- 1Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, Provincial Key Laboratory of Biotechnology of Shaanxi Province, The College of Life Sciences, Northwest University, Xi’an, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3BGI College and Henan Institute of Medical and Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, China
- 4Department of Burn Surgery, The First Affiliated Hospital of Naval Medical University, Shanghai, China
- 5BGI Research, Hangzhou, China
- 6BGI Research, Shenzhen, China
Burn injuries often leave behind a “stasis zone”, a region of tissue critically important for determining both the severity of the injury and the potential for recovery. To understand the intricate cellular and epigenetic changes occurring within this critical zone, we utilized single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) to profile over 31,500 cells from both healthy rat skin and the stasis zone at nine different time points after a burn injury. This comprehensive approach revealed 26 distinct cell types and the dynamic shifts in the proportions of these cell types over time. We observed distinct gene activation patterns in different cell types at various stages post-burn, highlighting key players in immune activation, tissue regeneration, and blood vessel repair. Importantly, our analysis uncovered the regulatory networks governing these genes, offering valuable insights into the intricate mechanisms orchestrating burn wound healing. This comprehensive cellular and molecular atlas of the stasis zone provides a powerful resource for developing targeted therapies aimed at improving burn injury recovery and minimizing long-term consequences.
1 Introduction
Burns are a significant global health issue that not only affect patients’ physiological functions but also lead to long-term psychosocial burdens (Jeschke and Herndon, 2014; Radzikowska-Büchner et al., 2023). The repair of skin damage following a burn is a complex process involving interactions among various cell types and molecular signaling pathways (Roshangar et al., 2019). Histologically, the burn area can be divided into three distinct zones: the necrotic zone, which is directly and irreversibly damaged by high temperatures; the stasis zone, which is damaged but has the potential for recovery; and the surrounding normal tissue (Tiwari, 2012; Burgess et al., 2022), which includes the epidermis, dermis, adipose tissue, and muscle layers (Malhotra et al., 2024). The stasis zone is crucial for burn treatment as it represents the transitional area between the wound margin and healthy tissue. With appropriate interventions, this tissue has the potential to be salvaged, thus facilitating the overall healing process (Guo et al., 2021; Lu et al., 2022).
Research indicates that burns heal through epithelial regeneration (Li B. et al., 2021; Boyce and Kagan, 2023), progressing through four stages, hemostasis, inflammation, proliferation, and remodeling (G El Baassiri et al., 2023; Siddique et al., 2023). The management and protection of the stasis zone directly influence inflammatory responses, tissue regeneration, and scar formation during these stages (Guo et al., 2015). First is the early stage after burn, a rapid inflammatory response occurs in the stasis zone, with elevated levels of cytokines such as TNF-α and the IL-1 family in the body. The main cell types involved include monocytes, macrophages, and mast cells, which function in hemostasis, pathogen defense, and necrotic tissue clearance, laying the groundwork for the subsequent proliferation and remodeling phases to promote wound repair (Eming et al., 2007; Burgess et al., 2022). Therefore, the inflammatory response is crucial for balancing apoptosis and proliferation (Ren et al., 2022). As the burn enters the proliferation and remodeling phases, the survival capacity of endothelial cells and fibroblasts significantly increases, primarily to promote angiogenesis and collagen deposition (Huang et al., 2015; Shi et al., 2023). With the formation of granulation tissue at the wound site, epidermal basal cells demonstrate the ability to exceed their self-renewal capacity, gradually differentiating and migrating to the site of injury (Gross-Amat et al., 2020). This process is influenced by growth factors (such as EGF, TGF-β, VEGF, and FGF), extracellular matrix components, moisture, and skin appendages (Burgess et al., 2022; Fan et al., 2024). Migrating epithelial cells also facilitate the formation of hair follicles and sweat glands. Ultimately, the proliferative migratory basal cells generate new epithelial cells, restoring the skin’s barrier function after a burn (Sukmawati et al., 2020). Reports indicate that skin injury induces significant changes in the epigenetic mechanisms of the skin, establishing unique epigenetic demands that influence inflammation and tissue repair. Specifically, this involves the production of damage-stress cytokines via epigenetic alterations during inflammation, and the recruitment and differentiation of resident stem cells following skin injury through epigenetic modifications, such as histone methylation and acetylation during re-epithelialization (Plikus et al., 2015; Ran et al., 2023; Rolim et al., 2024). These findings emphasize the complex physiological and biochemical processes involved in burn healing and reveal the importance of chromatin accessibility in regulating the rapid cellular response to injury (Lander et al., 2017; Nascimento-Filho et al., 2020; Cheng et al., 2024). ScATAC-seq data hold promise for providing a more detailed understanding of the role of the stasis zone in burn healing. However, there is still a lack of comprehensive datasets for rat burn.
Our study aims to fill this gap by providing an extensive scATAC-seq atlas of chromatin accessibility in rat skin at various time points following burn injuries. We analyzed 31,503 cells from normal tissue (ctrl) and the burn stasis zone at 0 h (0 h), 12 h (12 h), Day 1 (D1), Day 3 (D3), Day 7 (D7), Day 11 (D11), Day 15 (D15), and Day 19 (D19) post-burn, identifying 26 distinct cell types and their associated regulatory elements. This detailed characterization of chromatin accessibility and cell type-specific gene score will enhance our understanding of the molecular mechanisms underlying burn wound healing and provide valuable resources for future research in mammalian skin regeneration.
2 Materials and methods
2.1 Tissue collection
This study was approved by the Affiliated Hospital of the Naval Medical University (license number: SYXK (Hu) 2020–0033) and the BGI Technology Ethics Committee (approval number: BGI-IRB A23034). Experiments were conducted on SPF-grade SD male rats, 8 weeks old. A total of 9 rats were used in the study. In the modeling experiment, a 1 × 2 cm sponge was used to uniformly create 8 burn wounds. At each time point, the stasis zone of two wounds from one rat was collected for scATAC-seq experiments (Figure 1B). The specific modeling steps are as follows: Rats were depilated and prepared the day before the experiment to fully expose the back area. Following anesthesia, the rats were secured on a heated pad. A sponge pad, was heated in boiling water for 10 min, reaching approximately 100°C. The sponge was quickly dried and applied evenly to the rat’s back, ensuring even heating. The burn wound was then covered with gauze and bandaged. Tissue samples from the normal tissue (ctrl) and burn stasis zone were collected at 0h, 12h, D1, D3, D7, D11, D15, and D19 post-burn, preserved in a tissue storage solution, and processed within 48 h (Figures 1A, B).
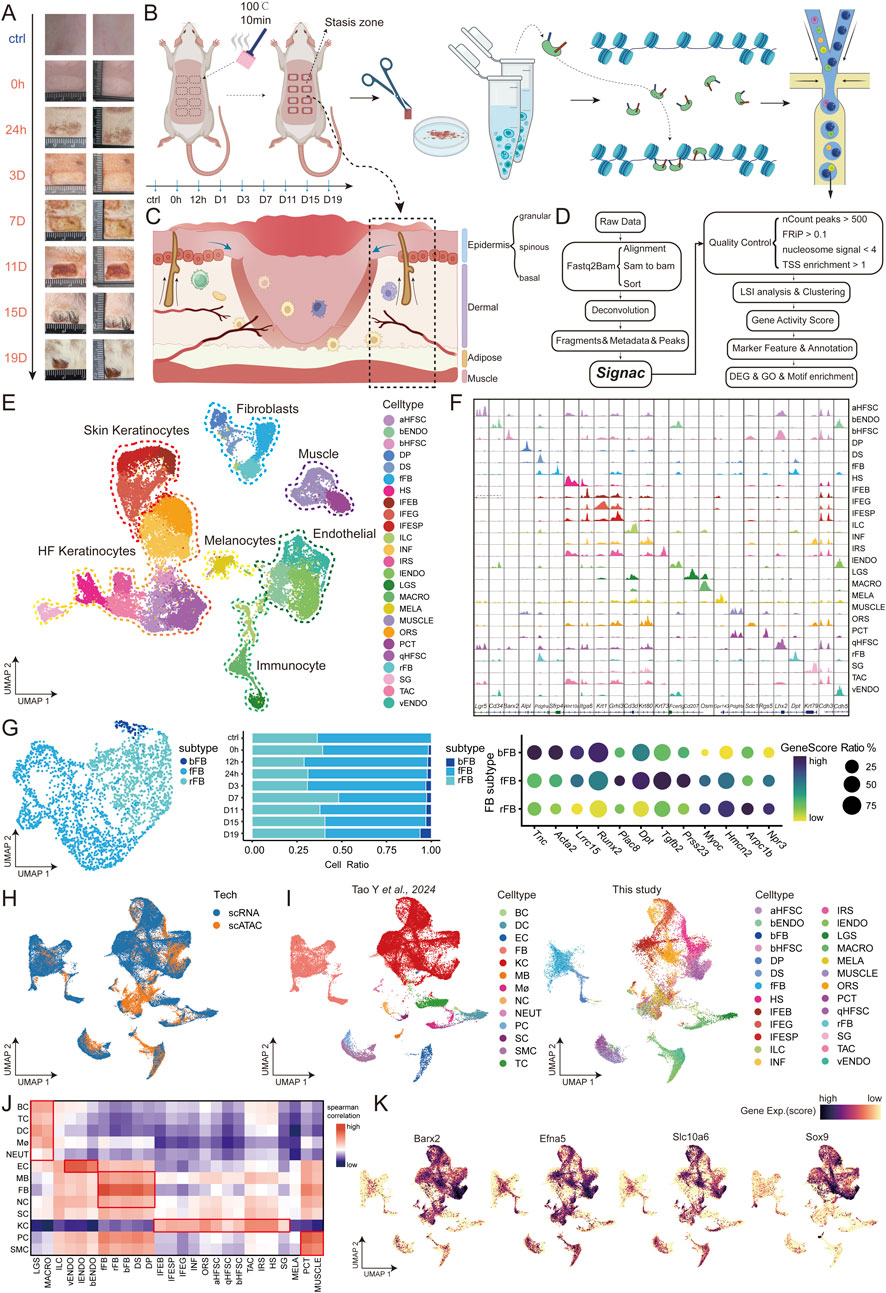
Figure 1. Identification of scATAC data cell types in burn stasis area of rats (A) Images of rat burn samples at each time point. (B) Experimental workflow: Rat skin samples were collected at nine time points (ctrl, 0h, 12h, D1, D3, D7, D11, D15, D19) for scATAC-seq profiling. (C) Diagram of skin burn pattern, including the structure and state of the necrotic area, the stasis area, and the normal skin. (D) The analysis workflow for scATAC-seq profiles. (E) UMAP visualization of 26 cell types identified by gene score in scATAC-seq data. aHFSC, active hair follicle stem cell; bENDO, burn specific endothelial; bHFSC, burn specific hair follicle stem cells; DP, dermal papilla; DS, dermal sheath; fFB, fascial fibroblast; HS, hair shaft; IFEB, interfollicular epidermis basal; IFEG, interfollicular epidermis granular; IFESP, interfollicular epidermis spinous; ILC, innate lymphoid cell; INF, infundibular; IRS, Inner root sheath; lENDO, lymphatic endothelial; LGS, langerhans; MACRO, macrophage; MELA, melanocyte; MUSCLE, muscle; ORS, outter root sheath; PCT, pericyte; qHFSC, quiescent hair follicle stem cell; rFB, reticular fibroblast; SG, sebaceous gland; TAC, transit amplifying cell; vENDO, vascular endothelium. (F) Cell type-specific genome browser views of scATAC-seq signal for the dynamically identified peaks. (G) Subpopulations of skin fibroblasts, marker genes, and their proportions over time. (H) UMAP shows cell clusters from two datasets. (I) Integration of scRNA data from rat skin with radiation injury by Tao Y et al. and distribution of cell types in the two datasets. (J) Correlation analysis between cell types in this study and public datasets, with red indicating high correlation and blue indicating low correlation. (K) FeaturePlot of marker gene expression for bHFSC in the integrated dataset, with darker colors indicating higher expression.
2.2 Dissociation of skin tissue
Skin tissues were placed in PBS (GIBCO), washed, and transferred to a 2 mL tube with 1 mL of enzyme 1 (100 × P/S (XP Biomed), 0.25% trypsin (GIBCO)). The tissue was finely minced with scissors, transferred to a 50 mL tube, and washed with enzyme solution 1. The solution was sealed, placed in a PE glove to prevent leaks, and incubated at 37°C with agitation at 130 rpm for 25 min. The solution was then centrifuged at 500 g for 5 min, and the supernatant was carefully removed. Next, 10 mL of enzyme solution 2 (DMEM/F-12 (GIBCO), collagenase IV(BBI), collagenase I (SANGON), dispase II (SIGMA), P/S) was added, mixed, and incubated under the same conditions for 60 min. The cell suspension was filtered through a 70 μm filter, washed with DPBS containing 2% FBS, and filtered again using a 30 μm filter. Cells were centrifuged at 300 g for 5 min at 4°C, resuspended in PBS (0.04% BSA (SIGMA)), and assessed for viability using trypan blue staining (INVITROGEN) dye with a Countstar cell analyzer.
2.3 scATAC-seq library construction and sequencing
For scATAC-seq library preparation, we used the DNBelab C4 (Yu et al., 2021) Series Single-Cell ATAC Library Prep Set (MGI, #1000012554) to generate 17 libraries across 9 time points, with 2 libraries for each post-burn time point and 1 control. The process involved cell fixation, nuclear extraction, nuclear counting, transposition, droplet generation, emulsion breaking, PCR amplification, purification, fragment selection, circular library construction, and sequencing on the DNBSEQ-T1 platform with read lengths of 115 bp for read 1, 69 bp for read 2, and 10 bp for the sample index sequencing scheme of the China National GeneBank (CNGB) (Huang et al., 2017).
2.4 scATAC-seq raw data processing
The scATAC-seq data processing followed these steps: raw reads were demultiplexed into insertions and barcodes, then filtered using PISA (v 1.1) with a minimum sequencing quality of 20. The filtered reads were aligned to the rat genome using BWA (v 0.7.17-r1188) (Li and Durbin, 2009), and BAM files were further processed with bap2 (v 0.6.2) (Lareau et al., 2019) to assign barcodes to individual cells.
2.5 Quality control (QC) of the scATAC-seq downstream analysis
Low-quality cells were filtered using Signac (v1.6.0) (Stuart et al., 2021) with the following criteria: nCount peaks >500, fraction of reads in peaks (FRiP) > 0.1, nucleosome signal <4, and transcription start site (TSS) enrichment >1 (Figure 1D). Doublets were identified and removed using the scDblFinder (v1.12.0) package.
2.6 Integration and clustering of scATAC-seq data
In Signac (Stuart et al., 2021), use the reduce function to merge multiome peaks sets from scATAC-seq libraries. LSI dimensionality reduction was performed with the RunSVD function, and sample integration was done using the IntegrateEmbeddings with Canonical Correlation Analysis (CCA). Clustering was carried out with FindClusters function at a resolution of 0.7, and cluster-specific peaks were identified using the FindAllMarkers function.
2.7 scATAC-seq gene activity scores
Gene activity scores were computed using Signac’s GeneActivity function, with the RNA matrix created using the CreateAssayObject function. Due to the sparsity of scATAC-seq data, we employed MAGIC (Markov Affinity-based Graph Imputation of Cells) (van Dijk et al., 2018) for data smoothing and imputation by constructing an affinity graph and applying Markov diffusion.
2.8 Gene ontology (GO) enrichment analysis
GO enrichment analysis was conducted on gene sets for each cell type using the enrichGO function in the clusterProfiler (v 4.6.0) (parameters: OrgDb = org. Rn.e.g.,.db, ont = ‘ALL’) (Wu et al., 2021). Input genes (p < 0.01, log2FC > 0.5) were obtained from the gene activity matrix using FindAllMarkers.
2.9 Motif enrichment analysis
Motif enrichment was performed with Signac’s getMatrixSet function (tax_group = “vertebrates”) to retrieve motif position frequency matrices. The AddMotifs function (genome = “BSgenome.Rnorvegicus.UCSC.rn7”) was used to add motif information, and a heatmap was generated to display the top 4 of cell type-specific motifs.
2.10 Soft clustering of peaks varying with burn time points
To perform clustering analysis using Mfuzz (v 2.58.0) (Kumar and E Futschik, 2007), first input the peak matrix, where rows represent different peaks and columns represent different time points or cells. Use the standardise function to standardize the data, then estimate the fuzzification parameter (m) with the mestimate function. Next, execute the fuzzy C-means clustering analysis with the mfuzz function. The clustering results indicate the temporal trends of each peak.
2.11 Monocle 2 pseudotime analysis
Analyzing the differentiation trajectory of bHFSC and the genes influencing the trajectory using monocle2 (v2.22.0) (Qiu et al., 2017). The top specific peaks calculated by the Seurat FindAllMarkers function will be input into monocle2 to identify cell differentiation trajectories. After selecting the keratinocyte lineage, dimensionality reduction will be performed using the DDRTree method, and visualized with the plot_cell_trajectory function, followed by monocle2 pseudotime analysis. To identify peaks influencing the generation of the IFE lineage from bHFSC, the BEAM function will be used to identify lineage branch-enriched peaks (P-value < 0.01). Peaks will be annotated to genes using the annotatePeak function within regions 2 kb upstream and downstream of the transcription start site (TSS)., the chromatin accessibility of relevant genes along the lineage will be visualized using ComplexHeatmap (v2.14.0) (Gu et al., 2016).
3 Results
3.1 scATAC-seq quality control across burn time points
In this study, we collected samples of normal skin and stasis zone tissues from rats at nine time points, spanning from 0 h to 19 days post-burn. Nuclei were extracted from these tissues for scATAC-seq experiment. The raw sequencing data were processed through alignment and deconvolution to generate fragments, metadata, and peak files, yielding an average of over 23 million scATAC-seq reads per sample. Quality control was performed using the Signac workflow, with the following thresholds: nCount peaks >500, FRiP >0.1, nucleosome signal <4, and TSS enrichment >1 (see Methods). High-quality data were obtained, with each sample containing an average of 3,500 cells. Additionally, the mean unique cell fragments per sample were 11,623, and the average TSS enrichment was 3.78 (Supplementary Table S1), confirming a significant concentration of reads around TSS regions (Buenrostro et al., 2013) (Supplementary Figures S1A–C). To ensure purity, doublets were removed from each library using scDblFinder (Supplementary Figure S1D). The final dataset comprised 31,503 single cells, offering a comprehensive chromatin landscape of stasis-zone tissues after burn injury.
3.2 Comprehensive chromatin accessibility map revealing multiple cell types in burned skin and its appendages
Data from different time points were effectively integrated, followed by visualization with Uniform Manifold Approximation and Projection (UMAP), revealing the heterogeneity and unity of cell types across various burn injury time points (Supplementary Figures S1E, F). Our chromatin accessibility map of burned skin identified 26 distinct cell types and their subtypes within the epidermis, dermis, and skin appendages (Figures 1E–G; Supplementary Table S2). Starting with the epidermis, a critical layer of the skin, we identified various cell types, including basal cells of the interfollicular epidermis (IFEB) (Itga6, Tp63, Wn3), granular cells (IFEG) (Krt1, Spink5, Krt10), and spinous cells (IFESP) (Grhl3, Pkp1, Dsc3) (Mazzone et al., 2016; Ågren et al., 2022). Specialized cells within the epidermis include melanocytes (MELA) (Gpr143, Tyr, Mlana) (Carney et al., 2024), which are essential for pigmentation, and sebaceous gland cells (SG) (Krt79, Acsbg1) (Lanz et al., 2011), which contribute to lipid production. Epidermal lineage-related hair follicle stem cells, including quiescent (qHFSC) (Lhx2, Krt15, Itgb8), activated (aHFSC) (Lgr5, Itgb8), and burn-specific states (bHFSC) (Barx2, Itgb8) (da Silva-Diz et al., 2013), were also identified. In the dermal compartment (Figures 1E, F), we identified multiple fibroblast populations, fascial fibroblasts (fFB) (Sfrp4, Prss23, Tgfb2), reticular fibroblasts (rFB) (Dpt, Npr3, Arpc1b) and burn-specific fibroblasts (bFB) (Tnc, Acta2, Lrrc15) (Figure 1G) (Gay et al., 2020; Correa-Gallegos et al., 2023). The hair follicle structures within the dermis include dermal papilla (DP) (Alpl, Col23a1, Runx3), dermal sheath (DS) (Pdgfra, Entpd1, Fbn1) (Kim et al., 2024). The hair shaft (HS) (Wnt10a, Wnt3, Krt27) and surrounding layers, such as the inner root sheath (IRS) (Krt73, Krt25, Krt27) and outer root sheath (ORS) (Sdc1, Krt17), were also detected. Additionally, infundibular cells (INF) (Krt80, Krt17, Itgb8)and transit amplifying cells (TAC) (Cdh3, Lef1, Bmp7) play key roles in hair follicle biology, while muscle cells (MUSCLE) (Pdgfrb, Mylk, Rgs5) and pericyte (PCT) (Rgs5,Tagln) reflect the multifunctionality of the skin (Takahashi et al., 2020). The immune system within the skin includes innate lymphoid cells (ILC) (Cd3d, Ptprc, Runx3), macrophages (MACRO) (Osm, Itgax, Cd86) (Wang et al., 2019), and Langerhans cells (LGS) (Cd207, Itgax) (Hochgerner et al., 2022). Finally, the vascular and lymphatic systems were represented by burn-specific endothelial cells (bENDO) (Pecam1,Cd34,Aqp1), lymphatic endothelial cells (lENDO) (Fcerlg, Il1r1, Flt4), and vascular endothelium (vENDO) (Cdh5, Cd93, Pecaml) (Figure 1F; Supplementary Figure S2A; Supplementary Table S2) (Hou et al., 2020; Li et al., 2020). As burn injury progresses over time, there is a significant shift in the proportion of various cell types. For instance, MACRO and IENDO cells dominate the early stages at 0h and 12 h post-burn, which is likely associated with the inflammatory response in the stasis zone. Conversely, the proportions of other cell types such as vENDO and the hair follicle components HS, IRS, and ORS decrease at 0h and 12 h post-burn, potentially due to cell death caused by the burn injury. Subsequently, the ratios of vENDO, qHFSC, and the stratified cells of IFE gradually increase, which may indicate the process of tissue regeneration and remodeling (Supplementary Figure S1G). This diverse cellular composition reflects the complex structural and function of the skin and its response to burn injury.
3.3 Integration and comparative analysis of cell types in skin burn and radiation injury healing processes
Our dataset was integrated with the public scRNA-seq dataset of radiation-induced skin injury (GSE193564). Single-cell RNA-Seq analysis of molecular changes during radiation-induced skin injury: the involvement of Nur77 and various cell types were well integrated (Figures 1H, I). We reproduced the cell type-specific marker genes provided in the public article for annotation (Supplementary Figure S1F, I), and the results showed good consistency between our cell type annotations and the public data. Next, we calculated the Spearman correlation between the two datasets and compared the RNA matrices of cell types from both datasets. The results showed that the cell types mainly fell into five major categories: immune cells, endothelial cells, fibroblasts, keratinocytes, and muscle cells, all of which exhibited high correlation (Figure 1J). Additionally, the openness of marker genes Barx2, Efna5, Slc10a6, and Sox9 for bHFSC in our dataset showed consistent trends with their expression in the scRNA data (Figure 1K) (Tierney et al., 2024). In summary, the chromatin accessibility revealed by our scATAC-seq data showed a high correlation with the mRNA expression of the same cell types in the public scRNA-seq data, demonstrating the completeness of our burn dataset time points and the high reliability of our data.
However, some differences were also observed. For example, the bENDO and MELA cell types we identified were rarely found in the radiation injury dataset (Figure 1I). By comparing sampling photos of the two injury types, we found that the radiation injury site showed signs of healing after 19 days, while the burn stasis zone had fully recovered, but the necrotic area was still scabbing (Figure 1A) (Yan et al., 2024). This suggests that burns may cause a more intense inflammatory response and prolong the inflammation phase, leading to an increase in bENDO cells, which delays the proliferation and remodeling phase of the skin and may result in more pigment deposition during burn healing (Burgess et al., 2022).
3.4 Gene functions and motif enrichment analysis reveals key transcription factors in burn skin cell types
To explore the underlying mechanisms of chromatin accessibility profiles, we performed peak differential analysis, gene scoring, and motif enrichment analysis on 163,441 peaks. We identified potential cis-regulatory elements (Figure 2A), transcription factors (TFs), genes, and their functions closely associated with the 26 cell types. For example, qHFSC are enriched for cell proliferation and adhesion-related functions (Supplementary Figure S2B). The gene Trabd2b is involved in regulating the Wnt signaling pathway, which controls stem cell self-renewal (Figure 2B). Additionally, Cux1 motifs are highly enriched, maintaining the balance of cell proliferation and differentiation in epithelial tissues (Figure 2C) (Ren et al., 2023). aHFSC are enriched for functions involved in hair follicle development and hair growth cycle (Figure 2D). The gene Lgr5 is involved in maintaining hair follicle stem cell self-renewal (Figure 2B) (da Silva-Diz et al., 2013). Furthermore, Fox family motifs (Foxf1, Foxp1) are enriched, indicating their active proliferation to support follicle growth. (Yang et al., 2022). The Fox family genes are known to play crucial roles in stem cell maintenance, differentiation, and follicle cycle regulation (Figure 2C) (Wang et al., 2016). bHFSC are associated with cell movement and neurotransmitter transmission (Figure 2D). The gene Anxa8 related to dynamic changes in the cell membrane and cell migration (Figure 2B) (Yuan et al., 2021), with the Snai1 motif regulating cell migration and survival (Figure 2C), bENDO cells are enriched for functions related to the clearance and phagocytosis of apoptotic cells (Figure 2D). The gene Snai1 enhances cell anti-apoptotic ability and promotes cell survival (Figure 2B) (Freihen et al., 2020). With enrichment for STAT family motifs (Stat3, Stat1). Stat3 promotes cell survival, proliferation, and differentiation by activating growth-promoting and apoptosis-inhibiting genes (Chou et al., 2021), while Stat1 is involved in regulating immune responses (Figure 2C) (Xu et al., 2019). bFB cells are enriched for functions related to collagen fiber tissue remodeling (Figure 2D). The gene Bicc1 plays an important role in fibrosis in the kidney and other tissues (Figure 2B) (Leal-Esteban et al., 2018), promoting the generation of myofibroblasts, with enrichment for Tead4 motifs, which bind to TEA/ATTS elements on DNA and activate genes such as α-smooth muscle actin (α-SMA) (Figure 2C) (Mesrouze et al., 2017). These results list the key motifs associated with burn skin remodeling, including both previously reported and newly discovered burn-specific TFs. This demonstrates the comprehensiveness and accuracy of our cell type data, strongly suggesting that the scATAC-seq profiles of burn rats precisely identify accessible chromatin regions within the rat genome, providing valuable data resources and theoretical foundations for studying mammalian burn healing.
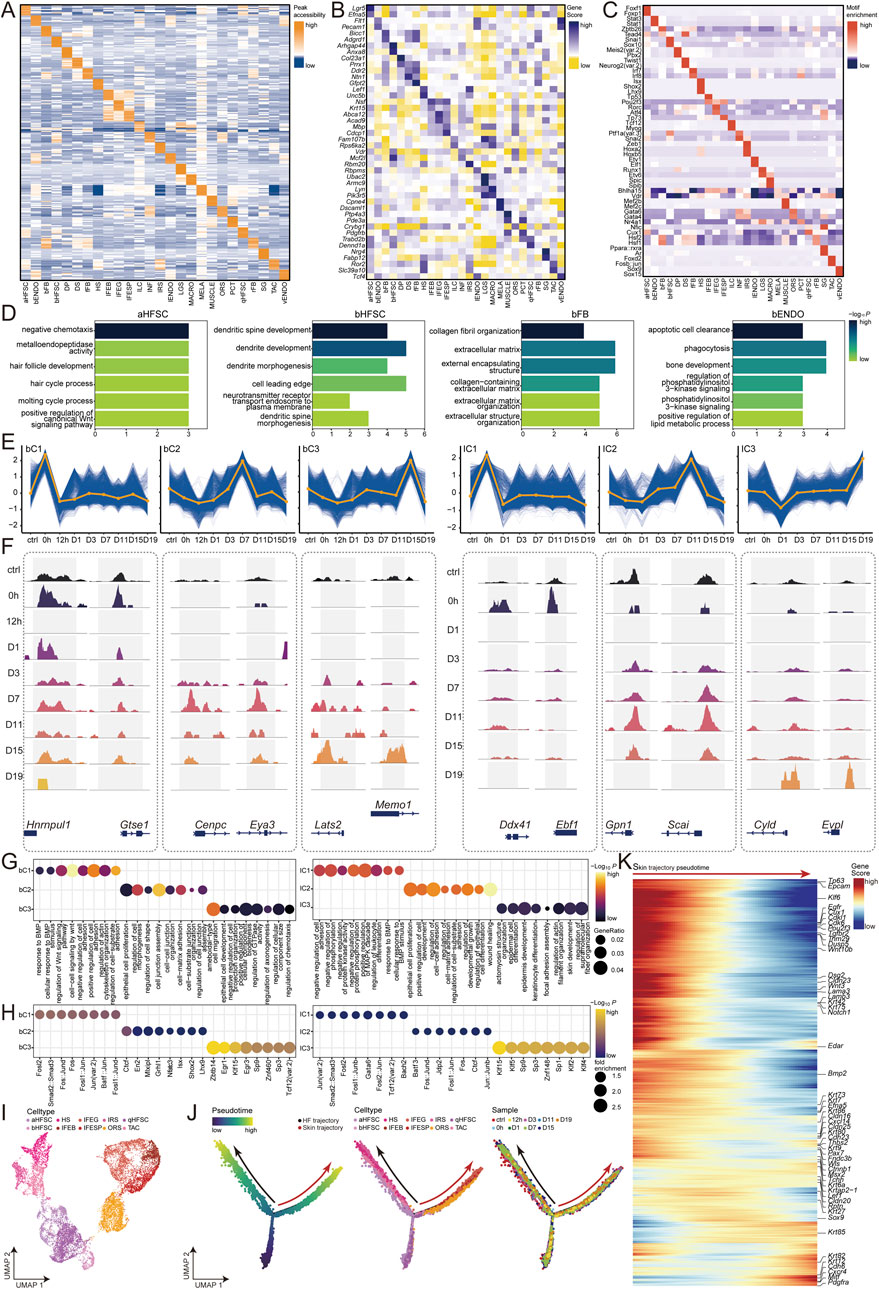
Figure 2. Temporal and pseudotime analyses of gene regulation, cell type dynamics, and the integration of single-cell RNA data. (A) Cell type-specific peaks. (B) Cell type-specific genes. (C) Cell type-specific motifs. (D) Enriched Gene Ontology (GO) Terms for aHFSC, bHFSC, bFB and bENDO. (E) Standardized fuzzy clustering of scATAC signals for bHFSC and IFEB, with blue lines representing individual loci and orange lines representing cluster center values. (F) Genomic browser view of dynamic peaks of bHFSC and IFEB signals. (G) GO enrichment of clusters for bHFSC and IFEB, with dot size representing the proportion of genes in the GO term and color representing the -Log10 (P-value). (H) Motif enrichment of clusters for bHFSC and IFEB, with dot size representing the fold. enrichment and color representing the -Log10 (P-value). (I) UMAP of cell types constructed in pseudotime. (J) Pseudotime analysis of post-injury epithelial lineage of HFSC and normal hair follicle differentiation using Monocle2. (K) Peaks associated with burn-specific epithelial temporal changes will be annotated to genes, with red coloration indicating high scores and blue coloration indicating low scores.
3.5 Temporal dynamics of chromatin accessibility during burn wound healing
The skin samples collected from the burn stasis zone of rats from 0 to 19 days post-burn constitute a comprehensive scATAC-seq dataset (Figure 1A), which can elucidate which cell types are most sensitive to burns and the molecular mechanisms behind healing. We found that the cell types in the skin niche mainly exhibit two trends with the change in burn time points: one increases in the early stages of the burn, producing burn stress responses and immune responses, including bHFSC, MACRO, MELA, fFB, and PCT. The other increases in the later stages of the burn, participating in skin remodeling, including IFEB, IFESP, IFEG, and ORS (Supplementary Figure S3A). To explore the molecular mechanisms of epithelialization after burns, we focused on bHFSC, which increases in the early stages, and IFEB, which increases in the later stages. We performed normalized time point fuzzy clustering on peaks and selected three significant trends for each cell type, showing the categories at different stages of burn healing (Supplementary Tables S4, S5): (1) bHFSC cluster 1 (bC1, n = 11773) accessible immediately after injury at 0h, indicating stress response; (2) bHFSC cluster 2 (bC2, n = 8,844) accessible during stem cell differentiation and proliferation from D3-D11; (3) bHFSC cluster 3 (bC3, n = 8,568) accessible during skin remodeling from D11-D19; (4) IFEB cluster 1 (IC1, n = 11399) accessible immediately after injury at 0h, indicating cell damage response; (5) IFEB cluster 2 (IC2, n = 10574) accessible during epithelial cell proliferation from D7-D15; (6) IFEB cluster 3 (IC3, n = 3,516) accessible during skin remodeling from D15-D19 (Figure 2C). These clusters highlight the chromatin changes during wound healing, with burns causing rapid cell damage and immune responses, clearing necrotic cells to prepare for later skin healing. The dynamics of chromatin accessibility provide new insights into cell fate determination during burn healing.
We identified significantly changed chromatin accessibility peaks in response to injury stress from bC1. By associating them with genes, we found that they mainly play roles in functions related to BMP and Wnt signaling pathways (Figure 1G). For example, Gtse1 is involved in cell cycle regulation and DNA damage response, affecting the cells response to signals (Lei et al., 2020). Hnrnpul1 can influence the cells response to DNA damage by regulating the function of the p53 protein (Figure 1F) (Sharma et al., 2015). The enriched motif is a heterodimer composed of Fos and Jun family proteins, which can respond to cellular stress and regulate gene expression (Figure 1H) (Papachristou et al., 2006). bC2 describes the roles of cells in epithelial cell proliferation, morphogenesis, cell junction, and adhesion regulation. For example, the Cenpc gene encodes centromere protein C, which is involved in chromosome segregation and cell division (Figure 1G) (Fellmeth et al., 2023); the Eya3 gene encodes a transcription factor that plays an important role in cell development and differentiation (Figure 1F) (Wiedner et al., 2023). Grhl1 and Nfatc3 motifs are involved in these processes (Figure 1H) (de la Garza et al., 2013). bC3 functions in migration, epithelial cell development, GTPase activity regulation, and cell component size regulation (Figure 1G). For example, the Lats2 gene encodes a protein that is a key regulator of the Hippo signaling pathway, involved in the regulation of cell proliferation, apoptosis, and morphogenesis (Guo et al., 2019); the Memo1 gene encodes a protein that plays an important role in cell migration and signal transduction (Figure 1F) (Labrecque et al., 2021). Gr1 and Egr3 motifs are involved in the regulation of cell migration and epithelial cell development (Figure 1H) (Shaw et al., 2015; Nishimura et al., 2024). IC1 functions in the regulation and response to cell death and immune response (Figure 1G). For example, the Ddx41 gene encodes a protein involved in RNA metabolism and innate immune response (Kim K. et al., 2023); the Ebf1 gene encodes a transcription factor that plays an important role in B cell development and differentiation (Figure 1F) (Kim E. E. et al., 2023). Bach2 and Gata6 motifs are involved in the regulation of cell death and B cell development and immune response (Figure 1H) (Jayakumar et al., 2022; Weng et al., 2024). IC2 describes the regulatory functions of cells in epithelial cell proliferation, development, adhesion, differentiation, and wound healing (Figure 1G). For example, the Gpn1 gene encodes a protein involved in the nuclear localization and stability of RNA polymerase, with its main function related to transcription regulation (Huang et al., 2021); the Scai gene encodes a protein that plays an important role in cell migration and invasion, particularly by regulating the cytoskeleton and adhesion molecules (Figure 1F) (Yi et al., 2019). Fos::Jund, Fos, Fosl1::Jun, and Jun::Junb motifs are involved in the regulation of epithelial cell proliferation, development, adhesion, differentiation, and wound healing (Figure 1H) (Sarate et al., 2024). IC3 describes the remodeling functions of cells in actin-myosin structure, cell junction, epithelial cell differentiation, and skin development (Figure 1G). For example, the Cyld gene encodes a deubiquitinating enzyme involved in regulating cell signaling, cell cycle, and apoptosis (Li Y. et al., 2021); the Evpl gene encodes a structural protein in the epidermis and other epithelial tissues, involved in the organization of the cytoskeleton and stabilization of cell junctions (Figure 1F) (Laky et al., 2023). Klf5 and Klf4 motifs are involved in the regulation of cell junction stabilization and epithelial cell differentiation (Figure 1H) (Laky et al., 2023). In summary, our comprehensive analysis of chromatin accessibility dynamics in response to burn injury provides valuable insights into the molecular mechanisms and cellular processes involved in wound healing, highlighting the distinct roles of various cell types and signaling pathways at different stages of the healing process.
3.6 Putative cell lineage transition in stasis zone of burn healing skin
The skin contains various epidermal stem cell populations, among which HFSC are crucial for the cyclical growth of hair (Kloepper et al., 2015). Previous studies have reported that HFSC undergo a change in stem cell identity and mobilize to participate in epidermal repair when the skin is scratched or abraded (Tierney et al., 2024). However, little is known about the lineage changes of HFSC during the burn process (Babakhani et al., 2020). To investigate the relationship between HFSC and burn skin healing, we applied the Monocle2 algorithm to scATAC-seq data to predict the fate transitions of HFSC after burns (Qiu et al., 2017). Our analysis clearly revealed the burn-specific epithelialization lineage and the lineage of HFSC in normal hair follicle growth. Specifically, we found that qHFSC transition into bHFSC after burns, subsequently producing IFESP and IFEG through IFEB. Additionally, we predicted that qHFSC activate into aHFSC in normal conditions, which then differentiate into ORS and TAC, with TAC further generating IRS and HS. (Figures 2I, J).
In the proliferation, differentiation, and formation of basal cells of the skin after burns, which subsequently generate the spinous and granular layers, the following genes play key roles: First, during the proliferation of qHFSC, stem cell genes with high gene scores, such as Tp63, Klf6, Pou2f3, and Tgfbr2 are crucial in maintaining stem cell characteristics (Paris et al., 2012; Tomela et al., 2021; Li et al., 2022). Subsequently, during the transition to bHFSC, stem cell and cell migration genes with high gene scores, such as Dsg2, Cldn23, Wnt3, and Notch1 promote cell proliferation and migration (Kishimoto et al., 2000; Brennan et al., 2007). Next, bHFSC differentiate into IFEB, basal layer genes with highly gene scores, such as Bmp2, Krt7, Krt73, Krt80, Krt9, and Pax7 playing important roles in basal cell differentiation (Gosselet et al., 2007; Kim et al., 2016). Following this, basal cells continue to proliferate and begin to differentiate into the spinous and granular layers, keratinization genes with high gene scores, such as Krt6a, Krtap2-1, Krt27, Krt85, Krt82, and Krt12 being crucial for the formation and maturation of the keratin layer (Suárez-Fariñas et al., 2015; Morgan et al., 2020; Erjavec et al., 2022). Additionally, transcription factors such as Lef1, Msx2, Sox9, and Mitf also play regulatory roles in this process, coordinating cell differentiation and keratin layer formation (Figure 2K) (Yeh et al., 2009; Phan et al., 2020; Wang et al., 2022). Our lineage analysis uncovered the time point-specific transitions and changes in chromatin accessibility during the burn process.
4 Conclusion
This study identified 26 different cell types, including various cell types in the epidermis, dermis, and skin appendages, and analyzed their dynamic changes during the burn process. By integrating and comparing burn data with scRNA-seq data of radiation injury, it was found that burns may cause a more intense inflammatory response and prolong the inflammation phase, leading to an increase in bENDO cells, delaying the skin proliferation and remodeling phases, and potentially resulting in more pigment deposition during burn healing. Through motif enrichment analysis, key transcription factors and gene functions related to burn skin cell types were identified, such as cell adhesion, Epidermal development, cell movement, phagocytosis, and collagen fiber tissue remodeling. The study revealed dynamic changes in chromatin accessibility after burns, indicating that burns cause rapid cell damage and immune responses, clearing necrotic cells to prepare for subsequent skin healing. The study predicted the fate transitions of HFSCs in normal hair follicle growth and burn-specific epithelialization lineages, revealing time point-specific transitions and changes in chromatin accessibility during the burn process. These findings provide important theoretical foundations and data resources for understanding the molecular mechanisms of burn skin repair and offer new insights for developing targeted therapies for burn treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. All raw data have been submitted to the CNGB (Accession ID, CNP0006143).
Ethics statement
The animal studies were approved by This study was approved by the Affiliated Hospital of the Naval Medical University [license number: SYXK (Hu) 2020-0033] and the BGI Technology Ethics Committee (approval number: BGI-IRB A23034). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
RL: Data curation, Formal Analysis, Visualization, Writing–original draft, Writing–review and editing. JLi: Data curation, Writing–original draft. SL: Conceptualization, Writing–review and editing. XG: Investigation, Validation, Writing–review and editing. JLu: Conceptualization, Investigation, Writing–review and editing. TW: Investigation, Writing–original draft. JC: Conceptualization, Data curation, Writing–review and editing. YZ: Investigation, Writing–original draft. YY: Investigation, Validation, Writing–review and editing. JD: Software, Writing–original draft. BZ: Data curation, Investigation, Writing–original draft. XW: Writing–original draft, Writing–review and editing. PG: Supervision, Writing–review and editing. LL: Resources, Supervision, Writing–review and editing. XX: Resources, Supervision, Writing–review and editing, Conceptualization. XD: Methodology, Supervision, Writing–review and editing. RH: Conceptualization, Resources, Validation, Writing–review and editing. XL: Supervision, Writing–review and editing. XH: Supervision, Writing–review and editing. SW: Supervision, Writing–review and editing. SJ: Writing–review and editing, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 31770152), the Shaanxi Fundamental Science Research Project for Chemistry and Biology (Grant No. 22JHZ008) and Guangdong Genomics Data Center (2021B1212100001).
Acknowledgments
We would like to thank DCS Cloud (https://cloud.stomics.tech/) for providing the computational resources and software support necessary for this study. This work is supported by China National GeneBank (CNGB) for providing sequencing service.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1519926/full#supplementary-material
References
Ågren, M. S., Litman, T., Eriksen, J. O., Schjerling, P., Bzorek, M., and Gjerdrum, L. M. R. (2022). Gene expression linked to reepithelialization of human skin wounds. Int. J. Mol. Sci. 23, 15746. doi:10.3390/ijms232415746
Babakhani, A., Nobakht, M., Pazoki Torodi, H., Dahmardehei, M., Hashemi, P., Mohajer Ansari, J., et al. (2020). Effects of hair follicle stem cells on partial-thickness burn wound healing and tensile strength. Iran. Biomed. J. 24, 99–109. doi:10.29252/ibj.24.2.99
Boyce, S. T., and Kagan, R. J. (2023). Composition and performance of autologous engineered skin substitutes for repair or regeneration of excised, full-thickness burns. J. Burn Care Res. 44, S50–S56. doi:10.1093/jbcr/irac107
Brennan, D., Hu, Y., Joubeh, S., Choi, Y. W., Whitaker-Menezes, D., O’Brien, T., et al. (2007). Suprabasal Dsg2 expression in transgenic mouse skin confers a hyperproliferative and apoptosis-resistant phenotype to keratinocytes. J. Cell Sci. 120, 758–771. doi:10.1242/jcs.03392
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y., and Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. doi:10.1038/nmeth.2688
Burgess, M., Valdera, F., Varon, D., Kankuri, E., and Nuutila, K. (2022). The immune and regenerative response to burn injury. Cells 11, 3073. doi:10.3390/cells11193073
Carney, B. C., Simbulan-Rosenthal, C. M., Rosenthal, D. S., and Shupp, J. W. (2024). A nude mouse model of xenografted hypertrophic scar cells to test therapeutics in the skin. Front. Biosci. Landmark Ed. 29, 230. doi:10.31083/j.fbl2906230
Cheng, X. C., Tong, W. Z., Rui, W., Feng, Z., Shuai, H., and Zhe, W. (2024). Single-cell sequencing technology in skin wound healing. Burns Trauma 12, tkae043. doi:10.1093/burnst/tkae043
Chou, J.-C., Lieu, F.-K., Ho, D. M.-T., Shen, H.-Y., Lin, P.-H., Hu, S., et al. (2021). Regulation of extracellular and intracellular prolactin on cell proliferation and survival rate through GHR/JAK2/STAT3 pathway in NSCLC. Chemosphere 264, 128604. doi:10.1016/j.chemosphere.2020.128604
Correa-Gallegos, D., Ye, H., Dasgupta, B., Sardogan, A., Kadri, S., Kandi, R., et al. (2023). CD201+ fascia progenitors choreograph injury repair. Nature 623, 792–802. doi:10.1038/s41586-023-06725-x
da Silva-Diz, V., Solé-Sánchez, S., Valdés-Gutiérrez, A., Urpí, M., Riba-Artés, D., Penin, R. M., et al. (2013). Progeny of Lgr5-expressing hair follicle stem cell contributes to papillomavirus-induced tumor development in epidermis. Oncogene 32, 3732–3743. doi:10.1038/onc.2012.375
de la Garza, G., Schleiffarth, J. R., Dunnwald, M., Mankad, A., Weirather, J. L., Bonde, G., et al. (2013). Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Invest. Dermatol. 133, 68–77. doi:10.1038/jid.2012.269
Eming, S. A., Krieg, T., and Davidson, J. M. (2007). Inflammation in wound repair: molecular and cellular mechanisms. J. Invest. Dermatol. 127, 514–525. doi:10.1038/sj.jid.5700701
Erjavec, S. O., Gelfman, S., Abdelaziz, A. R., Lee, E. Y., Monga, I., Alkelai, A., et al. (2022). Whole exome sequencing in Alopecia Areata identifies rare variants in KRT82. Nat. Commun. 13, 800. doi:10.1038/s41467-022-28343-3
Fan, R., Zhang, C., Li, F., Li, B., McCarthy, A., Zhang, Y., et al. (2024). Hierarchically assembled nanofiber scaffolds with dual growth factor gradients promote skin wound healing through rapid cell recruitment. Adv. Sci. Weinh. Baden-Wurtt. Ger. 11, e2309993. doi:10.1002/advs.202309993
Fellmeth, J. E., Jang, J. K., Persaud, M., Sturm, H., Changela, N., Parikh, A., et al. (2023). A dynamic population of prophase CENP-C is required for meiotic chromosome segregation. PLoS Genet. 19, e1011066. doi:10.1371/journal.pgen.1011066
Freihen, V., Rönsch, K., Mastroianni, J., Frey, P., Rose, K., Boerries, M., et al. (2020). SNAIL1 employs β-Catenin-LEF1 complexes to control colorectal cancer cell invasion and proliferation. Int. J. Cancer 146, 2229–2242. doi:10.1002/ijc.32644
Gay, D., Ghinatti, G., Guerrero-Juarez, C. F., Ferrer, R. A., Ferri, F., Lim, C. H., et al. (2020). Phagocytosis of wnt inhibitor SFRP4 by late wound macrophages drives chronic wnt activity for fibrotic skin healing. Sci. Adv. 6, eaay3704. doi:10.1126/sciadv.aay3704
G El Baassiri, M., Dosh, L., Haidar, H., Gerges, A., Baassiri, S., Leone, A., et al. (2023). Nerve growth factor and burn wound healing: update of molecular interactions with skin cells. Burns J. Int. Soc. Burn Inj. 49, 989–1002. doi:10.1016/j.burns.2022.11.001
Gosselet, F. P., Magnaldo, T., Culerrier, R. M., Sarasin, A., and Ehrhart, J.-C. (2007). BMP2 and BMP6 control p57(Kip2) expression and cell growth arrest/terminal differentiation in normal primary human epidermal keratinocytes. Cell. Signal. 19, 731–739. doi:10.1016/j.cellsig.2006.09.006
Gross-Amat, O., Guillen, M., Salmon, D., Nataf, S., and Auxenfans, C. (2020). Characterization of a topically testable model of burn injury on human skin explants. Int. J. Mol. Sci. 21, 6956. doi:10.3390/ijms21186956
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma. Oxf. Engl. 32, 2847–2849. doi:10.1093/bioinformatics/btw313
Guo, C., Liang, C., Yang, J., Hu, H., Fan, B., and Liu, X. (2019). LATS2 inhibits cell proliferation and metastasis through the Hippo signaling pathway in glioma. Oncol. Rep. 41, 2753–2761. doi:10.3892/or.2019.7065
Guo, S., Fang, Q., Chen, L., Yu, M., Chen, Y., Li, N., et al. (2021). Locally activated mitophagy contributes to a “built-in” protection against early burn-wound progression in rats. Life Sci. 276, 119095. doi:10.1016/j.lfs.2021.119095
Guo, S. X., Jin, Y. Y., Fang, Q., You, C. G., Wang, X. G., Hu, X. L., et al. (2015). Beneficial effects of hydrogen-rich saline on early burn-wound progression in rats. PloS One 10, e0124897. doi:10.1371/journal.pone.0124897
Hochgerner, M., Bauer, T., Zyulina, V., Glitzner, E., Warsi, S., Konkel, J. E., et al. (2022). BMPR1a is required for the optimal TGFβ1-dependent CD207+ langerhans cell differentiation and limits skin inflammation through CD11c+ cells. J. Invest. Dermatol. 142, 2446–2454.e3. doi:10.1016/j.jid.2022.02.014
Hou, H., Li, J., Zhou, L., Liang, J., Wang, J., Li, J., et al. (2020). An effective method of isolating microvascular endothelial cells from the human dermis. Cell Biol. Int. 44, 2588–2597. doi:10.1002/cbin.11448
Huang, F., Huffman, K. E., Wang, Z., Wang, X., Li, K., Cai, F., et al. (2021). Guanosine triphosphate links MYC-dependent metabolic and ribosome programs in small-cell lung cancer. J. Clin. Invest. 131, e139929. doi:10.1172/JCI139929
Huang, J., Liang, X., Xuan, Y., Geng, C., Li, Y., Lu, H., et al. (2017). A reference human genome dataset of the BGISEQ-500 sequencer. GigaScience 6, 1–9. doi:10.1093/gigascience/gix024
Huang, T., Zhang, K., Sun, L., Xue, X., Zhang, C., Shu, Z., et al. (2015). Body protective compound-157 enhances alkali-burn wound healing in vivo and promotes proliferation, migration, and angiogenesis in vitro. Drug Des. devel. Ther. 9, 2485–2499. doi:10.2147/DDDT.S82030
Jayakumar, P., Laganson, A., and Deng, M. (2022). GATA6+ peritoneal resident macrophage: the immune custodian in the peritoneal cavity. Front. Pharmacol. 13, 866993. doi:10.3389/fphar.2022.866993
Jeschke, M. G., and Herndon, D. N. (2014). Burns in children: standard and new treatments. Lancet lond. Engl. 383, 1168–1178. doi:10.1016/S0140-6736(13)61093-4
Kim, D., Hossain, M. Z., Nieves, A., Gu, L., Ratliff, T. S., Mi Oh, S., et al. (2016). To control site-specific skin gene expression, autocrine mimics paracrine canonical wnt signaling and is activated ectopically in skin disease. Am. J. Pathol. 186, 1140–1150. doi:10.1016/j.ajpath.2015.12.030
Kim, E. E., Shekhar, A., Ramachandran, J., Khodadadi-Jamayran, A., Liu, F.-Y., Zhang, J., et al. (2023a). The transcription factor EBF1 non-cell-autonomously regulates cardiac growth and differentiation. Dev. Camb. Engl. 150, dev202054. doi:10.1242/dev.202054
Kim, K., Ong, F., and Sasaki, K. (2023b). Current understanding of DDX41 mutations in myeloid neoplasms. Cancers 15, 344. doi:10.3390/cancers15020344
Kim, Y., Lee, J. O., Lee, J. M., Lee, M.-H., Kim, H.-M., Chung, H.-C., et al. (2024). Low molecular weight collagen peptide (LMWCP) promotes hair growth by activating the wnt/GSK-3β/β-catenin signaling pathway. J. Microbiol. Biotechnol. 34, 17–28. doi:10.4014/jmb.2308.08013
Kishimoto, J., Burgeson, R. E., and Morgan, B. A. (2000). Wnt signaling maintains the hair-inducing activity of the dermal papilla. Genes Dev. 14, 1181–1185. doi:10.1101/gad.14.10.1181
Kloepper, J. E., Baris, O. R., Reuter, K., Kobayashi, K., Weiland, D., Vidali, S., et al. (2015). Mitochondrial function in murine skin epithelium is crucial for hair follicle morphogenesis and epithelial-mesenchymal interactions. J. Invest. Dermatol. 135, 679–689. doi:10.1038/jid.2014.475
Kumar, L., and E Futschik, M. (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7. doi:10.6026/97320630002005
Labrecque, C. L., Hilton, C. N., Airas, J., Blake, A., Rubenstein, K. J., Parish, C. A., et al. (2021). Identification of phenazine-based MEMO1 small-molecule inhibitors: virtual screening, fluorescence polarization validation, and inhibition of breast cancer migration. ChemMedChem 16, 1163–1171. doi:10.1002/cmdc.202000797
Laky, K., Kinard, J. L., Li, J. M., Moore, I. N., Lack, J., Fischer, E. R., et al. (2023). Epithelial-intrinsic defects in TGFβR signaling drive local allergic inflammation manifesting as eosinophilic esophagitis. Sci. Immunol. 8, eabp9940. doi:10.1126/sciimmunol.abp9940
Lander, J. M., Supp, D. M., He, H., Martin, L. J., Chen, X., Weirauch, M. T., et al. (2017). Analysis of chromatin accessibility in human epidermis identifies putative barrier dysfunction-sensing enhancers. PloS One 12, e0184500. doi:10.1371/journal.pone.0184500
Lanz, C., Ledermann, M., Slavík, J., and Idle, J. R. (2011). The production and composition of rat sebum is unaffected by 3 gy gamma radiation. Int. J. Radiat. Biol. 87, 360–371. doi:10.3109/09553002.2010.537432
Lareau, C. A., Duarte, F. M., Chew, J. G., Kartha, V. K., Burkett, Z. D., Kohlway, A. S., et al. (2019). Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 37, 916–924. doi:10.1038/s41587-019-0147-6
Leal-Esteban, L. C., Rothé, B., Fortier, S., Isenschmid, M., and Constam, D. B. (2018). Role of Bicaudal C1 in renal gluconeogenesis and its novel interaction with the CTLH complex. PLoS Genet. 14, e1007487. doi:10.1371/journal.pgen.1007487
Lei, X., Du, L., Zhang, P., Ma, N., Liang, Y., Han, Y., et al. (2020). Knockdown GTSE1 enhances radiosensitivity in non-small-cell lung cancer through DNA damage repair pathway. J. Cell. Mol. Med. 24, 5162–5167. doi:10.1111/jcmm.15165
Li, B., Tang, H., Bian, X., Ma, K., Chang, J., Fu, X., et al. (2021a). Calcium silicate accelerates cutaneous wound healing with enhanced re-epithelialization through EGF/EGFR/ERK-mediated promotion of epidermal stem cell functions. Burns Trauma 9, tkab029. doi:10.1093/burnst/tkab029
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinforma. Oxf. Engl. 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Li, J., Wei, C., Yang, Y., Gao, Z., Guo, Z., and Qi, F. (2022). Apoptotic bodies extracted from adipose mesenchymal stem cells carry microRNA-21-5p to induce M2 polarization of macrophages and augment skin wound healing by targeting KLF6. Burns J. Int. Soc. Burn Inj. 48, 1893–1908. doi:10.1016/j.burns.2021.12.010
Li, K.-C., Wang, C.-H., Zou, J.-J., Qu, C., Wang, X.-L., Tian, X.-S., et al. (2020). Loss of Atg7 in endothelial cells enhanced cutaneous wound healing in a mouse model. J. Surg. Res. 249, 145–155. doi:10.1016/j.jss.2019.12.004
Li, Y., Shi, F., Hu, J., Xie, L., Zhao, L., Tang, M., et al. (2021b). Stabilization of p18 by deubiquitylase CYLD is pivotal for cell cycle progression and viral replication. NPJ Precis. Oncol. 5, 14. doi:10.1038/s41698-021-00153-8
Lu, M., Zhao, J., Wang, X., Zhang, J., Shan, F., and Jiang, D. (2022). Research advances in prevention and treatment of burn wound deepening in early stage. Front. Surg. 9, 1015411. doi:10.3389/fsurg.2022.1015411
Malhotra, H., Sharma, P., Kamal, M. A., Kaushik, P., and Rani, N. (2024). Concise review on scientific approaches to burns and scars. Curr. Drug Saf. 19, 191–199. doi:10.2174/1574886318666230509143017
Mazzone, L., Pratsinis, M., Pontiggia, L., Reichmann, E., and Meuli, M. (2016). Successful grafting of tissue-engineered fetal skin. Pediatr. Surg. Int. 32, 1177–1182. doi:10.1007/s00383-016-3977-z
Mesrouze, Y., Meyerhofer, M., Bokhovchuk, F., Fontana, P., Zimmermann, C., Martin, T., et al. (2017). Effect of the acylation of TEAD4 on its interaction with co-activators YAP and TAZ. Protein Sci. Publ. Protein Soc. 26, 2399–2409. doi:10.1002/pro.3312
Morgan, H. J., Benketah, A., Olivero, C., Rees, E., Ziaj, S., Mukhtar, A., et al. (2020). Hair follicle differentiation-specific keratin expression in human basal cell carcinoma. Clin. Exp. Dermatol. 45, 417–425. doi:10.1111/ced.14113
Nascimento-Filho, C. H. V., Silveira, E. J. D., Goloni-Bertollo, E. M., de Souza, L. B., Squarize, C. H., and Castilho, R. M. (2020). Skin wound healing triggers epigenetic modifications of histone H4. J. Transl. Med. 18, 138. doi:10.1186/s12967-020-02303-1
Nishimura, F. G., Sampaio, B. B., do Couto, G. O., da Silva, A. D., da Silva, W. J., Peronni, K. C., et al. (2024). The transcriptome of BT-20 breast cancer cells exposed to curcumin analog NC2603 reveals a relationship between EGR3 gene modulation and cell migration inhibition. Mol. Basel Switz. 29, 1366. doi:10.3390/molecules29061366
Papachristou, D., Pirttiniemi, P., Kantomaa, T., Agnantis, N., and Basdra, E. K. (2006). Fos- and Jun-related transcription factors are involved in the signal transduction pathway of mechanical loading in condylar chondrocytes. Eur. J. Orthod. 28, 20–26. doi:10.1093/ejo/cji101
Paris, M., Rouleau, M., Pucéat, M., and Aberdam, D. (2012). Regulation of skin aging and heart development by TAp63. Cell Death Differ. 19, 186–193. doi:10.1038/cdd.2011.181
Phan, Q. M., Fine, G. M., Salz, L., Herrera, G. G., Wildman, B., Driskell, I. M., et al. (2020). Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. eLife 9, e60066. doi:10.7554/eLife.60066
Plikus, M. V., Guerrero-Juarez, C. F., Treffeisen, E., and Gay, D. L. (2015). Epigenetic control of skin and hair regeneration after wounding. Exp. Dermatol. 24, 167–170. doi:10.1111/exd.12488
Qiu, X., Mao, Q., Tang, Y., Wang, L., Chawla, R., Pliner, H. A., et al. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. doi:10.1038/nmeth.4402
Radzikowska-Büchner, E., Łopuszyńska, I., Flieger, W., Tobiasz, M., Maciejewski, R., and Flieger, J. (2023). An overview of recent developments in the management of burn injuries. Int. J. Mol. Sci. 24, 16357. doi:10.3390/ijms242216357
Ran, Y., Yan, Z., Huang, M., Zhou, S., Wu, F., Wang, M., et al. (2023). Severe burn injury significantly alters the gene expression and m6A methylation tagging of mRNAs and lncRNAs in human skin. J. Pers. Med. 13, 150. doi:10.3390/jpm13010150
Ren, H., Zhao, F., Zhang, Q., Huang, X., and Wang, Z. (2022). Autophagy and skin wound healing. Burns Trauma 10, tkac003. doi:10.1093/burnst/tkac003
Ren, S., Wu, D., Shen, X., Wu, Q., Li, C., Xiong, H., et al. (2023). Deciphering the role of extrachromosomal circular DNA in adipose stem cells from old and young donors. Stem Cell Res. Ther. 14, 341. doi:10.1186/s13287-023-03575-2
Rolim, L. S. A., Nascente, P. da S., Castilho, R. M., and Squarize, C. H. (2024). Feeling the heat. Mapping the epigenetic modifications of histone during burn wound healing. J. Burn Care Res. 45, 499–507. doi:10.1093/jbcr/irad187
Roshangar, L., Soleimani Rad, J., Kheirjou, R., Reza Ranjkesh, M., and Ferdowsi Khosroshahi, A. (2019). Skin burns: review of molecular mechanisms and therapeutic approaches. Wounds Compend. Clin. Res. Pract. 31, 308–315.
Sarate, R. M., Hochstetter, J., Valet, M., Hallou, A., Song, Y., Bansaccal, N., et al. (2024). Dynamic regulation of tissue fluidity controls skin repair during wound healing. Cell 187, 5298–5315.e19. doi:10.1016/j.cell.2024.07.031
Sharma, V., Khurana, S., Kubben, N., Abdelmohsen, K., Oberdoerffer, P., Gorospe, M., et al. (2015). A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 16, 1520–1534. doi:10.15252/embr.201540437
Shaw, A. K., Pickup, M. W., Chytil, A., Aakre, M., Owens, P., Moses, H. L., et al. (2015). TGFβ signaling in myeloid cells regulates mammary carcinoma cell invasion through fibroblast interactions. PloS One 10, e0117908. doi:10.1371/journal.pone.0117908
Shi, Z., Yao, C., Shui, Y., Li, S., and Yan, H. (2023). Research progress on the mechanism of angiogenesis in wound repair and regeneration. Front. Physiol. 14, 1284981. doi:10.3389/fphys.2023.1284981
Siddique, R., Mehmood, M. H., Hussain, L., Malik, A., Sethi, A., Farrukh, M., et al. (2023). Role of medicinal herbs and phytochemicals in post burn management. Inflammopharmacology 31, 1695–1714. doi:10.1007/s10787-023-01246-5
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A., and Satija, R. (2021). Single-cell chromatin state analysis with signac. Nat. Methods 18, 1333–1341. doi:10.1038/s41592-021-01282-5
Suárez-Fariñas, M., Ungar, B., Noda, S., Shroff, A., Mansouri, Y., Fuentes-Duculan, J., et al. (2015). Alopecia areata profiling shows TH1, TH2, and IL-23 cytokine activation without parallel TH17/TH22 skewing. J. Allergy Clin. Immunol. 136, 1277–1287. doi:10.1016/j.jaci.2015.06.032
Sukmawati, D., Eryani, A., and Damayanti, L. (2020). Silver sulfadiazine’s effect on keratin-19 expression as stem cell marker in burn wound healing. BioMedicine 10, 5–11. doi:10.37796/2211-8039.1014
Takahashi, R., Grzenda, A., Allison, T. F., Rawnsley, J., Balin, S. J., Sabri, S., et al. (2020). Defining transcriptional signatures of human hair follicle cell states. J. Invest. Dermatol. 140, 764–773. doi:10.1016/j.jid.2019.07.726
Tierney, M. T., Polak, L., Yang, Y., Abdusselamoglu, M. D., Baek, I., Stewart, K. S., et al. (2024). Vitamin A resolves lineage plasticity to orchestrate stem cell lineage choices. Science 383, eadi7342. doi:10.1126/science.adi7342
Tiwari, V. K. (2012). Burn wound: how it differs from other wounds? Indian J. Plast. Surg. 45, 364–373. doi:10.4103/0970-0358.101319
Tomela, K., Karolak, J. A., Ginter-Matuszewska, B., Kabza, M., and Gajecka, M. (2021). Influence of TGFBR2, TGFB3, DNMT1, and DNMT3A knockdowns on CTGF, TGFBR2, and DNMT3A in neonatal and adult human dermal fibroblasts cell lines. Curr. Issues Mol. Biol. 43, 276–285. doi:10.3390/cimb43010023
van Dijk, D., Sharma, R., Nainys, J., Yim, K., Kathail, P., Carr, A. J., et al. (2018). Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729. doi:10.1016/j.cell.2018.05.061
Wang, E. C. E., Dai, Z., Ferrante, A. W., Drake, C. G., and Christiano, A. M. (2019). A subset of TREM2+ dermal macrophages secretes oncostatin M to maintain hair follicle stem cell quiescence and inhibit hair growth. Cell Stem Cell 24, 654–669. doi:10.1016/j.stem.2019.01.011
Wang, J., Xie, X., Deng, Y., Yang, H., Du, X., Liu, P., et al. (2022). SOX9 in keratinocytes regulates claudin 2 transcription during skin aging. Contrast Media Mol. Imaging 2022, 6884308. doi:10.1155/2022/6884308
Wang, L., Siegenthaler, J. A., Dowell, R. D., and Yi, R. (2016). Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science 351, 613–617. doi:10.1126/science.aad5440
Weng, X., Zheng, M., Liu, Y., and Lou, G. (2024). The role of Bach2 in regulating CD8 + T cell development and function. Cell Commun. Signal. CCS 22, 169. doi:10.1186/s12964-024-01551-8
Wiedner, H. J., Blue, R. E., Sadovsky, M., Mills, C. A., Wehrens, X. H. T., Herring, L. E., et al. (2023). RBFOX2 regulated EYA3 isoforms partner with SIX4 or ZBTB1 to control transcription during myogenesis. iScience 26, 108258. doi:10.1016/j.isci.2023.108258
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov. Camb. Mass 2, 100141. doi:10.1016/j.xinn.2021.100141
Xu, H., Jiang, Y., Xu, X., Su, X., Liu, Y., Ma, Y., et al. (2019). Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 20, 1621–1630. doi:10.1038/s41590-019-0542-7
Yan, T., Yang, P., Bai, H., Song, B., Liu, Y., Wang, J., et al. (2024). Single-cell RNA-Seq analysis of molecular changes during radiation-induced skin injury: the involvement of Nur77. Theranostics 14, 5809–5825. doi:10.7150/thno.100417
Yang, Q., Zhang, J., Bao, Q., Zhong, J., Wang, X., Tao, Y., et al. (2022). Foxp1 and Foxp4 deletion causes the loss of follicle stem cell niche and cyclic hair shedding by inducing inner bulge cell apoptosis. Stem Cells Dayt. Ohio 40, 843–856. doi:10.1093/stmcls/sxac045
Yeh, J., Green, L. M., Jiang, T.-X., Plikus, M., Huang, E., Chang, R. N., et al. (2009). Accelerated closure of skin wounds in mice deficient in the homeobox gene Msx2. Wound Repair Regen. Off. Publ. Wound heal. Soc. Eur. Tissue Repair Soc. 17, 639–648. doi:10.1111/j.1524-475X.2009.00535.x
Yi, T., Zhou, X., Sang, K., Zhou, J., and Ge, L. (2019). MicroRNA-1270 modulates papillary thyroid cancer cell development by regulating SCAI. Biomed. Pharmacother. Biomedecine Pharmacother. 109, 2357–2364. doi:10.1016/j.biopha.2018.08.150
Yu, Y., Wei, X., Deng, Q., Lan, Q., Guo, Y., Han, L., et al. (2021). Single-nucleus chromatin accessibility landscape reveals diversity in regulatory regions across distinct adult rat cortex. Front. Mol. Neurosci. 14, 651355. doi:10.3389/fnmol.2021.651355
Keywords: rat burn injuries, stasis zone, chromatin accessibility, wound healing, transcription factor
Citation: Li R, Li J, Liu S, Guo X, Lu J, Wang T, Chen J, Zheng Y, Yuan Y, Du J, Zhu B, Wei X, Guo P, Liu L, Xu X, Dai X, Huang R, Liu X, Hu X, Wang S and Ji S (2025) A scATAC-seq atlas of stasis zone in rat skin burn injury wound process. Front. Cell Dev. Biol. 12:1519926. doi: 10.3389/fcell.2024.1519926
Received: 30 October 2024; Accepted: 13 December 2024;
Published: 07 January 2025.
Edited by:
Kyoko Yokomori, University of California, Irvine, United StatesCopyright © 2025 Li, Li, Liu, Guo, Lu, Wang, Chen, Zheng, Yuan, Du, Zhu, Wei, Guo, Liu, Xu, Dai, Huang, Liu, Hu, Wang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shizhao Ji, c2hpemhhb2ppMjAyMkAxNjMuY29t; Shiwei Wang, d2FuZ3N3QG53dS5lZHUuY24=; Xiaoyan Hu, MTM4MTcxOTQ5NDNAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ruikang Li
Ruikang Li Jiashan Li
Jiashan Li Shuai Liu3†
Shuai Liu3† Yue Yuan
Yue Yuan Xiaoyu Wei
Xiaoyu Wei Xun Xu
Xun Xu Xi Dai
Xi Dai Runzhi Huang
Runzhi Huang Xin Liu
Xin Liu Xiaoyan Hu
Xiaoyan Hu Shiwei Wang
Shiwei Wang Shizhao Ji
Shizhao Ji