
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Chem. Eng., 16 October 2023
Sec. Biochemical Engineering
Volume 5 - 2023 | https://doi.org/10.3389/fceng.2023.1256334
This article is part of the Research Topic3D-Engineered Organoids for Modelling Tissue Development and Precision MedicineView all 5 articles
Gastrointestinal organ failure, from congenital or postnatally acquired pathologies, is a major cause of death across countries of all income levels. Organoids and engineered tissues have been widely investigated as tools to model organ functions and treat pathologies. In this review we aim to describe the progress in human organoid engineering applied to the gastrointestinal tract (namely esophagus, stomach, and intestine). Starting from the onset of the organoid culture technique, we illustrate genetic engineering, stem cell niche engineering, bioprinting, and microfluidics approaches used to integrate mechano-physiological parameters with human organoids. Thanks to these improvements, organoid technology allows disease modelling of patient-specific pathologies, and personalized treatment screening, also offering a cell source for autologous transplantation. We further present an overview of the advances of tissue engineering in animal systems, concerning novel materials and scaffolds to be combined with a variety of cell types to reconstitute a viable surrogate for implantation. The effort in this field sets organoids as an important tool in personalized and regenerative medicine. Their application combined with the advances in tissue engineering holds great potential for translational application.
The gastrointestinal (GI) tract is the primary organ system responsible for digestive functions, which involve nutrient, water and electrolyte absorption, as well as the elimination of waste to maintain homeostasis. Numerous pathologies can disrupt the proper functioning of the human GI tract. These include strictures, esophageal atresia, Barret’s esophagus, gastroesophageal reflux disease, and inflammatory bowel disease. The need for more accessible human models to study these conditions is growing, whether it be for discovering precision medicine approaches or creating disease models to aid in their understanding (Figure 1).
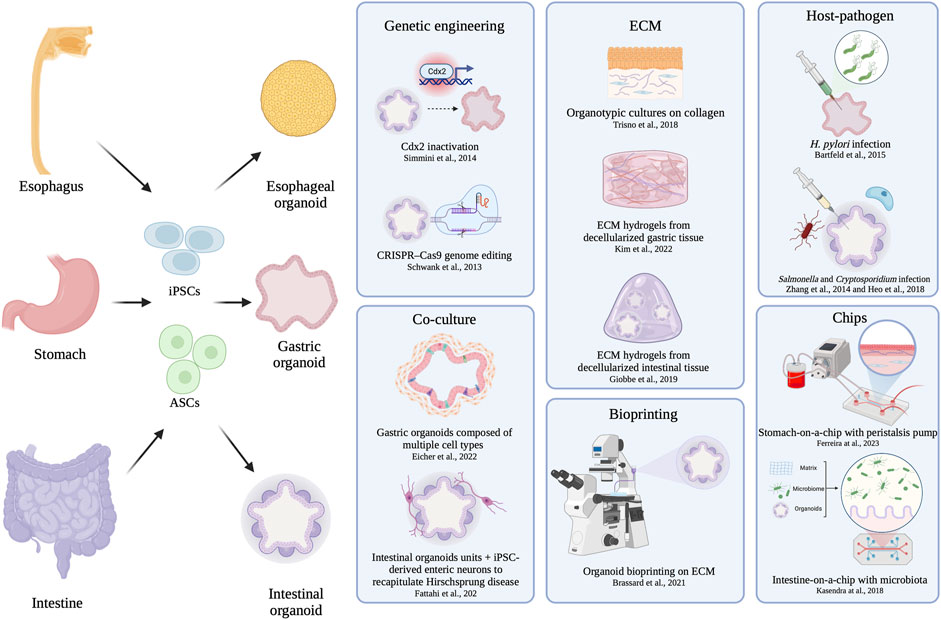
FIGURE 1. Schematic overview of organoid and tissue engineering. Schematics of organoid derivation: after cell derivation from patient, the obtained organoids can be engineered by genetic approaches, niche modification introducing other cell type, ECM modulation, niche modification by pathogen interaction, or more complicated systems involving chips and bioprinting.
In this scenario, organoids are considered a game changing technology for the study of human physiology and disease, providing systems to investigate medical research, drug screening/development, personalized medicine, generating interest both in academia and industry.
In order to replicate the functions of the various organs within the (GI) tract in vitro, it is crucial to accurately reproduce the complexity of the epithelial cell types, with organoids being a promising solution to this. Significant progress has been made in this field over the past decade through the use of three-dimensional (3D) cell culture. The initial breakthroughs occurred in the epithelium of the intestine, where multipotent crypt stem cells were isolated from patient biopsies. These stem cells can be expanded and differentiated into all different cell types found in the intestinal epithelium, when cultured in 3D on an appropriate extracellular matrix that mimics the natural microenvironment (Barker et al., 2007; Sato et al., 2009). Since then, additional compartments of the GI tract have been investigated to establish in vitro cultures derived from primary adult stem cells (Barker et al., 2010; Sato et al., 2011). However, organoids often lack specialized cell types and fail to recapitulate the complexity of native organs (Jensen and Little, 2023). This deficiency comes from the absence of key components such as mesenchymal compartment, vascularization, and microbiome. An alternative approach involves the guided differentiation of human pluripotent stem cells (PSC), either induced pluripotent stem cells (iPSC) or embryonic stem cells (ESC). Endoderm-derived epithelial organoids can be obtained by culturing PSCs in 3D matrices and supplying the appropriate biochemical stimuli. By harnessing the natural molecular patterning that naturally happens along the rostro-caudal axis of the GI tract, it is possible to differentiate PSC-derived organoids into specific cell-types that recapitulate the unique epithelial composition of each organ (Spence et al., 2010; McCracken et al., 2014; 2017; Múnera et al., 2017; Sinagoga et al., 2018; Broda et al., 2019). In contrast to adult stem cell (ASC) derived-organoids, PSC-organoids possess the advantage of incorporating a mesenchymal layer, which facilitates the process of differentiation. However, once established, PSC-derived organoids pose challenges in terms of propagation (Kim et al., 2020a), and their development is reliant of the stochastic nature of the differentiation process. Consequently, this can result in organoid heterogeneity (Brassard and Lutolf, 2019).
Discovering the optimal organoid culture conditions and engineering techniques is imperative due to the increasingly prominent role that organoids are acquiring in tissue engineering. To this extent, substantial efforts have been dedicated to developing alternative scaffolds for the replacement of damaged tissues. Numerous biomaterials have been identified, including various types of three-dimensional matrices that mimic the extracellular matrix. Additionally, the use of decellularized scaffolds has been introduced. The integration of these structures with organoids has been experimentally tested in several animal models, paving the way to a new organ source (Figure 2).
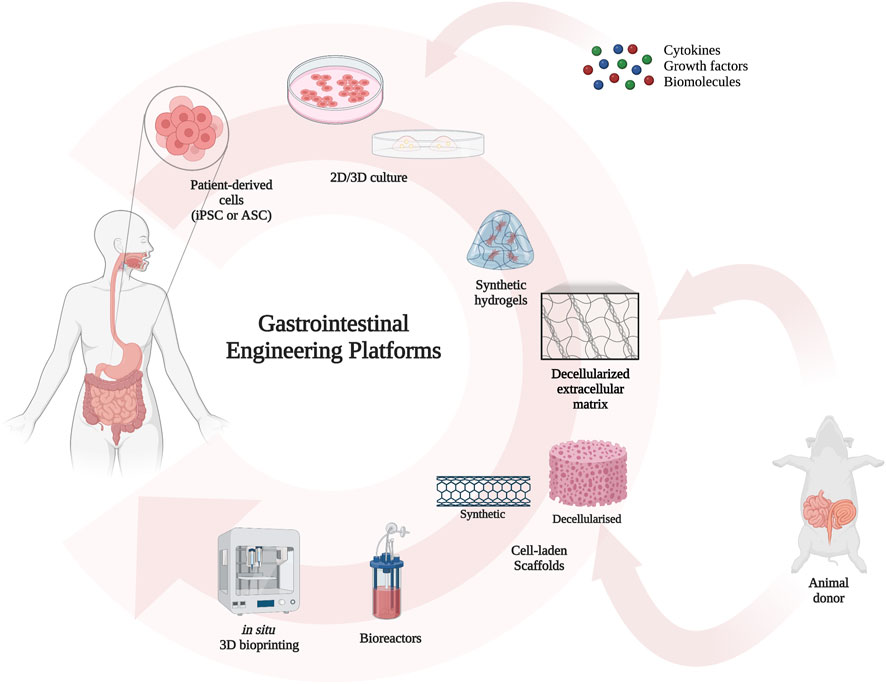
FIGURE 2. Schematics of tissue engineering workflow: after cell derivation from the patient itself, the cells are grown into organoids which can be engineered as mentioned. Matrices and decellularized scaffolds can be obtained from appropriate animals. The repopulation and growth in bioreactor can produce a source for organ transplantation.
For these reasons, this review aims to provide an overview of the available approaches for engineer GI tract organoids. Specifically, we discuss the engineering details for esophagus, stomach, and intestine. Starting from genetic engineering, we move to dissecting niche modifications, which span from medium composition to ECM composition, and microarchitecture bioprinting. Furthermore, we delve into the advancements made in microfluidics devices and organ-on-a-chip systems. Additionally, due to the new frontier of organoids as a cell source for tissue engineering, a critical analysis of the most relevant tissue engineering publications from recent years will be presented to illustrate the emerging role of organoids in the field.
The first section of this review discusses the advancement of esophageal engineering. Esophageal diseases, such as gastroesophageal reflux disease (GERD) and eosinophilic esophagitis (EoE), are characterized by dysfunctional esophageal epithelium. It is possible to derive 3D organoids from stem cells isolated from esophageal biopsies (Busslinger et al., 2021) (Table 1). There is not yet consensus on the specific molecular inputs in the culture medium required to simulate the original niche factors. Consequently, further research is needed to optimize this organoid model.
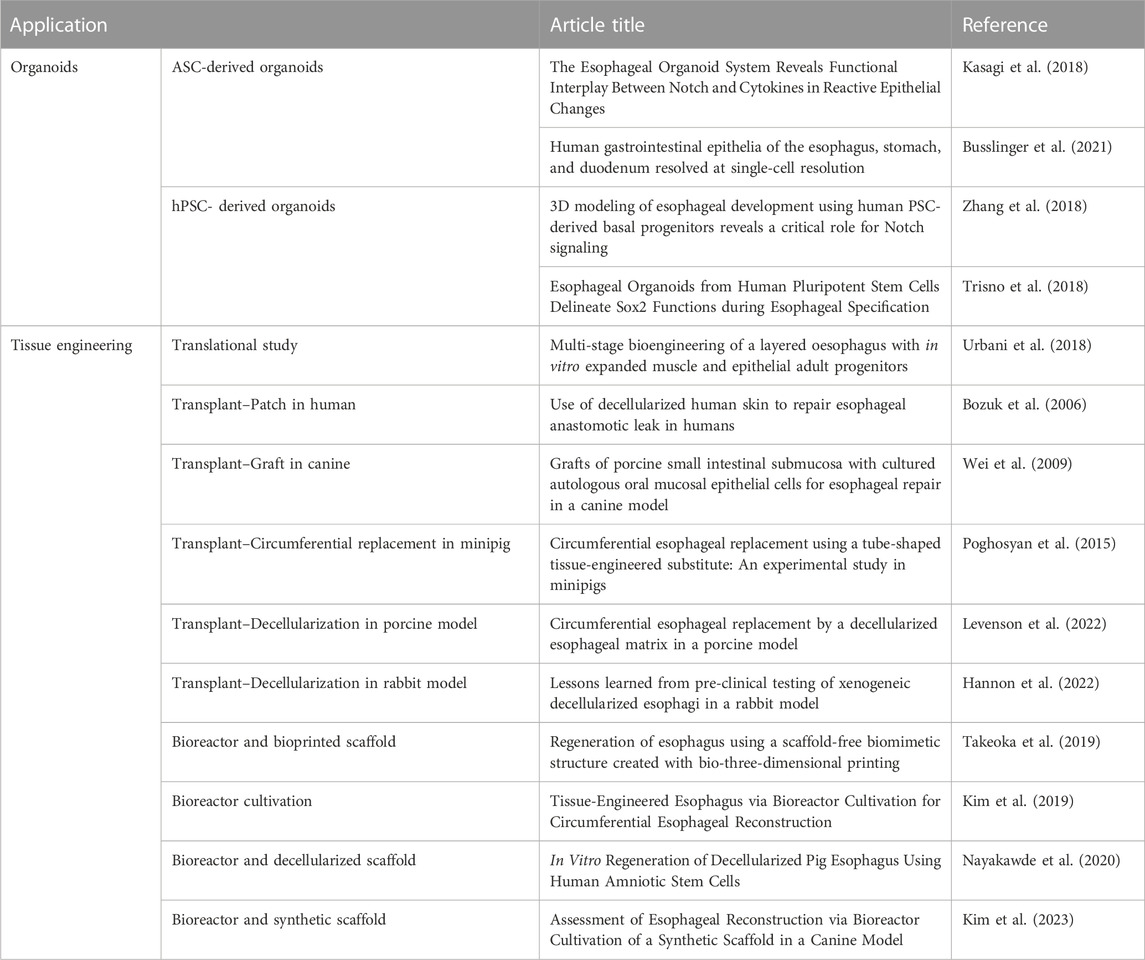
TABLE 1. Summary of esophageal derived organoid models and advances in esophageal tissue engineering.
To date, patient-derived esophageal organoids have been employed to investigate Notch-dependent regulatory mechanisms by either genetic or pharmacological manipulation, allowing for comparison between healthy and EoE conditions (Kasagi et al., 2018). On the other hand, esophageal epithelial progenitors (EPCs) have been derived from hPSCs, to be used to model and study the regulatory mechanism underlying the human esophagus development (Zhang et al., 2018). Another model of esophageal development was established on hPSCs by modulating BMP, Wnt, and RA signaling pathways. This approach allowed the patterning of definitive endoderm into foregut, anterior foregut, and dorsal anterior foregut. The latter, in a 3D matrix formed human esophageal organoids (HEOs). HEOs cells could be transitioned into two-dimensional cultures and grown as esophageal organotypic rafts, which could develop into a stratified squamous epithelium (Trisno et al., 2018).
While these newly developed HEO systems are well-suited for studying the esophageal epithelium, they lack other primary components critical for normal esophageal function. As a result, the range of diseases that can be modelled using these systems, such as motility defects or EoE, is limited. To this purpose, further investigation is required on human systems, drawing insights from preliminary studies conducted on animal models. Combining epithelial organoids with tissue engineering novel techniques would lead to a more comprehensive model.
In recent years, tissue engineering has emerged as a valuable approach for constructing esophageal scaffolds, providing a new method for esophageal repair and reconstruction. Several technologies have been developed to introduce either patches or whole-ring organ substitutes. The first reported application in humans involved the repair of a thoracic anastomotic esophageal leak using Alloderm, a decellularized human skin product (Bozuk et al., 2006). A successful animal approach involved the use of patches of acellular porcine small intestinal submucosa employed for esophagoplasticity. In canine model, it was demonstrated that combining such patches with a cellular component (autologous oral epithelial cells), resulted in superior reconstruction of esophageal defects (Wei et al., 2009). While tissue patches are beneficial for defect correction, there are many conditions that require full circumference transplantation. In a porcine model, a circumferential replacement of the cervical esophagus was achieved using a tubularized small intestine acellular matrix, recellularized with autologous skeletal myoblasts. This structure was further covered with a human amniotic membrane, previously seeded with autologous oral epithelial cells (Poghosyan et al., 2015). In another study conducted on a porcine model, allogenic decellularized esophagus recellularized with autologous bone marrow mesenchymal stromal cells was used for transplantation (Levenson et al., 2022). Both these studies used the great momentum as a natural in vivo bioreactor for cell maturation. In a rabbit model, esophageal replacement was achieved by using decellularized porcine esophagus with a vascularizing muscle flap (Hannon et al., 2022). The engineered organ was grown in static chamber to allow cell maturation. Although the anastomosis was successful and early vascularization was present, the long-term survival was limited due to the fragility of the animal model. Prior work has demonstrated the feasibility of transplanting circumferential grafts using repopulated autologous decellularized esophagus in rats. Here, muscle progenitors and fibroblasts were used as cellular components (Urbani et al., 2018). The esophageal muscles achieved organized maturation after functional integration with neural crest stem cells and dynamic culture in a bioreactor. Perfusion-rotation bioreactors have been shown to be the most effective in vitro method for recellularizing decellularized esophagus (Nayakawde et al., 2020; Kim et al., 2023).
Furthermore, bioprinting techniques have been tested in rat models. For instance, three-dimensional printed esophageal grafts were used for muscle regeneration and reepithelialisation of circumferential esophageal defects, while being grown in a bioreactor (Kim et al., 2019). The technology advanced further when a scaffold-free structure with a mixture of cell types was developed using three-dimensional bioprinting, which was transplanted in rat (Takeoka et al., 2019).
The second section of our review focuses on the innovation in gastric engineering. The stomach is a vital organ which plays a crucial role in human physiology. Its epithelium is divided into three regions: fundus and corpus, comprised of acid-secreting parietal cells, and antrum, containing hormone-secreting cells (McCracken et al., 2017). Three-dimensional (3D) gastric organoid models were developed to recapitulate the mucosa cell-type composition and architecture, providing a promising tool to study gastric tissue regeneration, infections, and cancer (Table 2).
Initially, gastric organoids were generated from Lgr5+ adult stem cells (ASCs) from antrum glands of a mouse stomach (Barker et al., 2010). These cells were embedded in a laminin-rich matrix (Matrigel) and cultured with a gastric-specific growth media, resulting in the formation of complex 3D self-organized structures that resembled the stomach epithelium. This pioneering study served as basis for the subsequent generation of gastric organoids from human stomach tissue (Bartfeld et al., 2015).
The ASC-derived gastric organoids consist of gastric epithelial cells only. In contrast, a human pluripotent stem cell (hPSC) system was used for the generation of gastric organoids containing both epithelial and mesenchymal stem cells (McCracken et al., 2014). The presence of a mesenchymal compartment allows for its interaction with gastric epithelial, favoring the differentiation of mature cell types.
However, these organoid models lack crucial specialized cell types such as neural, muscle and immune cells, thus limiting their ability to fully replicate native gastric development and functionality. Consequently, significant effort was dedicated to integrating other cell types to engineer a more comprehensive gastric niche. A recent advancement was achieved by co-culturing antral and fundic gastric organoids with human PSC-derived splanchnic mesenchyme and enteric neural crest cells (ENCCs). The so engineered gastric tissue model contained cells from all 3 germ layers and formed organoids containing innervated epithelial glands and several layers of functional smooth muscle, enabling the modeling of muscle contraction (Eicher et al., 2022).The organoid units can be engineered to modify their cell-cell interactions, the surrounding micro-environment, and the intrinsic properties of the different cell types, enabling better modeling of the tissue. One approach is the genetic manipulation of organoids. Upon inactivation of the intestinal-specific transcription factor Cdx2 in human intestinal organoids, a loss of intestinal identity was observed, with organoids giving rise to gastric pyloric organoids (Simmini et al., 2014). Niche engineering is another important strategy for evaluating the self-renewal and differentiation capabilities of the gastric organoids. Several tissue ECM hydrogels were investigated as an alternative to Matrigel. ECM hydrogels were derived from various tissues sources, including stomach and small intestine. The study demonstrated that tissue ECM hydrogels are a viable option for GI organoid engineering and disease modelling (Kim et al., 2022).
Alternatively, human gastric organoids have been used to study host-pathogen interactions and gain insights into the mechanisms of pathogens colonization and disease development. For example, gastric organoids were used to investigate H. pylori bacterial infection, which is associated with gastric cancer (Schlaermann et al., 2016). Microinjection of H. pylori into hASC-derived organoid’s lumen resulted in increased expression of inflammatory genes, indicating an immune response activation (Bartfeld et al., 2015). This system was also used to unveil the tropism of H. pylori, demonstrating its preferential binding to differentiated pit cells (DPCs) in hASC-derived GO through a chemotactic attraction towards urea (Aguilar et al., 2022). New insights into the mechanisms involved in infection will offer new treatment options. Therefore, these organoid models hold important implications for the development of novel treatments for gastric ulcers and stomach cancers caused by H. pylori infection.
Furthermore, gastric organoids have been used to investigate the susceptibility of the stomach to SARS-Cov-2 infection. Researchers investigated the infection of fetal, pediatric and adult gastric organoids with SARS-CoV-2, revealing specific gene expression changes, including the upregulation of inflammatory genes, indicating that the stomach serves as an additional site of SARS-CoV-2 replication (Giobbe et al., 2021).
However, such in vitro gastric organoid models lack the presence of physiological stimuli, hence not being able to fully recapitulate the biomechanical and physiological features of the native organ. To address this challenge, microfluidics platforms have emerged as valuable tools for growing gastric organoids, generating “organ-on-a-chip” technologies that better simulate the gastric physiological microenvironment of the stomach. A “stomach on-a-chip” (SoC) system was used to introduce luminal flow through hPSC-derived antral gastric organoids. This bioengineered platform incorporated a membrane within the chip, to create peristaltic-like contractions, and fibroblasts were incorporated to simulate the biochemical environment (Kug Lee et al., 2018). Likewise, Ferreira and others developed a SoC model to encase layers of gastric fibroblasts and epithelial cells, exposing them to peristalsis-like movement and intraluminal flow (Ferreira et al., 2023). The bioengineered device mimicked the architecture and function of the native stomach’s innermost mucosa layers, providing a more physiologically relevant environment for studying gastric physiology. Another study by Jeong and co-workers described a micro-physiological system that combined human antral organoids and gastric mesenchymal stromal cells (gMSCs) to study the dynamic mucosal mechanism of the stomach. The constant fluid flow in this system enhanced epithelial-mesenchymal interactions, creating a functional gastric mucosal barrier (Jeong et al., 2022).
The integration of microfluidics systems with organoid technologies holds great promise for developing optimal in vitro models of the stomach, to be grown in a controlled and reproducible environment.
The tissue engineering of the entire stomach is still limited, given its shape and scale. For this reason, current studies focus on generating tissue patches. These grafts have been implanted into preclinical models to investigate their effectiveness and healing properties in treating stomach defects.
One promising approach for delivering autologous stem cells involves the use of hydrogels, which aim to promote healing in wounded areas. For example, a 3D alginate hydrogel was used to develop a gastric mucosa model containing human gastric mesenchymal stem cells (MSCs). Gastric epithelial cells were seeded on top of this cell-containing hydrogel, enabling the reproduction of the physiological conditions of the gastric barrier. The stomach fibroblasts appeared to maintain a mucosal architecture and produce extracellular matrix in this model (Lourenço et al., 2018). Additionally, murine MSCs were seeded onto alginate-gelatin scaffolds and placed onto the stomach lumen of a gastroparesis mouse model. The MSCs successfully penetrated the intramuscular region, which is typically depleted of Interstitial Cells of Cajal (ICCs) during gastroparesis. Furthermore, the MSCs differentiated into an ICC phenotype (Joddar et al., 2018). However, safe and non-invasive methods to deliver the hydrogels still require further exploration.
Studies have also focused on developing protocols for decellularizing regions of the gastrointestinal tract. Zambaiti and others produced decellularized rat gastric tissue with preserved micro-structure, serving as a natural scaffold that enabled new cell growth while maintaining a native ECM structure (Zambaiti et al., 2019). Moreover, decellularized scaffolds derived from porcine Small Intestinal Submucosa (SIS) were combined with MSCs to regenerate stomach defects in rat models, resulting in improved repopulation of the seeded scaffold, even when cells were not directly derived from these progenitors (Nakatsu et al., 2015).
Furthermore, alternative engineering technologies have been employed to study stomach physiology in vitro. Habib and co-workers designed a continuous bioreactor system composed of multiple compartments to mimic the digestive tract and facilitate the growth of different microbial communities. This dynamic model can maintain a complex yet controlled microbial environment to investigate the bacterial response to specific metabolites naturally secreted by the host into the gut (Habib et al., 2021).
In situ bioprinting technique has been used to deposit bioink layers directly onto a defective site to create or repair living tissues. A micro printer connected to an endoscope could reach the inside of a synthetic stomach model and perform 3D in situ bioprinting on the injury site. Bioprinting of two-layer tissue scaffolds was carried out using bioinks containing a gelatin-alginate hydrogel mix, along with human gastric epithelial and smooth muscle cells, to simulate gastric ulcer injury repair (Zhao and Xu, 2020).
This third and last section of the review focuses on the advancement of intestinal engineering. The large bowel epithelium is composed of a simple columnar epithelium with a thin brush border, arranged throughout tube-like glands. On the other hand, the small intestine mucosa has also a simple columnar epithelium, but it is organized in the typical crypt-villus architecture. Overall, the whole intestinal mucosa is characterized by an extensive cell-type complexity, essential for the organ to carry out its functions effectively. However, due to the variegated nature of the epithelial intestinal cell-type, it is challenging to replicate it in vitro. As previously mentioned, the groundbreaking research conducted by Clevers Lab played a crucial role in understanding cell-type differentiation within the crypt-villus architecture. This research identified Lgr5+ stem cells as key players responsible for epithelial regeneration (Barker et al., 2007). This discovery paved the way for the development of small intestinal organoids from patient-derived ASCs, capable of mimicking the crypt-villus pattern in vitro (Sato et al., 2009; Sato et al., 2011). Similarly, to what previously mentioned for the other GI tract organs, investigation on hPSC-derived intestinal organoids has been performed, which were proved to be able to engraft in vivo (Watson et al., 2014). This approach has become an alternative when access to gut tissue is somewhat limited. This method promotes the presence of mesenchymal cells that provide support within the niche (Spence et al., 2010; Múnera et al., 2017; Takahashi et al., 2018).
The use of patient derived intestinal organoids has enabled the modelling of diseases in vitro (Table 3). This approach allowed researchers to study and understand various pathological conditions in vitro, opening the possibility of functional repair to correct genetic issues within these diseases. As an example, intestinal primary organoids derived from cystic fibrosis patients were engineered with CRISPR/Cas9 technology to correct the genetic abnormality. By comparing the intestinal epithelium between healthy and diseased organoids, researchers were able to investigate their differences (Schwank et al., 2013).
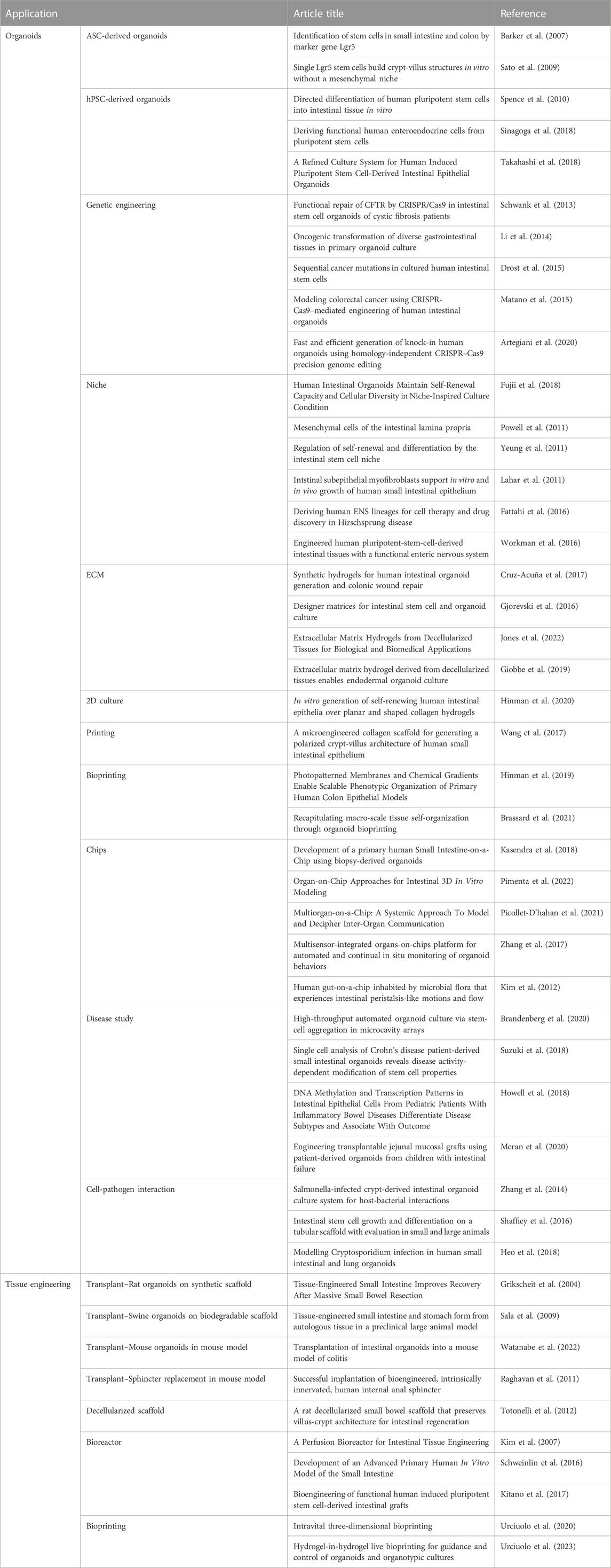
TABLE 3. Summary of intestinal derived organoid models and advances in intestinal tissue engineering.
Given the significant incidence of intestinal and colorectal cancer, extensive research has focused on exploring the establishment of oncogenic transformation. CRISPR/Cas9 has been instrumental, as it allowed for the sequential introduction of mutations to simulate the process of cancer development (Li et al., 2014; Drost et al., 2015; Matano et al., 2015). Homology-independent CRISPR/Cas9 has been used to generate knock-in human organoids (Artegiani et al., 2020). This technique offered further opportunities for studying specific genetic alterations and their effects in organoids.
As part of the engineering process, organoids need to be modulated by acting on the niche in which they grow. Attempt to replicate the paracrine effect exerted by the mesenchymal cells in the natural niche proved to be effective (Sato et al., 2009; Fujii et al., 2018).
The intestinal lamina propria comprises various types of mesenchymal cells and primary intestinal organoids can be co-cultured with stromal cells to better mimic the in vivo environment (Powell et al., 2011). Intestinal subepithelial myofibroblasts (ISEMFs) provided pivotal cues to the stem cell niche by secreting growth factors (Yeung et al., 2011). By co-culturing ASC-derived intestinal organoids with ISEMFs, it was possible to grow organoids without the presence of some growth factors in the medium. This co-culture approach has been successful in promoting engraftment and proliferation upon transplantation (Lahar et al., 2011).
The enteric nervous system (ENS) is another essential component involved in intestinal functions. However, both ASC-derived and PSC-derived intestinal organoids lack ENS direction. Efforts have been undertaken to differentiate PSCs into vagal neural crest cells, to be integrated into the organoids, thereby improving their functionality (Fattahi et al., 2016; Workman et al., 2016).
In addition to niche modulation, the choice of matrix used for the 3D culture of organoids also plays a significant role in providing stimuli. To avoid tumor-derived matrices, efforts have been made to develop biocompatible synthetic hydrogels to support the cultures. One example is the four-armed maleimide-terminated poly (ethylene glycol) macromer, which was shown to support robust and reproducible in vitro growth of human intestinal organoids (Gjorevski et al., 2016; Cruz-Acuña et al., 2017). As an alternative, for the establishment of a Good-Manufacturing-Practice (GMP)-compliant system for organoids expansion, ECM hydrogels derived from decellularized tissues can provide an environment capable of directing cell growth. When generated from porcine small intestine mucosa, these hydrogels proved to possess the biochemical signature of the intestinal-specific ECM (Giobbe et al., 2019; Jones et al., 2022).
Nevertheless, while current organoid models have made significant progress in replicating aspects of intestinal architecture and lineage compartmentalization, they still do not fully mimic the complexity of the in vivo environment. Therefore, researchers have explored various strategies to enhance the spatial organization and functionality of this model. One approach involved the use of micropatterned collagen scaffolds with suitable stiffness, to generate in vitro self-renewing human small intestinal epithelium crypt-villus architecture. This system allowed for the establishment of proper supply of chemical gradients, which in turn led to appropriate cell compartmentalization along the axis (Wang et al., 2017; Hinman et al., 2020). Another explored solution was the generation of a standardized well-plate platform with photopatterned porous membrane for the administration of an array of chemical gradients along the intestinal crypt axis (Hinman et al., 2019). By using tissue engineering and the intrinsic self-organization properties of cells, intestinal stem cells formed tube-shaped epithelia with an accessible lumen and similar spatial arrangement of crypt- and villus-like domains (Nikolaev et al., 2020). To further advance the spatial organization of human organoids, merging biofabrication techniques with organoid technology has shown promising data using mouse cells. The proposed system utilizes 3D bioprinting to control the spatial deposition of cells at microscale, with organoid-forming stem cells used as building blocks that can be directly deposited into an ECM conducive to their spontaneous self-organization (Brassard et al., 2021).
The use of organs-on-a-chip systems provides another system for integrating different intestinal cell types and architectural complexities. Organs-on-a-chip enabled the study of cell-cell interaction within the same organ (Kasendra et al., 2018; Pimenta et al., 2022). Moreover, they were used to simulate crosstalk between cells from different organs, mimicking more complex interactions (Picollet-D’hahan et al., 2021). The advantage of organ-on-a-chip systems is their ability to incorporate specific sensors within the chip itself, enabling measurements of physiological parameters otherwise challenging to achieve (Zhang et al., 2017). Organ-on-a-chip is also a valuable tool to study the physiological microbiota or host-pathogen interaction. This technique was used for recreating specific scenarios such as oxygen gradients (Kim et al., 2020b), or simulate peristaltic movements (Kim et al., 2012), thus enabling investigations of these factors on cell behavior.
Besides, simpler systems that utilize organoids alone can be employed to study specific cell features. For instance, coculturing intestinal organoids with commensal bacteria such as Lactobacillus showed an increased organoid proliferation and differentiation into Paneth cells (Shaffiey et al., 2016). In addition, pathogens such as Salmonella (Zhang et al., 2014), or Cryptosporidium (Heo et al., 2018) were incorporated into intestinal organoids to investigate their interaction with the intestinal epithelium. Overall, organ-on-a-chip systems offer a versatile platform for studying cell-cell interactions, complex organ interactions, physiological parameters, microbiota-host interactions, and pathogen interaction, providing valuable information on human intestinal biology and disease.
Intestinal tissue engineering for transplantation purposes has been largely investigated. Early trials involved applying organoid units onto a polymer scaffold for massive small bowel resection in rats (Grikscheit et al., 2004). Intestinal organoids were also applied onto biodegradable scaffold tubes in the swine model (Sala et al., 2009). Subsequently, the use of decellularized scaffold gained attention. Decellularized small bowel scaffolds preserved villus-crypt architecture and was successfully applied in rat and mice (Totonelli et al., 2012; Watanabe et al., 2022). Although human applications for transplantation are still at early stages, human tissues have been used in animal model research. For example, human internal anal sphincter innervated with fetal enteric neurons was successfully implanted in mice (Raghavan et al., 2011). Also, patient-derived jejunal organoids were seeded onto decellularized human intestinal (small intestine or colon) matrix with intact nanotopography, resulting in grafts exhibiting physiological jejunal functions and forming a lumen after transplantation into subcutaneous pockets in mice (Meran et al., 2020). These tissue structures were cultured in costume-designed bioreactors, which allowed the perfusion of the culture medium through the engineered tissue, either the whole cylinder (Kim et al., 2007; Schweinlin et al., 2016) or graft (Kitano et al., 2017).
Recent advancements focused on fabricating 3D functional tissues directly in live animals using minimally invasive approaches. Cell-laden photosensitive polymer hydrogels were bioprinted across and within the tissue of live mice (Urciuolo et al., 2020). This material proved to be suitable for intestinal organoid guidance and differentiation into crypt/villus domain (Urciuolo et al., 2023).
These developments highlight progress made in creating complex tissue structure and functional tissue transplantation purposes, with the ultimate goal of providing viable solutional for intestinal repair and replacement.
Gastrointestinal organoid engineering holds great promise in the fields of disease modelling and personalized medicine. The discussed review provides an overview of the available approaches for engineering GI tract, focusing on esophageal, gastric, and intestinal organoids systems. The ability to model a specific patient’s disease, considering their unique genetic background, opens numerous possibilities, from drug screening to innovative personalized therapeutic approaches. At the macroscale level, tissue engineering allows the autologous engineering and transplantation of artificially produced grafts.
Various studies have used tissue engineering techniques for esophageal repair and reconstruction in animal models (Wei et al., 2009; Poghosyan et al., 2015; Urbani et al., 2017; Nayakawde et al., 2020; Levenson et al., 2022; Kim et al., 2023). A further improvement would be the use of esophageal organoids (either derived from biopsies or iPSCs) in tissue engineering. This would offer clear advantages for the treatment of diseases requiring esophagus reconstruction, such as esophageal atresia, or epidermolysis bullosa, by taking advantage of a gene therapy approach (Shacham-Silverberg and Wells, 2020).
Similarly, gastric organoids are particularly relevant for investigating genetic diseases. Bioengineered patient-derived stomach organoids allow for unique profiling and mechanistic studies of patient pathophysiology in vitro. For instance, gastric and intestinal organoids generated from iPSCs derived from two patients with unique homozygous mutations in PDX1 were used to identify the multiple GI pathologies that were caused by the PDX1 mutations, and thus appropriately tailor the patients’ clinical needs according to the pathologies observed (Krishnamurthy et al., 2022). Moreover, extreme conditions of these pathologies showing non-functional epithelium might benefit from organ engineering.
Gastric tissue engineering holds great potential in cases where stomach tissue is lost due to trauma, resection (following tumor or bariatric surgery) or congenital conditions such as microgastria. By using synthetic hydrogels or decellularized tissue-derived -ECM hydrogels and scaffolds, researchers aim to create culture systems that are more biologically relevant and can better mimic the natural environment of the intestines, enhancing the growth and functionality of organoids (Giobbe et al., 2019).
Intestinal organoid engineering is comparably beneficial for a multitude of pathologies. For instance, micro-engineered cell culture devices allow high-throughput screening of anticancer drugs using GI organoids cultured in suspension in the absence of matrix within a polymer-hydrogel substrate (Brandenberg et al., 2020), enabling personalized treatments. Patient-derived organoids can also recapitulate congenital or genetic diseases in vitro, facilitating the study of biological features of the pathology or evaluating therapeutic approaches. Examples include Crohn’s disease (CD) patient-derived small intestinal organoids, which reveal modifications of stem cell properties due to the inflammatory environment (Suzuki et al., 2018). Alternatively, intestinal bowel disease (IBD) patient-derived intestinal organoids provided information on the pathological state, and evidence of transcriptional and methylation alterations (Howell et al., 2018).
Advancements in biofabrication and spatial control techniques have the potential to enhance the structural and functional complexity of human organoids, leading to better representation of in vivo tissue architecture and functionality. Organ-on-a-chip platforms offer opportunities to study the complex interplay between different cell types, mechanical forces, and physiological stimuli, resulting in more accurate and physiologically relevant models for tissue regeneration research. On another level, the GI tract is colonized by various microbes, each with its specific microbiota composition. The study of the interaction between these microbes (either commensal or pathogens) and epithelial cells provides insight into the patient’s metabolic features.
Although there are still challenges to overcome, such complexity, reproducibility, and scalability to maximize their translational relevance, their potential is clear. The combination of organoid engineering with tissue engineering will pave the way to future personalized medicine. This will be achieved by the combination of the two elements discussed in this review. On one hand, the production of autologous organoid either from hPSC or ASC, which can be engineered to correct eventual anomalies and be used to generate an epithelium with mature and differentiated cell types. On the other hand, these cells would be seeded on a support of appropriate size, which could be either artificial material, tissue derived ECM, or decellularized scaffolds. The consequent output will be a new source of biological material for autologous implantation, according to the patient’s needs.
GB: Data curation, Investigation, Writing–original draft. BF: Data curation, Writing–original draft. PD: Funding acquisition, Resources, Writing–review and editing. Giovanni GG: Conceptualization, Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the OAK Foundation Award W1095/OCAY-14-191; The Great Ormond Street Hospital (GOSH) Children’s Charity; and the National Institute for Health Research Great Ormond Street Hospital Biomedical Research Centre (NIHR GOSH BRC). GB is supported by the OAK Foundation. PD is supported by National Institute for Health Research Professorship and the GOSH Children’s Charity. GG is supported by the NIHR GOSH BRC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author PC declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Aguilar, C., Pauzuolis, M., Pompaiah, M., Vafadarnejad, E., Arampatzi, P., Fischer, M., et al. (2022). Helicobacter pylori shows tropism to gastric differentiated pit cells dependent on urea chemotaxis. Nat. Commun. 13, 5878. doi:10.1038/S41467-022-33165-4
Artegiani, B., Hendriks, D., Beumer, J., Kok, R., Zheng, X., Joore, I., et al. (2020). Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 22 (3), 321–331. doi:10.1038/s41556-020-0472-5
Barker, N., Huch, M., Kujala, P., van de Wetering, M., Snippert, H. J., van Es, J. H., et al. (2010). Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. doi:10.1016/j.stem.2009.11.013
Barker, N., Van Es, J. H., Kuipers, J., Kujala, P., Van Den Born, M., Cozijnsen, M., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. doi:10.1038/NATURE06196
Bartfeld, S., Bayram, T., Van De Wetering, M., Huch, M., Begthel, H., Kujala, P., et al. (2015). In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136.e6. doi:10.1053/j.gastro.2014.09.042
Bozuk, M. I., Fearing, N. M., and Leggett, P. L. (2006). Use of decellularized human skin to repair esophageal anastomotic leak in humans. JSLS 10 (1), 83–85.
Brandenberg, N., Hoehnel, S., Kuttler, F., Homicsko, K., Ceroni, C., Ringel, T., et al. (2020). High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 4, 863–874. doi:10.1038/S41551-020-0565-2
Brassard, J. A., and Lutolf, M. P. (2019). Engineering stem cell self-organization to build better organoids. Cell Stem Cell 24, 860–876. doi:10.1016/J.STEM.2019.05.005
Brassard, J. A., Nikolaev, M., Hübscher, T., Hofer, M., and Lutolf, M. P. (2021). Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater 20, 22–29. doi:10.1038/S41563-020-00803-5
Broda, T. R., McCracken, K. W., and Wells, J. M. (2019). Generation of human antral and fundic gastric organoids from pluripotent stem cells. Nat. Protoc. 14, 28–50. doi:10.1038/S41596-018-0080-Z
Busslinger, G. A., Weusten, B. L. A., Bogte, A., Begthel, H., Brosens, L. A. A., and Clevers, H. (2021). Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Rep. 34, 108819. doi:10.1016/j.celrep.2021.108819
Cruz-Acuña, R., Quirós, M., Farkas, A. E., Dedhia, P. H., Huang, S., Siuda, D., et al. (2017). Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326–1335. doi:10.1038/ncb3632
Drost, J., Van Jaarsveld, R. H., Ponsioen, B., Zimberlin, C., Van Boxtel, R., Buijs, A., et al. (2015). Sequential cancer mutations in cultured human intestinal stem cells. Nature 521 (7550), 43–47. doi:10.1038/nature14415
Eicher, A. K., Kechele, D. O., Sundaram, N., Berns, H. M., Poling, H. M., Haines, L. E., et al. (2022). Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell Stem Cell 29, 36–51.e6. doi:10.1016/J.STEM.2021.10.010
Fattahi, F., Steinbeck, J. A., Kriks, S., Tchieu, J., Zimmer, B., Kishinevsky, S., et al. (2016). Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 531, 105–109. doi:10.1038/NATURE16951
Ferreira, D. A., Conde, J. P., Rothbauer, M., Ertl, P., Granja, P. L., and Oliveira, C. (2023). Bioinspired human stomach-on-a-chip with in vivo like function and architecture. Lab. Chip 23, 495–510. doi:10.1039/D2LC01132H
Fujii, M., Matano, M., Toshimitsu, K., Takano, A., Mikami, Y., Nishikori, S., et al. (2018). Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23, 787–793.e6. doi:10.1016/J.STEM.2018.11.016
Giobbe, G. G., Bonfante, F., Jones, B. C., Gagliano, O., Luni, C., Zambaiti, E., et al. (2021). SARS-CoV-2 infection and replication in human gastric organoids. Nat. Commun. 12, 6610–6614. doi:10.1038/s41467-021-26762-2
Giobbe, G. G., Crowley, C., Luni, C., Campinoti, S., Khedr, M., Kretzschmar, K., et al. (2019). Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 10, 5658. doi:10.1038/s41467-019-13605-4
Gjorevski, N., Sachs, N., Manfrin, A., Giger, S., Bragina, M. E., Ordóñez-Morán, P., et al. (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539 (7630), 560–564. doi:10.1038/nature20168
Grikscheit, T. C., Siddique, A., Ochoa, E. R., Srinivasan, A., Alsberg, E., Hodin, R. A., et al. (2004). Tissue-engineered small intestine improves recovery after massive small bowel resection. Ann. Surg. 240, 748–754. doi:10.1097/01.SLA.0000143246.07277.73
Habib, S., Swaby, A. M., Gaisawat, M. B., Kubow, S., and Agellon, L. B. (2021). A novel, scalable, and modular bioreactor design for dynamic simulation of the digestive tract. Biotechnol. Bioeng. 118, 4338–4346. doi:10.1002/BIT.27902
Hannon, E., Pellegrini, M., Scottoni, F., Durkin, N., Shibuya, S., Lutman, R., et al. (2022). Lessons learned from pre-clinical testing of xenogeneic decellularized esophagi in a rabbit model. iScience 25, 105174. doi:10.1016/J.ISCI.2022.105174
Heo, I., Dutta, D., Schaefer, D. A., Iakobachvili, N., Artegiani, B., Sachs, N., et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823. doi:10.1038/S41564-018-0177-8
Hinman, S. S., Wang, Y., and Allbritton, N. L. (2019). Photopatterned membranes and chemical gradients enable scalable phenotypic organization of primary human colon epithelial models. Anal. Chem. 91, 15240–15247. doi:10.1021/acs.analchem.9b04217
Hinman, S. S., Wang, Y., Kim, R., and Allbritton, N. L. (2020). In vitro generation of self-renewing human intestinal epithelia over planar and shaped collagen hydrogels. Nat. Protoc. 16 (1), 352–382. doi:10.1038/s41596-020-00419-8
Howell, K. J., Kraiczy, J., Nayak, K. M., Gasparetto, M., Ross, A., Lee, C., et al. (2018). DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 154, 585–598. doi:10.1053/J.GASTRO.2017.10.007
Jensen, K. B., and Little, M. H. (2023). Organoids are not organs: sources of variation and misinformation in organoid biology. Stem Cell Rep. 18, 1255–1270. doi:10.1016/J.STEMCR.2023.05.009
Jeong, H.-J., Park, J.-H., Kang, J. H., Kong, S.-H., and Park, T.-E. (2022). Organoid-based human stomach micro-physiological system to recapitulate the dynamic mucosal defense mechanism. bioRxiv. doi:10.1101/2022.03.02.482603
Joddar, B., Tasnim, N., Thakur, V., Kumar, A., McCallum, R. W., and Chattopadhyay, M. (2018). Delivery of mesenchymal stem cells from gelatin–alginate hydrogels to stomach lumen for treatment of gastroparesis. Bioengineering 5, 12. doi:10.3390/BIOENGINEERING5010012
Jones, B. C., Elvassore, N., De Coppi, P., and Giobbe, G. G. (2022). Extracellular matrix hydrogels from decellularized tissues for biological and biomedical applications. Multifunct. Hydrogels Biomed. Appl., 1–22. doi:10.1002/9783527825820.ch1
Kasagi, Y., Chandramouleeswaran, P. M., Whelan, K. A., Tanaka, K., Giroux, V., Sharma, M., et al. (2018). The esophageal organoid system reveals functional interplay between Notch and cytokines in reactive epithelial changes. CMGH 5, 333–352. doi:10.1016/j.jcmgh.2017.12.013
Kasendra, M., Tovaglieri, A., Sontheimer-Phelps, A., Jalili-Firoozinezhad, S., Bein, A., Chalkiadaki, A., et al. (2018). Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 8 (1), 2871–2914. doi:10.1038/s41598-018-21201-7
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab. Chip 12, 2165–2174. doi:10.1039/C2LC40074J
Kim, I. G., Wu, Y., Choi, J. S., Kwon, S. K., Choi, S. H., Jung, K. C., et al. (2023). Assessment of esophageal reconstruction via bioreactor cultivation of a synthetic scaffold in a canine model. Clin. Exp. Otorhinolaryngol. 16, 165–176. doi:10.21053/CEO.2022.01522
Kim, I. G., Wu, Y., Park, S. A., Cho, H., Choi, J. J., Kwon, S. K., et al. (2019). Tissue-engineered esophagus via bioreactor cultivation for circumferential esophageal reconstruction. Tissue Eng. Part A 25, 1478–1492. doi:10.1089/TEN.TEA.2018.0277
Kim, J., Koo, B. K., and Knoblich, J. A. (2020a). Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584. doi:10.1038/S41580-020-0259-3
Kim, R., Attayek, P. J., Wang, Y., Furtado, K. L., Tamayo, R., Sims, C. E., et al. (2020b). An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 12, 015006. doi:10.1088/1758-5090/ab446e
Kim, S., Min, S., Choi, Y. S., Jo, S. H., Jung, J. H., Han, K., et al. (2022). Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 13, 1692. doi:10.1038/S41467-022-29279-4
Kim, S. S., Penkala, R., and Abrahimi, P. (2007). A perfusion bioreactor for intestinal tissue engineering. J. Surg. Res. 142, 327–331. doi:10.1016/J.JSS.2007.03.039
Kitano, K., Schwartz, D. M., Zhou, H., Gilpin, S. E., Wojtkiewicz, G. R., Ren, X., et al. (2017). Bioengineering of functional human induced pluripotent stem cell-derived intestinal grafts. Nat. Commun. 8 (1), 765–813. doi:10.1038/s41467-017-00779-y
Krishnamurthy, M., Kechele, D. O., Broda, T., Zhang, X., Enriquez, J. R., McCauley, H. A., et al. (2022). Using human induced pluripotent stem cell-derived organoids to identify new pathologies in patients with PDX1 mutations. Gastroenterology 163, 1053–1063.e7. doi:10.1053/J.GASTRO.2022.06.083
Kug Lee, K., McCauley, H. A., Broda, T. R., Kofron, M. J., Wells bcd, J. M., and Hong, C. I. (2018). Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab. Chip. 18, 3079–3085. doi:10.1039/c8lc00910d
Lahar, N., Lei, N. Y., Wang, J., Jabaji, Z., Tung, S. C., Joshi, V., et al. (2011). Intestinal subepithelial myofibroblasts support in vitro and in vivo growth of human small intestinal epithelium. PLoS One 6, e26898. doi:10.1371/JOURNAL.PONE.0026898
Levenson, G., Berger, A., Demma, J., Perrod, G., Domet, T., Arakelian, L., et al. (2022). Circumferential esophageal replacement by a decellularized esophageal matrix in a porcine model. Surgery 171, 384–392. doi:10.1016/J.SURG.2021.07.009
Li, X., Nadauld, L., Ootani, A., Corney, D. C., Pai, R. K., Gevaert, O., et al. (2014). Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20 (7), 769–777. doi:10.1038/nm.3585
Lourenço, B. N., dos Santos, T., Oliveira, C., Barrias, C. C., and Granja, P. L. (2018). Bioengineering a novel 3D in vitro model of gastric mucosa for stomach permeability studies. Acta Biomater. 82, 68–78. doi:10.1016/J.ACTBIO.2018.10.007
Matano, M., Date, S., Shimokawa, M., Takano, A., Fujii, M., Ohta, Y., et al. (2015). Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 21 (3), 256–262. doi:10.1038/nm.3802
McCauley, H. A., and Wells, J. M. (2017). Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 144, 958–962. doi:10.1242/DEV.140731
McCracken, K. W., Aihara, E., Martin, B., Crawford, C. M., Broda, T., Treguier, J., et al. (2017). Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature 541, 182–187. doi:10.1038/NATURE21021
McCracken, K. W., Catá, E. M., Crawford, C. M., Sinagoga, K. L., Schumacher, M., Rockich, B. E., et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. doi:10.1038/NATURE13863
Meran, L., Massie, I., Campinoti, S., Weston, A. E., Gaifulina, R., Tullie, L., et al. (2020). Engineering transplantable jejunal mucosal grafts using patient-derived organoids from children with intestinal failure. Nat. Med. 26, 1593–1601. doi:10.1038/s41591-020-1024-z
Múnera, J. O., Sundaram, N., Rankin, S. A., Hill, D., Watson, C., Mahe, M., et al. (2017). Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 21, 51–64.e6. doi:10.1016/j.stem.2017.05.020
Nakatsu, H., Ueno, T., Oga, A., Nakao, M., Nishimura, T., Kobayashi, S., et al. (2015). Influence of mesenchymal stem cells on stomach tissue engineering using small intestinal submucosa. J. Tissue Eng. Regen. Med. 9, 296–304. doi:10.1002/TERM.1794
Nayakawde, N. B., Methe, K., Banerjee, D., Berg, M., Premaratne, G. U., and Olausson, M. (2020). In vitro regeneration of decellularized pig esophagus using human amniotic stem cells. Biores Open Access 9, 22–36. doi:10.1089/BIORES.2019.0054
Nikolaev, M., Mitrofanova, O., Broguiere, N., Geraldo, S., Dutta, D., Tabata, Y., et al. (2020). Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585 (7826), 574–578. doi:10.1038/s41586-020-2724-8
Picollet-D’hahan, N., Zuchowska, A., Lemeunier, I., and Le Gac, S. (2021). Multiorgan-on-a-Chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 39, 788–810. doi:10.1016/J.TIBTECH.2020.11.014
Pimenta, J., Ribeiro, R., Almeida, R., Costa, P. F., da Silva, M. A., and Pereira, B. (2022). Organ-on-Chip approaches for intestinal 3D in vitro modeling. Cell Mol. Gastroenterol. Hepatol. 13, 351–367. doi:10.1016/J.JCMGH.2021.08.015
Poghosyan, T., Sfeir, R., Michaud, L., Bruneval, P., Domet, T., Vanneaux, V., et al. (2015). Circumferential esophageal replacement using a tube-shaped tissue-engineered substitute: an experimental study in minipigs. Surgery 158, 266–277. doi:10.1016/J.SURG.2015.01.020
Powell, D. W., Pinchuk, I. V., Saada, J. I., Chen, X., and Mifflin, R. C. (2011). Mesenchymal cells of the intestinal lamina propria. Annu. Rev. Physiol. 73, 213–237. doi:10.1146/ANNUREV.PHYSIOL.70.113006.100646
Raghavan, S., Gilmont, R. R., Miyasaka, E. A., Somara, S., Srinivasan, S., Teitelbaum, D. H., et al. (2011). Successful implantation of bioengineered, intrinsically innervated, human internal anal sphincter. Gastroenterology 141, 310–319. doi:10.1053/J.GASTRO.2011.03.056
Sala, F. G., Kunisaki, S. M., Ochoa, E. R., Vacanti, J., and Grikscheit, T. C. (2009). Tissue-engineered small intestine and stomach form from autologous tissue in a preclinical large animal model. J. Surg. Res. 156, 205–212. doi:10.1016/J.JSS.2009.03.062
Sato, T., Stange, D. E., Ferrante, M., Vries, R. G. J., Van Es, J. H., Van Den Brink, S., et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. doi:10.1053/j.gastro.2011.07.050
Sato, T., Vries, R. G., Snippert, H. J., Van De Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi:10.1038/NATURE07935
Schlaermann, P., Toelle, B., Berger, H., Schmidt, S. C., Glanemann, M., Ordemann, J., et al. (2016). A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 65, 202–213. doi:10.1136/GUTJNL-2014-307949
Schwank, G., Koo, B. K., Sasselli, V., Dekkers, J. F., Heo, I., Demircan, T., et al. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658. doi:10.1016/J.STEM.2013.11.002
Schweinlin, M., Wilhelm, S., Schwedhelm, I., Hansmann, J., Rietscher, R., Jurowich, C., et al. (2016). Development of an advanced primary human in vitro model of the small intestine. Tissue Eng. Part C Methods 22, 873–883. doi:10.1089/TEN.TEC.2016.0101
Shacham-Silverberg, V., and Wells, J. M. (2020). Generation of esophageal organoids and organotypic raft cultures from human pluripotent stem cells. Methods Cell Biol. 159, 1–22. doi:10.1016/BS.MCB.2020.04.009
Shaffiey, S. A., Jia, H., Keane, T., Costello, C., Wasserman, D., Quidgley, M., et al. (2016). Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen. Med. 11, 45–61. doi:10.2217/RME.15.70
Simmini, S., Bialecka, M., Huch, M., Kester, L., Van De Wetering, M., Sato, T., et al. (2014). Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat. Commun. 5, 5728. doi:10.1038/NCOMMS6728
Sinagoga, K. L., McCauley, H. A., Muńera, J. O., Reynolds, N. A., Enriquez, J. R., Watson, C., et al. (2018). Deriving functional human enteroendocrine cells from pluripotent stem cells. Development 145, dev165795. doi:10.1242/DEV.165795
Spence, J. R., Mayhew, C. N., Rankin, S. A., Kuhar, M. F., Vallance, J. E., Tolle, K., et al. (2010). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470 (7332), 105–109. doi:10.1038/nature09691
Suzuki, K., Murano, T., Shimizu, H., Ito, G., Nakata, T., Fujii, S., et al. (2018). Single cell analysis of Crohn’s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J. Gastroenterol. 53, 1035–1047. doi:10.1007/S00535-018-1437-3
Takahashi, Y., Sato, S., Kurashima, Y., Yamamoto, T., Kurokawa, S., Yuki, Y., et al. (2018). A refined culture system for human induced pluripotent stem cell-derived intestinal epithelial organoids. Stem Cell Rep. 10, 314–328. doi:10.1016/J.STEMCR.2017.11.004
Takeoka, Y., Matsumoto, K., Taniguchi, D., Tsuchiya, T., Machino, R., Moriyama, M., et al. (2019). Regeneration of esophagus using a scaffold-free biomimetic structure created with bio-three-dimensional printing. PLoS One 14, e0211339. doi:10.1371/JOURNAL.PONE.0211339
Totonelli, G., Maghsoudlou, P., Garriboli, M., Riegler, J., Orlando, G., Burns, A. J., et al. (2012). A rat decellularized small bowel scaffold that preserves villus-crypt architecture for intestinal regeneration. Biomaterials 33, 3401–3410. doi:10.1016/J.BIOMATERIALS.2012.01.012
Trisno, S. L., Philo, K. E. D., McCracken, K. W., Catá, E. M., Ruiz-Torres, S., Rankin, S. A., et al. (2018). Esophageal organoids from human pluripotent stem cells delineate Sox2 functions during esophageal specification. Cell Stem Cell 23, 501–515.e7. doi:10.1016/J.STEM.2018.08.008
Urbani, L., Camilli, C., Phylactopoulos, D. E., Crowley, C., Natarajan, D., Scottoni, F., et al. (2018). Multi-stage bioengineering of a layered oesophagus with in vitro expanded muscle and epithelial adult progenitors. Nat. Commun. 9, 4286. doi:10.1038/S41467-018-06385-W
Urbani, L., Maghsoudlou, P., Milan, A., Menikou, M., Hagen, C. K., Totonelli, G., et al. (2017). Long-term cryopreservation of decellularised oesophagi for tissue engineering clinical application. PLoS One 12, e0179341. doi:10.1371/JOURNAL.PONE.0179341
Urciuolo, A., Giobbe, G. G., Dong, Y., Michielin, F., Brandolino, L., Magnussen, M., et al. (2023). Hydrogel-in-hydrogel live bioprinting for guidance and control of organoids and organotypic cultures. Nat. Commun. 14 (1), 3128. doi:10.1038/s41467-023-37953-4
Urciuolo, A., Poli, I., Brandolino, L., Raffa, P., Scattolini, V., Laterza, C., et al. (2020). Intravital three-dimensional bioprinting. Nat. Biomed. Eng. 4, 901–915. doi:10.1038/s41551-020-0568-z
Wang, Y., Gunasekara, D. B., Reed, M. I., DiSalvo, M., Bultman, S. J., Sims, C. E., et al. (2017). A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55. doi:10.1016/J.BIOMATERIALS.2017.03.005
Watanabe, S., Kobayashi, S., Ogasawara, N., Okamoto, R., Nakamura, T., Watanabe, M., et al. (2022). Transplantation of intestinal organoids into a mouse model of colitis. Nat. Protoc. 17, 649–671. doi:10.1038/S41596-021-00658-3
Watson, C. L., Mahe, M. M., Múnera, J., Howell, J. C., Sundaram, N., Poling, H. M., et al. (2014). An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 20, 1310–1314. doi:10.1038/NM.3737
Wei, R. Q., Tan, B., Tan, M. Y., Luo, J. C., Deng, L., Chen, X. H., et al. (2009). Grafts of porcine small intestinal submucosa with cultured autologous oral mucosal epithelial cells for esophageal repair in a canine model. Exp. Biol. Med. (Maywood) 234, 453–461. doi:10.3181/0901-RM-5
Workman, M. J., Mahe, M. M., Trisno, S., Poling, H. M., Watson, C. L., Sundaram, N., et al. (2016). Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23 (1), 49–59. doi:10.1038/nm.4233
Yeung, T. M., Chia, L. A., Kosinski, C. M., and Kuo, C. J. (2011). Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell Mol. Life Sci. 68, 2513–2523. doi:10.1007/S00018-011-0687-5
Zambaiti, E., Scottoni, F., Rizzi, E., Russo, S., Deguchi, K., Eaton, S., et al. (2019). Whole rat stomach decellularisation using a detergent-enzymatic protocol. Pediatr. Surg. Int. 35, 21–27. doi:10.1007/S00383-018-4372-8
Zhang, Y. G., Wu, S., Xia, Y., and Sun, J. (2014). Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol. Rep. 2, e12147. doi:10.14814/PHY2.12147
Zhang, Y. S., Aleman, J, Shin, S. R., Kilic, T., Kim, D., Mousavi Shaegh, S. A., et al. (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U. S. A. 114, E2293–E2302. doi:10.1073/pnas.1612906114
Zhang, Y., Yang, Y., Jiang, M., Huang, S. X., Zhang, W., Al Alam, D., et al. (2018). 3D modeling of esophageal development using human PSC-derived basal progenitors reveals a critical role for Notch signaling. Cell Stem Cell 23, 516–529.e5. doi:10.1016/J.STEM.2018.08.009
Keywords: human organoids, tissue engineering, GI tract, esophagus, stomach, intestine
Citation: Benedetti G, Fournon Berodia B, De Coppi P and Giobbe GG (2023) Human gastro-intestinal organoid engineering: a state of the art. Front. Chem. Eng. 5:1256334. doi: 10.3389/fceng.2023.1256334
Received: 10 July 2023; Accepted: 02 October 2023;
Published: 16 October 2023.
Edited by:
Onelia Gagliano, University of Padua, ItalyReviewed by:
Yongfei Xue, Central South University of Forestry and Technology, ChinaCopyright © 2023 Benedetti, Fournon Berodia, De Coppi and Giobbe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giovanni Giuseppe Giobbe, Zy5naW9iYmVAdWNsLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.