- 1Division of Clinical Medicine, School of Medicine and Population Health, The Medical School, University of Sheffield, Sheffield, United Kingdom
- 2Centre of Preclinical Science Studies, Faculty of Dentistry, University Teknologi MARA (UiTM), Sg. Buloh, Selangor, Malaysia
- 3Institute of Veterinary Medicine and Animal Sciences, Estonian University of Life Sciences, Tartu, Estonia
- 4Department of Pathophysiology, Institute of Biomedicine and Translational Medicine, Faculty of Medicine, University of Tartu, Tartu, Estonia
- 5Department of Hormonal Action Mechanisms, Institute of Animal Reproduction and Food Research, Polish Academy of Sciences, Olsztyn, Poland
- 6Department of Animal Science, Faculty of Agriculture, University of Peradeniya, Peradeniya, Sri Lanka
The growing understanding of the role of extracellular vesicles (EVs) in embryo-maternal communication has sparked considerable interest in their therapeutic potential within assisted reproductive technology, particularly in enhancing implantation success. However, the major obstacle remains the large-scale production of EVs, and there is still a gap in understanding how different culture systems affect the characteristics of the EVs. In the current study, trophoblast analogue human chorionic carcinoma cell line was cultivated in both conventional monolayer culture (2D) and as spheroids in suspension culture (3D) and how the cell growth environment affects the physical, biochemical and cellular signalling properties of EVs produced by them was studied. Interestingly, the 3D system was more active in secreting EVs compared to the 2D system, while no significant differences were observed in terms of morphology, size, and classical EV protein marker expression between EVs derived from the two culture systems. There were substantial differences in the proteomic cargo profile and cellular signalling potency of EVs derived from the two culture systems. Notably, 2D EVs were more potent in inducing a cellular response in endometrial epithelial cells (EECs) compared to 3D EVs. Therefore, it is essential to recognize that the biological activity of EVs depends not only on the cell of origin but also on the cellular microenvironment of the parent cell. In conclusion, caution is warranted when selecting an EV production platform, especially for assessing the functional and therapeutic potential of EVs through in vitro studies.
1 Introduction
Extracellular vesicles (EVs) have emerged as important mediators of intercellular communication in both humans and animals (Pitt et al., 2016; Gurunathan et al., 2022). The inherent properties of EVs in cellular communication hold great potential for diagnostic and therapeutic applications across a spectrum of diseases, including cancer (Bell et al., 2016), regenerative disorders (Ramasubramanian et al., 2019), cardio vascular (Chong et al., 2019), and infectious diseases (Kumar et al., 2020) etc. The growing recognition of the role of EVs in the process of embryo implantation has presented promising prospects for their potential utility in improving pregnancy rates in assisted reproduction technologies (ARTs) (Poh et al., 2023).
Embryo implantation is a highly co-ordinated process that initiates with molecular signalling from the embryo, followed by its apposition, attachment, and invasion in to the receptive uterine epithelium (Kim and Kim, 2017). Despite rapid advances in ARTs and reproductive medicine, the inability of the embryo to implant remains the bottleneck to successful pregnancy (Ma et al., 2023). Evidently, success of embryo implantation depends on three main factors, namely, the quality of the embryo (della Ragione et al., 2007; Ahlstrom et al., 2011; Oron et al., 2014), the endometrial receptivity (Achache and Revel, 2006) and the synchronized crosstalk between the two (Idelevich and Vilella, 2020). Despite the presence of a good quality embryo and an optimally receptive endometrium, inadequate communication between these two can result in pregnancy failure. This reciprocal communication between conceptus and endometrium is now widely believed to be facilitated by paracrine signalling (Guzeloglu-Kayisli et al., 2009; Yockey and Iwasaki, 2018). While soluble factors such as cytokines and growth factors have been critical in this interplay, the role of EVs is also starting to come to light (Godakumara et al., 2021; Hart et al., 2023; Muhandiram et al., 2023). EVs are a heterogeneous group of nano-sized particles (30–5,000 nm in size) secreted by almost all cell types into the extracellular environment. They play vital functions, including the maintainance of cellular homeostasis (Takahashi et al., 2017; Raposo and Stahl, 2019), and intercellular signalling in different physiological and pathological conditions. EVs carry a range of bioactive cargoes including proteins, lipids, and nucleic acids, to neighbouring or distant recipient cells (Raposo and Stahl, 2019; Cocks et al., 2021). EV molecular cargo is known to exert various functions within recipient cells. Recent studies propose that EVs secreted by the embryo might alter the uterine epithelium rendering it more receptive (Latifi et al., 2018; Kurian and Modi, 2019; Godakumara et al., 2021; Hart et al., 2022) and endometrial cell derived EVs in turn can affect the growth and development of the embryo (Mishra et al., 2021; Segura-Benítez et al., 2022; Aguilera et al., 2024). These unique characteristics of EVs hold great potential for both diagnostic and therapeutic applications in contemporary assisted reproductive practices, encompassing both human and livestock (Dissanayake et al., 2020).
Investigating the in vivo human embryo implantation process poses significant challenges due to the inaccessibility to the real in vivo environment and ethical concerns related to obtaining biological samples. The outer most layer of the pre-implanting embryo (blastocyst) consists with a layer of trophectoderm cells (responsible for blastocyst hatching, endometrial attachment and placentation) and the quality of these cells are known to correlate highly with the pregnancy progression in humans (Garcia-Belda et al., 2024). The EVs derived from trophoblast cells have shown to support endometrial receptivity and embryo attachment (Es-Haghi et al., 2019; Godakumara et al., 2021; Muhandiram et al., 2023). Trophoblastic EVs also play a role in facilitating epithelial-to-mesenchymal transition of endometrial cells and modulating inflammation during the pre-implantation period (by interacting with immune cells to mitigate maternal rejection of the embryo) (Shi et al., 2021). On the other hand, EV-encapsulation with specific molecules can be utilized to exert diverse effects in the endometrium (Taravat et al., 2023). Therefore, the characteristics of EVs derived from trophectoderm cells in their natural form or as engineered vesicles are of interest in investigating potential clinical interventions for implantation failure. Despite the substantial interest in EVs as a therapeutic and diagnostic modality, the large-scale generation of natural EVs remains a significant challenge (Syromiatnikova et al., 2022).
Cell lines are an attractive option for producing trophoblastic EVs on a large scale due to their ease of manipulation and ready availability. One of the most commonly utilized immortalized cell lines serving as an analogue of trophoblast cells is the human choriocarcinoma JAr cell line (Weber et al., 2021). JAr cells demonstrates significant similarities to human trophoblast cells in terms of hormone production and their morphology in general. The cell line is known to produce placental human chronic gonadodrophin (hCG), estrogen, and progesterone hormones in both monolayer and spheroid culture mimicking a pre-implanting embryo (White et al., 1988; King et al., 2000). For studies involving EVs, cells can be cultivated using three different culture techniques: 2D monolayer culture, 3D culture with scaffolds, and 3D culture without scaffolds (Langhans, 2018; Chen and Wang, 2020). The choice of cell culture method depends on the specific application. The trophectoderm of the pre-implantation embryo can be easily mimicked using simple spheroids generated from JAr cells (3D) in suspension culture (Ho et al., 2013; Weimar et al., 2013; Green et al., 2015; Li et al., 2017). In general, the scalability of 3D culture makes it easily adaptable for large-scale production of EVs as well. The 3D cultures are also known to better mimic cell-cell and cell-extracellular-matrix (ECM) interactions in vivo while replicating nutrient and waste disposal in cells in a spatio-temporal manner (Huh et al., 2011; Syromiatnikova et al., 2022). The main differences between 2D and 3D cell culture systems can be seen in cell shapes, nutrient distribution, formation of cellular junctions, cell proliferation rates, responsiveness to stimuli, and gene or protein expression profiles. Moreover, variations can occur in the culture duration and reproducibility (Vantangoli et al., 2015). These differences can affect the cellular functions as well. On the other hand, EVs also exhibit remarkable heterogeneity, with their molecular characteristics predominantly shaped by the originating cells, cellular microenvironment, and the metabolic state of the parent cells (Riazifar et al., 2017; Kalluri and LeBleu, 2020). Hence, it is critical to exam how the culture conditions (static or flow culture), fluid flow dynamics in suspension cultures, and the pattern of cell-matrix interactions affect the production of EVs. These factors may influence not only the cells themselves but also the EVs they produce, ultimately affecting their ability to modulate recipient cell functions. To study this, we first evaluated the impact of 2D and 3D cell culture micro-environment on the physico-chemical characteristics and proteome composition of EVs released by trophoblast cells.
However, there are no proper indicators to evaluate the functionality of trophoblastic EVs in embryo implantation. In a previous study, we demonstrated that increased secretion of Milk fat globule-EGF factor 8 protein (MFGE8 or human lactadherin) serves as a cellular response of EECs to trophoblast-derived EVs (Muhandiram et al., 2023), thereby functioning as a marker for EV signalling during embryo implantation. MFGE8 is a glycoprotein rich in cysteine (Wang, 2014). Interestingly, it displays changes in expression in the endometrium during the menstrual cycle and appears to be associated with endometrial receptivity (Bocca et al., 2012). MFGE8 in EECs is also known as required for embryo attachment to the endometrium demonstrating its functional significance in embryo implantation (Schmitz et al., 2014). Therefore, we used MFGE8 to examine the impact of 2D and 3D EVs on cellular signalling in EECs. Hence, in the current study we further evaluated the biological implications of JAr-EVs produced in 2D and 3D culture systems and their differences in function in an in vitro embryo implantation model.
2 Materials and methods
2.1 Cell culture and spheroid formation
The human choriocarcinoma cell line (JAr) and human endometrial adenocarcinoma cell line (RL95-2) were purchased from the American Type Culture Collection (HTB-144™, Teddington, United Kingdom). JAr cell line was originally established from the trophoblastic tumour of the placenta of a 24 year old woman and the cell line is from a male fetus (passage 55). JAr cells (HTB-144™, Teddington, UK) were cultured in T75 flasks with RPMI 1640 medium supplemented with 10% Fetal Bovine serum (FBS), 1% Penicillin Streptomycin (P/S) and 1% L-glutamine in 5% CO2 at 37°C. The media was changed every second day until the cells reached 80% confluency. For 2D monolayer culture, cells were harvested and 1 × 106 cells were cultured in 5 mL of supplemented RPMI 1640 medium in 60 mm petri dishes for 48 h. Next, the cells were washed with Dulbecco’s phosphate-buffered saline without Ca+ 2 and Mg+ 2 (DPBS, Verviers, Belgium) and changed to FBS free medium. Cell culture conditioned medium (CM) was collected after 6 h.
JAr spheroids were formed as described previously with some modifications (Es-Haghi et al., 2019). In brief, JAr cells at 80% confluency were washed with DPBS, harvested using trypsin-EDTA (Gibco® Trypsin, New York, United States) and pelleted by centrifugation at 250 g for 5 min. Next, 1 × 106 cells were cultured in 5 mL of supplemented RPMI 1640 medium in 60 mm petri dishes on a gyratory shaker (Biosan PSU-2 T, Riga, Latvia), set at 295 rotations per minute (rpm) at 5% CO2 at 37 °C for 48 h to form spheroids (Supplementary Figure S1A). Then, spheroids were washed, replaced with FBS free media and CM was collected after 6 h. The spheroid viability was confirmed using Live/dead® viability/cytotoxicity assay kit (Molecular Probes, Eugene, Oregon, United States), according to the manufacturer’s instructions (Supplementary Figure S1B).
RL95-2 cells were used as an analogue of receptive endometrial epithelial cells (EECs). Cells were maintained in Dulbecco’s Modified Eagles medium F12 (DMEM 12-604F, Lonza, Verviers, Belgium) supplemented with 10% FBS (Gibco™, 10500064), 1% P/S (Gibco™, 15140122, Bleiswijk, Netherlands), and 5 μg/mL insulin (human recombinant insulin, Gibco™, Invitrogen, Denmark) in 5% CO2 at 37°C.
2.2 EV isolation and characterization
EV isolation was carried out using methods described previously (Es-Haghi et al., 2019; Godakumara et al., 2021). EVs were harvested from CM collected from both 2D monolayer culture and 3D JAr spheroids (100 mL each). Soon after collection, CM was centrifuged at 400 g for 10 min. The resulting supernatant was further centrifuged at 4,000 g for 10 min and thereafter at 10,000 g for 10 min to remove cell debris and apoptotic bodies. Samples were then directly processed for EV isolation. CM was concentrated to 500 µL using Amicon® Ultra-15 centrifugal filter devices (10 kDa cut-off). Next, EVs were purified using size exclusion chromatography (SEC). A gel filtration medium consisting of 4%–6% agarose matrix was used in 15 cm length columns to separate EV fractions from contaminating proteins. Fractions 7–10 were collected (each fraction was 500 µL in volume) and concentrated again to a total volume of 500 µL using a Amicon® Ultra centrifugal filter device with a 10 kDa cut-off and stored in −80°C until further analysis. Characterisation of the isolated EVs was carried out using methods described in detail elsewhere (Es-Haghi et al., 2019; Midekessa et al., 2020; Godakumara et al., 2021). In summary, the nanoparticle size and concentration in EV fractions were measured using Nano Particle Tracking Analyser (NTA) (Particle Metrix GmbH, Inning am Ammersee, Germany). Transmission electron microscopy (TEM) was used for the physical characterization of EVs. Enrichment of EV protein markers was confirmed by label free proteomic analysis of 2D and 3D EVs. EVs derived from JAr 2D monolayer are referred to as 2D EVs. EVs from JAr spheroids are referred to as 3D EVs.
2.3 Fluorescence–nanoparticle tracking analysis (FL-NTA)
FL-NTA of 3D and 2D JAr EVs was carried out to measure the size and concentration of EVs following staining them with the flouroscent stain CMG (CellMask™ Green Plasma Membrane Staining, Thermo Fisher Scientific, Waltham, MA, United States), as described previously, with some modifications (Midekessa et al., 2021). In summary, JAr EVs purified in SEC were diluted separately in 1 × PBS to a particle concentration of about 1 × 1010 particles/mL. Before incubating EVs with CMG dye molecules, 1 µL of 5 mg/mL CMG stock (CellMask™ Green Plasma Membrane Staining, Thermo Fisher Scientific, Waltham, MA, United States) was added to 50 µL of PBS. Then, 1 µL of CMG in 1 × PBS was added to 10 µL diluted EVs and incubated at RT for an hour on a shaker at 350 rpm. All experimental tubes were covered with aluminium foil during incubation. After the incubation, the incubated samples were added to 990 µL of 1 × PBS suspension medium to achieve a final volume of 1 mL with a pH value of 7.2. The size and concentration profiles of both 2D and 3D JAr EVs were measured in the fluorescence mode using a ZetaView PMX 120 V4.1 instrument (Particle Metrix GmbH, Ammersee, Bavaria, Germany). Before the measurements, auto-alignment of the Instrument was performed using a known concentration of 100 nm polystyrene (PS) and fluorescent Yellow Green (YG) nanoparticles (Applied Microspheres B.V., Leusden, Utrecht, The Netherland). Subsequently, the particle concentration and size distribution were measured in triplicate with the following settings; Sensitivity:72, shutter: 100 and frames per cycle: 11. Number of particles were reported in both scatter mode (t-NPs) and florescent mode (Fl-NPs). PBS buffer-only and dye control were used to detect labelling artefacts before measurements.
2.4 Measurement of Zeta Potential (ZP)
EVs carry a net negative charge on the surface, and the ZP is a measure of the surface potential. Importantly, ZP is an indicator of colloidal stability and nanoparticle stability which are important parameters for EV pharmacokinetics. The zeta potential of EV preparations was measured using methods described previously with slight modifications (Midekessa et al., 2020, 2021). The zeta potential (ZP) of EVs was measured using ZetaView PMX 110 V3.0 instrument (Particle Metrix GmbH, Germany), three times at 25°C under the following scatter and fluorescent settings: Sensitivity was set at 72, shutter value at 100, and frame rate at 30 frames per second. For the size and ZP measurement of fluorescently labelled EVs, the sensitivity was set at 90. Data were analyzed by ZetaView NTA software. PBS buffer control was used to eliminate artefacts.
2.5 Sample preparation for protein quantification with liquid chromatography-tandem mass spectrometry (LC-MS/MS)
EV or conditioned media samples were precipitated with trichloroacetic acid deoxycholate (TCA-DOC) precipitation overnight. Approximate protein quantities were estimated based on the size of the pellets. The pellets were then solubilized in 7 M urea, 2 M thiourea, 100 mM ammonium bicarbonate (ABC), 20 mM methylamine buffer. Protein reduction was performed with 5 mM dithiothreitol (DTT) by incubating 1 h at room temperature. Protein alkylation was performed with 10 mM chloroacetamide by incubating 1 h at room temperature in the dark. Next, protease LysC (Wako) was added to an enzyme:substrate ratio (E:S) of 1:50, and the samples were incubated for 1 h at room temperature. Samples were then diluted five times with 100 mM ABC, trypsin (Sigma Aldrich) was added to 1:50 E:S ratio and incubated overnight at room temperature. After digestion, samples were acidified with trifluoroacetic acid (TFA) to a concentration of 1%, and samples were desalted using in-house made C18 StageTips. Samples were reconstituted in 0.5% TFA and peptide concentrations were determined with a Pierce colorimetric peptide assay (Thermo Fisher Scientific).
For MS analysis, 1 µg of EV peptides was injected to an Easy-nLC 1000 system (Thermo Scientific). The sample was eluted at 250 nL/min from the trap to a 75 µm ID ×50 cm emitter-column (New Objective) packed with C18 material (3 μm, 300 Å particles, Dr Maisch). The separating gradient was 2%–35% B 60 min and 40%–100% B 5 min [A: 0.1% formic acid (FA), B: 80% ACN + 0.1% FA]. Eluted peptides were sprayed onto a Q Exactive Plus (Thermo Fisher Scientific) quadrupole-orbitrap mass spectrometer (MS) using nano-electrospray ionization at 2.4 kV (applied through liquid-junction). The MS was operated with a top-5 data-dependent acquisition strategy. Briefly, one 350–1,400 m/z MS scan at a resolution setting of R = 70,000 at 200 m/z was followed by five higher-energy collisional dissociation fragmentation (normalized collision energy of 26) of five most intense ions (z: +2 to +6) at R = 17,500. MS and MS/MS ion target values were 3e6 and 5e4 with 50 m injection time. Dynamic exclusion was limited to 40 s.
2.6 Database searching and protein identification
Mass spectrometric raw files were processed using the MaxQuant software package (versions 1.6.15.0 and 2.0.3.0) to identify proteins with their respective label-free quantification values. Methionine oxidation, asparagine and glutamine deamidation and protein N-terminal acetylation were set as variable modifications, while cysteine carbamidomethylation was defined as a fixed modification. Label-free protein quantification (LFQ) was enabled with LFQ and protein minimum ratio count was set to 1. The search was performed against Homo sapiens, Bos taurus reference proteomes, using the tryptic digestion rule. Peptide-spectrum match and protein false discovery rate (FDR) were kept below 1% using a target-decoy approach. All other parameters were set to default. The mass spectrometry data are available in the ProteomeXchange Consortium via the PRIDE with the dataset identifier PXD048789.
Differential protein analysis on the identified proteins was carried out in LFQ-analyst (Shah et al., 2020) platform. Data were normalized based on the assumption that the majority of proteins do not change between conditions. In summary, contaminated proteins, proteins identified “only by site” and reverse sequences were filtered out. Proteins identified only by a single peptide and those not consistently identified/quantified in the same condition were also removed. The LFQ protein intensity values were log2 transformed, and missing values were imputed using “missing not at random” method. Protein-wise linear models, combined with empirical Bayes statistics were used for the differential expression analyses. A cutoff of the adjusted p-value of 0.05 (Benjamini-Hochberg method), along with a log2 fold change of 1 was applied to determine significantly up and downregulated proteins between 3D and 2D EVs. Differentially expressed proteins in the heatmap were clustered using k-means clustering.
2.7 Functional annotation and pathway enrichment analysis
Biological significance of differentially expressed protein list was determined by performing functional annotation, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) online platform (Huang et al., 2009; Sherman et al., 2022). DAVID determined the proportion of genes with a specific GO or KEGG functional annotation in the differentially expressed gene list relative to the proportion of those genes in the genome. All relevant differentially enriched gene IDs were submitted to the DAVID Bioinformatics platform for GO and KEGG pathway enrichment analysis. Gene set enrichment analysis of KEGG pathways were performed using the clusterProfiler package in R (Yu et al., 2012; Wu et al., 2021). A complete protein list with log2 fold change and FDR was submitted to clusterProfiler to perform gene set enrichment analysis and visualization. FDR significance cut off was ≤ 0.05.
2.8 MFGE8 enzyme linked immunosorbent assay
Cell culture supernatants (1 mL of media) were centrifuged at 400 g for 10 min to remove any contaminating cells, followed by centrifugation at 4000 g for 10 min and 10,000 g for 10 min to remove other cellular debris and apoptotic bodies. Samples were snap frozen in liquid nitrogen before storing in −80°C. MFGE8 protein concentration was measured in cell culture supernatants using a commercially available ELISA kit (Human MFGE8 Quantikine ELISA Kit, R&D systems) according to the manufacturer’s instructions. The optical density of each well was measured using a microplate reader set to 450 nm (Multiskan FC microplate photometer, Life Technologies, China).
2.9 Preparation of EV depleted cell culture media
EV depletion in FBS was carried out using methods described previously (Godakumara et al., 2021). FBS was filtered using Amicon ultra-15 centrifugal filters (100 kDa, MERCK KGAA, Darmstadt, Germany) at 3,000 g for 55 min and then used as a 10% supplementation to prepare RL95-2 cell culture media.
2.10 Statistical analysis
The size and concentration of EVs were expressed as mean ± SD. Statistical analysis was performed using One-way ANOVA (for multiple comparisons) or unpaired t-test. p-values < 0.05 were considered statistically significant marked with an asterisk (*) symbol. All the experiments were performed in three biological replicates.
2.11 Experimental design
2.11.1 Determining the EV secretion from 2D vs 3D JAr culture
JAr cells were seeded in 1 × 106 cells in 5 mL of supplemented RPMI 1640 medium in 60 mm petri dishes. Cells were kept on a gyratory shaker (Biosan PSU-2 T, Riga, Latvia), set at 295 rotations per minute (rpm) at 5% CO2 in 37°C for 48 h to form spheroids (3D culture). Similarly JAr cells were grown monolayer culture by growing 1 × 106 cells in 5 mL of supplemented RPMI 1640 medium in 60 mm Petri dishes (2D culture) at 5% CO2 in 37°C for 48 h. AAfter 48 h, the JAr spheroids and monolayer cultures were washed with FBS-free complete medium. Next, the cell culture medium was replaced with FBS-free medium, and cells or spheroids were incubated for another 6 h before collecting the cell culture conditioned medium. Finally, cell culture supernatants were collected (100 mL each) and EVs were isolated. Particle concentration in EV samples were determined using nanoparticle tracking analysis.
2.11.2 Determining the physico-chemical characteristics of the EVs derived from 2D vs 3D culture
EVs from JAr cells grown in 3D and 2D microenvironments were isolated using sequential centrifugation followed by SEC. Then the physical and biochemical characteristics (size, concentration, zeta potential) of the EVs were further analysed using NTA and LC-MS/MS. The protein cargo of 2D EVs was compared with 3D EVs using LC-MS/MS.
2.11.3 Determining the MFGE8 secretion from receptive endometrial epithelial cells in response to 2D and 3D JAr EVs
MFGE8 secretion from EECs in response to 2D and 3D JAr EVs was determined using RL95-2 cells. The cells were seeded in 12 well plates (1 × 105 cells/mL) and grown until they reached 85% confluency. After reaching the desired confluency, the cells were washed with DPBS, and EVs derived from 2D and 3D JAr cells were supplemented at a concentration of 1 × 109 particles/mL in EV depleted medium and incubated for 24 h. Cell culture supernatants were collected at both 0 h and 24 h, and MFGE8 protein secretion from RL95-2 cells in response to 2D and 3D JAr EVs within 24 h was determined.
2.11.4 Determining the functional implications of 2D and 3D JAr EVs
Finally protein cargo of 3D and 2D JAr EVs were compared with proteins secreted by human embryos (proteins in human blastocoels) prior to implantation (Poli et al., 2015) and proteins related to embryo implantation (Díaz-Gimeno et al., 2011; Enciso et al., 2018).
3 Results
3.1 Isolation and characterization of 3D and 2D EVs from human trophoblast cells
EVs from 3D and 2D trophoblast cells were isolated and characterized using the methods described above. Total particle count was significantly higher in 3D EVs compared to 2D EVs (Figure 1A). The normalized EV size distribution graph illustrated that the majority of EVs produced by the two culture systems were predominantly in the 100–200 nm size range (Figure 1B). However, mean particle size was higher in 3D EVs compared to 2D EVs (Figure 1C). Next, membrane labelling of the nanoparticles was performed with CMG dye to characterize the EV population more specifically. The lipophilic membrane dye labelled a total of 55.81% of nanoparticles in the 3D EV samples and 53.77% in the 2D EV samples, respectively (Figure 1D). However, the average size of the labelled particles did not exhibit significant differences between 3D and 2D EVs (Figure 1E). The zeta potential of the 3D and 2D sample nanoparticles were similar. Interestingly, the zeta potential of the fluorescently labelled nanoparticles in 3D vs. 2D EVs was significantly different with 3D EVs having a more negative zeta potential (Figure 1F). TEM analysis demonstrated that both 3D and 2D nanoparticles were less than 200 nm in diameter and uniform in nature with typical EV characteristics such as dual membrane, cup shape, and circular cross section (Figures 1G,H). Mass spectrometry analysis of EV samples and their respective conditioned media samples showed that classical EV protein markers such as CD9, CD81, CD63, FLOT1, and TSG101 were enriched in both 3D and 2D EVs samples compared to their respective CM samples (Supplementary Table S1). Enrichment of EV protein markers in 3D and 2D EVs and CM samples were semi quantitatively visualized in the heat map (Figure 1I). In total 11 EV protein markers and 1 purity marker (LDHA) were shown in the current analysis, confirming that our EV samples are enriched with the majority of known EV proteins. LDHA did not show significant enrichment, suggesting the EV preparations mainly consists of EVs, with limited non EV protein contaminations. EV protein markers were selected according to the five-component framework of MISEV guidelines for reporting protein composition of EVs (Welsh et al., 2024).
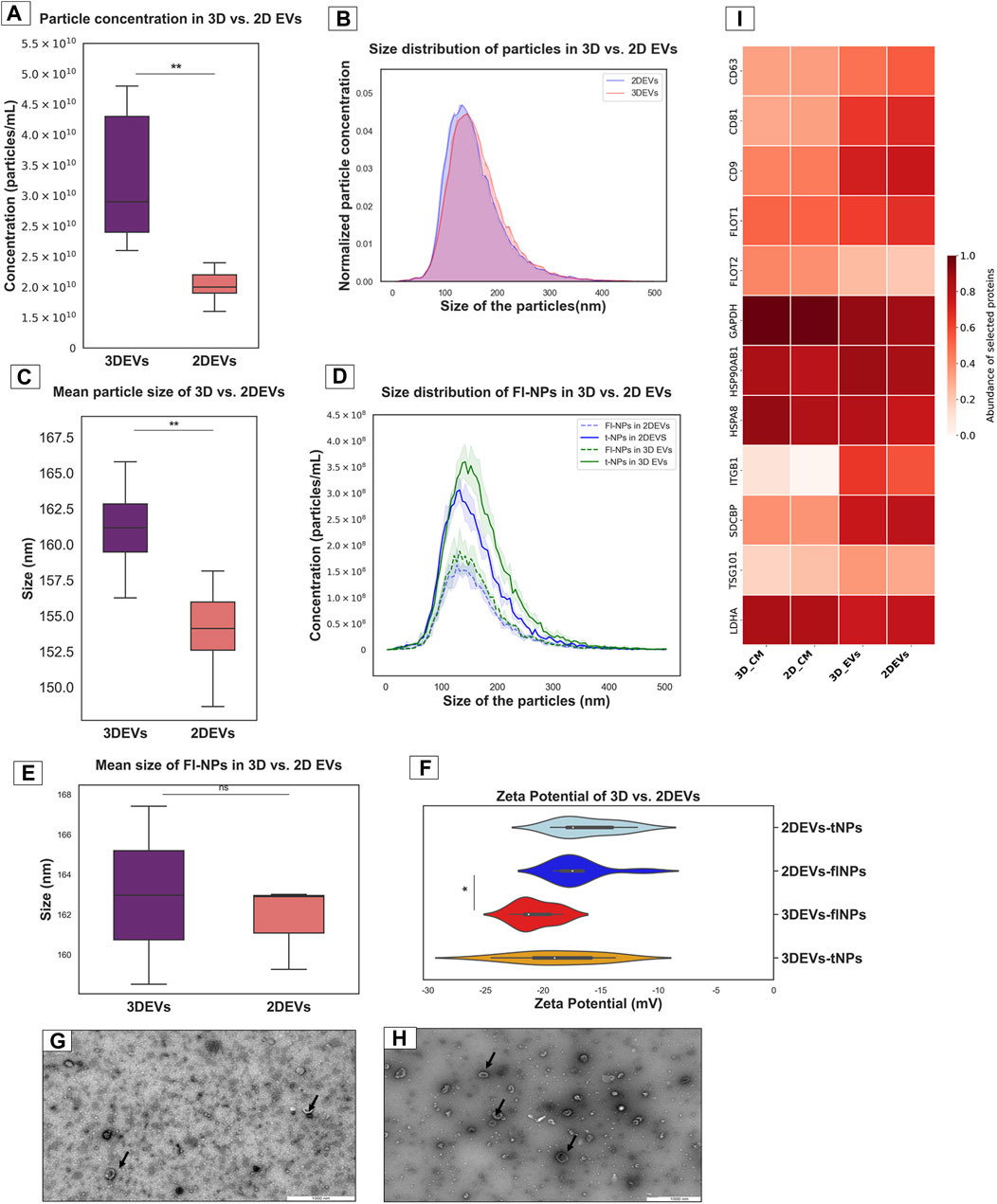
Figure 1. Characterization of nanoparticles derived from trophoblast cells grown in 3D and 2D cell culture systems as EVs (A). Total particle count of nanoparticles released by 3D vs. 2D EVs (B). The number and size profile of both 3D vs. 2D EVs exhibited a typical distribution of particles mainly less than 200 nm in size (C). Mean size of nanoparticles released by 3D vs. 2D EVs (D). Distribution of size and concentration of fluorescent nanoparticles released by 3D vs. 2D EVs (E). Mean size of fluorescent nanoparticles released by 3D vs. 2D EVs. t-NPs: total nanoparticles, fl-NPs: florescent nanoparticles. (F). Zeta potential of nanoparticles and fluorescently labelled nanoparticles in 3D vs. 2D EVs. tNPs: total nanoparticles, flNPs: florescent nanoparticles. EV morphology was assessed using TEM images of isolated EVs secreted from (G). 2D and (H). 3D cell culture systems. Scale bar: 1,000 nm. Black arrow shows EVs with typical characteristics (I). The heat map illustrates the presence and enrichment of specific proteins in 2D EVs compared to 2D CM (2D_CM) and 3D EVs compared to 3D CM (3D_CM). The proteins reported in the heat map show enrichment of standard EV markers in 2D and 3D EV preparations. The mean LFQ value for each protein corresponding to each sample type was log transformed and re-scaled before visualization in the heat map. All the experiments were conducted in biological triplicate (n = 3), data were presented as mean ± SD, and p < 0.05 considered statistically significant.
3.2 Proteomic composition of 3D vs 2D EVs
To gain insight in to whether the cell culture growing conditions affected the protein expression in 3D and 2D cell derived EVs, we analysed their protein cargo profile using label-free mass spectrometry. A total of 1,313 proteins were confidently detected with FDR< 0.01 in at least one of the three replicates of the 3D and 2D EV groups, where 1,204 proteins were detected in 3D EV and 1,159 proteins in 2D EV group, respectively. The principle component analysis and hierarchical clustering separated EVs derived from specific cell types, indicating global proteomic changes based on cell culture conditions (Figures 2A,B). Total of 1102 proteins were found to be common to both EV types. Interestingly, 102 proteins were uniquely detected in 3D EVs, whereas 57 proteins were uniquely detected in the 2D EV group. Of the 1,102 proteins that were common to both EV types, 80 were common with the top 100 EV proteins reported in Vesiclepedia database (Figure 2C). The 3D and 2D EV proteomic profile shared significant similarity with previously reported proteins profile of human trophectoderm stem cell derived EVs as well (Figure 2D). The commonly shared proteins included antioxidants (PRDX2, PRDX6), adhesion molecules (ITGB1, ITGB5) and cytoskeleton regulators (RAC1 and RHOA) etc. These proteins can play roles in antioxidant defences, cell adhesion and signal transduction processes which are considered important in embryo implantation (Poh et al., 2021). Differential expression analysis of the protein composition between 3D and 2D EVs resulted in 153 differentially expressed proteins (DEP). Of the 153 proteins (Supplementary Table S2), 120 proteins were upregulated, and 33 proteins downregulated. Among the 153 protein changes, 11 proteins are known to be classical EV proteins according to vesiclepedia (ANXA2, GNB1, SLC3A2, BSG, GNAS, RAB5C, RAC1, GNB2, ATP1A1, RAP1B, and RALA) (Figures 2E,F).
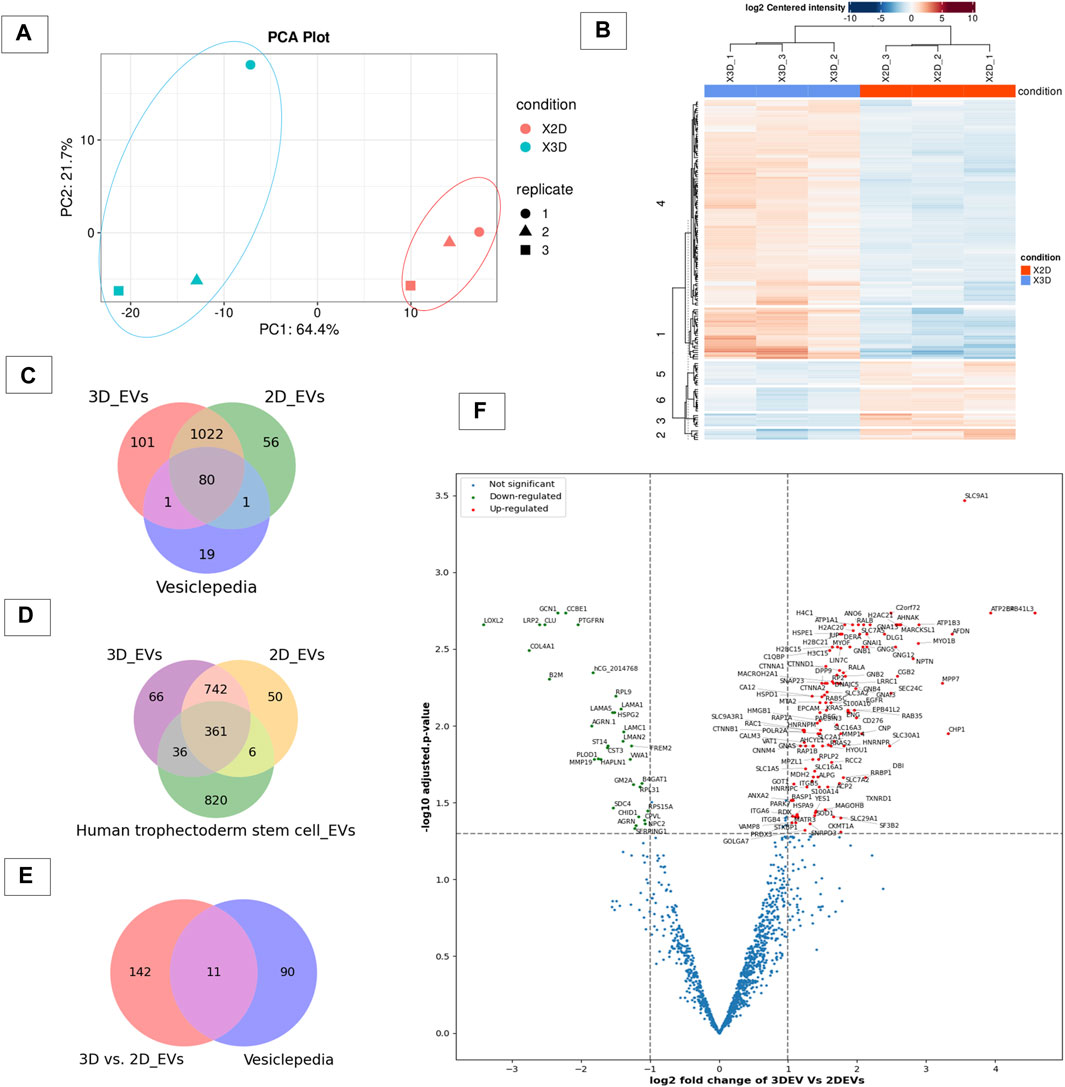
Figure 2. Proteomic profile of 3D vs 2D EVs. (A) Principle component analysis of protein composition of 3D vs. 2D EVs showing distinctive proteomic profiles. (B) Heat map of clustering among samples based on differentially enriched proteins between 3D and 2D EVs. (C) Venn diagram displaying the number of proteins identified in EVs derived from cells grown in 3D and 2D conditions and vesiclepedia most common proteins (top 100 proteins). (D) Venn diagram displaying the number of proteins identified in EVs derived from JAr cells grown in 3D and 2D conditions and proteins enriched in human trophectoderm stem cell derived EVs (Poh et al., 2021). (E) Venn diagram of EV proteins differentially expressed between 3D and 2D culture conditions and vesiclepedia top most common proteins (top 100 proteins). (F) Volcano plot showing differential expressed proteins between 3D and 2D EVs.
GO and KEGG pathway analysis provided insight in to the biological background of the DEP (Figures 3A,B). The significantly enriched (FDR< 0.05) GO biological terms (BP) revealed that cell-cell adhesion, protein localization to plasma membrane, and cell migration were enriched in EVs from 3D cells compared to 2D cells. For molecular function (MF), pathways such as GDP binding, cytoskeleton protein binding, and cadherin binding were enriched. The differentially expressed protein list was mapped to the KEGG database, and it was found that the identified proteins were enriched in 25 pathways (FDR< 0.05). Taken as a whole, pathways like ECM receptor interaction, adherens junctions, Ras signalling pathway, focal adhesion, regulation of actin cytoskeleton, and P13K-Akt signalling pathways were highly enriched in 3D EVs compared to 2D EVs (Supplementary Tables S3, S4).
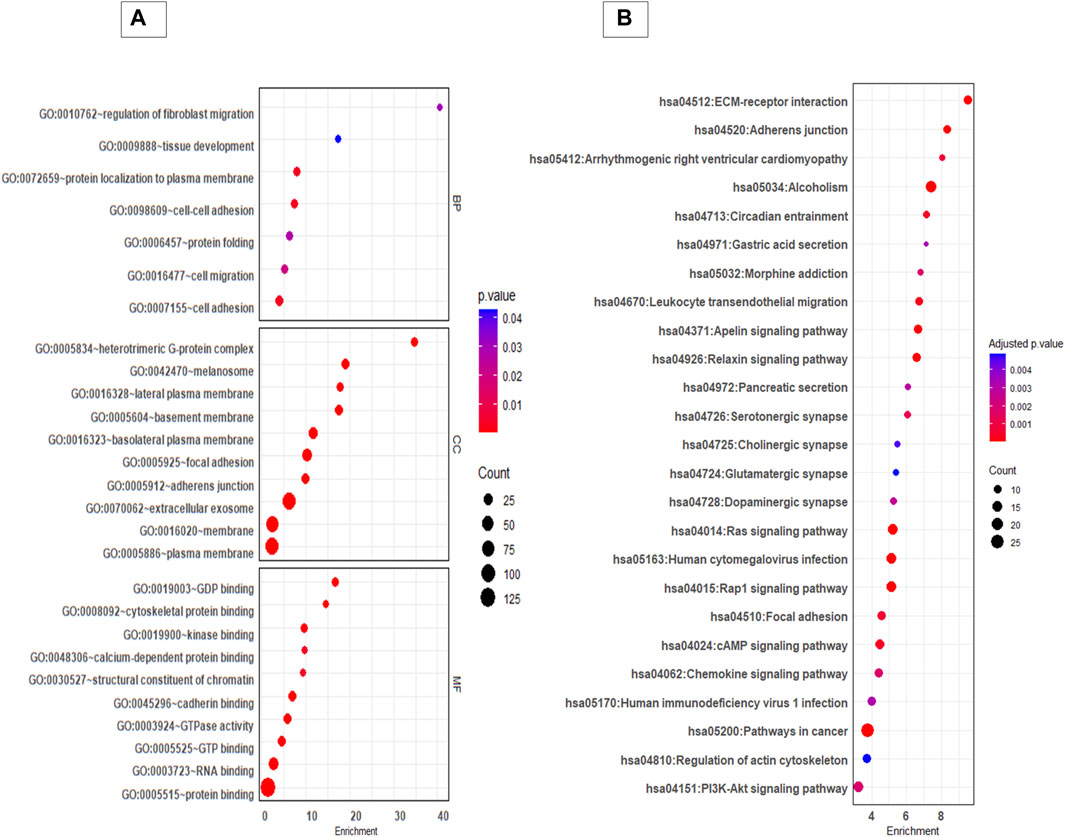
Figure 3. GO and KEGG pathway analysis of differentially expressed proteins between 3D and 2D EVs. (A) GO analysis of biological pathways (BP), cellular components (CC) and molecular function (MF). (B) KEGG enrichment pathways of differentially expressed proteins between 3D and 2D EVs. The colour of the bubble chart show the adjusted p. value and gene count in each term is represented by bubble size.
The gene set enrichment analysis of KEGG pathways (Supplementary Table S5) revealed pathways such as regulation of actin cytoskeleton, adherent junctions, Ras signalling and Rap signalling were profoundly upregulated in 3D EVs compared to 2D EVs, whereas pathways like cholesterol metabolism and galactose metabolism were downregulated (Figure 4).
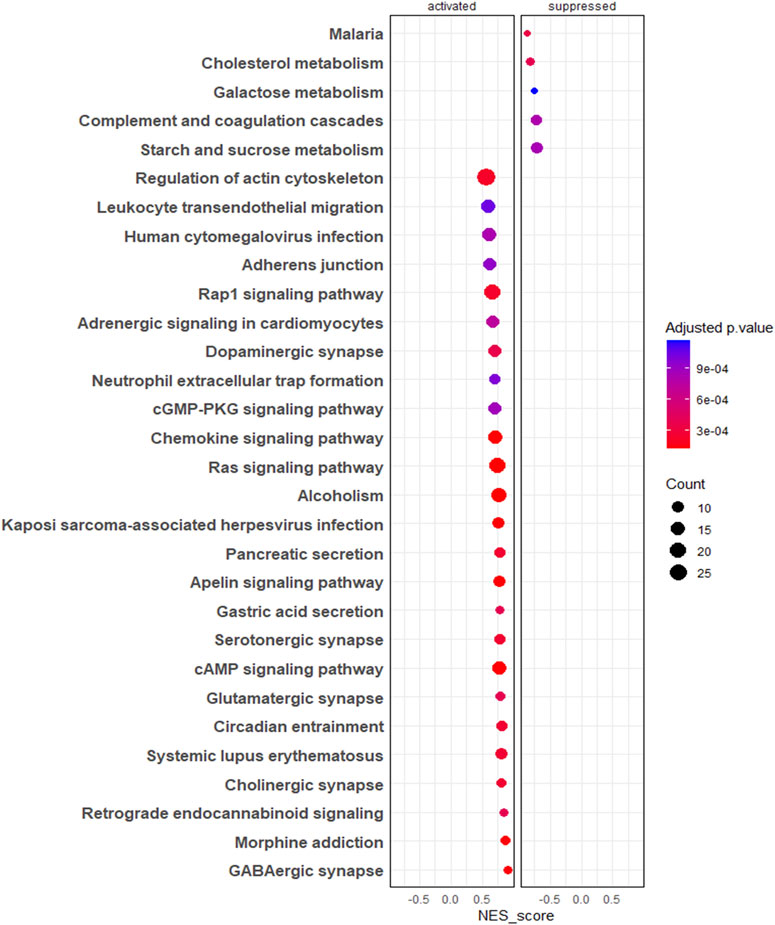
Figure 4. Gene set enrichment analysis (GSEA) of KEGG pathways. The colour of the bubble chart show the adjusted p. value and gene count in each term is represented by bubble size.
3.3 Endometrial epithelial cells sense the trophoblastic EV cargo changes
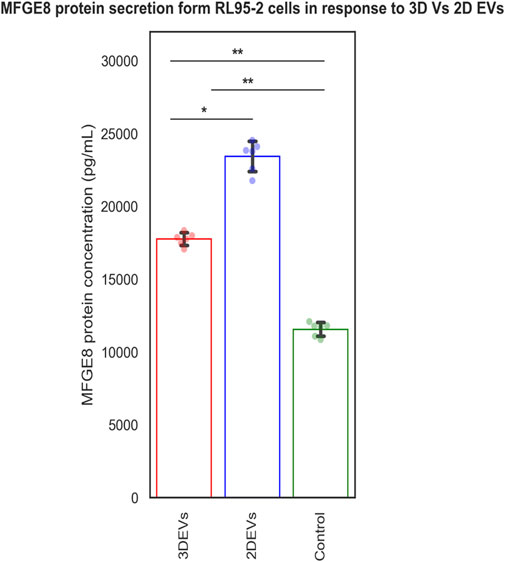
The MFGE8 secretion was increased in both 3D and 2D EV treated RL95-2 cells compared to its control. Increased MFGE8 secretion was seen in 2D EV treated RL95-2 cells compared to 3D EVs suggesting that 3D and 2D EVs have different potencies in trophoblast EV mediated cellular signalling (Figure 5).
3.4 Trophoblast cell derived extracellular vesicles may have the potential to prepare endometrium for embryo implantation
Trophoblasts cells are critical cellular players of embryo implantation. Here, we investigated whether the trophoblast analogue JAr cell derived EVs resemble the embryo secretions in humans (blastocoel proteome from embryos produced for in vitro fertilization). We identified around 115 proteins that are commonly found within embryo secretions and EVs secreted by trophoblastic cells (Supplementary Table S6). However, a total of 120 proteins were shared between 3D EVs and embryo secretions, whereas 117 proteins were shared between 2D EVs and embryo secretions. (Figure 6A). Among them, 115 proteins common to 2D EVs, 3D EVs and human embryo secretions. Some of these proteins were known to play roles in endometrial receptivity (ANXA2, CALR, STMN1) (Cheng et al., 2009; Gou et al., 2015; Yoshie et al., 2021), antioxidant defence (PRDX1, PRDX2) (Wu et al., 2017), trophoblast attachment and invasion by actin polymerization (TAGLN2) (Liang et al., 2019) etc. (Figure 6B).
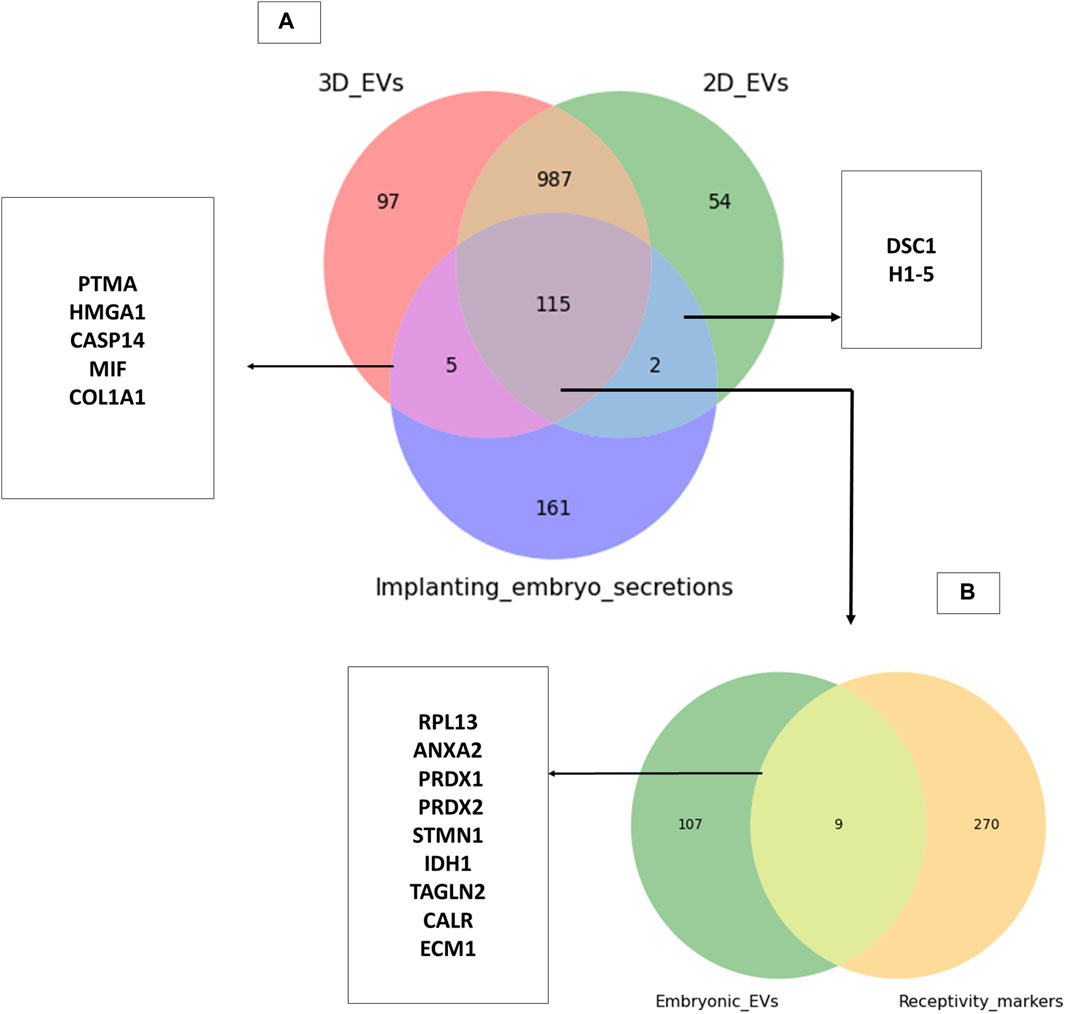
Figure 6. Common proteins between trophoblastic EVs and human embryo secretions. (A) Comparison of 3D and 2D trophoblast cell derived EVs with secretions of human embryos prior to implantation (Poli et al., 2015). (B) Comparison of proteins common to trophoblast EVs and human embryo secretions to known endometrial receptivity markers (Díaz-Gimeno et al., 2011; Enciso et al., 2018).
Through manual searching of the list of proteins (115) that were common to both trophoblast EVs and human embryo secretions, we discovered additional proteins that may play a role in the process of embryo implantation (Table 1). The common proteins for EVs and embryo secretions have been indicated in the process of embryo implantation and hence may have a therapeutic value in preparing the endometrium for embryo implantation.
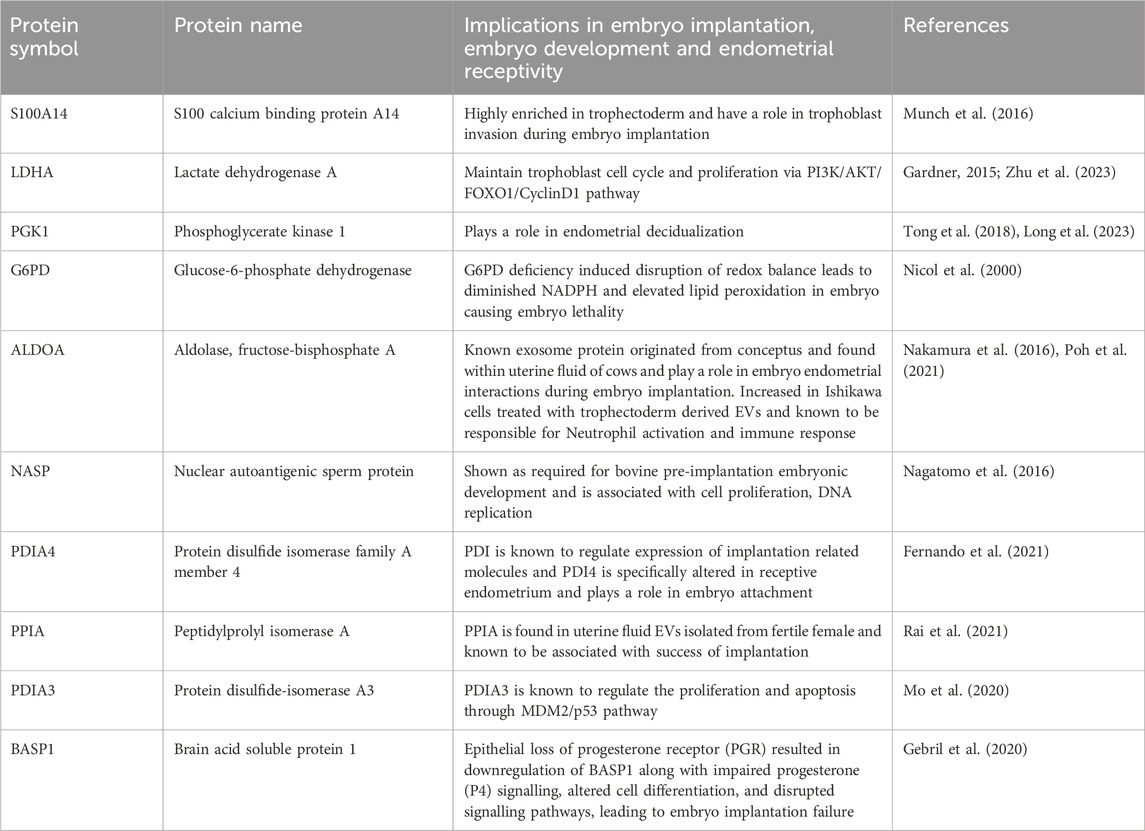
Table 1. Proteins commonly shared among pre-implanting single embryo secretions, 3D and 2D trophoblast EVs and their known role in the process of embryo implantation.
4 Discussion
Both 3D and 2D cultures are recognized as having applications in cancer research, drug discovery and in understanding physiological and pathological processes within biological systems. Three dimensional cultures are becoming more attractive in the field of drug discovery and tissue engineering owing to their ability to better mimic the in vivo cellular microenvironment. While the majority of EV studies have been conducted using EVs from 2D cell cultures, there is evidence suggesting that 2D and 3D cell secretomes and EV compositions differ. With the increasing interest in EVs as a therapeutic modality to improve the embryo implantation process, standardized production of EVs remains a challenge. While 3D cultures offer opportunities to scale up EV production, how the cellular micro-environment could affect EV production, bioactivity, and molecular composition remain elusive. Therefore, we studied whether the composition of EVs derived from cells grown in 3D and 2D microenvironment is different and how this affects the EV mediated cellular communication using an embryo implantation model. In the current study, we show that EV signalling can be tuned based on the proteomic composition of the EVs, which seems to depend on the cellular microenvironment where the donor cells were grown.
There are limited availability in primary tissues to isolate fresh trophoblast cells, therefore cell line derived from normal placenta, embryonal carcinomas and malignant tissue with evidence of trophoblast differentiation are imperative for EV production and functional studies. Some cell lines used as mimics of trophoblast cells are JAR, BeWo, and JEG-3, and these cell lines produce hormones and show cell growth similar to normal trophoblast cells (Grummer et al., 1994). These cell lines can be far from perfect, however, they can be used to generate significant interventional targets for clinics that can be later validated for more appropriate source materials or used for EV engineering purposes. Additionally, obtaining enough material for EV isolation from cell culture is challenging. EV studies typically employ three cell culture techniques, which includes, monolayer 2D culture, scaffold-based 3D culture, and scaffold-free 3D culture (Langhans, 2018; Chen and Wang, 2020). EV production can be scaled up using cell growth in bioreactors, induction of EV secretion from cells using stressors (by modulation of culture conditions, or by using physical and chemical stimulants) and by cell fragmentation methods (Syromiatnikova et al., 2022). The choice of cell culture method mostly depend on the specific application. In 2D monolayer cultures, cells grow flat in shape and are in contact with their neighbouring cells, cell culture media, and the culture vessel. However, they lack spatial polarization and cell-extracellular matrix (ECM) interactions (Kapałczyńska et al., 2016). In scaffold based 3D cultures, cells are basically in contact with the neighbouring cells, scaffold and cell culture media. In scaffold-free 3D cultures, cells are in contact with neighbouring cells and the culture medium. Non-scaffold-based 3D cultures are suspension cultures grown non-adherently to the culture surface such as hanging drop cultures, magnetic levitation and rotary cell cultures. Scaffold-based 3D cultures have concerns regarding biocompatibility and utility in clinical applications as they use non-human material. The cells in scaffold free culture self-assemble and secrete their own ECM, therefore allow more accurate cell-cell interactions, special organization and physiological responses (Vu et al., 2021). In general 3D culture can be easily scaled up for the large production of EVs, and rotary-shaker base suspension culture is an easy way of producing trophoblast spheroids for EV isolation (Huh et al., 2011; Syromiatnikova et al., 2022).
In the current study, we isolated EVs from trophoblast CM, comparing cells grown as a monolayer and spheroids from rotary shaker-based suspension culture. EVs can be isolated from source materials using different EV isolation techniques such as SEC, ultracentrifugation (UC), filtration, and precipitation methods (Welsh et al., 2024). Methods such as filtration and precipitation result in more protein contaminants and non EV particles in EV preparations. Therefore, the SEC and UC are common methods of EV isolation and can separate EVs from non EV particles and proteins to greater extent. Emerging evidence suggest that EVs produced by SEC has more intact biophysical properties and higher functionality compared to EVs from UC (Mol et al., 2017), therefore we used SEC to isolate EVs. Then, the physical and biochemical properties of the 3D and 2D EVs were characterized according to the guidelines of the Minimal information of Studies of EVs (Théry et al., 2018; Welsh et al., 2024). The secretion dynamics of EVs secreted by 2D vs 3D cells showed that 3D cells secreted more EVs compared to 2D cells, when the similar number cells and time in culture were used for producing EVs. The observed discrepancy in EV production rates between the two culture systems may stem from differences in cell proliferation or how cells are distributed within each culture model. These findings are consistent with the previous reports that suggested active secretion of EVs from 3D culture compared to 2D culture (Villasante et al., 2016; Rocha et al., 2019; Sun et al., 2023). This suggest the possibility of scaling up 3D cell culture for standardized and effective EV production (Guerreiro et al., 2018). While studying real-time EV secretion over extended periods could offer a more comprehensive understanding of EV production efficiency, current technological limitations hinder such investigations. Moreover, spheroids in culture for extended periods of time may start to develop a necrotic core that pose the risk of dying cells releasing EVs mainly consisting of apoptotic bodies rather than exosomes or microvesicles (Abdollahi, 2021). Majority of the EVs derived from both culture systems fell within the size range of 100–200 nm that is typical for small EVs, and TEM images showed that the EVs had a typical cup-shaped morphology. There were slight differences in the size of the particles derived from 2D culture vs 3D culture, where 3D-derived particles were slightly larger than the 2D derived particles. Previous reports also suggest that there can be size differences in EVs of 2D and 3D cell origin when measured with NTA (Thippabhotla et al., 2019; Clément et al., 2022). NTA used to measure the EV size and concentration is based on the principle of light scattering and Brownian motion; however, it is incapable of avoiding any non EV particles from their analysis. Techniques to more specifically label EVs based on antibodies or lipid membrane dyes have been emerged during recent years. To differentiate between EVs and non-EV particles, EVs can be labelled with lipid membrane dyes and then detected under fluorescent mode of the NTA (Midekessa et al., 2021). In contrast to the NTA results, fluorescent NTA showed that the size of the nanoparticles of EV origin derived from both 2D and 3D cells were not different. However, it is also important to keep in mind that this techniques can introduce labelling artefacts leading to over or underestimating the real EV numbers (Welsh et al., 2024). The zeta potential of fluorescent-labelled nanoparticles were different between 2D and 3D EVs, where 3D EVs showed a more negative charge. ZP is one of the most useful tool to investigate the colloidal stability of the nanoparticles, and this characteristics is important in nanoparticle stability and in understanding their pharmacokinetics while preparing nanomedicines (Midekessa et al., 2020). The EVs generated from 3D cells seem more stable compared to 2D EVs, further encouraging their suitability as an EV bio-manufacturing platform. Both 2D EVs and 3D EVs were enriched with classical EV protein markers such as CD9, CD81, CD63, and ITGB. Altogether, this confirmed that EVs could be isolated from both culture conditions and both EV types were of a typical cup shaped with similar mean size range and enriched with classical EV markers.
We next examined the effect of cell culture growth conditions on EV protein cargo composition. The cell growth conditions impacted the proteomic composition of EV cargo. Overall protein secretion was more active in 3D culture compared to 2D culture (more proteins identified in 3D EVs compared to 2D EVs), which was consistent with previous findings (Kusuma et al., 2022). The 2D and 3D EV proteome were clearly distinguishable, with around 153 proteins differentially expressed in 3D EVs compared to 2D EVs. Among the proteins identified, classical EVs proteins were also found. Both the 3D and 2D EV proteomic profiles shared proteins with those previously identified in human trophectoderm stem cell-derived EVs, indicating that the samples were indeed enriched with EVs. Among the differentially expressed proteins, 11 proteins were among the top most common EV proteins, suggesting that, EV cargo protein loading was different in 3D EVs compared to 2D EVs. Interestingly, among these proteins ANXA2 (Garrido-Gómez et al., 2012; Wang and Shao, 2020), SLC3A2 (Poh et al., 2021), and RAC1 (Tu et al., 2016) were identified as critical for embryo adhesiveness to the human endometrium. The key processes of proteins specially enriched in 3D EVs compared to 2D EVs were involved in cell-cell adhesion, cell migration, ECM receptor interaction, regulation of actin-cytoskeleton and the Ras signalling pathway among others. These pathways were previously highlighted as enriched in 3D culture derived EVs (Kusuma et al., 2022). There is no clear consensus on the exact mechanisms of EV secretion, however, cytoskeleton components play important role in EV secretion (Holliday et al., 2019). EV formation and release from a cell involve changes in actin polymerization followed by changes in contraction proteins, and fusion machineries (Rädler et al., 2023). Therefore, we can postulate that morphological changes in cells in spheroid form compared to a monolayer might affect cytoskeleton proteins, leading to changes in EV protein cargo as well. The characteristics of the cellular microenvironment can vary based on several factors, including the properties of the extracellular matrix, the nature and physical properties of the cells themselves (whether they are of the same origin or not), the presence of bioactive molecules like chemokines, cytokines and growth factors secreted by cells, and mechanical forces exerted by the surrounding fluid (Barthes et al., 2014). Consequently, cells cultivated in 2D and 3D environments are expected to exhibit different physiochemical properties. For instance, in 3D rotary culture, the movement of cell culture media generates variations in fluid shear stress on cells compared to the static environment of 2D culture. Fluid shear stress can be transmitted to the cell plasma membrane, potentially influencing tension at cell-cell junctions and subsequently altering the proteins associated with these junctions (White and Frangos, 2007). The spheroid formation take few steps. In the first step cadherin-cadherin and integrin binding to ECM form a loose cellular aggregate which is further compacted over time. After the cellular re-organization period cell aggregate morphologically transit in to spheroids and this is mainly mediated by cadherins (Egger et al., 2018). Interestingly the trophoblast EVs generated in 3D culture compared to 2D culture also seems to reflect these changes as shown in the pathway analysis.
The functional effects of 2D and 3D EVs were assessed in relation to EV-mediated embryo-maternal communication. We previously identified that the MFGE8 protein secretion increases in receptive endometrial epithelial cells in response to trophoblast-derived EVs, suggesting it as a potential marker of EV mediated intercellular communication between the embryo and the endometrium. To gain insight into the effects of molecular cargo differences on EV signalling, we assessed MFGE8 secretion from EEC in response to 2D and 3D EVs, respectively. MFGE8 secretion was increased in EECs treated with both 2D and 3D EVs. Nevertheless, the data suggested that there are distinct differences in the activity of 2D and 3D EVs in tuning MFGE8 secretion from endometrial cells. Interestingly, the effects of 2D EVs on the increase in MFGE8 secretion was higher than that of 3D EVs showing differences in EV potency. Increased potency of 2D EVs compared to 3D EVs has been reported previously in mesenchymal stem cell derived EVs as well (Sun et al., 2023). It can be postulated that the tuning of paracrine EV signalling depends up on their parent cell characteristics and microenvironment in which they were grown. MFGE8 protein expression is required for embryo attachment (Schmitz et al., 2014). Additionally, MFGE8 can influence endometrial stromal cell apoptosis, thereby facilitating embryo invasion (Riggs et al., 2012). Therefore it can be postulated that 2D and 3D EVs may have different potencies in mediating embryo implantation process. Previous literature suggest that molecular changes between 3D and 2D EV preparations may be implicated in other cellular processes such as the immunological processes, extracellular matrix or membrane re-organization or cell proliferation and migration processes (Thippabhotla et al., 2019). However, there are also instances where the molecular cargo differences between 2D and 3D EVs do not translate into functional disparities (Kusuma et al., 2022). So more studies are required in this context for different cell types and culture conditions.
Finally, the 2D and 3D EV protein cargo composition was compared with secretions from the single human embryo prior to implantation and with known endometrial receptivity markers. The majority of those proteins common to trophoblast EVs and human embryo secretions were implicated to be involved in the processes of embryo development, endometrial receptivity, and implantation suggesting the potential value of trophoblastic EVs as a therapeutic tool to facilitate embryo implantation process. Trophoblast cells undergo a transformation into a significant portion of the placenta, an essential fetal organ that facilitates the connection between the fetus and the maternal bloodstream. Within this structure, they release a substantial number of vesicles into the maternal bloodstream (Tong and Chamley, 2015). This vesicle transfer plays a crucial role in facilitating feto-maternal communication, aiding in fetal development, mediating maternal immune tolerance, and sustaining the pregnancy (Buca et al., 2020; Martin et al., 2022).
Less standardized protocols for EV production hinders progress in their clinical translation. Although 3D culture is recognized for better mimicking the in vivo cellular status compared to 2D culture, there remains a gap in understanding EV characteristics, composition, and signalling in relation to cell growth conditions. According to previous reports multiple factors can affect EV production from cells. For example, mesenchymal stem cells cultured in both 2D and 3D environments cell culture conditions exhibited variations in cell secretome and EVs, thereby impacting cellular functionality (Carter et al., 2019; Kusuma et al., 2022). Additionally, cell culture media composition (such as presence of FBS, EV depleted FBS or serum starved media) may also affect the EV production and their characteristics (Lehrich et al., 2021). Interestingly, bidirectional fluidic flow in primary liver cell cultures has been shown to alter cellular metabolic activity significantly compared to 2D static culture, potentially due shear stress sensing, increased metabolic and gas exchange, or through accumulation of growth factors (Esch et al., 2015). Therefore, it is crucial to consider factors such as culture status (static or flow culture) or composition, fluid flow rate, fluid flow direction and pattern of cell-matrix interactions when producing EVs, as these can impact not only the cells but also the EVs they generate, leading to changes in cellular signalling. In conclusion, this study provides compelling evidence of how changes in the cellular microenvironment are reflected in the protein cargo of EVs, subsequently affecting EV-mediated intercellular communication. Nevertheless, the EV characteristics, composition and potency may also vary depending on the cell type from which they originate. Therefore, it would be valuable to investigate these aspects in primary trophoblast cells or other cell lines of trophoblastic origin. Future studies should focus on investigating the potential effects of each of the above factors on EV production in different culture systems to establish standardized protocols for EV production for translational or other purposes.
5 Conclusion
In conclusion, the role of EVs in embryo maternal communication is beginning to be unveiled, opening new avenues for the development of diagnostics and therapeutics. However, this is largely limited by a lack of standardized bio-manufacturing methods for EV mass production. While 3D cultures offers opportunities to scale-up EV production, how the cellular microenvironment could affect EV production, bioactivity and molecular composition remained elusive. Our comparison of EVs generated by 2D and 3D cells showed a remarkable variation between the trophoblast EVs cargo proteome and functionality within the embryo implantation model. The findings suggest that there is an effect of cellular architecture on EVs generated from them, which then probably has an effect on the potency of EV signalling at the recipient cell level. As a whole, caution is warranted when selecting an EV manufacturing platform, especially for assessing the functional and therapeutic potential of EVs.
Data availability statement
The data presented in this study are available in the ProteomeXchange Consortium via the PRIDE with the dataset identifier PXD048789.
Author contributions
NA: Conceptualization, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. SM: Conceptualization, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. KD: Formal Analysis, Investigation, Methodology, Writing–review and editing. KG: Formal Analysis, Methodology, Supervision, Writing–review and editing. GM: Formal Analysis, Investigation, Methodology, Writing–review and editing. AA: Formal Analysis, Methodology, Supervision, Writing–review and editing. PH: Conceptualization, Project administration, Resources, Supervision, Writing–review and editing. SK: Formal Analysis, Methodology, Supervision, Writing–review and editing. AH: Formal Analysis, Methodology, Writing–review and editing. AF: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by European Union’s Horizon 2020 research and innovation programme under grant agreement No 857418 COMBIVET and The European Union through Horizon coordination and support actions under grant agreement No. 101079349 OH-Boost.
Acknowledgments
We express our gratitude to Prof. Tiziana A.L. Brevini for her constructive feedback on the content of this manuscript, which significantly contributed to enhancement of its quality.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1382552/full#supplementary-material
References
Abdollahi, S. (2021). Extracellular vesicles from organoids and 3D culture systems. Biotechnol. Bioeng. 118, 1029–1049. doi:10.1002/bit.27606
Achache, H., and Revel, A. (2006). Endometrial receptivity markers, the journey to successful embryo implantation. Hum. Reprod. Update 12, 731–746. doi:10.1093/humupd/dml004
Aguilera, C., Wong, Y. S., Gutierrez-Reinoso, M. A., Velásquez, A. E., Melo-Báez, B., Cabezas, J., et al. (2024). Embryo-maternal communication mediated by extracellular vesicles in the early stages of embryonic development is modified by in vitro conditions. Theriogenology 214, 43–56. doi:10.1016/j.theriogenology.2023.10.005
Ahlstrom, A., Westin, C., Reismer, E., Wikland, M., and Hardarson, T. (2011). Trophectoderm morphology: an important parameter for predicting live birth after single blastocyst transfer. Hum. Reprod. 26, 3289–3296. doi:10.1093/humrep/der325
Barthes, J., Özçelik, H., Hindié, M., Ndreu-Halili, A., Hasan, A., and Vrana, N. E. (2014). Cell microenvironment engineering and monitoring for tissue engineering and regenerative medicine: the recent advances. Biomed. Res. Int. 2014, 921905. doi:10.1155/2014/921905
Bell, B. M., Kirk, I. D., Hiltbrunner, S., Gabrielsson, S., and Bultema, J. J. (2016). Designer exosomes as next-generation cancer immunotherapy. Nanomedicine Nanotechnol. Biol. Med. 12, 163–169. doi:10.1016/j.nano.2015.09.011
Bocca, S. M., Anderson, S., Amaker, B., Swanson, R. J., Franchi, A., Lattanzio, F., et al. (2012). Milk fat globule epidermal growth factor 8 (MFG-E8): a novel protein in the mammalian endometrium with putative roles in implantation and placentation. Placenta 33, 795–802. doi:10.1016/j.placenta.2012.06.015
Buca, D., Bologna, G., D’Amico, A., Cugini, S., Musca, F., Febbo, M., et al. (2020). Extracellular vesicles in feto–maternal crosstalk and pregnancy disorders. Int. J. Mol. Sci. 21, 2120. doi:10.3390/ijms21062120
Carter, K., Lee, H. J., Na, K., Fernandes-Cunha, G. M., Blanco, I. J., Djalilian, A., et al. (2019). Characterizing the impact of 2D and 3D culture conditions on the therapeutic effects of human mesenchymal stem cell secretome on corneal wound healing in vitro and ex vivo. Acta Biomater. 99, 247–257. doi:10.1016/j.actbio.2019.09.022
Chen, Q., and Wang, Y. (2020). The application of three-dimensional cell culture in clinical medicine. Biotechnol. Lett. 42, 2071–2082. doi:10.1007/s10529-020-03003-y
Cheng, S.-Q., He, J.-L., Dong, Y.-L., Liu, X.-Q., Ding, Y.-B., Gao, R.-F., et al. (2009). Characterization of calreticulin expression in mouse endometrium during embryo implantation. Biol. Res. 42, 505–516. doi:10.4067/S0716-97602009000400012
Chong, S. Y., Lee, C. K., Huang, C., Ou, Y. H., Charles, C. J., Richards, A. M., et al. (2019). Extracellular vesicles in cardiovascular diseases: alternative biomarker sources, therapeutic agents, and drug delivery carriers. Int. J. Mol. Sci. 20, 3272. doi:10.3390/ijms20133272
Clément, V., Roy, V., Paré, B., Goulet, C. R., Deschênes, L. T., Berthod, F., et al. (2022). Tridimensional cell culture of dermal fibroblasts promotes exosome-mediated secretion of extracellular matrix proteins. Sci. Rep. 12, 19786. doi:10.1038/s41598-022-23433-0
Cocks, A., Martinez-Rodriguez, V., Del Vecchio, F., Schukking, M., Broseghini, E., Giannakopoulos, S., et al. (2021). Diverse roles of EV-RNA in cancer progression. Semin. Cancer Biol. 75, 127–135. doi:10.1016/j.semcancer.2020.11.022
della Ragione, T., Verheyen, G., Papanikolaou, E. G., Van Landuyt, L., Devroey, P., and Van Steirteghem, A. (2007). Developmental stage on day-5 and fragmentation rate on day-3 can influence the implantation potential of top-quality blastocysts in IVF cycles with single embryo transfer. Reprod. Biol. Endocrinol. 5, 2. doi:10.1186/1477-7827-5-2
Díaz-Gimeno, P., Horcajadas, J. A., Martínez-Conejero, J. A., Esteban, F. J., Alamá, P., Pellicer, A., et al. (2011). A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil. Steril. 95, 50–60. doi:10.1016/j.fertnstert.2010.04.063
Dissanayake, K., Nõmm, M., Lättekivi, F., Ressaissi, Y., Godakumara, K., Lavrits, A., et al. (2020). Individually cultured bovine embryos produce extracellular vesicles that have the potential to be used as non-invasive embryo quality markers. Theriogenology 149, 104–116. doi:10.1016/j.theriogenology.2020.03.008
Egger, D., Tripisciano, C., Weber, V., Dominici, M., and Kasper, C. (2018). Dynamic cultivation of mesenchymal stem cell aggregates. Bioengineering 5, 48. doi:10.3390/bioengineering5020048
Enciso, M., Carrascosa, J. P., Sarasa, J., Martínez-Ortiz, P. A., Munné, S., Horcajadas, J. A., et al. (2018). Development of a new comprehensive and reliable endometrial receptivity map (ER Map/ER Grade) based on RT-qPCR gene expression analysis. Hum. Reprod. 33, 220–228. doi:10.1093/humrep/dex370
Esch, M. B., Prot, J.-M., Wang, Y. I., Miller, P., Llamas-Vidales, J. R., Naughton, B. A., et al. (2015). Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab. Chip 15, 2269–2277. doi:10.1039/C5LC00237K
Es-Haghi, M., Godakumara, K., Häling, A., Lättekivi, F., Lavrits, A., Viil, J., et al. (2019). Specific trophoblast transcripts transferred by extracellular vesicles affect gene expression in endometrial epithelial cells and may have a role in embryo-maternal crosstalk. Cell Commun. Signal. 17, 146. doi:10.1186/s12964-019-0448-x
Fernando, S. R., Kottawatta, K. S. A., Jiang, L., Chen, X., Cheng, K. W., Wong, B. P. C., et al. (2021). Differential expression of protein disulfide isomerase (PDI) in regulating endometrial receptivity in humans. Reprod. Biol. 21, 100498. doi:10.1016/j.repbio.2021.100498
Garcia-Belda, A., Cairó, O., Martínez-Moro, Á., Cuadros, M., Pons, M. C., de Mendoza, M. V. H., et al. (2024). Considerations for future modification of the Association for the Study of Reproductive Biology embryo grading system incorporating time-lapse observations. Reprod. Biomed. Online 48, 103570. doi:10.1016/j.rbmo.2023.103570
Gardner, D. K. (2015). Lactate production by the mammalian blastocyst: manipulating the microenvironment for uterine implantation and invasion? BioEssays 37, 364–371. doi:10.1002/bies.201400155
Garrido-Gómez, T., Dominguez, F., Quiñonero, A., Estella, C., Vilella, F., Pellicer, A., et al. (2012). Annexin A2 is critical for embryo adhesiveness to the human endometrium by RhoA activation through F-actin regulation. FASEB J. 26, 3715–3727. doi:10.1096/fj.12-204008
Gebril, M., Hirota, Y., Aikawa, S., Fukui, Y., Kaku, T., Matsuo, M., et al. (2020). Uterine epithelial progesterone receptor governs uterine receptivity through epithelial cell differentiation. Endocrinology 161, bqaa195–16. doi:10.1210/endocr/bqaa195
Godakumara, K., Ord, J., Lättekivi, F., Dissanayake, K., Viil, J., Boggavarapu, N. R., et al. (2021). Trophoblast derived extracellular vesicles specifically alter the transcriptome of endometrial cells and may constitute a critical component of embryo-maternal communication. Reprod. Biol. Endocrinol. 19, 115. doi:10.1186/s12958-021-00801-5
Gou, J., Jia, J., Zhao, X., Yi, T., and Li, Z. (2015). Identification of stathmin 1 during peri-implantation period in mouse endometrium by a proteomics-based analysis. Biochem. Biophys. Res. Commun. 461, 211–216. doi:10.1016/j.bbrc.2015.02.171
Green, C. J., Fraser, S. T., and Day, M. L. (2015). Insulin-like growth factor 1 increases apical fibronectin in blastocysts to increase blastocyst attachment to endometrial epithelial cells in vitro. Hum. Reprod. 30, 284–298. doi:10.1093/humrep/deu309
Grummer, R., Hohn, H.-P., Mareel, M. M., and Denker, H.-W. (1994). Adhesion and invasion of three human choriocarcinoma cell lines into human endometrium in a three-dimensional organ culture system. Placenta 15, 411–429. doi:10.1016/0143-4004(94)90008-6
Guerreiro, E. M., Vestad, B., Steffensen, L. A., Aass, H. C. D., Saeed, M., Øvstebø, R., et al. (2018). Efficient extracellular vesicle isolation by combining cell media modifications, ultrafiltration, and size-exclusion chromatography. PLoS One 13, e0204276. doi:10.1371/journal.pone.0204276
Gurunathan, S., Kang, M.-H., Song, H., Kim, N. H., and Kim, J.-H. (2022). The role of extracellular vesicles in animal reproduction and diseases. J. Anim. Sci. Biotechnol. 13, 62. doi:10.1186/s40104-022-00715-1
Guzeloglu-Kayisli, O., Kayisli, U., and Taylor, H. (2009). The role of growth factors and cytokines during implantation: endocrine and paracrine interactions. Semin. Reprod. Med. 27, 062–079. doi:10.1055/s-0028-1108011
Hart, A. R., Khan, N. L. A., Dissanayake, K., Godakumara, K., Andronowska, A., Eapen, S., et al. (2023). The extracellular vesicles proteome of endometrial cells simulating the receptive menstrual phase differs from that of endometrial cells simulating the non-receptive menstrual phase. Biomolecules 13, 279. doi:10.3390/biom13020279
Hart, A. R., Khan, N. L. A., Godakumara, K., Dissanayake, K., Piibor, J., Muhandiram, S., et al. (2022). The role of extracellular vesicles in endometrial receptivity and their potential in reproductive therapeutics and diagnosis. Reprod. Biol. 22, 100645. doi:10.1016/j.repbio.2022.100645
Ho, H., Singh, H., Heng, S., Nero, T. L., Paule, S., Parker, M. W., et al. (2013). Small molecule proprotein convertase inhibitors for inhibition of embryo implantation. PLoS One 8, e81380. doi:10.1371/journal.pone.0081380
Holliday, L. S., Faria, L. P. de, and Rody, W. J. (2019). Actin and actin-associated proteins in extracellular vesicles shed by osteoclasts. Int. J. Mol. Sci. 21, 158. doi:10.3390/ijms21010158
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Huh, D., Hamilton, G. A., and Ingber, D. E. (2011). From 3D cell culture to organs-on-chips. Trends Cell Biol. 21, 745–754. doi:10.1016/j.tcb.2011.09.005
Idelevich, A., and Vilella, F. (2020). Mother and embryo cross-communication. Genes (Basel) 11, 376. doi:10.3390/genes11040376
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367, eaau6977. doi:10.1126/science.aau6977
Kapałczyńska, M., Kolenda, T., Przybyła, W., Zajączkowska, M., Teresiak, A., Filas, V., et al. (2016). 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Arch. Med. Sci. 14, 910–919. doi:10.5114/aoms.2016.63743
Kim, S. M., and Kim, J. (2017). A review of mechanisms of implantation. Dev. Reprod. 21, 351–359. doi:10.12717/DR.2017.21.4.351
King, A., Thomas, L., and Bischof, P. (2000). Cell culture models of trophoblast II: trophoblast cell lines-A workshop report. Placenta 21, S113–S119. doi:10.1053/plac.1999.0526
Kumar, S., Zhi, K., Mukherji, A., and Gerth, K. (2020). Repurposing antiviral protease inhibitors using extracellular vesicles for potential therapy of COVID-19. Viruses 12, 486. doi:10.3390/v12050486
Kurian, N. K., and Modi, D. (2019). Extracellular vesicle mediated embryo-endometrial cross talk during implantation and in pregnancy. J. Assist. Reprod. Genet. 36, 189–198. doi:10.1007/s10815-018-1343-x
Kusuma, G. D., Li, A., Zhu, D., McDonald, H., Inocencio, I. M., Chambers, D. C., et al. (2022). Effect of 2D and 3D culture microenvironments on mesenchymal stem cell-derived extracellular vesicles potencies. Front. Cell Dev. Biol. 10, 819726. doi:10.3389/fcell.2022.819726
Langhans, S. A. (2018). Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 9, 6–14. doi:10.3389/fphar.2018.00006
Latifi, Z., Fattahi, A., Ranjbaran, A., Nejabati, H. R., and Imakawa, K. (2018). Potential roles of metalloproteinases of endometrium-derived exosomes in embryo-maternal crosstalk during implantation. J. Cell. Physiol. 233, 4530–4545. doi:10.1002/jcp.26259
Lehrich, B. M., Liang, Y., and Fiandaca, M. S. (2021). Foetal bovine serum influence on in vitro extracellular vesicle analyses. J. Extracell. Vesicles 10, e12061. doi:10.1002/jev2.12061
Li, H.-W. R., Li, Y.-X., Li, T.-T., Fan, H., Ng, E. H.-Y., Yeung, W. S.-B., et al. (2017). Effect of ulipristal acetate and mifepristone at emergency contraception dose on the embryo-endometrial attachment using an in vitro human trophoblastic spheroid and endometrial cell co-culture model. Hum. Reprod. 32, 2414–2422. doi:10.1093/humrep/dex328
Liang, X., Jin, Y., Wang, H., Meng, X., Tan, Z., Huang, T., et al. (2019). Transgelin 2 is required for embryo implantation by promoting actin polymerization. FASEB J. 33, 5667–5675. doi:10.1096/fj.201802158RRR
Long, J., Li, W., Chen, M., Ding, Y., Chen, X., Tong, C., et al. (2023). Uterine deficiency of Dnmt3b impairs decidualization and causes consequent embryo implantation defects. Cell Biol. Toxicol. 39, 1077–1098. doi:10.1007/s10565-021-09664-3
Ma, J., Gao, W., and Li, D. (2023). Recurrent implantation failure: a comprehensive summary from etiology to treatment. Front. Endocrinol. (Lausanne) 13, 1061766–1061821. doi:10.3389/fendo.2022.1061766
Martin, C., Bergamelli, M., Malnou, C. E., and D’Angelo, G. (2022). Placental extracellular vesicles in maternal-fetal communication during pregnancy. Biochem. Soc. Trans. 50, 1785–1795. doi:10.1042/BST20220734
Midekessa, G., Godakumara, K., Dissanayake, K., Hasan, M. M., Reshi, Q. U. A., Rinken, T., et al. (2021). Characterization of extracellular vesicles labelled with a lipophilic dye using fluorescence nanoparticle tracking analysis. Membr. (Basel) 11, 779. doi:10.3390/membranes11100779
Midekessa, G., Godakumara, K., Ord, J., Viil, J., Lättekivi, F., Dissanayake, K., et al. (2020). Zeta potential of extracellular vesicles: toward understanding the attributes that determine colloidal stability. ACS Omega 5, 16701–16710. doi:10.1021/acsomega.0c01582
Mishra, A., Ashary, N., Sharma, R., and Modi, D. (2021). Extracellular vesicles in embryo implantation and disorders of the endometrium. Am. J. Reprod. Immunol. 85, e13360. doi:10.1111/aji.13360
Mo, H.-Q., Tian, F.-J., Ma, X.-L., Zhang, Y.-C., Zhang, C.-X., Zeng, W.-H., et al. (2020). PDIA3 regulates trophoblast apoptosis and proliferation in preeclampsia via the MDM2/p53 pathway. Reproduction 160, 293–305. doi:10.1530/REP-20-0156
Mol, E. A., Goumans, M. J., Doevendans, P. A., Sluijter, J. P. G., and Vader, P. (2017). Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine Nanotechnol. Biol. Med. 13, 2061–2065. doi:10.1016/j.nano.2017.03.011
Muhandiram, S., Dissanayake, K., Orro, T., Godakumara, K., Kodithuwakku, S., and Fazeli, A. (2023). Secretory proteomic responses of endometrial epithelial cells to trophoblast-derived extracellular vesicles. Int. J. Mol. Sci. 24, 11924. doi:10.3390/ijms241511924
Munch, E. M., Sparks, A. E., Gonzalez Bosquet, J., Christenson, L. K., Devor, E. J., and Van Voorhis, B. J. (2016). Differentially expressed genes in preimplantation human embryos: potential candidate genes for blastocyst formation and implantation. J. Assist. Reprod. Genet. 33, 1017–1025. doi:10.1007/s10815-016-0745-x
Nagatomo, H., Kohri, N., Akizawa, H., Hoshino, Y., Yamauchi, N., Kono, T., et al. (2016). Requirement for nuclear autoantigenic sperm protein mRNA expression in bovine preimplantation development. Anim. Sci. J. 87, 457–461. doi:10.1111/asj.12538
Nakamura, K., Kusama, K., Bai, R., Sakurai, T., Isuzugawa, K., Godkin, J. D., et al. (2016). Induction of IFNT-stimulated genes by conceptus-derived exosomes during the attachment period. PLoS One 11, e0158278. doi:10.1371/journal.pone.0158278
Nicol, C. J., Zielenski, J., Tsui, L., and Wells, P. G. (2000). An embryoprotective role for glucose-6-phosphate dehydrogenase in developmental oxidative stress and chemical teratogenesis. FASEB J. 14, 111–127. doi:10.1096/fasebj.14.1.111
Oron, G., Son, W. Y., Buckett, W., Tulandi, T., and Holzer, H. (2014). The association between embryo quality and perinatal outcome of singletons born after single embryo transfers: a pilot study. Hum. Reprod. 29, 1444–1451. doi:10.1093/humrep/deu079
Pitt, J. M., Kroemer, G., and Zitvogel, L. (2016). Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J. Clin. Invest. 126, 1139–1143. doi:10.1172/JCI87316
Poh, Q. H., Rai, A., Carmichael, I. I., Salamonsen, L. A., and Greening, D. W. (2021). Proteome reprogramming of endometrial epithelial cells by human trophectodermal small extracellular vesicles reveals key insights into embryo implantation. Proteomics 21, 2000210. doi:10.1002/pmic.202000210
Poh, Q. H., Rai, A., Pangestu, M., Salamonsen, L. A., and Greening, D. W. (2023). Rapid generation of functional nanovesicles from human trophectodermal cells for embryo attachment and outgrowth. Proteomics 12, e2300056. doi:10.1002/pmic.202300056
Poli, M., Ori, A., Child, T., Jaroudi, S., Spath, K., Beck, M., et al. (2015). Characterization and quantification of proteins secreted by single human embryos prior to implantation. EMBO Mol. Med. 7, 1465–1479. doi:10.15252/emmm.201505344
Rädler, J., Gupta, D., Zickler, A., and Andaloussi, S. E. L. (2023). Exploiting the biogenesis of extracellular vesicles for bioengineering and therapeutic cargo loading. Mol. Ther. 31, 1231–1250. doi:10.1016/j.ymthe.2023.02.013
Rai, A., Poh, Q. H., Fatmous, M., Fang, H., Gurung, S., Vollenhoven, B., et al. (2021). Proteomic profiling of human uterine extracellular vesicles reveal dynamic regulation of key players of embryo implantation and fertility during menstrual cycle. Proteomics 21, e2000211–e2000217. doi:10.1002/pmic.202000211
Ramasubramanian, L., Kumar, P., and Wang, A. (2019). Engineering extracellular vesicles as nanotherapeutics for regenerative medicine. Biomolecules 10, 48. doi:10.3390/biom10010048
Raposo, G., and Stahl, P. D. (2019). Extracellular vesicles: a new communication paradigm? Nat. Rev. Mol. Cell Biol. 20, 509–510. doi:10.1038/s41580-019-0158-7
Riazifar, M., Pone, E. J., Lötvall, J., and Zhao, W. (2017). Stem cell extracellular vesicles: extended messages of regeneration. Annu. Rev. Pharmacol. Toxicol. 57, 125–154. doi:10.1146/annurev-pharmtox-061616-030146
Riggs, R. M., Bocca, S., Anderson, S., Franchi, A., Rhavi, B. S., and Oehninger, S. (2012). Epithelial cell protein milk fat globule-epidermal growth factor 8 and human chorionic gonadotropin regulate stromal cell apoptosis in the human endometrium. Fertil. Steril. 98, 1549–1556. doi:10.1016/j.fertnstert.2012.07.1127
Rocha, S., Carvalho, J., Oliveira, P., Voglstaetter, M., Schvartz, D., Thomsen, A. R., et al. (2019). 3D cellular architecture affects microRNA and protein cargo of extracellular vesicles. Adv. Sci. 6, 1800948. doi:10.1002/advs.201800948
Schmitz, C., Yu, L., Bocca, S., Anderson, S., Cunha-Filho, J. S., Rhavi, B. S., et al. (2014). Role for the endometrial epithelial protein MFG-E8 and its receptor integrin αvβ3 in human implantation: results of an in vitro trophoblast attachment study using established human cell lines. Fertil. Steril. 101, 874–882. doi:10.1016/j.fertnstert.2013.12.015
Segura-Benítez, M., Bas-Rivas, A., Juárez-Barber, E., Carbajo-Garcia, M. C., Faus, A., Pellicer, A., et al. (2022). Human blastocysts uptake extracellular vesicles secreted by primary endometrial epithelial cells containing miRNAs related to implantation and early embryo development. Fertil. Steril. 118, e73–e74. doi:10.1016/j.fertnstert.2022.08.226
Shah, A. D., Goode, R. J. A., Huang, C., Powell, D. R., and Schittenhelm, R. B. (2020). LFQ-Analyst: an easy-to-use interactive web platform to analyze and visualize label-free proteomics data preprocessed with MaxQuant. J. Proteome Res. 19, 204–211. doi:10.1021/acs.jproteome.9b00496
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi:10.1093/nar/gkac194
Shi, S., Tan, Q., Liang, J., Cao, D., Wang, S., Liang, J., et al. (2021). Placental trophoblast cell-derived exosomal microRNA-1290 promotes the interaction between endometrium and embryo by targeting LHX6. Mol. Ther. Nucleic Acids 26, 760–772. doi:10.1016/j.omtn.2021.09.009
Sun, L., Ji, Y., Chi, B., Xiao, T., Li, C., Yan, X., et al. (2023). A 3D culture system improves the yield of MSCs-derived extracellular vesicles and enhances their therapeutic efficacy for heart repair. Biomed. Pharmacother. 161, 114557. doi:10.1016/j.biopha.2023.114557
Syromiatnikova, V., Prokopeva, A., and Gomzikova, M. (2022). Methods of the large-scale production of extracellular vesicles. Int. J. Mol. Sci. 23, 10522. doi:10.3390/ijms231810522
Takahashi, A., Okada, R., Nagao, K., Kawamata, Y., Hanyu, A., Yoshimoto, S., et al. (2017). Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 8, 15287. doi:10.1038/ncomms15287
Taravat, M., Asadpour, R., Jozani, R. J., Fattahi, A., and Khordadmehr, M. (2023). Enhanced anti-inflammatory effect of Rosmarinic acid by encapsulation and combination with the exosome in mice with LPS-induced endometritis through suppressing the TLR4-NLRP3 signaling pathway. J. Reprod. Immunol. 159, 103992. doi:10.1016/j.jri.2023.103992
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750. doi:10.1080/20013078.2018.1535750
Thippabhotla, S., Zhong, C., and He, M. (2019). 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep. 9, 13012. doi:10.1038/s41598-019-49671-3
Tong, J., Zhao, W., Lv, H., Li, W., Chen, Z., and Zhang, C. (2018). Transcriptomic profiling in human decidua of severe preeclampsia detected by RNA sequencing. J. Cell. Biochem. 119, 607–615. doi:10.1002/jcb.26221
Tong, M., and Chamley, L. W. (2015). Feto-maternal communication. Cold Spring Harb. Perspect. Med. 5, 1–17. doi:10.1101/cshperspect.a023028
Tu, Z., Wang, Q., Cui, T., Wang, J., Ran, H., Bao, H., et al. (2016). Uterine RAC1 via Pak1-ERM signaling directs normal luminal epithelial integrity conducive to on-time embryo implantation in mice. Cell Death Differ. 23, 169–181. doi:10.1038/cdd.2015.98
Vantangoli, M. M., Madnick, S. J., Huse, S. M., Weston, P., and Boekelheide, K. (2015). MCF-7 human breast cancer cells form differentiated microtissues in scaffold-free hydrogels. PLoS One 10, e0135426. doi:10.1371/journal.pone.0135426
Villasante, A., Marturano-Kruik, A., Ambati, S. R., Liu, Z., Godier-Furnemont, A., Parsa, H., et al. (2016). Recapitulating the size and cargo of tumor exosomes in a tissue-engineered model. Theranostics 6, 1119–1130. doi:10.7150/thno.13944
Vu, B., Souza, G. R., and Dengjel, J. (2021). Scaffold-free 3D cell culture of primary skin fibroblasts induces profound changes of the matrisome. Matrix Biol. Plus 11, 100066. doi:10.1016/j.mbplus.2021.100066
Wang, B., and Shao, Y. (2020). Annexin A2 acts as an adherent molecule under the regulation of steroids during embryo implantation. Mol. Hum. Reprod. 26, 825–836. doi:10.1093/molehr/gaaa065
Weber, M., Weise, A., Vasheghani, F., Göhner, C., Fitzgerald, J. S., Liehr, T., et al. (2021). Cytogenomics of six human trophoblastic cell lines. Placenta 103, 72–75. doi:10.1016/j.placenta.2020.10.011
Weimar, C. H. E., Post Uiterweer, E. D., Teklenburg, G., Heijnen, C. J., and Macklon, N. S. (2013). In-vitro model systems for the study of human embryo-endometrium interactions. Reprod. Biomed. Online 27, 461–476. doi:10.1016/j.rbmo.2013.08.002
Welsh, J. A., Goberdhan, D. C. I., O’Driscoll, L., Buzas, E. I., Blenkiron, C., Bussolati, B., et al. (2024). Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J. Extracell. Vesicles 13, e12404. doi:10.1002/jev2.12404
White, C. R., and Frangos, J. A. (2007). The shear stress of it all: the cell membrane and mechanochemical transduction. Philos. Trans. R. Soc. B Biol. Sci. 362, 1459–1467. doi:10.1098/rstb.2007.2128
White, T. E., Saltzman, R. A., Di Sant’agnese, P. A., Keng, P. C., Sutherland, R. M., and Miller, R. K. (1988). Human choriocarcinoma (JAr) cells grown as multicellular spheroids. Placenta 9, 583–598. doi:10.1016/0143-4004(88)90002-1
Wu, F., Tian, F., Zeng, W., Liu, X., Fan, J., Lin, Y., et al. (2017). Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis. 8, e2908–e2918. doi:10.1038/cddis.2017.301
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141. doi:10.1016/j.xinn.2021.100141
Yockey, L. J., and Iwasaki, A. (2018). Interferons and proinflammatory cytokines in pregnancy and fetal development. Immunity 49, 397–412. doi:10.1016/j.immuni.2018.07.017
Yoshie, M., Kusama, K., Tanaka, R., Okubo, T., Kojima, J., Takaesu, Y., et al. (2021). Possible roles of calreticulin in uterine decidualization and receptivity in rats and humans. Int. J. Mol. Sci. 22, 10505. doi:10.3390/ijms221910505
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16, 284–287. doi:10.1089/omi.2011.0118
Keywords: three dimensional, two dimensional, cell culture systems, extracellular vesicles, embryo implantation, potency
Citation: Khan NLA, Muhandiram S, Dissanayake K, Godakumara K, Midekessa G, Andronowska A, Heath PR, Kodithuwakku S, Hart AR and Fazeli A (2024) Effect of 3D and 2D cell culture systems on trophoblast extracellular vesicle physico-chemical characteristics and potency. Front. Cell Dev. Biol. 12:1382552. doi: 10.3389/fcell.2024.1382552
Received: 05 February 2024; Accepted: 30 April 2024;
Published: 21 May 2024.
Edited by:
Shiro Suetsugu, Nara Institute of Science and Technology (NAIST), JapanReviewed by:
José Martin Murrieta-Coxca, University Hospital Jena, GermanyRishi Man Chugh, University of Kansas Medical Center, United States
Ananth Kumar Kammala, University of Texas Medical Branch, United States
Copyright © 2024 Khan, Muhandiram, Dissanayake, Godakumara, Midekessa, Andronowska, Heath, Kodithuwakku, Hart and Fazeli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alireza Fazeli, ZmF6ZWxpQGVtdS5lZQ==
†These authors have contributed equally to this work and share first authorship
 Norhayati Liaqat Ali Khan
Norhayati Liaqat Ali Khan Subhashini Muhandiram
Subhashini Muhandiram Keerthie Dissanayake
Keerthie Dissanayake Kasun Godakumara3,4
Kasun Godakumara3,4 Aneta Andronowska
Aneta Andronowska Paul R. Heath
Paul R. Heath Suranga Kodithuwakku
Suranga Kodithuwakku Amber Rose Hart
Amber Rose Hart Alireza Fazeli
Alireza Fazeli