin based on the prototypical model of YadA from Yersinia enterocolitica, with the domains predicted by the domain annotation of trimeric autotransporter adhesins (daTAA) program. The residues numbers for each domain of the YadA protein are showed. For the head and a part of the stalk the 3D ribbon structures are available and showed with the respective PDB ID. The 3D structure of the YadA anchor has not been solved yet and the monomer model was constructed using the automated Swiss model Workspace (http://www.ebi.ac.uk/newt/; Arnold et al., 2006) with 3EMO as a template. Finally, a predicted 3D model of the trimeric YadA anchor domain (12-stranded beta-barrel) was generated using Cluspro 2.0 software (http://cluspro.bu.edu/home.php; Comeau et al., 2004).
Several TAAs have been characterized in terms of function and structure within a large number of bacterial pathogens, including, among others, YadA from Y. enterocolitica (Nummelin et al., 2004), Adhesin A from Bartonella henselae (Riess et al., 2004), NadA from Neisseria meningitidis (Capecchi et al., 2005), Hia from Haemophilus influenza (Meng et al., 2006), an IgD-binding protein from Moraxella catarrhalis (Riesbeck et al., 2006), AipA and TaaP from Proteus mirabilis (Alamuri et al., 2010), BpaA from Burkholderia pseudomallei (Edwards et al., 2010), Sad A from Salmonella enterica (Raghunathan et al., 2011), and Cha from Haemophilus cryptic genospecies (Sheets and St Geme III, 2011). YadA from Y. enterocolitica has been one of the most extensively studied and is found to display a multifaceted activity during host–pathogen interaction (reviewed in Linke et al., 2006).
Herein, as a first approach, we have conducted an in silico analysis in completed genomes of Bcc members aiming to identify TAA-encoding sequences. The proteins selected were studied through sequence similarity, phylogeny, and synteny conservation data. Then, we focused our analysis on the epidemic strain B. cenocepacia J2315 in which seven TAAs were annotated. Among those, we particularly focused our attention on three clustered TAAs-encoding genes (BCAM0219, BCAM0223, BCAM0224) that are strong candidates for multifunctional pathogenic factors. Finally, we review here recent results arising from the functional analysis of BCAM0224 as a model locus.
We used as a starting point the Bcc protein sequences predicted by the domain annotation of trimeric autotransporter adhesins (daTAA) program1 (Szczesny and Lupas, 2008). These sequences were used to search the 15 Bcc genomes [(9) finished and (6) unfinished] available in Burkholderia genome database2 and integrated microbial genomes (IMG) system3 searching for other putative TAAs. Next, to confirm the results, we used BLASTP 2.0 against Bcc genomes available at National Center for Biotechnology Information (NCBI) and all proteins identified were verified with daTAA program to confirm the presence of the membrane anchor [Pfam YadA domain (PF03895)] and at least another characteristic domain of TAAs. The following Bcc genomes were analyzed: B. ambifaria MC40-6, B. ambifaria MEX-5, B. ambifaria IOP40-10, B. cenocepacia AU 1054, B. cenocepacia HI2424, B. cenocepacia J2315, B. cenocepacia MC0-3, B. cenocepacia PC184, B. cepacia AMMD, B. dolosa AUO158, B. lata 383, B. multivorans ATCC 17616, B. multivorans CGD1, B. ubonensis Bu, B. vietnamiensis G4.
In total, our analysis revealed the existence of 74 putatively TAA-encoding sequences. Compared with other bacterial species, some Bcc genomes contain a large number of TAAs, which probably reflects their large multireplicon genome sizes and may have been acquired by insertions of transposable elements and/or bacteriophages or through horizontal transfer of DNA fragments. Furthermore, the higher number of TAAs found on Bcc genomes also suggests a high genome plasticity that ultimately may be relevant in their capacity to adhere and colonize human hosts as well as other environments. The absolute numbers and the density of TAAs are variable among the Bcc genomes under study. The genome of B. cenocepacia MC0-3 contains the highest number (8) and density and B. vietnamiensis G4 the lowest number (2) and density. Whether these TAAs have redundant or unique functions is an important question that needs to be answered. In addition, we also calculated the density of TAAs encoding genes relative to non-Burkholderia genomes available in the data database. To date, there are only five bacterial genera showing higher TAA gene density than those of Bcc genomes, namely, Fusobacterium, Bartonella, Haemophilus, Moraxella, and Xylella.
To analyze the phylogenetic relationship between the full-length amino acid sequences of the TAAs, we first created a multiple sequence alignment with ClustalW 2.0.12 (Thompson et al., 1994). Despite the conservation in domain architecture, different lengths, and a low overall sequence identity were observed across all TAAs. However, as expected, the C-terminal translocator domains are highly conserved.
The unrooted phylogenetic tree prepared with the 74 TAAs reveals that the sequences have been found to fall into at the least eight clusters with different evolutionary lineages (Figure 2). The tree based on the sequences of the C-terminal translocator conserved domain (data not shown) gave essentially the same result as the tree constructed from the entire sequences. The clusters I, V, VII, and VIII, respectively with 12, 12, 14, and 15 members were the tree most representative groups. The clusters I, II, III appear to be related, thereby suggesting that they were derived from a single ancestral. In contrast, clusters V and VI have clearly different lineages that may represent separate evolutionary histories (Figure 2). The evolutionary related clusters VII and VIII include only those TAAs with serine-rich repeats; cluster VII is formed with proteins containing only one serine-rich repeat domain whereas cluster VIII grouped the proteins with several extensive serine-rich repeats (Figures 2 and 3). As far as we know, these serine-rich repeat proteins have been found only in Gram-positive bacteria where they appear to play a decisive role in colonization; these include, among others, the glycoproteins Fap1 of Streptococcus parasanguinis, which mediates bacterial adhesion to saliva-coated hydroxyapatite (Wu et al., 2007), GspB of S. gordonii M99, which mediates bacterial binding of human platelets (Bensing et al., 2004) and SrpA of S. cristatus which mediates bacterial adhesion in oral biofilms (Handley et al., 2005). A common mechanism involved in these interactions is recognition of surface-associated host sialoglycoconjugates via the hydroxyl groups of S or T residues (O-glycosylation; Zhou and Wu, 2009).
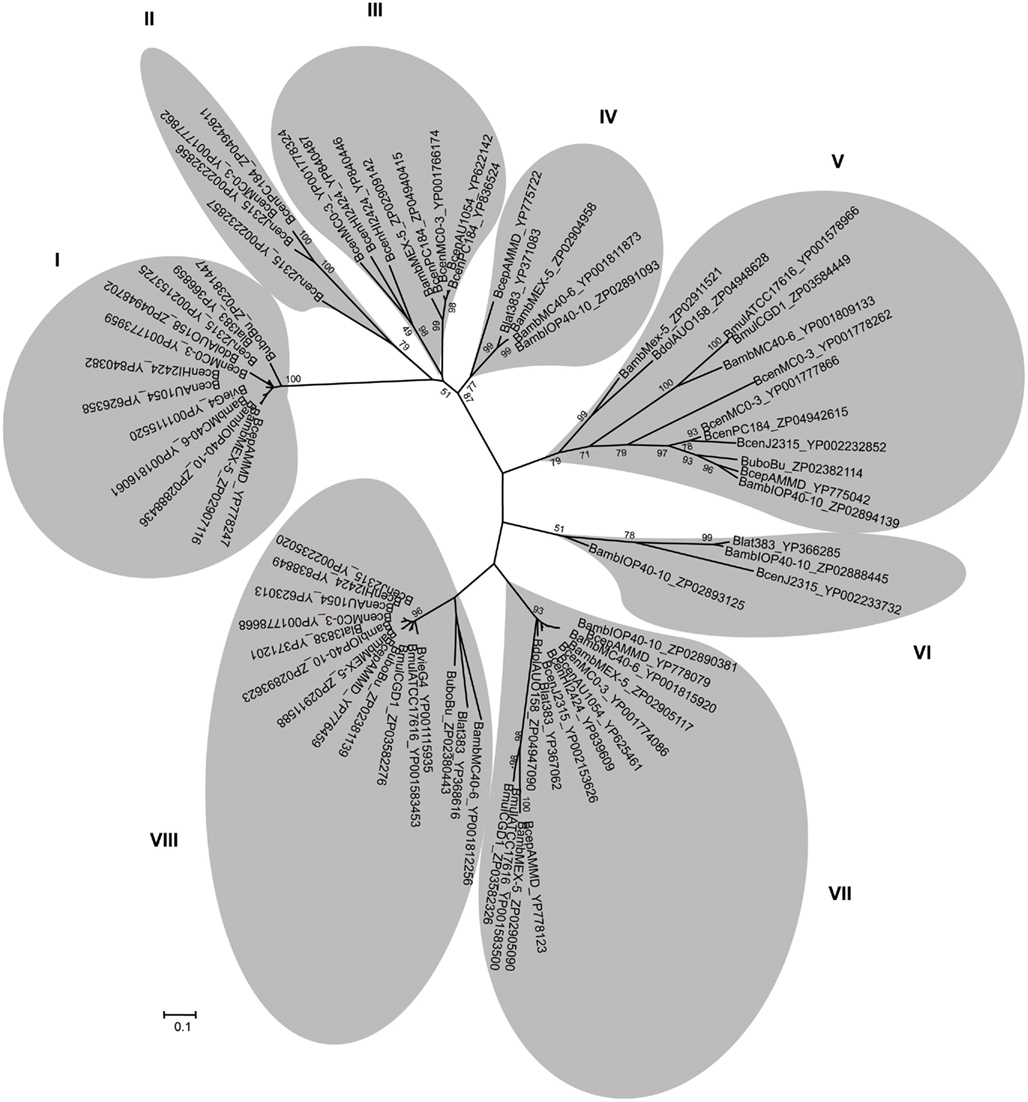
Figure 2. Phylogenetic tree of the 74 TAA sequences, performed with MEGA5 (Tamura et al., 2011), using the neighbor-joining method (Perriere and Gouy, 1996). The phylogenetic tree was constructed based on the result of the global alignment of the 74 sequences performed by ClustalW 2.0.12 (Thompson et al., 1994). A branch length of 0.1 substitution/site is given to phylogenetic distances. The values adjacent to a node indicate the percentage of 1000 bootstrap trees that contain the node. Each sequence is identified using the strain abbreviated name composed by the first letter of the genus, the three first letters of the specie and the strain code, followed by the GenBank accession number.
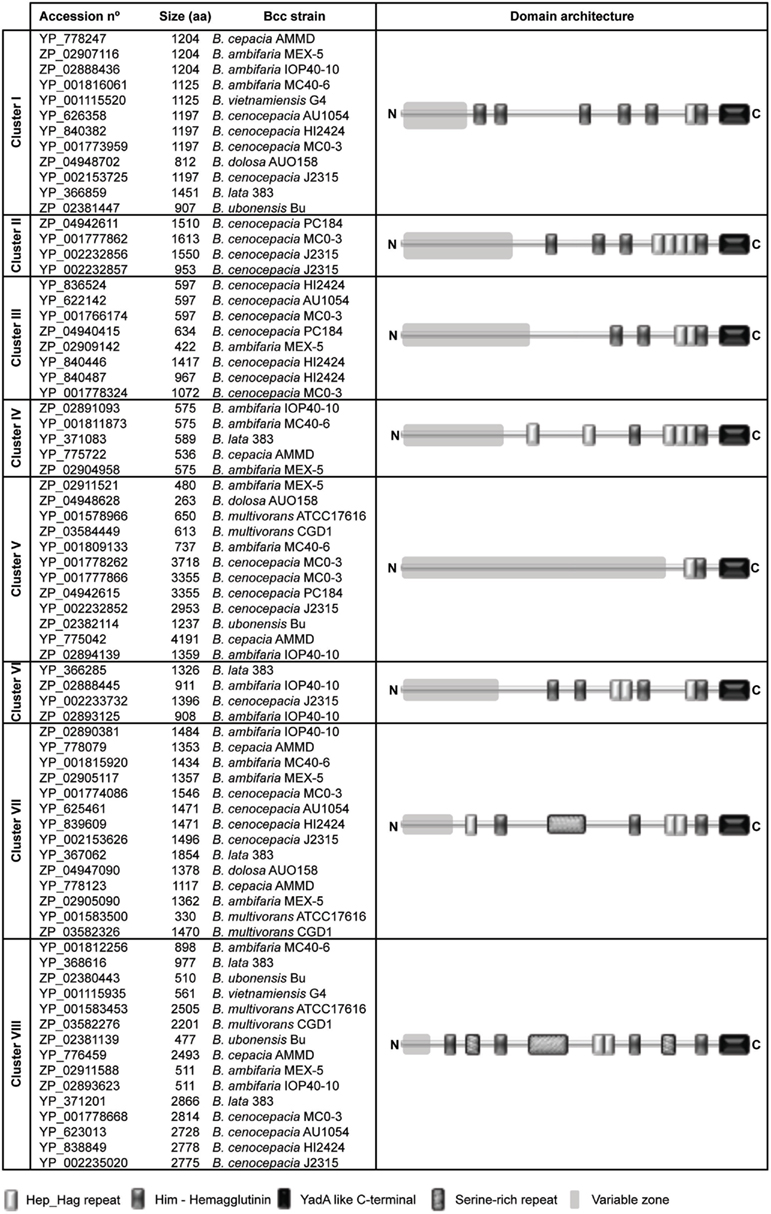
Figure 3. Domain architecture of TAA proteins being conserved throughout the tree clusters defined in Figure 2. The Pfam database (Finn et al., 2010) was used to obtain the details of domain organization. Keys for the Pfam domains are shown in the bottom.
We further analyzed the domain architecture representative of each cluster using the Pfam protein family database (Finn et al., 2010). Each of them contains an identical C-terminal YadA domain and a variable number of the other typical domains found in TAAs, such as the HIM and Hep_Hag domains. In addition, they have variable regions (in size and sequence) that are not conserved between the defined tree clusters (Figure 3).
We next analyzed the distribution of the TAAs across the eight clusters defined in the topology of the tree. A general finding was that the TAAs are under-represented in three Bcc species, namely B. multivorans, B. dolosa, and B. vietnamiensis (Figure 4). Since B. multivorans is one of the most prevalent Bcc species in CF patients, the results showed in Figure 4 render a clear difficulty to draw a logical distribution of TAAs across the Bcc species understudy. Furthermore, two other interesting findings emerged from our analysis: (i) TAAs included in the clusters I, VII, and VIII are the most representatives within the Bcc species; (ii) although the distribution of TAAs is not species specific, the representatives included in the clusters II and III are almost exclusive found in the genomes of the B. cenocepacia strains (Figure 4). Further work is needed to characterize the functions and specificities of these proteins, and the role they play in the pathogenesis of Bcc species.
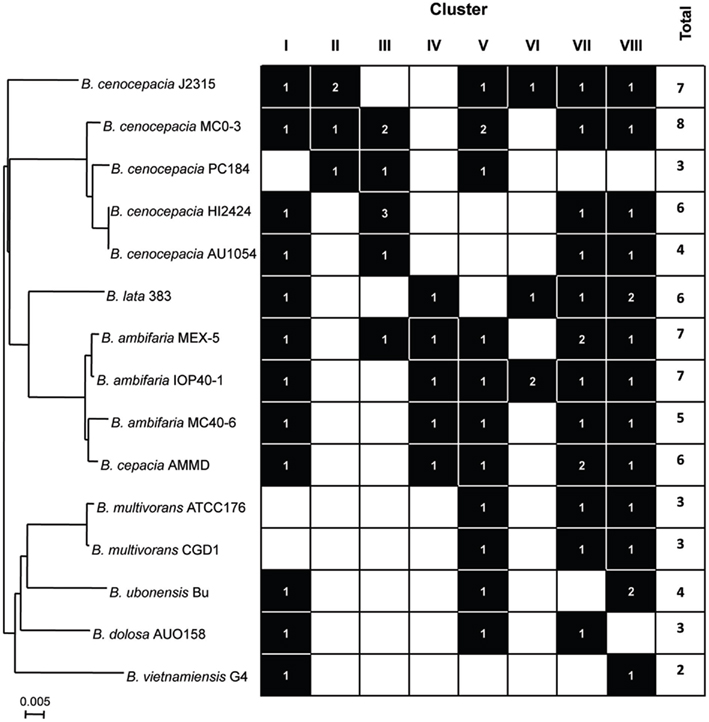
Figure 4. Relationship between Bcc phylogeny (based on recA sequence) and the presence/absence of the TAAs across the eight tree clusters defined in Figure 2. Phylogenetic distances were estimated using the neighbor-joining method (Saitou and Nei, 1987), applying a distance matrix and visualized with NJplot (Perriere and Gouy, 1996). A branch length of 0.005 substitution/site is given to phylogenetic distances.
Finally, all of the TAA sequences identified were examined in terms of the chromosomal arrangement and annotation of their neighboring genes. As shown in Figure 5, we concluded that each defined tree cluster is likely to represent distinct conserved genetic organizations and ultimately can reflect functional relationships between genes. Interestingly, in the genetic organizations defined for clusters I, II, and VII, our analysis reveals the existence of genes encoding a sensor (histidine kinase or TonB like) and one or more response-regulator proteins in the vicinity of the TAA-encoding gene. This finding suggests a possible two-component signal transduction system, where the periplasmic sensor histidine kinase is responsible for sensing stimuli and a second component regulates the virulence effector, namely the TAA gene (Figure 5). The bacterial prototype for this system is the Bordetella pertussis BvgAS two-component regulatory system which is involved in the expression of many adhesins and toxins (Jones et al., 2005). It is now important to obtain experimental data in order to validate the hypothesis raised by this in silico analysis.
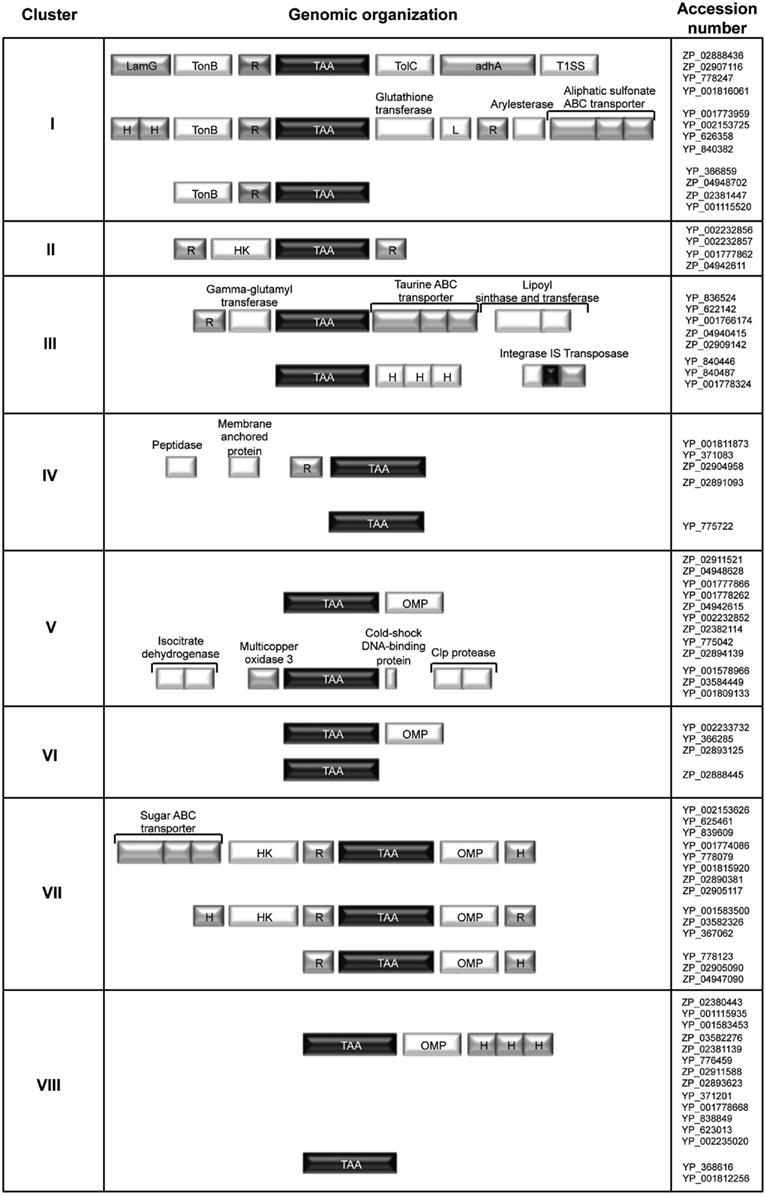
Figure 5. Conserved synteny blocks around the TAA genes in the Bcc genomes. Each syntenic block is representative of each defined cluster of the phylogenetic tree showed in Figure 2. The conserved chromosomal arrangement and annotation of their neighboring genes are presented. (TonB, TonB-dependent siderophore receptor; HK, histidine kinase; R, regulator; OMP, outer membrane protein; TAA, trimeric autotransporter adhesin; L, lipoprotein; LamG, laminin G domain of extracellular proteins; TolC, TolC outer membrane channel; adhA, cable pili-associated adhesin; T1SS, type I secretion system; H, hypothetical protein).
Furthermore, the analysis of synteny between the tree clusters V, VI, VII, and VIII, reveals the existence of a conserved gene encoding an outer membrane protein (OMP) in the vicinity of the TAA-encoding gene (Figure 5). It is noteworthy that recent biochemical and structural studies have raised pertinent questions about the traditional paradigm of TAA biogenesis as self-contained secretion system. In fact, several evidences now support that many passenger domains are transported across the outer membrane by exogenous auxiliary proteins, such as OMPs and periplasmic chaperones (Ieva and Bernstein, 2009; Ruiz-Perez et al., 2009; Wagner et al., 2009).
We further focused our analysis on the epidemic strain B. cenocepacia J2315 in which seven TAAs were annotated (Figure 6A). Of the seven TAA-encoding genes, five were located on chromosome 2 (BCAM0219, BCAM0223, BCAM0224, BCAM2418, BCAM1115) and two on chromosome 3 (BCAS0236, BCAS0335; Figure 6B). These observations are consistent with the fact that the chromosomes 2 and 3 contain a large number of virulence genes, whereas chromosome 1, the largest replicon, carries the majority of the core functions (Holden et al., 2009).
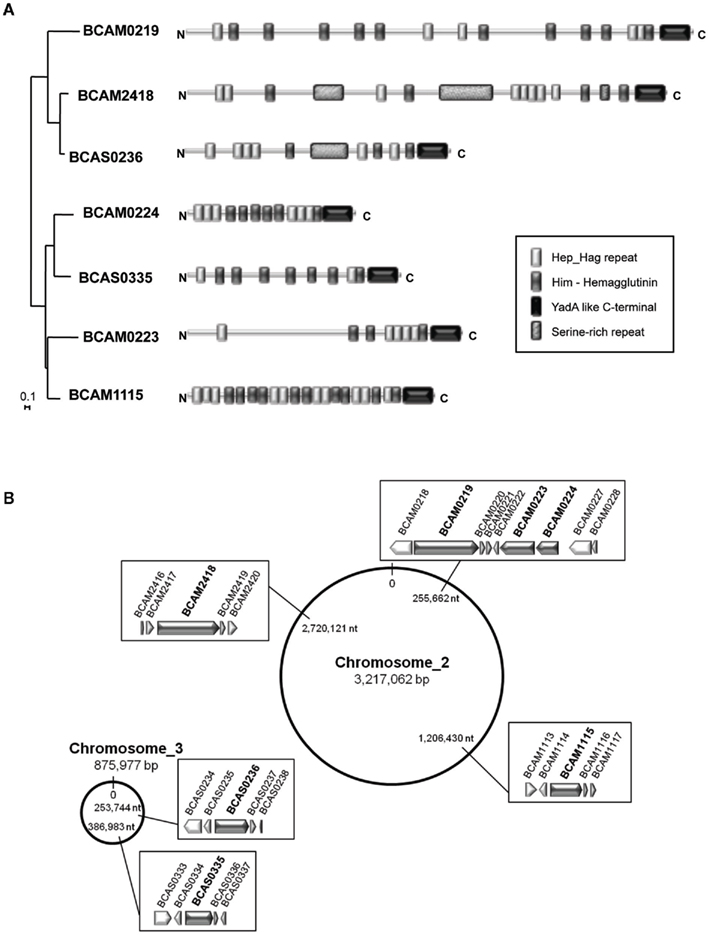
Figure 6. (A) Phylogenetic relationships and domain organization of the seven TAAs from the epidemic strain B. cenocepacia J2315. Phylogenetic distances of the C-terminal (70–71 aa) of the seven TAAs from B. cenocepacia J2315, were estimated using the neighbor-joining method (Saitou and Nei, 1987), applying a distance matrix and visualized with NJplot (Perriere and Gouy, 1996). A branch length of 0.1 substitution/site is given to phylogenetic distances. Domain organization is defined based on Pfam analysis (Finn et al., 2010). Keys for the Pfam domains are shown in the bottom. (B) Chromosomal location of the seven TAA-encoding genes in the genome of the epidemic strain B. cenocepacia J2315.
As previously stated in Figure 4, these seven TAAs are scattered over the phylogenetic tree, as follows: one member in cluster I, V, VI, VII, and VIII and two members in cluster II. Analysis of these TAA proteins reveals the existence of common complex architectures represented by multi-modular and polyfunctional domains, despite only weak sequence conservation (Figure 6A). Given the importance of TAAs in the virulence of Gram-negative pathogens, it is likely that these multifunctional proteins may play decisive roles in B. cenocepacia virulence.
Of the seven TAAs identified, three (BCAM0219, BCAM0223, BCAM0224) are described by Mil-Homens et al. (2010) and form part of a 24-kb cluster located downstream of the B. cenocepacia cci pathogenicity island (Baldwin et al., 2004). This cluster has a unique gene arrangement composed by three TAA-encoding genes (BCAM0219, BCAM0223, BCAM0224), one lipoprotein (BCAM0220), two sensor histidine kinases (BCAM0218, BCAM0227), and three response-regulator genes (BCAM0221, BCAM0222, and BCAM0228; Mil-Homens et al., 2010). Recently, McCarthy et al. (2010) demonstrated the involvement of the BCAM0227 sensor kinase in the perception of a cell–cell signaling molecule known as the Burkholderia diffusible signal factor (BDSF).
Quantitative real-time PCR analysis reveals that these TAAs clustered genes are overexpressed under certain environmental conditions such as, high osmolarity, oxygen limited conditions, and oxidative stress (Mil-Homens et al., 2010). Further, Mil-Homens et al. (2010) have developed a series of PCR-based assays to verify the presence of the selected TAA genes in 47 genomes representing the 17 species of the Bcc (Mil-Homens et al., 2010). It is noteworthy that a PCR test targeting the TAA-encoding gene BCAM0224 has been proved to be specific for epidemic B. cenocepacia strains belong to the ET-12 lineage (Mil-Homens et al., 2010). Aiming to prove the usefulness of this PCR assay as a genetic marker to discriminate epidemic strains of the ET-12 lineage, Mil-Homens et al. (2010) also assessed the use of the BCESM marker (Mahenthiralingam et al., 1997) across the same panel of Bcc strains. The results obtained have shown that the BCAM0224 sequence was exclusively detected in members of the ET-12 lineage whereas the BCESM sequence was found in ET-12 isolates as well as in some other epidemic and non-epidemic B. cenocepacia isolates (Mil-Homens et al., 2010). Thereby, we consider that this novel PCR-based assay may serve as a valuable tool to aid in Bcc strain identification.
The deduced protein encoded by BCAM0224 is composed of 953 amino acids, with a calculated molecular mass of 85 kDa and a pI of 4.02 (Mil-Homens et al., 2010). Analysis of the amino acid composition revealed that the protein contained 7.1, 3.8, 41.6, and 58.4% acidic, basic, polar, and hydrophobic residues, respectively. The presence of an extended signal sequence with a predicted cleavage site between amino acids 1 and 43 was identified. The deduced amino acid sequence encoded by BCAM0224 showed a head–stalk–anchor modular structure composed by seven clusters of Hep_Hag (Pfam domain PF05658), six clusters of HIM (Pfam domain PF05662), and two collagen-binding domains (Pfam domain PF01391; Figure 6A; Mil-Homens et al., 2010).
In order to investigate the contribution of BCAM0224 for virulence, Mil-Homens et al. (2010) have constructed a knockout mutant and tested its ability to adhere to ECM components and to kill the larvae Galleria mellonella, used as a model host to study Burkholderia pathogenesis (Mil-Homens et al., 2010). Overall, the TAA BCAM0224 protein showed adhesive properties to collagen type I, one of the most abundant components of the ECM. Furthermore, the same authors also examined adhesion of the Escherichia coli BL21 cells expressing the gene of interest. A significant difference was found between the recombinant and the vector control, confirming that BCAM0224 has collagen-binding properties. Finally, Mil-Homens et al. (2010) used the insect Galleria mellonella as a model of infection to analyze whether BCAM0224 is a virulence determinant in the pathogenesis of B. cenocepacia. At 72 h post-infection, compared to the wild-type B. cenocepacia K56-2, the BCAM0224-mutant exhibited attenuated (10%) killing ability in comparison to the wild-type (Mil-Homens et al., 2010). Collectively, these results strongly suggest that BCAM0224 is important for adhesion and virulence of B. cenocepacia cells.
Over the last years, important advances have been made in the study of TAAs, as novel virulence factors produced by Gram-negative bacteria, where their main function is to act as adhesins. To initiate infection Bcc species must be able to colonize the respiratory epithelium by binding to specific host macromolecules. This process has only begun to be studied and remains to be fully characterized. With these aspects in mind, here we present an in silico approach to identify TAA-encoding sequences in Bcc pathogenic strains. As a result, 74 TAA-encoding genes potential implicated in functional aspects associated to Bcc pathogenicity, such as cell adhesion, were predicted and classified by phylogenetic analysis. Among the candidates, we review experimental data supporting that the BCAM0224 from B. cenocepacia J2315 represents a collagen-binding TAA with an important role in cellular adhesion and virulence. Overall, the TAA proteins identified in this study are promising targets for future experimental analysis and could represent a valuable resource for unveiling mechanisms underlying the pathogenesis of Bcc bacteria.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported in part by Fundação para a Ciência e a Tecnologia (FCT), Portugal (grant PTDC/EBB-BIO/100326/2008) and Ph.D. fellowship to D. Mil-Homens.
Alamuri, P., Lower, M., Hiss, J. A., Himpsl, S. D., Schneider, G., and Mobley, H. L. T. (2010). Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect. Immun. 78, 4882–4894.
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201.
Baldwin, A., Sokol, P. A., Parkhill, J., and Mahenthiralingam, E. (2004). The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72, 1537–1547.
Bensing, B. A., Gibson, B. W., and Sullam, P. M. (2004). The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J. Bacteriol. 186, 638–645.
Capecchi, B., Adu-Bobie, J., Di Marcello, F., Ciucchi, L., Masignani, V., Taddei, A., Rappuoli, R., Pizza, M., and Aricò, B. (2005). Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55, 687–698.
Comeau, S. R., Gatchell, D. W., Vajda, S., and Camacho, C. J. (2004). ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50.
Cotter, S. E., Surana, N. K., and St Geme, J. W. III. (2005). Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13, 199–205.
Drevinek, P., and Mahenthiralingam, E. (2010). Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16, 821–830.
Edwards, T. E., Phan, I., Abendroth, J., Dieterich, S. H., Masoudi, A., Guo, W., Hewitt, S. N., Kelley, A., Leibly, D., Brittnacher, M. J., Staker, B. L., Miller, S. I., Van Voorhis, W. C., Myler, P. J., and Stewart, L. J. (2010). Structure of a Burkholderia pseudomallei trimeric autotransporter adhesin head. PLoS ONE 5, e12803. doi:10.1371/journal.pone.0012803
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., Gavin, O. L., Gunasekaran, P., Ceric, G., Forslund, K., Holm, L., Sonnhammer, E. L. L., Eddy, S. R., and Bateman, A. (2010). The Pfam protein families database. Nucleic Acids Res. 38, 211–222.
Handley, P. S., Correia, F. F., Russell, K., Rosan, B., and DiRienzo, J. M. (2005). Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol. Immunol. 20, 131–140.
Heise, T., and Dersch, P. (2006). Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc. Natl. Acad. Sci. U.S.A. 103, 3375–3380.
Holden, M. T. G., Seth-Smith, H. M. B., Crossman, L. C., Sebaihia, M., Bentley, S. D., Cerdeño-Tárraga, A. M., Thomson, N. R., Bason, N., Quail, M. A., Sharp, S., Cherevach, I., Churcher, C., Goodhead, I., Hauser, H., Holroyd, N., Mungall, K., Scott, P., Walker, D., White, B., Rose, H., Iversen, P., Mil-Homens, D., Rocha, E. P. C., Fialho, A. M., Baldwin, A., Dowson, C., Barrell, B. G., Govan, J. R., Vandamme, P., Hart, C. A., Mahenthiralingam, E., and Parkhill, J. (2009). The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191, 261–277.
Ieva, R., and Bernstein, H. D. (2009). Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 19120–19125.
Jones, A. M., Boucher, P. E., Williams, C. L., Stibitz, S., and Cotter, P. A. (2005). Role of BvgA phosphorylation and DNA binding affinity in control of Bvg-mediated phenotypic phase transition in Bordetella pertussis. Mol. Microbiol. 58, 700–713.
Kline, K. A., Fälker, S., Dahlberg, S., Normark, S., and Henriques-Normark, B. (2009). Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592.
Koretke, K. K., Szczesny, P., Gruber, M., and Lupas, A. N. (2006). Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struct. Biol. 155, 154–161.
Lehr, U., Schütz, M., Oberhettinger, P., Ruiz-Perez, F., Donald, J. W., Palmer, T., Linke, D., Henderson, I. R., and Autenrieth, I. B. (2010). C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol. Microbiol. 78, 932–946.
Linke, D., Riess, T., Autenrieth, I. B., Lupas, A., and Kempf, V. A. J. (2006). Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14, 264–270.
Mahenthiralingam, E., Simpson, D. A., and Speert, D. P. (1997). Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35, 808–816.
McCarthy, Y., Yang, L., Twomey, K. B., Sass, A., Tolker-Nielsen, T., Mahenthiralingam, E., Dow, J. M., and Ryan, R. P. (2010). A sensor kinase recognizing the cell–cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol. Microbiol. 77, 1220–1236.
McClean, S., and Callaghan, M. (2009). Burkholderia cepacia complex: epithelial cell-pathogen confrontations and potential for therapeutic intervention. J. Med. Microbiol. 58, 1–12.
Meng, G., Surana, N. K., St Geme, J. W., and Waksman, G. (2006). Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 25, 2297–2304.
Mil-Homens, D., Rocha, E. P. C., and Fialho, A. M. (2010). Genome-wide analysis of DNA repeats in Burkholderia cenocepacia J2315 identifies a novel adhesin-like gene unique to epidemic-associated strains of the ET-12 lineage. Microbiology 156, 1084–1096.
Müller, J. E. N., Papic, D., Ulrich, T., Grin, I., SchÃ1/4tz, M., Oberhettinger, P., Tommassen, J., Linke, D., Dimmer, K. S., Autenrieth, I. B., and Rapaport, D. (2011a). Mitochondria can recognize and assemble fragments of a beta-barrel structure. Mol. Biol. Cell 22, 1638–1647.
Müller, N. F., Kaiser, P. O., Linke, D., Schwarz, H., Riess, T., Schäfer, A., Eble, J. A., and Kempf, V. A. J. (2011b). Trimeric autotransporter adhesin-dependent adherence of Bartonella henselae, Bartonella quintana and Yersinia enterocolitica to matrix components and endothelial cells under static and dynamic flow conditions. Infect. Immun. 79, 2544–2553.
Nummelin, H., Merckel, M. C., Leo, J. C., Lankinen, H., Skurnik, M., and Goldman, A. (2004). The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23, 701–711.
Perriere, G., and Gouy, M. (1996). WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369.
Raghunathan, D., Wells, T. J., Morris, F. C., Shaw, R. K., Bobat, S., Peters, S. E., Paterson, G. K., Jensen, K. T., Leyton, D. L., Blair, J. M. A., Browning, D. F., Pravin, J., Flores-Langarica, A., Hitchcock, J. R., Moraes, C. T. P., Piazza, R. M. F., Maskell, D. J., Webber, M. A., May, R. C., MacLennan, C. A., Piddock, L. J., Cunningham, A. F., and Henderson, I. R. (2011). SadA, a trimeric autotransporter from Salmonella enterica serovar Typhimurium, can promote biofilm formation and provides limited protection against infection. Infect. Immun. 79, 4342–4352.
Riesbeck, K., Tan, T., and Forsgren, A. (2006). MID and UspA1/A2 of the human respiratory pathogen Moraxella catarrhalis, and interactions with the human host as basis for vaccine development. Acta Biochim. Pol. 53, 445–456.
Riess, T., Andersson, S. G. E., Lupas, A., Schaller, M., Schäfer, A., Kyme, P., Martin, J., Wälzlein, J.-H., Ehehalt, U., Lindroos, H., Schirle, M., Nordheim, A., Autenrieth, I. B., and Kempf, V. A. J. (2004). Bartonella adhesin a mediates a proangiogenic host cell response. J. Exp. Med. 200, 1267–1278.
Ruiz-Perez, F., Henderson, I. R., Leyton, D. L., Rossiter, A. E., Zhang, Y., and Nataro, J. P. (2009). Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J. Bacteriol. 191, 6571–6583.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sajjan, U., Liu, L., Lu, A., Spilker, T., Forstner, J., and LiPuma, J. J. (2002). Lack of cable pili expression by cblA-containing Burkholderia cepacia complex. Microbiology 148, 3477–3484.
Sajjan, U., Wu, Y., Kent, G., and Forstner, J. (2000). Preferential adherence of cable-piliated Burkholderia cepacia to respiratory epithelia of CF knockout mice and human cystic fibrosis lung explants. J. Med. Microbiol. 49, 875–885.
Serruto, D., Spadafina, T., Scarselli, M., Bambini, S., Comanducci, M., Höhle, S., Kilian, M., Veiga, E., Cossart, P., Oggioni, M. R., Savino, S., Ferlenghi, I., Taddei, A. R., Rappuoli, R., Pizza, M., Masignani, V., and Aricò, B. (2009). HadA is an atypical new multifunctional trimeric coiled-coil adhesin of Haemophilus influenzae biogroup aegyptius, which promotes entry into host cells. Cell. Microbiol. 11, 1044–1063.
Sheets, A. J., and St Geme, J. W. III. (2011). Adhesive activity of the Haemophilus cryptic genospecies cha autotransporter is modulated by variation in tandem peptide repeats. J. Bacteriol. 193, 329–339.
Szczesny, P., and Lupas, A. (2008). Domain annotation of trimeric autotransporter adhesins – daTAA. Bioinformatics 24, 1251–1256.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Tiyawisutsri, R., Holden, M., Tumapa, S., Rengpipat, S., Clarke, S., Foster, S., Nierman, W., Day, N., and Peacock, S. (2007). Burkholderia Hep_Hag autotransporter (BuHA) proteins elicit a strong antibody response during experimental glanders but not human melioidosis. BMC Microbiol. 7, 19. doi:10.1186/1471-2180-7-19
Wagner, J. K., Heindl, J. E., Gray, A. N., Jain, S., and Goldberg, M. B. (2009). Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J. Bacteriol. 191, 815–821.
Wu, H., Zeng, M., and Fives-Taylor, P. (2007). The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect. Immun. 75, 2181–2188.
Keywords: Burkholderia cepacia complex, trimeric autotransporter adhesins, virulence factors
Citation: Mil-Homens D and Fialho AM (2011) Trimeric autotransporter adhesins in members of the Burkholderia cepacia complex: a multifunctional family of proteins implicated in virulence. Front. Cell. Inf. Microbio. 1:13. doi: 10.3389/fcimb.2011.00013
Received: 15 September 2011;
Accepted: 15 November 2011;
Published online: 07 December 2011.
Edited by:
Joanna Goldberg, University of Virginia Health System, USAReviewed by:
Odile Tresse, French National Institute for Agricultural Research/Nantes-Atlantic National College of Veterinary Medicine, FranceCopyright: © 2011 Mil-Homens and Fialho. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Arsénio M. Fialho, Department of Bioengineering, Instituto Superior Técnico, Technical University of Lisbon, Av. Rovisco Pais, Lisbon 1049-001, Portugal. e-mail:YWZpYWxob0Bpc3QudXRsLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.