- 1Tianjin Key Laboratory of Organic Solar Cells and Photochemical Conversion, School of Chemistry and Chemical Engineering, Tianjin University of Technology, Tianjin, China
- 2Tianjin Key Laboratory of Drug Targeting and Bioimaging, School of Chemistry and Chemical Engineering, Tianjin University of Technology, Tianjin, China
CRISPR-associated Cas9 endonuclease (CRISPR/Cas9) systems are widely used to introduce precise mutations, such as knocking in/out at targeted genomic sites. Herein, we successfully disrupted the transcription of multiple genes in Bacillus pumilus LG3145 using a series of unspecific guide RNAs (gRNAs) and UgRNA:Cas9 system-assisted cre-box editing. The bases used as gRNAs shared 30–70% similarity with a consensus sequence, a cis-acting element (cre-box) mediating carbon catabolite repression (CCR) of many genes in Bacillus. This triggers trans-crRNA:Cas9 complex wobble cleavage up/downstream of cre sites in the promoters of multiple genes (up to 7), as confirmed by Sanger sequencing and next-generation sequencing (NGS). LG3145 displayed an obvious CCR release phenotype, including numerous secondary metabolites released into the culture broth, ∼ 1.67 g/L white flocculent protein, pigment overflow causing orange-coloured broth (absorbance = 309 nm), polysaccharide capsules appearing outside cells, improved sugar tolerance, and a two-fold increase in cell density. We assessed the relationship between carbon catabolite pathways and phenotype changes caused by unspecific UgRNA-directed cre site wobble editing. We propose a novel strategy for editing consensus targets at operator sequences that mediates transcriptional regulation in bacteria.
Introduction
Bacillus pumilus, a Gram-positive soil bacterium, is considered a potential host strain for the production of chemicals, heterologous proteins and antimicrobial materials (Westers et al., 2004; Su et al., 2020). However, in Bacillus, there is a general regulatory mechanism regulating many catabolic genes and operons through repression by favoured carbon sources such as glucose, fructose or mannose, termed carbon catabolite repression (CCR) (Stülke and Hillen, 2000). CCR helps to maintain carbon catabolic/metabolic network balance according to metabolic capacities, and can act as a serious barrier preventing the use of Bacillus as cell factories. Thus, alleviating CCR is an established area of research (Chen et al., 2020). Three common regulatory components have been identified that silence multiple operons: the catabolite responsive element cre-box, a promoter proximal cis-acting conserved sequence that negatively regulates multiple genes in Bacillus species; the histidine phosphocarrier protein Hpr, an intermediate of the phosphoenolpyruvate:sugar transport system (PTS); and the catabolite control protein CcpA, a trans-acting factor that binds cre-box through Hpr-Ser-p to control target gene transcription (Peng et al., 2020; Langa et al., 2021; Neira et al., 2021).
Accumulating evidence indicates that cre-boxes are globally distributed in the genome of Bacillus, hence simultaneous point mutations at these cre-boxes may trigger global regulation network rearrangement (Zhang et al., 2018; Han et al., 2020). In the present study, we wished to manipulate this trigger in Bacillus pumilus to disrupt the transcriptional regulation of multiple genes simultaneously to overcome CCR limitations, enhance sugar resistance, and improve secretion. The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system, a primitive adaptive immunity system in bacteria that defends against foreign genetic contamination, is capable of editing multiple genes simultaneously (Jinek et al., 2012; Cong et al., 2013). Specific editing using the CRISPR/Cas9 system cleaves target sites in the genome, but the approach suffers from genome-wide off-target effects (Guilinger et al., 2014; Wu et al., 2019). However, the off-target effects may be utilised to edit the consensus sequence at multiple-gene loci such as cre-boxes. Thus, an unspecific CRISPR/Cas9 system was constructed to control guide RNA (gRNA):Cas9 complex off-target effects. Specifically, three 20 nt oligonucleotides sharing 30–70% similarity with target sites were synthesised to serve as single gRNAs involved in targeting seven genes (listed in Supplementary Table S1).
Herein, we used a pCas9 derivate plasmid to assist multiple cre-box editing. The plasmid encodes Cas9 nuclease and a double-stranded DNA (dsDNA) endonuclease that uses a CRISPR RNA (crRNA) guide to specify the site of cleavage (Jiang et al., 2013). Spacers (gRNA) can be inserted into the crRNA array between BsaI sites using annealed oligonucleotides. We reprogramed three unspecific gRNAs sharing 30–70% similarity with the target fragments (as shown in Supplementary Table S2) and reconstructed three plasmids (pCas9-Cax, pCas9-Cac and pCas9-Cpt). Simultaneous use of three unspecific gRNAs enabled wobble editing up/downstream of cre-box sites in the seven target genes. Sanger sequencing and next-generation sequencing (NGS) were used to confirm mutagenesis near the cre-boxes (-10 and -35 domains falling in the range of ± 100 bp). The unspecific CRISPR/Cas9 system led to pleiotropic genome mutations in B. pumilus, resulting in strain LG3145, which exhibited remarkable phenotypic changes, such as smoother colonies, altered colour pigments, polysaccharide capsules outside cells, and elevated secretion. It is possible that CCR release of numerous genes may be responsible for the observed phenotypical changes.
Materials and Methods
Strains and Reagents
Previously, we isolated a B. pumilus wild-type strain from soil, and the genotype was the same as that of B. pumilus SH-B9 (NCBI Accession no: NZ_CP011007.1) according to whole-genome sequencing. The other strains used in this study are listed in Supplementary Table S3. Escherichia coli DH5α (the host for pCas9) and Escherichia coli Top10, both purchased from Thermo Fisher Scientific (Waltham, MA, United States), were used for plasmid construction and amplification, and cultured in Luria-Bertani medium containing 25 μg/ml chloromycetin. All chemicals, a DNA gel purification kit, and a plasmid extraction kit were purchased from TIANGEN Biotech (Beijing, China). Restriction enzymes and DNA ligase were purchased from New England Biolabs (Ipswich, MA, United States). Taq DNA polymerase and oligonucleotides used for PCR experiments were purchased from Ruibio Biotech (Beijing, China).
Plasmids and Primers
The pCas9 plasmid and sequences used in this study (Supplementary Table S4) are available from the Addgene nonprofit plasmid repository (Cat: 42,876). The pCas9 plasmid is 9,326 bp based on the low-copy backbone of pACYC184 (ATCC 37033), which endows resistance to chloramphenicol (25 μg/ml). The other elements of this plasmid include tracrRNA, Cas9, and an array of crRNAs with two BsaI sites for spacers (Figure 1). The plasmids pCas9-Cax, pCas9-Cac and pCas9-Cpt were constructed for cre-box editing of seven target genes. The spacer oligonucleotides for cloning into pCas9 and the primers used for PCR are listed in Supplementary Table S5.
Unspecific gRNA Array Construction and Multiple cre Sites Editing Strategy
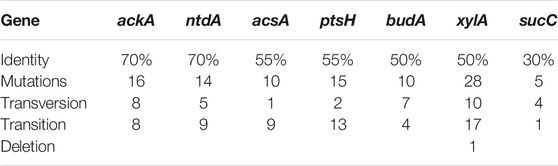
For editing multiple cre-box sites at the seven operons and simultaneously controlling the range of gRNA:Cas9 complex off-targets, oligonucleotides of gRNAs must direct the Cas9 protein to recognize multiple target sites. Because the cre-boxes of B. pumilus have the consensus sequence WWGNAANCGNWNNCW (Supplementary Table S1), we chose NNG for the PAM and cre proximal sequences for target sequences designed using the Zhang Lab website (http://crispor.tefor.net/), and the results (Supplementary Table S2) include partial or complete cre sequences (Supplementary Figure S2). In order to better describe the seven target operons, we continued using the same gene symbols of the type strain B. subtilis168 (NCBI Accession no: NC_000964.3) where annotation was consistent. The seven target sequences were divided into three groups with the highest (70%), moderate (70–50%), and lowest (50–30%) identity with the three unspecific gRNAs (Cac, Cpt, and Cax).
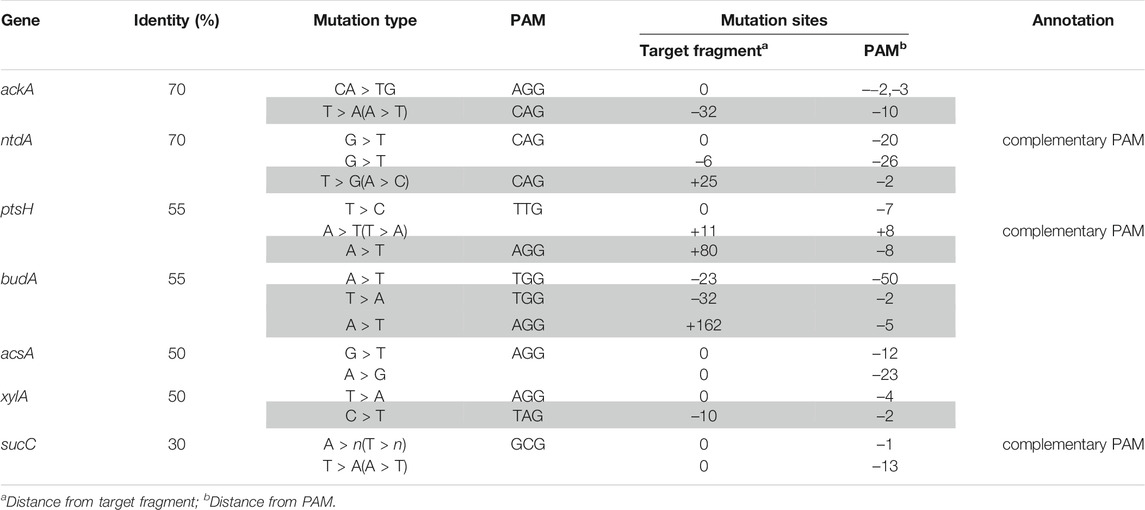
Among them, ackA and ntdA share the highest identity (70%) with the unique gRNA Cac and the common gRNA Cpt , both of which share >50% identity with ptsH and budA. The ackA encoding acetate kinase that converts pyruvate into acetate and is positively regulated by CcpA protein, ntdA encoding an aminotransferase responsible for converting dehydroglucose-6-phosphate to kanosamine-6-phosphate, ptsH encoding phosphocarrier protein Hpr that is also repressed by CcpA, and budA encoding acetolactate decarboxylase that converts pyruvate to acetoin and is also activated by CcpA. The target site sharing only 30% identity is the sucC operon encoding succinyl-CoA synthetase, which converts succinyl-CoA to succinate in the tricarboxylic acid (TCA) cycle and is negatively regulated by CcpA, and shares the gRNA Cax with two other operons (acsA and xylA) but they share more than 50% identity. Acetyl-CoA synthase, encoded by acsA, converts pyruvate into propanoate, which can be activated by CcpA and xylA encoding a xylose isomerase responsible for converting D-glucose into D-fructose. The three unspecific gRNAs (Cac, Cpt, and Cax) were inserted into the BsaI sites of pCas9 as the CRISPR spacers after annealing at 95°C for 5 min and 37°C for 30 min using T4 ligase according to the pCas9 protocol (Huang et al., 2016). The strategy of multiple editing at cre sites used in this study is depicted in Figure 2.
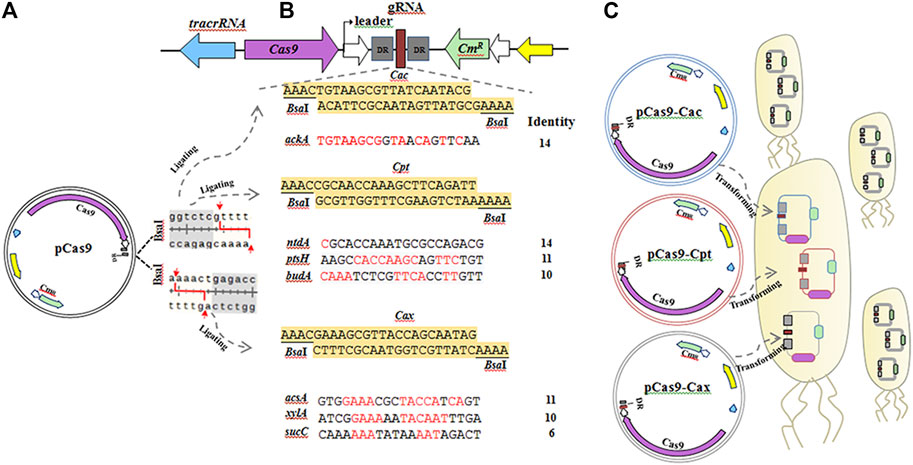
FIGURE 2. The strategy for multiple editing at cre sites in seven operons using three unspecific gRNA:Cas9 systems in B. pumilus. (A) The double BsaI restriction sites of pCas9 allow the gRNA to be inserted. (B) The three unspecific gRNAs used in this study contain 24 bp oligonucleotides and two BsaI sites at both ends, in which Cax has the highest identity (70%) with 14 bp shared with the target sequences of ackA, Cac and Cpt (moderate, 50–70%), or lower identity gRNAs (30–55%) with 10–14 bp shared with ntdA, ptsH and budA, and 6–10 bp shared with acsA, xylA and sucC. (C) After annealing, Cax, Cac and Cpt were inserted into pCas9 to construct three plasmids (pCas9-Cax, pCas9-Cac and pCas9-Cpt) using T4 DNA ligase, which were transformed into B. pumilus wild-type (WT) cells using the protoplast transformation method. Chloromycetin was added into the CMR media used for isolating transformants.
UgRNA:Cas9 System-Assisted Multiple cre Sites Editing
E. coli Top10 cells were used to construct and amplify plasmids. Ligations of gRNA-annealed oligos and BsaI-digested pCas9 were gently mixed with competent E. coli Top10 cells at a ratio of 1:200 and incubated on ice for 30 min, then heated at 42°C in a water bath for 90 s, and chilled for 1–2 min on ice. The cells were then incubated in preheated LB medium (150 μl) at 37°C for 45 min, then plated onto 25 μg/ml chloramphenicol selective media and incubated overnight at 37°C. The following morning, transformants were picked and incubated in LB medium. After cell growth, plasmids were extracted using an appropriate kit and the pCas9-Cac, pCas9-Cpt and pCas9-Cax constructs were verified by DNA sequencing.
Protoplasts of B. pumilus wild-type (WT) were transformed with plasmid DNA using a modified method described by Chang and Cohen (1979). A single colony of B. pumilus WT was inoculated into 5 ml of LB medium and cultured overnight at 37°C in a shaker (200 rpm). The overnight culture was diluted 50-fold into 5 ml of fresh LB medium and grown for 8 h at 37°C (200 rpm). Cells were harvested by centrifugation (9,000 × g, 10 min, 4°C). The supernatant was removed and cells were resuspended in 1 ml of SMMP solution (equal volumes of 2× SMM buffer and 4× Penassay broth). Lysozyme powder was added to 0.4 mg/ml, mixed, and incubated at 37°C for 45 min (100 rpm) to prepare protoplasts (globular appearance). Protoplasts were harvested by centrifugation (5,000 × g, 10 min, RT), the supernatant was poured off, and the cell pellet was resuspended gently in 0.05 ml SMMP. Plasmid DNA (0.2 μg) was mixed with 0.05 ml of protoplasts and added to a microtube containing 150 μl of polyethylene glycol (PEG) 6,000. After gentle mixing, the sample was incubated at 37°C for 2 min in a water bath shaker to allow plasmids to enter protoplasts, and 0.5 ml of SMMP was added for termination. Protoplasts were harvested by centrifuging (10,000 × g, 7 min) and the supernatant was discarded. SMMP (300 μl) was added and incubated for 60–90 min at 37°C in a water bath shaker (100 rpm) to allow for expression of the antibiotic resistance marker. Because there is no B. pumilus replication site in pCas9, the pCas9-Cac, pCas9-Cpt and pCas9-Cax plasmids were integrated into the genome. Cells were plated on CMR-selective regeneration medium containing chloramphenicol to select transformants. The three plasmids were successively transformed into B. pumilus WT three times. We engineered the B. pumilus LG3145 strain from eight transformants through DNA sequencing.
Fermentation and Morphological Measurements
To analyse the effects of multiple cre sites editing on phenotype, WT and mutant strains were grown in high-glucose fermentation medium (GYN; 10–30% glucose, 2% yeast extract, 0.05% MgSO4•7H2O, 1% urea) in triplicate at 37°C with shaking at 220 rpm. A 1 ml sample of culture was diluted 3-fold every 2 h and the optical density at 600 nm (OD600) was measured using a T6 UV spectrophotometer using a Purkinje General Instrument (Beijing, China) to plot the growth curve. Cells were harvested by centrifugation at 10,000 × g after culturing for 12 h, washed three times with sterile water, and diluted 100-fold for morphological assay. A 3 μl sample of cell diluent was placed in the centre of a 13 mm × 18 mm piece of mica, freeze-dried for 8 h, and observed by scanning electron microscopy (SEM) using a Zeiss Merlin Compact instrument (Jena, Thuringia, Germany). Cells for atomic force microscopy (AFM) analysis must be washed repeatedly and diluted at appropriate times. A sample of 3–5 μl was placed on a 25 mm × 25 mm fresh, sterile mica sheet and freeze-dried for 8 h before analysis by a Bruker ICON instrument (Berlin, Germany).
We selected the ScanAsyst-air scanning mode to image cells of each strain. For the probe, a silicon tip on a nitride lever was employed, with a cantilever length of 115 μm and a light spring constant of 0.4 N/m. The morphology of colonies on LB agar plates was observed by stereo microscopy using a Motic str6 instrument (Xiamen, China) with a silicon tip on a nitride lever.
Analysis of Extracellular Pigments and Proteins
The supernatants of LG3145 and WT strains were collected from LB medium cultures by centrifugation (6,000 × g, 20 min, 4°C) and mixed with precooled 12.5% trichloroacetic acid. Following freezing (−20°C for 12 h), the supernatant was centrifuged at 4°C for 5 min to collect proteins. The precipitate was washed 3–5 times with 80% precooled acetone to remove residual trichloroacetic acid and stood for 30 min to completely evaporate acetone. Some precipitates were dialysed for 5 h and freeze-dried for 8 h, then observed using an Oxford X-max20 energy dispersive spectrometer (EDS) and by SEM; other precipitates were dissolved in 1 ml ddH2O and mixed with 5 ml Folin-phenol reagent A and 0.5 ml reagent B, both purchased from Lanji technology (Shanghai, China). After standing at ambient temperature for 30 min, the mixture was analyzed using a spectrophotometer at 700 nm (Wan et al., 2009; Han et al., 2019). The protein concentration was calculated using the equation y = 0.2823x + 0.1011 (y-axis = absorbance, x-axis = protein concentration).
For pigment analysis, minimal medium (MM; 3.48 g KH2PO4, 1.5 g Na2HPO4.12H2O, 3.96 g (NH4)2SO4, 0.7 g MgSO4.7H2O, 0.01 g yeast extract, 1 L ddH2O, pH 7–7.5) supplemented with 2.5% (w/v) glycerol or glucose was used to culture each strain at 35°C with shaking at 220 rpm. The absorbance of extracellular pigments was measured from 200 to 800 nm by a Shimadzu UV-3600plus spectrophotometer (Kyoto, Japan). All experiments were repeated in triplicate.
Results and Discussion
UgRNA:Cas9 Edits Multiple cre Sites
The cre sites of seven target genes were edited by transforming UgRNA:Cas9 expression plasmids pCas9-Cax, pCas9-Cac and pCas9-Cpt into WT strain protoplast following the strategy shown in Figure 2. Chloromycetin (25 μg/ml) was added to the CMR media used for selecting transformants harbouring the UgRNA:Cas9 system.
The promoter regions, 500 bp upstream fragments of target genes ackA, ntdA, ptsH, budA, acsA, xylA and sucC, were amplified by PCR (Supplementary Figure S1) and mutations of the seven target genes were confirmed by DNA sequencing (Supplementary Figure S2). Surprisingly, there were eight mutants with the same genotype among the 10 transformants (success rate = 80%).
The mutation maps of each gene (top of Supplementary Figure S2) show that all point mutations occurred around the target fragments, and the closer the mutations to the centre, the denser they are, and most are distributed along the direction from PAM to target fragment. The number of mutations was significantly positively correlated with the identity of UgRNAs shown in Table 1 (except for xylA). Most of the mutations are transitions, with almost no deletions. Thus, a UgRNA:Cas9 system with more than 50% identity can edit multiple genes at the same time. In fact, the genomes of the mutants were determined by next-generation sequencing (NGS) and the results indicate that most of the mutation sites occurred around the seven target fragments, but some mutation sites occurred at other sites. The occurrence of off-target sites editing appeared to correlate with the similarities of UgRNAs. These relationships will be investigated in future work.
According to editing characteristics, the DNA strand around the target may be twisted and rolled, especially the DNA strand in the binding direction of UgRNAs, and the higher the similarity between UgRNA and target fragment, the larger the curl range. Because the UgRNAs were partially complementary to the target fragments, Cas9 endonuclease seems to wobble within this region and recognise multiple PAM sites for cleaving, which can be verified by the locations and the corresponding PAMs of mutation points shown in Table 2. The results indicate that all mutations occurred in the promoters, which include 1–2 bp mutations at target fragments according to the preset PAM, especially the edited sites of ptsH, acsA and sucC which coincided exactly with cre sites. There were many off-target mutation points with an efficient PAM nearby, such as the transversion T > A(A > T) near the -10 domain of ackA, with a CAG after10 nt; the transversions T > A and A > T of budA, also with TGG and AGG after two and five nucleotides respectively; the point mutation T > G(A > C) of ntdA with a CAG after 2 nt; and the mutation C > T before cre of xylA with an efficient PAM ‘TAG’. All data are marked with a grey shadow in Table 2. However, there were a few distinct off-target mutations in sucC, and only one base exactly edited at the target fragment, possibly because the lower the similarity of UgRNAs, the fewer the number of DNA fragments involved. This phenomenon further proves that UgRNA:Cas9 may cause swing editing of Cas9, and the swing range is affected by UgRNA identity.
UgRNA:Cas9 Mediates Carbon Catabolism
The DNA sequencing analysis described above showed that multiple cre sites of B. pumilus LG3145 were edited successfully. Additionally, various phenotypes of LG3145 were markedly altered, including resistance to sugar, capsules outside cells, and white flocculent protein floating on the medium upper layer. We subsequently analysed changes in carbon catabolism, and explored the relationships between mutations and phenotypes.
Peptidoglycan Synthesis is Enhanced in B. pumilus LG3145
In Bacillus, raising the amount of carbon in vivo can lead to carbon catabolite repression (CCR) and inhibit the absorption of glucose. Herein, we compared the growth patterns of the WT and the multiple cre editing LG3145 strain in high sugar broth. The results indicate that LG3145 cells were strongly resistant to sugar after editing around the cre site (Figure 3A). LG3145 cells entered a vigorous exponential growth phase (lasting ∼48 h) after 6 h of cultivation, during which the cell density (OD600) was >6, whereas the WT strain displayed a shorter (14 h) exponential growth phase with an OD600 < 2. Furthermore, AFM images revealed a large number of secretions outside the bacteria, believed to be bacterial glycocalyx, that help to protect against environmental stresses (Su et al., 2012; Meng et al., 2017) (Supplementary Figure S3C). The glycocalyx outside LG3145 cells may be related to the level of glucose catabolites because when we used 10% glucose GYN the glycocalyx became slimmer and flimsier (Supplementary Figure S3A). Even in LB broth, we observed a thicker polysaccharide capsule outside the cells of the LG3145 strain in TEM and AFM images (Supplementary Figures S4B, S4C, black arrow) compared with WT cells.
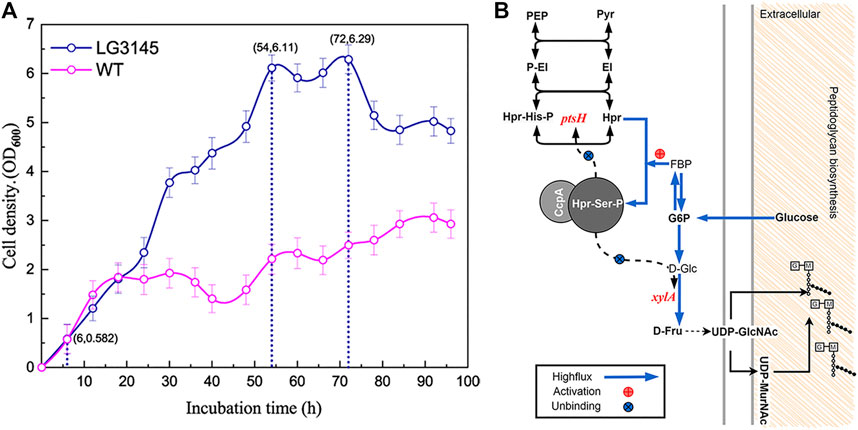
FIGURE 3. (A) Comparison of the growth patterns of LG3145 mutant and WT cells. Periodic (2 h) optical density (OD) readings were taken to assess cell growth of strains in GYN plus 30% (w/v) glucose at 37°C with shaking at 220 rpm. (B) Peptidoglycan synthesis-related pathways.
To further demonstrate this phenomenon, traditional negative staining using Indian ink and safranin was used to observe the presence of capsules by microscopy, and halos clearly surrounded each LG3145 cell, as expected (Supplementary Figure S4A). We speculate that the main components of the capsule are peptidoglycan because cre site mutation of xylA encoding xylose isomerase resulted in the CcpA∼(Hpr-Ser-p) complex that was unable to bind its promoter, regardless of how much glucose was supplied, and xylose isomerase was expressed normally (Dahl and Hillen, 1995). Additionally, the ptsH gene, encoding the key HPr factor associated with CCR, was also mutated at the cre site. In the LG3145 strain, more glucose was transported into cells, and expression of Hpr and Hpr-Ser-p was elevated, but this did not repress ptsH and xylA (Figure 3B) (Kraus et al., 1994). Thus, when glucose was increased, D-Fru was produced in greater quantities, the flux from D-Fru to UDP-GlcNac and UDP-MurNAc was enhanced, and more peptidoglycan was synthesised and secreted from cells to form the capsules that protect cells against dehydration in high-permeability glucose medium (Lovering et al., 2012). This explains why LG3145 cells could survive better than WT cells (Supplementary Figure S3B).
The glycocalyx is believed to be made from polypeptide, polysaccharide, or both, and secreted from cells (Li et al., 2018; Scarff et al., 2018; Rausch et al., 2019; Sun and Zhang, 2021). Coincidentally, we observed a white flocculent biofilm on the surface of the medium when LG3145 cells were incubated in a test tube (Supplementary Figure S5B). The biofilm substance was isolated by dialysis and observed by SEM, which revealed porous and viscous properties of protein (Figure 4A). When culturing, we could see that some cells were embedded (red circle in Figure 4B). The biofilm substance was found to contain C, N, O and S elements, the main elements of amino acids (Figure 4C), according to the results of EDS analysis. Total extracellular protein levels reached 1.908 g/L after 96 h, as determined by the Folin-Phenol method, but there was little protein in vitro with the WT strain (Figure 4D). Protease activity was also assessed using a two-layer skim-milk plate (Supplementary Figure S5C). This indicated that editing cre sites caused an increase in extracellular proteolytic activity at the late exponential phase. All experiments were repeated in triplicate and the same phenomenon was observed for LG3145.
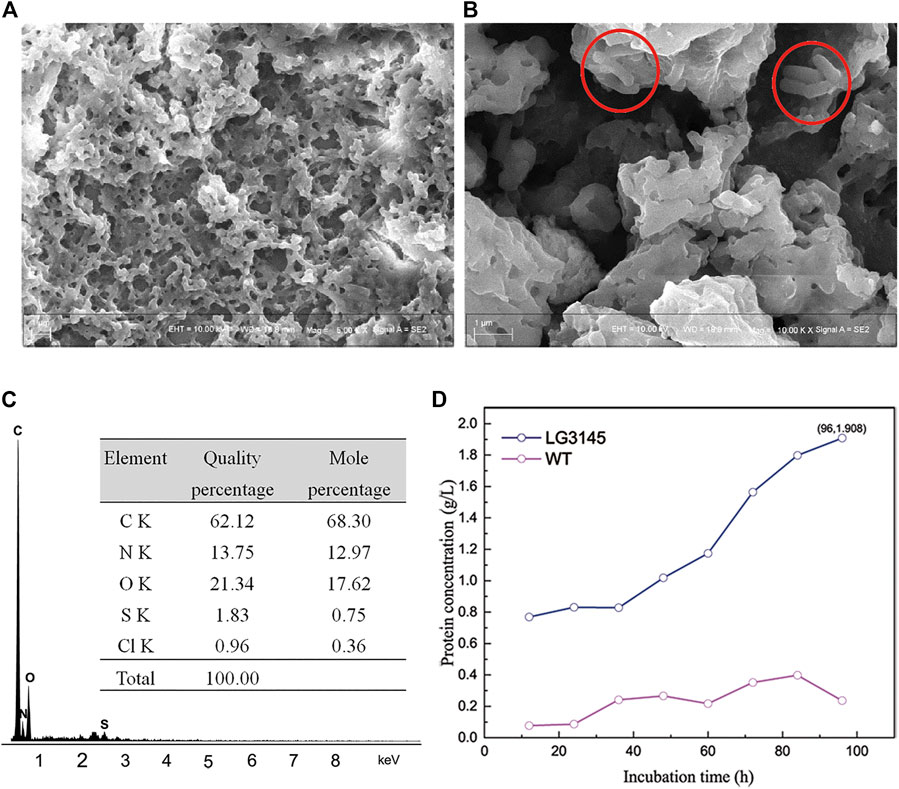
FIGURE 4. (A) Extracellular protein analysis and (B) detailed SEM images. (C) Sample energy dispersive spectrum (EDS) showing the elemental ratios. (D) Comparison of extracellular protein concentrations between LG3145 and WT cells.
Cytochrome Synthesis is Enhanced in B. pumilus LG3145
Since previous studies on CCR in Bacillus showed that cre negatively regulates genes related to carbohydrate catabolism (Sousa et al., 2019), mutation of cre could potentially disrupt secondary pathways, and lead to a significant increase in extracellular excretion. When we incubated the mutant LG3145 strain on LB agar, colonies were more adhesive and pigmented under the naked eye, with fuzzy edges, whereas WT colonies appeared smooth, white, and with distinct edges (Supplementary Figures S6A, 6B). To further explore the relationship between pigmentation and carbon source, MM with different carbon sources was used to culture each strain. Amazingly, an orange-coloured suspension of LG3145 cells was evident on day 3 in MM supplemented with 2.5% (w/v) glycerol or glucose after culturing at 35°C. In particular, with glycerol as the carbon source, the pigmentation of the suspension was more intense than with the preferred carbon source (glucose), while the broth of WT cells was colourless (Supplementary Figures S6C, 6D). These results imply that mutation of cre sites of target genes disrupted the carbon catabolism network, specifically by increasing the flux of secondary pathways, when utilising carbon sources other than glucose (Chen et al., 2019). As shown in Figure 5A, cre site mutation of ptsH resulted in overexpression of the Hpr protein, and the amount of Hpr-His-p and Hpr-Ser-p was also increased. Glycerol as a carbon source is transported by the transporter GlpF, then converted into glycerol-3-phosphate by glycerol kinase GlpK. The activity of GlpK is induced by Hpr-P-His through the promotion of GlpK phosphorylation. Thus, in LG3145 cells, more Hpr-His-P was produced, more GlpK was converted to p-GlpK, and more glycerol-3-phosphate was present in cells. In the next step, glycerol-3-phosphate is converted to dihydroxyacetone phosphate (DHAP), which enters the glycolytic pathway, and the flux from glycerol to the glycolytic pathway is enhanced following mutation of ptsH. However, the overflow pathways from pyruvate to acetate, acetoin and propanoate were repressed, as shown in Figure 5A, because cre site mutation of ackA, acsA and budA must be activated by CcpA∼Hpr-Ser-p complex binding to cre sites, which differs from the CCR mechanism of other genes (Turinsky et al., 2000; Moir-Blais et al., 2001). Thus, glycerol metabolic flux is diverted to the pentose phosphate pathway or to secondary metabolism, and pigments such as riboflavin or carotene accumulate, resulting in a colour change. The results of UV-Vis spectral analysis revealed maximal peaks shifting from 255 to 309 nm when LG3145 cells were cultured with glycerol as a carbon source (Figure 5B), and the culture emitted 450–480 nm blue-green phosphorescence upon UV excitation. The profiles resembled the features indicative of some cytochromes, but the molecular organisation of the pigments remains to be determined in future studies.
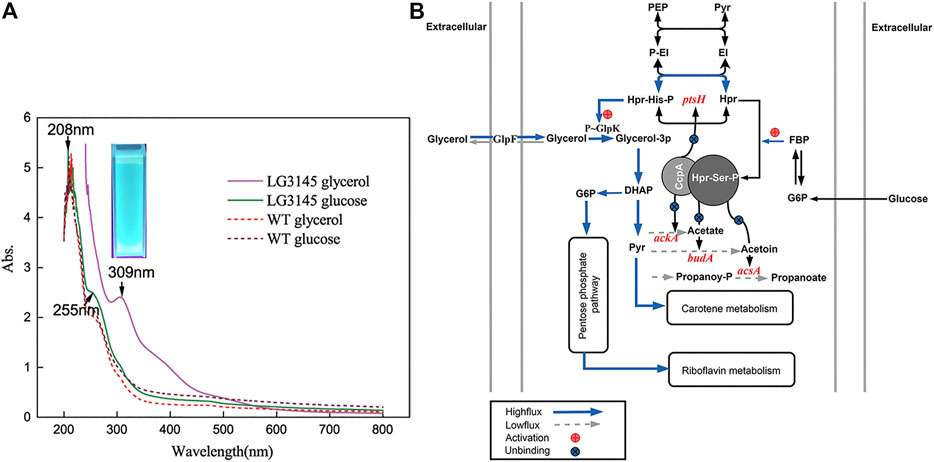
FIGURE 5. (A) Cytochrome synthesis-related pathways. (B) UV-Vis spectral analysis of LG3145 and WT cells incubated in MM supplemented with glycerol or glucose and cultured at 37°C. Images were obtained using a Chemidoc XRS + system (trans-UV 302 nm, BioRad, United States).
Secondary Metabolism is Enhanced in B. pumilus LG3145
For cytochrome experiments, liquid chromatography-mass spectrometry (LC-MS) was employed to probe molecular organisation. Unexpectedly, we found an amino sugar antibiotic, kanosamine (3-amino-3-deoxy-D-glucose), in the extracellular secretions of LG3145 cells incubated in MM supplemented with glucose. The mass spectrum of the metabolite (Figure 6A) resembled the kanamycin standard spectrum in the mzCloud MS database. Kanosamine, a component of kanamycin, can be produced in a three-step pathway from glucose 6-phosphate in Bacillus spp. (Prasertanan and Palmer, 2019).
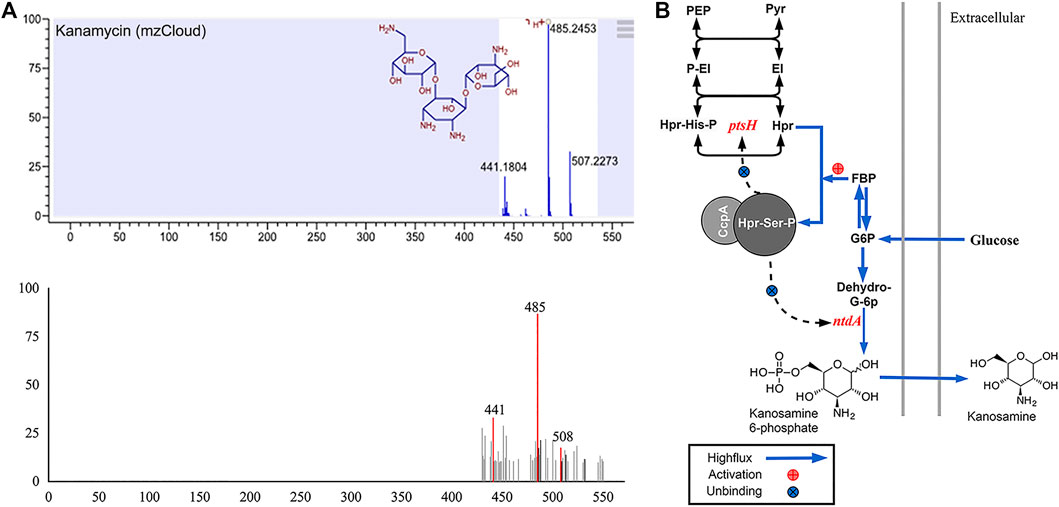
FIGURE 6. (A) MS spectrum showing kanamycin among LG3145 metabolites and the standard spectrum in mzCloud. (B) Kanosamine synthesis-related pathways in B. pumilus.
As shown in Figure 6B, glucose is phosphorylated after entering cells, and the more glucose 6-phosphate, the more CcpA∼(Hpr-Ser-p) complex in vivo. However, in LG3145, cre site mutation at ntdA encoding a sugar aminotransferase (Van Straaten et al., 2013) resulted in the CcpA∼(Hpr-Ser-p) complex that was unable to bind the promoter, regardless of how much glucose was included in the medium. Additionally, ptsH encoding the key HPr factor related to CCR, was also mutated at the cre site, and the more glucose transported into the cells, the greater the expression of Hpr and Hpr-Ser-p, but this did not repress expression of ptsH and ntdA. When glucose is increased, the kanosamine synthesis pathway is enhanced and secretion is elevated.
Conclusion
Herein, a novel CRISPR gene editing strategy was used to edit multi-target cre-boxes and relieve CCR in B. pumilus. We constructed a series of unspecific gRNAs as crRNAs according to similarity in the CRISPR/Cas9 system. Using seven metabolic genes as the editing targets, wobble editing occurred around cre sites, and this positively correlated with similarity. In addition, the phenotypes of the LG3145 mutant strain were markedly altered, and this was correlated with the seven edited genes. These phenotypes demonstrated that mutation at multiple cre sites caused target gene transcription changes and disruption of the carbon central pathway, which resulted in an increase in secondary metabolites such as extracellular pigments, proteins, and kanosamine, and the formation of polysaccharide capsules. The B. pumilus LG3145 strain has great potential as a host organism for secretion in cell factory processes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YW designed and performed experiments, and wrote the first draft of the manuscript; LC, MB, SW, MC, and XC participated in experiment design and analysis of experiment results; MY and LH designed and supervised the project, and guided the experimental design, data analysis, manuscript writing and revision. All authors read and agreed to the final text.
Funding
The publication fees for open access can be supported by National Natural Science Foundation of China (42077212) and Tianjin Technical Expert Project (Grant No.14JCTPJC00479).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We greatly appreciate Tianjin Academy of Agricultural Sciences of China for providing wild type strain of B. pumilus.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.717609/full#supplementary-material
References
Chen, B., Wen, J., Zhao, X., Ding, J., and Qi, G. (2020). Surfactin: A Quorum-sensing Signal Molecule to Relieve CCR in Bacillus Amyloliquefaciens. Front. Microbiol. 11, 631. doi:10.3389/fmicb.2020.00631
Chen, C., Wang, L., Lu, Y., Yu, H., and Tian, H. (2019). Comparative Transcriptional Analysis of Lactobacillus Plantarum and its ccpA-Knockout Mutant under Galactooligosaccharides and Glucose Conditions. Front. Microbio. 10, 1584. doi:10.3389/fmicb.2019.01584
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. doi:10.1126/science.1231143
Dahl, M. K., and Hillen, W. (1995). Contributions of Xy1R, CcpA and HPr to Catabolite Repression of the Xyl Operon inBacillus Subtilis. FEMS Microbiol. Lett. 132, 79–83. doi:10.1111/j.1574-6968.1995.tb07814.x
Guilinger, J. P., Thompson, D. B., and Liu, D. R. (2014). Fusion of Catalytically Inactive Cas9 to FokI Nuclease Improves the Specificity of Genome Modification. Nat. Biotechnol. 32, 577–582. doi:10.1038/nbt.2909
Han, L.-L., Liu, Y.-C., Miao, C.-C., and Feng, H. (2019). Disruption of the Pleiotropic Gene scoC Causes Transcriptomic and Phenotypical Changes in Bacillus Pumilus BA06. BMC Genomics 20, 327. doi:10.1186/s12864-019-5671-8
Han, L., Liu, K., Ma, W., Jiang, Y., Hou, S., Tan, Y., et al. (2020). Redesigning Transcription Factor Cre1 for Alleviating Carbon Catabolite Repression in Trichoderma Reesei. Synth. Syst. Biotechnol. 5, 230–235. doi:10.1016/j.synbio.2020.07.002
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided Editing of Bacterial Genomes Using CRISPR-Cas Systems. Nat. Biotechnol. 31, 233–239. doi:10.1038/nbt.2508
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. doi:10.1126/science.1225829
Kraus, A., Hueck, C., Gärtner, D., and Hillen, W. (1994). Catabolite Repression of the Bacillus Subtilis Xyl Operon Involves a Cis Element Functional in the Context of an Unrelated Sequence, and Glucose Exerts Additional xylR-dependent Repression. J. Bacteriol. 176, 1738–1745. doi:10.1128/jb.176.6.1738-1745.1994
Langa, S., Peirotén, Á., Arqués, J. L., and Landete, J. M. (2021). Catabolite Responsive Elements as a Strategy for the Control of Heterologous Gene Expression in Lactobacilli. Appl. Microbiol. Biotechnol. 105, 225–233. doi:10.1007/s00253-020-11010-2
Li, K., Zhao, L.-S., Sun, H.-M., Tang, R.-C., Chen, Z.-H., Su, H.-N., et al. (2018). Capsular Polysaccharide Production from Zunongwangia Profunda SM-A87 Monitored at Single Cell Level by Atomic Force Microscopy. Deep Sea Res. Part Topical Stud. Oceanography 155, 44–49. doi:10.1016/j.dsr2.2017.08.009
Lovering, A. L., Safadi, S. S., and Strynadka, N. C. J. (2012). Structural Perspective of Peptidoglycan Biosynthesis and Assembly. Annu. Rev. Biochem. 81, 451–478. doi:10.1146/annurev-biochem-061809-112742
Meng, J., Zhao, X., Tang, X., Xia, Y., Ma, X., and Gao, D. (2017). Surface Characterization of ICF Capsule by AFM-Based Profilometer. High Pow Laser Sci. Eng. 5, e21. doi:10.1017/hpl.2017.20
Moir-Blais, T. R., Grundy, F. J., and Henkin, T. M. (2001). Transcriptional Activation of the Bacillus Subtilis ackA Promoter Requires Sequences Upstream of the CcpA Binding Site. J. Bacteriol. 183 (7), 2389–2393. doi:10.1128/jb.183.7.2389-2393.2001
Neira, J. L., Cámara-Artigas, A., Hernández-Cifre, J. G., and Ortore, M. G. (2021). The Histidine Phosphocarrier Kinase/phosphorylase from Bacillus Subtilis Is an Oligomer in Solution with a High thermal Stability. Ijms 22, 3231. doi:10.3390/ijms22063231
Peng, Q., Zhao, X., Wen, J., Huang, M., Zhang, J., and Song, F. (2020). Transcription in the Acetoin Catabolic Pathway Is Regulated by AcoR and CcpA in Bacillus Thuringiensis. Microbiol. Res. 235, 126438. doi:10.1016/j.micres.2020.126438
Prasertanan, T., and Palmer, D. R. J. (2019). The Kanosamine Biosynthetic Pathway in Bacillus Cereus UW85: Functional and Kinetic Characterization of KabA, KabB, and KabC. Arch. Biochem. Biophys. 676, 108139. doi:10.1016/j.abb.2019.108139
Rausch, M., Deisinger, J. P., Ulm, H., Müller, A., Li, W., Hardt, P., et al. (2019). Coordination of Capsule Assembly and Cell wall Biosynthesis in Staphylococcus aureus. Nat. Commun. 10, 1404. doi:10.1038/s41467-019-09356-x
Scarff, J. M., Seldina, Y. I., Vergis, J. M., Ventura, C. L., and O'Brien, A. D. (2018). Expression and Contribution to Virulence of Each Polysaccharide Capsule of Bacillus Cereus Strain G9241. PLoS ONE 13, e0202701. doi:10.1371/journal.pone.0202701
Sousa, J., Westhoff, P., Methling, K., and Lalk, M. (2019). The Absence of Pyruvate Kinase Affects Glucose-dependent Carbon Catabolite Repression in Bacillus Subtilis. Metabolites 9, 216. doi:10.3390/metabo9100216
Stülke, J., and Hillen, W. (2000). Regulation of Carbon Catabolism inBacillusSpecies. Annu. Rev. Microbiol. 54, 849–880. doi:10.1146/annurev.micro.54.1.849
Su, H.-N., Chen, Z.-H., Liu, S.-B., Qiao, L.-P., Chen, X.-L., He, H.-L., et al. (2012). Characterization of Bacterial Polysaccharide Capsules and Detection in the Presence of Deliquescent Water by Atomic Force Microscopy. Appl. Environ. Microbiol. 78, 3476–3479. doi:10.1128/aem.00207-12
Su, Y., Liu, C., Fang, H., and Zhang, D. (2020). Bacillus Subtilis: A Universal Cell Factory for Industry, Agriculture, Biomaterials and Medicine. Microb. Cel Factories 19, 173. doi:10.1186/s12934-020-01436-8
Sun, X., and Zhang, J. (2021). Bacterial Exopolysaccharides: Chemical Structures, Gene Clusters and Genetic Engineering. Int. J. Biol. Macromolecules 173, 481–490. doi:10.1016/j.ijbiomac.2021.01.139
Turinsky, A. J., Moir-Blais, T. R., Grundy, F. J., and Henkin, T. M. (2000). Bacillus Subtilis ccpA Gene Mutants Specifically Defective in Activation of Acetoin Biosynthesis. J. Bacteriol. 182 (19), 5611–5614. doi:10.1128/jb.182.19.5611-5614.2000
Van Straaten, K. E., Ko, J. B., Jagdhane, R., Anjum, S., Palmer, D. R. J., and Sanders, D. A. R. (2013). The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus Subtilis, Reveals a New Subclass of Aminotransferases. J. Biol. Chem. 288, 34121–34130. doi:10.1074/jbc.m113.500637
Wan, M.-Y., Wang, H.-Y., Zhang, Y.-Z., and Feng, H. (2009). Substrate Specificity and Thermostability of the Dehairing Alkaline Protease from Bacillus Pumilus. Appl. Biochem. Biotechnol. 159, 394–403. doi:10.1007/s12010-008-8497-4
Westers, L., Westers, H., and Quax, W. J. (2004). Bacillus Subtilis as Cell Factory for Pharmaceutical Proteins: a Biotechnological Approach to Optimize the Host Organism. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1694, 299–310. doi:10.1016/j.bbamcr.2004.02.011
Wu, W., Yang, Y., and Lei, H. (2019). Progress in the Application of CRISPR: From Gene to Base Editing. Med. Res. Rev. 39, 665–683. doi:10.1002/med.21537
Keywords: Bacillus pumilus, UgRNA:Cas9, cre-box, CCR, multiple gene wobble editing
Citation: Wang Y, Cao L, Bi M, Wang S, Chen M, Chen X, Ying M and Huang L (2021) Wobble Editing of Cre-box by Unspecific CRISPR/Cas9 Causes CCR Release and Phenotypic Changes in Bacillus pumilus. Front. Chem. 9:717609. doi: 10.3389/fchem.2021.717609
Received: 01 June 2021; Accepted: 30 July 2021;
Published: 09 August 2021.
Edited by:
Youhui Lin, Xiamen University, ChinaReviewed by:
Jiantao Guo, University of Nebraska-Lincoln, United StatesParipok Phitsuwan, King Mongkut’s University of Technology Thonburi, Thailand
Copyright © 2021 Wang, Cao, Bi, Wang, Chen, Chen, Ying and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Ying, eW1AdGp1dC5lZHUuY24=; Lei Huang, aHVhbmdsZWlAdGp1dC5lZHUuY24=
 Yingxiang Wang
Yingxiang Wang Linfeng Cao
Linfeng Cao Meiying Bi
Meiying Bi Sicheng Wang
Sicheng Wang Meiting Chen
Meiting Chen Xingyu Chen
Xingyu Chen Ming Ying
Ming Ying Lei Huang
Lei Huang