
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 31 May 2024
Sec. Cancer Cell Biology
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1416697
This article is part of the Research TopicNew Strategies for Treating Fusion-Driven SarcomasView all 6 articles
Capicua (CIC)-rearranged sarcomas are an aggressive subset of undifferentiated round cell sarcomas. CIC::DUX4, the proto-typical CIC fusion oncoprotein is associated with rapid clinical progression and chemotherapy resistance leading to poor clinical outcomes. Recent studies have identified additional CIC fusions (CIC::NUTM1, CIC::FOXO4, and CIC::LEUTX) that largely retain CIC-binding specificity but leverage C-terminal binding partners (NUTM1, FOXO4, and LEUTX) to potentially activate transcriptional programs that drive oncogenesis. Moreover, the recent development of preclinical models to study CIC::DUX4 sarcoma have advanced our understanding of the underlying biological mechanisms and uncovered key dependencies that can be translated into rational therapies. In this review, we will highlight these recent advancements in CIC-rearranged sarcoma biology with a vision for clinical translation to improve patient outcomes.
Capicua (CIC)-rearranged sarcomas represent a rare and highly aggressive subset of undifferentiated small round cell sarcomas. The most frequently observed CIC-rearranged protein involves a fusion between the transcriptional repressor, CIC, and the double homeobox 4 gene, DUX4. Subsequent to their initial description, the World Health Organization (WHO) classified CIC::DUX4 sarcoma (CDS) as a “Ewing sarcoma-like” tumor based primarily on their morphological similarities to Ewing sarcoma (ES). Consequently, the clinical management of CDS often mimics ES, albeit with worse clinical outcomes relative to the latter. Since this initial classification, emerging data now indicates that CIC::DUX4 drives sarcomagenesis through a distinct transcriptional program compared to ES and other small round cell sarcomas. Moreover, there are divergent clinical manifestations and outcomes associated with CDS patients that support the notion that CDS is a distinct disease entity that warrants a well-defined clinical and molecular distinction from other small round cell sarcomas.
Here, we will review the molecular characteristics of CIC-rearranged proteins including CIC::DUX4, CIC::NUTM1, CIC::FOXO4, and CIC::LEUTX with a focus on fusion structure, function, and transcriptional targets. Moreover, we will report on the preclinical models that have been developed to better understand CIC fusion oncoproteins in the context of cancer and how these insights are fueling treatment strategies. Thereafter, we aim to compare and contrast the clinical manifestations between CIC fusions with an emphasis on the anatomical distribution and clinical outcomes among patients. These molecular and clinical observations will potentially provide a framework to develop consensus therapies for this underserved subset of human sarcoma.
Native wild-type (WT) Capicua (CIC) was initially discovered as a high-mobility-group (HMG) box transcriptional repressor in Drosophila melanogaster, where it operates as a MAPK-ERK signaling sensor to coordinate embryonic pattern formation (Jiménez et al., 2000). The mechanism by which CIC confers transcriptional repression in mammals has recently emerged through mass spectrometry (MS) analysis. Specifically, MS has delineated physical interactions with the SIN3-HDAC corepressor complex at CIC transcriptional target sites (Weissmann et al., 2018; Hwang et al., 2020). These studies have also suggested a mechanistic link between the CIC-SIN3/HDAC complex and SWI/SNF recruitment to CIC DNA foci (Hwang et al., 2020; Takemon et al., 2023). These early studies collectively suggest that CIC cooperates with SIN3/HDAC and the mSWI/SNF repressor complexes to transcriptionally silence target genes in mammalian systems. Upon recruitment and repressor complex assembly at CIC DNA binding sites, CIC silences highly conserved MAPK-ERK target genes through the recognition of TGAATGAA-like DNA-motifs (Forés et al., 2017). Upon MAPK activation, CIC is functionally suppressed through ERK–mediated degradation leading to the derepression of MAPK-ERK responsive genes. The underlying mechanism of CIC DNA binding and sequence recognition is thought to be regulated by its conserved HMG box and C1 domains, which enables high sequence specificity (Ajuria et al., 2011; Forés et al., 2017; Webb et al., 2022). The most well studied CIC targets include the PEA3 family of transcription factors, namely ETV1, ETV4, and ETV5. In the context of cancer, genetic loss or functional suppression of CIC derepresses ETV1, ETV4, and/or ETV5 in multiple subsets of human cancer such as lung, gastric, breast, and colorectal cancer, resulting in enhanced tumor proliferation, self-renewal, and invasion/metastasis (Okimoto et al., 2017; Lee et al., 2020; Yoe et al., 2020).
Despite its predominant function as a transcriptional repressor, the role of CIC dysregulation in cancer was first described through its activating capacity as a fusion oncoprotein in 2006 (Kawamura-Saito et al., 2006). Specifically, using in vitro and in vivo transformation models, Takuro Nakamura and colleagues functionally characterized the first patient-derived CIC rearrangement, namely the CIC::DUX4 fusion oncoprotein. Structurally, the CIC::DUX4 fusion almost always retains >90% of native CIC, including the HMG box and C1 domain, and is conjoined with the C-terminal trans-activation domain of the double homeobox four gene DUX4 as a result of translocations t (4; 19) (q35; q13) or t (10; 19) (q26; q13) (Kawamura-Saito et al., 2006; Specht et al., 2014; Antonescu et al., 2017) (Figure 1). Through this fusion, the C-terminal end of CIC is replaced with the DUX4 transactivating domain that interacts with the p300/CBP complex. Thus, it is reasonable to hypothesize that these structural changes shift the interaction from the SIN3-HDAC and SWI/SNF repressor complexes to a p300/CBP dominant interaction, which results in transcriptional activation of CIC target genes. Thus, it is reasonable to conceive that CIC::DUX4 attains transcriptional activating capacity through a conserved DUX4 recruitment of p300/CBP, which induces histone H3 acetylation to enable target gene activation (Choi et al., 2016; Bosnakovski et al., 2021). Consistent with this, we and others have observed that global CIC::DUX4 binding overlaps with H3K27ac marks in patient derived CIC::DUX4 cell lines, again suggesting that p300/CBP may in part confer activating capacity (Thomas et al., 2023; Bakaric et al., 2024). Collectively, CIC co-opts the activating capacity of the p300/CBP associated transcription factor, DUX4, to upregulate key target genes including PEA3 family members, cell cycle genes such as CCND2 and CCNE1, and negative MAPK regulators including DUSP4 and DUSP6 that drive malignant phenotypes in aggressive undifferentiated round cell sarcomas (Kawamura-Saito et al., 2006; Yoshimoto et al., 2017; Okimoto et al., 2019; Ponce et al., 2022). Collectively, these structure-function studies reveal that CIC::DUX4 largely retains CIC DNA-binding specificity while transforming its native repressor function (SIN3-HDAC and SWI/SNF) into a potent oncogene in part through DUX4 mediated p300/CBP recruitment.
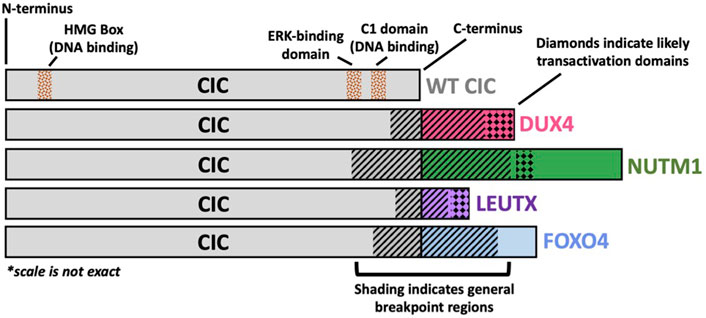
Figure 1. Structural Schematic of CIC Fusion Oncoproteins. Structural schematic of CIC fusions highlighting the highly conserved wild-type (WT) CIC region with variability in the fusion break point and c-terminal binding partner. The CIC::DUX4 fusion oncoprotein has been shown to retain WT CIC DNA binding specificity through its HMG box and C1 domain but acquires transcriptional activating capacity via DUX4-mediated p300 recruitment to CIC binding sites. The C-terminal bindings partners, NUTM1, LEUTX, and FOXO4 have previously been reported to interact with p300/CBP.
Since the initial discovery of CIC::DUX4, other CIC fusions have been reported including CIC::NUTM1, CIC::FOXO4, and CIC::LEUTX. With some variability in the 3’ CIC breakpoint, these CIC fusions largely retain the majority of WT CIC, including its HMG box domain, but replace the C-terminal end of CIC with components of another transcriptional regulator, including NUT midline carcinoma family member 1 (NUTM1), forkhead box O4 (FOXO4), or leucine twenty homeobox (LEUTX). Intriguingly, these CIC binding partners are well-defined developmentally regulated transcription factors that interact with p300/CBP (van der Heide and Smidt, 2005; Shiota et al., 2018; Gawriyski et al., 2023). Thus, it is reasonable to anticipate that these CIC-rearranged proteins can potentially operate in a similar fashion to CIC::DUX4 fusion oncoproteins.
CIC::NUTM1 was first reported in 2016 in a molecular analysis of primitive neuroectodermal tumors of the central nervous system (CNS), where two patients harbored a fusion between CIC exon 16 and NUTM1 exon 4 (Sturm et al., 2016). Since then, CIC::NUTM1 fusions have been identified in at least an additional one CNS patient and twenty sarcoma patients, where the chimera occurs between CIC exons 16, 17, 18, and 20 and NUTM1 exons 2, 3, 4, 5, and 6 (Mangray et al., 2018; Schaefer et al., 2018; Watson et al., 2018; Le Loarer et al., 2019; Biederman et al., 2022; Yang et al., 2022; Ma et al., 2023; Sievers et al., 2023) (Figure 1). While functional studies illuminating molecular pathology are limited, non-CIC::NUTM1 fusions have been implicated in NUTM1 midline carcinoma, specifically the BRD4::NUTM1 fusion, with p300 recruitment and subsequent histone acetylation to be a hypothesized mechanism of oncogenic activation (Reynoird et al., 2010; French, 2018). Structurally, CIC::NUTM1 fusions retain the CIC HMG box domain, but in contrast to CIC::DUX4, CIC::NUTM1 fusions appear to not universally harbor the CIC C1 domain, which has been previously linked to DNA-binding and target gene specificity (Forés et al., 2017). Interestingly, transcriptional profiling of patient-derived CIC::DUX4 and CIC::NUTM1 fusions demonstrated that these two groups shared some similarity but slightly diverged at the distal hierarchical branches (Watson et al., 2018). These findings suggest that while CIC::DUX4 and CIC::NUTM1 may share transcriptional targets, there may be key distinctions that molecularly differentiate these two fusion oncoproteins. Thus, a deeper and more comprehensive analysis may reveal key molecular distinctions that uniquely associate with each fusion. For example, if and/or how the C1 domain of CIC contributes to target gene specificity in the context of CIC fusions (CIC::DUX4 retains the C1 domain; CIC::NUTM1 does not retain the C1 domain). Additionally, this comparison could highlight key distinctions between how CIC binding partners, including DUX4 and NUTM1, impact fusion specific target gene regulation. Future comparative studies aimed at addressing these questions is warranted as we believe that the molecular underpinnings of how these fusions mechanistically regulate cellular transformation may reveal key insights to better understand both the clinical features and therapeutic approaches for these patients.
The more rare CIC::FOXO4 fusion has been reported in a total of four patients, with breakpoints identified in three patients occurring between CIC exons 19 or 20 and FOXO4 exons 2 or 3 (Brohl et al., 2014; Sugita et al., 2014; Babkoff et al., 2023) (Figure 1). Thus, similar to CIC::DUX4 fusions, CIC::FOXO4 oncoproteins structurally share CIC DNA-binding elements (HMG-box and C1 domains), suggesting that CIC DNA-binding specificity and target gene regulation may be conserved relative to CIC::DUX4. While little is known about the molecular mechanisms of CIC::FOXO4 fusion oncoproteins, FOXO4 (also known as AFX) and FOXO1 (also known as FKHR) chimeras have been identified in MLL-rearranged acute leukemia (AFX::MLL) and alveolar rhabdomyosarcoma (PAX3::FOXO1) (Borkhardt et al., 1997; Sorensen et al., 2002). Similar to DUX4 and NUTM1, p300 has been shown to interact with the FOXO family of transcription factors (van der Heide and Smidt, 2005). Not only was FOXO4 depicted to co-immunoprecipitate with p300 in a peroxide-induced, redox-dependent manner, but it was also recently shown that a single cysteine in the activation domain of PAX3::FOXO1 is important for the recruitment of p300/CBP to chromatin, which was also conserved in other members of the FOXO family, including FOXO4 (Dansen et al., 2009; Asante et al., 2023). We fully anticipate that with improved CIC::FOXO4 models, we will better understand the molecular mechanisms of CIC::FOXO4 fusions and how they contribute to sarcomagenesis.
A fourth CIC fusion has been identified in sarcoma and non-sarcoma cancers—CIC::LEUTX. Published patient case reports have identified the CIC::LEUTX chimera to occur between CIC exon 20 and LEUTX exon 3 across the different cancers of renal sarcoma, spinal cord sarcoma, angiosarcoma, central nervous system embryonal tumors, and pediatric-type high-grade neuroepithelial tumors (Huang et al., 2016; Hu et al., 2020; Lake et al., 2020; Song et al., 2022; Sievers et al., 2023; Tang et al., 2023) (Figure 1). Interestingly, LEUTX is a developmentally regulated transcriptional factor that is expressed in early human embryonic development but subsequently silenced in somatic tissues. Importantly, DUX4, which is a key early embryonic genome activation factor, can transcriptionally upregulate LEUTX expression (Gawriyski et al., 2023). Additionally, WT LEUTX has been shown to physically interact with and depend on p300/CBP mediated acetylation for target gene activation (Gawriyski et al., 2023). Notably, LEUTX interacts with p300 through a 9 amino-acid transactivating domain in its C-terminus, which is conserved in the context of the CIC::LEUTX fusion (Sievers et al., 2023). Thus, we suspect that CIC::LEUTX fusion oncoproteins may in part share some overlap in their molecular targets and mode of transcriptional activation with CIC::DUX4. A recent study by Sievers et al. leveraged DNA methylation patterns to differentiate tumors harboring CIC::LEUTX fusions from other CIC-rearranged (non-CIC::LEUTX) central nervous system (CNS) sarcomas (Sievers et al., 2023). Their analysis revealed that CIC::LEUTX bearing CNS tumors segregated into its own group characterized by a pediatric-type high-grade neuroepithelial histology that was distinct from the CIC-rearranged CNS sarcoma group. Consistent with the comparison between CIC::DUX4 and CIC::NUTM1 tumors, these findings suggest that while CIC::LEUTX fusions may share some similarities with CIC::DUX4 tumors, there are likely key distinctions that differentiate between these molecular subsets. Further studies are clearly warranted to further elucidate both shared and divergent mechanisms between CIC fusions.
The first widely validated CIC::DUX4 patient-derived cell lines were established in 2017 by Tadashi Kondo and colleagues (Oyama et al., 2017). Specifically, a CIC::DUX4 fusion positive soft tissue tumor was derived from a 29 year-old female patient and was implanted into immunodeficient mice that was serially propagated three times to generate NCC-CDS1-X1 (cell line generated after first passage) and NCC-CDS1-X3 (cell line generated after third passage). Sanger sequencing of the explanted tumor identified an in-frame CIC::DUX4 transcript fusing CIC exon 20 to exon 1 of DUX4 with a junction nucleotide sequence 5′-GGGTGGAG-3′ (Oyama et al., 2017). Eventually, Kondo and colleagues established NCC-CDS2-C1 patient-derived cell lines using a surgically resected tumor tissue from another patient with CIC::DUX4 sarcoma (CDS) containing a similar CIC::DUX4 fusion transcript (Yoshimatsu et al., 2020). Furthermore, a third cell line named Kitra-SRS was generated from a soft tissue tumor derived from a 9 year-old girl with metastatic CDS. The Kitra-SRS cell line was generated through xenograft propagation following the excision of the primary tumor. Sequence profiling revealed a fusion between CIC exon 20 and DUX4 exon 1 with the 5′-GGGTGG-3′ nucleotide sequence identified at the junction between CIC and DUX4, similar to the NCC-CDS1-X1 and NCC-CDS1-X3 cells (Nakai et al., 2019). Notably, all these aforementioned models were histologically similar to the original resected CDS tumors, and Kitra-SRS tumor-bearing mice exhibited lung metastases, which recapitulated the aggressive disease pattern observed in the host from which it was derived.
While case reports identifying CIC::NUTM1, CIC::FOXO4, and CIC::LEUTX fusions in patient tumors have been increasing, there has yet to be a patient-derived cell line or xenograft generated for experimental use. We eagerly await these tools that will greatly increase our understanding of CIC-rearranged biology and oncogenesis.
Recently, Hendrickson et al. attempted to generate a transgenic CIC::DUX4 mouse model (Hendrickson et al., 2024). They engineered three CIC::DUX4 chimeric mouse models (Ch7CDS, Ai9CDS, and TOPCDS), which developed spontaneous CDS-like tumors associated with presumed widespread metastases in the lung and liver in the absence of Cre-recombinase. The authors speculate that the CIC::DUX4 fusion is a highly potent oncogene (similar to Kras) that leads to spontaneous tumor formation. These studies largely represent proof-of-principle experiments since all three chimeric models died prior to reproduction due to rapid spontaneous tumor formation and subsequent death. Despite these limitations, the authors were able to generate important molecular datasets and investigated underlying mechanisms of CIC::DUX4 mediated gene activation in mice. Collectively, their findings align with human CDS as they demonstrate multiple shared transcriptional targets including known PEA3 family members that were, in part, associated with p300 transcriptional activation. One interesting observation that is consistent with other findings is that CIC::DUX4 localizes and binds to GGAA-motif sequences on DNA (Kim et al., 2022; Bakaric et al., 2024). These findings indicate that WT CIC and CIC::DUX4 (among other CIC rearrangements) may regulate gene expression through non-consensus (non TGAATGAA-like) like motifs (Thomas et al., 2023). Future studies aimed at identifying and differentiating which genes are regulated by CIC::DUX4 at TGAATGAA versus GGAA-like DNA-motifs is highly warranted.
Through CIC::DUX4 transduction of mouse embryonic mesenchymal cells (eMC), Yoshimoto et al. generated an ex vivo CDS mouse model that closely recapitulates the histological and clinical features of human CDS (Yoshimoto et al., 2017). In this model, CIC::DUX4 expressing eMCs were transplanted into the subcutaneous soft tissue of immunodeficient nude mice. Recipient mice rapidly developed primary tumors at 100% penetrance with a mean latency period of 24 days. Serial transplantation of these CIC::DUX4 tumors resulted in spontaneous lung metastases in 28% of recipient mice. Correlative studies further identified conserved human CIC::DUX4 transcriptional targets including ETV1, ETV4, and CCND2. The authors subsequently leveraged this mouse model to test treatment strategies and noted tumor growth inhibition using genetic and pharmacologic approaches that block the CCND2-CDK4/6 complex.
In our series of CIC::DUX4 confirmed case reports median age of CDS diagnosis was 29.5 years old with a range from 11 to 82 years old and a male-to-female ratio of fourteen to eighteen (Machado et al., 2013; Bielle et al., 2014; Kajtár et al., 2014; Haidar et al., 2015; Tardío et al., 2015; Chebib and Jo, 2016; Bergerat et al., 2017; Krskova et al., 2017; Loke et al., 2017; Tsukamoto et al., 2017; Camille et al., 2018; Donahue et al., 2018; Mangray et al., 2018; Tang and Dodd, 2018; Lehane et al., 2019; Maloney et al., 2020; Ricker et al., 2020; Tamada et al., 2020; Yamada et al., 2020; Chen et al., 2021; Maejima et al., 2021; Maekawa et al., 2021; Vieira et al., 2021; Wu and He, 2022; Aranza et al., 2023; Ascione et al., 2023; Wu et al., 2023) (Table 1). Within the thirty-two CDS patients that we analyzed through case studies, ten tumors localized primarily in the soft tissue of lower extremities, with a significant fraction also presenting in the soft tissue of the upper extremities and kidney as well as pelvic cavity area. (Figure 2). In comparison, the median age of CIC::NUTM1 sarcomas is 15.5 years old (range 2–61 years old) and presents with an eight-to-six ratio of male to female incidence from the available clinical data (Table 1). Interestingly, a significant percentage of CIC::NUTM1 tumors localized to the CNS, with over 70% confined to the spine (cervical and thoracic levels) and brain (temporal and occipital areas of meninges, parenchyma, and sarcoma, trigone of the lateral ventricle, interventricular foramen, parietal lobes) with others infrequently observed in the lung and kidney soft tissue (Sturm et al., 2016; Mangray et al., 2018; Schaefer et al., 2018; Watson et al., 2018; Le Loarer et al., 2019; Biederman et al., 2022; Yang et al., 2022; Ma et al., 2023; Sievers et al., 2023) (Figure 2). The CNS tropism is consistent with prior reports (Watson et al., 2018). Thus far, only four patient case reports have been published on the CIC::FOXO4 fusion, with the median age of diagnosis of 36.5 years old (range 13–63 years old), and a three-to-one male-to-female ratio (Solomon et al., 2014; Sugita et al., 2014; Connolly et al., 2022; Babkoff et al., 2023) (Table 1). In these patients, primary tumors localized to the scalp, neck, and thigh (Figure 2).
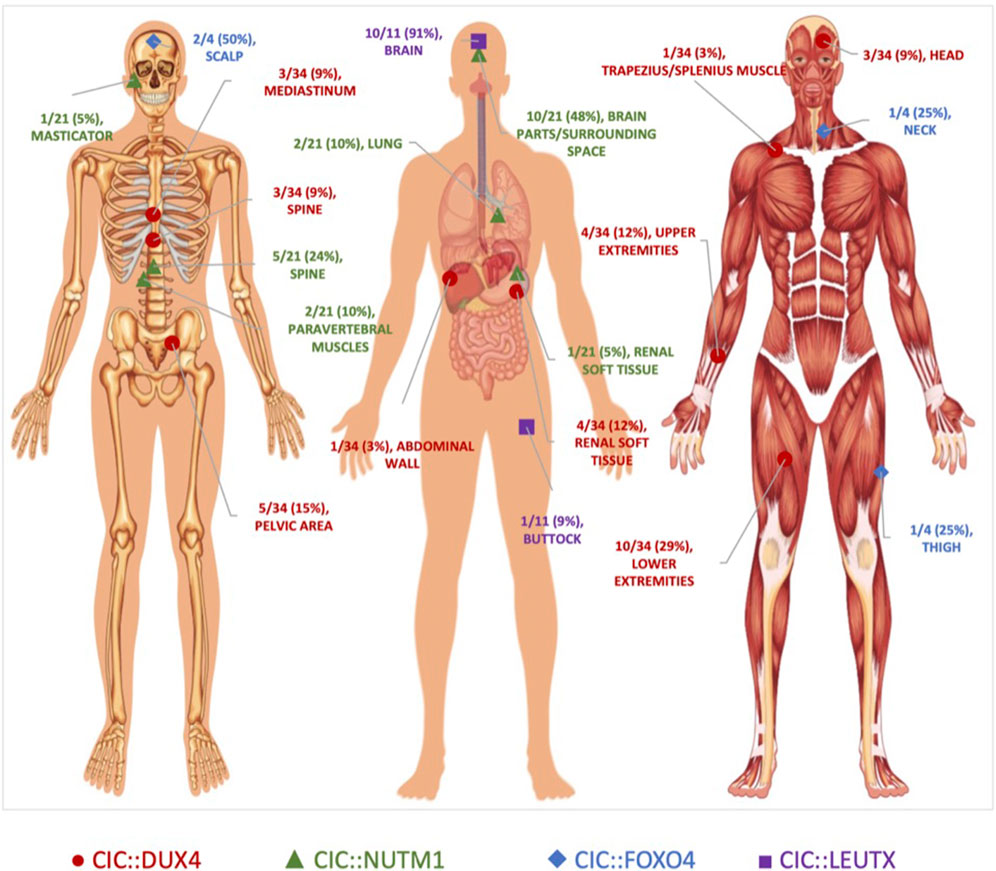
Figure 2. Anatomical Localization of CIC-rearranged Primary Tumors. Primary tumor location of CIC-fusion positive cases reported in the literature. CIC::DUX4 (red circles) tumors localized primarily in the soft tissue. CIC::NUTM1 (green triangles) tumors were predominantly observed in the central nervous system. CIC::LEUTX (purple squares) tumors were confined mostly in the brain, while CIC::FOXO4 (blue diamonds) tumors were interspersed in the scalp, neck, and thigh. Original unlabeled anatomical image from Vecton/Shutterstock.com.
While much less is known about the clinical features associated with CIC::LEUTX sarcoma, a recent study that profiled pediatric CNS tumors identified 9 patients with CIC::LEUTX fusions that all occurred within the supratentorial compartment (Sievers et al., 2023). The median age of onset is 7 years old (range 2–19) with a female predominance (three-to-six ratio of male to female) (Table 1). Clinical outcomes associated with CIC::LEUTX patients were available for a subset of patients (n = 6). Median progression-free survival was 13.5 months (range 6–16 months) with all patients developing relapse. Only one of the patients died of the disease during the follow-up period at 15 months. Treatment response was not available in this study. Another study identified two additional patients with CIC::LEUTX fusions and one was located in the midbrain/thalamus while the other was in the left buttock and associated with pulmonary metastases (Linos et al., 2023). Treatment response to radiation and temozolomide was noted in the patient with midbrain involvement, while chemotherapy (AIM) response was noted in the pulmonary metastases of the patient with the left buttock CIC::LEUTX sarcoma. Long-term clinical follow up and low numbers make it difficult to generalize these observations and more cases are needed to draw reliable conclusions about treatment response and/or anatomical distribution.
Of the four CIC-rearranged sarcomas, summary statistics of the available data within the case reports indicate CIC::DUX4 sarcoma to be associated with a relatively more aggressive clinical course, presenting with the highest rate of metastasis (76%; total n = 29) and a predilection for the lungs (Machado et al., 2013; Bielle et al., 2014; Kajtár et al., 2014; Haidar et al., 2015; Tardío et al., 2015; Chebib and Jo, 2016; Bergerat et al., 2017; Krskova et al., 2017; Loke et al., 2017; Tsukamoto et al., 2017; Camille et al., 2018; Donahue et al., 2018; Mangray et al., 2018; Tang and Dodd, 2018; Lehane et al., 2019; Maloney et al., 2020; Ricker et al., 2020; Tamada et al., 2020; Yamada et al., 2020; Chen et al., 2021; Maejima et al., 2021; Maekawa et al., 2021; Vieira et al., 2021; Wu and He, 2022; Aranza et al., 2023; Ascione et al., 2023; Wu et al., 2023). Sarcoma patients harboring the CIC::FOXO4 fusion also had a high rate of pulmonary metastasis (75%; total n = 4), while patients with the CIC::NUTM1 tumors had the lowest overall rate of metastasis (25%; total n = 12) with distant sites including the lungs, thyroid, bone, and brain (Solomon et al., 2014; Sugita et al., 2014; Mangray et al., 2018; Schaefer et al., 2018; Le Loarer et al., 2019; Biederman et al., 2022; Connolly et al., 2022; Yang et al., 2022; Babkoff et al., 2023; Ma et al., 2023; Sievers et al., 2023). Despite differences in metastatic pattern, at least half of the patients in each CIC rearrangement cohort, excluding CIC::LEUTX, died of their disease (Table 1).
There is no consensus treatment strategy for patients with CIC-rearranged sarcomas. As a result, CIC-rearranged sarcomas are treated with a similar paradigm to other more chemotherapy sensitive small round cell sarcomas including Ewing sarcoma (ES). The efficacy of these conventional systemic approaches along with clinical outcomes data of patients with CIC-rearranged tumors who undergo multimodal treatment strategies (surgery, radiation, and chemotherapy) have been extensively discussed elsewhere (Antonescu et al., 2017; Connolly et al., 2022; Brahmi et al., 2023; Murphy et al., 2024). Therefore, in this review we will only briefly mention the standard chemotherapy backbones and largely focus on the emerging mechanism-based approaches to target CIC-fused sarcomas. A short list of standard chemotherapy approaches include vincristine, doxorubicin, cyclophosphamide alternating with ifosfamide, and etoposide (VDC/IE) or doxorubicin plus ifosfamide with MESNA (AIM) (Table 2). It is important to emphasize that these treatment strategies were largely based on the histological similarities between CIC::DUX4 and ES, rather than the degree of tumor sensitivity to these chemotherapy regimens (Connolly et al., 2022).
Of the four CIC::FOXO4 sarcoma patients featured in the literature, two died of their disease despite utilization of surgical resection, adjuvant chemotherapy composed of common therapeutic combinations (e.g., VDC/IE), or use of kinase inhibitors (e.g., cabozantinib on clinical trial) (Solomon et al., 2014; Sugita et al., 2014; Connolly et al., 2022; Babkoff et al., 2023) (Table 2). Although CIC::NUTM1 has the lowest metastatic rate amongst the CIC-rearranged sarcomas, eight of the fourteen patients featured in the case reports died of their disease despite only two experiencing metastatic progression and amidst the use of surgical resection, chemotherapy, and radiotherapy in most of these patients (Le Loarer et al., 2019; Yang et al., 2022). One of the patients specifically followed the ES chemotherapeutic combination regimen of six cycles of VIDE (vincristine, ifosfamide, doxorubicin, and etoposide) but unfortunately continued to progress. Functional studies to elucidate the molecular pathogenesis of CIC::NUTM1 and CIC::FOXO4 sarcomas and identify therapeutic targets are warranted.
Although the CIC::DUX4 fusion was the first CIC-rearranged sarcoma identified in 2006, there are still inadequate chemotherapy regimens utilized for CDS. Recent studies have focused on understanding the mechanistic underpinnings of the CIC::DUX4 fusion oncoprotein, illuminating potential druggable targets (Table 2). Preclinical studies identified CIC::DUX4 to molecularly depend on cell cycle mediators CCNE1 and WEE1 (Okimoto et al., 2019; Ponce et al., 2022) with CCNE1 being established as a direct transcriptional target of CIC::DUX4 that drives tumor growth and survival through an acquired dependence on CCNE/CDK2 complex. Hyperactivation of the CCNE/CDK2 complex compromises the G1/S transition and confers sensitivity to CDK2 inhibitors in a relatively fusion specific manner (Okimoto et al., 2019). In order to prevent the accumulation of DNA damage in S phase and to subsequently prevent premature mitotic entry, CIC::DUX4 sarcomas depend on the G2/M checkpoint kinase WEE1 to delay mitotic entry and to ensure proper DNA repair and integrity prior to mitosis. Thus, WEE1 kinase inhibition with adavosertib has been shown to induce tumor regression in CIC::DUX4 tumor xenograft models (Ponce et al., 2022). These preclinical studies provide rationale to develop CDK2 and WEE1 inhibitor–based clinical trials for CIC::DUX4 patients. To this end, Blueprint Medicine is testing their CDK2 inhibitor (BLU-222) in patients with CIC-rearranged tumors through their ongoing clinical trial, NCT05252416 (Table 2). Additionally, based on the preclinical findings mentioned above, we and others are working to develop a future WEE1-directed strategy to test in patients with CIC-rearranged tumors.
Another mechanism-based approach to target CDS was identified through the development and study of the Kitra-SRS CIC::DUX4 positive cell line (Nakai et al., 2019), which was discussed above. Specifically, analysis of Kitra-SRS cells revealed autocrine activation of the insulin-like growth factor 1 (IGF-1)/IGF-1 receptor (IGF-1R) signaling pathway (Table 2). Thus, inhibiting this pathway with the IGF-1R inhibitor, linsitinib, limited IGF-1R/AKT signaling and reduced Kitra-SRS tumor growth both in vitro and in vivo.
Since MAPK-ERK signaling leads to WT CIC degradation, Lin et al. hypothesized that pharmacologic activation of ERK could potentially lead to direct degradation of the CIC::DUX4 oncoprotein, which retains the highly conserved ERK binding domain. Through a series of biochemical and genetic studies, they demonstrated that ERK activation through inhibition of the DUSP6 phosphatase (dephosphorylates ERK to decrease activity) could lead to CIC::DUX4 degradation. Moreover, they identify that CIC::DUX4 transcriptionally upregulates DUSP6 to silence ERK activity and sustain CIC::DUX4 oncoprotein expression in sarcoma (Lin et al., 2020). The use of DUSP6 inhibitors is not in clinical use largely due to the potential negative effects of activating ERK in non-CIC::DUX4 sarcoma (Table 2).
A broader approach to target CIC::DUX4 transcriptional activity was identified by Bosnakovski and colleagues. Leveraging biological insight of how WT DUX4 (and CIC::DUX4) interacts with p300, they rationalized that p300 inhibition could potentially overcome CIC::DUX4 sarcoma growth and survival. They demonstrated that a iP300w, a potent p300/CBP inhibitor, reversed CIC::DUX4 associated histone H3 acetylation marks and consequently suppressed CIC::DUX4 transcriptional activity while limiting tumor growth (Bosnakovski et al., 2021) (Table 2).
Finally, therapies that directly target components of the CIC::DUX4 fusion oncoprotein are emerging. Specifically, Targeted RNAi Molecules (TRiM) are being employed to intercept DUX4 in facioscapulohumeral muscular dystrophy (NCT06131983). Cell penetrating antibodies designed to target intracellular neoantigens including fusion oncoproteins are also in early development and could be of clinical interest in the near future (Herrmann et al., 2019). Anti-DUX4 strategies are of potential interest largely since WT DUX4 is typically silenced in adult somatic tissues (Karpukhina et al., 2021). Thus, RNAi-based technologies and/or cell penetrating antibodies that can effectively target the DUX4 in the context of the CIC::DUX4 fusion can open another therapeutic avenue directed at selective targeting of the CIC::DUX4 oncoprotein in human sarcoma (Table 2).
Collectively, we hope that future and ongoing studies aimed at identifying and exploiting key dependencies in CIC::DUX4 sarcoma will apply to other CIC-rearranged sarcomas to improve outcomes for these patients who currently have no therapeutic options.
CIC::DUX4 was first recognized in a subset of undifferentiated small round cell sarcomas that lacked conventional EWSR1 fusions that characterize ES. Improved model systems coupled with more advanced profiling studies have enabled a deeper understanding into the biological underpinnings of CIC::DUX4 mediated oncogenesis. Moreover, enhanced sequencing technologies and pathological stratification have led to the discovery of more CIC-rearranged proteins including CIC::NUTM1, CIC::FOXO4, and CIC::LEUTX. On the structural level, these fusions share a significant fraction of WT CIC including its DNA-binding domains and target gene specificity. Early data suggest that the developmentally regulated C-terminal binding partners (DUX4, NUTM1, FOXO4, and LEUTX) may confer activating capacity through the recruitment of the p300/CBP complex. However, further studies are needed to validate this hypothesis. Despite their commonalities in fusion structure, we anticipate transcriptional and perhaps functional divergence between CIC fusion types, which is supported through the aforementioned transcriptional studies comparing CIC::DUX4 to CIC::NUTM1 and/or CIC::LEUTX tumors which are closely but not identically clustered. These subtle molecular differences may provide insight into the key differences that differentiate CIC rearrangements in patients, including anatomical tropism, metastatic potential, and response to therapy.
Development of preclinical cell-based and animal models to further dissect the molecular and functional mechanisms of not only CIC::DUX4 but also the newly recognized CIC::NUTM1, CIC::FOXO4, and CIC::LEUTX chimeras will provide insight to improve treatment strategies for these patients. We applaud rare tumor programs that aim to develop new patient-derived models, such as CIC-rearranged sarcoma cell lines that can be subsequently shared with the scientific community to improve our understanding through mechanistic studies.
Due to its rarity, the development of new therapeutic strategies to target CIC::DUX4 sarcoma remains limited. Through deep mechanistic dissection of CIC::DUX4 biology we and others have identified preclinical targets that warrant clinical validation to refine the treatment paradigm for patients with CIC-rearranged sarcoma. We encourage clinical investigators to work together on a national or international level to enroll patients with CIC-rearranged sarcomas on ongoing and/or future mechanism-based clinical trials.
Another important barrier to overcome is within the clinical realm, calling for the unified differentiation by the sarcoma community to distinguish CIC-rearranged sarcomas from other small round cell sarcomas such as ES (Blay et al., 2016). This is of particular importance given the overall lack of therapeutic consensus and misinformed treatment strategies that patients with CIC fusions often face in the clinic. One strategy to overcome the barriers of enrolling patients with rare CIC-rearranged sarcomas onto clinical trials is to leverage the National Cancer Institute (NCI)-sponsored Experimental Therapeutics Clinical Trials Network (ETCTN). ETCTN participating sites throughout the United States can collaborate to conduct early trials that target CIC-rearranged sarcoma with NCI-IND agents, such as WEE1 inhibitors. Collaboration between the scientific and clinical community is essential in order to combat these rare but lethal subtypes of sarcoma with novel and effective molecular-based therapeutic interventions.
RP: Conceptualization, Data curation, Formal Analysis, Validation, Writing–original draft, Writing–review and editing. CL: Data curation, Methodology, Validation, Writing–review and editing. RO: Conceptualization, Funding acquisition, Supervision, Validation, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by a 2023 Conquer Cancer Medical Student Rotation from Underrepresented Populations Award to RKMP. Any opinions, findings, and conclusion expressed in this material are those of the author(s) and do not necessarily reflect those of the American Society of Clinical Oncology® or Conquer Cancer. RAO was funded by an NCI grant (R37 CA255453), the Children’s Cancer Research Fund (CCRF), and Cookies for Kids’ Cancer. CL acknowledges funding from the UCSF Discovery Fellows Program and NIGMS 1T32GM136547-01.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ajuria, L., Nieva, C., Winkler, C., Kuo, D., Samper, N., Andreu, M. J., et al. (2011). Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development 138 (5), 915–924. doi:10.1242/dev.057729
Antonescu, C. R., Owosho, A. A., Zhang, L., Chen, S., Deniz, K., Huryn, J. M., et al. (2017). Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: a clinicopathologic and molecular study of 115 cases. Am. J. Surg. Pathology 41 (7), 941–949. doi:10.1097/PAS.0000000000000846
Aranza, S., Roydhouse, C., Mitchell, C., Vissers, J. H. A., Lo, W. Y., Grimmond, S. M., et al. (2023). A rare diagnostically challenging case of CIC-DUX4 sarcoma arising in the neck. Pathology 55 (4), 568–571. doi:10.1016/j.pathol.2022.03.010
Asante, Y., Benischke, K., Osman, I., Ngo, Q. A., Wurth, J., Laubscher, D., et al. (2023). PAX3-FOXO1 uses its activation domain to recruit CBP/P300 and shape RNA Pol2 cluster distribution. Nat. Commun. 14 (1), 8361. doi:10.1038/s41467-023-43780-4
Ascione, A., Martino, G., Di Donato, F., Casini, B., Covello, R., and Ascani, S. (2023). CIC-rearranged sarcoma presenting with superior vena cava syndrome: case report. Pathologica 115 (2), 97–100. doi:10.32074/1591-951X-784
Babkoff, A., Berner-Wygoda, Y., Diment, J., Kustanovich, A., Zick, A., Katz, D., et al. (2023). First female patient with a rare CIC-FOXO4-Translocated sarcoma: a case report. Case Rep. Oncol. 16 (1), 954–962. doi:10.1159/000533519
Bakaric, A., Cironi, L., Praz, V., Sanalkumar, R., Broye, L. C., Favre-Bulle, K., et al. (2024). CIC-DUX4 chromatin profiling reveals new epigenetic dependencies and actionable therapeutic targets in CIC-rearranged sarcomas. Cancers (Basel) 16 (2), 457. doi:10.3390/cancers16020457
Bergerat, S., Barthelemy, P., Mouracade, P., Lang, H., Saussine, C., Lindner, V., et al. (2017). Primary CIC-DUX4 round cell sarcoma of the kidney: a treatment-refractory tumor with poor outcome. Pathol. Res. Pract. 213 (2), 154–160. doi:10.1016/j.prp.2016.11.015
Biederman, L. E., Lee, K., Yeager, N. D., Sribnick, E. A., and Shenoy, A. (2022). CIC::NUTM1 sarcoma mimicking primitive myxoid mesenchymal tumour of infancy: report of a case. Histopathology 81 (1), 131–133. doi:10.1111/his.14630
Bielle, F., Zanello, M., Guillemot, D., Gil-Delgado, M., Bertrand, A., Boch, A. L., et al. (2014). Unusual primary cerebral localization of a CIC–DUX4 translocation tumor of the Ewing sarcoma family. Acta Neuropathol. 128 (2), 309–311. doi:10.1007/s00401-014-1312-0
Blay, J. Y., Coindre, J. M., Ducimetière, F., and Ray-Coquard, I. (2016). The value of research collaborations and consortia in rare cancers. Lancet Oncol. 17 (2), e62–e69. doi:10.1016/S1470-2045(15)00388-5
Borkhardt, A., Repp, R., Haas, O. A., Leis, T., Harbott, J., Kreuder, J., et al. (1997). Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23). Oncogene 14 (2), 195–202. doi:10.1038/sj.onc.1200814
Bosnakovski, D., Ener, E. T., Cooper, M. S., Gearhart, M. D., Knights, K. A., Xu, N. C., et al. (2021). Inactivation of the CIC-DUX4 oncogene through P300/CBP inhibition, a therapeutic approach for CIC-DUX4 sarcoma. Oncogenesis 10 (10), 68. doi:10.1038/s41389-021-00357-4
Brahmi, M., Gaspar, N., Gantzer, J., Toulmonde, M., Boudou-Rouquette, P., Bompas, E., et al. (2023). Patterns of care and outcome of CIC-rearranged sarcoma patients: a nationwide study of the French sarcoma group. Cancer Med. 12 (7), 7801–7807. doi:10.1002/cam4.5539
Brohl, A. S., Solomon, D. A., Chang, W., Wang, J., Song, Y., Sindiri, S., et al. (2014). The genomic landscape of the ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 10 (7), e1004475. doi:10.1371/journal.pgen.1004475
Camille, A., Anne-Sophie, B., Cécile, P., Severine, B. C., Gaelle, P., Olivier, D., et al. (2018). Sarcoma with CIC-DUX4 gene fusion: case report of kidney tumor location in a 12-year-old boy. Pediatr. Dev. Pathology 21 (4), 406–410. doi:10.1177/1093526617706818
Chebib, I., and Jo, V. Y. (2016). Round cell sarcoma with CIC-DUX4 gene fusion: discussion of the distinctive cytomorphologic, immunohistochemical, and molecular features in the differential diagnosis of round cell tumors. Cancer Cytopathol. 124 (5), 350–361. doi:10.1002/cncy.21685
Chen, T., Wang, Y., Goetz, L., Corey, Z., Dougher, M. C., Smith, J. D., et al. (2021). Novel fusion sarcomas including targetable NTRK and ALK. Ann. Diagn Pathol. 54, 151800. doi:10.1016/j.anndiagpath.2021.151800
Choi, S. H., Gearhart, M. D., Cui, Z., Bosnakovski, D., Kim, M., Schennum, N., et al. (2016). DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44 (11), 5161–5173. doi:10.1093/nar/gkw141
Connolly, E. A., Bhadri, V. A., Wake, J., Ingley, K. M., Lewin, J., Bae, S., et al. (2022). Systemic treatments and outcomes in CIC-rearranged Sarcoma: a national multi-centre clinicopathological series and literature review. Cancer Med. 11 (8), 1805–1816. doi:10.1002/cam4.4580
Dansen, T. B., Smits, L. M. M., van Triest, M. H., de Keizer, P. L. J., van Leenen, D., Koerkamp, M. G., et al. (2009). Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 5 (9), 664–672. doi:10.1038/nchembio.194
Donahue, J. E., Yakirevich, E., Zhong, S., Treaba, D. O., Lakis, N. S., Ali, S. M., et al. (2018). Primary spinal epidural CIC-DUX4 undifferentiated sarcoma in a child. Pediatr. Dev. Pathology 21 (4), 411–417. doi:10.1177/1093526617707856
Forés, M., Simón-Carrasco, L., Ajuria, L., Samper, N., González-Crespo, S., Drosten, M., et al. (2017). A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 13 (3), e1006622. doi:10.1371/journal.pgen.1006622
French, C. A. (2018). NUT Carcinoma: clinicopathologic features, pathogenesis, and treatment. Pathol. Int. 68 (11), 583–595. doi:10.1111/pin.12727
Gawriyski, L., Jouhilahti, E. M., Yoshihara, M., Fei, L., Weltner, J., Airenne, T. T., et al. (2023). Comprehensive characterization of the embryonic factor LEUTX. iScience 26 (3), 106172. doi:10.1016/j.isci.2023.106172
Haidar, A., Arekapudi, S., DeMattia, F., Abu-Isa, E., and Kraut, M. (2015). High-grade undifferentiated small round cell sarcoma with t(4;19)(q35;q13.1) CIC-DUX4 fusion: emerging entities of soft tissue tumors with unique histopathologic features – a case report and literature review. Am. J. Case Rep. 16, 87–94. doi:10.12659/AJCR.892551
Hendrickson, P. G., Oristian, K. M., Browne, M. R., Luo, L., Ma, Y., Cardona, D. M., et al. (2024). Spontaneous expression of the CIC::DUX4 fusion oncoprotein from a conditional allele potently drives sarcoma formation in genetically engineered mice. Oncogene 43 (16), 1223–1230. doi:10.1038/s41388-024-02984-8
Herrmann, A., Nagao, T., Zhang, C., Lahtz, C., Li, Y. J., Yue, C., et al. (2019). An effective cell-penetrating antibody delivery platform. JCI Insight 4 (14), e127474. doi:10.1172/jci.insight.127474
Hu, W., Wang, J., Yuan, L., Zhang, X., Ji, Y., Song, C., et al. (2020). Case report: a unique case of pediatric central nervous system embryonal tumor harboring the CIC–LEUTX fusion, germline NBN variant and somatic TSC2 mutation: expanding the spectrum of CIC-rearranged neoplasia. Front. Oncol. 10, 598970. doi:10.3389/fonc.2020.598970
Huang, S. C., Zhang, L., Sung, Y. S., Chen, C. L., Kao, Y. C., Agaram, N. P., et al. (2016). Recurrent CIC gene abnormalities in angiosarcomas: a molecular study of 120 cases with concurrent investigation of PLCG1, kdr, myc, and FLT4 gene alterations. Am. J. Surg. Pathology 40 (5), 645–655. doi:10.1097/PAS.0000000000000582
Hwang, I., Pan, H., Yao, J., Elemento, O., Zheng, H., and Paik, J. (2020). CIC is a critical regulator of neuronal differentiation. JCI Insight 5 (9), e135826. doi:10.1172/jci.insight.135826
Jiménez, G., Guichet, A., Ephrussi, A., and Casanova, J. (2000). Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 14 (2), 224–231. doi:10.1101/gad.14.2.224
Kajtár, B., Tornóczky, T., Kálmán, E., Kuzsner, J., Hogendoorn, P. C. W., and Szuhai, K. (2014). CD 99-positive undifferentiated round cell sarcoma diagnosed on fine needle aspiration cytology, later found to harbour a CIC-DUX4 translocation: a recently described entity. Cytopathology 25 (2), 129–132. doi:10.1111/cyt.12079
Karpukhina, A., Tiukacheva, E., Dib, C., and Vassetzky, Y. S. (2021). Control of DUX4 expression in facioscapulohumeral muscular dystrophy and cancer. Trends Mol. Med. 27 (6), 588–601. doi:10.1016/j.molmed.2021.03.008
Kawamura-Saito, M., Yamazaki, Y., Kaneko, K., Kawaguchi, N., Kanda, H., Mukai, H., et al. (2006). Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 15 (13), 2125–2137. doi:10.1093/hmg/ddl136
Kim, J. W., Luck, C., Wu, W., Ponce, R. K., Lin, Y. K., Gupta, N., et al. (2022). Capicua suppresses YAP1 to limit tumorigenesis and maintain drug sensitivity in human cancer. Cell Rep. 41 (1), 111443. doi:10.1016/j.celrep.2022.111443
Krskova, L., Stejskalova, E., Kabickova, E., Mrhalova, M., and Kodet, R. (2017). A t(4;19) pediatric undifferentiated sarcoma with a novel variant of the CIC-DUX4 fusion transcript. Pathol. Res. Pract. 213 (3), 281–285. doi:10.1016/j.prp.2016.12.005
Lake, J. A., Donson, A. M., Prince, E., Davies, K. D., Nellan, A., Green, A. L., et al. (2020). Targeted fusion analysis can aid in the classification and treatment of pediatric glioma, ependymoma, and glioneuronal tumors. Pediatr. Blood Cancer 67 (1), e28028. doi:10.1002/pbc.28028
Lee, J. S., Kim, E., Lee, J., Kim, D., Kim, H., Kim, C. J., et al. (2020). Capicua suppresses colorectal cancer progression via repression of ETV4 expression. Cancer Cell Int. 20 (1), 42. doi:10.1186/s12935-020-1111-8
Lehane, F., Tsikleas, G., Bettington, A., Limarporn, K., Wilkinson, L., and Lehane, K. (2019). “Cyst” on the forearm of a 28-year-old female: case report of a CIC-rearranged sarcoma. J. Cutan. Pathol. 46 (8), 599–602. doi:10.1111/cup.13478
Le Loarer, F., Pissaloux, D., Watson, S., Godfraind, C., Galmiche-Rolland, L., Silva, K., et al. (2019). Clinicopathologic features of CIC-NUTM1 sarcomas, a new molecular variant of the family of CIC-fused sarcomas. Am. J. Surg. Pathology 43 (2), 268–276. doi:10.1097/PAS.0000000000001187
Lin, Y. K., Wu, W., Ponce, R. K., Kim, J. W., and Okimoto, R. A. (2020). Negative MAPK-ERK regulation sustains CIC-DUX4 oncoprotein expression in undifferentiated sarcoma. Proc. Natl. Acad. Sci. 117 (34), 20776–20784. doi:10.1073/pnas.2009137117
Linos, K., Dermawan, J. K., Bale, T., Rosenblum, M. K., Singer, S., Tap, W., et al. (2023). Expanding the molecular diversity of CIC-rearranged sarcomas with novel and very rare partners. Mod. Pathol. 36 (5), 100103. doi:10.1016/j.modpat.2023.100103
Loke, B. N., Lee, V. K. M., Sudhanshi, J., Wong, M. K., Kuick, C. H., Puhaindran, M., et al. (2017). Novel exon–exon breakpoint in CIC-DUX4 fusion sarcoma identified by anchored multiplex PCR (Archer FusionPlex Sarcoma Panel). J. Clin. Pathol. 70 (8), 697–701. doi:10.1136/jclinpath-2016-204247
Ma, Y., Feng, J., Ding, D., Tian, F., and Zhao, J. (2023). SMARCB1/INI1-deficient undifferentiated pancreatic carcinoma in a 13-year-old male patient: a case report. Pediatr. Blood Cancer 70 (9), e30038. doi:10.1002/pbc.30038
Machado, I., Cruz, J., Lavernia, J., Rubio, L., Campos, J., Barrios, M., et al. (2013). Superficial EWSR1-negative undifferentiated small round cell sarcoma with CIC/DUX4 gene fusion: a new variant of Ewing-like tumors with locoregional lymph node metastasis. Virchows Arch. 463 (6), 837–842. doi:10.1007/s00428-013-1499-9
Maejima, E., Mitsui, H., Ohnuma, T., Oishi, N., Odate, T., Deguchi, N., et al. (2021). Case of CIC-DUX4 sarcoma of the skin: histological simulant of epithelioid angiosarcoma. J. Dermatol 48 (12), e594–e595. doi:10.1111/1346-8138.16157
Maekawa, A., Matsunobu, T., Nabeshima, A., Fukushima, S., Makihara, K., Hisaoka, M., et al. (2021). Cardiac tamponade as an unusual initial clinical manifestation of CIC-DUX4 sarcoma. Am. J. Case Rep. 22, e929349. doi:10.12659/AJCR.929349
Maloney, N., Smith, S. M., Peters, S. B., Batistatou, A., Evangelou, Z., Harms, P. W., et al. (2020). Expanding the differential of superficial tumors with round-cell morphology: report of three cases of CIC -rearranged sarcoma, a potentially under-recognized entity. J. Cutan. Pathol. 47 (6), 535–540. doi:10.1111/cup.13639
Mangray, S., Kelly, D. R., LeGuellec, S., Fridman, E., Aggarwal, S., Shago, M., et al. (2018). Clinicopathologic features of a series of primary renal CIC-rearranged sarcomas with comprehensive molecular analysis. Am. J. Surg. Pathology 42 (10), 1360–1369. doi:10.1097/PAS.0000000000001098
Murphy, J., Resch, E. E., Leland, C., Meyer, C. F., Llosa, N. J., Gross, J. M., et al. (2024). Clinical outcomes of patients with CIC-rearranged sarcoma: a single institution retrospective analysis. J. Cancer Res. Clin. Oncol. 150 (3), 112. doi:10.1007/s00432-024-05631-7
Nakai, S., Yamada, S., Outani, H., Nakai, T., Yasuda, N., Mae, H., et al. (2019). Establishment of a novel human CIC-DUX4 sarcoma cell line, Kitra-SRS, with autocrine IGF-1R activation and metastatic potential to the lungs. Sci. Rep. 9 (1), 15812. doi:10.1038/s41598-019-52143-3
Okimoto, R. A., Breitenbuecher, F., Olivas, V. R., Wu, W., Gini, B., Hofree, M., et al. (2017). Inactivation of Capicua drives cancer metastasis. Nat. Genet. 49 (1), 87–96. doi:10.1038/ng.3728
Okimoto, R. A., Wu, W., Nanjo, S., Olivas, V., Lin, Y. K., Ponce, R. K., et al. (2019). CIC-DUX4 oncoprotein drives sarcoma metastasis and tumorigenesis via distinct regulatory programs. J. Clin. Investigation 129 (8), 3401–3406. doi:10.1172/JCI126366
Oyama, R., Takahashi, M., Yoshida, A., Sakumoto, M., Takai, Y., Kito, F., et al. (2017). Generation of novel patient-derived CIC- DUX4 sarcoma xenografts and cell lines. Sci. Rep. 7 (1), 4712. doi:10.1038/s41598-017-04967-0
Ponce, R. K. M., Thomas, N. J., Bui, N. Q., Kondo, T., and Okimoto, R. A. (2022). WEE1 kinase is a therapeutic vulnerability in CIC-DUX4 undifferentiated sarcoma. JCI Insight 7 (6), e152293. doi:10.1172/jci.insight.152293
Reynoird, N., Schwartz, B. E., Delvecchio, M., Sadoul, K., Meyers, D., Mukherjee, C., et al. (2010). Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 29 (17), 2943–2952. doi:10.1038/emboj.2010.176
Ricker, C. A., Berlow, N. E., Crawford, K. A., Georgopapadakos, T., Huelskamp, A. N., Woods, A. D., et al. (2020). Undifferentiated small round cell sarcoma in a young male: a case report. Mol. Case Stud. 6 (1), a004812. doi:10.1101/mcs.a004812
Schaefer, I., Dal Cin, P., Landry, L. M., Fletcher, C. D. M., Hanna, G. J., and French, C. A. (2018). CIC-NUTM1 fusion: a case which expands the spectrum of NUT -rearranged epithelioid malignancies. Genes Chromosom. Cancer 57 (9), 446–451. doi:10.1002/gcc.3
Shiota, H., Barral, S., Buchou, T., Tan, M., Couté, Y., Charbonnier, G., et al. (2018). Nut directs p300-dependent, genome-wide H4 hyperacetylation in male germ cells. Cell Rep. 24 (13), 3477–3487. doi:10.1016/j.celrep.2018.08.069
Sievers, P., Sill, M., Schrimpf, D., Abdullaev, Z., Donson, A. M., Lake, J. A., et al. (2023). Pediatric-type high-grade neuroepithelial tumors with CIC gene fusion share a common DNA methylation signature. NPJ Precis. Oncol. 7 (1), 30. doi:10.1038/s41698-023-00372-1
Solomon, D. A., Brohl, A. S., Khan, J., and Miettinen, M. (2014). Clinicopathologic features of a second patient with ewing-like sarcoma harboring CIC-FOXO4 gene fusion. Am. J. Surg. Pathology 38 (12), 1724–1725. doi:10.1097/PAS.0000000000000335
Song, K., Huang, Y., Xia, C., Zhu, H., and Wang, J. (2022). A case of CIC-rearranged sarcoma with CIC-LEUTX gene fusion in spinal cord. Neuropathology 42 (6), 555–562. doi:10.1111/neup.12850
Sorensen, P. H. B., Lynch, J. C., Qualman, S. J., Tirabosco, R., Lim, J. F., Maurer, H. M., et al. (2002). PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the Children’s Oncology group. J. Clin. Oncol. 20 (11), 2672–2679. doi:10.1200/JCO.2002.03.137
Specht, K., Sung, Y., Zhang, L., Richter, G. H. S., Fletcher, C. D., and Antonescu, C. R. (2014). Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion–positive round cell tumors compared to EWSR1 -rearranged ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosom. Cancer 53 (7), 622–633. doi:10.1002/gcc.22172
Sturm, D., Orr, B. A., Toprak, U. H., Hovestadt, V., Jones, D. T. W., Capper, D., et al. (2016). New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164 (5), 1060–1072. doi:10.1016/j.cell.2016.01.015
Sugita, S., Arai, Y., Tonooka, A., Hama, N., Totoki, Y., Fujii, T., et al. (2014). A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am. J. Surg. Pathology 38 (11), 1571–1576. doi:10.1097/PAS.0000000000000286
Takemon, Y., LeBlanc, V. G., Song, J., Chan, S. Y., Lee, S. D., Trinh, D. L., et al. (2023). Multi-omic analysis of CIC’s functional networks reveals novel interaction partners and a potential role in mitotic fidelity. Cancers (Basel) 15 (10), 2805. doi:10.3390/cancers15102805
Tamada, H., Kobayashi, M., Sano, K., Uehara, T., Matsumoto, Y., Tateishi, A., et al. (2020). Ultrastructure of CIC-DUX4 sarcoma: the first pathological report. Ultrastruct. Pathol. 44 (2), 237–244. doi:10.1080/01913123.2020.1737610
Tang, S., and Dodd, L. G. (2018). CIC-DUX4 sarcoma diagnosed by fine-needle aspiration cytology: a case report. Diagn Cytopathol. 46 (11), 958–963. doi:10.1002/dc.24027
Tang, Y., Lu, X., and Zhan, R. (2023). Renal CIC-LEUTX rearranged sarcoma with multiple pulmonary metastases: a case report and literature review. BMC Nephrol. 24 (1), 354. doi:10.1186/s12882-023-03404-x
Tardío, J. C., Machado, I., Navarro, L., Idrovo, F., Sanz-Ortega, J., Pellín, A., et al. (2015). Ewing-like sarcoma with CIC-DUX4 gene fusion in a patient with neurofibromatosis type 1. A hitherto unreported association. Pathol. Res. Pract. 211 (11), 877–882. doi:10.1016/j.prp.2015.08.003
Thomas, N. J., Luck, C., Shlimon, N., Ponce, R. K. M., Kosibaty, Z., and Okimoto, R. A. (2023). Mapping chromatin state and transcriptional response in CIC-DUX4 undifferentiated round cell sarcoma. bioRxiv. doi:10.1101/2023.10.11.561932
Tsukamoto, Y., Futani, H., Yoshiya, S., Watanabe, T., Kihara, T., Matsuo, S., et al. (2017). Primary undifferentiated small round cell sarcoma of the deep abdominal wall with a novel variant of t(10;19) CIC-DUX4 gene fusion. Pathol. Res. Pract. 213 (10), 1315–1321. doi:10.1016/j.prp.2017.06.008
van der Heide, L. P., and Smidt, M. P. (2005). Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem. Sci. 30 (2), 81–86. doi:10.1016/j.tibs.2004.12.002
Vieira, A. C., Xavier, C. B., Vieira, T. D., Carvalho, F. M., Scaranti, M., Munhoz, R. R., et al. (2021). CIC-DUX4 rearranged uterine cervix round-cell sarcoma exhibiting near-complete pathologic response following radiation and neoadjuvant chemotherapy: a case report. Gynecol. Oncol. Rep. 36, 100745. doi:10.1016/j.gore.2021.100745
Watson, S., Perrin, V., Guillemot, D., Reynaud, S., Coindre, J., Karanian, M., et al. (2018). Transcriptomic definition of molecular subgroups of small round cell sarcomas. J. Pathol. 245 (1), 29–40. doi:10.1002/path.5053
Webb, J., Liew, J., Gnann, A., Patterson, M., Paul, S., Forés, M., et al. (2022). Molecular basis of DNA recognition by the HMG-box-C1 module of Capicua. bioRxiv.doi:10.1101/2022.03.28.485992
Weissmann, S., Cloos, P. A., Sidoli, S., Jensen, O. N., Pollard, S., and Helin, K. (2018). The tumor suppressor CIC directly regulates MAPK pathway genes via histone deacetylation. Cancer Res. 78 (15), 4114–4125. doi:10.1158/0008-5472.CAN-18-0342
Wu, D. J., Murugan, P., and Skubitz, K. M. (2023). Patched homolog 1 (PTCH1) mutation in a CIC-rearranged sarcoma: lack of response to the smoothened (SMO) vismodegib. Cureus 15, e34281. doi:10.7759/cureus.34281
Wu, Q., and He, Y. (2022). A case report of CIC–DUX4 fusion-positive sarcoma in the pelvic cavity with targeted next-generation sequencing results. Front. Oncol. 15, 12. doi:10.3389/fonc.2022.1018992
Yamada, S., Muto, J., De Leon, J. C. A., Kumai, T., Ito, K., Murayama, K., et al. (2020). Primary spinal intramedullary Ewing-like sarcoma harboring CIC-DUX4 translocation: a similar cytological appearance as its soft tissue counterpart but no lobulation in association with desmoplastic stroma. Brain Tumor Pathol. 37 (3), 111–117. doi:10.1007/s10014-020-00366-y
Yang, S., Liu, L., Yan, Y., Jiang, L., Han, S., Shen, D., et al. (2022). CIC-NUTM1 sarcomas affecting the spine. Arch. Pathol. Lab. Med. 146 (6), 735–741. doi:10.5858/arpa.2021-0153-OA
Yoe, J., Kim, D., Kim, S., and Lee, Y. (2020). Capicua restricts cancer stem cell-like properties in breast cancer cells. Oncogene 39 (17), 3489–3506. doi:10.1038/s41388-020-1230-7
Yoshimatsu, Y., Noguchi, R., Tsuchiya, R., Kito, F., Sei, A., Sugaya, J., et al. (2020). Establishment and characterization of NCC-CDS2-C1: a novel patient-derived cell line of CIC-DUX4 sarcoma. Hum. Cell 33 (2), 427–436. doi:10.1007/s13577-019-00312-x
Keywords: CIC, CIC-DUX sarcomas, CIC-DUX4, CIC-NUTM1 sarcomas, CIC-rearranged sarcoma, Capicua (CIC)
Citation: Ponce RKM, Luck C and Okimoto RA (2024) Molecular and therapeutic advancements in Capicua (CIC)-rearranged sarcoma. Front. Cell Dev. Biol. 12:1416697. doi: 10.3389/fcell.2024.1416697
Received: 12 April 2024; Accepted: 14 May 2024;
Published: 31 May 2024.
Edited by:
Sean Bong Lee, Tulane University, United StatesReviewed by:
Le Su, HudsonAlpha Institute for Biotechnology, United StatesCopyright © 2024 Ponce, Luck and Okimoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ross A. Okimoto, cm9zcy5va2ltb3RvQHVjc2YuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.