- National Institutes for Food and Drug Control, Beijing, China
Objective: To establish a method for the determination of the chemical structure of vancomycin hydrochloride.
Methods: Nuclear magnetic resonance spectroscopy and mass spectrometry were conducted to analyze the chemical structure of vancomycin hydrochloride.
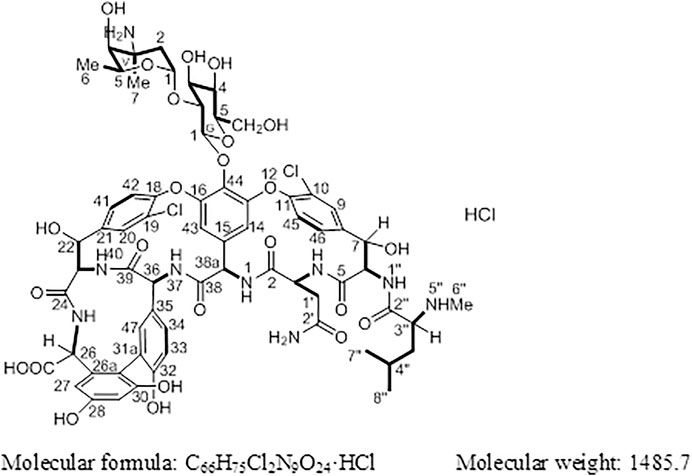
Results: In this study, the target compound (1) was identified as (Sα)-(3S, 6R, 7R, 22R, 23S, 26S, 36R, 38αR)-44-[[2-O-(3-amino-2, 3, 6-trideoxy-3-C-methyl-α-L-lyso-hexopyranosyl)-β-D-glucopyranosyl] oxy]-3-(carbamoylmethyl)-10, 19-dichloro-7, 22, 28, 30, 32-pentahydroxy-6-[[(2R)-4-methyl-2-(methylamino) pentanoyl] amino]-2, 5, 24, 38, 39-pentaoxo-2, 3, 4, 5, 6, 7, 23, 24, 25, 26, 36, 37, 38, 38α-tetradecahydro-22H-8, 11: 18, 21-dietheno-23, 36-(iminomethano)-13, 16: 31, 35-dimetheno-1H, 13H-[1, 6, 9] oxadiazacyclohexadecino [4, 5-m] [10, 2, 16]-benzoxadiazacyclotetracosine-26-carboxylic acid hydrochloride.
Conclusion: The method used in this study is accurate and can be used for the production and structural elucidation of vancomycin hydrochloride.
Introduction
Vancomycin hydrochloride (Figure 1) is a glycopeptide antibiotic that exhibits significant antibacterial activity against various Gram-positive bacteria, including cocci and bacilli (Mccormick et al., 1956). The mechanism of action of vancomycin hydrochloride is as follows: vancomycin binds to the alanine moiety of the precursor peptide present on the sensitive bacterial cell wall with high affinity, thereby preventing the synthesis of the polymer peptidoglycan that constitutes the bacterial cell wall, resulting in cell wall defects and bacterial death. Existing literature shows that vancomycin can selectively inhibit the synthesis of ribonucleic acid by altering the permeability of bacterial cell membranes (Wang et al., 2018). This property is listed in the Chinese Pharmacopoeia (2020 edition, Volume II), Japanese Pharmacopoeia (17th edition), United States Pharmacopoeia (43rd edition), and European Pharmacopoeia (10.0 edition).
China is an important supplier of the raw materials used in vancomycin synthesis. Presently, to the best of our knowledge, the structural analysis of vancomycin has not been reported. Moreover, due to its complex structure, the signals obtained in the nuclear magnetic resonance (NMR) spectrum of vancomycin tend to overlap, making it difficult to analyze and elucidate its structure. In this study, mass spectrometry (MS), one- (1H, 13C) and two-dimensional (1H-1H) correlation spectroscopy (COSY), and heteronuclear multiple quantum coherence (HMQC) and heteronuclear multiple bond coherence (HMBC)) NMR spectroscopy were comprehensively performed to analyze the structure on the basis of the spectral data of vancomycin hydrochloride. The obtained results will provide a scientific basis for the qualitative analysis of this compound.
Materials and Methods
Reagents
Formic acid (Fluka, GR), acetonitrile (Fisher, HPLC), Watsons purified deionized water (Xellia pharmaceuticals, batch number: A3250080), and vancomycin hydrochloride reference standard (National Institutes for Food and Drug Control, batch number: 130360-201302) were used in this study. Deuterated dimethyl sulfoxide (Merck) was used for NMR analysis of 1.
Instruments
Mass spectra were recorded using an AB Sciex 3200 Qtrap mass spectrometer (AB SCIEX, United States). NMR spectra were obtained using a Varian INOVA 600 MHz NMR spectrometer.
Results
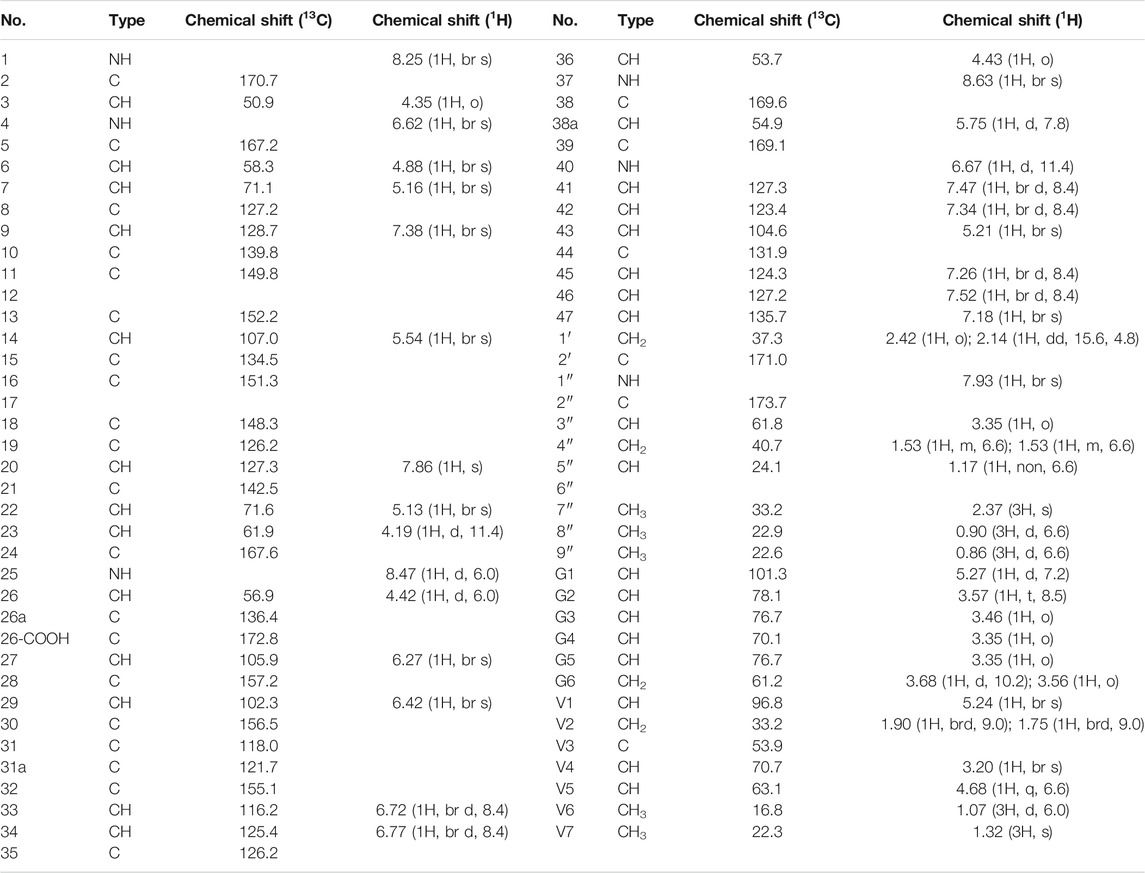
Compound 1 is a white solid. The positive ion electrospray ionization mass spectra [(+)-ESIMS] of 1 revealed a quasi-molecular ion peak at (m/z) 1448 [M+H]+. High-resolution (+)-ESIMS afforded the accurate mass-to-charge ratio of the quasi-molecular ion at m/z 1448.4380 [M+H]+, suggesting that the molecular composition of the test product was C66H75Cl2N9O24 (the calculated value was 1448.4375, and the corresponding molecular composition was C66H76Cl2N9O24). The signals in the 1H-NMR spectrum of 1 were assigned to five groups of aromatic proton as follows: 1) three groups of ABX-system aromatic proton: δH 7.38 (1H, br s, H-9), 7.52 (1H, brd, J = 8.4 Hz, H-46), and 7.26 (1H, brd, J = 8.4 Hz, H-45); δH 7.86 (1H, s, H-20), 7.47 (1H, brd, J = 8.4 Hz, H-41), and 7.34 (1H, brd, J = 8.4 Hz, H-42); δH 7.18 (1H, br s, H-47), 6.77 (1H, brd, J = 8.4 Hz, H-34), and 6.72 (1H, brd, J = 8.4 Hz, H-33); and 2) two groups of 3-substituted aromatic proton signals: δH 6.42 (1H, br s, H-29), 6.27 (1H, br s, H-27), and δH 5.54 (1H, br s, H-14), 5.21 (1H, br s, H-43)]. Moreover, three signals of methyl protons bonded to methylidyne [δH 0.90 (3H, d, J = 6.6 Hz, H3-8″), 0.86 (3H, d, J = 6.6 Hz, H3-9″), and 1.07 (3H, d, J = 6.0 Hz, H3-V6)], a signal of methyl protons bonded to a nitrogen atom [δH 2.37 (3H, s, H3-7″)], a signal of methyl protons bonded to a quaternary carbon [δH 1.32 (3H, s, H3-V7)], and 19 signals of protons bonded to various heteroatoms [δH 5.75 (1H, d, J = 7.8 Hz, H-38a), 5.27 (1H, d, J = 7.2 Hz, H-G1), 5.24 (1H, br s, H-V1), 5.16 (1H, br s, H-7), 5.13 (1H, br s, H-22), 4.88 (1H, br s, H-6), 4.68 (1H, q, J = 6.6 Hz, H-V5), 4.43 (1H, o, H-36), 4.42(1H, d, J = 6.0 Hz, H-26), 4.35(1H, o H-3), 4.19(1H, d, J = 11.4 Hz, H-23), 3.68(1H, d, J = 10.2 Hz, H-G6a), 3.57(1H, t, J = 8.5 Hz, H-G2), 3.56(1H, o, H-G6b), 3.46(1H, o, H-G3), 3.35(1H, o, H-G5), 3.35 (1H, o, H-G4), 3.35 (1H, o, H-3″), and 3.20 (1H, br s, H-V4)] were observed in the NMR spectra of 1. After deuterium exchange, the NMR spectra of 1 exhibited at least 16 exchangeable active proton signals [δH 9.44 (1H, br s, OH), 9.11 (1H, br s, OH), 8.63 (1H, br s, NH-37), 8.47 (1H, d, J = 6.0 Hz, NH-25), 8.25 (1H, br s, NH-1), 7.93 (1H, br s, NH-1″), 7.37 (2H, br s, CONH2), 6.91 (2H, br s, CONH2), 6.67 (1H, d, J = 11.4 Hz, NH-40), 6.62 (1H, br s, NH-4), 5.96 (1H, d, J = 4.2 Hz, OH-22), 5.83 (1H, br s, OH-7), 5.43 (1H, br s, OH-V4), 5.38 (1H, br s, OH-G3), 5.11 (1H, br s, OH-G4), and 4.05 (1H, br s, OH-G5)]. In addition, multiple aliphatic hydrogen signals were observed in the upfield region. The above spectral data show that 1 is a glycopeptide containing multiple amino acid fragments (Harris et al., 1983; Zhou et al., 2004).
The 1H and 13C NMR signals of 1 were accurately assigned using HSQC and 1H-1H COSY; the details are listed in Table 1. The 1H-1H COSY spectrum of 1 showed the cross-peak correlation between the protons. The arrangement of the structural fragments of 1 was determined on the basis of the proton couplings in the structure (represented by thick lines in Figure 5).
The fragments were then arranged based on the HMBC spectrum as follows:
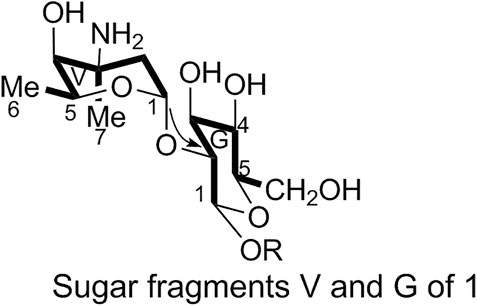
1) Linkage of sugar fragments:
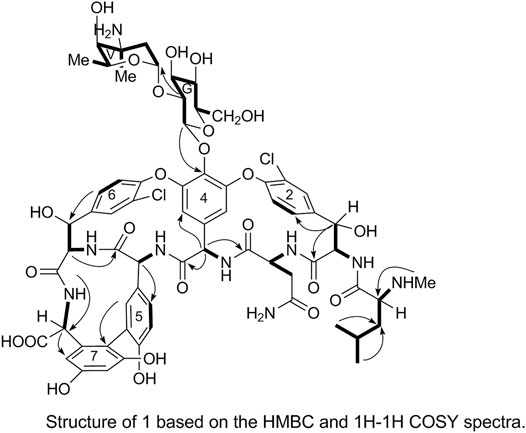
Two end group carbon signals were observed in the 1H-NMR and 13C-NMR spectra of 1. The structures and the linkage locations of the two sugar fragments (represented by V and G, respectively in Figure 2) were confirmed based on the HSQC, 1H-1H COSY, and HMBC spectra. Especially, in the HMBC spectrum, H-V1 can be attributed to C-G2, thereby confirming the α-L-vancosaminyl-(1→2)-O-β-D-glucosyl linkage between the two sugars.
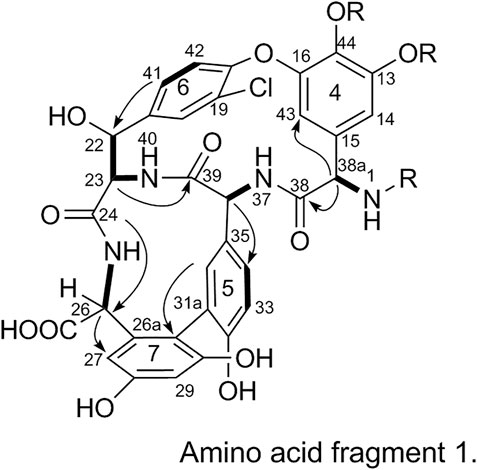
2) Linkage of amino acid fragments 4, 5, 6, and 7:
The substitution patterns of benzene rings 4, 5, 6, and 7 were determined using the δH, δC, and JH-H values observed in the 1H-NMR and 13C-NMR spectra of 1. In the HMBC spectrum of 1, H-38a corresponds to C-43 and C38, H-36 corresponds to C-34, H-41 corresponds to C-22, H-23 corresponds to C-39, and H-26 corresponds to C-24 and C-27. Thus, it can be inferred that 1 contains a 4-amino acid cyclic peptide structure (represented by thick lines in Figure 3).
3) Linkage of amino acid fragments 1, 2, and 3:
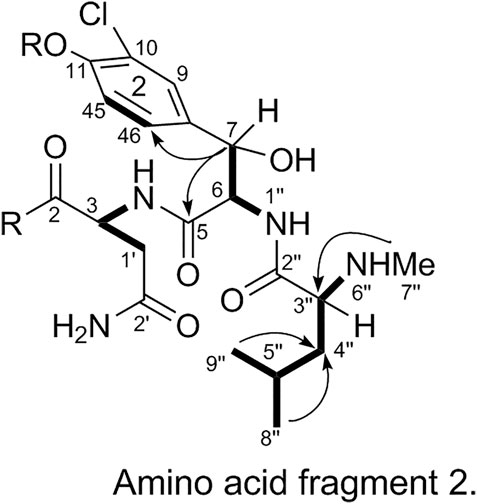
The substitution pattern of benzene ring 2 was confirmed using the δH, δC, and JH-H values observed in the 1H-NMR and 13C-NMR spectra of 1. In the HMBC spectrum, H3-7″ corresponds to C-3″, H-8″ and H-9″ correspond to C-1a, H-7 corresponds to C-11 and C-2, and H-3 corresponds to C-2. Therefore, it could be inferred that there was another 3-amino acid fragment linkage in 1 (represented by thick lines in Figure 4).
4) Linkage of amino acid and sugar fragments.
In the HMBC spectrum, H-G1 corresponds to C-44 and H-38a corresponds to C-2. These results confirmed the proposed arrangements of the above-mentioned three fragments (marked by arrows in Figure 5). The above analyses show that 1 is a glycopeptide antibiotic with a cyclic peptide core consisting of seven amino acids and two sugars linked by an oxygen glycosidic bond; its planar structure is identical to that of vancomycin (Mccormick et al., 1956).
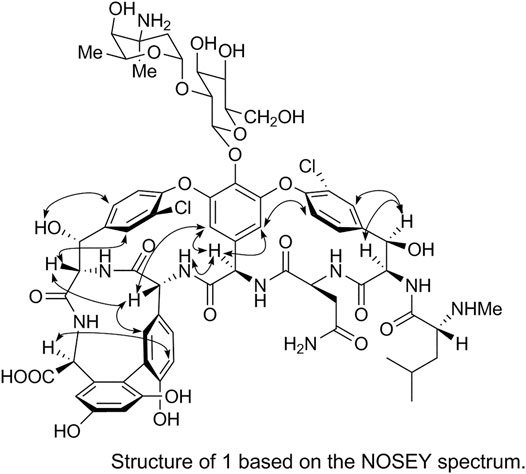
The spatial structure of 1 was determined by comparing its NOSEY and circular dichroism (CD) spectra with those of the reference standard. First, the relative configuration of 1 was determined using the NOSEY experimental data (indicated by arrows in Figure 6). Moreover, the δH, JH-H, and δC values and the NOSEY spectra of 1 were consistent with those of the reference standard. These results confirm that 1 has a configuration identical to that of the vancomycin hydrochloride reference standard.
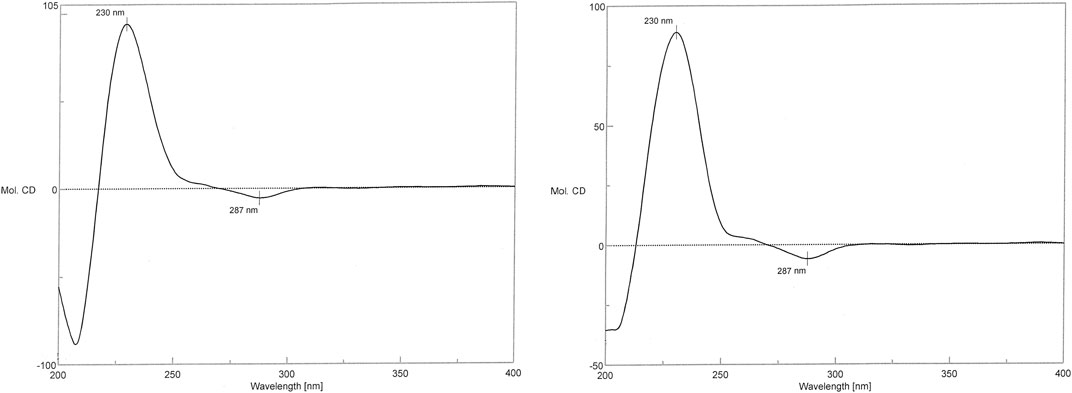
In the CD spectrum of 1, a negative Cotton effect was observed at 287 nm, while a positive Cotton effect was observed at 230 nm. The CD spectrum of 1 was identical to that of the vancomycin hydrochloride reference standard (2) (Figure 7). Therefore, 1 exhibited the three-dimensional structure identical to that of the vancomycin hydrochloride reference standard.
Discussion
Through MS and NMR spectroscopy, 1 was identified as vancomycin hydrochloride. Moreover, the 1H and 13C NMR signals were accurately assigned on the basis of the one- and two-dimensional NMR spectra of 1. Thus, this method can be effectively utilized for the structural and qualitative analysis of complex compounds.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found in the European Nucleotide Archive (ebi.ac.uk) under accession number PRJEB47449.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
Details of all funding sources should be provided, including grant numbers if applicable. Please ensure to add all necessary funding information, as after publication this is no longer possible.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.753060/full#supplementary-material
References
Harris, C. M., Kopecka, H., and Harris, T. M. (1983). Vancomycin: Structure and Transformation to CDP-I. J. Am. Chem. Soc. 105, 6915–6922. doi:10.1021/ja00361a029
Mccormick, M. H., Mcguire, J. M., Pittenger, G. E., Pittenger, R. C., and Stark, W. M. (1956). Vancomycin, a New Antibiotic. I. Chemical and Biologic Properties. Antibiot. Annu. 3, 606–611. doi:10.1021/ja00361a029
Wang, C., Wang, L. X., and Hu, C. Q. (2018). Quality Assessment of the Domestic Vancomycin Hydrochloride for Injection. Chin. J. Antibiot. 43, 262–268.
Keywords: NMR, MS, chemical shift, vancomycin, structure elucidation
Citation: Tian Y, Chong X, Yao S and Xu M (2021) Spectral Data Analysis and Identification of Vancomycin Hydrochloride. Front. Chem. 9:753060. doi: 10.3389/fchem.2021.753060
Received: 04 August 2021; Accepted: 08 September 2021;
Published: 20 September 2021.
Edited by:
Anna Napoli, University of Calabria, ItalyReviewed by:
Giuseppina De Luca, University of Calabria, ItalyWei-Lung Tseng, National Sun Yat-sen University, Taiwan
Copyright © 2021 Tian, Chong, Yao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangchen Yao, eXdfc2NAc2luYS5jb20=; Mingzhe Xu, dGlhbnllQG5pZmRjLm9yZy5jbg==
†These authors have contributed equally to this work
 Ye Tian
Ye Tian Xiaomeng Chong†
Xiaomeng Chong†