- 1Department of Chemistry, The Graduate Center, City University of New York, New York, NY, United States
- 2Department of Physics, City College of New York, New York, NY, United States
- 3Department of Physics, The Graduate Center, City University of New York, New York, NY, United States
Biological membranes are barriers to polar molecules, so membrane embedded proteins control the transfers between cellular compartments. Protein controlled transport moves substrates and activates cellular signaling cascades. In addition, the electrochemical gradient across mitochondrial, bacterial and chloroplast membranes, is a key source of stored cellular energy. This is generated by electron, proton and ion transfers through proteins. The gradient is used to fuel ATP synthesis and to drive active transport. Here the mechanisms by which protons move into the buried active sites of Photosystem II (PSII), bacterial RCs (bRCs) and through the proton pumps, Bacteriorhodopsin (bR), Complex I and Cytochrome c oxidase (CcO), are reviewed. These proteins all use water filled proton transfer paths. The proton pumps, that move protons uphill from low to high concentration compartments, also utilize Proton Loading Sites (PLS), that transiently load and unload protons and gates, which block backflow of protons. PLS and gates should be synchronized so PLS proton affinity is high when the gate opens to the side with few protons and low when the path is open to the high concentration side. Proton transfer paths in the proteins we describe have different design features. Linear paths are seen with a unique entry and exit and a relatively straight path between them. Alternatively, paths can be complex with a tangle of possible routes. Likewise, PLS can be a single residue that changes protonation state or a cluster of residues with multiple charge and tautomer states.
Introduction
Protons serve as substrate or product in many chemical and biological reactions. In proteins, protons often travel 10 Å or more from the surface to reach an active site. Proton gradients across the membranes of bacteria, mitochondria and chloroplasts contribute to the electrochemical gradients, ∆Ψ, used to store cellular energy (Mitchell, 1961; Rich, 2008; Nicholls, 2010; Gunner et al., 2013). The proton gradient can be generated by vectorial electron transfer, where reactants are oxidized and reduced on different sides of the membrane. Here the electrons cross the membrane, but the protons only move to or from the separated redox sites. In contrast, proton pumps transfer protons through the transmembrane proteins, requiring mechanisms to avoid downhill proton transfer.
Regardless of the mechanism a protein uses, it takes energy to build a proton gradient. The input energy is light in photosynthetic proteins (Cardona et al., 2012; Ge and Gunner, 2016; Cardona and Rutherford, 2019), redox reactions in the electron transfer chain (Kaila and Hummer, 2011), ATP hydrolysis (Vasanthakumar and Rubinstein, 2020) or the dissipation of the gradient of another ion (Fowler et al., 2015; Brandt, 2019). The protons flow down the electrochemical gradient then fuel processes such as ATP synthesis in F1/F0 ATPase (Walker et al., 1991; Futai et al., 2012; Yanagisawa and Frasch, 2017) and the active transport of ions and metabolites (Accardi and Picollo, 2010; Gunner et al., 2013).
To build the gradient, protons are transferred from the more negative, N-side of the membrane, where they are at lower concentration (higher pH) to the positive, P-side where they are at higher concentration (lower pH). The P-side is in the periplasm of bacteria, the outer membrane space of mitochondria and in the lumen on the inside of the chloroplast thylakoid membrane. The N-side is toward the bacterial cytoplasm, the mitochondrial matrix and the chloroplast stroma. The electrochemical gradient, ∆Ψ, is made up of the gradient of protons (the ∆pH) but also has contributions from other ions, adding to a voltage change, ∆V, across the membrane (Decoursey, 2003). The ∆Ψ across a given membrane determines the energy needed to push a proton uphill in the protein pumps described here or the energy liberated when protons run from P- to N-side as used for ATP synthesis.
Although we refer to “protons”, H+ does not travel alone. Rather it is associated with a water (hydronium, H3O+) or two water molecules as a Zundel cation (H5O2+) or as a larger, Eigen complex (H9O4+) (Agmon, 1995; Wraight, 2006; Farahvash and Stuchebrukhov, 2018). In proteins, the proton can also be bound to redox cofactors, to acidic or basic residues or trapped as a stabilized hydronium (Xu and Voth, 2006; Freier et al., 2011; Ikeda et al., 2017).
Protons move through a chain of oriented molecules by a Grotthuss proton transfer mechanism (Agmon, 1995; Cukierman, 2006; de Grotthuss, 2006; DeCoursey and Hosler, 2014). An active group in the middle of the chain is: 1) a hydrogen bond donor to the next group in the direction of proton transfer and 2) has a lone pair of electrons that is a hydrogen bond acceptor from the neighbor toward the proton input side. In the Grotthuss mechanism no proton moves more than one bond, as the proton acceptor takes ownership from the neighboring proton donor. However, overall the coupled transfers lead to a proton rapidly leaving the input side and appearing at the end of the chain. There are many reviews of the chemistry of proton transfer reactions as well as of proton transfer reactions in proteins (Hammes-Schiffer, 2001; Pomès and Roux, 2002; Blomberg and Siegbahn, 2006; Swanson et al., 2007; Knight and Voth, 2012; Ishikita and Saito, 2014; Miyake and Rolandi, 2015; Wikström et al., 2015; Sakashita et al., 2020).
Two requirements create barriers for Grotthuss proton transfers. First, the chain of hydrogen bonds between proton donors and acceptors needs to be pre-organized. Then, once the proton has transferred, the hydrogen bonds are arranged to return the proton back to the origin, not to move another proton in the same direction. The hydrogen bonded chain needs to fully reorient for the next proton to transfer, so overall proton flux is limited by this slow “hop and turn” process (Nagle and Morowitz, 1978).
Vectorial proton coupled electron transfer. Proteins such as PSII, cytochromes bc1 and b6f use vectorial electron transfer reactions where oxidation and reduction reactions are spatially separated to add to the proton gradient. Thus, oxidation occurs on the P-side, where protons are released because the loss of an electron lowers the oxidized product pKa below the pH of the nearby compartment. Reduction occurs on the N-side, where reduction shifts the product pKa to be higher than the compartment pH (Rich, 2008; Nicholls, 2010; Gunner et al., 2013; Gunner and Koder, 2017). Within the protein, a sequence of electron tunneling reactions pass the electrons 30 Å or more between the terminal electron donor and acceptor (Gray and Winkler, 2003; Moser et al., 2006). The interior electron transfer reactions are not coupled to gain or loss of protons. Thus, a proton gradient is generated without moving protons through the membrane by a redox loop mechanism as suggested by Mitchell (Mitchell, 1977). The intra-membrane, middle of these proteins are mostly non-polar side chains with few associated water molecules, so discourage proton transfer. However, as will be seen in the discussion of PSII and bRCs, the sites of final, proton coupled oxidation or reduction can be 10 Å or more from the surface, requiring long-range proton transfer to move the protons to the active site.
Proton pumps. The proton pumps include the well-studied, light-driven Bacteriorhodopsin (Balashov, 2000; Luecke, 2000), Complex I (Mathiesen and Hägerhäll, 2002; Hirst, 2013; Sazanov, 2014) and the heme copper oxidase (HuCuOx) family (Kaila et al., 2010; Lee and Ädelroth, 2013). To ensure protons move in the correct direction pumps require three elements. These are proton transfer paths, as found in vectorial electron transfer proteins. However, pumps need Proton Loading Sites, PLS, placed along the proton transfer path, and gates. PLS transiently change their proton affinity to load a proton when the gate is open to the N-side and releases it when it is open to the P-side. Pathway gating and proton loading must be synchronized to guard against energy dissipating proton transfer from P- to N-side.
This review will compare and contrast the residues that make up the proton transfer elements in three light activated proteins: Bacteriorhodopsin (bR) and the photosynthetic proteins, Photosystem II (PSII) and the purple non-sulfur photosynthetic bacterial reactions centers (bRCs) and in the proton pumps Complex I and Cytochrome c oxidase, which are the first and last protein in the erobic electron transfer chain.
Overview of Proton Transfer Paths
The role of the different residues in proton transfer paths. The review will describe the residues found along proton transfer paths. Water is the quintessential Grotthuss competent molecule so water filled channels through protein structures often trace the proton transfer paths. However, some side chains can be a part of a proton transfer chain. Hydroxyl residues are well established in proton transfer paths, such as in Green Fluorescent Protein, GFP, which has a Ser on the short proton transfer path (Brejc et al., 1997; Donati et al., 2018). A neutral His has a proton on Nε with a lone pair on Nδ (or the proton/lone pair swap positions). It can accept a proton from one side of the imidazole and donate a proton from the other side. His plays this role in proton transfer in the M2 proton channel (Wang et al., 1995) and in carbonic anhydrase (Tu et al., 1989). Histidine analogs have been used in synthetic electron coupled proton transfer chains that lead to the Grotthuss transfer of a proton over long distances (Odella et al., 2018; Odella et al., 2019). Ionized side chains cannot be both a hydrogen bond donor and acceptor as required for Grotthuss proton transfer (Ge and Gunner, 2016; Lazaridis and Hummer, 2017; Duster and Lin, 2019). Thus, deprotonated Asp- and Glu- are hydrogen bond acceptors but have no proton to donate, while protonated His+, Lys+ or Arg+ have no lone pairs to accept a proton. This review will show examples where acidic and basic residues are found as PLS in the proton transfer chain, serving as meta-stable intermediates that can cycle between loaded (protonated) and unloaded states. Polar residues such as Asn, Gln and Trp are found to anchor the hydrogen bond chain, but are unlikely be active elements in Grotthuss proton transfer chains (Hammes-Schiffer, 2001; Goings et al., 2020), while non-polar residue are insulators stopping water penetration and proton leaks.
Linear vs. complex proton transfer paths. As proton transfers have been investigated in different proteins, we have found they can take place via linear or complex paths. Linear paths, as defined here, have a single entry and exit and a well-defined road between them. There are limited branches, which never deviate far from the main path. Linear paths can often be identified in a protein structure that includes well resolved water molecules (Sharpe and Ferguson-Miller, 2008; Sazanov, 2015). Mutation of a single entry or exit residue can block proton transfer.
However, representative structures will be shown to reveal interior regions with tangled webs of polar and protonatable groups and many water molecules (Krammer et al., 2009; Cai et al., 2020; Khaniya et al., 2020). These complex proton transfer paths provide multiple choices for protons to follow. Here mutations of individual residue may lead to partial loss of activity, generating ambiguous results that neither fully confirm nor deny their role. The proteins reviewed here use linear and complex paths in different regions of the overall transfer of protons through the membrane.
Proton Loading Sites
Type of Residues That Can Serve as PLS
A successful pump takes a proton through the protein from the N-side to the P-side, even though it is thermodynamically unfavorable. A PLS must transiently hold protons with gates open to the N-side and to be released to the P-side, synchronized with a turnover time of microseconds to milliseconds (Balashov, 2000; Kaila et al., 2010). The PLS is thus a residue or cluster of residues whose proton affinity changes dramatically between different reaction intermediates (Supplementary Material S2). The carboxylic acids, Asp, Glu and heme propionic acids are the most common PLS components in the proteins described here. They are found as single site PLS as well as PLS clusters. His and Lys are more often found coupled to acidic residues in clusters. H3O+, trapped between several acidic residues has been suggested to be part of PLS clusters (Freier et al., 2011; Kovalevsky et al., 2011; Supekar et al., 2016). However, the pKa for Arg+ to lose a proton is as high as that of water or a hydroxyl side chain (Fitch et al., 2015). Thus, the protonated Arg can help stabilize the negative charge but is unlikely to lose a proton in a PLS.
PLS clusters. The PLS and complex proton transfer paths often have regions with many interacting, buried ionizable and polar residues (Lancaster et al., 1996; Kannt et al., 1998). For a PLS cluster with n protonatable residues there are n+1 charge states and 2n microstates, which identify the number and distribution of protons (Gunner et al., 2020). The charge ranges from -Nacids (the number of acids) (assuming all bases are neutral) to +Nbases (number of bases) (assuming all acids are neutral). Tautomers are protonation microstates with the same charge but different proton locations. With m protons distributed over n binding sites in a PLS there are:
tautomers. The relative energies of the different tautomers determine the proton positions within the loaded and unloaded clusters. This review will describe examples of mechanisms by which PLS can change their protein affinity to load and unload protons.
Model Systems
Green Fluorescent Protein (GFP). GFP provides a simple example of the role of side chains in and around the proton transfer path (Figure 1). It also shows how fast protons can transfer via a pre-organized Grotthuss competent chain (Brejc et al., 1997; Zimmer, 2009; Donati et al., 2018). GFP is well studied as it has revolutionized cell biology. When introduced into a genome it is co-expressed with a specific protein of interest and its characteristic florescence allows the targeted protein to be localized within a living cell. The chromophore in GFP is a photoacid that absorbs light in the near UV and emits in the green (Zhou and Han, 2018). The large Stokes shift results from the ground state absorption and excited state emission occurring from molecules with different charges. Thus, in the ground state the chromophore is the PLS, while Glu 222 is the proton acceptor when the chromophore proton affinity is diminished by excitation. In this system the path for rapid proton release must be ready to carry the proton away prior to relaxation of the chromophore (Chattoraj et al., 1996).
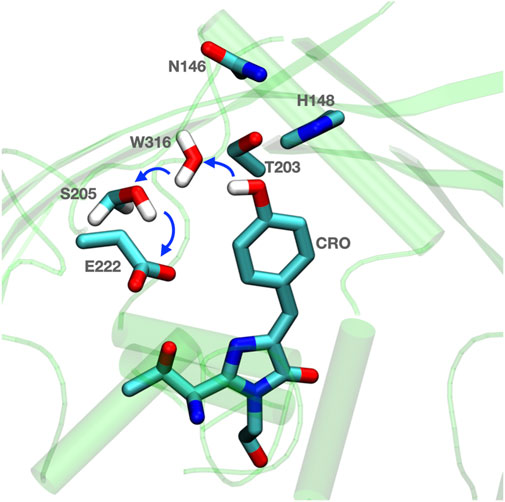
FIGURE 1. Residues are prearranged for Grotthuss proton transfer in GFP. Blue thick arrows show direction of proton transfer from chromophore (CRO) to E222 via water (W316) and S205. The surrounding H148 and N146 can help to anchor the active proton transfer path. The coordinates for Equorea victoria GFP are from PDB ID: 1EMA (Ormö et al., 1996).
The proton is transferred through bound water and Ser 205 to Glu 222. A nearby His 148, Thr 203 and Asn 146 are on the outskirts of the proton transfer wire (Stoner-Ma et al., 2005; Di Donato et al., 2011). These provide a polar residue fence to pre-orient the hydrogen bond network. As the protein is light activated experiments can synchronize the protein for kinetic measurements to follow changes in the hydrogen bonding network. The proton arrives on the Glu in less than 10 ps (Donati et al., 2018). Vibrational spectroscopy shows that there are rapid changes in hydrogen bond orientation that precede proton transfer, presumably to fine tune the hydrogen bond connections for Grotthuss proton transfer. The kinetic transients are distinguished as rearrangements, which do not have a kinetic isotope effect, and proton transfers, which do (Di Donato et al., 2011).
Gramicidin (gA). The gA channel is a proton and cation conducting channel that has been used to study proton transfer through a linear water wire, with no intervening side chains. gA is made of two short ß-strand peptides, capped on N- and C-terminal ends. The N-termini meet in the center of the membrane. In the ß-helix the side chains are to the outside, with sufficient space to hold a linear chain of ≈8 water molecules in the middle of the helix. gA is an antibiotic, allowing protons and other cations to cross, depolarizing the cell’s electrochemical gradient (Kelkar and Chattopadhyay, 2007; Li et al., 2016). The simplicity of this system has made it ideal for the experimental (Dorman and Jordan, 2004; Ryu et al., 2015; Paulino et al., 2020) and computational (Roux, 2002; Allen et al., 2004; Till et al., 2008; Lazaridis and Hummer, 2017; Zhang et al., 2020) studies of the Grotthuss proton transfer mechanism.
The water molecules in gA form hydrogen bonds with the two neighboring water molecules and with the amides of the surrounding peptides. The balance of the water-water and water-amide interactions determines the stability of the water wire, the stability of an excess proton within the wire and the barrier for flipping the wire orientation once a proton has translocated to transport another proton via the hop and turn mechanism. The rate determining step for transfer can be initially orienting the water molecules or flipping the oriented water dipoles to the correct direction (Pomès and Roux, 2002; Agmon et al., 2016; Bozdaganyan et al., 2019).
Recent computer simulations showed a rather substantial sensitivity of the water wire orientation to the force field and simulation method (Zhang et al., 2020). In molecular dynamics simulations, with a classical force field the water molecules are fully aligned within the channel with rare flips from one orientation to the other. In contrast, MD with a the Drude polarizable force field shows more disorganized water molecules. Monte Carlo sampling with a Continuum Electrostatic force field also show relatively disorganized water chain. Thus, the balance of the forces that determine the orientation of the water molecules are such that different simulation conditions induce different behavior.
Experiments have also supported a range of structures for water in the channel. The experimentally derived rate of proton translocation through the channel under a transmembrane voltage gradient appears to be diffusion limited (≈2 × 109 s−1) even at pH 0 (Cukierman, 2000; Decoursey, 2003). The reorientation of the water chain is likely to be the rate determining step in Grotthuss proton transfer (Pomès and Roux, 1998). In the MD simulations with a classical force field the water chain flips its direction at ≈4 × 108 s−1 and this process is faster with the Drude force field. Thus, the simulations are in general agreement with the measured rate of proton transfer. However, recent solid-state NMR studies show a well-organized water-wire with flip rates on the millisecond time scale, which would suggest very slow turnover for proton transfers. The NMR studies point to hydrogen bonds between water molecules and amides near the first and last turns of each ß-helix leading to this stability. Thus, despite the simplicity of its structure, gA remains a protein where our understanding of the channel water structure and the mechanism of proton transport remains incomplete.
Bacteriorhodopsin
bR is the simplest and best studied proton pump (Balashov, 2000; Baudry et al., 2001; Gunner et al., 2006; Lanyi, 2006; Lórenz-Fonfría et al., 2008; Clemens et al., 2011). The bacteriorhodopsin family uses retinal not chlorophyll based photoactivation to fuel proton or ion pumping (Figure 2). Absorption of a 568 nm photon initiates a reaction cycle that removes a proton from the cell interior (N-side) and releases one to the outside (P-side) adding to the proton gradient. The overall reaction is simply:
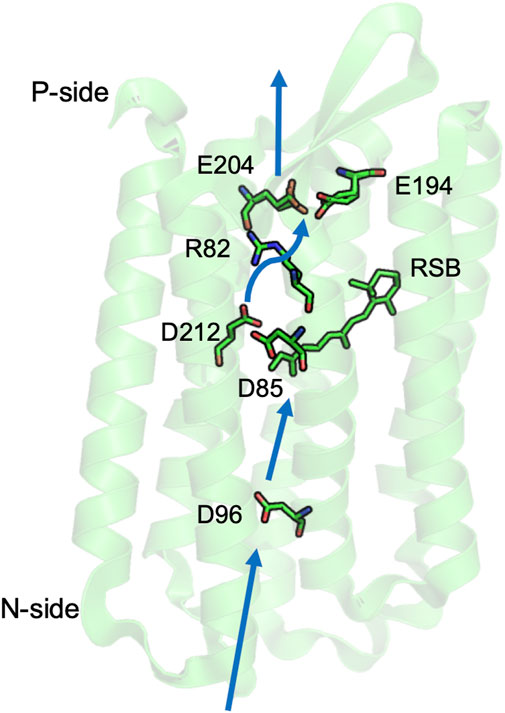
FIGURE 2. Key residues for proton transfer in bacteriorhodopsin. Three separated PLS are: the isolated D96; the central cluster D85, D212, and retinal Schiff base (RSB); and the exit cluster E194 and E204. Movement of R82 stabilizes unloading the central or exit clusters. Blue thick arrows show direction of proton transfer from N- to P-side by water mediated proton transfer paths. The coordinates for Halobacterium salinarum bR are from PDB ID: 5ZIM (Hasegawa et al., 2018).
The proton transfers are driven by the light induced conformational changes of the retinal from all trans to 13-cis, which is coupled to small changes in the helical packing in the protein core. There are three PLS in bR that illustrate different mechanisms to control PLS protonation and the connection to the proton transfer path.
Characterization of proton transfer intermediates. In light activated proteins such as GFP, bR, PSII and bRCs experiments can follow the sequential reaction mechanism since the flash of light used to start the reaction synchronizes the population. Time resolved visible and IR spectroscopy, combined with site directed mutations allow assignment of kinetic features to individual residues. bR intermediates were identified with the proton resting on acidic amino acids making up three well separated PLSs, each with metastable intermediates with different ionization states (Balashov, 2000; Lórenz-Fonfría and Kandori, 2009; Lórenz-Fonfría et al., 2011; Lórenz-Fonfría and Heberle, 2014). These intermediates clearly demonstrate the role of transient resting places for protons. Changes occur in times ranging from the picosecond transitions that trap the photon’s energy in the isomerized retinal to the milliseconds required to complete the full photocycle.
In bR, it is possible to crystallize protein trapped in different intermediates by a combination of mutation and temperature changes (Edmonds and Luecke, 2004; Hirai et al., 2009; Wickstrand et al., 2015). Simulations using these structures have shown that the calculated equilibrium proton distribution changes between intermediates as expected (Bashford and Gerwert, 1992; Spassov et al., 2001; Onufriev et al., 2003; Song et al., 2003; Song and Gunner, 2014). More recent time resolved crystal structures have been obtained using X-ray free electron lasers (XFEL) (Nogly et al., 2018; Wickstrand et al., 2019). These structures show many of the motions of water molecules and side chains and helices seen in earlier trapped structures. However, as the XFEL structures are not in deeply trapped intermediates, the dynamic structures provide additional information. However, as the transitions between photocycle intermediates are not all well separated in time, the XFEL structures each contain a mixture of states.
bR Demonstrates the Character of Simple and Cluster PLS
The central cluster tautomer shift. The central cluster consists of three residues: the retinal Schiff base (RSB), Asp 85 and Asp 212, which binds one proton on the RSB in the ground state (RSBH+: Asp 85-: Asp 212-). Light absorption leads to isomerization of the retinal, which rotates the RSB from facing the P-side to the N-side, leaving the proton on Asp 85 (M state: RSB: Asp85H: Asp 212-). This transition demonstrates a feature of a cluster PLS, as it moves between states with different proton distributions (tautomers) while retaining the same number of protons. The redistribution of the proton coupled to the retinal isomerization serves as a gate as it changes the direction of proton access. Thus, a proton will be passed from the trans-RSB to Asp85 toward the P-side, while later a proton is bound to the 13-cis RSB from the N-side (Bondar et al., 2007; Clemens et al., 2011; Wolter et al., 2013). The retinal returns to the P-side facing trans isomer only after it has bound the proton (Balashov, 2000).
A complex PLS can trap a proton on multiple sites. The complex exit cluster PLS, with Glu 194 and 204, has multiple tautomers for the proton loaded state. IR spectroscopy (Daldrop et al., 2018) and simulations (Bashford and Gerwert, 1992; Spassov et al., 2001; Phatak et al., 2008) support a protonated water stabilized by the two anionic glutamic acids, while the proton can also be trapped by a hydrogen bonded pair with one acid protonated and a water nearby (Song et al., 2003; Phatak et al., 2008). An advantage of using a cluster PLS is that it can use the multiple ways to store the proton to be less sensitive to mutation. If one of the Glu is mutated to an Asp the cluster is no longer properly positioned to trap a hydronium so the water cation IR signature is lost. The proton is now trapped on an acid, thereby retaining PLS function (Balashov, 2000; Gerwert et al., 2014).
An isolated acidic PLS requires hydration to lose its proton. Asp 96 on the N-side of bR plays a key role in proton transport (Miller and Oesterhelt, 1990). Asp 96 is an isolated PLS, as it is not in a cluster with other protonatable residues and has few hydrogen bonding opportunities to residues beyond Thr46 in the neighborhood. In the neutral, unloaded structure, there are few nearby water molecules and the acid is very stable in its neutral, loaded state (Gerwert et al., 2014; Wolf et al., 2014). A combination of time resolved IR and MD simulations show that isomerization of the retinal, 10 Å from Asp96, and the transfer of the proton from RSBH+ to the nearby Asp 85 leads to formation of a linear water chain on the N-side (Freier et al., 2011). The water molecules provide a proton transfer path, and also stabilize the negative charge on Asp 96 so that it can release a proton to the RSB near the end of the photocycle. Thus, a single site PLS cannot change its proton affinity by small movements of the polar and charged groups. Here flooding the site with water is required to both stabilize the charged Asp- and to open the gate for proton release by connecting it to the proton transfer path.
Photosystem II
PSII (Umena et al., 2011) is a multi-subunit protein pigment complex present in the thylakoid membrane of plants and cyanobacteria (Cox et al., 2013; Vinyard and Brudvig, 2017; Pantazis, 2018). The reaction is initiated by chlorophyll excitation with a 680 nm photon. Water is the primary electron donor and plastoquinone, PQ, is the final electron acceptor (Figure 3) (McEvoy and Brudvig, 2006). The overall reaction is:
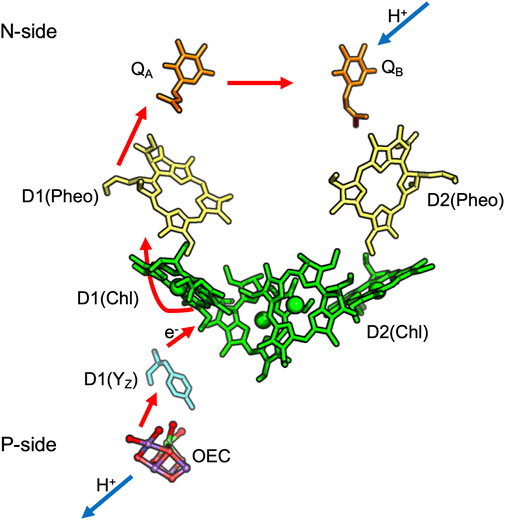
FIGURE 3. Cofactors in PSII: QA, QB are plastoquinones, Pheo is Pheophytin; Chl is Chlorophyll. D1 and D2 are the protein subunit that binds each cofactor. Red arrows show electron transfer from the OEC to QA and QB while blue lines show proton uptake from the stroma to QB and release from the OEC to the lumen. The coordinates for cyanobacteria Thermosynechococcus vulcanus are obtained from PDB ID: 3ARC.
The Oxygen Evolving Complex (OEC), an inorganic Mn4CaO5 cluster, catalyzes water oxidation following four sequential oxidations of the cluster (through five S-states) releasing O2 (Suga et al., 2019). The OEC is ≈20 Å from the surface, requiring paths for water entry and O2 and proton release. The region around the OEC is filled with water molecules that separate into three discrete water-filled channels moving to the lumen (Figures 4A,B) (Vassiliev et al., 2012; Vogt et al., 2015). On the electron acceptor side, QB in PSII is quite close to the stroma requiring only a short proton transfer path to bring in protons (Saito et al., 2013).
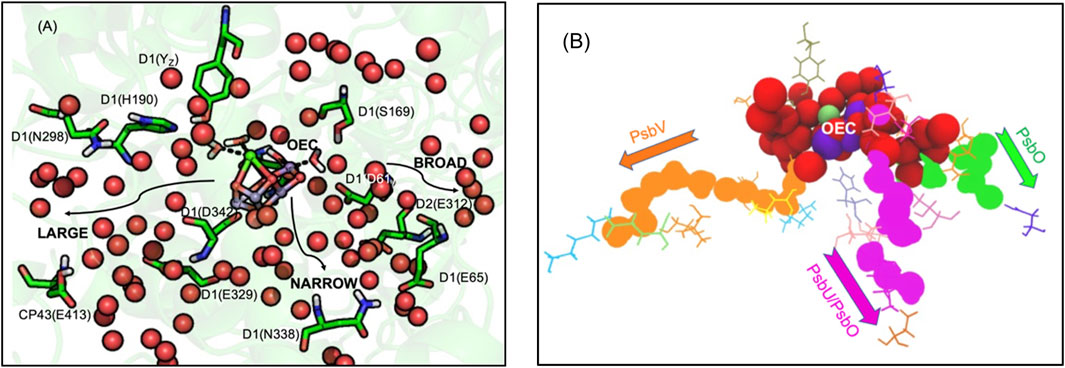
FIGURE 4. (A,B). (A) Water channels around the Oxygen Evolving Complex (OEC) of PSII. Water spheres (red) and amino acid residues highlighting the directions toward the large, broad and narrow channels (Vassiliev et al., 2012; Vogt et al., 2015). The notation for each residue is subunit-residue type, number. Residues in large channel: YZ, D1-H190, N298, E329, D342 (OEC primary ligand), CP43-E413. Narrow channel: D1-S169, N338. Broad channel: D1-D61, E65, D2-E312. (B) Waters extending from the OEC to the N-side lumen. Red spheres show highly interconnected water molecules extending ≈10 Å from the OEC. The structure then resolves to form three separated water filled channels: large (orange), narrow (magenta) and broad (green). Large channel extends from O1 of OEC to PsbV, narrow channel extends from O4 to PsbU/PsbO while broad channel extends from O5 to PsbO. See Supplementary Figure S1 for a more detailed view of the OEC. Coordinates from one MD snapshot initiated from Thermosynechococcus vulcanus PDB ID: 4UB6.
Proton Paths to the P-Side Near the OEC in PSII
Three water filled paths. Proton transfer paths combine Grotthuss competent water molecules and residues with protonatable groups to serve as transient proton loading sites. The proton release paths in PSII are dominated by water molecules, requiring minimal assistance from residues. There are three identified water filled channels leading from the OEC to the lumen (Figure 4A) (Vassiliev et al., 2012; Vogt et al., 2015). The narrow channel originates from one side of the Mn4 water ligands and extends through the PsbU/PsbO subunits (Figure 4B). The broad channel originates from the other side of Mn4, extending to the PsbO subunit, while the large channel originates from the OEC Ca water ligands leading to the PsbV subunit (Vassiliev et al., 2012; Vogt et al., 2015). These channels can provide paths for the transfer of the four product protons and O2 to the lumen and entry of the two substrate water molecules.
Which path does the proton take? With so many choices, an open question is which channel is best suited for proton transfer to the surface. Various simulation techniques have explored the nature of the water channels. Molecular dynamics investigations (Vassiliev et al., 2012) and quantum chemical studies (Retegan et al., 2016) favored the narrow channel for substrate water delivery. QM/MM studies (Saito et al., 2015) supported proton transport through the narrow channel. Continuum electrostatics calculations considered the proton affinity of residues lining the broad channel, finding increasing proton affinity, lowering the barrier for proton transfer, nearer the channel exit (Ishikita et al., 2006). Steered MD calculations (Vassiliev et al., 2012) found the large channel favorable for O2 transport and the narrow channel for substrate water delivery. However, other experimental and computational studies favored the large channel for proton transport (Chrysina et al., 2011; Nakamura et al., 2014; Sakamoto et al., 2017) or for substrate water delivery (McEvoy and Brudvig, 2004; Isobe et al., 2015; Shoji et al., 2015; Ugur et al., 2016; Kim and Debus, 2017). Thus, despite experimental and computational studies, a consensus for the role of each channel is yet to be established (Pantazis, 2018).
While earlier studies focused on individual linear paths, network analysis provides a somewhat different view of the connectivity of the water networks near the OEC (Kaur et al., 2021). These studies indicate that beyond ≈10–12 Å from the cofactor the three paths do become well separated as indicated by inspection of the structures. However, closer to the OEC all water molecules are highly interconnected. A proton from any of the Mn terminal water ligands or any of the oxygens that bridge the OEC Mn (except O2 and O3) can find its way to any of the three channels (Figure 4B) (Kaur et al., 2021). Comparing the proton affinity of H3O+ placed on individual water molecules in the three separated paths shows the broad channel as being more hospitable to the positive charge as suggested earlier (Ishikita et al., 2006; Bondar and Dau, 2012).
Mutations and time resolved IR difference spectroscopy support a complex proton transfer path near the OEC. The question is how to characterize a highly interconnected proton transfer path dominated by water molecules. FTIR-difference spectra followed through the cycle of reactions that lead to oxygen evolution shows changes in an extensive hydrogen bonding network around the cofactor. For example, FTIR-difference spectra shows a carboxylate peak (near 1,747 cm−1) whose proton affinity decreases in the step in the OEC oxidation cycle where a proton is not released to the lumen (S1 to S2) (Debus, 2015). This feature is lost when mutations are made of residues separated by ≈20 Å including D1-Asp61Ala, D1-Glu65Ala, D2-Glu312Ala, D1-Arg334Ala, D1-Glu329Gln (Figure 4). Mutation of each of these residues disrupts the hydrogen bond network and blocks or slows O2 evolution (Service et al., 2010; Debus, 2014). All of these residues are found in the network analysis that reveled the connections of all water molecules near the OEC (Kaur et al., 2021).
PLS used for proton coupled electron transfer near the OEC. PSII provides an example of the use of a PLS to stabilize the redox reactions of an intermediate on a longer electron transfer chain. Through the S-state cycle the redox active Tyr161, Yz, is the electron donor to the oxidized chlorophyll, P680•+. Yz•+ is then reduced by the OEC (Figure 5) (Lavergne and Junge, 1993). The pKa of an oxidized Tyr is -2 (Tommos and Babcock, 2000), so Yz•+ will lose its proton. D1-His 190 serves as a PLS, trapping the proton for the microseconds to several milliseconds that Yz is oxidized (Figure 5) (Rappaport et al., 1994). This His has a low enough proton affinity that it is neutral in the ground state, yet its proton affinity is higher than the oxidized Yz•. The protein must block the proton from being lost to the lumen from the His. A tight hydrogen bond between the Tyr and the His helps as does the presence of polar, but non-proton conducting residues such as D1-Asn 298 surrounding the pair (Saito et al., 2011). The proton is shuttled between the Tyr cofactor and its adjacent PLS, never moving in or out of the protein, while the electron is passed from the OEC to P680 via YZ (Figure 5) (Saito et al., 2011; Ishikita and Saito, 2014).
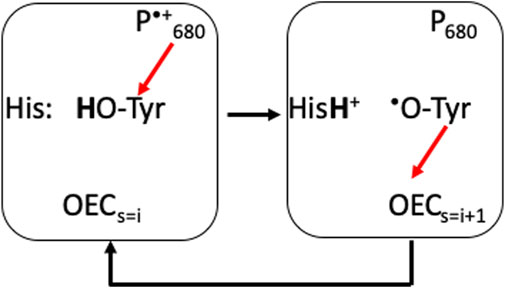
FIGURE 5. D1-His 190 is an example of a PLS supporting transient redox cycling of YZ (D1-Tyr 161), which is an intermediate electron donor/acceptor in the PSII electron transfer chain. Red arrows show electron transfers. YZ is an electron donor to the oxidized P680•+ (≈11 Å away) and acceptor from the OEC (≈5 Å away). The pKa of the oxidized Tyr is <0, while it is 9.8 when it is reduced. His 190 ensures that the proton is captured when the Tyr is oxidized and returned when the Tyr is reduced. As YZ and His 190 are hydrogen bonded together, the proton simply moves between the two residues and does not escape (Ishikita and Saito, 2014).
Gates in proton transfer pathways in proteins that are not pumps. A gate along proton transfer path guards against unwanted proton transfers. An inhibitory gate can be identified in PSII, where a chloride ion helps to keep the entrance to the broad channel open by preventing a salt bridge between D1-Asp 61 and D2-Lys 317. Experiments show chloride depletion blocks the advancement of S-state transition beyond S2 (Ono et al., 1986; Pokhrel and Brudvig, 2014). Simulations show depletion of chloride leads to formation of a salt bridge between D1-Asp 61 and D2-Lys 317 hindering proton loss (Rivalta et al., 2011; Amin et al., 2016; Kaur et al., 2019).
Fences support a proton path. The sides of the water filled channels contain residues such as Asn and Arg (e.g., D1-Asn 87 and CP43-Arg 357). These cannot participate in Grotthuss proton transfers, nor are their pKa in a range that would let them be PLS. Rather, these residues can anchor, the hydrogen bond connections, orienting the water molecules. One example that has been investigated is D1-Asn 298 near the OEC. Mutating this residue influences oxygen evolution (Kuroda et al., 2014) and the FTIR spectrum of the OEC network (Nagao et al., 2017; Chrysina et al., 2019). Simulations show the Asn changes the orientation of its side chain amide dipole in the transition from S2 to S3 leading to rearrangement of the hydrogen bond network (Chrysina et al., 2019).
Bacterial Reaction Center
The reaction center, bRC, of the purple non-sulfur bacteria is the first membrane protein whose structure was solved at atomic resolution (Deisenhofer et al., 1985). RCs are light activated proteins so as with GFP, bR and PSII, time resolved measurements allow individual steps in the series of electron and/or proton transfer reactions to be monitored by time-resolved spectroscopy, showing the individual steps in the reactions (Okamura et al., 2000; Wraight, 2006).
bRCs and PSII are Type II reaction centers where a fully reduced, QH2 is the final product. The quinol dissociates into the membrane to serve as the substrate of the b6f complex in oxygenic photosynthesis and the bc1 complex in bacteria (Cardona et al., 2012; Cardona and Rutherford, 2019). The D1 and D2 subunits of PSII are related to the L and M subunits in bRCs (Raymond and Blankenship, 2004). The bacterial systems use a photon, in the range of 860–960 nm. Thus, they do not have enough energy to carry out the PSII reaction, which uses a 680 nm photon to fuel the uphill transfer of electrons from water to quinone (Heathcote et al., 2002). The primary electron donor in bRCs is periplasmic (P-side) cytochrome c. The redox reactions of cytochrome c are not coupled to proton binding/release. The overall reaction is:
In contrast to PSII, which has a very short distance to the N-side, bRCs have an H subunit, capping the N-side of the protein, requiring a much longer path for the protons to reach the QB site (Figure 6).
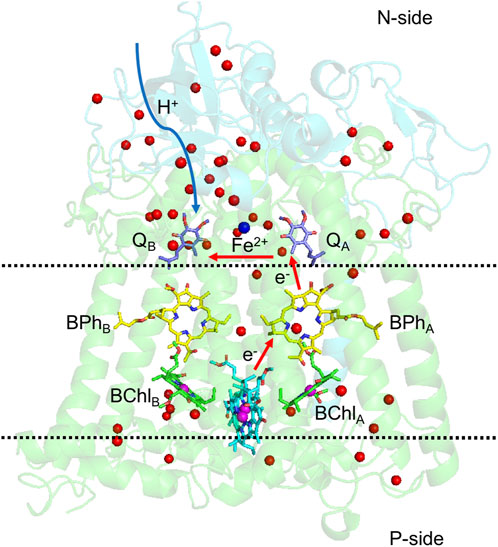
FIGURE 6. Structure of bacterial photosynthetic reaction centers of Rhodobacter sphaeroides. L and M subunits in green, H subunit in cyan. Red arrows follow the electron transfer path, and the blue arrow is the path protons travel from the N-side to QB. Water molecules in the structure are shown as red balls. The region between the two dashed lines has few water molecules or polar residues, which discourages proton transfer across the protein. The two quinones are still within the lipid membrane, but in a region of the protein with multiple polar residues and water molecules to transport protons. Coordinates are from PDB ID: 1AIG.
Quinones as a model redox coupled proton transfer reactant. Electrons move one at a time between cofactors in proteins generating free radical intermediates. While some cofactors such as chlorophylls, hemes, iron sulfur clusters and Tyr are stable one electron redox cofactors, unpaired electrons are often sources of toxic reactive oxygen intermediates (Weisz et al., 2017). Quinones function as single electron donors/acceptors within proteins, but accumulate two electrons and protons (Paddock et al., 2003; Müh et al., 2012). These lipid soluble cofactors thus transport electrons from many proteins including PSII, bRCs and complex I described here to the bc1 complex in mitochondria and bacteria or b6f complexes in chloroplasts as electrons move down the electron transfer chains.
As quinones cycle between oxidized quinone (Q), semiquinone (Q•-) and fully reduced quinol (QH2) their proton affinity changes. The quinone pKa is <0, it is <5 for the semiquinone (Zhu and Gunner, 2005; Gunner et al., 2008; Hasegawa et al., 2017). Hence, the anionic semiquinone is often the stable state within the protein. However, the generation of Q•- can trigger proton binding to nearby residues (Graige et al., 1996; Abresch et al., 1998; Alexov and Gunner, 1999; Paddock et al., 2003). The pKas for the fully reduced quinol is >10 so the second QB reduction is coupled to binding two protons to the cofactor. The difference in the hydrogen bonding pattern for the two quinone carbonyls and two quinol hydroxyls promote quinone dissociation in PSII (Shevela et al., 2012; Saito et al., 2013). As will be described below, in Complex I quinone reduction leads to a large conformational change that trigger proton pumping through distant pathways (Gupta et al., 2020; Gutiérrez-Fernández et al., 2020; Kampjut and Sazanov, 2020).
The quinone electrochemistry is modified within the protein, but the underlying proton affinity of each redox state in the isolated compound strongly influences the order in which protons are bound. The sequence of reactions in bRCs (Graige et al., 1996) and likely in PSII is: 1) QB is first reduced to the anionic semiquinone. The negative potential causes protons to be bound to nearby amino acids (Okamura and Feher, 1992; Wraight, 2006; Gunner et al., 2008); 2) uphill protonation of the semiquinone is the rate-determining step preceding the second reduction; 3) reduction is followed by binding a second proton and release of the quinol.
Proton transfer pathways to QBin bRCs. The proton transfer paths to QB have been well studied (Abresch et al., 1998; Okamura et al., 2000). These are different than those described above near the OEC, as residues play a much larger role in the network. However, they are similar in that there is a tangled complex of proton transfer paths. There is a large number of acidic and basic residues buried in the protein near QB that influence the electrochemistry of the quinone and provide paths for proton transfer (Figure 7) (Sebban et al., 1995; Lancaster et al., 1996; Abresch et al., 1998; Alexov and Gunner, 1999; Rabenstein et al., 2000). FTIR difference spectra obtained on QA and QB reduction shows broad features characteristic of a polarized, interconnected hydrogen bonded network of water molecules and amino acids around the two quinones (Breton and Nabedryk, 1998). L-Asp 210 and Asp 213 may share a proton in the ground state, serving as a PLS. Protonating one of the acids removes a negative charge, stabilizing the semiquinone QB•- and keeping a proton available for passage to the quinone itself (Lancaster et al., 1996; Alexov and Gunner, 1999; Ishikita et al., 2003). The mutants L-Asp213Asn and L-Ser223Ala slow the rate of this reaction, with the mutation of L-Asp 213 having a bigger impact (Paddock et al., 1994; Paddock et al., 1995). In the absence of L-Asp 213, H-Glu 173 may provide an alternative location for the proton in this extended PLS (Paddock et al., 2003). Thus, this web of acidic residues combines the functions of a cluster PLS and complex proton transfer path.
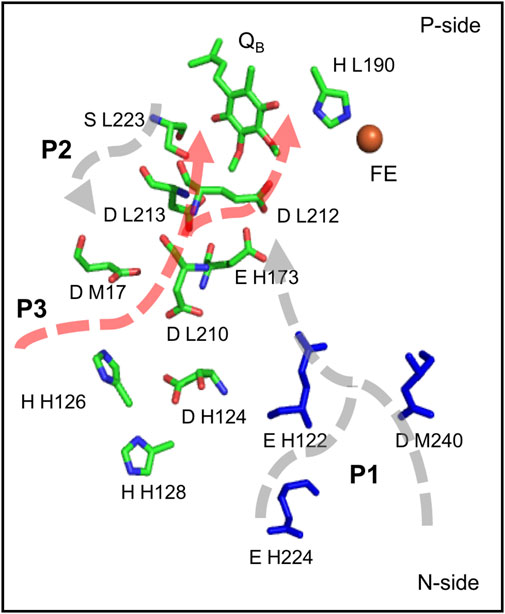
FIGURE 7. Three proposed paths for the two protons from the surface to the QB site (Abresch et al., 1998), showing key residues on each path including H-His 126, H-His 128, H-Asp 124, L-Asp 210, M-Asp 17, H-Glu 173, L-Asp 213, L-Ser 223 and L-Glu 212 (Okamura et al., 2000; Paddock et al., 2003; Wraight, 2004). Coordinates from Rb. sphaeroides bRCs PDB ID: 1AIG.
L-Glu 212, which is a protonated PLS in the ground state, provides the second proton to QB (Wraight, 2004). The pKa of the Glu is ≈10, trapping a proton near the quinone in the ground state ready when needed (Kleinfeld et al., 1984; Okamura and Feher, 1992). The mutant L-Glu212Gln does not affect the delivery of the first proton, supporting this site being neutral in the presence of QB-. However, the transfer of the second proton is totally blocked, indicating L-Glu 212 is a unique single site PLS (Paddock et al., 1989; Shinkarev et al., 1993; P. H.; McPherson et al., 1994; Okamura et al., 2000; Wraight, 2004).
Three possible paths. The complex web of acidic and other polar residues near QB leads to the question of what is the route for proton transfer from the N-side surface to the quinone binding site. The crystal structures reveal three likely paths (Figure 7) (Abresch et al., 1998). The longest path, P1, is ≈20 Å long. It enters the protein near H-Asp 224 or M-Asp 240 and passes to L-Glu 212, which provides the second proton to QB. P2, also ≈20 Å long, starts near M-Tyr 3 and moves via H-Glu 173 to L-Asp 213, which donates the first proton to QB. P3 is the shortest path, with only ≈7 Å between L-Asp 213 and the surface M-Asp 17 with one water molecule in the middle (Abresch et al., 1998).
Surface PLS as a proton collection site. RCs also have a well characterized external cluster near the entrance to P3 made up of H-Asp 124, H-His 126, H-His 128. The cluster is a proposed proton collection site (Utschig et al., 1998; Paddock et al., 1999; Okamura et al., 2000) (Figure 7). Zn2+ or Cd2+ bind here and slow proton transfer to QB. Clusters of protonatable groups near the surface of proton transfer paths are found in other proteins. A similar proton accumulation site is found in the D-channel (Cai et al., 2018). The broad channel in PSII exits to a cluster of surface acidic residues that can trap the released proton (Bondar and Dau, 2012; Kaur et al., 2021).
P3 appears to carry protons in wild-type bRCs. Mutation of L-Asp 210 and M-Asp 17, have a larger impact when Zn2+ or Cd2+ are present, showing an additive effect of multiple changes to this pathway. However even with P3 blocked, protons still enter to QB, indicating that other routes can serve as pathways, but with slower transfer rates (Okamura et al., 2000). Thus, in the tangled potential proton transfer network, multiple paths are possible, but some are preferred.
NADH-Ubiquinone Oxidoreductase (Complex I)
Complex I is the first and the largest protein in the erobic respiratory electron transfer chain of bacteria and mitochondria (Brandt et al., 2003; Hirst, 2013; Sazanov, 2015; Kaila, 2018). Within the protein electrons are transferred from NADH to a flavin and through a series of Iron Sulfur (FeS) complexes to a quinone (Verkhovskaya et al., 2008; Efremov et al., 2010; Hirst, 2013; Zickermann et al., 2015). The overall reaction is:
The quinone, Q, is often ubiquinone but can be menaquinone in bacteria such as Thermus thermophilus, the source of the protein for the first complete crystal structure (Baradaran et al., 2013). The transfer of the two electrons, which occurs in the N-side peripheral arm, leads to the pumping of four protons from the N- to P-side of the membrane embedded portion of the protein (Figure 8).
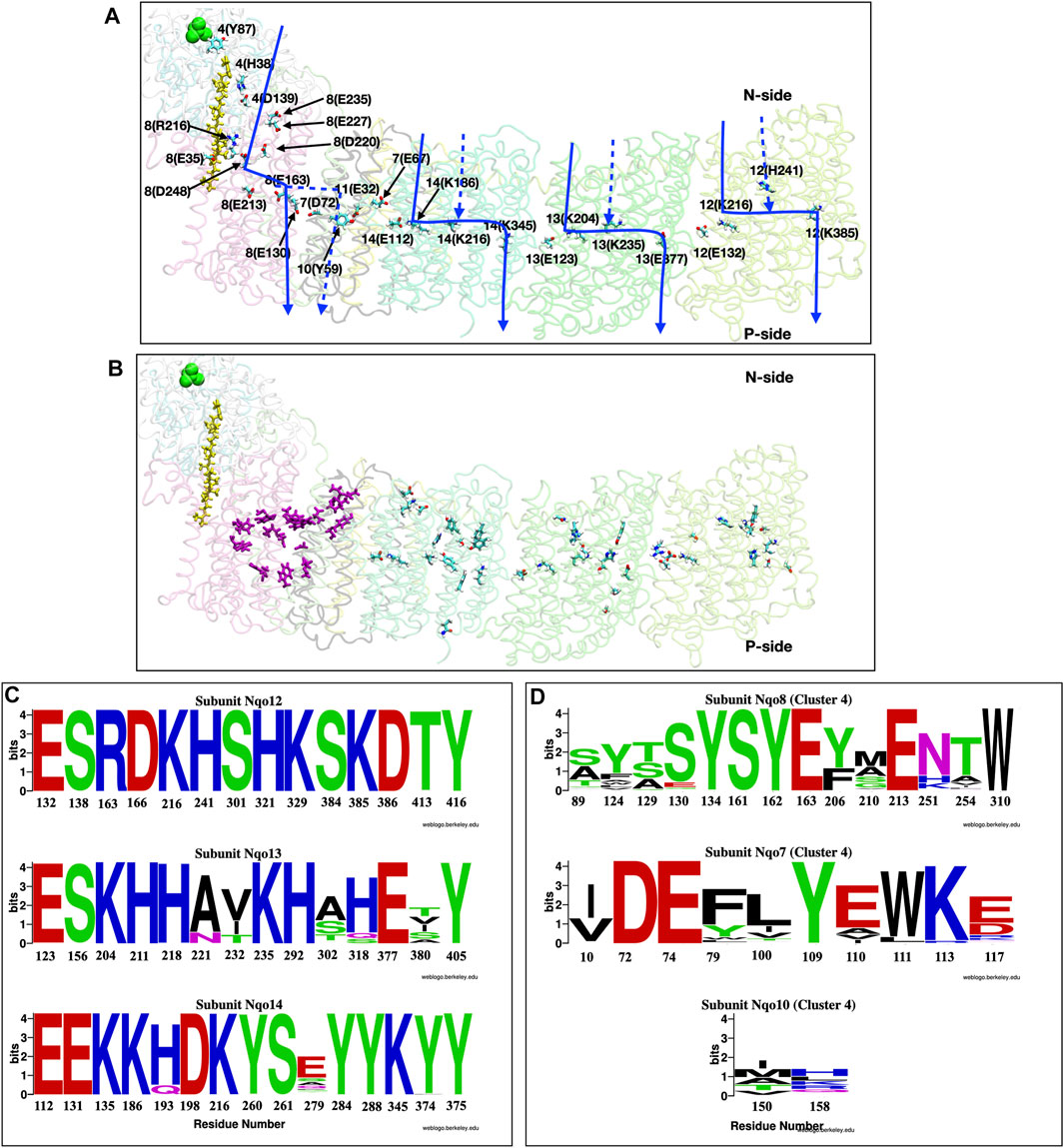
FIGURE 8. (A,B) Structure of the full membrane domain of complex I and only the Nqo4 subunit of the periplasmic domain from T. thermophilus [PDB ID: 4HEA (Baradaran et al., 2013)] highlighting proton transfer paths. (A) Residues that have been proposed to be important for proton pumping are shown as sticks and labeled as subunit number (one letter amino acid designation-residue number). Quinone is yellow and N2 of each Iron Sulfur cluster is green. Approximate locations of putative proton transfer paths are blue arrows and dashed lines show alternative, proposed paths. (B) Residues that are included in the conservation analysis. Purple sticks: E channel cluster in the center of the protein [Cluster 4 in (Khaniya et al., 2020)]; Sticks colored by atom type: antiporter residues (Baradaran et al., 2013; Di Luca et al., 2017; Haapanen and Sharma, 2017; Gutiérrez-Fernández et al., 2020). (C,D) Conservation analysis shown as Weblogo (Crooks, 2004) representation of multiple sequence alignment of 1,000 complex I sequences (Johnson et al., 2008; Sievers et al., 2011). (C) Residues in the three linear antiporter pathways in Nqo12 (Top), Nqo13 (Middle) and Nqo14 (Bottom) subunits. (D) Residues in the E-channel cluster 4. Residues from Nqo8 (Top), Nqo7 (Middle) and Nqo10 (Bottom).
Complex I combines elements of two disparate proteins. The long, peripheral arm is likely derived from a soluble hydrogenase and three of the proton channels are related to Mrp Na-H antiporters (denoted antiporter channels), however, the fourth, E-channel, is unique to Complex I and its close relatives (Efremov and Sazanov, 2012; Brandt, 2019). Complex I is remarkable, as the tightly coupled electron transfer and proton pumping elements are separated by as much as 300 Å from the NADH binding site, at the end of the peripheral arm, to the distal proton pumping subunit (Baradaran et al., 2013; Kaila, 2018). Quinone binding and reduction lead to a rotation of the soluble arm that connects the redox reactions to proton pumping in some way (Gupta et al., 2020; Gutiérrez-Fernández et al., 2020; Kampjut and Sazanov, 2020).
Antiporter: simple pathway. Complex I provides examples of both simple and complex proton transfer pathways. There are four proton paths, three through the antiporter subunits and one through the E-channel (Hirst, 2013; Ripple et al., 2013; Sazanov, 2015; Di Luca et al., 2017; Haapanen and Sharma, 2018; Saura and Kaila, 2019). The crystal structures show likely, linear paths through each antiporter subunit (Efremov and Sazanov, 2011; Baradaran et al., 2013; Zickermann et al., 2015) which have chain of well conserved acidic and basic residues in the center running parallel to the membrane (Figures 8A,B) (Fearnley and Walker, 1992; Torres-Bacete et al., 2007; Efremov and Sazanov, 2012). Recognizable water chains leading to the N- and P-sides are seen in computational studies (Kaila et al., 2014; Di Luca et al., 2017; Haapanen and Sharma, 2017; Röpke et al., 2020). Moving along each pathway from the N-side is a Glu/Lys pair then a central Lys followed by either a Lys or Glu (Baradaran et al., 2013; Kaila, 2018). Their protonation states change as the proton is handed from one ionizable residue to the next. Thus, the antiporter channels are simple linear proton transfer paths.
With a linear proton transfer path, it is often possible to identify a unique gating element. Simulations have been carried out to investigate the behavior of Complex I with different protonation states for these residues (Kaila et al., 2014; Di Luca et al., 2017; Haapanen and Sharma, 2017). Increasing the net charge in the interior leads to water molecules being brought into the protein in MD trajectories and they are expelled when the charges are neutralized (Kaila et al., 2014; Hummer and Wikström, 2016; Di Luca et al., 2017). These hydration/dehydration changes will gate proton transfer through the channels, similar to that described above for Asp96 in bR. They can be validated by seeing waters in different locations in structures trapped in different intermediates or by interpretation of IR spectroscopy (Lórenz-Fonfría et al., 2008). However, a buried charge will attract water in MD simulations so it is important that the residue protonation states be correctly assigned in the simulation (Hummer and Wikström, 2016).
Comparison of the structures of Complex I from different organisms shows conservation of the P-side proton release paths in the antiporter subunits. However, on the N-side MD studies (Kaila et al., 2014; Di Luca et al., 2017; Haapanen and Sharma, 2017) found a pathway similar to one identified in the crystal structure of Y. lipolytica (Zickermann et al., 2015) Complex I but different from the one proposed from the T. thermophilus (Sazanov, 2015) crystal structure. Thus, it is not known if the exit path is conserved. Similar changes in pathways through evolution are also found comparing A- and B-type CcO as will be described below.
E-channel: A complex proton transfer path. In contrast to the linear proton transfer path seen through the three antiporter subunits, the fourth proton travels through a path directly under the periplasmic arm denoted the E-channel. This region has a web of water molecules and polar and protonatable residues characteristic of a complex proton transfer path with several PLS clusters (Di Luca et al., 2017; Saura and Kaila, 2019; Gutiérrez-Fernández et al., 2020; Khaniya et al., 2020). There are several competing proposals for the proton transfer path through the E-channel. It has been suggested to use subunits Nqo10 and Nqo11 (Efremov and Sazanov, 2011; Zickermann et al., 2015) or subunit Nqo8 (Baradaran et al., 2013). Various computational studies also provide different answers (Kaila et al., 2014; Di Luca et al., 2017; Haapanen and Sharma, 2017). This uncertainty about the route is characteristic of complex proton transfer paths. There is a growing consensus that residues in subunits Nqo7, 8 10 and 11 are important for E-channel function (Figure 8A). Network analysis, which can accommodate complexity, has proposed a complete path through subunit Nqo4 and Nqo8 at the N-side entry, moving through subunit Nqo8 and Nqo7 in the center, and exiting through subunit Nqo10 and Nqo11 (Khaniya et al., 2020).
Role of quinone in Complex I as a gate. Complex I is able to couple the energy releasing redox reactions in the peripheral arm to the energy requiring proton pumping through four, distant well separated pumping sites (Baradaran et al., 2013; Kaila, 2018). The quinone binding site in complex I is ≈25–30 Å above the membrane surface, which is different from its location in any other quinone dependent membrane protein (Baradaran et al., 2013; Zickermann et al., 2015). Quinone binding leads to the rotation and tilt of the peripheral arm (Gutiérrez-Fernández et al., 2020). MD simulations (Gamiz-Hernandez et al., 2017; Warnau et al., 2018; Gupta et al., 2020) and Monte Carlo sampling (Khaniya et al., 2020) find changes in the connectivity of the hydrogen bond network that depend on the presence and redox state of the quinone. Movement of subunit Nqo4 and Nqo8 leads to changes in the E-channel hydrogen bond network that propagate into the first antiporter channel by a distortion near Nqo10 (Tyr 59) (Gutiérrez-Fernández et al., 2020). The shifts in hydrogen bond network and residue protonation initiated by the quinone reactions thus yield changes in the interaction between the key Glu/Lys residues in the very distant antiporter channels (Efremov and Sazanov, 2011; Kampjut and Sazanov, 2020). This web of long-range communication is not needed in smaller proteins such as CcO, described below, where the change in electrostatic potential due to the redox reactions can directly modify PLS proton affinity coupled to proton pumping.
MD trajectories show quinone binding influences the E-channel by enriching the number of hydrogen bonds near the N-side, which are proposed to open the channel for proton uptake (Gupta et al., 2020; Khaniya et al., 2020). However, when QH2 is bound the charge of conserved residues change leading to modification of the water wires in the proton transfer channels (Gamiz-Hernandez et al., 2017; Kaila, 2018). The E-channel central region has extended clusters of protonatable residues including Nqo7 (Asp 72), 8 (Glu 130), 8 (Glu 163), 8 (Glu 213) (Khaniya et al., 2020) [the nomenclature uses residue numbering from the T. thermophilus complex I in the form as Nqo subunit (residues)].
Beyond the central cluster of polar residues there is a hydrophobic barrier that blocks the proton transfer to the P-side in the E-channel. Thus, while there have been several studies of the proton entry, connections are rarely drawn from the center to the P-side (Baradaran et al., 2013; Kaila et al., 2014; Zickermann et al., 2015; Di Luca et al., 2017; Haapanen and Sharma, 2017). Network analysis of the hydrogen bonds made in MD trajectories suggests several paths that rely on transient wetting events (Khaniya et al., 2020). One lies near Nqo7 (Tyr 7) and 8 (Tyr 124). Another possibility is from Nqo8 (Glu 130) to 10 (Tyr 59) (Gutiérrez-Fernández et al., 2020; Steiner and Sazanov, 2020). However, these proposed bridging residues are not well conserved. Thus, what permits the proton to cross the hydrophobic barrier, and whether it conserved through Complex I evolution, is still an open question. A similar hydrophobic barrier is also seen in the voltage-sensing domain (VSD) of voltage-gated ion channels. Here mutations of the hydrophobic residues make the system leaky, showing the importance of non-polar residues to block uncontrolled proton transfers (Banh et al., 2019).
Residue conservation. The conservation and sensitivity of residues to mutation can provide evidence that there is a unique pathway for protons. The multisequence alignment of the residues in the linear paths through the three antiporter subunits (Baradaran et al., 2013; Di Luca et al., 2017; Haapanen and Sharma, 2017; Gutiérrez-Fernández et al., 2020) were compared with that found for the central cluster in the complex E-channel pathway (cluster 4 residues) (Khaniya et al., 2020). The WebLogo (Crooks, 2004) provides a graphical comparison of the results (Figures 8C,D). The residues along the linear antiporter paths are highly conserved. In contrast, the E-channel central cluster shows much weaker conservation, suggesting that, while the cluster as a whole must function, individual residues may not be uniquely important.
Mutation The sensitivity to mutation may also distinguish linear from complex proton transfer paths. Thus, mutation of residues along a linear path should severely impair activity. In contrast, a complex path may be less sensitive as there are multiple routes for the proton, though as shown above for bRCs, not all need be equally favorable. Many of the residues in the well-defined antiporter channels have been subjected to site-directed mutations. Mutation of the residues shown in (Figure 8A) severely reduce quinone oxidoreductase activity that is tightly coupled to proton transfer since there is no alternative paths for proton transport (Torres-Bacete et al., 2007; Euro et al., 2008; Michel et al., 2011). However, the E-channel is more complex and there is less consensus about the path. Mutations of proposed E-channel residues often modify but do not kill activity (Taylor et al., 2002; Yang et al., 2009).
Cytochrome c Oxidase
CcO is a proton pump belonging to the heme-copper oxidase superfamily (Kaila et al., 2010; Liang et al., 2017; Kaur et al., 2019). The energy for proton pumping comes from electrons from cytochrome c (Cyt c) reducing O2 to water in the binuclear (Heme & Cu) center (BNC), located in the protein center (Figure 9). The BNC is reduced stepwise, one at a time to store four electrons. O2 reduction takes place in one step in the fully reduced BNC (Kaila et al., 2010; Cai et al., 2020). O2 production likewise takes place in one step in the fully oxidized OEC of PSII. This mechanism protects against the release of toxic reactive oxygen intermediates. The overall reaction is:
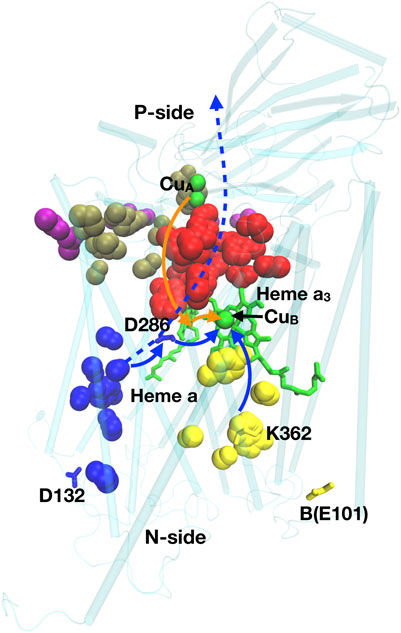
FIGURE 9. Residues of importance for proton pumping in CcO. Residues D132, K362, D286, B(E101) are shown; heme a and a3 are green sticks; CuA and CuB, are green spheres. Blue spheres: D-channel; red: P-side cluster PLS; yellow: K-channel; Brown: P-exit path; Purple: P-exit surface cluster. Solid blue lines: path of protons used for chemistry in the BNC (heme a2 and CuB), dashed blue line: path for pumped protons; orange line: electron transfer path. Structure of Rb. sphaeroides CcO from PDB ID:1M56.
Four electrons come from the cytochrome c on the P-side, and four protons from the N-side to the BNC for chemistry. m is the number of protons pumped across the membrane. There are several related classes of CcO, denoted A, B and C. The A-type CcO is found from bacteria to mammals, while B- and C-type are found in bacteria that live at low O2 levels. In the A-type CcO, m = 4. B- and C- type CcOs differ in the types of heme used and in the number of protons pumped/electron, with m generally less than four (Lee et al., 2012). CcO provides examples of simple and complex proton transfer paths and simple and cluster PLS as well as a gate generated by hydration/dehydration changes.
Proton transfer paths through CcO. In the A-type CcO two linear water filled channels (D- and K-channels) are seen. Each has an essential ionizable residue at the entry on the N-side and at the end near the BNC, but none within the channels. The D-channel has Asp 132 at the entry and the essential, isolated Glu 286 as the PLS (Wikström et al., 2000; Brändén et al., 2001). The K channel has Lys 362 near the BNC and B-Glu 101 near the entry (Ma et al., 1999; Brändén et al., 2002). The buried Glu and Lys are both isolated in hydrophobic parts of the protein. In the ground states, their proton affinity has shifted so both are neutral (Rb. sphaeroides CcO numbering used here).
The D-channel carries six of the eight protons in the A-type CcO reaction cycle, while the K-channel carries two. At the center of CcO is heme a and heme a3 and CuB of the active site BNC. As the retinal does in bR, the large cofactors may help to block proton transfer through the protein. The protons from the K channel exit into the BNC to be added to the reduced product water, while the D channel exits between the two hemes. The importance of these linear pathways were demonstrated by mutation of the residues at the beginning and end of the channels leading to loss of activity (Jünemann et al., 1997; Qian et al., 1997; Mills and Ferguson-Miller, 2002). Oddly, the D-channel is missing in B- and C-type CcOs, with only a K-channel remaining (Lee et al., 2012). Thus, as suggested in complex I, proton transfer paths may shift through evolution. In the B- and C- type CcOs it remains unclear how the pumped protons move around the active site to be delivered to the P-side.
Complex proton transfer pathways. The P-side of all CcOs has a tangled cluster of strongly interacting polar and protonatable residues that do not provide an obvious single exit path, although linear paths have been suggested (Popović and Stuchebrukhov, 2005; Björck et al., 2019). The hydrogen bond network on the P-side of A- and B-type CcO, was analyzed using Monte Carlo sampling and network analysis (Cai et al., 2018; Cai et al., 2020). Calculations were initiated with experimental crystal structures as well as with snapshots from MD trajectories carried out in different redox states of the hemes and protonation states of key residues. This analysis recognized a linear proton transfer path through the D-channel in the A-type CcO. A very large cluster of interconnected residues was identified as the P-side PLS (Figure 9). This cluster exits through several paths to a region near the cytochrome c binding site. Thus, there is an exit region not a unique exit for protons in this complex path.
Single residue PLS in CcO. Glu 286 is located at the top of the D-channel of A-type CcO (Kaila et al., 2010). It is isolated from other protonatable residues so forms a simple PLS. It plays an essential role, releasing a proton to the BNC for chemistry and to a P-side PLS cluster for pumping. The X-ray crystal structures show it is in a dry region and all simulation techniques give it a high proton affinity as there is nothing in the structure to stabilize an anionic residue (Hummer and Wikström, 2016). Measurements found a pKa of ≈10 for turnover that is assigned to Glu286 (Namslauer et al., 2003). However, MD simulations showed protonation of a propionic acid in the P-side PLS breaks a hydrogen bond and opens a cavity which then fills with water (Goyal et al., 2013; Son et al., 2017).
The hydration of the water cavity near Glu 286 also serves as a gate for the proton transfer pathway. In the crystal structures and in protein equilibrated in MD trajectories without the water cavity there is no exit found from the D-channel to the P-side PLS. Thus, the closed cavity blocks the backflow of protons (Cai et al., 2018). However, when the cavity is hydrated Glu 286 becomes well connected to the extensive PLS cluster on the P-side. Changes in hydration also help control the proton transfer from Glu 286 to the BNC (Wikström et al., 2003) Thus, hydration tunes the proton affinity of an isolated residue and serves as a gate in the proton transfer pathway, a pattern described for Asp 96 in bR and for the antiport subunits in complex I.
The role of tautomer shifts in a complex PLS. A proton pump must regulate the thermodynamics of PLS loading and unloading, and then change proton affinity when the reaction progresses. This requires tuning the free energy difference between the loaded and unloaded states at the pH of interest as well as the shift in this value as the protein goes through the reaction cycle (See fuller description in Supplementary Material S1). The PLS must remain in the appropriate loaded/unloaded state until the reaction has progressed and the accessibility of N- and P-side is modified by the gates opening/closing (Kim et al., 2007; Kim et al., 2009; Kim and Hummer, 2012; Stuchebrukhov, 2018; Stuchebrukhov, 2019). The challenge is to find the changes that can trap, hold and then release the proton. An analysis of the proton distribution in the PLS cluster in B-type CcO provides some insight into the atomic details of one mechanism.
On the P-side of the B-type CcO, an extended cluster of six residues was found to behave as a PLS (Figure 10) (Cai et al., 2020). The unloaded PLS has one proton bound (net charge -4) while the loaded cluster has two protons. The protein surrounding the PLS provides sufficient long-range positive potential to stabilize the cluster negative charge. There are six tautomers with one proton and 12 with two protons distributed over the six residues (Eq. 1). By investigating the proton affinity of different tautomers in snapshots derived from MD trajectories it was found that a shift in the hydrogen bond pattern changes the tautomer selected. This resulted in dynamic states where a loaded and unloaded state are close in energy so changes at the active site lead to proton binding or release. However, the BNC is ≈15 Å from the PLS and the change in the BNC do not shift the PLS proton affinity enough to fully load a proton to the PLS cluster. This behavior is seen Supplementary Figures S2A,B when there is only a small shift in proton affinity in a group with a pKa near the pH (Supplementary Figure S2C, middle titration). The problem of incomplete loading/unloading appears to be solved by moving the bound protons and rearranging the hydrogen bonding pattern, which is described as a tautomer trap. This leads to the PLS being trapped loaded or unloaded because the cluster proton affinity is either too high or too low (Supplementary Figure S2C, right-most titration) or unloaded (Supplementary Figure S2C, left-most titration).
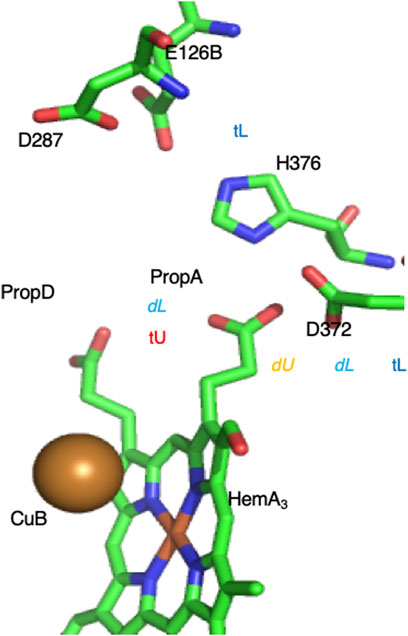
FIGURE 10. Complex PLS on the P-side of the B-type CcO. The proton moves between Prop A [trapped unloaded (tU), with very low proton affinity] to D372 [dynamic unloaded (dU)] where electron transfer to Heme A3 or CuB in the BNC active site 15 Å distant will lead to proton loading into the cluster (Cai et al., 2020). In the loaded state one proton is on D372. If the second proton is on PropA the system is dynamic (dL), so addition of a proton to the product water trapped in the BNC leads to the PLS unloading. If the second proton is on H386 the cluster proton affinity is too high to lose a proton (tL). The intra-cluster distances determine the relative energy of the loaded and unloaded tautomers. The crystal structure is likely trapped in the loaded state. Structure of Th. thermophilus CcO from PDB ID:3S8F.
The proton shift in the CcO complex PLS shows how a tautomer trap can solve the problem that a PLS, which is sensitive to changes in the protein, may not strongly trap the proton. Thus, in active structures, the change in proton affinity due to the BNC will lead to some changes in the PLS protonation state. But this shift in free energy is insufficient to reliably move the PLS between being fully loaded and unloaded (Supplementary Figure S2). Then a tautomer shift moves the PLS from the dynamic configuration (where the proton was bound or lost) to the stable, fully loaded or unloaded locked configuration. This mechanism may be similar to activation and inactivation process in voltage gated ion channels, where conformational changes occur when the channels are an active open state, then block the channel and transit to inactive state (Aldrich, 2001). A tautomer trap is only available to a PLS cluster, it is not possible in a single residue PLS.
Conclusion
The structure of multiple proton transfer paths in several proteins that add to the transmembrane electrochemical gradient show a range of motifs. Thus, they can be simple linear paths as found in the D- and K- channels of CcO and the antiporter subunits of complex I. They can also be complex paths as seen on the P-side of CcO and the E-channel of complex I. Paths can be filled with water so that a proton never needs to use a side chain as found around the OEC in PSII or to be handed through a mixture of side chains and water molecules as found in GFP and in bRCs. With simple, single site PLS, as at the exit from the D-channel and the N-side of bR, changes far from the PLS trigger water influx that leads to proton release and production of a water chain to ferry protons. A similar mechanism is used in the antiporter subunits of complex I. In contrast, in the PLS cluster on the proton release side of bR and CcO, small, local rearrangements of a cluster of strongly interacting residues leads to large changes in proton affinity to cause the PLS to load and unload.
Thus, the framework that proteins will have proton transfer paths, Proton Loading Sites (PLS) and gates allows the analysis of each of these proteins. However, the motifs vary in the residues that make up the needed elements and in their complexity. Each proton pump reviewed here use structures with different complexity for different parts of the proton transfer paths. The advantages of different motifs remain to be determined.
Author Contributions
DK, UK, YZ, and MG contributed to the draft preparation. DK, UK, and YZ contributed equally. MG reviewed and edited the manuscript.
Funding
The authors would like to acknowledge the funds from the Department of Energy Basic Energy Sciences grant number DE-SC0001423 and the National Science Foundation grant MCB-1519640.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Xiuhong Cai and Junjun Mao for many helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.660954/full#supplementary-material
References
Abresch, E. C., Paddock, M. L., Stowell, M. H. B., McPhillips, T. M., Axelrod, H. L., Soltis, S. M., et al. (1998). Identification of Proton Transfer Pathways in the X-ray crystal Structure of the Bacterial Reaction center from Rhodobacter Sphaeroides. Photosyn. Res. 55, 119–125. doi:10.1023/a:1006047519260
Accardi, A., and Picollo, A. (2010). CLC Channels and Transporters: Proteins with Borderline Personalities. Biochim. Biophys. Acta (Bba) - Biomembranes 1798, 1457–1464. doi:10.1016/j.bbamem.2010.02.022
Agmon, N., Bakker, H. J., Campen, R. K., Henchman, R. H., Pohl, P., Roke, S., et al. (2016). Protons and Hydroxide Ions in Aqueous Systems. Chem. Rev. 116, 7642–7672. doi:10.1021/acs.chemrev.5b00736
Agmon, N. (1995). The Grotthuss Mechanism. Chem. Phys. Lett. 244, 456–462. doi:10.1016/0009-2614(95)00905-J
Alexov, E. G., and Gunner, M. R. (1999). Calculated Protein and Proton Motions Coupled to Electron Transfer: Electron Transfer from QA-To QBin Bacterial Photosynthetic Reaction Centers†. Biochemistry 38, 8253–8270. doi:10.1021/bi982700a
Allen, T. W., Andersen, O. S., and Roux, B. (2004). Energetics of Ion Conduction through the Gramicidin Channel. Proc. Natl. Acad. Sci. 101, 117–122. doi:10.1073/pnas.2635314100
Amin, M., Pokhrel, R., Brudvig, G. W., Badawi, A., and Obayya, S. S. A. (2016). Effect of Chloride Depletion on the Magnetic Properties and the Redox Leveling of the Oxygen-Evolving Complex in Photosystem II. J. Phys. Chem. B 120, 4243–4248. doi:10.1021/acs.jpcb.6b03545
Balashov, S. P. (2000). Protonation Reactions and Their Coupling in Bacteriorhodopsin. Biochim. Biophys. Acta (Bba) - Bioenerg. 1460, 75–94. doi:10.1016/S0005-2728(00)00131-6
Banh, R., Cherny, V. V., Morgan, D., Musset, B., Thomas, S., Kulleperuma, K., et al. (2019). Hydrophobic Gasket Mutation Produces Gating Pore Currents in Closed Human Voltage-Gated Proton Channels. Proc. Natl. Acad. Sci. U.S.A. 116, 18951–18961. doi:10.1073/pnas.1905462116
Baradaran, R., Berrisford, J. M., Minhas, G. S., and Sazanov, L. A. (2013). Crystal Structure of the Entire Respiratory Complex I. Nature 494, 443–448. doi:10.1038/nature11871
Bashford, D., and Gerwert, K. (1992). Electrostatic Calculations of the pKa Values of Ionizable Groups in Bacteriorhodopsin. J. Mol. Biol. 224, 473–486. doi:10.1016/0022-2836(92)91009-e
Baudry, J., Tajkhorshid, E., Molnar, F., Phillips, J., and Schulten, K. (2001). Molecular Dynamics Study of Bacteriorhodopsin and the Purple Membrane. J. Phys. Chem. B 105, 905–918. doi:10.1021/jp000898e
Björck, M. L., Vilhjálmsdóttir, J., Hartley, A. M., Meunier, B., Näsvik Öjemyr, L., Maréchal, A., et al. (2019). Proton-transfer Pathways in the Mitochondrial S. cerevisiae Cytochrome C Oxidase. Sci. Rep. 9, 20207. doi:10.1038/s41598-019-56648-9
Blomberg, M. R. A., and Siegbahn, P. E. M. (2006). Quantum Chemistry Applied to the Mechanisms of Transition Metal Containing Enzymes-Cytochromec Oxidase, a Particularly Challenging Case. J. Comput. Chem. 27, 1373–1384. doi:10.1002/jcc.20448
Bondar, A.-N., and Dau, H. (2012). Extended Protein/water H-Bond Networks in Photosynthetic Water Oxidation. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 1177–1190. doi:10.1016/j.bbabio.2012.03.031
Bondar, A.-N., Suhai, S., Fischer, S., Smith, J. C., and Elstner, M. (2007). Suppression of the Back Proton-Transfer from Asp85 to the Retinal Schiff Base in Bacteriorhodopsin: A Theoretical Analysis of Structural Elements. J. Struct. Biol. 157, 454–469. doi:10.1016/j.jsb.2006.10.007
Bozdaganyan, M. E., Lokhmatikov, A. V., Voskoboynikova, N., Cherepanov, D. A., Steinhoff, H.-J., Shaitan, K. V., et al. (2019). Proton Leakage across Lipid Bilayers: Oxygen Atoms of Phospholipid Ester Linkers Align Water Molecules into Transmembrane Water Wires. Biochim. Biophys. Acta (Bba) - Bioenerg. 1860, 439–451. doi:10.1016/j.bbabio.2019.03.001
Brändén, M., Sigurdson, H., Namslauer, A., Gennis, R. B., Ädelroth, P., and Brzezinski, P. (2001). On the Role of the K-Proton Transfer Pathway in Cytochrome C Oxidase. Proc. Natl. Acad. Sci. 98, 5013–5018. doi:10.1073/pnas.081088398
Brändén, M., Tomson, F., Gennis, R. B., and Brzezinski, P. (2002). The Entry Point of the K-Proton-Transfer Pathway in CytochromecOxidase†. Biochemistry 41, 10794–10798. doi:10.1021/bi026093+
Brandt, U., Kerscher, S., Dröse, S., Zwicker, K., and Zickermann, V. (2003). Proton Pumping by NADH:ubiquinone Oxidoreductase. A Redox Driven Conformational Change Mechanism? FEBS Lett. 545, 9–17. doi:10.1016/S0014-5793(03)00387-9
Brandt, U. (2019). Adaptations of an Ancient Modular Machine. Science 363, 230–231. doi:10.1126/science.aaw0493
Brejc, K., Sixma, T. K., Kitts, P. A., Kain, S. R., Tsien, R. Y., Ormo, M., et al. (1997). Structural Basis for Dual Excitation and Photoisomerization of the Aequorea victoria green Fluorescent Protein. Proc. Natl. Acad. Sci. 94, 2306–2311. doi:10.1073/pnas.94.6.2306
Breton, J., and Nabedryk, E. (1998). Proton Uptake upon Quinone Reduction in Bacterial Reaction Centers: IR Signature and Possible Participation of a Highly Polarizable Hydrogen Bond Network. Photosyn. Res. 55, 301–307. doi:10.1023/a:1005972514425
Cai, X., Haider, K., Lu, J., Radic, S., Son, C. Y., Cui, Q., et al. (2018). Network Analysis of a Proposed Exit Pathway for Protons to the P-Side of Cytochrome C Oxidase. Biochim. Biophys. Acta (Bba) - Bioenerg. 1859, 997–1005. doi:10.1016/j.bbabio.2018.05.010
Cai, X., Son, C. Y., Mao, J., Kaur, D., Zhang, Y., Khaniya, U., et al. (2020). Identifying the Proton Loading Site Cluster in the Ba Cytochrome C Oxidase that Loads and Traps Protons. Biochim. Biophys. Acta (Bba) - Bioenerg. 1861, 148239. doi:10.1016/j.bbabio.2020.148239
Cardona, T., and Rutherford, A. W. (2019). Evolution of Photochemical Reaction Centres: More Twists? Trends Plant Sci. 24, 1008–1021. doi:10.1016/j.tplants.2019.06.016
Cardona, T., Sedoud, A., Cox, N., and Rutherford, A. W. (2012). Charge Separation in Photosystem II: a Comparative and Evolutionary Overview. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 26–43. doi:10.1016/j.bbabio.2011.07.012
Chattoraj, M., King, B. A., Bublitz, G. U., and Boxer, S. G. (1996). Ultra-Fast Excited State Dynamics in Green Fluorescent Protein: Multiple States and Proton Transfer. Proc. Natl. Acad. Sci. 93, 8362–8367. doi:10.1073/pnas.93.16.8362
Chrysina, M., Zahariou, G., Sanakis, Y., Ioannidis, N., and Petrouleas, V. (2011). Conformational Changes of the Intermediate of the S2 to S3 Transition in Photosystem II. J. Photochem. Photobiol. B: Biol. 104, 72–79. doi:10.1016/j.jphotobiol.2011.02.012
Chrysina, M., de Mendonça Silva, J. C., Zahariou, G., Pantazis, D. A., and Ioannidis, N. (2019). Proton Translocation via Tautomerization of Asn298 during the S2-S3 State Transition in the Oxygen-Evolving Complex of Photosystem II. J. Phys. Chem. B 123, 3068–3078. doi:10.1021/acs.jpcb.9b02317
Clemens, M., Phatak, P., Cui, Q., Bondar, A.-N., and Elstner, M. (2011). Role of Arg82 in the Early Steps of the Bacteriorhodopsin Proton-Pumping Cycle. J. Phys. Chem. B 115, 7129–7135. doi:10.1021/jp201865k
Cox, N., Pantazis, D. A., Neese, F., and Lubitz, W. (2013). Biological Water Oxidation. Acc. Chem. Res. 46, 1588–1596. doi:10.1021/ar3003249
Crooks, G. E. (2004). WebLogo: A Sequence Logo Generator. Genome Res. 14, 1188–1190. doi:10.1101/gr.849004
Cukierman, S. (2000). Proton Mobilities in Water and in Different Stereoisomers of Covalently Linked Gramicidin A Channels. Biophysical J. 78, 1825–1834. doi:10.1016/S0006-3495(00)76732-4
Cukierman, S. (2006). Et Tu, Grotthuss! and Other Unfinished Stories. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 876–885. doi:10.1016/j.bbabio.2005.12.001
Daldrop, J. O., Saita, M., Heyden, M., Lorenz-Fonfria, V. A., Heberle, J., and Netz, R. R. (2018). Orientation of Non-spherical Protonated Water Clusters Revealed by Infrared Absorption Dichroism. Nat. Commun. 9, 311. doi:10.1038/s41467-017-02669-9
de Grotthuss, C. J. T. (2006). Memoir on the Decomposition of Water and of the Bodies that it Holds in Solution by Means of Galvanic Electricity. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 871–875. doi:10.1016/j.bbabio.2006.07.004
Debus, R. J. (2014). Evidence from FTIR Difference Spectroscopy that D1-Asp61 Influences the Water Reactions of the Oxygen-Evolving Mn4CaO5Cluster of Photosystem II. Biochemistry 53, 2941–2955. doi:10.1021/bi500309f
Debus, R. J. (2015). FTIR Studies of Metal Ligands, Networks of Hydrogen Bonds, and Water Molecules Near the Active Site Mn4CaO5 Cluster in Photosystem II. Biochim. Biophys. Acta (Bba) - Bioenerg. 1847, 19–34. doi:10.1016/j.bbabio.2014.07.007
DeCoursey, T. E., and Hosler, J. (2014). Philosophy of Voltage-Gated Proton Channels. J. R. Soc. Interf. 11, 20130799. doi:10.1098/rsif.2013.0799
Decoursey, T. E. (2003). Voltage-Gated Proton Channels and Other Proton Transfer Pathways. Physiol. Rev. 83, 475–579. doi:10.1152/physrev.00028.2002
Deisenhofer, J., Epp, O., Miki, K., Huber, R., and Michel, H. (1985). Structure of the Protein Subunits in the Photosynthetic Reaction centre of Rhodopseudomonas Viridis at 3Å Resolution. Nature 318, 618–624. doi:10.1038/318618a0
Di Donato, M., van Wilderen, L. J. G. W., Van Stokkum, I. H. M., Stuart, T. C., Kennis, J. T. M., Hellingwerf, K. J., et al. (2011). Proton Transfer Events in GFP. Phys. Chem. Chem. Phys. 13, 16295. doi:10.1039/c1cp20387h
Di Luca, A., Gamiz-Hernandez, A. P., and Kaila, V. R. I. (2017). Symmetry-related Proton Transfer Pathways in Respiratory Complex I. Proc. Natl. Acad. Sci. U.S.A. 114, E6314–E6321. doi:10.1073/pnas.1706278114
Donati, G., Petrone, A., Caruso, P., and Rega, N. (2018). The Mechanism of a green Fluorescent Protein Proton Shuttle Unveiled in the Time-Resolved Frequency Domain by Excited State Ab Initio Dynamics. Chem. Sci. 9, 1126–1135. doi:10.1039/C7SC02803B
Dorman, V. L., and Jordan, P. C. (2004). Ionic Permeation Free Energy in Gramicidin: A Semimicroscopic Perspective. Biophysical J. 86, 3529–3541. doi:10.1529/biophysj.103.039214
Duster, A. W., and Lin, H. (2019). Tracking Proton Transfer through Titratable Amino Acid Side Chains in Adaptive QM/MM Simulations. J. Chem. Theor. Comput. 15, 5794–5809. doi:10.1021/acs.jctc.9b00649
Edmonds, B. W., and Luecke, H. (2004). Atomic Resolution Structures and the Mechanism of Ion Pumping in Bacteriorhodopsin. Front. Biosci. 9, 1556–1566. doi:10.2741/1264
Efremov, R. G., and Sazanov, L. A. (2011). Structure of the Membrane Domain of Respiratory Complex I. Nature 476, 414–420. doi:10.1038/nature10330
Efremov, R. G., and Sazanov, L. A. (2012). The Coupling Mechanism of Respiratory Complex I - A Structural and Evolutionary Perspective. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 1785–1795. doi:10.1016/j.bbabio.2012.02.015
Efremov, R. G., Baradaran, R., and Sazanov, L. A. (2010). The Architecture of Respiratory Complex I. Nature 465, 441–445. doi:10.1038/nature09066
Euro, L., Belevich, G., Verkhovsky, M. I., Wikström, M., and Verkhovskaya, M. (2008). Conserved Lysine Residues of the Membrane Subunit NuoM Are Involved in Energy Conversion by the Proton-Pumping NADH:ubiquinone Oxidoreductase (Complex I). Biochim. Biophys. Acta (Bba) - Bioenerg. 1777, 1166–1172. doi:10.1016/j.bbabio.2008.06.001
Farahvash, A., and Stuchebrukhov, A. (2018). Investigating the Many Roles of Internal Water in Cytochrome C Oxidase. J. Phys. Chem. B 122, 7625–7635. doi:10.1021/acs.jpcb.7b11920
Fearnley, I. M., and Walker, J. E. (1992). Conservation of Sequences of Subunits of Mitochondrial Complex I and Their Relationships with Other Proteins. Biochim. Biophys. Acta (Bba) - Bioenerg. 1140, 105–134. doi:10.1016/0005-2728(92)90001-I
Fitch, C. A., Platzer, G., Okon, M., Garcia-Moreno E., B. B., and McIntosh, L. P. (2015). Arginine: Its pKavalue Revisited. Protein Sci. 24, 752–761. doi:10.1002/pro.2647
Fowler, P. W., Orwick-Rydmark, M., Radestock, S., Solcan, N., Dijkman, P. M., Lyons, J. A., et al. (2015). Gating Topology of the Proton-Coupled Oligopeptide Symporters. Structure 23, 290–301. doi:10.1016/j.str.2014.12.012
Freier, E., Wolf, S., and Gerwert, K. (2011). Proton Transfer via a Transient Linear Water-Molecule Chain in a Membrane Protein. Proc. Natl. Acad. Sci. 108, 11435–11439. doi:10.1073/pnas.1104735108
Futai, M., Nakanishi-Matsui, M., Okamoto, H., Sekiya, M., and Nakamoto, R. K. (2012). Rotational Catalysis in Proton Pumping ATPases: From E. coli F-ATPase to Mammalian V-ATPase. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 1711–1721. doi:10.1016/j.bbabio.2012.03.015
Gamiz-Hernandez, A. P., Jussupow, A., Johansson, M. P., and Kaila, V. R. I. (2017). Terminal Electron-Proton Transfer Dynamics in the Quinone Reduction of Respiratory Complex I. J. Am. Chem. Soc. 139, 16282–16288. doi:10.1021/jacs.7b08486
Ge, X., and Gunner, M. R. (2016). Unraveling the Mechanism of Proton Translocation in the Extracellular Half-Channel of Bacteriorhodopsin. Proteins 84, 639–654. doi:10.1002/prot.25013
Gerwert, K., Freier, E., and Wolf, S. (2014). The Role of Protein-Bound Water Molecules in Microbial Rhodopsins. Biochim. Biophys. Acta (Bba) - Bioenerg. 1837, 606–613. doi:10.1016/j.bbabio.2013.09.006
Goings, J. J., Li, P., Zhu, Q., and Hammes-Schiffer, S. (2020). Formation of an Unusual Glutamine Tautomer in a Blue Light Using Flavin Photocycle Characterizes the Light-Adapted State. Proc. Natl. Acad. Sci. USA 117, 26626–26632. doi:10.1073/pnas.2016719117
Goyal, P., Lu, J., Yang, S., Gunner, M. R., and Cui, Q. (2013). Changing Hydration Level in an Internal Cavity Modulates the Proton Affinity of a Key Glutamate in Cytochrome C Oxidase. Proc. Natl. Acad. Sci. 110, 18886–18891. doi:10.1073/pnas.1313908110
Graige, M. S., Paddock, M. L., Bruce, J. M., Feher, G., and Okamura, M. Y. (1996). Mechanism of Proton-Coupled Electron Transfer for Quinone (QB) Reduction in Reaction Centers of Rb. Sphaeroides. J. Am. Chem. Soc. 118, 9005–9016. doi:10.1021/ja960056m
Gray, H. B., and Winkler, J. R. (2003). Electron Tunneling through Proteins. Quart. Rev. Biophys. 36, 341–372. doi:10.1017/S0033583503003913
Gunner, M. R., and Koder, R. (2017). The Design Features Cells Use to Build Their Transmembrane Proton Gradient. Phys. Biol. 14, 013001. doi:10.1088/1478-3975/14/1/013001
Gunner, M. R., Mao, J., Song, Y., and Kim, J. (2006). Factors Influencing the Energetics of Electron and Proton Transfers in Proteins. What Can Be Learned from Calculations. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 942–968. doi:10.1016/j.bbabio.2006.06.005
Gunner, M. R., Madeo, J., and Zhu, Z. (2008). Modification of Quinone Electrochemistry by the Proteins in the Biological Electron Transfer Chains: Examples from Photosynthetic Reaction Centers. J. Bioenerg. Biomembr. 40, 509–519. doi:10.1007/s10863-008-9179-1
Gunner, M. R., Amin, M., Zhu, X., and Lu, J. (2013). Molecular Mechanisms for Generating Transmembrane Proton Gradients. Biochim. Biophys. Acta (Bba) - Bioenerg. 1827, 892–913. doi:10.1016/j.bbabio.2013.03.001
Gunner, M. R., Murakami, T., Rustenburg, A. S., Işık, M., and Chodera, J. D. (2020). Standard State Free Energies, Not pKas, Are Ideal for Describing Small Molecule Protonation and Tautomeric States. J. Comput. Aided Mol. Des. 34, 561–573. doi:10.1007/s10822-020-00280-7
Gupta, C., Khaniya, U., Chan, C. K., Dehez, F., Shekhar, M., Gunner, M. R., et al. (2020). Charge Transfer and Chemo-Mechanical Coupling in Respiratory Complex I. J. Am. Chem. Soc. 142, 9220–9230. doi:10.1021/jacs.9b13450
Gutiérrez-Fernández, J., Kaszuba, K., Minhas, G. S., Baradaran, R., Tambalo, M., Gallagher, D. T., et al. (2020). Key Role of Quinone in the Mechanism of Respiratory Complex I. Nat. Commun. 11, 4135. doi:10.1038/s41467-020-17957-0
Haapanen, O., and Sharma, V. (2017). Role of Water and Protein Dynamics in Proton Pumping by Respiratory Complex I. Sci. Rep. 7, 7747. doi:10.1038/s41598-017-07930-1
Haapanen, O., and Sharma, V. (2018). A Modeling and Simulation Perspective on the Mechanism and Function of Respiratory Complex I. Biochim. Biophys. Acta (Bba) - Bioenerg. 1859, 510–523. doi:10.1016/j.bbabio.2018.04.001
Hammes-Schiffer, S. (2001). Theoretical Perspectives on Proton-Coupled Electron Transfer Reactions. Acc. Chem. Res. 34, 273–281. doi:10.1021/ar9901117
Hasegawa, R., Saito, K., Takaoka, T., and Ishikita, H. (2017). pK a of Ubiquinone, Menaquinone, Phylloquinone, Plastoquinone, and Rhodoquinone in Aqueous Solution. Photosynth Res. 133, 297–304. doi:10.1007/s11120-017-0382-y
Hasegawa, N., Jonotsuka, H., Miki, K., and Takeda, K. (2018). X-ray Structure Analysis of Bacteriorhodopsin at 1.3 Å Resolution. Sci. Rep. 8, 13123. doi:10.1038/s41598-018-31370-0
Heathcote, P., Fyfe, P. K., and Jones, M. R. (2002). Reaction Centres: the Structure and Evolution of Biological Solar Power. Trends Biochem. Sci. 27, 79–87. doi:10.1016/s0968-0004(01)02034-5
Hirai, T., Subramaniam, S., and Lanyi, J. K. (2009). Structural Snapshots of Conformational Changes in a Seven-helix Membrane Protein: Lessons from Bacteriorhodopsin. Curr. Opin. Struct. Biol. 19, 433–439. doi:10.1016/j.sbi.2009.07.009
Hirst, J. (2013). Mitochondrial Complex I. Annu. Rev. Biochem. 82, 551–575. doi:10.1146/annurev-biochem-070511-103700
Hummer, G., and Wikström, M. (2016). Molecular Simulation and Modeling of Complex I. Biochim. Biophys. Acta (Bba) - Bioenerg. 1857, 915–921. doi:10.1016/j.bbabio.2016.01.005
Ikeda, T., Saito, K., Hasegawa, R., and Ishikita, H. (2017). The Existence of an Isolated Hydronium Ion in the Interior of Proteins. Angew. Chem. 129, 9279–9282. doi:10.1002/ange.201705512
Ishikita, H., and Saito, K. (2014). Proton Transfer Reactions and Hydrogen-Bond Networks in Protein Environments. J. R. Soc. Interf. 11, 20130518. doi:10.1098/rsif.2013.0518
Ishikita, H., Morra, G., and Knapp, E.-W. (2003). Redox Potential of Quinones in Photosynthetic Reaction Centers from Rhodobacter Sphaeroides: Dependence on Protonation of Glu-L212 and Asp-L213. Biochemistry 42, 3882–3892. doi:10.1021/bi026781t
Ishikita, H., Saenger, W., Loll, B., Biesiadka, J., and Knapp, E.-W. (2006). Energetics of a Possible Proton Exit Pathway for Water Oxidation in Photosystem II. Biochemistry 45, 2063–2071. doi:10.1021/bi051615h
Isobe, H., Shoji, M., Shen, J.-R., and Yamaguchi, K. (2015). Strong Coupling between the Hydrogen Bonding Environment and Redox Chemistry during the S2 to S3 Transition in the Oxygen-Evolving Complex of Photosystem II. J. Phys. Chem. B 119, 13922–13933. doi:10.1021/acs.jpcb.5b05740
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a Better Web Interface. Nucleic Acids Res. 36, W5–W9. doi:10.1093/nar/gkn201
Jünemann, S., Meunier, B., Gennis, R. B., and Rich, P. R. (1997). Effects of Mutation of the Conserved Lysine-362 in CytochromecOxidase fromRhodobacter Sphaeroides†. Biochemistry 36, 14456–14464. doi:10.1021/bi971458p
Kaila, V. R. I., and Hummer, G. (2011). Energetics of Direct and Water-Mediated Proton-Coupled Electron Transfer. J. Am. Chem. Soc. 133, 19040–19043. doi:10.1021/ja2082262
Kaila, V. R. I., Verkhovsky, M. I., and Wikström, M. (2010). Proton-coupled Electron Transfer in Cytochrome Oxidase. Chem. Rev. 110, 7062–7081. doi:10.1021/cr1002003
Kaila, V. R. I., Wikstrom, M., and Hummer, G. (2014). Electrostatics, Hydration, and Proton Transfer Dynamics in the Membrane Domain of Respiratory Complex I. Proc. Natl. Acad. Sci. 111, 6988–6993. doi:10.1073/pnas.1319156111
Kaila, V. R. I. (2018). Long-range Proton-Coupled Electron Transfer in Biological Energy Conversion: towards Mechanistic Understanding of Respiratory Complex I. J. R. Soc. Interf. 15, 20170916. doi:10.1098/rsif.2017.0916
Kampjut, D., and Sazanov, L. A. (2020). The Coupling Mechanism of Mammalian Respiratory Complex I. Science 370, eabc4209. doi:10.1126/science.abc4209
Kannt, A., D. Lancaster, C. R., and Michel, H. (1998). The Role of Electrostatic Interactions for Cytochrome C Oxidase Function. J. Bioenerg. Biomembr 30, 81–87. doi:10.1023/A:1020563629032
Kaur, D., Cai, X., Khaniya, U., Zhang, Y., Mao, J., Mandal, M., et al. (2019). Tracing the Pathways of Waters and Protons in Photosystem II and Cytochrome C Oxidase. Inorganics 7, 14. doi:10.3390/inorganics7020014
Kaur, D., Zhang, Y., Reiss, K. M., Mandal, M., Brudvig, G. W., Batista, V. S., et al. (2021). Proton Exit Pathways Surrounding the Oxygen Evolving Complex of Photosystem II. Biochim. Biophys. Acta Bioenerg. 1862, 148446. doi:10.1016/j.bbabio.2021.148446
Kelkar, D. A., and Chattopadhyay, A. (2007). The Gramicidin Ion Channel: A Model Membrane Protein. Biochim. Biophys. Acta (Bba) - Biomembranes 1768, 2011–2025. doi:10.1016/j.bbamem.2007.05.011
Khaniya, U., Gupta, C., Cai, X., Mao, J., Kaur, D., Zhang, Y., et al. (2020). Hydrogen Bond Network Analysis Reveals the Pathway for the Proton Transfer in the E-Channel of T. thermophilus Complex I. Biochim. Biophys. Acta (Bba) - Bioenerg. 1861, 148240. doi:10.1016/j.bbabio.2020.148240
Kim, C. J., and Debus, R. J. (2017). Evidence from FTIR Difference Spectroscopy that a Substrate H2O Molecule for O2 Formation in Photosystem II Is provided by the Ca Ion of the Catalytic Mn4CaO5 Cluster. Biochemistry 56, 2558–2570. doi:10.1021/acs.biochem.6b01278
Kim, Y. C., and Hummer, G. (2012). Proton-pumping Mechanism of Cytochrome C Oxidase: A Kinetic Master-Equation Approach. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 526–536. doi:10.1016/j.bbabio.2011.09.004
Kim, Y. C., Wikström, M., and Hummer, G. (2007). Kinetic Models of Redox-Coupled Proton Pumping. Proc. Natl. Acad. Sci. 104, 2169–2174. doi:10.1073/pnas.0611114104
Kim, Y. C., Wikström, M., and Hummer, G. (2009). Kinetic Gating of the Proton Pump in Cytochrome C Oxidase. Proc. Natl. Acad. Sci. 106, 13707–13712. doi:10.1073/pnas.0903938106
Kleinfeld, D., Okamura, M. Y., and Feher, G. (1984). Electron Transfer in Reaction Centers of Rhodopseudomonas Sphaeroides. I. Determination of the Charge Recombination Pathway of D+QAQ−B and Free Energy and Kinetic Relations between Q−AQB and QAQ−B. Biochim. Biophys. Acta (Bba) - Bioenerg. 766, 126–140. doi:10.1016/0005-2728(84)90224-x
Knight, C., and Voth, G. A. (2012). The Curious Case of the Hydrated Proton. Acc. Chem. Res. 45, 101–109. doi:10.1021/ar200140h
Kovalevsky, A. Y., Hanson, B. L., Mason, S. A., Yoshida, T., Fisher, S. Z., Mustyakimov, M., et al. (2011). Identification of the Elusive Hydronium Ion Exchanging Roles with a Proton in an Enzyme at Lower pH Values. Angew. Chem. Int. Ed. 50, 7520–7523. doi:10.1002/anie.201101753
Krammer, E.-M., Till, M. S., Sebban, P., and Ullmann, G. M. (2009). Proton-Transfer Pathways in Photosynthetic Reaction Centers Analyzed by Profile Hidden Markov Models and Network Calculations. J. Mol. Biol. 388, 631–643. doi:10.1016/j.jmb.2009.03.020
Kuroda, H., Kodama, N., Sun, X.-Y., Ozawa, S.-i., and Takahashi, Y. (2014). Requirement for Asn298 on D1 Protein for Oxygen Evolution: Analyses by Exhaustive Amino Acid Substitution in the green Alga Chlamydomonas Reinhardtii. Plant Cel Physiol 55, 1266–1275. doi:10.1093/pcp/pcu073
Lancaster, C. R., Michel, H., Honig, B., and Gunner, M. R. (1996). Calculated Coupling of Electron and Proton Transfer in the Photosynthetic Reaction center of Rhodopseudomonas Viridis. Biophysical J. 70, 2469–2492. doi:10.1016/S0006-3495(96)79820-X
Lanyi, J. K. (2006). Proton Transfers in the Bacteriorhodopsin Photocycle. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 1012–1018. doi:10.1016/j.bbabio.2005.11.003
Lavergne, J. r. m., and Junge, W. (1993). Proton Release during the Redox Cycle of the Water Oxidase. Photosynth Res. 38, 279–296. doi:10.1007/BF00046752
Lazaridis, T., and Hummer, G. (2017). Classical Molecular Dynamics with Mobile Protons. J. Chem. Inf. Model. 57, 2833–2845. doi:10.1021/acs.jcim.7b00603
Lee, H. J., and Ädelroth, P. (2013). The Heme-Copper Oxidase Superfamily Shares a Zn2+-Binding Motif at the Entrance to a Proton Pathway. FEBS Lett. 587, 770–774. doi:10.1016/j.febslet.2013.01.069
Lee, H. J., Reimann, J., Huang, Y., and Ädelroth, P. (2012). Functional Proton Transfer Pathways in the Heme-Copper Oxidase Superfamily. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 537–544. doi:10.1016/j.bbabio.2011.10.007
Li, H., Anuwongcharoen, N., Malik, A., Prachayasittikul, V., Wikberg, J., and Nantasenamat, C. (2016). Roles of D-Amino Acids on the Bioactivity of Host Defense Peptides. IJMS 17, 1023. doi:10.3390/ijms17071023
Liang, R., Swanson, J. M. J., Wikström, M., and Voth, G. A. (2017). Understanding the Essential Proton-Pumping Kinetic gates and Decoupling Mutations in Cytochrome C Oxidase. Proc. Natl. Acad. Sci. USA 114, 5924–5929. doi:10.1073/pnas.1703654114
Lórenz-Fonfría, V. A., and Heberle, J. (2014). Channelrhodopsin Unchained: Structure and Mechanism of a Light-Gated Cation Channel. Biochim. Biophys. Acta (Bba) - Bioenerg. 1837, 626–642. doi:10.1016/j.bbabio.2013.10.014
Lórenz-Fonfría, V. A., and Kandori, H. (2009). Spectroscopic and Kinetic Evidence on How Bacteriorhodopsin Accomplishes Vectorial Proton Transport under Functional Conditions. J. Am. Chem. Soc. 131, 5891–5901. doi:10.1021/ja900334c
Lórenz-Fonfría, V. A., Furutani, Y., and Kandori, H. (2008). Active Internal Waters in the Bacteriorhodopsin Photocycle. A Comparative Study of the L and M Intermediates at Room and Cryogenic Temperatures by Infrared Spectroscopy†. Biochemistry 47, 4071–4081. doi:10.1021/bi7024063
Lórenz-Fonfría, V. A., Kandori, H., and Padrós, E. (2011). Probing Specific Molecular Processes and Intermediates by Time-Resolved Fourier Transform Infrared Spectroscopy: Application to the Bacteriorhodopsin Photocycle. J. Phys. Chem. B 115, 7972–7985. doi:10.1021/jp201739w
Luecke, H. (2000). Atomic Resolution Structures of Bacteriorhodopsin Photocycle Intermediates: the Role of Discrete Water Molecules in the Function of This Light-Driven Ion Pump. Biochim. Biophys. Acta (Bba) - Bioenerg. 1460, 133–156. doi:10.1016/S0005-2728(00)00135-3
Ma, J., Tsatsos, P. H., Zaslavsky, D., Barquera, B., Thomas, J. W., Katsonouri, A., et al. (1999). Glutamate-89 in Subunit II of Cytochromebo3fromEscherichia coliIs Required for the Function of the Heme−Copper Oxidase†. Biochemistry 38, 15150–15156. doi:10.1021/bi991764y
Mathiesen, C., and Hägerhäll, C. (2002). Transmembrane Topology of the NuoL, M and N Subunits of NADH:quinone Oxidoreductase and Their Homologues Among Membrane-Bound Hydrogenases and Bona Fide Antiporters. Biochim. Biophys. Acta (Bba) - Bioenerg. 1556, 121–132. doi:10.1016/S0005-2728(02)00343-2
McEvoy, J. P., and Brudvig, G. W. (2004). Structure-based Mechanism of Photosynthetic Water Oxidation. Phys. Chem. Chem. Phys. 6, 4754–4763. doi:10.1039/B407500E
McEvoy, J. P., and Brudvig, G. W. (2006). Water-splitting Chemistry of Photosystem II. Chem. Rev. 106, 4455–4483. doi:10.1021/cr0204294
McPherson, P. H., Schoenfeld, M., Paddock, M. L., Okamura, M. Y., and Feher, G. (1994). Protonation and Free Energy Changes Associated with Formation of QBH2 in Native and Glu-L212 .Fwdarw. Gln Mutant Reaction Centers from Rhodobacter Sphaeroides. Biochemistry 33, 1181–1193. doi:10.1021/bi00171a018
Michel, J., DeLeon-Rangel, J., Zhu, S., Van Ree, K., and Vik, S. B. (2011). Mutagenesis of the L, M, and N Subunits of Complex I from Escherichia coli Indicates a Common Role in Function. PLoS One 6, e17420. doi:10.1371/journal.pone.0017420
Miller, A., and Oesterhelt, D. (1990). Kinetic Optimization of Bacteriorhodopsin by Aspartic Acid 96 as an Internal Proton Donor. Biochim. Biophys. Acta (Bba) - Bioenerg. 1020, 57–64. doi:10.1016/0005-2728(90)90093-J
Mills, D. A., and Ferguson-Miller, S. (2002). Influence of Structure, pH and Membrane Potential on Proton Movement in Cytochrome Oxidase. Biochim. Biophys. Acta (Bba) - Bioenerg. 1555, 96–100. doi:10.1016/S0005-2728(02)00261-X
Mitchell, P. (1961). Coupling of Phosphorylation to Electron and Hydrogen Transfer by a Chemi-Osmotic Type of Mechanism. Nature 191, 144–148. doi:10.1038/191144a0
Mitchell, P. (1977). Vectorial Chemiosmotic Processes. Annu. Rev. Biochem. 46, 996–1005. doi:10.1146/annurev.bi.46.070177.005024
Miyake, T., and Rolandi, M. (2015). Grotthuss Mechanisms: from Proton Transport in Proton Wires to Bioprotonic Devices. J. Phys. Condens. Matter 28, 023001. doi:10.1088/0953-8984/28/2/023001
Moser, C. C., Farid, T. A., Chobot, S. E., and Dutton, P. L. (2006). Electron Tunneling Chains of Mitochondria. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 1096–1109. doi:10.1016/j.bbabio.2006.04.015
Müh, F., Glöckner, C., Hellmich, J., and Zouni, A. (2012). Light-induced Quinone Reduction in Photosystem II. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 44–65. doi:10.1016/j.bbabio.2011.05.021
Nagao, R., Ueoka-Nakanishi, H., and Noguchi, T. (2017). D1-Asn-298 in Photosystem II Is Involved in a Hydrogen-Bond Network Near the Redox-Active Tyrosine YZ for Proton Exit during Water Oxidation. J. Biol. Chem. 292, 20046–20057. doi:10.1074/jbc.M117.815183
Nagle, J. F., and Morowitz, H. J. (1978). Molecular Mechanisms for Proton Transport in Membranes. Proc. Natl. Acad. Sci. 75, 298–302. doi:10.1073/pnas.75.1.298
Nakamura, S., Nagao, R., Takahashi, R., and Noguchi, T. (2014). Fourier Transform Infrared Detection of a Polarizable Proton Trapped between Photooxidized Tyrosine YZand a Coupled Histidine in Photosystem II: Relevance to the Proton Transfer Mechanism of Water Oxidation. Biochemistry 53, 3131–3144. doi:10.1021/bi500237y
Namslauer, A., Aagaard, A., Katsonouri, A., and Brzezinski, P. (2003). Intramolecular Proton-Transfer Reactions in a Membrane-Bound Proton Pump: The Effect of pH on the Peroxy to Ferryl Transition in CytochromecOxidase†,⊥. Biochemistry 42, 1488–1498. doi:10.1021/bi026524o
Nicholls, D. G. (2010). Mitochondrial Ion Circuits. Essays Biochem. 47, 25–35. doi:10.1042/bse0470025
Nogly, P., Weinert, T., James, D., Carbajo, S., Ozerov, D., Furrer, A., et al. (2018). Retinal Isomerization in Bacteriorhodopsin Captured by a Femtosecond X-ray Laser. Science 361, eaat0094. doi:10.1126/science.aat0094
Odella, E., Mora, S. J., Wadsworth, B. L., Huynh, M. T., Goings, J. J., Liddell, P. A., et al. (2018). Controlling Proton-Coupled Electron Transfer in Bioinspired Artificial Photosynthetic Relays. J. Am. Chem. Soc. 140, 15450–15460. doi:10.1021/jacs.8b09724
Odella, E., Wadsworth, B. L., Mora, S. J., Goings, J. J., Huynh, M. T., Gust, D., et al. (2019). Proton-Coupled Electron Transfer Drives Long-Range Proton Translocation in Bioinspired Systems. J. Am. Chem. Soc. 141, 14057–14061. doi:10.1021/jacs.9b06978
Okamura, M. Y., and Feher, G. (1992). Proton Transfer in Reaction Centers from Photosynthetic Bacteria. Annu. Rev. Biochem. 61, 861–896. doi:10.1146/annurev.bi.61.070192.004241
Okamura, M. Y., Paddock, M. L., Graige, M. S., and Feher, G. (2000). Proton and Electron Transfer in Bacterial Reaction Centers. Biochim. Biophys. Acta (Bba) - Bioenerg. 1458, 148–163. doi:10.1016/S0005-2728(00)00065-7
Ono, T., Zimmermann, J. L., Inoue, Y., and Rutherford, A. W. (1986). EPR Evidence for a Modified S-State Transition in Chloride-Depleted Photosystem II. Biochim. Biophys. Acta (Bba) - Bioenerg. 851, 193–201. doi:10.1016/0005-2728(86)90125-8
Onufriev, A., Smondyrev, A., and Bashford, D. (2003). Proton Affinity Changes Driving Unidirectional Proton Transport in the Bacteriorhodopsin Photocycle. J. Mol. Biol. 332, 1183–1193. doi:10.1016/s0022-2836(03)00903-3
Ormö, M., Cubitt, A. B., Kallio, K., Gross, L. A., Tsien, R. Y., and Remington, S. J. (1996). Crystal Structure of the Aequorea victoria green Fluorescent Protein. Science 273, 1392–1395. doi:10.1126/science.273.5280.1392
Paddock, M. L., Rongey, S. H., Feher, G., and Okamura, M. Y. (1989). Pathway of Proton Transfer in Bacterial Reaction Centers: Replacement of Glutamic Acid 212 in the L Subunit by Glutamine Inhibits Quinone (Secondary Acceptor) Turnover. Pnas 86, 6602–6606. doi:10.1073/pnas.86.17.6602
Paddock, M. L., Rongey, S. H., McPherson, P. H., Juth, A., Feher, G., and Okamura, M. Y. (1994). Pathway of Proton Transfer in Bacterial Reaction Centers: Role of Aspartate-L213 in Proton Transfers Associated with Reduction of Quinone to Dihydroquinone. Biochemistry 33, 734–745. doi:10.1021/bi00169a015
Paddock, M. L., Feher, G., and Okamura, M. Y. (1995). Pathway of Proton Transfer in Bacterial Reaction Centers: Further Investigations on the Role of Ser-L223 Studied by Site-Directed Mutagenesis. Biochemistry 34, 15742–15750. doi:10.1021/bi00048a019
Paddock, M. L., Graige, M. S., Feher, G., and Okamura, M. Y. (1999). Identification of the Proton Pathway in Bacterial Reaction Centers: Inhibition of Proton Transfer by Binding of Zn2+ or Cd2+. Proc. Natl. Acad. Sci. 96, 6183–6188. doi:10.1073/pnas.96.11.6183
Paddock, M. L., Feher, G., and Okamura, M. Y. (2003). Proton Transfer Pathways and Mechanism in Bacterial Reaction Centers. FEBS Lett. 555, 45–50. doi:10.1016/S0014-5793(03)01149-9
Pantazis, D. A. (2018). Missing Pieces in the Puzzle of Biological Water Oxidation. ACS Catal. 8, 9477–9507. doi:10.1021/acscatal.8b01928
Paulino, J., Yi, M., Hung, I., Gan, Z., Wang, X., Chekmenev, E. Y., et al. (2020). Functional Stability of Water Wire-Carbonyl Interactions in an Ion Channel. Proc. Natl. Acad. Sci. U.S.A. 117, 11908–11915. doi:10.1073/pnas.2001083117
Phatak, P., Ghosh, N., Yu, H., Cui, Q., and Elstner, M. (2008). Amino Acids with an Intermolecular Proton Bond as Proton Storage Site in Bacteriorhodopsin. Proc. Natl. Acad. Sci. 105, 19672–19677. doi:10.1073/pnas.0810712105
Pokhrel, R., and Brudvig, G. W. (2014). Oxygen-evolving Complex of Photosystem II: Correlating Structure with Spectroscopy. Phys. Chem. Chem. Phys. 16, 11812–11821. doi:10.1039/C4CP00493K
Pomès, R., and Roux, B. (1998). Free Energy Profiles for H+ Conduction along Hydrogen-Bonded Chains of Water Molecules. Biophysical J. 75, 33–40. doi:10.1016/S0006-3495(98)77492-2
Pomès, R., and Roux, B. (2002). Molecular Mechanism of H+ Conduction in the Single-File Water Chain of the Gramicidin Channel. Biophysical J. 82, 2304–2316. doi:10.1016/s0006-3495(02)75576-8
Popović, D. M., and Stuchebrukhov, A. A. (2005). Proton Exit Channels in Bovine Cytochrome C Oxidase. J. Phys. Chem. B 109, 1999–2006. doi:10.1021/jp0464371
Qian, J., Shi, W., Pressler, M., Hoganson, C., Mills, D., Babcock, G. T., et al. (1997). Aspartate-407 inRhodobacter sphaeroidesCytochromecOxidase Is Not Required for Proton Pumping or Manganese Binding†. Biochemistry 36, 2539–2543. doi:10.1021/bi962721+
Rabenstein, B., Ullmann, G. M., and Knapp, E.-W. (2000). Electron Transfer between the Quinones in the Photosynthetic Reaction Center and its Coupling to Conformational Changes†. Biochemistry 39, 10487–10496. doi:10.1021/bi000413c
Rappaport, F., Blanchard-Desce, M., and Lavergne, J. (1994). Kinetics of Electron Transfer and Electrochromic Change during the Redox Transitions of the Photosynthetic Oxygen-Evolving Complex. Biochim. Biophys. Acta (Bba) - Bioenerg. 1184, 178–192. doi:10.1016/0005-2728(94)90222-4
Raymond, J., and Blankenship, R. E. (2004). The Evolutionary Development of the Protein Complement of Photosystem 2. Biochim. Biophys. Acta (Bba) - Bioenerg. 1655, 133–139. doi:10.1016/j.bbabio.2003.10.015
Retegan, M., Krewald, V., Mamedov, F., Neese, F., Lubitz, W., Cox, N., et al. (2016). A Five-Coordinate Mn(IV) Intermediate in Biological Water Oxidation: Spectroscopic Signature and a Pivot Mechanism for Water Binding. Chem. Sci. 7, 72–84. doi:10.1039/c5sc03124a
Rich, P. R. (2008). A Perspective on Peter Mitchell and the Chemiosmotic Theory. J. Bioenerg. Biomembr 40, 407–410. doi:10.1007/s10863-008-9173-7
Ripple, M. O., Kim, N., and Springett, R. (2013). Mammalian Complex I Pumps 4 Protons Per 2 Electrons at High and Physiological Proton Motive Force in Living Cells*. J. Biol. Chem. 288, 5374–5380. doi:10.1074/jbc.M112.438945
Rivalta, I., Amin, M., Luber, S., Vassiliev, S., Pokhrel, R., Umena, Y., et al. (2011). Structural-Functional Role of Chloride in Photosystem II. Biochemistry 50, 6312–6315. doi:10.1021/bi200685w
Röpke, M., Saura, P., Riepl, D., Pöverlein, M. C., and Kaila, V. R. I. (2020). Functional Water Wires Catalyze Long-Range Proton Pumping in the Mammalian Respiratory Complex I. J. Am. Chem. Soc. 142, 21758–21766. doi:10.1021/jacs.0c09209
Roux, B. (2002). Computational Studies of the Gramicidin Channel. Acc. Chem. Res. 35, 366–375. doi:10.1021/ar010028v
Ryu, H., Lee, H., Iwata, S., Choi, S., Ki Kim, M., Kim, Y.-R., et al. (2015). Investigation of Ion Channel Activities of Gramicidin A in the Presence of Ionic Liquids Using Model Cell Membranes. Sci. Rep. 5, 11935. doi:10.1038/srep11935
Saito, K., Shen, J.-R., Ishida, T., and Ishikita, H. (2011). Short Hydrogen Bond between Redox-Active Tyrosine YZand D1-His190 in the Photosystem II Crystal Structure. Biochemistry 50, 9836–9844. doi:10.1021/bi201366j
Saito, K., Rutherford, A. W., and Ishikita, H. (2013). Mechanism of Proton-Coupled Quinone Reduction in Photosystem II. Proc. Natl. Acad. Sci. 110, 954–959. doi:10.1073/pnas.1212957110
Saito, K., William Rutherford, A., and Ishikita, H. (2015). Energetics of Proton Release on the First Oxidation Step in the Water-Oxidizing Enzyme. Nat. Commun. 6, 8488. doi:10.1038/ncomms9488
Sakamoto, H., Shimizu, T., Nagao, R., and Noguchi, T. (2017). Monitoring the Reaction Process during the S2 → S3 Transition in Photosynthetic Water Oxidation Using Time-Resolved Infrared Spectroscopy. J. Am. Chem. Soc. 139, 2022–2029. doi:10.1021/jacs.6b11989
Sakashita, N., Ishikita, H., and Saito, K. (2020). Rigidly Hydrogen-Bonded Water Molecules Facilitate Proton Transfer in Photosystem II. Phys. Chem. Chem. Phys. 22, 15831–15841. doi:10.1039/D0CP00295J
Saura, P., and Kaila, V. R. I. (2019). Molecular Dynamics and Structural Models of the Cyanobacterial NDH-1 Complex. Biochim. Biophys. Acta (Bba) - Bioenerg. 1860, 201–208. doi:10.1016/j.bbabio.2018.11.010
Sazanov, L. A. (2014). The Mechanism of Coupling between Electron Transfer and Proton Translocation in Respiratory Complex I. J. Bioenerg. Biomembr. 46, 247–253. doi:10.1007/s10863-014-9554-z
Sazanov, L. A. (2015). A Giant Molecular Proton Pump: Structure and Mechanism of Respiratory Complex I. Nat. Rev. Mol. Cel Biol. 16, 375–388. doi:10.1038/nrm3997
Sebban, P., Maróti, P., Schiffer, M., and Hanson, D. K. (1995). Electrostatic Dominoes: Long Distance Propagation of Mutational Effects in Photosynthetic Reaction Centers of Rhodobacter Capsulatus. Biochemistry 34, 8390–8397. doi:10.1021/bi00026a021
Service, R. J., Hillier, W., and Debus, R. J. (2010). Evidence from FTIR Difference Spectroscopy of an Extensive Network of Hydrogen Bonds Near the Oxygen-Evolving Mn4Ca Cluster of Photosystem II Involving D1-Glu65, D2-Glu312, and D1-Glu329. Biochemistry 49, 6655–6669. doi:10.1021/bi100730d
Sharpe, M. A., and Ferguson-Miller, S. (2008). A Chemically Explicit Model for the Mechanism of Proton Pumping in Heme-Copper Oxidases. J. Bioenerg. Biomembr 40, 541–549. doi:10.1007/s10863-008-9182-6
Shevela, D., Eaton-Rye, J. J., Shen, J.-R., and Govindjee, G. (2012). Photosystem II and the Unique Role of Bicarbonate: A Historical Perspective. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 1134–1151. doi:10.1016/j.bbabio.2012.04.003
Shinkarev, V. P., Takahashi, E., and Wraight, C. A. (1993). Flash-induced Electric Potential Generation in Wild Type and L212EQ Mutant Chromatophores of Rhodobacter Sphaeroides: QBH2 Is Not Released from L212EQ Mutant Reaction Centers. Biochim. Biophys. Acta (Bba) - Bioenerg. 1142, 214–216. doi:10.1016/0005-2728(93)90106-P
Shoji, M., Isobe, H., and Yamaguchi, K. (2015). QM/MM Study of the S2 to S3 Transition Reaction in the Oxygen-Evolving Complex of Photosystem II. Chem. Phys. Lett. 636, 172–179. doi:10.1016/j.cplett.2015.07.039
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, Scalable Generation of High‐quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Son, C. Y., Yethiraj, A., and Cui, Q. (2017). Cavity Hydration Dynamics in Cytochrome C Oxidase and Functional Implications. Proc. Natl. Acad. Sci. U.S.A. 114, E8830–E8836. doi:10.1073/pnas.1707922114
Song, Y., and Gunner, M. R. (2014). Halorhodopsin Pumps Cl-And Bacteriorhodopsin Pumps Protons by a Common Mechanism that Uses Conserved Electrostatic Interactions. Proc. Natl. Acad. Sci. U.S.A. 111, 16377–16382. doi:10.1073/pnas.1411119111
Song, Y., Mao, J., and Gunner, M. R. (2003). Calculation of Proton Transfers in Bacteriorhodopsin bR and M Intermediates†. Biochemistry 42, 9875–9888. doi:10.1021/bi034482d
Spassov, V. Z., Luecke, H., Gerwert, K., and Bashford, D. (2001). p K a calculations suggest storage of an excess proton in a hydrogen-bonded water network in bacteriorhodopsin 1 1Edited by G. von Heijne. J. Mol. Biol. 312, 203–219. doi:10.1006/jmbi.2001.4902
Steiner, J., and Sazanov, L. (2020). Structure and Mechanism of the Mrp Complex, an Ancient Cation/proton Antiporter. eLife 9, e59407. doi:10.7554/eLife.59407
Stoner-Ma, D., Jaye, A. A., Matousek, P., Towrie, M., Meech, S. R., and Tonge, P. J. (2005). Observation of Excited-State Proton Transfer in Green Fluorescent Protein Using Ultrafast Vibrational Spectroscopy. J. Am. Chem. Soc. 127, 2864–2865. doi:10.1021/ja042466d
Stuchebrukhov, A. A. (2018). Redox-Driven Proton Pumps of the Respiratory Chain. Biophysical J. 115, 830–840. doi:10.1016/j.bpj.2018.07.022
Stuchebrukhov, A. A. (2019). Kinetics and Efficiency of Energy-Transducing Enzymes. J. Phys. Chem. B 123, 9456–9465. doi:10.1021/acs.jpcb.9b08191
Suga, M., Akita, F., Yamashita, K., Nakajima, Y., Ueno, G., Li, H., et al. (2019). An Oxyl/oxo Mechanism for Oxygen-Oxygen Coupling in PSII Revealed by an X-ray Free-Electron Laser. Science 366, 334–338. doi:10.1126/science.aax6998
Supekar, S., Gamiz-Hernandez, A. P., and Kaila, V. R. I. (2016). A Protonated Water Cluster as a Transient Proton-Loading Site in Cytochrome cOxidase. Angew. Chem. Int. Ed. 55, 11940–11944. doi:10.1002/anie.201603606
Swanson, J. M. J., Maupin, C. M., Chen, H., Petersen, M. K., Xu, J., Wu, Y., et al. (2007). Proton Solvation and Transport in Aqueous and Biomolecular Systems: Insights from Computer Simulations. J. Phys. Chem. B 111, 4300–4314. doi:10.1021/jp070104x
Taylor, R. W., Morris, A. A., Hutchinson, M., and Turnbull, D. M. (2002). Leigh Disease Associated with a Novel Mitochondrial DNA ND5 Mutation. Eur. J. Hum. Genet. 10, 141–144. doi:10.1038/sj.ejhg.5200773
Till, M. S., Essigke, T., Becker, T., and Ullmann, G. M. (2008). Simulating the Proton Transfer in Gramicidin A by a Sequential Dynamical Monte Carlo Method. J. Phys. Chem. B 112, 13401–13410. doi:10.1021/jp801477b
Tommos, C., and Babcock, G. T. (2000). Proton and Hydrogen Currents in Photosynthetic Water Oxidation. Biochim. Biophys. Acta (Bba) - Bioenerg. 1458, 199–219. doi:10.1016/S0005-2728(00)00069-4
Torres-Bacete, J., Nakamaru-Ogiso, E., Matsuno-Yagi, A., and Yagi, T. (2007). Characterization of the NuoM (ND4) Subunit in Escherichia coli NDH-1. J. Biol. Chem. 282, 36914–36922. doi:10.1074/jbc.M707855200
Tu, C. K., Silverman, D, N., Forsman, C., Jonsson, B. H., and Lindskog, S. (1989). Role of Histidine 64 in the Catalytic Mechanism of Human Carbonic Anhydrase II Studied with a Site-specific Mutant. Biochemistry 28, 7913–7918. doi:10.1021/bi00445a054
Ugur, I., Rutherford, A. W., and Kaila, V. R. I. (2016). Redox-coupled Substrate Water Reorganization in the Active Site of Photosystem II-The Role of Calcium in Substrate Water Delivery. Biochim. Biophys. Acta (Bba) - Bioenerg. 1857, 740–748. doi:10.1016/j.bbabio.2016.01.015
Umena, Y., Kawakami, K., Shen, J.-R., and Kamiya, N. (2011). Crystal Structure of Oxygen-Evolving Photosystem II at a Resolution of 1.9 Å. Nature 473, 55–60. doi:10.1038/nature09913
Utschig, L. M., Ohigashi, Y., Thurnauer, M. C., and Tiede, D. M. (1998). A New Metal-Binding Site in Photosynthetic Bacterial Reaction Centers that Modulates QAto QBElectron Transfer†. Biochemistry 37, 8278–8281. doi:10.1021/bi980395n
Vasanthakumar, T., and Rubinstein, J. L. (2020). Structure and Roles of V-type ATPases. Trends Biochem. Sci. 45, 295–307. doi:10.1016/j.tibs.2019.12.007
Vassiliev, S., Zaraiskaya, T., and Bruce, D. (2012). Exploring the Energetics of Water Permeation in Photosystem II by Multiple Steered Molecular Dynamics Simulations. Biochim. Biophys. Acta (Bba) - Bioenerg. 1817, 1671–1678. doi:10.1016/j.bbabio.2012.05.016
Verkhovskaya, M. L., Belevich, N., Euro, L., Wikström, M., and Verkhovsky, M. I. (2008). Real-time Electron Transfer in Respiratory Complex I. Proc. Natl. Acad. Sci. 105, 3763–3767. doi:10.1073/pnas.0711249105
Vinyard, D. J., and Brudvig, G. W. (2017). Progress toward a Molecular Mechanism of Water Oxidation in Photosystem II. Annu. Rev. Phys. Chem. 68, 101–116. doi:10.1146/annurev-physchem-052516-044820
Vogt, L., Ertem, M. Z., Pal, R., Brudvig, G. W., and Batista, V. S. (2015). Computational Insights on Crystal Structures of the Oxygen-Evolving Complex of Photosystem II with Either Ca2+ or Ca2+ Substituted by Sr2+. Biochemistry 54, 820–825. doi:10.1021/bi5011706
Walker, J. E., Lutter, R., Dupuis, A., and Runswick, M. J. (1991). Identification of the Subunits of F1F0-ATPase from Bovine Heart Mitochondria. Biochemistry 30, 5369–5378. doi:10.1021/bi00236a007
Wang, C., Lamb, R. A., and Pinto, L. H. (1995). Activation of the M2 Ion Channel of Influenza Virus: a Role for the Transmembrane Domain Histidine Residue. Biophysical J. 69, 1363–1371. doi:10.1016/S0006-3495(95)80003-2
Warnau, J., Sharma, V., Gamiz-Hernandez, A. P., Di Luca, A., Haapanen, O., Vattulainen, I., et al. (2018). Redox-coupled Quinone Dynamics in the Respiratory Complex I. Proc. Natl. Acad. Sci. U.S.A. 115, E8413–E8420. doi:10.1073/pnas.1805468115
Weisz, D. A., Gross, M. L., and Pakrasi, H. B. (2017). Reactive Oxygen Species Leave a Damage Trail that Reveals Water Channels in Photosystem II. Sci. Adv. 3, eaao3013. doi:10.1126/sciadv.aao3013
Wickstrand, C., Dods, R., Royant, A., and Neutze, R. (2015). Bacteriorhodopsin: Would the Real Structural Intermediates Please Stand up? Biochim. Biophys. Acta (Bba) - Gen. Subjects 1850, 536–553. doi:10.1016/j.bbagen.2014.05.021
Wickstrand, C., Nogly, P., Nango, E., Iwata, S., Standfuss, J., and Neutze, R. (2019). Bacteriorhodopsin: Structural Insights Revealed Using X-Ray Lasers and Synchrotron Radiation. Annu. Rev. Biochem. 88, 59–83. doi:10.1146/annurev-biochem-013118-111327
Wikström, M., Jasaitis, A., Backgren, C., Puustinen, A., and Verkhovsky, M. I. (2000). The Role of the D- and K-Pathways of Proton Transfer in the Function of the Haem-Copper Oxidases. Biochim. Biophys. Acta (Bba) - Bioenerg. 1459, 514–520. doi:10.1016/s0005-2728(00)00191-2
Wikström, M., Verkhovsky, M. I., and Hummer, G. (2003). Water-gated Mechanism of Proton Translocation by Cytochrome C Oxidase. Biochim. Biophys. Acta (Bba) - Bioenerg. 1604, 61–65. doi:10.1016/s0005-2728(03)00041-0
Wikström, M., Sharma, V., Kaila, V. R. I., Hosler, J. P., and Hummer, G. (2015). New Perspectives on Proton Pumping in Cellular Respiration. Chem. Rev. 115, 2196–2221. doi:10.1021/cr500448t
Wolf, S., Freier, E., and Gerwert, K. (2014). A Delocalized Proton-Binding Site within a Membrane Protein. Biophysical J. 107, 174–184. doi:10.1016/j.bpj.2014.05.019
Wolter, T., Welke, K., Phatak, P., Bondar, A.-N., and Elstner, M. (2013). Excitation Energies of a Water-Bridged Twisted Retinal Structure in the Bacteriorhodopsin Proton Pump: a Theoretical Investigation. Phys. Chem. Chem. Phys. 15, 12582. doi:10.1039/c3cp44280b
Wraight, C. A. (2004). Proton and Electron Transfer in the Acceptor Quinone Complex of Photosynthetic Reaction Centers from Rhodobacter Sphaeroides. Front. Biosci. 9, 309–337. doi:10.2741/1236
Wraight, C. A. (2006). Chance and Design-Proton Transfer in Water, Channels and Bioenergetic Proteins. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 886–912. doi:10.1016/j.bbabio.2006.06.017
Xu, J., and Voth, G. A. (2006). Free Energy Profiles for H+ Conduction in the D-Pathway of Cytochrome C Oxidase: A Study of the Wild Type and N98D Mutant Enzymes. Biochim. Biophys. Acta (Bba) - Bioenerg. 1757, 852–859. doi:10.1016/j.bbabio.2006.05.028
Yanagisawa, S., and Frasch, W. D. (2017). Protonation-dependent Stepped Rotation of the F-type ATP Synthase C-Ring Observed by Single-Molecule Measurements. J. Biol. Chem. 292, 17093–17100. doi:10.1074/jbc.M117.799940
Yang, J., Zhu, Y., Tong, Y., Chen, L., Liu, L., Zhang, Z., et al. (2009). Confirmation of the Mitochondrial ND1 Gene Mutation G3635A as a Primary LHON Mutation. Biochem. Biophysical Res. Commun. 386, 50–54. doi:10.1016/j.bbrc.2009.05.127
Zhang, Y., Haider, K., Kaur, D., Ngo, V. A., Cai, X., Mao, J., et al. (2020). Characterizing the Water Wire in the Gramicidin Channel Found by Monte Carlo Sampling Using Continuum Electrostatics and in Molecular Dynamics Trajectories with Conventional or Polarizable Force fields. J. Theor. Comput. Chem., 2042001. doi:10.1142/S0219633620420018
Zhou, P., and Han, K. (2018). Unraveling the Detailed Mechanism of Excited-State Proton Transfer. Acc. Chem. Res. 51, 1681–1690. doi:10.1021/acs.accounts.8b00172
Zhu, Z., and Gunner, M. R. (2005). Energetics of Quinone-dependent Electron and Proton Transfers inRhodobacter sphaeroidesPhotosynthetic Reaction Centers. Biochemistry 44, 82–96. doi:10.1021/bi048348k
Zickermann, V., Wirth, C., Nasiri, H., Siegmund, K., Schwalbe, H., Hunte, C., et al. (2015). Mechanistic Insight from the crystal Structure of Mitochondrial Complex I. Science 347, 44–49. doi:10.1126/science.1259859
Keywords: proton transfer pathways, bacteriorhodopsin, photosystem II, bacterial reaction center, complex I, cytochrome c oxidase, Grotthuss mechanism
Citation: Kaur D, Khaniya U, Zhang Y and Gunner MR (2021) Protein Motifs for Proton Transfers That Build the Transmembrane Proton Gradient. Front. Chem. 9:660954. doi: 10.3389/fchem.2021.660954
Received: 30 January 2021; Accepted: 31 May 2021;
Published: 15 June 2021.
Edited by:
Vivek Sharma, University of Helsinki, FinlandReviewed by:
Ville Kaila, Stockholm University, SwedenThomas E. DeCoursey, Rush University, United States
Copyright © 2021 Kaur, Khaniya, Zhang and Gunner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. R. Gunner, bWd1bm5lckBjY255LmN1bnkuZWR1
†These authors have contributed equally to this work and share first authorship
 Divya Kaur
Divya Kaur Umesh Khaniya
Umesh Khaniya Yingying Zhang
Yingying Zhang M. R. Gunner
M. R. Gunner