- 1Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, East China University of Science and Technology, Shanghai, China
- 2State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Zhangjiang Institute for Advanced Study, Shanghai Jiao Tong University, Shanghai, China
- 3State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China
Cyclic di-guanosine monophosphate (c-di-GMP) acts as a second messenger regulating bacterial behaviors including cell cycling, biofilm formation, adhesion, and virulence. Monitoring c-di-GMP levels is crucial for understanding these processes and designing inhibitors to combat biofilm-related antibiotic resistance. Here, we developed a genetically encoded biosensor, cdiGEBS, based on the transcriptional activity of the c-di-GMP-responsive transcription factor MrkH. Notably, cdiGEBS can detect both low and high cellular c-di-GMP levels, with a high fluorescence dynamic change of 23-fold. Moreover, it can detect subtle changes in c-di-GMP concentrations due to variations in the expression of c-di-GMP synthesis or degradation enzymes and can distinguish different synthesis activities among WspR mutants. These capabilities allow us to apply cdiGEBS for identifying new diguanylate cyclases and evaluating chemicals that modulate c-di-GMP levels, highlighting its potential as a high-throughput tool for screening inhibitors of c-di-GMP synthesis enzymes. Overall, cdiGEBS enhances the study of c-di-GMP-regulated functions and holds the potential for screening antimicrobials targeting c-di-GMP or its synthesis enzymes.
1 Introduction
C-di-GMP is a ubiquitous bacterial second messenger, regulating bacterial crucial behaviors and physiological functions, including biofilm, virulence, and the cell cycle (Römling et al., 2013; Ha and O’Toole, 2015; Kulasakara et al., 2006). Studies suggested that c-di-GMP plays a particularly pivotal role in governing the transition of bacteria from a planktonic state to a biofilm lifestyle (Jenal et al., 2017). Specifically, high c-di-GMP levels promote biofilm formation (Flemming et al., 2016), while low c-di-GMP levels could increase motility and facilitate biofilm dispersal (Rumbaugh and Sauer, 2020). Through biofilm formation, bacteria could defend against host immune responses and external environmental threats, including antimicrobials like colistin and chlorhexidine (Ciofu et al., 2015; Jenal et al., 2017; Wang Z. et al., 2023). To tightly regulate biofilm formation, bacteria have evolved sophisticated regulatory networks that maintain c-di-GMP homeostasis (Jenal and Malone, 2006; Park and Sauer, 2022).
Intracellular c-di-GMP levels are predominantly regulated by two types of antagonistic enzymes, diguanylate cyclases (DGCs) and phosphodiesterases (PDEs). DGCs that possess the GG(D/E)EF domain synthesize c-di-GMP by cyclizing two molecules of guanosine triphosphate (GTP) (Schirmer, 2016). In contrast, PDEs featuring either EAL or HD-GYP domains, catalyze the linearization of c-di-GMP into 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) or degrade c-di-GMP into two molecules of guanosine monophosphate (GMP) (Christen et al., 2005; Bellini et al., 2014). The c-di-GMP signaling network is characterized by numerous DGCs and PDEs, many of which contain putative sensory domains that perceive environmental cues (Almblad et al., 2021; Randall et al., 2022).
In order to gain deeper insights into c-di-GMP-mediated bacterial physiological functions and explore uncharacterized c-di-GMP signaling regulatory pathways, numerous tools have been developed to monitor intracellular c-di-GMP fluctuations (Petchiappan et al., 2020; McCaughey et al., 2024). Liquid chromatography coupled with gas chromatography-mass spectrometry (LC-GC/MS) is frequently employed to quantify c-di-GMP concentrations in extracted biological samples due to high accuracy and sensitivity (Simm et al., 2009; Irie and Parsek, 2014). However, the complex sample preparation and costly equipment required for these analytical tools limit their application. To overcome these limitations, genetically encoded fluorescent biosensors, typically incorporated into cells as plasmid DNA, have been extensively developed for monitoring the spatiotemporal dynamics of signaling molecules in living cells (Palmer et al., 2011; Greenwald et al., 2018; Wang et al., 2022). Specifically, for c-di-GMP detection in vivo, a serial of genetically encoded fluorescent biosensors have been established, including the fluorescence resonance energy transfer (FRET) - based biosensors (Christen et al., 2010; Petersen et al., 2019), the bimolecular fluorescence complementation (BiFC) - based biosensors (Halte et al., 2022), the sensor protein conformational change-based biosensors and others (Ho et al., 2013; Vrabioiu and Berg, 2022; Kaczmarczyk et al., 2024; McCaughey et al., 2024).
These biosensors have significantly advanced the study of c-di-GMP-mediated transcriptional pathways and bacterial behaviors. However, since these sensors typically require direct coupling of the c-di-GMP responsive element with a fluorescent reporter, amplifying the fluorescent signal and achieving a high fluorescence dynamic range remains challenging. In contrast, transcription factor (TF) - based biosensors, which rely on the transcriptional activity of the TF, can amplify the detection signal. These biosensors typically comprise a TF and a TF-recognized promoter fused to a fluorescent protein-encoding gene. Upon binding to the target molecule, the TF’s transcriptional activity is activated, leading to robust expression of the fluorescent protein reporter. Rybtke et al. developed a TF activity-based pCdrA::gfp reporter for c-di-GMP detection in Pseudomonas aeruginosa, significantly advancing the study of c-di-GMP-regulated cellular processes in this species (Rodesney et al., 2017; Wang L. et al., 2023; Hendrix et al., 2024; Kennelly et al., 2024). However, such a straightforward set up utilizing the ligand-regulated transcriptional activity of TF has not been established in E. coli. Current TF-based biosensors in E. coli rely on conformational changes in the TF upon c-di-GMP binding, as in the mScarlet I-mVenusNB-MrkH biosensor (Vrabioiu and Berg, 2022), or on TF dimerization induced by c-di-GMP binding, as in the BiFC-based CensYBL biosensor (Halte et al., 2022), resulting in a relatively limited dynamic range of fluorescence for c-di-GMP detection (0.9 to 3-fold).
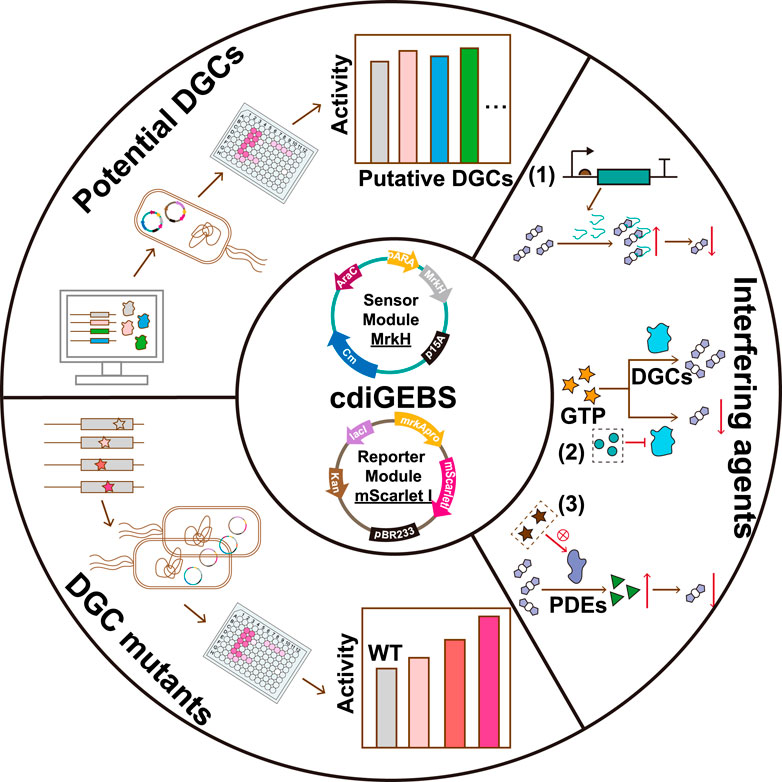
In this study, we introduced a TF transcriptional activity-based c-di-GMP biosensor (named cdiGEBS for c-di-GMP Genetically Encoded BioSensor), which utilized the c-di-GMP-responsive transcription factor MrkH from Klebsiella pneumonia and its corresponding promoter (PmrkA) as the sensing module, with the fluorescent protein mScarlet I as the reporter element. Our sensor demonstrates a fluorescence dynamic range of 23-fold, substantially improving the sensitivity for detecting both elevated and reduced intracellular c-di-GMP levels. Notably, cdiGEBS is capable of detecting subtle changes in c-di-GMP concentrations resulting from altered expression of its biogenesis or degradation enzymes. Additionally, it can distinguish the c-di-GMP synthesis activities of different mutants of the diguanylate cyclase WspR. These features encourage us to successfully apply cdiGEBS for identifying novel diguanylate cyclases and assessing the impact of various compounds on cellular c-di-GMP levels. Taken together, cdiGEBS not only serves as a versatile and easy-to-use tool for the functional analysis of c-di-GMP biogenesis enzymes but also shows promise for high-throughput screening of novel inhibitors targeting c-di-GMP synthesis for combating biofilm-associated bacterial infections.
2 Materials and methods
2.1 Plasmid constructions
Strains and plasmids used in this paper are listed in Table 1. Plasmid construction in this work follows the overlap extension PCR cloning procedure as described previously (Nelson and Fitch, 2011). E. coli strain Turbo (Tsingke Biotech) was used for competent cell preparation. The wspR, mrkA promoter, and mrkH sequences were synthesized by Tsingke Biotech, while the dgcT, dgcI, pdeH, and dgcF sequences were amplified from the genome of the E. coli MG1655 strain and the proC promoter and mScarlet I sequences were amplified from pEB2-mScarlet I-mVenusNB-MrkH vector (Addgene No. 182291).
The mrkH gene was inserted into the medium-copy plasmid pBAD33, generating the plasmid pBAD33-MrkH. Subsequently, the pdeH or wspR D70E gene, along with an additional ribosome binding site, was inserted downstream of the mrkH gene, resulting in the plasmids pBAD33-MrkH-PdeH and pBAD33-MrkH-WspR D70E, respectively. Plasmid pET28b(+)-PmrkA-mScarlet I was constructed by replacing the T7 promoter of the high-copy plasmid pET28b(+)-MCS with mrkA promoter and mScarlet I-encoding gene. For the plasmids pBAD33-PdeH-proC-MrkH and pBAD33-WspR D70E-proC-MrkH, which express PdeH or WspR D70E under the control of the araBAD promoter and constitutively express MrkH via the proC promoter respectively, were constructed by first inserting the PdeH or WspR D70E gene downstream of the araBAD promoter, followed by the insertion of the proC promoter, an additional ribosome binding site, and mrkH gene downstream of the pdeH or wspR D70E gene.
Quick change mutagenesis method was employed for introducing point mutations of the wspR, dgcT, dgcI, and dgcF genes (Liu and Naismith, 2008). All the oligonucleotides, promoters, and genes used in this work are listed in Supplementary Tables S1, S2.
2.2 Fluorescence microscopy
Overnight cultures of the strains harboring cdiGEBS and either PdeH or WspR D70E were diluted 1:100 into 4 mL of fresh LB medium supplemented with 100 μg mL−1 kanamycin, 34 μg mL−1 chloramphenicol, and 0.002% L-arabinose. Cells were then cultivated at 30°C for 6 h with a shaking speed of 220 rpm. Subsequently, 0.1 OD600 nm cells were collected and resuspended in 40 µL phosphate-buffered saline (PBS). A 5 µL aliquot was placed on a microscope slide, coated with 1% agarose, and covered with a coverslip. Fluorescence images were captured using a Zeiss LSM 880 Confocal Laser Scanning Microscope, with the excitation wavelength set to 568 nm.
2.3 Fluorescence measurement with a microplate reader
Overnight cultures of E. coli co-expressing cdiGEBS and PdeH, or cdiGEBS and each WspR mutant, or cdiGEBS and each putative DGC were inoculated at a 1:100 dilution-fold into 4 mL fresh LB medium supplemented with 100 μg mL−1 kanamycin, 34 μg mL−1 chloramphenicol, and 0.002% L-arabinose. After grown at 30°C for 6 h, 1 OD600 nm cells were collected and resuspended in 220 µL PBS for fluorescence measurements by an automated microplate reader (SynergyHTX hybrid reader, BioTek) with the excitation and the emission set at 540 ± 35 nm and 600 ± 40 nm, respectively.
2.4 Fluorescence intensity and fluorescence spectra detection by a fluorescence spectrometer
Overnight cultures of E. coli co-expressing cdiGEBS and various CSPs were inoculated at a 1:100 dilution-fold into 4 mL fresh LB medium supplemented with 100 μg mL−1 kanamycin, 34 μg mL−1 chloramphenicol, 100 μg mL−1 streptomycin, 100 μg mL−1 spectinomycin, and 0.002% L-arabinose. Cells were then cultured at 30°C, 220 rpm until the OD600 nm reached 0.3, followed by adding 0.1 mM isopropyl-β-D-thiogalactoside (IPTG) to induce the expression of 6 × His-MBP-CSP. The 6 × His-MBP tag was fused with various CSP to prevent cellular degradation of CSP (Hee et al., 2020). Following 6 h of 6 × His-MBP-CSP expression, cells equivalent to 4 OD600 nm units were harvested and resuspended in 2 mL of PBS buffer for fluorescence intensity and spectral measurements using a fluorescence spectrometer (FS5 Spectrofluorometer, Edinburgh Instruments), with the excitation wavelength set at 568 nm and an emission range of 575–650 nm.
2.5 Time-dependent fluorescence detection
Overnight cultures of E. coli harboring cdiGEBS were diluted 1:100 into fresh LB medium supplemented with 100 μg mL−1 kanamycin, 34 μg mL−1 chloramphenicol, and 0.002% L-arabinose. Then 200 μL cell cultures were transferred to each well of a 96-well plate containing different concentrations of tyrosol or ampicillin. Fluorescence intensity and cell density (OD600 nm) were monitored by using a Synergy HTX Multi-Mode Microplate Reader with a 15-min detection period for 21 h at 30°C. For fluorescence intensity measurement, the excitation and emission wavelength were set at 540 ± 35 nm and 600 ± 40 nm, respectively.
2.6 Western blots analysis
Cells were cultivated and induced for protein expression as described in the “Fluorescence Measurement by a Microplate Reader” Section. 1 OD600 nm cells were collected and resuspended in PBS to a final OD600 nm of 5.0. Then cell samples were mixed with 5 × protein loading buffer, and subjected to 12% SDS-PAGE after boiling for 10 min. For Western blot analysis, proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Merck) using a wet transfer apparatus (Tanon). Then the PVDF membranes were blocked with 5% non-fat milk in TBST (Tris buffer saline containing 0.1% Tween-20), followed by incubation with the primary antibodies: anti-6 × His from mouse or anti-trigger factor from rabbit (1:4,000, GenScript, Cat. No. A01329) in TBST containing 5% non-fat milk. Afterward, the membranes were incubated with fluorescently labeled IRDye 800 CW secondary antibodies (1:10,000, LI-COR Biosciences), and fluorescence images were recorded using the Odyssey Sa Infrared Imaging System (LI-COR Biosciences).
3 Results
3.1 Construction and characterization of a transcription factor-based c-di-GMP biosensor
The transcriptional regulator MrkH is a member of the PliZ family and exhibits a high affinity for c-di-GMP in the nanomolar range (Schumacher and Zeng, 2016; Wang F. et al., 2016). Upon c-di-GMP binding, MrkH undergoes a conformational change-induced activation (Figure 1A) (Schumacher and Zeng, 2016), allowing it to bind the mrkA promoter and highly upregulate the transcription of the downstream mrkABCDF operon (Wilksch et al., 2011). Based on this, we introduced the c-di-GMP sensing element MrkH under the regulation of the araBAD promoter and the fluorescent reporter element mScarlet I under the control of the mrkA promoter in E. coli to establish a genetically encoded fluorescent biosensor for c-di-GMP detection. Specifically, the mrkH gene was placed downstream of the araBAD promoter in the medium-copy number plasmid pBAD33 vector, while the mScarlet I gene was inserted downstream of the mrkA promoter (PmrkA) on the high-copy plasmid pET28b(+) backbone with the original T7 promoter removed (Figure 1B).
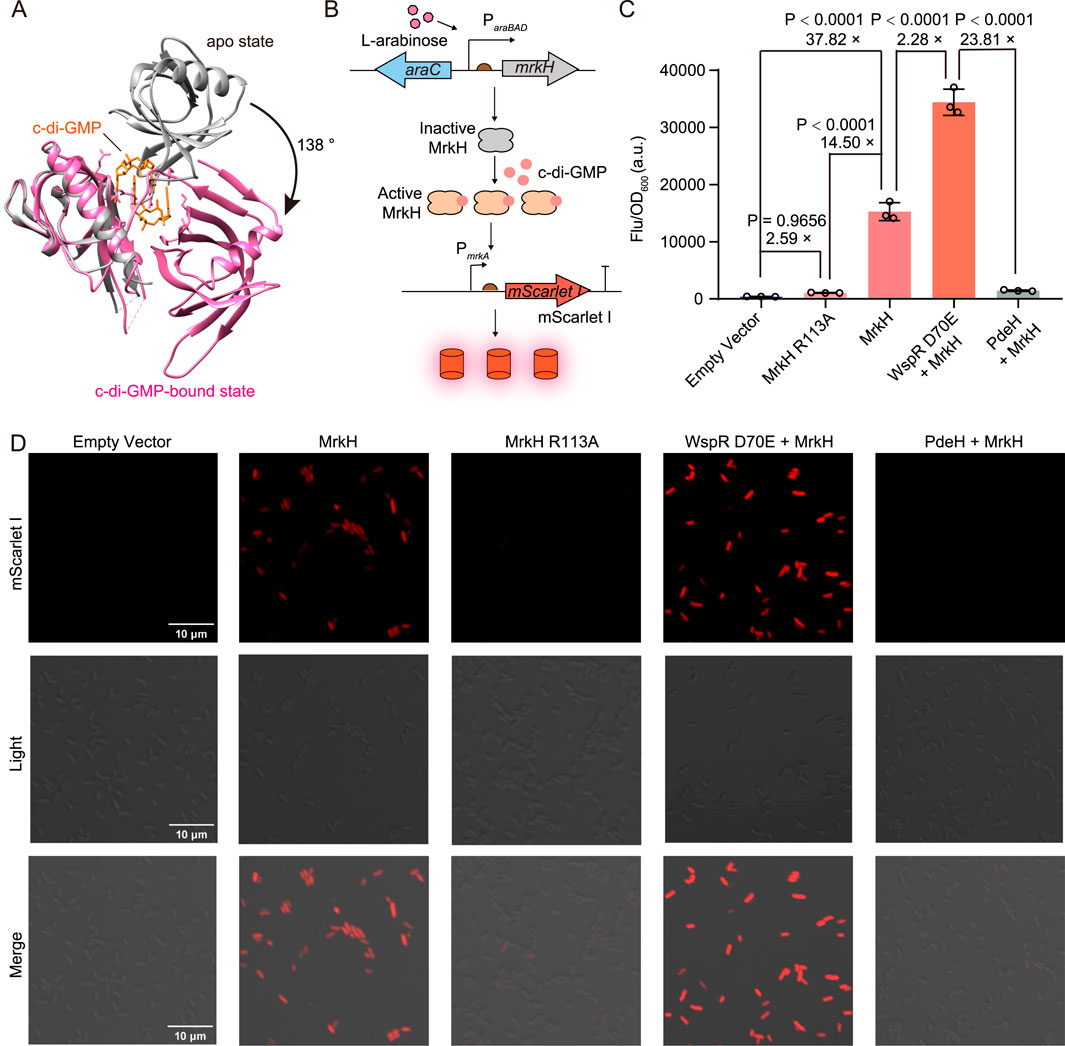
Figure 1. Construction and characterization of cdiGEBS. (A) Structural alignment of the apo (gray, PDB: 5KEC) and c-di-GMP-bound MrkH (pink, PDB: 5KGO). (B) Schematic illustration of the working mechanism of cdiGEBS. (C) Performance of cdiGEBS in detecting both high and low cellular c-di-GMP levels. Fluorescence was measured in strains co-harboring the PmrkA-mScarlet I plasmid and the pBAD33 plasmid expressing the indicated proteins. An additional ribosome binding site, fused with the gene encoding WspR D70E or PdeH, was cloned downstream of the mrkH sequence on the pBAD33 plasmid to enable co-expression of WspR D70E or PdeH with MrkH. The strain harboring both pBAD33-Empty and PmrkA-mScarlet I plasmids served as the “Empty vector” control. Samples were induced with 0.002% L-arabinose at 30°C for 6 h. Fluorescence was recorded by using a microplate reader with the excitation and emission wavelengths set at 540 ± 35 nm and 600 ± 40 nm, respectively. Individual data points (circles) and mean ± SD (n = 3) are shown. (D) Fluorescence microscopy images of strains expressing the corresponding proteins. Statistical analysis was performed with GraphPad Prism 8 using one-way ANOVA with Tukey’s multiple comparisons tests.
We observed a significant fluorescence increase when the strain containing the biosensor was induced with arabinose for MrkH expression, suggesting that the endogenous c-di-GMP in E. coli can substantially activate the transcriptional activity of MrkH and thus upregulate mScarlet I expression (Figure 1C). To further confirm that the observed increase in mScarlet I fluorescence was due to the transcriptional activation of MrkH upon binding c-di-GMP, we introduced the R113A mutation in MrkH to substantially diminish its c-di-GMP-binding ability (Vrabioiu and Berg, 2022). We found that the resultant strain expressing the MrkH R113A mutant exhibited a significantly decreased fluorescence intensity compared to the strain expressing the wild-type MrkH (Figure 1C), indicating that the fluorescence increase in the strain expressing wild-type MrkH was indeed mediated by the binding between MrkH and c-di-GMP.
To assess whether cdiGEBS can respond to varying intracellular levels of c-di-GMP, we co-expressed the biosensor with either a constitutively activated diguanylate cyclase WspR D70E or the phosphodiesterase PdeH to induce high or low c-di-GMP concentrations, respectively (Dippel et al., 2018). By optimizing the arabinose concentration, cultivation temperature, and induction time, we found that the biosensor could effectively distinguish between high and low c-di-GMP levels, exhibiting a remarkable fluorescence change readout of over 23-fold (Supplementary Figure S1 and Figure 1C). These findings were further supported by fluorescence imaging (Figure 1D), and Western blot analysis confirmed that the observed differences in fluorescence intensity were not due to variations in MrkH expression levels (Supplementary Figure S2). Taken together, these results indicate that cdiGEBS can respond to high and low c-di-GMP concentrations with highly dynamic fluorescence fold changes.
3.2 Distinguishing various cellular c-di-GMP levels with cdiGEBS
To further investigate the sensitivity of cdiGEBS in distinguishing varying cellular c-di-GMP levels, we maintained the mScarlet I reporter element and modified the sensing element as follows: 1) MrkH was constitutively expressed under the control of the proC promoter, and 2) WspR D70E or PdeH expression was induced by the araBAD promoter (Figure 2A). By titrating the resulting strains with different concentrations of L-arabinose, we were able to gradually adjust the levels of the c-di-GMP synthase WspR D70E or the degradation enzyme PdeH, leading to progressively increased or decreased cellular c-di-GMP concentrations, respectively.
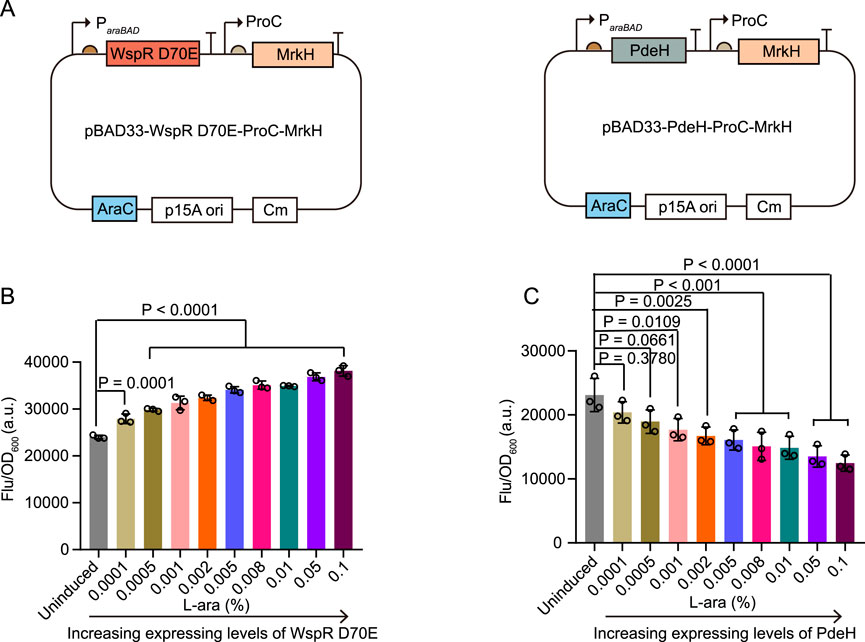
Figure 2. Sensitivity characterization of cdiGEBS. (A) Schematic illustrations of the modified biosensors for sensitivity characterization. (B, C) Normalized mScarlet I fluorescence intensities (Ex = 540 ± 35 nm, Em = 600 ± 40 nm, measured with a microplate reader) of strains co-expressing cdiGEBS with WspR D70E (B) or PdeH (C) under increasing concentrations of L-arabinose. Overnight cultures were inoculated into fresh LB culture at a 1:100 dilution fold. At the same time, various concentrations of L-arabinose were added to induce WspR D70E or PdeH expression at 30°C for 9 h. Subsequently, 1 OD cells were collected and used for fluorescence measurement. Individual data points (circles) and mean ± SD (n = 3) are shown. Statistical analysis was performed with GraphPad Prism 8 using one-way ANOVA with Tukey’s multiple comparisons tests.
We observed that the strain co-expressing MrkH and WspR D70E exhibited a gradual increase in mScarlet I fluorescence with increasing concentrations of L-arabinose (Figure 2B). In contrast, the strain co-expressing MrkH and PdeH showed a dose-dependent decrease in mScarlet I fluorescence intensity with L-arabinose (Figure 2C). Furthermore, by titrating the strain containing only the biosensor, without WspR D70E or PdeH, with different concentrations of L-arabinose, we excluded the possibility that these fluorescence changes were due to the effects of L-arabinose on the mScarlet I fluorophore or MrkH transcriptional activity (Supplementary Figure S3). Overall, these results demonstrate that cdiGEBS is sensitive and capable of distinguishing subtle variations in intracellular c-di-GMP levels, which result from finely tuned adjustments in the expression of c-di-GMP synthesis or degradation enzymes.
3.3 Assessment of diguanylate cyclase activities in vivo
The capability of cdiGEBS to distinguish varying c-di-GMP levels promoted us to explore its application in assessing the activity of diguanylate cyclases (DGCs). There are 29 genes in Escherichia coli K-12 participating in c-di-GMP biosynthesis or degradation (Hengge et al., 2016; Povolotsky and Hengge, 2016). Previous studies have shown that overexpression of DgcT resulted in a significant increase of intracellular c-di-GMP, implying that DgcT processed diguanylate cyclase activity (Jonas et al., 2008). Additionally, the uncharacterized membrane-associated proteins DgcI and DgcF have also been annotated as putative DGCs in E. coli since strains lacking DgcF or DgcI (ΔdgcF or ΔdgcI) display increased swimming motility (Sanchez-Torres et al., 2011; Povolotsky and Hengge, 2016). DGCs generally function as homodimers, with their GG(D/E)EF domains being responsible for the production of c-di-GMP (Dahlstrom and O’Toole, 2017). To gain structural insights into these putative DGCs, we predicted the potential dimeric structures of these proteins using AlphaFold 3 and identified the presence of typical GG(D/E)EF domains in all of them (Figure 3A) (Abramson et al., 2024).
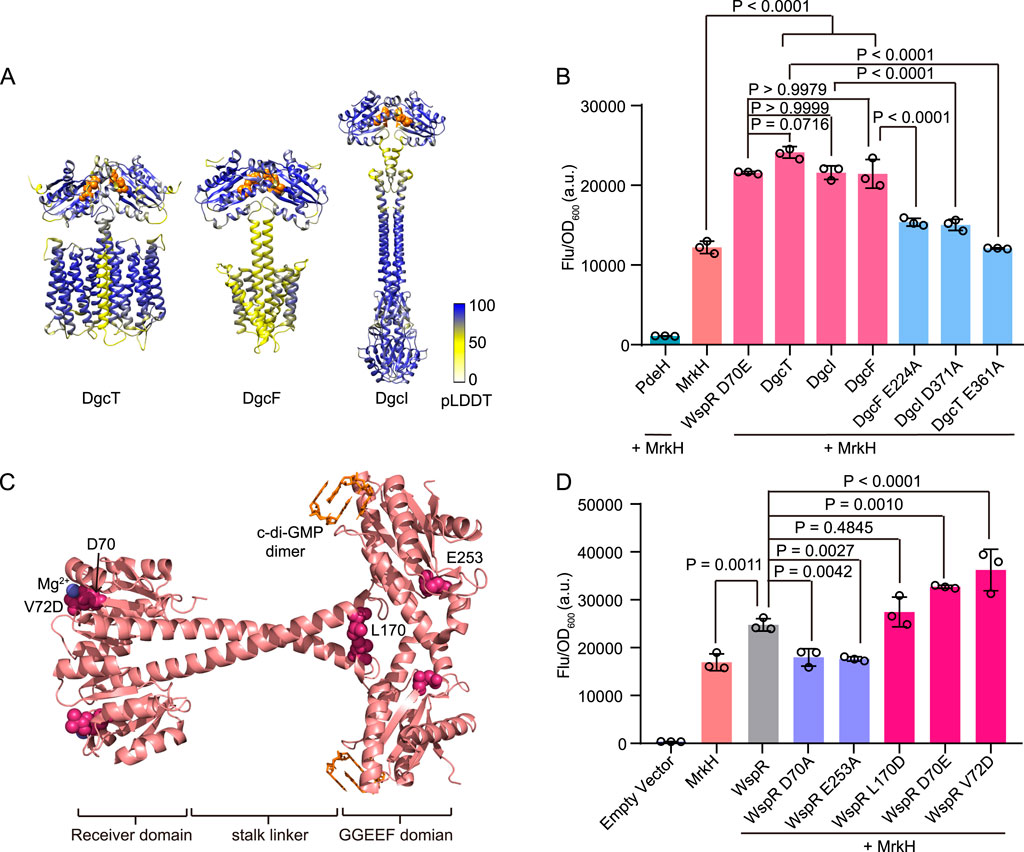
Figure 3. Diguanylate cyclase activity detection using cdiGEBS. (A) Homodimer structure prediction of DgcT, DgcF, and DgcI by AlphaFold 3. The white-to-blue color scale indicates pLDDT scores ranging from 0 to 100. The GG(D/E)EF domain of each structure was represented as orange spheres. (B) Diguanylate cyclase activity assessment of wild-type and mutants of the putative DGCs using cdiGEBS. All strains contain the PmrkA-mScarlet I reporter plasmid. PdeH, WspR D70E, DgcT, DgcI, DgcF, DgcT E361A, DgcI D371A, or DgcF E224A was co-expressed with MrkH under the control of the araBAD promoter on the pBAD33 plasmid. Samples were induced with 0.002% L-arabinose at 30°C for 6 h (C) WspR dimer structure (PDB: 3BRE) with each mutation site represented as a pink sphere. C-di-GMP dimer was colored orange and the magnesium ion was colored blue. (D) Distinguishing the activity differences of various WspR mutants using cdiGEBS. Wild-type WspR and its mutants were co-expressed with MrkH under the control of the araBAD promoter on the pBAD33 plasmid. The strain harboring both pBAD33-Empty and PmrkA-mScarlet I plasmids was served as the “Empty vector” control. For (B) and (D), samples were excited at 540 ± 35 nm and the fluorescence signals were recorded at 600 ± 40 nm by using a microplate reader. Individual data points (circles) and mean ± SD (n = 3) are shown. Statistical analysis was performed with GraphPad Prism 8 using one-way ANOVA with Tukey’s multiple comparisons tests.
We then investigated the in vivo diguanylate cyclase activities of these putative DGCs using cdiGEBS, with the constitutively active DGC WspR D70E serving as the positive control. Strains co-expressing the biosensor and any of the putative DGCs showed a substantial increase in mScarlet I fluorescence intensity, comparable to the strains co-expressing the biosensor and WspR D70E (Figure 3B). This result indicates that these three putative DGCs can indeed catalyze the synthesis of c-di-GMP in vivo. Moreover, to investigate the effect of the GG(D/E)EF domain of DgcT, DgcF, and DgcI on their DGC activity, we substituted the third residue of the GG(D/E)EF domain with alanine. We found that the resulting strains expressing the protein mutants (DgcT E361A, DgcI D371A, and DgcF E224A) showed significantly decreased fluorescence intensities compared to the strains expressing the wild-type proteins (Figure 3B). This suggests that DgcT, DgcI, and DgcF all catalyze the synthesis of c-di-GMP in a GG(D/E)EF domain-dependent manner.
The sensitive performance of our biosensor in tracking DGC activities drove us to further investigate whether it could discriminate the subtle differences in the enzymatic activities of various DGC mutants. To examine this, we characterized the activities of the canonical DGC WspR and its mutants with defined DGC activities in vivo using cdiGEBS (Figures 3C, D) (Huangyutitham et al., 2013). The D70A substitution disrupts the phosphorylation site, resulting in the loss of cyclase activity (De et al., 2009). Similarly, the E253A mutation inactivates WspR by impairing the active site (Malone et al., 2007). In contrast, the D70E mutation mimics the phosphorylated state, leading to constitutive activation (Wang X. C. et al., 2016). The V72D mutant also exhibits constitutive diguanylate cyclase activity, as identified in a random mutagenesis study of WspR (Huangyutitham et al., 2013). Additionally, studies have demonstrated that the tetrameric form of WspR displays high activity levels, with the WspR L170D variant enhancing activity by stabilizing this tetrameric form (De et al., 2008).
We found that strains co-expressing the biosensor and the wild-type WspR showed a significant increase in fluorescence intensity compared with the strain containing only the biosensor. In contrast, strains co-expressing the biosensor and the inactive mutants WspR D70A or WspR E253A exhibited low fluorescence intensities similar to the strain harboring the biosensor only, implying that these mutations nearly completely abolished DGC activities (Figure 3D). Consistent with the literature, we observed that strains co-expressing the biosensor and activity-enhanced mutants D70E, V72D, and L170D all showed a trend of increased fluorescence intensity compared to the strain co-expressing the biosensor and the wild-type WspR, although the increase in fluorescence intensity of the strain expressing the mutant L170D was not statistically significant (Figure 3D) (Huangyutitham et al., 2013). Additionally, Western blot analysis suggested that the observed fluorescence differences were not due to variations in the expression levels of MrkH and WspR mutants (Supplementary Figure S4). Taken together, these results suggested that cdiGEBS was sensitive and could be used for functional analysis and activity evaluation of DGCs in vivo.
3.4 Monitoring the interference of chemicals on the c-di-GMP levels in vivo
High c-di-GMP levels could stimulate biofilm formation, leading to a 10 to 1,000-fold increase in antibiotic resistance of bacteria in the sessile state than in the planktonic state (Dostert et al., 2019; Ciofu and Tolker-Nielsen, 2019; Dostert et al., 2021). Decreasing the cellular c-di-GMP levels is a promising strategy for combating biofilm-related antibiotic resistance and potential chronic infection. Therefore, researchers have developed a number of structurally diverse compounds to diminish c-di-GMP concentrations in bacteria, either by directly sequestering c-di-GMP or by enhancing PDE activity (Qvortrup et al., 2019; Hee et al., 2020). Encouraged by the sensitivity of cdiGEBS in monitoring c-di-GMP levels in vivo, we next sought to examine whether this sensor was capable of reflecting the effects of typical anti-biofilm chemicals on the cellular c-di-GMP level.
Hee et al. designed a serial of short c-di-GMP-sequestering peptides (CSP) which could directly bind c-di-GMP and thus reduce the concentration of free cellular c-di-GMP in Pseudomonas aeruginosa and consequently inhibit biofilm formation (Hee et al., 2020). We found that the strain co-expressing the biosensor and the peptide CSP2 which bound c-di-GMP tightest exhibited a substantial decrease in mScarlet I fluorescence intensity compared with the strain harboring the biosensor only (Figures 4A–C). In contrast, co-expressing the biosensor and the binding affinity attenuated peptides CSP3 or CSP2 R169A only led to a moderate fluorescence decrease of the resulting strains (Figures 4B, C).
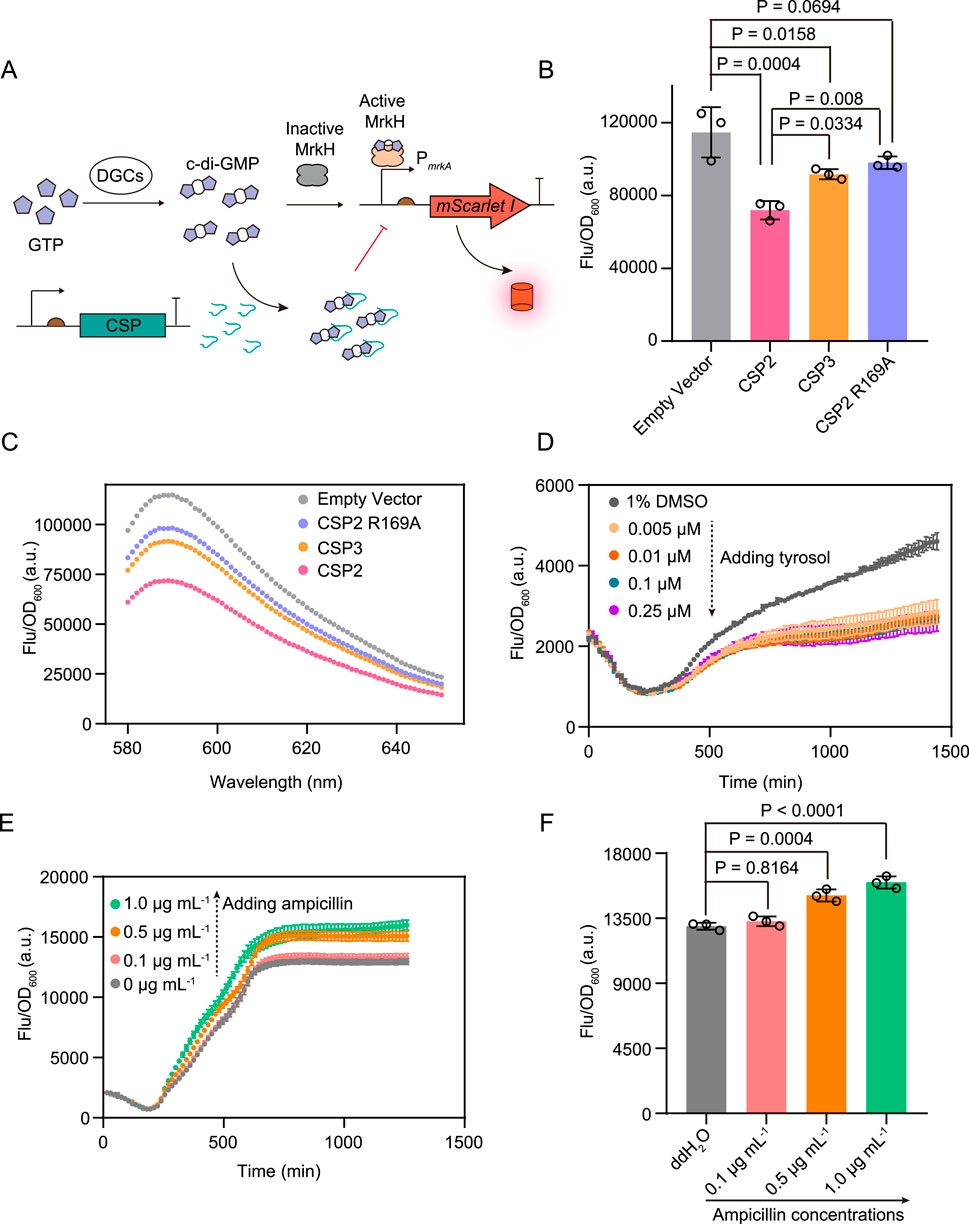
Figure 4. The application of cdiGEBS for investigating the effects of chemical compounds on cellular c-di-GMP levels. (A) Schematic illustration for detecting the effects of CSPs on cellular c-di-GMP levels by cdiGEBS. (B) Fluorescence intensities of strains co-expressing different CSPs and cdiGEBS. pCDFDuet vectors expressing each CSP were co-transformed with the cdiGEBS plasmids (pBAD33-MrkH, PmrkA-mScarlet I). The strain harboring the pBAD33-MrkH, PmrkA-mScarlet I, and pCDFDuet-Empty plasmids were used as the “Empty vector” control. (C) Fluorescence emission spectra of strains co-expressing different CSPs and cdiGEBS. Representative curves from three independent measurements are shown. For (B, C), fluorescence was recorded by using a fluorescence spectrometer with the excitation and emission wavelengths set at 568 nm and 589 nm, respectively. (D, E) Time-dependent fluorescence changes of strains expressing cdiGEBS in the presence of various concentrations of tyrosol (D) and ampicillin (E). Strains expressing cdiGEBS were cultured in 96-well microplates to allow continuous measurement of fluorescence and optical density using a microplate reader. (F) Fluorescence intensities of strains expressing cdiGEBS after 21 h of treatment with various concentrations of ampicillin. For (B, F), individual data points (circles) and mean ± SD (n = 3) are shown. Statistical analysis was performed with GraphPad Prism 8 using one-way ANOVA with Tukey’s multiple comparisons tests.
Furthermore, Choi et al. demonstrated that tyrosol could decrease c-di-GMP levels by activating PDE activity (Choi and Kim, 2024). We observed that treating the strain containing cdiGEBS with increasing concentrations of tyrosol resulted in decreased mScarlet I fluorescence intensity compared with the control strain which was treated with 1% DMSO (Figure 4D). In contrast to biofilm dispersal chemicals, studies have shown that subinhibitory concentrations of ampicillin can facilitate c-di-GMP production (Boehm et al., 2009; Ho et al., 2013; Gao et al., 2022). We also found that cdiGEBS could be used for monitoring the increasing effect of ampicillin on the cellular c-di-GMP levels (Figure 4F). Additionally, neither over-expressing the CSPs nor tyrosol treatment substantially hinders bacterial growth (Supplementary Figure S5). For ampicillin treatment, although bacterial growth was slightly reduced at 1.0 μg mL−1, the unaltered growth at 0.1–0.5 μg mL−1 confirms that the observed increase in fluorescence is genuine and not a result of signal normalization (Supplementary Figure S5). Furthermore, neither tyrosol nor ampicillin treatment affected the fluorescence of the strain constitutively expressing mScarlet I under the control of the J23100 promoter, indicating that these chemicals do not interfere with the mScarlet I fluorophore (Supplementary Figure S6). Taken together, these results demonstrate that cdiGEBS is capable of reflecting the effects of typical chemicals on cellular c-di-GMP levels and even has the potential to be used for validating the binding affinities between various chemicals and c-di-GMP in vivo.
Recent studies have emphasized the strategy of developing anti-biofilm compounds by reducing cellular c-di-GMP levels for the treatment of biofilm-associated diseases (Martin et al., 2021; Hultqvist et al., 2024). The high sensitivity of our sensor suggests strong potential for its use as a high-throughput screening platform to identify novel agents that reduce intracellular c-di-GMP concentrations. Future efforts focusing on screening compounds that inhibit DGC activity, activate PDE activity, or sequester free c-di-GMP by combining tools of our sensor and fluorescence-activated cell sorting (FACS) could offer new insights for combating biofilm-associated diseases.
4 Discussion
In this work, we developed a c-di-GMP biosensor (cdiGEBS) based on the transcriptional activity of the c-di-GMP-responsive activator MrkH (Yang et al., 2013), achieving a high fluorescence dynamic range of 23-fold. The moderate expression of the sensor module MrkH using the medium-copy plasmid pBAD33 and the overexpression of the mScarlet I reporter with the high-copy plasmid pET28b(+) likely contribute to this high fluorescence dynamic range. Remarkably, our sensor demonstrates the capability in detecting subtle intracellular c-di-GMP changes, enabling it to be used to identify novel DGCs, distinguish various c-di-GMP synthesis activities of DGC mutants, as well as assess the effects of chemical agents on cellular c-di-GMP levels. However, while our sensor, based on TF activity, is well-suited for detecting steady-state differences in the c-di-GMP concentration, it is not designed for tracking rapid fluctuations of c-di-GMP. Consistent with this, approximately 5 h were required to observe significant fluorescence changes reflecting differences in c-di-GMP levels between the strain expressing the c-di-GMP synthesis enzyme WspR D70E and the strain that does not (Supplementary Figure S7).
Biofilm formation enhances bacterial resistance to antibiotics and the human immune system, making it closely associated with the development of urinary tract infections, cystic fibrosis lung infections, and other diseases (Gomez and Waters, 2023; Hultqvist et al., 2024). Studies have shown that elevated levels of intracellular c-di-GMP play a crucial role in the bacterial transition to a biofilm state (Jenal et al., 2017; Römling et al., 2013). Recent studies have emphasized the strategy of developing anti-biofilm compounds by reducing cellular c-di-GMP levels for the treatment of biofilm-associated diseases (Hultqvist et al., 2024; Martin et al., 2021). Although TF activity-based sensors could not provide cellular insights at the single-cell level, the high sensitivity of our sensor suggests strong potential for its use as a high-throughput screening platform to identify novel agents that reduce intracellular c-di-GMP concentrations. Considering the more complex c-di-GMP-mediated networks in pathogens such as Pseudomonas aeruginosa and Vibrio cholerae compared to E. coli, more studies are needed to determine whether cdiGEBS can effectively capture dynamic changes in c-di-GMP concentrations in these bacteria. Nevertheless, future efforts focusing on screening compounds that inhibit DGC activity, activate PDE activity, or sequester free c-di-GMP in E. coli by combining tools of our sensor and fluorescence-activated cell sorting (FACS) could offer new insights for combating biofilm-associated diseases.
In conclusion, we present a novel genetically encoded c-di-GMP biosensor, cdiGEBS, which not only enables more straightforward quantification of cellular c-di-GMP compared to traditional phenotypic assays including Congo red staining and crystal violet assays but also exceeds the fluorescence dynamic range of most existing biosensors, i.e., a 21-fold increase compared with mScarlet I-mVenusNB-MrkH and a 20-fold increase over CensYBL. The application of cdiGEBS in investigating DGC and PDE activities, as well as the effects of chemical agents on cellular c-di-GMP levels, underscores its potential for high-throughput screening of c-di-GMP-targeting agents, offering significant implications for biomedical applications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
HL: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing–original draft. SQ: Conceptualization, Funding acquisition, Resources, Supervision, Writing–review and editing. WH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China, Grant Number: 2022YFF1102900; The National Natural Science Foundation of China (NSFC), Grant Numbers: 32222049, 32201043, and 32171269; The Natural Science Foundation of Shanghai, Grant Number: 23ZR1415300; The Shanghai Frontier Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Municipal Education Commission, Grant Number: 2021 Sci & Tech 03 28; Shanghai Municipal Science and Technology Major Project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2024.1528626/full#supplementary-material
References
Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., et al. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500. doi:10.1038/s41586-024-07487-w
Almblad, H., Randall, T. E., Liu, F., Leblanc, K., Groves, R. A., Kittichotirat, W., et al. (2021). Bacterial cyclic diguanylate signaling networks sense temperature. Nat. Commun. 12, 1986. doi:10.1038/s41467-021-22176-2
Bellini, D., Caly, D. L., McCarthy, Y., Bumann, M., An, S. Q., Dow, J. M., et al. (2014). Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol. Microbiol. 91, 26–38. doi:10.1111/mmi.12447
Boehm, A., Steiner, S., Zaehringer, F., Casanova, A., Hamburger, F., Ritz, D., et al. (2009). Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 72, 1500–1516. doi:10.1111/j.1365-2958.2009.06739.x
Choi, H. Y., and Kim, W. G. (2024). Tyrosol blocks E. coli anaerobic biofilm formation via YbfA and FNR to increase antibiotic susceptibility. Nat. Commun. 15, 5683. doi:10.1038/s41467-024-50116-3
Christen, M., Christen, B., Folcher, M., Schauerte, A., and Jenal, U. (2005). Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837. doi:10.1074/jbc.M504429200
Christen, M., Kulasekara, H. D., Christen, B., Kulasekara, B. R., Hoffman, L. R., and Miller, S. I. (2010). Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328, 1295–1297. doi:10.1126/science.1188658
Ciofu, O., and Tolker-Nielsen, T. (2019). Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front. Microbiol. 10, 913. doi:10.3389/fmicb.2019.00913
Ciofu, O., Tolker-Nielsen, T., Jensen, P., Wang, H., and Høiby, N. (2015). Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv. Drug Deliv. Rev. 85, 7–23. doi:10.1016/j.addr.2014.11.017
Dahlstrom, K. M., and O’Toole, G. A. (2017). A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu. Rev. Microbiol. 71, 179–195. doi:10.1146/annurev-micro-090816-093325
De, N., Navarro, M. V., Raghavan, R. V., and Sondermann, H. (2009). Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393, 619–633. doi:10.1016/j.jmb.2009.08.030
De, N., Pirruccello, M., Krasteva, P. V., Bae, N., Raghavan, R. V., and Sondermann, H. (2008). Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6, e67. doi:10.1371/journal.pbio.0060067
Dippel, A. B., Anderson, W. A., Evans, R. S., Deutsch, S., and Hammond, M. C. (2018). Chemiluminescent biosensors for detection of second messenger cyclic di-GMP. ACS Chem. Biol. 13, 1872–1879. doi:10.1021/acschembio.7b01019
Dostert, M., Belanger, C. R., and Hancock, R. E. W. (2019). Design and assessment of anti-biofilm peptides: steps toward clinical application. J. Innate Immun. 11, 193–204. doi:10.1159/000491497
Dostert, M., Trimble, M. J., and Hancock, R. E. W. (2021). Antibiofilm peptides: overcoming biofilm-related treatment failure. RSC Adv. 11, 2718–2728. doi:10.1039/d0ra09739j
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi:10.1038/nrmicro.2016.94
Gao, Y., Xu, Y., Li, Y., Chen, K., Wu, X., Liu, Y., et al. (2022). The first FRET-based RNA aptamer NanoKit for sensitively and specifically detecting c-di-GMP. Nano Lett. 22, 716–725. doi:10.1021/acs.nanolett.1c03970
Gomez, J. B., and Waters, C. M. (2023). Switching on cyclic di-GMP heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Microbiol. 8, 1380–1381. doi:10.1038/s41564-023-01428-5
Greenwald, E. C., Mehta, S., and Zhang, J. (2018). Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707–11794. doi:10.1021/acs.chemrev.8b00333
Ha, D. G., and O'Toole, G. A. (2015). C-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 3, 0003-2014. doi:10.1128/microbiolspec.MB-0003-2014
Halte, M., Wörmann, M. E., Bogisch, M., Erhardt, M., and Tschowri, N. (2022). BldD-based bimolecular fluorescence complementation for in vivo detection of the second messenger cyclic di-GMP. Mol. Microbiol. 117, 705–713. doi:10.1111/mmi.14876
Hee, C. S., Habazettl, J., Schmutz, C., Schirmer, T., Jenal, U., and Grzesiek, S. (2020). Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP. Proc. Natl. Acad. Sci. United States of America 117, 17211–17220. doi:10.1073/pnas.2001232117
Hendrix, H., Itterbeek, A., Longin, H., Delanghe, L., Vriens, E., Vallino, M., et al. (2024). PlzR regulates type IV pili assembly in Pseudomonas aeruginosa via PilZ binding. Nat. Commun. 15, 8717. doi:10.1038/s41467-024-52732-5
Hengge, R., Gründling, A., Jenal, U., Ryan, R., and Yildiz, F. (2016). Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J. Bacteriol. 198, 15–26. doi:10.1128/jb.00331-15
Ho, C. L., Chong, K. S., Oppong, J. A., Chuah, M. L., Tan, S. M., and Liang, Z. X. (2013). Visualizing the perturbation of cellular cyclic di-GMP levels in bacterial cells. J. Am. Chem. Soc. 135, 566–569. doi:10.1021/ja310497x
Huangyutitham, V., Güvener, Z. T., and Harwood, C. S. (2013). Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4, e00242–e00213. doi:10.1128/mBio.00242-13
Hultqvist, L. D., Andersen, J. B., Nilsson, C. M., Jansen, C. U., Rybtke, M., Jakobsen, T. H., et al. (2024). High efficacy treatment of murine Pseudomonas aeruginosa catheter-associated urinary tract infections using the c-di-GMP modulating anti-biofilm compound Disperazol in combination with ciprofloxacin. Antimicrob. Agents Chemother. 68, e0148123. doi:10.1128/aac.01481-23
Irie, Y., and Parsek, M. R. (2014). LC/MS/MS-based quantitative assay for the secondary messenger molecule, c-di-GMP. Methods Mol. Biol. 1149, 271–279. doi:10.1007/978-1-4939-0473-0_22
Jenal, U., and Malone, J. (2006). Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40, 385–407. doi:10.1146/annurev.genet.40.110405.090423
Jenal, U., Reinders, A., and Lori, C. (2017). Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284. doi:10.1038/nrmicro.2016.190
Jonas, K., Edwards, A. N., Simm, R., Romeo, T., Römling, U., and Melefors, O. (2008). The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70, 236–257. doi:10.1111/j.1365-2958.2008.06411.x
Kaczmarczyk, A., van Vliet, S., Jakob, R. P., Teixeira, R. D., Scheidat, I., Reinders, A., et al. (2024). A genetically encoded biosensor to monitor dynamic changes of c-di-GMP with high temporal resolution. Nat. Commun. 15, 3920. doi:10.1038/s41467-024-48295-0
Kennelly, C., Tran, P., and Prindle, A. (2024). Environmental purines decrease Pseudomonas aeruginosa biofilm formation by disrupting c-di-GMP metabolism. Cell Rep. 43, 114154. doi:10.1016/j.celrep.2024.114154
Kulasakara, H., Lee, V., Brencic, A., Liberati, N., Urbach, J., Miyata, S., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3'-5')-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. United States of America 103, 2839–2844. doi:10.1073/pnas.0511090103
Liu, H., and Naismith, J. H. (2008). An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91. doi:10.1186/1472-6750-8-91
Malone, J. G., Williams, R., Christen, M., Jenal, U., Spiers, A. J., and Rainey, P. B. (2007). The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153, 980–994. doi:10.1099/mic.0.2006/002824-0
Martin, I., Waters, V., and Grasemann, H. (2021). Approaches to targeting bacterial biofilms in cystic fibrosis airways. Int. J. Mol. Sci. 22, 2155. doi:10.3390/ijms22042155
McCaughey, C. S., Trebino, M. A., McAtamney, A., Isenberg, R. Y., Mandel, M. J., Yildiz, F. H., et al. (2024). A label-free approach for relative spatial quantitation of c-di-GMP in microbial biofilms. Anal. Chem. 96, 8308–8316. doi:10.1021/acs.analchem.3c04687
Nelson, M. D., and Fitch, D. H. (2011). Overlap extension PCR: an efficient method for transgene construction. Methods Mol. Biol. 772, 459–470. doi:10.1007/978-1-61779-228-1_27
Palmer, A. E., Qin, Y., Park, J. G., and McCombs, J. E. (2011). Design and application of genetically encoded biosensors. Trends Biotechnol. 29, 144–152. doi:10.1016/j.tibtech.2010.12.004
Park, S., and Sauer, K. (2022). Controlling biofilm development through cyclic di-GMP signaling. Adv. Exp. Med. Biol. 1386, 69–94. doi:10.1007/978-3-031-08491-1_3
Petchiappan, A., Naik, S. Y., and Chatterji, D. (2020). Tracking the homeostasis of second messenger cyclic-di-GMP in bacteria. Biophys. Rev. 12, 719–730. doi:10.1007/s12551-020-00636-1
Petersen, E., Mills, E., and Miller, S. I. (2019). Cyclic-di-GMP regulation promotes survival of a slow-replicating subpopulation of intracellular Salmonella Typhimurium. Proc. Natl. Acad. Sci. United States of America 116, 6335–6340. doi:10.1073/pnas.1901051116
Povolotsky, T. L., and Hengge, R. (2016). Genome-based comparison of cyclic di-GMP signaling in pathogenic and commensal Escherichia coli strains. J. Bacteriol. 198, 111–126. doi:10.1128/jb.00520-15
Qvortrup, K., Hultqvist, L. D., Nilsson, M., Jakobsen, T. H., Jansen, C. U., Uhd, J., et al. (2019). Small molecule anti-biofilm agents developed on the basis of mechanistic understanding of biofilm formation. Front. Chem. 7, 742. doi:10.3389/fchem.2019.00742
Randall, T. E., Eckartt, K., Kakumanu, S., Price-Whelan, A., Dietrich, L. E. P., and Harrison, J. J. (2022). Sensory perception in bacterial cyclic diguanylate signal transduction. J. Bacteriol. 204, e0043321. doi:10.1128/jb.00433-21
Rodesney, C. A., Roman, B., Dhamani, N., Cooley, B. J., Katira, P., Touhami, A., et al. (2017). Mechanosensing of shear by Pseudomonas aeruginosa leads to increased levels of the cyclic-di-GMP signal initiating biofilm development. Proc. Natl. Acad. Sci. United States of America 114, 5906–5911. doi:10.1073/pnas.1703255114
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. MMBR 77, 1–52. doi:10.1128/mmbr.00043-12
Rumbaugh, K. P., and Sauer, K. (2020). Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586. doi:10.1038/s41579-020-0385-0
Sanchez-Torres, V., Hu, H., and Wood, T. K. (2011). GGDEF proteins YeaI, YedQ, and YfiN reduce early biofilm formation and swimming motility in Escherichia coli. Appl. Microbiol. Biotechnol. 90, 651–658. doi:10.1007/s00253-010-3074-5
Schirmer, T. (2016). C-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J. Mol. Biol. 428, 3683–3701. doi:10.1016/j.jmb.2016.07.023
Schumacher, M. A., and Zeng, W. (2016). Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. Proc. Natl. Acad. Sci. United States of America 113, 10067–10072. doi:10.1073/pnas.1607503113
Simm, R., Morr, M., Remminghorst, U., Andersson, M., and Römling, U. (2009). Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 386, 53–58. doi:10.1016/j.ab.2008.12.013
Vrabioiu, A. M., and Berg, H. C. (2022). Signaling events that occur when cells of Escherichia coli encounter a glass surface. Proc. Natl. Acad. Sci. United States of America 119, e2116830119. doi:10.1073/pnas.2116830119
Wang, F., He, Q., Su, K., Gao, F., Huang, Y., Lin, Z., et al. (2016). The PilZ domain of MrkH represents a novel DNA binding motif. Protein and Cell 7, 766–772. doi:10.1007/s13238-016-0317-y
Wang, L., Wong, Y. C., Correira, J. M., Wancura, M., Geiger, C. J., Webster, S. S., et al. (2023). The accumulation and growth of Pseudomonas aeruginosa on surfaces is modulated by surface mechanics via cyclic-di-GMP signaling. NPJ Biofilms Microbiomes 9, 78. doi:10.1038/s41522-023-00436-x
Wang, L., Wu, C., Peng, W., Zhou, Z., Zeng, J., Li, X., et al. (2022). A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging. Nat. Commun. 13, 5363. doi:10.1038/s41467-022-32994-7
Wang, X. C., Wilson, S. C., and Hammond, M. C. (2016). Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res. 44, e139. doi:10.1093/nar/gkw580
Wang, Z., Song, L., Liu, X., Shen, X., and Li, X. (2023). Bacterial second messenger c-di-GMP: emerging functions in stress resistance. Microbiol. Res. 268, 127302. doi:10.1016/j.micres.2023.127302
Wilksch, J. J., Yang, J., Clements, A., Gabbe, J. L., Short, K. R., Cao, H., et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7, e1002204. doi:10.1371/journal.ppat.1002204
Keywords: c-di-GMP, genetically encoded fluorescent biosensor, transcription factor, MrkH, diguanylate cyclases
Citation: Li H, Quan S and He W (2025) A genetically encoded fluorescent biosensor for sensitive detection of cellular c-di-GMP levels in Escherichia coli. Front. Chem. 12:1528626. doi: 10.3389/fchem.2024.1528626
Received: 15 November 2024; Accepted: 24 December 2024;
Published: 10 January 2025.
Edited by:
Zhenhao Tian, Northwestern Polytechnical University, ChinaReviewed by:
Rigumula Wu, Genentech Inc., United StatesFangrui Zhong, Huazhong University of Science and Technology, China
Copyright © 2025 Li, Quan and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Quan, c2h1cXVhbkBzanR1LmVkdS5jbg==; Wei He, d2hlQG1haWwuZWN1c3QuZWR1LmNu
†ORCID: He Li, orcid.org/0009-0008-0209-9119; Shu Quan, orcid.org/0000-0002-6672-4947; Wei He, orcid.org/0000-0001-6263-5931
 He Li1†
He Li1† Shu Quan
Shu Quan Wei He
Wei He