- 1Development and Regeneration Key Laboratory of Sichuan Province, Department of Anatomy and Histology and Embryology, School of Basic Medical Sciences, Chengdu Medical College, Chengdu, China
- 2Department of Ultrasound, Luzhou People’s Hospital, Luzhou, China
- 3Department of Cardiology, Chengdu Seventh People’s Hospital, Chengdu, Sichuan, China
- 4Department of Neurology, The Second Affiliated Hospital of Chengdu Medical College, (China National Nuclear Corporation 416 Hospital), Chengdu, China
- 5Department of Pathology and Pathophysiology, School of Basic Medical Sciences, Chengdu Medical College, Chengdu, China
- 6Ministry of Education Key Laboratory of Child Development and Disorders, China International Science and Technology Cooperation Base of Child Development and Critical Disorders, National Clinical Research Center for Child Health and Disorders, Chongqing Key Laboratory of Pediatrics, Children’s Hospital of Chongqing Medical University, Chongqing, China
Cdon and boc are members of the cell adhesion molecule subfamily III Ig/fibronectin. Although they have been reported to be involved in muscle and neural development at late developmental stage, their early roles in embryonic development remain unknown. Here, we discovered that in zebrafish, cdon, but not boc, is expressed in dorsal forerunner cells (DFCs) and the epithelium of Kupffer’s vesicle (KV), suggesting a potential role for cdon in organ left-right (LR) patterning. Further data showed that liver and heart LR patterning were disrupted in cdon morphants and cdon mutants. Mechanistically, we found that loss of cdon function led to defect in DFCs clustering, reduced KV lumen, and defective cilia, resulting in randomized Nodal/spaw signaling and subsequent organ LR patterning defects. Additionally, predominant distribution of a cdon morpholino (MO) in DFCs caused defects in DFC clustering, KV morphogenesis, cilia number/length, Nodal/spaw signaling, and organ LR asymmetry, similar to those observed in cdon morphants and cdon−/− embryos, indicating a cell-autonomous role for cdon in regulating KV formation during LR patterning. In conclusion, our data demonstrate that during gastrulation and early somitogenesis, cdon is essential for proper DFC clustering, KV formation, and normal cilia, thereby playing a critical role in establishing organ LR asymmetry.
1 Introduction
The establishment of left-right (LR) patterning is a crucial event in early embryonic development. Organ asymmetry defects are closely related to the occurrence of various congenital diseases such as congenital heart disease and schizophrenia (Huang et al., 2014; Gabriel and Lo, 2020). Although the mechanisms for establishing organ LR patterning are complex and diverse across different animal models (Hamada and Tam, 2020; Onuma et al., 2020; Zhu et al., 2020), the role of the Kupffer’s vesicle (KV)/Node-Cilia axis in zebrafish and mice is highly conserved (Grimes and Burdine, 2017; Little and Norris, 2021; Forrest et al., 2022). In zebrafish, the normal development of KV precursor cells, called dorsal forerunner cells (DFCs), and KV morphogenesis/ciliogenesis play crucial roles in initiating left-sided signals in the embryonic lateral plate mesoderm (LPM) (Grimes and Burdine, 2017). During organ LR patterning, DFCs proliferate rapidly during gastrulation movement (Gokey et al., 2015; Abdel-Razek et al., 2023). After the bud stage, DFCs differentiate to form the KV and produce cilia, which oscillate to generate a counter-clockwise liquid flow and initiate specific asymmetric Nodal/spaw expression on the LPM (Bakkers et al., 2009; Liu et al., 2019). Furthermore, the left-sided Nodal/spaw and its downstream genes pitx2 and lft2 are amplified in the LPM. The embryonic midline (such as shh and ntl) also impedes the expansion of left-sided Nodal signals to the right side of the embryo, thus stabilizing the left-sided asymmetric Nodal signals (Burdine and Grimes, 2016). Finally, at the late stage of organ development, the left-sided Nodal directs an asymmetric migration of organ precursors to establish organ asymmetry patterning (Kajikawa et al., 2022).
The role of zebrafish KV morphogenesis/ciliogenesis and the underlying molecular mechanisms during organ LR patterning have received extensive attention. It has been found that many classical signaling pathways, such as Wnt and Fgf (Neugebauer et al., 2009; Xu et al., 2010; Caron et al., 2012; Zhang et al., 2012) and other genes, such as Rab5 (Kuhns et al., 2019) and FOR20 (Xie et al., 2019), participated in KV morphogenesis or ciliogenesis. Our previous studies found that during zebrafish gastrulation, inhibition of retinoic acid (RA) increases the expression of fgf8 in DFCs, leading to longer cilia and cilia motility disorders (Huang et al., 2011). More recently, we found that the chemokine receptor cxcr4a is highly expressed in DFCs, and that cxcr4a promotes CyclinD1 expression to regulate ciliogenesis by regulating the phosphorylation of ERK1/2 (Liu et al., 2019). Although many genes have been reported to play crucial roles during KV morphogenesis and ciliogenesis, the mechanisms underlying these processes are far from being elucidated.
Cell-adhesion molecule-related/downregulated by oncogenes (cdon) and its paralog boc [Brother of cdon (Kang et al., 2002)] are two members of the cell adhesion molecule subfamily III Ig/fibronectin (Sanchez-Arrones et al., 2012; Jeong et al., 2014). They have been reported to synergistically or individually regulate the development of different organs or diseases (Zhang et al., 2011; Sanchez-Arrones et al., 2012; Jeong et al., 2014; Jeong et al., 2017; Lencer et al., 2023). Regarding cdon, early reports showed that mouse cdon regulates skeletal muscle and central nervous system development via Shh and N-cadherin/cell adhesion (Cole et al., 2004; Zhang et al., 2006a; Zhang et al., 2006b; Jeong et al., 2014). More detailed studies have shown that cdon binds to N-cadherin and induces p38/MAPK signaling to guide cell differentiation and apoptosis during muscle formation and neural differentiation (Lu and Krauss, 2010). In mouse, Cdon has been reported to negatively regulate the Wnt signaling pathway during forebrain development, promoting neural differentiation and ventral cell fate determination via interaction with LRP6 (Jeong et al., 2014). Further, researchers found that the localization of Cx43 protein in cells was disorganized in cdon mutants, resulting in cardiac structural and functional lesions (Jeong et al., 2017). Regarding boc, early reports showed that it plays a role in axon guidance and neuron growth (Connor et al., 2005; Okada et al., 2006). Recently, Lencer et al. found that in zebrafish, cdon and boc double mutants display trunk neural crest cell (tNCC) migratory defects and loss of slow-twitch muscle, suggesting a non-cell-autonomous role for cdon and boc in regulating tNCC migration (Lencer et al., 2023). Although cdon and boc have been reported to play important roles in various contexts in mouse and zebrafish, their roles in early embryonic development remain unknown. Our data showed that during early embryonic development in zebrafish, cdon, but not boc, is expressed in DFCs during gastrulation movement and in the epithelial cells of KV at the early somitogenesis stage (Figure 1; Supplementry Figures S1). Additionally, cdon has been reported to control cell apoptosis and migration in various cell types (Sanchez-Arrones et al., 2012; Chapouly et al., 2020; Lencer et al., 2023). These findings suggest that cdon may be involved in regulating the clustering movement of DFCs and the subsequent organ LR patterning in zebrafish. Here, our data identify that cdon regulates organ LR patterning via the KV/Cilia-Nodal/spaw cascade during early embryonic development.
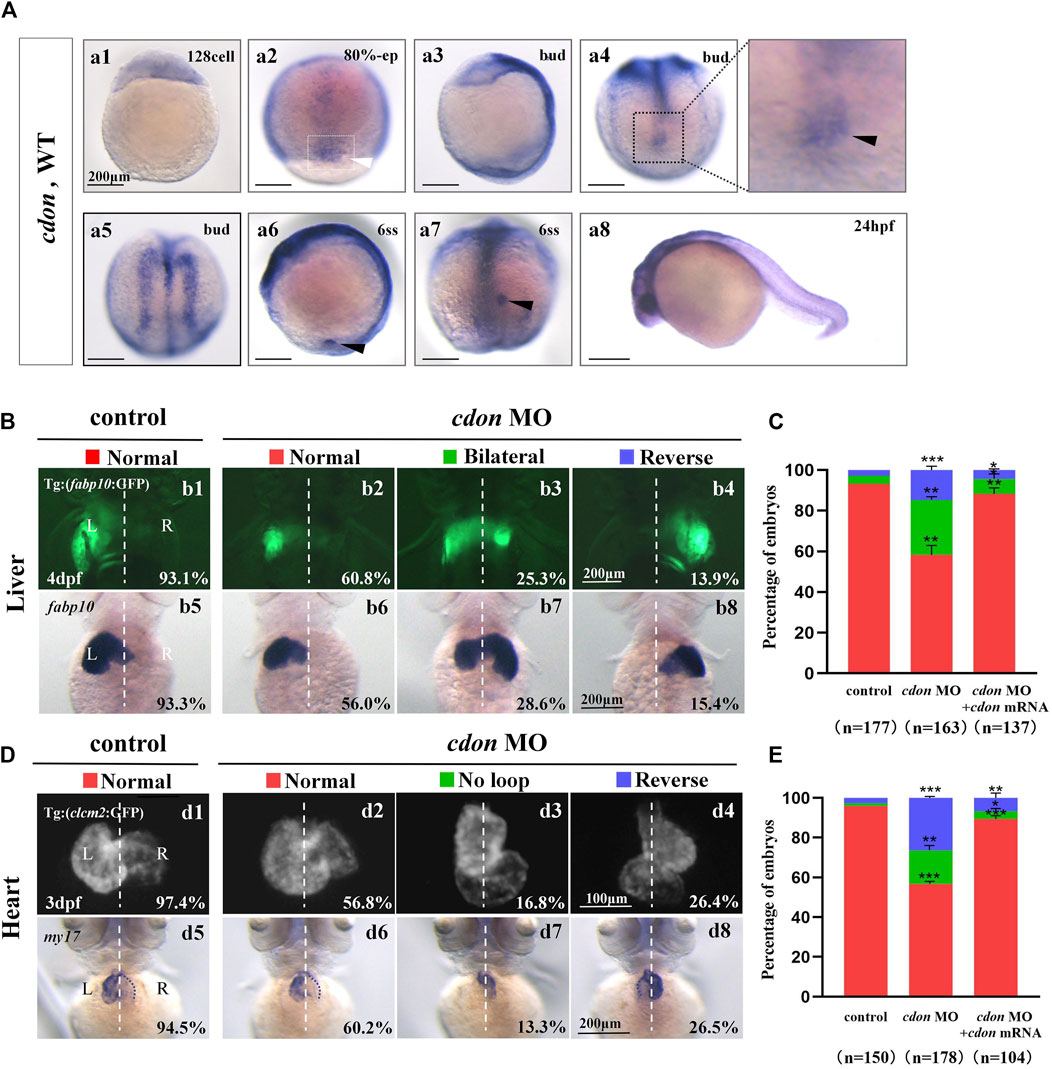
Figure 1. Organ left-right patterning defects in embryos injected with cdon MO at the 4-cell stage (A) The expression of cdon was examined using whole mount in situ hybridization from 128-cell stage to 24hpf. Cdon was expressed at 128-cell stage (Aa1). White arrowhead showed the expression of cdon in DFCs at 80% epiboly stage (Aa2). Black arrowhead showed the expression of cdon in DFCs at bud stage (Aa3-a4). Cdon was also expressed in midline and presumptive neural crest (Aa5). Black arrow head showed the expression of cdon in epithelial cell in KV at 6ss (Aa6-a7). Expression of cdon was examined at 24 hpf (Aa8). (B) Embryos injected with cdon MO displayed liver LR defects. b1, normal liver in Tg (fabp10:GFP) transgenic controls (93.1%, n = 102); b2, normal liver in embryos injected with cdon MO (60.8%, n = 79); b3, liver bifida in embryos injected with cdon MO (25.3%, n = 79); b4, reversed liver in embryos injected with cdon MO (13.9%, n = 79). b5, normal liver in wild type controls (93.3%, n = 75); b6-b8, embryos injected with cdon MO were examined at 4dpf using in situ experiments (n = 84). (C) Percentages of normal liver, liver bifida, and reversed liver in embryos used as control, embryos injected with cdon MO and embryos co-injected with cdon MO and cdon mRNA, in respectively. Embryos injected with cdon MO show a statistically significant difference compared to controls, and embryos co-injected with cdon MO and cdon mRNA show a statistically significant difference compared to those injected with cdon MO alone. (D) Embryos injected with cdon MO displayed heart LR defects. d1, normal-loop in Tg(cmlc2:GFP) transgenic controls (97.4%, n = 77); d2, normal-loop in embryos injected with cdon MO (56.8%, n = 95); d3, no loop in embryos injected with cdon MO (16.8%, n = 95); d4, reversed-loop in embryos injected with cdon MO (26.4%, n = 95); d5, normal-loop in wild type controls (94.5%, n = 73); d6-d8, wild-type embryos injected with cdon MO at the 4-cell stage were examined for cardiac looping at 72 hpf by WISH against my17 (n = 83). The blue dashed line indicates the atrial edge. (E) Percentages of normal looping, no looping, and reversed looping of the heart in embryos being as control, embryos injected with cdon MO, and embryos co-injcted with cdon MO and cdon mRNA. Cdon mRNA injection partially rescued the heart LR defect in embryos injected with cdon MO (E, the right column showing). Embryos injected with cdon MO show a statistically significant difference compared to controls, and embryos co-injected with cdon MO and cdon mRNA show a statistically significant difference compared to those injected with cdon MO alone. Notice: in “C” and “E,” all the transgenic embryos and wild type embryos were used together to calculate the percentage. Statistical analysis was performed using Student’s t-test. “*”p < 0.05, “**” p < 0.01, “***” p < 0.001. Notice: “control” refers to wild-type embryos that were not injected with cdon MO.
2 Methods and materials
2.1 Zebrafish
Zebrafish wild type (AB), Tg (cmcl2:GFP) (Liu et al., 2022), Tg (fabp10:GFP) (Liu et al., 2022) and Tg (sox17:EGFP) (Chung and Stainier, 2008) lines were maintained at 28.5°C. The embryonic stages were determined according to external morphology, based on the standard criteria described previously (Kimmel et al., 1995).
2.2 Whole-mount in situ hybridization (WISH)
Embryos used for whole-mount in situ hybridization (WISH) were collected at desired stages and fixed in 4% PFA at 4°C overnight. The embryos were then washed with PBST (2 times, 5 min each), dehydrated with 100% MeOH, and stored in MeOH at −20°C. The WISH procedure followed the experimental protocol described previously (Huang et al., 2011; Zhu et al., 2019). The antisense probes my17, fabp10, spaw, lefty1, lefty2, sox17, and sox32 were prepared as previously described (Zhu et al., 2019). To prepare the cdon antisense probe, the cDNA of cdon was amplified using PCR and then cloned into the pCS2 + vector. The detailed process can be found in the section “Plasmid Construction.” This construct was then used as the template to synthesize the cdon antisense probe according to the previously established protocol (Zhu et al., 2019).
2.3 Immunostaining
The immunostaining was performed as previously reported (Zhu et al., 2019). Briefly, the embryos were fixed with 4% PFA at 4°C overnight, then dehydrated with a methanol gradient and stored at −20°C. After methanol/PBST gradient rehydration, PBTN (4% BSA, 0.02% NaN3, PT) was added, and the embryos were incubated at 4°C for 3 h. Then, the α-tubulin antibody (Sigma, T7451, diluted with PBTN at 1:100) was added and incubated overnight on a shaker at 4°C. The next day, the embryos were washed with PT (0.3% Triton X-100 in 1X PBS) 4 times (30 min each), and the secondary antibody, Goat anti-Mouse IgG (H + L) Alexa Fluor 594 (Sigma, SAB4600105), was added (diluted with PBTN at 1:1,000) for overnight incubation in darkness. Finally, the embryos were rinsed with PT more than 6 times (30 min each), and imaging was performed.
2.4 Plasmids construction
Total RNA was extracted from zebrafish embryos at 24 hpf according to the reagent instructions (TRIzol, Ambion No.15596-026). cDNA was prepared using the RevertAid First Strand cDNA Synthesis Kit (Fermentas No. K1622) according to the manufacturer’s instructions. The cDNA of cdon was amplified using PCR (PrimeSTAR Max Premix, Takara No. R045 A) and cloned into the vector pCS2+ to generate the expression constructs (5x In-Fusion HD Enzyme Premix, Takara No. 639649). The primers for cloning were as follows:
PCS2+_F: 5′-CTCGAGCCTCTAGAACTATAGTG-3′
PCS2+_R: 5′-TGGTGTTTTCAAAGCAACGATATCG-3′
Cdon_F: 5′-TCTTTTTGCAGGATCCGTGAAACAGCGTCATGGAGGAC-3′
Cdon_R: 5′-GTTCTAGAGGCTCGAGTGTTCAGATCTCCTGCACGGT-3′
2.5 MO and mRNA injection
The antisense cdon ATG morpholino oligonucleotide (cdon MOatg) was synthesized (Gene Tools) and applied to block cdon mRNA translation. cdon MOatg: 5′-ATAATCTCAGGCCACCGTCCTCCAT-3′, 400 μM (Cardozo et al., 2014). Cdon MO was injected into the yolk of zebrafish embryos at the 1–4 cell stage to block the translation of cdon in whole embryos, or at the 256–512 cell stage to specifically block the translation of cdon mRNA in DFCs. The cdon mRNA was synthesized in vitro using the mMESSAGE Kit (AM1340, Ambion). In rescue experiments, the concentration for cdon mRNA injection was 15 ng/μL.
2.6 Generation of cdon−/− mutants
To generate cdon−/− mutants, we used the CRISPR design tool (http://crispr.mit.edu) to design CRISPR short guide RNAs (sgRNAs) targeting exon 3 of the cdon gene. Injection mixtures contained 500 pg Cas9 mRNA and 120 pg gRNA in each embryo. Embryos injected with sgRNA and Cas9 mRNA generated the F0 line with mosaic mutations. These F0 mosaic adult fish were outcrossed with wild-type fish to produce F1 heterozygous lines. In one F1 adult, the sequence “AAGGGC” in exon 3 of the cdon gene was changed to “TTGATGAATGGGG”, resulting in a truncated Cdon protein. This F1 heterozygote was outcrossed with wild-type fish to produce F2 heterozygous fish, then incrossed them to obtain F3 homozygous mutants. The homozygous embryos from this cdon−/− mutant line were used to perform experiments as required.
2.7 Imaging
Images of the whole-mount in situ hybridization (in 100% glycerol) were taken with an OLYMPUS SZX16 microscope at room temperature. Live embryos of the transgenic line Tg (sox17:EGFP) were placed in 1% low-point melting agarose (LPM agarose) and DFCs were photographed using a confocal microscope (OLYMPUS Fluview FV1000). To obtain images of cilia for immunostained embryos, the embryos were oriented correctly in 1% LPM agarose, and the cilia were photographed using a confocal microscope (OLYMPUS Fluview FV1000).
2.8 Statistical analysis
The data were statistically analyzed using GraphPad Prism 9 and ImageJ software. The length of the cilia was measured using a confocal microscope (OLYMPUS Fluview FV1000), and the OLYMPUS SZX16 microscope was used to measure the KV lumen area. The statistical results are presented as the mean ± SEM of three independent experiments. Statistical comparisons between two groups were performed using Student’s t-test. Statistical significance was defined as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
2.9 Ethics statement
The study was approved by the Institutional Review Board of Chengdu Medical College (SYXK(川)2015–196), and zebrafish were maintained in accordance with the Guidelines of Experimental Animal Welfare from the Ministry of Science and Technology of the People’s Republic of China (2006).
3 Results
3.1 Heart and liver LR patterning was disturbed in cdon morphants
In zebrafish, cdon and boc have been reported to be involved in trunk neural crest cell (NC) migration and correct proximo-distal patterning in eye development (Cardozo et al., 2014; Lencer et al., 2023), demonstrating the crucial role of cdon in zebrafish organ development. Since cdon and boc synergistically regulate NCs migration (Lencer et al., 2023), facial and eye development (Zhang et al., 2011), to study the role of cdon in more early embryonic development, we examined the detailed expression pattern of cdon and boc from 128-cell stage to 24 hpf. The data showed that cdon is a maternal factor that is distributed unequally in cells (Figure 1Aa1). During gastrulation, besides its expression in the presumptive neural crest and midline [Figure 1Aa5, (Jeong et al., 2014)], cdon was also highly expressed in DFCs (Figure 1Aa2–a4). At the 6-somite stage, cdon was enriched in the epithelium of the KV (Figures 1Aa6, a7). In contrast, although boc was maternally and ubiquitously expressed during gastrulation (Supplementry Figures S1A, B), it was not enriched in DFCs or the epithelium of the KV (Supplementry Figures S1E, F). Since disrupting DFC development and the sequential KV/ciliogenesis leads to organ LR patterning defects (Xie et al., 2019; Zhu et al., 2019), we proposed that cdon is involved in organ LR patterning. To preliminarily and rapidly evaluate this hypothesis, an ATG MO for cdon was used to downregulate the function of cdon (Jeong et al., 2014), and organ LR patterning was examined. The results showed that in Tg (fabp10:GFP) transgenic embryos and wild type embryos, injection of cdon MO did not lead to embryonic defects (Supplementary Figure S2), but gave rise to liver LR patterning defects (Figures 1Bb1–b8, C). Next, we examined whether injection of cdon MO led to heart looping defects. The results showed that injection of cdon MO caused heart LR patterning defects (Figures 1D, E); many cdon morphants displayed linear heart (Figures 1Dd3, d7, E) and reversed heart looping (Figures 1Dd4, d8, E). To further confirm the role of cdon in liver and heart LR patterning, we evaluated whether injection of cdon mRNA could rescue the organ LR patterning defects in cdon morphants. The data showed that injection of cdon mRNA partially restored organ LR patterning defects in embryos injected with cdon MO (Supplementary Figure S3; Figures 1C, E; the right columns show). These data indicate that cdon plays a critical role in organ LR patterning in zebrafish.
3.2 Left-sided Nodal/spaw cascade is randomized in cdon morphants
The crucial role of Nodal/spaw in LR patterning has been demonstrated in previous studies (Raya and Izpisua Belmonte, 2006; Speder et al., 2007; Kang et al., 2013). Disruption of Nodal/spaw leads to organ laterality defects (Huang et al., 2011). However, in some types of mutants with organ LR patterning defects, Nodal/spaw was not disturbed (Trinh and Stainier, 2004; Kurpios et al., 2008; Yin et al., 2010; Huang et al., 2014). To reveal how cdon regulates organ laterality, we examined whether left-sided Nodal/spaw was disturbed in embryos injected with cdon MO. The data showed that left-sided Nodal/spaw was disturbed in cdon morphants, displaying both-sided and right-sided(Figures 2Aa2–a4, B). Furthermore, we sequentially examined lefty1 and lefty2, the downstream genes of spaw in cdon morphants. The data showed that the left-sided expression patterns of lefty2 and lefty1 in the heart field were also perturbed (Figures 2Cc2–c4, D, Ee2–e4, F). Since cdon was also expressed in the midline [Figure 1Aa5, (Jeong et al., 2014)], we observed whether lefty1 expression in the midline was affected. The data showed that lefty1 expression in the midline was not affected (Figures 2Cc2–c4, red arrow). These data suggest that the left-sided Nodal/spaw cascade was disturbed in cdon morphants, which may lead to organ LR patterning defects in cdon morphants.
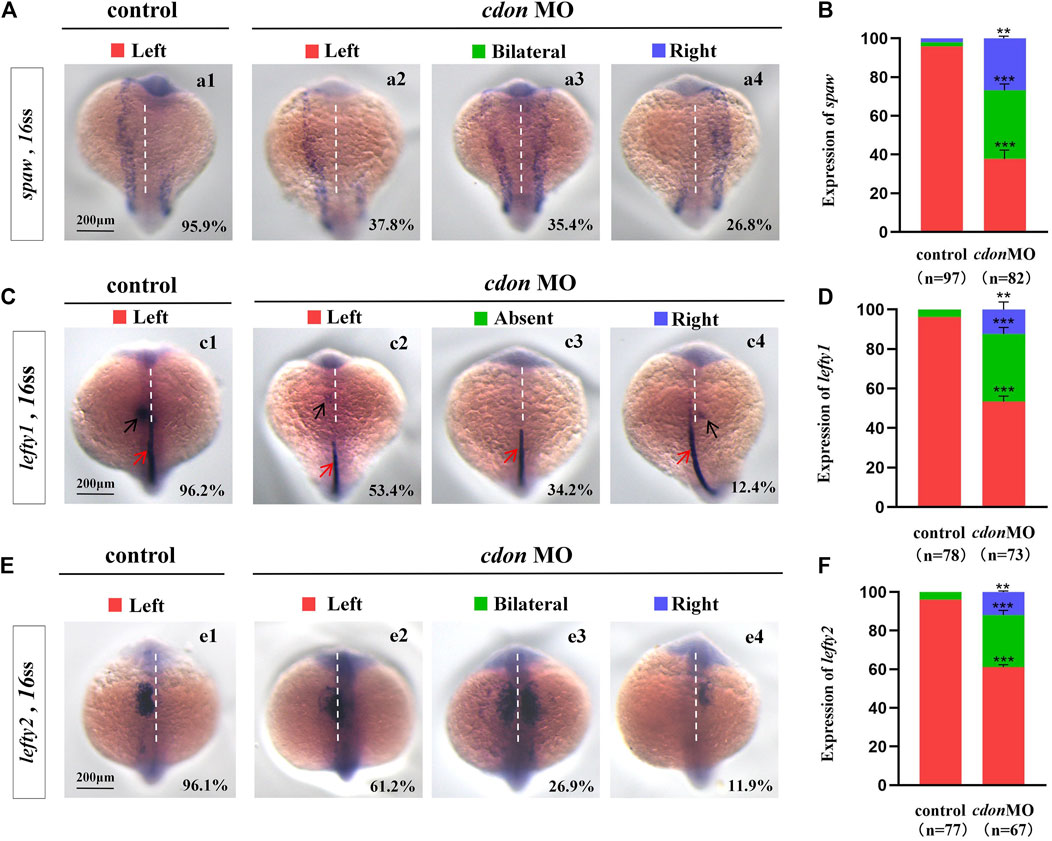
Figure 2. Expression of left-sided Nodal signaling in controls and embryos injected with cdon MO (A,B) Expression of spaw was evaluated in different groups of embryos. a1, left-sided spaw in controls (95.9%, n = 97); a2, left-sided spaw in embryos injected with cdon MO (37.8%, n = 82, p < 0.001); a3, bilateral spaw in embryos injected with cdon MO (35.4%, n = 82, p < 0.001); a4, right-sided spaw in embryos injected with cdon MO (26.8%, n = 82, p < 0.01). (C,D) Lefty1 is expressed in the heart field (black arrow) and trunk midline (red arrow) in controls and embryos injected with cdon MO. c1, left-sided lefty1 in embryos being as controls (96.2%, n = 78, black arrow showed); c2, left-sided lefty1 in embryos injected with cdon MO (53.4%, n = 73, p < 0.001, black arrow showed); c3, absent lefty1 in embryos injected with cdon MO (34.2%, n = 73, p < 0.001); c4, right-sided lefty1 in embryos injected with cdon MO (12.4%, n = 73, p < 0.01, black arrow showed) (E,F) Lefty2 is examined in control embryos and embryos injected with cdon MO. e1, left-sided lefty2 in controls (96.1%, n = 77); e2, left-sided lefty2 in embryos injected with cdon MO (61.2%, n = 67, p < 0.001); e3, bilateral expression of lefty2 in embryos injected with cdon MO (26.9%, n = 67, p < 0.001); e4, right-sided lefty2 in embryos injected with cdon MO (11.9%, n = 67, p < 0.01). Statistical analysis was performed using Student’s t-test. “**” p < 0.01, “***” p < 0.001. Notice: “control” refers to wild-type embryos that were not injected with cdon MO.
3.3 DFCs clustering, KV morphogenesis and cilia are disturbed in cdon morphants
Our current research showed that cdon was expressed in the DFCs during the gastrulation stage and in the epithelial cells of the KV (Figures 1Aa2–a7). In zebrafish, DFCs form the KV at the early somite stage. Defective KV morphogenesis or ciliogenesis can lead to disturbed left-sided Nodal/spaw expression and subsequent organ LR defects (Long et al., 2003; Essner et al., 2005; Wang et al., 2011). To determine whether cdon regulates DFC development, KV morphogenesis, or ciliogenesis, we analyzed the expression of DFC markers sox17 and sox32, as well as KV morphogenesis and ciliogenesis in cdon morphants. Compared with control embryos, at the 80% epiboly stage, the clustering DFC was disturbed in cdon morphants (Figures 3Aa2–a4, Aa6–a8, B), displaying linear expression (Figures 3Aa3, Aa7) and dispersed expression (Figures 3Aa4, Aa8). To further verify whether clustering DFC is affected, we injected cdon MO into Tg(sox17:EGFP) embryos. The results showed that at the 70% epiboly stage, DFCs remained as a tight cluster in the control embryos, while linear (Figure 3Cc3) or fragmented DFC clusters (Figure 3Cc4–c6) were detected in the cdon morphants (Figure 3C). Additionally, there was a significant decrease in the number of DFC cells at the 70% epiboly stage in the embryos injected with cdon MO compared to the control embryos (Figure 3D). Further, at the 10–13 somite stage (SS), the size of the KV lumen was smaller in the majority of cdon morphants (Figure 3Ee3, F, G), and a small portion of morphants displayed tiny or absent KV lumen (Figures 3Ee4, F, G). At the 10 somite stage, we used Tg (sox17:EGFP) transgenic zebrafish to examine the number of epithelium cells in KV. The results showed that, compared to the control, the number of epithelium cells in KV was significantly reduced in embryos injected with cdon MO at the 4-cell stage (Figures 3H, I). Furthermore, we examined cilia development and found that cdon morphants displayed slightly shorter cilia than controls (Figures 3Jj2–j4, K), and the number of cilia was also decreased in cdon morphants (Figures 3Jj2–j4, L). These results showed that normal expression of cdon is required for DFC development and KV morphogenesis, cdon MO injection also led to shorter cilia and decreased the number of cilia.
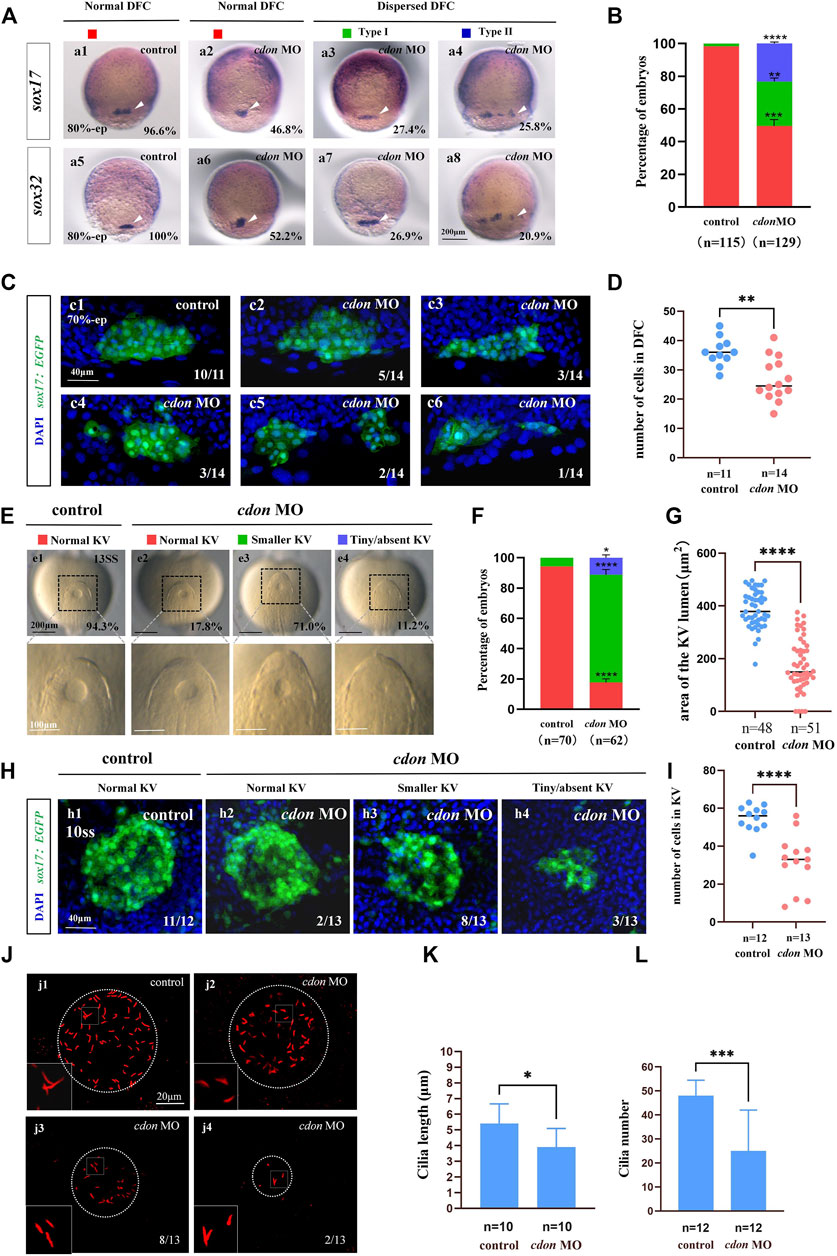
Figure 3. Cdon loss of function disturbs clustering DFCs, KV morphogenesis and cilia (A) Expression of sox17 and sox32 was examined using WISH at 80% epiboly (white arrow). a1, normal expression of sox17 in controls (96.6%, n = 58); a2, normal expression of sox17 in embryos injected with cdon MO at the 4-cell stage (46.8%, n = 62); a3, dispersed expression of sox17 (type I) in embryos injected with cdon MO at the 4-cell stage (27.4%, n = 62); a4, dispersed expression of sox17 (type II) in embryos injected with cdon MO at the 4-cell stage (25.8%, n = 62). a5, normal expression of sox32 in controls (100%, n = 57); a6, normal expression of sox32 in embryos injected with cdon MO at the 4-cell stage (52.2%, n = 67); a7, dispersed expression of sox32 (type I) in embryos injected with cdon MO at the 4-cell stage (26.9%, n = 67); a8, dispersed expression of sox32 (type II) in embryos injected with cdon MO at the 4-cell stage (20.9%, n = 67). (B) Statistical analysis was performed for the expression of sox17 and sox32 in controls and embryos injected with cdon MO. Here all the embryos staining with sox17 or sox32 were used together to calculate the percentage. Normal DFC and dispersed DFC show significant differences between control embryos and embryos injected with cdon MO at the 4-cell stage. (C) Using Tg (sox17:EGFP) transgenic zebrafish to detect DFC cell migration at the 70% epiboly stage in both control embryos and embryos injected with cdon MO at the 4-cell stage. c1, the DFCs in control embryos are tightly clustered (10/11, n = 11); c2, tightly clustered DFCs in embryos injected with cdon MO at the 4-cell stage (5/14, n = 14). c3, in embryos injected with cdon MO at the 4-cell stage, the DFCs are arranged in an elongated pattern with a reduced number of cells (3/14, n = 14). c4, mildly dispersed DFCs in embryos injected with cdon MO at the 4-cell stage (3/14, n = 14). c5, moderately dispersed DFCs in embryos injected with cdon MO at the 4-cell stage (2/14, n = 14). c6, in embryos injected with cdon MO at the 4-cell stage, the DFCs are dispersed, and the number of cells is significantly reduced (1/14, n = 14). (D) Number of cells in DFC in control and cdon MO-injected embryos. “n” represents the sample size. (E,F) KV morphology was evaluated. e1, normal KV in embryos in controls (94.3%, n = 70); e2, normal KV in embryos injected with cdon MO at the 4-cell stage (17.8%, n = 62, p < 0.001); e3, smaller KV in embryos injected with cdon MO at the 4-cell stage (71.0%, n = 62, p < 0.0001); e4, tiny/absent KV in embryos injected with cdon MO at the 4-cell stage (11.2%, n = 62, p < 0.05). “Normal KV” represents a KV lumen area greater than 300 μm2, “smaller KV” represents a KV lumen area between 100 and 300 μm2, and “tiny/absent KV” represents a KV lumen area less than 100 μm2. (G) Area of the KV lumen (µm2) in control and cdon MO-injected embryos. “n” represents the sample size. (H) The number of epithenium cells in KV were measured on confocal images. (I) Number of epithenium cells in KV in control and cdon MO-injected embryos. “n” represents the sample size. (J) Number and length of cilia were evaluated in controls and embryos injected with cdon MO. j1, cilia in embryos being as controls; j2-j4, cilia in embryos injected with cdon MO at the 4-cell stage. (K) Statistical chart for cilia length in KV. “n”represents the sample size. (L) Statistical chart for cilia number in KV. “n”represents the sample size. Statistical analysis was performed using Student’s t-test. “*” p < 0.05, “**” p < 0.01, “***” p < 0.001, “****” p < 0.0001. Notice: “control” refers to wild-type embryos that were not injected with cdon MO.
3.4 Cdon mutation leads to organ LR patterning defect
To confirm the role of cdon in organ LR patterning, we generated a cdon mutant line using the CRISPR-Cas9 method (Shankaran et al., 2017). To generate the cdon mutant, we selected a specific sequence in the exon 3 of cdon as the target sequence (Figure 4A). As a result, in F1 adults we screened out a frame shift mutation line (Figures 4B, C). In this mutation, the sequence “AAGGGC” in exon 3 of the cdon gene was changed to “TTGATGAATGGGG” (Figures 4B, C), resulting in a truncated Cdon protein (only 104 amino acids) (Figure 4C). In addition, even though we found that the expression of cdon mRNA was greatly downregulated in cdon−/− embryos at the 8-somite stage and 24 hpf (Supplementary Figures S4C, D), the cdon−/− embryos had no distinct external phenotype at different stages (Supplementary Figures S4A, B) and could grow to adulthood. Then we evaluated whether this frameshift mutation leads to liver and heart LR patterning defects using in situ experiments. The data showed that cdon−/− embryos, but not control embryos (Figures 4Dd1), displayed liver LR patterning defects: 71.2% of cdon−/− embryos displayed left-sided liver (Figures 4Dd2, E), 17.8% displayed bilateral liver (Figures 4Dd3, E), and 11.0% displayed right-sided liver (Figures 4Dd4, E). Similarly, no heart LR patterning defects were observed in control embryos (Figures 4Ff1, G), but cdon−/− embryos displayed no-loop heart (16.4%; Figures 4Ff3, G) and reversed-loop heart (10.9%; Figures 4Ff4, G). To further confirm the critical role of cdon in organ LR patterning, we examined whether injection of cdon mRNA could rescue liver and heart LR patterning defects in cdon−/− embryos. The data showed that injection of cdon mRNA partially restored liver and heart LR patterning (Figures 4E, G). All these data in cdon−/− embryos further demonstrate that cdon is essential for organ LR patterning.
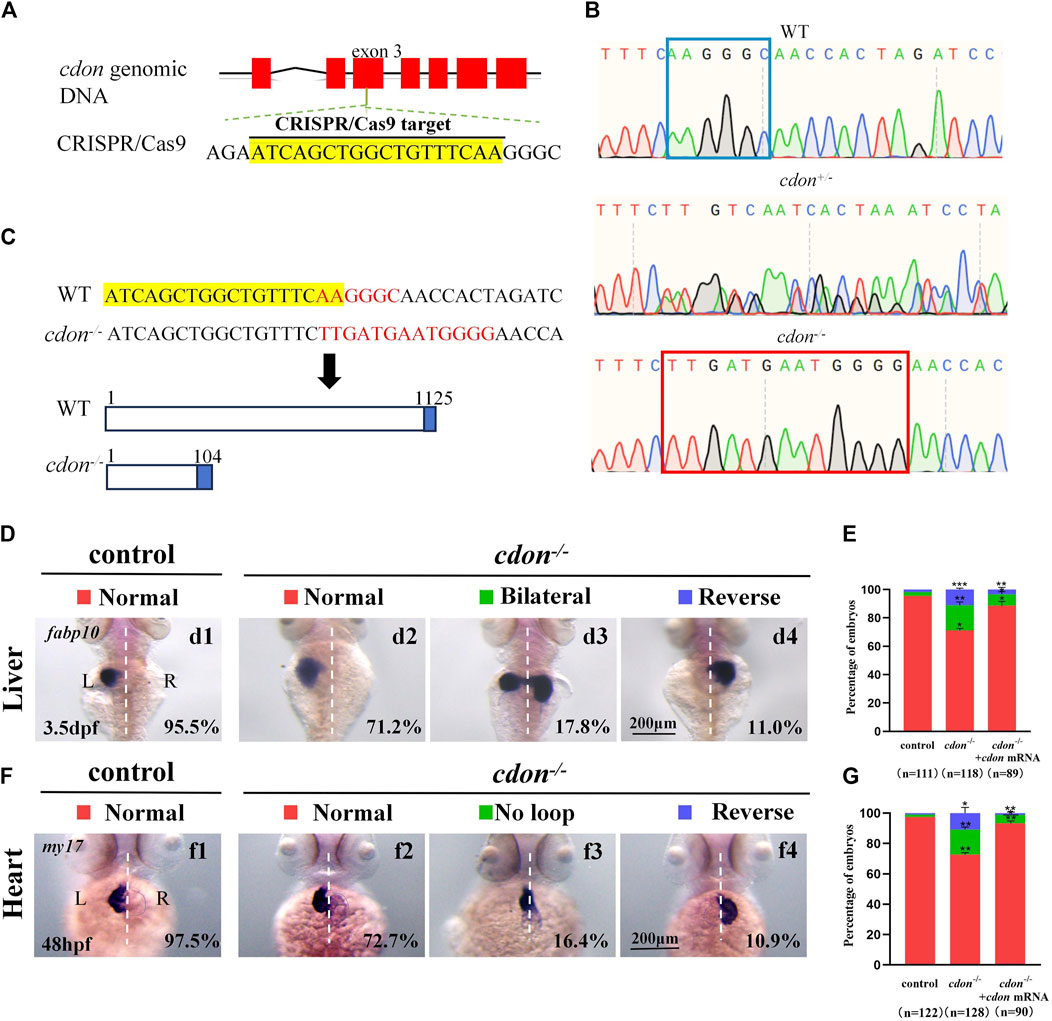
Figure 4. Organ LR patterning defects in cdon−/−embryos (A) CRISPR/Cas9 target was showed. The sequence in the exon 3 of cdon gene was chosen as the target (yellow highlighted). (B) Sequencing results for the genomic DNA in WT embryos, cdon+/− embryos and cdon−/− embryos. (C) Nucleotide sequences highlighted in yellow are CRISPR/Cas9 targets. In the cdon mutant, the sequence “AAGGGC” was changed to “TTGATGAATGGGG”. Cdon mutant harbors a frameshift mutation that is predicted to result in the production of truncated Cdon protein (104 amino acids). (D) Cdon−/− embryos were found to display liver LR defects using WISH. d1, left-sided liver in controls (95.5%, n = 111); d2, left-sided liver in cdon−/− embryos (71.2%, n = 118); d3, liver bifida in cdon−/− embryos (17.8%, n = 118); d4, right-sided liver in cdon−/− embryos (11.0%, n = 118). (E) Percentages of left-sided liver, liver bifida, and right-sided liver in cdon−/− embryos (n = 118), controls (n = 111) and cdon−/− embryos injected with cdon mRNA (n = 89), cdon−/− embryos show a statistically significant difference compared to controls, and cdon−/− embryos injected with cdon mRNA show a statistically significant difference compared to cdon−/− embryos. (F) Cdon−/− embryos displayed heart LR defects. f1, normal-loop heart in controls (97.5%, n = 122); f2, normal-loop heart in cdon−/− embryos (72.7%, n = 128); f3, no loop heart in cdon−/−embryos (16.4%, n = 128); f4, reversed loop heart in cdon−/− embryos (10.9%, n = 128). The blue dashed line indicates the atrial edge. (G) Percentages of normal looping, no looping, and reversed looping of the heart in cdon−/− embryos (n = 128), controls (n = 122) and cdon−/− embryos injected with cdon mRNA (n = 90), cdon−/− embryos show a statistically significant difference compared to controls, and cdon−/− embryos injected with cdon mRNA show a statistically significant difference compared to cdon−/− embryos. Statistical analysis was performed using Student’s t-test. “*” p < 0.05, “**” p < 0.01, “***” p < 0.001. Notice: “control” refers to wild-type embryos.
3.5 KV/cilia-Nodal/spaw cascade was also disturbed in cdon mutants
To further confirm the mechanism by which cdon regulates organ LR patterning, we also examined whether the KV/cilia-Nodal/spaw cascade was affected in cdon−/− embryos. First, we examined the expression of sox17 and sox32 at the 80% epiboly stage to evaluate whether the clustering DFC migration is disturbed in cdon−/− embryos. The data showed that 55.7% of cdon-/-embryos displayed dispersed expression of sox17 (Figures 5Aa3, a4, B), and 48.0% of cdon−/− embryos displayed dispersed expression of sox32 (Figures 5Aa7, a8, B). In contrast, the expression of sox17 and sox32 was normal in control embryos (Figures 5Aa1, Aa5, B). This data indicated that clustering DFC was disturbed in cdon−/− embryos. Then we evaluated whether KV morphogenesis and cilia development were affected in cdon−/− embryos. The data showed that in many cdon−/− embryos, the size of the KV lumen became smaller or absent (Figures 5Cc3, c4, D, E). Similar to cdon morphants, in cdon−/− embryos, the cilia length is slightly shorter (Figures 5Ff1–f4, G) and the cilia number is decreased (Figures 5Ff1–f4, H). Finally, we examined the expression pattern of spaw in control and cdon−/− embryos. The data showed that left-sided spaw expression was also disturbed in cdon−/− embryos, displaying left-sided spaw (Figures 5Ii2, J), bilateral spaw (Figures 5Ii3, J), and right-sided spaw (Figures 5Ii4, J). In addition, the expression of the Nodal/spaw downstream genes lefty1 and lefty2 was also disturbed in cdon−/− embryos (Supplementary Figures 5A–D). These data further confirm that the KV/cilia-Nodal/spaw cascade may mediate the regulation of heart and liver LR patterning by cdon during early development.
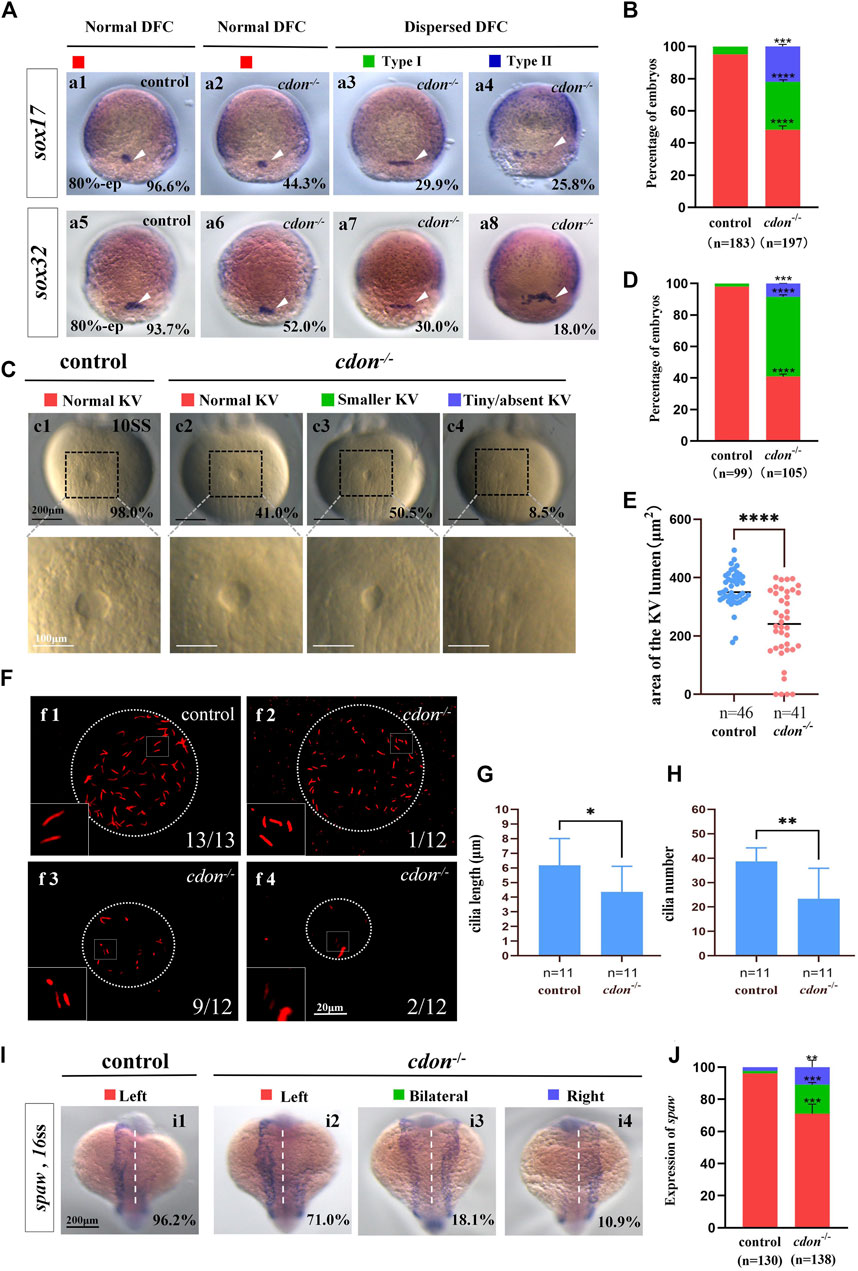
Figure 5. Cdon mutation gave rise to defects in KV formation and cilia (A) Expression of sox17 and sox32 was examined using WISH at 80% epiboly. a1, normal expression of sox17 in controls (96.6%, n = 88); a2, normal expression of sox17 in cdon−/− embryos (44.3%, n = 97); a3, dispersed expression of sox17 (type I) in cdon−/− embryos (29.9%, n = 97); a4, dispersed expression of sox17 (type II) in cdon−/− embryos (25.8%, n = 97). DFCs (white arrow). a5, normal expression of sox32 in controls (93.7%, n = 95); a6, normal expression of sox32 in cdon−/− embryos (52.0%, n = 100); a7, dispersed expression of sox32 (type I) in cdon−/− embryos (30%, n = 100); a8, dispersed expression of sox32 (type II) in cdon−/− embryos (18.0%, n = 100). DFCs (white arrow). (B) Statistical analysis was performed for DFCs clustering migration in controls (n = 183) and cdon−/− embryos (n = 197). Here all the embryos staining with sox17 or sox32 were used together to calculate the percentage. Normal DFC and dispersed DFC show significant differences between control embryos and cdon−/− embryos (C,D) Morphology of KV in different group of embryos at 10sss. c1, normal KV in controls (98.0%, n = 99); c2, normal KV in cdon−/− embryos (41.0%, n = 105, p < 0.0001); c3, smaller KV in cdon−/− embryos (50.5%, n = 105, p < 0.0001); c4, tiny/absent KV in cdon−/− embryos (8.5%, n = 105, p < 0.001). “Normal KV” represents a KV lumen area greater than 300 μm2, “smaller KV” represents a KV lumen area between 100 and 300 μm2, and “tiny/absent KV” represents a KV lumen area less than 100 μm2. (E) Area of the KV lumen (µm2) in control and cdon−/− embryos. “n” represents the sample size. (F) The number and the length of cilia were examined at 10 sss. f1, cilia in controls; f2-f4, cilia in cdon−/− embryos (G) Statistical chart for cilia length in KV. “n” represents the sample size. (H) Statistical chart for cilia number in KV. “n” represents the sample size. (I,J) Expression of spaw in controls and cdon−/− embryos. i1, left-sided spaw in controls (96.2%, n = 130); i2, left-sided spaw in cdon−/− embryos (71.0%, n = 138, p < 0.001); i3, bilateral spaw in cdon−/− embryos (18.1%, n = 138, p < 0.001); i4,right-sided spaw in cdon−/− embryos (10.9%, n = 138, p < 0.01). Statistical analysis was performed using Student’s t-test. “*” p < 0.05, “**” p < 0.01, “***” p < 0.001, “****” p < 0.0001. Notice: “control” refers to wild-type embryos.
3.6 Cdon loss of function in DFCs results in organ LR patterning defects
Our data have shown that cdon is expressed not only in DFCs and KV epithelial cells but also in other cells such as those in the midline and PSM (Figure 1A; Supplementary Figures S4C, D). To confirm whether cdon specifically regulates organ LR patterning via the DFCs-KV/cilia-Nodal/spaw cascade, we injected cdon MO at the 256–512 cell stage to predominantly block the translation of cdon mRNA in DFCs (Amack and Yost, 2004; Zhu et al., 2019), then evaluated whether organ LR patterning was disturbed. Indeed, in the embryos injected with cdon MO at the 256–512 cell stage, liver and heart LR patterning were disturbed (Figures 6A–D). In liver transgenic embryos Tg (fabp10:GFP), heart transgenic embryos Tg (cmlc2:GFP) and wild-type embryos, after predominantly down-regulating the function of cdon in DFCs, the liver (Figures 6A, B) and heart (Figures 6C, D) LR patterning are all disturbed. Next, we evaluated whether the DFCs-KV/cilia cascade was affected after predominantly down-regulating the function of cdon in DFCs. The data showed that clustering DFC was disturbed (Supplementary Figures S6Aa1-a8, B), the size of the KV lumen was smaller (Supplementary Figures S6Cc1-c4, D-E), the length of cilia was slightly shorter (Supplementary Figures S6Ff1-f4,G), and the number of cilia was also decreased (Supplementary Figures S6Ff1-f4, H). Finally, we evaluated whether Nodal/spaw signaling was disturbed in embryos injected with cdon MO at the 256–512 cell stage. As a result, the expression of left-sided Nodal/spaw and its downstream genes lefty1 and lefty2 was also randomized in embryos injected with cdon MO at the 256–512 cell stage (Figures 6E–J). Regarding the expression of spaw, 44.6% (Figures 6Ee2), 40.2% (Figures 6Ee3), and 15.2% (Figures 6Ee4) of embryos injected with cdon MO displayed left-sided spaw, both-sided spaw, and right-sided spaw, respectively, while 97.5% of control embryos displayed left-sided spaw (Figures 6Ee1). Regarding the expression of lefty1, 54.9% (Figures 6Gg2), 29.7% (Figures 6Gg3), and 15.4% (Figures 6Gg4) of embryos injected with cdon MO displayed left-sided lefty1, absent lefty1, and right-sided lefty1, respectively, while 96.5% of control embryos displayed left-sided lefty1. Regarding the expression of lefty2, 60.9% (Figures 6Ii2), 27.6% (Figures 6Ii3), and 11.5% (Figures 6Ii4) of embryos injected with cdon MO displayed left-sided lefty2, both-sided lefty2, and right-sided lefty2, respectively, while 97.2% of control embryos displayed left-sided lefty2 (Figures 6Ii1). These data indicate that left-sided Nodal/spaw signaling was randomized after down-regulating the function of cdon in DFCs. In conclusion, all these data suggest that cdon specifically regulates organ LR patterning via the DFCs-KV/cilia-Nodal/spaw cascade.
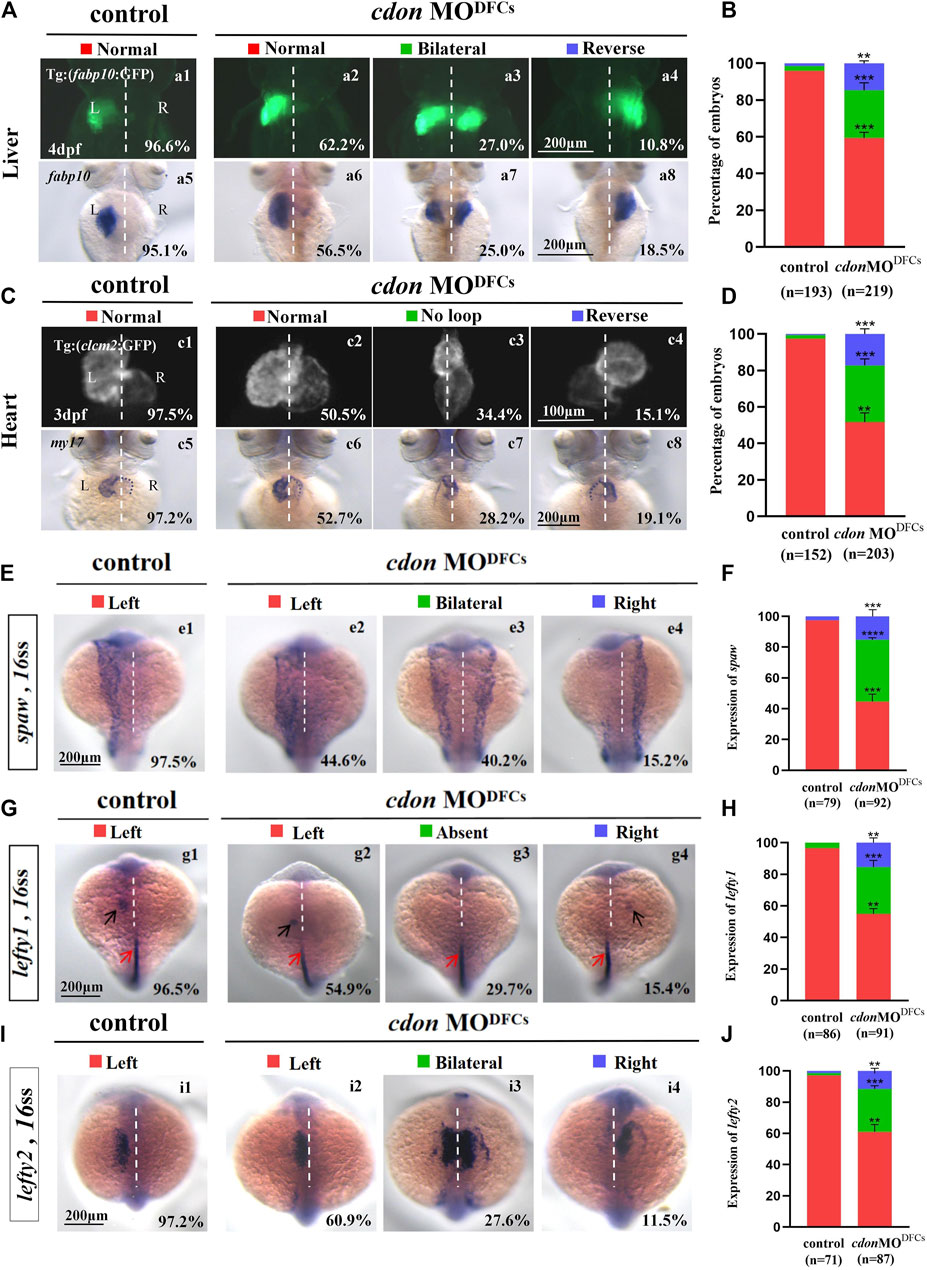
Figure 6. Organ left-right patterning defects in embryos injected with cdon MO at the 256-cell stage (A) Embryos injected with cdon MO at the 256-cell stage were found to cause liver LR defects. a1, normal liver in Tg(fabp10:GFP) transgenic controls (96.6%, n = 90); a2, normal liver in Tg(fabp10:GFP) transgenic embryos injected with cdon MO at the 256-cell stage (62.2%, n = 111); a3, liver bifida in Tg(fabp10:GFP) transgenic embryos injected with cdon MO at the 256-cell stage (27.0%, n = 111); a4, reversed liver in Tg(fabp10:GFP) transgenic embryos injected with cdon MO at the 256-cell stage (10.8%, n = 111). a5, normal liver in wild-type controls (95.1%, n = 103); a6-a8, wild-type embryos injected with cdon MO at the 256-cell stage were examined for liver laterality at 4 dpf by WISH against fabp10 probe (n = 108). (B) Percentages of normal liver, liver bifida, and reversed liver in control embryos and embryos injected with cdon MO at the 256-cell stage. Embryos injected with cdon MO at the 256-cell stage show a statistically significant difference compared to controls. (C) Heart morphogenesis in Tg(cmcl2:GFP) transgenic embryos and wild-type embryos injected with cdon MO at the 256-cell stage. c1, normal-loop in Tg(cmcl2:GFP) transgenic controls (97.5%, n = 80); c2, normal-loop in Tg(cmcl2:GFP) transgenic embryos injected with cdon MO at the 256-cell stage (50.5%, n = 93); c3, no loop in Tg(cmcl2:GFP) transgenic embryos injected with cdon MO at the 256-cell stage (34.4%, n = 93); c4, reversed-loop in Tg(cmcl2:GFP) transgenic embryos injected with cdon MO at the 256-cell stage (15.1%, n = 93); c5, normal-loop in wild type controls (97.2%, n = 72); c6-c8, wild-type embryos injected with cdon MO at 256-cell stage were examined for cardiac looping at 72 hpf by WISH against my17 (n = 110). The blue dashed line indicates the atrial edge. (D) Percentages of normal looping, no looping, and reversed looping of the heart in all the embryos injected with or without cdon MO at the 256-cell stage. Embryos injected with cdon MO at the 256-cell stage show a statistically significant difference compared to controls. (E,F) Expression of Nodal/spaw in controls (n = 79) and embryos injected with cdon MO at the 256-cell stage (n = 92). (G,H) Expression of lefty1 in controls (n = 86) and embryos injected with cdon MO at the 256-cell stage (n = 91). (I,J) Expression of lefty2 in controls (n = 71) and embryos injected with cdon MO at the 256-cell stage (n = 87). Statistical analysis was performed using Student’s t-test. “**” p < 0.01, “***” p < 0.001, “****” p < 0.0001. Notice: in “B” and “D”, all the transgenic embryos and wild-type embryos were used together to calculate the percentage. “Control” refers to wild-type embryos that were not injected with cdon MO.
4 Discussion
Mouse node/cilia [referred to as the “left-right organizer” (LRO)] was first identified to play a critical role during organ LR patterning (Sulik et al., 1994; Nonaka et al., 1998; Schneider et al., 1999; Kajikawa et al., 2022). Similar to mice, transient structures were identified in other vertebrate embryos such as zebrafish, suggesting a conserved cilia-based mechanism regulating LR patterning in zebrafish (Essner et al., 2002). Functional studies confirmed the presence of motile cilia and asymmetric fluid flow in Kupffer’s vesicle in zebrafish (Essner et al., 2005), and genetic or embryological perturbation of these ciliated structures disrupted asymmetric Nodal pathway expression and organ laterality (Essner et al., 2005; Amack, 2014; Kuhns et al., 2019; Liu et al., 2019). In zebrafish, the transgenic line Tg (sox17:EGFP) was developed to label the DFC/KV cells (Chung and Stainier, 2008), and several developmental steps have been identified to build a functional KV/Cilia. DFCs appear at mid-epiboly stage, migrate, proliferate, and then undergo a mesenchymal-to-epithelial transition to form the KV in early somite stage (Forrest et al., 2022). The KV develops directional fluid flow and establishes LR signaling, and then breaks down around 18 hpf when KV cells undergo an epithelial-to-mesenchymal transition (Amack, 2021) and migrate to incorporate into muscle and notochord (Ikeda et al., 2022). In the past decades, many genes and environmental elements have been reported to be involved in DFC clustering, migration (Ablooglu et al., 2010; Gao et al., 2011; Lai et al., 2012; Kajikawa et al., 2022; Liu et al., 2022), proliferation (Zhang et al., 2012; Gokey et al., 2015; Liu et al., 2019; Abdel-Razek et al., 2023), and the final KV formation and ciliogenesis. However, the mechanisms underlying this process are far from being completely elucidated.
Cdon is a cell surface glycoprotein that belong to a subgroup of the immunoglobulin (Ig) superfamily of cell adhesion molecules (Sanchez-Arrones et al., 2012). The role of cdon in organ development and function has been reported in many literature, including its role in neural differentiation, migration and survival (Jeong et al., 2014; Powell et al., 2015; Wang et al., 2017; Uluca et al., 2022; Kim et al., 2023), cardiac remodeling and fibrosis (Jeong et al., 2017) and myoblast fusion (Castiglioni et al., 2018). More recently, in zebrafish, cdon was reported to be involved in trunk neural crest cell migration, slow-twitch muscle development (Lencer et al., 2023), and limb growth (Echevarria-Andino et al., 2023). However, whether it plays a critical role in organ LR patterning at earlier stage has not been reported.
Here, we identified that cdon is expressed in DFCs during the gastrulation movement (Figures 1Aa2–a5) and in the epithelial cells of the KV at the early somitogenesis stage (Figures 1Aa6, a7). Further data suggested that cdon loss of function leads to defect in DFC clustering and decreases DFCs number. These defects are correlated with disturbances in KV formation, cilia number and the subsequent organ LR patterning. Therefore, during embryonic development, in addition to its role in regulating trunk neural crest cell migration (Lencer et al., 2023) and defining the correct proximo-distal patterning of eye development (Cardozo et al., 2014), we identified an earlier role of cdon in regulating LR patterning. Comparing our data in cdon morphants and cdon mutants, we discovered that the organ LR patterning defect in cdon morphants is stronger than that in cdon mutants. Given the well-known genetic compensation response in zebrafish (Ma et al., 2019), the possible reason is the mutation of cdon may upregulate other genes to compensate for the loss of cdon function. This possibility needs further work to elucidate.
Even our work identified the role of cdon in organ LR patterning, the detailed mechanism underlying was not elucidated. In our data, we found that the number of DFCs and epithelium cells in KV was decreased (Figures 3C, D, H, I), as well the cohesive migration was defect in cdon mutants (Figures 3A–D), so the genes being relative to cell migration, cell proliferation/differentiation or cell apoptosis should be affected in cdon mutants. In zebrafish, reduction of Wnt signaling leads to a disruption of LR patterning, shorter and fewer cilia (Caron et al., 2012), depletion of β-catenin 1 or β-catenin 2 in DFCs/KV leads to poor KV cell proliferation, abnormal cilia formation (Zhang et al., 2012). On the contrary, knockdown of Autotaxin/Lpar3 signaling activates β-catenin and also compromises DFC cohesive migration, KV formation and ciliogenesis (Lai et al., 2012). These reports suggested that both downregulation and upregulation of Wnt signaling would give rise to defect in KV morphogenesis, abnormal cilia formation and the sequential organ LR patterning. Importantly, early reports in mouse demonstrated that Cdon negatively regulates Wnt signaling during neural development and cardiac remodeling (Jeong et al., 2014; Jeong et al., 2017), the Cdo-deficient dorsal forebrain displays stronger Wnt signalling activity, increased cell proliferation and enhanced expression of the dorsal markers and Wnt targets (Jeong et al., 2014). These literature suggested the possibility that in zebrafish, loss of cdon function upregulates Wnt signaling, which compromises DFC cohesive migration, KV formation, ciliogenesis and the sequential organ LR patterning. While far more work is needed to clarify whether Wnt signaling lies downstream of cdon to regulate LR patterning.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Institutional Review Board of Chengdu Medical College (SYXK (川)2015-196). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZD: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing–original draft. QR: Conceptualization, Formal Analysis, Validation, Visualization, Writing–original draft, Methodology. WC: Data curation, Investigation, Validation, Visualization, Writing–original draft, Conceptualization, Formal Analysis. CL: Investigation, Validation, Visualization, Writing–original draft, Data curation. BL: Validation, Writing–original draft, Investigation, Visualization. SH: Data curation, Validation, Writing–original draft, Methodology. JH: Validation, Writing–original draft, Data curation, Formal Analysis. KZ: Investigation, Validation, Writing–original draft, Visualization. YL: Writing–original draft, Data curation, Investigation, Validation. XL: Methodology, Writing–original draft, Formal Analysis. ZG: Conceptualization, Data curation, Investigation, Project administration, Resources, Supervision, Validation, Writing–review and editing. SH: Conceptualization, Data curation, Funding acquisition, Investigation, Resources, Supervision, Validation, Visualization, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 32070805) and the Science and Technology Department of Sichuan Province (2021ZYD0074) and the Disciplinary Construction Innovation Team Foundation of Chengdu Medical College (CMC-XK-2102) and the Chengdu medical research project (2023433).
Acknowledgments
We would like to thank Qiang Wang for his advice on this work; we also would like to thank the members working in our fish facility for their help in taking care of all the fish lines in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1429782/full#supplementary-material
References
Abdel-Razek, O., Marzouk, A., MacKinnon, M., Guy, E. T. t., Pohar, S. A., Zhushma, E., et al. (2023). Calcium signaling mediates proliferation of the precursor cells that give rise to the ciliated left-right organizer in the zebrafish embryo. Front. Mol. Biosci. 10, 1292076. doi:10.3389/fmolb.2023.1292076
Ablooglu, A. J., Tkachenko, E., Kang, J., and Shattil, S. J. (2010). Integrin alphaV is necessary for gastrulation movements that regulate vertebrate body asymmetry. Development 137, 3449–3458. doi:10.1242/dev.045310
Amack, J. D. (2014). Salient features of the ciliated organ of asymmetry. Bioarchitecture 4, 6–15. doi:10.4161/bioa.28014
Amack, J. D. (2021). Cellular dynamics of EMT: lessons from live in vivo imaging of embryonic development. Cell. Commun. Signal 19, 79. doi:10.1186/s12964-021-00761-8
Amack, J. D., and Yost, H. J. (2004). The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr. Biol. 14, 685–690. doi:10.1016/j.cub.2004.04.002
Bakkers, J., Verhoeven, M. C., and Abdelilah-Seyfried, S. (2009). Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev. Biol. 330, 213–220. doi:10.1016/j.ydbio.2009.04.011
Burdine, R. D., and Grimes, D. T. (2016). Antagonistic interactions in the zebrafish midline prior to the emergence of asymmetric gene expression are important for left-right patterning. Philos. Trans. R. Soc. Lond B Biol. Sci. 371, 20150402. doi:10.1098/rstb.2015.0402
Cardozo, M. J., Sanchez-Arrones, L., Sandonis, A., Sanchez-Camacho, C., Gestri, G., Wilson, S. W., et al. (2014). Cdon acts as a Hedgehog decoy receptor during proximal-distal patterning of the optic vesicle. Nat. Commun. 5, 4272. doi:10.1038/ncomms5272
Caron, A., Xu, X., and Lin, X. (2012). Wnt/β-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer's vesicle. Development 139, 514–524. doi:10.1242/dev.071746
Castiglioni, I., Caccia, R., Garcia-Manteiga, J. M., Ferri, G., Caretti, G., Molineris, I., et al. (2018). The Trithorax protein Ash1L promotes myoblast fusion by activating Cdon expression. Nat. Commun. 9, 5026. doi:10.1038/s41467-018-07313-8
Chapouly, C., Hollier, P. L., Guimbal, S., Cornuault, L., Gadeau, A. P., and Renault, M. A. (2020). Desert hedgehog-driven endothelium integrity is enhanced by Gas1 (growth arrest-specific 1) but negatively regulated by cdon (cell adhesion molecule-related/downregulated by oncogenes). Arterioscler. Thromb. Vasc. Biol. 40, e336–e349. doi:10.1161/ATVBAHA.120.314441
Chung, W. S., and Stainier, D. Y. (2008). Intra-endodermal interactions are required for pancreatic beta cell induction. Dev. Cell. 14, 582–593. doi:10.1016/j.devcel.2008.02.012
Cole, F., Zhang, W., Geyra, A., Kang, J. S., and Krauss, R. S. (2004). Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Dev. Cell. 7, 843–854. doi:10.1016/j.devcel.2004.10.009
Connor, R. M., Allen, C. L., Devine, C. A., Claxton, C., and Key, B. (2005). BOC, brother of CDO, is a dorsoventral axon-guidance molecule in the embryonic vertebrate brain. J. Comp. Neurol. 485, 32–42. doi:10.1002/cne.20503
Echevarria-Andino, M. L., Franks, N. E., Schrader, H. E., Hong, M., Krauss, R. S., and Allen, B. L. (2023). CDON contributes to Hedgehog-dependent patterning and growth of the developing limb. Dev. Biol. 493, 1–11. doi:10.1016/j.ydbio.2022.09.011
Essner, J. J., Amack, J. D., Nyholm, M. K., Harris, E. B., and Yost, H. J. (2005). Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247–1260. doi:10.1242/dev.01663
Essner, J. J., Vogan, K. J., Wagner, M. K., Tabin, C. J., Yost, H. J., and Brueckner, M. (2002). Conserved function for embryonic nodal cilia. Nature 418, 37–38. doi:10.1038/418037a
Forrest, K., Barricella, A. C., Pohar, S. A., Hinman, A. M., and Amack, J. D. (2022). Understanding laterality disorders and the left-right organizer: insights from zebrafish. Front. Cell. Dev. Biol. 10, 1035513. doi:10.3389/fcell.2022.1035513
Gabriel, G. C., and Lo, C. W. (2020). Left-right patterning in congenital heart disease beyond heterotaxy. Am. J. Med. Genet. C Semin. Med. Genet. 184, 90–96. doi:10.1002/ajmg.c.31768
Gao, W., Xu, L., Guan, R., Liu, X., Han, Y., Wu, Q., et al. (2011). Wdr18 is required for Kupffer's vesicle formation and regulation of body asymmetry in zebrafish. PLoS One 6, e23386. doi:10.1371/journal.pone.0023386
Gokey, J. J., Dasgupta, A., and Amack, J. D. (2015). The V-ATPase accessory protein Atp6ap1b mediates dorsal forerunner cell proliferation and left-right asymmetry in zebrafish. Dev. Biol. 407, 115–130. doi:10.1016/j.ydbio.2015.08.002
Grimes, D. T., and Burdine, R. D. (2017). Left-right patterning: breaking symmetry to asymmetric morphogenesis. Trends Genet. 33, 616–628. doi:10.1016/j.tig.2017.06.004
Hamada, H., and Tam, P. (2020). Diversity of left-right symmetry breaking strategy in animals. F1000Res 9, F1000. doi:10.12688/f1000research.21670.1
Huang, S., Ma, J., Liu, X., Zhang, Y., and Luo, L. (2011). Retinoic acid signaling sequentially controls visceral and heart laterality in zebrafish. J. Biol. Chem. 286, 28533–28543. doi:10.1074/jbc.M111.244327
Huang, S., Xu, W., Su, B., and Luo, L. (2014). Distinct mechanisms determine organ left-right asymmetry patterning in an uncoupled way. Bioessays 36, 293–304. doi:10.1002/bies.201300128
Ikeda, T., Inamori, K., Kawanishi, T., and Takeda, H. (2022). Reemployment of Kupffer's vesicle cells into axial and paraxial mesoderm via transdifferentiation. Dev. Growth Differ. 64, 163–177. doi:10.1111/dgd.12774
Jeong, M. H., Ho, S. M., Vuong, T. A., Jo, S. B., Liu, G., Aaronson, S. A., et al. (2014). Cdo suppresses canonical Wnt signalling via interaction with Lrp6 thereby promoting neuronal differentiation. Nat. Commun. 5, 5455. doi:10.1038/ncomms6455
Jeong, M. H., Kim, H. J., Pyun, J. H., Choi, K. S., Lee, D. I., Solhjoo, S., et al. (2017). Cdon deficiency causes cardiac remodeling through hyperactivation of WNT/β-catenin signaling. Proc. Natl. Acad. Sci. U. S. A. 114, E1345–E1354. doi:10.1073/pnas.1615105114
Kajikawa, E., Miki, T., Takeda, M., Kiyonari, H., and Hamada, H. (2022). Left-right asymmetric expression of the Nodal-Lefty-Pitx2 module in developing turtle forebrain. Front. Cell. Dev. Biol. 10, 929808. doi:10.3389/fcell.2022.929808
Kang, J. S., Mulieri, P. J., Hu, Y., Taliana, L., and Krauss, R. S. (2002). BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 21, 114–124. doi:10.1093/emboj/21.1.114
Kang, Y., Kim, J., Anderson, J. P., Wu, J., Gleim, S. R., Kundu, R. K., et al. (2013). Apelin-APJ signaling is a critical regulator of endothelial MEF2 activation in cardiovascular development. Circ. Res. 113, 22–31. doi:10.1161/CIRCRESAHA.113.301324
Kim, S., An, S., Lee, J., Jeong, Y., You, C. L., Kim, H., et al. (2023). Cdon ablation in motor neurons causes age-related motor neuron degeneration and impaired sciatic nerve repair. J. Cachexia Sarcopenia Muscle 14, 2239–2252. doi:10.1002/jcsm.13308
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi:10.1002/aja.1002030302
Kuhns, S., Seixas, C., Pestana, S., Tavares, B., Nogueira, R., Jacinto, R., et al. (2019). Rab35 controls cilium length, function and membrane composition. EMBO Rep. 20, e47625. doi:10.15252/embr.201847625
Kurpios, N. A., Ibanes, M., Davis, N. M., Lui, W., Katz, T., Martin, J. F., et al. (2008). The direction of gut looping is established by changes in the extracellular matrix and in cell:cell adhesion. Proc. Natl. Acad. Sci. U. S. A. 105, 8499–8506. doi:10.1073/pnas.0803578105
Lai, S. L., Yao, W. L., Tsao, K. C., Houben, A. J., Albers, H. M., Ovaa, H., et al. (2012). Autotaxin/Lpar3 signaling regulates Kupffer's vesicle formation and left-right asymmetry in zebrafish. Development 139, 4439–4448. doi:10.1242/dev.081745
Lencer, E., Rains, A., Binne, E., Prekeris, R., and Artinger, K. B. (2023). Mutations in cdon and boc affect trunk neural crest cell migration and slow-twitch muscle development in zebrafish. Development 150, dev201304. doi:10.1242/dev.201304
Little, R. B., and Norris, D. P. (2021). Right, left and cilia: how asymmetry is established. Semin. Cell. Dev. Biol. 110, 11–18. doi:10.1016/j.semcdb.2020.06.003
Liu, J., Zhu, C., Ning, G., Yang, L., Cao, Y., Huang, S., et al. (2019). Chemokine signaling links cell-cycle progression and cilia formation for left-right symmetry breaking. PLoS Biol. 17, e3000203. doi:10.1371/journal.pbio.3000203
Liu, M., Zou, X., Fu, M., Bai, X., Zhao, Y., Chen, X., et al. (2022). Mild cold stress specifically disturbs clustering movement of DFCs and sequential organ left-right patterning in zebrafish. Front. Cell. Dev. Biol. 10, 952844. doi:10.3389/fcell.2022.952844
Long, S., Ahmad, N., and Rebagliati, M. (2003). The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130, 2303–2316. doi:10.1242/dev.00436
Lu, M., and Krauss, R. S. (2010). N-cadherin ligation, but not sonic hedgehog binding, initiates Cdo-dependent p38alpha/beta MAPK signaling in skeletal myoblasts. Proc. Natl. Acad. Sci. U. S. A. 107, 4212–4217. doi:10.1073/pnas.0908883107
Ma, Z., Zhu, P., Shi, H., Guo, L., Zhang, Q., Chen, Y., et al. (2019). PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263. doi:10.1038/s41586-019-1057-y
Neugebauer, J. M., Amack, J. D., Peterson, A. G., Bisgrove, B. W., and Yost, H. J. (2009). FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature 458, 651–654. doi:10.1038/nature07753
Nonaka, S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y., et al. (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 95, 829–837. doi:10.1016/s0092-8674(00)81705-5
Okada, A., Charron, F., Morin, S., Shin, D. S., Wong, K., Fabre, P. J., et al. (2006). Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444, 369–373. doi:10.1038/nature05246
Onuma, T. A., Hayashi, M., Gyoja, F., Kishi, K., Wang, K., and Nishida, H. (2020). A chordate species lacking Nodal utilizes calcium oscillation and Bmp for left-right patterning. Proc. Natl. Acad. Sci. U. S. A. 117, 4188–4198. doi:10.1073/pnas.1916858117
Powell, D. R., Williams, J. S., Hernandez-Lagunas, L., Salcedo, E., O'Brien, J. H., and Artinger, K. B. (2015). Cdon promotes neural crest migration by regulating N-cadherin localization. Dev. Biol. 407, 289–299. doi:10.1016/j.ydbio.2015.07.025
Raya, A., and Izpisua Belmonte, J. C. (2006). Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat. Rev. Genet. 7, 283–293. doi:10.1038/nrg1830
Sanchez-Arrones, L., Cardozo, M., Nieto-Lopez, F., and Bovolenta, P. (2012). Cdon and Boc: two transmembrane proteins implicated in cell-cell communication. Int. J. Biochem. Cell. Biol. 44, 698–702. doi:10.1016/j.biocel.2012.01.019
Schneider, A., Mijalski, T., Schlange, T., Dai, W., Overbeek, P., Arnold, H. H., et al. (1999). The homeobox gene NKX3.2 is a target of left-right signalling and is expressed on opposite sides in chick and mouse embryos. Curr. Biol. 9, 911–914. doi:10.1016/s0960-9822(99)80397-2
Shankaran, S. S., Dahlem, T. J., Bisgrove, B. W., Yost, H. J., and Tristani-Firouzi, M. (2017). CRISPR/Cas9-Directed gene editing for the generation of loss-of-function mutants in high-throughput zebrafish F(0) screens. Curr. Protoc. Mol. Biol. 119, 31.9.1–31.9.22. doi:10.1002/cpmb.42
Speder, P., Petzoldt, A., Suzanne, M., and Noselli, S. (2007). Strategies to establish left/right asymmetry in vertebrates and invertebrates. Curr. Opin. Genet. Dev. 17, 351–358. doi:10.1016/j.gde.2007.05.008
Sulik, K., Dehart, D. B., Iangaki, T., Carson, J. L., Vrablic, T., Gesteland, K., et al. (1994). Morphogenesis of the murine node and notochordal plate. Dev. Dyn. 201, 260–278. doi:10.1002/aja.1002010309
Trinh, L. A., and Stainier, D. Y. (2004). Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev. Cell. 6, 371–382. doi:10.1016/s1534-5807(04)00063-2
Uluca, B., Lektemur Esen, C., Saritas Erdogan, S., and Kumbasar, A. (2022). NFI transcriptionally represses CDON and is required for SH-SY5Y cell survival. Biochim. Biophys. Acta Gene Regul. Mech. 1865, 194798. doi:10.1016/j.bbagrm.2022.194798
Wang, G., Cadwallader, A. B., Jang, D. S., Tsang, M., Yost, H. J., and Amack, J. D. (2011). The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer's vesicle in zebrafish. Development 138, 45–54. doi:10.1242/dev.052985
Wang, L. C., Kennedy, T. E., and Almazan, G. (2017). A novel function of TBK1 as a target of Cdon in oligodendrocyte differentiation and myelination. J. Neurochem. 140, 451–462. doi:10.1111/jnc.13882
Xie, S., Jin, J., Xu, Z., Huang, Y., Zhang, W., Zhao, L., et al. (2019). Centrosomal protein FOR20 is essential for cilia-dependent development in zebrafish embryos. FASEB J. 33, 3613–3622. doi:10.1096/fj.201801235RR
Xu, P. F., Zhu, K. Y., Jin, Y., Chen, Y., Sun, X. J., Deng, M., et al. (2010). Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc. Natl. Acad. Sci. U. S. A. 107, 2521–2526. doi:10.1073/pnas.0914396107
Yin, C., Kikuchi, K., Hochgreb, T., Poss, K. D., and Stainier, D. Y. (2010). Hand2 regulates extracellular matrix remodeling essential for gut-looping morphogenesis in zebrafish. Dev. Cell. 18, 973–984. doi:10.1016/j.devcel.2010.05.009
Zhang, M., Zhang, J., Lin, S. C., and Meng, A. (2012). β-Catenin 1 and β-catenin 2 play similar and distinct roles in left-right asymmetric development of zebrafish embryos. Development 139, 2009–2019. doi:10.1242/dev.074435
Zhang, W., Hong, M., Bae, G. U., Kang, J. S., and Krauss, R. S. (2011). Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Dis. Model. Mech. 4, 368–380. doi:10.1242/dmm.005744
Zhang, W., Kang, J. S., Cole, F., Yi, M. J., and Krauss, R. S. (2006a). Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev. Cell. 10, 657–665. doi:10.1016/j.devcel.2006.04.005
Zhang, W., Yi, M. J., Chen, X., Cole, F., Krauss, R. S., and Kang, J. S. (2006b). Cortical thinning and hydrocephalus in mice lacking the immunoglobulin superfamily member CDO. Mol. Cell. Biol. 26, 3764–3772. doi:10.1128/MCB.26.10.3764-3772.2006
Zhu, C., Guo, Z., Zhang, Y., Liu, M., Chen, B., Cao, K., et al. (2019). Aplnra/b sequentially regulate organ left-right patterning via distinct mechanisms. Int. J. Biol. Sci. 15, 1225–1239. doi:10.7150/ijbs.30100
Keywords: cdon, left right patterning, DFCs, KV morphogenesis, cilia, nodal signaling
Citation: Deng Z, Ran Q, Chang W, Li C, Li B, Huang S, Huang J, Zhang K, Li Y, Liu X, Liang Y, Guo Z and Huang S (2024) Cdon is essential for organ left-right patterning by regulating dorsal forerunner cells clustering and Kupffer’s vesicle morphogenesis. Front. Cell Dev. Biol. 12:1429782. doi: 10.3389/fcell.2024.1429782
Received: 08 May 2024; Accepted: 02 August 2024;
Published: 22 August 2024.
Edited by:
Ashley Bruce, University of Toronto, CanadaReviewed by:
Jeffrey Amack, Upstate Medical University, United StatesHeidi Hehnly, Syracuse University, United States
Copyright © 2024 Deng, Ran, Chang, Li, Li, Huang, Huang, Zhang, Li, Liu, Liang, Guo and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sizhou Huang, aHVhbmd5dXkxMDI3QGNtYy5lZHUuY24=; Zhenhua Guo, Z3poMTI4MjAzQDE2My5jb20=; Yundan Liang, NDE0Nzc5NjkyQHFxLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Zhilin Deng
Zhilin Deng Qin Ran3†
Qin Ran3† Chengni Li
Chengni Li Ke Zhang
Ke Zhang Sizhou Huang
Sizhou Huang