
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Chem. Eng. , 19 January 2022
Sec. Biochemical Engineering
Volume 3 - 2021 | https://doi.org/10.3389/fceng.2021.790261
This article is part of the Research Topic Saccharomyces cerevisiae as Model Organism for Biochemical Engineering and Bioprocesses View all 5 articles
Squalene is an important polyunsaturated triterpene with wide applications in the food, cosmetics, and pharmaceutical industries. Currently, the main method for squalene production is extraction from oil-producing plants, but the scale is limited. The microbial fermentation with Saccharomyces cerevisiae still needs improvement to be economically viable. This study aimed to improve squalene production by metabolic engineering and random mutagenesis. First, the mevalonate (MVA) pathway was enhanced, by integrating tHMG1 and IDI1 into multi-copy site Ty2. Subsequently, the ACL gene from Yarrowia lipolytica, encoding citrate lyase was introduced and the β-oxidation pathway was enhanced with multiple copies of key genes. In addition, a high throughput screening strategy based on Nile red staining was established for high squalene-producer screening. After treatment with ARTP mutagenesis, a higher-producing mutant was obtained, with squalene production enhanced by 18.4%. A two-stage fermentation of this mutant in a 5 L bioreactor produced 8.2 g/L of squalene. These findings may facilitate the development of industrial squalene production by fermentation and potentially, other terpenoids.
Squalene is a natural product found in vegetable oils, fish oils, and human sebum. It is the main hydrocarbon component of human epidermal fats, accounting for about 10% by weight. Deep sea shark livers from the South Pacific, around Australia are the main source of squalene at present (Bhattacharjee et al., 2001). Squalene also exists in plant tissues, especially the oil in plant seeds, which is also an important industrial source of squalene (Gutierrez et al., 2014). Squalene is a lipophilic, acyclic triterpenoid compound formed from six isoprene units. It is the precursor of many biologically active compounds, including steroids and hopene (Katabami et al., 2015). Squalene itself also has useful applications, as a moisturizing ingredient in cosmetics (Kohno et al., 1995) and in pharmaceuticals (Reddy and Couvreur, 2009), based on its in anti-tumor (Rao et al., 1998), anti-fungal (Smith, 2000), and cholesterol-lowering (Kelly, 1999) effects. Currently, the main method of squalene production is extraction from plant materials, but the scale is limited by the long growing cycle of the plants (Lou-Bonafonte et al., 2018).
Compared with the conventional way of producing squalene from plant sources, microbial fermentation has the advantages in sustainability and greater environmental friendliness (Kim et al., 2019). For example, high titer squalene was obtained from the fungus, Yarrowia lipolytica by optimizing the fermentation medium (Tang et al., 2021), and 230 mg/L of squalene was obtained from Escherichia coli in flask culture by co-expressing mevalonate pathway with heterogeneous squalene synthase (Katabami et al., 2015). The cyanobacterium, Synechocystis, produced 5.1 mg/L of squalene, after introduction of a green algal squalene synthase (Pattanaik et al., 2020). However, few microorganisms possess a natural pathway for squalene synthesis. Enabling the production of squalene requires introduction of exogenous genes. Recently, the production of squalene with Saccharomyces cerevisiae has received considerable attention, because S. cerevisiae has a natural metabolic pathway for squalene synthesis, i.e., the mevalonate (MVA) pathway. In addition, S. cerevisiae is a well-developed eukaryotic producer microorganism and has been applied to produce many valuable plant natural products, including linalool, artemisinic acid and ginsenoside (Liu et al., 2013; Paramasivan and Mutturi, 2017a; Lian et al., 2018).
Many metabolic engineering strategies have been implemented to optimize squalene production by S. cerevisiae. The MVA pathway has been improved by overexpressing HMG-CoA reductase (HMGR) (Westfall et al., 2012). A truncated HMG-CoA reductase enzyme was overexpressed to eliminate feedback inhibition by ergosterol, (Polakowski et al., 1998). Metabolic flux optimization and NADH regeneration were achieved by co-overexpressing tHMG1 with POS5 (encoding NADH kinase), resulting in a 27.5-fold increase in squalene production (Paramasivan and Mutturi, 2017b). An engineered strain able to use ethanol as carbon source, with increased squalene synthesis, was obtained by overexpressing acetaldehyde dehydrogenase and alcohol dehydrogenase encoded by ADA and ADH2, respectively (Li et al., 2020b). Squalene production was also enhanced by increasing the storage capacity for hydrophobic compounds, by overexpressing DGA1, coding for diacylglycerol acyltransferase (Wei et al., 2018), and by cytoplasmic and peroxisomal engineering (Liu et al., 2020).
Practical industrial squalene production, however, still needs further enhancement of production by S. cerevisiae. Insufficient flux through the MVA pathway is an important limiting factor and the natural biosynthesis rate of the essential precursor, acetyl-CoA is insufficient for high squalene production. In this study, several strategies were implemented to enhance the production of squalene. To reduce the conversion of squalene into downstream compounds by squalene monooxygenase, 13 promoters were tested to replace the native promoter of the squalene monooxygenase gene, ERG1 (PERG1). To enhance the MVA pathway flux, IDI1 (isoprenoid diphosphate isomerase), and tHMG1 (HMG-CoA reductase, HMGR) were integrated at the multi-copy site Ty2 and NADH-HMG1 derived from Silicibacter pomeroyi was introduced. To supply more acetyl-CoA for squalene synthesis in the cytoplasm, TGL3, CIT2, FOX1, FOX2, FOX3, related to β-oxidation in peroxisomes, were overexpressed, and ACL from Y. lipolytica was heterologously expressed (Figure 1). In addition, a high-throughput screening method was established, to efficiently screen for high squalene-producing strains, resulting in a high-producing mutant, obtained by ARTP mutagenesis. The production of squalene was enhanced by 18.4%, permitting 8.2 g/L of squalene to be produced by a two-stage fermentation strategy, in a 5-L bioreactor.
S. cerevisiae C800 (CEN.PK2-1D; MATα; ura3-52; his3Δ1; trp1-289; leu2-3,112; MAL2-8C; SUC2; gal80:KanMX) (Gao et al., 2020) was used for modification. Escherichia coli JM109 was used for routine transformation and plasmid storage. The composition of synthetic complete medium (SC medium) was as follows: Yeast nitrogen base (Sangon Biotech, No. B540132), 20 g/L glucose, and 50 mg/L amino acid. The composition of YPD medium was as follows: 20 g/L glucose, 10 g/L yeast extract, and 20 g/L peptone. Standard squalene was from Sigma-Aldrich (St. Louis, MO, United States). Yeast genome extraction kits were from Takara (Dalian, China). Other chemicals were from Titan tech. (Shanghai, China).
Empty vectors were amplified from pY26 to construct gene expression cassettes. Promoters, terminators, and endogenous genes were all amplified from the genome of CENPK2-1D and assembled by Gibson assembly. The 20 nt of sgRNA plasmids were designed by Benchling (https://benchling.com/) and constructed with PCR and Gibson assembly. Donor DNAs were constructed with no less than each 400 bp upstream and downstream homologous arms. The templates were digested with DpnI. All vectors were cloned in E. coli JM109. DNA polymerase and DNA ligase were purchased from Vazyme (Nanjing, China). Genes were synthesized by GENEWIZ, Inc. (Suzhou, China). All constructed plasmids were sequencing by Saisofi Biotech. (Wuxi, China). Primers were from Sangon and their sequences are shown in Supplementary Table S1.
The yeast strains used in this study are listed in Supplementary Table S2. All plasmids and linearized DNAs were transformed into S. cerevisiae by using the Frozen-EZ-Yeast Transformation Ⅱ Kit™ (Irvine, CA, United States). SC-X solid medium (X represents the amino acids that need to be added) was used to screen transformants. Colony PCR was used to verify positive transformants. To carry out following gene editing, SC-5-FoA plate was used to eliminate sgRNA plasmids.
Thirteen promoters with different strengths were chosen to optimize ERG1 expression levels. The original promoter PERG1 was replaced by PTDH1, PCCW12, PTDH3p, PCIT1p, PPDC1p, PATP3p, PRPS5, PADE6, PARO7, PFAD1, PTRP1, PGPD2, and PHXT1 (listed in strength order from high to low) (Gao et al., 2020). Promoters with homologous arm list above were amplified from plasmids pUC19-x (x represents the promoter name). These PCR products along with sgRNA were then transformed into S. cerevisiae. SC-His-Leu-Trp agar plate was used to screen transformants. These agar plates were cultured at 30°C for 2–4 days and then incubated in liquid medium. After 4 days culture in shake-flasks, the best performing strain was selected as in Extraction and Quantification of Squalene.
Leucine degradation tags were connected to the integrated gene fragments and leucine-deficient YNB plates were used to screen high-copy strains of tHMG1 and IDI1. The first colony on the plate after the transformation was recorded, and 20 fast-growing, large, and round strains were selected for plate fermentation.
Shake flask condition: At least 10 colonies were grown in 50 ml flask containing 5 ml SC-X liquid medium at 30°C, 220 rpm for 24 h, and 2% (v/v) of the culture was reinoculated into 250 ml flask containing 25 ml YPD liquid medium, then cultured at 30°C, 220 rpm for 96 h.
Fed-batch fermentation condition: Bioreactor fermentation was carried out in a 5 L bioreactor containing 2.5 L of the YPD medium (containing 10 g/L CaCO3, which is used for maintaining the pH). The feeding medium was modified as described previously (Li et al., 2020a), including 400 g/L glucose, 10.24 g/L MgSO4·7H2O, 50 g/L (NH4)2SO4, 18 g/L KH2PO4, 0.56 g/L Na2SO4, 7 g/L K2SO4, 20 ml/L vitamin solution, and 24 ml/L trace metal solution. At least 10 colonies were grown in 250 ml flask containing 25 ml SC medium and cultured at 30°C and 220 rpm for 24 h, then the seed solution was reinoculated to a 250 ml flask containing 25 ml SC medium for another 24 h. The seed culture was then inoculated into a 5 L bioreactor containing YPD medium. The temperature was controlled at 30°C and the pH was maintained at 5.5 via automated addition of 50% ammonia. The concentration of dissolved oxygen (DO) was maintained at 50% by adjusting the stirring rate (250–800 rpm) up to 72 h, then at 30% after 72 h. The feeding medium was added at 6 ml/h from 14–24 h, then 8 ml/h from 24–72 h for maintaining the cell growth. In the second stage, the feeding medium was added at 5 ml/h from 72 to 120 h for accumulating squalene.
The harvested yeast cells were washed twice with PBS and diluted to a concentration of OD600 = 1.0. Then each diluted strain (1.0 ml) was stained with 0.5, 1, and 1.5 μg/ml Nile red for 30 min at room temperature, respectively. The Nile Red stock solution was prepared at 1.0 mg/ml in dimethyl sulfoxide (Choi et al., 2018). Stained sample (200 μL) was measured with a fluorescence microplate reader. The greatest fluorescence intensity was achieved at an excitation wavelength of 505 nm and an emission wavelength of 585 nm, so these wavelengths were used for the high-throughput screening. The wild-type strain stained with Nile Red was used as the control.
After fermentation, cell suspension (1.0 ml) was added to a tube of Lysing Matrix C and then centrifuged at 13,000 × g for 5 min. After removing the supernatant, beads and 1.0 ml of acetone were added to the tube. The samples were mechanically disrupted with FastPrep-24 (MP Biomedicals, Irvine, CA, United States) and the squalene acetone mixture was analyzed by HPLC. The HPLC system (Waters, Milford MA, United States) was equipped with a C18 column (30 m × 0.25 mm, 0.25 μm film thickness), maintained at 40°C. The squalene was separated by isocratic elution with acetonitrile (100%) of 1.6 ml/min for 15 min.
CRISPR/cas9-based approach was used to carry out mating-type change from “α” to “a” (Xie et al., 2018). Transformants with switched mating type were identified by colony PCR and sequencing. To realize the construction of diploids, two different mating type strains with different amino acid labels (one with LEU, another with TRP) were mixed in 25 ml of YPD medium in a 250 ml flask and cultured for 60 h at 30°C. SC-URA-HIS plate was used to screen diploids.
13 promoters of different strengths were chosen to replace the native promoter of ERG1 to weaken the expression of ERG1, thereby promoting the accumulation of squalene. All these promoters were chosen from a promoter library and had different translation levels. Most of the promoters with low transcription levels resulted in increased squalene accumulation. When promoter PERG1 was replaced by promoter PHXT1, the titer of squalene was highest from the corresponding engineered strain, reaching 826.5 mg/L at 96 h, 1.94-fold higher than the original strain, CP12, whereas cell growth was significantly lower (Figure 2).
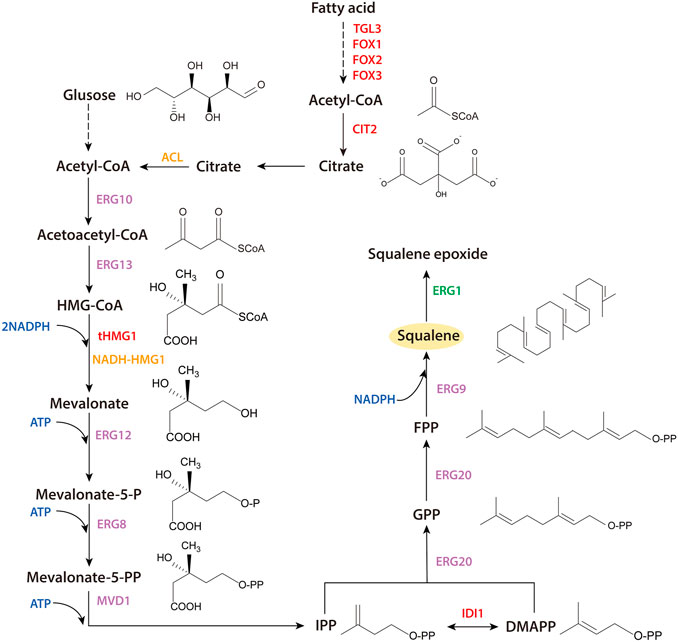
FIGURE 1. Engineered metabolic pathway of squalene production in S. cerevisiae. Heterologous expressed genes are highlighted in orange. Over-expressed genes are highlighted in red; downregulated genes are highlighted in green. HMG-CoA, 3-hydroxyl-3-methyl-glutaryl-CoA; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate.
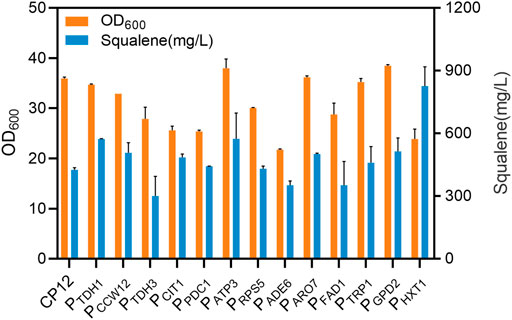
FIGURE 2. Biomass concentrations and squalene contents of constructed strains, resulting from promoter substitution, after 96 h of culture. The promoter of ERG1 (the squalene monooxygenase gene) was replaced by 13 different promoters: PTDH1, PCCW12, PTDH3, PCIT1, PPDC1, PATP3, PRPS5, PADE6, PARO7, PFAD1, PTRP1, PGPD2, and PHXT1. Error bars show SD values.
To investigate the influence of tHMG1 and IDI1 copy number on the enhancement of the MVA pathway, five strains were established with different copy numbers, namely wild-type strains C800, CP02 (1 tHMG1), CP05 (1 tHMG1 and 1 IDI1), CP08 (2 tHMG1 and 2 IDI1), and CP12 (3 tHMG1 and 3 IDI1), respectively. The accumulation of squalene was positively correlated with the copy number of tHMG1 and IDI1 (Figure 3A). Therefore, the multi-copy site Ty2 was selected for integration of tHMG1 and IDI1 to increase their copy numbers. After culturing in 24-well plates for 96 h, the production of squalene was enhanced by all these engineered strains, integrating different copy numbers, of which the highest-yielding strain (SQ10) could accumulate 781.2 mg/L of squalene (Figure 3B). The obtained engineered strain SQ10 could accumulate 703.7 mg/L of squalene for 96 h in shaking flasks, which was superior to CP12. To further improve HMGR activity without breaking of balance of redox status, an HMG-CoA reductase NADH-HMGR with high specificity for NADH from S. pomeroyi was introduced into SQ10 to produce strain SQ11. The titer of squalene with SQ11 was further increased, reaching 787.3 mg/L (Figure 3C).
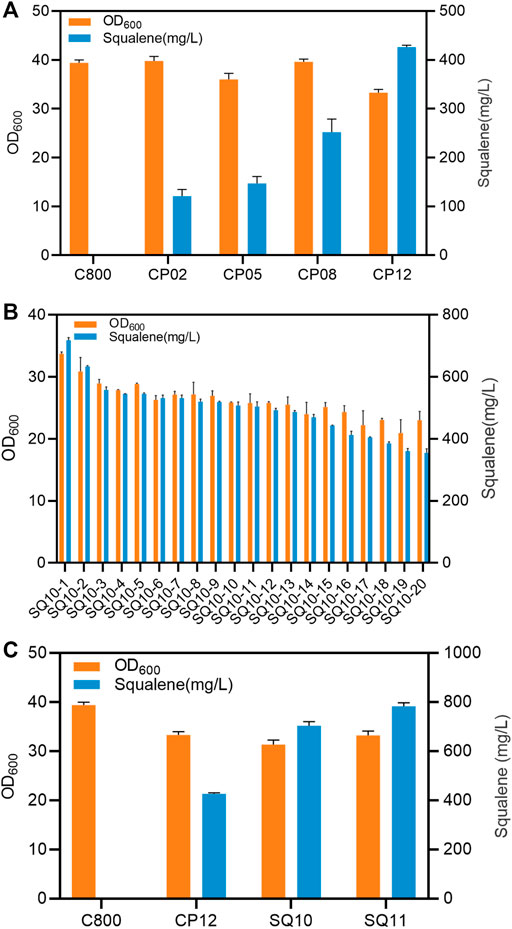
FIGURE 3. The influence of copy numbers of tHMG1/IDI1. (A) Growth and squalene accumulation of strains with different copy numbers of tHMG1 and IDI1 in shake-flasks at 96 h. (B) Growth and squalene accumulation of strains of integrating tHMG1 and IDI1 in multiple copy site Ty2 in 24-well plates at 96 h. (C) Growth and squalene accumulation of different strains in shake flasks at 96 h. Error bars show SD values.
Based on the above findings, a strain (SQ13) was obtained, by replacing the original promoter with PHXT1 and integrating tHMG1 and IDI1 at the Ty2 site. To increase the accumulation of acetyl-CoA in the cytoplasm, citrate lyase (encoded by ACL) from Yarrowia lipolytica was introduced. In addition, TGL3, CIT2, FOX1, FOX2, FOX3, related to β-oxidation in peroxisomes, were also overexpressed in different combinations. The squalene production of strain SQB1 that heterologous expressed ACL was increased by 257 mg/L compared with the original strain SQ13 (Figure 4). Compared with the heterologous expression of ACL alone, the expression of other β-oxidation-related genes had no obvious effect on squalene accumulation, while overexpression of FOX1 could slightly improve the squalene production to 293 mg/L (Figure 4). To enhance the resistance of strains to environmental and metabolic stress, another strain SQ6 (mating type a, PERG1 was replaced by promoter PHXT1) had been constructed. Two haploid strains SQB4 and SQ6, with different mating types, were hybridized, resulting in a diploid strain SQ9. There was a marked improvement in cell growth of SQ9, which was increased by 15.5% compared with that of SQB4.
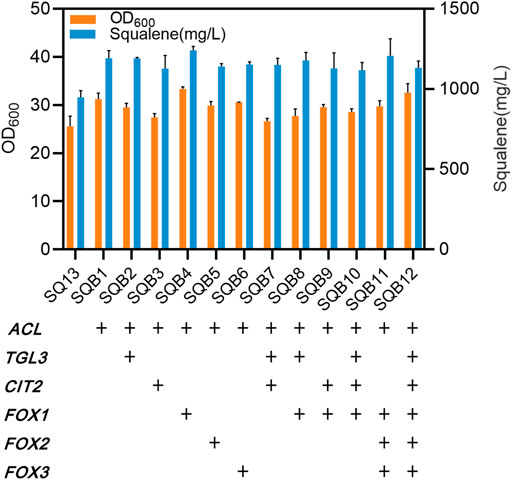
FIGURE 4. Biomass concentrations and squalene contents of engineered strains after 96 h of culture. SQB1-SQB12 were modified from SQ13. + signs signify genes are strengthened. Error bars show SD values.
To establish a method for rapid determination of the intracellular squalene concentration, samples of 3 different strains fermented for 72 h were coped with 3 different concentrations of Nile red (0.5, 1.0, and 1.5 μg/ml) to stain the cells. The fluorescence intensity of the stained cells was determined and correlated with the content of squalene determined by HPLC. A linear relationship between the squalene content and the fluorescence intensity was obtained (Figure 5). The best linear relationship was obtained at a Nile Red concentration of 1.0 μg/ml. The relationship between squalene content and the fluorescence intensity could be expressed as Y = 1057.8X-9945.5 (R2 = 0.9313), indicating that the staining method was suitable for the high-throughput screening procedure. The fatality curve showed that the fatality rate was 80.46% after 75 s of ARTP treatment. The strain SQ9 was treated with three rounds of ARTP mutagenesis and high-throughput screening (Figure 6) identified a mutant strain which accumulated 1470.9 mg/L of squalene, 18.4% higher than the initial strain. The mutant strain was tested for practical squalene production in a 5 L bioreactor, with a two-stage, high density cell fermentation, producing 8.2 g/L of squalene, and a final OD600 of 156.3 (Figure 7).
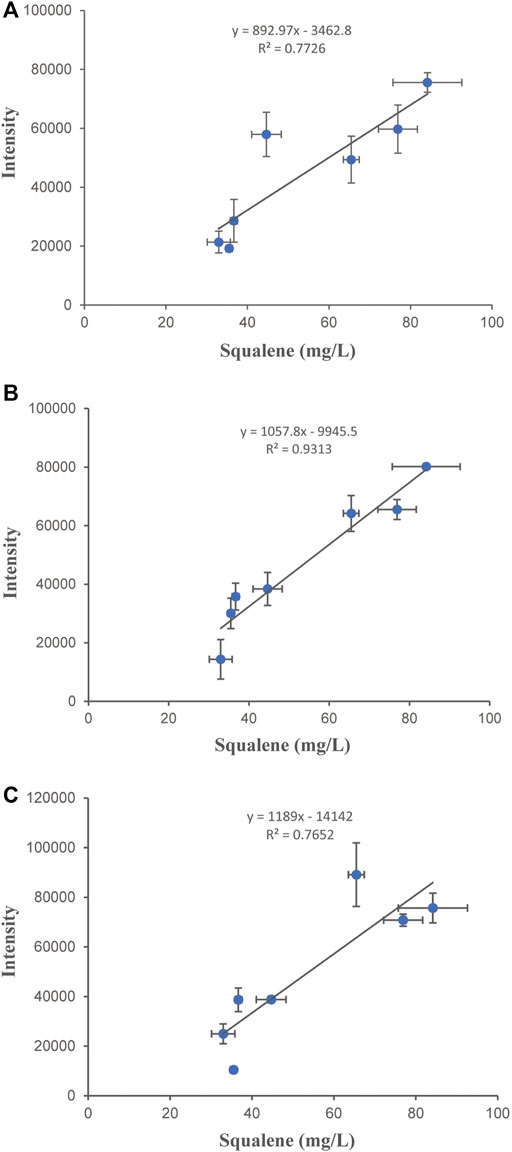
FIGURE 5. Establishment of the high-throughput intracellular squalene assay. Relationship between fluorescence intensity and squalene production at different Nile Red cell-staining concentrations: (A) 0.5 μg/ml; (B) 1 μg/ml; (C) 1.5 μg/ml. Error bars show SD values.
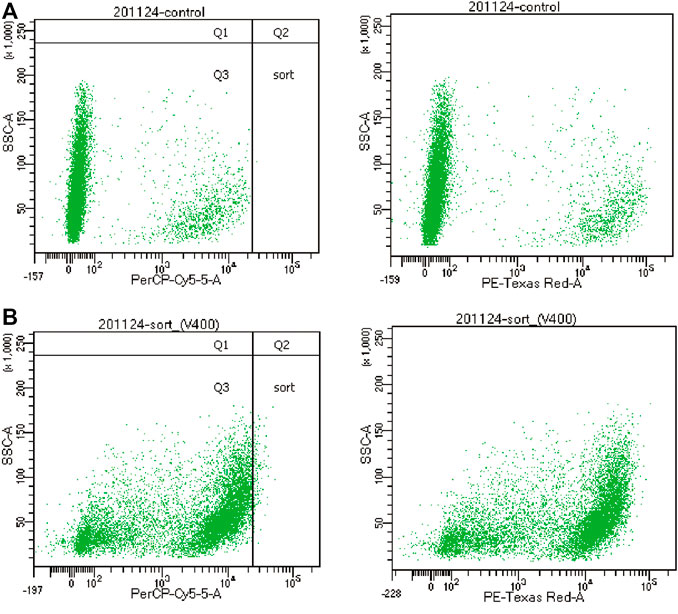
FIGURE 6. Cell populations under different fluorescence intensities of the flow cytometer. (A): Cell grouping under Nile Red staining of non-squalene-producing S. cerevisiae. (B): Cell grouping under Nile Red staining of squalene-producing S. cerevisiae. “Sort” represents the distribution of cells that we need to screen out.
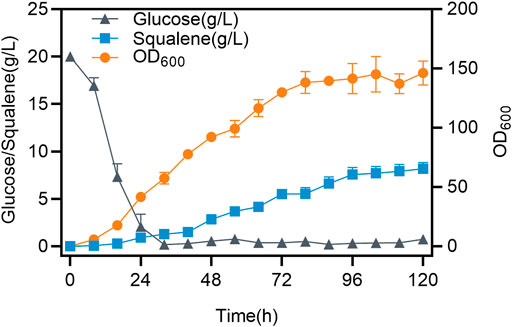
FIGURE 7. Time course of squalene accumulation in a 5-L bioreactor. Time courses showing changes in squalene production and cell growth by strain SQ9 in a 5 L bioreactor. Error bars indicate SD values.
S. cerevisiae is one of the most common microbial hosts for synthetic biology and metabolic engineering. It is a promising platform to produce squalene compared with natural resource extraction, or chemical synthesis (Polakowski et al., 1998; Thompson et al., 2014). In this study, the highest accumulation of squalene was achieved after the native promoter PERG1 was replaced by PHXT1. By randomly integrating tHMG1 and IDI1 into the multi-copy site Ty2, squalene production of 703.7 mg/L was obtained. Squalene production was further increased by 257 mg/L by increasing β-oxidation capacity. A high-throughput screening method was then established for screening high-efficiency squalene producers. After treatment by ARTP mutagenesis, a higher producing mutant strain was obtained, which accumulated 1470.9 mg/L squalene in a shake flask and 8.2 g/L in a 5 L fed-batch fermentation.
Weaker promoters of the squalene monooxygenase gene, ERG1, were tested to lower its expression level and minimize the conversion of squalene into downstream compounds. None of the weaker promoters resulted in significant squalene accumulation except PHXT1, which increased the squalene yield by 94.3% compared with the original strain. This indicated that the inducible promoter PHXT1 was important for regulation of key genes related to yeast growth. PHXT1 can also respond to the concentration of glucose (Ozcan and Johnston, 1995). During the growth stage of yeast, when the glucose concentration in the medium is relatively high, ERG1 is normally expressed, and the synthesis of sterols is sufficient to promote the rapid growth of the yeast cells. When the glucose in the culture medium is exhausted, expression of ERG1 is inhibited, thereby increasing the accumulation of squalene in the yeast cells (Zhao et al., 2017). Many reports have highlighted the importance of tHMG1 and IDI1 in increasing mevalonate (MVA) pathway flux and promoting squalene production (Liu et al., 2013). In this study, making various combinations of different copy numbers of tHMG1 and IDI1 showed that the production of squalene was proportional to their copy numbers. The integration of pathway genes in one or several copies, especially those encoding rate-limiting steps, is usual insufficient to maintain high metabolic fluxes (Maury et al., 2016). In addition, the expression of a multi-copy plasmid is unstable. The Ty2 element, with 34 insertions in the S. cerevisiae genome (Kim et al., 1998), was selected as the integration site for tHMG1 and IDI1.
Acetyl-CoA is a crucial precursor metabolite for terpene biosynthesis (Hellgren et al., 2020). In S. cerevisiae, acetyl-CoA metabolism is complex, because of its compartmentalized biosynthesis pathway and its involvement in various key cellular processes, including the tricarboxylic acid (TCA) cycle, biosynthesis of sterols, fatty acids, and polyketides (Nielsen, 2014). Strategies have been applied to enhance the supply of acetyl-CoA to the MVA pathway, such as introducing heterologous acetyl-CoA synthetase to increase acetyl-CoA (Shiba et al., 2007), removing bypass genes to eliminate competitive products (Lian et al., 2014), and establishing an efficient acetyl-CoA-producing bypass (Shiba et al., 2007; Li et al., 2020b). In addition, acetyl-CoA is a metabolite of fatty acid oxidation. In the peroxisome, it can be converted into citric acid and transported out of the peroxisome, through the glyoxylic acid cycle. The acetyl-CoA is converted to citric acid by citrate synthase (encoded by CIT2) and the resulting citric acid can be transported out of the peroxisomes to the cytoplasm (van Rossum et al., 2016). A heterologous citrate lyase, encoded by ACL1 and ACL2, was introduced, which converted citrate to acetyl-CoA, can promote the supply of acetyl-CoA to the cytoplasm. Genes involved in β-oxidation were also overexpressed. The citric acid generated by β-oxidation could be catalyzed to acetyl-CoA by the heterologous citrate lyase and further promoted squalene production. Furthermore, to enhance the growth of engineered strains, a diploid yeast was hybridized with two haploid, stress tolerant (e.g., to ethanol and starvation) yeasts, SQB4, and SQ6 (Tomova et al., 2019) (Supplementary Table S2).
The content of squalene in microorganisms is usually detected by high-performance liquid chromatography, which complicated and slow, because the cells must be disrupted, and extracted with organic solvent. It is, therefore, not suitable for high-throughput screening of high squalene-yield yeasts. Intracellular squalene is present as lipid droplets, which can be detected by Nile red staining (Greenspan et al., 1985; Chen et al., 2009; Sitepu et al., 2012; Rostron and Lawrence, 2017; Choi et al., 2018). After Nile red staining, the accumulation of squalene and lipid droplets in S. cerevisiae can be observed with a fluorescence microscope (Ta et al., 2012). This facilitated the establishment of a high-throughput screen, based on fluorescent Nile red staining of S. cerevisiae. The quantitative correlation between fluorescence intensity and squalene concentration from Nile red staining was analyzed with numerous cells of different squalene productivity. A high-throughput screening method was established, with a good linear relationship between squalene production and fluorescence intensity, by optimizing the concentration of Nile Red. Combined with the ARTP mutagenesis strategy and flow cytometry sorting technology (Figure 6), the screening efficiency was enhanced and a mutant strain was obtained, with squalene production increased by 18.4%.
Compared with previous reports, it appeared that the use of multi-copy site integration was an efficient way to increase MVA pathway flux and squalene production. However, the target sites for genomic integration have not been identified and there is a risk of DNA loss (Maury et al., 2016). A more accurate screening approach should be used after multi-copy site integration, such as whole genome sequencing. Furthermore, to promote the utilization of citric acid from β-oxidation, the glyoxylic acid cycle, which may be a competitive pathway, should be regulated to enhance squalene production. Except strengthening the supply of acetyl-CoA, overproduction of squalene can also be achieved by introducing a partial MVA pathway from acetyl-CoA to mevalonate in mitochondria (Zhu et al., 2021). It provides a new insight into the production of squalene and other terpenes in yeasts. Squalene distribution in yeast cells was visualized by Nile Red staining in this study, dense and large lipid droplets, spread over the whole cell were visualized by laser scanning confocal microscopy. Transporting squalene to the extracellular space appeared to be the main impediment to increased squalene accumulation. However, there are few reports on the transport of terpenoids in microorganisms, particularly hydrophobic terpenoids like squalene. The proteins most reported as being related to the transport of terpenoids are mainly ABC transporters, like SpTUR2 (Spirodela polyrrhiza) for the diterpene sclareol (Stukkens et al., 2005) and Snq2p for β-carotene (Sun et al., 2020). These findings indicated that the production of squalene could be further increased by discovering potential new transporters. Metabolic engineering has great potential to be applied to various cell factories to enhance the secretion of squalene.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
LX, WZ, and JZ designed the study and wrote the article. LX, WZ, and JZ critically revised the article. LX, YL, SL, SY, WZ, and JZ performed the experiments and analyzed the results. WZ and JZ designed and supervised the project. All authors discussed the results and commented on the article.
This work was supported by the National Key Research and Development Program of China (2019YFA0905300), the National Outstanding Youth Foundation (21822806), and the Qingdao Biomanufacturing Industry Joint Fund of Think Tank (QDSWZK202110).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceng.2021.790261/full#supplementary-material
Supplementary Table 1 | Primers used in this study.
Supplementary Table 2 | Plasmids and strains used in this study.
Bhattacharjee, P., Shukla, V. B., Singhal, R. S., and Kulkarni, P. R. (2001). Studies on Fermentative Production of Squalene. World J. Microbiol. Biotechnol. 17, 811–816. doi:10.1023/A:1013573912952
Chen, W., Zhang, C., Song, L., Sommerfeld, M., and Hu, Q. (2009). A High Throughput Nile Red Method for Quantitative Measurement of Neutral Lipids in Microalgae. J. Microbiol. Methods 77, 41–47. doi:10.1016/j.mimet.2009.01.001
Choi, S. Y., Sim, S. J., Choi, J.-I., and Woo, H. M. (2018). Identification of Small Droplets of Photosynthetic Squalene in Engineered Synechococcus Elongatus PCC 7942 Using TEM and Selective Fluorescent Nile Red Analysis. Lett. Appl. Microbiol. 66, 523–529. doi:10.1111/lam.12874
Entian, K.-D., and Kötter, P. (2007). “25 Yeast Genetic Strain and Plasmid Collections,” in Yeast Genetic Strain and Plasmid Collections. Yeast Gene Analysis. Editors I. Stansfield, and M.J.R. Stark. Second Edition (Elsevier), 629–666. doi:10.1016/s0580-9517(06)36025-4
Gao, S., Zhou, H., Zhou, J., and Chen, J. (2020). Promoter-Library-Based Pathway Optimization for Efficient (2S)-Naringenin Production from P-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 68, 6884–6891. doi:10.1021/acs.jafc.0c01130
Greenspan, P., Mayer, E. P., and Fowler, S. D. (1985). Nile Red: a Selective Fluorescent Stain for Intracellular Lipid Droplets. J. Cel Biol. 100, 965–973. doi:10.1083/jcb.100.3.965
Gutiérrez, M. C., Siles, J. A., Chica, A. F., and Martín, M. A. (2014). Kinetics of Biofuel Generation from Deodorizer Distillates Derived from the Physical Refining of Olive Oil and Squalene Recovery. Biomass Bioenergy 62, 93–99. doi:10.1016/j.biombioe.2014.01.010
Hellgren, J., Godina, A., Nielsen, J., and Siewers, V. (2020). Promiscuous Phosphoketolase and Metabolic Rewiring Enables Novel Non-oxidative Glycolysis in Yeast for High-Yield Production of Acetyl-CoA Derived Products. Metab. Eng. 62, 150–160. doi:10.1016/j.ymben.2020.09.003
Katabami, A., Li, L., Iwasaki, M., Furubayashi, M., Saito, K., and Umeno, D. (2015). Production of Squalene by Squalene Synthases and Their Truncated Mutants in Escherichia coli. J. Biosci. Bioeng. 119, 165–171. doi:10.1016/j.jbiosc.2014.07.013
Kim, J. M., Vanguri, S., Boeke, J. D., Gabriel, A., and Voytas, D. F. (1998). Transposable Elements and Genome Organization: A Comprehensive Survey of Retrotransposons Revealed by the Complete Saccharomyces cerevisiae Genome Sequence. Genome Res. 8, 464–478. doi:10.1101/gr.8.5.464
Kim, J.-E., Jang, I.-S., Son, S.-H., Ko, Y.-J., Cho, B.-K., Kim, S. C., et al. (2019). Tailoring the Saccharomyces cerevisiae Endoplasmic Reticulum for Functional Assembly of Terpene Synthesis Pathway. Metab. Eng. 56, 50–59. doi:10.1016/j.ymben.2019.08.013
Kohno, Y., Egawa, Y., Itoh, S., Nagaoka, S.-i., Takahashi, M., and Mukai, K. (1995). Kinetic Study of Quenching Reaction of Singlet Oxygen and Scavenging Reaction of Free Radical by Squalene in N-Butanol. Biochim. Biophys. Acta (Bba) - Lipids Lipid Metab. 1256, 52–56. doi:10.1016/0005-2760(95)00005-w
Li, G., Li, H., Lyu, Y., Zeng, W., and Zhou, J. (2020a). Enhanced Biosynthesis of Dihydromyricetin in Saccharomyces cerevisiae by Coexpression of Multiple Hydroxylases. J. Agric. Food Chem. 68, 14221–14229. doi:10.1021/acs.jafc.0c05261
Li, T., Liu, G.-S., Zhou, W., Jiang, M., Ren, Y.-H., Tao, X.-Y., et al. (2020b). Metabolic Engineering of Saccharomyces cerevisiae to Overproduce Squalene. J. Agric. Food Chem. 68, 2132–2138. doi:10.1021/acs.jafc.9b07419
Lian, J., Si, T., Nair, N. U., and Zhao, H. (2014). Design and Construction of Acetyl-CoA Overproducing Saccharomyces cerevisiae Strains. Metab. Eng. 24, 139–149. doi:10.1016/j.ymben.2014.05.010
Lian, J., Mishra, S., and Zhao, H. (2018). Recent Advances in Metabolic Engineering of Saccharomyces cerevisiae: New Tools and Their Applications. Metab. Eng. 50, 85–108. doi:10.1016/j.ymben.2018.04.011
Liu, J., Zhang, W., Du, G., Chen, J., and Zhou, J. (2013). Overproduction of Geraniol by Enhanced Precursor Supply in Saccharomyces cerevisiae. J. Biotechnol. 168, 446–451. doi:10.1016/j.jbiotec.2013.10.017
Liu, G.-S., Li, T., Zhou, W., Jiang, M., Tao, X.-Y., Liu, M., et al. (2020). The Yeast Peroxisome: A Dynamic Storage Depot and Subcellular Factory for Squalene Overproduction. Metab. Eng. 57, 151–161. doi:10.1016/j.ymben.2019.11.001
Lou-Bonafonte, J. M., Martínez-Beamonte, R., Sanclemente, T., Surra, J. C., Herrera-Marcos, L. V., Sanchez-Marco, J., et al. (2018). Current Insights into the Biological Action of Squalene. Mol. Nutr. Food Res. 62, 1800136. doi:10.1002/mnfr.201800136
Maury, J., Germann, S. M., Baallal Jacobsen, S. A., Jensen, N. B., Kildegaard, K. R., Herrgård, M. J., et al. (2016). EasyCloneMulti: A Set of Vectors for Simultaneous and Multiple Genomic Integrations in Saccharomyces cerevisiae. PLoS One 11, e0150394. doi:10.1371/journal.pone.0150394
Nielsen, J. (2014). Synthetic Biology for Engineering Acetyl Coenzyme A Metabolism in Yeast. mBio 5, 2153. doi:10.1128/mBio.02153-14
Ozcan, S., and Johnston, M. (1995). Three Different Regulatory Mechanisms Enable Yeast Hexose Transporter (HXT) Genes to Be Induced by Different Levels of Glucose. Mol. Cel. Biol. 15, 1564–1572. doi:10.1128/MCB.15.3.1564
Paramasivan, K., and Mutturi, S. (2017a). Progress in Terpene Synthesis Strategies through Engineering of Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 37, 974–989. doi:10.1080/07388551.2017.1299679
Paramasivan, K., and Mutturi, S. (2017b). Regeneration of NADPH Coupled with HMG-CoA Reductase Activity Increases Squalene Synthesis in Saccharomyces cerevisiae. J. Agric. Food Chem. 65, 8162–8170. doi:10.1021/acs.jafc.7b02945
Pattanaik, B., Englund, E., Nolte, N., and Lindberg, P. (2020). Introduction of a Green Algal Squalene Synthase Enhances Squalene Accumulation in a Strain of Synechocystis Sp. PCC 6803. Metab. Eng. Commun. 10, e00125. doi:10.1016/j.mec.2020.e00125
Polakowski, T., Stahl, U., and Lang, C. (1998). Overexpression of a Cytosolic Hydroxymethylglutaryl-CoA Reductase Leads to Squalene Accumulation in Yeast. Appl. Microbiol. Biotechnol. 49, 66–71. doi:10.1007/s002530051138
Rao, C., Newmark, H. L., and Reddy, B. S. (1998). Chemopreventive Effect of Squalene on Colon Cancer. Carcinogenesis 19, 287–290. doi:10.1093/carcin/19.2.287
Reddy, L. H., and Couvreur, P. (2009). Squalene: A Natural Triterpene for Use in Disease Management and Therapy. Adv. Drug Deliv. Rev. 61, 1412–1426. doi:10.1016/j.addr.2009.09.005
Rostron, K. A., and Lawrence, C. L. (2017). Nile Red Staining of Neutral Lipids in Yeast. Methods Mol. Biol. 1560, 219–229. doi:10.1007/978-1-4939-6788-9_16
Shiba, Y., Paradise, E. M., Kirby, J., Ro, D.-K., and Keasling, J. D. (2007). Engineering of the Pyruvate Dehydrogenase Bypass in Saccharomyces cerevisiae for High-Level Production of Isoprenoids. Metab. Eng. 9, 160–168. doi:10.1016/j.ymben.2006.10.005
Sitepu, I. R., Ignatia, L., Franz, A. K., Wong, D. M., Faulina, S. A., Tsui, M., et al. (2012). An Improved High-Throughput Nile Red Fluorescence Assay for Estimating Intracellular Lipids in a Variety of Yeast Species. J. Microbiol. Methods 91, 321–328. doi:10.1016/j.mimet.2012.09.001
Smith, T. J. (2000). Squalene: Potential Chemopreventive Agent. Expert Opin. Investig. Drugs 9, 1841–1848. doi:10.1517/13543784.9.8.1841
Stukkens, Y., Bultreys, A., Grec, S., Trombik, T., Vanham, D., and Boutry, M. (2005). NpPDR1, a Pleiotropic Drug Resistance-type ATP-Binding Cassette Transporter from Nicotiana Plumbaginifolia, Plays a Major Role in Plant Pathogen Defense. Plant Physiol. 139, 341–352. doi:10.1104/pp.105.062372
Sun, L., Atkinson, C. A., Lee, Y. G., and Jin, Y. S. (2020). High‐level β‐carotene Production from Xylose by Engineered Saccharomyces cerevisiae without Overexpression of a Truncated HMG1 (T HMG1 ). Biotechnol. Bioeng. 117, 3522–3532. doi:10.1002/bit.27508
Ta, M. T., Kapterian, T. S., Fei, W., Du, X., Brown, A. J., Dawes, I. W., et al. (2012). Accumulation of Squalene Is Associated with the Clustering of Lipid Droplets. FEBS J. 279, 4231–4244. doi:10.1111/febs.12015
Tang, W.-Y., Wang, D.-P., Tian, Y., Fan, X., Wang, C., Lu, X.-Y., et al. (2021). Metabolic Engineering of Yarrowia Lipolytica for Improving Squalene Production. Bioresour. Technol. 323, 124652. doi:10.1016/j.biortech.2020.124652
Thompson, A., Kwak, S., and Jin, Y. S. (2014). Squalene Production Using Saccharomyces cerevisiae. i-ACES 1, 1–2.
Tomova, A. A., Kujumdzieva, A. V., and Petrova, V. Y. (2019). Carbon Source influencesSaccharomyces Cerevisiaeyeast Cell Survival Strategies: Quiescence or Sporulation. Biotechnol. Biotechnol. Equip. 33, 1464–1470. doi:10.1080/13102818.2019.1674188
van Rossum, H. M., Kozak, B. U., Niemeijer, M. S., Duine, H. J., Luttik, M. A. H., Boer, V. M., et al. (2016). Alternative Reactions at the Interface of Glycolysis and Citric Acid Cycle inSaccharomyces Cerevisiae. FEMS Yeast Res. 16, fow017–21. doi:10.1093/femsyr/fow017
Wei, L.-J., Kwak, S., Liu, J.-J., Lane, S., Hua, Q., Kweon, D.-H., et al. (2018). Improved Squalene Production through Increasing Lipid Contents inSaccharomyces Cerevisiae. Biotechnol. Bioeng. 115, 1793–1800. doi:10.1002/bit.26595
Westfall, P. J., Pitera, D. J., Lenihan, J. R., Eng, D., Woolard, F. X., Regentin, R., et al. (2012). Production of Amorphadiene in Yeast, and its Conversion to Dihydroartemisinic Acid, Precursor to the Antimalarial Agent Artemisinin. Proc. Natl. Acad. Sci. 109, E111–E118. doi:10.1073/pnas.1110740109
Xie, Z.-X., Mitchell, L. A., Liu, H.-M., Li, B.-Z., Liu, D., Agmon, N., et al. (2018). Rapid and Efficient CRISPR/Cas9-Based Mating-type Switching of Saccharomyces cerevisiae. G3 (Bethesda) 8, 173–183. doi:10.1534/g3.117.300347
Zhao, J., Li, C., Zhang, Y., Shen, Y., Hou, J., and Bao, X. (2017). Dynamic Control of ERG20 Expression Combined with Minimized Endogenous Downstream Metabolism Contributes to the Improvement of Geraniol Production in Saccharomyces cerevisiae. Microb. Cel. Fact. 16, 17. doi:10.1186/s12934-017-0641-9
Keywords: Saccharomyces cerevisiae, promotor substitution, multi-copy site integration, high-throughput screening, β-oxidation
Citation: Xia L, Lv Y, Liu S, Yu S, Zeng W and Zhou J (2022) Enhancing Squalene Production in Saccharomyces cerevisiae by Metabolic Engineering and Random Mutagenesis. Front. Chem. Eng. 3:790261. doi: 10.3389/fceng.2021.790261
Received: 06 October 2021; Accepted: 20 December 2021;
Published: 19 January 2022.
Edited by:
Zongbao K. Zhao, Dalian Institute of Chemical Physics (CAS), ChinaReviewed by:
Hui Wu, East China University of Science and Technology, ChinaCopyright © 2022 Xia, Lv, Liu, Yu, Zeng and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weizhu Zeng, end6ZW5nQGppYW5nbmFuLmVkdS5jbg==; Jingwen Zhou, emhvdWp3MTk4MkBqaWFuZ25hbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.