
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 26 January 2024
Sec. Morphogenesis and Patterning
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1336308
The mechanism underlying the differentiation of the dorsal midbrain into two morphologically and functionally distinct compartments, the inferior colliculus (IC) and superior colliculus (SC), which process auditory and visual information, respectively, remains largely unexplored. By using null and conditional alleles, we uncover the roles of a homeodomain transcription factor Dbx1 in the regulation of IC and SC differentiation. We show that Dbx1 regulates GABAergic neuron development in the dorsal midbrain. In the absence of Dbx1 function, the dorsal-most m1-m2 progenitor domains in the midbrain fail to activate GABAergic neuron-specific gene expression and instead switch to a glutamatergic phenotype. These results identify Dbx1 as a dorsal midbrain-specific GABAergic determinant that regulates the selector genes, Helt, Gata2, and Tal2. Furthermore, we demonstrate that maturation of the dorsal midbrain into the IC and SC is dependent on Dbx1. Null mutation of Dbx1 impairs the identity and fate of IC and SC neurons. Surprisingly, Dbx1 is required for preventing IC into SC fate switch and thus Dbx1-deficient IC neurons undergo acquisition of SC identity. Conditional inactivation of Dbx1 at late developmental phase leads to alteration in the identity and fate of the IC, but not the SC. These results suggest that SC differentiation is dependent on the early function of Dbx1, and that the IC requires the prolonged action for its normal formation. Furthermore, we uncover that Tcf7l2 acts downstream of Dbx1 selectively to promote IC differentiation. Altogether, our study identifies a molecular mechanism underlying spatial and temporal control of dorsal midbrain development.
Early growth and patterning of the dorsal midbrain are tightly linked to the key signaling molecules of the isthmic organizer (Shamim et al., 1999; Joyner et al., 2000; Wurst and Bally-Cuif, 2001; Chi et al., 2003). FGF signaling functions in a dose- and time-dependent manner to control growth, specification and survival of the dorsal midbrain (Chi et al., 2003; Basson et al., 2008; Dee et al., 2016). Engrailed, Pax and Meis2 play differential roles in forming the dorsal midbrain (Urbánek et al., 1994; Matsunaga et al., 2001; Sgaier et al., 2007; Agoston and Schulte, 2009), and Otx2 contributes to the control of multiple aspects of dorsal midbrain development (Broccoli et al., 1999; Puelles et al., 2003; Di Giovannantonio et al., 2014). Although previous works have greatly broadened our understanding of the early phase of dorsal midbrain development, the molecular basis underlying maturation of the dorsal midbrain at the late embryonic and postnatal stages remains unexplored.
Following the induction of the midbrain during early embryogenesis, the territories of midbrain progenitors are specified into the roof plate, alar plate, basal plate and floor plate along the dorso-ventral axis by Shh and Bmp/Wnt signals (Zervas et al., 2005; Puelles, 2007; Dessaud et al., 2008; Le Dréau and Martí, 2012). These midbrain progenitors are further assigned into seven subdivisions (m1-m7) with specific combinations of gene expression codes (Nakatani et al., 2007; Kala et al., 2009; Lahti et al., 2013). The most dorsal midbrain neuronal populations derived from m1-m2 domains are clustered in the superior colliculus (SC) and inferior colliculus (IC) along the anteroposterior axis. The SC forms its stereotypical laminated structure and differentiates into a number of layers that can be grouped into the superficial layers receiving inputs related to vision and the deeper layers responding to nonvisual multiple modalities (McLaughlin and O’Leary, 2005). The IC located caudal to the SC functions as an acoustic integration center, and develops into the central nucleus (core region) receiving auditory inputs and the lateral and dorsal cortex (shell region) receiving nonauditory projections (Oliver and Morest, 1984; Cant and Benson, 2006). Despite the functional significance of the IC and SC, how the dorsal midbrain differentiates into these two distinct compartments is largely unknown.
Distinct subtypes of GABAergic neurons are formed at different locations and times during midbrain development. GABAergic neurogenesis can be detected first in the ventral midbrain, where GABAergic neuronal markers are expressed at E10.5∼E11.5. By contrast, in the dorsal midbrain, GABAergic neurogenesis begins 1 day later and persists longer (Achim et al., 2012). Basic HLH factors Helt (also known as Megane or Heslike), Ascl1, Tal2 and zinc finger protein Gata2 have been shown to be indispensable for specifying GABAergic identity in the midbrain (Guimera et al., 2006; Nakatani et al., 2007; Kala et al., 2009; Peltopuro et al., 2010; Achim et al., 2013; Wende et al., 2015). These observations suggest that GABAergic neuronal differentiation requires different dorsoventral molecular mechanisms. However, region-specific determinants or selector genes that govern genetic program remain to be determined.
Recently, we showed that a homeodomain transcription factor Dbx1 (Developing brain homeobox 1) functions as a key regulator that controls genetic programs for postnatal survival of the IC (Tran et al., 2023). Loss of Dbx1 causes apoptotic cell death by upregulating c-Jun and pro-apoptotic BH3 only factors and leads to specific loss of the IC. Furthermore, we found that the function of Dbx1 as a survival factor is mediated by Tcf7l2 and Ap-2δ transcription factors. In the present study, we assess the role of Dbx1 in regulating developmental acquisition of dorsal midbrain neuron identity. We show that Dbx1 functions as a dorsal midbrain-specific GABAergic determinant by regulating the selector genes, Helt, Gata2, and Tal2. Furthermore, we uncover that Dbx1 is required to partition the dorsal midbrain into the IC and SC.
All animal procedures were carried out in accordance with the guidelines and protocols approved by the Kyung Hee University Institutional Animal Care and Use Committee. The generation of Dbx1loxp/loxp (JAX028223), Tcf7l2loxp/loxp (JAX031436), Ascl1−/+ (JAX012881), and EIIa-Cre mice (JAX003314) obtained from The Jackson Laboratory, Bar Harbor, ME.was described previously (Lakso et al., 1996; Leung et al., 2007; Angus-Hill et al., 2011; Sokolowski et al., 2015).
Embryonic brains were fixed in 4% formaldehyde in PBS, pH7.4 for 2–3 h at 4°C, immersed in 30% sucrose, cryosectioned at 25 µm by Leica CM3050 Cryostat, and placed on glass slides. Sections were blocked with 2% normal sheep serum (Sigma) in PBS for 1 h, incubated in primary antibody made up in 1% sheep serum, 0.4% Triton X-100 in PBS overnight. The following primary antibodies were used: anti-phospho-Histone H3 (Upstate Biotechnology, 06–570; 1:1000), and anti-BrdU (Sigma, B8434; 1:1000). Following incubation with primary antibody, sections were washed three times with PBS, pH7.4 and incubated with species-specific secondary antibodies conjugated to Alexa Fluor (Molecular Probes, A11034, A11003, A11010, A11029; 1:200). After three washes in PBS, sections were mounted on slides with Fluoromount G (Electron Microscopy Sciences).
BrdU labeling was performed as described previously (Miller and Nowakowski, 1988), with some modifications. Timed pregnant female mice were injected subcutaneously with 50 mg/kg of 5-bromo-2′-deoxyuridine (Sigma). For pulse labeling experiments, mice were sacrificed after 2–24 h. For birth dating experiments, mice were sacrificed at E18.5. Cryostat sections were permeabilized in 0.4% Triton X-100, treated with 50% formamide/2x SSC for 2 h at 65°C, 2x SSC for 10 min, 2 M HCl for 1 h at 37°C with, washed twice for 10-min in 0.1 M sodium borate, and stained with anti-BrdU antibody.
For quantitation of proliferation and BrdU birth dating, images of stained sections were captured using a Leica DMI6000B digital camera (Leica Microsystems), and the number of cells was scored from captured images. Counting grids were set at 200 μm × 200 μm in the SC, and at 100 μm × 100 μm in the IC. In most cases, 20–30 counting sites were evaluated in one section for the region of interest. All data were obtained from at least three samples of each genotype, and tested for significance using a two sample Student's t-test with unequal variances.
Whole-mount and section in situ hybridization were performed using digoxigenin-UTP-labeled riboprobes essentially as described previously (Tran et al., 2020). Embryonic brains were collected from timed pregnant females, and fixed in 4% paraformaldehyde at 4°C for overnight. Brain tissues were embedded in Tissue-Tek O.C.T. compound, and 25 μm frozen sections were collected. Sections were rehydrated in PBS, and immersed in pre-hybridization buffer (1% SDS, 5x SSC, 50% formamide) for 1 h at 65°C. For riboprobes, template DNAs for in vitro transcription by T7 RNA polymerase were generated by using PCR primers, of which sequences were obtained from the Allen Brain Atlas or designed against unique regions of transcripts to avoid cross-reactivity (Supplementary Table S1). The promoter sequence for T7 RNA polymerase (5′-TGT AAT ACG ACT CAC TAT AGG GC-3′) was placed at the 5′ end of reverse primers. DIG-labeled RNA probe was then added in hybridization buffer (1% SDS, 5x SSC, 50% formamide, 200 mg/mL yeast tRNA, 5% heparin). Incubation overnight at 65°C followed washing for 30 min in 1x SSC/50% formamide at 65°C. Slides were then treated with RNase A (20 mg/mL) in a buffer containing 10 mM Tris pH 7.5, 500 mM NaCl, 1 mM EDTA at 37°C for 30 min, washed in 2x SSC for 20 min at 65°C, and then twice for 20 min each in 0.2x SSC. After incubation overnight with anti-DIG AP antibody (1:2500, Roche), slides were washed in MABT and equilibrated in NTM for 10 min. Color detection was performed using BM purple (Roche). At least three independent experiments were performed for any given riboprobes, which showed highly reproducible gene expression data.
All mutant phenotypes reported in Ascl1-/-, Dbx1del, Dbx1cko, and Tcf7l2del embryos were completely penetrant. The number of independent values in each experiment were as follows: Figure 1: n = 3 for E10.5, E11,5 and E12.5 for ISH coronal and sagittal sections. Figure 2: n = 3 for E12.5 ISH. Figure 3: n = 3 for E12.5 and E16.5 coronal ISH. Figure 4: n = 3 for E12.5 ISH and IHC with PH3. Three independent biological replicates for BrdU labelling at E12.5 with 2 and 24 h of incubation. Three sections of three embryos from each genotype were selected to count pH3+ and BrdU+. Figure 5: n = 3 for E15.5 and E18.5 ISH. Figure 6: n = 3 for E18.5 ISH. Figure 7: three independent biological replicates for BrdU birth-dating tracing from E11.5, E12.5, E13.5 and E14.5. For counting BrdU+ cells, 10–20 counting site (counting grid for SC: 200 μm × 200 μm; for IC: 100 μm × 100 μm) were evaluated in one section. The value was calculated from five sections of six embryos for each genotype. Figure 8: n = 3 for E18.5 ISH. Figure 9: n = 3 for E18.5 ISH.
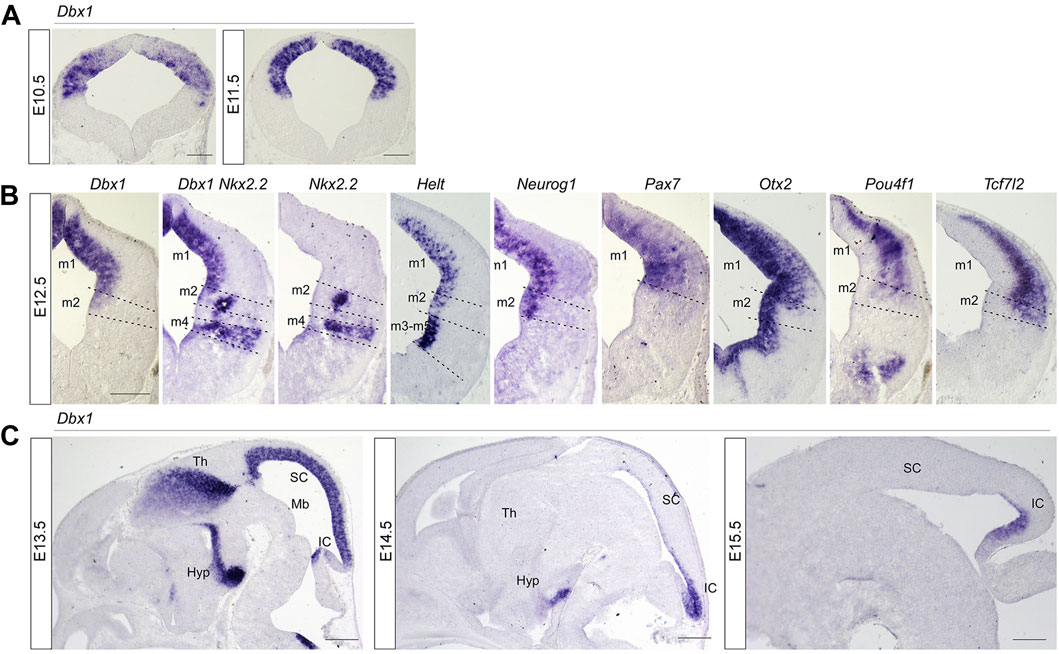
FIGURE 1. Spatial and temporal pattern of Dbx1 expression in embryonic mouse midbrain compared with expression of neural subtype markers. (A) Representative transverse midbrain sections of wild-type mouse embryos hybridized with Dbx1 riboprobe at E10.5 and E11.5. (B) In situ hybridization (ISH) with Dbx1, Nkx2.2, Helt, Neurog1, Pax7, Otx2, Pou4f1, and Tcf7l2 probes on transverse sections of E12.5 wild-type embryos. (C) ISH with Dbx1 probe on sagittal sections of wild-type embryos at E13.5 –E15.5. m1-m5, dorsoventral midbrain domains 1-5. Hyp, hypothalamus; IC inferior colliculus; Mb, midbrain; SC, superior colliculus, Th, thalamus. Scale bars, A = 100 μm, B = 200 μm, C = 400 μm.
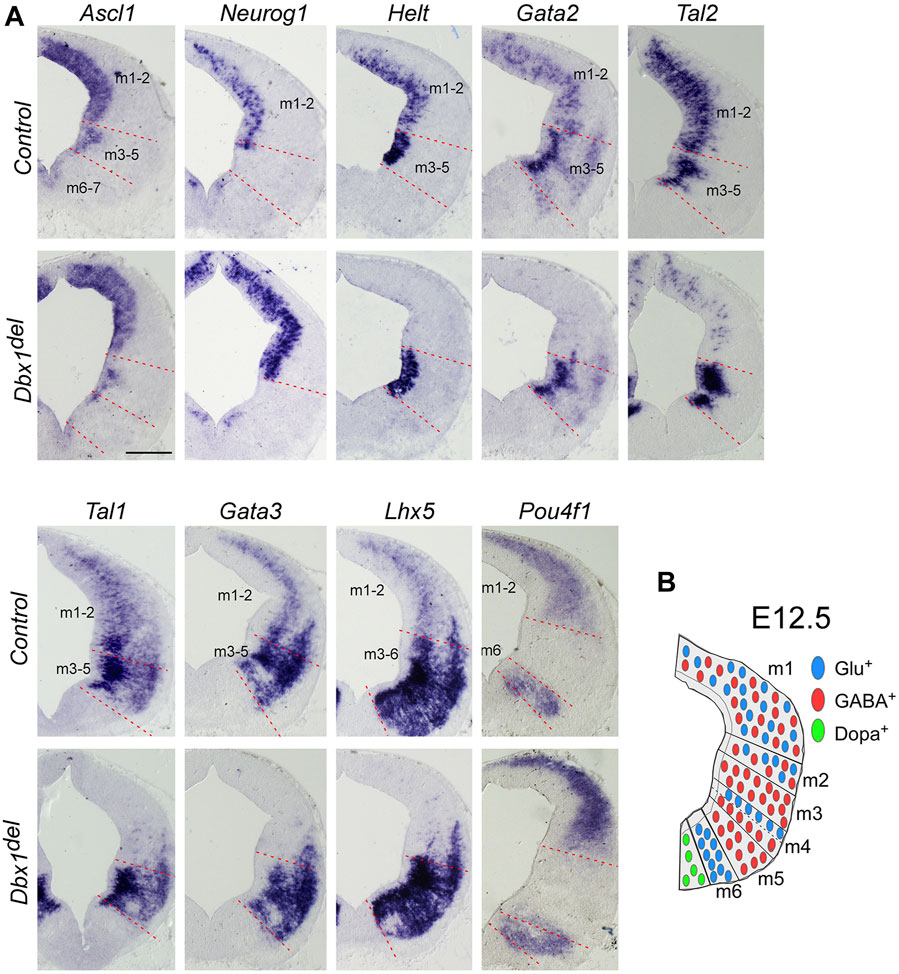
FIGURE 2. Dbx1 is required for selection of GABAergic over glutamatergic neuronal fate in the m1 and m2 dorsoventral midbrain domains (A) ISH on midbrain transverse sections of control and Dbx1del embryos at E12.5. Ascl1, Neurog1, and Helt were expressed in the ventricular progenitor region, while Gata2 and Tal2 expression were observed in the ventricular and intermediate zones. Tal1, Gata3, Lhx5, and Pou4f1 were detectable in the intermediate and postmitotic marginal zones. Loss of Dbx1 resulted in severe downregulation of GABAergic lineage genes Helt, Gata2, Tal2, Tal1, Gata3, and Lhx5, while glutamatergic lineage genes Neurog1 and Pou4f1 were upregulated. (B) Schematic view of E12.5 midbrain depicting domains m1– m7 and the distribution of glutamatergic (blue circles), GABAergic (red circles), and dopaminergic (green circles) neurons. Scale bar, A = 200 μm.
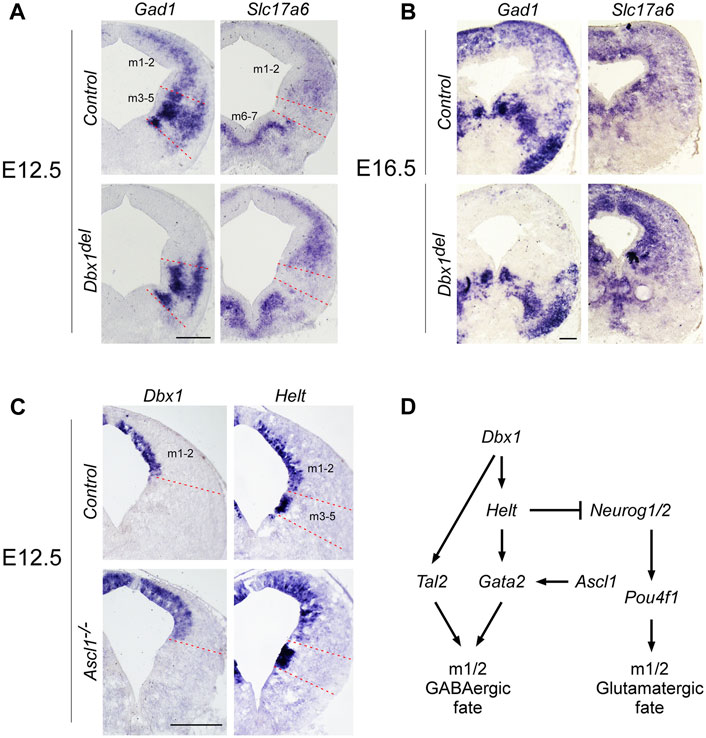
FIGURE 3. Distinct regulatory pathways control GABAergic neuronal fate determination in the m1 and m2 domains (A,B) ISH on midbrain transverse sections of control and Dbx1del embryos at E12.5 (A) and E16.5 (B). In control embryos, Gad1 and Slc17a6 expression were commonly detectable in m1 and m2 domains of the midbrain. Inactivation of Dbx1 downregulated Gad1 staining selectively in m1-m2 domains, but not in m3-m5 domains. By contrast, Slc17a6 expression was upregulated in m1-m2 domains of Dbx1del mutants. (C) Representative transverse midbrain sections of wild-type and Ascl1−/− mouse embryos hybridized with Dbx1 and Helt riboprobes. (D) Schematic showing genetic cascades that control GABAergic neuronal specification in the dorsal midbrain. Scale bars, A, C = 200 μm, B = 400 μm.
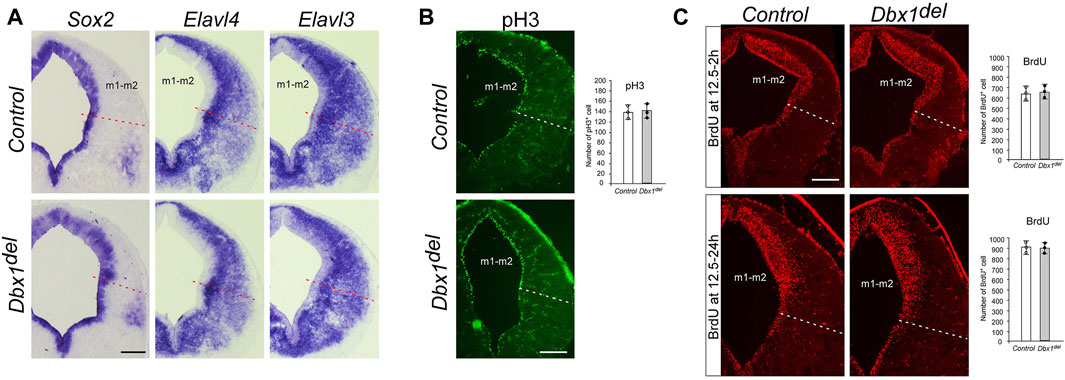
FIGURE 4. Unaltered progenitor cell proliferation and neurogenesis in Dbx1del midbrain (A) ISH of Sox2 and Elavl3/4 (HuC/D) on transverse sections of E12.5 embryos demonstrates no obvious difference between control and Dbx1del midbrains. (B) Immunohistochemistry (IHC) staining for phospho-histone H3 (pH3) on transverse sections of E12.5 midbrain. The number of pH3-positive cells counted from the ventricular zone of the m1-m2 domain (three sections from three embryos for each genotype) (C) Incorporation of BrdU in E12.5 control and Dbx1del midbrain during a 2–24 h labeling pulse detected by anti-BrdU IHC (three sections from three embryos for each genotype). Scale bars, 200 μm.
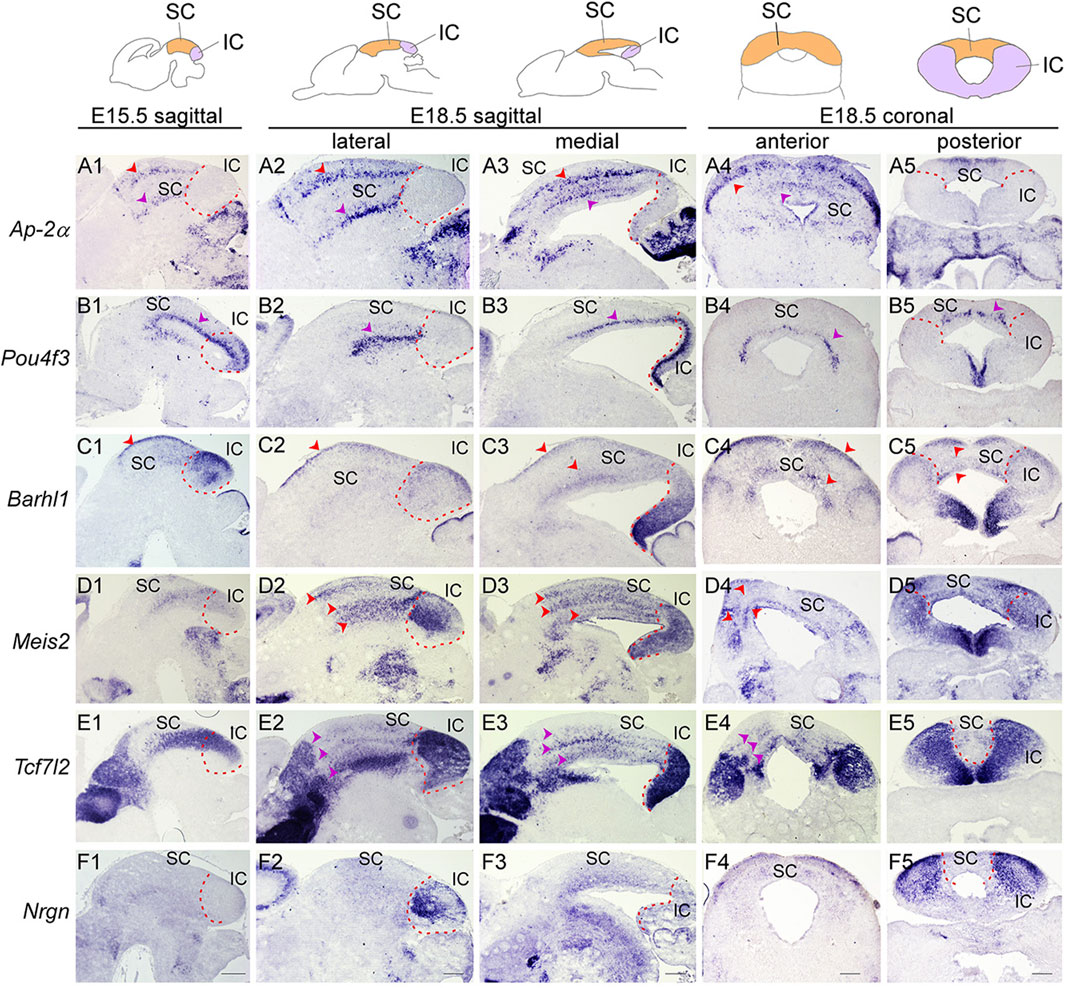
FIGURE 5. Molecular organization of the developing SC and IC (Top) Schematic view of E15.5 and E18.5 brain sections showing the superior colliculus (SC, orange) and inferior colliculus (IC, purple). (A–F) ISH with Ap-2α (A), Pou4f3 (B), Barhl1 (C), Meis2 (D), Tcf7l2 (E) and Nrgn (F) probes on sagittal sections of E15.5 wild-type embryos, and sagittal and transverse sections of E18.5 wild-type embryos. Multiple cellular layers of the SC can be identified by the expression of Ap-2α, Barhl1, Pou4f3, Meis2, and Tcf7l2 (arrowheads). Meis2 and Tcf7l2 expression are present in the almost entire region of the IC, while Barhl1, Pou4f3, and Nrgn are confined to discrete regions of the IC. Dashed curves delimit the IC region. Scale bars, 400 μm.
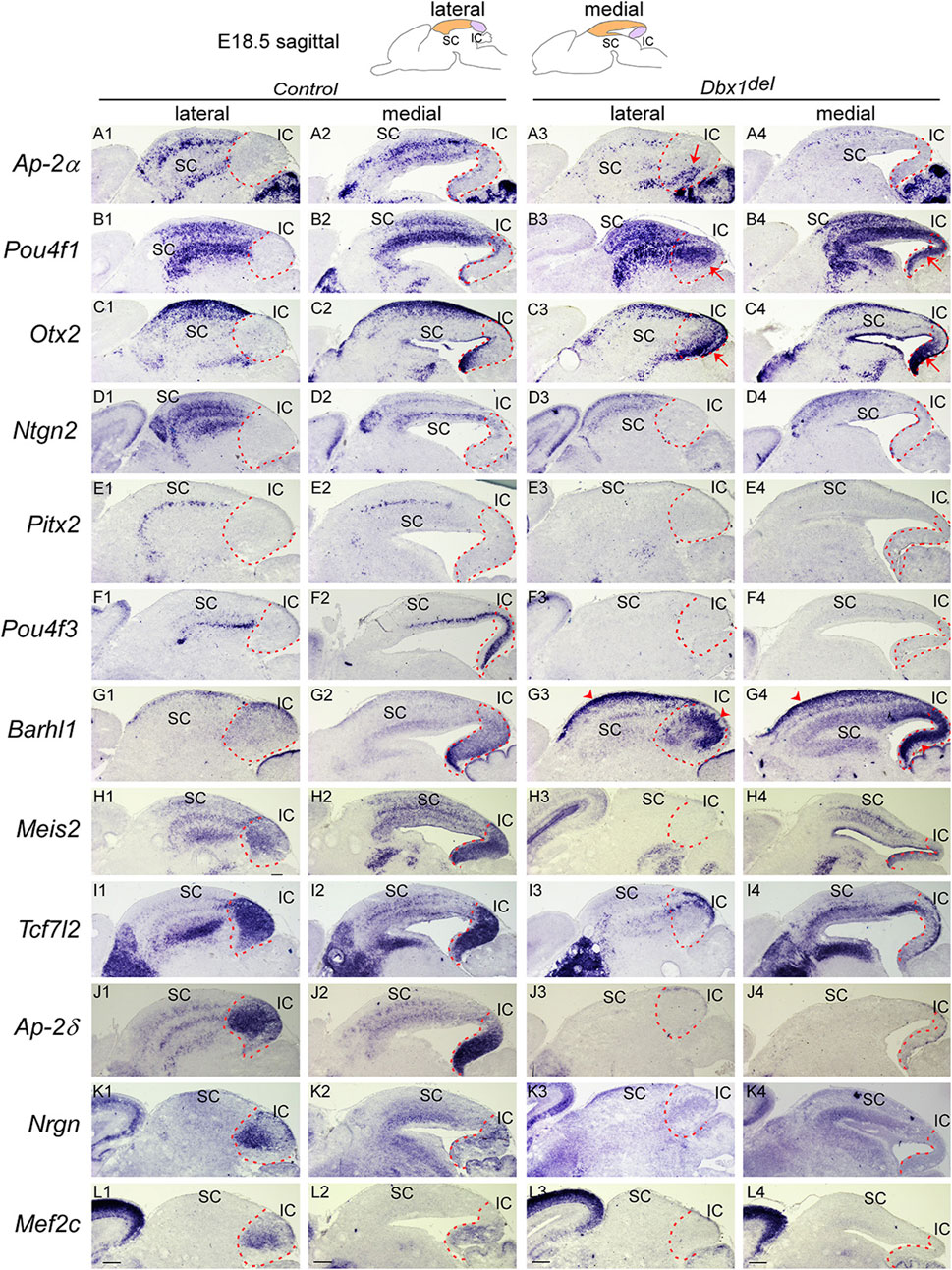
FIGURE 6. Loss of Dbx1 remarkably affects identity and differentiation of the superior and inferior colliculi (A–L) Section ISH on midbrain sagittal sections of control and Dbx1del embryos at E18.5 with Ap-2α (A), Pou4f1 (B), Otx2 (C), Ntng2 (D), Pitx2 (E), Pou4f3 (F), Barhl1 (G), Meis2 (H), Tcf7l2 (I), Ap-2δ (J), Nrgn (K), Mef2c (L). Red arrows indicate ectopic upregulation of Otx2, Pou4f1, and Ap-2α in the IC. Dashed curves delimit the IC region. Scale bars, 400 μm.
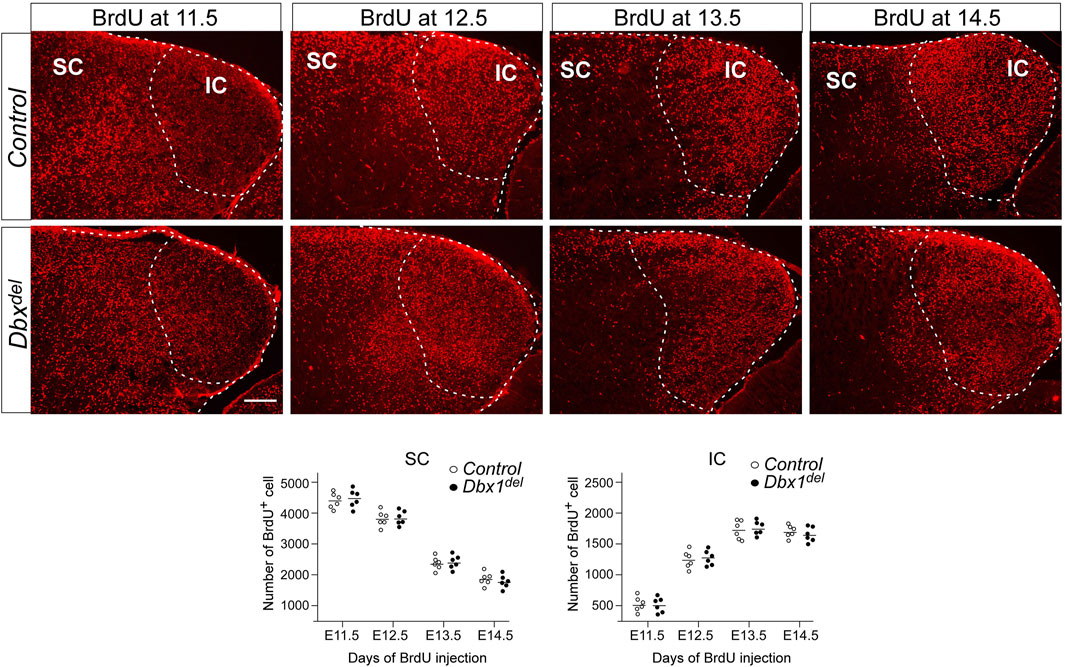
FIGURE 7. Birthdating of IC and SC neurons Pregnant mice were injected with BrdU at E11.5-E14.5, and brains were harvested at E18.5. For counting of BrdU+ cells, 10–20 counting sites (counting grid for SC, 200 μm × 200 μm; for IC, 100 μm × 100 μm) were evaluated in one section. The values were calculated from five sections of six embryos for each genotype. Scale bars, 200 μm.
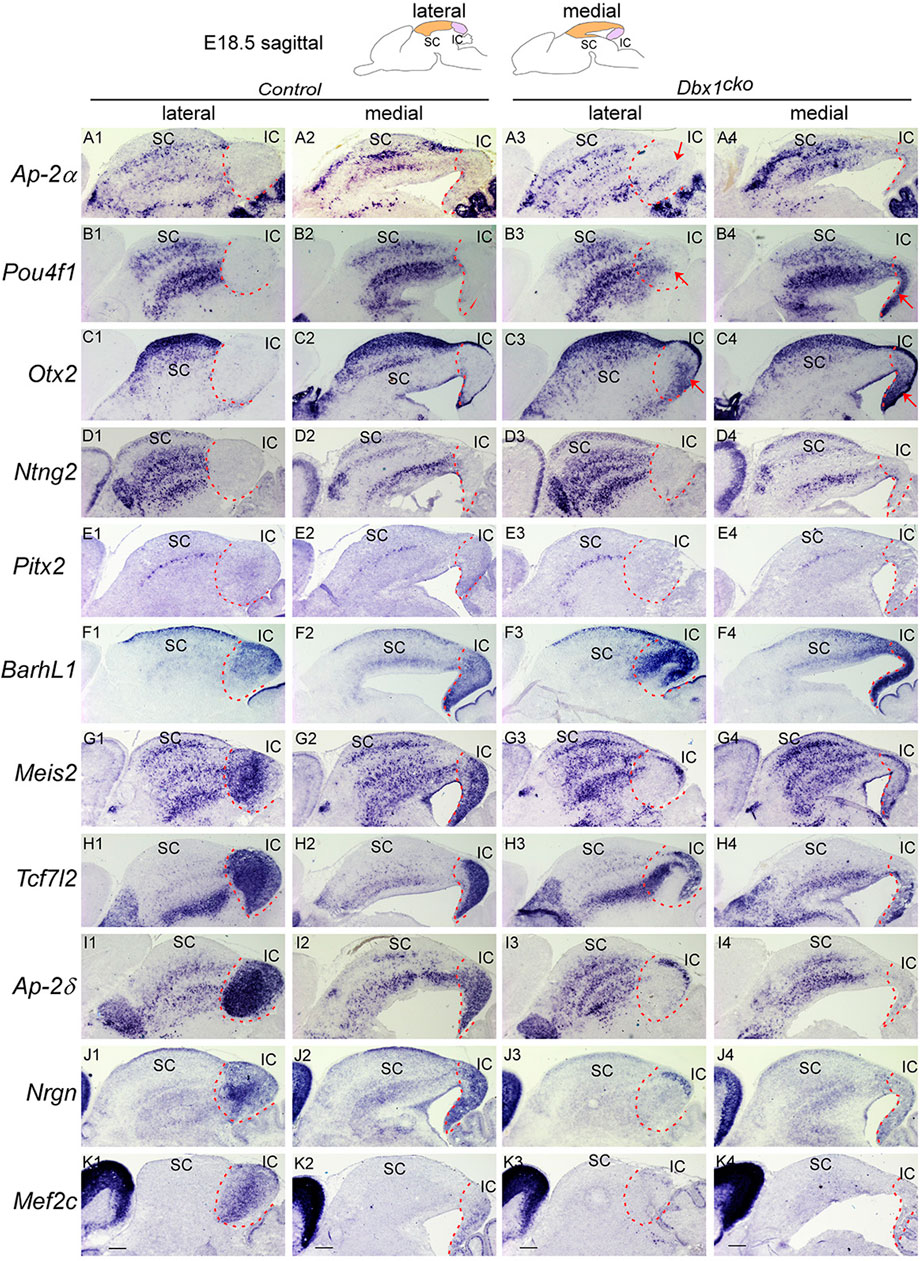
FIGURE 8. Dbx1 is required during distinct temporal windows for development of different structures in the dorsal midbrain Section ISH on midbrain sagittal sections of control and Dbx1cko embryos at E18.5 with Ap-2α (A), Pou4f1 (B), Otx2 (C), Ntng2 (D), Pitx2 (E), Barhl1 (F), Meis2 (G), Tcf7l2 (H), Ap-2δ (I), Nrgn (J), Mef2c (K). Red arrows indicate ectopic upregulation of Otx2, Pou4f1, and Ap-2α in the IC. Dashed curves delimit the IC region. Scale bars, 400 μm.
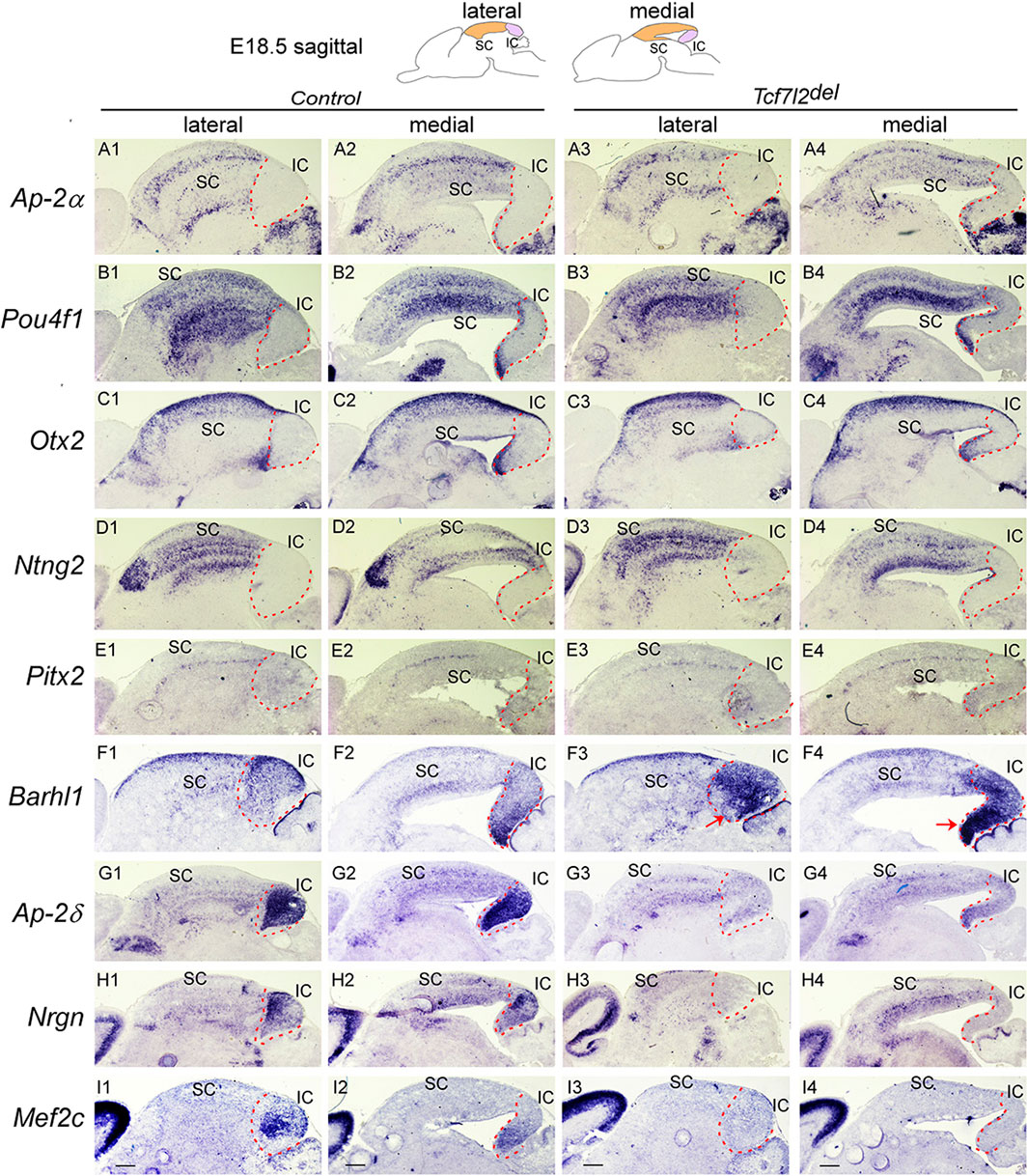
FIGURE 9. IC differentiation is impaired in the absence of Tcf7l2 Section ISH on midbrain sagittal sections of control and Tcf7l2del embryos at E18.5 with Ap-2α (A), Pou4f1 (B), Otx2 (C), Ntng2 (D), Pitx2 (E), Barhl1 (F), Ap-2δ (G), Nrgn (H), Mef2c (I). Red arrows (F3,F4) indicate ectopic upregulation of Barhl1 in the IC. Dashed curves delimit the IC region. Scale bars, 400 μm.
In order to understand the role of Dbx1 in mesencephalic neuronal differentiation, we first compared the distribution pattern of Dbx1 with those of transcription factors potentially involved in the specification of mesencephalic progenitors and derived neurons. Dbx1 expression was detectable in the dorsolateral wall of the mesencephalic neuroepithelium at E9.5 ∼ E10.5 (Lu et al., 1992; Shoji et al., 1996) (Figure 1A), and subsequently, at E11.5, was limited to the ventricular zone (Figure 1A). At E12.5-E13.5, Dbx1 was co-expressed in the m1-m2 dorsoventral progenitor domains of the dorsal midbrain with Ascl1, Helt, Neurog1, Pax7, and Otx2 (Figure 1B; Figures 2A, B). At E14.5-E15.5, there was a marked reduction in Dbx1 expression in the SC, but its expression persisted in the IC (Figure 1C).
The results of our gene expression experiments demonstrated that Dbx1 is actively expressed in the dorsal midbrain progenitors at the time when their dorsoventral identity is being established. Thus we intended to determine whether Dbx1 is involved in early regional patterning and neurogenesis in the dorsal midbrain. To achieve these goals, we analyzed Dbx1del mutants in which Dbx1 is inactivated at the pre-implantation stage (Tran et al., 2023). Recent studies demonstrated that GABAergic and glutamatergic neurons derived from the progenitor domains of the developing midbrain are regulated by combinations of multiple transcription factors acting as fate selectors. GABAergic fate determination requires the function of Ascl1 and Helt in the progenitor region and then subsequent activation of the post-mitotic selectors Gata2 and Tal2 (Miyoshi et al., 2004; Guimera et al., 2006; Nakatani et al., 2007; Kala et al., 2009; Peltopuro et al., 2010; Achim et al., 2013; Wende et al., 2015), whereas the glutamatergic lineage fate is initiated by progenitors expressing Neurog1, which is required for post-mitotic activation of Pou4f1 (Brn3a) (Guimera et al., 2006; Nakatani et al., 2007). In Dbx1-deficient embryos, Ascl1 expression was apparently normal, whereas the expression of Helt, Gata2, and Tal2 was lost specifically in the m1-m2 dorsoventral domains despite normal expression in the ventral domains (Figure 2A). Consistently, the post-mitotic GABAergic neuronal markers Gata3, Tal1, and Lhx5 were not detectable in the m1-m2 domain of Dbx1del mutants (Figure 2A). By contrast, loss of Dbx1 resulted in upregulation of the glutamatergic lineage markers, Neurog1 and Pou4f1 in the m1-m2 domain despite normal expression in the ventral region. These defects in GABAergic differentiation were similarly detected also in the posterior-most region of the midbrain (Supplementary Figure S1). Next, we examined the neurotransmitter profile in Dbx1-deficient embryos. GABAergic neurons express genes encoding the glutamate decarboxylase enzyme, Gad1/2, while glutamatergic neurons express the vesicular glutamate transporter Slc17a6. Compared to controls, in Dbx1del mutants, there was an almost complete loss of Gad1 staining in the m1-m2 domain, while Slc17a6 expression was increased (Figures 3A, B; Supplementary Figure S2).
To determine a genetic hierarchy, we analyzed Dbx1 and Helt expression in Ascl1 mutants. Loss of Ascl1 did not alter Dbx1 or Helt expression, although the progenitor domain expanded due to a failure in cell cycle exit (Peltopuro et al., 2010; Wende et al., 2015) (Figure 3C), implicating two parallel (Dbx1 vs. Ascl1) regulatory strategies for GABAergic fate determination in the dorsal midbrain (Figure 3D). Notably, as Tal2 is regulated by Dbx1 (Figure 2A), but not by Ascl1, Helt or Gata2 in the dorsal midbrain (Kala et al., 2009; Peltopuro et al., 2010; Wende et al., 2015), these observations demonstrate that Dbx1 regulates GABAergic fate via two different downstream cascades, Helt-Gata2 vs. Tal2 (Figure 3D).
Since Dbx1 is selectively expressed in the ventricular progenitor region and the onset of its transcription precedes neurogenesis, we examined the general properties of m1-m2 progenitor cells in Dbx1 mutant embryos. During neural development, Sox2 acts to maintain an undifferentiated cell state, and its expression becomes restricted to early progenitor and neural stem cells (Bylund et al., 2003; Graham et al., 2003; Cavallaro et al., 2008; Hagey and Muhr, 2014). The neuron-specific RNA binding proteins HuC/D encoded by Elavl3/4 genes are restricted to post-mitotic neurons (Szabo et al., 1991; Okano and Darnell, 1997). In Dbx1 mutant embryos, we detected no major difference in the thickness of the Sox2+ ventricular zone or the Elavl3/4+ (HuC/D) marginal zone (Figure 4A). Consistently, there was no major changes in the numbers of phosphor-histone H3+ mitotic cells or BrdU+ S-phase cells (Figures 4B, C). Thus, these results suggest that loss of Dbx1 does not impair generic neurogenic properties.
Early failure in GABAergic fate determination in the dorsal midbrain of Dbx1 mutants led us to ask whether Dbx1 is also required for morphological maturation of the dorsal midbrain. We first characterized molecular features of the dorsal midbrain at E15.5, when the IC and SC, two morphologically distinct compartments of the dorsal midbrain, are detectable, and the SC forms multiple cellular and fibrous layers. Ap-2α expression was restricted to the SC and not detectable in the IC (Figure 5A1). Pou4f3, Barhl1, Meis2, and Tcf7l2 expression were continuous between the SC and IC at E15.5 (Figures 5B1–E1). At E18.5, the overall patterns of Ap-2α and Barhl1 expression were maintained (Figures 5A2, A3, C2, C3), but the lateral region of the IC was completely devoid of Pou4f3 expression (Figures 5B2). Meis2 and Tcf7l2 displayed discontinuous and discrete expression pattern between the SC and IC (Figures 5D2, D3, E2, E3), and Nrgn expression was strongly detected in the lateral region of the IC (Figures 5F2, F5). The SC develops a layered structure with a number of layers that varies by species. The SC layers can be grouped into the superficial layers (zonal, superficial gray, and optic layers) and the deeper layers (intermediate gray, intermediate white, deep gray, and deep white layers). Ap-2α, Barhl1, Meis2, and Tcf7l2 expression were detectable in the superficial and deeper layers (Figures 5A2–A4, C2–C4, D2–D4, E2–E4), but Pou4f3 expression was specific for the deep gray layer (Figures 5B2–B4). Besides, molecularly distinct regions of the IC can be identified by differential gene expression of Pou4f3, Barhl1, Meis2, Tcf7l2 and Nrgn (Figures 5B5–F5). The spatial distribution patterns of these genes observed at E18.5 were maintained until postnatal or adult stages (Allen Brain Atlas, https://mouse.brain-map.org/), indicating that the molecular organization of the SC and IC are established during a narrow window of embryonic development (E15.5–E18.5).
To investigate whether Dbx1 regulates differentiation of the SC and IC, we analyzed Dbx1del mutants at E18.5, when morphological maturation is established. In the SC of Dbx1del embryos, Ap-2α expression was severely downregulated despite strong staining in the cerebellum (Figures 6A3, A4). Pou4f1 (Brn3a) was expressed in multiple layers of control SC (Figures 6B1, B2), but its striped pattern was disrupted in the SC of Dbx1del embryos (Figures 6B3, B4). Otx2 expression, which was confined to the superficial layers of control SC, was downregulated in the absence of Dbx1 (Figures 6C3, C4). In control embryos, Ntng2 expression was detected in both superficial and deep layers of the SC (Figures 6D1, D2), while in the SC of Dbx1del embryos, the striped pattern of Ntng2 expression was abolished (Figures 6D3, D4). Pitx2 and Pou4f3, which normally mark the intermediate grey and deep grey layers, respectively (Figures 6E1, E2, F1, F2), were not detected in the SC of Dbx1del embryos (Figures 6E3, E4, F3, F4). Notably, Pitx2 is identified as a target of Gata2 and required for GABAergic neuron differentiation in the SC (Kala et al., 2009; Waite et al., 2011). Barhl1, which marks the zonal layer, was strongly upregulated in Dbx1del mutants (Figures 6G3, G4, arrowheads). Targeted deletion of Barhl1 have been shown to disrupt the formation of the SC layers (Martin et al., 2004; Li and Xiang, 2006; Masullo et al., 2019). Meis2, Tcf7l2, and Ap-2δ, which are normally expressed in multiple layers of the SC, were almost absent or downregulated in Dbx1del mutants (Figures 6H3, H4, I3, I4, J3, J4). A previous report demonstrated that loss of Ap-2δ results in a diminished IC due to abnormal apoptotic cell death (Hesse et al., 2011). Together, these observations indicate that Dbx1 is critical for normal SC laminar organization.
In the IC of Dbx1del embryos, expression of Pou4f3, Meis2, Tcf7l2, Ap-2δ, Nrgn, and Mef2c genes were completely absent or severely downregulated (Figures 6F3, F4, H3, H4, I3, I4, J3, J4, K3, K4, L3, L4). By contrast, Barhl1 expression was strongly upregulated in the IC of Dbx1del embryos (Figures 6G3, G4, arrowheads). Surprisingly, loss of Dbx1 resulted in strong ectopic activation of SC-specific genes such as Ap-2α, Otx2 and Pou4f1 in the IC (Figures 6A3, A4, B3, B4, C3, C4, arrows), indicating that Dbx1 inactivation de-represses a SC-like fate in the IC. Therefore, it is likely that one of the crucial functions of Dbx1 is to prevent IC from acquiring SC identity.
We next examined whether abnormalities observed in the SC and IC might be associated with a defect in neuronal migration. To estimate the relationships between birthdates and destinations of the dorsal midbrain neurons, we labeled newly born cells with BrdU at E11.5 ∼ E14.5, when neurogenesis in the dorsal midbrain occurs (Cowan et al., 1968; Edwards et al., 1986; Tan et al., 2002). Harvesting embryos at E18.5, we counted labeled cells, having divided the SC into 20 equal zones and the IC into 10 equal zones. In control embryos, more of the earliest generated cells were situated in the SC than in the IC, while the majority of the latest born cells were situated in the IC (Figure 7). These observations were in agreement with previous findings that SC neurons are born earlier than IC neurons (Cowan et al., 1968; Altman and Bayer, 1981b; 1981a; Edwards et al., 1986). We did not observe significant differences in the distribution of BrdU+ cells in the SC and IC between controls and Dbx1del mutants (Figure 7), suggesting that loss of Dbx1 does not affect cell migration in the developing dorsal midbrain.
Given that Dbx1 expression persists at high levels in the dorsal midbrain during late gestation, our observations raise an intriguing possibility that Dbx1 has a continued role in IC and SC neuron development during embryogenesis. To determine whether the dependency of dorsal midbrain development on Dbx1 is regulated temporally, we analyzed Dbx1cko mutants in which Dbx1 function in the midbrain is inactivated at E12.5 (Tran et al., 2023). In contrast to Dbx1del embryos, Dbx1cko mutants displayed no major alteration in expression of SC-specific genes such as Ap-2α, Pou4f1, Otx2, Ntng2 and Pitx2 (Figures 8A3, A4, B3, B4, C3, C4, D3, D3, E3, E4). Compared to controls, we observed no significant change in Barhl1 expression in the SC of Dbx1cko mutants, whereas its expression in the IC was strongly upregulated in a pattern similar to Dbx1del embryos (Figures 8F1–F4). In additions, expression of Meis2, Tcf7l2, and Ap-2δ genes were significantly lost in the IC of Dbx1cko mutants despite similar expression in the SC (Figures 8G1–G4, H1–H4, I1–I4). Like in Dbx1del IC, the IC-specific markers Nrgn and Mef2c expression were almost absent in the Dbx1cko IC (Figures 8J3, J4, K3, K4). Moreover, we observed ectopic activation of Ap-2α, Pou4f1, and Otx2 in the IC of Dbx1cko mutants in a pattern similar to those in Dbx1del embryos (Figures 8A3, B3, B4, C3, C4), indicating that persistent expression of Dbx1 is required for preventing IC into SC fate switch. Therefore, conditional loss of Dbx1 at later stages did not affect SC differentiation, but caused IC phenotypes with striking similarity to those of Dbx1del mutants. These data suggest that SC differentiation is dependent on the early function of Dbx1, and the IC requires early and prolonged action for its normal specification and morphological maturation.
We also examined early regional patterning and neuronal specification in the dorsal midbrain of Dbx1cko mutant embryos. Compared to controls, we observed no substantial changes in the expression of Helt, Gata2, Tal2, Tal1, Gad1, and Slc17a6 in the developing midbrain of Dbx1cko mutants (Supplementary Figure S3), indicating that specification of GABAergic neuronal identity is dependent on early Dbx1 activity.
Previously, we showed that Tcf7l2 mediates the function of Dbx1 in the regulation of IC survival (Tran et al., 2023). To address whether Tcf7l2 is involved in embryonic midbrain patterning and IC/SC differentiation, we first examined the temporal and spatial pattern of Tcf7l2 expression in the developing midbrain. Tcf7l2 transcription in the midbrain was detectable after E10.5, and limited to m1-m2 post-mitotic regions (Supplementary Figure S4). Tcf7l2 expression then persisted in the SC and IC during later embryonic and adult stages (Supplementary Figure S4). To address whether Tcf7l2 is involved in regulating GABAergc neuronal fate in the dorsal midbrain, we analyzed Tcf7l2del mutants in which Tcf7l2 is inactivated at the pre-implantation stage (Tran et al., 2023). In Tcf7l2del mutants, there was no significant alteration in the expression of GABAergic lineage markers such as Ascl1, Helt, Gata2/3, Tal1/2, and Gad1 (Supplementary Figure S5), indicating that Tcf7l2 is not required for specifying GABAergic neuronal identity. To determine whether Tcf7l2 regulates maturation of the dorsal midbrain into SC and IC, we next examined the expression of molecular markers for the IC and SC. We observed no major difference in expression of Ap-2α, Pou4f1, Otx2, Ntng2, and Pitx2 in the SC of Tcf7l2del mutants compared with that in controls (Figures 9A1–A4, B1–B4, C1–C4, D1–D4, E1–E4). Barhl1 expression was similarly detected in the SC of control and Tcf7l2del mutants, while its expression was strongly upregulated in the IC of Tcf7l2del mutants (Figures 9F1–F4). Furthermore, expression of IC markers such as Ap-2δ, Nrgn, and Mef2c were severely downregulated in Tcf7l2del mutants (Figures 9G1–G4, H1–H4, I1–I4), indicating that loss of Tcf7l2 leads to a phenotype reminiscent of Dbx1cko mutants. These data suggest that Tcf7l2 is selectively required for differentiation of the IC, but not the SC. However, unlike Dbx1 mutants, there was no ectopic activation of SC-specific markers in the IC of in Tcf7l2del mutants (Figures 9A3, A4, B3, B4, C3, C4). Therefore, the function of Dbx1 to promote IC differentiation is not likely to be mediated entirely by Tcf7l2 gene.
The present study identifies Dbx1 as a master regulator of GABAergic neuron fate in the dorsal midbrain. Moreover, our results show that in the m1-m2 domains, Dbx1 functions upstream of Helt, Gata2, and Tal2, and that Dbx1 and Ascl1 act in parallel. Inactivation of Helt or Ascl1 resulted in complete loss of GABAergic neurons in m1-m2 domains, whereas GABAergic neurons in m3-m5 domains were still generated (Guimera et al., 2006; Nakatani et al., 2007; Peltopuro et al., 2010). However, combined deletion of Helt and Ascl1 abrogates induction of GABAergic neurons in all of the m1-m5 domains. Therefore, these observations suggest that GABAergic progenitor specification employs distinct molecular mechanisms depending on the location of the cells’ birth (Wende et al., 2015). Interestingly, loss of Ascl1 resulted in delayed cell cycle exit and onset of differentiation (Peltopuro et al., 2010), whereas deletion of Dbx1 or Helt did not affect that process (Guimera et al., 2006; Nakatani et al., 2007) (Figure 4), suggesting that neurogenesis and subtype specification including neurotransmitter selection may be separate processes. After cell cycle exit, Gata2 and Tal2 function as post-mitotic selectors between GABAergic and glutamatergic neurons. Gata2 expression was dependent on Helt and Ascl1, whereas Tal2 activation required none of those factors except for in the m5 domain (Kala et al., 2009; Peltopuro et al., 2010; Wende et al., 2015). Notably, loss of Dbx1 abrogated Tal2 expression in the m1-m2 domains, indicating that Dbx1 functions to promote GABAergic fate determination at least by controlling two different regulatory axes, the Helt/Gata2-dependent and Tal2-dependent cascades (Figure 3D).
By studying the sequential genetic changes underlying the role of Dbx1, we demonstrated that Dbx1 functions for distinct lengths of time in the development of different dorsal structures of the midbrain. A question arises why the SC require the early activity of Dbx1, and IC development is dependent on persistent expression of Dbx1. Previous studies showed that growth and expansion of the dorsal midbrain occur in an anterior-to-posterior direction so that the posterior region of the dorsal midbrain continues to divide even after the anterior region ceases proliferation and undergoes neurogenesis (Cowan et al., 1968; Altman and Bayer, 1981b; 1981a; Edwards et al., 1986). Consistently, our BrdU fate mapping experiments also demonstrated that SC neurons are born earlier with peak generation around E11.5 ∼ E12.5, whereas IC neuron generation peaks around E12.5 ∼ E14.5. In agreement with these results, Dbx1 expression persisted in the IC even after it was excluded from the SC (Figure 1C). These observations together may explain why IC, but not SC requires the prolonged activity of Dbx1 for normal differentiation.
Dbx1 plays important roles in the generation of specific subclasses of neurons in a region-specific manner by regulating the expression of effector genes, which work together to control the formation of V0 interneurons in the spinal cord, Pre-Bötzinger complex neurons in the hindbrain, orexigenic neurons in the hypothalamus (Pierani et al., 2001; Bouvier et al., 2010; Gray et al., 2010; Sokolowski et al., 2015)). In this study, we demonstrate that Tcf7l2 functions as an effector gene to mediate the activity of Dbx1 for IC differentiation. During embryogenesis, Dbx1 and Tcf7l2 expression are restricted to m1-m2 domains, which differentiate into the SC and IC. Loss of Dbx1 resulted in downregulation of Tcf7l2 in the SC and IC. However, unlike Dbx1, loss of Tcf7l2 did not affect SC differentiation. A question arises why the IC requires Tcf7l2 function for its development, and SC development is not dependent on Tcf7l2. Wnt/β-catenin signaling controls multiple processes during early midbrain development and dopaminergic (mDA) neuron differentiation. All Wnts that use canonical signaling converge on β-catenin, which activates target genes via Tcf/Lef factors (Liu et al., 2022). Vertebrates encode four Tcf/Lef genes (Tcf7, Lef1, Tcf7l1, Tcf7l2), which play important roles in mediating the diverse functions of Wnt/β-catenin signaling in different tissues at different stages. The overlapping expression patterns and functions of Tcf/Lef factors preclude an understanding of the specificity of individual factor involvement in neuron development in vivo. All four Tcf/Lef factors are expressed in the developing midbrain at early stages. Interestingly, later, Tcf7l2 is the only factor expressed in the IC (Allen Brain Atlas). Therefore, it is likely that functional redundancy may exist during SC development (Cadigan and Waterman, 2012).
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
The animal study was approved by the Kyung Hee University Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
H-NT: Formal Analysis, Writing–review and editing, Data curation, Methodology, Investigation, Validation. Q-HN: Data curation, Formal Analysis, Methodology, Writing–review and editing. YJ: Formal Analysis, Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing, Investigation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea Ministry of Science and ICT (Nos 2019R1F1A1042941 and 2022R1F1A1064555).
The authors would like to thank Drs. Kwanghee Baek and Jaeseung Yoon Labs at Kyung Hee University for the use of fluorescent microscope.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1336308/full#supplementary-material
Achim, K., Peltopuro, P., Lahti, L., Li, J., Salminen, M., and Partanen, J. (2012). Distinct developmental origins and regulatory mechanisms for GABAergic neurons associated with dopaminergic nuclei in the ventral mesodiencephalic region. Development 139, 2360–2370. doi:10.1242/dev.076380
Achim, K., Peltopuro, P., Lahti, L., Tsai, H.-H., Zachariah, A., Åstrand, M., et al. (2013). The role of Tal2 and Tal1 in the differentiation of midbrain GABAergic neuron precursors. Biol. Open 2, 990–997. doi:10.1242/bio.20135041
Agoston, Z., and Schulte, D. (2009). Meis2 competes with the Groucho co-repressor Tle4 for binding to Otx2 and specifies tectal fate without induction of a secondary midbrain-hindbrain boundary organizer. Development 136, 3311–3322. doi:10.1242/dev.037770
Altman, J., and Bayer, S. A. (1981a). Time of origin of neurons of the rat inferior colliculus and the relations between cytogenesis and tonotopic order in the auditory pathway. Exp. Brain Res. 42, 411–423. doi:10.1007/BF00237506
Altman, J., and Bayer, S. A. (1981b). Time of origin of neurons of the rat superior colliculus in relation to other components of the visual and visuomotor pathways. Exp. Brain Res. 42, 424–434. doi:10.1007/BF00237507
Angus-Hill, M. L., Elbert, K. M., Hidalgo, J., and Capecchi, M. R. (2011). T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc. Natl. Acad. Sci. 108, 4914–4919. doi:10.1073/pnas.1102300108
Basson, M. A., Echevarria, D., Petersen Ahn, C., Sudarov, A., Joyner, A. L., Mason, I. J., et al. (2008). Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development 135, 889–898. doi:10.1242/dev.011569
Bouvier, J., Thoby-Brisson, M., Renier, N., Dubreuil, V., Ericson, J., Champagnat, J., et al. (2010). Hindbrain interneurons and axon guidance signaling critical for breathing. Nat. Neurosci. 13, 1066–1074. doi:10.1038/nn.2622
Broccoli, V., Boncinelli, E., and Wurst, W. (1999). The caudal limit of Otx2 expression positions the isthmic organizer. Nature 401, 164–168. doi:10.1038/43670
Bylund, M., Andersson, E., Novitch, B. G., and Muhr, J. (2003). Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6, 1162–1168. doi:10.1038/nn1131
Cadigan, K. M., and Waterman, M. L. (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 4, a007906. doi:10.1101/cshperspect.a007906
Cant, N. B., and Benson, C. G. (2006). Organization of the inferior colliculus of the gerbil (Meriones unguiculatus): differences in distribution of projections from the cochlear nuclei and the superior olivary complex. J. Comp. Neurol. 495, 511–528. doi:10.1002/cne.20888
Cavallaro, M., Mariani, J., Lancini, C., Latorre, E., Caccia, R., Gullo, F., et al. (2008). Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 135, 541–557. doi:10.1242/dev.010801
Chi, C. L., Martinez, S., Wurst, W., and Martin, G. R. (2003). The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130, 2633–2644. doi:10.1242/dev.00487
Cowan, W. M., Martin, A. H., and Wenger, E. (1968). Mitotic patterns in the optic tectum of the chick during normal development and after early removal of the optic vesicle. J. Exp. Zool. 169, 71–92. doi:10.1002/jez.1401690110
Dee, A., Li, K., Heng, X., Guo, Q., and Li, J. Y. H. (2016). Regulation of self-renewing neural progenitors by FGF/ERK signaling controls formation of the inferior colliculus. Development 143, 3661–3673. doi:10.1242/dev.138537
Dessaud, E., McMahon, A. P., and Briscoe, J. (2008). Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489–2503. doi:10.1242/dev.009324
Di Giovannantonio, L. G., Di Salvio, M., Omodei, D., Prakash, N., Wurst, W., Pierani, A., et al. (2014). Otx2 cell-autonomously determines dorsal mesencephalon versus cerebellum fate independently of isthmic organizing activity. Development 141, 377–388. doi:10.1242/dev.102954
Edwards, M. A., Caviness, V. S., and Schneider, G. E. (1986). Development of cell and fiber lamination in the mouse superior colliculus. J. Comp. Neurol. 248, 395–409. doi:10.1002/cne.902480308
Graham, V., Khudyakov, J., Ellis, P., and Pevny, L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765. doi:10.1016/S0896-6273(03)00497-5
Gray, P. A., Hayes, J. A., Ling, G. Y., Llona, I., Tupal, S., Picardo, M. C. D., et al. (2010). Developmental origin of preBötzinger complex respiratory neurons. J. Neurosci. 30, 14883–14895. doi:10.1523/JNEUROSCI.4031-10.2010
Guimera, J., Weisenhorn, D. V., and Wurst, W. (2006). Megane/Heslike is required for normal GABAergic differentiation in the mouse superior colliculus. Development 133, 3847–3857. doi:10.1242/dev.02557
Hagey, D. W., and Muhr, J. (2014). Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell. Rep. 9, 1908–1920. doi:10.1016/j.celrep.2014.11.013
Hesse, K., Vaupel, K., Kurt, S., Buettner, R., Kirfel, J., and Moser, M. (2011). AP-2δ is a crucial transcriptional regulator of the posterior midbrain. PLoS One 6, e23483. doi:10.1371/journal.pone.0023483
Joyner, A. L., Liu, A., and Millet, S. (2000). Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell. Biol. 12, 736–741. doi:10.1016/s0955-0674(00)00161-7
Kala, K., Haugas, M., Lilleväli, K., Guimera, J., Wurst, W., Salminen, M., et al. (2009). Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development 136, 253–262. doi:10.1242/dev.029900
Lahti, L., Achim, K., and Partanen, J. (2013). Molecular regulation of GABAergic neuron differentiation and diversity in the developing midbrain. Acta Physiol. (Oxf). 207, 616–627. doi:10.1111/apha.12062
Lakso, M., Pichel, J. G., Gorman, J. R., Sauer, B., Okamoto, Y., Lee, E., et al. (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93, 5860–5865. doi:10.1073/pnas.93.12.5860
Le Dréau, G., and Martí, E. (2012). Dorsal-ventral patterning of the neural tube: a tale of three signals. Dev. Neurobiol. 72, 1471–1481. doi:10.1002/dneu.22015
Leung, C. T., Coulombe, P. A., and Reed, R. R. (2007). Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 10, 720–726. doi:10.1038/nn1882
Li, S., and Xiang, M. (2006). Barhl1 is required for maintenance of a large population of neurons in the zonal layer of the superior colliculus. Dev. Dyn. 235, 2260–2265. doi:10.1002/dvdy.20858
Liu, J., Xiao, Q., Xiao, J., Niu, C., Li, Y., Zhang, X., et al. (2022). Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 7, 3. doi:10.1038/s41392-021-00762-6
Lu, S., Bogarad, L. D., Murtha, M. T., and Ruddle, F. H. (1992). Expression pattern of a murine homeobox gene, Dbx, displays extreme spatial restriction in embryonic forebrain and spinal cord. Proc. Natl. Acad. Sci. U. S. A. 89, 8053–8057. doi:10.1073/pnas.89.17.8053
Martin, D. M., Skidmore, J. M., Philips, S. T., Vieira, C., Gage, P. J., Condie, B. G., et al. (2004). PITX2 is required for normal development of neurons in the mouse subthalamic nucleus and midbrain. Dev. Biol. 267, 93–108. doi:10.1016/j.ydbio.2003.10.035
Masullo, L., Mariotti, L., Alexandre, N., Freire-Pritchett, P., Boulanger, J., Tripodi, M., et al. (2019). Genetically defined functional modules for spatial orienting in the mouse superior colliculus. Curr. Biol. 29, 2892–2904. doi:10.1016/j.cub.2019.07.083
Matsunaga, E., Araki, I., and Nakamura, H. (2001). Role of Pax3/7 in the tectum regionalization. Development 128, 4069–4077. doi:10.1242/dev.128.20.4069
McLaughlin, T., and O’Leary, D. D. M. (2005). Molecular gradients and development of retinotopic maps. Annu. Rev. Neurosci. 28, 327–355. doi:10.1146/annurev.neuro.28.061604.135714
Miller, M. W., and Nowakowski, R. S. (1988). Use of bromodeoxyuridine-immunohistochemistry to examine the proliferation, migration and time of origin of cells in the central nervous system. Brain Res. 457, 44–52. doi:10.1016/0006-8993(88)90055-8
Miyoshi, G., Bessho, Y., Yamada, S., and Kageyama, R. (2004). Identification of a novel basic helix-loop-helix gene, Heslike, and its role in GABAergic neurogenesis. J. Neurosci. 24, 3672–3682. doi:10.1523/JNEUROSCI.5327-03.2004
Nakatani, T., Minaki, Y., Kumai, M., and Ono, Y. (2007). Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development 134, 2783–2793. doi:10.1242/dev.02870
Okano, H. J., and Darnell, R. B. (1997). A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 17, 3024–3037. doi:10.1523/JNEUROSCI.17-09-03024.1997
Oliver, D. L., and Morest, D. K. (1984). The central nucleus of the inferior colliculus in the cat. J. Comp. Neurol. 222, 237–264. doi:10.1002/cne.902220207
Peltopuro, P., Kala, K., and Partanen, J. (2010). Distinct requirements for Ascl1 in subpopulations of midbrain GABAergic neurons. Dev. Biol. 343, 63–70. doi:10.1016/j.ydbio.2010.04.015
Pierani, A., Moran-Rivard, L., Sunshine, M. J., Littman, D. R., Goulding, M., Jessell, T. M., et al. (2001). Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron 29, 367–384. doi:10.1016/s0896-6273(01)00212-4
Puelles, E. (2007). Genetic control of basal midbrain development. J. Neurosci. Res. 85, 3530–3534. doi:10.1002/jnr.21363
Puelles, E., Acampora, D., Lacroix, E., Signore, M., Annino, A., Tuorto, F., et al. (2003). Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nat. Neurosci. 6, 453–460. doi:10.1038/nn1037
Sgaier, S. K., Lao, Z., Villanueva, M. P., Berenshteyn, F., Stephen, D., Turnbull, R. K., et al. (2007). Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development 134, 2325–2335. doi:10.1242/dev.000620
Shamim, H., Mahmood, R., Logan, C., Doherty, P., Lumsden, A., Mason, I., et al. (1999). Sequential roles for Fgf4, En1 and Fgf8 in specification and regionalisation of the midbrain. Development 126, 945–959. doi:10.1242/dev.126.5.945
Shoji, H., Ito, T., Wakamatsu, Y., Hayasaka, N., Ohsaki, K., Oyanagi, M., et al. (1996). Regionalized expression of the Dbx family homeobox genes in the embryonic CNS of the mouse. Mech. Dev. 56, 25–39. doi:10.1016/0925-4773(96)00509-6
Sokolowski, K., Esumi, S., Hirata, T., Kamal, Y., Tran, T., Lam, A., et al. (2015). Specification of select hypothalamic circuits and innate behaviors by the embryonic patterning gene dbx1. Neuron 86, 403–416. doi:10.1016/j.neuron.2015.03.022
Szabo, A., Dalmau, J., Manley, G., Rosenfeld, M., Wong, E., Henson, J., et al. (1991). HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 67, 325–333. doi:10.1016/0092-8674(91)90184-z
Tan, S.-S., Valcanis, H., Kalloniatis, M., and Harvey, A. (2002). Cellular dispersion patterns and phenotypes in the developing mouse superior colliculus. Dev. Biol. 241, 117–131. doi:10.1006/dbio.2001.0505
Tran, H.-N., Nguyen, Q.-H., Jeong, J., Loi, D.-L., Nam, Y. H., Kang, T. H., et al. (2023). The embryonic patterning gene Dbx1 governs the survival of the auditory midbrain via Tcf7l2-Ap2δ transcriptional cascade. Cell. Death Differ. 30, 1563–1574. doi:10.1038/s41418-023-01165-6
Tran, H.-N., Park, W., Seong, S., Jeong, J.-E., Nguyen, Q.-H., Yoon, J., et al. (2020). Tcf7l2 transcription factor is required for the maintenance, but not the initial specification, of the neurotransmitter identity in the caudal thalamus. Dev. Dyn. 249, 646–655. doi:10.1002/dvdy.146
Urbánek, P., Wang, Z. Q., Fetka, I., Wagner, E. F., and Busslinger, M. (1994). Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79, 901–912. doi:10.1016/0092-8674(94)90079-5
Waite, M. R., Skidmore, J. M., Billi, A. C., Martin, J. F., and Martin, D. M. (2011). GABAergic and glutamatergic identities of developing midbrain Pitx2 neurons. Dev. Dyn. 240, 333–346. doi:10.1002/dvdy.22532
Wende, C.-Z., Zoubaa, S., Blak, A., Echevarria, D., Martinez, S., Guillemot, F., et al. (2015). Hairy/Enhancer-of-Split MEGANE and proneural MASH1 factors cooperate synergistically in midbrain GABAergic neurogenesis. PLoS One 10, e0127681. doi:10.1371/journal.pone.0127681
Wurst, W., and Bally-Cuif, L. (2001). Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2, 99–108. doi:10.1038/35053516
Keywords: Dbx1, GABAergic neuron, inferior colliculus, superior colliculus, Tcf7l2, dorsal midbrain
Citation: Tran H-N, Nguyen Q-H and Jeong Y (2024) Dbx1 is a dorsal midbrain-specific determinant of GABAergic neuron fate and regulates differentiation of the dorsal midbrain into the inferior and superior colliculi. Front. Cell Dev. Biol. 12:1336308. doi: 10.3389/fcell.2024.1336308
Received: 10 November 2023; Accepted: 15 January 2024;
Published: 26 January 2024.
Edited by:
Pin-Xian Xu, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Eduardo Puelles, Miguel Hernández University of Elche, SpainCopyright © 2024 Tran, Nguyen and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongsu Jeong, eW9uZ3N1QGtodS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.