
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Chem. , 26 July 2022
Sec. Nanoscience
Volume 10 - 2022 | https://doi.org/10.3389/fchem.2022.957572
This article is part of the Research Topic Recent Progress in Drug Delivery Technologies for Advanced Pharmaceutics View all 6 articles
 Faranak Hejabi1†
Faranak Hejabi1† Mohammad Sadegh Abbaszadeh2†
Mohammad Sadegh Abbaszadeh2† Shirinsadat Taji3
Shirinsadat Taji3 Andrew O’Neill4
Andrew O’Neill4 Fatemeh Farjadian5
Fatemeh Farjadian5 Mohammad Doroudian1*
Mohammad Doroudian1*In recent decades, clustered regularly interspaced short palindromic repeat/CRISPR-associated protein (CRISPR/Cas) has become one of the most promising genome-editing tools for therapeutic purposes in biomedical and medical applications. Although the CRISPR/Cas system has truly revolutionized the era of genome editing, the safe and effective delivery of CRISPR/Cas systems represents a substantial challenge that must be tackled to enable the next generation of genetic therapies. In addition, there are some challenges in the in vivo delivery to the targeted cells/tissues. Nanotechnology-based drug delivery systems can be employed to overcome this issue. This review discusses different types and forms of CRISPR/Cas systems and the current CRISPR/Cas delivery systems, including non-viral carriers such as liposomes, polymeric, and gold particles. The focus then turns to the viral nanocarriers which have been recently used as a nanocarrier for CRISPR/Cas delivery.
Genome-editing technologies are crucial in the field of genetic engineering primarily due to the development of tools that make the manipulation of genetic material possible (Smyth, 2022; Nambiar et al., 2022). Although numerous genome-editing methods have been employed, the CRISPR-Cas approach has opened a new era for genetic modification as a result of its notable simplicity and versatility (Liu et al., 2022). The CRISPR/Cas system was first discovered in 1987 (Ishino et al., 1987) and was later found to be present in approximately 90% of archaea and 40% of bacteria. In 2005, the CRISPR/Cas system was revealed to be a component of the defensive system of prokaryotic cells against phages. In a landmark 2012 study, CRISPR/Cas systems were employed for gene-editing purposes (Jinek et al., 2012). Compared with other similar genome-editing tools, the CRISPR/Cas system has notable benefits including low cost and the fact that the system is relatively easy to use (Yang et al., 2022). In addition, the CRISPR/Cas system can overcome current genome-editing limits of gene overexpression or gene shut-down. Furthermore, CRISPR/Cas systems have been used successfully in a wide range of disease indications including bacterial infections, viral infections, and also cancers (Figure 1) (Chen et al., 2019; Uddin et al., 2020). As mentioned previously, the CRISPR/Cas system is part of the adaptive immune system of bacteria and archaea, playing a key role in the defensive immune response to a variety of stimuli including bacteriophages and mobile genetic elements (MGEs) by cleaving their genetic material. CRISPR/Cas systems are highly diverse across organisms and can be exchanged via horizontal gene transfer (Hasanzadeh et al., 2022). The CRISPR system is composed of two parts: single-guide RNA (sgRNA) and an endonuclease protein known as CRISPR-associated protein (Cas). The sgRNA identifies a precise genomic target sequence and leads the Cas nuclease to precisely cleave the detected DNA sequence. Induced double-stranded breaks are repaired through non-homologous end joining to generate insertions, deletions, or homology-directed repair for gene modifications (Petassi et al., 2020; Vo et al., 2021). Delivery of the CRISPR/Cas system is an attractive strategy because it is the most direct and rapid method for gene-editing purposes and has been shown to have fewer off-target effects and lower immunogenicity than other similar technologies (Wei et al., 2020). There still exist numerous challenges with the technology, with the development of improved delivery systems to enable effective systemic delivery of the CRISPR/Cas system a key challenge. Nanotechnology is a multidisciplinary field that has been increasingly capitalized for a variety of applications (Doroudian et al., 2019). Reducing the administered dose enhances the efficiency of drugs or active substances by specifically generating a synergistic effect and utilizing the multivalency of nanomaterial (Aghasadeghi et al., 2013; Doroudian et al., 2020). Furthermore, nanotechnology can also convert traditional delivery systems into smart carriers that are not only specifically transported to targeted tissues or cells, but also sustainably release payload in response to the microenvironmental stimuli like pH and temperature at targeted sites (Doroudian et al., 2021a). Protecting cargo against degradation by digestive elements is of significant importance for nucleic structures as they are highly exposed to the nucleases once entering the body. Therefore, nano-based delivery systems provide a solution to a number of key challenges in the context of therapeutic delivery (Doroudian et al., 2021b).
FIGURE 1. The CRISPR/Cas9 system has a wide diversity of applications for the treatment of different diseases.
Different types of CRISPR have been developed in recent years which are divided into various groups based on their functions and structures (Figure 2) (Nidhi et al., 2021). Sequence-similarity–based clustering and phylogenetic analysis of conserved Cas proteins are the main factors for the group arrangement/classification (Pinilla-Redondo et al., 2021). There are two main classes of CRISPR/Cas systems with each of them consisting of further subclasses. Class 1 consists of types I, III, and IV, while class 2 includes types II, V, and VI types (Makarova et al., 2020; Mohanraju et al., 2022). Class 1 systems are found in bacteria and archaea (Birkeland et al., 2017), while class 2 are only detected in bacteria that are not hyperthermophiles. The structure of proteins in class 2 systems is also less complex than those found in class I systems (Shmakov et al., 2017; Makarova et al., 2020). Different Cas proteins have different roles in the CRISPR‐Cas system. The Cas1 protein is a well-known integrase enzyme required for the specific breaking of a CRISPR array to insert a newly identified spacer in prokaryotic DNA (Givens et al., 2018; Roma-Rodrigues et al., 2020). The role of the Cas2 protein has not been identified thus far. However, this protein has RNase and DNase activities and is essential for the adaptation phase in Escherichia coli (E. coli) (Guzmán et al., 2021). The most frequently used subtype of CRISPR/Cas systems is the type II CRISPR/Cas9, employed for gene editing in eukaryotic cells (Table 1) (Makarova et al., 2020; Ramachandran et al., 2020).
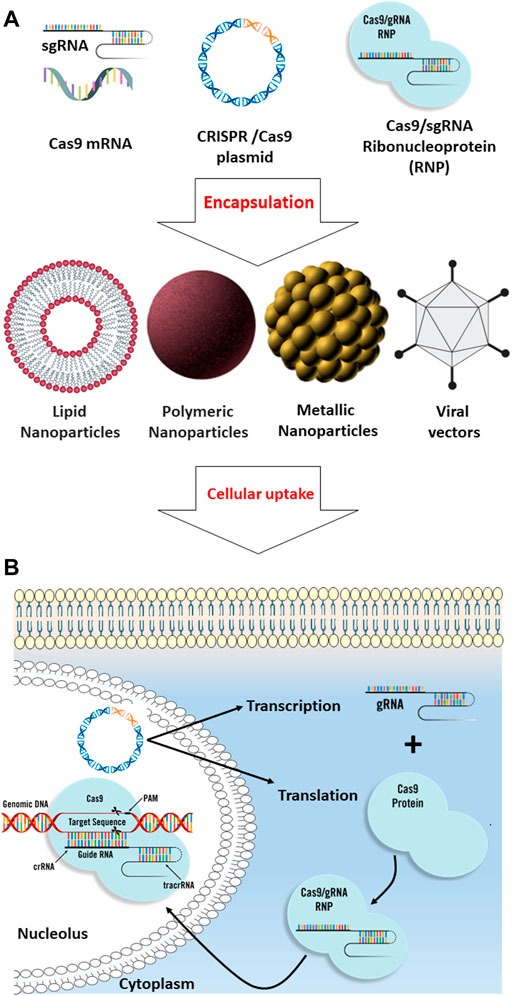
FIGURE 2. Comparison of different biomolecular CRISPR/Cas9 formats. (A) CRISPR/Cas9 systems include Cas9 plasmid, Cas9 mRNA, and sgRNA, and Cas9/sgRNA ribonucleoprotein can be encapsulated into different types of nanoparticles such as lipid-based, polymeric, metal-based, and viral nanodrug delivery systems for efficient intracellular uptake. (B) After cellular uptake through endocytosis, the CRISPR/Cas9 systems are released from the nanoparticles. For plasmid DNA delivery, the plasmid should be delivered into the nucleus, where the transcription mechanism must be employed to transcribe the gene into gRNA and Cas9 mRNA; in the cytoplasm, the Cas9 mRNA will be translated into the Cas9, and then, the gRNA and Cas9 protein will be transported back into the nucleus where the CRISPR mechanism can apply its effect on the targeted genomic DNA. For the Cas9 mRNA delivery strategy, the cargo should be released in the cytosol to allow the mRNA translation to Cas9 protein. Ribonucleoprotein (RNP) delivery is instantaneous compared to other strategies since the translation and transcription processes can be skipped, resulting in the immediate onset of gene editing.
There are various formats for the CRISPR/Cas9 system, including 1) ribonucleoprotein complexes containing Cas9 endonuclease protein and sgRNA molecule, 2) a plasmid that encodes both Cas9 and sgRNA, 3) two separate plasmids each encoding Cas9 and sgRNA, and 4) Cas9 mRNA and sgRNA molecule (Table 2) (Deng et al., 2019; Cheng et al., 2021).
Using a plasmid is a more stable and cost-effective mechanism in comparison to Cas9 mRNA and protein. The use of a plasmid form also further extends the production of Cas9 nuclease for constant gene editing (Ahmadi Danesh et al., 2012; Eoh and Gu, 2019). One of the advantages of the plasmid format is its ability to pass through cellular and nuclear membranes which is essential for the expression of the Cas9 nuclease protein in eukaryotic cells (Zhang et al., 2022). Therefore, the risk of off-target effects, random integration of the plasmid into the genome, and host immunologic responses may increase the continuous expression of plasmid in mammalian cells (Chen et al., 2020; Yang et al., 2021).
Cas9 mRNA has attractive features such as effective nuclear entry, fast onset of gene editing, and the removal of the need for DNA transcription to mRNA. Moreover, encoding Cas9 in mRNA, which is translated in the cytoplasm, results in rapid protein expression and reduced off-target effects and insertion into the genome (Salehi Chaleshtori et al., 2012; Doroudian et al., 2013).
The least complex approach is to transfer Cas protein with sgRNA specifically. This strategy offers the only temporal expression time and allows fast gene editing which removes the requirement for mRNA transcription and translation (Zhang et al., 2021; Yan et al., 2022). Furthermore, using the ribonucleoprotein strategy offers safe delivery to protect endonuclease Cas9 and protect RNA from rapid degradation. However, some challenges must be considered such as low efficiency for cellular uptake, high risk of endotoxin contamination, and high immunogenicity (Chen et al., 2020; Yang et al., 2021).
Biomacromolecular therapeutic agents, including DNAs and RNAs, are often degraded in biological fluids (Doroudian et al., 2021b; Azhdari et al., 2022). Moreover, they are ineffective at crossing the cell‐membrane barrier due to their hydrophilicity and negative charge (Hanjani et al., 2022). Therefore, nanocarriers are frequently applied to overcome these limitations and these are frequently used to deliver CRISPR/Cas9 to target cells (Wan et al., 2021). Nano delivery systems enhance the stability of biomacromolecular therapeutic agents and protect them from premature degradation and rapid clearance in vivo, thereby enabling the delivery of their drug content to the target area (Eoh and Gu, 2019; Roma-Rodrigues et al., 2020; Kazemian et al., 2022). Nano delivery systems present a promising strategy for overcoming current challenges with CRISPR/Cas9 delivery. In this context, nanoparticles are used as carriers for the direct delivery of therapeutic components to the nuclei of targeted cells (Wang et al., 2017; Doroudian et al., 2021b). Nano delivery systems improve gene delivery by covering the negative charge of DNA, therefore preventing the cargo from intracellular nuclease degradation (Li et al., 2022). This is achieved by pressing DNA through electrostatic interactions between anionic DNA and polycations, encapsulating it with biodegradable polymers, or adsorbing it (Ibraheem et al., 2014; Doroudian et al., 2017). Chemical transfection transfers cargo into target cells using chemical vectors such as lipid vesicles and polymer-based chemicals. The chemical vectors work as a carrier system for encapsulating genetic-editing plasmid DNA and mRNA (Duan et al., 2021; Han et al., 2022; Mahmoud et al., 2022). Inorganic formats of nucleic acid delivery, such as gold nanoparticles, carbon nanotubes, and graphene, also produce significant performance. Protein and other biomolecules can also be delivered using chemical transfection (He et al., 2020; Hosseinkazemi et al., 2022). Polymeric carriers are chemically diverse and versatile. Polyethylenimine (PEI), chitosan, and poly (l-lysine) have been the most common polymeric DNA vectors (PLL) (Wang et al., 2017; Ashok et al., 2021).
Liposomes are promising nanodrug delivery systems owing to their similarity to the plasma membrane. Lipid-based nanoparticles are one of the most widely used nanoparticles for nucleic acid delivery (Filipczak et al., 2020). The lipid layer enhanced the ability of the CRISPR cargo to cross the cell membrane and also to preserve it from RNases, enzymatic degradation, and immune responses (Liu et al., 2018a; Liu et al., 2018b). Furthermore, Lipofectamine, RNAiMAX, TurboFectTM, and StemfectTM are commercially available lipid-based nanoparticles that have been employed for CRISPR plasmids, mRNA, gRNA, and Cas9-sgRNA RNP delivery (Mout et al., 2017; Givens et al., 2018; Chen et al., 2020). Liposomes have been shown to be promising carriers for CRISPR/Cas9 delivery. The lipid bilayer protects protein and nucleic acid from decomposition in the blood circulatory system (Qiu et al., 2021). It can also increase endosomal escape via fusing with the endosomal membrane (Quagliarini et al., 2021). It has been shown that CRISPR/sgRNA RNPs carried by cationic liposome could detect 65% of reporter gene disruption in U2OS EGFP cells, a human osteosarcoma cell line. It was also identified that Cas9/sgRNA RNP delivery via cationic liposomes results in an approximately 13% degradation of the reporter gene near the injection site of the inner ear in a murine in vivo study. Furthermore, it has been shown that cationic liposomes can be an effective vehicle for genome-editing platform delivery (Zuris et al., 2015). A temporary cytosolic delivery system for correct Cas9 ribonucleoprotein is critical in the CRISPR/Cas9 target specificity. The improvement of the delivery system remains the main challenge due to the large size of the Cas9 protein and a negative-strand RNA (Hou et al., 2021). To address this issue, a novel nanocarrier was employed, which was a size-controlled lipopeptide-based nanosome structure derived from the blood–brain barrier–permeable dNP2 peptide to deliver a hyper-accurate Cas9 RNP complex (HypaRNP) into human cells for gene editing (Lin et al., 2022). The use of encapsulated HypaRNP in an in vivo model resulted in rapid nuclear localization without causing cytotoxicity. Thus, the HypaRNP effectively silenced endogenous eGFP in human embryonic kidney cells and glioblastoma cells (Saadati and Hosseini, 2012; Thach et al., 2019). This strategy was also used to modify the dipeptidyl peptidase-4 gene (DPP-4) to upregulate the glucagon-like peptide 1 in a murine model of type 2 diabetes mellitus (T2DM) db/db mice. The nanocarrier, loaded with the Cas9 ribonucleoprotein complex effectively blocked the expression of the DPP-4 gene in T2DM mice. Normalized blood glucose levels followed the decrease in DPP-4 enzyme activity and improved insulin response with less liver and kidney damage. These effects were similar to those of sitagliptin, a new chemical DPP-4 inhibitor that requires repeated dosing. This demonstrates that a nano-liposomal carrier system containing therapeutic Cas9 ribonucleoprotein has enormous potential as a tool for improving gene therapy for human liver disorders (Cho et al., 2019). To deliver Cas9 mRNA and sgRNA, a liposome (LNP-INT01) was developed with biodegradable ionizable lipid, and PEG-DMG. LNP-INT01 half-life cycle in the liver was just around 6 h. As a result, it was safer than other forms of non-degradable liposomes used in treatment. LNP-INT01 consisting of Cas9 mRNA and sgRNA was supplied to CD-1 mice at various doses to downregulate serum TTR levels. Using a single dose at the peak of three mpk, serum TTR levels were reduced by more than 97 percent for more than 12 months compared to controls. Furthermore, it was confirmed that the majority of the LNP-INT01 was accumulated in the liver. Thus, approximately 70% of the resulting DNA editing was found in the liver. Furthermore, the spleen and kidney were shown to have very poor levels of editing, indicating organ specificity. As a result, this type of LNP-INT01 is intended to be used in clinical trials for liver-based genetic disorders (Table 3) (Finn et al., 2018).
Polymers are extensively used in drug delivery applications (Kanduri et al., 2021; Lin et al., 2022). The homology-independent targeted integration (HITI) approach, which has demonstrated improved site-specific transgene integration frequencies, facilitates successful CRISPR/Cas9-mediated knock-in of therapeutic genes in non-dividing cells in vivo. Supramolecular nanoparticle (SMNP) carriers were used in a study to deliver two DNA plasmids, the CRISPR/Cas9 genome-editing framework and a therapeutic gene, Retinoschisin 1 (RS1), allowing CRISPR/Cas9 knock-in of the RS1 gene with HITI (Chou et al., 2020). In another study, Xiao-Yan He et al. demonstrated that efficient reversal of tumor immunosuppression is vital in cancer therapy. A nano delivery system for CRISPR/Cas9 plasmid was developed to reverse tumor immunosuppression by silencing the β-catenin gene. As a targeted nano delivery system, it was decorated with aptamer-conjugated hyaluronic acid and peptide-conjugated hyaluronic acid. This strategy resulted in a dramatically increased plasmid accumulation in malignant cell nuclei. The genome-editing system can effectively induce β-catenin knockout and inhibit the Wnt/β-catenin pathway, resulting in substantially downregulated proteins associated with tumor growth and immunosuppression (He et al., 2020). Cationic polymer polyethyleneimine-β-cyclodextrin (PC), which is proven helpful for small plasmid transfection, can efficiently deliver Cas9 and sgRNA plasmids. However, PC can compact and capture large plasmids with a high N/P ratio but plasmids show results in effective editing at two genome loci, namely, hemoglobin subunit beta (19.1%) and rhomboid five homologs 1 (RHBDF1) (7.0%). Furthermore, Sanger sequencing indicates that genome editing at these loci was effective (Zhang et al., 2019). In another study, Ami Jo and colleagues developed a nanoparticle carrier using poly lactic-co-glycolic acid (PLGA) to deliver a CRISPR–Cas9 plasmid into the bone marrow. In this system, the authors engineered a PLGA-based nano delivery system composed of several copies of plasmid which have been fluorescently labeled with TIPS pentacene fluorophore which were then encapsulated by the PLGA polymer. Following 24-h of treatment, most DNA including two to three copies of plasmid per nanoparticle were released and in vitro screening results demonstrated a majority of TIPS pentacene positive cells and the detection of Cas9 proteins inside the cells (Jo et al., 2020). Rohiwal et al. also attempted to characterize nanoparticles used to deliver plasmid DNA of the CRISPR/Cas system in vitro to eukaryotic cells. Magnetic nanoparticles (MNPs) were synthesized implementing CRISPR/Cas9-complexed polyethyleneimine (PEI). They evaluated effective homology-directed repair (HDR) and non-homologous end-joining (NHEJ) events following nanoparticle transfection using a robust HEK-293 cell line expressing the traffic light reporter (TLR-3) system. MNPs were generated through the co-precipitation process, with an average particle size of about 20 nm. The dynamic light scattering and zeta potential measurements revealed that the NPs had a small size range and adequate colloidal stability. This method investigated the use of non-viral delivery of CRISPR/Cas9 and a DNA template to execute HDR and NHEJ in the same assay. The results show that PEI-MNPs are an effective delivery vehicle for plasmids form of CRISPR/Cas9 and template DNA in comparison with the standard lipofectamine transfection. Moreover, this type of delivery system has the potential to increase the safety and efficiency of genetic modifications (Rohiwal et al., 2020). Further research has also documented a sequence of reducible branched ester-amine quadpolymers (rBEAQs) that were generated and tested for their ability to co-encapsulate and deliver DNA plasmids and RNA oligonucleotides. The goal of using these engineered rBEAQs was to use polymer branching, reducibility, and hydrophobicity to efficiently co-encapsulate DNA and RNA in nanoparticles at low polymer-to–nucleic acid w/w ratios while enabling high distribution efficiency. Moreover, they also evaluated the development of a safe DNA–siRNA co-delivery and also non-viral CRISPR-mediated gene editing using Cas9 DNA and sgRNA co-delivery (Rui et al., 2019).
Gold is among the most commonly used metals in biomedicine as a result of its anti-inflammatory and antibiotic properties (Ko et al., 2022). In recent years, various applications of gold nanoparticles as vectors in therapeutic strategies have been investigated (Trigueros et al., 2019; He et al., 2021; Jahangiri-Manesh et al., 2022). Coelho et al. recently designed a drug delivery nanosystem based on pegylated gold nanoparticles with anthracycline and tyrosine kinase inhibitor doxorubicin, a small anti-cancer molecule, and varlitinib, an inhibitor of the tyrosine kinases, for a combination strategy against pancreatic cancer cells (Coelho et al., 2019). In a recent study, gold nanoparticles were used for combined gene and antimicrobial therapy by binding antimicrobial peptides with cationic gold nanoparticles for gene delivery to mesenchymal stem cells (Peng et al., 2016). While CRISPR/Cas-based fluorescent biosensors have been produced recently, they still need amplification for measurement. The first CRISPR/Cas12a-based nucleic acid amplification-free fluorescent biosensor has been newly produced by Jin-Ha Choi’s team to detect cfDNA through the metal-enhanced fluorescence phenomenon (MEF) using DNA-functionalized gold nanoparticles. Moreover, MEF occurred with color shifts from purple to red-purple after activating the CRISPR/Cas12a complex with the target cfDNA and corresponding single-strand DNA (ssDNA) degradation between gold nanoparticles and fluorophore. Breast cancer gene-1 (BRCA-1) can be diagnosed with high sensitivity in 30 min using this method. This fast and highly selective sensor can be used to quantify other nucleic acid biomarkers, including viral DNA, in a field-deployable and point-of-care testing (POCT) framework (Imani et al., 2011; Jahangiri-Manesh et al., 2022). Generally, the research by Rabiee and his collaborators emphasizes the role of various parameters in developing a non-viral vector in gene delivery carriers. Work is ongoing in the context of 2D crystals, precisely layered double hydroxides, because of their considerable adjustability and cost-effective preparation process. Hence, the relationship between various physicochemical properties parameters of LDH, such as pH, scale, zeta potential, and synthesis process, was evaluated and adjusted for CRISPR/Cas9 delivery and reverse fluorescence response to EGFP. ZnAl LDH and ZnAl HMTA LDH were produced, characterized, and used to deliver CRISPR/Cas9 to the HEK-293 cell line throughout this study. The analysis demonstrated adequate binding potential with the DNA, making it an effective vector for CRISPR/Cas9 delivery (Rabiee et al., 2020). Furthermore, another independent study showed the advantages of biomimetic cancer cell–coated zeolitic imidazolate frameworks (ZIFs) for cells-specific delivery of genome-editing systems. This study found that coating ZIF-8 with a cancer cell membrane and encapsulating CRISPR/Cas9 (CC-ZIF) resulted in evenly coated cell membrane–localized C3-ZIF. Furthermore, when C3-ZIFMCF was incubated with MCF-7, HeLa, HDFn, and aTC cell lines, MCF-7 cells presented the highest uptake, while non-cancerous cells showed minimal uptake. In terms of genome editing, MCF-7 was transfected with C3-ZIFMCF, which resulted in 3-fold repression of EGFP expression compared to 1-fold repression of EGFP expression when MCF-7 was transfected with C3-ZIFHELA. Alyami et al. described in vivo testing confirmed C3-ZIFMCF’s selectivity to aggregate in MCF-7 tumor cells (Alyami et al., 2020).
Viral vectors have been developed over the last 30 years for gene therapy to deliver various nucleic acid-based therapeutics, and some have been approved for clinical use. Despite their safety concerns and the possibility of introducing undesired mutations, viral delivery systems have one of the most efficient strategies to deliver plasmid-based nucleic acids to mammalian cells in vitro and in vivo thus far (Bulcha et al., 2021). Adenovirus and lentivirus have been employed frequently among all other viruses (Zheng et al., 2018). Adeno-associated viruses (AAVs) are single-stranded DNA viruses that contain two genes (Berns and Giraud, 1996). Recombinant gene delivery vector systems (rAAVs) were first generated based on AAV serotype 2 in the 1980s. Currently, rAAVs are the leading platform for in vivo delivery of gene therapies (Duncan, 2022). Nano-sized viral delivery systems offer numerous advantages, including the highest level of biological safety with limited pathogenic properties. In addition, rAAV provides efficient gene delivery and sustained transgene expression in multiple tissues, including muscle, lung, liver, and central nervous system. Furthermore, rAAV has enjoyed some success in human clinical trials. AAV is the most popular viral vector to treat cancer due to its oncolytic therapy ability (Senís et al., 2014; Ibraheim et al., 2018; Lino et al., 2018; Wang et al., 2020). For example, in vivo as well as ex vivo genome editing on KRAS, p53, and LKB1, using AAV, lentivirus-, or particle-mediated delivery of guide RNA in neurons, immune cells, and endothelial cells has been successfully accomplished due to simultaneously modeled dynamics of the top three significantly mutated genes in lung adenocarcinoma (Platt et al., 2014). In the viral delivery method, the adenovirus is the most important and most commonly used due to its broad range of serotype specificity, non-pathogenicity, and less immunogenicity (Chew et al., 2016; Li et al., 2019; Yu and Wu, 2019; Wang et al., 2020). In 2012, the EU approved the first AAV-based gene therapy drug, Glybera, for patients with lipoprotein lipase deficiency, suggesting the great promise of AAV for gene therapy. Recombined rAAV can reduce the off-targeting rate due to its capability to identify the target cell more efficiently (Gaj et al., 2016; Bak and Porteus, 2017; Hanlon et al., 2019; Krooss et al., 2020). In the long term, it is important that we investigate further the integration of the CRISPR/Cas9 system in host DNA (Li et al., 2015; Heck and Brault, 2018; Uchida et al., 2021).
The CRISPR/Cas9 system is a practical, low cost, and highly efficient tool, recently used for genome editing in comparison with other genome-editing tools such as TALEN and ZFN. Although, choosing a suitable carrier for delivery is still challenging, the presence of a wide range of delivery materials is resulting in improved access. Furthermore, physical, chemical, and biological transfer methods have their own unique advantages and disadvantages. In this article, we have focused on these very advantages and disadvantages to highlight the recent advances in the field to provide a balanced outlook on current developments. As for future perspectives, one of the most challenging parts of genomic editing technology, particularly in clinical applications, is to develop active targeting nanodrug delivery systems. This strategy provides efficient and effective tools for the manipulation of certain cells/tissues. Smart nanodrug delivery systems, including targeted nanodrug carriers and stimuli-responsive nanoparticles, are promising novel approaches that can be employed to tackle these challenges. These novel strategies would provide a near-term future in which the therapy of genetic disorders is within our reach.
FH: Writing—original draft. MA: Writing—original draft. ST: review and editing. AO’n: Writing—review and editing. FF: Writing—review and editing. Mohammad Doroudian: Project administration, writing—original draft, review, and editing, artwork. The first two authors contributed equally to this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aghasadeghi, M. R., Heidari, H., M Sadat, S., Irani, S., Amini, S., D Siadat, S., et al. (2013). Lamivudine-PEGylated chitosan: A novel effective nanosized antiretroviral agent. Curr. HIV Res. 11 (4), 309–320. doi:10.2174/1570162x113119990043
Ahmadi Danesh, H., Saadati, M., Doroudian, M., Yakhchali, B., Karkhaneh, A. A., Hosseini, S. M., et al. (2012). Cloning virG gene and generation mutant construct of pGEM∆ virG in order to induction recombination in native Shigella dysenteriae. Med. Sci. 22 (3), 184–190.
Alyami, M. Z., Alsaiari, S. K., Li, Y., Qutub, S. S., Aleisa, F. A., Sougrat, R., et al. (2020). Cell-type-specific CRISPR/Cas9 delivery by biomimetic metal organic frameworks. J. Am. Chem. Soc. 142 (4), 1715–1720. doi:10.1021/jacs.9b11638
Ashok, B., Peppas, N. A., and Wechsler, M. E. (2021). Lipid-and polymer-based nanoparticle systems for the delivery of CRISPR/Cas9. J. Drug Deliv. Sci. Technol. 65, 102728. doi:10.1016/j.jddst.2021.102728
Azhdari, M. H., Goodarzi, N., Doroudian, M., and MacLoughlin, R. (2022). Molecular insight into the therapeutic effects of stem cell-derived exosomes in respiratory diseases and the potential for pulmonary delivery. Int. J. Mol. Sci. 23 (11), 6273. doi:10.3390/ijms23116273
Bak, R. O., and Porteus, M. H. (2017). CRISPR-mediated integration of large gene cassettes using AAV donor vectors. Cell Rep. 20 (3), 750–756. doi:10.1016/j.celrep.2017.06.064
Berns, K., and Giraud, C. (1996). Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218, 1–23.
Birkeland, N-K., Schönheit, P., Poghosyan, L., Fiebig, A., and Klenk, H. P. (2017). Complete genome sequence analysis of Archaeoglobus fulgidus strain 7324 (DSM 8774), a hyperthermophilic archaeal sulfate reducer from a North Sea oil field. Stand. Genomic Sci. 12 (1), 79. doi:10.1186/s40793-017-0296-5
Bulcha, J. T., Wang, Y., Ma, H., Tai, P. W. L., and Gao, G. (2021). Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 6 (1), 53. doi:10.1038/s41392-021-00487-6
Chen, F., Alphonse, M., and Liu, Q. (2020). Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 12 (3), e1609. doi:10.1002/wnan.1609
Chen, M., Mao, A., Xu, M., Weng, Q., Mao, J., and Ji, J. (2019). CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett. 447, 48–55. doi:10.1016/j.canlet.2019.01.017
Cheng, H., Zhang, F., and Ding, Y. (2021). CRISPR/Cas9 delivery system engineering for genome editing in therapeutic applications. Pharmaceutics 13 (10), 1649. doi:10.3390/pharmaceutics13101649
Chew, W. L., Tabebordbar, M., Cheng, J. K., Mali, P., Wu, E. Y., Ng, A. H., et al. (2016). A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Methods 13 (10), 868–874. doi:10.1038/nmeth.3993
Cho, E. Y., Ryu, J-Y., Lee, H. A. R., Hong, S. H., Park, H. S., Hong, K. S., et al. (2019). Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes. J. Nanobiotechnology 17 (1), 19. doi:10.1186/s12951-019-0452-8
Chou, S. J., Yang, P., Ban, Q., Yang, Y. P., Wang, M. L., Chien, C. S., et al. (2020). Dual supramolecular nanoparticle vectors enable CRISPR/Cas9-Mediated knockin of Retinoschisin 1 gene—a potential nonviral therapeutic solution for X-linked juvenile retinoschisis. Adv. Sci. (Weinh). 7 (10), 1903432. doi:10.1002/advs.201903432
Coelho, S. C., Reis, D. P., Pereira, M. C., and Coelho, M. A. N. (2019). Doxorubicin and varlitinib delivery by functionalized gold nanoparticles against human pancreatic adenocarcinoma. Pharmaceutics 11 (11), 551. doi:10.3390/pharmaceutics11110551
Deng, H., Huang, W., and Zhang, Z. (2019). Nanotechnology based CRISPR/Cas9 system delivery for genome editing: Progress and prospect. Nano Res. 12 (10), 2437–2450. doi:10.1007/s12274-019-2465-x
Doroudian, M., Azhdari, M. H., Goodarzi, N., O’Sullivan, D., and Donnelly, S. C. (2021b). Smart nanotherapeutics and lung cancer. Pharmaceutics 13 (11), 1972. doi:10.3390/pharmaceutics13111972
Doroudian, M., MacLoughlin, R., Poynton, F., Prina-Mello, A., and Donnelly, S. C. (2019). Nanotechnology based therapeutics for lung disease. Thorax 74 (10), 965–976. doi:10.1136/thoraxjnl-2019-213037
Doroudian, M., O'Neill, A., O'Reilly, C., Tynan, A., Mawhinney, L., McElroy, A., et al. (2020). Aerosolized drug-loaded nanoparticles targeting migration inhibitory factors inhibit Pseudomonas aeruginosa-induced inflammation and biofilm formation. Nanomedicine 15 (30), 2933–2953. doi:10.2217/nnm-2020-0344
Doroudian, M., O’Neill, A., Mac Loughlin, R., Prina-Mello, A., Volkov, Y., and Donnelly, S. C. (2021a). Nanotechnology in pulmonary medicine. Curr. Opin. Pharmacol. 56, 85–92. doi:10.1016/j.coph.2020.11.002
Doroudian, M., Poynton, F., O'Reilly, C., Crosbie-Staunton, K., MacLoughlin, R., Prina-Mello, A., et al. (2017). Aerosolized nanodrug delivery system to inhibit Macrophage migration inhibitory factor in order to treat lung cancer. Dublin: Future Medicine. 20(1):729-735.
Doroudian, M., Saadati, M., Ghahroudi, A., Jahantigh, D., Salehi, A. R., Hosseini, M., et al. (2013). Generation mutant construct of pgem-7zf::ΔIcsa (catr) for attenuation of Shigella flexneri 2A. Iran. J. Infect. Dis. Trop. Med. 17 (59), 41–47.
Duan, L., Ouyang, K., Xu, X., Xu, L., Wen, C., Zhou, X., et al. (2021). Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front. Genet. 12, 673286. doi:10.3389/fgene.2021.673286
Duncan, G. A. (2022). Integrative approaches to enhance adeno-associated viral gene delivery. J. Control. Release 341, 44–50. doi:10.1016/j.jconrel.2021.11.013
Eoh, J., and Gu, L. (2019). Biomaterials as vectors for the delivery of CRISPR–Cas9. Biomater. Sci. 7 (4), 1240–1261. doi:10.1039/c8bm01310a
Filipczak, N., Pan, J., Yalamarty, S. S. K., and Torchilin, V. P. (2020). Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 156, 4–22. doi:10.1016/j.addr.2020.06.022
Finn, J. D., Smith, A. R., Patel, M. C., Shaw, L., Youniss, M. R., van Heteren, J., et al. (2018). A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 22 (9), 2227–2235. doi:10.1016/j.celrep.2018.02.014
Gaj, T., Epstein, B. E., and Schaffer, D. V. (2016). Genome engineering using adeno-associated virus: Basic and clinical research applications. Mol. Ther. 24 (3), 458–464. doi:10.1038/mt.2015.151
Givens, B. E., Naguib, Y. W., Geary, S. M., Devor, E. J., and Salem, A. K. (2018). Nanoparticle-based delivery of CRISPR/Cas9 genome-editing therapeutics. AAPS J. 20 (6), 108. doi:10.1208/s12248-018-0267-9
Guzmán, N. M., Esquerra-Ruvira, B., and Mojica, F. J. M. (2021). Digging into the lesser-known aspects of CRISPR biology. Int. Microbiol. 24 (4), 473–498. doi:10.1007/s10123-021-00208-7
Han, J. P., Kim, M., Choi, B. S., Lee, J. H., Lee, G. S., Jeong, M., et al. (2022). In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy. Sci. Adv. 8 (3), eabj6901. doi:10.1126/sciadv.abj6901
Hanjani, N. A., Esmaelizad, N., Zanganeh, S., Gharavi, A. T., Heidarizadeh, P., Radfar, M., et al. (2022). Emerging role of exosomes as biomarkers in cancer treatment and diagnosis. Crit. Rev. Oncology/Hematology 169, 103565. doi:10.1016/j.critrevonc.2021.103565
Hanlon, K. S., Kleinstiver, B. P., Garcia, S. P., Zaborowski, M. P., Volak, A., Spirig, S. E., et al. (2019). High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 10 (1), 4439. doi:10.1038/s41467-019-12449-2
Hasanzadeh, A., Noori, H., Jahandideh, A., Haeri Moghaddam, N., Kamrani Mousavi, S. M., Nourizadeh, H., et al. (2022). Smart strategies for precise delivery of CRISPR/Cas9 in genome editing. ACS Appl. Bio Mat. 5 (2), 413–437. doi:10.1021/acsabm.1c01112
He, J-S., Liu, S-J., Zhang, Y-R., Chu, X-D., Lin, Z-B., Zhao, Z., et al. (2021). The application of and strategy for gold nanoparticles in cancer immunotherapy. Front. Pharmacol. 12, 687399. doi:10.3389/fphar.2021.687399
He, X. Y., Ren, X. H., Peng, Y., Zhang, J. P., Ai, S. L., Liu, B. Y., et al. (2020). Aptamer/peptide-functionalized genome-editing system for effective immune restoration through reversal of PD-L1-mediated cancer immunosuppression. Adv. Mat. 32 (17), 2000208. doi:10.1002/adma.202000208
Heck, M., and Brault, V. (2018). Targeted disruption of aphid transmission: A vision for the management of crop diseases caused by luteoviridae members. Curr. Opin. Virol. 33, 24–32. doi:10.1016/j.coviro.2018.07.007
Hosseinkazemi, H., Samani, S., O’Neill, A., Soezi, M., Moghoofei, M., Azhdari, M. H., et al. (2022). Applications of iron oxide nanoparticles against breast cancer. J. Nanomater. 2022, 12. doi:10.1155/2022/6493458
Hou, X., Zaks, T., Langer, R., and Dong, Y. (2021). Lipid nanoparticles for mRNA delivery. Nat. Rev. Mat. 6 (12), 1078–1094. doi:10.1038/s41578-021-00358-0
Ibraheem, D., Elaissari, A., and Fessi, H. (2014). Gene therapy and DNA delivery systems. Int. J. Pharm. X. 459 (1-2), 70–83. doi:10.1016/j.ijpharm.2013.11.041
Ibraheim, R., Song, C-Q., Mir, A., Amrani, N., Xue, W., and Sontheimer, E. J. (2018). All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 19 (1), 137. doi:10.1186/s13059-018-1515-0
Imani, S., Zagari, Z., Rezaei-Zarchi, S., Zand, A. M., Dorodiyan, M., Bariabarghoyi, H., et al. (2011). Antibacterial effect of CrO and CoFe2O4 nanoparticles upon Staphylococcus aureus. JABS 1 (3), 175–181.
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 (12), 5429–5433. doi:10.1128/jb.169.12.5429-5433.1987
Jahangiri-Manesh, A., Mousazadeh, M., Taji, S., Bahmani, A., Zarepour, A., Zarrabi, A., et al. (2022). Gold nanorods for drug and gene delivery: An overview of recent advancements. Pharmaceutics 14 (3), 664. doi:10.3390/pharmaceutics14030664
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Jo, A., Ringel-Scaia, V. M., McDaniel, D. K., Thomas, C. A., Zhang, R., Riffle, J. S., et al. (2020). Fabrication and characterization of PLGA nanoparticles encapsulating large CRISPR–Cas9 plasmid. J. Nanobiotechnology 18 (1), 16. doi:10.1186/s12951-019-0564-1
Kanduri, V., LaVigne, D., and Larsen, J. (2021). Current advances toward the encapsulation of Cas9. ACS Macro Lett. 10 (12), 1576–1589. doi:10.1021/acsmacrolett.1c00538
Kazemian, P., Yu, S-Y., Thomson, S. B., Birkenshaw, A., Leavitt, B. R., and Ross, C. J. (2022). Lipid-nanoparticle-based delivery of CRISPR/Cas9 genome-editing components. Mol. Pharm. 19, 1669–1686. doi:10.1021/acs.molpharmaceut.1c00916
Ko, W-C., Wang, S-J., Hsiao, C-Y., Hung, C-T., Hsu, Y-J., Chang, D-C., et al. (2022). Pharmacological role of functionalized gold nanoparticles in disease applications. Molecules 27 (5), 1551. doi:10.3390/molecules27051551
Krooss, S. A., Dai, Z., Schmidt, F., Rovai, A., Fakhiri, J., Dhingra, A., et al. (2020). Ex vivo/in vivo gene editing in hepatocytes using “all-in-one” CRISPR-adeno-associated virus vectors with a self-linearizing repair template. iScience 23 (1), 100764. doi:10.1016/j.isci.2019.100764
Li, A., Lee, C. M., Hurley, A. E., Jarrett, K. E., De Giorgi, M., Lu, W., et al. (2019). A self-deleting AAV-CRISPR system for in vivo genome editing. Mol. Ther. - Methods & Clin. Dev. 12, 111–122. doi:10.1016/j.omtm.2018.11.009
Li, L., He, Z-Y., Wei, X-W., Gao, G-P., and Wei, Y-Q. (2015). Challenges in CRISPR/CAS9 delivery: Potential roles of nonviral vectors. Hum. Gene Ther. 26 (7), 452–462. doi:10.1089/hum.2015.069
Li, Q., Lv, X., Tang, C., and Yin, C. (2022). Co-delivery of doxorubicin and CRISPR/Cas9 or RNAi-expressing plasmid by chitosan-based nanoparticle for cancer therapy. Carbohydr. Polym. 287, 119315. doi:10.1016/j.carbpol.2022.119315
Lin, Y., Wagner, E., and Lächelt, U. (2022). Non-viral delivery of the CRISPR/cas system: DNA versus RNA versus RNP. Biomater. Sci. 10 (5), 1166–1192. doi:10.1039/d1bm01658j
Lino, C. A., Harper, J. C., Carney, J. P., and Timlin, J. A. (2018). Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. (Lond). 25 (1), 1234–1257. doi:10.1080/10717544.2018.1474964
Liu, K., Cui, J-J., Zhan, Y., Ouyang, Q-Y., Lu, Q-S., Yang, D-H., et al. (2022). Reprogramming the tumor microenvironment by genome editing for precision cancer therapy. Mol. Cancer 21 (1), 98. doi:10.1186/s12943-022-01561-5
Liu, Q., Zhu, H., Liu, Y., Musetti, S., and Huang, L. (2018b). BRAF peptide vaccine facilitates therapy of murine BRAF-mutant melanoma. Cancer Immunol. Immunother. 67 (2), 299–310. doi:10.1007/s00262-017-2079-7
Liu, Q., Zhu, H., Tiruthani, K., Shen, L., Chen, F., Gao, K., et al. (2018a). Nanoparticle-mediated trapping of Wnt family member 5A in tumor microenvironments enhances immunotherapy for B-Raf proto-oncogene mutant melanoma. ACS Nano 12 (2), 1250–1261. doi:10.1021/acsnano.7b07384
Mahmoud, L. M., Kaur, P., Stanton, D., Grosser, J. W., and Dutt, M. (2022). A cationic lipid mediated CRISPR/Cas9 technique for the production of stable genome edited citrus plants. Plant methods 18 (1), 33. doi:10.1186/s13007-022-00870-6
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 18 (2), 67–83. doi:10.1038/s41579-019-0299-x
Mohanraju, P., Saha, C., van Baarlen, P., Louwen, R., Staals, R. H. J., and van der Oost, J. (2022). Alternative functions of CRISPR–Cas systems in the evolutionary arms race. Nat. Rev. Microbiol. 20 (6), 351–364. doi:10.1038/s41579-021-00663-z
Mout, R., Ray, M., Tonga, G. Y., Lee, Y-W., Tay, T., Sasaki, K., et al. (2017). Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 11 (3), 2452–2458. doi:10.1021/acsnano.6b07600
Nambiar, T. S., Baudrier, L., Billon, P., and Ciccia, A. (2022). CRISPR-based genome editing through the lens of DNA repair. Mol. Cell 82 (2), 348–388. doi:10.1016/j.molcel.2021.12.026
Nidhi, S., Anand, U., Oleksak, P., Tripathi, P., Lal, J. A., Thomas, G., et al. (2021). Novel CRISPR–cas systems: An updated review of the current achievements, applications, and future research perspectives. Int. J. Mol. Sci. 22 (7), 3327. doi:10.3390/ijms22073327
Peng, L-H., Huang, Y-F., Zhang, C-Z., Niu, J., Chen, Y., Chu, Y., et al. (2016). Integration of antimicrobial peptides with gold nanoparticles as unique non-viral vectors for gene delivery to mesenchymal stem cells with antibacterial activity. Biomaterials 103, 137–149. doi:10.1016/j.biomaterials.2016.06.057
Petassi, M. T., Hsieh, S-C., and Peters, J. E. (2020). Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. Cell 183 (7), 1757–1771.e18. e1718. doi:10.1016/j.cell.2020.11.005
Pinilla-Redondo, R., Russel, J., Mayo-Muñoz, D., Shah Shiraz, A., Garrett Roger, A., Nesme, J., et al. (2021). CRISPR-Cas systems are widespread accessory elements across bacterial and archaeal plasmids. Nucleic Acids Res. 50 (8), 4315–4328. doi:10.1093/nar/gkab859
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159 (2), 440–455. doi:10.1016/j.cell.2014.09.014
Qiu, M., Li, Y., Bloomer, H., and Xu, Q. (2021). Developing biodegradable lipid nanoparticles for intracellular mRNA delivery and genome editing. Acc. Chem. Res. 54 (21), 4001–4011. doi:10.1021/acs.accounts.1c00500
Quagliarini, E., Renzi, S., Digiacomo, L., Giulimondi, F., Sartori, B., Amenitsch, H., et al. (2021). Microfluidic formulation of DNA-loaded multicomponent lipid nanoparticles for gene delivery. Pharmaceutics 13 (8), 1292. doi:10.3390/pharmaceutics13081292
Rabiee, N., Bagherzadeh, M., Ghadiri, A. M., Salehi, G., Fatahi, Y., and Dinarvand, R. (2020). ZnAl nano layered double hydroxides for dual functional CRISPR/Cas9 delivery and enhanced green fluorescence protein biosensor. Sci. Rep. 10 (1), 20672. doi:10.1038/s41598-020-77809-1
Ramachandran, A., Summerville, L., Learn, B. A., DeBell, L., and Bailey, S. (2020). Processing and integration of functionally oriented prespacers in the Escherichia coli CRISPR system depends on bacterial host exonucleases. J. Biol. Chem. 295 (11), 3403–3414. doi:10.1074/jbc.ra119.012196
Rohiwal, S., Dvorakova, N., Klima, J., Vaskovicova, M., Senigl, F., Slouf, M., et al. (2020). Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci. Rep. 10 (1), 4619. doi:10.1038/s41598-020-61465-6
Roma-Rodrigues, C., Rivas-García, L., Baptista, P. V., and Fernandes, A. R. (2020). Gene therapy in cancer treatment: Why go nano? Pharmaceutics 12 (3), 233. doi:10.3390/pharmaceutics12030233
Rui, Y., Wilson, D. R., Sanders, K., and Green, J. J. (2019). Reducible branched ester-amine quadpolymers (rBEAQs) codelivering plasmid DNA and RNA oligonucleotides enable CRISPR/Cas9 genome editing. ACS Appl. Mat. Interfaces 11 (11), 10472–10480. doi:10.1021/acsami.8b20206
Saadati, M., and Hosseini, S. M. (2012). Engineered and construction of pDS132::∆ virG as suicide vector for targeted gene deletion of virG from Shigella flexneri 2a in order to generation a live attenuated Shigella vaccine. JABS 1 (4), 197–205.
Salehi Chaleshtori, A., Saadati, M., Ahsani Arani, Y., Bahador, A., Hosseini, M., Hesaraki, M., et al. (2012). Evaluation of live attenuated Shigella dysenteriae type 1 strain as a candidate vaccine. J. Police Med. 1 (1), 31–39.
Senís, E., Fatouros, C., Große, S., Wiedtke, E., Niopek, D., Mueller, A. K., et al. (2014). CRISPR/Cas9-mediated genome engineering: An adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 9 (11), 1402–1412. doi:10.1002/biot.201400046
Shmakov, S., Smargon, A., Scott, D., Cox, D., Pyzocha, N., Yan, W., et al. (2017). Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 15 (3), 169–182. doi:10.1038/nrmicro.2016.184
Smyth, S. J. (2022). Contributions of genome editing technologies towards improved nutrition, environmental sustainability and poverty reduction. Front. Genome Ed. 15, 863193. doi:10.3389/fgeed.2022.863193
Thach, T. T., Bae, D. H., Kim, N. H., Kang, E. S., Lee, B. S., Han, K., et al. (2019). Lipopeptide-based nanosome-mediated delivery of hyperaccurate CRISPR/Cas9 ribonucleoprotein for gene editing. Small 15 (46), 1903172. doi:10.1002/smll.201903172
Trigueros, S., B Domènech, E., Toulis, V., and Marfany, G. (2019). In vitro gene delivery in retinal pigment epithelium cells by plasmid dna-wrapped gold nanoparticles. Genes (Basel). 10 (4), 289. doi:10.3390/genes10040289
Uchida, N., Drysdale, C. M., Nassehi, T., Gamer, J., Yapundich, M., DiNicola, J., et al. (2021). Cas9 protein delivery non-integrating lentiviral vectors for gene correction in sickle cell disease. Mol. Ther. - Methods & Clin. Dev. 21, 121–132. doi:10.1016/j.omtm.2021.02.022
Uddin, F., Rudin, C. M., and Sen, T. (2020). CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 10, 1387. doi:10.3389/fonc.2020.01387
Vo, P. L. H., Ronda, C., Klompe, S. E., Chen, E. E., Acree, C., Wang, H. H., et al. (2021). CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 39 (4), 480–489. doi:10.1038/s41587-020-00745-y
Wan, F., Draz, M. S., Gu, M., Yu, W., Ruan, Z., and Luo, Q. (2021). Novel strategy to combat antibiotic resistance: A sight into the combination of CRISPR/cas9 and nanoparticles. Pharmaceutics 13 (3), 352. doi:10.3390/pharmaceutics13030352
Wang, D., Zhang, F., and Gao, G. J. C. (2020). CRISPR-Based therapeutic genome editing: Strategies and in vivo delivery by AAV vectors. Cell 181 (1), 136–150. doi:10.1016/j.cell.2020.03.023
Wang, H-X., Li, M., Lee, C. M., Chakraborty, S., Kim, H-W., Bao, G., et al. (2017). CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 117 (15), 9874–9906. doi:10.1021/acs.chemrev.6b00799
Wei, T., Cheng, Q., Min, Y-L., Olson, E. N., and Siegwart, D. J. (2020). Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11 (1), 3232. doi:10.1038/s41467-020-17029-3
Yan, J., Kang, D. D., Turnbull, G., and Dong, Y. (2022). Delivery of CRISPR-Cas9 system for screening and editing RNA binding proteins in cancer. Adv. Drug Deliv. Rev. 180, 114042. doi:10.1016/j.addr.2021.114042
Yang, P., Lee, A. Y-T., Xue, J., Chou, S-J., Lee, C., Tseng, P., et al. (2022). Nano-vectors for CRISPR/Cas9-mediated genome editing. Nano Today 44, 101482. doi:10.1016/j.nantod.2022.101482
Yang, Y., Xu, J., Ge, S., and Lai, L. (2021). CRISPR/Cas: Advances, limitations, and applications for precision cancer research. Front. Med. 8, 649896. doi:10.3389/fmed.2021.649896
Yu, W., and Wu, Z. (2019). “Use of AAV vectors for CRISPR-mediated in vivo genome editing in the retina,” in Adeno-associated virus vectors (Maryland: Springer), 123–139.
Zhang, Z., Wan, T., Chen, Y., Chen, Y., Sun, H., Cao, T., et al. (2019). Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid Commun. 40 (5), 1800068. doi:10.1002/marc.201800068
Zhang, S., Shen, J., Li, D., and Cheng, Y. (2021). Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 11 (2), 614–648. doi:10.7150/thno.47007
Zhang, X., Jin, H., Huang, X., Chaurasiya, B., Dong, D., Shanley, T. P., et al. (2022). Robust genome editing in adult vascular endothelium by nanoparticle delivery of CRISPR-Cas9 plasmid DNA. Cell Rep. 38 (1), 110196. doi:10.1016/j.celrep.2021.110196
Zheng, C-X., Wang, S-M., Bai, Y-H., Luo, T-T., Wang, J-Q., Dai, C-Q., et al. (2018). Lentiviral vectors and adeno-associated virus vectors: Useful tools for gene transfer in pain research. Anat. Rec. Hob. 301 (5), 825–836. doi:10.1002/ar.23723
Keywords: nanocarriers, CRISPR/Cas9, gene therapy, nanomedicine, non-viral vector
Citation: Hejabi F, Abbaszadeh MS, Taji S, O’Neill A, Farjadian F and Doroudian M (2022) Nanocarriers: A novel strategy for the delivery of CRISPR/Cas systems. Front. Chem. 10:957572. doi: 10.3389/fchem.2022.957572
Received: 31 May 2022; Accepted: 04 July 2022;
Published: 26 July 2022.
Edited by:
Ahmad Gholami, Shiraz University of Medical Sciences, IranReviewed by:
Lin Li, Northwestern Polytechnical University, ChinaCopyright © 2022 Hejabi, Abbaszadeh, Taji, O’Neill, Farjadian and Doroudian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Doroudian, ZG9yb3VkaWFuQGtodS5hYy5pcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.