- 1Department of Biological Repositories, Human Genetic Resources Preservation Center of Hubei Province, Hubei Key Laboratory of Urological Diseases, Zhongnan Hospital of Wuhan University, Wuhan, China
- 2Taihe Skills Training Center, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, China
- 3Department of Urology, Taihe Hospital, Hubei University of Medicine, Shiyan, Hubei, China
- 4Department of Urology, The First Affiliated Hospital of Yangtze University, Jingzhou, Hubei, China
- 5Department of Urology, Laboratory of Precision Medicine, Zhongnan Hospital of Wuhan University, Wuhan, China
Cell-free DNA (cfDNA), a burgeoning class of molecular biomarkers, has been extensively studied across a variety of biomedical fields. As a key component of liquid biopsy, cfDNA testing is gaining prominence in disease detection and management due to the convenience of sample collection and the abundant wealth of genetic information it provides. However, the broader clinical application of cfDNA is currently impeded by a lack of standardization in the preanalytical procedures for cfDNA analysis. A number of fundamental challenges, including the selection of appropriate preanalytical procedures, prevention of short cfDNA fragment loss, and the validation of various cfDNA measurement methods, remain unaddressed. These existing hurdles lead to difficulties in comparing results and ensuring repeatability, thereby undermining the reliability of cfDNA analysis in clinical settings. This review discusses the crucial preanalytical factors that influence cfDNA analysis outcomes, including sample collection, transportation, temporary storage, processing, extraction, quality control, and long-term storage. The review provides clarification on achievable consensus and offers an analysis of the current issues with the goal of standardizing preanalytical procedures for cfDNA analysis.
1 Introduction
Cell-free DNA (cfDNA) was first discovered in the peripheral blood of healthy individuals in 1948 (Mandel and Metais, 1948). Subsequent research demonstrated elevated levels of cfDNA in patients with cancer (Song et al., 2022; Wu et al., 2022) and inflammation (Schneck et al., 2017), indicating that cfDNA analysis could be a valuable tool for health monitoring (Hayashi et al., 2019). Originating from apoptotic or necrotic events and active release mechanisms in the presence of intracellular circulating nucleases (Qi et al., 2023), cfDNA, with its modal size around 166 base pairs (bp) in plasma, has been linked to nucleosomal structures (Lo et al., 2021). Due to the stability of cfDNA molecules in body fluids (Polini et al., 2019), their wealth of genetic and epigenetic information (Wu et al., 2019; Xiao et al., 2022), and the noninvasive or minimally invasive nature of body fluid collection for cfDNA analysis (Ellervik and Vaught, 2015), cfDNA is considered an ideal biomarker for disease prevention, diagnosis, treatment, and prognosis (Szilagyi et al., 2020). Fetal noninvasive prenatal testing (NIPT) based on cfDNA levels was the first successful application of cfDNA in health monitoring (Schmid et al., 2018). Recent studies have explored the role of cfDNA in various areas, including NIPT (Zhu et al., 2021), cancer (Nabet et al., 2020), diabetes (Humardani et al., 2023), cardiovascular diseases (Polina et al., 2020), organ transplantation (Wolf-Doty et al., 2021), autoimmune diseases (Mondelo-Macia et al., 2021), and sepsis (Lenz et al., 2022). However, the progress of most related studies remains in the preliminary stage, likely due to the challenges presented by preanalytical variables.
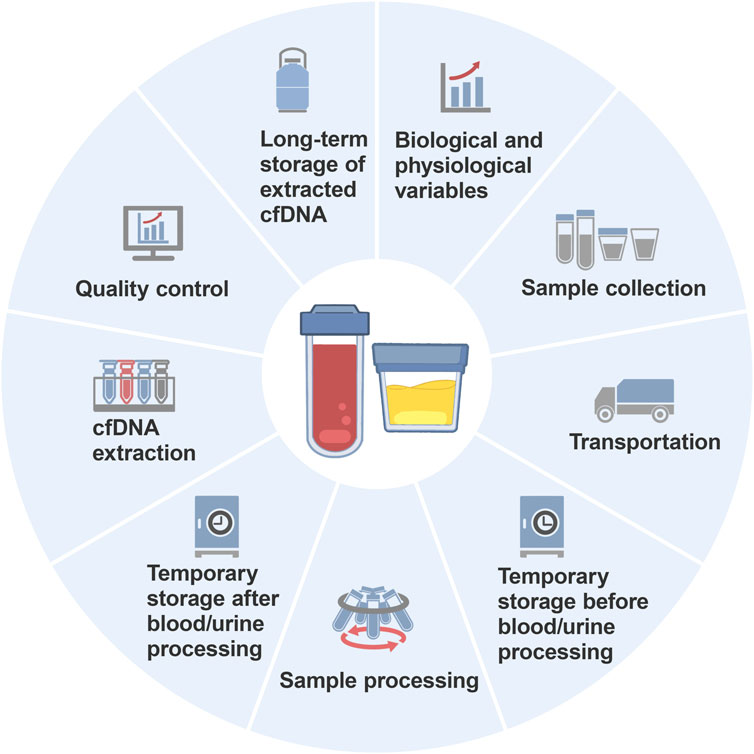
The journey from sample collection to cfDNA analysis is intricate and involves several steps, such as preparation, collection, transportation, temporary storage, processing, extraction, quality control, and long-term storage (Figure 1). Each step involves numerous conditions or details, and the variables interact with each other. Moreover, many studies inadequately describe the preanalytical variables for cfDNA analysis in their Materials and Methods sections (Campbell et al., 2015; Wolf-Doty et al., 2021; Shen et al., 2022), leading to questionable credibility of analytical results and inefficiency in method verification. Diao et al. surveyed the quality assurance (the questionnaire included preanalysis, postanalysis and performance validation for mNGS) of metagenomic next-generation sequencing (mNGS) used for detecting microbial cfDNA in blood samples across 80 laboratories in China and found significant variation in the mNGS workflow among the laboratories (Diao et al., 2022). Specifically, the sequencing platforms used in the mNGS laboratories included 49 Illumina laboratories, 16 Beijing Genomics Institute laboratories, 13 Ion Torrent laboratories and 2 Nanopore sequencing laboratories, and the interpretation standards for the mNGS results were inconsistent among the laboratories. Consequently, establishing widely applicable standards and consensuses presents a formidable challenge.
Blood and urine samples are valuable resources in biomedical research. Over the past few years, progress has been made in some effective methods (e.g., EDTA tubes, specialized collection tubes and specialized kits) (Salvianti et al., 2020; Ungerer et al., 2020; Deger et al., 2021; Wever et al., 2022) and in establishing preliminary guidelines for preanalytical variables (Meddeb et al., 2019). However, with the advancement of technology and new insights into unresolved issues, the existing consensus on preanalytical variables for blood-derived cfDNA analysis needs to be updated. cfDNA in urine has shown great potential in noninvasive diagnosis. Urine is an ideal biomaterial for the study of urological diseases due to its direct contact with the urinary system and convenient collection of sufficient volume (Ruppert et al., 2023). Research has shown significantly higher levels of urine cfDNA (ucfDNA) in patients with urinary tumors compared to healthy individuals (Nikanjam et al., 2022). Nevertheless, the clinical application of ucfDNA is relatively rare, possibly due to its sensitivity to environmental conditions (e.g., temperature and pH level of preservation solution) (Kim et al., 2023; Ruppert et al., 2023), which makes it easier to degrade rapidly, resulting in inadequate concentrations for downstream analysis (Nel et al., 2023) compared to blood-derived cfDNA. Therefore, the weaker stability makes ucfDNA analysis more susceptible to complex preanalytical variables, highlighting the importance of establishing a consensus on ucfDNA experimental procedures.
Clear documentation of the key details and preanalytical variables in experimental procedures is important, as it forms the basis for discussion and analytical results. Such detailed information is of paramount importance for consensus building. In this review, we delve into the preanalytical variables affecting cfDNA analysis. We clarify the achievable consensus in preanalytical variables and analyze existing challenges with the aim of standardizing preanalytical procedures for cfDNA analysis.
2 Preanalytical variables affecting cfDNA analysis
2.1 Biological and physiological variables prior to sample collection
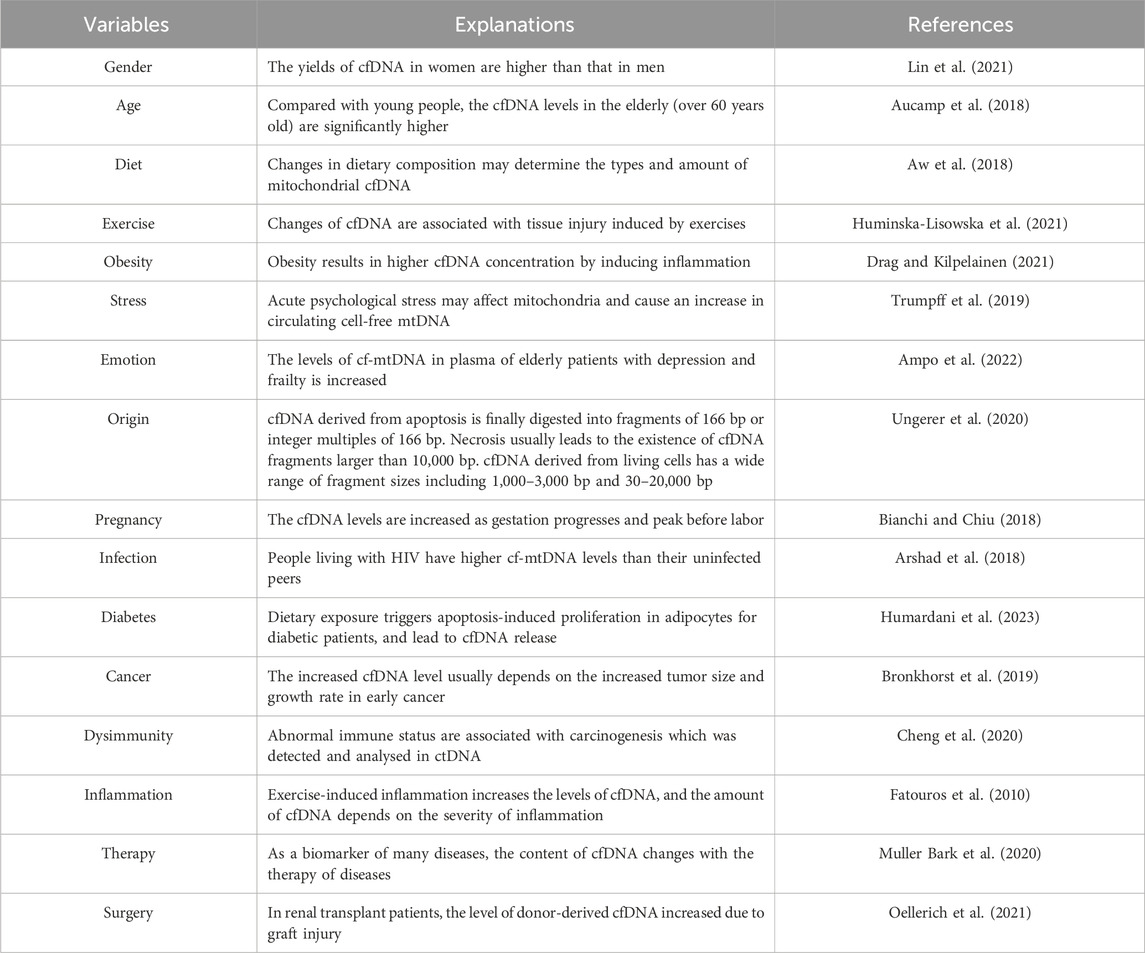
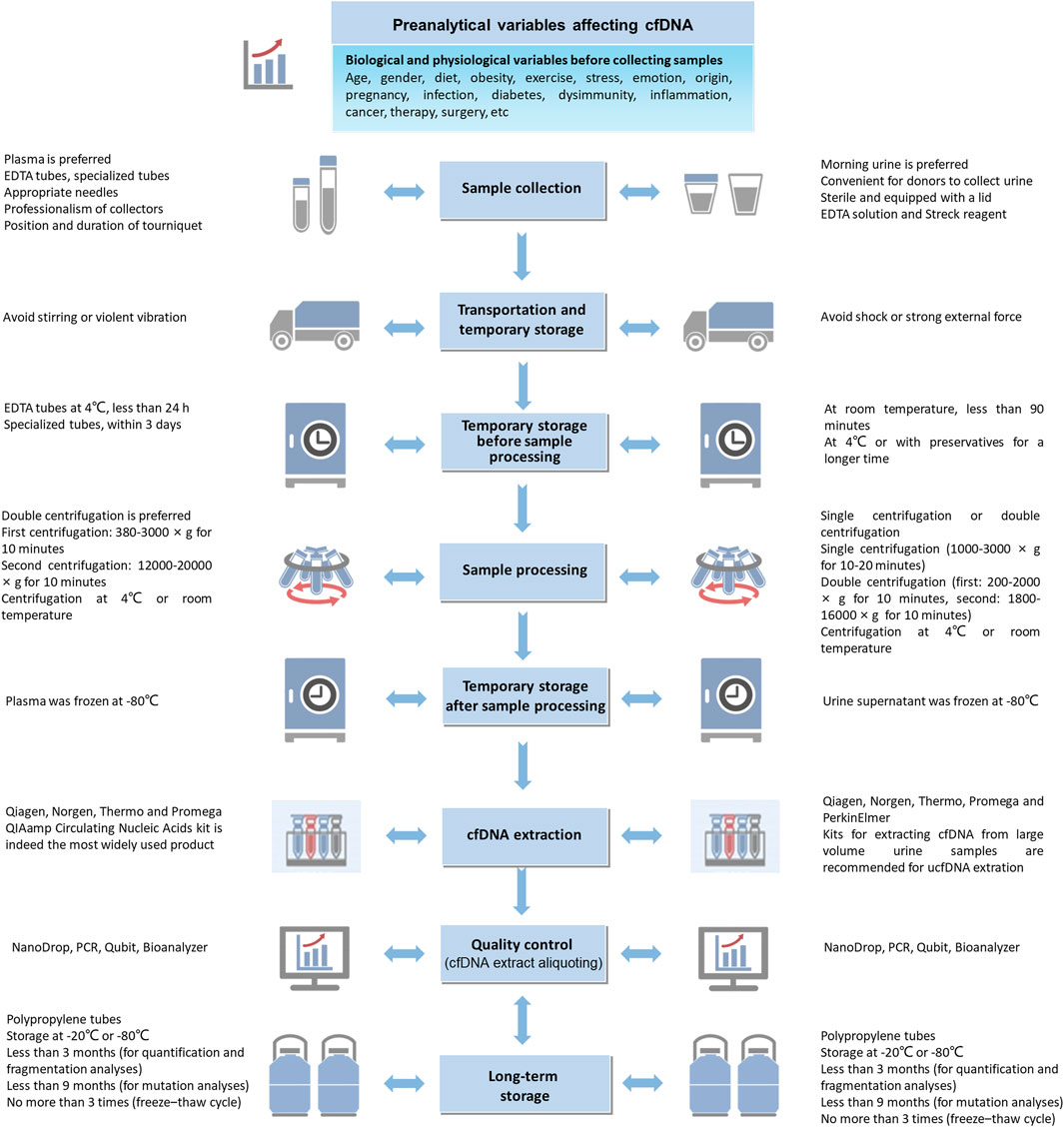
The characteristics of cfDNA in biospecimens are influenced by many biological and physiological variables before collection. These variables are often interrelated and subject to significant intra- and inter-individual differences (Ungerer et al., 2020). However, few of these variables have been individually studied, resulting in a limited understanding of their specific impacts on cfDNA characteristics. Potential biological and physiological variables (Table 1) that may affect cfDNA characteristics mainly include demographic differences (e.g., age and gender) (Aucamp et al., 2018; Lin et al., 2021), living habits (e.g., diet and exercise) (Aw et al., 2018; Huminska-Lisowska et al., 2021), psychophysical state (e.g., obesity, stress and emotion) (Trumpff et al., 2019; Drag and Kilpelainen, 2021; Ampo et al., 2022), origin (Stejskal et al., 2023), physiological process (e.g., menstruation and pregnancy) (Bianchi and Chiu, 2018; Yuwono et al., 2022), infection (Arshad et al., 2018), pathological diseases (e.g., diabetes, cancer, dysimmunity, and inflammation) (Fatouros et al., 2010; Bronkhorst et al., 2019; Cheng et al., 2020; Humardani et al., 2023), therapy (Muller Bark et al., 2020) and surgery (Oellerich et al., 2021). To shed light on how these variables influence cfDNA characteristics, representative examples, such as cfDNA origin mechanisms and cfDNA in cancer, are discussed below.
cfDNA originates from various sources, and its characteristics vary greatly, often identifiable based on DNA fragment length. Currently, several major mechanisms of cfDNA origin are recognized, including passive release from apoptotic (Handayani et al., 2023) and necrotic cells (Jahr et al., 2001) and active release from living cells (Thakur et al., 2014). During apoptosis, nucleosomes, composed of histone octamers and double-stranded DNA wrapped around the protein complex, are released into the blood and sheared by various nucleases to form cfDNA (Duplessis et al., 2018; Fedyuk et al., 2023). Consequently, cfDNA fragments resulting from apoptosis are typically 160–180 bp or 360 bp in length, consistent with the length of the nucleosome (Jahr et al., 2001; Markus et al., 2022). In contrast, cfDNA fragments from necrotic cells are usually larger than 10,000 bp (Fujihara et al., 2021). Furthermore, living cells can actively secrete cfDNA in various forms of extracellular vesicles, containing cfDNA fragments ranging from 150 to 6,000 bp (Thakur et al., 2014; Fernando et al., 2017) and even up to two million bp (Vagner et al., 2018).
Despite different origin mechanisms, the cfDNA fragment ranges in blood and urine are largely regular due to metabolic equilibrium. Plasma cfDNA is predominantly split into 166 bp fragments, as confirmed by precise sequencing technologies (Jiang et al., 2015; Hudecova et al., 2022). ucfDNA, filtered through the renal barrier or directly released into urine following apoptosis and necrosis of urogenital cells (Cimmino et al., 2021), displays a wider range of lengths (Jain et al., 2019). Large molecular fragments, usually more than 1,000 bp, mainly originate from the necrosis of exfoliated uroepithelial cells or leukocytes (Streleckiene et al., 2018). Conversely, small molecular fragments, usually 10–150 bp and 150–200 bp (Melkonyan et al., 2008), mainly originate from apoptotic cells in the bloodstream.
Circulating tumor DNA (ctDNA), a subtype of cfDNA, is released by necrotic or apoptotic tumor cells and carries a wealth of genetic information related to tumorigenesis and progression (Weng et al., 2020). Increased ctDNA levels typically correlate with increased tumor size and growth rate in early-stage cancer (Fiala and Diamandis, 2018). Generally, cancer patients exhibit higher plasma ctDNA levels than healthy individuals (Xu et al., 2021). Apart from presenting significant intraindividual and interindividual variation, ctDNA levels in malignant tumors are significantly greater than those in nonmalignant tumors (Thierry et al., 2016). In summary, ctDNA levels vary greatly across different cancer stages and can be efficiently used to detect alterations in cancer-related genes (Bettegowda et al., 2014), which is highly important for the early detection of cancers (Song et al., 2022).
2.2 Sample collection procedure
2.2.1 Blood collection
As vital biological materials, blood samples are most frequently collected for cfDNA analysis due to the body’s reliance on blood circulation for metabolism. To achieve more reliable results, the choice between serum or plasma as the cfDNA analysis matrix is crucial (Kumar et al., 2018). Research suggests a preference for plasma in cfDNA analysis, as it helps circumvent the effects of genomic DNA (gDNA) released by leukocyte lysis on the concentration and purity of cfDNA (Martignano, 2019; Pittella-Silva et al., 2020). Although several studies reported higher cfDNA concentrations in serum than in plasma due to DNA degradation and contamination of gDNA from white blood cells (Wong et al., 2016; Li et al., 2017; Huang et al., 2020), cfDNA in plasma has proven to be more stable over time (Board et al., 2008). Moreover, a study aimed at evaluating the positive rate of epidermal growth factor receptor (EGFR) mutations in cfDNA revealed greater sensitivity when using plasma rather than serum (Vallee et al., 2013), underscoring the reliability of plasma-derived cfDNA.
Collection tubes with superior performance are preferred for blood collection. Anticoagulants, key components of blood collection tubes, can impact the quality and integrity of cfDNA (Luo et al., 2022). Widely used anticoagulants such as EDTA, citrate, and heparin exhibit different functional characteristics (Akat et al., 2019). Previously, EDTA tubes were commonly considered the standard for cfDNA analysis because EDTA inhibits DNase (Barra et al., 2015) and demonstrates better storage effects than heparin or citrate for delayed blood processing (Lam et al., 2004). Evidence suggests that plasma samples collected in EDTA tubes and processed within 6 h are most suitable for ctDNA analysis (Kang et al., 2016). However, when blood processing is delayed due to long-distance transportation or other unavoidable circumstances, these collection tubes may not preserve samples efficiently. This has led to the development of specialized blood collection tubes designed to preserve samples for extended periods (Sorber et al., 2020).
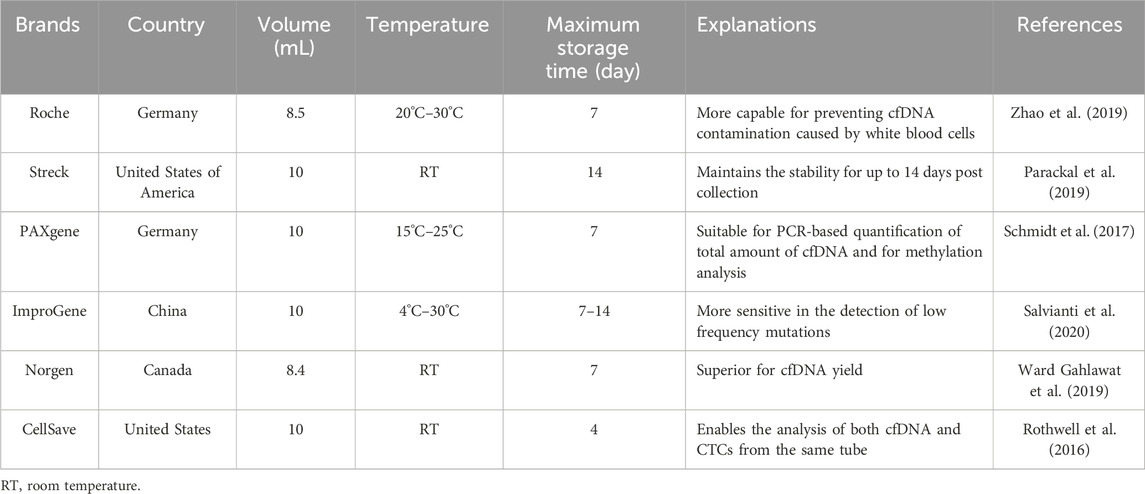
Recently, various specialized collection tubes with different properties have been widely utilized for blood collection for cfDNA analysis (Alidousty et al., 2017; Ungerer et al., 2020). While these specialized collection tubes (Table 2) vary slightly in specifications (e.g., volume and shape), they extend the storage time of blood samples without the need for controlled environmental conditions (Rothwell et al., 2016; Schmidt et al., 2017; Parackal et al., 2019; Ward Gahlawat et al., 2019; Zhao et al., 2019; Salvianti et al., 2020). To ascertain the tubes’ ability to maintain cfDNA levels during transportation or temporary storage, studies have compared them with each other or with EDTA tubes. Overall, the specialized tubes outperform EDTA tubes in preventing gDNA contamination and extending storage time (van Ginkel et al., 2017; de Kock et al., 2019). Furthermore, these specialized tubes maintain the quality of cfDNA samples within 3 days equally well, but their storage effects reportedly differ slightly after more than 7 days (Zhao et al., 2019).
Some often overlooked variables during blood sample collection can impact cfDNA analysis. Proper needle selection is necessary, as excessively thin needles can cause hemolysis (Mouser et al., 2017). When collecting blood samples from children, professional collectors can enhance the efficiency of blood collection and minimize discomfort to the participants (Simundic et al., 2018). To prevent hemolysis, the tourniquet should be correctly positioned and not left in place too long during blood drawing (Phelan et al., 2018). The recommended duration for tourniquet application is generally within 1 min (Wall et al., 2014), as prolonged tourniquet use can lead to blood sample concentration and hemolysis (Jacob et al., 2021). Even though these operational details affecting cfDNA quality have not been fully investigated or described in the current literature, they should be considered during blood collection.
2.2.2 Urine collection
Collecting urine samples is a completely noninvasive process typically carried out by the donors themselves. Urine collection is more convenient than blood collection, provided that there is good communication with donors beforehand (Itoh et al., 2013). Different types of urine samples, such as 24-h, morning, and random samples, are collected for various purposes (24-h and random urine for the urinary biochemical parameters, and morning urine for the extraction of tumor markers) (Cook et al., 2000; Shojaei-Far et al., 2017; Zhang et al., 2018). Morning urine is frequently preferred for cfDNA analysis due to its more stable total cfDNA content compared to that of other urine types (Zhou et al., 2021). This is because factors that might alter cfDNA content, such as the donor intentionally drinking excessive water before collection, are hard to control. Therefore, morning urine collection tends to be less affected.
The choice of suitable collection containers is also crucial, as they need to be user-friendly for donors. Ideally, these containers should be sterile (Sigdel et al., 2013; Cheng et al., 2017) and have a lid to prevent leakage. The donor’s gender, age, and physical condition should be considered when selecting an appropriate container. In addition, it is also recommended to immediately divide the collected urine into multiple portions to avoid freeze-thaw cycles that could affect the quality of the ucfDNA (Jordaens et al., 2023).
The question of whether to add protective agents to the collected urine samples is important. Taking cues from the practice of prolonging the storage time of blood samples collected in tubes with protective agents, adding additives to urine may enhance the stability of cfDNA (Murugesan et al., 2019). EDTA is most commonly used as a protective agent added to urine for optimal cfDNA storage outcomes (Lee et al., 2020). In a study comparing the extraction methods of short cfDNA fragments from urine, a 10 mmol/L EDTA solution was added to the collected urine to enhance analytical result accuracy (Oreskovic et al., 2019). Besides EDTA, the Streck reagent has also been used as a urine preservative for cfDNA protection (Murugesan et al., 2019). However, numerous studies have not clearly outlined whether protective agents were added to the collected urine (Table 4). Further research is necessary to enhance the effectiveness of preservatives in preserving ucfDNA. It is possible to find inspiration from the key components present in specialized collection tubes designed for the extended preservation of blood cfDNA. Moreover, the methods of long-term preservation of DNA might be beneficial to develop more effective preservatives for preserving ucfDNA.
2.3 Sample transportation before processing
Due to a lack of necessary equipment or well-trained professionals, samples often cannot be processed immediately after collection and have to be transported. Unpredictable conditions, such as violent shaking during transportation, prolonged transportation, or high temperature during temporary storage, may negatively impact sample quality and cfDNA analysis.
2.3.1 Blood transportation
During transportation, stirring or violent vibration may cause blood sample hemolysis, resulting in the release of cell metabolites that inhibit the Taq enzyme activity and decrease PCR amplification efficiency (Ellervik and Vaught, 2015). As reviewed in El Messaoudi et al. (2013), cfDNA concentration slightly increased in blood samples stirred for 3 h at room temperature (El Messaoudi et al., 2013). Hence, significant or prolonged vibration should be avoided during blood sample transportation, especially at room temperature (the duration of stable cfDNA level is much longer in EDTA tubes when stored at 4°C than at room temperature) (Hidestrand et al., 2012).
In addition to blood collection tubes, the time and temperature of blood transportation postcollection should be controlled. A study comparing three collection tubes (K2EDTA, Roche, and Streck) revealed that Roche and Streck tubes were similarly effective in preventing gDNA release after 7 days of storage at room temperature, while K2EDTA tubes resulted in significant gDNA release (Zhao et al., 2019). Therefore, specialized tubes should be used when processing steps have to be delayed (Zhang et al., 2022). Hidestrand et al. reported that the samples should avoid being exposed to extreme temperatures during transportation by comparing the total cfDNA in plasma of EDTA samples and BCT samples at room temperature and 4°C (Hidestrand et al., 2012). As indicated in Table 3, the recommended transportation temperature is 4°C for EDTA tubes or room temperature for specialized tubes.
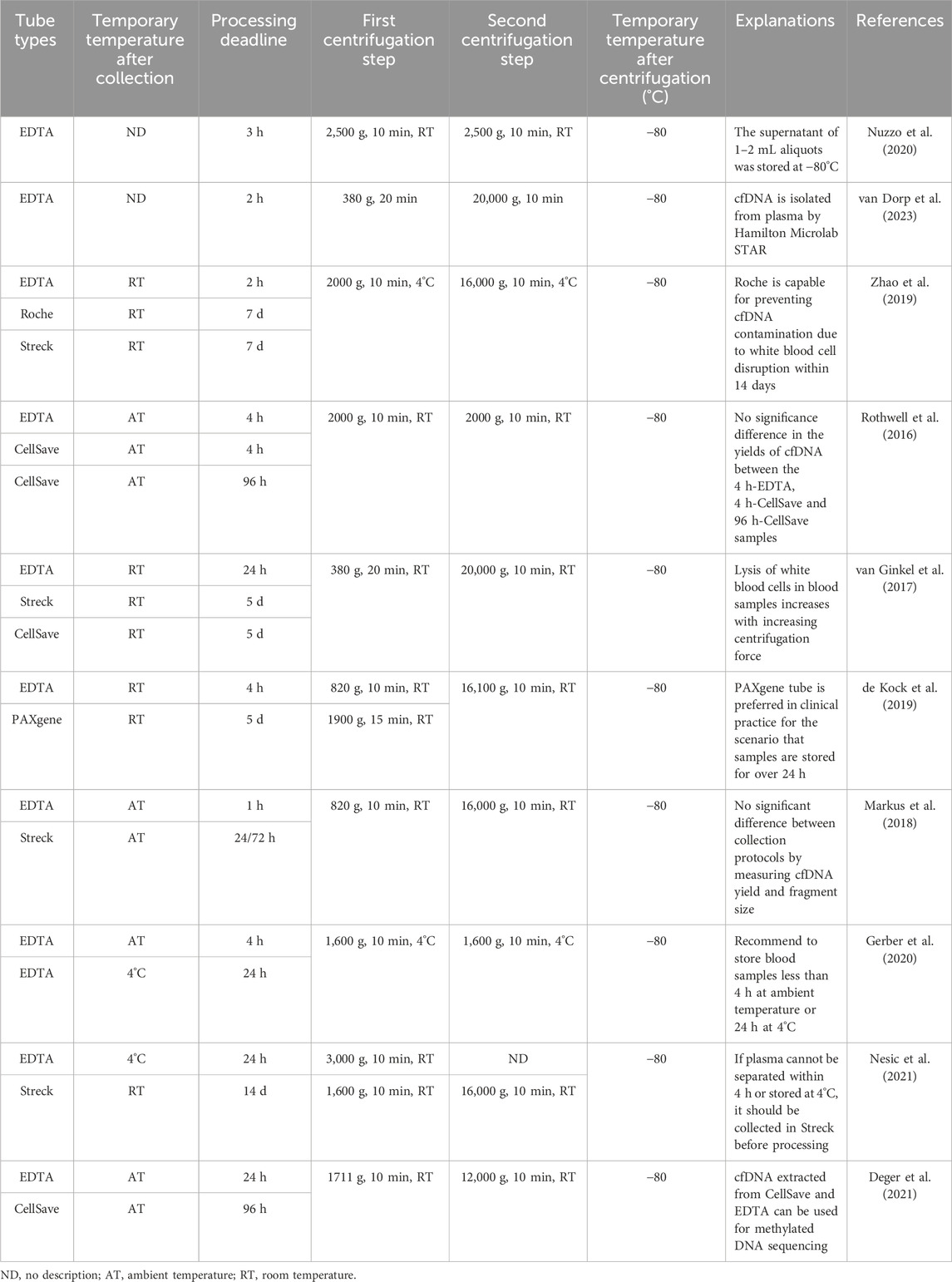
Table 3. Summary of used/recommended temporary storage conditions and different centrifugal procedures after collecting blood for cfDNA analysis.
2.3.2 Urine transportation
The urine sample transportation temperature should be regulated based on different transportation distances. For distances that allow urine samples to be processed within 90 min, samples can be stored at room temperature for transportation (Eisinger et al., 2013; Dolscheid-Pommerich et al., 2016). For longer distances, measures such as using ice packs to maintain the urine samples at approximately 4°C or adding preservatives to urine samples (Ercan et al., 2015) should be implemented to prevent changes in physical and chemical properties of urine or the degradation of cfDNA fragments. The long-distance transportation may lead to the instability of cfDNA level in urine, which is equivalent to prolonging the temporary storage time before sample processing. Therefore, reliable transportation methods (e.g., equipping with enough ice packs or adding suitable preservatives) should be prioritized, especially when transferring rare disease samples. Additionally, it is necessary to prevent the rupture of the urine collection tube due to shock or external force during transportation, which might lead to urine sample leakage.
2.4 Temporary storage before sample processing
2.4.1 Blood temporary storage
Research has shown that the storage temperature and delay time during temporary storage before sample processing significantly impact cfDNA concentration, fragments, and purity (Gerber et al., 2020). The concentration of cfDNA slightly increases with time during temporary storage (Bronkhorst et al., 2015; Bhangu et al., 2017). However, the unified delay time has not yet been determined.
Previous studies have investigated blood samples collected in EDTA tubes and processed within specific times, such as 1 h (Markus et al., 2018), 2 h (Zhao et al., 2019), 3 h (Nuzzo et al., 2020), 4 h (Gerber et al., 2020), 6 h (El Messaoudi et al., 2013) or 24 h (Nesic et al., 2021). However, a consensus on the maximum permissible delay time for processing has not been reached. Table 3 shows that the storage temperature of blood correlates with the delayed processing time, suggesting that blood samples should be processed within the delayed processing time corresponding to the specific storage temperature as soon as possible to ensure the optimal quality of cfDNA. Otherwise, it is recommended that blood samples collected in EDTA tubes should be stored at 4°C, and the delay time should not exceed 24 h (Van Paemel et al., 2021). The blood samples collected by specialized collection tubes are temporarily stored for a longer time than EDTA tubes, and the results are summarized in Tables 2, 3.
2.4.2 Urine temporary storage
The delay time after urine collection is also different in previous studies, including 0 h (Janovicova et al., 2023), 1 h at 4°C (Cheng et al., 2017), 3 h (no description of temperature) (Nuzzo et al., 2020) and 24–72 h (no description of temperature) (Wever et al., 2022). The temporary storage conditions of urine samples after collection were usually neglected, so detailed descriptions of the variables were not provided in most related studies (Table 4). Due to the lack of comparative studies on the temporary storage conditions of urine samples, it is difficult to establish standard operational consensuses. The concentration of ucfDNA is greatly dependent on the addition of urine preservatives (Lee et al., 2020) during temporary storage. Table 4 shows that urine samples with EDTA can be temporarily stored for a longer period of time. Adding preservatives after urine collection is extremely important for sample stability (Pages et al., 2022), which makes the temporary storage conditions more variable and flexible.
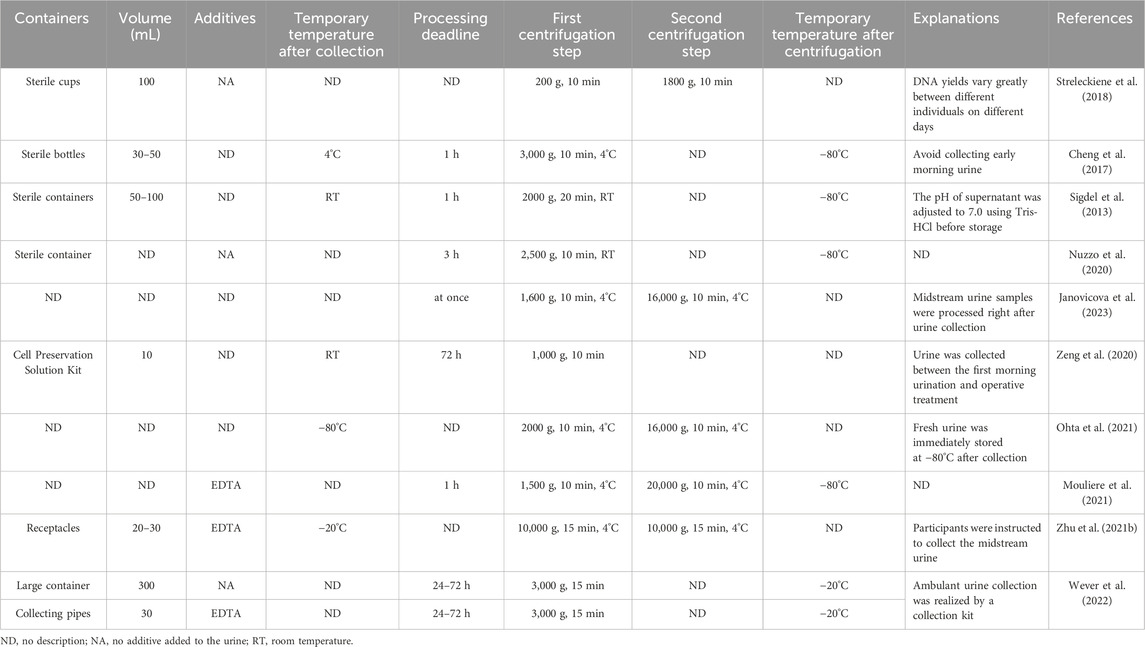
Table 4. Summary of used/recommended temporary storage conditions and centrifugal procedures for urine cfDNA analysis.
2.5 Sample processing procedure
Ensuring that plasma and urine supernatant used for extracting cfDNA are free from cellular components, various specific protocols for processing blood and urine samples have been developed (Tables 3, 4). The main factors that may result in DNA contamination during sample processing include centrifugation speeds, centrifugal temperature, single or double centrifugation steps, and the duration of the centrifugation steps (Ungerer et al., 2020; Shin et al., 2022). These preanalytical variables can affect the effectiveness of blood and urine processing to varying degrees. However, no consensus has been reached regarding these preanalytical variables in the current sample processing protocols. Therefore, there is an urgent need to develop a unified and effective centrifugation protocol for processing blood and urine samples.
2.5.1 Blood processing
Whole blood samples are primarily processed through centrifugation steps to remove cellular components, thus avoiding gDNA contamination of cfDNA (Martignano, 2019). The parameters involved in these steps, such as centrifugal force, temperature, number of centrifugations, and duration of centrifugation, can potentially introduce sample contamination. Therefore, it is crucial to determine the optimal parameters for a centrifugation scheme that can be universally applied to blood processing. Previous studies have indicated that the number of centrifugation steps and the centrifugal force are the key parameters in developing a centrifugation scheme for blood processing (de Kock et al., 2019; Zhao et al., 2019). These parameters should be given more attention than the temperature or duration of centrifugation steps to prevent sample contamination. Currently, a well-established approach for obtaining cell-free plasma fractions during blood processing involves an initial centrifugation step with lower centrifugal force followed by a subsequent second centrifugation step with higher centrifugal force. Moreover, the yield of plasma cfDNA did not differ after the third centrifugation step at 16,000 g compared to the second centrifugation step. Therefore, double centrifugation is widely recognized and used in current blood sample processing protocols to achieve satisfactory cfDNA analysis results (Volckmar et al., 2018; Sorber et al., 2019).
Centrifugal parameters for blood samples vary significantly and are summarized in Table 3. In the first centrifugation step, a slow centrifugal force, primarily ranging from 380–3,000×g for 10 min, is used to remove a large number of cell components (Nesic et al., 2021; van Dorp et al., 2023). In the second centrifugation step, a faster centrifugal force, mainly ranging from 12,000–20,000×g for 10 min, is usually performed to remove cellular residues and debris (Deger et al., 2021; van Dorp et al., 2023). These centrifugation steps are generally carried out at 4°C (Zhao et al., 2019; Gerber et al., 2020) or room temperature (Markus et al., 2018; de Kock et al., 2019).
2.5.2 Urine processing
ucfDNA degrades more easily than blood-derived cfDNA due to urinary nucleases and contaminants (Yao et al., 2016); therefore, collected urine samples should be processed as soon as possible. Similar to blood samples, collected urine typically undergoes single or double centrifugation to remove cellular components or cell debris (Casadio and Salvi, 2019). However, the range of centrifugal force for urine samples is much larger than that for blood samples, as summarized in Table 4. Single centrifugation usually takes 10–20 min at speeds ranging from 1,000–3,000×g (Cheng et al., 2017; Zeng et al., 2020). The double centrifugation procedure consists of a first centrifugation step at 200–2000×g for 10 min, followed by a faster second centrifugation step at 1800–16,000×g for 10 min (Streleckiene et al., 2018; Mouliere et al., 2021; Ohta et al., 2021). These centrifugation steps are also carried out at 4°C (Cheng et al., 2017; Zhu et al., 2021) or room temperature (Sigdel et al., 2013; Nuzzo et al., 2020).
In some studies, urine samples were directly frozen at −20°C or −80°C after collection without a centrifugation step (Kim et al., 2022; Janovicova et al., 2023). The frozen samples must be thawed for subsequent processing or analysis, which can result in cell lysis in urine during the freeze-thaw cycle (Luo et al., 2018). In a study by Oreskovic A et al. on the diagnostic accuracy of a tuberculosis cfDNA test using sequence-specific purification of ucfDNA, the collected urine samples underwent several stages, including freezing at −20°C at the collection point, transportation on dry ice, freezing at −80°C, and thawing at 37°C before centrifugation (Oreskovic et al., 2021). However, the study did not describe or discuss whether the urine samples were immediately processed or not, nor the potential effect of the above steps on the urine. All preanalytical variables may affect the quality and final analysis of cfDNA, which should be clearly described in each study.
2.6 Temporary storage between sample processing and extraction
Many studies do not immediately proceed to cfDNA extraction after centrifuging the collected blood or urine samples. This delay is often due to specific experimental purposes or the need for centralized cfDNA extraction. The conditions of temporary storage, such as the duration and temperature between sample processing and extraction, are vital variables that could impact cfDNA quality. Cellular components and cell debris are removed from the samples during centrifugation, suggesting that changes in cfDNA likely result from DNA fragment degradation during temporary storage (Ellervik and Vaught, 2015). However, temporary storage conditions are not yet standardized.
After centrifugation, the majority of the collected blood and urine samples were frozen at −80°C until DNA extraction, as summarized in Tables 3, 4. One study showed that cfDNA concentration increased slightly when the centrifuged plasma samples were stored at room temperature for varying lengths of time, ranging from 0 to 4 h, before extraction (El Messaoudi et al., 2013). Another study revealed that cfDNA fragmented gradually over 3 months when centrifuged plasma was stored at −20°C (Bronkhorst et al., 2015). The plasma used for detecting specific DNA sequences can be stored at −80°C for up to 10 years, while samples for quantitative analysis can only be stored at −80°C for 9 months (Diao et al., 2022).
Specialized kits allow the collected urine to be temporarily stored for a longer period of time before extraction. Zeng et al. reported that the Cell Preservation Solution Kit was used to collect urine samples and allowed the samples to be transferred to the laboratory for processing within 72 h at room temperature (Zeng et al., 2020). In another more detailed report, urine samples were collected using the specialized kits including a large collection container (300 mL) and three 30 mL collection tubes and then transported to the Department of Pathology of Amsterdam UMC (Wever et al., 2022). Importantly, 2 mL of 0.6 M EDTA as a preservative agent in the collection tubes allowed the samples to be processed within 24–72 h. Except for urine samples collected by specialized kits stored at −20°C after a single centrifugation or samples without relevant storage descriptions, the other collected urine samples were frozen at −80°C after single or double centrifugation until DNA extraction (Table 4). Studies specifically related to the temporary storage of collected urine samples are scarce. Nonetheless, the conditions summarized above for plasma samples are also applicable to urine samples that have undergone single or double centrifugation during temporary storage.
2.7 cfDNA extraction procedure
Efficient cfDNA extraction is essential for ensuring the accuracy and reliability of downstream analytical results. However, the extracted cfDNA can often be too fragmented or too low in content, possibly leading to regrettable analytical data or failed application. Thus, finding a way to efficiently and cost-effectively separate cfDNA from samples has been a central issue for researchers.
A wide array of extraction methods, including traditional (liquid-phase-based or solid-phase-based DNA isolation methods) (Janku et al., 2021), improved (methods for separating cfDNA mainly based on chromatographic columns or magnetic beads) (Lin et al., 2021), and novel (methods for separating cfDNA using new technologies or materials) (Liu et al., 2022) technologies, as well as manual (Wang et al., 2021) or automatic (Lee et al., 2018) methods, have been employed to extract cfDNA. These methods vary in terms of recovery efficiency, fragment discrimination, and reproducibility (Ungerer et al., 2020), making it challenging to select the optimal method for cfDNA isolation. Factors such as the efficiency of extracting low-content DNA (Lee et al., 2018), purity (Uwiringiyeyezu et al., 2022), repeatability (Letendre and Goggs, 2017), and cost (Diefenbach et al., 2018) are usually considered when applying extraction protocols. Commercial specialized kits based on current optimized technologies seem to offer clear advantages for cfDNA extraction (Janovicova et al., 2020) and are routinely used in many studies.
Currently, innovative technologies based on magnetic particles (van der Leest et al., 2022) or spin columns (Diefenbach et al., 2018) are the most common methods in specialized commercial kits for cfDNA extraction. A comparative study of a series of commercial kits analyzing artificially added DNA fragments showed that the Qiagen QIAamp circulating nucleic acid kit, based on a spin column, was the most stable kit (Diefenbach et al., 2018). However, the Qiagen QIAamp kits have a significant shortcoming: some short DNA fragments are lost during extraction and purification, resulting in a decrease in cfDNA yield (Kemp et al., 2014). Comparatively, the kits based on magnetic particles for cfDNA isolation have a higher recovery rate for short cfDNA fragments (50–250 bp) than those based on silica membranes (Markus et al., 2018; Ungerer et al., 2020).
2.7.1 Kits for extracting blood-derived cfDNA
Several manufacturers, such as Qiagen, Norgen, Thermo, and Promega, produce specialized commercial kits for extracting blood-derived cfDNA (Diefenbach et al., 2018; van Dessel et al., 2019; Huebner et al., 2021; Wang et al., 2021). Among these, Qiagen’s systematic kits are the most commonly used (Vermeulen et al., 2017; Jain et al., 2019; Jiang et al., 2020; Kallionpaa et al., 2021), However, comparing the performance of these kits is challenging due to variations in sample collection, processing, and analysis procedures (Table 5). For example, when PCR is used to quantify specific genes, the sensitivity may decrease or even vanish as cfDNA fragments become shorter (Ungerer et al., 2020). Nonetheless, a few of the few studies have compared these kits under identical conditions. Devonshire et al. compared the extraction efficiency of four commercial kits (QIAamp circulating nucleic acid kit, NucleoSpin Plasma XS kit, FitAmp plasma/serum DNA isolation kit, and QIAamp DNA blood mini kit) using quantitative PCR measurements of seven different reference genes (Devonshire et al., 2014). They found that the extraction efficiency of the kits was in the following order: QIAamp circulating nucleic acid kit > QIAamp DNA blood mini kit > NucleoSpin Plasma XS kit > FitAmp plasma/serum DNA isolation kit. The QIAamp circulating nucleic acid kit and NucleoSpin Plasma XS kit were more efficient in extracting short DNA fragments than the QIAamp DNA blood mini kit.
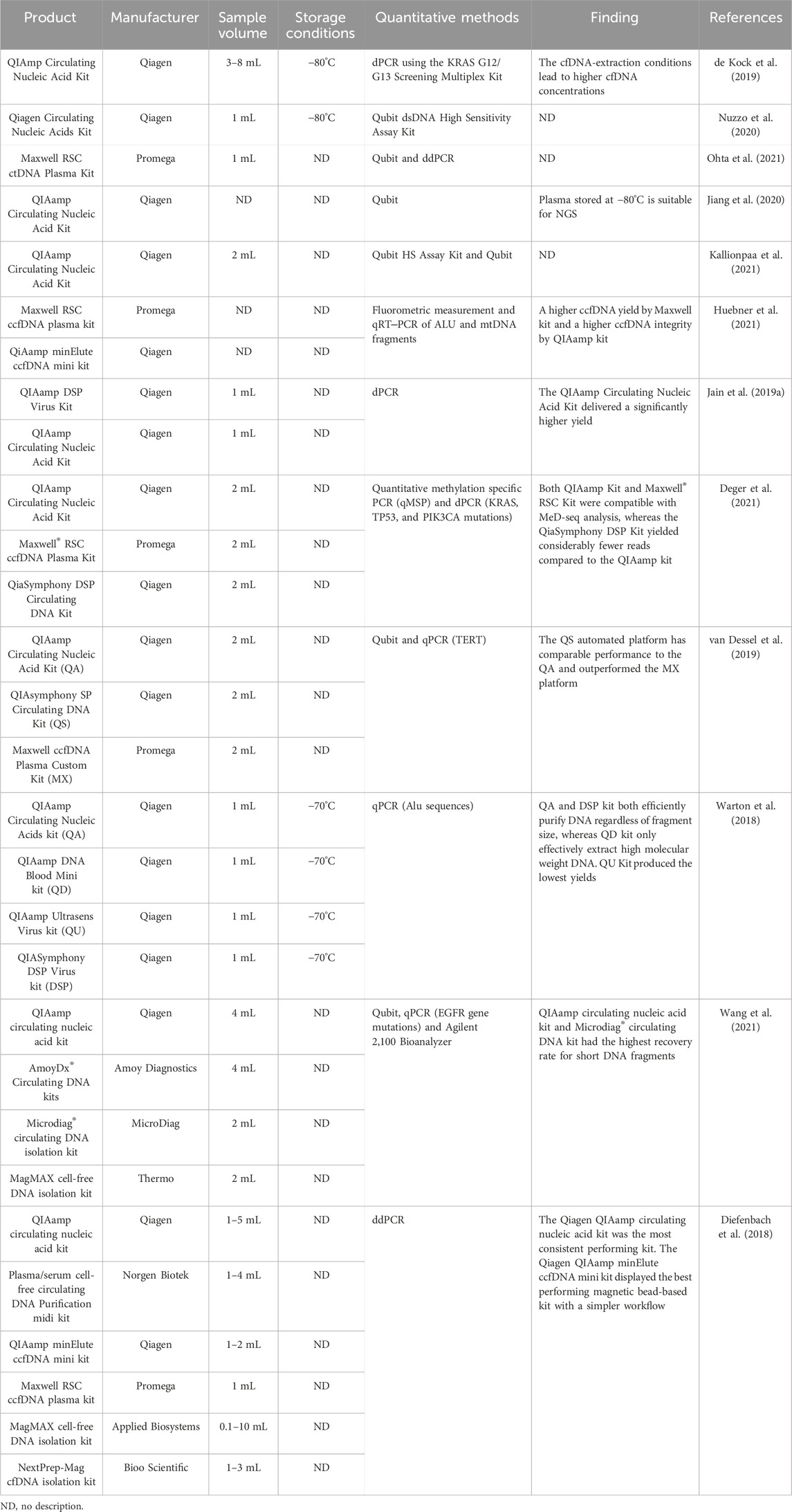
Table 5. Summary of kits for blood-derived cfDNA extraction and storage conditions of extracted cfDNA.
Warton K et al. evaluated four commercial DNA purification kits (QIAamp Circulating Nucleic Acids kit, QIAamp Ultrasens Virus kit, QIAamp DNA Blood Mini kit, and QIASymphony DSP Virus kit) for the extraction of low- (115 base) and high-molecular-weight DNA (247 base) from plasma by qPCR quantification of endogenous Alu sequences (Warton et al., 2018). The study revealed that both the Circulating Nucleic Acids kit and the QIASymphony DSP Virus kit efficiently extracted DNA from plasma regardless of the size of DNA fragments, while the DNA Blood Mini kit only effectively extracted high-molecular-weight DNA. Overall, the QIAamp Circulating Nucleic Acids kit is the most widely used product with better performance than other kits for cfDNA analysis.
In summary, although the Qiagen Company kits lost short DNA fragments during the extraction procedure, they showed relatively higher efficiency and recovery rates (Wang et al., 2021). Additionally, long duration of storage before sample processing may cause cell lysis, resulting in a higher total amount of cfDNA. Therefore, the selection of kits should be based on the specific analytical targets (Table 5).
2.7.2 Kits for extracting urine-derived cfDNA
The extraction protocols of cfDNA in many studies were originally developed mainly for extracting high-integrity gDNA from blood or virus particles rather than highly fragmented cfDNA (Repiska et al., 2013). Therefore, how to efficiently extract cfDNA from urine is usually neglected. Apart from cfDNA originating from exfoliated urothelial cells, the peak length of urine-derived cfDNA depends on glomerular filtration, which requires further degradation of all cfDNA fragments before entering the urine (Yao et al., 2016). While the peak length of plasma cfDNA is 160–167 bp, most urine cfDNA fragments are less than 100 bp (Burnham et al., 2018). Therefore, kits designed for blood-derived cfDNA extraction may not be suitable for urine-derived cfDNA isolation (Oreskovic et al., 2019). Currently, specialized commercial kits for extracting urine-derived cfDNA are offered by manufacturers such as Qiagen, Norgen, Thermo, Promega, and PerkinElmer (Table 6). Lee EY et al. compared the efficiency of four commercial kits (Urine Cell-Free Circulating DNA Purification Midi Kit, Quick-DNA™ Urine Kit, QIAamp Circulating Nucleic Acid Kit, and MagMAX™ Cell-Free DNA Isolation Kit) by an Agilent 2,100 Bioanalyzer for ucfDNA isolation and found that the QIAamp Circulating Nucleic Acid Kit and the MagMAX™ Cell-Free DNA Isolation Kit had the highest cfDNA yield within the 50–300 bp fragment range, while the MagMAX™ Cell-Free DNA Isolation Kit and the Urine Cell-Free Circulating DNA Purification Midi Kit had the highest cfDNA yield within the 50–100 bp fragment range (Lee et al., 2020). Another study by Oreskovic A et al. compared three commercial kits (Norgen, QIAamp, and MagMAX) for extracting short cfDNA fragments from urine (Oreskovic et al., 2019). The study showed that the Norgen kit exhibited a high recovery rate for short cfDNA fragments, although the kit resulted in PCR inhibition, while the other two kits had the lowest recovery rate for short cfDNA fragments. In summary, each kit for urine-derived cfDNA extraction has its own characteristics and advantages, emphasizing the importance of selecting a suitable cfDNA isolation kit based on the specific research purpose. In addition, for ucfDNA extraction, kits allowing the extraction of cfDNA from large-volume urine samples (usually greater than or equal to 10–15 mL) are recommended, as this facilitates obtaining a sufficient amount of ucfDNA (Casadio and Salvi, 2019; Martignano, 2019; Oreskovic et al., 2019; Janovicova et al., 2020).
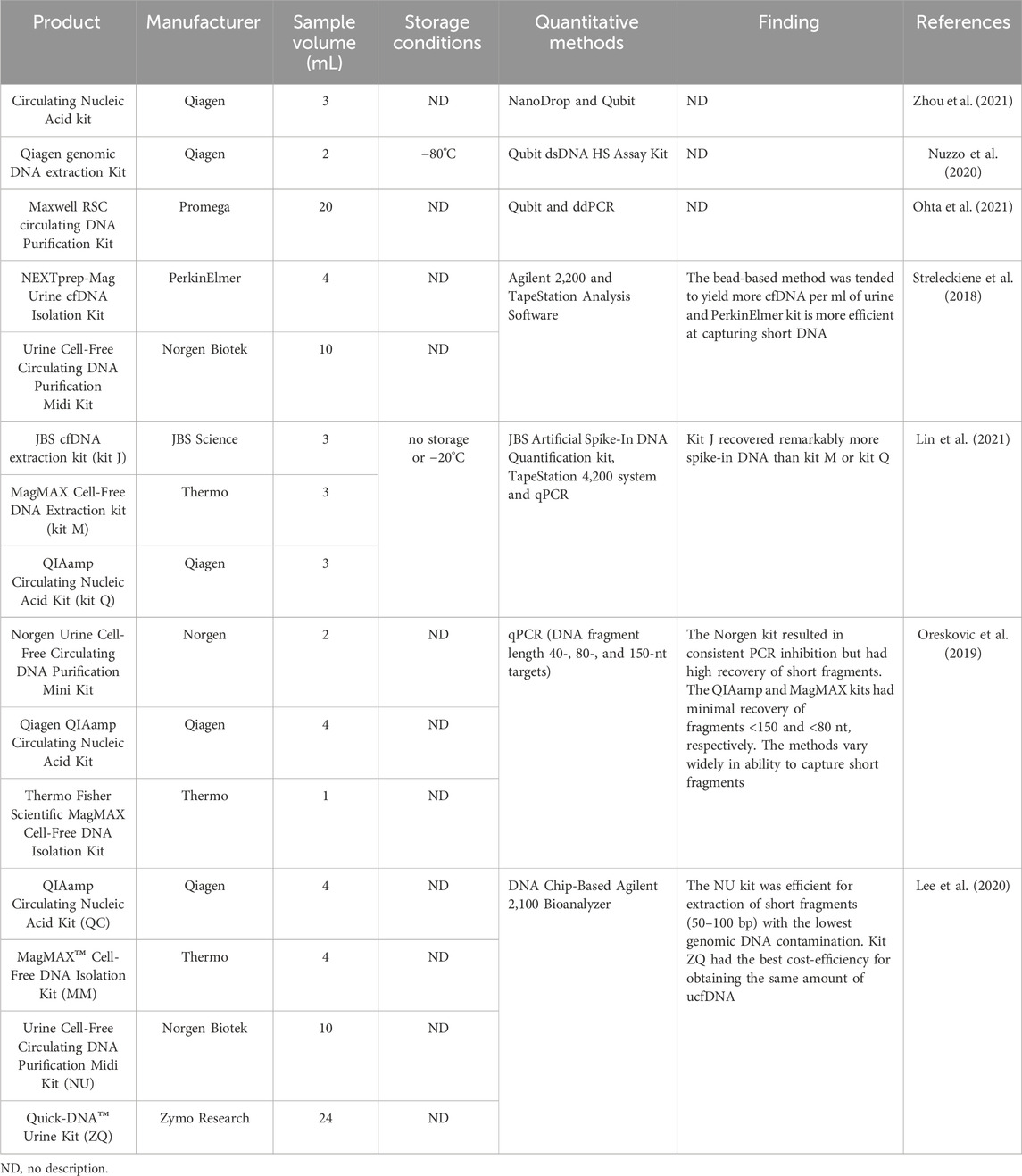
Table 6. Summary of kits for urine-derived cfDNA extraction and storage conditions of extracted cfDNA.
2.8 Quality control of extracted cfDNA
Quality evaluation of extracted cfDNA prior to analysis is critical. This involves assessing concentration, fragment size, and DNA Integrity Number (DIN). The quality control results may indicate the reliability of preanalytical procedures. Unexpected results often suggest potential issues with these procedures. Therefore, sensitive and accurate methods are required for cfDNA quantification.
qPCR-based techniques are also commonly used to quantify cfDNA.With their high sensitivity (Pan et al., 2017), accuracy (Leung et al., 2021), and low false positive rate (Yin et al., 2022), these methods can be used to measure trace nucleic acids effectively (Pan et al., 2017) and analyze cfDNA for known mutations (Hatipoglu et al., 2022). By detecting housekeeping genes (Aucamp et al., 2016) or noncoding repetitive sequences (Hussein et al., 2019) in cfDNA and fitting the standard curve with a reference substance (Tang et al., 2020), absolute cfDNA concentrations can be quantified using PCR-based methods. Frequently used reference genes include TERT (Akuta et al., 2020), GAPDH (Salinas-Sanchez et al., 2021), EGFR (Sugimoto et al., 2023), KRAS (Berchuck et al., 2022), and ALU (Shi et al., 2020). However, the lack of unified reference genes results in significant variations in the quantitative results of PCR-based methods, hindering efficient comparisons across different studies (Devonshire et al., 2014). Moreover, these methods are easily interfered with by compounds. Yokota et al. reported that heparin in plasma could inhibit Taq reaction in PCR analysis (Yokota et al., 1999). The use of cfDNA extracted from jaundice plasma occasionally interfered with PCR reaction, suggesting that a compound in jaundice plasma is not conducive to PCR analysis (Meddeb et al., 2019).
NGS, which greatly reduces sequencing costs and improves accuracy, can analyze millions of ctDNA molecules simultaneously and has been applied to ctDNA detection (Chen and Zhao, 2019). Although PCR-based methods are sensitive, inexpensive and do not require complex information, they are only able to detect known sequences (Taly et al., 2017). In contrast, NGS is high-throughput and can identify new genetic information, but the method is time-consuming and depends on complex data analysis (Postel et al., 2018).
Considering a wide range of applications and advanced technologies, Qubit and Bioanalyzer are currently the optimal methods for cfDNA quantification (Kumar et al., 2018). The Qubit fluorometer offers excellent analytical sensitivity (Burnham et al., 2018) and can simultaneously detect up to eight samples (Parackal et al., 2019). Compared to NanoDrop and qPCR-based methods, the Qubit fluorometer is a suitable compromise considering measurement precision, processing time, and cost simultaneously (Burnham et al., 2018; Khetan et al., 2019). The Bioanalyzer system from Agilent Technology provides detailed information on cfDNA fragment size and level (Lapin et al., 2018), and automatically provides DIN values ranging from 1 (highly degraded) to 10 (extremely intact) to quantitatively evaluate DNA integrity (Truszewska et al., 2020). This system is ideal for quality control of cfDNA samples utilized in NGS (Yu et al., 2021) and qPCR workflows (Hussing et al., 2018). The Cell-free DNA ScreenTape assay can be used to analyze cfNDA samples from 50 bp to 700 bp and detect high molecular weight DNA contaminations (Terp et al., 2024). However, Femtopulse, another Agilent Technology, is a powerful and effective pulsed field capillary electrophoresis system with high sensitivity (Hashem et al., 2020). The system can run for up to 88 samples for cfDNA analysis on a gel simultaneously and provides results in as little as 1.5 h, which is quicker and more economical than the use of a bioanalyzer.
A new technique that can detect cfDNA directly in plasma without prior DNA extraction was developed in 2018. The developers used this technology to analyze cfDNA and found that the measured cfDNA concentrations correlated with those measured by digital PCR (Andriamanampisoa et al., 2018). Further study demonstrated that the analytical performance of the technology is equivalent to that obtained after purification and concentration, with a precision of ∼1% for size features (Boutonnet et al., 2023). In addition, several emerging technologies, including sophisticated cellular biosensors (Cooper et al., 2023), electrochemical biosensors (Wang et al., 2022) and fluorescence-enhancing all-dielectric metasurface biosensors (Iwanaga et al., 2023), can detect the content of cfDNA without complicated processing. These methods with high sensitivity have advantages in low-content detection, but they are not suitable for more detailed analysis of DNA fragments, so they are not generally used for quality control of cfDNA analysis.
Overall, each method has advantages and disadvantages. PCR can accurately and sensitively detect trace amounts of DNA, but it is susceptible to interference from certain compounds. Bioanalyzer and Femtopulse are often used to analyze the fragments and concentration of cfDNA. A bioanalyzer cannot detect larger fragments that can be analyzed by qPCR, while some samples that cannot be amplified by qPCR can be detected by a bioanalyzer (Krasic et al., 2021). In summary, the comparison of cfDNA yields obtained by different quantitative methods is infeasible, and non-PCR methods can compensate for the deficiency of PCR methods (Akbariqomi et al., 2019).
2.9 Long-term storage of extracted cfDNA
If not immediately analyzed, extracted cfDNA should be stored at low temperatures. Long-term preservation is crucial to ensure effective downstream applications. Shorter cfDNA fragments may yield poor quantitative results (Cook et al., 2018), so factors causing DNA degradation or breakage during storage should be mitigated.
Chemical degradation poses the main threat to DNA preservation, so nuclease contamination should be avoided during sample processing and extraction (Ellervik and Vaught, 2015). Factors impacting cfDNA quality during long-term storage include storage temperature and duration, repeated freeze-thaw cycles, and storage tubes (Ungerer et al., 2020).
Long-term storage of cfDNA is typically performed at −20°C or −80°C (Nuzzo et al., 2020; Lin et al., 2021). The appropriate temperature depends on the requirements of subsequent applications. As reviewed in El Messaoudi et al. (2013), cfDNA should be stored at −20°C for less than 3 months for quantification and fragmentation analyses, while it can be stored at −20°C or −80°C for up to 9 months for mutation analyses (El Messaoudi et al., 2013). Low concentrations of cfDNA are more prone to degradation (Martignano, 2019), reducing the storage time.
Polypropylene tubes are recommended for storing cfDNA before freezing as they absorb less DNA (Meddeb et al., 2019). The walls of LoBind tubes may absorb cfDNA, leading to lower sample concentrations (Ungerer et al., 2020). After freezing, extracted cfDNA should not undergo more than three freeze-thaw cycles. Shao et al. (Shao et al., 2012) found that increasing freeze-thaw cycles accelerates DNA degradation, with larger DNA fragments degrading most readily. Increasing DNA concentration can reduce degradation caused by repeated freeze-thaw cycles.
3 Conclusion and future directions
Achieving consensus on the various variables in preanalytical procedures is critical for ensuring the reliability and repeatability of cfDNA measurements. Notable progress, including the development of analytical methods and specialized products, has been made in recent years. However, many variables associated with preanalytical procedures remain undefined or inconsistent or interact with each other, particularly in the case of urine samples. The guidelines for preanalytical variables of blood samples have been developed and summarized in recent years (Meddeb et al., 2019; Greytak et al., 2020; Lampignano et al., 2020), offering valuable insights and inspiration for the standardization of preanalytical procedures for urine-derived cfDNA analysis. For instance, the addition of EDTA to collected samples (Markus et al., 2021) and timely processing of samples (Zhang et al., 2022) is beneficial to enhance cfDNA quality, and these protocols are also applicable to urine-derived cfDNA.
This review discusses and summarizes the crucial variables in each preanalytical stage for analyzing blood-derived and urine-derived cfDNA (Figure 2). However, it is undeniable that achieving perfect coordination in the implementation of standardized preanalytical procedures is challenging due to objective conditions such as potential differences in funding and equipment resources among institutions or laboratories. Nevertheless, the following points can be explored further: (1) further verification of the impact of preanalytical variables on ucfDNA analysis; (2) development of multifunctional kits that efficiently extract short DNA fragments while preventing gDNA contamination; and (3) comparison and verification of measurement results from different internal reference genes when using PCR-based technologies for cfDNA quantification. These future directions will not only help address existing gaps in cfDNA preanalytical procedure standardization but also facilitate the broader and more accurate application of cfDNA analysis in clinical diagnostics and research.
Author contributions
HP: Writing–original draft, Writing–review and editing. MP: Writing–original draft, Writing–review and editing. ZZ: Writing–original draft. CC: Writing–original draft. XX: Writing–original draft. SC: Writing–original draft. SZ: Writing–original draft. HZ: Writing–original draft, Writing–review and editing. KQ: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund, Project CXPY2020031 and ZLYNXM20200213.
Acknowledgments
The authors would like to thank Dr. Yuruo Chen, a diagram expert at Chinese Academy of Sciences, for editing the figure.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akat, K. M., Lee, Y. A., Hurley, A., Morozov, P., Max, K. E., Brown, M., et al. (2019). Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight 5 (9), e127317. doi:10.1172/jci.insight.127317
Akbariqomi, M., Heidari, R., Gargari, S. S., Omrani, M. D., Rigi, G., Sanikhani, N. S., et al. (2019). Evaluation and statistical optimization of a method for methylated cell-free fetal DNA extraction from maternal plasma. J. Assist. Reprod. Genet. 36 (5), 1029–1038. doi:10.1007/s10815-019-01425-w
Akuta, N., Suzuki, F., Kobayashi, M., Fujiyama, S., Kawamura, Y., Sezaki, H., et al. (2020). Detection of TERT promoter mutation in serum cell-free DNA using wild-type blocking PCR combined with Sanger sequencing in hepatocellular carcinoma. J. Med. Virol. 92, 3604–3608. doi:10.1002/jmv.25724
Alidousty, C., Brandes, D., Heydt, C., Wagener, S., Wittersheim, M., Schafer, S. C., et al. (2017). Comparison of blood collection tubes from three different manufacturers for the collection of cell-free DNA for liquid biopsy mutation testing. J. Mol. Diagn 19 (5), 801–804. doi:10.1016/j.jmoldx.2017.06.004
Ampo, E., Mendes-Silva, A. P., Goncalves, V., Bartley, J. M., Kuchel, G. A., and Diniz, B. S. (2022). Increased levels of circulating cell-free mtDNA in the plasma of subjects with late-life depression and frailty: a preliminary study. Am. J. Geriatr. Psychiatry 30 (3), 332–337. doi:10.1016/j.jagp.2021.07.012
Andriamanampisoa, C. L., Bancaud, A., Boutonnet-Rodat, A., Didelot, A., Fabre, J., Fina, F., et al. (2018). BIABooster: online DNA concentration and size profiling with a limit of detection of 10 fg/μL and application to high-sensitivity characterization of circulating cell-free DNA. Anal. Chem. 90 (6), 3766–3774. doi:10.1021/acs.analchem.7b04034
Arshad, O., Gadawska, I., Sattha, B., Cote, H. C. F., and Hsieh, A. Y. Y. (2018). Elevated cell-free mitochondrial DNA in filtered plasma is associated with HIV infection and inflammation. J. Acquir Immune Defic. Syndr. 78 (1), 111–118. doi:10.1097/QAI.0000000000001650
Aucamp, J., Bronkhorst, A. J., Badenhorst, C. P. S., and Pretorius, P. J. (2018). The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol. Rev. Camb Philos. Soc. 93 (3), 1649–1683. doi:10.1111/brv.12413
Aucamp, J., Bronkhorst, A. J., Wentzel, J. F., and Pretorius, P. J. (2016). A quantitative assessment of cell-free DNA utilizing several housekeeping genes: measurements from four different cell lines. Adv. Exp. Med. Biol. 924, 101–103. doi:10.1007/978-3-319-42044-8_19
Aw, W. C., Towarnicki, S. G., Melvin, R. G., Youngson, N. A., Garvin, M. R., Hu, Y., et al. (2018). Genotype to phenotype: diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 14 (11), e1007735. doi:10.1371/journal.pgen.1007735
Barra, G. B., Santa Rita, T. H., de Almeida Vasques, J., Chianca, C. F., Nery, L. F., and Santana Soares Costa, S. (2015). EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin. Biochem. 48 (15), 976–981. doi:10.1016/j.clinbiochem.2015.02.014
Berchuck, J. E., Facchinetti, F., DiToro, D. F., Baiev, I., Majeed, U., Reyes, S., et al. (2022). The clinical landscape of cell-free DNA alterations in 1671 patients with advanced biliary tract cancer. Ann. Oncol. 33 (12), 1269–1283. doi:10.1016/j.annonc.2022.09.150
Bettegowda, C., Sausen, M., Leary, R. J., Kinde, I., Wang, Y., Agrawal, N., et al. (2014). Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6 (224), 224ra24. doi:10.1126/scitranslmed.3007094
Bhangu, J. S., Taghizadeh, H., Braunschmid, T., Bachleitner-Hofmann, T., and Mannhalter, C. (2017). Circulating cell-free DNA in plasma of colorectal cancer patients - a potential biomarker for tumor burden. Surg. Oncol. 26 (4), 395–401. doi:10.1016/j.suronc.2017.08.001
Bianchi, D. W., and Chiu, R. W. K. (2018). Sequencing of circulating cell-free DNA during pregnancy. N. Engl. J. Med. 379 (5), 464–473. doi:10.1056/NEJMra1705345
Board, R. E., Williams, V. S., Knight, L., Shaw, J., Greystoke, A., Ranson, M., et al. (2008). Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann. N. Y. Acad. Sci. 1137, 98–107. doi:10.1196/annals.1448.020
Boutonnet, A., Pradines, A., Mano, M., Kreczman-Brun, M., Mazières, J., Favre, G., et al. (2023). Size and concentration of cell-free DNA measured directly from blood plasma, without prior DNA extraction. Anal. Chem. 95 (24), 9263–9270. doi:10.1021/acs.analchem.3c00998
Bronkhorst, A. J., Aucamp, J., and Pretorius, P. J. (2015). Cell-free DNA: preanalytical variables. Clin. Chim. Acta 450, 243–253. doi:10.1016/j.cca.2015.08.028
Bronkhorst, A. J., Ungerer, V., and Holdenrieder, S. (2019). Early detection of cancer using circulating tumor DNA: biological, physiological and analytical considerations. Crit. Rev. Clin. Lab. Sci. 57 (4), 253–269. doi:10.1080/10408363.2019.1700902
Burnham, P., Dadhania, D., Heyang, M., Chen, F., Westblade, L. F., Suthanthiran, M., et al. (2018). Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 9 (1), 2412. doi:10.1038/s41467-018-04745-0
Campbell, G., Alderson, P., Smith, A. F., and Warttig, S. (2015). Warming of intravenous and irrigation fluids for preventing inadvertent perioperative hypothermia. Cochrane Database Syst. Rev. 2015 (4), CD009891. doi:10.1002/14651858.CD009891.pub2
Casadio, V., and Salvi, S. (2019). Urinary cell-free DNA: isolation, quantification, and quality assessment. Methods Mol. Biol. 1909, 211–221. doi:10.1007/978-1-4939-8973-7_16
Chen, M., and Zhao, H. (2019). Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum. Genomics 13 (1), 34. doi:10.1186/s40246-019-0220-8
Cheng, H., Luo, G., Jin, K., Fan, Z., Huang, Q., Gong, Y., et al. (2020). Kras mutation correlating with circulating regulatory T cells predicts the prognosis of advanced pancreatic cancer patients. Cancer Med. 9 (6), 2153–2159. doi:10.1002/cam4.2895
Cheng, T. H. T., Jiang, P., Tam, J. C. W., Sun, X., Lee, W. S., Yu, S. C. Y., et al. (2017). Genomewide bisulfite sequencing reveals the origin and time-dependent fragmentation of urinary cfDNA. Clin. Biochem. 50 (9), 496–501. doi:10.1016/j.clinbiochem.2017.02.017
Cimmino, I., Bravaccini, S., and Cerchione, C. (2021). Urinary biomarkers in tumors: an overview. Methods Mol. Biol. 2292, 3–15. doi:10.1007/978-1-0716-1354-2_1
Cook, J. D., Caplan, Y. H., LoDico, C. P., and Bush, D. M. (2000). The characterization of human urine for specimen validity determination in workplace drug testing: a review. J. Anal. Toxicol. 24 (7), 579–588. doi:10.1093/jat/24.7.579
Cook, L., Starr, K., Boonyaratanakornkit, J., Hayden, R., Sam, S. S., and Caliendo, A. M. (2018). Does size matter? Comparison of extraction yields for different-sized DNA fragments by seven different routine and four new circulating cell-free extraction methods. J. Clin. Microbiol. 56 (12), e01061. doi:10.1128/JCM.01061-18
Cooper, R. M., Wright, J. A., Ng, J. Q., Goyne, J. M., Suzuki, N., Lee, Y. K., et al. (2023). Engineered bacteria detect tumor DNA. Science 381 (6658), 682–686. doi:10.1126/science.adf3974
Deger, T., Boers, R. G., de Weerd, V., Angus, L., van der Put, M. M. J., Boers, J. B., et al. (2021). High-throughput and affordable genome-wide methylation profiling of circulating cell-free DNA by methylated DNA sequencing (MeD-seq) of LpnPI digested fragments. Clin. Epigenetics 13 (1), 196. doi:10.1186/s13148-021-01177-4
de Kock, R., Deiman, B., Kraaijvanger, R., and Scharnhorst, V. (2019). Optimized (pre) analytical conditions and workflow for droplet digital PCR analysis of cell-free DNA from patients with suspected lung carcinoma. J. Mol. Diagn 21 (5), 895–902. doi:10.1016/j.jmoldx.2019.05.003
Devonshire, A. S., Whale, A. S., Gutteridge, A., Jones, G., Cowen, S., Foy, C. A., et al. (2014). Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal. Bioanal. Chem. 406 (26), 6499–6512. doi:10.1007/s00216-014-7835-3
Diao, Z. L., Zhang, R., and Li, J. M. (2022). Analysis of the methods and quality assurance of metagenomic next-generation sequencing to detect the microbial cfDNA from blood samples in China. Zhonghua Yi Xue Za Zhi 102 (15), 1114–1118. doi:10.3760/cma.j.cn112137-20220104-00017
Diefenbach, R. J., Lee, J. H., Kefford, R. F., and Rizos, H. (2018). Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 228-229, 21–27. doi:10.1016/j.cancergen.2018.08.005
Dolscheid-Pommerich, R. C., Klarmann-Schulz, U., Conrad, R., Stoffel-Wagner, B., and Zur, B. (2016). Evaluation of the appropriate time period between sampling and analyzing for automated urinalysis. Biochem. Med. Zagreb. 26 (1), 82–89. doi:10.11613/BM.2016.008
Drag, M. H., and Kilpelainen, T. O. (2021). Cell-free DNA and RNA-measurement and applications in clinical diagnostics with focus on metabolic disorders. Physiol. Genomics 53 (1), 33–46. doi:10.1152/physiolgenomics.00086.2020
Duplessis, C., Gregory, M., Frey, K., Bell, M., Truong, L., Schully, K., et al. (2018). Evaluating the discriminating capacity of cell death (apoptotic) biomarkers in sepsis. J. Intensive Care 6, 72. doi:10.1186/s40560-018-0341-5
Eisinger, S. W., Schwartz, M., Dam, L., and Riedel, S. (2013). Evaluation of the BD Vacutainer Plus Urine C&S Preservative Tubes compared with nonpreservative urine samples stored at 4°C and room temperature. Am. J. Clin. Pathol. 140 (3), 306–313. doi:10.1309/AJCP5ON9JHXVNQOD
Ellervik, C., and Vaught, J. (2015). Preanalytical variables affecting the integrity of human biospecimens in biobanking. Clin. Chem. 61 (7), 914–934. doi:10.1373/clinchem.2014.228783
El Messaoudi, S., Rolet, F., Mouliere, F., and Thierry, A. R. (2013). Circulating cell free DNA: preanalytical considerations. Clin. Chim. Acta 424, 222–230. doi:10.1016/j.cca.2013.05.022
Ercan, M., Akbulut, E. D., Abusoglu, S., Yilmaz, F. M., Oguz, E. F., Topcuoglu, C., et al. (2015). Stability of urine specimens stored with and without preservatives at room temperature and on ice prior to urinalysis. Clin. Biochem. 48 (13-14), 919–922. doi:10.1016/j.clinbiochem.2015.05.016
Fatouros, I. G., Jamurtas, A. Z., Nikolaidis, M. G., Destouni, A., Michailidis, Y., Vrettou, C., et al. (2010). Time of sampling is crucial for measurement of cell-free plasma DNA following acute aseptic inflammation induced by exercise. Clin. Biochem. 43 (16-17), 1368–1370. doi:10.1016/j.clinbiochem.2010.08.020
Fedyuk, V., Erez, N., Furth, N., Beresh, O., Andreishcheva, E., Shinde, A., et al. (2023). Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics. Nat. Biotechnol. 41 (2), 212–221. doi:10.1038/s41587-022-01447-3
Fernando, M. R., Jiang, C., Krzyzanowski, G. D., and Ryan, W. L. (2017). New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One 12 (8), e0183915. doi:10.1371/journal.pone.0183915
Fiala, C., and Diamandis, E. P. (2018). Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med. 16 (1), 166. doi:10.1186/s12916-018-1157-9
Fujihara, J., Takinami, Y., Kawai, Y., Kimura-Kataoka, K., and Takeshita, H. (2021). Comparison of serum cell-free DNA between postmortem and living samples. Clin. Chim. Acta 519, 255–259. doi:10.1016/j.cca.2021.05.013
Gerber, T., Taschner-Mandl, S., Saloberger-Sindhoringer, L., Popitsch, N., Heitzer, E., Witt, V., et al. (2020). Assessment of pre-analytical sample handling conditions for comprehensive liquid biopsy analysis. J. Mol. Diagn 22 (8), 1070–1086. doi:10.1016/j.jmoldx.2020.05.006
Greytak, S. R., Engel, K. B., Parpart-Li, S., Murtaza, M., Bronkhorst, A. J., Pertile, M. D., et al. (2020). Harmonizing cell-free DNA collection and processing practices through evidence-based guidance. Clin. Cancer Res. 26 (13), 3104–3109. doi:10.1158/1078-0432.CCR-19-3015
Handayani, N., Aubry, D., Boediono, A., Wiweko, B., Sirait, B., Sini, I., et al. (2023). The origin and possible mechanism of embryonic cell-free DNA release in spent embryo culture media: a review. J. Assist. Reprod. Genet. 40 (6), 1231–1242. doi:10.1007/s10815-023-02813-z
Hashem, V., Tiwari, A., Bewick, B., Teive, H. A. G., Moscovich, M., Schuele, B., et al. (2020). Pulse-Field capillary electrophoresis of repeat-primed PCR amplicons for analysis of large repeats in Spinocerebellar Ataxia Type 10. PLoS One 15 (3), e0228789. doi:10.1371/journal.pone.0228789
Hatipoglu, T., Esmeray Sonmez, E., Hu, X., Yuan, H., Danyeli, A. E., Seyhanli, A., et al. (2022). Plasma concentrations and cancer-associated mutations in cell-free circulating DNA of treatment-naive follicular lymphoma for improved non-invasive diagnosis and prognosis. Front. Oncol. 12, 870487. doi:10.3389/fonc.2022.870487
Hayashi, Y., Fujita, K., Matsuzaki, K., Matsushita, M., Kawamura, N., Koh, Y., et al. (2019). Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci. 110 (5), 1771–1779. doi:10.1111/cas.14000
Hidestrand, M., Stokowski, R., Song, K., Oliphant, A., Deavers, J., Goetsch, M., et al. (2012). Influence of temperature during transportation on cell-free DNA analysis. Fetal Diagn Ther. 31 (2), 122–128. doi:10.1159/000335020
Huang, Y., Mu, J., Qi, L., Ge, W., Fang, X., Song, Y., et al. (2020). Diverse fragment lengths dismiss size selection for serum cell-free DNA: a comparative study of serum and plasma samples. Clin. Chem. Lab. Med. 58 (9), 1451–1459. doi:10.1515/cclm-2020-0078
Hudecova, I., Smith, C. G., Hansel-Hertsch, R., Chilamakuri, C. S., Morris, J. A., Vijayaraghavan, A., et al. (2022). Characteristics, origin, and potential for cancer diagnostics of ultrashort plasma cell-free DNA. Genome Res. 32 (2), 215–227. doi:10.1101/gr.275691.121
Huebner, H., Lubrich, H., Blum, S., Antoniadis, S., Lermann, J., Ekici, A., et al. (2021). Comparison of methods for isolation and quantification of circulating cell-free DNA from patients with endometriosis. Reprod. Biomed. Online 43 (5), 788–798. doi:10.1016/j.rbmo.2021.08.004
Humardani, F. M., Mulyanata, L. T., and Dwi Putra, S. E. (2023). Adipose cell-free DNA in diabetes. Clin. Chim. Acta 539, 191–197. doi:10.1016/j.cca.2022.12.008
Huminska-Lisowska, K., Mieszkowski, J., Kochanowicz, A., Stankiewicz, B., Niespodzinski, B., Brzezinska, P., et al. (2021). cfDNA changes in maximal exercises as a sport adaptation predictor. Genes (Basel) 12 (8), 1238. doi:10.3390/genes12081238
Hussein, N. A., Mohamed, S. N., and Ahmed, M. A. (2019). Plasma ALU-247, ALU-115, and cfDNA integrity as diagnostic and prognostic biomarkers for breast cancer. Appl. Biochem. Biotechnol. 187 (3), 1028–1045. doi:10.1007/s12010-018-2858-4
Hussing, C., Kampmann, M. L., Mogensen, H. S., Borsting, C., and Morling, N. (2018). Quantification of massively parallel sequencing libraries - a comparative study of eight methods. Sci. Rep. 8 (1), 1110. doi:10.1038/s41598-018-19574-w
Itoh, M., Sako, Y., Itoh, S., Ishikawa, Y., Akabane, H., Nakaya, K., et al. (2013). Immunodiagnosis of alveolar echinococcosis using urine samples. Parasitol. Int. 62 (6), 514–516. doi:10.1016/j.parint.2013.07.007
Iwanaga, M., Hironaka, T., Ikeda, N., Sugasawa, T., and Takekoshi, K. (2023). Metasurface biosensors enabling single-molecule sensing of cell-free DNA. Nano Lett. 23 (12), 5755–5761. doi:10.1021/acs.nanolett.3c01527
Jacob, E., Jacob, A., Davies, H., Jacob, D., Jenkins, M., Husain, M., et al. (2021). The impact of blood sampling technique, including the use of peripheral intravenous cannula, on haemolysis rates: a cohort study. J. Clin. Nurs. 30 (13-14), 1916–1926. doi:10.1111/jocn.15744
Jahr, S., Hentze, H., Englisch, S., Hardt, D., Fackelmayer, F. O., Hesch, R. D., et al. (2001). DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61 (4), 1659–1665.
Jain, M., Balatsky, A. V., Revina, D. B., and Samokhodskaya, L. M. (2019a). Direct comparison of QIAamp DSP Virus Kit and QIAamp Circulating Nucleic Acid Kit regarding cell-free fetal DNA isolation from maternal peripheral blood. Mol. Cell Probes 43, 13–19. doi:10.1016/j.mcp.2018.12.006
Jain, S., Lin, S. Y., Song, W., and Su, Y. H. (2019b). Urine-based liquid biopsy for nonurological cancers. Genet. Test. Mol. Biomarkers 23 (4), 277–283. doi:10.1089/gtmb.2018.0189
Janku, F., Huang, H. J., Pereira, D. Y., Kobayashi, M., Chiu, C. H., Call, S. G., et al. (2021). A novel method for liquid-phase extraction of cell-free DNA for detection of circulating tumor DNA. Sci. Rep. 11 (1), 19653. doi:10.1038/s41598-021-98815-x
Janovicova, L., Kmetova, K., Tothova, L., Vlkova, B., and Celec, P. (2023). DNA in fresh urine supernatant is not affected by additional centrifugation and is protected against deoxyribonuclease. Mol. Cell Probes 68, 101900. doi:10.1016/j.mcp.2023.101900
Janovicova, L., Konecna, B., Vlkova, B., and Celec, P. (2020). Isolation and quantification of extracellular DNA from biofluids. Bio Protoc. 10 (16), e3726. doi:10.21769/BioProtoc.3726
Jiang, P., Chan, C. W., Chan, K. C., Cheng, S. H., Wong, J., Wong, V. W., et al. (2015). Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. U. S. A. 112 (11), E1317–E1325. doi:10.1073/pnas.1500076112
Jiang, X., Li, H., Liu, J., Sun, H., Zhang, L., Li, W., et al. (2020). Feasibility analysis of cell-free DNA derived from plasma of lung cancer patients for next-generation sequencing. Biopreserv Biobank 18 (2), 117–121. doi:10.1089/bio.2019.0115
Jordaens, S., Arora, A., MacDonald, K. W., Wood, C., Hendrickx, J. O., Zwaenepoel, K., et al. (2023). UAS-A urine preservative for oncology applications. Cancers (Basel) 15 (12), 3119. doi:10.3390/cancers15123119
Kallionpaa, R. A., Ahramo, K., Aaltonen, M., Pennanen, P., Peltonen, J., and Peltonen, S. (2021). Circulating free DNA in the plasma of individuals with neurofibromatosis type 1. Am. J. Med. Genet. A 185 (4), 1098–1104. doi:10.1002/ajmg.a.62081
Kang, Q., Henry, N. L., Paoletti, C., Jiang, H., Vats, P., Chinnaiyan, A. M., et al. (2016). Comparative analysis of circulating tumor DNA stability in K(3)EDTA, Streck, and CellSave blood collection tubes. Clin. Biochem. 49 (18), 1354–1360. doi:10.1016/j.clinbiochem.2016.03.012
Kemp, B. M., Winters, M., Monroe, C., and Barta, J. L. (2014). How much DNA is lost? Measuring DNA loss of short-tandem-repeat length fragments targeted by the PowerPlex 16(R) system using the Qiagen MinElute Purification Kit. Hum. Biol. 86 (4), 313–329. doi:10.13110/humanbiology.86.4.0313
Khetan, D., Gupta, N., Chaudhary, R., and Shukla, J. S. (2019). Comparison of UV spectrometry and fluorometry-based methods for quantification of cell-free DNA in red cell components. Asian J. Transfus. Sci. 13 (2), 95–99. doi:10.4103/ajts.AJTS_90_19
Kim, A. K., Hamilton, J. P., Lin, S. Y., Chang, T. T., Hann, H. W., Hu, C. T., et al. (2022). Urine DNA biomarkers for hepatocellular carcinoma screening. Br. J. Cancer 126 (10), 1432–1438. doi:10.1038/s41416-022-01706-9
Kim, A. K., Lin, S. Y., Wang, Z., Luu, H., Hamilton, J. P., Song, W., et al. (2023). Impact of cell-debris and room-temperature storage on urine circulating tumor DNA from hepatocellular carcinoma. J. Mol. Diagn 25 (12), 913–920. doi:10.1016/j.jmoldx.2023.08.006
Krasic, J., Abramovic, I., Vrtaric, A., Nikolac Gabaj, N., Kralik-Oguic, S., Katusic Bojanac, A., et al. (2021). Impact of preanalytical and analytical methods on cell-free DNA diagnostics. Front. Cell Dev. Biol. 9, 686149. doi:10.3389/fcell.2021.686149
Kumar, M., Choudhury, Y., Ghosh, S. K., and Mondal, R. (2018). Application and optimization of minimally invasive cell-free DNA techniques in oncogenomics. Tumour Biol. 40 (2), 1010428318760342. doi:10.1177/1010428318760342
Lam, N. Y., Rainer, T. H., Chiu, R. W., and Lo, Y. M. (2004). EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin. Chem. 50 (1), 256–257. doi:10.1373/clinchem.2003.026013
Lampignano, R., Neumann, M. H. D., Weber, S., Kloten, V., Herdean, A., Voss, T., et al. (2020). Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (Pre)analytical work flows. Clin. Chem. 66 (1), 149–160. doi:10.1373/clinchem.2019.306837
Lapin, M., Oltedal, S., Tjensvoll, K., Buhl, T., Smaaland, R., Garresori, H., et al. (2018). Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 16 (1), 300. doi:10.1186/s12967-018-1677-2
Lee, E. Y., Lee, E. J., Yoon, H., Lee, D. H., and Kim, K. H. (2020). Comparison of four commercial kits for isolation of urinary cell-free DNA and sample storage conditions. Diagn. (Basel) 10 (4), 234. doi:10.3390/diagnostics10040234
Lee, H., Na, W., Park, C., Park, K. H., and Shin, S. (2018). Centrifugation-free extraction of circulating nucleic acids using immiscible liquid under vacuum pressure. Sci. Rep. 8 (1), 5467. doi:10.1038/s41598-018-23766-9
Lenz, M., Maiberger, T., Armbrust, L., Kiwit, A., Von der Wense, A., Reinshagen, K., et al. (2022). cfDNA and DNases: new biomarkers of sepsis in preterm neonates-A pilot study. Cells 11 (2), 192. doi:10.3390/cells11020192
Letendre, J. A., and Goggs, R. (2017). Measurement of plasma cell-free DNA concentrations in dogs with sepsis, trauma, and neoplasia. J. Vet. Emerg. Crit. Care (San Ant.) 27 (3), 307–314. doi:10.1111/vec.12592
Leung, E., Han, K., Zou, J., Zhao, Z., Zheng, Y., Wang, T. T., et al. (2021). HPV sequencing facilitates ultrasensitive detection of HPV circulating tumor DNA. Clin. Cancer Res. 27 (21), 5857–5868. doi:10.1158/1078-0432.CCR-19-2384
Li, J., Dittmar, R. L., Xia, S., Zhang, H., Du, M., Huang, C. C., et al. (2017). Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol. Oncol. 11 (8), 1099–1111. doi:10.1002/1878-0261.12077
Lin, S. Y., Luo, Y., Marshall, M. M., Johnson, B. J., Park, S. R., Wang, Z., et al. (2021). A new method for improving extraction efficiency and purity of urine and plasma cell-free DNA. Diagn. (Basel) 11 (4), 650. doi:10.3390/diagnostics11040650
Liu, Y., Cheng, L., Wang, G., Lv, J., He, Y., Shao, P. L., et al. (2022). A nano-magnetic size selective cfDNA extraction platform for liquid biopsy with enhanced precision. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 1199, 123236. doi:10.1016/j.jchromb.2022.123236
Lo, Y. M. D., Han, D. S. C., Jiang, P., and Chiu, R. W. K. (2021). Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 372 (6538), eaaw3616. doi:10.1126/science.aaw3616
Luo, J., Wang, S., Zhang, S., He, Y., Li, S., Han, J., et al. (2022). Performance of ImproGene cell-free DNA tubes for stabilization and analysis of cfDNA in blood samples. Fetal Pediatr. Pathol. 41 (5), 771–780. doi:10.1080/15513815.2021.1979143
Luo, X., Gu, X., and Li, L. (2018). Development of a simple and efficient method of harvesting and lysing adherent mammalian cells for chemical isotope labeling LC-MS-based cellular metabolomics. Anal. Chim. Acta 1037, 97–106. doi:10.1016/j.aca.2017.11.054
Mandel, P., and Metais, P. (1948). Nuclear acids in human blood plasma. C R. Seances Soc. Biol. Fil. 142 (3-4), 241–243.
Markus, H., Chandrananda, D., Moore, E., Mouliere, F., Morris, J., Brenton, J. D., et al. (2022). Refined characterization of circulating tumor DNA through biological feature integration. Sci. Rep. 12 (1), 1928. doi:10.1038/s41598-022-05606-z
Markus, H., Contente-Cuomo, T., Farooq, M., Liang, W. S., Borad, M. J., Sivakumar, S., et al. (2018). Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci. Rep. 8 (1), 7375. doi:10.1038/s41598-018-25810-0
Markus, H., Zhao, J., Contente-Cuomo, T., Stephens, M. D., Raupach, E., Odenheimer-Bergman, A., et al. (2021). Analysis of recurrently protected genomic regions in cell-free DNA found in urine. Sci. Transl. Med. 13 (581), eaaz3088. doi:10.1126/scitranslmed.aaz3088
Martignano, F. (2019). Cell-free DNA: an overview of sample types and isolation procedures. Methods Mol. Biol. 1909, 13–27. doi:10.1007/978-1-4939-8973-7_2
Meddeb, R., Dache, Z. A. A., Thezenas, S., Otandault, A., Tanos, R., Pastor, B., et al. (2019). Quantifying circulating cell-free DNA in humans. Sci. Rep. 9 (1), 5220. doi:10.1038/s41598-019-41593-4
Meddeb, R., Pisareva, E., and Thierry, A. R. (2019). Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin. Chem. 65 (5), 623–633. doi:10.1373/clinchem.2018.298323
Melkonyan, H. S., Feaver, W. J., Meyer, E., Scheinker, V., Shekhtman, E. M., Xin, Z., et al. (2008). Transrenal nucleic acids: from proof of principle to clinical tests. Ann. N. Y. Acad. Sci. 1137, 73–81. doi:10.1196/annals.1448.015
Mondelo-Macia, P., Castro-Santos, P., Castillo-Garcia, A., Muinelo-Romay, L., and Diaz-Pena, R. (2021). Circulating free DNA and its emerging role in autoimmune diseases. J. Pers. Med. 11 (2), 151. doi:10.3390/jpm11020151
Mouliere, F., Smith, C. G., Heider, K., Su, J., van der Pol, Y., Thompson, M., et al. (2021). Fragmentation patterns and personalized sequencing of cell-free DNA in urine and plasma of glioma patients. EMBO Mol. Med. 13 (8), e12881. doi:10.15252/emmm.202012881
Mouser, A., Uettwiller-Geiger, D., Plokhoy, E., Berube, J., Ahuja, A. J., and Stankovic, A. K. (2017). Evaluation of pain and specimen quality by use of a novel 25-gauge blood collection set with ultra-thin wall cannula and 5-bevel tip design. J. Appl. Lab. Med. 2 (2), 201–210. doi:10.1373/jalm.2017.023564
Muller Bark, J., Kulasinghe, A., Chua, B., Day, B. W., and Punyadeera, C. (2020). Circulating biomarkers in patients with glioblastoma. Br. J. Cancer 122 (3), 295–305. doi:10.1038/s41416-019-0603-6
Murugesan, K., Hogan, C. A., Palmer, Z., Reeve, B., Theron, G., Andama, A., et al. (2019). Investigation of preanalytical variables impacting pathogen cell-free DNA in blood and urine. J. Clin. Microbiol. 57 (11), e00782. doi:10.1128/JCM.00782-19
Nabet, B. Y., Esfahani, M. S., Moding, E. J., Hamilton, E. G., Chabon, J. J., Rizvi, H., et al. (2020). Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 183 (2), 363–376. doi:10.1016/j.cell.2020.09.001
Nel, I., Munch, C., Shamkeeva, S., Heinemann, M. L., Isermann, B., and Aktas, B. (2023). The challenge to stabilize, extract and analyze urinary cell-free DNA (ucfDNA) during clinical routine. Diagn. (Basel) 13 (24), 3670. doi:10.3390/diagnostics13243670
Nesic, M., Bodker, J. S., Terp, S. K., and Dybkaer, K. (2021). Optimization of preanalytical variables for cfDNA processing and detection of ctDNA in archival plasma samples. Biomed. Res. Int. 2021, 5585148. doi:10.1155/2021/5585148
Nikanjam, M., Kato, S., and Kurzrock, R. (2022). Liquid biopsy: current technology and clinical applications. J. Hematol. Oncol. 15 (1), 131. doi:10.1186/s13045-022-01351-y
Nuzzo, P. V., Berchuck, J. E., Korthauer, K., Spisak, S., Nassar, A. H., Abou Alaiwi, S., et al. (2020). Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med. 26 (7), 1041–1043. doi:10.1038/s41591-020-0933-1
Oellerich, M., Sherwood, K., Keown, P., Schutz, E., Beck, J., Stegbauer, J., et al. (2021). Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury. Nat. Rev. Nephrol. 17 (9), 591–603. doi:10.1038/s41581-021-00428-0
Ohta, R., Yamada, T., Sonoda, H., Matsuda, A., Shinji, S., Takahashi, G., et al. (2021). Detection of KRAS mutations in circulating tumour DNA from plasma and urine of patients with colorectal cancer. Eur. J. Surg. Oncol. 47 (12), 3151–3156. doi:10.1016/j.ejso.2021.07.017
Oreskovic, A., Brault, N. D., Panpradist, N., Lai, J. J., and Lutz, B. R. (2019). Analytical comparison of methods for extraction of short cell-free DNA from urine. J. Mol. Diagn 21 (6), 1067–1078. doi:10.1016/j.jmoldx.2019.07.002
Oreskovic, A., Panpradist, N., Marangu, D., Ngwane, M. W., Magcaba, Z. P., Ngcobo, S., et al. (2021). Diagnosing pulmonary tuberculosis by using sequence-specific purification of urine cell-free DNA. J. Clin. Microbiol. 59 (8), e0007421. doi:10.1128/JCM.00074-21
Pages, M., Rotem, D., Gydush, G., Reed, S., Rhoades, J., Ha, G., et al. (2022). Liquid biopsy detection of genomic alterations in pediatric brain tumors from cell-free DNA in peripheral blood, CSF, and urine. Neuro Oncol. 24 (8), 1352–1363. doi:10.1093/neuonc/noab299
Pan, X., Wang, J., Zhang, Y., Dong, P., Li, C., and Liang, X. (2017). Detection of trace amounts of target DNA from massive background of nucleic acids by using the LM-PCR-based preamplification method. Biotechnol. Appl. Biochem. 64 (6), 879–887. doi:10.1002/bab.1545
Parackal, S., Zou, D., Day, R., Black, M., and Guilford, P. (2019). Comparison of Roche Cell-Free DNA collection Tubes ® to Streck Cell-Free DNA BCT ® s for sample stability using healthy volunteers. Pract. Lab. Med. 16, e00125. doi:10.1016/j.plabm.2019.e00125
Phelan, M. P., Reineks, E. Z., Schold, J. D., Hustey, F. M., Chamberlin, J., and Procop, G. W. (2018). Preanalytic factors associated with hemolysis in emergency department blood samples. Arch. Pathol. Lab. Med. 142 (2), 229–235. doi:10.5858/arpa.2016-0400-OA
Pittella-Silva, F., Chin, Y. M., Chan, H. T., Nagayama, S., Miyauchi, E., Low, S. K., et al. (2020). Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin. Chem. 66 (7), 946–957. doi:10.1093/clinchem/hvaa103
Polina, I. A., Ilatovskaya, D. V., and DeLeon-Pennell, K. Y. (2020). Cell free DNA as a diagnostic and prognostic marker for cardiovascular diseases. Clin. Chim. Acta 503, 145–150. doi:10.1016/j.cca.2020.01.013
Polini, B., Carpi, S., Romanini, A., Breschi, M. C., Nieri, P., and Podesta, A. (2019). Circulating cell-free microRNAs in cutaneous melanoma staging and recurrence or survival prognosis. Pigment. Cell Melanoma Res. 32 (4), 486–499. doi:10.1111/pcmr.12755
Postel, M., Roosen, A., Laurent-Puig, P., Taly, V., and Wang-Renault, S. F. (2018). Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev. Mol. Diagn 18 (1), 7–17. doi:10.1080/14737159.2018.1400384
Qi, T., Pan, M., Shi, H., Wang, L., Bai, Y., and Ge, Q. (2023). Cell-free DNA fragmentomics: the novel promising biomarker. Int. J. Mol. Sci. 24 (2), 1503. doi:10.3390/ijms24021503
Repiska, G., Sedlackova, T., Szemes, T., Celec, P., and Minarik, G. (2013). Selection of the optimal manual method of cell free fetal DNA isolation from maternal plasma. Clin. Chem. Lab. Med. 51 (6), 1185–1189. doi:10.1515/cclm-2012-0418
Rothwell, D. G., Smith, N., Morris, D., Leong, H. S., Li, Y., Hollebecque, A., et al. (2016). Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Mol. Oncol. 10 (4), 566–574. doi:10.1016/j.molonc.2015.11.006
Ruppert, T., Roth, A., Kollmeier, J., Mairinger, T., and Frost, N. (2023). Cell-free DNA extraction from urine of lung cancer patients and healthy individuals: evaluation of a simple method using sample volume up-scaling. J. Clin. Lab. Anal. 37 (21-22), e24984. doi:10.1002/jcla.24984
Salinas-Sanchez, A. S., Garcia-Olmo, D. C., Martinez-Sanchiz, C., Picazo-Martinez, M. G., Gimenez-Bachs, J. M., Flores-Bautista, A. B., et al. (2021). Clinical value of perioperative levels of DNA and mRNA in plasma of patients with renal cell carcinoma. Transl. Oncol. 14 (2), 100999. doi:10.1016/j.tranon.2020.100999
Salvianti, F., Gelmini, S., Costanza, F., Mancini, I., Sonnati, G., Simi, L., et al. (2020). The pre-analytical phase of the liquid biopsy. N. Biotechnol. 55, 19–29. doi:10.1016/j.nbt.2019.09.006
Schmid, M., White, K., Stokowski, R., Miller, D., Bogard, P. E., Valmeekam, V., et al. (2018). Accuracy and reproducibility of fetal-fraction measurement using relative quantitation at polymorphic loci with microarray. Ultrasound Obstet. Gynecol. 51 (6), 813–817. doi:10.1002/uog.19036
Schmidt, B., Reinicke, D., Reindl, I., Bork, I., Wollschlager, B., Lambrecht, N., et al. (2017). Liquid biopsy - performance of the PAXgene® Blood ccfDNA Tubes for the isolation and characterization of cell-free plasma DNA from tumor patients. Clin. Chim. Acta 469, 94–98. doi:10.1016/j.cca.2017.03.031
Schneck, E., Samara, O., Koch, C., Hecker, A., Padberg, W., Lichtenstern, C., et al. (2017). Plasma DNA and RNA differentially impact coagulation during abdominal sepsis-an explorative study. J. Surg. Res. 210, 231–243. doi:10.1016/j.jss.2016.11.044
Shao, W., Khin, S., and Kopp, W. C. (2012). Characterization of effect of repeated freeze and thaw cycles on stability of genomic DNA using pulsed field gel electrophoresis. Biopreserv Biobank 10 (1), 4–11. doi:10.1089/bio.2011.0016
Shen, Q., Cen, H., Jiang, J., Cong, Z., Zhou, Y., Huang, X., et al. (2022). The level and integrity of plasma circulating cell-free DNA in patients with primary multiple myeloma. Transl. Cancer Res. 11 (11), 4137–4147. doi:10.21037/tcr-22-2416
Shi, H., Li, Q., Liao, J., Bai, L., Wu, S., Xie, J., et al. (2020). Different primers for diagnosing circulating cell-free DNA of colorectal cancer. Transl. Cancer Res. 9 (5), 3435–3442. doi:10.21037/tcr-19-2017
Shin, K. H., Lee, S. M., Park, K., Choi, H., Kim, I. S., Yoon, S. H., et al. (2022). Effects of different centrifugation protocols on the detection of EGFR mutations in plasma cell-free DNA. Am. J. Clin. Pathol. 158 (2), 206–211. doi:10.1093/ajcp/aqac024
Shojaei-Far, Z., Razi, F., Bandarian, F., Rambod, C., and Qorbani, M. (2017). A detailed comparison of morning and random urine specimen levels with 24 hour urinary excretion levels of seven biochemical parameters with a proposed formula. Ann. Clin. Lab. Sci. 47 (2), 201–207.
Sigdel, T. K., Vitalone, M. J., Tran, T. Q., Dai, H., Hsieh, S. C., Salvatierra, O., et al. (2013). A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 96 (1), 97–101. doi:10.1097/TP.0b013e318295ee5a
Simundic, A. M., Bolenius, K., Cadamuro, J., Church, S., Cornes, M. P., van Dongen-Lases, E. C., et al. (2018). Joint EFLM-COLABIOCLI Recommendation for venous blood sampling. Clin. Chem. Lab. Med. 56 (12), 2015–2038. doi:10.1515/cclm-2018-0602
Song, P., Wu, L. R., Yan, Y. H., Zhang, J. X., Chu, T., Kwong, L. N., et al. (2022). Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 6 (3), 232–245. doi:10.1038/s41551-021-00837-3
Sorber, L., Zwaenepoel, K., Jacobs, J., De Winne, K., Goethals, S., Reclusa, P., et al. (2019). Circulating cell-free DNA and RNA analysis as liquid biopsy: optimal centrifugation protocol. Cancers (Basel) 11 (4), 458. doi:10.3390/cancers11040458
Sorber, L., Zwaenepoel, K., Jacobs, J., De Winne, K., Van Casteren, K., Augustus, E., et al. (2020). Specialized blood collection tubes for liquid biopsy: improving the pre-analytical conditions. Mol. Diagn Ther. 24 (1), 113–124. doi:10.1007/s40291-019-00442-w
Stejskal, P., Goodarzi, H., Srovnal, J., Hajduch, M., van 't Veer, L. J., and Magbanua, M. J. M. (2023). Circulating tumor nucleic acids: biology, release mechanisms, and clinical relevance. Mol. Cancer 22 (1), 15. doi:10.1186/s12943-022-01710-w
Streleckiene, G., Reid, H. M., Arnold, N., Bauerschlag, D., and Forster, M. (2018). Quantifying cell free DNA in urine: comparison between commercial kits, impact of gender and inter-individual variation. Biotechniques 64 (5), 225–230. doi:10.2144/btn-2018-0003
Sugimoto, A., Matsumoto, S., Udagawa, H., Itotani, R., Usui, Y., Umemura, S., et al. (2023). A large-scale prospective concordance study of plasma- and tissue-based next-generation targeted sequencing for advanced non-small cell lung cancer (LC-SCRUM-Liquid). Clin. Cancer Res. 29 (8), 1506–1514. doi:10.1158/1078-0432.CCR-22-1749
Szilagyi, M., Pos, O., Marton, E., Buglyo, G., Soltesz, B., Keseru, J., et al. (2020). Circulating cell-free nucleic acids: main characteristics and clinical application. Int. J. Mol. Sci. 21 (18), 6827. doi:10.3390/ijms21186827
Taly, V., Pekin, D., Benhaim, L., Kotsopoulos, S. K., Le Corre, D., Li, X., et al. (2017). Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 59 (12), 1722–1731. doi:10.1373/clinchem.2013.206359
Tang, Y., Zou, B., Wang, R., Luo, N., Qi, X., Zhou, G., et al. (2020). Multiplex-invasive reaction-assisted qPCR for quantitatively detecting the abundance of EGFR exon 19 deletions in cfDNA. Anal. Methods 12 (26), 3344–3350. doi:10.1039/d0ay00897d
Terp, S. K., Pedersen, I. S., and Stoico, M. P. (2024). Extraction of cell-free DNA: evaluation of efficiency, quantity, and quality. J. Mol. Diagn 26 (4), 310–319. doi:10.1016/j.jmoldx.2024.01.008
Thakur, B. K., Zhang, H., Becker, A., Matei, I., Huang, Y., Costa-Silva, B., et al. (2014). Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24 (6), 766–769. doi:10.1038/cr.2014.44
Thierry, A. R., El Messaoudi, S., Gahan, P. B., Anker, P., and Stroun, M. (2016). Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 35 (3), 347–376. doi:10.1007/s10555-016-9629-x
Trumpff, C., Marsland, A. L., Sloan, R. P., Kaufman, B. A., and Picard, M. (2019). Predictors of ccf-mtDNA reactivity to acute psychological stress identified using machine learning classifiers: a proof-of-concept. Psychoneuroendocrinology 107, 82–92. doi:10.1016/j.psyneuen.2019.05.001
Truszewska, A., Wirkowska, A., Gala, K., Truszewski, P., Krzemien-Ojak, L., Perkowska-Ptasinska, A., et al. (2020). Cell-free DNA profiling in patients with lupus nephritis. Lupus 29 (13), 1759–1772. doi:10.1177/0961203320957717
Ungerer, V., Bronkhorst, A. J., and Holdenrieder, S. (2020). Preanalytical variables that affect the outcome of cell-free DNA measurements. Crit. Rev. Clin. Lab. Sci. 57 (7), 484–507. doi:10.1080/10408363.2020.1750558
Uwiringiyeyezu, T., El Khalfi, B., Saile, R., Belhachmi, J., and Soukri, A. (2022). Comparability of CMV DNA extraction methods and validation of viral load. Methods Protoc. 5 (1), 6. doi:10.3390/mps5010006
Vagner, T., Spinelli, C., Minciacchi, V. R., Balaj, L., Zandian, M., Conley, A., et al. (2018). Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 7 (1), 1505403. doi:10.1080/20013078.2018.1505403
Vallee, A., Marcq, M., Bizieux, A., Kouri, C. E., Lacroix, H., Bennouna, J., et al. (2013). Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 82 (2), 373–374. doi:10.1016/j.lungcan.2013.08.014
van der Leest, P., Ketelaar, E. M., van Noesel, C. J. M., van den Broek, D., van Boerdonk, R. A. A., Deiman, B., et al. (2022). Dutch national round robin trial on plasma-derived circulating cell-free DNA extraction methods routinely used in clinical Pathology for molecular tumor profiling. Clin. Chem. 68 (7), 963–972. doi:10.1093/clinchem/hvac069
van Dessel, L. F., Vitale, S. R., Helmijr, J. C. A., Wilting, S. M., van der Vlugt-Daane, M., Oomen-de Hoop, E., et al. (2019). High-throughput isolation of circulating tumor DNA: a comparison of automated platforms. Mol. Oncol. 13 (2), 392–402. doi:10.1002/1878-0261.12415
van Dorp, J., Pipinikas, C., Suelmann, B. B. M., Mehra, N., van Dijk, N., Marsico, G., et al. (2023). High- or low-dose preoperative ipilimumab plus nivolumab in stage III urothelial cancer: the phase 1B NABUCCO trial. Nat. Med. 29 (3), 588–592. doi:10.1038/s41591-022-02199-y
van Ginkel, J. H., van den Broek, D. A., van Kuik, J., Linders, D., de Weger, R., Willems, S. M., et al. (2017). Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med. 6 (10), 2297–2307. doi:10.1002/cam4.1184
Van Paemel, R., De Koker, A., Caggiano, C., Morlion, A., Mestdagh, P., De Wilde, B., et al. (2021). Genome-wide study of the effect of blood collection tubes on the cell-free DNA methylome. Epigenetics 16 (7), 797–807. doi:10.1080/15592294.2020.1827714
Vermeulen, C., Geeven, G., de Wit, E., Verstegen, M., Jansen, R. P. M., van Kranenburg, M., et al. (2017). Sensitive monogenic noninvasive prenatal diagnosis by targeted haplotyping. Am. J. Hum. Genet. 101 (3), 326–339. doi:10.1016/j.ajhg.2017.07.012
Volckmar, A. L., Sultmann, H., Riediger, A., Fioretos, T., Schirmacher, P., Endris, V., et al. (2018). A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Genes Chromosom. Cancer 57 (3), 123–139. doi:10.1002/gcc.22517
Wall, P. L., Coughlin, O., Rometti, M. R. P., Birkholz, S., Gildemaster, Y., Grulke, L., et al. (2014). Tourniquet pressures: strap width and tensioning system widths. J. Spec. Oper. Med. 14 (4), 19–29. doi:10.55460/IT3C-9I89
Wang, K., Peng, Z., Lin, X., Nian, W., Zheng, X., and Wu, J. (2022). Electrochemical biosensors for circulating tumor DNA detection. Biosens. (Basel) 12 (8), 649. doi:10.3390/bios12080649
Wang, M., Huang, X., Li, X., Guo, Q., Xu, W., Zhao, M., et al. (2021). Performance comparison of commercial kits for isolating and detecting circulating tumor DNA. Scand. J. Clin. Lab. Invest. 81 (4), 276–281. doi:10.1080/00365513.2020.1821394
Ward Gahlawat, A., Lenhardt, J., Witte, T., Keitel, D., Kaufhold, A., Maass, K. K., et al. (2019). Evaluation of storage tubes for combined analysis of circulating nucleic acids in liquid biopsies. Int. J. Mol. Sci. 20 (3), 704. doi:10.3390/ijms20030704
Warton, K., Graham, L. J., Yuwono, N., and Samimi, G. (2018). Comparison of 4 commercial kits for the extraction of circulating DNA from plasma. Cancer Genet. 228-229, 143–150. doi:10.1016/j.cancergen.2018.02.004
Weng, J., Atyah, M., Zhou, C., and Ren, N. (2020). Prospects and challenges of circulating tumor DNA in precision medicine of hepatocellular carcinoma. Clin. Exp. Med. 20 (3), 329–337. doi:10.1007/s10238-020-00620-9
Wever, B. M. M., Bach, S., Tibbesma, M., Ter Braak, T. J., Wajon, D., Dickhoff, C., et al. (2022). Detection of non-metastatic non-small-cell lung cancer in urine by methylation-specific PCR analysis: a feasibility study. Lung Cancer 170, 156–164. doi:10.1016/j.lungcan.2022.06.013
Wolf-Doty, T. K., Mannon, R. B., Poggio, E. D., Hinojosa, R. J., Hiller, D., Bromberg, J. S., et al. (2021). Dynamic response of donor-derived cell-free DNA following treatment of acute rejection in kidney allografts. Kidney360 2 (4), 729–736. doi:10.34067/KID.0000042021
Wong, F. C., Sun, K., Jiang, P., Cheng, Y. K., Chan, K. C., Leung, T. Y., et al. (2016). Cell-free DNA in maternal plasma and serum: a comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin. Biochem. 49 (18), 1379–1386. doi:10.1016/j.clinbiochem.2016.09.009
Wu, J., Dai, W., Wu, L., Li, W., Xia, X., and Wang, J. (2019). Decoding genetic and epigenetic information embedded in cell free DNA with adapted SALP-seq. Int. J. Cancer 145 (9), 2395–2406. doi:10.1002/ijc.32206
Wu, Z., Yu, L., Hou, J., Cui, L., Huang, Y., Chen, Q., et al. (2022). Plasma cfDNA for the diagnosis and prognosis of colorectal cancer. J. Oncol. 2022, 9538384. doi:10.1155/2022/9538384
Xiao, Y., Ju, L., Qian, K., Jin, W., Wang, G., Zhao, Y., et al. (2022). Non-invasive diagnosis and surveillance of bladder cancer with driver and passenger DNA methylation in a prospective cohort study. Clin. Transl. Med. 12 (8), e1008. doi:10.1002/ctm2.1008
Xu, F., Yu, S., Han, J., Zong, M., Tan, Q., Zeng, X., et al. (2021). Detection of circulating tumor DNA methylation in diagnosis of colorectal cancer. Clin. Transl. Gastroenterol. 12 (8), e00386. doi:10.14309/ctg.0000000000000386
Yao, W., Mei, C., Nan, X., and Hui, L. (2016). Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene 590 (1), 142–148. doi:10.1016/j.gene.2016.06.033
Yin, J., Xia, L., Zou, Z., Zhuang, J., and Mu, Y. (2022). A direct and multiplex digital PCR chip for EGFR mutation. Talanta 250, 123725. doi:10.1016/j.talanta.2022.123725
Yokota, M., Tatsumi, N., Nathalang, O., Yamada, T., and Tsuda, I. (1999). Effects of heparin on polymerase chain reaction for blood white cells. J. Clin. Lab. Anal. 13 (3), 133–140. doi:10.1002/(sici)1098-2825(1999)13:3<133::aid-jcla8>3.0.co;2-0
Yu, Y., Qian, J., Shen, L., Ji, W., and Lu, S. (2021). Distinct profile of cell-free DNA in malignant pleural effusion of non-small cell lung cancer and its impact on clinical genetic testing. Int. J. Med. Sci. 18 (6), 1510–1518. doi:10.7150/ijms.52306
Yuwono, N. L., Alonso, A., Abbott, J., Houshdaran, S., Henry, C. E., Rodgers, R., et al. (2022). Circulating cell-free endometrial DNA level is unaltered during menstruation and in endometriosis. Hum. Reprod. 37 (11), 2560–2569. doi:10.1093/humrep/deac198
Zeng, S., Ying, Y., Xing, N., Wang, B., Qian, Z., Zhou, Z., et al. (2020). Noninvasive detection of urothelial carcinoma by cost-effective low-coverage whole-genome sequencing from urine-exfoliated cell DNA. Clin. Cancer Res. 26 (21), 5646–5654. doi:10.1158/1078-0432.CCR-20-0401
Zhang, H., He, B., Cui, J., Zhao, M., and Zhang, Z. (2018). Comparison of circulating DNA from plasma and urine for EGFR mutations in NSCLC patients. Cancer Biomark. 23 (3), 427–436. doi:10.3233/CBM-181511
Zhang, S., Zhou, D., Li, S., Bai, Y., Huang, B., Han, J., et al. (2022). Performance of ImproGene cfDNA blood collection tubes for mutation analysis in cancer patients. Scand. J. Clin. Lab. Invest. 82 (5), 378–384. doi:10.1080/00365513.2022.2100272
Zhao, Y., Li, Y., Chen, P., Li, S., Luo, J., and Xia, H. (2019). Performance comparison of blood collection tubes as liquid biopsy storage system for minimizing cfDNA contamination from genomic DNA. J. Clin. Lab. Anal. 33 (2), e22670. doi:10.1002/jcla.22670
Zhou, Q., Liu, F., Guo, L., Chen, R., Yuan, X., Li, C., et al. (2021). A novel urine cell-free DNA preservation solution and its application in kidney transplantation. Nephrol. Carlt. 26 (8), 684–691. doi:10.1111/nep.13884
Zhu, J., Hui, F., Mao, X., Zhang, S., Qi, H., and Du, Y. (2021a). cfDNA deconvolution via NIPT of a pregnant woman after bone marrow transplant and donor egg IVF. Hum. Genomics 15 (1), 14. doi:10.1186/s40246-021-00311-w
Keywords: cell-free DNA (cfDNA), preanalytical variables, standardization, blood, urine
Citation: Peng H, Pan M, Zhou Z, Chen C, Xing X, Cheng S, Zhang S, Zheng H and Qian K (2024) The impact of preanalytical variables on the analysis of cell-free DNA from blood and urine samples. Front. Cell Dev. Biol. 12:1385041. doi: 10.3389/fcell.2024.1385041
Received: 11 February 2024; Accepted: 22 April 2024;
Published: 09 May 2024.
Edited by:
Maria Rachele Ceccarini, University of Perugia, ItalyReviewed by:
Filippo Martignano, Prevention and Clinical Network (ISPRO), ItalyIvana Kurelac, University of Bologna, Italy
Aurelien Bancaud, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2024 Peng, Pan, Zhou, Chen, Xing, Cheng, Zhang, Zheng and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Zheng, emgtdXJvbG9neUB3aHUuZWR1LmNu; Kaiyu Qian, cWt5MTAwOUB3aHUuZWR1LmNu
†These authors have contributed equally to this work
 Hongwei Peng
Hongwei Peng Ming Pan2†
Ming Pan2† Hang Zheng
Hang Zheng Kaiyu Qian
Kaiyu Qian