
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 08 June 2022
Sec. Medicinal and Pharmaceutical Chemistry
Volume 10 - 2022 | https://doi.org/10.3389/fchem.2022.911232
 Jun-Jie Ke
Jun-Jie Ke Jing Lin
Jing Lin Xin Zhang
Xin Zhang Xiao-Zheng Wu
Xiao-Zheng Wu Ying-Ying Zheng
Ying-Ying Zheng Chun-Mei Hu
Chun-Mei Hu Yu Kang
Yu Kang Kun Zhang
Kun Zhang Zhuang Xiong*
Zhuang Xiong* Zhi-Qiang Ma*
Zhi-Qiang Ma*A series of benzylidene analogs of oleanolic acid 4a∼4s were synthesized and assessed for their α-glucosidase and α-amylase inhibitory activities. The results presented that all synthesized analogs exhibited excellent-to-moderate inhibitory effects on α-glucosidase and α-amylase. Analog 4i showed the highest α-glucosidase inhibition (IC50: 0.40 μM), and analog 4o presented the strongest α-amylase inhibition (IC50: 9.59 μM). Inhibition kinetics results showed that analogs 4i and 4o were reversible and mixed-type inhibitors against α-glucosidase and α-amylase, respectively. Simulation docking results demonstrated the interaction between analogs and two enzymes. Moreover, analogs 4i and 4o showed a high level of safety against 3T3-L1 and HepG2 cells.
Type 2 diabetes mellitus (T2DM) is a metabolic disease characterized by hyperglycemia resulting from insulin resistance and insufficient insulin secretion by pancreatic β-cells (Silva et al., 2016; Ahmed et al., 2020). T2DM can also bring about complications such as hepatic, cardiac, and renal disorders (Forouhi and Wareham, 2010; Mollica et al., 2018). In addition, the aggravation of insulin resistance and pancreatic β-cell dysfunction can be ascribed to genetic predisposition, increasing age, and obesity (Kokil et al., 2015; Mollica et al., 2017). It is conservatively estimated that T2DM will affect approximately 500 million people worldwide by 2030, and the mortality rate for the disease and its associated complications is one death every 6 s as of now (Kumar et al., 2021). The effective way to reduce blood glucose levels in the treatment and prevention of T2DM and its complications is the clinical use of oral hypoglycemic agents such as sulfonylureas (Liang et al., 2020; Ling et al., 2021), biguanides (Zheng T.-L. et al., 2021; Kathuria et al., 2021), thiazolidinedione-derived drugs (Long et al., 2021; Liu et al., 2021), dipeptidyl-peptidase IV inhibitors (Carullo et al., 2021; Cheng et al., 2022), α-glucosidase inhibitors (Hossain et al., 2020), sodium-glucose cotransporter-2 inhibitors (Provenzano et al., 2021), and glucagon-like peptide-1 (GLP-1) receptor agonists (Furer et al., 2021; Saxena et al., 2021). Among these, α-glucosidase inhibitors are widely used as clinical drugs including acarbose, voglibose, miglitol, and emiglitate, and there are reports on α-amylase inhibitors derived from the natural product. In addition, it is well-known that starch is hydrolyzed by α-amylases into disaccharides or oligosaccharides in the mouth and small intestine, respectively, followed by the further hydrolysis of α-glucosidases into glucose units in the small intestinal lumen. The inhibitors against α-glucosidases or α-amylases can combine with the active units of these two enzymes to form complexes with stronger affinity than that of the carbohydrate–enzyme complex, thereby realizing the inhibition against α-glucosidases or α-amylases. (Proença et al., 2021).
Oleanolic acids (OA) is a pentacyclic oleanane-type triterpenoid with a broad spectrum of bioactivities in which the potential application in the management of T2DM and its associated comorbidities, resulting from their antihyperglycemic, antihyperlipidemic, antiatherogenic, antioxidant, and anti-inflammatory action, has attracted much attention (Liu, 2005; Pollier and Goossens, 2012; Loza-Rodriguez et al., 2020). It is worth noting that OA can reduce postprandial hyperglycemia in diabetic people by inhibiting α-glucosidase and the pancreatic and salivary α-amylase without apparent hepatotoxicity in the experimental studies (Castellano et al., 2013). Given that, much effort has been focused on the modification of OA in order to improve its potential druggability. So far, OA derivatives are mainly obtained by means of OA semi-synthetic pathways: 1) esterification, glycosylation, or oxidization at C-3 position (Jensen et al., 1997; Wu, et al., 2021; Zhong et al., 2021); 2) amidation or esterification at C-28 position (Tang et al., 2014; Kazakova et al., 2021); 3) lactonization between C-12 and C-28 positions (Zhong et al., 2019); and 4) condensation with various aldehydes at C-2 position (Zhong et al., 2019); all these derivatizations toward OA can improve α-glucosidase or α-amylase inhibitory activity. In addition, the benzylidene group is reported to be one key substituent in many pharmacological compounds, such as chalcones and cinnamic acids(Csuk, et al., 2012; Kim et al., 2014; Gupta et al., 2017), and the introduction of the benzylidene group can increase the inhibitory activity against α-glucosidase or α-amylase inhibitory activity in our previous work (Deng et al., 2022).
In view of these findings, the strategy to incorporate the benzylidene side chain into the OA skeleton was adopted to enhance the bioavailability of OA. Herein, the synthesis of benzylidene analogs 3a–s of OA by Claisen–Schmidt condensation was developed, followed by reduction at the C-3 position to obtain OA analogs 4a–4s. The α-glucosidase and α-amylase inhibitory activity of OA analogs has been evaluated in vitro. The results showed that most compounds revealed a better inhibitory effect than OA. On this basis, the inhibitory mechanism that OA analogs interacted with these two enzymes has been probed into by biochemical and computational assays.
OA was used as a starting material to obtain benzylidene analogs of oleanolic acid according to the synthetic route shown in Scheme 1. First, OA (1) was oxidized to 3-oxo-olean-12-en-28-oic acid (2) by Jones reagent, further reacting with substituted aromatic aldehydes to produce intermediate 3. Then, target analogs 4a–4s were prepared through reduction reaction of intermediate 3. All synthetic analogs were identified by 1H NMR, 13C NMR, and HRMS.
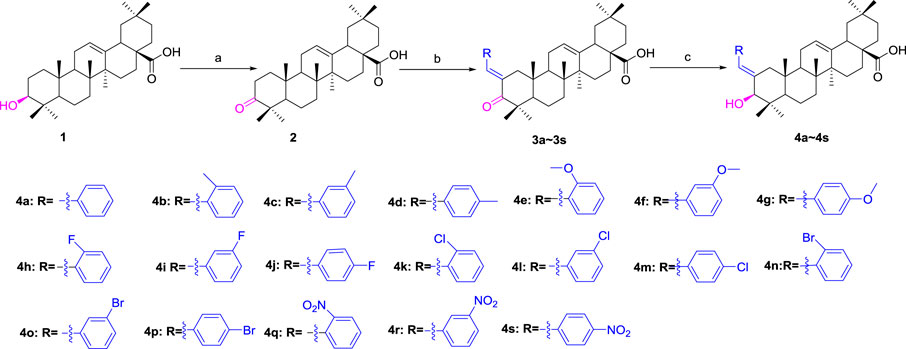
SCHEME 1. Synthesis of benzylidene analogs of oleanolic acid 4a–4s. Reagent and condition: (a) Jones reagent, acetone, 0°C; (b) Substituted aromatic aldehydes, EtOH, KOH, room temperature, and overnight; (c) NaBH4, DCM, MeOH, 2h, 0 C—room temperature
OA analogs 4a–4s were first assessed for their α-glucosidase inhibitory activities, and the results are listed in Table 1. All synthesized OA analogs (4a–4s) exhibited potent α-glucosidase inhibitory activity (IC50: 0.40–3.96 μM), which was higher than that of OA (IC50: 4.09 μM). Analog 4i showed the strongest α-glucosidase inhibition (IC50: 0.40 μM), which was ∼1,663 times stronger than that of acarbose (IC50: 665.56 μM). The results showed that the modification of OA with benzylidene could improve its α-glucosidase inhibitory activity. From the α-glucosidase inhibitory activities of OA analogs (4a–4s), it could be seen that introduction of the donating groups (methyl and methoxy) at the para-position of substituted aryl aldehydes could reduce inhibitory activities, while the introduction of the withdrawing group (fluorine, chlorine, bromine, and nitro) at the para-position could improve the inhibition. In addition, it was surprisingly found that the inhibition of compounds with substituents at meta-position was mostly superior to that of compounds with substituents at ortho-position, and the inhibition of compounds with substituents at the ortho-position was better than that of compounds with substituents at para-position. Finally, we found that analog 4i with the withdrawing group (fluorine) at the meta-position of substituted aryl aldehydes showed the highest α-glucosidase inhibition (IC50: 0.40 μM).
Subsequently, OA analogs (4a–4s) were evaluated for their α-amylase inhibitory activities (Table 1). It was found that all OA analogs presented obvious α-amylase inhibitory activities with IC50 values of 9.59–65.78 μM, compared with those of OA (IC50: 94.10 μM) and analog 4o had the strongest α-amylase inhibitory activity (IC50: 9.59 μM), which was ∼10 times higher than that of acarbose (IC50: 100.01 μM). The modification of OA with benzylidene could also contribute to the improvement of α-amylase inhibitory activity. But, the compound’s inhibitory activity has no relation with the electrical properties of substituents. Compared with the unsubstituted compound 4a, introduction of functional groups (fluorine, chlorine, and bromine) at the C2, C3, and C4 position of the substituted aldehydes, functional groups (methyl and methoxy) at the C2 position, and nitro group at the C3 position of benzene group improved the inhibitory activity. But, the introduction of the donating groups (methyl and methoxy) at the C3 and C4 position and nitro group at the para-position of substituted aryl aldehydes resulted in lower inhibitory activity.
As could be seen from the aforementioned results, all OA analogs (4a–4s) displayed bifunctionality against α-glucosidase and α-amylase. All OA analogs displayed dual-inhibitory activities against α- glucosidase and α-amylase, which were better than those of acarbose.
To decide the inhibition kinetics of OA analogs (4a–4s) against α-glucosidase and α-amylase, analogs 4i and 4o with the strongest α-glucosidase and α-amylase inhibitory activity, respectively, were selected as the representative analogs. As shown in Figure 1A, each trend line of the enzymatic reaction rate vs. α-glucosidase concentration with or without the presence of analog 4i passed through the origin, showing that the inhibition of analog 4i on α-glucosidase was reversible. Its inhibition kinetics parameters were assayed using the Lineweaver–Burk plots (Figure 1B). It could be seen that each trend line of the enzymatic reaction rate vs. substrate concentration with or without the presence of analog 4i intersected in the second quadrant, indicating that analog 4i was a mixed-type inhibitor against α-glucosidase.
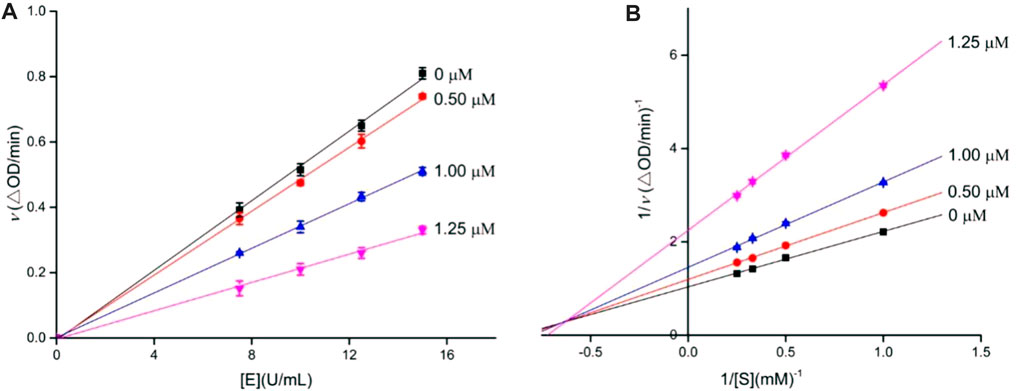
FIGURE 1. Inhibition kinetics of analog 4i against α-glucosidase. (A) Plots of the enzymatic reaction rate vs. α-glucosidase concentration with or without the presence of analog 4i; (B) Lineweaver–Burk plots of enzymatic reaction rate vs. substrate concentration with or without the presence of analog 4i.
Similarly, the inhibition kinetics of analog 4o against α-amylase was studied. The plots of the enzymatic reaction rate vs. α-amylase concentration passed through the origin (Figure 2A), and Lineweaver–Burk plots of enzymatic reaction rate vs. substrate concentration intersected in the first quadrant (Figure 2B). The results showed that analog 4o functioned as a reversible and mixed-type inhibitor against α-amylase.
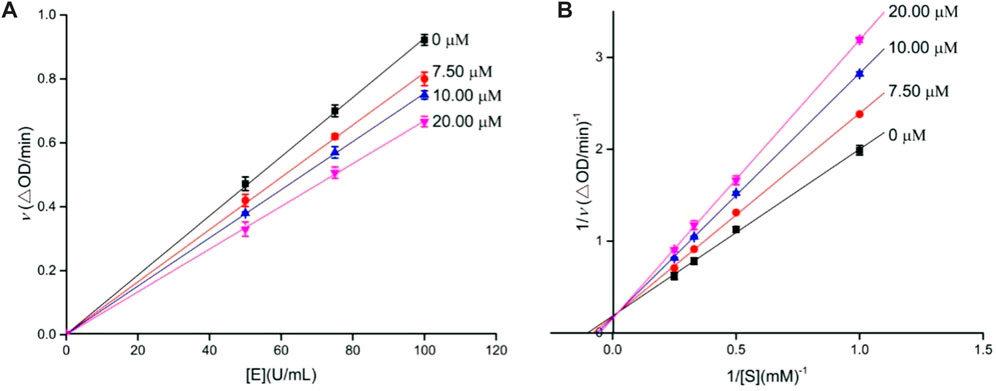
FIGURE 2. Inhibition kinetics of analog 4o against α-amylase. (A) Plots of enzymatic reaction rate vs. α-amylase concentration with or without the presence of analog 4o; (B) Lineweaver–Burk plots of enzymatic reaction rate vs. substrate concentration with or without the presence of analog 4o.
Molecular docking simulations using SYBYL software for analogs 4i and 4o were carried out targeting α-glucosidase and α-amylase, respectively. On the basis of docking results, the binding interactions were analyzed. The docking simulation between analog 4i and homology model α-glucosidase is shown in Figure 3. Benzylidene and the hydroxyl moiety of analog 4i bind at the entrance of the α-glucosidase pocket and carboxyl moiety positioned in the interior of the pocket (Figures 3A,B). Detailed analysis (Figures 3C,D) presented that the carboxyl moiety of analog 4i formed a hydrogen bond with Arg 439 (2.4 Å). The fluorine substituent of benzylidene formed a hydrogen bond with Asn 412 and two halogen bonds with Phe 157 and Asp 408. The benzylidene formed a π–π stacking with Phe 311 (5.0 Å). Furthermore, analog 4i formed hydrophobic interactions with Tyr 71, Phe 177, His 279, and Phe 300.
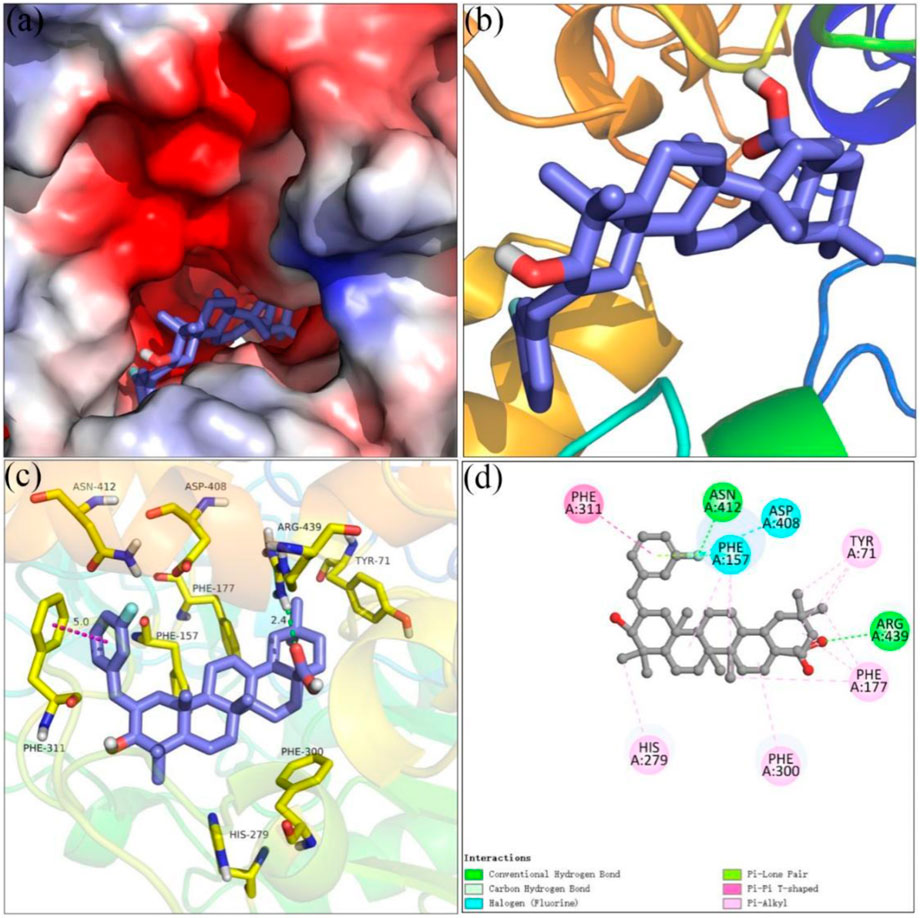
FIGURE 3. Molecular docking of analog 4i with α-glucosidase. (A) 4i in the electrostatics active pocket; (B) 4i in the active pocket; (C) 3D view of 4i with α-glucosidase; (D) 2D view of 4i with α-glucosidase.
Figure 4 showed the docking results of analog 4o and α-amylase. It could be seen that benzylidene and the hydroxyl moiety of analog 4o docked at the interior of the α-glucosidase pocket and carboxyl moiety positioned at the entrance of the pocket (Figures 4A,B). Detailed binding results (Figures 4C,D) pointed that analog 4o formed a hydrogen bond with Asp 300 (2.0 Å), a π–π stacking with Trp 59 (4.6 Å), and hydrophobic interactions with Leu 162 and His 305.
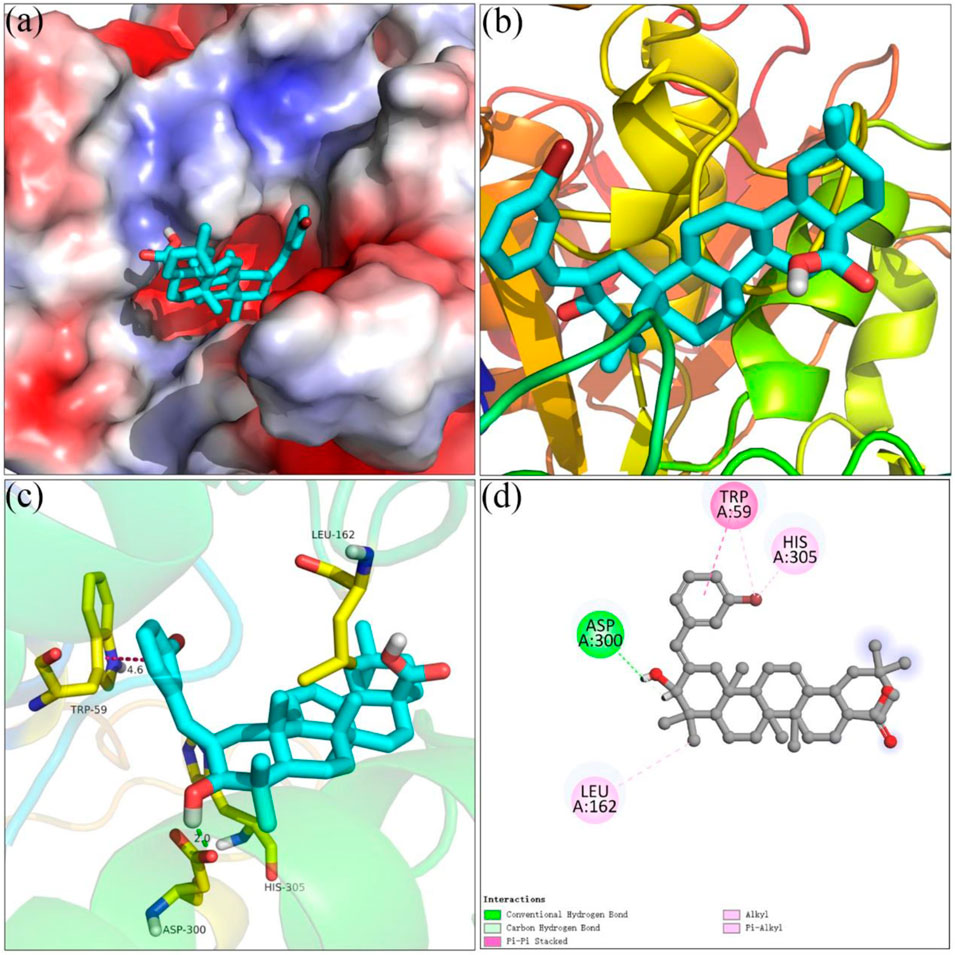
FIGURE 4. Molecular docking of analog 4o and α-amylase. (A) 4o in the electrostatics active pocket; (B) 4o in the active pocket; (C) 3D view of 4o and α-amylase; (D) 2D view of 4o and α-amylase.
The cell cytotoxicity of strongest inhibitory activity analogs 4i and 4o was evaluated against HepG2 and 3T3-L1 cells. The results showed that analogs 4i and 4o had a non-cytotoxic effect to HepG2 and 3T3-L1 cells under a concentration of 100 μM (Figure 5).
In summary, benzylidene analogs of oleanolic acid 4a–4s were synthesized for finding potential α-glucosidase and α-amylase inhibitors. All synthesized analogs displayed bifunctionality against α-glucosidase and α-amylase. Analog 4i showed the highest α-glucosidase inhibition (IC50: 0.40 μM), and analog 4o presented the strongest α-amylase inhibition (IC50: 9.59 μM). Inhibition kinetics results showed that analogs 4i and 4o were reversible and had mixed-type inhibitors against α-glucosidase and α-amylase, respectively. Moreover, analogs 4i and 4o showed a high level of safety against HepG2 and 3T3-L1 cells, which provided strong support for the further in vivo assay. In viewpoint of all these experimental data, benzylidene analogs of oleanolic acid have the potential to be developed into the leading compound in the management of T2D.
OA was purchased from Energy Chemical Company, China. α-glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20), α-amylase from hog pancreas (EC 3.2.1.1), and p-nitrophenyl-α-D-galactopyranoside (p-NPG) were provided by Sigma-Aldrich. Water-soluble starch was obtained from Shanghai Yuanye Biological Technology Co., Ltd. 3T3-L1 cells and HepG2 cells were supplied by ATCC. Other reagents were purchased from commercial suppliers. All the compounds were dissolved in DMSO, and DMSO working concentration in the enzyme inhibition test was 5%. The NMR spectra were recorded on a Bruker AM spectrometer (500 MHz). High-resolution mass spectral analysis (HRMS) was carried out on the Apex II by means of the ESI technique. Melting points were detected on a WRS-2C micro melting point instrument.
To a solution of oleanolic acid (1.0 mmol) in acetone (10 ml) was added freshly prepared Jones reagent at 0°C for 1 h. The mixture was quenched with methanol, then extracted with ethyl acetate, dried over Na2SO4, and concentrated in vacuo. Then, the crude product was purified by silica gel column chromatography to yield compound 2. Then, compound 2 (0.5 mmol) was condensed with substituted aromatic aldehydes (0.75 mmol) at room temperature overnight. After the completion of the reaction, the mixture was adjusted to pH = 1 with 1N diluted hydrochloric acid, then extracted with ethyl acetate, washed with water, dried by Na2SO4, and evaporated to dryness. Compounds 3a–3s were obtained through the purification of silica gel column chromatography.
Last, NaBH4 (1.25 mmol) was added to the solution of 3a–3s (0.25 mmol) in methanol (2 ml) at 0 C. The mixture was warmed to room temperature and stirred for 1–2 h. The mixture was quenched with cold water, extracted with ethyl acetate, dried by Na2SO4, and separated by silica gel column chromatography to the desired product 4a–4s.
(4a, C37H52O3). White solid; Yield: 75%; mp: 183–184 °C; 1H NMR (500 MHz, chloroform-d) δ 7.34–7.29 (m, 2H), 7.22 (d, J = 7.8 Hz, 3H), 6.69 (s, 1H), 5.22 (t, J = 3.6 Hz, 1H), 3.87 (d, J = 2.0 Hz, 1H), 2.92 (d, J = 12.9 Hz, 1H), 2.79 (dd, J = 13.8, 4.5 Hz, 1H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.82–1.52 (m, 9H), 1.47 (td, J = 12.4, 3.8 Hz, 1H), 1.43–1.17 (m, 5H), 1.14 (s, 4H), 1.13 (s, 4H), 1.11–1.03 (m, 2H), 0.91 (s, 3H), 0.89 (s, 3H), 0.78 (s, 3H), 0.75 (s, 3H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.19, 143.63, 140.33, 138.13, 128.96, 128.26, 126.20, 122.67, 122.56, 81.26, 55.90, 47.02, 46.65, 45.95, 41.78, 41.74, 41.69, 41.07, 40.36, 39.57, 33.88, 33.21, 32.58, 32.53, 30.80, 28.76, 27.79, 26.04, 23.71, 23.46, 22.95, 18.51, 17.12, 15.78, and 15.61. HRMS (ESI-MS) m/z: [M + Na]+ calculated for C37H52O3Na: 567.3809; found: 567.3809.
(4b, C38H54O3). White solid; Yield: 65%; mp: 210–211 °C; 1H NMR (500 MHz, chloroform-d) δ 7.15 (ddt, J = 17.2, 5.9, 3.3 Hz, 3H), 7.04 (dd, J = 6.5, 2.3 Hz, 1H), 6.64 (s, 1H), 5.16 (t, J = 3.2 Hz, 1H), 3.92 (d, J = 1.9 Hz, 1H), 2.76 (dd, J = 13.8, 4.5 Hz, 1H), 2.60 (d, J = 12.8 Hz, 1H), 2.24 (s, 3H), 1.93 (td, J = 13.4, 4.0 Hz, 1H), 1.78–1.49 (m, 8H), 1.49–1.23 (m, 6H), 1.21–1.00 (m, 10H), 0.89 (d, J = 3.2 Hz, 7H), 0.78 (s, 3H), 0.66 (s, 3H), and 0.63 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.20, 143.55, 139.85, 137.27, 136.55, 129.84, 129.42, 126.53, 125.42, 122.56, 122.12, 81.17, 55.76, 47.09, 46.64, 45.94, 42.14, 41.70, 41.47, 41.08, 40.07, 39.54, 33.86, 33.21, 32.56, 30.78, 28.68, 27.74, 26.05, 23.69, 23.44, 22.90, 20.20, 18.55, 17.04, 15.80, and 15.32. HRMS (ESI-MS) m/z: [M + H]+ calculated for C38H55O3: 559.4146; found: 559.4148.
(4c, C38H54O3). White solid; Yield: 71%; mp: 128–129 °C; 1H NMR (500 MHz, chloroform-d) δ 7.20 (t, J = 7.5 Hz, 1H), 7.06–7.00 (m, 3H), 6.66 (s, 1H), 5.23 (t, J = 3.6 Hz, 1H), 3.86 (d, J = 2.1 Hz, 1H), 2.94 (d, J = 12.9 Hz, 1H), 2.80 (dd, J = 13.9, 4.6 Hz, 1H), 2.34 (s, 3H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.87–1.52 (m, 9H), 1.52–1.18 (m, 6H), 1.17–1.01 (m, 9H), 0.90 (d, J = 7.9 Hz, 7H), 0.80 (s, 3H), 0.74 (s, 3H), and 0.69 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.27, 143.68, 140.10, 138.03, 137.70, 129.76, 128.13, 126.95, 125.96, 122.68, 122.58, 81.29, 55.93, 47.00, 46.66, 45.96, 41.81, 41.75, 41.67, 41.06, 40.35, 39.57, 33.89, 33.20, 32.58, 32.53, 30.79, 28.75, 27.81, 26.04, 23.70, 23.43, 22.96, 21.64, 18.50, 17.14, 15.78, and 15.67. HRMS (ESI-MS) m/z: [M + Na]+ calculated for C38H54O3Na: 581.3965; found: 581.3959.
(4d, C38H54O3). White solid; Yield: 73%; mp: 146–147 °C; 1H NMR (500 MHz, chloroform-d) δ 7.12 (s, 4H), 6.65 (s, 1H), 5.22 (d, J = 3.7 Hz, 1H), 3.86 (d, J = 2.0 Hz, 1H), 2.94 (d, J = 12.9 Hz, 1H), 2.79 (dd, J = 13.8, 4.6 Hz, 1H), 2.35 (s, 3H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.86–1.51 (m, 9H), 1.51–1.43 (m, 1H), 1.38 (t, J = 8.2 Hz, 2H), 1.34–1.26 (m, 4H), 1.23–1.18 (m, 1H), 1.13 (d, J = 1.7 Hz, 6H), 1.05 (td, J = 13.1, 12.5, 2.8 Hz, 2H), 0.91 (s, 3H), 0.89 (s, 3H), 0.78 (s, 3H), 0.74 (s, 3H), and 0.69 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.10, 143.62, 139.69, 135.75, 135.10, 128.99, 128.84, 122.58, 122.47, 81.31, 55.92, 47.02, 46.65, 45.95, 41.79, 41.75, 41.66, 41.09, 40.36, 39.57, 33.89, 33.21, 32.59, 32.53, 30.80, 29.84, 28.74, 27.80, 26.04, 23.71, 23.49, 22.96, 21.30, 18.51, 17.11, 15.77, and 15.61. HRMS (ESI-MS) m/z: [M + H]+ calculated for C38H55O3: 559.4146; found: 559.4149.
(4e, C38H54O4). White solid; Yield: 72%; mp: 200–201°C; 1H NMR (500 MHz, chloroform-d) δ 7.22 (td, J = 7.9, 1.7 Hz, 1H), 7.14 (dd, J = 7.6, 1.6 Hz, 1H), 6.90 (td, J = 7.5, 1.1 Hz, 1H), 6.87 (dd, J = 8.2, 1.0 Hz, 1H), 6.65 (s, 1H), 5.19 (t, J = 3.7 Hz, 1H), 3.89 (d, J = 2.0 Hz, 1H), 3.80 (s, 3H), 2.77 (dd, J = 13.8, 4.6 Hz, 1H), 2.72 (d, J = 12.9 Hz, 1H), 1.95 (td, J = 13.5, 4.0 Hz, 1H), 1.78–1.69 (m, 3H), 1.68–1.52 (m, 5H), 1.49–1.23 (m, 5H), 1.22–1.00 (m, 10H), 0.89 (d, J = 7.0 Hz, 7H), 0.76 (d, J = 9.7 Hz, 6H), and 0.67 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.04, 157.38, 143.57, 140.11, 130.12, 127.72, 127.04, 122.63, 120.16, 118.57, 110.58, 81.39, 55.91, 55.55, 47.05, 46.65, 45.95, 42.21, 41.75, 41.58, 41.08, 40.18, 39.56, 33.88, 33.20, 32.62, 32.52, 30.79, 28.71, 27.79, 26.04, 23.71, 23.46, 22.96, 18.51, 17.12, 15.80, and 15.36. HRMS (ESI-MS) m/z: [M + Na]+ calculated for C38H54O4Na: 597.3914; found: 597.3925.
(4f, C38H54O4). White solid; Yield: 62%; mp: 200–201°C; 1H NMR (500 MHz, chloroform-d) δ 7.15 (d, J = 8.7 Hz, 2H), 6.86 (d, J = 8.7 Hz, 2H), 6.62 (s, 1H), 5.23 (t, J = 3.6 Hz, 1H), 3.85 (d, J = 2.0 Hz, 1H), 3.82 (s, 3H), 2.92 (d, J = 12.9 Hz, 1H), 2.79 (dd, J = 13.9, 4.6 Hz, 1H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.86–1.70 (m, 2H), 1.70–1.52 (m, 6H), 1.47 (td, J = 12.3, 4.0 Hz, 1H), 1.40–1.32 (m, 3H), 1.31–1.24 (m, 4H), 1.23–1.18 (m, 1H), 1.13 (d, J = 3.2 Hz, 6H), 1.10–1.01 (m, 2H), 0.91 (s, 3H), 0.89 (s, 3H), 0.78 (s, 3H), 0.73 (s, 3H), and 0.69 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.06, 159.49, 143.69, 140.54, 139.51, 129.23, 122.56, 122.53, 121.55, 114.09, 112.13, 81.27, 55.94, 55.34, 47.01, 46.65, 45.96, 41.87, 41.76, 41.72, 41.06, 40.36, 39.57, 33.89, 33.20, 32.59, 32.52, 30.80, 28.76, 27.81, 26.04, 23.70, 23.50, 22.98, 18.50, 17.14, 15.80, and 15.70. HRMS (ESI-MS) m/z: [M + Na]+ calculated for C38H54O4Na: 597.3914; found: 597.3915.
(4g, C38H54O4). White solid; Yield: 70%; mp: 296–297°C; 1H NMR (500 MHz, chloroform-d) δ 7.15 (d, J = 8.7 Hz, 2H), 6.86 (d, J = 8.7 Hz, 2H), 6.62 (s, 1H), 5.23 (t, J = 3.6 Hz, 1H), 3.85 (d, J = 2.0 Hz, 1H), 3.82 (s, 3H), 2.92 (d, J = 12.9 Hz, 1H), 2.79 (dd, J = 13.9, 4.6 Hz, 1H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.86–1.70 (m, 2H), 1.70–1.52 (m, 6H), 1.47 (td, J = 12.3, 4.0 Hz, 1H), 1.40–1.32 (m, 3H), 1.31–1.24 (m, 4H), 1.23–1.18 (m, 1H), 1.13 (d, J = 3.2 Hz, 6H), 1.10–1.01 (m, 2H), 0.91 (s, 3H), 0.89 (s, 3H), 0.78 (s, 3H), 0.73 (s, 3H), and 0.69 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.16, 157.96, 143.64, 139.12, 130.56, 130.05, 122.57, 122.03, 113.69, 81.31, 55.91, 55.37, 47.00, 46.65, 45.95, 41.75, 41.62, 41.08, 40.32, 39.57, 33.88, 33.21, 32.59, 32.53, 30.80, 29.84, 28.73, 27.79, 26.04, 23.71, 23.50, 22.96, 18.50, 17.11, 15.77, and 15.60. HRMS (ESI-MS) m/z: [M + H]+ calculated for C38H55O4: 575.4095; found: 575.4072.
(4h, C37H51FO3). White solid; Yield: 75%; mp: 165–166 °C; 1H NMR (500 MHz, chloroform-d) δ 7.26–7.17 (m, 2H), 7.13–7.03 (m, 2H), 6.63 (s, 1H), 5.21 (t, J = 3.6 Hz, 1H), 3.93 (d, J = 1.9 Hz, 1H), 2.79 (dd, J = 13.9, 4.6 Hz, 1H), 2.65 (d, J = 12.9 Hz, 1H), 1.97 (td, J = 13.5, 4.1 Hz, 1H), 1.75 (qd, J = 9.1, 8.5, 5.8 Hz, 3H), 1.70–1.53 (m, 5H), 1.53–1.43 (m, 2H), 1.41–1.19 (m, 7H), 1.19–1.04 (m, 8H), 0.92 (s, 3H), 0.91 (s, 3H), 0.79 (s, 3H), 0.73 (s, 3H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ184.15, 161.29, 159.34, 143.16, 130.85, 128.14, 125.80, 123.67, 122.49, 115.88, 115.46, 81.20, 55.78, 47.05, 46.64, 45.94, 42.37, 41.73, 41.69, 41.06, 40.21, 39.55, 33.86, 33.20, 32.53, 30.78, 29.85, 28.72, 27.76, 26.04, 23.70, 23.41, 22.94, 18.54, 17.05, 15.74, and 15.32.HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52FO3: 563.3895; found: 563.3904.
(4i, C37H51FO3). White solid; Yield: 69%; mp: 157–158 °C; 1H NMR (500 MHz, chloroform-d) δ 7.28 (d, J = 7.9 Hz, 1H), 6.97 (d, J = 7.7 Hz, 1H), 6.91 (d, J = 9.4 Hz, 2H), 6.66 (s, 1H), 5.25–5.18 (m, 1H), 3.87 (d, J = 2.0 Hz, 1H), 2.89–2.75 (m, 2H), 1.95 (td, J = 13.5, 4.0 Hz, 1H), 1.83–1.51 (m, 8H), 1.51–1.17 (m, 9H), 1.13 (d, J = 2.6 Hz, 7H), 0.90 (d, J = 8.2 Hz, 7H), 0.76 (s, 3H), 0.74 (s, 3H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.38, 163.77, 161.83, 143.68, 141.52, 140.47, 129.62, 124.69, 122.11, 115.66, 113.04, 81.14, 55.82, 47.00, 46.64, 45.94, 41.77, 41.75, 41.72, 41.05, 40.40, 39.55, 33.87, 33.19, 32.52, 30.78, 29.83, 28.74, 27.77, 26.02, 23.69, 23.41, 22.92, 18.49, 17.09, 15.76, and 15.59. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52FO3: 563.3895; found: 563.3879.
(4j, C37H51FO3). White solid; Yield: 57%; mp: 226–227°C; 1H NMR (500 MHz, chloroform-d) δ 7.18–7.13 (m, 2H), 7.00 (t, J = 8.5 Hz, 2H), 6.65 (s, 1H), 5.24–5.20 (m, 1H), 3.86 (d, J = 1.9 Hz, 1H), 2.80 (dd, J = 16.4, 11.2 Hz, 2H), 1.95 (td, J = 13.5, 4.1 Hz, 1H), 1.85–1.51 (m, 10H), 1.50–1.16 (m, 7H), 1.13 (s, 6H), 1.10–1.01 (m, 2H), 0.91 (s, 3H), 0.89 (s, 3H), 0.75 (s, 3H), 0.73 (s, 3H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.15, 161.38, 143.69, 140.42, 134.08, 130.41, 122.47, 121.69, 115.14, 81.17, 55.83, 47.01, 46.64, 45.95, 41.75, 41.69, 41.67, 41.08, 40.35, 39.57, 33.88, 33.20, 32.56, 32.53, 30.80, 28.75, 27.78, 26.04, 23.71, 23.45, 22.94, 18.50, 17.10, 15.77, and 15.57. HRMS (ESI-MS) m/z: [M + Na]+ calculated for C37H52FO3: 563.3895; found: 563.3899.
(4k, C37H51ClO3). White solid; Yield: 58%; mp: 219–220 °C; 1H NMR (500 MHz, chloroform-d) δ7.38 (dd, J = 7.3, 1.8 Hz, 1H), 7.20–7.13 (m, 3H), 6.67 (s, 1H), 5.17 (d, J = 3.1 Hz, 1H), 3.92 (d, J = 1.9 Hz, 1H), 2.76 (dd, J = 14.0, 4.5 Hz, 1H), 2.58 (d, J = 12.8 Hz, 1H), 1.94 (td, J = 13.5, 4.1 Hz, 1H), 1.78–1.50 (m, 10H), 1.44 (td, J = 14.2, 12.6, 9.8 Hz, 2H), 1.38–1.27 (m, 3H), 1.22–1.17 (m, 1H), 1.14 (d, J = 14.9 Hz, 6H), 1.09–1.02 (m, 3H), 0.89 (s, 3H), 0.89 (s, 3H), 0.80 (s, 3H), 0.68 (s, 3H), and 0.64 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.20, 143.64, 141.64, 136.67, 134.09, 130.93, 129.40, 127.79, 126.31, 122.50, 120.78, 81.16, 55.82, 47.09, 46.64, 45.95, 42.22, 41.77, 41.72, 41.08, 40.28, 39.56, 33.86, 33.21, 32.57, 32.52, 30.79, 28.69, 27.76, 26.05, 23.70, 23.42, 22.92, 18.55, 17.07, 15.72, and 15.18. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52ClO3: 579.3599; found: 579.3577.
(4l, C37H51ClO3). White solid; Yield: 69%; mp: 242–243 °C; 1H NMR (500 MHz, chloroform-d) δ 7.27–7.15 (m, 3H), 7.08 (d, J = 7.5 Hz, 1H), 6.64 (s, 1H), 5.24 (d, J = 3.6 Hz, 1H), 3.87 (d, J = 2.0 Hz, 1H), 2.81 (dd, J = 19.1, 11.1 Hz, 2H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.83–1.53 (m, 8H), 1.51–1.40 (m, 2H), 1.41–1.26 (m, 4H), 1.29–1.15 (m, 2H), 1.13 (d, J = 3.2 Hz, 7H), 1.10–1.01 (m, 2H), 0.90 (d, J = 8.1 Hz, 6H), 0.75 (d, J = 12.9 Hz, 6H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.11, 143.69, 141.76, 140.05, 134.06, 129.49, 128.97, 127.12, 126.29, 122.50, 121.60, 81.14, 55.83, 47.01, 46.64, 45.95, 41.79, 41.77, 41.75, 41.06, 40.45, 39.57, 33.88, 33.20, 32.53, 30.80, 28.77, 27.79, 26.04, 23.70, 23.41, 22.94, 18.51, 17.12, 15.79, and 15.65. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52ClO3: 579.3599; found: 579.3572.
(4m, C37H51ClO3). White solid; Yield: 57%; mp: 196–197 °C; 1H NMR (500 MHz, chloroform-d) δ 7.28 (d, J = 8.4 Hz, 2H), 7.13 (d, J = 8.3 Hz, 2H), 6.64 (s, 1H), 5.23 (t, J = 3.7 Hz, 1H), 3.86 (d, J = 2.1 Hz, 1H), 2.81 (t, J = 13.1 Hz, 2H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.82–1.52 (m, 8H), 1.55–1.17 (m, 8H), 1.13 (s, 7H), 1.06 (ddd, J = 17.3, 10.9, 4.1 Hz, 2H), 0.90 (d, J = 10.2 Hz, 6H), 0.74 (d, J = 7.3 Hz, 6H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.13, 143.70, 141.12, 136.60, 131.86, 130.26, 128.42, 122.42, 121.66, 81.16, 55.81, 47.02, 46.63, 45.94, 41.74, 41.72, 41.06, 40.41, 39.57, 33.87, 33.20, 32.54, 32.52, 30.79, 28.75, 27.77, 26.04, 23.71, 23.48, 22.93, 18.50, 17.08, 15.78, and 15.58. HRMS (ESI-MS) m/z: [M + K]+ calculated for C37H51ClO3K: 617.3158; found: 617.3146.
(4n, C37H51BrO3). White solid; Yield: 58%; mp: 234–235 °C; 1H NMR (500 MHz, chloroform-d) δ 7.57 (d, J = 7.9 Hz, 1H), 7.24 (d, J = 7.6 Hz, 1H), 7.13 (d, J = 7.6 Hz, 1H), 7.09 (t, J = 7.8 Hz, 1H), 6.62 (s, 1H), 5.17 (d, J = 3.0 Hz, 1H), 3.91 (d, J = 1.9 Hz, 1H), 2.76 (dd, J = 13.9, 4.5 Hz, 1H), 2.57 (d, J = 12.8 Hz, 1H), 1.94 (td, J = 13.6, 4.1 Hz, 1H), 1.78–1.61 (m, 5H), 1.55 (dd, J = 22.2, 13.7 Hz, 4H), 1.49–1.23 (m, 5H), 1.08 (dd, J = 50.0, 12.8 Hz, 11H), 0.89 (d, J = 4.4 Hz, 6H), 0.81 (s, 3H), 0.69 (s, 3H), and 0.64 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 183.95, 143.63, 141.30, 138.47, 132.57, 131.01, 128.00, 126.95, 124.65, 123.04, 122.51, 81.11, 55.81, 47.10, 46.63, 45.94, 42.17, 41.84, 41.72, 41.10, 40.32, 39.56, 33.86, 33.21, 32.58, 32.52, 30.79, 28.70, 27.75, 26.04, 23.70, 23.42, 22.92, 18.55, 17.08, 15.81, and 15.21. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52BrO3: 623.3094; found: 623.3067.
(4o, C37H51BrO3). White solid; Yield: 60%; mp: 197–198 °C; 1H NMR (500 MHz, chloroform-d) δ 7.38–7.31 (m, 2H), 7.17 (t, J = 7.8 Hz, 1H), 7.12 (d, J = 7.7 Hz, 1H), 6.64 (s, 1H), 5.26–5.21 (m, 1H), 3.87 (d, J = 1.9 Hz, 1H), 2.88–2.74 (m, 2H), 1.95 (td, J = 13.5, 4.1 Hz, 1H), 1.83–1.51 (m, 8H), 1.51–1.24 (m, 7H), 1.23–1.17 (m, 1H), 1.16–1.01 (m, 9H), 0.90 (d, J = 8.0 Hz, 6H), 0.75 (d, J = 13.9 Hz, 6H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.31, 143.69, 141.80, 140.35, 131.86, 129.77, 129.17, 127.55, 122.49, 122.33, 121.50, 81.14, 55.84, 47.01, 46.64, 45.95, 41.79, 41.74, 41.04, 40.46, 39.57, 33.88, 33.20, 32.53, 30.79, 29.83, 28.77, 27.79, 23.70, 23.39, 22.93, 18.50, 17.12, 15.79, and 15.65. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52BrO3: 623.3094; found: 623.3087.
(4p, C37H51BrO3). White solid; Yield: 66%; mp: 224–225 °C; 1H NMR (500 MHz, chloroform-d) δ7.43 (d, J = 8.3 Hz, 2H), 7.07 (d, J = 8.2 Hz, 2H), 6.62 (s, 1H), 5.34–5.09 (m, 1H), 3.86 (d, J = 2.0 Hz, 1H), 2.81 (t, J = 12.5 Hz, 2H), 1.96 (td, J = 13.5, 4.1 Hz, 1H), 1.85–1.51 (m, 7H), 1.51–1.17 (m, 9H), 1.13 (s, 9H), 0.91 (s, 3H), 0.89 (s, 3H), 0.74 (s, 3H), 0.73 (s, 3H), and 0.68 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.11, 143.69, 141.22, 137.09, 131.38, 130.63, 122.44, 121.70, 120.01, 81.18, 55.82, 47.04, 46.64, 45.95, 41.75, 41.73, 41.08, 40.43, 39.58, 33.88, 33.21, 32.55, 32.53, 30.80, 29.85, 28.76, 27.78, 26.05, 23.72, 23.50, 22.94, 18.52, 17.09, 15.79, and 15.60. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H52BrO3: 623.3094; found: 623.3067.
(4q, C37H51NO5). White solid; Yield: 55%; mp: 197–198 °C; 1H NMR (500 MHz, chloroform-d) δ 8.02 (dd, J = 8.2, 1.3 Hz, 1H), 7.54 (td, J = 7.5, 1.4 Hz, 1H), 7.46–7.35 (m, 1H), 7.24 (d, J = 7.7 Hz, 1H), 6.89 (s, 1H), 5.15 (d, J = 2.7 Hz, 1H), 3.90 (d, J = 1.8 Hz, 1H), 2.74 (dd, J = 13.9, 4.5 Hz, 1H), 2.41 (d, J = 12.8 Hz, 1H), 1.94 (td, J = 13.2, 4.1 Hz, 1H), 1.71 (td, J = 13.9, 4.5 Hz, 1H), 1.56 (dt, J = 15.8, 7.4 Hz, 6H), 1.44 (d, J = 13.0 Hz, 2H), 1.37–1.24 (m, 5H), 1.19–1.01 (m, 10H), 0.88 (d, J = 1.8 Hz, 7H), 0.82 (s, 3H), and 0.63 (d, J = 4.2 Hz, 6H). 13C NMR (126 MHz, CDCl3) δ 183.70, 148.59, 143.64, 141.68, 133.92, 132.66, 132.31, 127.42, 124.63, 122.18, 119.76, 80.98, 55.62, 46.89, 46.50, 45.84, 42.32, 41.81, 41.64, 40.95, 40.09, 39.43, 33.74, 33.07, 32.43, 32.33, 30.66, 28.62, 27.63, 25.89, 23.58, 23.25, 22.85, 18.45, 16.92, 15.56, and 14.94. HRMS (ESI-MS) m/z: [M + K]+ calculated for C37H51NO5K: 628.3399; found: 628.3420.
(4r, C35H51NO5). White solid; Yield: 64%; mp: 191–192 °C; 1H NMR (500 MHz, chloroform-d) δ8.11 (d, J = 2.0 Hz, 1H), 8.07 (dt, J = 7.7, 1.8 Hz, 1H), 7.54–7.46 (m, 2H), 6.76 (s, 1H), 5.22 (d, J = 3.6 Hz, 1H), 3.90 (d, J = 2.0 Hz, 1H), 2.85–2.74 (m, 2H), 1.96 (td, J = 13.5, 4.0 Hz, 1H), 1.82–1.65 (m, 4H), 1.63–1.53 (m, 4H), 1.47 (d, J = 12.6 Hz, 2H), 1.38–1.28 (m, 3H), 1.25 (d, J = 2.9 Hz, 1H), 1.14 (d, J = 9.3 Hz, 7H), 1.11–0.97 (m, 3H), 0.89 (d, J = 7.5 Hz, 7H), 0.77 (d, J = 17.2 Hz, 6H), and 0.69 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 183.73, 148.21, 143.60, 143.21, 139.79, 135.02, 129.08, 123.51, 122.26, 121.09, 120.74, 80.95, 55.68, 46.92, 46.51, 45.83, 41.84, 41.65, 41.59, 40.94, 40.53, 39.47, 33.76, 33.07, 32.42, 32.39, 30.68, 28.69, 27.67, 25.91, 23.57, 23.27, 22.83, 18.39, 16.99, 15.70, and 15.56. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H51NO5: 590.3840; found: 590.3849.
(4s, C34H51NO5). White solid; Yield: 50%; mp: 219–220 °C; 1H NMR (500 MHz, chloroform-d) δ 8.19–8.16 (m, 2H), 7.37–7.31 (m, 2H), 6.77 (s, 1H), 5.22 (t, J = 3.6 Hz, 1H), 3.91 (d, J = 1.9 Hz, 1H), 2.86–2.75 (m, 2H), 1.95 (td, J = 13.5, 4.1 Hz, 1H), 1.80–1.40 (m, 12H), 1.39–1.17 (m, 5H), 1.14 (d, J = 8.4 Hz, 6H), 1.11–1.00 (m, 2H), 0.90 (d, J = 9.0 Hz, 6H), 0.74 (d, J = 3.9 Hz, 6H), and 0.67 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 184.06, 146.10, 145.44, 144.30, 143.81, 129.58, 123.68, 122.18, 121.45, 81.06, 55.71, 47.06, 46.59, 45.92, 42.02, 41.92, 41.74, 41.05, 40.72, 39.59, 33.85, 33.18, 32.50, 30.77, 29.81, 28.78, 27.75, 26.02, 23.68, 23.46, 22.89, 18.51, 17.04, 15.82, and 15.59. HRMS (ESI-MS) m/z: [M + H]+ calculated for C37H51NO5: 590.3840; found: 590.3849.
The α-glucosidase inhibition assay of analogs 4a–4s was carried out as in previously reported methods, with minor modifications (Xu et al., 2020). Then, 10 μL α-glucosidase and 10 μL test sample (dissolved in DMSO) were added into 130 μL PBS (50 mM phosphate saline buffer, pH 6.8) and incubated for 10 min at 37°C. After adding 50 μL pNPG, the absorbance change of the mixture at 405 nm was monitored. Then, the percent inhibition was calculated (Ademiluyi and Oboh, 2013). All experiments were carried out in quadruplicate.
The α-amylase inhibitory activity was performed according to methods followed in previous reports, with minor modification (Hameed et al., 2019). Then, 10 μL α-amylase (final concentration 0.25 U/mL), 10 μL test compound, and 80 μL phosphate buffer (20 mM, pH 6.9) were mixed and incubated for 10 min at 37°C. Then, 100 μL starch solution (final concentration 0.5%) was added into the mixture and further incubated for 10 min. After 100 μL DNS (containing 1 M potassium sodium tartrate and 48 mM 3,5-dinitrosalicylic acid) was added, the mixture was boiled for 15 min. After diluting using 900 μL distilled water, the absorbance was measured at 540 nm. Then, the percent inhibition was calculated (Aispuro-Perez et al., 2020). All experiments were carried out in quadruplicate.
The inhibition kinetics of 4i and 4o against α-glucosidase and α-amylase, respectively, was investigated using a similar aforementioned inhibition assay method. The enzyme inhibitory kinetics was obtained by the plots of the enzymatic reaction rate vs. enzyme concentration with or without the inhibitor, and the substrate inhibitory kinetics was measured using the Lineweaver–Burk plot of the enzymatic reaction rate vs. substrate concentration with or without the inhibitor.
Molecular docking was performed using SYBYL software to investigate the interaction between inhibitors and target protein (Zheng P.-F. et al., 2021; Hu, et al., 2021). The crystal structure of Saccharomyces cerevisiae α-glucosidase was not resolved so far. The homology model of α-glucosidase was constructed using the protocol reported earlier (Wang, et al., 2017; Wang et al., 2017). In brief, the structure of Saccharomyces cerevisiae isomaltase (PDB ID: 3AJ7) was selected as a template, the sequence in FASTA format of α-glucosidase was obtained from UniProt (access code P53341), and the homology model was prepared using modeler 10.1 software. The quality of the homology model was verified by Ramachandran plot (Yamamoto et al., 2010). Porcine pancreatic α-amylase (PDB: 3BAJ) was retrieved from the Protein Data Bank (Rafique et al., 2020). The target protein was optimized by removing water molecules, adding hydrogen atoms, adding charge, and repairing end residues, followed by the generation of an active pocket. The compounds were charged with Gasteiger–Hückle charges and prepared by an energy minimization program. Thus, the docking between the compounds and target protein was operated in the default format, and the results were visualized by PyMol and Discover studio software.
All data were presented as mean ± SD. One-way ANOVA was performed to evaluate the difference between groups. p < 0.05 was considered significant.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
J-JK, JL, and XZ contributed to the synthesis and purification of compounds; J-JK, X-ZW, and Y-YZ contributed to the characterization of compounds and interpretation of data for the work. J-JK, X-ZW, Y-YZ, C-MH, and YK contributed to the enzyme inhibition assay and enzyme kinetics assay; J-JK performed the molecular docking simulation. KZ and Z-QM conceived and designed the experiments. ZX supervised the work and prepared the manuscript.
This work was financially supported by the Department of Education of Guangdong Province (Nos. 2019KZDXM035, 2021KTSCX135, and 2021KCXTD044) and Jiangmen City Science and Technology Basic Research Project (No.2020030101030005457).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.911232/full#supplementary-material
Ademiluyi, A. O., and Oboh, G. (2013). Aqueous Extracts of Roselle (Hibiscus sabdariffa Linn.) Varieties Inhibit α-Amylase and α-Glucosidase Activities In Vitro. J. Med. Food 16, 88–93. doi:10.1089/jmf.2012.0004
Ahmed, A. A., Mollica, A., Stefanucci, A., Tayrab, E., Ahmed, H., and Adam Essa, M. E. (2020). Gum Arabic Improves the Reproductive Capacity through Upregulation of Testicular Glucose Transporters (GLUTs) mRNA Expression in Alloxan Induced Diabetic Rat. Bioact. Carbohydrates Diet. Fibre 22, 100218. doi:10.1016/j.bcdf.2020.100218
Aispuro-Pérez, A., López-Ávalos, J., García-Páez, F., Montes-Avila, J., Picos-Corrales, L. A., Ochoa-Terán, A., et al. (2020). Synthesis and Molecular Docking Studies of Imines as α-glucosidase and α-amylase Inhibitors. Bioorg. Chem. 94, 103491. doi:10.1016/j.bioorg.2019.103491
Carullo, G., Mazzotta, S., Vega-Holm, M., Iglesias-Guerra, F., Vega-Pérez, J. M., Aiello, F., et al. (2021). GPR120/FFAR4 Pharmacology: Focus on Agonists in Type 2 Diabetes Mellitus Drug Discovery. J. Med. Chem. 64, 4312–4332. doi:10.1021/acs.jmedchem.0c01002
Castellano, J. M., Guinda, A., Delgado, T., Rada, M., and Cayuela, J. A. (2013). Biochemical Basis of the Antidiabetic Activity of Oleanolic Acid and Related Pentacyclic Triterpenes. Diabetes 62, 1791–1799. doi:10.2337/db12-1215
Cheng, F., Duan, D.-S., Jiang, L.-M., Li, B.-S., Wang, J.-X., Zhou, Y.-J., et al. (2022). Copper-Catalyzed Asymmetric Ring-Opening Reaction of Cyclic Diaryliodonium Salts with Imides. Org. Lett. 24, 1394–1399. doi:10.1021/acs.orglett.2c00247
Csuk, R., Stark, S., Nitsche, C., Barthel, A., and Siewert, B. (2012). Alkylidene Branched Lupane Derivatives: Synthesis and Antitumor Activity. Eur. J. Med. Chem. 53, 337–345. doi:10.1016/j.ejmech.2012.04.023
Deng, X.-Y., Ke, J.-J., Zheng, Y.-Y., Li, D.-L., Zhang, K., Zheng, X., et al. (2022). Synthesis and Bioactivities Evaluation of Oleanolic Acid Oxime Ester Derivatives as α-glucosidase and α-amylase Inhibitors. J. Enzyme Inhibition Med. Chem. 37, 451–461. doi:10.1080/14756366.2021.2018682
Forouhi, N. G., and Wareham, N. J. (2010). Epidemiology of Diabetes. Medicine 38, 602–606. doi:10.1016/j.mpmed.2010.08.007
Furer, N., Kaushansky, N., and Shlush, L. I. (2021). The Vicious and Virtuous Circles of Clonal Hematopoiesis. Nat. Med. 27, 949–950. doi:10.1038/s41591-021-01396-5
Gupta, N., Rath, S. K., Singh, J., Qayum, A., Singh, S., and Sangwan, P. L. (2017). Synthesis of Novel Benzylidene Analogues of Betulinic Acid as Potent Cytotoxic Agents. Eur. J. Med. Chem. 135, 517–530. doi:10.1016/j.ejmech.2017.04.062
Hameed, S., KanwalSeraj, F., Seraj, F., Rafique, R., Chigurupati, S., Wadood, A., et al. (2019). Synthesis of Benzotriazoles Derivatives and Their Dual Potential as α-amylase and α-glucosidase Inhibitors In Vitro: Structure-Activity Relationship, Molecular Docking, and Kinetic Studies. Eur. J. Med. Chem. 183, 111677. doi:10.1016/j.ejmech.2019.111677
Hossain, U., Das, A. K., Ghosh, S., and Sil, P. C. (2020). An Overview on the Role of Bioactive α-glucosidase Inhibitors in Ameliorating Diabetic Complications. Food Chem. Toxicol. 145, 111738. doi:10.1016/j.fct.2020.111738
Hu, C.-M., Wang, W.-J., Ye, Y.-N., Kang, Y., Lin, J., Wu, P.-P., et al. (2021). Novel Cinnamic Acid Magnolol Derivatives as Potent α-glucosidase and α-amylase Inhibitors: Synthesis, In Vitro and In Silico Studies. Bioorg. Chem. 116, 105291. doi:10.1016/j.bioorg.2021.105291
Jensen, H. E., Christensen, J. P., Bisgaard, M., and Nielsen, O. L. (1997). Immunohistochemistry for the Diagnosis of Aspergillosis in Turkey Poults. Avian Pathol. 26, 5–18. doi:10.1080/03079459708419189
Kathuria, D., Raul, A. D., Wanjari, P., and Bharatam, P. V. (2021). Biguanides: Species with Versatile Therapeutic Applications. Eur. J. Med. Chem. 219, 113378. doi:10.1016/j.ejmech.2021.113378
Kazakova, O., Rubanik, L., Smirnova, I., Poleschuk, N., Petrova, A., Kapustsina, Y., et al. (2021). Synthesis and In Vitro Activity of Oleanolic Acid Derivatives against Chlamydia trachomatis and Staphylococcus aureus. Med. Chem. Res. 30, 1408–1418. doi:10.1007/s00044-021-02741-6
Kim, H. R., Lee, H. J., Choi, Y. J., Park, Y. J., Woo, Y., Kim, S. J., et al. (2014). Benzylidene-linked Thiohydantoin Derivatives as Inhibitors of Tyrosinase and Melanogenesis: Importance of the β-phenyl-α,β-unsaturated Carbonyl Functionality. Med. Chem. Commun. 5, 1410–1417. doi:10.1039/c4md00171k
Kokil, G. R., Veedu, R. N., Ramm, G. A., Prins, J. B., and Parekh, H. S. (2015). Type 2 Diabetes Mellitus: Limitations of Conventional Therapies and Intervention with Nucleic Acid-Based Therapeutics. Chem. Rev. 115, 4719–4743. doi:10.1021/cr5002832
Kumar, K., Suebsuwong, C., Wang, P., Garcia-Ocana, A., Stewart, A. F., and DeVita, R. J. (2021). DYRK1A Inhibitors as Potential Therapeutics for β-Cell Regeneration for Diabetes. J. Med. Chem. 64, 2901–2922. doi:10.1021/acs.jmedchem.0c02050
Liang, H., Gou, A., Gao, Z., Lei, L., Wang, B., Yu, L., et al. (2020). A New Strategy for the Synthesis of Tertiary Amides via a Copper-Catalyzed Decyanation Reaction of N,N-Disubstituted 2-Aminomalononitriles. Acta Chim. Sin. 78, 1064–1068. doi:10.6023/a20070296
Ling, S., Zaccardi, F., Lawson, C., Seidu, S. I., Davies, M. J., and Khunti, K. (2021). Glucose Control, Sulfonylureas, and Insulin Treatment in Elderly People with Type 2 Diabetes and Risk of Severe Hypoglycemia and Death: An Observational Study. Diabetes Care 44, 915–924. doi:10.2337/dc20-0876
Liu, J. (2005). Oleanolic Acid and Ursolic Acid: Research Perspectives. J. Ethnopharmacol. 100, 92–94. doi:10.1016/j.jep.2005.05.024
Liu, S., Cui, M., Wang, B., Hu, C., Zheng, Y., Li, J., et al. (2021). Triethyl Amine-Promoted Cyclization Reaction between Cyclopropenone and α-Halogenated Hydroxamate for the Synthesis of Polysubstituted 6H-1,3-Oxazin-6-One. Chin. J. Org. Chem. 41, 1622–1630. doi:10.6023/cjoc202010041
Long, N., Le Gresley, A., and Wren, S. P. (2021). Thiazolidinediones: An In-Depth Study of Their Synthesis and Application to Medicinal Chemistry in the Treatment of Diabetes Mellitus. ChemMedChem 16, 1717–1736. doi:10.1002/cmdc.202100177
Loza-Rodríguez, H., Estrada-Soto, S., Alarcón-Aguilar, F. J., Huang, F., Aquino-Jarquín, G., Fortis-Barrera, Á., et al. (2020). Oleanolic Acid Induces a Dual Agonist Action on PPARγ/α and GLUT4 Translocation: A Pentacyclic Triterpene for Dyslipidemia and Type 2 Diabetes. Eur. J. Pharmacol. 883, 173252. doi:10.1016/j.ejphar.2020.173252
Mollica, A., Stefanucci, A., Zengin, G., Locatelli, M., Macedonio, G., Orlando, G., et al. (2018). Polyphenolic Composition, Enzyme Inhibitory Effects Eex-Vvivo and Iin-Vvivo Studies on Two Brassicaceae of North-Central Italy. Biomed. Pharmacother. 107, 129–138. doi:10.1016/j.biopha.2018.07.169
Mollica, A., Zengin, G., Locatelli, M., Stefanucci, A., Mocan, A., Macedonio, G., et al. (2017). Anti-diabetic and Anti-hyperlipidemic Properties of Capparis Spinosa L.: In Vivo and In Vitro Evaluation of its Nutraceutical Potential. J. Funct. Foods 35, 32–42. doi:10.1016/j.jff.2017.05.001
Pollier, J., and Goossens, A. (2012). Oleanolic Acid. Phytochemistry 77, 10–15. doi:10.1016/j.phytochem.2011.12.022
Proença, C., Ribeiro, D., Freitas, M., and Fernandes, E. (2021). Flavonoids as Potential Agents in the Management of Type 2 Diabetes through the Modulation of α-amylase and α-glucosidase Activity: a Review. Crit Rev Food Sci Nutr, 62. 3137. doi:10.1080/10408398.2020.1862755
Provenzano, M., Pelle, M. C., Zaffina, I., Tassone, B., Pujia, R., Ricchio, M., et al. (2021). Sodium-Glucose Co-transporter-2 Inhibitors and Nephroprotection in Diabetic Patients: More Than a Challenge. Front. Med. 8, 654557. doi:10.3389/fmed.2021.654557
Rafique, R., Khan, K. M., Arshia, K., Kanwal, S., Chigurupati, S., Wadood, A., et al. (2020). Synthesis of New Indazole Based Dual Inhibitors of α-glucosidase and α-amylase Enzymes, Their In Vitro, In Silico and Kinetics Studies. Bioorg. Chem. 94, 103195. doi:10.1016/j.bioorg.2019.103195
Saxena, A. R., Gorman, D. N., Esquejo, R. M., Bergman, A., Chidsey, K., Buckeridge, C., et al. (2021). Danuglipron (PF-06882961) in Type 2 Diabetes: a Randomized, Placebo-Controlled, Multiple Ascending-Dose Phase 1 Trial. Nat. Med. 27 (6), 1079–1087. doi:10.1038/s41591-021-01391-w
Silva, F. S. G., Oliveira, P. J., and Duarte, M. F. (2016). Oleanolic, Ursolic, and Betulinic Acids as Food Supplements or Pharmaceutical Agents for Type 2 Diabetes: Promise or Illusion? J. Agric. Food Chem. 64, 2991–3008. doi:10.1021/acs.jafc.5b06021
Tang, C., Zhu, L., Chen, Y., Qin, R., Mei, Z., Xu, J., et al. (2014). Synthesis and Biological Evaluation of Oleanolic Acid Derivative-Chalcone Conjugates as α-glucosidase Inhibitors. RSC Adv. 4, 10862–10874. doi:10.1039/c3ra46492j
Wang, G., Wang, J., Xie, Z., Chen, M., Li, L., Peng, Y., et al. (2017). Discovery of 3,3-Di(indolyl)indolin-2-One as a Novel Scaffold for α-glucosidase Inhibitors: In Silico Studies and SAR Predictions. Bioorg. Chem. 72, 228–233. doi:10.1016/j.bioorg.2017.05.006
Wu, P., He, H., Ma, H., Tu, B., Li, J., Guo, S., et al. (2021). Oleanolic Acid Indole Derivatives as Novel α-glucosidase Inhibitors: Synthesis, Biological Evaluation, and Mechanistic Analysis. Bioorg. Chem. 107, 104580. doi:10.1016/j.bioorg.2020.104580
Xu, X.-T., Deng, X.-Y., Chen, J., Liang, Q.-M., Zhang, K., Li, D.-L., et al. (2020). Synthesis and Biological Evaluation of Coumarin Derivatives as α-glucosidase Inhibitors. Eur. J. Med. Chem. 189, 112013. doi:10.1016/j.ejmech.2019.112013
Yamamoto, K., Miyake, H., Kusunoki, M., and Osaki, S. (2010). Crystal Structures of Isomaltase from Saccharomyces cerevisiae and in Complex with its Competitive Inhibitor Maltose. FEBS J. 277, 4205–4214. doi:10.1111/j.1742-4658.2010.07810.x
Zheng, P.-F., Xiong, Z., Liao, C.-y., Zhang, X., Feng, M., Wu, X.-Z., et al. (2021b). In Vitro and In Silico Studies of Bis (Indol-3-yl) Methane Derivatives as Potential α-glucosidase and α-amylase Inhibitors. J. Enzyme Inhibition Med. Chem. 36, 1938–1951. doi:10.1080/14756366.2021.1971976
Zheng, T.-L., Liu, S.-Z., Huo, C.-Y., Li, J., Wang, B.-W., Jin, D.-P., et al. (2021a). Au-Catalyzed 1,3-Acyloxy Migration/Cyclization Cascade: A Direct Strategy toward the Synthesis of Functionalized Abietane-type Diterpenes. CCS Chem. 3, 2795–2802. doi:10.31635/ccschem.020.202000582
Zhong, Y.-Y., Chen, H.-S., Wu, P.-P., Zhang, B.-J., Yang, Y., Zhu, Q.-Y., et al. (2019). Synthesis and Biological Evaluation of Novel Oleanolic Acid Analogues as Potential α-glucosidase Inhibitors. Eur. J. Med. Chem. 164, 706–716. doi:10.1016/j.ejmech.2018.12.046
Keywords: oleanolic acid, α-amylase, α-glucosidase, enzyme inhibitor, docking
Citation: Ke J-J, Lin J, Zhang X, Wu X-Z, Zheng Y-Y, Hu C-M, Kang Y, Zhang K, Xiong Z and Ma Z-Q (2022) Synthesis of Benzylidene Analogs of Oleanolic Acid as Potential α-Glucosidase and α-Amylase Inhibitors. Front. Chem. 10:911232. doi: 10.3389/fchem.2022.911232
Received: 04 April 2022; Accepted: 21 April 2022;
Published: 08 June 2022.
Edited by:
Xi Zheng, Rutgers, The State University of New Jersey, United StatesReviewed by:
Xiping Cui, Guangdong University of Technology, ChinaCopyright © 2022 Ke, Lin, Zhang, Wu, Zheng, Hu, Kang, Zhang, Xiong and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuang Xiong, d3l1Y2hlbXh6QDEyNi5jb20=; Zhi-Qiang Ma, Y2V6cW1hQHNjdXQuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.