
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 08 July 2022
Sec. Organic Chemistry
Volume 10 - 2022 | https://doi.org/10.3389/fchem.2022.948687
This article is part of the Research Topic Bioactive Natural Products-Oriented Synthetic and Functional Studies View all 24 articles
Quorum sensing plays a necessary role in the production of virulence factors and the formation of biofilm on Pseudomonas aeruginosa. Thus, the development of inhibition of quorum sensing is one of the most promising methods to control bacterial infection and antibiotic resistance. In this work, nine novel AHL analogs were designed, synthesized, and evaluated as potential quorum sensing inhibitors. The results depicted that structural modifications have significant effects on quorum sensing inhibition activity of AHL molecules. Without inhibiting the growth of P. aeruginosa, 2-(4-bromophenyl)-N-(2-oxotetrapyridinefuran-3-yl) butanamide (compound no.10) showed the excellent performance in inhibiting biofilm formation and virulence factor production among all the compounds through robustly suppressing the expression of QS related genes. In a molecular docking study, compound no.10 exhibited a higher affinity toward LasR than other AHL analogs. In addition, compound no.10 also exhibits the best inhibition effect on virulence production in the Caenorhabditis elegans infection model.
Pseudomonas aeruginosa as an important nosocomial pathogen could threaten those who are long-term intubated and immunocompromised, and patients suffering from cystic fibrosis, traumatized cornea, burns, Gustilo open fractures, and lead to lethal infections (Hogardt and Heesemann, 2013; Moradali et al., 2017). A previous study reported that biofilm formation and virulence factor production are the major causes of P. aeruginosa infections (Schuster and Greenberg, 2006). At present, antibiotics such as tobramycin, meropenem, or ciprofloxacin, are considered the first choice in treating infections of P. aeruginosa. However, the wild use of antibiotics to treat infections of P. aeruginosa has led to the generation of multidrug-resistant pathogens (Abbas et al., 2017). Thus, combating P. aeruginosa infections need a new target.
Quorum sensing (QS) is a bacteria-to-bacteria communication system. Also, QS is moderated by the production of different kinds of signal molecules known as autoinducers, which are usually N-acyl-homoserine lactones (AHLs) in Gram-negative bacteria (Ng and Bassler, 2009). In P. aeruginosa, the QS system is composed of four interconnected systems (Las, Rhl, PQS, and IQS), among them, the LasI/LasR system is the most representative and primary (Hançer Aydemir et al., 2018). In addition, each system utilizes a particular signal molecule such as N-(3-oxododecanoyl)-L-homoserine lactone (OdDHL), N-butanoyl-L-homoserine lactone (BHL), 2-heptyl-3-hydroxy-4-quinolone, and 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde (Hao et al., 2021). Typically, the accumulation of signal molecules interacts with a transcriptional activator protein (Abbas et al., 2017), and then the expression of QS-regulated genes were activated (Schuster and Greenberg, 2006).
It is reported that pathogenicity of P. aeruginosa is generally related to virulence production and biofilm formation (Bassler and Losick, 2006; Hançer Aydemir et al., 2018). As QS inhibitors (QSIs) had no influence on bacterial growth, they are believed to generate weaker selection for antibiotic resistance than conventional antibiotics. Thus, QSIs can be a novel and promising way to attenuate P. aeruginosa infection (Hançer Aydemir et al., 2018). Nowadays, numerous QS signal molecule analogs and plant-derived natural substances have been reported as QSIs to interfere with the QS system of P. aeruginosa (Qu et al., 2016; Abbas et al., 2020; Hao et al., 2021; Saqr et al., 2021). Unfortunately, its dose-dependent toxicity and a weak effect significantly limit clinical applications. Therefore, development of novel safe and effective QSIs is urgently needed for the treatment of P. aeruginosa infections.
In this study, nine novel AHL analogs were designed, synthesized, and evaluated as potential QSIs against P. aeruginosa. The effects of AHL analogs on biofilm formation and virulence factor production in vitro were evaluated. Furthermore, the mechanism was studied by related gene expression and molecular docking.
Pseudomonas aeruginosa PAO1 and Escherichia coli OP50 were stored in our laboratory. Caenorhabditis elegans N2 were obtained from the Caenorhabditis Genetics Center (CGC). P. aeruginosa PAO1 was cultured using Luria–Bertani broth (LB) at 37 °C and maintained on a nutrient agar plate (Solarbio, Beijing, China) at 4 °C. C. elegans was cultured using the nematode growth medium (NGM) at 20 °C and E. coli OP50 was used as the standard food source. 4-Br-PHL (compound no.1) and other AHL analogs (compound no.2 to compound no.10) were dissolved in DMSO before treatment (Solarbio, Beijing, China).
Based on the structures of OdDHL and BHL (Figure 1), we synthesized a series of novel AHL-based QSIs, in which the natural homoserine lactone ring is present but the natural acyl group was substituted. The structures of analogs were modified from three aspects: 1) the acyl group, 2) the length of the acyl side chain, and 3) the type and position of the substituents on the benzene ring. The synthesis method is shown in Figure 2. Those AHL analogs used in this study were synthesized by us and characterized by NMR, and MS with a purity of ≥95% (Supplementary Table S1). To circumvent the issues of different assessment results, we also synthesized N-(4-bromophenylacetanoyl)-L-homoserine lactone (4-Br-PHL, no. 1) as a positive control (Geske et al., 2005).
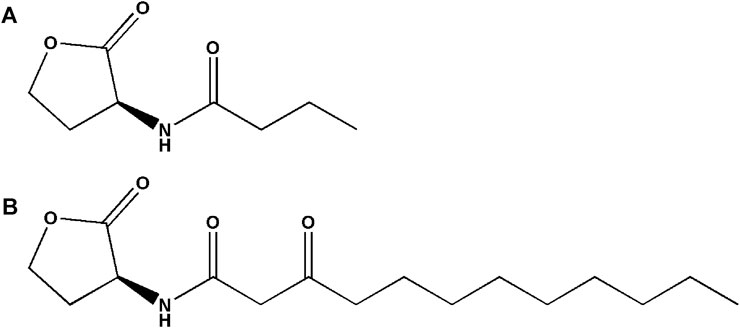
FIGURE 1. Chemical structure of P. aeruginosa PAO1 natural signal molecule. (A) Structure of BHL and (B) structure of OdDHL.
To initial screen compounds with QS-inhibited activity, QS-regulated biofilm formation was tested as the screening indicator. As described previously (Luo et al., 2017), biofilm mass was measured using a crystal violet assay.
The planktonic cell growth assay was slightly modified (Qu et al., 2016). Overnight cultures of P. aeruginosa PAO1 were diluted in LB broth. The suspension was supplemented with two compounds (no.3 and no.10) with concentrations ranging from 10 to 400 μM and incubated at 37 °C. Then, 1 ml samples were obtained every 2 h, and the turbidity was recorded at 600 nm using a spectrophotometer (Thermo scientific, Finland), then a growth curve was generated.
In this assay, the inhibitory effects of compound no.3 and compound no.10 on biofilm formation were further identified at different concentrations ranging from 50 to 400 μM. The method of this assay is similar to the method of initial screening of analogs. In addition, cover slides were incubated statically with or without 200 μM compound no.3 and compound no.10 at 37 °C to facilitate cell attachment and biofilm formation. Also, compound no.1 was conducted as a positive control. Finally, the structures of biofilm were observed using an upright microscope (BX53, Olympus, Japan) at ×20 magnification.
The swimming assay method was modified from Rashid and Kornberg (2000). Swimming agar plates (10 g/L peptone, 5 g/L NaCl, and 0.3% agarose) with compound no.3 and no.10 (200 μM) and blank control plates were center stabbed with P. aeruginosa PAO1 using sterile toothpick. Also, compound no.1 was conducted as a positive control. After incubating at 30 °C for 12–14 h, swimming zones were determined by measuring the diameter of circular expansion.
The swarming plate was composed of nutrient agar (8 g/L) and glucose (5.0 g/L). Then, swarming plates with and without supplementation of 200 μM compound no.3 and no.10 were added 5 μl overnight culture of P. aeruginosa PAO1. In addition, compound no.1 was conducted as a positive control. Then, the swarming plates were placed at 30 °C and incubated for 18–24 h (Kumar et al., 2013), and swarming zones were determined by measuring the diameter of circular expansion.
Twitching motility of P. aeruginosa PAO1 was modified from Saqr et al. (2021). A measure of 1% of LB agar with or without compounds (compound no.3 and compound no.10, 200 μM) stabbed with a toothpick up to bottom of the Petri dish from overnight culture P. aeruginosa PAO1 in order to evaluate twitching motility inhibition. Compound no.1 was conducted as a positive control, the agar was removed after incubation at 37 °C for 48 h, and then the plates were dried in air and stained with crystal violet. The dye was washed off with sterile water and the twitching zones were measured.
The assay of the inhibitory effect of compound no.3 and compound no.10 on pyocyanin production was conducted using previous methods (O'Loughlin et al., 2013). Overnight, P. aeruginosa PAO1 cultures were subcultured into LB broth with the compound no.3 and compound no.10 at the concentration of 200 μM for endpoint assays following 24 h of aerobic growth with shaking at 37 °C. Also, compound no.1 was conducted as a positive control. Finally, the production of pyocyanin was measured by recording the absorbance of this pink layer at 520 nm.
The skim milk agar method (Hao et al., 2021) was used to determine the effect of compound no.3 and compound no.10 on elastase activity. Also, compound no.1 was conducted as a positive control. After overnight culturing, P. aeruginosa PAO1 with or without compound no.3 and compound no.10 were incubated at 37 °C for 24 h at 180 rpm. After 24 h , the cultures were centrifuged at 10,000 rpm for 15 min. A measure of 100 μl of supernatant was added to the wells of skim milk agar and then plates were incubated for 24 h at 37 °C. The elastase activity was determined by measuring the diameter of clear zone.
To further investigate the anti-virulence effect of compound no.3 and compound no.10, we used a C. elegans life span assay. Also, compound no.1 was conducted as a positive control. This assay was modified from a previous study (Cezairliyan et al., 2013; O'Loughlin et al., 2013). After synchronization, C. elegans was grown to the L4 stage at 20 °C. In addition, an equivalent volume of compound no.1, compound no.3, and compound no.10 were mixed to the P. aeruginosa PAO1 cultures during growth and DMSO was added as a blank control. Then, an overnight culture of P. aeruginosa PAO1 was poured on Petri plates, containing PGS agar (1% peptone, 1% NaCl, 1% glucose, 150 mM sorbitol, and 1.7% agar) and kept for 24 h incubation at 37 °C. After 8–24 h, 25–30 L4 stage C. elegans were seeded with each plate. Then, live or dead worms were scored every 3 h at 25 °C and again at 24 h.
According to the previous study (Qu et al., 2016), the expression of key QS regulatory genes (lasI, lasR, rhlI, rhlR, and mvfR) of P. aeruginosa PAO1 was monitored in the presence of compound no.3 and compound no.10. Also, compound no.1 was conducted as a positive control. The primers for the genes are shown in Supplementary Table S2. The 16S rRNA housekeeping gene was served as an internal control. Total RNA was isolated with an Ultrapure RNA kit (CWBIO, Beijing, China), and the first-strand cDNA was synthesized with a Hifair® Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR (Yeasen, Beijing, China) according to the manufacturer’s instructions. The reaction was carried out with Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (Yeasen, Beijing, China) also according to the manufacturer. The relative gene expression was calculated using the 2−ΔΔCt method.
Molecular docking was conducted to analyze the interaction of compound no.3 and compound no.10 with the LasR receptor of P. aeruginosa PAO1. The crystal structure of P. aeruginosa PAO1 LasR was obtained from the Protein Data Bank (PDB ID:2UV0) (Bottomley et al., 2007). Hydrogen atoms were added according to the hydrogen network optimization and then minimized with an Amber19SB force field with UCSF Chimera. Finally, the protein structure was converted to a pdbqt file by prepare_receptor4.py script using AutoDockTools, and all the hydrogen atoms were kept. The ligands structures were built with UCSF Chimera, minimized with the GAFF force field, and converted to a pdbqt format by using the prepare_ligand4.py script with all hydrogen atoms. The docking site was determined by the P. aeruginosa PAO1 binding pocket. Finally, the molecular docking was performed by watvina (https://www.github.com/biocheming/watvina), which was optimized with new scorning function, with van der Waals, hydrogen bond, polar repulsion, and weak hydrogen bonds contribution. Conformation searching was performed by a simplified genetic algorithm and simulated annealing mixed with BFGS local optimization. Finally, the docking pose was optimized while the protein was kept fixed.
All experiments were conducted in triplicate to validate the reproducibility. All values are presented as the mean ± standard error. One-way ANOVA was performed using SPSS 23.0 software. The significance was accepted when the p-value was less than 0.05. Graphs were constructed using GraphPad Prism 7.0 software.
A series of novel AHL-based QS inhibitors are presented in Table 1. All compounds were characterized by NMR, MS with a purity of ≥95% (Supplementary Table S1).
The biofilm formation inhibition activity of compounds 1–10 was assessed. As shown in Table 1, all compounds showed inhibitory activity against biofilm formation of P. aeruginosa PAO1, except for compound no.6. Among all analogs, compound no.3 and compound no.10 exhibited the best inhibition activity and were identified as the potential QS-inhibitor to further evaluate in the following experiments.
The growth curves were drawn to identify the influence on the growth of P. aeruginosa PAO1. As shown in Figure 3, compound no.3 and compound no.10 had no inhibition effect on growth of P. aeruginosa PAO1 at the concentrations ranging from 10 to 400 μM.
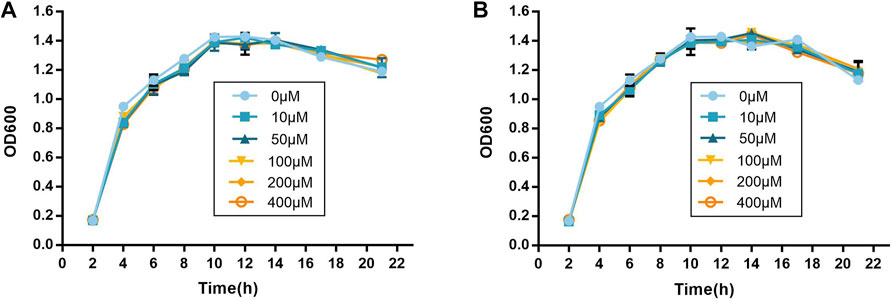
FIGURE 3. Inhibitory effect of compound no.3 and compound no.10 on (A) pyocyanin and (B) elastase productions of P. aeruginosa PAO1.
As shown in Figures 4A–C, inhibitory activities of compound no.3 and compound no.10 on biofilm formation were dose-dependent. Compound no.1 had slightly reduced biofilm formation at the concentration of 200 μM. By contrast, compound no.3 and compound no.10 had more obvious inhibitory effect on biofilm formation at the concentrations ranging from 200 to 400 μM. In the presence of compound no.3, the biofilm formation of P. aeruginosa PAO1 was decreased to nearly 35% at the concentration ranging from 50 to 400 μM. Remarkedly, compound no.10 inhibited more than 60% of biofilm formation at above 200 μM.
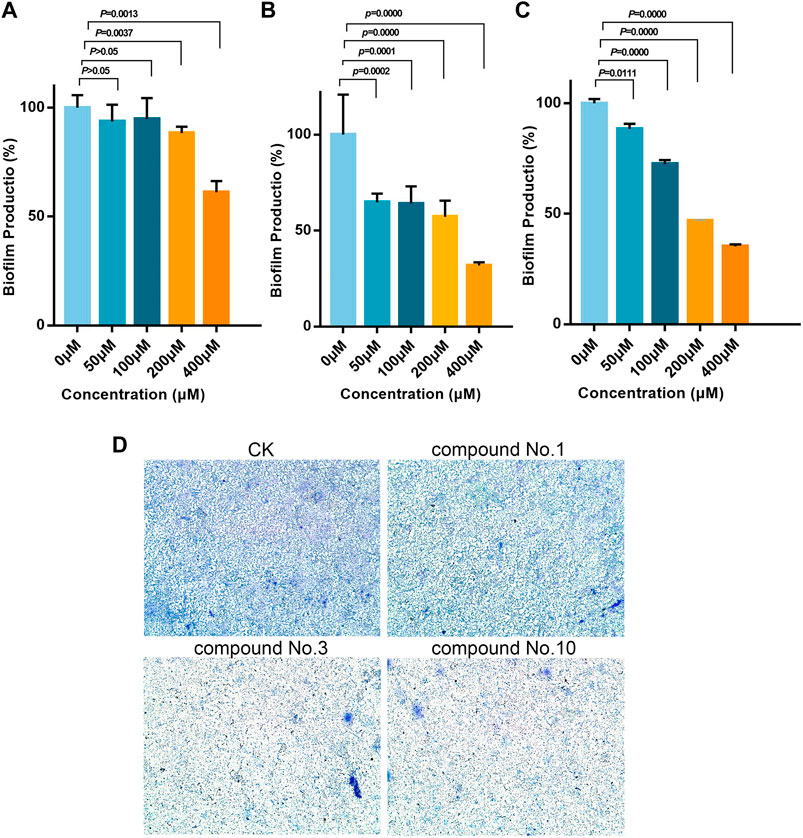
FIGURE 4. Inhibitory effect of compound no.3 and compound no.10 on P. aeruginosa biofilm formation. Biofilm was stained by crystal violet and measured as absorbance at 570 nm. (A) Compound no.1, (B) compound no.3, (C) and compound no.10. (D) Representative microscope images of treated with compounds at 200 μM.
The aforementioned results were further revealed under a microscope. From Figure 4D, reduction of biofilm attached to the surface of plates was observed in treated groups. Compared with compound no.1, the reduction of biofilms was more obvious in compound no.3 and compound no.10 treated groups.
As shown in Figure 5, in the absence of compounds, the swimming, swarming, and twitching zones for P. aeruginosa PAO1 are 2.53 ± 0.08 cm, 1.88 ± 0.20 cm, and 5.62 ± 0.16 cm, respectively. Compound no.1 exerted a weak inhibitory effect that the swimming, swarming, and twitching zones are 1.5 ± 0.05 cm, 1.58 ± 0.13 cm, and 4.32 ± 0.28 cm. In the presence of compound no.3, it resulted in a significant decrease in motility zones to 0.87 ± 0.07 cm, 1.35 ± 0.13 cm, and 2.38 ± 0.16 cm. Moreover, compound no.10 also reduce the motility zones to 0.98 ± 0.10 cm, 1.40 ± 0.05 cm, and 3.43 ± 0.16 cm, respectively.
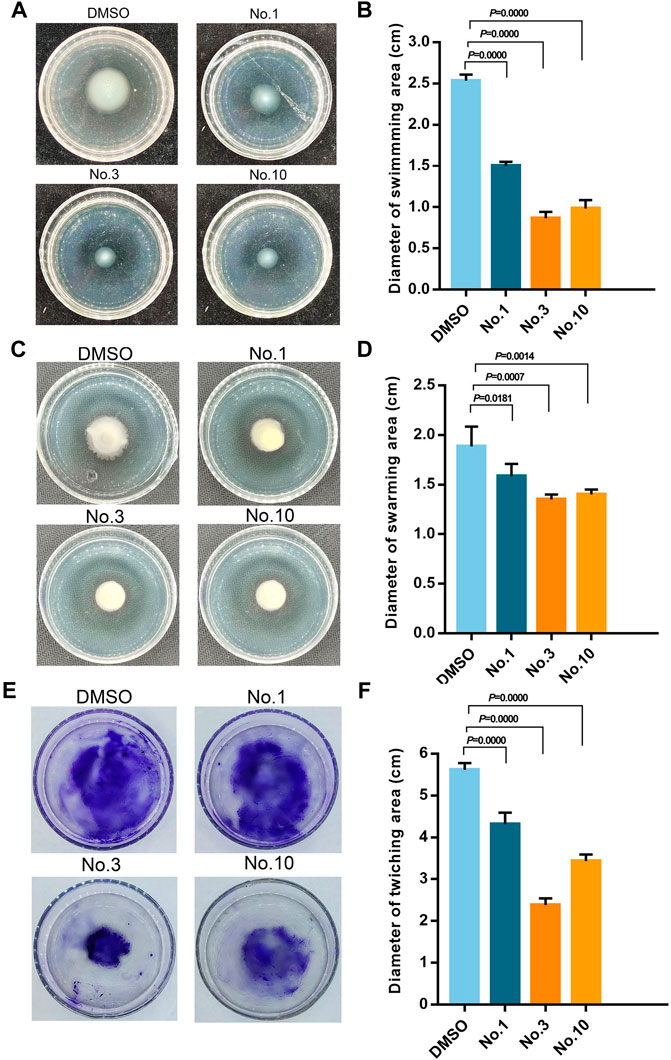
FIGURE 5. Inhibitory effect of compound no.3 and compound no.10 on P. aeruginosa motility. (A) and (B) represent swimming motility of P. aeruginosa PAO1, (C) and (D) represent swarming motility of P. aeruginosa PAO1, (E) and (F) represent twitching motility of P. aeruginosa PAO1. DMSO and compound no.1 represent negative and positive control, respectively.
Pyocyanin is a green exotoxin generated by P. aeruginosa governed by the QS system. The effect of compounds on pyocyanin production was studied using the chloroform extraction assay. As shown in Figure 6A, compound no.1 and compound no.3 exhibited a weak inhibitory effect. By contrast, compound no.10 robustly lowered pyocyanin production by 65.29%.
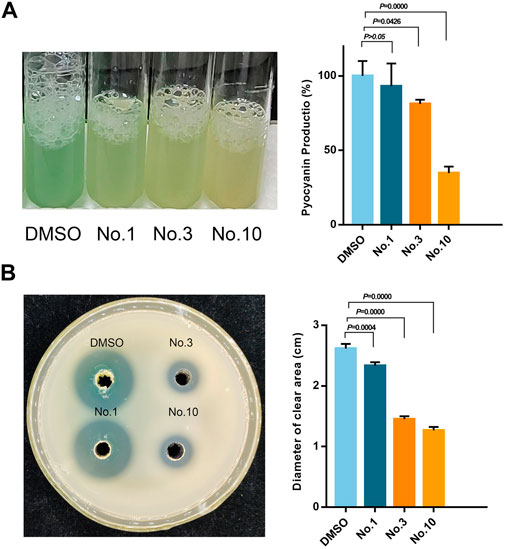
FIGURE 6. Inhibitory effect of compound no.3 and compound no.10 on (A) pyocyanin and (B) elastase productions of P. aeruginosa PAO1.
The effect of compounds on elastase activity was assessed with the skim milk agar method. As shown in Figure 6B, all the compounds can effectively inhibit the activity of elastase compared with the control group. Compared with compound no.1, significant reduction in the diameter of the clear zone was produced in compound no.3 and no.10 treated groups on a skim milk plate.
As shown in Figure 7, at 0–6 h, compound no.3 and compound no.10 exhibited an excellent effect on the survival rate compared with compound no.1. Then, the effect of compound no.10 was more obvious in improving the survival rate of infected C. elegans than no.3 at the 6–24 h. Compared with the untreated group, the survival rate of C. elegans of the compound no.10 group reaches nearly 80% within 6 h of incubation and can still be reached to 52% within 24 h.
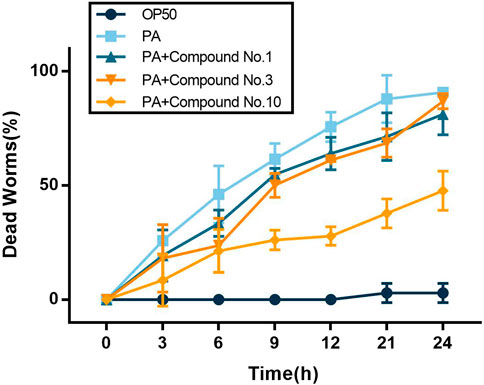
FIGURE 7. Compound no.3 and compound no.10 decreased toxicity of P. aeruginosa PAO1 in the C. elegans model.
To further determine the inhibition mechanism of compounds on P. aeruginosa PAO1 virulence and biofilm formation, the expression level of key QS-related genes was assessed using qPCR. From Figure 8, it shows that compound no.3 and no.10 downregulated the expression of lasI, lasR, rhlI, rhlR, and mvfR. Compared with the untreated group, compound no.1 slightly inhibited the expression of lasI, rhlI, and rhlR. The relative expression of QS regulatory gene treatment with compound no.3 decreased by 87% for lasI, 59% for lasR, 66% for rhlI, 80% for rhlR, and 17% for mvfR, respectively, and compound no.10 decreased 82% for lasI, 77% for lasR, 57% for rhlI, 91% for rhlR, and 61% for mvfR, respectively.
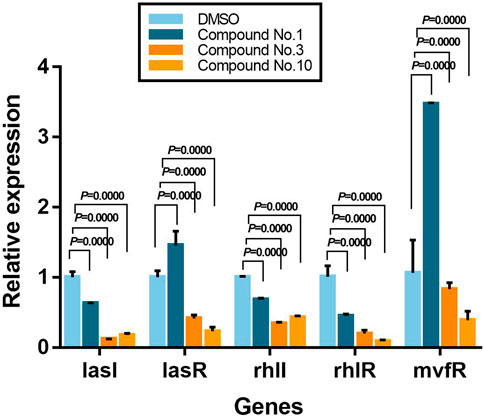
FIGURE 8. Relative expression levels of QS-regulated genes in the presence of compound no.1, compound no.3, and compound no.10 as determined by real-time quantitative PCR.
To further explore the binding mode of compounds to the LasR, molecular docking analysis of compounds no.1, no.3, and no.10 was performed. As shown in Figure 9, the interaction between compound no.1 and LasR by formation of H-bonding with Try 56, Trp 60, and Asp 73. It may be the reason that compound no.1 had weaker effects on biofilm formation, motility, and virulence production of P. aeruginosa PAO1. By contrast, the interaction between compounds (no.3 and no.10) and LasR receptor by formation of H-bonding with the Try 56, Trp 60, Asp 73, and Ser 129, is consistent with the binding sites of OdDHL (Abbas et al., 2017). The Lavina scores indicated a higher affinity of compound no.10 to bind with LasR than compound no.3 resulting in a more considerable inhibition activity of biofilm formation and virulence production.
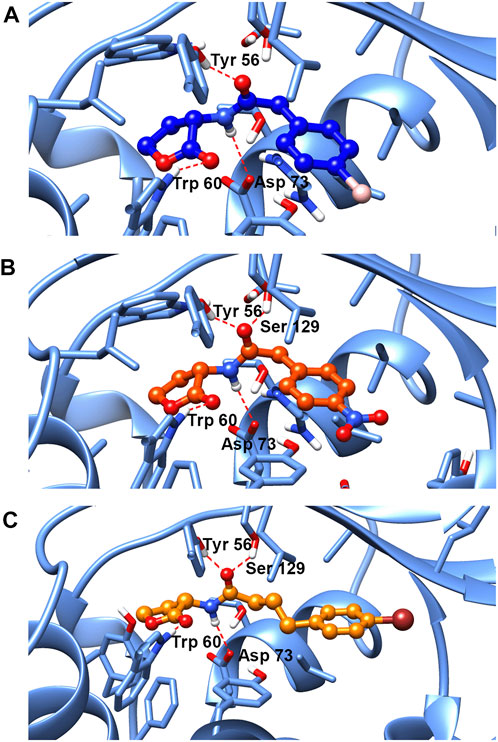
FIGURE 9. Molecular docking of (A) compound no.1, (B) compound no.3, and (C) compound no.10 into the active site of LasR. Both compound no.3 and compound no.10 interacted with LasR by formation of H-bonding with the Try 56, Trp 60, Asp 73, and Ser 129. Compound no.1 interacted with LasR with Try 56, Trp 60, and Asp 73 by H-bonding.
It is known that the pathogenicity of P. aeruginosa is contributed to the virulence factor production and the biofilm formation, which could lead to lethal infection and antibiotics defense (Costerton, 2001; Clatworthy et al., 2007; Zhao and Liu, 2010). Research evidence indicated that QS of P. aeruginosa was closely related to these phenomena (Lee and Zhang, 2015; Hegazy et al., 2021). Indeed, the structures of natural autoinducers have often served as an intermediate for the synthesis of the QS inhibitor (Galloway et al., 2011; O'Loughlin et al., 2013). In this study, we designed and synthesized nine novel AHL analogs and evaluated their inhibition activities and revealed their mechanism in the QS system of P. aeruginosa.
As described previously, subtle alterations to substituents and their placement on the AHL acyl group dramatically influenced analog inhibitory activity (Hodgkinson et al., 2012). Also then, the inhibitory activity of AHL analogs is highly dependent on the structure and position of substituents in the benzene group (Geske et al., 2008; Abbas et al., 2017). In this study, the chemical modification of AHL analogs is initiated by the replacement of the hydrocarbon chain length and by replacing substituents on the benzene ring, then nine novel AHL analogs were designed and synthesized.
In the initial screening assay, by using the QS-regulated production of biofilm as the screening indicator, compound no.3 and compound no.10 were selected to conduct further investigation. Consistent with the biofilm inhibition results, compound no.3 and compound no.10 also significantly inhibited the swimming, swarming, and twitching ability compared with compound no.1 at the concentration of 200 μM. In a previous study, the biofilm formation and adhesion to tissue related to the motility of bacteria were confirmed (Landry et al., 2006). Furthermore, it is reported that thin and dispersed biofilm was formed by QS deficient P. aeruginosa (Lee et al., 2018; Mayer et al., 2020). Therefore, we supposed that the inhibition in motility is one of the reasons for biofilm formation inhibition by compound no.3 and compound no.10.
P. aeruginosa is armed by an armory rich in virulence factors that are regulated by QS and pyocyanin and elastase are representative virulence of P. aeruginosa (Chatterjee et al., 2016). After being treated with compound no.3 and compound no.10, the production of pyocyanin was reduced by nearly 19 and 66%, respectively. The activity of elastase was also inhibited by 44 and 52%. Compound no.10 had a more obvious effect on inhibiting the virulence production of PAO1 in vitro. To further illustrate the inhibitory effect of those compound no.1, compound no.3, and compound no.10 on virulence production, C. elegans life span assays were conducted (Utari and Quax, 2013). Compared with compound no.3, compound no.10 had a more continuous and significant effect on improving the survival rate of infected C. elegans.
Moreover, compound no.10 exhibited more inhibition activity on pyocyanin production compared with other AHL analogs with a non-native tail region and native head group reported by previous studies (Morkunas et al., 2012). Chlorolactone (CL) was synthesized by Swem et al. (2009), and it exhibited 50% QS inhibition activity against C. violaceum at a concentration of 295 nM. However, O'Loughlin et al. (2013) proved that it had a weak QS inhibition effect in P. aeruginosa (Swem et al., 2009; O'Loughlin et al., 2013). Compared with 1,3-benzoxazol-2(3 H)-one derivatives, compound no.10 was able to achieve the similar QS-inhibition activity at lower concentrations (Miandji et al., 2012).
Based on aforementioned results, we supposed that the phenotype of QS system inhibition was attributed to the interaction between compounds and LasR. Molecular docking results have shown that compound no.1 interacted with LasR by formation of hydrogen bonds with three sites: Try 56, Trp 60, and Asp 73. While both the compound no.3 and compound no.10 interacted with receptors at four sites: Try 56, Trp 60, Asp 73, and Ser 129, consistent with the binding site of the natural ligand (Abbas et al., 2017). A previous study has shown that the absence of the hydrogen bonds donated by Ser 129 in the LasR ligand-binding pocket may result in the reduction of QS inhibition activity of ligands (Manson et al., 2020). It was proved that Ser 129 of LasR may be an essential site for QS inhibitor binding to LasR. Moreover, the Lavina scores of compound no.3 and no.10 are −9 and −7, respectively, which indicated a high affinity of compound no.10 to bind with LasR, and it could contribute to its anti-QS activity.
In a previous study, biofilm formation related to the LasI/R system and RhlI/R system was confirmed, which is responsible for biofilm differentiation and optimal biofilm formation, respectively (Favre-Bonté et al., 2003). While virulence factors, such as pyocyanin and elastase, also were controlled by PQS and RhlI/R system (Oliveira et al., 2016; McCready et al., 2019). To further explore the compounds’ mechanism, the relative expressions of QS-regulated (lasI, lasR, rhlI, rhlR, and mvfR) genes were selected to be tested (Kim et al., 2015). It has shown that compound no.3 and compound no.10 effectively downregulate lasI, lasR, rhlI, rhlR, and mvfR, while compound no.1 slightly inhibited the expression of lasI, rhlI, and rhlR. These results illustrated the possible causes that compound no.3 and compound no.10 had more significant bioactivity of anti-biofilm and anti-virulence at the gene level. In addition, compound no.10 has shown the more significant effect on downregulating the expression of QS-regulated genes than compound no.3, especially reduction in the expression of mvfR, which is the most important regulator of virulence production (Cao et al., 2001).
Based on aforementioned results, the structure–activity relationship trends influencing inhibitory activity are provided as follows: 1) AHL analog aromatic functionality with electron-withdrawing groups had stronger QS inhibition activity. 2) AHL analogs with electron-withdrawing and lipophilic substituents in the 4th position on the phenyl group displayed the stronger antagonistic activities than other analogs. 3) AHL analogs with an elastic spacer flexible carbon spacer of at least one carbon between the homoserine lactone ring and the benzene ring showed optimal inhibition activity.
In addition, our work proved that the substitutes of natural acyl group can be used as the strategy for synthesizing QS inhibitor. Moreover, our compounds had no stress on neither bacterial nor normal mammalian cell growth (Supplementary Figure S1); it is safe and can avoid the development of drug resistance.
In conclusion, 2-(4-bromophenyl)-N-(2-oxotetrapyridinefuran-3-yl) butanamide (compound no.10) could be considered as the most promising anti-QS compound against P. aeruginosa. Nevertheless, further experiments are conducted to evaluate the effect of compound no.10 on clinical drug-resistant strains and assess the therapeutic effect of compound no.10 on the infected animal model. In addition, further experiments are conducted to explore the QS inhibition mechanism of compound no.10.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Conceptualization, ZW and YL; methodology, TL, YG, EW, and WL; formal analysis, QZ; funding acquisition, HL; grammar correction, XW. All authors have read and agreed to the published version of the manuscript.
This research was funded by the National Natural Science Foundation of China (No.31571989, No.31772147), Science and Technology Cooperation Program of Gansu Academy of Sciences (grant number 2019HZ-02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are grateful to Ximing Xu, PhD (Ocean University of China) for providing assistance of the molecular docking.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.948687/full#supplementary-material
Abbas, H. A., Elsherbini, A. M., and Shaldam, M. A. (2017). Repurposing Metformin as a Quorum Sensing Inhibitor in Pseudomonas A. Afr. Health Sci. 17 (3), 808–819. doi:10.4314/ahs.v17i3.24
Abbas, H. A., Shaldam, M. A., and Eldamasi, D. (2020). Curtailing Quorum Sensing in Pseudomonas A by Sitagliptin. Curr. Microbiol. 77 (6), 1051–1060. doi:10.1007/s00284-020-01909-4
Bassler, B. L., and Losick, R. (2006). Bacterially Speaking. Cell 125 (2), 237–246. doi:10.1016/j.cell.2006.04.001
Bottomley, M. J., Muraglia, E., Bazzo, R., and Carfì, A. (2007). Molecular Insights into Quorum Sensing in the Human Pathogen Pseudomonas A from the Structure of the Virulence Regulator LasR Bound to its Autoinducer. J. Biol. Chem. 282 (18), 13592–13600. doi:10.1074/jbc.M700556200
Cao, H., Krishnan, G., Goumnerov, B., Tsongalis, J., Tompkins, R., and Rahme, L. G. (2001). A Quorum Sensing-Associated Virulence Gene of Pseudomonas A Encodes a LysR-Like Transcription Regulator with a Unique Self-Regulatory Mechanism. Proc. Natl. Acad. Sci. U. S. A. 98 (25), 14613–14618. doi:10.1073/pnas.251465298
Cezairliyan, B., Vinayavekhin, N., Grenfell-Lee, D., Yuen, G. J., Saghatelian, A., and Ausubel, F. M. (2013). Identification of Pseudomonas Aeruginosa Phenazines that Kill Caenorhabditis Elegans. PLoS Pathog. 9 (1), e1003101. doi:10.1371/journal.ppat.1003101
Chatterjee, M., Anju, C. P., Biswas, L., Anil Kumar, V., Gopi Mohan, C., and Biswas, R. (2016). Antibiotic Resistance in Pseudomonas A and Alternative Therapeutic Options. Int. J. Med. Microbiol. 306 (1), 48–58. doi:10.1016/j.ijmm.2015.11.004
Clatworthy, A. E., Pierson, E., and Hung, D. T. (2007). Targeting Virulence: A New Paradigm for Antimicrobial Therapy. Nat. Chem. Biol. 3 (9), 541–548. doi:10.1038/nchembio.2007.24
Costerton, J. W. (2001). Cystic Fibrosis Pathogenesis and the Role of Biofilms in Persistent Infection. Trends Microbiol. 9 (2), 50–52. doi:10.1016/s0966-842x(00)01918-1
Favre-Bonte, S., Köhler, T., and Van Delden, C. (2003). Biofilm Formation by Pseudomonas A: Role of the C4-HSL Cell-To-Cell Signal and Inhibition by Azithromycin. J. Antimicrob. Chemother. 52 (4), 598–604. doi:10.1093/jac/dkg397
Galloway, W. R. J. D., Hodgkinson, J. T., Bowden, S. D., Welch, M., and Spring, D. R. (2011). Quorum Sensing in Gram-Negative Bacteria: Small-Molecule Modulation of AHL and AI-2 Quorum Sensing Pathways. Chem. Rev. 111 (1), 28–67. doi:10.1021/cr100109t
Geske, G. D., O'Neill, J. C., Miller, D. M., Wezeman, R. J., Mattmann, M. E., Lin, Q., et al. (2008). Comparative Analyses of N-Acylated Homoserine Lactones Reveal Unique Structural Features that Dictate Their Ability to Activate or Inhibit Quorum Sensing. Chembiochem 9 (3), 389–400. doi:10.1002/cbic.200700551
Geske, G. D., Wezeman, R. J., Siegel, A. P., and Blackwell, H. E. (2005). Small Molecule Inhibitors of Bacterial Quorum Sensing and Biofilm Formation. J. Am. Chem. Soc. 127 (37), 12762–12763. doi:10.1021/ja0530321
Hançer Aydemir, D., Çifci, G., Aviyente, V., and Boşgelmez-Tinaz, G. (2018). Quorum-sensing Inhibitor Potential of Trans-Anethole Aganist Pseudomonas A. J. Appl. Microbiol. 125 (3), 731–739. doi:10.1111/jam.13892
Hao, S., Yang, D., Zhao, L., Shi, F., Ye, G., Fu, H., et al. (2021). EGCG-Mediated Potential Inhibition of Biofilm Development and Quorum Sensing in Pseudomonas A. Int. J. Mol. Sci. 22 (9), 4946. doi:10.3390/ijms22094946
Hegazy, W. A. H., Khayat, M. T., Ibrahim, T. S., Youns, M., Mosbah, R., and Soliman, W. E. (2021). Repurposing of Antidiabetics as Serratia M Virulence Inhibitors. Braz J. Microbiol. 52 (2), 627–638. doi:10.1007/s42770-021-00465-8
Hodgkinson, J. T., Galloway, W. R. J. D., Wright, M., Mati, I. K., Nicholson, R. L., Welch, M., et al. (2012). Design, Synthesis and Biological Evaluation of Non-Natural Modulators of Quorum Sensing in Pseudomonas A. Org. Biomol. Chem. 10 (30), 6032–6044. doi:10.1039/c2ob25198a
Hogardt, M., and Heesemann, J. (2013). Microevolution of Pseudomonas A to a Chronic Pathogen of the Cystic Fibrosis Lung. Curr. Top. Microbiol. Immunol. 358, 91–118. doi:10.1007/82_2011_199
Kim, H.-S., Lee, S.-H., Byun, Y., and Park, H.-D. (2015). 6-Gingerol Reduces Pseudomonas A Biofilm Formation and Virulence via Quorum Sensing Inhibition. Sci. Rep. 5, 8656. doi:10.1038/srep08656
Kumar, L., Chhibber, S., and Harjai, K. (2013). Zingerone Inhibit Biofilm Formation and Improve Antibiofilm Efficacy of Ciprofloxacin against Pseudomonas A PAO1. Fitoterapia 90, 73–78. doi:10.1016/j.fitote.2013.06.017
Landry, R. M., An, D., Hupp, J. T., Singh, P. K., and Parsek, M. R. (2006). Mucin-Pseudomonas aeruginosa Interactions Promote Biofilm Formation and Antibiotic Resistance. Mol. Microbiol. 59 (1), 142–151. doi:10.1111/j.1365-2958.2005.04941.x
Lee, D. J., Jo, A. R., Jang, M. C., Nam, J., Choi, H. J., Choi, G.-W., et al. (2018). Analysis of Two Quorum Sensing-Deficient Isolates of Pseudomonas A. Microb. Pathog. 119, 162–169. doi:10.1016/j.micpath.2018.04.014
Lee, J., and Zhang, L. (2015). The Hierarchy Quorum Sensing Network in Pseudomonas A. Protein Cell 6 (1), 26–41. doi:10.1007/s13238-014-0100-x
Luo, J., Dong, B., Wang, K., Cai, S., Liu, T., Cheng, X., et al. (2017). Baicalin Inhibits Biofilm Formation, Attenuates the Quorum Sensing-Controlled Virulence and Enhances Pseudomonas A Clearance in a Mouse Peritoneal Implant Infection Model. PLoS One 12 (4), e0176883. doi:10.1371/journal.pone.0176883
Manson, D. E., O’Reilly, M. C., Nyffeler, K. E., and Blackwell, H. E. (2020). Design, Synthesis, and Biochemical Characterization of Non-Native Antagonists of the Pseudomonas A Quorum Sensing Receptor LasR with Nanomolar IC(50) Values. ACS Infect. Dis. 6 (4), 649–661. doi:10.1021/acsinfecdis.9b00518
Mayer, C., Muras, A., Parga, A., Romero, M., Rumbo-Feal, S., Poza, M., et al. (2020). Quorum Sensing as a Target for Controlling Surface Associated Motility and Biofilm Formation in Acinetobacter Baumannii ATCC(®) 17978(TM). Front. Microbiol. 11, 565548. doi:10.3389/fmicb.2020.565548
McCready, A. R., Paczkowski, J. E., Cong, J.-P., and Bassler, B. L. (2019). An Autoinducer-Independent RhlR Quorum-Sensing Receptor Enables Analysis of RhlR Regulation. PLoS Pathog. 15 (6), e1007820. doi:10.1371/journal.ppat.1007820
Miandji, A., Ulusoy, S., Dündar, Y., Özgen, S., Onurdağ, F., Boşgelmez-Tınaz, G., et al. (2012). Synthesis and Biological Activities of Some 1,3-Benzoxazol-2(3H)-One Derivatives as Anti-Quorum Sensing Agents. Arzneimittelforschung 62 (7), 330–334. doi:10.1055/s-0032-1312590
Moradali, M. F., Ghods, S., and Rehm, B. H. A. (2017). Pseudomonas A Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 7, 39. doi:10.3389/fcimb.2017.00039
Morkunas, B., Galloway, W. R. J. D., Wright, M., Ibbeson, B. M., Hodgkinson, J. T., O'Connell, K. M. G., et al. (2012). Inhibition of the Production of the Pseudomonas A Virulence Factor Pyocyanin in Wild-Type Cells by Quorum Sensing Autoinducer-Mimics. Org. Biomol. Chem. 10 (42), 8452–8464. doi:10.1039/c2ob26501j
Ng, W.-L., and Bassler, B. L. (2009). Bacterial Quorum-Sensing Network Architectures. Annu. Rev. Genet. 43, 197–222. doi:10.1146/annurev-genet-102108-134304
Oliveira, B. D. Á., Rodrigues, A. C., Cardoso, B. M. I., Ramos, A. L. C. C., Bertoldi, M. C., Taylor, J. G., et al. (2016). Antioxidant, Antimicrobial and Anti-Quorum Sensing Activities of Rubus Rosaefolius Phenolic Extract. Industrial Crops Prod. 84, 59–66. doi:10.1016/j.indcrop.2016.01.037
O’Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., and Bassler, B. L. (2013). A Quorum-Sensing Inhibitor Blocks Pseudomonas A Virulence and Biofilm Formation. Proc. Natl. Acad. Sci. U. S. A. 110 (44), 17981–17986. doi:10.1073/pnas.1316981110
Qu, L., She, P., Wang, Y., Liu, F., Zhang, D., Chen, L., et al. (2016). Effects of Norspermidine on Pseudomonas A Biofilm Formation and Eradication. Microbiologyopen 5 (3), 402–412. doi:10.1002/mbo3.338
Rashid, M. H., and Kornberg, A. (2000). Inorganic Polyphosphate is Needed for Swimming, Swarming, and Twitching Motilities of Pseudomonas A. Proc. Natl. Acad. Sci. U. S. A. 97 (9), 4885–4890. doi:10.1073/pnas.060030097
Saqr, A. A., Aldawsari, M. F., Khafagy, E.-S., Shaldam, M. A., Hegazy, W. A. H., and Abbas, H. A. (2021). A Novel Use of Allopurinol as A Quorum-Sensing Inhibitor in Pseudomonas A. Antibiotics 10 (11), 1385. doi:10.3390/antibiotics10111385
Schuster, M., and Greenberg, E. P. (2006). A Network of Networks: Quorum-Sensing Gene Regulation in Pseudomonas A. Int. J. Med. Microbiol. 296 (2-3), 73–81. doi:10.1016/j.ijmm.2006.01.036
Swem, L. R., Swem, D. L., O'Loughlin, C. T., Gatmaitan, R., Zhao, B., Ulrich, S. M., et al. (2009). A Quorum-Sensing Antagonist Targets Both Membrane-Bound and Cytoplasmic Receptors and Controls Bacterial Pathogenicity. Mol. Cell 35 (2), 143–153. doi:10.1016/j.molcel.2009.05.029
Utari, P. D., and Quax, W. J. (2013). Caenorhabditis E Reveals Novel Pseudomonas A Virulence Mechanism. Trends Microbiol. 21 (7), 315–316. doi:10.1016/j.tim.2013.04.006
Keywords: AHL analogs, quorum sensing, Pseudomonas aeruginosa, anti-virulence, antibiofilm
Citation: Wei Z, Li T, Gu Y, Zhang Q, Wang E, Li W, Wang X, Li Y and Li H (2022) Design, Synthesis, and Biological Evaluation of N-Acyl-Homoserine Lactone Analogs of Quorum Sensing in Pseudomonas aeruginosa. Front. Chem. 10:948687. doi: 10.3389/fchem.2022.948687
Received: 20 May 2022; Accepted: 09 June 2022;
Published: 08 July 2022.
Edited by:
Xuetao Xu, Wuyi University, ChinaReviewed by:
Xin Li, Henan University of Science and Technology, ChinaCopyright © 2022 Wei, Li, Gu, Zhang, Wang, Li, Wang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Li, bGlfeWFuZ0BsenUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.