
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem., 31 August 2022
Sec. Catalytic Reactions and Chemistry
Volume 10 - 2022 | https://doi.org/10.3389/fchem.2022.978668
This article is part of the Research TopicHot Topic: Chemical Reactions & Catalysis for a Sustainable FutureView all 6 articles
 Pritam Biswas1
Pritam Biswas1 Aniruddha Adhikari2
Aniruddha Adhikari2 Uttam Pal3
Uttam Pal3 Susmita Mondal2
Susmita Mondal2 Dipanjan Mukherjee2
Dipanjan Mukherjee2 Ria Ghosh3
Ria Ghosh3 Rami J. Obaid4
Rami J. Obaid4 Ziad Moussa5
Ziad Moussa5 Sudeshna Shyam Choudhury1*
Sudeshna Shyam Choudhury1* Saleh A. Ahmed4,6*
Saleh A. Ahmed4,6* Ranjan Das7*
Ranjan Das7* Samir Kumar Pal2,3*
Samir Kumar Pal2,3*Enzyme function can be altered via modification of its amino acid residues, side chains and large-scale domain modifications. Herein, we have addressed the role of residue modification in catalytic activity and molecular recognition of an enzyme alpha-chymotrypsin (CHT) in presence of a covalent cross-linker formalin. Enzyme assay reveals reduced catalytic activity upon increased formalin concentration. Polarization gated anisotropy studies of a fluorophore 8-Anilino-1-naphthalenesulfonic acid (ANS) in CHT show a dip rise pattern in presence of formalin which is consistent with the generation of multiple ANS binding sites in the enzyme owing to modifications of its local amino acid residues. Molecular docking study on amino acid residue modifications in CHT also indicate towards the formation of multiple ANS binding site. The docking model also predicted no change in binding behavior for the substrate Ala-Ala-Phe-7-amido-4-methylcoumarin (AMC) at the active site upon formalin induced amino acid cross-linking.
Covalent modifications introduce addition or removal of groups from the amino acid residues of a protein altering its structure and function. Formalin, simplest aldehyde is flammable and highly reactive and polymerizes readily under normal temperature and pressure. Formalin is known to react with a plethora of functional groups in small peptides and other substrates to generate covalently modified products (Fraenkel-Conrat and Olcott, 1948; Hoffman et al., 2015) which can include both intramolecular and intermolecular cross-linked species. Formalin due to its high reactivity and permeability into cells and tissues has led to its use in numerous applications in medicine and medical biotechnology. (Hoffman et al., 2015). From a chemical standpoint, formalin can react with biological nucleophiles such as protein and DNA and facilitates in vitro formation of intra-strand and DNA-protein cross links. (Brutlag et al., 1969; Metz et al., 2004; Hoffman et al., 2015). Formalin related reactions in cells are likely responsible for its toxic/carcinogenic effects, however the precise chemistry of such reactions and their cellular prevalence is still not clearly defined. (Lu et al., 2010). Studies have indicated that formalin causes crosslinking of proteins, involving the formation of a methylene bridge between two proximal amino acids. (Feldman, 1973; Metz et al., 2006). Formalin is widely used as a fixative, as it chemically modifies proteins irreversibly, although the first stages of such reactions are reversible, such as the reaction of an aldehyde with a free thiol to form a thioacetal, RS-C(OH)H2, or with an amine to form a Schiff-base adduct > C=N-. (Wait et al., 2011). Pioneering works by McGhee and Von Hippel (McGhee and Von Hippel, 1977) and a recent study by Kamps et al. (Kamps et al., 2019) as well Tayri-Wilk et al. (2020) on the reactions of nucleosides/nucleotides and amino acids with formalin have envisaged to understand the chemistry of such reactions.
To have a better understanding how formalin influence health and diseases, it is important to investigate the correlation between formalin induced structural modification and function of biological macromolecules. Previous studies on interaction of formalin with biological macromolecules were mostly restricted to residue modification studies. (Metz et al., 2004; Metz et al., 2006; Lu et al., 2008; Metz et al., 2020). These studies, however, revealed very little information regarding residue modification and its correlation to protein function and molecular recognition. In the present study, residue modification of a common digestive enzyme a-chymotrypsin (CHT; EC 3.4.21.1) by formalin was correlated with its catalytic activity and molecular recognition of a fluorescence probe (ANS) using steady state and pico-second resolved fluorescence spectroscopy, circular dichroism (CD), Dynamic light scattering (DLS). Here, we also utilize structural bioinformatics and develop a molecular docking model for understanding the effect of residue modification on molecular recognition.
α-Chymotrypsin (CHT) (Bostaurus) (≥40 U/mg), Ala-Ala-Phe-7-amido-4-methylcoumarin (AMC) (≥98%), 8-Anilino-1-naphthalenesulfonic acid (ANS) (≥97%) were purchased from Sigma (Saint Louis, United States). Formaldehyde (37%) was purchased from Merck. a-Chymotrypsin, AMC and ANS solutions were prepared in phosphate buffer (10mM, pH 7.0) using water from Millipore system.
Formalin modified CHT was prepared by incubating the enzyme at different formalin (1%, 2%, 4%) concentrations at 4°C for 5 h, which was dialysed before experimentation.
ANS is a well-known protein probe, and is a drug mimic. The ANS-CHT complex was prepared by mixing ANS (0.5 µM) with CHT (5 µM) in phosphate buffer (pH 7.0). The mix was stirred continuously for 5 h at 4°C, which was filtered extensively to remove the free probe. (Pal et al., 2002).
Circular dichroism (CD) spectroscopy analysis of CHT was conducted with 2 µM CHT solution incubated with different concentrations of formalin (1%, 2%, 4%) using a quartz cuvette of 1 mm path length in a JASCO 815 spectrometer. The deconvolution of the CD signals into relevant secondary structure was done by CDNN software. (Böhm et al., 1992).
Dynamic light scattering (DLS) measurements was done with 1.5 µM CHT solution incubated with different concentrations of formalin using a Nano S Malvern instrument employing a 4 mW He-Ne laser (λex = 632 nm).
Enzymatic activity of CHT was conducted with 0.5 µM of CHT at different formalin concentrations (1%, 2%, 4%). The substrate Ala-Ala-Phe-AMC (λmax = 325 nm) was cleaved to produce 7-amido-4-methyl-coumarin (λmax = 370 nm), the absorbance of which was monitored in Shimadzu Model UV-2600 spectrophotometer.
Picosecond-resolved fluorescence spectroscopic study were carried out using a commercial time correlated single photon counting (TCSPC) from Edinburgh instruments. The picosecond excitation pulses from the picoquant diode laser were used at 375 nm for the excitation of ANS with an instrument response function (IRF) of 80 ps at different formalin concentration. Fluorescence photons were detected by a microchannel-plate-photomultiplier tube (MCP-PMT; Hamamatsu Photonics, Kyoto, Japan). The time-resolved instrument provided a time resolution of 20 ps, which is one-fourth of the instrument response (IRF) for the detection of time constants after de-convolution of the IRF. For all decays, the emission polarizer was set at 55° (magic angle) with respect to the polarization axis of the emission beam. The observed fluorescence transients were fitted using a nonlinear least-squares fitting procedure. The details of which can be found elsewhere. (Sardar et al., 2015; Bagchi et al., 2016).
For the anisotropy measurements (r(t)), the emission polarizer was adjusted to be parallel and perpendicular to that of the excitation. Time-resolved anisotropy, r(t), was determined from the following equation:
where IVV(t) and IVH(t) are parallel and perpendicular polarized fluorescence decays of the dye, respectively, recorded using a vertically polarized excitation light. G is an instrument grating factor, which is an instrument and wavelength dependent correction factor to compensate for the polarization bias of the detection system and its magnitude was obtained by a long tail matching technique. (Schröder et al., 2005; O'Connor, 2012)
Solvent accessible surface area (SASA) is a measure of formation of contacts between the atoms on the surface of the protein and the solvent molecules. Thus, solvent accessibility provides a better insight on the amino acid residues of the enzyme (CHT) vulnerable to formalin induced modification. The validation and selection of the appropriate protein data bank (PDB) structure of CHT was made according to Chakraborty et al. (Chakraborty et al., 2020). Solvent accessible surface area (SASA) of CHT (PDB ID: 1CGJ) is computed using the PyMOL software (Educational version). The catalytic and residues of ANS binding site were identified as the prime targets for the SASA analysis.
Solvent exposed residues susceptible to formalin modification were modified followed by minimization of the modified residues using Schrodinger Maestro 2018-1 (Academic release). All covalent modifications were conducted according to Kamps et al. (2019) Targeted molecular docking studies of the modified amino acid residues were performed using AutoDock Vina (Trott and Olson, 2010) to study the influence of formalin induced covalent modification on substrate and molecular recognition of ANS by CHT. Protein and ligand preparation were done using AutoDock Tools 1.5.6. Polar hydrogens were added, the bond order and the subsequent renumbering of the newly added polar hydrogen were done. To add partial charge to the CHT molecule Gasteiger charges were added accordingly. The docking grid was localized on the substrate and ANS binding site. Furthermore, SASA analyses of the bound ANS were also performed according to the methodology described in the previous section.
α-Chymotrypsin (CHT) is a proteolytic enzyme of the class serine protease (EC 3.4.21.1) which is associated with hydrolysis of peptide bonds in the mammalian digestive system. The catalytic triad of CHT is located in the hydrophobic S1 pocket of CHT, (Ma et al., 2005; Andersson et al., 2017). Figure 1 shows the 3D structure of CHT. Figure 1A highlights the catalytic triad comprising the residues histidine-57; aspartate-102 and serine-195. The active site residue serine-195 serves as probe binding site (Banerjee and Pal, 2008). While, the site at cys-1-122 disulfide bond (Figure 1B) also serves as one of the probe (ANS) binding sites and is located almost opposite to the catalytic site (Johnson et al., 1979).
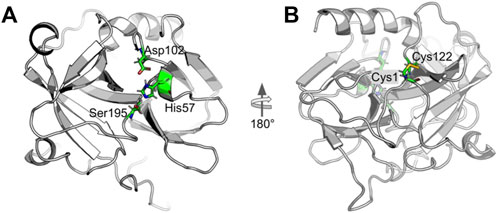
FIGURE 1. Cartoon representation of alpha-Chymotrypsin (CHT) [PDB ID:1CGJ]. (A) The coloured stick representation of the active site catalytic triad; showing the active site residues [His-57; Asp-102; Ser-195]. (B) At the N-terminal, the coloured stick representation [Cys-1; Cys-122] shows the molecular recognition site [External ANS binding site]. This site is located almost opposite to the catalytic site.
The catalytic activity of CHT was measured with different AMC (substrate) concentrations (5–400 µM) at varying formalin concentrations (1%–4%). To ensure the reaction between the enzyme and formalin, CHT was kept for 5 h at 4°C at the different ascribed concentrations of formalin, followed by dialysis of the enzyme before experimentation. Formalin is a chemical modification agent which causes irreversible covalent inhibition of CHT. CHT belongs to the class of serine protease that catalyzes hydrolysis of AMC to 7-amido-4-methy-coumarin (Figures 2A,B). Figures 2C,D illustrate the effects of increasing formalin concentration on the product formation and the apparent catalytic efficiency. A reduction in the apparent catalytic efficiency was observed (Table 1) with increasing formalin concentration reflecting on reduced catalytic activity which likely originates from a possible modification of the substrate binding site. A reduction in the apparent catalytic efficiency (kcat/Km) with increasing formalin concentration indicates a reduced catalytic activity which likely originates from a possible modification of the substrate binding site as indicated from previous reports. (Marini and Martin, 1971; Martin et al., 1972). however, it did not escape our attention that the reduced catalytic activity could also be caused due to enzyme aggregation and precipitation due to formalin modification. Hence, a structural analysis was conducted.
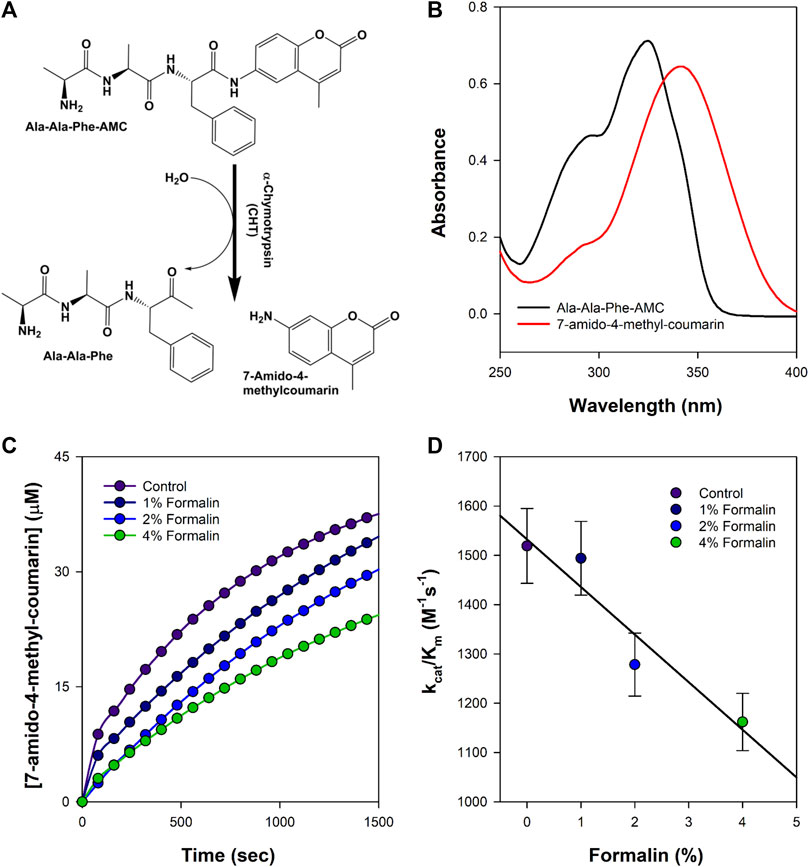
FIGURE 2. Schematic representation of the formalin treated enzyme catalysis. (A) Simple reaction outline catalysed by CHT; the substrate AMC hydrolysed to produce 7-amido-4-methyl-coumarin. (B) Absorbance spectra of the substrate AMC and product 7-amido-4-methyl-coumarin during catalysis. (C) Product formation at different representative formalin concentration; diminishing product formation with increased formalin concentration. (D) Apparent catalytic efficiency (kcat/Km) at different representative formalin concentration; retarding catalytic efficiency is consistent with decreasing product formation with increasing formalin concentration (Experiments were performed in triplicate).
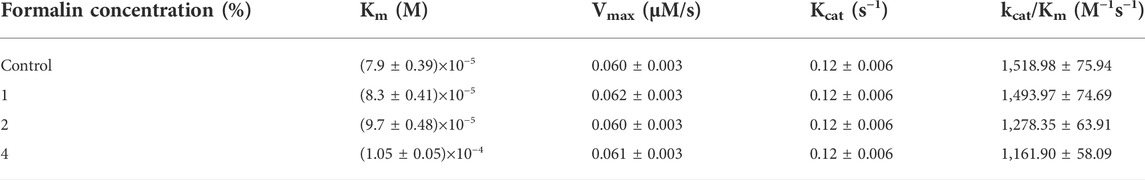
TABLE 1. Kinetics of CHT catalyzed hydrolysis of Ala-Ala-Phe-AMC at different formalin concentration.
Close analysis of the 3D structure of CHT (Figure 1) shows that the enzyme consists of anti-parallel ß-sheets which are highly distorted, forming very short irregular strands. Such conformation can lead to a shift in the negative band from the ideal ß-sheet position (210–220 nm) towards 200 nm. The effect of formalin induced modification on CHT was quantified by circular dichroism (CD) spectroscopy in the far UV region. The far UV spectra of CHT were characterized by a minimum at 202 nm with no positive band (Figure 3). The spectral data for all formalin concentration (1%, 2%, and 4%) was recorded. Since, 4% formalin corresponds to 1.3 M, the reliability of the CD spectra can be questioned. So, we recorded the CD voltage for all the corresponding CD spectra and found the CD voltage to be < 500 V, which was well within the acceptable range (<700 V) and hence, the CD spectral data was considered reliable. Upon de-convolution, a 4% increase in anti-parallel ß-sheet content was observed along with a nominal decrease in α-helix and insignificant changes in parallel ß-sheet, turns and random coils (Table 2). These results indicate an insignificant change in the secondary structure of the enzyme reflecting on its structural stability. The effect of formalin on the globular tertiary structure of CHT was monitored by dynamic light scattering (DLS). The DLS study (Figure 3A Inset) reveals the hydrodynamic diameter of CHT to be ∼7 nm, which agrees with previous studies (Shaw et al., 2007; Banerjee and Pal, 2008). Upon treatment with 4% formalin, a decrease in the hydrodynamic diameter of CHT from ∼7 nm (control) to ∼5 nm was noted (Supplementary Table S1) indicating no CHT aggregation due formalin treatment within the experimental conditions.
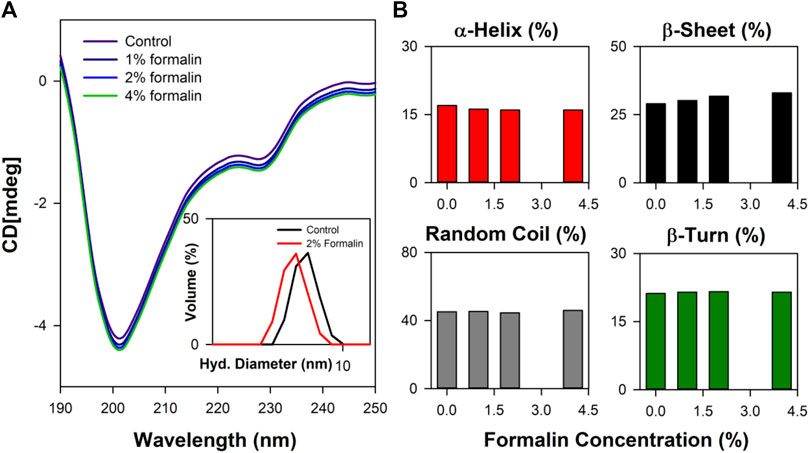
FIGURE 3. Far UV CD spectroscopy study of CHT. (A) The far UV CD spectra representing structural integrity of formalin treated CHT. The inset shows the DLS spectra of CHT at representative formalin concentration. (B) Percentage composition of secondary structure shown as bar plots. Analysis of secondary structure indicated structural stability within the experimental setup.
In water fluorescence of ANS decays rapidly with a time constant of 0.25 ns. (Collini et al., 2003). The fluorescence transients of ANS in CHT at different formalin concentrations (Figure 4) are characterized by three-time constants (Table 3) indicating its binding to the enzyme. The faster component
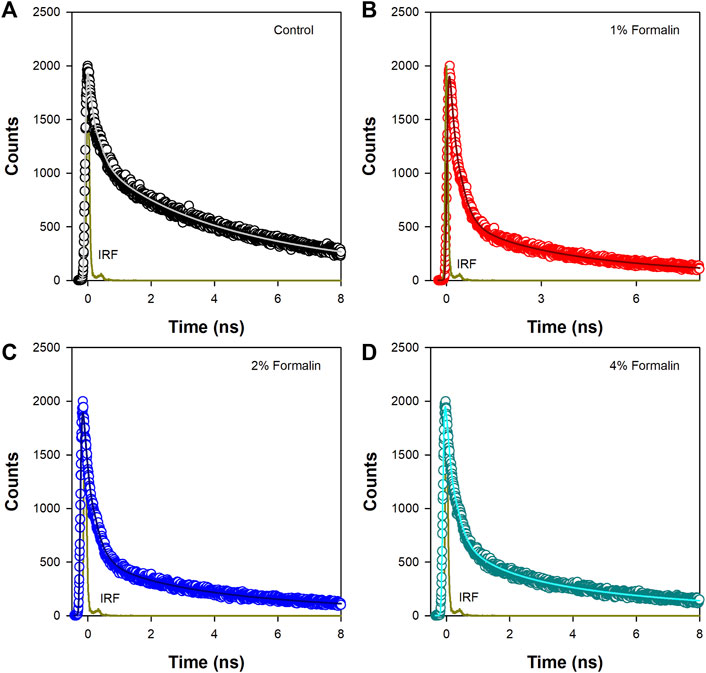
FIGURE 4. Time resolved fluorescence transients of ANS in (A) Pure CHT (control) and at varying formalin concentrations (B–D).
Polarization-gated fluorescence anisotropy of ANS in CHT was measured (Figure 5) at different formalin concentrations to monitor changes around the ANS binding sites of the enzyme. The anisotropy decay in CHT (Figure 5; control) is characterized by three rotational time constants of ∼56 ps (θ1), ∼712 ps (θ2) and 30 ns (θ3), respectively, (Table 4). The faster (θ1) and the relatively slower correlation time (θ2) is attributed to orientational motion of the dye in water (Pal et al., 2002) and that (ANS) bound to an external binding site in CHT (Cys-1-122) (Johnson et al., 1979), respectively. On the other hand, the much longer time constant (θ3) originates from rotational motion of the ANS/CHT complex where the probe is strongly bound to an internal binding site of the enzyme so that it rotates with the enzyme as a protein/dye complex. (Johnson et al., 1979). Upon interaction with formalin the fluorescence anisotropy decay of ANS in CHT is remarkably modified and a dip-rise pattern becomes evident (Figure 5) owing to heterogeneous distribution of ANS in different microenvironments with different fluorescence lifetimes and associated rotational correlation times. (Sinha et al., 2008; Paul and Guchhait, 2010; Lakowicz, 2013). When a fluorophore exists in different microenvironments, its anisotropy decay, r(t) can be expressed as: (Broos et al., 1995; Paul et al., 2015):
where the fractional intensity of the mth component at any time t is given by
αm is the amplitude corresponding to the fluorescence lifetime
where θm is the single rotational correlation time of the mth component and r0m is corresponding initial anisotropy.
The analysis of the dip and rise anisotropy decay (Figure 5) using equation (2) shows presence of three rotational correlation times (Table 4) where the fastest (θ1) and the longest (θ3) rotational correlation times remain almost unchanged with a variation in the formalin content. The longer rotational relaxation constant is not an arbitrary one. In order to investigate the longer time constants which corresponds to the global tumbling motion of the entire CHT-ANS complex, we have employed modified Stokes-Einstein-Debye equation where τr is defined as:
Here,
However, a gradual decrease in the intermediate correlation time (θ2) from 712 to 244 ps with increasing formalin content is observed which may be attributed to covalent modification of the amino acid residues in the enzyme by formalin. As a result of such residue modification the external binding site of ANS in the enzyme likely becomes more exposed to water giving rise to faster rotational relaxation of the dye (ANS).
Figure 6 shows solvent accessible surface area of CHT (1CGJ). It becomes evident that several amino acid residues of CHT are exposed to solvent and hence, are vulnerable to formalin induced modification. However, we have restricted our study to the substrate and the probe (ANS) binding sites in the enzyme. At the catalytic S1 pocket of CHT which also serves as the buried internal site of ANS binding, Ser-195 is solvent exposed and contains a free hydroxyl group making it susceptible to formalin induced modification. (Kamps et al., 2019). Furthermore, study of the external ANS binding, which is at Cys-1-122 (Johnson et al., 1979) site indicates Cys-1 to be vulnerable to modification by formalin owing to its high extent of solvent exposure and the presence of a free amino (-NH2) group. In contrary, Cys-122 has no free amino (NH2) group making it almost immune to formalin induced modification. This study also reveals the presence of susceptible residues Asn-48 and Ser-119 within 3 Å of the ANS binding site, making them suitable candidates for our study.
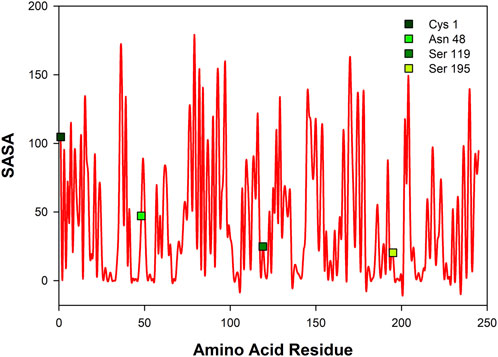
FIGURE 6. Solvent accessible surface area (SASA) of CHT [PDB ID: 1CGJ]. Four different residues corresponding to the catalytic site and molecular recognition were selected [Cys-1; Asn-48; Ser-119; Ser-195] for formalin induced modification.
To study the influence of residue modification on the binding of substrate (AMC) and its ability to recognize molecular binding partner (ANS) a molecular docking model was designed, the residues selected from the SASA study (Ser-195; Cys-1; Asn-48; Ser-119) were subjected to residue modification according to Kamps et al. (2019) Figure 7 shows various possible modifications due to reaction with formalin. From Figure 7A it becomes evident that serine has three possible modified outcomes. The free hydroxyl (OH) group undergoes modification forming a methylol adduct (Ser’), which can undergo internal cyclization, forming oxazolidone adduct (Ser’‘) or forming a crosslink with the nearby His-57 residue (Ser’’’). Similarly, Cys-1 and Asn-48, undergo modifications forming a methylol adduct at the free thiol (-SH) or amino (-NH2) group (Figures 7B,C). However, due to the formation of a disulfide bond between Cys-1 and Cys-122, no thiol group is available for formalin induced modification. Cys-1 being the N-terminal amino acid, contains a free amino group (-NH2) which reacts with the formalin.
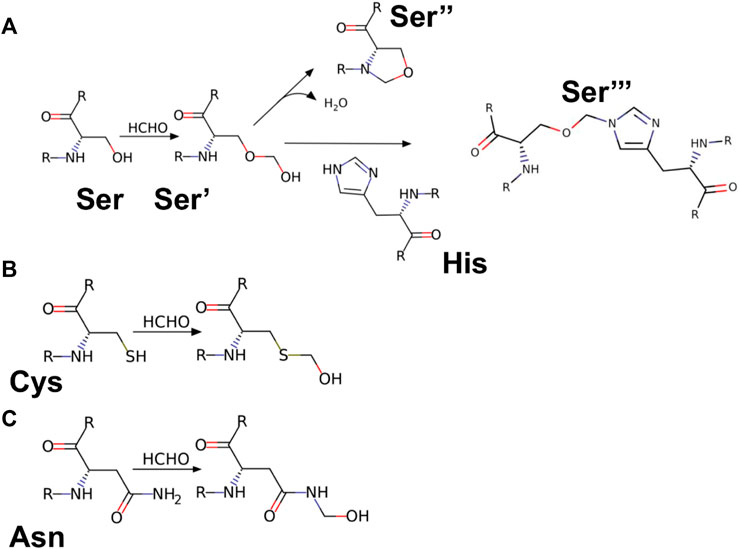
FIGURE 7. Reaction products of HCHO with different amino acids. (A) Amino acid Serine (Ser) reacting with HCHO form methylol derivative [Ser’]; which undergo cyclisation to form oxazolidine adduct [Ser’‘]; the methylol derivative can react with nearby amino acid [case in point amino acid Histidine (His)] to form a crosslink derivative [Ser’’‘]. (B,C) Reaction of HCHO with amino acids Cysteine (Cys) and Asparagine (Asn) forming their respective methylol derivative.
Targeted molecular docking was performed on the modified CHT structure. It should be noted that only the lowest energy binding poses were selected for the study. Figures 8–10 illustrate different ligand bound structures. It becomes evident from the docking studies that residue modification at the catalytic site has no influence on binding of CHT to the substrate (AMC) (Figure 8). Furthermore, a study of the van der Waals surface indicated no major structural distortions of the binding pocket (Supplementary Figure S3). The possible formation of Ser-195-His-57 crosslink upon formalin treatment may cause a disruption of the serine protease activity at the catalytic triad; thus, impairing the product formation and release, leading to reduced catalytic activity.
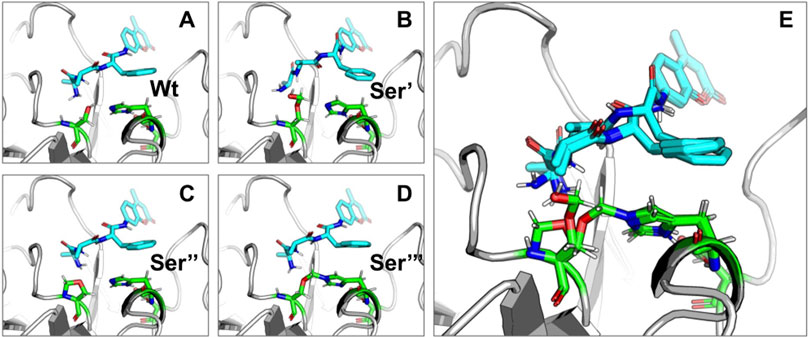
FIGURE 8. Binding of substrate (AMC) with Wt and HCHO modified CHT as obtained by molecular docking simulations. (A–D) Different modes of AMC binding with CHT on modification of the active site Serine residue (Ser-195) as mentioned previously [Wt indicates the wild type Ser-195 residue]. (E) Overlapping view of the different binding interactions; residue modification causes no major change in binding mode of AMC with CHT.
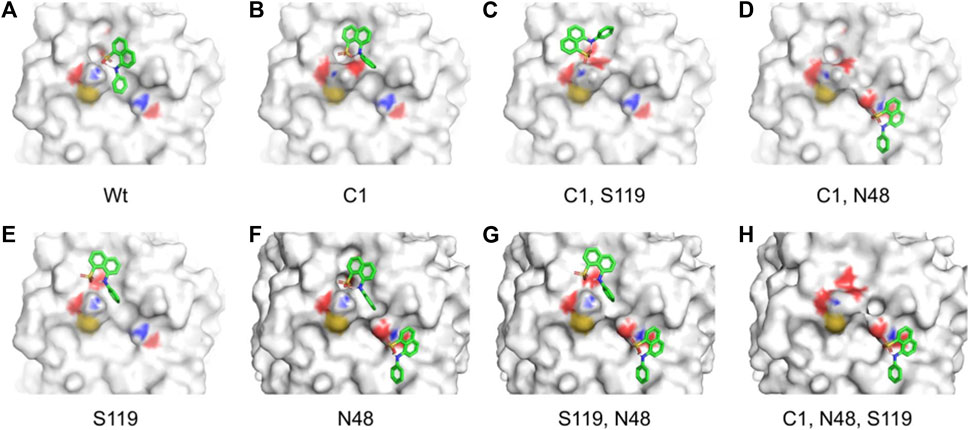
FIGURE 9. Different binding modes of ANS with CHT (external site) as obtained by molecular docking simulation. (A–H) Best binding modes of ANS on residue modification [Cys-1; Asn-48; Ser-119] as mentioned previously; extensive change in binding modes resulting in heterogeneity of ANS binding is evident.
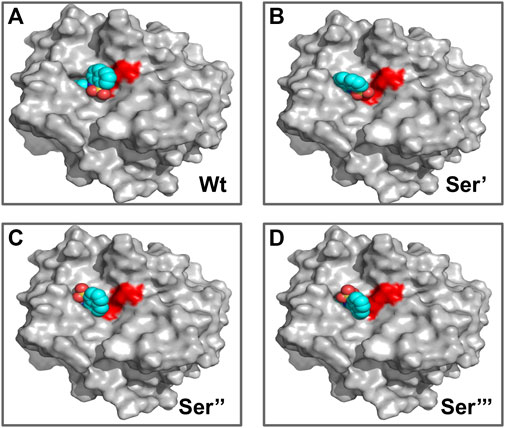
FIGURE 10. Different binding modes of ANS with CHT (internal site) as obtained by molecular docking simulation. (A–D) ANS binding modes with CHT on active site serine (Ser-195) modification as previously mentioned. [Wt indicates the wild type/unmodified CHT].
Our time resolved studies indicate the presence of two probe binding sites-internal binding site (buried), external binding site (exposed); which was described in previous studies (Johnson et al., 1979; Banerjee and Pal, 2008). Targeted molecular docking of the probe ANS and CHT was performed at the two binding sites, namely, the buried internal site at the S1 pocket and an external binding site at Cys-1-122 di-sulfide bond (Johnson et al., 1979; Banerjee and Pal, 2008). It becomes evident from Figure 9 that, formalin induced residue modification at the external binding site of ANS may lead to the generation of multiple probe (ANS) binding sites and a heterogeneous probe binding distribution which is indeed reflected by the dip-rise pattern in the anisotropy decay (Figure 5). Docking simulations of CHT modified at the external binding site Cys-1 and its nearby susceptible residues Ser-119, Asn-48 generated binding models (Figures 9B–H) which deviate from the unmodified CHT (Figure 9A). Although molecular docking does not provide pronounced solvent effects in protein/ligand interactions, however, by probing the solvent accessibility of the bound ligand (ANS), solvent effects in protein-ligand interactions can be elucidated from molecular docking studies. SASA analysis of the bound probe found a diverse range of solvent effects upon formalin modification of the external site. The varied solvent effects ranged from mimicking the unmodified ANS binding site [modification at Cys-1; 1,244 Å2 and Ser-119; 1,236 Å2] and lesser degree of solvent accessibility [modifications at Asn-48; 1150Å2] to experiencing higher solvent effects [modifications in conjunction at Cys-1 Asn-48; (1374Å2), Cys-1 Ser-119; (1402Å2), Ser-119 Asn-48; (1369Å2), Cys-1 Asn-48 Ser-119; (1360Å2)] (Table 5) signifying the bound probe to be more exposed and susceptible to solvent effects.
Similar to the binding interactions of substrate (AMC), residue modification at the internal ANS binding site in CHT (Ser-195) has no effect on binding of the probe (Figure 10). SASA analysis of ANS at the internal binding site of CHT suggests no major changes in solvent exposure of ANS (Table 6) suggesting that the bound probe is shielded from the surrounding solvent even after residue modification. [The calculated binding energies and cluster of poses are provided in the Supplementary Material]
CHT causes the catalytic hydrolysis of AMC to 7-amido 4-methyl-coumarin. (Biswas and Pal, 2004). The effect of formalin induced modification was monitored at different concentrations of formalin (1%–4%). We have observed a gradual decrease (Figure 2) in product formation with an increase in formalin concentration as well as an apparent fall in catalytic efficiency (Table 1). The Lineweaver-Burk plot (Supplementary Figure S1) and Table 1 indicates formalin exhibiting competitive inhibition (no change in Vmax and increasing Km) which may be inaccurate. Since formalin covalently modifies the active site residues causing irreversible covalent inhibition and can cause crosslinks (Marini and Martin, 1971; Martin et al., 1972; Tayri-Wilk et al., 2020) understanding the kinetic parameters of CHT using Lineweaver-Burk plot (Michealis-Menten kinetics) may not be accurate. A crucial assumption of the Michaelis–Menten model is that the total enzyme concentration does not change over time. As the active enzyme concentration changes due to formalin induced modification, the apparent kcat and Km values cannot be used to categorize the formalin mediated enzyme inhibition. Hence, whether formalin modification causes impairment of substrate binding at the active site cannot be discerned from Michaelis-Menten kinetics study of CHT under formalin modification. However, chymotrypsin goes through burst kinetics, we observed a longer initial phase for the formalin treated CHT (Supplementary Figure S2), suggesting retardation in the de-acylation of Ser-195 due to residue modification upon formalin treatment, which was also echoed in previous literature (Marini and Martin, 1971). Stability of the secondary and tertiary structure indicated to residue modification at the catalytic S1 pocket as the possible architect in reduction of catalytic activity of the enzyme in presence of formalin (Figure 3), further any change to the mass of the enzyme due to crosslinking would have reflected in its hydrodynamic diameter, which we found no evidence of in our DLS study. To support our hypothesis, we developed a molecular docking model to predict the cross-linking effects of active site residues on the binding of substrate. Previous studies by Martin et al. (1972), Zhang et al. (2014) found direct correlation between modification of the catalytic site residues and activity. Ser-195 at the catalytic triad was solvent exposed. Ser-195 modification and molecular docking showed no change in substrate (AMC) binding at the catalytic triad (Figures 7,8). Furthermore, no major structural distortions in the binding pocket were observed (Supplementary Figure S2). The analysis implied a stable enzyme-substrate formation; however, disruption in the proton transfer cascade from Ser-195 to His-57 to Asp-102 due to serine-histidine cross-linking by formalin impairs product formation and release leading to the reduced catalytic activity. Molecular docking simulations further support that modifications of amino acid residues are solely responsible for the impaired catalysis.
Time resolved fluorescence studies of ANS in CHT manifest heterogeneity in binding of ANS upon formalin induced modification (Figures 4,5). As a result of formalin induced structural modification in the enzyme, ANS bound to an external binding site (Cys-1-122) (Figure 9) becomes increasingly exposed to water thereby leading to a decreased fluorescence lifetime and rotational relaxation time (Tables 3,4).
To understand the effects of amino acid residue modification on substrate and probe binding SASA calculations were performed. From our SASA calculations we identified four amino acid residues [Ser-195; Cys-1; Asn-48 & Ser-119] as possible centers residue modification for our study (Figure 6). Using the SASA analysis a molecular docking model was designed to understand the substrate and probe binding behavior upon formalin modification. Targeted molecular docking analysis of external probe binding site (Cys-1) indicated generation of new ANS binding sites having different solvent accessibility on formalin modification (Figure 9). However, no change in binding was observed for the internal binding site (Ser-195) of ANS in CHT. The SASA of bound ANS suggested the bound probe to be buried and shielded from the surrounding solvent (Tables 5,6). Our solvent effect and docking simulation studies of the bound probe was found to corroborate with the time resolved fluorescence anisotropy studies manifesting heterogeneity in ANS binding to CHT following formalin induced modification.
The present work primarily focuses on the influence of residue modification on the catalytic activity and molecular recognition of CHT. The study found changes in the catalytic activity and molecular recognition of the enzyme (CHT) upon formalin induced chemical modification of its amino acid residues. The structural and molecular docking analysis provide evidence of no impediment to the formation of a stable enzyme-substrate complex due to formalin induced modifications; rather we assume the impairment of the proton transfer cascade in the catalytic triad due to Ser-195-His-57 cross-linking leads to reduced catalytic activity. Picosecond resolved fluorescence studies reveal generation of multiple binding sites of a fluorophore ANS in CHT, and upon treatment with formalin a reduction in fluorescence lifetime and rotational relaxation time of the probe is observed when it is bound to an external binding site. Subsequent molecular docking studies and SASA analysis provide evidence towards heterogeneity of ligand (ANS) interaction with the enzyme (CHT) upon amino acid residues. The present findings may in the future offer background regarding drug-target interaction, molecular recognition, macromolecular modification to generate new binding sites for enhanced ligand binding through protein engineering.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Conceptualization: PB, AA, UP, SSC, SKP. Methodology: PB, AA, UP, SM, RG, DM, ZM. Investigation: PB, AA, UP, SM, RG, DM. Funding acquisition: SKP, SAA, ZM. Project administration: SKP, SSC, RD. Supervision: SKP, RD, SSC, SAA, ZM, RJO. Writing—original draft: PB, AA, UP, SM, DM. Writing—review and editing: SKP, SSC, RD, SAA.
SKP thanks the Indian National Academy of Engineering (INAE) for the Abdul Kalam Technology Innovation National Fellowship, INAE/121/AKF. The authors would like to acknowledge the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant code: 22UQU4320545DSR22. Dr. ZM is grateful to the United Arab Emirates University (UAEU) of Al-Ain and the Research Office for supporting the research developed in his laboratory (Grant no. G00003291).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.978668/full#supplementary-material
Andersson, C. A., Martinez, N., Zeller, D., Rondahl, S. H., Koza, M. M., Frick, B., et al. (2017). Changes in dynamics of α-chymotrypsin due to covalent inhibitors investigated by elastic incoherent neutron scattering. Phys. Chem. Chem. Phys. 19 (37), 25369–25379. doi:10.1039/c7cp04041e
Bagchi, D., Ghosh, A., Singh, P., Dutta, S., Polley, N., Althagafi, I. I., et al. (2016). Allosteric inhibitory molecular recognition of a photochromic dye by a digestive enzyme: dihydroindolizine makes α-chymotrypsin photo-responsive. Sci. Rep. 6 (1), 34399–34411. doi:10.1038/srep34399
Banerjee, D., and Pal, S. K. (2008). Conformational dynamics at the active site of α-chymotrypsin and enzymatic activity. Langmuir 24 (15), 8163–8168. doi:10.1021/la8010184
Biswas, R., and Pal, S. K. (2004). Caging enzyme function: α-chymotrypsin in reverse micelle. Chem. Phys. Lett. 387 (4-6), 221–226. doi:10.1016/j.cplett.2004.02.018
Böhm, G., Muhr, R., and Jaenicke, R. (1992). Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. Des. Sel. 5 (3), 191–195. doi:10.1093/protein/5.3.191
Broos, J., Visser, A. J. W. G, Engbersen, J. F. J., Verboom, W., van Hoek, A., and Reinhoudt, D. N. (1995). Flexibility of enzymes suspended in organic solvents probed by time-resolved fluorescence anisotropy. Evidence that enzyme activity and enantioselectivity are directly related to enzyme flexibility. J. Am. Chem. Soc. 117 (51), 12657–12663. doi:10.1021/ja00156a001
Brutlag, D., Schlehuber, C., and Bonner, J. (1969). Properties of formaldehyde-treated nucleohistone. Biochemistry 8 (8), 3214–3218. doi:10.1021/bi00836a013
Chakraborty, B., Sengupta, C., Pal, U., and Basu, S. (2020). Probing the hydrogen bond involving Acridone trapped in a hydrophobic biological nanocavity: Integrated spectroscopic and docking analyses. Langmuir 36 (5), 1241–1251. doi:10.1021/acs.langmuir.9b03506
Collini, M., D'Alfonso, L., Molinari, H., Ragona, L., Catalano, M., and Baldini, G. (2003). Competitive binding of fatty acids and the fluorescent probe 1-8-anilinonaphthalene sulfonate to bovine β-lactoglobulin. Protein Sci. 12 (8), 1596–1603. doi:10.1110/ps.0304403
Feldman, M. Y. (1973). Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog. Nucleic Acid. Res. Mol. Biol. 13, 1–49. doi:10.1016/s0079-6603(08)60099-9
Fraenkel-Conrat, H., and Olcott, H. S. (1948). Reaction of formaldehyde with proteins VI. cross-linking of amino groups with phenol, imidazole, or indole groups. J. Biol. Chem. 174 (3), 827–843. doi:10.1016/s0021-9258(18)57292-6
Hoffman, E. A., Frey, B. L., Smith, L. M., and Auble, D. T. (2015). Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 290 (44), 26404–26411. doi:10.1074/jbc.r115.651679
Johnson, J. D., El-Bayoumi, M. A., Weber, L. D., and Tulinsky, A. (1979). Interaction of. alpha.-chymotrypsin with the fluorescent probe 1-anilinonaphthalene-8-sulfonate in solution. Biochemistry 18 (7), 1292–1296. doi:10.1021/bi00574a027
Kamps, J. J. A. G., Hopkinson, R. J., Schofield, C. J., and Claridge, T. D. W. (2019). How formaldehyde reacts with amino acids. Commun. Chem. 2 (1), 126–214. doi:10.1038/s42004-019-0224-2
Lakowicz, J. R. (2013). Principles of fluorescence spectroscopy. New York, NY: Springer science & business media.
Lu, K., Boysen, G., Gao, L., Collins, L. B., and Swenberg, J. A. (2008). Formaldehyde-induced histone modifications in vitro. Chem. Res. Toxicol. 21 (8), 1586–1593. doi:10.1021/tx8000576
Lu, K., Ye, W., Zhou, L., Collins, L. B., Chen, X., Gold, A., et al. (2010). Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132 (10), 3388–3399. doi:10.1021/ja908282f
Ma, W., Tang, C., and Lai, L. (2005). Specificity of trypsin and chymotrypsin: loop-motion-controlled dynamic correlation as a determinant. Biophysical J. 89 (2), 1183–1193. doi:10.1529/biophysj.104.057158
Marini, M. A., and Martin, C. J. (1971). Chymotrypsin catalysis in the presence of formaldehyde: the pH-dependence of ester hydrolysis. Eur. J. Biochem. 19 (2), 153–161. doi:10.1111/j.1432-1033.1971.tb01299.x
Martin, C. J., Oza, N. B., and Marini, M. A. (1972). Formaldehyde as an active site label of α-chymotrypsin. Can. J. Biochem. 50 (10), 1114–1121. doi:10.1139/o72-152
McGhee, J. D., and Von Hippel, P. H. (1977). Formaldehyde as a probe of DNA structure. 3. Equilibrium denaturation of DNA and synthetic polynucleotides. Biochemistry 16 (15), 3267–3276. doi:10.1021/bi00634a001
Metz, B., Kersten, G. F. A., Hoogerhout, P., Brugghe, H. F., Timmermans, H. A. M., De Jong, A., et al. (2004). Identification of formaldehyde-induced modifications in proteins reactions with model peptides. J. Biol. Chem. 279 (8), 6235–6243. doi:10.1074/jbc.m310752200
Metz, B., Kersten, G. F. A., Baart, G. J. E., de Jong, A., Meiring, H., ten Hove, J., et al. (2006). Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjug. Chem. 17 (3), 815–822. doi:10.1021/bc050340f
Metz, B., Michiels, T., Uittenbogaard, J., Danial, M., Tilstra, W., Meiring, H. D., et al. (2020). Identification of formaldehyde-induced modifications in diphtheria toxin. J. Pharm. Sci. 109 (1), 543–557. doi:10.1016/j.xphs.2019.10.047
Pal, S. K., Peon, J., and Zewail, A. H. (2002). Ultrafast surface hydration dynamics and expression of protein functionality: α-Chymotrypsin. Proc. Natl. Acad. Sci. U. S. A. 99 (24), 15297–15302. doi:10.1073/pnas.242600399
Paul, B. K., and Guchhait, N. (2010). Modulated photophysics of an ESIPT probe 1-hydroxy-2-naphthaldehyde within motionally restricted environments of liposome membranes having varying surface charges. J. Phys. Chem. B 114 (39), 12528–12540. doi:10.1021/jp1048138
Paul, B. K., Ghosh, N., and Mukherjee, S. (2015). Interplay of multiple interaction forces: Binding of norfloxacin to human serum albumin. J. Phys. Chem. B 119 (41), 13093–13102. doi:10.1021/acs.jpcb.5b08147
Sardar, S., Chaudhuri, S., Kar, P., Sarkar, S., Lemmens, P., and Pal, S. K. (2015). Direct observation of key photoinduced dynamics in a potential nano-delivery vehicle of cancer drugs. Phys. Chem. Chem. Phys. 17 (1), 166–177. doi:10.1039/c4cp03749a
Schröder, G. F., Alexiev, U., and Grubmüller, H. (2005). Simulation of fluorescence anisotropy experiments: probing protein dynamics. Biophysical J. 89 (6), 3757–3770. doi:10.1529/biophysj.105.069500
Shaw, A. K., Sarkar, R., Banerjee, D., Hintschich, S., Monkman, A., and Pal, S. K. (2007). Direct observation of protein residue solvation dynamics. J. Photochem. Photobiol. A Chem. 185 (1), 76–85. doi:10.1016/j.jphotochem.2006.05.014
Sinha, S. S., Mitra, R. K., and Pal, S. K. (2008). Temperature-dependent simultaneous ligand binding in human serum albumin. J. Phys. Chem. B 112 (16), 4884–4891. doi:10.1021/jp709809b
Tayri-Wilk, T., Slavin, M., Zamel, J., Blass, A., Cohen, S., Motzik, A., et al. (2020). Mass spectrometry reveals the chemistry of formaldehyde cross-linking in structured proteins. Nat. Commun. 11 (1), 3128–3129. doi:10.1038/s41467-020-16935-w
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Wait, A. F., Brandmayr, C., Stripp, S. T., Cavazza, C., Fontecilla-Camps, J. C., Happe, T., et al. (2011). Formaldehyde-- A rapid and reversible inhibitor of hydrogen production by [FeFe]-Hydrogenases. J. Am. Chem. Soc. 133 (5), 1282–1285. doi:10.1021/ja110103p
Keywords: amino acid modification, chymotrypsin, formalin, cross-linking, spectroscopy, molecular docking
Citation: Biswas P, Adhikari A, Pal U, Mondal S, Mukherjee D, Ghosh R, Obaid RJ, Moussa Z, Choudhury SS, Ahmed SA, Das R and Pal SK (2022) A combined spectroscopic and molecular modeling Study on structure-function-dynamics under chemical modification: Alpha-chymotrypsin with formalin preservative. Front. Chem. 10:978668. doi: 10.3389/fchem.2022.978668
Received: 26 June 2022; Accepted: 01 August 2022;
Published: 31 August 2022.
Edited by:
Dhenadhayalan Namasivayam, National Taiwan University, TaiwanReviewed by:
Cassandra Fleming, Auckland University of Technology, New ZealandCopyright © 2022 Biswas, Adhikari, Pal, Mondal, Mukherjee, Ghosh, Obaid, Moussa, Choudhury, Ahmed, Das and Pal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sudeshna Shyam Choudhury, c3VkZXNobmEucy5jaG91ZGh1cnlAc3hjY2FsLmVkdQ==; Saleh A. Ahmed, c2FhaG1lZEB1cXUuZWR1LnNh; Ranjan Das, cmFuamFuLmRhczY4QGdtYWlsLmNvbQ==; Samir Kumar Pal, c2twYWxAYm9zZS5yZXMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.