
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem., 05 February 2020
Sec. Nanoscience
Volume 8 - 2020 | https://doi.org/10.3389/fchem.2020.00051
This article is part of the Research TopicBio-Nano Interfaces: From Biosensors to NanomedicineView all 7 articles
Protein aggregation, involving the formation of dimers, oligomers, and fibrils, is associated with many human diseases. Type 2 diabetes is one of the common amyloidosis and linked with the aggregation of human islet amyloid polypeptide (hIAPP). A series of nanoparticles are reported to be able to interact with proteins and enhance/inhibit protein aggregation. However, the effects of C60 (a model system of hydrophobic nanoparticle) and C60(OH)8 (a hydroxylated fullerene) on hIAPP aggregation remain unknown. In this study, we investigate the influences of pristine fullerene C60 and hydroxylated C60 on the dimerization of hIAPP using molecular dynamics (MD) simulations. Extensive replica exchange molecular dynamics (REMD) simulations show that isolated hIAPP dimers adopt β-sheet structure containing the amyloid-precursor (β-hairpin). Both C60 and C60(OH)8 notably inhibit the β-sheet formation of hIAPP dimer and induce the formation of collapsed disordered coil-rich conformations. Protein—nanoparticle interaction analyses reveal that the inhibition of hIAPP aggregation by C60 is mainly via hydrophobic and aromatic-stacking interactions, while the prevention of hIAPP aggregation by C60(OH)8 is mostly through collective hydrogen bonding and aromatic-stacking interactions. Conventional MD simulations indicate that both C60 and C60(OH)8 weaken the interactions within hIAPP protofibril and disrupt the β-sheet structure. These results provide mechanistic insights into the possible inhibitory mechanism of C60 and C60(OH)8 toward hIAPP aggregation, and they are of great reference value for the screening of potent amyloid inhibitors.
Human islet amyloid polypeptide (hIAPP) is an intrinsically disordered protein and plays a significant role in the progression of type 2 diabetes (Cooper et al., 1987). hIAPP has a high propensity to form amyloid aggregates (Larson and Miranker, 2004; Brender et al., 2010). Amyloid deposits derived from hIAPP are observed in human islet extracellular space in type 2 diabetes and the formation of intracellular hIAPP oligomers may conduce to β-cell loss in Type 2 diabetes (Haataja et al., 2008). Inhibition of hIAPP aggregation and destabilization of preformed hIAPP fibrils are considered as two major therapeutic strategies for treating Type 2 diabetes. Finding an effective inhibitor of hIAPP aggregation is a crucial step for reducing islet β-cells death and the development of drugs against Type 2 diabetes. Researchers have made great efforts to search for inhibitors against hIAPP aggregation. Increasing experimental studies show that peptides (Yan et al., 2006; Abedini et al., 2007; Saunders et al., 2016), and natural small molecules (Cao and Raleigh, 2012; Palhano et al., 2013; Young et al., 2015; Pithadia et al., 2016) can modulate hIAPP aggregation and drive the peptides into disordered off-pathway aggregates which almost has no toxicity. Very recently, Ke et al. demonstrated that nanomaterials can inhibit hIAPP aggregation and reduce the toxicity in silico, in vitro, and in vivo (Wang et al., 2018; Faridi et al., 2019; Ke et al., 2019).
Carbon nanoparticles including graphene, carbon nanotube, fullerene, and its derivatives (especially hydroxylated fullerenes) have also been of great concern due to their excellent physicochemical properties (Mahmoudi et al., 2013) [such as high capacity to cross biological barriers (Tsuchiya et al., 1996; Sumner et al., 2010), low biotoxicity (Zhu et al., 2007), and high solubility (Da Ros and Prato, 1999; Maciel et al., 2011)]. Experimental studies have demonstrated that fullerenes and their derivatives can prevent the aggregation of amyloid proteins. For example, pristine fullerenes, carboxyfullerenes, and hydroxylated fullerene, strongly inhibit the aggregation of Aβ and Aβ fragments (Dugan et al., 1997; Kim and Lee, 2003; Podolski et al., 2007; Bobylev et al., 2011). Hydroxylated carbon nanotubes can significantly impede the aggregation of hIAPP (Mo et al., 2018). Graphene quantum dots are able to prevent the aggregation of hIAPP and reduce the toxicity in vivo (Wang et al., 2018). On the computational side, researchers investigated the interactions of amyloid proteins and carbon nanoparticles at atomic level of details with an attempt to uncover the underlying inhibitory mechanisms. By atomistic replica exchange molecular dynamics (REMD) simulations, Li et al. found that carbon nanotube can significantly suppress the formation of β-sheet rich Aβ16−22 oligomers (Li et al., 2011). Using the same simulation method, Xie et al. explored the effect of different size of fullerenes on the aggregation of Aβ16−22. Their simulations showed that fullerene C180, albeit with a smaller surface area than 3C60, exhibits an unexpectedly more effective inhibition of β-sheet formation. The stronger inhibition of β-sheet formation by C180 is due to the stronger hydrophobic and aromatic-stacking interactions between the fullerene hexagonal rings and the Phe rings than that between the pentagonal rings and the Phe rings (Xie et al., 2014). MD simulations revealed that C60 can destabilize Aβ protofibrils by disrupting the D23–K28 salt bridge (Andujar et al., 2012; Zhou et al., 2014). Guo et al. explored the influences of graphene, carbon nanotube, and C60 on oligomerization of IAPP22−28 fragment and found that these carbon nanoparticles inhibit the formation of the β-sheet-rich oligomers (Guo et al., 2013). However, questions remain to be addressed. For example, can pristine C60 inhibit the aggregation of full length hIAPP and disrupt hIAPP protofibrils? If yes, what is the inhibitory mechanism and how different is it from that of hydroxylated C60?
In this work, we conducted extensive explicit solvent replica-exchange molecular dynamics (REMD) simulations on hIAPP dimer with and without four C60/C60(OH)8 nanoparticles. Our aim is to explore the effects of pristine and hydroxylated C60 nanoparticles on full-length hIAPP aggregation. REMD simulations showed that both C60 and C60(OH)8 display a strong inhibition of β-sheet formation. The nanoparticle—peptide interactions analyses revealed that the strong β-sheet inhibition results from the strong binding of C60/C60(OH)8 to hIAPP. C60 preferentially binds to the hydrophobic residues and aromatic residues, while C60(OH)8 has a relatively high probability to bind to hydrophilic residues and aromatic residues. In addition, to examine whether C60/C60(OH)8 nanoparticles can disrupt the preformed protofibril, we carried out conventional MD simulations for hIAPP protofibril in the absence and presence of C60/C60(OH)8. The MD simulations revealed that both C60 and C60(OH)8 can disrupt the β-sheet structure and destabilize hIAPP protofibril.
The hIAPP dimer with/without C60/C60(OH)8 nanoparticles, were simulated, and they were denoted as hIAPP-dimer, hIAPP-dimer + C60 and hIAPP-dimer + C60(OH)8. hIAPP has 37 amino acid residues (with sequence KCNTATCATQ10 RLANFLVHSS20 NNFGAILSST30 NVGSNTY) with an amidated C-terminus and a disulfide bond forming between Cys2 and Cys7. In accordance with previous experimental studies (Nanga et al., 2011), the N-terminus, the side chains of Lys1 and Arg11 were protonated (NH3+, Lys+, and Arg+). And the sidechain of H18 was uncharged to mimic the experimental conditions with pH of ~7.3 (Goldsbury et al., 2000). It is true that the protonation state of His will change along with the local environment changes. As done recently by other groups (Dupuis et al., 2009, 2011; Deng et al., 2013; Qiao et al., 2013), we neglected the pKa shift in all of our simulations as the involvement of pKa calculation in MD simulations makes it very computationally expensive. Every hIAPP dimer was put in the center of a cubic box with a side length of 6.7 nm. Four C60 or four C60(OH)8 molecules were displaced in the solvated peptide system at random thus nanoparticle: peptide is 2:1 at molar ratio, which were consistent with previous simulation studies (Bai et al., 2019). The partial charges of oxygen and hydrogen atoms in hydroxyl groups were −0.8 and +0.3, and that of carbon atoms bonded with hydroxyl groups in C60(OH)8 was +0.5, while other carbon atoms were uncharged (Goldsbury et al., 2000). Six counter ions (Cl−) were added to the three systems for neutralization. More details about the system preparation could be found in the Supporting Information section.
The hIAPP protofibril with/without C60/C60(OH)8 nanoparticles, are simulated, and they are denoted as hIAPP-protofibril, hIAPP-protofibril + C60 and hIAPP-protofibril + C60(OH)8 systems. Kindly provided by Professor Tycko (Luca et al., 2007), the initial structure of hIAPP protofibril is a hIAPP decamer including two rotationally symmetric protofibrillar pentamers, Each pentamer contains two anti-parallel β-strand, β1 (spanning residues 8–17) and β2 (spanning residues 28–37), and a “loop” (spanning residues 18–27). The three regions together formed a U-shaped structure. There are 20 C60/C60(OH)8 nanoparticles in hIAPP-protofibril + C60/C60(OH)8 systems (nanoparticle: peptide = 2:1 in molar ratio). Thirty counter ions (Cl−) are added to the systems for neutralization. C60/C60(OH)8 nanoparticles are randomly distributed in the simulation box. The dimensions of the simulation box are 9.5 × 9.5 × 9.5 nm3 for all the three systems.
Both REMD and MD simulations are performed in the isothermal-isobaric (NPT) ensemble at a pressure of 1 bar using GROMACS−4.5.3 software package (Sugita and Okamoto, 1999; Nadler and Hansmann, 2008). We choose OPLS force field TIP4P water molecules, consistent with previous computational studies of hIAPP (Qi et al., 2014; Mo et al., 2016; Bai et al., 2019). hIAPP molecules are kept from the water box at least for 1.0 nm. There are 48 replicas for each system in the REMD simulations, at different temperatures exponentially distributed from 306 to 409 K. Every 1,000 integration steps, two adjacent replicas attempt to exchange with an average acceptance ratios of ~15% for each system. The integration time step is 2 fs. Each replica was simulated for 360 ns, and thus the accumulative simulation time period for each system was 17.28 μs. For MD simulations, we perform two individual 300 ns simulations for each of the three systems: hIAPP-protofibril and hIAPP-protofibril + C60 and hIAPP-protofibril + C60(OH)8.
The tools implemented in GROMACS software package and our in-house developed codes were both used to analyze the trajectories. For REMD simulations, we chose the last 160 ns simulation data for analysis as the first 200 ns data of each replica may have bias of the initial structures. Daura method was used for cluster analysis with a Cα-root-mean-square deviation (Cα-RMSD) cutoff of 0.35 nm (Daura et al., 1999). We analyzed the REMD trajectories by calculating the secondary structure propensities by the DSSP program, number of hydrogen bonds (H-bonds), percentage of β-strand length, free energy landscape (or potential of mean force), hIAPP–nanoparticles binding probabilities, and hIAPP–nanoparticles contact surface area (CSA), pairwise residue contact probabilities of both main chain-main chain (MC-MC) contact and side chain-side chain (SC-SC) contact. For MD simulations, H-bond number within hIAPP protofibril and between hIAPP and nanoparticles, secondary structure probabilities of hIAPP and the hIAPP–nanoparticle binding probability were calculated. The VMD (Humphrey et al., 1996) and Pymol (Schrodinger, 2015) programs were used for graphical structure analysis and trajectory visualization.
For each system at 310 K, we examined the data convergence within two different time intervals (200–280 and 280–360 ns) before analyzing the REMD simulation data by comparing four parameters as followed. These parameters include the probability density functions (PDF) of the radius of gyration (Rg) and the hydrogen bond (H-bond), the probabilities of coil, β-sheet and helix structure of each amino acid residue. As it can be seen in Figures S2–S5, the simulation data from the two independent time periods coincide very well in terms of all these parameters, demonstrating that the last 160 ns REMD simulations nicely converged. Unless specified, all the REMD simulation results presented below are based on the last 160 ns (t = 200–360 ns) simulation data generated at 310 K.
We first examined the percentages of different types of secondary structure formed by hIAPP dimer in each REMD system and the results were listed in Table 1. For the isolated hIAPP dimer, the probabilities of coil and β-sheet are 40.5 and 10.6%, respectively. The secondary structure propensities are in good agreement with previous circular dichroism (CD) studies (Kayed et al., 1999; Goldsbury et al., 2000) and with our recent REMD simulation results using AMBER99SB-ILDN force field (Lao et al., 2019). In comparison of the secondary structure content of isolated hIAPP dimer, the β-sheet contents of hIAPP dimer with C60 or C60(OH)8 are reduced from 10.6% (hIAPP-dimer system) to 1.8% (hIAPP-dimer + C60 system) or 4.2% (hIAPP-dimer + C60(OH)8 system) and the probability of coil increases from 40.5 to 46.7% or 48.2%. The influences of C60/C60(OH)8 on the probabilities of helix, turn, bend, and β-bridge structures are minor. The dominant secondary structure (β-sheet and coil) propensities of each residue of hIAPP dimer in the three systems are presented in Figures 1A,B. Figure 1A shows that residues in Q10–L16, S20–S29, and T30–T36 regions have the highest probabilities (7.0–22.7%) to form β-sheets. The three regions are reported to be the amyloidogenic regions in many experimental studies (Jaikaran et al., 2001; Nielsen et al., 2009; Bedrood et al., 2012; Zhang et al., 2013; Weirich et al., 2016). With the addition of C60, those β-sheet rich regions all display a reduced β-sheet probability (0.01–4.9%) (the dark cyan bars in Figure 1A). In hIAPP-dimer + C60(OH)8 system, except for polar residues N21, N22, S28, and S29, other residues including hydrophobic residues F23–L27 located in the primary amyloid core region (S20–S29) (Goldsbury et al., 2000; Brender et al., 2008; Dupuis et al., 2011) exhibit a dramatically decreased β-sheet probability (the orange bars in Figure 1A). The coil propensity of all these residues increases modestly in the presence of C60 or C60(OH)8. As shown in Figure 1C, nanoparticles also affect the probability distribution of β-sheet length. Upon addition of C60 and C60(OH)8, the probabilities of long β-sheets almost disappear. The probabilities of short β-sheets also decrease dramatically in the presence of C60, while their change becomes less prominent in hIAPP dimer + C60(OH)8 system. Taken together, these data demonstrate that C60 and C60(OH)8 substantially suppress hIAPP β-sheet formation, especially in the amyloidogenic core region, S20–S29.
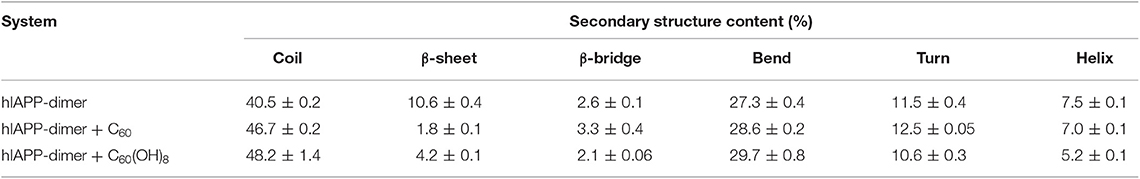
Table 1. Secondary structure probabilities of hIAPP dimer in the absence or presence of carbon nanoparticles.
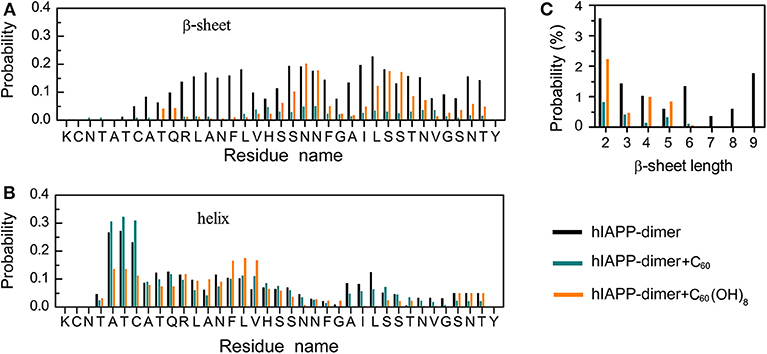
Figure 1. Influence of C60 and C60(OH)8 on the secondary structures of hIAPP dimer. Probabilities of β-sheet (A) and coil (B) as a function of amino acid residue. (C) The probability distribution of β-sheet length.
We first performed a RMSD-based cluster analysis for each REMD system at 310 K using a Cα-root-mean-square deviation (Cα-RMSD) cutoff of 0.35 nm to investigate the three-dimensional (3D) conformations of hIAPP dimer in the three systems. The conformations of hIAPP dimer in the three systems were separated into 85, 73, and 68 clusters, respectively. Figure 2 showed the centers of the top six most populated clusters, which represent 42.5, 44.0, and 61.5% of all conformations, respectively, for the three systems. As shown in Figure 2A, hIAPP dimer transiently adopts a three-stranded antiparallel β-sheet structure with a β-hairpin, which was considered to be hIAPP amyloidogenic precursor (Dupuis et al., 2009, 2011; Qiao et al., 2013). In hIAPP dimer + C60 system, this β-hairpin structure disappears and both intra- and inter-chain β-sheet content dramatically reduced, leading to collapsed disordered coil-rich conformations (Figure 2B). In hIAPP dimer + C60(OH)8 systems, inter-chain β-sheets are significantly reduced while a few intra-chain β-sheets (including short β-hairpins) still exist.
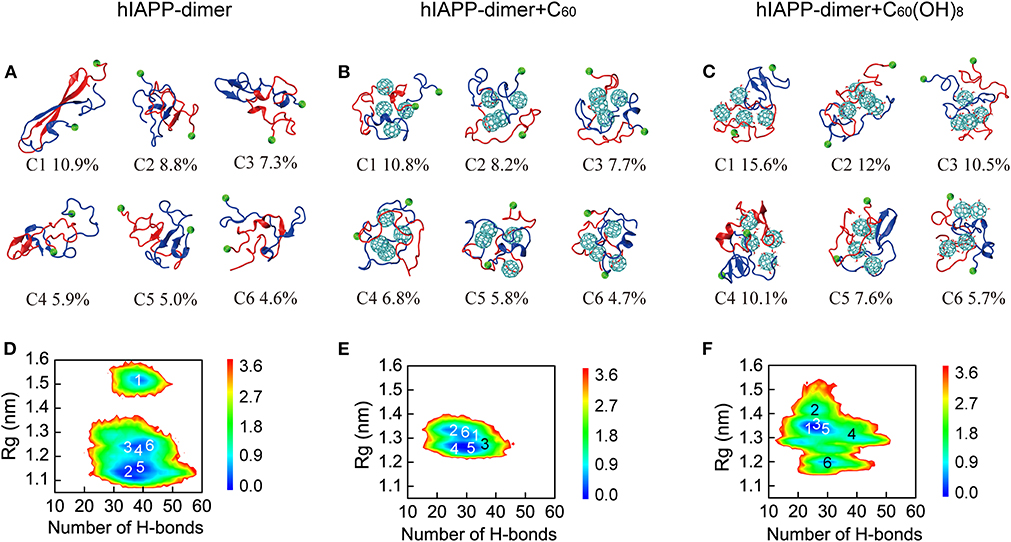
Figure 2. Analysis of 3D conformational properties and 2D free energy landscape of hIAPP dimer with and without C60/C60(OH)8. Representative conformations of the first six most-populated clusters for hIAPP dimer in hIAPP-dimer (A), hIAPP-dimer + C60 (B), and hIAPP-dimer + C60(OH)8 (C) systems. The corresponding population of each cluster is given below the snapshots. Free energy landscape (in kcal/mol) of hIAPP dimer as a function of the total number of H-bonds and Rg for three systems (D–F). The numbers in the PMF correspond to the cluster index. The green balls refer to the Cα atoms of the N-terminal residue K1.
To have an overall view of the effects of C60 and C60(OH)8 on the whole space of conformations of hIAPP dimer, we plotted the two-dimensional (2D) free energy landscape as a function of H-bond number and Rg. It can be seen from Figure 2D that there are three minimum-energy basins of the free energy surface of isolated hIAPP dimer, located at (H-bond number, Rg) values of (40.0, 1.52 nm), (40.0, 1.23 nm), and (38.0, 1.14 nm). The first basin with the largest Rg values corresponds to the three-stranded antiparallel β-sheet structure with a β-hairpin as mentioned earlier. In the presence of C60 (Figure 2E), hIAPP dimer has only one narrow and deep basin located at (30.0, 1.28 nm), corresponding to collapsed disordered dimers. It is noteworthy that the basin located at (40.0, 1.52 nm) disappears, indicating that the β-hairpin amyloid precursor of hIAPP is completely suppresses in presence of C60. With C60(OH)8 (Figure 2F), the free energy landscape becomes shallower than that of the isolated hIAPP dimer and has a basin centered at (28.0, 1.35 nm). The decreased number of H-bond and the increased range of Rg imply a collapsed and loosely packed coil-rich hIAPP dimer.
To explore the effects of C60 and C60(OH)8 on the hIAPP interactions, we plotted the pairwise residue inter-peptide and intra-peptide MC–MC (Figures 3A–C) and SC–SC (Figure S6) contact probabilities of hIAPP dimer in the three systems. The maps of contact probability in these three systems display distinct interaction patterns, suggesting that both inter-peptide and intra-peptide interactions are remarkably altered by C60 and C60(OH)8. As shown in Figure 3A, without nanoparticles, residues V17–L27 and C7–V17 present the dominant probabilities of inter-chain contact. The highest probabilities of intra-chain MC–MC contact show strong anti-diagonal contacts between A8–L16 and A25–G33, suggesting the appearance of the amyloid precursor β-hairpin structure that many studies previously reported (Jiang et al., 2009; Xu et al., 2009; Qi et al., 2014; Zhao et al., 2015; Qiao et al., 2016). In the presence of C60, the β-hairpin pattern disappears and both the inter- and intra-chain contact probabilities are observably reduced (Figure 3B). With C60(OH)8 molecules, the inter-chain contact probabilities greatly decrease, and the amyloid precursor β-hairpin structure pattern disappears. Other short β-hairpin structures, such as that formed between S28–N31 and S19–N22, were also observed (Figure 3C). The results above demonstrate that C60 can markedly block both intra- and inter-peptide interactions critical for hIAPP aggregation, while C60(OH)8 dramatically alters intra-peptide interaction patterns and weakens inter-peptide interactions.
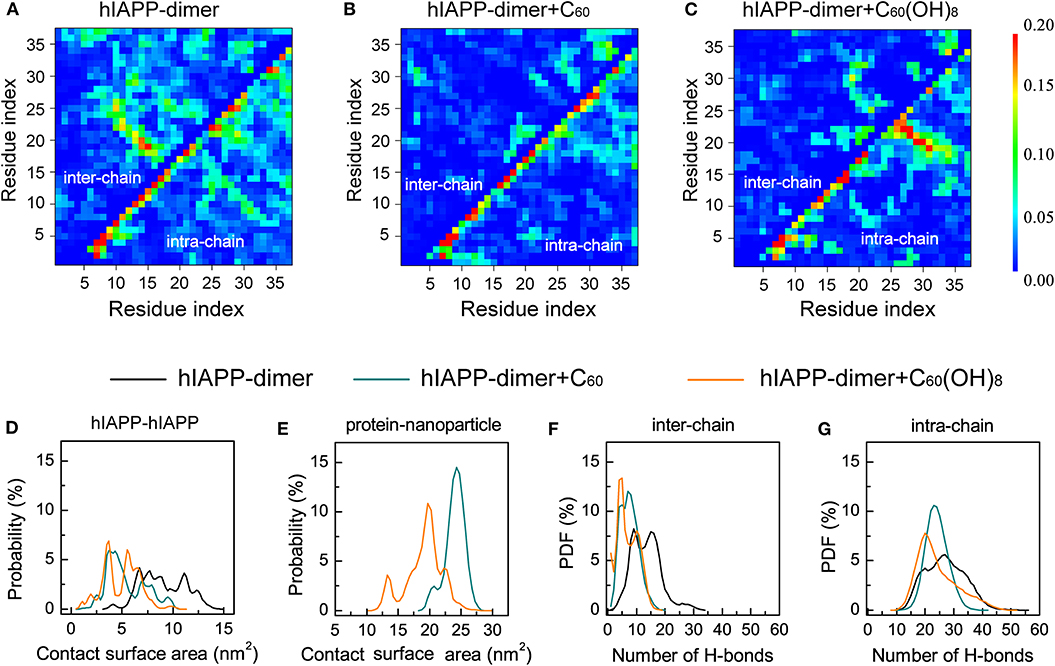
Figure 3. Analysis of the effects of C60/C60(OH)8 on the hIAPP interactions. Inter- and intra-peptide MC-MC contact probability maps for hIAPP dimer in the three different systems, hIAPP-dimer (A), hIAPP-dimer + C60 (B), and hIAPP-dimer + C60(OH)8 (C). PDF of protein–protein (D) and protein–nanoparticle (E) contact surface area (CSA). PDF of inter-chain (F) and intra-chain (G) H-bond number.
We also analyzed the contact surface area (CSA) probability between the two hIAPP monomers. without nanoparticles, the average inter-chain CSA value is 9.0 nm2. In the presence of C60/C60(OH)8 (Figure 3D), the average inter-chain CSA reduces to 5.3/4.9 nm2 while the CSA between hIAPP and nanoparticles is large (Figure 3E), indicating a strong binding between nanoparticles and hIAPP. We also find that hIAPP has a larger contact surface area with C60 than C60(OH)8, resulting in a more prominent inhibitory effect of C60. We further calculated the probability density function (PDF) of H-bond number formed within hIAPP dimer in the three systems. As shown in Figures 3F,G, in the presence of C60 or C60(OH)8, the numbers of inter-chain H-bonds are dramatically decreased as a result of the interactions between hIAPP and nanoparticles. These data suggest that the hIAPP-C60/C60(OH)8 interaction is stronger than hIAPP-hIAPP interaction, thus weaken hIAPP-hIAPP interactions and prevent hIAPP aggregation. Our result is consistent with recent studies showing that whether nanoparticles can inhibit or prevent peptide aggregation depends on the competition between peptide-peptide and peptide-nanoparticle interactions (Gladytz et al., 2016; Ke et al., 2019).
To identify the most favorable binding sites of nanoparticles, we calculated the contact probabilities of C60/C60(OH)8 with each amino acid residue of hIAPP. Figures 4A,B shows that C60 nanoparticles have a relatively high probability to bind with the hydrophobic residues L12, L16, V17, L27, V32, and aromatic residues F15, F23, and Y37, reflecting that both hydrophobic and aromatic interactions play an important role in inhibiting hIAPP dimerization. It is well-known that π-π stacking of aromatic residues is crucial to the amyloid fibril formation (Azriel and Gazit, 2001; Gazit, 2002; Porat et al., 2004). It can be seen from Figures 4A,B that C60 and C60(OH)8 both have high propensities to interact with F15, F23, and Y37, which are the only three aromatic residues and proposed to be of great importance in hIAPP aggregation (Padrick and Miranker, 2001; Marek et al., 2007). We also find that C60(OH)8 has a relatively high probability to bind with hydrophilic residues H18, N21, N31, and N35. The hydroxylation of C60 makes it more hydrophilic than pristine C60, which weakens its interactions with hydrophobic amino acids and enhances its interactions with hydrophilic amino acids. Therefore, C60 and C60(OH)8 display different binding sites on hIAPP.
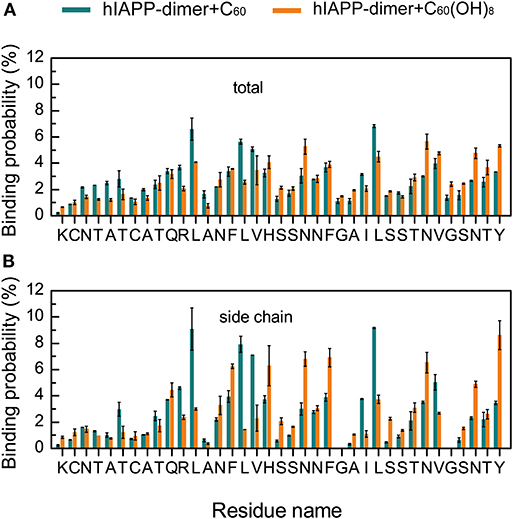
Figure 4. Binding probabilities of C60/C60(OH)8 with hIAPP dimer. Binding probabilities of C60/C60(OH)8 with all the atoms (A) and the side chain atoms (B) of each residue in hIAPP-dimer + C60 and hIAPP-dimer + C60(OH)8 systems.
Interestingly, we find that, the positively charged residues R11 in hIAPP dimer have relatively high binding probabilities with the hydrophobic C60. Thus, we calculated the minimum distance distribution between the atom NE of the side chain of R11 and the geometry center of each ring of C60. The distance distribution curve in Figure 5A shows that there exists a sharp peak centered at 0.40 nm, indicating strong cation-π interactions between R11 and C60 (Figure 5B). We can see from Figure 4 that C60 has a high contact probability with hydrophobic residue L12, this strong hydrophobic interaction might induce the cation-π interaction between R11 and C60.
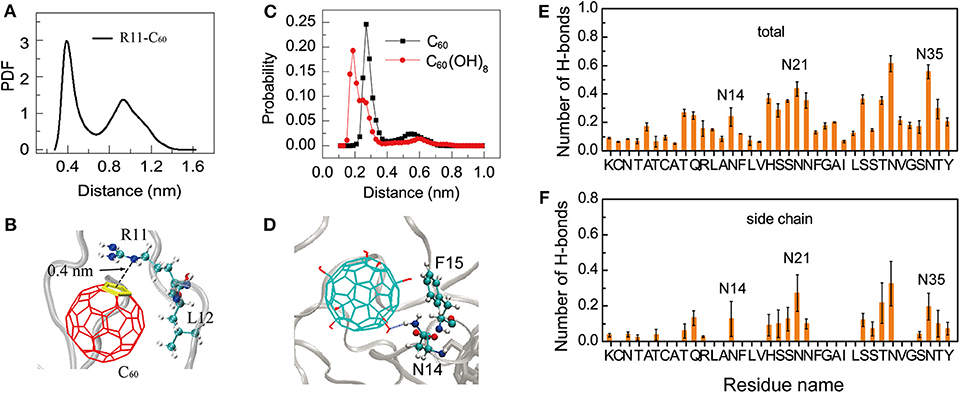
Figure 5. Binding mechanism of C60/C60(OH)8 on hIAPP dimer. (A) PDF of the minimum distance between NE atom of R11 and the geometry center of the C60 ring; (B) a representative snapshot showing the cation-π interaction between the atom NE in residue R11 and the ring of C60; (C) probability distribution of the minimum distance between N14/N21/N35 and C60/C60(OH)8. (D) A representative snapshot showing the collective hydrophilic and aromatic-stacking interactions between C60(OH)8 and hIAPP. (E) Total number of H-bonds between C60(OH)8 and each residue of hIAPP. (F) Number of H-bonds between C60(OH)8 and the side chain of each hIAPP residue.
Another interesting phenomenon is that C60 has stronger hydrophobicity than C60(OH)8, but C60(OH)8 exhibits much higher binding probability with the side chains of aromatic residues F15, H18, F23, and Y37 than C60 (Figure 4B). Meanwhile we noticed that C60(OH)8 displays high interacting probabilities with their neighboring residues N14, N21, and N35. To understand this observation at atomic level, we computed the minimum distance distribution between N14/N21/N35 and C60/C60(OH)8. The distance distribution curve in Figure 5C shows that the distance between C60(OH)8 and residue N14/N21/N35 is much shorter than that between C60 and N14/N21/N35. This result is probably attributed to the H-bond formation between the hydroxyl group of C60(OH)8 and Asn. Thus, we calculated the H-bond number between C60(OH)8 and each residue of hIAPP and found that residues N14, N21, and N35 all have high propensities to form H-bonds with C60(OH)8 (Figures 5E,F). These results suggest that the relatively high binding probability of C60(OH)8 with the aromatic residues F15, F23, and Y37 results from the cooperative contribution of aromatic interactions between F15, F23, Y37, and C60(OH)8, and hydrogen bonding interactions between their nearest neighboring residues N14, N21, N35, and C60(OH)8 (Figure 5D). We also find four consecutive hydrophilic residues S19, S20 N21, and N22 in the amyloid core region and four consecutive hydrophilic residues S28, S29, T30, and N31 in the C-terminal region that have high propensities to form H-bonds with C60(OH)8. It indicates that hydrogen bonding interaction between C60(OH)8 and hydrophilic residues of hIAPP also plays a role in inhibiting hIAPP aggregation.
We further performed MD simulations to probe into the effects of C60/C60(OH)8 on preformed hIAPP protofibrils. As shown in Figures 6A,B, compared to the β-sheet content in hIAPP-protofibril system (44.66%), β-sheet probabilities are reduced in both the hIAPP-protofibril + C60 (38.67%) and hIAPP-protofibril + C60(OH)8 (39.16%) systems, especially for the N- and C-terminal residues of the β-sheet regions (β1 and β2). It should be pointed out that the β-sheet disruption by C60/C60(OH)8 is less pronounced than β-sheet inhibition (β-sheet probability: 10.6% in hIAPP-dimer system, 1.8% in hIAPP-dimer + C60 system and 4.2% in hIAPP-dimer + C60(OH)8 system). The snapshots in Figure 6C shows that all the C60 and C60(OH)8 molecules bind to the surface of hIAPP protofibril. It can be seen in Figure 6F that the favorite binding sites of C60 and C60(OH)8 are the three aromatic amino acids, F15, F23, and Y37, revealing that the π-π staking may play a crucial role in the protein–nanoparticles interactions. F15 and F23 are located, respectively, in the C-terminal region of the β1 and the turn region of the protofibril, and Y37 is near the turn region in space. In addition, other residues with high binding probabilities are mostly located in the regions of the turn and the C-terminal of β1. These results demonstrate that both C60 and C60(OH)8 prefer to bind to the turn and the C-terminal of β1. The strong protein—C60/C60(OH)8 interactions significantly weaken and remodel the protein–protein interactions (Figures 6D,E, Figures S7A–F, S8). It is noted that C60(OH)8 displays a less prominent protofibril disruptive effect than C60, indicating the H-bonds (Figure 6G) formed between hydroxyl groups of C60(OH)8 and hIAPP might have limited disruptive effects on the protein–protein interactions. Taken together, both C60 and C60(OH)8 weaken the protein–protein interactions and disrupt the secondary structures.
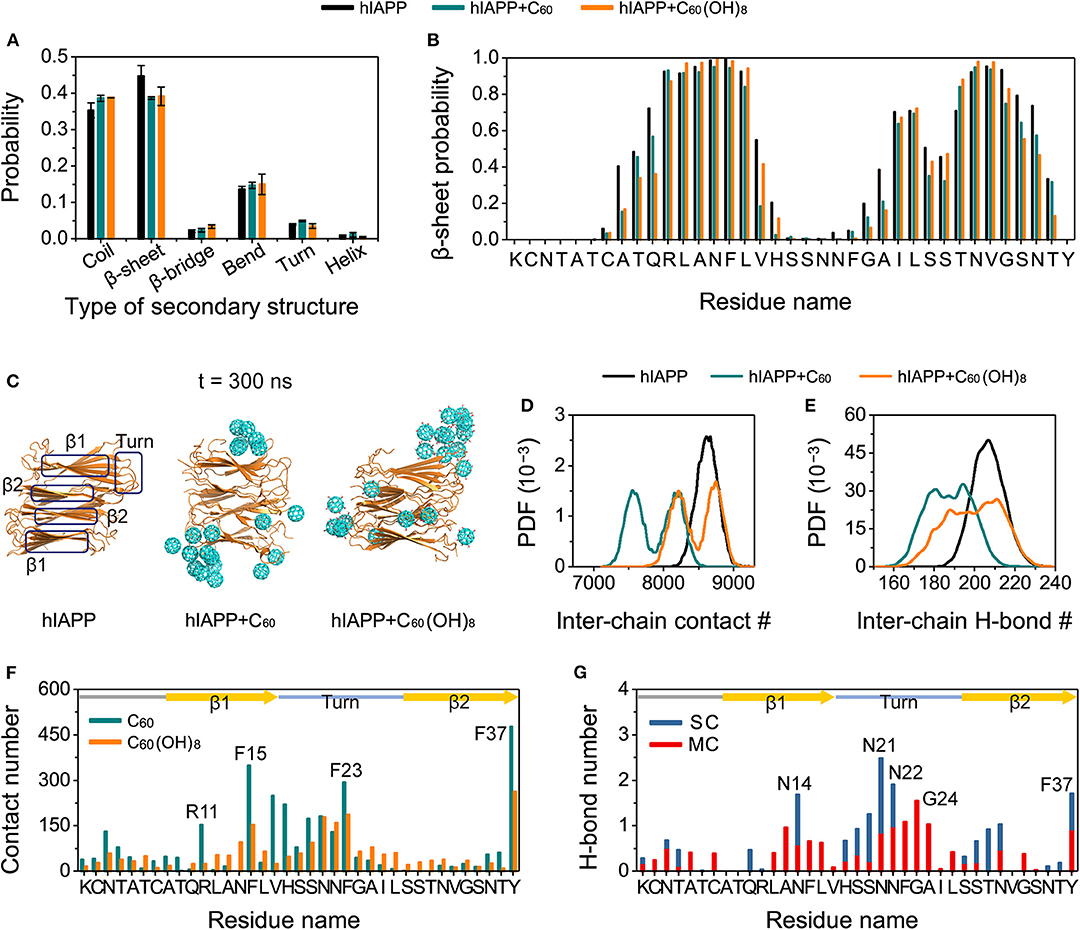
Figure 6. Influences of C60 and C60(OH)8 on the hIAPP protofibril. The average probability of each type of secondary structure (A), the β-sheet probability of each amino acid of hIAPP (B), the snapshots of hIAPP protofibril at t = 300 ns (C), the PDF of the inter-chain contact number (abbreviated as #) (D) and the inter-chain H-bond number (E) for hIAPP-fibril, hIAPP-fibril + C60, and hIAPP-fibril + C60(OH)8 systems, contact number between C60/C60(OH)8 and each amino acid of hIAPP (F), the number of H-bond formed between C60(OH)8 and the main-chain (MC) and side-chain (SC) atoms of each amino acid (G). The β1, β2, and turn regions are highlighted by rectangles in (C) and by yellow arrows and light blue line in (F,G).
In this study, we performed both REMD and MD simulations to study the effects of pristine and hydroxylated C60 on hIAPP aggregation. All-atom REMD simulations of hIAPP dimers reveal that C60 and C60(OH)8 can significantly suppress β-sheet formation of hIAPP. We found that, isolated hIAPP dimers adopt mostly disordered coil with a small proportion of short β-sheets. Interestingly, the previously proposed β-hairpin amyloidogenic precursor (Dupuis et al., 2009), contained in a three-stranded antiparallel β-sheet structure is also transiently populated. In the presence of C60 or C60(OH)8, the three-stranded antiparallel β-sheet structure with a β-hairpin completely disappears, resulting in disordered coil states. Protein-nanoparticle and protein-protein interaction analysis shows that C60 and C60(OH)8 both have strong binding with hIAPP and disrupt the peptide-peptide interactions responsible for hIAPP aggregation. These results indicate that both C60 and C60(OH)8 could slow down or hinder the aggregation of hIAPP. Further analyses reveal that the inhibition of hIAPP aggregation by C60 and C60(OH)8 is through different mechanism: hydrophobic and aromatic-stacking interactions for C60, and collective hydrogen bonding and aromatic-stacking interactions for C60(OH)8. MD simulations indicate that both C60 and C60(OH)8 are more likely to bind to the turn and the C-terminal of β1 via hydrophobic interactions, weaken the protein–protein interactions and disrupt the β-sheet of hIAPP protofibril. The obtained results are helpful for understanding the possible inhibitory mechanism of C60 and C60(OH)8 on hIAPP aggregation and provided valuable reference for the screening of potent amyloid inhibitors.
The β-sheet inhibition effect of C60/C60(OH)8 on amyloid proteins is sequence dependent. For example, this study together with our previous work shows that C60 can observably inhibit the β-sheet formation of Aβ16−22 (Xie et al., 2014) and hIAPP, while a recent MD study by Sun et al. reported that pristine C60 displays weak inhibitory impact on the aggregation of NACore of α-synuclein (Sun et al., 2019). Similarly, our REMD study demonstrates that C60 exhibits stronger inhibition capacity on hIAPP aggregation than C60(OH)8, whereas the work by Sun et al. shows that C60 has weaker inhibition ability on the aggregation of NACore of α-synuclein than C60(OH)8. It is noted that C60 and C60(OH)8 have poor water solubility, which limits their applications. This limitation can be overcome by increasing their extent of hydroxylation or their hydrophilicity through chemical modifications. Recently, it has been reported that graphene quantum dots and gold nanoparticles display excellent inhibition capacity against amyloidosis of hIAPP (Kim et al., 2018; Javed et al., 2019). Our results together with previous studies (Cabaleiro-Lago et al., 2012; Xie et al., 2014; Bednarikova et al., 2016; Nedumpully-Govindan et al., 2016; Kim et al., 2018) provide a better understanding of the inhibitory mechanism of nanomaterials targeting protein aggregation.
The datasets generated for this study are available on request to the corresponding author.
CB and GW conceived and designed the research. CB performed the simulations. CB, ZL, YC, and YT analyzed the simulation data. CB and GW wrote the paper and all authors approved the article.
This work was supported by the NSF of China (Grant No. 11674065) and National Key Research and Development Program of China (Grant No. 2016YFA0501702).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All simulations were performed using the high-performance computational facilities at the National High Performance Computing Center of Fudan University and at Guangzhou Supercomputer Center.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.00051/full#supplementary-material
Additional data are provided, including construction of initial structures of hIAPP dimer, parameters for simulation data analyses, temperature list, and eight supplementary figures. These figures present the initial states of the hIAPP dimer in our REMD simulations, the convergence check for the REMD runs of the three systems, inter-/intra-chain SC-SC contact probability maps, inter-chain contact number, inter-chain H-bond number, and the differentials of the inter-chain contact maps. In the figures of the convergence check, several parameters were used, including H-bonds number, Rg, the average probability of each types of secondary structure, secondary structure propensity of each residue at 310 K. We use two time periods, 200–280 and 280–360 ns for analysis in all the REMD systems.
Abedini, A., Meng, F., and Raleigh, D. P. (2007). A single-point mutation converts the highly amyloidogenic human islet amyloid polypeptide into a potent fibrillization inhibitor. J. Am. Chem. Soc. 129, 11300–11301. doi: 10.1021/ja072157y
Andujar, S. A., Lugli, F., Höfinger, S., Enriz, R. D., and Zerbetto, F. (2012). Amyloid-beta fibril disruption by C60-molecular guidance for rational drug design. Phys. Chem. Chem. Phys. 14, 8599–8607. doi: 10.1039/c2cp40680b
Azriel, R., and Gazit, E. (2001). Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. an experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 276, 34156–34161. doi: 10.1074/jbc.M102883200
Bai, C., Lin, D., Mo, Y., Lei, J., Sun, Y., Xie, L., et al. (2019). Influence of fullerenol on hIAPP aggregation: amyloid inhibition and mechanistic aspects. Phys. Chem. Chem. Phys. 21, 4022–4031. doi: 10.1039/C8CP07501H
Bednarikova, Z., Huy, P. D. Q., Mocanu, M. M., Fedunova, D., Li, M. S., and Gazova, Z. (2016). Fullerenol C-60(OH)(16) prevents amyloid fibrillization of a beta(40)–in vitro and in silico approach. Phys. Chem. Chem. Phys. 18, 18855–18867. doi: 10.1039/C6CP00901H
Bedrood, S., Li, Y., Isas, J. M., Hegde, B. G., Baxa, U., Haworth, I. S., et al. (2012). Fibril structure of human islet amyloid polypeptide. J. Biol. Chem. 287, 5235–5241. doi: 10.1074/jbc.M111.327817
Bobylev, A. G., Kornev, A. B., Bobyleva, L. G., Shpagina, M. D., Fadeeva, I. S., Fadeev, R. S., et al. (2011). Fullerenolates: metallated polyhydroxylated fullerenes with potent anti-amyloid activity. Org. Biomol. Chem. 9, 5714–5719. doi: 10.1039/c1ob05067b
Brender, J. R., Hartman, K., Nanga, R. P., Popovych, N., de la Salud Bea, R., Vivekanandan, S., et al. (2010). Role of zinc in human islet amyloid polypeptide aggregation. J. Am. Chem. Soc. 132, 8973–8983. doi: 10.1021/ja1007867
Brender, J. R., Lee, E. L., Cavitt, M. A., Gafni, A., Steel, D. G., and Ramamoorthy, A. (2008). Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the type-2-diabetes-related peptide. J. Am. Chem. Soc. 130, 6424–6429. doi: 10.1021/ja710484d
Cabaleiro-Lago, C., Szczepankiewicz, O., and Linse, S. (2012). The effect of nanoparticles on amyloid aggregation depends on the protein stability and intrinsic aggregation rate. Langmuir 28, 1852–1857. doi: 10.1021/la203078w
Cao, P., and Raleigh, D. P. (2012). Analysis of the inhibition and remodeling of islet amyloid polypeptide amyloid fibers by flavanols. Biochemistry 51, 2670–2683. doi: 10.1021/bi2015162
Cooper, G. J., Willis, A. C., Clark, A., Turner, R. C., Sim, R. B., and Reid, K. B. (1987). Purification and characterization of a peptide from amyloid-rich pancreases of type-2 diabetic-patients. Proc. Natl. Acad. Sci. U.S.A. 84, 8628–8632. doi: 10.1073/pnas.84.23.8628
Da Ros, T., and Prato, M. (1999). Medicinal chemistry with fullerenes and fullerene derivatives. Chem. Commun. 8, 663–669. doi: 10.1039/a809495k
Daura, X., Gademann, K., Jaun, B., Seebach, D., van Gunsteren, W. F., and Mark, A. E. (1999). Peptide folding: when simulation meets experiment. Angew. Chem. Int. Ed. 38, 236–240. doi: 10.1002/(SICI)1521-3773(19990115)38:1/2<236::AID-ANIE236>3.0.CO;2-M
Deng, Y., Qian, Z., Luo, Y., Zhang, Y., Mu, Y., and Wei, G. (2013). Membrane binding and insertion of a pHLIP peptide studied by all-atom molecular dynamics simulations. Int. J. Mol. Sci. 14, 14532–14549. doi: 10.3390/ijms140714532
Dugan, L. L., Turetsky, D. M., Du, C., Lobner, D., Wheeler, M., Almli, C. R., et al. (1997). Carboxyfullerenes as neuroprotective agents (vol 94, pg 9434, 1997). Proc. Natl. Acad. Sci. U.S.A. 94, 12241–12241. doi: 10.1073/pnas.94.22.12241-a
Dupuis, N. F., Wu, C., Shea, J. E., and Bowers, M. T. (2009). Human islet amyloid polypeptide monomers form ordered beta-hairpins: a possible direct amyloidogenic precursor. J. Am. Chem. Soc. 131, 18283–18292. doi: 10.1021/ja903814q
Dupuis, N. F., Wu, C., Shea, J. E., and Bowers, M. T. (2011). The amyloid formation mechanism in human IAPP: dimers have beta-strand monomer-monomer interfaces. J. Am. Chem. Soc. 133, 7240–7243. doi: 10.1021/ja1081537
Faridi, A., Sun, Y., Mortimer, M., Aranha, R. R., Nandakumar, A., Li, Y., et al. (2019). Graphene quantum dots rescue protein dysregulation of pancreatic beta-cells exposed to human islet amyloid polypeptide. Nano Res. 12, 2827–2834. doi: 10.1007/s12274-019-2520-7
Gazit, E. (2002). A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 16, 77–83. doi: 10.1096/fj.01-0442hyp
Gladytz, A., Abel, B., and Risselada, H. J. (2016). Gold-induced fibril growth: the mechanism of surface-facilitated amyloid aggregation. Angew. Chem. Int. Ed. 55, 11242–11246. doi: 10.1002/anie.201605151
Goldsbury, C., Goldie, K., Pellaud, J., Seelig, J., Frey, P., Müller, S. A., et al. (2000). Amyloid fibril formation from full-length and fragments of amylin. J. Struct. Biol. 130, 352–362. doi: 10.1006/jsbi.2000.4268
Guo, J., Li, J., Zhang, Y., Jin, X., Liu, H., and Yao, X. (2013). Exploring the influence of carbon nanoparticles on the formation of beta-sheet-rich oligomers of IAPP(2)(2)(-)(2)(8) peptide by molecular dynamics simulation. PLoS ONE 8:e65579. doi: 10.1371/journal.pone.0065579
Haataja, L., Gurlo, T., Huang, C. J., and Butler, P. C. (2008). Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endocr. Rev. 29, 303–316. doi: 10.1210/er.2007-0037
Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: Visual molecular dynamics. J. Mol. Graph. Model. 14, 33–38. doi: 10.1016/0263-7855(96)00018-5
Jaikaran, E. T., Higham, C. E., Serpell, L. C., Zurdo, J., Gross, M., Clark, A., et al. (2001). Identification of a novel human islet amyloid polypeptide beta-sheet domain and factors influencing fibrillogenesis. J. Mol. Biol. 308, 515–525. doi: 10.1006/jmbi.2001.4593
Javed, I., He, J. C., Kakinen, A., Faridi, A., Yang, W., Davis, T. P., et al. (2019). Probing the aggregation and immune response of human islet amyloid polypeptides with ligand-stabilized gold nanoparticles. ACS Appl. Mater. Interfaces. 11, 10462–10471. doi: 10.1021/acsami.8b19506
Jiang, P., Xu, W., and Mu, Y. (2009). Amyloidogenesis abolished by proline substitutions but enhanced by lipid binding. PLoS Comput. Biol. 5:e1000357. doi: 10.1371/journal.pcbi.1000357
Kayed, R., Bernhagen, J., Greenfield, N., Sweimeh, K., Brunner, H., Voelter, W., et al. (1999). Conformational transitions of islet amyloid polypeptide (IAPP) in amyloid formation in vitro. J. Mol. Biol. 287, 781–796. doi: 10.1006/jmbi.1999.2646
Ke, P. C., Pilkington, E. H., Sun, Y., Javed, I., Kakinen, A., Peng, G., et al. (2019). Mitigation of amyloidosis with nanomaterials. Adv. Mater. doi: 10.1002/adma.201901690. [Epub ahead of print].
Kim, D., Yoo, J. M., Hwang, H., Lee, J., Lee, S. H., Yun, S. P., et al. (2018). Graphene quantum dots prevent alpha-synucleinopathy in Parkinson's disease. Nat. Nanotechnol. 13, 812–818. doi: 10.1038/s41565-018-0179-y
Kim, J. E., and Lee, M. (2003). Fullerene inhibits beta-amyloid peptide aggregation. Biochem. Biophys. Res. Commun. 303, 576–579. doi: 10.1016/S0006-291X(03)00393-0
Lao, Z., Chen, Y., Tang, Y., and Wei, G. (2019). Molecular dynamics simulations reveal the inhibitory mechanism of dopamine against human islet amyloid polypeptide (hIAPP) aggregation and its destabilization effect on hIAPP protofibrils. ACS Chem. Neurosci. 10, 4151–4159. doi: 10.1021/acschemneuro.9b00393
Larson, J. L., and Miranker, A. D. (2004). The mechanism of insulin action on islet amyloid polypeptide fiber formation. J. Mol. Biol. 335, 221–231. doi: 10.1016/j.jmb.2003.10.045
Li, H., Luo, Y., Derreumaux, P., and Wei, G. (2011). Carbon nanotube inhibits the formation of beta-sheet-rich oligomers of the Alzheimer's amyloid-beta(16–22) peptide. Biophys. J. 101, 2267–2276. doi: 10.1016/j.bpj.2011.09.046
Luca, S., Yau, W. M., Leapman, R., and Tycko, R. (2007). Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry 46, 13505–13522. doi: 10.1021/bi701427q
Maciel, C., Fileti, E. E., and Rivelino, R. (2011). Assessing the solvation mechanism of C-60(OH)(24) in aqueous solution. Chem. Phys. Lett. 507, 244–247. doi: 10.1016/j.cplett.2011.03.080
Mahmoudi, M., Kalhor, H. R., Laurent, S., and Lynch, I. (2013). Protein fibrillation and nanoparticle interactions: opportunities and challenges. Nanoscale 5, 2570–2588. doi: 10.1039/c3nr33193h
Marek, P., Abedini, A., Song, B. B., Kanungo, M., Johnson, M. E., Gupta, R., et al. (2007). Aromatic interactions are not required for amyloid fibril formation by islet amyloid polypeptide but do influence the rate of fibril formation and fibril morphology. Biochemistry 46, 3255–3261. doi: 10.1021/bi0621967
Mo, Y., Brahmachari, S., Lei, J., Gilead, S., Tang, Y., Gazit, E., et al. (2018). The inhibitory effect of hydroxylated carbon nanotubes on the aggregation of human islet amyloid polypeptide revealed by a combined computational and experimental study. ACS Chem. Neurosci. 9, 2741–2752. doi: 10.1021/acschemneuro.8b00166
Mo, Y., Lei, J., Sun, Y., Zhang, Q., and Wei, G. (2016). Conformational ensemble of hIAPP dimer: insight into the molecular mechanism by which a green tea extract inhibits hIAPP aggregation. Sci. Rep. 6:33076. doi: 10.1038/srep33076
Nadler, W., and Hansmann, U. H. (2008). Optimized explicit-solvent replica exchange molecular dynamics from scratch. J. Phys. Chem. B 112, 10386–10387. doi: 10.1021/jp805085y
Nanga, R. P., Brender, J. R., Vivekanandan, S., and Ramamoorthy, A. (2011). Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochim. Biophys. Acta Biomemb. 1808, 2337–2342. doi: 10.1016/j.bbamem.2011.06.012
Nedumpully-Govindan, P., Gurzov, E. N., Chen, P. Y., Pilkington, E. H., Stanley, W. J., Litwak, S. A., et al. (2016). Graphene oxide inhibits hIAPP amyloid fibrillation and toxicity in insulin-producing NIT-1 cells. Phys. Chem. Chem. Phys. 18, 94–100. doi: 10.1039/C5CP05924K
Nielsen, J. T., Bjerring, M., Jeppesen, M. D., Pedersen, R. O., Pedersen, J. M., Hein, K. L., et al. (2009). Unique identification of supramolecular structures in amyloid fibrils by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 48, 2118–2121. doi: 10.1002/anie.200804198
Padrick, S. B., and Miranker, A. D. (2001). Islet amyloid polypeptide: identification of long-range contacts and local order on the fibrillogenesis pathway. J. Mol. Biol. 308, 783–794. doi: 10.1006/jmbi.2001.4608
Palhano, F. L., Lee, J., Grimster, N. P., and Kelly, J. W. (2013). Toward the molecular mechanism(s) by which EGCG treatment remodels mature amyloid fibrils. J. Am. Chem. Soc. 135, 7503–7510. doi: 10.1021/ja3115696
Pithadia, A., Brender, J. R., Fierke, C. A., and Ramamoorthy, A. (2016). Inhibition of IAPP aggregation and toxicity by natural products and derivatives. J. Diabetes Res. 2016:2046327. doi: 10.1155/2016/2046327
Podolski, I. Y., Podlubnaya, Z. A., Kosenko, E. A., Mugantseva, E. A., Makarova, E. G., Marsagishvili, L. G., et al. (2007). Effects of hydrated forms of C-60 fullerene on amyloid beta-peptide fibrillization in vitro and performance of the cognitive task. J. Nanosci. Nanotechnol. 7, 1479–1485. doi: 10.1166/jnn.2007.330
Porat, Y., Mazor, Y., Efrat, S., and Gazit, E. (2004). Inhibition of islet amyloid polypeptide fibril formation: a potential role for heteroaromatic interactions. Biochemistry 43, 14454–14462. doi: 10.1021/bi048582a
Qi, R., Luo, Y., Ma, B., Nussinov, R., and Wei, G. (2014). Conformational distribution and alpha-Helix to beta-Sheet transition of human amylin fragment dimer. Biomacromolecules 15, 122–131. doi: 10.1021/bm401406e
Qiao, Q., Bowman, G. R., and Huang, X. (2013). Dynamics of an intrinsically disordered protein reveal metastable conformations that potentially seed aggregation. J. Am. Chem. Soc. 135, 16092–16101. doi: 10.1021/ja403147m
Qiao, Q., Qi, R., Wei, G., and Huang, X. (2016). Dynamics of the conformational transitions during the dimerization of an intrinsically disordered peptide: a case study on the human islet amyloid polypeptide fragment. Phys. Chem. Chem. Phys. 18, 29892–29904. doi: 10.1039/C6CP05590G
Saunders, J. C., Young, L. M., Mahood, R. A., Jackson, M. P., Revill, C. H., Foster, R. J., et al. (2016). An in vivo platform for identifying inhibitors of protein aggregation. Nat. Chem. Biol. 12, 94–101. doi: 10.1038/nchembio.1988
Schrodinger, L. L. C. (2015). The PyMOL Molecular Graphics System, Version 1.8. Schrödinger. Available online at: https://pymol.org/2/
Sugita, Y., and Okamoto, Y. (1999). Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314, 141–151. doi: 10.1016/S0009-2614(99)01123-9
Sumner, S. C., Fennell, T. R., Snyder, R. W., Taylor, G. F., and Lewin, A. H. (2010). Distribution of carbon-14 labeled C60 ([14C]C60) in the pregnant and in the lactating dam and the effect of C60 exposure on the biochemical profile of urine. J. Appl. Toxicol. 30, 354–360. doi: 10.1002/jat.1503
Sun, Y., Kakinen, A., Zhang, C., Yang, Y., Faridi, A., Davis, T. P., et al. (2019). Amphiphilic surface chemistry of fullerenols is necessary for inhibiting the amyloid aggregation of alpha-synuclein NACore. Nanoscale 11, 11933–11945. doi: 10.1039/C9NR02407G
Tsuchiya, T., Oguri, I., Yamakoshi, Y. N., and Miyata, N. (1996). Novel harmful effects of [60]fullerene on mouse embryos in vitro and in vivo. FEBS Lett. 393, 139–145. doi: 10.1016/0014-5793(96)00812-5
Wang, M., Sun, Y., Cao, X., Peng, G., Javed, I., Kakinen, A., et al. (2018). Graphene quantum dots against human IAPP aggregation and toxicity in vivo. Nanoscale 10, 19995–20006. doi: 10.1039/C8NR07180B
Weirich, F., Gremer, L., Mirecka, E. A., Schiefer, S., Hoyer, W., and Heise, H. (2016). Structural characterization of fibrils from recombinant human islet amyloid polypeptide by solid-state NMR: the central FGAILS segment is part of the beta-sheet core. PLoS ONE 11:e0161243. doi: 10.1371/journal.pone.0161243
Xie, L., Luo, Y., Lin, D., Xi, W., Yang, X., and Wei, G. (2014). The molecular mechanism of fullerene-inhibited aggregation of Alzheimer's beta-amyloid peptide fragment. Nanoscale 6, 9752–9762. doi: 10.1039/C4NR01005A
Xu, W., Jiang, P., and Mu, Y. (2009). Conformation preorganization: effects of S20G mutation on the structure of human islet amyloid polypeptide segment. J. Phys. Chem. B 113, 7308–7314. doi: 10.1021/jp8106827
Yan, L. M., Tatarek-Nossol, M., Velkova, A., Kazantzis, A., and Kapurniotu, A. (2006). Design of a mimic of nonamyloidogenic and bioactive human islet amyloid polypeptide (IAPP) as nanomolar affinity inhibitor of IAPP cytotoxic fibrillogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 2046–2051. doi: 10.1073/pnas.0507471103
Young, L. M., Saunders, J. C., Mahood, R. A., Revill, C. H., Foster, R. J., Tu, L. H., et al. (2015). Screening and classifying small-molecule inhibitors of amyloid formation using ion mobility spectrometry-mass spectrometry. Nat. Chem. 7, 73–81. doi: 10.1038/nchem.2129
Zhang, S., Andreasen, M., Nielsen, J. T., Liu, L., Nielsen, E. H., Song, J., et al. (2013). Coexistence of ribbon and helical fibrils originating from hIAPP(20-29) revealed by quantitative nanomechanical atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 110, 2798–2803. doi: 10.1073/pnas.1209955110
Zhao, D. S., Chen, Y. X., and Li, Y. M. (2015). Rational design of an orthosteric regulator of hIAPP aggregation. Chem. Commun. 51, 2095–2098. doi: 10.1039/C4CC06739H
Zhou, X., Xi, W., Luo, Y., Cao, S., and Wei, G. (2014). Interactions of a water-soluble fullerene derivative with amyloid-beta protofibrils: dynamics, binding mechanism, and the resulting saltbridge disruption. J. Phys. Chem. B 118, 6733–6741. doi: 10.1021/jp503458w
Keywords: type 2 diabetes, hIAPP aggregation, inhibitory mechanism, replica exchange molecular dynamics simulations, C60
Citation: Bai C, Lao Z, Chen Y, Tang Y and Wei G (2020) Pristine and Hydroxylated Fullerenes Prevent the Aggregation of Human Islet Amyloid Polypeptide and Display Different Inhibitory Mechanisms. Front. Chem. 8:51. doi: 10.3389/fchem.2020.00051
Received: 31 October 2019; Accepted: 16 January 2020;
Published: 05 February 2020.
Edited by:
Ruhong Zhou, IBM Research, United StatesCopyright © 2020 Bai, Lao, Chen, Tang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanghong Wei, Z2h3ZWlAZnVkYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.