- 1Artemisinin Research Center, Institute of Chinese Meteria Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 2School of Graduate Students, Tianjin University of Traditional Chinese Medicine, Tianjin, China
Introduction: Artemisinin (1) is a milestone compound in malaria treatment, and it exhibits a broad scope of bioactivities. Herein, sequential chemo-reduction and biotransformation of artemisinin were undertaken to obtain a series of artemisinin derivatives.
Methods: First, 10-deoxyartemisinin (2) and 9-ene-10-deoxyartemisinin (3) were synthesized after simple handling with boron trifluoride/diethyl ether and sodium borohydride. Then, biotransformation of 10-deoxyartemisinin was conducted with Cunninghamella echinulata CGMCC 3.4879 and Cunninghamella elegans CGMCC 3.4832, and the transformed products were separated and identified. The antimalarial activity of these products was tested in vitro against Plasmodium falciparum 3D7.
Results: Fifteen metabolites (4–18), including seven novel compounds, were isolated and identified after cultivation. Compounds 2, 3, 13, 15, 16, and 18 displayed moderate-to-good antimalarial activity, with a half-maximal inhibitory concentration ranging from 6 to 223 nM.
Discussion: This work explored the combination of chemical and biological transformation to develop a co-environmental, efficient, and cost-efficiency synthetic methodology and applied it to synthesize novel derivatives of artemisinin. The association of the two strategies will hopefully provide an abundant source for the development of novel drugs with bioactivities.
1 Introduction
Artemisinin (also named qinghaosu, ART) is a legendary antimalarial agent. It was discovered from Artemisia annua L. by Tu et al. (1981) in 1972. In the last 50 years, millions of people suffering from malaria have been saved by artemisinin or its derivatives. Artemisinin-based combination therapy (ACT) is a recommended first-line treatment for malaria by the World Health Organization (especially for chloroquine-resistant malaria) (World Health Organization, 2021).
In recent years, parasite clearance for some patients in the Great Mekong Area and parts of Africa has been reported to be delayed after treatment with artesunate for 3 days (Uwimana et al., 2020). According to clinical studies, parasites can be cleared through 7-day treatment of artesunate, but the aforementioned delayed clearance has increased the anxiety about the possibility of resistance to artemisinin (Wang et al., 2019). To prevent the consequences of unpredictable drug resistance, there is an urgent need to explore antimalarial agents, including novel derivatives of artemisinin or other new chemical skeletons.
Microorganism-mediated modification of natural products and bioactive molecules is an efficient route for drug development (Cao et al., 2015). The abundant enzymes found in microorganisms enable hydroxylation, oxidation, reduction, and coupling reactions, with excellent chemo-, regio-, and even stereo-selectivities (Asha and Vidyavathi, 2009). In recent years, numerous microbial-based transformations of artemisinin have been attempted and various microbiological strains have been applied, including those of the genera Aspergillus, Streptomyces, Penicillium, and Cunninghamella (Parshikov et al., 2004a; Liu et al., 2006; Goswami et al., 2010; Ponnapalli et al., 2018). However, the number of compounds converted in a single transformation has been modest. In the latest research, four compounds from artemisinin were converted by Aspergillus niger in 2022 (Luo et al., 2022). Comparatively, plentiful metabolites were reported in this work.
Our research team has been engaged in the optimization of artemisinin (1) for the development of antimalarial drugs. In recent years, we have prepared a series of artemisinin derivatives through fungal-mediated transformations (Bai et al., 2019; Ma et al., 2019; Bai et al., 2021; Zhao et al., 2021). The labile lactone structure of artemisinin is widely perceived to contribute to diminishing the stability of the entire molecule. Herein, the lactone was reduced to methylene to offer 10-deoxyartemisinin (2), accompanied by a byproduct: 9-ene-10-deoxyartemisinin (3). The 10-deoxyartemisinin was modified by Cunninghamella species to obtain structurally divergent metabolites. The antimalarial activity of these generated products against P. falciparum (Pf.) 3D7 was examined to obtain potential lead compounds for drug development.
2 Materials and methods
2.1 General experimental procedures
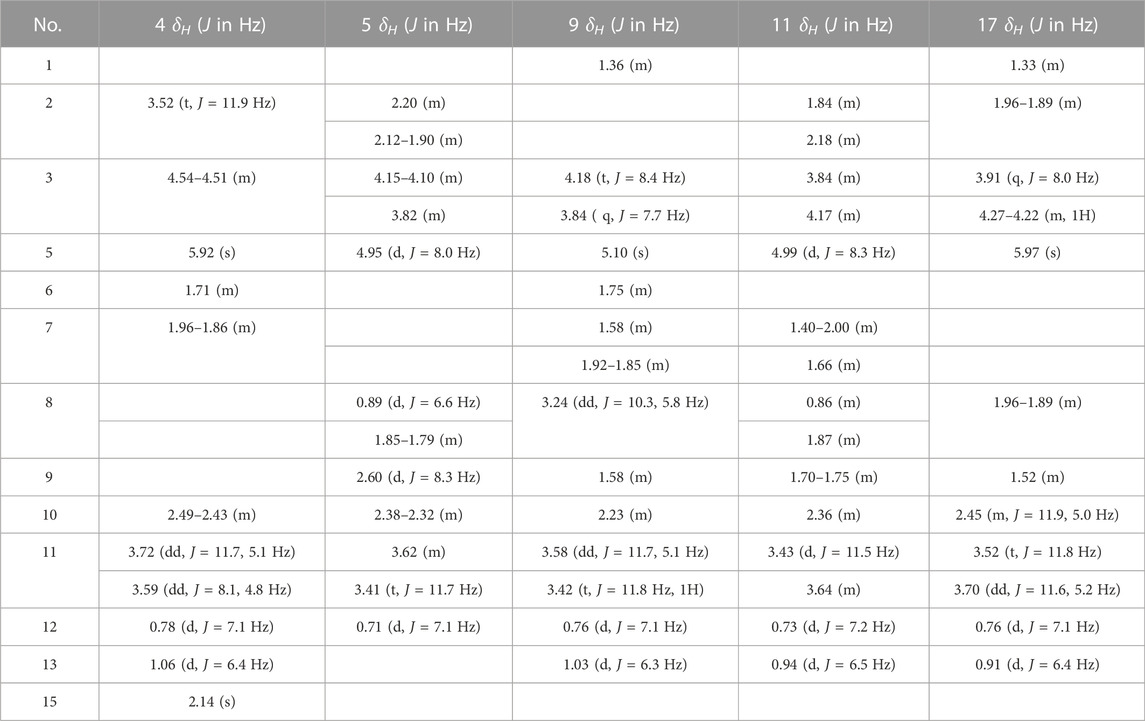
1H (600 MHz), 13C (150 MHz), and two-dimensional nuclear magnetic resonance (2D-NMR) spectroscopy were undertaken on a spectrometer (AV 600; Bruker, Billerica, MA, United States) with tetramethylsilane as an internal reference. Chemical shifts (δ) are given in ppm. Coupling constants (J) are given in hertz. X-ray diffraction was carried out using a diffractometer (D8 Venture; Bruker) with Cu Kα radiation. Column chromatography was performed with a silica flash column (330 g; Qingdao Marine Chemical Group, Qingdao, China), silica gel (200–300 mesh; Qingdao Marine Chemical Group), and a Chromatorex (FujiSilysia Chemicals, Kasugai, Japan) system. Analytical thin-layer chromatography was carried out on pre-coated silica-gel GF254 plates (Qingdao Marine Chemical Group). Water was prepared using a Milli-Q™ system operating at 18.2 MΩ (Millipore, Bedford, MA, United States). Unless stated otherwise, all chemicals were obtained from commercially available sources and were used without further purification.
2.2 Synthesis of 10-deoxyartemisinin and 9-ene-10-deoxyartemisinin
Artemisinin (99% by high-performance liquid chromatography (HPLC); batch number, C00120160) was purchased from Kunming Pharmaceutical Group (Kunming, China). Under an inert atmosphere, a solution of artemisinin (2 g) and boron trifluoride/diethyl ether (BF3/Et2O) (26.4 mL) in dry tetrahydrofuran (THF; 30 mL) at 0 °C was added dropwise to an ice-cooled solution of sodium tetrahydroborate (NaBH4; 0.6 g) in dry THF (30 mL). The reaction was carried out for 3 h at 0°C and then heated to reflux for 15 min. The synthetic route is shown in Figure 1. After cooling to room temperature, the reaction mixture was extracted thrice with ether (1:1, v/v). The combined organic phase was washed with saturated sodium chloride and dried over anhydrous sodium sulfate (NaSO4). The solvent was removed after evaporation in vacuo to obtain the crude product. The latter was purified by silica-gel column chromatography. The target product, 10-deoxyartemisinin (2, yield 50%), and the byproduct, 9-ene-10-deoxyartemisinin (3, yield 22%), were obtained with petroleum ether–ethyl acetate as the eluent. High-resolution-electrospray ionization-mass spectrometry (HR-ESI-MS) and 1H-NMR and 13C-NMR spectroscopy were used to identify structures. The NMR data of 10-deoxoartemisinin and 9-ene-10-deoxyartemisinin are shown in Table 1 and Table 2. The synthesis scheme is shown in Figure 1.
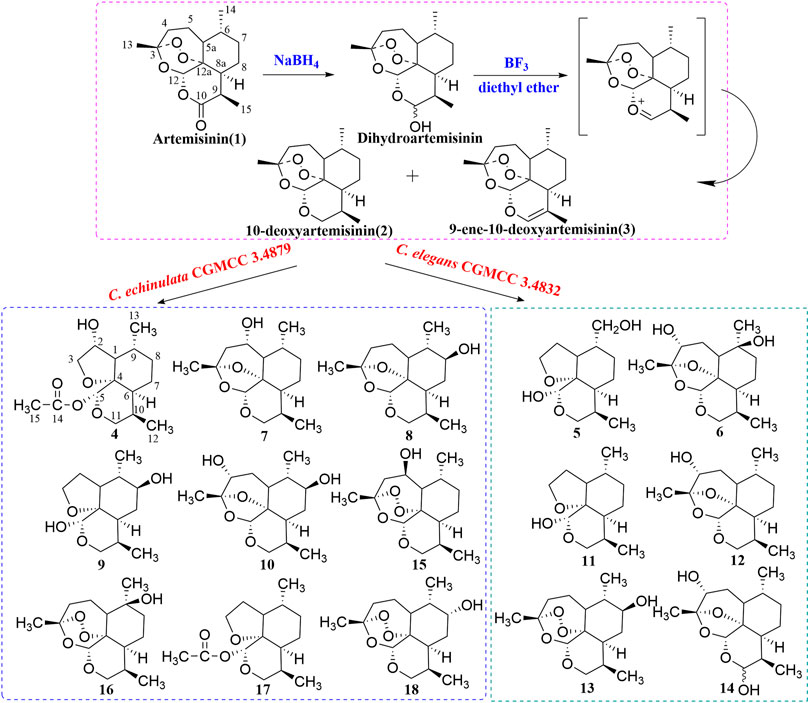
FIGURE 1. Synthesis route of 10-deoxyartemisinin and its metabolites by C. elegans and C. echinulata.
2.3 Preparative biotransformation, extraction, and isolation of metabolites
C. echinulata CGMCC 3.4879 and C. elegans CGMCC 3.4832 were obtained from the China General Microbiological Culture Collection Center (Beijing, China). Culture was conducted in a medium comprising sabouraud dextrose broth (20 g/L), peptone (10 g/L), and sucrose (15 g/L). Two-stage fermentation was carried out. The substrate (10-deoxyartemisinin) was dissolved in methanol (25 mg/mL) and added to each flask after the second fermentation to reach a final concentration of 0.5 mg/mL. Cultures were incubated at 28 °C and agitated at 180 rpm/min for 14 days. Then, they were filtered and extracted thrice with ethyl acetate at an equal volume. The extract was dried with anhydrous Na2SO4 and concentrated under a vacuum at 45 °C to provide a residue.
The residue from C. echinulata CGMCC 3.4879 was subjected to a silica-gel column chromatography by elution with petroleum ether/ethyl acetate to provide six subfractions (Fr.1–Fr.6). Fractions 1, 2, and 4 were separated by Chromatorex silica-gel column chromatography with petroleum ether/ethyl acetate to obtain compound 12 (200 mg), compound 9 (9 mg), and compound 13 (250 mg), respectively. Fraction 3 was purified by recrystallization from ethyl acetate to provide compound 11 (120 mg). Fractions 5 and 6 were purified by semi-preparative normal-phase HPLC (methanol/water) to gain compound 14 (7 mg) and compounds 4–6 (34, 12, and 6 mg), respectively.
The residue from C. elegans CGMCC 3.4832 was subjected to silica-gel column chromatography by elution with petroleum ether/ethyl acetate to provide seven fractions (Fr.1–Fr.7). Fractions 1, 5, and 7 were separated by Chromatorex silica-gel column chromatography with petroleum ether/ethyl acetate to afford compound 17 (50 mg), compound 18 (154 mg), and compound 9 (5 mg), respectively. Fraction 3 was separated by reverse-phase C18 column chromatography with methanol/water to obtain compound 15 (10 mg). Fraction 4 was separated by Chromatorex silica-gel column chromatography with petroleum ether/ethyl acetate with additional reverse-phase C18 column chromatography to obtain compound 7 (10 mg), compound 8 (4 mg), compound 16 (20 mg), and compound 10 (100 mg).
2.4 Identification of compounds
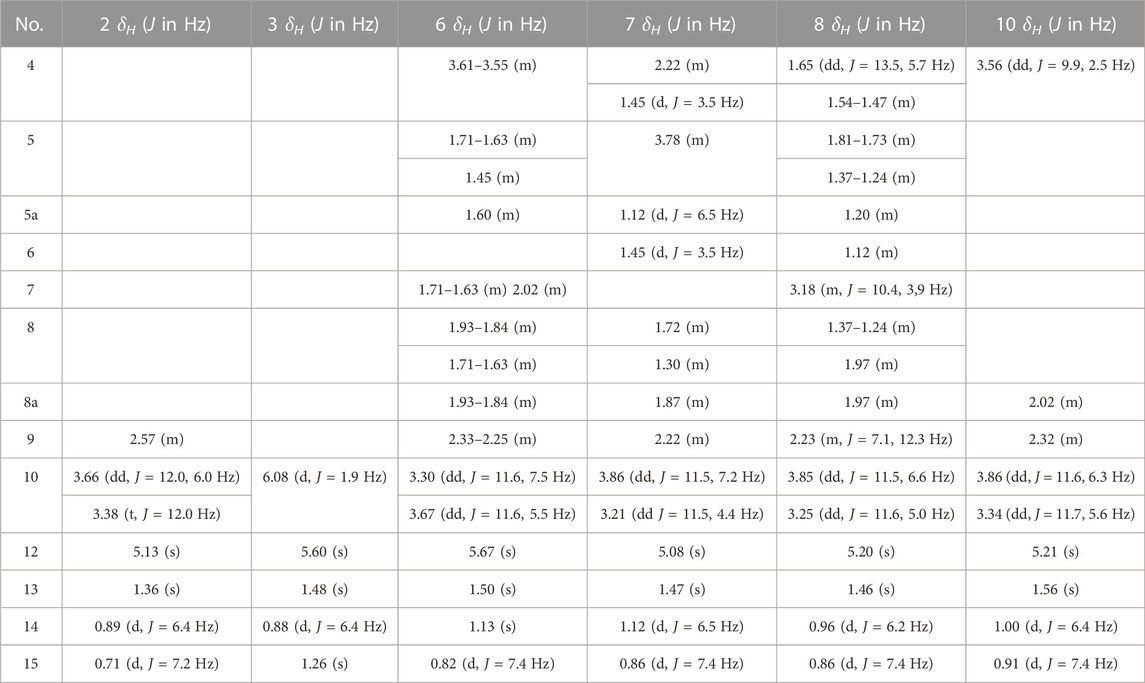
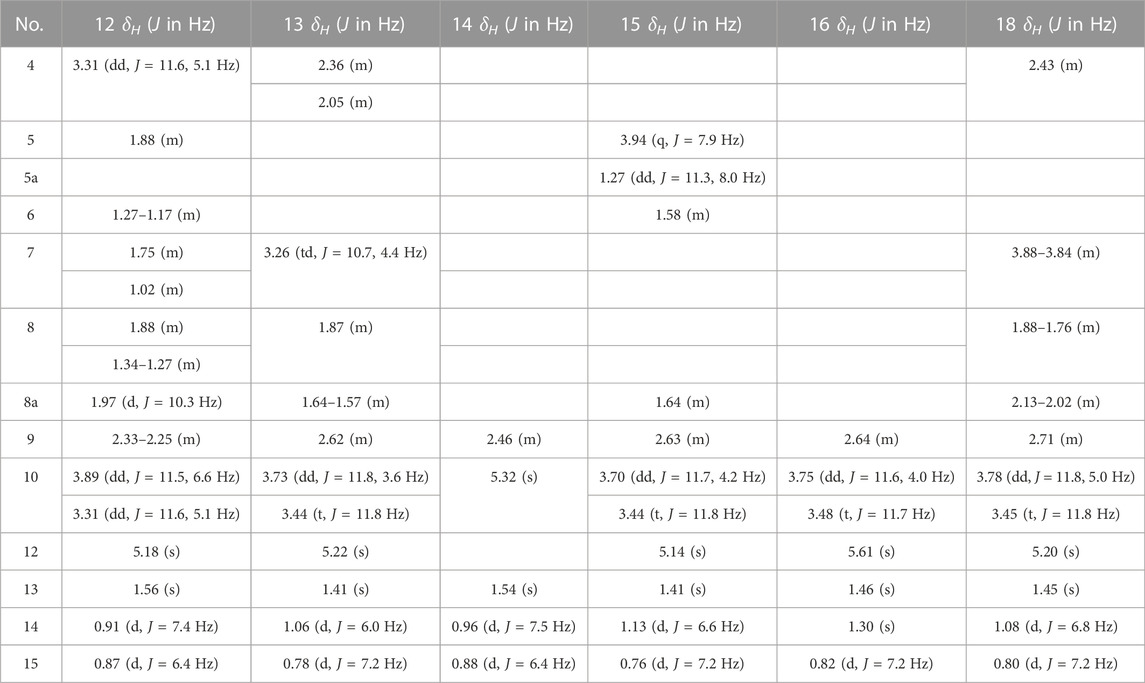
10-Deoxyartemisinin (2) (Jung et al., 1990): White acicular crystals (ethyl acetate). HR-ESI-MS m/z 291.1624 [M + Na]+ (calcd for C15H24O4, 268.1675). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 2.
9-ene-10-Deoxyartemisinin (3) (Xie et al., 2001): White powder (ethyl acetate). HR-ESI-MS m/z 289.1425 [M + Na]+ (calcd for C15H22O4, 266.1518). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 2.
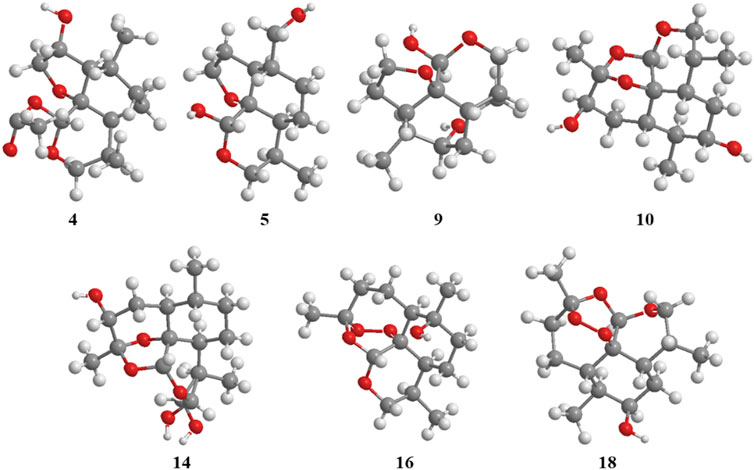
2α-Hydroxy-5α-acetoxy-artemethin-Ⅱ (4): Colorless, transparent, columnar crystals (ethyl acetate). HR-ESI-MS m/z 307.1520 [M + Na]+ (calcd for C15H24O5, 284.1624). Crystal data: C15H24O5, M = 284.34, monoclinic system, crystal size is 0.47 mm3 × 0.40 mm3 × 0.39 mm3, a = 13.2788 (6) Å, b = 14.9910 (7) Å, c = 15.8479 (8) Å; α = β = γ = 90.00°, V = 3154.7 (3) Å3, space group P212121 (NO. 19), T = 273.15 K, Z = 8, Z′ = 2, μ (Cu Kα) = 0.089 mm−1, wavelength/Å = 0.71073, R1 = 0.1017, wR (F2) = 0.1283. Flack parameter: 0.2 (2). Crystallographic data of compound 4 have been deposited to CCDC (www.ccdc.cam.ac.uk/, number = CCDC 2218207). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 4.
5α,13-Dihydroxy-artemethin-Ⅱ (5): Colorless, transparent, columnar crystals (petroleum ether and acetone). HR-ESI-MS m/z 265.1415 [M + Na]+ (calcd for C13H22O4, 242.1518). Crystal data: C13H22O4, M = 242.30, monoclinic system, crystal size is 0.47 mm3 × 0.40 mm3 × 0.39 mm3, a = 6.1074 (3) Å, b = 15.6846 (7) Å, c = 6.7830 (3) Å; α = γ = 90°, V = 634.54 (5) Å3, space group P21 (NO. 4), T = 273.15 K, Z = 2, μ (Cu Kα) = 0.092 mm−1, wavelength/Å = 0.71073, R1 = 0.0319 [I > 2σ (I)], wR (F2) = 0.0875. Flack parameter: 0.2 (2). Crystallographic data of compound 5 have been deposited to CCDC (www.ccdc.cam.ac.uk/, number = CCDC 2218208). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 4.
4α,6β-Dihydroxy-1-deoxy-10-deoxoartemisinin (6): White powder (ethyl acetate). HR-ESI-MS 307.1549 [M + Na]+ (calcd for C15H24O5, 284.1624). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 2.
5α-Hydroxy-1-deoxy-10-deoxoartemisinin (7): White powder (ethyl acetate). HR-ESI-MS 269.1756 [M + H]+ (calcd for C15H24O4, 268.1675). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 2.
7β-Hydroxy-1-deoxy-10-deoxoartemisinin (8): Yellow powder (ethyl acetate). HR-ESI-MS 269.1754 [M + H]+ (calcd for C15H24O4, 268.1675). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 2.
5α,8β-Dihydroxy-artemethin-Ⅱ (9): Colorless, transparent, needle crystals (ethyl acetate). HR-ESI-MS m/z 265.1416 [M + H]+ (calcd for C13H22O4, 242.1518). Crystal data: C13H22O4, M = 242.31, triclinic system, crystal size is 0.38 mm3 × 0.11 mm3 × 0.08 mm3, a = 9.5815 (4) Å, b = 10.7290 (5) Å, c = 13.8292 (3) Å; α = 79.138 (3)°, β = 72.174 (3)°, γ = 75.565 (3)°, V = 1300.94 (9) Å3, space group P1 (NO. 1), T = 111.6 (3) K, Z = 4, μ (Cu Kα) = 0.738 mm−1, R1 = 0.0436 (all data), wR (F2) = 0.1088. Flack parameter: 0.06 (11). Crystallographic data of compound 9 have been deposited to CCDC (www.ccdc.cam.ac.uk/, number = CCDC 2218214). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 4.
3α,7β-Dihydroxy-1-deoxy-10-deoxoartemisinin (10): Colorless, transparent, columnar crystals (ethyl acetate and acetone). HR-ESI-MS m/z 285.1690 [M + Na]+ (calcd for C15H24O5, 284.1624). Crystal data: C15H24O5, M = 283.94, triclinic system, crystal size is 0.46 mm3 × 0.31 mm3 × 0.27 mm3, a = 9.3731 (18) Å, b = 12.574 (3) Å, c = 15.842 (3) Å; α = 85.623 (6)°, β = 81.953 (6)°, γ = 89.302 (7)°, V = 89.302 (7) Å3, space group P1 (NO. 1), T = 273.15 K, Z = 5, Z' = 5 μ (Cu Kα) = 0.095 mm−1, wavelength/Å = 0.71073, R1 = 0.0463 [I > 2σ (I)], wR (F2) = 0.1078. Flack parameter: −0.05 (19). Crystallographic data of compound 10 have been deposited to CCDC (www.ccdc.cam.ac.uk/, number = CCDC 2218210). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 2.
5α-Hydroxy-artemethin-Ⅱ (11) (Gaur et al., 2014): White powder (ethyl acetate). HR-ESI-MS m/z 249.1467 [M + Na]+ (calcd for C13H22O3, 226.1569). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 4.
4α-Hydroxy-1-deoxy-10-deoxoartemisinin (12) (Parshikov et al., 2004b): White powder (ethyl acetate). HR-ESIMS m/z 269.1741 [M + H]+ (calcd for C15H24O4, 268.1675). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 3.
7β-Hydroxy-10-deoxoartemisinin (13) (Parshikov et al., 2004a): White powder (ethyl acetate). HR-ESI-MS 307.1521 [M + Na]+(calcd for C15H24O5, 284.1624). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 3.
4α-Hydroxy-1-deoxydihydroartemisinin (14) (Lee et al., 1990): Colorless, transparent, columnar crystals (ethyl acetate). HR-ESI-MS m/z 307.1526 [M + Na]+ (calcd for C15H24O5, 284.1624). Crystal data: C15H24O5, M = 284.34, monoclinic system, crystal size is 0.25 mm3 × 0.21 mm3 × 0.13 mm3, a = 5.7937 (4) Å, b = 8.5070 (4) Å, c = 29.3066 (17) Å; α = β = γ = 90°, V = 1444.43 (15) Å3, space group P212121 (NO. 19), T = 294.0 K, Z = 4, μ (Cu Kα) = 0.097 mm−1, wavelength/Å = 0.71073, R1 = 0.0585, wR (F2) = 0.1157. Flack parameter: 0.0 (12). Crystallographic data of compound 14 have been deposited to CCDC (www.ccdc.cam.ac.uk/. number = CCDC 2218211). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 3.
5β-Hydroxy-10-deoxoartemisinin (15) (Parshikov et al., 2004b): White powder (ethyl acetate). HR-ESI-MS 307.1519 [M + Na]+ (calcd for C15H24O5, 284.1624). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 3.
6β-Hydroxy-10-deoxoartemisinin (16) (Medeiros et al., 2002): Colorless, transparent, columnar crystals (ethyl acetate). HR-ESI-MS m/z 307.1522 [M + Na]+ (calcd for C15H24O5, 284.1624). Crystal data: C15H24O5, M = 284.34, monoclinic system, crystal size is 0.24 mm3 × 0.21 mm3 × 0.10 mm3, a = 7.1619 (10) Å, b = 12.1265 (17) Å, c = 16.910 (2) Å; α = β = γ = 90.00°, V = 1468.6 (3) Å3, space group P212121 (NO. 19), T = 273.15 K, Z = 4, Z′ = 1, μ (Cu Kα) = 0.095 mm−1, R1 = 0.0469 [I > 2σ (I)], wR (F2) = 0.1142. Flack parameter: −0.2 (4). Crystallographic data of compound 16 have been deposited to CCDC (www.ccdc.cam.ac.uk/, number = CCDC 2218212). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 3.
5α-Acetoxy-artemethin-Ⅱ (17) (Khalifa et al., 1995): Colorless, transparent, oily substance (ethyl acetate). HR-ESI-MS m/z 291.1573 [M + Na]+ (calcd for C15H24O4, 268.1675). 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 4.
7α-Hydroxy-10-deoxoartemisinin (18) (Khalifa et al., 1995): Colorless, transparent, columnar crystals (ethyl acetate). HR-ESI-MS m/z 307.1520 [M + Na]+ (calcd for C15H24O5, 284.1624). Crystal data: C15H24O5, M = 284.34, triclinic system, crystal size is 0.46 mm3 × 0.31 mm3 × 0.27 mm3, a = 10.2388 (17) Å, b = 14.937 (3) Å, c = 9.8826 (17) Å; α = 94.230 (6)°, β = 101.038 (6)°, γ = 90.123 (5)°, V = 1479.2 (4) Å3, space group P1 (NO. 1), T = 293 (2) K, Z = 4, μ (Cu Kα) = 0.095 mm−1, R1 = 0.1021 [I > 2σ (I)], wR (F2) = 0.2594. Flack parameter: −0.3 (15). Crystallographic data of compound 18 have been deposited to CCDC (www.ccdc.cam.ac.uk/, number = CCDC 2218213). The structure of a single crystal is shown in Figure 2. 13C-NMR data are shown in Table 1. 1H-NMR data are shown in Table 3.
2.5 Evaluation of antimalarial activity in vitro
Pf. 3D7 strains were obtained from Professor Chenqijun (Institute of Zoonosis, Jilin University, Jilin, China). Pf. 3D7 strains were grown under a gas mixture (5% CO2, 5% O2, and 90% N2). Human erythrocytes were grown at 2% hematocrit. Synchronization was carried out by treatment with 5% D-sorbitol when most parasites were in the “ring” stage. All compounds were prepared in dimethyl sulfoxide and diluted serially in culture medium (100 µL) across the columns of a 96-well tissue-culture plate. Artemisinin was used as a positive control drug. Then, 100 µL of a parasite suspension (1% ring-infected erythrocytes at 4% hematocrit) was added to each well. The plate was incubated under the gas mixture for 72 h at 37 °C. After incubation, 100 µL of lysis buffer (Tris-Cl (1 M), EDTA (0.5 M), 10% saponin, 0.08% Triton X-100, pH 7.5, SYBR™ Green 1, at the recommended dilution of the manufacturer) was added to each well. The plate was agitated for 1.5 h, and fluorescence was measured at an excitation wavelength of 485 nm and emission wavelength of 530 nm. The half-maximal inhibitory concentration (IC50) was used to evaluate the anti-malarial action of all compounds.
3 Results
3.1 Structural elucidation
The two chemically synthesized derivatives, 10-deoxyartemisinin (2) and 9-ene-10-deoxyartemisinin (3), were obtained following reduction, dehydration, and a second reduction from artemisinin. In addition, 10-deoxyartemisinin (2) was employed as the substrate for microbial transformation with C. elegans CGMCC 3.4832 and C. echinulata CGMCC 3.4879.
Seven new metabolites and eight known metabolites were isolated and characterized unambiguously by various spectroscopy methods (Figure 1). The biotransformation products were identified to be 2α-hydroxy-5α-acetoxy-artemethin-Ⅱ (4), 5α,13-dihydroxy-artemethin-Ⅱ (5), 4α,6β-dihydroxy-1-deoxy-10-deoxoartemisinin (6), 5α-hydroxy-1-deoxy-10-deoxoartemisinin (7), 7β-hydroxy-1-deoxy-10-deoxoartemisinin (8), 5α,8β-dihydroxy-artemethin-Ⅱ (9), and 4α,7β-dihydroxy-1-deoxy-10-deoxoartemisinin (10). The known compounds were determined to be 5α-hydroxy-artemethin-Ⅱ (11), 4α-hydroxy-1-deoxy-10-deoxoartemisinin (12), 7β-hydroxy-10-deoxoartemisinin (13), 4α-hydroxy-1-deoxydihydroartemisinin (14), 5β-hydroxy-10-deoxoartemisinin (15), 6β-hydroxy-10-deoxoartemisinin (16), 5α-acetoxy-artemethin-Ⅱ (17), and 7α-hydroxy-10-deoxoartemisinin (18).
Metabolite 4 had a molecular formula of C15H24O5, as deduced from HR-ESI-MS m/z of 307.1520 [M + Na]+. 13C-NMR data suggested one hydroxy carbon signal (δC 75.7) instead of an alkane carbon signal. The hydroxy group was at C-2 on the basis of the data for 5α-acetoxy-artemethin-II (Khalifa et al., 1995). The structure was confirmed by X-ray crystallography. The structure of a single crystal is shown in Figure 2. Thus, metabolite 4 was identified as 2α-hydroxy-5α-acetoxy-artemethin-Ⅱ.
Metabolite 5 had a molecular formula of C13H22O4, as deduced from its HR-ESI-MS m/z of 265.1415 [M + Na]+. 1H-NMR spectra showed −CH3 (δH 0.95, 3H) to be substituted by −CH2OH (δH 0.94, 2H). 13C-NMR spectra showed that δC 20.5 (C-13) shifted to δC 65.8 compared with 5α-hydroxy-artemethin-II (Gaur et al., 2014). Thus, the hydroxy group was at C-13. The structure was confirmed by X-ray crystallography. The structure of a single crystal is shown in Figure 2. Thus, metabolite 5 was identified as 5α,13-dihydroxy-artemethin-Ⅱ.
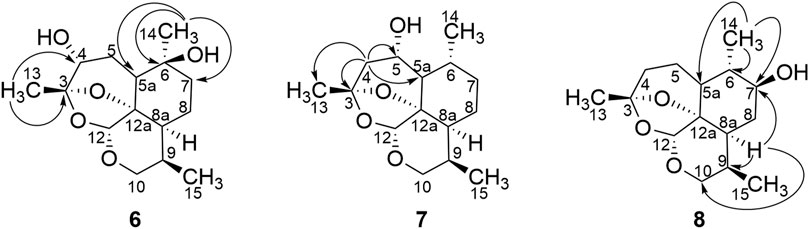
Metabolite 6 had a molecular formula of C15H24O5, as deduced from its HR-ESI-MS m/z of 307.1549 [M + Na]+. 13C-NMR (150 MHz, CDCl3) and distortionless enhancement by polarization transfer (DEPT) spectroscopy showed 15 carbon signals: three methyl, four methylene, five methine, and three quaternary carbon atoms. The low-field shift of C-3 (δC 106.4) together with C-12 (δC 94.9) and C-12a (δC 82.7) compared with those of 2 implied deoxidation of an endoperoxide bridge. Compared with 4α-hydroxy-1-deoxy-10-deoxoartemisinin (Parshikov et al., 2004a), a quaternary carbon signal δC 70.3 was found in place of a tertiary carbon signal. Together with a mass shift of 16 Da, the aforementioned information implied one more hydroxy group than that in 4α-hydroxy-1-deoxy-10-deoxoartemisinin. Heteronuclear multiple-bond coherence (HMBC) correlation from the methoxy proton (δH 1.13, H-14) to the quaternary carbon (δC 70.3, C-6) confirmed a hydroxyl group at C-6. HMBC correlation for metabolite 6 is shown in Figure 3. Thus, metabolite 6 was identified as 4α,6β-dihydroxy-1-deoxy-10-deoxoartemisinin.
Metabolite 7 had a molecular formula of C15H24O4, as determined by HR-ESI-MS at m/z of 269.1756 [M + H]+. 13C-NMR (150 MHz, CDCl3), and DEPT spectroscopy showed three methyl carbon signals (δ 22.7, 20.7, and 15.8), four methylene signals (δ 63.3, 44.3, 34.4, and 23.0), six tertiary carbon signals (δ 95.2, 67.9, 54.1, 39.0, 34.6, and 25.4), and two quaternary carbon signals (δ 105.0 and 81.7). Compared with 4α-hydroxy-1-deoxy-10-deoxoartemisinin (Parshikov et al., 2004b), metabolite 7 was indicated to be a monohydroxy of 1,10-deoxy-artemisinin. 1H-NMR spectroscopy (600 MHz, CDCl3) showed three methyl hydrogen signals: δH 1.47 (s, 3 H, H-13), 1.12 (d, J = 6.5 Hz, 3 H, H-14), and .86 (d, J = 7.4 Hz, 3 H, H-15), which implied that the hydroxyl group may be present at positions 5, 7, or 8. Based on HMBC spectroscopy, δH 2.22 (m, 1H, H-4α) and δH 1.45 (d, 1H, J = 3.5 Hz, H-4β) were remotely correlated to δC 105.0 (C-3), δC 15.8 (C-13), δC 53.1 (C-5a), and δC 67.9, and the hydroxyl group was suggested to be located at C-5. HMBC correlation for metabolite 7 is shown in Figure 3. Given the characterized hydroxylation C-5 of artemisinin, the structure of metabolite 7 was identified as 5α-hydroxy-1-deoxy-10-deoxoartemisinin.
Metabolite 8 had a molecular formula of C15H24O4, as deduced from its HR-ESI-MS m/z of 269.1754 [M + H]+. 13C-NMR (150 MHz, CDCl3), and DEPT spectroscopy showed three methyl signals (δ 22.9, 15.3, and 13.3), four methylene signals (δ 63.3, 33.1, 32.1, and 21.0), six tertiary carbon signals (δ 94.9, 73.8, 42.3, 41.5, 36.8, and 25.1), and two quaternary carbon atoms. Compared with 13C-NMR spectroscopy of 1-deoxy-10-deoxoartemisinin, δC 73.8 was predicted to be a hydroxyl carbon signal. Metabolite 8 was suggested to be hydroxyl 1-deoxy-10-deoxoartemisinin. HMBC spectroscopy showed a correlation from δH 0.96 (d, J = 6.2 Hz, 3H, H-14) to δC 42.3 (C-5a), δC 41.50 (C-6), and δC 73.8, and a correlation from δH 2.02–1.92 (m, 1 H, H-8a) to δC 25.1 (C-9), δC 63.3 (C-10), δC 32.1 (C-8), and δC 73.8. Thus, the hydroxyl group was suggested to be positioned at C-7. HMBC correlation for metabolite 8 is shown in Figure 3. Based on the distinction between 7α (δC 73.5) and 7β (δC 68.7) hydroxylated chemical-shift deviation of artemisinin C-7 (δC 33.6), metabolite 8 was finally identified as 7β-hydroxy-1-deoxy-10-deoxoartemisinin.
Metabolite 9 had a molecular formula of C13H22O4, as deduced from its HR-ESI-MS m/z of 265.1416 [M + H]+. 13C-NMR spectroscopy showed one more hydroxy carbon signal (δC 78.1) than that for 5α-hydroxy-artemethin-II. Combined with its molecular formula, metabolite 9 was predicted to be hydroxylated 5α-hydroxy-artemethin-II. The 1H-NMR spectrum showed a hydrogen signal δH 3.24 (dd, J = 10.3, 5.8 Hz, 1H), which indicated that the hydroxy group was at position C-8. The structure of metabolite 9 was confirmed by X-ray crystallography. The structure of a single crystal is shown in Figure 2. Thus, metabolite 9 was identified as 5α,8β-dihydroxy-artemethin-Ⅱ.
Metabolite 10 had a molecular formula of C15H24O5, as deduced from its HR-ESI-MS m/z of 285.1690 [M + Na]+. 13C-NMR spectroscopy indicated that metabolite 10 was a two hydroxy of 1-deoxy-10-deoxoartemisinin, of which hydroxy carbon signals were at δC 69.2 and δC 74.6. Compared with 4α-hydroxy-1-deoxy-10-deoxoartemisinin, 1H-NMR spectroscopy indicated that the hydroxy groups were located at C-4 and C-7. Finally, the structure of metabolite 10 was confirmed by X-ray crystallography. The structure of a single crystal is shown in Figure 2. Thus, metabolite 10 was identified as 4α,7β-dihydroxy-1-deoxy-10-deoxoartemisinin.
3.2 Antimalarial activity in vitro
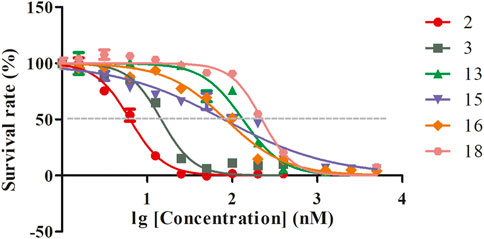
The positive control drug (artemisinin) exhibited in vitro antimalarial activity against Pf. 3D7, with an IC50 (50% inhibition concentration) value of 11 nM. The in vitro antimalarial activity against Pf. 3D7 of compounds 2, 3, 13, 15, 16, and 18 was indicated by IC50 values (nM) of 6, 15, 133, 79, 84, and 223 (Figure 4). The other compounds did not show activity against Pf. 3D7.
4 Discussion
Artemisinin is an unusual sesquiterpene lactone possessing an endoperoxide moiety. The sesquiterpene lactone endoperoxide structure of artemisinin is the pharmacophore characteristic element. The partial resistance of artemisinin refers to a delay in the clearance of malaria parasites from the bloodstream following treatment with ACT. This class of compounds with parent nucleus structures is characterized by a relatively complex metabolic process in vivo, which was considered to be partly responsible for the drug resistance phenomenon by some perspectives. Exploring the associated hydroxylation derivatives, especially hydroxylation, which is highly similar to the in vivo disposal process of artemisinin, is an effective approach to discover novel agents with higher activities.
Structural modification of artemisinin to improve its solubility, stability, and bioavailability has been a research “hotspot” in medicinal chemistry since its discovery. In recent years, the need for novel antimalarial drugs to prevent unpredictable drug tolerance has been urgent. 10-Deoxyartemisinin was first synthesized by Jung in 1989, and it was shown to be more stable in gastric acid and more efficacious than artemisinin. The synthesis of 10-deoxyartemisinin has been previously examined in detail, and in this work the reaction conditions were optimized, including temperature, duration, and solvent. As a result, a direct one-step reduction of the carbonyl function of artemisinin into 10-deoxyartemisinin was successfully achieved by NaBH4 in the presence of BF3/Et2O, with a yield of 50%. Moreover, 9-ene-10-deoxyartemisinin was isolated as a by-product, and it demonstrated similar antimalarial activity to that of artemisinin. Subsequently, 10-deoxyartemisinin was chosen as the substrate for microbial transformation.
Cunninghamella species are commonly used models for microbial transformation. Numerous research studies have confirmed that Cunninghamella functioned with outstanding hydroxylation ability, which was responsible for the cytochrome P450 activity of fungus (Wang et al., 2000; Asha and Vidyavathi, 2009). In addition, some research studies found that the cytochrome P450 in Cunninghamella species belongs to the CYP5I family, and the role of these enzymes was confirmed as the function of CYP3A4 enzyme in mammal metabolism (Dube et al., 2016). Thus, Cunninghamella was also employed to transform xenobiotics for the simulation of phase I (oxidative) and phase II (conjugative) metabolism (Zhang et al., 1996). In this work, two strains were chosen based on our previous study on the biotransformation of artemisinin (Ma et al., 2019) and dihydroartemisinin (unpublished data). The activity evaluation of the microbial transformation products showed that the antimalarial activity of the C-5 hydroxylated product was better than others. Though metabolites 13 and 18 were both hydroxylated products at the C-7 position, the antimalarial activity of the β-OH product exhibited two times better efficiency than the α-OH product. Unfortunately, hydroxylated products at C-5, C-6, and C-7 positions of 10-deoxyartemisinin led to attenuated antimalarial activities. Nevertheless, hydroxylation could improve the solubility of the compound, while also providing the possibility for further functionalization. Derivatives with a reduced peroxide bridge exhibited negligible antimalarial activity, a finding that is in accordance with previous reports (O’Neill et al., 2010). The synergistic effect of these products on malarial treatment and other bioactivities needs to be further studied.
5 Conclusion
In summary, seventeen artemisinin derivatives, including seven novel compounds (4–10) and ten known compounds, were isolated and identified through combined chemical and biological transformation. This protocol provided a highly efficient and divergent translation strategy for artemisinin. The pharmacological activities of the generated products were evaluated, and some derivatives displayed good antimalarial activity, which inspired us to conduct a comprehensive druggability study. The novel products with divergent structural moieties provided promising candidates for further bio-evaluation and drug development.
Data availability statement
The data presented in the study are deposited in the article/Supplementary Material. The crystallographic data presented in the study are deposited in www.ccdc.cam.ac.uk/, accession numbers CCDC 2218207 (M4), 2218208 (M5), 2218214 (M9), 2218210 (10), 2218211 (14), 2218212 (M16), 2218213 (M18).
Author contributions
Conceptualization and methodology: YM; investigation and original draft preparation: XG and YB; data analysis: DZ; software: YZ; writing—review: PS and HG; and funding acquisition: LY. All authors read and agreed to the published version of the manuscript.
Funding
This research was funded by the Beijing Natural Science Foundation (No. 7214292, China), the National Natural Science Foundation of China (Nos. 82104516 and 82141004, China), the Fundamental Research Funds for the Central Public Welfare Research Institutes of China Academy of Chinese Medical Sciences (Nos. ZZ14-YQ-054 and ZZ13-YQ-098), and the Innovation Project of China Academy of Chinese Medical Sciences (CI2021A05102 and CI2021A04002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.1089290/full#supplementary-material
References
Asha, S., and Vidyavathi, M. (2009). Cunninghamella-a microbial model for drug metabolism studies-a review. Biotechnol. Adv. 27 (1), 16–29. doi:10.1016/j.biotechadv.2008.07.005
Bai, Y., Zhang, D., Sun, P., Zhao, Y. F., Chang, X. Q., Ma, Y., et al. (2019). Evaluation of microbial transformation of 10-Deoxoartemisinin by UPLC-ESI-Q-TOF-MSE. Molecules 24 (21), 3874. doi:10.3390/molecules24213874
Bai, Y., Zhao, Y. F., Gao, X. N., Zhang, D., Ma, Y., Yang, L., et al. (2021). A novel antimalarial metabolite in erythrocyte from the hydroxylation of dihydroartemisinin by Cunninghamella elegans. Front. Chem. 10, 850133. doi:10.3389/fchem.2022.850133
Cao, H., Chen, X., Jassbi, A. R., Xiao, J., and Kumar, M. S. (2015). Microbial biotransformation of bioactive flavonoids Biotransformation of bromhexine by Cunninghamella elegans, C. echinulata and C. blakesleeana. Biotechnol. Adv.Braz. J. Microbiol. 3348 (12), 214259–223267. doi:10.1016/j.bjm.2016.11.003
Dube, A. K., and Kumar, M. S. (2016). Biotransformation of bromhexine by Cunninghamella elegans, C. echinulata and C. blakesleeana. Braz. J. Microbiol. 48 (2), 259–267. doi:10.1016/j.bjm.2016.11.003
Gaur, R., Patel, S., Verma, R. K., Mathur, A., and Bhakuni, R. S. (2014). Biotransformation of Artemisinin derivatives by glycyrrhiza glabra, lavandula officinalis, and panax quinquefolium. Med. Chem. Res. 23, 1202–1206. doi:10.1007/s00044-013-0726-x
Goswami, A., Saikia, P., Barua, N., Bordoloi, M., Yadav, A., Bora, T., et al. (2010). Biotransformation of artemisinin using soil microbe: Direct C-acetoxylation of artemisinin at C-9 by Penicillium simplissimum. Bioorg. Med. Chem. Lett. 20, 359–361. doi:10.1016/j.bmcl.2009.10.097
Jung, M., Li, X., Bustos, D. A., ElSohly, H. N., McChesney, J. D., and Milhous, W. K. (1990). Synthesis and antimalarial activity of (+)- Deoxoartemisinin. J. Med. Chem. 33, 1516–1518. doi:10.1021/jm00167a036
Khalifa, S. I., Baker, J. K., Jung, M., Mcchesney, J. D., and Hufford, C. D. (1995). Microbial and mammalian metabolism studies on the semisynthetic antimalarial, Deoxoartemisinin. Pharm. Res. 12, 1493–1498. doi:10.1023/a:1016239505506
Lee, I. S., Elsohly, H. N., and Hufford, C. D. (1990). Microbial metabolism studies of the antimalarial drug arteether. Pharm. Res. 7, 199–203. doi:10.1023/a:1015845306124
Liu, J. H., Chen, Y. G., Yu, B. Y., and Chen, Y. J. (2006). A novel ketone derivative of Artemisinin biotransformed by Streptomyces griseus ATCC 13273. Bioorg. Med. Chem. Lett. 16, 1909–1912. doi:10.1016/j.bmcl.2005.12.076
Luo, J., Mobley, R., Woodfine, S., Drijfhout, F., Horrocks, P., Ren, X. D., et al. (2022). Biotransformation of Artemisinin to a novel derivative via ring rearrangement by Aspergillus niger. Appl. Microbiol. Biot. 106 (7), 2433–2444. doi:10.1007/s00253-022-11888-0
Ma, Y., Sun, P., Zhao, Y., Wang, K., Chang, X., Bai, Y., et al. (2019). A microbial transformation model for simulating mammal metabolism of artemisinin. Molecules 24 (2), 315. doi:10.3390/molecules24020315
Medeiros, S. F., Avery, M. A., Avery, B., Leite, S., and Williamson, J. S. (2002). Biotransformation of 10-deoxoartemisinin to its 7β-hydroxy derivative by Mucor ramannianus. Biotechnol. Lett. 24, 937–941. doi:10.1023/A:1015516929682
O’Neill, P., Barton, V., and Ward, S. (2010). The molecular mechanism of action of artemisinin-the debate continues. Molecules 15, 1705–1721. doi:10.3390/molecules15031705
Parshikov, I. A., Muraleedharan, K. M., Avery, M. A., and Williamson, J. S. (2004a). Transformation of artemisinin by Cunninghamella elegans. Appl. Microbiol. Biot. 64 (6), 782–786. doi:10.1007/s00253-003-1524-z
Parshikov, I. A., Muraleedharan, K. M., Miriyala, B., Avery, M. A., and Williamson, J. S. (2004b). Hydroxylation of 10-deoxoartemisinin by Cunninghamella elegans. Nat. Prod. 67, 1595–1597. doi:10.1021/np040089c
Ponnapalli, M. G., Suea, M. B., Sudhakar, R., Govindarajalu, G., and Sijwali, P. S. (2018). Biotransformation of artemisinin to 14-hydroxydeoxyartemisinin: C-14 hydroxylation by Aspergillus flavus. J. Agric. Food. Chem. 66 (40), 10490–10495. doi:10.1021/acs.jafc.8b03573
Tu, Y. Y., Ni, M. Y., Zhong, Y. R., Li, L. N., Cui, S. L., Zhang, M. Q., et al. (1981). Studies on the constituents of Artemisia annua L. Acta. Pharm. Sin. B 16 (5), 366. doi:10.16438/j.0513-4870.1981.05.008
Uwimana, A., Legrand, E., Stokes, B. H., Ndikumana, J. L. M., Warsame, M., Umulisa, N., et al. (2020). Emergence and clonal expansion of in vitro Artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 26 (10), 1602–1608. doi:10.1038/s41591-020-1005-2
Wang, J. G., Xu, C., Liao, F. L., Jiang, T., Krishna, S., and Tu, Y. (2019). A temporizing solution to "artemisinin resistance. New. Engl. J. Med. 380 (22), 2087–2089. doi:10.1056/NEJMp1901233
Wang, R., Cao, W., Khan, A., Khan, A., and Cerniglia, C. (2000). Cloning, sequencing, and expression in Escherichia coli of a cytochrome P450 gene from Cunninghamella elegans. Fems. Microbiol. Lett. 188, 55–61. doi:10.1111/j.1574-6968.2000.tb09168.x
World Health Organization (2021). World malaria report 2021, Available at: https://apps.who.int/iris/handle/10665/350147.
Xie, X. T., Zheng, P., Zhang, G. Y., and Yang, M. (2001). Isolation and identification of new components in mother liquor of synthetic Dihydroartemisinin methyl ether. Chin. Traditional Herb. Drugs 32, 388.
Zhang, D., Yang, Y., Leakey, J., and Cerniglia, C. (1996). Phase I and phase II enzymes produced by Cunninghamella elegans for the metabolism of xenobiotics. Fems. Microbiol. Lett. 138, 221–226. doi:10.1111/j.1574-6968.1996.tb08161.x
Keywords: artemisinin, microbial transformation, Cunninghamella genus, anti-malarial activity, bioactive metabolites
Citation: Gao X, Bai Y, Sun P, Gao H, Yang L, Zhang D, Zhao Y and Ma Y (2023) Combined chemical transformation and biological transformation of artemisinin: A facile approach to diverse artemisinin derivatives. Front. Chem. 10:1089290. doi: 10.3389/fchem.2022.1089290
Received: 04 November 2022; Accepted: 31 December 2022;
Published: 24 January 2023.
Edited by:
Jawad Nasim, Saarland University, GermanyReviewed by:
Caijuan Zheng, Hainan Normal University, ChinaClemens Zwergel, Sapienza University of Rome, Italy
Copyright © 2023 Gao, Bai, Sun, Gao, Yang, Zhang, Zhao and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Ma, eW1hQGljbW0uYWMuY24=
 Xinna Gao
Xinna Gao Yue Bai
Yue Bai Peng Sun
Peng Sun Huimin Gao
Huimin Gao Lan Yang
Lan Yang Dong Zhang
Dong Zhang Yifan Zhao
Yifan Zhao Yue Ma
Yue Ma