- 1Biomaterials and Biomanufacturing Department, Sandia National Laboratories, Livermore, CA, United States
- 2Systems Biology Department, Sandia National Laboratories, Livermore, CA, United States
- 3Bioresource and Environmental Security Department, Sandia National Laboratories, Livermore, CA, United States
- 4Computational Biology and Biophysics Department, Sandia National Laboratories, Livermore, CA, United States
Naturally occurring microbial communities are able to decompose lignocellulosic biomass through the concerted production of a myriad of enzymes that degrade its polymeric components and assimilate the resulting breakdown compounds by members of the community. This process includes the conversion of lignin, the most recalcitrant component of lignocellulosic biomass and historically the most difficult to valorize in the context of a biorefinery. Although several fundamental questions on microbial conversion of lignin remain unanswered, it is known that some fungi and bacteria produce enzymes to break, internalize, and assimilate lignin-derived molecules. The interest in developing efficient biological lignin conversion approaches has led to a better understanding of the types of enzymes and organisms that can act on different types of lignin structures, the depolymerized compounds that can be released, and the products that can be generated through microbial biosynthetic pathways. It has become clear that the discovery and implementation of native or engineered microbial consortia could be a powerful tool to facilitate conversion and valorization of this underutilized polymer. Here we review recent approaches that employ isolated or synthetic microbial communities for lignin conversion to bioproducts, including the development of methods for tracking and predicting the behavior of these consortia, the most significant challenges that have been identified, and the possibilities that remain to be explored in this field.
Introduction
Lignocellulosic biomass is the most abundant source of renewable carbon on Earth, making it a top candidate for conversion to biofuels and bioproducts as part of the strategies to transition to a sustainable bioeconomy (Rajesh Banu et al., 2021). This carbon is stored in plants in the form of large, interlinked polymers: cellulose, hemicellulose, and lignin. These polymers have evolved to withstand physical, chemical, and biological degradation, and provide plants with rigidity and resistance to environmental aggressions (Bomble et al., 2017). Therefore, numerous efforts have been devoted to developing technologies that would enable the cheap, efficient, and sustainable deconstruction and upgrading of lignocellulosic biomass (Hassan et al., 2018; Bhatia et al., 2020).
Considerable progress has been made in the last two decades to convert cellulose and hemicellulose to monomeric sugars and ferment them to ethanol or other biofuels (Balan et al., 2013; Raud et al., 2019; Saravanan et al., 2022). Lignin, on the other hand, has proven to be the most recalcitrant component of lignocellulosic biomass. This is due to its heterogeneous chemical structure comprising large chains of phenolic rings interlinked through different types of chemical bonds, resulting in poor solubility in aqueous systems (Li et al., 2016). Although the relative amounts, chemical composition, and structure of each of the main components is known to be highly variable (Ralph et al., 2019), lignin valorization has been deemed essential to improve the economics of lignocellulosic refineries (Martinez-Hernandez et al., 2019).
Thermochemical approaches to lignin depolymerization such as pyrolysis, hydrolysis, gasification, and oxidative or reductive approaches have been effective to produce low molecular weight compounds, usually aromatic compounds (Liu et al., 2020). However, these procedures typically require high temperature and pressure conditions, which can be costly to implement and difficult to tune in order to obtain high amounts of specific products (Schutyser et al., 2018). On the other side, biological approaches to depolymerize lignin may be a cost- and energy-effective solution that involves utilizing microorganisms that secrete cocktails of lignin-depolymerizing enzymes (Martínez et al., 2018; Kamimura et al., 2019).
Early experiments on biological lignin conversion indicated that the white-rot fungi Phanerochaete chrysosporium and Coriolus versicolor can degrade 14C-labeled lignin and convert it to 14CO2 (Kirk et al., 1975). This led to the discovery of the first lignin-degrading enzyme, lignin peroxidase (LiP), an H2O2-dependent enzyme system that was detected in the extracellular medium of ligninolytic cultures with P. chrysosporium (Tien and Kirk, 1983). Other types of fungal peroxidases include manganese-dependent peroxidase (MnP) and versatile peroxidase (VP) that can work in synergy with LiP, which has a higher redox potential (Martínez et al., 2018). Some bacteria and fungi also produce dye-decolorizing peroxidases (DyP) with lignin-degrading activity. Although the bacterial DyPs are not efficient at degrading non-phenolic compounds and therefore unlikely to act on most of the native lignin molecule (Linde et al., 2021), they are proposed to contribute to the degradation of small phenolic compounds released by the attack of fungal enzymes on lignin. Lastly, laccases are known to play an important role in lignin biosynthesis and degradation in nature and are found in fungi, bacteria and plants. Since laccases do not require H2O2 and use oxygen as electron acceptor, they are truly green and industrially attractive enzymes; however, the production and purification of fungal laccases at high concentrations has proven challenging (Leynaud Kieffer Curran et al., 2022).
Enzymatic lignin-degradation strategies can be combined with biological lignin-valorization strategies that incorporate microbes that have evolved intracellular metabolic pathways for assimilation of lignin fragments produced during depolymerization (Kamimura et al., 2017; Lee et al., 2019). The use of microorganisms to funnel an array of depolymerized lignin compounds into selected metabolic intermediates has been explored during the last decade as a potential solution to counter the heterogeneity in depolymerized lignin solutions (Beckham et al., 2016). Some studies have shown a synergy between lignin-modifying enzymes and bacteria for lignin degradation. For example, cell growth and selective degradation of different functional groups was observed when the bacterium Rhodococcus opacus was incubated with lignin in the presence of a laccase (Zhao et al., 2016). The heterologous expression of a laccase in Pseudomonas putida also resulted in improved growth and lignin degradation (Cao et al., 2021). Regarding substrate conversion capabilities, R. opacus was found to consume lower-molecular weight compounds obtained from lignin, such as guaiacol (Ravi et al., 2019). Similarly, an engineered Amycolatopsis sp., strain was also able to consume guaiacol and produce cis,cis-muconic acid, a precursor of commercial plastics (Barton et al., 2018). In a separate work, a higher extent of lignin depolymerization was observed when P. putida was incubated with a fungal secretome and lignin (Salvachua et al., 2016). Engineered P. putida strains have also been constructed to produce cis,cis-muconic acid from depolymerized lignin such as catechol and other monomeric aromatics (Kohlstedt et al., 2018).
Although these developments represent only a few examples of the numerous achievements in microbial conversion of lignin in recent years (Weiland et al., 2022), some disadvantages have become evident. These include lignin repolymerization by the same enzymes involved in depolymerization, low solubility of lignin and lignin-derived compounds in water, and microbial toxicity caused by the depolymerized compounds (Bugg and Rahmanpour, 2015; Beckham et al., 2016). One of the biggest challenges is the difficulty in predicting which compounds can be produced from certain lignins and which of them are amenable to microbial conversion (Davis and Moon, 2020). In addition, an organism that has been engineered to overproduce a specific metabolite from a lignin-derived substrate will direct less carbon to biosynthetic pathways, which could limit its ability to grow and the productivity of a bioprocess. The metabolic burden of expressing different enzymes required for depolymerization and conversion of multiple lignin-derived compounds in a single organism could also significantly impact the growth rate of the engineered strains and limit their usefulness (Roell et al., 2019).
The use of microbial communities for valorization of lignocellulose has steadily gained interest as an alternative to overcome some of the current limitations in biological conversion processes because communities exhibit favorable traits such as increased resilience and metabolic diversity, compared to single organisms. However, more research is needed to understand the dynamics and tackle the complexity of systems comprised of a heterogeneous substrate such as lignin and multiple microorganisms. We provide a discussion of recent advances related to the development of lignin valorization processes using microbial consortia in the sections below.
Studies on natural microbial consortia
Beneficial relationships of microorganisms in the natural environment have been exploited for agriculture, food, medicine, and bioremediation purposes, with many examples available (Frey-Klett et al., 2011; Ijoma and Tekere, 2017). Different aerobic and anaerobic communities containing fungal and bacterial species were found to be able to degrade lignin-derived compounds (Chen et al., 1985a; 1985b; Jokela et al., 1985; Tuomela et al., 2000). Mixed bacterial cultures were also able to degrade lignin, and in some cases, auxiliary activity families and metabolic pathways of aromatic compounds were identified by metagenomic studies (Jiménez et al., 2016; Silva et al., 2021).
It has been suggested that bacteria can catabolize low molecular weight compounds produced by fungal lignin depolymerization. However, it remains unclear to what extent bacteria can depolymerize larger compounds, including high molecular mass lignin, and some studies indicate that bacteria have significantly less lignin-degradation ability than fungal systems (Bugg et al., 2011; Brown and Chang, 2014). If the prevailing paradigm is that both fungal and bacterial species are required for the complete mineralization of lignin, this means that it should be possible to identify metabolites that can only be consumed by specific members of a community and synergistic interactions that can be exploited to improve the conversion process. However, it remains difficult to draw such conclusions from data in the scientific literature, since variations in the source of lignin, organisms, and cultivation conditions dictate the composition of the resulting community and the extent of lignin depolymerization (Mendes et al., 2021; Azubuike et al., 2022). It has been demonstrated that certain microbes can degrade specific C-C and C-O bonds in lignin while also changing the content of H, G, and S lignin moieties (Bugg et al., 2020; Xu et al., 2022; Zhu et al., 2022). Similar in-depth structural characterizations of the effects of treatments with microbial consortia are currently lacking and future structural studies will be critical to progress in the field.
Typically, top-down approaches must be performed when working with natural consortia to narrow down the diversity of organisms and conversion pathways, as well as identifying community members that promote high lignin conversion (Puentes-Tellez and Falcao Salles, 2018; Gilmore et al., 2019). For example, Fang et al. (2018) performed enrichment of microbial consortia isolated from rotten wood by screening for microbes that can degrade guaiacol and a sequential transferring of the cultures to medium containing tree trimmings, which led to the isolation of a community able to degrade up to 28% of the lignin in the biomass. Complementary approaches such as “dilution-to-extinction” have also been applied to identify species in minimal active consortia for lignocellulose conversion (Diaz-Rodriguez et al., 2022). For comparison purposes, Table 1 lists the most prevalent microorganisms in consortia with high reported degrees of lignin conversion. A strong appeal of studying natural consortia is the possibility of discovering novel genes and organisms that can be used to rationally design pathways for degradation and utilization of lignin (Moraes et al., 2018; Díaz-García et al., 2020).
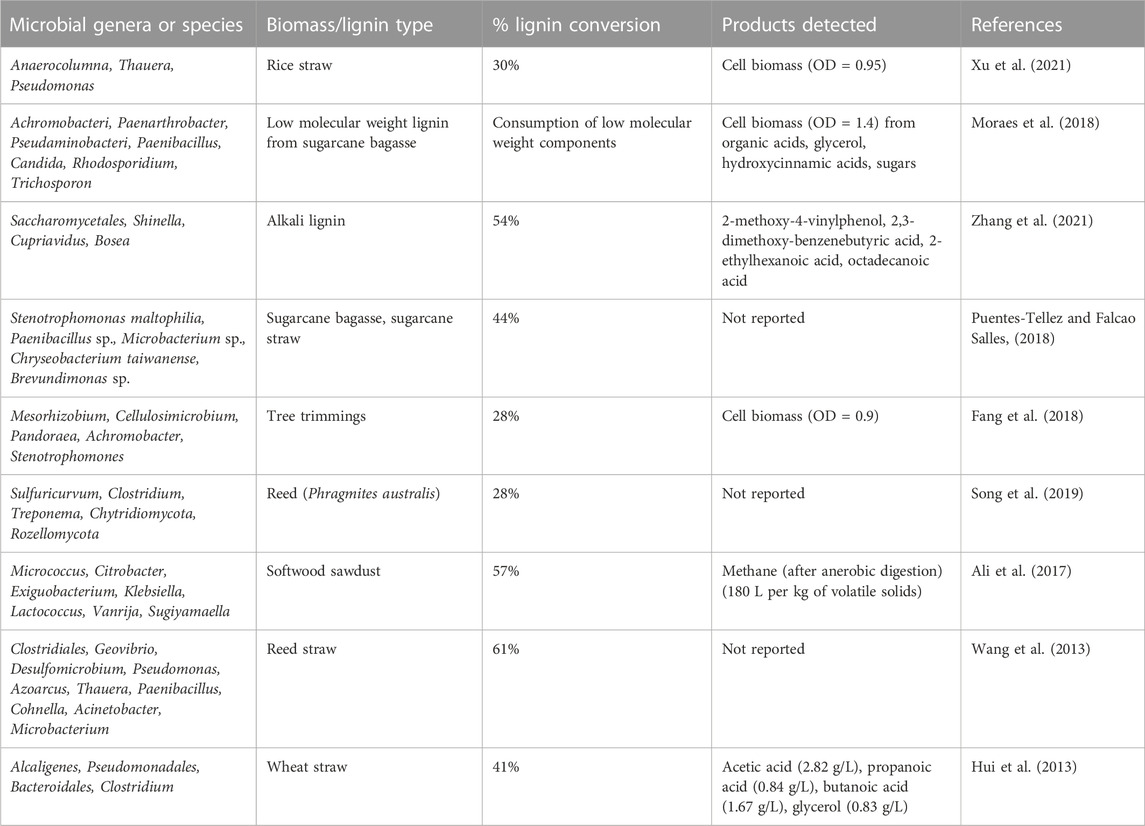
TABLE 1. Most abundant organisms present in microbial consortia with reported lignin conversion capabilities.
One important point of discussion is the feasibility of using natural communities to not just deconstruct but valorize lignin. Although they represent an untapped resource of enzymes and metabolic reactions, the added complexity of promoting carbon conversion to specific bioproducts is still an important barrier to overcome. For example, the number of organisms that interact in a specific ecosystem can be extremely large, and not all organisms can be cultivated in laboratory conditions. Moreover, only a small fraction of those that can be cultivated will have genetic engineering tools available, making it difficult to promote conversion of lignin-derived compounds into defined products. In this regard, anaerobic communities isolated from the rumen of herbivores have been found to degrade lignin in lignocellulosic substrates and convert it to methane (Ahring et al., 2015), expanding the possibilities for implementing lignin conversion processes with mixtures of anaerobic organisms (Podolsky et al., 2019). Although anaerobic digestion has the potential to improve the economics of a biorefinery, the extent of lignin degradation under these conditions remains low and, unless converted, lignin will have a negative impact on the process (Ali et al., 2018). Improvements in this area may require the implementation of oxidative pretreatment approaches (Khan and Ahring, 2019) coupled with efficient lignolytic consortia that include more specialized organisms with potentially impactful bioconversion capabilities (Liu et al., 2019; Lankiewicz et al., 2022).
Studies on synthetic or engineered consortia
Although the use of natural communities is a promising approach to degrade complex substrates, the stability and behavior of members of the community could be difficult to control. This has encouraged biologists to leverage systems-level knowledge on natural communities to assemble defined consortia of organisms using bottom-up approaches, which sacrifices diversity for a higher degree of control and the ability to perform specific tasks more efficiently (Lin 2022). Some examples of engineered communities for production of value-added compounds from lignocellulosic biomass involve the production of butanol from hemicellulose (Jiang et al., 2020), isobutanol from cellulose (Minty et al., 2013), and butyric acid from cellulose (Shahab et al., 2020). A multi-species consortium has also been used to consume not just the lignocellulosic substrates present in hydrolysates but also compounds that can be toxic to some species (Lee et al., 2021).
These examples highlight the value of using engineered consortia to produce biofuels and bioproducts; however, their specific application to lignin conversion is still an uncharted territory. One research group focused on building 65 synthetic communities with combinations of 18 strains found in a native consortium, to evaluate their potential to degrade polymeric sugars and lignin when fed with raw biomass (Puentes-Tellez and Falcao Salles, 2018). The authors found synergies and demonstrated that a minimal community of 5 organisms was able to degrade up to 44% of the lignin in the biomass. In a different work, synthetic bacterial-fungal consortia have been found to be capable of deconstructing and growing on wheat straw biomass (Wang et al., 2021). However, the microbial activity was determined as function of the total substrate weight loss and not the specific degree of lignin conversion, which can be difficult to measure accurately.
Several challenges need to be addressed to achieve microbial community-based lignin conversion to bioproducts. Firstly, different species of known degraders (i.e., bacteria and fungi) have different growth and substrate consumption rates, which are crucial parameters in a bioprocess. The reaction conditions (pH, temperature, media components) required for optimal growth or enzymatic lignin depolymerization could be vastly different for each organism or enzyme (Janusz et al., 2017), resulting in members of the community with compromised activity or stability. Another hurdle is the differential consumption of substrates and intermediates from members of the community and the difficulty of mapping and controlling these carbon fluxes. It is also difficult to predict the type of relationship that can exist if two or more species are cultivated together and recognize when antagonistic interactions may be present (Jimenez et al., 2020). One way to approach these issues could be to design a synthetic community where the tasks related to converting lignin to a bioproduct are divided among the members. One can envision this concept with organisms called cutters, likely white-rot fungi, which deconstruct lignin into smaller fragments that are captured by a scavenger, likely a bacterium or yeast (Figure 1). Synergism in lignin depolymerization and increased ligninase activity has been observed in co-cultures of white-rot fungi (Cui et al., 2021), signaling that fungal treatment via strong enzymes such as LiP, MnP, and VP could be a reasonable first step in the process. Bacteria or yeasts can then potentially contribute to the degradation of small oligomers with the help of etherases, laccases, DyP, and other oxidases and auxiliary enzymes (Bugg et al., 2020; Putra et al., 2022). Considering that microbes have the ability to scavenge different types of compounds, it should be possible to evaluate the performance of different organisms in a consortium to funnel lignin-derived compounds to specific metabolites. A database of microbial lignin catabolism such as eLignin could be an important resource when selecting scavengers for conversion experiments (Brink et al., 2019). Success in this task would allow for the introduction of specialized organisms (“manufacturers”) that are easy to engineer and can convert one or more of these secreted metabolites into a higher value bioproduct without the need to harbor lignin assimilation pathways.
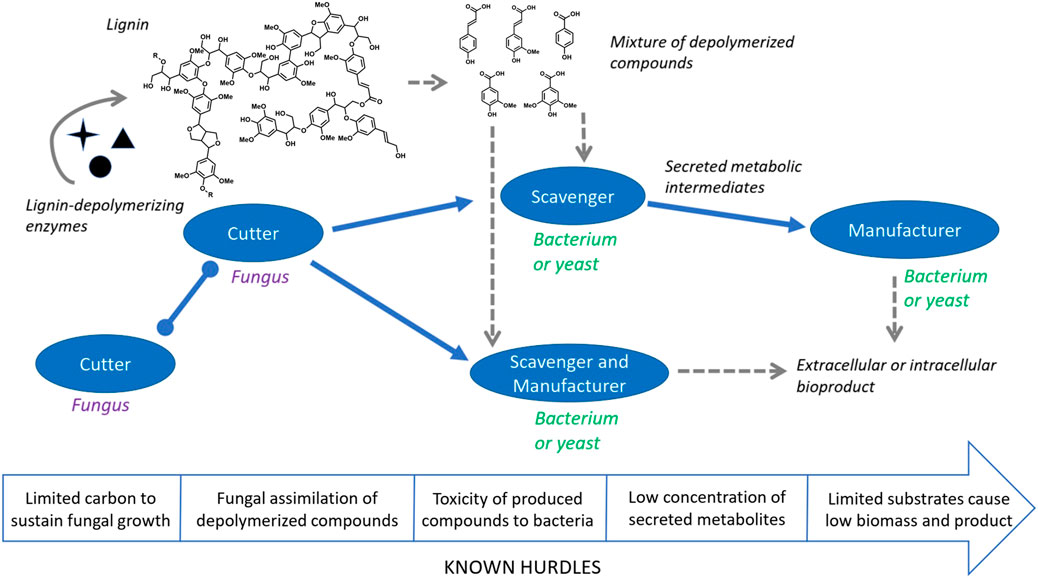
FIGURE 1. A concept for building a synthetic microbial community for conversion of lignin to valuable products. Cutters represent fungi that secrete lignin degrading enzymes, scavengers represent microbes that catabolize lignin fragments and potentially convert them to a shareable intermediate, and manufacturers represent engineered hosts that convert either lignin fragments or a shared intermediate to bioproducts. The arrow at the bottom depicts hurdles that may be present at various stages of the conversion process.
Simulation and optimization of synthetic communities
An important topic when utilizing synthetic communities is the development of tools and methods to guide their rational design, testing, and optimization, which involves considering the complexities of the interactions among community members that influence properties such as stability and productivity. It is evident that new developments in these areas will have an impact beyond lignin conversion, and methods for monitoring and predicting the dynamics of microbial communities have recently been published. For example, programmable interactions between organisms have been created using synthetic gene circuits (Kong et al., 2018). A pipeline to select organisms based on meta-data analysis of their microbial physiologies and subsequent screening has been reported for the production of biohydrogen (Ergal et al., 2021). Another work describes a high-throughput phenotypic screening using a droplets-based platform to rapidly assemble variable microbial communities (Kehe et al., 2019). Immobilization of community members as shown by Huang et al. (2019) can tackle some of the challenges associated with stability and toxicity. Finally, molecular methods that allow for systematic screening of lignin-related enzymes and products such as biosensors could be key to the discovery and implementation of robust microbial consortia (Gonçalves et al., 2020).
One of the areas with highest potential impact are studies on the structure of communities enriched on lignin and the interaction of their members. However, the development of consensus communities and consensus interactions among members can be difficult to understand because 1) the relationships between species (mutualism, commensalism, or facultative symbioses) are difficult to determine, and 2) interacting does not necessarily mean cause and effect. To this end, in silico simulation approaches provide a means for describing, understanding, and optimizing microbial systems in diverse environments. Approaches using individual-based modeling (IBM) in which microbes and their interactions are defined, and the temporal-spatial evolution of the system is simulated, are becoming increasingly powerful. For example, BacArena models individual microbes using flux-balance analysis and simulates their interactions and community evolution using IBM. This approach has been used to obtain insights into Pseudomonas aeruginosa biofilm formation and a seven species community of the human gut (Bauer et al., 2017). Martin et al. (2012), integrated Dynamic Energy Budget (DEB) theory into IBM to enable exploration of the evolution and dynamics of both individual microbes and their community as a function of the set of DEB parameters of each species and their interactions with their environment (Martin et al., 2013; García-Jiménez et al., 2021). Applying these IBM approaches to lignin-associated microbial communities may be an effective strategy towards optimizing synthetic microbial consortia for lignin valorization.
Discussion
In this article we argue that the isolation, enrichment, and de novo design of microbial communities can provide solutions to some of the issues associated with the structural diversity and recalcitrance of lignin for valorization purposes. While there are many publications on the use of microbial consortia for lignocellulose conversion, there are fewer reports on the use of natural or engineered consortia strictly for lignin conversion. This alone indicates significant challenges to obtain efficient processes for the microbial conversion of lignin, such as limitations in methods and tools available for systematically assembling and assessing microbial communities.
From the examples discussed here, it is evident that considerable progress has been made in terms of proving that microbial conversion of lignin is a feasible approach. Nevertheless, the different temporality in lignin breakdown (mostly performed by slow-growing fungi) and assimilation (ideally performed by fast growing bacteria) could be one of the main hurdles to overcome when testing and designing a bioprocess. For example, if one organism is engineered to achieve high yields of a product, its ability to grow efficiently in a challenging environment may be negatively impacted. In order to maximize carbon assimilation and conversion of lignin-derived compounds to a high-value product by a specific organism, it may be desirable to have organisms that do not consume the depolymerized fragments, which can prove challenging given their scavenging nature and diverse metabolism (del Cerro et al., 2021). This would add the requirement of having an additional carbon source for those “cutter” organisms and possibly use genetic engineering to inactivate some of their natural assimilation pathways. Therefore, biorefinery streams that are lignin-rich but also contain traces of sugars and other assimilable components could be a more compelling substrate than high purity lignins.
Finally, if multiple species of fungi are included in a consortium, it may be necessary to perform compatibility assays due to the known extensive secretion of potentially toxic metabolites and enzymes. Depending on the application, it will also be necessary to differentiate between a community that is able to share a number of metabolic intermediates and a co-culture of organisms where different species individually convert a substrate to different products. Considering the rising interest from the scientific community on the biological valorization of lignin and the important progress reported in recent years, we are confident that these hurdles will be overcome.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was supported by a Laboratory Directed Research and Development grant from Sandia National Laboratories with contract number 20-0590. Sandia National Laboratories is a multimission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC, a wholly-owned subsidiary of Honeywell International, Inc., for the United States Department of Energy’s National Nuclear Security Administration under contract DE-NA0003525. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the United States Department of Energy or the United States Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahring, B. K., Biswas, R., Ahamed, A., Teller, P. J., and Uellendahl, H. (2015). Making lignin accessible for anaerobic digestion by wet-explosion pretreatment. Bioresour. Technol. 175, 182–188. doi:10.1016/j.biortech.2014.10.082
Ali, S. S., Abomohra, A. E.-F., and Sun, J. (2017). Effective bio-pretreatment of sawdust waste with a novel microbial consortium for enhanced biomethanation. Bioresour. Technol. 238, 425–432. doi:10.1016/j.biortech.2017.03.187
Ali, S., Shah, T. A., Afzal, A., and Tabassum, R. (2018). Exploring lignocellulosic biomass for bio-methane potential by anaerobic digestion and its economic feasibility. Energy & Environ. 29, 742–751. doi:10.1177/0958305X18759009
Azubuike, C. C., Allemann, M. N., and Michener, J. K. (2022). Microbial assimilation of lignin-derived aromatic compounds and conversion to value-added products. Curr. Opin. Microbiol. 65, 64–72. doi:10.1016/j.mib.2021.10.014
Balan, V., Chiaramonti, D., and Kumar, S. (2013). Review of US and EU initiatives toward development, demonstration, and commercialization of lignocellulosic biofuels. Biofuels, Bioprod. Bioref. 7, 732–759. doi:10.1002/bbb.1436
Barton, N., Horbal, L., Starck, S., Kohlstedt, M., Luzhetskyy, A., and Wittmann, C. (2018). Enabling the valorization of guaiacol-based lignin: Integrated chemical and biochemical production of cis,cis-muconic acid using metabolically engineered Amycolatopsis sp ATCC 39116. Metab. Eng. 45, 200–210. doi:10.1016/j.ymben.2017.12.001
Bauer, E., Zimmermann, J., Baldini, F., Thiele, I., and Kaleta, C. (2017). BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. PloS Comput. Biol. 13, e1005544. doi:10.1371/journal.pcbi.1005544
Beckham, G. T., Johnson, C. W., Karp, E. M., Salvachúa, D., and Vardon, D. R. (2016). Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 42, 40–53. doi:10.1016/j.copbio.2016.02.030
Bhatia, S. K., Jagtap, S. S., Bedekar, A. A., Bhatia, R. K., Patel, A. K., Pant, D., et al. (2020). Recent developments in pretreatment technologies on lignocellulosic biomass: Effect of key parameters, technological improvements, and challenges. Bioresour. Technol. 300, 122724. doi:10.1016/j.biortech.2019.122724
Bomble, Y. J., Lin, C.-Y., Amore, A., Wei, H., Holwerda, E. K., Ciesielski, P. N., et al. (2017). Lignocellulose deconstruction in the biosphere. Curr. Opin. Chem. Biol. 41, 61–70. doi:10.1016/j.cbpa.2017.10.013
Brink, D. P., Ravi, K., Lidén, G., and Gorwa-Grauslund, M. F. (2019). Mapping the diversity of microbial lignin catabolism: Experiences from the eLignin database. Appl. Microbiol. Biotechnol. 103, 3979–4002. doi:10.1007/s00253-019-09692-4
Brown, M. E., and Chang, M. C. Y. (2014). Exploring bacterial lignin degradation. Curr. Opin. Chem. Biol. 19, 1–7. doi:10.1016/j.cbpa.2013.11.015
Bugg, T. D. H., Ahmad, M., Hardiman, E. M., and Singh, R. (2011). The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400. doi:10.1016/j.copbio.2010.10.009
Bugg, T. D. H., and Rahmanpour, R. (2015). Enzymatic conversion of lignin into renewable chemicals. Curr. Opin. Chem. Biol. 29, 10–17. doi:10.1016/j.cbpa.2015.06.009
Bugg, T. D. H., Williamson, J. J., and Rashid, G. M. M. (2020). Bacterial enzymes for lignin depolymerisation: New biocatalysts for generation of renewable chemicals from biomass. Curr. Opin. Chem. Biol. 55, 26–33. doi:10.1016/j.cbpa.2019.11.007
Cao, L., Lin, L., Sui, H., Wang, H., Zhang, Z., Jiao, N., et al. (2021). Efficient extracellular laccase secretion via bio-designed secretory apparatuses to enhance bacterial utilization of recalcitrant lignin. Green Chem. 23 (5), 2079–2094. doi:10.1039/d0gc04084c
Chen, W., Ohmiya, K., Shimizu, S., and Kawakami, H. (1985a). Degradation of dehydrodivanillin by anaerobic bacteria from cow rumen fluid. Appl. Environ. Microbiol. 49, 211–216. doi:10.1128/aem.49.1.211-216.1985
Chen, W., Supanwong, K., Ohmiya, K., Shimizu, S., and Kawakami, H. (1985b). Anaerobic degradation of veratrylglycerol-beta-guaiacyl ether and guaiacoxyacetic acid by mixed rumen bacteria. Appl. Environ. Microbiol. 50, 1451–1456. doi:10.1128/aem.50.6.1451-1456.1985
Cui, T., Yuan, B., Guo, H., Tian, H., Wang, W., Ma, Y., et al. (2021). Enhanced lignin biodegradation by consortium of white rot fungi: Microbial synergistic effects and product mapping. Biotechnol. Biofuels 14, 162. doi:10.1186/s13068-021-02011-y
Davis, K., and Moon, T. S. (2020). Tailoring microbes to upgrade lignin. Curr. Opin. Chem. Biol. 59, 23–29. doi:10.1016/j.cbpa.2020.04.001
Del Cerro, C., Erickson, E., Dong, T., Wong, A. R., Eder, E. K., Purvine, S. O., et al. (2021). Intracellular pathways for lignin catabolism in white-rot fungi. Proc. Natl. Acad. Sci. U. S. A. 118, e2017381118. doi:10.1073/pnas.2017381118
Díaz Rodríguez, C. A., Díaz-García, L., Bunk, B., Spröer, C., Herrera, K., Tarazona, N. A., et al. (2022). Novel bacterial taxa in a minimal lignocellulolytic consortium and their potential for lignin and plastics transformation. ISME Commun. 2 (1), 89. doi:10.1038/s43705-022-00176-7
Díaz-García, L., Bugg, T. D. H., and Jiménez, D. J. (2020). Exploring the lignin catabolism potential of soil-derived lignocellulolytic microbial consortia by a gene-centric metagenomic approach. Microb. Ecol. 80, 885–896. doi:10.1007/s00248-020-01546-1
Ergal, İ., Bochmann, G., Fuchs, W., and Rittmann, S. K.-M. (2021). Design and engineering of artificial microbial consortia for biohydrogen production. Curr. Opin. Biotechnol. 73, 74–80. doi:10.1016/j.copbio.2021.07.010
Fang, X., Li, Q., Lin, Y., Lin, X., Dai, Y., Guo, Z., et al. (2018). Screening of a microbial consortium for selective degradation of lignin from tree trimmings. Bioresour. Technol. 254, 247–255. doi:10.1016/j.biortech.2018.01.058
Frey-Klett, P., Burlinson, P., Deveau, A., Barret, M., Tarkka, M., and Sarniguet, A. (2011). Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 75, 583–609. doi:10.1128/MMBR.00020-11
García-Jiménez, B., Torres-Bacete, J., and Nogales, J. (2021). Metabolic modelling approaches for describing and engineering microbial communities. Comput. Struct. Biotechnol. J. 19, 226–246. doi:10.1016/j.csbj.2020.12.003
Gilmore, S. P., Lankiewicz, T. S., Wilken, S. E., Brown, J. L., Sexton, J. A., Henske, J. K., et al. (2019). Top-down enrichment guides in formation of synthetic microbial consortia for biomass degradation. ACS Synth. Biol. 8, 2174–2185. doi:10.1021/acssynbio.9b00271
Gonçalves, C. C., Bruce, T., Silva, C. O. G., Fillho, E. X. F., Noronha, E. F., Carlquist, M., et al. (2020). Bioprospecting microbial diversity for lignin valorization: Dry and wet screening methods. Front. Microbiol. 11, 1081. doi:10.3389/fmicb.2020.01081
Hassan, S. S., Williams, G. A., and Jaiswal, A. K. (2018). Emerging technologies for the pretreatment of lignocellulosic biomass. Bioresour. Technol. 262, 310–318. doi:10.1016/j.biortech.2018.04.099
Huang, J., Yang, X., Wu, Q., Mai, S., and Chi, H. (2019). Application of independent immobilization in benzo[a]pyrene biodegradation by synthetic microbial consortium. Environ. Sci. Pollut. Res. Int. 26, 21052–21058. doi:10.1007/s11356-019-05477-4
Hui, W., Jiajia, L., Yucai, L., Peng, G., Xiaofen, W., Kazuhiro, M., et al. (2013). Bioconversion of un-pretreated lignocellulosic materials by a microbial consortium XDC-2. Bioresour. Technol. 136, 481–487. doi:10.1016/j.biortech.2013.03.015
Ijoma, G. N., and Tekere, M. (2017). Potential microbial applications of co-cultures involving ligninolytic fungi in the bioremediation of recalcitrant xenobiotic compounds. Int. J. Environ. Sci. Technol. 14, 1787–1806. doi:10.1007/s13762-017-1269-3
Janusz, G., Pawlik, A., Sulej, J., Swiderska-Burek, U., Jarosz-Wilkolazka, A., and Paszczynski, A. (2017). Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 41, 941–962. doi:10.1093/femsre/fux049
Jiang, Y., Dong, W., Xin, F., and Jiang, M. (2020). Designing synthetic microbial consortia for biofuel production. Trends Biotechnol. 38, 828–831. doi:10.1016/j.tibtech.2020.02.002
Jiménez, D. J., de Lima Brossi, M. J., Schückel, J., Kračun, S. K., Willats, W. G. T., and van Elsas, J. D. (2016). Characterization of three plant biomass-degrading microbial consortia by metagenomics- and metasecretomics-based approaches. Appl. Microbiol. Biotechnol. 100, 10463–10477. doi:10.1007/s00253-016-7713-3
Jiménez, D. J., Wang, Y., Chaib de Mares, M., Cortes-Tolalpa, L., Mertens, J. A., Hector, R. E., et al. (2020). Defining the eco-enzymological role of the fungal strain Coniochaeta sp. 2T2.1 in a tripartite lignocellulolytic microbial consortium. FEMS Microbiol. Ecol. 96, fiz186. doi:10.1093/femsec/fiz186
Jokela, J., Pellinen, J., Salkinoja-Salonen, M., and Brunow, G. (1985). Biodegradation of two tetrameric lignin model compounds by a mixed bacterial culture. Appl. Microbiol. Biotechnol. 23, 38–46. doi:10.1007/BF02660116
Kamimura, N., Sakamoto, S., Mitsuda, N., Masai, E., and Kajita, S. (2019). Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 56, 179–186. doi:10.1016/j.copbio.2018.11.011
Kamimura, N., Takahashi, K., Mori, K., Araki, T., Fujita, M., Higuchi, Y., et al. (2017). Bacterial catabolism of lignin-derived aromatics: New findings in a recent decade: Update on bacterial lignin catabolism. Environ. Microbiol. Rep. 9, 679–705. doi:10.1111/1758-2229.12597
Kehe, J., Kulesa, A., Ortiz, A., Ackerman, C. M., Thakku, S. G., Sellers, D., et al. (2019). Massively parallel screening of synthetic microbial communities. Proc. Natl. Acad. Sci. U. S. A. 116, 12804–12809. doi:10.1073/pnas.1900102116
Khan, M. U., and Ahring, B. K. (2019). Lignin degradation under anaerobic digestion: Influence of lignin modifications -A review. Biomass Bioenergy 128, 105325. doi:10.1016/j.biombioe.2019.105325
Kirk, T. K., Connors, W. J., Bleam, R. D., Hackett, W. F., and Zeikus, J. G. (1975). Preparation and microbial decomposition of synthetic [14C]ligins. Proc. Natl. Acad. Sci. U. S. A. 72, 2515–2519. doi:10.1073/pnas.72.7.2515
Kohlstedt, M., Starck, S., Barton, N., Stolzenberger, J., Selzer, M., Mehlmann, K., et al. (2018). From lignin to nylon: Cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 47, 279–293. doi:10.1016/j.ymben.2018.03.003
Kong, W., Meldgin, D. R., Collins, J. J., and Lu, T. (2018). Designing microbial consortia with defined social interactions. Nat. Chem. Biol. 14, 821–829. doi:10.1038/s41589-018-0091-7
Lankiewicz, T. S., Lillington, S. P., and O’Malley, M. A. (2022). Enzyme discovery in anaerobic fungi (neocallimastigomycetes) enables lignocellulosic biorefinery innovation. Microbiol. Mol. Biol. Rev. 86, e0004122. doi:10.1128/mmbr.00041-22
Lee, J. A., Baugh, A. C., Shevalier, N. J., Strand, B., Stolyar, S., and Marx, C. J. (2021). Cross-feeding of a toxic metabolite in a synthetic lignocellulose-degrading microbial community. Microorganisms 9, 321. doi:10.3390/microorganisms9020321
Lee, S., Kang, M., Bae, J.-H., Sohn, J.-H., and Sung, B. H. (2019). Bacterial valorization of lignin: Strains, enzymes, conversion pathways, biosensors, and perspectives. Front. Bioeng. Biotechnol. 7, 209. doi:10.3389/fbioe.2019.00209
Leynaud Kieffer Curran, L. M. C., Pham, L. T. M., Sale, K. L., and Simmons, B. A. (2022). Review of advances in the development of laccases for the valorization of lignin to enable the production of lignocellulosic biofuels and bioproducts. Biotechnol. Adv. 54, 107809. doi:10.1016/j.biotechadv.2021.107809
Li, M., Pu, Y., and Ragauskas, A. J. (2016). Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 4, 45. doi:10.3389/fchem.2016.00045
Lin, L. (2022). Bottom-up synthetic ecology study of microbial consortia to enhance lignocellulose bioconversion. Biotechnol. Biofuels Bioprod. 15, 14. doi:10.1186/s13068-022-02113-1
Linde, D., Ayuso-Fernández, I., Laloux, M., Aguiar-Cervera, J. E., de Lacey, A. L., Ruiz-Dueñas, F. J., et al. (2021). Comparing ligninolytic capabilities of bacterial and fungal dye-decolorizing peroxidases and class-II peroxidase-catalases. Int. J. Mol. Sci. 22, 2629. doi:10.3390/ijms22052629
Liu, X., Bouxin, F. P., Fan, J., Budarin, V. L., Hu, C., and Clark, J. H. (2020). Recent advances in the catalytic depolymerization of lignin towards phenolic chemicals: A review. ChemSusChem 13, 4296–4317. doi:10.1002/cssc.202001213
Liu, Z.-H., Le, R. K., Kosa, M., Yang, B., Yuan, J., and Ragauskas, A. J. (2019). Identifying and creating pathways to improve biological lignin valorization. Renew. Sustain. Energy Rev. 105, 349–362. doi:10.1016/j.rser.2019.02.009
Martin, B. T., Jager, T., Nisbet, R. M., Preuss, T. G., and Grimm, V. (2013). Predicting population dynamics from the properties of individuals: A cross-level test of dynamic energy budget theory. Am. Nat. 181, 506–519. doi:10.1086/669904
Martin, B. T., Zimmer, E. I., Grimm, V., and Jager, T. (2012). Dynamic energy budget theory meets individual-based modelling: A generic and accessible implementation. Methods Ecol. Evol. 3, 445–449. doi:10.1111/j.2041-210X.2011.00168.x
Martínez, A. T., Camarero, S., Ruiz-Dueñas, F. J., and Martínez, M. J. (2018). “Chapter 8. Biological lignin degradation,” in Lignin valorization: Emerging approaches energy and environment series. Editor G. T. Beckham (Cambridge: Royal Society of Chemistry), 199–225. doi:10.1039/9781788010351-00199
Martinez-Hernandez, E., Cui, X., Scown, C. D., Amezcua-Allieri, M. A., Aburto, J., and Simmons, B. A. (2019). Techno-economic and greenhouse gas analyses of lignin valorization to eugenol and phenolic products in integrated ethanol biorefineries. Biofuels, Bioprod. Bioref. 13, 978–993. doi:10.1002/bbb.1989
Mendes, I. V., Garcia, M. B., Bitencourt, A. C. A., Santana, R. H., Lins, P. de C., Silveira, R., et al. (2021). Bacterial diversity dynamics in microbial consortia selected for lignin utilization. PloS ONE 16, e0255083. doi:10.1371/journal.pone.0255083
Minty, J. J., Singer, M. E., Scholz, S. A., Bae, C.-H., Ahn, J.-H., Foster, C. E., et al. (2013). Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc. Natl. Acad. Sci. U. S. A. 110, 14592–14597. doi:10.1073/pnas.1218447110
Moraes, E. C., Alvarez, T. M., Persinoti, G. F., Tomazetto, G., Brenelli, L. B., Paixão, D. A. A., et al. (2018). Lignolytic-consortium omics analyses reveal novel genomes and pathways involved in lignin modification and valorization. Biotechnol. Biofuels 11, 75. doi:10.1186/s13068-018-1073-4
Podolsky, I. A., Seppälä, S., Lankiewicz, T. S., Brown, J. L., Swift, C. L., and O’Malley, M. A. (2019). Harnessing nature’s anaerobes for biotechnology and bioprocessing. Annu. Rev. Chem. Biomol. Eng. 10, 105–128. doi:10.1146/annurev-chembioeng-060718-030340
Puentes-Téllez, P. E., and Falcao Salles, J. (2018). Construction of effective minimal active microbial consortia for lignocellulose degradation. Microb. Ecol. 76, 419–429. doi:10.1007/s00248-017-1141-5
Putra, F. J. N., Kahar, P., Kondo, A., and Ogino, C. (2022). Valorization of lignin and its derivatives using yeast. Processes 10, 2004. doi:10.3390/pr10102004
Rajesh Banu, J., PreethiKavitha, S., Tyagi, V. K., Gunasekaran, M., Karthikeyan, O. P., Kumar, G., et al. (2021). Lignocellulosic biomass based biorefinery: A successful platform towards circular bioeconomy. Fuel 302, 121086. doi:10.1016/j.fuel.2021.121086
Ralph, J., Lapierre, C., and Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi:10.1016/j.copbio.2019.02.019
Raud, M., Kikas, T., Sippula, O., and Shurpali, N. J. (2019). Potentials and challenges in lignocellulosic biofuel production technology. Renew. Sustain. Energy Rev. 111, 44–56. doi:10.1016/j.rser.2019.05.020
Ravi, K., Abdelaziz, O. Y., Nöbel, M., García-Hidalgo, J., Gorwa-Grauslund, M. F., Hulteberg, C. P., et al. (2019). Bacterial conversion of depolymerized Kraft lignin. Biotechnol. Biofuels 12, 56. doi:10.1186/s13068-019-1397-8
Roell, G. W., Zha, J., Carr, R. R., Koffas, M. A., Fong, S. S., and Tang, Y. J. (2019). Engineering microbial consortia by division of labor. Microb. Cell Fact. 18, 35. doi:10.1186/s12934-019-1083-3
Salvachúa, D., Katahira, R., Cleveland, N. S., Khanna, P., Resch, M. G., Black, B. A., et al. (2016). Lignin depolymerization by fungal secretomes and a microbial sink. Green Chem. 18, 6046–6062. doi:10.1039/C6GC01531J
Saravanan, A., Senthil Kumar, P., Jeevanantham, S., Karishma, S., and Vo, D.-V. N. (2022). Recent advances and sustainable development of biofuels production from lignocellulosic biomass. Bioresour. Technol. 344, 126203. doi:10.1016/j.biortech.2021.126203
Schutyser, W., Renders, T., Van den Bosch, S., Koelewijn, S. F., Beckham, G. T., and Sels, B. F. (2018). Chemicals from lignin: An interplay of lignocellulose fractionation, depolymerisation, and upgrading. Chem. Soc. Rev. 47, 852–908. doi:10.1039/c7cs00566k
Shahab, R. L., Brethauer, S., Davey, M. P., Smith, A. G., Vignolini, S., Luterbacher, J. S., et al. (2020). A heterogeneous microbial consortium producing short-chain fatty acids from lignocellulose. Science 369, eabb1214. doi:10.1126/science.abb1214
Silva, J. P., Ticona, A. R. P., Hamann, P. R. V., Quirino, B. F., and Noronha, E. F. (2021). Deconstruction of lignin: From enzymes to microorganisms. Molecules 26, 2299. doi:10.3390/molecules26082299
Song, N., Xu, H., Yan, Z., Yang, T., Wang, C., and Jiang, H.-L. (2019). Improved lignin degradation through distinct microbial community in subsurface sediments of one eutrophic lake. Renew. Energy 138, 861–869. doi:10.1016/j.renene.2019.01.121
Tien, M., and Kirk, T. K. (1983). Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium burds. Science 221, 661–663. doi:10.1126/science.221.4611.661
Tuomela, M., Vikman, M., Hataka, A., and Itavaara, M. (2000). Biodegradation of lignin in a compost environment: A review. Bioresour. Technol. 72, 169–183. doi:10.1016/S0960-8524(99)00104-2
Wang, Y., Elzenga, T., and van Elsas, J. D. (2021). Effect of culture conditions on the performance of lignocellulose-degrading synthetic microbial consortia. Appl. Microbiol. Biotechnol. 105 (20), 7981–7995. doi:10.1007/s00253-021-11591-6
Wang, Y., Liu, Q., Yan, L., Gao, Y., Wang, Y., and Wang, W. (2013). A novel lignin degradation bacterial consortium for efficient pulping. Bioresour. Technol. 139, 113–119. doi:10.1016/j.biortech.2013.04.033
Weiland, F., Kohlstedt, M., and Wittmann, C. (2022). Guiding stars to the field of dreams: Metabolically engineered pathways and microbial platforms for a sustainable lignin-based industry. Metab. Eng. 71, 13–41. doi:10.1016/j.ymben.2021.11.011
Xu, C., Su, X., Wang, J., Zhang, F., Shen, G., Yuan, Y., et al. (2021). Characteristics and functional bacteria in a microbial consortium for rice straw lignin-degrading. Bioresour. Technol. 331, 125066. doi:10.1016/j.biortech.2021.125066
Xu, T., Zong, Q.-J., Liu, H., Wang, L., Liu, Z.-H., Li, B.-Z., et al. (2022). Identifying ligninolytic bacteria for lignin valorization to bioplastics. Bioresour. Technol. 358, 127383. doi:10.1016/j.biortech.2022.127383
Zhang, W., Ren, X., Lei, Q., and Wang, L. (2021). Screening and comparison of lignin degradation microbial consortia from wooden antiques. Molecules 26, 2862. doi:10.3390/molecules26102862
Zhao, C., Xie, S., Pu, Y., Zhang, R., Huang, F., Ragauskas, A. J., et al. (2016). Synergistic enzymatic and microbial lignin conversion. Green Chem. 18, 1306–1312. doi:10.1039/C5GC01955A
Keywords: lignin, microbial communities, synthetic consortia, biomass valorization, white-rot fungi
Citation: Rodriguez A, Hirakawa MP, Geiselman GM, Tran-Gyamfi MB, Light YK, George A and Sale KL (2023) Prospects for utilizing microbial consortia for lignin conversion. Front. Chem. Eng. 5:1086881. doi: 10.3389/fceng.2023.1086881
Received: 01 November 2022; Accepted: 06 January 2023;
Published: 17 January 2023.
Edited by:
Daehwan Kim, Hood College, United StatesReviewed by:
Steven D. Karlen, University of Wisconsin-Madison, United StatesSandip Kumar Singh, University of Kansas, United States
Copyright © 2023 Rodriguez, Hirakawa, Geiselman, Tran-Gyamfi, Light, George and Sale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto Rodriguez, YWxiZXJ0b0BsYmwuZ292
 Alberto Rodriguez
Alberto Rodriguez Matthew P. Hirakawa
Matthew P. Hirakawa Gina M. Geiselman
Gina M. Geiselman Mary B. Tran-Gyamfi3
Mary B. Tran-Gyamfi3 Yooli K. Light
Yooli K. Light Kenneth L. Sale
Kenneth L. Sale