
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. Biol. , 01 March 2024
Sec. Molecular Sciences
Volume 3 - 2024 | https://doi.org/10.3389/fchbi.2024.1362147
This article is part of the Research Topic Biosynthesis of secondary metabolites in bacteria: genes, pathways, and evolution View all 7 articles
 Siraprapa Brooks1*
Siraprapa Brooks1* Jack A. Weaver2
Jack A. Weaver2 Anthikan Klomchit1
Anthikan Klomchit1 Shumukh A. Alharthi3
Shumukh A. Alharthi3 Thanyarat Onlamun4
Thanyarat Onlamun4 Rithika Nurani5
Rithika Nurani5 Thienthu Khanh Vong5
Thienthu Khanh Vong5 Fabrizio Alberti2
Fabrizio Alberti2 Claudio Greco3*
Claudio Greco3*Endophytic fungi constitute a rich source of secondary metabolites that can be manipulated to produce desirable novel analogs for combating current agricultural challenges for crop production, especially controlling plant disease. The endophytic fungus Daldinia eschscholtzii MFLUCC 19-0629, was newly isolated from tropical ancient plants, Oncosperma sp., and displays a broad-spectrum of antifungal and antibacterial activities against several plant pathogens including Ralstonia solanacearum, Fusarium oxysporum, Colletotrichum gloeosporioides, Colletotrichum acutatum, Stagonosporopsis cucurbitacearum, Corynespora cassiicola and Stemphylium spp. A high-quality genome sequence was obtained using Oxford nanopore technology, the accuracy and length of reads resulting in no need for Illumina or other sequencing techniques, for D. eschscholtzii MFLUCC 19-0629, resulting in a genome size of 37.56 Mb assembled over 11 contigs of significant size, likely to be at the chromosomal level. Bioinformatics analysis revealed that this strain is biosynthetically talented encoding 67 predicted biosynthetic gene clusters (BGCs). Only eight of the 67 BGCs matched or demonstrated high similarity to previously characterized BGCs linked to the production of known secondary metabolites. The high number of predicted unknown BGCs makes this strain a promising source of novel natural products. The discovery that D. eschscholtzii MFLUCC 19-0629 has a broad spectrum of antimicrobial activity against seven major plant pathogenic microorganisms relevant to crop production and its complete genome sequence carries immense importance in the advancement of novel microbial biocontrol agents (MBCAs). This also unveils the prospect of uncovering new compounds that could be utilized for sustainable agriculture and pharmaceutical purposes.
Vegetables are affordable sources of vitamins, minerals, and antioxidants which are vital for human health. Thus, vegetables are essential for world food security, and culinary diversity. The most dominant vegetables that provide positive economic impacts are tomatoes, cucurbit, and chilies (Rajasree and Pugalendhi, 2021). Several factors can have an impact on vegetable production such as soil (soil type, soil pH, soil fertility, soil salinity) and environment (humidity, light, and temperature). In addition, plant diseases caused by pathogenic organisms, especially fungi and bacteria have tremendous negative impacts on vegetable production. For instance, 20–100% of yield losses in Solanaceae family including tomato, chili, and hot peppers are due to infection of bacterial soil-borne pathogens, Ralstonia solanacearum, that causes bacterium wilt disease, and fungal soil-borne pathogens, Fusarium oxysporum and Colletotrichum spp. that cause Fusarium wilt and anthracnose disease, respectively (Saxena et al., 2016; Ma et al., 2023; Wang et al., 2023). Leaf spot and late blight that are caused by pathogenic fungi, Corynespora cassiicola and Stemphylium spp., also significantly impact the yield of tomato crops, which is commonly a susceptible host, by causing severe leaf chlorosis, necrosis followed by leaf defoliation (Farr and Rossman, 2013; Bessadat et al., 2020). As for cucurbit, several diseases threaten the yield of this family, gummy stem blight caused by Stagonosporopsis cucurbitacearum (syn. Dimymella bryoniae) is one of the major devastating diseases, especially for watermelon, cantaloupe, cucumber, pumpkin and other melons (Seblani et al., 2023).
Chemical pesticides have been applied to control those pathogenic organisms; however excessive applications have simultaneously increased concern about negative effects on human health and the environment (Aktar et al., 2009; Tudi et al., 2021). Sustainable agriculture is increasingly recognized as a green strategy to manage plant diseases with less concerns for pollution from inorganic residuals. Interestingly, fungi are an excellent source of bioactive molecules, especially secondary metabolites with antimicrobial properties. Microbial biological control agents (MBCAs) from fungi have become increasingly recognized as solutions to disease management in sustainable agriculture. Various species of Trichoderma spp., Clonostachys spp., Coniothyrium sp., and Phoma sp. are either commonly used as MBCAs or well-studied on their biocontrol potential (Cao et al., 2009; Hamid et al., 2013; Park et al., 2019; Sood et al., 2020; Thambugala et al., 2020; Srisuksam et al., 2021). The genus Daldinia within the Xylariaceae family also has been widely reported as a rich source of various bioactive compounds (Stadler et al., 2014; Li et al., 2021). In particular, D. eschscholtzii species have been documented to produce a great number of secondary metabolites (SMs), (Wang et al., 2015; Shylaja et al., 2018; Yang et al., 2018; Zhang et al., 2019; Liao et al., 2019; Lin et al., 2019; ZHOU et al., 2019), even when compared to other Daldinia species including D. concentrica (Quang et al., 2002), D. hawksworthii (Pažoutová et al., 2013) and D. childiae (Stadler et al., 2014).
Although, MBCAs have been proven to be effective agents in sustainable agriculture, at present there is a great need for novel sources of antimicrobials that could prevent or control a broad range of diseases. In our ongoing bioactivity screening against crop pathogens, the newly isolated fungal strain Daldinia eschscholtzii MFLUCC 19-0629 showed an excellent broad range inhibition against relevant plant pathogens, Ralstonia solanacearum, Fusarium oxysporum, Colletotrichum spp., Corynespora cassiicola, Stemphylium spp. and Stagonosporopsis cucurbitacearum. This suggested the huge potential of this strain to develop high-efficiency MBCAs.
Here, we addressed its molecular phylogeny and fully annotated the high-quality, uncontaminated genome of this novel strain to further understand and facilitate research into the function of BGCs associated with valuable SMs. More importantly, this will be a major step in developing effective MBCAs that provide a broad range of protection against major diseases in vegetable production which will indirectly increase crop yields while contributing to sustainable and eco-friendly farming practices.
Fresh and healthy samples of nibung palm (Oncosperma spy.) were collected from Suratthani province, Thailand (8° 37′ 47″ N, 99° 20′ 35″ E). Plant materials were transferred to the laboratory in sterile bags and stored at 4 °C until processed. The surfaces of plant material were washed with running tap water to remove surface microbes and sterilized with 70% ethanol for 3 minutes followed by 5% sodium hypochlorite for 3 minutes and rinsed with sterilized water for 3 minutes in aseptic conditions twice. The sterile plant tissues were dried on sterile filter paper, cut to obtain small tissue sections (0.5 × 0.5 cm2) and transferred to potato dextrose agar (PDA) plates. The plates were incubated for 2–10 days at 28°C. Growing hyphal tips from the plant tissue sections were transferred to fresh PDA plates and incubated at 28 °C to obtain pure cultures. Pure cultures were preserved in separate 1.5 tubes of sterilized 15% glycerol, distilled water, and PDA for further use.
Sixty-two endophytic strains isolated from the native palm Oncosperma were grown on potato dextrose agar (PDA, Himedia, India) at 28°C for 7 days. Then, five fungal mycelial plugs (5 mm in diameter) were inoculated into 1,000 mL Erlenmeyer flask containing 500 mL of PDB (Himedia, India). The cultures were grown in an incubator at 28 °C with shaking at 100 rpm on a reciprocal shaker. After 3 weeks, the cultures were filtrated by Whatman’s No. 1 paper to collect the supernatant. After that, the culture filtrate of endophytic fungi was extracted twice with an equal volume of ethyl acetate (1:1 v/v). The upper solvent phase was collected and evaporated using a rotary evaporator. The crude extract of each strain was stored at −20°C for further experiments.
All seven pathogenic fungi used in this study were kindly provided by Chia Tai Co., Ltd. (Bangkok, Thailand). Three pathogenic fungi, Fusarium oxysporum MFLUCC BK-0012, Corynespora cassiicola MFLUCC TSM-EB11, Stemphylium spp. MFLUCC TSM-LB13, Alternaria solani MFLUCC AS0050 were isolated from tomato plants with Fusarium wilt, leaf spot, late blight, and early blight respectively. Two strains of Colletotrichum spp., C. gloeosporioides MFLUCC CK-0032 and C. acutatum MFLUCC CC-0036, were isolated from chili with anthracnose disease. Stagonosporopsis cucurbitacearum MFLUCC BB-001 was isolated from honeydew melon. All of them were cultured on PDA at 28 C.
The poisoned culture technique was performed to investigate the antagonistic activity of crude extracts from the 62 isolated fungal strains following a protocol modified from the previous protocol (Schmitz, 1930). The effect of dimethylsulfoxide (DMSO) and recommended fungicides, Captan and Mancozeb were pre-evaluated on the growth of all pathogenic strains that were tested. The most effective fungicide was used as positive control, namely, Captan with F. oxysporum MFLUCC BK-0012, C. gloeosporioides MFLUCC CK-0032, C. acutatum MFLUCC CC-0036 and S. cucurbitacearum MFLUCC BB-001; while Mancozeb was used with Corynespora cassiicola MFLUCC TSM-EB11 and Stemphylium spp. MFLUCC TSM-LB13. Since DMSO did not result in hyphal growth inhibition in any of the plant pathogenetic strains, PDA mixed with sterile water was used as a negative control plate. The fungal crude extract solution was prepared by dissolving it in DMSO, then the fungal crude extract was added to the autoclaved PDA media to obtain a final concentration of 1 mg/mL. The experiments were each carried out with triplicates for each sample and the whole experiment was repeated three separate times. Briefly, a 7 mm dish from a week-old culture of pathogenic fungi on PDA medium was inoculated at the center of PDA mixed with fungal crude extract, fungicides and sterile water. All plates were incubated at 28 C for 7–10 days before measuring radial mycelial growth. The percentage of inhibition (PI%) was calculated as follows:
where PI% is inhibition of radial mycelial growth; C is radial growth measurement of the pathogen in the negative control; T is the radial growth of the pathogen in the presence of fungal crude extract.
Three bacterial strains of Ralstonia solanacearum, including BWTM-01, BWEP-05, and BWHP-03 that cause bacterial wilt in eggplant (BWTM-01), tomato (BWEP-05), and hot pepper (BWHP-03), and one strain of Acidovorax citrulli JT-0003 that causes bacterial fruit blotch in the cucurbits family, were kindly provided from Chai Tai Co., Ltd. Antimicrobial activities of fungal crude extract were determined by the broth microdilution technique using 96-well microplates (Sarker et al., 2007). All four strains were cultured in Nutrient Broth (NB) at 37 °C for overnight, and bacterial suspensions were adjusted to 0.5 McFarland standard turbidity (1×108 CFU/mL). The stock concentration of fungal crude extract (10 mg/mL) was prepared by weighing 10 mg of dry crude extract and dissolved in 1 mL of 10% methanol. Each well in a sterile microdilution plate was filled with 100 µL NB and 100 µL of each treatment was added (sterile distilled water and 10% methanol as a negative control; Ampicillin as a positive control; fungal crude extract with the final well concentration was 1 mg/mL). Then, 10 µL of bacterial suspension was added to all wells and mixed thoroughly. After overnight incubation at 37°C, 15 µL of 0.01% resazurin was added to all wells and then incubated for 3 h before the absorbance was measured at 600 nm. The reduction percentage was calculated as follows:
The 10-day-old pure cultures of D. eschscholtzii strain MFLUCC 19-0629 on PDA were used for DNA extraction. The mycelia were scraped off from pure cultures and genomic DNA was extracted using a PureDire X genomic DNA isolation kit (Plant) (bio-helix. Co., Ltd, United States) following the manufacturer’s instructions. The quantity and quality of the genomic DNA were evaluated by a Thermo Scientific NanoDrop spectrophotometer, Qubit Fluorometer and by visual observation through 1.5% agarose gel electrophoresis.
The molecular identification of the endophytic fungi was performed with four sets of primers. The internal transcribed spacer regions (ITS4/5) (White et al., 1990) and partial sequences of the large subunit of the rDNA (LR7/LROR) (Vilgalys and Hester, 1990; Bunyard et al., 1994), RNA polymerase II (RPB2-5F/RPB2-7C) (Liu et al., 1999), and beta-tubulin (T1/T22) (O’Donnell and Cigelnik, 1997). PCR reactions were carried out in a total volume of 25 μL, containing 2 μL of genomic DNA, 1.25 μL of MgCl2, 0.5 μL of deoxynucleoside triphosphate (dNTP), 2 μL of 10x buffer, 1 μL of each primer, 0.25 μL of Taq DNA polymerase and 17 μL Milli-Q waters in ram1x reaction buffer. The PCR amplifications were performed following the thermal cycling conditions mentioned in Supplementary Table S1. The PCR products were checked on 1.5% agarose gel electrophoresis for 30 min at 100 V. PCR purification and DNA sequencing of PCR products were carried out at Biogenomed Co., Ltd, Thailand.
The BLAST search engine of the National Centre for Biotechnology Information (NCBI) was used for the preliminary identification of DNA sequences of the new isolates. Sequence data of the closely related taxa to our isolates were retrieved from GenBank based on the BLAST search results and recent publications (Supplementary Table S2). Sequences of the individual loci were aligned with MAFFT v. 7 online versions using default settings. BioEdit v. 7.0.5.2 software was used to refine the alignments manually where necessary. Maximum likelihood (ML) trees were generated using CIPRES Science Gateway v. 3.3; http://www.phylo.org/portal2/. Parameters of maximum likelihood were set to 1,000 bootstrap replicates with the GTRGAMMA+I model of nucleotide evolution. The best fit nucleotide substitution model for the dataset was separately determined using MrModeltest v. 2.2. The Bayesian analysis was conducted with MrBayes v. 3.2.2 to evaluate posterior probabilities (PP) by Markov Chain Monte Carlo sampling (MCMC). Two independent runs of six simultaneous Markov chains were run for 1,000,000 generations and trees were sampled every 100th generation and 10,000 trees were obtained. The first 20% of trees, representing the burn-in phase of the analyses, were discarded. The remaining 80% of trees were used to calculate PP in the majority rule consensus tree. Trees were visualized with FigTree v. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) (Rambaut, 2014).
A genomic DNA library was prepared using kit SQK-LSK109 from Oxford Nanopore Technology (ONT). The manufacturers protocol was modified for DNA repair and end-prep stages by increasing the incubation time and temperature after ethanol washing to 15 min minimum at 37°C. Native barcode ligation took place at 37°C for 30 min. The long fragment buffer was used to enrich DNA fragments of 3 kb or longer. 1.2 μg of genomic DNA was loaded at the beginning of library preparation. The library was sequenced on a MinION Mk1C (ONT) with a FLO-MIN-106 R9.4 flow-cell (ONT).
The raw data produced was base-called and demultiplexed using Guppy v6.0.1. Guppy used config file dna_r9.4.1_450bps_hac.cfg, all other parameters were left on default. Outputs were merged into a single.fastq file that was inputted into Flye version 2.9 using the nano-hq setting to assemble the draft genome (Kolmogorov et al., 2019). The draft genome was polished by aligning to the raw reads with Minimap2 v2.24 (Li, 2018) and correcting errors using Racon v1.4.20 (Vaser et al., 2017), the values given to bases were as follows; 8 for matches, −6 for mismatches, −8 for gaps, with a 500 base window size. The output from Racon was entered back into Minimap2 and the process repeated for 4 total rounds of polishing. Medaka v1.5.0 (ONT) was used to make further corrections for the genome against model r941_min_high_g360. The polished genome was analyzed using BUSCO v5.0.0 against the sordariomycete_odb10. Functional annotation was performed using the Funannotate version 1.8.9 pipeline (Palmer and Stajich, 2020). The prediction and annotation of functional elements were done using Neurospora crassa as the seed species and the BUSCO sordariomycetes database, minimum training models were lowered to 100, and the Augustus optimization setting was called. Unless specified all settings were left on their default parameters. CMscan (Burge et al., 2013) was used with StructRNAfinder (Arias-Carrasco et al., 2018) for screening for non-coding RNA, with the Rfam database (Griffiths-Jones, 2004) as the input.
The genomes of D. eschscholtzii strains IFB-TL01 (GCA_001951055.1) (Fang et al., 2012), UM1020 (GCA_000261445.1) (Ng et al., 2012), FL0578 (GCA_022478725.1) (Franco et al., 2022) were downloaded from NCBI. The annotated genome of D. eschscholtzii MFLUCC 19-0629 and the other genomes were analyzed using antiSMASH fungal version 7.1.0, the detection strictness was set to ‘relaxed’ and the extract features ‘KnownClusterBlast’, ‘ClusterBlast’, ‘SubClusterBlast’, ‘MIBiG cluster comparison’, ‘ActiveSiteFinder’, ‘REFinder’ and ‘Cluster Pfam analysis’ were set on (Blin et al., 2023). The genes predicted in BGCs were further searched using NCBI BLAST. BGCs comparison across strains and to the MIBiG database was done using BiG-SCAPE (Navarro-Muñoz et al., 2020).
The 62 fungal endophytes were isolated from healthy plant tissue of the Nibung palm (Oncosperma sp.) an ancient plant aged over 50 years. The fungal crude extracts of these endophytes were evaluated for their antagonistic activity against six plant pathogenic strains by poison food assay. In general, fungicides that were used as positive control exhibited a higher percentage of mycelium inhibition against all tested pathogens than fungal crude extracts (Figures 1, 2). While antagonistic activities were observed in at least 20 fungal crude extract (data not shown), only fungal crude extract from strain MFLUCC 19-0629 displayed varying high degrees of inhibitory activity against six pathogenic fungi except for Alternaria solani MFLUCC AS0050. Consequently, this strain was selected for further investigation. The highest inhibition of fungal crude extract was observed towards the pathogen that causes gummy stem blight disease in cucurbits as shown by a 92.88 ± 3.28% inhibition which is the same level inhibition of Captan (92.97 ± 0.92% inhibition; Figure 1). While the potence of fungal crude extract toward C. acutatum MFLUCC CC-0036, Corynespora cassiicola MFLUCC TSM-EB11 and Stemphylium spp. MFLUCC TSM-LB13 was slightly lower than the recommended fungicide (Figure 1). The lowest effect of fungal crude extract of strain MFLUCC 19-0629 was observed in F. oxysporum MFLUCC BK-0012 as shown in only 38.84 ± 6.38% mycelium growth inhibition while Captan provide 82.12+3.56% inhibition (Figure 1).
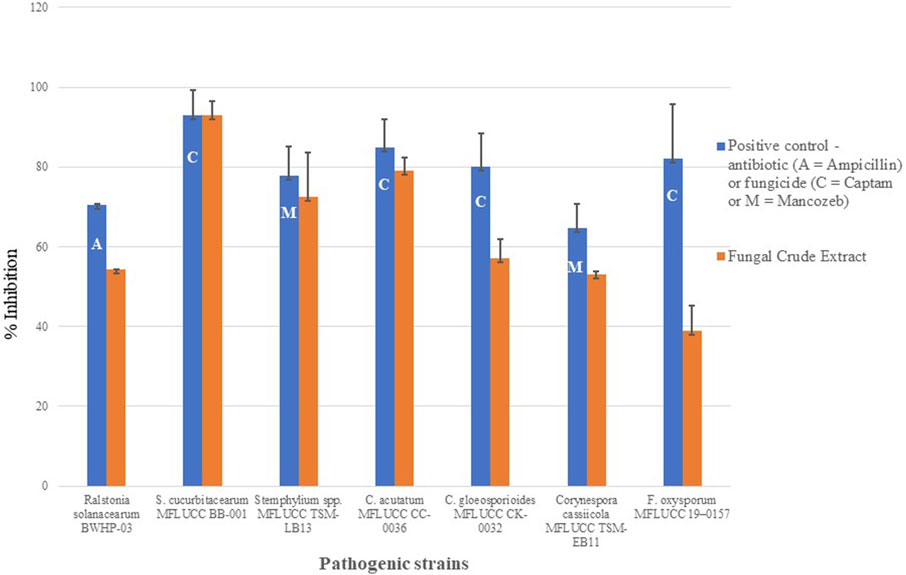
FIGURE 1. Bioactivity screening against plant pathogens. The percent of inhibition of antibiotic, recommended fungicide (Captan, Mancozeb) or antibacterial (ampicillin) (shown in blue), and fungal crude extract of D. eschscholtzii MFLUCC 19-0629 (shown in orange) towards inhibition of bacterial strain, R. solanacearum BWHP-03 and mycelium growth of six strains of fungal pathogens F. oxysporum MFLUCC BK-0012, C. gloeosporioides MFLUCC CK-0032, C. acutatum MFLUCC CC-0036, S. cucurbitacearum MFLUCC BB-001, Corynespora cassiicola MFLUCC TSM-EB11 and Stemphylium spp. MFLUCC TSM-LB13. Standard deviation (±SD) is calculated from the nine replications.
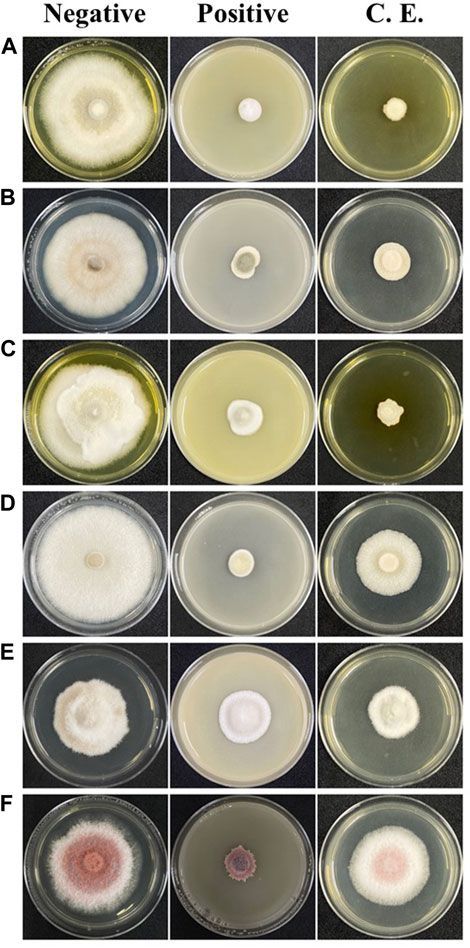
FIGURE 2. Poison food assay plates. (A) S. cucurbitacearum MFLUCC BB-001; (B) C. acutatum MFLUCC CC-0036; (C) Stemphylium spp. MFLUCC TSM-LB13 (D) C. gloeosporioides MFLUCC CK-0032; (E) Corynespora cassiicola MFLUCC TSM-EB11; (F) F. oxysporum MFLUCC BK-0012; inoculated on PDA (negative control; left column), recommended fungicides (positive control; middle column), and fungal crude extract of D. eschscholtzii MFLUCC 19-0629 (crude extract = C. E., right column).
The crude extract from this strain was also used to screen for antibacterial activity against three strains of R. solanacearum and one strain of Acidovorax citrulli. The antibiotic ampicillin, used as positive control, had the highest inhibitory percentage against all strains of tested bacteria (percentage of reduction 44.762 ± 0.047 to 70.580 ± 0.010%), while the negative control, methanol, did not present any growth inhibition. The MFLUCC 19-0629 crude extract showed no inhibitory effect against Acidovorax citrulli and weak inhibitory effect (percentage of reduction of less than 5%) on two strains of pathogenic bacteria (R. solanacearum BWTM-01 and BWEP-05); however, potent inhibition was observed against R. solanacearum BWHP-03 with a reduction percentage of up to 54% (Figure 1).
Fungus culture MFLUCC 19-0629 was identified based on both morphology and molecular means. The morphology is matched to the previous report of Daldinia eschscholtzii (Figure 3) (Rehm, 1904). The growth rate of colonies grown on PDA for 2 weeks ranged from 3 to 4 mm (diameter)/day. Early hyphae were showing fairly rapid growth, at first whitish hyphae becoming form a felty, azonate mycelium and become greenish in patches, finally turning mouse grey in the rings of white: reverse green/yellow and become dull green (with age resulting). Asexual morph: Mycelium composed of septate, branched hyphae. Conidiophores hyaline with virgariella-like to nodulisporium-like branching patterns, di- or trichotomously branched, with two to three conidiogenous cells arising from each terminus. Conidiogenous cells are hyaline, cylindrical, holoblastic, terminal or intercalary. Conidia size and shape were 4.49–6.24 µm × 2–3.7 µm (x = 5.2 × 2.7, n = 30), aseptate, hyaline, smooth, obovoid to ellipsoid.
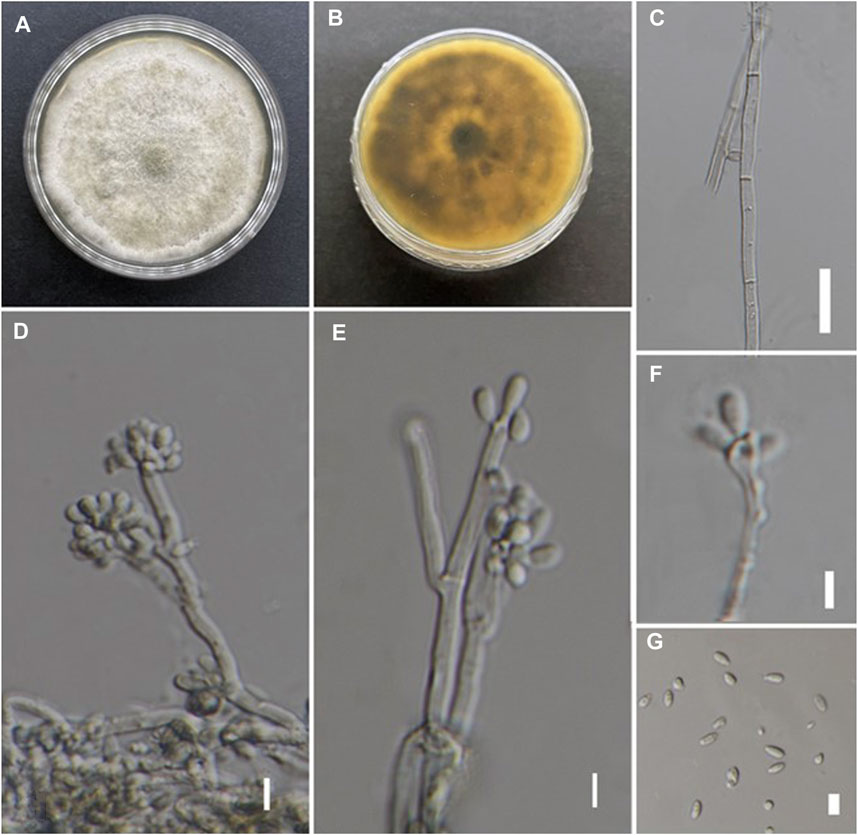
FIGURE 3. Morphological analysis of D. eschscholtzii MFLUCC 19-0629. (A,B) Colonies on PDA after 10 days. (C) Mycelium with septa. (D,E) Conidiophores with di–or trichotomously branching, showing nodulisporium-like branching pattern in the mycelium. (F) Conidiogenous cell with conidia. (G) Conidia. Scale bar: c = 20 μm, d–g = 5 μm.
Molecular characterization was performed through PCR amplification and Sanger sequencing of four target conserved sequences, which were compared with available sequences from NCBI. Based on the analysis of the homology of the ITS rDNA sequences, partial sequences of the large subunit of the rDNA, RNA polymerase II, and beta-tubulin regions, a phylogenetic relationship was constructed by RAxML analyses (Figure 4). The maximum composite likelihood analysis of MFLUCC 19-0629 sequences alignment revealed that the D. eschscholtzii species form a monophyletic group in a consensus tree. The bootstrap value of 90% supported the MFLUCC19-0629 sequence analysis, thus inferred that the isolate belongs to the D. eschscholtzii species.
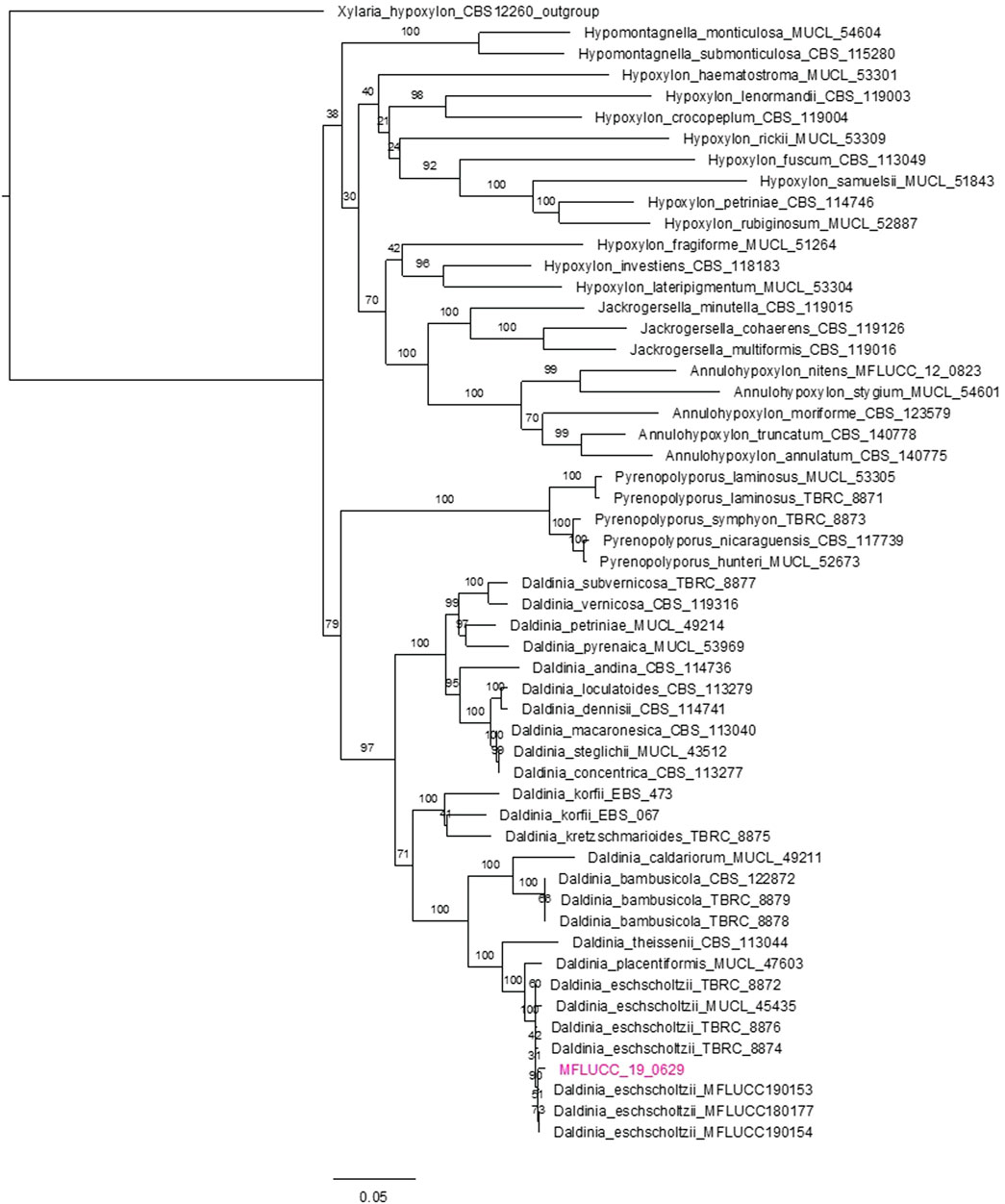
FIGURE 4. Phylogenetic tree based on the nucleotide sequences of rDNA-ITS. Partial sequences of the large subunit of the rDNA, RNA polymerase II, and beta-tubulin regions involved 56 nucleotide sequences. The isolates and their phylogenetic position are based on the ITS region according to the RAxML analyses. Bootstrap support values for ML equal to or greater than 90%.
Genomic DNA from Daldinia eschscholtzii MFLUCC 19-0629 was sequenced harnessing Oxford nanopore sequencing. This led to approximately 2,779,445,032 bases from the nanopore data. As our assembly of D. eschscholtzii has a 37.56 Mb genome, this gives a coverage of 73.9 reads per base. The assembly presented here is of similar size to D. eschscholtzii genome analysis carried out by other researchers (35 Mb) (Chan et al., 2015). Assembly statistics shown in Table 1 show this assembly has 11 contigs of significant size, these are likely to be assembled at the chromosomal level although this requires further investigation to determine.
BUSCO (Manni et al., 2021) was used to determine the quality of the genomes by quantifying the completeness of core conserved genes in D. eschscholtzii that would be expected across the sordariomycetes class. Table 1 shows the conserved genes identified across the scaffold of this assembly, while Supplementary Table S3 shows the conserved proteins identified in the predicted proteome for this species. Predicted non-coding RNA can be found in Supplementary Table S4, generated using StructRNAfinder, this data is publicly available at NCBI (Bioproject PRJNA1053602).
The predicted proteome generated using the Funannotate package was run through Eggnog-mapper v2.1.7. Where 80.3% of the 10,466 protein coding genes were able to be assigned a COG (Clusters of Orthologous Groups) functional category. The distribution of genes across these categories is shown in Supplementary Figure S1. It is important to note that some genes are assigned multiple COG categories. Eggnog-mapper was also capable of assigning gene ontology terms, frequently with multiple terms assigned to individual predicted proteins, enzyme commission numbers, matches across multiple KEGG databases, and BRITE hierarchies are also included (Supplementary Figure S2). A small number of predicted proteins (1.7%) could be assigned to matches in the CAZy database for carbohydrate active enzymes, 1.3% were found to match with BiGG IDs. 74% of genes were matched to the pfam database. All the Eggnog-mapper results are included in Dataset Supplementary Figure S2.
The genome of D. eschscholtzii was analyzed using antiSMASH fungal version 7.1.0. This analysis revealed that there are 67 putative biosynthetic gene clusters (BGCs). Polyketide synthase (PKS) containing BGCs were the most prevalent with 23, followed by non-ribosomal peptide synthetase (NRPS) and NRPS-like BGCs with 15 (6 and 9 respectively), ribosomally synthesized and post-translationally modified peptides (RiPPs) with 13, terpenes with 10, indole with 3 and one each for hybrid PKS/NRPS, indole, betalactone and the newly reported isocyanide. Analysis of the PKS domain architecture determined that they can be subdivided into 19 highly-reducing Type I PKS, two non-reducing Type I PKS, one partially-reducing Type I PKS and one Type III PKS (Cox, 2007). Out of 67 predicted BGCs, only eight matched or have close similarity with known clusters. A complete match was observed to the BGCs for mellein 1 (Zhang et al., 2019), naphthalenes 2-3 (Chooi et al., 2015), 1-(2,6-dihydroxyphenyl)but-2-en-1-one (PBEO) 4 (Fang et al., 2012) and ergotamine 5 (Lorenz et al., 2007) deposited in the MiBiG database (Figure 5A). Other strains of D. eschscholtzii have been previously reported to produce mellein (Zhang et al., 2019), PBEO and naphthalene-derived compounds (Fang et al., 2012). Comparison with MiBig database identified BGCs with high similarity to cytochalasin E 6 (Qiao et al., 2011), trichoxide 7 (Liu et al., 2019), solanapyrone D 8 (Kasahara et al., 2010) (Figure 5B). Other D. eschscholtzii strains have been reported to produce cytochalasins (Yang et al., 2018).
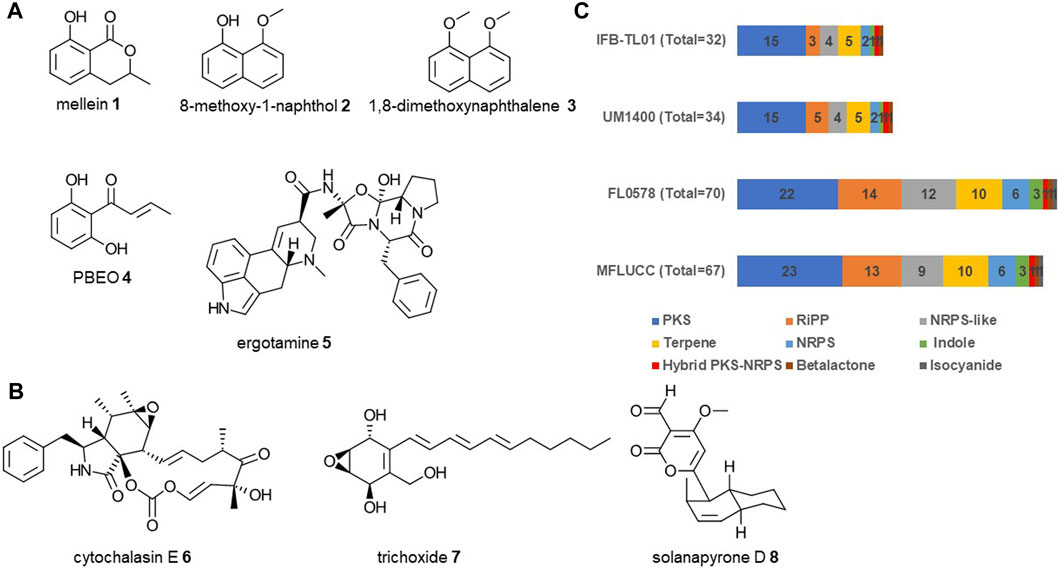
FIGURE 5. Overview of the secondary metabolites and BGCs from D. eschscholtzii MFLUCC 19-0629. (A) Compounds that could be produced by MFLUCC 19-0629 based on a complete match to characterized BGCs. (B) Compounds that could be produced because they share a high similarity to known BGCs. (C) Comparison of the distribution of BGCs by types of secondary metabolites for D. eschscholtzii MFLUCC 19-0629 and three other D. eschscholtzii strains that have their genomes available.
The biosynthetic potential of this strain was compared to other D. eschscholtzii strains that had their genome available on NCBI. They are strain IFB-TL01 isolated from mantis gut (Wang et al., 2015), strain UM1020 isolated from human blood (Ng et al., 2012) and FL0578 isolated from lichen thallus of Cladonia evansii (Franco et al., 2022). Their genomes were analyzed using fungiSMASH, which revealed a huge variation in biosynthetic potential across different D. eschscholtzii strains (Figure 5C). The strains isolated from gut or blood sample (IFB-TL01 and UM1020) encoded only for 32 and 34 BGCs respectively. While the strain (FL0578) isolated from lichen encoded for 70 BGCs, which is slightly higher than the strain from this study.
Various mechanisms are employed to maintain a harmonious equilibrium among the endophyte, host, and pathogen, involving strategies like mycoparasitism, the generation of lytic enzymes or antibiotics, and the secretion of secondary metabolites (Hassan et al., 2013; Cheong et al., 2017; Yan et al., 2018). This study particularly assesses the efficiency of secondary metabolites derived from 62 endophytic fungi isolated from Oncosperma in managing destructive pathogenic bacteria and fungi in vegetable crops. Among the tested pathogens, Fusarium oxysporum MFLUCC BK-0012, Corynespora cassiicola MFLUCC TSM-EB11, Stemphylium spp. MFLUCC TSM-LB13, Alternaria solani MFLUCC AS0050, Colletotrichum spp., C. gloeosporioides MFLUCC CK-0032, and C. acutatum MFLUCC CC-0036, Stagonosporopsis cucurbitacearum MFLUCC, and bacterial pathogens Ralstonia solanacearum BWTM-01, BWEP-05, BWHP-03, and Acidovorax citrulli JT-0003 were tested against the crude extracts from these fungal endophytes. One strain, D. eschscholtzii MFLUCC 19-0629, exhibited a notably broad spectrum of microbial activity by displaying antagonistic effects against seven out of the eleven tested pathogenic microorganisms. This strain showed notably high antagonistic activity, especially against C. acutatum MFLUCC CC-0036, S. cucurbitacearum MFLUCC BB-001, and Stemphylium spp. MFLUCC TSM-LB13, inhibiting their mycelium growth by 72.44–92.88% in this study.
Presently, research developments regarding the utilization of endophytic fungi and the production of biocontrol agents (BCAs) are progressing. Nevertheless, their efficacy remains limited in terms of the range of pathogens they can effectively control. For instance, the endophytic fungus Phoma sp. L28 that colocalized with mangrove was reported to produce macrosporin that displayed the ability to control F. oxysporum, C. gloeosporioides, and A. solani (Huang et al., 2017). Similarly, an alkaloid compound, penochalasin, secreted from Penicillium chrysogenum V11 was able to inhibit the growth of plant pathogenic fungi (C. gloeosporioides) and bacteria (Rhizoctonia solani) (Zhu et al., 2017). Highlighting the significance of this study, we successfully identified a fungal endophyte strain, D. eschscholtzii MFLUCC 19-0629, whose crude extract displayed a broad-spectrum activity against seven major pathogenic strains in vegetable crop production. The crude extract bioactivity against a range of pathogens suggests the presence of other metabolites contributing to antifungal and antibacterial effects. The genome was sequenced using Oxford Nanopore Technology, which resulted in a very high-quality genome assembly, which highlights the advancement of this sequencing technology and its application for sequencing fungal genomes. Previous flow cells which are used to collect the long-read data generated by nanopore sequencing were not of sufficient accuracy to assemble a fungal genome by themselves, requiring a supplementary round of polishing by aligning to short read sequencing as shown in previous work (Tamizi et al., 2022). This is no longer the case and Oxford Nanopore Technology flow cells are now capable of generating reads of an accuracy on par with that of other high throughput yet shorter read length sequencing (Ni et al., 2023). Bioinformatics analysis uncovered 67 putative BGCs within this fungus strain, indicating the potential to produce many SMs. Furthermore, four out of 67 BGCs had a complete match to characterized ones, while another four had high homology to previously mined BGCs, suggesting that similar molecules to these may be produced. Some of these predicted compounds are, for example, mellein 1 which has been reported to have broad-spectrum antimicrobial activity against several bacterial and fungal plants and human pathogens (Morales-Sánchez et al., 2021; Saraswathi et al., 2022), 8-Methoxy-1-naphthol 2 which has shown antifungal activity against Athelia rolfsii responsible for blight disease on tomatoes (Tanapichatsakul et al., 2020), the naphthalene derivative 1,8-dimethoxynaphthalene 3, which proved superior anti-fungal activity against several pathogens that threaten agroeconomic security (Santra and Banerjee, 2022), ergotamine, which is the main ingredient to treat migraines and cluster headaches (Tfelt-Hansen, 2000) and cytochalasins, a class of compounds which have been reported to improve the efficacy of chemotherapeutics (Trendowski et al., 2015). In conclusion, the high number of unknown BGCs encoded in the genome of D. eschscholtzii MFLUCC 19-0629 and the broad range of bioactivities against plant pathogens make this newly isolated fungal strain an excellent candidate to mine novel bioactive natural products with applications in agriculture and medicine. Future work will combine metabolomics studies, bioactivity screening and fungal engineering to increase the production of new secondary metabolites and identify potential bioactive compounds to develop effective MBCAs from this strain.
The genome sequence can be found in NCBI under the Bioproject accession number PRJNA1053602. The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
SB: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. JW: Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing–original draft, Writing–review and editing. AK: Data curation, Formal Analysis, Investigation, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. SA: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft, Writing–review and editing. TO: Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. RN: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. TV: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. FA: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing–original draft, Writing–review and editing. CG: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. CG was supported by the BBSRC BB/V005723/2. SA’s PhD Scholarship was supported by Taif University. JW was supported by a scholarship from the Engineering and Physical Sciences Research Council and the Biotechnology and Biological Sciences Research Council (EP/L016494/1) through the Centre for Doctoral Training in Synthetic Biology (SynBioCDT). FA was supported by a UKRI Future Leaders Fellowship (MR/V022334/1). SB was supported by Mae Fah Luang University under the grant name “Reinventing University 2021” and Minister of Higher Education, Science, Research and Innovation under grant name “Talent Mobility 2023 (TM65005).”
The authors would like to acknowledge Miss Machima Saengket for collecting fungal materials.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchbi.2024.1362147/full#supplementary-material
Aktar, M. W., Sengupta, D., and Chowdhury, A. (2009). Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip. Toxicol. 2, 1–12. doi:10.2478/v10102-009-0001-7
Arias-Carrasco, R., Vásquez-Morán, Y., Nakaya, H. I., and Maracaja-Coutinho, V. (2018). StructRNAfinder: an automated pipeline and web server for RNA families prediction. BMC Bioinforma. 19, 55. doi:10.1186/s12859-018-2052-2
Bessadat, N., Hamon, B., Bataillé-Simoneau, N., Chateau, C., Mabrouk, K., and Simoneau, P. (2020). Occurrence of leaf spot disease caused by Alternaria crassa (sacc.) rands on jimson weed and potential additional host plants in Algeria. Plant Pathol. J. 36, 179–184. doi:10.5423/PPJ.NT.01.2020.0003
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50. doi:10.1093/nar/gkad344
Bunyard, B. A., Nicholson, M. S., and Royse, D. J. (1994). A systematic assessment of Morchella using RFLP analysis of the 28S ribosomal RNA gene. Mycologia 86, 762–772. doi:10.1080/00275514.1994.12026481
Burge, S. W., Daub, J., Eberhardt, R., Tate, J., Barquist, L., Nawrocki, E. P., et al. (2013). Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232. doi:10.1093/nar/gks1005
Cao, R., Liu, X., Gao, K., Mendgen, K., Kang, Z., Gao, J., et al. (2009). Mycoparasitism of endophytic fungi isolated from reed on soilborne phytopathogenic fungi and production of cell wall-degrading enzymes in vitro. Curr. Microbiol. 59, 584–592. doi:10.1007/s00284-009-9477-9
Chan, C. L., Yew, S. M., Ngeow, Y. F., Na, S. L., Lee, K. W., Hoh, C.-C., et al. (2015). Genome analysis of Daldinia eschscholtzii strains UM 1400 and UM 1020, wood-decaying fungi isolated from human hosts. BMC Genomics 16, 966. doi:10.1186/s12864-015-2200-2
Cheong, S. L., Cheow, Y. L., and Ting, A. S. Y. (2017). Characterizing antagonistic activities and host compatibility (via simple endophyte-calli test) of endophytes as biocontrol agents of Ganoderma boninense. Biol. Control 105, 86–92. doi:10.1016/j.biocontrol.2016.12.002
Chooi, Y.-H., Krill, C., Barrow, R. A., Chen, S., Trengove, R., Oliver, R. P., et al. (2015). An in planta -expressed polyketide synthase produces (R)-Mellein in the wheat pathogen parastagonospora nodorum. Appl. Environ. Microbiol. 81, 177–186. doi:10.1128/AEM.02745-14
Cox, R. J. (2007). Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 5, 2010. doi:10.1039/b704420h
Fang, W., Ji, S., Jiang, N., Wang, W., Zhao, G. Y., Zhang, S., et al. (2012). Naphthol radical couplings determine structural features and enantiomeric excess of dalesconols in Daldinia eschscholzii. Nat. Commun. 3, 1039. doi:10.1038/ncomms2031
Farr, D. F., and Rossman, A. Y. (2013). “Fungal databases,” in Systematic mycology and microbiology laboratory, ARS, USDA. Available at: http://nt.ars-grin.gov/fungaldatabases/
Franco, M. E. E., Wisecaver, J. H., Arnold, A. E., Ju, Y. M., Slot, J. C., Ahrendt, S., et al. (2022). Ecological generalism drives hyperdiversity of secondary metabolite gene clusters in xylarialean endophytes. New Phytol. 233, 1317–1330. doi:10.1111/NPH.17873
Griffiths-Jones, S. (2004). Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124. doi:10.1093/nar/gki081
Hamid, M. I., Zeng, F., Cheng, J., Jiang, D., and Fu, Y. (2013). Disruption of heat shock factor 1 reduces the formation of conidia and thermotolerance in the mycoparasitic fungus Coniothyrium minitans. Fungal Genet. Biol. 53, 42–49. doi:10.1016/j.fgb.2012.12.002
Hassan, S. E., Hijri, M., and St-Arnaud, M. (2013). Effect of arbuscular mycorrhizal fungi on trace metal uptake by sunflower plants grown on cadmium contaminated soil. New Biotechnol. 30, 780–787. doi:10.1016/j.nbt.2013.07.002
Huang, S., Xu, J., Li, F., Zhou, D., Xu, L., and Li, C. (2017). IDENTIFICATION and antifungal activity of metabolites from the mangrove fungus phoma sp. L28. Chem. Nat. Compd. 53, 237–240. doi:10.1007/s10600-017-1961-z
Kasahara, K., Miyamoto, T., Fujimoto, T., Oguri, H., Tokiwano, T., Oikawa, H., et al. (2010). Solanapyrone synthase, a possible diels–alderase and iterative type I polyketide synthase encoded in a biosynthetic gene cluster from Alternaria solani. ChemBioChem 11, 1245–1252. doi:10.1002/cbic.201000173
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi:10.1038/s41587-019-0072-8
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi:10.1093/bioinformatics/bty191
Li, J., Li, L., Zhu, R., Lim, V., Sun, Q., and Fang, L. (2021). Natural products from the genus Daldinia and their bioactivities. Med. Res. 5, 210005. doi:10.21127/yaoyimr20210005
Liao, H.-X., Zheng, C.-J., Huang, G.-L., Mei, R.-Q., Nong, X.-H., Shao, T.-M., et al. (2019). Bioactive polyketide derivatives from the mangrove-derived fungus Daldinia eschscholtzii HJ004. J. Nat. Prod. 82, 2211–2219. doi:10.1021/acs.jnatprod.9b00241
Lin, L., Jiang, N., Wu, H., Mei, Y., Yang, J., and Tan, R. (2019). Cytotoxic and antibacterial polyketide-indole hybrids synthesized from indole-3-carbinol by Daldinia eschscholzii. Acta Pharm. Sin. B 9, 369–380. doi:10.1016/j.apsb.2018.09.011
Liu, L., Tang, M. C., and Tang, Y. (2019). Fungal highly reducing polyketide synthases biosynthesize salicylaldehydes that are precursors to epoxycyclohexenol natural products. J. Am. Chem. Soc. 141, 19538–19541. doi:10.1021/jacs.9b09669
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi:10.1093/oxfordjournals.molbev.a026092
Lorenz, N., Wilson, E. V., Machado, C., Schardl, C. L., and Tudzynski, P. (2007). Comparison of ergot alkaloid biosynthesis gene clusters in claviceps species indicates loss of late pathway steps in evolution of C. fusiformis. Appl. Environ. Microbiol. 73, 7185–7191. doi:10.1128/AEM.01040-07
Ma, P., Liu, E., Zhang, Z., Li, T., Zhou, Z., Yao, W., et al. (2023). Genetic variation in <i>ZmWAX2</i> confers maize resistance to Fusarium verticillioides. Plant Biotechnol. J. 21, 1812–1826. doi:10.1111/PBI.14093
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi:10.1093/molbev/msab199
Morales-Sánchez, V., Díaz, C. E., Trujillo, E., Olmeda, S. A., Valcarcel, F., Muñoz, R., et al. (2021). Bioactive metabolites from the endophytic fungus Aspergillus sp. SPH2. J. Fungi 7, 109. doi:10.3390/jof7020109
Navarro-Muñoz, J. C., Selem-Mojica, N., Mullowney, M. W., Kautsar, S. A., Tryon, J. H., Parkinson, E. I., et al. (2020). A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68. doi:10.1038/s41589-019-0400-9
Ng, K. P., Ngeow, Y. F., Yew, S. M., Hassan, H., Soo-Hoo, T. S., Na, S. L., et al. (2012). Draft genome sequence of Daldinia eschscholzii isolated from blood culture. Eukaryot. Cell 11, 703–704. doi:10.1128/EC.00074-12
Ni, Y., Liu, X., Simeneh, Z. M., Yang, M., and Li, R. (2023). Benchmarking of Nanopore R10.4 and R9.4.1 flow cells in single-cell whole-genome amplification and whole-genome shotgun sequencing. Comput. Struct. Biotechnol. J. 21, 2352–2364. doi:10.1016/j.csbj.2023.03.038
O’Donnell, K., and Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the FungusFusariumAre nonorthologous. Mol. Phylogenetics Evol. 7, 103–116. doi:10.1006/mpev.1996.0376
Palmer, J. M., and Stajich, J. (2020). Funannotate V1.8.1: eukaryotic genome annotation. Available at: https://funannotate.readthedocs.io/en/latest/.
Park, Y.-H., Chandra Mishra, R., Yoon, S., Kim, H., Park, C., Seo, S.-T., et al. (2019). Endophytic Trichoderma citrinoviride isolated from mountain-cultivated ginseng (Panax ginseng) has great potential as a biocontrol agent against ginseng pathogens. J. Ginseng Res. 43, 408–420. doi:10.1016/j.jgr.2018.03.002
Pažoutová, S., Follert, S., Bitzer, J., Keck, M., Surup, F, Šrůtka, P., et al. (2013). A new endophytic insect-associated Daldinia species, recognised from a comparison of secondary metabolite profiles and molecular phylogeny. Fungal Divers. doi:10.1007/s13225-013-0238-5
Qiao, K., Chooi, Y.-H., and Tang, Y. (2011). Identification and engineering of the cytochalasin gene cluster from Aspergillus clavatus NRRL 1. Metab. Eng. 13, 723–732. doi:10.1016/j.ymben.2011.09.008
Quang, D. N., Hashimoto, T., Tanaka, M., Baumgartner, M., Stadler, M., and Asakawa, Y. (2002). Constituents of the ascomycete Daldinia concentrica. J. Nat. Prod. doi:10.1021/np020301h
Rajasree, V., and Pugalendhi, L. (2021). “Breeding vegetables for nutritional security,” in Veganism - a fashion trend or food as a medicine (IntechOpen). doi:10.5772/intechopen.95349
Rambaut, A. (2014). FigTree v1.4.2. Available at: http://beast.bio.ed.ac.uk/figtree.
Santra, H. K., and Banerjee, D. (2022). Broad-spectrum antimicrobial action of cell-free culture extracts and volatile organic compounds produced by endophytic fungi curvularia eragrostidis. Front. Microbiol. 13, 920561. doi:10.3389/fmicb.2022.920561
Saraswathi, M., Meshram, S. H., Siva, B., Misra, S., and Suresh Babu, K. (2022). Isolation, purification and structural elucidation of Mellein from endophytic fungus Lasiodiplodia theobromae strain (SJF-1) and its broad-spectrum antimicrobial and pharmacological properties. Lett. Appl. Microbiol. 75, 1475–1485. doi:10.1111/lam.13813
Sarker, S. D., Nahar, L., and Kumarasamy, Y. (2007). Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods. doi:10.1016/j.ymeth.2007.01.006
Saxena, A., Raghuwanshi, R., Gupta, V. K., and Singh, H. B. (2016). Chilli anthracnose: the epidemiology and management. Front. Microbiol. 7, 1527. doi:10.3389/fmicb.2016.01527
Schmitz, H. X. (1930). A suggested toximetric method for wood preservatives. Industrial Eng. Chem. Anal. Ed. 2, 361–363. doi:10.1021/ac50072a004
Seblani, R., Keinath, A. P., and Munkvold, G. (2023). Gummy stem blight: one disease, three pathogens. Mol. Plant Pathol. 24, 825–837. doi:10.1111/mpp.13339
Shylaja, G., Sasikumar, K., and Sathiavelu, A. (2018). Antimycobacterial and anti-oxidant potential of the bioactive metabolite isolated from the endophytic fungus Daldinia eschscholtzii. Bangladesh J. Pharmacol. 13, 330–332. doi:10.3329/bjp.v13i4.38593
Sood, M., Kapoor, D., Kumar, V., Sheteiwy, M. S., Ramakrishnan, M., Landi, M., et al. (2020). Trichoderma: the “secrets” of a multitalented biocontrol agent. plants 9, 762. doi:10.3390/plants9060762
Srisuksam, C., Yodpanan, P., Suntivich, R., Tepboonrueng, P., Wattananukit, W., Jongsareejit, B., et al. (2022). The fungus Phoma multirostrata is a host-specific pathogen and a potential biocontrol agent for a broadleaf weed. Fungal Biol. 126, 162–173. doi:10.1016/j.funbio.2021.11.008
Stadler, M., Læssøe, T., Fournier, J., Decock, C., Schmieschek, B., Tichy, H.-V., et al. (2014). A polyphasic taxonomy of Daldinia (Xylariaceae)1. Stud. Mycol. 77, 1–143. doi:10.3114/sim0016
Tamizi, A.-A., Mat-Amin, N., Weaver, J. A., Olumakaiye, R. T., Akbar, M. A., Jin, S., et al. (2022). Genome sequencing and analysis of Trichoderma (hypocreaceae) isolates exhibiting antagonistic activity against the papaya dieback pathogen, erwinia mallotivora. J. Fungi 8, 246. doi:10.3390/jof8030246
Tanapichatsakul, C., Pansanit, A., Monggoot, S., Brooks, S., Prachya, S., Kittakoop, P., et al. (2020). Antifungal activity of 8-methoxynaphthalen-1-ol isolated from the endophytic fungus Diatrype palmicola MFLUCC 17-0313 against the plant pathogenic fungus Athelia rolfsii on tomatoes. PeerJ 8, e9103. doi:10.7717/peerj.9103
Tfelt-Hansen, P. (2000). Ergotamine in the acute treatment of migraine: a review and European consensus. Brain 123, 9–18. doi:10.1093/brain/123.1.9
Thambugala, K. M., Daranagama, D. A., Phillips, A. J. L., Kannangara, S. D., and Promputtha, I. (2020). Fungi vs fungi in biocontrol: an overview of fungal antagonists applied against fungal plant pathogens. Front. Cell Infect. Microbiol. 10, 604923. doi:10.3389/fcimb.2020.604923
Trendowski, M., Christen, T. D., Acquafondata, C., and Fondy, T. P. (2015). Effects of cytochalasin congeners, microtubule-directed agents, and doxorubicin alone or in combination against human ovarian carcinoma cell lines in vitro. BMC Cancer 15, 632. doi:10.1186/s12885-015-1619-9
Tudi, M., Daniel Ruan, H., Wang, L., Lyu, J., Sadler, R., Connell, D., et al. (2021). Agriculture development, pesticide application and its impact on the environment. Int. J. Environ. Res. Public Health 18, 1112. doi:10.3390/ijerph18031112
Vaser, R., Sović, I., Nagarajan, N., and Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi:10.1101/gr.214270.116
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi:10.1128/jb.172.8.4238-4246.1990
Wang, G., Fan, J., Zhang, W., Hua, C., Chen, C., Yan, W., et al. (2015). Polyketides from Mantis-associated fungus Daldinia eschscholzii IFB-TL01. Chem. Biodivers. 12, 1349–1355. doi:10.1002/cbdv.201400414
Wang, Z., Luo, W., Cheng, S., Zhang, H., Zong, J., and Zhang, Z. (2023). Ralstonia solanacearum – a soil borne hidden enemy of plants: research development in management strategies, their action mechanism and challenges. Front. Plant Sci. 14, 1141902. doi:10.3389/fpls.2023.1141902
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and application (New York: Academic Press Inc), 315–322.
Yan, L., Zhao, H., Zhao, X., Xu, X., Di, Y., Jiang, C., et al. (2018). Production of bioproducts by endophytic fungi: chemical ecology, biotechnological applications, bottlenecks, and solutions. Appl. Microbiol. Biotechnol. 102, 6279–6298. doi:10.1007/s00253-018-9101-7
Yang, L.-J., Liao, H.-X., Bai, M., Huang, G.-L., Luo, Y.-P., Niu, Y.-Y., et al. (2018). One new cytochalasin metabolite isolated from a mangrove-derived fungus Daldinia eschscholtzii HJ001. Nat. Prod. Res. 32, 208–213. doi:10.1080/14786419.2017.1346641
Zhang, A. H., Jiang, N., Wang, X. Q., and Tan, R. X. (2019a). Galewone, an anti-fibrotic polyketide from Daldinia eschscholzii with an undescribed carbon skeleton. Sci. Rep. 9, 14316. doi:10.1038/s41598-019-50868-9
Zhang, P., Zhou, S., Wang, G., An, Z., Liu, X., Li, K., et al. (2019b). Two transcription factors cooperatively regulate DHN melanin biosynthesis and development in Pestalotiopsis fici. Mol. Microbiol. 112, 649–666. doi:10.1111/MMI.14281
Zhou, Z.-Z., Zhu, H.-J., Yang, C.-L., Liu, Y.-J., Jiang, N., Xiao, Y.-S., et al. (2019). Dalestones A and B, two anti-inflammatory cyclopentenones from Daldinia eschscholzii with an edited strong promoter for the global regulator LaeA-like gene. Chin. J. Nat. Med. 17, 387–393. doi:10.1016/S1875-5364(19)30045-7
Keywords: secondary metabolites, fungal endophytes, biosynthetic gene clusters, microbial biocontrol agents, Daldinia, genome
Citation: Brooks S, Weaver JA, Klomchit A, Alharthi SA, Onlamun T, Nurani R, Vong TK, Alberti F and Greco C (2024) Unveiling the potential of Daldinia eschscholtzii MFLUCC 19-0629 through bioactivity and bioinformatics studies for enhanced sustainable agriculture production. Front. Chem. Biol 3:1362147. doi: 10.3389/fchbi.2024.1362147
Received: 27 December 2023; Accepted: 15 February 2024;
Published: 01 March 2024.
Edited by:
Natalia Miguel Vior, John Innes Centre, United KingdomReviewed by:
Wei Yan, Nanjing Agricultural University, ChinaCopyright © 2024 Brooks, Weaver, Klomchit, Alharthi, Onlamun, Nurani, Vong, Alberti and Greco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siraprapa Brooks, c2lyYXByYXBhLmJyb0BtZnUuYWMudGg=; Claudio Greco, Y2xhdWRpby5ncmVjb0Bzd2Fuc2VhLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.