- 1Department of Chemistry, Faculty of Applied Sciences, Umm Al-Qura University, Makkah, Saudi Arabia
- 2Department of Chemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
- 3Department of Chemistry, College of Science, United Arab Emirates University, Al Ain, Abu Dhabi, United Arab Emirates
- 4Department of Epidemic Disease Research, Institute for Research & Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, 31441, Saudi Arabia
- 5Department of Health Informatics, College of Public Health and Health Informatics, Qassim University, Al Bukayriyah, Saudi Arabia
- 6Research on Advanced BioMedical Solutions Pvt Ltd, KPHB, Hyderabad, 500071, India
- 7Department of Chemistry, Faculty of Science, Assiut University, 71516, Assiut, Egypt
- 8Research Laboratories Unit, Faculty of Applied Science, Umm Al-Qura University, 21955, Makkah, Saudi Arabia
A molecular modeling assisted rational design and synthesis of naphthalene diimide linked bis-naphthalimides as potential DNA interactive agents is described. Chemical templates incorporating naphthalene diimide as a linker in bis-naphthalimide motif were subjected to molecular docking analysis at specific intercalation and telomeric DNA G-quadruplex sites. Excellent results were obtained, which were better than the standards. A short and convenient synthetic route was employed to access these hybrids experimentally, followed by evaluation of their ability to cause thermal denaturation of DNA and cytotoxic properties along with ADME predictions. The obtained results provided useful insights and two potential molecules were identified for further development.
Introduction
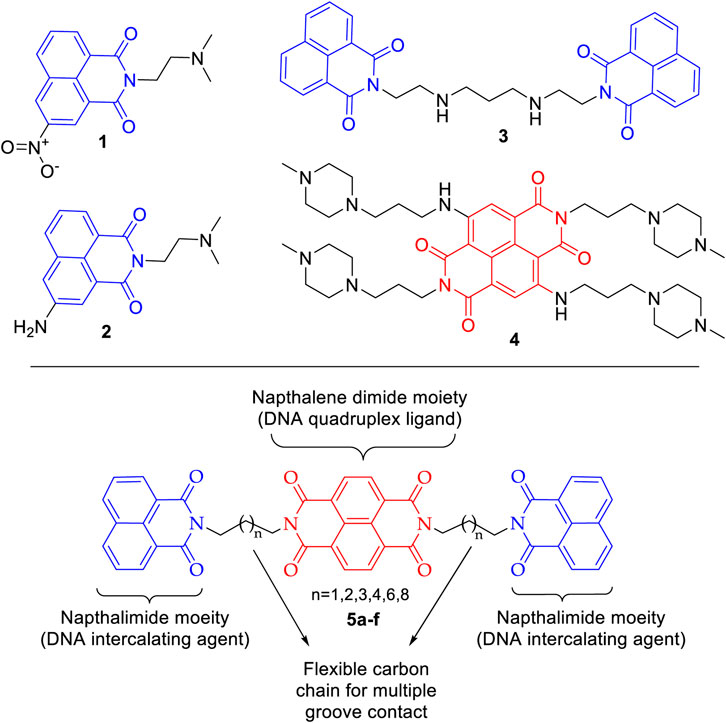
DNA, the carrier of genetic information, is a prominent target in anticancer drug discovery programs. The drugs interact with the DNA and its associated proteins in varied manners resulting in cessation of the unbridled cell growth in cancer (Hurley, 2002; Izbicka and Tolcher, 2004; Delgado et al., 2018). Many DNA targeting drugs are currently in clinical use for treatment of different forms of cancer. Intercalation in DNA base pairs accompanied by interfering with DNA-protein interaction is one of the many mechanisms through which DNA interactive compounds exert cytotoxicity (Mukherjee and Sasikala, 2013). 1,8-Naphthalimide derivatives have emerged as important DNA intercalating class of compounds (Banerjee et al., 2013; Chen et al., 2018; Tomczyk and Walczak, 2018). Mitonafide (1) and amonafide (2) exhibited good DNA intercalation properties and were taken up for clinical studies with limited success (Casado et al., 1996; Allen and Lundberg, 2011). To further improve the DNA binding ability and the overall therapeutic profile of naphthalimides, the concept of bifunctional intercalating group was introduced. The designed bis-naphthalimides exhibited enhanced DNA binding properties and are integral area of research in DNA intercalators (Banerjee et al., 2013; Tandon et al., 2017). One such compound is elinafide (LU79553) (3), which is a lead molecule in the bis-naphthalimide class of compounds. Elinafide (3) exhibited encouraging antitumor activity in phase I clinical trial, though some cumulative toxicity issues were observed (Figure 1) (Villalona-Calero et al., 2001). On the other hand, in recent decades G-quadruplex DNA has emerged as an exciting target in cancer research (Hänsel-Hertsch et al., 2017). The G-quadruplex arrangement of 3′ end of human telomere inhibits the telomerase activity and can be exploited in the selective inhibition of cancer cell. For this purpose, small molecules that stabilize the telomeric end into a G-quadruplex structure are being developed (Monchaud and Teulade-Fichou, 2008). Naphthalene diimides (NDIs) scaffold is increasingly being harnessed as a suitable moiety in the design of quadruplex binding ligands because of its large planar surface (Ohnmacht et al., 2015; Pirota et al., 2019). In an interesting report, the NDI with N-methyl-piperazine side chain BMSG-SH-3 (4) bound exclusively to the 3′ surface of the DNA quadruplex, thus providing impetus to the design of new NDIs based molecules (Collie et al., 2012; Hampel et al., 2013). In addition to medicinal application, the NDIs have potential applications in molecular sensors, organic optoelectronics, photovoltaic cells, and flexible displays. These varied applications of NDIs stem from their characteristic high electron affinity, charge carrier capability, and profound oxidative as well as thermal stabilities (Kobaisi et al., 2016).
Cancer, being a disease of our own cells, is lethal with high rate of incidence and mortality. Extensive anticancer drug development programs are underway to develop newer drugs with enhanced efficacy and safety. The importance of bis-naphthalimides and naphthalene diimides in the design of new DNA interactive agents is well established with considerable success. Here, we envisaged chemical templates (5a–f) in which NDI motif is harnessed as a linker in a bis-naphthalimide scaffold. Further, to enhance the affinity of this template toward DNA intercalation and DNA quadruplex, the flexibility is regulated by six different carbon chain spacers. This could help in multiple groove contacts with DNA base pairs. The naphthalene diimide linked bis-naphthalimide templates were subjected to molecular docking to understand their DNA intercalating and telomeric G-quadruplex binding potential. This was followed by chemical synthesis of these compounds and in vitro evaluation of their DNA thermal denaturation property and cytotoxic potency against selected cancer cell lines along with ADME analysis.
Materials and Methods
Molecular Modeling
Chemical Templates Preparation
The chemical structures of the selected compounds were drawn using ChemDraw 19.1.0.8. Using chemical canonical SMILES IDs of the selected compounds, the 3D structures were obtained by CORINA classic 3D structure generator server (https://www.mn-am.com/online_demos/corina_demo). Discovery Studio Visualizer, 2019 was used to implement CHARMM force field to complete energy minimization process for the generated 3D structures of selected compounds (Vanommeslaeghe et al., 2010).
DNA Molecules Preparation
The 3D structures of DNA molecules (PDB:1CX3 and PDB:3SC8) were downloaded from Protein Data Bank (PDB) (www.rcsb.org). Further HETATOM and water molecules were removed from the native PDB 3D structure files. For energy minimization CHARMM force field was applied using Discovery Studio Visualizer, 2019.
Computational Docking Studies
In silico docking was performed between compounds (5a–f) and DNA molecules (PDB:1CX3 and PDB:3SC8) using AutoDock 4.2 software. AutoDock runs Lamarckian Genetic Algorithm (LGA) and empirical binding free energy function as a scoring function for the ligand-receptor interaction (Morris et al., 1998; Morris et al., 2009). The docking was executed implementing the default AutoDock parameters; however, to cover the maximum area 60 × 60 × 60 Å grid box was used and the grid center point coordinates X, Y, and Z were set as 0.739, −0.12, −0.024 for PDB:1CX3 and −12.145, −15.576, 4.968 for PDB:3SC8, respectively, with the default value of grid points spacing 0.375 Å. After successful completion of docking steps, the results were analyzed and the graphics generation was performed using Discovery Studio Visualizer, 2019.
Chemical Synthesis
Reactions were monitored by thin layer chromatography (silica gel glass plates containing 60 F-254) and visualization was achieved by UV light or iodine indicator. Infrared (IR) spectra were recorded on Perkin-Elmer model 683 or 1,310 spectrometers with sodium chloride optics. 1H NMR spectra were recorded on Gemini (200 MHz) (Varian Inc., Palo Alto, CA, United States) and chemical shifts (d) were reported in ppm, downfield from internal TMS standard. ESI spectra were recorded on Micromass Quattro LC using ESIþ software with capillary voltage of 3.98 kV and ESI mode positive ion trap detector. Elemental analyses were performed on an elemental analyzer (Model: VARIO EL, Elementar, Hanau, Germany). Starting materials and reagents were purchased from Sigma-Aldrich and Alfa-Aesar.
General Procedure for Synthesis of N-Alkyl Azido Naphthalimide Derivatives 9a–f
A mixture of naphthalimide 6 (1 equiv) and dibromoalkanes 7a–f (3 equiv) was dissolved in acetonitrile, K2CO3 (5 equiv) was added, and the resulting solution was refluxed at 80°C for about 3–4 h. The reaction was monitored using TLC and after completion of reaction, the mixture was filtered through sintered funnel. Then the resulting filtrate was concentrated in vacuo and the residue was washed with petroleum ether to remove the unreacted dibromoalkane providing the crude product 8a–f, which was used for azidation reaction. A mixture of N-3-bromopropyl naphthalimide 8a–f (1 equiv) and sodium azide (3 equiv) was dissolved in DMF and the resulting mixture was stirred at room temperature. The reaction was monitored using TLC and after the completion of reaction, the reaction mixture was added to ice-cold water and the resulting solution was extracted with ethyl acetate. Then the resulting filtrate was concentrated in vacuo and the products 9a–f were isolated using column chromatography.
2-(3-Azidopropyl)-2,3-dihydro-1H-benzo[de]isoquinoline-1,3-dione (9a)
It is prepared by following the general procedure of 9a–f from naphthalimide 6 (200 mg, 1.015 mmol), 1,3 dibromopropane 7a (612 mg, 3.045 mmol), and K2CO3 (700 mg, 5.076 mmol) in first step to provide 8a. This is followed by azidation of 8a (200 mg, 0.628 mmol) using sodium azide (122 mg, 1.886 mmol) to afford 160 mg of 9a as gummy solid. Yield: 94%; IR (KBr): ν 2,090, 1,685, 1,662, 1,155 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.58–2.12 (m, 4H), 3.28–3.52 (m, 2H), 7.62–8.33 (m, 6H). MS (ESI): m/z 280 (M+). Anal. Calcd for C15H12N4O2: C, 64.28; H, 4.32; N, 19.99 Found: C, 64.02; H, 4.28; N, 19.56%.
2-(4-Azidobutyl)-2,3-dihydro-1H-benzo[de]isoquinoline-1,3-dione (9b)
It is prepared by following the general procedure of 9a–f from naphthalimide 6 (200 mg, 1.015 mmol), 1,4-dibromobutane 7b (654 mg, 3.045 mmol), and K2CO3 (700 mg, 5.076 mmol) in first step to provide 8b. This is followed by azidation of 8b (200 mg, 0.602 mmol) using sodium azide (117 mg, 1.807 mmol) to afford 165 mg of 9b as gummy solid. Yield: 93%; IR (KBr): ν 2,103, 1,682, 1,659, 1,162 cm−1.1H NMR (200 MHz, CDCl3): δ ppm 1.23–2.03 (m, 6H), 3.28–3.48 (m, 2H), 7.57–8.21 (m, 6H). MS (ESI): m/z 294 (M+). Anal. Calcd for C16H14N4O2: C, 65.30; H, 4.79; N, 19.04 Found: C, 65.27; H, 4.68; N, 18.89%.
2-(5-Azidopentyl)-2,3-dihydro-1H-benzo[de]isoquinoline-1,3-dione (9c)
It is prepared by following the general procedure of 9a–f from naphthalimide 6 (200 mg, 1.015 mmol), 1,5-dibromopentane 7c (697 mg, 3.045 mmol), and K2CO3 (700 mg, 5.076 mmol) in first step to provide 8c. This is followed by azidation of 8c (200 mg, 0.578 mmol) using sodium azide (112 mg, 1.734 mmol) to afford 162 mg of 9c as gummy solid. Yield: 91%; IR (KBr): ν 2,085, 1,678, 1,659, 1,162 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.16–2.11 (m, 8H), 3.19–3.42 (m, 2H), 7.37–8.19 (m, 6H). MS (ESI): m/z 308 (M+). Anal. Calcd for C17H16N4O2: C, 66.22; H, 5.23; N, 18.17 Found: C, 65.98; H, 5.06; N, 18.20%.
2-(6-Azidohexyl)-2,3-dihydro-1H-benzo[de]isoquinoline-1,3-dione (9d)
It is prepared by following the general procedure of 9a–f from naphthalimide 6 (200 mg, 1.015 mmol), 1,6-dibromohexane 7d (739 mg, 3.045 mmol), and K2CO3 (700 mg, 5.076 mmol) in first step to provide 8d. This is followed by azidation of 8d (200 mg, 0.555 mmol) using sodium azide (108 mg, 1.66 mmol) to afford 170 mg of 9d as gummy solid. Yield: 94%; IR (KBr): ν 2,095, 1,689, 1,665, 1,162 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.21–2.19 (m, 10H), 3.21–3.38 (m, 2H), 7.29–8.21 (m, 6H). MS (ESI): m/z 322 (M+). Anal. Calcd for C18H18N4O2: C, 67.07; H, 5.63; N, 17.38 Found: C, 67.01; H, 5.56; N, 17.22%.
2-(8-Azidooctyl)-2,3-dihydro-1H-benzo[de]isoquinoline-1,3-dione (9e)
It is prepared by following the general procedure of 9a–f from naphthalimide 6 (200 mg, 1.015 mmol), 1,8-dibromooctane 7e (828 mg, 3.045 mmol), and K2CO3 (700 mg, 5.076 mmol) in first step to provide 8e. This is followed by azidation of 8e (200 mg, 0.515 mmol) using sodium azide (100 mg, 1.546 mmol) to afford 168 mg of 9e as gummy solid. Yield: 94%; IR (KBr): ν 2,102, 1,688, 1,663, 1,156 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.29–2.02 (m, 14H), 3.19–3.28 (m, 2H), 7.33–8.29 (m, 6H). MS (ESI): m/z 350 (M+). Anal. Calcd for C20H22N4O2: C, 68.55; H, 6.33; N, 15.99 Found: C, 68.50; H, 6.25; N, 15.92%.
2-(10-Azidodecyl)-2,3-dihydro-1H-benzo[de]isoquinoline-1,3-dione (9f)
It is prepared by following the general procedure of 9a–f from naphthalimide 6 (200 mg, 1.015 mmol), 1,10-dibromodecane 7f (913 mg, 3.045 mmol), and K2CO3 (700 mg, 5.076 mmol) in first step to provide 8f. This is followed by azidation of 8f (200 mg, 0.480 mmol) using sodium azide (93 mg, 1.442 mmol) to afford 166 mg of 9f as gummy solid. Yield: 92%; IR (KBr): ν 2095, 1,689, 1,659, 1,152 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.21–2.12 (m, 18H), 3.15–3.29 (m, 2H), 7.31–8.32 (m, 6H). MS (ESI): m/z 378 (M+). Anal. Calcd for C22H26N4O2: C, 69.82; H, 6.92; N, 14.80 Found: C, 69.78; H, 6.88; N, 14.75%.
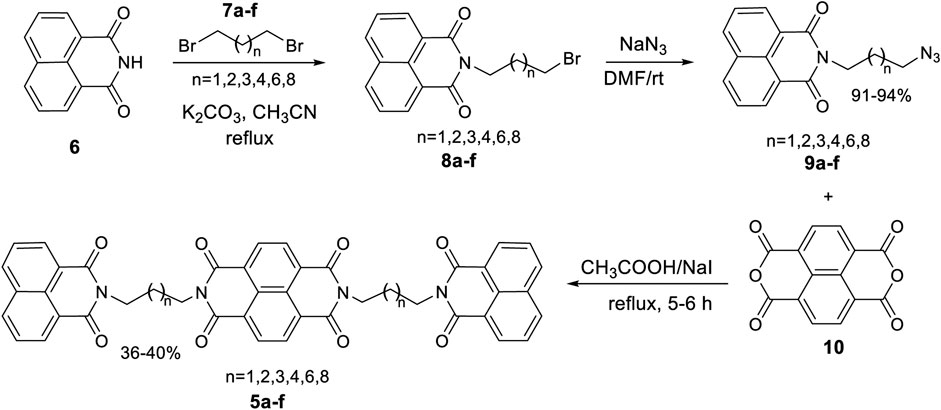
General Procedure for Synthesis of Naphthalene Diimide Linked Bis-Naphthalimides 5a–f
A mixture of N-3-azidopropyl naphthalimide 9a–f (1 equiv) and naphthalic dianhydride 10 (2 equiv) was dissolved in acetic acid. Sodium iodide (6 equiv) was added and the resulting mixture was stirred at reflux temperature for 12 h. After the reaction time, ethyl acetate was added and stirred for 10 min and the organic layer was washed with aq. NaHCO3 followed by aq. Na2S2O3 solution. The organic layer was then concentrated under reduced pressure to obtain crude product which was purified using preparative TLC using hexane:ethyl acetate as solvent system to afford desired compounds 5a–f as white solid.
2,7-Di[3-(1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-2-yl)propyl]-1,2,3,6,7,8-hexahydrobenzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone (5a)
It is prepared by following the general procedure of 5a–f from N-3-azidopropyl naphthalimide 9a (100 mg, 0.357 mmol), naphthalic dianhydride 10 (191 mg, 0.710 mmol), and NaI (321 mg, 2.142 mmol). 105 mg of product 5a was obtained as white solid. Yield: 40%; mp 141°C; IR (KBr): ν 2,950, 1,696, 1,664, 1,160 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 2.23–2.45 (m, 4H), 3.38–3.61 (m, 8H), 7.62–7.91 (m, 4H), 8.11–8.31 (m, 8H), 8.58 (d, 4H). 13C NMR (75 MHz, CDCl3): δ ppm 161.1, 136.3, 134.3, 131.8, 130.7, 127.49, 125.3, 125.1, 42.1, 37.4. MS (ESI): m/z 763 (M+23). Anal. Calcd. for C44H28N4O8: C, 71.35; H, 3.81; N, 7.56 Found: C, 71.30; H, 3.78; N, 7.54%.
2,7-Di[4-(1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-2-yl)butyl]-1,2,3,6,7,8-hexahydrobenzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone (5b)
It is prepared by following the general procedure of 5a–f from N-3-azidopropyl naphthalimide 9b (100 mg, 0.340 mmol), naphthalic dianhydride 10 (182 mg, 0.680 mmol), and NaI (306 mg, 2.04 mmol). 94 mg of product 5b was obtained as white solid, yield 36%; mp 151°C. IR (KBr): ν 2,948, 1,697, 1,666, 1,155 cm−1.1H NMR (200 MHz, CDCl3): δ ppm 1.42–1.85 (m, 8H), 3.37–3.63 (m, 8H), 7.61–7.93 (m, 4H), 8.12–8.29 (m, 8H), 8.58 (d, 4H). MS (ESI): m/z 768 (M+). Anal. Calcd for C46H32N4O8: C, 71.87; H, 4.20; N, 7.29 Found: C, 71.80; H, 4.16; N, 7.25%.
2,7-Di[5-(1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-2-yl)pentyl]-1,2,3,6,7,8-hexahydrobenzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone (5c)
It is prepared by following the general procedure of 5a–f from N-3-azidopropyl naphthalimide 9c (100 mg, 0.308 mmol), naphthalic dianhydride 10 (173 mg, 0.649 mmol), and NaI (277 mg, 1.848 mmol). 100 mg of product 5c was obtained as white solid, yield 39%; mp 156°C. IR (KBr): ν 2,954, 1700, 1,670, 1,060 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.44–1.88 (m, 12H), 3.36–3.69 (m, 8H), 7.59–7.82 (m, 4H), 8.11–8.31 (m, 8H), 8.58 (d, J = 7.38 Hz, 4H). 13C NMR (75 MHz, CDCl3): δ ppm 161.1, 136.3, 134.3, 131.8, 130.7, 127.49, 125.4, 125.1, 42.1, 28.9, 28.4, 27.6. MS (ESI): m/z 819 (M+23). Anal. Calcd for C48H36N4O8: C, 72.35; H, 4.55; N, 7.03 Found: C, 72.30; H, 4.51; N, 7.00%.
2,7-Di[6-(1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-2-yl)hexyl]-1,2,3,6,7,8-hexahydrobenzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone (5d)
It is prepared by following the general procedure of 5a–f from N-3-azidopropyl naphthalimide 9d (100 mg, 0.289 mmol), naphthalic dianhydride 10 (155 mg, 0.579 mmol), and NaI (260 mg, 1.734 mmol). 94 mg of product 5d was obtained as white solid, yield 37%; mp 162°C. IR (KBr): ν 2,948, 1,697, 1,666, 1,075 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.43–1.88 (m, 16H), 3.34–3.62 (m, 8H), 7.55–7.79 (m, 4H), 8.13–8.30 (m, 8H), 8.56 (d, 4H). MS (ESI): m/z 874 (M+23). Anal. Calcd for C50H40N4O8: C, 72.80; H, 4.89; N, 6.79 Found C, 72.80; H, 4.89; N, 6.79%.
2,7-Di[8-(1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-2-yl)octyl]-1,2,3,6,7,8-hexahydrobenzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone (5e)
It is prepared by following the general procedure of 5a-f from N-3-azidopropyl naphthalimide 9e (100 mg, 0.285 mmol), naphthalic dianhydride 10 (153 mg, 0.571 mmol), and NaI (256 mg, 1.71 mmol). 95 mg of product 5e was obtained as white solid, yield 38%; mp 172°C. IR (KBr): ν 2,948, 1706, 1,660, 1,068 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.41–1.87 (m, 24H), 3.39–3.60 (m, 8H), 7.63–7.84 (m, 4H), 8.10–8.30 (m, 8H), 8.58 (d, 4H). MS (ESI): m/z 880 (M+). Anal. Calcd for C54H48N4O8: C, 73.62; H, 5.49; N, 6.36 Found: C, 73.60; H, 5.44; N, 6.34%.
2,7-Di[10-(1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-2-yl)decyl]-1,2,3,6,7,8-hexahydrobenzo[lmn][3,8]phenanthroline-1,3,6,8-tetraone (5f)
It is prepared by following the general procedure of 5a–f from N-3-azidopropyl naphthalimide 9f (100 mg, 0.264 mmol), naphthalic dianhydride 10 (141 mg, 0.529 mmol), and NaI (238 mg, 1.584 mmol). 88 mg of product 5f was obtained as white solid, yield 36%; mp 184°C. IR (KBr): ν 2,951, 1,694, 1,670, 1,074 cm−1. 1H NMR (200 MHz, CDCl3): δ ppm 1.43–1.98 (m, 32H), 3.38–3.61 (m, 8H), 7.62–7.82 (m, 4H), 8.11–8.31 (m, 8H), 8.58 (d, 4H). MS (ESI): m/z 960 (M+23). Anal. Calcd for C58H56N4O8: C, 74.34; H, 6.02; N, 5.98 Found: C, 74.34; H, 6.02; N, 5.98%.
Biological Assays
Thermal Denaturation Studies
Compounds 5a–f were subjected to thermal denaturation studies with duplex-form CT-DNA using reported method (Kamal et al., 2002). Working solutions in aqueous buffer (10 mM NaH2PO4/Na2HPO4, 1 mM Na2EDTA, pH 7.00 ± 0.01) containing CT-DNA (100 μM in phosphate) and the 5a–f (20 μM) were prepared by addition of concentrated 5a–f solutions in DMSO to obtain a fixed [5a–f ]/[DNA] molar ratio of 1:5. The DNA–5a–f solutions were incubated at 37°C for 0, 18, and 36 h prior to analysis. Samples were monitored at 260 nm using a BECKMAN COULTER DU 800 spectrophotometer fitted with high performance temperature controller, and heating was applied at 1°C min−1 in the 40–90°C range. DNA helix→coil transition temperature (Tm) has been obtained from the maxima in the d(A260)/dT derivative plots. Results have been given as the mean ± standard deviation from three determinations and are corrected for the effects of DMSO cosolvent using a linear correction term. Drug-induced alterations in DNA melting behavior have been calculated by ∆Tm =Tm (DNA+ 5a–f)-Tm (DNA alone), where the Tm value for the [5a–f]-free CT-DNA is 69.2 ± 0.01°C.
Cytotoxicity Assay
In vitro anticancer screening assay was performed according to NCI procedures in triplicate. A panel of ten cancer lines belonging to seven different cancer types, prostrate (DU145, PC-3), colon (colo-205, HCT-15, 502713), breast (MCF-7), neuroblastoma (IMR-32), liver (Hep-2), ovary (OVCAR-5), and lung cancers (A549), was selected. The human tumor cell lines of the cancer screening panel were grown in RPMI 1640 medium containing 5% fetal bovine serum and 2 mM l-glutamine. For a typical screening experiment, cells were inoculated into 96-well microtiter plates in 100 μL at plating densities ranging from 5,000 to 40,000 cells/well depending on the doubling time of individual cell lines. After cell inoculation, the microtiter plates were incubated at 37°C, 5% CO2, 95% air, and 100% relative humidity for 24 h prior to addition of experimental drugs. Experimental drugs were solubilized in dimethyl sulfoxide at 400-fold the desired final maximum test concentration and stored frozen prior to use. Following drug addition, the plates were incubated for an additional 48 h at 37°C, 5% CO2, 95% air, and 100% relative humidity. For adherent cells, the assay was terminated by the addition of cold trichloro acetic acid (TCA). Sulforhodamine B (SRB) solution (100 μL) at 0.4% (w/v) in 1% acetic acid was added to each well, and plates are incubated for 10 min at room temperature. After staining, unbound dye is removed by washing five times with 1% acetic acid and the plates were air-dried. Bound stain was subsequently solubilized with 10 mM trizma base, and the absorbance was read on an automated plate reader at a wavelength of 515 run. Using the absorbance measurements [time zero, (Tz), control growth in DMSO without drug, (C), and test growth in the presence of drug at the different concentration levels (Ti)], the percentage growth was calculated at each of the drug concentrations levels.
ADME Prediction
ADME profiling of compounds was carried out using computational approach. ADME properties of compounds 5a–5f and standards, elinafide (3) and BMSG-SH-3 (4), were calculated using preADMET and swissADME predictor online programs (Lee et al., 2003; Daina et al., 2017). The properties like BBB, PPB, Caco2, P-gp interaction, and TPSA were analyzed to understand the ADME properties of these compounds.
Results and Discussions
Molecular Modeling
The designed chemical entities incorporating the DNA interacting bis-naphthalimides and naphthalene diimides moieties were subjected to DNA interaction analysis at molecular level using AutoDock software. First, the compounds 5a–f were studied for their DNA intercalating ability at d(atgcat)2 site of DNA and compared with the standard 3 (Figure 2). The results revealed that the compounds exhibited excellent DNA intercalating property compared to the standard 3 (Table 1). The binding affinity of most of the compounds was better than 3 with compound 5a exhibiting exceptional binding affinity score of −10.93 kcal/mol and inhibition constant of 9.69 nM. The interaction of 5a with the DNA molecule resulted through the formation of three hydrogen bonds with bond lengths of 2.67–3.31 Å and other noncovalent interactions. In comparison the standard 3 exhibited binding affinity of −6.90 kcal/mol and inhibition constant of 8.73 uM with only single hydrogen bond formation. The compound 5b interacted with the DNA site with two hydrogen bonds and a binding energy of −9.45 kcal/mol. Similarly, in case of the compounds 5c and 5f also the binding affinity was better than 3 with values of −9.69 and −9.01 kcal/mol resulting from one and three hydrogen bonds’ formation, respectively, with 5c showing an inhibition constant of 79.37 nM. Comparatively, low binding affinities were observed in compounds 5d and 5e with formation of one and two hydrogen bonds and a resulting binding energy of −6.32 and −7.63 kcal/mol, respectively.
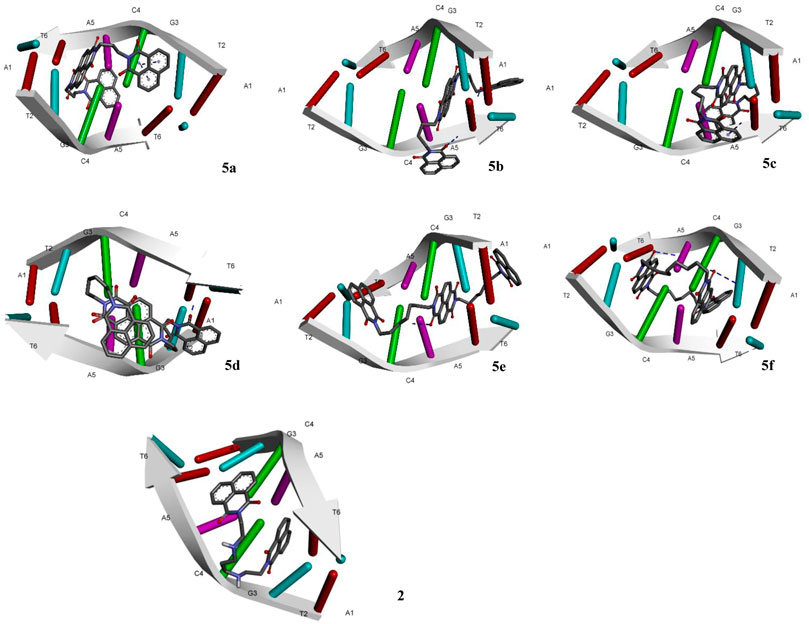
FIGURE 2. Molecular docking of compounds 5a–f and standard elinafide 2 at d(atgcat)2 site of DNA revealing intercalation ability.
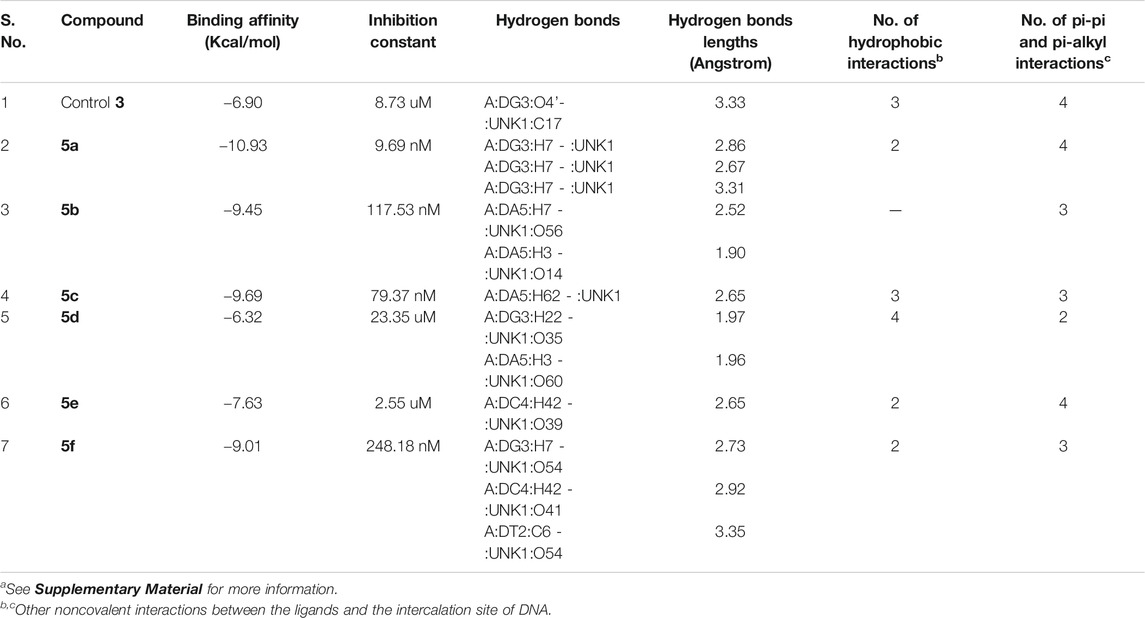
TABLE 1. Molecular docking analysis of some critical parameters of compounds 5a–f and elinafide 3 at d(atgcat)2 site of DNAa.
In addition to the hydrogen bonding interactions, some other noncovalent interactions such as hydrophobic interactions and pi–pi and pi-alkyl interactions were also observed. The compound 5d exhibited most hydrophobic interactions with the DNA helix, followed by compound 5c that showed three interactions similar to standard 3. Two hydrophobic interactions were seen in other compounds and compound 5b exhibited no such interaction. Other noncovalent interactions (pi-pi and pi-alkyl) were also observed and compound 5a as well as 5e showed maximum of these interactions, similar to standard 3 (see Supplementary Material).
The molecular docking of the compounds 5a–f for DNA intercalating ability exhibited excellent results and encouraged us to study their interaction with telomeric G-quadruplex, a promising cancer target. The tetrameric naphthalene diimide derivative BMSG-SH-3 (4) was used as a control (Figure 3). All the compounds 5a-f showed binding affinity in the range of −4.15 to −7.82 kcal/mol, which were better than the standard 3 (−3.70 kcal/mol) as shown in Table 2. The compound 5b was the best among the series with a binding affinity score of −7.82 kcal/mol resulting from formation of four hydrogen bonds with the nitrogen base pairs of quadruplex DNA and an inhibition constant of 1.86 uM. This was higher than three hydrogen bonds’ formation exhibited by the standard 3. Similarly to 5b, the compound 5d also exhibited good binding with telomeric G-quadruplex via four hydrogen bonds’ formation with binding energy and inhibition constant scores of −7.47 kcal/mol and 3.34 uM, respectively. This was followed by compound 5c, which exhibited a binding affinity of −6.42 kcal/mol with the formation of three hydrogen bonds with nitrogen base pairs. The binding energies of −5.33 and −4.81 kcal/mol were shown by compounds 5a and 5e, respectively, with one hydrogen bond’s formation. On the other hand, the affinity of compound 5f was the least with binding energy of −4.15kca/mol with three hydrogen bonds’ formation. In addition to the hydrogen bonding interactions, the docking analysis revealed the presence of other interactions such as hydrophobic interactions and pi–pi and pi-alkyl interactions. In telomeric quadruplex docking also the compound 5d exhibited most hydrophobic interactions (six) with the DNA base pairs, followed by four interactions seen in compound 5f. All the compounds exhibited similar or higher number of hydrogen interactions compared to standard 4 with compounds 5b and 5c displaying three hydrophobic interactions. Other noncovalent interactions (pi-pi and pi-alkyl) were also observed and most of the compounds exhibited similar or higher number of pi-pi and pi-alkyl interactions compared to standard 4 (see Supplementary Material).
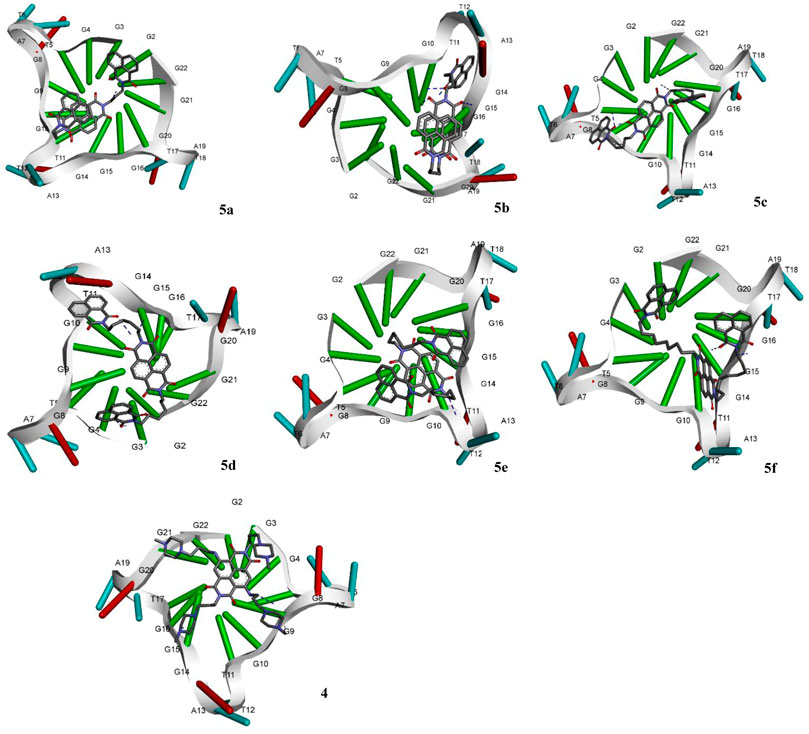
FIGURE 3. Molecular docking of compounds 5a–f and standard BMSG-SH-3 (4) with telomeric G-quadruplex, a promising cancer target.
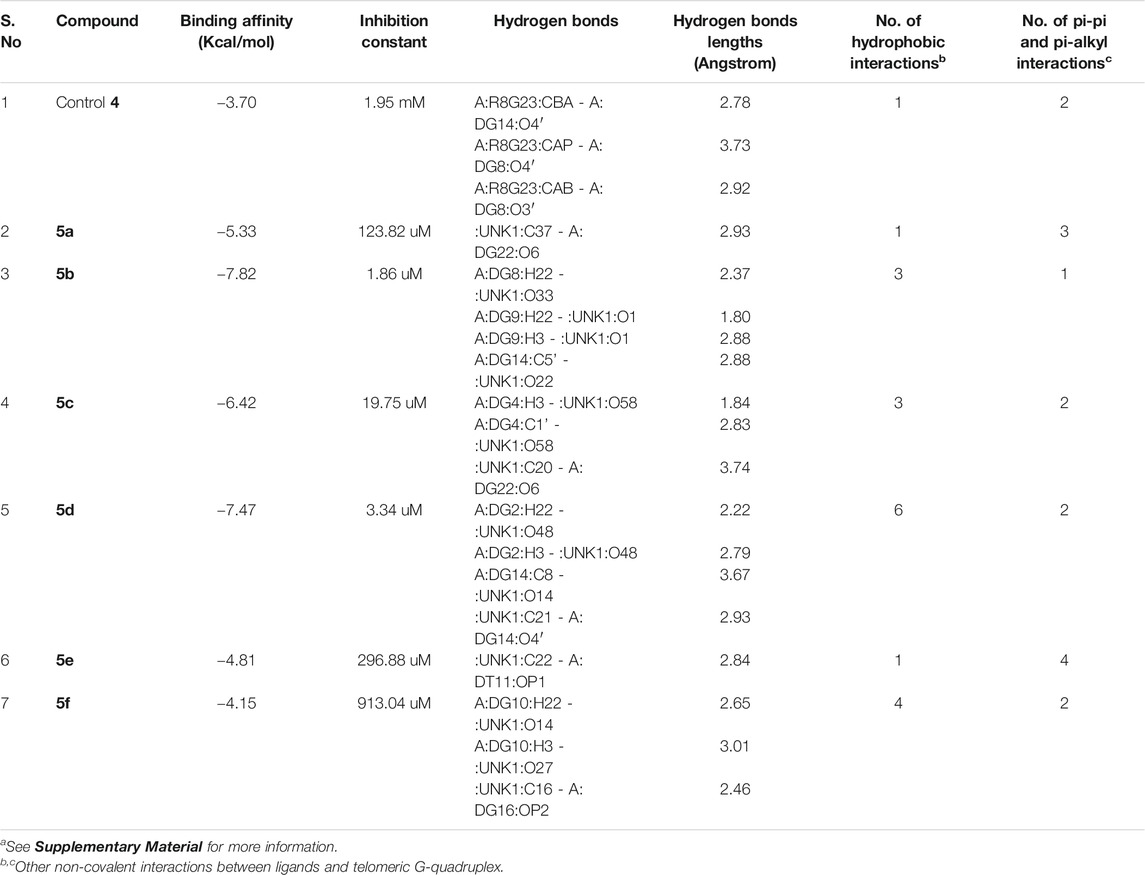
TABLE 2. Molecular docking analysis of some critical parameters of compounds 5a–f and BMSG-SH-3 (4) with telomeric G-quadruplexa.
Chemical Synthesis
Molecular docking analysis of the designed naphthalene diimide linked bis-naphthalimides on DNA intercalation and telomeric DNA quadruplex sites provided excellent results compared to the standard. Therefore, we proceeded to synthesize the compounds and study whether the promising docking results could be reflected on the actual DNA denaturation and cytotoxic properties of the compounds. For this purpose, we designed a convenient synthetic strategy to access the desired molecules 5a–f (Scheme 1). 1,8-Naphthalimide 6 was used as the starting material and was subjected to alkylation employing dibromoalkanes 7a–f with varying carbon chain length. The reaction was carried out in the presence of a base with acetonitrile as solvent under refluxing condition to afford N-alkyl naphthalimides 8a–f. The azidation of these N-alkyl naphthalimides 8a–f was done by employing sodium azide in dimethyl formamide as solvent to yield the key intermediates N-alkyl azido naphthalimides 9a–f. The key intermediates 9a–f were then treated with 1,4,5,8 naphthalic dianhydride in the presence of excess sodium iodide with acetic acid as solvent under reflux condition, to yield the desired naphthalene diimide linked bis-naphthalimides 5a–f.
Biological Evaluation
The synthesized naphthalene diimide linked bis-naphthalimides 5a–f were evaluated for their in vitro DNA binding ability to substantiate the molecular docking results, followed by assessment of cytotoxic properties against a panel of human cancer cell lines.
Thermal Denaturation Studies
Chemical entities that bind preferentially to DNA duplex are known to elevate the melting temperature (Tm) of DNA by stabilizing the double helix. This thermal denaturation analysis is a straightforward way to demonstrate the binding of drug to the DNA. The DNA binding ability for the compounds 5a–f was determined by conducting thermal denaturation studies using duplex-form calf thymus (CT) DNA (Kamal et al., 2002). These studies were carried out at 1:5 molar ratio of compounds 5a–f/DNA and all the compounds elevated the helix melting temperature of CT-DNA (Table 3). These compounds showed a change in melting temperature (∆Tm) in the range of 0.3–1.0°C at 0, 18, and 36 h time intervals (Table 3). No significant change in Tm was observed after 36 h incubation of compounds with DNA. Among the series, compounds 5b and 5c displayed better ∆Tm than the naturally occurring DC-81, which is usually used as a standard in thermal denaturation studies. The compounds 5b and 5c exhibited ∆Tm of 0.7°C at 18 h of incubation time; however after 36 h of incubation ∆Tm was better than standard DC-81. The compound 5b was the best that exhibited ∆Tm of 1.0°C and a change in thermal denaturation of 0.8°C was observed with compound 5c. It is interesting to note that among all the compounds, 5b and 5c exhibited best binding affinities toward the DNA helices in the docking studies. Moreover, the results validate the promising DNA interactive properties of the compounds 5b and 5c.
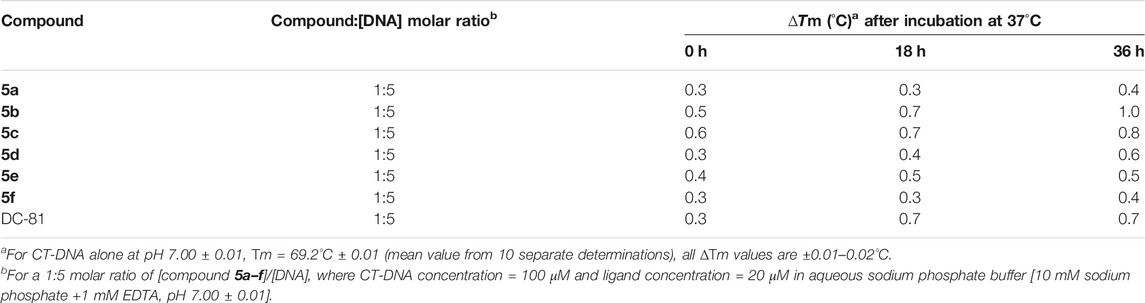
TABLE 3. Thermal denaturation studies of compounds 5a–f using duplex-form calf thymus (CT) DNA at 1:5 molar ratio.
Cytotoxicity
The computational and DNA thermal denaturation studies revealed broadly that naphthalene diimide linked bis-naphthalimides with the short chain spacers exhibit better DNA interactive ability. We then proceeded to evaluate whether the DNA interactive ability of the 5a–f could induce cytotoxic effects against human cancer cell line. The compound 5b was selected and evaluated for anticancer screening at National Cancer Institute (NCI), United States. It was screened against a panel of 60 human tumor cell lines organized in subpanels representing melanoma, leukemia, and cancers of breast, prostate, lung, colon, ovary, CNS, and kidney by using sulforhodamine B (SRB) method at single dose concentration of 10−5 M. Compounds which reduced the growth of the cell lines to 50% or less compared with untreated control cells at this concentration were considered as active against the corresponding cell lines. The compound 5b exhibited significant activity selectively against non-small cell lung cancer and leukemia cell lines as shown in Table 4. A percent growth of 45.42 was seen in HOP-92 lung cancer cell line and in case of leukemia, percent growth of 23.93 and 44.91 was observed in cell lines MOLT-4 and SR, respectively.

TABLE 4. Growth percent of compounds 5b in selected cell lines in National Cancer Institute single dose screeninga (10−5 M).
In addition to this, all the compounds 5a–f were tested against ten human tumor cell lines organized in subpanels representing cancers of breast, prostate, lung, colon, ovary, and neuroblastoma at two different concentrations, i.e.,10−5 M and 10−6 M, and the results are shown in Table 5. All the tested compounds exhibited little activity against cell lines of prostrate, neuroblastoma, liver, and ovary cancer. Some activity was observed in MCF-7 breast cancer cell line and HCT-15 colon cancer cell lines. The compounds 5a–f exhibited some promising activity against colon-205 and A549 lung cancer cell lines. Among the series, compounds 5a and 5b with short chain spacers displayed good growth inhibition of around 45% against A459 lung cancer at 10 μM concentration. On the other hand, compounds 5e and 5f showed 40% and 41% growth inhibitions against A549 and colo-205, respectively.
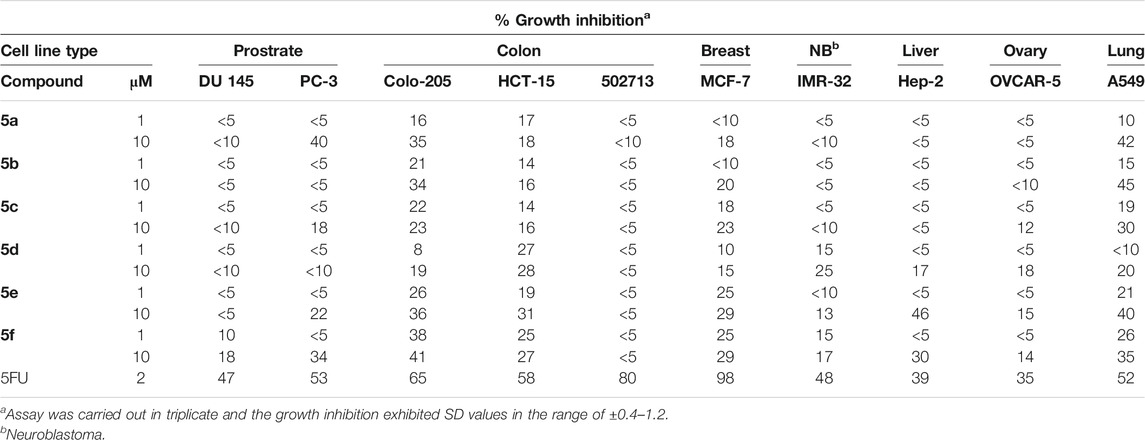
TABLE 5. Cytotoxicity assay of compounds 5a–f on selected cancer cell lines employing sulforhodamine B (SRB) assay at 10−5 M and 10−6 M concentrations.
ADME Computational Analysis
The pharmacokinetic and pharmacodynamic properties of the compounds reveal whether a chemical compound has potential to be developed as a clinical drug. In silico ADME predictions provide an insight into these critical properties prior to conducting the actual in vitro and in vivo studies. The druglikeness and ADME properties of compounds 5a–f were predicted by using preADMET and SwissADME online tools and compared with standard 3, which reached clinical studies (Table 6). The predicted Caco2 values revealed that all the compounds along with the standard exhibited similar biological barrier crossing ability and were not blood brain barrier permeant. However, the compounds acted as inhibitor of P-gp, a transporter protein, that could result in decreased bioavailability. This was further validated by high TPSA and low bioavailability scores. iLOGP is a critical physiochemical parameter in drug design and is one of the factors contributing to the drug availability. Interestingly, the compounds 5a and 5b exhibited iLOGP values comparable to the standard 3. Both the compounds are promising candidate to be taken further for development by structural diversification.
Conclusion
Bis-naphthalimide and naphthalene diimide motifs are important pharmacophores in anticancer drug design. A molecular modeling assisted rational design of new naphthalene diimide linked bis-naphthalimides as potential DNA interactive anticancer agents was undertaken. These compounds were subjected to molecular docking for their DNA intercalating and telomeric G-quadruplex binding potential. Compounds 5a and 5b provided excellent results for DNA intercalation and G-quadruplex binding, respectively, compared to the standards. This encouraged us to synthesize these compounds and study their in vitro DNA interactive ability and cytotoxicity. The compound 5b stabilized the CT-DNA better than the standard and exhibited promising anticancer activity against HOP-92, A549 (lung cancer), MOLT-4, and SR (leukemia). Similarly, 5a also exhibited decent activity against few cancer cell lines. A prediction of ADME properties was undertaken to understand the exhibited cytotoxicity of the compounds and thereby identify a potential molecule for further development. Based on all the studies, it was found that naphthalene diimide linked bis-naphthalimides with short chain spacers have potential for further development. Notwithstanding the excellent computational and promising experimental results suggesting DNA binding ability, the lower cytotoxicity of these compounds against cancer cell lines could be attributed to decreased bioavailability as shown in ADME predictions. However, a more focused chemical diversification of compounds 5a and 5b to overcome the bioavailability issues could lead to more potent molecules.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by grant code 19-SCI-1-01-0019.
Conflict of Interest
Thokhir B. Shaik was employed by Research on Advanced BioMedical Solutions.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.630357/full#supplementary-material.
References
Allen, S. L., and Lundberg, A. S. (2011). Amonafide: a potential role in treating acute myeloid leukemia. Expet Opin. Invest. Drugs 20 (7), 995–1003. doi:10.1517/13543784.2011.585756
Banerjee, S., Veale, E. B., Phelan, C. M., Murphy, S. A., Tocci, G. M., Gillespie, L. J., et al. (2013). Recent advances in the development of 1,8-naphthalimide based DNA targeting binders, anticancer and fluorescent cellular imaging agents. Chem. Soc. Rev. 42 (4), 1601–1618. doi:10.1039/c2cs35467e
Casado, A., Rosell, R., García-Gómez, R., Díaz-Rubio, E., Pérez-Manga, G., Font, A., et al. (1996). Phase II study of mitonafide in non-small cell lung cancer (NSCLC). Invest. N. Drugs 14 (4), 415–417. doi:10.1007/BF00180820
Chen, Z., Xu, Y., and Qian, X. (2018). Naphthalimides and analogues as antitumor agents: a review on molecular design, bioactivity and mechanism of action. Chin. Chem. Lett. 29 (12), 1741–1756. doi:10.1016/j.cclet.2018.09.020
Collie, G. W., Promontorio, R., Hampel, S. M., Micco, M., Neidle, S., and Parkinson, G. N. (2012). Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 134 (5), 2723–2731. doi:10.1021/ja2102423
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7 (1), 42717. doi:10.1038/srep42717
Delgado, J. L., Hsieh, C. M., Chan, N. L., and Hiasa, H. (2018). Topoisomerases as anticancer targets. Biochem. J. 475 (2), 373–398. doi:10.1042/BCJ20160583
Hampel, S. M., Pepe, A., Greulich-Bode, K. M., Malhotra, S. V., Reszka, A. P., Veith, S., et al. (2013). Mechanism of the antiproliferative activity of some naphthalene diimide G-quadruplex ligands. Mol. Pharmacol. 83 (2), 470–480. doi:10.1124/mol.112.081075
Hänsel-Hertsch, R., Di Antonio, M., and Balasubramanian, S. (2017). DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 18 (5), 279–284. doi:10.1038/nrm.2017.3
Hurley, L. H. (2002). DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer 2 (3), 188–200. doi:10.1038/nrc749
Izbicka, E., and Tolcher, A. W. (2004). Development of novel alkylating drugs as anticancer agents. Curr. Opin. Invest. Drugs 5 (6), 587–591.
Kamal, A., Ramesh, G., Laxman, N., Ramulu, P., Srinivas, O., Neelima, K., et al. (2002). Design, synthesis, and evaluation of new noncross-linking pyrrolobenzodiazepine dimers with efficient DNA binding ability and potent antitumor activity. J. Med. Chem. 45 (21), 4679–4688. doi:10.1021/jm020124h
Kobaisi, M. A., Bhosale, S. V., Latham, K., Raynor, A. M., and Bhosale, S. V. (2016). Functional naphthalene diimides: synthesis, properties, and applications. Chem. Rev. 116 (19), 11685–11796. doi:10.1021/acs.chemrev.6b00160
Lee, S. K., Lee, I. H., Kim, H. J., Chang, G. S., Chung, J. E., and No, K. T. (2003). “The PreADME Approach: web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties”, in EuroQSAR 2002 designing drugs and crop protectants: processes, problems and solutions. (London: Blackwell Publishing), 418–420.
Monchaud, D., and Teulade-Fichou, M. P. (2008). A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 6 (4), 627–636. doi:10.1039/b714772b
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30 (16), 2785–2791. doi:10.1002/jcc.21256
Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., et al. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19 (14), 1639–1662. doi:10.1002/(sici)1096-987x(19981115)19:14<1639::aid-jcc10>3.0.co;2-b
Mukherjee, A., and Sasikala, W. D. (2013). Drug-DNA intercalation: from discovery to the molecular mechanism. Adv Protein Chem Struct Biol 92, 1–62. doi:10.1016/B978-0-12-411636-8.00001-8
Ohnmacht, S. A., Marchetti, C., Gunaratnam, M., Besser, R. J., Haider, S. M., Di Vita, G., et al. (2015). A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 5, 11385. doi:10.1038/srep11385
Pirota, V., Nadai, M., Doria, F., and Richter, S. N. (2019). Naphthalene diimides as multimodal G-quadruplex-selective ligands. Molecules 24 (3), 426. doi:10.3390/molecules24030426
Tandon, R., Luxami, V., Kaur, H., Tandon, N., and Paul, K. (2017). 1,8-Naphthalimide: a potent DNA intercalator and target for cancer therapy. Chem. Rec. 17 (10), 956–993. doi:10.1002/tcr.201600134
Tomczyk, M. D., and Walczak, K. Z. (2018). l,8-Naphthalimide based DNA intercalators and anticancer agents. A systematic review from 2007 to 2017. Eur. J. Med. Chem. 159, 393–422. doi:10.1016/j.ejmech.2018.09.055
Vanommeslaeghe, K., Hatcher, E., Acharya, C., Kundu, S., Zhong, S., Shim, J., et al. (2010). CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31 (4), 671–690. doi:10.1002/jcc.21367
Keywords: bis-naphthalimides, naphthalene diimide, molecular modeling, DNA interactive, anticancer agents
Citation: Malik MS, Farooq Adil S, Moussa Z, Altass HM, Althagafi II, Morad M, Ansari MA, Sajid Jamal QM, Obaid RJ, Al-Warthan AA, Shaik TB and Ahmed SA (2021) Rational Design and Synthesis of Naphthalene Diimide Linked Bis-Naphthalimides as DNA Interactive Agents. Front. Chem. 9:630357. doi: 10.3389/fchem.2021.630357
Received: 17 November 2020; Accepted: 07 January 2021;
Published: 10 March 2021.
Edited by:
Stalin Thambusamy, Alagappa University, IndiaReviewed by:
Narayana Nagesh, Centre for Cellular & Molecular Biology (CCMB), IndiaTimothy Andrew Barendt, University of Birmingham, United Kingdom
Copyright © 2021 Malik, Farooq Adil, Moussa, Altass, Althagafi, Morad, Ansari, Sajid Jamal, Obaid, Al-Warthan, Shaik and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Shaheer Malik, msmalik@uqu.edu.sa; Saleh A. Ahmed, saahmed@uqu.edu.sa
 M. Shaheer Malik
M. Shaheer Malik Syed Farooq Adil2
Syed Farooq Adil2 Mohammad Azam Ansari
Mohammad Azam Ansari Qazi Mohammad Sajid Jamal
Qazi Mohammad Sajid Jamal Thokhir B. Shaik
Thokhir B. Shaik Saleh A. Ahmed
Saleh A. Ahmed