- 1Department of Molecular and Medical Genetics, Oregon Health and Science University, Portland, OR, United States
- 2Division of Reproductive and Developmental Sciences, Oregon National Primate Research Center, Beaverton, OR, United States
- 3Department of Obstetrics and Gynecology, Oregon Health and Science University, Portland, OR, United States
- 4Department of Biomedical Engineering, Oregon Health and Science University, Portland, OR, United States
Mammalian preimplantation embryos often contend with aneuploidy that arose either by the inheritance of meiotic errors from the gametes, or from mitotic mis-segregation events that occurred following fertilization. Regardless of the origin, mis-segregated chromosomes become encapsulated in micronuclei (MN) that are spatially isolated from the main nucleus. Much of our knowledge of MN formation comes from dividing somatic cells during tumorigenesis, but the error-prone cleavage-stage of early embryogenesis is fundamentally different. One unique aspect is that cellular fragmentation (CF), whereby small subcellular bodies pinch off embryonic blastomeres, is frequently observed. CF has been detected in both in vitro and in vivo-derived embryos and likely represents a response to chromosome mis-segregation since it only appears after MN formation. There are multiple fates for MN, including sequestration into CFs, but the molecular mechanism(s) by which this occurs remains unclear. Due to nuclear envelope rupture, the chromosomal material contained within MN and CFs becomes susceptible to double stranded-DNA breaks. Despite this damage, embryos may still progress to the blastocyst stage and exclude chromosome-containing CFs, as well as non-dividing aneuploid blastomeres, from participating in further development. Whether these are attempts to rectify MN formation or eliminate embryos with poor implantation potential is unknown and this review will discuss the potential implications of DNA removal by CF/blastomere exclusion. We will also extrapolate what is known about the intracellular pathways mediating MN formation and rupture in somatic cells to preimplantation embryogenesis and how nuclear budding and DNA release into the cytoplasm may impact overall development.
Introduction
Chromosomal instability (CIN) is a common form of genomic variability that causes changes in chromosome number and/or structure (Bielski and Taylor, 2021). It is characterized by either whole or segmental duplications/deletions of individual chromosomes or produces complex abnormalities that impact multiple chromosomes simultaneously. Losses and/or gains of entire chromosomes is termed aneuploidy and results in copy number variation (CNV) between cells. Besides containing a different number of chromosomes than their chromosomally normal (euploid) counterparts, aneuploid cells will also likely experience gene dosage differences that manifests as genetic variation (Kojima and Cimini, 2019; Stamoulis et al., 2019). While thought to be hallmarks of tumor transformation and cancer, CIN and aneuploidy are also frequently observed in mammalian preimplantation embryos (Vanneste et al., 2009). Aneuploidy can arise in the gametes during meiosis or after fertilization from the mitotic divisions of early embryogenesis. Unlike mitotic divisions in somatic cells, however, cleavage-stage embryos increase in cell number without a change in overall size. This largely occurs in the absence of new transcription until the major wave of embryonic genome activation (EGA) around the ∼4–8 cell stage in several mammals (Braude et al., 1988; Dobson et al., 2004; Vassena et al., 2011; Asami et al., 2022). Thus, the first three mitotic divisions are the most susceptible to chromosome mis-segregation events. Depending on the stage at which it arises, a mitotic error can be just as detrimental as a meiotic error and induce embryo arrest or be surmounted in subsequent development. Even if an aneuploid embryo reaches the blastocyst stage, however, it can still fail to implant or result in spontaneous miscarriage following implantation (Hassold et al., 1980; Schaeffer et al., 2004).
Our knowledge of mitotic chromosome mis-segregation largely derives from somatic cells and includes relaxed cell cycle checkpoints, premature loss or prolonged chromosome cohesion, defective spindle attachments, and abnormal centrosome number (Ganem et al., 2009; Soto et al., 2019) (Figure 1A). However, only the mitotic checkpoint complex (MCC) has been investigated in greater detail within the context of embryogenesis (Wei et al., 2011; Bolton et al., 2016; Vazquez-Diez et al., 2019a; Brooks et al., 2022). At the zygote stage, replication stress due to incomplete DNA replication and early entry into mitosis also contributes to chromosome mis-segregation (Palmerola et al., 2022). Following a mis-segregation event, the chromosome(s) will become encapsulated in nuclear envelope (NE) and form abnormal intracellular structures known as micronuclei (MN) that are spatially distinct from the main nucleus (Chavez et al., 2012; Daughtry et al., 2019)
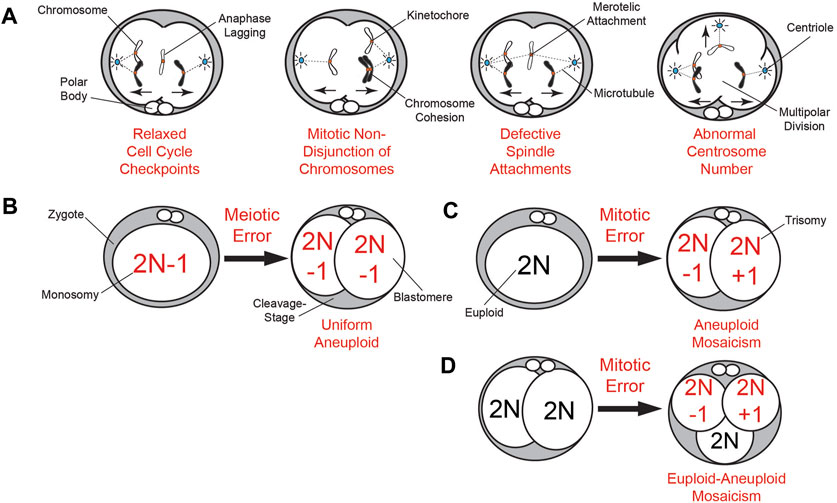
FIGURE 1. Summary of meiotic and mitotic chromosome mis-segregation mechanisms and embryonic aneuploidy outcomes. (A) Known mechanisms of mitotic mis-segregation from dividing somatic cells that have also been detected to some extent in cleavage-stage embryos from humans (Coonen et al., 2004), NHPs (Daughtry et al., 2019), and livestock (Yao et al., 2018; Brooks et al., 2019; Brooks et al., 2022). This includes relaxed cell cycle checkpoints that permit aberrations such as anaphase lagging of chromosomes, non-disjunction events due to premature loss or prolonged chromosome cohesion, defective spindle attachments of chromosomes to microtubules, and abnormal centrosome number leading to multipolar divisions. (B) The inheritance of a meiotic segregation error (a monosomy in this case) from the oocyte results in a 2-cell embryo with uniform aneuploidy, whereby each daughter blastomere has the same karyotype (2N−1). (C) Mitotic chromosome mis-segregation in a zygote during the first cleavage division can be just as detrimental as the inheritance of a meiotic error, producing a 2-cell embryo with blastomeres exhibiting different chromosomal losses (2N−1; monosomy) or gains (2N + 1; trisomy) known as aneuploid mosaicism. (D) Mitotic errors occurring later in development beyond the 2-cell stage produce embryos with euploid-aneuploid mosaicism, or those containing a mixture of both euploid and aneuploid blastomeres.
Meiotic versus mitotic origin(s) of aneuploidy in mammalian preimplantation embryos
Using whole-genome approaches, it was shown that 50%–80% of even high-quality cleavage-stage human embryos are aneuploid to some extent (Vanneste et al., 2009; Johnson et al., 2010b; Chavez et al., 2012; Chow et al., 2014; McCoy et al., 2015), which is major contributor to in vitro fertilization (IVF) failure. Estimates of aneuploidy in human blastocysts based on TE biopsy are less than the cleavage-stage, but still quite high at 20%–40% (Johnson et al., 2010b; Fiorentino et al., 2014; Popovic et al., 2018). By inferring chromosome dosage from single-cell RNA-seq data, an analysis of the morula-to-blastocyst transition suggested that the aneuploidy frequency at this stage of development is probably closer to 80% (Starostik et al., 2020). Indeed, a recent study used single-cell DNA-seq (scDNA-seq) of human blastocysts to demonstrate numerical and structural chromosome abnormalities in 82% of the embryos (Chavli et al., 2024). A similarly high percentage of aneuploidy is thought to occur in cleavage-stage embryos from rhesus monkeys (Daughtry et al., 2019), but to a lesser extent in bovine (Destouni et al., 2016; Hornak et al., 2016; Tsuiko et al., 2017) and porcine embryos (Hornak et al., 2015), and 1%–4% of murine embryos (Macaulay et al., 2015; Treff et al., 2016). To date, only DNA-Fluorescence In Situ Hybridization of a subset of chromosomes has been used to assess the frequency of aneuploidy in cleavage-stage equine embryos (Rambags et al., 2005). However, like bovine oocytes, equine oocytes are typically matured in vitro, which is known to increase aneuploidy by itself (Treff et al., 2016). Chromosomal mis-segregation in oocytes that arises during meiosis is one of the primary sources of aneuploidy and estimated to be around ∼20% in young women, but this percentage increases with maternal age (Hou et al., 2013; Gruhn et al., 2019; Charalambous et al., 2022) (Figure 1B). However, there is evidence that these meiotic errors can be corrected during oocyte maturation by the extrusion of reciprocal aneuploid polar bodies (PBs) (Hassold and Chiu, 1985; Forman et al., 2013; Treff et al., 2016). In contrast, sperm typically exhibit an aneuploidy frequency of ∼1–2% that is not influenced by paternal age (Lu et al., 2012; Bell et al., 2020). Although specific factors to meiotic mis-segregation have been identified, much of our knowledge of mitotic mis-segregation comes from somatic cells during tumorigenesis and cancer progression. This is in spite of findings that mitotic errors in cleavage-stage embryos are just as, or more, frequent as meiotic errors and appear independent of both maternal age or fertility status (Vanneste et al., 2009; Chavez et al., 2012; McCoy et al., 2015) (Figures 1C, D).
Mechanisms of chromosome mis-segregation during gametogenesis and embryogenesis
Non-disjunction, or the failure of homologous chromosomes (meiosis I) or sister chromatids (meiosis II) to properly separate, is the most common cause of chromosome mis-segregation in oocytes (Wartosch et al., 2021) (Figure 1B). While non-disjunction can occur during both meiotic stages, non-disjunction events leading to major aneuploidy issues such as uniform monosomy or trisomy originate primarily from meiosis I (Ghosh et al., 2009; Herbert et al., 2015). Mitotic non-disjunction is also observed in embryos following fertilization, but the failure of parental chromosomes to properly migrate to the spindle poles during anaphase is thought to be the most frequent mechanism for aneuploidy to arise during cleavage divisions (Coonen et al., 2004; Daughtry et al., 2019; Brooks et al., 2022) (Figure 1A). However, anaphase lagging of chromosomes in somatic cells is often indicative of defects in kinetochore-microtubule attachments and therefore, these are not necessarily mutually exclusive events (Thompson and Compton, 2011). There is also evidence that incomplete DNA replication during the S phase of the cell cycle and premature entry into mitosis results in subsequent whole and segmental chromosome errors at the zygote stage (Palmerola et al., 2022).
By monitoring the bipolar attachment of spindle microtubules to kinetochores, the MCC prevents activation of the anaphase promoting complex/cyclosome and delays mitotic progression until stable attachments are established. The main components of the MCC are evolutionarily conserved and include CDC20, as well as the serine/threonine kinases, BUB1B/BUBR1, BUB3, and MAD2. Unlike somatic cells, which require all three kinases to prevent aneuploidy, gametes and embryos primarily rely on BUBR1/BUB1B to ensure chromosome fidelity (Jeganathan and van Deursen, 2006; Touati et al., 2015; Vazquez-Diez et al., 2019b; Brooks et al., 2022). Moreover, because Bub1B/BubR1 has been shown to be maternally inherited in Drosophila (Perez-Mongiovi et al., 2005) and early cleavage divisions are largely under the control of maternal protein and RNA until EGA (Braude et al., 1988; Dobson et al., 2004; Vassena et al., 2011; Asami et al., 2022), BUBR1/BUB1B and other cell cycle associated factors derived from the oocyte likely contribute to maternal age related fertility decline (Qiu et al., 2018). Sperm also provide a small number of transcripts at fertilization, but whether these RNAs participate in EGA or other processes during the oocyte-to-embryo transition is still under investigation (Sendler et al., 2013; Corral-Vazquez et al., 2021). However, it has been shown that certain sperm borne microRNAs regulate early mitotic timing in human embryos and are required for the first cleavage division at least in mice (Liu et al., 2012; Shi et al., 2020). The centrosome for the first mitotic division(s) is also paternally inherited from the sperm in most mammalian species except rodents and can still cause aneuploidy via abnormal divisions (Sathananthan et al., 1991; Schatten et al., 1991) (Figure 1A).
CF is likely a response to chromosome mis-segregation and micronuclei formation
Even though bipolar divisions are more likely to produce euploid embryos that reach the blastocyst stage (Figure 2A), a segregation error from anaphase lagging or another mechanism during mitosis can still result in the formation of MN (Figure 2B). MN are frequently detected in early cleavage-stage embryos from humans (Chavez et al., 2012), nonhuman primates (NHPs) (Daughtry et al., 2019), horses (Brooks et al., 2019), and cattle (Yao et al., 2018; Brooks et al., 2022; Yao et al., 2022), but not in murine embryos at comparable levels until the morula stage (Vazquez-Diez et al., 2016; Vazquez-Diez et al., 2019b). Once formed, MN can sustain one of multiple fates, including persistence in subsequent divisions, fusion with the primary nucleus from which it arose, or relocation to an adjacent cell via a chromatin bridge during anaphase (Vazquez-Diez et al., 2016; Brooks et al., 2022). The appearance of MN in embryos is likely a precursor to CF since MN may be detected in the absence of CF, but not vice versa (Daughtry et al., 2019). Analogous to aneuploidy, the incidence of CF varies across mammalian species, with ∼50% of human, NHP, equine, and porcine embryos exhibiting CF (Alikani et al., 1999; Antczak and Van Blerkom, 1999; Tremoleda et al., 2003; Mateusen et al., 2005; Daughtry et al., 2019), ∼15% of bovine embryos undergoing CF (Somfai et al., 2010; Sugimura et al., 2010), and murine embryos rarely displaying CF unless experimentally induced (Dozortsev et al., 1998; Chavez et al., 2014). CF has been shown to occur in vivo in several mammals, including humans (Pereda and Croxatto, 1978; Buster et al., 1985; Gjorret et al., 2003; Mateusen et al., 2005), indicating it is not an artifact of in vitro culture, and is distinct from the cell death-induced DNA fragmentation that can arise late in preimplantation development (Hardy et al., 2001; Xu et al., 2001). Despite indications that CF is evolutionary shared among some mammals, comparative studies focused on the mechanistic details of CF and its relationship to aneuploidy are still lacking.
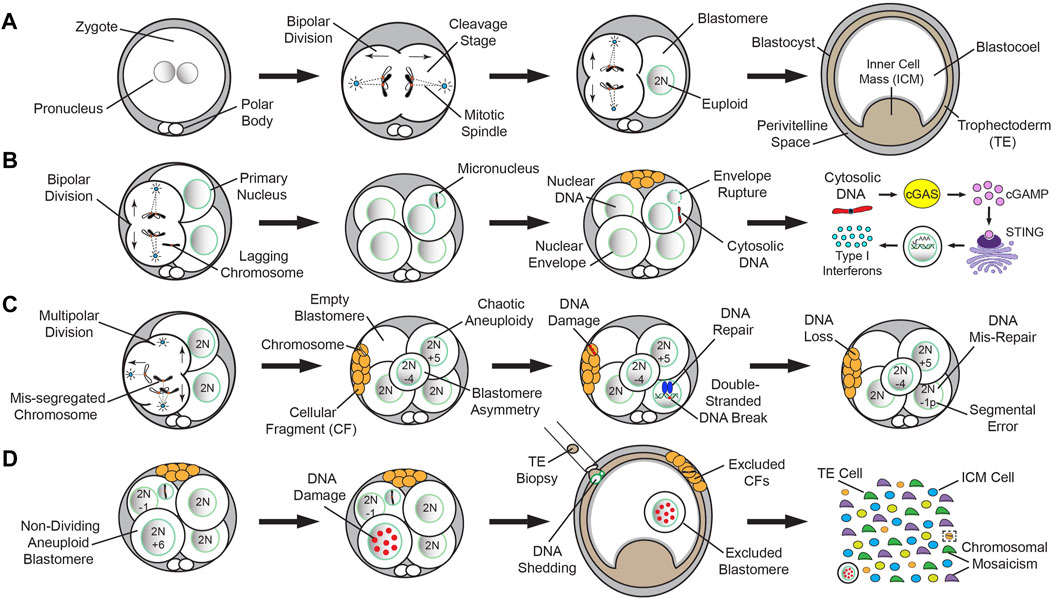
FIGURE 2. Normal versus abnormal mitotic chromosome segregation and different mechanisms of DNA loss from embryos. A simplified model of normal mitotic chromosome segregation in mammalian cleavage-stage embryos and mis-segregation induced DNA elimination up to the blastocyst stage in humans (Chavez et al., 2012; Domingo-Muelas et al., 2023), NHPs (Daughtry et al., 2019), horses (Brooks et al., 2019), and cattle (Yao et al., 2018; Brooks et al., 2022; Yao et al., 2022). (A) Normal mitotic chromosome segregation events from bipolar cleavage divisions are more likely to produce euploid embryos that reach the blastocyst stage. (B) A segregation error from anaphase lagging or another mechanism during mitosis can still result in the formation of MN. The appearance of MN in embryos is likely a precursor to CF since MN may be detected in the absence of CF, but not vice versa. MN are fragile and their nuclear envelope often ruptures, leading to the release of DNA into the cytoplasm. In other biological contexts, cytosolic DNA activates the cGAS-STING DNA-sensing pathway, which in turn, induces an immune response through transcription of Type I Interferons (IFNs) and other cytokines. (C) Multipolar divisions at the cleavage-stage often result in blastomere asymmetry, chaotic aneuploidy, and/or one or more cells lacking a nucleus (empty blastomere). Although there are likely other mechanisms, CF during or following the abnormal division may capture whole chromosomes or chromosomal segments lost from blastomeres. The chromosome(s) contained within CFs lack a nuclear envelope and undergo DNA damage, resulting in eventual DNA loss. In contrast to whole chromosomal mis-segregation, segmental errors most often emerge from improperly repaired double-stranded DNA breaks. (D) Despite the presence of MN, CF, and/or non-dividing aneuploid blastomeres, certain cleavage-stage embryos will still successfully reach the blastocyst stage. However, these entities are often subjected to extensive DNA damage. Upon blastocyst formation, chromosome-containing CFs may be expelled to the perivitelline space, while the arrested blastomeres are excluded into the blastocoel cavity. Blastocyst expansion mechanically constrains the TE layer, causing nuclear budding and the shedding of DNA into the cytoplasm, which is exacerbated by TE biopsy. Single-cell analyses of disassembled blastocysts and co-isolation of DNA and RNA from the same individual cell will assist in determining the impact of chromosome loss on the embryo and the extent of chromosomal mosaicism between the TE and ICM when cell identity is confirmed by gene expression.
Whole chromosomes and/or segmental DNA may be contained within CFs
While CF was first described in the 1980s as a morphological indicator of an embryo with low implantation potential (Trounson and Sathananthan, 1984; Alikani et al., 1999), the significance of CF and how it impacted embryogenesis was only recently determined. In 1999, the timing and pattern of CF in human embryos was examined and the authors concluded that the earlier in preimplantation development CF occurred, the worse the outcome (Antczak and Van Blerkom, 1999). Four basic patterns of CF were also proposed: (1) a single monolayer of small CFs on the surface of one blastomere with no apparent reduction in cell size, (2) the presence of multiple layers of CFs accompanied by a significant reduction in blastomere size, (3) complete disintegration of one or more blastomeres into CFs, and (4) a limited number of small CFs scattered over several blastomeres with no indication of which cell(s) they came from. The findings also suggested that the release of large fragments at the early cleavage-stage may deprive blastomeres of vital proteins and organelles, hindering further development. Initial attempts to selectively remove CFs by microsurgery suggested that this procedure improved blastocyst formation and IVF outcomes (Eftekhari-Yazdi et al., 2006; Keltz et al., 2006), but other studies reported no benefit from fragment removal (Halvaei et al., 2016). Ultrastructure analyses of both in vitro and in vivo derived embryos revealed that mitochondria were the most abundant organelle in CFs (Pereda and Croxatto, 1978; Halvaei et al., 2016), and immunostaining suggested that there was DNA in some fragments (Chavez et al., 2012). It was not until the contents of CFs were examined by next-generation sequencing and the entire embryo reconstructed at a single-cell level, however, that this was confirmed. Using rhesus macaque embryos as a model for human preimplantation development, our group demonstrated that fragments may encapsulate whole chromosomes or chromosomal segments lost from blastomeres (Daughtry et al., 2019) (Figure 2C). There was no preferential sequestering of certain chromosomes as both small and large chromosomes were affected, and high variability in the size of DNA segments, with a slightly greater propensity for maternal chromosomes to be encapsulated. However, while ∼18% of the embryos had chromosome-containing fragments, only ∼6% of all CFs contained DNA detectable by sequencing. Given that most embryos with CF exhibited non-reciprocal chromosome losses not found in other cells or fragments (Daughtry et al., 2019), this suggested that CF is not efficient at removing mis-segregated chromosomes or there is an alternative fate for MN.
Time-lapse imaging uncovers the mechanism(s) of chromosome encapsulation by CFs
Although many aneuploid embryos will arrest at the cleavage-stage, some will still form blastocysts and may even appear morphologically indistinguishable from euploid embryos, especially when assessed at static time points (Magli et al., 2000; Dupont et al., 2010). Thus, the use of time-lapse imaging to monitor preimplantation embryogenesis in real-time has enabled the tracking of morphological events indicative of developmental potential such as the timing, polarity, and symmetry of mitotic divisions, as well as the fate of CFs (Daughtry and Chavez, 2018). Time-lapse imaging has also shown that a fragment can be reabsorbed by the original blastomere from which it arose, providing an opportunity to restore euploidy (Chavez et al., 2012). If a chromosome-containing fragment fuses with a neighboring blastomere that is euploid, however, this could have detrimental effects on the embryo (Hardarson et al., 2002). When we evaluated whether there were imaging parameters indicative of chromosome sequestration by CFs in our study, there was a clear correlation between this event and multipolar divisions at the zygote or 2-cell stage (Daughtry et al., 2019). Consequently, the multipolar division often resulted in blastomere asymmetry, chaotic aneuploidy, and/or one or more cells lacking a nucleus (Figure 2C). In extreme cases of chaotic aneuploidy, the multipolar division produced embryos where every cell had multiple chromosomes affected in what appeared to be a random pattern. Not all embryos with chromosome-containing CFs exhibited multipolar divisions, however, suggesting that there are other mechanisms leading to chromosome encapsulation and potential loss.
Mitotic mis-segregation events often lead to chromosomal mosaicism
Unlike mitotic errors at the zygote stage (Figure 1C), a mitotic mis-segregation event that arises in 2-cell embryos or later in development can produce cells with diverse karyotypes known as chromosomal mosaicism (Figure 1D). Euploid-aneuploid mosaic embryos containing a mixture of both chromosomally normal and abnormal cells are the most common (Chuang et al., 2020), but there are also reports of embryos with mixoploidy, whereby cells differ according to whether they are haploid, diploid, or polyploid (Destouni et al., 2016; Carson et al., 2018; Daughtry et al., 2019; De Coster et al., 2022). However, these types of errors typically go undetected and/or are classified as euploid unless parental DNA is inputted to assign chromosomes as either maternal or paternal. While it has been known for quite some time that the smaller chromosomes are more susceptible to mis-segregation events (Smith et al., 1998), recent studies suggest that mosaicism disproportionately impacts large chromosomes (Chuang et al., 2020). Chromosomal rearrangements also often emerge that were produced from improper repair of double-stranded DNA breaks (Burssed et al., 2022) (Figure 2C). This mis-repair leads to DNA damage and segmental errors that may still impact implantation potential, resulting in a higher likelihood of miscarriage and reduced live birth rate if transferred (Zore et al., 2019). Several studies have now shown that segmental aneuploidy is more often mitotic in origin, mostly paternally-derived, and tends to occur within distinct chromosomal regions (Chavez et al., 2012; Babariya et al., 2017; McCarty et al., 2022; Palmerola et al., 2022). Regardless of whether whole and/or partial chromosomes are affected, the DNA will become subjected to further damage due to the fragility of the NE that encapsulates the mis-segregated material, which can have significant consequences for embryo development.
MN rupture and the release of cytosolic DNA is irreversible and potentially catastrophic
Studies in cancer cells have shown that the NE of MN is quite fragile for a variety of reasons, including premature chromatin condensation in MN, asynchrony in the timing and rate of DNA replication between MN and the primary nucleus, and the failure of MN to import key proteins such as nuclear pore complexes necessary to maintain NE integrity (Xu et al., 2011; Crasta et al., 2012; Liu et al., 2018). This eventually results in MN rupture (Figure 2B), leading to the irreversible loss of nuclear-cytoplasmic compartmentalization and a greater propensity for chromothripsis to occur (Hatch et al., 2013). Chromothripsis is a catastrophic mutagenesis process, whereby chromosomes are shattered and randomly reassembled, promoting somatic cell tumorigenesis and cancer genome evolution (Cortes-Ciriano et al., 2020; Shoshani et al., 2021). Similar events are also known to arise in the germline of patients with certain congenital disorders and there are strong indications of a paternal bias in its origin (Kloosterman and Cuppen, 2013). While hallmarks of this process have been observed during early embryogenesis (Pellestor, 2014; Pellestor et al., 2014), whether preimplantation embryos are equally afflicted by chromothripsis is difficult to determine given the depth of genome coverage and large amplicon size needed to accurately call such small structural variants.
Upon nuclear rupture in other biological contexts, the genetic material contained within MN is released into the cytoplasm, resulting in the activation of the cyclic GMP-AMP (cGAS)-cyclic GMP-AMP receptor stimulator of interferon genes (STING) DNA-sensing pathway. Although cGAS typically resides in the cytoplasm, it can also be observed in the nucleus and plasma membrane (Barnett et al., 2019; Volkman et al., 2019), suggesting that its subcellular location may confer specificity. STING is normally present in the endoplasmic reticulum, but transferred to the Golgi after activation, where it activates the TANK-binding kinase 1 (TBK1) (Mukai et al., 2016; Ogawa et al., 2018). In turn, TBK1 phosphorylates interferon regulatory factor 3 and nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) (Yum et al., 2021), which translocate from the cytoplasm to the nucleus and induce transcription of type I Interferons and other cytokines (Sun et al., 2013; Wu et al., 2013). As part of the innate immune response, cGAS-STING typically provide protection from the invasion of pathogenic DNA during viral or microbial infections (Mackenzie et al., 2017; Yu and Liu, 2021). However, the cGAS-STING pathway can also be activated by the presence of endogenous DNA produced from chromosomal instability in tumorigenesis and cancer progression (Bakhoum and Cantley, 2018; Bakhoum et al., 2018). Given the high incidence of MN observed in cleavage-stage embryos from most mammals, it seems likely that cGAS-STING would serve as a surveillance mechanism for DNA release in this context as well, but this is still speculative (Figure 2B). Additional research suggested that the cGAS-STING pathway may also play a role in autophagy, senescence, and apoptosis (Yang et al., 2017; Yang et al., 2019; Zierhut et al., 2019), processes that also frequently occur during preimplantation development.
DNA is eliminated from the embryo by multiple mechanisms at the blastocyst stage
Despite the presence of MN, CF, and/or aneuploidy, cleavage-stage embryos may continue to divide and still successfully reach the blastocyst stage. This suggests that embryonic chromosomal instability can be overcome, but the frequency and underlying mechanism(s) of such events are unknown. Our group demonstrated that chromosome-containing CFs which persisted throughout preimplantation embryogenesis are often expelled to the perivitelline space prior to blastocyst formation (Daughtry et al., 2019) (Figure 2D). In addition, we also observed that non-dividing blastomeres from the early cleavage-stage may be excluded to the blastocoel cavity during the morula-to-blastocyst transition. Following CNV analysis, we determined that these excluded blastomeres were highly chaotic, with multiple chromosome losses and gains, and immunostaining revealed abnormal nuclear morphology with extensive DNA damage (Daughtry et al., 2019). A previous report showed that E-cadherin expression is absent or altered in the excluded blastomeres of human embryos with no or abnormal compaction, suggesting a disruption in cell-cell adhesion (Alikani, 2005). Another study with reconstituted mouse embryos containing a mixture of control and hyperploid blastomeres demonstrated that the latter exhibited slower cell cycle progression and frequent DNA fragmentation beginning at the 16-cell stage (Pauerova et al., 2020). However, we observed selection against blastomeres as early as the 2- to 8-cell stage when embryos are most susceptible to chromosome mis-segregation events, which indicates that these previously observed changes were the consequence rather than the cause of aneuploidy. Nevertheless, the extruded fragments and blastomeres would presumably be left behind upon embryo hatching from the zona pellucida and prevented from participating in further development.
A recent study also showed that blastocyst expansion mechanically constrains the TE layer, causing nuclear budding and the shedding of DNA into the cytoplasm, and that this was exacerbated by biopsying 5–10 TE cells for preimplantation genetic testing of aneuploidy (Domingo-Muelas et al., 2023) (Figure 2D). Thus, similar to fragment removal (Halvaei et al., 2016), TE biopsy may provide no benefit especially if the biopsied cells are not representative of the embryo overall (Gleicher et al., 2017; Wu et al., 2021; Ren et al., 2022), or actually harmful for development by causing further DNA loss. Whether preferential allocation of aneuploid cells to the TE layer makes these cells more susceptible to DNA elimination even in the absence of TE biopsy remains to be determined, but preliminary evidence in human blastocysts suggests that there is no difference in the frequency of aneuploidy in relation to cell lineage by scDNA-seq (Chavli et al., 2024). However, the study did observe that complex chromosomal abnormalities are more commonly observed in TE cells than the inner cell mass (ICM). For this work, the TE and ICM were separated by biopsy prior to disaggregation into single cells, which can result in cross contamination between cell types due to cytoplasmic strings that connect the ICM to TE cells (Ebner et al., 2020; Joo et al., 2023). Thus, additional studies are needed to confirm each lineage by isolating both DNA and RNA from the same individual cell for scDNA-seq and gene expression (Macaulay et al., 2015; Macaulay et al., 2016), respectively.
Conclusion and future directions
CIN and aneuploidy are remarkably common in preimplantation embryos, but difficult to accurately detect due to differences in the mechanism(s) of meiotic versus mitotic errors, extent of chromosomal mosaicism, and the response to chromosome mis-segregation through MN formation and the elimination of DNA by one of the above-described processes. Further work is needed to connect the intracellular dynamics of chromosome loss with measurable morphological events such as mitotic timing, MN formation, CF incidence, and blastomere asymmetry to noninvasively assess embryo potential. Ultimately, the goal will be to identify possible therapeutic targets to prevent or alleviate aneuploidy altogether, which may be difficult to accomplish if CIN is inherent to natural conception and aneuploidy occurs at a similar frequency in vitro and in vivo.
Author contributions
JB: Conceptualization, Data curation, Investigation, Writing–original draft, Writing–review and editing. SC: Conceptualization, Data curation, Investigation, Writing–original draft, Writing–review and editing, Funding acquisition, Project administration, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. JB was supported by the “Program in Molecular and Cellular Biosciences” (T32GM071338) and the NSF Graduate Research Fellowship Program (GRFP). This work was supported by the NIH/NICHD (R01HD086073-A1 and R01HD105320-A1 to SC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alikani, M. (2005). Epithelial cadherin distribution in abnormal human pre-implantation embryos. Hum. Reprod. 20, 3369–3375. doi:10.1093/humrep/dei242
Alikani, M., Cohen, J., Tomkin, G., Garrisi, G. J., Mack, C., and Scott, R. T. (1999). Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil. Steril. 71, 836–842. doi:10.1016/s0015-0282(99)00092-8
Antczak, M., and Van Blerkom, J. (1999). Temporal and spatial aspects of fragmentation in early human embryos: possible effects on developmental competence and association with the differential elimination of regulatory proteins from polarized domains. Hum. Reprod. 14, 429–447. doi:10.1093/humrep/14.2.429
Asami, M., Lam, B. Y. H., Ma, M. K., Rainbow, K., Braun, S., VerMilyea, M. D., et al. (2022). Human embryonic genome activation initiates at the one-cell stage. Cell. Stem Cell. 29, 209–216.e4. doi:10.1016/j.stem.2021.11.012
Babariya, D., Fragouli, E., Alfarawati, S., Spath, K., and Wells, D. (2017). The incidence and origin of segmental aneuploidy in human oocytes and preimplantation embryos. Hum. Reprod. 32, 2549–2560. doi:10.1093/humrep/dex324
Bakhoum, S. F., and Cantley, L. C. (2018). The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 174, 1347–1360. doi:10.1016/j.cell.2018.08.027
Bakhoum, S. F., Ngo, B., Laughney, A. M., Cavallo, J. A., Murphy, C. J., Ly, P., et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472. doi:10.1038/nature25432
Barnett, K. C., Coronas-Serna, J. M., Zhou, W., Ernandes, M. J., Cao, A., Kranzusch, P. J., et al. (2019). Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell. 176, 1432–1446. doi:10.1016/j.cell.2019.01.049
Bell, A. D., Mello, C. J., Nemesh, J., Brumbaugh, S. A., Wysoker, A., and McCarroll, S. A. (2020). Insights into variation in meiosis from 31,228 human sperm genomes. Nature 583, 259–264. doi:10.1038/s41586-020-2347-0
Bielski, C. M., and Taylor, B. S. (2021). Homing in on genomic instability as a therapeutic target in cancer. Nat. Commun. 12, 3663. doi:10.1038/s41467-021-23965-5
Bolton, H., Graham, S. J. L., Van der Aa, N., Kumar, P., Theunis, K., Fernandez Gallardo, E., et al. (2016). Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat. Commun. 7, 11165. doi:10.1038/ncomms11165
Braude, P., Bolton, V., and Moore, S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459–461. doi:10.1038/332459a0
Brooks, K. E., Daughtry, B. L., Davis, B., Yan, M. Y., Fei, S. S., Shepherd, S., et al. (2022). Molecular contribution to embryonic aneuploidy and karyotypic complexity in initial cleavage divisions of mammalian development. Development 149, dev198341. doi:10.1242/dev.198341
Brooks, K. E., Daughtry, B. L., Metcalf, E. S., Masterson, K., Battaglia, D., Gao, L., et al. (2019). Assessing equine embryo developmental competency by time-lapse image analysis. Reprod. Fertil. Dev. 31 (12), 1840–1850.
Burssed, B., Zamariolli, M., Bellucco, F. T., and Melaragno, M. I. (2022). Mechanisms of structural chromosomal rearrangement formation. Mol. Cytogenet. 15, 23. doi:10.1186/s13039-022-00600-6
Buster, J. E., Bustillo, M., Rodi, I. A., Cohen, S. W., Hamilton, M., Simon, J. A., et al. (1985). Biologic and morphologic development of donated human ova recovered by nonsurgical uterine lavage. Am. J. Obstet. Gynecol. 153, 211–217. doi:10.1016/0002-9378(85)90116-4
Carson, J. C., Hoffner, L., Conlin, L., Parks, W. T., Fisher, R. A., Spinner, N., et al. (2018). Diploid/triploid mixoploidy: a consequence of asymmetric zygotic segregation of parental genomes. Am. J. Med. Genet. A 176, 2720–2732. doi:10.1002/ajmg.a.40646
Charalambous, C., Webster, A., and Schuh, M. (2022). Aneuploidy in mammalian oocytes and the impact of maternal ageing. Nat. Rev. Mol. Cell. Biol. 24, 27–44. doi:10.1038/s41580-022-00517-3
Chavez, S. L., Loewke, K. E., Han, J., Moussavi, F., Colls, P., Munne, S., et al. (2012). Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat. Commun. 3, 1251. doi:10.1038/ncomms2249
Chavez, S. L., McElroy, S. L., Bossert, N. L., De Jonge, C. J., Rodriguez, M. V., Leong, D. E., et al. (2014). Comparison of epigenetic mediator expression and function in mouse and human embryonic blastomeres. Hum. Mol. Genet. 23, 4970–4984. doi:10.1093/hmg/ddu212
Chavli, E. A., Klaasen, S. J., Van Opstal, D., Laven, J. S., Kops, G. J., and Baart, E. B. (2024). Single-cell DNA sequencing reveals a high incidence of chromosomal abnormalities in human blastocysts. J. Clin. Investig., e174483. Epub ahead of print. doi:10.1172/JCI174483
Chow, J. F., Yeung, W. S. B., Lau, E. Y. L., Lee, V. C. Y., Ng, E. H. Y., and Ho, P. C. (2014). Array comparative genomic hybridization analyses of all blastomeres of a cohort of embryos from young IVF patients revealed significant contribution of mitotic errors to embryo mosaicism at the cleavage stage. Reprod. Biol. Endocrinol. 12, 105. doi:10.1186/1477-7827-12-105
Chuang, T. H., Chang, Y. P., Lee, M. J., Wang, H. L., Lai, H. H., and Chen, S. U. (2020). The incidence of mosaicism for individual chromosome in human blastocysts is correlated with chromosome length. Front. Genet. 11, 565348. doi:10.3389/fgene.2020.565348
Coonen, E., Derhaag, J. G., Dumoulin, J. C. M., van Wissen, L. C. P., Bras, M., Janssen, M., et al. (2004). Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Hum. Reprod. 19, 316–324. doi:10.1093/humrep/deh077
Corral-Vazquez, C., Blanco, J., Aiese Cigliano, R., Sarrate, Z., Rivera-Egea, R., Vidal, F., et al. (2021). The RNA content of human sperm reflects prior events in spermatogenesis and potential post-fertilization effects. Mol. Hum. Reprod. 27, gaab035. doi:10.1093/molehr/gaab035
Cortes-Ciriano, I., Lee, J. J. K., Xi, R., Jain, D., Jung, Y. L., Yang, L., et al. (2020). Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 52, 331–341. doi:10.1038/s41588-019-0576-7
Crasta, K., Ganem, N. J., Dagher, R., Lantermann, A. B., Ivanova, E. V., Pan, Y., et al. (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58. doi:10.1038/nature10802
Daughtry, B. L., and Chavez, S. L. (2018). Time-lapse imaging for the detection of chromosomal abnormalities in primate preimplantation embryos. Methods Mol. Biol. 1769, 293–317. doi:10.1007/978-1-4939-7780-2_19
Daughtry, B. L., Rosenkrantz, J. L., Lazar, N. H., Fei, S. S., Redmayne, N., Torkenczy, K. A., et al. (2019). Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 29, 367–382. doi:10.1101/gr.239830.118
De Coster, T., Masset, H., Tšuiko, O., Catteeuw, M., Zhao, Y., Dierckxsens, N., et al. (2022). Parental genomes segregate into distinct blastomeres during multipolar zygotic divisions leading to mixoploid and chimeric blastocysts. Genome Biol. 23, 201. doi:10.1186/s13059-022-02763-2
Destouni, A., Zamani Esteki, M., Catteeuw, M., Tšuiko, O., Dimitriadou, E., Smits, K., et al. (2016). Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res. 26, 567–578. doi:10.1101/gr.200527.115
Dobson, A. T., Raja, R., Abeyta, M. J., Taylor, T., Shen, S., Haqq, C., et al. (2004). The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 13, 1461–1470. doi:10.1093/hmg/ddh157
Domingo-Muelas, A., Skory, R. M., Moverley, A. A., Ardestani, G., Pomp, O., Rubio, C., et al. (2023). Human embryo live imaging reveals nuclear DNA shedding during blastocyst expansion and biopsy. Cell. 186, 3166–3181. doi:10.1016/j.cell.2023.06.003
Dozortsev, D., Ermilov, A., El-Mowafi, D. M., and Diamond, M. (1998). The impact of cellular fragmentation induced experimentally at different stages of mouse preimplantation development. Hum. Reprod. 13, 1307–1311. doi:10.1093/humrep/13.5.1307
Dupont, C., Segars, J., DeCherney, A., Bavister, B. D., Armant, D. R., and Brenner, C. A. (2010). Incidence of chromosomal mosaicism in morphologically normal nonhuman primate preimplantation embryos. Fertil. Steril. 93, 2545–2550. doi:10.1016/j.fertnstert.2009.06.040
Ebner, T., Sesli, Ö., Kresic, S., Enengl, S., Stoiber, B., Reiter, E., et al. (2020). Time-lapse imaging of cytoplasmic strings at the blastocyst stage suggests their association with spontaneous blastocoel collapse. Reprod. Biomed. Online 40, 191–199. doi:10.1016/j.rbmo.2019.11.004
Eftekhari-Yazdi, P., Valojerdi, M. R., Ashtiani, S. K., Eslaminejad, M. B., and Karimian, L. (2006). Effect of fragment removal on blastocyst formation and quality of human embryos. Reprod. Biomed. Online 13, 823–832. doi:10.1016/s1472-6483(10)61031-0
Fiorentino, F., Bono, S., Biricik, A., Nuccitelli, A., Cotroneo, E., Cottone, G., et al. (2014). Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum. Reprod. 29 (12), 2802–2813. doi:10.1093/humrep/deu277
Forman, E. J., Treff, N. R., Stevens, J. M., Garnsey, H. M., Katz-Jaffe, M. G., Scott, R. T., et al. (2013). Embryos whose polar bodies contain isolated reciprocal chromosome aneuploidy are almost always euploid. Hum. Reprod. 28, 502–508. doi:10.1093/humrep/des393
Ganem, N. J., Godinho, S. A., and Pellman, D. (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. doi:10.1038/nature08136
Ghosh, S., Feingold, E., and Dey, S. K. (2009). Etiology of Down syndrome: evidence for consistent association among altered meiotic recombination, nondisjunction, and maternal age across populations. Am. J. Med. Genet. A 149A, 1415–1420. doi:10.1002/ajmg.a.32932
Gjorret, J. O., Knijn, H. M., Dieleman, S. J., Avery, B., Larsson, L. I., and Maddox-Hyttel, P. (2003). Chronology of apoptosis in bovine embryos produced in vivo and in vitro. Biol. Reprod. 69, 1193–1200. doi:10.1095/biolreprod.102.013243
Gleicher, N., Metzger, J., Croft, G., Kushnir, V. A., Albertini, D. F., and Barad, D. H. (2017). A single trophectoderm biopsy at blastocyst stage is mathematically unable to determine embryo ploidy accurately enough for clinical use. Reprod. Biol. Endocrinol. 15, 33. doi:10.1186/s12958-017-0251-8
Gruhn, J. R., Zielinska, A. P., Shukla, V., Blanshard, R., Capalbo, A., Cimadomo, D., et al. (2019). Chromosome errors in human eggs shape natural fertility over reproductive life span. Science 365, 1466–1469. doi:10.1126/science.aav7321
Halvaei, I., Khalili, M. A., Esfandiari, N., Safari, S., Talebi, A. R., Miglietta, S., et al. (2016). Ultrastructure of cytoplasmic fragments in human cleavage stage embryos. J. Assist. Reprod. Genet. 33, 1677–1684. doi:10.1007/s10815-016-0806-1
Hardarson, T., Löfman, C., Coull, G., Sjögren, A., Hamberger, L., and Edwards, R. G. (2002). Internalization of cellular fragments in a human embryo: time-lapse recordings. Reprod. Biomed. Online 5, 36–38. doi:10.1016/s1472-6483(10)61594-5
Hardy, K., Spanos, S., Becker, D., Iannelli, P., Winston, R. M., and Stark, J. (2001). From cell death to embryo arrest: mathematical models of human preimplantation embryo development. Proc. Natl. Acad. Sci. U. S. A. 98, 1655–1660. doi:10.1073/pnas.98.4.1655
Hassold, T., and Chiu, D. (1985). Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum. Genet. 70, 11–17. doi:10.1007/BF00389450
Hassold, T., Chen, N., Funkhouser, J., Jooss, T., Manuel, B., Matsuura, J., et al. (1980). A cytogenetic study of 1000 spontaneous abortions. Ann. Hum. Genet. 44 (2), 151–164. doi:10.1111/j.1469-1809.1980.tb00955.x
Hatch, E. M., Fischer, A. H., Deerinck, T. J., and Hetzer, M. W. (2013). Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 154, 47–60. doi:10.1016/j.cell.2013.06.007
Herbert, M., Kalleas, D., Cooney, D., Lamb, M., and Lister, L. (2015). Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb. Perspect. Biol. 7, a017970. doi:10.1101/cshperspect.a017970
Hornak, M., Jeseta, M., Hanulakova, S., and Rubes, J. (2015). A high incidence of chromosome abnormalities in two-cell stage porcine IVP embryos. J. Appl. Genet. 56, 515–523. doi:10.1007/s13353-015-0280-y
Hornak, M., Kubicek, D., Broz, P., Hulinska, P., Hanzalova, K., Griffin, D., et al. (2016). Aneuploidy detection and mtDNA quantification in bovine embryos with different cleavage onset using a next-generation sequencing-based protocol. Cytogenet Genome Res. 150, 60–67. doi:10.1159/000452923
Hou, Y., Fan, W., Yan, L., Li, R., Lian, Y., Huang, J., et al. (2013). Genome analyses of single human oocytes. Cell. 155, 1492–1506. doi:10.1016/j.cell.2013.11.040
Jeganathan, K. B., and van Deursen, J. M. (2006). Differential mitotic checkpoint protein requirements in somatic and germ cells. Biochem. Soc. Trans. 34, 583–586. doi:10.1042/BST0340583
Johnson, D. S., Cinnioglu, C., Ross, R., Filby, A., Gemelos, G., Hill, M., et al. (2010a). Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol. Hum. Reprod. 16 (12), 944–949. doi:10.1093/molehr/gaq062
Johnson, D. S., Gemelos, G., Baner, J., Ryan, A., Cinnioglu, C., Banjevic, M., et al. (2010b). Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum. Reprod. 25, 1066–1075. doi:10.1093/humrep/dep452
Joo, K., Nemes, A., Dudas, B., Berkes-Bara, E., Murber, A., Urbancsek, J., et al. (2023). The importance of cytoplasmic strings during early human embryonic development. Front. Cell. Dev. Biol. 11, 1177279. doi:10.3389/fcell.2023.1177279
Keltz, M. D., Skorupski, J. C., Bradley, K., and Stein, D. (2006). Predictors of embryo fragmentation and outcome after fragment removal in in vitro fertilization. Fertil. Steril. 86, 321–324. doi:10.1016/j.fertnstert.2006.01.048
Kloosterman, W. P., and Cuppen, E. (2013). Chromothripsis in congenital disorders and cancer: similarities and differences. Curr. Opin. Cell. Biol. 25, 341–348. doi:10.1016/j.ceb.2013.02.008
Kojima, S., and Cimini, D. (2019). Aneuploidy and gene expression: is there dosage compensation? Epigenomics 11, 1827–1837. doi:10.2217/epi-2019-0135
Liu, S., Kwon, M., Mannino, M., Yang, N., Renda, F., Khodjakov, A., et al. (2018). Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 561, 551–555. doi:10.1038/s41586-018-0534-z
Liu, W. M., Pang, R. T. K., Chiu, P. C. N., Wong, B. P. C., Lao, K., Lee, K. F., et al. (2012). Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. U. S. A. 109, 490–494. doi:10.1073/pnas.1110368109
Lu, S., Zong, C., Fan, W., Yang, M., Li, J., Chapman, A. R., et al. (2012). Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338, 1627–1630. doi:10.1126/science.1229112
Macaulay, I. C., Haerty, W., Kumar, P., Li, Y. I., Hu, T. X., Teng, M. J., et al. (2015). G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522. doi:10.1038/nmeth.3370
Macaulay, I. C., Teng, M. J., Haerty, W., Kumar, P., Ponting, C. P., and Voet, T. (2016). Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat. Protoc. 11, 2081–2103. doi:10.1038/nprot.2016.138
Mackenzie, K. J., Carroll, P., Martin, C. A., Murina, O., Fluteau, A., Simpson, D. J., et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465. doi:10.1038/nature23449
Magli, M. C., Jones, G. M., Gras, L., Gianaroli, L., Korman, I., and Trounson, A. O. (2000). Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum. Reprod. 15, 1781–1786. doi:10.1093/humrep/15.8.1781
Mateusen, B., Van Soom, A., Maes, D. G. D., Donnay, I., Duchateau, L., and Lequarre, A. S. (2005). Porcine embryo development and fragmentation and their relation to apoptotic markers: a cinematographic and confocal laser scanning microscopic study. Reproduction 129, 443–452. doi:10.1530/rep.1.00533
McCarty, K. J., Haywood, M. E., Lee, R., Henry, L., Arnold, A., McReynolds, S., et al. (2022). Segmental aneuploid hotspots identified across the genome concordant on reanalysis. Mol. Hum. Reprod. 29, gaac040. doi:10.1093/molehr/gaac040
McCoy, R. C., Demko, Z. P., Ryan, A., Banjevic, M., Hill, M., Sigurjonsson, S., et al. (2015). Evidence of selection against complex mitotic-origin aneuploidy during preimplantation development. PLoS Genet 11 (10), e1005601. doi:10.1371/journal.pgen.1005601
Mukai, K., Konno, H., Akiba, T., Uemura, T., Waguri, S., Kobayashi, T., et al. (2016). Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7, 11932. doi:10.1038/ncomms11932
Ogawa, E., Mukai, K., Saito, K., Arai, H., and Taguchi, T. (2018). The binding of TBK1 to STING requires exocytic membrane traffic from the ER. Biochem. Biophys. Res. Commun. 503, 138–145. doi:10.1016/j.bbrc.2018.05.199
Palmerola, K. L., Amrane, S., De Los Angeles, A., Xu, S., Wang, N., de Pinho, J., et al. (2022). Replication stress impairs chromosome segregation and preimplantation development in human embryos. Cell. 185, 2988–3007. doi:10.1016/j.cell.2022.06.028
Pauerova, T., Radonova, L., Kovacovicova, K., Novakova, L., Skultety, M., and Anger, M. (2020). Aneuploidy during the onset of mouse embryo development. Reproduction 160, 773–782. doi:10.1530/REP-20-0086
Pellestor, F., Gatinois, V., Puechberty, J., Genevieve, D., and Lefort, G. (2014). Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil. Steril. 102, 1785–1796. doi:10.1016/j.fertnstert.2014.09.006
Pellestor, F. C. (2014). How does such a catastrophic event impact human reproduction? Hum. Reprod. 29, 388–393. doi:10.1093/humrep/deu003
Pereda, J., and Croxatto, H. B. (1978). Ultrastructure of a seven-cell human embryo. Biol. Reprod. 18, 481–489. doi:10.1095/biolreprod18.3.481
Perez-Mongiovi, D., Malmanche, N., Bousbaa, H., and Sunkel, C. (2005). Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster. Development 132, 4509–4520. doi:10.1242/dev.02028
Popovic, M., Dheedene, A., Christodoulou, C., Taelman, J., Dhaenens, L., and Nieuwerburgh, F. V. (2018). Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing?. Hum. Reprod. 33 (7), 1342–1354. doi:10.1093/humrep/dey106
Qiu, D., Hou, X., Han, L., Li, X., Ge, J., and Wang, Q. (2018). Sirt2-BubR1 acetylation pathway mediates the effects of advanced maternal age on oocyte quality. Aging Cell. 17, e12698. doi:10.1111/acel.12698
Rambags, B. P., Krijtenburg, P. J., Drie, H. F. v., Lazzari, G., Galli, C., Pearson, P. L., et al. (2005). Numerical chromosomal abnormalities in equine embryos produced in vivo and in vitro. Mol. Reprod. Dev. 72, 77–87. doi:10.1002/mrd.20302
Ren, Y., Yan, Z., Yang, M., Keller, L., Zhu, X., Lian, Y., et al. (2022). Regional and developmental characteristics of human embryo mosaicism revealed by single cell sequencing. PLoS Genet. 18, e1010310. doi:10.1371/journal.pgen.1010310
Sathananthan, A. H., Kola, I., Osborne, J., Trounson, A., Ng, S. C., Bongso, A., et al. (1991). Centrioles in the beginning of human development. Proc. Natl. Acad. Sci. U. S. A. 88, 4806–4810. doi:10.1073/pnas.88.11.4806
Schaeffer, A. J., Chung, J., Heretis, K., Wong, A., Ledbetter, D. H., and Martin, C. L. (2004). Comparative genomic hybridization–array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am. J. Hum. Genet. 74 (6), 1168–1174. doi:10.1086/421250
Schatten, G., Simerly, C., and Schatten, H. (1991). Maternal inheritance of centrosomes in mammals? Studies on parthenogenesis and polyspermy in mice. Proc. Natl. Acad. Sci. U. S. A. 88, 6785–6789. doi:10.1073/pnas.88.15.6785
Sendler, E., Johnson, G. D., Mao, S., Goodrich, R. J., Diamond, M. P., Hauser, R., et al. (2013). Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 41, 4104–4117. doi:10.1093/nar/gkt132
Shi, S., Shi, Q., and Sun, Y. (2020). The effect of sperm miR-34c on human embryonic development kinetics and clinical outcomes. Life Sci. 256, 117895. doi:10.1016/j.lfs.2020.117895
Shoshani, O., Brunner, S. F., Yaeger, R., Ly, P., Nechemia-Arbely, Y., Kim, D. H., et al. (2021). Chromothripsis drives the evolution of gene amplification in cancer. Nature 591, 137–141. doi:10.1038/s41586-020-03064-z
Smith, S. E., Toledo, A. A., Massey, J. B., and Kort, H. I. (1998). Simultaneous detection of chromosomes X, Y, 13, 18, and 21 by fluorescence in situ hybridization in blastomeres obtained from preimplantation embryos. J. Assist. Reprod. Genet. 15, 314–319. doi:10.1023/a:1022504829854
Somfai, T., Inaba, Y., Aikawa, Y., Ohtake, M., Kobayashi, S., Konishi, K., et al. (2010). Relationship between the length of cell cycles, cleavage pattern and developmental competence in bovine embryos generated by in vitro fertilization or parthenogenesis. J. Reprod. Dev. 56, 200–207. doi:10.1262/jrd.09-097a
Soto, M., Raaijmakers, J. A., and Medema, R. H. (2019). Consequences of genomic diversification induced by segregation errors. Trends Genet. 35, 279–291. doi:10.1016/j.tig.2019.01.003
Stamoulis, G., Garieri, M., Makrythanasis, P., Letourneau, A., Guipponi, M., Panousis, N., et al. (2019). Single cell transcriptome in aneuploidies reveals mechanisms of gene dosage imbalance. Nat. Commun. 10, 4495. doi:10.1038/s41467-019-12273-8
Starostik, M. R., Sosina, O. A., and McCoy, R. C. (2020). Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism. Genome Res. 30, 814–825. doi:10.1101/gr.262774.120
Sugimura, S., Akai, T., Somfai, T., Hirayama, M., Aikawa, Y., Ohtake, M., et al. (2010). Time-lapse cinematography-compatible polystyrene-based microwell culture system: a novel tool for tracking the development of individual bovine embryos. Biol. Reprod. 83, 970–978. doi:10.1095/biolreprod.110.085522
Sun, L., Wu, J., Du, F., Chen, X., and Chen, Z. J. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. doi:10.1126/science.1232458
Thompson, S. L., and Compton, D. A. (2011). Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl. Acad. Sci. U. S. A. 108, 17974–17978. doi:10.1073/pnas.1109720108
Touati, S. A., Buffin, E., Cladière, D., Hached, K., Rachez, C., van Deursen, J. M., et al. (2015). Mouse oocytes depend on BubR1 for proper chromosome segregation but not for prophase I arrest. Nat. Commun. 6, 6946. doi:10.1038/ncomms7946
Treff, N. R., Krisher, R. L., Tao, X., Garnsey, H., Bohrer, C., Silva, E., et al. (2016). Next generation sequencing-based comprehensive chromosome screening in mouse polar bodies, oocytes, and embryos. Biol. Reprod. 94, 76. doi:10.1095/biolreprod.115.135483
Tremoleda, J. L., Stout, T. A. E., Lagutina, I., Lazzari, G., Bevers, M. M., Colenbrander, B., et al. (2003). Effects of in vitro production on horse embryo morphology, cytoskeletal characteristics, and blastocyst capsule formation. Biol. Reprod. 69, 1895–1906. doi:10.1095/biolreprod.103.018515
Trounson, A., and Sathananthan, A. H. (1984). The application of electron microscopy in the evaluation of two-to four-cell human embryos cultured in vitro for embryo transfer. J Vitro Fert Embryo Transf 1, 153–165. doi:10.1007/BF01139208
Tsuiko, O., Catteeuw, M., Zamani Esteki, M., Destouni, A., Bogado Pascottini, O., Besenfelder, U., et al. (2017). Genome stability of bovine in vivo-conceived cleavage-stage embryos is higher compared to in vitro-produced embryos. Hum. Reprod. 32, 2348–2357. doi:10.1093/humrep/dex286
Vanneste, E., Voet, T., Le Caignec, C., Ampe, M., Konings, P., Melotte, C., et al. (2009). Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 15, 577–583. doi:10.1038/nm.1924
Vassena, R., Boué, S., González-Roca, E., Aran, B., Auer, H., Veiga, A., et al. (2011). Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138, 3699–3709. doi:10.1242/dev.064741
Vazquez-Diez, C., Paim, L. M. G., and FitzHarris, G. (2019a). Cell-size-independent spindle checkpoint failure underlies chromosome segregation error in mouse embryos. Curr. Biol. 29, 865–873. doi:10.1016/j.cub.2018.12.042
Vazquez-Diez, C., Paim, L. M. G., and FitzHarris, G. (2019b). Cell-size-independent spindle checkpoint failure underlies chromosome segregation error in mouse embryos. Curr. Biol. 29, 865–873. doi:10.1016/j.cub.2018.12.042
Vazquez-Diez, C., Yamagata, K., Trivedi, S., Haverfield, J., and FitzHarris, G. (2016). Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. Proc. Natl. Acad. Sci. U. S. A. 113, 626–631. doi:10.1073/pnas.1517628112
Volkman, H. E., Cambier, S., Gray, E. E., and Stetson, D. B. (2019). Tight nuclear tethering of cGAS is essential for preventing autoreactivity. Elife 8, e47491. doi:10.7554/eLife.47491
Wartosch, L., Schindler, K., Schuh, M., Gruhn, J. R., Hoffmann, E. R., McCoy, R. C., et al. (2021). Origins and mechanisms leading to aneuploidy in human eggs. Prenat. Diagn 41, 620–630. doi:10.1002/pd.5927
Wei, Y., Multi, S., Yang, C. R., Ma, J., Zhang, Q. H., Wang, Z. B., et al. (2011). Spindle assembly checkpoint regulates mitotic cell cycle progression during preimplantation embryo development. PLoS One 6, e21557. doi:10.1371/journal.pone.0021557
Wu, J., Sun, L., Chen, X., Du, F., Shi, H., Chen, C., et al. (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. doi:10.1126/science.1229963
Wu, L., Jin, L., Chen, W., Liu, J. M., Hu, J., Yu, Q., et al. (2021). The true incidence of chromosomal mosaicism after preimplantation genetic testing is much lower than that indicated by trophectoderm biopsy. Hum. Reprod. 36, 1691–1701. doi:10.1093/humrep/deab064
Xu, B., Sun, Z., Liu, Z., Guo, H., Liu, Q., Jiang, H., et al. (2011). Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS One 6, e18618. doi:10.1371/journal.pone.0018618
Xu, J., Cheung, T., Chan, S. T., Ho, P., and Yeung, W. S. (2001). The incidence of cytoplasmic fragmentation in mouse embryos in vitro is not affected by inhibition of caspase activity. Fertil. Steril. 75, 986–991. doi:10.1016/s0015-0282(01)01687-9
Yang, H., Wang, H., Ren, J., Chen, Q., and Chen, Z. J. (2017). cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. U. S. A. 114, E4612–E4620. doi:10.1073/pnas.1705499114
Yang, J., Tang, X., Nandakumar, K. S., and Cheng, K. (2019). Autophagy induced by STING, an unnoticed and primordial function of cGAS. Cell. Mol. Immunol. 16, 683–684. doi:10.1038/s41423-019-0240-2
Yao, T., Suzuki, R., Furuta, N., Suzuki, Y., Kabe, K., Tokoro, M., et al. (2018). Live-cell imaging of nuclear-chromosomal dynamics in bovine in vitro fertilised embryos. Sci. Rep. 8, 7460. doi:10.1038/s41598-018-25698-w
Yao, T., Ueda, A., Khurchabilig, A., Mashiko, D., Tokoro, M., Nagai, H., et al. (2022). Micronucleus formation during early cleavage division is a potential hallmark of preimplantation embryonic loss in cattle. Biochem. Biophys. Res. Commun. 617, 25–32. doi:10.1016/j.bbrc.2022.05.075
Yu, L., and Liu, P. (2021). Cytosolic DNA sensing by cGAS: regulation, function, and human diseases. Signal Transduct. Target Ther. 6, 170. doi:10.1038/s41392-021-00554-y
Yum, S., Li, M., Fang, Y., and Chen, Z. J. (2021). TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. U. S. A. 118, e2100225118. doi:10.1073/pnas.2100225118
Zierhut, C., Yamaguchi, N., Paredes, M., Luo, J. D., Carroll, T., and Funabiki, H. (2019). The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell. 178, 302–315. doi:10.1016/j.cell.2019.05.035
Keywords: cellular fragmentation, DNA shedding, excluded blastomere, micronuclei, mitosis, nuclear budding, preimplantation embryo, aneuploidy
Citation: Budrewicz J and Chavez SL (2024) Insights into embryonic chromosomal instability: mechanisms of DNA elimination during mammalian preimplantation development. Front. Cell Dev. Biol. 12:1344092. doi: 10.3389/fcell.2024.1344092
Received: 25 November 2023; Accepted: 15 January 2024;
Published: 05 February 2024.
Edited by:
Judith Yanowitz, Magee-Womens Research Institute, United StatesReviewed by:
Sue Hammoud, University of Michigan, United StatesSatoshi Sugimura, Tokyo University of Agriculture and Technology, Japan
Miguel Abraham Velazquez, Newcastle University, United Kingdom
Copyright © 2024 Budrewicz and Chavez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shawn L. Chavez, Y2hhdmVzaEBvaHN1LmVkdQ==
 Jacqueline Budrewicz
Jacqueline Budrewicz Shawn L. Chavez
Shawn L. Chavez