
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem., 17 February 2022
Sec. Chemical Biology
Volume 10 - 2022 | https://doi.org/10.3389/fchem.2022.815788
This article is part of the Research TopicExpansion of the Genetic Code: Unnatural Amino Acids and their ApplicationsView all 10 articles
The expansion of the genetic code beyond a single type of noncanonical amino acid (ncAA) is hindered by inefficient machinery for reassigning the meaning of sense codons. A major obstacle to using directed evolution to improve the efficiency of sense codon reassignment is that fractional sense codon reassignments lead to heterogeneous mixtures of full-length proteins with either a ncAA or a natural amino acid incorporated in response to the targeted codon. In stop codon suppression systems, missed incorporations lead to truncated proteins; improvements in activity may be inferred from increased protein yields or the production of downstream reporters. In sense codon reassignment, the heterogeneous proteins produced greatly complicate the development of screens for variants of the orthogonal machinery with improved activity. We describe the use of a previously-reported fluorescence-based screen for sense codon reassignment as the first step in a directed evolution workflow to improve the incorporation of a ncAA in response to the Arg AGG sense codon. We first screened a library with diversity introduced into both the orthogonal Methanocaldococcus jannaschii tyrosyl tRNA anticodon loop and the cognate aminoacyl tRNA synthetase (aaRS) anticodon binding domain for variants that improved incorporation of tyrosine in response to the AGG codon. The most efficient variants produced fluorescent proteins at levels indistinguishable from the E. coli translation machinery decoding tyrosine codons. Mutations to the M. jannaschii aaRS that were found to improve tyrosine incorporation were transplanted onto a M. jannaschii aaRS evolved for the incorporation of para-azidophenylalanine. Improved ncAA incorporation was evident using fluorescence- and mass-based reporters. The described workflow is generalizable and should enable the rapid tailoring of orthogonal machinery capable of activating diverse ncAAs to any sense codon target. We evaluated the selection based improvements of the orthogonal pair in a host genomically engineered for reduced target codon competition. Using this particular system for evaluation of arginine AGG codon reassignment, however, E. coli strains with genomes engineered to remove competing tRNAs did not outperform a standard laboratory E. coli strain in sense codon reassignment.
The genetically-encoded introduction of noncanonical amino acids (ncAAs) is a powerful tool for increasing the chemical diversity of proteins because it enables the precise placement of desired side chain functionality within a growing peptide chain. Two general approaches to expand the genetic code have been widely employed: nonsense suppression and residue specific reassignment (Figure 1) (Wiltschi and Budisa, 2007; Ngo and Tirrell, 2011). Advances in both technologies have increased the efficiency with which a single ncAA can be genetically encoded. Nonsense suppression technology has been improved through genome modifications, enabling the elimination of the release factor that typically competes to read the amber stop codon as a termination signal (Mukai et al., 2010; Johnson et al., 2011; Lajoie et al., 2013). Amino acid-specific reassignment has been updated by breaking the degeneracy of the genetic code to enable the reassignment of individual sense codons (Kwon et al., 2003; Bohlke and Budisa, 2014; Zeng et al., 2014; Lee et al., 2015; Mukai et al., 2015; Ho et al., 2016; Kwon and Choi, 2016; Wang and Tsao, 2016). Improvements in both methods have focused primarily on genetic additions and deletions rather than on functional improvements of the evolved orthogonal translation components central to both technologies.
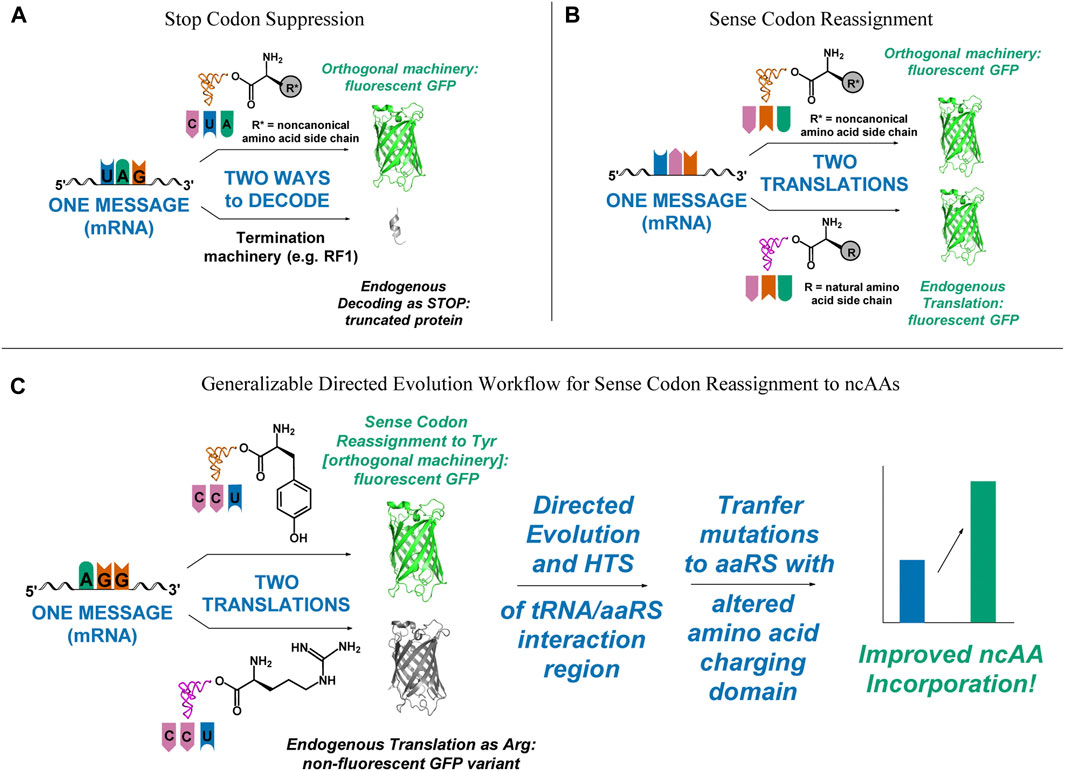
FIGURE 1. Visual representation of outcomes of successful and unsuccessful selection schemes, depicting the need for a fast, functional screen for improving sense codon reassignment. (A) In stop codon suppression, only successful incorporation of the ncAA leads to functional full-length protein. Missed incorporations lead to truncated products. (B) In sense codon reassignment, incorporation of either the desired ncAA or a canonical amino acid lead to full-length, functional protein. (C) A directed evolution pathway that incorporates improvement of sense codon reassignment using a readily screenable reporter, followed by transplantation of the mutations that gave rise to the improved reassignment efficiency onto an orthogonal pair that incorporates a ncAA would lower one of the major barriers to utilizing sense codon reassignment for expansion of the genetic code to 22 amino acids and beyond.
In vivo incorporation of ncAAs by either nonsense suppression or sense codon reassignment (SCR) requires the introduction of an orthogonal aminoacyl tRNA synthetase (aaRS) engineered to recognize and attach a ncAA onto its cognate, orthogonal tRNA (Furter, 1998; Wang et al., 2001). The orthogonal tRNA is not recognized by the set of endogenous aaRSs, and the orthogonal aaRS does not recognize the complement of endogenous tRNAs. The vast majority of the ribosomally incorporated ncAAs have been introduced using derivatives of only two orthogonal pairs: the tyrosine tRNA/aaRS pair from Methanocaldococcus jannaschii (M. jannaschii) and the pyrrolysine tRNA/aaRS pair from Methanosarcina species. (Wang et al., 2001; Polycarpo et al., 2006). Variants of these pairs that recognize and aminoacylate over 150 different ncAAs have been developed (Liu and Schultz, 2010; Wan et al., 2014; Dumas et al., 2015). Continuing efforts to find or engineer new orthogonal pairs have identified several additional potential orthogonal systems. These systems, however, have not yet found widespread use in genetic code expansion (Cervettini et al., 2020; Ding et al., 2020; Zhao et al., 2021).
Here, our previously-developed fluorescence-based screen is utilized as the first step in a directed evolution workflow to improve the incorporation of a ncAA in response to the Arg AGG sense codon (Kuhn et al., 2010; Biddle et al., 2015). The fluorescence-based screen takes advantage of the absolute requirement of tyrosine in the central position of a Thr-Tyr-Gly triad for mature GFP fluorophore formation. We first screened a library with diversity introduced into both the orthogonal M. jannaschii tyrosyl tRNA anticodon loop and the cognate aaRS anticodon binding domain for variants that improved incorporation of tyrosine in response to the AGG codon. The most efficient variants produced fluorescent protein at levels indistinguishable from the Escherichia coli (E. coli) translation machinery decoding Tyr codons, and reassignment efficiency improved from 56.9 ± 2.4% to 98.6 ± 4.7%. Mutations to the M. jannaschii aaRS that were found to improve tyrosine incorporation were transplanted onto a M. jannaschii aaRS evolved for the incorporation of para-azidophenylalanine (pAzF) (Chin et al., 2002). The same suite of mutations improved incorporation of pAzF in response to the AGG codon from 29.5 ± 0.1% to 50.1 ± 1.1% as measured via fluorescence, an approximately 1.7 fold improvement for both amino acids. The improved para-azidophenylalanine incorporating tRNA/aaRS pair was evaluated in strains from which competition for the AGG codon was reduced by deletion of the gene for the primary endogenous tRNA that reads the AGG codon (Lee et al., 2015). Improvements in sense codon reassignment efficiency due to reduced competition did not appear to combine with improvements selected through directed evolution. We have previously quantified the reassignment efficiency of several sense codons by both the natural, tyrosine-incorporating M. jannaschii and an evolved tyrosine-incorporating M. barkeri orthogonal pair (Schmitt et al., 2018; Schwark et al., 2020; Schwark et al., 2021). This workflow should be generalizable for improving incorporation of other ncAAs in response to other sense codon targets.
Nonsense (stop codon) suppression has been the most widely used method for introduction of noncanonical amino acids into the genetic code. In E. coli, the amber stop codon is employed both because it is a termination signal and because it is the least frequently used codon. Nonsense suppression utilizes orthogonal aaRS variants to aminoacylate >150 different ncAAs onto essentially an identical cognate tRNA with a CUA anticodon. Variation across orthogonal systems that incorporate different ncAAs primarily resides within the amino acid recognition domain of the aaRS. tRNA/aaRS recognition is typically not altered across variants. The kinetic efficiencies of aaRSs evolved to recognize ncAAs are generally poor. In vitro kinetic aminoacylation efficiencies for these enzymes are on the order of 1% that of natural enzymes (Guo et al., 2014; Amiram et al., 2015; Rauch et al., 2016). The measured efficiency of ncAA incorporation is dependent on systemic variables beyond the kinetics of aminoacylation (e.g. ncAA, tRNA and aaRS concentrations), and single site incorporation efficiencies of 10–30% are readily achievable. For systems in which a single amber stop codon is targeted for ncAA incorporation, this strategy and enzyme function level are sufficient to produce modified proteins in usable quantities. The expected protein yield drops as the nth power of the single site incorporation efficiency (where n is the number of incorporations attempted). Even for a highly efficient (e.g. 50%) ncAA-incorporating tRNA/aaRS system, the expected protein yield for a protein containing 3 amber stop codons would be 12.5% of the yield expected if the amber codon were a typical canonical amino acid (Schwark et al., 2018).
Advances in the technology that underpins both amber stop codon suppression and sense codon reassignment by breaking the degeneracy of the genetic code have focused primarily on extrinsic system improvements, such as increasing the expression level of the orthogonal pair, improving the interactions of the orthogonal tRNA with the endogenous translation components (e.g. EF-Tu), or reducing endogenous competition for the targeted codon. The orthogonal translation components are orthogonal because they are taken from a phylogenetically distant species, and the efficiency of incorporation could potentially be increased by improved assimilation of the orthogonal components into the host organism. A large amount of “orthogonal space” in the vicinity of each orthogonal system appears to exist, as mutually orthogonal variants are readily evolvable (Neumann et al., 2010; Willis and Chin, 2018; Cervettini et al., 2020). Orthogonal machinery levels have been adjusted by expressing both tRNAs and aaRSs from cassettes with different promoters in plasmids with different copy numbers (Ryu and Schultz, 2006; Young et al., 2010; Chatterjee et al., 2013). A handful of attempts to improve amber stop codon suppression via tRNA mutation have focused on changes to tRNA sequences that modulate interactions with elongation factor Tu (EF-Tu) (Guo et al., 2009; Schrader et al., 2011; Mittelstaet et al., 2013; Fan et al., 2015; Maranhao and Ellington, 2017). The extent to which tRNA mutations that improved incorporation of one ncAA in response to amber codons functioned when transferred between aaRSs specific for different ncAAs suggested that the extent of transfer is idiosyncratic, although in general, improved variants were identified (Guo et al., 2009). Reducing the competition from endogenous translation components has been the major mode of improving genetic code expansion systems. Three separate approaches involving different combinations of organismal genomic modifications enabled the deletion of the gene for the release factor that typically reads the amber stop codon as a termination signal (Mukai et al., 2010; Johnson et al., 2011; Lajoie et al., 2013). Additional genome rewriting projects seek to generate multiple “free” codons for ncAA incorporation (Ostrov et al., 2016; Wang et al., 2016; Fredens et al., 2019). Attempts to improve sense codon reassignment have involved deletion of competing tRNA genes and antisense RNAs to reduce competition between orthogonal and host components reading the AGG arginine codon (Zeng et al., 2014; Lee et al., 2015).
An alternative strategy for improving the efficiency of orthogonal tRNA/aaRS pairs targets interactions intrinsic to the cognate pair itself. The anticodon is often an important identity element that allows a specific aaRS to recognize its appropriate tRNA (Giege et al., 1998). Changing the anticodon of the M. jannaschii tRNA, even to CUA for amber suppression, is known to strongly affect the efficiency of aminoacylation (Fechter et al., 2001). Increasing the recognition between the orthogonal tRNA and the anticodon binding domain of the aaRS is expected to lead to a higher concentration of aminoacylated tRNA and better kinetic competition against endogenous tRNAs capable of decoding the targeted codon. Improvements of the interactions between the components of the orthogonal translation machinery should facilitate improved incorporation efficiency with less dependence upon the system characteristics into which orthogonal translation system is placed. Several general selection strategies have been developed to improve the function of stop codon suppressing orthogonal pair systems, although the maturation of initially selected aaRSs utilized for ncAA incorporation is not commonly performed (Pott et al., 2014; Wang et al., 2015; Rauch et al., 2016; Maranhao and Ellington, 2017; Owens et al., 2017). Directed evolution of M. jannaschii tRNA/aaRS pair variants for improved reassignment of the amber stop codon as well as Lys AAG and His CAU sense codons have been described (Takimoto et al., 2009; Kuhn et al., 2010; Mittelstaet et al., 2013; Biddle et al., 2015; Wang et al., 2015; Biddle et al., 2016).
A major obstacle to using directed evolution to improve the efficiency of sense codon reassignment is that fractional sense codon reassignments lead to heterogeneous mixtures of full-length proteins with either a ncAA or a natural amino acid incorporated in response to the targeted codon. In stop codon suppression systems, missed incorporations lead to truncated proteins and improvements in activity can be inferred from increased protein yields or the production of downstream reporters (Figure 1A). In sense codon reassignment, heterogeneous mixtures of full-length proteins greatly complicate the development of screens for variants with improved activity (Figure 1B). Implementation of a general workflow that allows selection of orthogonal pair variants that reassign a particular sense codon first by evaluating variants with a fast, facile screen (e.g. fluorescence-activated cell sorting, FACS) followed by subsequent transfer of the mutations to a version of that orthogonal pair engineered to recognize a ncAA, for which screening would be more challenging, would lower the barrier to wider utilization of sense codon reassignment for genetic code expansion (Figure 1C).
The Supplementary Materials file includes general reagents and materials, as well as detailed experimental protocols for site-directed mutagenesis, the fluorescence-based screen, and protein expression and isolation for reassignment efficiency analysis and mass spectrometry (Kunkel, 1985; Gao et al., 2003; Clackson and Lowman, 2004). This file also includes a graphical representation of the directed evolution workflow (Supplementary Figure S1), an image of a representative protein gel (Supplementary Figure S2), and a representative optical density vs. time plot for reassigning systems (Supplementary Figure S3). Finally, further experimental information, including a list of the oligonucleotide primers used for library creation, cell strain information, and preparation of electrocompetent cells are provided (Sambrook and Russell, 2001).
We selected the arginine AGG codon for initial evaluation of this directed evolution workflow because AGG has been the most common target for sense codon reassignment using variants of both the orthogonal pyrrolysyl and M. jannaschii tyrosyl tRNA/aaRS pairs (Krishnakumar et al., 2013; Zeng et al., 2014; Lee et al., 2015; Mukai et al., 2015; Wang and Tsao, 2016). Zeng et al. reported approximately 90% efficient reassignment using a variant of the Methanosarcina pyrrolysyl tRNA/aaRS pair and media in which the concentration of arginine was controlled (Zeng et al., 2014). Mukai et al. mutated the 38 AGG codons found in essential E. coli genes to other arginine codons and subsequently removed the gene for endogenous tRNACCU, argW, from the genome (Mukai et al., 2015). These genetic transformations allowed incorporation of a close structural analogue of arginine at very high levels using a variant of the Methanosarcina pyrrolysyl tRNA/aaRS pair. Lee et al. described reassignment of the AGG codon using a variant of the M. jannaschii tyrosyl tRNA/aaRS pair (Lee et al., 2015). Their strategy also utilized an argW knockout strain, however the distribution of AGG codons in the genome was not adjusted. The fact that argW knockout strains are viable and exhibit only slightly reduced growth suggests that the remaining E. coli tRNAUCU is able to read AGG codons to some extent via an expected G/U wobble interaction. Double knockouts of both E. coli tRNAUCU and tRNACCU are not viable (Mukai et al., 2015).
The efficiency of AGG reassignment was quantified in E. coli DH10B expressing the M. jannaschii tyrosyl aaRS, its cognate tRNAOpt with a CCU anticodon (M. jannaschii tRNACCU/TyrRS), and a GFP reporter protein with an arginine AGG codon specifying the central fluorophore position (Young et al., 2010; Chatterjee et al., 2013; Biddle et al., 2015). The gene sequence of the GFP reporter uses a reduced codon set such that instances of the 20 sense codons decoded through wobble interactions and 10 additional least frequently used sense codons are eliminated or greatly reduced from the gene (Schmitt et al., 2018). No AGG codons are present in the aaRS gene. Changing the anticodon of the M. jannaschii tRNAOpt to Watson—Crick base pair with the AGG codon results in 56.9 ± 2.4% reassignment efficiency. Significantly, this high efficiency is evident in rich media without any modifications to the E. coli genome or alterations to the orthogonal tRNA and aaRS beyond the anticodon change. Reassignment efficiency is quantified by normalizing the observed fluorescence with a test codon at the fluorophore position to a “100% fluorescence” reference value determined by expressing sfGFP with a tyrosine UAC codon at position 66. A “0% fluorescence” reference is determined by expressing sfGFP with a non-tyrosine codon specifying the fluorophore. Both fluorescence reference systems include a plasmid expressing the orthogonal machinery to maintain a similar metabolic burden on each system.
A library of M. jannaschii tRNA anticodon loop and aaRS anticodon binding domain variants was constructed and screened for improved efficiency of reassignment of the AGG codon to Tyr (Figure 2). Co-evolution of the tRNA anticodon loop and the aaRS anticodon binding domain was hypothesized to increase the recognition and aminoacylation of the tRNA by the aaRS, which should, in turn, increase the effective concentration of the aminoacylated tRNA and lead to better competition against endogenous tRNAs for decoding the targeted codon. Furthermore, the positions within the M. jannaschii Tyr aaRS that contact the cognate tRNA anticodon are spatially distant from and within a separate domain relative to the amino acid binding pocket, suggesting that mutations which lead to improvement of incorporation for one amino acid may be transferable to a variant of the same enzyme that recognizes and aminoacylates a different amino acid (Kobayashi et al., 2003).
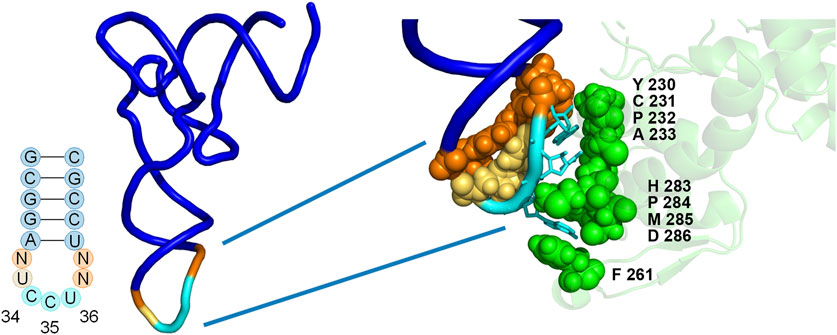
FIGURE 2. Positions of library mutations mapped onto the co-crystal structure of the M. jannaschii tRNA/aaRS pair (PDB 1J1U). Positions 32, 37, and 38 within the tRNA anticodon loop were allowed to vary in conjunction with 9 amino acids in the anticodon binding domain of the aaRS. The diversity available at each position in the aaRS is described both in the text and in Table 1.
The library included diversity at 9 amino acid positions in the aaRS and 3 of 4 nucleotides in the tRNA anticodon loop outside of the anticodon (32, 37, and 38) (Biddle et al., 2015). The varied amino acid positions were chosen based on the proximity to the nucleotides of the anticodon in the co-crystal structure of the M. jannaschii tyrosyl tRNA/aaRS orthogonal pair (PDB 1J1U) (Figure 2) (Kobayashi et al., 2003). Amino acid residues with side chains within 5Å of anticodon nucleobases were varied. Amino acid diversity at each of the positions was limited in order to maintain a manageable library size, and degenerate nucleotides were selected to include the parent M. jannaschii aaRS amino acid at each position (Table 1). Amino acids 230-233 were allowed to vary between four amino acids. Greater diversity was included at the remaining 5 positions; diversity at positions 261 and 283-286 varied between 9 and 15 different amino acids. Three of the four nucleotides flanking the anticodon in the tRNA were allowed to vary; the universally conserved U33 was preserved (Figure 2; Table 1). The library had a theoretical diversity of 4.1 × 1010 at the DNA level, 4.1 × 109 at the expressed level.
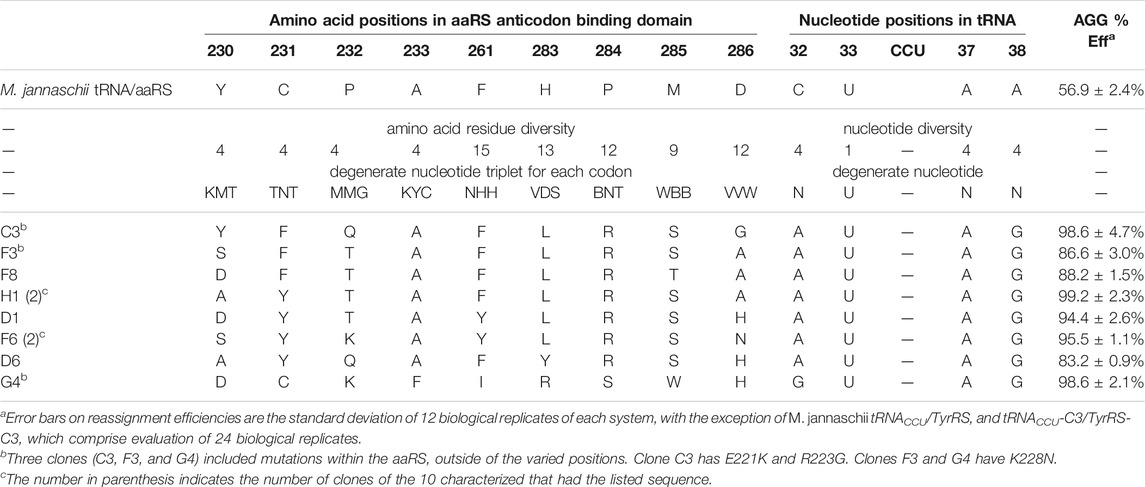
TABLE 1. Sequences and AGG reassignment efficiencies for M. jannaschii tyrosyl tRNA/aaRS variants selected from the library.
The constructed library contained 3 × 108 unique transformants, which were allowed to multiply 25-fold (∼109 cells). Of these, approximately 107 cells were screened using fluorescence activated cell sorting (FACS), and the brightest 2% of cells were collected. Following amplification, the population was resorted, and the top 1% of fluorescent cells were collected. After amplification and a third round FACS, a portion of the collected cells were plated for single clone analysis (Supplementary Figure S1 in Supplementary Material). 75 of 84 visibly green clones showed increased fluorescence relative to the starting M. jannaschii tRNACCU/TyrRS. The clones displayed a range of reassignment efficiencies, with the vast majority reassigning the AGG codon at greater than 75% efficiency. 10 of the 84 clones were selected for further analysis. Sequencing revealed eight unique sets of M. jannaschii tRNACCU/TyrRS variants (Table 1). Because false positives resulting from reporter fluorophore revertants or mutations increasing the expression of GFP have been observed in other aaRS selections, the orthogonal translation machinery plasmids were isolated and retransformed into cells with unselected GFP reporter plasmids. Analysis of the eight identified M. jannaschii tRNACCU/TyrRS variants suggested that the mutations within the tRNA and aaRS were responsible for the improved incorporation of tyrosine in response to the AGG codon. Sense codon reassignment efficiencies ranged from 83.2 ± 0.9% to 99.2 ± 2.3% (measurements from twelve colonies of each variant).
Strong consensus was apparent across the sequences of the selected M. jannaschii tRNACCU/TyrRS variants (Table 1). In the case of the tRNA anticodon loop, the nucleotides identified at the three varied positions were identical for seven of the eight aaRS variants. The predominantly selected tRNA sequence was 5′-auCCUag-3′, where loop positions are written in lower case and anticodon nucleotides in upper case. Positions 32 and 38 were different than the starting M. jannaschii tRNACCU: 5′-cuCCUaa-3’. While the predominant tRNA variant included purine nucleotides at the 3 varied positions, the majority of E. coli tRNAs include a pyrimidine at position 32 and a purine at 38 as in the starting sequence. No E. coli tRNA has a purine in both positions 32 and 38. A purine/pyrimidine relationship also exists for tRNA positions 33/37, with the universally-conserved uridine at position 33. In E. coli, the choice of purine at position 37 is strongly dependent on the nucleotide at position 36. The majority of E. coli tRNAs feature adenosine (or a modified adenosine) at position 37; only a few of the tRNAs with G36 anticodons have a modified guanosine at position 37 (Machnicka et al., 2013).
The seven aaRS sequences (from nine characterized variants) that were selected along with the single tRNA sequence have strikingly similar amino acid sequences (Table 1). Alanine, the amino acid present at position 233 in the original aaRS, was present in each of the seven sequences. Despite having the greatest opportunity to vary, position 261 maintained a similar amino acid to the wild type aaRS, with all seven sequences either remaining Phe or mutating to Tyr. The changes selected in the remaining seven varied positions modified the amino acid size and polarity relative to the amino acid present in the starting aaRS. In all seven selected sequences, position 284 mutated from Pro to Arg. Six of seven sequences included H283L and M285S. For both Leu 283 and Ser 285, the multiple codons for each amino acid available as a result of degenerate codon choice were present at the DNA level. All seven sequences included an aromatic residue at position 231. Less of a consensus was apparent at positions 232 and 286. Position 232 universally changed from the proline found in the starting aaRS with a weak consensus for Thr (4/7 sequences). Position 286 showed a weak consensus for alanine (3/7 sequences) or histidine (2/7 sequences). The amino acids identified at position 230 were evenly distributed between Tyr, Ala, Asp, and Ser, the 4 amino acids available at that position. The single selected tRNA with a different anticodon loop sequence (5′-guCCUag-3′) was co-selected with an aaRS with a unique suite of mutations (clone G4 tRNA/aaRS pair). The amino acids selected at six of nine varied positions in clone G4 were not identified in any of the other variants.
Three of the ten clones, including the highly efficient co-evolved tRNA/aaRS pair C3, had additional, spontaneous mutations to the aaRS sequence. Clone C3 had two mutations: E221K and R223G. Clones F3 and G4 had a K228N mutation. All three of these mutations occur within an α-helix near the anticodon binding domain. The E221K and R223G mutations in C3 occur near the end of the α-helix furthest removed from the tRNA. The K228N mutation was also observed in three of ten characterized clones identified from a similar tRNA anticodon loop/aaRS anticodon binding domain library targeting improvement of Lys AAG codon reassignment (Biddle et al., 2015). The anticodons required to Watson—Crick base pair with Lys AAG (CUU) and Arg AGG (CCU) differ only at position 35. That the same mutations arise in selections for improving the function of two very similar anticodon loops is not surprising.
Importantly, the orthogonality of the M. jannaschii tRNACCU-C3/TyrRS-C3 and M. jannaschii tRNACCU-G4/TyrRS-G4 variants was maintained. Incorporation of tyrosine in response to an AGG codon using orthogonal translation machinery vectors from which the tRNA was removed was below the limit of detection of the fluorescence-based screen (0.2%, 2 of every 1000 incorporation events in response to the AGG codon in the reporter). The anticodon is often an important identity element for aaRSs, and the M. jannaschii TyrRS is known to utilize the anticodon during the process of aminoacylation. The most likely way in which orthogonality of the pair might be broken after mutation of the aaRS anticodon binding domain to better recognize the tRNACCU would be aminoacylation of Tyr onto the E. coli tRNACCU. The absence of detectable fluorescence in vectors expressing either TyrRS-C3 or TyrRS-G4 without their cognate tRNA suggests that is not a frequent occurrence. These two aaRS variants were selected for the verification of orthogonality because they represent the 2 broad sequence spaces identified in the characterized clones.
As with our previous reports of sense codon reassignment using the fluorescence-based screen, the fluorescence measurements from the in cell assay were compared to fluorescence measurements from isolated protein. GFP protein production across the different AGG reassigning systems and non-reassigning controls is remarkably similar (Supplementary Figure S2). In this report, clarified lysate fluorescence measurements were normalized to protein amounts determined from image quantification of SDS-PAGE gels. Reassignment efficiency of AGG to tyrosine was identical within error using both types of measurement. Analysis of clarified lysates showed that M. jannaschii tRNACCU/TyrRS was 60.7 ± 2.4% efficient and that M. jannaschii tRNACCU-C3/TyrRS-C3 was 96.0 ± 2.0% efficient. The protein reassignment efficiencies from clarified lysates comprise at least 4 biological replicates of each system.
Reassignment efficiencies are calculated using the optical density-corrected fluorescence over a 4 h period of cell growth in order to eliminate biases from differences in the growth profiles of different codon reassigning systems. Bacterial cells are largely tolerant of induced missense mutations. We have previously discussed the average impact of high level codon reassignment on cellular fitness (Schmitt et al., 2018; Schwark et al., 2020). Using these orthogonal translation machinery and reporter vectors, the average fitness reduction for 20 different sense codon reassigning systems is about 2.5x the fitness reduction imposed by using 2 antibiotics to maintain 2 different protein expressing plasmids in the cells. The cell strain used for sense codon reassignment evaluation in the majority of our prior work was SB3930. Our recent reports have utilized the more commonly-available laboratory strain DH10B (Schwark et al., 2021). Codon reassignments have similar impacts on cell health in both strains.
Reassignment of Arg AGG using M. jannaschii tRNACCU/TyrRS results in reduced carrying capacity and instantaneous doubling times in DH10B relative to an “empty” translation machinery vector from which both the tRNA and aaRS gene cassettes have been deleted. E. coli DH10B expressing M. jannaschii no-tRNA/no-aaRS and the GFP reporter vector with a Tyr codon in the fluorophore have an instantaneous doubling time of 40.0 ± 3.0 min, while those reassigning the AGG codon to tyrosine have an instantaneous doubling time of 45.8 ± 2.3 min. The relative system fitness of the AGG reassigning system is ∼87% that of the “empty” translation machinery control. Interestingly, expression of M. jannaschii tRNACCU-C3/TyrRS-C3 restores cell fitness back to that of the “empty” translational machinery vector control. E. coli DH10B reassigning the AGG codon with M. jannaschii tRNACCU-C3/TyrRS-C3 double every 36.7 ± 2.2 min in the exponential phase, for a relative system fitness of ∼109%. The tolerance toward high level reassignment of Arg AGG codons is expected to partly be the result of the low usage of the AGG codon throughout the E. coli genome. A representative optical density vs. time cell growth graph is provided as Supplementary Figure S3.
As part of a general process for directed evolution of sense codon reassigning systems, the fluorescence-based screen with the tyrosine-incorporating aaRSs would be utilized as a stand in for aaRS variants that activate ncAAs. A group of clones with different sequences (C3, H1, and G4) exhibited very high, near quantitative reassignment of AGG to Tyr using the fluorescence-based screen. The reassignment efficiencies for each of these systems were all within error of each other, based on evaluation of 12 or 24 biological replicates of each system. In order to evaluate the transferability of improvements in tRNA/aaRS interactions between aaRS variants that activate different amino acids, the tRNA anticodon loop and aaRS anticodon binding domain mutations present in clone C3 were transferred to an aaRS variant that activates para-azidophenylalanine (pAzFRS) and evaluated both using both the fluorescence-based screen and mass spectrometry (Chin et al., 2002; Kirshenbaum et al., 2002). The C3 mutations improved AGG reassignment to Tyr from 56.9 ± 2.4% to 98.6 ± 4.7% (Figure 3A).
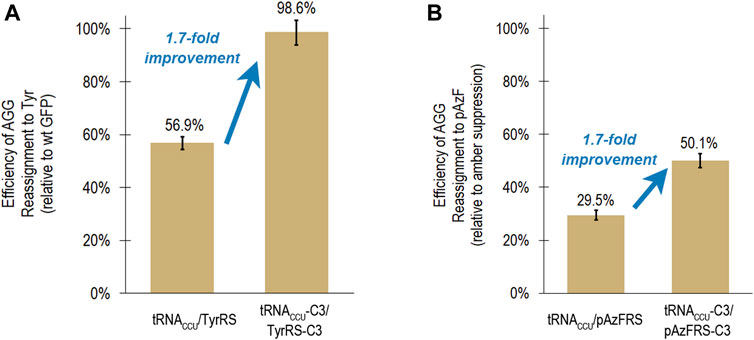
FIGURE 3. Transferability of mutations that led to increased reassignment of the AGG codon to tyrosine to an aaRS capable of incorporating a ncAA (para-azidophenylalanine). In both cases, the same suite of mutations to the tRNA anticodon loop and aaRS anticodon binding domain resulted in a 1.7-fold improvement in reassignment efficiency. (A) Efficiency of AGG sense codon reassignment to tyrosine by two orthogonal pairs: M. jannaschii tRNACCU/TyrRS and tRNACCU-C3/TyrRS-C3. Reported efficiencies are the mean and standard deviation for 24 biological replicates of each system evaluated in several independent iterations of the fluorescence-based screen. Reassignment efficiencies are corroborated by fluorescence analysis of the GFP protein mixtures isolated from these cells. (B) Efficiency of AGG sense codon reassignment to a ncAA, pAzF, by two orthogonal pairs: M. jannaschii tRNACCU/pAzFRS and tRNACCU-C3/pAzFRS-C3. Reported efficiencies are the mean and standard deviation of at least 4 biological replicates of each system quantified using the fluorescence per protein of GFP mixtures isolated from these cells. The “100% pAzF incorporation” control in each experiment is based on the fluorescence per protein analysis of full length GFP proteins produced after suppression of an amber stop codon in the fluorophore position 66.
The M. jannaschii pAzF incorporating variant has been employed to direct incorporation of pAzF in response to both amber stop and arginine AGG codons (Chin et al., 2002; Lee et al., 2015). pAzF has been widely used for protein crosslinking via photolysis and bioorthogonal derivatization via copper-catalyzed Huisgen cyclization with alkynes. pAzF is one of several ncAAs that produce fluorescent proteins when introduced at the fluorophore tyrosine position in GFP (Wang et al., 2003a; Kajihara et al., 2005). Introduction of pAzF into GFP results in a protein with less intense fluorescence and blue shifted maximum relative to sfGFP. The apparent brightness (quantum yield*extinction coefficient) of sfGFP with pAzF in the fluorophore is approximately 10% of wild type sfGFP (Wang et al., 2012; Reddington et al., 2013). The actual fluorescent species in GFP with pAzF substitution is the reduced form of pAzF, para-aminophenylalanine. Reports differ on the extent to which spontaneous reduction of the azide to an amine occurs in GFP (Wang et al., 2003a; Morris et al., 2013; Reddington et al., 2013). In biological contexts, azides are often subject to rapid reduction by common dithiols (Bayley et al., 1978; Staros et al., 1978).
The fluorescence of cells expressing both the AGG reassignment reporter and either the M. jannaschii tRNACCU/pAzFRS or M. jannaschii tRNACCU-C3/pAzFRS-C3 was monitored in both the presence and absence of pAzF. Meaningful in vivo fluorescence was not observed for these cultures, possibly due to the lower inherent brightness of para-aminophenylalanine-containing GFP, the absence of spontaneous reduction of the azide, or obfuscation of protein fluorescence by fluorescent components in the rich medium. A slight increase in fluorescence was observed after irradiation of the cell cultures with UV light; however, controlled irradiation of several cell cultures was challenging. Reduction of the azide side chain following protein isolation proved to be more controllable.
In order to use fluorescence to quantify ncAA incorporation efficiency, the differences in the photophysical properties of GFP with pAmF (reduced pAzF) in the fluorophore need to be included in the measurements. The 100% fluorescence control for reassignment to pAzF was the per protein fluorescence of pAzF incorporated in response to an amber stop codon in the fluorophore of GFP. Although the efficiencies of introduction of ncAAs in response to the amber stop codon using M. jannaschii tRNA/aaRS pair variants are typically in the 10–30% range relative to sense codons (Young et al., 2010), no protein is produced in the absence of ncAA. The protein isolated and subsequently quantified contains only the ncAA. The amount of protein was determined from SDS-PAGE gels using GFP standards to calibrate the visualization. The observed fluorescence was normalized to the protein amounts to derive a value for fluorescence per protein.
Based on fluorescence analysis of isolated proteins, M. jannaschii tRNACCU/pAzFRS incorporated pAzF in response to the AGG codon in the fluorophore of GFP with 29.5 ± 0.1% efficiency in rich media. The M. jannaschii tRNACCU-C3/pAzFRS-C3 orthogonal pair incorporated pAzF in response to the AGG codon with 50.1 ± 1.1% efficiency, a 1.7-fold improvement over the original M. jannaschii tRNACCU/pAzFRS (Figure 3B). This same relative improvement was observed between M. jannaschii tRNACCU/TyrRS (56.9 ± 2.4%) and M. jannaschii tRNACCU-C3/TyrRS-C3 (98.6 ± 4.7%) (Figure 3A). The improved reassignment efficiency observed for the C3 variant is expected to be the result of increased effective concentration of aminoacylated tRNA due to improved recognition between the anticodon of the tRNA and the aaRS. In this instance, improvements made to the M. jannaschii tRNA/aaRS pair for incorporation one amino acid, Tyr, also increased the efficiency of incorporating another amino acid, pAzF, in response to AGG codons.
Electrospray ionization mass spectrometry (ESI-MS) was used to evaluate the levels of incorporation of arginine, tyrosine, and pAzF in response to a single AGG codon in the Z domain of protein A (Braisted and Wells, 1996). The Z domain is a small, soluble, 8.3 kDa, three-helix protein that has been employed as a reporter for stop codon suppression (Xie et al., 2007). The phenylalanine codon at position 5 in the native Z domain, a known permissive site, was mutated to AGG. The AGG codon containing Z domain variant was expressed in DH10B cells also expressing the M. jannaschii machinery to reassign AGG codons to tyrosine or pAzF (tRNACCU/TyrRS, tRNACCU-C3/TyrRS-C3, tRNACCU/pAzFRS, or tRNACCU-C3/pAzFRS-C3). Z domain proteins were isolated and analyzed using ESI-MS of the intact protein followed by deconvolution of the mass spectra using the Maximum Entropy algorithm (MassHunter Software, Agilent Technologies).
The calculated mass of the Z domain protein with arginine at the single AGG codon is 8308 Da, and the +14 Da peak observed for all parent peaks is likely due to a methylation of the Z domain (Stock et al., 1987; Apostol et al., 1995). The Z domain protein has been reported to be variously modified (Wang et al., 2003b; Zhang et al., 2003). Incorporation of tyrosine rather than arginine in response to the AGG codon results in a +7 Da shift. Incorporation of pAzF rather than arginine in response to the AGG codon results in a either a +6 or +32 Da shift (for the reduced amine or azide form of the amino acid, respectively).
Reassignment of the AGG codon to either tyrosine or pAzF was not detectable using the original forms of the M. jannaschii translation machinery where only the anticodon of the orthogonal tRNA was modified (tRNACCU/TyrRS or tRNACCU/pAzFRS). Masses corresponding to arginine incorporation were observed (Figures 4A,B). AGG reassignment is apparent in the deconvoluted mass spectra of cells expressing either tRNACCU-C3/TyrRS-C3 or tRNACCU-C3/pAzFRS-C3 (Figures 4A,C). In the case of protein isolated from cells expressing tRNACCU-C3/TyrRS-C3, the peaks corresponding to arginine incorporation (8308 and 8322 Da) shrink, and peaks corresponding to tyrosine incorporation (8315 and 8329 Da) become evident. A similar trend is observed for proteins isolated from cells expressing tRNACCU-C3/pAzFRS-C3 in the presence of pAzF.
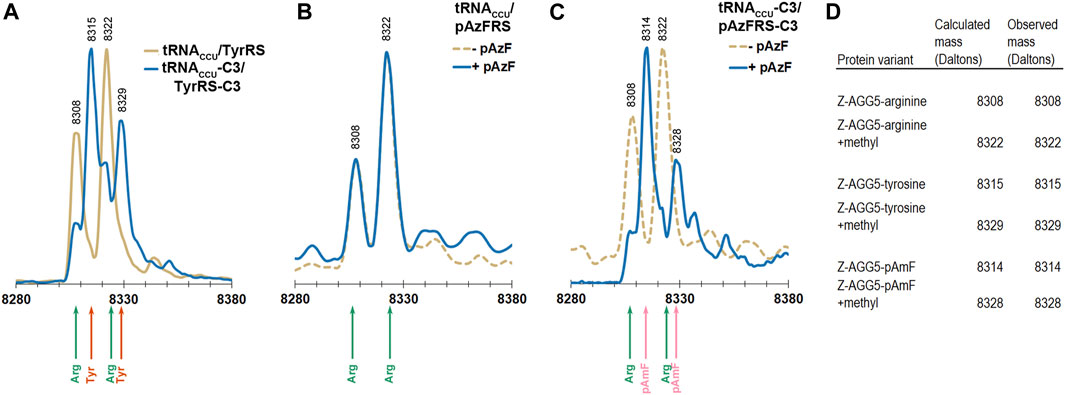
FIGURE 4. ESI-MS of purified Z domain proteins for identification of amino acids incorporated in response to an AGG codon at position 5 in the gene. The expected mass for incorporation of Arg in response to AGG is 8308 Da. The expected mass for incorporation of Tyr in response to AGG is 8315 Da. The expected mass for incorporation of reduced pAzF (pAmF) in response to AGG is 8314 Da. In all 3 instances, an additional mass at +14 Da relative to the parent mass is observed and likely corresponds to methylation of the Z domain. (A) Mass spectra for proteins produced using the tyrosine-incorporating M. jannaschii tRNACCU/TyrRS pair (gold line) and M. jannaschii tRNACCU-C3/TyrRS-C3 (blue line) variants. In the case of the original M. jannaschii tRNACCU/TyrRS pair, only peaks for arginine incorporation are apparent (8308, 8322 Da, green arrows). Peaks at 8315 and 8329 Da (orange arrows) corresponding to tyrosine incorporation in response to AGG are readily apparent in the Z domain expressed in the presence of the M. jannaschii tRNACCU-C3/TyrRS-C3 variant. (B) Mass spectra for proteins produced using the para-azidophenylalanine-incorporating M. jannaschii tRNACCU/pAzFRS machinery in the presence (blue line) and absence (gold dotted line) of ncAA. In both cases, incorporation of Arg in response to AGG is the only set of masses detected (green arrows). (C) Mass spectra for proteins produced using the para-azidophenylalanine-incorporating M. jannaschii tRNACCU-C3/pAzFRS-C3 machinery in the presence (blue line) and absence (gold dotted line) of ncAA. Only in the presence of para-azidophenylalanine are peaks corresponding to incorporation of ncAA observed at 8314 and 8328 Da (pink arrows). In the absence of ncAA, only arginine incorporation is detected (green arrows). (D) Table of calculated and observed masses (Daltons) for each protein.
The apparent difference in reassignment efficiencies in the Z domain protein as measured by ESI-MS and the GFP fluorescence-based screen is consistent with large observed variations in the efficiency of incorporation measured at different positions within the same protein and between various reporter proteins (Young et al., 2010). The efficiency of stop codon reassignment is thought to be sequence dependent based largely on differential interactions of release factors and suppressor tRNAs with regions outside the codon, codon context effects, additionally modulated by mRNA structure (Bossi and Roth 1980; Chevance et al., 2014; Gamble et al., 2016; Schwark et al., 2018). The extent to which codon context affects the rate and efficiency of sense codon reading and reassignment has not been widely investigated. While the absolute reassignment efficiencies are decidedly different in the two reporter protein systems, the trends in both suggest that tRNA/aaRS variants selected from a library that improve reassignment of AGG codons to tyrosine are transferable and increase the reassignment of AGG codons to pAzF.
The increase in sense codon reassignment efficiency that results from modifying interactions between the orthogonal tRNA/aaRS pair is expected to be independent from and potentially combinable with improvements that result from removing competition from endogenous tRNAs through genomic engineering. As near-quantitative reassignment of the AGG codon to tyrosine was observed using the M. jannaschii tRNACCU-C3/TyrRS-C3, genomic engineering was not expected to markedly improve this system. Our evaluation of the effect of host strain genomic engineering on sense codon reassignment began instead with M. jannaschii tRNACCU/TyrRS, which reassigns AGG to tyrosine with 56.9 ± 2.4% efficiency in E. coli DH10B.
One of the simplest genome modifications that may affect decoding the AGG codon is removal of the gene which encodes the E. coli rare arginine tRNACCU, argW. The argW knockout of E. coli DH10B was prepared via the lambda red recombinase method (DS-dArgW) (Datsenko and Wanner, 2000). The strain mimics the previously-reported BS01 strain (DH10B ΔargW) evaluated for reassignment of the AGG codon (Lee et al., 2015). An argW/argA double knockout named DS-dArgWArgA was also evaluated. The secondary argA knockout renders the cells auxotrophic for arginine. Sanger sequencing of the products of PCR amplifications of relevant segments of the chromosome and tRNA amplification tests indicated that the genomic knockouts were successful.
Eliminating competition for the AGG codon by removing the endogenous tRNA with a CCU anticodon resulted in a 1.6-fold decrease in measured reassignment efficiency. The M. jannaschii tRNACCU/TyrRS reassigned AGG to tyrosine with 37.3 ± 0.3% efficiency in E. coli DS-dArgWArgA (12 biological replicates), compared to 56.9 ± 2.4% in E. coli DH10B. The observation that knocking out the competing tRNA was detrimental, as opposed to neutral or even beneficial, to sense codon reassignment efficiency was surprising. Cells from the original report (BS02, DH10B ΔargW ΔargA) were obtained for comparison to the DS-dArgWArgA strain and yielded similar results with our orthogonal machinery (Lee et al., 2016).
The finding that expression of the minor E. coli tRNACCU is not a major factor in allowing high level sense codon reassignment at the AGG codon is consistent with several other reports. Zeng et al. attempted to reduce the effective concentration of E. coli tRNACCU by introducing an antisense RNA and found that it had a no effect on sense codon reassignment efficiency (Zeng et al., 2014). Mukai et al. reported cells that E. coli cells were viable after knocking out argW, but were unable to survive after knocking out the genes for both the E. coli tRNACCU and E. coli tRNAUCU, which is able to read both the Arg AGA and AGG codons (Mukai et al., 2015). Supplemental information in Lee et al. suggests that very high levels of AGG codon reassignment are present in argW+ parent cells consistent with the AGG reassignment efficiencies measured by the GFP screen (Lee et al., 2016). Direct comparisons between these systems are difficult as the vectors employed to produce both the orthogonal machinery and reporter proteins differ significantly across the reports. Preliminary evaluation of combinations of the variously evolved versions of orthogonal machinery expressed from different plasmid vectors (e.g. origin of replication, antibiotic resistance marker, and the promoters driving expression of the aaRS and tRNA) in the wildtype and knockout cell lines utilizing both fluorescence and mass reporters produced confounding results. The general conclusion that can be drawn from these experiments is that the systemic context in which a set of improvements in orthogonal system performance is selected is important. Combinations of improvements selected under different conditions do not necessarily add in predictable ways.
We have described a general directed evolution pipeline for tailoring the tRNA/aaRS interactions of already-evolved ncAA-activating aaRSs for improved sense codon reassignment. Our pipeline exploits an easily screenable, tyrosine-activating aaRS as a stand in for ncAA-activating orthogonal aaRSs without readily screenable side chain chemistries. In the case of the ∼100 ncAA activating M. jannaschii orthogonal aaRS variants, the stand in aaRS is the natural, parent, tyrosine-activating M. jannaschii aaRS. This workflow should be applicable to ncAA-activating variants of the M. barkeri pyrrolysine aaRS as well, as we recently reported the evolution of a highly efficient tyrosine-incorporating variant of that pair (Schwark et al., 2021). We demonstrate the utility of the pipeline in improving the efficiency of para-azidophenylalanine incorporation in response to the arginine AGG codon. We selected variants that could more efficiently translate the AGG codon placed at the fluorophore tyrosine position in sfGFP from a library of M. jannaschii Tyr tRNA and aaRSs with focused mutations in the tRNA anticodon loop and aaRS tRNA anticodon binding domain. Translation of the codon at position 66 in a GFP reporter as either tyrosine or the amino acid typically specified by the target codon results in fluorescence signals proportional to the extent of successful reassignment. Screening large combinatorial libraries for improved variants is accomplished quickly via FACS. We showed that mutations that led to improved AGG codon reassignment identified using a tyrosine-activating aaRS also improved the efficiency of pAzF incorporation in response to the arginine AGG codon.
We expect that this directed evolution workflow will be readily applicable to improving incorporation of ncAAs in response to other targeted sense codons exploiting orthogonal tRNA/aaRS pairs already-evolved to recognize ncAAs and target suppression of an amber stop codon. Although decreasing competition from the endogenous translation machinery via genomic alterations proved effective for improving reassignment of the Arg AGG codon in other reports, the genomic alteration strategy is not necessarily readily transferrable to other attractive sense codon targets. Given the idiosyncrasies of in vivo protein translation and of the orthogonal pairs transplanted into it, improvements for one amino acid may not result in the same magnitude of improvement for another amino acid each time. However, we expect this approach to serve as a reasonable starting point for rapid identification of positional mutants with the likelihood of improving tRNA/aaRS recognition across aaRSs evolved for many amino acids.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Conceptualization: WB and JF; Funding acquisition: MS and JF; Investigation, WB, DS, and MS; Methodology, WB, DS, and MS; Supervision: JF; Visualization, WB and MS; Writing–original draft, WB; Writing–review and editing, MS and JF.
This research was funded by the National Science Foundation, grant number CHE-1507055 to JF. The APC was funded by University of Colorado Denver.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Tae Hyeon Yoo for the generous gift of the pSPEL plasmid for expression of the Z domain and their laboratory’s genomically-engineered BS-02 E. coli for comparison with our strain. We thank two facilities for maintaining FACS instruments and expertise: Leslie Armstrong-Lea and the Colorado State University Proteomics and Metabolomics Facility and the Colorado State University Flow Cytometry Facility. Mass spectrometry analysis was performed in the Chemistry Instrumentation Facility at Colorado State University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.815788/full#supplementary-material
Amiram, M., Haimovich, A. D., Fan, C., Wang, Y.-S., Aerni, H.-R., Ntai, I., et al. (2015). Evolution of Translation Machinery in Recoded Bacteria Enables Multi-Site Incorporation of Nonstandard Amino Acids. Nat. Biotechnol. 33 (12), 1272–1279. doi:10.1038/nbt.3372
Apostol, I., Aitken, J., Levine, J., Lippincott, J., Davidson, J. S., and Abbott-Brown, D. (1995). Recombinant Protein Sequences Can Trigger Methylation of N-Terminal Amino Acids inEscherichia Coli. Protein Sci. 4 (12), 2616–2618. doi:10.1002/pro.5560041219
Bayley, H., Standring, D. N., and Knowles, J. R. (1978). Propane-1,3-dithiol: A Selective Reagent for the Efficient Reduction of Alkyl and Aryl Azides to Amines. Tetrahedron Lett. 19, 3633–3634. doi:10.1016/s0040-4039(01)95015-4
Biddle, W., Schmitt, M. A., and Fisk, J. D. (2015). Evaluating Sense Codon Reassignment with a Simple Fluorescence Screen. Biochemistry 54 (50), 7355–7364. doi:10.1021/acs.biochem.5b00870
Biddle, W., Schmitt, M. A., and Fisk, J. D. (2016). Modification of Orthogonal tRNAs: Unexpected Consequences for Sense Codon Reassignment. Nucleic Acids Res. 44 (21), 10042–10050. doi:10.1093/nar/gkw948
Bohlke, N., and Budisa, N. (2014). Sense Codon Emancipation for Proteome-wide Incorporation of Noncanonical Amino Acids: Rare Isoleucine Codon AUA as a Target for Genetic Code Expansion. FEMS Microbiol. Lett. 351 (2), 133–144. doi:10.1111/1574-6968.12371
Bossi, L., and Roth, J. R. (1980). The Influence of Codon Context on Genetic Code Translation. Nature 286 (5769), 123–127. doi:10.1038/286123a0
Braisted, A. C., and Wells, J. A. (1996). Minimizing a Binding Domain from Protein A. Proc. Natl. Acad. Sci. 93 (12), 5688–5692. doi:10.1073/pnas.93.12.5688
Cervettini, D., Tang, S., Fried, S. D., Willis, J. C. W., Funke, L. F. H., Colwell, L. J., et al. (2020). Rapid Discovery and Evolution of Orthogonal Aminoacyl-tRNA Synthetase-tRNA Pairs. Nat. Biotechnol. 38 (8), 989–999. doi:10.1038/s41587-020-0479-2
Chatterjee, A., Sun, S. B., Furman, J. L., Xiao, H., and Schultz, P. G. (2013). A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia C. Biochemistry 52 (10), 1828–1837. doi:10.1021/bi4000244
Chevance, F. F. V., Le Guyon, S., and Hughes, K. T. (2014). The Effects of Codon Context on In Vivo Translation Speed. Plos Genet. 10 (6), e1004392. doi:10.1371/journal.pgen.1004392
Chin, J. W., Santoro, S. W., Martin, A. B., King, D. S., Wang, L., and Schultz, P. G. (2002). Addition of P-Azido-L-Phenylalanine to the Genetic Code of Escherichia C. J. Am. Chem. Soc. 124 (31), 9026–9027. doi:10.1021/ja027007w
Clackson, T., and Lowman, H. B. (2004). “Phage Display a Practical Approach,” in Practical Approach Series. Editors T. Clackson, and H. B. Lowman (Oxford: Oxford University Press).
Datsenko, K. A., and Wanner, B. L. (2000). One-Step Inactivation of Chromosomal Genes in Escherichia C K-12 Using PCR Products. Proc. Natl. Acad. Sci. 97 (12), 6640–6645. doi:10.1073/pnas.120163297
Ding, W., Zhao, H., Chen, Y., Zhang, B., Yang, Y., Zang, J., et al. (2020). Chimeric Design of Pyrrolysyl-tRNA Synthetase/tRNA Pairs and Canonical Synthetase/tRNA Pairs for Genetic Code Expansion. Nat. Commun. 11 (1), 3154. doi:10.1038/s41467-020-16898-y
Dumas, A., Lercher, L., Spicer, C. D., and Davis, B. G. (2015). Designing Logical Codon Reassignment - Expanding the Chemistry in Biology. Chem. Sci. 6 (1), 50–69. doi:10.1039/c4sc01534g
Fan, C., Xiong, H., Reynolds, N. M., and Söll, D. (2015). Rationally Evolving tRNA(Pyl) for Efficient Incorporation of Noncanonical Amino Acids. Nucleic Acids Res. 43 (22), e156. doi:10.1093/nar/gkv800
Fechter, P., Rudinger-Thirion, J., Tukalo, M., and Giege, R. (2001). Major Tyrosine Identity Determinants in Methanococcus Jannaschii and Saccharomyces C tRNA(Tyr) Conserved but Expressed Differently. Eur. J. Biochem. 268 (3), 761–767. doi:10.1046/j.1432-1327.2001.01931.x
Fredens, J., Wang, K., de la Torre, D., Funke, L. F. H., Robertson, W. E., Christova, Y., et al. (2019). Total Synthesis of Escherichia coli with a Recoded Genome. Nature 569 (7757), 514–518. doi:10.1038/s41586-019-1192-5
Furter, R. (1998). Expansion of the Genetic Code: Site-Directed P-Fluoro-Phenylalanine Incorporation in Escherichia coli. Protein Sci. 7 (2), 419–426. doi:10.1002/pro.5560070223
Gamble, C. E., Brule, C. E., Dean, K. M., Fields, S., and Grayhack, E. J. (2016). Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell 166 (3), 679–690. doi:10.1016/j.cell.2016.05.070
Gao, X., Yo, P., Keith, A., Ragan, T. J., and Harris, T. K. (2003). Thermodynamically Balanced Inside-Out (TBIO) PCR-Based Gene Synthesis: A Novel Method of Primer Design for High-Fidelity Assembly of Longer Gene Sequences. Nucleic Acids Res. 31 (22), e143. doi:10.1093/nar/gng143
Giege, R., Sissler, M., and Florentz, C. (1998). Universal Rules and Idiosyncratic Features in tRNA Identity. Nucleic Acids Res. 26 (22), 5017–5035. doi:10.1093/nar/26.22.5017
Guo, J., Melançon, C. E., Lee, H. S., Groff, D., and Schultz, P. G. (2009). Evolution of Amber Suppressor tRNAs for Efficient Bacterial Production of Proteins Containing Nonnatural Amino Acids. Angew. Chem. Int. Edition 48 (48), 9148–9151. doi:10.1002/anie.200904035
Guo, L.-T., Wang, Y.-S., Nakamura, A., Eiler, D., Kavran, J. M., Wong, M., et al. (2014). Polyspecific Pyrrolysyl-tRNA Synthetases from Directed Evolution. Proc. Natl. Acad. Sci. USA 111 (47), 16724–16729. doi:10.1073/pnas.1419737111
Ho, J. M., Reynolds, N. M., Rivera, K., Connolly, M., Guo, L.-T., Ling, J., et al. (2016). Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia coli. ACS Synth. Biol. 5 (2), 163–171. doi:10.1021/acssynbio.5b00197
Johnson, D. B. F., Xu, J., Shen, Z., Takimoto, J. K., Schultz, M. D., Schmitz, R. J., et al. (2011). RF1 Knockout Allows Ribosomal Incorporation of Unnatural Amino Acids at Multiple Sites. Nat. Chem. Biol. 7 (11), 779–786. doi:10.1038/nchembio.657
Kajihara, D., Hohsaka, T., and Sisido, M. (2005). Synthesis and Sequence Optimization of GFP Mutants Containing Aromatic Non-natural Amino Acids at the Tyr66 Position. Protein Eng. Des. Selection 18 (6), 273–278. doi:10.1093/protein/gzi033
Kirshenbaum, K., Carrico, I. S., and Tirrell, D. A. (2002). Biosynthesis of Proteins Incorporating a Versatile Set of Phenylalanine Analogues. ChemBioChem 3 (2-3), 235–237. doi:10.1002/1439-7633(20020301)3:2/3<235:AID-CBIC235>3.0.CO;2-7
Kobayashi, T., Nureki, O., Ishitani, R., Yaremchuk, A., Tukalo, M., Cusack, S., et al. (2003). Structural Basis for Orthogonal tRNA Specificities of Tyrosyl-tRNA Synthetases for Genetic Code Expansion. Nat. Struct. Mol. Biol. 10 (6), 425–432. doi:10.1038/nsb934
Krishnakumar, R., Prat, L., Aerni, H.-R., Ling, J., Merryman, C., Glass, J. I., et al. (2013). Transfer RNA Misidentification Scrambles Sense Codon Recoding. ChemBioChem 14 (15), 1967–1972. doi:10.1002/cbic.201300444
Kuhn, S. M., Rubini, M., Fuhrmann, M., Theobald, I., and Skerra, A. (2010). Engineering of an Orthogonal Aminoacyl-tRNA Synthetase for Efficient Incorporation of the Non-natural Amino Acid O-Methyl-L-Tyrosine Using Fluorescence-Based Bacterial Cell Sorting. J. Mol. Biol. 404 (1), 70–87. doi:10.1016/j.jmb.2010.09.001
Kunkel, T. A. (1985). Rapid and Efficient Site-Specific Mutagenesis without Phenotypic Selection. Proc. Natl. Acad. Sci. 82 (2), 488–492. doi:10.1073/pnas.82.2.488
Kwon, I., and Choi, E. S. (2016). Forced Ambiguity of the Leucine Codons for Multiple-Site-Specific Incorporation of a Noncanonical Amino Acid. PLoS One 11 (3), e0152826. doi:10.1371/journal.pone.0152826
Kwon, I., Kirshenbaum, K., and Tirrell, D. A. (2003). Breaking the Degeneracy of the Genetic Code. J. Am. Chem. Soc. 125 (25), 7512–7513. doi:10.1021/ja0350076
Lajoie, M. J., Rovner, A. J., Goodman, D. B., Aerni, H.-R., Haimovich, A. D., Kuznetsov, G., et al. (2013). Genomically Recoded Organisms Expand Biological Functions. Science 342 (6156), 357–360. doi:10.1126/science.1241459
Lee, B. S., Shin, S., Jeon, J. Y., Jang, K.-S., Lee, B. Y., Choi, S., et al. (2015). Incorporation of Unnatural Amino Acids in Response to the AGG Codon. ACS Chem. Biol. 10 (7), 1648–1653. doi:10.1021/acschembio.5b00230
Lee, K. B., Hou, C. Y., Kim, C.-E., Kim, D.-M., Suga, H., and Kang, T. J. (2016). Genetic Code Expansion by Degeneracy Reprogramming of Arginyl Codons. ChemBioChem 17 (13), 1198–1201. doi:10.1002/cbic.201600111
Liu, C. C., and Schultz, P. G. (2010). Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 79, 413–444. doi:10.1146/annurev.biochem.052308.105824
Machnicka, M. A., Milanowska, K., Osman Oglou, O., Purta, E., Kurkowska, M., Olchowik, A., et al. (2013). MODOMICS: A Database of RNA Modification Pathways-2013 Update. Nucleic Acids Res. 41 (D1), D262–D267. doi:10.1093/nar/gks1007
Maranhao, A. C., and Ellington, A. D. (2017). Evolving Orthogonal Suppressor tRNAs To Incorporate Modified Amino Acids. ACS Synth. Biol. 6 (1), 108–119. doi:10.1021/acssynbio.660014510.1021/acssynbio.6b00145
Mittelstaet, J., Konevega, A. L., and Rodnina, M. V. (2013). A Kinetic Safety Gate Controlling the Delivery of Unnatural Amino Acids to the Ribosome. J. Am. Chem. Soc. 135 (45), 17031–17038. doi:10.1021/ja407511q
Morris, J. L., Reddington, S. C., Murphy, D. M., Jones, D. D., Platts, J. A., and Tippmann, E. M. (2013). Aryl Azide Photochemistry in Defined Protein Environments. Org. Lett. 15 (4), 728–731. doi:10.1021/ol3028779
Mukai, T., Hayashi, A., Iraha, F., Sato, A., Ohtake, K., Yokoyama, S., et al. (2010). Codon Reassignment in the Escherichia coli Genetic Code. Nucleic Acids Res. 38 (22), 8188–8195. doi:10.1093/nar/gkq707
Mukai, T., Yamaguchi, A., Ohtake, K., Takahashi, M., Hayashi, A., Iraha, F., et al. (2015). Reassignment of a Rare Sense Codon to a Non-Canonical Amino Acid inEscherichia Coli. Nucleic Acids Res. 43 (16), 8111–8122. doi:10.1093/nar/gkv787
Neumann, H., Slusarczyk, A. L., and Chin, J. W. (2010). De Novo Generation of Mutually Orthogonal Aminoacyl-tRNA Synthetase/tRNA Pairs. J. Am. Chem. Soc. 132 (7), 2142–2144. doi:10.1021/ja9068722
Ngo, J. T., and Tirrell, D. A. (2011). Noncanonical Amino Acids in the Interrogation of Cellular Protein Synthesis. Acc. Chem. Res. 44 (9), 677–685. doi:10.1021/ar200144y
Ostrov, N., Landon, M., Guell, M., Kuznetsov, G., Teramoto, J., Cervantes, N., et al. (2016). Design, Synthesis, and Testing toward a 57-Codon Genome. Science 353 (6301), 819–822. doi:10.1126/science.aaf3639
Owens, A. E., Grasso, K. T., Ziegler, C. A., and Fasan, R. (2017). Two‐Tier Screening Platform for Directed Evolution of Aminoacyl-tRNA Synthetases with Enhanced Stop Codon Suppression Efficiency. ChemBioChem 18 (12), 1109–1116. doi:10.1002/cbic.201700039
Polycarpo, C. R., Herring, S., Bérubé, A., Wood, J. L., Söll, D., and Ambrogelly, A. (2006). Pyrrolysine Analogues as Substrates for Pyrrolysyl-tRNA Synthetase. FEBS Lett. 580 (28-29), 6695–6700. doi:10.1016/j.febslet.2006.11.028
Pott, M., Schmidt, M. J., and Summerer, D. (2014). Evolved Sequence Contexts for Highly Efficient Amber Suppression with Noncanonical Amino Acids. ACS Chem. Biol. 9 (12), 2815–2822. doi:10.1021/cb5006273
Rauch, B. J., Porter, J. J., Mehl, R. A., and Perona, J. J. (2016). Improved Incorporation of Noncanonical Amino Acids by an Engineered tRNA(Tyr) Suppressor. Biochemistry 55 (3), 618–628. doi:10.1021/acs.biochem.5b01185
Reddington, S. C., Rizkallah, P. J., Watson, P. D., Pearson, R., Tippmann, E. M., and Jones, D. D. (2013). Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence. Angew. Chem. Int. Ed. 52 (23), 5974–5977. doi:10.1002/anie.201301490
Ryu, Y., and Schultz, P. G. (2006). Efficient Incorporation of Unnatural Amino Acids into Proteins in Escherichia coli. Nat. Methods 3 (4), 263–265. doi:10.1038/nmeth864
Sambrook, J., and Russell, D. W. (2001). “Molecular Cloning : a Laboratory Manual,” in Cold Spring Harbor (N.Y., Cold Spring Harbor: Cold Spring Harbor Laboratory Press).
Schmitt, M. A., Biddle, W., and Fisk, J. D. (2018). Mapping the Plasticity of the Escherichia coli Genetic Code with Orthogonal Pair-Directed Sense Codon Reassignment. Biochemistry 57, 2762–2774. doi:10.1021/acs.biochem.8b00177
Schrader, J. M., Chapman, S. J., and Uhlenbeck, O. C. (2011). Tuning the Affinity of Aminoacyl-tRNA to Elongation Factor Tu for Optimal Decoding. Proc. Natl. Acad. Sci. 108 (13), 5215–5220. doi:10.1073/pnas.1102128108
Schwark, D., Schmitt, M., and Fisk, J. (2018). Dissecting the Contribution of Release Factor Interactions to Amber Stop Codon Reassignment Efficiencies of the Methanocaldococcus Jannaschii Orthogonal Pair. Genes 9 (11), 546. doi:10.3390/genes9110546
Schwark, D. G., Schmitt, M. A., Biddle, W., and Fisk, J. D. (2020). The Influence of Competing tRNA Abundance on Translation: Quantifying the Efficiency of Sense Codon Reassignment at Rarely Used Codons. ChemBioChem 21, 2274–2286. doi:10.1002/cbic.202000052
Schwark, D. G., Schmitt, M. A., and Fisk, J. D. (2021). Directed Evolution of the Methanosarcina Barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential. Int. J. Mol. Sci. 22 (2), 895. doi:10.3390/ijms22020895
Staros, J. V., Bayley, H., Standring, D. N., and Knowles, J. R. (1978). Reduction of Aryl Azides by Thiols: Implications for the Use of Photoaffinity Reagents. Biochem. Biophysical Res. Commun. 80 (3), 568–572. doi:10.1016/0006-291x(78)91606-6
Stock, A., Schaeffer, E., Koshland, D. E., and Stock, J. (1987). A Second Type of Protein Methylation Reaction in Bacterial Chemotaxis. J. Biol. Chem. 262 (17), 8011–8014. doi:10.1016/s0021-9258(18)47518-7
Takimoto, J. K., Adams, K. L., Xiang, Z., and Wang, L. (2009). Improving Orthogonal tRNA-Synthetase Recognition for Efficient Unnatural Amino Acid Incorporation and Application in Mammalian Cells. Mol. Biosyst. 5 (9), 931–934. doi:10.1039/b904228h
Wan, W., Tharp, J. M., and Liu, W. R. (2014). Pyrrolysyl-tRNA Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1844 (6), 1059–1070. doi:10.1016/j.bbapap.2014.03.002
Wang, Y., and Tsao, M.-L. (2016). Reassigning Sense Codon AGA to Encode Noncanonical Amino Acids inEscherichia Coli. ChemBioChem 17 (23), 2234–2239. doi:10.1002/cbic.201600448
Wang, L., Brock, A., Herberich, B., and Schultz, P. G. (2001). Expanding the Genetic Code of Escherichia coli. Science 292 (5516), 498–500. doi:10.1126/science.1060077
Wang, L., Xie, J., Deniz, A. A., and Schultz, P. G. (2003a). Unnatural Amino Acid Mutagenesis of Green Fluorescent Protein. J. Org. Chem. 68 (1), 174–176. doi:10.1021/jo026570u
Wang, L., Zhang, Z., Brock, A., and Schultz, P. G. (2003b). Addition of the Keto Functional Group to the Genetic Code of Escherichia coli. Proc. Natl. Acad. Sci. 100 (1), 56–61. doi:10.1073/pnas.0234824100
Wang, F., Niu, W., Guo, J., and Schultz, P. G. (2012). Unnatural Amino Acid Mutagenesis of Fluorescent Proteins. Angew. Chem. Int. Ed. 51 (40), 10132–10135. doi:10.1002/anie.201204668
Wang, N., Ju, T., Niu, W., and Guo, J. (2015). Fine-Tuning Interaction between Aminoacyl-tRNA Synthetase and tRNA for Efficient Synthesis of Proteins Containing Unnatural Amino Acids. ACS Synth. Biol. 4 (3), 207–212. doi:10.1021/sb500195w
Wang, K., Fredens, J., Brunner, S. F., Kim, S. H., Chia, T., and Chin, J. W. (2016). Defining Synonymous Codon Compression Schemes by Genome Recoding. Nature 539 (7627), 59–64. doi:10.1038/nature20124
Willis, J. C. W., and Chin, J. W. (2018). Mutually Orthogonal Pyrrolysyl-tRNA Synthetase/tRNA Pairs. Nat. Chem 10 (8), 831–837. doi:10.1038/s41557-018-0052-5
Wiltschi, B., and Budisa, N. (2007). Natural History and Experimental Evolution of the Genetic Code. Appl. Microbiol. Biotechnol. 74 (4), 739–753. doi:10.1007/s00253-006-0823-6
Xie, J., Supekova, L., and Schultz, P. G. (2007). A Genetically Encoded Metabolically Stable Analogue of Phosphotyrosine in Escherichia coli. ACS Chem. Biol. 2 (7), 474–478. doi:10.1021/cb700083w
Young, T. S., Ahmad, I., Yin, J. A., and Schultz, P. G. (2010). An Enhanced System for Unnatural Amino Acid Mutagenesis in E. coli. J. Mol. Biol. 395 (2), 361–374. doi:10.1016/j.jmb.2009.10.030
Zeng, Y., Wang, W., and Liu, W. R. (2014). Towards Reassigning the Rare AGG Codon inEscherichia Coli. ChemBioChem 15 (12), 1750–1754. doi:10.1002/cbic.201400075
Zhang, Z., Smith, B. A. C., Wang, L., Brock, A., Cho, C., and Schultz, P. G. (2003). A New Strategy for the Site-Specific Modification of Proteins In Vivo. Biochemistry 42 (22), 6735–6746. doi:10.1021/bi0300231
Keywords: genetic code expansion, sense codon reassignment, noncanonical amino acid, synthetic biology, directed evolution, protein engineering
Citation: Biddle W, Schwark DG, Schmitt MA and Fisk JD (2022) Directed Evolution Pipeline for the Improvement of Orthogonal Translation Machinery for Genetic Code Expansion at Sense Codons. Front. Chem. 10:815788. doi: 10.3389/fchem.2022.815788
Received: 15 November 2021; Accepted: 24 January 2022;
Published: 17 February 2022.
Edited by:
Jiantao Guo, University of Nebraska-Lincoln, United StatesReviewed by:
Shixian Lin, Zhejiang University, ChinaCopyright © 2022 Biddle, Schwark, Schmitt and Fisk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John D. Fisk, am9obi5maXNrQHVjZGVudmVyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.