- Freie Universität Berlin, Department of Physics, Theoretical Molecular Biophysics Group, Berlin, Germany
Membrane-bound proteins that change protonation during function use specific protein groups to bind and transfer protons. Knowledge of the identity of the proton-binding groups is of paramount importance to decipher the reaction mechanism of the protein, and protonation states of prominent are studied extensively using experimental and computational approaches. Analyses of model transporters and receptors from different organisms, and with widely different biological functions, indicate common structure-sequence motifs at internal proton-binding sites. Proton-binding dynamic hydrogen-bond networks that are exposed to the bulk might provide alternative proton-binding sites and proton-binding pathways. In this perspective article I discuss protonation coupling and proton binding at internal and external carboxylate sites of proteins that use proton transfer for function. An inter-helical carboxylate-hydroxyl hydrogen-bond motif is present at functionally important sites of membrane proteins from archaea to the brain. External carboxylate-containing H-bond clusters are observed at putative proton-binding sites of protonation-coupled model proteins, raising the question of similar functionality in spike protein S.
Introduction
Proton transfer reactions are fundamental to cells from all branches of life. Among proteins that use proton binding and proton transfer for biological function, one of the best studied is bacteriorhodopsin, a small light-driven proton pump of the halophile archaeon Halobacterium salinarium. The first three-dimensional structure of bacteriorhodopsin, solved in 1975 by Henderson and Unwin using cryo-Electron Microscopy (cryo-EM), identified the seven transmembrane helices of the protein at a resolution of 7Å (Henderson and Unwin, 1975). In recent years, structures of bacteriorhodopsin solved with time-resolved serial femtosecond crystallography provided an atomic-resolution movie of structural changes during the proton-transfer reaction cycle (Nango et al., 2016; Weinert et al., 2019). Knowledge derived from simple model systems such as bacteriorhodopsin and other microbial rhodopsins can inform and guide studies of significantly more complex proteins and protein complexes that use protonation change and proton transfer during their reaction cycles (Ernst et al., 2014; Govorunova et al., 2017; Bondar and Lemieux, 2019). An example are the ASIC-like ion channels thought to participate in a cellular pH-sensing mechanism of specialized neurons (Jalalvand et al., 2016; Jalalvand et al., 2018). During ischemia and traumatic brain injury, a drop in extracellular pH and release of glutamate associate with hyperactive N-methyl-D-aspartate (NMDA) receptors and neuronal cell death (Regan et al., 2019).
pH sensing by proteins is thought to involve the chemical reaction of protonation change of a relatively small number of the titratable groups, typically carboxylate and histidine sidechains (Damaghi et al., 2013; Schönichen et al., 2013). Histidine sidechains, with their nominal pKa of 6.54 ± 0.4, can titrate at physiological values of cytosolic pH (Schönichen et al., 2013). The pKa value of an amino acid residue may, however, depend strongly on the environment (Thurlkill et al., 2006a). For example, whereas the nominal pKa values for aspartic and glutamic acids in solution are, respectively, 3.67 ± 0.4 and 4.25 ± 0.5 (Thurlkill et al., 2006b), pKa values as low as 1.56 ± 0.14 and as high as seven were determined with Nuclear Magnetic Resonance (NMR) for aspartic groups of the soluble protein PsbO (Gerland et al., 2020). In this perspective article, common proton-binding and H-bonding motifs are discussed with examples from simple model systems to more complex proteins.
An Inter-helical Carboxylate-Hydroxyl H-Bond Motif Common to Membrane Transporters and Receptors
Membrane insertion of Asp/Glu groups is energetically costly, with apparent free energy penalties of ∼4–5 kcal/mol (Xie et al., 2007), and indeed there is only a relatively small probability that Asp and Glu locate in the hydrophobic core of α-helical membrane proteins (Ulmschneider and Sansom, 2001). Presence of carboxylates in transmembrane regions of membrane transporters and receptors is thus of direct interest to evaluate structure-function relationships, particularly for proteins in which these carboxylate groups could be involved in proton binding and proton transfer. Examples discussed here suggest that internal carboxylates used by membrane proteins for proton binding might be part of common inter-helical H-bond motifs; this could guide studies of other, more complex proteins for which proton-binding groups are yet to be identified.
A prominent example of a membrane transporter that uses internal carboxylates to bind protons is bacteriorhodopsin, which for decades has served as model system to study proton pumps (Lanyi, 1999; Herzfeld and Tongue, 2000; Balashov and Ebrey, 2001; Kandori, 2004; Kouyama et al., 2004; Gerwert et al., 2014). The primary proton acceptor group of bacteriorhodopsin is D85 (Metz et al., 1992), a carboxylate within one helical turn of T89, whose sidechain can serve as intermediate carrier for the proton transferred from the Schiff base proton donor to D85 (Bondar et al., 2004; Nango et al., 2016; Ni et al., 2018) (Figure 1A). Adjacent to T89, T90 has an inter-helical H-bond with D115, a carboxylate that stays protonated throughout the entire reaction cycle (Metz et al., 1992), and an intra-helical H-bond to the backbone carbonyl of W86 (del Val et al., 2014a). In the structure of the resting state of bacteriorhodopsin (Luecke et al., 1999), the cytoplasmic proton donor D96 (Gerwert et al., 1989) is within inter-helical H-bond distance from the T46 hydroxyl, which is within H-bond distance from the backbone carbonyl of F42 (Figure 1A); later during the reaction cycle, rearrangements of hydrophobic sidechains associate with formation of a water chain from D96 to the retinal Schiff base (Weinert et al., 2019). That is, at two internal sites where protons bind, bacteriorhodopsin can have inter-helical carboxylate-hydroxyl H-bonding and intra-helical H-bonding of the Thr hydroxyl group to a backbone carbonyl group (del Val et al., 2014a). The fact that only D96 changes protonation, whereas D115 remains protonated, suggests local protein environment other than with the direct H-bond partner impacts protonation change (del Val et al., 2014a). At the extracellular proton-release side, H-bonding between S193 and E204 depends on the protonation state (Gerwert et al., 2014).
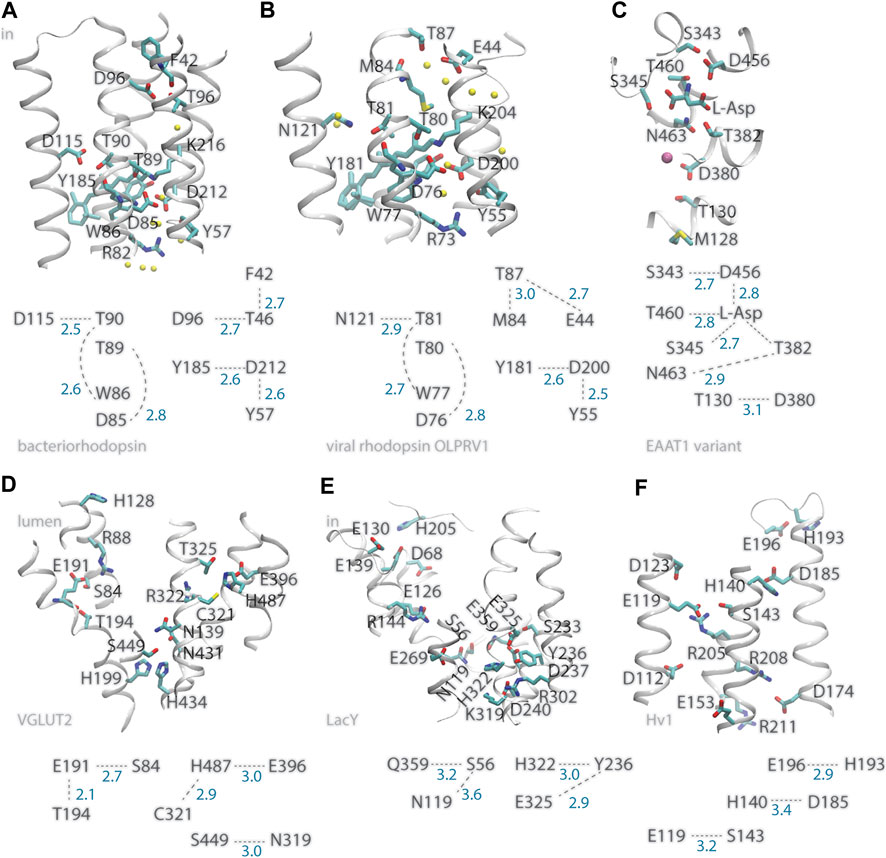
FIGURE 1. Illustration of inter-helical carboxylate-hydroxyl H-bond motifs. For clarity, only selected backbone groups are shown. Numbers in blue represent H-bond distances in Å. (A) The bacteriorhodopsin proton pump, PDB ID:5ZIM (Hasegawa et al., 2018). (B) The viral ion channel OLPVR1, PDB ID:7AKY (Zabelskii et al., 2021). (C) EAAT1 bound to L-Asp substrate, PDB ID:5LLU (Canul-Tec et al., 2017). (D) The glutamate transporter VGLUT2, PDB ID:64VD (Li et al., 2020). (E) The LacY symporter, PDB ID:2V8N. (F) The HV1 proton channel, PDB ID:5OKK, molecule #7 (Bayrhuber et al., 2019). All molecular graphics were prepared with Visual Molecular Dynamics, VMD (Humphrey et al., 1996).
Similar inter-helical carboxylate-hydroxyl motifs were identified in structures of other membrane transporters, such as the C1C2 channelrhodopsin chimera and the sodium pump rhodopsin KR2 (del Val et al., 2014b; Bondar and Smith, 2017; Siemers et al., 2019), the SERCA calcium pump ATPase, and the multidrug transporter AcrB (del Val et al., 2014a; Bondar and Lemieux, 2019), and can be observed in the recently solved structure of viral rhodopsin OLPVR1, a sodium/potassium ion channel (Zabelskii et al., 2021) (Figure 1B). Likewise, recently solved structures of glutamate transporters VGLUT2 (Li et al., 2020) and EAAT1 (Canul-Tec et al., 2017), which are members of families of neurotransmitter transporters essential for the functioning of the brain (Münster-Wandowski et al., 2016; Martineau et al., 2017), indicate inter-helical carboxylate-hydroxyl motifs at functionally important sites of both transporters: In the structure of VGLUT2, the putative pH sensor E191 (Li et al., 2020) H-bonds to S84 and T194 (Figure 1D); in EAAT1, D380 is within H-bond distance from T130 and thus close to M128, a group whose mutation to Arg inhibits glutamate transport (Chivukala et al., 2020); D456 H-bonds to the L-Asp substrate and to S343—which was noted as a conserved group whose mutation in wild-type EAAT1 inhibits transport (Canul-Tec et al., 2017); L-Asp H-bonds to three Ser/Thr hydroxyl groups (Figure 1C).
G-Protein Coupled Receptors (GPCRs) are membrane proteins that mediate communication between cells and their external environments. Protonation change might occur at carboxylate D2.50 (Zhang et al., 2015; Vickery et al., 2018), a carboxylate group part of the sodium-binding site in many class-A GPCRs (Katritch et al., 2014). H-bond networks conserved among static structures of GPCRs show D2.50 paired with S7.46, that is, a carboxylate-hydroxyl pair is present at a functionally important site (Bertalan et al., 2020) [the Ballesteros and Weinstein numbering scheme is used for GPCR groups (Ballesteros and Weinstein, 1995)].
Inter-helical H-bonding between Asp/Glu and Ser/Thr could provide an efficient mechanism to couple protonation change with local protein dynamics (Bondar and Smith 2017). Hydrophobic contacts of non-polar segments of these H-bond partners can reduce H-bond dynamics and strengthen H-bonding (Nemethy et al., 1963; Fernandez and Scheraga, 2003), and alteration of the pKa of an internal carboxylate might require local protein conformational change enabling water access to the carboxylate (Szaraz et al., 1994). In bacteriorhodopsin, the T90-D115 H-bond (Figure 1A) is in a local environment with more non-polar contacts than T46-D96, and its H-bond distance experiences smaller-amplitude fluctuations at room temperature (del Val et al., 2014a); the lesser hydrophobic packing near T46-D96 associates with water molecules entering rapidly the vicinity of D96 when the protein is mutated elsewhere (del Val et al., 2014a), and during the reaction cycle of the wild-type pump a file of waters assembles to bridge D96 to the Schiff base proton acceptor (Freier et al., 2011; Weinert et al., 2019).
Protonation change at an inter-helical carboxylate-hydroxyl pair might couple to altered local dynamics of the helix hosting the hydroxyl group: Ser/Thr hydroxyl groups can compete with backbone amide groups for H-bonding to a carbonyl group (Gray and Matthews, 1984; Presta and Rose, 1988), which increases local fluctuations of the helix (del Val et al., 2012). Alternatively, altered helix dynamics might impact local dynamics and hydration of the hydroxyl-carboxylate H-bond (Figure 2A).
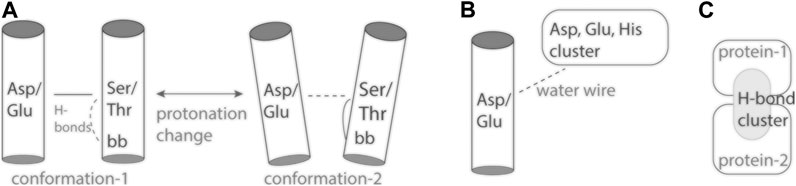
FIGURE 2. Cartoon representation of inter-helical carboxylate-hydroxyl H-bonds and H-bond clusters. (A) The inter-helical carboxylate-hydroxyl H-bond may couple protonation change with changes in local protein dynamics (Bondar and Smith, 2017). (B) An internal carboxylate group and an external proton antenna cluster communicate via a water wire. (C) An interface H-bond cluster, as observed for spike protein S bound to ACE2 (Karathanou et al., 2020).
Other H-Bond Motifs at Internal Proton-Binding Sites of Membrane Transporters
Local H-bonding and hydrophobic packing are likely key mechanisms used by proteins to control protonation of a proton-binding group–and different solutions as to which sidechain serves as H-bond partner for proton-transfer sidechains of membrane transporters appear to exist.
In the plasma membrane proton pump AHA2, which is a P-type ATPase, the primary proton donor/acceptor D684 (Buch-Pedersen and Palmgren, 2003) makes an inter-helical H-bond with an Asn (Pedersen et al., 2007; Guerra and Bondar, 2015; Focht et al., 2017); Likewise, OLPRV1, EAAT1, VGLUT2, and LacY, have at functionally important sites inter-helical bonds between Ser/Thr and Asn instead of Asp (Figure 1B–E).
In bacteriorhodopsin D212, which might functions as an intermediate proton carrier (Dioumaev et al., 1999; Phatak et al., 2009) has inter-helical H-bonds with Y57 and Y185 (Figure 1A), both important for optimal control of H-bond dynamics in the wild-type pump: mutating either Tyr group alters the kinetics of the reaction cycle (Rath et al., 1993; Govindjee et al., 1995), and Y57F has weak Y185-D212 H-bonding (del Val et al., 2014a). In the Escherichia coli proton-galactopyranoside symporter LacY E325, which deprotonates during the reaction cycle (Sahin-Toh and Kaback, 2001; Guan and Kaback, 2006), is part of a local H-bond network with Tyr and His sidechains (Figure 1E). Deprotonation of E325 causes protein structure changes that depend on the composition of the surrounding lipid bilayer (Andersson et al., 2012).
Hv1, also known as HVCN1 (human voltage-gated proton channel) is a proton-selective ion channel whose action early after ischemia contributes to brain damage by providing the cancer cell with protons to compensate for the electron-transfer action of phagocytic cell NADPH oxidase (Wu et al., 2012). Proton selectivity of Hv1 requires a carboxylate group at D112 (Figure 1F), as some mutants with a neutral group instead Asp or Glu at this position loose proton specificity and became anion selective, and proton specificity could not be recovered when mutating D112 to His (Musset et al., 2011). That Hv1 becomes predominantly anion selective when its proton sensor is neutralized by mutation (Musset et al., 2011) resembles the behavior of bacteriorhodopsin, which from an outward proton pump becomes an inward chloride pump when the primary proton acceptor D85 is mutated to Thr (Sasaki et al., 1995), and underlines the need to decipher the role of electrostatic interactions–of static protein groups and of mobile ions, that govern charge states along the reaction cycle of ion transporters (Song and Gunner, 2014). The recent solution NMR structure of Hv1 reports for the sidechains of D112, R208, and R211 (Figure 1F) coordinates modeled by considering steric packing (Bayrhuber et al., 2019), and interactions of internal arginine sidechains can change in the presence of membrane voltage (Geragotelis et al., 2020). This suggests Hv1 has a rather dynamical internal H-bond network.
H-Bond Networks of Histidine Sidechains at Proton-Binding Sites of Membrane Transporters and Receptors
Internal histidine groups can be central to the reaction mechanisms of protonation-coupled transporters and receptors. Examples include 1) H322 of LacY is part of an inter-helical H-bond motif with E325 at a proton-binding site (Figure 1D); 2) H128 of VGLU2 is thought to pick up a proton at the lumen side of the membrane, and connect to the E119 proton sensor via a water-filled pore (Li et al., 2020) (Figure 1); 3) H140 of Hv1 is a putative binding site for Zn2+ (DeCoursey et al., 2016; Bayrhuber et al., 2019), and its inter-helical H-bond partner D185 might be involved in ΔpH-gating (DeCoursey, 2018); 4) A pair of two H37 groups of the influenza channel M2 could participate in a proton-conductance mechanism that involves low-barrier H-bonding (Fu et al., 2020); 5) A cluster of histidine sidechains was proposed to ensure proton-sensing by the ovarian cancer GPCR 1, known as OGR1 or GPR68 (Ludwig et al., 2003; Holzer, 2009). OGR1, and other members of the proton-sensing GPCR subgroup, are activated when the extracellular pH becomes acidic, and histidine protonation couples with protein conformational change (Radu et al., 2004; Wang et al., 2004).
Taken together, the examples above suggest that internal histidine sidechains implicated in proton binding tend to be part of local H-bond clusters with carboxylate and/or other histidine sidechains. An Asp-His pair is also involved in pH sensing by the soluble protein hemoglobin (Srivastava et al., 2007).
Carboxylate Clusters as Proton Antennas on Protein Surfaces
Proton antennas consist of clusters of closely spaced carboxylate, or of carboxylate and histidine groups, which can capture protons and prolong the time protons spend bound at the surface of the protein (Gutman and Nachliel, 1990; Sacks et al., 1998; Ädelroth and Brzezinski, 2004), and deliver a proton to a proton-entry site (Checover et al., 2001) (Figure 2B). Putative proton antenna clusters have been identified for bacteriorhodopsin (Checover et al., 2001), cytochrome c oxidase (Ädelroth and Brzezinski, 2004), and for the PsbO subunit of photosystem II (Shutova et al., 2007; Lorch et al., 2015; Kemmler et al., 2019). Surface carboxylates could be part of a pH-dependent conformational switch of PsbO (Bommer et al., 2016), and of a local buffering mechanism used by photosystem II to respond to acidification of the lumen (Gerland et al., 2020). The two carboxylate groups thought to bind the proton are part of a dynamic protein-water H-bond network that extends across the interface between PsbO and another protein subunit of photosystem II (Kemmler et al., 2019).
H-Bond Networks of SARS-CoV-2 Protein S
The acidic environment of the endosome activates membrane fusion of coronaviruses, but it remains unclear whether acidity is required for proteases that cleave protein S to activate fusion, or for the spike protein itself (Heald-Sargent and Gallagher, 2012; Li, 2016). Membrane fusion might also be assisted by calcium binding to conserved negatively charged groups of SARS-CoV spike protein S (Millet and Whittaker, 2018).
Sequences of corona spike proteins S tend to carry significant net negative charges and contain patches rich in carboxylate, or carboxylate and histidine groups (Karathanou et al., 2020), but a discussion of a putative involvement of these patches in ion binding would be speculative. From graph-based analyses we discovered that titratable groups can participate in H-bond clusters that tend to have three-fold compositional symmetry when SARS-CoV-2 protein S is closed (Karathanou et al., 2020). Loss of this three-fold symmetry in the pre-fusion conformation could contribute to conformational selection of one protein S protomer to bind to the ACE2 receptor (Karathanou et al., 2020). An H-bond network that extends across the interface between the Receptor Binding Domain (RBD) of protein S and ACE2 contains N501 (Karathanou et al., 2020), which is mutated to Tyr in the United Kingdom virus variant (Leung et al., 2021). E484, which is mutated to Lys in the South African variant (Tegally et al., 2021), is close in sequence to RBD-Y449, which H-bonds to ACE2-D38 (Karathanou et al., 2020).
Elsewhere on the surface of protein S, intra-molecular H-bonding of D614 would be lost in the D614G virus variant (Korber et al., 2020); this mutation favors enhanced conformational dynamics of the spike protein, which could enable ACE2 binding (Benton et al., 2021). At the C-terminal region of the ectodomain of the closed protein S, a symmetrical H-bond cluster of no fewer than 33 protein groups includes six carboxylates and six histidine sidechains (Karathanou et al., 2020). Close to the R815 proteolytic cleavage site, E819 H-bonds to three Ser/Thr groups in all protomers of the closed and open conformations, and contributes to a large H-bond cluster in a pre-fusion protomer (Karathanou et al., 2020). Cryo-EM data suggest D830 and D843 are part of a pH-dependent switch that enables protein S to adopt, at low pH, a conformation incompatible with binding of particular antibodies (Zhou et al., 2020). Future experiments and computation are needed to evaluate any pH-dependent conformational changes at the interface between protein S and ACE2 (Figure 2C).
Conclusion
Proteins across all branches of life rely on proton binding and proton transfer to exert their biological function. As three-dimensional structures of proteins and macro-molecular protein complexes involved in human physiology and disease become available, reasonable predictions of likely proton-binding sites and proton-transfer events are of interest to understand reaction mechanisms and potentially guide drug design. Simpler model proteins inform on H-bonding motifs common at proton-binding sites, and on structural elements that can shape the energetics of proton transfers in heterogeneous protein environments. Examples discussed here (Figure 1) illustrate an inter-helical H-bond carboxylate-hydroxyl motif that can ensure coupling between carboxylate protonation, protein conformational dynamics, and local water accessibility (Figure 2A). On the surface of the protein, carboxylate clusters may transiently bind a proton to then deliver it to an internal proton-binding site (Figure 2B) or for buffering, and participate in pH-dependent conformational switching that could impact protein interactions (Figure 2C). Dissecting the functional role of surface proton-binding clusters would need to account for local water interactions and protein conformational dynamics.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: Protein Data Bank, with accession codes as indicated in the article.
Author Contributions
A-NB wrote the manuscript and prepared the figures.
Funding
European Union’s Horizon 2020 research and innovation program, Marie Sklodowska-Curie grant agreement No 860592, Innovative Training Network “Proton and proton-coupled transport”. German Cancer Research Foundation (DFG) Collaborative Research Center SFB 1078, project C4.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research reviewed here was supported in part by the German Research Foundation, DFG, Collaborative Research Center SFB1078 Protonation Dynamics in Protein Function, SFB1078, Project C4. Research supported in part by the Innovative Training Network PROTON founded by the European Commission. I acknowledge support from the Open Access Publication Initiative of the Freie Universität Berlin.
References
Ädelroth, P., and Brzezinski, P. (2004). Surface-mediated Proton-Transfer Reactions in Membrane-Bound Proteins. Biochim. Biophys. Acta (Bba) - Bioenerg. 1655, 102–115. doi:10.1016/j.bbabio.2003.10.018
Andersson, M., Bondar, A.-N., Freites, J. A., Tobias, D. J., Kaback, H. R., and White, S. H. (2012). Proton-coupled Dynamics in Lactose Permease. Structure 20, 1893–1904. doi:10.1016/j.str.2012.08.021
Balashov, S. P., and Ebrey, T. G. (2001). Trapping and Spectroscopic Identification of the Photointermediates of Bacteriorhodopsin at Low Temperatures. Photochem. Photobiol. 73, 453–462. doi:10.1562/0031-8655(2001)0730453TASIOT2.0.CO2
Ballesteros, J. A., and Weinstein, H. (1995). Integrated Methods for the Construction of Three-Dimensional Models and Computational Probing of Structure-Function Relations in G Protein-Coupled Receptors. Methods Neurosci. 25, 366–428. doi:10.1016/s1043-9471(05)80049-7
Bayrhuber, M., Maslennikov, I., Kwiatkowski, W., Sobol, A., Wierschem, C., Eichmann, C., et al. (2019). Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel. Biochemistry 58, 4017–4027. doi:10.1021/acs.biochem.9b00471
Benton, D. J., Wroberl, A. G., Roustan, C., Borg, A., Xu, P., Martin, S. R., et al. (2021). The Effect of the D614G Substitution on the Structure of the Spike Glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. 118, e2022586118. doi:10.1073/pnas.2022586118
Bertalan, É., Lešnik, S., Bren, U., and Bondar, A.-N. (2020). Protein-water Hydrogen-Bond Networks of G Protein-Coupled Receptors: Graph-Based Analyses of Static Structures and Molecular Dynamics. J. Struct. Biol. 212, 107634. doi:10.1016/j.jsb.2020.107634
Bommer, M., Bondar, A.-N., Zouni, A., Dobbek, H., and Dau, H. (2016). Crystallographic and Computational Analysis of the Barrel Part of the PsbO Protein of Photosystem II: Carboxylate-Water Clusters as Putative Proton Transfer Relays and Structural Switches. Biochemistry 55, 4626–4635. doi:10.1021/acs.biochem.6b00441
Bondar, A.-N., Elstner, M., Suhai, S., Smith, J. C., and Fischer, S. (2004). Mechanism of Primary Proton Transfer in Bacteriorhodopsin. Structure 12, 1281–1288. doi:10.1016/j.str.2004.04.016
Bondar, A.-N., and Lemieux, M. J. (2019). Reactions at Biomembrane Interfaces. Chem. Rev. 119, 6162–6183. doi:10.1021/acs.chemrev.8b00596
Bondar, A.-N., and Smith, J. C. (2017). Protonation-state-Coupled Conformational Dynamics in Reaction Mechanisms of Channel and Pump Rhodopsins. Photochem. Photobiol. 93, 1336–1344. doi:10.1111/php.12790
Buch-Pedersen, M. J., and Palmgren, M. G. (2003). Conserved Asp684 in Transmembrane Segment M6 of the Plant Plasma Membrane P-type Proton Pump AHA2 Is a Molecular Determinant of Proton Translocation. J. Biol. Chem. 278, 17845–17851. doi:10.1074/jbc.m212729200
Canul-Tec, J. C., Assal, R., Cirri, E., Legrand, P., Brier, S., Chamot-Rooke, J., et al. (2017). Structure and Allosteric Inhibition of Excitatory Amino Acid Transporter 1. Nature 544, 446–451. doi:10.1038/nature22064
Checover, S., Marantz, Y., Nachliel, E., Gutman, M., Pfeiffer, M., Tittor, J., et al. (2001). Dynamics of the Proton Transfer Reaction on the Cytoplasmic Surface of Bacteriorhodopsin†. Biochemistry 40, 4281–4292. doi:10.1021/bi002574m
Chivukala, A. S., Suslova, M., Kortzak, D., Kovermann, P., and Fahlke, C. (2020). Functional Consequences of SLC1A3 Mutations Associated with Episodic Ataxia 6. Hum. Mutat , 1–14. doi:10.1002/humu.24089
Damaghi, M., Wojktowiak, J. W., and Gillies, R. J. (2013). pH Sensing and Regulation in Cancer. Front. Physiol. 4, 1–10. doi:10.3389/fphys.2013.00370
DeCoursey, T. E., Morgan, D., Musset, B., and Cherny, V. V. (2016). Insights into the Structure and Function of Hv1 from a Meta-Analysis of Mutation Studies. J. Gen. Physiol. 148, 97–118. doi:10.1085/jgp.201611619
DeCoursey, T. E. (2018). Voltage and pH Sensing by the Voltage-Gated Proton Channel, Hv1. J. R. Soc. Interf. 15, 20180108. doi:10.1098/rsif.2018.0108
Del Val, C., Bondar, L., and Bondar, A.-N. (2014a). Coupling between Inter-helical Hydrogen Bonding and Water Dynamics in a Proton Transporter. J. Struct. Biol. 186, 95–111. doi:10.1016/j.jsb.2014.02.010
Del Val, C., Royuela-Flor, J., Milenkovic, S., and Bondar, A.-N. (2014b). Channelrhodopsins: A Bioinformatics Perspective. Biochim. Biophys. Acta - Bioenerg. 1837, 643–655. doi:10.1016/j.bbabio.2013.11.005
Del Val, C., White, S. H., and Bondar, A.-N. (2012). Ser/Thr Motifs in Transmembrane Proteins: Conservation Patterns and Effects on Local Protein Structure and Dynamics. J. Membr. Biol 245, 717–730. doi:10.1007/s00232-012-9452-4
Dioumaev, A. K., Brown, L. S., Needleman, R., and Lanyi, J. K. (1999). Fourier Transform Infrared Spectra of a Late Intermediate of the Bacteriorhodopsin Photocycle Suggest Transient Protonation of Asp-212. Biochemistry 38, 10070–10078. doi:10.1021/bi990873+
Ernst, O. P., Lodowski, D. T., Elstner, M., Hegemann, P., Brown, L. S., and Kandori, H. (2014). Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 114, 126–163. doi:10.1021/cr4003769
Fernandez, A., and Scheraga, H. A. (2003). Insufficiently Dehydrated Hydrogen Bonds as Determinants of Protein Interactions. Proc. Natl. Acad. Sci. 100, 113–118. doi:10.1073/pnas.0136888100
Focht, D., Croll, T. I., Pedersen, B. P., and Nissen, P. (2017). Improved Model of Proton Pump crystal Structure Obtained by Interactive Molecular Dynamics Flexible Fitting Expands the Mechanistic Model for Proton Translocation in P-type ATPases. Front. Physiol 8, 202. doi:10.3389/fphys.2017.00202
Freier, E., Wolf, S., and Gerwert, K. (2011). Proton Transfer via a Transient Linear Water-Molecule Chain in a Membrane Protein. Proc. Natl. Acad. Sci. 108, 11435–11439. doi:10.1073/pnas.1104735108
Fu, R., Miao, Y., Qin, H., and Cross, T. A. (2020). Observation of the Imidazole-Imidazolium Hydrogen Bonds Responsible for Selective Proton Conductance in the Influenza A M2 Channel. J. Am. Chem. Soc. 142, 2115–2119. doi:10.1021/jacs.9b09985
Geragotelis, A. D., Wood, M. L., Göddeke, H., Hong, L., Webster, P. D., Wong, E. K., et al. (2020). Voltage-dependent Structural Models of the Human Hv1 Proton Channel from Long-Timescale Molecular Dynamics Simulations. Proc. Natl. Acad. Sci. USA 117, 13490–13498. doi:10.1073/pnas.1920943117
Gerland, L., Friedrich, D., Hopf, L., Donovan, E. J., Wallmann, A., Erdmann, N., et al. (2020). pH‐Dependent Protonation of Surface Carboxylate Groups in PsbO Enables Local Buffering and Triggers Structural Changes. ChemBioChem 21, 1597–1604. doi:10.1002/cbic.201900739
Gerwert, K., Freier, E., and Wolf, S. (2014). The Role of Protein-Bound Water Molecules in Microbial Rhodopsins. Biochim. Biophys. Acta - Bioenerg. 1837, 606–613. doi:10.1016/j.bbabio.2013.09.006
Gerwert, K., Hess, B., Soppa, J., and Oesterhelt, D. (1989). Role of Aspartate-96 in Proton Translocation by Bacteriorhodopsin. Proc. Natl. Acad. Sci. 86, 4943–4947. doi:10.1073/pnas.86.13.4943
Govindjee, R., Kono, M., Balashov, S. P., Imasheva, E., Sheves, M., and Ebrey, T. G. (1995). Effects of Substitution of Tyrosine 57 with Asparagine and Phenylalanine on the Properties of Bacteriorhodopsin. Biochemistry 34, 4828–4838. doi:10.1021/bi00014a040
Govorunova, E. G., Sineshchekov, O. A., Li, H., and Spudich, J. L. (2017). Microbial Rhodopsins: Diversity, Mechanisms, and Optogenetic Applications. Annu. Rev. Biochem. 86, 845–872. doi:10.1146/annurev-biochem-101910-144233
Gray, T. M., and Matthews, B. W. (1984). Intrahelical Hydrogen Bonding of Serine, Threonine and Cysteine Residues within α-helices and its Relevance to Membrane-Bound Proteins. J. Mol. Biol. 175, 75–81. doi:10.1016/0022-2836(84)90446-7
Guan, L., and Kaback, H. R. (2006). Lessons from Lactose Permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67–91. doi:10.1146/annurev.biophys.35.040405.102005
Guerra, F., and Bondar, A.-N. (2015). Dynamics of the Plasma Membrane Proton Pump. J. Membr. Biol 248, 443–453. doi:10.1007/s00232-014-9732-2
Gutman, M., and Nachliel, E. (1990). The Dynamic Aspects of Proton Transfer Processes. Biochim. Biophys. Acta (Bba) - Bioenerg. 1015, 391–414. doi:10.1016/0005-2728(90)90073-d
Hasegawa, N., Jonotsuka, H., Miki, K., and Takeda, K. (2018). X-ray Structure Analysis of Bacteriorhodopsin at 1.3Å Resolution. Sci. Rep. 8, 13123. doi:10.1038/s41598-018-31370-0
Heald-Sargent, T., and Gallagher, T. (2012). Ready, Set, Fuse! the Coronavirus Spike Protein and Acquisition of Fusion Competence. Viruses 4, 557–580. doi:10.3390/v4040557
Henderson, R., and Unwin, P. N. T. (1975). Three-dimensional Model of Purple Membrane Obtained by Electron Microscopy. Nature 257, 28–32. doi:10.1038/257028a0
Herzfeld, J., and Tongue, B. (2000). NMR Probes Vectoriality in the Proton-Motive Photocycle of Bacteriorhodopsin: Evidence for an ‘Electrostatic Steering’ Mechanism. Biochim. Biophys. Acta 1460, 95–105. doi:10.1016/s0005-2728(00)00132-8
Holzer, P. (2009). Acid-sensitive Ion Channels and Receptors. Handb. Exp. Pharmacol. 194, 283–332. doi:10.1007/978-3-540-79090-7_9
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: Visual Molecular Dynamics. J. Mol. Graphics 14, 33–38. doi:10.1016/0263-7855(96)00018-5
Jalalvand, E., Robertson, B., Tostivint, H., Löw, P., Wallén, P., and Grillner, S. (2018). Cerebrospinal Fluid-Contacting Neurons Sense pH Changes and Motion in the Hypothalamus. J. Neurosci. 38, 7713–7724. doi:10.1523/jneurosci.3359-17.2018
Jalalvand, E., Robertson, B., Wallén, P., and Grillner, S. (2016). Ciliated Neurons Lining the central Canal Sense Both Fluid Movement and pH through ASIC3. Nat. Comm 7, 10002. doi:10.1038/ncomms10002
Kandori, H. (2004). Role of Internal Water Molecules in Bacteriorhodopsin. Biochim. Biophys. Acta 1460, 177–191. doi:10.1016/s0005-2728(00)00138-9
Karathanou, K., Lazaratos, M., Bertalan, É., Siemers, M., Buzar, K., Schertler, G. F. X., et al. (2020). A Graph-Based Approach Identifies Dynamic H-Bond Communication Networks in Spike Protein S of SARS-CoV-2. J. Struct. Biol. 212, 107617. doi:10.1016/j.jsb.2020.107617
Katritch, V., Fenalti, G., Abola, E. E., Roth, B. L., Cherezov, V., and Stevens, R. C. (2014). Allosteric Sodium in Class A GPCR Signaling. Trends Biochem. Sci. 39, 233–244. doi:10.1016/j.tibs.2014.03.002
Kemmler, L., Ibrahim, M., Dobbek, H., Zouni, A., and Bondar, A.-N. (2019). Dynamic Water Bridging and Proton Transfer at a Surface Carboxylate Cluster of Photosystem II. Phys. Chem. Chem. Phys. 21, 25449–25466. doi:10.1039/c9cp03926k
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H. W., Theiler, J., Abfalterer, W., et al. (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 1–16. doi:10.1016/j.cell.2020.06.043
Kouyama, T., Nishikawa, T., Tokuhisa, T., and Okumura, H. (2004). Crystal Structure of the L Intermediate of Bacteriorhodopsin: Evidence for Vertical Translocation of a Water Molecule during the Proton Pumping Cycle. J. Mol. Biol. 335, 531–546. doi:10.1016/j.jmb.2003.10.068
Lanyi, J. K. (1999). Bacteriorhodopsin. Int. Rev. Cytol. 187, 161–202. doi:10.1016/s0074-7696(08)62418-3
Leung, K., Shum, M. H. H., Leung, G. M., Lam, T. T. Y., and Wu, J. T. (2021). Early Transmissibility Assessment of the N501Y Mutant Strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 26, 2002106. doi:10.2807/1560-7917.es.2020.26.1.2002106
Li, F., Eriksen, J., Finer-Moore, J., Chang, R., Nguyen, P., Bowen, A., et al. (2020). Ion Transport and Regulation in a Synaptic Vesicle Glutamate Transporter. Science 368, 893–897. doi:10.1126/science.aba9202
Li, F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 3, 237–261. doi:10.1146/annurev-virology-110615-042301
Lorch, S., Capponi, S., Pieront, F., and Bondar, A.-N. (2015). Dynamic Carboxylate/water Networks on the Surface of the PsbO Subunit of Photosystem II. J. Phys. Chem. B 119, 12172–12181. doi:10.1021/acs.jpcb.5b06594
Ludwig, M.-G., Vanek, M., Guerini, D., Gasser, J. A., Jones, C. E., Junker, U., et al. (2003). Proton-sensing G-Protein-Coupled Receptors. Nature 425, 93–98. doi:10.1038/nature01905
Luecke, H., Schobert, B., Richter, H.-T., Cartailler, J.-P., and Lanyi, J. K. (1999). Structure of Bacteriorhodopsin at 1.55 Å Resolution. J. Mol. Biol. 291, 899–911. doi:10.1006/jmbi.1999.3027
Martineau, M., Guzman, R. E., Fahlke, C., and Klingauf, J. (2017). VGLUT1 Functions as a Glutamate/proton Exchanger with Chloride Channel Activity in Hippocampal Glutamergic Synapses. Nat. Comm 8, 2279. doi:10.1038/s41467-017-02367-6
Metz, G., Siebert, F., and Engelhard, M. (1992). Asp85 Is the Only Internal Aspartic Acid that Gets Protonated in the M Intermediate and the Purple-To-Blue Transition of Bacteriorhodopsin. A Solid-State 13C CP-MAS NMR Investigation. FEBS Lett. 303, 237–241. doi:10.1016/0014-5793(92)80528-o
Millet, J. K., and Whittaker, G. R. (2018). Physiological and Molecular Triggers for SARS-CoV Membrane Fusion and Entry into Host Cells. Virology 517, 3–8. doi:10.1016/j.virol.2017.12.015
Münster-Wandowski, A., Zander, J.-F., Richter, K., and Ahnert-Hilger, G. (2016). Co-existence of Functionally Different Vesiculr Neurotransmitter Transporters. Front. Synaptic Neurosci. 8. doi:10.3389/fnsyn.2016.00004
Musset, B., Smith, S. M. E., Rajan, S., Morgan, D., Cherny, V. V., and DeCoursey, T. E. (2011). Aspartate 112 Is the Selectivity Filter of the Human Voltage-Gated Proton Channel. Nature 480, 273–277. doi:10.1038/nature10557
Nango, E., Royant, A., Kubo, M., Nakane, T., Wickstrand, C., Kimura, T., et al. (2016). A Three-Dimensional Movie of Structural Changes in Bacteriorhodopsin. Science 354, 1552–1557. doi:10.1126/science.aah3497
Némethy, G., Steinberg, I. Z., and Scheraga, H. A. (1963). Influence of Water Structure and of Hydrophobic Interactions on the Strength of Side-Chain Hydrogen Bonds in Proteins. Biopolymers 1, 43–69. doi:10.1002/bip.360010107
Ni, Q. Z., Can, T. V., Daviso, E., Griffin, R. G., and Herzfeld, J. (2018). Primary Transfer Step in the Light-Driven Ion Pump Bacteriorhodopsin: An Irreversible U-Turn Revealed by Dynamic Nuclear Polarization-Enhanced Magic Angle Spinning NMR. J. Am. Chem. Soc. 140, 4085–4091. doi:10.1021/jacs.8b00022
Pedersen, B. P., Buch-Pedersen, M. J., Preben Morth, J., Palmgren, M. G., and Nissen, P. (2007). Crystal Structure of the Plasma Membrane Proton Pump. Nature 450, 1111–1114. doi:10.1038/nature06417
Phatak, P., Frähmcke, J. S., Wanko, M., Hoffmann, M., Strodel, P., Smith, J. C., et al. (2009). Long-distance Proton Transfer with a Break in the Bacteriorhodopsin Active Site. J. Am. Chem. Soc. 131, 7064–7078. doi:10.1021/ja809767v
Presta, L., and Rose, G. (1988). Helix Signals in Proteins. Science 240, 1632–1641. doi:10.1126/science.2837824
Radu, C. G., Nijagal, A., Mclaughlin, J., Wang, L., and Witte, O. N. (2004). Differential Proton Sensitivity of Related G Protein-Coupled Receptors T Cell Death-Associated Gene 8 and G2A Expressed in Immune Cells. Proc. Natl. Acad. Sci. U S A. 102, 1632–1637. doi:10.1073/pnas.0409415102
Rath, P., Krebs, M. P., He, Y., Khorana, H. G., and Rothschild, K. J. (1993). Fourier Transform Raman Spectroscopy of the Bacteriorhodopsin Mutant Tyr-185 Phe: Formation of a Stable O-like Species during Light Adaptation and Detection of its Transient N-like Photoproduct. Biochemistry 32, 2272–2281. doi:10.1021/bi00060a020
Regan, M. C., Zhu, Z., Yuan, H., Myers, S. J., Menaldino, D. S., Tahirovic, Y. A., et al. (2019). Structural Elemets of a pH-Sensitive Inhibitor Binding Site in NMDA Receptors. Nat. Comm 10, 321. doi:10.1038/s41467-019-08291-1
Sacks, V., Marantz, Y., Aagaard, A., Checover, S., Nachliel, E., and Gutman, M. (1998). The Dynamic Feature of the Proton Collecting Antenna of a Protein Surface. Biochim. Biophys. Acta - Bioenerg. 1365, 232–240. doi:10.1016/s0005-2728(98)00073-5
Sahin-Toh, M., and Kaback, H. R. (2001). Arg-302 Facilitates Deprotonation of Glu-325 in the Transport Mechanism of the Lactose Permease from Escherichia coli. Proc. Natl. Acad. Sci. 98, 6068–6073. doi:10.1073/pnas.111139698
Sasaki, J., Brown, L., Chon, Y., Kandori, H., Maeda, A., Needleman, R., et al. (1995). Conversion of Bacteriorhodopsin into a Chloride Ion Pump. Science 269, 73–75. doi:10.1126/science.7604281
Schönichen, A., Webb, B. A., Jacobson, M. P., and Barber, D. L. (2013). Considering Protonation as a Posttranslational Modification Regulating Protein Structure and Function. Annu. Rev. Biophys. 42, 289–314. doi:10.1146/annurev-biophys-050511-102349
Shutova, T., Klimov, V. V., Andersson, B., and Samuelsson, G. (2007). A Cluster of Carboxylic Groups in PsbO Protein Is Involved in Proton Transfer from the Water Oxidizing Complex of Photosystem II. Biochim. Biophys. Acta - Bioenerg. 1767, 434–440. doi:10.1016/j.bbabio.2007.01.020
Siemers, M., Lazaratos, M., Karathanou, K., Guerra, F., Brown, L. S., and Bondar, A.-N. (2019). Bridge: A Graph-Based Algorithm to Analyze Dynamic H-Bond Networks in Membrane Proteins. J. Chem. Theor. Comput. 15, 6781–6798. doi:10.1021/acs.jctc.9b00697
Song, Y., and Gunner, M. R. (2014). Halorhodopsin Pumps Cl− and Bacteriorhodopsin Pumps Protons by a Common Mechanism that Uses Conserved Electrostatic Interactions. Proc. Natl. Acad. Sci. USA 111, 16377–16382. doi:10.1073/pnas.1411119111
Srivastava, J., Barber, D. L., and Jacobson, M. P. (2007). Intracellular pH Sensors: Design Principles and Functional Significance. Physiology 22, 30–39. doi:10.1152/physiol.00035.2006
Száraz, S., Oesterhelt, D., and Ormos, P. (1994). pH-Induced Structural Changes in Bacteriorhodopsin Studied by Fourier Transform Infrared Spectroscopy. Biophysical J. 67, 1706–1712. doi:10.1016/s0006-3495(94)80644-7
Tegally, H., Wilkinson, E., Giovanetti, M., Iranzadeh, A., Fonseca, V., Giandhari, J., et al. (2021). Emergence of Rapid Spread of a New Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-S) Lineage with Multiple Spike Mutations in South Africa. [Epub ahead of print]. 10.1101/2020.12.21.20248640
Thurlkill, R. L., Grimsley, G. R., Scholtz, J. M., and Pace, C. N. (2006a). Hydrogen Bonding Markedly Reduces the pK of Buried Carboxyl Groups in Proteins. J. Mol. Biol. 362, 594–604. doi:10.1016/j.jmb.2006.07.056
Thurlkill, R. L., Grimsley, G. R., Scholtz, J. M., and Pace, C. N. (2006b). pK Values of the Ionizable Groups of Proteins. Protein Sci. 15, 1214–1218. doi:10.1110/ps.051840806
Ulmschneider, M. B., and Sansom, M. S. P. (2001). Amino Acid Distributions in Integral Membrane Protein Structures. Biochim. Biophys. Acta - Biomembranes 1512, 1–14. doi:10.1016/s0005-2736(01)00299-1
Vickery, O. N., Carvalheda, C. A., Zaidi, S. A., Pisliakov, A. V., Katritch, V., and Zachariae, U. (2018). Intracellular Transfer of Na+ in an Active-State G-Protein-Coupled Receptor. Structure 26, 171–180. doi:10.1016/j.str.2017.11.013
Wang, J.-Q., Kon, J., Mogi, C., Tobo, M., Damirin, A., Sato, K., et al. (2004). TDAG8 Is a Proton-Sensing and Psychosine-Sensitive G-Protein-Coupled Receptor. J. Biol. Chem. 279, 45626–45633. doi:10.1074/jbc.m406966200
Weinert, T., Skopintsev, P., James, D., Dworkowski, F., Panepucci, E., Kekilli, D., et al. (2019). Proton Uptake Mechanism in Bacteriorhodopsin Captured by Serial Synchrothron Crystallography. Science 365. 61–65. doi:10.1126/science.aaw8634
Wu, L.-J., Wu, G., Sharif, M. R. A., Baker, A., Jia, Y., Fahey, F. H., et al. (2012). The Voltage-Gated Proton Channel Hv1 Enhances Brain Damage from Ischemic Stroke. Nat. Neurosci. 15, 565–573. doi:10.1038/nn.3059
Xie, K., Hessa, T., Seppälä, S., Rapp, M., Von Heijne, G., and Dalbey, R. E. (2007). Features of Transmembrane Segments that Promote the Lateral Release from the Translocase into the Lipid Phase. Biochemistry 46, 15153–15161. doi:10.1021/bi701398y
Zabelskii, D., Alekseev, A., Kovalev, K., Rankovic, V., Balandin, T., Soloviov, D., et al. (2021). Viral Rhodopsins 1 Are an Unique Family of Light-Gated Cation Channels. Nat. Commun. 11, 5707. doi:10.1038/s41467-020-19457-7
Zhang, X. C., Cao, C., Zhou, Y., and Zhao, Y. (2015). Proton Transfer-Mediated GPCR Activation. Protein Cell 6, 12–17. doi:10.1007/s13238-014-0106-4
Zhou, T., Tsybovsky, Y., Gorman, J., Rapp, M., Cerutti, G., Chuang, G.-Y., et al. (2020). Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host Microbe 28, 867–879. doi:10.1016/j.chom.2020.11.004
Keywords: hydrogen-bonding, proton transfer, proton antenna, membrane transporter, spike protein S
Citation: Bondar A-N (2021) Proton-Binding Motifs of Membrane-Bound Proteins: From Bacteriorhodopsin to Spike Protein S. Front. Chem. 9:685761. doi: 10.3389/fchem.2021.685761
Received: 26 March 2021; Accepted: 18 May 2021;
Published: 31 May 2021.
Edited by:
Petra Hellwig, Université de Strasbourg, FranceReviewed by:
Steffen Wolf, University of Freiburg, GermanyAlexei Stuchebrukhov, University of California, United States
Copyright © 2021 Bondar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana-Nicoleta Bondar, bmJvbmRhckB6ZWRhdC5mdS1iZXJsaW4uZGU=
 Ana-Nicoleta Bondar
Ana-Nicoleta Bondar