
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 05 December 2024
Sec. Stem Cell Research
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1510746
This article is part of the Research TopicAdvancements in Molecular and Cellular Mechanisms of Stem Cells in Tissue Development and RegenerationView all 7 articles
 Xuanran Feng1†
Xuanran Feng1† Xue Du1†
Xue Du1† Xiaoyu Yang1
Xiaoyu Yang1 Changqi Chen1
Changqi Chen1 Zhanping Liang1
Zhanping Liang1 Xiaonan Xu1
Xiaonan Xu1 Yi Wang2
Yi Wang2 Jialin C. Zheng1,3,4,5,6,7*
Jialin C. Zheng1,3,4,5,6,7* Xiaohuan Xia1,3,4,5,6*
Xiaohuan Xia1,3,4,5,6* Jianhui Liu1,3*
Jianhui Liu1,3*Background: MicroRNAs (miRNAs) have emerged as an essential regulator of the cell fate commitment of neural stem/progenitor cells (NPCs), although the impacts of certain miRNAs on NPCs remain vague. The aim of this study is to investigate the regulatory effects of miR-185-5p on the cell fate commitment of NPCs.
Methods: We investigated the impact of miR-185-5p on the proliferation and differentiation capacities of primary NPCs by manipulating the expression of miR-185-5p using specific mimics and inhibitors. The effects of miR-185-5p on NPCs was confirmed in vivo through stereotactic injection of miR-185-5p antagonists to the brains of mice at postnatal day 1 (P1).
Results: The expression levels of miR-185-5p kept increasing in the differentiation process of NPCs in vivo and in vitro. Perturbation of miR-185-5p’s function showed that miR-185-5p inhibited NPCs’ proliferation and promoted embryonic NPCs to differentiate more favorably to the glial lineage. We then validated the anti-proliferation and pro-glial roles of miR-185-5p using NPCs isolated from P1 mouse brains. In vivo study further showed enlarged NPCs pools and inhibited gliogenesis in the brains of P1 mice after animals received antagomir-185-5p.
Conclusion: Our study suggests miR-185-5p as an important regulator for the proliferation and glial fate commitment of NPCs.
During the development of the vertebrate central nervous system (CNS), multipotent neural stem/progenitor cells (NPCs) give rise to various types of neurons and glial cells in a spatially and temporally conserved pattern (Okano and Temple, 2009). Emerging evidence from multiple approaches suggests that the maintenance and differentiation of NPCs are regulated by dynamic interplay among transcription factors, epigenetic factors, non-coding RNAs (ncRNAs), and cell-extrinsic signals from the microenvironment in which NPCs reside. Among them, the involvement of ncRNAs, particular miRNAs, in NPCs regulation has received increasing attentions in the past decade (Ayana et al., 2017; D'Anca et al., 2022; Sharma et al., 2024; Wakabayashi et al., 2014). Among these ncRNAs, miRNAs are the mostly investigated one.
MicroRNAs (miRNAs) are evolutionary conserved ncRNAs that were first reported in 1993 (Lee et al., 1993; Blakaj and Lin, 2008). Being the smallest type of ncRNAs, miRNAs are typically 22–24 nucleotides in length. They bind to partially complementary sequences in the 3′ untranslated region (3′UTR) of target transcripts and regulate their expression at post-transcriptional level (Carthew and Sontheimer, 2009). miRNAs have the ability to play important roles in stem cell fate commitment, therefore controlling the developmental timing and regeneration capacity of the CNS (Sempere et al., 2004; Coolen and Bally-Cuif, 2009; Lopez-Ramirez and Nicoli, 2014; Ghibaudi et al., 2017). Notably, different miRNAs exhibit diverse impacts on cell fate commitment of NPCs, for instance, members of let-7 and miR-106b miRNA families regulate the proliferation of NPCs (Rybak et al., 2008; Xia et al., 2019a; Xia and Ahmad, 2016), while miR-9, miR-21, and miR-124 are important for neuronal induction (Zhao et al., 2009; Visvanathan et al., 2007; Xia et al., 2019b; Ma et al., 2019; Yuan et al., 2021). Our previous study has identified miR-185-5p as one differentially expressed miRNA during the differentiation process of retinal stem/progenitor cells (RPCs), NPC-like cells in the retina (Xia et al., 2019b). However, the expression patterns and functions of miR-185-5p in NPCs during brain development remain vague (Liu et al., 2019; Değerli et al., 2020). Our study found that the expression of miR-185-5p increased with embryonic development, suggesting that miR-185-5p may be involved in the regulation of cellular processes in NPCs. Therefore, it is essential to explore new functions of miR-185-5p to reveal new factors in the regulation of neural development.
In this study, we found that the expression levels of miR-185-5p in mouse brains increased from embryonic stage to adult one. The upregulation of miR-185-5p expression was observed in NPCs differentiation process in vitro, suggesting the involvement of miR-185-5p in neurogenesis. We then investigated the effect of miR-185-5p on mouse embryonic NPCs. Our in vitro studies showed that miR-185-5p is critical for inhibiting the proliferation of NPCs and promoting the differentiation of NPCs. We further verified that miR-185-5p is a key regulator in controlling the balance of NPCs proliferation and differentiation by administering antagomir-185-5p in vivo. Thus, our observations suggest miR-185-5p as a promotor of NPCs differentiation, regardless of the neuronal or glial lineage, shifting fate of NPCs from maintenance to differentiation.
NPCs were isolated from mouse fetal brain tissue as described previously (Ma et al., 2019). Briefly, cortical tissue was isolated from embryonic day 14 (E14) C57BL/6J mice and dissociated into single cells. Single cells were cultured for 3–4 days to form neurospheres in NPCs proliferation medium, which contained NeuroCult® NSC Basal Medium (Stem Cell Technologies), NeuroCult® NSC Proliferation Supplements (Stem Cell Technologies), 20 ng/mL bFGF (BioWalkersville), 20 ng/mL EGF (BioWalkersville), 2 μg/mL heparin (Sigma), N2 supplement, 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Primary neurospheres were collected, centrifuged at low speed to remove individual cells, dissociated into single cells by accutase (Sigma) at 37°C for 5 min, and a second round of neurospheres was formed in suspension culture. Enriched NPCs were harvested after three rounds of neurosphere formation for NPCs proliferation experiments.
Differentiation of NPCs was performed as described previously (Ma et al., 2019). Briefly, NPCs were seeded on Matrigel-coated coverslips in 24-well plates, 2 × 105 per well, which were supplemented with DMEM/F12 (Gibco) and supplemented with 1 × N2 supplement (Gibco), 1 × B27 supplement (ThermoFisher), 1.0 mM GlutaMAX (ThermoFisher), 10 ng/mL brain-derived neurotrophic factor (BDNF) (Peprotech), 10 ng/mL glial cell line-derived neurotrophic factor (GDNF) (Peprotech), 100 U/mL penicillin, and 100 μg/mL streptomycin. The medium was changed every 3 days.
Astrocyte differentiation medium consisted of DMEM/F12 (Gibco) and supplemented with 1 × N2 supplement (Gibco), 10% FBS (Gibco), 1.0 mM GlutaMAX (ThermoFisher), 100 U/mL penicillin, and 100 μg/mL streptomycin.
The mimic negtive control, miR-185-5p mimic, inhibitor negtive control, and miR-185-5p inhibitor were purchased from GenePharma (GenePharma Co., Ltd., Shanghai). Transfection was performed using Lipofectamine 2000 Reagent (Invitrogen) according to the manufacturer’s instructions.
The antagomir negative control and antagomiR-185-5p were purchased from GenePharma (GenePharma Co., Ltd., Shanghai). Mice were administered 2 μL of 100 μM antagomirs (antagomir-negative control or antagomiR-185-5p) bilaterally in the lateral ventricles.
The miRNA and mRNA were isolated from tissue or cell samples using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA was removed and cDNA was synthesized using the DNase I Digestion Kit (Qiagen) and miScript II Reverse Transcription Kit (Qiagen), respectively. Transcripts were amplified using gene-specific primers (Supplementary Table S1) and SYBR Green PCR kit (Qiagen) with ABI7500 (Applied Biosystems). All RT-qPCR results were measured in triplicate for each sample, and negative controls were used without template blanks. Amplification curves and gene expression were normalized to the housekeeping genes Gapdh (mRNA) and U6 snRNA (miRNA).
For cellular experiments, samples were fixed in 4% formaldehyde for 20 min at room temperature and then washed 3 times with PBS. For animal experiments, mouse brain tissue was cryosectioned and the brain slices were washed 3 times with PBS. Cell samples or brain slices were permeabilized with 0.4% Triton X-100 in PBS for 15 min, blocked with 5% BSA and 10% goat serum in PBS for 1 h at room temperature, and then incubated with primary antibody at 4°C overnight. Samples were washed 3 times with PBS and incubated with secondary antibody for 1 h at room temperature, blocked, and then images were acquired.
All results are shown as the means of at least three independent experiments ± S.D. The statistical difference between two independent groups was analyzed with the unpaired Student’s t-test. Data from multiple groups were statistically assessed by one-way ANOVA followed by Tukey post hoc tests. Significance was considered when p < 0.05.
To determine the temporal expression patterns of miR-185-5p during the differentiation process of NPCs, we first examined miR-185-5p expression levels in the cortical tissues of embryonic day 14 (E14), embryonic day 18 (E18), postnatal day 1 (P1), and adult mice (Figure 1A). We found that the expression levels of miR-185-5p increased with brain development from E14 and reached the maximum levels at P1 (Figure 1B). The expression level of miR-185-5p in the cortical tissues of adult mice was reduced compared to the P1 stage (Figure 1B). This trend showed a negative correlation with the mRNA levels of Nestin (Figure 1C), the expressed transcript corresponding to NPCs, and Ki67 (Figure 1D), the proliferation marker, and showed a positive correlation with the mRNA levels of the neuronal markers’ transcripts Tubb3 and Map2 (Figures 1E, F), and the astrocyte marker Gfap (Figure 1G). We further examined the expression level of miR-185-5p in the hippocampus and found that the expression levels of miR-185-5p in the mouse hippocampi gradually increased from E18 to adulthood (Figure 1H). This trend was opposite to that of Nestin (Figure 1I) and Ki67 (Figure 1J), but not completely consistent with that of Tubb3 (Figure 1K) and Map2 (Figure 1L). We found that the expression trend of miR-185-5p in the hippocampus was the same as Gfap (Figure 1M). The above results suggest that miR-185-5p may be related to the differentiation of NPCs.
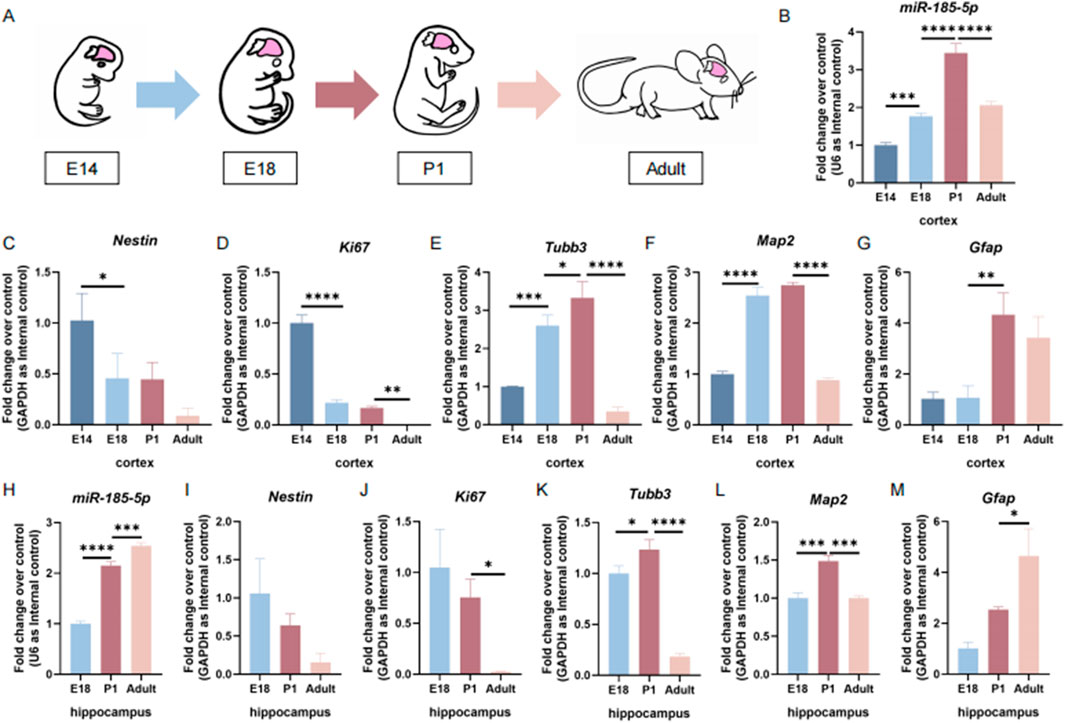
Figure 1. The temporal expression pattern of miR-185-5p corresponds to NPCs differentiation in vivo. (A) A schematic representation of the sample collection during brain development. (B) The temporal expression levels of miR-185-5p in cortical tissues were determined by RT-qPCR assay. (C–G) The expression levels of Nestin/Ki67/Tubb3/Map2/Gfap transcripts in cortical tissues were determined by RT-qPCR analysis. (H) The temporal expression levels of miR-185-5p in hippocampal tissues were determined by RT-qPCR assay. (I–M) The expression levels of Nestin/Ki67/Tubb3/Map2/Gfap transcripts in hippocampal tissues was determined by RT-qPCR analysis, respectively. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates.
Next, we determined the expression pattern of miR-185-5p in NPCs and differentiated cells in vitro (Figure 2A). E14 cortical dissociates cultured in the presence of EGF and FGF2 produced neurospheres enriched with Ki67/Nestin/Sox2 cells 3 days after inoculation, indicating an enrichment of NPCs (Figure 2B). NPCs were cultured under differentiation conditions and differentiated into β3-Tubulin-positive/Map2-positive neurons and GFAP+ glial cells (Figures 2B, C). The qPCR results showed that miR-185-5p was lowly expressed in NPCs but highly expressed in differentiated cells, and miR-185-5p expression gradually increased with the increase of differentiation days (Figure 2D), which had the same expression pattern as the transcript corresponding to Gfap and was different from Ki67, Tubb3, and Map2 (Figures 2E–H). Thus, in vivo and in vitro studies demonstrated a corresponding positive correlation between miR-185-5p expression and the differentiation of NPCs, especially with astrocytes, suggesting that its function is involved in the regulation of NPCs differentiation.
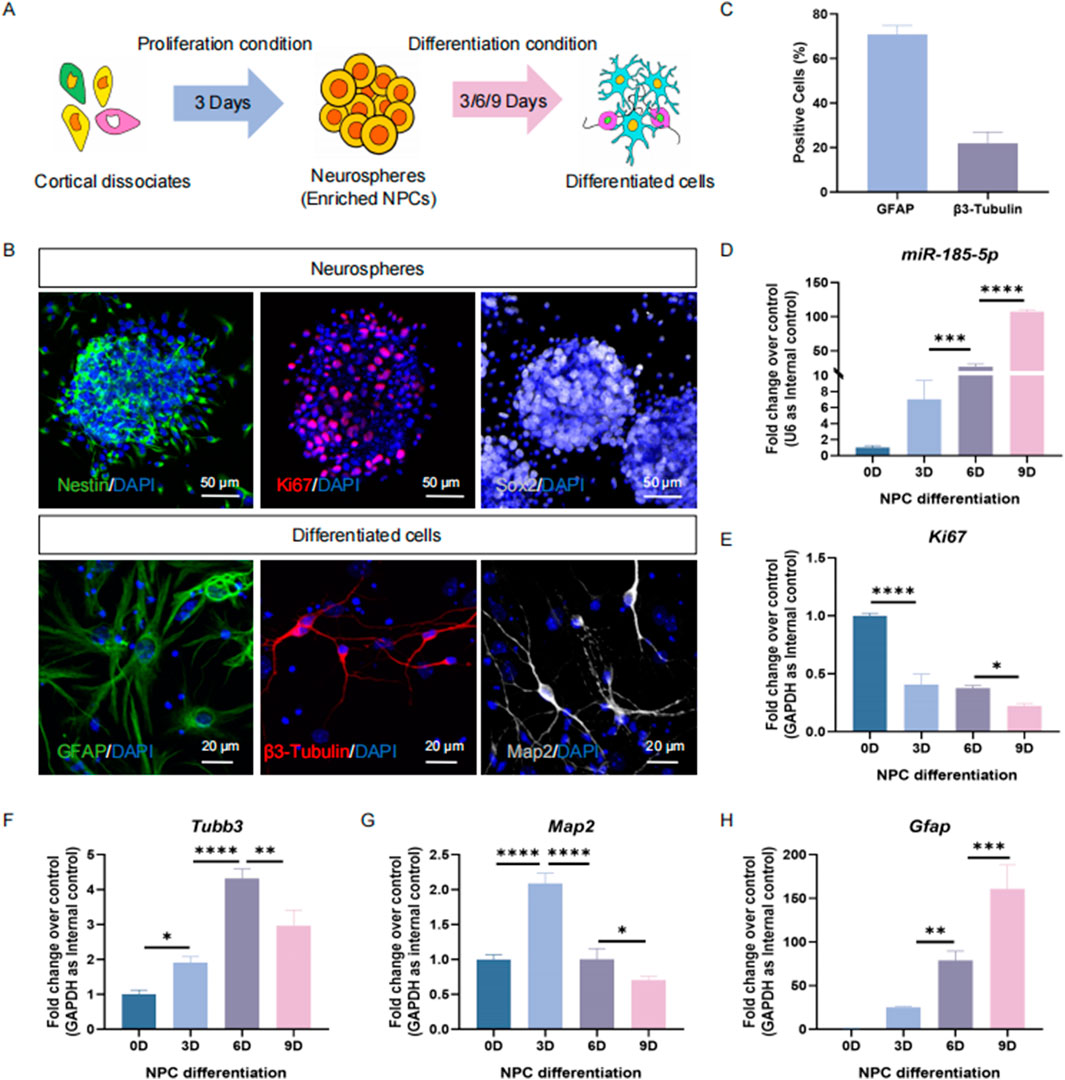
Figure 2. The temporal expression pattern of miR-185-5p corresponds to NPCs differentiation in vitro. (A) A schematic representation of the enrichment and differentiation of NPCs. (B) The enrichment and differentiation of NPCs was confirmed by immunoreactivities corresponding to protein markers of NPCs (β3-Tubulin-positive/Ki67/Sox2) and differentiated cells (GFAP/β3-Tubulin/Map2), respectively. Scale bar: 50 μm and 20 μm. (C) Number of GFAP-positive cells and β3-Tubulin-positive cells were quantified using Image J. (D) The expression levels of miR-185-5p during NPCs’ differentiation were determined by RT-qPCR analysis. (E–H) The expression levels of Gfap/Tubb3/Map2/Ki67 transcripts during NPCs’ differentiation were determined by RT-qPCR analysis. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates.
To understand the roles of miR-185-5p in the regulation of NPCs, we first investigated the involvement of miR-185-5p in the proliferation of NPCs. miR-185-5p loss-of-function (LOF) and gain-of-function (GOF) assays were performed using miR-185-5p-specific inhibitors and mimics, respectively (Figures 3A, I). In the miR-185-5p LOF assay, NPCs were transfected with either miR-185-5p inhibitor or inhibitor negative control (nc), and cultured under proliferative conditions for 3 days (Figure 3A). After 3 days’ culture, a significant increase in the number and size of neurospheres (Figures 3B–D) was observed, indicating enhanced proliferation and self-renewal of NPCs in the miR-185-5p inhibitor group compared to the inhibitor nc group. Transfection efficiency was verified by RT-qPCR (Figure 3E). The results revealed significantly reduced miR-185-5p expression levels in the miR-185-5p inhibitor group, compared with controls (Figure 3E). RT-qPCR analysis further showed that the expression levels of transcripts corresponding to Nestin, Sox2, and Ki67 were significantly increased in the miR-185-5p inhibitor group versus controls (Figure 3F). Moreover, the proportion of immunoreactive cells of Ki67, Nestin, and Sox2 in the miR-185-5p inhibitor group was significantly increased, compared with the inhibitor nc group (Figures 3G, H), confirming that miR-185-5p LOF promotes the proliferation of NPCs.
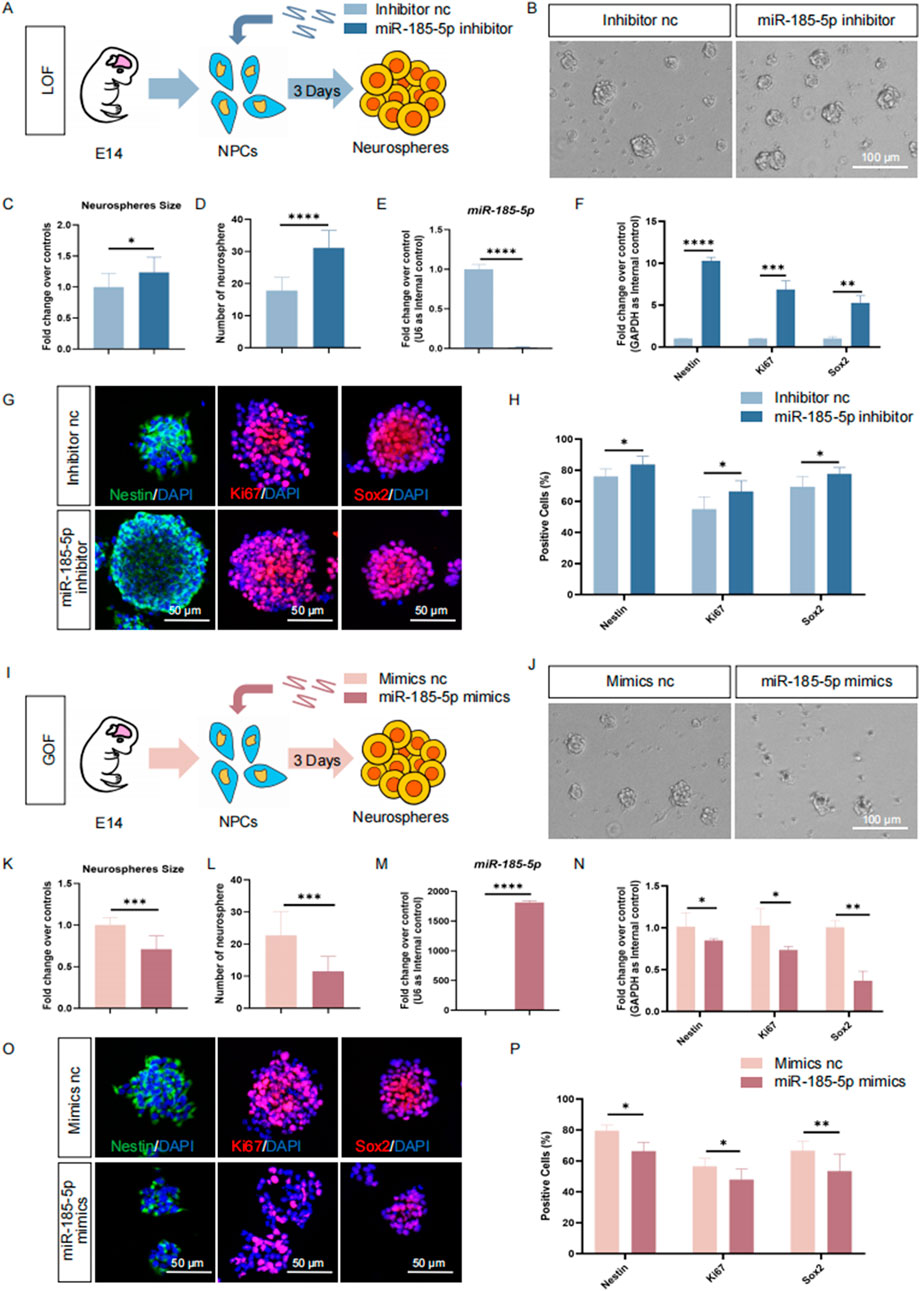
Figure 3. miR-185-5p inhibited the proliferation of NPCs. (A) A schematic representation of miR-185-5p LOF approach. (B) Bright field images of neurospheres in miR-185-5p LOF and control groups. (C, D) The number (C) and size (D) of neurospheres were quantified in the miR-185-5p LOF group, compared with controls. (E, F) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of NPCs (Nestin/Sox2) and proliferating cells (Ki67) in the miR-185-5p LOF groups, compared with controls. (G) Immunofluorescence analysis of transduced cells displaying NPCs (Nestin/Sox2) and proliferating cells (Ki67) specific immunoreactivities in LOF experiments. Scale bar: 50 μm. (H) Quantification of cells displaying immunoreactivities corresponding to NPCs and proliferating cells in the miR-185-5p LOF group, compared with controls. (I) A schematic representation of miR-185-5p GOF approach. (J) Bright field images of neurospheres in miR-185-5p LOF and control groups. (K, L) The number (K) and size (L) of neurospheres were quantified in the miR-185-5p GOF group, compared to controls. (M, N) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of NPCs (Nestin/Sox2) and proliferating cells (Ki67) in the miR-185-5p GOF groups, compared with controls. (O) Immunofluorescence analysis of transduced cells displaying NPCs (Nestin/Sox2) and proliferating cells (Ki67) specific immunoreactivities in GOF experiments. Scale bar: 50 μm. (P) Quantification of cells displaying immunoreactivities corresponding to NPCs and proliferating cells in the miR-185-5p GOF group, compared with controls. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation.
Next, we performed GOF using the same strategy as miR-185-5p LOF, in which NPCs were transfected with either miR-185-5p mimics or mimics negative control (nc) and cultured under proliferative conditions for 3 days (Figure 3I). We noticed a significant increase in the number and size of neurospheres in the group transfected with the miR-185-5p mimic, indicating enhanced proliferation and self-renewal of NPCs (Figures 3J–L). RT-qPCR analysis showed a significant increase in miR-185-5p levels (Figure 3M), validating the transfection efficiency. Compared with the mimics nc group, the miR-185-5p mimics group showed downregulation of Nestin, Sox2 and Ki67 transcript levels (Figure 3N), as well as a reduced proportion of cells with Ki67/Nestin/Sox2 specific immunoreactivity (Figures 3O, P), suggesting inhibition of proliferation and self-renewal of NPCs. Thus, our results suggest that miR-185-5p negatively regulates the proliferation of NPCs.
Next, we investigated the role of miR-185-5p in the differentiation of NPCs. Similar to previous studies, we performed LOF and GOF approaches to explore the role of miR-185-5p under differentiation conditions. We transfected NPCs with miR-185-5p inhibitor or inhibitor nc and cultured them under differentiation conditions for 3 days (Figure 4A). RT-qPCR analysis showed a significant decrease in miR-185-5p levels (Figure 4B), validating the transfection efficiency. We observed a reduction in the transcript expression levels of the astrocyte-specific markers Gfap and GS (Figures 4C, D), and of the neuron-specific marker Map2 compared to controls, with no significant difference in Tubb3 (Figures 4E, F). Immunofluorescence analysis showed a significant reduction in the percentage of immunoreactivity for GFAP (Figures 4G, H) and the percentage of immunoreactivity for β3-Tubulin showed a decreasing trend but no significant difference (Figure 4I), confirming that miR-185-5p LOF inhibited the differentiation of NPCs, especially their differentiation to astrocytes. In miR-185-5p GOF analysis, NPCs were transfected with miR-185-5p mimics or negative controls and cultured for 3 days under differentiation conditions (Figures 4J, K). Ectopic expression of miR-185-5p significantly increased the differentiation of glial cells compared to the results obtained by the LOF method, which was confirmed by a significant increase in the transcript levels corresponding to cell type-specific markers (Figures 4L–O) and in the number of cells displaying cell type-specific immunoreactivity (Figures 4P–R). Thus, both LOF and GOF studies suggest that miR-185-5p positively regulates the differentiation of NPCs, especially their differentiation to astrocytes.
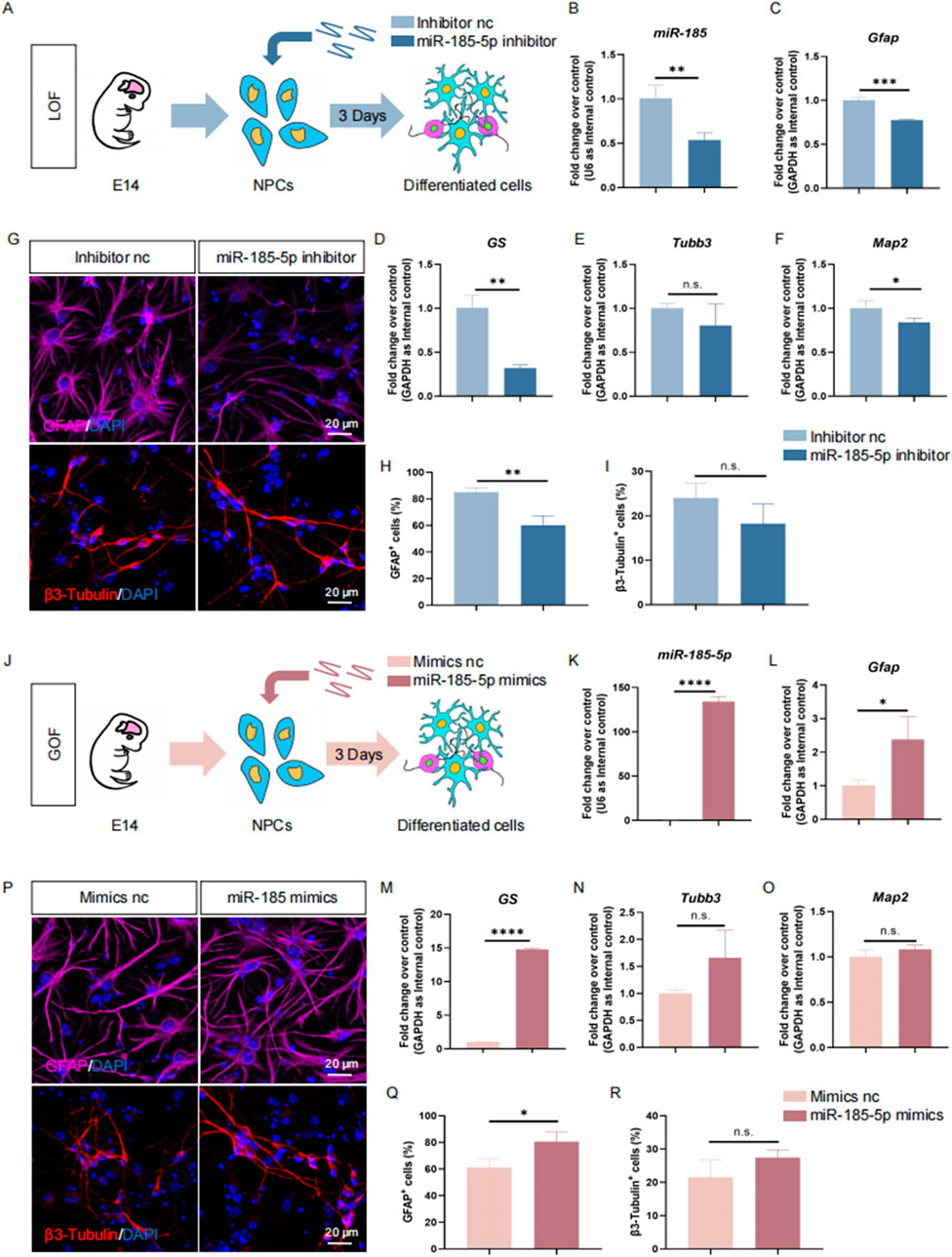
Figure 4. miR-185-5p promoted the differentiation of NPCs. (A) A schematic representation of miR-185-5p LOF approach. (B–F) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of differentiated cells (Gfap/GS/Tubb3/Map2) in the miR-185-5p LOF groups, compared with controls. (G) Representative images of immunofluorescence staining for the astrocyte-specific marker GFAP and the neuron-specific marker β3-Tubulin in LOF experiments. Scale bar: 20 μm. (H) Quantitative analysis of the number of GFAP-positive cells in LOF experiments. (I) Quantitative analysis of the number of β3-Tubulin-positive cells in LOF experiments. (J) A schematic representation of miR-185-5p GOF approach. (K–O) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of differentiated cells (Gfap/GS/Tubb3/Map2) in the miR-185-5p GOF groups, compared with controls. (P) Representative images of immunofluorescence staining for the astrocyte-specific marker GFAP and the neuron-specific marker β3-Tubulin in GOF experiments. Scale bar: 20 μm. (Q) Quantitative analysis of the number of GFAP-positive cells in GOF experiments. (R) Quantitative analysis of the number of β3-Tubulin-positive cells in GOF experiments. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation.
To further confirm the positive effects of miR-185-5p on NPCs’ differentiation to astrocyte, we isolated mouse NPCs in P1, a stage when gliogenesis occupies a dominant position versus neurogenesis (Dwyer et al., 2016; Guerra, 2014). We also performed proliferation experiments in P1 mouse-derived NPCs, and the results of our LOF assay (Figures 5A–H) and GOF assay (Figures 5I–P) indicated that miR-185-5p inhibited the proliferation and self-renewal of NPCs, which is consistent with the results we obtained in E14-derived NPCs. We induced these P1 NPCs to differentiate into astrocytes by culturing them in astrocyte differentiation medium for 3 days (Figures 6A–C). RT-qPCR analysis detected a significant increase in miR-185-5p expression after differentiation of NPCs to astrocytes (Figure 6D). Afterwards, both miR-185-5p LOF and GOF were conducted on P1 NPCs to determine the effects of miR-185-5p on astrocyte differentiation (Figures 6E, K). In the LOF assay, we verified the transfection efficiency of the miR-185-5p inhibitor (Figure 6F) and found that the mRNA levels of Gfap and GS were reduced in the miR-185-5p inhibitor group (Figures 6G, H) and the number of GFAP-positive cells was decreased in the immunofluorescence analysis compared to the inhibitor nc group (Figures 6I, J). In the GOF assay, we similarly verified the transfection efficiency of the miR-185-5p mimics (Figure 6L) and found increased mRNA levels of Gfap and GS in the miR-185-5p mimics group compared to the mimics nc group (Figures 6M, N), as verified by an increase in the number of GFAP-positive cells by immunofluorescence analysis (Figures 6O, P). The above results suggest that miR-185-5p is capable of promoting the differentiation of NPCs to astrocytes.
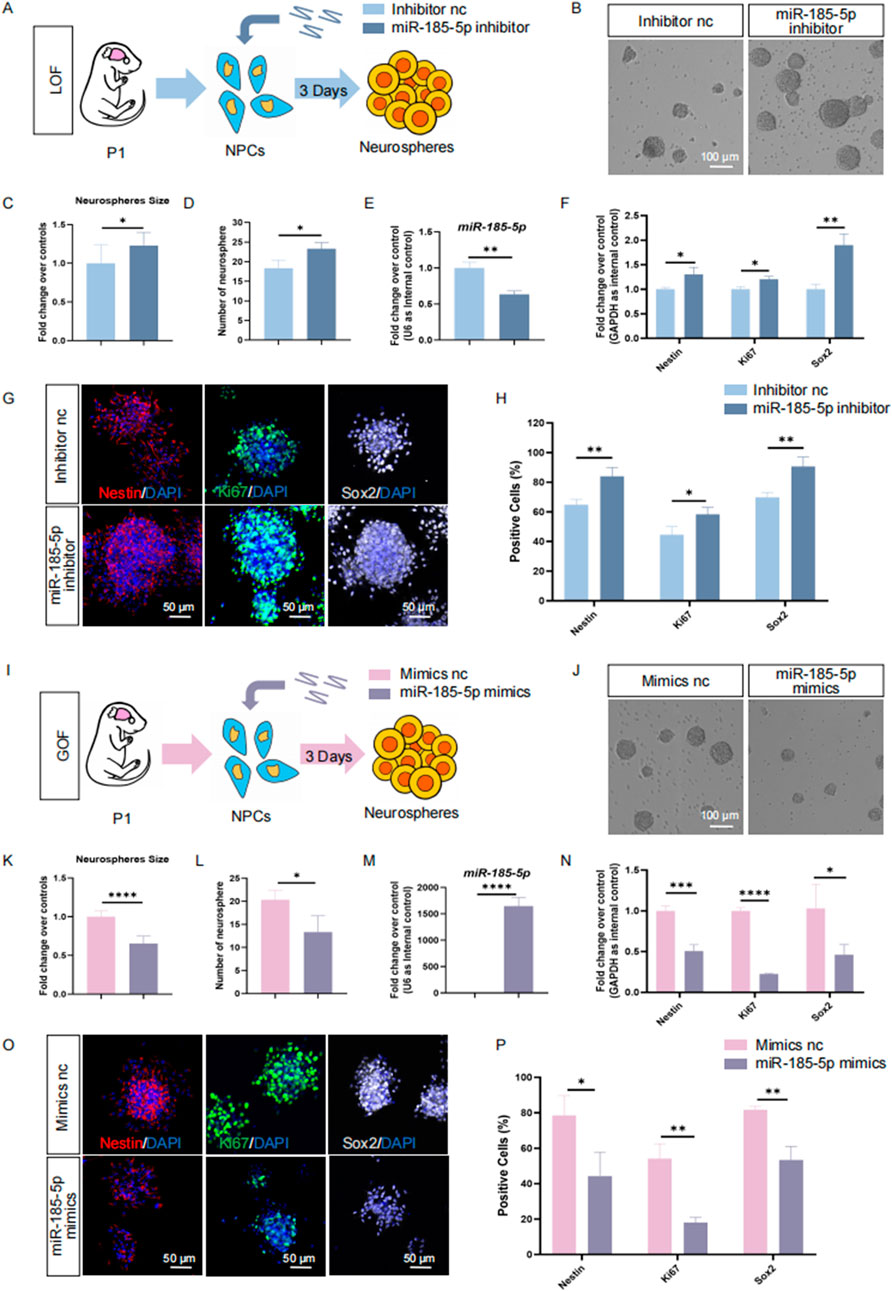
Figure 5. miR-185-5p inhibits proliferation of P1 NPCs. (A) A schematic representation of miR-185-5p LOF approach. (B) Bright field images of neurospheres in miR-185-5p GOF and control groups. (C, D) The number (C) and size (D) of neurospheres were quantified in the miR-185-5p LOF group, compared with controls. (E, F) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of NPCs (Nestin/Sox2) and proliferating cells (Ki67) in the miR-185-5p LOF groups, compared with controls. (G) Immunofluorescence analysis of transduced cells displaying NPCs (Nestin/Sox2) and proliferating cells (Ki67) specific immunoreactivities in LOF experiments. Scale bar: 50 μm. (H) Quantification of cells displaying immunoreactivities corresponding to NPCs and proliferating cells in the miR-185-5p LOF group, compared with controls. (I) A schematic representation of miR-185-5p GOF approach. (J) Bright field images of neurospheres in miR-185-5p LOF and control groups. (K, L) The number (K) and size (L) of neurospheres were quantified in the miR-185-5p GOF group, compared with controls. (M, N) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of NPCs (Nestin/Sox2) and proliferating cells (Ki67) in the miR-185-5p GOF groups, compared with controls. (O) Immunofluorescence analysis of transduced cells displaying NPCs (Nestin/Sox2) and proliferating cells (Ki67) specific immunoreactivities in GOF experiments. Scale bar: 50 μm. (P) Quantification of cells displaying immunoreactivities corresponding to NPCs and proliferating cells in the miR-185-5p GOF group, compared with controls. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation.
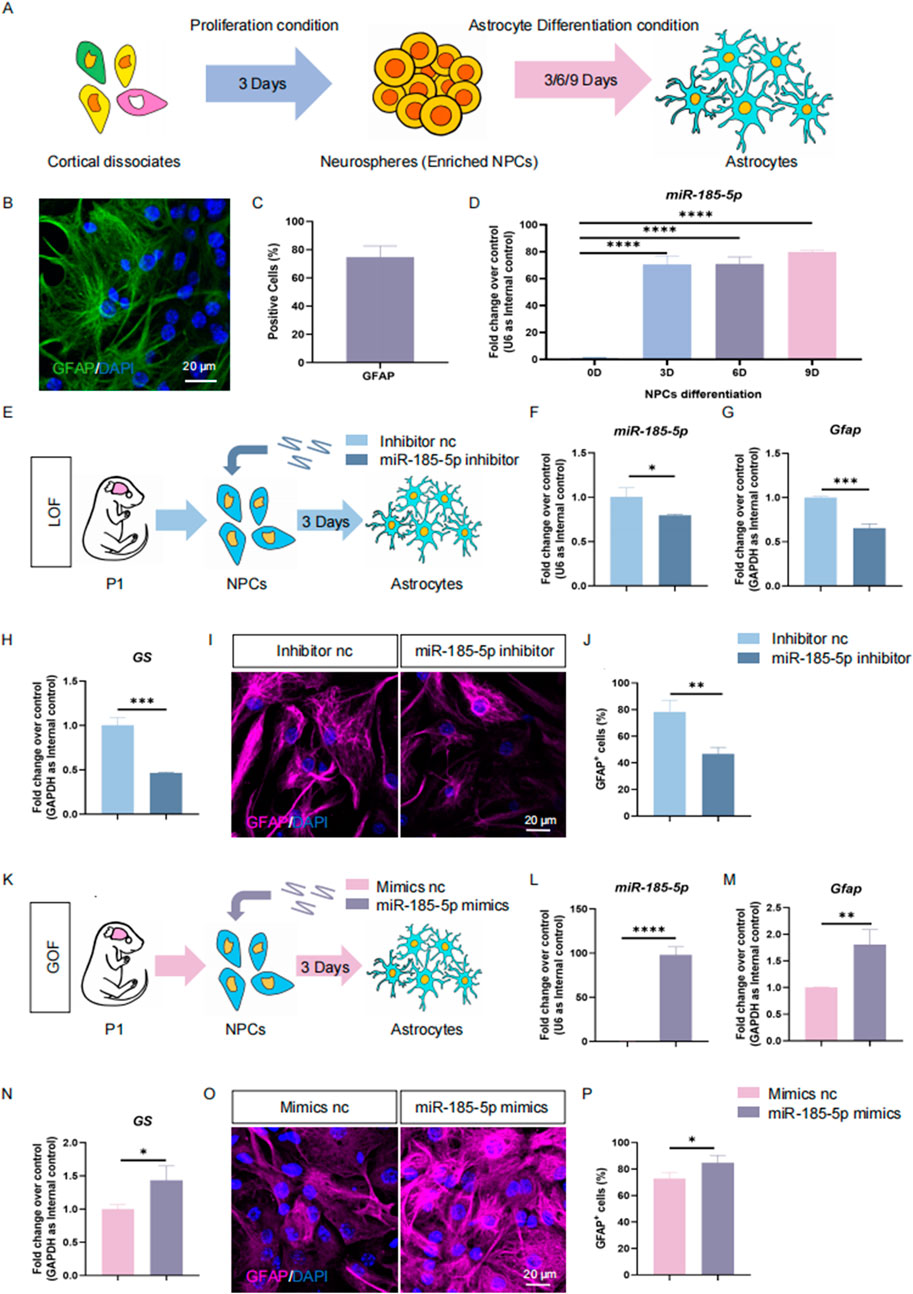
Figure 6. miR-185-5p promotes directed differentiation of P1 NPCs. (A) A schematic representation of the enrichment and astrocytes differentiation of NPCs. (B) Number of GFAP-positive cells were quantified using Image J. (C) Immunofluorescence staining for the astrocyte-specific marker GFAP identifies differentiation of NPCs to astrocytes. Scale bar: 20 μm. (D) RT-qPCR analysis showed that the expression levels of miR-185-5p were elevated in the differentiation of NPCs to astrocytes. (E) A schematic representation of miR-185-5p LOF approach. (F–H) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of astrocytes (Gfap/GS) in the miR-185-5p LOF groups, compared with controls. (I) Representative images of immunofluorescence staining for the astrocyte-specific marker GFAP in LOF experiments. Scale bar: 20 μm. (J) Quantitative analysis of the number of GFAP-positive cells in LOF experiments. (K) A schematic representation of miR-185-5p GOF approach. (L–N) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of astrocytes (Gfap/GS) in the miR-185-5p GOF groups, compared with controls. (O) Representative images of immunofluorescence staining for the astrocyte-specific marker GFAP in GOF experiments. Scale bar: 20 μm. (P) Quantitative analysis of the number of GFAP-positive cells in GOF experiments. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation.
Our in vitro studies demonstrated the critical importance of miR-185-5p in promoting the differentiation of NPCs, especially astrocytes. To further validate our observations, we performed miR-185-5p knockdown in P1 C57BL/6J mice. The lateral ventricles of P1 mice were injected with antagomir-185–5p (=LOF group) or antagomir negative control (NC), and the animals were executed 72 h later (Figure 7A). The expression of miR-185-5p was suppressed in the brain tissues of mice in the miR-185-5p LOF group compared with the antagomir NC group (Figure 7B), indicating that miR-185-5p was successfully knocked down in vivo. RT-qPCR results showed that the expression of transcripts corresponding to Gfap and GS was significantly reduced in the miR-185-5p LOF group compared with the antagomir NC group (Figures 7C, D). The proportion of cells showing GFAP immunoreactivity was significantly decreased in the miR-185-5p LOF group compared to the antagomir NC group in both the subgranular zone (SGZ) of the hippocampal dentate gyrus (Figures 7E, F) and the subventricular zone (SVZ) of the lateral ventricles, two main regions where NPCs exist in the mammalian brain throughout adulthood (Figures 7H, I). In addition, miR-185-5p LOF enhanced the proliferation of NPCs compared to the antagomir NC group, which was determined by the increase in the number of Ki67-positive cells in the miR-185-5p LOF group in the SGZ (Figures 7E,G) and SVZ (Figures 7H, J). Thus, our results suggest that miR-185-5p plays an essential role in promoting the differentiation of NPCs in vivo.
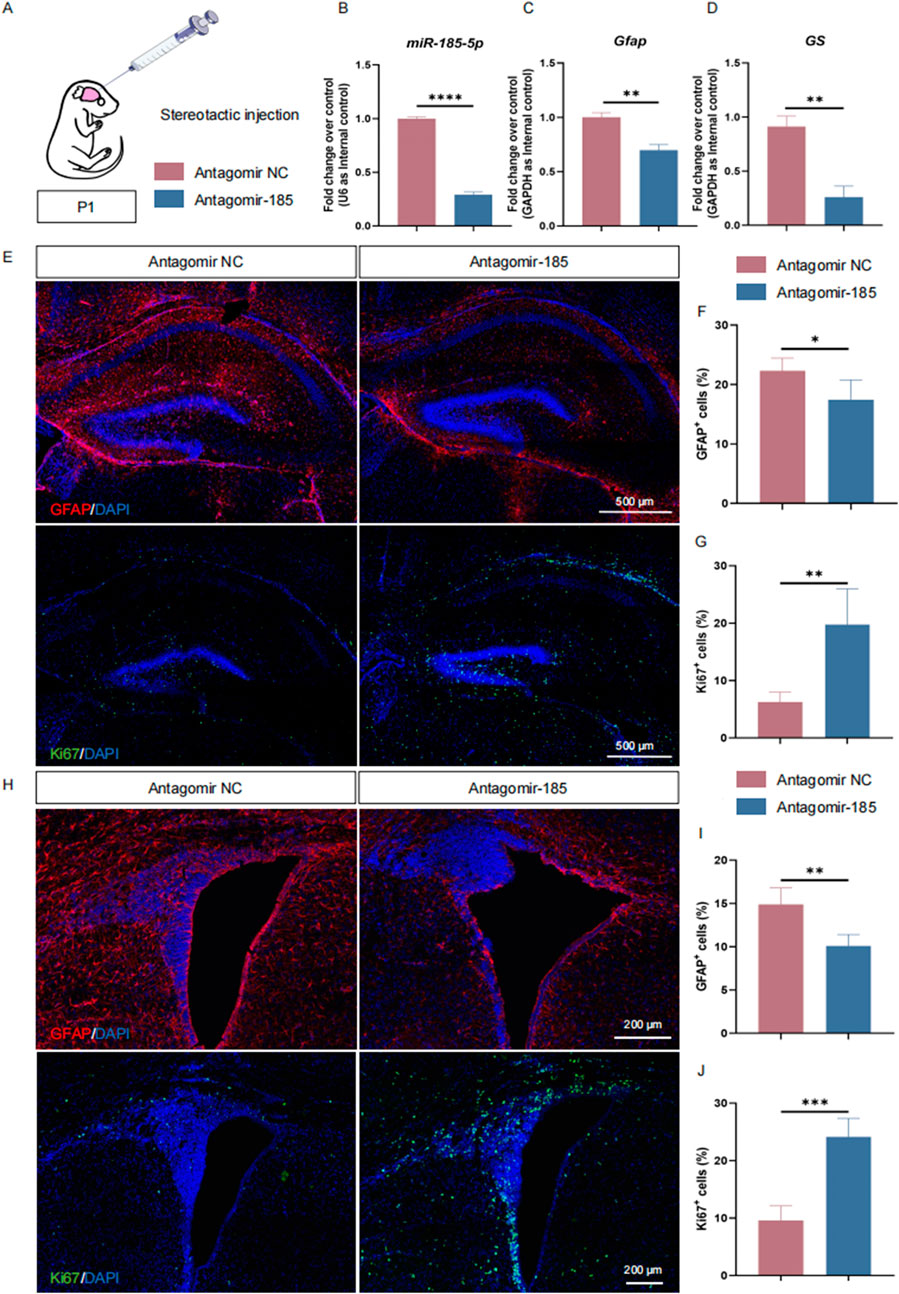
Figure 7. miR-185-5p regulates the proliferation and differentiation of NPCs in vivo. (A) A schematic representation of miR-185-5p LOF approach in vivo. (B–D) RT-qPCR analysis of expression levels of miR-185-5p and transcripts corresponding to markers of astrocytes (Gfap/GS) in the miR-185-5p LOF groups, compared with controls (n = 3). (E–G) Immunofluorescence analysis and quantification of transduced cells displaying GFAP and Ki67 specific immunoreactivities in the SGZ (n = 3). (H–J) Immunofluorescence analysis and quantification of transduced cells displaying GFAP and Ki67 specific immunoreactivities in the SVZ (n = 3). Scale bar: 500 μm and 200 μm. Data are mean ± S.D. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vivo perturbation.
The development of the vertebrate CNS is a highly conserved and dynamic process, characterized by distinct stages that begin with the formation of the neural tube (Rakic, 2009; Jessell and Sanes, 2000). The cell fate commitment of NPCs, including self-renewal and differentiation into diverse lineages, is crucial throughout these stages (Thor, 2024; Yoon et al., 2018). Deregulation of NPCs during CNS development leads to significant developmental abnormalities, including malformations and intellectual disabilities (Jessell and Sanes, 2000). Identifying the critical factors involved in the regulation of NPCs is essential for understanding CNS development, and could be of great advantage to the prevention and treatment of developmental defects.
In recent years, miRNAs have been found to regulate both the proliferation and differentiation capacities of NPCs in vitro and in vivo. In this study, we found that the expression levels of miR-185-5p increased as neurodevelopment proceeds in mouse cortical and hippocampal tissues. We also extracted primary NPCs to confirm the positive correlation of the expression levels of miR-185-5p with NPCs differentiation, and the negative correlation with NPCs proliferation. We then implemented LOF and GOF approaches, and found that miR-185-5p inhibits the proliferation of E14 NPCs and promotes the differentiation of NPCs into astrocytes in vitro. These effects of miR-185-5p on NPCs were further validated by perturbing the expression of miR-185-5p in NPCs isolated from mouse brains at P1, the time point when miR-185-5p expression reaches the maximum levels. More importantly, the lateral ventricle administration of antagomir-185-5p repressed the generation of astrocytes and enhanced cell proliferation in the SVZ and SGZ, two key brain regions where NPCs reside in the adulthood (Doetsch et al., 1997), matching with our observations in vitro.
Prior to our study, multiple studies of miR-185-5p have shown that miR-185-5p suppress tumor cell division, suggesting miR-185-5p an anti-tumor (anti-proliferation) miRNA (Wang et al., 2021; Tang et al., 2014). Afterwards, studies revealed that miR-185-5p also functions as an anti-proliferation miRNAs in many types of normal cells, including endothelial cells (Feng et al., 2022), aortic vascular smooth muscle cells (Wang et al., 2017), and mesangial cells (Wang et al., 2017). In our study, miR-185-5p significantly suppresses the proliferation of NPCs derived from embryonic and postnatal mouse brains. Thus, our results, together with others’ findings, indicate that miR-185-5p as a general blocker of proliferation not only in the CNS, but also in other types of cells under physiological and pathological conditions. Notably, the mechanisms of miR-185-5p -mediated proliferation inhibition of NPCs remains unclear. Inspiringly, researches on other types of cells can provide us with many insights. For example, miR-185-5p inhibit the proliferation of tumor cells by targeting multiple genes, including ROCK2 (Niu and Tang, 2019), IGF2 (Zhuang et al., 2020), RAGE (Yin et al., 2018), HMGA2 (Lu et al., 2022), BCL2 (Değerli et al., 2020), BCL2L1 (Değerli et al., 2020; Ostadrahimi et al., 2018), and KLF7 (Zhao et al., 2019). Our previous study has identified HMGA2, a key chromatin-associated protein, as a key gene enhancing proliferation of RPCs (Xia and Ahmad, 2016; Parameswaran et al., 2014). Besides, the inhibition of either ROCK (Zhang et al., 2016), IGF2 (Yang et al., 2015), or RAGE (Li et al., 2014) pathways has also been reported to suppress the proliferation of NSCs. Hence, aforementioned may regulate proliferation of NPCs via inhibiting aforementioned pathways, which needs to be investigated in the future studies.
Besides the anti-proliferation effect, we also found that miR-185-5p plays a key role in gliogenesis in vitro and in vivo. The development of the CNS follows a strict sequence, and one of the well-established aspects is that neurogenesis precedes gliogenesis. There are multiple miRNAs that are found to regulate neurogliogenesis decision including miR-9, miR-124, and miR-29 (Xia et al., 2019b; Åkerblom and Jakobsson, 2014; Coolen et al., 2013). For example, we have found the miR-9 enhances neurogenesis and inhibits gliogenesis by targeting HES1, a downstream transcription factor of NOTCH signaling (Yuan et al., 2021). Moreover, miR-9 also enhances neurogenesis through MCPIP1 (Yang et al., 2013), ELAVL3 (Coolen et al., 2012), TLX (Madelaine et al., 2017), and ONECUTs (Madelaine et al., 2017). Another well-recognized pro-neural miRNA is miR-124, which promotes neurogenesis via targeting NEUROD1 (Liu et al., 2011) and AAK1 (Song et al., 2023). Besides, our recent study also identified miR-29 as a regulator of neuroligogenesis by down-regulating REST expression (Xia et al., 2019b). Here, we identified miR-185-5p as a novel miRNA for promoting gliogenesis, although the underlying molecular mechanisms remains unknown. Based on the information from online database Targetscan.org, miR-185-5p does not directly binds to the well-known miR-9/124/29 target transcripts like HES1, TLX, and REST, suggesting that miR-185-5p regulates NPCs proliferation and differentiation through distinct mechanisms. Interestingly, in addition to regulating proliferation, HMGA2 also has the functions of enhancing neurogenesis and suppressing gliogenesis through regulating HES5, a NOTCH effector and well-established node that facilitates the astrocytic fate commitment in the CNS (Bansod et al., 2017; Patterson et al., 2014). These findings imply HMGA2 as a core downstream target of miR-185-5p that mediates the anti-proliferative and pro-glial effects of miR-185-5p on NPCs. Another possible target of miR-185-5p in the regulation of gliogenesis is IGF2 pathway (Zhuang et al., 2020), which controls the timing of neurogliogenesis during brain development (Balzer et al., 2010). Future investigations that aim to clarify the molecular mechanisms of miR-185-5p-mediated astrocytic commitment are urgently needed.
It is worth noting that, besides the regulatory effects on NPCs and brain development, miR-185-5p also participate in the modulation of neurological disease progression. For example, we detected reduced miR-185-5p levels in the serum of Alzheimer’s disease (AD) patients (Ding et al., 2022). We further found that miR-185-5p negatively regulates the expression of amyloid precursor protein (APP) through the direct binding to the APP 3′UTR (Ding et al., 2022). Therefore, exosomal miR-185-5p in the microenvironment of the CNS functions as a blocker for APP expression, and the loss of miR-185-5p in exosomes releases APP mRNAs from degradation or translational inhibition in the recipient cells to promote Aβ production (Ding et al., 2022). Downregulation of miR-185-5p has also been identified as a common pathogenic event in 22q11.2 deletion syndrome-related and idiopathic schizophrenia (Sabaie et al., 2022). Moreover, reduced miR-185-5p levels were found in glioma tissues, indicating that miR-185-5p was associated with a poor outcome in glioma patients (Tang et al., 2012). Thus, these observations, together with our findings, imply a great potential of miR-185-5p as a biomarker for neurological diseases, particularly development disorders and brain tumors that directly associate with dysregulated proliferation of normal or cancer cells in the brains. For instance, lowered circulating miR-185-5p has been reported as a predictive biomarker for diagnosis and prognosis of glioma (Tabibkhooei et al., 2020; Tang et al., 2015). Besides, as anti-proliferative effects of miR-185-5p have been reported in brain tumor (Guan et al., 2022), miR-185-5p may also serve as a potential drug for brain tumors, which will be investigated in our future works.
In conclusion, we found that miR-185-5p expression levels increase during both the development of mouse brains and differentiation process of NPCs. The upregulation of miR-185-5p expression suppresses proliferation of NPCs and facilitates astrocyte differentiation in vitro and in vivo. Thus, our study identified miR-185-5p as a novel player in the cell fate commitment of NPCs, suggesting the complexity of miRNA-mediated regulation in the brain development.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was approved by The Institutional Animal Care and Use Committee of Tongji University School of Medicine. The study was conducted in accordance with the local legislation and institutional requirements.
XF: Data curation, Formal Analysis, Methodology, Project administration, Writing–original draft. XD: Data curation, Formal Analysis, Writing–original draft. XY: Data curation, Formal Analysis, Writing–original draft. CC: Data curation, Formal Analysis, Writing–original draft. ZL: Data curation, Writing–original draft. XNX: Data curation, Writing–original draft. YW: Methodology, Writing–review and editing. JZ: Conceptualization, Project administration, Supervision, Validation, Writing–review and editing. XHX: Conceptualization, Formal Analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing–review and editing. JL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by research grants from the National Natural Science Foundation of China (Nos. 82171194 and 82371204 to JL).
We thank Qihui Wu, Jie Zhu, Yanyan Zhang, Lu Ding and Zhihao Sheng for technical assistance and proofreading the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1510746/full#supplementary-material
NPCs, neural stem/progenitor cells; miRNA, microRNA; 3′UTR, 3′ untranslated region; CNS, central nervous system; E14, embryonic day 14; E18, embryonic day 18; P1, postnatal day 1; SD, standard deviation; APP, amyloid precursor protein; AD, Alzheimer’s disease.
Åkerblom, M., and Jakobsson, J. (2014). MicroRNAs as neuronal fate determinants. Neuroscientist 20 (3), 235–242. doi:10.1177/1073858413497265
Ayana, R., Singh, S., and Pati, S. (2017). Decoding crucial LncRNAs implicated in neurogenesis and neurological disorders. Stem Cells Dev. 26 (8), 541–553. doi:10.1089/scd.2016.0290
Balzer, E., Heine, C., Jiang, Q., Lee, V. M., and Moss, E. G. (2010). LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 137 (6), 891–900. doi:10.1242/dev.042895
Bansod, S., Kageyama, R., and Ohtsuka, T. (2017). Hes5 regulates the transition timing of neurogenesis and gliogenesis in mammalian neocortical development. Development 144 (17), 3156–3167. doi:10.1242/dev.147256
Blakaj, A., and Lin, H. (2008). Piecing together the mosaic of early mammalian development through microRNAs. J. Biol. Chem. 283 (15), 9505–9508. doi:10.1074/jbc.R800002200
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136 (4), 642–655. doi:10.1016/j.cell.2009.01.035
Coolen, M., and Bally-Cuif, L. (2009). MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol. 19 (5), 461–470. doi:10.1016/j.conb.2009.09.006
Coolen, M., Katz, S., and Bally-Cuif, L. (2013). miR-9: a versatile regulator of neurogenesis. Front. Cell Neurosci. 7, 220. doi:10.3389/fncel.2013.00220
Coolen, M., Thieffry, D., Drivenes, Ø., Becker, T. S., and Bally-Cuif, L. (2012). miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell 22 (5), 1052–1064. doi:10.1016/j.devcel.2012.03.003
D'Anca, M., Buccellato, F. R., Fenoglio, C., and Galimberti, D. (2022). Circular RNAs: emblematic players of neurogenesis and neurodegeneration. Int. J. Mol. Sci. 23 (8), 4134. doi:10.3390/ijms23084134
Değerli, E., Torun, V., and Cansaran-Duman, D. (2020). miR-185-5p response to usnic acid suppresses proliferation and regulating apoptosis in breast cancer cell by targeting Bcl2. Biol. Res. 53 (1), 19. doi:10.1186/s40659-020-00285-4
Ding, L., Yang, X., Xia, X., Li, Y., Wang, Y., Li, C., et al. (2022). Exosomes mediate APP dysregulation via APP-miR-185-5p Axis. Front. Cell Dev. Biol. 10, 793388. doi:10.3389/fcell.2022.793388
Doetsch, F., García-Verdugo, J. M., and Alvarez-Buylla, A. (1997). Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 17 (13), 5046–5061. doi:10.1523/JNEUROSCI.17-13-05046.1997
Dwyer, N. D., Chen, B., Chou, S. J., Hippenmeyer, S., Nguyen, L., and Ghashghaei, H. T. (2016). Neural stem cells to cerebral cortex: emerging mechanisms regulating progenitor behavior and productivity. J. Neurosci. 36 (45), 11394–11401. doi:10.1523/JNEUROSCI.2359-16.2016
Feng, N., Wang, Z., Wu, Y., Zheng, H., Jiang, X., Wang, Z., et al. (2022). ADAMTS9-AS2 promotes angiogenesis of brain microvascular endothelial cells through regulating miR-185-5p/IGFBP-2 Axis in ischemic stroke. Mol. Neurobiol. 59 (4), 2593–2604. doi:10.1007/s12035-021-02641-1
Ghibaudi, M., Boido, M., and Vercelli, A. (2017). Functional integration of complex miRNA networks in central and peripheral lesion and axonal regeneration. Prog. Neurobiol. 158, 69–93. doi:10.1016/j.pneurobio.2017.07.005
Guan, Y., Yang, W., Zhang, F., Zhang, L., and Wang, L. (2022). CircPOSTN competes with KIF1B for miR-185-5p binding sites to promote the tumorigenesis of glioma. Brain Res. Bull. 180, 86–96. doi:10.1016/j.brainresbull.2021.12.014
Guerra, M. (2014). Neural stem cells: are they the hope of a better life for patients with fetal-onset hydrocephalus? Fluids Barriers CNS 11, 7. doi:10.1186/2045-8118-11-7
Jessell, T. M., and Sanes, J. R. (2000). Development. The decade of the developing brain. Curr. Opin. Neurobiol. 10 (5), 599–611. doi:10.1016/s0959-4388(00)00136-7
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Li, M., Sun, L., Luo, Y., Xie, C., Pang, Y., and Li, Y. (2014). High-mobility group box 1 released from astrocytes promotes the proliferation of cultured neural stem/progenitor cells. Int. J. Mol. Med. 34 (3), 705–714. doi:10.3892/ijmm.2014.1820
Liu, C., Cai, L., and Li, H. (2019). miR-185 regulates the growth of osteosarcoma cells via targeting Hexokinase 2. Mol. Med. Rep. 20 (3), 2774–2782. doi:10.3892/mmr.2019.10534
Liu, K., Liu, Y., Mo, W., Qiu, R., Wang, X., Wu, J. Y., et al. (2011). MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 39 (7), 2869–2879. doi:10.1093/nar/gkq904
Lopez-Ramirez, M. A., and Nicoli, S. (2014). Role of miRNAs and epigenetics in neural stem cell fate determination. Epigenetics 9 (1), 90–100. doi:10.4161/epi.27536
Lu, W., Huang, Z., Wang, J., and Liu, H. (2022). Long non-coding RNA DANCR accelerates colorectal cancer progression via regulating the miR-185-5p/HMGA2 axis. J. Biochem. 171 (4), 389–398. doi:10.1093/jb/mvab011
Ma, Y., Li, C., Huang, Y., Wang, Y., Xia, X., and Zheng, J. C. (2019). Exosomes released from neural progenitor cells and induced neural progenitor cells regulate neurogenesis through miR-21a. Cell Commun. Signal 17 (1), 96. doi:10.1186/s12964-019-0418-3
Madelaine, R., Sloan, S. A., Huber, N., Notwell, J. H., Leung, L. C., Skariah, G., et al. (2017). MicroRNA-9 couples brain neurogenesis and angiogenesis. Cell Rep. 20 (7), 1533–1542. doi:10.1016/j.celrep.2017.07.051
Niu, Y., and Tang, G. (2019). miR-185-5p targets ROCK2 and inhibits cell migration and invasion of hepatocellular carcinoma. Oncol. Lett. 17 (6), 5087–5093. doi:10.3892/ol.2019.10144
Okano, H., and Temple, S. (2009). Cell types to order: temporal specification of CNS stem cells. Curr. Opin. Neurobiol. 19 (2), 112–119. doi:10.1016/j.conb.2009.04.003
Ostadrahimi, S., Abedi Valugerdi, M., Hassan, M., Haddad, G., Fayaz, S., Parvizhamidi, M., et al. (2018). miR-1266-5p and miR-185-5p promote cell apoptosis in human prostate cancer cell lines. Asian Pac J. Cancer Prev. 19 (8), 2305–2311. doi:10.22034/APJCP.2018.19.8.2305
Parameswaran, S., Xia, X., Hegde, G., and Ahmad, I. (2014). Hmga2 regulates self-renewal of retinal progenitors. Development 141 (21), 4087–4097. doi:10.1242/dev.107326
Patterson, M., Gaeta, X., Loo, K., Edwards, M., Smale, S., Cinkornpumin, J., et al. (2014). let-7 miRNAs can act through notch to regulate human gliogenesis. Stem Cell Rep. 3 (5), 758–773. doi:10.1016/j.stemcr.2014.08.015
Rakic, P. (2009). Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10 (10), 724–735. doi:10.1038/nrn2719
Rybak, A., Fuchs, H., Smirnova, L., Brandt, C., Pohl, E. E., Nitsch, R., et al. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10 (8), 987–993. doi:10.1038/ncb1759
Sabaie, H., Gharesouran, J., Asadi, M. R., Farhang, S., Ahangar, N. K., Brand, S., et al. (2022). Downregulation of miR-185 is a common pathogenic event in 22q11.2 deletion syndrome-related and idiopathic schizophrenia. Metab. Brain Dis. 37 (4), 1175–1184. doi:10.1007/s11011-022-00918-5
Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5 (3), R13. doi:10.1186/gb-2004-5-3-r13
Sharma, H., Kaushik, M., Goswami, P., Sreevani, S., Chakraborty, A., Ashique, S., et al. (2024). Role of miRNAs in brain development. Microrna 13 (2), 96–109. doi:10.2174/0122115366287127240322054519
Song, Y., Shi, R., Liu, Y., Cui, F., Han, L., Wang, C., et al. (2023). M2 microglia extracellular vesicle miR-124 regulates neural stem cell differentiation in ischemic stroke via AAK1/NOTCH. Stroke 54 (10), 2629–2639. doi:10.1161/STROKEAHA.122.041611
Tabibkhooei, A., Izadpanahi, M., Arab, A., Zare-Mirzaei, A., Minaeian, S., Rostami, A., et al. (2020). Profiling of novel circulating microRNAs as a non-invasive biomarker in diagnosis and follow-up of high and low-grade gliomas. Clin. Neurol. Neurosurg. 190, 105652. doi:10.1016/j.clineuro.2019.105652
Tang, H., Liu, P., Yang, L., Xie, X., Ye, F., Wu, M., et al. (2014). miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol. Cancer Ther. 13 (12), 3185–3197. doi:10.1158/1535-7163.MCT-14-0243
Tang, H., Liu, Q., Liu, X., Ye, F., Xie, X., Xie, X., et al. (2015). Plasma miR-185 as a predictive biomarker for prognosis of malignant glioma. J. Cancer Res. Ther. 11 (3), 630–634. doi:10.4103/0973-1482.146121
Tang, H., Wang, Z., Liu, X., Liu, Q., Xu, G., Li, G., et al. (2012). LRRC4 inhibits glioma cell growth and invasion through a miR-185-dependent pathway. Curr. Cancer Drug Targets 12 (8), 1032–1042. doi:10.2174/156800912803251180
Thor, S. (2024). Indirect neurogenesis in space and time. Nat. Rev. Neurosci. 25 (8), 519–534. doi:10.1038/s41583-024-00833-x
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21 (7), 744–749. doi:10.1101/gad.1519107
Wakabayashi, T., Hidaka, R., Fujimaki, S., Asashima, M., and Kuwabara, T. (2014). MicroRNAs and epigenetics in adult neurogenesis. Adv. Genet. 86, 27–44. doi:10.1016/B978-0-12-800222-3.00002-4
Wang, S., Tang, L., Zhou, Q., Lu, D., Duan, W., Chen, C., et al. (2017). miR-185/P2Y6 Axis inhibits angiotensin II-induced human aortic vascular smooth muscle cell proliferation. DNA Cell Biol. 36 (5), 377–385. doi:10.1089/dna.2016.3605
Wang, X., Zhu, X., and Zhao, Y. (2021). Targeting miR-185-3p inhibits head and neck squamous cell carcinoma by modulating RAB25. Front. Oncol. 11, 721416. doi:10.3389/fonc.2021.721416
Xia, X., and Ahmad, I. (2016). let-7 microRNA regulates neurogliogenesis in the mammalian retina through Hmga2. Dev. Biol. 410 (1), 70–85. doi:10.1016/j.ydbio.2015.12.010
Xia, X., Lu, H., Li, C., Huang, Y., Wang, Y., Yang, X., et al. (2019a). miR-106b regulates the proliferation and differentiation of neural stem/progenitor cells through Tp53inp1-Tp53-Cdkn1a axis. Stem Cell Res. Ther. 10 (1), 282. doi:10.1186/s13287-019-1387-6
Xia, X., Teotia, P., and Ahmad, I. (2019b). miR-29c regulates neurogliogenesis in the mammalian retina through REST. Dev. Biol. 450 (2), 90–100. doi:10.1016/j.ydbio.2019.03.013
Yang, L., Chao, J., Kook, Y. H., Gao, Y., Yao, H., and Buch, S. J. (2013). Involvement of miR-9/MCPIP1 axis in PDGF-BB-mediated neurogenesis in neuronal progenitor cells. Cell Death Dis. 4 (12), e960. doi:10.1038/cddis.2013.486
Yang, M., Yang, S. L., Herrlinger, S., Liang, C., Dzieciatkowska, M., Hansen, K. C., et al. (2015). Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development 142 (9), 1616–1627. doi:10.1242/dev.120543
Yin, C., Zhang, G., Sun, R., Pan, X., Wang, X., Li, H., et al. (2018). miR-185-5p inhibits F-actin polymerization and reverses epithelial mesenchymal transition of human breast cancer cells by modulating RAGE. Mol. Med. Rep. 18 (3), 2621–2630. doi:10.3892/mmr.2018.9294
Yoon, K. J., Vissers, C., Ming, G. L., and Song, H. (2018). Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence. J. Cell Biol. 217 (6), 1901–1914. doi:10.1083/jcb.201802117
Yuan, P., Ding, L., Chen, H., Wang, Y., Li, C., Zhao, S., et al. (2021). Neural stem cell-derived exosomes regulate neural stem cell differentiation through miR-9-hes1 Axis. Front. Cell Dev. Biol. 9, 601600. doi:10.3389/fcell.2021.601600
Zhang, C., Wu, J. M., Liao, M., Wang, J. L., and Xu, C. J. (2016). The ROCK/GGTase pathway are essential to the proliferation and differentiation of neural stem cells mediated by simvastatin. J. Mol. Neurosci. 60 (4), 474–485. doi:10.1007/s12031-016-0811-y
Zhao, C., Sun, G., Li, S., and Shi, Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16 (4), 365–371. doi:10.1038/nsmb.1576
Zhao, L., Zhang, Y., Liu, J., Yin, W., Jin, D., Wang, D., et al. (2019). miR-185 inhibits the proliferation and invasion of non-small cell lung cancer by targeting KLF7. Oncol. Res. 27 (9), 1015–1023. doi:10.3727/096504018X15247341491655
Keywords: miR-185-5p, neural stem/progenitor cells, proliferation, differentiation, astrocytes
Citation: Feng X, Du X, Yang X, Chen C, Liang Z, Xu X, Wang Y, Zheng JC, Xia X and Liu J (2024) miR-185-5p regulates the proliferation and differentiation of neural stem/progenitor cells. Front. Cell Dev. Biol. 12:1510746. doi: 10.3389/fcell.2024.1510746
Received: 13 October 2024; Accepted: 20 November 2024;
Published: 05 December 2024.
Edited by:
Finosh Thankam, Western University of Health Sciences, United StatesReviewed by:
Sini Sunny, University of Alabama at Birmingham, United StatesCopyright © 2024 Feng, Du, Yang, Chen, Liang, Xu, Wang, Zheng, Xia and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jialin C. Zheng, amlhbGluemhlbmdAdG9uZ2ppLmVkdS5jbg==; Xiaohuan Xia, eGlhb2h1YW5feGlhMUAxNjMuY29t; Jianhui Liu, amlhbmh1aWxpdV8xMjQ2QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.