- 1Shenzhen Key Laboratory of Marine Bioresource and Eco-Environmental Science, Shenzhen Engineering Laboratory for Marine Algal Biotechnology, Guangdong Provincial Key Laboratory for Plant Epigenetics, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, College of Optoelectronic Engineering, Shenzhen University, Shenzhen, China
- 3BASIS International School Park Lane Harbour, Shenzhen, China
With the continuous growth of the world’s population and the increasing development of industrialization, the demand for energy by human beings has been expanding, resulting in an increasingly severe energy crisis. Microalgae are considered the most potential alternatives to traditional fossil fuels due to their many advantages, like fast growth rate, strong carbon sequestration capacity, and low growth environment requirements. Euglena can use carbon sources such as glucose, ethanol, and others for heterotrophic growth. Moreover, Euglena is highly adaptable to the environment and has a high tolerance to various environmental stresses, such as salinity, heavy metals, antibiotics, etc. Different treatments of Euglena cells could affect their growth and the accumulation of bioactive substances, especially fatty acids. To expand the industrial application of Euglena as a potential biodiesel candidate, we determine the physiological responses of Euglena against environmental stresses (antibiotics, heavy metals, salinity) or carbon resources (glucose and ethanol), and evaluate the potential for higher quality and yield of fatty acid with a high growth rate. Adding glucose into the culture media increases cell biomass and fatty acid production with high-quality biodiesel characters. The transcriptome analysis helped explore the possible regulation and biosynthesis of fatty acids under different treatments and exploited in the improvement of biodiesel production. This study provides insights for further improvement and various culture treatments for Euglena-based biodiesel and jet fuels.
1 Introduction
Environmental problems and energy crises are becoming increasingly severe; therefore, it is imperative to discover novel, renewable, and environmentally friendly energy sources (Xia et al., 2019; Luo et al., 2021). Many microalgae, such as Chlorella, Spirulina, and Dunaliella, are cultured on a large scale and used in industrial applications (Gilmour, 2019). The microalga Euglena gracilis has characteristics of both plants and animals, with secondary endosymbiotic chloroplasts without cell walls. Its cells contain many high-value bioproducts, including vitamins, amino acids, unsaturated fatty acids, and paramylon (Grimm et al., 2015; Zakryś et al., 2017). E. gracilis is of considerable environmental importance and biotechnological value: it can survive a variety of carbon resources, toxic chemicals, and adverse environmental conditions such as heavy metals, antibiotics, acid, salinity, and high levels of ionizing radiation, and can be observed in most waters such as ponds, fish farms and small rivers (Rodríguez-Zavala et al., 2007; Kottuparambil et al., 2012; Mukaida et al., 2016; Moreno-Sánchez et al., 2017; He et al., 2021). Indeed, E. gracilis exhibits remarkable metabolic diversity, blooming as photosynthetic autotrophy, heterotrophy, and photoheterotrophy (Zakryś et al., 2017). Thus, Euglena species are highly flexible to different nutrients and tolerant to adverse environments, which makes Euglena a model microorganism for environmental assessments, wastewater remediation, and sources of numerous bioproducts.
Environmental factors such as temperature, nutrients, carbon source, heavy metals, trace elements, antibiotics, and organic matter affect the growth of microalgae (Béchet et al., 2013; Gao et al., 2021; Maltsev et al., 2021). In this study, biomass, fatty acid (FA) contents, compositions, and biodiesel quality under the addition of paromomycin (PRM), ethanol, glucose, CdCl2, and NaCl with continuously light cultivation were evaluated.
In recent years, with the development of next-generation sequencing platforms, transcriptome sequencing (RNA sequencing, RNA-seq) has been gradually applied to microalgae research, with significant results (Khan et al., 2018). The first de novo transcriptome study revealed unexpected metabolic capabilities for carbohydrate and natural product biochemistry in Euglena (O’neill et al., 2015). Another comparative transcriptome analysis investigated Euglena’s response to anaerobic conditions, focusing on paramylon and wax ester metabolic pathways (Yoshida et al., 2016). Five available E. gracilis transcriptome data, under fermentative, mixotrophic, heterotrophic, and phototrophic culture conditions, were well summarized (Geimer et al., 2009; Ebenezer et al., 2017, 2019; Cordoba et al., 2021). Thus, research on microalgae transcriptomes helps us characterize microalgal diversity better. The application of this technology will enable us to understand the mechanisms by which E. gracilis responds to different carbon sources and environmental stresses. However, no data were collected for transcriptome investigation in Euglena under environmental stresses.
In this study, we used RNA-seq technology and de novo assembly to perform analysis of the E. gracilis transcriptome under selected treatments as addition of ethanol (E1.0, 1.0% vol/vol), glucose (G1.0, 1.0% weight), NaCl (NaCl1, 1.0%) Cd0.5 (CdCl2 0.5 mM), and PRM5.0 (5 μg/ml). Differential gene expression and related metabolic pathways of E. gracilis in response to various treatments were investigated. Euglena cells showed both shared and distinct responses to these treatments at the transcriptional level. CdCl2 stress significantly down-regulated thousands of genes, some of them related to amino acid metabolism and N-glycan biosynthesis. NaCl inhibited cell growth but did not change gene expression much at the transcriptional level. Different carbon sources such as ethanol and glucose promoted cell growth and altered the expression of genes related to photosynthesis, carbon fixation, nucleotide biosynthetic processes, component of the plasma membrane, ABC transporters, and the mRNA surveillance pathway. GO and KEGG pathway enrichments provide insights into the mechanisms of differential responses of E. gracilis to different treatments, especially the FA contents and composition changes.
2 Materials and Methods
2.1 Strain and Cultural Conditions
E. gracilis 1224/5Z was obtained from the Culture Collection of Algae and Protozoa (https://www.ccap.ac.uk/). Aliquots of cells (1–3 × 105) were incubated in 5 ml of fresh culture medium. The algal cells were grown in photosynthetic medium (1.8 g/L NH4Cl, 0.6 g/L KH2PO4, 0.6 g/L MgSO4, 60 mg/L urea, 0.02 g/L CaCl2, 0.48 mg/L Na2EDTA, 2 mg/L Fe2(SO4)3, 60 μL HCl, 0.01 mg/L Vb1, 0.0005 mg/L Vb12, 20 mg/L CuSO4·5H2O, 0.4 g/L ZnSO4·7H2O, 1.3 g/L Co(NH3)·H2O, and 1.6 g/L MnCl2·4H2O) under a light intensity of approximately 100 μmol/m2/s in an illuminating incubator at 26 °C until algal cells reached the stationary phase (Afiukwa et al., 2007; Wang et al., 2018).
2.2 Cultivation and Stress Treatments
E. gracilis cells were cultured for 6 days in a photosynthetic medium, then 1×106 cells/mL were centrifuged at 5,000×g for 3 min and transferred into an equal medium volume. Treatments were applied, including supplementation with PRM (PRM1, 5, 25 as 1, 5, 25 μg/ml), ethanol (E0.5, 1.0, 1.5 as 0.5, 1, 1.5% vol/vol), glucose (G0.5, G1.0, G1.5 as 0.5, 1.0, 1.5% weight), CdCl2 (Cd0.5, 1, 1.5 as CdCl2 of 0.5, 1.0, 1.5 mM), or NaCl1.0 (NaCl as 1% weight); After 6 days, samples were collected and used for future experiments. The concentrations were selected based on previous references (Kirk, 1962; Gonzalez-Moreno et al., 1997; Sánchez-Thomas et al., 2016; Ji et al., 2018) and our primarily experimental results.
2.3 Growth Biomass
The dry weight of 107–108 cells was measured using the oven-drying method (Edmunds, 1965). The total algal chlorophyll was extracted with 95% ethanol, and the content was spectrophotometrically assayed according to the method (Harris, 2009). Triplicates of each treatment were conducted.
2.4 Fatty Acids and FAME Analyses
After 6 days’ growth, E. gracilis samples were taken separately and centrifuged at 3,000 × g for 10 min to collect algal cells. The samples are freeze-dried for 48 h and weighed. Take 10 mg dry algae powder, add C19: 0 fatty acid methyl ester as internal standard, 0.01% butylated hydroxytoluene (BHT) methanol solution as an antioxidant, and refer to Zeng et al. (2016) for processing. The composition and content of fatty acids are detected by gas chromatography-mass spectrometry GC-MS (United States, 7890A-5975C). The column model is VF-23 ms (0.32 mm × 0.15 μm × 60 m), high-purity helium with a purity greater than 99.999% is used as the carrier gas, the injection volume is 1 μL, the solvent is delayed for 5 min, the inlet temperature is 240°C, constant flow mode, and the split ratio is 10: 1. For calculating the fatty acid content, the internal standard curve method is used to calculate the absolute content and then convert it into the percentage content.
One-way ANOVA is used for the statistics of different treatment samples. The statistical software is SPSS20. The quality of Euglena fatty acids derived biodiesels, key indexes like saponification (SN), iodine values (IN), and cetane number (CN) were also investigated (Lu et al., 2012). The saponification value (SN) is the mass of sodium hydroxide required for the complete saponification of 1 g grease, which can reflect the relative molecular weight of grease, and SN = Σ(560 × Pi)/MWi. The iodine value (IN) indicates the degree of grease unsaturation. The iodine value reflects the number of double bonds in FA and IN = Σ(254 × D × Pi)/MWi. CN is a standard to measure the combustion performance of biodiesel in engines, and CN = 46.3 + 5458/SN - 0.225 × IN. This study combines GC-MS component analysis and gets three main performance parameters of FA in E. gracilis as biodiesel under different treatments (Lu et al., 2012).
2.5 RNA Isolation and RNA-Seq Analyses
The cDNA library construction and double-ended PE125 Illumina sequencing were performed by Real Omics (Biotech) Co., Ltd (Shenzhen, China). Approximately 106–107 cells were harvested by centrifugation at 5,000×g for 5 min at 4°C and used for total RNA isolation with a Trizol Kit (Invitrogen, United States). For RNA-seq, triplicates of each sample were used.
Because E. gracilis has no reference genome, Trinity software was used to assemble the clean reads, Corset was used to perform hierarchical clustering, and Benchmarking Universal Single-Copy Orthologs (BUSCO) was used to evaluate the completeness and integrity of the transcriptome assembly. Seven NCBI databases were used to annotate the assembled transcripts: NCBI non-redundant protein sequences (Nr), NCBI nucleotide sequences (Nt), Protein family (Pfam), euKaryotic Ortholog Groups (KOG) Clusters of Orthologous Groups of proteins (COG), Swiss-Prot manually annotated and reviewed protein sequence database, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO). The input data for the differential expression analysis were read counts obtained from the analysis of gene expression data. The analysis of samples with biological replicates was performed in DESeq2 based on a negative binomial distribution.
The KEGG Orthology Based Annotation System (KOBAS) was used to perform KEGG pathway enrichment analysis for each comparison with an automatic annotation server e-value of 1e−10. GO annotations were obtained from the annotations of the top 10 Nr blast hits using Blast2GO (Conesa et al., 2005) and from the Pfam database using InterPro scan (Anders and Huber, 2010; Kimbrel et al., 2011). The Seq method was used to identify pathways enriched in up-regulated and down-regulated differential gene sets for each group. When more genes are up-regulated than down-regulated in a selected pathway, the pathway would be marked up-regulated under this condition (Campanaro et al., 2007).
2.6 Statistical Analysis
Significant differences in growth biomass, fatty acids, and biodiesel fuel properties were tested using Dunnett’s t-test. All data were obtained and averaged from at least three independent experiments, and standard errors were calculated and displayed as error bars.
3 Results
3.1 Biomass and Chlorophyll Contents Under Different Treatments
Biomass differed significantly under different environmental treatments (Figure 1A). The PRM treatment caused no significant change in biomass, and NaCl and CdCl2 treatment significantly reduced biomass (p < 0.001) by 18.18 and 18.72% relative to the control, respectively. All ethanol and glucose concentrations significantly increased biomass (p < 0.001) relative to the control by 64.75–107.66% and 216.47%, respectively. Glucose treatments showed higher biomass increases under all current concentrations in this case.
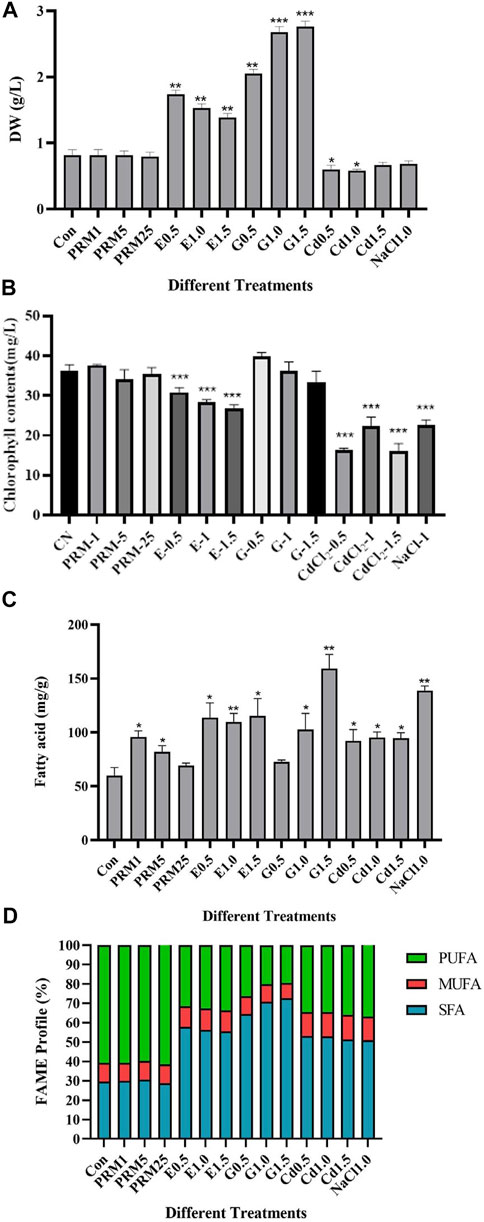
FIGURE 1. Physiological changes of E. gracilis under different treatments. Dried weights (A), chlorophyll contents (B), variations of fatty acids contents (C) and FAME profile (D) of E. gracilis under different treatments. Asterisks “*”, “**” indicate significant differences between treatments and control treatment (Con) at p < 0.05 and p < 0.01, respectively. Treatments include supplementation with paromomycin (PRM1, 5, 25 as 1, 5, 25 μg/ml), ethanol (E0.5, 1.0, 1.5 as 0.5, 1, 1.5% vol/vol), glucose (G0.5, G1.0, G1.5 as 0.5, 1.0, 1.5% weight), CdCl2 (Cd0.5, 1.0, 1.5 as CdCl2 of 0.5, 1.0, 1.5 mM), or NaCl1.0 (NaCl as 1% weight).
Chlorophylls in microalgae are sensitive to environmental stress. The addition of ethanol (all concentrations) CdCl2 (all doses), and NaCl decreased the chlorophyll content of E. gracilis significantly (Figure 1B), whereas PRM and Glucose had no significant effect.
3.2 Fatty Acid Contents, Compositions, and FAME
The total fatty acid (TFA) content of E. gracilis under all treatments increases to varying degrees compared with the control, and some treated groups showed concentration-dependent manners, such as paromomycin (PRMs) and glucose (G) treatments (Figure 1C). The TFA content in PRM groups tends to decrease as the concentration increases. A low concentration of PRM (PRM1) has a higher effect on accumulating total fatty acids. However, in glucose-treated groups (G) TFA tends to increase with concentration, with the highest in G1.5 by about 132% increases compared to the control (Con). The second high TFA content was detected in the NaCl group, around 140 mg/g. The addition of ethanol (E0.5, E1.0, E1.5, as 0.5–1.5% vol/vol) nearly doubled the TFA compared to Con, with no significant difference among different concentrations. This study also showed a similar TFA increase by CdCl2 at different concentrations.
Investigation of FA composition, namely SFA, MUFA, and PUFA, indicated interesting differences under treatments (Figure 1D and Supplementary Table S1). Compared with the control, the FA composition of E. gracilis under paromomycin treatments (PRM1, -5, -25) changed little. In contrast, the FAME profile under CdCl2, ethanol, glucose, and NaCl treatment changed significantly.
The general trend of FA composition changes was almost the same, both which SFA proportion increases and PUFA proportion decreases, indicating more saturated fatty acids under these treatments. MUFA proportion changes little, and the most significant change was observed in the G1.5 treatment, whose SFA proportion increased from 29.51% of the control group to 72.72%. The PUFA proportion decreases from 60.85% of the control group to 19.49%. The change patterns in the other groups, Cd0.5, 1.0, 1.5, E0.5, 1.0, 1.5, and NaCl, were similar with no apparent dose-dependent pattern.
3.3 Evaluation of Biodiesel Fuel Properties of E. gracilis TFA
Biodiesel fuel characters were evaluated for E. gracilis TFA under different consumption methods. All the data are obtained and averaged from at least three independent experiments. As shown in the figure, compared with the control, the changes of SN (a), IN (b), and CN (c) of E. gracilis under different treatments were generally different (Figure 2).
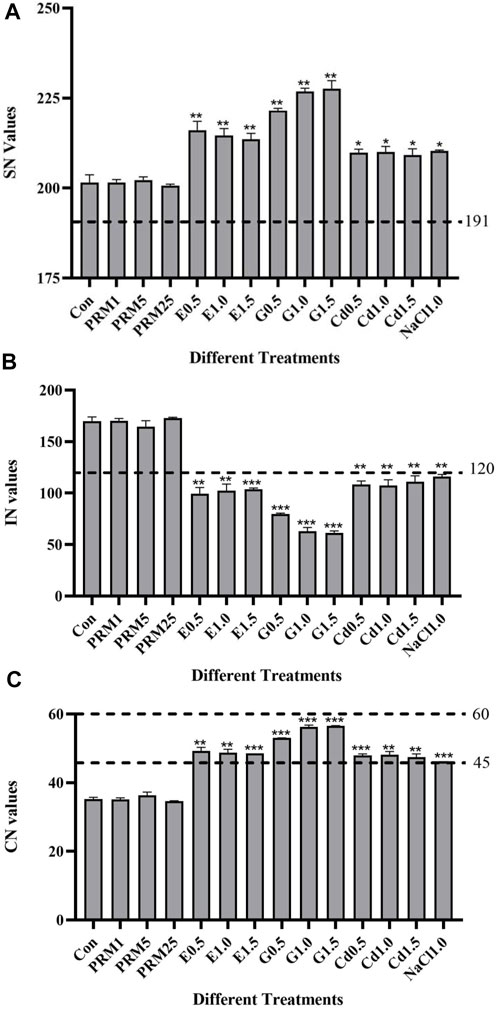
FIGURE 2. Biodiesel fuel properties of E. gracilis TFA under different treatments at the sixth day. (A) saponification value (SN), (B) iodine value (IN), (C) cetane number (CN). Asterisks “*”, “**” indicate significant differences between treatments and control treatment (Con) at p < 0.05 and p < 0.01, respectively. Lines indicate the standard threshold values for each biodiesel fuel property (SN, IN, or CN).
The SN value of E. gracilis TFA slightly or significantly increased under different treatments than the control, except for no change under antibiotics treatments. The SN value of the G-1.5 treatment is the largest compared with the control, which increases by about 13%. The German EN 14214 standard requires that the SN value of biodiesel should be 187–191 mg/g. The SN value of each group in this study was larger than the standard (Figure 2A).
The minimum IN value standard in various countries’ biodiesel production standards is <125 g/100 g (Arguelles and Martinez-Goss, 2021). The IN values of E. gracilis TFA in the control and paromomycin treated group are higher than the minimum standard by 35.92–36.02%, which is not suitable for direct use as biodiesel feedstock. In addition, the IN values of E. gracilis TFA under CdCl2, ethanol, glucose, and NaCl treatments are significantly lower than those in control (Figure 2B). With the highest IN level (116.15), NaCl treated group has already met the standards of China, Australia, and the European Union (<120 g/100 g) and could be used as an ideal choice for biodiesel feedstock. The IN values of CdCl2, ethanol, and glucose treatments (107.44–111.03, 99.22–103.62, 61.21–79.63) all reach the strict standards of biodiesel production in various countries, like the biodiesel production standards <115 g/100 g in Germany (EN 14214) and France, with excellent combustibility (Lu et al., 2012; Arguelles and Martinez-Goss, 2021). Among the low IN value treatments, the G1.5 treatment has the lowest compared with the control, with a significant reduction of about 64%.
Compared with the control, CN was improved under all treatments except for PRM (Figure 2C). The minimum requirement of CN in biodiesel production standards of various countries is higher than 45. The CN value of control and PRM groups is still 22.87–22.96%, respectively, lower than the minimum production standards of biodiesel. It could not be used directly as the raw material for biodiesel. The CN value of the E0.5 treatment is already 0.04% higher than the Chinese GB/T 20,828 standard and has the potential as a biodiesel feedstock. The CN values of G0.5, G1.0, and G1.5 treatment groups (53.02, 56.22, 56.51) have all reached the most demanding EU 14124 standard (> 51). The ideal treatment is the addition of glucose under different concentrations, about 51–60% higher than the control.
3.4 RNA-Seq Analyses
3.4.1 Summary of RNA-Seq and Differentially Expressed Genes
Our biomass and TFA results showed that treatments of E1.0, G1.0, NaCl1.0, Cd0.5, PRM25 caused significant differences compared to control samples, thus, these conditions were used for further transcriptome analyses. An average of 47,452,424 reads was generated for each sample. After removing low-quality reads, an average of 46,542,906 clean reads was obtained for each sample. The average GC content was less than 60%, the Q20 and Q30 values were 97 and 92%, respectively, and the sequencing error rate was 3%.
Compared with the control, overall gene expression changes were the most obvious in the CdCl2 treatment, with 3,040 and 1,284 up-regulated and down-regulated genes. The heavy metal Cd2+, therefore, appeared to have a significant effect on the transcriptome of E. gracilis (Table 1). The gene expression changes were less evident in the ethanol and glucose treatments. Under ethanol addition (E1.0), there were 56 and 1,040 up- and down-regulated genes, i.e., about 20 times more down-regulated genes than up-regulated ones. Similarly, in the glucose treatment (G1), there were 1,043 down-regulated genes and only 37 up-regulated genes.
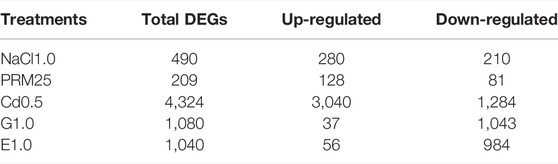
TABLE 1. Differentially expressed genes (DEGs) based on RNA-seq under different treatments compared with the control. Treatments include supplementation with paromomycin (PRM25, 25 μg/ml), ethanol (E1.0, 1.0% vol/vol), glucose (G1.0, 1.0% weight), CdCl2 (Cd0.5, 0.5 mM), or NaCl1.0 (NaCl as 1.0% weight).
Compared with thousands of genes altered under the addition of Cd2+, glucose, and ethanol treatments, PRM and NaCl treatments had relatively modest changes on the overall transcriptome, causing up-regulation of 128–280 genes and down-regulation of 81–210 genes (Table 1).
3.4.2 Gene Expression Patterns Related to Reactive Oxygen Species, Fatty Acid Metabolism and Chlorophyll Biosynthesis
Reactive oxygen species (ROS) scavenging genes, such as APX, L-ascorbate peroxidase; FTRC, Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic; GPX, glutathione peroxidase; SOD1, Superoxide dismutase [Fe]; SOD2, Superoxide dismutase [Mn]; SOD3, Superoxide dismutase [Zn]; CAT1,2, catalase1, 2; trxB, Thioredoxin reductase, were screened for differential gene expression analysis (Table 2). Very obviously, 13 of 18 selected genes were induced by addition of CdCl2, while most genes were down-regulated with PRM25 (13/18) and NaCl (11/18) (Table 2).
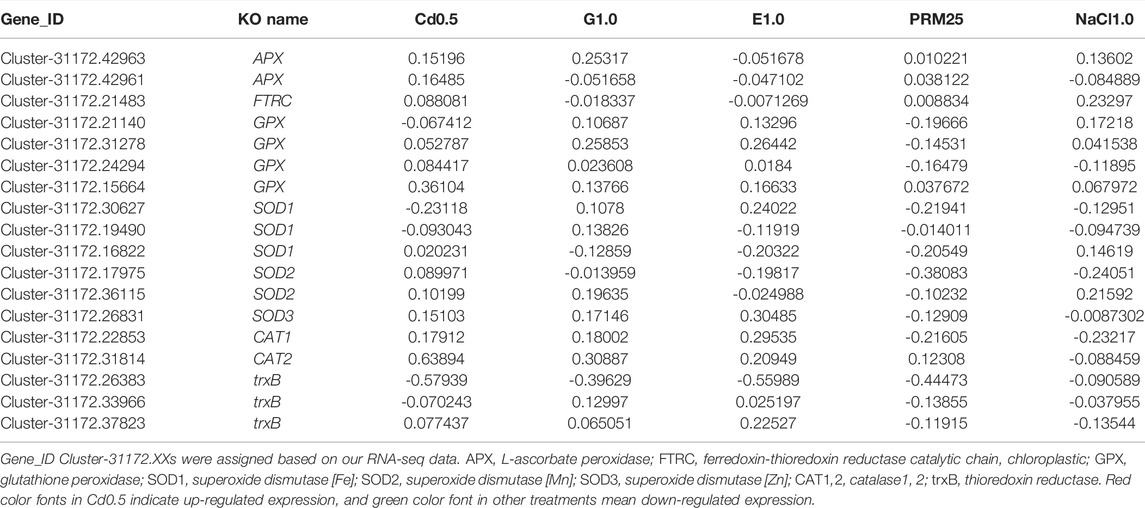
TABLE 2. The Log2FC (fold change) of selected Reactive Oxygen Species (ROS) scavenging genes under different treatments, treatment vs. control.
According to FA metabolism, total 31 genes were identified and no significant changes with more than 1.5 or 2 fold. With slightly up-regulation, most FA metabolism related genes were correlated with E1.0 (26/31 genes) and G1.0 (26/31 genes). NaCl treatment did not cause much up-regulation instead (9/31 genes) (Supplementary Table S2).
Based on chlorophyll synthesis and metabolism genes, mostly down-regulated gene numbers were identified in treatments of NaCl1.0 (17/24 genes) and Cd0.5 (10/24 genes) (Table 3). Again, most changes of these genes were less than 1.0 fold.
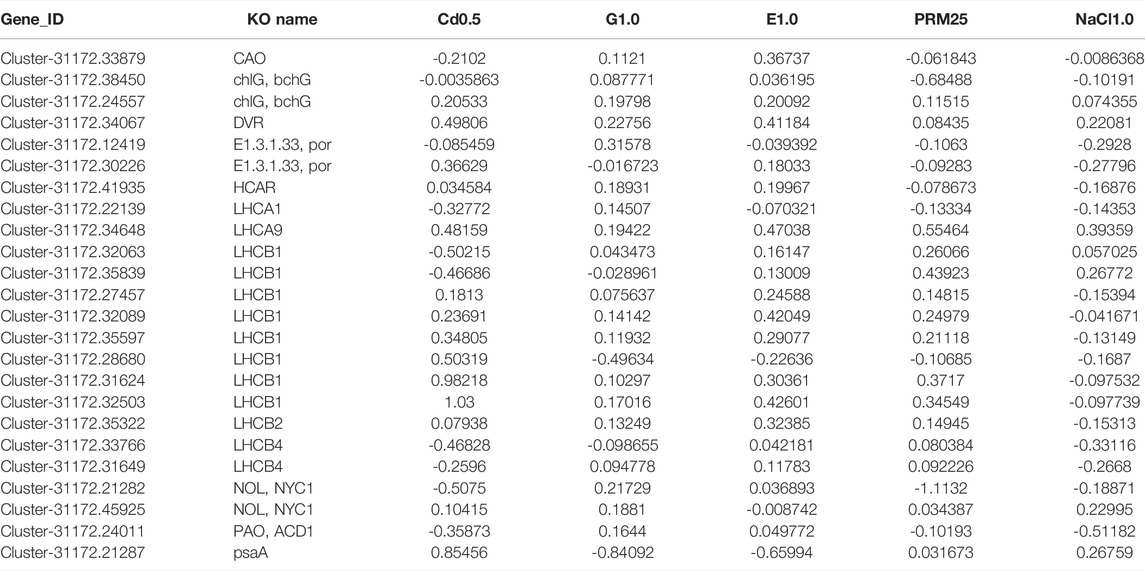
TABLE 3. The Log2FC (fold change) of selected chlorophyll related genes under different treatments, treatment vs. control. Gene_ID Cluster-31172.XXs were assigned based on our RNA-seq data.
3.4.3 GO and KEGG Pathway Enrichments
GO Pathways enriched under different treatments showed various and similar patterns under our selected treatments. Pathways down-regulated in response to 1.0% ethanol (E1) included several nucleotide biosynthetic or metabolic processes, signal transduction, and cell communication; integral and intrinsic component of the plasma membrane (Supplementary Figure S1 E1.0 GO). Similarly, adding glucose reduced nucleotide biosynthetic or metabolic processes, signal transduction, and the component of plasma membranes (Supplementary Figure S1 G1.0 GO). NaCl treatment caused up-regulation of genes associated with the arginine biosynthetic process, argininosuccinate synthase activity, and down-regulation of genes in the monosaccharide binding pathway (Supplementary Figure S1 NaCl GO). In response to the antibiotic PRM, genes in the pyruvate, proline, and arginine metabolic processes, sodium ion transmembrane transport, and sodium ion export were up-regulated. Still, genes involved with oxaloacetate decarboxylase activity were down-regulated (Supplementary Figure S1 PRM GO). The heavy metal treatment is an exception in this case: more pathways were up-regulated than down-regulated. Most significantly, Na + transport, organic transmembrane transports, non-membrane-bounded and plasma membranes, and heterocyclic and organic cyclic compound binding pathways were increased against Cd ions (Supplementary Figure S1 Cd GO).
KEGG pathways enriched under E1 are involved in the down-regulation of photosynthesis, carbon fixation in photosynthetic organisms, glyoxylate and dicarboxylate metabolism, and ABC transporters. Upregulated pathways like spliceosome, cysteine, and methionine metabolism were detected under E-1 (Supplementary Figure S2 E-1 KEGG). Photosynthesis, glyoxylate and dicarboxylate metabolism, glycerophospholipid, fatty acid degradation, and lipid metabolism pathways were down-regulated with glucose (G1). In contrast, sulfur metabolism, N-glycan biosynthesis, mRNA surveillance pathway, endocytosis, and cysteine and methionine metabolism were up-regulated (Supplementary Figure S2 G1 KEGG). Similar changes were also detected in NaCl treatment. Under NaCl treatment, photosynthesis, oxidative phosphorylation, glycerophospholipid, glycerolipid, and ether lipid metabolisms were down-regulated. In contrast, selenocompound metabolism, plant-pathogen interaction, plant hormone signal transduction, endocytosis, and cysteine and methionine metabolism were up-regulated (Supplementary Figure S2 NaCl KEGG). There are relatively few pathways changed by PRM treatment, with down-regulated purine, porphyrin, chlorophyll metabolisms, and up-regulated glycolysis/gluconeogenesis and endocytosis (Supplementary Figure S2 PRM KEGG). More complicated changes were disturbed by CdCl2, with many pathways such as down-regulated spliceosome, protein processing, nucleotide excision repair, N-glycan biosynthesis, basal transcription factors, and up-regulated ribosome, glycolysis/gluconeogenesis, and fatty acid biosynthesis and ether lipid metabolism pathways (Supplementary Figure S2 Cd KEGG).
4 Discussion
Cd2+ is a serious environmental pollutant. It is highly toxic to living organisms because it is an unnecessary ion, even at low concentrations. Not only Cd2+accumulates in Euglena, but other heavy metals such as Hg2+, Cu2+, Zn2 +, Pb2+, Tc7+, and Cr6+, thereby contributing to the remediation of waste and polluted water (Mendoza-Cózatl et al., 2006). E. gracilis is considered an efficient and reliable bioremediation microorganism with the potential to remediate heavy metal environments (Sánchez-Thomas et al., 2016). In this study, E. gracilis growth was slightly reduced due to a low concentration of 0.5 mM CdCl2. A previous study showed that Chlorella also had some tolerance to cadmium. Its growth was unaffected by 0.5 and 1 mg/L 3CdSO4•8H2O, although concentrations of 3 and 5 mg/L inhibited growth, and 7 mg/L completely blocked growth (Cheng et al., 2016). Based on the thousands of genes differentially expressed in response to CdCl2 treatment and the observed reductions in biomass, we can conclude that CdCl2 has a significant impact on Euglena cells.
NaCl treatment produced similar results, reducing biomass by 18.18%, consistent with previous studies of NaCl in Scenedesmus and E. gracilis (Ji et al., 2018). This may reflect that NaCl promotes the degradation of chlorophyll a, reduces photosynthetic capacity, and ultimately inhibits microalgae growth. This is verified through our chlorophyll assessments and related gene expression analysis.
The biochemical effects of antibiotics paromomycin (PRM) have seldom been studied in E. gracilis and other microalgae. PRM and streptomycin are aminoglycoside antibiotics, and streptomycin has a known inhibitory effect on E. gracilis growth (Kirk, 1962). However, this study showed that low concentrations (1–25 μg/ml) of PRM had little effect on E. gracilis growth, suggesting that E. gracilis is tolerant to low PRM concentrations, consistent with previous reports on Euglena antibiotic tolerance (Kirk, 1962; He et al., 2021). E. gracilis could grow mixotrophically; that is, it can first use organic carbons in the medium or field waters instead of CO2 via photosynthesis under the light. Transcriptome analyses indicated down-regulated photosynthesis with the addition of ethanol or glucose. Ethanol has been reported to induce cell division and promote the growth of E. gracilis (Horrum and Schwartzbach, 1980; Venugopal et al., 2006). Likewise, studies have shown that adding glucose to the culture medium can promote the growth of E. gracilis and Scenedesmus quadriceps (Taylor, 1960; Nicolas et al., 1980). Here, we also found out that ethanol and glucose significantly promoted the growth of E. gracilis, and the effect of glucose was significant under selected concentrations in this study.
All treatments with low doses of ethanol (all doses), glucose (1.0, 1.5%), CdCl2, NaCl, and PRM (1–5 μg/ml) can promote the accumulation of fatty acids of E. gracilis. Also, FA compositions showed some interesting patterns under different treatments.
Coleman’s research shows that the fatty acid synthesis rate of E. gracilis with an additional carbon source is higher than that of photosynthetic carbon fixation. In this study, adding ethanol and glucose significantly increases the fatty acid contents of E. gracilis compared with the control, of which G1.5 treatment has the highest FA content and the total lipid content of E. gracilis can reach 16.59% (p < 0.001). When E. gracilis is cultivated in ethanol-containing media, its total lipid content increases (Coleman et al., 1988; Thuillier-Bruston et al., 1990). When treated with different ethanol concentrations, the total lipid content can also be increased by 82.59–146.96% (p < 0.01). The glucose-rich medium can increase the lipid content in Scenedesmus quadricauda (Kandimalla et al., 2016), and glucose (35 mM) as a carbon source can promote the accumulation of lipid in Chromochloris zofingiensis (Zhang et al., 2019). The addition of carbon sources into E. gracilis culture medium alters the FA compositions a lot. The transcriptome data indicated very similar responses to ethanol and glucose. For instance, nucleotide biosynthetic or metabolic processes such as signal transduction, integral and intrinsic components of plasma membrane pathways were down-regulated under both treatments. Characters of plasma membrane depend on the composition of membrane lipids based on the saturated ratio. With the concentrations in this study, glucose increased significantly higher FA contents than ethanol. The reduced fatty acid degradation detected in the G1.0 treatment may contribute partially to higher FA production under glucose addition.
Our transcriptome analysis showed up-regulated ROS related genes in CdCl2 treatment group. Alho’s research shows that Cd has a more significant impact on the biochemical composition of Raphidocelis subcapitata, increasing total lipid content (7.2 times), total carbohydrates (3.5 times), and ROS production (3.7 times) (Alho et al., 2019). Addition of CdCl2 into E. gracilis culture medium caused the up-regulation of sodium ion and organic transmembrane transports to pump out Cd2+. At the same time, CdCl2 treatment increased glycolysis, fatty acid biosynthesis, and ether lipid metabolism changed non-membrane bound and plasma membranes with increased SFA and reduced PUFA. Thus, increased FA content and changed FA composition of E. gracilis might be the defense mechanism of E. gracilis when exposed to Cd2+, which helps reduce the metal damage to photosynthesis (Alho et al., 2019).
As we all know, under environmental pressure, FA accumulates as energy reserves, allowing algae to survive (De Carvalho and Caramujo, 2018). Increased intracellular lipid can maintain redox homeostasis because these biomolecules act as electron absorbers and help resist oxidative damage caused by ROS (Sun et al., 2018). Our transcriptome data showed NaCl also inhibited photosynthesis and changed glycerophospholipid, glycerolipid, and ether lipid metabolism. In the end, NaCl treatment resulted in the FA content increasing and FA composition alternating with a high SFA content ratio.
PRM could promote FA accumulation with no change of FA composition under low concentrations. Based on our transcriptome data, porphyrin and chlorophyll metabolisms were down-regulated while glycolysis was up-regulated under PRM treatment. We propose that the decreased chlorophyll inhibits photosynthesis, similar to NaCl, ethanol, or glucose treatments, and promotes FA accumulation. There is no alternation of any membrane metabolism among GO and KEGG pathways enriched from our RNA-seq data. This observation is consistent with no FA composition change under PRM treatment.
Interestingly, FA compositions showed significantly different and similar patterns under different treatments. Except for PRM treatments, all treatments in this study increased saturated rates 1.7∼2.4 fold compared to the control. The lipids in green cells of E. gracilis grown in the light differ from those in etiolated cells (with no photosynthesis) grown in the dark. The FA of the photosynthesizing green E. gracilis cells is mainly of the unsaturated variety (Rosenberg, 1963). An early study showed a complex lipid pattern of Euglena: phosphatidylglycerol sulfolipid, and several glycolipids appeared to be the dominant lipids during photosynthesis (Helmy et al., 1967). E. gracilis was cultured under heterotrophic or phototrophic growth conditions using ethanol, glucose, or CO2 as the primary carbon source. TFA analyses indicated that ethanol produced more highly unsaturated acids than glucose (Reitz and Moore, 1972). In E. gracilis synthesis, the more significant part of the cellular unsaturated fatty acids accompanies photosynthesis. Although it is not known whether unsaturated fatty acids participate directly in the photosynthetic mechanism, their synthesis is an essential factor in the economy of the photosynthesizing cells. Comparing the relative proportions of fatty acid species in a chloroplast fraction of green E. gracilis cells showed a significantly increased ratio of unsaturated to saturated homologs in the chloroplast (Rosenberg, 1963). The effects of ethanol and glucose on fatty acid profile showed significantly higher SFA accumulation (ΣSFA/(ΣMUFA+ΣPUFA) with glucose compared to ethanol (2.5 vs. 0.9) (Barsanti et al., 2000). In our previous report, in fully green photosynthetic E. gracilis cells under light conditions, desaturase activity was enhanced to generate a set of MUFAs and PUFAs (Zeng et al., 2016). Transcriptome analyses provide some clues about FA composition changes under treatments. Down-regulated photosynthesis was detected in E1, G1, and NaCl treatments for KEGG enriched pathways, which correlates with the significantly increased saturated FA in these treated cells.
The increase of SFA and the decrease of PUFA proportion mean that the saturation of biodiesel increases. It helps to change some indicators related to the combustion performance, like increasing the CN and lowering IN of biodiesel. This study finds that using suitable conditions to treat E. gracilis could improve not only the yield of E. gracilis FA can but alsobiodiesel quality. Based on the SN, IN, and CN, E. gracilis FA was evaluated as biodiesel. According to the biodiesel production standard, the performance of E. gracilis FA in the glucose treatment group is the best. Among them, both IN and CN of E. gracilis FA of G1.5 treatment reach the highest standards for biodiesel production in various countries. However, E. gracilis FA in all groups has the same shortcoming. The SN is too high to meet biodiesel standards in many countries.
5 Conclusion
The total fatty acids and related components of E. gracilis have increased under various treatment conditions with various TFA saturated rates and biodiesel fuel properties. Both iodine value and cetane number have reached the highest standard of current international biodiesel production under some treatments. Among them, TFA with glucose treatment is excellent, which can be used as an in-depth research object in biodiesel. The transcriptome analyses indicate that high-quality biodiesel could be achieved in E. gracilis by carefully designed cultivation medium and conditions, andbalancing the growth and photosynthesis.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA714494.
Author Contributions
JW conceived the concept, and JH, YC, YL, JZ conducted the experiments and collected the data. MD analyzed the data. JW wrote the manuscript draft. AL and WF helped to collect materials and finish the manuscript. AL, JW, JZ, and MD offered help to this project and/or revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was partially supported by China’s National Key R&D Programs (2018YFA0902500; 2020YFA0908703; 2021YFA0910800) and the National Natural Science Foundation of China (41876188).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2022.884451/full#supplementary-material
Supplementary Figure S1 | Functional categorization of up- (red color) and down-regulated (blue color) genes in E. gracilis under different treatments based on gene ontology (GO) annotations. BP, biological process, CC, cellular components, MP, molecular functions.
Supplementary Figure S2 | Bubble plots of DEGs for visualizing KEGG pathway enrichments. Bubble size indicates the log size of the KEGG term, enrichment of the down-regulated genes (down KEGG), and up-regulated genes (up KEGG).
Supplementary Table S1 | The contents of fatty acids in the unit cell of E. gracilis under different treatment conditions (pg/cell). Treatments include supplementation with paromomycin (PRM1, 5, 25 as 1, 5, 25 µg/mL), ethanol (E0.5, 1.0, 1.5 as 0.5, 1, 1.5% vol/vol), glucose (G0.5, G1.0, G1.5 as 0.5, 1.0, 1.5% weight), CdCl2 (Cd0.5, 1.0, 1.5 as CdCl2 of 0.5, 1.0, 1.5 mM), or NaCl1.0 (NaCl as 1% weight), CN, control.
Supplementary Table S2 | The Log2FC (fold change) of selected fatty acid metabolism genes under different treatments, treatment vs. control.
References
Afiukwa, C. A., and Ogbonna, J. C. (2007). Effects of Mixed Substrates on Growth and Vitamin Production by Euglena gracilis. Afr. J. Biotechnol. 6 (22), 2612–2615.
Alho, L. D. O. G., Gebara, R. C., Paina, K. D. A., Sarmento, H., and Melão, M. D. G. G. (2019). Responses of Raphidocelis Subcapitata Exposed to Cd and Pb: Mechanisms of Toxicity Assessed by Multiple Endpoints. Ecotoxicol. Environ. Saf. 169, 950–959. doi:10.1016/j.ecoenv.2018.11.087
Anders, S., and Huber, W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11, R106. doi:10.1186/gb-2010-11-10-r106
Arguelles, E. D. L. R., and Martinez-Goss, M. R. (2021). Lipid Accumulation and Profiling in Microalgae Chlorolobion Sp. (BIOTECH 4031) and Chlorella Sp. (BIOTECH 4026) during Nitrogen Starvation for Biodiesel Production. J. Appl. Phycol. 33, 1–11. doi:10.1007/s10811-020-02126-z
Barsanti, L., Bastianini, A., Passarelli, V., Tredici, M. R., and Gualtieri, P. (2000). Fatty Acid Content in Wild Type and WZSL Mutant of Euglena Gracilis - Effects of Carbon Source and Growth Conditions. J. Appl. Phycol. 12, 515–520. doi:10.1023/a:1008187514624
Béchet, Q., Shilton, A., and Guieysse, B. (2013). Modeling the Effects of Light and Temperature on Algae Growth: State of the Art and Critical Assessment for Productivity Prediction during Outdoor Cultivation. Biotechnol. Adv. 31, 1648–1663.
Campanaro, S., Picelli, S., Torregrossa, R., Colluto, L., Ceol, M., Del Prete, D., et al. (2007). Genes Involved in TGFβ1-Driven Epithelial-Mesenchymal Transition of Renal Epithelial Cells Are Topologically Related in the Human Interactome Map. BMC Genomics 8, 383. doi:10.1186/1471-2164-8-383
Cheng, J., Qiu, H., Chang, Z., Jiang, Z., and Yin, W. (2016). The Effect of Cadmium on the Growth and Antioxidant Response for Freshwater Algae Chlorella Vulgaris. Springerplus 5, 1290. doi:10.1186/s40064-016-2963-1
Coleman, L. W., Rosen, B. H., and Schwartzbach, S. D. (1988). Environmental-control of Carbohydrate and Lipid-Synthesis in Euglena. Plant Cell Physiology 29, 423–432. doi:10.1016/s0022-2275(20)38565-5
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21, 3674–3676. doi:10.1093/bioinformatics/bti610
Cordoba, J., Perez, E., Van Vlierberghe, M., Bertrand, A. R., Lupo, V., Cardol, P., et al. (2021). De Novo transcriptome Meta-Assembly of the Mixotrophic Freshwater Microalga Euglena Gracilis. Genes (Basel) 12. doi:10.3390/genes12060842
De Carvalho, C. C. C. R., and Caramujo, M. J. (2018). The Various Roles of Fatty Acids. Molecules 23, 2583. doi:10.3390/molecules23102583
Ebenezer, T. E., Carrington, M., Lebert, M., Kelly, S., and Field, M. C. (2017). Euglena Gracilis Genome and Transcriptome: Organelles, Nuclear Genome Assembly Strategies and Initial Features. Adv. Exp. Med. Biol. 979, 125–140. doi:10.1007/978-3-319-54910-1_7
Ebenezer, T. E., Zoltner, M., Burrell, A., Nenarokova, A., Novák Vanclová, A. M. G., Prasad, B., et al. (2019). Transcriptome, Proteome and Draft Genome of Euglena Gracilis. BMC Biol. 17, 11. doi:10.1186/s12915-019-0626-8
Edmunds, L. N. (1965). Studies on Synchronously Dividing Cultures ofEuglena Gracilis Klebs (Strain Z). II. Patterns of Biosynthesis during the Cell Cycle. J. Cell. Comp. Physiol. 66, 159–181. doi:10.1002/jcp.1030660205
Gao, P., Guo, L., Zhao, Y., Jin, C., She, Z., and Gao, M. (2021). Enhancing Microalgae Growth and Product Accumulation with Carbon Source Regulation: New Perspective for the Coordination between Photosynthesis and Aerobic Respiration. Chemosphere 278, 130435. doi:10.1016/j.chemosphere.2021.130435
Geimer, S., Belicová, A., Legen, J., Sláviková, S., Herrmann, R. G., and Krajčovič, J. (2009). Transcriptome Analysis of the Euglena Gracilis Plastid Chromosome. Curr. Genet. 55, 425–438. doi:10.1007/s00294-009-0256-8
Gilmour, D. J. (2019). Microalgae for Biofuel Production. Adv. Appl. Microbiol. 109, 1–30. doi:10.1016/bs.aambs.2019.10.001
Gonzalez-Moreno, S., Gomez-Barrera, J., Perales, H., and Moreno-Sanchez, R. (1997). Multiple Effects of Salinity on Photosynthesis of the Protist Euglena Gracilis. Physiol. Plant 101, 777–786. doi:10.1111/j.1399-3054.1997.tb01063.x
Grimm, P., Risse, J. M., Cholewa, D., Müller, J. M., Beshay, U., Friehs, K., et al. (2015). Applicability of Euglena Gracilis for Biorefineries Demonstrated by the Production of α-tocopherol and Paramylon Followed by Anaerobic Digestion. J. Biotechnol. 215, 72–79. doi:10.1016/j.jbiotec.2015.04.004
Harris, E. H. (2009). The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and its Laboratory Use. 2nd Edn. San Diego, CA: Academic Press, 119267–158274.
He, J., Liu, C., Du, M., Zhou, X., Hu, Z., Lei, A., et al. (2021). Metabolic Responses of a Model Green Microalga Euglena Gracilis to Different Environmental Stresses. Front. Bioeng. Biotechnol. 9, 662655. doi:10.3389/fbioe.2021.662655
Helmy, F. M., Hack, M. H., and Yaeger, R. G. (1967). Comparative Lipid Biochemistry-VI. Lipids of Green and Etiolated Euglena Gracilis and of Blastocrithidia Culicis. Comp. Biochem. Physiology 23, 565–567. doi:10.1016/0010-406x(67)90408-2
Horrum, M. A., and Schwartzbach, S. D. (1980). Nutritional Regulation of Organelle Biogenesis in Euglena. Plant Physiol. 65, 382–386. doi:10.1104/pp.65.2.382
Ji, X., Cheng, J., Gong, D., Zhao, X., Qi, Y., Su, Y., et al. (2018). The Effect of NaCl Stress on Photosynthetic Efficiency and Lipid Production in Freshwater Microalga-Scenedesmus Obliquus XJ002. Sci. Total Environ. 633, 593–599. doi:10.1016/j.scitotenv.2018.03.240
Kandimalla, P., Desi, S., and Vurimindi, H. (2016). Mixotrophic Cultivation of Microalgae Using Industrial Flue Gases for Biodiesel Production. Environ. Sci. Pollut. Res. 23, 9345–9354. doi:10.1007/s11356-015-5264-2
Khan, A. Z., Shahid, A., Cheng, H., Mahboob, S., Al-Ghanim, K. A., Bilal, M., et al. (2018). Omics Technologies for Microalgae-Based Fuels and Chemicals: Challenges and Opportunities. Ppl 25, 99–107. doi:10.2174/0929866525666180122100722
Kimbrel, J. A., Di, Y., Cumbie, J. S., and Chang, J. H. (2011). RNA-seq for Plant Pathogenic Bacteria. Genes 2, 689–705. doi:10.3390/genes2040689
Kirk, J. T. (1962). Effect of Streptomycin on Greening and Biosynthesis in Euglena Gracilis. Biochimica Biophysica Acta 56, 139–151. doi:10.1016/0006-3002(62)90534-6
Kottuparambil, S., Shin, W., Brown, M. T., and Han, T. (2012). UV-B Affects Photosynthesis, ROS Production and Motility of the Freshwater Flagellate, Euglena Agilis Carter. Aquat. Toxicol. 122-123, 206–213. doi:10.1016/j.aquatox.2012.06.002
Lu, S., Wang, J., Niu, Y., Yang, J., Zhou, J., and Yuan, Y. (2012). Metabolic Profiling Reveals Growth Related FAME Productivity and Quality of Chlorella Sorokiniana with Different Inoculum Sizes. Biotechnol. Bioeng. 109, 1651–1662. doi:10.1002/bit.24447
Luo, Y., Zhang, X., Huang, C., Han, X., Jiang, Q., Zhou, T., et al. (2021). Zn0.8Cd0.2S Hollow Spheres with a Highly Dispersed Ni Dopant for Boosting Photocatalytic Hydrogen Generation. ACS omega 6, 13544–13553. doi:10.1021/acsomega.0c06038
Maltsev, Y., Maltseva, K., Kulikovskiy, M., and Maltseva, S. (2021). Influence of Light Conditions on Microalgae Growth and Content of Lipids, Carotenoids, and Fatty Acid Composition. Biology 10, 1060. doi:10.3390/biology10101060
Mendoza-Cózatl, D. G., Rangel-González, E., and Moreno-Sánchez, R. (2006). Simultaneous Cd2+, Zn2+, and Pb2+ Uptake and Accumulation by Photosynthetic Euglena Gracilis. Archives Environ. Contam. Toxicol. 51, 521–528.
Moreno-Sánchez, R., Rodríguez-Enríquez, S., Jasso-Chávez, R., Saavedra, E., and García-García, J. D. (2017). Biochemistry and Physiology of Heavy Metal Resistance and Accumulation in Euglena. Adv. Exp. Med. Biol. 979, 91–121.
Mukaida, S., Ogawa, T., Ohishi, K., Tanizawa, Y., Ohta, D., and Arita, M. (2016). The Effect of Rapamycin on Biodiesel-Producing Protist Euglena Gracilis. Biosci. Biotechnol. Biochem. 80, 1223–1229. doi:10.1080/09168451.2016.1141040
Nicolas, P., Freyssinet, G., and Nigon, V. (1980). Effect of Light on Glucose Utilization by Euglena Gracilis. Plant Physiol. 65, 631–634. doi:10.1104/pp.65.4.631
O'Neill, E. C., Trick, M., Hill, L., Rejzek, M., Dusi, R. G., Hamilton, C. J., et al. (2015). The Transcriptome of Euglena Gracilis Reveals Unexpected Metabolic Capabilities for Carbohydrate and Natural Product Biochemistry. Mol. Biosyst. 11, 2808–2820. doi:10.1039/c5mb00319a
Reitz, R. C., and Moore, G. S. (1972). Effects of Changes in the Major Carbon Source on the Fatty Acids ofEuglena Gracilis. Lipids 7, 217–220. doi:10.1007/bf02533068
Rodríguez-Zavala, J. S., García-García, J. D., Ortiz-Cruz, M. A., and Moreno-Sánchez, R. (2007). Molecular Mechanisms of Resistance to Heavy Metals in the Protist Euglena Gracilis. J. Environ. Sci. Health A Tox Hazard Subst. Environ. Eng. 42, 1365–1378.
Rosenberg, A. (1963). A Comparison of Lipid Patterns in Photosynthesizing and Nonphotosynthesizing Cells of Euglena Gracilis*. Biochemistry 2, 1148–1154. doi:10.1021/bi00905a042
Sánchez-Thomas, R., Moreno-Sánchez, R., and García-García, J. D. (2016). Accumulation of Zinc Protects against Cadmium Stress in Photosynthetic Euglena Gracilis. Environ. Exp. Bot. 131, 19–31.
Sun, X.-M., Ren, L.-J., Zhao, Q.-Y., Ji, X.-J., and Huang, H. (2018). Microalgae for the Production of Lipid and Carotenoids: a Review with Focus on Stress Regulation and Adaptation. Biotechnol. Biofuels 11, 272. doi:10.1186/s13068-018-1275-9
Taylor, F. J. (1960). The Absorption of Glucose by Scenedesmus Quadricauda. I. Some Kinetic Aspects. Proc. R. Soc. Lond B Biol. Sci. 151, 400–418. doi:10.1098/rspb.1960.0006
Thuillier-Bruston, F., Briand, J., and Laval-Martin, D. (1990). Effects of a First Exposure to Ethanol on the Compositions of Neutral and Polar Lipids in Euglena Gracilis Z, Taken as a Hepatic Cell Model: Equilibration by Citrulline-Malate. Biochem. Med. Metabolic Biol. 44, 159–174. doi:10.1016/0885-4505(90)90057-8
Venugopal, V., Prasanna, R., Sood, A., Jaiswal, P., and Kaushik, B. D. (2006). Stimulation of Pigment Accumulation inAnabaena Azollae Strains: Effect of Light Intensity and Sugars. Folia Microbiol. 51, 50–56. doi:10.1007/bf02931450
Wang, Y., Seppänen-Laakso, T., Rischer, H., and Wiebe, M. G. (2018). Euglena gracilis Growth and Cell Composition Under Different Temperature, Light and Trophic Conditions. PLoS One 13 (4), e0195329.
Xia, K., Chi, Y., Fu, J., Zhu, Z., Zhang, H., Du, C., et al. (2019). A Triboelectric Nanogenerator Based on Cosmetic Fixing Powder for Mechanical Energy Harvesting. Microsyst. Nanoeng. 5, 26. doi:10.1038/s41378-019-0066-1
Yoshida, Y., Tomiyama, T., Maruta, T., Tomita, M., Ishikawa, T., and Arakawa, K. (2016). De Novo assembly and Comparative Transcriptome Analysis of Euglena Gracilis in Response to Anaerobic Conditions. BMC Genomics 17, 182. doi:10.1186/s12864-016-2540-6
Zakryś, B., Milanowski, R., and Karnkowska, A. (2017). Evolutionary Origin of Euglena. Adv. Exp. Med. Biol. 979, 3–17. doi:10.1007/978-3-319-54910-1_1
Zeng, M., Hao, W., Zou, Y., Shi, M., Jiang, Y., Xiao, P., et al. (2016). Fatty Acid and Metabolomic Profiling Approaches Differentiate Heterotrophic and Mixotrophic Culture Conditions in a Microalgal Food Supplement 'Euglena'. BMC Biotechnol. 16, 49. doi:10.1186/s12896-016-0279-4
Keywords: Euglena gracilis, fatty acids, biodiesel, transcriptome, environmental stresses, carbon sources
Citation: He J, Du M, Chen Y, Liu Y, Zhang J(, Fu W, Lei A and Wang J (2022) Fatty Acid Accumulations and Transcriptome Analyses Under Different Treatments in a Model Microalga Euglena gracilis. Front. Chem. Eng. 4:884451. doi: 10.3389/fceng.2022.884451
Received: 26 February 2022; Accepted: 22 June 2022;
Published: 26 July 2022.
Edited by:
He Huang, Nanjing Normal University, ChinaCopyright © 2022 He, Du, Chen, Liu, Zhang, Fu, Lei and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangxin Wang, anh3YW5nQHN6dS5lZHUuY24=
 Jiayi He1,2
Jiayi He1,2 Ming Du
Ming Du Anping Lei
Anping Lei Jiangxin Wang
Jiangxin Wang