- 1Central Laboratory, Guangzhou Panyu Central Hospital, Guangzhou, China
- 2Rehabilitation Medicine Institute of Panyu District, Guangzhou, China
- 3Department of General Surgery, Guangzhou Panyu Central Hospital, Guangzhou, China
- 4Department of Gynaecology and Obstetrics, Guangzhou Panyu Central Hospital, Guangzhou, China
- 5Clinical Laboratory, Guangzhou Panyu Central Hospital, Guangzhou, China
- 6He Xian Memorial Hospital, Southern Medical University, Guangzhou, China
Cell death is ubiquitous during development and throughout life and is a genetically determined active and ordered process that plays a crucial role in regulating homeostasis. Cell death includes regulated cell death and non-programmed cell death, and the common types of regulatory cell death are necrosis, apoptosis, necroptosis, autophagy, ferroptosis, and pyroptosis. Apoptosis, Necrosis and necroptosis are more common than autophagy, ferroptosis and pyroptosis among cell death. Non-coding RNAs are regulatory RNA molecules that do not encode proteins and include mainly microRNAs, long non-coding RNAs, and circular RNAs. Non-coding RNAs can act as oncogenes and tumor suppressor genes, with significant effects on tumor occurrence and development, and they can also regulate tumor cell autophagy, ferroptosis, and pyroptosis at the transcriptional or post-transcriptional level. This paper reviews the recent research progress on the effects of the non-coding RNAs involved in autophagy, ferroptosis, and pyroptosis on tumorigenesis, tumor development, and treatment, and looks forward to the future direction of this field, which will help to elucidate the molecular mechanisms of tumorigenesis and tumor development, as well as provide a new vision for the treatment of tumors.
Background
Cell death is an irreversible process that plays an important role in normal development and inhibits the rapid growth of tumor cells (Mahapatra et al., 2021; Patra et al., 2022). Cell death includes regulatory cell death and non-programmed cell death. Regulatory cell death is ubiquitous during development and is an active and orderly process determined by genes that plays an important role in maintaining homeostasis (Chen L. et al., 2021). Non-coding RNAs are RNA molecules that do not encode proteins, including mainly microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). Non-coding RNAs participate in protein, DNA, and RNA interactions and have roles in a variety of cellular activities, including gene activation and silencing, RNA splicing, modification, and editing, and protein translation (Bahreini et al., 2021). An increasing number of studies have demonstrated that non-coding RNAs regulate tumor cell death by acting directly on cell death-related proteins or acting indirectly on downstream targets or pathways. In recent years, tremendous progress has been made in the study of the molecular mechanisms underlying the regulation of tumor cell death by non-coding RNAs, which is of great significance for elucidating the mechanisms of tumorigenesis and tumor development and guiding the treatment and intervention of tumors based on cell death. This article reviews the progress of research from this aspect.
Classification and regulatory mechanisms of cell death
At present, the common types of regulatory cell death are autophagy, ferroptosis, and pyroptosis (Buchser et al., 2012; Green, 2019). The differences in these processes are shown in Supplementary Table S1 (Additional File S1). Autophagy is a highly conserved self-digestion process in eukaryotic cells that extensively regulates cell growth, development, senescence, and death. In some cases, autophagy acts on cell death as autophagic cell death. Autophagy is the process by which cells use lysosomes to degrade damaged organelles and macromolecular substances to maintain homeostasis. The cells first form a monolayer or bilayer membrane that develops into a vesicle-like autophagosome, which then fuses with a lysosome to form an autolysosome, leading to the lysosomal hydrolase degradation of the contents and the recycling of the products (New and Tooze, 2019; Chen et al., 2022), as shown in Supplementary Figure S1 (Additional File S2).
Ferroptosis is a newly identified form of regulatory cell death caused by the abnormal accumulation of lipid reactive oxygen species (ROS). Ferroptosis and iron metabolism, glutathione metabolism, and lipid peroxidation are closely related, Thus, a large number of molecules have roles in ferroptosis, including transferrin receptor 1, ferritin, cystine/glutamate reverse transporter, glutathione peroxidase 4 (GPX4), and lipoxygenase (Rochette et al., 2022). The regulatory mechanisms of ferroptosis are described in Supplementary Figure S2 (Additional File S3).
Pyroptosis is a novel form of programmed cell death that depends mainly on the cleavage and activation of gasdermin protein by the inflammasome-activating caspase family of proteins, which then translocates to the membrane to form holes, leading to cell swelling, cytosolic outflow, and cell membrane rupture (Zhang et al. 2021c). The regulatory mechanism of pyroptosis is shown in Supplementary Figure S3 (Additional File S4).
Features and functions of non-coding RNAs
Non-coding RNAs are transcribed, but are not translated into a protein, and include mainly miRNAs, lncRNAs, and circRNAs (Bahreini et al., 2021). The features and functions of miRNAs, lncRNAs, and circRNAs are summarized in Supplementary Table S2 (Additional File S1). miRNAs are an endogenous class of RNA molecules that are 21–25 nucleotides in length. miRNAs regulate various metabolic pathways at the transcription and translation levels and have key roles in regulating tumor cell growth, migration, invasion, and chemoresistance (Ambros, 2001). lncRNAs are RNA molecules greater than 200 nucleotides in length, with or without an open reading frame, that are involved in transcriptional silencing and activation, chromosome modification, and nuclear transport (Anastasiadou et al., 2018). circRNAs are non-coding RNA molecules formed by reverse splicing that are stable and not easy to degrade. circRNAs are derived from introns or exons, can act as RNA sponges, and can also be combined with proteins, so as to participate in the regulation of gene expression and affect protein function (Zheng et al., 2017).
Role of miRNAs in tumor cell death for tumorigenesis, development, and treatment
miRNAs and autophagy
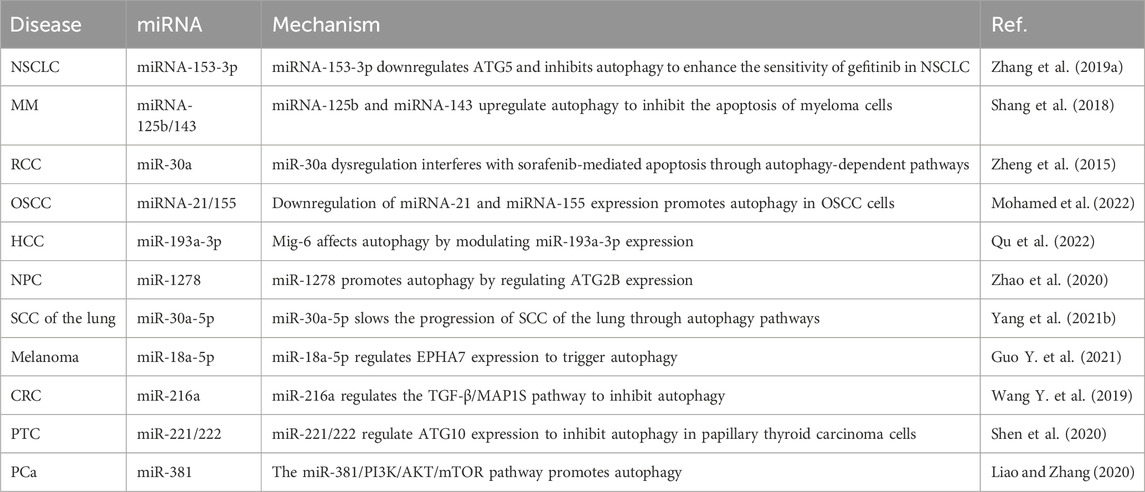
In recent years, miRNAs have been shown to be involved mainly in influencing the role of autophagy in lung cancer, multiple myeloma, kidney cancer, oral cancer, liver cancer, nasopharyngeal carcinoma (NPC), melanoma, thyroid cancer, and prostate cancer (PCa), as summarized in Table 1. For example, miRNA-153-3p expression is low in gefitinib-resistant non-small cell lung cancer (NSCLC) cell lines, and its expression level is negatively correlated with that of the autophagic activity marker tubulin 1 light chain 3. miRNA-153-3p overexpression inhibits the occurrence of autophagy and enhances sensitivity to gefitinib by downregulating the expression of autophagy-related 5 homolog (ATG5) (Zhang et al., 2019a). The expression levels of miRNA-125b and miRNA-143 are significantly correlated with the concentrations of beta-2-microglobulin, albumin, and hemoglobin. The upregulation of miRNA-125b and downregulation of miRNA-143 both promote autophagy, thus inhibiting apoptosis in myeloma cells (Shang et al., 2018). miR-30a inhibits autophagy in renal cancer cells by downregulating the expression of Beclin-1 and interferes with sorafenib-mediated cell apoptosis through an autophagy-dependent pathway (Zheng et al., 2015). Isoliquiritigenin promotes apoptosis and autophagy in oral squamous cell carcinoma (OSCC) cells by downregulating the expression of miRNA-21 and miRNA-155, which provides new targets for the treatment of OSCC (Mohamed et al., 2022). Mitogen-inducible gene 6 (Mig-6) promotes apoptosis and autophagy in hepatocellular carcinoma (HCC) cells by regulating the expression of miR-193a-3p (Qu et al., 2022). miR-1278 specifically regulates the expression of ATG2B to promote autophagy in NPC cells, thereby increasing the chemical resistance of NPC cells to cisplatin; miR-1278 may become a novel therapeutic target for the treatment of NPC (Zhao et al., 2020). miR-30a-5p inhibits ATG5-mediated autophagy in lung SCC cells and slows the progression of lung SCC through the autophagy pathway (Yang et al., 2021b). miR-18a-5p expression is significantly increased in melanoma tissues and cell lines, and the expression of ephrin type-A receptor 7 (EPHA7) is negatively regulated to promote the proliferation of melanoma cells and inhibits apoptosis and autophagy, which provide clues to reveal the pathogenesis of miRNA-mediated melanoma (Guo Y. et al., 2021). miR-216a expression is downregulated in colorectal cancer (CRC) tissues, and autophagy is inhibited by the transforming growth factor beta 1 (TGF-β)/microtubule-associated protein 1S (MAP1S) pathway (Wang Y. et al., 2019). The high expression of miR-221/222 is associated with the regional lymph node and distant metastasis stage of papillary thyroid carcinoma, and miR-221/222 target ATG10 expression, thus inhibiting autophagy and apoptosis in papillary thyroid carcinoma cells (Shen et al., 2020). In PCa cells, the upregulation of miR-381 expression suppresses the expression of recombinant reelin and then suppresses the activation of the PI3K/AKT/mammalian target of rapamycin (mTOR) signaling pathway to promote apoptosis and autophagy (Liao and Zhang, 2020).
miRNAs and ferroptosis
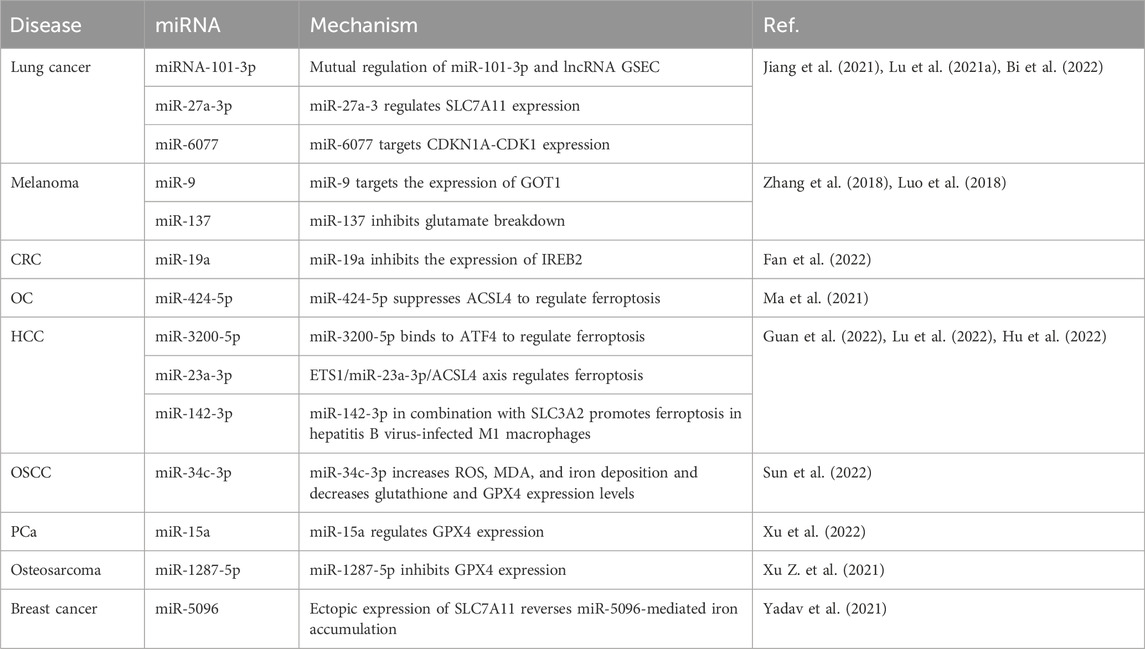
Studies of miRNAs involved in the effects of ferroptosis on the occurrence, development, and treatment of lung cancer, melanoma, CRC, ovarian cancer, HCC, OSCC, PCa, osteosarcoma, and breast cancer are summarized in Table 2. For example, miRNA-101-3p is downregulated in lung adenocarcinoma, and miR-101-3p and the lncRNA G-tetra-chain body formation sequence (GSEC) regulate each other to promote ferroptosis in lung adenocarcinoma cells (Jiang et al., 2021). miR-27a-3p directly targets the expression of solute carrier family 7 member 11 (SLC7A11) in NSCLC cells to regulate ferroptosis (Lu et al., 2021a). miR-6077 regulates KEAP1-NRF2(nuclear factor erythroid 2-related factor)-SLC7A11-NQO1 (NAD (P)H quinone dehydrogenase 1)-mediated ferroptosis to protect lung adenocarcinoma cells from cisplatin/pemetrexed-induced cell death, thereby increasing their resistance to chemotherapeutic drugs (Bi et al., 2022). In melanoma cells, miR-9 acts as an important regulator of ferroptosis, and its overexpression inhibits glutamic-oxaloacetic transaminase 1 (GOT1) expression and reduces ferroptosis induced by erastin and RAS-selective lethal 3 (RSL3), an inhibitor of GPX4 (Zhang et al., 2018). miR-137 negatively regulates ferroptosis by directly targeting the binding glutamine transporter SLC1A5 in melanoma cells, pointing the way for potential therapies for melanoma (Luo et al., 2018). miR-19a promotes the proliferation and invasion of CRC cells and suppresses ferroptosis by inhibiting the expression of iron-response element-binding protein 2 (IREB2) (Fan et al., 2022). Upregulation of miR-424-5p suppresses acyl-CoA synthetase long chain family member 4 (ACSL4) expression, thereby reducing ferroptosis induced by erastin and RSL3, while downregulation of miR-424-5p increases the sensitivity of ovarian cancer cells to erastin and RSL3 (Ma et al., 2021). The expression of miR-3200-5p, which binds to activated transcription factor 4 (ATF4), regulates ferroptosis and inhibits the proliferation and metastasis of HCC cells (Guan et al., 2022). The ETS proto-oncogene 1, transcription factor (ETS1)/miR-23a-3p/ACSL4 axis increases the resistance of liver cancer cells to sorafenib by regulating ferroptosis, and miR-23a-3p may be a potential target for improving the sensitivity of HCC to sorafenib (Lu et al., 2022). miR-142-3p binds to SLC7A5 and promotes ferroptosis in hepatitis B virus-infected M1 macrophages, thereby accelerating the progression of HCC (Hu et al., 2022). In the OSCC SCC-25 cell line, miR-34c-3p overexpression suppresses cell proliferation and increases ROS, malondialdehyde (MDA), and iron concentrations, and decreases glutathione and GPX4 expression levels to promote ferroptosis (Sun et al., 2022). miR-15a overexpression and decreased GPX4 expression inhibit the proliferation of PCa cells and increase intracellular iron and ROS accumulation, and miR-15a induces ferroptosis by regulating GPX4 expression (Xu et al., 2022). The upregulation of miR-1287-5p expression increases the sensitivity of osteosarcoma cells to cisplatin chemotherapy by inhibiting GPX4 expression (Xu Z. et al., 2021). The ectopic expression of SLC7A11 partially reverses miR-5096-mediated ROS, lipid peroxidation, iron accumulation, GSH, hydroxyl radicals, mitochondrial membrane potential, and colony formation in breast cancer cells (Yadav et al., 2021).
miRNAs and pyroptosis
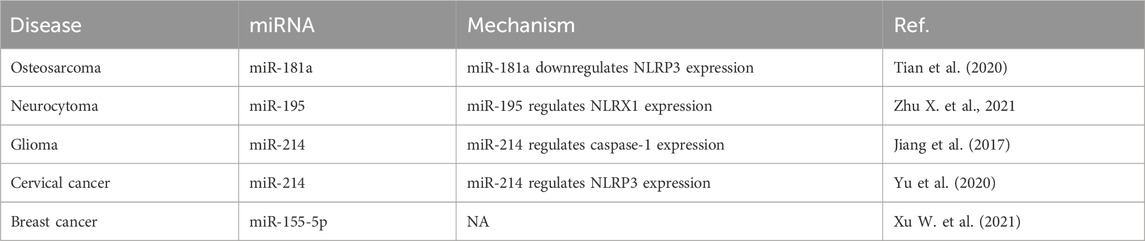
Studies of the roles of miRNAs in pyroptosis for the development of osteosarcoma, neurocytoma, glioma, cervical cancer, and breast cancer are summarized in Table 3. For example, the downregulation of miR-181a expression in osteosarcoma cells activates the expression of nucleotide-binding oligomerization domain-like receptor pyrin domain-containing 3 (NLRP3) to promote pyroptosis and inhibit tumor growth (Tian et al., 2020). miR-195 expression is significantly downregulated in neuroblastoma SH-SY5Y cells infected with enterovirus A71, and miR-195 binds to NLR family member X1 and NLRs to regulate the pyroptosis induced by enterovirus A71 infection (Zhu X. et al., 2021). In glioma tissues as well as the glioblastoma cell lines U87 and T98G, caspase-1 expression is increased while miR-214 expression is significantly downregulated, and miR-214 regulates caspase 1-mediated pyroptosis to inhibit cell proliferation and migration, providing a novel intervention for glioma (Jiang et al., 2017). miR-214 expression is significantly downregulated in cervical cancer cell lines and regulates NLRP3 expression, which induces pyroptosis (Yu et al., 2020). miRNA-155-5p expression is upregulated in triple-negative breast cancer cells, while downregulation of its expression induces pyroptosis and enhances the role of cetuximab in the MDA-MB-468 xenograft model (Xu W. et al., 2021).
Role of lncRNAs in tumor cell death for tumorigenesis, development, and therapy
lncRNAs and autophagy
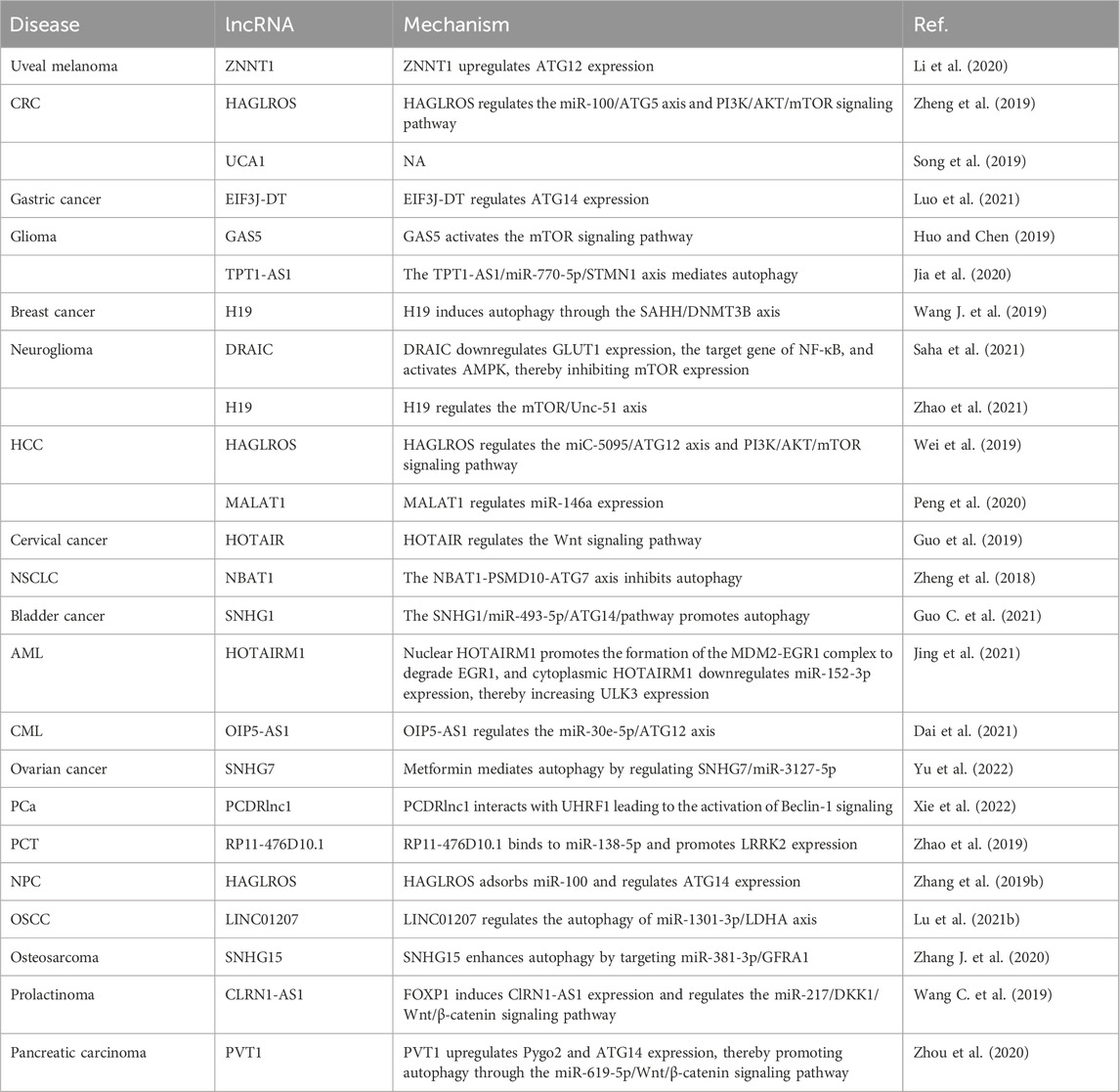
lncRNAs are involved mainly in autophagy affecting uveal melanoma, CRC, gastric cancer, glioma, breast cancer, glioblastoma, glioma, hepatoma, HCC, NSCLC, bladder cancer, acute myelogenous leukemia (AML), chronic myelogenous leukemia (CML), ovarian cancer, PCa, thyroid papillary carcinoma, NPC, OSCC, osteosarcoma, prolactinoma, and pancreatic cancer, as summarized in Table 4. For example, the lncRNA ZNF706 neighboring transcript 1 (ZNNT1) is expressed at a low level in uveal melanoma cells, and ZNNT1 promotes autophagy by upregulating ATG12 expression, and thus has a tumor suppressive effect (Li et al., 2020).
The high expression of the lncRNA HAGLR complementary chain (HAGLROS) is associated with the shortened survival time of patients with CRC; HAGLROS induces CRC cell apoptosis and suppresses autophagy through the miR-100/ATG5 and PI3K/AKT/mTOR pathways (Zheng et al., 2019). Downregulation of lncRNA urothelial cancer-associated 1 (UCA1) expression suppresses CRC cell proliferation and autophagy (Song et al., 2019). EIF3J divergent transcript (EIF3J-DT) is highly expressed in gastric cancer cells with chemotherapeutic drugs, and EIF3J-DT specifically regulates ATG14 expression, activates autophagy, and induces drug resistance in gastric cancer cells (Luo et al., 2021). The lncRNA growth arrest-specific 5 (GAS5) is expressed in the U138 and LN18 cell lines, where it induces cisplatin sensitivity through the activation of mTOR signaling and thereby inhibits autophagy (Huo and Chen, 2019). The expression of tumor protein translation control 1 antisense RNA 1 (TPT1-AS1) is upregulated in glioma cells, and the TPT1-AS1/miR-770-5p/stathmin 1 (STMN1) axis mediates autophagy in glioma cells, which is important for the targeted treatment of glioma (Jia et al., 2020). Downregulation of H19 imprinted maternally expressed transcript (H19) expression inhibits autophagy in tamoxifen-resistant MCF7 cells; H19 induces autophagy through the S-adenosyl-L-homocysteine hydrolase (SAHH)/DNA methyltransferase 3 beta (DNMT3B) axis, which helps to elucidate the molecular mechanism of tamoxifen resistance in breast cancer (Wang J. et al., 2019). Downregulated RNA in cancer (DRAIC). Inhibits the invasion of glioblastoma-derived cell lines and activates adenosine monophosphate-activated protein kinase (AMPK) by downregulating the expression of glucose transporter 1 (GLUT1), thereby inhibiting the expression of mTOR, and eventually leading to increased autophagy (Saha et al., 2021). Overexpression of H19 promotes the proliferation and migration of glioma cells, and H19 promotes the proliferation and autophagy of glioma cells through the mTOR/Unc-51-like autophagy-activating kinase 1 (ULK1) pathway (Zhao et al., 2021). HAGLROS is highly expressed in HCC, and its high expression may be related to both the miC-5095/ATG12 axis and the PI3K/AKT/mTOR signaling pathway, which are involved in apoptosis and autophagy (Wei et al., 2019). Downregulation of the expression of metastasis-related lung adenocarcinoma transcript 1 (MALAT1) promotes apoptosis and autophagy in HCC cells and regulates miR-146a expression, affecting the progression of HCC (Peng et al., 2020). Upregulation of HOX transcript antisense RNA (HOTAIR) expression promotes the progression of cervical cancer, while its downregulation reduces autophagy and reverses epithelial stromal transformation by inhibiting the Wnt signaling pathway, thereby enhancing the sensitivity of cervical cancer to radiotherapy (Guo et al., 2019). Downregulation of neuroblastoma-associated transcript 1 (NBAT1) expression inhibits autophagy in NSCLC cells. Furthermore, the interaction of NBAT1 with proteasome 26S subunit, non-ATPase 10 (PSMD10) promotes autophagic degradation (Zheng et al., 2018). High small nucleolar RNA host gene 1 (SNHG1) expression is positively correlated with bladder cancer cell invasion, proliferation, and autophagy, and SNHG1 functions through the miR-493-5p/ATG14 pathway (Guo C. et al., 2021). HOXA transcript antisense RNA, myeloid-specific 1 (HOTAIRM1) promotes the proliferation and autophagy of AML cells; nuclear HOTAIRM1 promoted early growth response 1 (EGR1) degradation by serving as a scaffold to facilitate MDM2 (Murine double minute 2)-EGR1 complex formation, while cytoplasmic HOTAIRM1 acted as a sponge for miR-152-3p to increase ULK3 (Unc-51 like kinase 3) expression (Jing et al., 2021). In CML K562 cells, upregulation of opa interacting protein 5 antisense RNA 1 (OIP5-AS1) expression enhances autophagy, while its downregulation suppresses autophagy and enhances sensitivity to imatinib, and OIP5-AS1 promotes autophagy-related imatinib resistance in CML cells through the miR-30e-5p/ATG12 axis (Dai et al., 2021). Metformin inhibits the viability of paclitaxel ovarian cancer cells, increases the expression of SNHG7, and promotes autophagy in ovarian cancer cells; Metformin enhances the sensitivity of ovarian cancer cells to paclitaxel by regulating the SNHG7/miR-3127-5p axis to mediate autophagy (Yu et al., 2022). Decreasing PCa docetaxel resistance-associated lncRNA1 (PCDRlnc1) expression significantly inhibits autophagy in PCa cells; PCDRlnc1 interacts with ubiquitin-like with plant homeodomain and ring finger domain 1 (UHRF1) and promotes its transcription in PCa cells, leading to the activation of autophagy-related Beclin-1 protein (Xie et al., 2022). Silencing of lncRNA RP11-476D10.1 expression enhances apoptosis and autophagy in papillary thyroid carcinoma cells; lncRNA RP11-476D10.1 binds to miR-138-5p and promotes leucine-rich repeat kinase 2 (LRRK2) expression (Zhao et al., 2019). Silencing of HAGLROS expression promotes NPC cell apoptosis and inhibits autophagy in NPC cells; HAGLROS affects NPC progression by regulating ATG14 expression by adsorbing miR-100 (Zhang et al., 2019b). LINC01207 expression is upregulated in OSCC cells to promote apoptosis and autophagy; the LINC01207/miR-1301-3p/lactate dehydrogenase A (LDHA) regulatory axis promotes the proliferation of OSCC cells (Lu et al., 2021b). SNHG15 is upregulated in doxorubicin-resistant cell lines, and knockdown of SNHG15 expression inhibits the proliferation and autophagy of osteosarcoma cells; SNHG15 targets miR-381-3p/GDNF family receptor alpha 1 (GFRA1) expression to promote autophagy and enhances the resistance of osteosarcoma cells to doxorubicin (Zhang J. et al., 2020). CLRN1 antisense RNA 1 (CLRN1-AS1) inhibits the proliferation and autophagy of prolactinoma cells; forkhead box protein P1 (FOXP1) induces the expression of CLRN1-AS1, which adsorbs miR-217 and affects the biological function of pituitary prolactinoma cells through the Dickkopf WNT signaling pathway inhibitor 1 (DKK1)/Wnt/β-catenin signaling pathway (Wang C. et al., 2019). Pvt1 oncogene (PVT1) is upregulated in gemcitabine-resistant pancreatic cancer cell lines, and PVT1 promotes Pygopus family PHD finger 2 (Pygo2) and ATG14 expression; the PVT1/miR-619-5p/Wnt/β-catenin axis promotes cellular autophagy and attenuates resistance to gemcitabine (Zhou et al., 2020).
lncRNAs and ferroptosis
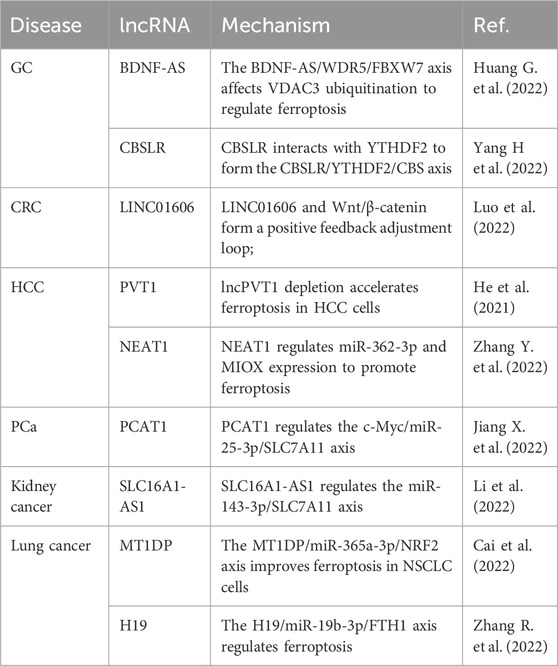
Studies examining the involvement of lncRNAs in ferroptosis for the occurrence, development, and metallurgical treatment of HCC, gastric cancer, colon cancer, PCa, kidney cancer, and lung cancer are summarized in Table 5. For example, the high expression of brain-derived neurotrophic factor antisense RNA (BDNF-AS) is positively correlated with the progression and poor prognosis of gastric cancer (Huang G. et al., 2022). BDNF-AS overexpression protects gastric cancer cells from ferroptosis and promotes the progression of gastric cancer. The BDNF-AS/WD repeat domain 5 (WDR5)/F-box and WD repeat domain-containing 7 (FBXW7) axis regulates ferroptosis in gastric cancer cells by affecting the ubiquitination of voltage-dependent anion channel 3 (VDAC3) (Huang G. et al., 2022). CBS mRNA stabilizing lncRNA (CBSLR) interacts with YTH domain linker protein 2 (YTHDF2) to form the CBSLR/YTHDF2/cystathionine beta-synthase (CBS) signal axis, which reduces the stability of CBS mRNA by enhancing the binding of YTHDF2 to the m6A-modified coding sequence of CBS mRNA. This protects gastric cancer cells from iron death and leads to resistance to chemotherapy drugs (Yang H. et al., 2022). LINC01606 protects colon cancer cells from ferroptosis by reducing the concentration of iron, lipid ROS, and mitochondrial superoxide, and by increasing mitochondrial membrane potential, while LINC01606 and Wnt/β-catenin form a positive feedback regulatory loop that further inhibits ferroptosis (Luo et al., 2022). lncPVT1 depletion accelerates ferroptosis in HCC cells, and decreased miR-214-3p expression and increased GPX4 expression reverse this effect and promote 4-ketamine-induced ferroptosis (He et al., 2021). In HCC cells, overexpression of nuclear paraspeckle assembly transcript 1 (NEAT1) enhances ferroptosis and the antitumor activity of erastin and RSL3; NEAT1 plays a role in ferroptosis by regulating the expression of miR-362-3p and myo-inositol oxygenase (MIOX) (Zhang Y. et al., 2022). Prostate cancer-associated transcript 1 (PCAT1) suppresses ferroptosis and enhances docetaxel resistance in PCa cells; PCAT1 inhibits the expression of miR-25-3p, thus promoting SLC7A11 expression, while transcription factor AP-2 γ activates PCAT1 expression, and finally reduces ferroptosis and enhances resistance to chemotherapeutic agents (Jiang X. et al., 2022). SLC16A1-antisense nucleic acid 1 (AS1) is highly expressed in kidney cancer, and is associated with overall survival; knockdown of SLC16A1-AS1 expression suppresses the proliferation and migration of kidney cancer cells, and SLC16A1-AS1 induces ferroptosis in kidney cancer through the miR-143-3p/SLC7A11 axis (Li et al., 2022). The metallothionein 1D pseudogene (MT1DP) downregulates miR-365a-3p expression by stabilizing the expression of nuclear factor-erythroid 2 p45-related factor 2, and further promotes ferroptosis in lung cancer cells (Gai et al., 2020). Overexpression of lncRNA H19 eliminates the anticancer effects of curcumin in lung cancer cells, whereas knockdown of H19 expression promotes curcumin-induced ferroptosis; lncRNA H19 acts as a competing endogenous RNA for miR-19b-3p, thereby enhancing the transcriptional activity of its endogenous target, ferritin heavy chain 1 (FTH1) (Zhang R. et al., 2022).
lncRNAs and pyroptosis
At present, research on the participation of lncRNAs in pyroptosis in tumors focuses mainly on using bioinformatics technology to predict the role of lncRNAs associated with pyroptosis in tumor diagnosis, treatment, and prognosis. However, molecular biology experiments are needed to verify their potential functions. For example, investigators downloaded the sequencing results from a total of 454 lung adenocarcinoma samples from The Cancer Genome Atlas (TCGA) database and identified 19 prognostic lncRNAs related to pyroptosis (Huang H. et al., 2022). Based on RNA sequencing data from the TCGA database, researchers analyzed 14 lncRNAs associated with pyroptosis that may be prognostic markers and promising therapeutic targets in head and neck SCC (Zhu W. et al., 2021). Pyroptosis-associated lncRNAs are associated with the tumor immune microenvironment, which may provide a new indicator for the selection of patients with ovarian cancer for immunotherapy (Zhang et al., 2021e).
According to analyses of the TCGA and China Glioma Genome Atlas databases, a pyroptosis-related lncRNA model was established by using consensus clustering and weighted gene co-expression network analysis, and glioblastoma patients with a high pyroptosis-related lncRNA score were found to have a richer immune infiltrate, higher immune checkpoint gene expression, and better response to immunotherapy, but worse response to chemotherapy (Xing et al., 2022). In gastric adenocarcinoma, HAND2-AS1, LINC01354, RP11-276H19.1, and PGM5-AS1 were suggested to be involved in pyroptosis, which could be used to guide effective patient prognosis and to provide evidence for the development of molecular-targeted therapies associated with pyroptosis (Wang Z. et al., 2022). lncRNA-XIST knockdown triggers pyroptosis mediated by the miR-335/superoxide dismutase 2/ROS signaling pathway, thereby inhibiting the progression of NSCLC (Liu et al., 2019). An lncRNA profile including ELFN1-AS1, PCAT6, TNRC6C-AS1, and ZEB1-AS1 related to pyroptosis could accurately predict the prognosis of CRC patients (Chen S. et al., 2021).
An investigator-constructed risk model containing 10 lncRNAs associated with pyroptosis that were identified as independent predictors of overall survival in breast cancer patients, with RP11-459E5.1, RP11-1070N10.3, and RP11-817J15.3 downregulation, was significantly associated with worse overall survival (Yang et al., 2021d). Eight pyroptosis-associated lncRNAs were identified by using a co-expression network of genes and lncRNAs and by further screening with univariate Cox regression analysis, which may be potential molecular markers and therapeutic targets in patients with bladder cancer (Lia et al., 2022). A novel risk score for pyroptosis-related lncRNAs could be used as a promising prognostic biomarker for HCC patients and may help to guide precision drugs and immunotherapy (Wang T. et al., 2022). Based on transcriptomic data, miRNA sequencing data, and related clinical information downloaded from the TCGA database, and constructed a competing endogenous RNA regulatory network that included 132 lncRNAs and 7 miRNAs, and established 11 lncRNA risk models related to pyroptosis, which has good prognostic value and can predict the immunotherapy outcome of colon adenocarcinoma (Tan et al., 2022).
Role of circRNAs in tumor cell death for tumorigenesis, development, and treatment
circRNAs and autophagy
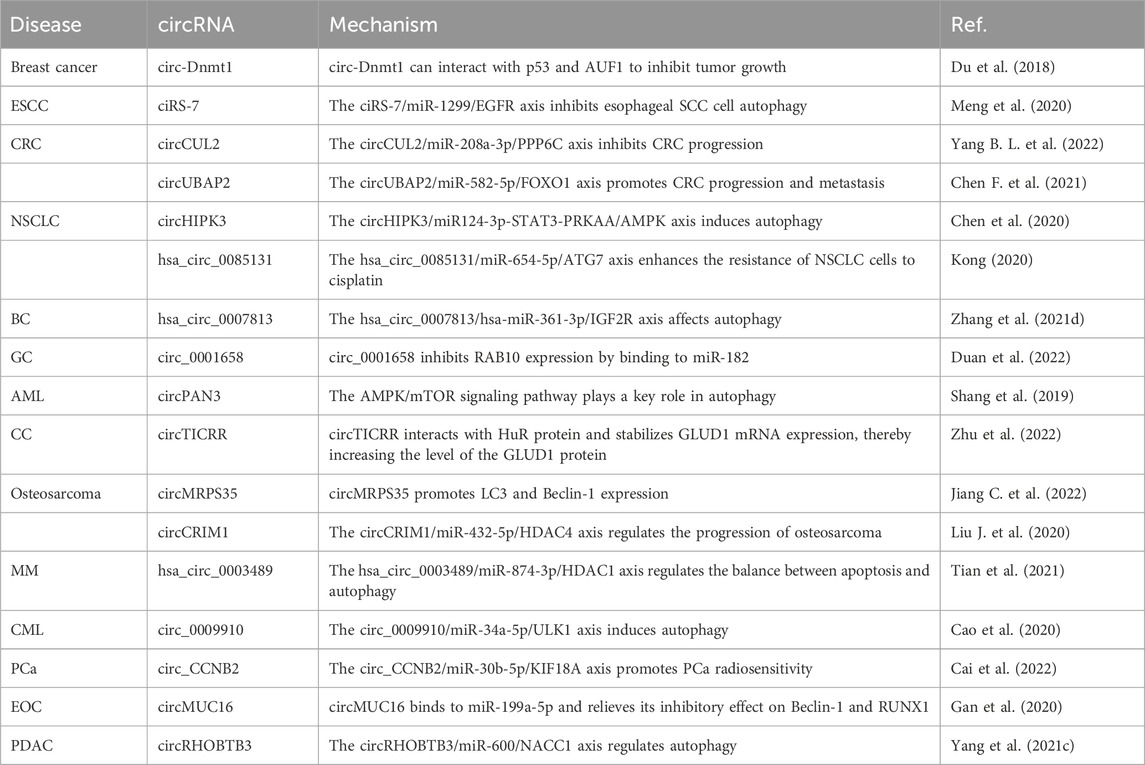
CircRNAs are involved mainly in the influence of autophagy on the occurrence, development, and treatment of breast cancer, SCC, NSCLC, bladder cancer, gastric cancer, AML, cervical carcinoma, osteosarcoma, Bozzolo’s disease, PCa, epithelial ovarian cancer, and pancreatic ductal adenocarcinoma, as summarized in Table 6. For example, circDNMT1 increases the proliferation of breast cancer cells by promoting cell autophagy (Du et al., 2018). Ectopically expressed circDNMT1 promotes the nuclear translocation of p53 and AU-rich element RNA-binding factor 1 (AUF1); the nuclear translocation of p53 induces autophagy, while the nuclear translocation of AUF1 reduces the instability of Dnmt1, ultimately increasing DNMT1 translation (Du et al., 2018). While ciRS-7 is highly expressed in triple-negative breast cancer, it suppresses rapamycin-induced autophagy in esophageal SCC, and miR-1299 promotes rapamycin-induced autophagy in esophageal SCC; ciRS-7 interacts with miR-1299 and regulates epidermal growth factor receptor (EGFR) expression to inhibit autophagy in esophageal SCC cells (Meng et al., 2020). circCUL2 overexpression inhibits the proliferation and autophagy of CRC cells and inhibits CRC progression through the miR-208a-3p/PPP6C signaling pathway (Yang B. L. et al., 2022). circUBAP2 induces autophagy in CRC cells in vitro and in vivo; circUBAP2 interacts directly with miR-582-5p and regulates the expression of forkhead box protein O1 (FOXO1) to promote CRC progression and metastasis (Chen F. et al., 2021). Silencing of circHIPK3 expression significantly inhibits the proliferation of lung cancer cells and induces autophagy; circHIPK3 has potential clinical value in evaluating the prognosis of lung cancer (Chen et al., 2020). High hsa_circ_0085131 expression is associated with the recurrence of NSCLC; hsa_circ_0085131 interacts with miR-654-5p to promote the expression of ATG7, thus enhancing the resistance of NSCLC cells to cisplatin (Kong, 2020). Reduced hsa_circ_0007813 expression inhibits the proliferation, migration, and autophagy of bladder cancer cells in vitro and in vivo; hsa_circ_0007813 binds to hsa-miR-361-3p and regulates the expression of insulin-like growth factor 2 receptor, thereby affecting bladder cancer progression (Zhang et al., 2021d). Silencing of circ_0001658 expression reduces the viability and autophagy of gastric cancer cells, and circ_0001658 acts by binding miR-182 and inhibiting the expression of member RAS oncogene family (RAB10) (Duan et al., 2022). circRNA-poly (A)-nuclease deadenylation complex subunit 3 (circPAN3) increases the drug resistance of AML cells by regulating autophagy, and the AMPK/mTOR signaling pathway plays a key role in this process (Shang et al., 2019). Knockdown of circRNA TOPBP1-interacting checkpoint and replication regulator (circTICRR) expression activates autophagy in cervical cancer cells; circTICRR binds to F287/F289 in the ribonucleotide reductase regulatory subunit M3 domain of human antigen R (HuR) and increases the expression of glutamate dehydrogenase 1 (GLUD1) protein, ultimately promoting autophagy (Zhu et al., 2022). circMRPS35 increases microtubule associated protein 1 light chain 3 (LC3) and Beclin-1 expression to promote autophagy in osteosarcoma cells (Jiang C. et al., 2022). After downregulation in osteosarcoma cells, circCRIM1 inhibits cell proliferation, promotes cell autophagy, and acts on the miR-432-5p/HDAC4 axis to inhibit osteosarcoma growth (Liu J. et al., 2020). The hsa_circ_0003489/miR-874-3p/HDAC1 axis balances apoptosis and autophagy in multiple myeloma cells, silences the expression of hsa_circ_0003489, inhibits autophagy, and enhances the sensitivity of multiple myeloma cells to bortezomib (Tian et al., 2021).
Knockdown of circ_0009910 expression inhibits CML cell proliferation and autophagy; the circ_0009910/miR-34a-5p/ULK1 axis induces autophagy in CML cells and accelerates the development of resistance to imatinib (Cao et al., 2020). Knockdown of circ_CCNB2 expression inhibits autophagy in PCa cells and promotes their sensitivity to radiotherapy through the miR-30b-5p/kinesin family member 18A (KIF18A) axis (Cai et al., 2022). circMUC16-mediated autophagy accelerates the invasion and metastasis of epithelial ovarian cancer, and circMUC16 can bind directly to miR-199a-5p and deinhibit the target genes Beclin-1 and Runt-related transcription factor 1 (RUNX1) to exert its effects (Gan et al., 2020). circRHOBTB3 is highly expressed in pancreatic ductal cancer cells, binds directly to miR-600 and regulates the expression of nucleus accumbens-associated 1 (NACC1), thereby promoting the autophagy response mediated by the Akt/mTOR pathway (Yang et al., 2021c).
circRNAs and ferroptosis
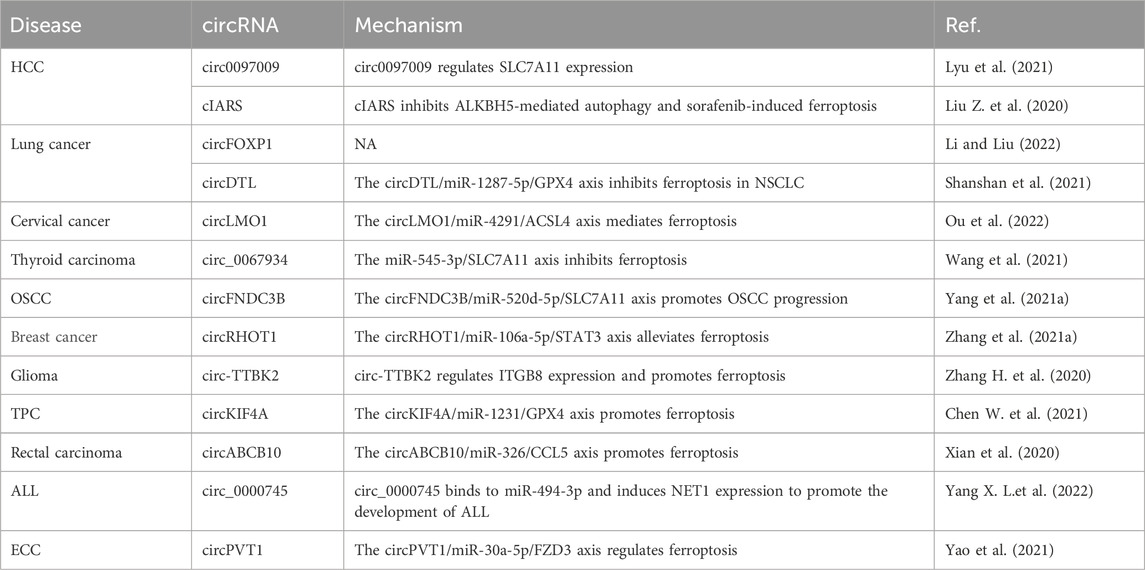
The influence of circRNAs on ferroptosis in the occurrence, development, and treatment of HCC, lung cancer, cervical cancer, thyroid carcinoma, OSCC, breast cancer, glioma, thyroid papillary carcinoma, rectal carcinoma, and acute lymphoblastic leukemia (ALL) is summarized in Table 7. For example, knockdown of circ0097009 expression inhibits the proliferation and invasion of HCC cells; circ0097009, as a competing endogenous RNA, adsorbs miR-1261 and regulates the expression of SLC7A11, a key regulator of ferroptosis, to affect the occurrence and development of liver cancer (Lyu et al., 2021). The expression of cIARS (hsa_circ_0008367) is significantly upregulated in HCC cells after treatment with sorafenib. Downregulation of cIARS expression inhibits ferroptosis, and cIARS inhibits AlkB homolog 5, RNA demethylase (ALKBH5)-mediated autophagy and inhibits sorafenib-induced ferroptosis (Liu Z. et al., 2020). circFOXP1 expression is significantly upregulated in lung cancer tissues, and knockdown of circFOXP1 expression inhibits the viability of lung cancer cells and promotes lung cancer progression by inhibiting ferroptosis (Li and Liu, 2022). circRNA denticleless E3 ubiquitin protein ligase homolog (circDTL) expression is upregulated in NSCLC cells, and knockdown of its expression promotes apoptosis and ferroptosis in NSCLC cells; circDTL functions through the miR-1287-5p/GPX4 axis, which provides a potential target for NSCLC treatment (Shanshan et al., 2021). circLMO1 overexpression in vitro and in vivo suppresses the growth and metastasis of cervical cancer, and circLMO1 inhibits cervical cancer progression through ferroptosis mediated by the miR-4291/ACSL4 axis (Ou et al., 2022). Knockdown of circ_0067934 expression in vitro and in vivo induces the proliferation and apoptosis of thyroid cancer cells; circ_0067934 inhibits ferroptosis in thyroid cancer cells through the miR-545-3p/SLC7A11 axis (Wang et al., 2021). Reduced circFNDC3B expression inhibits the growth and ferroptosis of OSCC cells, and the circFNDC3B/miR-520d-5p/SLC7A11 axis promotes the progression of OSCC (Yang et al., 2021a). circRHOT1 acts via the miR-106a-5p/signal transducer and activator of transcription 3 (STAT3) axis to promote the malignant progression of breast cancer and alleviates ferroptosis, providing new insights into the molecular mechanisms underlying the development of breast cancer (Zhang et al., 2021a). In glioma tissues and cells, the expression of the circRNA tau tubulin kinase 2 (circTTBK2) and integrin subunit beta 8 (ITGB8) is upregulated, and circTTBK2 targets ITGB8 expression to promote glioma cell proliferation, invasion, and ferroptosis (Zhang H. Y. et al., 2020). Knockdown of circRNA kinesin family member 4A (circKIF4A) expression inhibits the growth and migration of papillary thyroid cells; the circKIF4A-miR-1231-GPX4 axis plays an important role in promoting the proliferation and ferroptosis of papillary thyroid cancer cells (Chen W. et al., 2021). In rectal cancer tissues, the expression of circRNA ATP-binding box subfamily B member 10 (circABCB10) and C-C motif chemokine ligand 5 (CCL5) is upregulated, while miR-326 is knocked down to promote ferroptosis; the circABCB10/miR-326/CCL5 axis affects rectal cancer cell ferroptosis (Xian et al., 2020). Knockdown of circ_0000745 expression in an ALL cell line suppresses cell cycle progression and glycolysis, and triggers apoptosis and ferroptosis; circ_0000745 binding to miR-494-3p induces neuroepithelial cell transforming 1 (NET1) expression and promotes the development of ALL (Yang X. et al., 2022). circPVT1 enhances the sensitivity of esophageal SCC cells to 5-fluorouracil and regulates the Wnt/β-catenin pathway and ferroptosis via the miR-30a-5p/frizzled class receptor 3 (FZD3) axis (Yao et al., 2021).
circRNAs and pyroptosis
Pyroptosis is an important natural immune response of the body that plays a key role in fighting infection (Deng et al., 2022). circRNAs belong to a class of abundant non-coding RNAs, with stability and specificity, and thus have great potential in cancer treatment (Zhang et al., 2021b). The involvement of circRNAs in the role of pyroptosis in tumors has not been studied in depth. Investigators selected 1,875 differentially expressed circRNAs in unirradiated and irradiated lung cancer A549 cells (Zhang T. et al., 2022). Knockdown of circRNA Nei-like DNA glycosylase 3 (circNEIL3) expression was shown to promote radiation-induced pyroptosis. However, circNEIL3 overexpression had the opposite effect; by binding to miR-1184, circNEIL3 released the inhibitory effect of miR-1184 on PIF1 5′-to-3′ DNA helicase (PIF1) to induce DNA damage and trigger the activation of the absent in melanoma 2 inflammasome. The circNEIL3/miR-1184/PIF1 axis may be a new and promising clinical treatment strategy for lung cancer (Zhang T. et al., 2022).
Conclusion and future directions
Clinical and basic research into cancer, as a profound public health issue worldwide, has made many breakthroughs in recent years, but we still lack the ability to control tumor morbidity and mortality. How to kill tumor cells accurately and protect normal cells is the last and most important focus of tumor research. With our deepening understanding of intracellular molecules, an increasing number of molecules have been identified with roles in tumor progression, and the participation of non-coding RNAs in cell death plays a key role in tumor occurrence and development. However, under different biological backgrounds, regulated cell death may play very different biological roles. The functions of most cell death-related genes in tumors have not been studied thoroughly, and the signal of regulated cell death in tumors is also undecipherable. Therefore, revealing the regulatory mechanisms of non-coding RNAs involved in cell death in cancer, identifying potential therapeutic cell death targets of cancer, and developing novel immunotherapies based on the non-coding RNAs involved in cell death will have great and far-reaching significance for conquering cancer.
Author contributions
ZH: Writing–original draft. WL: Writing–review and editing. JS: Data curation, Writing–review and editing. FX: Data curation, Writing–review and editing. JL: Data curation, Writing–review and editing. XY: Data curation, Writing–review and editing. TP: Data curation, Writing–review and editing. YLv: Data curation, Writing–review and editing. YLi: Supervision, Writing–review and editing. XT: Funding acquisition, Writing–review and editing. JH: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Guangdong Provincial Medical Science and Technology Research Fund Projects (Nos A2023216, A2022524, and A2020304), Basic and Applied Basic Research Foundation of Guangdong Province (No. 2022A1515220217), Science and Technology Program of Guangzhou (Nos 202201010840, 202201010810, 202102080532, 202002030032, 202002020023, and 20211A011116), Health Commission Program of Guangzhou (Nos 20212A010025 and 20201A010085), Science and Technology Project of Panyu, Guangzhou (Nos 2022-Z04-009, 2022-Z04-090, 2022-Z04-072, 2021-Z04-053, 2020-Z04-052, 2020-Z04-026, and 2019-Z04-02), and Scientific Research Project of Guangzhou Panyu Central Hospital (Nos PY-2023-001, PY-2023-002, PY-2023-003, PY-2023-004, PY-2023-005, 2022Y002; 2021Y004, and 2021Y002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1284934/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Schematic representation of the regulatory mechanism of autophagy.
SUPPLEMENTARY FIGURE S2 | Schematic representation of the regulatory mechanisms of ferroptosis.
SUPPLEMENTARY FIGURE S3 | Schematic representation of the regulatory mechanisms of autophagy.
References
Ambros, V. (2001). microRNAs: tiny regulators with great potential. Cell 107 (7), 823–826. doi:10.1016/s0092-8674(01)00616-x
Anastasiadou, E., Faggioni, A., Trivedi, P., and Slack, F. J. (2018). The nefarious nexus of noncoding RNAs in cancer. Int. J. Mol. Sci. 19 (7), 2072. doi:10.3390/ijms19072072
Bahreini, F., Jabbari, P., Gossing, W., Aziziyan, F., Frohme, M., and Rezaei, N. (2021). The role of noncoding RNAs in pituitary adenoma. Epigenomics 13 (17), 1421–1437. doi:10.2217/epi-2021-0165
Bi, G., Liang, J., Zhao, M., Zhang, H., Jin, X., Lu, T., et al. (2022). miR-6077 promotes cisplatin/pemetrexed resistance in lung adenocarcinoma via CDKN1A/cell cycle arrest and KEAP1/ferroptosis pathways. Mol. Ther. Nucleic acids 28, 366–386. doi:10.1016/j.omtn.2022.03.020
Buchser, W. J., Laskow, T. C., Pavlik, P. J., Lin, H. M., and Lotze, M. T. (2012). Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 72 (12), 2970–2979. doi:10.1158/0008-5472.CAN-11-3396
Cai, F., Li, J., Zhang, J., and Huang, S. (2022). Knockdown of circ_CCNB2 sensitizes prostate cancer to radiation through repressing autophagy by the miR-30b-5p/KIF18A Axis. Cancer biotherapy Radiopharm. 37 (6), 480–493. doi:10.1089/cbr.2019.3538
Cao, H. X., Miao, C. F., Sang, L. N., Huang, Y. M., Zhang, R., Sun, L., et al. (2020). Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 243, 117255. doi:10.1016/j.lfs.2020.117255
Chen, F., Guo, L., Di, J., Li, M., Dong, D., and Pei, D. (2021). Circular RNA ubiquitin-associated protein 2 enhances autophagy and promotes colorectal cancer progression and metastasis via miR-582-5p/FOXO1 signaling. J. Genet. genomics = Yi chuan xue bao 48 (12), 1091–1103. doi:10.1016/j.jgg.2021.07.017
Chen, L., Niu, X., Qiao, X., Liu, S., Ma, H., Shi, X., et al. (2021). Characterization of interplay between autophagy and ferroptosis and their synergistical roles on manipulating immunological tumor microenvironment in squamous cell carcinomas. Front. Immunol. 12, 739039. doi:10.3389/fimmu.2021.739039
Chen, S., Zhu, J., and Zhi, X. (2021). A novel pyroptosis-associated long noncoding RNA signature to predict the prognosis of patients with colorectal cancer. Int. J. general Med. 14, 6111–6123. doi:10.2147/IJGM.S328842
Chen, W., Fu, J., Chen, Y., Li, Y., Ning, L., Huang, D., et al. (2021). Circular RNA circKIF4A facilitates the malignant progression and suppresses ferroptosis by sponging miR-1231 and upregulating GPX4 in papillary thyroid cancer. Aging 13 (12), 16500–16512. doi:10.18632/aging.203172
Chen, X., Mao, R., Su, W., Yang, X., Geng, Q., Guo, C., et al. (2020). Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy 16 (4), 659–671. doi:10.1080/15548627.2019.1634945
Chen, Y., Wang, K., Yang, J., Zhang, A., Dong, X., Zhou, Z., et al. (2022). Mechanism of ferroptosis in hypertensive nephropathy. Transl. Androl. urology 11 (5), 617–626. doi:10.21037/tau-22-276
Dai, H., Wang, J., Huang, Z., Zhang, H., Wang, X., Li, Q., et al. (2021). LncRNA OIP5-AS1 promotes the autophagy-related imatinib resistance in chronic myeloid leukemia cells by regulating miR-30e-5p/ATG12 Axis. Technol. Cancer Res. Treat. 20, 15330338211052150. Technology in cancer research and treatment. doi:10.1177/15330338211052150
Deng, H., Wei, Z., Qiu, S., Ye, D., Gu, S., Shen, Y., et al. (2022). Pyroptosis patterns and immune infiltrates characterization in head and neck squamous cell carcinoma. J. Clin. Lab. Anal. 36 (4), e24292. doi:10.1002/jcla.24292
Du, W. W., Yang, W., Li, X., Awan, F. M., Yang, Z., Fang, L., et al. (2018). A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 37 (44), 5829–5842. doi:10.1038/s41388-018-0369-y
Duan, X., Yu, X., and Li, Z. (2022). Circular RNA hsa_circ_0001658 regulates apoptosis and autophagy in gastric cancer through microRNA-182/Ras-related protein Rab-10 signaling axis. Bioengineered 13 (2), 2387–2397. doi:10.1080/21655979.2021.2024637
Fan, H., Ai, R., Mu, S., Niu, X., Guo, Z., and Liu, L. (2022). MiR-19a suppresses ferroptosis of colorectal cancer cells by targeting IREB2. Bioengineered 13 (5), 12021–12029. doi:10.1080/21655979.2022.2054194
Gai, C., Liu, C., Wu, X., Yu, M., Zheng, J., Zhang, W., et al. (2020). MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell death Dis. 11 (9), 751. doi:10.1038/s41419-020-02939-3
Gan, X., Zhu, H., Jiang, X., Obiegbusi, S. C., Yong, M., Long, X., et al. (2020). CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. cancer 19 (1), 45. doi:10.1186/s12943-020-01163-z
Green, D. R. (2019). The coming decade of cell death research: five riddles. Cell 177 (5), 1094–1107. doi:10.1016/j.cell.2019.04.024
Guan, L., Wang, F., Wang, M., Han, S., Cui, Z., Xi, S., et al. (2022). Downregulation of HULC induces ferroptosis in hepatocellular carcinoma via targeting of the miR-3200-5p/ATF4 Axis. Oxidative Med. Cell. Longev. 2022, 9613095. doi:10.1155/2022/9613095
Guo, C., Li, X., Xie, J., Liu, D., Geng, J., Ye, L., et al. (2021). Long noncoding RNA SNHG1 activates autophagy and promotes cell invasion in bladder cancer. Front. Oncol. 11, 660551. doi:10.3389/fonc.2021.660551
Guo, Y., Shi, W., and Fang, R. (2021). miR-18a-5p promotes melanoma cell proliferation and inhibits apoptosis and autophagy by targeting EPHA7 signaling. Mol. Med. Rep. 23 (1), 79. doi:10.3892/mmr.2020.11717
Guo, X., Xiao, H., Guo, S., Li, J., Wang, Y., Chen, J., et al. (2019). Long noncoding RNA HOTAIR knockdown inhibits autophagy and epithelial-mesenchymal transition through the Wnt signaling pathway in radioresistant human cervical cancer HeLa cells. J. Cell. physiology 234 (4), 3478–3489. doi:10.1002/jcp.26828
He, G. N., Bao, N. R., Wang, S., Xi, M., Zhang, T. H., and Chen, F. S. (2021). Ketamine induces ferroptosis of liver cancer cells by targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des. Dev. Ther. 15, 3965–3978. doi:10.2147/DDDT.S332847
Hu, Z., Yin, Y., Jiang, J., Yan, C., Wang, Y., Wang, D., et al. (2022). Exosomal miR-142-3p secreted by hepatitis B virus (HBV)-hepatocellular carcinoma (HCC) cells promotes ferroptosis of M1-type macrophages through SLC3A2 and the mechanism of HCC progression. J. Gastrointest. Oncol. 13 (2), 754–767. doi:10.21037/jgo-21-916
Huang, G., Xiang, Z., Wu, H., He, Q., Dou, R., Lin, Z., et al. (2022). The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int. J. Biol. Sci. 18 (4), 1415–1433. doi:10.7150/ijbs.69454
Huang, H., Shi, Z., Li, Y., Zhu, G., Chen, C., Zhang, Z., et al. (2022). Pyroptosis-related LncRNA signatures correlate with lung adenocarcinoma prognosis. Front. Oncol. 12, 850943. doi:10.3389/fonc.2022.850943
Huo, J. F., and Chen, X. B. (2019). Long noncoding RNA growth arrest-specific 5 facilitates glioma cell sensitivity to cisplatin by suppressing excessive autophagy in an mTOR-dependent manner. J. Cell. Biochem. 120 (4), 6127–6136. doi:10.1002/jcb.27900
Jia, L., Song, Y., Mu, L., Li, Q., Tang, J., Yang, Z., et al. (2020). Long noncoding RNA TPT1-AS1 downregulates the microRNA-770-5p expression to inhibit glioma cell autophagy and promote proliferation through STMN1 upregulation. J. Cell. physiology 235 (4), 3679–3689. doi:10.1002/jcp.29262
Jiang, C., Jiang, Z., and Zhang, X. (2022). Circular RNA circMRPS35 regulates progression and autophagy in osteosarcoma cells by recruiting KAT6B to govern FOXO3. Anti-cancer drugs 33 (7), 607–613. doi:10.1097/CAD.0000000000001276
Jiang, X., Guo, S., Xu, M., Ma, B., Liu, R., Xu, Y., et al. (2022). TFAP2C-Mediated lncRNA PCAT1 inhibits ferroptosis in docetaxel-resistant prostate cancer through c-Myc/miR-25-3p/SLC7A11 signaling. Front. Oncol. 12, 862015. doi:10.3389/fonc.2022.862015
Jiang, X., Yuan, Y., Tang, L., Wang, J., Zhang, D., and Duan, L. (2021). Systematic analysis and validation of the prognosis, immunological role and biology function of the ferroptosis-related lncRNA GSEC/miRNA-101-3p/CISD1 Axis in lung adenocarcinoma. Front. Mol. Biosci. 8, 793732. doi:10.3389/fmolb.2021.793732
Jiang, Z., Yao, L., Ma, H., Xu, P., Li, Z., Guo, M., et al. (2017). miRNA-214 inhibits cellular proliferation and migration in glioma cells targeting caspase 1 involved in pyroptosis. Oncol. Res. 25 (6), 1009–1019. doi:10.3727/096504016X14813859905646
Jing, Y., Jiang, X., Lei, L., Peng, M., Ren, J., Xiao, Q., et al. (2021). Mutant NPM1-regulated lncRNA HOTAIRM1 promotes leukemia cell autophagy and proliferation by targeting EGR1 and ULK3. J. Exp. Clin. cancer Res. CR 40 (1), 312. doi:10.1186/s13046-021-02122-2
Kong, R. (2020). Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol. Int. 44 (9), 1945–1956. doi:10.1002/cbin.11401
Li, H., and Liu, L. (2022). Zinc moderates circular RNA CircFOXP1 expression in order to regulate ferroptosis during lung adenocarcinoma. Chemico-biological Interact. 352, 109760. doi:10.1016/j.cbi.2021.109760
Li, P., He, J., Yang, Z., Ge, S., Zhang, H., Zhong, Q., et al. (2020). ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy 16 (7), 1186–1199. doi:10.1080/15548627.2019.1659614
Li, Y. Z., Zhu, H. C., Du, Y., Zhao, H. C., and Wang, L. (2022). Silencing lncRNA slc16a1-AS1 induced ferroptosis in renal cell carcinoma through miR-143-3p/slc7a11 signaling. Technol. cancer Res. Treat. 21, 15330338221077803. doi:10.1177/15330338221077803
Lia, T., Shao, Y., Regmi, P., and Li, X. (2022). Development and validation of pyroptosis-related lncRNAs prediction model for bladder cancer. Biosci. Rep. 42 (1). doi:10.1042/BSR20212253
Liao, W., and Zhang, Y. (2020). MicroRNA-381 facilitates autophagy and apoptosis in prostate cancer cells via inhibiting the RELN-mediated PI3K/AKT/mTOR signaling pathway. Life Sci. 254, 117672. doi:10.1016/j.lfs.2020.117672
Liu, J., Feng, G., Li, Z., Li, R., and Xia, P. (2020). Knockdown of CircCRIM1 inhibits HDAC4 to impede osteosarcoma proliferation, migration, and invasion and facilitate autophagy by targeting miR-432-5p. Cancer Manag. Res. 12, 10199–10210. doi:10.2147/CMAR.S253130
Liu, Z., Wang, Q., Wang, X., Xu, Z., Wei, X., and Li, J. (2020). Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell death Discov. 6, 72. doi:10.1038/s41420-020-00306-x
Liu, J., Yao, L., Zhang, M., Jiang, J., Yang, M., and Wang, Y. (2019). Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging 11 (18), 7830–7846. doi:10.18632/aging.102291
Lu, X., Chen, L., Li, Y., Huang, R., Meng, X., and Sun, F. (2021b). Long non-coding RNA LINC01207 promotes cell proliferation and migration but suppresses apoptosis and autophagy in oral squamous cell carcinoma by the microRNA-1301-3p/lactate dehydrogenase isoform A axis. Bioengineered 12 (1), 7780–7793. doi:10.1080/21655979.2021.1972784
Lu, X., Kang, N., Ling, X., Pan, M., Du, W., and Gao, S. (2021a). MiR-27a-3p promotes non-small cell lung cancer through slc7a11-mediated-ferroptosis. Front. Oncol. 11, 759346. doi:10.3389/fonc.2021.759346
Lu, Y., Chan, Y. T., Tan, H. Y., Zhang, C., Guo, W., Xu, Y., et al. (2022). Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J. Exp. Clin. cancer Res. CR 41 (1), 3. doi:10.1186/s13046-021-02208-x
Luo, M., Wu, L., Zhang, K., Wang, H., Zhang, T., Gutierrez, L., et al. (2018). miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell death Differ. 25 (8), 1457–1472. doi:10.1038/s41418-017-0053-8
Luo, Y., Huang, S., Wei, J., Zhou, H., Wang, W., Yang, J., et al. (2022). Long noncoding RNA LINC01606 protects colon cancer cells from ferroptotic cell death and promotes stemness by SCD1-Wnt/β-catenin-TFE3 feedback loop signalling. Clin. Transl. Med. 12 (4), e752. doi:10.1002/ctm2.752
Luo, Y., Zheng, S., Wu, Q., Wu, J., Zhou, R., Wang, C., et al. (2021). Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 17 (12), 4083–4101. doi:10.1080/15548627.2021.1901204
Lyu, N., Zeng, Y., Kong, Y., Chen, Q., Deng, H., Ou, S., et al. (2021). Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR-1261/SLC7A11 axis. Ann. Transl. Med. 9 (8), 675. doi:10.21037/atm-21-997
Ma, L. L., Liang, L., Zhou, D., and Wang, S. W. (2021). Tumor suppressor miR-424-5p abrogates ferroptosis in ovarian cancer through targeting ACSL4. Neoplasma 68 (1), 165–173. doi:10.4149/neo_2020_200707N705
Mahapatra, K. K., Mishra, S. R., Behera, B. P., Patil, S., Gewirtz, D. A., and Bhutia, S. K. (2021). The lysosome as an imperative regulator of autophagy and cell death. Cell. Mol. life Sci. CMLS 78 (23), 7435–7449. doi:10.1007/s00018-021-03988-3
Meng, L., Liu, S., Ding, P., Chang, S., and Sang, M. (2020). Circular RNA ciRS-7 inhibits autophagy of ESCC cells by functioning as miR-1299 sponge to target EGFR signaling. J. Cell. Biochem. 121 (2), 1039–1049. doi:10.1002/jcb.29339
Mohamed, L. A., El Bolok, A. H. M., Elgayar, S. F., and Fahmy, ANJOAMJ. M. S. (2022). miRNA-155 as a novel target for isoliquiritigenin to induce autophagy in oral squamous cell carcinoma. Open Access Maced. J. Med. Sci. 10 (A), 481–488. doi:10.3889/oamjms.2022.8278
New, M., and Tooze, S. (2019). The role of autophagy in pancreatic cancer-recent advances. Biology 9 (1), 7. doi:10.3390/biology9010007
Ou, R., Lu, S., Wang, L., Wang, Y., Lv, M., Li, T., et al. (2022). Circular RNA circLMO1 suppresses cervical cancer growth and metastasis by triggering miR-4291/ACSL4-mediated ferroptosis. Front. Oncol. 12, 858598. doi:10.3389/fonc.2022.858598
Patra, S., Praharaj, P. P., Klionsky, D. J., and Bhutia, S. K. (2022). Vorinostat in autophagic cell death: a critical insight into autophagy-mediated, -associated and -dependent cell death for cancer prevention. Drug Discov. today 27 (1), 269–279. doi:10.1016/j.drudis.2021.08.004
Peng, N., He, J., Li, J., Huang, H., Huang, W., Liao, Y., et al. (2020). Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis. Cancer cell Int. 20, 165. doi:10.1186/s12935-020-01231-w
Qu, L., Tian, Y., Hong, D., Wang, F., and Li, Z. (2022). Mig-6 inhibits autophagy in HCC cell lines by modulating miR-193a-3p. Int. J. Med. Sci. 19 (2), 338–351. doi:10.7150/ijms.66040
Rochette, L., Dogon, G., Rigal, E., Zeller, M., Cottin, Y., and Vergely, C. (2022). Lipid peroxidation and iron metabolism: two corner stones in the homeostasis control of ferroptosis. Int. J. Mol. Sci. 24 (1), 449. doi:10.3390/ijms24010449
Saha, S., Zhang, Y., Wilson, B., Abounader, R., and Dutta, A. (2021). The tumor-suppressive long noncoding RNA DRAIC inhibits protein translation and induces autophagy by activating AMPK. J. cell Sci. 134 (24), jcs259306. doi:10.1242/jcs.259306
Shang, J., Chen, W. M., Liu, S., Wang, Z. H., Wei, T. N., Chen, Z. Z., et al. (2019). CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leukemia Res. 85, 106198. doi:10.1016/j.leukres.2019.106198
Shang, J., Chen, Z. Z., Wang, Z. H., Wei, T. N., Wu, W. B., and Chen, W. M. (2018). Effect of expression levels of MiRNA-132, -125, -143 and -145 on autophagy and apoptosis of multiple myeloma cells. Zhongguo shi yan xue ye xue za zhi 26 (6), 1688–1694. doi:10.7534/j.issn.1009-2137.2018.06.018
Shanshan, W., Hongying, M., Jingjing, F., Yiming, Y., Yu, R., and Rui, Y. (2021). CircDTL functions as an oncogene and regulates both apoptosis and ferroptosis in non-small cell lung cancer cells. Front. Genet. 12, 743505. doi:10.3389/fgene.2021.743505
Shen, H., Lin, Z., Shi, H., Wu, L., Ma, B., Li, H., et al. (2020). MiR-221/222 promote migration and invasion, and inhibit autophagy and apoptosis by modulating ATG10 in aggressive papillary thyroid carcinoma. 3 Biotech. 10 (8), 339. doi:10.1007/s13205-020-02326-x
Song, F., Li, L., Liang, D., Zhuo, Y., Wang, X., and Dai, H. (2019). Knockdown of long noncoding RNA urothelial carcinoma associated 1 inhibits colorectal cancer cell proliferation and promotes apoptosis via modulating autophagy. J. Cell. physiology 234 (5), 7420–7434. doi:10.1002/jcp.27500
Sun, K., Ren, W., Li, S., Zheng, J., Huang, Y., Zhi, K., et al. (2022). MiR-34c-3p upregulates erastin-induced ferroptosis to inhibit proliferation in oral squamous cell carcinomas by targeting SLC7A11. Pathol. Res. Pract. 231, 153778. doi:10.1016/j.prp.2022.153778
Tan, Y., Lu, L., Liang, X., and Chen, Y. (2022). Identification of a pyroptosis-related lncRNA risk model for predicting prognosis and immune response in colon adenocarcinoma. World J. Surg. Oncol. 20 (1), 118. doi:10.1186/s12957-022-02572-8
Tian, B. G., Hua, Z., Wang, Z. J., and Li, J. (2020). Knockdown of microRNA-181a inhibits osteosarcoma cells growth and invasion through triggering NLRP3-dependent pyroptosis. Eur. Rev. Med. Pharmacol. Sci. 24 (3), 1030–1040. doi:10.26355/eurrev_202002_20153
Tian, F. Q., Chen, Z. R., Zhu, W., Tang, M. Q., Li, J. H., Zhang, X. C., et al. (2021). Inhibition of hsa_circ_0003489 shifts balance from autophagy to apoptosis and sensitizes multiple myeloma cells to bortezomib via miR-874-3p/HDAC1 axis. J. gene Med. 23 (9), e3329. doi:10.1002/jgm.3329
Wang, C., , Tan, C., Wen, Y., Zhang, D., Li, G., Chang, L., et al. (2019). FOXP1-induced lncRNA CLRN1-AS1 acts as a tumor suppressor in pituitary prolactinoma by repressing the autophagy via inactivating Wnt/β-catenin signaling pathway. Cell death Dis. 10 (7), 499. doi:10.1038/s41419-019-1694-y
Wang, Y., Zhang, S., Dang, S., Fang, X., and Liu, M. (2019). Overexpression of microRNA-216a inhibits autophagy by targeting regulated MAP1S in colorectal cancer. Onco Targets Ther. 12, 4621–4629. doi:10.2147/OTT.S196992
Wang, J., , Xie, S., Yang, J., Xiong, H., Jia, Y., Zhou, Y., et al. (2019). The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12 (1), 81. doi:10.1186/s13045-019-0747-0
Wang, H. H., Ma, J. N., and Zhan, X. R. (2021). Circular RNA Circ_0067934 attenuates ferroptosis of thyroid cancer cells by miR-545-3p/slc7a11 signaling. Front. Endocrinol. 12, 670031. doi:10.3389/fendo.2021.670031
Wang, T., Yang, Y., Sun, T., Qiu, H., Wang, J., Ding, C., et al. (2022). The pyroptosis-related long noncoding RNA signature predicts prognosis and indicates immunotherapeutic efficiency in hepatocellular carcinoma. Front. cell Dev. Biol. 10, 779269. doi:10.3389/fcell.2022.779269
Wang, Z., Cao, L., Zhou, S., Lyu, J., Gao, Y., and Yang, R. (2022). Construction and validation of a novel pyroptosis-related four-lncRNA prognostic signature related to gastric cancer and immune infiltration. Front. Immunol. 13, 854785. doi:10.3389/fimmu.2022.854785
Wei, H., Hu, J., Pu, J., Tang, Q., Li, W., Ma, R., et al. (2019). Long noncoding RNA HAGLROS promotes cell proliferation, inhibits apoptosis and enhances autophagy via regulating miR-5095/ATG12 axis in hepatocellular carcinoma cells. Int. Immunopharmacol. 73, 72–80. doi:10.1016/j.intimp.2019.04.049
Xian, Z. Y., Hu, B., Wang, T., Cai, J. L., Zeng, J. Y., Zou, Q., et al. (2020). CircABCB10 silencing inhibits the cell ferroptosis and apoptosis by regulating the miR-326/CCL5 axis in rectal cancer. Neoplasma 67 (5), 1063–1073. doi:10.4149/neo_2020_191024N1084
Xie, J., Chen, X., Wang, W., Guan, Z., Hou, J., and Lin, J. (2022). Long non-coding RNA PCDRlnc1 confers docetaxel resistance in prostate cancer by promoting autophagy. J. Cancer 13 (7), 2138–2149. doi:10.7150/jca.65329
Xing, Z., Liu, Z., Fu, X., Zhou, S., Liu, L., Dang, Q., et al. (2022). Clinical significance and immune landscape of a pyroptosis-derived LncRNA signature for glioblastoma. Front. cell Dev. Biol. 10, 805291. doi:10.3389/fcell.2022.805291
Xu, P., Wang, Y., Deng, Z., Tan, Z., and Pei, X. (2022). MicroRNA-15a promotes prostate cancer cell ferroptosis by inhibiting GPX4 expression. Oncol. Lett. 23 (2), 67. doi:10.3892/ol.2022.13186
Xu, W., Song, C., Wang, X., Li, Y., Bai, X., Liang, X., et al. (2021). Downregulation of miR-155-5p enhances the anti-tumor effect of cetuximab on triple-negative breast cancer cells via inducing cell apoptosis and pyroptosis. Aging 13 (1), 228–240. doi:10.18632/aging.103669
Xu, Z., Chen, L., Wang, C., Zhang, L., and Xu, W. (2021). MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through inhibiting GPX4. Free Radic. Res. 55 (11-12), 1119–1129. doi:10.1080/10715762.2021.2024816
Yadav, P., Sharma, P., Sundaram, S., Venkatraman, G., Bera, A. K., and Karunagaran, D. (2021). SLC7A11/xCT is a target of miR-5096 and its restoration partially rescues miR-5096-mediated ferroptosis and anti-tumor effects in human breast cancer cells. Cancer Lett. 522, 211–224. doi:10.1016/j.canlet.2021.09.033
Yang, J., Cao, X. H., Luan, K. F., and Huang, Y. D. (2021a). Circular RNA FNDC3B protects oral squamous cell carcinoma cells from ferroptosis and contributes to the malignant progression by regulating miR-520d-5p/slc7a11 Axis. Front. Oncol. 11, 672724. doi:10.3389/fonc.2021.672724
Yang, J., Rao, S., Cao, R., Xiao, S., Cui, X., and Ye, L. (2021b). miR-30a-5p suppresses lung squamous cell carcinoma via ATG5 - mediated autophagy. Aging 13 (13), 17462–17472. doi:10.18632/aging.203235
Yang, T., Shen, P., Chen, Q., Wu, P., Yuan, H., Ge, W., et al. (2021c). FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J. Exp. Clin. cancer Res. CR 40 (1), 261. doi:10.1186/s13046-021-02063-w
Yang, X., Weng, X., Yang, Y., and Jiang, Z. (2021d). Pyroptosis-related lncRNAs predict the prognosis and immune response in patients with breast cancer. Front. Genet. 12, 792106. doi:10.3389/fgene.2021.792106
Yang, B. L., Liu, G. Q., Li, P., and Li, X. H. (2022). Circular RNA CUL2 regulates the development of colorectal cancer by modulating apoptosis and autophagy via miR-208a-3p/PPP6C. Aging 14 (1), 497–508. doi:10.18632/aging.203827
Yang, H., Hu, Y., Weng, M., Liu, X., Wan, P., Hu, Y., et al. (2022). Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J. Adv. Res. 37, 91–106. doi:10.1016/j.jare.2021.10.001
Yang, X., Li, Y., Zhang, Y., and Liu, J. (2022). Circ_0000745 promotes acute lymphoblastic leukemia progression through mediating miR-494-3p/NET1 axis. Hematol. Amst. Neth. 27 (1), 11–22. doi:10.1080/16078454.2021.2008590
Yao, W., Wang, J., Meng, F., Zhu, Z., Jia, X., Xu, L., et al. (2021). Circular RNA CircPVT1 inhibits 5-fluorouracil chemosensitivity by regulating ferroptosis through MiR-30a-5p/FZD3 Axis in esophageal cancer cells. Front. Oncol. 11, 780938. doi:10.3389/fonc.2021.780938
Yu, S., Zhao, N., He, M., Zhang, K., and Bi, X. (2020). MiRNA-214 promotes the pyroptosis and inhibits the proliferation of cervical cancer cells via regulating the expression of NLRP3. Cell. Mol. Biol. (Noisy-le-Grand, France) 66 (6), 59–64. doi:10.14715/cmb/2020.66.6.11
Yu, Z., Wang, Y., Wang, B., and Zhai, J. (2022). Metformin affects paclitaxel sensitivity of ovarian cancer cells through autophagy mediated by long noncoding RNASNHG7/miR-3127-5p Axis. Cancer biotherapy Radiopharm. 37 (9), 792–801. doi:10.1089/cbr.2019.3390
Zhang, H., Ge, Z., Wang, Z., Gao, Y., Wang, Y., and Qu, X. (2021a). Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging 13 (6), 8115–8126. doi:10.18632/aging.202608
Zhang, K., Wu, L., Zhang, P., Luo, M., Du, J., Gao, T., et al. (2018). miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Mol. Carcinog. 57 (11), 1566–1576. doi:10.1002/mc.22878
Zhang, S., Sun, J., Gu, M., Wang, G., and Wang, X. (2021b). Circular RNA: a promising new star for the diagnosis and treatment of colorectal cancer. Cancer Med. 10 (24), 8725–8740. doi:10.1002/cam4.4398
Zhang, X., Wei, X., Wang, Y., Wang, S., Ji, C., Yao, L., et al. (2021c). Pyroptosis regulators and tumor microenvironment infiltration characterization in clear cell renal cell carcinoma. Front. Oncol. 11, 774279. doi:10.3389/fonc.2021.774279
Zhang, Z., Mou, Z., Xu, C., Wu, S., Dai, X., Chen, X., et al. (2021d). Autophagy-associated circular RNA hsa_circ_0007813 modulates human bladder cancer progression via hsa-miR-361-3p/IGF2R regulation. Cell death Dis. 12 (8), 778. doi:10.1038/s41419-021-04053-4
Zhang, Z., Xu, Z., and Yan, Y. (2021e). Role of a pyroptosis-related lncRNA signature in risk stratification and immunotherapy of ovarian cancer. Front. Med. 8, 793515. doi:10.3389/fmed.2021.793515
Zhang, H. Y., Zhang, B. W., Zhang, Z. B., and Deng, Q. J. (2020). Circular RNA TTBK2 regulates cell proliferation, invasion and ferroptosis via miR-761/ITGB8 axis in glioma. Eur. Rev. Med. Pharmacol. Sci. 24 (5), 2585–2600. doi:10.26355/eurrev_202003_20528
Zhang, J., Rao, D., Ma, H., Kong, D., Xu, X., and Lu, H. (2020). LncRNA SNHG15 contributes to doxorubicin resistance of osteosarcoma cells through targeting the miR-381-3p/GFRA1 axis. Open life Sci. 15 (1), 871–883. doi:10.1515/biol-2020-0086
Zhang, R., Pan, T., Xiang, Y., Zhang, M., Xie, H., Liang, Z., et al. (2022). Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact. Mater. 13, 23–36. doi:10.1016/j.bioactmat.2021.11.013
Zhang, T., Wu, D. M., Luo, P. W., Liu, T., Han, R., Deng, S. H., et al. (2022). CircNEIL3 mediates pyroptosis to influence lung adenocarcinoma radiotherapy by upregulating PIF1 through miR-1184 inhibition. Cell death Dis. 13 (2), 167. doi:10.1038/s41419-022-04561-x
Zhang, Y., Luo, M., Cui, X., O'Connell, D., and Yang, Y. (2022). Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell death Differ. 29 (9), 1850–1863. doi:10.1038/s41418-022-00970-9
Zhang, W., Dong, Y. Z., Du, X., Peng, X. N., and Shen, Q. M. (2019a). MiRNA-153-3p promotes gefitinib-sensitivity in non-small cell lung cancer by inhibiting ATG5 expression and autophagy. Eur. Rev. Med. Pharmacol. Sci. 23 (6), 2444–2452. doi:10.26355/eurrev_201903_17391
Zhang, W., Zhang, Y., and Xi, S. (2019b). Upregulation of lncRNA HAGLROS enhances the development of nasopharyngeal carcinoma via modulating miR-100/ATG14 axis-mediated PI3K/AKT/mTOR signals. Artif. cells, nanomedicine, Biotechnol. 47 (1), 3043–3052. doi:10.1080/21691401.2019.1640233
Zhao, W., Lin, X., Han, H., Zhang, H., Li, X., Jiang, C., et al. (2021). Long noncoding RNA H19 contributes to the proliferation and autophagy of glioma cells through mTOR/ULK1 pathway. Neuroreport 32 (5), 352–358. doi:10.1097/WNR.0000000000001602
Zhao, Y., Wang, P., and Wu, Q. (2020). miR-1278 sensitizes nasopharyngeal carcinoma cells to cisplatin and suppresses autophagy via targeting ATG2B. Mol. Cell. probes 53, 101597. doi:10.1016/j.mcp.2020.101597
Zhao, Y., Zhao, L., Li, J., and Zhong, L. (2019). Silencing of long noncoding RNA RP11-476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2. J. Cell. physiology 234 (11), 20980–20991. doi:10.1002/jcp.28702
Zheng, B., Zhu, H., Gu, D., Pan, X., Qian, L., Xue, B., et al. (2015). MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem. biophysical Res. Commun. 459 (2), 234–239. doi:10.1016/j.bbrc.2015.02.084
Zheng, J., Liu, X., Xue, Y., Gong, W., Ma, J., Xi, Z., et al. (2017). TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J. Hematol. Oncol. 10 (1), 52. doi:10.1186/s13045-017-0422-2
Zheng, T., Li, D., He, Z., Feng, S., and Zhao, S. (2018). Long noncoding RNA NBAT1 inhibits autophagy via suppression of ATG7 in non-small cell lung cancer. Am. J. cancer Res. 8 (9), 1801–1811.
Zheng, Y., Tan, K., and Huang, H. (2019). Long noncoding RNA HAGLROS regulates apoptosis and autophagy in colorectal cancer cells via sponging miR-100 to target ATG5 expression. J. Cell. Biochem. 120 (3), 3922–3933. doi:10.1002/jcb.27676
Zhou, C., Yi, C., Yi, Y., Qin, W., Yan, Y., Dong, X., et al. (2020). LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. cancer 19 (1), 118. doi:10.1186/s12943-020-01237-y
Zhu, T., Cen, Y., Chen, Z., Zhang, Y., Zhao, L., Wang, J., et al. (2022). Oncogenic circTICRR suppresses autophagy via binding to HuR protein and stabilizing GLUD1 mRNA in cervical cancer. Cell death Dis. 13 (5), 479. doi:10.1038/s41419-022-04943-1
Zhu, W., Ye, Z., Chen, L., Liang, H., and Cai, Q. (2021). A pyroptosis-related lncRNA signature predicts prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int. Immunopharmacol. 101, 108268. doi:10.1016/j.intimp.2021.108268
Zhu, X., , Wu, T., Chi, Y., Ge, Y., Jiao, Y., Zhu, F., et al. (2021). MicroRNA-195 suppresses enterovirus A71-induced pyroptosis in human neuroblastoma cells through targeting NLRX1. Virus Res. 292, 198245. doi:10.1016/j.virusres.2020.198245
Glossary
Keywords: non-coding RNAs, autophagy, ferroptosis, pyroptosis, tumorigenesis, tumor progression, treatment
Citation: Han Z, Luo W, Shen J, Xie F, Luo J, Yang X, Pang T, Lv Y, Li Y, Tang X and He J (2024) Non-coding RNAs are involved in tumor cell death and affect tumorigenesis, progression, and treatment: a systematic review. Front. Cell Dev. Biol. 12:1284934. doi: 10.3389/fcell.2024.1284934
Received: 29 August 2023; Accepted: 08 January 2024;
Published: 28 February 2024.
Edited by:
Zhe-Sheng Chen, St. John’s University, United StatesReviewed by:
Tatsushi Yoshida, Kyoto Prefectural University of Medicine, JapanLeli Zeng, Sun Yat-sen University, China
Copyright © 2024 Han, Luo, Shen, Xie, Luo, Yang, Pang, Lv, Li, Tang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhua He, MzMyNTE4NTc5QHFxLmNvbQ==; Xingkui Tang, dGFuZ3hpbmdrdWlAMTYzLmNvbQ==; Yuguang Li, bHlnX3B5QDEyNi5jb20=
†These authors have contributed equally to this work
 Zeping Han1,2†
Zeping Han1,2† Jian Shen
Jian Shen Fangmei Xie
Fangmei Xie Xiang Yang
Xiang Yang Yubing Lv
Yubing Lv Yuguang Li
Yuguang Li Xingkui Tang
Xingkui Tang Jinhua He
Jinhua He