- 1Graduate School of Science, Technology and Innovation, Kobe University, Kobe, Japan
- 2Technology Research Association of Highly Efficient Gene Design (TRAHED), Kobe, Japan
- 3Engineering Biology Research Center, Kobe University, Kobe, Japan
- 4Department of Chemical Science and Engineering, Graduate School of Engineering, Kobe University, Kobe, Japan
- 5RIKEN Center for Sustainable Resource Science, Yokohama, Japan
Flavonoids, a major group of secondary metabolites in plants, are promising for use as pharmaceuticals and food supplements due to their health-promoting biological activities. Industrial flavonoid production primarily depends on isolation from plants or organic synthesis, but neither is a cost-effective or sustainable process. In contrast, recombinant microorganisms have significant potential for the cost-effective, sustainable, environmentally friendly, and selective industrial production of flavonoids, making this an attractive alternative to plant-based production or chemical synthesis. Structurally and functionally diverse flavonoids are derived from flavanones such as naringenin, pinocembrin and eriodictyol, the major basic skeletons for flavonoids, by various modifications. The establishment of flavanone-producing microorganisms can therefore be used as a platform for producing various flavonoids. This review summarizes metabolic engineering and synthetic biology strategies for the microbial production of flavanones. In addition, we describe directed evolution strategies based on recently-developed high-throughput screening technologies for the further improvement of flavanone production. We also describe recent progress in the microbial production of structurally and functionally complicated flavonoids via the flavanone modifications. Strategies based on synthetic biology will aid more sophisticated and controlled microbial production of various flavonoids.
1 Introduction
Flavonoids are a major group of secondary metabolites in plants that shares a diphenylpropane backbone (C6-C3-C6) in which three methylene units link two aromatic rings. Flavonoids are divided into several subclasses based on modifications to the backbone: flavanone, flavone, dihydroflavonol, flavonol, flavane-3,4-diol, anthocyanidin, flavane-3-ol, isoflavone and the derivatives of these aglycones such as flavonoid glycoside and prenylated flavonoid (Figure 1). To date, approximately ten thousand types of flavonoids have been identified from plants (Dixon and Pasinetti, 2010) and are promising as pharmaceuticals and food supplements due to their health-promoting biological activities (Harborne and Williams, 2000; Fraga et al., 2019).
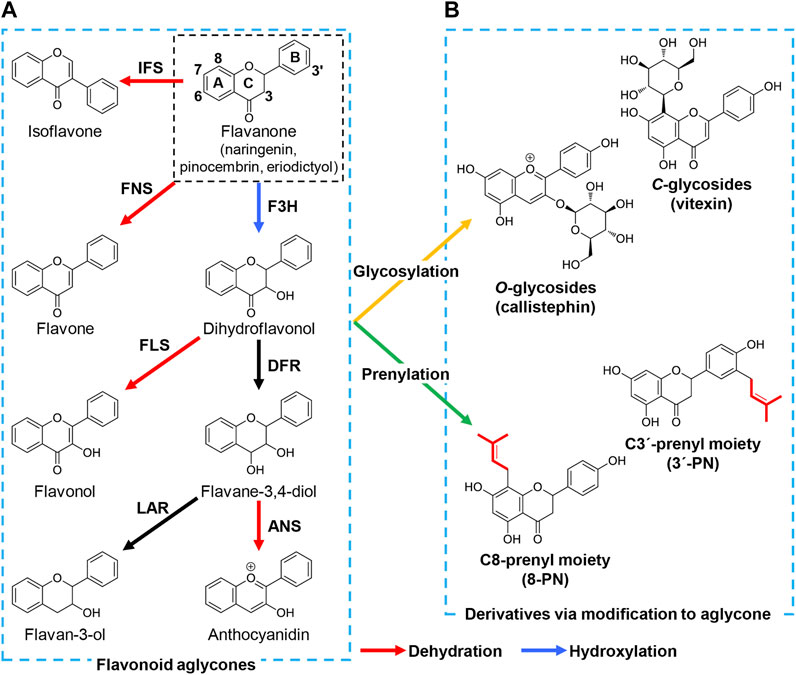
FIGURE 1. Structure and biosynthesis of major flavonoid aglycones and their derivatives. The (A) represents the core structures and biosynthetic pathway of flavonoid aglycones: flavanone, flavone, dihydroflavonol, flavonol, flavane-3,4-diol, anthocyanidin, flavane-3-ol, and isoflavone. In brief, flavanone is hydroxylated to dihydroflavonol by F3H. FLS, and DFR catalyze the dehydration and hydroxylation of dihydroflavonol, yielding flavonol, and flavane-3,4-diol, respectively. Flavane-3,4-diol is converted to anthocyanidin by ANS and flavane-3-ol by LAR. Flavone and isoflavone are also synthesized from naringenin by FNS and IFS, respectively. The (B) shows the formation of derivatives via modification to flavonoid aglycones. Sugar and prenyl moieties are attached to the hydroxyl group(s) and carbon atom(s) of flavonoid aglycones to form flavonoid glycosides and prenylated flavonoids, such as callistephin (pelargonidin-3-O-glucoside, one of anthocyanins) and vitexin (apigenin-8-C-glycoside), 8-prenylnaringenin (8-PN) and 3′-prenylnaringenin (3′-PN). Red, blue, orange, and green arrows indicate enzymatic reactions of dehydration, hydroxylation, glycosylation, and prenylation respectively. F3H, flavanone-3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; LAR, leucoanthocyanidin reductase; FNS, flavone synthase; IFS, isoflavone synthase.
Flavonoids are primarily isolated from plants or generated by organic synthesis; however, neither plant-based isolation nor chemical synthesis are cost-effective and sustainable because 1) plants produce small quantities of flavonoids and large expanses of land and longtime are required to grow these plants, making the isolation of flavonoids, including harvest, extraction and purification, arduous and expensive, and 2) organic synthesis is often challenging, given the structural complexity of flavonoids. As an alternative, flavonoids can be produced using recombinant microbes in a cost-effective and sustainable process due to their selective production of valuable compounds from inexpensive substrates (Liu et al., 2017b).
Most botanical flavonoids are biosynthesized from flavanone by various enzymatic modifications, such as hydroxylation, dehydration, methylation, glycosylation, and prenylation (Figure 1). The establishment of flavanone-producing microorganisms can therefore provide a platform for producing structurally complicated flavonoids. Flavonoid biosynthesis occurs in two steps: flavanone biosynthesis, and its modification(s) (Winkel, 2006; Tohge et al., 2017). Naringenin, the most common flavanone, is synthesized through the aromatic amino acids phenylalanine and tyrosine by sequential enzymatic reactions in plants (Figure 2). First, phenylalanine ammonia lyase (PAL) catalyzes the deamination of phenylalanine to form trans-cinnamic acid, then trans-cinnamic acid is hydroxylated by cinnamate-4-hydroxylase (C4H, cytochrome P450) coupled with cytochrome P450 reductase (CPR) to yield p-coumaric acid. Alternatively, p-coumaric acid is synthesized by direct deamination of tyrosine by the enzymatic reaction of tyrosine ammonia lyase (TAL). Then, 4-coumaroyl-CoA ligase (4CL) catalyzes the ligation of CoA to p-coumaric acid to form p-coumaroyl-CoA. Chalcone synthase (CHS) subsequently catalyzes the Claisen-type condensation of p-coumaroyl-CoA with three molecules of malonyl-CoA to yield naringenin chalcone, then chalcone isomerase (CHI) converts naringenin chalcone to naringenin by formation of the C-ring. PAL has been identified in plants, whereas TAL has not. Alternatively, plant uses a bifunctional phenylalanine/tyrosine ammonia-lyase (PTAL) for p-coumaric acid synthesis from tyrosine, which catalyzes the deamination of both phenylalanine and tyrosine (Barros and Dixon, 2020). The biosynthesis of flavanones uses a common route with the naringenin biosynthesis except for the difference in phenylpropanoyl-CoA precursors (i.e., p-coumaroyl-CoA for naringenin) for chalcone formation. In brief, pinocembrin and eriodictyol are produced from trans-cinnamic acid and caffeic acid via the phenylpropanoyl-CoA (trans-cinnamoyl-CoA and caffeoyl-CoA, respectively) by the enzymatic reactions of 4CL, CHS and CHI (Figure 2). Since flavanones share thus the same precursors, amino acids (phenylalanine and tyrosine) and malonyl-CoA, metabolic engineering strategies and synthetic biology approaches have been employed to increase the intracellular availability of these precursors in microbial production of flavonoids especially de novo synthesis of them.
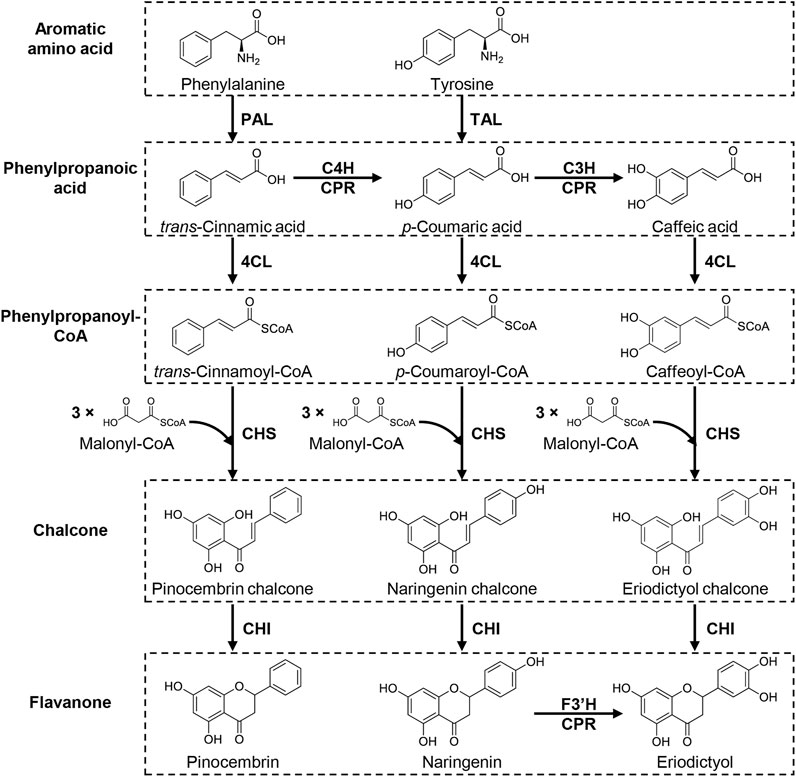
FIGURE 2. Biosynthetic pathway of flavanones. Flavanones are synthesized from aromatic amino acids, phenylalanine and tyrosine, via four enzymatic steps: deamination of aromatic amino acid to phenylpropanoic acid, ligation of CoA with phenylpropanoic acid to yield phenylpropanoyl-CoA, the Claisen-type condensation of phenylpropanoyl-CoA with three molecules of malonyl-CoA to form chalcone and isomerization of chalcone to form the C-ring. In this pathway, p-coumaric acid is produced from phenylalanine by a combination of deamination-oxygenation by PAL and C4H (CPR) or directly synthesized from tyrosine via TAL-catalyzed deamination. p-Coumaric acid is further hydroxylated by C3H (CPR) to form caffeic acid, a precursor for eriodictyol. Eriodictyol can be produced via hydroxylation of C3′ in the B-ring of naringenin by F3′H (CPR). PAL, phenylalanine ammonia lyase; TAL, tyrosine ammonia lyase; C4H, tans-cinnamate 4-hydroxylase; C3H, p-coumaric acid 3-hydroxylase; CPR, cytochrome P450 reductase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3′H, flavonoid 3′-hydroxylase.
The modification enzymes attach various functional groups to the flavanone core structure to form diverse flavonoids. The modification steps are categorized into two types: variation in the heterocyclic C-ring by hydroxylation and dehydration, and attachment of diverse side chains such as glycosylation and prenylation (Figure 1). For example, flavone synthase (FNS) catalyzes the dehydration of the C-ring in flavanone to flavone, whereas flavanone-3-hydroxylase (F3H) attaches a hydroxyl group onto C3 in the C-ring of flavanone to yield dihydroflavonol, which is then converted to flavonol via dehydration by flavanol synthase (FLS). Dihydroflavanol is also converted to anthocyanidin or flavane-3-ol by sequential modifications by dihydroflavonol 4-reductase (DFR) along with anthocyanidin synthase (ANS) or leucoanthocyanidin reductase (LAR). Several carbon atoms in the B-ring of naringenin are hydroxylated to yield structurally different compounds during these modification steps such as hydroxylation of C3′ or both C3′ and C5′ in the B-ring of flavonoids by flavanone-3′-hydroxylase (F3′H) and flavanone-3′5′-hydroxylase (F3′5′H) respectively. For example, eriodictyol can be synthesized by hydroxylating C3′ of naringenin (Amor et al., 2010) as well as synthesized from caffeic acid described above (Figure 2). Similarly, myricetin, one of flavonol, is biosynthesized from naringenin by combination of F3H, FLS, and F3′5′H (Leonard et al., 2006). Additionally, isoflavone synthase (IFS), a P450 monooxygenase, catalyzes the transposition of the B-ring from C2 to C3 in the C-ring.
Glycosylation and prenylation convert naringenin-derived flavonoids into more complex flavonoids in plants (Figure 1). Glycosylation increases the solubility and bioactivity of flavonoids, with the increased solubility contributing to their utilization upon eating the plants. Plant glycosyltransferase produces flavonoid glycosides mainly by linking the activated sugar moiety to the oxygen atom of the flavonoid backbone (O-glycosylation) or by directly forming a C-C linkage between the sugar and the flavonoid (C-glycosylation). An alternative modification to glycosylation, prenylation, attaches one or more prenyl groups, such as dimethylallyl (C5) and geranyl (C10) moieties, to the flavonoid backbone. Prenylation increases the liposolubility of flavonoids, contributing to the maintenance of these compounds in cell membranes compared with non-prenylated flavonoids (Mukai et al., 2012). Additionally, the attachment position and chain length of the prenyl moieties, and further modification after prenylation, can alter and enhance the biological activity of flavonoids. These modifications are essential for the various biological activities of flavonoids and their structural diversity.
In this review, we summarize current engineering strategies for improving the microbial production of plant flavonoids. First, we describe metabolic engineering strategies, including the screening of host organisms and plant-derived enzymes, and pathway designs using these organisms and enzymes mainly focusing on naringenin. Next, we describe evolutionary strategies utilizing high-throughput screening techniques to identify cells with improved production of naringenin and its derivatives. Moreover, we explain how genetic circuits that regulate pathway gene expression improve the production of flavonoids and their precursors. Finally, we describe recent progress in the microbial production of structurally complicated flavonoids using various modification enzymes.
2 Flavanones Production in Microorganisms
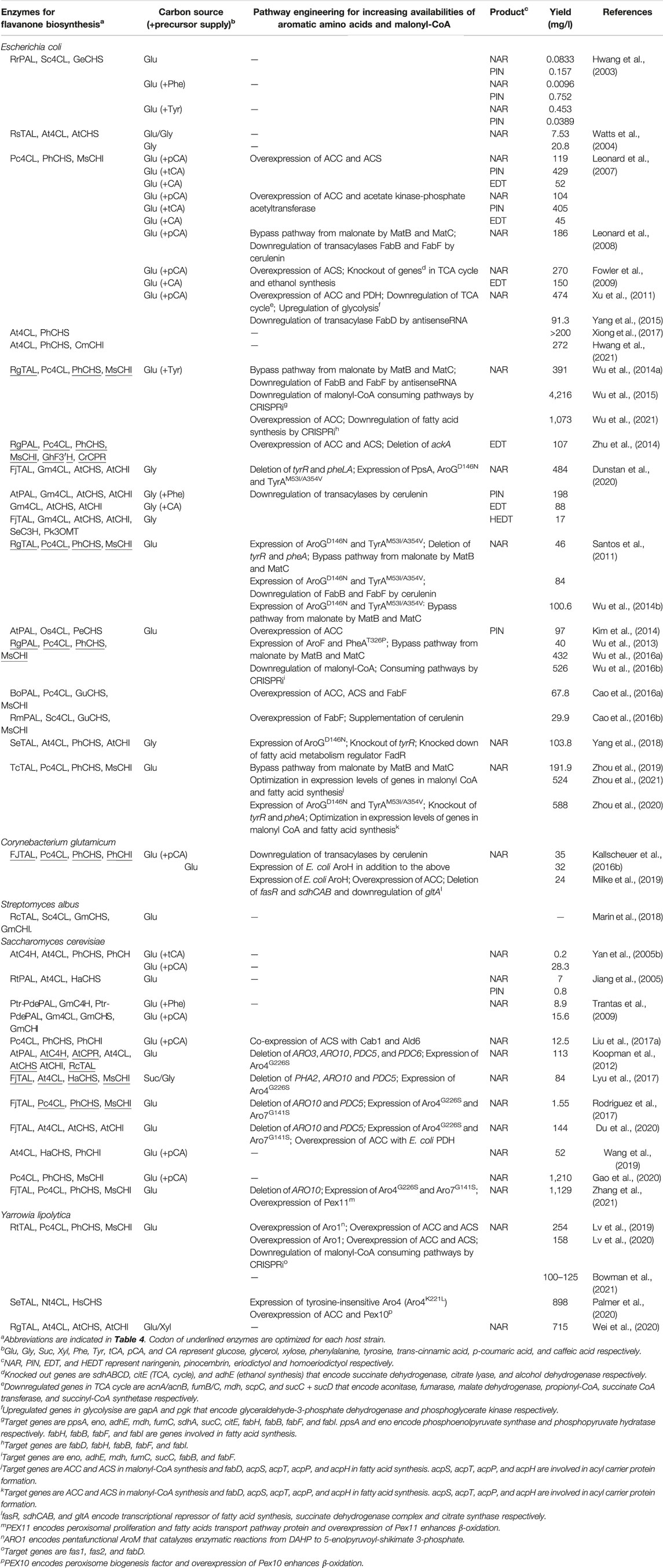
Most structurally and functionally diverse flavonoids are biosynthesized from flavanones such as naringenin, pinocembrin and eriodictyol (Figure 1). Since naringenin is the most basic skeleton for flavanone, there have been many studies on reconstructing naringenin biosynthetic pathways in microorganisms. In this section, we introduce various aspects of microbial flavanone production, including metabolic engineering strategies and synthetic biology approaches, with a focus mainly on naringenin as well as some studies on pinocembrin and eriodictyol (Tables 1, 2).
2.1 Production Host
Bacteria and yeasts, especially Escherichia coli and Saccharomyces cerevisiae, have generally been used for the microbial production of flavonoids (Tables 1–3). Differences between these hosts significantly influence the strategy for reconstructing flavanone biosynthetic pathways. This section summarizes the characteristics of these host strains and examples of their development with respect to flavanone production.
2.1.1 Bacteria
In general, bacteria grow faster than yeasts. In addition, heterogeneous genes can be expressed in a polycistronic manner in bacteria, decreasing the complexity of designing genetic systems. Furthermore, some bacteria represented by E. coli have extraordinarily powerful expression systems for proteins, which often leads to the clear phenotype change by the introduced metabolic enzymes. In some cases, however, functional expression of eukaryotic enzymes is difficult in bacteria. For example, Watts et al. (2004) has reported to fail a functional expression of the plant C4H enzyme, a cytochrome P450 monooxygenase, in E. coli, resulting in the accumulation of trans-cinnamic acid. The lack of C4H enzyme significantly influenced the conversion efficiency of phenylalanine to p-coumaric acid when only PAL from the yeast Rhodotorula mucilaginosa, which catalyzes the deamination of phenylalanine and tyrosine, was expressed, resulting in low naringenin production compared with tyrosine supplementation (Hwang et al., 2003). Reconstruction of the naringenin biosynthetic pathway in E. coli thus utilized microbial TAL (usually a yeast enzyme), bacterial/plant 4CL, and plant CHS and CHI using tyrosine (Watts et al., 2004; Wu et al., 2014a; Wu et al., 2014b; Wu et al., 2015; Yang et al., 2018; Zhou et al., 2019; Zhou et al., 2020; Zhou et al., 2021). Expressing these heterogonous genes by engineering precursor metabolic pathways provided the highest levels of naringenin production in E. coli to date: 1,073 mg/L from tyrosine (Wu et al., 2021) and 588 mg/L from glucose (Zhou et al., 2020). As well as the naringenin production, expressing 4CL, CHS and CHI enzymes enabled pinocembrin production from trans-cinnamic acid (Leonard et al., 2007). An additional expression of PAL allowed to produce pinocembrin from glucose (Hwang et al., 2003; Wu et al., 2013; Kim et al., 2014; Cao et al., 2016a; Wu et al., 2016a; Cao et al., 2016b; Wu et al., 2016b) or phenylalanine (Dunstan et al., 2020). Similarly, the engineered E. coli produced eriodictyol from caffeic acid by expressing 4CL, CHS and CHI (Leonard et al., 2007; Fowler et al., 2009; Dunstan et al., 2020) or tyrosine by additional expression of TAL, F3′H and CPR with those pathway enzymes (Zhu et al., 2014).
Recently, several studies employed Corynebacterium glutamicum as the host strain instead of E. coli (Kallscheuer et al., 2016b; Milke et al., 2019). C. glutamicum is used for the several million ton-scale industrial production of amino acids such as glutamate and lysine (Koffas et al., 2003; Becker et al., 2011), and therefore might have a great potential to supply the precursor tyrosine through its endogenous pathway. Kallscheuer et al. (2016a) have previously found the potential of this organism to grow on several phenylpropanoids and identified the phenylpropanoid degradation pathway (Phd pathway), reporting an engineered C. glutamicum that produces 35 mg/L of naringenin from p-coumaric acid with deletion of the pathways for degradation and unspecific conversion of the phenylpropanoid substrates (Kallscheuer et al., 2016b). They also achieved 32 mg/L of naringenin from glucose followed by increasing the intracellular availability of precursors (tyrosine and malonyl-CoA) by additional engineering of the shikimate pathway and supplementation of cerulenin, an inhibitor of fatty acid synthesis (see Section 2.4) (Kallscheuer et al., 2016b). In a subsequent study by the same group, the authors modulated gene expressions of the fatty acid synthesis and the tricarboxylic acid (TCA) cycle of the host strain to enhance the malonyl-CoA supply without the supplementation of any inhibitors, achieving 24 mg/L of naringenin from glucose (Milke et al., 2019).
Furthermore, some cyanobacteria species, such as Synechocistis and Synechococcus, have been used as production host for plant secondary metabolites including flavonoids and their pathway intermediates (Xue et al., 2014; Ni et al., 2016), achieving 7.1 mg/L of naringenin with cerulenin supplementation (Ni et al., 2016). Although further improvement in productivity and pathway design is required, cyanobacterium is an attractive host for flavonoid production owing to following features (Jin et al., 2021). They can use carbon dioxides (CO2) and sunlight as carbon and energy sources, respectively, contributing sustainable and low-cost microbial production of flavonoids. They can also supply large quantities of energies and reductants, ATP and NADPH, by photosynthetic system, facilitating the reaction steps of energy consumption (e.g., 4CL) and P450 enzymes (e.g., C4H, F3H and IFS) in the biosynthesis of flavonoids.
2.1.2 Yeasts
Leveraging the eukaryotic nature of yeasts facilitates the functional expression of plant-derived genes. For example, heterogeneous production of naringenin in S. cerevisiae has been achieved by introducing a five- or six-step pathway from phenylalanine (Trantas et al., 2009; Koopman et al., 2012) or a four-step pathway from tyrosine (Lyu et al., 2017; Rodriguez et al., 2017; Du et al., 2020). Furthermore, additional expression of microbial TAL in yeast harboring the naringenin biosynthetic pathway from phenylalanine improved the p-coumaric acid supply, leading to a 1.5-fold improvement in naringenin production (Koopman et al., 2012). To our knowledge, the highest reported production of naringenin from phenylalanine and glucose in S. cerevisiae is 8.9 mg/L (Trantas et al., 2009) and 1,129 mg/L (Zhang et al., 2021), respectively. Furthermore, pinocembrin production in S. cerevisiae was achieved by expressing PAL, 4CL and CHS (Jiang et al., 2005) and additional expression of CHI (Liu et al., 2020).
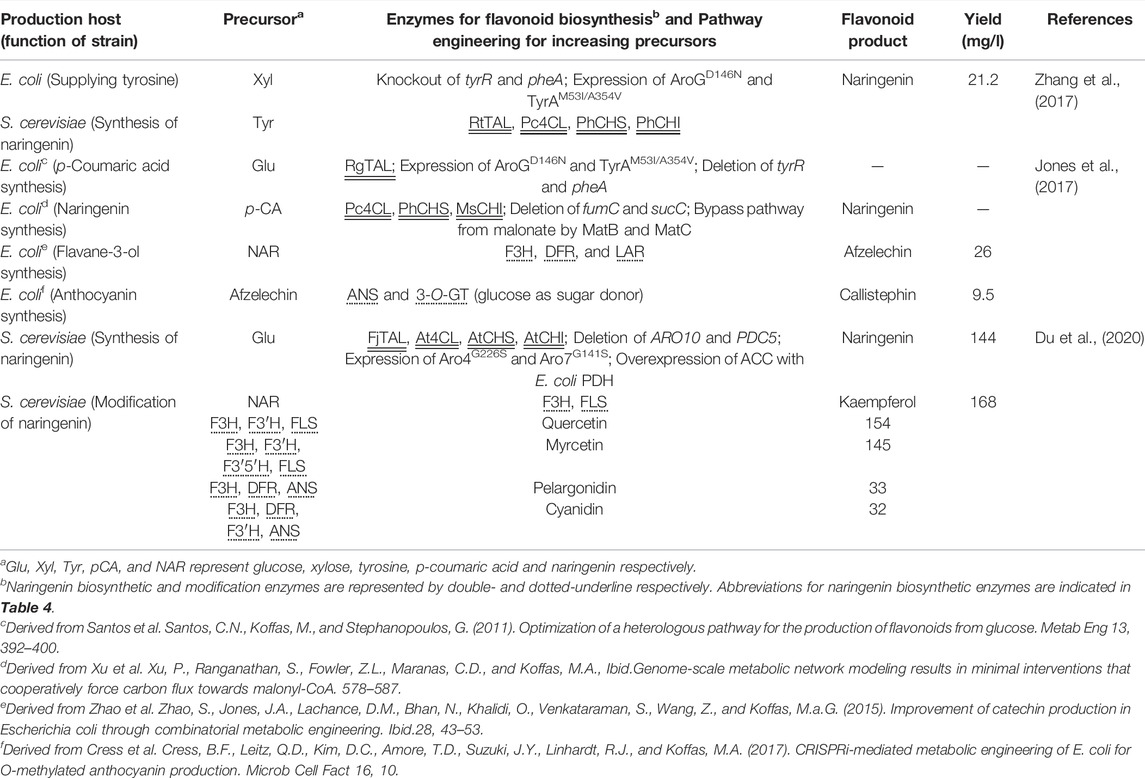
Recent research used the non-conventional yeast Yarrowia lipolytica as a host strain instead of S. cerevisiae. Although the genetic background of Y. lipolytica is less well understood in terms of gene manipulation than laboratory yeast, this yeast strain is expected to have superior malonyl-CoA cell availability, with the potential to enhance naringenin biosynthesis (Beopoulos et al., 2009). Indeed, the production of naringenin by Y. lipolytica was achieved by reconstructing the naringenin biosynthetic pathway and enhancing precursor supply (Lv et al., 2019; Palmer et al., 2020; Wei et al., 2020). The engineered Y. lipolytica generated 898 mg/L of naringenin from glucose (Palmer et al., 2020). Nonetheless, the introduction of several genes into yeasts to construct a multi-step pathway is laborious due to the monocistronic nature of their gene expression; thus, recent studies employed the four-step pathway from tyrosine, a simpler genetic design compared with that from phenylalanine (Lyu et al., 2017; Rodriguez et al., 2017; Du et al., 2020). To address this drawback, Zhang et al. reported a co-culture system of S. cerevisiae and E. coli, in which the former is engineered to efficiently produce naringenin from tyrosine and the latter is engineered to supply the precursor tyrosine from d-xylose to the former. This system resulted in the production of 21.16 mg/L naringenin (Zhang et al., 2017). More recently, a co-culture system of two different S. cerevisiae strains was designed to efficiently produce naringenin derivatives (Du et al., 2020) (Table 2).
In addition to the above-mentioned bacteria and yeasts, the bacteria Lactococcus lactis (Solopova et al., 2019) and Streptomyces albus (Marin et al., 2018) and the non-conventional yeast Pichia pastoris (Komagataella phaffii) (Wang et al., 2016) have recently been used as alternative hosts for flavonoid production, but further engineering is necessary to improve the productivity and design pathways (Tables 1, 3).
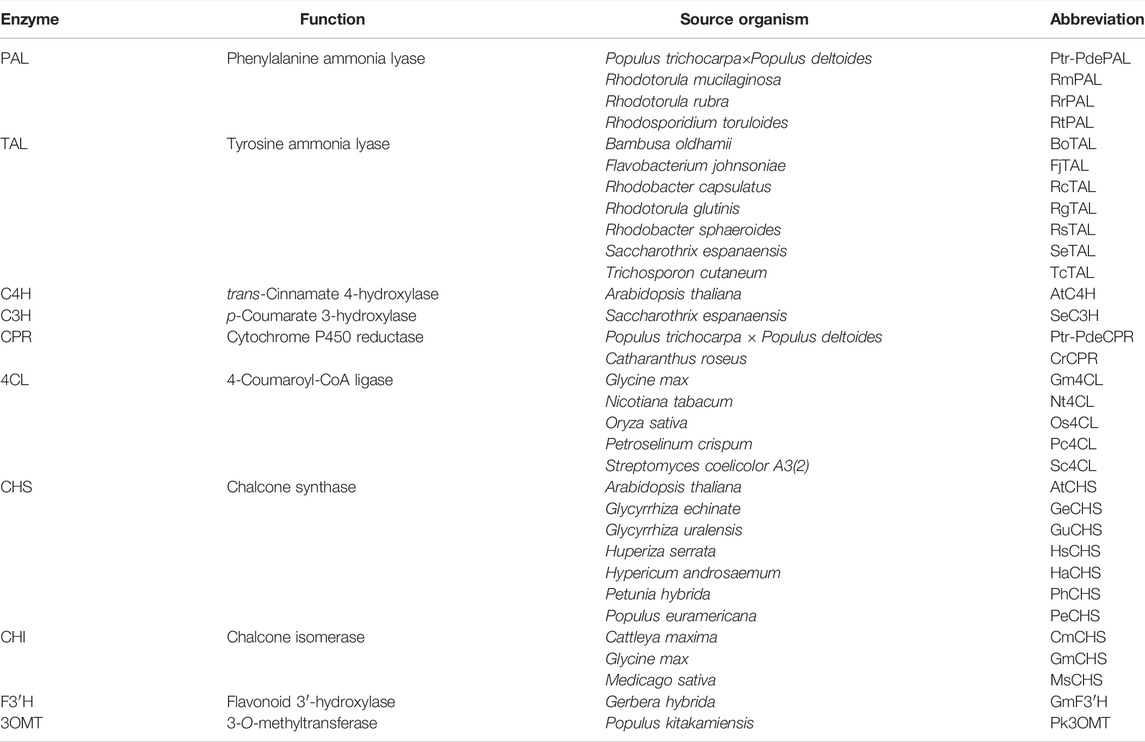
2.2 Heterologous Genes for Flavanone Biosynthesis
Various plants produce flavonoids as secondary metabolites with different productivities and diversities. It is thus likely that genes from high-producing organisms encode pathway enzymes (Figure 2) with high catalytic activity. However, genes from a limited number of plants have been tested to date, as listed in Table 4 due to a lack of genetic information on these natural producers. Because of this situation, Arabidopsis thaliana, one of the well-characterized plant species, has been widely used as the source organism for naringenin and flavonoid biosynthesis genes despite of its lower productivity. Recently, Mark et al. (2019) demonstrated that using 4CL and CHS from different plants such as Medicago truncatula and Vitis vinifera instead of A. thaliana resulted in the increased production of naringenin. In addition, expressing PAL from Bambusa oldhamii and 4CL from Petroselinum crispum instead of those from Rhodotorula mucilaginosa and Streptomyces coelicolor A3(2) increased intracellular trans-cinnamic acid, resulting in a 3.5-fold increase in pinocembrin production (Cao et al., 2016a). Similarly, Dunstan et al. (2020) evaluated the different combinations of three TAL, five 4CL and two CHS genes from various plants, from which they identified the pathway constructs that enabled efficient flavanone production in E. coli; 484 mg/L of naringenin and 17 mg/L of homoeriodictyol from glycerol, 198 mg/L of pinocembrin from phenylalanine and 88 mg/L of eriodictyol from caffeic acid with cerulenin supplementation, respectively. Taken together, reconstruction of the flavanone biosynthetic pathways with different combinations of pathway genes screened from various plants will be promising for increasing flavanone production by engineered microorganisms.
2.3 Biosynthesis From Pathway Intermediates
Supplementation of precursor aromatic amino acids and their derivative phenylpropanoids (e.g., tyrosine and p-coumaric acid) to the culture medium increases their intracellular availability in engineered microorganisms, improving flavanone production without modulating their biosynthetic pathways. For example, phenylalanine supplementation increased p-coumaric acid production in S. cerevisiae, enabling the production of 8.9 mg/L naringenin from phenylalanine (Trantas et al., 2009). Moreover, the addition of tyrosine and phenylalanine to the culture medium resulted in a five-fold increase in production of naringenin and pinocembrin in E. coli, respectively (Hwang et al., 2003). More recent research reported naringenin production using engineered E. coli expressing the genes responsible for naringenin biosynthesis from tyrosine (i.e., TAL, 4CL, CHS, and CHI) upon tyrosine supplementation, achieving 1,073 mg/L naringenin from tyrosine (Wu et al., 2014b; Wu et al., 2015; Wu et al., 2021). Furthermore, supplementation of trans-cinnamic acid or p-coumaric acid improved naringenin production in both E. coli (Leonard et al., 2007; Fowler et al., 2009; Xu et al., 2011) and S. cerevisiae (Yan et al., 2005b; Trantas et al., 2009). Similarly, supplementation of trans-cinnamic acid enabled pinocembrin production in E. coli expressing 4CL, CHS, and CHI (Leonard et al., 2007), while eriodictyol production in E. coli was achieved by addition of tyrosine (Zhu et al., 2014) or caffeic acid (Leonard et al., 2007; Fowler et al., 2009; Dunstan et al., 2020), respectively. The addition of these pathway intermediates can decrease the necessity of introducing the genes responsible for intermediate biosynthesis, therefore simplifying the expression system. However, the use of these intermediates often negatively influences the microbial production of flavanone due to the increase in precursor cost, the toxicity (e.g., acidic nature) of these compounds, and the low solubility of these intermediates.
2.4 Pathway Design for De Novo Synthesis From a Sole Carbon Source
De novo synthesis from a simple sole carbon source such as glucose is the most cost-effective approach for microbial flavanone production, but would lead to an insufficient supply of precursors (the aromatic amino acids phenylalanine and tyrosine, and malonyl-CoA) due to tight regulation of the biosynthetic pathways for these compounds in a host microorganism. In the following subsections, we summarize efforts to date to overcome these limitations by the metabolic engineering of biosynthetic pathways for precursors (Table 1) (Santos et al., 2011; Koopman et al., 2012; Wu et al., 2014b; Kim et al., 2014; Zhu et al., 2014; Cao et al., 2016a; Wu et al., 2016a; Cao et al., 2016b; Wu et al., 2016b; Rodriguez et al., 2017; Yang et al., 2018; Zhou et al., 2019; Du et al., 2020; Dunstan et al., 2020; Zhou et al., 2020; Zhou et al., 2021).
2.4.1 Biosynthesis of Aromatic Amino Acids
Phenylalanine and tyrosine are biosynthesized from phosphoenolpyruvate (PEP) and d-erythrose-4-phosphate (E4P) via the shikimate pathway (Figures 3A,B). 3-Deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase catalyzes the first step of the shikimate pathway by condensing PEP and E4P to form DAHP. Next, DAHP is converted to chorismite through shikimate via two-step enzymatic reactions. Then, chorismate mutase catalyzes the rearrangement of chorismate to prephenate, which is the last step in the shikimate pathway. Finally, prephenate is converted to tyrosine and phenylalanine via dehydrogenation and transamination in parallel pathways. The biosynthesis of phenylalanine and tyrosine via chorismate (the shikimate pathway) is tightly regulated at both the biochemical and genetic levels. DAHP synthases are feedback-inhibited by phenylalanine, tyrosine, and tryptophan, whereas chorismate mutases are subjected to feedback inhibition by phenylalanine and tyrosine. In addition, the expression of these biosynthetic enzymes is regulated in response to these aromatic amino acids (Braus, 1991; Rodriguez et al., 2014). These regulations of the pathway enzymes inhibit the overproduction of phenylalanine and tyrosine in the cell, leading to insufficient supply of these compounds. Therefore, removing the negative regulation of aromatic amino acid biosynthesis is a promising approach for enhancing the endogenous biosynthesis of these precursors.
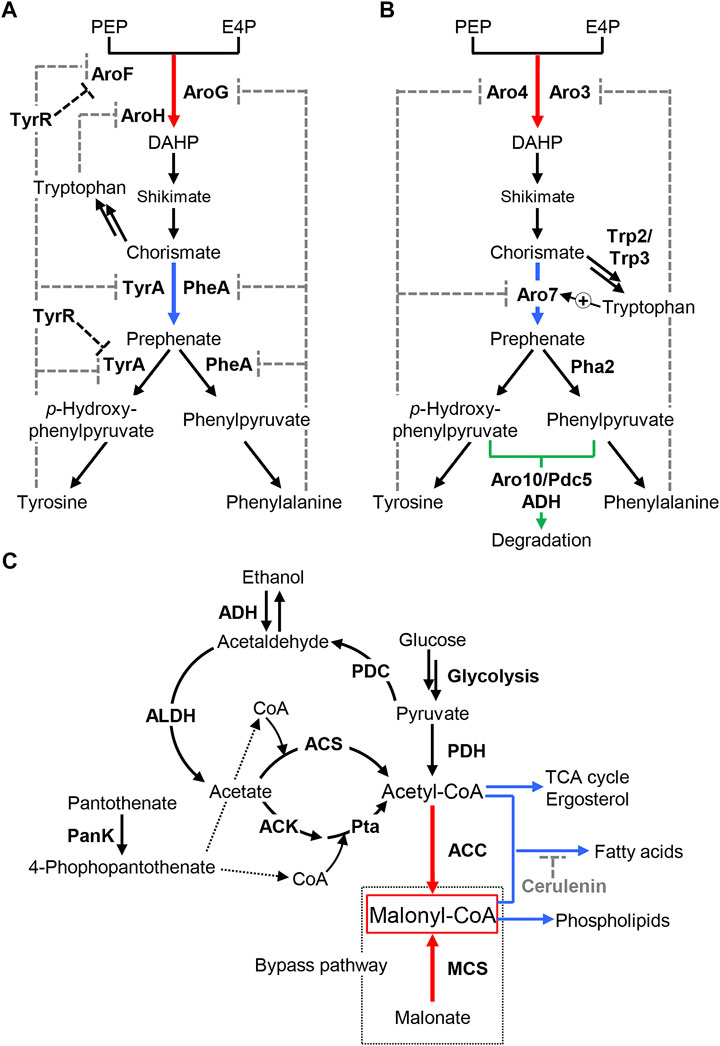
FIGURE 3. Metabolic engineering of endogenous biosynthetic pathways for precursors. (A,B) Biosynthesis of aromatic amino acids in E. coli (A) and S. cerevisiae (B). Phenylalanine and tyrosine are synthesized via the shikimate pathway. DAHP synthase and chorismate mutase, which catalyze the first and last step of the shikimate pathway, respectively, are subjected to feedback regulation by aromatic amino acids (indicated by gray-dashed lines). Chorismate is bypassed to tryptophan. Red and blue arrows indicate the enzymatic reactions catalyzed by DAHP synthase and chorismate mutase, respectively. (A) The transcriptional regulator TyrR represses the expression of genes involved in the biosynthesis of phenylalanine and tyrosine, such as aroF and tyrA (indicated by black-dashed lines). (B) The enzymatic activity of Aro7 is activated by tryptophan. Green arrow indicates degradation of phenylpyruvate and p-hydroxyphenylpyruvate, precursors of phenylalanine and tyrosine, respectively, to the corresponding alcohols by decarboxylases, Aro10, and Pdc5, and alcohol dehydrogenase (ADH). PEP: phosphoenolpyruvate, E4P: d-erythrose-4-phosphate, DAHP: 3-deoxy-d-arabino-heptulosonate 7-phosphate. (C) Biosynthesis of malonyl-CoA. Endogenous malonyl-CoA is synthesized from acetyl-CoA by ACC. MCS catalyzes the formation of malonyl-CoA from malonate, which is used as the bypass pathway in malonyl-CoA synthesis, in addition to the natural metabolic pathway. Acetyl-CoA is synthesized in the PDH complex from pyruvate and by ACS from acetate in S. cerevisiae. The Ack-Pta pathway from acetate is also used in addition to these biosynthetic pathways in E. coli. PanK catalyzes the phosphorylation of pantothenate to 4-phospho pantothenate, a precursor of CoA. Red arrows represent two key enzymatic reactions for directly increasing intracellular malonyl-CoA, which are catalyzed by ACC ant MCS. Whereas, blue arrows indicate pathways for acetyl-CoA consumption by glycolysis, the TCA cycle, and fatty acid synthesis, and malonyl-CoA consumption by the synthesis of fatty acids and phospholipids. Cerulenin inhibits fatty acid synthesis as indicated by grey dotted line. Pyruvate, a precursor for acetyl-CoA, is catabolized to acetate, another precursor of acetyl-CoA, and ethanol via acetaldehyde. The reaction catalyzed by ADH is bi-directional; thus, overexpression of ADH that prefers to convert ethanol to acetaldehyde, such as Adh2 in S. cerevisiae, was used for increasing acetate availability. ACC, acetyl-CoA carboxylase; MCS, malonyl-CoA synthetase; PDH, pyruvate dehydrogenase; ACS, acetyl-CoA synthetase; Ack, acetate kinase; Pta, phosphate acetyltransferase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase; ALDH, acetaldehyde dehydrogenase; PanK, pantothenate kinase.
In E. coli, there are three isozymes of DAHP synthase (AroF, AroG, and AroH) and two isoforms of chorismate mutase (TyrA, and PheA) (Figure 3A). tyrA and pheA encode the bifunctional enzyme that catalyzes the conversion of chorismate to prephenate (chorismate mutase) and the dehydrogenation of prephenate (prephenate dehydrogenase). AroF, AroG, and AroH are feedback-inhibited by tyrosine, phenylalanine and tryptophan, respectively, whereas the enzymatic activities of TyrA and PheA are subjected to feedback inhibition by tyrosine and phenylalanine, respectively. Previous studies described AroG and TyrA mutants that are insensitive to such feedback inhibition (Kikuchi et al., 1997; Lutke-Eversloh and Stephanopoulos, 2005). Since AroG contributes 80% of total cellular DAHP synthase activity, the expression of these AroG and TyrA mutants (AroGD146N and TyrAM53I/A354V) increases intracellular tyrosine in E. coli even in the presence of tyrosine-sensitive AroF (Lütke-Eversloh and Stephanopoulos, 2007). Introducing the aroG and tyrA mutants (AroGD146N, TyrAM53I/A354V) resulted in increased tyrosine supply in E. coli, enabling the production of over 80 mg/L of naringenin from glucose (Santos et al., 2011; Wu et al., 2014b; Yang et al., 2018). Because tyrosine biosynthesis in E. coli is transcriptionally regulated by TyrR (Pittard et al., 2005), the removal of downregulation by TyrR also increases intracellular tyrosine (Kim et al., 2018), contributing to the increase in naringenin production (Yang et al., 2018). Furthermore, Dunston et al. (2020) demonstrated that deletion of pheA together with the introduction of ppsA that catalyzes the generation of PEP from pyruvate enabled 484 mg/L of naringenin production from glucose in combination with above mentioned strategies (i.e., the introduction of aroG and tyrA mutants and the knockout of tyrR). In contrast, the similar approach was employed to increase intracellular phenylalanine availability for heterologous pinocembrin production from phenylalanine via trans-cinnamic acid. Previous study reported that co-expression of PheA mutant (PheAT326P) that is insensitive to feedback inhibition by phenylalanine with the wild-type AroF increased intracellular phenylalanine pool (Zhou et al., 2010). Introducing the wild-type aroF and the pheA mutant resulted in 40 mg/L of pinocembrin production from glucose (Wu et al., 2013), subsequently achieving 432 mg/L along with optimization of PAL expression level by modifying its mRNA secondary structure (Wu et al., 2016a).
In S. cerevisiae, two DAHP synthases, Aro3 and Aro4, are subjected to feedback inhibition by phenylalanine and tyrosine, respectively, while one chorismate mutase, Aro7, is inhibited by tyrosine but activated by tryptophan (Figure 3B) (Schnappauf et al., 1997; Helmstaedt et al., 2005). Koopman et al. (2012) deleted the ARO3 gene and introduced the tyrosine-insensitive ARO4 mutant (Aro4G226S) (Hartmann et al., 2003) into engineered S. cerevisiae to enhance the flux from glucose to phenylalanine. They also deleted genes encoding decarboxylase (Aro10, Pdc5, and Pdc6) to alleviate the loss of phenylpyruvate, a precursor of phenylalanine. Next, they introduced both PAL and TAL to the resultant strain to form p-coumaric acid from phenylalanine and tyrosine, enabling the production of 113 mg/L of naringenin from glucose. Later, tyrosine-insensitive Aro4 and/or Aro7 mutants (Aro4K229L and Aro7G141S) were used to increase naringenin production together with the heterologous expression of TAL in S. cerevisiae (Lyu et al., 2017; Rodriguez et al., 2017; Du et al., 2020). This strategy would improve the production of chorismate and prephenate; however, these compounds are also converted to tryptophan (by Trp2/Trp3) and phenylalanine (by Pha2), respectively, resulting in the loss of tyrosine. Lyu et al. (2017) revealed that weakening PHA2 expression resulted in a 20% improvement in naringenin production due to enhanced tyrosine availability. Additionally, p-hydroxy-phenylpyruvate, a precursor of tyrosine, is degraded by endogenous decarboxylases, such as Aro10 and Pdc5. Deletion of these decarboxylases aided the accumulation of intracellular tyrosine (Rodriguez et al., 2015; Rodriguez et al., 2017), resulting in an approximately two-fold improvement in naringenin production (Lyu et al., 2017; Du et al., 2020).
2.4.2 Biosynthesis of Malonyl-CoA
The metabolic engineering of pathways responsible for malonyl-CoA biosynthesis is another effective strategy for enhancing naringenin production (Figure 3C). Endogenous synthesis of malonyl-CoA depends on the conversion from acetyl-CoA by acetyl-CoA carboxylase (ACC) using adenosine triphosphate (ATP) (Milke and Marienhagen, 2020). Additional expression (overexpression) of ACC is therefore the most common way to enhance the intracellular malonyl-CoA pool and thus increase productivities of naringenin (Leonard et al., 2007; Xu et al., 2011; Du et al., 2020; Wu et al., 2021), pinocembrin (Leonard et al., 2007; Kim et al., 2014; Cao et al., 2016a), and eriodictyol (Leonard et al., 2007; Zhu et al., 2014). Another strategy is the addition of a synthetic bypass pathway for malonyl-CoA synthesis. For example, malonyl-CoA synthetase (MCS) encoded by matB can directly synthesize malonyl-CoA from malonate (An and Kim, 1998). Co-expression of MatB and the malonate carrier protein MatC in engineered E. coli would enhance intracellular malonyl-CoA by supplementing malonate (Wu et al., 2014a; Wu et al., 2014b), resulting in between a 1.6–2.7-fold increase in naringenin production (Leonard et al., 2008; Santos et al., 2011). Similarly, expression of bypass pathway from malonate was employed to increase malonyl-CoA supply for pinocembrin production (Wu et al., 2013; Wu et al., 2016a). Further improvement of the intracellular malonyl-CoA pool can be accomplished by increasing acetyl-CoA availability and deleting the branching pathway from malonyl-CoA to fatty acids and phospholipids. In S. cerevisiae, acetyl-CoA is synthesized by the pyruvate dehydrogenase (PDH) complex from pyruvate and by acetyl-CoA synthetase (ACS) from acetate using ATP, whereas acetate kinase-phosphate acetyltransferase catalyzes the formation of acetyl-CoA in addition to PDH and ACS pathways in E. coli. Therefore, the overexpression of these enzymes can increase acetyl-CoA availability. For example, PDH (Xu et al., 2011), ACS (Leonard et al., 2007; Fowler et al., 2009), and ACC and acetate kinase-phosphate acetyltransferase (Ack-Pta) (Leonard et al., 2007) were overexpressed in engineered E. coli. More recently, Zhu et al. (2014) overexpressed the genes encoding endogenous ACS (acs) and heterologous ACC (accBC and dtsR1 from C. glutamicum), and removed the acetate competition pathway by deletion of acetate kinase gene (ackA), achieving 107 mg/L of eriodictyol production from tyrosine in E. coli (Zhu et al., 2014). Similarly, a heterologous ACS mutant (AcsL641P from Salmonella enteric) was co-expressed with endogenous acetaldehyde dehydrogenase (Ald6) to bypass acetyl-CoA synthesis from PDH pathway, producing 2.9 mg/L of naringenin from p-coumaric acid in engineered S. cerevisiae. They also achieved 10.5 mg/L of naringenin followed by increasing CoA availability by additional expression of endogenous pantothenate kinase (PanK encoded by CAB1). Pantothenate supplementation to this yeast led to another 19% increase in naringenin production (12.5 mg/L) (Liu et al., 2017). In the subsequent studies, Adh2, an enzyme converting ethanol to acetaldehyde, or Adh2 and ACC were additionally expressed to further increase malonyl-CoA availability in engineered S. cerevisiae which produce derivatives of naringenin and pinocembrin (see Section 3.1 and 3.2) (Liu et al., 2018; Liu et al., 2020). Additionally, the downregulation of acetyl-CoA and pyruvate consuming pathways enhanced acetyl-CoA availability by, for example, deleting the genes involved in the TCA cycle and ethanol synthesis in engineered E. coli (Fowler et al., 2009) and deleting or downregulating ergosterol biosynthetic enzymes in engineered S. cerevisiae (Du et al., 2020). Furthermore, downregulation of the TCA cycle combined with the upregulation of glycolysis and malonyl-CoA synthesis achieved a four-fold increase in intracellular malonyl-CoA, enabling the production of 474 mg/L of naringenin by engineered E. coli (Xu et al., 2011). Interception of malonyl-CoA from fatty acid synthesis was also reported to increase malonyl-CoA availability in cells. Transacylases that catalyze the condensation of acyl-CoA (acetyl-CoA and malonyl-CoA) with acyl carrier protein were downregulated by the inhibitor cerulenin (Leonard et al., 2008; Cao et al., 2016b; Dunstan et al., 2020), antisense RNA (Wu et al., 2014a; Yang et al., 2015) and knockdown of the regulator (Yang et al., 2018) in engineered E. coli. This strategy was also adopted for optimizing malonyl-CoA allocation in Y. lipolytica using CRISPRi-based genetic circuits (see Section 2.6) (Lv et al., 2020). Furthermore, CRISPRi-based downregulation of the various pathways for malonyl-CoA consumption, TCA cycle, ethanol synthesis and fatty acid synthesis, increased intracellular malonyl-CoA supply in E. coli, achieving the production of naringenin up to 1,073 mg/L from tyrosine (Wu et al., 2015; Lv et al., 2020; Wu et al., 2021) and 526 mg/L of pinocembrin from glucose (Wu et al., 2016b). These engineering efforts resulted in a 1.14- to 24-fold increased production of naringenin, 4- to 12-fold of pinocembrin and 2- to 3-fold of eriodictyol compared with the parental strains of E. coli, S. cerevisiae and Y. lipolytica, respectively. In addition, these engineering strategies were employed to modulate intracellular availability in C. glutamicum, resulting in the production of 24 mg/L of naringenin by the overexpression of ACC and downregulation of the TCA cycle (Milke et al., 2019). More recently, overexpression of peroxisomal proliferation and free fatty-acid transport pathway protein (Pex11) increased acetyl-CoA availability via β-oxidation, enabling the production of 1,129 mg/l of naringenin in S. cerevisiae (Zhang et al., 2021). The similar approach enabled the production of 898 mg/l of naringenin from glucose in Y. lipolytica (Palmer et al., 2020).
2.5 Directed Evolution/Pathway Tuning
The expression of heterogonous genes such as flavonoid biosynthetic genes in microorganisms often utilizes an inducible or constitutive expression system, but the resulting high-level expression of heterogonous genes often causes metabolic flux imbalance, a reduction in cell growth, an accumulation of (toxic) intermediates, and the synthesis of undesired by-products, resulting in reduced carbon influx into flavonoid biosynthesis. To overcome this problem, fine-tuning of the expression level of pathway gene(s) and directed evolution of pathway enzyme(s) have been used to optimize the introduced biosynthetic pathway (Jones et al., 2015).
The expression of pathway genes can be efficiently balanced by constructing strains with different expression levels of each pathway gene, followed by screening for high naringenin production. Zhou et al. (2019) established a high-throughput method to evaluate naringenin production using ultraviolet spectrophotometry–fluorescence spectrophotometry. In this method, cells are incubated in the presence of aluminum ion (Al3+) to allow the binding of Al3+ to naringenin, and the resulting Al3+-bound naringenin is measured using fluorescence excitation at 382 nm and emission at 505 nm and/or UV absorbance at 373 nm. This approach was used to fine-tune the pathway enzymes to optimize promoter strength for naringenin pathway genes in both E. coli (Zhou et al., 2019) and S. cerevisiae (Gao et al., 2020). Another example utilized the naringenin-inducible transcription factor FdeR and its cognate binding site (fdeO) to construct a naringenin-responsive transcription switch in S. cerevisiae (Wang et al., 2019). This biosensor enabled the high-throughput screening of as many as 972 combinations of expression cassette with different promoter-terminator pairs and plasmid vectors for three naringenin biosynthetic genes 4CL, CHS, and CHI, leading to identify the yeast strain producing 52.0 mg/L naringenin. Additionally, naringenin-responsive riboswitches have been developed (Jang et al., 2017; Xiu et al., 2017) and utilized for fine-tuning the expression of multiple enzymes in E. coli, enabling a 3-fold improvement in naringenin production (Hwang et al., 2021).
The high-throughput screening approaches using genetically encoded biosensors described above can be used not only to balance the expression of pathway enzymes, but also to perform directed evolution of the catalytic activity of these enzymes. To this end, several genetically encoded biosensors that translate intracellular concentrations of naringenin and malonyl-CoA into reporter gene expression levels have been described. For example, the transcription factor TtgR from Pseudomonas putida and its target promoter have been evolutionarily engineered in E. coli to respond to resveratrol, which is synthesized from p-coumaroyl-CoA by stilbene synthase (resveratrol synthase) catalysis (Xiong et al., 2017). The resultant biosensor was used to screen 4CL1 (A. thaliana 4CL) mutants with improved activity from a randomly mutagenized 4CL1 library, leading to the enhanced production of naringenin and resveratrol with p-coumaric acid supplementation. Furthermore, the repressor QdoR from Bacillus subtilis responsive for downstream flavonoids (flavonols; quercetin and kaempferol, see Section 3.1 for more information on these compounds) can be used for the high-throughput screening of functional FLS mutants that convert dihydrokaempferol into kaempferol with different efficiencies (Siedler et al., 2014).
As mentioned above, biosensor-assisted metabolic engineering is a powerful technique to obtain high production of flavonoids, but this strategy requires well-optimized biosensors for each host species. Both rational and irrational engineering toolboxes have been described in detail for E. coli and S. cerevisiae, but not for non-conventional microbes such as Y. lipolytica. Recently, Bowman et al. (2021) addressed this problem by developing a droplet-based high-throughput screening technique that enables the co-encapsulation of two different microbes in a single droplet: one is a mutant cell that produces naringenin, and the other is a sensor cell that translates the naringenin concentration inside the droplet into an altered level of gene expression. Different flavonoids can be monitored by simply changing the sensor cells using this method.
2.6 Dynamic Regulation for Optimizing the Allocation of Precursors in Flavonoid Synthesis
Since flavonoids share the same precursors as lipids during biosynthesis, reallocation of the precursors could increase flavonoid production. To this end, metabolite sensors that respond to flavonoids or their precursors have been assembled into genetic circuits to dynamically control the expression of branching pathway enzymes. Lv et al. (2020) constructed CRISPRi-based, fatty acid-responsive genetic circuits in Y. lipolytica to improve naringenin production by optimizing malonyl-CoA allocation. They used a synthetic, fatty acid inducible promoter, based on the regulatory components from natural fatty acids inducible promoter, POX2 (Shabbir Hussain et al., 2017), to regulate the expression of three gRNAs that repress three genes responsible for fatty acids synthesis [FAS1, FAS2, and YALI0E18590g (equivalent to bacterial fabD or yeast MCT1)], together with constitutively expressed dCas9. This approach repressed fatty acid synthesis in response to excessive fatty acid synthesis, enabling an increase in naringenin production by up to 74.8%. The expression of pathway genes can be regulated in response not only to intracellular metabolite concentrations but also to cell density. For example, Dinh and Prather created an autonomous genetic circuit that leads to the activation of TAL and 4CL gene expression along with the CRISPRi-based downregulation of malonyl-CoA consuming pathway genes (fabF and sucC) in response to high cell density in E. coli (Dinh and Prather, 2019). This dynamic regulation resulted in a 6-fold increase in naringenin production compared with the control strain without the dynamic control and the CRISPRi-based downregulation even in a bioreactor fermentation.
In addition to pathway gene regulation, metabolite sensors can be used for the selective growth of cells producing target metabolites to increase productivity. Lv et al. (2020) created a naringenin-responsive transcription switch based on the transcription factor FdeR and its binding sites (fdeO) fused to the TEF1 promoter, without an upstream activation sequence controlling the LEU2 gene. In this system, only cells producing sufficient naringenin to induce LEU2 expression can grow in medium lacking leucine. This strategy, together with the aforementioned fatty-acid responsive CRISPRi system, enabled stable naringenin production by up to 324 generations.
Despite the wide utility of metabolite sensors described above, the sensors are often saturated by the metabolite to be sensed. In naringenin production, the FdeR-based naringenin sensor could not detect naringenin over 40 mg/L. Boada et al. (2020) addressed this problem by indirectly detecting large amounts of naringenin by converting a small fraction of the naringenin into kaempferol (see Section 3.1 for more information on this compound), which can be detected using the kaempferol-responsive transcription factor QdoR.
3 Microbial Production of Diversified Flavonoids
In plants, structurally complicated flavonoids are synthesized using flavanones as starting compounds (Figure 1) (Winkel, 2006). Introducing plant-derived genes encoding modification enzymes into microbes allows microbial production of these complicated flavonoids. This section introduces the microbial production of structurally complicated flavonoids derived from naringenin, pinocembrin and eriodictyol using various types of modifications (Table 3).
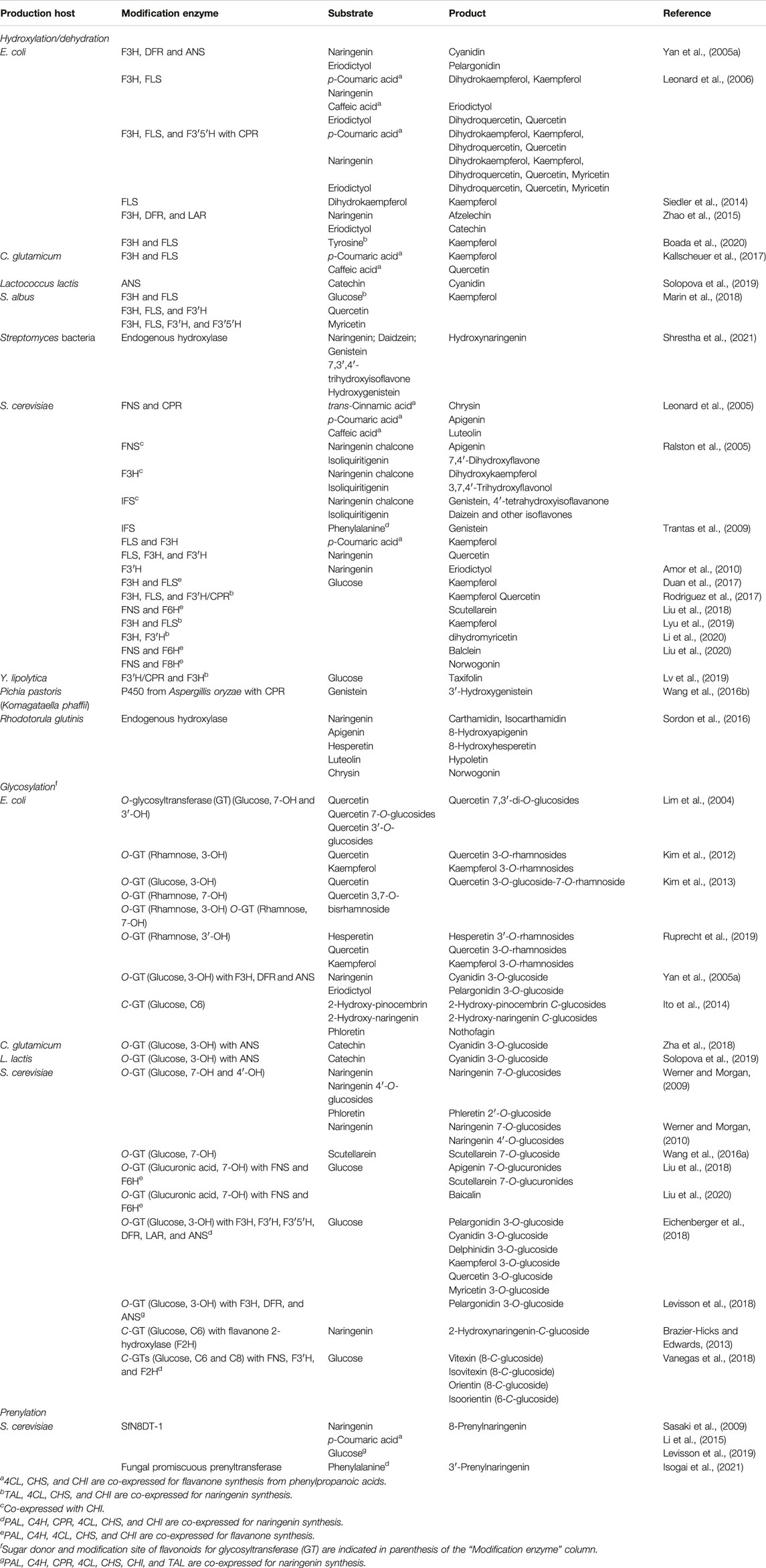
3.1 Hydroxylation/Dehydration
Flavone, dihydroflavonol, flavonol, anthocyanidin, flavane-3-ol, and isoflavone can be synthesized via flavanone (Figure 1). This has prompted many studies on the bioconversion of naringenin and eriodictyol by the expression of one or several modification enzymes or the partial reconstruction of the biosynthetic pathway in E. coli (Yan et al., 2005a; Leonard et al., 2006; Zhao et al., 2015) and S. cerevisiae (Leonard et al., 2005; Ralston et al., 2005; Trantas et al., 2009; Amor et al., 2010). In E. coli, kaempferol, a flavonol, was produced by expressing FLS (FLS1) from A. thaliana to convert dihydrokaempferol, a dihydroflavonol. The concentration of kaempferol produced was monitored by a QdoR-based metabolite sensor responsive to kaempferol and quercetin (flavonols) but not to dihydrokaempferol (see Section 2.5) (Siedler et al., 2014). Furthermore, kaempferol was produced in E. coli from tyrosine by expressing F3H and FLS in addition to the naringenin biosynthetic pathway via feedback control of CHS expression (downregulation) using a genetic circuit incorporating the QdoR-based kaempferol biosensor (see Section 2.6) (Boada et al., 2020). Recently, the production of these derivatives using bacteria and yeasts other than E. coli and S. cerevisiae has been reported: kaempferol and quercetin production from phenylpropanoic acids by C. glutamicum expressing F3H and FLS together with 4CL, CHS and CHI (Kallscheuer et al., 2017), hydroxylation of genistein (an isoflavone) by P. pastoris (Wang et al., 2016), and conversion of catechin (a flavane-3-ol) to cyanidin (a anthocyanidin) by L. lactis expressing ANS (Solopova et al., 2019). This engineered L. lactis further glycosylated cyanidin to anthocyanin (see Section 3.2).
The de novo synthesis of various naringenin derivatives, including kaempferol and quercetin, was recently achieved by expressing suitable modification enzymes in S. cerevisiae carrying the reconstructed naringenin biosynthetic pathway, namely, the production of kaempferol (Duan et al., 2017; Rodriguez et al., 2017; Lyu et al., 2019) and quercetin (Rodriguez et al., 2017) by expression of FLS, and dihydromyricetin (a dihydroflavonol) by multiple hydroxylases, including F3H (Li et al., 2020). In addition, Liu et al. (2018) identified P450 enzymes responsible for hydroxylation of C6 (F6H) and C8 (F8H) of flavanone backbone. Expression of these hydroxylases with FNS achieved the production of hydroxylated flavones from glucose in engineered S. cerevisiae: expression of F6H, FNS and naringenin biosynthetic enzymes produced scutellarein, whereas that of F6H or F8H with FNS and genes responsible for pinocembrin biosynthesis enabled the production of balclein and norwogonin respectively (Liu et al., 2020). The engineered S. cerevisiae further glycosylated these flavones by additional expression of glycosyltransferase (see section 3.2). Furthermore, engineered S. albus and Y. lipolytica de novo synthesized flavonols (myricetin, kaempferol and quercetin) (Marin et al., 2018) and taxifolin (a dihydroflavonol) (Lv et al., 2019), respectively. More recently, the co-culture system of two different S. cerevisiae strains has been developed, in which one strain synthesized naringenin and the other converted it to its derivatives (Table 2) (Du et al., 2020). This platform enabled the production of different flavonols and anthocyanidins simply by exchanging naringenin-converting strain with that expressing different modification enzymes. Another approach for the hydroxylation of flavonoids involves the bioconversion of flavonoids using yeast or Streptomyces bacteria without the use of heterogeneous genes (Sordon et al., 2016; Shrestha et al., 2021).
3.2 Glycosylation
Several studies reported the bioconversion of flavonoid scaffolds to O-glycosylated derivatives using E. coli (Lim et al., 2004; Kim et al., 2012; Kim et al., 2013; Ruprecht et al., 2019) and S. cerevisiae (Werner and Morgan, 2009, 2010; Wang et al., 2016) by expressing botanical O-glycosyltransferase (Figure 1). E. coli (Ito et al., 2014) and S. cerevisiae (Brazier-Hicks and Edwards, 2013) expressing C-glycosyltransferase converted flavone and dihydrochalcone to the corresponding C-glycosylated derivatives. Moreover, anthocyanins, which are pigments synthesized via the glycosylation of anthocyanidins, were biosynthesized from flavanone (eriodictyol and naringenin) in engineered E. coli expressing O-glycosyltransferase with F3H, DFR, ANS (Yan et al., 2005a), and from catechin by C. glutamicum (Zha et al., 2018) and L. lactis (Solopova et al., 2019) expressing O-glycosyltransferase and ANS.
More recent studies reported the de novo synthesis of anthocyanins by the expression of O-glycosyltransferase, F3H, DFR, and ANS with reconstruction of the naringenin biosynthetic pathway in engineered S. cerevisiae (Eichenberger et al., 2018; Levisson et al., 2018). Additionally, a co-culture system comprising four different E. coli strains was developed for the de novo synthesis of anthocyanin. Each E. coli strain was designed to provide one of the following modules: synthesis of phenylpropanoic acids from glucose, formation of flavanones, conversion of flavanones to flavane-3-ols, and production of anthocyanins from flavane-3-ols (Table 2) (Jones et al., 2017). Furthermore, the de novo synthesis of flavonoid-7-O-glucuronides, such as apigenin-7-O-glucuronide, scutellarin and baicalin, were reported by the expression of 7-O-glucuronosyltransferase with FNS in engineered S. cerevisiae that produce naringenin (Liu et al., 2018) or pinocembrin (Liu et al., 2020). Similarly, C-glycosylated flavones were de novo synthesized using C-glycosyltransferase together with the reconstruction of the naringenin biosynthetic pathway in yeast (Vanegas et al., 2018).
3.3 Prenylation
Flavonoid prenyltransferases, a class of enzymes responsible for flavonoid prenylation, are membrane-bound and strictly control the stereoselectivity of prenyl transfer (Figure 1). The prenyltransferase SfN8DT-1, isolated from Sophora flavescens, is responsible for the C8-prenylation of naringenin. Expression of this enzyme in S. cerevisiae resulted in the bioconversion of naringenin to 8-prenylnaringenin (Sasaki et al., 2009). Moreover, SfN8DT expression in naringenin-producing S. cerevisiae resulted in the synthesis of 8-prenylnatingenin from glucose (Levisson et al., 2019), phenylalanine (Isogai et al., 2021) and p-coumaric acid (Li et al., 2015).
However, to date only 13 flavonoid prenyltransferases have been identified from plants, limiting the microbial production of prenylated flavonoids. For example, three kinds of prenylnaringenins have been isolated from plants: 8-, 6-, and 3′-prenylnaringenin carrying the dimethylallyl moiety at C8 or C6 in the A-ring, and at C3′ in the B-ring, respectively. However, only the prenyltransferase SfN8DT-1 was identified, and enzymes responsible for the C6- and C3′-prenylation of naringenin remain unidentified. Recently, the in vivo production of 3′-prenylnaringenin was achieved by the expression of fungal prenyltransferase with broad substrate specificity (Isogai et al., 2021). Such microbial promiscuous enzyme may also be able to catalyze prenylation of other positions of flavonoids, for which specific enzymes have not yet been identified.
4 Conclusion and Future Perspectives
Flavonoids are promising as pharmachemicals and food supplements due to their various biological activities; however, their supply is not stable or environmentally friendly. Therefore, the development of microbial production platform for flavonoids is desired. In this review, we focused on the strategies to increase microbial production of flavanones, specifically naringenin, pinocembrin, and eriodictyol, that serve as a starting compound for diverse flavonoids. The strategies are mainly the pathway engineering for the biosynthesis of flavanones and their precursors (aromatic amino acids and malonyl-CoA) by selecting host organism, gene source and optimizing enzyme expression levels.
As described in Section 2.6, several recent studies have demonstrated that genetically encoded biosensors for flavonoids can be useful for improving naringenin production by dynamically controlling the expression of pathway enzymes in response to cellular conditions. However, such strategies largely depend on the specifications of the biosensors, thereby necessitating the reconstruction of biosensors for functional fine-tuning or improvement. To this end, both rational and evolutionary methodologies for engineering synthetic promoters (Snoek et al., 2020; Cazier and Blazeck, 2021; Tominaga et al., 2021) can be adapted. The assembly of well-optimized biosensors into genetic circuits to control the important pathway enzymes will further sophisticate the flavonoids production as well as balancing their expression levels via translational control using a ribosome binding site (RBS) library (Wang et al., 2021).
We also described recent progress in the microbial production of structurally and functionally diverse flavonoids via flavanones such as naringenin, some of which have complex structures. Diverse flavonoids can be efficiently produced by introducing modification enzymes into flavanone-producing microbes. Although thousands of flavonoids are known to be produced in plants, there are few reports describing the microbial production of such flavonoids, and these were summarized in this review. Progress faces two possible bottlenecks: only a few modification enzymes from plants have been identified, and these enzymes are membrane-bound, complicating their functional expression in bacteria. Methodologies for the rapid identification and microbial expression of these enzymes need to be developed. The massive increase in genomic information due to the expansion of next-generation sequencing will help to uncover novel microbial enzymes, as well as botanical enzymes suitable for various modification steps (Unamba et al., 2015; Medema and Osbourn, 2016). Alternatively, microbial promiscuous enzymes can be used not only as modification enzymes but also as easy-to-express counterparts of botanical enzymes. Altogether, advanced methodologies combining metabolic engineering and synthetic biology will enable microbial production of an incredibly diverse range of plant flavonoids.
Author Contributions
SI: conceptualization, investigation, original draft preparation, visualization, writing and editing, MT: conceptualization, investigation, original draft preparation, visualization, writing and editing, AK: conceptualization, supervision and JI: conceptualization, investigation, editing.
Funding
This work was partially supported by the Commission for Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterials from the Ministry of Economy, Trade and Industry (METI), the CREST program (JPMJCR21N2) and the JST-Mirai Program (JPMJMI17EJ) from the Japan Science and Technology Agency (JST), JSPS KAKENHI (Grant Number 18K14374), and the Intensive Support for Young Promising Researchers program (JPNP20004) from the New Energy and Industrial Technology Development Organization (NEDO).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amor, I. L., Hehn, A., Guedone, E., Ghedira, K., Engasser, J. M., Chekir-Ghedrira, L., et al. (2010). Biotransformation of Naringenin to Eriodictyol by Saccharomyces Cerevisiea Functionally Expressing Flavonoid 3' Hydroxylase. Nat. Prod. Commun. 5, 1893–1898. doi:10.1177/1934578x1000501211
An, J. H., and Kim, Y. S. (1998). A Gene Cluster Encoding Malonyl-CoA Decarboxylase (MatA), Malonyl-CoA Synthetase (MatB) and a Putative Dicarboxylate Carrier Protein (MatC) in Rhizobium Trifolii . Cloning, Sequencing, and Expression of the Enzymes in Escherichia coli. Eur. J. Biochem. 257, 395–402. doi:10.1046/j.1432-1327.1998.2570395.x
Barros, J., and Dixon, R. A. (2020). Plant Phenylalanine/Tyrosine Ammonia-Lyases. Trends Plant Sci. 25, 66–79. doi:10.1016/j.tplants.2019.09.011
Becker, J., Zelder, O., Häfner, S., Schröder, H., and Wittmann, C. (2011). From Zero to Hero-Design-Based Systems Metabolic Engineering of Corynebacterium Glutamicum for L-Lysine Production. Metab. Eng. 13, 159–168. doi:10.1016/j.ymben.2011.01.003
Beopoulos, A., Cescut, J., Haddouche, R., Uribelarrea, J.-L., Molina-Jouve, C., and Nicaud, J.-M. (2009). Yarrowia Lipolytica as a Model for Bio-Oil Production. Prog. Lipid Res. 48, 375–387. doi:10.1016/j.plipres.2009.08.005
Boada, Y., Vignoni, A., Picó, J., and Carbonell, P. (2020). Extended Metabolic Biosensor Design for Dynamic Pathway Regulation of Cell Factories. iScience 23, 101305. doi:10.1016/j.isci.2020.101305
Bowman, E. K., Wagner, J. M., Yuan, S. F., Deaner, M., Palmer, C. M., D'oelsnitz, S., et al. (2021). Sorting for Secreted Molecule Production Using a Biosensor-In-Microdroplet Approach. Proc. Natl. Acad. Sci. U. S. A. 118. doi:10.1073/pnas.2106818118
Braus, G. H. (1991). Aromatic Amino Acid Biosynthesis in the Yeast Saccharomyces cerevisiae: a Model System for the Regulation of a Eukaryotic Biosynthetic Pathway. Microbiol. Rev. 55, 349–370. doi:10.1128/mr.55.3.349-370.1991
Brazier-Hicks, M., and Edwards, R. (2013). Metabolic Engineering of the Flavone-C-Glycoside Pathway Using Polyprotein Technology. Metab. Eng. 16, 11–20. doi:10.1016/j.ymben.2012.11.004
Cao, W., Ma, W., Wang, X., Zhang, B., Cao, X., Chen, K., et al. (2016a). Enhanced Pinocembrin Production in Escherichia coli by Regulating Cinnamic Acid Metabolism. Sci. Rep. 6, 32640. doi:10.1038/srep32640
Cao, W., Ma, W., Zhang, B., Wang, X., Chen, K., Li, Y., et al. (2016b). Improved Pinocembrin Production in Escherichia coli by Engineering Fatty Acid Synthesis. J. Ind. Microbiol. Biotechnol. 43, 557–566. doi:10.1007/s10295-015-1725-3
Cazier, A. P., and Blazeck, J. (2021). Advances in Promoter Engineering: Novel Applications and Predefined Transcriptional Control. Biotechnol. J. 16, e2100239. doi:10.1002/biot.202100239
Dinh, C. V., and Prather, K. L. J. (2019). Development of an Autonomous and Bifunctional Quorum-sensing Circuit for Metabolic Flux Control in Engineered Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 116, 25562–25568. doi:10.1073/pnas.1911144116
Dixon, R. A., and Pasinetti, G. M. (2010). Flavonoids and Isoflavonoids: from Plant Biology to Agriculture and Neuroscience. Plant Physiol. 154, 453–457. doi:10.1104/pp.110.161430
Du, Y., Yang, B., Yi, Z., Hu, L., and Li, M. (2020). Engineering Saccharomyces cerevisiae Coculture Platform for the Production of Flavonoids. J. Agric. Food Chem. 68, 2146–2154. doi:10.1021/acs.jafc.9b07916
Duan, L., Ding, W., Liu, X., Cheng, X., Cai, J., Hua, E., et al. (2017). Biosynthesis and Engineering of Kaempferol in Saccharomyces cerevisiae. Microb. Cell. Fact. 16, 165. doi:10.1186/s12934-017-0774-x
Dunstan, M. S., Robinson, C. J., Jervis, A. J., Yan, C., Carbonell, P., Hollywood, K. A., et al. (2020). Engineering Escherichia coli towards De Novo Production of Gatekeeper (2S)-Flavanones: Naringenin, Pinocembrin, Eriodictyol and Homoeriodictyol. Synth. Biol. (Oxf) 5, ysaa012. doi:10.1093/synbio/ysaa012
Eichenberger, M., Hansson, A., Fischer, D., Dürr, L., and Naesby, M. (2018). De Novo biosynthesis of Anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 18. doi:10.1093/femsyr/foy046
Fowler, Z. L., Gikandi, W. W., and Koffas, M. A. G. (2009). Increased Malonyl Coenzyme A Biosynthesis by Tuning the Escherichia coli Metabolic Network and its Application to Flavanone Production. Appl. Environ. Microbiol. 75, 5831–5839. doi:10.1128/aem.00270-09
Fraga, C. G., Croft, K. D., Kennedy, D. O., and Tomás-Barberán, F. A. (2019). The Effects of Polyphenols and Other Bioactives on Human Health. Food Funct. 10, 514–528. doi:10.1039/c8fo01997e
Gao, S., Zhou, H., Zhou, J., and Chen, J. (2020). Promoter-Library-Based Pathway Optimization for Efficient (2S)-Naringenin Production from P-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 68, 6884–6891. doi:10.1021/acs.jafc.0c01130
Harborne, J. B., and Williams, C. A. (2000). Advances in Flavonoid Research since 1992. Phytochemistry 55, 481–504. doi:10.1016/s0031-9422(00)00235-1
Hartmann, M., Schneider, T. R., Pfeil, A., Heinrich, G., Lipscomb, W. N., and Braus, G. H. (2003). Evolution of Feedback-Inhibited β/α Barrel Isoenzymes by Gene Duplication and a Single Mutation. Proc. Natl. Acad. Sci. U.S.A. 100, 862–867. doi:10.1073/pnas.0337566100
Helmstaedt, K., Strittmatter, A., Lipscomb, W. N., and Braus, G. H. (2005). Evolution of 3-deoxy- D - Arabino -Heptulosonate-7-Phosphate Synthase-Encoding Genes in the Yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 102, 9784–9789. doi:10.1073/pnas.0504238102
Hwang, E. I., Kaneko, M., Ohnishi, Y., and Horinouchi, S. (2003). Production of Plant-specific Flavanones by Escherichia coli Containing an Artificial Gene Cluster. Appl. Environ. Microbiol. 69, 2699–2706. doi:10.1128/aem.69.5.2699-2706.2003
Hwang, H. G., Noh, M. H., Koffas, M. A. G., Jang, S., and Jung, G. Y. (2021). Multi-level Rebalancing of the Naringenin Pathway Using Riboswitch-Guided High-Throughput Screening. Metab. Eng. 67, 417–427. doi:10.1016/j.ymben.2021.08.003
Isogai, S., Okahashi, N., Asama, R., Nakamura, T., Hasunuma, T., Matsuda, F., et al. (2021). Synthetic Production of Prenylated Naringenins in Yeast Using Promiscuous Microbial Prenyltransferases. Metab. Eng. Commun. 12, e00169. doi:10.1016/j.mec.2021.e00169
Ito, T., Fujimoto, S., Shimosaka, M., and Taguchi, G. (2014). Production of C-Glucosides of Flavonoids and Related Compounds by Escherichia coli Expressing Buckwheat C-Glucosyltransferase. Plant Biotechnol. 31, 519–524. doi:10.5511/plantbiotechnology.14.1016a
Jang, S., Jang, S., Xiu, Y., Kang, T. J., Lee, S.-H., Koffas, M. A. G., et al. (2017). Development of Artificial Riboswitches for Monitoring of Naringenin In Vivo. ACS Synth. Biol. 6, 2077–2085. doi:10.1021/acssynbio.7b00128
Jiang, H., Wood, K. V., and Morgan, J. A. (2005). Metabolic Engineering of the Phenylpropanoid Pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 2962–2969. doi:10.1128/aem.71.6.2962-2969.2005
Jin, H., Wang, Y., Zhao, P., Wang, L., Zhang, S., Meng, D., et al. (2021). Potential of Producing Flavonoids Using Cyanobacteria as a Sustainable Chassis. J. Agric. Food Chem. 69, 12385–12401. doi:10.1021/acs.jafc.1c04632
Jones, J. A., Vernacchio, V. R., Collins, S. M., Shirke, A. N., Xiu, Y., Englaender, J. A., et al. (2017). Complete Biosynthesis of Anthocyanins Using E. coli Polycultures. mBio 8, e00621–00617. doi:10.1128/mBio.00621-17
Jones, J. A., Toparlak, Ö. D., and Koffas, M. A. (2015). Metabolic Pathway Balancing and its Role in the Production of Biofuels and Chemicals. Curr. Opin. Biotechnol. 33, 52–59. doi:10.1016/j.copbio.2014.11.013
Kallscheuer, N., Vogt, M., Bott, M., and Marienhagen, J. (2017). Functional Expression of Plant-Derived O -methyltransferase, Flavanone 3-hydroxylase, and Flavonol Synthase in Corynebacterium Glutamicum for Production of Pterostilbene, Kaempferol, and Quercetin. J. Biotechnol. 258, 190–196. doi:10.1016/j.jbiotec.2017.01.006
Kallscheuer, N., Vogt, M., Kappelmann, J., Krumbach, K., Noack, S., Bott, M., et al. (2016a). Identification of the Phd Gene Cluster Responsible for Phenylpropanoid Utilization in Corynebacterium Glutamicum. Appl. Microbiol. Biotechnol. 100, 1871–1881. doi:10.1007/s00253-015-7165-1
Kallscheuer, N., Vogt, M., Stenzel, A., Gätgens, J., Bott, M., and Marienhagen, J. (2016b). Construction of a Corynebacterium Glutamicum Platform Strain for the Production of Stilbenes and (2S)-Flavanones. Metab. Eng. 38, 47–55. doi:10.1016/j.ymben.2016.06.003
Kikuchi, Y., Tsujimoto, K., and Kurahashi, O. (1997). Mutational Analysis of the Feedback Sites of Phenylalanine-Sensitive 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase of Escherichia coli. Appl. Environ. Microbiol. 63, 761–762. doi:10.1128/aem.63.2.761-762.1997
Kim, B.-G., Kim, H. J., and Ahn, J.-H. (2012). Production of Bioactive Flavonol Rhamnosides by Expression of Plant Genes in Escherichia coli. J. Agric. Food Chem. 60, 11143–11148. doi:10.1021/jf302123c
Kim, B., Binkley, R., Kim, H. U., and Lee, S. Y. (2018). Metabolic Engineering ofEscherichia Colifor the Enhanced Production Ofl‐tyrosine. Biotechnol. Bioeng. 115, 2554–2564. doi:10.1002/bit.26797
Kim, B. G., Lee, H., and Ahn, J.-H. (2014). Biosynthesis of Pinocembrin from Glucose Using Engineered escherichia Coli. J. Microbiol. Biotechnol. 24, 1536–1541. doi:10.4014/jmb.1406.06011
Kim, H. J., Kim, B.-G., and Ahn, J.-H. (2013). Regioselective Synthesis of Flavonoid Bisglycosides Using Escherichia coli Harboring Two Glycosyltransferases. Appl. Microbiol. Biotechnol. 97, 5275–5282. doi:10.1007/s00253-013-4844-7
Koffas, M. A. G., Jung, G. Y., and Stephanopoulos, G. (2003). Engineering Metabolism and Product Formation in Corynebacterium Glutamicum by Coordinated Gene Overexpression. Metab. Eng. 5, 32–41. doi:10.1016/s1096-7176(03)00002-8
Koopman, F., Beekwilder, J., Crimi, B., Van Houwelingen, A., Hall, R. D., Bosch, D., et al. (2012). De Novo production of the Flavonoid Naringenin in Engineered Saccharomyces cerevisiae. Microb. Cell. Fact. 11, 155. doi:10.1186/1475-2859-11-155
Leonard, E., Lim, K.-H., Saw, P.-N., and Koffas, M. A. G. (2007). Engineering Central Metabolic Pathways for High-Level Flavonoid Production in Escherichia coli. Appl. Environ. Microbiol. 73, 3877–3886. doi:10.1128/aem.00200-07
Leonard, E., Yan, Y., Fowler, Z. L., Li, Z., Lim, C.-G., Lim, K.-H., et al. (2008). Strain Improvement of Recombinant Escherichia coli for Efficient Production of Plant Flavonoids. Mol. Pharm. 5, 257–265. doi:10.1021/mp7001472
Leonard, E., Yan, Y., and Koffas, M. (2006). Functional Expression of a P450 Flavonoid Hydroxylase for the Biosynthesis of Plant-specific Hydroxylated Flavonols in Escherichia coli. Metab. Eng. 8, 172–181. doi:10.1016/j.ymben.2005.11.001
Leonard, E., Yan, Y., Lim, K. H., and Koffas, M. A. G. (2005). Investigation of Two Distinct Flavone Synthases for Plant-specific Flavone Biosynthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 8241–8248. doi:10.1128/aem.71.12.8241-8248.2005
Levisson, M., Araya-Cloutier, C., De Bruijn, W. J. C., Van Der Heide, M., Salvador López, J. M., Daran, J.-M., et al. (2019). Toward Developing a Yeast Cell Factory for the Production of Prenylated Flavonoids. J. Agric. Food Chem. 67, 13478–13486. doi:10.1021/acs.jafc.9b01367
Levisson, M., Patinios, C., Hein, S., De Groot, P. A., Daran, J.-M., Hall, R. D., et al. (2018). Engineering De Novo Anthocyanin Production in Saccharomyces cerevisiae. Microb. Cell. Fact. 17, 103. doi:10.1186/s12934-018-0951-6
Li, G., Li, H., Lyu, Y., Zeng, W., and Zhou, J. (2020). Enhanced Biosynthesis of Dihydromyricetin in Saccharomyces cerevisiae by Coexpression of Multiple Hydroxylases. J. Agric. Food Chem. 68, 14221–14229. doi:10.1021/acs.jafc.0c05261
Li, H., Ban, Z., Qin, H., Ma, L., King, A. J., and Wang, G. (2015). A Heteromeric Membrane-Bound Prenyltransferase Complex from Hop Catalyzes Three Sequential Aromatic Prenylations in the Bitter Acid Pathway. Plant Physiol. 167, 650–659. doi:10.1104/pp.114.253682
Lim, E.-K., Ashford, D. A., Hou, B., Jackson, R. G., and Bowles, D. J. (2004). Arabidopsis Glycosyltransferases as Biocatalysts in Fermentation for Regioselective Synthesis of Diverse Quercetin Glucosides. Biotechnol. Bioeng. 87, 623–631. doi:10.1002/bit.20154
Liu, W., Zhang, B., and Jiang, R. (2017a). Improving Acetyl-CoA Biosynthesis in Saccharomyces cerevisiae via the Overexpression of Pantothenate Kinase and PDH Bypass. Biotechnol. Biofuels 10, 41. doi:10.1186/s13068-017-0726-z
Liu, X., Cheng, J., Zhang, G., Ding, W., Duan, L., Yang, J., et al. (2018). Engineering Yeast for the Production of Breviscapine by Genomic Analysis and Synthetic Biology Approaches. Nat. Commun. 9, 448. doi:10.1038/s41467-018-02883-z
Liu, X., Cheng, J., Zhu, X., Zhang, G., Yang, S., Guo, X., et al. (2020). De Novo Biosynthesis of Multiple Pinocembrin Derivatives in Saccharomyces cerevisiae. ACS Synth. Biol. 9, 3042–3051. doi:10.1021/acssynbio.0c00289
Liu, X., Ding, W., and Jiang, H. (2017b). Engineering Microbial Cell Factories for the Production of Plant Natural Products: from Design Principles to Industrial-Scale Production. Microb. Cell. Fact. 16, 125. doi:10.1186/s12934-017-0732-7
Lütke-Eversloh, T., and Stephanopoulos, G. (2005). Feedback Inhibition of Chorismate Mutase/prephenate Dehydrogenase (TyrA) of Escherichia coli: Generation and Characterization of Tyrosine-Insensitive Mutants. Appl. Environ. Microbiol. 71, 7224–7228. doi:10.1128/AEM.71.11.7224-7228.2005
Lütke-Eversloh, T., and Stephanopoulos, G. (2007). L-tyrosine Production by Deregulated Strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75, 103–110. doi:10.1007/s00253-006-0792-9
Lv, Y., Gu, Y., Xu, J., Zhou, J., and Xu, P. (2020). Coupling Metabolic Addiction with Negative Autoregulation to Improve Strain Stability and Pathway Yield. Metab. Eng. 61, 79–88. doi:10.1016/j.ymben.2020.05.005
Lv, Y., Marsafari, M., Koffas, M., Zhou, J., and Xu, P. (2019). Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 8, 2514–2523. doi:10.1021/acssynbio.9b00193
Lyu, X., Ng, K. R., Lee, J. L., Mark, R., and Chen, W. N. (2017). Enhancement of Naringenin Biosynthesis from Tyrosine by Metabolic Engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 65, 6638–6646. doi:10.1021/acs.jafc.7b02507
Lyu, X., Zhao, G., Ng, K. R., Mark, R., and Chen, W. N. (2019). Metabolic Engineering of Saccharomyces cerevisiae for De Novo Production of Kaempferol. J. Agric. Food Chem. 67, 5596–5606. doi:10.1021/acs.jafc.9b01329
Marín, L., Gutiérrez-del-Río, I., Entrialgo-Cadierno, R., Villar, C. J., and Lombó, F. (2018). De Novo biosynthesis of Myricetin, Kaempferol and Quercetin in Streptomyces Albus and Streptomyces Coelicolor. PLoS One 13, e0207278. doi:10.1371/journal.pone.0207278
Mark, R., Lyu, X., Ng, K. R., and Chen, W. N. (2019). Gene Source Screening as a Tool for Naringenin Production in Engineered Saccharomyces cerevisiae. ACS Omega 4, 12872–12879. doi:10.1021/acsomega.9b00364
Medema, M. H., and Osbourn, A. (2016). Computational Genomic Identification and Functional Reconstitution of Plant Natural Product Biosynthetic Pathways. Nat. Prod. Rep. 33, 951–962. doi:10.1039/c6np00035e
Milke, L., Ferreira, P., Kallscheuer, N., Braga, A., Vogt, M., Kappelmann, J., et al. (2019). Modulation of the Central Carbon Metabolism of Corynebacterium Glutamicum Improves malonyl‐CoA Availability and Increases Plant Polyphenol Synthesis. Biotechnol. Bioeng. 116, 1380–1391. doi:10.1002/bit.26939
Milke, L., and Marienhagen, J. (2020). Engineering Intracellular Malonyl-CoA Availability in Microbial Hosts and its Impact on Polyketide and Fatty Acid Synthesis. Appl. Microbiol. Biotechnol. 104, 6057–6065. doi:10.1007/s00253-020-10643-7
Mukai, R., Horikawa, H., Fujikura, Y., Kawamura, T., Nemoto, H., Nikawa, T., et al. (2012). Prevention of Disuse Muscle Atrophy by Dietary Ingestion of 8-prenylnaringenin in Denervated Mice. PLoS One 7, e45048. doi:10.1371/journal.pone.0045048
Ni, J., Tao, F., Wang, Y., Yao, F., and Xu, P. (2016). A Photoautotrophic Platform for the Sustainable Production of Valuable Plant Natural Products from CO2. Green Chem. 18, 3537–3548. doi:10.1039/c6gc00317f
Palmer, C. M., Miller, K. K., Nguyen, A., and Alper, H. S. (2020). Engineering 4-Coumaroyl-CoA Derived Polyketide Production in Yarrowia Lipolytica through a β-oxidation Mediated Strategy. Metab. Eng. 57, 174–181. doi:10.1016/j.ymben.2019.11.006
Pittard, J., Camakaris, H., and Yang, J. (2005). The TyrR Regulon. Mol. Microbiol. 55, 16–26. doi:10.1111/j.1365-2958.2004.04385.x
Ralston, L., Subramanian, S., Matsuno, M., and Yu, O. (2005). Partial Reconstruction of Flavonoid and Isoflavonoid Biosynthesis in Yeast Using Soybean Type I and Type II Chalcone Isomerases. Plant Physiol. 137, 1375–1388. doi:10.1104/pp.104.054502
Rodriguez, A., Kildegaard, K. R., Li, M., Borodina, I., and Nielsen, J. (2015). Establishment of a Yeast Platform Strain for Production of P-Coumaric Acid through Metabolic Engineering of Aromatic Amino Acid Biosynthesis. Metab. Eng. 31, 181–188. doi:10.1016/j.ymben.2015.08.003
Rodriguez, A., Martnez, J. A., Flores, N., Escalante, A., Gosset, G., and Bolivar, F. (2014). Engineering Escherichia coli to Overproduce Aromatic Amino Acids and Derived Compounds. Microb. Cell. Fact. 13, 126. doi:10.1186/s12934-014-0126-z
Rodriguez, A., Strucko, T., Stahlhut, S. G., Kristensen, M., Svenssen, D. K., Forster, J., et al. (2017). Metabolic Engineering of Yeast for Fermentative Production of Flavonoids. Bioresour. Technol. 245, 1645–1654. doi:10.1016/j.biortech.2017.06.043
Ruprecht, C., Bönisch, F., Ilmberger, N., Heyer, T. V., Haupt, E. T. K., Streit, W. R., et al. (2019). High Level Production of Flavonoid Rhamnosides by Metagenome-Derived Glycosyltransferase C in Escherichia coli Utilizing Dextrins of Starch as a Single Carbon Source. Metab. Eng. 55, 212–219. doi:10.1016/j.ymben.2019.07.002
Santos, C. N. S., Koffas, M., and Stephanopoulos, G. (2011). Optimization of a Heterologous Pathway for the Production of Flavonoids from Glucose. Metab. Eng. 13, 392–400. doi:10.1016/j.ymben.2011.02.002
Sasaki, K., Tsurumaru, Y., and Yazaki, K. (2009). Prenylation of Flavonoids by Biotransformation of Yeast Expressing Plant Membrane-Bound Prenyltransferase SfN8DT-1. Biosci. Biotechnol. Biochem. 73, 759–761. doi:10.1271/bbb.80729
Schnappauf, G., Sträter, N., Lipscomb, W. N., and Braus, G. H. (1997). A Glutamate Residue in the Catalytic Center of the Yeast Chorismate Mutase Restricts Enzyme Activity to Acidic Conditions. Proc. Natl. Acad. Sci. U.S.A. 94, 8491–8496. doi:10.1073/pnas.94.16.8491
Shabbir Hussain, M., Wheeldon, I., and Blenner, M. A. (2017). A Strong Hybrid Fatty Acid Inducible Transcriptional Sensor Built from Yarrowia Lipolytica Upstream Activating and Regulatory Sequences. Biotechnol. J. 12. doi:10.1002/biot.201700248
Shrestha, L., Marasini, B. P., Pradhan, S. P., Shrestha, R. K., Shrestha, S., Regmi, K. P., et al. (2021). Biotransformation of Daidzein, Genistein, and Naringenin by Streptomyces Species Isolated from High-Altitude Soil of Nepal. Int. J. Microbiol. 2021, 9948738. doi:10.1155/2021/9948738
Siedler, S., Stahlhut, S. G., Malla, S., Maury, J., and Neves, A. R. (2014). Novel Biosensors Based on Flavonoid-Responsive Transcriptional Regulators Introduced into Escherichia coli. Metab. Eng. 21, 2–8. doi:10.1016/j.ymben.2013.10.011
Snoek, T., Chaberski, E. K., Ambri, F., Kol, S., Bjørn, S. P., Pang, B., et al. (2020). Evolution-guided Engineering of Small-Molecule Biosensors. Nucleic Acids Res. 48, e3. doi:10.1093/nar/gkz954
Solopova, A., Van Tilburg, A. Y., Foito, A., Allwood, J. W., Stewart, D., Kulakauskas, S., et al. (2019). Engineering Lactococcus Lactis for the Production of Unusual Anthocyanins Using Tea as Substrate. Metab. Eng. 54, 160–169. doi:10.1016/j.ymben.2019.04.002
Sordon, S., Madej, A., Popłoński, J., Bartmańska, A., Tronina, T., Brzezowska, E., et al. (2016). Regioselective Ortho-Hydroxylations of Flavonoids by Yeast. J. Agric. Food Chem. 64, 5525–5530. doi:10.1021/acs.jafc.6b02210
Tohge, T., De Souza, L. P., and Fernie, A. R. (2017). Current Understanding of the Pathways of Flavonoid Biosynthesis in Model and Crop Plants. J. Exp. Bot. 68, 4013–4028. doi:10.1093/jxb/erx177
Tominaga, M., Nozaki, K., Umeno, D., Ishii, J., and Kondo, A. (2021). Robust and Flexible Platform for Directed Evolution of Yeast Genetic Switches. Nat. Commun. 12, 1846. doi:10.1038/s41467-021-22134-y
Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic Engineering of the Complete Pathway Leading to Heterologous Biosynthesis of Various Flavonoids and Stilbenoids in Saccharomyces cerevisiae. Metab. Eng. 11, 355–366. doi:10.1016/j.ymben.2009.07.004
Unamba, C. I. N., Nag, A., and Sharma, R. K. (2015). Next Generation Sequencing Technologies: The Doorway to the Unexplored Genomics of Non-model Plants. Front. Plant Sci. 6, 1074. doi:10.3389/fpls.2015.01074
Vanegas, K. G., Larsen, A. B., Eichenberger, M., Fischer, D., Mortensen, U. H., and Naesby, M. (2018). Indirect and Direct Routes to C-Glycosylated Flavones in Saccharomyces cerevisiae. Microb. Cell. Fact. 17, 107. doi:10.1186/s12934-018-0952-5
Wang, H., Yang, Y., Lin, L., Zhou, W., Liu, M., Cheng, K., et al. (2016a). Engineering Saccharomyces cerevisiae with the Deletion of Endogenous Glucosidases for the Production of Flavonoid Glucosides. Microb. Cell. Fact. 15, 134. doi:10.1186/s12934-016-0535-2
Wang, R., Cress, B. F., Yang, Z., Hordines, J. C., Zhao, S., Jung, G. Y., et al. (2019). Design and Characterization of Biosensors for the Screening of Modular Assembled Naringenin Biosynthetic Library in Saccharomyces cerevisiae. ACS Synth. Biol. 8, 2121–2130. doi:10.1021/acssynbio.9b00212
Wang, T. Y., Tsai, Y. H., Yu, I. Z., and Chang, T. S. (2016b). Improving 3′-Hydroxygenistein Production in Recombinant Pichia pastoris Using Periodic Hydrogen Peroxide-Shocking Strategy. J. Microbiol. Biotechnol. 26, 498–502. doi:10.4014/jmb.1509.09013
Wang, Y., Cheng, H., Liu, Y., Liu, Y., Wen, X., Zhang, K., et al. (2021). In-situ Generation of Large Numbers of Genetic Combinations for Metabolic Reprogramming via CRISPR-Guided Base Editing. Nat. Commun. 12, 678. doi:10.1038/s41467-021-21003-y
Watts, K. T., Lee, P. C., and Schmidt-Dannert, C. (2004). Exploring Recombinant Flavonoid Biosynthesis in Metabolically Engineered Escherichia coli. Chembiochem 5, 500–507. doi:10.1002/cbic.200300783
Wei, W., Zhang, P., Shang, Y., Zhou, Y., and Ye, B.-C. (2020). Metabolically Engineering of Yarrowia Lipolytica for the Biosynthesis of Naringenin from a Mixture of Glucose and Xylose. Bioresour. Technol. 314, 123726. doi:10.1016/j.biortech.2020.123726
Werner, S. R., and Morgan, J. A. (2010). Controlling Selectivity and Enhancing Yield of Flavonoid Glycosides in Recombinant Yeast. Bioprocess Biosyst. Eng. 33, 863–871. doi:10.1007/s00449-010-0409-7
Werner, S. R., and Morgan, J. A. (2009). Expression of a Dianthus Flavonoid Glucosyltransferase in Saccharomyces cerevisiae for Whole-Cell Biocatalysis. J. Biotechnol. 142, 233–241. doi:10.1016/j.jbiotec.2009.05.008
Winkel, B. S. J. (2006). “The Biosynthesis of Flavonoids,” in The Science of Flavonoids. Editor E. Grotewold (New York, NY: Springer New York), 71–95. doi:10.1007/978-0-387-28822-2_3
Wu, J., Du, G., Chen, J., and Zhou, J. (2015). Enhancing Flavonoid Production by Systematically Tuning the Central Metabolic Pathways Based on a CRISPR Interference System in Escherichia coli. Sci. Rep. 5, 13477. doi:10.1038/srep13477
Wu, J., Du, G., Zhou, J., and Chen, J. (2013). Metabolic Engineering of Escherichia coli for (2S)-Pinocembrin Production from Glucose by a Modular Metabolic Strategy. Metab. Eng. 16, 48–55. doi:10.1016/j.ymben.2012.11.009
Wu, J., Yu, O., Du, G., Zhou, J., and Chen, J. (2014a). Fine-tuning of the Fatty Acid Pathway by Synthetic Antisense RNA for Enhanced (2 S )-Naringenin Production from L -Tyrosine in Escherichia coli. Appl. Environ. Microbiol. 80, 7283–7292. doi:10.1128/aem.02411-14
Wu, J., Zhang, X., Dong, M., and Zhou, J. (2016a). Stepwise Modular Pathway Engineering of Escherichia coli for Efficient One-step Production of (2S)-Pinocembrin. J. Biotechnol. 231, 183–192. doi:10.1016/j.jbiotec.2016.06.007
Wu, J., Zhang, X., Zhou, J., and Dong, M. (2016b). Efficient Biosynthesis of (2S)-Pinocembrin from D-Glucose by Integrating Engineering Central Metabolic Pathways with a pH-Shift Control Strategy. Bioresour. Technol. 218, 999–1007. doi:10.1016/j.biortech.2016.07.066
Wu, J., Zhou, L., Duan, X., Peng, H., Liu, S., Zhuang, Q., et al. (2021). Applied Evolution: Dual Dynamic Regulations-Based Approaches in Engineering Intracellular Malonyl-CoA Availability. Metab. Eng. 67, 403–416. doi:10.1016/j.ymben.2021.08.004
Wu, J., Zhou, T., Du, G., Zhou, J., and Chen, J. (2014b). Modular Optimization of Heterologous Pathways for De Novo Synthesis of (2S)-Naringenin in Escherichia coli. PLoS One 9, e101492. doi:10.1371/journal.pone.0101492
Xiong, D., Lu, S., Wu, J., Liang, C., Wang, W., Wang, W., et al. (2017). Improving Key Enzyme Activity in Phenylpropanoid Pathway with a Designed Biosensor. Metab. Eng. 40, 115–123. doi:10.1016/j.ymben.2017.01.006
Xiu, Y., Jang, S., Jones, J. A., Zill, N. A., Linhardt, R. J., Yuan, Q., et al. (2017). Naringenin-responsive Riboswitch-Based Fluorescent Biosensor Module for Escherichia coli Co-cultures. Biotechnol. Bioeng. 114, 2235–2244. doi:10.1002/bit.26340
Xu, P., Ranganathan, S., Fowler, Z. L., Maranas, C. D., and Koffas, M. A. G. (2011). Genome-scale Metabolic Network Modeling Results in Minimal Interventions that Cooperatively Force Carbon Flux towards Malonyl-CoA. Metab. Eng. 13, 578–587. doi:10.1016/j.ymben.2011.06.008
Xue, Y., Zhang, Y., Cheng, D., Daddy, S., and He, Q. (2014). Genetically Engineering Synechocystis Sp. Pasteur Culture Collection 6803 for the Sustainable Production of the Plant Secondary Metabolite P -coumaric Acid. Proc. Natl. Acad. Sci. U.S.A. 111, 9449–9454. doi:10.1073/pnas.1323725111
Yan, Y., Chemler, J., Huang, L., Martens, S., and Koffas, M. A. G. (2005a). Metabolic Engineering of Anthocyanin Biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 71, 3617–3623. doi:10.1128/aem.71.7.3617-3623.2005
Yan, Y., Kohli, A., and Koffas, M. A. G. (2005b). Biosynthesis of Natural Flavanones in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71, 5610–5613. doi:10.1128/aem.71.9.5610-5613.2005
Yang, D., Kim, W. J., Yoo, S. M., Choi, J. H., Ha, S. H., Lee, M. H., et al. (2018). Repurposing Type III Polyketide Synthase as a Malonyl-CoA Biosensor for Metabolic Engineering in Bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 9835–9844. doi:10.1073/pnas.1808567115
Yang, Y., Lin, Y., Li, L., Linhardt, R. J., and Yan, Y. (2015). Regulating Malonyl-CoA Metabolism via Synthetic Antisense RNAs for Enhanced Biosynthesis of Natural Products. Metab. Eng. 29, 217–226. doi:10.1016/j.ymben.2015.03.018
Zha, J., Zang, Y., Mattozzi, M., Plassmeier, J., Gupta, M., Wu, X., et al. (2018). Metabolic Engineering of Corynebacterium Glutamicum for Anthocyanin Production. Microb. Cell. Fact. 17, 143. doi:10.1186/s12934-018-0990-z
Zhang, Q., Yu, S., Lyu, Y., Zeng, W., and Zhou, J. (2021). Systematically Engineered Fatty Acid Catabolite Pathway for the Production of (2S)-Naringenin in Saccharomyces cerevisiae. ACS Synth. Biol. 10, 1166–1175. doi:10.1021/acssynbio.1c00002
Zhang, W., Liu, H., Li, X., Liu, D., Dong, X.-T., Li, F.-F., et al. (2017). Production of Naringenin from D-Xylose with Co-culture ofE. coliandS. Cerevisiae. Eng. Life Sci. 17, 1021–1029. doi:10.1002/elsc.201700039
Zhao, S., Jones, J. A., Lachance, D. M., Bhan, N., Khalidi, O., Venkataraman, S., et al. (2015). Improvement of Catechin Production in Escherichia coli through Combinatorial Metabolic Engineering. Metab. Eng. 28, 43–53. doi:10.1016/j.ymben.2014.12.002
Zhou, H., Liao, X., Wang, T., Du, G., and Chen, J. (2010). Enhanced L-Phenylalanine Biosynthesis by Co-expression of pheAfbr and aroFwt. Bioresour. Technol. 101, 4151–4156. doi:10.1016/j.biortech.2010.01.043
Zhou, S., Hao, T., and Zhou, J. (2020). Fermentation and Metabolic Pathway Optimization to De Novo Synthesize (2S)-Naringenin in Escherichia coli. J. Microbiol. Biotechnol. 30, 1574–1582. doi:10.4014/jmb.2008.08005
Zhou, S., Lyu, Y., Li, H., Koffas, M. A. G., and Zhou, J. (2019). Fine‐tuning the (2 S )‐naringenin Synthetic Pathway Using an Iterative High‐throughput Balancing Strategy. Biotechnol. Bioeng. 116, 1392–1404. doi:10.1002/bit.26941
Zhou, S., Yuan, S.-F., Nair, P. H., Alper, H. S., Deng, Y., and Zhou, J. (2021). Development of a Growth Coupled and Multi-Layered Dynamic Regulation Network Balancing Malonyl-CoA Node to Enhance (2S)-Naringenin Biosynthesis in Escherichia coli. Metab. Eng. 67, 41–52. doi:10.1016/j.ymben.2021.05.007
Keywords: microbial production, flavonoid, naringenin, metabolic engineering, synthetic biology, directed evolution, biosensor
Citation: Isogai S, Tominaga M, Kondo A and Ishii J (2022) Plant Flavonoid Production in Bacteria and Yeasts. Front. Chem. Eng. 4:880694. doi: 10.3389/fceng.2022.880694
Received: 21 February 2022; Accepted: 09 June 2022;
Published: 08 July 2022.
Edited by:
Shang-Tian Yang, The Ohio State University, United StatesReviewed by:
Jingwen Zhou, Jiangnan University, ChinaYu Wang, Tianjin Institute of Industrial Biotechnology (CAS), China
Copyright © 2022 Isogai, Tominaga, Kondo and Ishii. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Ishii, anVuanVuQHBvcnQua29iZS11LmFjLmpw
†Present address: Shota Isogai, Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, Ikoma, Japan
 Shota Isogai
Shota Isogai Masahiro Tominaga1,3
Masahiro Tominaga1,3 Akihiko Kondo
Akihiko Kondo Jun Ishii
Jun Ishii