- Biology, The Life Science Center, School of Science and Technology, Örebro University, Örebro, Sweden
Background: The classical concept of brain sex differentiation suggests that steroid hormones released from the gonads program male and female brains differently. However, several studies indicate that steroid hormones are not the only determinant of brain sex differentiation and that genetic differences could also be involved.
Methods: In this study, we have performed RNA sequencing of rat brains at embryonic days 12 (E12), E13, and E14. The aim was to identify differentially expressed genes between male and female rat brains during early development.
Results: Analysis of genes expressed with the highest sex differences showed that Xist was highly expressed in females having XX genotype with an increasing expression over time. Analysis of genes expressed with the highest male expression identified three early genes, Sry2, Eif2s3y, and Ddx3y.
Discussion: The observed sex-specific expression of genes at early development confirms that the rat brain is sexually dimorphic prior to gonadal action on the brain and identifies Sry2 and Eif2s3y as early genes contributing to male brain development.
1 Introduction
Sexual dimorphism, including maternal care, sexual behavior, brain function, structure, and susceptibility to neurological disorders is evident in humans as well as in nonhuman species. Studies of human male and female brains have revealed sex differences in connectome, methylome, and transcriptome profiles (Ingalhalikar et al., 2014; Xu et al., 2014). Despite extensive advancement in neuroscience, the molecular regulation of these sex differences remains unclear.
The classical model of brain sex differentiation that placed gonadal steroid hormones as the main drivers in establishing male and female neural networks was derived from earlier studies (Phoenix et al., 1959; Arnold, 2009). This model states that the chromosomal constitution (XX or XY) determines the gonadal sex and that the hormone secreted by the gonads programs the brain neural network differently (Phoenix et al., 1959; Arnold, 2009).
The initiation of sex differentiation is governed by the sex determining region Y (Sry) master regulator gene located on the Y chromosome, that signals for the activation of the male sex differentiation pathway and the formation of testes (Koopman, 2005). The earliest gonadal expression of Sry is at around E10.5 in mouse, and peaks at E11.5 to initiate testis differentiation (Sim et al., 2008). It has also been observed that Sry is present in the mouse brain at E11 (Mayer et al., 2000). In rats, multiple Sry copies were identified by Turner and co-workers (Turner et al., 2007). They observed that among the different Sry homologues, Sry2 had the highest expression in rat testis and adrenal glands at 15 weeks of age. In a separate study, it was observed that the expression of Sry2 in rat gonads began from E11, as revealed by transcriptional analysis (Prokop et al., 2020). In humans the expression of Sry is observed first at E41 to peak on E44 (Hanley et al., 2000). Studies using the four-core mouse model indicate that Sry may be needed for proper masculinization of the animal, both gonadal and neuronal (Arnold, 2009). A study in chicken also indicate that male and female brains are sexually dimorphic prior to gonadal development (Lee et al., 2009).
Studies on zebra finch has been instrumental in revealing the involvement of genetic elements in neuronal cell sex differentiation. In a study on zebra finch, Gahr and Metzdorf, showed that androgen treatment of female birds did not fully masculinize the song center (Gahr and Metzdorf, 1999). Furthermore, an involvement of genetic factors in brain sex development was evident from a study of a gynandromorphic zebra finch (Arnold, 2003). Despite the whole brain being under the influence of the same gonadal hormones, the zebra finch still developed histologically identifiable song centers in the male side of the brain while the female side of the brain remained feminine (Arnold, 2003). In addition, aromatase inhibitor treatment, that induced testicular tissue in the genetic female zebra finch, failed to masculinize the song system that remained feminine (Wade and Arnold, 1996).
To identify genes involved in brain sexual development Dewing and coworkers performed a study using embryonic mouse heads and microarray analysis (Dewing et al., 2003) They observed sex differences in gene expression at embryonic day 10.5 (E10.5) were observed in the head region. DEAD-box RNA helicase y (Ddx3y; Dby) and eukaryotic translation initiation factor 2 subunit 3y (Eif2s3y) were identified as having the highest male biased gene expression at 10.5 dpc (days post coitum), while X-inactive specific transcript (Xist) showed the highest female bias. While studies on human brain sex differences are limited and generally obtained from mid-gestational developmental stages, they do provide information on brain sex differences (Reinius and Jazin, 2009; Johansson et al., 2016). Analysis of gene expression of sex-chromosome linked genes showed 11 Y-chromosome linked genes that were upregulated in comparison to their X-chromosome homologues in females. Further analysis of two of these genes (Pcdh11y and Nlgn4y) showed that these genes were predominantly expressed in different glial and neuronal cell populations in the CNS. In line with this it has also been shown that the microglia cell displays sex differences with males exhibiting higher microglia count than females throughout the neonatal period (Bordt et al., 2020).
To further elucidate the mechanisms leading to differential transcription in the developing male and female brain, and to identify genes showing sex dependent regulation in the developing rodent brain, we performed a transcriptomic analysis of rat brain during the period of gonadal activation. We selected three embryonic stages to determine sex differences in gene expression in the brain. These were stage E12 prior to gonadal activation (Prokop et al., 2020), E13 at the time of gonadal activation, and at E14 following gonadal activation (Val et al., 2003).
The temporal expression from E12 to E14 revealed two Y-chromosome genes with the highest expression levels at E12, Sry2, and Eif2s3y. As Sry2 showed sex biased gene expression at all three developmental stages, locally expressed Sry cannot be excluded as a regulator of gene expression in the brain prior to testis differentiation. Our results support that there are genetic sex differences in developing brains prior to hormone action and suggest that these differences may be involved in the differential development of male and female brains.
2 Materials and methods
2.1 Sample processing, genotyping, and RNA sequencing
Brain samples from Sprague Dawley rats (Rattus norvegicus) were obtained from Brain Bits (United States). The Spraque Dawley rats represent an outbred strain that was selected to allow for determination of responses related to a normal population of the species. Following sampling the brains were maintained in Hibernate™-E Medium (ThermoFisher Scientific, United States) and were shipped on ice. Directly following receiving the samples they were transferred to −80°C. Three males and three females were used to obtain brain samples from each developmental stage. For genotyping, tissue samples from the body were extracted using a DNA isolation kit (Zymo Research, United States). DNA was quantified using NanoDrop (Denovix, United States) and qPCR was performed for Xist and Sry, to confirm the sex of the sample and to validate brain specific expression. The primer sequences used for Sry and Xist are listed in Table 1. The qPCR reaction conditions were as follows: 95°C for 5 min followed by 35 cycles of 95°C for 10 s, 55°C for 15 s, and 72°C for 1 min. The PCR product was run on 1% agarose gel. Following genotyping, three individuals per sex and stage were used for RNA sequencing.
The samples were homogenized in Tri Reagent (Sigma) and RNA extraction was performed using Directzol RNA extraction kit (Zymo Research, United States). RNA samples were quantified using NanoDrop (Denovix, United States), and the quality was analyzed using RNA denaturing gel. RNA at a concentration of 50 ng/μL with a OD 260/280 between 1.8 and 2.0, OD 260/230 between 2.0 and 2.2 were sent to GATC Biotech/Eurofins for RNA sequencing. RNA samples with a RNA integrity number equal to or exceeding eight were used for sequencing, Sequencing was performed using Illumina platform to generate 2 × 51 bp reads.
2.2 Data analysis
The raw data files were first analyzed for sequence quality using pre-alignment QA/QC. Reads were trimmed from three prime ends based on quality score and the average Phred score for the reads was determined for all replicates. The reads were aligned to the rat genome (Rnor_6.0) using BWA-MEM alignment algorithm followed by quantification to alignment model using Partek genomic software (Partek Inc., St. Louis, United States). BWA was chosen due to its well described alignment accuracy and handling of false negative results which is usually observed by other algorithms with short sequence reads (Wu et al., 2019; Alganmi and Abusamra, 2023). The gene expression counts were normalized using the Counts Per Million reads (CPM) method, followed by 1.0e-4 addition. This adjustment is particularly important for genes located on the Y chromosome, as they do not exhibit any expression in female samples. Lowest gene coverage of 10 was employed to filter out low expressed genes. The resulting normalized counts were used to identify differentially regulated genes using DESeq2 (Anders and Huber, 2010). Analysis of the number of reads of Y chromosome genes was performed by calculating reads per million using Integrated Genomics Viewer version 2.12 (Robinson et al., 2011). Manual inspection of the reads of Sry genes was performed to ascertain localization at the correct position and ensured that the low read counts were not merely noise.
Hierarchical clustering and heatmaps were generated for each stage with the average linkage cluster distance metric, in conjunction with the Euclidean point distance metric (https://www.bioinformatics.com.cn/en). Standardized gene expression values (z-scores) were used to visualize each stage. Principal component analysis (PCA) was utilized to visualize relationships between various embryonic stages (Partek Inc., St. Louis, United States). To ensure uniform feature contribution in the construction of the PCA, 18 principal components (normalized counts) underwent a logarithmic transformation with a base of two and a transform offset of 1.
Venn diagrams were individually created using jvenn (https://jvenn.toulouse.inrae.fe/app/index.html) for male and female rats at each developmental stage, illustrating the unique gene sets for each sex. Furthermore, the overlapping genes across different stages were highlighted. Key Y-chromosome gene expression profiles were derived through the utilization of counts per million, allowing for the visualization of gene expression patterns across successive developmental stages.
Bubble plots were created to highlight GO annotations and KEGG pathway analysis for each embryonic stage by employing identified differentially expressed genes (DEGs) using an online data analysis and visualization platform (https://www.bioinformatics.com.cn/en). Terms and pathways were ranked based on their enrichment score and the top 10 enriched terms and pathways are presented. A color gradient was employed to indicate the percentage of sex ratios for the given term. Pathways that were overrepresented in males and females were indicated by red and a green coloring, respectively.
Relationships between proteins encoded by the DEGs at the embryonic stages were visualized using STRING database version 12.0 (Szklarczyk et al., 2019). Networks were individually created for each developmental stage. Interactions were predicted based on a confidence score computed by combining the probabilities from experimental, co-expression, co-occurrence, neighborhood, text mining, and database evidence corrected for the probability of randomly observing an interaction. Interactions with a medium confidence score (0.4) are shown. The thickness of the connecting lines indicates the strength of the interaction. Proteins were assigned to one of three clusters according to their global interaction score. Clusters were created with the KMEANS algorithm. Interactions between clusters are shown by dotted lines. Analysis was performed using False Discovery Rate (FDR <0.1) without limiting to 1.5 times fold change. Gene ontology and pathway enrichment was done in Reactome using Human orthologues (Fabregat et al., 2018) to determine whether the genes were separated into specific categories.
2.3 Statistical analysis
Candidate DEGs were initially identified based on a p-value threshold of less than 0.05. To control for multiple testing, the FDR step-up method was utilized, with a criterion of an FDR step-up value less than or equal to 0.1. Additionally, a fold change requirement of less than −1.5 or greater than 1.5 was applied to identify DEGs. This approach ensured the selection of genes that are both statistically significant and show substantial changes in expression levels. Significant interactions among gene sets were determined at p ≤ 0.05. Statistical analysis was performed using One way ANOVA followed by Tukey’s multiple comparison post-test using the GraphPad Prism eight software (GraphPad software, Boston, United States). The differences were considered significant when the p-value was <0.05 (∗p < 0.05; ∗∗p < 0.01).
3 Results
3.1 Alignment results
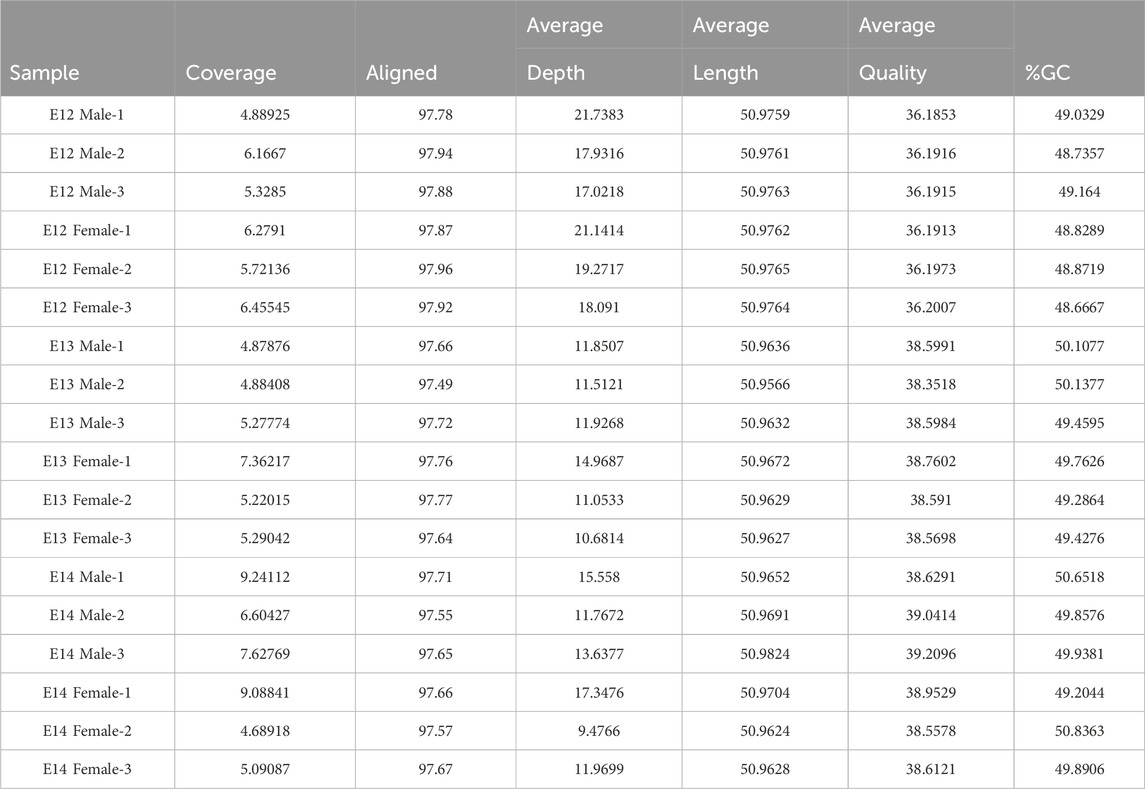
Comprehensive QA/QC results for samples used in this study are shown in Table 2. Key metrics, including coverage, alignment, average coverage depth, average sequence length, average sequence quality, and %GC content, have been summarized. High alignment percentages around 97% indicate robust alignment to the reference genome, ensuring the reliability of the sequencing data. Moreover, the stable average sequence length at approximately 50.97 base pairs emphasized the uniformity of sequenced fragments, that allowed robust downstream data analysis. The average sequence quality scores, ranging from 36.19 to 39.21, reflect the accuracy of base calling, with higher scores indicating lower sequencing errors. Furthermore, the %GC content, consistently ranging from 48.54% to 50.84%, suggested a homogeneous base composition among the samples.
3.2 Differentially expressed genes in rat brain
Analysis of differentially expressed genes in rat brains from E12 to E14 was performed to identify sex differences during early embryogenesis. The number of genes with higher read counts in male brains compared to female brains increased from E12 to E14 (Figure 1; Supplementary Figure S1). At E12, only 37 genes had higher read counts in males, and this number increased to 184 at E13 and 1,655 genes at E14 (Supplementary Table S2). Of these genes, six Y-chromosome genes were differentially expressed at all three developmental stages. In addition, there is an overlap of five DEGs between stages E13 and E14. Furthermore, a set of four genes exhibits differential expression at both E12 and E14. It was interesting to note that there were no overlapping genes between E12 and E13 except for the six Y-chromosome genes.
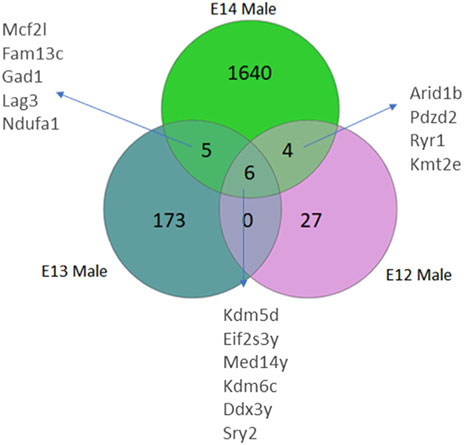
Figure 1. Venn diagrams illustrate the number of differentially expressed genes (DEGs) in male rats, categorized by their developmental stage. DEGs were identified based on an adjusted p-value <0.05, following False Discovery Rate (FDR) correction. DEGs common to multiple stages are also annotated.
The gene expression profile in female brains followed a similar pattern with the numbers of female biased genes increasing from E12 to E14 (Figure 2; Supplementary Figure S1). Here, four X-chromosome genes, and one autosomal gene, had significantly higher read counts in female brain at all three developmental stages. Moreover, an overlap of three genes was observed between E12 and E13, and 22 DEGs were common between E13 and E14. Notably, there were no common DEGs between E12 and E14 except for the four X chromosome genes, and one autosomal gene, that overlapped in all three stages.
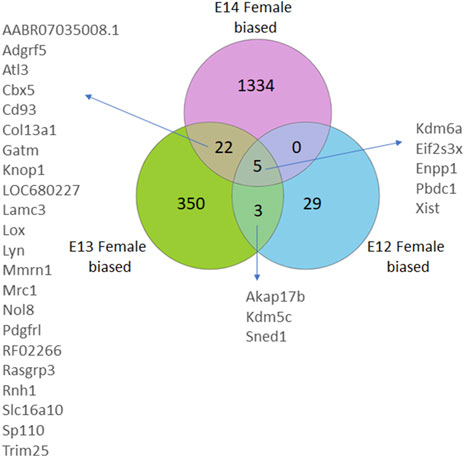
Figure 2. Venn diagrams illustrate the number of differentially expressed genes (DEGs) in female rats, categorized by their developmental stage. DEGs were identified based on an adjusted p-value <0.05, following False Discovery Rate (FDR) correction. DEGs common to multiple stages are also annotated.
Analysis of the overall gene expression pattern clearly grouped the brains into male and female (Figure 3A). A separation between the sexes at the later stages is apparent from the PCA analysis where the two sexes are clearly separated at E14 (Figure 3B).
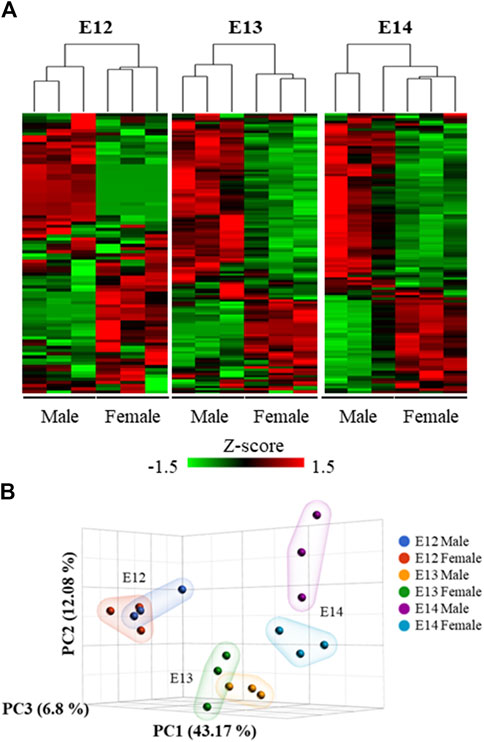
Figure 3. Hierarchical clustering and heat map of the differentially expressed genes. (A) The expression patterns at the three studied developmental stages were visualized using standardized gene expression values (z-scores). (B) 3D scatter plot illustrates the results of a Principal Component Analysis (PCA) performed on gene expression data from male and female embryos at different developmental stages.
Analysis of abundance and changes in Y chromosome genes was performed by calculating reads per million instead of fold change as these genes are not present in females (Supplementary Figure S2). Manual inspection of the Sry reads located at the correct position ensured that the low read counts were not merely noise. The highest expression of the Sry genes was observed for Sry2 (Supplementary Figure S2A). The expression levels of Sry2 were highest at E12 (Figure 4A). The brain specific expression of Sry genes was verified by qPCR, confirming that Sry expression was highest at E12 (Supplementary Figure S3). The remaining Sry genes showed much lower expression and no stage specific differences (Supplementary Figure S2B-I). It is interesting to note that Sry2 is located within an Kdm5d gene intron in opposite direction of Kdm5d (Supplementary Figure S2J). The other Y-chromosome genes that were overlapping between all three stages are shown in Figure 4. Of these, Eif2s3y showed a similar expression pattern as Sry2, with downregulation from E12 to E14 (Figure 4B). Ddx3y (Figure 4C), Kdm5d (Figure 4D), and Kdm6c (Figure 4E) showed statistically significant upregulation from E12 to E14. The Med14y gene was upregulated from E13 to E14 (Figure 4F), together with the X-chromosome linked Med14 gene.
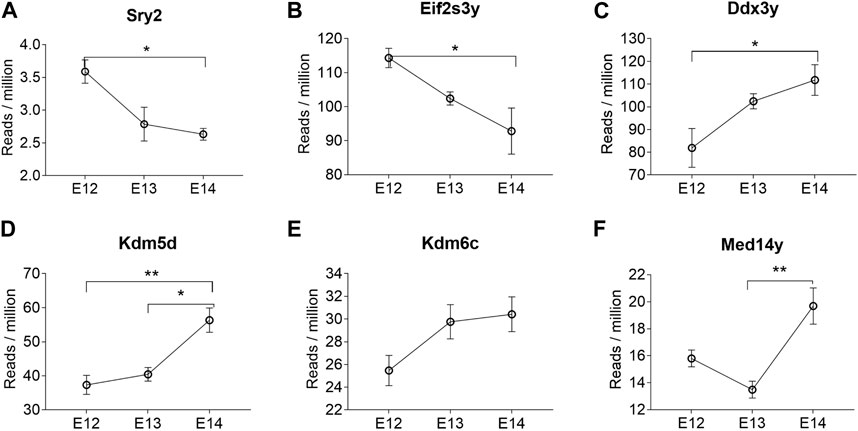
Figure 4. Y chromosome genes differentially expressed at all three developmental stages in male rat brains. The reads per million of DEGs at all three developmental stages are shown. Statistical analysis was performed using One way ANOVA followed by Tukey’s multiple comparison post-test (*p < 0.05 and **p < 0.01) (n = 3, mean ± SEM).
Four X-chromosome linked genes, Kdm6a, Eifs3x, Pbdc1 and Xist were present at all developmental stages with higher expression in females than in males (Figure 5). Of these, Xist was upregulated during embryogenic development in female brain and showed no expression in male brain (Figure 5A). The brain specific expression of Xist was verified by qPCR, confirming the upregulation from E12 to E14 (Supplementary Figure S2). Kdm6a was only downregulated in female brains at E14 (Figure 5B). Eif2s3x, and Pbdc1 were downregulated in both male and female brains (Figures 5C, E). Also present at all developmental stages was Enpp1, coding for an enzyme involved in ATP cleavage, located on chromosome 1. Enpp1 showed higher expression in female brains at E12 and E13 (Figure 5D). At E14 Enpp1 was downregulated in both sexes and no sex difference remained. While Kdm6a was upregulated at all three developmental stages, the Kdm5c gene showed female biased expression at E12 and E13. Both the X-linked Akap17b gene and the Sned1 gene (chromosome 9) showed female biased expression at E12 and E13. There were no common autosomal DEGs between all three developmental stages. However, several autosomal genes associated with proper neuronal functions had higher read counts in male brains at E12 (Supplementary Table S1). These include neuregulin 1 (Nrg1, normal development of nervous system), amyloid beta precursor protein (App, neural plasticity, and synapse formation), DnaJ Heat Shock Protein Family (Hsp40) Member A3 (Dnaja3, associated with Alzheimer disease), interleukin one receptor type 1 (Il1r1, neuron migration), and neuroligin 1 (Nlgn1, synapse function). In addition, ethanolamine kinase 2 (Etnk2) which is involved in choline metabolism also had higher read counts in the male rat brain at E12. In female brains, three genes stood out as highly expressed at E12. These were Somatostatin (Sst), Cytochrome P450 26b1 (Cyp26b1) and (Cyp2j4).
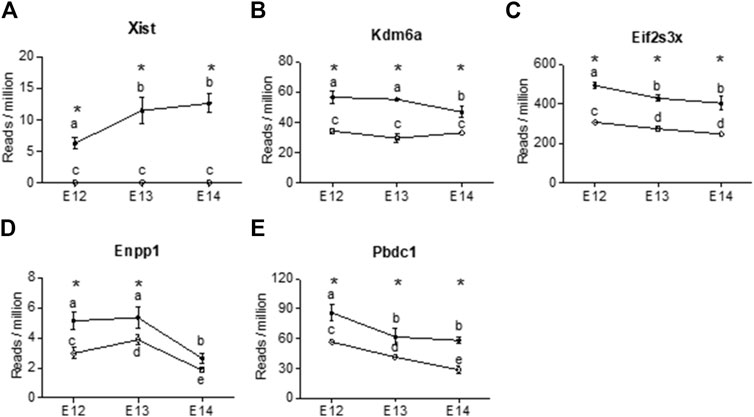
Figure 5. X chromosome genes were differentially expressed at all three developmental stages in female rat brain (closed circles). The reads per million of DEGs at all three developmental stages are shown For male brains (open circles) only three of the genes showed differences in reads per million between developmental stages. (A) Xist, (B) Kdm6a, (C) Eifs2s3x, (D) Enpp1, (E) Pbdc2. Statistical analysis was performed using One way ANOVA followed by Tukey’s multiple comparison post-test. Different letters were used to denote the level of significance between stages, and asterisk was used to denote statistical differences between sexes at p < 0.05 (n = 3, mean ± SEM).
3.3 Sex and stage specific effects on biological processes
Gene Ontology (GO) enrichment analysis showed consistent and evolving patterns across the three distinct embryonic stages (Figure 6). Biological processes (BP) crucial to development and regulation, such as developmental process, regulation of nervous system development, and neuron differentiation, exhibited sustained enrichment. However, the interplay of sex-specific influences within these enriched processes emerged when sex ratios were considered for the pathways. At the E12 stage, genes associated with the enriched processes displayed a higher expression in males, emphasizing a potential male gene dominance in these developmental pathways at this point. Interestingly, this dynamic shifted during the E13 stage, with a notable trend towards female gene regulation, suggesting a transitional period in the influence of different sexes. Finally, by the E14 stage, there was a reversion to a male influence, indicating the complexity and variability of sex-specific contributions to development across different stages.
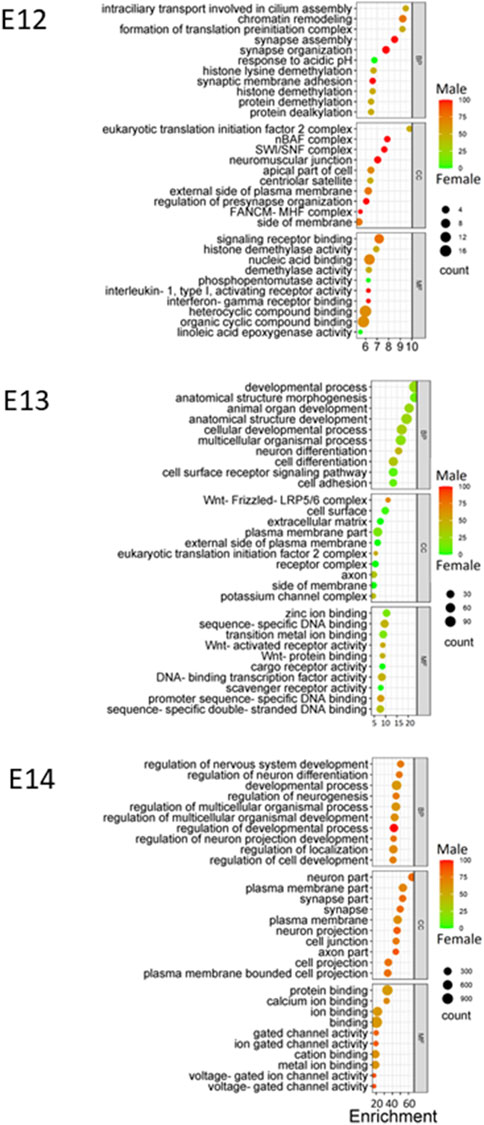
Figure 6. GO enrichment at E12 to E14. Bubble plots were constructed to visually represent Gene Ontology (GO) annotations for each embryonic stage utilizing DEGs identified at each stage, providing a comprehensive overview of the functional categories that are enriched during development. Enriched biological processes (BP), cellular components (CC), and molecular functions (MF) are presented using color gradient to represent the percentage of sex bias for given terms. Red color indicates male dominated enrichment while green color indicates female dominated enrichment.
Throughout the embryonic stages, certain cellular components (CC) were consistently enriched. The plasma membrane part, synapse part, and neuron part emerged as recurrently enriched components (Figure 6). Moreover, molecular functions (MF) that encompass binding and receptor activity, including zinc ion binding, sequence-specific DNA binding, and signaling receptor binding, were also consistently enriched across stages. Additionally, the variations in sex percentage involvement across processes, components, and functions provide an intriguing opportunity for further investigation into the interplay between genetics, sex-specific influences, and developmental outcomes.
3.4 Brain sex development leads to differentially enriched pathways
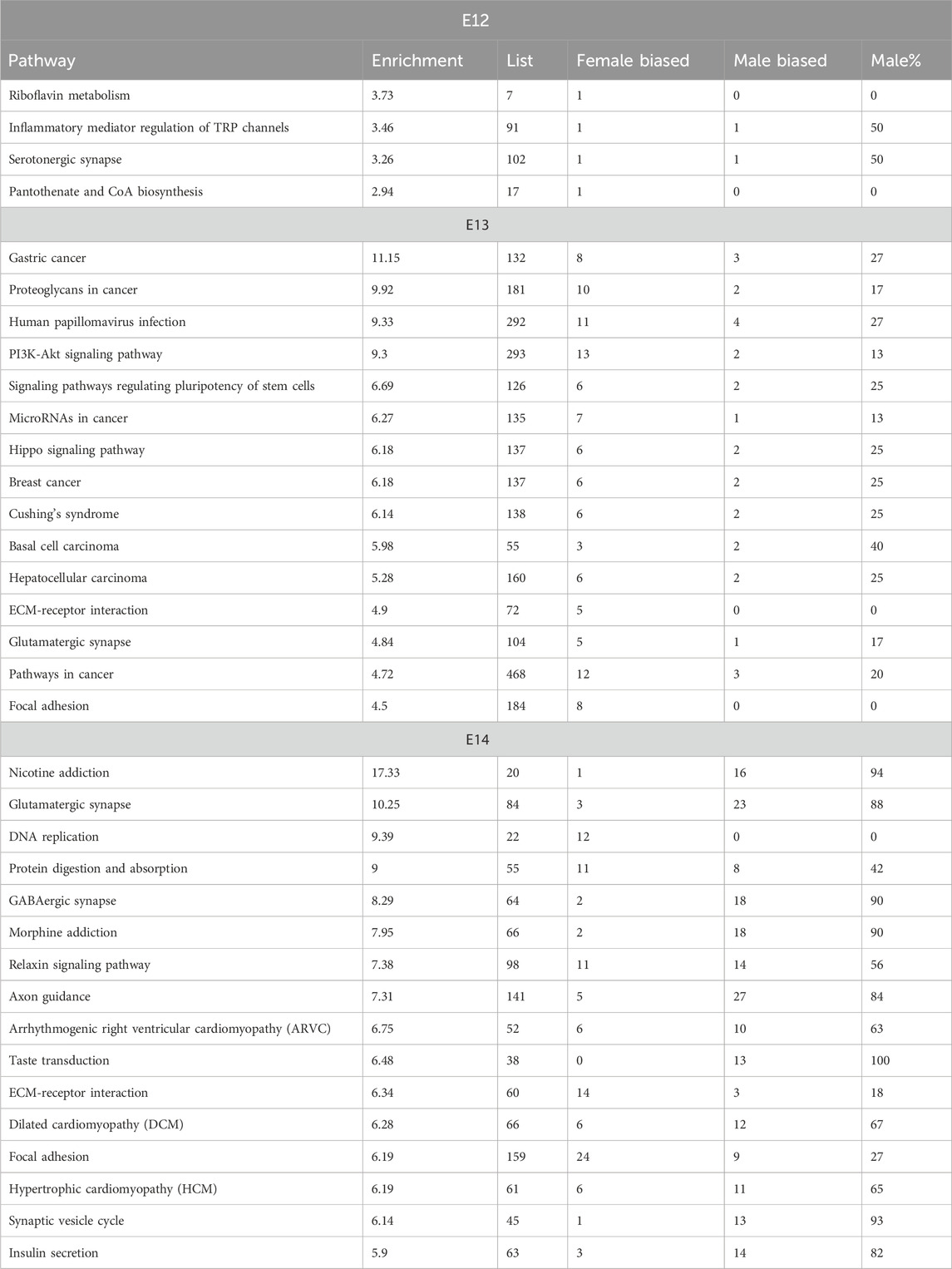
The analysis of enriched pathways in the KEGG database showed significant sex differences across the embryonic stages, which is in line with the dynamic nature of embryonic development (Table 3; Supplementary Figure S4). The p-values associated with these pathways provided a measure of statistical significance. Some pathways, particularly at later stages, displayed exceptionally low p-values, suggesting high statistical significance. In contrast, pathways at E12, such as riboflavin metabolism, exhibited relatively higher p-values, indicating less statistically significant associations. The analysis revealed that certain pathways are particularly associated with specific embryonic stages. Notably, embryonic stage E13 was characterized by pathways related to disease and viral infections such as gastric cancer, proteoglycans in cancer, and human papillomavirus infection. In contrast, at embryonic stage E14, neural and cellular processes dominated, with nicotine addiction and glutamatergic synapse showing noteworthy significance. The number of genes associated with these pathways varied significantly between stages and they increased in number as the development of the embryo progressed (Table 3). This was in line with the increasing number of DEGs for consecutive stages. Although the p-values and gene counts differed, several pathways, such as extracellular matrix (ECM)-receptor interaction and Focal adhesion, were enriched in both E13 and E14 indicating that these pathways play significant roles in both stages of embryonic development. At embryonic stage E14, a distinct cluster of pathways related to cardiac function and signaling was observed. Pathways such as Hypertrophic cardiomyopathy and Arrhythmogenic right ventricular cardiomyopathy indicated a critical period for the development of the cardiac system during this stage (Table 3).
3.5 Predicted stage specific functional protein networks during sex differentiation of the brain
Functional gene network of neural development genes showed significant associations/interactions among differentially expressed genes in the three developmental stages (Supplementary Figure S5). In each stage, three distinct clusters were identified. There were also interactions among protein products within or between neighboring clusters. Interacting proteins contribute to a shared function, an indication of biological interactions. In all developmental stages, Y chromosome genes showed a distinct network and interacted with X-linked genes. At embryonic stage E12, multiple interactions were observed among autosomal proteins involved in signal transduction and neural development and function. In E13, highly expressed genes involved in the development of the reproductive system, produced a distinct network. At embryonic stage E14, a large network of highly expressed genes involved in neural development and gene expression was observed. The reactome analysis furthermore indicates pathway similarities and differences between the three stages (Supplementary Figure S6). At E12 and E13 both developmental biology and signal transduction pathways are enriched, while at E14 developmental and extracellular matrix organization is augmented.
4 Discussion
In the present study our aim was to further the understanding of early key events in the process of sex differentiation of the brain. Earlier studies on zebra finch (Agate et al., 2003), mouse (Dewing et al., 2003), and chicken (Lee et al., 2009) suggest involvement of genetic signals in the development of male and female brain neuronal networks. By performing a temporal analysis of gene expression patterns in the early developing brain, around the time of gonadal sex differentiation we were able to determine temporal the regulation of differentially expressed genes. The gene expression pattern gave insight into the increased specialization of the brain, occurring from E12 to E14 and revealed sex biased enriched pathways related to nicotine and morphine addiction.
From experiments using the four-core mouse model it is apparent that Sry is needed for sex differentiation of both gonadal and somatic cells (Arnold, 2009). However, until now it has been unclear if Sry is expressed in neuronal cells during sex differentiation of the brain. RNA sequencing of rat embryonic brains revealed the presence of Sry2 with the highest expression already at the first studied embryonal stage, E12. The expression of Sry in rat gonads is detectable at E11 with the expression of Sry2 followed by the upregulation of other Sry1, Sry3C and Sry4a at E12 (Prokop et al., 2020). At later stages, Sry4a appears to be the main form expressed in rat testis (Prokop et al., 2020). Sequence analysis of the different Sry isoforms in rat shows that they differ in amino acid sequence with Sry2 having two point mutations (H4Q and R21H) in the first helix of the HMG box, leading to reduced nuclear translocation (Prokop et al., 2013; Prokop et al., 2016). This has led to questioning the ability of Sry2 to translocate to the nucleus and activate gene transcription. However, it has been shown that transfection of Chinese hamster ovarian (CHO) cell lines with the Sry2 transcript results in activation of downstream genes (Milsted et al., 2010; Prokop et al., 2016). In the earlier study Sry2 was compared to Sry1 and Sry3 for its ability to regulate the renin-angiotensin system (Milsted et al., 2010). While Sry1 and Sry3 were more potent than Sry2 it remains that all three isoforms could regulate the renin-angiotensin system. In the later study expression of the Sry1, Sry2 and Sry3 isoforms in CHO cells resulted in translocation to the nucleus all three isoforms, with Sry2 showing equal distribution between nucleus and cytosol (Prokop et al., 2016). Thus, while Sry2 appears to be less efficient at nuclear translocation and activation of downstream gene regulation it still is functional.
In mice testis, Sry first appears at 10.5 dpc, peaks at 11.5 dpc, and by 12.5 dpc the expression is undetectable (Larney et al., 2014). Sry has previously been reported to be expressed in adult mouse and human male brains (Mayer et al., 1998; Vawter et al., 2004; Dewing et al., 2006; Czech et al., 2012). In male rat gonads, Sry expression peaks at E12 (Prokop et al., 2020). However, at this stage the gonad is still underdeveloped (Val et al., 2003). Hence, it can be assumed that steroid secretion is still not initiated from the testis at E13. Thus, it can be concluded that the differential gene expression profiles observed for rat brains at E12 occur prior to hormonal actions on the brain.
In mice brain at E10.5 it was observed that Eif2s3y and Dby (Ddx3y) had the highest fold differential expression in males (Dewing et al., 2003). The remaining genes indicated to be differentially expressed were not located on the Y chromosome (Dewing et al., 2003). In the present study, we identified an increasing number of male biased genes from E12 to E14. There were six Y-chromosome genes showing differential expression at all developmental stages. Besides Sry2, we also identified both Eif2s3y and Ddx3y as early upregulated genes in the male brain. Of these two genes, Eif2s3y showed a similar expression profile as Sry2, with the highest expression at E12. Overexpression of Eif2s3y in mouse neurons has been shown to lead to autism-like behavior in male mice (Zhang et al., 2021). Several autosomal genes (Nrg1, App, Dnaja3, Il1r1, Arid1b, and Nlgn1b) that were upregulated at E12 are also associated with neuronal functions (Mucke et al., 1996; Stefansson et al., 2004; Tabarean et al., 2006; Martinez-Mir et al., 2013; Ka et al., 2016; Zhou et al., 2020). These results show that the early gene expression pattern in male mice is closely connected to neuronal outcomes in adult males.
In the present study we identified four Y chromosome genes, expressed at all three stages, with homologues on the X chromosome that escape X chromosome inactivation. These were ddx3y, eif2s3y, kdm5d, and kdm6c. In female brain the X chromosome homologues to three of these genes, eif2s3x, kdm5c, and kdm6a showed female biased expression, suggesting that the Y chromosome homologues could complement the expression of the X chromosome genes. However, it is worth noting that for all four Y chromosome genes it has been established that their functions are overlapping, but not identical, to their X chromosome counterpart (Mayfour et al., 2019; Shen et al., 2022; Dicke et al., 2023; Li et al., 2023; Liu et al., 2023; Rock et al., 2023). Thus, the sex differences in regulation of these genes may lead to sex specific functions. Ddx3y showed increasing expression in brain from E12 to E14. Ddx3y and Kdm6c, are both located in the AZFa region of the Y-chromosome. The AZFa, b, and c regions on the Y-chromosome have been identified to be required for normal spermatogenesis (Vog et al., 1996). In the AZFa region, it has been suggested that Ddx3y is the main gene responsible for infertility (Foresta et al., 2000). In male gonads, Ddx3y is expressed in spermatogonia before meiosis and Ddx3x is expressed in spermatids (Rauschendorf et al., 2014). Deletion of Ddx3y disrupts germ cell development and leads to infertility in males (Ramathal et al., 2015; Dicke et al., 2023). The increased expression of Ddx3y from E12 to E14 in rat brain indicate that this gene has key functions in male brain development.
Kdm5d and Kdm6c were both present at all three developmental stages with Kdm5d being upregulated from E12 to E14. The functions of both genes have been shown to differ from their X chromosome homologues (Meyfour et al., 2019; Rock et al., 2023). These genes are active demethylases, acting on Lys 27 of histone H3 (Shpargel et al., 2012; Walport et al., 2014). Kdm6c shows >88% similarity with Kdm6a and has recently been shown to be an active demethylase, demethylating H3K27, but with lower activity than KDM6A (Walport et al., 2014). Kdm6a regulation of H3K27me3 has been indicated to be an important regulator in embryonic stem cells (Welstead et al., 2012). It was also shown that Kdm6c complemented Kdm6a during cellular differentiation. Both genes have been demonstrated to be important for male neurogenesis (Pottmeier et al., 2020).
While few Y-chromosome genes overlapping in males between all three stages, the number of X-chromosome and autosomal genes increased over time in male brains. At E12, Foxo4 was the only X-chromosome gene upregulated in the male brain. Foxo4 is a member of the FOXO family that regulates genes involved in metabolism (Liu et al., 2020). In a study on triple knock-out mice, the loss of Foxo4, together with Foxo1 and Foxo3, was shown to reduce insulin responses in male, but not female mice (Penniman et al., 2019). In pathway enrichment analysis, Foxo4 and T-box transcription factor 20 (Tbx20) enriched carcinogenesis developmental process in E12, indicating a crucial role of the genes in the early heart development in male. While FOXO4 along with NKX2-5, and MEF2C bind the promoter of the MYOCD (Myocardin) gene, TBX20 and ISL1:LDB1 complex bind the anterior heart field enhancer of the MEF2C gene during cardiogenesis (Takeuchi et al., 2005; Creemers et al., 2006). The role of Foxo4 in male brain development is of interest for further studies. In female brains Xist showed the highest expression with an upregulation from 12 to E13. In addition to Xist, Kdm5c also showed increased expression at E13. The Y chromosome homologue, kdm5d, showed increased expression first at E14. It is worth noting that Kdm5c, but not Kdm5d, has been shown to be involved in the activation of Xist (Samanta et al., 2022).
Transcriptome analysis revealed that the progression from E12 to E14 resulted in increased specialization of the brain. Following male biased gene expression at E12 there was a shift toward female biased expression at E13 and again a shift back to male biased expression at E14 (Figure 6). A possible rational for this may be that the reprograming of the male brain at E12 requires more time to activate the new downstream genes compared to a continued differentiation of female neurons. Although it has not been explicitly demonstrated, research suggests that cell proliferation rates in the amygdala of rats may be higher in females compared to males (Krebs-Kraft et al., 2010). The GO enrichment analysis reveals dynamic patterns across embryonic stages, highlighting the role of sex in shaping developmental pathways. The consistent enrichment of certain cellular components and molecular functions underscores their importance in embryonic development. The enrichment of biological processes crucial to development and regulation, such as developmental process, regulation of nervous system development, and neuron differentiation, aligns with the understanding that these processes are fundamental to embryogenesis (Zhang et al., 2019).
Analysis of sex biased enriched pathways showed a strong male bias for nicotine and morphine addiction as well as GABAergic and glutamateric synapses at E14. The neuronal signalling is overlapping for these systems, and it has earlier been shown that they exhibit male biased expression in adults (D’Souza and Markou, 2013; Morgan and Ataras, 2022; Medrano et al., 2023). Nicotine exposure can lead to sex-dependent regulation of signalling pathways (Lee et al., 2021). This suggests that the sex biased effects on adults are established already during early development.
The Reactome pathway analysis indicated that the expression of Rac1 and Rock1 resulted in enriched semaphorin 4D. Semaphorin 4D axon guidance molecule influence cell migration and axon guidance which are essential for the proper formation and function of the nervous system (Jongbloets et al. 2014). These results show that axon guidance in nervous system development begin at E12. No neuronal development pathways were enriched during E13. We also observed enriched RHOH GTPase cycle, which regulates the proliferation, survival, migration, and engraftment of hematopoietic progenitor cells and specifically T cell development (Li et al., 2002; Yi et al., 2005). Rho GTPase activation is also associated with increased cancer cell invasiveness and associated with the development and progression of various cancers, including breast cancer, lung cancer, and skin cancer (Murray et al., 2014; Butera et al., 2020).
At E14, NCAM signaling for neurite out-growth and the L1CAM interaction axon guidance pathways were significantly overrepresented by DEGs. Three of the 4 DEGs in the NCAM signaling for neurite out-growth, Cacna1i, Cacna1d, and Cacna1h, were upregulated at E14. Similarly, four of the 5 DEGs, Dlg3, Dcx, Ank3, and L1cam, that enriched the L1CAM interactions were upregulated. The NCAM and L1CAMs are involved in the formation and maintenance of the nervous system. These neural adhesion molecules act as coreceptors for integrins, growth factors and axon guidance/axon pathfinding receptorsduring nervous system development (Schmid and Maness, 2008; Russell and Bashaw, 2018).
In the present study, we have mapped temporal expression patterns in male and female rat brains. The result shows that a limited number of Y-chromosome and X-chromosome genes are involved in all three studied embryonic stages, forming a cluster of interacting genes. In contrast, the expression of autosomal genes was highly stage specific with no gene involved in all three stages. This suggests that once the signaling for sex determination has been activated, there is a progression in functions that will be developed. In addition, the shift from male to female and back to male gene bias from E12 to E14, indicates that the shift from female to male brain requires more time than the direct development of the female brain. The present study reveals that one of the Sry genes, Sry2, peaked at E12 or earlier, together with Eif2s3y. This is followed by the upregulation of Ddx3y and Kdm5d. The results suggest that Sry2 is involved in the early sex differentiation cascade for the male brain.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1046089.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
BP: Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing–original draft, Writing–review and editing. SP: Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. YB: Formal Analysis, Investigation, Methodology, Writing–review and editing. P-EO: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was financed by the Swedish Research Council (Grant number 2019-04455) and Örebro University.
Acknowledgments
We are grateful to Dr. Carina Modig for proofreading the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1343800/full#supplementary-material
References
Agate, R. J., Grisham, W., Wade, J., Mann, S., Wingfield, J., Schanen, C., et al. (2003). The gender of the voice within: the neural origin of sex differences in the brain. Curr. Opin. Neurobiol. 13 (6), 759–764. doi:10.1016/j.conb.2003.10.005
Alganmi, N., and Abusamra, H. (2023). Evaluation of an optimized germline exomes pipeline using BWA-MEM2 and Dragen-GATK tools. PLOS ONE 18, e0288371. doi:10.1371/journal.pone.0288371
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11 (10), R106. doi:10.1186/gb-2010-11-10-r106
Arnold, A. P. (2009). The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm. Behav. 55 (5), 570–578. doi:10.1016/j.yhbeh.2009.03.011
Bordt, E. A., Ceasrine, A. M., and Bilbo, S. D. (2020). Microglia and sexual differentiation of the developing brain: a focus on ontogeny and intrinsic factors. Glia 68 (6), 1085–1099. doi:10.1002/glia.23753
Butera, A., Cassandri, M., Rugolo, F., Agostini, M., and Melino, G. (2020). The ZNF750–RAC1 axis as potential prognostic factor for breast cancer. Cell Death Discov. 6 (1), 135. doi:10.1038/s41420-020-00371-2
Creemers, E. E., Sutherland, L. B., McAnally, J., Richardson, J. A., and Olson, E. N. (2006). Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development 133 (21), 4245–4256. doi:10.1242/dev.02610
Czech, D. P., Lee, J., Sim, H., Parish, C. L., Vilain, E., and Harley, V. R. (2012). The human testis-determining factor SRY localizes in midbrain dopamine neurons and regulates multiple components of catecholamine synthesis and metabolism. J. Neurochem. 122 (2), 260–271. doi:10.1111/j.1471-4159.2012.07782.x
Dewing, P., Chiang, C. W., Sinchak, K., Sim, H., Fernagut, P. O., Kelly, S., et al. (2006). Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 16 (4), 415–420. doi:10.1016/j.cub.2006.01.017
Dewing, P., Shi, R. T., Horvath, S., and E. Vilain, E. (2003). Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res. Mol. Brain Res. 118 (1-2), 82–90. doi:10.1016/s0169-328x(03)00339-5
Dicke, A. K., Pilatz, A., Wyrwoll, M. J., Punab, M., Ruckert, C., Nagirinaja, L., et al. (2023). Ddx3y is likely the key spermatogenic factor in the AZFa region that contributes to human non-obstructive azoospermia. Commun. Biol. 6, 350. doi:10.1038/s42003-023-04714-4
D’Sousa, M. S., and Markou, A. (2013). The “Stop” and “go” of nicotine dependence: role of GABA and Glutamate. Cold Spring Harb. Perspect. Med. 3, a012146. doi:10.1101/cshperspect.a012146
Fabregat, A., Jupe, S., Matthews, L., Sidiropoulos, K., Gillespie, M., Garapati, P., et al. (2018). The reactome pathway knowledgebase. Nucleic Acids Res. 46 (D1), D649–D655. doi:10.1093/nar/gkx1132
Foresta, C., Ferlin, A., and Moro, E. (2000). Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum. Mol. Genet. 9 (8), 1161–1169. doi:10.1093/hmg/9.8.1161
Gahr, M., and Metzdorf, R. (1999). The sexually dimorphic expression of androgen receptors in the song nucleus hyperstriatalis ventrale pars caudale of the zebra finch develops independently of gonadal steroids. J. Neurosci. 19 (7), 2628–2636. doi:10.1523/JNEUROSCI.19-07-02628.1999
Hanley, N. N., Hagan, D. M., Clement-Jones, M., Ball, S. G., Srachan, T., Salas-Cortés, L., et al. (2000). Sry, Sox9, and Dax1 expression patterns during human sex determination and gonadal development. Mech. Dev. 91, 403–407. doi:10.1016/s0925-4773(99)00307-x
Ingalhalikar, M., Smith, A., Parker, D., Satterthwaite, T. D., Elliott, M. A., Ruparel, K., et al. (2014). Sex differences in the structural connectome of the human brain. PNAS 111 (2), 823–828. doi:10.1073/pnas.1316909110
Johansson, M. M., Lundin, E., Qian, X., Mirzazadeh, M., Halvardson, J., Darj, E., et al. (2016). Spatial sexual dimorphism of X and Y homolog gene expression in the human central nervous system during early male development. Biol. Sex. Diff. 7, 5. doi:10.1186/s13293-015-0056-4
Jongbloets, B. C., Jeroen, R., and Pasterkamp, R. J. (2014). Semaphorin signalling during development. Development 141, 3292–3297. doi:10.1242/dev.105544
Ka, M., Chopra, D. A., Dravid, M. S., and Kim, W. Y. (2016). Essential roles for ARID1B in dendritic arborization and spine morphology of developing pyramidal neurons. J. Neurosci. 36 (9), 2723–2742. doi:10.1523/JNEUROSCI.2321-15.2016
Koopman, P. (2005). Sex determination: a tale of two Sox genes. Trends Genet. 21 (7), 367–370. doi:10.1016/j.tig.2005.05.006
Krebs-Kraft, D. L., Hill, M. N., Hillard, C. J., and McCarthy, M. M. (2010). Sex difference in cell proliferation in developing rat amygdala mediated by endocannabinoids has implications for social behavior. PNAS 107 (47), 20535–20540. doi:10.1073/pnas.1005003107
Larney, C., Bailey, T. L., and P. Koopman, P. (2014). Switching on sex: transcriptional regulation of the testis-determining gene Sry. Development 141 (11), 2195–2205. doi:10.1242/dev.107052
Lee, A. M., Mansuri, M. S., Wilson, R. S., Lam, T. T., Nairn, A. C., and Picciotto, M. R. (2021). Sex differences in the ventral tegmental area and nucleus accumbens proteome at baseline and following nicotine exposure. Front. Mol. Neurosci. 14 (14), 657064. doi:10.3389/fnmol.2021.657064
Lee, S. I., Lee, W. K., Shin, J. H., Han, B. K., Moon, S., Cho, S., et al. (2009). Sexually dimorphic gene expression in the chick brain before gonadal differentiation. Poult. Sci. 88 (5), 1003–1015. doi:10.3382/ps.2008-00197
Li, X., Bu, X., Lu, B., Avraham, H., Flavell, R. A., and Lim, B. (2002). The hematopoiesis-specific GTP-binding protein RhoH is GTPase deficient and modulates activities of other Rho GTPases by an inhibitory function. Mol. Cell Biol. 22 (4), 1158–1171. doi:10.1128/mcb.22.4.1158-1171.2002
Li, Y., Wu, W., Xu, W., Wang, Y., Wan, S., Cjen, W., et al. (2023). Eif2s3y alleviated LPS-induced damage to mouse testis and maintained spermatogenesis by negatively regulating Adamts5. Theriogenology 211, 65–75. doi:10.1016/j.theriogenology.2023.08.003
Liu, W., Li, N., Zhang, M., Arisha, A. H., and Hua, J. (2023). The role of Eif2s3y in mouse spermatogenesis and ESC. Curr. Stem Cell Res. Ther. 17, 750–755. doi:10.2174/1574888X16666211102091513
Liu, W., Yong, L., and Luo, B. (2020). Current perspective on the regulation of FOXO4 and its role in disease progression. Cell Mol. Life Sci. 77, 651–663. doi:10.1007/s00018-019-03297-w
Martinez-Mir, A., González-Pérez, A., Gayán, J., Antúnez, C., Marín, J., Boada, J. M., et al. (2013). Genetic study of neurexin and neuroligin genes in Alzheimer's disease. J. Alzheimers Dis. 35 (2), 403–412. doi:10.3233/JAD-122257
Mayer, A., Lahr, G., Swaab, D. F., Pilgrim, C., and Reisert, I. (1998). The Y-chromosomal genes SRY and ZFY are transcribed in adult human brain. Neurogenetics 1 (4), 281–288. doi:10.1007/s100480050042
Mayer, A., Mosler, G., Just, W., Pilgrim, C., and Reisert, I. (2000). Developmental profile of Sry transcripts in mouse brain. Neurogenetics 3 (1), 25–30. doi:10.1007/s100480000093
Mayfour, A., Pahlavan, S., Ansari, H., Baharvand, H., and Salekdeh, G. H. (2019). Down-regulation of a male-specific H3K4 demethylase, kdm5d, impairs cardiomyocyte differentiation. J. Proteome 18, 4277–4282. doi:10.1021/acs.jproteome.9b00395
Medrano, M. C., Darlot, F., Cador, M., and Caille, S. (2023). Poor inhibitory control predicts sex-specific vulnerability to nicotine rewarding properties in mice. Psycopharmacology 240, 1973–1986. doi:10.1007/s00213-023-06418-3
Milsted, A., Underwood, A. C., Dunmire, J., DelPuerto, H. L., Martins, A. S., Ely, D. L., et al. (2010). Regulation of multiple renin-angiotensin system genes by Sry. J. Hypertens. 28 (19), 59–64. doi:10.1097/HJH.0b013e328332b88d
Morgan, M. M., and Ataras, K. (2022). Sex differences in the impact of pain, morphine administration and morphine withdrawal on quality of life in rats. Pharmacol. Biochem. Behav. 219, 173451. doi:10.1016/j.pbb.2022.173451
Mucke, L., Abraham, C. R., and Masliah, E. (1996). Neurotrophic and neuroprotective effects of hAPP in transgenic mice. Ann. N. Y. Acad. Sci. 17 (777), 82–88. doi:10.1111/j.1749-6632.1996.tb34405.x
Murray, D. W., Didier, S., Chan, A., Paulino, V., Van Aelst, L., Ruggieri, R., et al. (2014). Guanine nucleotide exchange factor Dock7 mediates HGF-induced glioblastoma cell invasion via Rac activation. Br. J. Cancer 110 (5), 1307–1315. doi:10.1038/bjc.2014.39
Penniman, C. M., Suarez Beltran, P. A., Bhardwaj, G., Junck, T. L., Jena, J., Poro, K., et al. (2019). Loss of FoxOs in muscle reveals sex-based differences in insulin sensitivity but mitigates diet-induced obesity. Mol. Metab. 30, 203–220. doi:10.1016/j.molmet.2019.10.001
Phoenix, C. H., Goy, R. W., Gerall, A. A., and Young, W. C. (1959). Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female Guinea pig. Endocrinol 65, 369–382. doi:10.1210/endo-65-3-369
Pottmeier, P., Doszyn, O., Peuckert, C., and Jazin, E. (2020). Increased expression of Y-encoded demethylases during differentiation of human male neural stem cells. Stem Cells Dev. 29 (23), 1497–1509. doi:10.1089/scd.2020.0138
Prokop, J. W., Chhetri, S. B., van Veen, J. E., Chen, X., Underwood, A. C., Uhl, K., et al. (2020). Transcriptional analysis of the multiple Sry genes and developmental program at the onset of testis differentiation in the rat. Biol. Sex. Differ. 11 (1), 28–8. doi:10.1186/s13293-020-00305-8
Prokop, J. W., Tsaih, S.-W., Faber, A. B., Boehme, S., Underwood, A. C., Troyer, S., et al. (2016). The phenotypic impact of the male-specific region of chromosome-Y in inbred mating: the role of genetic variants and gene duplications in multiple inbred rat strains. Biol. Sex. Diff. 7, 10. doi:10.1186/s13293-016-0064-z
Prokop, J. W., Underwood, A. C., Turner, M. E., Miller, N., Pietrzak, D., Scott, S., et al. (2013). Analysis of Sry duplications on the Rattus norvegicus Y-chromosome. BMC Genomics 14, 792. doi:10.1186/1471-2164-14-792
Ramathal, C., Angulo, B., Sukhwani, M., Cui, J., Durruthy-Durruthy, J., Fang, F., et al. (2015). DDX3Y gene rescue of a Y chromosome AZFa deletion restores germ cell formation and transcriptional programs. Sci. Rep. 5, 15041. doi:10.1038/srep15041
Rauschendorf, M. A., Zimmer, J., Ohnmacht, C., and Vogt, P. H. (2014). DDX3X, the X homologue of AZFa gene DDX3Y, expresses a complex pattern of transcript variants only in the male germ line. Mol. Hum. Reprod. 20 (12), 1208–1222. doi:10.1093/molehr/gau081
Reinius, B., and Jazin, E. (2009). Prenatal sex differences in the human brain. Mol. Psychiatry 14, 988–989. doi:10.1038/mp.2009.79
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Gad Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi:10.1038/nbt.1754
Rock, K. D., Folts, L. M., Zierden, H. C., Marx-Rattner, R., Leu, N. A., Nugent, B. M., et al. (2023). Developmental transcriptomic patterns can be altered by transgenic overexpression of Uty. Sci. Rep. 13, 21082. doi:10.1038/s41598-023-47977-x
Russell, S. A., and Bashaw, G. J. (2018). Axon guidance pathways and the control of gene expression. Dev. Dyn. 247, 571–580. doi:10.1002/dvdy.24609
Samanta, M. K., Gayen, S., Harris, C., Maclary, E., Murata-Nakamura, Y., Malcore, R. M., et al. (2022). Activation of Xist by an evolutionarily conserved function of kdm5c demethylase. Nat. Commun. 13, 2602. doi:10.1038/s41467-022-30352-1
Schmid, R. S., and Maness, P. F. (2008). L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 18, 245–250. doi:10.1016/j.conb.2008.07.015
Shen, H., Yanas, A., Owens, M. C., Zhang, C., Fritsch, C., Fare, C. M., et al. (2022). Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol. Cell 82, 2588–2603.e9. doi:10.1016/j.molcel.2022.04.022
Shpargel, K. B., Sengoku, T., Yokohama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLOS Genet. 8 (9), e1002964. doi:10.1371/journal.pgen.1002964
Sim, H., Argentaro, A., and Harley, V. R. (2008). Boys, girls and shuttling of SRY and SOX9. Trends Endocrinol. Metab. 19 (6), 213–222. doi:10.1016/j.tem.2008.04.002
Stefansson, H., Steinthorsdottir, V., Thorgeirsson, T. E., Gulcher, J. R., and Stefansson, K. (2004). Neuregulin 1 and schizophrenia. Ann. Med. 36 (1), 62–71. doi:10.1080/07853890310017585
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613. doi:10.1093/nar/gky1131
Tabarean, I. V., Korn, H., and Barfai, T. (2006). Interleukin-1beta induces hyperpolarization and modulates synaptic inhibition in preoptic and anterior hypothalamic neurons. Neuroscience 141, 1685–1695. doi:10.1016/j.neuroscience.2006.05.007
Takeuchi, J. K., Mileikovskaia, M., Koshiba-Takeuchi, K., Heidt, A. B., Mori, A. D., Arruda, E. P., et al. (2005). Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132 (10), 2463–2474. doi:10.1242/dev.01827
Turner, M. E., Martin, C., Martins, A. S., Dunmire, J., Farkas, J., Ely, D. L., et al. (2007). Genomic and expression analysis of multiple Sry loci from a single Rattus norvegicus Y chromosome. BMC Genet. 8 (11), 11. doi:10.1186/1471-2156-8-11
Val, P., Lefrançois-Martinez, A. M., Veyssière, G., and Martinez, A. (2003). SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl. Recept 1 (1), 8. doi:10.1186/1478-1336-1-8
Vawter, M. P., Evans, S., Choudary, P., Tomita, H., Meador-Woodruff, J., Molnar, M., et al. (2004). Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology 29 (2), 373–384. doi:10.1038/sj.npp.1300337
Vog, P., Edelmann, A., Kirsch, S., Henegariu, O., Hirschmann, P., Kiesewetter, F., et al. (1996). Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum. Mol. Genet. 5 (7), 933–943. doi:10.1093/hmg/5.7.933
Wade, J., and Arnold, A. P. (1996). Functional testicular tissue does not masculinize development of the zebra finch song system. PNAS 93 (11), 5264–5268. doi:10.1073/pnas.93.11.5264
Walport, L. J., Hopkinson, R. J., Vollmar, M., Madden, S. K., Gileadi, C., Oppermann, U., et al. (2014). Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J. Biol. Chem. 289 (26), 18302–18313. doi:10.1074/jbc.M114.555052
Welstead, G. G., Creyghton, M.-P., Bilodeau, S., Cheng, A. W., Markoulaki, S., Young, R. A., et al. (2012). X-lin ked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. PNAS 109 (32), 13004–13009. doi:10.1073/pnas.1210787109
Wu, X., Heffelfinger, C., Zhao, H., and Dellaporta, S. L. (2019). Benchmarking variant identification tools for plant diversity discovery. BMC Genomics 20, 701. doi:10.1186/s12864-019-6057-7
Xu, H., Wang, F., Liu, Y., Yu, Y., Gelernter, J., and Zhang, H. (2014). Sex-biased methylome and transcriptome in human prefrontal cortex. Hum. Mol. Genet. 23 (5), 1260–1270. doi:10.1093/hmg/ddt516
Yi, G., Jasti, A. C., Jansen, M., and Siefring, J. E. (2005). RhoH, a hematopoietic-specific Rho GTPase, regulates proliferation, survival, migration, and engraftment of hematopoietic progenitor cells. Blood 105 (4), 1467–1475. doi:10.1182/blood-2004-04-1604
Zhang, M., Zhou, Y., Jiang, Y., Lu, Z., Xiao, X., Ning, J., et al. (2021). Profiling of sexually dimorphic genes in neural cells to identify Eif2s3y, whose overexpression causes autism-like behaviors in male mice. Front. Cell Dev. Biol. 9, 669798. doi:10.3389/fcell.2021.669798
Zhang, X., Gan, Y., Zou, G., Guan, J., and Zhou, S. (2019). Genome-wide analysis of epigenetic dynamics across human developmental stages and tissues. BMC Genomics 20 (Suppl. 2), 221. doi:10.1186/s12864-019-5472-0
Keywords: neuronal, sex chromosome, RNA sequencing, sexual dimorphism, differentiation
Citation: Paylar B, Pramanik S, Bezabhe YH and Olsson P-E (2024) Temporal sex specific brain gene expression pattern during early rat embryonic development. Front. Cell Dev. Biol. 12:1343800. doi: 10.3389/fcell.2024.1343800
Received: 24 November 2023; Accepted: 05 June 2024;
Published: 19 June 2024.
Edited by:
Dagmar Wilhelm, The University of Melbourne, AustraliaReviewed by:
Rasoul Godini, Monash University, AustraliaFrancis Poulat, Université de Montpellier, France
Joohyung Lee, Monash University, Australia
Copyright © 2024 Paylar, Pramanik, Bezabhe and Olsson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Per-Erik Olsson, cGVyLWVyaWsub2xzc29uQG9ydS5zZQ==
†These authors share first authorship
‡Present addresses: Subrata Pramanik, Jyoti and Bhupat Mehta School of Health Sciences and Technology, Indian Institute of Technology Guwahati, Guwahati, Assam 781039, India; Centre for Nanotechnology, Indian Institute of Technology Guwahati, Guwahati 781039, Assam, India
 Berkay Paylar
Berkay Paylar Subrata Pramanik†‡
Subrata Pramanik†‡ Yared H. Bezabhe
Yared H. Bezabhe Per-Erik Olsson
Per-Erik Olsson