- 1Department of Epidemiology and Health Statistics, School of Public Health, Qingdao University, Qingdao, Shandong, China
- 2Qingdao Municipal Centre for Disease Control and Prevention, Qingdao, Shandong, China
- 3Qingdao Institute of Preventive Medicine, Qingdao, Shandong, China
Background: The decline in muscle strength and function with aging is well recognized, but remains poorly characterized at the molecular level. Here, we report the epigenetic relationship between genome-wide DNA methylation and handgrip strength (HGS) among Chinese monozygotic (MZ) twins.
Methods: DNA methylation (DNAm) profiling was conducted in whole blood samples through Reduced Representation Bisulfite Sequencing method. Generalized estimating equation was applied to regress the DNAm of each CpG with HGS. The Genomic Regions Enrichment of Annotations Tool was used to perform enrichment analysis. Differentially methylated regions (DMRs) were detected using comb-p. Causal inference was performed using Inference about Causation through Examination of Familial Confounding method. Finally, we validated candidate CpGs in community residents.
Results: We identified 25 CpGs reaching genome-wide significance level. These CpGs located in 9 genes, especially FBLN1, RXRA, and ABHD14B. Many enriched terms highlighted calcium channels, neuromuscular junctions, and skeletal muscle organ development. We identified 21 DMRs of HGS, with several DMRs within FBLN1, SLC30A8, CST3, and SOCS3. Causal inference indicated that the DNAm of 16 top CpGs within FBLN1, RXRA, ABHD14B, MFSD6, and TYW1B might influence HGS, while HGS influenced DNAm at two CpGs within FBLN1 and RXRA. In validation analysis, methylation levels of six CpGs mapped to FLBN1 and one CpG mapped to ABHD14B were negatively associated with HGS weakness in community population.
Conclusion: Our study identified multiple DNAm variants potentially related to HGS, especially CpGs within FBLN1 and ABHD14B. These findings provide new clues to the epigenetic modification underlying muscle strength decline.
1 Introduction
Muscle strength is a crucial factor in healthy aging and an important predictor of several adverse health outcomes, which decline from midlife and throughout life (McLeod et al., 2016; Balogun et al., 2019; Alcazar et al., 2020). HGS, a simple measure of upper body muscle strength in clinical practice (Lauretani et al., 2003), is influenced by both environmental and genetic factors (Tian et al., 2017). The correlation between HGS and various factors, encompassing nutritional parameters and levels of physical activity, has been extensively explored through epidemiological studies (Rondanelli et al., 2016; Mendes et al., 2017; Weng et al., 2022). Moreover, previous genome-wide association studies (GWAS) have elucidated the influence of genetics on HGS, revealing heritability estimates within the range of 40%–65% (Tiainen et al., 2004; Matteini et al., 2010; Matteini et al., 2016). Recent emerging GWASs have facilitated the identification of human genetic variants associated with HGS (Matteini et al., 2016; Luo et al., 2022). Nevertheless, the single nucleotide-level polymorphisms (SNPs) reported in these studies only explained a small portion of heritability of HGS, suggesting that additional gene-regulatory mechanisms are involved in muscle maintenance or decline (He et al., 2020).
Emerging evidence indicated that epigenetic processes play a crucial role in the development of complex human traits or diseases (Godfrey et al., 2007). Epigenetic modifications serve as potential mediators for the impact of external factors on muscle maintenance or decline, providing a mechanistic framework that integrates environmental and genetic influences. DNA methylation, a well-studied epigenetic modification, provides a novel perspective on the variability of muscle strength. It is widely recognized that DNA methylation is a fundamental mechanism underlying myogenesis (Carrió et al., 2015; Laker and Ryall, 2016). Previous research has suggested that DNAm regulates skeletal muscle differentiation and proliferation by modulating the expression of genes from the myocyte enhancer and myogenic pathway (Bharathy et al., 2013). Several studies have reported a hypermethylated state in skeletal muscle tissue in older subjects compared to younger subjects (Zykovich et al., 2014; Turner et al., 2020). There are multiple epigenome-wide association studies (EWAS) of sarcopenia or muscle mass and strength among people of European ancestry (Livshits et al., 2016; Voisin et al., 2021; Antoun et al., 2022). These studies have identified many CpG loci and differentially methylated regions (DMRs) that may be associated with muscle strength. Nevertheless, three EWASs have tested associations between whole-blood DNAm level and HGS, but no significant association between methylation levels at single CpG and HGS was observed (Bell et al., 2012; Marioni et al., 2015; Soerensen et al., 2019).
The trait-discordant monozygotic twins design is a valuable and powerful design for EWAS that can link epigenetic modification to traits, as the genetic component is perfectly matched within monozygotic twin pairs (Tan et al., 2015). This design also provides an opportunity to identify different DNA methylation patterns triggered by environmental factors. Due to difference in environmental exposures, genetic backgrounds, and lifestyles among different ethnicities, DNAm patterns are likely to exhibit distinctions. Hence, it is necessary to conduct comparable studies using samples from different ethnic groups. Currently, relevant investigation has not been reported in the Chinese population.
2 Materials and methods
2.1 Participants and study procedures
The study population was a subsample of the Qingdao Twin Registry System. For detailed information on recruitment procedures, please refer to our previous work (Xu et al., 2017). Participants were excluded if they suffered from stroke, cardiovascular disease, and/or tumor, or were unable to complete the examination. Twin pairs with intra-pair HGS difference ≥0.1 kg were chosen for the trait-discordant monozygotic twin design based on our previous experience with similar studies (Wang W. et al., 2021; Wang T. et al., 2021; Wang et al., 2023). A total of 66 HGS-discordant monozygotic twin pairs were included. The Regional Ethics Committee of the Institutional Review Committee of Qingdao CDC approved this study. This study followed the Helsinki Declaration and all participants provided informed consent.
After a fasting period of 10–12 h, the participants completed a questionnaire and physical examination, and a venous blood sample of 10 mL was collected. Handgrip strength was measured using a hand-held dynamometer (WCS-100, Nantong, China). Participants were asked to forcefully squeeze the dynamometer three times for each hand, with the maximum value being used for subsequent analysis.
2.2 DNA methylation data
Total DNA was extracted from venous blood samples for reduced representation bisulfite sequencing experiments. In summary, the initial step involved digestion of genomic DNA to produce shorter fragments. Subsequently, the CpG-rich DNA fragments underwent bisulfite conversion. Finally, the resulting cDNA library was sequenced. The resulting raw methylation data encompassed 551,447 CpGs throughout the genome of each individual. The raw reads were mapped to the human Genome Reference Consortium Human Build 37 using Bismark (Krueger and Andrews, 2011), and methylation levels were determined by smoothing the data with the BiSeq package in R software. We maintained control over the coverage, ensuring it was within the 90% quantile. CpGs with an average methylation β-value below 0.01 or those with more than 10 missing observations were excluded. The methylation β-values were transformed to M-values by log2 transformation (Wang T. et al., 2021).
2.3 Cell-type composition estimate
DNAm patterns vary across different cell types. To address the potential confounding effect of cell-type composition on DNAm analysis in whole blood, we used ReFACTor, a reference-free, unsupervised method that utilizes principal component analysis to estimate and adjust for cellular heterogeneity (Jaffe and Irizarry, 2014; Rahmani et al., 2016). In our study, we utilized the top five components identified by ReFACTor to control the potential effects of cell-type heterogeneity in EWAS analysis.
2.4 Gene expression data preparation
A subsample of 12 MZ twin pairs were included in the gene expression analysis. Briefly, total mRNA was extracted from whole peripheral blood. Then the RNA-Seq library was constructed and sequenced to obtain the sequenced data, which was mapped to the human genome by TopHat2 (Kim et al., 2013). The gene expression level was evaluated by FPKM value through Cufflinks.
2.5 Statistical analysis
2.5.1 Epigenome-wide association analysis
For single CpG analysis, generalized estimating equation (GEE) model was applied to estimate the relationship between DNAm and HGS, with taking the correlation within each twin pair into account and adjusting for sex, age, and cell-type composition. We conducted this analysis using the geeglm function of geepack package in R software (version 4.1.0). The false discovery rate (FDR) was calculated to correct for multiple testing and genome-wide significance was defined as FDR <0.05. We annotated the identified genomic CpGs to the nearest genes by biomaRt package in R software. Causal inference.
2.5.2 Causal inference
The potential causal relationships between genome-wide significant CpGs and HGS were estimated using the ICE FALCON method, a regression-based method for causal inference in twin pair or family design (Li et al., 2005; Li et al., 2020). Estimations of βself, βco-twin and β’self, β’co-twin were calculated by the GEE model, where βco-twin is the estimation of family confounding proportion; βself represents the overall correlation including family confounding and casual proportion; and β’co-twin and β’self were the estimations of the full model. If the absolute difference between βself and β’self is greater than the absolute difference between βco-twin and β’co-twin (ratio >1.5), it suggests a causal relationship. However, if the association is a result of family confounding, the absolute difference between βself and β’self would be similar to the absolute difference between βco-twin and β’co-twin.
2.5.3 Differentially methylated region (DMR) analysis
Considering that DNAm of adjacent CpGs may be functionally or spatially linked, exploring genomic regions containing biologically linked DNAm may provide new perspectives compared to the analysis of single CpGs. Hence, we explored the DMRs potentially related to HGS by comb-p (Pedersen et al., 2012). The significantly enriched DMRs were identified using the Stouffer–Liptak–Kechris (slk) correction method with a significance threshold of p < 0.05.
2.5.4 Biological pathway analysis
We performed genomic region enrichment analyses using the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010). The CpGs that were identified with a p < 0.05 were submitted to the GREAT online platform for ontology enrichment. Annotation was conducted based on Genome Reference Consortium Human Build 37, employing the default “basal plus extension” association rule. Enrichment items with FDR<0.05 were deemed statistically significant.
2.5.5 EWAS power estimation
According to a recent computer simulation study on the power of EWAS based on twin designs, for traits with a heritability of 0.6 or higher, the maximal sample size of 63 pairs is required for statistical power exceeding 80% in utilizing trait-discordant twin design (Li et al., 2018; Wang et al., 2023). Compared to ordinary case-control design, twin design can significantly reduce the required sample size. Therefore, our sample size could meet the criteria for statistical power exceeding 80%.
2.5.6 DNA methylation and gene expression analysis
The correlation between top CpGs methylation level and corresponding gene expression level was calculated using Spearman’s rank correlation. In addition, we also queried the position of significant CpGs in NCBI to illustrate the gene structures where these CpGs are located.
2.5.7 Quantitative methylation analysis of FBLN1 and ABHD14B
To validate CpGs located at FBLN1 and ABHD14B, we recruited 117 participants with low HGS (defined as male<28 kg, female<18 kg) and 117 healthy controls from the community. The method for HGS testing is consistent with the aforementioned approach. Participants attended interviews and venous blood samples were collected for DNAm level measurements. Primers for the FBLN1 and ABHD14B genes were designed to cover the majority of the CpGs identified in EWAS. The cleavage products were analyzed using MALDI-TOF mass spectrometry based on MassARRAY System (Bio Miao Biological Technology, Beijing, China). Subsequently, the resulting spectra were processed using the MassARRAY EpiTYPER software (Agena Bioscience, San Diego, California) to determine the methylation ratio. The Wilcoxon rank-sum test was used to compare the DNAm levels of GpGs between the two groups. The association between each CpG and low HGS was evaluated by logistic regression, with adjusting for age, sex, and BMI. The significance level was defined as p < 0.05.
3 Results
The epigenome-wide association study included 66 MZ with a median HGS of 32.0 kg (95% range 16.7, 57.0) and the median of absolute values of intra-pair HGS was 3.9 kg (95% range 0.3, 13.3). Significant correlations were observed between various clinical indicators within twin pairs, such as height, weight, BMI, fat rate, systolic and diastolic blood pressure, glucose, serum uric acid, cholesterol, triglycerides, HDL cholesterol, and LDL cholesterol, suggesting that utilizing a trait-discordant monozygotic twin design provided significant advantages (Supplementary Table S1).
3.1 Epigenome-wide association analysis
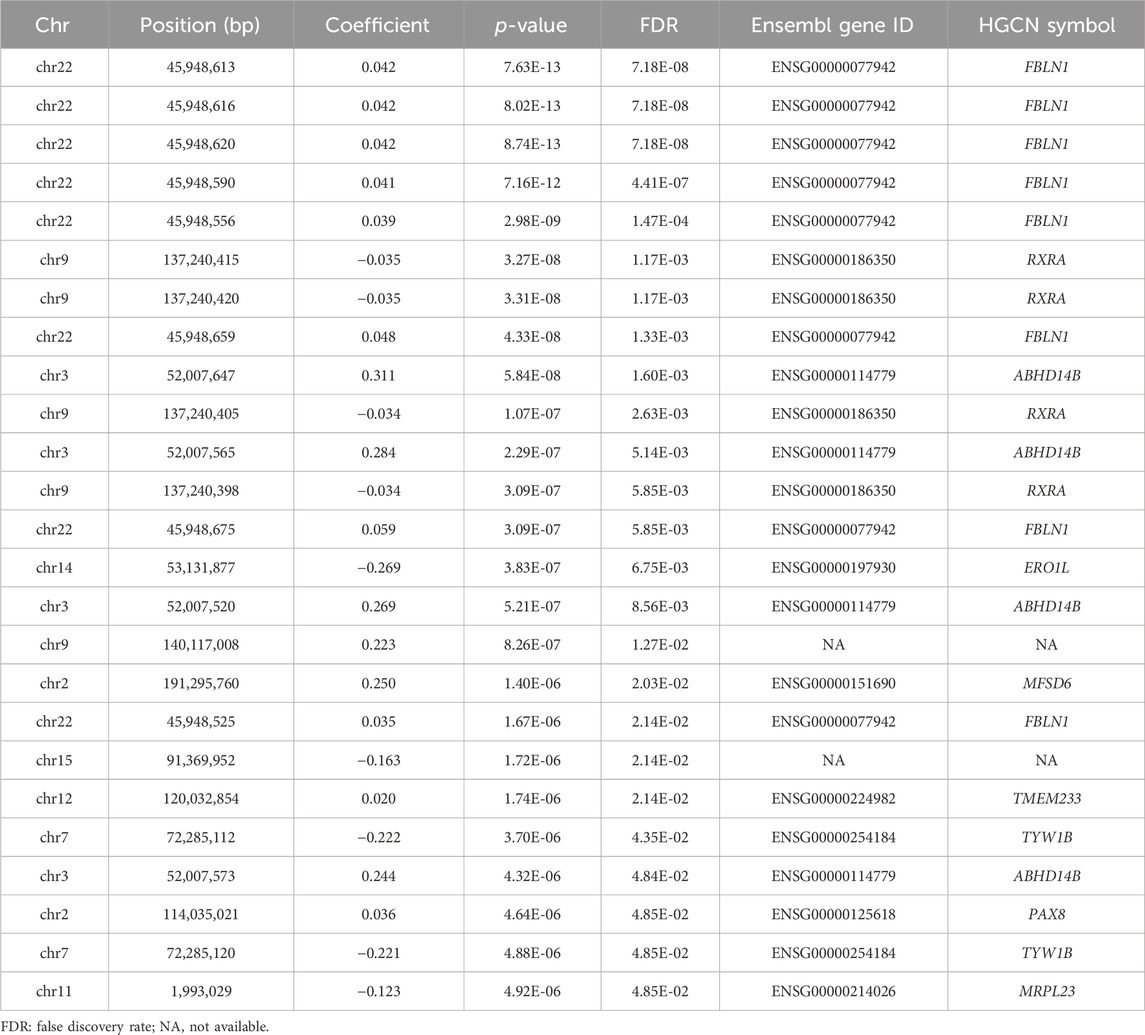
Figure 1 illustrates the Manhattan plot of the EWAS. A total of 25 CpGs were found reaching genome-wide significance level (FDR <0.05). Specifically, eight CpGs (chr3: 45,948,525–45,948,675 bp) located at FBLN1; four CpGs (chr9: 137,240,398–137,240,420 bp) located at RXRA; four CpGs (chr3: 52,007,520–52,007,647 bp) located at ABHD14B, and the remaining nine CpGs located at six different genes, including ERO1L, MFSD6, TMEM233, TYW1B, PAX8, and MRPL23. Methylation level of 16 CpGs (located at FBLN1, ABHD14B, MFSD6, TMEM233, and PAX8 genes) were positively associated with HGS, indicating that hypermethylation of these CpGs in twins with higher HGS. While nine CpGs (located at RXRA, ERO1L, TYW1B, and MRPL23 genes) were negatively associated with HGS, indicating that hypomethylation of these CpGs in twins with higher HGS (Table 1).
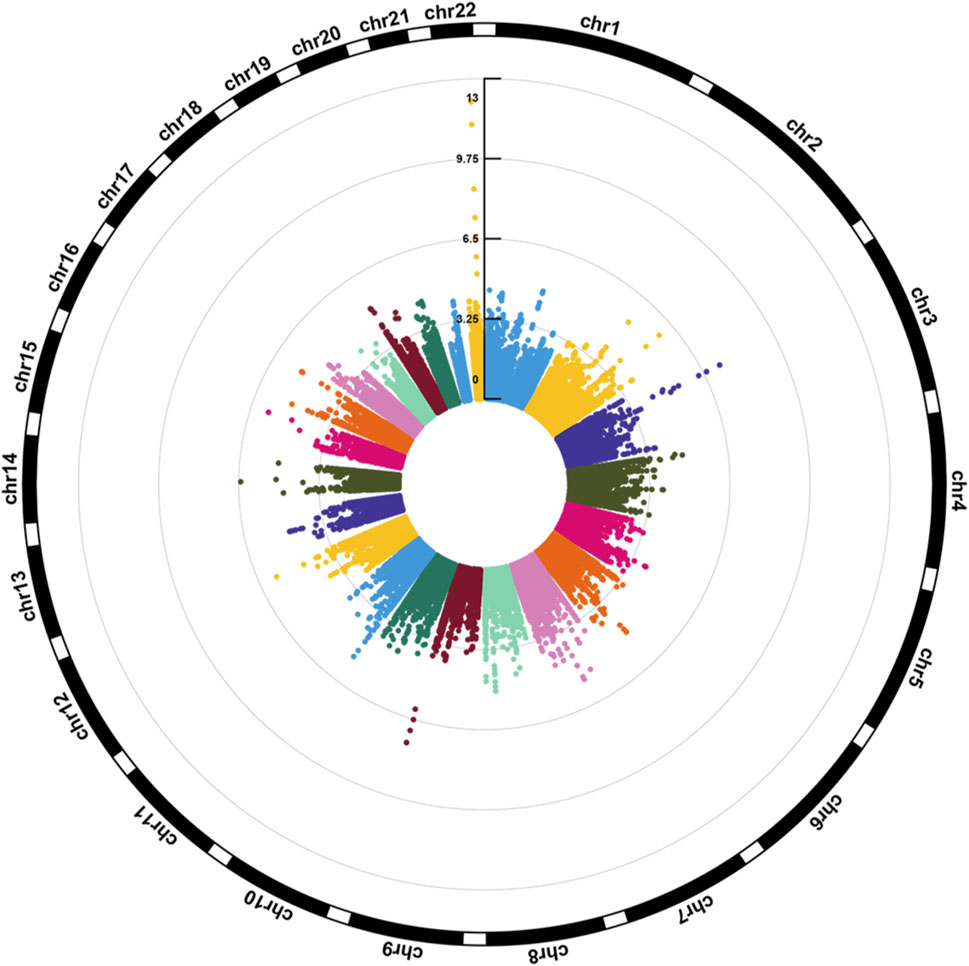
Figure 1. Circular Manhattan plot for the epigenome-wide association study of HGS. The number of chromosomes and the -log10 of p-values for statistical significance are shown. The dots represent the observed CpGs.
3.2 Causal inference analysis
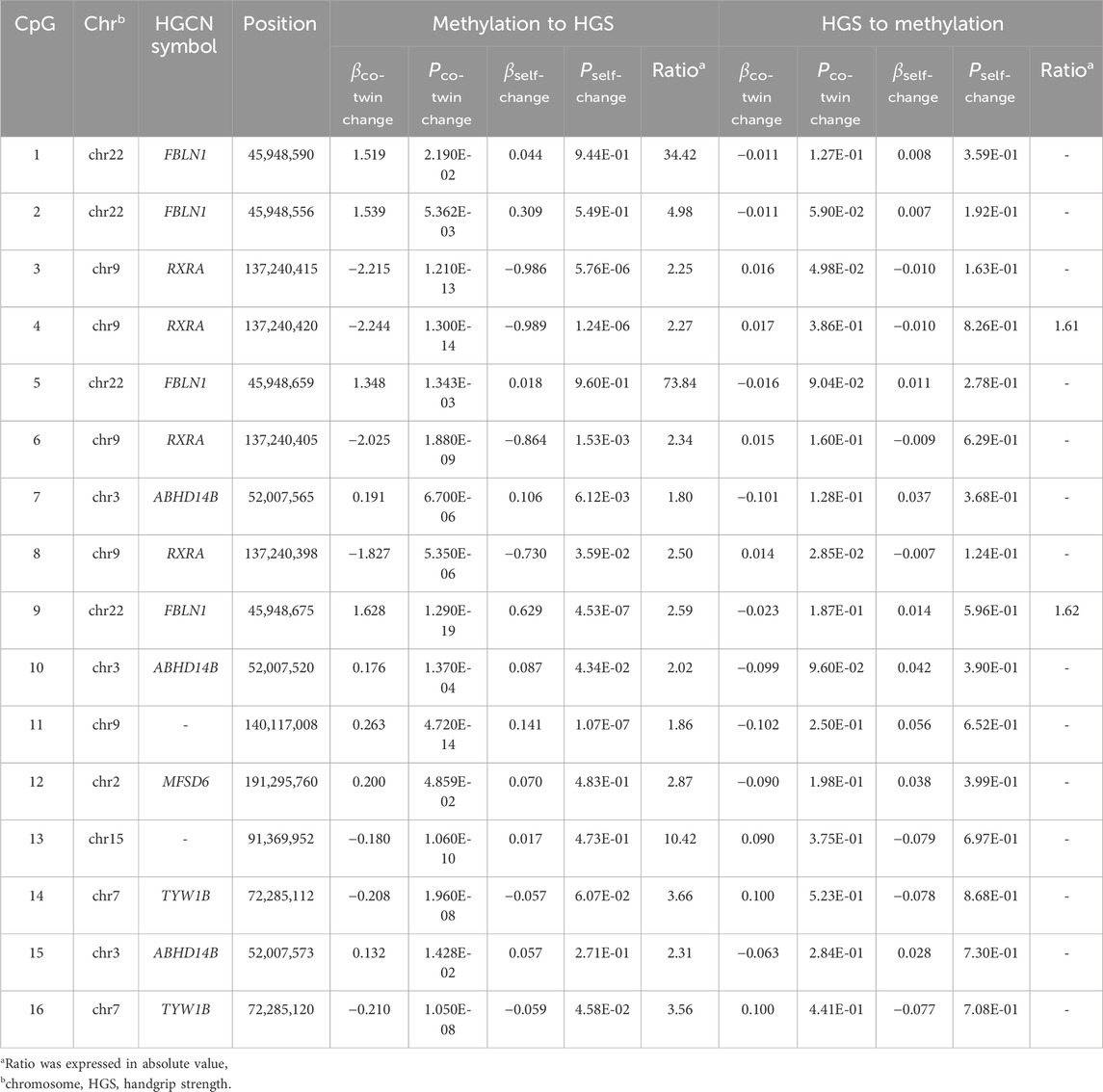
Table 2 presents the estimation of the causal inference between genome-wide significant CpGs and HGS. In the causal inference from DNAm to HGS, the causal effect was supported for 16 CpGs located at FBLN1, RXRA, ABHD14B, MFSD6, and TYW1B. The causal effects from HGS to DNAm were also observed among two CpGs located at FBLN1 and RXRA.
3.3 Region-based analysis
A total of 21 DMRs were detected for HGS (Table 3). Notably, among these DMRs, 11 DMRs (A, C, E, F-J, L, S, and T) indicated a positive correlation with HGS, whereas five (B, D, Q, R, and U) near/at CPLX1, PLEC, AKR1D1, WNK2, and ZNF597 exhibited a negative association. In the case of five DMRs (K, M-P), the methylation levels were inconclusive (Figure 2).
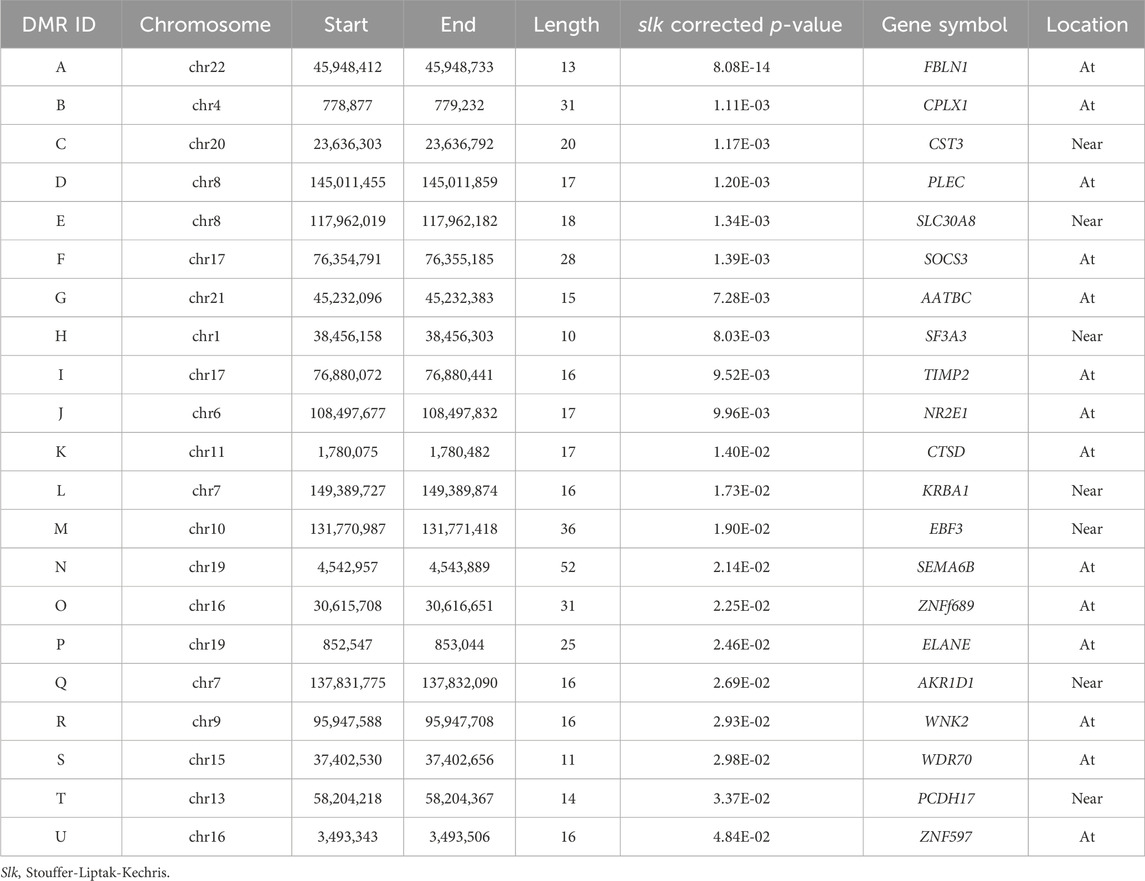
Table 3. The results of annotation to significant differentially methylated regions (DMRs) (slk corrected p < 0.05).
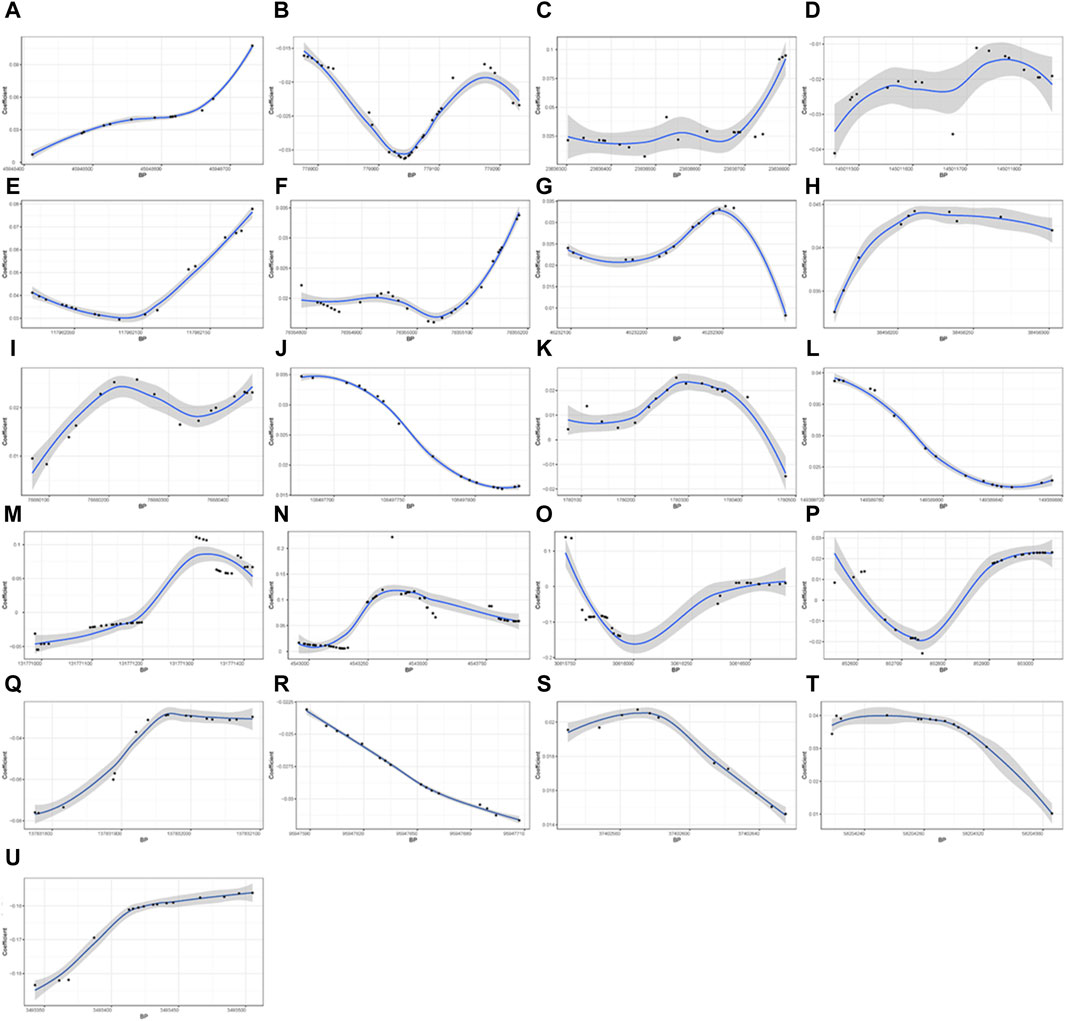
Figure 2. Differential methylation patterns for the identified DMRs. 11 DMRs (A, C, E–J, L, S, and T) indicated a positive correlation with HGS, whereas five (B, D, Q, R, and U) exhibited a negative association The vertical axis shows the coefficient for the association of each CpG with HGS, and the horizontal axis shows the chromosome positions with the black points indicating each CpG. The black line presents the methylation pattern for each DMR. BP, base pair; chr, chromosome.
3.4 Biological pathway analysis
The GO-enriched terms mainly highlighted calcium channels, neuromuscular junctions, and skeletal muscle organ development. Moreover, the pathway terms associated with HGS were significantly enriched, including calcium transport, choline biosynthesis, genes involved in the integration of energy metabolism, PKC-catalyzed phosphorylation of inhibitory phosphoprotein of myosin phosphatase, and the Notch signaling pathway (Table 4). Some items are related to skeletal muscle function in previous studies and have shown a potential role of epigenetic changes in the denervation of calcium channels and neuromuscular junctions.
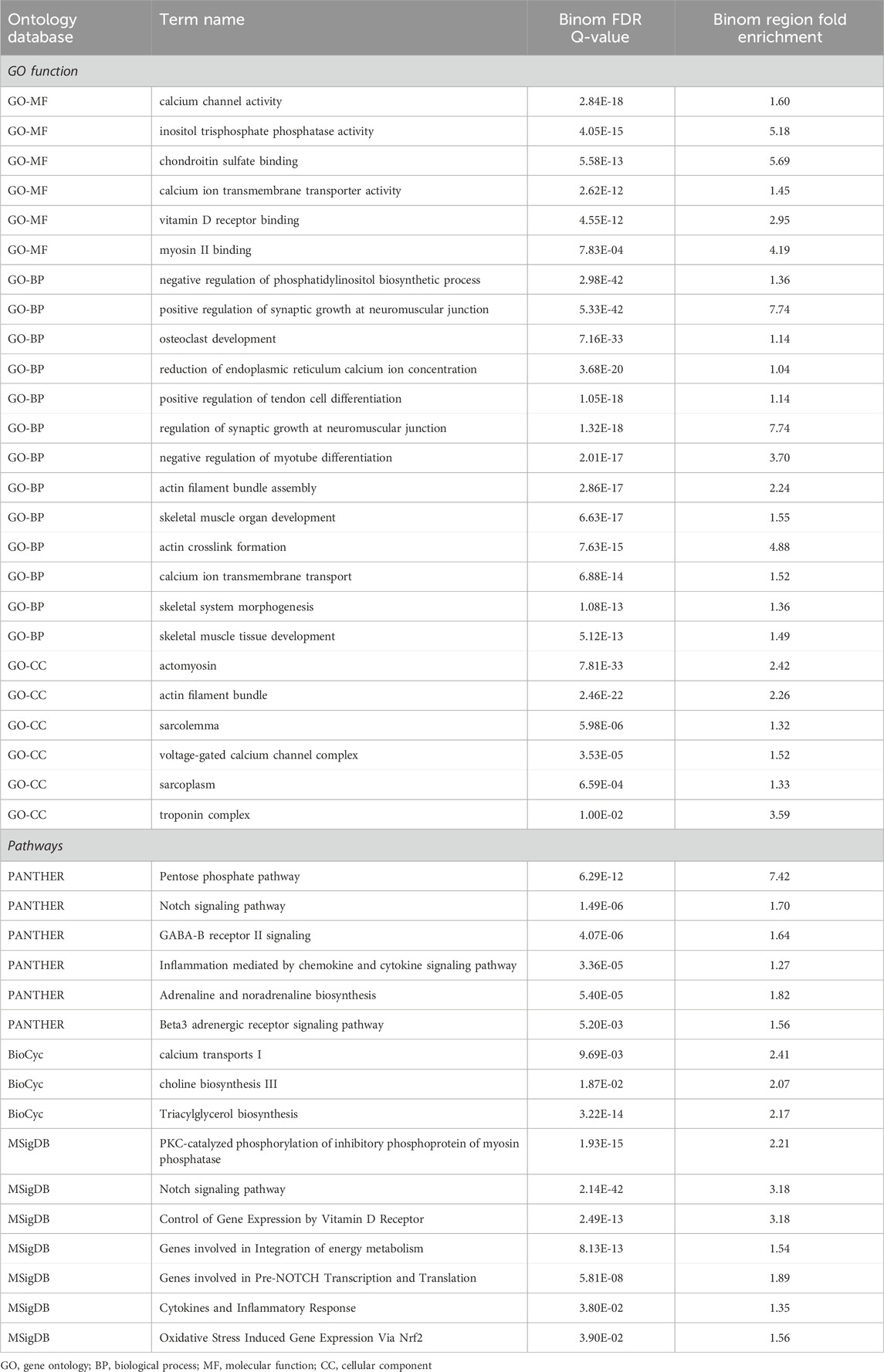
Table 4. The top GREAT ontology enrichment for regions potentially related to handgrip strength by using a binomial test.
3.5 DNA methylation and gene expression analysis
A total of 12 twin pairs with a median age of 53 years (95% range 43–65) and a median HGS of 36.9 kg (95% range 18.4–61.3) were included in the analysis. Supplementary Table S2 presented the results of DNA methylation and gene expression analysis. Methylation level of CpGs located at FBLN1, RXRA, and MRPL23 gene were positively associated with corresponding gene expression level. Methylation level of CpGs located at ABHD14B were negatively associated with gene expression. As shown in Supplementary Figure S1, all of these CpGs are located within gene bodies.
3.6 Quantitative methylation analysis of FBLN1 and ABHD14B
We quantified nine CpGs (FDR< 0.05) mapped to FBLN1 using the Sequenom MassARRAY platform. Six CpGs were negatively associated with HGS weakness (Supplementary Table S3). Four out of the five CpGs mapped on ABHD14B were quantified and one CpG (chr3: 52,007,647) were not detected in the validation experiment. Just one CpG (chr3: 52,007,595) was validated to be hypomethylated in the low HGS group (Supplementary Table S3), although this CpG did not reach genome-wide significance in the EWAS analysis (FDR = 0.054).
4 Discussion
In the present study, we detected epigenetic variants of handgrip strength using EWAS based on monozygotic twins. Our analysis identified several CpGs, genes, DMRs, and pathways that might elucidate the mechanism of muscle strength change. As additional validation, several candidates CpGs mapped to FBLN1 and ABHD14B were quantified and validated.
Research has indicated that the effects of DNA methylation are associated with its occurrence at different positions in the genome and DNA methylation occurring in gene bodies may be related to gene expression (Aquino et al., 2018). Similarly, we found that the methylation levels of CpGs within FBLN1, RXRA, and MRPL23 are positively correlated with gene expression. The methylation of CpGs in FBLN1 is also positively associated with HGS. Fibulin-1, encoded by FBLN1, is an important protein in the structure of elastic fibers and basement membranes of various tissues. Previous studies suggested that Fibulin-1 implicated in tissue organogenesis during myotome development, bone formation, ossification, and digits of the development limbs (de Vega et al., 2009; Bohlega et al., 2014; Cooley et al., 2014). Methylation levels of CpGs located at RXRA and MRPL23 were negatively associated with HGS in EWAS. RXRA is a member of the nuclear receptor superfamily involved in lipid, glucose, energy, and hormone metabolism. Evidence from animal experiments indicates that RXRA promotes adipogenesis and accelerate fat accumulation (Pan et al., 2023). Previous research reported that the DNAm level at four CpGs located at RXRA (chr 9: 136,355,569–136,355,885) was negatively associated with bone mass, which might affect muscle strength (Harvey et al., 2014; Kim et al., 2018). The mechanism underlying the relationship between MRPL23 and muscle strength remains unclear at present and awaits further investigation in future studies. The methylation levels of CpGs located within the gene body of ABHD14B is positively associated with HGS and negatively correlated with gene expression. A previous study found that cortical ABHD14B levels were negatively associated with motor resilience, which reflects muscle endurance and motor function in the elderly (Buchman et al., 2023).
The present study also identified 21 DMRs in genomic regions located near or at 21 genes, of which FBLN1, SLC30A8, CST3, and SOCS3 have potential biological functions related to muscle strength. Sprouse et al. reported that rs13266634 in SLC30A8 is associated with higher muscle strength and larger arm skeletal muscle volume (Sprouse et al., 2014). The protein encoded by CST3 has been implicated as a biomarker for sarcopenia, as measured by the creatinine-to-cystatin C ratio (Tabara et al., 2020; Tabara et al., 2021). PLEC encodes a cytoskeletal protein that maintains tissue integrity by regulating intracellular signaling in response to mechanical stimulation (Rice et al., 2019). SOCS3 has been identified as a crucial mediator in myogenesis. SOCS3 overexpression during myogenesis enhanced myogenin and α-actin mRNA expression (Caldow et al., 2011). The protein encoded by CTSD maintains cellular protein homeostasis and is regarded as a bone-related biomarker of osteoporosis (Deng et al., 2021; Wang et al., 2022; Wu et al., 2022).
Our findings provide evidence of causation underlying DNAm-HGS. We observed HGS might be a response to the methylation level of several CpGs located at different genes. The potential function of FBLN1, RXRA, and ABHD14B in muscle strength has been discussed above. Recent research has indicated that the methylation level of RXRA promoters could change in response to the external stimulus of mechanical loading (Krstic et al., 2022), which might explain the reason for the DNAm response to HGS changes. Additionally, studies have reported an association between lower plasma vitamin D levels and elevated RXRA gene methylation (Curtis et al., 2019; Krstic et al., 2022). Considering the role of vitamin D in muscle strength, the changes in HGS-induced RXRA DNA methylation alterations may also be attributed to the potential confounding effect of vitamin D.
The strengths and advantages of the current study are as follows. First, our study included monozygotic twin pairs, which controlled the genetic background and enhanced the credibility of the results. Second, we investigated the causal relationship between DNAm and HGS, providing a deeper understanding of the effect of epigenetic modifications on HGS. Thirdly, our study is the first EWAS of HGS in the Asian population. Given the diverse genetic backgrounds and environmental exposures among different ethnic groups, our research illuminates the intrinsic physiological mechanisms contributing to the variation in HGS.
There are limitations of our study that must be acknowledged. First, our study did not consider physical activity and exercise due to a lack of relevant information, which can directly affect HGS or DNA methylation. The second limitation lies in the use of DNA methylation data derived from blood rather than the more intuitively relevant muscle tissue. Despite the challenges in obtaining muscle tissue, we acknowledge the tissue specificity of DNA methylation. Previous study has also indicated differences in the epigenetic feature between muscle tissue and blood (Slieker et al., 2013). However, a study conducted by Pilling et al. identified potential gene expression characteristics related to muscle strength in the blood and revealed several genes previously reported to be associated with muscle strength (Pilling et al., 2016). Additionally, some studies suggest that while the use of whole blood DNA methylation data may only reflect changes in blood, it can still provide information about variations in muscle mass (Livshits et al., 2016). Therefore, the use of blood samples for epigenetic studies on muscle strength may be a feasible approach (Soerensen et al., 2019). Finally, we recognize that HGS serves as an indicator of upper limb strength rather than a comprehensive measure of whole-body strength. Hence, future research should employ more representative indicators to validate our findings.
5 Conclusion
In summary, our study utilizing monozygotic twin pairs detected many DNAm variants that may be associated with handgrip strength, particularly the loci within FBLN1 and ABHD14B. Our findings provide new clues to the epigenetic modification underlying muscle strength.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: We are preparing the raw data (fastq files for RRBS libraries) for uploading to the public website, and until then the data used or analyzed during the current study are available from the corresponding author upon reasonable request. Requests to access these datasets should be directed to DZ, emhhbmdkZjE5NjFAMTI2LmNvbQ==.
Ethics statement
The studies involving humans were approved by the Regional Ethics Committee of the Qingdao CDC Institutional Review Boards. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JLu: Conceptualization, Formal Analysis, Methodology, Software, Visualization, Writing–original draft. WW: Methodology, Software, Validation, Writing–review and editing. JLi: Data curation, Software, Writing–original draft. HD: Data curation, Investigation, Writing–original draft. CX: Data curation, Investigation, Writing–original draft. XT: Data curation, Investigation, Writing–original draft. DZ: Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the National Natural Science Foundation of China (82073641). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The authors would like to thank all the participants.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1378680/full#supplementary-material
Abbreviations
MZ, Monozygotic; EWAS, Epigenome-Wide Association Study; GEE, Generalized estimating equation; ICE FALCON, Inference about Causation through Examination of Familial Confounding method; FDR, False discovery rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DMRs, Differentially methylation regions; HGS, Handgrip strength; DNAm, DNA methylation; GWAS, Genome-wide association study; GREAT, Genomic Regions Enrichment of Annotations Tool.
References
Alcazar, J., Aagaard, P., Haddock, B., Kamper, R. S., Hansen, S. K., Prescott, E., et al. (2020). Age- and sex-specific changes in lower-limb muscle power throughout the lifespan. journals gerontology Ser. A, Biol. Sci. Med. Sci. 75 (7), 1369–1378. doi:10.1093/gerona/glaa013
Antoun, E., Garratt, E. S., Taddei, A., Burton, M. A., Barton, S. J., Titcombe, P., et al. (2022). Epigenome-wide association study of sarcopenia: findings from the Hertfordshire Sarcopenia Study (HSS). J. cachexia, sarcopenia muscle 13 (1), 240–253. doi:10.1002/jcsm.12876
Aquino, E. M., Benton, M. C., Haupt, L. M., Sutherland, H. G., Griffiths, L. R., and Lea, RAJOG (2018). Current understanding of DNA methylation and age-related disease. OBM Genet. 2 (2), 1–16. doi:10.21926/obm.genet.1802016
Balogun, S., Winzenberg, T., Wills, K., Scott, D., Jones, G., Callisaya, M. L., et al. (2019). Prospective associations of low muscle mass and strength with health-related quality of life over 10-year in community-dwelling older adults. Exp. Gerontol. 118, 65–71. doi:10.1016/j.exger.2019.01.008
Bell, J. T., Tsai, P. C., Yang, T. P., Pidsley, R., Nisbet, J., Glass, D., et al. (2012). Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 8 (4), e1002629. doi:10.1371/journal.pgen.1002629
Bharathy, N., Ling, B. M., and Taneja, R. (2013). Epigenetic regulation of skeletal muscle development and differentiation. Sub-cellular Biochem. 61, 139–150. doi:10.1007/978-94-007-4525-4_7
Bohlega, S., Al-Ajlan, H., and Al-Saif, A. (2014). Mutation of fibulin-1 causes a novel syndrome involving the central nervous system and connective tissues. Eur. J. Hum. Genet. 22 (5), 640–643. doi:10.1038/ejhg.2013.210
Buchman, A. S., Yu, L., Klein, H. U., Zammit, A. R., Oveisgharan, S., Grodstein, F., et al. (2023). Proteome-wide discovery of cortical proteins that may provide motor resilience to offset the negative effects of pathologies in older adults. journals gerontology Ser. A, Biol. Sci. Med. Sci. 78 (3), 494–503. doi:10.1093/gerona/glac105
Caldow, M. K., Steinberg, G. R., and Cameron-Smith, D. (2011). Impact of SOCS3 overexpression on human skeletal muscle development in vitro. Cytokine 55 (1), 104–109. doi:10.1016/j.cyto.2011.03.012
Carrió, E., Díez-Villanueva, A., Lois, S., Mallona, I., Cases, I., Forn, M., et al. (2015). Deconstruction of DNA methylation patterns during myogenesis reveals specific epigenetic events in the establishment of the skeletal muscle lineage. Stem cells Dayt. Ohio 33 (6), 2025–2036. doi:10.1002/stem.1998
Cooley, M. A., Harikrishnan, K., Oppel, J. A., Miler, S. F., Barth, J. L., Haycraft, C. J., et al. (2014). Fibulin-1 is required for bone formation and Bmp-2-mediated induction of Osterix. Bone 69, 30–38. doi:10.1016/j.bone.2014.07.038
Curtis, E. M., Krstic, N., Cook, E., D'Angelo, S., Crozier, S. R., Moon, R. J., et al. (2019). Gestational vitamin D supplementation leads to reduced perinatal RXRA DNA methylation: results from the MAVIDOS trial. J. bone mineral Res. 34 (2), 231–240. doi:10.1002/jbmr.3603
Deng, Y. X., He, W. G., Cai, H. J., Jiang, J. H., Yang, Y. Y., Dan, Y. R., et al. (2021). Analysis and validation of hub genes in blood monocytes of postmenopausal osteoporosis patients. Front. Endocrinol. 12, 815245. doi:10.3389/fendo.2021.815245
de Vega, S., Iwamoto, T., and Yamada, Y. (2009). Fibulins: multiple roles in matrix structures and tissue functions. Cell. Mol. life Sci. 66 (11-12), 1890–1902. doi:10.1007/s00018-009-8632-6
Godfrey, K. M., Lillycrop, K. A., Burdge, G. C., Gluckman, P. D., and Hanson, M. A. (2007). Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr. Res. 61 (5), 5R–10R. doi:10.1203/pdr.0b013e318045bedb
Harvey, N. C., Sheppard, A., Godfrey, K. M., McLean, C., Garratt, E., Ntani, G., et al. (2014). Childhood bone mineral content is associated with methylation status of the RXRA promoter at birth. J. bone mineral Res. 29 (3), 600–607. doi:10.1002/jbmr.2056
He, L., Khanal, P., Morse, C. I., Williams, A., and Thomis, M. (2020). Associations of combined genetic and epigenetic scores with muscle size and muscle strength: a pilot study in older women. J. cachexia, sarcopenia muscle 11 (6), 1548–1561. doi:10.1002/jcsm.12585
Jaffe, A. E., and Irizarry, R. A. (2014). Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15 (2), R31. doi:10.1186/gb-2014-15-2-r31
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14 (4), R36. doi:10.1186/gb-2013-14-4-r36
Kim, K. M., Lim, S., Oh, T. J., Moon, J. H., Choi, S. H., Lim, J. Y., et al. (2018). Longitudinal changes in muscle mass and strength, and bone mass in older adults: gender-specific associations between muscle and bone losses. journals gerontology Ser. A, Biol. Sci. Med. Sci. 73 (8), 1062–1069. doi:10.1093/gerona/glx188
Krstic, N., Bishop, N., Curtis, B., Cooper, C., Harvey, N., Lilycrop, K., et al. (2022). Early life vitamin D depletion and mechanical loading determine methylation changes in the RUNX2, RXRA, and osterix promoters in mice. Genes & Nutr. 17 (1), 7. doi:10.1186/s12263-022-00711-0
Krueger, F., and Andrews, S. R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinforma. Oxf. Engl. 27 (11), 1571–1572. doi:10.1093/bioinformatics/btr167
Laker, R. C., and Ryall, J. G. (2016). DNA methylation in skeletal muscle stem cell specification, proliferation, and differentiation. Stem cells Int. 2016, 5725927. doi:10.1155/2016/5725927
Lauretani, F., Russo, C. R., Bandinelli, S., Bartali, B., Cavazzini, C., Di Iorio, A., et al. (2003). Age-associated changes in skeletal muscles and their effect on mobility: an operational diagnosis of sarcopenia. J. Appl. physiology (1985) 95 (5), 1851–1860. doi:10.1152/japplphysiol.00246.2003
Li, S., Wong, E. M., Bui, M., Nguyen, T. L., Joo, J. E., Stone, J., et al. (2005)2019). Inference about causation between body mass index and DNA methylation in blood from a twin family study. Int. J. Obes. 43 (2), 243–252. doi:10.1038/s41366-018-0103-4
Li, W., Baumbach, J., Larsen, M. J., Mohammadnejad, A., Lund, J., Christensen, K., et al. (2020). Differential long noncoding RNA profiling of BMI in twins. Epigenomics 12 (17), 1531–1541. doi:10.2217/epi-2020-0033
Li, W., Christiansen, L., Hjelmborg, J., Baumbach, J., and Tan, Q. (2018). On the power of epigenome-wide association studies using a disease-discordant twin design. Bioinforma. Oxf. Engl. 34 (23), 4073–4078. doi:10.1093/bioinformatics/bty532
Livshits, G., Gao, F., Malkin, I., Needhamsen, M., Xia, Y., Yuan, W., et al. (2016). Contribution of heritability and epigenetic factors to skeletal muscle mass variation in United Kingdom twins. J. Clin. Endocrinol. metabolism 101 (6), 2450–2459. doi:10.1210/jc.2016-1219
Luo, J., Zhang, T., Wang, W., and Zhang, D. (2022). Genome-wide association study of handgrip strength in the Northern Chinese adult twins. Connect. tissue Res. 64, 117–125. doi:10.1080/03008207.2022.2104160
Marioni, R. E., Shah, S., McRae, A. F., Ritchie, S. J., Muniz-Terrera, G., Harris, S. E., et al. (2015). The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 44 (4), 1388–1396. doi:10.1093/ije/dyu277
Matteini, A. M., Fallin, M. D., Kammerer, C. M., Schupf, N., Yashin, A. I., Christensen, K., et al. (2010). Heritability estimates of endophenotypes of long and health life: the Long Life Family Study. journals gerontology Ser. A, Biol. Sci. Med. Sci. 65 (12), 1375–1379. doi:10.1093/gerona/glq154
Matteini, A. M., Tanaka, T., Karasik, D., Atzmon, G., Chou, W. C., Eicher, J. D., et al. (2016). GWAS analysis of handgrip and lower body strength in older adults in the CHARGE consortium. Aging Cell 15 (5), 792–800. doi:10.1111/acel.12468
McLean, C. Y., Bristor, D., Hiller, M., Clarke, S. L., Schaar, B. T., Lowe, C. B., et al. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28 (5), 495–501. doi:10.1038/nbt.1630
McLeod, M., Breen, L., Hamilton, D. L., and Philp, A. (2016). Live strong and prosper: the importance of skeletal muscle strength for healthy ageing. Biogerontology 17 (3), 497–510. doi:10.1007/s10522-015-9631-7
Mendes, J., Amaral, T. F., Borges, N., Santos, A., Padrão, P., Moreira, P., et al. (2017). Handgrip strength values of Portuguese older adults: a population based study. BMC Geriatr. 17 (1), 191. doi:10.1186/s12877-017-0590-5
Pan, Z., Li, X., Wu, D., Chen, X., Zhang, C., Jin, S., et al. (2023). The duck RXRA gene promotes adipogenesis and correlates with feed efficiency. Animals open access J. MDPI. 13 (4), 680. doi:10.3390/ani13040680
Pedersen, B. S., Schwartz, D. A., Yang, I. V., and Kechris, K. J. (2012). Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinforma. Oxf. Engl. 28 (22), 2986–2988. doi:10.1093/bioinformatics/bts545
Pilling, L. C., Joehanes, R., Kacprowski, T., Peters, M., Jansen, R., Karasik, D., et al. (2016). Gene transcripts associated with muscle strength: a CHARGE meta-analysis of 7,781 persons. Physiol. genomics 48 (1), 1–11. doi:10.1152/physiolgenomics.00054.2015
Rahmani, E., Zaitlen, N., Baran, Y., Eng, C., Hu, D., Galanter, J., et al. (2016). Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat. methods 13 (5), 443–445. doi:10.1038/nmeth.3809
Rice, S. J., Tselepi, M., Sorial, A. K., Aubourg, G., Shepherd, C., Almarza, D., et al. (2019). Prioritization of PLEC and GRINA as osteoarthritis risk genes through the identification and characterization of novel methylation quantitative trait loci. Arthritis & rheumatology (Hoboken, NJ) 71 (8), 1285–1296. doi:10.1002/art.40849
Rondanelli, M., Klersy, C., Terracol, G., Talluri, J., Maugeri, R., Guido, D., et al. (2016). Whey protein, amino acids, and vitamin D supplementation with physical activity increases fat-free mass and strength, functionality, and quality of life and decreases inflammation in sarcopenic elderly. Am. J. Clin. Nutr. 103 (3), 830–840. doi:10.3945/ajcn.115.113357
Slieker, R. C., Bos, S. D., Goeman, J. J., Bovée, J. V., Talens, R. P., van der Breggen, R., et al. (2013). Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics chromatin 6 (1), 26. doi:10.1186/1756-8935-6-26
Soerensen, M., Li, W., Debrabant, B., Nygaard, M., Mengel-From, J., Frost, M., et al. (2019). Epigenome-wide exploratory study of monozygotic twins suggests differentially methylated regions to associate with hand grip strength. Biogerontology 20 (5), 627–647. doi:10.1007/s10522-019-09818-1
Sprouse, C., Gordish-Dressman, H., Orkunoglu-Suer, E. F., Lipof, J. S., Moeckel-Cole, S., Patel, R. R., et al. (2014). SLC30A8 nonsynonymous variant is associated with recovery following exercise and skeletal muscle size and strength. Diabetes 63 (1), 363–368. doi:10.2337/db13-1150
Tabara, Y., Kohara, K., Okada, Y., Ohyagi, Y., and Igase, M. (2020). Creatinine-to-cystatin C ratio as a marker of skeletal muscle mass in older adults: J-SHIPP study. Clin. Nutr. Edinb. Scotl. 39 (6), 1857–1862. doi:10.1016/j.clnu.2019.07.027
Tabara, Y., Okada, Y., Ochi, M., Ohyagi, Y., and Igase, M. (2021). Association of creatinine-to-cystatin C ratio with myosteatosis and physical performance in older adults: the Japan shimanami health promoting program. J. Am. Med. Dir. Assoc. 22 (11), 2366–2372.e3. doi:10.1016/j.jamda.2021.03.021
Tan, Q., Christiansen, L., von Bornemann Hjelmborg, J., and Christensen, K. (2015). Twin methodology in epigenetic studies. J. Exp. Biol. 218 (1), 134–139. doi:10.1242/jeb.107151
Tiainen, K., Sipilä, S., Alen, M., Heikkinen, E., Kaprio, J., Koskenvuo, M., et al. (2004). Heritability of maximal isometric muscle strength in older female twins. J. Appl. physiology (1985) 96 (1), 173–180. doi:10.1152/japplphysiol.00200.2003
Tian, X., Xu, C., Wu, Y., Sun, J., Duan, H., Zhang, D., et al. (2017). Genetic and environmental influences on pulmonary function and muscle strength: the Chinese twin study of aging. Twin Res. Hum. Genet. 20 (1), 53–59. doi:10.1017/thg.2016.97
Turner, D. C., Gorski, P. P., Maasar, M. F., Seaborne, R. A., Baumert, P., Brown, A. D., et al. (2020). DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: the role of HOX genes and physical activity. Sci. Rep. 10 (1), 15360. doi:10.1038/s41598-020-72730-z
Voisin, S., Jacques, M., Landen, S., Harvey, N. R., Haupt, L. M., Griffiths, L. R., et al. (2021). Meta-analysis of genome-wide DNA methylation and integrative omics of age in human skeletal muscle. J. cachexia, sarcopenia muscle 12 (4), 1064–1078. doi:10.1002/jcsm.12741
Wang, T., Wang, W., Li, W., Duan, H., Xu, C., Tian, X., et al. (2021b). Genome-wide DNA methylation analysis of pulmonary function in middle and old-aged Chinese monozygotic twins. Respir. Res. 22 (1), 300. doi:10.1186/s12931-021-01896-5
Wang, W., Li, W., Wu, Y., Tian, X., Duan, H., Li, S., et al. (2021a). Genome-wide DNA methylation and gene expression analyses in monozygotic twins identify potential biomarkers of depression. Transl. psychiatry 11 (1), 416. doi:10.1038/s41398-021-01536-y
Wang, W., Yao, J., Li, W., Wu, Y., Duan, H., Xu, C., et al. (2023). Epigenome-wide association study in Chinese monozygotic twins identifies DNA methylation loci associated with blood pressure. Clin. epigenetics 15 (1), 38. doi:10.1186/s13148-023-01457-1
Wang, X., Pei, Z., Hao, T., Ariben, J., Li, S., He, W., et al. (2022). Prognostic analysis and validation of diagnostic marker genes in patients with osteoporosis. Front. Immunol. 13, 987937. doi:10.3389/fimmu.2022.987937
Weng, W. H., Cheng, Y. H., Yang, T. H., Lee, S. J., Yang, Y. R., and Wang, R. Y. (2022). Effects of strength exercises combined with other training on physical performance in frail older adults: a systematic review and meta-analysis. Archives gerontology geriatrics 102, 104757. doi:10.1016/j.archger.2022.104757
Wu, P. H., Glerup, R. I., Svensson, M. H. S., Eriksson, N., Christensen, J. H., Linde, T., et al. (2022). Osteoprotegerin predicts cardiovascular events in patients treated with haemodialysis. Nephrol. Dial. Transplant. 37 (6), 1162–1170. doi:10.1093/ndt/gfab192
Xu, C., Zhang, D., Tian, X., Wu, Y., Pang, Z., Li, S., et al. (2017). Genetic and environmental basis in phenotype correlation between physical function and cognition in aging Chinese twins. Twin Res. Hum. Genet. 20 (1), 60–65. doi:10.1017/thg.2016.98
Keywords: epigenetics, DNA methylation, monozygotic twins, handgrip strength, causal inference
Citation: Luo J, Wang W, Li J, Duan H, Xu C, Tian X and Zhang D (2024) Epigenome-wide association study identifies DNA methylation loci associated with handgrip strength in Chinese monozygotic twins. Front. Cell Dev. Biol. 12:1378680. doi: 10.3389/fcell.2024.1378680
Received: 30 January 2024; Accepted: 22 March 2024;
Published: 03 April 2024.
Edited by:
Wieslawa Lesniak, Polish Academy of Sciences, PolandReviewed by:
Magdeldin Elgizouli, University of Zurich, SwitzerlandLiam C. Hunt, Rhodes College, United States
Copyright © 2024 Luo, Wang, Li, Duan, Xu, Tian and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongfeng Zhang, emhhbmdkZjE5NjFAMTI2LmNvbQ==
 Jia Luo
Jia Luo Weijing Wang
Weijing Wang Jingxian Li
Jingxian Li Haiping Duan
Haiping Duan Chunsheng Xu
Chunsheng Xu Xiaocao Tian
Xiaocao Tian Dongfeng Zhang
Dongfeng Zhang