
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. , 19 January 2017
Sec. Chemical Biology
Volume 5 - 2017 | https://doi.org/10.3389/fchem.2017.00001
This article is part of the Research Topic Antimicrobial and Anticancer Peptides View all 15 articles
 Wenyi Li1,2
Wenyi Li1,2 Zhe Sun3,4
Zhe Sun3,4 Neil M. O'Brien-Simpson3,4
Neil M. O'Brien-Simpson3,4 Laszlo Otvos5
Laszlo Otvos5 Eric C. Reynolds3,4
Eric C. Reynolds3,4 Mohammed A. Hossain1,2*
Mohammed A. Hossain1,2* Frances Separovic2,4*
Frances Separovic2,4* John D. Wade1,2*
John D. Wade1,2*In vivo pharmacokinetics studies have shown that the proline-rich antimicrobial peptide, A3-APO, which is a discontinuous dimer of the peptide, Chex1-Arg20, undergoes degradation to small fragments at positions Pro6-Arg7 and Val19-Arg20. With the aim of minimizing or abolishing this degradation, a series of Chex1-Arg20 analogs were prepared via Fmoc/tBu solid phase peptide synthesis with D-arginine or, in some cases, peptide backbone Nα-methylated arginine, substitution at these sites. All the peptides were tested for antibacterial activity against the Gram-negative bacterium Klebsiella pneumoniae. The resulting activity of position-7 substitution of Chex1-Arg20 analogs showed that arginine-7 is a crucial residue for maintaining activity against K. pneumoniae. However, arginine-20 substitution had a much less deleterious effect on the antibacterial activity of the peptide. Moreover, none of these peptides displayed any cytotoxicity to HEK and H-4-II-E mammalian cells. These results will aid the development of more effective and stable PrAMPs via judicious amino acid substitutions.
The increasing widespread onset of bacterial multi-drug resistance, associated with major clinical pathogenic infections, has resulted in calls for the development new antimicrobial agents (Laxminarayan et al., 2013). Due to their broad-spectrum activities and multi-modal actions against pathogens, antimicrobial peptides (AMPs) (also known host-defense peptides), are considered as attractive potential candidates for new antibiotics (Hilchie et al., 2013; Lam et al., 2016). Importantly, these peptides have also attracted considerable attention as alternative means of plant disease control to conventional treatments that are polluting and hazardous to both human health and the environment (Datta et al., 2015, 2016). Among these peptides, the class of proline-rich AMPs (PrAMPs) possess a unique multi-modal mechanism of action against pathogens and display potent activity against Gram-negative bacteria (Otvos et al., 2005; Czihal et al., 2012; Guida et al., 2015). These actions include membrane rupture (Li et al., 2014), inhibition of the bacterial shock heat protein DnaK (Kragol et al., 2001; Scocchi et al., 2009), blockade of bacterial ribosomal protein expression (Krizsan et al., 2014; Roy et al., 2015; Seefeldt et al., 2015, 2016; Goldbach et al., 2016), and immunostimulatory activity (Ostorhazi et al., 2011). Recently, a PrAMP and other AMPs were impregnated into nanofibers or hydrogels for the potential treatment of skin injuries in general and battlefield burns (Mateescu et al., 2015; Sebe et al., 2016).
The peptide, Chex1-Arg20, was de novo designed based on native PrAMPs with additional sequence optimization to enhance bacterial membrane penetration (Otvos et al., 2005; Noto et al., 2008; Rozgonyi et al., 2009). It has been shown that multimerization of Chex-Arg20 to a discontinuous dimer or tetramer results in an alteration of its mechanism of interaction with the Escherichia coli membrane (Li et al., 2015a). These observations were further confirmed on investigation of Chex1-Arg20 and its multimers with model membranes (Li et al., 2016). Additionally, specific C-terminal chemical modifications of the Chex1-Arg20 monomer were shown to expand both its activity and spectrum of Gram-negative bacterial action (Li et al., 2015b). These observations led to the development of a series of tetrameric Chex1-Arg20 bearing a C-terminal hydrazide that were shown to possess a more compact structure and potent and broadened activity against Gram-negative nosocomial pathogens (Li et al., 2017).
The discontinuous dimer of Chex1-Arg20, A3-APO, was shown in in vivo pharmacokinetic studies to undergo degradation at positions Pro6-Arg7 and Val19-Arg20, as well as to produce the major metabolite, Chex1-Arg20 (Noto et al., 2008). A key goal is to undertake chemical modifications at these labile sites to confer significant improvement in peptide stability in serum without undue effect on their activity (Otvos and Wade, 2014). D-amino acid substitution in AMPs has previously been shown to be a successful strategy (Hong et al., 1999). This suggests that partial D-amino acid substitutions within Chex1-Arg20 might be a useful means to improve its activity and stability. Furthermore, backbone N-methylation of peptide bonds can also confer high stability against proteases and improved pharmacological bioavailability (Di Gioia et al., 2016). Therefore, we undertook to incorporate the unnatural D-amino acid and Nα-methyl-amino acid into two key points within the peptide sequence to determine the effect on activity against Gram-negative bacterium K. pneumoniae.
Nine-Fluorenylmethoxylcarbonyl (Fmoc)-L-amino acids, 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylamonium hexafluorophosphate (HCTU), and 1-[Bis(dimethylamino) methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid (HATU) were from GL Biochem (Shanghai, China). TentaGel-MB-RAM-resin was from Rapp Polymere (Tubingen, Germany). Nα-Fmoc-Nα-methyl-L-arginine(Nω-Pbf), and Nα-Fmoc-D-arginine(D-Pbf) were purchased from Novabiochem (Sydney, Australia). N,N-Diisopropylethylamine (DIPEA), dimethylformamide (DMF), and trifluoroacetic acid (TFA) were obtained from Auspep (Melbourne, Australia). Piperidine, triisopropylsilane (TIPS), anisole, and acetonitrile (CH3CN) were all obtained from Sigma (Sydney, Australia).
The peptides were synthesized by Fmoc/tBu solid-phase methods (Fields and Noble, 1990) using a CEM Liberty microwave-assisted synthesizer and TentaGel-MB-RAM-resin as previously described (Li et al., 2015a). Standard Fmoc-chemistry was used throughout with a 4-fold molar excess of the Fmoc-protected amino acids in the presence of 4-fold HCTU and 8-fold DIPEA. For the arginine derivative substitution, 1.5-fold of amino acid coupling was used together with 1.5 equivalents HATU and 3 equivalents of DIPEA. After synthesis, the peptides were cleaved from the solid support resin with TFA in the presence of anisole and TIPS as scavengers (95:3:2, v/v) for 2 h at room temperature. After filtration to remove the resin, the filtrate was concentrated under a stream of nitrogen and the peptide products were precipitated in ice-cold diethyl ether and washed three times. The peptides were then purified by reversed-phase high performance liquid chromatography (RP-HPLC) in water and acetonitrile containing 0.1% TFA using a gradient of 10–40% (acetonitrile) in 40 min. Due to the variation in hydrophobicity between the different analogs, the final products were characterized by RP-HPLC using a gradient of either 0–40% (acetonitrile) in 40 min or 10–40% (acetonitrile) in 30 min. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was also used for characterization.
An antibacterial assay was undertaken to determine the minimal inhibitory concentration (MIC) as described previously (Li et al., 2015b). The Gram-negative nosocomial bacterium, K. pneumoniae ATCC13883, was selected for testing the antibacterial activities of the Chex1-Arg20 analogs using 2.5 × 105 cells/ml in Mueller Hinton broth (MHB) at 37°C immediately prior to the determination of MIC.
The proliferation of HEK-293 (ATCC® CRL-1573™) and H-4-II-E (ATCC® CRL-1548™) cells were tested with the Chex1-Arg20 analogs using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) as described previously (Li et al., 2015b).
Peptide 1 was prepared as described in a previous report (Li et al., 2015b) and 2–8 were prepared on TentaGel-MB-RAM-resin via standard Fmoc/tBu solid-phase methods. Unnatural amino acid incorporation was achieved in presence of HATU instead of HCTU (Table 1) which produced better quality products. Each Chex1-Arg20 analog was obtained in an overall yield of ca. ~15% relative to the crude cleaved starting material. Each analog was then subjected to comprehensive chemical characterization including analytical RP-HPLC and MALDI-TOF MS to confirm their purity (Figure 1).
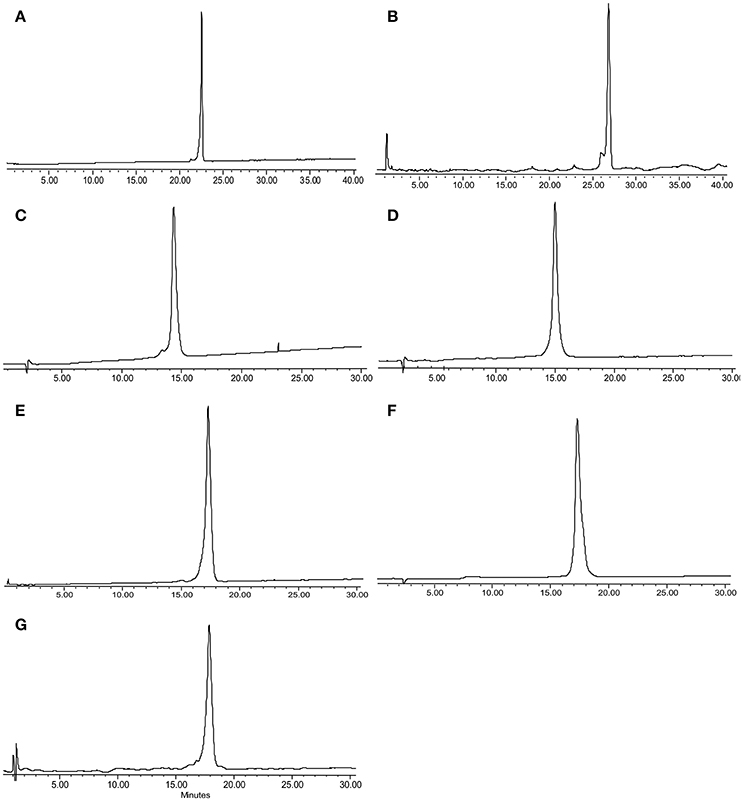
Figure 1. RP-HPLC and MALDI-TOF/ESI MS for peptide analogs 2–8, respectively: (A) 2, DR7; (B) 3, DR7(1–19); (C) 4, DR7(7–19); (D) 5, Chex1-Val19; (E) 6, DR20; (F) 7, mR20; (G) 8, reverse. Analysis condition: Phenomenex C18 column (WIDEPORE 3.6 μ XB-C18, 150 × 4.6 nm); buffer A, 0.1% aq. TFA; buffer B, 0.1% TFA in acetonitrile; gradient, buffer B 0–40% in 40 min for (A) 2 and (B) 3, and 10–40% in 30 min for (C) 4–(G) 8.
Each Chex1-Arg20 analog was assayed against the nosocomial Gram-negative bacterium K. pneumoniae ATCC 13883. The results are shown in Table 2 in comparison with analog 1, Chex1-Arg20. Replacement of arginine at position 7 with the D-form (analog 2) resulted in substantial loss of activity. This highlighted the importance of arginine-7 and its native L-configuration for characteristic antimicrobial activity. Curiously, truncation of the C-terminal Arg20 from analog 2 to produce analog 5 partially restored activity. Compared with analog 5, the N-terminal shortened analogs 2–4 containing a D-arginine substitution at position 7 showed a drastic loss of activity against this pathogen in MHB. In contrast, replacement of position Arg20 with either the D-arginine or Nα-methylated-arginine (analogs 6–7) led to a maintenance of significant activity of the native Chex1-Arg20 which indicates that this residue is more tolerant to modification to improve its in vivo stability to degradation. Finally, the reverse sequence (analog 8) was also evaluated and, as expected, it showed no activity against K. pneumoniae which confirmed the necessity of the native sequence for antibacterial action.

Table 2. Antibacterial activity, MIC (μM), of Chex1-Arg20 analogs against Gram-negative pathogen K. pneumoniae ATCC 13883.
In vitro cytotoxicity was also measured via the Promega CellTiter 96 AqueousNon-Radioactive Cell Proliferation Assay (Li et al., 2015a) using the mammalian cell lines HEK-293 (ATCC CRL 1573) and H-4-II-E (ATCC CRL-1548). None of the Chex1-Arg20 analogs showed any toxicity against either mammalian cell line at the highest tested concentration (100 μM) (Table 3).

Table 3. Cytoxocity (μM) of Chex1-Arg20 analogs against mammalian cell lines, H-4-II-E (ATCC® CRL-1573™) and H-4-II-E (ATCC® CRL-1548™), in which >100 μM or >50 indicated there was no cytotoxicity at the highest tested concentration 100 μM or 50 μM.
In summary, a series of D-amino acid substituted analogs of the PrAMP, Chex1-Arg20, were prepared by standard Fmoc/tBu solid phase peptide synthesis. These analogs were tested against the Gram-negative bacterium K. pneumoniae for antibacterial activity. In this study, the activity of D-arginine Chex1-Arg20 showed the replacement of arginine at position seven led to drastic loss of activity. The short fragments, Arg2-Val19 and Arg7-Val19, also displayed no antibacterial activity. However, substitution at position 20 with either D-arginine or Nα-methyl-arginine did not greatly affect the activity against K. pneumoniae. Moreover, none of these peptides showed any cytotoxicity to HEK and H-4-II-E mammalian cells. Such findings will assist the development of more effective and stable Chex1-Arg20 and A3-APO analogs with further substitution at position 20.
WL performed chemical syntheses, antibacterial assay and drafted the manuscript; ZS performed cytotoxicity test; NO, LO, ER, MH, FS, and JW took part in experimental design. All authors worked on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge support of the studies undertaken in the authors' laboratory by ARC Discovery Project grants (DP150103522) to JW and MH, and NHMRC Project grants (APP1029878) to NMOBS and (APP1008106) to ER and NMOBS. JW is an NHMRC (Australia) Principal Research Fellow. WL is the recipient of an MIRS PhD award and Dr Albert Shimmins Postgraduate Writing-Up award (University of Melbourne). Research at the FINMH was also supported by the Victorian Government's Operational Infrastructure Support Program.
Czihal, P., Knappe, D., Fritsche, S., Zahn, M., Berthold, N., Piantavigna, S., et al. (2012). Api88 is a novel antibacterial designer peptide to treat systemic infections with multidrug-resistant gram-negative pathogens. ACS Chem. Biol. 7, 1281–1291. doi: 10.1021/cb300063v
Datta, A., Bhattacharyya, D., Singh, S., Ghosh, A., Schmidtchen, A., Malmsten, M., et al. (2016). Role of aromatic amino acids in lipopolysaccharide and membrane interactions of antimicrobial peptides for use in plant disease control. J. Biol. Chem. 291, 13301–13317. doi: 10.1074/jbc.M116.719575
Datta, A., Ghosh, A., Airoldi, C., Sperandeo, P., Mroue, K. H., Jiménez-Barbero, J., et al. (2015). Antimicrobial peptides: insights into membrane permeabilization, lipopolysaccharide fragmentation and application in plant disease control. Sci. Rep. 5:11951. doi: 10.1038/srep11951
Di Gioia, M. L., Leggio, A., Malagrinò, F., Romio, E., Siciliano, C., and Liguori, A. (2016). N-Methylated alpha-amino acids and peptides: synthesis and biological activity. Mini Rev. Med. Chem. 16, 683–690. doi: 10.2174/1389557516666160322152457
Fields, G. B., and Noble, R. L. (1990). Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 35, 161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x
Goldbach, T., Knappe, D., Reinsdorf, C., Berg, T., and Hoffmann, R. (2016). Ribosomal binding and antibacterial activity of ethylene glycol-bridged apidaecin Api137 and oncocin Onc112 conjugates. J. Pept. Sci. 22, 592–599. doi: 10.1002/psc.2905
Guida, F., Benincasa, M., Zahariev, S., Scocchi, M., Berti, F., Gennaro, R., et al. (2015). Effect of size and N-terminal residue characteristics on bacterial cell penetration and antibacterial activity of the proline-rich peptide Bac7. J. Med. Chem. 58, 1195–1204. doi: 10.1021/jm501367p
Hilchie, A. L., Wuerth, K., and Hancock, R. E. (2013). Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 9, 761–768. doi: 10.1038/nchembio.1393
Hong, S. Y., Oh, J. E., and Lee, K.-H. (1999). Effect of D-amino acid substitution on the stability, the secondary structure, and the activity of membrane-active peptide. Biochem. Pharmacol. 58, 1775–1780. doi: 10.1016/S0006-2952(99)00259-2
Kragol, G., Lovas, S., Varadi, G., Condie, B. A., Hoffmann, R., and Otvos, L. Jr. (2001). The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of dnak and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026. doi: 10.1021/bi002656a
Krizsan, A., Volke, D., Weinert, S., Sträter, N., Knappe, D., and Hoffmann, R. (2014). Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew. Chem. Int. Ed. Engl. 53, 12236–12239. doi: 10.1002/anie.201407145
Lam, S. J., O'Brien-Simpson, N. M., Pantarat, N., Sulistio, A., Wong, E. H. H., Chen, Y.-Y., et al. (2016). Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 1:16162. doi: 10.1038/nmicrobiol.2016.162
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K. M., Wertheim, H. F. L., Sumpradit, N., et al. (2013). Antibiotic resistance - the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/S1473-3099(13)70318-9
Li, W., O'Brien-Simpson, N. M., Yao, S., Tailhades, J., Reynolds, E. C., Dawson, R. M., et al. (2017). C-Terminal modification and multimerization increase the efficacy of a proline-rich antimicrobial peptide. Chem. Eur. J. doi: 10.1002/chem.201604172
Li, W., O'Brien-Simpson, N., Tailhades, J., Pantarat, N., Dawson, R., Otvos, L. Jr., et al. (2015a). Multimerization of a proline-rich antimicrobial peptide, Chex-Arg20, alters its mechanism of interaction with the Escherichia coli membrane. Chem. Biol. 22, 1250–1258. doi: 10.1016/j.chembiol.2015.08.011
Li, W., Sani, M.-A., Jamasbi, E., Otvos L., Jr, Hossain, M. A., Wade, J. D., et al. (2016). Membrane interactions of proline-rich antimicrobial peptide, Chex1-Arg20, multimers. Biochim. Biophys. Acta 1858, 1236–1243. doi: 10.1016/j.bbamem.2016.02.035
Li, W., Tailhades, J., Hossain, M. A., O'Brien-Simpson, N. M., Reynolds, E. C., Otvos, L., et al. (2015b). C-Terminal modifications broaden activity of the proline-rich antimicrobial peptide, Chex1-Arg20. Aust. J. Chem. 68, 1373–1378. doi: 10.1071/CH15169
Li, W., Tailhades, J., O'Brien-Simpson, N., Separovic, F., Otvos, L. Jr., Hossain, M. A., et al. (2014). Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids 46, 2287–2294. doi: 10.1007/s00726-014-1820-1
Mateescu, M., Baixe, S., Garnier, T., Jierry, L., Ball, V., Haikel, Y., et al. (2015). Antibacterial peptide-based gel for prevention of medical implanted-device infection. PLoS ONE 10:e0145143. doi: 10.1371/journal.pone.0145143
Noto, P. B., Abbadessa, G., Cassone, M., Mateo, G. D., Agelan, A., Wade, J. D., et al. (2008). Alternative stabilities of a proline-rich antibacterial peptide in vitro and in vivo. Protein Sci. 17, 1249–1255. doi: 10.1110/ps.034330.108
Ostorhazi, E., Holub, M. C., Rozgonyi, F., Harmos, F., Cassone, M., Wade, J. D., et al. (2011). Broad-spectrum antimicrobial efficacy of peptide A3-APO in mouse models of multidrug-resistant wound and lung infections cannot be explained by in vitro activity against the pathogens involved. Int. J. Antimicrob. Agents 37, 480–484. doi: 10.1016/j.ijantimicag.2011.01.003
Otvos, L. Jr., and Wade, J. D. (2014). Current challenges in peptide-based drug discovery. Front. Chem. 2:62. doi: 10.3389/fchem.2014.00062
Otvos, L. Jr., Wade, J. D., Lin, F., Condie, B. A., Hanrieder, J., and Hoffmann, R. (2005). Designer antibacterial peptides kill fluoroquinolone-resistant clinical isolates. J. Med. Chem. 48, 5349–5359. doi: 10.1021/jm050347i
Roy, R. N., Lomakin, I. B., Gagnon, M. G., and Steitz, T. A. (2015). The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Mol. Biol. 22, 466–469. doi: 10.1038/nsmb.3031
Rozgonyi, F., Szabo, D., Kocsis, B., Ostorhazi, E., Abbadessa, G., Cassone, M., et al. (2009). The antibacterial effect of a proline-rich antibacterial peptide A3-APO. Curr. Med. Chem. 16, 3996–4002. doi: 10.2174/092986709789352295
Scocchi, M., Lüthy, C., Decarli, P., Mignogna, G., Christen, P., and Gennaro, R. (2009). The proline-rich antibacterial peptide Bac7 binds to and inhibits in vitro the molecular chaperone DnaK. Int. J. Pept. Res. Ther. 15, 147–155. doi: 10.1007/s10989-009-9182-3
Sebe, I., Ostorhazi, E., Fekete, A., Kovacs, K., Zelko, R., Kovalszky, I., et al. (2016). Polyvinyl alcohol nanofiber formulation of the designer antimicrobial peptide APO sterilizes Acinetobacter baumannii-infected skin wounds in mice. Amino Acids 48, 203–211. doi: 10.1007/s00726-015-2080-4
Seefeldt, A. C., Graf, M., Pérébaskine, N., Nguyen, F., Arenz, S., Mardirossian, M., et al. (2016). Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 44, 2429–2438. doi: 10.1093/nar/gkv1545
Keywords: A3-APO, Chex1-Arg20, D-arginine, Gram-negative bacteria, K. pneumoniae, backbone Nα-methylation, proline-rich antimicrobial peptide
Citation: Li W, Sun Z, O'Brien-Simpson NM, Otvos L, Reynolds EC, Hossain MA, Separovic F and Wade JD (2017) The Effect of Selective D- or Nα-Methyl Arginine Substitution on the Activity of the Proline-Rich Antimicrobial Peptide, Chex1-Arg20. Front. Chem. 5:1. doi: 10.3389/fchem.2017.00001
Received: 24 November 2016; Accepted: 04 January 2017;
Published: 19 January 2017.
Edited by:
Maria Luisa Mangoni, Sapienza University of Rome, ItalyCopyright © 2017 Li, Sun, O'Brien-Simpson, Otvos, Reynolds, Hossain, Separovic and Wade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammed A. Hossain, YWtodGVyLmhvc3NhaW5AZmxvcmV5LmVkdS5hdQ==
Frances Separovic, ZnNAdW5pbWVsYi5lZHUuYXU=
John D. Wade, am9obi53YWRlQGZsb3JleS5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.