- 1College of Animal Science and Technology, Qingdao Agricultural University, Qingdao, Shandong, China
- 2Animal Products Quality and Safety Center of Shandong Province, Jinan, Shandong, China
- 3Provincial Animal Husbandry Station of Shandong Province, Jinan, Shandong, China
As a crucial component of the male reproductive system, the epididymis plays multiple roles, including sperm storage and secretion of nutritive fluids for sperm development and maturation. The acquisition of fertilization capacity by sperm occurs during their transport through the epididymis. Compared with the testis, little has been realized about the importance of the epididymis. However, with the development of molecular biology and single-cell sequencing technology, the importance of the epididymis for male fertility should be reconsidered. Recent studies have revealed that different regions of the epididymis exhibit distinct functions and cell type compositions, which are likely determined by variations in gene expression patterns. In this research, we primarily focused on elucidating the cellular composition and region-specific gene expression patterns within different segments of the epididymis and provided detailed insights into epididymal function in male fertility.
1 Introduction
The epididymis, primarily composed of an epithelial membrane and a thin annular muscle, exhibits secretory functions that contribute to sperm nutrition and maturation. Species variations are observed in the length and morphology of the epididymis. Notably, mice possess an epididymal tube exceeding 1 m in length (approximately 100 times longer than the epididymis itself), while rats have 3 m and humans have 6 m (Von and Neuhaeuser, 1964; Turner et al., 1990; Jiang et al., 1994; Stoltenberg et al., 1998). As a component of the male reproductive tract, the development of the epididymis is dependent on androgens and originates from the Wolffian duct (Shaw and Renfree, 2014). Studies indicated that Hoxa10 and Hoxa11 genes, belonging to the homeobox (Hox) gene family, played crucial roles in epididymal development (Branford et al., 2000; Snyder et al., 2010).
The function of the epididymis is closely related to the sperm maturation. It has been established that mammalian spermatozoa undergo a process of “ripening” as they traverse the epididymis (Westfalewicz et al., 2017) While human sperm transport typically takes an average of 12 days (Rowley et al., 1970). In addition to these maturation steps, the epididymis provides a sophisticated microenvironment that allows for stationary storage of sperm during epididymal transit, avoiding premature sperm activation (Acott and Carr, 1984). Fundamentally, the four primary functions of the epididymis encompass storing, transporting, protecting, and eliminating sperm.
The earliest report on the epididymis was published in the 18th century (National Library of Medicine, 1859): discutient application to the indurated epididymis, focusing on discutient application to treat indurated epididymis. Early understanding of the epididymis stemmed from various pathological studies (Rockwell, 1888; Bryant, 1892; Some Diseases of the Male Genital System, 1908) and investigations into its anatomical structures and functions (Griffiths, 1893; Watson, 1902). Since the 1950s, a significant number of research papers had emerged in this field. Over the past 70 years, extensive exploration had been conducted on different aspects of the epididymis. Initially, pioneers elucidated its anatomy in horses (Goglia, 1954), guinea pigs (Goglia and Magli, 1957) and humans (Montagna, 1952). Subsequently, cellular and chemical analyses gained prominence with cytochemical research and histochemical study of mouse epididymis (Allen, 1961; Allen and Slater, 1961; Birnbaum et al., 1961). Since the 1960s, continuous research efforts were dedicated to study the epididymis as evidenced by an increasing number of annual publications depicted in Figure 1A. With advancements in sequencing technology came a greater emphasis on understanding detailed functions of the epididymis. Consequently, its crucial role in sperm maturation was discovered (Salisbury et al., 1963; Salisbury and Graves, 1963). This led to further investigations into the proteome, transcriptome, and genetics of this organ resulting in numerous reported findings (Dacheux et al., 2009; Wong et al., 2020; Zheng et al., 2021). Transcriptomic and proteomic techniques facilitated progress in studying gene expression specific to the epididymis along with their associated functions (Lye and Hinton, 2004; Légaré and Sullivan, 2020). It is now well-established that each segment within this highly segmented structure expresses distinct genes as well as the related proteins (Domeniconi et al., 2016). Figure 1B illustrated that research pertaining to region-specific gene expression patterns began appearing since 1983 with an upward trend observed annually.
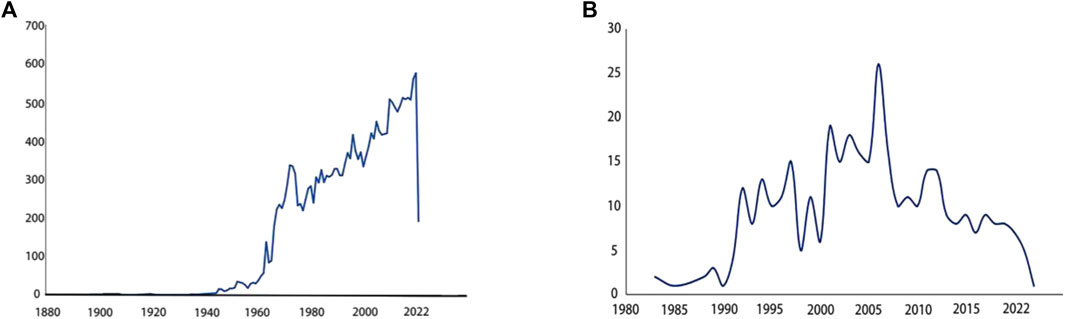
Figure 1. Paper counts published annually related to specific keywords (A) Paper counts published annually on epididymis. Search for epididymis keywords in PubMed. The number of research papers published each year is shown in the line graph. From 1960 to 2022, research papers about epididymis published in PubMed every year. More and more research papers are published every year, and the trend is rising in a straight line. (B) Paper counts published annually on region specific-gene expression in epididymis. Search for epididymis region specific gene expression keywords in PubMed. The number of research papers published each year is shown in the line graph. It is found that relevant research papers only appeared in 1983. From 1983 to 2022, the trend first rose and then declined.
It had been reported that the environment of the epididymal duct might influence on the steps of sperm maturation (Shimazaki et al., 1976). During the process of mammalian sperm transport through the epididymis, the sperm encountered intraluminal fluid with disparate protein compositions, which interacted with the sperm surface and confers fertilization capabilities to the male gamete (Dacheux et al., 2005). Numerous studies had shown that a majority of sperm in the cauda region of epididymis were matured. As sperm passed through the epididymis, they undergo capacitation, acquire fertility, and could be used for in vitro fertilization (IVF) or artificial insemination (AI) (Axnér et al., 1999; Tsutsui et al., 2003; Toyonaga et al., 2011).
2 Relevant advancements Applied to research of epididymis
Rats and mice had been the most commonly used research models for studying the epididymis. In recent decades, advancements in technology, such as gene-knockout approaches (Eddy et al., 1996), transcriptome sequencing (Wu et al., 2021a), microarray analysis (Jervis and Robaire, 2003), proteomics (Fouchécourt et al., 2000), cell lines (Masuda et al., 2022), and single cell RNA-seq methods for transcriptome analysis (Rinaldi et al., 2020; Lang et al., 2022), had significantly enhanced our understanding of epididymal composition. Numerous knockout mouse models have exhibited phenotypes that impact male reproductive function. For instance, sperm from Clgn, Adam1a, Adam2, and Adam3 knockout mice were unable to migrate to the fallopian tubes (Cho et al., 1998; Ikawa et al., 2001; Nishimura et al., 2004). However, the dynamic functioning of these genes remained largely unknown. Both in vivo and in vitro models had been employed to investigate epididymal function. Currently, culturing techniques for epididymal cells had become well-established (Klinefelter et al., 1982; Chen et al., 1998; Lee et al., 2007). A rat caput epididymal cell line had demonstrated characteristics similar to epithelial principal cells found in vivo (Araki et al., 2002; Dufresne et al., 2005). The advent of RNA sequencing technologies had enabled researchers to analyze the transcriptomes of specific groups or even individual cells (Vazquez et al., 2019). Correlative studies had revealed novel distribution patterns of mitochondria and key genes that might be as-sociated with initial and subsequent sperm waves (Shi et al., 2021).
At present, there are many achievements in the application of single cell technology in the field of epididymis research. Human nasal, bronchial and epididymal epithelial cells were compared by single cell analysis. It has been found that when secretory cells, ciliated cells and basal cells are located in different anatomical locations, their transcriptome features are different (Paranjapye et al., 2022). Eight clusters of cell types have been identified in the proximal human epididymis, including secretory and basal epithelial cells, as well as rare clear cells, all of which have overlapping functions with bronchial and nasal epithelial cells (Paranjapye et al., 2022). Aged principal cells showed a variety of functional gene expression changes related to acrosome response and sperm maturation, suggesting that sperm activation and maturation is an asynchronous process during epididymal transport (Zhuang et al., 2023). At the same time, pathway alterations associated with aging were found in immune cells, particularly “cell chemotaxis” in Cx3cr1Hi epididymal dendritic cells (Zhuang et al., 2023). The monocyte specific expression of chemokine Ccl8 increased with age (Zhuang et al., 2023). Single-cell analysis of zebrafish testis revealed thousands of new marker genes specific to cell types, and through ligand receptor (LR) analysis, it was found that Zebrafish stromal cells had a stronger paracrine effect on germ cells than sertolith cells (Qian et al., 2022). The regulatory network, upstream transcription factors and downstream pathways of human epididymal fragment specific miRNA-mRNA were revealed, which provided a basis for further study of epididymal fragment specific function (Chen et al., 2023). Human epididymis was analyzed by single-cell RNA sequencing to further characterize efferent tubes and model systems (Leir et al., 2023).
The RNA-Seq dataset revealed a substantial number of genes expressed in different regions of the epididymis and the gene expression patterns exhibited significant differences among these regions. Notably, principal cells displayed diverse subsets that were distributed throughout the caput, corpus, and cauda of the epididymis (Rinaldi et al., 2020). However, numbers of genes could be used to distinguish the principal cells from different regions of the epididymal tube. RNase10, Cst11, Lcn2 and Mfge8 genes exhibited high expression levels in caput principal cells. Lcn5, Rnase9 and Plac8 genes were expressed in corpus principal cells (Rinaldi et al., 2020). Additionally, Gpx3, Hint1, Spink10 and Crisp1 showed significant expression in cauda principal cells (Rinaldi et al., 2020). Clear cells were identified by expression of the V-ATPase encoding marker genes such as Atp6v0c, Atpv1e1, and Atp6v1a (Rinaldi et al., 2020). Three clusters of basal cells were identified based on the shared expression of marker genes including Itga6 and Krt14 (Rinaldi et al., 2020). Another study employing single-cell analysis discovered eight subsets within the population of principal cells (Shi et al., 2021). According to GO analysis, two of these subsets were found to be enriched with genes associated with cilia organization and assembly, microtubule-based movement, and cilia-dependent cell movement (Shi et al., 2021). Other major subsets of epididymal epithelial cells were also examined. In detail, four cell subtypes were identified in cluster of basal cells, clear cells, and halo cells (Shi et al., 2021). In this study, the principal cells were characterized by the expression of Aqp9 and Cftr as marker genes, while Krt5 and Cldn1 served as marker genes for basal cells. Clear cells were identified by the presence of Atp6v1b1 and Foxil as marker genes (Shi et al., 2021). Additionally, novel region-specific genes were discovered in principal cells: Lcp1, Pemt, and Ucp2 exhibited high expression levels in the caput; Ccdc198, Ramp3, and Srgn showed high expression levels in the corpus; Atf3 and Zfp36 displayed high expression levels in the cauda of the epididymis (Shi et al., 2021). As new marker genes continue to be discovered, there are slight differences with marker genes in different cell types of different research. It was found that the number of mitochondria in the corpus and cauda of the mice was significantly higher than that in the caput of the epididymis (Shi et al., 2021). Mitochondria are a very important and abundant organelle type in the cytoplasm, involved in the production of adenosine triphosphate (ATP), the establishment of developmental capacity, the maintenance of calcium homeostasis, the regulation of apoptosis and other processes (Shi et al., 2021). The high mitochondrial count and MT% in the epididymis may be required to provide enough energy to synthesize and secrete thousands of molecules that are necessary for various sperm functions (Shi et al., 2021). Over the past 2 years, spatial transcriptomics had provided a wealth of quantitative gene expression data regarding mRNA distribution within tissue slices. These emerging technologies offered new opportunities for bioinformatics analyses in both research and diagnosis (Ståhl et al., 2016). In this paper, we provided an overview of various techniques utilized in epididymis research. Table 1 listed representative research papers showcasing applications of these new technologies within this field. It had been acknowledged that variations in gene and protein expression exist among distinct regions of the epididymis (Johnston et al., 2005), indicating diverse roles or functions in sperm maturation across these regions.
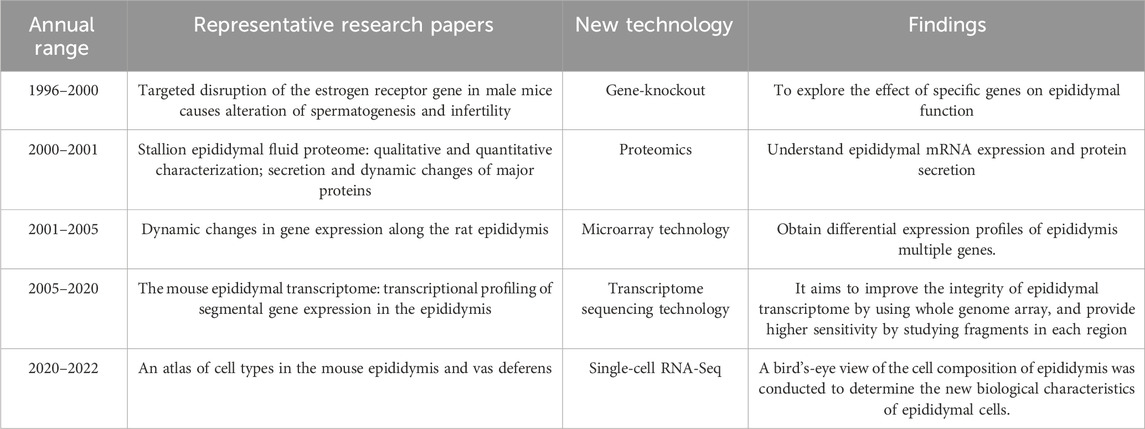
Table 1. Representative research papers on the application of new technologies in the field of epididymis.
3 Regional segmentation and functional differentiation in epididymis
In previous studies, the epididymis had been traditionally classified into three regions–caput, corpus, and cauda. However, in rodents, the epididymis was further divided into four anatomical regions-initial segment, caput, corpus, and cauda (Gervasi and Visconti, 2017). While these divisions were well-established in rodents, they had not been clearly defined in the human epididymis. Previous reports had primarily used the caput, corpus, and cauda as boundaries for analyzing gene and protein expression patterns within the epididymis (Jervis and Robaire, 2001; Chauvin and Griswold, 2004). The caput, corpus, and cauda of the epididymis were proximal to distal, relative to the testis (Figure 2A).
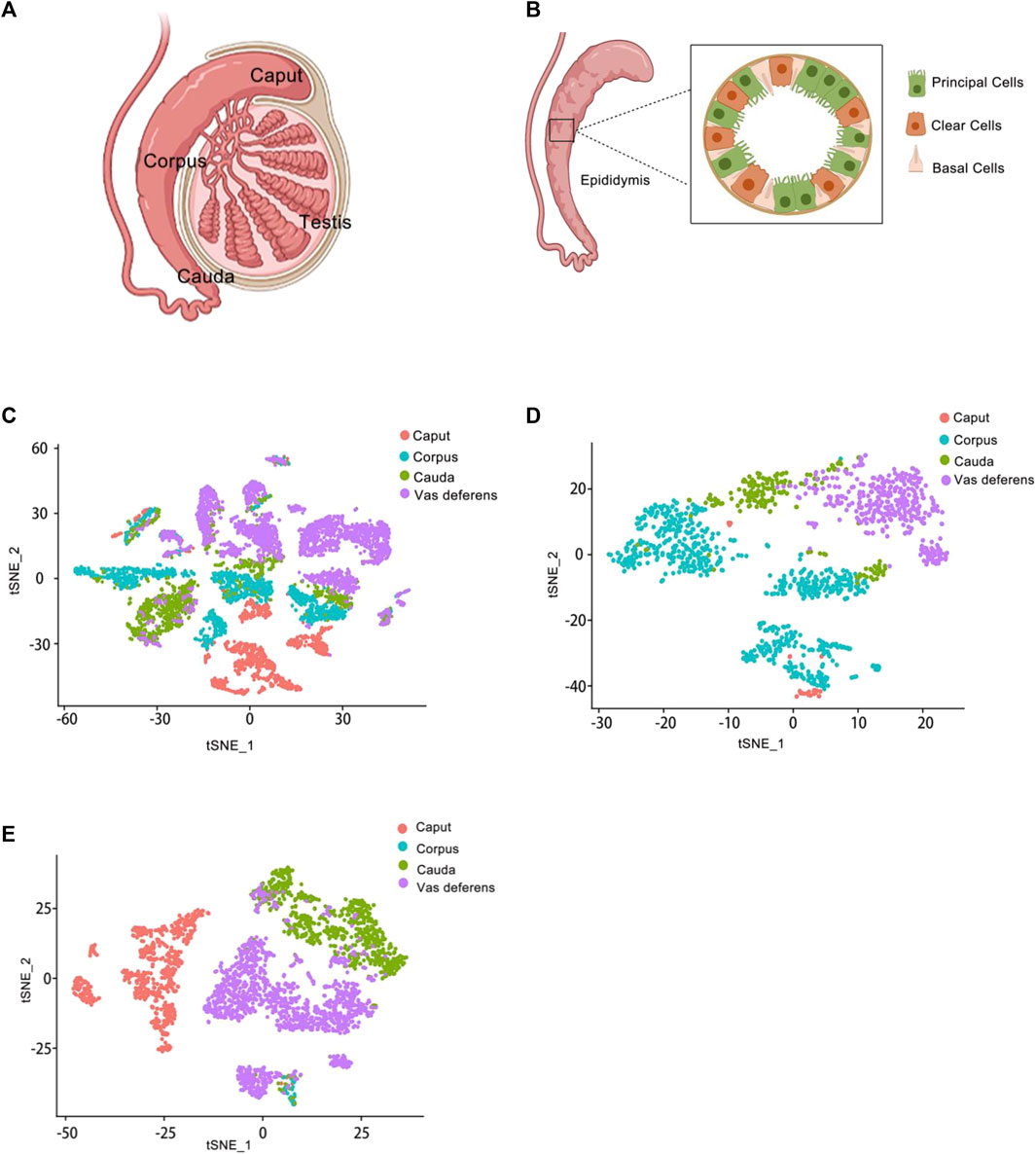
Figure 2. Epididymis atlas from anatomy to single cell level (A) Anatomy of the epididymis. The epididymis is attached to the testis. From top to bottom, it is the caput of the epididymis, the corpus of the epididymis, and the cauda of the epididymis. (B) Schematic representation of the main cell types in the epididymis. Several cell types make up the epididymal epithelium. It mainly consists of principal cells, basal cells and clear cells. Principal cells are the most abundant cells, which responsible for the absorption and secretion of substances into the epididymal cavity. Basal cells have a different morphological appearance, and they have the property of extending elongated body projections into the lumen between adjacent epithelial cells. Clear cells express proton-pumping ATPase in their apical membrane. (C) Single-cell cluster, clustered by t-SNE and annotated according to the four anatomical regions (caput, corpus, cauda and vas deferens). The data of epididymal caput, carpus, cauda and vas deferens were analyzed jointly. This is the t-SNE cluster diagram of four anatomical regions. Each color represents a cluster of cells in an anatomical region. The red dot represents the epididymis caput cells; The blue dot represents the epididymis corpus cells; The green dot represents the epididymis cauda cells; The purple dot represents the epididymis vas deference cells. (D) Reclustering of extracted basal cells, visualized by t-SNE and annotated according to the four anatomical regions. This figure shows the t-SNE cluster diagram of basal cells in the caput, carpus, cauda and vas deferens of epididymis regions. Each color represents a cluster of cells in an anatomical region. The red dots represent the epididymis caput basal cells; The blue dots represent the epididymis corpus basal cells; The green dots represent the epididymis cauda basal cells; The purple dots represent the epididymis vas deference basal cells. (E) Reclustering of extracted principal cells, visualized by t-SNE and annotated according to the four anatomical regions. This figure shows the t-SNE cluster diagram of principal cells in the caput, carpus, cauda and vas deferens of epididymis regions. Each color represents a cluster of cells in an anatomical region. The red dots represent the epididymis caput principal cells; The blue dots represent the epididymis corpus principal cells; The green dots represent the epididymis cauda principal cells; The purple dots represent the epididymis vas deference principal cells.
Anatomical analysis of the human epididymis based on histology, proteomics data and 3D reconstruction had led to a subdivision of the caput, corpus and cauda into four six and ten regions respectively (Zhao et al., 2020). This research revealed significant variations in protein composition within different luminal regions of the epididymis (Dacheux et al., 2006). It should be noted that regional divisions of the epididymis also differ among other species. For instance, the mouse epididymis consisted of ten distinct segments while rats comprised fourteen segments (Adamali et al., 1999; Johnston et al., 2005). Furthermore, variations could be observed in size and shape of specific segments acrossing different species (Jelinsky et al., 2007).
The function of the epididymal corpus and cauda were found to be similar. Specifically, immune-related biological processes were enriched in both regions. During transport between the corpus and cauda of the epididymis, sperm acquired fertilization capacity. Currently, it had been demonstrated that sperm in the epididymal corpus area possess a similar ability to cauda sperm, including capacitation, oocyte fertilization, and survival during cryopreservation (Kunkitti et al., 2015; Kunkitti et al., 2016a; Kunkitti et al., 2016b). The cauda of the epididymis served as the final storage site for matured sperm prior to ejaculation (Souza et al., 2020). In many animal species, sperm could maintain fertility for several weeks within this environment (Esponda and Bedford, 1986). These findings highlighted the crucial role of processing and modification in each region of the epididymis for maintaining sperm function.
The biosynthesis of secreted proteins in the cauda of the epididymis, similar to that in the caput, was also regulated by androgen (Foldesy and Bedford, 1982; Wong et al., 1982; Esponda and Bedford, 1986; Regalado et al., 1993). In comparison to other mammals, human sperm production was relatively lower. Furthermore, the functional significance of temperature variation within the epididymis remained unknown (Bedford, 2015). In summary, sperm maturation was intricately linked to the unique regional functions within the epididymis, enabling successful fertilization.
4 Distinct cellular compositions across different epididymal regions
4.1 Epididymal epithelial and cellular phenotypes
In the past two decades, extensive research had been dedicated to comprehending the contribution of epididymal epithelial cells. The epididymal epithelium exhibited region-specific characteristics and functional diversity in the caput, corpus, and cauda regions. Each of these regions played a distinct role in ensuring that sperm acquired both motility and fertility functions. Furthermore, the epithelial lining primarily consisted of principal and basal cells (Arrighi, 2014). During the undifferentiated stage of epididymis development, small undifferentiated columnar cells formed the composition of epithelial cells (Sun and Flickinger, 1979). Subsequently, these undifferentiated short columnar cells differentiated into basal cells as well as high columnar/narrow cells which further develop into principal cells, clear cells, narrow cells, and apical cells (Dufresne et al., 2022).
Epididymal epithelial cells had been successfully isolated and cultured in many species, such as bull, mouse, rabbit, as well as human (Orgebin-Crist et al., 1984; Joshi, 1985; Moore and Hartman, 1986; Bongso and Trounson, 1996; Castellón and Huidobro, 1999). Several studies indicated that epithelial-mesenchymal interactions were important in epididymal morphogenesis (Higgins et al., 1989). The epididymal mesenchymal interactions, such as the phenotypic effects of androgens, were proved as the result of interactions between specific androgenic de-pending cells (De Gendt and Verhoeven, 2012). Besides, epididymal epithelium provided an optimal acidic liquid microenvironment in epididymal lumen (Hinton and Palladino, 1995; Zuo et al., 2011). Different regions of the epididymis contained a variety of epithelial cell types that served distinct functions.
Earlier studies had identified four types of epididymal epithelial cells: principal cells, narrow cells, clear cells, and basal cells (Jervis and Robaire, 2001). Subsequently, the classification was expanded to include six different cell types: principal cells, narrow cells, apical cells, clear cells, halo cells, and basal cells (Cornwall, 2009). Furthermore, numerous studies had categorized epididymal epithelial cells into five types: principal cell, narrow cell, clear cell, apical cell and basal cell (Shum et al., 2009). The presence of various cell types varies across species; for instance, apical cells, narrow cells, clear cells, and halo cells (Arrighi, 2014). Recently, single cell RNA-sequencing samples had enabled the identification of multiple subtypes within the epididymis (Leir et al., 2020; Rinaldi et al., 2020). In 2020, the study by Leir et al. (Leir et al., 2020) obtained 1876 cells, 1309 cells, and 2114 cells from human epididymis aged 32, 57 and 32 years, respectively. Eight distinct proximal epididymal cell types were identified in humans including principal cells, basal cells, clear cells, stomal cells, apical/narrow cells, immune cells, sperm, and efferent ducts (Leir et al., 2020). The identification of specific marker genes allowed for the division of each cell type into multiple subtypes. In the study, a total of nine principal cell sub-types and three basal cell subtypes were analyzed from the dataset (Rinaldi et al., 2020). Further investigation would be required to elucidate the contribution of different epididymal epithelial cell subtypes to sperm maturation.
4.2 Principal cells
The most abundant epididymis epithelial cells were the principal cells, which dis-tributed throughout the whole epididymis. According to the analysis of epididymis, the principal cells accounted from 65% to 80% of the epididymal epithelium. It is reported that the number of principal cells was at least three times more than the sum of other cell types (Hermo et al., 1994; Joseph et al., 2011). The cells were mainly responsible for the absorption and the secretion of substances into the epididymal cavity, so it had significantly secretory activity (Cornwall, 2009). In addition, the principal cells were the places where liquid, ions, antioxidants and exons were produced and released (Sullivan and Saez, 2013).
The epididymal junction complex between adjacent principal cells comprised apical adhesions and tight junctions, leading to the formation of the blood-epididymal barrier (Cyr et al., 1995). The blood-epididymis barrier limited the molecular exchange between blood and lumen, providing a guarantee for the stability of the epididymal lumen environment. Consequently, specific microenvironments were established within the epididymal lumen, which played a crucial role in sperm maturation. Principal cells secreted proteins that bound to maturing spermatozoa and regulated their maturational process (Cyr et al., 2007). Dysfunction of epididymis principal cells could potentially impact sperm motility quality and fertilization capacity. Notably, Occludin (OCLN), a tight junction protein, localized at the apical junction of proximal epididymis epithelium’s principal cells. Deletion mutation of OCLN had been shown to cause male infertility in mice (Liu et al., 2022a).
4.3 Basal cells
Basal cells were present in the columnar pseudostratified epithelium that covered the mammalian epididymis (Arrighi, 2014). These cells could be found in the epididymal epithelium of all species, including caput, corpus, and cauda regions (Hermo et al., 1994; Arrighi, 2014). These cells were located in the basal layer and extend long and slender cytoplasmic projections to the lumen. It was generally believed that there were more basal cells in the epididymal corpus and fewer in the cauda (Arrighi et al., 1991; Schon and Blottner, 2009). Initially, basal cells were thought to have macrophage-like qualities and were associated with epididymal immunity (Yeung et al., 1994; Seiler et al., 1998). Basal cells might scan and perceive the lumen environment of pseudostratified epithelium and regulate epithelial function through the mechanism of crosstalk with other epithelial cells (Shum et al., 2008). In addition to their role in regulating epithelial function through crosstalk with other cell types, basal cells also had a capacity to monitor the fluid environment within the epididymis.
Early studies showed that basal cells of epididymis could be detected as stem cells (Mandon et al., 2015; Mou et al., 2016). Furthermore, several signaling pathways related to stem cell functions such as Notch1, Hedgehog, p63, and Wnt were enriched in basal cells (Yalcin-Ozuysal et al., 2010), some of which were known to be involved in sperm function. Specifically, Wnt ligands had been shown to promote stability of sperm proteome and enhanced sperm motility by orchestrating post-transcriptional sperm maturation programs (Koch et al., 2015). The relationship between stem cell-related signaling pathways within basal cells and sperm function warranted further investigation. Primary cilia were observed on both surfaces of basal cells as well as within intercellular spaces. Damage to these primary cilia on basal cells lead to imbalanced apoptosis rates among epididymal epithelial cells, thus highlighting their role as guardians for maintaining homeostasis within pseudostratified epithelia (Girardet et al., 2022).
4.4 Other cells
Apical cells were mainly located in the initial segment of the epididymal epithelium and had endocytosis activity. Narrow cells also existed only within the initial segment, as their name suggested, being narrower than the adjacent principal cells. Both of apical cells and narrow cells had been shown to secrete H+ into the epididymal lumen and are responsible for endocytosis (Cornwall, 2009). Clear cells were large endocytotic cells scattered among the principal cells in the caput, corpus and cauda regions. In addition to secretion, endocytosis of lumen proteins was the main function of epithelial clear cells (Moore and Bedford, 1979). Narrow, apical and clear cells were strongly expressing vacuolar proton pump ATPase (V-ATPase) in their apical membranes, which was considered to be the reason for proton secretion into the lumen (Da Silva et al., 2007; Kujala et al., 2007; Belleannée et al., 2010).
In the epididymis, intraepithelial cells were believed to perform distinct and integrated functions. Recent studies had demonstrated that basal cells regulated electrolyte transport in principal cells by releasing paracrine factors (Cheung et al., 2005). It was concluded that cell–cell interaction underlied the formation of the epididymal environment in maturing spermatozoa. Therefore, the intricate interplay between different epithelial cell types contributed to the activation of luminal acidification in the epididymis, which was essential for sperm maturation and storage.
5 Different gene expression patterns in different epididymis regions
5.1 Specially expressed genes and gene families
Regional specificity of gene expression in epididymis had been proposed for a long time (Douglass et al., 1991; Sullivan et al., 2011). Some genes were predominantly expressed in the epididymis, which suggested that these genes might possess epididymis-specific functions. To gain insights into this phenomenon, the mRNA of the caput, corpus and cauda of the epididymis had been sequenced by transcriptome sequencing. Specific mRNA synthesis supported the concept that the region-specific expression pattern of epididymal tubule transcription was the main molecular basis of the region-specific expression pattern of tubule lumen proteins (Douglass et al., 1991). Furthermore, each gene had distinct spatial expression and regulation in the epididymis. The genes expressed in the epididymis played an important role in sperm maturation and motility, and changed in epididymal-specific gene expression may lead to infertility. In the following, we would specify the expression of representative epididymal genes.
The mouse epididymis transcripts were analyzed based on a 17,000 oligonucleotides microarray. These genes were up or downregulated more than four times between at least two different fragments of mouse epididymis (Ståhl et al., 2016). The expression patterns of these genes identify distinct patterns of segmental regulation. Analysis based on rat microarray technology found that the number of genes detected in each region of the epididymis varied greatly. Notably, 53% of the genes with the highest expression were expressed in the cauda (626 out of 1176 genes) (Jervis and Robaire, 2001). Recently, the region-specific ex-pression genes in sheep were analyzed by GO and KEGG analysis to screen the key genes related to sperm maturation. A total of 129, 54 and 99 specific genes were obtained in the caput, corpus and cauda, respectively (Wu et al., 2021a). The heatmap showed that six genes of the LCN family (the lipocalin (Lcn) family was a hydrophobic ligand binding protein) were highly expressed in the sheep epididymal caput. RNase10 (ribonuclease, RNase A family, 10, predicted to enable nucleic acid binding activity) was also highly specifically expressed in caput. It was the same for other species for which RNase10 had been identified, such as humans, mice and pigs (Castella et al., 2004; Devor et al., 2004; Krutskikh et al., 2012).
5.2 scRNA-seq showed gene expression patterns in the epididymis
We reanalyzed the data provided in previous research (Rinaldi et al., 2020) with the Seurat software package, collecting the data of the mouse epididymal caput, corpus, cauda and vas deferens. The organ was divided into four regions: caput, corpus, cauda, and vas deferens. The single-cell RNA sequencing library was prepared using the Chrome single-cell 3′kit V2 (10X Genomics). Using single-cell data uploaded to the GEO (Gene Expression Omnibus) website combined with our own mouse epididymis data, we visualized the expression of several genes in basal cells and principal cells.
Finally, we obtained a total of 9167 individual cells in a single assay (Figure 2C). Within the dataset, 1619 cells were collected from the caput of the epididymis, 2345 cells from the corpus of the epididymis, 1658 cells from the cauda of the epididymis, and up to 3545 cells were collected from the vas deferens. To visualize these four regions of data, we employed t-distributed stochastic neighbor embedding (t-SNE), a dimensionality reduction technique that enables visualization of data pointed on a two-dimensional map. In the t-SNE map, genes with similar functions tended to cluster together and form distinct groups within a coordinate system. Each region of the epididymal sample is represented by a different color (Figure 2C). Subsequently, principal cell and basal cell populations representing two major cell types in the epididymis were individually extracted and clustered. By employing t-SNE analysis and annotating them based on their respective regions, we successfully identified basal cells (Figure 2D) and principal cells (Figure 2E). There were 1410 cells within the basal cluster (Figure 2D) and 2099 cells in the principal cluster (Figure 2E). Furthermore, numerous genes were selected for examination regarding their distribution across different regions within the epididymis (Figure 3).
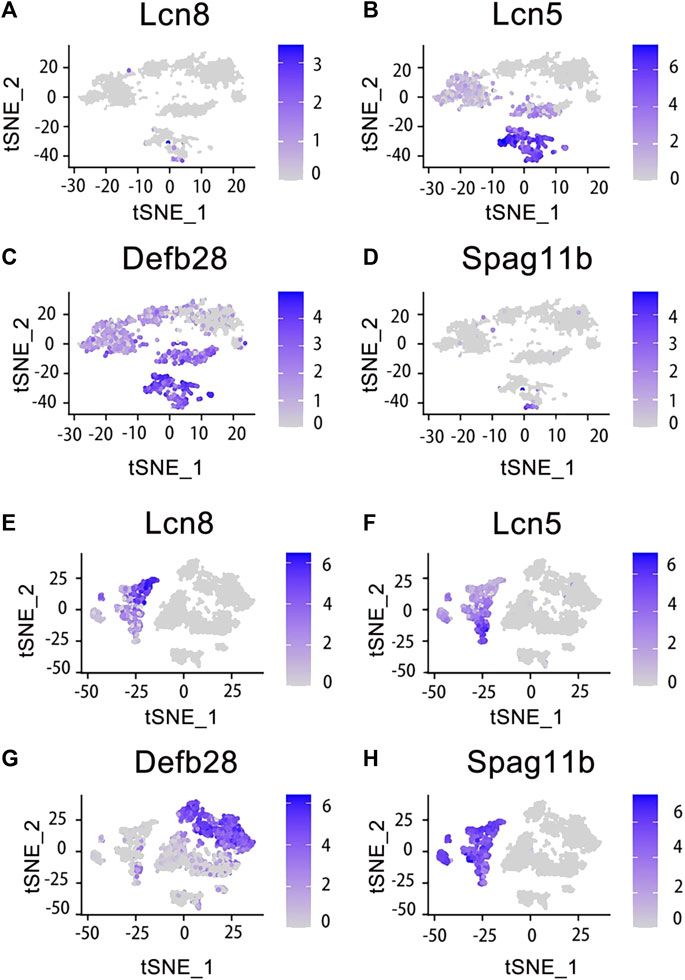
Figure 3. Expression of specific genes across basal cells and principal cells. Those genes were selected and looked at their epididymal distribution in different parts respectively. The tSNE cluster map shows the differential expression of genes in basal cells. The darker blue indicates the higher expression level. (A) Lcn8 is highly expressed in epididymis caput basal cells. (B) Lcn5 is highly expressed in epididymis caput and corpus basal cells. (C) Defb28 is highly expressed in corpus, cauda and vas deferens of epididymis basal cells. (D) Spag11b is highly expressed in epididymis caput basal cells. (E) Lcn8 is highly expressed in epididymis caput principal cells. (F) Lcn5 is highly expressed in epididymis caput principal cells. (G) Defb28 is highly expressed in epididymis cauda principal cells. (H) Spag11b is highly expressed in epididymis caput principal cells.
6 Discussion
The epididymis comprises convoluted tubules that connect the testis and vas deferens within the male reproductive system. It plays a pivotal role in regulating sperm maturation and fertilization processes. Sperm maturation necessitated epididymal processing, encompassing regional alterations in spermatozoa structure and molecular composition (Jones et al., 2007). Additionally, it involves immune and metabolic physiological mechanisms. Notably, numerous macrophages were present within the epididymal lumen and were capable of recognizing and eliminating suboptimal spermatozoa. Consequently, the epididymis assumed a critical function in both sperm maturation and the acquisition of fertilization competence (Elbashir et al., 2021).
With the advancement of sequencing technologies (Wong et al., 2020; Wu et al., 2021a; Zhao et al., 2021a; Liu et al., 2022b), a greater understanding of the molecular mechanisms underlying epididymal function has been achieved. The significance of the epididymis in male reproductive biology has garnered increasing attention. Furthermore, distinct regional disparities within the epididymis have been elucidated (Belleannée et al., 2012; Cyr et al., 2018; Wu et al., 2021b; Zhao et al., 2021b). For instance, the caput region exhibited heightened activity and accounted for approximately 70%–80% of total protein secretion (Cornwall, 2009). The corpus of the epididymis plays a pivotal role in immune functions. The comprehension of the epididymis evolved from its initial morphological and structural characterization to encompass diverse functional aspects across different regions (Martin et al., 1902; Sullivan et al., 2019; James et al., 2020; Zhao et al., 2020). Notably, unique gene expression patterns were identified in the caput, corpus, and cauda regions through techniques such as microarray or single-cell RNA sequencing analysis. In summary, while the caput region secretes nutrients and androgens to facilitate sperm maturation, sperm traverse through the corpus region towards the cauda, which acts as a barrier against autoimmune reactions, additionally serving as a conduit for transporting sperm to the vas deferens. Consequently, functional discrepancies exist among various regions within the epididymis due to differential gene expression profiles that drive specific functions associated with each respective region.
The male reproductive system exhibited significant interspecies variability. In the epididymis of rats and mice, distinct regions displayed pronounced differences (Abou-Haïla and Fain-Maurel, 1984). Due to their shorter life cycle than humans, rodents have become the most common animal model in research. Recently, dogs have emerged as viable biological models for studying the molecular functions of the epididymis (Kirchhoff, 2002). However, extrapolating reproductive characteristics of other mammals based solely on a specific species might lead to biased conclusions in terms of reproductive biology.
The cell classifications of the epididymal epithelium in each region of the epididymis varied (Chen et al., 2021). Among them, the principal cells constituted 80% of the epithelial cells in the epididymis tubule. Further analysis revealed distinct gene expression patterns for the same cell type across different regions of the epididymis. In this study, we reanalyzed and visualized gene expression data from the mouse epididymis and vas deferens based on previous findings and publicly available single-cell datasets. Marker genes specific to caput principal cells included RNase10, Cst11, and Lcn2. Similarly, the Lcn5, RNase9, and Plac8 genes distinguished corpus principal cells, while the Gpx3, Klk1b27, Hint1, and Gstm2 genes characterized cauda principal cells. Basal cells were identified by the Itga6 and Krt14 genes. Interestingly, our analysis revealed differential mRNA expression levels between principal and basal cells for several genes, including Lcn5, which showed high expression in caput principal cells but was significantly detected in both caput and corpus basal cells; Defb28 exhibited high expression in cauda principal cells but was significantly detected in epididymis basal cells as well (Rinaldi et al., 2020). The results demonstrated the essential role of Rnase10 in sperm adhesion within the epididymis, a critical characteristic for mouse sperm transport in the female vagina (Krutskikh et al., 2012). Combined with the data analysis on Lcn5 in the principal cells, we indicated that the caput of the mouse epididymis was related to lipid metabolism and adhesion. Similarly, basal cells exhibited significant enrichment of genes related to membrane transport and lipid metabolism (Rinaldi et al., 2020). These findings suggested that different cell types share similarities, contributing to an optimal environment for sperm maturation within the epididymal cavity. Additionally, certain coregulatory factors displayed fragment-specific enrichment patterns during their expression processes, suggesting their potential contribution to specific responses (Browne et al., 2016). Each region of the epididymis possesses a unique transcriptomic signature, which regulates luminal composition and ultimately influences male sperm maturation.
7 Conclusion
The intricate patterns of gene expression and cellular composition in the caput, corpus, and cauda regions of the epididymis contribute to distinct functions in sperm maturation. However, interspecies variations exist in gene expression within the epididymis. Exploring the functional diversity across different regions of the epididymis presents an exciting avenue for research. It is anticipated that advancements in sequencing technologies, such as spatial transcriptomics, would play a pivotal role in unraveling the complexities of epididymal biology.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
M-ML: Data curation, Formal Analysis, Software, Writing–original draft, Data curation, Formal Analysis, Software, Writing–original draft. X-LF: Data curation, Visualization, Writing–review and editing, Data curation, Visualization, Writing–review and editing. CQ: Methodology, Validation, Visualization, Writing–review and editing, Methodology, Validation, Visualization, Writing–review and editing. S-EZ: Data curation, Formal Analysis, Methodology, Resources, Visualization, Writing–review and editing, Data curation, Formal Analysis, Methodology, Resources, Visualization, Writing–review and editing. G-LZ: Funding acquisition, Investigation, Project administration, Writing–review and editing, Funding acquisition, Investigation, Project administration, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (31902158), Key Technology R&D Program of Shandong Province (2021LZGC001), Shandong Modern Agricultural Industry Technology System (SDAIT-08-16), Shaanxi Modern Agricultural Industry Technology System (K3031222128), China Postdoctoral Science Foundation (2019M662293S), Shandong Pro-vincial Natural Science Foundation (ZR2020QC099), and Research Foundation for Advanced Talents of Qingdao Agricultural University (665/1119013).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AI, artificial insemination; DEFB, defensin, beta; Hox, homeobox; IVF, in vitro fertilization; LCN, lipocalin; OCLN, occludin; Rnase10, ribonuclease A family member 10 (inactive); t-SNE, t-distributed stochastic neighbor embedding.
References
Abou-Haïla, A., and Fain-Maurel, M. A. (1984). Regional differences of the proximal part of mouse epididymis: morphological and histochemical characterization. Anat. Rec. 209 (2), 197–208. doi:10.1002/ar.1092090207
Acott, T. S., and Carr, D. W. (1984). Inhibition of bovine spermatozoa by caudal epididymal fluid: II. Interaction of pH and a quiescence factor. Biol. Reprod. 30 (4), 926–935. doi:10.1095/biolreprod30.4.926
Adamali, H. I., Somani, I. H., Huang, J. Q., Mahuran, D., Gravel, R. A., Trasler, J. M., et al. (1999). I. Abnormalities in cells of the testis, efferent ducts, and epididymis in juvenile and adult mice with β-hexosaminidase A and B deficiency. J. Androl. 20 (6), 779–802. doi:10.1002/j.1939-4640.1999.tb03385.x
Allen, J. M. (1961). The histochemistry of glucose-6-phosphatase in the epididymis of the mouse. J. Histochem Cytochem 9, 681–689. doi:10.1177/9.6.681
Allen, J. M., and Slater, J. J. (1961). A cytochemical study of Golgi associated thiamine pyrophosphatase in the epididymis of the mouse. J. Histochem Cytochem 9, 418–423. doi:10.1177/9.4.418
Araki, Y., Suzuki, K., Matusik, R. J., Obinata, M., and Orgebin-Crist, M. C. (2002). Immortalized epididymal cell lines from transgenic mice overexpressing temperature-sensitive simian virus 40 large T-antigen gene. J. Androl. 23 (6), 854–869. doi:10.1002/j.1939-4640.2002.tb02344.x
Arrighi, S. (2014). Are the basal cells of the mammalian epididymis still an enigma? Reprod. Fertil. Dev. 26 (8), 1061–1071. doi:10.1071/RD13301
Arrighi, S., Romanello, M. G., and Domeneghini, C. (1991). Morphological examination of epididymal epithelium in the mule (E. hinnus) in comparison with parental species (E. asinus and E. caballus). Histol. Histopathol. 6 (3), 325–337.
Axnér, E., Linde-Forsberg, C., and Einarsson, S. (1999). Morphology and motility of spermatozoa from different regions of the epididymal duct in the domestic cat. Theriogenology 52 (5), 767–778. doi:10.1016/S0093-691X(99)00170-3
Bedford, J. M. (2015). Human spermatozoa and temperature: the elephant in the room. Biol. Reprod. 93 (4), 97. doi:10.1095/biolreprod.115.130658
Belleannée, C., Da Silva, N., Shum, W. W. C., Brown, D., and Breton, S. (2010). Role of purinergic signaling pathways in V-ATPase recruitment to apical membrane of acidifying epididymal clear cells. Am. J. Physiol. Cell Physiol. 298 (4), C817–C830. doi:10.1152/ajpcell.00460.2009
Belleannée, C., Thimon, V., and Sullivan, R. (2012). Region-specific gene expression in the epididymis. Cell Tissue Res. 349 (3), 717–731. doi:10.1007/s00441-012-1381-0
Birnbaum, D., Hall, T., and Lee, R. (1961). The zinc content of rat sperm cells from ejaculate, vas, epididymis and testis. Proc. Soc. Exp. Biol. Med. 108, 321–324. doi:10.3181/00379727-108-26926
Bongso, A., and Trounson, A. (1996). Evaluation of motility, freezing ability and embryonic development of murine epididymal sperm after coculture with epididymal epithelium. Hum. Reprod. 11 (7), 1451–1456. doi:10.1093/oxfordjournals.humrep.a019418
Branford, W. W., Benson, G. V., Ma, L., Maas, R. L., and Potter, S. S. (2000). Characterization of Hoxa-10/Hoxa-11 transheterozygotes reveals functional redundancy and regulatory interactions. Dev. Biol. 224 (2), 373–387. doi:10.1006/dbio.2000.9809
Browne, J. A., Yang, R., Leir, S. H., Eggener, S. E., and Harris, A. (2016). Expression profiles of human epididymis epithelial cells reveal the functional diversity of caput, corpus and cauda regions. Mol. Hum. Reprod. 22 (2), 69–82. doi:10.1093/molehr/gav066
Bryant, T. (1892). Case of torsion of the spermatic cord, with strangulation of the epididymis and testicle in an incompletely descended organ. Med. Chir. Trans. 75, 247–256. doi:10.1177/095952879207500115
Castella, S., Fouchécourt, S., Teixeira-Gomes, A. P., Vinh, J., Belghazi, M., Dacheux, F., et al. (2004). Identification of a member of a new RNase a family specifically secreted by epididymal caput epithelium. Biol. Reprod. 70 (2), 319–328. doi:10.1095/biolreprod.103.022459
Castellón, E. A., and Huidobro, C. C. (1999). Androgen regulation of glycosidase secretion in epithelial cell cultures from human epididymis. Hum. Reprod. 14 (6), 1522–1527. doi:10.1093/humrep/14.6.1522
Chauvin, T. R., and Griswold, M. D. (2004). Androgen-regulated genes in the murine epididymis. Biol. Reprod. 71 (2), 560–569. doi:10.1095/biolreprod.103.026302
Chen, H., Alves, M. B. R., and Belleannée, C. (2021). Contribution of epididymal epithelial cell functions to sperm epigenetic changes and the health of progeny. Hum. Reprod. Update 28 (1), 51–66. doi:10.1093/humupd/dmab029
Chen, T., Yao, L., Liu, W., Luan, J., Wang, Y., Yang, C., et al. (2023). Epididymal segment-specific miRNA and mRNA regulatory network at the single cell level. Cell Cycle 22 (19), 2194–2209. doi:10.1080/15384101.2023.2280170
Chen, Y. C., Bunick, D., Bahr, J. M., Klinefelter, G. R., and Hess, R. A. (1998). Isolation and culture of epithelial cells from rat ductuli efferentes and initial segment epididymidis. Tissue Cell 30 (1), 1–13. doi:10.1016/s0040-8166(98)80002-5
Cheung, K. H., Leung, G. P. H., Leung, M. C. T., Shum, W. W. C., Zhou, W. L., and Wong, P. Y. D. (2005). Cell-cell interaction underlies formation of fluid in the male reproductive tract of the rat. J. Gen. Physiol. 125 (5), 443–454. doi:10.1085/jgp.200409205
Cho, C., Bunch, D. O., Faure, J. E., Goulding, E. H., Eddy, E. M., Primakoff, P., et al. (1998). Fertilization defects in sperm from mice lacking fertilin beta. Science 281 (5384), 1857–1859. doi:10.1126/science.281.5384.1857
Cornwall, G. A. (2009). New insights into epididymal biology and function. Hum. Reprod. Update 15 (2), 213–227. doi:10.1093/humupd/dmn055
Cyr, D. G., Dufresne, J., and Gregory, M. (2018). Cellular junctions in the epididymis, a critical parameter for understanding male reproductive toxicology. Reprod. Toxicol. 81, 207–219. doi:10.1016/j.reprotox.2018.08.013
Cyr, D. G., Gregory, M., Dubé, E., Dufresne, J., Chan, P. T. K., and Hermo, L. (2007). Orchestration of occludins, claudins, catenins and cadherins as players involved in maintenance of the blood-epididymal barrier in animals and humans. Asian J. Androl. 9 (4), 463–475. doi:10.1111/j.1745-7262.2007.00308.x
Cyr, D. G., Robaire, B., and Hermo, L. (1995). Structure and turnover of junctional complexes between principal cells of the rat epididymis. Microsc. Res. Tech. 30 (1), 54–66. doi:10.1002/jemt.1070300105
Dacheux, J. L., Belghazi, M., Lanson, Y., and Dacheux, F. (2006). Human epididymal secretome and proteome. Mol. Cell Endocrinol. 250 (1-2), 36–42. doi:10.1016/j.mce.2005.12.022
Dacheux, J. L., Belleannée, C., Jones, R., Labas, V., Belghazi, M., Guyonnet, B., et al. (2009). Mammalian epididymal proteome. Mol. Cell Endocrinol. 306 (1-2), 45–50. doi:10.1016/j.mce.2009.03.007
Dacheux, J. L., Castella, S., Gatti, J. L., and Dacheux, F. (2005). Epididymal cell secretory activities and the role of proteins in boar sperm maturation. Theriogenology 63 (2), 319–341. doi:10.1016/j.theriogenology.2004.09.015
Da Silva, N., Shum, W. W., and Breton, S. (2007). Regulation of vacuolar proton pumping ATPase-dependent luminal acidification in the epididymis. Asian J. Androl. 9 (4), 476–482. doi:10.1111/j.1745-7262.2007.00299.x
De Gendt, K., and Verhoeven, G. (2012). Tissue- and cell-specific functions of the androgen receptor revealed through conditional knockout models in mice. Mol. Cell Endocrinol. 352 (1-2), 13–25. doi:10.1016/j.mce.2011.08.008
Devor, E. J., Moffat-Wilson, K. A., and Galbraith, J. J. (2004). LOC 390443 (RNase 9) on chromosome 14q11.2 is related to the RNase A superfamily and contains a unique amino-terminal preproteinlike sequence. Hum. Biol. 76 (6), 921–935. doi:10.1353/hub.2005.0016
Domeniconi, R. F., Souza, A. C. F., Xu, B., Washington, A. M., and Hinton, B. T. (2016). Is the epididymis a series of organs placed side by side? Biol. Reprod. 95 (1), 10. doi:10.1095/biolreprod.116.138768
Douglass, J., Garrett, S. H., and Garrett, J. E. (1991). Differential patterns of regulated gene expression in the adult rat epididymis. Ann. N. Y. Acad. Sci. 637, 384–398. doi:10.1111/j.1749-6632.1991.tb27324.x
Dufresne, J., Gregory, M., Pinel, L., and Cyr, D. G. (2022). Differential gene expression and hallmarks of stemness in epithelial cells of the developing rat epididymis. Cell Tissue Res. 389 (2), 327–349. doi:10.1007/s00441-022-03634-9
Dufresne, J., St-Pierre, N., Viger, R. S., Hermo, L., and Cyr, D. G. (2005). Characterization of a novel rat epididymal cell line to study epididymal function. Endocrinology 146 (11), 4710–4720. doi:10.1210/en.2004-1634
Eddy, E. M., Washburn, T. F., Bunch, D. O., Goulding, E. H., Gladen, B. C., Lubahn, D. B., et al. (1996). Targeted disruption of the estrogen receptor gene in male mice causes alteration of spermatogenesis and infertility. Endocrinology 137 (11), 4796–4805. doi:10.1210/endo.137.11.8895349
Elbashir, S., Magdi, Y., Rashed, A., Henkel, R., and Agarwal, A. (2021). Epididymal contribution to male infertility: an overlooked problem. Andrologia 53 (1), e13721. doi:10.1111/and.13721
Esponda, P., and Bedford, J. M. (1986). The influence of body temperature and castration on the protein composition of fluid in the rat cauda epididymidis. J. Reprod. Fertil. 78 (2), 505–514. doi:10.1530/jrf.0.0780505
Foldesy, R. G., and Bedford, J. M. (1982). Biology of the scrotum. I. Temperature and androgen as determinants of the sperm storage capacity of the rat cauda epididymidis. Biol. Reprod. 26 (4), 673–682. doi:10.1095/biolreprod26.4.673
Fouchécourt, S., Métayer, S., Locatelli, A., Dacheux, F., and Dacheux, J. L. (2000). Stallion epididymal fluid proteome: qualitative and quantitative characterization; secretion and dynamic changes of major proteins. Biol. Reprod. 62 (6), 1790–1803. doi:10.1095/biolreprod62.6.1790
Gervasi, M. G., and Visconti, P. E. (2017). Molecular changes and signaling events occurring in spermatozoa during epididymal maturation. Andrology 5 (2), 204–218. doi:10.1111/andr.12320
Girardet, L., Cyr, D. G., and Belleannée, C. (2022). Arl13b controls basal cell stemness properties and Hedgehog signaling in the mouse epididymis. Cell Mol. Life Sci. 79 (11), 556. doi:10.1007/s00018-022-04570-1
Goglia, G. (1954). Histochemical research on the ductus epididymis of the horse. Boll. Soc. Ital. Biol. Sper. 30 (8-11), 1151–1154.
Goglia, G., and Magli, G. (1957). Histochemical research on secretory processes of coni vasculosi and epididymis canal in the Guinea pig. Boll. Soc. Ital. Biol. Sper. 33 (4), 418–421.
Griffiths, J. (1893). Observations on the appendix of the testicle, and on the cysts of the epididymis, the vasa efferentia, and the rete testis. J. Anat. Physiol. 28 (1), 107–116.
Hermo, L., Oko, R., and Morales, C. R. (1994). Secretion and endocytosis in the male reproductive tract: a role in sperm maturation. Int. Rev. Cytol. 154, 106–189.
Higgins, S., Young, P., Brody, J. R., and Cunha, G. R. (1989). Induction of functional cytodifferentiation in the epithelium of tissue recombinants. I. Homotypic seminal vesicle recombinants. Homotypic Semin. vesicle Recomb. 106 (2), 219–234. doi:10.1242/dev.106.2.219
Hinton, B. T., and Palladino, M. A. (1995). Epididymal epithelium: its contribution to the formation of a luminal fluid microenvironment. Microsc. Res. Tech. 30 (1), 67–81. doi:10.1002/jemt.1070300106
Ikawa, M., Nakanishi, T., Yamada, S., Wada, I., Kominami, K., Tanaka, H., et al. (2001). Calmegin is required for fertilin alpha/beta heterodimerization and sperm fertility. Dev. Biol. 240 (1), 254–261. doi:10.1006/dbio.2001.0462
James, E. R., Carrell, D. T., Aston, K. I., Jenkins, T. G., Yeste, M., and Salas-Huetos, A. (2020). The role of the epididymis and the contribution of epididymosomes to mammalian reproduction. Int. J. Mol. Sci. 21 (15), 5377. doi:10.3390/ijms21155377
Jelinsky, S. A., Turner, T. T., Bang, H. J., Finger, J. N., Solarz, M. K., Wilson, E., et al. (2007). The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol. Reprod. 76 (4), 561–570. doi:10.1095/biolreprod.106.057323
Jervis, K. M., and Robaire, B. (2001). Dynamic changes in gene expression along the rat epididymis. Biol. Reprod. 65 (3), 696–703. doi:10.1095/biolreprod65.3.696
Jervis, K. M., and Robaire, B. (2003). Effects of caloric restriction on gene expression along the epididymis of the Brown Norway rat during aging. Exp. Gerontol. 38 (5), 549–560. doi:10.1016/s0531-5565(03)00034-2
Jiang, F. X., Temple-Smith, P., and Wreford, N. G. (1994). Postnatal differentiation and development of the rat epididymis: a stereological study. Anat. Rec. 238 (2), 191–198. doi:10.1002/ar.1092380205
Johnston, D. S., Jelinsky, S. A., Bang, H. J., DiCandeloro, P., Wilson, E., Kopf, G. S., et al. (2005). The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol. Reprod. 73 (3), 404–413. doi:10.1095/biolreprod.105.039719
Jones, R. C., Dacheux, J. L., Nixon, B., and Ecroyd, H. W. (2007). Role of the epididymis in sperm competition. Asian J. Androl. 9 (4), 493–499. doi:10.1111/j.1745-7262.2007.00284.x
Joseph, A., Shur, B. D., and Hess, R. A. (2011). Estrogen, efferent ductules, and the epididymis. Biol. Reprod. 84 (2), 207–217. doi:10.1095/biolreprod.110.087353
Joshi, M. S. (1985). Isolation and cell culture of the epithelial cells of cauda epididymidis of the bull. Biol. Reprod. 33 (1), 187–200. doi:10.1095/biolreprod33.1.187
Kirchhoff, C. (2002). The dog as a model to study human epididymal function at a molecular level. Mol. Hum. Reprod. 8 (8), 695–701. doi:10.1093/molehr/8.8.695
Klinefelter, G. R., Amann, R. P., and Hammerstedt, R. H. (1982). Culture of principal cells from the rat caput epididymidis. Biol. Reprod. 26 (5), 885–901. doi:10.1095/biolreprod26.5.885
Koch, S., Acebron, S. P., Herbst, J., Hatiboglu, G., and Niehrs, C. (2015). Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell 163 (5), 1225–1236. doi:10.1016/j.cell.2015.10.029
Krutskikh, A., Poliandri, A., Cabrera-Sharp, V., Dacheux, J. L., Poutanen, M., and Huhtaniemi, I. (2012). Epididymal protein Rnase10 is required for post-testicular sperm maturation and male fertility. Faseb J. 26 (10), 4198–4209. doi:10.1096/fj.12-205211
Kujala, M., Hihnala, S., Tienari, J., Kaunisto, K., Hästbacka, J., Holmberg, C., et al. (2007). Expression of ion transport-associated proteins in human efferent and epididymal ducts. Reproduction 133 (4), 775–784. doi:10.1530/rep.1.00964
Kunkitti, P., Axnér, E., Bergqvist, A. S., and Sjunnesson, Y. (2016a). In vitro fertilization using frozen-thawed feline epididymal spermatozoa from corpus and cauda regions. Theriogenology 86 (6), 1403–1408. doi:10.1016/j.theriogenology.2016.04.085
Kunkitti, P., Bergqvist, A. S., Sjunnesson, Y., and Axnér, E. (2015). The ability of feline spermatozoa in different epididymal regions to undergo capacitation and acrosome reaction. Anim. Reprod. Sci. 161, 64–74. doi:10.1016/j.anireprosci.2015.08.005
Kunkitti, P., Bergqvist, A. S., Sjunnesson, Y., Johannisson, A., and Axnér, E. (2016b). The tolerance of feline corpus and cauda spermatozoa to cryostress. Theriogenology 85 (3), 502–508. doi:10.1016/j.theriogenology.2015.09.032
Lang, X., Adjei, M., Wang, C., Chen, X., Li, C., Wang, P., et al. (2022). RNA-Seq reveals the functional specificity of epididymal caput, corpus, and cauda genes of cattleyak. Anim. Sci. J. 93 (1), e13732. doi:10.1111/asj.13732
Lee, G. Y., Kenny, P. A., Lee, E. H., and Bissell, M. J. (2007). Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4 (4), 359–365. doi:10.1038/nmeth1015
Légaré, C., and Sullivan, R. (2020). Differential gene expression profiles of human efferent ducts and proximal epididymis. Andrology 8 (3), 625–636. doi:10.1111/andr.12745
Leir, S. H., Paranjapye, A., and Harris, A. (2023). Functional genomics of the human epididymis: further characterization of efferent ducts and model systems by single-cell RNA sequencing analysis. Andrology 2023. doi:10.1111/andr.13477
Leir, S. H., Yin, S., Kerschner, J. L., Cosme, W., and Harris, A. (2020). An atlas of human proximal epididymis reveals cell-specific functions and distinct roles for CFTR. Life Sci. Alliance 3 (11), e202000744. doi:10.26508/lsa.202000744
Liu, B. Y., Zhang, B. L., Gao, D. Y., Li, Q., Xu, X. Y., and Shum, W. (2022a). Epididymal epithelial degeneration and lipid metabolism impairment account for male infertility in occludin knockout mice. Front. Endocrinol. (Lausanne) 13, 1069319. doi:10.3389/fendo.2022.1069319
Liu, J., Yao, T., Weng, X., Yao, R., Li, W., Xie, L., et al. (2022b). Antioxidant properties and transcriptome of cauda epididymis with different levels of fertility in Hu lambs. Theriogenology 182, 85–95. doi:10.1016/j.theriogenology.2022.01.035
Lye, R. J., and Hinton, B. T. (2004). Technologies for the study of epididymal-specific genes. Mol. Cell Endocrinol. 216 (1-2), 23–30. doi:10.1016/j.mce.2003.10.072
Mandon, M., Hermo, L., and Cyr, D. G. (2015). Isolated rat epididymal basal cells share common properties with adult stem cells. Biol. Reprod. 93 (5), 115. doi:10.1095/biolreprod.115.133967
Martin, E., Carnett, J. B., Levi, J. V., and Pennington, M. E. (1902). The surgical treatment of sterility due to obstruction at the epididymis; together with a study of the morphology of human spermatozoa. Univ. Pa Med. Bull. 15 (1), 2–15.
Masuda, K., Watanabe, K., Ueno, T., Nakazawa, Y., Tanabe, Y., Ushiki-Kaku, Y., et al. (2022). Novel cell line development strategy for monoclonal antibody manufacturing using translational enhancing technology. J. Biosci. Bioeng. 133 (3), 273–280. doi:10.1016/j.jbiosc.2021.11.010
Montagna, W. (1952). Some cytochemical observations on human testes and epididymides. Ann. N. Y. Acad. Sci. 55 (4), 629–642. doi:10.1111/j.1749-6632.1952.tb26583.x
Moore, H. D., and Bedford, J. M. (1979). Short-term effects of androgen withdrawal on the structure of different epithelial cells in the rat epididymis. Anat. Rec. 193 (2), 293–311. doi:10.1002/ar.1091930209
Moore, H. D., and Hartman, T. D. (1986). In-vitro development of the fertilizing ability of hamster epididymal spermatozoa after co-culture with epithelium from the proximal cauda epididymidis. J. Reprod. Fertil. 78 (2), 347–352. doi:10.1530/jrf.0.0780347
Mou, H., Vinarsky, V., Tata, P. R., Brazauskas, K., Choi, S. H., Crooke, A. K., et al. (2016). Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 19 (2), 217–231. doi:10.1016/j.stem.2016.05.012
National Library of Medicine (1859). Discutient application to the indurated epididymis. Atlanta Med. Surg. J. 4 (8), 515.
Nishimura, H., Kim, E., Nakanishi, T., and Baba, T. (2004). Possible function of the ADAM1a/ADAM2 Fertilin complex in the appearance of ADAM3 on the sperm surface. J. Biol. Chem. 279 (33), 34957–34962. doi:10.1074/jbc.M314249200
Orgebin-Crist, M. C., Jonas-Davies, J., Storey, P., and Olson, G. E. (1984). Effect of D-valine and cytosine arabinoside on [3H]thymidine incorporation in rat and rabbit epididymal epithelial cell cultures. Vitro 20 (1), 45–52. doi:10.1007/BF02633331
Paranjapye, A., Leir, S. H., Huang, F., Kerschner, J. L., and Harris, A. (2022). Cell function and identity revealed by comparative scRNA-seq analysis in human nasal, bronchial and epididymis epithelia. Eur. J. Cell Biol. 101 (3), 151231. doi:10.1016/j.ejcb.2022.151231
Qian, P., Kang, J., Liu, D., and Xie, G. (2022). Single cell transcriptome sequencing of zebrafish testis revealed novel spermatogenesis marker genes and stronger leydig-germ cell paracrine interactions. Front. Genet. 13, 851719. doi:10.3389/fgene.2022.851719
Regalado, F., Esponda, P., and Nieto, A. (1993). Temperature and androgens regulate the biosynthesis of secretory proteins from rabbit cauda epididymidis. Mol. Reprod. Dev. 36 (4), 448–453. doi:10.1002/mrd.1080360407
Rinaldi, V. D., Donnard, E., Gellatly, K., Rasmussen, M., Kucukural, A., Yukselen, O., et al. (2020). An atlas of cell types in the mouse epididymis and vas deferens. Elife 9, e55474. doi:10.7554/eLife.55474
Rockwell, F. W. (1888). Case of removal of both testicles for recurrent carcinoma of the epididymis. Ann. Surg. 8 (6), 446–449. doi:10.1097/00000658-188807000-00167
Rowley, M. J., Teshima, F., and Heller, C. G. (1970). Duration of transit of spermatozoa through the human male ductular system. Fertil. Steril. 21 (5), 390–396. doi:10.1016/s0015-0282(16)37502-1
Salisbury, G. W., and Graves, C. N. (1963). substrate-free epididymal-like bovine spermatozoa. J. Reprod. Fertil. 6, 351–359. doi:10.1530/jrf.0.0060351
Salisbury, G. W., Graves, C. N., Nakabayashi, N. T., and Cragle, R. G. (1963). Observations on the aerobic metabolism of bull and goat epididymal spermatozoa. J. Reprod. Fertil. 6, 341–349. doi:10.1530/jrf.0.0060341
Schon, J., and Blottner, S. (2009). Seasonal variations in the epididymis of the roe deer (Capreolus capreolus). Anim. Reprod. Sci. 111 (2-4), 344–352. doi:10.1016/j.anireprosci.2008.03.008
Seiler, P., Wenzel, I., Wagenfeld, A., Yeung, C. H., Nieschlag, E., and Cooper, T. G. (1998). The appearance of basal cells in the developing murine epididymis and their temporal expression of macrophage antigens. Int. J. Androl. 21 (4), 217–226. doi:10.1046/j.1365-2605.1998.00116.x
Shaw, G., and Renfree, M. B. (2014). Wolffian duct development. Sex. Dev. 8 (5), 273–280. doi:10.1159/000363432
Shi, J., Fok, K. L., Dai, P., Qiao, F., Zhang, M., Liu, H., et al. (2021). Spatio-temporal landscape of mouse epididymal cells and specific mitochondria-rich segments defined by large-scale single-cell RNA-seq. Cell Discov. 7 (1), 34. doi:10.1038/s41421-021-00260-7
Shimazaki, J., Yamanaka, H., Taguchi, I., and Shida, K. (1976). Free amino acids in the caput and the cauda epididymis of adult rats. Endocrinol. Jpn. 23 (2), 149–156. doi:10.1507/endocrj1954.23.149
Shum, W. W., Da Silva, N., Brown, D., and Breton, S. (2009). Regulation of luminal acidification in the male reproductive tract via cell-cell crosstalk. J. Exp. Biol. 212 (11), 1753–1761. doi:10.1242/jeb.027284
Shum, W. W., Da Silva, N., McKee, M., Smith, P. J. S., Brown, D., and Breton, S. (2008). Transepithelial projections from basal cells are luminal sensors in pseudostratified epithelia. Cell 135 (6), 1108–1117. doi:10.1016/j.cell.2008.10.020
Snyder, E. M., Small, C. L., Bomgardner, D., Xu, B., Evanoff, R., Griswold, M. D., et al. (2010). Gene expression in the efferent ducts, epididymis, and vas deferens during embryonic development of the mouse. Dev. Dyn. 239 (9), 2479–2491. doi:10.1002/dvdy.22378
Some Diseases of the Male Genital System (1908). Some Diseases of the male genital system: VIII. Tuberculous disease of the testis and epididymis. Hosp. (Lond 1886) 43 (1120), 526.
Souza, A. P. B., Lopes, T. N., da Silva, A. F. T., Santi, L., Beys-da-Silva, W. O., Yates, J. R., et al. (2020). Changes in porcine cauda epididymal fluid proteome by disrupting the HPT axis: unveiling potential mechanisms of male infertility. Mol. Reprod. Dev. 87 (9), 952–965. doi:10.1002/mrd.23408
Ståhl, P. L., Salmén, F., Vickovic, S., Lundmark, A., Navarro, J. F., Magnusson, J., et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353 (6294), 78–82. doi:10.1126/science.aaf2403
Stoltenberg, M., Therkildsen, P., Andreasen, A., Jensen, K. B., Juhl, S., Ernst, E., et al. (1998). Computer-assisted visualization of the rat epididymis: a methodological study based on paraffin sections autometallographically stained for zinc ions. Histochem J. 30 (4), 237–244. doi:10.1023/a:1003255705503
Sullivan, R., Légaré, C., Lamontagne-Proulx, J., Breton, S., and Soulet, D. (2019). Revisiting structure/functions of the human epididymis. Andrology 7 (5), 748–757. doi:10.1111/andr.12633
Sullivan, R., Legare, C., Thabet, M., and Thimon, V. (2011). Gene expression in the epididymis of normal and vasectomized men: what can we learn about human sperm maturation? J. Androl. 32 (6), 686–697. doi:10.2164/jandrol.110.012575
Sullivan, R., and Saez, F. (2013). Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction 146 (1), R21–R35. doi:10.1530/REP-13-0058
Sun, E. L., and Flickinger, C. J. (1979). Development of cell types and of regional differences in the postnatal rat epididymis. Am. J. Anat. 154 (1), 27–55. doi:10.1002/aja.1001540104
Toyonaga, M., Sato, Y., Sasaki, A., Kaihara, A., and Tsutsui, T. (2011). Artificial insemination with cryopreserved sperm from feline epididymides stored at 4 °C. Theriogenology 76 (3), 532–537. doi:10.1016/j.theriogenology.2011.03.005
Tsutsui, T., Wada, M., Anzai, M., and Hori, T. (2003). Artificial insemination with frozen epididymal sperm in cats. J. Vet. Med. Sci. 65 (3), 397–399. doi:10.1292/jvms.65.397
Turner, T. T., Gleavy, J. L., and Harris, J. M. (1990). Fluid movement in the lumen of the rat epididymis: effect of vasectomy and subsequent vasovasostomy. J. Androl. 11 (5), 422–428. doi:10.1002/j.1939-4640.1990.tb00170.x
Vazquez, J., Ong, I. M., and Stanic, A. K. (2019). Single-cell technologies in reproductive immunology. Am. J. Reprod. Immunol. 82 (3), e13157. doi:10.1111/aji.13157
Von, L., and Neuhaeuser, G. (1964). Morphometric analysis of the human epididymis. Z Anat. Entwicklungsgesch 124, 126–152.
Watson, J. H. (1902). Some observations on the origin and nature of the so-called hydatids of morgagni found in men and women, with especial reference to the fate of the müllerian duct in the epididymis. J. Anat. Physiol. 36 (2), 147–161.
Westfalewicz, B., Dietrich, M. A., Mostek, A., Partyka, A., Bielas, W., Niżański, W., et al. (2017). Identification and functional analysis of bull (Bos taurus) cauda epididymal fluid proteome. J. Dairy Sci. 100 (8), 6707–6719. doi:10.3168/jds.2016-12526
Wong, J., Damdimopoulos, A., Damdimopoulou, P., Gasperoni, J. G., and Grommen, S. V. H. (2020). Transcriptome analysis of the epididymis from Plag1 deficient mice suggests dysregulation of sperm maturation and extracellular matrix genes. Dev. Dyn. 249 (12), 1500–1513. doi:10.1002/dvdy.254
Wong, P. Y., Au, C. L., and Bedford, J. M. (1982). Biology of the scrotum. II. Suppression by abdominal temperature of transepithelial ion and water transport in the cauda epididymidis. Biol. Reprod. 26 (4), 683–689. doi:10.1095/biolreprod26.4.683
Wu, C., Wang, C., Zhai, B., Zhao, Y., Zhao, Z., Yuan, Z., et al. (2021a). Study on the region-specific expression of epididymis mRNA in the rams. PLoS One 16 (1), e0245933. doi:10.1371/journal.pone.0245933
Wu, C., Wang, C., Zhai, B., Zhao, Y., Zhao, Z., Yuan, Z., et al. (2021b). Study of microRNA expression profile in different regions of ram epididymis. Reprod. Domest. Anim. 56 (9), 1209–1219. doi:10.1111/rda.13978
Yalcin-Ozuysal, O., Fiche, M., Guitierrez, M., Wagner, K. U., Raffoul, W., and Brisken, C. (2010). Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 17 (10), 1600–1612. doi:10.1038/cdd.2010.37
Yeung, C. H., Nashan, D., Sorg, C., Oberpenning, F., Schulze, H., Nieschlag, E., et al. (1994). Basal cells of the human epididymis-antigenic and ultrastructural similarities to tissue-fixed macrophages. Biol. Reprod. 50 (4), 917–926. doi:10.1095/biolreprod50.4.917
Zhao, J., Zhai, X., Ma, Y., Zhang, T., Wang, Z., and Chong, T. (2020). Anatomic characteristics of epididymis based on histology, proteomic, and 3D reconstruction. Andrology 8 (6), 1787–1794. doi:10.1111/andr.12842
Zhao, W., Ahmed, S., Ahmed, S., Yangliu, Y., Wang, H., and Cai, X. (2021a). Analysis of long non-coding RNAs in epididymis of cattleyak associated with male infertility. Theriogenology 160, 61–71. doi:10.1016/j.theriogenology.2020.10.033
Zhao, W., Hussain Solangi, T., Wu, Y., Yang, X., Xu, C., Wang, H., et al. (2021b). Comparative rna-seq analysis of region-specific miRNA expression in the epididymis of cattleyak. Reprod. Domest. Anim. 56 (4), 555–576. doi:10.1111/rda.13893
Zheng, W., Zhang, Y., Sun, C., Ge, S., Tan, Y., Shen, H., et al. (2021). A multi-omics study of human testis and epididymis. Molecules 26 (11), 3345. doi:10.3390/molecules26113345
Zhuang, J., Li, X., Yao, J., Sun, X., Liu, J., Nie, H., et al. (2023). Single-cell RNA sequencing reveals the local cell landscape in mouse epididymal initial segment during aging. Immun. Ageing 20 (1), 21. doi:10.1186/s12979-023-00345-9
Keywords: epididymis, regional, cell composition, gene expression pattern, male fertility
Citation: Liu M-M, Feng X-L, Qi C, Zhang S-E and Zhang G-L (2024) The significance of single-cell transcriptome analysis in epididymis research. Front. Cell Dev. Biol. 12:1357370. doi: 10.3389/fcell.2024.1357370
Received: 10 January 2024; Accepted: 12 March 2024;
Published: 21 March 2024.
Edited by:
Charles Bradley Shuster, New Mexico State University, United StatesReviewed by:
Michael C. Golding, Texas A and M University, United StatesMehmet Özbek, Mehmet Akif Ersoy University, Türkiye
Copyright © 2024 Liu, Feng, Qi, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-Liang Zhang, MjAxOTAxMDIyQHFhdS5lZHUuY24=
 Meng-Meng Liu1
Meng-Meng Liu1 Guo-Liang Zhang
Guo-Liang Zhang