- 1School of Chemistry and Chemical Engineering, Guangxi University, Nanning, China
- 2Guangxi Research Institute of Chemical Industry Co., Ltd., Nanning, China
- 3School of Public Health, Guangxi Medical University, Nanning, China
- 4Guangxi Colleges and Universities Key Laboratory of Prevention and Control of Highly Prevalent Diseases, Guangxi Medical University, Nanning, China
A series of novel menthone derivatives bearing pyrimidine and urea moieties was designed and synthesized to explore more potent natural product-derived antitumor agents. The structures of the target compounds were confirmed by FTIR, NMR, and HRMS. The in vitro antitumor activity was tested by standard methyl thiazolytetrazolium assay and showed that 4i, 4g, 4s, and 4m are the best compounds with IC50 values of 6.04 ± 0.62µM, 3.21 ± 0.67µM, 19.09 ± 0.49µM, and 18.68 ± 1.53µM, against Hela, MGC-803, MCF-7, and A549, respectively. The results of the preliminary action mechanism studies showed that compound 4i, the representative compound, could induce cell apoptosis in Hela cells in a dose-dependent manner and might arrest the cell cycle in the G2/M phase. Furthermore, the results of network pharmacology prediction and Western blot experiments indicated that compound 4i might inhibit Hela cells through inhibit PI3K/Akt/mTOR signaling pathway. The binding modes and the binding sites interactions between compound 4i and the target proteins were predicted preliminarily by the molecular docking method.
Introduction
Cancer is a life-threatening disease with a high mortality rate and a major public health problem worldwide (Siegel et al., 2018). Although many new significant therapeutic methods were developed during the past decades, chemotherapy remains the main method for cancer treatment (Bukowski et al., 2020). However, the drug resistance of tumors restricted the applications of many chemotherapeutic drugs in treatments (Kumar et al., 2015; Gao et al., 2019). The tumors could be resistant to some chemotherapeutic drugs intrinsically, and show acquired drug resistance after chemotherapy, which makes the tumors become insensitive to similar drugs later. Therefore, the development of new drugs to overcome the resistance is still an important mission for medicinal chemists.
PI3K/Akt/mTOR pathway has been reported as an important cell growth signaling pathway. This pathway is excessively activated in many tumor cells and facilitates the cancer cells resistance to chemotherapy (Chen et al., 2010; Zhang and Yang, 2020). In addition, there is evidence that the sensitivity of tumor cells to therapeutic drugs could be restored by inhibiting PI3K/Akt/mTOR pathway, which also prompts the apoptosis of tumor cells (Papadimitrakopoulo, 2012). Thus, many small molecule inhibitors against PI3K/Akt/mTOR signaling pathway have been studied for antitumor use (Alzahrani, 2019).
Natural product is a prolific source with diverse structures and biological properties (Seca and Pinto 2018). Many plants have been used as traditional medicine and the leading compounds for the development of antitumor agents (Agarwal et al., 2019). L-menthone is a naturally occurring monocyclic monoterpenone found in many essential oils (Tahghighi et al., 2019; Kikowska et al., 2020; Montenegro et al., 2020), especially in peppermint essential oil. It can also be conveniently prepared by oxidation of L-menthol, which is the dominant component of peppermint essential oil. L-menthone and its derivatives have been reported to have a wide range of biological activities, such as antiviral (He et al., 2013), anti-inflammatory (Sun et al., 2014), antidepressant (Xue et al., 2015), antifungal (Boni et al., 2016), and anticancer activities (Bhalla et al., 2013; Chen et al., 2014; Mosbah et al., 2018; Qi et al., 2018). Herein, L-menthone has received growing interest from medicinal chemists due to their wide range of biological activities.
Many pyrimidine derivatives showed various biological activities such as antibacterial (Xiong et al., 2012), antifungal (Zhang et al., 2019), and anti-inflammatory (Horishny et al., 2021) activities. Especially, pyrimidine derivatives have been reported to show antitumor effect against different human cancers with various mechanisms (Chen N.-Y. et al., 2020; Wang et al., 2020). Meanwhile, many substituted pyrimidines displayed enzymes inhibitory activity (Heffron et al., 2010; Ni et al., 2011), such as PI3K inhibitor (Han et al., 2015), Akt inhibitor (Liu et al., 2017), PI3K and mTOR dual inhibitor (Han et al., 2017), and others. These reports inspire the development of novel pyrimidine-containing antitumor compounds.
On the other hand, urea derivatives are widely applied in medicinal synthesis due to their various biological activities, such as anti-inflammatory (Drexel et al., 2019), antitumor activities (Kang et al., 2019; Kilic-Kurta et al., 2020), and antimicrobial (Lafzi et al., 2021). Recent reports showed that urea derivatives could inhibit cell signaling transduction, such as RAS-RAFMEK-ERK signaling pathway and PI3K/Akt/mTOR pathway (Li et al., 2019), and inhibit protein activity, such as mTORC1 and mTORC2 (Gao et al., 2018). Thus, to design new compounds with urea moiety to explore chemotherapy agents is a feasible strategy.
In addition, it is an effective strategy in drug development to integrate various pharmacophores to obtain novel molecules with synergistic effect. In continuation of our interest in natural product-based antitumor compounds (Li et al., 2017; Wang et al., 2018; Chen Y. et al., 2020; Li et al., 2020; Zhu et al., 2020; Wang et al., 2021), a series of novel menthone-derived compounds containing pyrimidine and urea moieties were designed and synthesized according to the strategy, which was shown in Figure 1. The in vitro antitumor activities of the menthone derivatives against the four tumor cell lines were evaluated. The action mechanisms of the target compounds against tumor cell line were studied using a representative compound against Hela cell line by the tests of cell cycle and cell apoptosis, and further predicted by network pharmacology method and molecular docking. Western blot experiments were chosen to confirm the predicted result of the key signaling pathway regulated by this representative compound.
Materials and Methods
General
The melting points were recorded on an MP420 automatic melting point apparatus (Hanon Instruments Co., Ltd., Jinan, China) without calibration. Optical rotations were recorded at 20°C on precision scientific WZZ-1 digital automatic polarimeter (Shanghai Precision Technology Instrument Co., Ltd., Shanghai, China). FT-IR spectra were carried out using a Nicolet iS50 FT-IR spectrometer (Thermo Scientific Co., Ltd., United States) by KBr pellet method. NMR spectra were recorded on Bruker Avance III HD 500MHz/600MHz spectrometer (Bruker Co., Ltd., Switzerland) with TMS (tetramethylsilane) as an internal standard in CDCl3. High resolution mass spectrometry was determined on ESQUIRE HCT (Bruker Daltonics Co., Ltd., United States) equipped with a TOF/TOF/Ultimate 3000 Nano HPLC. GC analysis was conducted on an Agilent 6890GC (Agilent Technologies Inc., United States) equipped with column HP-1 (30m, 0.530 mm, 0.88μm) and FID. HPLC analysis was conducted on Waters 1525 HPLC instrument (Waters Corp., United States) equipped with Waters 2998 PDA detector and column C18 5μm (4.6mm × 150mm). L-Menthone (GC purity 98%) was provided by Shanghai Macklin Biochemical Co., Ltd., China. Routine thin-layer chromatography (TLC) was performed on silica gel plates (silica gel GF254 from Qingdao Haiyang Chemical Co., Ltd., Qingdao, China). Preparative flash column chromatography was performed on the 200–300 mesh silica gel (Qingdao Haiyang Chemical Co. Ltd.). Other reagents were purchased from commercial suppliers and used as received.
General Procedure for Compound 2
L-Menthone (3.50ml, 20mmol), KOH (1.40g, 25mmol), and anhydrous DMSO (15ml) were mixed and stirred at 20°C for 30min. Then, the mixed solution of benzaldehyde (2.50g, 22mmol) and anhydrous DMSO (10ml) was added dropwise to the reaction system, and the reaction temperature was controlled not to exceed 20°C. After continuous stirring for 4h, the reaction solution was poured into 250ml 4% acetic acid ice-water mixture and then extracted by EtOAc three times. The extracts were combined and washed with saturated sodium chloride and deionized water three times, respectively. Then, the resulting solution was dried with anhydrous Na2SO4 and concentrated in vacuum. The crude product was purified by silica gel column chromatography (EtOAc-petroleum ether = 1:60, v/v) to obtain L-menthone-derived α,β-unsaturated ketone, (3R,6S)-2-((E)-benzylidene)-6-isopropyl-3-methylcyclohexan-1-one, compound 2, as a pale yellow liquid in the yield of 93% and HPLC purity of 98.8%. [α]D20 = - 68.8°(c = 0.20, CH2Cl2); 1H NMR (600MHz, CDCl3) δ (ppm): 7.38 (d, J = 7.2 Hz, 4H), 7.32 (t, J = 6.8 Hz, 1H), 7.13 (s, 1H), 3.43 (dt, J = 10.8, 6.9 Hz, 1H), 2.58 (m, 1H), 2.26 (ddd, J = 11.5, 7.2, 3.5 Hz, 1H), 1.90 (td, J = 13.2, 7.6 Hz, 3H), 1.80 (m, 1H), 1.21 (d, J = 7.2 Hz, 3H), 0.99 (d, J = 7.1 Hz, 3H), 0.93 (d, J = 6.8 Hz, 3H); 13C NMR (151MHz, CDCl3) δ (ppm): 205.18, 145.00, 136.07, 132.66, 129.61, 128.55, 128.21, 56.07, 31.79, 30.66, 26.74, 20.54, 19.98, 18.85, 17.97; IR (KBr) v: 3438, 3199, 2959, 2939, 1691, 1614, 1488, 1458cm−1.
General Procedure for Compound 3
The mixed solution of compound 2 (1.20 g, 5mmol), guanidine hydrochloride (0.95g, 10mmol), K2CO3 (4.15g, 30mmol), distilled water (1.20g), and ethanol (30ml) were stirred and refluxed for 9h. After the reaction was completed, the reaction solution was diluted with water and then extracted by EtOAc three times. The extracts were combined and washed with saturated brine to neutrality. The resulting solution was dried over Na2SO4 and concentrated in vacuum. The crude product was purified by silica gel column chromatography (EtOAc-petroleum ether = 1:6, v/v) to obtain L-menthone-derived pyrimidine, (5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-amine, compound 3, as a white solid in the yield of 28.6% and HPLC purity of 98.2%; [α]D20 = + 31.3°(c = 0.20, CH2Cl2); m.p. 144.5–145.6°C; 1H NMR (500MHz, CDCl3) δ (ppm): 7.47–7.38 (m, 5H), 4.87 (s, 2H), 3.04 (td, J = 7.0, 3.9 Hz, 1H), 2.92 (m, 1H), 2.70 (td, J = 8.8, 3.9 Hz, 1H), 1.76 (d, J = 8.0 Hz, 2H), 1.70–1.65 (m, 2H), 1.08 (d, J = 7.1 Hz, 3H), 0.85 (d, J = 7.0 Hz, 3H), 0.75 (d, J = 6.8 Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 170.04, 167.30, 160.58, 139.35, 128.36, 128.30, 127.99, 123.97, 46.41, 29.59, 28.96, 27.56, 21.45, 20.78, 16.71, 16.44; IR (KBr) v: 3438, 3199, 2959, 2939, 1691, 1614, 1488, 1458cm−1; HRMS m/z: Calcd for C18H23N3 [M+H]+: 282.1965, found: 282.1954.
General Procedure for the Title Compounds 4a-4s
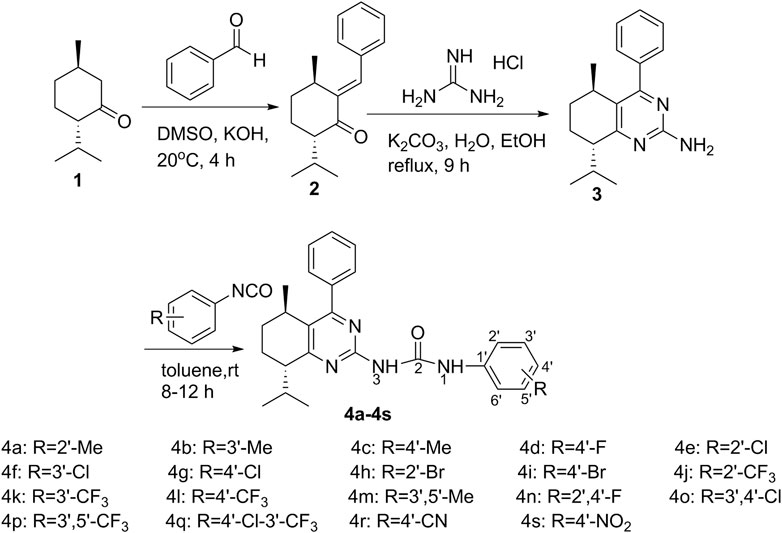
The mixture of compound 3 (0.28 g, 1mmol), substituted phenyl isocyanate (1.3mmol), and toluene (10ml) were stirred at room temperature for 8–12h. After compound 3 was fully reacted, the precipitate was filtered, and dissolved in EtOAc. The resulting solution was concentrated in vacuum. The crude product was purified by silica gel column chromatography (EtOAc-petroleum ether = 1:10, v/v) to afford L-menthone-derived pyrimidine-urea, compounds 4a-4s, as a white solid.
1-(2′-Methylphenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4a). White solid, Yield: 80%; HPLC purity: 95.3%; [α]D20 = + 10.2°(c = 0.11, CHCl3); m.p. 191.2–193.3°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.08 (s, 1H), 8.08 (d, J = 7.9 Hz, 1H), 7.57–7.46 (m, 4H), 7.45–7.40 (m, 2H), 7.21 (t, J = 7.8 Hz, 1H), 7.07 (d, J = 7.5 Hz, 1H), 6.99 (t, J = 7.4 Hz, 1H), 3.12 (m, 1H), 2.93 (ddd, J = 13.9, 8.9, 5.4 Hz, 1H), 2.83 (tt, J = 6.0, 3.2 Hz, 1H), 1.89 (s, 1H), 1.84–1.80 (m, 3H), 1.77 (s, 1H), 1.77–1.73 (m, 1H), 1.68 (d, J = 8.5 Hz, 1H), 1.10 (d, J = 7.1 Hz, 3H), 0.92 (d, J = 7.0 Hz, 3H), 0.77 (d, J = 6.8 Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 155.06, 152.21, 138.18, 136.92, 131.04, 130.18, 128.97, 128.89, 128.62, 128.46, 128.25, 128.04, 127.77, 126.63, 123.70, 46.62, 29.84, 28.66, 27.80, 21.30, 20.67, 18.03, 16.80, 16.33; IR (KBr) v: 3203, 3122, 3045, 2960, 2941, 2866, 1692, 1614, 1590, 1561, 1543, 1489, 1458, 1376cm−1; HRMS m/z: Calcd for C26H30N4O [M+H]+: 415.2492, found: 415.2492.
1-(3′-Methylphenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4b). White solid, Yield: 83%; HPLC purity: 97.2%; [α]D20 = + 11.0°(c = 0.10, CHCl3); m.p. 170.8–173.7°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.52 (s, 1H), 7.48 (d, J = 3.5Hz, 5H), 7.37–7.28 (m, 3H), 7.19 (t, J = 7.8Hz, 1H), 6.88 (d, J = 7.5Hz, 1H), 3.25 (m, 1H), 2.99 (tt, J = 10.6, 5.4Hz, 1H), 2.85 (s, 1H), 2.33 (s, 3H), 1.90–1.80 (m, 3H), 1.79–1.75 (m, 1H), 1.15 (d, J = 7.0Hz, 3H), 0.89 (d, J = 7.0Hz, 3H), 0.80 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.98, 151.94, 138.91, 138.57, 138.31, 129.15, 128.97, 128.59, 128.12, 127.76, 124.22, 120.39, 116.68, 46.82, 29.85, 28.66, 27.69, 21.69, 21.46, 20.75, 16.73, 16.24; IR (KBr) v: 3203, 3116, 3054, 2958, 2923, 2869, 1687, 1614, 1566, 1543, 1486, 1379cm−1; HRMS m/z: Calcd for C26H30N4O [M+H]+: 415.2492, found: 415.2487.
1-(4′-Methylphenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4c). White solid, Yield: 88%; HPLC purity: 96.8%; [α]D20 = + 12.6°(c = 0.11, CHCl3); m.p. 179.8–181.2°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.47 (s, 1H), 7.50 (d, J = 3.9Hz, 5H), 7.41 (d, J = 8.4Hz, 3H), 7.13 (d, J = 8.0Hz, 2H), 3.27 (q, J = 6.3, 5.5Hz, 1H), 3.04–2.94 (m, 1H), 2.86 (td, J = 8.6, 8.0, 4.0Hz, 1H), 2.33 (s, 3H), 1.92–1.81 (m, 3H), 1.28 (s, 1H), 1.16 (d, J = 7.0Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.9Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 155.00, 152.01, 138.32, 136.04, 132.93, 129.61, 129.11, 128.58, 128.09, 127.68, 119.75, 46.78, 29.83, 28.65, 27.68, 21.45, 20.93, 20.75, 16.73, 16.27; IR (KBr) v: 3197, 3119, 3033, 2956, 2921, 1694, 1610, 1562, 1544, 1514, 1499, 1371cm−1; HRMS m/z: Calcd for C26H30N4O [M+H]+: 415.2492, found: 415.2485.
1-(4′-Fluorophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4d). White solid, Yield: 86%; HPLC purity: 98.0%; [α]D20 = + 5.7°(c = 0.11, CHCl3); m.p. 191.5–192.9°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.56 (s, 1H), 7.54–7.43 (m, 8H), 7.01 (t, J = 8.5Hz, 2H), 3.27 (m, 1H), 2.96 (m, 1H), 2.86 (td, J = 8.5, 7.5, 4.1Hz, 1H), 1.85 (td, J = 12.8, 11.7, 3.8Hz, 3H), 1.80 (s, 1H), 1.16 (d, J = 7.0Hz, 3H), 0.90 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 159.94, 158.02, 154.91, 152.12, 138.27, 134.68, 129.19, 128.62, 128.07, 127.85, 121.23, 121.17, 115.77, 115.59, 46.80, 29.85, 28.63, 27.68, 21.44, 20.76, 16.76, 16.28; IR (KBr) v): 3197, 3116, 3042, 2960, 2929, 2870, 1689, 1614, 1565, 1542, 1510, 1376cm−1; HRMS m/z: Calcd for C25H27FN4O [M+H]+: 419.2242, found: 419.2235.
1-(2′-Chlorophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4e). White solid, Yield: 78%; HPLC purity: 95.6%; [α]D20 = + 50.2°(c = 0.11, CHCl3); m.p. 177.5–180.5°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.66 (s, 1H), 8.39 (d, J = 8.2Hz, 1H), 7.56–7.46 (m, 4H), 7.43 (dd, J = 6.8, 2.9Hz, 2H), 7.29–7.23 (m, 2H), 6.98 (t, J = 7.6Hz, 1H), 3.13 (m, 1H), 2.98 (m, 1H), 2.87 (dt, J = 9.4, 4.7Hz, 1H), 1.81 (t, J = 9.2Hz, 3H), 1.77–1.72 (m, 1H), 1.10 (d, J = 7.0Hz, 3H), 0.93 (d, J = 7.0Hz, 3H), 0.75 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.78, 152.01, 138.03, 136.01, 129.12, 128.87, 128.39, 128.18, 128.15, 127.44, 123.85, 123.49, 122.14, 46.48, 30.00, 28.63, 27.84, 21.20, 20.70, 16.76, 16.33; IR (KBr) v: 3203, 3140, 3036, 2958, 2876, 1689, 1592, 1557, 1538, 1498, 1442, 1381cm−1; HRMS m/z: Calcd for C25H27ClN4O [M+H]+: 435.1946, found: 435.1945.
1-(3′-Chlorophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4f). White solid, Yield: 80%; HPLC purity: 98.2%; [α]D20 = + 25.2°(c = 0.12, CHCl3); m.p. 170.8–173.7°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.74 (s, 1H), 7.61 (s, 1H), 7.51 (q, J = 8.1, 7.0Hz, 6H), 7.43–7.31 (m, 1H), 7.22 (t, J = 8.0Hz, 1H), 7.04 (d, J = 7.9Hz, 1H), 3.33–3.24 (m, 1H), 2.98 (m, 1H), 2.88 (dd, J = 9.9, 4.4Hz, 1H), 1.92–1.82 (m, 3H), 1.79 (dd, J = 12.2, 4.1Hz, 1H), 1.18 (d, J = 7.1Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.82 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.80, 151.85, 139.91, 138.20, 134.71, 130.08, 129.26, 128.65, 128.08, 128.01, 123.34, 119.68, 117.45, 46.83, 29.91, 28.62, 27.69, 21.45, 20.72, 16.74, 16.24; IR (KBr) v: 3215, 3134, 3057, 2959, 2870, 1694, 1595, 1560, 1541, 1480, 1421, 1379cm−1; HRMS m/z: Calcd for C25H27ClN4O [M+H]+: 435.1946, found: 435.1942.
1-(4′-Chlorophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4g). White solid, Yield: 70%; HPLC purity: 96.1%; [α]D20 = + 18.8°(c = 0.11, CHCl3); m.p. 183.6–185.5°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.67 (s, 1H), 7.61 (s, 1H), 7.54–7.44 (m, 7H), 7.27 (d, J = 8.5Hz, 2H), 3.28 (q, J = 6.3, 5.7Hz, 1H), 3.00–2.92 (m, 1H), 2.87 (m, 1H), 1.85 (td, J = 12.9, 12.0, 3.9Hz, 3H), 1.75 (dd, J = 14.0, 6.6Hz, 1H), 1.16 (d, J = 7.0Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.84, 151.99, 138.23, 137.30, 129.21, 129.06, 128.60, 128.24, 128.19, 128.06, 127.92, 120.77, 46.77, 29.87, 28.60, 27.66, 21.42, 20.74, 16.75, 16.26; IR (KBr) v: 3203, 3108, 3030, 2957, 2932, 2864, 1697, 1611, 1602, 1583, 1560, 1542, 1493, 1371cm−1; HRMS m/z: Calcd for C25H27ClN4O [M+H]+: 435.1946, found: 435.1940.
1-(2′-Bromophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4h). White solid, Yield: 76%; HPLC purity: 96.7%; [α]D20 = + 19.2°(c = 0.11, CHCl3); m.p. 184.2–187.1°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.50 (s, 1H), 8.32 (d, J = 8.2Hz, 1H), 7.52 (s, 1H), 7.50–7.44 (m, 4H), 7.42 (dd, J = 6.7, 3.1Hz, 2H), 7.30 (d, J = 8.0Hz, 1H), 6.92 (q, J = 7.3Hz, 1H), 3.15–3.06 (m, 1H), 2.98 (m, 1H), 2.87 (dt, J = 9.6, 4.6Hz, 1H), 1.82–1.73 (m, 3H), 1.65 (s, 1H), 1.10 (d, J = 7.1Hz, 3H), 0.93 (d, J = 7.0Hz, 3H), 0.75 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.74, 152.09, 138.07, 137.24, 132.69, 132.46, 128.86, 128.56, 128.39, 128.26, 128.18, 127.97, 124.51, 122.91, 46.46, 29.99, 28.64, 27.85, 21.22, 20.68, 16.74, 16.30; IR (KBr) v: 3212, 3137, 3033, 2960, 2923, 2870, 1690, 1581, 1569, 1556, 1536, 1498, 1438, 1421, 1380cm−1; HRMS m/z: Calcd for C25H27BrN4O [M+H]+: 479.1441, found: 479.1462.
1-(4′-Bromophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4i). White solid, Yield: 82%; HPLC purity: 95.8%; [α]D20 = + 20.2°(c = 0.11, CHCl3); m.p. 188.8–190.1°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.66 (s, 1H), 7.53 (s, 1H), 7.52–7.47 (m, 4H), 7.42 (d, J = 9.5 Hz, 5H), 3.32–3.23 (m, 1H), 2.96 (m, 1H), 2.86 (m, 1H), 1.86–1.78 (m, 3H), 1.67–1.61 (m, 1H), 1.16 (d, J = 7.0Hz, 3H), 0.90 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.81, 151.90, 138.22, 137.80, 132.04, 129.24, 128.64, 128.07, 127.98, 121.16, 115.85, 46.82, 29.89, 28.62, 27.69, 21.44, 20.75, 16.76, 16.27; IR (KBr) v: 3209, 3114, 3030, 2958, 2926, 2870, 1696, 1607, 1591, 1583, 1559, 1542, 1488, 1371cm−1; HRMS m/z: Calcd for C25H27BrN4O [M+H]+: 479.1441, found: 479.1446.
1-(2′-(Trifluoromethyl)phenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4j). White solid, Yield: 80%; HPLC purity: 95.2%; [α]D20 = + 54.7°(c = 0.14, CHCl3); m.p. 171.2–174.2°C; 1H NMR (500 MHz, CDCl3) δ (ppm): 11.29 (s, 1H), 8.04 (d, J = 8.2Hz, 1H), 7.57 (h, J = 9.6, 8.7Hz, 3H), 7.47 (dd, J = 5.3, 1.9Hz, 3H), 7.41 (dd, J = 6.8, 2.9Hz, 2H), 7.22 (t, J = 7.7Hz, 1H), 3.13 (m, 1H), 2.96–2.75 (m, 2H), 1.81–1.72 (m, 3H), 1.69–1.61 (m, 1H), 1.07 (d, J = 6.8Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.74 (d, J = 6.7 Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.81, 152.58, 137.94, 135.65, 132.49, 128.81, 128.34, 128.26, 128.05, 126.77, 126.09, 126.04, 126.00, 124.40, 122.45, 46.26, 30.03, 28.60, 27.82, 21.14, 20.63, 16.76, 16.32; IR (KBr) v: 3197, 3143, 3066, 3042, 2961, 2938, 2876, 1684, 1620, 1588, 1566, 1535, 1481, 1455, 1371cm−1; HRMS m/z: Calcd for C26H27F3N4O [M+H]+: 469.2210, found: 469.2211.
1-(3′-(Trifluoromethyl)phenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4k). White solid, Yield: 76%; HPLC purity: 97.4%; [α]D20 = + 16.7°(c = 0.11, CHCl3); m.p. 175.4–176.3°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.92 (s, 1H), 7.87 (s, 1H), 7.66 (s, 1H), 7.52 (d, J = 6.8Hz, 5H), 7.43 (dd, J = 9.8, 6.3Hz, 2H), 7.32 (d, J = 7.8Hz, 1H), 3.30 (d, J = 8.4Hz, 1H), 2.99 (m, 1H), 2.88 (q, J = 7.2, 4.9Hz, 1H), 1.85 (td, J = 13.3, 9.6Hz, 3H), 1.66–1.59 (m, 1H), 1.18 (d, J = 7.1Hz, 3H), 0.92 (d, J = 7.0Hz, 3H), 0.83 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.78, 151.92, 139.29, 138.18, 131.28, 129.76, 129.31, 128.84, 128.68, 128.12, 128.07, 125.18, 123.01, 122.49, 119.83, 116.11, 116.08, 46.88, 29.85, 28.62, 27.70, 21.48, 20.64, 16.69, 16.21; IR (KBr) v: 3206, 3120, 3063, 2963, 2926, 2876, 1696, 1614, 1566, 1541, 1486, 1373cm−1; HRMS m/z: Calcd for C26H27F3N4O [M+H]+: 469.2210, found: 469.2211.
1-(4′-(Trifluoromethyl)phenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4l). White solid, Yield: 70%; HPLC purity: 98.3%; [α]D20 = + 11.2°(c = 0.10, CHCl3); m.p. 204.7–206.2°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.89 (s, 1H), 7.64–7.43 (m, 10H), 3.33–3.22 (m, 1H), 2.97 (m, 1H), 2.87 (s, 1H), 1.87–1.80 (m, 3H), 1.61 (d, J = 3.2Hz, 1H), 1.17 (d, J = 7.0Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.9Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.74, 151.89, 141.85, 138.18, 129.32, 128.68, 128.18, 128.07, 126.40, 123.33, 119.14, 46.86, 29.85, 28.61, 27.71, 21.44, 20.76, 16.77, 16.27; IR (KBr) v: 3206, 3137, 3111, 3039, 2960, 2929, 2870, 1697, 1603, 1564, 1541, 1504, 1488, 1376cm−1; HRMS m/z: Calcd for C26H27F3N4O [M+H]+: 469.2210, found: 469.2202.
1-(3′,5′-Dimethylphenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4m). White solid, Yield: 80%; HPLC purity: 95.9%; [α]D20 = + 17.2°(c = 0.11, CHCl3); m.p. 166.9–169.1°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.54 (s, 1H), 7.51 (s, 5H), 7.36 (s, 1H), 7.18 (s, 2H), 6.74 (s, 1H), 3.28 (d, J = 7.3Hz, 1H), 3.03 (m, 1H), 2.87 (q, J = 7.3, 5.1Hz, 1H), 2.31 (s, 6H), 1.91–1.82 (m, 3H), 1.69 (s, 1H), 1.18 (d, J = 7.1Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.83 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.99, 151.89, 138.73, 138.49, 138.32, 129.12, 128.96, 128.56, 128.13, 127.70, 125.10, 117.38, 46.80, 29.84, 28.66, 27.67, 21.57, 21.46, 20.72, 16.71, 16.19; IR (KBr) v: 3206, 3131, 3021, 2957, 2920, 2867, 1692, 1617, 1566, 1548, 1485, 1388cm−1; HRMS m/z: Calcd for C27H32N4O [M+H]+: 429.2649, found: 429.2647.
1-(2′,4′-Difluorophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4n). White solid, Yield 75%; Purity 97.0%; [α]D20 = + 25.2°(c = 0.11, CHCl3); m.p. 179.7–182.2°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.61 (s, 1H), 8.31 (td, J = 9.0, 5.9 Hz, 1H), 7.54–7.45 (m, 6H), 6.86 (td, J = 13.3, 11.2, 7.1Hz, 2H), 3.23 (m, 1H), 3.00–2.92 (m, 1H), 2.88 (dt, J = 8.9, 4.4Hz, 1H), 1.86–1.80 (m, 3H), 1.68–1.64 (m, 1H), 1.12 (d, J = 7.0Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.77 (d, J = 6.9Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 159.10 (d, J = 11.1Hz), 157.15 (d, J = 11.3Hz), 154.81, 153.79 (d, J = 11.7Hz), 151.99, 138.04, 129.05, 128.51, 128.12, 123.44 (dd, J = 10.8, 4.0Hz), 122.70 (d, J = 9.2Hz), 111.37–110.73 (m), 104.02–102.92 (m), 46.59, 29.86 (d, J = 5.0Hz), 28.58, 27.75, 21.23, 20.75, 16.77, 16.34; IR (KBr) v: 3209, 3152, 3117, 3027, 2960, 2926, 2870, 1694, 1611, 1565, 1542, 1503, 1371cm−1; HRMS m/z: Calcd for C25H26F2N4O [M+H]+: 437.2147, found: 437.2149.
1-(3′,4′-Dichlorophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4o). White solid, Yield: 72%; HPLC purity: 95%; [α]D20 = + 10.2°(c = 0.12, CHCl3); m.p. 178.7–180.8°C; 1H NMR (500MHz, CDCl3) δ (ppm): 11.81 (s, 1H), 7.71 (s, 1H), 7.55–7.43 (m, 6H), 7.35 (d, J = 6.6 Hz, 2H), 3.29 (dq, J = 10.1, 6.0 Hz, 1H), 2.95 (m, 1H), 2.87 (m, 1H), 1.84 (dt, J = 15.8, 6.3Hz, 3H), 1.64–1.61 (m, 1H), 1.17 (d, J = 7.1Hz, 3H), 0.91 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.69, 151.78, 138.29, 138.16, 132.78, 130.61, 129.34, 128.69, 128.17, 128.06, 126.36, 121.17, 118.68, 46.86, 29.92, 28.60, 27.69, 21.46, 20.72, 16.75, 16.24; IR (KBr) v: 3215, 3146, 3030, 2959, 2932, 2870, 1694, 1592, 1566, 1538, 1476, 1371cm−1; HRMS m/z: Calcd for C25H26Cl2N4O [M+H]+: 469.1556, found: 469.1556.
1-(3′,5′-Bis(trifluoromethyl)phenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4p). White solid, Yield: 80%; HPLC purity: 96.5%; [α]D20 = + 26.2°(c = 0.10, CHCl3); m.p. 160.5–162.0°C; 1H NMR (500MHz, CDCl3): δ (ppm) 12.30 (s, 1H), 8.02 (s, 2H), 7.58–7.49 (m, 7H), 3.33 (s, 1H), 3.04–2.95 (m, 1H), 2.91 (d, J = 10.8Hz, 1H), 1.86 (dt, J = 14.0, 6.0Hz, 3H), 1.62 (d, J = 3.4Hz, 1H), 1.19 (d, J = 7.1Hz, 3H), 0.93 (d, J = 7.0Hz, 3H), 0.83 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.58, 151.81, 140.28, 138.06, 132.57, 132.30, 132.03, 129.47, 128.75, 128.46, 128.03, 126.61, 124.44, 122.27, 120.10, 118.97, 116.47, 46.93, 30.05, 28.58, 27.71, 21.50, 20.57, 16.67, 16.15; IR (KBr) v: 3224, 3164, 3108, 2962, 2873, 1706, 1625, 1583, 1565, 1545, 1473, 1450, 1389cm−1; HRMS m/z: Calcd for C27H26F6N4O [M+H]+: 537.2085, found: 537.2084.
1-(4′-Chloro-3′-(trifluoromethyl)phenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4q). White solid, Yield: 76%; HPLC purity: 95.2%; [α]D20 = + 19.2°(c = 0.11, CHCl3); m.p. 192.1–193.5°C; 1H NMR (500MHz, CDCl3) δ (ppm): 12.00 (s, 1H), 7.92 (s, 1H), 7.63 (s, 1H), 7.55–7.47 (m, 6H), 7.44 (d, J = 8.8Hz, 1H), 3.31 (d, J = 7.2Hz, 1H), 2.97 (m, 1H), 2.88 (m, 1H), 1.84 (dt, J = 13.3, 6.1Hz, 3H), 1.65 (s, 1H), 1.17 (d, J = 7.0Hz, 3H), 0.92 (d, J = 7.0Hz, 3H), 0.82 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.66, 151.89, 138.11, 137.71, 132.20, 129.36, 128.83, 128.71, 128.58, 128.26, 128.02, 125.61, 123.85, 123.31, 121.68, 119.51, 118.21 (d, J = 5.5Hz), 46.87, 29.99, 28.59, 27.70, 21.47, 20.63, 16.68, 16.20; IR (KBr) v: 3206, 3137, 3102, 3042, 2961, 2935, 2873, 1695, 1603, 1560, 1540, 1482, 1371cm−1; HRMS m/z: Calcd for C26H26ClF3N4O [M+H]+: 503.1820, found: 503.1824.
1-(4′-Cyanophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4r). White solid, Yield: 80%; HPLC purity: 95.5%; [α]D20 = + 16.2°(c = 0.11, CHCl3); m.p. 216.4–218.4°C; 1H NMR (500MHz, CDCl3) δ (ppm): 12.03 (s, 1H), 7.60 (d, J = 15.8Hz, 5H), 7.55–7.48 (m, 5H), 3.28 (t, J = 8.1 Hz, 1H), 3.01–2.92 (m, 1H), 2.87 (t, J = 6.8 Hz, 1H), 1.89–1.81 (m, 3H), 1.70–1.63 (m, 1H), 1.16 (d, J = 7.0Hz, 3H), 0.91 (d, J = 7.1Hz, 3H), 0.81 (d, J = 6.7Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.60, 151.79, 142.90, 138.09, 133.39, 129.38, 128.69, 128.34, 128.03, 119.37, 119.25, 106.11, 46.84, 29.83, 28.55, 27.69, 21.41, 20.74, 16.78, 16.26; IR (KBr) v: 3209, 3155, 3036, 2960, 2929, 2873, 2227, 1698, 1593, 1568, 1538, 1510, 1500, 1489, 1372cm−1; HRMS m/z: Calcd for C26H26ClF3N4O [M+H]+: 426.2288, found: 426.2288.
1-(4′-Nitrophenyl)-3-((5R,8S)-8-isopropyl-5-methyl-4-phenyl-5,6,7,8-tetrahydroquinazolin-2-yl)urea (4s). White solid, Yield: 82%; HPLC purity: 97.3%; [α]D20 = + 19.2°(c = 0.10, CHCl3); m.p. 221.4–224.0°C; 1H NMR (500MHz, CDCl3) δ (ppm): 12.20 (s, 1H), 8.19 (d, J = 8.6Hz, 2H), 7.64 (d, J = 12.8Hz, 3H), 7.56–7.46 (m, 5H), 3.30 (m, 1H), 2.97 (m, 1H), 2.92–2.82 (m, 1H), 1.92–1.83 (m, 3H, C3-H), 1.82–1.77 (m, 1H), 1.17 (d, J = 7.0Hz, 3H), 0.92 (d, J = 7.0Hz, 3H), 0.81 (d, J = 6.8Hz, 3H); 13C NMR (126MHz, CDCl3) δ (ppm): 154.55, 151.75, 144.83, 142.96, 138.06, 129.44, 128.73, 128.47, 128.17, 128.04, 125.28, 118.79, 46.87, 29.98, 28.55, 27.71, 21.41, 20.75, 16.80, 16.26; IR (KBr) v: 3206, 3120, 3039, 2958, 2923, 2870, 1701, 1614, 1600, 1561, 1543, 1506, 1371cm−1; HRMS m/z: Calcd for C26H26ClF3N4O [M+H]+: 446.2187, found: 446.2188.
Cytotoxicity Assay
The antitumor activity of the target compounds and the positive control, 5-FU, were evaluated against the human cancer cell lines A549, Hela, MCF-7, and MGC-803 by MTT assay. The cell lines were grown on 96-well micro-plates at a density of 5 × 103 cells per well in DMEM (Dulbecco’s modified eagle medium) with 10% FBS (fetal bovine serum). The plates were incubated and maintained at 37°C with 5% CO2. The cells were then exposed to different concentrations of the synthesized compounds and 5-FU and incubated for another 48h. Control wells were formed by culture media with the maximum concentration of DMSO used in each assay (1‰) (Ruvinsky and Meyuhas, 2006). A total of 20µL of MTT was added and incubated for about 4h. The medium was thrown away and replaced by 150ml DMSO. The absorbance (OD) value was measured at 490nm using an enzyme labeling instrument. Each experiment was repeated at least three times to obtain the mean values (Hafez and EL-Gazzar, 2017).
Cell Cycle Analysis
The Hela cell line was treated with different concentrations of compound 4i. After incubation for 48h, the treated cells were washed twice with ice-cold phosphate buffer saline (PBS), fixed, and permeabilized with ice-cold 70% ethanol (−20°C) overnight. The cells were treated with 100µg/ml rnase A at 37°C for 30min, then washed with ice-cold PBS, and finally stained with 1mg/ml propidium iodide (PI) (BD, Pharmingen) in the dark at 4°C for 30min. The data were analyzed by the system software (Cell Quest; BD Biosciences, Becton Dickinson FACSAriaIII, New York, United States).
Apoptosis Analysis
Hela cells were seeded at a concentration of 2 × 106 cells/mL of the DMEM medium with 10% FBS on 6-well plates to the final volume of 2ml, and then treated with compound 4i at different concentrations for 48h. After 48h, the cells were collected and washed twice with PBS, then resuspended in 100µL 1 × binding buffer. The cells were subjected to 5µL of FITC Annexin V and 5µL propidium iodide (PI) staining using an annexin-V FITC apoptosis kit (BD, Pharmingen) and incubated for 30min at RT (25°C) in the dark. Then, 100µL 1× binding buffer was added. The apoptosis ratio was quantified by system software (Cell Quest; BD Biosciences, Becton Dickinson FACSAriaIII, New York, United States).
Network Pharmacology Analysis
Targets Prediction of 4i
The structure of 4i was imported into the database of PharmMapper Server (http://www.lilab-ecust.cn/pharmmapper/) for prediction (Yi et al., 2017). The results, whose scores of Norm Fit were greater than zero, were chosen as the predicted targets of 4i. The targets, including the gene names and gene ID, were further extracted using UniProtKB (http://www.uniprot.org) (Wang et al., 2017).
Collection of Potential Targets for Cervical Cancer Hela Cell Line
“Hela human cervical cancer Hela cell line” were used as keywords to retrieve cervical cancer Hela cell line associated targets in Online Mendelian Inheritance in Man database (OMIM, https://omim.org) (Bateman et al., 2021) and Genecards database (https://www.genecards.org) (Hamosh et al., 2005). On mapping cervical cancer Hela cell line associated targets and predicted targets of 4i, the potential targets of 4i for treatment of cervical cancer Hela cell line were finally obtained.
Construction and Analysis of Protein-Protein Interaction Network
The potential targets gene list was imported into the STRING database (https://string-db.org) (Stelzer et al., 2016), with the species as ‘‘Homo sapiens” and a confidence score greater than 0.4, to obtain the data of PPI and the visualized PPI network. The topological parameters of the PPI network were analyzed with the Network Analyzer tool of Cytoscape. The targets whose degrees were greater than 1.9 times median were selected as the core targets.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis
The biological functions of the target genes were assessed through gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis. The R package, “DOSE”, “clusterProfiler”, “enrichplot”, and “org.Hs.eg.db”, were used to perform the enrichment analyses. The bar graphs of results were drawn by the R package “ggplot2”.
Molecular Docking Analysis
Molecular docking was performed using AutoDock 4.2.6 software (ADT) according to the report (Szklarczyk et al., 2019). The structures of the key targets were downloaded from RCSB PDB database (http://www.rcsb.org/). All the PDB files of the targets were cleaned by removing small molecules, ions, and native ligand in the crystal using PyMOL software. The resulting PDB files were prepared by ADT to adding hydrogen atoms, calculating and adding Gasteigar charges, merging non-polar hydrogen atoms, and setting rotatable torsion bonds to afford protein files for docking. The structure of 4i was drawn and minimized energy using Chem3D 19.1. The torsional bonds of 4i were automatically set by the AUTOTORS module in ADT. The docked sites of the protein were set in the place of the native ligand. The binding energy between 4i and the targets was calculated using the AutoGrid program with a grid spacing of 0.375Å by the Lamarckian genetic algorithm as a searching method. The interaction between 4i and the amino acid residues of the targets was found by LigPlot+ 2.1 and visualized by PyMOL software. The reliability of the docking method was confirmed when the RMSD value, which is obtained from the docking result between native ligand and the target, is less than 2Å. The accessible surface area (ASA) was calculated from Accessible Surface Area and Accessibility Calculation for Protein (ver. 1.2) (http://cib.cf.ocha.ac.jp/bitool/ASA/). The loss in ASA (ΔASA) was calculated as follows: ΔASA = ASAUnbound protein-ASA Protein−ligand complex. If the ΔASA loses more than 10Å2, the residue is involved in binding (Miao et al., 2019).
Western Blot Analysis of PI3K/Akt Signaling Pathway Inhibitory Activity of 4i
Hela cells were seeded at a density of 3 × 106 cells per dish and attached for 8h, and then treated with compounds in concentrations of 0, 5, 10, and 20μM for 24h. After that, cells were collected, washed with cold PBS, and lysed with lysis buffer (100mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol) on ice for 30min. Protein concentrations were detected using the bicinchoninc acid procedure (BCA) method (Beyotime Institute of Biotechnology, Haimen, China). Proteins were electrophoresed using sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE Bio-Rad, California, United States) and transferred electrophoretically to membranes. The membranes were blocked with 5% non-fat milk at room temperature for 1h and were incubated with primary antibodies overnight at 4°C. The next day, membranes were washed and incubated with the appropriate fluorescence-conjugated secondary antibodies at room temperature for 1h. Finally, membranes were washed and developed by the addition of enhanced chemiluminescence (ECL) substrate (Thermo Fisher Scientific, Rockford, United States). Proteins were visualized using the intelligent gel imaging system IBright FL1000 (Thermofisher, Rockford, United States). Primary antibodies were as follows: anti-phospho-4EBP1, anti-4EBP1, anti-mTOR, anti-phospho-mTOR, anti-Akt, anti-phospho-Akt, anti-PI3K(p85), anti-phospho-PI3K(p85), and anti-α-tubulin, which were purchased from Cell Signaling Technology (Cell Signaling Technology, Boston, MA, United States).
Statistical Analyses
Statistical analysis was conducted on SPSS 22.0 (SPSS, Chicago, IL, United States), with a data-processing method of independent-sample t test. Comparisons between groups were statistically analyzed, and p < 0.05 was considered to be statistically significant.
Results and Discussion
Chemistry
The synthetic approaches adopted to afford the target compounds are outlined in Scheme 1, L-menthone-derived α, β-unsaturated ketone 2 was prepared by Claisen-Schmidt condensation reaction of L-menthone 1 with benzaldehyde. Then, L-menthone-derived pyrimidine 3 was prepared by cyclization of compound 2 with guanidine hydrochloride in the presence of K2CO3 in ethanol. Finally, a series of L-menthone-derived pyrimidine-urea compounds 4a-4s were synthesized by nucleophilic addition reaction of compound 3 with substituted phenyl isocyanates.
The structures of all the target compounds were characterized by FT-IR, 1H NMR, 13C NMR, and HRMS. The spectra can be found in the Supplementary Material. In the FT-IR spectra of the target compounds, the absorption bands at about 3200cm−1 were attributed to the stretching vibrations of N-H. The characteristic absorption bands at about 1690cm−1 were assigned to the stretching vibrations of C=O. The characteristic absorption bands at about 1581–1620cm−1 were assigned to C=N in the pyrimidine moiety. In the 1H-NMR spectra, the protons of a benzene ring showed signals at 6.88–8.19ppm. The N–H protons exhibited signals at 11.08–12.30ppm and 7.51–8.13ppm, respectively. The protons of the menthone moiety displayed signals in the range of 0.77–3.12ppm. The 13C NMR spectra of the target compounds showed peaks for the carbon atom of C=O at about 155.00 ppm, carbon atoms of the benzene ring, and the unsaturated carbons at 102.92–158.02ppm. The other saturated carbons of menthone displayed signals in the region of 16.15–46.93ppm. Their molecular weights were confirmed by HRMS.
Cytotoxicity Measurement
The in vitro antitumor activity of L-menthone-derived pyrimidine-urea compounds 4a-4s against four human cancer cell lines (A549 human lung adenocarcinoma cell, Hela human cervical cancer cell line, MCF-7 human breast cancer cell line, andMGC-803 human gastric cancer cell line) was evaluated by MTT (methyl thiazolytetrazolium) assay and compared with the positive control 5-fluorouracil (5-FU) in each panel. The results are shown in Table 1.
It was found that 7, 16, 14, and 9 compounds exhibited better antitumor activity than the positive control, 5-FU, against A549, Hela, MCF-7, and MGC-803, respectively, while the target compounds showed more effective inhibitory activity against MGC-803 than other tested cell lines, which were inhibited by the target compounds in a similar level. Compounds 4m, 4i, 4s, and 4g showed the best IC50 values of 18.68 ± 1.53µM, 6.04 ± 0.62µM, 19.09 ± 0.49µM, and 3.21 ± 0.67µM against A549, Hela, MCF-7, and MGC-803, respectively. Compounds 4i and 4g showed good dual antitumor activity against Hela and MGC-803.
In addition, the compounds with para substituent showed good activity in the greater probability, especially the halogen substituted compounds. It is indicated that the lone electron pair on the para substituent might act as an important role in tumor inhibition. For the further action mechanism study, compound 4i was employed as a representative.
Cell Cycle Analysis
To study the possible role of compound 4i in the tumor growth inhibition, different concentrations of compound 4i were treated with Hela cells for 48h. The cell cycle distribution was investigated by flow cytometric analysis following the staining of DNA with propidium iodide (PI). It was observed that the G0/G1 phase cells were gradually decreased from 72.46% (0μM) to 67.05% (10μM, p = 0.043), and the S phase cells changed unobviously, while the G2/M phase cells gradually increased from 18.45% (0μM) to 22.83% (10μM) (Figure 2). Thus, these results suggested that the target compound 4i might arrest the Hela cells in the G2/M phase.
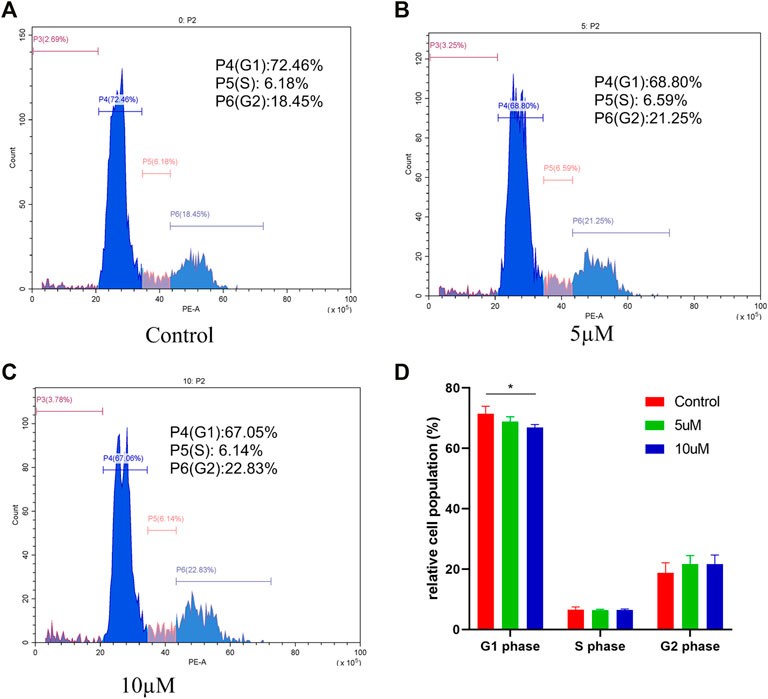
FIGURE 2. Effect of compound 4i (0°μM (A), 5°μM (B), and 10°μM (C)) on the cell cycle of human cervical cancer Hela cell. The graph (D) presents cells in the G0/G1, S, and G2/M phases in the Hela cell line using PI staining after exposure to compound 4i at (0°μM, 5°μM, and 10°μM) for 48 h. Data are expressed as the mean and SD from three independent experiments; *p < 0.05.
Compound 4i Induces Apoptosis in Hela Cells
To investigate whether compound 4i could induce cell apoptosis, Hela cells were treated with compound 4i in different concentrations for 48h. The Annexin V-FITC/PI dual staining assay was carried out by flow cytometry. The results are illustrated in Figure 3. The percentage of all apoptotic cells (early and late apoptotic cells, lower right quadrant and upper right quadrant, AV+/PI, respectively) significantly increased from 0.07% (0μM, early, 0.00%; later, 0.07%) to 26.22% (5μM, early, 16.57%, p = 0.003; later 9.65%, p = 0.007) and 53.78% (10μM, early, 37.81%, p = 0.004; later 15.97%, p = 0.001), while the increase in the 10µM treatment group was much greater than that in the 5µM treatment group (early, p = 0.015; later, p = 0.024). These results clearly confirmed that compound 4i effectively induced apoptosis in Hela cells in a dose-dependent manner.
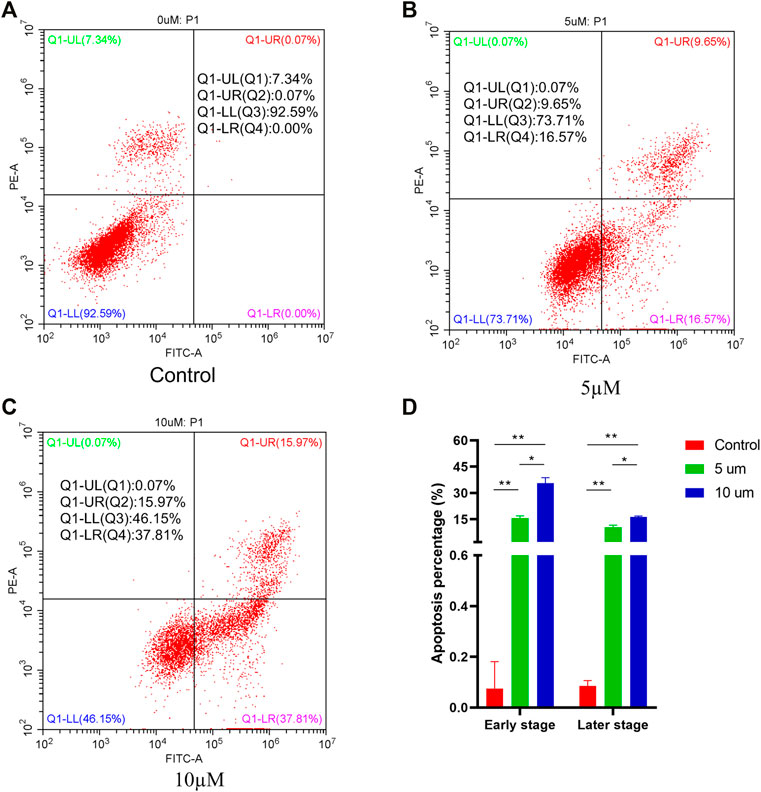
FIGURE 3. Apoptosis ratio detection of compound 4i (0°μM (A), 5°μM (B), and 10 (C) μM) by flow cytometry. The graph (D) presents cell apoptosis in an early stage and a later stage in the Hela cell line using PI staining after exposure to compound 4i at (0°μM, 5°μM, and 10°μM) for 48 h. Data are expressed as the mean and SD from three independents experiments; *P < 0.05, **P < 0.01.
Preliminary Exploration of the Mechanism of Compound 4i Against Hela Cells Based on Network Pharmacology and Molecular Docking Technology
Potential Targets of 4i Against Hela Cell Line
A total of 293 targets of compound 4i (Supplementary Table S1) were obtained by PharmMapper Server. A total of 834 Hela cell line associated targets (Supplementary Table S2) were collected by OMIM database and Genecards database after eliminating duplicates. Finally, 59 potential targets were obtained by overlapping the two sets (Supplementary Table S3).
Construction and Analysis of Protein-Protein Interaction Network
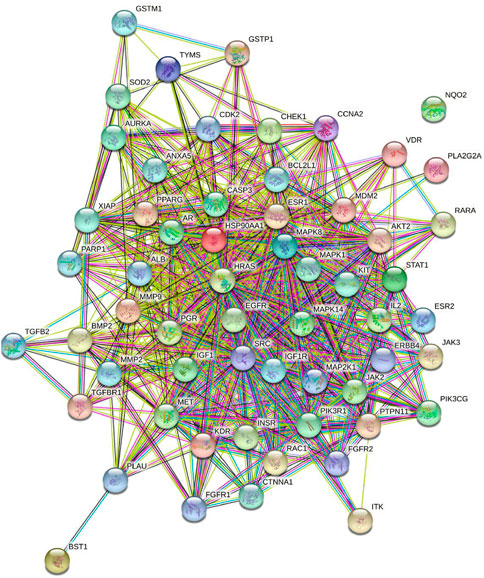
A total of 59 targets was imported into the STRING database to obtain the data of PPI and the visualized PPI network. As shown in Figure 4, the nodes in the network represent the potential targets, and the edges represent the interaction between the targets. The network was comprised of 59 nodes and 708 edges, and the average node degree was 24.4. The targets with the degree greater than 1.9 times median, EGFR, HRAS, and MAPK1, were selected as the core targets (Table 2).
Results of GO and KEGG Pathway Enrichment Analysis
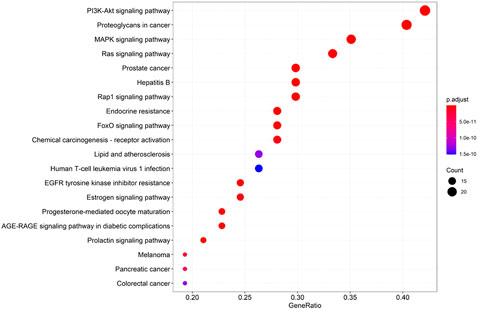
GO and KEGG pathway enrichment analysis were performed using the 59 potential targets in R packages, and the GO terms and KEGG pathways with p-value < 0.05 were significantly enriched (Supplementary Tables S4 and S5). The top 20 GO terms of biological process, molecular function, and cellular components were shown in Figure 5. The KEGG pathway enrichment analysis indicated that 54 of the 59 targets (91.52%) were enriched in 141 pathways. The top 20 pathways were shown in Figure 6. There are 6 cancer pathways, 5 signal transduction pathways, 3 endocrine system pathways, 2 antineoplastic drug resistance pathways, 2 viral infectious pathways, 1 endocrine and metabolic disease pathway, and 1 cardiovascular disease pathway (Supplementary Table S6). This result indicated that compound 4i against cancer may through regulating these pathways.
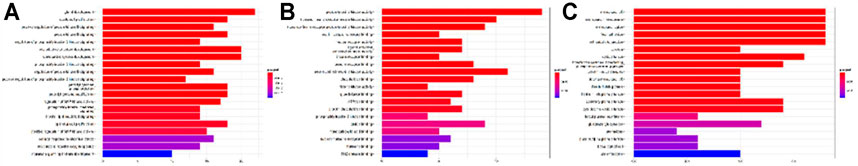
FIGURE 5. GO enrichment of 4i targets. (A) Biological process. (B) Molecular function. (C) Cellular components.
Furthermore, there are 12 proteins involved in the top 20 pathways more than 15 times, including MAP2K1(19), KDR (19), IGF1R (19), HRAS (19), IGF1 (19), FGFR1 (19), Akt2 (19), ESR1 (19), ERBB4(19), EGFR (19), CASP3(19), and MAPK1(16), suggesting that these targets may be the key proteins in the enriched pathways.
Molecular Docking Analysis
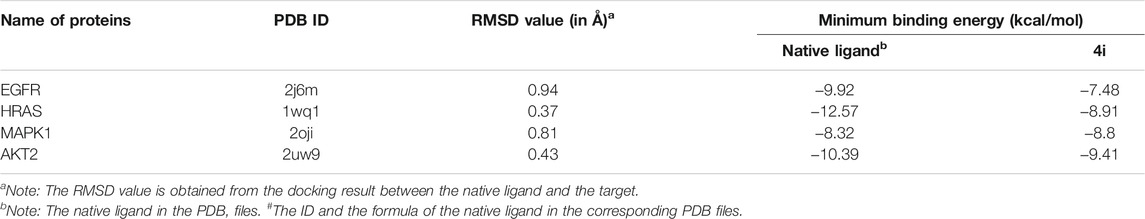
Proteins, EGFR, HRAS, and MAPK1, are the overlapping targets of the core targets in PPI network analysis and the key proteins in the top 20 pathways. Activated EGFR, HRAS, and MAPK1 can stimulate the activation of PI3K/Akt signaling pathway. Akt2 is a member protein in the pathway of PI3K-Akt signaling pathway which is the first pathway in the KEGG pathway enrichment. Thus, these four proteins, EGFR, HRAS, MAPK1, and Akt2, were studied in molecular docking. All the RMSD value in the docking resulted in less than 2Å, which is considered a good prediction for computed ligand-protein conformation and indicated the docking method is believable (García-Godoy et al., 2016).
Many reports show that the overexpression or the mutational activation of these four targets was found in cancer cells and provides strong growth and survival signals to cancer cells (Vajdos et al., 2001; Yun et al., 2007; Zhu et al., 2011; Rampias et al., 2014; Sharma et al., 2019; Jiang et al., 2021). For regulating the expression of the targets, some small molecules which could bind with the specific region at the target proteins were found as inhibitors for cancer therapy (Roskoski, 2018; Alzahrani, 2019). Thus, in this work, the ATP-binding sites, which contain the native ligand as inhibitor of the targets, were chosen as docking sites.
In the results, the predicted minimum binding energies of compound 4i with the target proteins are less than or equal to −7.48kcal/mol. Particularly, the minimum binding energy of compound 4i with MAPK1 is lower than that of the native ligand. These low binding energies indicated that compound 4i may form stable binding with the docking sites and might act as a well inhibitor against EGFR, HRAS, MAPK1, and Akt2, respectively (Table 3).
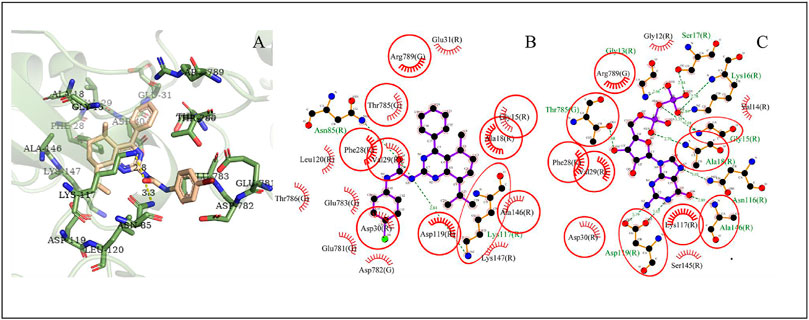
The possible interaction mode between the residues of EGFR and compound 4i were shown in Figure 7. Five of 12 residues (41.67%) are overlapping with the interacting residues of the native ligand, including an overlapping residue, Asp855, forming a hydrogen bond with urea moiety at N atom, which is also forming a hydrogen bond with binding site residue Asp855. Arg841 with the maximum ΔASA (48.158Å2) is seen as the key interacting residue (Supplementary Table S7).
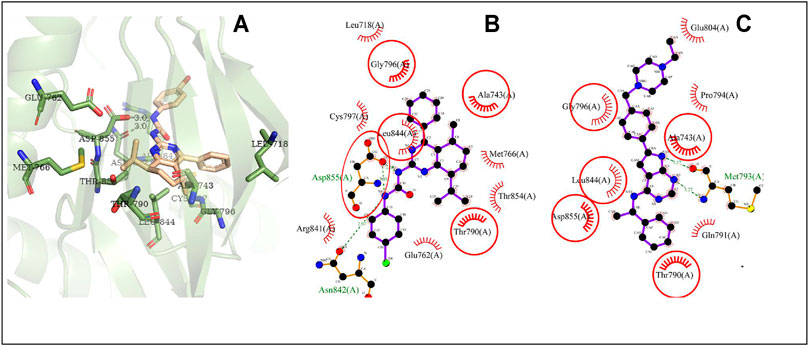
FIGURE 7. The conformation of 4i docked in EGFR. Note (A) 3D docking conformation; (B) 2D docking conformation; (C) 2D conformation of native ligand and residues, the same as below.
The possible interaction mode between the residues of HRAS and compound 4i were shown in Figure 8. Ten of 18 residues (55.56%) are overlapping with the interacting residues of the native ligand, including an overlapping residue, Lys117, forming a hydrogen bond with urea moiety at O atom, which is also forming a hydrogen bond with binding site residue Asn85. Lys117 with the maximum ΔASA (48.206Å2) is seen as the key interacting residue.
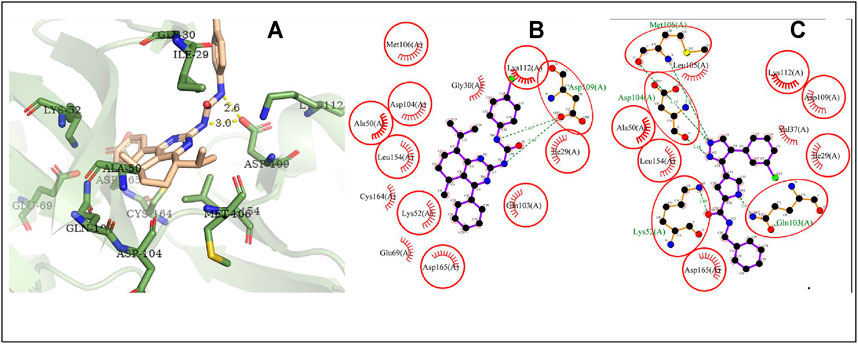
The possible interaction mode between the residues of MAPK1 and compound 4i were shown in Figure 9. Ten of 13 residues (76.92%) are overlapping with the interacting residues of the native ligand, including an overlapping residue, Asp109, forming two hydrogen bonds with the 2N atoms of urea moiety. Ile29 with the maximum ΔASA (46.449Å2) is seen as the key interacting residue.
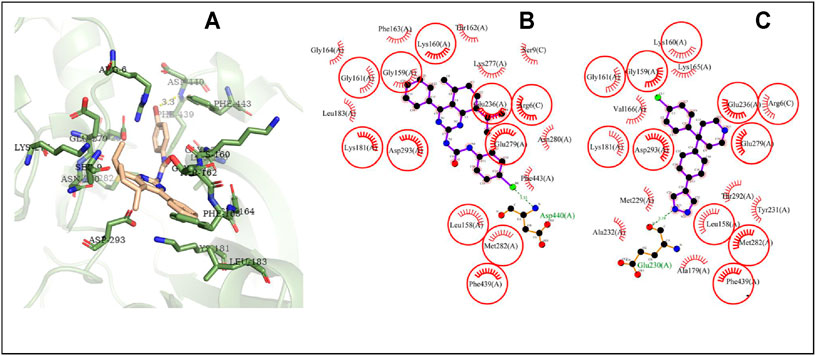
The possible interaction mode between the residues of Akt2 and compound 4i were shown in Figure 10. Eleven of 20 residues (55.00%) are overlapping with the interacting residues of the native ligand. The binding site residue Asp440 formed a hydrogen bond with the Br atom. Asp293 with the maximum ΔASA (48.293Å2) is seen as the key interacting residue.
The interaction mode study showed that the higher overlapping ratio of compound 4i to the native ligand interacting residues and the higher average ΔASA of compound 4i might relate to low minimum binding energy. A hydrogen bond is also a positive factor for ligand-protein accommodation. In the docking results, the hydrogen bonds’ interaction almost associates to urea moiety, indicating that introduction of urea moiety is favorable to form new compounds with good protein-binding ability. In addition, the Br atom of compound 4i could form a hydrogen bond with the Akt2 binding site residue that might be a factor of the para halogen substituted compounds showing good antitumor activity.
PI3K/Akt/mTOR Signaling Pathway Inhibitory Activity of 4i
The inhibitory activity of compound 4i against PI3K/Akt/mTOR signaling pathway, the first pathway in the KEGG pathway enrichment analysis and its downstream protein, was investigated. The proteins’ expressions of PI3K(p85), p-PI3K(p85), AKT, p-AKT, mTOR, p-mTOR, 4EBP1, p-4EBP1, and α-tubulin in Hela cells were detected by Western blot method. The result is shown in Figure 11. In the Hela cells treated by compound 4i, the total PI3K(p85), AKT, mTOR, 4EBP1, and α-tubulin were almost not affected by the treatments, while the expression of the phosphorylated proteins, p-PI3K(p85), p-AKT, p-mTOR, and p-4EBP1 suffered significant decrease.
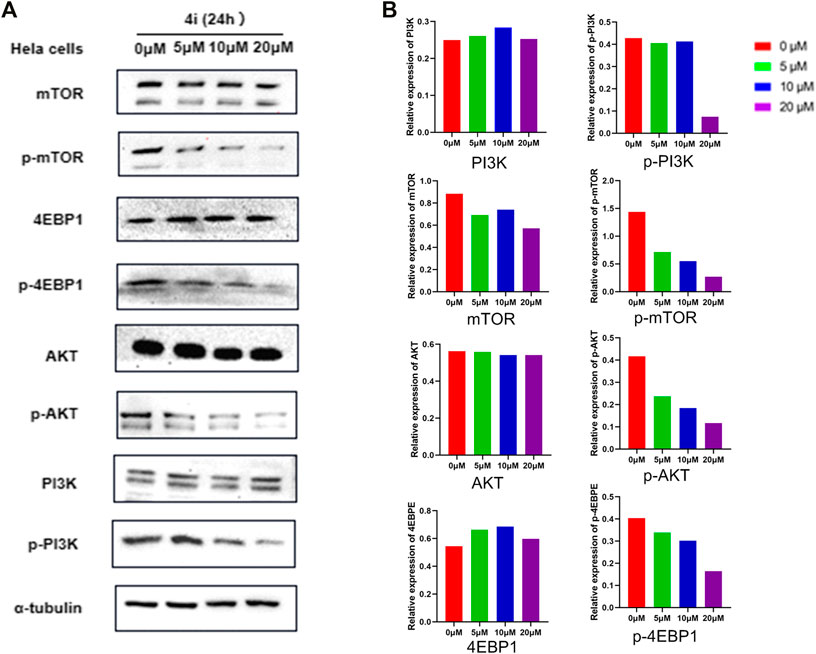
FIGURE 11. Inhibitory effects of compound 4i in the Hela cell line according to Western blot analysis. (A) The expression levels of the PI3K,p-PI3K, mTOP, p-mTOR, AKT, p-AKT, 4EBPE, and p-4EBPE in the HeLa cell which were treated with (0°μM, 5°μM, 10°μM, and 20°μM of 4i) for 24 h,were examined by Western Blot (B).
In PI3K/Akt/mTOR signaling pathway, the phosphorylated PI3K (p-PI3K), some of which were constructed by a catalytic subunit (p110) and a regulatory subunit (p85), could phosphorylate PIP2 to convert it into PIP3. PIP3 could activate Akt through phosphorylation, and further activate the mTOR as phosphorylated mTOR. The activated mTOR could deactivate its downstream effectors 4EBP1, which could bind with eIF4E to inhibit the cells’ proliferation, growth, and protein synthesis, through phosphorylation (Condon and Sabatini, 2019; Rehan and Bajouh, 2019; Sun et al., 2019). Thus, the decreased expression of the phosphorylated PI3K, Akt, mTOR, and downstream effectors 4EBP1 indicated that compound 4i could inhibit PI3K/Akt/mTOR signaling pathways significantly.
Conclusion
In summary, a series of novel menthone derivatives bearing pyrimidine and urea moieties were synthesized and identified by FTIR, NMR, and HRMS. All the synthesized compounds were evaluated for their cytotoxic effects against A549, Hela, MCF-7, and MGC-803 cell lines by standard MTT assay. The results revealed that most of the target compounds exhibited better antitumor activity than the positive control, 5-FU. Compounds 4m, 4i, 4s, and 4g showed the best IC50 values against A549, Hela, MCF-7, and MGC-803, respectively. In particular, compounds 4i and 4g showed good dual antitumor activity against Hela and MGC-803. The action mechanistic studies, using compound 4i as a representative, revealed that compound 4i could markedly induce Hela cell apoptosis in a dose-dependent manner and arrested them in the G2/M phase. Network pharmacology prediction showed that compound 4i might against Hela cells through regulating the expression and activation of the core targets, EGFR, HRAS, MAPK1, and Akt2. In KEGG pathway enrichment, PI3K/Akt signaling pathway was evaluated as the first pathway for compound 4i against Hela cells. The Western blot assay confirmed that compound 4i could inhibit PI3K/Akt signaling pathway, including the downstream protein mTOR and downstream effectors 4EBP1, significantly. Molecular docking shows that compound 4i was able to interact efficiently with the ATP-binding site of EGFR, HRAS, MAPK1, and Akt2. The urea moiety and para substituent halogen acted as important roles for ligand-protein stabling. These results indicated that the synthetic strategy in this study is feasible. The compound 4i might serve as a leading compound for potent antitumor agents such as PI3K/Akt/mTOR signaling pathway inhibitor.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
MH, WD, and GL carried out design, synthesis, characterization, and analyzed the data. NC and XW conducted the in vitro anticancer studies. NC conducted the cell cycle, cell apoptosis, and Western blot assay. WD and GL provided overall supervision and guidance. All the authors discussed all the results and helped with writing.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31870556) and Guangxi Natural Science Foundation (No. 2019GXNSFAA245057).
Conflict of Interest
MH was employed by Guangxi Research Institute of Chemical Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.815531/full#supplementary-material
References
Agarwal, G., Carcache, P. J. B., Addo, E. M., and Kinghorn, A. D. (2020). Current Status and Contemporary Approaches to the Discovery of Antitumor Agents from Higher Plants. Biotechnol. Adv. 38, 107337–107399. doi:10.1016/j.biotechadv.2019.01.004
Alzahrani, A. S. (2019). PI3K/Akt/mTOR Inhibitors in Cancer: At the Bench and Bedside. Semin. Cancer Biol. 59, 125–132. doi:10.1016/j.semcancer.2019.07.009
Bateman, A., Martin, M., Orchard, S., Magrane, M., Agivetova, R., Ahmad, S., et al. (2021). UniProt: the Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi:10.1093/nar/gkaa1100
Bhalla, Y., Gupta, V. K., and Jaitak, V. (2013). Anticancer Activity of Essential Oils: A Review. J. Sci. Food Agric. 93, 3643–3653. doi:10.1002/jsfa.6267
Boni, G. C., Feiria, S. N. B. D., Santana, P. D. L., Anibal, P. C., Boriollo, M. F. G., Buso-Ramos, M. M., et al. (2016). Antifungal and Cytotoxic Activity of Purified Biocomponents as Carvone, Menthone, Menthofuran and Pulegone from Mentha Spp. Afr. J. Plant Sci. 10, 203–210. doi:10.5897/AJPS2016.1454
Bukowski, K., Kciuk, M., and Kontek, R. (2020). Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Ijms 21, 3233–3257. doi:10.3390/ijms21093233
Chen, D., Sun, J., Lian, S., Liu, Z., Sun, K., Liu, W., et al. (2014). Dual pH-Responsive and CD44 Receptor Targeted Multifunctional Nanoparticles for Anticancer Intracellular Delivery. J. Nanopart. Res. 16, 2743–2479. doi:10.1007/s11051-014-2743-1
Chen, N.-Y., Xie, Y.-L., Lu, G.-D., Ye, F., Li, X.-Y., Huang, Y.-W., et al. (2020a). Synthesis and Antitumor Evaluation of (Aryl)methyl-amine Derivatives of Dehydroabietic Acid-Based B Ring-Fused-Thiazole as Potential PI3K/AKT/mTOR Signaling Pathway Inhibitors. Mol. Divers. 25, 967–979. doi:10.1007/s11030-020-10081-7
Chen, S., Rehman, S. K., Zhang, W., Wen, A., Yao, L., and Zhang, J. (2010). Autophagy Is a Therapeutic Target in Anticancer Drug Resistance. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1806, 220–229. doi:10.1016/j.bbcan.2010.07.003
Chen, Y., Yang, L., Qiao, H., Cheng, Z., Xie, J., Zhou, W., et al. (2020b). Discovery of New Thieno[3,2-D]pyrimidine Derivatives Targeting EGFRL858R/T790M NSCLCs by the Conformation Constrained Strategy. Eur. J. Med. Chem. 199, 112388. doi:10.1016/j.ejmech.2020.112388
Condon, K. J., and Sabatini, D. M. (2019). Nutrient Regulation of mTORC1 at a Glance. J. Cel Sci. 132, jcs222570–222575. doi:10.1242/jcs.222570
Drexel, M., Kirchmair, J., and Santos-Sierra, S. (2019). INH14, a Small-Molecule Urea Derivative, Inhibits the IKKα/β-dependent TLR Inflammatory Response. ChemBioChem 20, 710–717. doi:10.1002/cbic.201800647
Gao, F., Zhang, X., Wang, T., and Xiao, J. (2019). Quinolone Hybrids and Their Anti-cancer Activities: an Overview. Eur. J. Med. Chem. 165, 59–79. doi:10.1016/j.ejmech.2019.01.017
Gao, X., Ren, Y., Huang, J., and Pan, A. (2018). Molecular Modeling Studies of Urea-Morpholinopyrimidine Analogues as Dual Inhibitors of mTORC1 and mTORC2 Using 3D-QSAR, Topomer CoMFA and Molecular Docking Simulations. Lddd 15, 1026–1035. doi:10.2174/1570180815666180607090956
García-Godoy, M., López-Camacho, E., García-Nieto, J., Nebro, A., and Aldana-Montes, J. (2016). Molecular Docking Optimization in the Context of Multi-Drug Resistant and Sensitive EGFR Mutants. Molecules 21, 1575–1588. doi:10.3390/molecules21111575
Hafez, H. N., and EGazzar, A.-R. B. A. (2017). Synthesis and Evaluation of Antitumor Activity of New 4-substituted Thieno[3,2-D]pyrimidine and Thienotriazolopyrimidine Derivatives. Acta Pharmaceut 67, 527–542. doi:10.1515/acph-2017-0039
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A., and Mckusick, V. A. (2005). Online Mendelian Inheritance in Man (OMIM), a Knowledgebase of Human Genes and Genetic Disorders. Nucleic Acids Res. 33, D514–D517. doi:10.1093/nar/gki033
Han, F., Lin, S., Liu, P., Tao, J., Yi, C., and Xu, H. (2017). Synthesis and Structure-Activity Relationships of PI3K/mTOR Dual Inhibitors from a Series of 2-Amino-4-Methylpyrido[2,3-D]pyrimidine Derivatives. Bioorg. Med. Chem. Lett. 24, 4538–4541. doi:10.1016/j.bmcl.2014.07.073
Han, F., Lin, S., Liu, P., Liu, X., Tao, J., Deng, X., et al. (2015). Discovery of a Novel Series of Thienopyrimidine as Highly Potent and Selective PI3K Inhibitors. ACS Med. Chem. Lett. 6, 434–438. doi:10.1021/ml5005014
He, T., Tang, Q., Gou, L., Liu, J. W., Yang, J., Yu, L., et al. (2013). Study on Effect and Mechanism of Volatile Oil of Schizonepetae Herba and its Essential Components against Influenza Virus. Cjcmm 38, 1772–1777. doi:10.4268/cjcmm20131125
Heffron, T. P., Berry, M., Castanedo, G., Chang, C., Chuckowree, I., Dotson, J., et al. (2010). Identification of GNE-477, a Potent and Efficacious Dual PI3K/mTOR Inhibitor. Bioorg. Med. Chem. Lett. 20, 2408–2411. doi:10.1016/j.bmcl.2010.03.046
Horishny, V. Y., Zadorozhnii, P. V., Horishnia, I. V., and Matiychuk, V. S. (2020). Synthesis, Anti-inflammatory Activity and Molecular Docking Studies of 1,4,5,6-Tetrahydropyrimidine-2-Carboxamides. Pharm. Sci. 27, 353–365. doi:10.34172/PS.2020.100
Jiang, T., Xia, Y., Lv, J., Li, B., Li, Y., Wang, S., et al. (2021). A Novel Protein Encoded by circMAPK1 Inhibits Progression of Gastric Cancer by Suppressing Activation of MAPK Signaling. Mol. Cancer 20, 66–83. doi:10.1186/s12943-021-01358-y
Kang, S. J., Lee, J. W., Chung, S. H., Jang, S. Y., Choi, J., Suh, K. H., et al. (20192019). Synthesis and Anti-tumor Activity of Imidazopyrazines as TAK1 Inhibitors. Eur. J. Med. Chem. 163, 660–670. doi:10.1016/j.ejmech.2018.12.025
Kikowska, M., Kalemba, D., Dlugaszewska, J., and Thiem, B. (2020). Chemical Composition of Essential Oils from Rare and Endangered Species-eryngium Maritimum L. And E. Alpinum L. Plants 9, 417–431. doi:10.3390/plants9040417
Kilic-Kurt, Z., Aka, Y., and Kutuk, O. (2020). Novel Pyrrolopyrimidine Derivatives Induce P53-independent Apoptosis via the Mitochondrial Pathway in colon Cancer Cells. Chemico-Biological Interactions 330, 109236–109247. doi:10.1016/j.cbi.2020.109236
Kumar, R., Shin, W. S., Sunwoo, K., Kim, W. Y., Koo, S., Bhuniya, S., et al. (2015). Small Conjugate-Based Theranostic Agents: an Encouraging Approach for Cancer Therapy. Chem. Soc. Rev. 44, 6670–6683. doi:10.1039/c5cs00224a
Lafzi, F., Kilic, D., Yildiz, M., and Saracoglu, N. (2021). Design, Synthesis, Antimicrobial Evaluation, and Molecular Docking of Novel Chiral Urea/thiourea Derivatives Bearing Indole, Benzimidazole, and Benzothiazole Scaffolds. J. Mol. Struct. 1241, 130566–130579. doi:10.1016/j.molstruc.2021.130566
Li, F.-Y., Wang, X., Duan, W.-G., and Lin, G.-S. (2017). Synthesis and In Vitro Anticancer Activity of Novel Dehydroabietic Acid-Based Acylhydrazones. Molecules 22, 1087–1096. doi:10.3390/molecules22071087
Li, F., Huang, L., Zhou, X., Li, Q., Ma, X., Duan, W., et al. (2020). Synthesis and Cytotoxicity Evaluation of Dehydroabietic Acid Derivatives Bearing Nitrate Moiety. Chin. J. Org. Chem. 40, 2845–2854. doi:10.6023/cjoc202003062
Li, W., Chu, J., Fan, T., Zhang, W., Yao, M., Ning, Z., et al. (2019). Design and Synthesis of Novel 1-Phenyl-3-(5-(pyrimidin-4-Ylthio)-1,3,4-Thiadiazol- 2-yl)urea Derivatives with Potent Anti-CML Activity throughout PI3K/AKT Signaling Pathway. Bioorg. Med. Chem. Lett. 29, 1831–1835. doi:10.1016/j.bmcl.2019.05.005
Liu, Y., Yin, Y., Zhang, Z., Li, C. J., Zhang, H., Zhang, D., et al. (2017). Structural Optimization Elaborates Novel Potent Akt Inhibitors with Promising Anticancer Activity. Eur. J. Med. Chem. 138, 543–551. doi:10.1016/j.ejmech.2017.06.067
Miao, X.-d., Zheng, L.-j., Zhao, Z.-z., Su, S.-l., Zhu, Y., Guo, J.-m., et al. (2019). Protective Effect and Mechanism of Boswellic Acid and Myrrha Sesquiterpenes with Different Proportions of Compatibility on Neuroinflammation by LPS-Induced BV2 Cells Combined with Network Pharmacology. Molecules 24, 3946–3461. doi:10.3390/molecules24213946
Montenegro, I., Said, B., Godoy, P., Besoain, X., Parra, C., Díaz, K., et al. (2020). Antifungal Activity of Essential Oil and Main Components from Mentha Pulegium Growing Wild on the Chilean central Coast. Agronomy 10, 254–260. doi:10.3390/agronomy10020254
Mosbah, H., Chahdoura, H., Mannai, A., Snoussi, M., Aouadi, K., Abreu, R. M. V., et al. (2018). Biological Activities Evaluation of Enantiopure Isoxazolidine Derivatives: In Vitro, In Vivo and In Silico Studies. Appl. Biochem. Biotechnol. 187, 1113–1130. doi:10.1007/s12010-018-2868-2
Ni, Y., Gopalsamy, A., Cole, D., Hu, Y., Denny, R., Ipek, M., et al. (2011). Identification and SAR of a New Series of Thieno[3,2-D]pyrimidines as Tpl2 Kinase Inhibitors. Bioorg. Med. Chem. Lett. 21, 5952–5956. doi:10.1016/j.bmcl.2011.07.069
Papadimitrakopoulou, V. (2012). Development of PI3K/AKT/mTOR Pathway Inhibitors and Their Application in Personalized Therapy for Non-smalL-cell Lung Cancer. J. Thorac. Oncol. 7, 1315–1326. doi:10.1097/JTO.0b013e31825493eb
Qi, M., Zou, S., Guo, C., Wang, K., Yu, Y., Zhao, F., et al. (2018). Enhanced In Vitro and In Vivo Anticancer Properties by Using a Nanocarrier for Co-delivery of Antitumor Polypeptide and Curcumin. J. Biomed. Nanotechnol. 14, 139–149. doi:10.1166/jbn.2018.2479
Rampias, T., Giagini, A., Siolos, S., Matsuzaki, H., Sasaki, C., Scorilas, A., et al. (2014). RAS/PI3K Crosstalk and Cetuximab Resistance in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 20, 2933–2946. doi:10.1158/1078-0432.ccr-13-2721
Rehan, M., and Bajouh, O. S. (2019). Virtual Screening of Naphthoquinone Analogs for Potent Inhibitors against the Cancer‐signaling PI3K/AKT/mTOR Pathway. J. Cel. Biochem. 120, 1328–1339. doi:10.1002/jcb.27100
Roskoski, R. (2018). Small Molecule Inhibitors Targeting the EGFR/ErbB Family of Protein-Tyrosine Kinases in Human Cancers. Pharmacol. Res. 139, 395–411. doi:10.1016/j.phrs.2018.11.014
Ruvinsky, I., and Meyuhas, O. (2006). Ribosomal Protein S6 Phosphorylation: from Protein Synthesis to Cell Size. Trends Biochem. Sci. 31, 342–348. doi:10.1016/j.tibs.2006.04.003
Seca, A., and Pinto, D. (2018). Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Ijms 19, 263–284. doi:10.3390/ijms19010263
Sharma, P. C., Bansal, K. K., Sharma, A., Sharma, D., and Deep, A. (2019). Thiazole-containing Compounds as Therapeutic Targets for Cancer Therapy. Eur. J. Med. Chem. 188, 112016–112116. doi:10.1016/j.ejmech.2019.112016
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer Statistics, 2018. CA: A Cancer J. Clinicians 68, 7–30. doi:10.3322/caac.21442
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinformatics 54 (1), 30133–3033. doi:10.1002/cpbi.5
Sun, X., Wei, W., Ren, J., Liang, Y., Wang, M., Gui, Y., et al. (2019). Inhibition of 4E-BP1 Phosphorylation Promotes Tubular Cell Escaping from G2/M Arrest and Ameliorates Kidney Fibrosis. Cell. Signal. 62, 109331. doi:10.1016/j.cellsig.2019.05.016
Sun, Z., Wang, H., Wang, J., Zhou, L., and Yang, P. (2014). Chemical Composition and Anti-inflammatory, Cytotoxic and Antioxidant Activities of Essential Oil from Leaves of Mentha Piperita Grown in china. Plos One 9, e114767–e114781. doi:10.1371/journal.pone.0114767
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-wide Experimental Datasets. Nucleic Acids Res. 47, D607–D613. doi:10.1093/nar/gky1131
Tahghighi, A., Maleki-Ravasan, N., Dinparast Djadid, N., Alipour, H., Ahmadvand, R., Karimian, F., et al. (2019). GC-MS Analysis and Anti-mosquito Activities of Juniperus Virginiana Essential Oil against Anopheles stephensi (Diptera: Culicidae). Asian Pac. J. Trop. Biomed. 9, 168–175. doi:10.4103/2221-1691.256730
Vajdos, F. F., Ultsch, M., Schaffer, M. L., Deshayes, K. D., Liu, J., Skelton, N. J., et al. (2001). Crystal Structure of Human Insulin-like Growth Factor-1: Detergent Binding Inhibits Binding Protein Interactions. Biochemistry 40, 11022–11029. doi:10.1021/bi0109111
Wang, X., Duan, W., Lin, G., Li, B., Chen, M., and Lei, F. (2021). Synthesis, 3D-QSAR and Molecular Docking Study of nopoL-Based 1,2,4-Triazole-Thioether Compounds as Potential Antifungal Agents. Front. Chem. 9, 757584. doi:10.3389/fchem.2021.757584
Wang, X., Pang, F.-H., Huang, L., Yang, X.-P., Ma, X.-L., Jiang, C.-N., et al. (2018). Synthesis and Biological Evaluation of Novel Dehydroabietic Acid-Oxazolidinone Hybrids for Antitumor Properties. Ijms 19, 3116–3132. doi:10.3390/ijms19103116
Wang, X., Shen, Y., Wang, S., Li, S., Zhang, W., Liu, X., et al. (2017). PharmMapper 2017 Update: a Web Server for Potential Drug Target Identification with a Comprehensive Target Pharmacophore Database. Nucleic Acids Res. 45, W356–W360. doi:10.1093/nar/gkx374
Wang, Y., Chen, X., Yan, Y., Zhu, X., Liu, M., and Liu, X. (2020). Discovery and SARs of 5-Chloro-N4-Phenyl-N2-(pyridin-2-Yl)pyrimidine-2,4-Diamine Derivatives as Oral Available and Dual CDK 6 and 9 Inhibitors with Potent Antitumor Activity. J. Med. Chem. 63, 3327–3347. doi:10.1021/acs.jmedchem.9b02121
Xiong, Q., Lin, X., Liu, J., Bi, L., and Bao, X. (2012). Synthesis and Bioactivities of Novel 1,2,4-Triazolo[1,5-A]pyrimidine Derivatives Containing 1,2,4-Triazole-5-Thione Schiff Base Unit. Chin. J. Org. Chem. 32, 1255–1260. doi:10.6023/cjoc1112231
Xue, J. S., Li, H. Y., Fu, Q., and Qiang, S. P. (2015). Antidepressant-like Effects of L-Menthone in Depressed Mice and its Pos-Sible, Mechanisms. Pharm. Eut. Clin. Res. 23, 238–241. doi:10.13664/j.cnki.pcr.2015.03.008
Yi, Q. Y., Wan, D., Tang, B., Wang, Y. J., Zhang, W. Y., Du, F., et al. (2017). Synthesis, Characterization and Anticancer Activity In Vitro and In Vivo Evaluation of an Iridium (III) Polypyridyl Complex. Eur. J. Med. Chem. 145, 338–349. doi:10.1016/j.ejmech.2017.11.091
Yun, C.-H., Boggon, T. J., Li, Y., Woo, M. S., Greulich, H., Meyerson, M., et al. (2007). Structures of Lung Cancer-Derived EGFR Mutants and Inhibitor Complexes: Mechanism of Activation and Insights into Differential Inhibitor Sensitivity. Cancer Cell 11, 217–227. doi:10.1016/j.ccr.2006.12.017
Zhang, P., Guan, A., Xia, X., Sun, X., Wei, S., Yang, J., et al. (2019). Design, Synthesis, and Structure-Activity Relationship of New Arylpyrazole Pyrimidine Ether Derivatives as Fungicides. J. Agric. Food Chem. 67, 11893–11900. doi:10.1021/acs.jafc.9b05185
Zhang, X., and Yang, H. (2020). Research Progress on Long Non-coding RNAs and Drug Resistance of Breast Cancer. Clin. Breast Cancer 20, 275–282. doi:10.1016/j.clbc.2019.11.001
Zhu, C., Qi, X., Chen, Y., Sun, B., Dai, Y., and Gu, Y. (2011). PI3K/Akt and MAPK/ERK1/2 Signaling Pathways Are Involved in IGF-1-Induced VEGF-C Upregulation in Breast Cancer. J. Cancer Res. Clin. Oncol. 137, 1587–1594. doi:10.1007/s00432-011-1049-2
Keywords: menthone, pyrimidine, urea, antitumor activity, synthesis, PI3K/AKT/mTOR
Citation: Huang M, Duan W, Chen N, Lin G and Wang X (2022) Synthesis and Antitumor Evaluation of Menthone-Derived Pyrimidine-Urea Compounds as Potential PI3K/Akt/mTOR Signaling Pathway Inhibitor. Front. Chem. 9:815531. doi: 10.3389/fchem.2021.815531
Received: 15 November 2021; Accepted: 31 December 2021;
Published: 03 February 2022.
Edited by:
Maria Valeria Raimondi, University of Palermo, ItalyReviewed by:
Omaima AboulWafa, Alexandria University, EgyptHamid Morjani, Université de Reims Champagne-Ardenne, France
Copyright © 2022 Huang, Duan, Chen, Lin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wengui Duan, d2dkdWFuQGd4dS5lZHUuY24=; Naiyuan Chen, Y2hlbm5haXl1YW5AZ3htdS5lZHUuY24=
 Mei Huang1,2
Mei Huang1,2 Wengui Duan
Wengui Duan Xiu Wang
Xiu Wang