- Laboratorio de Enzimología Molecular, Instituto de Agrobiotecnología Del Litoral (CONICET—UNL), Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional Del Litoral, Santa Fe, Argentina
The phosphorolysis of cello-oligosaccharides is a critical process played in the rumen by Ruminococcus albus to degrade cellulose. Cellodextrins, made up of a few glucosyl units, have gained lots of interest by their potential applications. Here, we characterized a cellobiose phosphorylase (RalCBP) and a cellodextrin phosphorylase (RalCDP) from R. albus 8. This latter was further analyzed in detail by constructing a truncated mutant (Ral∆N63CDP) lacking the N-terminal domain and a chimeric protein by fusing a CBM (RalCDP-CBM37). RalCBP showed a typical behavior with high activity on cellobiose. Instead, RalCDP extended its activity to longer soluble or insoluble cello-oligosaccharides. The catalytic efficiency of RalCDP was higher with cellotetraose and cellopentaose as substrates for both reaction directions. Concerning properties of Ral∆N63CDP, results support roles for the N-terminal domain in the conformation of the homo-dimer and conferring the enzyme the capacity to catalyze the phosphorolytic reaction. This mutant exhibited reduced affinity toward phosphate and increased to glucose-1-phosphate. Further, the CBM37 module showed functionality when fused to RalCDP, as RalCDP-CBM37 exhibited an enhanced ability to use insoluble cellulosic substrates. Data obtained from this enzyme’s binding parameters to cellulosic polysaccharides agree with the kinetic results. Besides, studies of synthesis and phosphorolysis of cello-saccharides at long-time reactions served to identify the utility of these enzymes. While RalCDP produces a mixture of cello-oligosaccharides (from cellotriose to longer oligosaccharides), the impaired phosphorolytic activity makes Ral∆N63CDP lead mainly toward the synthesis of cellotetraose. On the other hand, RalCDP-CBM37 remarks on the utility of obtaining glucose-1-phosphate from cellulosic compounds.
1 Introduction
Cellulose, the quantitatively highest biopolymer on Earth, plays a critical role as a structural polymer, source of sugars and material for industrial applications (Moon et al., 2011; Huber et al., 2012; Nakajima M et al., 2017). Cellulose is a linear polymer of a hundred to a thousand glucosyl units linked through β-(1 → 4)-glycosidic bonds. Cellulose oligomers, or cellodextrins, made up of a few glucosyl units, have gained some interest because of their properties (Kluge et al., 2019; Ávila et al., 2021). These materials have potential applications for non-digestible dietary fiber products (Adharis et al., 2018), novel bio-based surfactants (Billès et al., 2016), hybrid nanomaterials (Enomoto-Rogers et al., 2010; 2011), and scaffold candidates for tissue engineering (Wang et al., 2017). Cellodextrins are interesting feed ingredients in livestock animals, showing potential prebiotic and health-promoting properties (Sybesma et al., 2015).
Cello-oligosaccharides have been produced from the degradation of natural cellulose or synthetic pathways via chemical or enzymatic reactions (Billès et al., 2017; Kluge et al., 2019). Soluble cellodextrins were proven to be beneficial dietary fibers and prebiotics that stimulate the growth of many healthy human gut bacteria more efficiently than inulin, trans-galacto-oligosaccharides, and cellobiose alone (Zhong et al., 2020). One way to generate cellodextrins is through chemical (e.g., acid-catalyzed cellulose degradation) or enzymatic hydrolysis of cellulose. However, there are important concerns about chemical degradation because of the generation of oligosaccharides and unwanted by-products. The enzymatic hydrolysis, meanwhile, requires highly pure enzymes (e.g., cellulases, β-glucosidases) and gives a mixture of cellodextrins (Zhong et al., 2019). Furthermore, the isolation of the cellodextrins from hydrolysis mixtures needs significant efforts in downstream processing (Zhang and Lynd, 2003). An alternative approach is to produce cellodextrins through bottom-up synthesis to overcome cellulose degradation issues. Since chemical synthesis involves multistep procedures, difficulties appear in controlling glycosidic bond formation’s regioselectivity and stereospecificity during polymerization and obtaining high molecular weight polysaccharides (Xiao and Grinstaff, 2017). Enzymatic routes constitute convenient tools for overcoming such critical troubles (Billès et al., 2017). In vitro enzymatic synthesis of cello-oligosaccharides provides some advantages compared to the previous methods. For example, well-controlled structures of products are obtained in a one-step polymerization owing to the high regio-, enantio-, chemo-, and stereo-selectivity of the enzymes. In addition, the production of controlled product composition eliminates the need for posterior purification steps (Zhong et al., 2020). Moreover, enzymes are non-toxic compounds that catalyze the reaction under ecologic environments, and they are isolated from sustainable resources or produced as recombinant proteins (Shoda et al., 2016; Fodor et al., 2017).
Ruminococcus albus 8 is a critical ruminal bacterium involved in the digestion of dietary cellulose. This microorganism degrades cello-oligosaccharides mainly through phosphorolysis catalyzed by cellobiose phosphorylase (RalCBP; EC 2.4.1.2) and cellodextrin phosphorylase (RalCDP; EC 2.4.1.49) rather than hydrolysis catalyzed by β-glucosidase (Hamura et al., 2012; Sawano et al., 2013). Glycoside phosphorylases belong to the glycoside hydrolase family 94 (GH94) and play an important role in the metabolism of long-chain cellodextrins (Ubiparip et al., 2021). They catalyze a glycoside’s reversible phosphorolysis to produce the corresponding sugar 1-phosphate (Eq. 1), enabling efficient synthesis of a diversity of oligosaccharides with sugar 1-phosphate as the donor and a suitable carbohydrate acceptor in the synthetic reaction with strict regioselectivity (Luley-Goedl and Nidetzky, 2010; Nakai et al., 2010; O’Neill and Field, 2015; Puchart, 2015; Zhong et al., 2019; Gabrielli et al., 2021).
Cellodextrin phosphorylases have been extensively used in the enzymatic synthesis of polysaccharides (Nakai et al., 2013; O’Neill and Field, 2015; Zhong et al., 2019; Zhong and Nidetzky, 2020; Nidetzky and Zhong, 2021). They have the broadest acceptor and donor substrate specificity of all GH94 enzymes, making them good tools for suitable biocatalytic production of diverse carbohydrates and related molecules. Still, the average degree of polymerization (DP) of cellodextrin produced by CDP ranged from 8 to 10 glucose units, suggesting that these enzymes cannot elongate high DP cellodextrin chains (Ubiparip et al., 2021). This lack of capacity may result from the decrease in cellodextrin solubility as their DP increases, leading to reduced access of the soluble enzyme to the longer substrates. To overcome this issue, enzymes from different GH families, usually contain a catalytic module and one or more non-catalytic carbohydrate-binding modules (CBMs) (Boraston et al., 2004). CBMs can enrich enzymes on the surface of solid substrates through affinity adsorption to enhance their activity (Shoseyov et al., 2006). Remarkably, in addition to the catalytic domain, most glycosyl hydrolases from R. albus display different motifs involved in substrate binding and/or cell adhesion (Christopherson et al., 2014; Storani et al., 2020; Yeoman et al., 2021). Among them, CBM from family 37 (CBM37), exclusive from this bacterium, exhibits an affinity to broad substrates like cellulose, xylans, and other oligosaccharides (Ezer et al., 2008). Unlike many glycoside hydrolases, no report deals with glycoside phosphorylase containing a CBM. From the crystal structure of Clostridium thermocellum CDP, it was identified that this protein had a unique N-terminal extension involved in homodimer assembly and contributed to the substrate binding site conformation (O’Neill et al., 2017). We identified that multiple GH94 enzymes have a very conserved N-terminal region that could be involved in homodimer assembly and/or binding site conformation.
The present work describes the structural and functional characterization of two members of the family GH94 (CDP and CBP) from R. albus 8. Here, we analyzed the putative function of the first 63 N-terminal amino acids conserved in RalCDP as a putative CBM. On the other hand, we hypothesized that adding a CBM to a cellodextrin phosphorylase catalytic domain could improve enzyme access to insoluble substrates, thus enhancing its activity. To achieve this goal, we explored how the fusion of a CBM37_2 from R. albus 8 Cel5G affects its activity on insoluble substrates.
2 Materials and methods
2.1 Bacteria, plasmids, primers, and reagents
Escherichia coli Top 10 F′ and E. coli BL21 (DE3) (Invitrogen) served as hosts for cloning purposes and expression of the genes cloned into pRSFDuet and pETDuet vectors (Novagen). DNA manipulations, E. coli cultures, and transformations were performed according to standard protocols (Sambrook and Russell, 2001). Supplementary Table S1 details all the primers (obtained from GenBiotech) utilized in this work. Carboxymethyl cellulose (CMC), glucose-1-phosphate (Glc-1P), cellobiose (C2) and glucose (Glc) were from Sigma-Aldrich. Cellotriose (C3), cellotetraose (C4) and cellopentaose (C5) were from CarboSynth. All other reagents were of the highest purity available. Phosphoric acid swollen cellulose (PASC) was prepared from Cellulose Powder (ICN) after water slurring, cellulose dissolution in ortho-phosphoric acid, and regeneration in water (Percival Zhang et al., 2006). Insoluble PASC was prepared in concentrated phosphoric acid with partial hydrolysis in an ice bath for 1 h (Zhang and Lynd, 2004).
2.2 Construction of CDP variants
The DNA sequences coding for RalCBP (GenBank accession number: OP866768) and RalCDP (GenBank accession number: OP866769), as reported in the elucidated genome assembly from R. albus 8 (GCA_000178155.2), were codon-optimized for expression in E. coli based on RefSeq: WP_002848762.1 and RefSeq: WP_002848532.1, respectively. Sequences were de novo synthesized by Bio Basic (Canada) and provided in pUC57/ralcdp and pUC57/ralcbp, respectively. The synthetic genes were cloned into the pETDuet vector (Novagen) between the SacI and SalI sites to produce the recombinant proteins fused to a His6-tag at the N-terminus.
For CBM37 fusion, the DNA sequence coding for CBM37 from R. albus 8 Cel5G was amplified using primers FowCBM37 and RevCBM37, respectively (Supplementary Table S1), containing BamHI and NotI restriction sites for cloning the PCR fragment into a designed cassette. This cassette allows cloning CBM37 to either N- or C- terminal of the previously cloned gene fused by a glycine linker (Gly X9) into pETDuet or pRSFDuet expression vectors between SacI and SalI restriction sites. To construct Ral∆N63CDP, the DNA region (first 189 nucleotides) coding for the putative CBM was removed by PCR from pUC57/ralcdp. Primers FowGHCDP and RevGHCDP (Supplementary Table S1), respectively containing SacI and SalI restriction sites, were used for cloning the PCR fragment into the pRSFDuet expression vector.
2.3 Protein expression and purification
The constructs [pETDuet/RalCBP] [pETDuet/RalCDP] [pRSFDuet/Ral∆N63CDP] and [pETDuet/RalCDP-CBM37] were used to transform E. coli BL21 (DE3) cells (Invitrogen). To produce the recombinant proteins, we inoculated 1 L of LB medium (supplemented with 50 μg mL−1 kanamycin or ampicillin) with a 1/100 dilution of an overnight culture. Cells were grown at 37°C and 180 rpm in an orbital shaker until OD600nm was ∼0.6. Gene expression was induced with the addition of 0.2 mM isopropyl-β-D-1-thiogalactopyranoside and further growing at 22°C overnight. The cells were harvested by centrifugation at 5,000 ×g at room temperature for 15 min and kept at −20°C until use.
The cell pellet was suspended in 25 mL of Buffer A [25 mM Tris-HCl pH 8.0, 300 mM NaCl, 5% (v/v) glycerol, 10 mM imidazole], and disrupted by sonication. The resulting suspension was centrifuged twice at 20,000 ×g at 4°C for 10 min. The supernatant was loaded onto a 1 mL HisTrap column connected to an ÄKTA Explorer 100 purification system (GE Healthcare), previously equilibrated with Buffer A. After washing the column with 10 mL of Buffer A, the recombinant protein was eluted with a linear gradient of imidazole (10–300 mM, 50 mL). The fractions containing the enzymes of interest were collected and concentrated with Amicon™ Ultra Centrifugal Filter (Merk). The enzyme pool was loaded onto a HiLoad 16/600 Superdex 200 column (GE Healthcare), previously equilibrated with Buffer G [50 mM HEPES-NaOH pH 8.0, 100 mM NaCl, 0.1 mM EDTA]. The pool of the active fractions was concentrated, and protein preparations were supplemented with 5% (v/v) glycerol and stored at −80°C until use. Under these conditions, the purified enzymes remained stable and active for at least 6 months.
2.4 Native molecular mass determination
The native molecular mass of the purified recombinant proteins was determined by using a Superdex 200 10/300 column (GE Healthcare), equilibrated with Buffer G. A calibration curve was constructed by plotting the Kav values versus log10 of the molecular mass of standard proteins (ribonuclease, 13.7 kDa; carbonic anhydrase, 29 kDa; ovalbumin, 43 kDa; conalbumin, 75 kDa; aldolase, 158 kDa; ferritin, 440 kDa; and thyroglobulin, 669 kDa). The Kav was calculated as (Ve-V0)/(Vt-V0), where Ve is the elution volume of the protein, V0 is the elution volume of Dextran Blue, and Vtis the total volume of the column.
2.5 Enzyme activity assay and determination of kinetic constants
Cellobiose and cellodextrin phosphorylase activities were determined for phosphorolysis and synthetic directions. The reaction of cello-oligosaccharides synthesis was measured following the inorganic phosphate (Pi) released from the glucosyl donor using a highly sensitive colorimetric method (Fusari et al., 2006). The standard reaction media contained 100 mM sodium acetate buffer pH 6.0, 2 mM Glc-1P, 20 mM of different glucosyl acceptors (glucose, cellobiose, cellotriose, cellotetraose or cellopentaose), and a proper dilution of the enzyme in a final volume of 50 μL. Reactions were incubated for 10 min at 45°C and terminated with the addition of the Malachite Green reagent. The complex formed with Pi was measured at 630 nm in a Multiskan GO microplate reader (Thermo Scientific). To determine phosphorolytic activity, the reaction mixture (50 μL) contained 50 mM sodium acetate buffer (pH 6.0), 20 mM cellobiose, 50 mM Pi (potassium salt), and an appropriate amount of enzyme. After 10 min incubation at 45°C, the reaction was stopped by heating at 100°C for 10 min. The glucose oxidase-peroxidase method measured D-glucose liberated from cellobiose (Ambade et al., 1998).
When an oligosaccharide (n > 2) was used as a substrate, arsenolysis was measured instead of phosphorolysis by incubating the substrate (20 mM or 10 mg/mL) in a reaction mixture of 50 μL containing 50 mM sodium acetate buffer (pH 6.0), 50 mM arsenate (sodium salt) and a proper enzyme dilution for 10 min at 45°C (Eis and Nidetzky, 1999). D-glucose released from arsenolysis (since glucose 1-arsenate is unstable) was determined by the glucose oxidase-peroxidase method (Ambade et al., 1998). Comparative determination between phosphorolysis and arsenolysis reactions was performed with cellobiose as substrate (Supplementary Figure S1).
In all the assays, the substrate consumption was maintained below 10% as a control to guarantee the proper determination of initial velocity (vo). One unit (U) of enzyme activity equals 1 μmol of glucose/Pi released per minute under the respective assay conditions specified above.
Kinetic assays were performed using specified concentrations and conditions for all reaction mixture components. Saturation curves were carried out by assaying the respective enzyme activity at the saturating level of a fixed substrate and different concentrations of the variable substrate. The experimental data were plotted as enzyme activity (U.mg-1) versus substrate concentration (mM or mg/mL), and kinetic constants were determined by fitting the data to the Michaelis-Menten equation (Eq. 2). kcat was calculated with the enzyme concentration ([E]) from Vmax as kcat = Vmax/[E]. All kinetic constants are the mean of at least three data sets, reproducible within ±10%.
2.6 Carbohydrate binding assays
Enzymes’ binding capacity to cellulosic polysaccharides was determined as follows. The enzyme (10 µM) was mixed with 5 mg PASC in 0.25 mL of 50 mM sodium acetate buffer (pH 6.0) and incubated at 25°C for 1 h with stirring (1,500 xg). Stability control was performed by setting the protein in the same conditions without the polysaccharide. After centrifugation at 5,000 xg for 5 min to precipitate the polysaccharide, the supernatant was separated, and the unbound enzyme was quantified by the Bradford procedure (Bradford, 1976). The insoluble polysaccharide was washed thrice with sodium acetate buffer; then bound protein was eluted with 0.25 mL sodium dodecyl sulfate (SDS) 10% (w/v) at 90°C for 10 min and analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE).
Assays quantifying the capacity of enzyme binding to saccharides were carried out with 0.3–10 µM protein with 5 mg of polysaccharide in 0.25 mL of 50 mM sodium acetate buffer (pH 6.0). The maximum adsorption capacity (Amax) and dissociation constant (Kd) were determined by regressing binding isotherm data to a modified Langmuir-type binding model as described previously (Hong et al., 2007).
2.7 Polysaccharides synthesis and phosphorolysis
The analysis of cello-oligosaccharides synthesized by carrying out reactions (triplicated) at 45°C under agitation at a rate of 800 rpm in a Thermo Mixer HCM100-Pro (DLab Scientific). The reaction mixture contained 10 mM cellobiose, 100 nM purified enzyme in sodium acetate buffer (50 mM, pH 6.0) and different Glc-1P concentrations (0, 10, 30 and 50 mM) in a total volume of 0.05 mL. The reactions were stopped at different times by incubating the mixture at 100°C for 10 min. The samples were centrifuged at 20,000 xg, and the supernatant was separated from the pellet. The supernatant was analyzed for soluble cellodextrins using thin-layer chromatography (TLC) with a mobile phase of ethyl acetate: acetic acid: water (4:3:3, by volume). Staining was performed with orcinol 0.25% reagent in ethanol: sulfuric acid solution (95:5, v/v) at 95°C.
The soluble oligosaccharides were additionally analyzed by high-performance liquid chromatography (HPLC) on a Shimadzu LC-2050 system (Merck, Darmstadt, Germany) using a Luna Omega 3 μm NH2 column (100 Å, 150 × 4.6 mm; Phenomenex, Aschaffenburg, Germany) operated at 30°C. Acetonitrile: water (65: 35, by volume) was used as eluent at a flow rate of 1 mL/min. Refractive index detection was used to quantify cello-oligosaccharides and cellobiose. Calibration was performed with cellobiose, cellotriose, cellotetraose, and cellopentaose as standards.
The capacity for deconstructing high DP cello-oligosaccharides of the enzyme was measured. PASC arsenolysis was performed in a reaction mixture of 50 μL containing 1 mg/mL PASC, 50 mM sodium acetate buffer pH 5, 100 mM sodium arsenate, and 5 nM enzyme. The glucose oxidase-peroxidase method (Ambade et al., 1998) determined D-glucose released from arsenolysis. PASC phosphorolysis was performed in the same conditions (a reaction mixture of 50 μL containing 1 mg/mL PASC, 50 mM sodium acetate buffer pH 5, 50 mM Pi, and 5 nM enzyme), but Glc-1P, released from the reaction, was determined enzymatically using a continuous coupled enzyme assay (Eis and Nidetzky, 1999).
2.8 Other assays
Protein concentration was determined following the procedure described by Bradford (Bradford, 1976), using BSA as the standard. Polyacrylamide gel electrophoresis was carried out under denaturing conditions (SDS-PAGE) to determine the purity of the enzymes (Laemmli, 1970). The average DP of PASC and CMC was determined by exhaustive hydrolysis by phenol-sulfuric method and determining reducing ends by Somogyi-Nelson (Zhang and Lynd, 2005).
3 Results
3.1 Sequence alignment and structure analysis
Cellobiose and cellodextrin phosphorylases belong to family GH94 together with other glycosyl phosphorylases like laminaribiose phosphorylases (LBPs), chitobiose phosphorylases (ChBPs), and cellobionic acid phosphorylases (CBAPs). They are commonly homodimers, and each subunit is composed of three distinct domains: an N-terminal β-sandwich domain that forms most of the dimer interface, a helical linker, and an (α/α)6-barrel catalytic domain (Hidaka et al., 2004, 2006; Bianchetti et al., 2011; Nakajima H et al., 2017; Kuhaudomlarp et al., 2019). Interestingly, the length and conformation of catalytic, adjacent, and opposite loops were identified as the determining features that restrict the size of the acceptor binding site differentiating between CBPs, CDPs, and ChBPs (Hidaka et al., 2004; 2006; De Groeve et al., 2010; Bianchetti et al., 2011; O’Neill et al., 2017; Gabrielli et al., 2021).
We recognized two putative glycosyl phosphorylases from the GH94 family in the proteome of R. albus 8 project ID UP000004259, identified with entry codes E9SB92 and E9SAK5. Protein sequence E9SB92 is entirely identical to CBP from R. albus NE1 (Hamura et al., 2012). It presents high identity (67% and 56%, respectively) to the enzymes from Clostridium thermocellum (PDB: 3qde) (Bianchetti et al., 2011) and Cellvibrio gilvus (PDB: 2cqs) (Hidaka et al., 2006). The protein derived from E9SAK5 aligned with the same sequences presenting much lower identity and similarity (30% and 45%, respectively). The alignment of E9SAK5 with members of the GH94 family showed higher sequence similarities with Vibrio proteolyticus ChBP, and Vibrio furnissii CBP, with an identity of 37% and a similarity of 68% for both proteins. Sequence alignments showed the conservation of critical amino acids involved in the phosphate-binding site and other residues conforming substrate-binding subsites −1, +1, and +2 (Figure 1).

FIGURE 1. Multiple alignment of partial sequences from GH94 family enzymes. Color lines over the sequences delimit opposite (orange), adjacent (blue), and catalytic (magenta) loops.
Structure comparison performed between V. proteolyticus ChBP (PDB code: 1v7x) (Hidaka et al., 2004), C. thermocellum CBP (PDB code: 3qde) (Bianchetti et al., 2011), CtCDP (PDB code: 5nz8) (O’Neill et al., 2017), and a model of the E9SAK5 protein designed by AlphaFold2 (Jumper et al., 2021) revealed significant structural differences in catalytic, adjacent, and opposite loops length (Figures 2A, B). Based on these dissimilarities, we assumed that the protein sequence from entry E9SAK5 corresponded to R. albus 8 cellodextrin phosphorylase (RalCDP), despite its relatively low sequence similarity with other CDPs, like Thermoclostridium stercorarium (53%), and Thermotoga neapolitana (49%). In addition, multiple sequences analysis of several glycosyl phosphorylases from the family GH94 revealed that the N-terminal domain presents a highly conserved region (amino acids 1–63). This region was predicted to be involved in substrate binding, homodimer interaction, and to be a putative CBM by the SMART tool (Letunic and Bork, 2018; Letunic et al., 2021). As far as we know, no other glycosyl phosphorylase from this family was reported to possess a CBM.
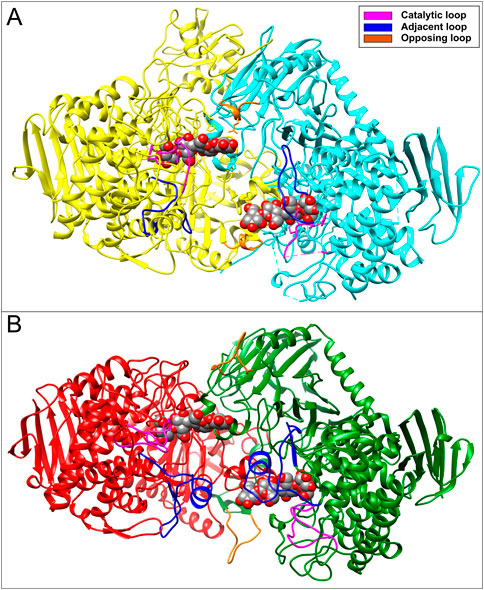
FIGURE 2. (A) Crystal structure of CtCDP (PDB code: 5zn8) and (B) 3D-model of RalCDP homodimer structure performed by Alphafold2. Both structures show the interaction with cellotetraose displayed as colored spheres.
3.2 Production of recombinant enzymes
Ruminococcus albus 8 CBP and four variants of CDP were constructed and produced in E. coli, as described in Figure 3. Thus, the constructs were designed to produce RalCBP (the E9SB92 protein), and RalCDP (the E9SAK5 protein) and its truncated version lacking the firsts sixty-three N-terminal amino acids (Ral∆N63CDP). Also, we joined the CBM37 by a flexible glycine linker to RalCDP and Ral∆N63CDP to construct the fusion proteins RalCDP-CBM37 and Ral∆N63CDP-CBM37 (Figure 3A). Flexible linkers are the most often employed since they allow freedom of mobility and interaction between domains (Martins et al., 2020). The applied strategy enables fusing the CBM to the enzymes’ N- or the C-extreme. Still, all resulting enzymes were inactive when the CBM was linked to the N- terminal. As also illustrated in Figure 2A, all the recombinant proteins were produced with a His-tag fused to its C-terminal and purified by Ni2+-affinity (Ni2+-IDA) chromatography.
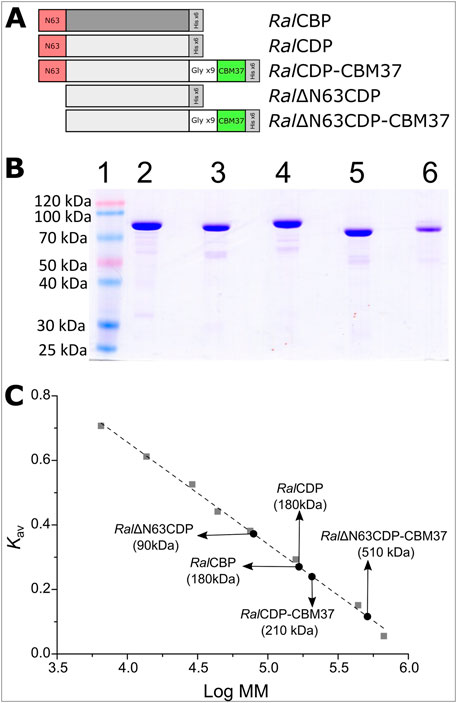
FIGURE 3. Schematic representation of CDP variants (A). SDS-Page of purified proteins: Molecular mass marker (1), RalCBP (2), RalCDP (3), RalCDP-CBM37 (4), Ral∆N63CDP (5) and Ral∆N63CDP-CBM37 (6) (B). Quaternary structure determination for each enzyme variant by size exclusion chromatography (C).
Figure 3B shows that recombinant proteins were produced and purified to a high electrophoretic degree. RalCBP has 829 amino acids and a theoretical molecular mass of 92.3 kDa, while RalCDP has 809 amino acids and 90.2 kDa. Analysis by SDS-PAGE of the purified proteins exposed a band of approximately 90 kDa. Gel filtration chromatography on Superdex 200 indicated that both enzymes are homodimers with a molecular mass of 180 kDa (Figure 3C). These data are in good agreement with that reported for other enzymes of the GH94 family previously characterized (Kim et al., 2002; Krishnareddy et al., 2002; Nakai et al., 2010; Bianchetti et al., 2011). Meanwhile, the mutant Ral∆N63CDP eluted as a single peak with an elution volume corresponding to a monomer. This latter result suggests a loss of interaction between the subunits consequent with the absence of the conserved region on the N-terminal. Conversely, adding the CBM37 to the C-extreme of RalCDP did not disturb the homodimer conformation. On the other hand, the CBM37 fusion to the C-terminal of Ral∆N63CDP produced a protein oligomer that eluted in a volume corresponding with a homo-pentamer (Figure 3C).
We measured the enzymatic activity of each elusion peak by assaying the respective enzyme in the synthesis sense of catalysis. RalCBP and RalCDP presented a specific activity of 57 U/mg and 98 U/mg, with glucose and cellobiose as substrates, respectively. For the truncated version Ral∆N63CDP, this value was 31 U/mg, despite its conformational change from a homodimer to a monomer. Different results were observed with the fusion of the CBM to these enzymes. RalCDP-CBM37 showed a specific activity of 45 U/mg; meanwhile, Ral∆N63CDP-CBM37 was completely inactive, probably due to its oligomeric state. These results show the importance of the N-terminal extreme of RalCDP, as the absence of such a region prevents the protein’s correct conformation as a homodimer, with a parallel decrease in enzyme activity. In addition, the mere presence of the His-tag or the CBM fusion at the N-terminal demolished the enzymatic activity.
3.3 Study of physicochemical properties
We examined the activity of the recombinant enzymes under study at different pH and temperature ranges since the modifications made to their sequences could alter the protein’s sensitivity to pH or temperature. No optimum pH and temperature differences were evident between any enzyme’s phosphorolytic and synthetic activities. RalCDP exhibited maximum activity at pH range 5–7, but the proteins RalCBP, Ral∆N63CDP, and RalCDP-CBM37 showed a narrower maximum activity peak at pH 6–7. At pH 5, RalCBP and Ral∆N63CDP displayed a reduction of 60% in their respective catalytic activity compared with RalCDP and RalCDP-CBM37. In the same way, a slight difference (∼20%) was detected for the mutant enzymes at pH 8.0 (Figure 4A). Regarding the proteins’ stability at different pH conditions, RalCDP and RalCDP-CBM37 conserved 80% of their enzymatic activity after incubating for 1 h in the pH range 4–9. RalCBP only preserved this activity level when incubated in the pH range of 4–6. Meanwhile, in the rest of the conditions tested, RalCBP showed around 50%–60% of its catalytic activity. Ral∆N63CDP was the enzyme with the least stability, with 40% residual activity after incubation with sodium citrate buffer (pH 4–5) or potassium phosphate (pH 5–9). However, this enzyme conserved 70% residual activity when incubated in sodium acetate buffer (between pH 4–6). Also, all enzymes showed shallow stability when incubated in sodium citrate buffer at pH 3.0 for 1 h (Figure 4B).
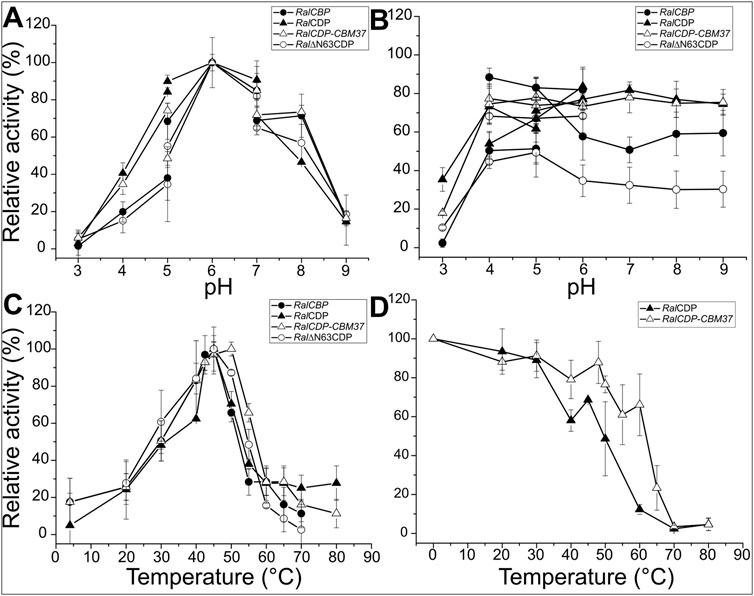
FIGURE 4. Enzymatic activity of RalCBP (black circles), RalCDP (black triangles), RalCDP-CBM37 (white triangles) and Ral∆N63CDP (white circles) at different pHs (A). Residual enzymatic activity of RalCBP (black circles), RalCDP (black triangles), RalCDP-CBM37 (white triangles), and Ral∆N63CDP (white circles) after being incubated for 1 h in different buffer solutions (B). Enzymatic activity of RalCBP (black circles), RalCDP (black triangles), RalCDP-CBM37 (white triangles) and Ral∆N63CDP (white circles) at different temperatures(C). Residual enzymatic activity of RalCDP (black triangles), RalCDP-CBM37 (white triangles) after 1 h incubations at different temperatures (D).
Determination of optimal temperature for catalytic activity showed that RalCBP, RalCDP, and Ral∆N63CDP presented a maximum at 45°C. However, RalCDP activity decreased considerably at temperatures slightly lower or higher than its optimal temperature (60% and 40% of its optimal activity at 40°C and 55°C, respectively). It was noticed that the fusion of the CBM37 domain to RalCDP improved the enzyme´s behavior toward temperature. Thus, RalCDP-CBM37 presented a maximum activity at 50°C with a reduction to 80% and over 60% values at 40°C and 55°C, respectively (Figure 4C). To analyze the proteins’ thermal stability, both enzymes were incubated for 1 h at different temperatures to measure the residual enzymatic activity later. RalCDP-CBM37 retained more than 60% of its catalytic capacity up to 60°C, while at this temperature, the RalCDP activity reduced by 90% (Figure 4D). The global of protein stability was reinforced by experiments determining melting temperatures for these enzymes shown in Supplementary Figure S2.
3.4 Kinetic characterization of Ruminococcus albus CBP and CDP variants
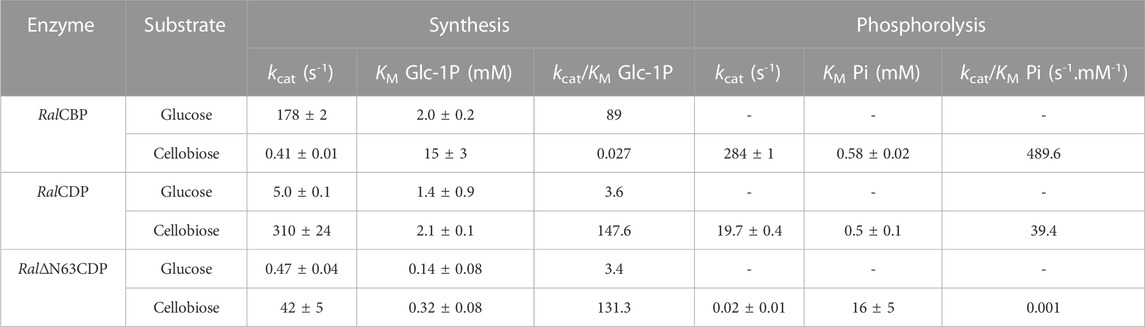
We further characterized the recombinant phosphorylases determining their kinetic properties when using different substrates in both senses of catalysis, with results detailed in Table 1. We also analyzed if the CBM addition improves enzyme activity on long-chain oligosaccharides or if the quaternary structure change (due to the lack of the N-terminal region in the protein) could modify the active site conformation, opening it to broader substrates. We tested the phosphorolytic and synthetic activities of RalCBP and the active CDP forms (RalCDP, Ral∆N63CDP, and RalCDP-CBM37) with substrates with different DP like glucose, cellobiose, cellotriose, cellotetraose, cellopentaose, and the two cellulosic oligosaccharides CMC and PASC.
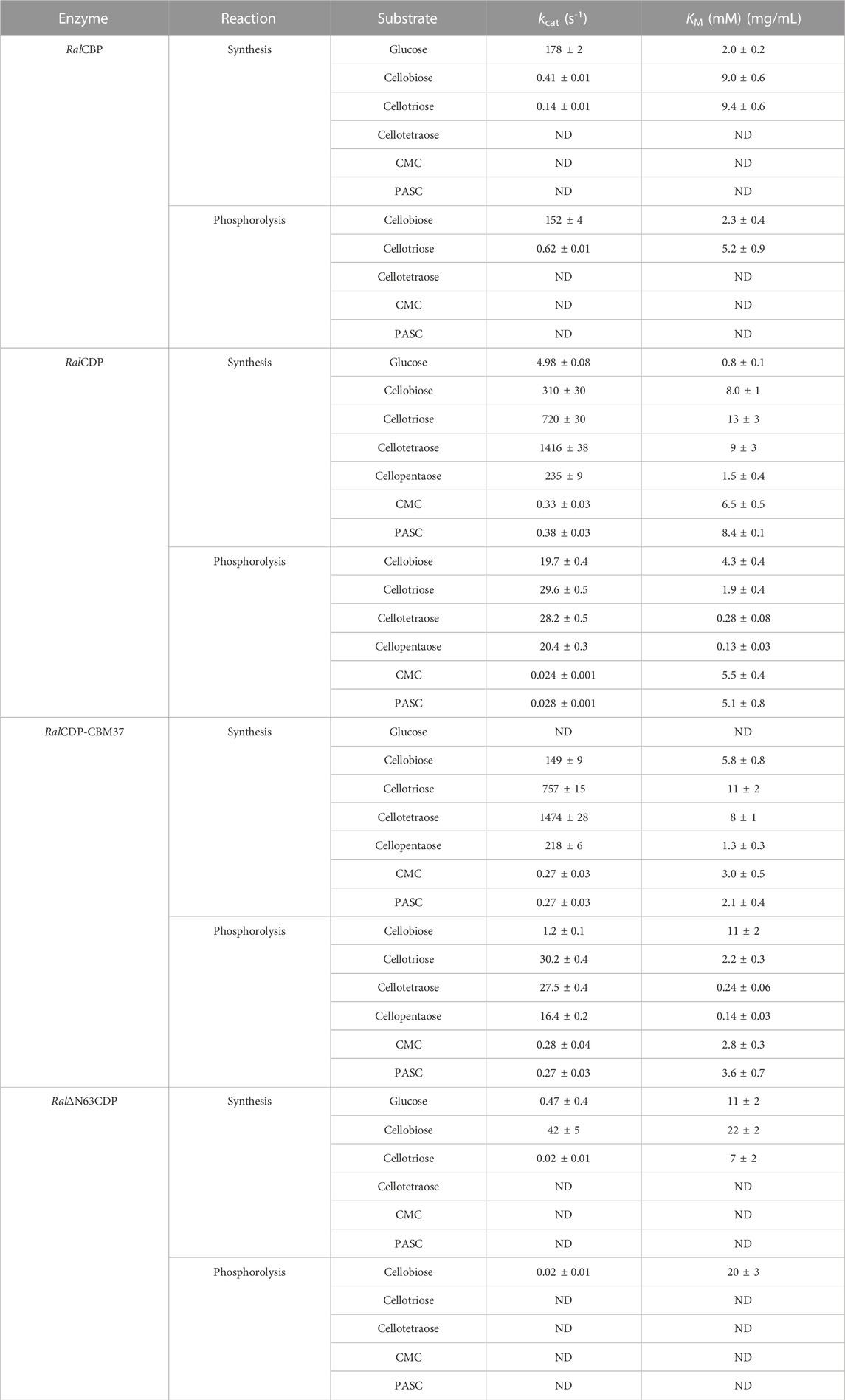
TABLE 1. Kinetic parameters from the different enzyme variants with substrates with different DP. aunits for substrates CMC and PASC ND: Not detected activity even at a 50-fold increase in enzyme concentration.
Table 1 shows that RalCBP exhibited phosphorolytic activity only with cellobiose and cellotriose. The kinetic parameters indicated a higher (by more than two orders of magnitude) catalytic performance of the enzyme with cellobiose (Figure 5). Indeed, the major difference was that the kcat value with cellotriose was 250-fold lower than that obtained with the disaccharide, although both KM values were similar (only 2-fold lower for cellobiose) (Table 1). Concerning synthetic activity using Glc-1P as the donor substrate, RalCBP exhibited maximum catalytic efficiency with glucose as the acceptor (Figure 5). The DP of the acceptor substrate affected considerably RalCBP catalytic ability. Compared with glucose, decreases of 450- and 1,100-fold in kcat were determined with cellobiose and cellotriose, respectively, but KM values exhibited increases of ∼5-fold (Table 1). Overall, the kinetic behavior of RalCBP agrees with its identification of cellobiose phosphorylase.
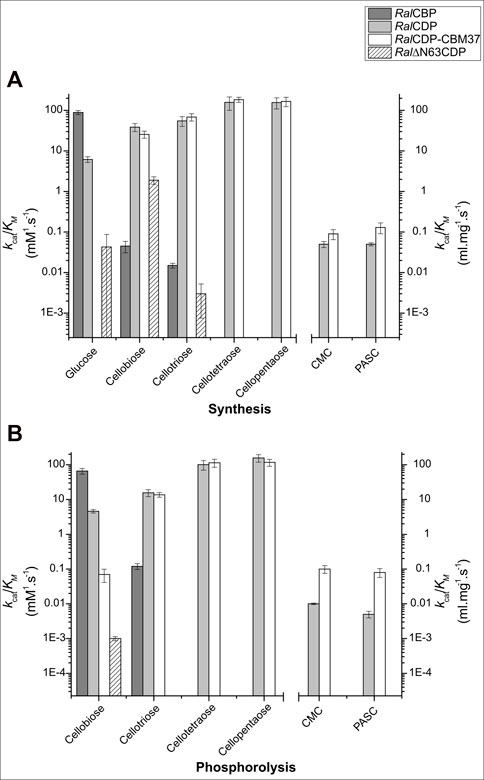
FIGURE 5. Enzymatic efficiencies of RalCBP (dark gray), RalCDP (light gray), RalCDP-CBM37 (white), and Ral∆N63CDP (striped) with cello-oligosaccharides of different DP for synthesis (A) and phosphorolysis (B).
Data obtained in the RalCDP characterization as a cellodextrin phosphorylase were markedly different from the above described for RalCBP. Thus, RalCDP showed synthetic activity with multiple acceptors, like glucose, cellobiose, cellotriose, cellotetraose, cellopentaose, and, less efficiently, the polysaccharides PASC and CMC (Table 1). The catalytic efficiency with the monosaccharide was considerably lower than with the other soluble substrates or RalCBP (Figure 5). Indeed, RalCDP exhibited maximum catalytic efficiency with cellotetraose and cellopentaose substrates and values 3-, 4- and 25-fold lower for cellotriose, cellobiose and glucose, respectively. On the other hand, the enzyme’s catalytic capacity for using the polysaccharides CMC and PASC was significantly low (Table 1; Figure 5). These results indicate the preference of this enzyme for oligosaccharides over the use of di- or monosaccharides as acceptors. However, the activity decreased when the DP increased considerably, like CMC and PASC polysaccharides, as described for other CDPs [54]. Similarly, RalCDP achieved maximum catalytic efficiencies in the phosphorolysis direction with cellopentaose and cellotetraose, diminishing in one order of magnitude by cellotriose and cellobiose (Figure 5). This decrease in catalytic efficiency was due to increased KM values as it decreased the substrate’s polymerization degree. In contrast, the kcat values were similar with all cello-oligosaccharides. Otherwise, phosphorolytic activity (in terms of kcat) with polysaccharides CMC and PASC was 1,000-fold lower (Table 1).
The N-terminal region removal provided notably different kinetic parameters to Ral∆N63CDP compared with the wild-type RalCDP. As shown in Table 1, the truncated enzyme exhibited a ∼1,000-fold reduction in its phosphorolytic capacity with cellobiose and was inactive with other disaccharides or oligosaccharides. Besides, the analysis of the synthetic reaction showed a 10-fold lower kcat for the elongation of cellobiose with Glc-1P. Other differences in kinetic behavior observed between Ral∆N63CDP and RalCDP for catalysis in the synthesis and phosphorolysis directions refer to affinity toward the substrates Glc-1P and phosphate (Table 2). Ral∆N63CDP showed a ∼30-fold reduction in phosphate affinity, leading to a significant decrease in its catalytic efficiency for phosphorolysis. However, the capacity for catalyzing the synthetic reaction with cellobiose remains at the same level as the wild-type RalCDP.
On the other hand, the fusion of the CBM37 to Ral∆N63CDP and RalCDP had quite varied effects on the catalytic properties. Ral∆N63CDP-CBM37 was completely inactive, while RalCDP-CBM37 showed similar kinetic parameters to RalCDP when using soluble oligosaccharides as substrates (Table 1). However, for the polysaccharides CMC and PASC, the CBM-tagged protein enhanced the kcat for phosphorolysis by one order of magnitude. It exhibited a 2-fold reduction in KM for CMC and PASC for both reaction directions (Table 2). This increased enzyme affinity for polysaccharides substrates enhances by ∼20-fold the catalytic efficiency for phosphorolysis compared to RalCDP (Figure 5).
3.5 Carbohydrate binding assay
The binding capacity of RalCDP, Ral∆N63CDP, and RalCDP-CBM37 to insoluble cellulosic polysaccharides was studied by incubating the enzymes with PASC. After incubation, the remaining soluble and bound fractions were analyzed by SDS-PAGE. As shown in Figure 6A, RalCDP could not bind to the insoluble polysaccharide PASC, remaining mainly in the soluble fraction after incubation. The same result was obtained for RalΔN63CDP, indicating that the N-terminal region, predicted as CBM, lacks a carbohydrate-binding function, at least to cellulosic substrates. However, the addition of CBM37, exemplified by RalCDP-CBM37, remarkably improved its binding capacity to this polysaccharide. The increment supports this result in enzyme affinity to polysaccharides observed in the kinetics parameters determined for this enzyme.
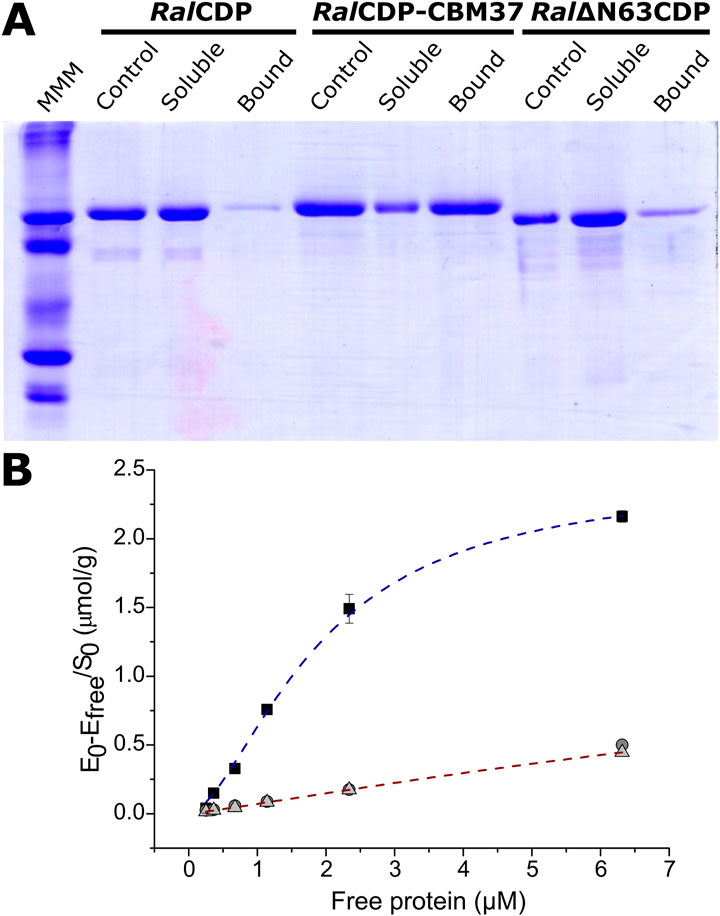
FIGURE 6. Analysis of CDP variants binding capacity to PASC. SDS-PAGE of the different fractions after incubation with the substrate (Control: enzyme incubated without substrate) (A). Binding isotherms for RalCDP (circles), RalCDP-CBM37 (squares), and Ral∆N63CDP (triangles) (B).
We calculated the respective binding parameters from the isotherm of the interaction betweenRalCDP-CBM37 and PASC, shown in Figure 5B. The fusion protein presented a dissociation constant (Kd) of 1.73 ± 0.06 µM and an adsorption capacity (Amax) of 2.36 ± 0.05 μmol/g. This adsorption capacity is 5-fold higher than that of enzymes lacking a CBM. These results indicate that CBM37 effectively recognizes the insoluble substrate PASC, also reinforcing the absence of functionality of the protein’s N-terminal domain for binding to the polysaccharide, as supported by results obtained with the complete RalCDP (Figure 6A). The increment in substrate binding affinity to the insoluble substrate was also reflected in the kinetic parameters of RalCDP-CBM37, which presents a decrease in KM with PASC compared to RalCDP (Table 1).
3.6 Evaluation of polysaccharides synthesis and degradation
The synthesis of polysaccharides by RalCDP and RalΔN63CDP was monitored utilizing cellobiose (10 mM) and different concentrations of Glc-1P (10, 30, and 50 mM) as substrates. Reaction products were analyzed by TLC (Figures 7A, B). In the RalCDP reactions, glucose can be observed as a result of the phosphorolytic activity of this enzyme, mainly at low Glc-1P concentrations (Figure 7A). However, this did not occur in reactions catalyzed by RalΔN63CDP for any of the Glc-1P concentrations tested (Figure 7B). The absence of glucose in RalΔN63CDP reactions is a consequence of its low affinity for Pi, as was shown in its kinetic characterization (Table 2). In addition, at low Glc-1P concentration (10 mM), we observed the synthesis of higher DP saccharides with the catalysis of RalΔN63CDP than that exerted by RalCDP, reflecting reaction displacement towards polysaccharides synthesis.
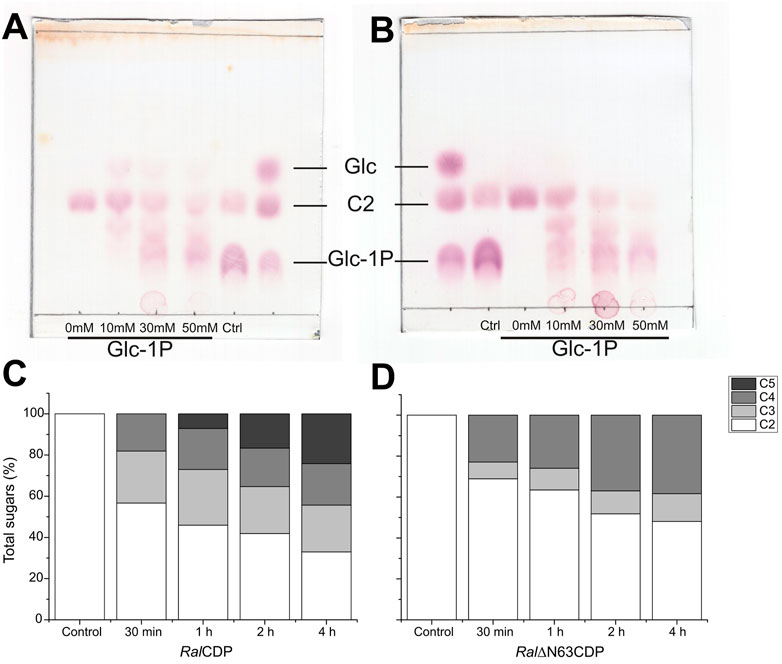
FIGURE 7. TLC analysis of RalCDP (A) and RalΔN63CDP (B) reactions at different Glc-1P: cellobiose ratios. HPLC analysis of cello-oligosaccharides synthesized by RalCDP (C) and RalΔN63CDP (D).
The use of HPLC allowed further analysis of the reaction products of both enzymes. The reactions were performed from 30 min to 4 h using Glc-1P and cellobiose in a 3:1 ratio. As illustrated by Figure 7C, RalCDP produced cellotriose, cellotetraose (∼25 and ∼20%, respectively), and increasing amounts of cellopentaose over the reaction time course (7, 17, and 24% for 1, 2, and 4 h, respectively). Meanwhile, in the reactions catalyzed by Ral∆N63CDP, only cellotriose and cellotetraose were produced (Figure 7D). The percentage of cellotriose detected was lower than those from RalCDP reactions. However, increasing amounts of cellotetraose were produced over time (23%–38%), being higher than those from RalCDP (18%–20%). These differences in reaction products could be attributed to the fact that Ral∆N63CDP cannot extend the oligosaccharide chain beyond cellotetraose, as shown in its kinetic characterization (Table 1), giving rise to the accumulation of this oligosaccharide. While, RalCDP has a high enzymatic activity with cellotetraose and cellopentaose. For these reasons, the RalCDP reaction would channel cellotetraose toward synthesizing oligosaccharides with a higher DP.
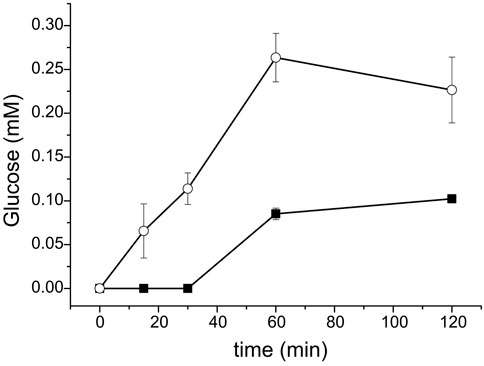
The higher catalytic efficiency of RalCDP-CBM37 with cellulosic polysaccharides could be an advantage for its application in different processes. Its higher phosphorolytic capacity could be useful for the production of Glc-1P from high available polysaccharides like cellulose. Thus, we followed the arsenolysis of PASC at different reaction times catalyzed by RalCDP and RalCDP-CBM37, the two enzyme versions exhibiting higher phosphorolytic activity. The strategy of replacing phosphate with arsenate was previously useful for measuring phosphorolytic activity on the insoluble polysaccharide PASC (Eis and Nidetzky, 1999; Zhang and Lynd, 2005). Figure 8 shows that RalCDP-CBM37 presented a ∼4-fold higher activity than RalCDP (0.71 vs. 0.16 µmol glucose/µmol enzyme. min). A maximum glucose yield of 0.26 mM was obtained after a 1 h reaction with RalCDP-CBM37, while RalCDP presented a maximum glucose yield of 0.1 mM after 2 h. This higher rate of phosphorolysis seems to be caused by the enhanced adsorption capacity, increasing the effective local enzyme concentration on the insoluble substrate due to the addition of the CBM.
4 Discussion
CDPs are carbohydrate-active enzymes with outstanding potential for the biocatalytic bottom-up synthesis of β-glucans as major bioactive compounds. The donor substrate α-Glc-1P serves as a cheaper version of the UDP-Glc required by the corresponding “Leloir glycosyltransferases” and can be conveniently prepared through the phosphorolysis of cheap and abundant resources like starch or cellulose (Ubiparip et al., 2021). They have the broadest acceptor and donor substrate specificity of all GH94 enzymes, making them good tools for suitable biocatalytic production of diverse carbohydrates and related molecules. However, only a few CDPs were biochemically characterized (Sawano et al., 2013; O’Neill et al., 2017; Wu et al., 2017; Ubiparip et al., 2021).
In this work, we identify a GH94 glycosyl phosphorylase from R. albus that showed relatively low sequence similarity with other CDPs, like T. stercorarium (53%), and T. neapolitana (49%). In addition, structure comparison with members of the GH94 family like V. proteolyticus ChBP (PDB code: 1v7x) (Hidaka et al., 2004), C. thermocellum CBP (PDB code: 3qde) (Bianchetti et al., 2011) and CtCDP (PDB code: 5nz8) (O’Neill et al., 2017), revealed significant sequence and structural differences in catalytic, adjacent, and opposite loops, which are highly involved in active site conformation. (O’Neill et al., 2017; Gabrielli et al., 2021). Here, we noticed that these differences led to an increase in the active site accessibility for long-chain oligosaccharides in RalCDP, as shown in Figure 2.
As was hypothesized from structural analysis and confirmed by 3D-models in Figure 2, RalCDP showed remarkable kinetic differences from other CDPs (detailed in Supplementary Table S2). This enzyme was active with substrates of different DP, ranging from monosaccharides to long-chain cellulosic oligosaccharides. Previously, TaCDP/CBP showed activity on both monosaccharides and cello-oligosaccharides (Wu et al., 2017), but only CtCDP showed activity on long-chain oligosaccharides (Ye et al., 2011). RalCDP activity with monosaccharides was similar to that of TaCDP/CBP but showed a 3- and 9-fold higher activity than CtCDP, for PASC phosphorolysis and synthesis, respectively (Supplementary Table S2).
Despite CDPs being active on cello-oligosaccharides, they are single modular proteins with no apparent CBM. However, we noticed that RalCDP presents a highly conserved N-terminal region, which is predicted to be involved in substrate binding, homodimer interaction, and/or a putative CBM. To analyze in depth the properties of these GH94 family proteins, we constructed two variants of RalCDP, one truncated mutant (RalΔN63CDP) lacking the conserved N-terminal region and another one (RalCDP-CBM37) fused to a CBM of family 37, unique from R. albus (Xu et al., 2004; Ezer et al., 2008). The exact mechanism of the CBM37 in the R. albus cellulase system is still unknown, but its members recognize and bind strongly to cellulose and numerous other polysaccharides (Xu et al., 2004).
Characterization of the recombinant proteins reveals that the conserved N-terminal region is critical to homodimer assembly since the construction on the truncated mutant RalΔN63CDP lead to a monomeric enzyme. This was also predicted in crystal structures of other GH94 enzymes (Hidaka et al., 2004; 2006; Bianchetti et al., 2011; Kuhaudomlarp et al., 2019). In addition, we identify that this modification of the quaternary structure decreased the enzyme capacity to catalyze the phosphorolysis of cellodextrins. Although, the activity in the direction of synthesis was only slightly affected in the truncated mutant. The kinetic analysis proved that differences in the enzymatic activity of monomeric RalΔN63CDP resulted from its affinity decrease toward phosphate and increase for Glc-1P. Kinetics parameters for Glc-1P and Pi of RalCDP showed that enzyme relative affinity for these substrates was 80- and 360-fold higher than those reported for R. albus NE1 CDP (Sawano et al., 2013) (Supplementary Table S2). These results indicate that the minimal differences observed in the sequence of the loops conforming to the active site lead to a significant difference in substrate recognition.
The reduced phosphorolytic capacity observed inRalΔN63CDP has been exploited for the synthesis of soluble cello-oligosaccharides from cellobiose and Glc-1P at lower donor: acceptor ratios than those reported from other CDPs wild-type versions (Zhong et al., 2019; Zhong and Nidetzky, 2020; Nidetzky and Zhong, 2021). RalΔN63CDP produced increasing amounts of cellotetraose over the reaction time course (Supplementary Figure S3). This confirms that this enzyme cannot extend the polysaccharide chain over four glucose units, as previously noticed in its kinetic characterization. These features of RalΔN63CDP make it an excellent molecular tool for synthesizing short-chain cello-oligosaccharides (Ubiparip, 2020; Zhong et al., 2020). Such soluble cellodextrins were proven to be useful dietary fibers and prebiotics that stimulate the growth of many healthy human gut bacteria more efficiently than inulin, trans-galacto-oligosaccharides, and cellobiose alone (Zhong et al., 2020). In addition, producing such controlled product composition eliminates the need for subsequent organic solvent purification steps.
Worthy, CDPs could also be employed to degrade β-glucans by phosphorolysis and produce high-value Glc-1P from a cheap and easily accessible substrate like cellulose. Kinetic characterization of RalCDP showed higher catalytic efficiency for the synthetic rather than the phosphorolytic reaction with cellulosic polysaccharides PASC and CMC. However, the fusion of the CBM37 to the C-terminal of RalCDP improved both the kcat for phosphorolysis and the enzyme affinity to insoluble cellulosic substrates (without affecting homodimer assembly). The increase in phosphorolytic activity here achieved by the fusion of the CBM37 to RalCDP was 13.5-fold higher than that from the fusion of CBM9 to CtCDP (Ye et al., 2011) (Supplementary Table S3). Thus, RalCDP-CBM37 would constitutes an improved molecular tool to degrade cellulosic polysaccharides generating Glc-1P. Also, the fusion of CBM37 increased the synthetic reaction’s catalytic efficiency, mainly reducing the enzyme KM (Supplementary Table S2). These effects on KM and catalysis with insoluble substrates were reported in studies fusing CBMs to different single-domain enzymes like cellulases, xylanases, or laccases. (Ravalason et al., 2009; Zhang et al., 2013; Dai et al., 2021).
The analysis of the CDPs binding capacity suggests that RalCDP and RalΔN63CDP could not bind to insoluble polysaccharides (PASC). However, the fusion of the CBM37 to RalCDP enhanced its adsorption capacity by 5-fold. The adsorption parameters obtained for this construct were similar to those achieved by the fusion of CBM9 to C. thermocellum CDP (Ye et al., 2011) (Supplementary Table S4). In the same way, CBM9 fusion did not modify the kinetic parameters with soluble substrates but improved enzyme activity on insoluble cellulose. Despite the poorly understood CBM37 binding mechanism to cellulose, our results suggest that this domain may direct the enzyme towards cellulose chain ends increasing the RalCDP-CBM37 activity and binding capacity to insoluble polysaccharides in the same level. Furthermore, a slight increment in optimum temperature and thermal stability was also observed after fusing CBM37 to RalCDP (Figure 4D; Supplementary Figure S2). This stabilization effect was previously noticed by the fusion of CBM3 and CBM9 to GH5 cellulase (Maharjan et al., 2018) and by linking CBM72 to Umcel9A (Duan et al., 2017). In the same way, the removal of the CBM from the thermostable endoglucanase from Phialophora sp. G5 observed a decrease in the enzyme’s thermal stability (Zhao et al., 2012).
As a concluding remark, we proved that RalCDP has activity with a wide range of substrates, from monosaccharides to long-chain polysaccharides. Also, we achieved two RalCDP versions with valuable features. Ral∆N63CDP catalyzes the reaction with higher efficiency in the synthetic direction under low Glc-1P:cellobiose ratios, an advantageous characteristic for the tightly controlled synthesis of short-chain cello-oligosaccharides. Conversely, RalCDP-CBM37 has enhanced activity on polysaccharides, thus enabling the prospect of producing sugars-1P from highly available cellulosic substrates or synthesizing long-chain oligosaccharides.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/OP866768.1/, OP866768; https://www.ncbi.nlm.nih.gov/nuccore/OP866769.1/, OP866769.
Author contributions
AS, SG, and AI conceived and designed research. AS conducted experiments. AS, SG, and AI analyzed data and wrote the manuscript. All authors read and approved the manuscript.
Funding
AS is a doctoral fellow from CONICET. AAI and SAG are investigator members from CONICET. This work was supported by grants from CONICET (PUE 2016-0040 to IAL), ANPCyT (PICT-2019-0349 to SAG, and PICT-2018-00929 and PICT-2020-03326 to AAI).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1176537/full#supplementary-material
References
Adharis, A., Petrovic, D. M., Ozdamar, I., Woortman, A. J., and Loos, K. (2018). Environmentally friendly pathways towards the synthesis of vinyl-based oligocelluloses. Carbohydr. Polym. 193, 196–204. doi:10.1016/j.carbpol.2018.03.098
Ambade, V. N., Sharma, Y., and Somani, B. (1998). Methods for estimation of blood glucose: A comparative evaluation. Med. J. Armed Forces India 54 (2), 131–133. doi:10.1016/S0377-1237(17)30502-6
Ávila, P. F., Silva, M. F., Martins, M., and Goldbeck, R. (2021). Cello-oligosaccharides production from lignocellulosic biomass and their emerging prebiotic applications. World J. Microbiol. Biotechnol. 37 (5), 73–11. doi:10.1007/s11274-021-03041-2
Bianchetti, C. M., Elsen, N. L., Fox, B. G., and Phillips, G. N. (2011). Structure of cellobiose phosphorylase from Clostridium thermocellum in complex with phosphate. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67 (11), 1345–1349. doi:10.1107/S1744309111032660
Billès, E., Coma, V., Peruch, F., and Grelier, S. (2017). Water-soluble cellulose oligomer production by chemical and enzymatic synthesis: A mini-review. Polym. Int. 66 (9), 1227–1236. doi:10.1002/pi.5398
Billès, E., Onwukamike, K. N., Coma, V., Grelier, S., and Peruch, F. (2016). Cellulose oligomers production and separation for the synthesis of new fully bio-based amphiphilic compounds. Carbohydr. Polym. 154, 121–128. doi:10.1016/j.carbpol.2016.07.107
Boraston, A. B., Bolam, D., Gilbert, H., and Davies, G. (2004). Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 382 (3), 769–781. doi:10.1042/BJ20040892
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 (1–2), 248–254. doi:10.1016/0003-2697(76)90527-3
Christopherson, M. R., Dawson, J. A., Stevenson, D. M., Cunningham, A. C., Bramhacharya, S., Weimer, P. J., et al. (2014). Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genomics 15 (1), 1066. doi:10.1186/1471-2164-15-1066
Dai, L., Qu, Y., Huang, J. W., Hu, Y., Hu, H., Li, S., et al. (2021). Enhancing PET hydrolytic enzyme activity by fusion of the cellulose–binding domain of cellobiohydrolase I from Trichoderma reesei. J. Biotechnol. 334, 47–50. doi:10.1016/j.jbiotec.2021.05.006
De Groeve, M. R. M., Remmery, L., Van Hoorebeke, A., Stout, J., Desmet, T., Savvides, S. N., et al. (2010). Construction of cellobiose phosphorylase variants with broadened acceptor specificity towards anomerically substituted glucosides. Biotechnol. Bioeng. 107 (3), 413–420. doi:10.1002/bit.22818
Duan, C. J., Huang, M. Y., Pang, H., Zhao, J., Wu, C. X., and Feng, J. X. (2017). Characterization of a novel theme C glycoside hydrolase family 9 cellulase and its CBM-chimeric enzymes. Appl. Microbiol. Biotechnol. 101 (14), 5723–5737. doi:10.1007/s00253-017-8320-7
Eis, C., and Nidetzky, B. (1999). Characterization of trehalose phosphorylase from Schizophyllum commune. Biochem. J. 341 (2), 385–393. doi:10.1042/bj3410385
Enomoto-Rogers, Y., Kamitakahara, H., Yoshinaga, A., and Takano, T. (2010). Radially oriented cellulose triacetate chains on gold nanoparticles. Cellulose 17 (5), 923–936. doi:10.1007/s10570-010-9437-3
Enomoto-Rogers, Y., Kamitakahara, H., Yoshinaga, A., and Takano, T. (2011). Water-soluble low-molecular-weight cellulose chains radially oriented on gold nanoparticles. Cellulose 18 (4), 929–936. doi:10.1007/s10570-011-9535-x
Ezer, A., Matalon, E., Jindou, S., Borovok, I., Atamna, N., Yu, Z., et al. (2008). Cell surface enzyme attachment is mediated by family 37 carbohydrate-binding modules, unique to Ruminococcus albus. J. Bacteriol. 190 (24), 8220–8222. doi:10.1128/JB.00609-08
Fodor, C., Golkaram, M., Woortman, A. J. J., van Dijken, J., and Loos, K. (2017). Enzymatic approach for the synthesis of biobased aromatic–aliphatic oligo-/polyesters. Polym. Chem. 8 (44), 6795–6805. doi:10.1039/C7PY01559C
Fusari, C., Demonte, A. M., Figueroa, C. M., Aleanzi, M., and Iglesias, A. A. (2006). A colorimetric method for the assay of ADP-glucose pyrophosphorylase. Anal. Biochem. 352 (1), 145–147. doi:10.1016/j.ab.2006.01.024
Gabrielli, V., Munoz-Garcia, J. C., Pergolizzi, G., Andrade, P., Khimyak, Y. Z., Field, R. A., et al. (2021). Molecular recognition of natural and non-natural substrates by cellodextrin phosphorylase from ruminiclostridium thermocellum investigated by NMR spectroscopy. Chem. – A Eur. J. 27 (63), 15688–15698. doi:10.1002/CHEM.202102039
Hamura, K., Saburi, W., Abe, S., Morimoto, N., Taguchi, H., Mori, H., et al. (2012) ‘Enzymatic characteristics of cellobiose phosphorylase from Ruminococcus albus NE1 and kinetic mechanism of unusual substrate inhibition in reverse phosphorolysis’, Biosci. Biotechnol. Biochem., 8451, pp. 812–818. doi:10.1271/bbb.110954
Hidaka, M., Honda, Y., Kitaoka, M., Nirasawa, S., Hayashi, K., Wakagi, T., et al. (2004). Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (α/α)6 barrel fold. Structure 12 (6), 937–947. doi:10.1016/j.str.2004.03.027
Hidaka, M., Kitaoka, M., Hayashi, K., Wakagi, T., Shoun, H., and Fushinobu, S. (2006). Structural dissection of the reaction mechanism of cellobiose phosphorylase. Biochem. J. 398 (1), 37–43. doi:10.1042/BJ20060274
Hong, J., Ye, X., and Zhang, Y.-H. P. (2007). Quantitative determination of cellulose accessibility to cellulase based on adsorption of a nonhydrolytic fusion protein containing CBM and GFP with its applications. Langmuir 23 (25), 12535–12540. doi:10.1021/la7025686
Huber, T., Mussig, J., Curnow, O., Pang, S., Bickerton, S., and Staiger, M. P. (2012). A critical review of all-cellulose composites. J. Mater Sci. 47 (3), 1171–1186. doi:10.1007/s10853-011-5774-3
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kim, Y.-K., Kitaoka, M., Krishnareddy, M., Mori, Y., and Hayashi, K. (2002). Kinetic studies of a recombinant Cellobiose Phosphorylase (CBP) of the Clostridium thermocellumYM4 strain expressed in Escherichia coli. J. Biochem. 132 (2), 197–203. doi:10.1093/oxfordjournals.jbchem.a003210
Kluge, S., Bonhage, B., Viell, J., Granstrom, M., Kindler, A., and Spiess, A. C. (2019). Enzymatic production of cello-oligomers with endoglucanases. Cellulose 26 (7), 4279–4290. doi:10.1007/s10570-019-02390-4
Krishnareddy, M., Kim, Y. K., Kitaoka, M., Mori, Y., and Hayashi, K. (2002). Cellodextrin Phosphorylase from Clostridium thermocellum YM4 strain expressed in Escherichia coli. J. Appl. Glycosci. 49 (1), 1–8. doi:10.5458/jag.49.1
Kuhaudomlarp, S., Stevenson, C. E. M., Lawson, D. M., and Field, R. A. (2019). The structure of a GH149 β-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-β-(1 → 3)-glucose (laminaribiose) phosphorylase. Proteins Struct. Funct. Bioinforma. 87 (10), 885–892. doi:10.1002/prot.25745
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature 227 (5259), 680–685. doi:10.1038/227680a0
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46 (D1), D493–D496. doi:10.1093/nar/gkx922
Letunic, I., Khedkar, S., and Bork, P. (2021). Smart: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49 (D1), D458–D460. doi:10.1093/nar/gkaa937
Luley-Goedl, C., and Nidetzky, B. (2010). Carbohydrate synthesis by disaccharide phosphorylases: Reactions, catalytic mechanisms and application in the glycosciences. Biotechnol. J. 5 (12), 1324–1338. doi:10.1002/biot.201000217
Maharjan, A., Alkotaini, B., and Kim, B. S. (2018). Fusion of carbohydrate binding modules to bifunctional cellulase to enhance binding affinity and cellulolytic activity. Biotechnol. Bioprocess Eng. 23 (1), 79–85. doi:10.1007/s12257-018-0011-4
Martins, M., Dinamarco, T. M., and Goldbeck, R. (2020). Recombinant chimeric enzymes for lignocellulosic biomass hydrolysis. Enzyme Microb. Technol. 140 (80), 109647. doi:10.1016/j.enzmictec.2020.109647
Moon, R. J., Martini, A., Nairn, J., Simonsen, J., and Youngblood, J. (2011). Cellulose nanomaterials review: Structure, properties and nanocomposites. Chem. Soc. Rev. 40, 3941–3994. doi:10.1039/c0cs00108b
Nakai, H., Hachem, M. A., Petersen, B. O., Westphal, Y., Mannerstedt, K., Baumann, M. J., et al. (2010). Efficient chemoenzymatic oligosaccharide synthesis by reverse phosphorolysis using cellobiose phosphorylase and cellodextrin phosphorylase from Clostridium thermocellum. Biochimie 92 (12), 1818–1826. doi:10.1016/j.biochi.2010.07.013
Nakai, H., Kitaoka, M., Svensson, B., and Ohtsubo, K. (2013). Recent development of phosphorylases possessing large potential for oligosaccharide synthesis. Curr. Opin. Chem. Biol. 17 (2), 301–309. doi:10.1016/j.cbpa.2013.01.006
Nakajima, H., Dijkstra, P., and Loos, K. (2017). The recent developments in biobased polymers toward general and engineering applications: Polymers that are upgraded from biodegradable polymers, analogous to petroleum-derived polymers, and newly developed. Polymers 9 (12), 523. doi:10.3390/polym9100523
Nakajima, M., Tanaka, N., Furukawa, N., Nihira, T., Kodutsumi, Y., Takahashi, Y., et al. (2017). Mechanistic insight into the substrate specificity of β-1,2-oligoglucan phosphorylase from Lachnoclostridium phytofermentans. Sci. Rep. 7 (1), 42671. doi:10.1038/srep42671
Nidetzky, B., and Zhong, C. (2021). Phosphorylase-catalyzed bottom-up synthesis of short-chain soluble cello-oligosaccharides and property-tunable cellulosic materials. Biotechnol. Adv. 51, 107633. doi:10.1016/j.biotechadv.2020.107633
O’Neill, E. C., and Field, R. A. (2015). Enzymatic synthesis using glycoside phosphorylases. Carbohydr. Res. 403, 23–37. doi:10.1016/j.carres.2014.06.010
O’Neill, E. C., Pergolizzi, G., Stevenson, C. E., Lawson, D. M., Nepogodiev, S. A., and Field, R. A. (2017). Cellodextrin phosphorylase from Ruminiclostridium thermocellum: X-Ray crystal structure and substrate specificity analysis. Carbohydr. Res. 451, 118–132. doi:10.1016/j.carres.2017.07.005
Puchart, V. (2015). Glycoside phosphorylases: Structure, catalytic properties and biotechnological potential. Biotechnol. Adv. 33 (2), 261–276. doi:10.1016/j.biotechadv.2015.02.002
Ravalason, H., Herpoel-Gimbert, I., Record, E., Bertaud, F., Grisel, S., de Weert, S., et al. (2009). Fusion of a family 1 carbohydrate binding module of Aspergillus niger to the Pycnoporus cinnabarinus laccase for efficient softwood kraft pulp biobleaching. J. Biotechnol. 142 (3–4), 220–226. doi:10.1016/j.jbiotec.2009.04.013
Sambrook, J., and Russell, D. W. (2001). Molecular cloning. New York: A Laboratory Manual Cold Spring Horbor laboratory Press’.
Sawano, T., Saburi, W., Hamura, K., Matsui, H., and Mori, H. (2013). Characterization of Ruminococcus albus cellodextrin phosphorylase and identification of a key phenylalanine residue for acceptor specificity and affinity to the phosphate group. FEBS J. 280 (18), 4463–4473. doi:10.1111/febs.12408
Shoda, S. I., Hiroshi, U., Jun-ichi, K., Shunsaku, K., and Shiro, K. (2016). Enzymes as green catalysts for precision macromolecular synthesis. Chem. Rev. Am. Chem. Soc. 1, 2307–2413. doi:10.1021/acs.chemrev.5b00472
Shoseyov, O., Shani, Z., and Levy, I. (2006). Carbohydrate binding modules: Biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70 (2), 283–295. doi:10.1128/MMBR.00028-05
Storani, A., Guerrero, S. A., and Iglesias, A. A. (2020). On the functionality of the N-terminal domain in xylanase 10A from Ruminococcus albus 8. Enzyme Microb. Technol. 142, 109673. doi:10.1016/j.enzmictec.2020.109673
Sybesma, W., Kort, R., and Lee, Y.-K. (2015). Locally sourced probiotics, the next opportunity for developing countries? Trends Biotechnol. 33 (4), 197–200. doi:10.1016/j.tibtech.2015.01.002
Ubiparip, Z., De Doncker, M., Beerens, K., Franceus, J., and Desmet, T. (2021). β-Glucan phosphorylases in carbohydrate synthesis. Appl. Microbiol. Biotechnol. 105 (10), 4073–4087. doi:10.1007/s00253-021-11320-z
Ubiparip, Z. (2020). Engineering of cellobiose phosphorylase for the defined synthesis of cellotriose. Appl. Microbiol. Biotechnol., 1, 8327–8337. doi:10.1007/s00253-020-10820-8
Wang, J., Niu, J., Sawada, T., Shao, Z., and Serizawa, T. (2017). A bottom-up synthesis of vinyl-cellulose nanosheets and their nanocomposite hydrogels with enhanced strength. Biomacromolecules 18 (12), 4196–4205. doi:10.1021/acs.biomac.7b01224
Wu, Y., Mao, G., Fan, H., Song, A., Zhang, Y. H. P., and Chen, H. (2017). Biochemical properties of GH94 cellodextrin phosphorylase THA_1941 from a thermophilic eubacterium Thermosipho africanus TCF52B with cellobiose phosphorylase activity. Sci. Rep. 7 (1), 4849–4912. doi:10.1038/s41598-017-05289-x
Xiao, R., and Grinstaff, M. W. (2017). Chemical synthesis of polysaccharides and polysaccharide mimetics. Prog. Polym. Sci. 74, 78–116. doi:10.1016/j.progpolymsci.2017.07.009
Xu, Q., Morrison, M., Nelson, K. E., Bayer, E. A., Atamna, N., and Lamed, R. (2004). A novel family of carbohydrate-binding modules identified with Ruminococcus albus proteins. FEBS Lett. 566 (1–3), 11–16. doi:10.1016/j.febslet.2004.04.005
Ye, X., Zhu, Z., Zhang, C., and Zhang, Y. H. P. (2011). Fusion of a family 9 cellulose-binding module improves catalytic potential of Clostridium thermocellum cellodextrin phosphorylase on insoluble cellulose. Appl. Microbiol. Biotechnol. 92 (3), 551–560. doi:10.1007/s00253-011-3346-8
Yeoman, C. J., Fields, C. J., Lepercq, P., Ruiz, P., Forano, E., White, B. A., et al. (2021). In vivo competitions between Fibrobacter succinogenes, Ruminococcus flavefaciens, and Ruminoccus albus in a gnotobiotic sheep model revealed by multi-omic analyses. mBio 12 (2), 035333–e3616. doi:10.1128/mBio.03533-20
Zhang, P., Lynd, L. R., and Kuang, L. R. (2006). A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: Evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7 (2), 644–648. doi:10.1021/BM050799C
Zhang, Y.-H. P., and Lynd, L. R. (2005). Determination of the number-average degree of polymerization of cellodextrins and cellulose with application to enzymatic hydrolysis. Biomacromolecules 6 (3), 1510–1515. doi:10.1021/bm049235j
Zhang, Y.-H. P., and Lynd, L. R. (2004). Toward an aggregated understanding of enzymatic hydrolysis of cellulose: Noncomplexed cellulase systems. Biotechnol. Bioeng. 88 (7), 797–824. doi:10.1002/bit.20282
Zhang, Y. H. P., and Lynd, L. R. (2003). Cellodextrin preparation by mixed-acid hydrolysis and chromatographic separation. Anal. Biochem. 322 (2), 225–232. doi:10.1016/j.ab.2003.07.021
Zhang, Y., Wang, L., Chen, J., and Wu, J. (2013). Enhanced activity toward PET by site-directed mutagenesis of Thermobifida fusca cutinase–CBM fusion protein. Carbohydr. Polym. 97 (1), 124–129. doi:10.1016/J.CARBPOL.2013.04.042
Zhao, J., Shi, P., Huang, H., Li, Z., Yuan, T., Yang, P., et al. (2012). A novel thermoacidophilic and thermostable endo-β-1,4-glucanase from Phialophora sp. G5: Its thermostability influenced by a distinct β-sheet and the carbohydrate-binding module. Appl. Microbiol. Biotechnol. 95 (4), 947–955. doi:10.1007/s00253-011-3807-0
Zhong, C., Luley-Goedl, C., and Nidetzky, B. (2019). Product solubility control in cellooligosaccharide production by coupled cellobiose and cellodextrin phosphorylase. Biotechnol. Bioeng. 116 (9), 2146–2155. doi:10.1002/bit.27008
Zhong, C., and Nidetzky, B. (2020). Three-enzyme phosphorylase cascade for integrated production of short-chain cellodextrins. Biotechnol. J. 15 (3), 1900349. doi:10.1002/biot.201900349
Keywords: cellobiose phosphorylase, cellodextrin phosphorylase, cello-oligosaccharides, CBM, enzymatic catalysis
Citation: Storani A, Guerrero SA and Iglesias AA (2023) Insights to improve the activity of glycosyl phosphorylases from Ruminococcus albus 8 with cello-oligosaccharides. Front. Chem. 11:1176537. doi: 10.3389/fchem.2023.1176537
Received: 28 February 2023; Accepted: 30 March 2023;
Published: 07 April 2023.
Edited by:
Heng Zhang, Guizhou University, ChinaReviewed by:
Yimin Zhu, Penn State Altoona, United StatesJian He, Jishou University, China
Yu-Cai He, Changzhou University, China
Copyright © 2023 Storani, Guerrero and Iglesias. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto A. Iglesias, aWdsZXNpYXNAZmJjYi51bmwuZWR1LmFy
 Alem Storani
Alem Storani Sergio A. Guerrero
Sergio A. Guerrero Alberto A. Iglesias
Alberto A. Iglesias