
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 11 April 2022
Sec. Organic Chemistry
Volume 10 - 2022 | https://doi.org/10.3389/fchem.2022.851674
This article is part of the Research Topic DNA-encoded Library and Chemistry View all 4 articles
C-N bond formation is one of the most commonly used reactions in medicinal chemistry. Herein, we report an efficient Pd-promoted hydroamination reaction between DNA-conjugated aryl alkenes and a wide scope of aliphatic amines. The described reactions are demonstrated in good to excellent conversions to furnish C (sp3)–N bonds on DNA. This DNA-compatible transformation has strong potentials for the application into DNA-encoded library synthesis.
DNA-encoded library technology (DELT), first conceptualized by Brenner and Lerner in 1992 Brenner and Lerner. (1992), has evolved into one of major platforms for small-molecule drug discovery. DNA-encoded libraries are built with combinatorial chemistries in a split-pool manner. Each individual compound in a DNA-encoded library is covalently attached to a unique DNA sequence, which serves as a barcode. The libraries, typically consisting of millions to billions of DNA-encoded molecules, can be pooled together and conveniently screened against biological targets with pharmaceutical interests in a single experiment. It has been recognized as a powerful and economic tool for hit identifications in both industry and academia (Gironda-Martínez et al., 2021).
One of the challenges in the area of DELT is to develop DNA-compatible chemistry for the synthesis of drug-like and diverse chemical entities as starting points for medicinal chemistry research (Zhu et al., 2019; Franzini and Randolph, 2016; Dickson and Kodadek, 2019; Götte et al., 2020). Most DNA-encoded library synthesis are reported to perform on unprotected DNA substrates (Shi et al., 2021; Fair et al., 2021); thus, the reaction conditions must be mild to preserve DNA information integrity, and consequently to remain its viability in PCR. In addition, chemical reactions used in DEL synthesis should have potentials to construct diverse encoded molecules by introducing multitudinous building blocks (BBs) through suitable functional groups on DNA (Zhang and Clark, 2021; Kalliokoski, 2015; Zabolotna et al., 2021), mainly represented by carboxylic acids, aldehydes, amines, aryl halides, etc.
C-N bond formation is of the most used chemistry transformations in medicinal chemistry due to the prevalence of nitrogen-containing motifs in approved drugs and bioactive compounds. C-N bond formation is extremely appealing for DEL synthesis because amines (or aldehydes) as BBs are easily accessible synthetic reagents (Zabolotna et al., 2021) for generating library diversity. Several on-DNA C-N bond formations, exemplified as C-N cross coupling (Buchwald coupling (Xu et al., 2019; de Pedro Beato et al., 2019; Chen et al., 2020; Lu et al., 2017), Ullmann coupling (Lu et al., 2017; Ruff and Berst, 2018), SNAr (Ding et al., 2018; Clark et al., 2009; Wang et al., 2019), and reductive amination (Flood et al., 2019; Ding et al., 2016), have been already reported for the synthesis of DELs. As depicted in Figure 1, these transformations require specific functional groups on DNA, such as DNA-tagged aryl halides, aldehydes, and amines. Herein, we describe the development of unprecedented Pd-promoted hydroamination reactions between DNA-conjugated aryl alkenes and a wide scope of aliphatic amines.
The headpiece-tagged 4-vinylpyridine 1a was used as a prototypical on-DNA substrate for the optimization of the hydroamination reactions in aqueous solutions (see headpiece structures in Supporting Information). After extensive reaction optimizations, the 2-(pyridin-4-yl)ethan-1-amine 3a was obtained in 88% conversion (Table 1, entry 1). The reaction was dramatically promoted by the palladium catalyst. There was only trace amounts of 3a detected under the condition without Pd(OAc)2 and most of the starting material 1a was inert (entry 2). Other palladium catalysts, such as PdCl2 and Pd(PPh3)4, which are frequently employed in cross coupling reactions, led to diminished conversions (entries 3 and 4). We then presumed that the conversion was maximized by introducing large excesses of isopropyl amine 2a. By comparison, 500 equivalents of 2a in the standard condition gave better conversions than 200 equivalents in entry 5 and 100 equivalents in entry 6. This is in line with the kinetic mechanism for developing on-DNA intermolecular transformations. We finally found pH value of the final reaction solution played vital roles on conversions. As depicted in entry 7, the conversion was decreased to 55% without adding CsOH. In a reverse way, excess CsOH led to the final pH of the reaction solution to 12 that gave a diminished conversion (entry 8). Finally, we performed the reaction in 100 nmol scale under the optimal condition and found that it gave a comparable conversion as the small-scale reaction (Table 1, entry 9).
With the optimal conditions in hand, we further studied substrate scopes of amines against headpiece-conjugated 4-vinylpyridine 1a (Scheme 1). Under the conditions, we found that most primary alkyl amines were well tolerated, giving moderate to excellent conversions (3a-3h). The conversions were decreased when increasing steric hindrance of amines (3i-3j). Various types of secondary amines were also studied. The results showed that cyclic amines led to excellent conversions, such as monocyclic amines (3k-3n), spirocyclic amines (3o-3p), bicyclic amines (3q-3r), and fused cyclic amines (3s). However, non-cyclic secondary amines, especially with bulky groups (3v), resulted in low conversions (3t-3v). Interestingly, we found that carboxylic acids (3w-3x), Boc-protected diamines (3y-3z), and aryl halides (3aa-3ab), which were frequently used for diversity derivations, were well tolerated under the optimal conditions. The tolerance of these functional groups provided a straightforward pathway for further creating library diversities. Unfortunately, aromatic amines were not tolerated with these conditions. Only trace amounts of desired products were detected, with most starting material remained (3ac-3af).
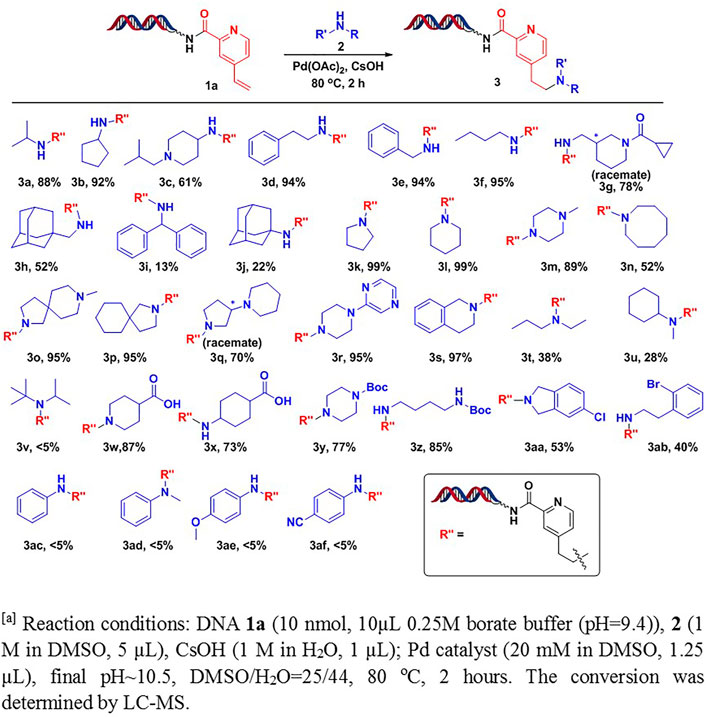
SCHEME 1. Scope study on amines Reaction conditions: DNA 1a [10 nmol, 10 μL, 0.25 M borate buffer (pH = 9.4)], 2 (1 M in DMSO, 5 μL), CsOH (1 M in H2O, 1 μL); Pd catalyst (20 mM in DMSO, 1.2 μL), pH∼10.5, DMSO/H2O = 25/44, 80°C, 2 h. The conversion was determined by LC-MS.
Various DNA-conjugated alkenes (Scheme 2) and aliphatic amines were subjected to the optimal reaction condition for a comprehensive substrate scope study. As outlined in Scheme 2, we first investigated the effects of DNA-tagged alkenes on the conversions. Two types of on-DNA alkenes were prepared. We found both styrene analogs (3b-3ao) and acrylamide analogs (3ap-3at) delivered decent conversions to desired products. To our delight, except mono-substituted alkenes, many disubstituted alkenes (3ao-3at) also worked well under the condition. Furthermore, DNA-tagged alkenes containing heterocyclic moieties, such as pyridine (3 aL-3am, 3aq), oxazole (3ah, 3au, 3bd, 3bi), imidazo [1,2-b]pyridazine (3aj, 3av, 3ax, 3bf, 3bg), pyrazolo [3,4-b]pyridine (3ak, 3aw, 3bb), isoxazole (3ar, 3ay, 3ba), pyrimidine (3ai, 3bc), and pyrazine (3 ag, 3be, 3bh), afforded the desired products in moderate-to-good conversions.
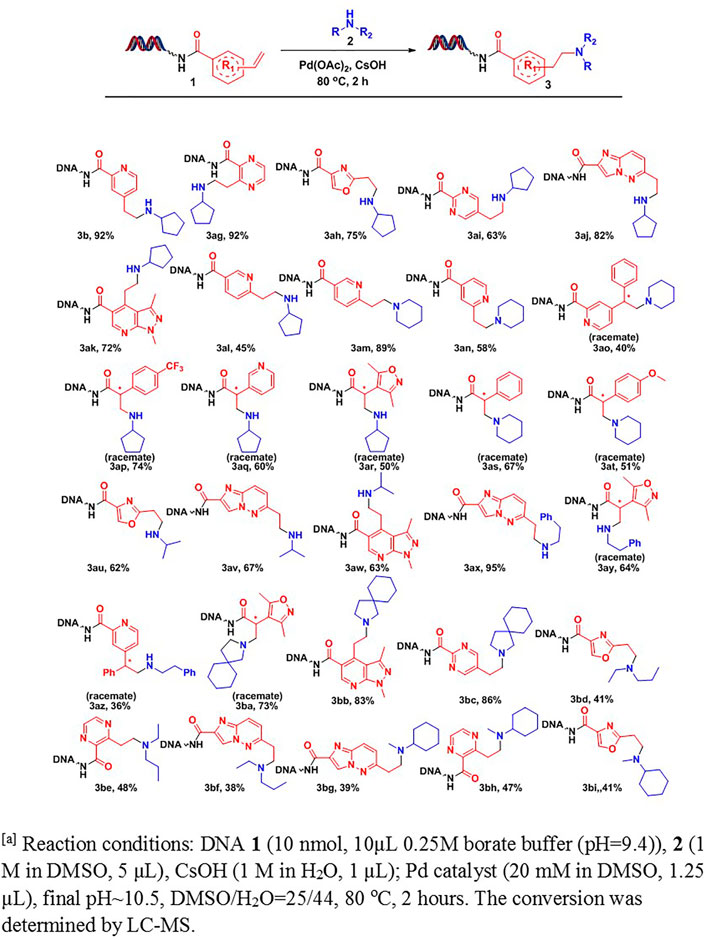
SCHEME 2. Scope study on DNA-conjugated aryl ethylene Reaction conditions: DNA 1 [10 nmol, 10 μL, 0.25 M borate buffer (pH = 9.4)], 2 (1 M in DMSO, 5 μL), CsOH (1 M in H2O, 1 μL); Pd catalyst (20 mM in DMSO, 1.2 μL), pH∼10.5, DMSO/H2O = 25/44, 80°C, 2 h. The conversion was determined by LC-MS.
To further confirm structures of the proposed products, co-injection experiments were carried out as shown in Scheme 3 (see details in Supporting Information). Small molecule 4, which was structurally confirmed by 1HNMR, was conjugated with DNA via acylation reactions. The obtained conjugate 5 was compared with on-DNA synthesized conjugate 3bj by co-injection experiments. Two samples gave identical m/z and HPLC trace, which indicated characterization of desired on-DNA product 3bj. Besides, the off-DNA small-molecule synthesis with established on-DNA condition was conducted, and the results indicated that the desired structure was generated with good regioselectivity (see details in Supporting Information, Supplementary Figure S17–S18).
A control experiment was also performed under our optimal conditions by employing a benzoyl-substituted DNA 6 as a substrate (Scheme 4). The purpose of the experiment was to confirm that the conjugated small molecule rather than the DNA backbone underwent the reaction. As expected, quantitative recovery of starting material 6 was observed, thereby lending credence to our contention that the C-N bond formation did not take place on the DNA backbone.
To demonstrate the compatibility of this reaction with DNA chain and its suitability for subsequent DEL construction, the C-N bond formation reaction on long-chain DNA (headpiece-primer-code 1) followed by enzymatic ligation and quantitative PCR (qPCR) was evaluated (see detailed information in Supporting Information). The conversion on long-chain DNA with 33-bp dsDNA was found to be consistent with that on short-chain DNA under the same reaction condition. No evidence of DNA decomposition was detected by LC-MS or gel electrophoresis. Subsequent qPCR experiments indicated no obvious DNA damage on either case.
In conclusion, we developed an efficient method of an efficient palladium-mediated hydroamination reaction to construct C[sp3]–N bond between DNA-conjugated aryl ethylenes and amines. The methodology was demonstrated with moderate-to-excellent product conversions under mild conditions. It was verified in terms of no obvious damage to the DNA substrate, indicating the suitability of the DNA encoded library production.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Authors KC, YR, WS, SG, JL, JW, and GL were employed by HitGen Inc.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge Rongfeng Wu at HitGen Inc. for testing repeatability of the reaction between conjugate 1a and amine 2a using the optimal reaction conditions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.851674/full#supplementary-material
Brenner, S., and Lerner, R. A. (1992). Encoded Combinatorial Chemistry. Proc. Natl. Acad. Sci. 89 (12), 5381–5383. doi:10.1073/pnas.89.12.5381
Chen, Y.-C., Faver, J. C., Ku, A. F., Miklossy, G., Riehle, K., Bohren, K. M., et al. (2020). C-N Coupling of DNA-Conjugated (Hetero)aryl Bromides and Chlorides for DNA-Encoded Chemical Library Synthesis. Bioconjug. Chem. 31 (3), 770–780. doi:10.1021/acs.bioconjchem.9b00863
Clark, M. A., Acharya, R. A., Arico-Muendel, C. C., Belyanskaya, S. L., Benjamin, D. R., Carlson, N. R., et al. (2009). Design, Synthesis and Selection of DNA-Encoded Small-Molecule Libraries. Nat. Chem. Biol. 5, 647–654. doi:10.1038/nchembio.211
de Pedro Beato, E., Priego, J., Gironda-Martínez, A., González, F., Benavides, J., Blas, J., et al. (2019). Mild and Efficient Palladium-Mediated C-N Cross-Coupling Reaction between DNA-Conjugated Aryl Bromides and Aromatic Amines. ACS Comb. Sci. 21 (2), 69–74. doi:10.1021/acscombsci.8b00142
Dickson, P., and Kodadek, T. (2019). Chemical Composition of DNA-Encoded Libraries, Past Present and Future. Org. Biomol. Chem. 17, 4676–4688. doi:10.1039/C9OB00581A
Ding, Y., Chai, J., Centrella, P. A., Gondo, C., DeLorey, J. L., and Clark, M. A. (2018). Development and Synthesis of DNA-Encoded Benzimidazole Library. ACS Comb. Sci. 20 (5), 251–255. doi:10.1021/acscombsci.8b00009
Ding, Y., Franklin, G. J., DeLorey, J. L., Centrella, P. A., Mataruse, S., Clark, M. A., et al. (2016). Design and Synthesis of Biaryl DNA-Encoded Libraries. ACS Comb. Sci. 18 (10), 625–629. doi:10.1021/acscombsci.6b00078
Fair, R. J., Walsh, R. T., and Hupp, C. D. (2021). The Expanding Reaction Toolkit for DNA-Encoded Libraries. Bioorg. Med. Chem. Lett. 51, 128339. doi:10.1016/j.bmcl.2021.128339
Flood, D. T., Asai, S., Zhang, X., Wang, J., Yoon, L., Adams, Z. C., et al. (2019). Expanding Reactivity in DNA-Encoded Library Synthesis via Reversible Binding of DNA to an Inert Quaternary Ammonium Support. J. Am. Chem. Soc. 141 (25), 9998–10006. doi:10.1021/jacs.9b03774
Franzini, R. M., and Randolph, C. (2016). Chemical Space of DNA-Encoded Libraries. J. Med. Chem. 59 (14), 6629–6644. doi:10.1021/acs.jmedchem.5b01874
Gironda-Martínez, A., Donckele, E. J., Samain, F., and Neri, D. (2021). DNA-encoded Chemical Libraries: A Comprehensive Review with Succesful Stories and Future Challenges. ACS Pharmacol. Transl. Sci. 4 (4), 1265–1279. doi:10.1021/acsptsci.1c00118
Götte, K., Chines, S., and Brunschweiger, A. (2020). Reaction Development for DNA-Encoded Library Technology: From Evolution to Revolution? Tetrahedron Lett. 61 (22), 151889. doi:10.1016/j.tetlet.2020.151889
Kalliokoski, T. (2015). Price-focused Analysis of Commercially Available Building Blocks for Combinatorial Library Synthesis. ACS Comb. Sci. 17 (10), 600–607. doi:10.1021/acscombsci.5b00063
Lu, X., Roberts, S. E., Franklin, G. J., and Davie, C. P. (2017). On-DNA Pd and Cu Promoted C-N Cross-Coupling Reactions. Med. Chem. Commun. 8, 1614–1617. doi:10.1039/c7md00289k
Ruff, Y., and Berst, F. (2018). Efficient Copper-Catalyzed Amination of DNA-Conjugated Aryl Iodides under Mild Aqueous Conditions. Med. Chem. Commun. 9, 1188–1193. doi:10.1039/C8MD00185E
Shi, Y., Wu, Y.-r., Yu, J.-q., Zhang, W.-n., and Zhuang, C.-l. (2021). DNA-encoded Libraries (DELs): a Review of On-DNA Chemistries and Their Output. RSC Adv. 11, 2359–2376. doi:10.1039/D0RA09889B
Wang, D.-Y., Wen, X., Xiong, C.-D., Zhao, J.-N., Ding, C.-Y., Meng, Q., et al. (2019). Non-transition Metal-Mediated Diverse Aryl-Heteroatom Bond Formation of Arylammonium Salts. iScience 15, 307–315. doi:10.1016/j.isci.2019.04.038
Xu, H., Ma, F., Wang, N., Hou, W., Xiong, H., Lu, F., et al. (2019). DNA‐Encoded Libraries: Aryl Fluorosulfonates as Versatile Electrophiles Enabling Facile On‐DNA Suzuki, Sonogashira, and Buchwald Reactions. Adv. Sci. 6 (23), 1901551. doi:10.1002/advs.201901551
Zabolotna, Y., Volochnyuk, D. M., Ryabukhin, S. V., Horvath, D., Gavrilenko, K. S., Marcou, G., et al. (2021). A Close-Up Look at the Chemical Space of Commercially Available Building Blocks for Medicinal Chemistry. J. Chem. Inf. Model. in press. doi:10.1021/acs.jcim.1c00811 https://pubs.acs.org/doi/10.1021/acs.jcim.1c00811
Zhang, Y., and Clark, M. A. (2021). Design Concepts for DNA-Encoded Library Synthesis. Bioorg. Med. Chem. 41, 116189. doi:10.1016/j.bmc.2021.116189
Keywords: DNA-encoded library, palladium catalysis, C-N bond formation, DNA-conjugated aryl ethylene, hydroamination
Citation: Cai K, Ran Y, Sun W, Gao S, Li J, Wan J and Liu G (2022) Palladium-Mediated Hydroamination of DNA-Conjugated Aryl Alkenes. Front. Chem. 10:851674. doi: 10.3389/fchem.2022.851674
Received: 10 January 2022; Accepted: 15 February 2022;
Published: 11 April 2022.
Edited by:
Jörg Scheuermann, ETH Zürich, SwitzerlandReviewed by:
Qinheng Zheng, University of California, United StatesCopyright © 2022 Cai, Ran, Sun, Gao, Li, Wan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guansai Liu, Z3MubGl1QGhpdGdlbi5jb20=; Wenbo Sun, d2Iuc3VuQGhpdGdlbi5jb20=; Sen Gao, c2VuLmdhb0BoaXRnZW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.