- 1Institute of Integrated Traditional Chinese and Western Medicine, Chinese Evidence-based Medicine Center, West China Hospital, Sichuan University, Chengdu, China
- 2Department of Orthopedics, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 3Clinical School of Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 4Department of Orthopedics, The Second Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, China
Background: Osteosarcoma is the most prevalent among primary bone malignancies, and its standard intervention involves neoadjuvant chemotherapy - surgical adjuvant chemotherapy (MAP regimen) with adriamycin, cisplatin, and high-dose methotrexate. Early-stage osteosarcoma can be effectively treated with surgical resection along with chemotherapy or radiotherapy. However, as the cancer progresses, the efficacy of chemo- and radiotherapy decreases, and the associated problems increase. The current understanding of osteosarcoma development, diagnosis, and treatment does not meet clinical demands. More recently, there has been a significant increase in exosome-associated osteosarcoma research, potentially opening up novel possibilities for osteosarcoma research.
Purpose: We comprehensively evaluated and analyzed the advancement of preclinical research related to exosome-osteosarcoma. We aimed to establish a practical, theoretical foundation for future research initiatives.
Study design: The selected design was a systematic review and meta-analysis.
Methods: Scientific databases, such as PubMed, Embase, The Cochrane Library, and Web of Science, were extensively screened for exosome and osteosarcoma articles. Two highly trained investigators separately reviewed the literature, extracted relevant information, and assessed study quality. Subsequently, we conducted a meta-analysis using Review Manager 5.4.
Results: In total, 25 animal-based randomized controlled trials (RCTs) were selected for analysis. Among them, 13 studies provided strong evidence of cellular exosomes regulating osteosarcoma development from bone marrow mesenchymal stem cells, osteosarcoma cells, and macrophages. In addition, 12 studies demonstrated the therapeutic potential of exosomes in managing osteosarcoma, among which 7 studies transplanted transfected exosomes directly into animals as drugs, and five studies employed exosomes as drug carriers, which were next transplanted into animals.
Conclusion: Based on our meta-analysis, macrophages strongly modulate osteosarcoma development, and engineered exosomes provide the most effective exosome-based osteosarcoma treatment.
1 Introduction
Osteosarcoma ranks first among all bone-related primary malignant tumors, with a reported incidence of about 4.8 per million, mainly in children and adolescents (Pingping et al., 2019; Lancia et al., 2019), and third among all cancers among cancers among children aged <20 years (Belayneh et al., 2021; Simpson and Brown, 2018). Statistically, approximately 2% of all tumors among children <14 years of age and 3% among children between 14–19 years is osteosarcoma (Siegel et al., 2022; Mirabello et al., 2009). Owing to its mesenchymal tissue origin, Osteosarcoma features malignant spindle cells that produce aberrant bone-like tissue (Ji et al., 2015). There are multiple reported risk factors for osteosarcoma, including high stature, elevated birth weight, certain inherited cancer susceptibility syndromes, and common genetic variants (Gianferante et al., 2017). At present, the optimal standard of care is neoadjuvant chemotherapy - surgical adjuvant chemotherapy with adriamycin, cisplatin, and high-dose methotrexate (MAP regimen) (Ritter and Bielack, 2010). This management is efficacious for early-stage osteosarcoma. However, the chemo- and radiotherapeutic efficacies decline with disease progression (Smrke et al., 2021). Moreover, conventional chemotherapeutic agents, namely, methotrexate, cisplatin, adriamycin, and iso-cyclophosphamide, which are often employed in osteosarcoma management, are accompanied by numerous complications, such as unstable blood levels, rapid clearance, poor targeting, variable toxicity to normal cells, and drug resistance. As such, their application among osteosarcoma patients is restricted (Grünewald et al., 2020). Given these factors, the past few decades have seen no marked improvement in patient survival (Corre et al., 2020). Post-recurrence patient prognosis also remains poor, with only 23%–29% of patients surviving over 5 years following a second diagnosis (Simpson and Brown, 2018) and long-term survival being <20% (Carrle and Bielack, 2006; Ferrari et al., 2003). The consequences of osteosarcoma can be utterly devastating, affecting both the physical and mental health of patients and their families (Mortus et al., 2014). Therefore, a safe and effective therapy is urgently needed for appropriate osteosarcoma management.
Exosomes are nanoscale extracellular vesicles that are released by a wide variety of cells (Liu et al., 2021a). Its primary role is normal cellular metabolic maintenance and intercellular communication (Song et al., 2021). Based on transmission electron microscopy images, exosomes resemble “cups” or “discs,” which are molecularly recognized by expressing specific protein markers. These structures mediate cell-to-cell communication via the transfer of reactive molecules, namely, proteins, DNA, mRNAs, and non-coding RNAs, from 1 cell to another (Tkach and Théry, 2016). Exosomes are made up of lipid bilayers. Hence, they are readily taken up by neighboring or distant receptor cells. Exosomal contents (such as small RNAs and proteins) provide physiological activities, such as immunomodulation (Teng et al., 2015), autophagy (Baixauli et al., 2014), stem cell differentiation (Nair et al., 2014), and intercellular communication (Tkach and Théry, 2016), in a process termed the third mode of cellular communication. There is significant interest in the development of exosomes for the management of osteosarcoma. The Exosomal lipid barrier protects exosomal miRNAs from RNase degradation, enabling stable transfer between two separate tumors and involvement in tumor growth (Tkach and Théry, 2016). Exosomal miRNAs significantly influence the cellular capacities for migration, proliferation, invasion, apoptosis, and chemoresistance in many cancers, all closely associated with patient prognosis, including osteosarcoma patients (Santos et al., 2018). At present, researchers believe that exosomes have broad prospects for treating osteosarcoma (Zhongyu et al., 2024). Exosomes are crucial for the onset and progression of osteosarcoma. Exosomes can increase osteosarcoma lung metastasis and negatively impact prognosis by modulating MMP and other processes that alter the interstitial microenvironment (Zhan et al., 2022). Conversely, exosomes are also crucial for osteosarcoma’s drug resistance. Osteosarcoma cells produce tumor cells resistant to cisplatin by activating autophagy, breaking down cell components, and preserving intracellular balance through exosomes. However, the role of exosomes not only worsens the disease but also plays an advantage in treatment. For example, exosomes of sensitive cells treated with Luteolin can improve the adriamycin response in adriamycin-resistant cells by increasing miR-384 and targeting the PTN/β-catenin/MDR1 axis. It is a promising therapeutic agent for chemotherapy-resistant OS (Qin et al., 2022). As a result, exosomes may be considered a double-edged sword in regulating bone tumor formation, influencing carcinogenesis, angiogenesis, and metastasis while inhibiting tumor progression (Vakhshiteh et al., 2019). Therefore, investigating the role of exosomes in the progression of osteosarcoma is essential.
2 Materials and methods
The analysis and findings were reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Liberati et al., 2009). This study did not require patient permission and ethical approval because the data and results were derived from previously approved research. The protocol was entered into the PROSPERO database (CRD42024502574).
2.1 Inclusion criteria
2.1.1 Patients and diseases (P)
Any animal and osteosarcoma cell lines used to produce osteosarcoma xenograft models were included.
2.1.2 Intervention (I)
Collect any interventions involving extracellular vesicles that inhibit the development of animal osteosarcoma models.
2.1.3 Control (C)
Positive control: exosomes from various transfected cells or drug-carrying exosomes.
Negative control: blank, DMEM, PBS, saline.
2.1.4 Results (O)
Tumor weight (mg).
Tumor volume (mm3) (tumor volume = length × width2/2) (Li et al., 2024)
2.1.5 Type of study (S)
Control studies were included, with no restrictions on the blind method.
2.2 Exclusion criteria
1) Irrelevant animals: Animals with comorbidities; non-mouse animal model studies; ex vivo, in vitro, and computer models.
2) Irrelevant controls: No controls for osteosarcoma.
3) Irrelevant study types: Case, crossover, and studies without independent control groups.
4) Unavailable data: specific study details or study data (e.g., reviews or conference abstracts) were missing.
5) Unreported studies of (1) tumor weight; (2) tumor volume.
6) Non-English studies.
7) Non-peer-reviewed journal articles
2.3 Data selection
Owing to the diverse collection of contemporary preclinical studies involving animal models as test subjects, we analyzed data involving exosomes promoting osteosarcoma development and suppressing osteosarcoma progression. Regarding exosomes promoting osteosarcoma development, we generated subcategories based on varying cellular sources of exosomes. In the case of exosome suppression osteosarcoma progression, we generated subcategories based on the varying mechanisms of exosomal action.
2.4 Data source and screening
Relevant articles were selected via extensive screening of four major scientific databases, PubMed, Embase, Web of Science, and Cochrane, published from 2014.03.01 to 2024.03.01. Screening was completed using a combination of the following terminology: [(“Exosomes” OR “Extracellular Vesicles” OR “Extracellular Vesicle” OR “Vesicle, Vesicle. Extracellular” OR “Vesicles, Extracellular” OR “Exovesicles” OR “Exovesicle”) AND (Osteosarcoma OR Osteosarcomas OR “Osteosarcoma Tumor” OR “Osteosarcoma Tumors” OR “Tumor, Osteosarcoma” OR “Tumors,” Osteosarcoma’ OR “Sarcoma, Osteogenic” OR “Osteogenic Sarcomas” OR “Sarcomas, Osteogenic” OR “Osteogenic Sarcoma” OR “Sarcoma, Ewing” OR “Sarcoma, Ewing’s” OR “Sarcoma, Ewings” OR “Ewing’s Sarcoma” OR “Ewings Sarcoma” OR “Ewing’s Tumor” OR “Ewings Tumor” OR “Tumor, Ewing’s” OR “Ewing Sarcoma” OR “Ewing Tumor” OR “Tumor, Ewing” OR “Osteosarcoma, Juxtacortical” OR “Juxtacortical Osteosarcoma” OR “Juxtacortical Osteosarcomas’ OR ‘Osteosarcomas. Juxtacortical”)]
2.5 Literature screening and data acquisition
After removing duplicate studies, two highly trained investigators reviewed the eligible articles and extracted data based on the inclusion/exclusion criteria, cross-checking their selections. Any disagreements were resolved by a third investigator. Data acquisition was based on the pre-established full-text data sheets, which included (1) basic study characteristics: article title, authors, publication year, type of study, baseline features of mice, sample size, modeling method, exosome cell type and source, exosome size, and marker proteins; (2) critical elements of the bias risk assessment; and (3) outcome measures of tumor volume and weight. In the case of studies reporting data in image form only, we employed the GetData Graph Digitizer software for data extraction from images.
2.6 Bias risk between included studies
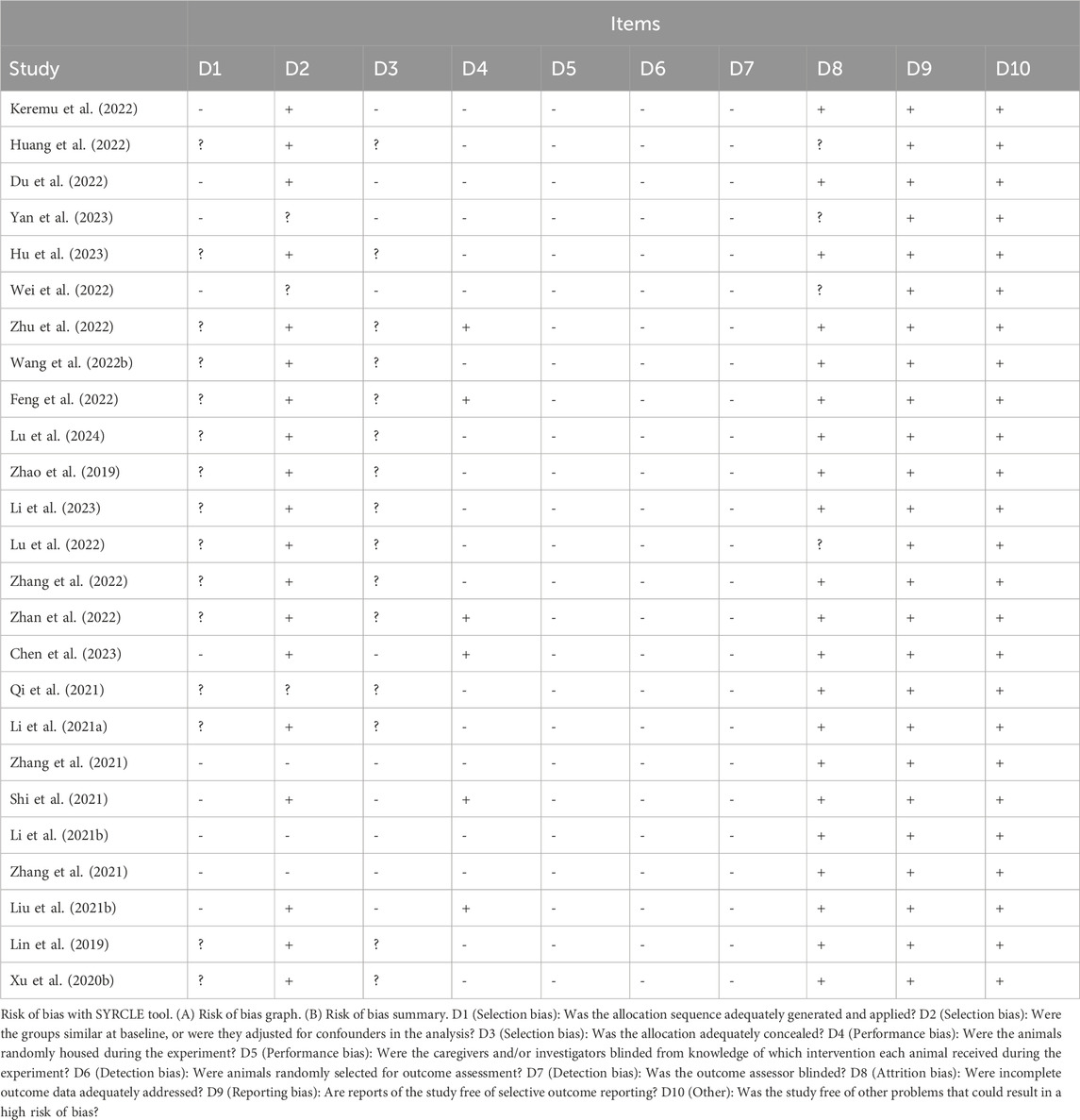
Two highly trained investigators evaluated and cross-checked the inherent bias risk of the selected studies based on the SYRCLE’s Risk of Bias in Animal Studies tool (Hooijmans et al., 2014). They covered selection bias, implementation bias, measurement bias, follow-up bias, reporting bias, and any other bias as part of a list of 10 questions or tools. Any disagreements were resolved via discussion with a third investigator. The evaluation questions were answered either with a “yes” (indicating low bias risk) or “no” (indicating high bias risk). Unclear items were classified as “not clear.”
2.7 Data analyses
Among the extracted data were tumor volume and weight. All data were analyzed using Review Manager 5.4 software. We conducted a sensitivity analysis to determine if excluding studies with a high risk of bias affected the estimated effect or heterogeneity of the outcome. Forest plots are depicted aggregated hazard ratios with their respective 95% confidence intervals. The inconsistency index (I2) was employed to assess statistical heterogeneity. Data exhibiting greater heterogeneity were analyzed using a random-effects model, while data demonstrating lesser heterogeneity were evaluated using a fixed-effects model. To address the uniqueness of animal research and to reduce bias from varying experimental designs, we select to use the mean difference to convey the impact of the intervention modality on the outcome measures. Std. Mean difference (SMD) is the quotient between the difference of two means and the combined standard deviation. It eliminates the influence of absolute value and measurement unit of a study, and is suitable for the analysis of numerical data with different units or large difference in means.
3 Results
3.1 Systematic screening
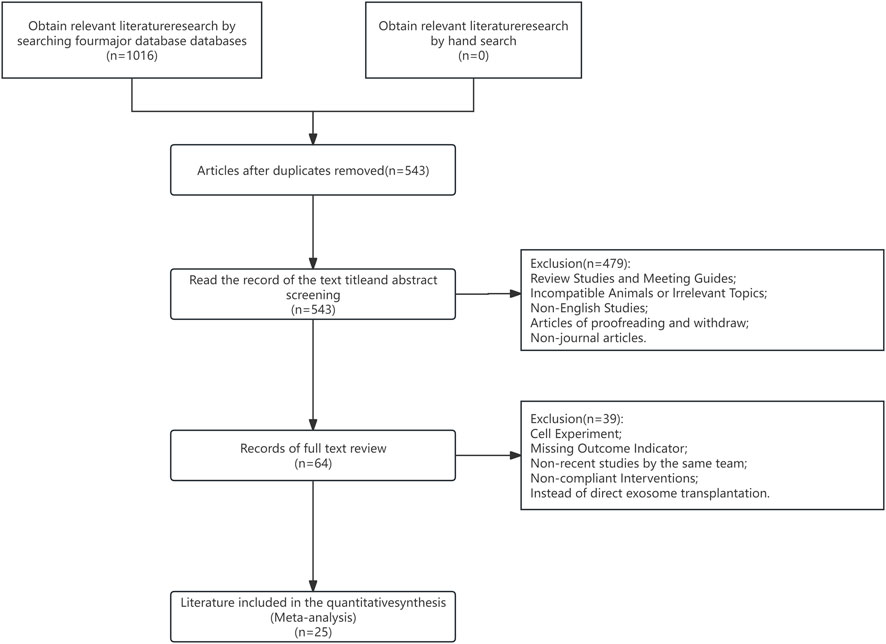
Overall, 1,016 articles were eligible for analysis. Among them, duplicate and irrelevant articles (e.g., subject inconsistency, reporting, etc.) were eliminated after reviewing study titles and abstracts. Subsequently, articles with inconsistent research methods (e.g., cellular experiments, endpoint indicators without tumor volume or tumor weight, and so on) were eliminated following the full-text review. Lastly, 25 preclinical studies were selected for meta-analysis. A PRISMA flowchart depicting our strict study inclusion criteria is provided in Figure 1.
3.2 Basic demographics of included studies
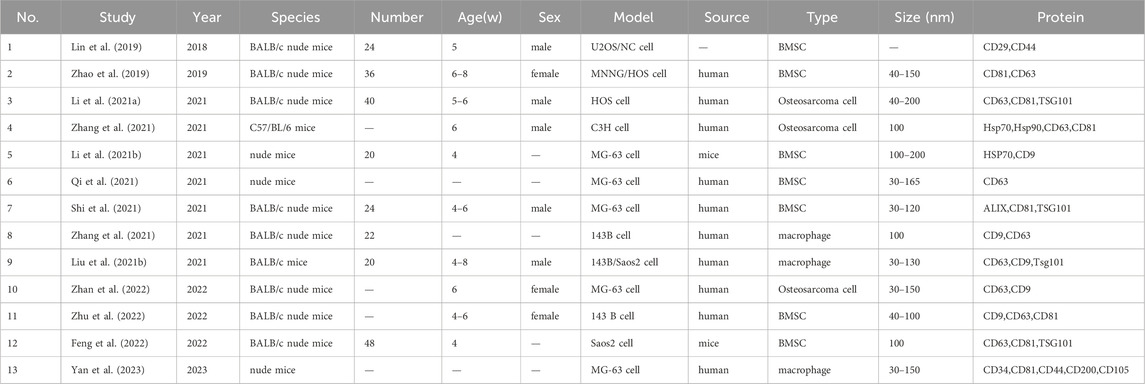
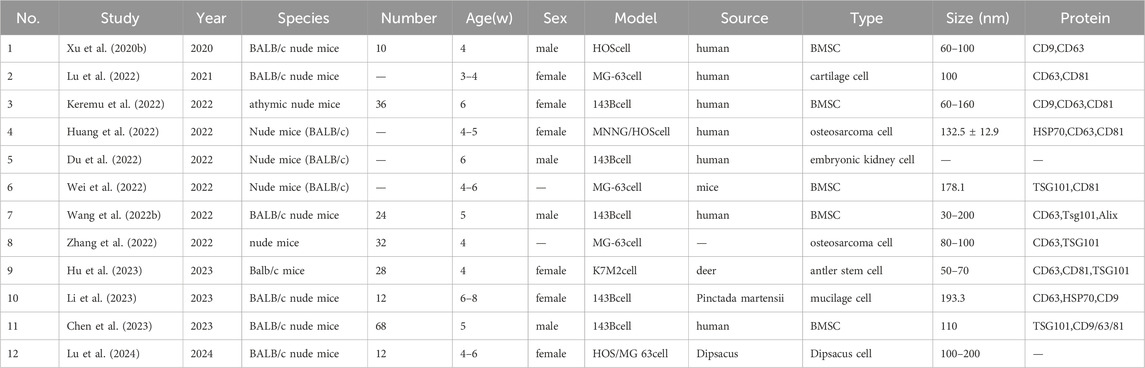
All selected studies were randomized controlled trials (RCTs), with the mouse as the examined animal. Overall, six studies failed to report the sex of mice. The age of model mice ranged between 3–8 weeks, and the sample size was between 10–68 mice. The model type was inoculated cells. Exosomes were derived from humans (17 studies), other sources (6 studies), and source unspecified (2 studies). Exosomal cell types were osteosarcoma cells (5 studies), bone marrow mesenchymal stem cells (12 studies), macrophages (3 studies), and other cells (5 studies). The exosomal diameter was between 30–200 nm. The exosome-specific surface proteins assessed were CD9, CD29, CD34, CD44, CD63, CD 81, CD105, CD200, HSP 70, Hsp 90, TSG101, and Alix. The information mentioned above is detailed in Tables 1, 2.
3.3 Bias risk assessment
In all, 25 studies reported baseline demographics of experimental mice, such as age, sex, and body weight, without significant bias. Six studies, meanwhile, reported placing animals at random during RCTs. We could not determine if researchers and/or animal handlers were blinded, whether animals were chosen randomly for outcome assessment, or whether researchers generated blinded results because of a lack of information. All of the papers selected reported the anticipated results despite the absence of a study protocol. All results from the offset risk evaluation of selected articles are summarized in Table 3.
3.4 Meta-analysis results
For exosome-mediated positive and negative control of osteosarcoma progression, we examined 25 RCTs. Based on the several exosome sources (cell types), we performed a subclass analysis to investigate the acceleration of osteosarcoma development. Based on the several modes of action, we performed subclass analyses while investigating the suppression of osteosarcoma advancement. Our observations were as follows:
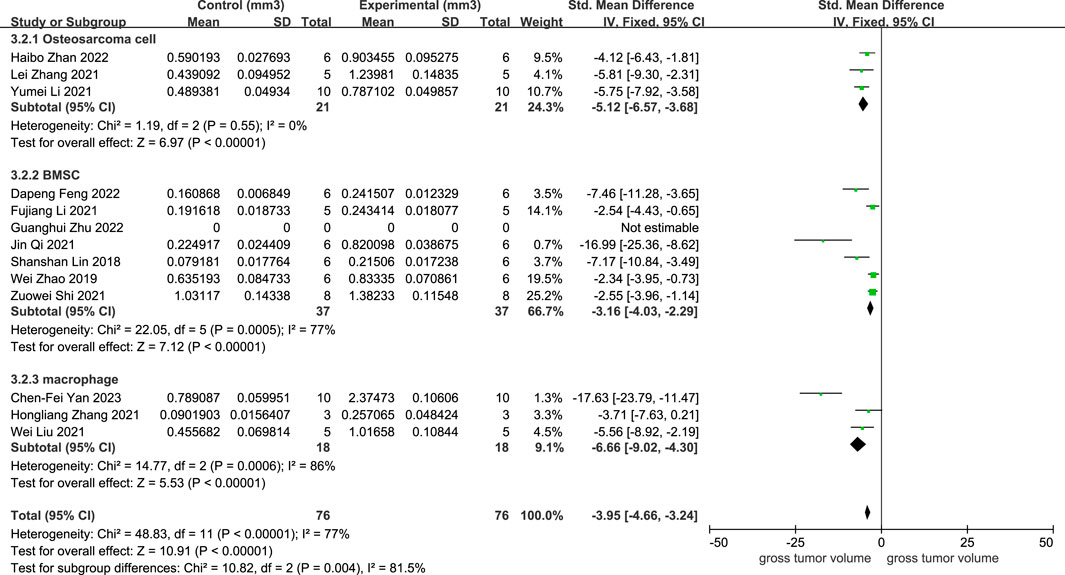
As illustrated in Figure 2, 12 studies revealed a rise in osteosarcoma volume following exosome transplantation, and the SMD value was −3.95 with a 95% confidence interval [-4.66, −3.24] under the fixed-effect model. At the same time, the heterogeneity test exhibited a P-value <0.00001, I2 = 77%. Subcategory evaluation revealed the following: the SMD value of OS cells was −5.12 with 95% confidence interval [-6.57, −3.68], the SMD value of BMSC cells was −3.16 with 95% confidence interval [-4.03, −2.29], and the SMD value of macrophage was −6.66 with 95% confidence interval [-9.02, −4.30].
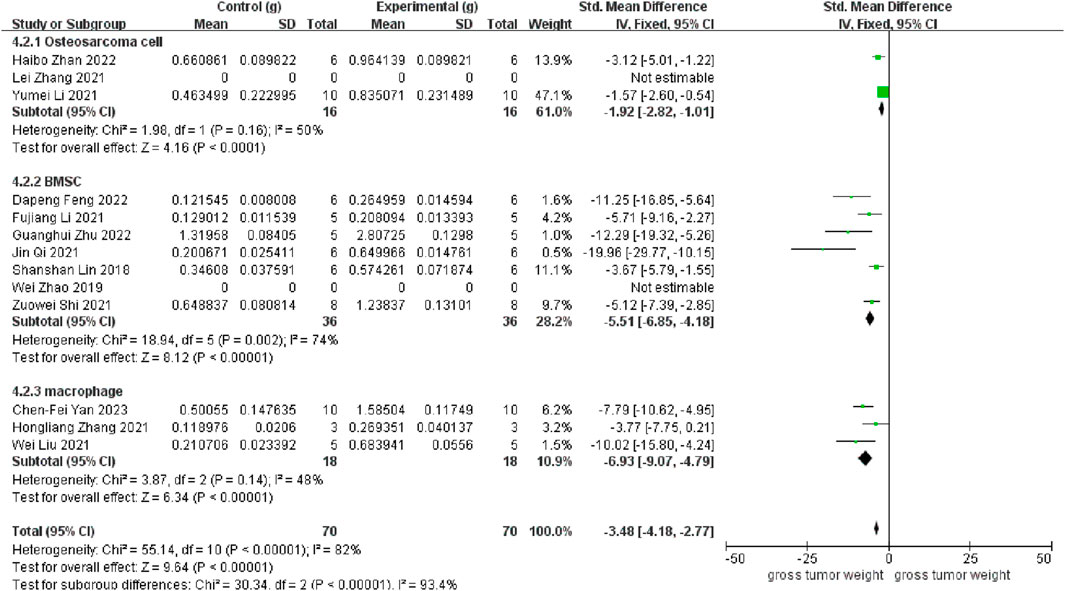
As depicted in Figure 3, 11 studies revealed a rise in osteosarcoma weight following exosome transplantation, and the corresponding SMD value was −3.48 with 95% confidence interval [-4.18, −2.77] under the fixed-effect model, and the heterogeneity test exhibited a P-value <0.00001 and I2 = 82%. Subcategory evaluation revealed the following: the SMD value of OS cells was −1.92 with 95% confidence interval [-2.82, −1.01], the SMD value of BMSC cells was −5.51 with 95% confidence interval [-6.85, −4.18], and the SMD value of macrophage was −6.93 with 95% confidence interval [-9.07, −4.79].
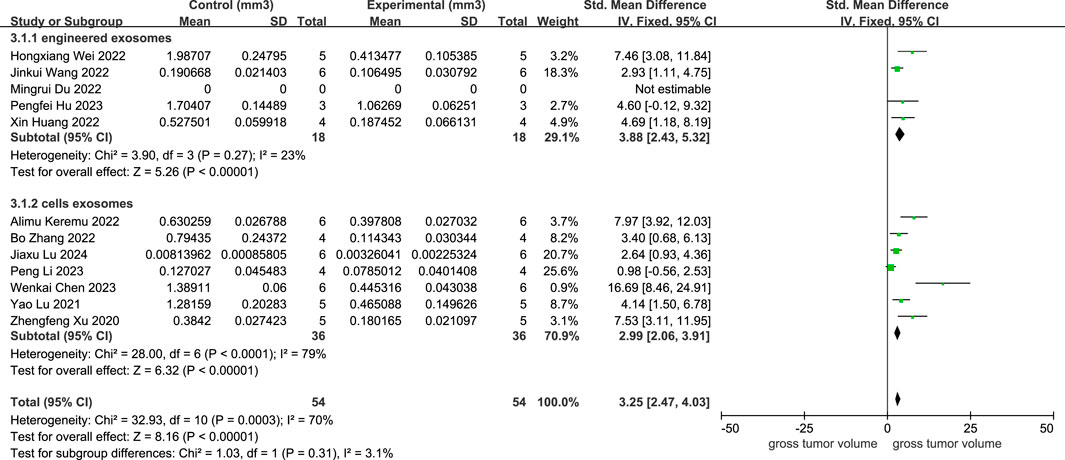
As provided in Figure 4, 11 studies depicted a decline in osteosarcoma volume following cellular exosome transplantation, and the corresponding SMD value was 3.25 with a 95% confidence interval [2.47, 4.03] under the fixed effect model, and the heterogeneity test exhibited a P-value equal to 0.0003, I2 = 70%. Based on the subcategory evaluation, the SMD value of engineered exosomes was 3.88 with a 95% confidence interval [2.43, 5.32], and the SMD value of cellular exosomes was 2.99 with a 95% confidence interval [2.06, 3.91].
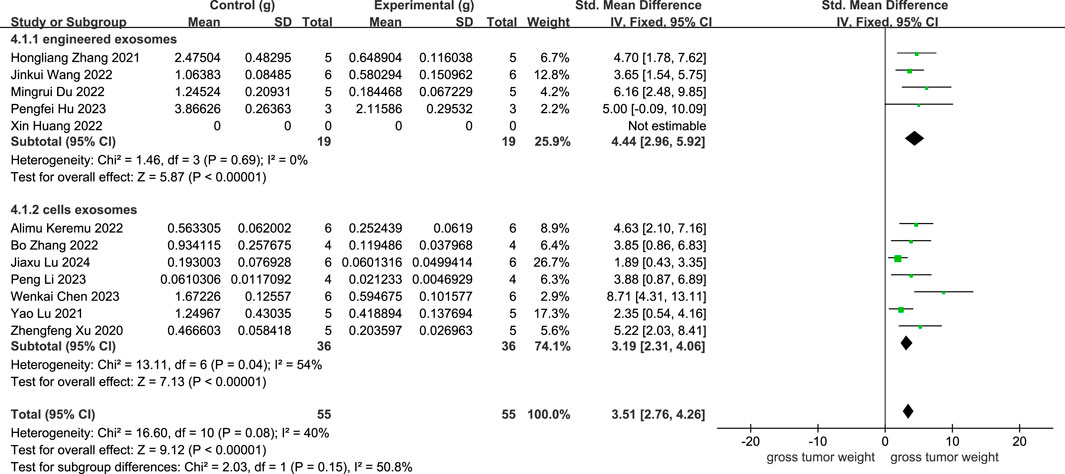
As revealed in Figure 5, 11 studies showed a rise in osteosarcoma weight following cellular exosome transplantation, with an SMD value of 3.51, 95% confidence interval of [2.76, 4.26], and a heterogeneity test of p = 0.08, I2 = 40% in the fixed-effects model. Subcategory analysis revealed the following: the SMD value of engineered exosomes was 4.44 with 95% confidence interval [2.96, 5.92], and the SMD value of cellular exosomes was 3.19 with 95% confidence interval [2.31, 4.06].
4 Discussion
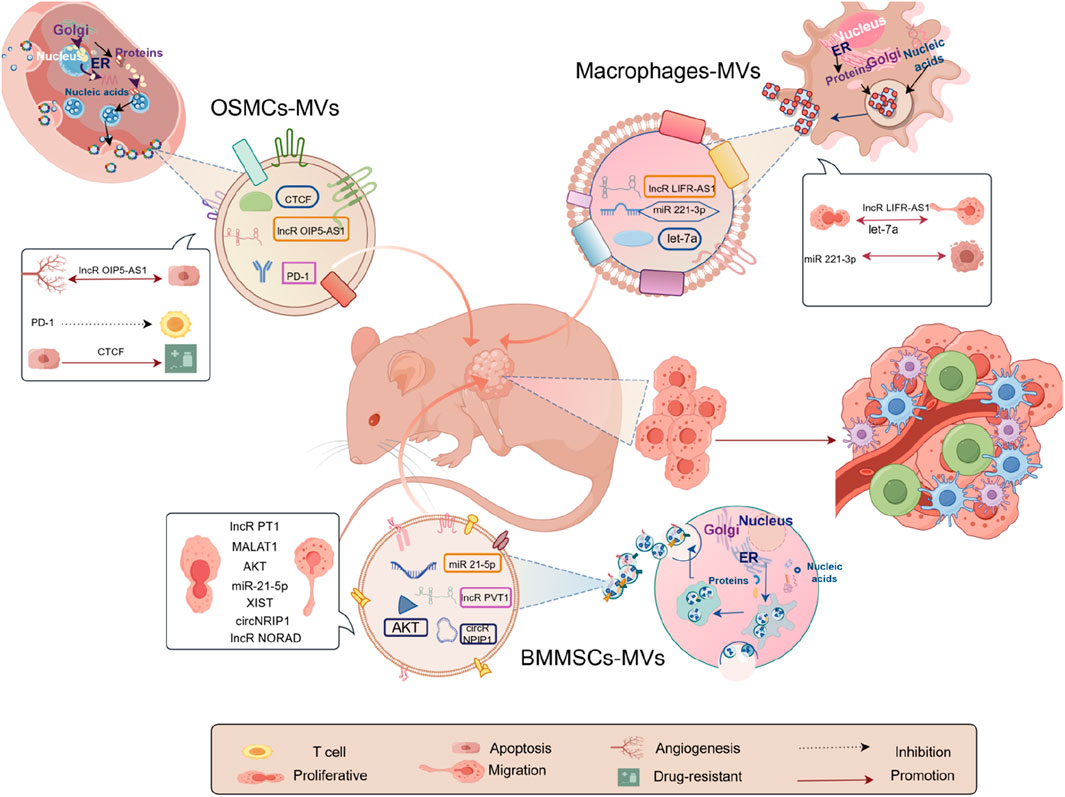
We examined 25 RCTs investigating exosome transplantation’s effect on animal osteosarcoma models. The replies we received from our data differed based on the exosomal design. This suggests that exosomes have promise for exploring the pathophysiology of osteosarcoma and for driving the development of focused diagnostic methods. Firstly, exosomes can help us explore osteosarcoma pathogenesis. For example, long non-coding RNAs PVT1 (Zhao et al., 2019), miR-21–5p (Qi et al., 2021), CircNRIP1 (Shi et al., 2021), LncRNA XIST (Zhu et al., 2022), lncRNA NORAD (Feng et al., 2022) encapsulated by exosomes derived from bone marrow mesenchymal stem cells are now known. Macrophage-derived lncRNA Livr-AS1 (Zhang et al., 2021), miRNA-221–3p (Liu et al., 2021b), let-7a (Yan et al., 2023); lncRNA OIP5-AS1 (Li et al., 2021a) CTCF (Zhan et al., 2022) derived from osteosarcoma cells can all participate in the progression of osteosarcoma, as shown in Figure 6. As depicted in Figure 6, based on our meta-analysis of 13 studies, 3 categories of cellular exosomes regulated osteosarcoma pathogenesis in mice, and their origins were bone marrow mesenchymal stem cells (BMMSCs), osteosarcoma cells (OSMCs), and macrophages, respectively. While bone marrow mesenchymal stem cells (BMMSCs) were the most commonly studied exosomes, our comparative analysis revealed that macrophage exosomes exerted a substantially higher effect on the model mice than BMMSCs and osteosarcoma cells (OSMCs), in terms of both tumor volume and weight. Recent years have seen a rise in studies on macrophages and their derivatives, particularly tumor-associated macrophages (TAMs), crucial immune cells. They constitute a major component of the tumor microenvironments and play a key in regulating TME. TAMs also critically regulate tumourigenesis and progression (Duan and Luo, 2021; Chen et al., 2019). It has been reported that TAMs can be polarized into either pro-tumor (M2 macrophages) or anti-tumor phenotype (M1 macrophages) (Xu et al., 2020a). As expected, TAMs also regulate osteosarcoma development (Li et al., 2024; Wang et al., 2022a), solidifying the macrophage-based exosomal role in osteosarcoma without any exception. Moreover, several studies demonstrated the pathogenesis and progression of tumors via exosomes (Zhang et al., 2021; Liu et al., 2021b; Yan et al., 2023). Given this evidence, the analyzed studies depicted a vital role of macrophages in exosome development (from all three sources) in osteosarcoma (Li et al., 2024; Wang et al., 2022a). Further, they verified that macrophage exosomes play a critical role in the process (Zhang et al., 2021; Liu et al., 2021b; Yan et al., 2023). Therefore, while the experimental findings indicated that the exosomes from the 3 cells mentioned above types modulate osteosarcoma in model organisms, the authors hypothesize that macrophages exert a more significant influence on osteosarcoma progression and may thus hold greater promise for advancing research and therapeutic strategies in osteosarcoma. Therefore, the considerable function of macrophages and their derivatives in the pathogenesis of osteosarcoma necessitates additional investigation to establish a theoretical foundation for future study and the development of accurate and effective diagnostic and therapeutic strategies for osteosarcoma.
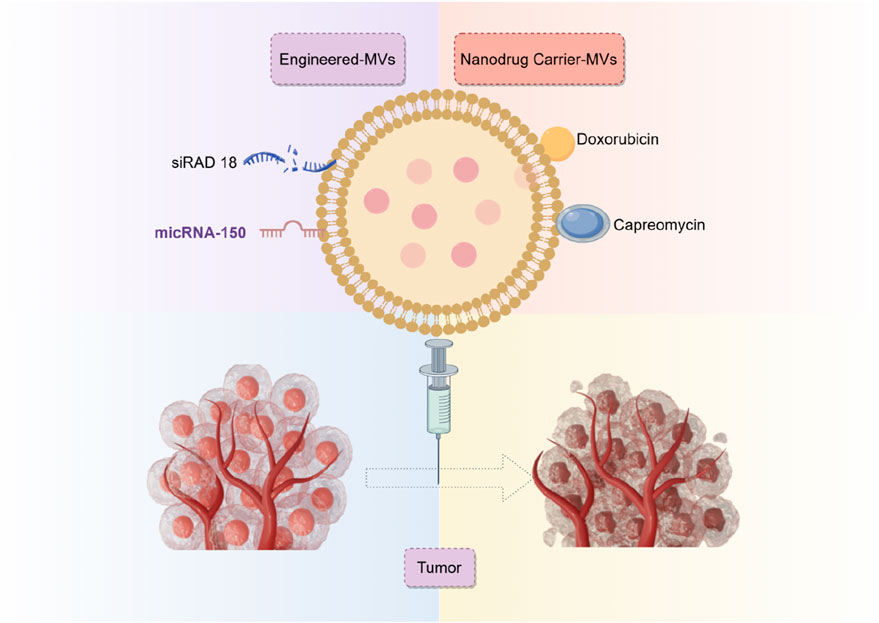
Secondly, it is equally crucial to ascertain the potential of biological or engineered exosomes in the direct treatment of osteosarcoma or as an adjunct therapy. Exo-miR-150 targets IGF2BP1 (Xu et al., 2020b), Exo-miR-195 targets KIF4A (Lu et al., 2022), Exo-miR-206 targets NRSN2 (Keremu et al., 2022), and RGD-Exo targets Rad18 (Du et al., 2022), along with other pathways that impede the evolution of osteosarcoma. This significantly expands potential opportunities for innovation in diagnosing and treating osteosarcoma (Figure 7). It has shown excellent therapeutic effects not only in osteosarcoma exosomes but also in other malignant tumors, such as colon cancer, hepatocellular carcinoma, breast cancer, etc (Zhao et al., 2020).
Overall, 12 analyzed studies reported the role of exosomes in mouse models. Seven employed exosomes directly transplanted from humans, shellfish, and sequential cell sources to mouse models to serve as therapeutic agents. Our timeline analysis indicates that specialists have prioritized the importance of human-derived exosomes in osteosarcoma-related exosome research. Exosomes derived from animal and plant sources have been utilized as exosomal technologies advance and mature, producing favorable results. Furthermore, five projects, predominantly from 2022, examined the application of exosomes as carriers for targeted drug delivery by generating modified exosomes. Emerging evidence revealed that engineered exosomes possess properties such as easy mass production, excellent therapeutic efficacy, strong targeting, and not elicit drug resistance (Du et al., 2022; Huang et al., 2022; Wei et al., 2022; Wang et al., 2022; Hu et al., 2023). Ultimately, we performed a detailed assessment of two types of exosomes: those derived from transfected cells and those designed for drug delivery. We demonstrated that the drug-loaded exosomes exhibited enhanced therapeutic efficacy compared to exosomes derived from transfected cells. The comparison results validate the effectiveness of modified exosomes in osteosarcoma treatment, indicating that the ideal cellular exosomes for this purpose necessitate additional investigation. In summary, the authors warrant accelerating engineered exosomal design and development to fast-track its application in clinical settings. In the meantime, researchers should also explore other sources of exosomes to elucidate more suitable exosomes for osteosarcoma management.
Exosomes are an extracellular component facilitating substantial information interchange, providing exceptional diagnostic and prognostic capacities for osteosarcoma and its associated complications, in addition to their previously mentioned roles. In this meta-analysis, we examined pertinent and contemporary literature. Exosomes, including various biomarkers, serve diagnostic purposes (Araki et al., 2023; Han et al., 2021). In addition to the primary foci, Osteosarcoma is notorious for its enhanced metastatic potential. In this regard, it was reported that differences in exosome sources modulate the metastatic potential of osteosarcoma (Roberts et al., 2022). Lung metastasis is the most frequent metastatic foci of osteosarcoma, is a major contributor to cancer-related death, and has recently received extensive attention from experts and scholars. Numerous reports revealed that cellular exosomes are critical in modulating osteosarcoma lung metastasis (Deng et al., 2024; Yu et al., 2023; Chang et al., 2023; Almeida et al., 2022; Zhong et al., 2021; Zhang et al., 2021; Cheng et al., 2021). In addition to metastasis, alterations in bone metabolism are a major complication of osteosarcoma. Activation of osteoclast cell activity with simultaneous inhibition of osteogenesis promotes pathological fracture and pain among osteosarcoma patients, which contributes to severe physical and psychological suffering. As before mentioned, exosomes from osteosarcoma cells possess a robust information interaction ability, which induces the formation of bone metabolic abnormalities within osteosarcoma patients (Ucci et al., 2021; Luo et al., 2021; Raimondi et al., 2020). Likewise, engineered exosomes promote bone formation in the osteosarcoma pathological state, thereby preventing bone destruction-mediated complications via precise targeting action (Gupta et al., 2023). Finally, exosomes also predict drug resistance, prognosis, and survival of osteosarcoma patients (Araki et al., 2023; Weinman et al., 2021; Wang et al., 2021; Wang et al., 2023).
We confirmed the crucial role of exosomes in the progression of osteosarcoma using meta-analysis and systematic review. We also found that exosomes generated from macrophages exceeded other sources in osteosarcoma regulation. Secondly, we identified the incredible potential of exosomes in osteosarcoma treatment and determined the efficacy of engineered exosomes and future research direction involving cellular exosomes based on our subcategory analysis. To expand our understanding of exosomes in osteosarcoma pathogenesis, progression, and treatment, we examined recent articles about the association between osteosarcoma and exosomes and osteosarcoma and related complication diagnostic and prognostic prediction.
4.1 Study strengths and weaknesses
Strengths of this study: (1) We were the first to extensively evaluate the significance of exosomes on osteosarcoma pathogenesis and in interventional therapy. We summarized the overall progression of the study and highlighted the challenges and the direction of future development. (2) Our hypotheses were entirely validated by a thorough exosome feature-based subcategory analysis that we created. Furthermore, we developed a solid research hypothesis for additional studies based on our subcategory analysis findings. (3) We reviewed recent investigations involving exosomes and osteosarcoma diagnosis and prognostic prediction, summarized overall research progress, and provided the basis for expanding ideas of related studies. (4) Using the SYRCLE risk of bias assessment method, we evaluated the intrinsic risk of bias in the investigated studies. Furthermore, we limited our selection to the most current findings from the same research organization within 5 years to reduce bias in those studies.
Limitations of this study: (1) Data was selected based on tumor size and weight in mouse models. We did not analyze other outcome indicators. This could lead to potential bias. Therefore, the reliability of our model requires further validation. (2) We could not accurately identify the heterogeneity source; thus, we employed SMD for data merging analysis, which produced more conservative conclusions. (3) We did not review grey literature and conference abstracts and opted for English-only databases, which may have introduced publication bias. (4) The quantity of examined studies and associated sample population were relatively small, potentially generating bias. (5) All included studies had an unclear blinding performance bias risk.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XHL: Writing–original draft, Writing–review and editing. JYY: Writing–review and editing. WLG: Conceptualization, Investigation, Methodology, Writing–review and editing. JQW: Data curation, Formal Analysis, Resources, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almeida, S. F. F., Fonseca, A., Sereno, J., Ferreira, H. R. S., Lapo-Pais, M., Martins-Marques, T., et al. (2022). Osteosarcoma-derived exosomes as potential PET imaging nanocarriers for lung metastasis. Small 18 (49), e2203999. doi:10.1002/smll.202203999
Araki, Y., Asano, N., Yamamoto, N., Hayashi, K., Takeuchi, A., Miwa, S., et al. (2023). A validation study for the utility of serum microRNA as a diagnostic and prognostic marker in patients with osteosarcoma. Oncol. Lett. 25 (6), 222. doi:10.3892/ol.2023.13808
Baixauli, F., López-Otín, C., and Mittelbrunn, M. (2014). Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 5, 403. doi:10.3389/fimmu.2014.00403
Belayneh, R., Fourman, M. S., Bhogal, S., and Weiss, K. R. (2021). Update on osteosarcoma. Curr. Oncol. Rep. 23 (6), 71. doi:10.1007/s11912-021-01053-7
Carrle, D., and Bielack, S. S. (2006). Current strategies of chemotherapy in osteosarcoma. Int. Orthop. 30 (6), 445–451. doi:10.1007/s00264-006-0192-x
Chang, X., Tan, Q., Xu, J., Wu, X., Wang, Y., Zhang, Y., et al. (2023). Tumor-derived exosomal linc00881 induces lung fibroblast activation and promotes osteosarcoma lung migration. Cancer Cell Int. 23 (1), 287. doi:10.1186/s12935-023-03121-3
Chen, W., Li, Z., Yu, N., Zhang, L., Li, H., Chen, Y., et al. (2023). Bone-targeting exosome nanoparticles activate Keap1/Nrf2/GPX4 signaling pathway to induce ferroptosis in osteosarcoma cells. J. Nanobiotechnology 21 (1), 355. doi:10.1186/s12951-023-02129-1
Chen, Y., Song, Y., Du, W., Gong, L., Chang, H., and Zou, Z. (2019). Tumor-associated macrophages: an accomplice in solid tumor progression. J. Biomed. Sci. 26 (1), 78. doi:10.1186/s12929-019-0568-z
Cheng, Z., Wang, L., Wu, C., Huang, L., Ruan, Y., and Xue, W. (2021). Tumor-derived exosomes induced M2 macrophage polarization and promoted the metastasis of osteosarcoma cells through tim-3. Arch. Med. Res. 52 (2), 200–210. doi:10.1016/j.arcmed.2020.10.018
Corre, I., Verrecchia, F., Crenn, V., Redini, F., and Trichet, V. (2020). The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells 9 (4), 976. doi:10.3390/cells9040976
Deng, C., Xu, Y., Chen, H., Zhu, X., Huang, L., Chen, Z., et al. (2024). Extracellular-vesicle-packaged S100A11 from osteosarcoma cells mediates lung premetastatic niche formation by recruiting gMDSCs. Cell Rep. 43 (2), 113751. doi:10.1016/j.celrep.2024.113751
Du, M., Gu, J., Liu, C., Liu, N., Yu, Z., Zhou, C., et al. (2022). Genome-wide CRISPR screen identified Rad18 as a determinant of doxorubicin sensitivity in osteosarcoma. J. Exp. Clin. Cancer Res. 41 (1), 154. doi:10.1186/s13046-022-02344-y
Duan, Z., and Luo, Y. (2021). Targeting macrophages in cancer immunotherapy. Signal Transduct. Target Ther. 6 (1), 127. doi:10.1038/s41392-021-00506-6
Feng, D., Li, Z., Yang, L., Liang, H., He, H., Liu, L., et al. (2022). BMSC-EV-derived lncRNA NORAD facilitates migration, invasion, and angiogenesis in osteosarcoma cells by regulating CREBBP via delivery of miR-877-3p. Oxid. Med. Cell Longev. 2022, 8825784. doi:10.1155/2022/8825784
Ferrari, S., Briccoli, A., Mercuri, M., Bertoni, F., Picci, P., Tienghi, A., et al. (2003). Postrelapse survival in osteosarcoma of the extremities: prognostic factors for long-term survival. J. Clin. Oncol. 21 (4), 710–715. doi:10.1200/JCO.2003.03.141
Gianferante, D. M., Mirabello, L., and Savage, S. A. (2017). Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 13 (8), 480–491. doi:10.1038/nrendo.2017.16
Grünewald, T. G., Alonso, M., Avnet, S., Banito, A., Burdach, S., Cidre-Aranaz, F., et al. (2020). Sarcoma treatment in the era of molecular medicine. EMBO Mol. Med. 12 (11), e11131. doi:10.15252/emmm.201911131
Gupta, S., Qayoom, I., Gupta, P., Gupta, A., Singh, P., Singh, S., et al. (2023). Exosome-functionalized, drug-laden bone substitute along with an antioxidant herbal membrane for bone and periosteum regeneration in bone sarcoma. ACS Appl. Mater Interfaces 15, 8824–8839. doi:10.1021/acsami.2c18308
Han, Z., Yi, J., Yang, Y., Li, D., Peng, C., Long, S., et al. (2021). SERS and MALDI-TOF MS based plasma exosome profiling for rapid detection of osteosarcoma. Analyst 146 (21), 6496–6505. doi:10.1039/d1an01163d
Hooijmans, C. R., Rovers, M. M., de Vries, R. B. M., Leenaars, M., Ritskes-Hoitinga, M., and Langendam, M. W. (2014). SYRCLE's risk of bias tool for animal studies. BMC Med. Res. Methodol. 14, 43. doi:10.1186/1471-2288-14-43
Hu, P., Ying, J., Wang, Y., Jiang, T., Pan, Z., Zhao, C., et al. (2023). Extracellular vesicles derived from 3D cultured antler stem cells serve as a new drug vehicle in osteosarcoma treatment. Cell Transpl. 32, 9636897231219830. doi:10.1177/09636897231219830
Huang, X., Wu, W., Jing, D., Yang, L., Guo, H., Wang, L., et al. (2022). Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J. Control Release 343, 107–117. doi:10.1016/j.jconrel.2022.01.026
Ji, G. R., Yu, N. c., Xue, X., and Li, Z. g. (2015). PERK-Mediated autophagy in osteosarcoma cells resists ER stress-induced cell apoptosis. Int. J. Biol. Sci. 11 (7), 803–812. doi:10.7150/ijbs.11100
Keremu, A., Aila, P., Tusun, A., Abulikemu, M., and Zou, X. (2022). Extracellular vesicles from bone mesenchymal stem cells transport microRNA-206 into osteosarcoma cells and target NRSN2 to block the ERK1/2-Bcl-xL signaling pathway. Eur. J. Histochem 66 (3), 3394. doi:10.4081/ejh.2022.3394
Lancia, C., Anninga, J. K., Sydes, M. R., Spitoni, C., Whelan, J., Hogendoorn, P. C. W., et al. (2019). A novel method to address the association between received dose intensity and survival outcome: benefits of approaching treatment intensification at a more individualised level in a trial of the European Osteosarcoma Intergroup. Cancer Chemother. Pharmacol. 83 (5), 951–962. doi:10.1007/s00280-019-03797-3
Li, C., Xiang, F., Gong, Y., Fu, Y., Chen, G., Wang, Z., et al. (2024). Tumor-derived microparticles promoted M2-like macrophages polarization to stimulate osteosarcoma progression. Int. J. Biochem. Cell Biol. 166, 106494. doi:10.1016/j.biocel.2023.106494
Li, F., Chen, X., Shang, C., Ying, Q., Zhou, X., Zhu, R., et al. (2021b). Bone marrow mesenchymal stem cells-derived extracellular vesicles promote proliferation, invasion and migration of osteosarcoma cells via the lncRNA MALAT1/miR-143/NRSN2/wnt/β-catenin Axis. Onco Targets Ther. 14, 737–749. doi:10.2147/OTT.S283459
Li, P., Zhan, W., Xie, D., Hong, G., Deng, M., Almeida, S. F. F., et al. (2023). Exosome-like nanovesicles derived from the mucilage of pinctada martensii exhibit antitumor activity against 143B osteosarcoma cells. ACS Appl. Mater Interfaces 15 (22), 26227–26240. doi:10.1021/acsami.2c21485
Li, Y., Lin, S., Xie, X., Zhu, H., Fan, T., and Wang, S. (2021a). Highly enriched exosomal lncRNA OIP5-AS1 regulates osteosarcoma tumor angiogenesis and autophagy through miR-153 and ATG5. Am. J. Transl. Res. 13 (5), 4211–4223.
Liberati, A., Altman, D. G., Tetzlaff, J., Mulrow, C., Gøtzsche, P. C., Ioannidis, J. P. A., et al. (2009). The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. Bmj 339, b2700. doi:10.1136/bmj.b2700
Lin, S., Zhu, B., Huang, G., Zeng, Q., and Wang, C. (2019). Microvesicles derived from human bone marrow mesenchymal stem cells promote U2OS cell growth under hypoxia: the role of PI3K/AKT and HIF-1α. Hum. Cell 32 (1), 64–74. doi:10.1007/s13577-018-0224-z
Liu, W., Long, Q., Zhang, W., Zeng, D., Hu, B., Liu, S., et al. (2021b). miRNA-221-3p derived from M2-polarized tumor-associated macrophage exosomes aggravates the growth and metastasis of osteosarcoma through SOCS3/JAK2/STAT3 axis. Aging (Albany NY) 13 (15), 19760–19775. doi:10.18632/aging.203388
Liu, Y., Xia, Y., Smollar, J., Mao, W., and Wan, Y. (2021a). The roles of small extracellular vesicles in lung cancer: molecular pathology, mechanisms, diagnostics, and therapeutics. Biochim. Biophys. Acta Rev. Cancer 1876 (1), 188539. doi:10.1016/j.bbcan.2021.188539
Lu, J., Chen, J., Ye, J., Shi, Z., Gao, X., Chen, P., et al. (2024). Dipsacus asperoides-derived exosomes-like nanoparticles inhibit the progression of osteosarcoma via activating P38/JNK signaling pathway. Int. J. Nanomedicine 19, 1097–1108. doi:10.2147/IJN.S446594
Lu, Y., Cao, G., Lan, H., Liao, H., Hu, Y., Feng, H., et al. (2022). Chondrocyte-derived exosomal miR-195 inhibits osteosarcoma cell proliferation and anti-apoptotic by targeting KIF4A in vitro and in vivo. Transl. Oncol. 16, 101289. doi:10.1016/j.tranon.2021.101289
Luo, T., Zhou, X., Jiang, E., Wang, L., Ji, Y., and Shang, Z. (2021). Osteosarcoma cell-derived small extracellular vesicles enhance osteoclastogenesis and bone resorption through transferring MicroRNA-19a-3p. Front. Oncol. 11, 618662. doi:10.3389/fonc.2021.618662
Mirabello, L., Troisi, R. J., and Savage, S. A. (2009). International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 125 (1), 229–234. doi:10.1002/ijc.24320
Mortus, J. R., Zhang, Y., and Hughes, D. P. (2014). Developmental pathways hijacked by osteosarcoma. Adv. Exp. Med. Biol. 804, 93–118. doi:10.1007/978-3-319-04843-7_5
Nair, R., Santos, L., Awasthi, S., von Erlach, T., Chow, L. W., Bertazzo, S., et al. (2014). Extracellular vesicles derived from preosteoblasts influence embryonic stem cell differentiation. Stem Cells Dev. 23 (14), 1625–1635. doi:10.1089/scd.2013.0633
Pingping, B., Yuhong, Z., Weiqi, L., Chunxiao, W., Chunfang, W., Yuanjue, S., et al. (2019). Incidence and mortality of Sarcomas in Shanghai, China, during 2002-2014. Front. Oncol. 9, 662. doi:10.3389/fonc.2019.00662
Qi, J., Zhang, R., and Wang, Y. (2021). Exosomal miR-21-5p derived from bone marrow mesenchymal stem cells promote osteosarcoma cell proliferation and invasion by targeting PIK3R1. J. Cell Mol. Med. 25 (23), 11016–11030. doi:10.1111/jcmm.17024
Qin, T., Zhu, W., Kan, X., Li, L., and Wu, D. (2022). Luteolin attenuates the chemoresistance of osteosarcoma through inhibiting the PTN/β-catenin/MDR1 signaling axis by upregulating miR-384. J. Bone Oncol. 34, 100429. doi:10.1016/j.jbo.2022.100429
Raimondi, L., De Luca, A., Gallo, A., Costa, V., Russelli, G., Cuscino, N., et al. (2020). Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis 41 (5), 666–677. doi:10.1093/carcin/bgz130
Ritter, J., and Bielack, S. S. (2010). Osteosarcoma. Ann. Oncol. 21 (Suppl. 7), vii320–5. doi:10.1093/annonc/mdq276
Roberts, B. K., Barnes, B., and Soffer, S. Z. (2022). Variable metastatic potential of osteosarcoma may Be due to differences in extracellular vesicles. J. Am. Coll. Surg. 235 (5), S195. doi:10.1097/01.xcs.0000894500.10236.5f
Santos, J. C., Lima, N. d. S., Sarian, L. O., Matheu, A., Ribeiro, M. L., and Derchain, S. F. M. (2018). Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 8 (1), 829. doi:10.1038/s41598-018-19339-5
Shi, Z., Wang, K., Xing, Y., and Yang, X. (2021). CircNRIP1 encapsulated by bone marrow mesenchymal stem cell-derived extracellular vesicles aggravates osteosarcoma by modulating the miR-532-3p/AKT3/PI3K/AKT Axis. Front. Oncol. 11, 658139. doi:10.3389/fonc.2021.658139
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Simpson, E., and Brown, H. L. (2018). Understanding osteosarcomas. Jaapa 31 (8), 15–19. doi:10.1097/01.JAA.0000541477.24116.8d
Smrke, A., Anderson, P. M., Gulia, A., Gennatas, S., Huang, P. H., and Jones, R. L. (2021). Future directions in the treatment of osteosarcoma. Cells 10 (1), 172. doi:10.3390/cells10010172
Song, H., Zhao, J., Cheng, J., Feng, Z., Wang, J., Momtazi-Borojeni, A. A., et al. (2021). Extracellular vesicles in chondrogenesis and cartilage regeneration. J. Cell Mol. Med. 25 (11), 4883–4892. doi:10.1111/jcmm.16290
Teng, X., Chen, L., Chen, W., Yang, J., Yang, Z., and Shen, Z. (2015). Mesenchymal stem cell-derived exosomes improve the microenvironment of infarcted myocardium contributing to angiogenesis and anti-inflammation. Cell Physiol. Biochem. 37 (6), 2415–2424. doi:10.1159/000438594
Tkach, M., and Théry, C. (2016). Communication by extracellular vesicles: where we are and where we need to go. Cell 164 (6), 1226–1232. doi:10.1016/j.cell.2016.01.043
Ucci, A., Cappariello, A., Ponzetti, M., Tennant, F., Loftus, A. E. P., Shefferd, K., et al. (2021). Anti-osteoblastogenic, pro-inflammatory and pro-angiogenic effect of extracellular vesicles isolated from the human osteosarcoma cell line MNNG/HOS. Bone 153, 116130. doi:10.1016/j.bone.2021.116130
Vakhshiteh, F., Atyabi, F., and Ostad, S. N. (2019). Mesenchymal stem cell exosomes: a two-edged sword in cancer therapy. Int. J. Nanomedicine 14, 2847–2859. doi:10.2147/IJN.S200036
Wang, B., Wang, X., Li, P., Niu, X., Liang, X., Liu, G., et al. (2022a). Osteosarcoma cell-derived exosomal ELFN1-AS1 mediates macrophage M2 polarization via sponging miR-138-5p and miR-1291 to promote the tumorgenesis of osteosarcoma. Front. Oncol. 12, 881022. doi:10.3389/fonc.2022.881022
Wang, J., Guo, W., Wang, X., Tang, X., Sun, X., and Ren, T. (2023). Circulating exosomal PD-L1 at initial diagnosis predicts outcome and survival of patients with osteosarcoma. Clin. Cancer Res. 29 (3), 659–666. doi:10.1158/1078-0432.CCR-22-2682
Wang, J., Li, M., Jin, L., Guo, P., Zhang, Z., Zhanghuang, C., et al. (2022b). Exosome mimetics derived from bone marrow mesenchymal stem cells deliver doxorubicin to osteosarcoma in vitro and in vivo. Drug Deliv. 29 (1), 3291–3303. doi:10.1080/10717544.2022.2141921
Wang, L., Wu, J., Song, S., Chen, H., Hu, Y., Xu, B., et al. (2021). Plasma exosome-derived sentrin SUMO-specific protease 1: a prognostic biomarker in patients with osteosarcoma. Front. Oncol. 11, 625109. doi:10.3389/fonc.2021.625109
Wei, H., Chen, F., Chen, J., Lin, H., Wang, S., Wang, Y., et al. (2022). Mesenchymal stem cell derived exosomes as nanodrug carrier of doxorubicin for targeted osteosarcoma therapy via SDF1-CXCR4 Axis. Int. J. Nanomedicine 17, 3483–3495. doi:10.2147/IJN.S372851
Weinman, M. A., Ramsey, S. A., Leeper, H. J., Brady, J. V., Schlueter, A., Stanisheuski, S., et al. (2021). Exosomal proteomic signatures correlate with drug resistance and carboplatin treatment outcome in a spontaneous model of canine osteosarcoma. Cancer Cell Int. 21 (1), 245. doi:10.1186/s12935-021-01943-7
Xu, F., Wei, Y., Tang, Z., Liu, B., and Dong, J. (2020a). Tumor-associated macrophages in lung cancer: friend or foe? (Review). Mol. Med. Rep. 22 (5), 4107–4115. doi:10.3892/mmr.2020.11518
Xu, Z., Zhou, X., Wu, J., Cui, X., Wang, M., Wang, X., et al. (2020b). Mesenchymal stem cell-derived exosomes carrying microRNA-150 suppresses the proliferation and migration of osteosarcoma cells via targeting IGF2BP1. Transl. Cancer Res. 9 (9), 5323–5335. doi:10.21037/tcr-20-83
Yan, C. F., Xia, J., Qun, W. S., Bing, W. Y., Guo, W. J., Yong, H. G., et al. (2023). Tumor-associated macrophages-derived exo-let-7a promotes osteosarcoma metastasis via targeting C15orf41 in osteosarcoma. Environ. Toxicol. 38 (6), 1318–1331. doi:10.1002/tox.23766
Yu, L., Fan, G., Wang, Q., Zhu, Y., Zhu, H., Chang, J., et al. (2023). In vivo self-assembly and delivery of VEGFR2 siRNA-encapsulated small extracellular vesicles for lung metastatic osteosarcoma therapy. Cell Death Dis. 14 (9), 626. doi:10.1038/s41419-023-06159-3
Zhan, H., Xiao, J., Wang, P., Mo, F., Li, K., Guo, F., et al. (2022). Exosomal CTCF confers cisplatin resistance in osteosarcoma by promoting autophagy via the IGF2-AS/miR-579-3p/MSH6 Axis. J. Oncol. 2022, 9390611. doi:10.1155/2022/9390611
Zhang, B., Yang, Y., Tao, R., Yao, C., Zhou, Z., and Zhang, Y. (2022). Exosomes loaded with miR-665 inhibit the progression of osteosarcoma in vivo and in vitro. Am. J. Transl. Res. 14 (10), 7012–7026.
Zhang, H., Yu, Y., Wang, J., Han, Y., Ren, T., Huang, Y., et al. (2021a). Macrophages-derived exosomal lncRNA LIFR-AS1 promotes osteosarcoma cell progression via miR-29a/NFIA axis. Cancer Cell Int. 21 (1), 192. doi:10.1186/s12935-021-01893-0
Zhang, L., Xue, L., Wu, Y., Wu, Q., Ren, H., and Song, X. (2021c). Exosomes loaded with programmed death ligand-1 promote tumor growth by immunosuppression in osteosarcoma. Bioengineered 12 (2), 9520–9530. doi:10.1080/21655979.2021.1996509
Zhang, Y., Liu, Z., Yang, X., Lu, W., Chen, Y., Lin, Y., et al. (2021b). H3K27 acetylation activated-COL6A1 promotes osteosarcoma lung metastasis by repressing STAT1 and activating pulmonary cancer-associated fibroblasts. Theranostics 11 (3), 1473–1492. doi:10.7150/thno.51245
Zhao, L., Gu, C., Gan, Y., Shao, L., Chen, H., and Zhu, H. (2020). Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control Release 318, 1–15. doi:10.1016/j.jconrel.2019.12.005
Zhao, W., Qin, P., Zhang, D., Cui, X., Gao, J., Yu, Z., et al. (2019). Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging (Albany NY) 11 (21), 9581–9596. doi:10.18632/aging.102406
Zhong, L., Liao, D., Li, J., Liu, W., Wang, J., Zeng, C., et al. (2021). Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. Signal Transduct. Target Ther. 6 (1), 59. doi:10.1038/s41392-020-00414-1
Zhongyu, X., Wei, X., Hongmei, Z., Xiaodong, G., Xiaojing, Y., Yuanpei, L., et al. (2024). Review of pre-metastatic niches induced by osteosarcoma-derived extracellular vesicles in lung metastasis: a potential opportunity for diagnosis and intervention. Biomed. Pharmacother. 178, 117203. doi:10.1016/j.biopha.2024.117203
Keywords: osteosarcoma, exosomes, preclinical studies, meta-analysis, chemotherapy
Citation: Liu X, Ye J, Guo W and Wang J (2024) Significance of exosomes in osteosarcoma research: a systematic review and meta-analysis of a singular clinical investigation. Front. Cell Dev. Biol. 12:1473044. doi: 10.3389/fcell.2024.1473044
Received: 30 July 2024; Accepted: 31 October 2024;
Published: 13 November 2024.
Edited by:
Jaime Villegas, Andres Bello University, ChileReviewed by:
Eszter Persa, Frédéric Joliot-Curie National Research Institute for Radiobiology and Radiohygiene, HungaryEmanuel Jeldes, University of Edinburgh, United Kingdom
Copyright © 2024 Liu, Ye, Guo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junqing Wang, d2FuZ2p1bnFpbmcwMDAxQDEyNi5jb20=
†These authors have contributed equally to this work
 Xuehong Liu
Xuehong Liu Jingyao Ye
Jingyao Ye Wenlong Guo
Wenlong Guo Junqing Wang4*
Junqing Wang4*