- Department of Chemistry, University of Connecticut, Storrs, CT, United States
Therapeutic nucleic acids hold immense potential in combating undruggable, gene-based diseases owing to their high programmability and relative ease of synthesis. While the delivery of this class of therapeutics has successfully entered the clinical setting, extrahepatic targeting, endosomal escape efficiency, and subcellular localization remain as major roadblocks. On the other hand, viruses serve as natural carriers of nucleic acids and have acquired a plethora of structures and mechanisms that confer remarkable transfection efficiency. Thus, understanding the structure and mechanism of viruses can guide the design of synthetic nucleic acid vectors. This review revisits relevant structural and mechanistic features of viruses as design considerations for efficient nucleic acid delivery systems. This article explores how viral ligand display and a metastable structure are central to the molecular mechanisms of attachment, entry, and viral genome release. For comparison, accounted for are details on the design and intracellular fate of existing nucleic acid carriers and nanostructures that share similar and essential features to viruses. The review, thus, highlights unifying themes of viruses and nucleic acid delivery systems such as genome protection, target specificity, and controlled release. Sophisticated viral mechanisms that are yet to be exploited in oligonucleotide delivery are also identified as they could further the development of next-generation nonviral nucleic acid vectors.
Introduction
Undruggable targets are disease-implicated proteins that lack easy-to-bind pockets where conventional therapeutics like small molecules can bind (Crews, 2010; Duffy and Crown, 2021). However, around 80% of the human proteome is difficult to reach or target (Verdine and Walensky, 2007). The past decade has shown enormous progress in targeting the previously thought to be unreachable sites such as growth factors, enzymes, defective genes, or nuclear transcription factors (Lazo and Sharlow, 2016). In particular, therapeutic nucleic acids such as small interfering RNAs (siRNAs), microRNAs (miRNAs), antisense oligonucleotides (ASOs), synthetic messenger RNAs (mRNAs), and CRISPR-Cas9-guide RNAs are programmable, easy to synthesize, and thus have the potential to treat previously undruggable diseases such as cancer and viral diseases (Dowdy, 2017). They hold great promise in treating the root cause of the disease rather than just treating the symptoms by targeting the mutated genes, mRNA, or proteins with high specificity and selectivity (Keefe et al., 2010; Damha 2019). The challenge lies in delivery (Juliano, 2016; Dowdy, 2017; Dowdy and Levy, 2018; Johannes and Lucchino, 2018; Juliano, 2018).
For billions of years, cells have evolved to keep genomic material on one side of the membrane. Thus, transfection by bare nucleic acids across an anionic lipid barrier is fundamentally prevented by the large size and density of negative charges (Dowdy, 2017; Dowdy and Levy, 2018; Johannes and Lucchino, 2018). Furthermore, medical translation necessitates a successful in vivo delivery. This is particularly challenging given the limited systemic stability of unmodified nucleic acids. Thus, an ideal delivery strategy should include nucleic acid protection from nuclease degradation and oxidation, prolonged systemic circulation, targeted delivery, efficient transfection across a membrane, facilitated access to the cytoplasm or nucleus, and little to no side effects (Zhu and Mahato, 2010). While progress has been made in designing and implementing safe, effective, and efficient nucleic acid delivery systems, realizing their therapeutic potential is, at present, challenged mainly by the lack of cellular target diversity and endosomal escape ability (Dowdy, 2017; Dowdy and Levy, 2018; Johannes and Lucchino, 2018; Juliano, 2018).
In contrast, viruses have evolved a diversity of enabling architectures for the infiltration of various host cells and controlled viral genome replication using the host cell machinery (Flint et al., 2015). While they have become longstanding models for engineering the transfection of therapeutic nucleic acids (Figure 1) (Ni et al., 2016), their delivery efficiency far outplays that of synthetic vectors (Ramamoorth and Narvekar, 2015). This underscores how our current molecular understanding of viral function and how this relates to nucleic acid transfection can be improved to achieve more effective translation to rational design.
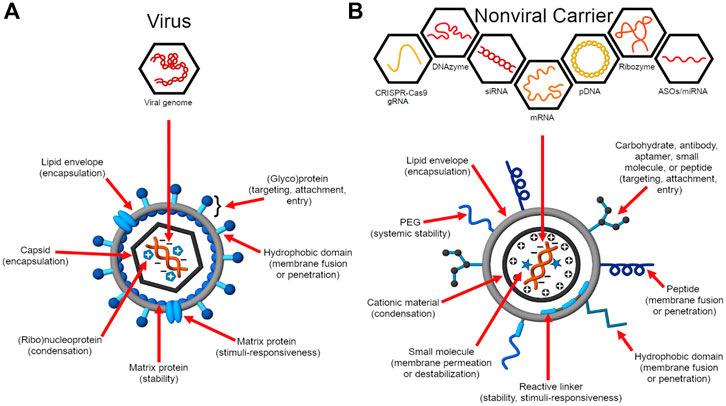
FIGURE 1. Virus structure and function inform the design of nucleic acid delivery systems. (A) Viruses evolve to deliver their genome efficiently to the host cell for replication (Flint et al., 2015). As such, their genome encodes proteins essential for genome protection, tropism, intracellular trafficking, controlled genome release, and replication. (B) Synthetic carriers are designed to deliver a diversity of therapeutic nucleic acid cargo including pDNA, siRNA, ASOs, miRNA, mRNA, CRISPR-Cas9 guide RNAs (gRNAs), ribozymes, and DNAzymes (Ni et al., 2016; Ni et al., 2019). Analogous to viruses, functional domains are embedded on the construct that enable a balance between nucleic acid protection and programmed, stimulus-induced release.
This review, therefore, details the structure and intracellular fate of existing nucleic acid delivery strategies whose designs are either directly inspired by viruses or their resulting formulation exhibits many similarities to that of viruses. Hence, relevant structural and mechanistic features of viruses as design considerations for viable nucleic acid delivery systems are examined. This article also explores how a dynamic and stimulus-responsive structure can play an important role in designing an effective nucleic acid carrier. Importantly, it also highlights how sophisticated ligand display is central to the molecular mechanisms of carrier trafficking and nucleic acid release.
General Structure of Nucleic Acid Carriers and Mechanism of Protection
An ideal carrier packs, stores, and protects nucleic acid cargo until it has reached the target site. In that regard, this section provides examples of select viruses and nonviral nucleic acid vectors and discusses their structural features relevant to the efficient packing and protection of nucleic acids. Figure 2 presents examples of common viruses to show that despite differences in sizes and shapes, viruses collectively protect their genome through condensation and encapsulation. In addition to these two mechanisms of nucleic acid protection, nonviral carriers also use chemical modifications, self-generated sterics, or a combination of these strategies to achieve the same effect.
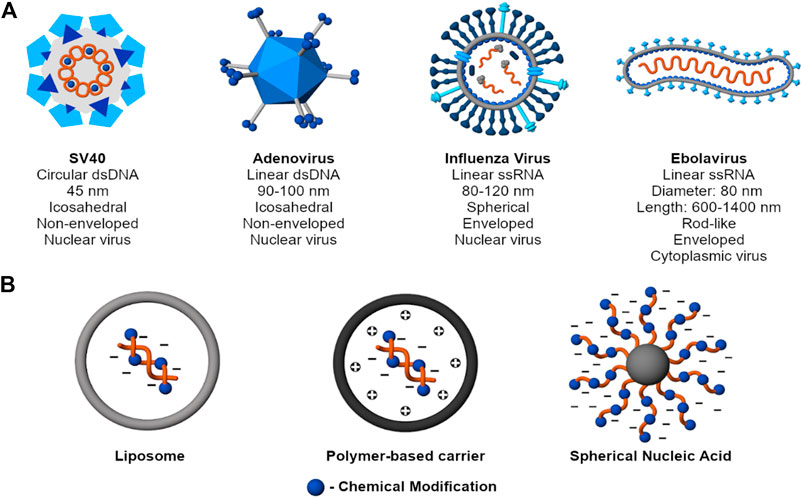
FIGURE 2. Mechanisms to protect nucleic acid cargo. (A) Examples of common viruses (SV40 - Martini et al. 2007; Adenovirus - Greber et al., 1997; Russel, 2009; Influenza Virus - James and Whitley, 2017; Ebolavirus - Beniac et al., 2012; Falasca et al., 2015). Despite structural diversity, viruses collectively protect their genome through charge condensation and encapsulation by a capsid and, for an enveloped virus, an outer lipid membrane. (B) Examples of nonviral nucleic acid delivery systems. Beyond condensation and encapsulation, nonviral carriers also use chemical modifications, self-generated sterics, or a combination of strategies to achieve the same purpose.
Structure of Viruses and Genome Protection
Viruses are obligate intracellular parasites (Gelderblom, 1996). They have evolved to transfect their DNA or RNA genome into the host cell for expression and subsequent production of more virus particles (Prasad and Schmid, 2011). At the core of virus structure are structural proteins that serve to protect the viral genome until it is delivered to the target site. These structural proteins assemble to form the viral capsid, which is the protein coat that wraps around the genome. The high degree of folding and dense packing of capsid proteins protect them from proteolytic digestion, making them stable carriers of nucleic acid cargo (Flint et al., 2015). Moreover, the viral genome is typically condensed by viral proteins through charge neutralization (Gelderblom, 1996), allowing confinement within the interior of the capsid. Enveloped viruses possess an outer lipid envelope that provides additional encapsulation and can fuse with the host plasma membrane during uptake or endosomal escape. The protein components encoded by the viral genome display highly specific and often, multiple, roles essential for structural integrity, attachment, and replication in the host cell (Flint et al., 2015).
For example, the main components of the influenza virus are the lipid bilayer, glycoprotein spikes hemagglutinin (HA) and neuraminidase (NA), matrix proteins (M1 and M2), the heterotrimeric RNA-dependent RNA polymerase (RdRP), the viral RNA segments, a nucleoprotein (NP), and two nonstructural proteins (NS1 and NS2 a.k.a. nuclear export protein or NEP). The outermost layer of the virus is a lipid membrane decorated with glycoproteins that, in turn, may be recognized by antibodies to protect the host against infection (James and Whitley, 2017). Thus, these glycoproteins are critical in both immune response and the development of therapeutics. Hemagglutinin, specifically its subunit HA1, is responsible for the targeting of and uptake by the host cells. HA1 binds to sialic acid functionalized cell surface receptors, resulting in receptor-mediated endocytosis. The lipid bilayer is stabilized by M1 on its cytoplasmic periphery and is spanned by M2, a proton ionophore. The core of the virion contains the viral genome as well as proteins essential for viral gene replication (RdRP), gene encapsulation (NP), and nuclear translocation (NEP). Each protein-coding ssRNA segment is coated by NPs and associated with an RdRP, forming a ribonucleoprotein (RNP) complex that is anchored to M1. The viral envelope of influenza virus has been used as a carrier for nucleic acids such as siRNA (de Jonge et al., 2006) and miRNA (Li J. et al., 2013). Particularly, the reconstituted influenza virus membrane envelope, called “virosome,” acts as an efficient carrier to target small nucleic acid such as siRNA in vitro as well as in vivo (de Jonge et al., 2006). As per this study, the functional integrity of HA viral protein helps in membrane fusion and efficient cytosolic delivery of siRNA.
Another example is the adenovirus (AdV), one of the largest (90–100 nm) non-enveloped double stranded linear DNA viruses. The icosahedral shaped capsid is made of many structural polypeptides. Most of the capsid coat (about 75%) is composed of a hexon protein, which is held together by protein IX. A unique feature of Adv capsid is that the vertices are made of a penton protein from which fiber knobs protrude out–both of which are essential for host cell entry. The viral genome is condensed by proteins V, VII and μ and is also covalently associated with the terminal protein. The cementing protein IIIa acts as capsid stabilizing protein by linking the facets of the icosahedron (Greber et al., 1997; Fay and Panté, 2015). Adenoviral vectors have been used for delivering shRNA, siRNA (Nayerossadat et al., 2012), and large sizes of DNA (up to 38 kb). However, unlike retroviruses, these cannot integrate the carried DNA into the host genome. Thus, the desired gene expression is limited. Also, the immunogenic response caused by adenoviral infection and low cell specificity limits the use of such viral vector only to few tissues such as lungs and liver (Vorburger and Hunt, 2002).
Despite the structural and mechanistic differences among viruses, all viral capsids are metastable, which means they are stable enough to protect the genome until they reach the target site to uncoat it. Thus, the virus construct is spring-loaded in that potential energy is stored during its assembly. Upon reaching the target site, a chemical trigger such as low pH or proteolytic enzymes overcome the energetic barrier, resulting in virus disassembly and uncoating of the genome. Metastability is achieved by the inherent symmetrical arrangement of identical capsid protein subunits that is stabilized by nonspecific noncovalent interactions. In this regard, many capsid proteins self-assemble into virus-like particles (VLPs) (Flint et al., 2015).
VLPs are non-infectious, multiprotein complexes that mimic the viral capsid assembly but are devoid of the genome. Their utility as experimental tools and as therapeutic carriers has been thoroughly reviewed elsewhere (Rohovie et al., 2017; Roldão et al., 2017). Recombinant versions with attenuated or inactivated antigens can also be reconstructed from complementary DNA of a viral genome. While VLPs are historically produced and extracted from the natural hosts themselves, nowadays they are primarily produced through various cell cultures (Roldão et al., 2017). The use of mammalian and non-mammalian cells, baculoviruses, and bacteria has been reported, but VLPs are commonly expressed in yeast cells due to the relative ease of protein expression, scalability, and lower production cost compared to mammalian and insect cells (Kim and Kim, 2017; Roldão et al., 2017).
Like viruses, VLPs have been successfully used in developing vaccines and vaccine adjuvants, and their use in gene therapy and immunotherapy has also been explored (Rohovie et al., 2017; Roldão et al., 2017). Some of those that have shown potential for nucleic acid delivery include bacteriophage-based MS2 (Pan et al., 2012a; Pan et al., 2012b), bacteriophage-based M13 (Yata et al., 2014), animal virus-based hepatitis B virus core (Brandenburg et al., 2005), and plant-based cowpea chlorotic mottle virus (Lam and Steinmetz, 2019).
Target specificity can be tailored by chemical conjugation or directly expressing targeting ligands on the protein coat (Rohovie et al., 2017). For example, Yata et al. (2014) demonstrated the use of a hybrid VLP/cationic polymer-based system for efficient gene transfer. The construct specifically used bacteriophage M13 that was genetically modified to express the RGD peptide on its surface for tumor targeting and was complexed with a cationic polymer for enhanced cellular uptake. Similarly, Lam and Steinmetz (2019) recently delivered siRNA for the knockdown of GFP and FOXA1 target genes using cowpea chlorotic mottle VLPs. With an SM(PEG)4 crosslinker, the VLPs were chemically labeled with m-lycotoxin, a cell-penetrating peptide, to enhance cellular uptake.
Strategies for Nucleic Acid Protection by Nonviral Carriers
While the ability of viruses and VLPs to efficiently encapsulate and transfect nucleic acids is remarkable, they are structurally more complex and, thus, typically require hosts for production and subsequent purification (Roldão et al., 2017), both of which may come at a high cost. Moreover, viruses and VLPs have a higher risk of triggering an immune response (Xue et al., 2015) and possess limited chemistry (Wagner, 2012). Therefore, tuning properties such as target specificity, particle stability, and subcellular localization is restricted, motivating the construction of non-viral vectors (Wagner, 2012). Beyond condensation and encapsulation, this section lists other strategies that have been employed for efficient protection of nucleic acid cargo such as chemical modifications and self-generated sterics. Furthermore, these strategies are often combined for enhanced protection.
Condensation by Cationic Materials
Viral assembly mainly involves electrostatic interactions between the capsid proteins and genomic cargo. Similarly, many first-generation designs of delivery agents relied on the electrostatic masking of the polyanionic backbone of nucleic acids for successful delivery into cells. Whereas viruses protect their nucleic acid cargo via capsid encapsulation, cationic materials such as natural and synthetic polymers, dendrimers, proteins, peptides, and cationic lipids as well as inorganic nanoparticles bearing a positive charge (to be discussed in Section Utility of Inorganic Nanoparticles) form an electrostatic interaction with the negative phosphate backbone of the nucleic acid cargo, providing protection from nuclease degradation (Ferrari et al., 1999; Moret et al., 2001; Thomas and Klibanov, 2003). This can be ascribed to the compaction of nucleic acids, which results in the blockage of enzymatic digestion sites, thereby conferring nuclease protection (Feng et al., 2015).
Electrostatic interactions also strengthen viral attachment to the surface of negatively—charged host cells. Thus, viruses such as the hepatitis C virus (Penin et al., 2001) and the influenza virus (Arinaminpathy and Grenfell, 2010) have conserved cationic regions in their glycoproteins that aid in membrane binding. In the same light, synthetic polycationic nucleic acid carriers not only allow compaction and protection from nuclease degradation but they also mediate cellular attachment and entry (Mislick and Baldeschwieler, 1996). However, this uptake mechanism is nonspecific, and polymeric materials tend to form aggregates with components of the blood such as serum proteins. For this reason, nonionic, hydrophilic polymers such as PEG are commonly added to confer stealth (Klibanov et al., 1990; Takemoto et al., 2014). Additionally, the structural flexibility of PEG makes its integration into different formulations very convenient. However, while PEG-ylation imparts blood compatibility and circulation longevity (Takemoto et al., 2014), it can compromise cellular uptake and/or endosomal escape (Fang et al., 2017).
To address this limitation, PEG-ylation typically involves responsive linkages that can be cleaved by cellular cues such as low pH or external stimuli such as temperature (Fang et al., 2017). An alternative way of using cleavable PEG was demonstrated by Li and co-workers (2013) where they used MMP-7-cleavable peptides as linkers. Matrix Metalloproteinase-7 (MMP-7) belongs to a class of zinc-dependent, extracellular proteases that are overexpressed on the surface of breast tumor cells. In their construct, the outer surface of the polymer-based siRNA-delivery vector was decorated with PEG attached to the core of the particle using a peptide substrate of MMP-7. When the peptide substrate came to contact with MMP-7, the PEG outer layer was cleaved off, revealing a highly cationic dimethylaminoethyl methacrylate core that then engages the membrane, facilitating uptake. Thus, the selective attachment and entry of the resulting construct is afforded through proximity activation by MMP-7.
Peptide-based vectors tend to rely on positive charge character to condense nucleic acids for packaging and protection. In particular, these consist of cationic amphiphilic peptides that are composed of a hydrophobic and a hydrophilic domain that form a well-defined nanoparticle (Kang et al., 2019). The hydrophobic region consists of non-polar neutral amino acids whereas the hydrophilic region has polar aliphatic residues. These peptides self-assemble to form a micellular structure. Small molecule drugs and DNA can be co-delivered using these multifunctional micelle-plexes, where each peptide plays a different role. For example, displaying a cell penetrating peptide on the surface facilitates binding and entry. Histidine residues cause endosomal escape while lysine residues condense DNA. These types of complexes have been used to deliver siRNA and plasmid DNA. Recent studies have also shown that the addition of stearyl, an alkyl chain, or cholesterol to the hydrophobic domain of self-assembled peptides further enhances DNA condensation and transfection efficiency (Kang et al., 2019).
In addition, highly branched polypeptides are used as hybrid-peptide based gene delivery vehicles. This is achieved by covalently joining multi-functional peptide sequences. Functional peptides are separated by spacers such as repeats of glycine residues that confer flexibility. Nucleic acids are also packed by condensation. Redox-active disulfide bonds can be used to connect peptides in a branched fashion, delivering genes more efficiently than linear counterparts. These disulfide bonds are then reduced in the cytoplasm by glutathione to liberate the nucleic acid cargo as well as to reduce cytotoxicity. Highly branched arginine-rich polypeptides are multivalent and flexible—attributes beneficial for nucleic acid compaction and cellular entry. Many of these reducible multibranched cationic polypeptides have the potential to be non-toxic, degradable vectors for gene delivery (Kang et al., 2019).
Among various polycationic formulations, materials based on synthetic polymers such as polymeric nanoparticles, dendrimers, polymer micelles, polymersomes, polyplexes, and lipopolyplexes have benefited from their chemical diversity, relatively simple design, and potential for multi-functionality (Takemoto et al., 2014; Yuan and Li, 2017). The chemistry, molecular weight, weight relative to the nucleic acid, and overall topology of the polymer determine its stability and transfection efficiency. Intracellularly cleavable linkages are typically inserted within the polymeric chain, affording a dynamic structure that reveals the nucleic acid payload in response to a site-specific stimulus (Troiber and Wagner, 2011).
In a similar sense, multiblock copolymers impart modularity and enable multifunctionality. As an example, polymeric carriers are often based on the electrostatic condensation and shielding by a cationic polymer such as polydimethylaminoethyl methacrylate (pDMAEA). pDMAEA can then be copolymerized with a second block of p(N-(3-(1H-imidazol-1-yl)propyl)acrylamide (pImPAA) and poly(butyl acrylate) (pBA) that mediates an acid-triggered endosomal escape. PImPAA and PBA were designed based on viral membranolytic peptides, and they disrupt the endosomal membrane synergistically through electrostatic and hydrophobic interactions, respectively (Truong et al., 2013; Gillard et al., 2014). Such cationic polymer-based carriers serve as valuable tools for assessing the potency of nucleic acids under study. At this time, structural heterogeneity, imprecise surface conjugation, lack of structure-function insights, and cytotoxicity at therapeutically effective formulations currently hamper their clinical utility (Lv et al., 2006; Troiber and Wagner 2011).
Encapsulation by Lipid-Based Vectors
Nucleic acid protection through charge neutralization and condensation by cationic materials may only provide partial nuclease resistance (Moret et al., 2001). Moreover, additional encapsulation by lipid membranes to form lipopolyplexes has been shown to enhance protection from nucleases and the overall therapeutic efficacy of nucleic acids (Yen et al., 2018). For this reason, lipid-based vectors such as liposomes and solid lipid nanoparticles are commonly explored as nucleic acid carriers (Barba et al., 2019). Compared to other nucleic acid delivery systems, lipid-based carriers offer ease of manufacturing and scalability. Their lipid formulation mimics the lipid bilayer, imparting biocompatibility and conveniently facilitating cellular uptake (Ghasemiyeh and Mohammadi-Samani, 2018).
Among these, liposomes have shown the most promise (Barba et al., 2019). They are spherical vesicles made of a lipid bilayer with an aqueous core (Kulkarni et al., 2018; Barba et al., 2019) and can be designed to carry both hydrophilic and lipophilic cargo (Ghasemiyeh and Mohammadi-Samani, 2018; Barba et al., 2019). The earliest work demonstrating liposome-mediated gene delivery was in 1980 by Fraley et al. (1980) when SV40 DNA was encapsulated and delivered using large unilamellar vesicles. They found that using PS exhibited the highest delivery efficiency. Felgner et al. (1987) then showed that using synthetic cationic lipids such as DOTMA resulted in a higher transfection efficiency. Since then, cationic lipids bearing different structure modifications such as DOTAP, DOSPA, DMRIE, and DL-cholesterol have been incorporated in liposome-based gene delivery systems (Zhi et al., 2013; Yin et al., 2014). For anionic cargo such as nucleic acids, the cationic head group permits condensation of the large biomolecule (Zhi et al., 2013). Moreover, polycationic head groups such as polyamines can be used to form polycationic liposomes. These combine the ability of cationic liposomes to complex nucleic acids and that of polycations to mediate endosomal escape via the proton sponge effect (Yamazaki et al., 2000; Sugiyama et al., 2004; Asai et al., 2011; Yonenaga et al., 2012). Nonionic lipids such as fusogenic DOPE and cholesterol can also be incorporated into the liposome to further enhance its stability and delivery efficiency (Wasungu and Hoekstra, 2006).
Modular release usually centers on the lipid formulation where the lipid envelope is destabilized either by an external stimulus such as temperature or an cellular stimulus such as low pH (Heidarli et al., 2017; Aghdam et al., 2019). As an example, Yatvin et al. (1978) introduced the idea that liposomes can preferentially release cargo at the diseased site in response to mild hyperthermic temperature (around 40°C). This was initially achieved using DPPC alone or with DSPC, which has a phase-transition temperature of 42–44°C, above which its membrane permeability increases (Kono et al., 2010; Aghdam et al., 2019). Among efforts that followed on the construction of heat-responsive liposomes (Matsumura and Maeda, 1986; Tomita et al., 1989; Maruyama et al., 1993; Gaber et al., 1995; Anyarambhatla and Needham, 1999; Needham et al., 2000), Anyarambhatla and Needham (1999) notably incorporated a lysolipid to DPPC to bring down the phase-transition temperature to a clinically achievable range (39–40°C) and initiate release within tens of seconds (Needham et al., 2000). As this design only achieved 50% cargo release within an hour at 42°C (Needham et al., 2000), succeeding studies focused on modulating the temperature-responsiveness of liposomes. One strategy is the incorporation of thermosensitive polymers that can impart a sharp and tunable phase transition temperature to the liposome. Upon heating, the polymeric components form hydrophobic domains that disrupt the lipid bilayer (Kono et al., 2010).
On the other hand, pH-sensitive liposomes exploit the differential acidification in the vicinity of malignant tumors or within endosomes for controlled release via membrane fusion or destabilization (Yatvin et al., 1980; Budker et al., 1996; Heidarli et al., 2017). Earlier anionic pH-responsive designs were constructed with a bilayer rich in PE that is stabilized by anionic lipids containing carboxylate head groups at physiological pH (Budker et al., 1996). PE typically forms an inverted hexagonal phase on its own (Chernomordik et al., 1995). Thus, when the anionic carboxylate head groups are protonated in a region of lower pH, the PE-rich bilayer is disrupted (Budker et al., 1996). While there were reports on using anionic liposomes for nucleic acid delivery (Wang and Huang 1989; Legendre and Szoka 1992), their negative charge limits both the efficient packing of polyanionic nucleic acids and interaction with the negatively charged cellular membrane. For this reason, cationic pH-sensitive liposomes were developed. These contain a weakly basic lipid component such as DOTAP and DODAP that have a pKa slightly below physiological pH (Budker et al., 1996; Sato et al., 2012).
Certain early formulations of lipid-based carriers were limited in part by toxicity and immunogenicity at high lipid concentrations, as well as by low bioavailability and low biodistribution (Zatsepin et al., 2016; Huggins et al., 2019). Overtime these formulations have been significantly improved. In addition, the ease of lipid synthesis and structural modifications permit thorough studies on structure-activity relationships and thus, enable a guided design of more efficient and safe delivery systems (Zhi et al., 2013). Furthermore, lipid-based carriers can be easily decorated with receptor ligands to target specific cell types such as tumor and angiogenic endothelial cells (Yonenaga et al., 2012). Such studies culminated in 2018 with the success of Patisiran (ONPATTRO®), a liposomal vector developed by Alnylam Pharmaceuticals, as the first US Food and Drug Administration approved synthetic carrier of siRNA into cells (Adams et al., 2018; Hoy, 2018; Wood, 2018).
Chemical Modifications
Chemical modifications may impart one or more of the following: in vivo stability, cellular delivery, reduced immunogenicity, and potency through enhanced target binding affinity (Judge et al., 2006; Corey, 2007; Whitehead et al., 2009). Such modifications may alter the phosphodiester backbone (phosphothiorates, boranophosphates, and locked nucleic acids), the ribose sugar (2′ modifications, 4′ thio), or the base (ribodifluorotoluyl nucleotide) (Corey 2007). In particular, 2′-O-modifications on siRNA impart nuclease resistance (Whitehead et al., 2009) and suppression of sequence-dependent immunostimulation by some sequences (Judge et al., 2005; Judge et al., 2006). Furthermore, Jackson et al. (Jackson et al., 2006) showed that by specifically modifying position two in the siRNA guide strand, off-target binding of other transcripts to the seed region is reduced. In addition, uncharged nucleic acid mimics such as peptide nucleic acids and morpholino oligomers present unique chemical properties and may improve biodistribution and efficacy. Details on the structure, properties, and applications of chemically modified nucleic acids and DNA/RNA mimics have been extensively reviewed elsewhere (Karkare and Bhatnagar, 2006; Summerton, 2006; Corey, 2007; Chery, 2016).
Utility of Inorganic Nanoparticles
Inorganic nanoparticles are emerging as appealing synthetic vectors for nucleic acid delivery owing to their unique properties such as tunable size and surface properties, multifunctional capabilities, chemical and thermal stability, and low inherent toxicity (Loh et al., 2015; Ding et al., 2014a). Incorporating nucleic acid cargo into inorganic nanoparticles can be accomplished using the following general strategies: complexation between negatively charged nucleic acid material and positively charged inorganic nanoparticle, direct conjugation of nucleic acid onto the inorganic particle with a stimuli-responsive linker, and addition of cationic amphiphilic polymer to facilitate the assembly formation between the inorganic nanoparticle and the nucleic acid (Loh et al., 2015).
Another approach to protect and deliver nucleic acid cargos is via encapsulation using metal-organic frameworks (MOFs) (Liang et al., 2015; Li Y. et al., 2019; Poddar et al., 2019; Tolentino et al., 2020). These are porous structures built from metal ions or metal clusters linked by organic ligands (Li G. et al., 2019). The nucleic acid can be accommodated in the MOF structure through electrostatic and coordination interactions. Such physical confinement and the characteristic positive surface charge of MOFs offer effective protection of nucleic acid cargo against enzymatic degradation, which is, in many ways, analogous to viral capsids (Li Y. et al., 2019; Poddar et al., 2019).
While viruses deliver their nucleic acid cargo mostly through vesical fusion with the aid of some membrane fusion proteins (Harrison, 2008), inorganic nanoparticles do so with more complexity and hence present some formidable challenges. To achieve intracellular response, the nucleic acid cargo preferably needs to disassemble from the inorganic nanoparticle construct and escape the endosome. The mechanism by which these events (cell internalization and endosomal escape) occur depends on the identity and properties of the inorganic core, chemistry of the conjugation technique utilized, and response of other nanoparticle components to cellular or external stimuli (Sokolova and Epple, 2008). For example, magnetic iron oxide (Fe3O4) nanoparticle, when utilized as a delivery vehicle, can be stimulated to produce oscillating magnetic fields which could then promote more efficient endocytosis (Fouriki and Dobson, 2014). Furthermore, the inclusion of cell penetrating peptides and cationic amphiphilic polymers (e.g. polyethylenimine) as transfecting components assists in the endosomal escape via membrane destabilization and osmotic swelling, respectively (Thomas and Klibanov, 2003; Dowaidar et al., 2017). On the other hand, biocompatible MOFs like Zeolithic Imidazolate Framework-8 (ZIF-8) possess a hydrophobic and positively charged surface (Zhuang et al., 2014), which enable them to interact with the cell membrane and enable internalization through endocytosis.
A promising use of a metal nanoparticle for nucleic acid delivery is exemplified by spherical nucleic acids (SNAs). SNAs radially display a high density of nucleic acids around a spherical nanoparticle. The introduction of high concentrations of salt masks the polyanionic backbone of the nucleic acids, permitting clustering around a very small surface area (Mirkin et al., 1996; Cutler et al., 2011; Cutler et al., 2012). Moreover, the attachment of nucleic acids to a scaffold enhances their target binding affinity to complementary nucleic acids by restricting their conformational flexibility, reducing the entropic cost of binding (Lytton-Jean and Mirkin, 2005). SNAs have low immunogenicity (Massich et al., 2009) and are readily taken up by cells (Cutler et al., 2011) via caveolin-dependent endocytosis (Choi et al., 2013), eliminating the need for potentially toxic transfection agents (Cutler et al., 2011; Cutler et al., 2012). Unlike the abovementioned examples of inorganic nanoparticles, SNAs do not rely on complexation nor encapsulation to protect their nucleic acid cargo (Mirkin et al., 1996; Cutler et al., 2011; Cutler et al., 2012). The mechanism by which they protect nucleic acids is discussed more in Section Self-Generated Sterics.
Self-Generated Sterics
The overall 3D architecture of spherical nucleic acids (SNAs) imparts nuclease resistance through steric-shielding and enhanced local ionic strength (Seferos et al., 2009). This sterics-based mechanism of nucleic acid protection has defined an entire class of nucleic acid delivery systems. These nucleic acid displaying nanomaterials or NADNs, have recently been reviewed by Gudipati et al. (2019). While the metallic gold core provides a means of sensing and tracking the intracellular fate of the nanoconstructs (Mirkin et al., 1996; Cutler et al., 2012), it has limited therapeutic use. Thus, later generations of SNAs that have been developed contain biocompatible cores such as such proteins (Brodin et al., 2015; Samanta et al., 2020) and liposomes (Banga et al., 2014).
Designed to build upon the successful properties of SNAs, NADNs utilize densely packed oligonucleotides around a scaffold, enhancing oligonucleotide stability and permitting scavenger-mediated endocytosis but are built upon biodegradable core materials. The scaffolds of reported NADNs are chemically diverse (Rush et al., 2013; Banga et al., 2014, 2017; Awino et al., 2017; Ding et al., 2018; Roloff et al., 2018; Ruan et al., 2018) and can be programmed for responsiveness to biochemical stimuli (Awino et al., 2017; Santiana et al., 2017). For example, our lab developed nucleic acid nanocapsules (NANs) comprised of nucleic acids photochemically tethered to the surface of stimuli-responsive, crosslinked micelles (Awino et al., 2017; Santiana et al., 2017).
Overall, this section underscores that virus particles are metastable machines built to protect the viral genome and that its overall responsiveness to the environment enables it to carry out its function as an infectious particle. In a similar fashion, nonviral synthetic carriers are designed to protect nucleic acid cargo and facilitate controlled release. Table 1 provides a summary of the structures and cellular trafficking of viral and nonviral carriers. Similar to viruses, functional components (as summarized in Table 2) are incorporated into the design of nonviral vectors that facilitate cellular entry (Section Cellular Targeting, Attachment, and Entry), endosomal escape (Section Cytosolic Delivery), and nuclear delivery (Section Nuclear Delivery).
Cellular Targeting, Attachment, and Entry
Tropism is the ability of viruses to target specific cell types by binding their surface protein or peptide ligands to specific host cell receptors. The elaborate means with which they make use of these ligands accounts for their cell target specificity and high uptake efficiency (Ni et al., 2016). Mechanisms governing the targeting and specific uptake of viruses and nonviral vectors alike rely on the use of electrostatic forces, multiple receptors for enhanced specificity, and multivalent interactions.
Receptor Ligands are Central to the Molecular Mechanisms of Targeting, Attachment, and Entry
Prior to entry, viruses often adhere to the cell surface via non-specific electrostatic interactions involving viral surface components (i.e. membrane glycoproteins) and negatively charged sugars (i.e. heparin sulfate) attached on the target cell surface (Grove and Marsh 2011; Mazzon and Marsh 2019). Though such interactions may lack specificity, they provide the virus an initial foothold on the cell before recruiting specific cell receptors and facilitating entry (Grove and Marsh 2011). Most viruses, which include influenza virus, coronavirus, reovirus and polyomavirus, utilize the sialic acid receptors on the host cell surface for initial attachment (Maginnis 2018). Taking inspiration from this virus behavior, a number of delivery methods have either functionalized nucleic acid cargo with sialic acid (St-Pierre et al., 2016) or encapsulated them in nanocarriers decorated with sialic acids on the surface (Tang et al., 2019). A notable example of the latter strategy is demonstrated in the work of Tang et al. (2019). In their study, they have successfully delivered reporter (luciferase) and functional (antitumor p53) mRNAs to cancer cells using a liposomal nanoparticle containing surface sialic acids. Other than sialic acids, viruses utilize a plethora of receptor ligands which are proteoglycans (i.e. cell adhesion molecules) and lipids (i.e. PS) by nature, to mediate cellular attachment and entry (Maginnis, 2018). On the other hand, synthetic vectors make use of a more chemically diverse array of ligands but mostly for targeting purposes.
Targeted delivery is desired for synthetic vectors as it confers safety, efficacy, and efficiency. It limits the release of the therapeutic to diseased cells or tissues, minimizing adverse off-target effects that could outweigh therapeutic benefits. Secondly, it enhances efficacy by localizing a high concentration of the drug to a specific site. Third, efficiency is achieved by providing access to sites such as certain cells or subcellular locations (e.g. nucleus) that are normally inaccessible to the therapeutic (Rohovie et al., 2017). Many non-viral strategies have derived targeting domains from viral ligands for specific cell or tissue targeting. For example, the adenovirus-derived RGD peptide has been used to direct the nucleic acid delivery of lipoplexes, dendriplexes, and polyplexes to tumor cells overexpressing integrin αvβ3 on the cell surface (Danhier et al., 2012). The successful delivery of RGD-conjugated ASOs to melanoma cells has also been demonstrated (Alam et al., 2008; Juliano et al., 2008; Kang et al., 2008; Juliano et al., 2011). An RGD-based polycationic liposome was also developed to specifically target cancer cells and angiogenic endothelial cells (Yonenaga et al., 2012).
Other ligands of non-viral origin also offer targeting properties. For example, monoclonal antibodies have a been highly effective at targeting delivery of cytotoxic drugs to cancer cells (Sievers et al., 2001; Krop et al., 2010; Younes et al., 2010). Their ability to specifically and avidly bind to cell-specific receptors makes them equally viable targeting domains for biologics such as therapeutic nucleic acids. Their use in directing nucleic acid carriers has been demonstrated in several studies (Palanca-Wessels et al., 2011; Ngamcherdtrakul et al., 2015; Moffett et al., 2017; Huggins et al., 2019; Nanna et al., 2020). They can be either directly conjugated to the nucleic acid (Huggins et al., 2019; Nanna et al., 2020) or to the vector (Palanca-Wessels et al., 2011; Ngamcherdtrakul et al., 2015; Moffett et al., 2017). Antibody-RNA conjugates (ARCs) are promising in that they overcome possible limitations of nanoparticle-based formulations such as poor diffusivity, toxicity, and immunogenicity while still significantly extending the half-life of the cargo (Nanna et al., 2020). Earlier conjugation methods for therapeutic attachment to antibodies involve nonselective conjugation to lysine or cysteine residues. Consequently, prior formulations suffer mainly from product heterogeneity (Huggins et al., 2019). Recently published works on ARC synthesis involved highly specific mechanisms for conjugation, giving a precise drug:antibody ratio of 2 (Huggins et al., 2019; Nanna et al., 2020).
Nucleic acid aptamers offer another promising approach in delivering nucleic acid cargos to specific cell-types (Dassie and Giangrande, 2013). Aptamers are short, chemically synthesized, single stranded oligonucleotides (DNA or RNA), which adopt a specific three-dimensional (3D) structure and bind to their ligands with high affinity (KDs in the pico-to nano-molar range) (Sun et al., 2014). Although aptamer-nucleic acid conjugates possess no innate mechanisms for endosomal escape on their own, aptamers can be conjugated on to nucleic acid carriers with endosomal escape activity as a way to improve cell specific targeting (Yan and Levy, 2018). For example, Zhao et al. (2011) designed a nanocomplex composed of a cationic PEI core endosomal escape component, CD30 RNA aptamer targeting lymphoma cells and siRNA that inhibits the expression of anaplastic lymphoma kinase (ALK). Such an assembly was proven to selectively bind lymphoma cells, deliver the siRNA intracellularly, silence ALK expression, and arrest the growth of lymphoma cells (Zhao et al., 2011).
Lastly, small molecules are commonly used as targeting ligands as they are easily synthesized at a modest cost. They are more stable than biological ligands such as aptamers and peptides, and their conjugation is often relatively simple. However, these molecules are often not the natural ligands of the target cell receptors and thus have lower affinity and specificity for a given receptor, the latter giving rise to off-target effects. Nevertheless, the relative structural simplicity and functional designability of small molecules make them attractive and viable targeting domains (Friedman et al., 2013).
For example, folate (Vitamin B9) is widely used for targeting folate receptor-positive cell lines, with a high affinity (KD = 1 nM) and minimal toxicity. Folate-functionalized vectors are typically internalized via receptor-mediated endocytosis, but reduced folate carriers, though having lower affinity, directly enter the cytosol. Folate-expressing imaging agents are currently in Phase I and Phase II clinical trials, but they are not yet clinically approved for targeting therapeutic nanoparticles (Sikorski et al., 2015).
Likewise, benzamides (anisamide, in particular) target sigma receptors that are upregulated in cancer cell lines. Benzamide analogues can also target dopamine receptors selectively. So far, these have been used to deliver small molecule drugs such as doxorubicin encapsulated in liposomes but have not been explored in gene-delivery yet (Banerjee et al., 2004; Mach et al., 2004).
Multivalent Interactions Facilitate Cellular Uptake
Multivalent interactions between the viral ligands and host cell surface receptors not only amplify the strength of the interaction but also promote viral entry. This is exemplified by the influenza virus where the interaction of multiple capsid protein trimers (2-4 per 100 nm2) with spatially concentrated sialic acid functionalities on the surface of the host cell (50–200 per 100 nm2) is necessary for effective attachment and uptake (Mammen et al., 1998). Apart from high surface density, the spatial arrangement of the ligands is equally important. For example, the internalization of the simian virus 40 (SV40) necessitates the pentameric presentation of its viral capsid protein one to successfully bind to the cell-surface GM1 receptors and facilitate endocytosis (Ewers et al., 2010).
This parallels with carbohydrate-based delivery systems such as siRNAs and ASOs conjugated to N-acetylgalactosamine (GalNAc) for hepatic targeting. GalNAc involves multi-site interactions with asioglycoprotein receptors (ASPGR) of hepatocytes, facilitating endocytosis. (Nair et al., 2014; Debacker et al., 2020). In 2019, Alnylam’s givosiran (GIVLAARI®) was the first US Food and Drug Administration approved GalNAc conjugate for acute hepatic porphyria, and other conjugates are underway (Debacker et al., 2020). ASPGR is a liver-specific receptor that has been targeted for hepatic-directed therapeutics. It is a heterooligomeric complex that is capable of interacting with multiple GalNAc molecules (Meier et al., 2000). The strong binding affinity of monomeric GalNAc with ASPGR is in the micromolar range, and the avidity of the interaction can be enhanced by 103 to 105, depending on the number and spacing of GalNAc units (Lee and Lee 2000). Specifically, the structure of ASPGR was found to optimally bind three divergent GalNAc residues (Lee and Lee 2000) spaced from a common branch point by 14–20 Å and separated from each other by 15–20 Å (Lee et al., 1983; Khorev et al., 2008).
Other synthetic vectors having multivalent interactions with cell receptors have been developed to mimic viral behavior and have shown an enhanced cellular uptake of the carriers or nucleic cargo. A prime example of this is the study of Nakagawa et al. (2010), wherein they delivered a splice switching antisense oligonucleotide (SSO) directly conjugated to anisamide, a sigma receptor present in plasma membranes, to tumor cells, and investigated their ability to modify the splicing of a reporter gene (luciferase). Mono-anisamide and tri-anisamide conjugates were synthesized, and it was demonstrated that the multivalent conjugate yielded a more enhanced receptor-specific cell uptake and biological effect (Nakagawa et al., 2010). Another study highlighting the beneficial effect of multivalency to nucleic acid cargo internalization is carried out by Kang et al. (2018). In their study, siRNA specific to Bcl2, an anti-apoptotic protein, was tethered to MUC-1- and nucleolin-targeting aptamers and delivered to cancer cells. Fluorescence microscopy revealed the positive correlation between aptamer valency (n = 1, 3, 9) and cellular internalization. Moreover, higher tumor accumulation was observed for multivalent aptamer conjugates compared to mono- and divalent conjugates. These studies underscore the critical need for multivalent interactions in designing delivery systems for nucleic acids.
Attachment to Multiple Receptors Confers Cell Target Specificity and Uptake Efficiency
Maginnis (2018) provides a comprehensive review of how virus interactions with host receptors govern pathogenicity. Worth noting are evolutionarily conserved mechanisms among viruses, redundancy in target primary receptors, and diversity of secondary receptors. One conserved mechanism is the conformational change involved in the sequential binding to multiple receptors that leads to fusion or endocytosis. For instance, the trimeric glycoprotein (GP) complex of the human immunodeficiency virus (HIV) is formed by the GP120/GP41 heterodimer and is necessary for cellular targeting and entry. GP120 binds CD4 on the surface of T-cells, T-cell precursors, macrophages, dendritic cells, and microglial cells. GP120 binding induces a conformational shift in the trimeric GP, revealing a GP120 binding domain specific for one of many chemokine coreceptors such as CXCR4 and CCR5. These coreceptors vary across different cells and thus mainly determine tropism (Fanales-Belasio et al., 2010; Wilen et al., 2012). The involvement of coreceptors form the basis of some anti-viral drugs such as Maraviroc, a US Food and Drug Administration and European Medicines Agency approved HIV/AIDS treatment. It acts by antagonizing CCR5, the secondary receptor of HIV in CD4+ T cells. In particular, maraviroc binding induces a change to the inactive conformer of CCR5 (López-Huertas et al., 2017).
In terms of redundant receptors, integrins are of particular interest because they are commonly involved in the internalization of viruses. Integrins are heterodimeric cell surface receptors that mediate cell adhesion, migration, differentiation, and tumor growth. The binding of a virus to a host induces the clustering and/or structural changes of integrins, resulting in intracellular cues that enhance binding affinity, drive structural changes in the cytoskeleton, and/or facilitate uptake. This is demonstrated by certain viruses such as the adenovirus whose secondary attachment to integrins initiates intracellular signals that ultimately lead to viral uptake (Stewart and Nemerow, 2007). For the human cytomegalovirus, the binding of its glycoproteins to both the epidermal growth factor receptors (EGFR) and integrin on the host cell brings EGFR and integrins into close proximity, eliciting signaling responses that facilitate cellular uptake and nuclear trafficking (Wang et al., 2005).
For synthetic vectors, engaging multiple receptors presents an opportunity for programming more specific and efficient nucleic acid delivery systems. The use of multiple ligands for enhanced specificity and uptake is guided by knowing which receptors are overexpressed in the tissue or region of interest. Just as integrins are often implicated in virus entry, they have become popular targets for drug and gene delivery for their natural abundance, efficient endocytosis, and differential expression on a number of tumor cells and angiogenic endothelial cells (Wang et al., 2010; Juliano et al., 2011). For instance, Nie et al. (2011) developed a synthetic dual-ligand targeted vector in which plasmid DNA is condensed by polyethylenimine (PEI). In this study, they conjugated PEG-ylated PEI-based polyplexes with peptides B6 and arginylglycylaspartic acid (RGD) that target transferrin and integrin, respectively. This strategy exploits the fact that tumor cells overexpress transferrin while vasculature that supply blood to these newly formed tumor cells overexpress integrins. Importantly, RGD-integrin binding stabilizes the B6-transferrin interaction. This design has shown to improve transfection efficiency and specificity. Thus, as illustrated in Figure 3, it demonstrates the power of mimicking the dual-receptor internalization of natural viruses such as the adenovirus, herpes simplex virus, and SV40 (Hussein et al., 2015).
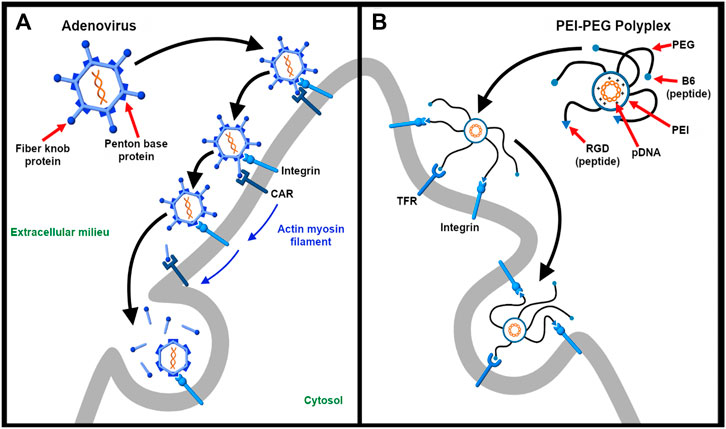
FIGURE 3. Targeting multiple receptors enhances cellular specificity and transfection efficiency. (A). The entry of adenovirus into the host cell occurs in a three-step process – binding, drifting, and shedding. First, the adenovirus binds to the Coxsackievirus and adenovirus receptor (CAR) of the host cell surface through fiber knobs jutting out the vertices of the icosahedral shaped viral capsid. Second, acto-myosin drifting of the virus-bound CAR receptor leads to internment of the penton base protein of the viral capsid by integrins expressed on the cell surface. Third, the slow drifting motion (0.1 μm/s) of the CAR receptor and the stable nature of binding causes mechanical stress onto the viral capsid, the first uncoating step in the capsid disassembling process. The protein VI of the inner capsid is exposed which makes lesions in the plasma membrane and undergoes integrin-dependent endocytosis (Burckhardt et al., 2011) (B). As described by Nie et al. (Nie et al., 2011), a synthetic dual-ligand targeted vector system was constructed using a cationic polymer PEI to deliver pDNA. PEG moieties were used to shield the charge of the polyplex. Inspired from natural viruses, the polyplex was conjugated with Transferrin receptor (TFR)-binding B6 peptide and integrin-recognizing RGD sequence for dual targeting purpose. The receptor specificity of the dual targeted polyplex shows increased gene transfection as compared to the single targeting peptide. The integrin receptor binding helps in cellular association and the vector is internalized via TFR-mediated endocytosis.
In another study, Dong et al. (2018) depict the dual targeting ability of RGDK peptide sequence. In this particular example, they designed a siRNA/amphiphilic dendrimer complex decorated with a dual targeting peptide RGDK. The design of the targeting peptide is such that it protects and stabilizes the siRNA-dendrimer complex by electrostatic interaction. Similar to Nie et al.’s study, the RGD part binds to target integrin receptors on tumor vasculature while the full length RGDK interacts with neuropilin-1 (Nrp-1), which is expressed on tumor cells, thereby enhancing cellular uptake.
Cytosolic Delivery
For a virus to deliver its genome to the cytosol or nucleus, it needs to penetrate either the cellular membrane or a subcellular membrane within the cytoplasm such as the endo-lysosomal membrane. This section talks about how viruses and synthetic carriers alike manage to bring their nucleic acid cargo into the host cell interior with mechanisms to overcome cellular barriers.
Direct Cytosolic Delivery
Some enveloped viruses such as HIV are able to directly translocate their genome into the cytosol via cell membrane fusion. As mentioned in Section Attachment to Multiple Receptors Confers Cell Target Specificity and Uptake Efficiency, the binding of the HIV glycoprotein to its primary receptor drives structural changes within the glycoprotein, facilitating a subsequent interaction with a coreceptor that then mediates viral entry (Wilen et al., 2012). Binding to two receptors enhances the strength of viral attachment (Grove and Marsh, 2011; Ni et al., 2016), and for HIV, this allows the N-terminal fusogenic peptide of GP41 to penetrate the membrane. The heptad repeats of GP41 interact to form a hairpin loop, facilitating the fusion of the viral and host cellular membranes (Chan et al., 1997; Fanales-Belasio et al., 2010).
For nonviral carriers, a particle can also be designed such that it directly transfects cargo to the cytosol. For instance, Motion et al., (2012) reported a promising phosphatase-triggered liposome carrier that was directly inspired by HIV. It incorporates an inactive phosphorylated version of the GP41 peptide that, when dephosphorylated, shifts to its fusogenic alpha-helical conformer. The phosphorylated form, on the other hand, has an increased random coil structure that is unable to interact with a lipid membrane. Since phosphates are overexpressed and secreted by diseased tissues, the fusogenic peptide is activated in a diseased cell, facilitating fusion with the plasma membrane and targeted cytosolic delivery. Such system has great potential as a nucleic acid carrier. Additionally, studies have shown that exogenous miRNA (Vickers et al., 2011) and siRNA (Shahzad et al., 2011; Ding et al., 2014b) can be directly delivered to the cytosol of target cells using endogenous or reconstituted high density lipoprotein by targeting scavenger receptor B1 (Shahzad et al. 2011).
In addition, siRNA (Jiang et al., 2015; Jiang et al., 2018) and CRISPR-Cas9 ribonucleoprotein (CRISPR-Cas9-RNP) (Mout et al., 2017) can be directly transfected across the cell membrane using nanoparticle-stabilized nanocapsules (NPSCs). Previously shown to mediate the direct cytosolic delivery of small molecules (Yang et al., 2011) and proteins (Tang et al., 2013), NPSCs are formed by assembling a preformed complex of nucleic acids and arginine-coated nanoparticles on the surface of an oil droplet (Jiang et al., 2015). The inorganic- and lipid-based hybrid construct efficiently delivered nucleic acid cargo to the cytosol with an siRNA knockdown efficiency of 90% (Jiang et al., 2015; Jiang et al., 2018) and to the nucleus with a CRISPR-Cas9-RNP gene editing efficiency of 30% (Mout et al., 2017). In vivo assays of spleen-directed siRNA loaded NPSCs showed good selectivity and immunomodulatory activity, demonstrating the potential for targeted delivery (Jiang et al., 2018).
Endosomal Escape
Most viruses and synthetic nucleic acid carriers are internalized via endocytosis. While viruses manage to escape into the cytosol efficiently, synthetic carriers pale in contrast (Ramamoorth and Narvekar, 2015), only having around 1–2% endosomal release (Gilleron et al., 2013). Thus, endosomal escape is the bottleneck of nucleic acid delivery and ultimately determines therapeutic efficiency (Gilleron et al., 2013; Shete et al., 2014; Selby et al., 2017).
While direct fusion with the plasma membrane may seem simpler, endocytosis offers several advantages—one being evasion of molecular crowding in the cytosol and microtubule-assisted shuttling to the nucleus or other subcellular locations (Barrow et al., 2013). Furthermore, as endocytosis is often linked to signaling cascades, the invading particle can influence its intracellular fate by targeting the appropriate receptor (Nemerow and Stewart, 1999; Marsh and Helenius, 2006). For viruses, endocytosis can lower the risk of triggering an immune response because rapid endocytotic uptake minimizes the exposure of viral immunogenic epitopes to the extracellular milieu (Miyauchi et al., 2009). Importantly, the physical integrity of the viral capsid is responsive to both chemical and mechanical stimuli brought about by interactions with the host. This provides a basis for disassembly once the genome has reached its target site (Greber 2016; Yamauchi and Greber, 2016). Similarly, endocytosis enables opportunities to embed responsiveness of a nonviral carrier to endolysosomal cues. For these reasons and the overwhelming tendency for nonviral carriers to undergo endocytotic entry, research efforts are more directed toward enhancing endosomal escape efficiency.
Cellular Cues Drive Endosomal Escape via Membrane Fusion or Penetration
Staring et al. (2018) provides an excellent discussion of how viruses carry out endosomal escape to avoid degradation or recycling. For their remarkable endosomal escape efficiency, viruses have served as templates for engineering the endosomal escape mechanism of non-viral vectors. A unifying theme is a conformational change in viral structural proteins that drives viral and endo-lysosomal membrane fusion for enveloped viruses or membrane penetration by nonenveloped viruses. These structural rearrangements are triggered by cellular cues such as low pH or acid-dependent proteolytic activity. Such viral proteins or peptides contain ionizable groups such as critical histidine residues whose imidazole groups (pKa∼6) are protonated as the pH drops in the endosome. These histidine residues act as pH sensors involved in pH-dependent structural changes of the protein or peptide as observed for the surface protein hemagglutinin (HA) glycoprotein (GP) of the influenza virus. Moreover, they also serve as internal buffers. This “proton sponge” effect leads to endosomal swelling and rupture. For this reason, histidine residues (5–20) are added to peptide domains (such as TAT) of nucleic acid carriers (Lo and Wang, 2008). A research study by Meng et al. (2016) has discussed a multifunctional peptide-based nanocarrier composed of different peptide fragments—a CPP segment (TAT) for cell penetration, an ELMD segment for endo-lysosomal membrane disruption, and stearyl moieties to improve hydrophobicity and cell membrane binding ability of the peptide-DNA complex. For the ELMD segment, six histidine resides were inserted to increase endosomal escape by “proton sponge” effect. All these amino acids were dextrorotatory to protect the DNA/peptide nanocarrier from proteolysis.
Membrane Fusion
For the endosomal escape of enveloped viruses, the influenza virus is a classic model (Figure 4A). The fusogenic HA has been used or mimicked as an endosomal escape domain. Following endocytosis, the acid-triggered proteolysis induces the conformational change of the viral GP spike. This exposes the hydrophobic subunit HA2 that facilitates the endosomal escape of the ribonucleoprotein contents into the cytosol (Pinto et al., 1992). Specifically, endosomal acidification induces a conformational change in HA that sequesters charged residues glutamate-15 and aspartate-19. This reveals a V-shaped HA conformer with a hydrophobic pocket that penetrates deeply into the endosomal membrane. The enhanced penetration increases the lateral pressure in the hydrophobic pocket and the surface tension at the interface of the viral and endosomal membranes. Altogether, these drive the hemifusion of the two lipid membranes (Han et al., 2001).
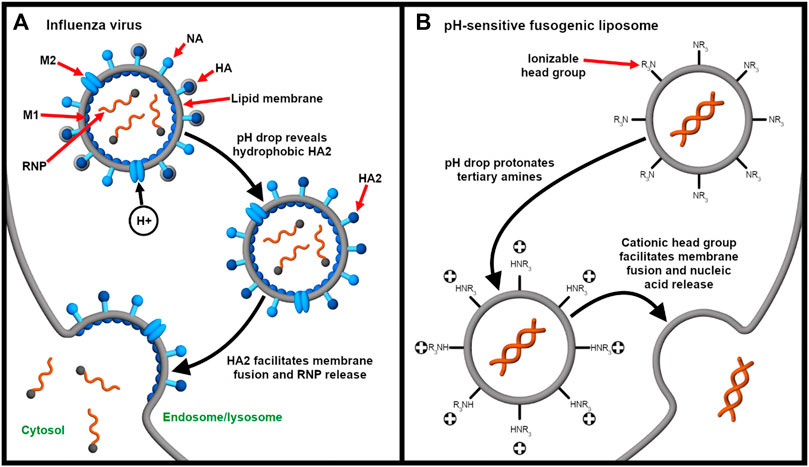
FIGURE 4. Endocytosis provides an opportunity for integrating stimulus-responsive nucleic acid release. (A) The influenza virus releases its genome (complexed with nucleoproteins, gray spheres) into the cytosol in a pH-dependent manner. Endosomal acidification drives the influx of protons through the Matrix Protein 2 (M2) ionophore. This liberates the ribonucleoprotein (RNP) complex from Matrix Protein 1 (M1) and exposes the fusogenic subunit HA2, which, in turn, facilitates fusion of the viral and endosomal membranes (Pinto et al., 1992). Neuraminidase (NA) enables release of the influenza virus from the host cell after replication (James and Whitley 2017). (B) On the other hand, pH-responsive fusogenic liposomes are composed of ionizable lipids with weakly basic head groups that are rapidly protonated as the pH drops in the endosomes. This enables the protonated lipids to promote fusion and nucleic acid release before lysosomal degradation (Budker et al., 1996; Kogure et al., 2008; Sato et al., 2012).
Synthetic HA2 analogs have demonstrated improved endosomal escape ability (Ye et al., 2012). Ye et al. (2012) developed and studied different types of fusogenic peptides (HA2, R8, TAT, TAT-HA2, and TAT-R8) by conjugating them to gelatin-silica nanoparticles (GSNPs). These GSNPs were used to deliver plasmid DNA and their endosomal escape efficiency was measured and compared. They concluded that the endosomal escape efficiency of TAT-HA2 conjugate was superior as compared to others. Moreover, the concentration of the peptide dictates the extent of its interaction with the membrane. While the peptide domains only engage the membrane electrostatically at low concentrations, pore formation is observed at higher concentrations.
The endosomal escape of the influenza virus can be largely ascribed to the sequestering of the hydrophilic cap of HA to reveal a hydrophobic domain HA2 that then engages the endosomal membrane. This mechanism has inspired Lönn et al. (2016) to develop endosomal escape domains (EEDS), which are hydrophobic peptides containing tryptophan and phenylalanine residues. For EED-TAT-siRNA conjugates, the presence of indole and/or phenyl rings at an optimal distance of six PEG units from the TAT domain is able to significantly enhance the endosomal escape of siRNA. Additionally, the concept of hydrophobic unmasking has also been exhibited by nucleic acid nanocapsules. Amphiphilic surfactant-DNA conjugates were constructed to mimic the disassembly products of the nanocapsule. The membrane permeating ability of these conjugates (Hartmann et al., 2018) suggests that the hydrophobic group revealed only after disassembly could facilitate the endosomal escape of the degradation products.
Similarly, pH-sensitive fusogenic liposomes (Figure 4B) have been developed to mimic the acid-triggered endosomal escape of viruses (Budker et al., 1996). Sato et al. (2012) described the delivery of siRNA for gene silencing using low pH-activatable cationic liposomes. The responsiveness to low pH is enabled by using a lipid containing a tertiary amine head group that is almost neutral at physiological pH but is cationic at low endosomal pH (Moriguchi et al., 2005; Kogure et al., 2008; Sato et al., 2012). The lipid also consists of two long linoleyl fatty acid chains, forming cone-shaped molecules that further mediate endosomal escape through membrane fusion. Because the apparent pK of the ionizable lipid is 6.5, rapid membrane fusion and siRNA release is induced in the endosomes before lysosomal degradation occurs (Sato et al., 2012; Sakurai et al., 2014).
Membrane Penetration
Unlike enveloped viruses that possess a lipid envelope capable of fusing with the plasma or endo-lysosomal membrane, nonenveloped viruses make use of membranolytic peptides to escape the endosome. While membrane penetration is not completely understood, the exact mechanism can range from temporary membrane destabilization to pore formation to complete disruption (Staring et al., 2018). The elegance of viral endosomal escape using membranolytic peptides is exemplified by the adenovirus. The mechanical stress caused by binding multiple receptors primes the shedding of the capsid coat (Burckhardt et al., 2011). This liberates membranolytic viral protein VI that then creates small lesions on the plasma membrane. As a response, the host secretes lipid hydrolase acid sphingomyelinase that catalyzes ceramide production for membrane repair. The increased level of ceramide enhances interaction of protein VI with the endosomal membrane, leading to endosomal rupture. This illustrates how the host cell’s natural response to membrane damage is exploited by a virus for it to escape the limiting vesicle (Staring et al., 2018). Moreover, a study by Ortega-Esteban et al. (2015) showed that upon virus maturation, the expansion of the genome stiffens virions. As in the case of the adenovirus, the rise in internal pressure renders the capsid more susceptible to disruption and, thus, contributes to the overall endosomal escape mechanism and eventual uncoating of the virus at the nuclear pore complex (Ortega-Esteban et al., 2015; Greber 2016).
Similarly, the Glutamic acid-Alanine-Leucine-Alanine (GALA) peptide is a targeting and endosomal escape peptide that has been used in siRNA delivery (Subbarao et al., 1987; Kusumoto et al., 2013; Kusumoto et al., 2014). GALA was originally designed to undergo an acid-triggered change from a random coil to a membrane-disrupting alpha helical structure (Subbarao et al., 1987). Later on it was found to target the sialic acid residues on lung endothelium (Kusumoto et al., 2013), making it a promising multifunctional ligand. On the other hand, KALA is a modified version of GALA with alanine to lysine substitutions and reduced glutamic acid content. These features allow DNA condensation, endo-lysosomal disruption, and nucleic acid release (Wyman et al., 1997; Shaheen et al., 2011). Miura et al. (2017) performed a complete study of KALA as a fusogenic peptide. They modified the surface of a DNA-encapsulating liposome with KALA peptide sequences. In this study, they found that as compared to the full-length KALA sequence (27 residues), the short-KALA3 peptide (14 residues) was the shortest KALA peptide to form a α-helical structure at physiological pH. Thus, short-KALA3 can be used to elicit transgene expression (Miura et al., 2017). KALA peptide has also been used before for the delivery of siRNA-PEG conjugates (Mok and Park 2008).
Small Molecules for Enhancing Endosomal Escape Efficiency
The fact that fusogenic or membranolytic peptides are often required to gain cytosolic access underscores the necessity for an endosomal escape component in a drug delivery system. This idea has been extended to various small molecules that can be used as tools to cross the endo-lysosomal membrane either through direct conjugation to or co-delivery with the nucleic acid cargo (Gilleron et al., 2015; Osborn et al., 2015; Maxfield 1982; Juliano et al., 2018; Joris et al., 2018; Du Rietz et al., 2020; Yang et al., 2015; Wang et al., 2017). For example, cationic amphiphilic drugs (CADS) have been shown to enhance siRNA delivery due to their ability to increase the permeability of the endo-lysosomal membrane (Joris et al., 2018; Du Rietz et al., 2020). On the other hand, oligonucleotide enhancing compounds (OECs) are small molecules covalently linked to siRNAs, ASOs, and single stranded oligonucleotides and have been screened for improved cytosolic and nuclear delivery without an external carrier (Yang et al., 2015; Wang et al., 2017). Through a set of structure-activity experiments, hydrophobic phenyl rings, the presence and relative placement of a tertiary amine, and carbamate modifications were identified as essential and tunable features for enhancing the therapeutic availability of the oligonucleotides. How OECs influence the intracellular redistribution of oligonucleotides is not yet clear but, similar to CADs, involves an increase in endomembrane permeability rather than complete disruption. Though the potency imparted by OECs holds great promise, the challenge of enhancing efficacy while minimizing cytotoxicity remains (Juliano et al., 2018).
Additionally, Orellana et al. (2019) reported the use of nigericin, a novel, small molecule endosomal escape agent, to enhance the cytosolic delivery of folate-conjugated miRNA. Nigericin is a proton ionophore that exchanges osmotically inactive protons inside the endosomes with potassium ions in the cytosol. The combined high concentration of sodium and potassium ions raises the osmotic pressure inside the endosomes, resulting in endosomal rupture and release of the miRNA payload.
Intracellular Receptor Targeting as a Potential Endosomal Escape Strategy
For effective host cell infection, the Lassa virus (LASV, Jae et al., 2014) and ebolavirus (EBOV, Carette et al., 2011; Côté et al., 2011; Wang et al., 2016a) escape the endosome via a critical switch from their extracellular receptor (involved in cellular attachment and entry) to an intracellular endo-lysosomal receptor to mediate membrane fusion (Jae and Brummelkamp 2015). This is commonly due to the pH drop in the endosome that primes the viral glycoprotein (GP) for a receptor switch (Staring et al., 2018).
In particular, LASV was found to bind mainly to α-dystroglycan (Cao et al., 1998) as well as TAM receptor tyrosine kinases, DC-SIGN of dendritic cells, and C-type lectins of liver and lymph nodes (Shimojima et al., 2012) and is taken up mainly through macropinocytosis (Oppliger et al., 2016). The trimeric LASV spike protein is composed of a receptor-binding domain (GP1), a fusion protein subunit (GP2), and a unique stable signal peptide (SSP) (Burri et al., 2012) that directs the polypeptide to the endoplasmic reticulum and also interacts with GP2 during membrane fusion (Nunberg and York 2012). Structural studies support an entry model wherein endo-lysosomal pH (5.0–6.0) induces a conformational change in GP1 that facilitates an intracellular receptor switch to LAMP1, a late endosomal/lysosomal protein (Cohen-Dvashi et al., 2015; Li et al., 2016). Further acidification in the lysosomes (pH 4.0) sheds GP1, exposing GP2 that mediates membrane fusion (Li et al., 2016). The pH-dependence of the conformational change is attributed to the pH-sensing histidine triad on the surface of the spike protein (Cohen-Dvashi et al., 2015; Cohen-Dvashi et al., 2016). Mutation of these His residues reveals that LAMP1 binding is not necessary for membrane fusion but greatly enhances viral infection efficiency (Cohen-Dvashi et al., 2016).
Similarly, attachment of EBOV to the host cell membrane facilitates internalization principally through macropinocytosis (Nanbo et al., 2010), with evidence that the virus is also taken up via clathrin-mediated endocytosis (Aleksandrowicz et al., 2011). Several cell membrane contact sites have been identified that seem to facilitate virus attachment such as β1-integrins and Tyro3 (TAM) family kinase receptors, but no sites for direct interaction with the EBOV GP have been identified yet. C-type lectins (L-SIGN, DC-SIGN, and hMGL) have also been shown to enhance adherence of the virus to the host cell membrane. Due to the broad tropism of EBOV across different cell types and different host organisms, it has been difficult to identify cell surface receptors that facilitate internalization (Hunt et al., 2012). So far, TIM-1 was determined to be the EBOV receptor for epithelial cells (Kondratowicz et al., 2011). Upon entry, endo-lysosomal acidification activates proteases cathepsin B and cathepsin L that cleave the EBOV GP. Proteolysis reveals the active conformer GP2, which then binds to Niemann-Pick C1 (NPC1), a cholesterol transporter embedded on the endo-lysosomal membrane. This interaction facilitates the fusion of the viral and lysosomal membranes, releasing the viral nucleocapsid into the cytosol (Carette et al., 2011).
Because NPC1 is involved in vesicular trafficking, it is even more interesting that it is responsible for limiting lipid nanoparticle-mediated siRNA delivery by shuttling the bulk of the lipid nanoparticles back to the outside of the cell after endocytosis (Sahay et al., 2013). Moreover, inhibition of NPC1 greatly increases the cytosolic delivery of the siRNA cargo (Wang et al., 2016b). A similar effect was observed when ESCRT-1, another endo-lysosomal protein involved in vesicular sorting, was knocked down to enhance the delivery of a therapeutic anti-miRNA (Wagenaar et al., 2015). Alternatively, the entrapment of oligonucleotides in the late endosomes can be exploited. Instead of inhibiting or knocking down endo-lysosomal-associated proteins such as NPC1, LAMP1, or ESCRT-1, a ligand that engages the intracellular receptor can be used to facilitate the cytosolic delivery of the cargo. This could potentially be applicable to lipid-based systems where membrane fusion precedes content release.
Nuclear Delivery
Unlike cytoplasmic viruses, nuclear viruses (such as SV40, adenovirus, influenza virus and HIV) need to travel further in order to replicate themselves in the nucleus of the host cell. They must cross a total of three cell barriers to reach the nucleus—the plasma membrane, cytosol and the nuclear membrane. Thus, they have evolved to use their structural features along with cellular transport machinery to hijack the well-protected nuclear import process. The size, structure, and composition of the viral proteins determines the mechanism by which it enters the nucleus. The structure and surface properties of nuclear viruses are also different from cytoplasmic viruses as the capsid of these viruses needs to be intact when they are traversing through the highly crowded cytosol but should breakdown in the perinuclear area (Cohen et al., 2011; Kobiler et al., 2012).
The nucleus is the main regulator of intracellular functions such as gene activation, cell division and proliferation, metabolism and protein production. As such, it is also considered as the most important target to deliver intact therapeutic exogenous oligonucleotides to treat diseases at the genetic level (Faustino et al., 2007; Pouton et al., 2007). However, cytosolic trafficking is a critical bottleneck for the efficient nuclear delivery of nucleic acids (Ni et al., 2019). Previous studies show that when a plasmid DNA is microinjected into the cytoplasm, the cellular enzymes degrade the DNA before it can reach the nucleus through Brownian motion (Cohen et al., 2009). Thus, it is necessary to protect as well as actively traffic the DNA to the perinuclear region.
To reach the nucleus, a number of different cytosolic trafficking strategies have been explored by nuclear viruses. Among these, the karyopherin-dependent and microtubule-assisted pathways have been extensively studied and mimicked for nucleic acid delivery (Bai et al., 2017). Thus, this section discusses these two common viral nuclear import mechanisms and how these pathways have inspired the development of nonviral vectors for therapeutic and diagnostic purposes (Cohen et al., 2011; Kobiler et al., 2012).
Karyopherin-Mediated Pathway
The nuclear trafficking of the viral ribonucleoproteins (vRNPs) is required for production and release of mature virions. To travel actively toward the nucleus, viruses use nuclear localization signals (NLSs) to mediate nucleus entry of the vRNPS. NLS sequences are short basic peptide motifs that are recognized by karyopherin proteins and are transported to the nucleus via karyopherin α/β-mediated pathway (Cros and Palese 2003). Detailed chemical and biophysical studies show that the influenza A virus, herpes simplex virus, and SV40 consist of these NLS sequences embedded in their viral proteins. These specific sequences interact with the α subunit of dimeric karyopherin α/β receptors with high specificity. The karyopherin α binding site classifies the type of NLS as either classical or nonclassical. The classical NLS (derived from SV40) binds to inner concave surface of the ARM domain of karyopherin α. On the other hand, nonclassical NLS are the viral peptides that bind specifically and exclusively to the minor groove of the karyopherin α (Xu et al., 2005). An example is the NLS obtained from influenza A virus Li G. et al. (2019). The trimeric karyopherin-NLS complex docks at the nuclear pore complex and is passaged across the nuclear envelope and released into the interior. This transport mechanism is based on nucleocytoplasmic gradient of the GTP bound form of Ran protein as the Ran-GTP/GDP ratio is high in the nucleus but low in the cytoplasm. This difference in concentration acts as the driving force to transport the trimeric complex inside the nucleus (Fay and Panté, 2015).
Miller and Dean (2009) summarized nuclear targeting ligands that can be used to deliver therapeutic nucleic acids. These ligands can be easily modified and conjugated to the surface of a nanoparticle or directly to the gene of interest. Variants of virus-derived NLS peptides are most commonly used as nuclear targeting ligands (Kim et al., 2017). Thus, carriers decorated with or nucleic acid cargo associated with the NLS peptide sequence also undergo nuclear uptake via the karyopherin α/β pathway (Pan et al., 2012; Ray et al., 2015; Zanta et al., 1999; Cartier and Reszka, 2002). One such example by Hu et al., 2012 has been discussed in detail in Figure 5 wherein the classical NLS peptide sequence derived from SV40 virus was used to deliver a plasmid DNA (pDNA) polyplex across the nuclear envelope via karyopherin-dependent pathway (Hu et al., 2012). Importantly, the unmasking of NLS peptide in case of SV40 and HIV virus only when it is needed reduces the off-target binding and increases the karyopherin-mediated uptake (Nakanishi et al., 2002; Fanales-Belasio et al., 2010). These kinds of smart techniques can be explored further as current synthetic carriers are designed to deliver the whole construct to the nucleus and not just the nucleic acid cargo (Hu et al., 2012; Favaro et al., 2018).
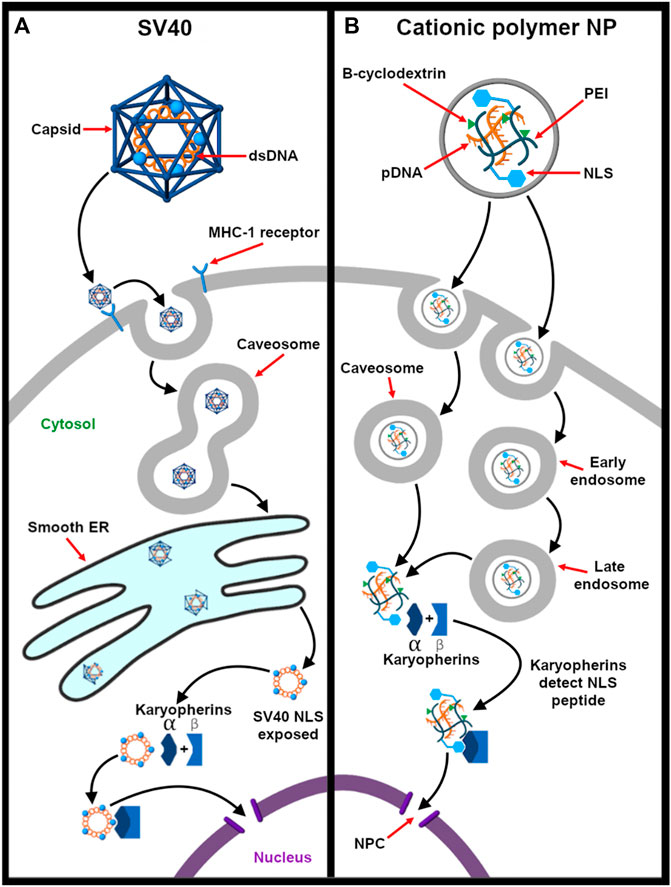
FIGURE 5. Karyopherin-mediated nuclear delivery of SV40 and of a synthetic nanovector. (A) SV40 binds to MHC-1 class receptors present on the host cell surface. This mediates the recruitment of caveolin-1 positive vesicles, and the virus is eventually taken up into caveosomes. These caveosomes undergo dynamic structural changes to form long tubular membrane extensions, which are then released from caveosomes and are transported to the smooth ER (Pelkmans et al., 2001). Once inside the ER lumen, the disassembly of viral capsid begins, and the partially disassembled capsid undergoes structural changes in the cytosol to expose the NLS embedded in the minor capsid. The NLS moiety is recognized by the karyopherin family, and the viral genome is transported to the nucleus as karyopherin cargo (Nakanishi et al., 2007; Toscano and de Haan 2018). (B) In this study by Hu et al. (2012), PEI conjugated to β-cyclodextrin (PC) was used to transfect pDNA. Results shows that it is internalized by caveolae- and clathrin-dependent pathways. To enhance the nuclear delivery of DNA, the NLS peptide inspired from SV40 virus was combined and conjugated to the PC backbone. Compared to PC/pDNA, PC/NLS/pDNA shows higher gene transfection efficiency.
Alternatively, the DNA nuclear-targeting sequence (DTS) is a 72 bp aptamer derived from SV40 and has innate affinity for NLS-tagged cytoplasmic proteins such as transcription factors (TFs) (Gaal et al., 2011). DTS-containing plasmids bind to one or more TFs, and the complex is shuttled into the nucleus. If cells are undergoing proliferation due to injury, the addition of DTS/NLS sequence shows limited effect in gene expression as the guard of the nuclear envelope breaks down (Miller and Dean, 2009). So far, DTS expressing plasmids have been delivered by electroporation or direct injection. Thus, it is possible to use DTS as a targeting ligand for gene vectors but not in vivo. In addition, plasmids complexed with proteins like HMG-1, histone H2B proteins, karyopherin receptors, and nucleoplasmin show increased transgene expression due to nuclear uptake (Miller and Dean, 2009).
Microtubule-Assisted Transport
Many viruses use microtubule (MT) facilitated transport to traverse the cytoplasmic medium. Viral proteins induce rearrangement of microfilaments and recruit molecular motors such as dynein and kinesin to traverse from the plus to the minus terminal of MTs (Döhner et al., 2005). The MT-organizing center nucleates the minus end of the MTs and is close to the nucleus. This is how the viral capsid is transported actively to reach nearby regions of the nucleus (Naghavi and Walsh, 2017). Viruses such as the adenovirus, adeno-associated virus (AAV), and influenza A virus are able to hijack the cellular microtubule transport system, intercepting traffic to the nucleus. Amongst these, the adenovirus and influenza A virus are released out of the endosome before traveling along the microtubule in a non-vesicle dependent manner. In contrast, AAV is transported while within the endosome and the endosomal vesicle ruptures near the nucleus. The ligands that attach the endosomal membrane to the MT system are still currently unknown (Cohen et al., 2011).
In an effort to mimic viruses, the dynein binding protein (DBP) is often used as a ligand for nuclear uptake as it can mediate the transport of cargo via the MT-assisted pathway (Favaro et al., 2014; Favaro et al., 2018). A review by Midoux et al., 2017 has listed the dynein binding viral proteins and selective peptide sequences that have been used for efficient nonviral gene delivery. These peptides help to actively deliver the nanovector to the centrosome wherein the dynein interacts dynamically with the nuclear envelope and rearranges the nuclear lamin protein filaments, thereby increasing the permeability of nucleus (Dalmau-Mena et al., 2018). Moreover, Cohen and Granek (2014) provided theoretical insights on the rational design of spherical nanocarriers that require active transport to the nucleus. One recent example using such pathway is a peptide vector synthesized by Favaro et al., 2018 (2018). In this study, a dynein binding protein (TRp3) was incorporated into the vector to enhance microtubule-assisted delivery of an encapsulated gene toward the nucleus of the cell (Figure 6).
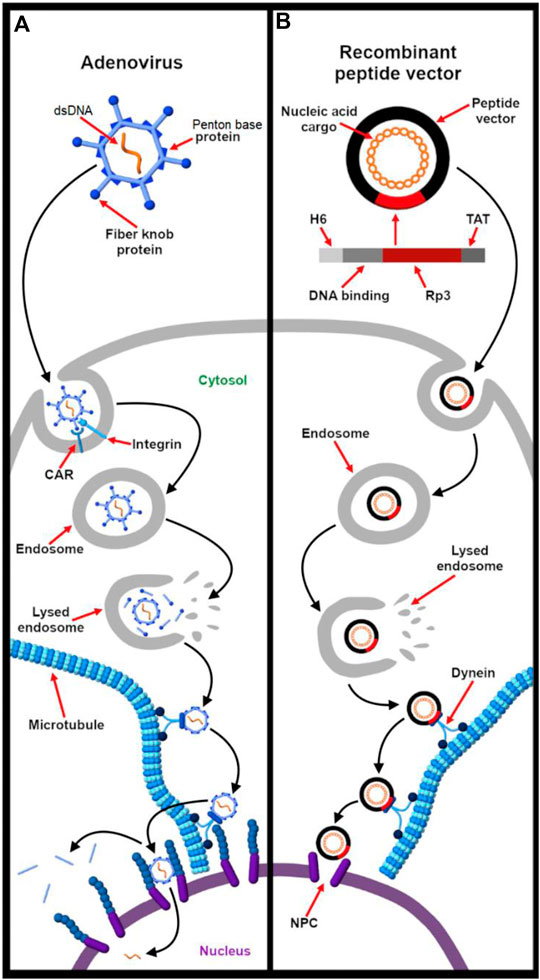
FIGURE 6. Microtubule (MT)-assisted nuclear delivery of adenovirus mimicked by a recombinant peptide vector. (A) Adenovirus undergoes receptor-mediated endocytosis by targeting CAR and integrin receptors present on the cell surface. Once inside the endosome, protein VI contains an N-terminal amphipathic helix that fragments the endosomal membrane. An adjacent peptide motif is also exposed which helps to drive the viral capsid out of the endosome (Flatt and Butcher 2019). After endosomal escape, the hexon facet of the viral capsid interacts with the kinesin light chain and cytoplasmic dynein protein. Thus, the virion hijacks the host’s dynein/dynactin motor proteins to hitchhike towards the nucleus. As the viral capsid docks onto the nuclear pore complex (NPC), the kinesin motor mediates a tug-of-war process for final uncoating of the viral capsid and release of the viral genome (Scherer and Vallee 2011). (B) To mimic this nuclear virus strategy, a peptide-based non-viral vector was synthesized by Favaro et al. (2018) wherein they used modular recombinant TRp3 protein (human dynein light chain) that interacts with dynein motor proteins and undergoes MT-assisted nuclear delivery. In addition to the MT-targeting protein, this peptide vector is composed of TAT for cell targeting, a DNA binding domain for electrostatic condensation of DNA and six histidine moieties for endosomal escape. Conclusively, this modular protein is able to efficiently deliver nucleic acid cargos including plasmid DNA, dsRNA and siRNA (Favaro et al., 2018).
Concluding Remarks
Evolution has honed viruses to be master hijackers of a broad range of host cells. They possess unique structural and mechanistic features wherein overarching themes such as capsid metastability, genome protection, stimuli-responsiveness, receptor duality, and synergistic ligand activity make them attractive templates for the design of non-viral nucleic acid carriers. Based on these outstanding characteristics of viruses, it is evident that an ideal carrier needs to find a balance between nucleic acid protection and release, two seemingly contradictory functions. A dynamic structure that responds to site-specific cues such as low pH or enzymatic activity help to control the release of nucleic acid cargo. These cues can vary with microenvironments within a cell, enabling biochemically controlled release mechanisms. Alternatively, the vector can be made sensitive to external stimuli such as light or temperature, which is more applicable to locally delivered formulations (Takemoto et al., 2014).
While therapeutic nucleic acids have made it to the clinical setting, extrahepatic targeting, endosomal escape, and controlled subcellular localization remain as major hurdles in their delivery (Juliano, 2016; Dowdy, 2017; Johannes and Lucchino, 2018; Juliano, 2018). Viruses commonly target multiple receptors for enhanced specificity and uptake, and this collective feature has been applied by synthetic carriers. Viral mimicry and the development of nucleic acid vectors iterate with our understanding of viral mechanism. Accordingly, advancements in techniques that identify viral ligands and corresponding host receptors, interrogate structure, and probe dynamics of ligand-receptor interactions may be translated to the design of more effective targeting domains for synthetic carriers.
In many ways, the outstanding difference in the transfection efficiency of viruses and synthetic vectors stems from the lack of a consensus of what drives endosomal escape. Escape from the endosome is influenced by a large range of factors such as nanoparticle properties (size, shape, and composition), mode of cellular uptake, and the type of cell (Selby et al., 2017). Moreover, mechanistic insights tend to be context-dependent as they are influenced by multiple factors such as the type of carrier, type of cell, and experimental conditions (LeCher et al., 2017). Structural studies on determinants of endosomal escape, while informative, often do not address the possible interplay of uptake route and intracellular trafficking. Moreover, uptake mechanisms are overlapping and poorly understood, making it difficult to determine the exact uptake mechanism of a particular construct (Nelemans and Gurevich, 2020). As uptake mechanisms typically involve signaling cascades, their relationship with intracellular trafficking are important considerations. Also, the implication of recycling pathways in viral and non-viral cytosolic access (Carette et al., 2011; Sahay et al., 2013; Wang et al., 2016a; Staring et al., 2018) suggests further studies on their exact role in therapeutic delivery. Filling such scientific gaps may help guide the design of more efficient nucleic acid delivery systems. Additionally, some viruses (such as the adenovirus) have been found to exploit cellular responses to membrane disruption concurrent with membrane fusion or penetration (Staring et al., 2018). In this light, future synthetic carriers may also be tailored to utilize host damage control to enhance therapeutic delivery. For this to be an effective strategy, it is imperative that the sensing of and response to invading particles by the host cell be exhaustively studied.
In summary, viruses can serve as a source of inspiration for chemists and materials scientists alike in the design considerations of non-viral vectors due to their efficient uptake and delivery of nucleic acid cargo. By designing nanoscale materials with stimuli-responsive properties and efficient targeting and internalization, therapeutic nucleic acids can be more rapidly brought forward for clinical application.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, D., Gonzalez-Duarte, A., O’Riordan, W. D., Yang, C.-C., Ueda, M., Kristen, A. V., et al. (2018). Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379 (1), 11–21. doi:10.1056/NEJMoa1716153
Aghdam, A., Marjan, R. B., Mosafer, J., Baradaran, B., Hashemzaei, M., Baghbanzadeh, A., et al. (2019). Recent advances on thermosensitive and PH-sensitive liposomes employed in controlled release. J. Control. Relea. 315, 1–22. doi:10.1016/j.jconrel.2019.09.018
Akinc, A., Querbes, W., De, S., Qin, J., and Frank-Kamenetsky, M. (2010). Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 18 (7), 1357–1364. doi:10.1038/mt.2010.85
Alam, M. R., Dixit, V., Kang, H., Li, Z. B., Chen, X., Trejo, J., et al. (2008). Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nuc. Acid. Res. 36 (8), 2764–2776. doi:10.1093/nar/gkn115
Aleksandrowicz, P., Marzi, A., Biedenkopf, N., Beimforde, N., Becker, S., Hoenen, T., et al. (2011). Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 204 (3), S957–S967. doi:10.1093/infdis/jir326
Alipour, M., Hosseinkhani, S., Sheikhnejad, R., and Cheraghi, R. (2017). Nano-biomimetic carriers are implicated in mechanistic evaluation of intracellular gene delivery. Sci. Rep. 7 (1), 41507. doi:10.1038/srep41507
Ämmälä, C., Drury, W. J., Knerr, L., Ahlstedt, I., Stillemark-Billton, P., Wennberg-Huldt, C., et al. (2018). Targeted delivery of antisense oligonucleotides to pancreatic β-cells. Sci. Adv. 4 (10), eaat3386. doi:10.1126/sciadv.aat3386
Anyarambhatla, G. R., and Needham, D. (1999). Enhancement of the phase transition permeability of DPPC liposomes by incorporation of mppc: a new temperature-sensitive liposome for use with mild hyperthermia. J. Liposome Res. 9 (4), 491–506. doi:10.3109/08982109909035549
Arana, L., Bayón-Cordero, L., Sarasola, L., Berasategi, M., and Ruiz, S. (2019). Solid lipid nanoparticles surface modification modulates cell internalization and improves chemotoxic treatment in an oral carcinoma cell line. Nanomaterials 9 (3), 464. doi:10.3390/nano9030464
Arinaminpathy, N., and Grenfell, B. (2010). Dynamics of glycoprotein charge in the evolutionary history of human influenza. PLoS One 5 (12), e15674. doi:10.1371/journal.pone.0015674
Asai, T., Matsushita, S., Kenjo, E., Tsuzuku, T., Yonenaga, N., Koide, H., et al. (2011). Dicetyl phosphate-tetraethylenepentamine-based liposomes for systemic SiRNA delivery. Bioconjug. Chem. 22 (3), 429–435. doi:10.1021/bc1004697
Ashley, C. E., Carnes, E. C., Phillips, G. K., Durfee, P. N., Lino, C. A., Padilla, D. P., et al. (2011). Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano 5 (7), 5729–5745. doi:10.1021/nn201397z
Awino, J. K., Gudipati, S., Hartmann, A. K., Santiana, J. J., and Cairns-Gibson, D. F. (2017). Nucleic acid nanocapsules for enzyme-triggered drug release. J. Am. Chem. Soc. 139 (18), 6278–6281. doi:10.1021/jacs.6b13087
Bai, H., Schiralli Lester, G. M., Petishnok, L. C., and Dean, D. A. (2017). Cytoplasmic transport and nuclear import of plasmid DNA. Biosci. Rep. 37, BSR20160616. doi:10.1042/BSR20160616
Banerjee, R., Tyagi, P., Li, S., and Huang, L. (2004). Anisamide-targeted stealth liposomes: a potent carrier for targeting doxorubicin to human prostate cancer cells. Int. J. Cancer 112 (4), 693–700. doi:10.1002/ijc.20452
Banga, R. J., Meckes, B., Narayan, S. P., Sprangers, A. J., Nguyen, S. T., and Mirkin, C. A. (2017). Cross-linked micellar spherical nucleic acids from thermoresponsive templates. J. Am. Chem. Soc. 139 (12), 4278–4281. doi:10.1021/jacs.6b13359
Banga, R. J., Chernyak, N., and Narayan, S. P. (2014). Liposomal spherical nucleic acids. J. Am. Chem. Soc. 136 (28), 9866–9869. doi:10.1021/ja504845f
Barba, A. A., Bochicchio, S., Dalmoro, A., and Lamberti, G. (2019). Lipid delivery systems for nucleic-acid-based-drugs: from production to clinical applications. Pharmaceutics 11 (8), 360. doi:10.3390/pharmaceutics11080360
Barrow, E., Nicola, A. V., and Liu, J. (2013). Multiscale perspectives of virus entry via endocytosis. Virol. J. 10, 177. doi:10.1186/1743-422X-10-177
Beniac, D. R., Melito, P. L., Devarennes, S. L., Hiebert, S. L., Rabb, M. J., and Lamboo, L. L. (2012). The organisation of ebola virus reveals a capacity for extensive, modular polyploidy. PLOS One 7 (1), e29608. doi:10.1371/journal.pone.0029608
Brandenburg, B., Stockl, L., Gutzeit, C., Roos, M., Lupberger, J., Schwartlander, R., et al. (2005). A novel system for efficient gene transfer into primary human hepatocytes via cell-permeable hepatitis B virus-like particle. Hepatology 42 (6), 1300–1309. doi:10.1002/hep.20950
Breuzard, G., Tertil, M., Gonçalves, C., Cheradame, H., Géguan, P., Pichon, C., et al. (2008). Nuclear delivery of NFkappaB-assisted DNA/polymer complexes: plasmid DNA quantitation by confocal laser scanning microscopy and evidence of nuclear polyplexes by FRET imaging. Nucleic Acids Res. 36 (12), e71. doi:10.1093/nar/gkn287
Brodin, J. D., Sprangers, A. J., McMillan, J. R., Mirkin, C. A., and Mirkin, C. A. (2015). DNA-mediated cellular delivery of functional enzymes. J. Am. Chem. Soc. 137 (47), 14838–14841. doi:10.1021/jacs.5b09711
Budker, V., Gurevich, V., Hagstrom, J. E., Bortzov, F., and Wolff, J. A. (1996). PH-sensitive, cationic liposomes: a new synthetic virus-like vector. Nat. Biotechnol. 14 (6), 760–764. doi:10.1038/nbt0696-760
Burckhardt, C. J., Suomalainen, M., Schoenenberger, P., Boucke, K., Hemmi, S., and Greber, U. F. (2011). Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host. Microbe. 10 (2), 105–117. doi:10.1016/j.chom.2011.07.006
Burger, N., Biswas, A., Barzan, D., Kirchner, A., Hosser, H., Hausmann, M., et al. (2014). A method for the efficient cellular uptake and retention of small modified gold nanoparticles for the radiosensitization of cells. Nanomedicine 10 (6), 1365–1373. doi:10.1016/j.nano.2014.03.011
Burgess, K. (2001). Solid-phase syntheses of beta-turn analogues to mimic or disrupt protein-protein interactions. Acc. Chem. Res. 34 (10), 826–835. doi:10.1021/ar9901523
Burri, D. J., da Palma, J. R., Kunz, S., and Pasquato, A. (2012). Envelope glycoprotein of arenaviruses. Viruses 4 (10), 2162–2181. doi:10.3390/v4102162
Cai, X., Chen, H. H., Wang, C. L., Chen, S. T., Lai, S. F., Chien, C. C., et al. (2011). Imaging the cellular uptake of tiopronin-modified gold nanoparticles. Anal. Bioanal. Chem. 401 (3), 809–816. doi:10.1007/s00216-011-4986-3
Callaway, E. (2020). The race for coronavirus vaccines: a graphical guide. Nature 580 (7805), 576–577. doi:10.1038/d41586-020-01221-y
Cao, W., Henry, M. D., Borrow, P., Yamada, H., Elder, J. H., Ravkov, E. V., et al. (1998). Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282 (5396), 2079–2081. doi:10.1126/science.282.5396.2079
Carette, J. E., Raaben, M., Wong, A. C., Herbert, A. S., Obernosterer, G., et al. (2011). Ebola virus entry requires the cholesterol transporter niemann-pick C1. Nature 477 (7364), 340–343. doi:10.1038/nature10348
Cartier, R., and Reszka, R. (2002). Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther. 9 (3), 157–167. doi:10.1038/sj.gt.3301635
Chan, D. C., Fass, D., Berger, J. M., and Kim, P. S. (1997). Core structure of Gp41 from the HIV envelope glycoprotein. Cell 89 (2), 263–273. doi:10.1016/S0092-8674(00)80205-6
Chang, K. L., Higuchi, Y., Kawakami, S., Yamashita, F., and Hashida, M. (2010). Efficient gene transfection by histidine-modified chitosan through enhancement of endosomal escape. Bioconjug. Chem. 21 (6), 1087–1095. doi:10.1021/bc1000609
Chernomordik, L., Kozlov, M. M., and Zimmerberg, J. (1995). Lipids in biological membrane fusion. J. Membr. Biol. 146 (1), 1–14. doi:10.1007/BF00232676
Chery, J. (2016). RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc. J.: A J. Postdoc. Res. Postdoc. Aff. 4 (7), 35–50. doi:10.14304/surya.jpr.v4n7.5
Choi, C. H. J., Hao, L., Narayan, S. P., Auyeung, E., and Mirkin, C. A. (2013). Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. 110 (19), 7625–7630. doi:10.1073/pnas.1305804110
Cohen, O., and Granek, R. (2014). Nucleus-targeted drug delivery: theoretical optimization of nanoparticles decoration for enhanced intracellular active transport. Nano Lett. 14 (5), 2515–2521. doi:10.1021/nl500248q
Cohen, R. N., van der Aavan der Aa, M. A., Macaraeg, N., Lee, A. P., and Szoka, F. C. (2009). Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Control. Relea. 135 (2), 166–174. doi:10.1016/j.jconrel.2008.12.016
Cohen, S., Au, S., and Panté, N. (2011). How viruses access the nucleus. Biochim. Biophys. Acta 1813 (9), 1634–1645. doi:10.1016/j.bbamcr.2010.12.009
Cohen-Dvashi, H., Cohen, N., Israeli, H., and Diskin, R. (2015). Molecular mechanism for LAMP1 recognition by Lassa virus. J. Virol. 89 (15), 7584–7592. doi:10.1128/JVI.00651-15
Cohen-Dvashi, H., Israeli, H., Shani, O., Katz, A., and Diskin, R. (2016). Role of LAMP1 binding and PH sensing by the spike complex of Lassa virus. J. Virol. 90 (22), 10329–10338. doi:10.1128/JVI.01624-16
Corbett, K. S., Edwards, D., Leist, S. R., Abiona, O. M., Boyoglu-Barnum, S., Gillespie, R. A., et al. (2020). SARS-CoV-2 MRNA vaccine development enabled by prototype pathogen preparedness. BioRxiv [Epub ahead of print]. doi:10.1101/2020.06.11.145920
Côté, M., Misasi, J., Ren, T., Bruchez, A., Lee, K., Filone, C. M., et al. (2011). Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477 (7364), 344–348. doi:10.1038/nature10380
Couch, R. B. (1996). “Orthomyxoviruses,” in Medical microbiology. Editors S. Baron (Galveston, TX: University of Texas Medical Branch).
Crews, C. M. (2010). Targeting the undruggable proteome: the small molecules of my dreams. Chem. Biol. 17 (6), 551–555. doi:10.1016/j.chembiol.2010.05.011
Cros, J. F., and Palese, P. (2003). Trafficking of viral genomic RNA into and out of the nucleus: influenza, thogoto and borna disease viruses. Virus. Res. 95 (1), 3–12. doi:10.1016/S0168-1702(03)00159-X
Cruz-Acuña, M., Halman, J. R., Afonin, K. A., and Dobson, J. (2018). Magnetic nanoparticles loaded with functional RNA nanoparticles. Nanoscale 10 (37), 17761–17770. doi:10.1039/C8NR04254C
Cutler, J. I., Auyeung, E., and Mirkin, C. A. (2012). Spherical nucleic acids. J. Am. Chem. Soc. 134 (3), 1376–1391. doi:10.1021/ja209351u
Cutler, J. I., Zhang, K., Zheng, D., Auyeung, E., Prigodich, A. E., and Mirkin, C. A. (2011). Polyvalent nucleic acid nanostructures. J. Am. Chem. Soc. 133 (24), 9254–9257. doi:10.1021/ja203375n
Cutler, J. I., Zheng, D., Xu, X., Giljohann, D. A., and Mirkin, C. A. (2010). Polyvalent oligonucleotide iron oxide nanoparticle “click” conjugates. Nano Lett. 10 (4), 1477–1480. doi:10.1021/nl100477m
Dalmau-Mena, I., del Pino, P., Pelaz, B., Cuesta-Geijo, M. Á., Galindo, I., Moros, M., et al. (2018). Nanoparticles engineered to bind cellular motors for efficient delivery. J. Nanobiotechnology 16 (1), 33. doi:10.1186/s12951-018-0354-1
Damha, M. J. (2019). Exciting times in the field of nucleic acid therapeutics. Trends Mol. Med. 25 (12), 1051–1052. doi:10.1016/j.molmed.2019.10.005
Danhier, F., Le Breton, A., and Préat, V. (2012). RGD-based strategies to target alpha(v) beta(3) integrin in cancer therapy and diagnosis. Mol. Pharm. 9, 2961–2971. doi:10.1021/mp3002733
Dassie, J. P., and Giangrande, P. H. (2013). Current progress on aptamer-targeted oligonucleotide therapeutics. Ther. Deliv. 4 (12), 1527–1546. doi:10.4155/tde.13.118
de Jonge, J., Holtrop, M., Wilschut, J., and Huckriede, A. (2006). Reconstituted influenza virus envelopes as an efficient carrier system for cellular delivery of small-interfering RNAs. Gene Ther. 13 (5), 400–411. doi:10.1038/sj.gt.3302673
Debacker, A. J., Voutila, J., Catley, M., Blakey, D., and Habib, N. (2020). Delivery of oligonucleotides to the liver with GalNAc: from research to registered therapeutic drug. Mol. Ther. 28 (8), 1759–1771. doi:10.1016/j.ymthe.2020.06.015
Ding, F., Gao, X., Huang, X., Ge, H., Xie, M., Qian, J., et al. (2020). Polydopamine-coated nucleic acid nanogel for SiRNA-mediated low-temperature photothermal therapy. Biomaterials 245, 119976. doi:10.1016/j.biomaterials.2020.119976
Ding, F., Mou, Q., Ma, Y., Pan, G., Guo, Y., Tong, G., et al. (2018). A crosslinked nucleic acid nanogel for effective SiRNA delivery and antitumor therapy. Angew. Chem. Int. Ed. Engl. 57 (12), 3064–3068. doi:10.1002/anie.201711242
Ding, F., Huang, X., Gao, X., Xie, M., Pan, G., Li, Q., et al. (2019). A non-cationic nucleic acid nanogel for the delivery of the CRISPR/Cas9 gene editing tool. Nanoscale 11 (37), 17211–17215. doi:10.1039/C9NR05233J
Ding, Y., Jiang, Z., Saha, K., Kim, C. S., Kim, S. T., Landis, R. F., et al. (2014a). Gold nanoparticles for nucleic acid delivery. Mol. Ther. 22 (6), 1075–1083. doi:10.1038/mt.2014.30
Ding, Y., Wang, Y., Zhou, J., Gu, X., Wang, W., Liu, C., et al. (2014b). Direct cytosolic SiRNA delivery by reconstituted high density lipoprotein for target-specific therapy of tumor angiogenesis. Biomaterials 35 (25), 7214–7227. doi:10.1016/j.biomaterials.2014.05.009
Döhner, K., Nagel, C.-H., and Sodeik, B. (2005). Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 13 (7), 320–327. doi:10.1016/j.tim.2005.05.010
Dong, Y., Yu, T., Ding, L., Laurini, E., Huang, Y., Zhang, M., et al. (2018). A dual targeting dendrimer-mediated SiRNA delivery system for effective gene silencing in cancer therapy. J. Am. Chem. Soc. 140 (47), 16264–16274. doi:10.1021/jacs.8b10021
Dowaidar, M., Hällbrink, M., Freimann, K., Kurrikoff, K., Zou, X., and Langel, Ü. (2017). Magnetic nanoparticle assisted self-assembly of cell penetrating peptides-oligonucleotides complexes for gene delivery. Sci. Rep. 7 (1), 9159. doi:10.1038/s41598-017-09803-z
Dowdy, S. F. (2017). Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 35 (3), 222–229. doi:10.1038/nbt.3802
Dowdy, S. F., and Levy, M. (2018). RNA therapeutics (almost) comes of age: targeting, delivery and endosomal escape. Nucleic Acid Ther. 28 (3), 107–108. doi:10.1089/nat.2018.29001.dow
Du Rietz, H., Hedlund, H., Wilhelmson, S., Nordenfelt, P., and Wittrup, A. (2020). Imaging small molecule-induced endosomal escape of SiRNA. Nat. Commun. 11 (1), 1809. doi:10.1038/s41467-020-15300-1
Duffy, M. J., and Crown, J. (2021). Drugging "undruggable" genes for cancer treatment: are we making progress?. Int. J. Cancer 148 (1), 8–17. doi:10.1002/ijc.33197
Elghanian, R., Storhoff, J. J., Mucic, R. C., Letsinger, R. L., and Mirkin, C. A. (1997). Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277 (5329), 1078–1081. doi:10.1126/science.277.5329.1078
Ewers, H., Römer, W., Smith, A. E., Bacia, K., Dmitrieff, S., Chai, W., et al. (2010). GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12 (1), 11–12. doi:10.1038/ncb1999
Falasca, L., Agrati, C., Petrosillo, N., Di Caro, A., Capobianchi, M. R., Ippolito, G., et al. (2015). Molecular mechanisms of ebola virus pathogenesis: focus on cell death. Cell Death Differ. 22 (8), 1250–1259. doi:10.1038/cdd.2015.67
Fan, Y., Yao, J., Du, R., Hou, L., Zhou, J., Lu, Y., et al. (2013). Ternary complexes with core-shell bilayer for double level targeted gene delivery: in vitro and in vivo evaluation. Pharm. Res. 30 (5), 1215–1227. doi:10.1007/s11095-012-0960-9
Fanales-Belasio, E., Raimondo, M., Suligoi, B., and Buttò, S. (2010). HIV virology and pathogenetic mechanisms of infection: a brief overview. Ann. Ist Super Sanita 46 (1), 5–14. doi:10.4415/ANN_10_01_02
Fang, Y., Xue, J., Gao, S., Lu, A., Yang, D., Jiang, H., et al. (2017). Cleavable PEGylation: a strategy for overcoming the "PEG dilemma" in efficient drug delivery. Drug Deliv. 24 (2), 22–32. doi:10.1080/10717544.2017.1388451
Faustino, R. S., Nelson, T. J., Terzic, A., and Perez-Terzic, C. (2007). Nuclear transport: target for therapy. Clin. Pharmacol. Ther. 81 (6), 880–886. doi:10.1038/sj.clpt.6100141
Favaro, M. T., de Toledo, M. A., Alves, R. F., Santos, C. A., Beloti, L. L., Janissen, R., et al. (2014). Development of a non-viral gene delivery vector based on the dynein light chain Rp3 and the TAT peptide. J. Biotechnol. 173, 10–18. doi:10.1016/j.jbiotec.2014.01.001
Favaro, M. T. P., Unzueta, U., de Cabo, M., Villaverde, A., Ferrer-Miralles, N., and Azzoni, A. R. (2018). Intracellular trafficking of a dynein-based nanoparticle designed for gene delivery. Eur. J. Pharm. Sci. 112, 71–78. doi:10.1016/j.ejps.2017.11.002
Fay, N., and Panté, N. (2015). Nuclear entry of DNA viruses. Front. Microbiol. 6, 467. doi:10.3389/fmicb.2015.00467
Felgner, P. L., Gadek, T. R., Holm, M., Roman, R., Chan, H. W., Wenz, M., et al. (1987). Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Proc Natl Acad. Sci. U.S.A. 84 (21), 7413–7417. doi:10.1073/pnas.84.21.7413
Feng, G., Chen, H., Li, J., Huang, Q., Gupte, M. J., Liu, H., et al. (2015). Gene therapy for nucleus pulposus regeneration by heme oxygenase-1 plasmid DNA carried by mixed polyplex micelles with thermo-responsive heterogeneous coronas. Biomaterials 52, 1–13. doi:10.1016/j.biomaterials.2015.02.024
Ferrari, S., Pettenazzo, A., Garbati, N., Zacchello, F., Behr, J. P., and Scarpa, M. (1999). Polyethylenimine shows properties of interest for cystic fibrosis gene therapy. Biochim. Biophys. Acta 1447 (2), 219–225. doi:10.1016/S0167-4781(99)00153-0
Flatt, J. W., and Butcher, S. J. (2019). Adenovirus flow in host cell networks. Open Biol. 9 (2), 190012. doi:10.1098/rsob.190012
Flint, J., Racaniello, V. R., Rall, G. F., Skalka, M., and Enquist, W. (2015). Principles of virology. 4th Edn. Hoboken, NJ: Wiley.
Fouriki, A., and Dobson, J. (2014). Oscillating magnet array-based nanomagnetic gene transfection of human mesenchymal stem cells. Nanomedicine 9 (7), 989–997. doi:10.2217/nnm.13.74
Fraley, R., Subramani, S., Berg, P., and Papahadjopoulos, D. (1980). Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 255 (21), 10431–10435. doi:10.1016/s0021-9258(19)70482-7
Friedman, A. D., Claypool, S. E., and Liu, R. (2013). The smart targeting of nanoparticles. Curr. Pharm. Des. 19 (35), 6315–6329. doi:10.2174/13816128113199990375
Fu, Y., and Li, J. (2016). A novel delivery platform based on bacteriophage MS2 virus-like particles. Virus. Res. 211, 9–16. doi:10.1016/j.virusres.2015.08.022
Gaal, E. V. B., Roel van Eijk, R. S. O., Bakowska, M., Feyen, D., Jan Kok, R., Hennink, W. E., et al. (2011). DNA nuclear targeting sequences for non-viral gene delivery. Pharm. Res. 28 (7), 1707–1722. doi:10.1007/s11095-011-0407-8
Gaber, M. H., Hong, K., Huang, S. K., and Papahadjopoulos, D. (1995). Thermosensitive sterically stabilized liposomes: formulation and in Vitro studies on mechanism of doxorubicin release by bovine serum and human plasma. Pharm. Res. 12 (10), 1407–1416. doi:10.1023/a:1016206631006
Galaway, F. A., and Stockley, P. G. (2013). MS2 viruslike particles: a robust, semisynthetic targeted drug delivery platform. Mol. Pharm. 10 (1), 59–68. doi:10.1021/mp3003368
Gelderblom, H. R. (1996). “Structure and classification of viruses,” in Medical microbiology. Editor S. Baron (Galveston, TX: University of Texas Medical Branch).
Ghasemiyeh, P., and Mohammadi-Samani, S. (2018). Solid lipid nanoparticles and nanostructured lipid carriers as novel drug delivery systems: applications, advantages and disadvantages. Res. Pharm. Sci. 13 (4), 288–303. doi:10.4103/1735-5362.235156
Gillard, M., Jia, Z., Hou, J. J., Song, M., Gray, P. P., Munro, T. P., et al. (2014). Intracellular trafficking pathways for nuclear delivery of plasmid DNA complexed with highly efficient endosome escape polymers. Biomacromolecules 15 (10), 3569–3576. doi:10.1021/bm5008376
Gilleron, J., Querbes, W., Zeigerer, A., Borodovsky, A., Marsico, G., Schubert, U., et al. (2013). Image-based analysis of lipid nanoparticle-mediated SiRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31 (7), 638–646. doi:10.1038/nbt.2612
Gilleron, J., Paramasivam, P., Zeigerer, A., Querbes, W., Marsico, G., Andree, C., et al. (2015). Identification of SiRNA delivery enhancers by a chemical library screen. Nucleic Acids Res. 43 (16), 7984–8001. doi:10.1093/nar/gkv762
Greber, U. F. (2016). Virus and host mechanics support membrane penetration and cell entry. J. Virol. 90 (8), 3802–3805. doi:10.1128/JVI.02568-15
Greber, U. F., Suomalainen, M., Stidwill, R. P., Boucke, K., and Ebersold, M. W. (1997). The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16 (19), 5998–6007. doi:10.1093/emboj/16.19.5998
Grove, J., and Marsh, M. (2011). The cell biology of receptor-mediated virus entry. J. Cell Biol. 195 (7), 1071–1082. doi:10.1083/jcb.201108131
Gudipati, S., Zhang, K., and Rouge, J. L. (2019). Towards self-transfecting nucleic acid nanostructures for gene regulation. Trends Biotechnol. 37 (9), 983–994. doi:10.1016/j.tibtech.2019.01.008
Hamid, F. B., Kim, J., and Shin, C.-G. (2015). Cellular and viral determinants of retroviral nuclear entry. Can. J. Microbiol. 62, 1. doi:10.1139/cjm-2015-0350
Han, X., Bushweller, J. H., Cafiso, D. S., and Tamm, L. K. (2001). Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8 (8), 715–720. doi:10.1038/90434
Hao, X., Li, Q., Guo, J., Ren, X., Feng, Y., Shi, C., et al. (2017). Multifunctional gene carriers with enhanced specific penetration and nucleus accumulation to promote neovascularization of HUVECs in Vivo. ACS Appl. Mater. Inter. 9 (41), 35613–35627. doi:10.1021/acsami.7b11615
Harrison, S. C. (2008). Viral membrane fusion. Nat. Struct. Mol. Biol. 15 (7), 690–698. doi:10.1038/nsmb.1456
Hartmann, A. K., Cairns-Gibson, D. F., Santiana, J. J., Tolentino, M. Q., and Barber, H. M. (2018). Enzymatically ligated DNA-surfactants: unmasking hydrophobically modified DNA for intracellular gene regulation. ChemBioChem 19 (16), 1734–1739. doi:10.1002/cbic.201800302
Hartmann, A. K., Gudipati, S., Pettenuzzo, A., Ronconi, L., and Rouge, J. L. (2020). Chimeric SiRNA-DNA surfactants for the enhanced delivery and sustained cytotoxicity of a gold(III) metallodrug. Bioconjug. Chem. 31 (4), 1063–1069. doi:10.1021/acs.bioconjchem.0c00047
Heidarli, E., Dadashzadeh, S., and Haeri, A. (2017). State of the art of stimuli-responsive liposomes for cancer therapy. Iran J. Pharm. Res. 16 (4), 1273–1304.
Corey, D. R. (2007). Chemical modification: the key to clinical application of RNA interference?. J. Clin. Invest. 117 (12), 3615–3622. doi:10.1172/JCI33483
Hoy, S. M. (2018). Patisiran: first global approval. Drugs 78 (15), 1625–1631. doi:10.1007/s40265-018-0983-6
Hu, Q., Wang, J., Shen, J., Liu, M., Jin, X., Tang, G., et al. (2012). Intracellular pathways and nuclear localization signal peptide-mediated gene transfection by cationic polymeric nanovectors. Biomaterials 33 (4), 1135–1145. doi:10.1016/j.biomaterials.2011.10.023
Huang, K., Ma, H., Liu, J., Huo, S., Kumar, A., Wei, T., et al. (2012). Size-dependent localization and penetration of ultrasmall gold nanoparticles in cancer cells, multicellular spheroids, and tumors in Vivo. ACS Nano 6 (5), 4483–4493. doi:10.1021/nn301282m
Huggins, I. J., Medina, C. A., van den Berg, A., and Jadhav, S. (2019). Site selective antibody-oligonucleotide conjugation via microbial transglutaminase. Molecules 24 (18), 3287. doi:10.3390/molecules24183287
Hunt, C. L., Lennemann, N. J., and Maury, W. (2012). Filovirus entry: a novelty in the viral fusion world. Viruses 4 (2), 258–275. doi:10.3390/v4020258
Huo, S., Jin, S., Ma, X., Xue, X., Yang, K., Kumar, A., et al. (2014). Ultrasmall gold nanoparticles as carriers for nucleus-based gene therapy due to size-dependent nuclear entry. ACS Nano 8 (6), 5852–5862. doi:10.1021/nn5008572
Hussein, H. A. M., Walker, L. R., Abdel-Raouf, U. M., Desouky, S. A., Montasser, A. K. M., Akula, S. A., et al. (2015). Beyond RGD: virus interactions with integrins. Arch. Virol. 160 (11), 2669–2681. doi:10.1007/s00705-015-2579-8
Jackson, A. L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., et al. (2006). Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12 (7), 1197–1205. doi:10.1261/rna.30706
Jae, L. T., Raaben, M., Herbert, A. S., Kuehne, A. I., Wirchnianski, A. S., Soh, T. K., et al. (2014). Lassa Virus Entry Requires a Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344 (6191), 1506–1510. doi:10.1126/science.1252480
Jae, L. T., and Brummelkamp, T. R. (2015). Emerging intracellular receptors for hemorrhagic fever viruses. Trends Microbiol. 23 (7), 392–400. doi:10.1016/j.tim.2015.04.006
James, S. H., and Whitley, R. J. (2017). “Influenza viruses,” in Infectious diseases (Amsterdam, Netherlands: Elsevier), 1465–1471. doi:10.1016/B978-0-7020-6285-8.00172-6
Jeffs, L. B., Palmer, L. R., Ambegia, E. G., Giesbrecht, C., Ewanick, S., and MacLachlan, I. (2005). A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 22 (3), 362–372. doi:10.1007/s11095-004-1873-z
Jiang, S., Eltoukhy, A. A., Love, K. T., Langer, R., and Anderson, D. G. (2013). Lipidoid-coated iron oxide nanoparticles for efficient DNA and SiRNA delivery. Nano Lett. 13 (3), 1059–1064. doi:10.1021/nl304287a
Jiang, Y., Tang, R., Duncan, B., Jiang, Z., Yan, B., Mout, R., et al. (2015). Direct cytosolic delivery of SiRNA using nanoparticle-stabilized nanocapsules. Angew. Chem. Int. Ed. Engl. 54 (2), 506–510. doi:10.1002/anie.201409161
Jiang, Y., Hardie, J., Liu, Y., Ray, M., Luo, X., Das, R., et al. (2018). Nanocapsule-mediated cytosolic SiRNA delivery for anti-inflammatory treatment. J. Control. Releas. 283, 235–240. doi:10.1016/j.jconrel.2018.06.001
Jin, R., Wu, G., Li, Z., Mirkin, C. A., and Schatz, G. C. (2003). What controls the melting properties of DNA-linked gold nanoparticle assemblies?. J. Am. Chem. Soc. 125 (6), 1643–1654. doi:10.1021/ja021096v
Johannes, L., and Lucchino, M. (2018). Current challenges in delivery and cytosolic translocation of therapeutic RNAs. Nucleic Acid Ther. 28 (3), 178–193. doi:10.1089/nat.2017.0716
Joris, F., De Backer, L., Van de Vyver, T., Bastiancich, C., De Smedt, S. C., and Raemdonck, K. (2018). Repurposing cationic amphiphilic drugs as adjuvants to induce lysosomal SiRNA escape in nanogel transfected cells. J. Control Releas. 269, 266–276. doi:10.1016/j.jconrel.2017.11.019
Judge, A. D., Bola, G., and Lee, A. C. (2006). Design of noninflammatory synthetic SiRNA mediating potent gene silencing in Vivo. Mol. Ther. 13 (3), 494–505. doi:10.1016/j.ymthe.2005.11.002
Judge, A. D., Sood, V., Shaw, J. R., and Fang, D. (2005). Sequence-dependent stimulation of the mammalian innate immune response by synthetic SiRNA. Nat. Biotechnol. 23 (4), 457–462. doi:10.1038/nbt1081
Juliano, R. L. (2016). The delivery of therapeutic oligonucleotides. Nucl. Acids Res. 44 (14), 6518–6548. doi:10.1093/nar/gkw236
Juliano, R. L. (2018). Intracellular trafficking and endosomal release of oligonucleotides: what we know and what we don't. Nucleic Acid Ther. 28 (3), 166–177. doi:10.1089/nat.2018.0727
Juliano, R. L., Ming, X., Nakagawa, O., Xu, R., and Yoo, H. (2011). Integrin targeted delivery of gene therapeutics. Theranostics 1, 211–219. doi:10.7150/thno/v01p0211
Juliano, R. L., Wang, L., Tavares, F., Brown, E. G., James, L., Ariyarathna, Y., et al. (2018). Structure-activity relationships and cellular mechanism of action of small molecules that enhance the delivery of oligonucleotides. Nucleic Acids Res. 46 (4), 1601–1613. doi:10.1093/nar/gkx1320
Juliano, R., Alam, M. R., Dixit, V., and Kang, H. (2008). Mechanisms and strategies for effective delivery of antisense and SiRNA oligonucleotides. Nucleic Acids Res. 36 (12), 4158–4171. doi:10.1093/nar/gkn342
Kang, H., Alam, M. R., Dixit, V., Fisher, M., and Juliano, R. L. (2008). Cellular delivery and biological activity of antisense oligonucleotides conjugated to a targeted protein carrier. Bioconjug. Chem. 19 (11), 2182–2188. doi:10.1021/bc800270w
Kang, Y. Y., Song, J., Jung, H. S. G., Kwak, G., Yu, J.-H., Kim, S. H., et al. (2018). Implication of multivalent aptamers in DNA and DNA-RNA hybrid structures for efficient drug delivery in vitro and in vivo. J. Ind. Eng. Chem. 60, 250–258. doi:10.1016/j.jiec.2017.11.011
Kang, Z., Meng, Q., and Liu, K. (2019). Peptide-based gene delivery vectors. J. Mater. Chem. B 7 (11), 1824–1841. doi:10.1039/C8TB03124J
Karimi, M., Mirshekari, H., Moosavi Basri, S. M., Bahrami, S., Moghoofei, M., and Hamblin, M. R. (2016). Bacteriophages and phage-inspired nanocarriers for targeted delivery of therapeutic cargos. Adv. Drug Deliv. Rev. 106, 45–62. doi:10.1016/j.addr.2016.03.003
Karkare, S., and Bhatnagar, D. (2006). Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA, and morpholino. Appl. Microbiol. Biotechnol. 71 (5), 575–586. doi:10.1007/s00253-006-0434-2
Keefe, A. D., Pai, S., and Ellington, A. (2010). Aptamers as therapeutics. Nat. Rev. Drug Discov. 9 (7), 537–550. doi:10.1038/nrd3141
Khan, N., Halcrow, P. W., Lakpa, K. L., Afghah, Z., Miller, N. M., Dowdy, S. F., et al. (2020). Two‐pore channels regulate Tat endolysosome escape and Tat‐mediated HIV‐1 LTR transactivation. FASEB j. 34 (3), 4147–4162. doi:10.1096/fj.201902534R
Khorev, O., Stokmaier, D., Schwardt, O., Cutting, B., and Ernst, B. (2008). Trivalent, gal/GalNAc-containing ligands designed for the asialoglycoprotein receptor. Bioorg. Med. Chem. 16 (9), 5216–5231. doi:10.1016/j.bmc.2008.03.017
Kim, A., Shin, T. H., Shin, S. M., Pham, C. D., Choi, D. K., Kwon, M. H., et al. (2012). Cellular internalization mechanism and intracellular trafficking of filamentous M13 phages displaying a cell-penetrating transbody and TAT peptide. PLOS One 7 (12), e51813. doi:10.1371/journal.pone.0051813
Kim, H. J., and Kim, H. J. (2017). Yeast as an expression system for producing virus-like particles: what factors do we need to consider?. Lett. Appl. Microbiol. 64 (2), 111–123. doi:10.1111/lam.12695
Kim, Y. H., Han, M.-E., and Oh, S.-O. (2017). The molecular mechanism for nuclear transport and its application. Anat. Cell Biol. 50 (2), 77. doi:10.5115/acb.2017.50.2.77
Klibanov, A. L., Maruyama, K., and Torchilin, V. P. (1990). Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 268 (1), 235–237. doi:10.1016/0014-5793(90)81016-H
Kobiler, O., Drayman, N., Butin-Israeli, V., and Oppenheim, A. (2012). Virus strategies for passing the nuclear envelope barrier. Nucleus 3 (6), 526–539. doi:10.4161/nucl.21979
Kogure, K., Akita, H., Yamada, Y., and Harashima, H. (2008). Multifunctional envelope-type nano device (MEND) as a non-viral gene delivery system. Adv. Drug Deliv. Rev. 60 (4), 559–571. doi:10.1016/j.addr.2007.10.007
Kondratowicz, A. S., Lennemann, N. J., Sinn, P. L., Davey, R. A., Hunt, C. L., Moller-Tank, S., et al. (2011). T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for zaire ebolavirus and lake victoria marburgvirus. Proc. Natl. Acad. Sci. U.S.A. 108 (20), 8426–8431. doi:10.1073/pnas.1019030108
Kono, K., Ozawa, T., Yoshida, T., Ozaki, F., Ishizaka, Y., Maruyama, K., et al. (2010). Highly temperature-sensitive liposomes based on a thermosensitive block copolymer for tumor-specific chemotherapy. Biomaterials 31 (27), 7096–7105. doi:10.1016/j.biomaterials.2010.05.045
Krop, I. E., Beeram, M., Modi, S., Jones, S. F., Holden, S. N., Yu, W., et al. (2010). Phase I study of trastuzumab-DM1, an HER2 antibody-drug conjugate, given every 3 Weeks to patients with HER2-positive metastatic breast cancer. J. Clin. Oncol. 28 (16), 2698–2704. doi:10.1200/JCO.2009.26.2071
Kulkarni, J. A., Cullis, P. R., and van der Meel, R. (2018). Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 28 (3), 146–157. doi:10.1089/nat.2018.0721
Kusumoto, K., Akita, H., Ishitsuka, T., Matsumoto, Y., Nomoto, T., Furukawa, R., et al. (2013). Lipid envelope-type nanoparticle incorporating a multifunctional peptide for systemic SiRNA delivery to the pulmonary endothelium. ACS Nano 7 (9), 7534–7541. doi:10.1021/nn401317t
Kusumoto, K., Akita, H., Santiwarangkool, S., and Harashima, H. (2014). Advantages of ethanol dilution method for preparing GALA-modified liposomal SiRNA carriers on the in Vivo gene knockdown efficiency in pulmonary endothelium. Int. J. Pharm. 473 (1–2), 144–147. doi:10.1016/j.ijpharm.2014.07.007
Lam, P., and Steinmetz, N. F. (2019). Delivery of SiRNA therapeutics using cowpea chlorotic mottle virus-like particles. Biomater. Sci. 7 (8), 3138–3142. doi:10.1039/C9BM00785G
Lam, P., and Steinmetz, N. F. (2018). Plant viral and bacteriophage delivery of nucleic acid therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotech. 10 (1), e1487. doi:10.1002/wnan.1487
Lazo, J. S., and Sharlow, E. R. (2016). Drugging undruggable molecular cancer targets. Annu. Rev. Pharmacol. Toxicol. 56 (1), 23–40. doi:10.1146/annurev-pharmtox-010715-103440
Lechardeur, D., Sohn, K. J., Haardt, M., Joshi, P. B., Monck, M., Graham, R. W., et al. (1999). Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 6 (4), 482–497. doi:10.1038/sj.gt.3300867
LeCher, J. C., Nowak, S. J., and McMurry, J. L. (2017). Breaking in and busting out: cell-penetrating peptides and the endosomal escape problem. Biomol. Concepts 8 (3–4), 131–141. doi:10.1515/bmc-2017-0023
Lee, Y. C., Townsend, R. R., Hardy, M. R., Lönngren, J., Arnarp, J., Haraldsson, M., et al. (1983). Binding of synthetic oligosaccharides to the hepatic gal/GalNAc lectin. Dependence on fine structural features. J. Biol. Chem. 258 (1), 199–202. doi:10.1016/s0021-9258(18)33240-x
Lee, Y. C., and Lee, R. T. (2000). “Interactions of oligosaccharides and glycopeptides with hepatic carbohydrate receptors,” in Carbohydrates in chemistry and biology (New York, NY: John Wiley & Sons, Ltd), 549–561. doi:10.1002/9783527618255.ch72
Legendre, J. Y., and Szoka, F. C. (1992). Delivery of plasmid DNA into mammalian cell lines using PH-sensitive liposomes: comparison with cationic liposomes. Pharm. Res. 9 (10), 1235–1242. doi:10.1023/a:1015836829670
Li, G., Qi, X., Hu, Z., and Tang, Q. (2019). Mechanisms mediating nuclear trafficking involved in viral propagation by DNA viruses. Viruses 11 (11), 1035. doi:10.3390/v11111035
Li, Y., Zhang, K., Liu, P., Chen, M., Zhong, Y., Ye, Q., et al. (2019). Encapsulation of plasmid DNA by nanoscale metal-organic frameworks for efficient gene transportation and expression. Adv. Mater. 31 (29), e1901570. doi:10.1002/adma.201901570
Li, H., Yu, S. S., Miteva, M., Nelson, C. E., Werfel, T., Giorgio, T. D., et al. (2013). Matrix metalloproteinase responsive, proximity-activated polymeric nanoparticles for SiRNA delivery. Adv. Funct. Mater. 23 (24), 3040–3052. doi:10.1002/adfm.201202215
Li, J., Arévalo, M. T., and Zeng, M. (2013). Engineering influenza viral vectors. Bioengineered 4 (1), 9–14. doi:10.4161/bioe.21950
Li, H., Zhang, B., Lu, X., Tan, X., Jia, F., Xiao, Y., et al. (2018). Molecular spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 115 (17), 4340–4344. doi:10.1073/pnas.1801836115
Li, J., Yu, M., Zheng, W., and Liu, W. (2015). Nucleocytoplasmic shuttling of influenza A virus proteins. Viruses 7 (5), 2668–2682. doi:10.3390/v7052668
Li, S., Sun, Z., Pryce, R., Parsy, M. L., Fehling, S. K., Schlie, K., et al. (2016). Acidic PH-induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. Plos Pathog. 12 (2), e1005418. doi:10.1371/journal.ppat.1005418
Li, W. (2015). The hepatitis B virus receptor. Annu. Rev. Cell Dev. Biol. 31 (1), 125–147. doi:10.1146/annurev-cellbio-100814-125241
Liang, K., Ricco, R., Doherty, C. M., Styles, M. J., Bell, S., Kirby, N., et al. (2015). Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules. Nat. Commun. 6 (1), 7240. doi:10.1038/ncomms8240
Lo, S. L., and Wang, S. (2008). An endosomolytic tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials 29 (15), 2408–2414. doi:10.1016/j.biomaterials.2008.01.031
Lobovkina, T., Jacobson, G. B., Gonzalez-Gonzalez, E., Hickerson, R. P., Leake, D., Kaspar, R. L., et al. (2011). In Vivo sustained release of SiRNA from solid lipid nanoparticles. ACS Nano 5 (12), 9977–9983. doi:10.1021/nn203745n
Loh, X. J., Lee, T. C., Dou, Q., and Deen, G. (2015). Utilising inorganic nanocarriers for gene delivery. Biomater. Sci. 4 (1), 70–86. doi:10.1039/C5BM00277J
Lönn, P., Kacsinta, A. D., Cui, X.-S., Hamil, A. S., Kaulich, M., Gogoi, K., et al. (2016). Enhancing endosomal escape for intracellular delivery of macromolecular biologic therapeutics. Sci. Rep. 6 (1), 32301. doi:10.1038/srep32301
López-Huertas, M. R., Jiménez-Tormo, L., Madrid-Elena, N., Gutiérrez, C., Rodríguez-Mora, S., Coiras, M., et al. (2017). The CCR5-antagonist maraviroc reverses HIV-1 latency in Vitro alone or in combination with the PKC-agonist bryostatin-1. Sci. Rep. 7, 2385. doi:10.1038/s41598-017-02634-y
Lv, H., Zhang, S., Wang, B., Cui, S., and Yan, J. (2006). Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control Release 114 (1), 100–109. doi:10.1016/j.jconrel.2006.04.014
Lytton-Jean, A. K., and Mirkin, C. A. (2005). A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J. Am. Chem. Soc. 127 (37), 12754–12755. doi:10.1021/ja052255o
Mach, R. H., Huang, Y., Freeman, R. A., Wu, L., Vangveravong, S., and Luedtke, R. R. (2004). Conformationally-flexible benzamide analogues as dopamine D3 and sigma 2 receptor ligands. Bioorg. Med. Chem. Lett. 14 (1), 195–202. doi:10.1016/j.bmcl.2003.09.083
Maginnis, M. S. (2018). Virus-receptor interactions: the key to cellular invasion. J. Mol. Biol. 430 (17), 2590–2611. doi:10.1016/j.jmb.2018.06.024
Mammen, M., Choi, S. K., and Whitesides, G. M. (1998). Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37 (20), 2754–2794. doi:10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3
Marsh, M., and Helenius, A. (2006). Virus entry: open sesame. Cell 124 (4), 729–740. doi:10.1016/j.cell.2006.02.007
Maruyama, K., Unezaki, S., Takahashi, N., and Iwatsuru, M. (1993). Enhanced delivery of doxorubicin to tumor by long-circulating thermosensitive liposomes and local hyperthermia. Biochim. Biophys. Acta 1149 (2), 209–216. doi:10.1016/0005-2736(93)90203-C
Massich, M. D., Giljohann, D. A., Seferos, D. S., Ludlow, L. E., Horvath, C. M., and Mirkin, C. A. (2009). Regulating immune response using polyvalent nucleic acid-gold nanoparticle conjugates. Mol. Pharm. 6 (6), 1934–1940. doi:10.1021/mp900172m
Matsumura, Y., and Maeda, H. (1986). A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 46 (12), 6387–6392.
Maxfield, F. R. (1982). Weak bases and ionophores rapidly and reversibly raise the PH of endocytic vesicles in cultured mouse fibroblasts. J. Cell Biol. 95 (2), 676–681. doi:10.1083/jcb.95.2.676
Mazzon, M., and Marsh, M. (2019). Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Res 8, 1628. doi:10.12688/f1000research.19694.1
McBain, S. C., Griesenbach, U., Xenariou, S., Keramane, A., Batich, C. D., Alton, E. W., et al. (2008). Magnetic nanoparticles as gene delivery agents: enhanced transfection in the presence of oscillating magnet arrays. Nanotechnology 19 (40), 405102. doi:10.1088/0957-4484/19/40/405102
Meier, M., Bider, M. D., Malashkevich, V. N., Spiess, M., and Burkhard, P. (2000). Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor. J. Mol. Biol. 300 (4), 857–865. doi:10.1006/jmbi.2000.3853
Mendes, R., Fernandes, A. R., and Baptista, P. V. (2017). Gold nanoparticle approach to the selective delivery of gene silencing in cancer-the case for combined delivery?. Genes 8 (3), 94. doi:10.3390/genes8030094
Meng, Z., Luan, L., Kang, Z., Feng, S., Meng, Q., and Liu, K. (2017). Histidine-enriched multifunctional peptide vectors with enhanced cellular uptake and endosomal escape for gene delivery. J. Mater. Chem. B 5 (1), 74–84. doi:10.1039/C6TB02862D
Midoux, P., Pigeon, L., Gonçalves, C., and Pichon, C. (2017). Peptides mediating DNA transport on microtubules and their impact on non-viral gene transfer efficiency. Biosci. Rep. 37 (5), BSR20170995. doi:10.1042/BSR20170995
Miller, A. M., and Dean, D. A. (2009). Tissue-specific and transcription factor-mediated nuclear entry of DNA. Adv. Drug Deliv. Rev. 61 (7–8), 603–613. doi:10.1016/j.addr.2009.02.008
Mirkin, C. A., Letsinger, R. L., Mucic, R. C., and Storhoff, J. J. (1996). A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382 (6592), 607–609. doi:10.1038/382607a0
Mislick, K. A., and Baldeschwieler, J. D. (1996). Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc. Natl. Acad. Sci. U.S.A. 93 (22), 12349–12354. doi:10.1073/pnas.93.22.12349
Miura, N., Tange, K., Nakai, Y., Yoshioka, H., Harashima, H., and Akita, H. (2017). Identification and evaluation of the minimum unit of a KALA peptide required for gene delivery and immune activation. J. Pharm. Sci. 106 (10), 3113–3119. doi:10.1016/j.xphs.2017.05.014
Miyauchi, K., Kim, Y., Latinovic, O., Morozov, V., and Melikyan, G. B. (2009). HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137 (3), 433–444. doi:10.1016/j.cell.2009.02.046
Moffett, H. F., Coon, M. E., Radtke, S., Stephan, S. B., McKnight, L., Lambert, A., et al. (2017). Hit-and-Run programming of therapeutic cytoreagents using MRNA nanocarriers. Nat. Commun. 8 (1), 389. doi:10.1038/s41467-017-00505-8
Mok, H., and Park, T. G. (2008). Self-crosslinked and reducible fusogenic peptides for intracellular delivery of SiRNA. Biopolymers 89 (10), 881–888. doi:10.1002/bip.21032
Moon, J. S., Kim, W. G., Kim, C., Park, G. T., Heo, J., Yoo, S. Y., et al. (2015). M13 bacteriophage-based self-assembly structures and their functional capabilities. Mini Rev. Org. Chem. 12 (3), 271–281. doi:10.2174/1570193X1203150429105418
Moret, I., Esteban Peris, J., Guillem, V. M., Benet, M., Revert, F., Dasí, F., et al. (2001). Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline PH, heparin and serum. J. Control. Releas. 76 (1–2), 169–181. doi:10.1016/s0168-3659(01)00415-1
Moriguchi, R., Kogure, K., Akita, H., Futaki, S., Miyagishi, M., Taira, K., et al. (2005). A multifunctional envelope-type nano device for novel gene delivery of SiRNA plasmids. Int. J. Pharm. 301 (1), 277–285. doi:10.1016/j.ijpharm.2005.05.021
Motion, J. P., Nguyen, J., and Szoka, F. C. (2012). Phosphatase-triggered fusogenic liposomes for cytoplasmic delivery of cell-impermeable compounds. Angew. Chem. Int. Ed. Engl. 51 (36), 9047–9051. doi:10.1002/anie.201204198
Mout, R., Ray, M., Yesilbag Tonga, G., Lee, Y. W., Tay, T., Sasaki, K., et al. (2017). Direct cytosolic delivery of CRISPR/Cas9-Ribonucleoprotein for efficient gene editing. ACS Nano 11 (3), 2452–2458. doi:10.1021/acsnano.6b07600
Mukherjee, S., Pfeifer, C. M., Johnson, J. M., Liu, J., and Zlotnick, A. (2006). Redirecting the coat protein of a spherical virus to assemble into tubular nanostructures. J. Am. Chem. Soc. 128 (8), 2538–2539. doi:10.1021/ja056656f
Naghavi, M. H., and Walsh., D. (2017). Microtubule regulation and function during virus infection. J. Virol. 91 (16), e00538-17. doi:10.1128/JVI.00538-17
Nair, J. K., Willoughby, J. L., Chan, A., Charisse, K., Alam, M. R., Wang, Q., et al. (2014). Multivalent N-Acetylgalactosamine-Conjugated SiRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136 (49), 16958–16961. doi:10.1021/ja505986a
Nakagawa, O., Ming, X., Huang, L., and Juliano, R. L. (2010). Targeted intracellular delivery of antisense oligonucleotides via conjugation with small-molecule ligands. J. Am. Chem. Soc. 132 (26), 8848–8849. doi:10.1021/ja102635c
Nakanishi, A., Li, P. P., Qu, Q., Jafri, Q. H., and Kasamatsu, H. (2007). Molecular dissection of nuclear entry-competent SV40 during infection. Virus. Res. 124 (1–2), 226–230. doi:10.1016/j.virusres.2006.10.001
Nakanishi, A., Shum, D., Morioka, H., Otsuka, E., and Kasamatsu, H. (2002). Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. J. VIROL. 76, 9368. doi:10.1128/jvi.76.18.9368-9377.2002
Nanbo, A., Imai, M., Watanabe, S., Noda, T., Takahashi, K., Neumann, G., et al. (2010). Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. Plos Pathog. 6 (9), e1001121. doi:10.1371/journal.ppat.1001121
Nanna, A. R., Kel’in, A. V., Theile, C., Zhi Xiang, V., Garg, A., Nair, J. K., Maier, M. A., et al. (2020). Generation and validation of structurally defined antibody-siRNA conjugates. Nucleic Acids Res. 48 (10), 5281–5293. doi:10.1093/nar/gkaa286
Nayerossadat, N., Maedeh, T., and Ali, P. A. (2012). Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 1, 27. doi:10.4103/2277-9175.98152
Needham, D., Anyarambhatla, G., Kong, G., and Dewhirst, M. W. (2000). A new temperature-sensitive liposome for use with mild hyperthermia: characterization and testing in a human tumor xenograft model. Cancer Res. 60 (5), 1197–1201. doi:10.1093/analys/60.1.13
Nelemans, L. C., and Gurevich, L. (2020). Drug delivery with polymeric nanocarriers-cellular uptake mechanisms. Materials 13 (2), 366. doi:10.3390/ma13020366
Nemerow, G. R., and Stewart, P. L. (1999). Role of αv integrins in adenovirus cell entry and gene delivery. Microbiol. Mol. Biol. Rev. 63 (3), 725–734. doi:10.1128/mmbr.63.3.725-734.1999
Neshatian, M., Chung, S., Yohan, D., Yang, C., and Chithrani, D. B. (2014). Determining the size dependence of colloidal gold nanoparticle uptake in a tumor-like interface (hypoxic). Colloids Interf. Sci. Commun. 1, 57–61. doi:10.1016/j.colcom.2014.07.004
Neumann, G., Castrucci, M. R., and Kawaoka, Y. (1997). Nuclear import and export of influenza virus nucleoprotein. J. Virol. 71 (12), 9690–9700. doi:10.1128/JVI.71.12.9690-9700.1997
Ngamcherdtrakul, W., Morry, J., Gu, S., Castro, D. J., Goodyear, S. M., Sangvanich, T., et al. (2015). Cationic polymer modified mesoporous silica nanoparticles for targeted SiRNA delivery to HER2+ breast cancer. Adv. Funct. Mater. 25 (18), 2646–2659. doi:10.1002/adfm.201404629
Ni, R., Zhou, J., Hossain, N., and Chau, Y. (2016). Virus-inspired nucleic acid delivery system: linking virus and viral mimicry. Adv. Drug Deliv. Rev. 106, 3–26. doi:10.1016/j.addr.2016.07.005
Ni, R., Feng, R., and Chau, Y. (2019). Synthetic approaches for nucleic acid delivery: choosing the right carriers. Life 9 (3), 59. doi:10.3390/life9030059
Nie, Y., Schaffert, D., Rödl, W., Ogris, M., Wagner, E., and Günther, M. (2011). Dual-targeted polyplexes: one step towards a synthetic virus for cancer gene therapy. J. Control. Release 152 (1), 127–134. doi:10.1016/j.jconrel.2011.02.028
Norkin, L. C., Anderson, H. A., Wolfrom, S. A., and Oppenheim, A. (2002). Caveolar endocytosis of Simian virus 40 is followed by Brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76 (10), 5156–5166. doi:10.1128/JVI.76.10.5156-5166.2002
Nunberg, J. H., and York, J. (2012). The curious case of arenavirus entry, and its inhibition. Viruses 4 (1), 83–101. doi:10.3390/v4010083
Oppliger, J., Torriani, G., Herrador, A., and Kunz, S. (2016). Lassa virus cell entry via dystroglycan involves an unusual pathway of macropinocytosis. J. Virol. 90 (14), 6412–6429. doi:10.1128/JVI.00257-16
Orellana, E. A., Abdelaal, A. M., Rangasamy, L., Tenneti, S., Myoung, S., Low, P. S., et al. (2019). Enhancing MicroRNA activity through increased endosomal release mediated by nigericin. Mol. Ther. Nucleic Acids 16, 505–518. doi:10.1016/j.omtn.2019.04.003
Orellana, E. A., Tenneti, S., Rangasamy, L., Lyle, L. T., Low, P. S., and Kasinski, A. L. (2017). FolamiRs: ligand-targeted, vehicle-free delivery of MicroRNAs for the treatment of cancer. Sci. Transl. Med. 9 (401). doi:10.1126/scitranslmed.aam9327
Ortega-Esteban, A., Condezo, G. N., Pérez-Berná, A. J., Chillón, M., Flint, S., Reguera, D., et al. (2015). Mechanics of viral chromatin reveals the pressurization of human adenovirus. ACS Nano 9 (11), 10826–10833. doi:10.1021/acsnano.5b03417
Osborn, M. F., Alterman, J. F., Nikan, M., Cao, H., Didiot, M. C., Hassler, M. R., et al. (2015). Guanabenz (Wytensin™) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Res. 43 (18), 8664–8672. doi:10.1093/nar/gkv942
Palanca-Wessels, M. C., Convertine, A. J., Richelle, C.-S., Booth, G. C., Lee, F., and Berguig, G. Y. (2011). Anti-CD22 antibody targeting of pH-responsive micelles enhances small interfering RNA delivery and gene silencing in lymphoma cells. Mol. Ther. 19 (8), 1529–1537. doi:10.1038/mt.2011.104
Pan, L., He, Q., Liu, J., Chen, Y., Ma, M., Zhang, L., et al. (2012). Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J. Am. Chem. Soc. 134 (13), 5722–5725. doi:10.1021/ja211035w
Pan, Y., Jia, T., Zhang, Y., Zhang, K., Zhang, R., Li, J., et al. (2012a). MS2 VLP-based delivery of MicroRNA-146a inhibits autoantibody production in lupus-prone mice. Int. J. Nanomedicine 7, 5957–5967. doi:10.2147/IJN.S37990
Pan, Y., Zhang, Y., Jia, T., Zhang, K., Li, J., and Wang, L. (2012b). Development of a MicroRNA delivery system based on bacteriophage MS2 virus-like particles. FEBS J. 279 (7), 1198–1208. doi:10.1111/j.1742-4658.2012.08512.x
Passaretti, P., Sun, Y., Dafforn, T. R., and Oppenheimer, P. G. (2020). Determination and characterisation of the surface charge properties of the bacteriophage M13 to assist bio-nanoengineering. RSC Adv. 10 (42), 25385–25392. doi:10.1039/D0RA04086J
Pelkmans, L., Kartenbeck, J., and Helenius, A. (2001). Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3 (5), 473–483. doi:10.1038/35074539
Peng, S., Bie, B., Sun, Y., Liu, M., Cong, H., Zhou, W., et al. (2018). Metal-organic frameworks for precise inclusion of single-stranded DNA and transfection in immune cells. Nat. Commun. 9 (1), 1293. doi:10.1038/s41467-018-03650-w
Penin, F., Combet, C., Germanidis, G., Frainais, P. O., Deléage, G., and Pawlotsky, J. M. (2001). Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75 (12), 5703–5710. doi:10.1128/JVI.75.12.5703-5710.2001
Pinto, L. H., Holsinger, L. J., and Lamb, R. A. (1992). Influenza virus M2 protein has ion channel activity. Cell 69 (3), 517–528. doi:10.1016/0092-8674(92)90452-I
Poddar, A., Conesa, J. J., Liang, K., Dhakal, S., Reineck, P., Bryant, G., et al. (2019). Encapsulation, visualization and expression of genes with biomimetically mineralized zeolitic imidazolate framework-8 (ZIF-8). Small 15 (36), e1902268. doi:10.1002/smll.201902268
Pouton, C., Wagstaff, K., Roth, D., Moseley, G., and Jans, D. (2007). “Targeted Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 59 (8), 698–717. doi:10.1016/j.addr.2007.06.010
Prasad, B. V. V., and Schmid, M. F. (2011). Principles of virus structural organization. Adv. Exp. Med. Biol. 726, 17–47. doi:10.1007/978-1-4614-0980-9_3
Prel, A., Caval, V., Gayon, R., Ravassard, P., Duthoit, C., Payen, E., et al. (2015). Highly efficient in Vitro and in Vivo delivery of functional RNAs using new versatile MS2-chimeric retrovirus-like particles. Mol. Ther. Methods Clin. Dev. 2, 15039. doi:10.1038/mtm.2015.39
Pretto, C., and van Hest, J. C. M. (2019). Versatile reversible cross-linking strategy to stabilize CCMV virus like particles for efficient SiRNA delivery. Bioconjug. Chem. 30 (12), 3069–3077. doi:10.1021/acs.bioconjchem.9b00731
Ramamoorth, M., and Narvekar, A. (2015). Non viral vectors in gene therapy- an overview. J. Clin. Diagn. Res. 9 (1), GE01–GE06. doi:10.7860/JCDR/2015/10443.5394
Ray, M., Tang, R., Jiang, Z., and Rotello, V. M. (2015). Quantitative tracking of protein trafficking to the nucleus using cytosolic protein delivery by nanoparticle-stabilized nanocapsules. Bioconjug. Chem. 26 (6), 1004–1007. doi:10.1021/acs.bioconjchem.5b00141
Rinaldi, S.-H., Zhi, D.-F., Zhao, Y.-N, Chen, H.-Y., Meng, Y., and Zhang, C.-M. (2016). Cationic lioposomes with folic acid as targeting ligand for gene delivery. Bioorg. Med. Chem. Lett. 26 (16), 4025–4029. doi:10.1016/j.bmcl.2016.06.085
Rohovie, M. J., Nagasawa, M., and Swartz, J. R. (2017). Virus-like particles: next-generation nanoparticles for targeted therapeutic delivery. Bioeng. Translational Med. 2 (1), 43–57. doi:10.1002/btm2.10049
Roldão, A., Silva, A. C., Mellado, M. C. M., Alves, P. M., and Carrondo, M. J. T. (2017). Viruses and virus-like particles in biotechnology: fundamentals and applications ☆. Compr. Biotechnol., 633–656. doi:10.1016/B978-0-12-809633-8.09046-4
Roloff, A., Nelles, D. A., Thompson, M. P., Yeo, G. W., and Gianneschi, N. C. (2018). Self-transfecting micellar RNA: modulating nanoparticle cell interactions via high density display of small molecule ligands on micelle coronas. Bioconjug. Chem. 29 (1), 126–135. doi:10.1021/acs.bioconjchem.7b00657
Rosi, N. L., Giljohann, D. A., Thaxton, C. S., Lytton-Jean, A. K. R., Han, M. S., and Mirkin, C. A. (2006). Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312, 1027–1030. doi:10.1126/science.1125559
Rouge, J. L., Sita, T. L., Hao, L., Fotini, M. K., WilliamStegh, E. B., and Alexander, H. (2015). Ribozyme-spherical nucleic acids. J. Am. Chem. Soc. 137 (33), 10528–10531. doi:10.1021/jacs.5b07104
Ruan, W., Zheng, M., An, Y., Liu, Y., Lovejoy, D. B., Hao, M., et al. (2018). DNA nanoclew templated spherical nucleic acids for SiRNA delivery. Chem. Commun. 54 (29), 3609–3612. doi:10.1039/C7CC09257A
Rush, A. M., Thompson, M. P., Tatro, E. T., and Gianneschi, N. C. (2013). Nuclease-resistant DNA via high-density packing in polymeric micellar nanoparticle coronas. ACS Nano 7 (2), 1379–1387. doi:10.1021/nn305030g
Russell, W. C. (2009). Adenoviruses: update on structure and function. J. Gen. Virol. 90 (1), 1–20. doi:10.1099/vir.0.003087-0
Sahay, G., Querbes, W., Alabi, C., Eltoukhy, A., Sarkar, S., Zurenko, C., et al. (2013). Efficiency of SiRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 31 (7), 653–658. doi:10.1038/nbt.2614
Sakurai, Y., Hatakeyama, H., Sato, Y., Hyodo, M., Akita, H., Ohga, N., et al. (2014). RNAi-mediated gene knockdown and anti-angiogenic therapy of RCCs using a cyclic RGD-modified liposomal-SiRNA system. J. Control. Release 173, 110–118. doi:10.1016/j.jconrel.2013.10.003
Samanta, D., Ebrahimi, S. B., Kusmierz, C. D., Cheng, H. F., and Mirkin, C. A. (2020). Protein spherical nucleic acids for live-cell chemical analysis. J. Am. Chem. Soc. 142 (31), 13350–13355. doi:10.1021/jacs.0c06866
Santiana, J. J., Sui, B., Gomez, N., and Rouge, J. L. (2017). Programmable peptide-cross-linked nucleic acid nanocapsules as a modular platform for enzyme specific cargo release. Bioconjug. Chem. 28 (12), 2910–2914. doi:10.1021/acs.bioconjchem.7b00629
Sato, Y., Hatakeyama, H., Sakurai, Y., Hyodo, M., Akita, H., and Harashima, H. (2012). A PH-sensitive cationic lipid facilitates the delivery of liposomal SiRNA and gene silencing activity in Vitro and in Vivo. J. Controll. Release 163 (3), 267–276. doi:10.1016/j.jconrel.2012.09.009
Scherer, J., and Vallee, R. B. (2011). Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to PH-primed hexon. Viruses 3 (8), 1417–1431. doi:10.3390/v3081417
Seferos, D. S., Prigodich, A. E., Giljohann, D. A., Pinal, C., and Mirkin, C. A. (2009). Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 9 (1), 308–311. doi:10.1021/nl802958f
Selby, L. I., Cortez-Jugo, C. M., Such, G. K., and Johnston, A. P. R. (2017). Nanoescapology: progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnolnanomed. Nanobiotech. 9 (5), e1452. doi:10.1002/wnan.1452
Semple, S. C., Akinc, A., Chen, J., Sandhu, A. P., Mui, B. L., Cho, C. K., et al. (2010). Rational design of cationic lipids for SiRNA delivery. Nat. Biotechnol. 28 (2), 172–176. doi:10.1038/nbt.1602
Seth, P. P., Tanowitz, M., and Bennett, C. (2019). Selective tissue targeting of synthetic nucleic acid drugs. J. Clin. Invest. 129 (3), 915–925. doi:10.1172/JCI125228
Shaheen, S. M., Akita, H., Nakamura, T., Takayama, S., Futaki, S., Yamashita, A., et al. (2011). KALA-modified multi-layered nanoparticles as gene carriers for MHC class-I mediated antigen presentation for a DNA vaccine. Biomaterials 32 (26), 6342–6350. doi:10.1016/j.biomaterials.2011.05.014
Shahzad, M. M., Mangala, L. S., Han, H. D., Lu, C., Bottsford-Miller, J., Nishimura, M., et al. (2011). Targeted delivery of small interfering RNA using reconstituted high-density lipoprotein nanoparticles. Neoplasia 13 (4), 309–319. doi:10.1593/neo.101372
Shete, H. K., Prabhu, R. H., and Patravale, V. B. (2014). Endosomal escape: a bottleneck in intracellular delivery. J. Nanosci Nanotechnol 14 (1), 460–474. doi:10.1166/jnn.2014.9082
Shi, Q., Nguyen, A. T., Angell, Y., Deng, D., Na, C. R., Burgess, K., et al. (2010). A combinatorial approach for targeted delivery using small molecules and reversible masking to bypass nonspecific uptake in Vivo. Gene Ther. 17 (9), 1085–1097. doi:10.1038/gt.2010.55
Shimojima, M., Ströher, U., Ebihara, H., Feldmann, H., and Kawaoka, Y. (2012). Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J. Virol. 86 (4), 2067–2078. doi:10.1128/JVI.06451-11
Sievers, E. L., Larson, R. A., Stadtmauer, E. A., Estey, E., Löwenberg, B., Dombret, H., et al. (2001). Efficacy and safety of gemtuzumab ozogamicin in patients with CD33-positive acute myeloid leukemia in first relapse. J. Clin. Oncol. 19 (13), 3244–3254. doi:10.1200/JCO.2001.19.13.3244
Sikorski, A. F., Toporkiewicz, M., Meissner, J., Matusewicz, L., and Czogalla, A. (2015). Toward a magic or imaginary bullet? Ligands for drug targeting to cancer cells: principles, hopes, and challenges, Int. J. Nanomedicine 10, 1399. doi:10.2147/IJN.S74514
Simeoni, F. (2003). Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of SiRNA into mammalian cells. Nucleic Acids Res. 31 (11), 2717–2724. doi:10.1093/nar/gkg385
Sokolova, V., and Epple, M. (2008). Inorganic nanoparticles as carriers of nucleic acids into cells. Angew. Chem. Int. Ed. 47 (8), 1382–1395. doi:10.1002/anie.200703039
Springer, A. D., and Dowdy, S. F. (2018). GalNAc-SiRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 28 (3), 109–118. doi:10.1089/nat.2018.0736
St-Pierre, G., Pal, S., Østergaard, M. E., Zhou, T., Yu, J., Tanowitz, M., et al. (2016). Synthesis and biological evaluation of sialyl-oligonucleotide conjugates targeting leukocyte B trans-membranal receptor CD22 as delivery agents for nucleic acid drugs. Bioorg. Med. Chem. 24 (11), 2397–2409. doi:10.1016/j.bmc.2016.03.047
Staring, J., Raaben, M., and Brummelkamp, T. R. (2018). Viral escape from endosomes and host detection at a glance. J. Cell Sci. 131 (15), jcs.216259. doi:10.1242/jcs.216259
Stewart, P. L., and Nemerow, G. R. (2007). Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol. 15 (11), 500–507. doi:10.1016/j.tim.2007.10.001
Subbarao, N. K., Parente, R. A., Szoka, F. C., Nadasdi, L., and Pongracz, K. (1987). PH-dependent bilayer destabilization by an amphipathic peptide. Biochemistry 26 (11), 2964–2972. doi:10.1021/bi00385a002
Sugiyama, M., Matsuura, M., Takeuchi, Y., Kosaka, J., Nango, M., and Oku, N. (2004). Possible mechanism of polycation liposome (PCL)-Mediated gene transfer. Biochim. Biophys. Acta 1660 (1–2), 24–30. doi:10.1016/j.bbamem.2003.10.012
Summerton, J. E. (2006). “Morpholinos and PNAs compared,” in Peptide nucleic acids, morpholinos and related antisense biomolecules. Editors C. G. Janson, and M. J. During (Boston, MA: Medical Intelligence), 89–113. doi:10.1007/0-387-32956-0_6
Sun, H., Zhu, X., Lu, P. Y., Rosato, R. R., Tan, W., and Zu, Y. (2014). Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 3, e182. doi:10.1038/mtna.2014.32
Sun, P., Li, Z., Wang, J., Gao, H., Yang, X., Wu, S., et al. (2018). Transcellular delivery of messenger RNA payloads by a cationic supramolecular MOF platform. Chem. Commun. 54 (80), 11304–11307. doi:10.1039/C8CC07047D
Sun, Y., Zheng, L., Yang, Y., Qian, X., Fu, T., Li, X., et al. (2020). Metal-organic framework nanocarriers for drug delivery in biomedical applications. Nano-micro Lett. 12 (1), 103. doi:10.1007/s40820-020-00423-3
Takemoto, H., Miyata, K., Nishiyama, N., and Kataoka, K. (2014). “Bioresponsive polymer-based nucleic acid carriers,” in Advances in genetics. Editors L. Huang, D. Liu, and E. Wagner (Cambridge, MA: Academic Press), 88, 289–323. doi:10.1016/B978-0-12-800148-6.00010-9Adv. Genet.
Tang, Q., Liu, J., Jiang, Y., Zhang, M., Mao, L., and Wang, M. (2019). Cell-selective messenger RNA delivery and CRISPR/Cas9 genome editing by modulating the interface of phenylboronic acid-derived lipid nanoparticles and cellular surface sialic acid. ACS Appl. Mater. Inter. 11 (50), 46585–46590. doi:10.1021/acsami.9b17749
Tang, R., Kim, C. S., and Solfiell, D. J. (2013). Solfiell, Subinoy Rana, Rubul Mout, Elih M. Velázquez-Delgado, Apiwat Chompoosor, et al.Direct Delivery of Functional Proteins and Enzymes to the Cytosol Using Nanoparticle-Stabilized Nanocapsules. ACS Nano 7 (8), 6667–6673. doi:10.1021/nn402753y
Tatsis, N., and Ertl, H. C. (2004). Adenoviruses as vaccine vectors. Mol. Ther. 10 (4), 616–629. doi:10.1016/j.ymthe.2004.07.013
Teplensky, M. H., Fantham, M., Poudel, C., Hockings, C., Lu, M., Guna, A., et al. (2019). A highly porous metal-organic framework system to deliver payloads for gene knockdown. Chem 5 (11), 2926–2941. doi:10.1016/j.chempr.2019.08.015
Thomas, M., and Klibanov, A. M. (2003). Non-viral gene therapy: polycation-mediated DNA delivery. Appl. Microbiol. Biotechnol. 62 (1), 27–34. doi:10.1007/s00253-003-1321-8
Tian, Y., Wu, M., Liu, X., Liu, Z., Zhou, Q., Niu, Z., et al. (2015). Probing the endocytic pathways of the filamentous bacteriophage in live cells using ratiometric PH fluorescent indicator. Adv. Healthc. Mater. 4 (3), 413–419. doi:10.1002/adhm.201400508
Tolentino, M. Q., Hartmann, A. K., Loe, D. T., and Rouge, J. L. (2020). Controlled release of small molecules and proteins from DNA-surfactant stabilized metal organic frameworks. J. Mater. Chem. B 8, 5627. doi:10.1039/D0TB00767F
Tomar, R. S., Matta, H., and Chaudhary, P. M. (2003). Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene 22 (36), 5712–5715. doi:10.1038/sj.onc.1206733
Tomita, T., Watanabe, M., Takahashi, T, Kumai, K., Tadakuma, T., and Yasuda, T. (1989). Temperature-sensitive release of adriamycin, an amphiphilic antitumor agent, from dipalmitoylphosphatidylcholine-cholesterol liposomes. Biochim. Biophys. Acta 978 (2), 185–190. doi:10.1016/0005-2736(89)90113-2
Toscano, M. G., and de Haan, P. (2018). How simian virus 40 hijacks the intracellular protein trafficking pathway to its own benefit and ours and ours. Front. Immunol. 9, 1160. doi:10.3389/fimmu.2018.01160
Troiber, C., and Wagner, E. (2011). Nucleic acid carriers based on precise polymer conjugates. Bioconjug. Chem. 22 (9), 1737–1752. doi:10.1021/bc200251r
Truong, N. P., Gu, W., Prasadam, I., Jia, Z., Crawford, R., Xiao, Y., et al. (2013). An influenza virus-inspired polymer system for the timed release of SiRNA. Nat. Commun. 4 (1), 1902. doi:10.1038/ncomms2905
Tsukuda, S., and Watashi, K. (2020). Hepatitis B virus biology and life cycle. Antivir. Res. 182, 104925. doi:10.1016/j.antiviral.2020.104925
Urie, R., and Rege, K. (2015). Nanoscale inorganic scaffolds as therapeutics and delivery vehicles. Curr. Opin. Chem. Eng. 7, 120–128. doi:10.1016/j.coche.2014.12.004
Venkatakrishnan, B., and Zlotnick, A. (2016). The structural biology of hepatitis B virus: form and function. Annu. Rev. Virol. 3 (1), 429–451. doi:10.1146/annurev-virology-110615-042238
Verdine, G. L., and Walensky, L. D. (2007). The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 13 (24), 7264–7270. doi:10.1158/1078-0432.CCR-07-2184
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell. Biol. 13 (4), 423–433. doi:10.1038/ncb2210
Villagrana-Escareño, M. V., Reynaga-Hernández, E., Galicia-Cruz, O. G., Durán-Meza, A. L., De la Cruz-González, V., Hernández-Carballo, C. Y., et al. (2019). VLPs derived from the CCMV plant virus can directly transfect and deliver heterologous genes for translation into mammalian cells. Biomed. Res. Int. 2019, 4630891. doi:10.1155/2019/4630891
Volpers, C., and Kochanek, S. (2004). Adenoviral vectors for gene transfer and therapy. J. Gene Med. 6 (S1), S164–S171. doi:10.1002/jgm.496
Vorburger, S. A., and Hunt, K. K. (2002). Adenoviral gene therapy. Oncologist 7, 46–59. doi:10.1634/theoncologist.7-1-46
Wagenaar, T. R., Tolstykh, T., Shi, C., Jiang, L., Zhang, J., Li, Z., et al. (2015). Identification of the endosomal sorting complex required for transport-I (ESCRT-I) as an important modulator of anti-MiR uptake by cancer cells. Nucleic Acids Res. 43 (2), 1204–1215. doi:10.1093/nar/gku1367
Wagner, E. (2012). Polymers for SiRNA delivery: inspired by viruses to Be targeted, dynamic, and precise. Acc. Chem. Res. 45 (7), 1005–1013. doi:10.1021/ar2002232
Wang, C. Y., and Huang, L. (1989). Highly efficient DNA delivery mediated by PH-sensitive immunoliposomes. Biochemistry 28 (24), 9508–9514. doi:10.1021/bi00450a039
Wang, H., Shi, Y., Song, J., Qi, J., Lu, G., Yan, J., et al. (2016a). Ebola viral glycoprotein bound to its endosomal receptor niemann-pick C1. Cell 164 (1), 258–268. doi:10.1016/j.cell.2015.12.044
Wang, H., Tam, Y. Y., Chen, S., Zaifman, J., van der Meel, R., Ciufolini, M. A., et al. (2016b). The niemann-pick C1 inhibitor NP3.47 enhances gene silencing potency of lipid nanoparticles containing SiRNA. Mol. Ther. 24 (12), 2100–2108. doi:10.1038/mt.2016.179
Wang, L., Ariyarathna, Y., Ming, X., Yang, B., James, L. I., Kreda, S. M., et al. (2017). A novel family of small molecules that enhance the intracellular delivery and pharmacological effectiveness of antisense and splice switching oligonucleotides. ACS Chem. Biol. 12 (8), 1999–2007. doi:10.1021/acschembio.7b00242
Wang, X., Huang, D. Y., Huong, S.-M., and Huang, E.-S. (2005). Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 11 (5), 515–521. doi:10.1038/nm1236
Wang, Z., Chui, W. K., and Ho, P. C. (2010). Integrin targeted drug and gene delivery. Expert Opin. Drug Deliv. 7 (2), 159–171. doi:10.1517/17425240903468696
Wasungu, L., and Hoekstra, D. (2006). Cationic lipids, lipoplexes and intracellular delivery of genes. J. Controlled Release 116 (2), 255–264. doi:10.1016/j.jconrel.2006.06.024
Wheeler, J. J., Palmer, L., Ossanlou, M., MacLachlan, I., Graham, R. W., Zhang, Y. P., et al. (1999). Stabilized plasmid-lipid particles: construction and characterization. Gene Ther. 6 (2), 271–281. doi:10.1038/sj.gt.3300821
Whitehead, K. A., Langer, R., and Anderson, D. G. (2009). Knocking down barriers: advances in SiRNA delivery. Nat. Rev. Drug Discov. 8 (2), 129–138. doi:10.1038/nrd2742
Wilen, C. B., Tilton, J. C., and Doms, R. W. (2012). Molecular mechanisms of HIV entry. Adv. Exp. Med. Biol. 726, 223–242. doi:10.1007/978-1-4614-0980-9_10
Wilner, S. E., Wengerter, B., Maier, K., de Lourdes Borba Magalhães, , Del Amo, D. S., Pai, S., et al. (2012). An RNA alternative to human transferrin: a new tool for targeting human cells. Mol. Ther. Nucleic Acids 1 (5), e21. doi:10.1038/mtna.2012.14
Wood, H. (2018). FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat. Rev. Neurol. 14 (10), 570. doi:10.1038/s41582-018-0065-0
Wyman, T. B., Nicol, F., Zelphati, O., Scaria, P. V., Plank, C., and Szoka, F. C. (1997). Design, synthesis, and characterization of a cationic peptide that binds to nucleic acids and permeabilizes bilayers. Biochemistry 36 (10), 3008–3017. doi:10.1021/bi9618474
Xiang, B., Jia, X. L., Qi, J. L., Yang, L. P., Sun, W. H., Yan, X., et al. (2017). Enhancing SiRNA-based cancer therapy using a new PH-responsive activatable cell-penetrating peptide-modified liposomal system. Int. J. Nanomedicine 12, 2385–2405. doi:10.2147/IJN.S129574
Xie, X., Liao, J., Shao, X., Li, Q., and Lin, Y. (2017). The effect of shape on cellular uptake of gold nanoparticles in the forms of stars, rods, and triangles. Sci. Rep. 7 (1), 3827. doi:10.1038/s41598-017-04229-z
Xu, D., McCarty, D., Fernandes, A., Fisher, M., Samulski, R. J., and Juliano, R. L. (2005). Delivery of MDR1 small interfering RNA by self-complementary recombinant adeno-associated virus vector. Mol. Ther. 11 (4), 523–530. doi:10.1016/j.ymthe.2004.12.019
Xue, H. Y., Guo, P., Wen, W. C., and Wong, H. L. (2015). Lipid-based nanocarriers for RNA delivery. Curr. Pharm. Des. 21 (22), 3140–3147. doi:10.2174/1381612821666150531164540
Yamauchi, Y., and Greber, U. F. (2016). Principles of virus uncoating: cues and the snooker ball. Traffic 17 (6), 569–592. doi:10.1111/tra.12387
Yamazaki, Y., Nango, M., Matsuura, M., Hasegawa, Y., Hasegawa, M., and Oku, N. (2000). Polycation liposomes, a novel nonviral gene transfer system, constructed from cetylated polyethylenimine. Gene Ther. 7 (13), 1148–1155. doi:10.1038/sj.gt.3301217
Yan, A. C., and Levy., M. (2018). Aptamer-mediated delivery and cell-targeting aptamers: room for improvement. Nucleic Acid Ther. 28 (3), 194–199. doi:10.1089/nat.2018.0732
Yang, B., Ming, X., Cao, C., Laing, B., Yuan, A., Porter, M. A., et al. (2015). High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides. Nucleic Acids Res. 43 (4), 1987–1996. doi:10.1093/nar/gkv060
Yang, X. C., Samanta, B., Agasti, S. S., Jeong, Y., Zhu, Z. J., Rana, S., et al. (2011). Drug delivery using nanoparticle-stabilized nanocapsules. Angew. Chem. Int. Ed. Engl. 50 (2), 477–481. doi:10.1002/anie.201005662
Yao, Y., Jia, T., Pan, Y., Gou, H., Li, Y., Sun, Y., et al. (2015). Using a novel MicroRNA delivery system to inhibit osteoclastogenesis. Int. J. Mol. Sci. 16 (4), 8337–8350. doi:10.3390/ijms16048337
Yata, T., Lee, K. Y., Dharakul, T., Songsivilai, S., Bismarck, A., Mintz, P. J., et al. (2014). Hybrid nanomaterial complexes for advanced phage-guided gene delivery. Mol. Ther. Nucleic Acids 3, e185. doi:10.1038/mtna.2014.37
Yatvin, M. B., Kreutz, W., Horwitz, B. A., and Shinitzky, M. (1980). PH-sensitive liposomes: possible clinical implications. Science 210 (4475), 1253–1255. doi:10.1126/science.7434025
Yatvin, M. B., Weinstein, J. N., Dennis, W. H., and Blumenthal, R. (1978). Design of liposomes for enhanced local release of drugs by hyperthermia. Science 202 (4374), 1290–1293. doi:10.1126/science.364652
Ye, S.-f., Tian, M.-M., Wang, T.-X., Ren, L., Wang, D., Shen, L.-H., et al. (2012). Synergistic effects of cell-penetrating peptide tat and fusogenic peptide HA2-enhanced cellular internalization and gene transduction of organosilica nanoparticles. Nanomed.: Nanotechno., Biol. Med. 8 (6), 833–841. doi:10.1016/j.nano.2011.10.003
Yen, A., Cheng, Y., Sylvestre, M., Gustafson, H. H., Puri, S., and Pun, S. H. (2018). Serum nuclease susceptibility of MRNA cargo in condensed polyplexes. Mol. Pharm. 15 (6), 2268–2276. doi:10.1021/acs.molpharmaceut.8b00134
Yin, H., Kanasty, R. L., Eltoukhy, A. A., Vegas, A. J., Dorkin, J., and Anderson, D. G. (2014). Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15 (8), 541–555. doi:10.1038/nrg3763
Yonenaga, N., Kenjo, E., Asai, T., Tsuruta, A., Shimizu, K., Dewa, T., et al. (2012). RGD-based active targeting of novel polycation liposomes bearing SiRNA for cancer treatment. J. Control Release 160 (2), 177–181. doi:10.1016/j.jconrel.2011.10.004
Younes, A., Bartlett, N. L., Leonard, J. P., and Kennedy, D. A. (2010). Brentuximab vedotin (SGN-35) for relapsed CD30-positive lymphomas. N. Engl. J. Med. 363 (19), 1812–1821. doi:10.1056/NEJMoa1002965
Young, K. L., Scott, A. W., Hao, L., Sarah, E. M., Guoliang, L., and Mirkin, C. A. (2012). Hollow spherical nucleic acids for intracellular gene regulation based upon biocompatible silica shells. Nano Lett. 12 (7), 3867–3871. doi:10.1021/nl3020846
Yuan, W., and Li, H. (2017). “Polymer-based nanocarriers for therapeutic nucleic acids delivery,” in Nanostructures for drug delivery. 445–460. doi:10.1016/B978-0-323-46143-6.00014-2
Zanta, M. A., Belguise-Valladier, P., and Behr, J. P. (1999). Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. U.S.A. 96 (1), 91–96. doi:10.1073/pnas.96.1.91
Zatsepin, T. S., Kotelevtsev, Y. V., and Koteliansky, V. (2016). Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int. J. Nanomedicine 11, 3077–3086. doi:10.2147/IJN.S106625
Zhang, J., Chen, R., Fang, X., Chen, F., Wang, Y., and Chen, M. (2015). Nucleolin targeting AS1411 aptamer modified PH-sensitive micelles for enhanced delivery and antitumor efficacy of paclitaxel. Nano Res. 8 (1), 201–218. doi:10.1007/s12274-014-0619-4
Zhao, N., Bagaria, H. G., Wong, M. S., and Zu, Y. (2011). A nanocomplex that is both tumor cell-selective and cancer gene-specific for anaplastic large cell lymphoma. J. Nanobiotechnology 9, 2. doi:10.1186/1477-3155-9-2
Zhi, D., Zhang, S., Cui, S., Zhao, Y., Wang, Y., and Zhao, D. (2013). The headgroup evolution of cationic lipids for gene delivery. Bioconjug. Chem. 24 (4), 487–519. doi:10.1021/bc300381s
Zhu, L., and Mahato, R. I. (2010). Lipid and polymeric carrier-mediated nucleic acid delivery. Expert Opin. Drug Deliv. 7 (10), 1209–1226. doi:10.1517/17425247.2010.513969
Keywords: nucleic acid delivery, viral mimicry, endosomal escape, nuclear targeting, nanoparticles
Citation: de la Fuente IF, Sawant SS, Tolentino MQ, Corrigan PM and Rouge JL (2021) Viral Mimicry as a Design Template for Nucleic Acid Nanocarriers. Front. Chem. 9:613209. doi: 10.3389/fchem.2021.613209
Received: 01 October 2020; Accepted: 06 January 2021;
Published: 10 March 2021.
Edited by:
Chun Xu, The University of Queensland, Brisbane, Qld, AustraliaReviewed by:
Chad A. Mirkin, Northwestern University, Evanston, IL, United StatesKai Ewert, University of California, Santa Barbara, CA, United States
Copyright © 2021 de la Fuente, Sawant, Tolentino, Corrigan and Rouge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica L. Rouge, amVzc2ljYS5yb3VnZUB1Y29ubi5lZHU=
 Ina F. de la Fuente
Ina F. de la Fuente Shraddha S. Sawant
Shraddha S. Sawant Mark Q. Tolentino
Mark Q. Tolentino Patrick M. Corrigan
Patrick M. Corrigan Jessica L. Rouge
Jessica L. Rouge