- 1Academy for Advanced Interdisciplinary Studies, Peking University, Beijing, China
- 2State Key Laboratory of North China Crop Improvement and Regulation, College of Plant Protection, Hebei Agricultural University, Baoding, China
- 3State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 4Guangdong Laboratory for Lingnan Modern Agriculture (Shenzhen Branch), Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
Plant-parasitic nematodes are a main limiting factor for worldwide agriculture. To reduce the global burden of nematode infections, chemical nematicides are still the most effective methods to manage nematodes. With the increasing resistance of nematodes, the development of new anti-nematicides drug is urgent. Nematode chitinases are found to play important roles in various physiological functions, such as larva moulting, hatching from eggshell, and host infection. Inhibition of nematode chitinase is considered a promising strategy for the development of eco-friendly nematicides. In this study, to develop novel nematode chitinase CeCht1 inhibitors, virtual screening of the ZINC database was performed using the pesticide-likeness rules, pharmacophore-based and docking-based approach in turn. Compounds HAU-4 and HAU-7 were identified as potent CeCht1 inhibitors with the IC50 values of 4.2 μM and 10.0 μM, respectively. Moreover, molecular dynamics simulations combined with binding free energy and free energy decomposition calculations were conducted to investigate the basis for the potency of the two inhibitors toward CeCht1. This work gives an insight into the future rational development of novel and potent nematode chitinase inhibitors.
Introduction
Nematodes occur as parasites in humans, animals and plants or as free-living forms in soil, fresh water, and marine environments (Traunspurger, 2000). Among them, plant-parasitic nematodes (PPNs) represent a significant limiting factor for global agriculture and cause up to $157 billion in economic losses worldwide each year (Abad et al., 2008; Chen J. et al., 2020). The most efficient approach of controlling PPNs is considered to be the use of chemical nematicides (Albonico et al., 1999; Ntalli et al., 2012). However, due to the widespread use of some traditional nematicides, the problem of nematode resistance has become increasingly serious (Abebew et al., 2022; Vanegas et al., 2022). Therefore, the development of green pesticides for novel nematode targets is of great significance for the control of plant-parasitic nematodes.
Chitin, a linear homopolymer of N-acetyl-D-glucosamine, is known to exist in the eggshell, microfilarial sheath, and pharynx of nematodes, but not in higher plants and mammals (Adam et al., 1996; Fanelli et al., 2005; Veronico et al., 2001; Zhang et al., 2005). The chitinase that catalyzes the degradation of nematode chitin is found to play an important role in various physiological functions, including larva moulting, hatching from eggshell, and host infection (Chen W. et al., 2020; Gao et al., 2002; Maeda et al., 2001). For example, using RNAi to silence the expression of Caenorhabditis elegans chitinase (CeCht1) led to hatching failure and eventual nematode death (Maeda et al., 2001). Down-regulating the expression level of Acanthocheilonema viteae chitinase (AvCht1) can affect nematode egg hatching and larval molting (Tachu et al., 2008). Downregulation of Bursaphelenchus xylophilus chitinase (BxCht1) led to hatching delay and spawning decrease (Ju et al., 2016). Accordingly, nematode chitinase may serve as a potential target for the development of novel nematicides (Chen W. et al., 2021; Chen Q. et al., 2021).
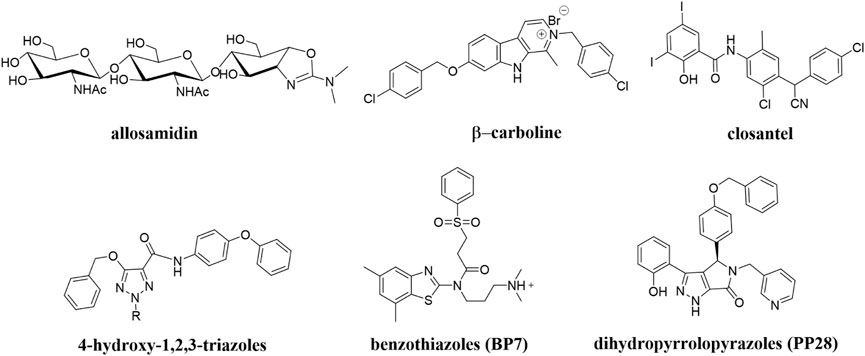
To date, a large number of chitinase inhibitors have been reported, and most of which show excellent application prospects in drug, insecticide, and fungicide (Chen J. et al., 2020; Gloeckner et al., 2010; Jiang et al., 2020). However, the development of chitinase inhibitors for nematicides is rarely mentioned. Only few nematode chitinase inhibitors have been reported, including allosamidin (Arnold et al., 1993), β-carboline (Gooyit et al., 2015), closantel (Gloeckner et al., 2010), 4-hydroxy-1,2,3-triazoles (Pippione et al., 2015), benzothiazoles (BP) (Chen Q. et al., 2021), and dihydropyrrolopyrazoles (PP) (Chen W. et al., 2021) (Figure 1). Among these inhibitors, allosamidin, carbolines, closantel, and 4-hydroxy-1,2,3-triazoles were reported to have good inhibitory effect on animal parasitic nematodes (e.g., Heligmosomoides polygyrus, Brugia malayi, Onchocerca volvulus). Benzothiazoles (BP) and dihydropyrrolopyrazole (PP) were recently found by Yang’s group to have excellent inhibitory potency against the model nematode Caenorhabditis elegans. Furthermore, the crystal structures of Caenorhabditis elegans chitinase CeCht1 and CeCht1-inhibitor complexes were obtained by Yang’s group (Chen W. et al., 2021; Chen Q. et al., 2021). The results showed that CeCht1 consists of a signal peptide, a catalytic domain, and two chitin-binding modules. The catalytic signature motif of CeCht1 is located in the loop between β4 and α4, and the key catalytic residues is composed of Asp175, Asp177, and Glu179 (Chen Q. et al., 2021). These crystallographic investigations on CeCht1 lay the solid foundation for the discovery of novel nematode chitinase inhibitors via structure-based virtual screening.
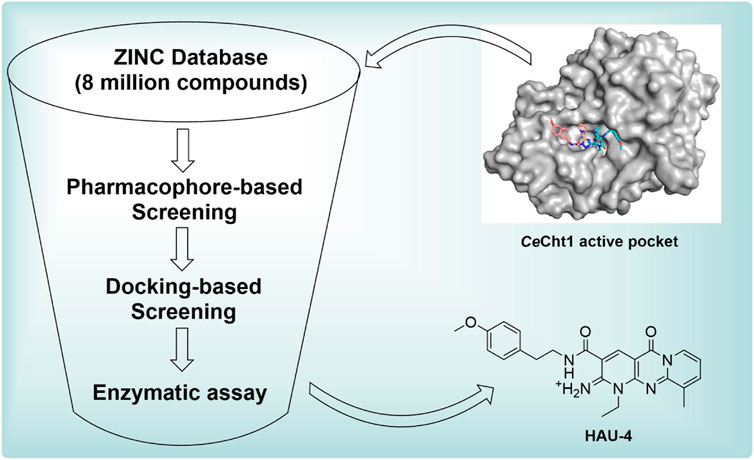
In silico virtual screening has been rapidly developed as a reliable, timesaving strategy to obtain novel inhibitors that target a given protein of interest (Dong et al., 2019; Gorgulla et al., 2020). In order to acquire more potent nematode chitinase inhibitors with novel chemical scaffolds, especially their application against plant-parasitic nematicides, the virtual screening targeting CeCht1 was carried out using both pharmacophore-based and docking-based methods in this study. In the subsequent steps, 15 compounds were selected for further enzymatic assay, and a molecule named 1-ethyl-3-((4-methoxyphenethyl) carbamoyl)- 10-methyl- 5-oxo- 1,5-dihydro- 2H-dipyrido [1,2-a:2′,3′-d] pyrimidin-2-iminium was identified as a novel type of CeCht1 inhibitor. Moreover, to analyze the possible inhibitory mechanisms of potent inhibitors with CeCht1, MD simulations combined with binding free energy and free energy decomposition calculations were performed. The results obtained in this work provide important insights into the future development of novel and potent plant-parasitic nematodes chitinase inhibitors.
Results and discussion
Pharmacophore-based virtual screening
To identify novel and potent nematode chitinase inhibitors, in this study, a total of 8 million molecules from ZINC library were used for virtual screening. Firstly, in order to improve the screening efficiency, the pesticide-likeness rules (Hao et al., 2011) were used to reasonably reduce the number of compounds and this yielded 3176361 molecules. Then, the reported co-crystal structure of CeCht1 with PP7 (PDB ID: 6LE8) (Chen W. et al., 2021) was selected as the template to generate the pharmacophore model in MOE. We focused on the H-bonding and hydrophobic interactions between ligand and residues Glu179, Trp138, and Trp394 at −1 and +1 subsists of CeCht1. As a result, a pharmacophore model was constructed with the features of a hydrogen bond donor at Glu179, aromatic center at Trp394, a hydrogen-bond acceptor and a hydrophobic center at Trp138 (Figure 2). Based on this pharmacophore model, 1376261 compounds were obtained.
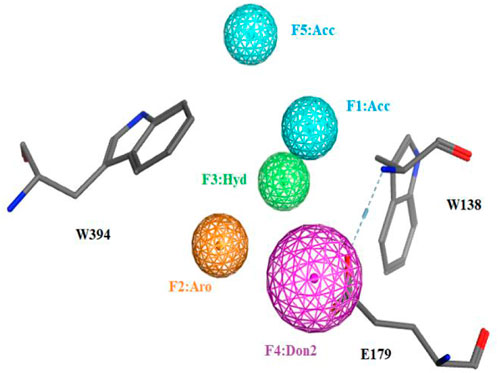
FIGURE 2. The pharmacophore model used for the virtual screening of CeCht1 inhibtors. Definitions: Don2, hydrogen-bond donor projection; Acc, hydrogen-bond acceptor; Aro, aromatic center; Hyd, hydrophobic centroid.
Docking-based virtual screening
To further reduce the number of the hits, these molecules were refined with docking-based virtual screening. The virtual screening was performed using the MOE modeling software, and the crystal structure of CeCht1 complexed with PP7 was selected as the template for molecular docking. The obtained poses were rank-ordered according to their binding scores (a higher negative score indicated a higher stable interaction). As a result, the top 500 compounds with score ranging from −9.75 to −9.00 were selected. These hits were further refined with visual inspection. Considering the interactions between CeCht1 and the corresponding inhibitors revealed from crystal complexes, only hits that mainly interacted with residues Trp138, Asp177, Asp179, Tyr247, Asp248 and Trp394 were considered. Finally, 15 representative compounds were purchased from Topscience Corporation and were used for biological activity assays. Structures, docking scores of the compounds are listed in Supplementary Table S1. The corresponding sequence of these steps is summarized in Figure 3.
Enzyme inhibition evaluation
After the 15 potential compounds were purchased, the corresponding inhibitory activities against CeCht1 were assayed at a concentration of 100 μM. As shown in Supplementary Table S1, compounds HAU-4, HAU-7, HAU-9, and HAU-11 exhibited good inhibitory potency against CeCht1 with inhibition rates >75% at 100 μM. Two compounds (namely, HAU-1 and HAU-3) showed moderate efficiency against CeCht1 with inhibition rates >50% at 100 μM. The remaining compounds showed relatively weak efficiency (inhibition rate <50% at 100 μM). These results preliminarily indicate that our virtual screening strategy is effective.
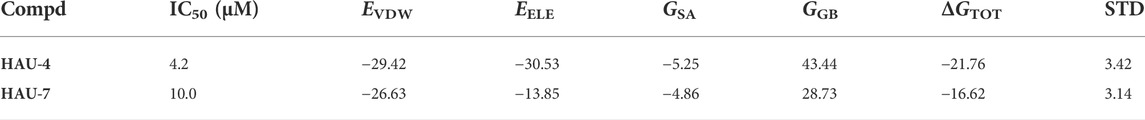
Seven compounds with inhibition rates >50% at the concentration of 100 μM were further assayed for their inhibitory activities. As shown in Table 1, compounds HAU-4 (ZINC09610803) and HAU-7 (ZINC12704597) exhibited excellent potency against CeCht1 with IC50 values of 4.2 μM and 10.0 μM, respectively (Supplementary Figure S1). The remaining compounds displayed inhibition rate <50% at 10 μM against CeCht1. Therefore, compounds HAU-4 and HAU-7 can be used as the novel nematode chitinase inhibitors, and the study of their inhibition mechanisms is of great significance for guiding the development of new nematicides.
Inhibitory mechanisms of HAU-4 and HAU-7 against CeCht1
To investigate the basis for the potency of compounds HAU-4 and HAU-7 toward CeCht1, the molecular docking studies were carried out. As shown in Supplementary Figure S2A,B,C, compounds HAU-4 and HAU-7 were all well-anchored in the substrate-binding pocket of CeCht1 at −1, +1, and +2 subsites, and stabilized by hydrophobic interactions and hydrogen bonds. Specifically, the dipyrido-pyrimidine moiety of HAU-4 was found to bind to the -1 and +1 subsites of CeCht1, and formed H-bonding interactions with residues Trp138 and Asp248 (Supplementary Figure S2B). The p-methoxyphenyl fragment of HAU-4 was located at the +2 subsite of the active pocket entrance consisting of Trp253 (Supplementary Figure S2B). For compound HAU-7, the pyrrole ring was shown to be inserted into the -1 subsite of CeCht1 via π-π stacking interactions with Trp394, and the amide linker can form H-bonding interactions with residues Trp138 and Asp248 (Supplementary Figure S2C). In addition, the naphthalene ring of HAU-7 extends to the +2 subsite (Supplementary Figure S2C).
To shed further light on the appropriate binding modes of compounds HAU-4 and HAU-7 with CeCht1 and in an effort to assay the stabilities of the potential compounds in the active pocket, 40 ns MD simulations were performed (Figure 4). As shown in Figure 4A, the two simulated systems all achieved dynamic equilibrium at approximately 25 ns and their root-mean-square deviation (RMSD) values ultimately maintained at 1.7 and 1.5 Å for protein-ligand complexes of compounds HAU-4 and HAU-7, respectively. These results indicated that these two systems underwent reasonable conformational changes.
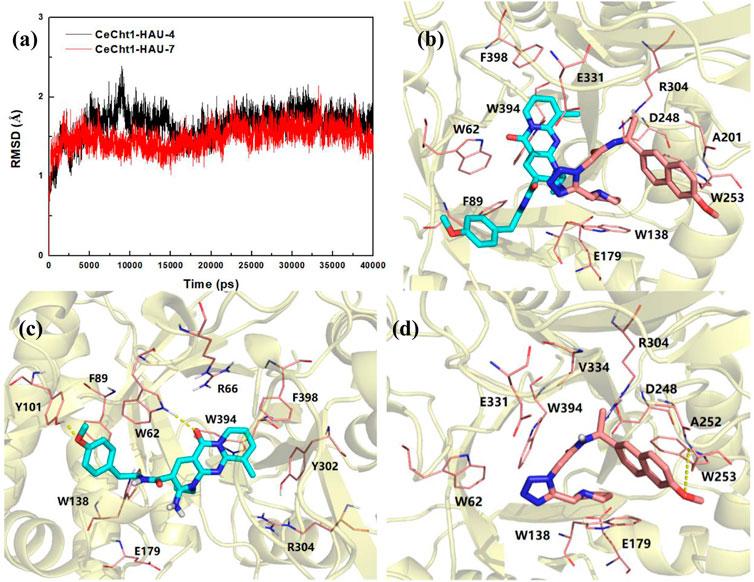
FIGURE 4. (A) RMSD changes of compounds HAU-4 and HAU-7 in complex with CeCht1. (B) Superimposition of the conformations of HAU-4 and HAU-7 in CeCht1 pocket at 40 ns MD simulations. Specific binding conformations of (C) CeCht1-HAU-4 and (D) CeCht1-HAU-7 systems revealed by MD simulations. Compound HAU-4 is shown in cyan; HAU-7 is shown in pink. Colored according to element.
Superimposition of the conformations of HAU-4 and HAU-7 with CeCht1 at 40 ns of MD simulations are shown in Figure 4B and Supplementary Figure S2D. Compound HAU-7 was found to be buried at the entrance of the CeCht1 active pocket, and mainly binds to the +1 and +2 subsites of CeCht1. In contrast, compound HAU-4 was found to enter deeper into the active pocket, and the dipyrido-pyrimidine ring was inserted into the -1 subsite of CeCht1 (Figure 4B and Supplementary Figure S2D).
The specific binding mode of HAU-4 with CeCht1 revealed by MD simulations is shown in Figure 4C. In comparison to the docking conformation (Supplementary Figure S2B), the dipyrido-pyrimidine ring of HAU-4 was demonstrated to move deeper into the CeCht1 pocket via 40 ns MD simulations, and the p-methoxyphenyl moiety was far from the +1 and +2 subsites (Supplementary Figure S2E). In detail, the dipyrido-pyrimidine moiety of HAU-4 stacked well with Trp394, Phe398, Tyr302, and Trp62 at −1 subsite. The oxygen atom on dipyrido-pyrimidine ring formed a hydrogen bond with Trp62. On the other hand, the p-methoxyphenyl fragment of HAU-4 bound in a small hydrophobic cave constructed by residues Tyr101, Phe89, Trp138, and it was further stabilized by forming a hydrogen bond with Tyr101. Furthermore, we found that the ethyl group on the dipyrido-pyrimidine points to the position of the catalytic residues Asp175, Asp177, and Glu179 (Chen Q. et al., 2021). This may suggest the introduction of suitable substituent (such as fragment with hydrogen bond donor) in the ethyl of HAU-4 may lead to increased activity. Thus, this binding pattern provided a clear molecular basis for the potent inhibitory activity of compound HAU-4 (IC50 = 4.2 μM) against CeCht1.
The conformations of compound HAU-7 before and after MD simulations in CeCht1 pocket are superimposed and shown in Supplementary Figure S2F. Compound HAU-7 was found to exhibit a great deal of folding and move to the entrance of the pocket relative to the conformation before MD simulations (Figure 4D, Supplementary Figure S2C,F). The specific binding mode of HAU-7 with CeCht1 revealed by MD simulations showed that the pyrrole moiety anchored in the hydrophobic pocket and formed π-π stacking interactions with Trp138, while the naphthalene ring interacted with Ala252 and Trp253 at +2 subsite of CeCht1. It is worth noting that HAU-7 could only form one hydrogen bond with Trp253, with reduced interactions in its docking mode (Figure 4D and Supplementary Figure S2C). Therefore, the affinity between HAU-7 with CeCht1 was determined to be relatively weak, resulting in a moderate inhibitory potency against CeCht1 (IC50 = 10.0 μM).
Binding free energy calculation
To further explore the cause of the inhibitory potency against CeCht1, binding free energy analyses of HAU-4 and HAU-7 were conducted by using MM/GBSA calculation methods. The predicted binding affinities of these complexes are summarized in Table 2. The results showed that the binding free energy (ΔGTOT) were in agreement with the experimental values of HAU-4 and HAU-7. Furthermore, both of the electrostatic (EELE) and the van der Waals (EVDW) contributions are essential for the two compounds when bound to CeCht1. For compound HAU-4, the electrostatic energy (EELE = −30.53 kcal/mol) was lower than its van der Waals energy (EVDM = −29.42 kcal/mol). On the contrary, the van der Waals interactions (EVDM = −26.63) of HAU-7 acted a more important role than the electrostatic interaction kcal/mol (EELE = −13.83 kcal/mol). Moreover, the nonpolar interactions (EVDM + GSA) of these two complexes were lower than the polar interactions (EELE + GGB), which suggested that the nonpolar interactions in these two systems were more beneficial for the binding of HAU-4 and HAU-7 to CeCht1.
Decomposition free energy calculation
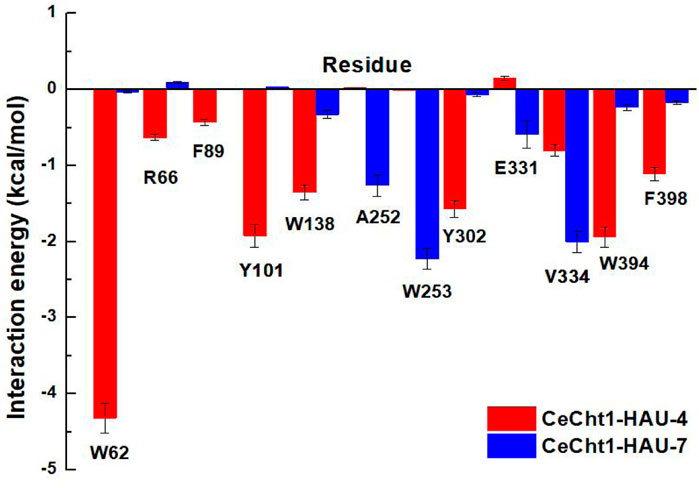
Free energy decomposition calculations were further performed by using MM/GBSA methods to investigate the detailed contributions of residues in the two systems. Residues near 5 Å of the ligands were selected to compute the decomposition free energy, and the main residue contributions are shown in Figure 5. Compound HAU-4 formed the strongest interaction with Trp62 (−4.32 kcal/mol) and formed moderate interactions with Trp394, Tyr101, Tyr302, and Trp138 (−1.94, −1.92, −1.57, and −1.35 kcal mol−1, respectively). This suggested that the hydrophobic interactions and hydrogen-bonding interactions at −1 subsite were the main reasons for the higher inhibitory potency of compound HAU-4. Compound HAU-7 mainly interacted with Trp253, Val334, and Ala252 at the entrance of the active pocket. This result further demonstrated the shallow binding mode of HAU-7 in the CeCht1 active pocket. It is noteworthy that neither compound bound to the catalytic residue Asp175, Asp177, and Glu179 (Chen Q. et al., 2021), illustrating that the efficiency of compounds HAU-4 and HAU-7 is in the micromolar range.
Conclusion
In conclusion, in order to develop novel nematicides with new mechanisms of action, in this study, we conducted pharmacophore-based and docking-based virtual screening methodology to identify novel and potent inhibitors targeting the model nematode chitinase CeCht1. As a result, 15 representative compounds from the ZINC database were selected for enzymatic evaluation, and HAU-4 and HAU-7 demonstrated the higher potency against CeCht1 with the IC50 values of 4.2 μM and 10.0 μM, respectively. Furthermore, MD simulations combined with binding free energy and free energy decomposition calculations of these two inhibitors with CeCht1 were carried out to investigate the basis for their potency against CeCht1. These computational study results also suggest that introducing a suitable substituent (such as fragment with hydrogen bond donor) on the ethyl group of dipyrido-pyrimidine ring of HAU-4 may result in additional bindings with catalytic residues of CeCht1 and ultimately improve the inhibitory activity. Taken in concert, the combined theoretical and experimental results in this manuscript may provide a new direction for the future discovery of potent nematodes chitinase inhibitors.
Materials and methods
Preparation of the Zinc database
The ZINC database containing 8 million molecules was used for our virtual screening targeting nematode chitinase CeCht1. Initially, the descriptors of all the molecules in the ZINC library were calculated via the Descriptor module in MOE software (version 2016). Secondly, these molecules were filtered based on “Hao’s pesticide-likeness” rules (Hao et al., 2011), which are MW < 435 Da, ClogP <6, HBA <6, HBD <2, ROB <9, ARB <17. In addition, the synthetic feasibility score in MOE of these compounds was considered. The synthetic feasibility score is the fraction of atoms of each new structure that ultimately appear in a retrosynthetic fragment, and a value of one means the molecule is likely to be synthesizable. Based on these criteria, the unsuitable molecules were gradually filtered out.
Pharmacophore-based screening
The complex crystal structure of CeCht1 (PDB ID: 6LE8) (Chen W. et al., 2021) was selected as the template to generate a pharmacophore model. The protein ligand interaction fingerprints (PLIF) tool in MOE was used to establish a pharmacophore model. The pharmacophore features take the spatial position and interactions between ligands and proteins into account, including hydrogen bond donor and acceptor, and hydrophobic interaction characteristics. Then, virtual screening module in MOE was used to perform the pharmacophore-based screening.
Docking-based screening
Molecular docking was carried out by using the Dock module in MOE (Dong et al., 2021). The complex crystal structure of CeCht1 with PP7 (PDB ID: 6LE8) (Chen W. et al., 2021) was used as the template for the docking-based screening, and the position of the ligand was defined as the docking site. First, the Structure Preparation module in MOE was used to prepare the protein structure. The typical steps in this process included the geometry and electron-density checks, addition of hydrogen atoms, optimization of their position, and energy minimization. Then, the Triangle Matcher placement method and Rigid Receptor post-placement refinement method were used to generate the poses of ligand. The number of poses returned by each ligand placement were set to a default value of 30. The London dG scoring function was used to estimate their free energy of binding. The other parameters were set default values. The molecules were ranked lowest to highest according to their scores, and the pose with the lowest score was retained.
Enzymatic assays
Nematode chitinase CeCht1 (CeCht1-CAD) was overexpressed in Pichia pastoris GS115 (Invitrogen, Carlsbad, CA) and then purified as described previously (Chen Q. et al., 2021).
The target compounds were assayed for their inhibitory activities against CeCht1 in end-point experiments using 4-methylumbelliferyl -N, N′-diacetyl-β-D-chitobioside (4-MU-(GlcNAc)2, Sigma, St. Louis, MO) as the substrate. The CeCht1 was assayed in 20 mM sodium phosphate buffer (pH 6.0). In a final volume of 100 μl, the reaction mixture containing buffer, DMSO (1% (v/v)), CeCht1-CAD (10 nM), 4-MU- (GlcNAc)2 (4 μM) and inhibitor was incubated at 25°C for 20 min. Then, the reaction was stopped by the addition of sodium carbonate solution (0.5 M, 100 μl). The fluorescence of the liberated 4-methylumbelliferone was quantitated at an excitation of 366 nm and emission of 440 nm. Experiments were performed in triplicate. For determination of the IC50 values, the inhibitory activity of the compounds against CeCht1 was monitored by changing the concentrations of inhibitors.
Molecular dynamics simulations
Molecular Dynamics (MD) simulations were carried out to assess the validity of the binding interactions of the selected compounds and CeCht1 by using AMBER14 package. The ligands and protein were selected for GAFF force field (Duan et al., 2003) and AMBER03 force field (Wang et al., 2004), respectively. The protein-inhibitor system was immersed in a radius truncated octahedral box with TIP3P water molecules extended 10 Å from the complex, and the system was neutralized by adding counterions (Na+). Then, two-step energy minimization was performed using the first 2,500 steps steepest-descent and last 2,500 steps conjugated gradient algorithms in Sander module. Next, the system was gradually heated from 0 to 300 K in the NVT ensemble and the temperature was maintained at 300 K during 100 ps with a restrain force constant of 5 kcal/mol/Å2. The SHAKE algorithm was used to constrain the hydrogen atoms (Ryckaert et al., 1977), and the particle mesh Ewald algorithm was applied to calculate the long-range electrostatic interactions with default cutoff of 8.0 Å under periodic boundary conditions (Darden et al., 1993). Finally, 40 ns MD simulations were carried out at the constant temperature and pressure (300 K and 105 Pa) using the PMEMD module in the AMBER14.
MM/GBSA calculations
The molecular mechanics/generalized Born surface area (MM-GBSA) molecular method was performed to calculate the binding free energy (ΔGbind) in the post-processing trajectory analysis in Amber14. The last 5 ns simulations were selected as the binding equilibrium conformation to calculate binding free energy. The ΔGbind of CeCht1-inhibitor complexes was evaluated by energy minimization of the ligand, receptor, and the complex structure as follows:
where Gcomplex, Greceptor, and Gligand represent the free energy of complex, CeCht1, and ligand, respectively. ΔGbind was evaluated by gasphase interaction energy (ΔEMM), solvation energy term (ΔGsolvation) and entropy term (TΔS) between the candidate molecules and CeCht1. ΔEMM is the gasphase interaction energy and it contains internal (ΔEinternal), electrostatic (ΔEelectrostatic), and van der Waals energies (ΔEVDW). ΔGsolvation is the solvation free energy, which is the sum of the polar solvation contribution and nonpolar solvation contributions. TΔS shows the change of conformational entropy upon ligand binding. The energy decomposition was carried out to evaluate the contribution of each residue of the CeCht1 via the MM-GBSA method in AMBER14.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
LD designed and guided this investigation. SS performed this study and wrote this paper. XJ, QY, BD, and MY implemented the modification of this paper in order to improve its quality. All authors have given approval to the final version of the manuscript.
Acknowledgments
We acknowledge the financial support by the National Natural Science Foundation (32102247), the Key Research and Development Project of Hebei Province (22326508D), State Key Laboratory of North China Crop Improvement and Regulation (NCCIR2022ZZ-18), the Research Project of Basic Scientific Research Business Expenses of Hebei Provincial Universities (KY2021066).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.1021295/full#supplementary-material
References
Abad, P., Gouzy, J., Aury, J.-M., Castagnone-Sereno, P., Danchin, E. G. J., Deleury, E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. doi:10.1038/nbt.1482
Abebew, D., Sayedain, F. S., Bode, E., and Bode, H. B. (2022). Uncovering nematicidal natural products from xenorhabdus bacteria. J. Agric. Food Chem. 70, 498–506. doi:10.1021/acs.jafc.1c05454
Adam, R., Kaltmann, B., Rudin, W., Friedrich, T., Marti, T., and Lucius, R. (1996). Identification of chitinase as the immunodominant filarial antigen recognized by sera of vaccinated rodents. J. Biol. Chem. 271 (3), 1441–1447. doi:10.1074/jbc.271.3.1441
Albonico, M., Crompton, D. W., and Savioli, L. (1999). Control strategies for human intestinal nematode infections. Adv. Parasitol. 42, 277–341. doi:10.1016/s0065-308x(08)60151-7
Arnold, K., Brydon, L. J., Chappell, L. H., and Gooday, G. W. (1993). Chitinolytic activities in Heligmosomoides polygyrusand their role in egg hatching. Mol. Biochem. Parasitol. 58 (2), 317–323. doi:10.1016/0166-6851(93)90054-2
Chen, J., Li, Q. X., and Song, B. (2020). Chemical nematicides: Recent research progress and outlook. J. Agric. Food Chem. 68, 12175–12188. doi:10.1021/acs.jafc.0c02871
Chen, W., Jiang, X., and Yang, Q. (2020). Glycoside hydrolase family 18 chitinases: The known and the unknown. Biotechnol. Adv. 43, 107553. doi:10.1016/j.biotechadv.2020.107553
Chen, W., Chen, Q., Kumar, A., Jiang, X., Zhang, K. Y. J., and Yang, Q. (2021). Structure-based virtual screening of highly potent inhibitors of the nematode chitinase CeCht1. J. Enzyme Inhib. Med. Chem. 36, 1198–1204. doi:10.1080/14756366.2021.1931862
Chen, Q., Chen, W., Kumar, A., Jiang, X., Janezic, M., Zhang, K. Y. J., et al. (2021). Crystal structure and structure-based discovery of inhibitors of the nematode chitinase CeCht1. J. Agric. Food Chem. 69 (11), 3519–3526. doi:10.1021/acs.jafc.1c00162
Darden, T., York, D., and Pedersen, L. (1993). Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98 (12), 10089–10092. doi:10.1063/1.464397
Dong, L., Shen, S., Chen, W., Xu, D., Yang, Q., Lu, H., et al. (2019). Discovery of novel inhibitors targeting human O-GlcNAcase: Docking-based virtual screening, biological evaluation, structural modification, and molecular dynamics simulation. J. Chem. Inf. Model 59, 4374–4382. doi:10.1021/acs.jcim.9b00479
Dong, L., Shen, S., Xu, Y., Wang, L., Yang, Q., Zhang, J., et al. (2021). Identification of novel insect β-N-acetylhexosaminidase OfHex1 inhibitors based on virtual screening, biological evaluation, and molecular dynamics simulation. J. Biomol. Struct. Dyn. 39, 1735–1743. doi:10.1080/07391102.2020.1743758
Duan, Y., Wu, C., Chowdhury, S., Lee, M. C., Xiong, G., Zhang, W., et al. (2003). A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012. doi:10.1002/jcc.10349
Fanelli, E., Di Vito, M., Jones, J. T., and De Giorgi, C. (2005). Analysis of chitin synthase function in a plant parasitic nematode, Meloidogyne artiellia, using RNAi. Gene 349, 87–95. doi:10.1016/j.gene.2004.11.045
Gao, B., Allen, R., Maier, T., McDermott, J. P., Davis, E. L., Baum, T. J., et al. (2002). Characterisation and developmental expression of a chitinase gene in Heterodera glycines. Int. J. Parasitol. 32, 1293–1300. doi:10.1016/S0020-7519(02)00110-8
Gloeckner, C., Garner, A. L., Mersha, F., Oksov, Y., Tricoche, N., Eubanks, L. M., et al. (2010). Repositioning of an existing drug for the neglected tropical disease Onchocerciasis. Proc. Natl. Acad. Sci. U. S. A. 107 (8), 3424–3429. doi:10.1073/pnas.0915125107
Gooyit, M., Tricoche, N., Javor, S., Lustigman, S., and Janda, K. D. (2015). Exploiting the polypharmacology of ß-carbolines to disrupt O. Volvulus molting. ACS Med. Chem. Lett. 6 (3), 339–343. doi:10.1021/ml500516r
Gorgulla, C., Boeszoermenyi, A., Wang, Z.-F., Fischer, P. D., Coote, P. W., Padmanabha Das, K. M., et al. (2020). An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668. doi:10.1038/s41586-020-2117-z
Hao, G., Dong, Q., and Yang, G. (2011). A comparative study on the constitutive properties of marketed pesticides. Mol. Inf. 30, 614–622. doi:10.1002/minf.201100020
Jiang, X., Kumar, A., Motomura, Y., Liu, T., Zhou, Y., Moro, K., et al. (2020). A series of compounds bearing a dipyrido-pyrimidine scaffold acting as novel human and insect pest chitinase inhibitors. J. Med. Chem. 63, 987–1001. doi:10.1021/acs.jmedchem.9b01154
Ju, Y., Wang, X., Guan, T., Peng, D., and Li, H. (2016). Versatile glycoside hydrolase family 18 chitinases for fungi ingestion and reproduction in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 46, 819–828. doi:10.1016/j.ijpara.2016.08.001
Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. (2001). Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11, 171–176. doi:10.1016/s0960-9822(01)00052-5
Ntalli, N. G., and Caboni, P. (2012). Botanical nematicides: A review. J. Agric. Food Chem. 60, 9929–9940. doi:10.1021/jf303107j
Pippione, A. C., Dosio, F., Ducime, A., Federico, A., Martina, K., Sainas, S., et al. (2015). Substituted 4-hydroxy-1, 2, 3-triazoles: Synthesis, characterization and first drug design applications through bioisosteric modulation and scaffold hopping approaches. MedChemComm 6 (7), 1285–1292. doi:10.1039/C5MD00182J
Ryckaert, J. P., Ciccotti, G., and Berendsen, H. J. C. (1977). Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341. doi:10.1016/0021-9991(77)90098-5
Tachu, B., Pillai, S., Lucius, R., and Pogonka, T. (2008). Essential role of chitinase in the development of the filarial nematode Acanthocheilonema viteae. Infect. Immun. 76, 221–228. doi:10.1128/iai.00701-07
Traunspurger, W. (2000). The biology and ecology of lotic nematodes. Freshw. Biol. 44, 29–45. doi:10.1046/j.1365-2427.2000.00585.x
Vanegas, J. A. G., Pacule, H. B., Capitao, R. M., Correia, C. R. D., Terra, W. C., Campos, V. P., et al. (2022). Methyl esters of (E)-Cinnamic acid: Activity against the plant-parasitic nematode Meloidogyne incognita and in silico interaction with histone deacetylase. J. Agric. Food Chem. 70 (22), 6624–6633. doi:10.1021/acs.jafc.1c08142
Veronico, P., Gray, L. J., Jones, J. T., Bazzicalupo, P., Arbucci, S., Cortese, M. R., et al. (2001). Nematode chitin synthases: Gene structure, expression and function in Caenorhabditis elegans and the plant parasitic nematode Meloidogyne artiellia. Mol. Genet. Genomics 266, 28–34. doi:10.1007/s004380100513
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., and Case, D. A. (2004). development and testing of a general amber force fieldJournal of computational chemistry. J. Comput. Chem. 26, 1157–1174. doi:10.1002/jcc.20145
Keywords: nematicide, chitinase, inhibitor, CeCht1, virtual screening, inhibitory mechanism
Citation: Shen S, Ding B, Jiang X, Yang M, Yang Q and Dong L (2022) Discovery of novel inhibitors targeting nematode chitinase CeCht1: Virtual screening, biological evaluation, and molecular dynamics simulation. Front. Chem. 10:1021295. doi: 10.3389/fchem.2022.1021295
Received: 17 August 2022; Accepted: 18 October 2022;
Published: 03 November 2022.
Edited by:
Renjith Thomas, Mahatma Gandhi University, IndiaReviewed by:
Xiuhai Gan, Guizhou University, ChinaSanja Josip Armakovic, University of Novi Sad, Serbia
Copyright © 2022 Shen, Ding, Jiang, Yang, Yang and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Yang, cWluZ3lhbmdAZGx1dC5lZHUuY24=; Lili Dong, bGlsaWRvbmdAaGViYXUuZWR1LmNu
 Shengqiang Shen1
Shengqiang Shen1 Qing Yang
Qing Yang Lili Dong
Lili Dong