- 1School of Life Science and Technology, ShanghaiTech University, Shanghai, China
- 2Zhenhai Lianhua Hospital, Ningbo City, Zhejiang, China
- 3Department of Cardiothoracic Surgery, Lihuili Hospital Affiliated to Ningbo University, Ningbo City, Zhejiang, China
Neural stem cells (NSCs) exhibit self-renewing and multipotential properties. Adult NSCs are located in two neurogenic regions of adult brain: the ventricular-subventricular zone (V-SVZ) of the lateral ventricle and the subgranular zone of the dentate gyrus in the hippocampus. Maintenance and differentiation of adult NSCs are regulated by both intrinsic and extrinsic signals that may be integrated through expression of some key factors in the adult NSCs. A number of transcription factors have been shown to play essential roles in transcriptional regulation of NSC cell fate transitions in the adult brain. Epigenetic regulators have also emerged as key players in regulation of NSCs, neural progenitor cells and their differentiated progeny via epigenetic modifications including DNA methylation, histone modifications, chromatin remodeling and RNA-mediated transcriptional regulation. This minireview is primarily focused on epigenetic regulations of adult NSCs during adult neurogenesis, in conjunction with transcriptional regulation in these processes.
Introduction
Adult neurogenesis is a process that generates functional neurons and glial cells from adult neural stem cells (NSCs) (Ming and Song, 2011; Hsieh and Zhao, 2016; Kuhn et al., 2018; Cope and Gould, 2019; Bond et al., 2021). There are two neurogenic regions in adult mouse brain, the ventricular-subventricular zone (V-SVZ) located in the lateral ventricle and the subgranular zone (SGZ) located in the dentate gyrus (DG) of hippocampus (Bond et al., 2021; Kobayashi and Kageyama, 2021). Adult NSCs at the DG niche often adopt a radial glial morphology and thus they are also called radial glia-like neural stem cells (RGLs), whereas those at the V-SVZ niche are termed B cells (Bond et al., 2021). The adult NSCs in V-SVZ have embryonic origin and enter a quiescence state during embryonic development. They are reactivated by both intrinsic and extrinsic signals before they give rises to neurons and small populations of glial cells (Delgado et al., 2021). First, they generate intermediate progenitor cells (IPCs), which undergo further differentiation to become immature neurons called neuroblasts (Lim and Alvarez-Buylla, 2016). Neuroblasts are precursors of neural cells. They migrate through the rostral migratory stream (RMS) to the olfactory bulb (OB) where they turn into mature inhibitory interneurons that are essential for olfaction. Occasionally adult NSCs located in the V-SVZ region can also give rise to oligodendrocytes which subsequently migrate to the corpus callosum and striatum where they further differentiate into myelinated or unmyelinated oligodendrocytes. The adult NSCs in the SGZ region are located in the granular cell layer and hilus of the dentate gyrus (Vicidomini et al., 2020; Kobayashi and Kageyama, 2021). These adult NSCs are released from the quiescent state in response to neural activity and environmental factors surrounding the niche of adult NSCs. They enter dentate migratory stream (DMS) to cross the granular cell layer radially before they give rise to IPCs, which in turn become neuroblasts and undergo further differentiation in the CA3 region of the hippocampus. They are integrated into the existing neural circuits to induce plasticity with important functions in cognition such as learning and memory (Goncalves et al., 2016). A small number of NSCs in SGZ can also migrate to the hilus and granular layers of hippocampus to generate oligodendrocytes and astrocytes. In summary, adult NSCs may remain quiescent to maintain a pool of stem cells in the V-SVZ and SGZ regions. They may undergo proliferation, differentiation, migration and integration into existing neural circuits to function as mature neurons. Besides neurons, NSCs may also give rise to glial cells (Figure 1; Figure 2).
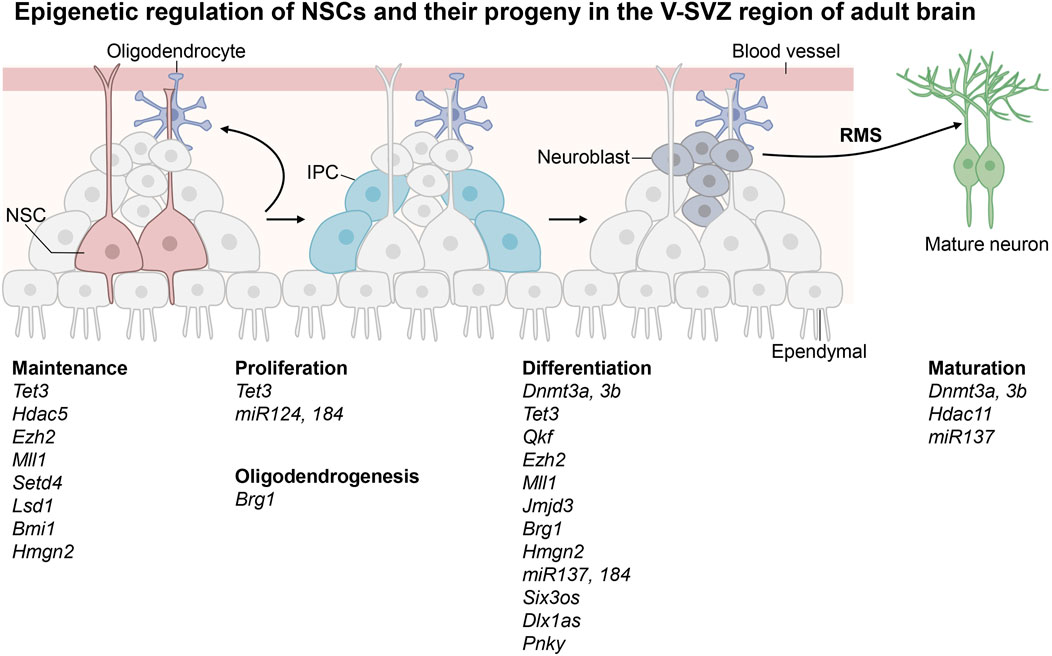
FIGURE 1. Epigenetic regulation of the neural stem cells in the ventricular-subventricular zone (V-SVZ) of adult brain. Adult neural stem cells (NSCs) in the V-SVZ give rise to intermediate progenitor cells (IPCs) before occurrence of neuroblasts. The immature neuroblasts enter rostral migratory stream (RMS) before they turn into mature neurons in the olfactory bulb. The epigenetic regulators shown in this figure are known to play important roles in maintenance, proliferation and differentiation of adult NSCs in the V-SVZ region.
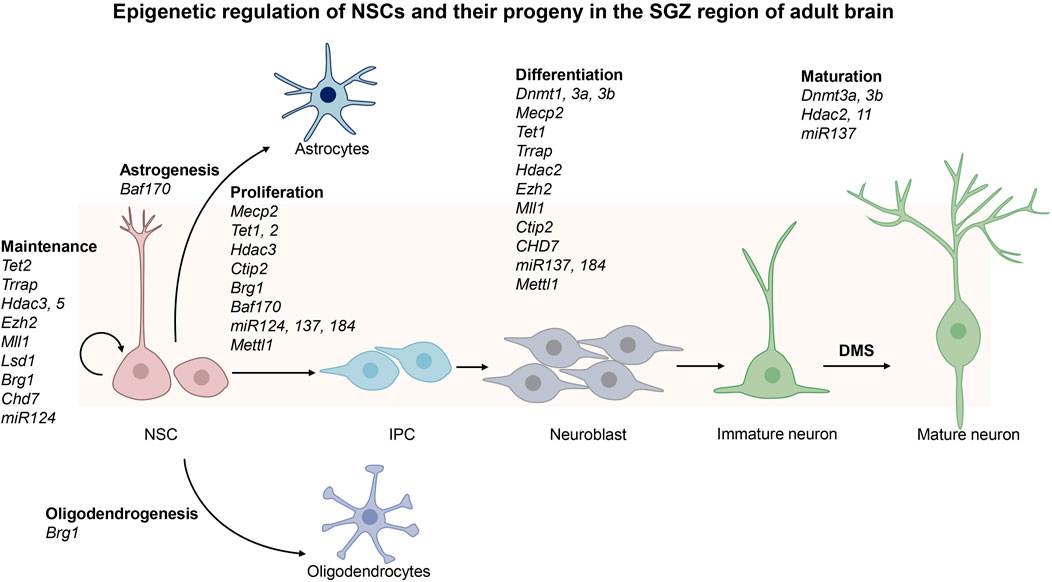
FIGURE 2. Epigenetic regulation of the neural stem cells in the subgranular zone (SGZ) of the adult brain. SGZ is located in the dentate gyrus (DG) of hippocampus. Upon activation, adult neural stem cells (NSCs) in SGZ give rise to intermediate progenitor cells (IPCs), astrocytes and oligodendrocytes. IPCs become neuroblasts, and then immature neurons before they further differentiate into mature neurons after they enter dentate migratory stream (DMS). The epigenetic regulators involved in these processes are shown in the figure.
Balancing maintenance and differentiation of adult NSCs is controlled by both intrinsic and extrinsic signals. Intrinsic signals include some key transcription factors produced in the NSCs and IPCs that are important for maintenance and/or differentiation of these cells, whereas extrinsic signals generally refer to growth factors and neurotrophins secreted in the surrounding niche (Covic et al., 2010; Matsubara et al., 2021). There are a lot of studies indicating the important roles of transcription factors in NSCs and their neural development (Harada et al., 2021; Fong et al., 2022; Guo et al., 2022; Li et al., 2022; Qin et al., 2022; Fan et al., 2023). Epigenetic modifications in response to both kinds of signals are crucial for maintaining the quiescence state of NSCs and dictating their cell lineage differentiation by spatial and temporal regulation in expression of some key factors in the NSCs (Yao et al., 2016). These epigenetic regulations are mediated through DNA methylation, histone modifications, chromatin remodeling, and non-coding RNAs, etc. This review primarily focuses on epigenetic and transcriptional regulations of adult NSCs located in the V-SVZ and SGZ of the adult mouse brain, together with some referenced studies in mouse embryos or cell culture in support of the findings and conclusions in adult mice (see below).
DNA methylation
Cytosine DNA methylation is essential for mammalian development by regulating lineage commitment in cell differentiation in embryos or stem cells (Li and Zhang, 2014; Dor and Cedar, 2018; Zeng and Chen, 2019; Chen and Zhang, 2020). It is altered in many human diseases including neurological diseases (Hamidi et al., 2015; Xie et al., 2023). It occurs primarily at the CpG sites in mammals, with S-adenyl methionine (SAM) as the donor (Zeng and Chen, 2019; Chen and Zhang, 2020). In the human and mouse brains, DNA methylation also occurs at the CpH sites (H = A, C or T), with generally much lower frequencies compared with those of CpG methylation (Guo et al., 2014).
DNA methylation is catalyzed by DNA methyltransferase (DNMT). The mammalian DNMT family mainly consists of DNMT1, DNMT3A and DNMT3B. Among them, DNMT3A and 3B are the major DNMTs for de novo DNA methylation, while DNMT1 is primarily involved in the maintenance DNA methylation (Chen and Zhang, 2020). DNMT1 is highly expressed in the central nervous system (CNS) of both mouse embryos and postnatal mice. It is crucial for neurogenesis as well as survival of newly generated neurons in SGZ although it does not appear to be required for the existing mature neurons (Noguchi et al., 2015; Cui and Xu, 2018). Dnmt3a is expressed in the NSCs of the SVZ of the mouse embryonic brain from E10.5 until E17.5, whereas Dnmt3b expression is detected in the ventricular zone of the brain in mouse embryos from E10.5 to E13.5 (Feng et al., 2005). Expression of both genes decreases postnatally. Nevertheless, loss of DNMT3A results in reduced number of newborn neurons in the SVZ and SGZ regions of the postnatal mouse brain (Wu et al., 2010). DNMT3A is shown to bind to the intergenic regions as well as across the transcribed regions or gene bodies of lowly expressed genes in newborn pups, in which DNA methylation occurs at the CpA sites of these regions that is required for fine-tuning of neuronal subtype-specific transcription in the adult brain (Stroud et al., 2017). DNMT3A and DNMT3B are also associated with the enhancers and gene bodies of the neuronal target genes in adult neurogenesis to establish neuron-specific methylomes and gene expression patterns that are essential for maturation and integration of newborn neurons in the adult brain (Zocher et al., 2021). They do not appear to affect proliferation or cell fate specification of newborn neurons in the adult hippocampus though. Interestingly, growth of dendrites and synaptogenesis are impaired when both DNMT3A and DNMT3B are ablated in adult NSCs, which causes learning and memory defects in the hippocampus (Zocher et al., 2021). In addition, phosphorylation of DNA methylation binding protein MeCP2 by Aurora kinase B is required for balancing proliferation and differentiation of NSCs in the adult brain through NOTCH signaling pathway (Li et al., 2014).
Ten-eleven translocation (TET) proteins are α-ketoglutarate-dependent and Fe2+-dependent dioxygenases that catalyze conversion of 5-methylcytosine (5 mC) to 5-hydroxymethylcytosine (5hmC), which ultimately leads to DNA demethylation (He et al., 2011; Ito et al., 2011; Wu and Zhang, 2017; Xu and Bochtler, 2020). There are three TET proteins in mammals, namely TET1, TET2 and TET3. Although it is much less abundant than 5 mC in most cell types, 5hmC is relatively enriched in the brain (Santiago et al., 2014; MacArthur and Dawlaty, 2021). TET1 may participate in SGZ neurogenesis by regulating NSC proliferation and differentiation as well as cognition in adult mice (Zhang et al., 2013). Knockdown of Tet1 causes promoter hypermethylation in the Dll3 (Delta-like 3) and Notch1 genes that leads to inhibition of NOTCH signaling pathway and results in decreased NSC proliferation (Chen et al., 2021). Similarly, TET1 can also affect neurogenesis in the adult hippocampus by modulating miR-124 expression (Choi et al., 2019). Hippocampal aging is accompanied by reduction in the TET2 protein and 5hmC levels, and Tet2 knockdown causes impairment of neural regeneration and cognitive function. In contrast, restoration of TET2 in the mature adult hippocampus can rescue these brain defects in the hippocampus (Gontier et al., 2018). Loss of TET3 causes proliferation and differentiation defects in neural progenitor cells. Intriguingly, TET3 has been shown to maintain the stem cell pool of the V-SVZ region by binding directly to the Snrpn gene and represses its transcription from the paternal allele (Montalban-Loro et al., 2019).
In mammals, DNA methylation plays an important role in genomic imprinting characterized by parent-of-origin-dependent mono-allelic expression (Li, 2013; Barlow and Bartolomei, 2014; Monk et al., 2019; Tucci et al., 2019; Bartolomei et al., 2020). Genomic imprinting has been implicated in regulating dosage-sensitive gene expression in the neurogenic niche (Montalban-Loro et al., 2015; Perez et al., 2016; Lozano-Urena et al., 2017). So far, about 200 known imprinted genes have been discovered to exhibit parent-of-origin–dependent monoallelic expression patterns (Tucci et al., 2019; Xu et al., 2022). Tissue-specific imprinting has been observed for some imprinted genes such as the Ube3a imprinted gene at the Snrpn imprinted region that shows mono-allelic expression in the brain but not in the other organs (Hsiao et al., 2019; Jiang et al., 2021). It is not expressed in the embryos either. Normally, Dlk1 is almost exclusively expressed from the paternal allele in mouse embryos, whereas it is bi-allelically expressed in the NSCs and niche astrocytes in the adult V-SVZ and SGZ regions (Ferron et al., 2011; Montalban-Loro et al., 2021). The Dlk1 gene expresses multiple transcript isoforms encoding membrane-bound and secreted proteins of DLK1. It is reported that the secreted DLK1 by niche astrocytes interacts with its membrane-bound DLK1 present on NSCs, which is important for neurogenesis and cognition (Montalban-Loro et al., 2021). The imprinted Igf2 gene is expressed solely from the paternal allele in the NSCs of SGZ that encodes an autocrine factor to prevent NSCs from apoptosis. In contrast, biallelic expression of Igf2 is observed in the cerebrospinal fluid and endothelial cells of V-SVZ (Ferron et al., 2015). It is possible that dosage-sensitive transcriptional regulation of Igf2 through its imprinted expression may be crucial for adult neurogenesis. Interestingly, IGF2 is also important for terminal differentiation of NSCs into neurons, astrocytes and oligodendrocytes through regulation of another imprinted gene Cdkn1c encoding a cell-cycle inhibitor p57 (Lozano-Urena et al., 2023). Thus, DNA methylation plays diverse and important roles in the maintenance, proliferation and differentiation of NSCs.
Histone post-translational modification
Histone post-translational modifications (PTMs) refer to the covalent chemical modification on histone proteins, including acetylation, methylation, ubiquitination, phosphorylation, ribosylation, and SUMOylation, etc. (Bannister and Kouzarides, 2011; Suganuma and Workman, 2018; Talbert et al., 2019; Millan-Zambrano et al., 2022). Many transcription factors are known to be important for CNS development and adult neurogenesis. Histone PTMs facilitate recruitment of these transcription factors to their binding sites on chromatin to promote transcriptional activation or repression in the brain (Adam and Harwell, 2020; Chen and Zhang, 2020). Histone acetylation catalyzed by histone acetyltransferases (HATs) is a marker of transcriptional activation, whereas deacetylation catalyzed by histone deacetylases (HDACs) inhibits transcription. QKF, a member of the MYST family of HAT, is highly expressed in the adult V-SVZ. Its loss causes reduced neuroblast migration through RMS, and accordingly fewer interneurons can reach OB (Merson et al., 2006; Sheikh et al., 2012). TRRAP-mediated histone acetylation regulates SP1-dependent transcription in adult neurogenesis, and TRRAP deletion inhibits self-renewal and differentiation potentials of adult NSCs (Yin et al., 2023). Furthermore, activation of adult NSCs is shown to be dependent on regulation of histone acetylation via ATP-citrate lyase (Liu et al., 2023b). In contrast, HDACs remove acetyl groups from histones to repress transcription (Chen and Zhang, 2020; Li et al., 2020). There are eighteen known HDACs that are classified into four groups (Class I, Class II, Class III and Class IV). HDAC2 and HDAC3 belong to class I HDACs. HDAC2 is shown to be required for transcriptional silencing during neuronal differentiation in adult neurogenesis, and its loss results in death of neurons at the maturation stages during adult neurogenesis (Jawerka et al., 2010). HDAC3 regulates proliferation and cell cycle progression in adult neurogenesis mainly through acetylation of G2/M cyclin-dependent kinase 1 (CDK1) (Jiang and Hsieh, 2014). HDAC5 belongs to class II HDACs and it is reported to interact with the orphan nuclear receptor TLX, a transcription factor necessary for NSC proliferation and self-renewal in cell culture (Sun et al., 2007). This interaction causes transcriptional repression of some TLX target genes in cell cycle regulation, including the cyclin-dependent kinase inhibitor p21 and the tumor suppressor gene pten in the NSC cell culture experiments (Sun et al., 2007). Class III HDACs consist of sirtuins such as SIRT1 and SIRT5 that are required for cell fate determination of adult NSCs and maintenance of the nervous system during aging, although the specific mechanism is still unclear (Rafalski and Brunet, 2011; Herskovits and Guarente, 2014; Santos et al., 2021). HDAC11 is the only member of Class IV HDAC family that is currently known to act in neuronal maturation by regulating dendritic length and complexity (Nunez-Alvarez and Suelves, 2022).
Histone methylation modification is catalyzed by histone methyltransferases (HMTs), with methyl group added onto the lysine (K) and arginine (R) residues. These may be either transcriptional activation marks or repression marks (Bedford and Clarke, 2009; Guccione and Richard, 2019; Jambhekar et al., 2019; Morgan and Shilatifard, 2020). Polycomb (PcG) proteins are important for formation of some repressive chromatin marks. There are two types of PcG complexes: polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2) (Almeida et al., 2020; Blackledge and Klose, 2021). Both PRC complexes regulate chromatin accessibility and expression of region-specific transcription factors in NPC differentiation and brain regionalization (Eto and Kishi, 2021). EZH2 is a key member of PRC2 that catalyzes H3K27me3 formation. It is expressed in the V-SVZ and SGZ of adult mice, and loss of EZH2 in these regions affects both differentiation and maintenance of neural stem cells and progenitor cells (Rhodes et al., 2018). MLL1 is an HMT of the Trithorax (TrxG) complex, and it is required for neurogenesis in V-SVZ by regulating transcription factor DLX2 (Lim et al., 2009). MLL1-deficient NSCs in SGZ can survive and proliferate. And they can differentiate into glial cell lineages, but their neuronal differentiation potential is severely impaired. MLL1 is also required for maintaining NSC positional identity through regulation of region-specific transcription factors such as NKX2-1 (Delgado et al., 2020). Without MLL1, expression of some dorsal identity genes increases in the neurons derived from adult NSCs. SETD4, an H4K20me3 writer, maintains the NSC population in the adult brain, and loss of SETD4 leads to depletion of adult NSCs in the mutant mice (Cai et al., 2022). In contrast, lysine-specific demethylase 1 (LSD1) that catalyzes histone lysine demethylation is required for NSC proliferation. It is recruited by TLX to suppress expression of its target genes after removal of activation methylation marks (Sun et al., 2010). JMJD3, a histone H3 lysine 27 (H3K27) demethylase, regulates the Dlx2 enhancer during NSC differentiation (Park et al., 2014).
Other histone modifications also play crucial roles in neurogenesis. BMI1 that is a member of the PcG family proteins and a component of polycomb repressive complex 1 (PRC1), cooperates with RING1, another component of PRC1, in ubiquitination of K119 of H2A. Loss of Bmi1 affects self-renewal of NSCs in the SVZ of adult mice partly through deregulation of cell cycle inhibitors such as p16Ink4a and P19Arf (Pardal et al., 2005). There are some well-documented studies regarding the important functions of other components of PRC1 such as RYBP, YAF2, PCGF5 and PCGF6 in neural differentiation from mouse embryonic stem cells (ESCs) or from human induced pluripotent stem cells (iPSCs) (Yao et al., 2018; Lan et al., 2022; Liu et al., 2023a). It is worth noting that RING1 (also called PCGF1) is required for neuronal subtype specification in the enteric nervous system of adult mouse (Putra et al., 2023). Nevertheless, it awaits further investigation if they may play similar roles in adult NSCs.
Taken together, histone modifications catalyzed by different enzymes are important for adult neurogenesis that can either activate or repress key target genes required for proper neurogenesis.
Chromatin remodeling
Chromatin-remodeling complexes contain ATPases to modulate chromatin structure and gene expression (Becker and Workman, 2013; Hota and Bruneau, 2016; Ahmad et al., 2022). In mammals, they can be divided into four major subfamilies based on the characteristics of their ATPase catalytic domains: BAF (SWI/SNF), ISWI, CHD/NuRD and INO80/SWR (Hota and Bruneau, 2016). Bcl11b/Ctip2 encodes a subunit of BAF regulating survival, differentiation as well as circuit integration of the granule neurons generated from SGZ. Loss of Ctip2 expression in the adult hippocampus and dentate gyrus results in reduced proliferation and differentiation of NSCs (Simon et al., 2012; Simon et al., 2016). Deletion of BRG1 in the BAF family, which directly interacts with the transcription factor PAX6, causes differentiation of adult NSCs into ependymal lineages in the V-SVZ, whereas migrating neuroblasts become glial cells in the RMS during their migration to OB (Ninkovic et al., 2013). BRG1 is also involved in the maintenance and proliferation of hippocampal progenitor cells through regulation of the p53-p21 axis (Petrik et al., 2015). Loss of BAF170, another subunit of the BAF complex, results in premature differentiation of NSCs into astrocytes in the SGZ that causes depletion of NSCs (Tuoc et al., 2017). CHD7, a member of the CHD family, maintains the NSC quiescent status in the hippocampal SGZ through induced expression of Hes5, a target gene of NOTCH signaling (Jones et al., 2015). It is required for expression of SOX4 and SOX11, two transcription factors necessary for neuronal differentiation (Feng et al., 2013). Taken together, chromatin-remodeling complexes are required for neurogenesis by modulating chromatin structure and gene expression.
Nucleosome and chromatin organization
Nucleosome positioning affects transcription. It is also important for neural development. Indeed, high-mobility group nucleosomal binding domain 2 (HMGN2) was shown to be expressed in the SVZ and SGZ regions of adult mouse brain and loss of HMGN2 caused reduced self-renewal and increased differentiation of adult NSCs, which resulted in microcephaly (Gao et al., 2020). There was correlation between nucleosome occupancy and histone modifications in the genome, and there was also an increase in average length of the nucleosomes in the differentiated neuronal cells derived from mouse ESCs (Teif et al., 2012). Nucleosome positioning may be also important for neural differentiation from human iPSCs, with an increase in the number of positioned nucleosomes as well as repositioning of nucleosomes upon differentiation of human iPSCs (Harwood et al., 2019). CTCF is an important regulator in enhancer-promoter interactions and 3D genome organization necessary for proper gene expression (Ghirlando and Felsenfeld, 2016; Chen and Long, 2023). It is required for embryonic neural development and neural differentiation from mouse and human ESCs (Bonev et al., 2017; Modrek et al., 2017; Arzate-Mejia et al., 2018; Pekowska et al., 2018; Kubo et al., 2021; Dong et al., 2022). However, it remains to be tested if similar nucleosome positioning and CTCF-mediated 3D chromatin organization effects may be observed in the process of neural differentiation of the adult NSCs.
Non-coding RNAs
Non-coding RNA (ncRNA) does not appear to encode any long peptide in its sequence (Chen and Rechavi, 2022; Nojima and Proudfoot, 2022; van Zonneveld et al., 2023). However, some ncRNAs have been shown to display important biological functions in many cellular processes including neural development (Soutschek and Schratt, 2023). Huge numbers of ncRNAs have been discovered and millions of ncRNAs may exist in the mammalian genome. There are a few known kinds of ncRNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs).
One of the first discovered ncRNAs is miRNA that contains a small single-stranded RNA molecule with approximately 22 nucleotides in length (Shang et al., 2023). More than one thousand miRNAs have been discovered in mouse or humans. In mammals miRNA usually targets the 3′UTR of mRNAs with imperfect complementary to cause translational inhibition, whereas it may trigger transcriptional repression or RNA degradation with nearly perfect complementation to its targets in plants (Chen and Rechavi, 2022). miR124 is one of highly abundant miRNAs present in the brain (Sun et al., 2013). It targets Sox9, a key transcription factor in neural development, to regulate neural regeneration in the V-SVZ region during the transition from IPCs to neuroblasts (Cheng et al., 2009). EZH2 is targeted by miR137, another miRNA, to regulate NSC proliferation and differentiation which is under the control of MeCP2 and SOX2 (Szulwach et al., 2010). It has been shown that miR137 plays a role in neuronal maturation and dendritic morphogenesis by targeting MIB1 in the ubiquitin-regulated pathway (Smrt et al., 2010). Another miRNA miR184 which is regulated by MBD1 promotes proliferation but inhibits differentiation of NSCs (Liu et al., 2010). A cluster of six miRNAs have been shown to be required for NSC proliferation at the expense of oligodendrocytes (Favaloro et al., 2022).
Generally, lncRNAs are more than 200 nucleotides in length, with no obviously translated protein product (Yao et al., 2019; Andergassen and Rinn, 2022; Nojima and Proudfoot, 2022; Mattick et al., 2023). There are hundreds of thousands of lncRNAs in mammals. The imprinted H19 gene product is the first discovered lncRNA (Brannan et al., 1990; Bartolomei et al., 1991). Xist involved in mammalian X chromosome inactivation is another well-known founding member of lncRNAs (Loda et al., 2022). LncRNAs may regulate gene expression in a tissue-specific pattern (Ernst and Morton, 2013; Zhang et al., 2019; Statello et al., 2021). They are prevalently expressed in the brain that contains the highest number of tissue-specific lncRNAs (Francescatto et al., 2014; Washietl et al., 2014; Ninou et al., 2021). Depletion of two lncRNAs, Six3os and Dlx1as, in the NPCs of the adult V-SVZ region results in increased astrocyte differentiation at the expense of neurons (Ramos et al., 2013). The lncRNA Pnky interacts with PTBP1 to regulate neural differentiation from NSCs in vivo, and Pnky knockdown increases neural commitment in differentiation of NSCs (Ramos et al., 2015). However, further studies are needed to elucidate the molecular mechanisms of lncRNAs in transcriptional regulation of adult NSCs.
RNA methylation
There are already more than 100 different kinds of known RNA modifications. RNA methylation is among the most common RNA modifications that may play important roles in neural development (Yoon et al., 2018). Without an rRNA methyltransferase FBL, neural differentiation and neuronal progression from NSCs is inhibited in mouse embryos because there is reduced translation of EZH2 and KDM6b (Wu et al., 2022). YTHDF2, an m6A reader, is required for self-renewal and neural differentiation of embryonic NSCs in mouse through RNA degradation (Li et al., 2018). It has also been shown that FMRP is a reader for m6A modified mRNAs and promotes their nuclear export in order to fulfill their roles in cell cycle progression and maintenance of neural progenitors derived from NSCs in mouse embryos (Edens et al., 2019). These findings may need to be tested and confirmed in adult NSCs. Interestingly, ablation of METTL1 inhibits m7G RNA methylation and causes reduced hippocampal neurogenesis from NSCs in adult mice (Li et al., 2023). Therefore, at least some RNA methylation seems to be important for adult NSCs.
Perspectives
The stem cell state of adult NSCs is maintained by both intrinsic factors and extrinsic signals. Through spatial and temporal regulation of expression of key transcription factors and signaling pathway modulators, many epigenetic regulators have already been shown to be required for maintenance of NSCs in the adult brain. They also play important roles in balancing proliferation and differentiation of NSCs in adult neural development. Interestingly, the epigenetic modifications established during early development may exert significant influence on neurogenesis in the adult brain. Consistent with this, adult NSCs are thought to be derived from embryonic radial glial (RG) cells and reversibly enter the quiescent state after they exit cell cycle. This also implies that the impact of epigenetic modifications established during early development may need to be taken into account in analyses of the mechanisms underlying maintenance, proliferation and differentiation of adult NSCs.
DNA and RNA methylation, histone modifications, chromatin remodeling, nucleosome positioning, 3D chromatin organization as well as non-coding RNAs may function in distinct pathways to ensure adult neurogenesis to progress in an orderly fashion. As shown in other cells and model systems, these epigenetic modifications do not act alone and may indeed function synergistically in order for adult NSCs to attain various cellular states. The cross-talk among different epigenetic modifications may be important for integration of intrinsic and extrinsic factors in cell fate transition of adult NSCs. Furthermore, there are a lot more epigenetic modifications that have yet to be discovered, and undoubtably some of them may be involved in regulation of adult NSCs. It remains to be explored how other new epigenetic modifications may modulate adult NSCs. It is also important to examine how many transcription factors and epigenetic regulators may share their functions in the adult NSCs in the V-SVZ and SGZ regions (Figure 1; Figure 2). It remains to be tested if these findings may be applicable to adult NSCs in human brains.
Despite much progresses in adult NSCs over the last few decades, it is not that clear how adult NSCs maintain their quiescent cellular state and how external signals and intrinsic factors drive them to re-enter cell cycle and give rise to different cell lineages before integration into the neural circuits. Application of high-throughput technologies in epigenetic research such as RNA-seq, WGBS, ChIP-seq and ATAC-seq, in combination with single cell analyses, will help us to better understand dynamic transcriptional regulation of adult NSCs in their cell fate transition and specification. It is still in the infancy stage to uncover epigenetic regulators and modifications in adult NSCs and their roles in adult neurogenesis and neural plasticity.
Author contributions
JS: Writing–original draft, Writing–review and editing. ZiW: Writing–review and editing. ZhW: Writing–review and editing. GS: Funding acquisition, Writing–review and editing. XL: Writing–review and editing, Conceptualization, Funding acquisition, Supervision, Writing–original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work in the authors’ laboratories has been supported by the grant from Science and Technology Commission of Shanghai Municipality (22WZ2503400) to XL, the grant from Ministry of Science and Technology of the People’s Republic of China (G2023013054) to XL and the major key project of Ningbo medical and health team (2022030107) to GS.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
XL has declared that he is an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adam, M. A., and Harwell, C. C. (2020). Epigenetic regulation of cortical neurogenesis; orchestrating fate switches at the right time and place. Curr. Opin. Neurobiol. 63, 146–153. doi:10.1016/j.conb.2020.03.012
Ahmad, K., Henikoff, S., and Ramachandran, S. (2022). Managing the steady state chromatin landscape by nucleosome dynamics. Annu. Rev. Biochem. 91, 183–195. doi:10.1146/annurev-biochem-032620-104508
Almeida, M., Bowness, J. S., and Brockdorff, N. (2020). The many faces of Polycomb regulation by RNA. Curr. Opin. Genet. Dev. 61, 53–61. doi:10.1016/j.gde.2020.02.023
Andergassen, D., and Rinn, J. L. (2022). From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo. Nat. Rev. Genet. 23, 229–243. doi:10.1038/s41576-021-00427-8
Arzate-Mejia, R. G., Recillas-Targa, F., and Corces, V. G. (2018). Developing in 3D: the role of CTCF in cell differentiation. Development 145, dev137729. doi:10.1242/dev.137729
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi:10.1038/cr.2011.22
Barlow, D. P., and Bartolomei, M. S. (2014). Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, a018382. doi:10.1101/cshperspect.a018382
Bartolomei, M. S., Oakey, R. J., and Wutz, A. (2020). Genomic imprinting: an epigenetic regulatory system. PLoS Genet. 16, e1008970. doi:10.1371/journal.pgen.1008970
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature 351, 153–155. doi:10.1038/351153a0
Becker, P. B., and Workman, J. L. (2013). Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 5, a017905. doi:10.1101/cshperspect.a017905
Bedford, M. T., and Clarke, S. G. (2009). Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13. doi:10.1016/j.molcel.2008.12.013
Blackledge, N. P., and Klose, R. J. (2021). The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833. doi:10.1038/s41580-021-00398-y
Bond, A. M., Ming, G. L., and Song, H. (2021). Ontogeny of adult neural stem cells in the mammalian brain. Curr. Top. Dev. Biol. 142, 67–98. doi:10.1016/bs.ctdb.2020.11.002
Bonev, B., Mendelson Cohen, N., Szabo, Q., Fritsch, L., Papadopoulos, G. L., Lubling, Y., et al. (2017). Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572. doi:10.1016/j.cell.2017.09.043
Brannan, C. I., Dees, E. C., Ingram, R. S., and Tilghman, S. M. (1990). The product of the H19 gene may function as an RNA. Mol. Cell Biol. 10, 28–36. doi:10.1128/mcb.10.1.28
Cai, S. L., Yang, Y. S., Ding, Y. F., Yang, S. H., Jia, X. Z., Gu, Y. W., et al. (2022). SETD4 cells contribute to brain development and maintain adult stem cell reservoir for neurogenesis. Stem Cell Rep. 17, 2081–2096. doi:10.1016/j.stemcr.2022.07.017
Chen, L. F., and Long, H. K. (2023). Topology regulatory elements: from shaping genome architecture to gene regulation. Curr. Opin. Struct. Biol. 83, 102723. doi:10.1016/j.sbi.2023.102723
Chen, W., Liu, N., Shen, S., Zhu, W., Qiao, J., Chang, S., et al. (2021). Fetal growth restriction impairs hippocampal neurogenesis and cognition via Tet1 in offspring. Cell Rep. 37, 109912. doi:10.1016/j.celrep.2021.109912
Chen, X., and Rechavi, O. (2022). Plant and animal small RNA communications between cells and organisms. Nat. Rev. Mol. Cell Biol. 23, 185–203. doi:10.1038/s41580-021-00425-y
Chen, Z., and Zhang, Y. (2020). Role of mammalian DNA methyltransferases in development. Annu. Rev. Biochem. 89, 135–158. doi:10.1146/annurev-biochem-103019-102815
Cheng, L. C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408. doi:10.1038/nn.2294
Choi, C., Kim, T., Chang, K. T., and Min, K. T. (2019). DSCR1-mediated TET1 splicing regulates miR-124 expression to control adult hippocampal neurogenesis. EMBO J. 38, e101293. doi:10.15252/embj.2018101293
Cope, E. C., and Gould, E. (2019). Adult neurogenesis, glia, and the extracellular matrix. Cell Stem Cell 24, 690–705. doi:10.1016/j.stem.2019.03.023
Covic, M., Karaca, E., and Lie, D. C. (2010). Epigenetic regulation of neurogenesis in the adult hippocampus. Hered. (Edinb) 105, 122–134. doi:10.1038/hdy.2010.27
Cui, D., and Xu, X. (2018). DNA methyltransferases, DNA methylation, and age-associated cognitive function. Int. J. Mol. Sci. 19, 1315. doi:10.3390/ijms19051315
Delgado, A. C., Maldonado-Soto, A. R., Silva-Vargas, V., Mizrak, D., von Kanel, T., Tan, K. R., et al. (2021). Release of stem cells from quiescence reveals gliogenic domains in the adult mouse brain. Science 372, 1205–1209. doi:10.1126/science.abg8467
Delgado, R. N., Mansky, B., Ahanger, S. H., Lu, C., Andersen, R. E., Dou, Y., et al. (2020). Maintenance of neural stem cell positional identity by mixed-lineage leukemia 1. Science 368, 48–53. doi:10.1126/science.aba5960
Dong, X., Guo, R., Ji, T., Zhang, J., Xu, J., Li, Y., et al. (2022). YY1 safeguard multidimensional epigenetic landscape associated with extended pluripotency. Nucleic Acids Res. 50, 12019–12038. doi:10.1093/nar/gkac230
Dor, Y., and Cedar, H. (2018). Principles of DNA methylation and their implications for biology and medicine. Lancet 392, 777–786. doi:10.1016/S0140-6736(18)31268-6
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m(6)a-dependent mRNA nuclear export. Cell Rep. 28, 845–854. doi:10.1016/j.celrep.2019.06.072
Ernst, C., and Morton, C. C. (2013). Identification and function of long non-coding RNA. Front. Cell Neurosci. 7, 168. doi:10.3389/fncel.2013.00168
Eto, H., and Kishi, Y. (2021). Brain regionalization by Polycomb-group proteins and chromatin accessibility. Bioessays 43, e2100155. doi:10.1002/bies.202100155
Fan, W., Jurado-Arjona, J., Alanis-Lobato, G., Peron, S., Berger, C., Andrade-Navarro, M. A., et al. (2023). The transcriptional co-activator Yap1 promotes adult hippocampal neural stem cell activation. EMBO J. 42, e110384. doi:10.15252/embj.2021110384
Favaloro, F., DeLeo, A. M., Delgado, A. C., and Doetsch, F. (2022). miR-17-92 exerts stage-specific effects in adult V-SVZ neural stem cell lineages. Cell Rep. 41, 111773. doi:10.1016/j.celrep.2022.111773
Feng, J., Chang, H., Li, E., and Fan, G. (2005). Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79, 734–746. doi:10.1002/jnr.20404
Feng, W., Khan, M. A., Bellvis, P., Zhu, Z., Bernhardt, O., Herold-Mende, C., et al. (2013). The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell 13, 62–72. doi:10.1016/j.stem.2013.05.002
Ferron, S. R., Charalambous, M., Radford, E., McEwen, K., Wildner, H., Hind, E., et al. (2011). Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 475, 381–385. doi:10.1038/nature10229
Ferron, S. R., Radford, E. J., Domingo-Muelas, A., Kleine, I., Ramme, A., Gray, D., et al. (2015). Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat. Commun. 6, 8265. doi:10.1038/ncomms9265
Fong, B. C., Chakroun, I., Iqbal, M. A., Paul, S., Bastasic, J., O'Neil, D., et al. (2022). The Rb/E2F axis is a key regulator of the molecular signatures instructing the quiescent and activated adult neural stem cell state. Cell Rep. 41, 111578. doi:10.1016/j.celrep.2022.111578
Francescatto, M., Vitezic, M., Heutink, P., and Saxena, A. (2014). Brain-specific noncoding RNAs are likely to originate in repeats and may play a role in up-regulating genes in cis. Int. J. Biochem. Cell Biol. 54, 331–337. doi:10.1016/j.biocel.2014.06.014
Gao, X. L., Tian, W. J., Liu, B., Wu, J., Xie, W., and Shen, Q. (2020). High-mobility group nucleosomal binding domain 2 protects against microcephaly by maintaining global chromatin accessibility during corticogenesis. J. Biol. Chem. 295, 468–480. doi:10.1074/jbc.RA119.010616
Ghirlando, R., and Felsenfeld, G. (2016). CTCF: making the right connections. Genes Dev. 30, 881–891. doi:10.1101/gad.277863.116
Goncalves, J. T., Schafer, S. T., and Gage, F. H. (2016). Adult neurogenesis in the Hippocampus: from stem cells to behavior. Cell 167, 897–914. doi:10.1016/j.cell.2016.10.021
Gontier, G., Iyer, M., Shea, J. M., Bieri, G., Wheatley, E. G., Ramalho-Santos, M., et al. (2018). Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981. doi:10.1016/j.celrep.2018.02.001
Guccione, E., and Richard, S. (2019). The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657. doi:10.1038/s41580-019-0155-x
Guo, J. U., Su, Y., Shin, J. H., Shin, J., Li, H., Xie, B., et al. (2014). Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222. doi:10.1038/nn.3607
Guo, N., McDermott, K. D., Shih, Y. T., Zanga, H., Ghosh, D., Herber, C., et al. (2022). Transcriptional regulation of neural stem cell expansion in the adult hippocampus. Elife 11, e72195. doi:10.7554/eLife.72195
Hamidi, T., Singh, A. K., and Chen, T. (2015). Genetic alterations of DNA methylation machinery in human diseases. Epigenomics 7, 247–265. doi:10.2217/epi.14.80
Harada, Y., Yamada, M., Imayoshi, I., Kageyama, R., Suzuki, Y., Kuniya, T., et al. (2021). Cell cycle arrest determines adult neural stem cell ontogeny by an embryonic Notch-nonoscillatory Hey1 module. Nat. Commun. 12, 6562. doi:10.1038/s41467-021-26605-0
Harwood, J. C., Kent, N. A., Allen, N. D., and Harwood, A. J. (2019). Nucleosome dynamics of human iPSC during neural differentiation. EMBO Rep. 20, e46960. doi:10.15252/embr.201846960
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. doi:10.1126/science.1210944
Herskovits, A. Z., and Guarente, L. (2014). SIRT1 in neurodevelopment and brain senescence. Neuron 81, 471–483. doi:10.1016/j.neuron.2014.01.028
Hota, S. K., and Bruneau, B. G. (2016). ATP-dependent chromatin remodeling during mammalian development. Development 143, 2882–2897. doi:10.1242/dev.128892
Hsiao, J. S., Germain, N. D., Wilderman, A., Stoddard, C., Wojenski, L. A., Villafano, G. J., et al. (2019). A bipartite boundary element restricts UBE3A imprinting to mature neurons. Proc. Natl. Acad. Sci. U. S. A. 116, 2181–2186. doi:10.1073/pnas.1815279116
Hsieh, J., and Zhao, X. (2016). Genetics and epigenetics in adult neurogenesis. Cold Spring Harb. Perspect. Biol. 8, a018911. doi:10.1101/cshperspect.a018911
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi:10.1126/science.1210597
Jambhekar, A., Dhall, A., and Shi, Y. (2019). Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625–641. doi:10.1038/s41580-019-0151-1
Jawerka, M., Colak, D., Dimou, L., Spiller, C., Lagger, S., Montgomery, R. L., et al. (2010). The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 6, 93–107. doi:10.1017/S1740925X10000049
Jiang, W., Shi, J., Zhao, J., Wang, Q., Cong, D., Chen, F., et al. (2021). ZFP57 dictates allelic expression switch of target imprinted genes. Proc. Natl. Acad. Sci. U. S. A. 118, e2005377118. doi:10.1073/pnas.2005377118
Jiang, Y., and Hsieh, J. (2014). HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc. Natl. Acad. Sci. U. S. A. 111, 13541–13546. doi:10.1073/pnas.1411939111
Jones, K. M., Saric, N., Russell, J. P., Andoniadou, C. L., Scambler, P. J., and Basson, M. A. (2015). CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33, 196–210. doi:10.1002/stem.1822
Kobayashi, T., and Kageyama, R. (2021). Lysosomes and signaling pathways for maintenance of quiescence in adult neural stem cells. FEBS J. 288, 3082–3093. doi:10.1111/febs.15555
Kubo, N., Ishii, H., Xiong, X., Bianco, S., Meitinger, F., Hu, R., et al. (2021). Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 28, 152–161. doi:10.1038/s41594-020-00539-5
Kuhn, H. G., Toda, T., and Gage, F. H. (2018). Adult hippocampal neurogenesis: a coming-of-age story. J. Neurosci. 38, 10401–10410. doi:10.1523/JNEUROSCI.2144-18.2018
Lan, X., Ding, S., Zhang, T., Yi, Y., Li, C., Jin, W., et al. (2022). PCGF6 controls neuroectoderm specification of human pluripotent stem cells by activating SOX2 expression. Nat. Commun. 13, 4601. doi:10.1038/s41467-022-32295-z
Li, E., and Zhang, Y. (2014). DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 6, a019133. doi:10.1101/cshperspect.a019133
Li, H., Zhong, X., Chau, K. F., Santistevan, N. J., Guo, W., Kong, G., et al. (2014). Cell cycle-linked MeCP2 phosphorylation modulates adult neurogenesis involving the Notch signalling pathway. Nat. Commun. 5, 5601. doi:10.1038/ncomms6601
Li, L., Medina-Menendez, C., Garcia-Corzo, L., Cordoba-Beldad, C. M., Quiroga, A. C., Calleja Barca, E., et al. (2022). SoxD genes are required for adult neural stem cell activation. Cell Rep. 38, 110313. doi:10.1016/j.celrep.2022.110313
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19, 69. doi:10.1186/s13059-018-1436-y
Li, P., Ge, J., and Li, H. (2020). Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 17, 96–115. doi:10.1038/s41569-019-0235-9
Li, Q., Liu, H., Li, L., Guo, H., Xie, Z., Kong, X., et al. (2023). Mettl1-mediated internal m(7)G methylation of Sptbn2 mRNA elicits neurogenesis and anti-alzheimer's disease. Cell Biosci. 13, 183. doi:10.1186/s13578-023-01131-2
Li, X. (2013). Genomic imprinting is a parental effect established in mammalian germ cells. Curr. Top. Dev. Biol. 102, 35–59. doi:10.1016/B978-0-12-416024-8.00002-7
Lim, D. A., and Alvarez-Buylla, A. (2016). The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harb. Perspect. Biol. 8, a018820. doi:10.1101/cshperspect.a018820
Lim, D. A., Huang, Y. C., Swigut, T., Mirick, A. L., Garcia-Verdugo, J. M., Wysocka, J., et al. (2009). Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533. doi:10.1038/nature07726
Liu, C., Teng, Z. Q., Santistevan, N. J., Szulwach, K. E., Guo, W., Jin, P., et al. (2010). Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444. doi:10.1016/j.stem.2010.02.017
Liu, Y., Hu, G., Yang, S., Yao, M., Liu, Z., Yan, C., et al. (2023a). Functional dissection of PRC1 subunits RYBP and YAF2 during neural differentiation of embryonic stem cells. Nat. Commun. 14, 7164. doi:10.1038/s41467-023-42507-9
Liu, Y., Wang, M., Guo, Y., Wang, L., and Guo, W. (2023b). D-2-hydroxyglutarate dehydrogenase governs adult neural stem cell activation and promotes histone acetylation via ATP-citrate lyase. Cell Rep. 42, 112067. doi:10.1016/j.celrep.2023.112067
Loda, A., Collombet, S., and Heard, E. (2022). Gene regulation in time and space during X-chromosome inactivation. Nat. Rev. Mol. Cell Biol. 23, 231–249. doi:10.1038/s41580-021-00438-7
Lozano-Urena, A., Lazaro-Carot, L., Jimenez-Villalba, E., Montalban-Loro, R., Mateos-White, I., Duart-Abadia, P., et al. (2023). IGF2 interacts with the imprinted gene Cdkn1c to promote terminal differentiation of neural stem cells. Development 150, dev200563. doi:10.1242/dev.200563
Lozano-Urena, A., Montalban-Loro, R., Ferguson-Smith, A. C., and Ferron, S. R. (2017). Genomic imprinting and the regulation of postnatal neurogenesis. Brain Plast. 3, 89–98. doi:10.3233/BPL-160041
MacArthur, I. C., and Dawlaty, M. M. (2021). TET enzymes and 5-hydroxymethylcytosine in neural progenitor cell biology and neurodevelopment. Front. Cell Dev. Biol. 9, 645335. doi:10.3389/fcell.2021.645335
Matsubara, S., Matsuda, T., and Nakashima, K. (2021). Regulation of adult mammalian neural stem cells and neurogenesis by cell extrinsic and intrinsic factors. Cells 10, 1145. doi:10.3390/cells10051145
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447. doi:10.1038/s41580-022-00566-8
Merson, T. D., Dixon, M. P., Collin, C., Rietze, R. L., Bartlett, P. F., Thomas, T., et al. (2006). The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 26, 11359–11370. doi:10.1523/JNEUROSCI.2247-06.2006
Millan-Zambrano, G., Burton, A., Bannister, A. J., and Schneider, R. (2022). Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 23, 563–580. doi:10.1038/s41576-022-00468-7
Ming, G. L., and Song, H. (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702. doi:10.1016/j.neuron.2011.05.001
Modrek, A. S., Golub, D., Khan, T., Bready, D., Prado, J., Bowman, C., et al. (2017). Low-grade astrocytoma mutations in IDH1, P53, and ATRX cooperate to block differentiation of human neural stem cells via repression of SOX2. Cell Rep. 21, 1267–1280. doi:10.1016/j.celrep.2017.10.009
Monk, D., Mackay, D. J. G., Eggermann, T., Maher, E. R., and Riccio, A. (2019). Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 20, 235–248. doi:10.1038/s41576-018-0092-0
Montalban-Loro, R., Lassi, G., Lozano-Urena, A., Perez-Villalba, A., Jimenez-Villalba, E., Charalambous, M., et al. (2021). Dlk1 dosage regulates hippocampal neurogenesis and cognition. Proc. Natl. Acad. Sci. U. S. A. 118, e2015505118. doi:10.1073/pnas.2015505118
Montalban-Loro, R., Domingo-Muelas, A., Bizy, A., and Ferron, S. R. (2015). Epigenetic regulation of stemness maintenance in the neurogenic niches. World J. Stem Cells 7, 700–710. doi:10.4252/wjsc.v7.i4.700
Montalban-Loro, R., Lozano-Urena, A., Ito, M., Krueger, C., Reik, W., Ferguson-Smith, A. C., et al. (2019). TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat. Commun. 10, 1726. doi:10.1038/s41467-019-09665-1
Morgan, M. A. J., and Shilatifard, A. (2020). Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 52, 1271–1281. doi:10.1038/s41588-020-00736-4
Ninkovic, J., Steiner-Mezzadri, A., Jawerka, M., Akinci, U., Masserdotti, G., Petricca, S., et al. (2013). The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell 13, 403–418. doi:10.1016/j.stem.2013.07.002
Ninou, E., Michail, A., and Politis, P. K. (2021). Long non-coding RNA lacuna regulates neuronal differentiation of neural stem cells during brain development. Front. Cell Dev. Biol. 9, 726857. doi:10.3389/fcell.2021.726857
Noguchi, H., Kimura, A., Murao, N., Matsuda, T., Namihira, M., and Nakashima, K. (2015). Expression of DNMT1 in neural stem/precursor cells is critical for survival of newly generated neurons in the adult hippocampus. Neurosci. Res. 95, 1–11. doi:10.1016/j.neures.2015.01.014
Nojima, T., and Proudfoot, N. J. (2022). Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 23, 389–406. doi:10.1038/s41580-021-00447-6
Nunez-Alvarez, Y., and Suelves, M. (2022). HDAC11: a multifaceted histone deacetylase with proficient fatty deacylase activity and its roles in physiological processes. FEBS J. 289, 2771–2792. doi:10.1111/febs.15895
Pardal, R., Molofsky, A. V., He, S., and Morrison, S. J. (2005). Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb. Symp. Quant. Biol. 70, 177–185. doi:10.1101/sqb.2005.70.057
Park, D. H., Hong, S. J., Salinas, R. D., Liu, S. J., Sun, S. W., Sgualdino, J., et al. (2014). Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 8, 1290–1299. doi:10.1016/j.celrep.2014.07.060
Pekowska, A., Klaus, B., Xiang, W., Severino, J., Daigle, N., Klein, F. A., et al. (2018). Gain of CTCF-anchored chromatin loops marks the exit from naive pluripotency. Cell Syst. 7, 482–495. doi:10.1016/j.cels.2018.09.003
Perez, J. D., Rubinstein, N. D., and Dulac, C. (2016). New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu. Rev. Neurosci. 39, 347–384. doi:10.1146/annurev-neuro-061010-113708
Petrik, D., Latchney, S. E., Masiulis, I., Yun, S., Zhang, Z., Wu, J. I., et al. (2015). Chromatin remodeling factor Brg1 supports the early maintenance and late responsiveness of nestin-lineage adult neural stem and progenitor cells. Stem Cells 33, 3655–3665. doi:10.1002/stem.2215
Putra, B. P., Ito, K., Cirillo, C., Sunardi, M., Koseki, H., Uesaka, T., et al. (2023). Pcgf1 gene disruption reveals primary involvement of epigenetic mechanism in neuronal subtype specification in the enteric nervous system. Dev. Growth Differ. 65, 461–469. doi:10.1111/dgd.12880
Qin, S., Yuan, Y., Huang, X., Tan, Z., Hu, X., Liu, H., et al. (2022). Topoisomerase IIA in adult NSCs regulates SVZ neurogenesis by transcriptional activation of Usp37. Nucleic Acids Res. 50, 9319–9338. doi:10.1093/nar/gkac731
Rafalski, V. A., and Brunet, A. (2011). Energy metabolism in adult neural stem cell fate. Prog. Neurobiol. 93, 182–203. doi:10.1016/j.pneurobio.2010.10.007
Ramos, A. D., Andersen, R. E., Liu, S. J., Nowakowski, T. J., Hong, S. J., Gertz, C., et al. (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. doi:10.1016/j.stem.2015.02.007
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K. Y., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi:10.1016/j.stem.2013.03.003
Rhodes, C. T., Zunino, G., Huang, S. A., Cardona, S. M., Cardona, A. E., Berger, M. S., et al. (2018). Region specific knock-out reveals distinct roles of chromatin modifiers in adult neurogenic niches. Cell Cycle 17, 377–389. doi:10.1080/15384101.2018.1426417
Santiago, M., Antunes, C., Guedes, M., Sousa, N., and Marques, C. J. (2014). TET enzymes and DNA hydroxymethylation in neural development and function - how critical are they? Genomics 104, 334–340. doi:10.1016/j.ygeno.2014.08.018
Santos, S. S., Moreira, J. B., Costa, M., Rodrigues, R. S., Sebastião, A. M., Xapelli, S., et al. (2021). The mitochondrial antioxidant Sirtuin3 cooperates with lipid metabolism to safeguard neurogenesis in aging and depression. Cells 11, 90. doi:10.3390/cells11010090
Shang, R., Lee, S., Senavirathne, G., and Lai, E. C. (2023). microRNAs in action: biogenesis, function and regulation. Nat. Rev. Genet. 24, 816–833. doi:10.1038/s41576-023-00611-y
Sheikh, B. N., Dixon, M. P., Thomas, T., and Voss, A. K. (2012). Querkopf is a key marker of self-renewal and multipotency of adult neural stem cells. J. Cell Sci. 125, 295–309. doi:10.1242/jcs.077271
Simon, R., Baumann, L., Fischer, J., Seigfried, F. A., De Bruyckere, E., Liu, P., et al. (2016). Structure-function integrity of the adult hippocampus depends on the transcription factor Bcl11b/Ctip2. Genes Brain Behav. 15, 405–419. doi:10.1111/gbb.12287
Simon, R., Brylka, H., Schwegler, H., Venkataramanappa, S., Andratschke, J., Wiegreffe, C., et al. (2012). A dual function of Bcl11b/Ctip2 in hippocampal neurogenesis. EMBO J. 31, 2922–2936. doi:10.1038/emboj.2012.142
Smrt, R. D., Szulwach, K. E., Pfeiffer, R. L., Li, X., Guo, W., Pathania, M., et al. (2010). MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070. doi:10.1002/stem.431
Soutschek, M., and Schratt, G. (2023). Non-coding RNA in the wiring and remodeling of neural circuits. Neuron 111, 2140–2154. doi:10.1016/j.neuron.2023.04.031
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Stroud, H., Su, S. C., Hrvatin, S., Greben, A. W., Renthal, W., Boxer, L. D., et al. (2017). Early-life gene expression in neurons modulates lasting epigenetic states. Cell 171, 1151–1164. doi:10.1016/j.cell.2017.09.047
Suganuma, T., and Workman, J. L. (2018). Chromatin and metabolism. Annu. Rev. Biochem. 87, 27–49. doi:10.1146/annurev-biochem-062917-012634
Sun, A. X., Crabtree, G. R., and Yoo, A. S. (2013). MicroRNAs: regulators of neuronal fate. Curr. Opin. Cell Biol. 25, 215–221. doi:10.1016/j.ceb.2012.12.007
Sun, G., Alzayady, K., Stewart, R., Ye, P., Yang, S., Li, W., et al. (2010). Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell Biol. 30, 1997–2005. doi:10.1128/MCB.01116-09
Sun, G., Yu, R. T., Evans, R. M., and Shi, Y. (2007). Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl. Acad. Sci. 104, 15282–15287. doi:10.1073/pnas.0704089104
Szulwach, K. E., Li, X., Smrt, R. D., Li, Y., Luo, Y., Lin, L., et al. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141. doi:10.1083/jcb.200908151
Talbert, P. B., Meers, M. P., and Henikoff, S. (2019). Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat. Rev. Genet. 20, 283–297. doi:10.1038/s41576-019-0105-7
Teif, V. B., Vainshtein, Y., Caudron-Herger, M., Mallm, J. P., Marth, C., Hofer, T., et al. (2012). Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 19, 1185–1192. doi:10.1038/nsmb.2419
Tucci, V., Isles, A. R., Kelsey, G., Ferguson-Smith, A. C., and Erice Imprinting, G. (2019). Genomic imprinting and physiological processes in mammals. Cell 176, 952–965. doi:10.1016/j.cell.2019.01.043
Tuoc, T., Dere, E., Radyushkin, K., Pham, L., Nguyen, H., Tonchev, A. B., et al. (2017). Ablation of BAF170 in developing and postnatal dentate gyrus affects neural stem cell proliferation, differentiation, and learning. Mol. Neurobiol. 54, 4618–4635. doi:10.1007/s12035-016-9948-5
van Zonneveld, A. J., Zhao, Q., Rotmans, J. I., and Bijkerk, R. (2023). Circulating non-coding RNAs in chronic kidney disease and its complications. Nat. Rev. Nephrol. 19, 573–586. doi:10.1038/s41581-023-00725-w
Vicidomini, C., Guo, N., and Sahay, A. (2020). Communication, cross talk, and signal integration in the adult hippocampal neurogenic niche. Neuron 105, 220–235. doi:10.1016/j.neuron.2019.11.029
Washietl, S., Kellis, M., and Garber, M. (2014). Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 24, 616–628. doi:10.1101/gr.165035.113
Wu, H., Coskun, V., Tao, J., Xie, W., Ge, W., Yoshikawa, K., et al. (2010). Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–448. doi:10.1126/science.1190485
Wu, Q., Shichino, Y., Abe, T., Suetsugu, T., Omori, A., Kiyonari, H., et al. (2022). Selective translation of epigenetic modifiers affects the temporal pattern and differentiation of neural stem cells. Nat. Commun. 13, 470. doi:10.1038/s41467-022-28097-y
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534. doi:10.1038/nrg.2017.33
Xie, J., Xie, L., Wei, H., Li, X. J., and Lin, L. (2023). Dynamic regulation of DNA methylation and brain functions. Biol. (Basel) 12, 152. doi:10.3390/biology12020152
Xu, G. L., and Bochtler, M. (2020). Reversal of nucleobase methylation by dioxygenases. Nat. Chem. Biol. 16, 1160–1169. doi:10.1038/s41589-020-00675-5
Xu, Z., Shi, J., Zhang, Y., Liu, Y., Zhao, J., Chen, Q., et al. (2022). Zfp57 exerts maternal and sexually dimorphic effects on genomic imprinting. Front. Cell Dev. Biol. 10, 784128. doi:10.3389/fcell.2022.784128
Yao, B., Christian, K. M., He, C., Jin, P., Ming, G. L., and Song, H. (2016). Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537–549. doi:10.1038/nrn.2016.70
Yao, M., Zhou, X., Zhou, J., Gong, S., Hu, G., Li, J., et al. (2018). PCGF5 is required for neural differentiation of embryonic stem cells. Nat. Commun. 9, 1463. doi:10.1038/s41467-018-03781-0
Yao, R. W., Wang, Y., and Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551. doi:10.1038/s41556-019-0311-8
Yin, B. K., Lazaro, D., and Wang, Z. Q. (2023). TRRAP-mediated acetylation on Sp1 regulates adult neurogenesis. Comput. Struct. Biotechnol. J. 21, 472–484. doi:10.1016/j.csbj.2022.12.024
Yoon, K. J., Vissers, C., Ming, G. L., and Song, H. (2018). Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence. J. Cell Biol. 217, 1901–1914. doi:10.1083/jcb.201802117
Zeng, Y., and Chen, T. (2019). DNA methylation reprogramming during mammalian development. Genes (Basel) 10, 257. doi:10.3390/genes10040257
Zhang, R. R., Cui, Q. Y., Murai, K., Lim, Y. C., Smith, Z. D., Jin, S., et al. (2013). Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245. doi:10.1016/j.stem.2013.05.006
Zhang, X., Wang, W., Zhu, W., Dong, J., Cheng, Y., Yin, Z., et al. (2019). Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 20, 5573. doi:10.3390/ijms20225573
Keywords: neural stem cell (NSC), epigenetic, transcription, adult brain, DNA methylation
Citation: Shi J, Wang Z, Wang Z, Shao G and Li X (2024) Epigenetic regulation in adult neural stem cells. Front. Cell Dev. Biol. 12:1331074. doi: 10.3389/fcell.2024.1331074
Received: 31 October 2023; Accepted: 12 January 2024;
Published: 31 January 2024.
Edited by:
Noriaki Sasai, Nara Institute of Science and Technology (NAIST), JapanReviewed by:
Yuyao Tian, Massachusetts General Hospital and Harvard Medical School, United StatesKaimeng Huang, Dana–Farber Cancer Institute, United States
Copyright © 2024 Shi, Wang, Wang, Shao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Wang, MTYzNTI5MTIyNkBxcS5jb20=; Guofeng Shao, c2dmMTk1OEBzaW5hLmNvbQ==; Xiajun Li, bGl4ajFAc2hhbmdoYWl0ZWNoLmVkdS5jbg==
 Jiajia Shi
Jiajia Shi Zilin Wang
Zilin Wang Zhijun Wang2*
Zhijun Wang2* Guofeng Shao
Guofeng Shao Xiajun Li
Xiajun Li