- 1Institute for Control of Chinese Traditional Medicine and Ethnic Medicine, National Institutes for Food and Drug Control, Beijing, China
- 2Xinjiang Uygur Autonomous Region Drug Inspection and Research Institute NMPA Key Laboratory for Quality Control of Traditional Chinese Medicine (Uyghur) Medicine Urumqi, Urumqi, China
Introduction: Polygoni Multiflori Radix (PMR) is a type of Chinese herbal medicine with rich chemical composition and pharmacological activity used widely in medicine and food. However, in recent years, there have been increasing numbers of negative reports about its hepatotoxicity. Identification of its chemical constituents for quality control and safe use is very important.
Methods: Three solvents of different polarities (water, 70% ethanol, and 95% ethanol solution) were used to extract the compounds from PMR. Extracts were analyzed and characterized by ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-Q-ToF MS/MS) in the negative-ion mode.
Results: 152 compounds were detected and identified: 50 anthraquinones, 33 stilbene derivatives, 21 flavonoids, seven naphthalene compounds, and 41 other compounds. Eight other compounds were reported for the first time in the PMR-related literature, and eight other compounds were potentially new compounds.
Discussion: This study lays a solid foundation for the screening of toxicity and quality-control indicators of PMR.
1 Introduction
In recent years, traditional Chinese medicine (TCM) has attracted increasing attention from researchers due to its diverse medicinal uses, mild efficacy, and few side effects. TCM-related products have been applied efficiently in medical treatment, food, cosmetics, and other fields.
Polygoni Multiflori Radix (He Shou Wu) (PMR) is derived from the dried root tubers of Polygonum multiflorum Thunb., which is a medicinal plant of the Polygonaceae family. It is used widely in medicines, food, and healthcare products. PMR can remove toxicity to eliminate carbuncles, prevent malaria, and moisten the bowel. It can also be used to treat pruritus caused by rubella, body deficiency due to malaria, and bowel dryness due to constipation (State Pharmacopoeia Commission. 2020).
PMR contains stilbene derivatives, anthraquinones, flavonoids, and polyphenols, which have significant effects against aging and tumors, protect nerves, and improve vascular function (Lin et al., 2015; Zhou et al., 2020; Ning et al., 2021; Teka et al., 2021; Wang et al., 2021). However, in recent years, adverse reactions after the clinical use of PMR (related mainly to hepatotoxicity using a normal dose) have focused attention on the safe use and quality control of PMR (Xia et al., 2017; Gong et al., 2019; Yang et al., 2021; Kong et al., 2022). The main compounds thought to cause the hepatotoxicity of PMR are anthraquinones, dianthraones, stilbene glycosides, and tannins (Yu et al., 2017; Kang et al., 2022; Yang et al., 2022).
PMR, as a type of TCM formulation, has multiple components, multiple signaling pathways, and multiple targets. Therefore, before carrying out an in-depth analysis of the toxic or active components of PMR, a comprehensive understanding of its chemical constituents is crucial. Previously, we showed that the order of hepatotoxicity of PMR extracts using different polar solvents in a zebrafish model was 70% ethanol extract >95% ethanol extract > water (Yang et al., 2018). Therefore, to comprehensively analyze and compare the components in different solvents, water, 70% ethanol, and 95% ethanol were used to extract the ingredients in PMR, and then an efficient method of ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-Q-ToF MS/MS) was established to analyze them.
2 Material and methods
2.1 Chemicals and reagents
Formic acid used for LC-MS (LC-MS grade) was purchased from Honeywell (Shanghai, China). Methanol (LC-MS grade) was obtained from MilliporeSigma (Burlington, MA, United States). Ultra-pure water was sourced from a water purification system (Alpha-Q; Millipore, Waltham, MA, United States). A microporous membrane (0.22 μm) was purchased from Dikma Technologies (Foothill Ranch, CA, United States). All the other reagents were analytically pure.
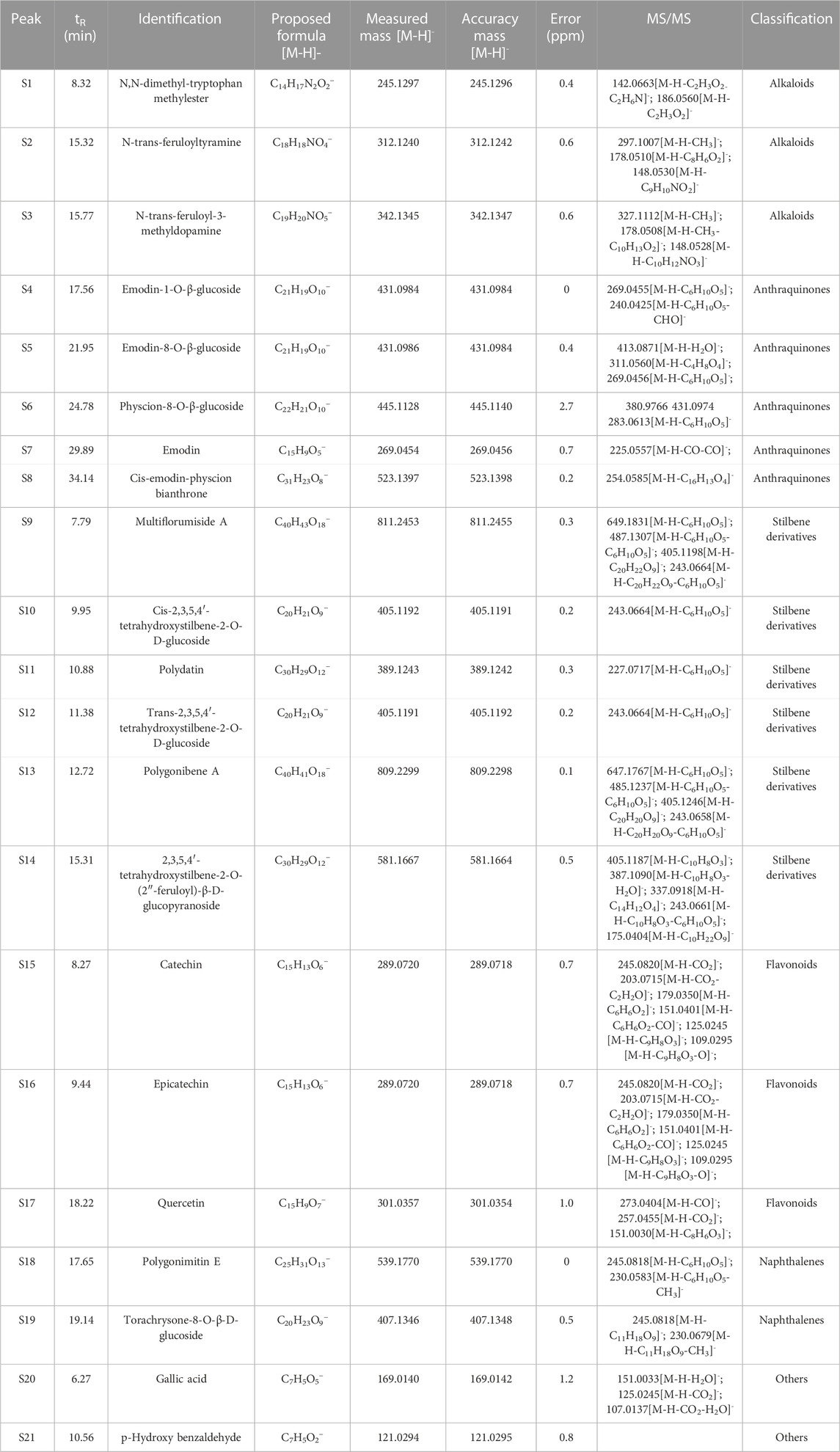
N,N-dimethyl-tryptophan methylester (S1), N-trans-feruloyltyramine (S2), N-trans-feruloyl-3-methyldopamine (S3), cis-emodin-physcion bianthrone (S8), multiflorumiside A (S9), polygonibene A (S13), 2,3,5,4′-tetrahydroxystilbene-2-O-(2″-feruloyl)-β-D-glucopyranoside (S14), polygonimitin E (S18), torachrysone-8-O-β-D-glucoside (S19), and p-hydroxy benzaldehyde (S21) were isolated by our research team. The purity of these compounds was >98%.
Emodin (S7; lot: 110758–201913), polydatin (S11; 111575–201603), trans-2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside (S12; 110844–201915), catechin (S15; 110878–201703), epicatechin (S16; 110877–202005), quercetin (S17; 100818–201610), and gallic acid (S20; 110831–201906) were purchased from China National Institute for Food and Drug Control (Beijing, China). Emodin-1-O-β-D-glucoside (S4; S28HB196292), emodin-8-O-β-glucoside (S5; ST10890126), physcion-8-O-β-D-glucoside (S6; ST23870105), and cis-2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside (S10; M31GB150098) were sourced from Shanghai Yuanye Biotechnology (Shanghai, China). The purity of these compounds was >95%.
2.2 Plant materials and apparatus
The dried roots of Polygonum multiflorum Thunb. were acquired from Deqing County in Zhaoqing City (Guangdong Province, China). We used a UPLC-QTOF-MS/MS system (1290 series) obtained from Agilent Technologies (Santa Clara, CA, United States). A 1/10,000 electronic analytical balance (XPE105) and a 1/1000 electronic analytical balance (Me 303/02) were obtained from Mettler Toledo (Geneva, Switzerland). An ultrasonic cleaner (Kq-300da) was obtained from Kunshan Ultrasonic Instruments (Beijing, China). A Lingsheng multi-functional grinder (C800) was obtained from Yongkang Hongsun Electromechanical (Yongkang, China). The MassHunter™ Workstation (version B.04.00) was obtained from Agilent Technologies.
2.3 Preparation of control samples and standard samples
Before extraction, dried crude medicinal materials were pulverized and screened through a 65-mesh sieve. Then, pulverized PMR (1 g) was weighed accurately in triplicate using an electronic analytical balance. Then, 20 mL of solvent (water, 70% ethanol, or 95% ethanol) was added. The mixture underwent ultrasound treatment for 20 min and was filtered with a polytetrafluoroethylene membrane (0.22 μm). PMR samples extracted with different solvents were obtained.
The 22 standard reference substances mentioned in Section 2.1 were dissolved in methanol separately and prepared into reference solutions of 0.5 mg/mL. According to the category of the compound, S1–S3 reference solutions were mixed in equal proportions to prepare a mixed reference solution containing alkaloids. In the same way, S4–S9 reference solutions, S10–S15 reference solutions, and S16–S22 reference solutions were mixed separately to prepare reference solutions containing anthraquinones, stilbene derivatives, and other compounds, respectively.
2.4 LC-MS conditions
LC was undertaken on a UHPLC system (1290; Agilent Technologies) equipped with a ZORBAX SB-C18 RRHD column (2.1 mm × 100 mm, 1.8 µm; Agilent Technologies) at 35°C. The mobile phase comprised methanol (A) and water containing 0.1% formic acid (B). The gradient program was as follows: 0–5 min, 25%–30% A; 5–15 min, 30%–40% A; 15–18 min, 40%–50% A; 18–23 min, 50%–55% A; 23–30 min, 55%–65% A; 30–35 min, 65%–85% A; 35–40 min, 85% A; 40–50 min, 85%–90% A; 50–60 min, 90% A; 60–61 min, 90%–100% A; 61–70 min, 100% A. The flow rate was 0.25 mL/min. The injection volume was 1 µL.
MS was undertaken on an Accurate-Mass Q-TOF/MS system (6520; Agilent Technologies) with an electrospray ionization source. Mass spectra were acquired in negative mode. The parameters were as follows: mass range = 100–1500 m/z; scanning rate = 3 spectra/s; capillary voltage = 3.5 kV; nozzle voltage = 1 kV; fragmentor voltage = 150 V; cone voltage = 65 V; drying temperature = 350°C; flow rate = 0.3 mL/min. Data were obtained by data-dependent MS/MS (i.e., in each cycle, the top two precursor ions were chosen for fragmentation at a collision energy of 0, 10, and 20 V). Data were recorded and processed using the MassHunter Workstation.
2.5 Data analyses
The MassHunter software was used to extract the base peak chromatogram (BPC) of a compound. MS data provided the precise molecular weight, molecular formula, isotope abundance ratio, and error range for chromatography. MS/MS data provided abundant information on fragment ions.
To evaluate the chemical components in PMR systematically and rapidly, some analytical methods were used. First, the cracking pathways of reference substances were analyzed and summarized by MS, which is useful for structural speculations of compounds of the same type. Second, our in-house MS database and MS/MS databases that include the chemical names, molecular formulae, accurate molecular mass, chemical structures, and fragment ions were established by searching the related literature and compound databases (ScienceDirect (Elsevier), ChemSpider, PubMed, Chinese National Knowledge Infrastructure, SciFinder, and PubChem). Based on the reference substance and our in-house database, compounds were identified with mass errors of <5 ppm for the precursor and one or more products. Feature extraction was used to identify compounds with a weak response signal and confluence.
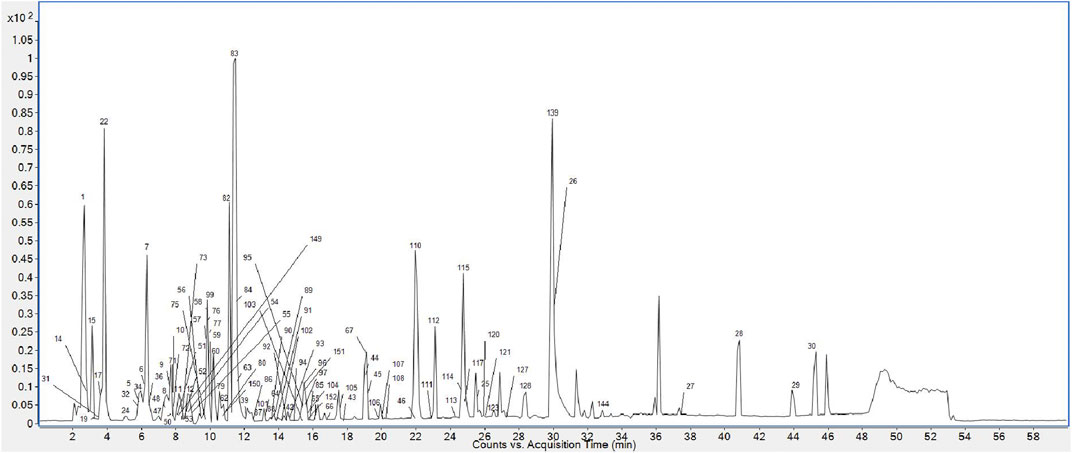
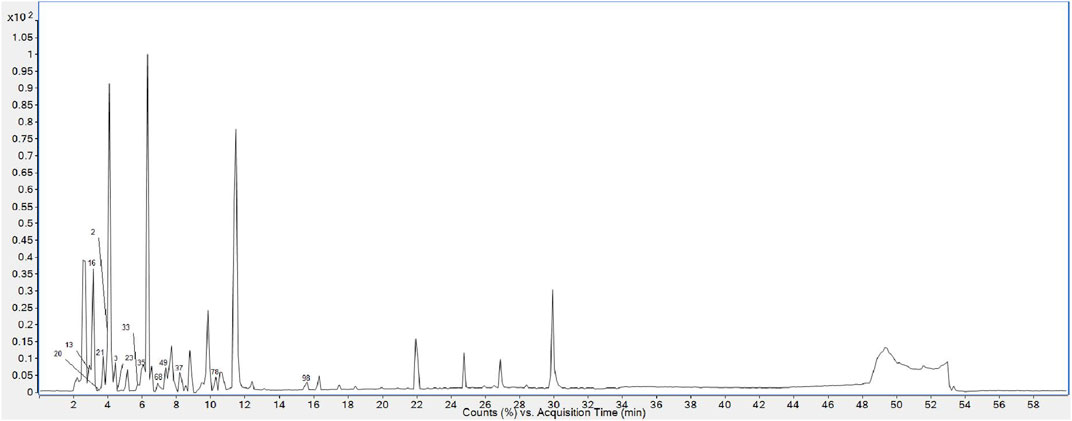
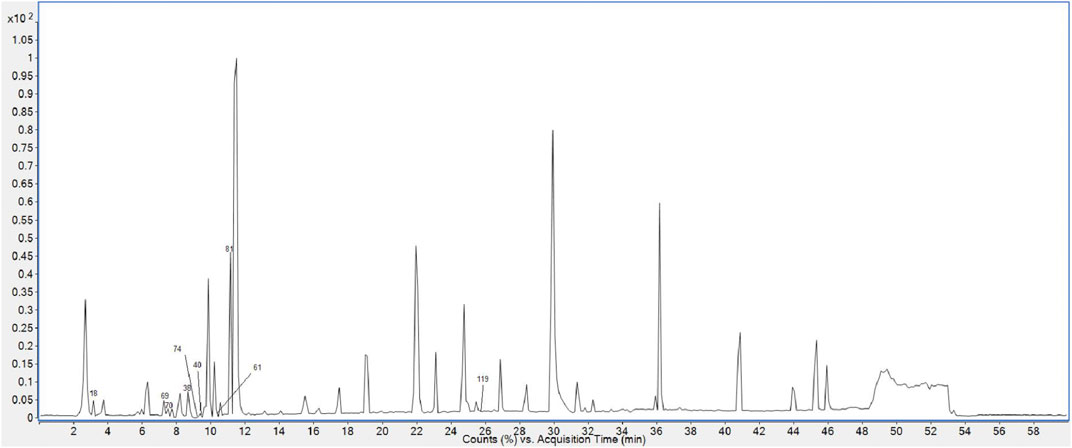
Finally, 158 compounds were identified: 50 anthraquinones, 31 stilbene derivatives, 21 flavonoids, seven naphthalene compounds, and 49 other compounds. The BPCs of different sample solutions are shown in Figures 1–3. Feature-extraction chromatograms and BPCs of mixed reference solutions are revealed in Supplementary Materials. The specific information of reference substances and identified compounds is given in Table 1 and Supplementary Table S1.
3 Results
3.1 Analysis of flavonoids
In a broad sense, flavonoids are formed by two benzene rings connected to each other by three carbon atoms (C6–C3–C6). Flavonoids have antioxidant and free radical-scavenging activity (Aziz et al., 2022; Shen et al., 2022). Depending on the substitution patterns and position of two benzene rings and the oxidation state of the heterocyclic-C3 ring, flavonoids can be classified into seven major subclasses: flavan-3-ols, flavones, flavonols, flavanones, anthocyanins, chalcones, and isoflavonoids (Dretcanu et al., 2022). Here, we also discuss proanthocyanidins as oligomers of (epi) catechin units.
We obtained three reference substances of flavonoids, namely, catechin, epicatechin, and quercetin. Catechin and epicatechin were optical isomers with the structural framework of flavanols. Catechin and epicatechin were eluted at 8.266 min and 9.441 min, respectively. They possessed the same precursor ions at m/z 289.0720 (C15H13O6) and the same main characteristic fragment ions at m/z 151 (C8H7O3) and m/z 125 (C6H5O3), which were produced by RDA (retro Diels–Alder reaction) rearrangement and heterocyclic cracking, along with 138 Da (C7H6O3) and 164 Da (C9H8O3) neutral loss (March and Brodbelt, 2008). So, fragment ions at m/z 289.0720 (C15H13O6), m/z 151 (C8H7O3), and m/z 125 (C6H5O3) could be taken as diagnostic ions of flavanols. Differently, the quercetin reference substance had a flavonol skeleton, which has an additional carbon–carbon double bond than the flavanones. It was eluted at 18.216 min, and its [M-H]− was at m/z 301.0357 (C15H9O7). In addition to forming the same daughter ion at m/z 151 (C8H7O3) as (epi) catechin, quercetin reference substance also produced other characteristic fragment ions at m/z 273 (C14H9O6) and m/z 257 (C14H9O5) by losing CO (28 Da) and CO2 (44 Da). The typical cracking pathway of flavonoids also involved the loss of CH3 (15Da) or H2O (18 Da). For other flavonoids, such as flavonoid glycosides or galloyl-flavonoids, fragmentation was characterized by the loss of glucose or galloyl along with the production of many skeleton fragment ions (March and Brodbelt, 2008; Vukics and Guttman, 2010; Aziz et al., 2022).
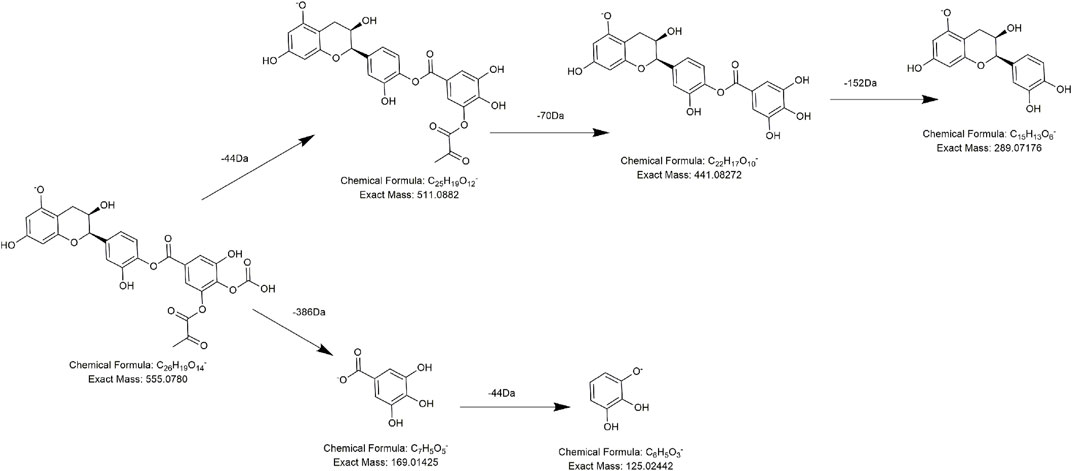
According to the retention time and MS/MS information, compounds FL6 and FL10 were identical to the catechin standard substance and epicatechin standard substance, respectively. Compounds FL14, FL15, and FL16 had precursor ions at m/z 441.0829 (C22H17O10), m/z 555.0756 (C24H19O14), and m/z 693.1825 (C35H33O15), respectively, and were presumed to be catechin derivatives because they all produced fragment ions at m/z 289 (C15H13O6) and m/z 125 (C6H5O3). Taking compound FL14 (which was eluted at 10.3 min) as an example, with a precursor ion at m/z 441.0829, it not only lost 152 Da (C8H8O3 ) to produce a fragment ion at m/z 289 (C15H13O6) but also possessed a daughter ion at m/z 169 (C7H5O5), which is a typical fragment of a galloyl substituent. Therefore, we presumed that FL14 was formed from the combination of (epi) catechin and galloyl substituents, that is, catechin galloyl ester (Wang et al., 2021). Similarly, except for the typical diagnostic ions of flavonoids (i.e., at m/z 289, m/z 125, and m/z 169), compound FL15 also generated the fragment ion at m/z 511.0865 (C23H19O12) by losing 44 Da (CO2), which suggested a carboxyl substituent. Also, in its subsequent collision, a neutral loss of 70 Da emerged to generate the fragment ion at m/z 441.0834 (C20H17O10), which implied a malonyl (C3H2O2) group. We preliminarily speculated that FL15 consisted of an (epi) catechin nucleus and a molecule of carboxyl and a molecule of malonyl substitution. Tentative speculation of its structure and the lytic pathway is stated in Figure 4, which was conjectured to be a potential new compound. Compound FL16 exhibited [M-H]− at m/z 693.1824 (C35H33O15) with a retention time of 10.81 min, and its primary fragment ions were at m/z 289 (C15H13O6) and m/z 405 (C29H23O10). Hence, it was identified as a polygonflavanol A (C35H34O15): a combination of a tetrahydroxystilbene glycoside (C29H24O10) and (epi) catechin (C15H14O6) formed by opening the ene bond and forming a ring. (Huang et al., 2018). As for FL1, its excimer ion (m/z 305) was 16 Da higher than that of (epi) catechin, and one of the typical fragment ions m/z 125 was also generated, so it is reasonable to deem that it has one more hydroxyl group than (epi) catechin. According to the literature, compound FL1 was speculated to be gallocatechin A (Wang et al., 2017).
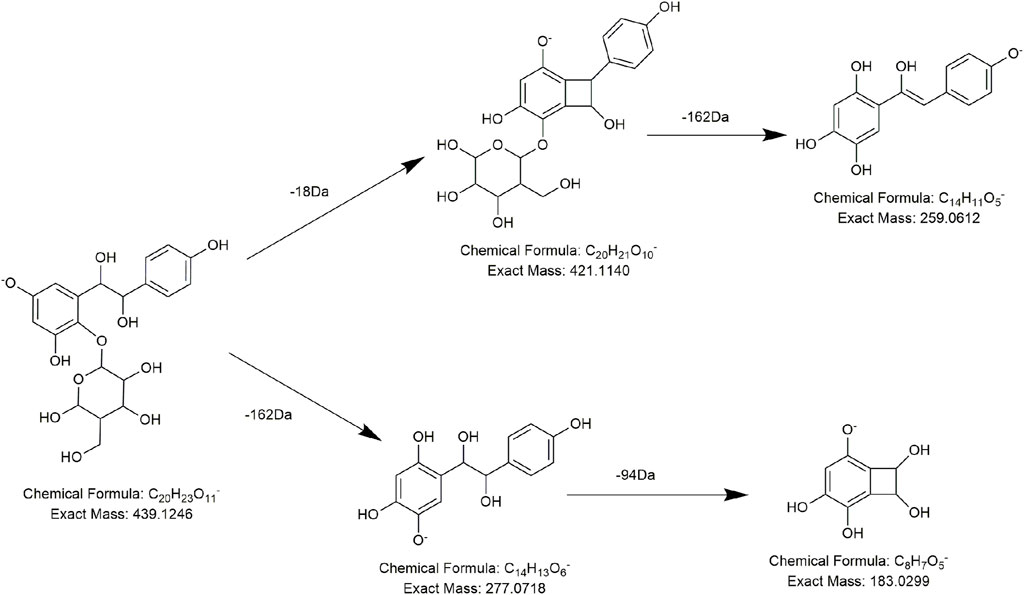
In a similar manner, compound FL18 was assumed to be an isomer of quercetin upon comparison with the eluting time and MS/MS information of quercetin’s standard substance. Meanwhile, several compounds share similar nuclear factors with quercetin. Take compound FL11 as an example, after losing a molecule of hexose substitution (162 Da, C6H10O5), a unit of H2O (18 Da) was lost immediately, and FL11 produced the fragment ions at m/z 287 and m/z 269. The two fragment ions were 14 Da and 32 Da lower than the typical diagnostic ions m/z 301 of quercetin, respectively. So, we believe that FL11 has one less hydroxide radical (16 Da) and one unsaturation (−2 Da) more than quercetin, so it most likely processes flavonol structures with one less phenolic hydroxide radical than quercetin. Because of the aforementioned reasons, FL11 was identified as dihydrokaempferol-3-glucoside (Huang et al., 2018).
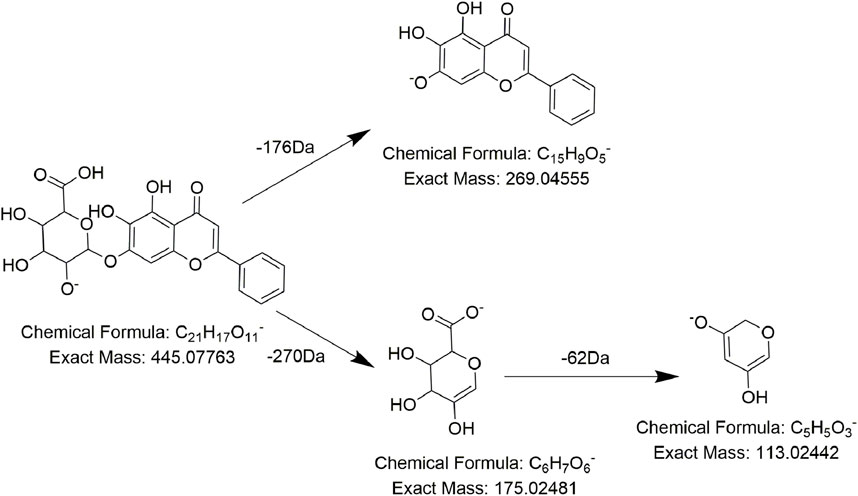
Analogously, compound FL20 displayed a precursor ion at m/z 444.0766 and presented typical fragment ions at m/z 269 after a 177 Da (C6H8O6) neutral loss. Combining with the daughter ions at m/z 113 and m/z 175, we believed that there is a glucuronic-group substitution (C6H8O6). So, m/z 269 was reasonably inferred to be the nucleus of compound FL20, which was the flavone nucleus. According to the literature (Wu et al., 2005), it was speculated to be baicalin (Figure 5), which was reported for the first time in PMR-related literature. Compound FL19 was eluted at 16.1 min with the precursor ion at m/z 447.40493, and a comparison with compound FL20 revealed its molecular weight to be 2 Da lower. The same as compound FL20, after losing 177 Da (C6H8O6), it generated the fragment ion at m/z 271 (269 Da +2 Da) and had fragment ions at m/z 175 and m/z 113. According to the literature, it was considered to be naringenin-7-O-glucuronide (Rui et al., 2020).
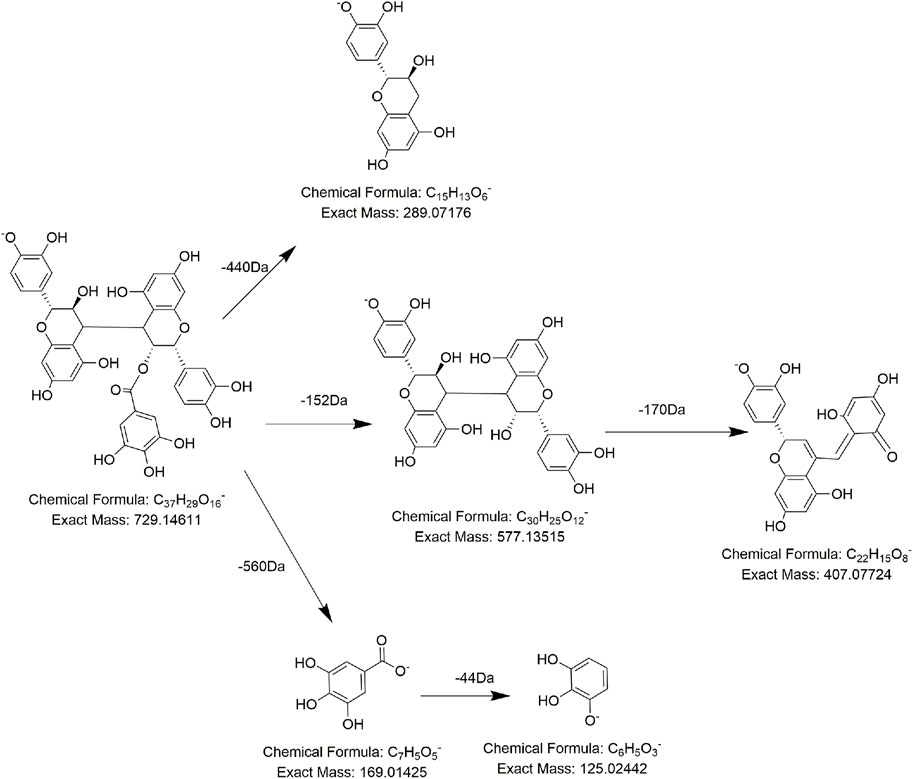
As for procyanidins, which are made up of two units of flavanol, they tend to break the C–C bond, losing one unit of nuclear flavanols. During this procedure, 288 Da (C15H12O6) neutral loss and characteristic ions at m/z 289 (C15H13O6) are generated. Compounds FL2, FL3, FL5, FL7, and FL8 not only presented the characteristic daughter ion at m/z 125 but also exhibited a typical neutral loss of 288 Da (C15H12O6). So, they were tentatively regarded as procyanidin derivatives. For example, compound FL5 was detected at 8.11 min, with the precursor ion at m/z 729.1454 (C37H29O16). Under the collision energy in MS/MS, compound FL5 lost 152 Da (C8H8O3) and 288 Da (C15H12O6) to produce fragment ions at m/z 577 (C30H25O12) and m/z 289 (C15H13O6) successively, which could be regarded as a decrease of one unit of galloyl and (epi) catechin structure. Therefore, according to the literature, compound FL5 was speculated to be 3-O-galloyl-procyanidin B (Figure 6) (Qiu et al., 2013). Analogously, compound FL2 and compound FL9 displayed [M-H]− at m/z 577.1349 (C30H25O12) and m/z 881.1571 (C44H33O20), of which one was 152-Da (C8H8O3) lower and one was 152-Da (C8H8O3) higher than the precursor ion of compound FL5. In addition, these three compounds shared the same daughter ions at m/z 729 (C37H29O16), m/z 577 (C30H25O12), and m/z 169 (C7H5O5). Hence, it was reasonable to assume that the three compounds differed by only one unit of a galloyl substituent (152 Da, C8H8O3) and that compound FL2 and compound FL9 might be procyanidin B and di-galloyl-procyanidin B, respectively (Qiu et al., 2013; Zhao et al., 2021). Compound FL3 was predicted to be the catechin trimer procyanidin C1 by the same method (Zhao et al., 2021). The MS behavior of FL7 and FL8 was identical to that of FL3 and FL5, except for the retention time, so we presumed them to be isomers of 3-O-galloyl-procyanidin B and procyanidin C1, respectively(Zhao et al., 2021).
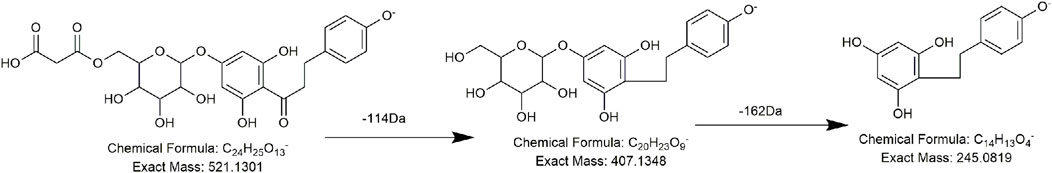
The precursor ions of compounds FL4 and FL21 were at m/z 519.1121 (C24H23O13) and m/z 521.1300 (C24H25O13), respectively, and in both of their MS/MS spectra, 114 Da (C4H2O4) and 162 Da (C6H10O5) were lost successively. So, we presumed that FL4 and FL21 process the same substitution, and during the MS/MS procedure, one unit of the malonyl group (86 Da, C3H2O3) plus one unit of CO (28 Da) and one unit of hexose (162 Da, C6H10O5) were lost in sequence. The difference in molecular weight (2 Da) might be due to the unsaturation of the parent nucleus. The proposed structure and pathway of compounds FL4 and FL21 are shown in Figures 7, 8, respectively. Compounds FL12, FL13, and FL17 had the same molecular weight and fragment ions as compound FL4 but not an identical retention time, so they were presumed to be conformational isomers of compound FL4. Compounds FL4, FL12, FL13, FL17, and FL21 were reported for the first time in the PMR-related literature.
3.2 Analysis of stilbene derivatives
Stilbene derivatives are typical components in PMR. They have antibacterial, anti-inflammatory, anticancer, antioxidant, neuroprotective, and immunomodulatory effects (Lin et al., 2015; De Filippis et al., 2017; Lee et al., 2019). Stilbene derivatives in PMR that have been isolated so far include tetrahydroxystilbene and its glycoside, substituted tetrahydroxystilbene glycoside, a dimer of tetrahydroxystilbene and its glycoside, and a small amount of trihydroxy stilbene glycoside derivatives (Zhang and Cui, 2016; Yuan et al., 2017; Li et al., 2018; Nguyen et al., 2020).
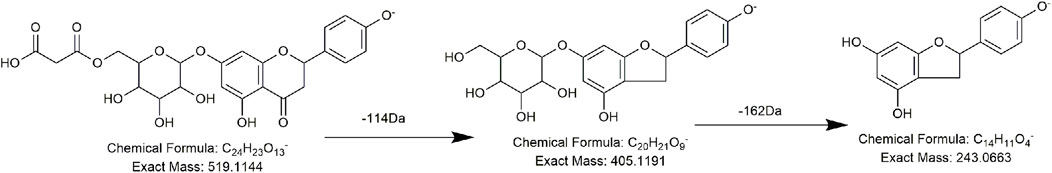
Reference substances cis-2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside (cis-THSG) and trans-2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside (trans-THSG) were isomers with a retention time of 9.98 min and 11.5 min, respectively. Except for the retention time, their MS behavior was essentially identical with the precursor ion at m/z 405.1191 (C20H21O9) and the primary fragment ion at m/z 243 (C14H11O4), which was obtained by losing one unit of hexose (162 Da, C14H11O4) (Wang et al., 2021). Among substituted stilbene derivatives, the reference substance 2,3,5,4′-tetrahydroxystilbene-2-O-(2″-feruloyl)-β-D-glucopyranoside aided further identification of homologous series. Under the action of collision energy, 2,3,5,4′-tetrahydroxystilbene-2-O-(2″-feruloyl)-β-D-glucopyranoside yielded fragments at m/z 405.1191 (C20H21O9) and m/z 243 (C14H11O4), which corresponded with a tetrahydroxystilbene-glucoside nucleus. Other debris, including those at m/z 193.0506 (C10H9O4) and m/z 175.0401 (C10H7O3), corresponded to the feruloyl group. Reference substances multiflorumiside A and polygonibene A showed two different patterns of tetrahydroxystilbene dimers; they are both polymerized by forming a ring, but the ring atoms are in different positions. Multiflorumiside A was a dimer of THSG with a retention time of 7.86 min and a precursor ion at m/z 811.2455 (C40H43O18). After appropriate interaction with collision energy, the polymerization C–C bond was broken, a unit of THSG was removed (406 Da, C20H22O9), and a daughter ion at m/z 405 (C20H21O9) was generated, followed by the loss of one unit of hexose to produce another characteristic fragment ion at m/z 243 (C14H11O4). Reference substance polygonibene A was also a dimer of THSG but formed by the C–O bond and C–C bond. It was detected at 12.49 min and possessed the parent ion at m/z 809.2298 (C40H41O18). Apart from having the same cleavage pathway as multiflorumiside A, the glycosidic bond of polygonibene A was easier to break and produced fragment ions at m/z 647 (C34H31O13) and m/z 485 (C28H21O8), along with the shedding of two units of hexose (162 Da, C14H11O4). Different from all of the previously mentioned references, the reference substance polydatin was a trihydroxy stilbene glycoside and was composed of one unit of resveratrol and one unit of glucose. Hence, the representative rupture was identical to that of tetrahydroxystilbene-glucoside: loss of a glycosidic bond (162 Da, C6H10O5) to produce a fragment ion at m/z 227.0713 (C14H11O3).
Compared with the previously stated standard substance, compounds D12 and D18 were identified as cis-2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside and trans-2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside, respectively, by comparing molecular weight, retention time, and MS information. Similarly, compounds D32 and D33 were presumed to be tetrahydroxystilbene-O-(feruloyl)-hexose and its isomer. Compounds D14, D22, D25, and D27 were identified as being isomers of polygonibene A. Compounds D9, D11, and D19 were assumed to be isomers of multiflorumiside A. Compounds D16 and D26 were identified as polydatin and resveratrol, respectively.
According to the MS fragments of compound D1, we found its major fragment ion at m/z 259.0614, which was 16 Da higher than the characteristic fragment ion of stilbene derivatives (243 Da, C14H11O4). Combined with the consecutive neutral loss of 18 Da (H2O) and 162 Da (C6H10O5) from m/z 439.1246 to m/z 259.0614, we speculated that compound D1 might be an oxidized derivative of tetrahydroxystilbene glucoside, which lost one molecule of water and one molecule of hexose in succession in the collision pool. Moreover, there was another lytic pathway, in which a unit of hexose and a unit of phenol were consecutively lost. In addition, specific elucidation of the two proposed cleavage methods is shown in Figure 9.
Similar to polygonibene A, compound D3, with a retention time of 6.87 min and precursor ion at m/z 827.2396 (C40H43O19), also generated the characteristic ion at m/z 485 (C28H21O8) under the effect of collision energy. Its molecular weight of 486 Da (C28H22O8) can be viewed as the addition of two molecules of tetrahydroxystilbene (243 Da, C14H11O4), and MS fragment information displayed that it lost two molecules of glucose in succession to generate m/z 485. Hence, in accordance with the literature, compound D3 was preliminarily identified as the dimer of THSG formed in different ways: polygonumside C/D, which polymerizes by one unit of the C–C bond (Wang et al., 2017). Compounds D4, D5, D6, and D8, which had the same precursor ions and fragment ions but were eluted at different times, were referred to as isomers of polygonumside C/D.
Compounds D10, D17, D20, D21, D23, D24, D28, D29, D30, D31, D32, and D33 produced the same typical fragment ions of THSG at m/z 405 (C20H21O9) by different types of neutral loss in MS/MS. Hence, it was rational to speculate that they were THSG derivatives formed by combining THSG with different substituents. For example, compounds D10, D17, and D20 shared the same [M-H]− at m/z 557.1299 (C27H25O13), and all lost 152 Da (C7H4O4) to produce the characteristic ion at m/z 405 (C20H21O9), so they were assumed to be THSG replaced by a galloyl group (152 Da, C7H4O4) (Xu et al., 2006; Qiu et al., 2013). For the same reason, compounds D21 and D23 with [M-H]− at m/z 447.1297 (C22H3O10) were presumed to be THSG substituted by an acetyl group (42 Da, C2H2O) as its major neutral loss of 42 Da coincided perfectly with the molecular weight of the acetyl group (42 Da, C2H2O) (Liang et al., 2018); Compound D24 and compound D29, with precursor ions at m/z 567.1506 (C26H31O14) and m/z 491.1191 (C23H23O12), respectively, were identified as THSG substituted by a caffeyl group (162 Da, C6H10O5) and malonyl group (162 Da, C6H10O5) for the typical 162 Da and 86 Da neutral loss (Qiu et al., 2013; Liang et al., 2018). Compound D31, with a precursor ion at m/z 551.1559 (C29H27O11), was identified tentatively as THSG substituted by a coumaroyl group (146 Da, C9H6O2), according to the 146 Da neutral loss (Wang et al., 2017; Yang et al., 2019); specific information is shown in Supplementary Table S1.
3.3 Analysis of anthraquinones
Anthraquinones are the most important and controversial ingredients in PMR. They show high-profile hepatotoxicity and widely reported pharmacological (anti-inflammatory, anti-cancer, antidepressant, and resistance to microorganisms) activities (Lin et al., 2015; Yang et al., 2018; Campora et al., 2021; Yang et al., 2022). Anthraquinone compounds reported in PMR so far mainly include anthraquinone derivatives of different degrees of reduction and dimers, such as anthraquinone, anthrone, and dianthrone (Li and Jiang, 2018). On the basis of whether they combine with sugars, anthraquinones in PMR can also be divided into conjugated anthraquinones and free anthraquinones. In addition, in view of its nucleus, there is an emodin nucleus and a physcion nucleus in anthraquinones.
The peak of the emodin reference substance was detected at 29.889 min, and [M-H]− was at m/z 269.0454 (C15H9O5). After crazing in the collision pool, a unit of CO (28 Da) was lost, and the characteristic fragment ion at m/z 241 (C14H9O4) was generated. With an increase in collision energy, the debris also lost 16 Da (O) to produce another characteristic fragment ion at m/z 225.0560 (C14H9O3). So, we believed that the typical fragments generated by anthraquinone compounds with emodin as the parent nucleus in MS were at m/z 269 (C15H9O5) and m/z 241 (C14H9O4) or m/z 240 (C14H8O4, radical ions) and m/z 225 (C14H9O3) (Qiu et al., 2013). For example, the standard substances emodin-1-O-β-glucoside and emodin-8-O-glucoside were the isomers formed by combining glucose on the first hydroxy group and eighth hydroxy group of the emodin nucleus, which were eluted at 17.418 min and 21.949 min, respectively. Their precursor ions at m/z 431.0984 (C21H19O10) produced characteristic fragment ions at m/z 269 (C15H9O5) by breaking the glycosidic bond and losing one glucose unit (162 Da, C6H10O5) under the action of collision energy.
Another anthraquinone reference substance, physcion-8-O-glucopyranoside, was observed at 24.78 min upon MS, and its primary ion was at m/z 445.1128 (C22H21O10). Similar to emodin-1-O-β-glucoside, glycosidic bonds were broken, along with the loss of one unit of glucoside (162 Da, C6H10O5) and the generation of the characteristic fragment ion at m/z 283 (C14H9O4), matching with the excimer ion of physcion.
In addition, the reference substance cis-emodin–physcion dianthrone is a typical example to explain the cracking tendency of dianthrone compounds. It comprises a parent nucleus of emodin and physcion, with a retention time of 9.952 min and [M-H]− at m/z 523.1397 (C31H23O8). After experiencing a certain amount of collision energy, the C10–C10’ bond connecting the two parent nuclei broke, and the characteristic fragment at m/z 254 (C14H9O4) was generated. In accordance with the literature, we assumed that dianthrones, such as emodin–physcion bianthrone, emodin–physcion bianthrone, and physcion–physcion bianthrone, tended to suffer breakage of the glycosidic bond and the C10–C10’ bond under the impact of collision energy (Yang et al., 2018).
By comparing the retention time and MS behavior with the reference substances, compounds A7, A12, A17, A41, and A47 were determined to be emodin-1-O-glucoside, emodin-8-O-β-glucoside, physcion-8-O-glucopyranoside, emodin, and cis-emodin–physcion bianthrone, respectively.
According to the rules summarized by the previously stated reference substances, compounds A6, A10, A13, A14, A15, A23, and A30 were speculated to be anthraquinones with a framework of emodin based on the characteristic fragment ion at m/z 269 (C15H9O5) or m/z 225 (C14H9O3). Taking compound A14 as an example, its [M-H]− ion was 517.0988 (C24H21O13), and it possessed the main fragment ions at m/z 473 (C23H21O11), m/z 431 (C21H19O10), and m/z 269 (C15H9O5), which were presumed to be generated after losing a molecule of CO2 (44 Da), a malonyl group (86 Da, C3H2O3), and one glucose unit (162 Da, C6H10O5) successively. The neutral loss of glucose (162 Da) occurred mainly after the shedding of a malonyl group (86 Da), so we speculated that the malonyl group was connected to glucose, and, by comparison with the literature, compound A14 was identified as emodin-O-(malonyl)-glucopyranoside (Liang et al., 2018). In a similar way, first, compounds A10 and A13 were preliminarily speculated as emodin derivatives according to the characteristic fragment ion at m/z 269 (C15H9O5). On the other hand, they lost 44 Da and 42 Da neutral segments separately to produce diagnostic ions at m/z 269, by which we deduced that there was a carboxyl group and an acetyl group, respectively. Ultimately, compounds A10 and A13 were inferred as acetyl-emodin-glucopyranoside and endocrocin, respectively. The primary and secondary fragment ions of compound A15 and compound A10 were identical, except for different retention times, so we speculated them to be isomers of acetyl-emodin-glucopyranoside. As for compound A6, it presented a relatively rare neutral loss (80 Da, SO3) and the typical diagnostic ion of emodin derivatives (m/z 269). By comparing with the literature, we identified it as emodin-8-O-β-hexose-sulfate (Wang et al., 2017).
Compared to the aforementioned compounds, compounds A9, A19, A23, A29, and A30 had a comparatively special way of cracking. As for compounds A29 and A30, they both generated the main fragment ion at m/z 283, one of the diagnostic ions of physcion. Moreover, they were both more likely to lose 28 Da (CO) in the collision pool, which was also consistent with anthraquinones. By searching relevant literature, they were tentatively taken for emodin methyl ester and acetyl-emodin (Wang et al., 2017; Huang et al., 2018; Wang et al., 2021). In the same way, compound A23 was speculated to be an isomer of acetyl emodin. Compared with compound A29, we could find that the molecular weight of compound A9 was 15 Da higher, but the neutral loss (28 Da and 15 Da) was the same as that of compound A29. So, we presumed that compound A9 was composed of emodin methyl ester plus methylene and was mostly like questinol (Qiu et al., 2013). Similarly, the molecular weight of compound A19 was 2 Da higher than that of compound A29 and lost 28 Da and 16 Da in succession in the collision pool; we deduced that it could be an emodin methyl ester analog. We preliminarily speculated it as citreorosein (Huang et al., 2018).
Other atypical anthraquinone derivatives were also present in PMR. Compounds A1 and A3 were anthranones and produced a fragment ion at m/z 255 (C14H10O4) upon MS/MS. The primary ion of compound A1 at m/z 479.1189 (C22H23O12) shattered into fragment ions at m/z 299 (C16H11O6) and m/z 255 (C14H10O4) by losing 180 Da (glucose and H2O) and 44 Da (CO2). As a result, compound A1 was presumed to be a compound ion of emodin acid-hexose (Qiu et al., 2013). Compound A3 had [M-H]− at m/z 417.1195 (C21H21O9), which produced a characteristic fragment ion at m/z 255 (C14H10O4) by losing 162 Da (hexose) and was speculated to be cassialoin (Luo et al., 2017). The [M-H]− of compound A5 was at m/z 447.0933 (C14H8O5), and a fragment ion at m/z 285 (C15H9O6) was generated after 162 Da (hexose) had been lost in the collision pool, so we speculated it to be citreorosein-8-O-glucopyranoside (Wang et al., 2015).
In view of the relatively higher molecular weight, successive breakage of glycosidic bonds and characteristic fragment ions at m/z 254 and m/z 269, compounds A2, A8, A11, A16, A18, A20, A22 A24–A28, A31–A40, A42–A45, and A48–A50 were speculated to be dianthranones (Yang et al., 2019), and information on specific compounds is provided in Supplementary Table S1.
3.4 Analysis of naphthalene compounds
Naphthalene compounds are important polyphenols in PMR. So far, three types of naphthalene compounds have been isolated from PMR: torachrysones (e.g., torachrysone-8-O-β-D-glucopyranoside), hydroxymusizins (e.g., 6-hydroxy-8-o-glucoside), and naphthoquinones (e.g., 2-methoxy-6-acetyl-7-methyljuglone) (Reddy. 2016; Luo et al., 2017; Yang et al., 2020). Different mother nucleus structures tend to generate different characteristic fragment ions, which guide the structural analysis of compounds.
The standard substances torachrysone-8-O-β-D-glucopyranoside and polygonimitin E were formed by combining one or two units of hexose, respectively, with a torachrysone skeleton. In LC-MS, torachrysone-8-O-β-D-glucopyranoside and polygonimitin E, with [M-H]− at m/z 407.1346 (C20H23O9) and m/z 539.1770 (C25H31O13), were eluted successively at 19.2 min and 17.7 min, respectively. Under the effect of collision energy, they all broke the glycosidic bond and lost one or two units of hexose (162 Da, C6H10O5), resulting in the characteristic fragment ion at m/z 245 (C14H13O4). Then, the methyl radical was lost in rapid sequence to produce another characteristic ion at m/z 230 (C13H10O4).
Six naphthalene-based components were resolved, among which compounds N4 and N6 were determined to be polygonimitin E and torachrysone-8-O-glucoside, respectively, by comparing the excimer ions, fragment ions, and retention time. Compound N5 was eluted at 19.1 min, with [M-H]− at m/z 245.0821 (C14H13O4), which was 162 Da (C6H10O5) lower than torachrysone-8-O-β-glucoside. In addition, combining with the characteristic fragment ion at m/z 230 (C13H10O4), compound N5 was speculated to be torachrysone. Similarly, the molecular weight of compound N7 was 42-Da (C2H2O) greater than that of the reference substance torachrysone-8-O-β-glucoside. Moreover, it was easy to lose 42 Da (C2H2O) to generate a fragment ion at m/z 405 (C20H21O9) followed by m/z 230 in MS/MS. The 42 Da (C2H2O) could be regarded as acetyl substitution (C2H2O); therefore, compound N7 was speculated to be torachrysone-8-O-(6′-O-acetyl)-D-glucopyranoside (Wang et al., 2017).
We found that hydroxymusizins and torachrysones differ only by one unit of methyl substitution (15 Da, CH3) and may have similar cleavage pathways and fragment ions. Taking compounds N1 and N3 as examples, the molecular weight of compounds N3 and N1 was 14 Da (CH2) lower than that of the reference substance torachrysone-8-O-glucoside, and the fragment ion at m/z 231 (C13H11O4) was generated after losing 162 Da (C6H10O5) in MS/MS. Therefore, we assumed that compounds N1 and N3 were hydroxymusizin-O-glucopyranoside and their isomers (Jin et al., 2007). Similarly, compound N2 was presumed to be hydroxymusizin-o-glucose-o-xylose.
3.5 Analysis of other compounds
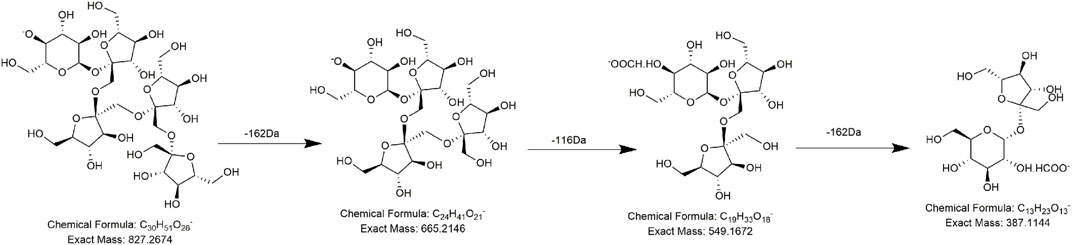
Alkaloids, fatty acids, phospholipids, amino acids, and sugars have also been found in PMR (Lin et al., 2015; Yang et al., 2020; Yang et al., 2021). We identified several compounds for the first time in PMR: three amino acids, two saccharides, and two fatty acids. Taking compound AM7 as an example, it generated the main fragment ion at m/z 257.1143 after losing 162 Da (C6H10O5) upon MS. The main fragment ion at m/z 257.1143 and its additional debris was consistent with the MS behavior of the identified compound gamma-L-glutamyl-L-pipecolic-acid reported in the literature (Huang et al., 2022). Hence, AM7 was thought to be gamma-L-glutamyl-L-pipecolic-acid glucopyranoside (Figure 10). Similarly, two saccharides, 1F-fructofuranosylnystose (G4, Figure 11) and maltose (G5), were deduced by the continuous loss of hexose molecules and the previously reported compounds raffinose and cephulac (Wang et al., 2017). Compound AM9 was tentatively identified as 1-piperidinepentanoic acid, α-amino-δ-oxo-, (S)- by the TCM library of Agilent Technology, and the proposed cracking pathway is shown in Figure 12.
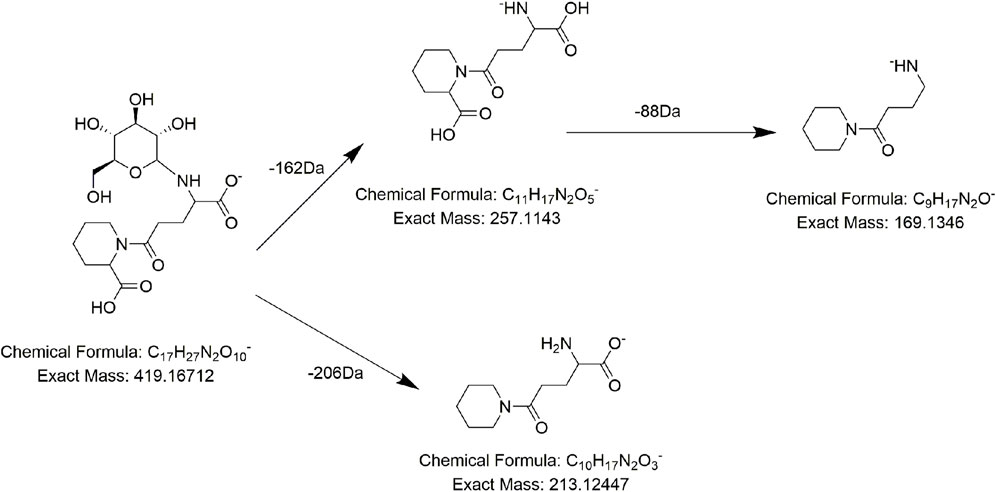
FIGURE 10. Proposed fragmentation pathways of AM7 (gamma-L-glutamyl-L-pipecolic-acid glucopyranoside).
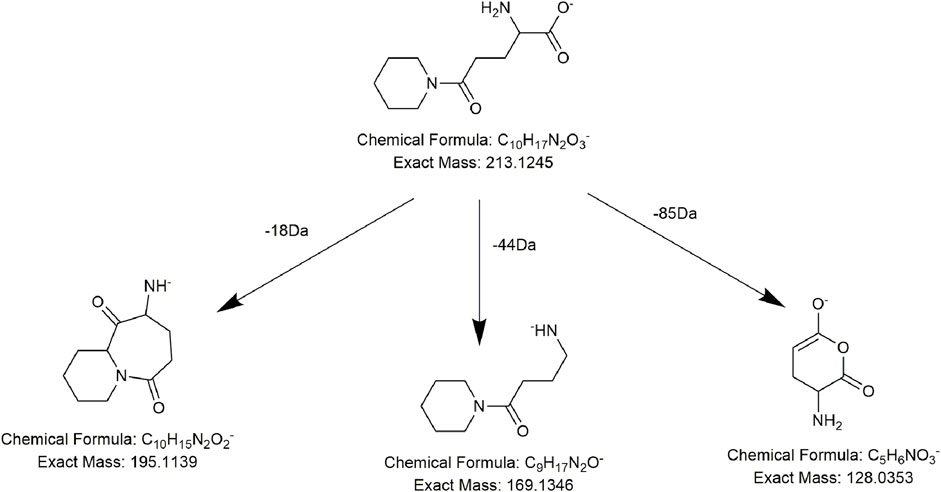
FIGURE 12. Proposed fragmentation pathways of AM9 (1-piperidinepentanoic acid, α-amino-δ-oxo-, (S)-).
4 Discussion
In recent years, discussion of the hepatotoxicity of PMR has increased, and our research team has also explored the hepatotoxic components of PMR (Yang et al., 2018; Li et al., 2020; Gao et al., 2022; Song et al., 2022). We showed that solutions of PMR extracted with different solvents showed different percentages of hepatotoxicity, and the difference in toxicity is mostly caused by chemical differences in each solvent (Yang et al., 2018). A comprehensive identification and analysis of the chemical constituents of PMR are of far-reaching significance to clarify its material basis and toxic mechanism.
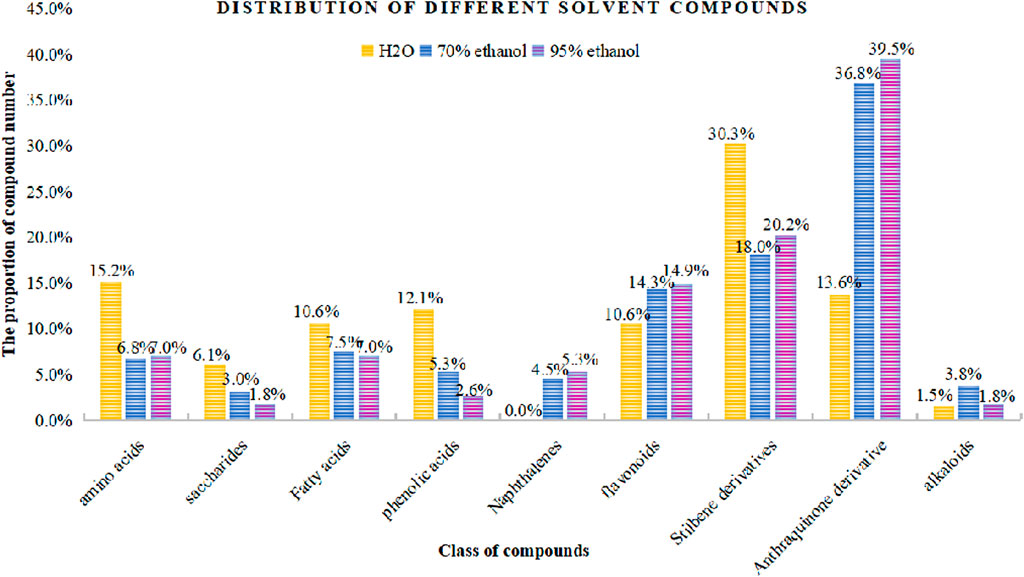
In the present study, a UPLC-Q-ToF-MS/MS method was established to systematically characterize and compare the components in three extracts of PMR. A total of 152 compounds were identified, of which 66, 133, and 114 compounds were identified in aqueous solution, 70% ethanol solution, and 95% ethanol solution, respectively. Simultaneously, we found that amino acids and stilbene derivatives were the most important compounds in the aqueous extract of PMR and that extracts in 70% ethanol and 95% ethanol contained more anthraquinones. Figure 13 shows the distribution of specific compounds. Moreover, 16 components show relatively higher content in the extract of 70% ethanol, including three alkaloids, one amino acid, four anthraquinones, one fatty acid, two polyphenols, and two stilbene derivatives. For these 16 compounds, the comparing histograms are shown in Figures 14,Figures 15Figures 15 A statistical table of the peak area is attached in the Supplementary Material.
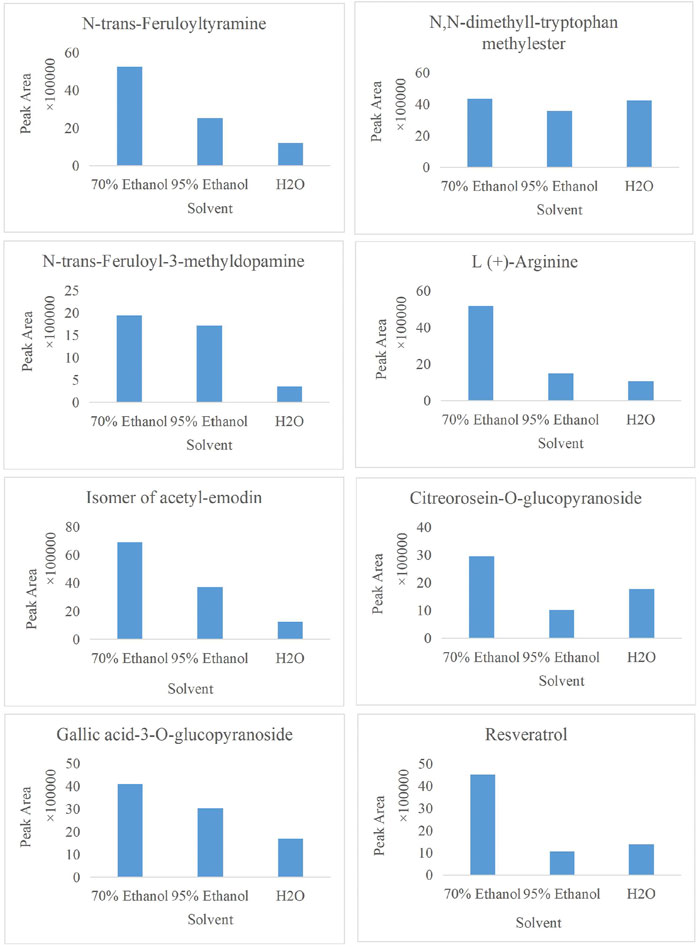
FIGURE 14. Histogram of peak area of compound N,N-dimethyl-tryptophan methylester, N-trans-feruloyltyramine, N-trans-feruloyl-3-methyldopamine, L (+)-arginine, the isomer of acetyl-emodin, citreorosein-O-glucopyranoside, gallic acid-3-O-glucopyranoside, and resveratrol in three solvents.
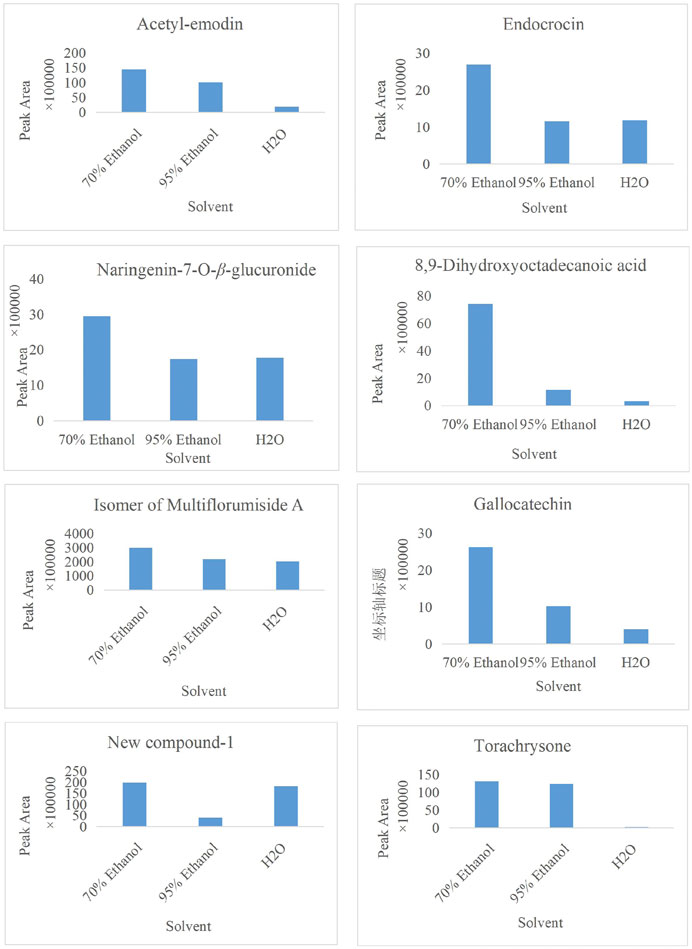
FIGURE 15. Histogram of peak area of compound acetyl-emodin, endocrocin, naringenin-7-O-β-glucuronide, 8,9-dihydroxyoctadecanoic acid, the isomer of multiflorumiside A, gallocatechin, new compound-1, and torachrysone in three solvents.
A few works of literature have reported the biological activity and toxicity of three alkaloidal constituents. (Ghosal et al. (1972) have reported that the total alkaloid fraction of E. variegata, including N,N-dimethyl-L-tryptophan methylester, has multiple effects: neuromuscular blocking, smooth muscle relaxant, central nervous system depressant, hydrocholeretic, and anticonvulsant effects. Regarding N-trans-feruloyltyramine, another alkaloid, there are some academic papers that demonstrate its antibacterial activity, α-glucosidase inhibitory activity, and antioxidant activity (Yokozawa et al., 2001; Panidthananon et al., 2018; Gao et al., 2019). However, its cytotoxicity is worth noting, which is normally involved in host defense mechanisms such as anti-bacterial (mainly G-bacteria), antiviral, and anti-parasitic but can destroy the body’s own cells, leading to tissue injury and disease in some pathological cases (Hsieh et al., 2001; Zhao et al., 2018; Gao et al., 2019). Similar to N-trans-feruloyltyramine, compound N-trans-feruloyl-3-methyldopamine also causes cytotoxicity (Zhao et al., 2018). Regarding the four anthraquinones, endocrocin, citreorosein-O-glucopyranoside, and acetyl-emodin and its isomer all belong to emodin derivatives, and the differences are the substituent groups that are attached. Endocrocin is mainly used as a pigment, and a small amount of literature has reported its biological activities, such as antifungal, antitumor, antioxidant, and pest-control effects (Liu et al., 2012; Wu, 2022). Zhang reported the neuroprotective effect of 2-acetyl-emodin, whose activity and toxicity need more exploration. As for citreorosein-O-glucopyranoside, the metabolites citreorosein and its isomer, aloe-emodin, have received more attention. Molecular docking experiment results showed that citreorosein has a strong binding ability with CYP1A1, CYP1A2, CYP2E1, CYP2B6, CYP2C9, and CYP3A4, and it is highly likely to induce liver toxicity by inhibiting CYP450 isoenzyme activity (Huang et al., 2021). Wang found that mild accumulation of aloe emodin and citreorosein was presented with the increase in dosage (Wang et al., 2022). Moreover, compared with the solvent control group, citreorosein 6.25, 12.5, and 25 μg·mL-1 groups significantly increased the mutation rate of the PIG-A gene in vitro (Wang et al., 2022). Thus, combined with our research results, more attention should be paid to citreorosein-O-glucopyranoside, which could be a potential hepatotoxic component. Gallocatechin and naringenin-7-O-β-glucuronide belong to flavonoids. Flavonoids are widely recognized as a group of active components that have anti-inflammatory, antioxidant, detoxification, hypoglycemic, and anticancer effects, and there have been no reports on their toxicity (Wang et al., 2019; Yuan et al., 2019; Dong et al., 2022). Similarly, resveratrol, as a compound that has been well-studied, processes many biological activities, including neuroprotective, antitumor, anti-inflammatory, and hepatoprotective effects (Yin et al., 2022; Ping, 2023; Yan et al., 2023). Multiflorumiside A, a dimer of tetrahydroxy stilbene glycoside, was first isolated and reported for its suppressive effects against NO production in lipopolysaccharide-stimulated RAW 264.7 cells (Li et al., 2018). More clinical value and risk assessment should be carried out.
These compounds deserve additional attention in further hepatotoxicity studies of PMR. Our study lays a solid foundation for the subsequent screening of toxicity and quality-control indicators of PMR. There are still some points in this study that need further improvement. On one hand, only one ionization mode was used to analyze the samples in our experiment, so some compounds that easily ionize hydroxide ions may not be adequately identified. On the other hand, more batches of samples are needed to verify the content distribution of 16 components in three types of solutions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author contributions
JY and XW designed the study. XW drafted the manuscript. FW and SM revised the manuscript. SM, FW, JY, and XC were responsible for collecting the samples. HG, YS, and YW provided technical support and advice for the study. All authors read and approved the final manuscript.
Funding
The work was financial supported by the National Natural Science Foundation of China (Grant Nos 81973476 and 81773874).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2023.1158717/full#supplementary-material
References
Avula, B., Khan, M. N., Wang, Y. H., Smillie, T. J., and Khan, I. A. (2009). Determination of shikimic acid in fruits of illicium species and various other plant samples by LC–UV and LC–ESI–MS. Chromatographia 69, 307–314. doi:10.1365/s10337-008-0884-z
Aziz, N., Khan, M. N., Ali, A., Khadim, A., Muhsinah, A. B., Uddin, J., et al. (2022). Rapid analysis of flavonoids based on spectral library development in positive ionization mode using LC-HR-ESI-MS/MS. Arabian J. Chem. 15, 103734. doi:10.1016/j.arabjc.2022.103734
Campora, M., Francesconi, V., Schenone, S., Tasso, B., and Tonelli, M. (2021). Journey on naphthoquinone and anthraquinone derivatives: New insights in alzheimer's disease. Pharm. (Basel) 14, 33. doi:10.3390/ph14010033
De Filippis, B., Ammazzalorso, A., Fantacuzzi, M., Giampietro, L., Maccallini, C., and Amoroso, R. (2017). Anticancer activity of stilbene-based derivatives. ChemMedChem 12, 558–570. doi:10.1002/cmdc.201700045
Dong, J., Zhu, W., and Si, Y. (2022). Study of Naringin against myocardial cytotoxicity of H9C2 induced by epirubicin. Chin. J. Hosp. Pharm. 42, 1204–1206. doi:10.13286/j.1001-5213.2022.12.05
Dretcanu, G., Stirbu, I., Leoplold, N., Cruceriu, D., Danciu, C., Stănilă, A., et al. (2022). Chemical structure, sources and role of bioactive flavonoids in cancer prevention: A review. Plants (Basel) 11, 1117. doi:10.3390/plants11091117
Gao, H., Yang, J., Wang, X., Song, Y., Cheng, X., Wei, F., et al. (2022). Exploratory quality control study for Polygonum multiflorum Thunb. Using dinuclear anthraquinones with potential hepatotoxicity. Molecules 27, 6760. doi:10.3390/molecules27196760
Gao, X., Wang, C., Chen, Z., Chen, Y., Santhanam, R. K., Xue, Z., et al. (2019). Effects of N-trans-feruloyltyramine isolated from laba garlic on antioxidant, cytotoxic activities and H(2)O(2)-induced oxidative damage in HepG2 and L02 cells. Food Chem. Toxicol. 130, 130–141. doi:10.1016/j.fct.2019.05.021
Ghosal, S., Dutta, S. K., and Bhattacharya, S. K. (1972). Erythrina—chemical and pharmacological evaluation II: Alkaloids of <em>Erythrina variegata</em> L. J. Pharm. Sci. 61, 1274. doi:10.1002/jps.2600610821
Gong, X., Liu, M., Gong, L., Li, Y., and Peng, C. (2019). Study on hepatotoxicity of different dosages of Polygoni multiflori radix praeparata in rats by metabolomics based on UPLC-Q-TOF-MS. J. Pharm. Biomed. Anal. 175, 112760. doi:10.1016/j.jpba.2019.07.008
Hsieh, T. J., Chang, F. R., and Chia, Y. C. (2001). Cytotoxic constituents of the fruits of Cananga odorata. J. Nat. Prod. 64, 616–619. doi:10.1021/np0005208
Huang, H., Xiong, W., and Tang, X. (2021). Screening of hepatotoxic components and CYP450 inhibitors of Polygonum multiflorum based on network toxicology and molecular docking. Genomics Appl. Biol. 40, 2863–2873. doi:10.13417/j.gab.040.002863
Huang, J., Zhang, J.-P., Bai, J.-Q., Wei, M. J., Huang, Z. H., et al. (2018). Chemical profiles and metabolite study of raw and processed Polygoni Multiflori Radix in rats by UPLC-LTQ-Orbitrap MS n spectrometry. Chin. J. Nat. Med. 16, 375–400. doi:10.1016/s1875-5364(18)30070-0
Huang, M. Q., Sun, L. N., and Dong, Z. Y. (2022). Comparative study on chemical components of Polygonum multiflorum between integrated processing and traditional processing. Chin. Traditional Herb. Drugs 53, 5293–5304. doi:10.7501/j.issn.0253-2670.2022.17.005
Jin, W., Wang, Y. F., Ge, R. L., Shi, H. M., Jia, C. Q., and Tu, P. F. (2007). Simultaneous analysis of multiple bioactive constituents in Rheum tanguticum Maxim. ex Balf. by high-performance liquid chromatography coupled to tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 2351–2360. doi:10.1002/rcm.3086
Kang, L., Li, D., Jiang, X., Zhang, Y., Pan, M., Hu, Y., et al. (2022). Hepatotoxicity of the major anthraquinones derived from Polygoni multiflori radix based on bile acid homeostasis. Front. Pharmacol. 13, 878817. doi:10.3389/fphar.2022.878817
Kong, W. S., Zhou, G., Xu, L. W., Wang, K., Feng, Y. M., Tao, L. Y., et al. (2022). Beware of the potential risks for Polygoni multiflori caulis-induced liver injury. Front. Pharmacol. 13, 868327. doi:10.3389/fphar.2022.868327
Lee, Y. H., Chen, Y. Y., Yeh, Y. L., Wang, Y. J., and Chen, R. J. (2019). Stilbene compounds inhibit tumor growth by the induction of cellular senescence and the inhibition of telomerase activity. Int. J. Mol. Sci. 20, 2716. doi:10.3390/ijms20112716
Li, H. Y., Yang, J. B., Li, W. F., Qiu, C. X., Hu, G., Wang, S. T., et al. (2020). In vivo hepatotoxicity screening of different extracts, components, and constituents of Polygoni Multiflori Thunb. in zebrafish (Danio rerio) larvae. Biomed. Pharmacother. 131, 110524. doi:10.1016/j.biopha.2020.110524
Li, S. G., Huang, X. J., Li, M. M., Liu, Q., Liu, H., Wang, Y., et al. (2018). Multiflorumisides A-G, dimeric stilbene glucosides with rare coupling patterns from the roots of Polygonum multiflorum. J. Nat. Prod. 81, 254–263. doi:10.1021/acs.jnatprod.7b00540
Li, Y., and Jiang, J. G. (2018). Health functions and structure-activity relationships of natural anthraquinones from plants. Food Funct. 9, 6063–6080. doi:10.1039/c8fo01569d
Liang, L., Xu, J., Zhou, W. W., Brand, E., Chen, H. B., and Zhao, Z. Z. (2018). Integrating targeted and untargeted metabolomics to investigate the processing Chemistry of Polygoni multiflori radix. Front. Pharmacol. 9, 934. doi:10.3389/fphar.2018.00934
Lin, L., Lin, H., Zhang, M., Ni, B., Yin, X., Qu, C., et al. (2015). A novel method to analyze hepatotoxic components in Polygonum multiflorum using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J. Hazard Mater 299, 249–259. doi:10.1016/j.jhazmat.2015.06.014
Lin, L., Ni, B., Lin, H., Zhang, M., Li, X., Yin, X., et al. (2015). Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb.: A review. J. Ethnopharmacol. 159, 158–183. doi:10.1016/j.jep.2014.11.009
Liu, Y., Wang, Q., and Jiang, M. (2012). Screening method of tyrosinase inhibitors in rhubarb based on spectral activity - structure - activity relationship. Chin. Tradit. Herb. Drugs 43, 2120–2126.
Luo, Y. Y., Cai, Z. Q., and Liu, X. H. (2017). Analysis of chemical constituents in Polygoni multiflori radix and Polygoni multifori caulis from the same origin based on UPLC-triple TOF- MS/MS. Nat. Prod. Res. Dev. 29. doi:10.16333/j.1001-6880.2017.12.012
March, R., and Brodbelt, J. (2008). Analysis of flavonoids: Tandem mass spectrometry, computational methods, and NMR. J. Mass Spectrom. 43, 1581–1617. doi:10.1002/jms.1480
Nguyen, T. A., Ha, M. T., Park, S. E., Choi, J. S., Min, B. S., and Kim, J. A. (2020). Stilbenes with potent protein tyrosine phosphatase-1B inhibitory activity from the roots of Polygonum multiflorum. J. Nat. Prod. 83, 323–332. doi:10.1021/acs.jnatprod.9b00777
Ning, F., Chen, L., Chen, L., Liu, X., Zhu, Y., Hu, J., et al. (2021). Combination of Polygoni multiflori radix praeparata and acori tatarinowii rhizoma alleviates learning and memory impairment in scopolamine-treated mice by regulating synaptic-related proteins. Front. Pharmacol. 12, 679573. doi:10.3389/fphar.2021.679573
Panidthananon, W., Chaowasku, T., Sritularak, B., and Likhitwitayawuid, K. (2018). A new benzophenone C-glucoside and other constituents of pseuduvaria fragrans and their α-glucosidase inhibitory activity. Molecules 23, 1600. doi:10.3390/molecules23071600
Ping, Y. (2023). Research progress of resveratrol in the treatment of knee osteoarthritis by regulating signal pathway. Chin. Tradit. Herb. Drugs 54, 1311–1320. doi:10.7501/j.issn.0253-2670.2023.04.032
Qiu, X., Zhang, J., Huang, Z., Zhu, D., and Xu, W. (2013). Profiling of phenolic constituents in Polygonum multiflorum Thunb. by combination of ultra-high-pressure liquid chromatography with linear ion trap-Orbitrap mass spectrometry. J. Chromatogr. A 1292, 121–131. doi:10.1016/j.chroma.2012.11.051
Reddy, M. V. B. (2016). A new naphthoquinone isolated from Polygonum multiflorum (Polygonaceae). Int. J. Pharm. Pharm. Sci. 8, 387–389.
Rui, W., Xia, W., Zhao, W., Li, B., Li, J., Feng, Y., et al. (2020). Differential constituents in roots, stems and leaves of Polygonum multiflorum Thunb. Screened by UPLC/ESI-Q-TOF-MS and multivariate statistical analysis. J. Chromatogr. Sci. 58, 136–143. doi:10.1093/chromsci/bmz086
Shen, N., Wang, T., Gan, Q., Liu, S., Wang, L., and Jin, B. (2022). Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 383, 132531. doi:10.1016/j.foodchem.2022.132531
Song, Y., Yang, J., Hu, X., Gao, H., Wang, P., Wang, X., et al. (2022). A stepwise strategy integrating metabolomics and pseudotargeted spectrum-effect relationship to elucidate the potential hepatotoxic components in Polygonum multiflorum. Front. Pharmacol. 13, 935336. doi:10.3389/fphar.2022.935336
State Pharmacopoeia Commission (2020). Pharmacopoeia of the people's Republic of China. Beijing: China Medical And Technology Press.
Teka, T., Wang, L., Gao, J., Mou, J., Pan, G., Yu, H., et al. (2021). Polygonum multiflorum: Recent updates on newly isolated compounds, potential hepatotoxic compounds and their mechanisms. J. Ethnopharmacol. 271, 113864. doi:10.1016/j.jep.2021.113864
Vukics, V., and Guttman, A. (2010). Structural characterization of flavonoid glycosides by multi-stage mass spectrometry. Mass Spectrom. Rev. 29, 1–16. doi:10.1002/mas.20212
Wang, L., Sang, M., Liu, E., Banahene, P. O., Zhang, Y., Wang, T., et al. (2017). Rapid profiling and pharmacokinetic studies of major compounds in crude extract from Polygonum multiflorum by UHPLC-Q-TOF-MS and UPLC-MS/MS. J. Pharm. Biomed. Anal. 140, 45–61. doi:10.1016/j.jpba.2017.03.016
Wang, M., Meng, N., and Wen, Y. (2019). Naringin inhibits lipopolysaccharide-induced inflammatory response in HaCaT cells through the P38 MAPK/NF-κB pathway. Drug Eval. study 42, 1081–1086. doi:10.7501/j.issn.1674-6376.2019.06.005
Wang, Q., Yang, J., and Wang, Y. (2022). Study on Toxicity kinetics of emodin type monoanthrone in rats. J. Pharm. Anal. 42, 1720–1728. doi:10.7501/j.issn.1674-6376.2022.07.003
Wang, S., Sun, X., An, S., Sang, F., Zhao, Y., and Yu, Z. (2021). High-throughput identification of organic compounds from Polygoni multiflori radix praeparata (zhiheshouwu) by UHPLC-Q-exactive orbitrap-MS. Molecules 26, 3977. doi:10.3390/molecules26133977
Wang, T. H., Zhang, J., Qiu, X. H., Bai, J. Q., Gao, Y. H., and Xu, W. (2015). Application of ultra-high-performance liquid chromatography coupled with LTQ-orbitrap mass spectrometry for the qualitative and quantitative analysis of Polygonum multiflorum thumb. And its processed products. Molecules 21, E40. doi:10.3390/molecules21010040
Wang, Y., Qian, Y., and Wang, X. (2022). Risk assessment of emodin type anthraquinone mutagenesis based on Pig-a gene mutation assay in vitro. Drug Eval. study 45, 1233–1239.
Wheelan, P., Zirrolli, J. A., and Murphy, R. C. (1995). Analysis of hydroxy fatty acids as pentafluorobenzyl ester, trimethylsilyl ether derivatives by electron ionization gas chromatography/mass spectrometry. J. Am. Soc. Mass Spectrom. 6, 40–51. doi:10.1016/1044-0305(94)00090-m
Wu, Y. (2022). Inhibitory effect of polyketones on enzyme activity in Filamentum filamentum. China: Jilin University.
Wu, S., Sun, A., and Liu, R. (2005). Separation and purification of baicalin and wogonoside from the Chinese medicinal plant Scutellaria baicalensis Georgi by high-speed counter-current chromatography. J. Chromatogr. A 1066, 243–247. doi:10.1016/j.chroma.2005.01.054
Xia, X. H., Yuan, Y. Y., and Liu, M. (2017). The assessment of the chronic hepatotoxicity induced by Polygoni multiflori radix in rats: A pilot study by using untargeted metabolomics method. J. Ethnopharmacol. 203, 182–190. doi:10.1016/j.jep.2017.03.046
Xu, M. L., Zheng, M. S., Lee, Y. K., Moon, D. C., Lee, C. S., Woo, M. H., et al. (2006). A new stilbene glucoside from the roots of Polygonum multiflorum Thunb. archives pharmacal Res. 29, 946–951. doi:10.1007/BF02969276
Yan, S., An, G., and Li, L. (2023). Research progress of resveratrol on liver diseases. Mod. Chin. Dr. 61, 137–140. doi:10.1007/bf02835739
Yang, J. B., Gao, H. Y., Song, Y. F., Liu, Y., Wang, Q., Wang, Y., et al. (2021). Advances in understanding the metabolites and metabolomics of Polygonum multiflorum Thunb: A mini-review. Curr. Drug Metab. 22, 165–172. doi:10.2174/1389200221666201201091345
Yang, J. B., Gao, H. Y., and Wang, X. T. (2021). A new lignanamide isolated from roots of Polygonum multiflorum. China Tradi Herb Drugs 52, 5475–5481. doi:10.7501/j.issn.0253-2670.2021.18.001
Yang, J. B., Gao, W. B., and Sun, H. (2022). Progress in basic research on hepatotoxic substances of Polygonum multiflorum. Chin. J. Pharmacovigil. 19, 611–614. doi:10.19803/j.1672-8629.2022.06.06
Yang, J. B., Li, W. F., Liu, Y., Wang, Q., Cheng, X. L., Wei, F., et al. (2018). Acute toxicity screening of different extractions, components and constituents of Polygonum multiflorum Thunb. on zebrafish (Danio rerio) embryos in vivo. Biomed. Pharmacother. 99, 205–213. doi:10.1016/j.biopha.2018.01.033
Yang, J. B., Liu, Y., Wang, Q., Wang, A. G., Cheng, X. L., et al. (2019). Characterization and identification of the chemical constituents of Polygonum multiflorum Thunb. by high-performance liquid chromatography coupled with ultraviolet detection and linear ion trap FT-ICR hybrid mass spectrometry. J. Pharm. Biomed. Anal. 172, 149–166. doi:10.1016/j.jpba.2019.03.049
Yang, J. B., Sun, B. W., and Sun, H. (2022). Progress in basic research on hepatotoxic substances of Polygonum multiflorum Thunb. Chin. J. Pharmacovigil. 19, 610–614. doi:10.19803/j.1672-8629.2022.06.06
Yang, J., Sun, H., Ma, J., Song, Y. f., Liu, Y., Wang, Q., et al. (2020). New phenolic constituents obtained from Polygonum multiflorum. Chin. Herb. Med. 12, 342–346. doi:10.1016/j.chmed.2020.02.001
Yang, J., Yan, Z., Dai, Z., Ma, S., Wang, A., Ren, J., et al. (2018). Polygonumnolides A1–B3, minor dianthrone derivatives from the roots of Polygonum multiflorum Thunb. Archives Pharmacal Res. 41, 617. doi:10.1007/s12272-016-0816-7
Yin, H., Cui, S., and Li, Z. (2022). Protective mechanism of resveratrol against retinal ganglion cell damage induced by high sugar through Nrf2/HO-1 pathway. World Sci. Technol. - TCM Mod. 24, 4681–4689. doi:10.11842/wst.20211125001
Yokozawa, T., Tanaka, T., and Kimura, T. (2001). Examination of the nitric oxide production-suppressing component in Tinospora tuberculata. Biol. Pharm. Bull. 24, 1153–1156. doi:10.1248/bpb.24.1153
Yu, Q., Jiang, L.-L., Luo, N., Fan, Y. X., Ma, J., Li, P., et al. (2017). Enhanced absorption and inhibited metabolism of emodin by 2, 3, 5, 4′-tetrahydroxystilbene-2-O-β-D-glucopyranoside: Possible mechanisms for Polygoni Multiflori Radix-induced liver injury. Chin. J. Nat. Med. 15, 451–457. doi:10.1016/s1875-5364(17)30067-5
Yuan, W., Gao, Z. P., and Yang, J. B. (2017). Study on chemical constituents of Polygoni multiflori radix. Chin. Herb. Med. 48, 631–634. doi:10.7501/j.issn.0253-2670.2017.04.002
Yuan, Y., Liu, H., and Yan, H. (2019). Antioxidant activity of foodborne naringin and its protective effect on furan-induced liver and kidney injury in mice. Chin. J. Food Sci. 19, 13–20. doi:10.16429/j.1009-7848.2019.09.002
Zhang, J. X., and Cui, Y. M. (2016). Study on chemical constituents of Polygonum multiflorum. J. Chin. Materia Medica 41, 3252–3255. doi:10.4268/cjcmm20161721
Zhao, C., Ying, Z., Tao, X., Jiang, M., and Yang, G. (2018). A new lactam alkaloid from Portulaca oleracea L. and its cytotoxity. Nat. Prod. Res. 32, 1548–1553. doi:10.1080/14786419.2017.1385022
Zhao, Y., Chu, S., Gui, S., Qin, Y., Xu, R., Shan, T., et al. (2021). Tissue-specific metabolite profiling of Fallopia multiflora (Heshouwu) and Fallopia multiflora var. angulata by mass spectrometry imaging and laser microdissection combined with UPLC-Q/TOF-MS. J. Pharm. Biomed. Anal. 200, 114070. doi:10.1016/j.jpba.2021.114070
Keywords: Polygoni Multiflori Radix, mass spectrometry, hepatotoxicity, identification, chemical components
Citation: Wang X, Yang J, Cheng X, Wang Y, Gao H, Song Y, Wei F and Ma S (2023) On-line identification of the chemical constituents of Polygoni Multiflori Radix by UHPLC-Q-ToF MS/MS. Front. Chem. 11:1158717. doi: 10.3389/fchem.2023.1158717
Received: 15 February 2023; Accepted: 05 April 2023;
Published: 21 April 2023.
Edited by:
Konstantinos M. Kasiotis, Benaki Phytopathological Institute, GreeceReviewed by:
Miaomiao Jiang, Tianjin University of Traditional Chinese Medicine, ChinaXiaowei Shi, Hebei Medical University, China
Copyright © 2023 Wang, Yang, Cheng, Wang, Gao, Song, Wei and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Wei, d2VpZmVuZ0BuaWZkYy5vcmcuY24=; Shuangcheng Ma, bWFzY0BuaWZkYy5vcmcuY24=
†These authors have contributed equally to this work
 Xueting Wang
Xueting Wang Jianbo Yang
Jianbo Yang Xianlong Cheng1
Xianlong Cheng1 Ying Wang
Ying Wang Huiyu Gao
Huiyu Gao Yunfei Song
Yunfei Song Feng Wei
Feng Wei Shuangcheng Ma
Shuangcheng Ma