- 1Integrative Program for Biological and Genome Sciences, The University of North Carolina at Chapel Hill, Chapel Hill, NC, Unites States
- 2Department of Cell Biology and Physiology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, Unites States
- 3Department of Biology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, Unites States
The molecular mechanisms that govern the metabolic commitment to reproduction, which often occurs at the expense of somatic reserves, remain poorly understood. We identified the Caenorhabditis elegans F-box protein FBXL-5 as a negative regulator of maternal provisioning of vitellogenin lipoproteins, which mediate the transfer of intestinal lipids to the germline. Mutations in fbxl-5 partially suppress the vitellogenesis defects observed in the heterochronic mutants lin-4 and lin-29, both of which ectopically express fbxl-5 at the adult developmental stage. FBXL-5 functions in the intestine to negatively regulate expression of the vitellogenin genes; and consistently, intestine-specific over-expression of FBXL-5 is sufficient to inhibit vitellogenesis, restrict lipid accumulation, and shorten lifespan. Our epistasis analyses suggest that fbxl-5 functions in concert with cul-6, a cullin gene, and the Skp1-related gene skr-3 to regulate vitellogenesis. Additionally, fbxl-5 acts genetically upstream of rict-1, which encodes the core mTORC2 protein Rictor, to govern vitellogenesis. Together, our results reveal an unexpected role for a SCF ubiquitin-ligase complex in controlling intestinal lipid homeostasis by engaging mTORC2 signaling.
Introduction
Reproduction is a metabolically expensive process that, in many metazoans, is supported by resources stored in somatic cells. In particular, reallocation of lipids from the soma to the germline, a process called vitellogenesis, is crucial for fertilization, embryonic development, and offspring resiliency (Perez et al., 2017; Perez and Lehner, 2019; Geens et al., 2023; Wang et al., 2023). In the nematode Caenorhabditis elegans, synthesis of vitellogenin lipoproteins occurs in the intestine where they mediate the assembly and secretion of triglyceride-rich low density-like lipoprotein (LDL-like) particles that constitute the maternal yolk (Kimble and Sharrock, 1983). The circulating yolk is captured and endocytosed by the RME-2 receptor expressed in mature oocytes (Grant and Hirsh, 1999). Deletion of rme-2 severely impairs yolk uptake, reduces progeny production by over 20-fold, and extends maternal lifespan (Grant and Hirsh, 1999; Dowen, 2019), which underscores the metabolic trade-off described by the disposable soma theory–that reproductive fitness is inversely related to longevity due to a limited amount of resources that must be balanced between somatic and germline functions (Kirkwood, 1977).
Expression of intestinal vitellogenin lipoproteins is coordinated by a suite of developmental, reproductive, nutritional, and metabolic signals (Perez and Lehner, 2019), which together define the temporal expression and the abundance of vitellogenin lipoproteins. We previously demonstrated that vitellogenesis is regulated by several developmental timing factors that are temporally expressed in the C. elegans hypodermis (Dowen et al., 2016). These include the heterochronic genes lin-4, let-7, and lin-29, which when mutated result in reiteration of larval cell divisions and retardation of subsequent developmental events (Chalfie et al., 1981; Ambros and Horvitz, 1984; Reinhart et al., 2000). Expression of LIN-29, a zinc finger transcription factor, in the hypodermis at the larva-to-adult transition is required for maximal vitellogenin production (Ambros, 1989; Dowen et al., 2016), and mutations that impair lin-29 expression (i.e., lin-4 and let-7) confer similar vitellogenesis defects. How these factors engage developmental or metabolic signaling pathways in the intestine is poorly understood.
Developmental and environmental regulation of vitellogenesis is coordinated by several pro-growth and homeostatic signaling pathways that function in the intestine. Some of the pathways that control vitellogenin transcription, including the TGF-β and the insulin/insulin-like growth factor signaling (IIS) pathways, are likely to respond to neuronally-secreted ligands (i.e., insulin-like peptides or the TGF-β family member DBL-1) that are modulated by environmental or nutritional conditions (Goszczynski et al., 2016; Perez and Lehner, 2019). In contrast, other intestinal signaling factors respond to developmental or reproductive signals from hypodermal or germline tissues, respectively (Balklava et al., 2016; Dowen et al., 2016; Goszczynski et al., 2016). We have previously demonstrated that the hypodermal heterochronic pathway engages intestinal mTOR (mechanistic target of rapamycin) signaling to promote vitellogenin gene expression (Dowen et al., 2016). The mTOR kinase resides in two multiprotein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which contain the complex-specific scaffolding proteins DAF-15/Raptor and RICT-1/Rictor, respectively. Loss of either RICT-1 or SGK-1 (serum/glucocorticoid-regulated kinase 1), a downstream target of mTORC2, severely disrupts vitellogenin expression (Dowen et al., 2016). While mTORC2 is necessary for maximal vitellogenin expression, it is more difficult to assess whether mTORC1 is required for vitellogenesis, as complete loss of DAF-15 results in developmental arrest. Although it is well established that insulin/IGF signaling stimulates mTORC2 activity in mammals, it is yet not clear whether mTORC2 is regulated by additional morphogens or pro-growth signaling pathways during development (Oh and Jacinto, 2011).
The factors that mediate crosstalk between the hypodermal heterochronic pathway and the intestinal mTORC2 signaling pathway are unknown. Here, we identify a previously uncharacterized F-box protein, FBXL-5, as an intestinal modulator of vitellogenin expression and intestinal lipid homeostasis in C. elegans. Mutations in fbxl-5 partially suppress the vitellogenin expression defects observed in lin-29 and lin-4 mutants, suggesting that FBXL-5 negatively regulates vitellogenesis. This function is performed in concert with the Skp1-related protein SKR-3 and Cullin protein CUL-6, which together form a canonical SCF ubiquitin-ligase complex. Moreover, we demonstrate that FBXL-5 acts within the mTORC2 signaling pathway to regulate intestinal homeostasis. Although FBXL-5 and mTORC2 function together to regulate a broad set of genes, we also found that FBXL-5 acts via a mTORC2-independent mechanism to regulate vitellogenesis and aging pathways. Thus, our work identifies a new cell-autonomous regulator of mTORC2 signaling that couples developmental timing events in the hypodermis to metabolic homeostasis in the intestine.
Materials and methods
Caenorhabditis elegans strains
All C. elegans strains were cultured at 20°C (unless specified otherwise) on NGM media seeded with E. coli OP50 as previously described (Brenner, 1974). To synchronize growth of C. elegans animals, embryos were isolated by bleaching, washed in M9 buffer, and incubated overnight at room temperature. The resulting synchronized L1s were dropped on seeded plates and grown until adulthood. For auxin treatment, gravid adults were allowed to lay eggs on NGM plates containing 4 mM Naphthaleneacetic Acid (K-NAA, PhytoTech) and the progeny were grown to the L4 stage, picked to new plates containing auxin, and grown for an additional 24 h prior to imaging. The strains used in this study are listed in Supplementary Table S1.
Generation and imaging of transgenic animals
The high-copy mgIs70[Pvit-3::GFP] and single-copy rhdSi42[Pvit-3::mCherry] transgenes have been previously described (Dowen et al., 2016; Torzone et al., 2023). The fbxl-5 rescue constructs were generated by fusing either the col-10 promoter (chromosome V: 9,166,416–9,165,291; WS284) or the vha-6 promoter (chromosome II: 11,439,355–11,438,422; WS284) to a mCherry::his-58::SL2::fbxl-5 cassette (fbxl-5 gene: 2,330 bp ORF and 176 bp of 3′UTR) within the pCFJ151 plasmid via Gibson assembly to generate plasmids pRD99 and pRD98, respectively (Frøkjær-Jensen et al., 2008; Gibson et al., 2009). The sequence-verified plasmids were microinjected into lin-4(e912); fbxl-5(rhd43); mgIs70[Pvit-3::GFP] animals at 20 ng/μL, along with 2.5 ng/μL pCFJ90(Pmyo-2::mCherry) and 77.5 ng/μL of 2-Log DNA ladder (New England BioLabs), to generate the DLS327 and DLS330 strains. To image these strains, L1 animals expressing Pmyo-2::mCherry were hand-picked on a SMZ-18 Stereo microscope with fluorescence, grown to adulthood, and were then randomly selected for fluorescence imaging under bright field using a standard Stereo microscope. To generate rhdIs2 and rhdIs4, wild-type animals carrying the extrachromosomal array Ex[Pvha-6::mCherry::his-58::SL2::fbxl-5 + Pmyo-2::mCherry] were gamma-irradiated using a137Cs source (∼3000 rad), resulting in random integration of the array into the genome. The resulting two independent lines were backcrossed at least three times. The fbxl-5 promoter (chromosome V: 7,746,887–7,748,867; WS288) was amplified by PCR and fused to a mCherry::unc-54 3′UTR fragment by PCR fusion (Hobert, 2002) to yield a Pfbxl-5::mCherry::unc-54 3′UTR PCR fragment, which was microinjected into wild-type animals at 75 ng/μL, along with 5 ng/μL Pmyo-3::GFP and 20 ng/μL of 2-Log DNA ladder (New England BioLabs), to generate the DLS344 strain. All strains carrying the mgIs70 or rhdSi42 transgenes were imaged with a Nikon SMZ-18 Stereo microscope equipped with a DS-Qi2 monochrome camera, while the DLS344 strain was imaged on a Nikon Eclipse E800 microscope.
Reporter imaging and quantification
Strains carrying either the mgIs70 or rhdSi42 reporter were grown asynchronously, L4s were picked to new plates, and animals were imaged 24 h later. Alternatively, synchronized animals were grown for 72 h before imaging. Day 1 adults were mounted on a 2% agarose pad with 25 mM levamisole and imaged with a Nikon SMZ-18 Stereo microscope equipped with a DS-Qi2 monochrome camera. The worm bodies were outlined in the brightfield channel and the mean intensities (i.e., gray values) were calculated using the GFP (mgIs70) or mCherry (rhdSi42) channel using the Fiji 2.14.0 software (Schindelin et al., 2012). Body size measurements (pixels/worm) were simultaneously performed, and the data were converted to mm2 based on the known imaging settings. The fluorescence and body size data were plotted in Prism 10 and a one-way ANOVA with a Bonferroni correction was performed to calculate p values. All tabular data and the associated statistical analyses can be found in Supplementary Table S5.
CRISPR/Cas9 gene editing
Genomic edits were performed by microinjection of Cas9:crRNA:tracrRNA complexes (Integrated DNA Technologies) into the germline as previously described (Ghanta and Mello, 2020). The crRNA guide sequences are listed in Supplementary Table S2. To generate missense, nonsense, or deletion mutations, single-stranded oligodeoxynucleotides were used as donor molecules. The deletion edits employed two sgRNAs positioned at each end of the deletion site and an oligo that bridged the predicted repair junction. To insert the 3xFLAG::AID cassette into the 5′ end of the fbxl-5 gene, dsDNA donor molecules with ∼40 bp homology arms were prepared by PCR using Q5 DNA Polymerase (New England BioLabs) and purified using HighPrep PCR Clean-up beads (MagBio) according to the manufacturers’ instructions. The melted and reannealed DNA repair templates were prepared and microinjected into the germline as previously described (Ghanta and Mello, 2020).
RNAi experiments
E. coli HT115 (DE3) strains carrying plasmids for fbxl-5 or lin-29 RNAi knockdown (Ahringer RNAi library), or the control strain containing the empty L4440 plasmid, were grown for ∼19 h at 37°C in Luria–Bertani media containing ampicillin (100 μg/mL). The bacteria were concentrated by 20–30x via centrifugation, seeded on NGM plates containing 5 mM isopropyl-β-D-thiogalactoside (IPTG) and 100 μg/mL ampicillin, and maintained at room temperature overnight to induce dsRNA expression. Synchronized L1s were dropped on RNAi plates, grown at 20°C until they were day 1 adults (∼72 h), and imaged. For the skr/cul RNAi screen, we screened cul-1,2,3,4,5,6 and skr-1,2,3,5,7,11,12,13,14,16,17,19,20,21 using existing clones from the Ahringer RNAi library and assessed their ability to confer any detectable increase in Pvit-3::GFP reporter expression (relative to the L4440 control) in the DLS447 strain. The skr-4,6,8,9,10,15,18 genes were not screened due to a lack of a sequence-verified RNAi clone in our collection. RNAi conditions that prevented development to the adult stage (e.g., skr-1 RNAi) were not included in the analysis.
Lifespan and brood size assays
Animals were grown on standard NGM E. coli OP50 plates for at least two generations without starvation prior to lifespan and brood size assays. Longitudinal lifespan assays were performed at 25°C in the presence of 50 μM FUDR as previously described (Torzone et al., 2023). Approximately 150 animals were assayed for each genotype and a log-rank test was used to determine significance. Lifespan assays were repeated at least twice with similar results. For brood size assays (20°C), L4 animals (n = 11 per genotype) were picked to individual plates and transferred daily throughout the reproductive period. Progeny were counted as L3, L4s, or as adults. The lowest brood size for each genotype was dropped from the analysis due to potential injury and the data were reported as the mean +/- the standard deviation. The tabular data and statistical analyses can be found in Supplementary Table S5.
Oil Red O staining
Animals were fixed in 60% isopropanol and stained for 7 h with 0.3% Oil Red O exactly as previously described (Torzone et al., 2023). Next, animals were mounted on 2% agarose pads and imaged with either a Nikon SMZ-18 Stereo microscope equipped with a DS-Qi2 monochrome camera (intensity quantification) or a Nikon Ti2 widefield microscope equipped with a DS-Fi3 color camera (representative color images). Quantification of Oil Red O staining was performed in Fiji 2.14.0. Animals were manually outlined, the mean gray value was measured in the worm area, and the value was subtracted from 65,536 (maximum gray value for 16-bit images). The data were plotted in Prism 10 and a one-way ANOVA with a Bonferroni correction was performed to calculate p values. The tabular data and statistical analyses are shown in Supplementary Table S5.
Quantitative PCR
Synchronized day 1 adult animals were harvested in M9 buffer, washed, and flash frozen. Isolation of total RNA was performed using Trizol Reagent (Thermo Fisher) with chloroform extraction, followed by isopropanol precipitation. Synthesis of cDNA was performed by oligo (dT) priming using the SuperScript IV VILO Master Mix with ezDNase Kit per the manufacturer’s instructions (Thermo Fisher). Quantitative PCR was performed exactly as previously described (Dowen, 2019). The qPCR primer sequences are listed in Supplementary Table S3. Data from at least three independent experiments were plotted in Prism 10 as the mean fold change relative to wild-type with the standard error of the mean (SEM). The individual Ct values and the statistical analyses can be found in Supplementary Table S5.
Western blot analyses
Synchronized L1 animals expressing the 3xFLAG::AID::FBXL-5 protein (DLS874 or DLS889) were grown at 20°C for either 36 h (L3s), 48 h (L4s), 72 h (day 1 adults), or 96 h (day 2 adults) before harvesting them in M9 buffer. The animals were washed three times and pellets were frozen in liquid nitrogen. Whole cell lysates and protein concentrations were determined exactly as previously described (Torzone et al., 2023). The protein samples (50 μg) were resolved by SDS-PAGE, transferred to a PVDF membrane, blocked in 5% nonfat dry milk (BioRad), and probed with either anti-FLAG M2 (F1804, Sigma) or anti-Actin (ab3280, Abcam) antibodies. The developmental time course experiment was performed twice with similar results.
EMS mutagenesis
The GR2123 strain was mutagenized with ethyl methanesulfonate (M0880, Sigma-Aldrich) exactly as previously described (Dowen et al., 2016). Suppressor mutants displaying increased mgIs70[Pvit-3::GFP] reporter expression were selected from ten distinct mutagenesis pools and were singled to individual plates. Mutations that suppressed the egg-laying defects of lin-4(e912) were presumed to disrupt lin-14 function and were not selected for subsequent studies.
Suppressor strains were crossed to the GR2122 strain (mgIs70[Pvit-3::GFP]) and F2 recombinants that displayed the characteristic egl and lon phenotypes of the lin-4(e912) mutant, and that also showed elevated mgIs70 expression, were singled. Starved animals from the individual plates were then pooled and the genomic DNA was prepared with the Qiagen Gentra Puregene Tissue Kit (Doitsidou et al., 2010). The DNA-Seq libraries were prepared using the NEBNext DNA library preparation kit according to the manufacturer’s instructions (E6040, New England Biolabs) and the libraries were sequenced on an Illumina HiSeq 4000 instrument. Identification of candidate suppressor mutations was performed using in-house scripts according to previously described methods (Minevich et al., 2012).
mRNA sequencing
Wild-type and mutant animals were reared on 10 cm NGM agarose plates (5000 animals/plate) until they reached the first day of adulthood. RNA was isolated using Trizol Reagent as described above (see quantitative PCR). The mRNA-Seq libraries were prepared with 1 μg of total RNA using the TruSeq RNA Library Prep Kit v2 per the manufacturer’s instructions (Illumina). Three independent biological replicates were prepared for each condition and libraries were sequenced on an Illumina HiSeq 4000 instrument (single-end, 50 bp) at the High Throughput Genomic Sequencing Facility at the University of North Carolina at Chapel Hill. The reads were aligned to the C. elegans genome (WS260), and the feature read counts were compiled exactly as previously described (Dowen, 2019). RPKM values and differentially expressed genes (1% FDR) were determined using the DESeq2 algorithm (Love et al., 2014). Lists of differentially expressed genes and RPKM values can be found in Supplementary Table S4. Tissue-specific enrichment for differential expression was calculated as the number of observed vs. expected differentially expressed genes using previously generated gene lists for each major tissue (Kaletsky et al., 2018). The gene ontology analysis was performed using WormCat (Holdorf et al., 2020). All scatter and violin plots displaying differential expression were generated using the DESeq2 RPKM values and the data were plotted in Prism 10. All tabular data and associated statistical analyses can be found in Supplementary Table S5. The raw and processed mRNA-Seq data have been deposited in GEO (GSE248602).
Results
Mutations in T05B11.1/fbxl-5 suppress lin-4 mutant defects in vitellogenesis
The heterochronic gene lin-4 encodes a miRNA that governs developmental timing by controlling larval cell divisions in the C. elegans hypodermis (Lee et al., 1993). Furthermore, lin-4 acts non-cell-anonymously to license expression of the vitellogenin genes (vit-1 through vit-6) in the intestine at the L4 larval to adult transition (Dowen et al., 2016). To identify genes that act in the lin-4 vitellogenesis pathway, we performed an EMS mutagenesis screen to identify mutations that suppress the vitellogenesis defects observed in the lin-4(e912) mutant using a Pvit-3::GFP vitellogenesis reporter as a readout (Supplementary Figure S1A). We selected lin-4 suppressor mutants that displayed elevated Pvit-3::GFP expression but that also maintained the egg-laying (egl) and long body length (lon) defects of the lin-4(e912) mutant, which selects against known lin-4 suppressors (e.g., lin-14 alleles). After backcrossing to remove unlinked background mutations, we performed whole genome sequencing of the lin-4 suppressor strains and identified candidate causative mutations using established bioinformatic approaches (Minevich et al., 2012). This resulted in the identification of two putative loss-of-function mutations in the uncharacterized T05B11.1 gene, which were recovered in two genetically distinct suppressor strains (Supplementary Figure S1B).
The T05B11.1 protein contains a F-box domain at the amino terminus and leucine-rich repeats (LRRs) that span most of the protein (Paysan-Lafosse et al., 2023). Thus, we will subsequently refer to the T05B11.1 gene as fbxl-5 (F-box leucine rich repeat containing protein 5). F-box proteins canonically function in protein ubiquitination, typically in concert with the Skp1 and Cullin proteins (the SCF complex), which together catalyze E3 ubiquitin ligase activity (Skaar et al., 2013). The F-box domain, and in some cases the LRRs as well, specifically recognizes target substrates and mediates poly-ubiquitination of proteins, resulting in 26S proteasomal degradation (K11/K48 linkages) or altered protein function/localization (K63 linkages). Although there is no clear mammalian orthologue of FBXL-5, highly similar proteins are found in other Caenorhabditis species, and it is possible that functional orthologues may exist in other species.
To characterize the role of FBXL-5 in vitellogenesis, we first compared expression levels of the Pvit-3::GFP reporter between the lin-4(e912) single mutant and the lin-4(e912); fbxl-5(rhd43) double mutant, finding that while reporter expression is increased in lin-4(e912) animals upon loss of fbxl-5 it does not reach that of wild-type (Figure 1A). Notably, the rhd43 lesion is a nonsense mutation and is thus predicted to be a strong loss-of-function or null allele. Consistently, knock-down of fbxl-5 by RNAi partially suppressed the Pvit-3::GFP expression defects observed in the lin-4 mutant (Supplementary Figure S1C). To rule out the possibility that the high-copy Pvit-3::GFP reporter lacks the sensitivity to robustly detect the fbxl-5 suppression phenotype, we employed a single-copy Pvit-3::mCherry reporter, which closely reflects endogenous vit-3 levels (Torzone et al., 2023), and generated large deletions within the fbxl-5 locus using CRISPR/Cas9 genomic editing (Supplementary Table S2; Supplementary Figure S1B). While each of the three fbxl-5 deletion alleles tested suppressed the lin-4(e912) mutant defects in Pvit-3::mCherry expression, they did not restore levels to those seen in wild-type animals (Figures 1B, C). Interestingly, all fbxl-5 deletion mutations also reduced the body size of the lin-4(e912) mutant (Figure 1D). Consistently, the fbxl-5(rhd43) mutation conferred smaller body sizes in otherwise wild-type animals (Supplementary Figure S2), suggesting that FBXL-5 may regulate additional developmental or metabolic processes outside of vitellogenesis.
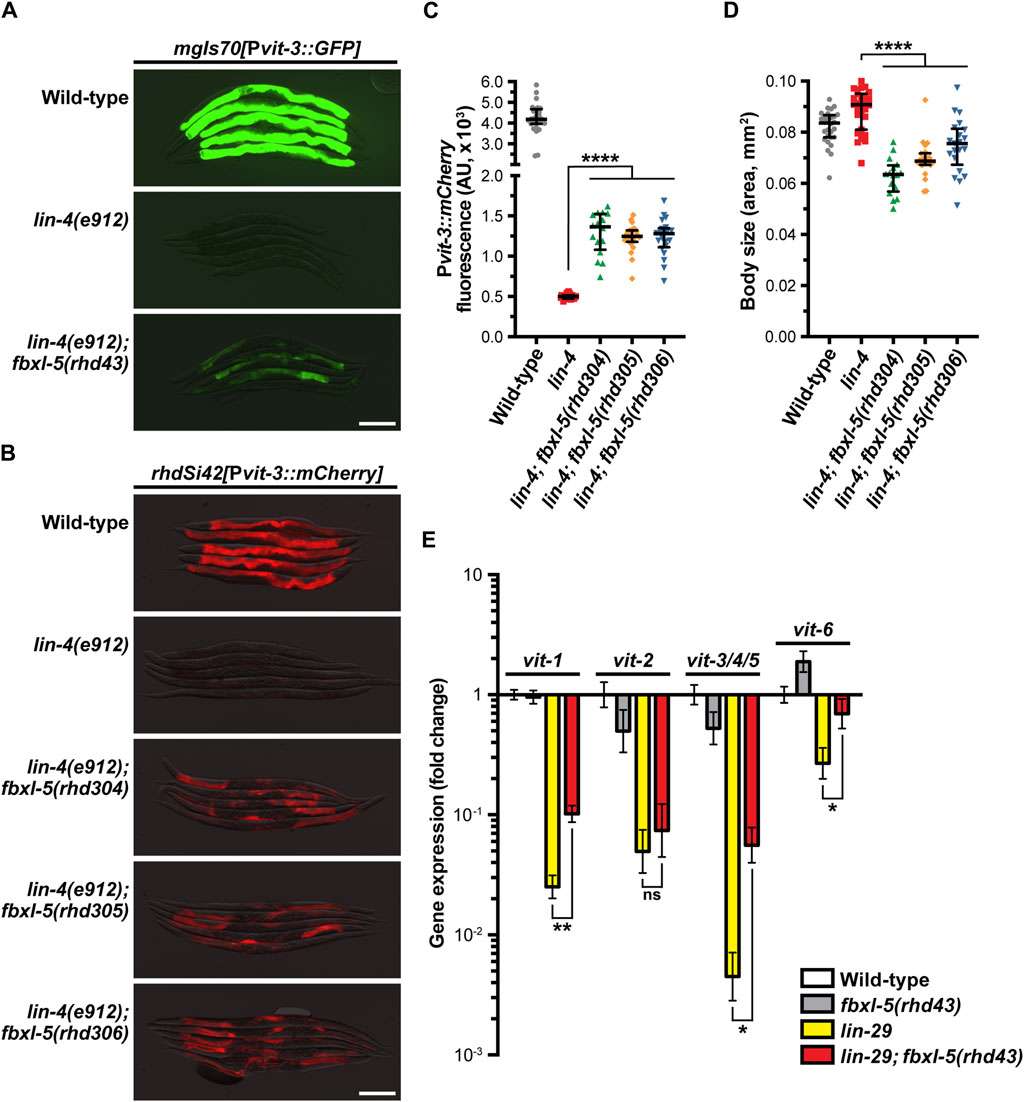
Figure 1. The F-box protein FBXL-5 negatively regulates vitellogenesis in the Caenorhabditis elegans heterochronic mutants. Overlaid DIC and fluorescence images of day 1 adult wild-type, lin-4, and lin-4; fbxl-5 animals expressing (A) a high-copy Pvit-3::GFP reporter (vit-3 promoter fused to GFP) or (B) a single-copy Pvit-3::mCherry reporter (scale bars, 200 μm). The fbxl-5(rhd43) allele is a nonsense mutation, while the rhd304-rhd306 are large fbxl-5 deletion alleles. Quantification of (C) Pvit-3::mCherry fluorescence and (D) body size of the strains shown in (B) at the day 1 adult stage. Data are displayed as the median and interquartile range (****, p < 0.0001, one-way ANOVA). (E) Expression of the vit genes as measured by RT-qPCR in wild-type, fbxl-5(rhd43), lin-29(n333), and lin-29(n333); fbxl-5(rhd43) day one adult animals (mean +/- SEM; ns, not significant, *, p < 0.05, **, p < 0.01, t-test).
Impairment of hypodermal lin-4 alters the expression of other developmental timing genes resulting in heterochrony. In particular, lin-4 mutants fail to properly express lin-29 at the larval to adult transition. Thus, we reasoned that loss of fbxl-5 may suppress the vit expression defects seen in the lin-29(n333) mutant. Using RT-qPCR to measure the endogenous vit transcripts, we found that mutation of fbxl-5 also partially suppressed the vitellogenesis defects in lin-29(n333) animals; however, loss of fbxl-5 on its own is not sufficient to increase vit transcripts in otherwise wild-type animals (Figure 1E). These data suggest that FBXL-5 may negatively regulate vitellogenesis only when developmental timing is impaired. Together, our results indicate that fbxl-5 acts genetically downstream or in parallel to lin-4/lin-29, possibly in the intestinal cells, to govern vitellogenin expression.
FBXL-5 governs specific aspects of cellular homeostasis
Although loss of fbxl-5 only partially suppressed the vitellogenesis defects in the lin-29 mutant, it is possible that FBXL-5 plays a broader role in regulating cellular metabolism or lipid allocation in the heterochronic mutants. Taking an unbiased approach, we employed mRNA sequencing (mRNA-Seq) to resolve transcriptome-level differences between the lin-29 single mutant and the lin-29; fbxl-5 double mutant (Supplementary Table S4). Remarkably, loss of fbxl-5 had no global impact on the expression levels of the 6,265 genes that are differentially expressed in the lin-29 mutant (R2 = 0.927, Figure 2A). Expression of the vit-3-5 genes, as well as a handful of other downregulated genes, did increase in the lin-29; fbxl-5 double mutant; however, their expression did not return to wild-type. Thus, FBXL-5 has very specific and limited effects on gene expression in the lin-29 heterochronic mutant.
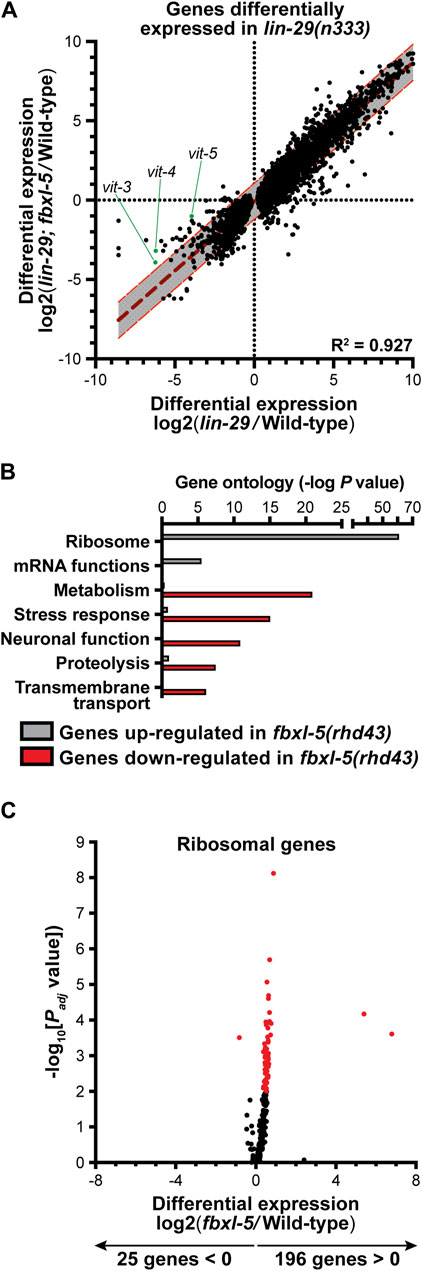
Figure 2. Mutation of fbxl-5 suppresses only a portion of the transcriptional defects caused by the lin-29 mutation and prompts a global upregulation of ribosomal genes. (A) A scatter plot of the mRNA-Seq differential expression values comparing the lin-29(n333) mutant to the lin-29(n333); fbxl-5(rhd43) double mutant for all 6,265 genes differentially expressed in lin-29(n333) animals (1% FDR). A linear regression analysis (dark red dashed line) is displayed and the 95% prediction bands for the best-fit line are shown in gray. (B) A bar plot showing the p values for the top functional categories derived from a gene ontology analysis of the genes differentially expressed in the fbxl-5(rhd43) mutant (1% FDR). (C) A volcano plot showing the differential expression values for all annotated ribosomal genes in the fbxl-5(rhd43) mutant (red dots indicate differentially expressed genes, 1% FDR).
To further define the role of FBXL-5 in wild-type animals, we performed mRNA-Seq on the fbxl-5(rhd43) single mutant. This analysis led to the identification of 828 genes that were differentially expressed upon loss of fbxl-5 (Supplementary Table S4). Mutation of fbxl-5 resulted in both increased and decreased mRNA levels for genes expressed in each of the major C. elegans tissues, which likely represents both the cell-autonomous and non-cell-autonomous effects of mutating fbxl-5 (Supplementary Figure S3). A gene ontology analysis revealed that the upregulated genes are enriched in ribosomal and mRNA function, while the downregulated genes are annotated to act in cellular metabolism, stress response pathways, or proteolysis (Figure 2B). Remarkably, expression of 196 of the 221 C. elegans ribosomal genes was increased in the fbxl-5 mutant (Figure 2C), suggesting that translation is globally upregulated upon loss of fbxl-5. Thus, FBXL-5 functions broadly in maintaining cellular energy and protein homeostasis, possibly by governing the activity of a core homeostatic regulator that senses metabolic or developmental signals to tune protein synthesis.
Heterochronic mutants ectopically express FBXL-5 at adulthood
Since loss of fbxl-5 in an otherwise wild-type background fails to upregulate vit gene expression (Figure 1E), we reasoned that FBXL-5 levels may be specifically altered in the lin-4 and lin-29 heterochronic mutants. To test this hypothesis, we first measured fbxl-5 transcript levels at the L4 and adult stages in the lin-4(e912) mutant by RT-qPCR, finding that fbxl-5 expression is specifically upregulated in lin-4 days 1 adult animals (Figure 3A). Consistently, adult expression of fbxl-5 is also elevated in the lin-29 mutant relative to wild-type animals (Supplementary Figure S4A). To measure FBXL-5 protein levels, we inserted a 3xFLAG::AID cassette into the 5’ end of the fbxl-5 locus by CRISPR/Cas9 editing, which did not disrupt FBXL-5 function and enabled detection by Western blotting (Supplementary Figure S4B). This strain, however, did not respond to auxin treatment (Supplementary Figure S4B), possibly because the auxin-inducible degron (AID) tag on FBXL-5 is buried in a larger protein complex that was inaccessible to TIR1. We assessed FBXL-5 expression levels across development by Western blot, probing lysates from wild-type and lin-4(e912) animals isolated at different developmental stages with an anti-FLAG antibody. Consistent with our gene expression measurements, the lin-4 mutant ectopically expresses FBXL-5 at the first day of adulthood, which persists into the second day of adulthood (Figure 3B; Supplementary Figure S4C). We observed a similar FBXL-5 expression pattern for animals subjected to lin-29 RNAi (Supplementary Figure S4D). While wild-type animals had the highest amount of FBXL-5 expression at the L4 stage, FBXL-5 protein was reproducibly detected at the L3s stage (Figure 3B; Supplementary Figure S4C), suggesting that fbxl-5 may be upregulated at the L3-L4 transition and may serve a specific role during the L4 developmental stage. Together, these data indicate that heterochronic programs control fbxl-5 expression, suggesting that the heterochrony of the lin-4 and lin-29 mutants may be, at least in part, attributed to improper levels of FBXL-5 during development.
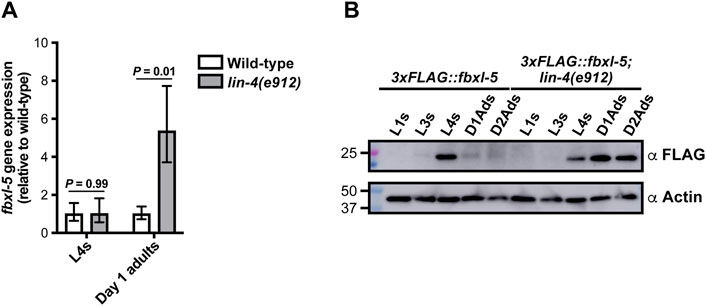
Figure 3. The FBXL-5 protein is misexpressed at the adult stage in the lin-4 heterochronic mutant. (A) mRNA expression levels of fbxl-5 in wild-type and lin-4(e912) animals at the L4 and day 1 adult stage as determined by RT-qPCR (mean ± SEM; t-test). (B) A Western blot analysis of lysates from wild-type or lin-4 mutants expressing an endogenously-tagged 3xFLAG::FBXL-5 protein at the indicated developmental stages (replicate experiment shown in Supplementary Figure S4C). An actin blot is included as a loading control.
FBXL-5 acts cell-autonomously to govern vitellogenin expression and intestinal homeostasis
Since loss of fbxl-5 resulted in altered transcription of genes expressed in a variety of tissues, we reasoned that FBXL-5 could act either cell-autonomously or non-cell-autonomously to control vitellogenin expression in the intestine. Importantly, LIN-29 functions in the hypodermis to regulate vitellogenesis in a non-cell-autonomous fashion (Dowen et al., 2016), and it is possible that ectopic expression of fbxl-5 in the hypodermis is partially responsible for the impaired vitellogenin production that is observed in the lin-29 mutant. To investigate the tissue-specificity of FBXL-5, we first generated a transgenic strain expressing a Pfbxl-5::mCherry transcriptional reporter, comprised of a ∼2 kb fragment of the fbxl-5 promoter driving the mCherry gene, and inspected animals for mCherry expression by fluorescence microscopy. This analysis revealed that fbxl-5 is indeed weakly expressed in the intestine of wild-type young adults; however, it is also expressed in a small set of neurons in the head and in the body wall muscle (Figure 4A; Supplementary Figure S5A), which is consistent with the widespread transcriptional changes that we observed. Surprisingly, we did not observe Pfbxl-5::mCherry reporter expression in the hypodermis despite finding that hypodermal genes were downregulated in the fbxl-5 mutant (Supplementary Figure S3). However, it is possible that fbxl-5 hypodermal expression is relatively low compared to other tissues, a hypothesis that is supported by existing tissue-specific mRNA-Seq data (Kaletsky et al., 2018). Intriguingly, in wild-type animals intestinal Pfbxl-5::mCherry reporter expression continued to increase throughout adulthood while FBXL-5 protein levels decreased (Figure 3B; Supplementary Figure S5B), suggesting that fbxl-5 may be post-transcriptionally or post-translationally regulated; however, the long-term stability of mCherry protein may at least partially contribute to this elevated signal in older animals.
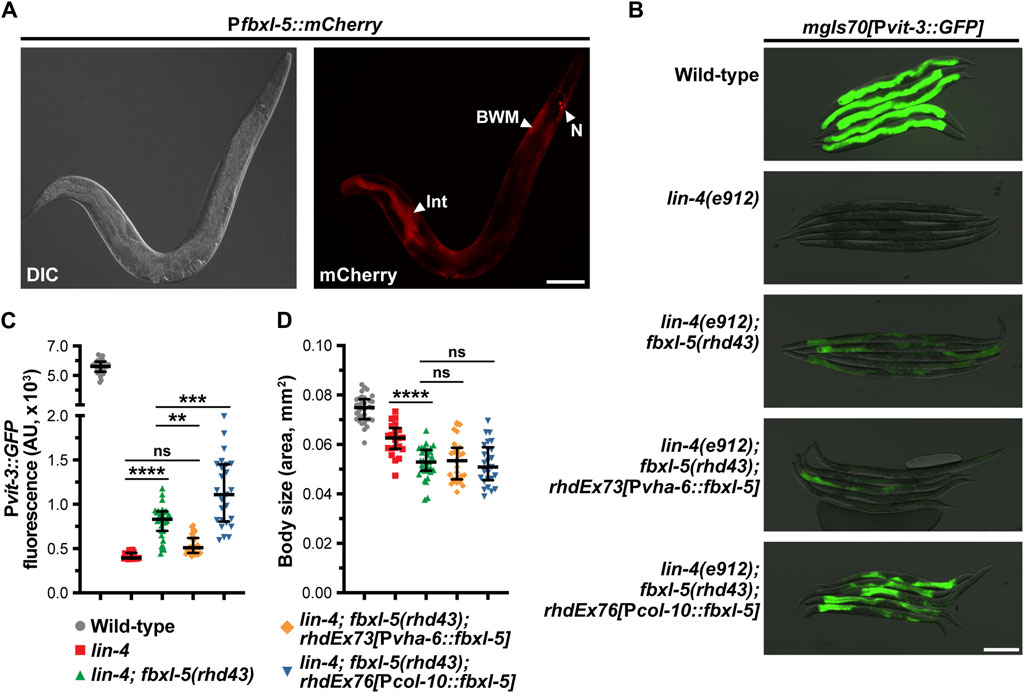
Figure 4. The FBXL-5 protein acts cell-autonomously to control vitellogenesis. (A) A representative DIC and mCherry fluorescence image of an animal expressing a Pfbxl-5::mCherry transcriptional reporter (Int, intestine; BWM, body wall muscle; N, neurons; scale bar, 100 μm). (B) Representative overlaid DIC and Pvit-3::GFP fluorescence images (scale bar, 200 μm), (C) Pvit-3::GFP reporter quantification, and (D) body sizes of day 1 adult wild-type, lin-4(e912), and lin-4(e912); fbxl-5(rhd43) animals, as well as the indicated lin-4(e912); fbxl-5(rhd43) tissue-specific rescue strains (Pvha-6, intestinal-specific rescue; Pcol-10, hypodermal-specific rescue). (C,D) Data are plotted as the median and interquartile range (ns, not significant, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001, one-way ANOVA).
Despite being expressed in several tissues, we reasoned that FBXL-5 likely acts in the intestine or in the hypodermis (the site of lin-29 action) to regulate vitellogenesis. Thus, we generated intestine-specific and hypodermis-specific fblx-5 rescue constructs to test whether the fblx-5(rhd43) mutation could be rescued by tissue-specific expression of fblx-5. Rescue of fblx-5 in the intestine, but not in the hypodermis, reversed the suppressive effects of fblx-5(rhd43) in the lin-4 mutant background back to baseline Pvit-3::GFP levels (Figures 4B, C). Surprisingly, hypodermal fblx-5 over-expression further enhanced the suppressive effects of the rhd43 allele, suggesting that FBXL-5 might promote vitellogenin expression when expressed outside of the intestine. While the vitellogenesis phenotype was rescued by intestinal expression of fbxl-5, the small body size phenotype that we observed in the lin-4(e912); fblx-5(rhd43) double mutant was not rescued by intestinal or hypodermal fbxl-5 expression (Figure 4D), suggesting that FBXL-5 may act non-cell-autonomously to control body size.
Since fbxl-5 is ectopically expressed in the lin-4 and lin-29 heterochronic mutants, we reasoned that the consequences of FBXL-5 misexpression should be mirrored by constitutive over-expression of fbxl-5 in the intestine of otherwise wild-type animals. Thus, we integrated a Pvha-6::fbxl-5 extrachromosomal array into the genome via gamma irradiation and crossed the resulting strains to our vitellogenesis reporter. Consistent with our fbxl-5 rescue experiments, constitutive over-expression of fbxl-5 in the intestine is sufficient to abrogate Pvit-3::GFP expression (Figures 5A, B). Furthermore, fbxl-5 over-expression prevented the accumulation of intestinal lipids and restricted body size (Figures 5C, D), suggesting that FBXL-5 might impair nutrient absorption or stimulate lipid catabolism pathways. Lipid levels are reduced both at the L4 larval stage and during reproduction by fbxl-5 over-expression (Figure 5E), suggesting that FBXL-5 does not act exclusively on a larval-specific metabolic regulator.
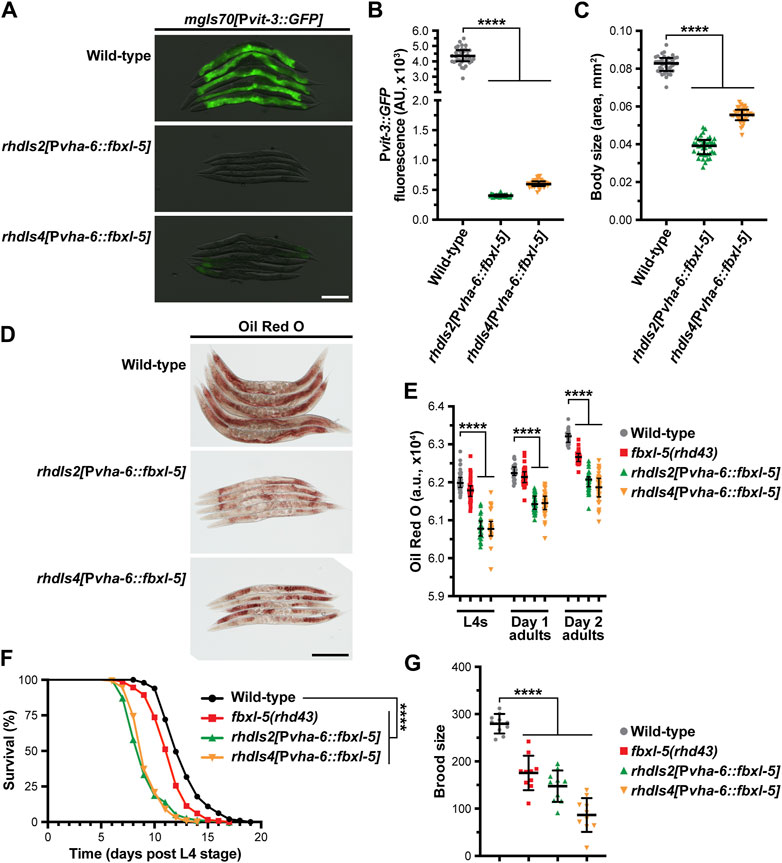
Figure 5. Constitutive overexpression of fbxl-5 in the intestine is sufficient to impair lipid homeostasis and shorten lifespan. (A) Overlaid DIC and Pvit-3::GFP fluorescence images (scale bar, 200 μm), (B) Pvit-3::GFP reporter quantification, (C) body size, and (D) representative images (scale bar, 200 μm) of Oil Red O-stained wild-type or fbxl-5 intestinal overexpression animals (Pvha-6::fbxl-5, two independently integrated array lines). (E) Quantification of Oil Red O staining for wild-type, fbxl-5(rhd43), and fbxl-5 overexpression animals at the L4, day one adult, and day 2 adult stages reveals that FBXL-5 restricts lipid accumulation. (F) Longitudinal lifespans (25°C with FUDR; ****, p < 0.0001, Log-rank test) and (G) brood size (20°C; mean +/- SD; ****, p < 0.0001, one-way ANOVA) of wild-type, fbxl-5(rhd43), and fbxl-5 overexpression animals. (B,C,E) Data are plotted as the median and interquartile range (****, p < 0.0001, one-way ANOVA).
Impaired vitellogenin production can indirectly extend organismal lifespan by stimulating intestinal autophagy and lipid catabolism (Murphy et al., 2003; Seah et al., 2016). Thus, we tested whether fbxl-5 over-expression, which severely reduces vitellogenin synthesis, increases longevity. Surprisingly, over-expression of fbxl-5 in the intestine reduced the lifespan of otherwise wild-type animals (Figure 5F), which is consistent with FBXL-5 playing a more widespread role in metabolic regulation. Moreover, the fbxl-5(rhd43) mutant also displayed a modest, but statistically significant, reduction in lifespan, which is consistent with the modest reduction in lipid levels and brood size that we observed in these mutant animals (Figures 5E, G). Predictably, intestinal over-expression of fbxl-5 reduced progeny production (Figure 5G), likely a consequence of the impaired vitellogenin production. Together, these data suggest that FBXL-5 can function broadly in the intestine to alter metabolic, reproduction, and longevity programs.
FBXL-5 functions in a canonical SCF complex
One canonical function of F-box proteins is to mediate poly-ubiquitination and proteasomal degradation of their substrates, which requires coordination with the Skp1 and Cullin proteins. While C. elegans expresses ∼520 F-box proteins, the genome only contains 21 Skp1-related (skr) genes and 6 Cullin (cul) genes, suggesting that these factors have less functional redundancy than the F-box family (Nayak et al., 2002; Thomas, 2006). To determine if FBXL-5 functions in concert with the SKR and CUL proteins, we performed a small-scale RNAi screen to identify the skr/cul genes that are required for reducing Pvit-3::GFP reporter expression in fbxl-5 over-expression animals. We screened all six cul genes, as well as a majority of the skr genes (14/21), for vitellogenesis phenotypes and excluded any clones that caused lethality or prevented larval development. Knock-down of skr-3 by RNAi modestly increased GFP levels in the reporter strain, which was recapitulated using the skr-3(ok365) mutant allele (Figure 6A; Supplementary Figure S6). Although we also identified cul-6 as a candidate in our RNAi screen, the cul-6(ok1614) mutation failed to replicate this finding; however, mutation of both skr-3 and cul-6 strongly increased Pvit-3::GFP expression in fbxl-5 over-expression animals relative to the control (Figures 6A, B; Supplementary Figure S6). Further mutation of skr-4 or skr-5, which both share significant similarity to skr-3 (Nayak et al., 2002), failed to dramatically enhance the skr-3(ok365); cul-6(ok365) double mutant. Together, these data suggest that FBXL-5 functions in concert with SKR-3 and CUL-6 to restrict vitellogenin production; however, it is likely that FBXL-5 acts with other redundantly-acting factors, as Pvit-3::GFP expression levels are only partially restored to wild-type levels in skr-3; cul-6 double mutant. Alternatively, high levels of FBXL-5 over-expression may facilitate interactions with other SKR/CUL proteins that are not normally preferred when FBXL-5 is expressed at endogenous levels, which may limit the suppressive effects of the skr-3 and cul-6 mutations in this experiment.
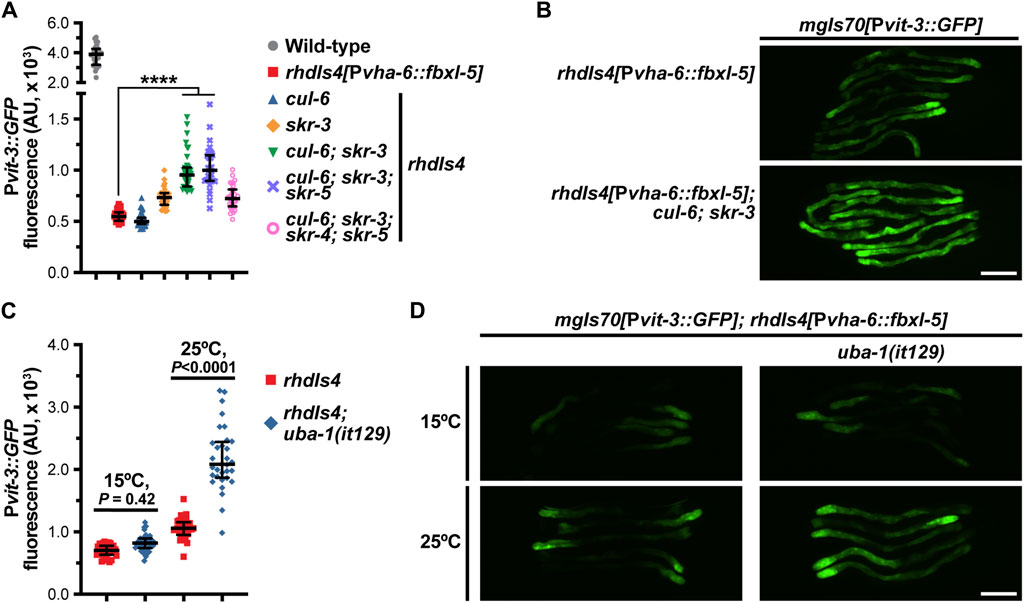
Figure 6. FBXL-5 acts in a SCF complex to restrict vitellogenesis likely via canonical ubiquitin-mediated proteasomal degradation. (A) Pvit-3::GFP fluorescence quantification and (B) representative Pvit-3::GFP images of fbxl-5 intestinal overexpression animals (rhdIs4) carrying the indicated skr (Skp1 related) or cul (Cullin) mutations suggesting that SKR-3 and CUL-6 act with FBXL-5 in a SCF complex. The full set of representative Pvit-3::GFP images can be found in Supplementary Figure S6. (C) Pvit-3::GFP fluorescence quantification and (D) representative Pvit-3::GFP images of rhdIs4 or rhdIs4; uba-1(it129) animals. The uba-1(it129) mutation is a temperature sensitive loss-of-function allele at 25°C. (A,C) Data are plotted as the median and interquartile range (****, p < 0.0001, one-way ANOVA). (B,D) Scale bars are 200 μm.
To further examine the role of poly-ubiquitination in the FBXL-5-dependent regulation of vitellogenesis, we introduced the uba-1(it129ts) temperature-sensitive mutation into the fbxl-5 over-expression strain. UBA-1 is the sole E1 ubiquitin-activating enzyme in C. elegans and its activity is required for the ubiquitin proteolytic pathway (Kulkarni and Smith, 2008). Consistent with our previous results, the uba-1(it129ts) mutation partially suppressed the effects of fbxl-5 over-expression at the restrictive, but not permissive, temperature (Figures 6C, D). Together, our results suggest that FBXL-5 acts in a SCF complex, likely with SKR-3 and CUL-6, to promote poly-ubiquitination-mediated proteasomal degradation. However, we are unable to eliminate the possibility that FBXL-5 associates with other SKR/CUL proteins in the intestine, or potentially a different set of SKR/CUL proteins in the muscle, to carry out some of its homeostatic functions.
FBXL-5 acts in concert with mTORC2 to govern intestinal homeostasis
We have previously shown that hypodermal lin-29 acts genetically upstream of intestinal mTORC2 signaling to regulate vitellogenesis (Dowen et al., 2016). We hypothesized that FBXL-5 might be responsible for modulating the signaling between LIN-29 and mTORC2, which we tested by performing genetic epistasis analyses. While knock-down of fbxl-5 by RNAi increased Pvit-3::GFP expression 3.3-fold in lin-4(e912) and 3.9-fold in lin-29(n333) mutant animals, the suppressive effects seen in rict-1(mg360) and sgk-1(ok538) animals were less striking (1.6- and 0.9-fold, respectively; Figure 7A; Supplementary Figure S7A). The mg360 allele is a missense mutation in rict-1, and thus, this mutant could retain some rict-1 function, which complicates the interpretation of our epistasis analysis. Therefore, we generated a complete rict-1 deletion mutant by CRISPR/Cas9 editing, removing the entire sequence between the start and stop codons, including the pqn-32 gene that is positioned between exons 10 and 11, and measured Pvit-3::GFP reporter expression. Consistent with the mg360 allele, complete deletion of rict-1 resulted in a robust decrease in Pvit-3::GFP levels, while further mutation of fbxl-5 resulted in a modest, but significant, 1.1-fold increase in reporter expression (Figure 7B). This weak suppression phenotype was not limited to vitellogenesis, as the fbxl-5(rhd43) mutation also partially suppressed the short lifespan of the rict-1(mg360) mutant (Figure 7C). To eliminate the possibility that FBXL-5 non-specifically restricts vitellogenin expression in the intestine, we performed fbxl-5 RNAi on daf-2(e1370) mutants, which also have reduced Pvit-3::GFP reporter expression (Dowen et al., 2016). Knock-down of fbxl-5 had no impact on vitellogenin expression in daf-2 mutant animals (Supplementary Figure S7B). Together, our results suggest that fbxl-5 acts genetically upstream of rict-1 and sgk-1 to regulate vitellogenesis; however, it is also likely that fbxl-5 acts simultaneously in a pathway parallel to rict-1 to regulate lifespan and vitellogenesis via a mTORC2-independent mechanism.
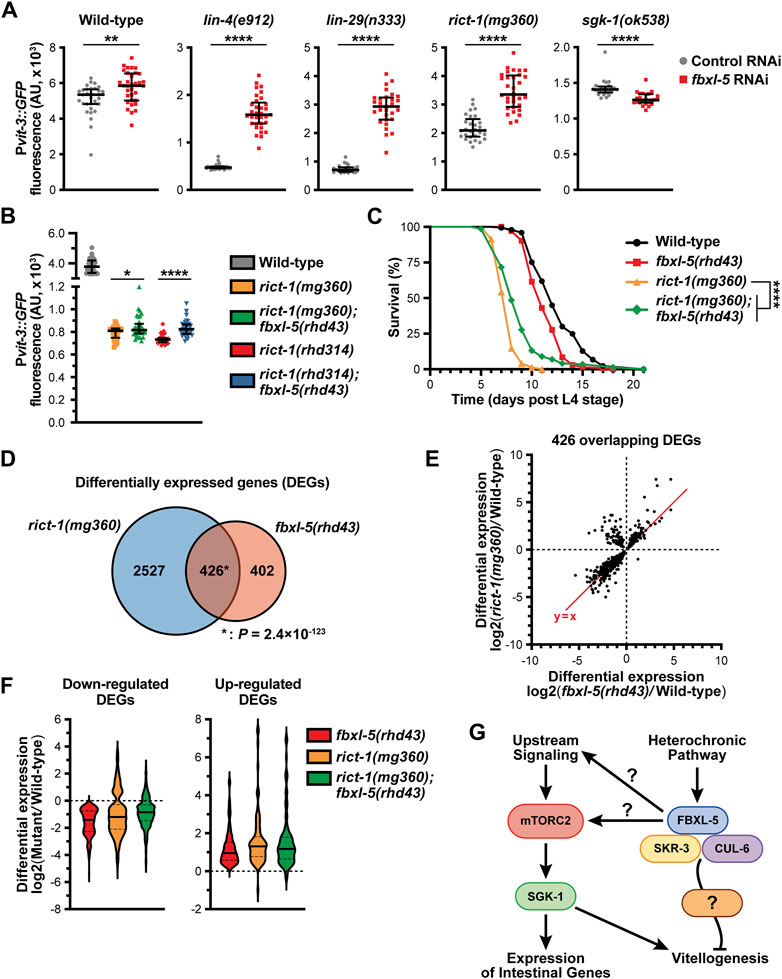
Figure 7. FBXL-5 functions in the mTORC2 pathway. (A) Quantification of Pvit-3::GFP fluorescence in the indicated heterochronic (lin-4, lin-29) or mTORC2 (rict-1, sgk-1) mutants following treatment with either control or fbxl-5 RNAi (median and interquartile range; ****, p < 0.0001, t-test). (B) Pvit-3::GFP fluorescence quantification of strains carrying either the rict-1(mg360) missense mutation or the rict-1(rhd314) null mutation, which lacks the entire rict-1 locus (median and interquartile range; *, p < 0.05, ****, p < 0.0001, t-test). The fbxl-5(rhd43) mutation modestly suppresses both rict-1 mutations. (C) Longitudinal lifespan assays of the indicated strains reared at 25°C in the presence of FUDR demonstrating that the fbxl-5(rhd43) mutation extends the lifespan of short-lived rict-1(mg360) animals (****, p < 0.0001, Log-rank test). (D) A Venn diagram showing the overlap of genes differentially expressed in the rict-1(mg360) and fbxl-5(rhd43) mutants relative to wild-type (1% FDR; *, hypergeometric p-value). (E) A scatter plot of the differential expression values of the 426 genes co-regulated by RICT-1 and FBXL-5 indicating that most genes are similarly regulated. (F) Violin plots comparing the differential expression values of the 426 RICT-1/FBXL-5 co-regulated genes for the fbxl-5(rhd43) and rict-1(mg360) single mutants, as well as the rict-1(mg360); fbxl-5(rhd43) double mutant, showing that loss of both genes fails to produce an additive effect. (G) A proposed model of FBXL-5 action in the Caenorhabditis elegans intestine.
To further define the relationship between FBXL-5 and RICT-1 signaling, we performed mRNA-Seq on the rict-1(mg360) mutant and compared the differentially expressed genes (DEGs) between the rict-1 and fbxl-5 mutants (Supplementary Table S4). There was a striking overlap between these datasets, with 51% of the fbxl-5 DEGs also misexpressed in the rict-1 mutant (Figure 7D). The vast majority of these overlapping DEGs showed similar directionalities and degrees of mis-expression between the two mutants; however, a small cluster of genes was upregulated in the rict-1(mg360) mutant but downregulated in fbxl-5(rhd43) animals (Figure 7E). This could be a result of FBXL-5 having different roles within the intestine or having unique functions in different tissues. With many genes similarly altered in rict-1 and fbxl-5 mutants, we performed mRNA sequencing on the rict-1(mg360); fbxl-5(rhd43) double mutant and calculated the differential expression values for the 426 co-regulated genes. Simultaneous loss of both rict-1 and fbxl-5 did not further alter gene expression levels compared to the single mutants (Figure 7F), suggesting that these genes are regulated by FBXL-5 and RICT-1 in a linear pathway. Together, our results indicate that FBXL-5 regulates gene expression via mTORC2-dependent and independent mechanisms (Figure 7G).
Discussion
Here, we demonstrate that the previously uncharacterized FBXL-5 protein functions in the mTORC2 pathway to govern lipid homeostasis. While much of FBXL-5’s function aligns with the hypothesis that it is a positive regulator of mTORC2 signaling, we have also shown that FBXL-5 acts in a mTORC2-independent pathway to confer modest effects on vitellogenesis and organismal lifespan (Figure 7G). It is likely that FBXL-5 functions in concert with SKR-3 and CUL-6 to mediate poly-ubiquitination-dependent proteasomal degradation of at least one target protein; however, it is not surprising that loss of fbxl-5 yields pleiotropic effects, as it is also equally possible that FBXL-5 acts on multiple targets and/or pathways. Alternatively, the effects of loss of fbxl-5 on mTORC2 signaling may have an indirect effect on other pathways, including the mTORC1 pathway. Notably, the mTORC1 complex shares many of the same proteins as mTORC2, which may be redistributed to mTORC1 upon loss of FBXL-5 (Nukazuka et al., 2011). Nonetheless, our results demonstrate a role for a FBXL-5-containing SCF complex in the regulation vitellogenesis, which has not been previously described.
Mutation of the lin-4 gene results in heterochrony due to cell lineage defects primarily in the larval hypodermis (Chalfie et al., 1981; Ambros and Horvitz, 1984). While there is significant evidence that lin-4 functions in a cell-autonomous fashion in several tissues (Zhang and Fire, 2010), loss of lin-4 or lin-29 impairs intestinal vitellogenin gene expression in a non-cell-autonomous manner (Dowen et al., 2016). This response is partially governed by intestinal FBXL-5. Consistent with these observations, migration of hermaphrodite-specific neurons is controlled non-cell-autonomously by hypodermal factors, which promote the expression of receptors or cell-adhesion molecules that maintain neuron-hypodermal contacts (Pedersen et al., 2013; Salzberg et al., 2013). High levels of FBXL-5, as modeled in the fbxl-5 over-expression animals, could impair communication between hypodermal and intestinal cells by restricting an inter-tissue signal pathway that in turn couples to intestinal mTORC2 signaling. The lin-4 and lin-29 mutants both display ectopic expression of fbxl-5 in adults, which could be inhibiting an adult-specific signal from the hypodermis that is required for initiating vitellogenesis; however, additional studies are needed to resolve what types of developmental signals are released from the hypodermis to coordinate intestinal metabolism.
The C. elegans genome encodes ∼520 F-box-containing proteins that serve as adaptors for SCF-mediated, ubiquitin-dependent proteolysis. In contrast, the human genome encodes ∼68 F-box proteins (Jin et al., 2004), suggesting that F-box proteins may serve redundant or additional roles in C. elegans, which may include responding to foreign or pathogenic proteins that nematodes commonly encounter in the wild (Thomas, 2006), performing nematode-specific developmental or reproductive functions (Nayak et al., 2002; Fielenbach et al., 2007; Gao et al., 2008), or providing tissue-specific responses to a ubiquitous signaling molecule. Importantly, only a handful of F-box proteins have clearly defined roles in C. elegans, although mutation of either the ubiquitin-activating enzyme (uba-1, E1), the ubiquitin-conjugating enzymes (ubc genes, E2s), the Skp1-related (skr) genes, or the Cullin (cul) genes results in a wide array of developmental phenotypes (Kipreos, 2005). Intriguingly, CUL-6, along with the redundantly acting SKR-3, SKR-4, and SKR-5 proteins, provide a modest amount of protection against Nematocida parisii (Bakowski et al., 2014), a microsporidia intracellular pathogen, and are strongly required for thermotolerance following heat shock (Panek et al., 2020). SKR-5 is functionally the most important for promoting thermotolerance (Panek et al., 2020). We find a role for cul-6 in vitellogenin regulation but only when skr-3 is also deleted, suggesting that other Cullin proteins may act redundantly with CUL-6. Regardless, SKR-3 is functionally the most important of the SKR-3,4,5 family for vitellogenesis. Thus, the intestinal SKR preference during heat shock tolerance differs from that during vitellogenesis, which in turn likely dictates the F-box adapter. Indeed, FBXA-75 and FBXA-158 promote thermotolerance in response to heat stress while FBXL-5 governs ubiquitin-mediated metabolic regulation (Panek et al., 2020).
In mammals, the mTOR signaling pathways are regulated by ubiquitination in several different contexts (Jiang et al., 2019). To our knowledge, FBXL-5 represents the first F-box protein to function in mTOR signaling outside of mammals, although it remains unclear whether FBXL-5 directly acts on a component of the mTORC2 complex. Mutation of fbxl-5 largely mirrors the transcriptional response to loss of mTORC2; however, the fbxl-5 mutation is unlikely to also impair mTORC1 activity, as loss of both complexes would be predicated to result in severe developmental defects. It is possible that FBXL-5 mediates switching of factors between the two mTOR complexes. This phenomena has been described in mammals, whereby the TRAF2 E3 ligase mediates K63-linked poly-ubiquitination of GβL/LST8, a shared component of both mTORC1 and mTORC2, which promotes mTOR-Raptor binding and dissociation of Sin1 from mTORC2, resulting in elevated mTORC1 signaling and reduced mTORC2 signaling (Wang et al., 2017). Alternatively, loss of FBXL-5 may directly impact the stability or activity of a core mTORC2 component; however, this hypothesis argues that ubiquitination would activate mTORC2 or that FBXL-5 inhibits a negative regulator of mTORC2 signaling. Consistent with the latter hypothesis, proteosomal degradation of Deptor, which itself inhibits both mTORC1 and mTORC2, promotes the activity of both complexes in vertebrates (Duan et al., 2011; Gao et al., 2011; Zhao et al., 2011); however, a functional homologue of Deptor has yet to be identified in C. elegans. In the future, it will be crucial to identify the target of FBXL-5 to gain mechanistic insight into how ubiquitination of mTORC2 signaling components may govern intestinal homeostasis.
Loss of fbxl-5 resulted in an increase in the transcription of most ribosomal protein genes. Interestingly, the expression of the cytosolic ribosomal proteins is reduced in the germline-less glp-1(e2141) mutant in a daf-16-dependent manner, suggesting that DAF-16/FOXO may be a transcriptional repressor of ribosomal protein gene expression (Hemphill et al., 2022). Moreover, reduced protein translation from impaired mTOR/DAF-16 signaling is likely to contribute to lifespan extension in multiple contexts (Hansen et al., 2007; Pan et al., 2007). It is therefore possible that the increase in ribosomal protein gene expression may contribute to the shorter lifespan that we observed in the fbxl-5 mutant. Paradoxically, over-expression of fbxl-5 in the intestine dramatically reduces lifespan and the fbxl-5 loss-of-function mutation extends the lifespan of the rict-1 mutant, suggesting that FBXL-5 may function in multiple pathways or in different tissues to regulate aging. Intriguingly, fbxl-5 is upregulated in the intestine of aging adults, which requires the homeodomain transcription factor UNC-62 (Van Nostrand et al., 2013). It is possible that a post-reproductive function of FBXL-5 may be to downregulate vitellogenesis by simultaneously restricting vitellogenin transcription and translation by reducing the expression of the vit genes and ribosomal protein genes, respectively.
The FBXL-5 protein acts in a SCF complex to govern ubiquitin-mediated proteasome degradation of a component within the mTORC2 pathway. We find that levels of FBXL-5 are controlled by the hypodermal heterochronic pathway. Our study elucidates a new mechanism by which intestinal metabolic regulation is coupled to a developmental timing pathway. Conceivably, dynamic upregulation of fbxl-5 in response to adverse environmental conditions, which could include reduced food availability or pathogen infection, might tune vitellogenin expression levels to match the energetic resources that are available. Alternatively, FBXL-5 might purely act as a developmental regulator that responds to a variety of inter-tissue signals from different tissues; however, a systematic analysis of the signaling pathways that govern vitellogenesis will be needed to determine whether fbxl-5 expression is under the control of additional developmental circuits. Importantly, identification of the target(s) of FBXL-5 could reveal a new upstream regulator of mTORC2 signaling or a novel ubiquitination-dependent mechanism of mTORC2 activation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE248602.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
PB: Data curation, Formal Analysis, Investigation, Writing–review and editing. KK: Data curation, Formal Analysis, Investigation, Writing–review and editing. MN: Data curation, Formal Analysis, Investigation, Writing–review and editing. RD: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Project administration, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Institute of General Medical Sciences grants R35GM137985 to RD and R25GM089569 to KK (UNC PREP in the Biomedical Sciences).
Acknowledgments
The Caenorhabditis Genetics Center provided some of the strains used in this study and is supported by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Yasmine Ackall and Monica Macharios for their assistance in carrying out some of the experiments described in this study. A preprint of this manuscript can be found on the bioRxiv server (Breen et al., 2024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1389077/full#supplementary-material
SUPPLEMENTARY DATA SHEET 1 | A file containing Supplementary Tables S1-S3 and Supplementary Figures S1-S7.
SUPPLEMENTARY TABLE 1 | A file containing processed data, including lists of differentially expressed genes, from mRNA-Seq experiments.
SUPPLEMENTARY TABLE 2 | A file containing all tabular data from the experiments shown in the article.
References
Ambros, V. (1989). A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 57, 49–57. doi:10.1016/0092-8674(89)90171-2
Ambros, V., and Horvitz, H. R. (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416. doi:10.1126/science.6494891
Bakowski, M. A., Desjardins, C. A., Smelkinson, M. G., Dunbar, T. L., Lopez-Moyado, I. F., Rifkin, S. A., et al. (2014). Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 10, e1004200. doi:10.1371/journal.ppat.1004200
Balklava, Z., Rathnakumar, N. D., Vashist, S., Schweinsberg, P. J., and Grant, B. D. (2016). Linking gene expression in the intestine to production of gametes through the phosphate transporter PITR-1 in Caenorhabditis elegans. Genetics 204, 153–162. doi:10.1534/genetics.116.188532
Breen, P. C., Kanakanui, K. G., Newman, M. A., and Dowen, R. H. (2024). The F-box protein FBXL-5 governs vitellogenesis and lipid homeostasis in C. elegans. bioRxiv 2024, 2024.04.18.590113. doi:10.1101/2024.04.18.590113
Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi:10.1093/genetics/77.1.71
Chalfie, M. (1981). Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 24, 59–69. doi:10.1016/0092-8674(81)90501-8
Doitsidou, M., Poole, R. J., Sarin, S., Bigelow, H., and Hobert, O. (2010). C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5, e15435. doi:10.1371/journal.pone.0015435
Dowen, R. H. (2019). CEH-60/PBX and UNC-62/MEIS coordinate a metabolic switch that supports reproduction in C. elegans. Dev. Cell. 49, 235–250. doi:10.1016/j.devcel.2019.03.002
Dowen, R. H., Breen, P. C., Tullius, T., Conery, A. L., and Ruvkun, G. (2016). A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes. Dev. 30, 1515–1528. doi:10.1101/gad.283895.116
Duan, S., Skaar, J. R., Kuchay, S., Toschi, A., Kanarek, N., Ben-Neriah, Y., et al. (2011). mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol. Cell. 44, 317–324. doi:10.1016/j.molcel.2011.09.005
Fielenbach, N., Guardavaccaro, D., Neubert, K., Chan, T., Li, D., Feng, Q., et al. (2007). DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev. Cell. 12, 443–455. doi:10.1016/j.devcel.2007.01.018
Frøkjær-Jensen, C., Wayne Davis, M., Hopkins, C. E., Newman, B. J., Thummel, J. M., Olesen, S.-P., et al. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383. doi:10.1038/ng.248
Gao, D., Inuzuka, H., Tan, M.-K. M., Fukushima, H., Locasale, J. W., Liu, P., et al. (2011). mTOR drives its own activation via SCFβTrCP-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell. 44, 290–303. doi:10.1016/j.molcel.2011.08.030
Gao, M. X., Liao, E. H., Yu, B., Wang, Y., Zhen, M., and Derry, W. B. (2008). The SCF FSN-1 ubiquitin ligase controls germline apoptosis through CEP-1/p53 in C. elegans. Cell. Death Differ. 15, 1054–1062. doi:10.1038/cdd.2008.30
Geens, E., Van De Walle, P., Caroti, F., Jelier, R., Steuwe, C., Schoofs, L., et al. (2023). Yolk-deprived Caenorhabditis elegans secure brood size at the expense of competitive fitness. Life Sci. Alliance 6, e202201675. doi:10.26508/lsa.202201675
Ghanta, K. S., and Mello, C. C. (2020). Melting dsDNA donor molecules greatly improves precision genome editing in Caenorhabditis elegans. Genetics 216, 643–650. doi:10.1534/genetics.120.303564
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi:10.1038/nmeth.1318
Goszczynski, B., Captan, V. V., Danielson, A. M., Lancaster, B. R., and McGhee, J. D. (2016). A 44 bp intestine-specific hermaphrodite-specific enhancer from the C. elegans vit-2 vitellogenin gene is directly regulated by ELT-2, MAB-3, FKH-9 and DAF-16 and indirectly regulated by the germline, by daf-2/insulin signaling and by the TGF-β/Sma/Mab pathway. Dev. Biol. 413, 112–127. doi:10.1016/j.ydbio.2016.02.031
Grant, B., and Hirsh, D. (1999). Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. MBoC 10, 4311–4326. doi:10.1091/mbc.10.12.4311
Hansen, M., Taubert, S., Crawford, D., Libina, N., Lee, S.-J., and Kenyon, C. (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 6, 95–110. doi:10.1111/j.1474-9726.2006.00267.x
Hemphill, C., Pylarinou-Sinclair, E., Itani, O., Scott, B., Crowder, C. M., and Van Gilst, M. R. (2022). Daf-16 mediated repression of cytosolic ribosomal protein genes facilitates a hypoxia sensitive to hypoxia resistant transformation in long-lived germline mutants. PLoS Genet. 18, e1009672. doi:10.1371/journal.pgen.1009672
Hobert, O. (2002). PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728–730. doi:10.2144/02324bm01
Holdorf, A. D., Higgins, D. P., Hart, A. C., Boag, P. R., Pazour, G. J., Walhout, A. J. M., et al. (2020). WormCat: an online tool for annotation and visualization of Caenorhabditis elegans genome-scale data. Genetics 214, 279–294. doi:10.1534/genetics.119.302919
Jiang, Y., Su, S., Zhang, Y., Qian, J., and Liu, P. (2019). Control of mTOR signaling by ubiquitin. Oncogene 38, 3989–4001. doi:10.1038/s41388-019-0713-x
Jin, J., Cardozo, T., Lovering, R. C., Elledge, S. J., Pagano, M., and Harper, J. W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes. Dev. 18, 2573–2580. doi:10.1101/gad.1255304
Kaletsky, R., Yao, V., Williams, A., Runnels, A. M., Tadych, A., Zhou, S., et al. (2018). Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14, e1007559. doi:10.1371/journal.pgen.1007559
Kimble, J., and Sharrock, W. J. (1983). Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev. Biol. 96, 189–196. doi:10.1016/0012-1606(83)90322-6
Kipreos, E. T. (2005). Ubiquitin-mediated pathways in C. elegans. WormBook, 1–24. doi:10.1895/wormbook.1.36.1
Kulkarni, M., and Smith, H. E. (2008). E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 4, e1000131. doi:10.1371/journal.pgen.1000131
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75, 843–854. doi:10.1016/0092-8674(93)90529-y
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Minevich, G., Park, D. S., Blankenberg, D., Poole, R. J., and Hobert, O. (2012). CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269. doi:10.1534/genetics.112.144204
Murphy, C. T., McCarroll, S. A., Bargmann, C. I., Fraser, A., Kamath, R. S., Ahringer, J., et al. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283. doi:10.1038/nature01789
Nayak, S., Santiago, F. E., Jin, H., Lin, D., Schedl, T., and Kipreos, E. T. (2002). The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12, 277–287. doi:10.1016/s0960-9822(02)00682-6
Nukazuka, A., Tamaki, S., Matsumoto, K., Oda, Y., Fujisawa, H., and Takagi, S. (2011). A shift of the TOR adaptor from Rictor towards Raptor by semaphorin in C. elegans. Nat. Commun. 2, 484. doi:10.1038/ncomms1495
Oh, W. J., and Jacinto, E. (2011). mTOR complex 2 signaling and functions. Cell. Cycle 10, 2305–2316. doi:10.4161/cc.10.14.16586
Pan, K. Z., Palter, J. E., Rogers, A. N., Olsen, A., Chen, D., Lithgow, G. J., et al. (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 6, 111–119. doi:10.1111/j.1474-9726.2006.00266.x
Panek, J., Gang, S. S., Reddy, K. C., Luallen, R. J., Fulzele, A., Bennett, E. J., et al. (2020). A cullin-RING ubiquitin ligase promotes thermotolerance as part of the intracellular pathogen response in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 117, 7950–7960. doi:10.1073/pnas.1918417117
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–D427. doi:10.1093/nar/gkac993
Pedersen, M. E., Snieckute, G., Kagias, K., Nehammer, C., Multhaupt, H. A. B., Couchman, J. R., et al. (2013). An epidermal MicroRNA regulates neuronal migration through control of the cellular glycosylation state. Science 341, 1404–1408. doi:10.1126/science.1242528
Perez, M. F., Francesconi, M., Hidalgo-Carcedo, C., and Lehner, B. (2017). Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature 552, 106–109. doi:10.1038/nature25012
Perez, M. F., and Lehner, B. (2019). Vitellogenins - yolk gene function and regulation in Caenorhabditis elegans. Front. Physiol. 10, 1067. doi:10.3389/fphys.2019.01067
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi:10.1038/35002607
Salzberg, Y., Díaz-Balzac, C. A., Ramirez-Suarez, N. J., Attreed, M., Tecle, E., Desbois, M., et al. (2013). Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell. 155, 308–320. doi:10.1016/j.cell.2013.08.058
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Seah, N. E., de Magalhaes Filho, C. D., Petrashen, A. P., Henderson, H. R., Laguer, J., Gonzalez, J., et al. (2016). Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy 12, 261–272. doi:10.1080/15548627.2015.1127464
Skaar, J. R., Pagan, J. K., and Pagano, M. (2013). Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell. Biol. 14, 369, 381. doi:10.1038/nrm3582
Thomas, J. H. (2006). Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 16, 1017–1030. doi:10.1101/gr.5089806
Torzone, S. K., Park, A. Y., Breen, P. C., Cohen, N. R., and Dowen, R. H. (2023). Opposing action of the FLR-2 glycoprotein hormone and DRL-1/FLR-4 MAP kinases balance p38-mediated growth and lipid homeostasis in C. elegans. PLoS Biol. 21, e3002320. doi:10.1371/journal.pbio.3002320
Van Nostrand, E. L., Sánchez-Blanco, A., Wu, B., Nguyen, A., and Kim, S. K. (2013). Roles of the developmental regulator unc-62/homothorax in limiting longevity in Caenorhabditis elegans. PLoS Genet. 9, e1003325. doi:10.1371/journal.pgen.1003325
Wang, B., Jie, Z., Joo, D., Ordureau, A., Liu, P., Gan, W., et al. (2017). TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature 545, 365–369. doi:10.1038/nature22344
Wang, W., Sherry, T., Cheng, X., Fan, Q., Cornell, R., Liu, J., et al. (2023). An intestinal sphingolipid confers intergenerational neuroprotection. Nat. Cell. Biol. 25, 1196–1207. doi:10.1038/s41556-023-01195-9
Zhang, H., and Fire, A. Z. (2010). Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Dev. Biol. 344, 603–610. doi:10.1016/j.ydbio.2010.05.018
Keywords: development, vitellogenesis, f-box, mTORC2, rict-1, lipid, SCF
Citation: Breen PC, Kanakanui KG, Newman MA and Dowen RH (2024) The F-box protein FBXL-5 governs vitellogenesis and lipid homeostasis in C. elegans. Front. Cell Dev. Biol. 12:1389077. doi: 10.3389/fcell.2024.1389077
Received: 20 February 2024; Accepted: 22 May 2024;
Published: 14 June 2024.
Edited by:
Maria Teresa Bengoechea Alonso, Hamad Bin Khalifa University, QatarReviewed by:
Nuno Miguel Luis, UMR7288 Institut de Biologie du Développement de Marseille (IBDM), FranceSpencer Gang, Colorado College, United States
Copyright © 2024 Breen, Kanakanui, Newman and Dowen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert H. Dowen, ZG93ZW5AZW1haWwudW5jLmVkdQ==
 Peter C. Breen1
Peter C. Breen1 Robert H. Dowen
Robert H. Dowen