- 1Canadian Wheat Improvement Flagship Program, National Research Council Canada (NRC-CNRC), Saskatoon, SK, Canada
- 2Saskatoon Research and Development Centre, Agriculture and Agri-Food Canada, Saskatoon, SK, Canada
- 3Department of Global Agricultural Sciences, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 4Institute of Crop Science (NICS), National Agriculture and Food Research Organization (NARO), Tsukuba, Japan
- 5Agrobioversity Research Area, International Center for Tropical Agriculture (CIAT), Cali, Colombia
Abiotic stresses such as, drought, heat, salinity, and flooding threaten global food security. Crop genetic improvement with increased resilience to abiotic stresses is a critical component of crop breeding strategies. Wheat is an important cereal crop and a staple food source globally. Enhanced drought tolerance in wheat is critical for sustainable food production and global food security. Recent advances in drought tolerance research have uncovered many key genes and transcription regulators governing morpho-physiological traits. Genes controlling root architecture and stomatal development play an important role in soil moisture extraction and its retention, and therefore have been targets of molecular breeding strategies for improving drought tolerance. In this systematic review, we have summarized evidence of beneficial contributions of root and stomatal traits to plant adaptation to drought stress. Specifically, we discuss a few key genes such as, DRO1 in rice and ERECTA in Arabidopsis and rice that were identified to be the enhancers of drought tolerance via regulation of root traits and transpiration efficiency. Additionally, we highlight several transcription factor families, such as, ERF (ethylene response factors), DREB (dehydration responsive element binding), ZFP (zinc finger proteins), WRKY, and MYB that were identified to be both positive and negative regulators of drought responses in wheat, rice, maize, and/or Arabidopsis. The overall aim of this review is to provide an overview of candidate genes that have been identified as regulators of drought response in plants. The lack of a reference genome sequence for wheat and non-transgenic approaches for manipulation of gene functions in wheat in the past had impeded high-resolution interrogation of functional elements, including genes and QTLs, and their application in cultivar improvement. The recent developments in wheat genomics and reverse genetics, including the availability of a gold-standard reference genome sequence and advent of genome editing technologies, are expected to aid in deciphering of the functional roles of genes and regulatory networks underlying adaptive phenological traits, and utilizing the outcomes of such studies in developing drought tolerant cultivars.
Introduction
Wheat is one of the important staple food crops supplying 20% of calories globally (Lobell and Gourdji, 2012; Shiferaw et al., 2013). Currently, two major wheat species, hexaploid bread wheat (Triticum aestivum; 2n = 6x = 42) and tetraploid durum wheat (Triticum durum; 2n = 4x = 28), are commercially important. Food and Agriculture Organization (FAO) of the United Nations has estimated 739.9 million tons of wheat production in 2017 (http://www.fao.org/worldfoodsituation/csdb/en/). Global wheat yields have increased at a mere 1.0% per year in the past two decades (Manes et al., 2012). Wheat crop is sensitive to heat and drought stresses mainly at the flowering and grain development stages, which negatively impact the yield and grain quality (lower 1,000 grain weight and change in protein quality). Annual production variability estimated at ~40% was mainly due to heat waves and drought situations in major wheat producing belts throughout the world (Zampieri et al., 2017). Demand for wheat is estimated to increase by 60% by 2050, but production might go down by 29% as a result of climate change imposed environmental stresses (Manickavelu et al., 2012). These predictions indicate that improving abiotic stress tolerance in wheat is paramount for global food security in the near future. Continued wheat genetic improvement is thus critically important as it has direct impact on economic development, food security, and international grain trade.
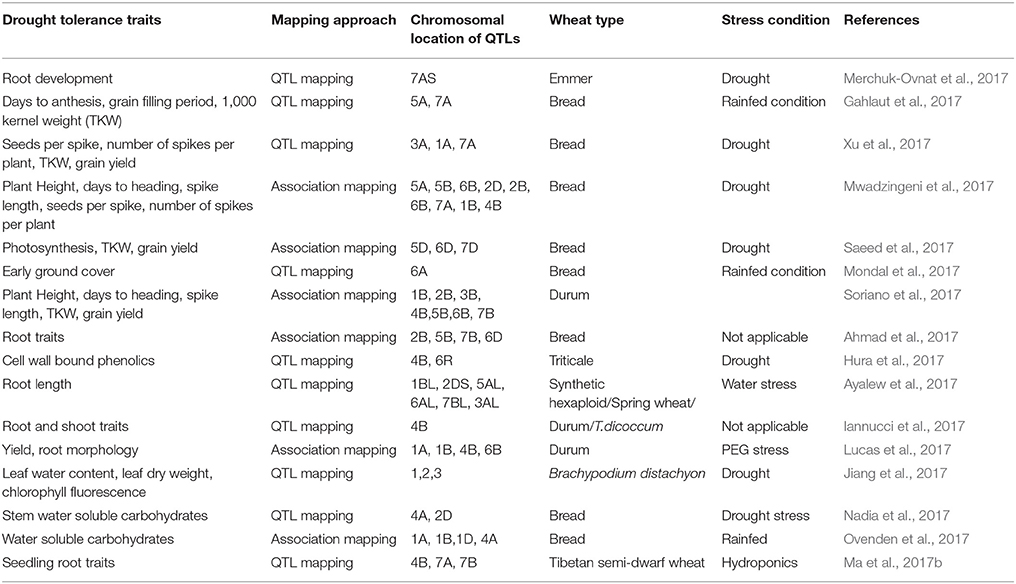
Most of the stress tolerance traits in wheat are polygenic and hence complex to understand at the physiological and molecular levels. Approaches like quantitative trait locus (QTL) mapping, marker assisted breeding, and introgression from wild gene pool are being employed to improve drought tolerance (Mwadzingeni et al., 2016). A summary of recent efforts in QTL and association mapping for drought tolerance associated traits is provided in Table 1. Success in QTL and association mapping approaches in wheat has unraveled value of these methods as a starting point for fine mapping and identification of genes affecting drought tolerance (Gupta et al., 2017). The recent advances in high-throughput genotyping and phenotyping methods are enabling more advanced approaches such as, genomic selection that allow analysis of the architecture of complex traits.
In the recent past, progress has been made in identifying key regulators of drought tolerance in wheat using transgenic approaches. Microarray and RNA sequencing-based gene expression analyses have also been used as important tools in the past to understand wheat responses to varoius abiotic stresses including drought stress. A recent field study looking at the effect of drought on wheat transcriptome changes during reproductive stages detected over 300 differentially expressed genes involved in many critical processes including floral development, photosynthetic activity, and stomatal movement (Ma et al., 2017a). A common response to drought stress involves differential expression of cytochrome P450, heat shock proteins, dehydrins, glutathione transferase, proteinase inhibitors, and regulatory proteins including transcription factors. Several transcription factors, such as, bHLH, bZIP, ERF, HD-ZIP, NAC, and WRKY were differentially expressed in a drought tolerant wheat genotype compared to susceptible genotype (Ergen et al., 2009). Transcriptomic and proteomic analyses of a pale green durum wheat mutant under drought stress indicated expression modulation of several genes encoding antioxidant enzymes, photosystem components, and enzymes representing carbohydrate metabolism and the tricarboxylic acid cycle that may be valuable in addressing drought resistance in wheat (Peremarti et al., 2014). Similarly, a number of other transcriptome and proteome profiling, and genetic manipulation studies have identified candidate genes with potential roles in drought tolerance mechanisms. A summary of promising candidates identified through such studies is provided in Table 2.
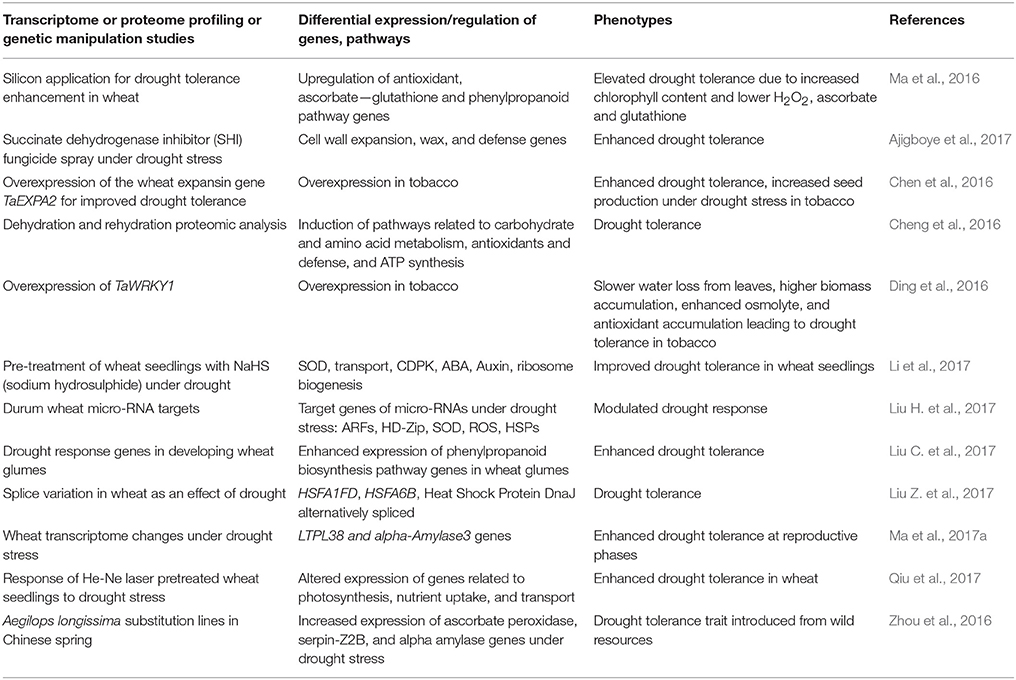
Table 2. Identification of candidate genes for drought tolerance through transcriptome and proteome profiling, and genetic manipulation.
Wheat cultivars have adapted various drought tolerance mechanisms, which include formation of deeper roots, accumulation of higher biomass, exertion of better stomatal control over transpiration (Chipilski et al., 2012), enhancement of osmoprotective and antioxidant response (Huseynova, 2012; Loutfy et al., 2012), and importantly a better coordination of positive and negative regulation of gene expression.
Developmental response of plants to drought stress is manifested through enhanced root growth and suppressed shoot growth resulting in increased root: shoot ratio (Sharp et al., 2004; Yamaguchi and Sharp, 2010; Xu et al., 2013). A combination of 20% faster root descent and more efficient roots can result in more effective water extraction from sub-soil (roots below 60 cm) and provide yield benefits of 0.32–0.44 t/ha in wheat (Lilley and Kirkegaard, 2011). In addition, Rauf et al. (2007) observed up to 50% increase in wheat root: shoot ratio in response to drought stress. Elevated abscisic acid (ABA) levels is shown to function as a promoter of root growth and simultaneously a repressor of shoot growth (Sharp et al., 2004; Xu et al., 2013). Crop's ability to extract water from larger soil volume is critical for yield stability under depleting soil moisture in rainfed production systems. Thus, deep root systems contribute to greater yield potential under drought conditions (Reynolds et al., 2009; Pask and Reynolds, 2013). DEEPER ROOTING 1 (DRO1) gene in rice and related genes in Arabidopsis and Prunus species, which will be discussed in detail in the following sections, have been shown to alter root architecture for drought avoidance and improved use of water resources (Uga et al., 2013; Guseman et al., 2017). In addition to improved root traits, greater crop transpiration efficiency (TE) is also critical for yield protection in the agro-ecological regions with limited soil moisture availability (Condon et al., 2004).
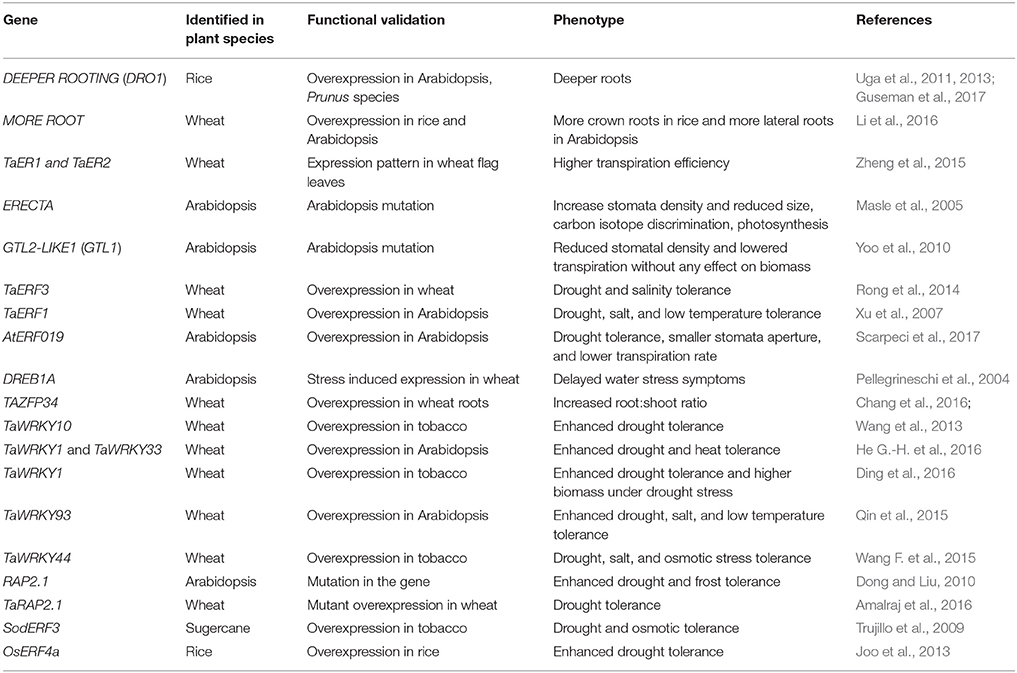
This review highlights the progress made in physiological and molecular traits in important crops to maintain yield stability under depleting soil moisture conditions. Here, we focus only on a set of important genes (DRO1, ERECTA) and transcription regulators (AP2/ERF, ZFPs, WRKY, and MYB) that are functionally characterized for their role in drought tolerance (Tables 3, 4).
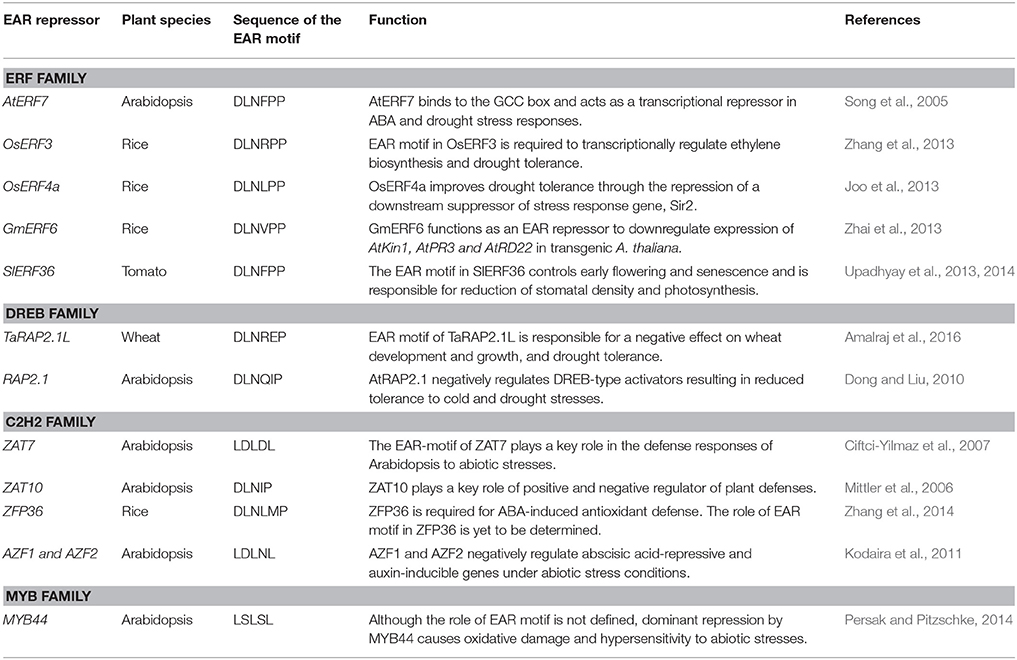
Table 4. Examples of EAR-motif containing transcriptional repressors involved in modulation of drought response.
Enhanced Root Growth for Drought Tolerance
Root system architecture (RSA) has been the target of wheat research and breeding to develop drought tolerant cultivars. Through QTL analysis, DRO1 was identified as a regulator of RSA by modulating root growth angle in rice (Uga et al., 2011, 2013). Kinandang Patong (a japonica upland rice) containing a full-length DRO1 copy exhibits deeper RSA; whereas, IR64 (an indica lowland rice) carrying a truncated copy, due to an insertion of 1 bp deletion within exon 4 resulting in introduction of a pre-mature stop codon of DRO1, exhibits shallower roots (Uga et al., 2013). The deep rooting was found to be beneficial in rice for not only drought tolerance but higher harvest index, nitrogen uptake, and flux of cytokinin from root to shoot during grain filling (Arai-Sanoh et al., 2014). These findings confirmed the positive contribution of root depth to drought avoidance facilitated by the ability to access moisture from deeper soil layers, better photosynthesis, and grain filling under drought conditions.
DRO1 orthologs are found in many different plants, including both dicot and monocot species (Guseman et al., 2017). Consistent with the findings in rice, orthologs of DRO1 in Arabidopsis and Prunus species were also found to influence the RSA, as evident from deeper rooting phenotypes in Arabidopsis and Prunus lines overexpressing DRO1 (Guseman et al., 2017). The wheat genome harbors three copies of DRO1 orthologs (TRIAE_CS42_5AL_TGACv1_374418_AA1199770, TRIAE_CS42_5BL_TGACv1_405332_AA1325250, and TRIAE_CS42_5DL_TGACv1_433409_AA1412320).
Notably, the wheat DRO1 orthologs share 76% identitity with rice DRO1, suggesting the likelihood of functional similarity and potential applications in altering RSA for drought avoidance in wheat.
Harmonal regulation of root development in wheat is another interesting area that has attracted attension of wheat researchers. The recent functional characterization of ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES DOMAIN (AS2/LBD) genes in wheat identified a transcription factor involved in root architecture enhancement. MORE ROOT from the D-genome of wheat (TaMOR-D), an auxin responsive transcription factor in the LBD family, when over-expressed in rice and Arabidopsis resulted in lateral root enhancement in Arabidopsis, and more crown roots, longer panicles and higher grain yield in rice (Li et al., 2016). It will be interesting to assess the response of these overexpression lines from Arabidopsis and rice to drought stress. As deeper and more effective root systems improve the capture of water from soil, further characterization of DRO1 orthologs and TaMOR genes would provide an effective strategy for improvement of drought tolerance.
Stomatal Traits for Enhanced Drought Tolerance
Stomata is the above ground control point for the entry of carbon dioxide (CO2) for photosynthesis and exit of water from plants via transpiration (Shahinnia et al., 2016). Stomatal closure as a response to stress leads to decreased leaf water potential, reduced carbon assimilation, oxidative stress, and increased canopy temperature (Ludlow and Muchow, 1990; Yokota et al., 2002). Maintaining better stomatal control over transpiration is critical for combating photosynthesis inhibition under drought stress (Bota et al., 2004). Stomatal pore area per leaf is determined by stomatal density and stomatal size. Significant genetic variation for stomatal size and density has been reported in wheat (Baloch et al., 2013; Shahinnia et al., 2016). Molecular understanding of genes regulating stomatal patterning and size in wheat is very important as this knowledge could be successfully employed to improve TE under drought stress. Smaller stomata size and higher density in wheat flag leaves were found to be associated with drought tolerance in wheat varieties (Baloch et al., 2013; Shahinnia et al., 2016). At least 40 genes in Arabidopsis are known to regulate stomatal development and patterning (Pillitteri and Torii, 2012). Interestingly, larger stomata and smaller density leads to better TE in Arabidopsis (Masle et al., 2005) but smaller stomata and higher density promotes higher TE in wheat (Baloch et al., 2013; Shahinnia et al., 2016). Molecular genetic understanding of genes and networks for stomatal patterning, size, and density regulation in wheat will enable modulation of stomatal index in wheat and improve TE under drought stress.
Transpiration efficiency is measured as biomass produced per unit of water transpired by a plant (Condon et al., 2004). Transpiration efficiency is connected to deeper root system and hence these two traits need to be simultaneously improved. Crop varieties that extract moisture from deeper zones (60–120 cm), maintain higher stomatal conductance and are able to maintain cooler canopy temperature (Pask and Reynolds, 2013). The stacking of deep root biomass and TE traits in wheat varieties will enhance protection from drought stress.
One of the classical examples of genes regulating TE is the ERECTA from Arabidopsis (Masle et al., 2005), a putative leucine-rich repeat receptor-like kinase (LRR-RLK) known to regulate stomatal density, epidermal cell expansion and patterning, mesophyll cell proliferation and cell-to-cell contact. Mutation of ERECTA in Arabidopsis increased stomatal density (resulting in increased stomatal conductance), decreased epidermal cell size and also decreased TE (by around 20%). Interestingly, ERECTA gene mutation can increase stomatal density without changing stomatal index. ERECTA thus regulates both photosynthesis ability as well as stomatal control over TE (Masle et al., 2005).
Two homologs of ERECTA in the wheat genome TaER1 and TaER2 were recently characterized (Zheng et al., 2015). Analysis of the expression patterns of TaER1 and TaER2 revealed a stronger negative correlation with carbon isotope discrimination, stomatal density, and transpiration rate but a positive correlation with flag leaf area, instant water use efficiency, biomass, and yield per plant, suggesting TaERs are involved in TE related traits and yield in bread wheat (Zheng et al., 2015). Interestingly, QTLs for stomatal density have been mapped to chromosome 7A on which one of the TaER2 homoeologues resides (Huang et al., 2013; Zheng et al., 2015; Shahinnia et al., 2016). Further characterization of allelic diversity in TaER genes is essential prior to their future deployment in improving both drought tolerance and agronomic traits in wheat.
Stomata are key regulators of internal plant water status and carbon assimilation. Stomatal pores control both uptake of CO2 and water use through controlling transpiration rate, thus has a major role in photosynthesis as well as TE. Arabidopsis mutants with reduced stomatal density and increased stomata size showed better drought tolerance through reduced transpiration and higher biomass accumulation under drought stress (Pillitteri and Torii, 2012). Other environmental factors such as, temperature, water availability, and humidity also could be important modulators of photosynthesis. In contrast to ERECTA gene in Arabidopsis, GTL2-LIKE1 (GTL1) controls stomata density, transpiration, and water use efficiency by repressing STOMATAL DENSITY AND DISTRIBUTION 1 (SDD1) gene (Yoo et al., 2010). GTL1 mutation reduced stomatal density and lowered transpiration without negative impact on CO2 assimilation and biomass production (Yoo et al., 2010) to improve photosynthesis efficiency in Arabidopsis.
Stomata pores cover only around 5% of the leaf area but contribute to around 70% water loss by plants (Hetherington and Woodward, 2003). Both stomatal conductance and stomatal index have an influence on carbon isotope discrimination (Δ13C). For example, Australian wheat varieties Drysdale and Rees developed for low Δ13C exhibit higher TE and around 10% better yield under dry and hot climate (Passioura, 2006; Richards, 2006). In environments where crops were able to maintain better water status, the genotypes exhibit a positive relationship between Δ13C and grain yields (Araus et al., 1998). Stomatal density and size in the complex wheat genome are dependent on ploidy level. Khazaei et al. (2010) compared Triticum monococcum (diploid), T. durum (tetraploid), and T. aestivum (hexaploid) for stomatal density and size, and observed a significant genetic variation for stomatal density. The high heritability of this trait enables dissection at molecular level (Bhagwat and Bhatia, 1993). Recently, Shahinnia et al. (2016) mapped QTLs for stomatal traits (size and density) on chromosomes 1A, 1B, 2B, and 7A in wheat. Interestingly, the 7A QTL co-localized with other QTLs for yield contributing traits such as, seeds per head, harvest index, and yield. Flag leaf may contribute up to 30–50% of the assimilates during grain filling (Sylvester-Bradley et al., 1990), hence flag leaf stomatal features are important to cope drought episodes.
Simultaneous improvements to root architecutre and depth for enhanced access to water from deeper soil layers and TE to preserve above ground water would be a valuable strategy that can be adopted to improve drought tolerance. A heat and drought tolerant wheat variety RAC875 had smaller and numerous stomata on flag leaf as compared to drought susceptible cultivar Kukri (Shahinnia et al., 2016). Root anatomical features such as, smaller central metaxylem (CMX) vessels and stele area complement RAC875 variety stomatal features by restricting root hydraulic conductivity under stress, another possible mechanism to reduce transpirational loss (Schoppach et al., 2014). Deeper roots enhance moisture uptake (Uga et al., 2013), whereas transcription factors from LBD gene family improve root architecture (Li et al., 2016). Drought tolerant spring wheat cultivars Anmol, Moomal, Bhittai, Sarsabz have smaller stomata size, lower stomatal conductance and are able to maintain higher relative water content under drought stress (Baloch et al., 2013). Based on these observations, we present a drought-tolerant wheat ideotype (Figure 1) with integration of adaptive root and stomatal traits including, deeper roots, narrower CMX, high moisture use and transpiration efficiency, and lower carbon discrimination and canopy temperature. Future research in wheat focused on understanding of genetic regulation of these complex traits would enable improvement of TE and MUE simultaneously.
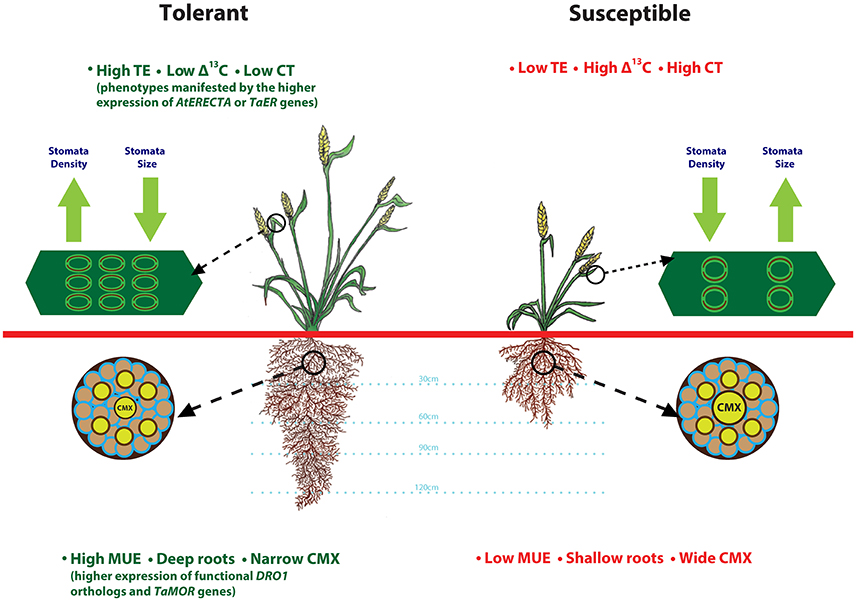
Figure 1. Root and stomatal traits that define drought tolerant and susceptible wheat plant ideotypes. This illustration is prepared based on the findings from various articles cited throughout the manuscript. Plant tolerance to drought stress relies on favorable root anatomical features such as, deeper roots and smaller central metaxylem (CMX) that contribute to improved moisture uptake-efficiency (MUE), and stomatal features such as high density and smaller size that contribute to lower canopy temperature (CT) and reduced carbon isotope discrimination.
Regulation of Gene Expression Under Drought Stress
Plants show remarkable transcriptional plasticity that allows them to thrive under harsh environmental conditions. Regulation of gene expression occurring at both the transcriptional and post-transcriptional level has a central role in plant's adaptation to environmental changes (Lopez-Maury et al., 2008). Gene regulation at the transcriptional level is coordinated by a complex network of functionally diverse regulatory proteins, including activators, repressors, co-activators, co-repressors, and chromatin modifiers. Positive regulation of gene expression in plants in response to drought stress has been well studied and extensively reviewed (Gahlaut et al., 2016; Joshi et al., 2016). Here, we highlight a few interesting discoveries of transcription factors that positively regulate adaptation responses to drought stress in wheat (summarized in Table 3).
Positive Regulation of Gene Expression under Drought Stress
AP2/ERF family transcription factors are well documented in many crop species for their role as mediators of both abiotic and biotic stress responses in plants (Licausi et al., 2013). AP2/ERF transcription factors are classified into four sub-families in wheat, including DREB, ERF, AP2, and RAV (Zhuang et al., 2011). ERFs are rapidly upregulated in response to stresses (He et al., 2011), and have been the subject of various overexpression studies to assess their usefulness in improving drought tolerance. Overexpression of TaERF3 in wheat resulted in enhancement of drought as well as salinity tolerance (Rong et al., 2014), potentially due to the increase in accumulation of proline as well as chlorophyll content as compared to non-transformed lines and activation of a number of downstream genes by binding to the GCC-box cis-elements present in the promoter regions of target genes (Rong et al., 2014). Overexpression of another wheat ERF, TaERF1, activated stress-related genes, including PR and COR/RD genes, under normal growth conditions, and improved drought, cold, and salt tolerance in transgenic plants (Xu et al., 2007). A recent study of AtERF019 indicated its role in drought tolerance with a phenotype of delayed flowering and maturity under drought stress suggesting overexpression of its orthologs could be used for obtaining enhanced drought tolerance in wheat without any compensation of the seed set (Scarpeci et al., 2017). ABA and ethylene are important phytohormones related to stress response regulation in plants. Induction of TaERF3 by ethylene (Zhang et al., 2007) and ABA (Rong et al., 2014), and the presence of GCC boxes (ethylene response elements) as well as ACGT and ABRE (response elements for ABA) in the promoter confirms well established complex hormone-gene interplay mechanisms in abiotic stress tolerance. As ERFs are involved in multiple stresses, it would be useful to identify the conserved domains/motifs in their sequences and analyze their correspondence to different stress tolerance mechanisms. DREB and HSFs are key regulators of complex heat-drought stress genetic networks in wheat. Stress induced expression of DREB1A from Arabidopsis enhanced drought tolerance in wheat, indicating a promising role for DREBs in improving wheat adaptation to drought stress (Pellegrineschi et al., 2004).
Zinc Finger Proteins (ZFPs) with a QALGGH conserved domain are known to be associated with modulation of gene expression under drought stress (Cheuk and Houde, 2016). Role of ZFPs in drought tolerance in wheat (Cheuk and Houde, 2016), rice (Zhang et al., 2014), and Arabidopsis (Hichri et al., 2014) has been well established. A number of ZFPs, such as ZFP182 and ZFP252 (rice), ZZ1 (soybean), ZFP1 (Glycine soja) and AZF1, AZF2, ZAT10, and ZAT11 (Arabidopsis) are known to be having a positive role in drought stress tolerance (Huang et al., 2012; Luo et al., 2012). TaZFP22, TaZFP34, and TaZFP46 are root expressed and drought induced Q-type C2H2 zinc finger transcriptional repressors in wheat (Chang et al., 2016). TaZFP34 was found to be up-regulated in response to multiple abiotic stresses, including heat, dehydration, salinity, and cold. Overexpressing this gene in wheat roots enhanced root:shoot ratio by reducing shoot growth while maintaining root elongation (Chang et al., 2016).
WRKY transcription factors are also important in critical plant developmental and physiological functions (Phukan et al., 2016). This gene family has been extensively studied for its role in abiotic stress tolerance in many plant species, including wheat (Ding et al., 2016), soybean (Wang F. et al., 2015), canola (He Y. et al., 2016), and common bean (Wu et al., 2017). A direct role of WRKY transcription factors in drought tolerance is evident from their up-regulation at the protein level in response to drought stress (Tripathi et al., 2014). Wheat WRKY transcription factors, TaWRKY44 and TaWRKY93, were identified to be critical response factors under drought stress (Qin et al., 2015; Wang X. et al., 2015). A functional proof of the role of WRKY in drought tolerance of wheat was evident from overexpression of TaWRKY10 in tobacco which enhanced drought tolerance response in transgenic tobacco lines with a suggested role as a negative regulator of antioxidant accumulation (Wang et al., 2013). Some of the wheat TaWRKYs (TaWRKY16, 24, 59, 61, and 82) were found to be differentially expressed in both leaf and root tissues under drought stress (Okay et al., 2014). A recent study identified TaWRKY1 and TaWRKY33 as candidates putatively involved in drought tolerance (He G.-H. et al., 2016); these genes when overexpressed in Arabidopsis lead to enhanced drought and heat tolerance. A positive role of TaWRKY93 in drought, heat, salt tolerance as well as root growth enhancement was also evident from enhanced cellular membrane stability in Arabidopsis overexpression lines (Qin et al., 2015). Similarly, overexpression of an ABA dependent TaWRKY1 in tobacco enhanced drought tolerance via stomatal closure to reduce water loss and altered osmotic adjustment to accumulate higher biomass (Ding et al., 2016). The role of TaWRKY44 in drought and salinity tolerance via activating reactive oxygen species or antioxidant pathways from overexpression studies in tobacco provides a promising example of multiple roles of transcription factors and their possible use in developing wheat lines with tolerance to drought, salt, and osmotic stresses (Wang X. et al., 2015). Above studies also indicate the importance of studying genome wide variation in various transcription factors and exploitation of the allelic variation to enhance drought tolerance.
Negative Regulation of Gene Expression under Drought Stress
Although the positive control of transcriptional activation has been well studied, until recently relatively little was known about the negative regulation of gene expression in response to environmental changes. In the past decade, notable progress has been made in elucidating the molecular nature and functions of transcriptional repressors (Payankaulam et al., 2010). A few active transcriptional repression motifs, including EAR (ethylene-responsive element binding factor-associated amphiphilic repression motif; Ohta et al., 2001), TLLLFR (Matsui et al., 2008), R/KLFGV (Ikeda and Ohme-Takagi, 2009), and LxLxPP (Paponov et al., 2009), have been identified to facilitate recruitment of transcriptional co-repressors and chromatin modifiers to abate gene expression (Kagale and Rozwadowski, 2011). The EAR motif is the most pre-dominant form of active transcriptional repression motif identified in plants so far and is highly conserved across evolutionarily diverse plant species (Kagale and Rozwadowski, 2010, 2011; Kagale et al., 2010; Sherif et al., 2013; Upadhyay et al., 2014; Dong et al., 2015; Amalraj et al., 2016; Ma et al., 2017). Here, we will focus on EAR motif containing transcriptional repressors and their potential modes of action.
The EAR motif, defined by the consensus sequence patterns LxLxL or DLNxxP, is found in numerous transcriptional repressors in plants that negatively regulate genes involved in developmental, hormonal, and stress signaling pathways (Kagale et al., 2010). It was initially identified in a subset of class-II ERFs and TFIIIA-type ZFPs (Ohta et al., 2001). Interestingly, when this motif was tethered to transcriptional activators, they functioned as dominant repressors (Hiratsu et al., 2003). The Arabidopsis EAR repressome comprises 219 transcriptional regulators belonging to 21 different families (Kagale et al., 2010). EAR repressors suppress the expression of target genes through chromatin modification via physically interacting with co-repressors such as, SAP18, known to directly interact with a histone deacetylase (HDA19) potentially forming a repression complex, or TOPLESS (TPL) which is also known to function in conjunction with HDA19 (Szemenyei et al., 2008; Pauwels et al., 2010; Kagale and Rozwadowski, 2011; Oh et al., 2014; Ma et al., 2017).
EAR-mediated transcriptional repression has emerged as one of the principal mechanisms for negative regulation of gene expression in response to abiotic stresses in plants. A number of EAR-motif containing transcriptional repressors from various plant species have been functionally characterized and their role in drought stress responses has been established (Table 4). The Arabidopsis gene RAP2.1, a DREB gene with an EAR motif, is strongly induced by drought and cold stresses (Dong and Liu, 2010). Overexpression of RAP2.1 leads to enhanced sensitivity to drought and cold stresses; whereas, T-DNA insertion mutant alleles of this gene exhibit enhanced tolerance to drought. Stress-induced expression of RD/COR genes was repressed in RAP2.1 overexpressing lines but increased in mutant lines (Dong and Liu, 2010). Similarly, overexpression of wheat ortholog of RAP2.1 (TaRAP2.1) led to dwarfism and frost sensitivity; whereas overexpression of its EAR-motif inactivated variant enhanced its ability to survive drought and frost (Amalraj et al., 2016). Overexpression of several other EAR motif-containing ERF/DREB proteins also results in decreased drought tolerance via reduced expression of stress defense genes (Song et al., 2005; Pan et al., 2010; Zhang et al., 2013; Upadhyay et al., 2014; Amalraj et al., 2016; Scarpeci et al., 2017). The complete deletion or mutation of conserved residues within the EAR motif in various transcriptional repressors has been shown to alter or even reverse their functions (Dong and Liu, 2010; Pan et al., 2010; Zhang et al., 2013; Amalraj et al., 2016). Remarkably, overexpression of an ERF protein from sugarcane (SodERF3) in tobacco increased tolerance to drought and osmotic stresses (Trujillo et al., 2009). The EAR motif in SodERF3 is different from other ERF proteins, such that the highly conserved proline residue in the DLNxxP type of EAR motifs found in various ERFs is replaced by leucine in SodERF3 (Trujillo et al., 2009). It may be speculated that this proline to leucine change in SodERF3 renders its EAR-motif inactive thereby converting it into a transcriptional activator.
Sustained activation of stress responses is metabolically expensive, and therefore plants have evolved negative gene regulation mechanisms to keep it under tight control during normal growth and development and also during stress conditions (Kazan, 2006). It is speculated that the EAR-motif containing ERF repressors perform “capping” role by limiting the upper levels of stress-responsive genes and thereby exerting strong regulatory control over stress responses (Dong and Liu, 2010; Amalraj et al., 2016).
Interestingly, a few EAR-motif containing transcriptional regulators are shown to function as positive regulators of drought responses in plants. For example, OsERF4a (an EAR motif containing ERF) when overexpressed in rice results in increased drought tolerance (Joo et al., 2013). Intriguingly, DRO1 in rice and its orthologs in other plant species also contain a conserved motif (IVLEI) in the C-terminal region, which is required for DRO1 overexpression phenotypes of deeper root in Arabdiopsis and Prunus species (Guseman et al., 2017) and is very similar to the EAR motif. It is possible that the EAR-motifs in the positive regulators enhance stress tolerance by suppressing the expression of other negative regulators of stress responses. Indeed, OsERF4a is shown to improve drought tolerance through the repression of another downstream suppressor of stress response gene, Sir2 (Joo et al., 2013). In summary, EAR-motif containing repressors despite being negative and/or positive regulators of gene expression play a key role in modulating plant stress responses.
Notably, the past efforts to characterize both positive and negative regulators of drought tolerance in wheat and other species have involved their genetic manipulation by transgenic approaches. The commercialization of transgenic wheat has been problematic due to regulatory hurdles in Europe and Africa. Thus, the future deployment of transcriptional regulators in wheat breeding programs should involve screening of natural allelic variation through EcoTilling approaches or generation of variation through non-transgenic gene editing approaches. The availability of a new gold-standard genome sequence of wheat produced by the International Wheat Genome Sequencing Consortium (IWGSC) and the recent advances made in genome editing technologies provide valuable resources and technologies for non-transgenic manipulation of the wheat genome to improve drought-related traits.
Concluding Remarks
Crop growth and development are greatly affected by changes in morphological and physiological responses resulting from the lack of soil moisture. So far only a small number of genes, such as, TaMOR, TaERs, and a few transcriptional regulators, affecting drought-adaptive traits in wheat have been fully investigated. Comparatively, the drought tolerance mechanisms have been extensively studied in other model species and crop plants, including Arabidopsis, rice, and maize. Functional validation of the wheat orthologs of drought tolerance genes identified in other species is essential for their deployment in breeding drought tolerant cultivars.
Combining deep roots capability with superior anatomical features, such as, smaller xylem diameter and increased stomatal density helps wheat to extract and use water to maintain normal photosynthesis under depleting soil moisture conditions. In addition, a tightly linked interaction between xylem-stomatal physiologies is a must to cope with drought episodes. The comprehensive gene expression analyses combined with both forward and reverse genetic approaches have identified several target genes that are useful in manipulating an acclimation response to drought stress in plants. Future elucidation of the precise mode of action of several uncharacterized genes by analyzing phenotypic effects of their mutant variants would significantly advance our understanding of the regulatory networks underlying the complex drought tolerance mechanisms and enable their utilization in developing drought tolerant cultivars.
Recent sequencing of the wheat genome has opened up tremendous opportunities to understand the complex architecture of drought tolerance mechanisms. Wheat harbors a very large and complex allohexaploid genome of 17 Gb with ~80% of repetitive elements and an estimated 124,201 annotated genes (International Wheat Genome Sequencing Consortium, 2014). Similar to other neopolyploid species, such as, brassicas, cotton, and maize, homoeolog expression bias which refers to the preferential or altered expression of one homeolog over the other is observed in wheat as about 55% of the wheat genes were reported to be expressed only from one or two homoeologs of the genome. Transcriptomic or microarray based gene expression analysis has been used as an important tool to understand transcript modulation under stress. The wheat genome sequence combined with transcriptome, proteome, and metabolome profiling of genes associated with various traits for drought tolerance will aid in overcoming the challenges posed by genome complexity and facilitate the analysis of genetic basis of drought tolerance in wheat. Furthermore, it will help in integration of phenotypic, biochemical, and genomics-assisted selection strategies for breeding drought tolerant wheat cultivars.
Author Contributions
MK, RS, and SK discussed and prepared the outline. MK did the literature search and prepared the first draft. RS and SK revised, edited and contributed to writing the manuscript. SO, YU, and MS contributed to discussions and critical revision of the manuscript.
Funding
This work was supported by National Research Council Canada through the Canadian Wheat Improvement Flagship to SK, MK and from Agriculture and Agri-Food Canada to RS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with several of the authors MK and SK.
Acknowledgments
We would like to thank Drs. Faouzi Bekkaoui and Arun Shunmugam for critical reading of the manuscript.
References
Ahmad, I., Ali, N., and Ahmad, H. (2017). “Association mapping of root traits for drought tolerance in bread wheat,” in Wheat Improvement, Management and Utilization, eds R. Wanyera and J. Owuoche (London: InTech.), 39–57.
Ajigboye, O. O., Lu, C., Murchie, E. H., Schlatter, C., Swart, G., and Ray, R. V. (2017). Altered gene expression by sedaxane increases PSII efficiency, photosynthesis and growth and improves tolerance to drought in wheat seedlings. Pestic. Biochem. Physiol. 137, 49–61. doi: 10.1016/j.pestbp.2016.09.008
Amalraj, A., Luang, S., Kumar, M. Y., Sornaraj, P., Eini, O., Kovalchuk, N., et al. (2016). Change of function of the wheat stress-responsive transcriptional repressor TaRAP2.1L by repressor motif modification. Plant Biotechnol. J. 14, 820–832. doi: 10.1111/pbi.12432
Arai-Sanoh, Y., Takai, T., Yoshinaga, S., Nakano, H., Kojima, M., Sakakibara, H., et al. (2014). Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Sci. Rep. 4:5563. doi: 10.1038/srep05563
Araus, J., Amaro, T., Casadesus, J., Asbati, A., and Nachit, M. (1998). Relationships between ash content, carbon isotope discrimination and yield in durum wheat. Funct. Plant Biol. 25, 835–842. doi: 10.1071/PP98071
Ayalew, H., Liu, H., and Yan, G. (2017). Identification and validation of root length QTLs for water stress resistance in hexaploid wheat (Titicum aestivum L.). Euphytica 213:126. doi: 10.1007/s10681-017-1914-4
Baloch, M. J., Dunwell, J., Khan, N. U., Jatoi, W. A., Khakhwani, A. A., Vessar, N. F., et al. (2013). Morpho-physiological characterization of spring wheat genotypes under drought stress. Int. J. Agric. Biol. 15, 945–950.
Bhagwat, S., and Bhatia, C. (1993). Selection for flag leaf stomatal frequency in bread wheat. Plant Breed. 110, 129–136. doi: 10.1111/j.1439-0523.1993.tb01224.x
Bota, J., Medrano, H., and Flexas, J. (2004). Is photosynthesis limited by decreased Rubisco activity and RuBP content under progressive water stress? New Phytol. 162, 671–681. doi: 10.1111/j.1469-8137.2004.01056.x
Chang, H., Chen, D., Kam, J., Richardson, T., Drenth, J., Guo, X., et al. (2016). Abiotic stress upregulated TaZFP34 represses the expression of type-B response regulator and SHY2 genes and enhances root to shoot ratio in wheat. Plant Sci. 252, 88–102. doi: 10.1016/j.plantsci.2016.07.011
Chen, Y., Han, Y., Zhang, M., Zhou, S., Kong, X., and Wang, W. (2016). Overexpression of the wheat expansin gene TaEXPA2 improved seed production and drought tolerance in transgenic tobacco plants. PLoS ONE 11:e0153494. doi: 10.1371/journal.pone.0153494
Cheng, L., Wang, Y., He, Q., Li, H., Zhang, X., and Zhang, F. (2016). Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration. BMC Plant Biol. 16:188. doi: 10.1186/s12870-016-0871-8
Cheuk, A., and Houde, M. (2016). Genome wide identification of C1-2i zinc finger proteins and their response to abiotic stress in hexaploid wheat. Mol. Genet. Genomics 291, 873–890. doi: 10.1007/s00438-015-1152-1
Chipilski, R. R., Kocheva, K. V., Nenova, V. R., and Georgiev, G. I. (2012). Physiological responses of two wheat cultivars to soil drought. Z. Naturforschung C 67, 181–186. doi: 10.5560/ZNC.2012.67c0181
Ciftci-Yilmaz, S., Morsy, M. R., Song, L., Coutu, A., Krizek, B. A., Lewis, M. W., et al. (2007). The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 282, 9260–9268. doi: 10.1074/jbc.M611093200
Condon, A. G., Richards, R., Rebetzke, G., and Farquhar, G. (2004). Breeding for high water-use efficiency. J. Exp. Bot. 55, 2447–2460. doi: 10.1093/jxb/erh277
Ding, W., Fang, W., Shi, S., Zhao, Y., Li, X., and Xiao, K. (2016). Wheat WRKY type transcription factor gene TaWRKY1 is essential in mediating drought tolerance associated with an ABA-dependent pathway. Plant Mol. Biol. Rep. 34, 1111–1126. doi: 10.1007/s11105-016-0991-1
Dong, C. J., and Liu, J. Y. (2010). The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 10:47. doi: 10.1186/1471-2229-10-47
Dong, L., Cheng, Y., Wu, J., Cheng, Q., Li, W., Fan, S., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Ergen, N. Z., Thimmapuram, J., Bohnert, H. J., and Budak, H. (2009). Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct. Integr. Genomics 9, 377–396. doi: 10.1007/s10142-009-0123-1
Gahlaut, V., Jaiswal, V., Kumar, A., and Gupta, P. K. (2016). Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 129, 2019–2042. doi: 10.1007/s00122-016-2794-z
Gahlaut, V., Jaiswal, V., Tyagi, B. S., Singh, G., Sareen, S., Balyan, H. S., et al. (2017). QTL mapping for nine drought-responsive agronomic traits in bread wheat under irrigated and rain-fed environments. PLoS ONE 12:e0182857. doi: 10.1371/journal.pone.0182857
Gupta, P. K., Balyan, H. S., and Gahlaut, V. (2017). QTL analysis for drought tolerance in wheat: present status and future possibilities. Agronomy 7:5. doi: 10.3390/agronomy7010005
Guseman, J. M., Webb, K., Srinivasan, C., and Dardick, C. (2017). DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J. 89, 1093–1105. doi: 10.1111/tpj.13470
He, G.-H., Xu, J.-Y., Wang, Y.-X., Liu, J.-M., Li, P.-S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16:116. doi: 10.1186/s12870-016-0806-4
He, Y., Li, W., Lv, J., Jia, Y., Wang, M., and Xia, G. (2011). Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 63, 1511–1522. doi: 10.1093/jxb/err389
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016). Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS ONE 11:e0157558. doi: 10.1371/journal.pone.0157558
Hetherington, A. M., and Woodward, F. I. (2003). The role of stomata in sensing and driving environmental change. Nature 424:901. doi: 10.1038/nature01843
Hichri, I., Muhovski, Y., ŽiŽková, E., Dobrev, P. I., Franco-Zorrilla, J. M., Solano, R., et al. (2014). The Solanum lycopersicum Zinc Finger2 cysteine-2/histidine-2 repressor-like transcription factor regulates development and tolerance to salinity in tomato and Arabidopsis. Plant Physiol. 164, 1967–1990. doi: 10.1104/pp.113.225920
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313X.2003.01759.x
Huang, J., Sun, S., Xu, D., Lan, H., Sun, H., Wang, Z., et al. (2012). A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol. 80, 337–350. doi: 10.1007/s11103-012-9955-5
Huang, L. Z., Yasir, T. A., Phillips, A. L., and Hu, Y. G. (2013). Isolation and characterization of ERECTA genes and their expression patterns in common wheat (Triticum aestivum L.). Aust. J. Crop Sci. 7, 381–390.
Hura, T., Tyrka, M., Hura, K., Ostrowska, A., and Dziurka, K. (2017). QTLs for cell wall-bound phenolics in relation to the photosynthetic apparatus activity and leaf water status under drought stress at different growth stages of triticale. Mol. Genet. Genomics 292, 415–433. doi: 10.1007/s00438-016-1276-y
Huseynova, I. M. (2012). Photosynthetic characteristics and enzymatic antioxidant capacity of leaves from wheat cultivars exposed to drought. Biochim. Biophys. Acta Bioenerg. 1817, 1516–1523. doi: 10.1016/j.bbabio.2012.02.037
Iannucci, A., Marone, D., Russo, M. A., De Vita, P., Miullo, V., Ferragonio, P., et al. (2017). Mapping QTL for Root and shoot morphological traits in a durum wheat × T. dicoccum segregating population at seedling stage. Int. J. Genomics 2017:17. doi: 10.1155/2017/6876393
Ikeda, M., and Ohme-Takagi, M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975. doi: 10.1093/pcp/pcp048
International Wheat Genome Sequencing Consortium (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
Jiang, Y., Wang, X., Yu, X., Zhao, X., Luo, N., Pei, Z., et al. (2017). Quantitative trait loci associated with drought tolerance in Brachypodium distachyon. Front. Plant Sci. 8:811. doi: 10.3389/fpls.2017.00811
Joo, J., Choi, H. J., Lee, Y. H., Kim, Y. K., and Song, S. I. (2013). A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta 238, 155–170. doi: 10.1007/s00425-013-1880-6
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Kagale, S., Links, M. G., and Rozwadowski, K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109–1134. doi: 10.1104/pp.109.151704
Kagale, S., and Rozwadowski, K. (2010). Small yet effective: the ethylene responsive element binding factor-associated amphiphilic repression (EAR) motif. Plant Signal. Behav. 5, 691–694. doi: 10.4161/psb.5.6.11576
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Kazan, K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11, 109–112. doi: 10.1016/j.tplants.2006.01.004
Khazaei, H., Monneveux, P., Hongbo, S., and Mohammady, S. (2010). Variation for stomatal characteristics and water use efficiency among diploid, tetraploid and hexaploid Iranian wheat landraces. Genet. Resour. Crop Evol. 57, 307–314. doi: 10.1007/s10722-009-9471-x
Kodaira, K. S., Qin, F., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2011). Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 157, 742–756. doi: 10.1104/pp.111.182683
Li, B., Liu, D., Li, Q., Mao, X., Li, A., Wang, J., et al. (2016). Overexpression of wheat gene TaMOR improves root system architecture and grain yield in Oryza sativa. J. Exp. Bot. 67, 4155–4167. doi: 10.1093/jxb/erw193
Li, H., Li, M., Wei, X., Zhang, X., Xue, R., Zhao, Y., et al. (2017). Transcriptome analysis of drought-responsive genes regulated by hydrogen sulfide in wheat (Triticum aestivum L.) leaves. Mol. Genet. Genomics 292, 1091–1110. doi: 10.1007/s00438-017-1330-4
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lilley, J., and Kirkegaard, J. (2011). Benefits of increased soil exploration by wheat roots. Field Crops Res. 122, 118–130. doi: 10.1016/j.fcr.2011.03.010
Liu, C., Mi, H., Liu, H., Xie, S., Wu, Y., Ning, P., et al. (2017). Response to water deficit in glume of wheat: expression profiling by microarray analysis. Euphytica 213:26. doi: 10.1007/s10681-016-1827-7
Liu, H., Able, A. J., and Able, J. A. (2017). Water-deficit stress-responsive microRNAs and their targets in four durum wheat genotypes. Funct. Integr. Genomics 17, 237–251. doi: 10.1007/s10142-016-0515-y
Liu, Z., Qin, J., Tian, X., Xu, S., Wang, Y., Li, H., et al. (2017). Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum asetivum L.). Plant Biotechnol. J. doi: 10.1111/pbi.12822. [Epub ahead of print].
Lobell, D. B., and Gourdji, S. M. (2012). The influence of climate change on global crop productivity. Plant Physiol. 160, 1686–1697. doi: 10.1104/pp.112.208298
Lopez-Maury, L., Marguerat, S., and Bahler, J. (2008). Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 9, 583–593. doi: 10.1038/nrg2398
Loutfy, N., El-Tayeb, M. A., Hassanen, A. M., Moustafa, M. F., Sakuma, Y., and Inouhe, M. (2012). Changes in the water status and osmotic solute contents in response to drought and salicylic acid treatments in four different cultivars of wheat (Triticum aestivum). J. Plant Res. 125, 173–184. doi: 10.1007/s10265-011-0419-9
Lucas, S. J., Salantur, A., Yazar, S., and Budak, H. (2017). High-throughput SNP genotyping of modern and wild emmer wheat for yield and root morphology using a combined association and linkage analysis. Funct. Integr. Genomics 17, 667–685. doi: 10.1007/s10142-017-0563-y
Ludlow, M., and Muchow, R. (1990). A critical evaluation of traits for improving crop yields in water-limited environments. Adv. Agron. 43, 107–153. doi: 10.1016/S0065-2113(08)60477-0
Luo, X., Bai, X., Zhu, D., Li, Y., Ji, W., Cai, H., et al. (2012). GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta 235, 1141–1155. doi: 10.1007/s00425-011-1563-0
Ma, D., Sun, D., Wang, C., Qin, H., Ding, H., Li, Y., et al. (2016). Silicon application alleviates drought stress in wheat through transcriptional regulation of multiple antioxidant defense pathways. J. Plant Growth Regul. 35, 1–10. doi: 10.1007/s00344-015-9500-2
Ma, H., Duan, J., Ke, J., He, Y., Gu, X., Xu, T. H., et al. (2017). A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci. Adv. 3:e1601217. doi: 10.1126/sciadv.1601217
Ma, J., Li, R., Wang, H., Li, D., Wang, X., Zhang, Y., et al. (2017a). Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant Sci. 8:592. doi: 10.3389/fpls.2017.00592
Ma, J., Luo, W., Zhang, H., Zhou, X.-H., Qin, N.-N., Wei, Y.-M., et al. (2017b). Identification of quantitative trait loci for seedling root traits from Tibetan semi-wild wheat (Triticum aestivum ssp. tibetanum). Genome 25, 1–8. doi: 10.1139/gen-2017-0097
Manes, Y., Gomez, H., Puhl, L., Reynolds, M., Braun, H., and Trethowan, R. (2012). Genetic yield gains of the CIMMYT international semi-arid wheat yield trials from 1994 to 2010. Crop Sci. 52, 1543–1552. doi: 10.2135/cropsci2011.10.0574
Manickavelu, A., Kawaura, K., Oishi, K., Shin-I, T., Kohara, Y., Yahiaoui, N., et al. (2012). Comprehensive functional analyses of expressed sequence tags in common wheat (Triticum aestivum). DNA Res. 19, 165–177. doi: 10.1093/dnares/dss001
Masle, J., Gilmore, S. R., and Farquhar, G. D. (2005). The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436:866. doi: 10.1038/nature03835
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Merchuk-Ovnat, L., Fahima, T., Ephrath, J. E., Krugman, T., and Saranga, Y. (2017). Ancestral QTL alleles from wild emmer wheat enhance root development under drought in modern wheat. Front. Plant Sci. 8:703. doi: 10.3389/fpls.2017.00703
Mittler, R., Kim, Y., Song, L., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Mondal, B., Singh, A., Yadav, A., Tomar, R. S. S., Vinod, G. P. S., and Prabhu, K. V. (2017). QTL mapping for early ground cover in wheat (Triticum aestivum L.) under drought stress. Curr. Sci. 112, 1266–1271. doi: 10.18520/cs/v112/i06/1266-1271
Mwadzingeni, L., Shimelis, H., Dube, E., Laing, M. D., and Tsilo, T. J. (2016). Breeding wheat for drought tolerance: progress and technologies. J. Integr. Agric. 15, 935–943. doi: 10.1016/S2095-3119(15)61102-9
Mwadzingeni, L., Shimelis, H., Rees, D. J. G., and Tsilo, T. J. (2017). Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS ONE 12:e0171692. doi: 10.1371/journal.pone.0171692
Nadia, K., Chang, X., and Jing, R. (2017). Genetic dissection of stem water-soluble carbohydrates and agronomic traits in wheat under different water regimes. J. Agric. Sci. 9:42. doi: 10.5539/jas.v9n3p42
Oh, E., Zhu, J. Y., Ryu, H., Hwang, I., and Wang, Z. Y. (2014). TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 5:4140. doi: 10.1038/ncomms5140
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/tpc.13.8.1959
Okay, S., Derelli, E., and Unver, T. (2014). Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol. Genet. Genomics 289, 765–781. doi: 10.1007/s00438-014-0849-x
Ovenden, B., Milgate, A., Wade, L. J., Rebetzke, G. J., and Holland, J. B. (2017). Genome-wide associations for water-soluble carbohydrate concentration and relative maturity in wheat using SNP and DArT marker arrays. Genes Genomes Genet. 7, 2821–2830. doi: 10.1534/g3.117.039842
Pan, I. C., Li, C. W., Su, R. C., Cheng, C. P., Lin, C. S., and Chan, M. T. (2010). Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta 232, 1075–1086. doi: 10.1007/s00425-010-1235-5
Paponov, I. A., Teale, W., Lang, D., Paponov, M., Reski, R., Rensing, S. A., et al. (2009). The evolution of nuclear auxin signalling. BMC Evol. Biol. 9:126. doi: 10.1186/1471-2148-9-126
Pask, A., and Reynolds, M. (2013). Breeding for yield potential has increased deep soil water extraction capacity in irrigated wheat. Crop Sci. 53, 2090–2104. doi: 10.2135/cropsci2013.01.0011
Passioura, J. (2006). Increasing crop productivity when water is scarce—from breeding to field management. Agric. Water Manage. 80, 176–196. doi: 10.1016/j.agwat.2005.07.012
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Perez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Payankaulam, S., Li, L. M., and Arnosti, D. N. (2010). Transcriptional repression: conserved and evolved features. Curr. Biol. 20, R764–R771. doi: 10.1016/j.cub.2010.06.037
Pellegrineschi, A., Reynolds, M., Pacheco, M., Brito, R. M., Almeraya, R., Yamaguchi-Shinozaki, K., et al. (2004). Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47, 493–500. doi: 10.1139/g03-140
Peremarti, A., Marè, C., Aprile, A., Roncaglia, E., Cattivelli, L., Villegas, D., et al. (2014). Transcriptomic and proteomic analyses of a pale-green durum wheat mutant shows variations in photosystem components and metabolic deficiencies under drought stress. BMC Genomics 15:125. doi: 10.1186/1471-2164-15-125
Persak, H., and Pitzschke, A. (2014). Dominant repression by Arabidopsis transcription factor MYB44 causes oxidative damage and hypersensitivity to abiotic stress. Int. J. Mol. Sci. 15, 2517–2537. doi: 10.3390/ijms15022517
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Pillitteri, L. J., and Torii, K. U. (2012). Mechanisms of stomatal development. Annu. Rev. Plant Biol. 63, 591–614. doi: 10.1146/annurev-arplant-042811-105451
Qin, Y., Tian, Y., and Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 464, 428–433. doi: 10.1016/j.bbrc.2015.06.128
Qiu, Z., Yuan, M., He, Y., Li, Y., and Zhang, L. (2017). Physiological and transcriptome analysis of He-Ne laser pretreated wheat seedlings in response to drought stress. Sci. Rep. 7:6108. doi: 10.1038/s41598-017-06518-z
Rauf, M., Munir, M., ul Hassan, M., Ahmad, M., and Afzal, M. (2007). Performance of wheat genotypes under osmotic stress at germination and early seedling growth stage. Afr. J. Biotechnol. 6, 971–975. doi: 10.5897/AJB2007.000-2119
Reynolds, M., Foulkes, M. J., Slafer, G. A., Berry, P., Parry, M. A., Snape, J. W., et al. (2009). Raising yield potential in wheat. J. Exp. Bot. 60, 1899–1918. doi: 10.1093/jxb/erp016
Richards, R. A. (2006). Physiological traits used in the breeding of new cultivars for water-scarce environments. Agric. Water Manage. 80, 197–211. doi: 10.1016/j.agwat.2005.07.013
Rong, W., Qi, L., Wang, A., Ye, X., Du, L., Liang, H., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Saeed, I., Chen, X., Bachir, D. G., Chen, L., and Hu, Y.-G. (2017). Association mapping for photosynthesis and yield traits under two moisture conditions and their drought indices in winter bread wheat (‘Triticum aestivum’L.) using SSR markers. Aust. J. Crop Sci. 11:248. doi: 10.21475/ajcs.17.11.03.pne252
Scarpeci, T. E., Frea, V. S., Zanor, M. I., and Valle, E. M. (2017). Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J. Exp. Bot. 68, 673–685. doi: 10.1093/jxb/erw429
Schoppach, R., Wauthelet, D., Jeanguenin, L., and Sadok, W. (2014). Conservative water use under high evaporative demand associated with smaller root metaxylem and limited trans-membrane water transport in wheat. Funct. Plant Biol. 41, 257–269. doi: 10.1071/FP13211
Shahinnia, F., Le Roy, J., Laborde, B., Sznajder, B., Kalambettu, P., Mahjourimajd, S., et al. (2016). Genetic association of stomatal traits and yield in wheat grown in low rainfall environments. BMC Plant Biol. 16:150. doi: 10.1186/s12870-016-0838-9
Sharp, R. E., Poroyko, V., Hejlek, L. G., Spollen, W. G., Springer, G. K., Bohnert, H. J., et al. (2004). Root growth maintenance during water deficits: physiology to functional genomics. J. Exp. Bot. 55, 2343–2351. doi: 10.1093/jxb/erh276
Sherif, S., El-Sharkawy, I., Paliyath, G., and Jayasankar, S. (2013). PpERF3b, a transcriptional repressor from peach, contributes to disease susceptibility and side branching in EAR-dependent and -independent fashions. Plant Cell Rep. 32, 1111–1124. doi: 10.1007/s00299-013-1405-6
Shiferaw, B., Smale, M., Braun, H.-J., Duveiller, E., Reynolds, M., and Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Soriano, J. M., Malosetti, M., Roselló, M., Sorrells, M. E., and Royo, C. (2017). Dissecting the old Mediterranean durum wheat genetic architecture for phenology, biomass and yield formation by association mapping and QTL meta-analysis. PLoS ONE 12:e0178290. doi: 10.1371/journal.pone.0178290
Sylvester-Bradley, R., Scott, R., and Wright, C. (1990). Physiology in the Production and Improvement of Cereals. London: Home Grown Cereals Authority, NHBS Ltd.
Szemenyei, H., Hannon, M., and Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. doi: 10.1126/science.1151461
Tripathi, P., Rabara, R. C., and Rushton, P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. doi: 10.1007/s00425-013-1985-y
Trujillo, L. E., Menéndez, C., Ochogavía, M. E., Hernández, I., Borrás, O., Rodríguez, R., et al. (2009). Engineering drought and salt tolerance in plants using SodERF3, a novel sugarcane ethylene responsive factor. Biotecnol. Appl. 26, 168–171.
Uga, Y., Okuno, K., and Yano, M. (2011). Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62, 2485–2494. doi: 10.1093/jxb/erq429
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Upadhyay, R. K., Gupta, A., Ranjan, S., Singh, R., Pathre, U. V., Nath, P., et al. (2014). The EAR motif controls the early flowering and senescence phenotype mediated by over-expression of SlERF36 and is partly responsible for changes in stomatal density and photosynthesis. PLoS ONE 9:e101995. doi: 10.1371/journal.pone.0101995
Upadhyay, R. K., Soni, D. K., Singh, R., Dwivedi, U. N., Pathre, U. V., Nath, P., et al. (2013). SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 64, 3237–3247. doi: 10.1093/jxb/ert162
Wang, C., Deng, P., Chen, L., Wang, X., Ma, H., Hu, W., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 8:e65120. doi: 10.1371/journal.pone.0065120
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., Sun, T., et al. (2015). Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6:615. doi: 10.3389/fpls.2015.00615
Wu, J., Chen, J., Wang, L., and Wang, S. (2017). Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 8:380. doi: 10.3389/fpls.2017.00380
Xu, W., Jia, L., Shi, W., Liang, J., Zhou, F., Li, Q., et al. (2013). Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 197, 139–150. doi: 10.1111/nph.12004
Xu, Y.-F., Li, S.-S., Li, L.-H., Ma, F.-F., Fu, X.-Y., Shi, Z.-L., et al. (2017). QTL mapping for yield and photosynthetic related traits under different water regimes in wheat. Mol. Breed. 37:34. doi: 10.1007/s11032-016-0583-7
Xu, Z.-S., Xia, L.-Q., Chen, M., Cheng, X.-G., Zhang, R.-Y., Li, L.-C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. doi: 10.1007/s11103-007-9237-9
Yamaguchi, M., and Sharp, R. E. (2010). Complexity and coordination of root growth at low water potentials: recent advances from transcriptomic and proteomic analyses. Plant Cell Environ. 33, 590–603. doi: 10.1111/j.1365-3040.2009.02064.x
Yokota, A., Kawasaki, S., Iwano, M., Nakamura, C., Miyake, C., and Akashi, K. (2002). Citrulline and DRIP-1 protein (ArgE homologue) in drought tolerance of wild watermelon. Ann. Bot. 89, 825–832. doi: 10.1093/aob/mcf074
Yoo, C. Y., Pence, H. E., Jin, J. B., Miura, K., Gosney, M. J., Hasegawa, P. M., et al. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22, 4128–4141. doi: 10.1105/tpc.110.078691
Zampieri, M., Ceglar, A., Dentener, F., and Toreti, A. (2017). Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ. Res. Lett. 12:064008. doi: 10.1088/1748-9326/aa723b
Zhai, Y., Li, J. W., Li, X. W., Lei, T. T., Yan, F., Zhao, Y., et al. (2013). Isolation and characterization of a novel transcriptional repressor GmERF6 from soybean. Biol. Plant. 57, 26–32. doi: 10.1007/s10535-012-0146-7
Zhang, H., Liu, Y., Wen, F., Yao, D., Wang, L., Guo, J., et al. (2014). A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 65, 5795–5809. doi: 10.1093/jxb/eru313
Zhang, H., Zhang, J., Quan, R., Pan, X., Wan, L., and Huang, R. (2013). EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 237, 1443–1451. doi: 10.1007/s00425-013-1852-x
Zhang, Z., Yao, W., Dong, N., Liang, H., Liu, H., and Huang, R. (2007). A novel ERF transcription activator in wheat and its induction kinetics after pathogen and hormone treatments. J. Exp. Bot. 58, 2993–3003. doi: 10.1093/jxb/erm151
Zheng, J., Yang, Z., Madgwick, P. J., Carmo-Silva, E., Parry, M. A., and Hu, Y.-G. (2015). TaER expression is associated with transpiration efficiency traits and yield in bread wheat. PLoS ONE 10:e0128415. doi: 10.1371/journal.pone.0128415
Zhou, J., Ma, C., Zhen, S., Cao, M., Zeller, F. J., Hsam, S. L., et al. (2016). Identification of drought stress related proteins from 1Sl (1B) chromosome substitution line of wheat variety Chinese Spring. Bot. Stud. 57:20. doi: 10.1186/s40529-016-0134-x
Keywords: wheat, drought, root traits, transpiration efficiency, transcriptional regulation, EAR motif
Citation: Kulkarni M, Soolanayakanahally R, Ogawa S, Uga Y, Selvaraj MG and Kagale S (2017) Drought Response in Wheat: Key Genes and Regulatory Mechanisms Controlling Root System Architecture and Transpiration Efficiency. Front. Chem. 5:106. doi: 10.3389/fchem.2017.00106
Received: 31 August 2017; Accepted: 07 November 2017;
Published: 05 December 2017.
Edited by:
Raju Datla, National Research Council Canada (NRC-CNRC), CanadaReviewed by:
Rupesh Kailasrao Deshmukh, Laval University, CanadaNoemi Tel Zur, Ben-Gurion University of the Negev, Beersheba, Israel
Copyright © 2017 Kulkarni, Soolanayakanahally, Ogawa, Uga, Selvaraj and Kagale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sateesh Kagale, U2F0ZWVzaC5LYWdhbGVAbnJjLWNucmMuZ2MuY2E=
 Manoj Kulkarni
Manoj Kulkarni Raju Soolanayakanahally
Raju Soolanayakanahally Satoshi Ogawa
Satoshi Ogawa Yusaku Uga4
Yusaku Uga4 Michael G. Selvaraj
Michael G. Selvaraj Sateesh Kagale
Sateesh Kagale