
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 21 March 2017
Sec. Chemical Biology
Volume 5 - 2017 | https://doi.org/10.3389/fchem.2017.00010
This article is part of the Research Topic Natural and artificially-engineered nucleic acid aptamers: design, characterization and technological applications View all 6 articles
Riboswitches are RNA elements that bind specific metabolites in order to regulate the gene expression involved in controlling the cellular concentration of the respective molecule or ion. Ligand recognition is mostly facilitated by Mg2+ mediated pre-organization of the riboswitch to an active tertiary fold. To predict these specific Mg2+ induced tertiary interactions of the btuB riboswitch from E. coli, we here report Mg2+ binding pockets in its aptameric part in both, the ligand-free and the ligand-bound form. An ensemble of weak and strong metal ion binding sites distributed over the entire aptamer was detected by terbium(III) cleavage assays, Tb3+ being an established Mg2+ mimic. Interestingly many of the Mn+ (n = 2 or 3) binding sites involve conserved bases within the class of coenzyme B12-binding riboswitches. Comparison with the published crystal structure of the coenzyme B12 riboswitch of S. thermophilum aided in identifying a common set of Mn+ binding sites that might be crucial for tertiary interactions involved in the organization of the aptamer. Our results suggest that Mn+ binding at strategic locations of the btuB riboswitch indeed facilitates the assembly of the binding pocket needed for ligand recognition. Binding of the specific ligand, coenzyme B12 (AdoCbl), to the btuB aptamer does however not lead to drastic alterations of these Mn+ binding cores, indicating the lack of a major rearrangement within the three-dimensional structure of the RNA. This finding is strengthened by Tb3+ mediated footprints of the riboswitch's structure in its ligand-free and ligand-bound state indicating that AdoCbl indeed induces local changes rather than a global structural rearrangement.
Riboswitches are a class of non-coding RNA that bind small molecules, i.e., metabolites, with high affinity and specificity (Mironov et al., 2002; Nahvi et al., 2002; Winkler et al., 2002). The two functional domains of the riboswitch, the aptamer, and the downstream situated expression platform undergo concomitant structural changes upon ligand binding to the aptamer region (Mandal and Breaker, 2004). This conformational switch ultimately regulates gene expression mainly at the transcriptional or translational level although other regulatory mechanisms have been proposed involving splicing, self-cleavage of the RNA and trans regulation (Kubodera et al., 2003; Winkler and Breaker, 2003; Soukup and Soukup, 2004; Winkler et al., 2004; Loh et al., 2009; Bastet et al., 2011).
The btuB riboswitch from E. coli belongs to the class of coenzyme B12 riboswitches recognizing and regulating the pool of coenzyme B12 (AdoCbl) inside the cell by altering the expression of the outer membrane B12 transporter BtuB (Reynolds et al., 1980; Gudmundsdottir et al., 1988; Nou and Kadner, 2000; Nahvi et al., 2004). The secondary structure of the btuB aptamer consists of a four way junction common with the other B12 riboswitches and a varying peripheral region (Nahvi et al., 2004). The two recently solved crystal structures of AdoCbl riboswitches from thermophiles have established the role of the four way junction in constituting the ligand binding pocket/the receptor core. The peripheral elements on the other hand further stabilize the binding pocket upon ligand binding and are crucial for specificity toward AdoCbl (Johnson et al., 2012; Peselis and Serganov, 2012).
In many riboswitches the aptamer is pre-organized to facilitate the interaction with its cognate ligand as was observed for the purine, lysine, FMN, SAM I, glucosamine-6 phosphate, and the preQ1 binding riboswitches (Hampel and Tinsley, 2006; Klein and Ferré-D'Amaré, 2006; Noeske et al., 2007; Garst et al., 2008; Montange and Batey, 2008; Baird and Ferré-D'Amaré, 2010; Heppell et al., 2011; Suddala et al., 2013). Due to their physiological abundance, divalent metal ions like Mg2+ help to assemble the active tertiary structure of the polyanionic RNA not only by shielding the negatively charged sugar-phosphate backbone but also by facilitating specific tertiary interactions within nucleobases (Misra and Draper, 1998; Pyle, 2002; Woodson, 2005). These metal ion interactions are mediated either through an inner sphere or an outer sphere co-ordination where the contacts to the RNA are made directly or via water molecules, respectively (Erat and Sigel, 2011; Schnabl et al., 2011).
Our earlier studies (Choudhary and Sigel, 2014) have given a strong indication that the btuB riboswitch is prefolded by Mg2+ already in the absence of its ligand AdoCbl. We also observed that AdoCbl does not switch the RNA conformation in the absence of Mg2+, indicating the essential role of Mg2+ in the pre-organization of the aptamer to enable ligand binding. However, the specific tertiary interactions induced by Mg2+ that constitute the ligand-free state of the riboswitch are still not defined. So far there was only little structural insight on the ligand-free fold of the B12-riboswitches. All secondary structural models were and still are based on the B12-bound state, and also the crystallized coenzyme B12 riboswitches represent the ligand-bound structure (Nahvi et al., 2004; Johnson et al., 2012; Peselis and Serganov, 2012). In order to validate the Mg2+ aided pre-organization of the btuB riboswitch in the absence of AdoCbl it is therefore crucial to locate the specific Mg2+ binding sites of this RNA.
A well-established method to map metal ion binding sites in large nucleic acids makes use of lanthanide(III) probes (Ciesiolka et al., 1989; Dorner and Barta, 1999; Sigel et al., 2000; Walter et al., 2000; Kaye et al., 2002; Sigel and Pyle, 2003; Waldsich and Pyle, 2008). Lanthanide(III) ions, specifically Tb3+ has been used as the closest mimic for Mg2+ due to its similar properties (Sigel et al., 2000; Sigel and Pyle, 2003). The ionic radius of hydrated Tb3+ (0.91 Å) is in the same range as hydrated Mg2+ (0.71 Å) and both ions have similar preferences for co-ordination to oxygen ligands (Saito and Suga, 2002). It has been shown that Tb3+ can replace Mg2+ at its specific binding sites without the rupture of the tertiary structure of the RNA (Sigel et al., 2000). Application of Tb3+ has the decisive advantage, that this ion can be applied at much lower concentrations and in parallel to Mg2+, still yielding a clear cleavage pattern at physiological pH. This is due to the lower pKa (~7.9) of the Tb3+ aqua species [Tb(H2O)n]3+ compared to the one of hydrated Mg2+ (pKa ~ 11.4) (Sigel et al., 2000; Harris and Walter, 2003).
Our current studies describe the specific Mn+ (n = 2 or 3) binding sites mapped on the btuB riboswitch of E. coli, both in the ligand-free btuB aptamer as well as in the AdoCbl-bound form, in order to establish the role of the Mg2+-assisted tertiary interactions for ligand binding. Furthermore, we compared our data to the consensus sequence (Nahvi et al., 2004) as well as to the sequences of the two crystallized AdoCbl riboswitches (Johnson et al., 2012; Peselis and Serganov, 2012) to see if there is a common set of Mg2+-mediated tertiary interactions crucial for the organization of the AdoCbl aptamer.
Nucleoside 5′ triphosphates (ATP, GTP, CTP, and UTP) were purchased from GE Healthcare and Sigma-Aldrich, respectively. Homemade T7 RNA polymerase (Gallo et al., 2005) was used for in-vitro RNA transcription. RNase T1 1000 U/μL was purchased from Fermentas and was diluted to 1 U/μL in a buffer containing 50 mM Tris-HCl (pH 7.4) and 50% (v/v) glycerol. Coenzyme B12 (Sigma-Aldrich) and anhydrous TbCl3 (Sigma-Aldrich) were used without any further purification. Denaturing polyacrylamide gels were prepared using Long Ranger™ gel solution, Lonza, Rockland ME (USA). All the buffers, salt solutions and gel solutions were filtered through 0.2 μm filters. All other chemicals were at least puriss p.a. and were purchased from Sigma-Aldrich. Gels were scanned by Storm860 PhosphoImager and analyzed by ImageQuant software (GE Healthcare).
The btuB RNA was obtained by in vitro transcription from the plasmid pPC1 by homemade T7 RNA polymerase (Gallo et al., 2005). The plasmid pPC1 is based on the plasmid pSG2 (Gallo, 2009) that contains the natural 202 nucleotide btuB aptamer sequence. In addition, pPC1 contains a newly introduced GGA sequence at the 5′-end of the btuB aptamer sequence for efficient transcription (Gallo et al., 2005) and a GAGCUCG sequence at 3′-end stemming from digestion with EcoR1 (Promega). These additional nucleotides still render the aptamer active (Supplementary Figure 5). After in-vitro transcription of the plasmid DNA, the btuB RNA was purified by 10% denaturing PAGE, electroeluted, precipitated with ethanol and concentrated with Vivaspin concentrator (5000 MWCO). The RNA was stored in water at −20°C.
TbCl3 stock solutions were prepared as previously described (Sigel et al., 2000). In a total volume of 10 μL, 1 μM of unlabeled RNA along with 10 nM of 32P-5′-labeled RNA was denatured at 90°C for 45 s in a reaction buffer containing 25 mM MOPS, pH 7.0 and 100 mM KCl followed by the addition of 20 mM MgCl2 and incubation at 37°C for 15 min. For the samples with ligand, AdoCbl was added to the RNA sample folded in 20 mM MgCl2 and the samples were further incubated at 37°C for 30 min. For the competition experiments with MgCl2, the RNA was folded in various MgCl2 concentrations after denaturation and before the addition of TbCl3. The cleavage reactions were performed for 1 h on ice. Subsequently quenching, buffer [80% formamide (v/v), 10 mM EDTA] was added to the samples to stop the reaction. The intensity of cleavage products did not change much even when the reaction was carried out for 1.5 and 2 h on ice (data not shown). The samples were precipitated with 2.5 volumes of ethanol, resuspended in 5 μL of formamide loading buffer [80% formamide (v/v), 10 mM EDTA; pH 8.0 at 20°C, 2% Bromophenol blue, 2% Xylene Cyanol] and the cleavage products were separated by 10% denaturing PAGE. RNase T1 ladder and alkaline hydrolysis ladder were prepared as described (Regulski and Breaker, 2008). The relative cleavage intensities of the nucleotides with respect to cleavage intensities at 0 mM Tb3+ concentration are calculated as described (Choudhary et al., 2014).
Tb3+ cleavage was carried out with the ligand free btuB aptamer pre-folded in 20 mM MgCl2 and varying the Tb3+ concentration. As shown in Figure 1, Tb3+ at concentrations up to 0.1 mM is unable to cleave the RNA indicating that the apparent KD of Tb3+ to any of the binding sites is higher than 100 μM under these conditions. As soon as the concentration of Tb3+ is raised to 0.25 mM, backbone cleavage becomes evident (Figure 1). Usually Tb3+ mediated cleavage at micromolar concentration indicates specific metal ion binding sites (Harris and Walter, 2003; Sigel and Pyle, 2003). Hence, the difference in cleavage intensities at 0.5 mM Tb3+ compared to the cleavage intensities at 0 mM Tb3+ were used to map the specific metal ion binding sites (Figure 2A). This comparison shows that the ligand-free btuB aptamer offers Mn+-binding sites of various strengths that are evenly distributed over its whole secondary structure (Figure 2B).
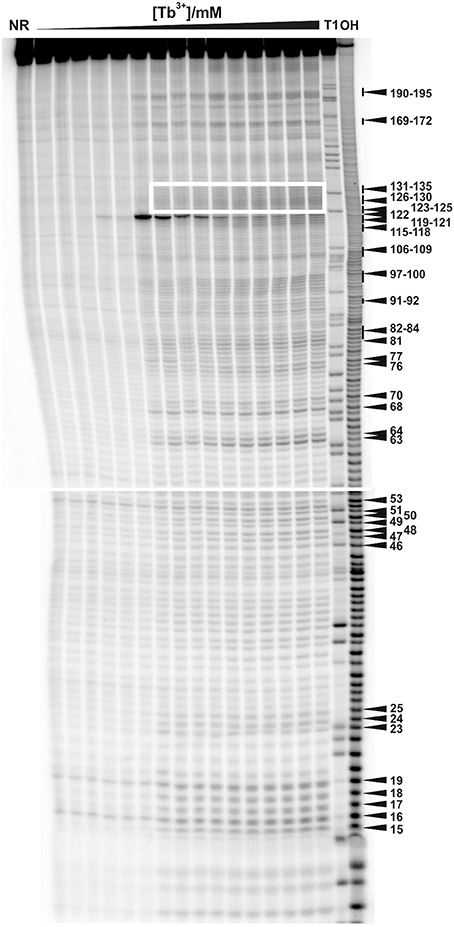
Figure 1. Tb3+ cleavage of the ligand-free and pre-folded btuB riboswitch of E. coli incubated with increasing concentrations of TbCl3 from left to right (0.0, 0.025, 0.05, 0.075, 0.1, 0.25, 0.5, 0.75, 1.0, 2.5, 5.0, 7.5, 10, 15, 20 mM). The nucleotides undergoing a distinct cleavage in the presence of TbCl3 are indicated on the right side. The white box indicates the supposedly helical region of P12 encompassing nucleotides 123–135. NR, Non-reacted RNA; T1, RNase T1 ladder; OH, alkaline hydrolysis ladder. The Mg2+ concentration was held constant at 20 mM.
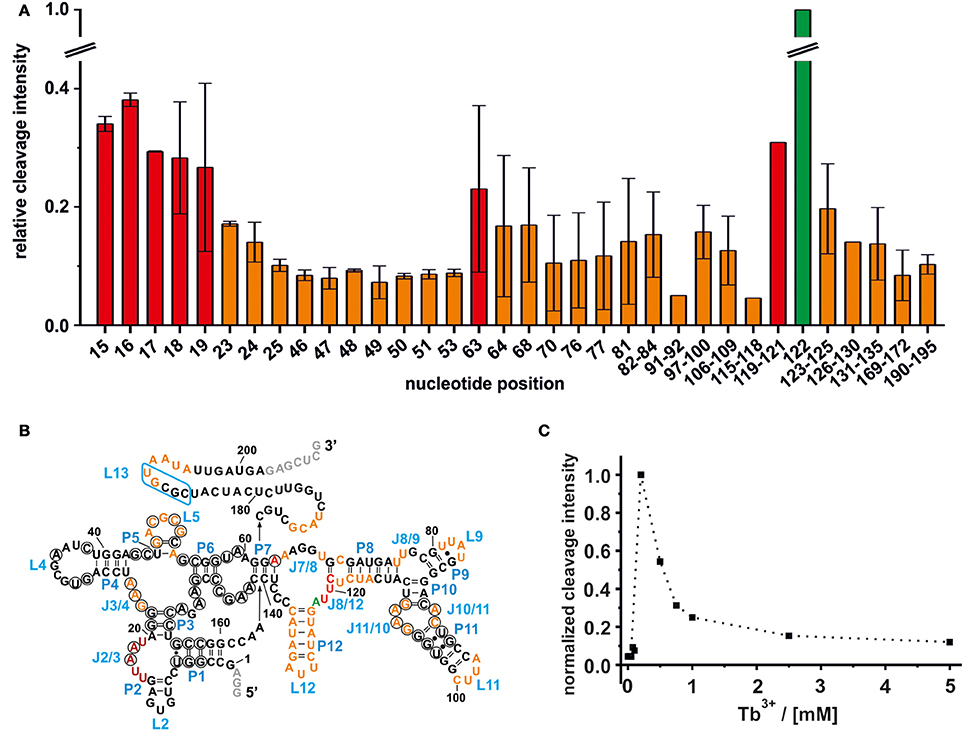
Figure 2. Mapping of the Mn+ binding sites on the ligand free btuB riboswitch of E. coli. (A) The relative cleavage intensity at the indicated nucleotides at 0.5 mM TbCl3 and 20 mM Mg2+ by nucleotide is displayed. Sites not showing any intensity change are not displayed. Shown is the average of two gels with the respective range while the cleavage intensity was normalized to the strongest cleavage found at A122. Weak cleavage sites (up to 0.2 relative cleavage intensity) are shown in orange, strong cleavage sites (up to 0.4 relative cleavage intensity) are shown in red, and the strongest cleavage site is shown in green. (B) Mapping of the Mn+ binding sites on the secondary structure of the btuB riboswitch. Nucleotides in black circles indicate the conserved bases from the consensus sequence of AdoCbl riboswitches (Nahvi et al., 2004). The blue box indicates the L13 region supposedly undergoing the kissing loop interaction with L5 upon AdoCbl binding. Nucleotides in gray were added to the original btuB sequence at both the 3′- and 5′-end to enhance the transcription yield (Choudhary and Sigel, 2014). (C) The change in cleavage intensity at A122 (J8/12) as a function of TbCl3 concentration derived from one representative experiment.
The intensity of a Tb3+ cleavage band depends on the geometry of the coordinating nucleotides within the RNA tertiary fold, i.e., on the accessibility of the 2′-hydroxyl group of their ribose (Harris and Walter, 2003). Weak Tb3+ cleavage was categorized as being below 20% cleavage intensity relative to the strongest cleavage observed at A122, and indicates a hindered accessibility due to geometrical constraints. Weak Tb3+ cleavage sites on the btuB riboswitch are located around the junctions J3/4, J7/8, and J8/9, the loops L9, L11, and L12, the internal loop region of J10/11 and J11/10 and within the base paired regions of P5, P8, and P12. Further, weak cleavage sites are situated proximal to nucleotides 169–172, on the kissing loop contributing L5 and around its counterpart at nucleotides190–195 (Figure 2B). Interestingly, some of the cleavage sites at J3/4, P5/L5, and at the internal loop region J11/10 and J10/11 involve nucleotides of conserved regions in the consensus sequence of the coenzyme B12 riboswitches (Nahvi et al., 2004). Comparison with the published crystal structures of the coenzyme B12 riboswitches from thermophiles (Johnson et al., 2012; Peselis and Serganov, 2012) indicates that the conserved nucleotides at J11/10 contact the nucleotides from the junction J6/3 helping to create one of the coaxial stems of the receptor core. Also metal ion binding at L5 might be important for the observed kissing loop (KL) interaction between L5 and L13 (Johnson et al., 2012; Peselis and Serganov, 2012), while metal ion binding at J3/4 could assist its supposed tertiary interactions to J6/3 and P3 (Peselis and Serganov, 2012).
The regions J2/3 and J8/12 as well as A63 situated in P7 harbor strong Tb3+ cleavage sites involving also the conserved nucleotides at J2/3 and A63 (Figure 2B). Metal ion binding at J2/3 could facilitate the supposed tertiary interactions to P1 as reported for the S. thermophilum AdoCbl riboswitch (Peselis and Serganov, 2012). Although strong cleavage is clustered at the above-mentioned sites, we want to draw the attention to the helical region of P12, which is just next to the strongest cleavage site found at A122 (Figures 1, 2B). Even though the nucleotides of P12 are proposed to undergo base pairing in the ligand-bound state (Nahvi et al., 2004) hampering Tb3+ accessibility, our experiments show partial cleavage in this region. The region P12 therefore appears to be at least to some extent in a dynamic or non-fully canonical conformation accessible for Tb3+ binding when in the ligand-free state. As stated before, nucleotide A122, which exhibits the strongest cleavage of all nucleotides, is situated next to this region of moderate Mn+ binding. The cleavage at nucleotide A122 decreases with increasing Tb3+ concentration, a trend completely opposite of any of the other cleavage sites (Figure 2C). We suggest that in contrast to the other sites, the geometry at A122 is altered at high Tb3+ concentrations, possibly by additional Tb3+ binding close-by, thereby inhibiting the cleavage.
Besides stem P12, all other proposed helical regions of the btuB aptamer were found to be unaffected from Tb3+-cleavage confirming their helical state also in the ligand-free form of the riboswitch. The lack of cleavage does however not exclude their involvement in metal ion binding since the helical regions often bind metal ions in the major groove without direct access to the 2′-hydroxyl group (Sigel et al., 2000), and therefore metal ion binding sites in such regions remain undetected.
Competing Tb3+ with an increasing Mg2+ concentration results in the decrease of Tb3+ cleavage intensity, ideally at nucleotides involved in Mg2+ binding, but also in general due to better charge screening by the increased M2+ concentration (Harris and Walter, 2003). Hence, not only specific metal ion binding sites are confirmed but also sites of non-specific Tb3+ binding are eliminated. In competition experiments with 500 μM Tb3+ and increasing concentrations of Mg2+, we observed a general decrease in the cleavage intensities at the mapped metal ion binding sites (Figure 3A and Supplementary Figure 1). In the absence of Mg2+, no specific Tb3+ cleavage is observed, as the riboswitch is apparently unfolded, not reaching a distinct three-dimensional structure. In the presence of Mg2+, i.e., when the riboswitch is folded correctly, nucleotides 82–84, 91–92, 97–100, and 106–109 exhibit a less pronounced competition with Tb3+ compared to the other sites, probably due to a smaller affinity toward Mg2+ ions (Figure 3A). These nucleotides belong to the peripheral structural elements (L9, J10/11, P11, J11/10) of the aptamer and in spite their accessibility as proposed by the comparison to the two solved crystal structures, this region shows a lower affinity to Mg2+ than the other, probably very structured part of the riboswitch. Interestingly, at all Mg2+ concentrations studied (20–100 mM), nucleotide A122 always exhibits the strongest cleavage by Tb3+ compared to the other mapped nucleotides. The evident strong cleavage as well as the distinctive decrease in cleavage intensity with increasing Mg2+ concentration suggests a strong Mg2+ binding site at or close to A122. The competition experiments thus confirm to a large extent the mapped metal ion binding sites as shown in Figures 1, 2.
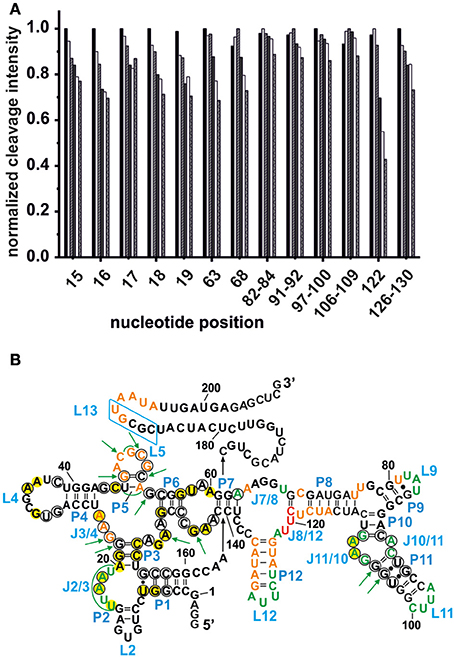
Figure 3. Competition between Mg2+ and Tb3+ for the identification of specific Mn+ binding sites. (A) The decrease in cleavage intensity at the mapped sites is shown as a function of Mg2+ while Tb3+ was held constant at 500 μM. The Mg2+ concentration increases from 20 to 90 mM (from left-to-right). (B) Comparison of the metal ion binding sites of the AdoCbl riboswitches from E. coli and S. thermophilum (Peselis and Serganov, 2012). Shown is the proposed secondary structure of the btuB riboswitch with circled nucleotides representing the conserved bases in the consensus sequence of AdoCbl riboswitches (Nahvi et al., 2004). Green nucleotides correspond to specific Mg2+ binding sites of the btuB riboswitch. The nucleotides marked by green arrows indicate the metal ion binding sites at conserved nucleotides as found in the S. thermophilum AdoCbl riboswitch while areas marked in green signal possible metal ion binding sites at non-conserved nucleotides. Bases marked in yellow could be involved in tertiary interactions as comparison with the S. thermophilum riboswitch suggests (Peselis and Serganov, 2012). The blue box indicates the L13 region supposedly undergoing the kissing loop interaction with L5 upon AdoCbl binding. This tertiary interaction is not visible in the crystallized riboswitch from S. thermophilum since it was truncated after P1. Orange and red nucleotides were shown to provide weak and strong Tb(III) binding sites respectively (see Figure 2).
In each class of riboswitches, the aptameric moiety is higher conserved than the expression platform ensuring a conserved three dimensional structure of the aptamer by tertiary contacts between conserved residues (Roth and Breaker, 2009). Finding a correlation between these conserved nucleotides and their affinity toward Mg2+ would provide a general scheme of important Mg2+ mediated tertiary interactions among different coenzyme B12 riboswitches. We therefore compared the metal ion binding behavior of the btuB riboswitch of E. coli to the crystallized AdoCbl riboswitch from S. thermophilum (Figure 3B) (Peselis and Serganov, 2012). Except for the two additional peripheral stems P9 and P12, the btuB riboswitch has a similar secondary structure not only as the S. thermophilum one, but also as the second crystallized AdoCbl riboswitch from T. tengcongensis. While the crystallized aptamer of the T. tengcongensis riboswitch includes stem P13 involved in the L5/L13 kissing-loop interaction, the crystallized construct of the S. thermophilum riboswitch was shortened on this site (Johnson et al., 2012; Peselis and Serganov, 2012). Nevertheless, it is highly likely that all three riboswitches share a respective tertiary structure of the aptamer region (Nahvi et al., 2004; Johnson et al., 2012; Peselis and Serganov, 2012).
The crystal structure of the AdoCbl riboswitch from S. thermophilum solved at 3.05 Å resolution (PDB ID: 4GXY; Peselis and Serganov, 2012) reveals two Mg2+ ions (Mg A310 and Mg A311) and seven iridium(III) hexammine complexes. The contact maps for the seven iridium(III) complexes indicate hydrogen bonds not only with O4 and O5 of pyrimidines as well as O6 and N7 of guanines but also with oxygen atoms from the phosphate backbone (Table 1). Since the contact maps for the two remaining Mg2+ ions are missing, we applied MINAS (Schnabl et al., 2011) to look for the nearest contacting nucleobases (Table 2). Presumably both Mg2+ ions interact with the predicted bases via outer sphere co-ordination since the distances of the contacts are more than 2.7 Å (Supplementary Figure 2 and Supplementary Table 1). One of the Mg2+ (A310) contacts a guanine quartet at L8 and P8 (Supplementary Figure 2A) whereas the other Mg2+ (A311) is placed in proximity to L5 (Supplementary Figure 2B). Interestingly, out of the 25 residues involved in metal ion binding in the crystal structure, 13 were found to be conserved in the consensus sequence of coenzyme B12 riboswitches (Nahvi et al., 2004). Furthermore, the binding of iridium(III) hexammine, to most non-conserved residues, namely to U34, U35, U38, G64, C72, U100, G101, G103, C104, involves the adjacent vicinity to the conserved bases at J2/3, J4/5, P5, and J10/11 (Peselis and Serganov, 2012).
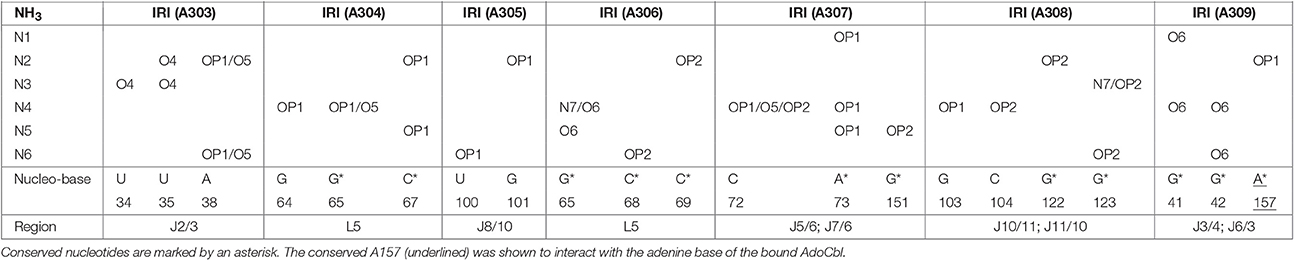
Table 1. Contact map for the ammine ligands (N1–N6) of the iridium(III) hexammine (IRI A303–A309) in the crystal structure of the AdoCbl riboswitch (S. thermophilum) (Peselis and Serganov, 2012).
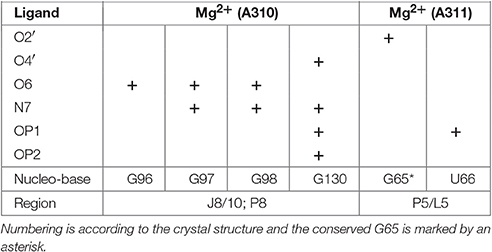
Table 2. Outer sphere coordination for Mg2+ ions in the crystal structure of the AdoCbl riboswitch (S. thermophilum; Peselis and Serganov, 2012) as calculated by MINAS (Schnabl et al., 2011).
Comparison of the specific Mg2+ binding sites of the ligand-free btuB riboswitch with the metal binding sites of the S. thermophilum riboswitch crystallized in its AdoCbl-bound form (Peselis and Serganov, 2012) show a good correlation at J2/3 and J11/10 (Figure 3B). Interestingly, these two regions involve conserved nucleotides potentially undergoing crucial tertiary contacts as found for the S. thermophilum riboswitch (Peselis and Serganov, 2012). It is therefore highly probable that the long range interaction of J11/10 to J5/6 and J6/3 as well as the P3-P1 stack are already formed in the ligand-free form of the riboswitch and that they are facilitated by Mg2+ binding. Other metal binding sites of the S. thermophilum riboswitch agree with some of the weaker metal binding sites found by Tb(III) cleavage, namely at J3/4 and P5/L5.
The btuB riboswitch is known to undergo a conformational change upon AdoCbl-binding mainly through the proposed pseudoknot formation between L5 and L13 disrupting thereby the anti-terminator stem (Nahvi et al., 2002; Gallo et al., 2008). We attempted to visualize this conformational change in terms of Mn+ binding properties. For this we compared its Mg2+ binding sites described above with the results of the ligand-bound aptamer. The btuB riboswitch was prefolded in 20 mM MgCl2 and then incubated with AdoCbl for 30 min before addition of 0.5 mM Tb3+. The incubation time of 30 min with AdoCbl was adequate for a structural switch of the RNA (Supplementary Figure 3), as indeed the conformational switch appears instantaneously upon addition of AdoCbl.
As already observed for the ligand-free RNA, cleavage of the AdoCbl-bound aptamer was again evident at Tb3+-concentrations higher than 0.25 mM (Figure 4). Interestingly, at Tb3+ concentrations between 0.25 and 1 mM, the strongest cleavage is again observed at nucleotide A122 (J8/12). However, in the presence of AdoCbl, the cleavage intensity of A122 achieves its maximum only at 1 mM Tb3+ and not at 0.25 mM as in the absence of the ligand (Figure 5A), showing a decreased affinity to Mn+ at this site upon ligand-binding. The overall cleavage pattern for the switched RNA (+AdoCbl) at higher Tb3+ concentrations (from 2.5 to 20 mM) remains the same as in the absence of AdoCbl except for the sites modulated by AdoCbl (Sites 1–8). Sites 1 (G23), 3 (G87), 4 (G106), and 8 (U183) undergo relative decrease in cleavage whereas sites 2 (U68), 5 (U110), and 7 (U167) exhibit increase in the cleavage intensity as expected (Figure 5B; Nahvi et al., 2002). However, site 2b (U77) displays an increase in cleavage intensity contrary to observation from the in-line probing experiments (Gallo et al., 2008). The AdoCbl dependent increase in the cleavage at site 2b is confirmed from the experiment where the btuB riboswitch was incubated with varying AdoCbl concentrations prior to cleavage by 0.5 mM Tb3+ (Supplementary Figure 4). This difference between in-line probing and Tb3+ mediated cleavage is probably due to the minor differences in binding between the two metal ions (Sigel and Pyle, 2003).
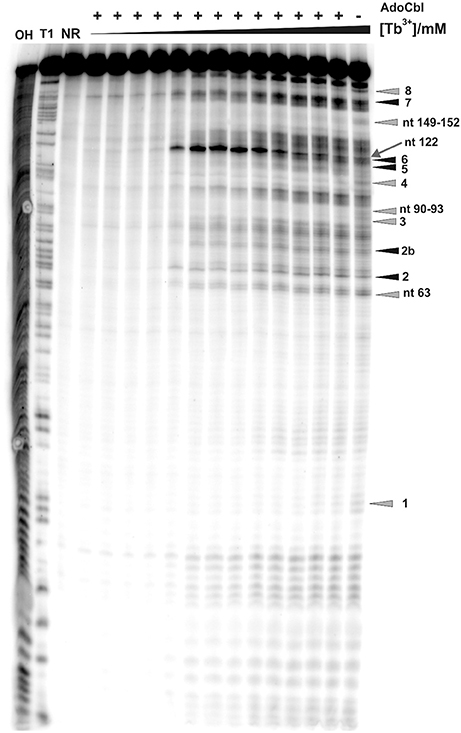
Figure 4. Tb3+ cleavage of the ligand bound btuB riboswitch. The RNA was prefolded in the presence of 100 μM AdoCbl and then incubated with increasing concentrations of TbCl3 (from left to right, 0.0, 0.025, 0.05, 0.1, 0.25, 0.5, 0.75, 1.0, 2.5, 5.0, 10, 15, 20 mM). The arrow shows nucleotide A122 displaying the strongest cleavage of all sites within the btuB riboswitch. Triangles in gray and black indicate the nucleotides with decreased and increased cleavage in the presence of AdoCbl (Supplementary Figure 4). The cleavage patterns of the two lanes on the right [20 mM Tb(III), ± AdoCbl] were used for the structural footprints shown in Figure 6. The numbered cleavage bands indicated on the right side correspond to the previously described in-line probing bands (Gallo et al., 2008). OH, Alkaline hydrolysis ladder; T1, RNase T1 ladder; NR, non-reacted RNA.
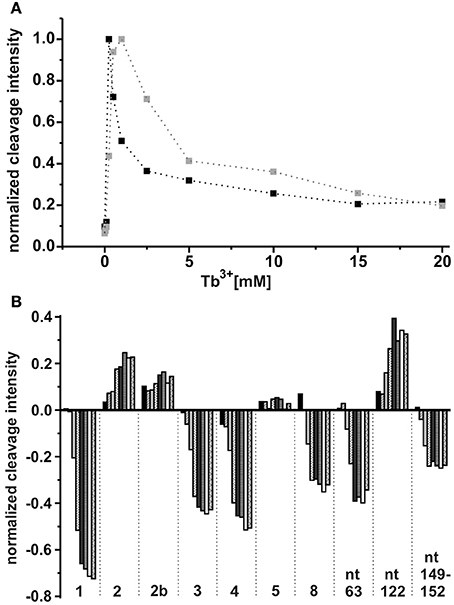
Figure 5. Tb3+ dependent changes in the ligand bound conformation of the btuB riboswitch. (A) TbCl3 concentration dependent changes at nucleotide A122 in the absence (black squares) and presence (gray squares) of 100 μM AdoCbl. The data points are connected by dotted lines for better visibility. (B) AdoCbl concentration dependent changes at the indicated nucleotides. The RNA was incubated with AdoCbl (0.05, 0.1, 0.5, 1, 5, 10, 50, 100 μM) prior to cleavage with 0.5 mM TbCl3.
There is a marked decrease in the cleavage intensity at nucleotides 63 (P7), 90–93 (P10–P11), and 149–152 (J6/3) along with the earlier reported sites modulated by AdoCbl (Figures 4, 5B). This could be due to AdoCbl induced tertiary interactions between P10–P11 and J6/3 in the btuB riboswitch similar to the one observed in the two reported crystal structures of AdoCbl riboswitches (Johnson et al., 2012; Peselis and Serganov, 2012).
Besides being a probe for Mg2+ binding sites, Tb3+ can also be used to investigate the native conformation of RNAs (Harris et al., 2004). At high millimolar concentration Tb3+ cleaves RNA additionally in a sequence-independent manner producing a foot printing pattern of single-stranded and non-Watson-Crick base paired elements (Sigel and Pyle, 2003; Harris et al., 2004). We compared the cleavage pattern of the btuB riboswitch at 20 mM Tb3+ in the absence and presence of AdoCbl (Figure 4, lanes 16 and 17) to get an insight into the arrangement of the riboswitch domains at native pH.
In the absence of AdoCbl (Figure 6A), the strongly cleaved regions belong to the proposed single stranded regions. These regions mainly involve the junctions (J2/3, J3/4, L5, J7/8, J8/12) and the loops (L9, L11, L12) along with the nucleotides 169–172 and 192–196 surrounding stem P13. The other proposed single stranded regions of the riboswitch, L4, J6/7, J7/8, and nucleotides 177–184 (surrounding stem P13) display relative protection from cleavage. For L4 and J6/7 this indicates their tertiary contact, fortifying the structural similarity of the btuB riboswitch to the other two crystallized AdoCbl riboswitches. The protection of the other two regions could be due to surface occlusion within the folded RNA, a finding however not supported by the two crystal structures (Johnson et al., 2012; Peselis and Serganov, 2012).
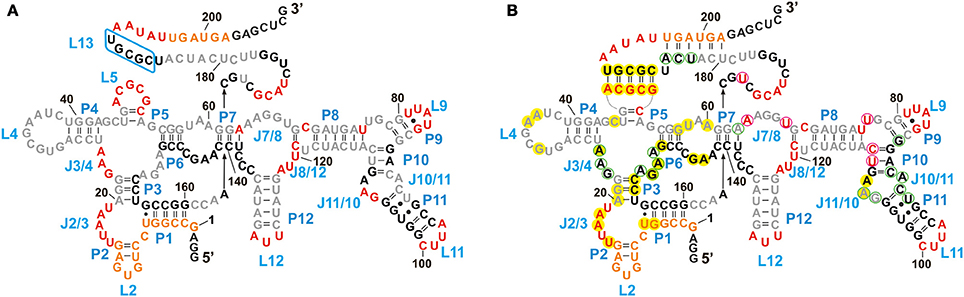
Figure 6. Structural footprint of the btuB aptamer obtained by Tb3+ cleavage in the absence (A) and presence (B) of AdoCbl. Nucleotides undergoing strong cleavage in the presence of 20 mM TbCl3 are shown in red, the ones with intermediate cleavage are marked in orange. Partially protected nucleotides are shown in gray and completely protected nucleotides are in black. Nucleotides circled in magenta and light green indicate an increased or decreased cleavage intensity, respectively, in the presence of 100 μM AdoCbl and 20 mM Tb(III). Bases marked in yellow could be involved in tertiary interactions as comparison with the S. thermophilum riboswitch suggests (Peselis and Serganov, 2012).
We observed a marked protection from cleavage at the conserved B12 box (140–160), P9–P11, J10/11, and J10/11 as well as at the regions encompassing nucleotides 164–168, 173–176, and 186–190. Out of these regions, only a part of the B12 box (140–141, 146–148, 153–160) and the peripheral face of the aptamer (at P8–P11) include proposed helical regions leaving the secluded position within the inner core the only explanation for the protection of the other parts. Apart from P9, which is missing in the two AdoCbl riboswitches from thermophiles, and of J3/13 (161–179), which is missing or not defined in the crystal structures, all of these protected areas match very well with the inner core as seen for the two crystallized AdoCbl riboswitches. Clear protection from cleavage is further evident for the regions encompassing nucleotides 20–22 and 26–62 that presumably constitute a part of the receptor core, again as seen for the crystallized AdoCbl riboswitches (Johnson et al., 2012; Peselis and Serganov, 2012).
Upon addition of AdoCbl, the overall global conformation of the aptamer appears similar to the one in the absence of the ligand except for the sites reported to be modulated by AdoCbl (Figure 6B).
Depending on the riboswitch class, the correct pre-organization of the aptameric part is a crucial step for ligand recognition and binding (Montange and Batey, 2008; Baird and Ferré-D'Amaré, 2010). Our earlier studies suggest that this fact is also true for the btuB riboswitch from E. coli to allow an interaction with its ligand AdoCbl (Choudhary and Sigel, 2014). The formation of this so-called binding-competent fold is mediated by Mg2+ binding to specific sites and thus facilitating tertiary interactions within the RNAs tertiary fold. The detection of these binding sites combined with the comparison with a riboswitch of similar structure, in our case the AdoCbl-sensing riboswitch of S. thermophilum (Peselis and Serganov, 2012), allowed for a quite adequate prediction of the btuB riboswitch tertiary structure. Furthermore, we compared the Mg2+ binding sites within the ligand-free and ligand-bound aptamer to reveal structural differences and to demonstrate if indeed there is a global rearrangement upon AdoCbl binding or only locally restricted structural changes.
Mapping the Mn+ binding sites of the btuB riboswitch we observed metal ion binding at/around a set of conserved residues (J2/3, J3/4, P5, L5, P7, J10/11, P11, and J11/10). Interestingly, these regions are very likely to undergo tertiary interactions crucial for the aptamer active structure, as was revealed by structural comparison with the two crystallized AdoCbl riboswitches from thermophiles (Johnson et al., 2012; Peselis and Serganov, 2012). Furthermore, the Mn+ binding sites of the btuB riboswitch show a partial match with the binding sites of the S. thermophilum riboswitch (Peselis and Serganov, 2012). We therefore propose a common set of Mg2+ mediated tertiary contacts for the aptameric part of the AdoCbl-binding subclass of the B12-riboswitches.
Both structures of the AdoCbl riboswitches crystallized in their ligand-bound form revealed complex tertiary structural elements such as coaxial stems (P3–P6 and P4–P6), a kissing loop (L5 and L13), a T loop-PK domain interaction between L4 and J6-7/J7-6 as well as a long range tertiary interaction between J10/11 and J6/3 (Johnson et al., 2012; Peselis and Serganov, 2012). In addition to these common features, the structure from S. thermophilum included a minor groove interaction of J2/3 with P1 and a zipper formation between J3/4, J6/3 and P3 (16). The formation of tertiary motifs like coaxial stems, kissing loops, and pseudoknots is largely dependent on Mg2+ for stabilization (Batey et al., 1999). The evident correlation in Mn+ binding sites of the btuB riboswitch to the two structurally known AdoCbl riboswitches leads to the conclusion that these three riboswitches share a corresponding tertiary fold within their aptameric part.
The observed cleavage pattern at stem P12, missing in the two crystallized riboswitches, indicates a partial dynamic geometry in this region. The biochemical data for AdoCbl riboswitches with and without stem P12 does not correlate to the affinity for AdoCbl (Nahvi et al., 2004). Therefore, we suggest that stem P12 is involved in stabilizing the overall binding pocket but may not necessarily contribute toward the specificity to AdoCbl. Located at the peripheral region of the btuB riboswitch is nucleotide A122 that appears to have the most suited geometry for Tb3+ binding. The strong cleavage at A122 is evident at low micromolar Tb3+ concentrations in both the ligand-free and the ligand-bound form. A122 is located at the J8/12 region of the btuB aptamer and since both of the crystallized AdoCbl riboswitches (Johnson et al., 2012; Peselis and Serganov, 2012) lack this peripheral part, it is difficult at present to predict the role of A122 in the overall structure of the btuB riboswitch of E. coli.
Our results show similar Tb3+ coordination in the ligand-free and ligand-bound forms of the btuB riboswitch of E. coli. The mapping experiments revealed that the crucial tertiary interactions involving the T loop pseudoknot, the long range interaction of J11/10–J5/6 and J6/3 and the KL-interaction supposed to form upon AdoCbl-binding are already formed in the absence of AdoCbl. Therefore, the binding pocket of the aptamer seems to be highly pre-organized by Mn+ already in the ligand-free form to allow optimal binding to AdoCbl. This is in strong agreement with our earlier studies (Choudhary and Sigel, 2014) indicating that the btuB riboswitch cannot be switched by AdoCbl in the absence of Mg2+.
This pre-assembly of the ligand binding pocket solely by Mg2+ is not peculiar for the btuB riboswitch but has been observed for the adenine riboswitch, the glmS ribozyme and for the lysine binding riboswitches (Garst et al., 2008; Baird and Ferré-D'Amaré, 2010; Fiegland et al., 2012). Mg2+ mediated pre-compaction of the RNA in the absence of the ligand has been further reported by SAXS studies for the glycine, the c-di-GMP, the TPP, the lysine, and the SAM I riboswitches (Baird and Ferré-D'Amaré, 2010). Therefore, Mg2+ mediated pre-organization of RNA to achieve an active tertiary structure of the ligand-free aptamer seems to be a common phenomenon in riboswitches and probably holds true for the AdoCbl riboswitches as well.
While the btuB riboswitch appears not to undergo a drastic structural change upon AdoCbl binding, two distinct local changes take place: First, similar intensity changes upon AdoCbl addition are observed in Tb3+ cleavage experiments as reported by in-line probing in earlier studies (Nahvi et al., 2002; Gallo et al., 2008). Second, the observed Mn+ binding to the two conserved adenosine residues of J11/10 upon AdoCbl binding which might aid its interaction with J6/3.
To summarize, we propose the presence of specific Mn+ binding sites and the pre-organization of the btuB aptamer in its ligand-free form encompassing Mn+ mediated tertiary interactions within the aptamer. These tertiary interactions could be common for all members of the AdoCbl-binding B12 riboswitches in order to facilitate the interaction with their complex ligand, AdoCbl. A pre-organized binding pocket benefits the riboswitch not only with a rapid recognition of the ligand but also with the prompt transmission of the regulatory signal (Baird and Ferré-D'Amaré, 2010).
PC performed all experimental work. PC and SG did the data analysis. RS designed the research. All authors wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Joachim Schnabl for providing the data related to the MINAS database. Financial support by the Swiss National Science Foundation, the University of Zurich, the European research Council (ERC Starting Grant to RS), and within the COST Action CM1105 is gratefully acknowledged.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fchem.2017.00010/full#supplementary-material
Baird, N. J., and Ferré-D'Amaré, A. R. (2010). Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16, 598–609. doi: 10.1261/rna.1852310
Bastet, L., Dubé, A., Massé, E., and Lafontaine, D. A. (2011). New insights into riboswitch regulation mechanisms. Mol. Microbiol. 80, 1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x
Batey, R. T., Rambo, R. P., and Doudna, J. A. (1999). Tertiary motifs in RNA structure and folding. Angew. Chem. 38, 2326–2343. doi: 10.1002/(SICI)1521-3773(19990816)38:16<2326::AID-ANIE2326>3.0.CO;2-3
Choudhary, P. K., Gallo, S., and Sigel, R. K. (2014). Monitoring global structural changes and specific metal binding sites in RNA by in-line probing and Tb(III) cleavage. Methods Mol. Biol. 1086, 143–158. doi: 10.1007/978-1-62703-667-2_8
Choudhary, P. K., and Sigel, R. K. O. (2014). Mg2+ induced conformational changes in the btuB riboswitch from E. coli. RNA 20, 36–45. doi: 10.1261/rna.039909.113
Ciesiolka, J., Marciniec, T., and Krzyzosiak, W. (1989). Probing the environment of lanthanide binding sites in yeast tRNA(Phe) by specific metal-ion-promoted cleavages. Eur. J. Biochem. 182, 445–450. doi: 10.1111/j.1432-1033.1989.tb14851.x
Dorner, S., and Barta, A. (1999). Probing ribosome structure by europium-induced RNA cleavage. Biol. Chem. 380, 243–251. doi: 10.1515/BC.1999.032
Erat, M. C., and Sigel, R. K. (2011). Methods to detect and characterize metal ion binding sites in RNA. Met. Ions Life Sci. 9, 37–100. doi: 10.1039/9781849732512-00037
Fiegland, L. R., Garst, A. D., Batey, R. T., and Nesbitt, D. J. (2012). Single-molecule studies of the lysine riboswitch reveal effector-dependent conformational dynamics of the aptamer domain. Biochemistry 51, 9223–9233. doi: 10.1021/bi3007753
Gallo, S. (2009). Investigations on the Interaction between Coenzyme B12 Derivatives and the btuB Riboswitch of E. coli. Ph.D Thesis, University of Zürich.
Gallo, S., Furler, M., and Sigel, R. K. O. (2005). In vitro transcription and purification of RNAs of different size. Chimia 59, 812–816. doi: 10.2533/000942905777675589
Gallo, S., Oberhuber, M., Sigel, R. K. O., and Kräutler, B. (2008). The corrin moiety of coenzyme B12 is the determinant for switching the btuB riboswitch of E. coli. Chembiochem 9, 1408–1414. doi: 10.1002/cbic.200800099
Garst, A. D., Héroux, A., Rambo, R. P., and Batey, R. T. (2008). Crystal structure of the lysine riboswitch regulatory mRNA element. J. Biol. Chem. 283, 22347–22351. doi: 10.1074/jbc.C800120200
Gudmundsdottir, A., Bradbeer, C., and Kadner, R. J. (1988). Altered binding and transport of vitamin B12 resulting from insertion mutations in the Escherichia coli btuB gene. J. Biol. Chem. 263, 14224–14230.
Hampel, K. J., and Tinsley, M. M. (2006). Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry 45, 7861–7871. doi: 10.1021/bi060337z
Harris, D. A., Tinsley, R. A., and Walter, N. G. (2004). Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 341, 389–403. doi: 10.1016/j.jmb.2004.05.074
Harris, D. A., and Walter, N. G. (2003). Probing RNA structure and metal-binding sites using terbium(III) footprinting. Curr. Protoc. Nucleic Acid Chem. Chapter 6:Unit 6.8. doi: 10.1002/0471142700.nc0608s13
Heppell, B., Blouin, S., Dussault, A. M., Mulhbacher, J., Ennifar, E., Penedo, J. C., et al. (2011). Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 7, 384–392. doi: 10.1038/nchembio.563
Johnson, J. E. Jr., Reyes, F. E., Polaski, J. T., and Batey, R. T. (2012). B(12) cofactors directly stabilize an mRNA regulatory switch. Nature 492, 133–137. doi: 10.1038/nature11607
Kaye, N. M., Zahler, N. H., Christian, E. L., and Harris, M. E. (2002). Conservation of helical structure contributes to functional metal ion interactions in the catalytic domain of ribonuclease P RNA. J. Mol. Biol. 324, 429–442. doi: 10.1016/S0022-2836(02)01094-X
Klein, D. J., and Ferré-D'Amaré, A. R. (2006). Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313, 1752–1756. doi: 10.1126/science.1129666
Kubodera, T., Watanabe, M., Yoshiuchi, K., Yamashita, N., Nishimura, A., Nakai, S., et al. (2003). Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5'-UTR. FEBS Lett. 555, 516–520. doi: 10.1016/S0014-5793(03)01335-8
Loh, E., Dussurget, O., Gripenland, J., Vaitkevicius, K., Tiensuu, T., Mandin, P., et al. (2009). A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139, 770–779. doi: 10.1016/j.cell.2009.08.046
Mandal, M., and Breaker, R. R. (2004). Gene regulation by riboswitches. Nat. Rev. Mol. Cell Bio. 5, 451–463. doi: 10.1038/nrm1403
Mironov, A. S., Gusarov, I., Rafikov, R., Lopez, L. E., Shatalin, K., Kreneva, R. A., et al. (2002). Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756. doi: 10.1016/S0092-8674(02)01134-0
Misra, V. K., and Draper, D. E. (1998). On the role of magnesium ions in RNA stability. Biopolymers 48, 113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y
Montange, R. K., and Batey, R. T. (2008). Riboswitches: emerging themes in RNA structure and function. Ann. Rev. Biophys. 37, 117–133. doi: 10.1146/annurev.biophys.37.032807.130000
Nahvi, A., Barrick, J. E., and Breaker, R. R. (2004). Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32, 143–150. doi: 10.1093/nar/gkh167
Nahvi, A., Sudarsan, N., Ebert, M. S., Zou, X., Brown, K. L., and Breaker, R. R. (2002). Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043–1049. doi: 10.1016/S1074-5521(02)00224-7
Noeske, J., Schwalbe, H., and Wöhnert, J. (2007). Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 35, 5262–5273. doi: 10.1093/nar/gkm565
Nou, X., and Kadner, R. J. (2000). Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc. Natl. Acad. Sci. U.S.A. 97, 7190–7195. doi: 10.1073/pnas.130013897
Peselis, A., and Serganov, A. (2012). Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat. Struct. Mol. Biol. 19, 1182–1184. doi: 10.1038/nsmb.2405
Pyle, A. M. (2002). Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7, 679–690. doi: 10.1007/s00775-002-0387-6
Regulski, E. E., and Breaker, R. R. (2008). In-line probing analysis of riboswitches. Methods Mol. Biol. 419, 53–67. doi: 10.1007/978-1-59745-033-1_4
Reynolds, P. R., Mottur, G. P., and Bradbeer, C. (1980). Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of BtuC and TonB. J. Biol. Chem. 255, 4313–4319.
Roth, A., and Breaker, R. R. (2009). The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334. doi: 10.1146/annurev.biochem.78.070507.135656
Saito, H., and Suga, H. (2002). Outersphere and innersphere coordinated metal ions in an aminoacyl-tRNA synthetase ribozyme. Nucleic Acids Res. 30, 5151–5159. doi: 10.1093/nar/gkf641
Schnabl, J., Suter, P., and Sigel, R. K. O. (2011). MINAS–a database of metal ions in nucleic acids. Nucleic Acids Res. 40, D434–D438. doi: 10.1093/nar/gkr920
Sigel, R. K., and Pyle, A. M. (2003). Lanthanide ions as probes for metal ions in the structure and catalytic mechanism of ribozymes. Met. Ions Biol. Syst. 40, 477–512.
Sigel, R. K., Vaidya, A., and Pyle, A. M. (2000). Metal ion binding sites in a group II intron core. Nat. Struct. Biol. 7, 1111–1116. doi: 10.1038/81958
Soukup, J. K., and Soukup, G. A. (2004). Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struc. Biol. 14, 344–349. doi: 10.1016/j.sbi.2004.04.007
Suddala, K. C., Rinaldi, A. J., Feng, J., Mustoe, A. M., Eichhorn, C. D., Liberman, J., et al. (2013). Single transcriptional and translational preQ1 riboswitches adopt similar pre-folded ensembles that follow distinct folding pathways into the same ligand-bound structure. Nucleic Acids Res. 41, 10462–10475. doi: 10.1093/nar/gkt798
Waldsich, C., and Pyle, A. M. (2008). A kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol. 375, 572–580. doi: 10.1016/j.jmb.2007.10.052
Walter, N. G., Yang, N., and Burke, J. M. (2000). Probing non-selective cation binding in the hairpin ribozyme with Tb(III). J. Mol. Biol. 298, 539–555. doi: 10.1006/jmbi.2000.3691
Winkler, W. C., and Breaker, R. R. (2003). Genetic control by metabolite-binding riboswitches. Chembiochem 4, 1024–1032. doi: 10.1002/cbic.200300685
Winkler, W. C., Nahvi, A., Roth, A., Collins, J. A., and Breaker, R. R. (2004). Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286. doi: 10.1038/nature02362
Winkler, W., Nahvi, A., and Breaker, R. R. (2002). Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956. doi: 10.1038/nature01145
Keywords: riboswitch, Tb3+, metal ion binding, tertiary interactions, coenzyme B12
Citation: Choudhary PK, Gallo S and Sigel RKO (2017) Tb3+-Cleavage Assays Reveal Specific Mg2+ Binding Sites Necessary to Pre-fold the btuB Riboswitch for AdoCbl Binding. Front. Chem. 5:10. doi: 10.3389/fchem.2017.00010
Received: 31 October 2016; Accepted: 14 February 2017;
Published: 21 March 2017.
Edited by:
J. Carlos Penedo, University of St Andrews, UKReviewed by:
Minkui Luo, Memorial Sloan Kettering Cancer Center, USACopyright © 2017 Choudhary, Gallo and Sigel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roland K. O. Sigel, cm9sYW5kLnNpZ2VsQGNoZW0udXpoLmNo
†Present Address: Pallavi K. Choudhary, Center for Molecular Biology of RNA, University of California at Santa Cruz, Santa Cruz, CA, USA
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.