- 1Department of Genetics, University of Cambridge, Cambridge, United Kingdom
- 2School of BioSciences, The University of Melbourne, Parkville, VIC, Australia
Genomic imprinting is an epigenetic process whereby genes are monoallelically expressed in a parent-of-origin-specific manner. Imprinted genes are frequently found clustered in the genome, likely illustrating their need for both shared regulatory control and functional inter-dependence. The Dlk1-Dio3 domain is one of the largest imprinted clusters. Genes in this region are involved in development, behavior, and postnatal metabolism: failure to correctly regulate the domain leads to Kagami–Ogata or Temple syndromes in humans. The region contains many of the hallmarks of other imprinted domains, such as long non-coding RNAs and parental origin-specific CTCF binding. Recent studies have shown that the Dlk1-Dio3 domain is exquisitely regulated via a bipartite imprinting control region (ICR) which functions differently on the two parental chromosomes to establish monoallelic expression. Furthermore, the Dlk1 gene displays a selective absence of imprinting in the neurogenic niche, illustrating the need for precise dosage modulation of this domain in different tissues. Here, we discuss the following: how differential epigenetic marks laid down in the gametes cause a cascade of events that leads to imprinting in the region, how this mechanism is selectively switched off in the neurogenic niche, and why studying this imprinted region has added a layer of sophistication to how we think about the hierarchical epigenetic control of genome function.
Introduction
Genomic imprinting in mammals
Genome function is regulated temporally and tissue specifically through the orchestrated interplay of regulatory factors, genomic features, and epigenetic states. Epigenetic modifications are dynamic during development and across the cell cycle. A hierarchy of successive epigenetic states, including DNA methylation, ensures the creation of healthy individuals. In mammals, extensive epigenetic reprogramming events occur during germ cell development, fertilization, and early embryogenesis (Smith and Meissner, 2013). Although DNA methylation is essential for normal mammalian development, there appear to be multiple ways in which it can regulate and maintain cell fate and function, which remain incompletely understood. Although intensively studied, the association between DNA methylation and transcription is often correlative, with little experimental evidence to support causal relationships.
One major process in which predictive and causal relationships between DNA methylation and gene expression is more comprehensively understood is that of mammalian genomic imprinting (Bartolomei and Ferguson-Smith, 2011; Ferguson-Smith, 2011). Genomic imprinting is an epigenetically regulated process causing genes to be expressed from one chromosome homolog according to the parent-of-origin. Imprinting is highly conserved in eutherian mammals, and mouse studies have provided insights into the repertoire of developmental and physiological pathways regulated by imprinted genes (Ferguson-Smith and Bourc’his, 2018; Cleaton et al., 2014). Failure to correctly establish and maintain imprints is associated with developmental syndromes, including growth abnormalities, neurological and metabolic disorders, and numerous forms of cancer (Uribe-Lewis et al., 2011; Ishida and Moore, 2013). Imprinted genes are also highly expressed in the developing and adult brain, and are implicated in numerous brain functions, including behavior (Keverne, 1997; Liu et al., 2010; Furutachi et al., 2013; Tsan et al., 2016). Several syndromes that result from dysregulation of imprinted loci involve brain dysfunction, such as Prader–Willi syndrome (PWS) (Aman et al., 2018), Angelman syndrome (Nicholls et al., 1998), Turner syndrome (Bondy, 2006; Lepage et al., 2013), autism (Flashner et al., 2013), bipolar depression (Pinto et al., 2011), and schizophrenia (Ingason et al., 2011; Isles et al., 2016).
Genomic imprinting in laboratory mice is a tractable model for studying epigenetic regulation as the two parentally inherited, genetically identical genomic regions within the same nucleus express a different repertoire of genes in a parental-origin-specific manner. Imprinting is established in the germ cells through the differential deposition of DNA methylation in the two parental germlines (Smallwood et al., 2011). Differential DNA methylation marks (DMRs) at imprinting control regions (ICRs) are maintained in the post-fertilization period and protected from the global methylation erasure in the early embryo by KRAB zinc finger proteins ZFP57 and ZFP445 in order to maintain the epigenetic memory of parental origin (Takahashi et al., 2019). However, the dynamic hierarchy of events initiated by the ICRs that leads to the long-range domain-wide temporal and tissue-specific behavior of imprinted genes is not fully understood.
Studies assessing global as well as locus-specific alterations to ICRs have emphasized that loss of imprinting results in reciprocal effects on imprinted genes with the biallelic expression of some genes within the cluster and biallelic repression at others (Tucker et al., 1996; Li et al., 2008; Demars and Gicquel, 2012; Azzi et al., 2014; Takahashi et al., 2019). Phenotypically, perturbations to individual imprinted genes exert effects in numerous developmental and physiological pathways (Kalish et al., 2014; Tucci et al., 2019). Together, this has led to the prevailing notion in the field that at least some imprinted genes are dosage-sensitive. Deletions or insertions of the genes themselves, and aberrations that disrupt the pattern of imprinted gene expression, like mutations in the ICR or uniparental disomy (UPD), contribute to tumor progression and disease. In addition, the balance between maternally and paternally inherited genes can modulate phenotypes. For instance, PWS patients with maternal UPD or with ICR deletions have increased maternal expression along with a loss of paternal gene expression. These individuals are far more associated with psychotic illnesses than PWS patients with individual paternal gene deletion genotypes (Nicholls et al., 1998; Tucci et al., 2019). Yet, because the intricate epigenetic control at imprinted clusters controls the parent-specific expression of multiple genes, it is difficult to assign the relative contribution of the individual gene dosage to the resulting physiological phenotypes. Utilizing a systemic set of mutants at a single imprinted domain allows us to dissect the relationship between allelic expression, dosage, epigenetic control, and phenotypical outcomes.
Genomic imprinting at the Dlk1-Dio3 domain
One of the largest imprinted clusters in mammals is the 1.2 Mb Dlk1-Dio3 domain. This region is conserved between mice and humans, and is one of the major developmentally regulated mammalian imprinted domains. Failure to correctly imprint genes in this cluster in humans leads to Temple or Kagami–Ogata syndromes, both of which exhibit neurological, developmental, and behavioral impairments (Ioannides et al., 2014; Kagami et al., 2015). In mice, both maternal and paternal UPDs containing this domain lead to prenatal lethality, further illustrating the developmental importance of the correct dosage of genes in the region.
Four protein-coding genes, Dlk1, Rtl1, Dio3, and one isoform of Begain (located upstream of Dlk1 beyond a large LINE1-rich region), are preferentially expressed from the paternal allele (Figure 1A) (Tierling et al., 2009). Dlk1 encodes delta like non-canonical Notch ligand 1. Paternal loss of the gene leads to partial neonatal lethality, and those animals that survive display post-natal growth retardation, increased adiposity, and skeletal defects (Moon et al., 2002). More recently, mice lacking Dlk1 have also been shown to be prone to anxiety-like behaviors (García-Gutiérrez et al., 2018). Rtl1 is a Ty3-gypsy retrotransposon-derived neogene that has evolved a function in placentation in eutherians. Loss of the paternal copy of Rtl1 causes placental retardation and, in some mouse strains, can cause delayed parturition (Youngson et al., 2005; Sekita et al., 2008; Ito et al., 2015; Kitazawa et al., 2020; Kitazawa et al., 2021). The most distal imprinted gene in the domain, Dio3, encodes type 3 iodothyronine deiodinase, which is a negative regulator of thyroid hormone metabolism (Tsai et al., 2002). Dio3 null mice show partial neonatal lethality and postnatal growth restriction. However, paternal loss of the gene leads to a much milder phenotype, reflecting the less stringent imprinting of this gene (Elena Martinez et al., 2014).
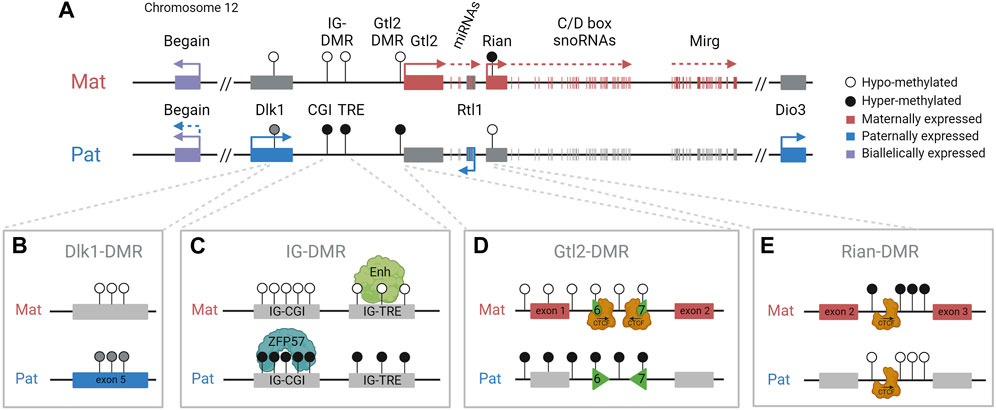
FIGURE 1. Dlk1-Dio3 imprinted domain and key regulatory features. (A). The paternally inherited chromosome expresses Dlk1, Rtl1, Dio3, and one isoform of Begain. The maternally inherited chromosome expresses Gtl2/Meg3, antiRtl1, and arrays of snoRNAs and miRNAs. The IG-DMR is methylated in sperm, is unmethylated in oocytes, and is the imprinting control region for the domain. DMRs are indicated by circles: black (methylated) and white (unmethylated). (B). The somatic Dlk1-DMR, in the last exon of the Dlk1 gene, is differentially methylated with partial methylation on the paternal chromosome. (C). The IG-DMR contains a CpG island (CGI) that binds ZFP57 on the methylated paternal copy and a transcriptional regulatory element (TRE) that has an enhancer-like function on the unmethylated maternal copy. (D). The Gtl2-DMR contains two differentially methylated CTCF binding sites (CTCF6 and 7) binding CTCF only on the unmethylated maternal chromosome. (E). The Rian-DMR, in the second intron of the Rian gene, is methylated in the reverse pattern as the IG-DMR and Gtl2-DMR, methylated on the maternal chromosome, and hypomethylated on the paternal chromosome.
The maternally inherited chromosome expresses multiple imprinted noncoding transcripts, including Gtl2 (also known as Meg3) (Figure 1). Gtl2 is a long non-coding RNA (lncRNA) that is downregulated or lost in numerous human cancers, including breast and colorectal cancers (Makoukji et al., 2016; Buccarelli et al., 2020). Gtl2 is also thought to form a long polycistronic transcript along with its associated transcripts Rian and Mirg, and acts as a host for multiple snoRNAs and miRNAs, including the miR-379/miR-410 cluster, all of which are driven by the Gtl2 promoter (Cavaillé et al., 2002; Seitz et al., 2004; Tierling et al., 2006). In humans, the miRNAs in this cluster have been shown to be downregulated in the pancreatic islets of donors with type 2 diabetes (Kameswaran et al., 2014). Neonatal mice with a maternal deletion of the entire miR-379/miR-410 cluster are hypoglycemic and show impaired transition from fetal to postnatal metabolism (Labialle et al., 2014). These mice have also been shown to have increased anxiety-related behaviors in adulthood (Marty et al., 2016). Therefore, appropriate expression of genes in this region is essential for the lifelong health of mammals, and understanding the epigenetic regulation of these genes has significant biomedical relevance for a diverse range of processes.
Imprinting at the Dlk1-Dio3 locus is regulated by the germline-derived intergenic DMR (IG-DMR) (Lin et al., 2003), which is required for regulating parent-specific expression in this locus (Rocha et al., 2008). This DMR is normally methylated in sperm and not methylated in oocytes, and maintains this parent-specific pattern throughout development (Figure 2A). After implantation, secondary somatic DMRs are established in the region: one at the promoter of the Gtl2 gene (the Gtl2-DMR) and another tissue-specific partial DMR over the fifth exon of Dlk1. Both of these somatic DMRs are also hypermethylated on the paternal chromosome (Takada et al., 2000). A third somatic DMR is located in the second intron of Rian (known as MEG8 in humans) (Zeng et al., 2014). This DMR is dependent on the IG-DMR, but in contrast to the other DMRs in the region, it gains secondary methylation on the maternally inherited chromosome (Figure 3). Here, we review the current knowledge on how differential epigenetic landscapes, genetic elements, and transcription are exquisitely coordinated to regulate genome function in this domain.
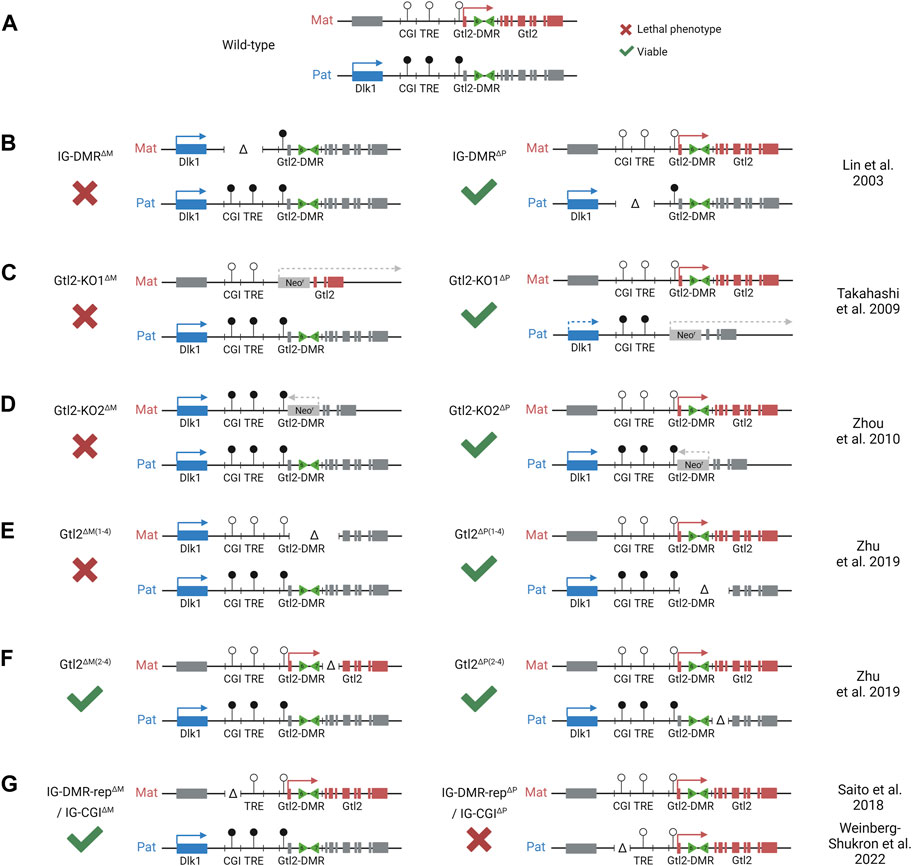
FIGURE 2. Integrated model depicting effects of different mice models with deletions at the Dlk1-Dio3 locus. Colored boxes represent expression from maternal (red) and paternal (blue) alleles. Gray boxes represent allelically repressed genes. Lollipops represent methylated (black) and unmethylated (white) regulatory elements. (A). WT pattern of expression at the Dlk1-Dio3 locus. (B). Maternal deletion of the entire IG-DMR results in a maternal-to-paternal epigenotype switch. (C). Maternal replacement of Gtl2 exons 1–7 with a forward-facing neomycin resistant cassette results in partial loss of the maternal gene expression. The same paternal substitution results in partial loss of the paternal gene expression. (D). Maternal replacement of Gtl2 exons 1–6 with a reverse facing neomycin resistant cassette results in loss of the maternal gene expression and activation of the maternal Dlk1 expression. The same paternal substitution had no effect on the expression of methylation in the region. (E). Maternal deletion of Gtl2 exons 1–4 (including part of the Gtl2-DMR) results in a maternal-to-paternal epigenotype switch, which is similar to the IG-DMR deletion. However, methylation at the IG-DMR is not affected by this deletion. Paternal deletion has no effect. (F). Neither maternal nor paternal deletion of Gtl2 exons 2–4 has an effect on the expression of methylation at the Dlk1-Dio3 locus, indicating that the Gtl2-DMR, not the Gtl2 gene, regulated imprinting at this region. (G). An isolated paternal deletion of the IG-CGI results in a paternal-to-maternal epigenotype switch. Although maternal deletion has no effect, indicating that the IG-CGI is the primary methylation mark on the paternal chromosome, it is dispensable from the maternal chromosome.
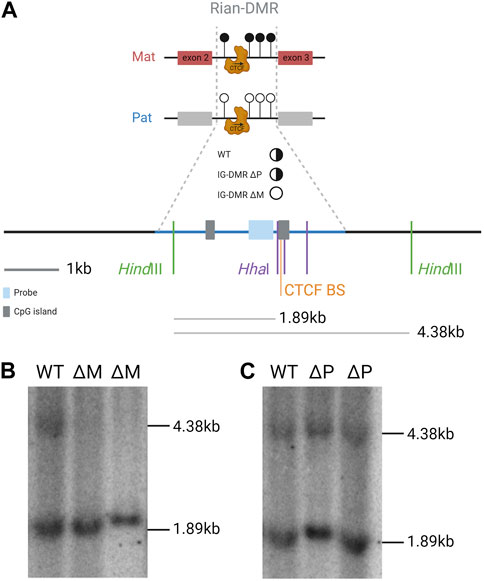
FIGURE 3. Rian-DMR is biallelically unmethylated in e16 embryos that inherit the IG-DMR deletion from their mother. (A). Scale summary of the Southern blot displayed in part b relative to sequence features of the region and the location of the hybridization probe and the digest fragments that are hybridized with the probe. Full black circle, fully methylated; half black half white circle, differentially methylated; white circle, unmethylated. Blue shade, probe; gray shade, CpG islands; blue line, Rian-DMR +/-1 kb. (B,C). Methylation-sensitive restriction-digested Southern blot of genomic DNA from e16 embryos hybridized with a Rian-DMR-specific probe. The genomic DNA was digested with HindIII in combination with HhaI in all lanes. WT, wild-type embryo; ΔM, IG-DMR maternally transmitted knockout embryo; ΔP, IG-DMR paternally transmitted knockout embryo.
Regulation and hierarchy of imprinting at the Dlk1-Dio3 region
The IG-DMR spans approximately 5 kb between Dlk1 and Gtl2. Maternal deletion of this ICR region leads to paternalization of the maternal chromosome, and mice die between e16.5 and birth (Lin et al., 2003; Lin et al., 2007). However, on paternal transmission, the deletion has no effect (Figure 2B). The IG-DMR contains a small CpG island comprising seven tandemly repeated sequences, five of which contain a ZFP57 binding motif (Takada et al., 2002). The sequence-specific zinc finger protein, ZFP57, only binds to the methylated paternal chromosome, where it interacts with TRIM28, which in turn recruits the repressive epigenetic machinery, including DNMTs and heterochromatin-associated proteins such as SETDB1 and HP1, thereby maintaining the methylation memory of the germline imprint in early development in an environment where most epigenetic modifications elsewhere are being erased (Quenneville et al., 2011; Messerschmidt et al., 2012).
Recently, the IG-DMR was shown to be a bipartite element comprising two distinct functional elements (Aronson et al., 2021). In addition to the CpG island (IG-CGI) described earlier, it also contains a transcriptional regulation element (IG-TRE) that can bind pluripotency transcription factors in mouse embryonic stem cells (mESCs) and exhibit active enhancer marks (H3K27ac) and nascent transcription (Danko et al., 2015; Luo et al., 2016) (Figure 1C). It was, therefore, suggested that the IG-TRE serves as a putative enhancer, driving the expression of the maternally inherited genes within the domain.
Furthermore, these data also indicate that the IG-CGI is required to inactivate the paternal IG-TRE and maintain a repressive chromatin landscape on the paternal chromosome (Stelzer et al., 2016; Kojima et al., 2022; Weinberg-Shukron et al., 2022). Surprisingly, in contrast to the full IG-DMR deletion (maternal to paternal epigenotype switch but no effect upon paternal transmission), an isolated paternally derived deletion of the IG-CGI results in the reciprocal paternal-to-maternal epigenotype switch, with the IG-TRE becoming hypomethylated on the paternal chromosome (Saito et al., 2018) (Figure 2G).
Together, these findings indicate that the key element regulating imprinted expression on the maternal chromosome is the IG-TRE, which promotes activity from the maternally inherited non-coding RNAs, with the unmethylated IG-CGI being irrelevant for that function. In addition, the key element on the paternally inherited chromosome is a germline-methylated IG-CGI that is required for methylation and repression of the IG-TRE.
Some ICRs, including the IG-DMR, have also been shown to bind AFF3, a component of the super elongation complex-like 3 (SEC-L3), on the methylated allele in ESCs where it is thought to interact with the ZFP57/TRIM28 complex (Luo et al., 2016). However, the function of this interaction is not clear as depletion of AFF3 in ESCs leads to decreased expression of the maternally expressed genes in the Dlk1-Dio3 region, demonstrating that it does not cooperate in protecting the IG-DMR from demethylation. Intriguingly, AFF3 also binds to a second region at the 3’ side of the IG-DMR downstream of the IG-TRE. Here, AFF3 is co-bound with ZFP281 but only on the unmethylated maternal copy (Wang et al., 2017). ZFP281 is a zinc finger protein that has previously been reported to act as both a transcriptional activator and a repressor (Wang et al., 2008). In the Dlk1-Dio3 locus, depletion of ZFP281 from ESCs leads to decreased AFF3 binding at this downstream region, but not at the methylated IG-CGI. As depletion of AFF3 leads to decreased expression of Gtl2, Mirg, and Rian, this suggests that the downstream bound region is relevant for AFF3 function and that it also acts as an enhancer for maternally expressed genes (Wang et al., 2017). Whether this second region is acting in concert with the IG-TRE to control the expression of Gtl2 and its associated transcripts remains to be established.
The combined results from the IG-DMR and the IG-CGI deletion indicate that the paternal IG-CGI is required to inactivate the paternal IG-TRE and maintain a repressive chromatin landscape on the paternal chromosome; this same element is dispensable on the maternal chromosome that is normally not methylated. On the other hand, the IG-TRE is dominant over the IG-CGI on the maternal chromosome where it is required to establish maternal gene expression and prevent methylation at the Gtl2-DMR. Together, the paradoxical effects imposed by distinct deletions within the IG-DMR represent an attractive experimental framework for dissecting the impact of changes in gene dosage on embryonic phenotypes. Synthesizing the result of the two genetic models shows that normal development cannot occur with biallelic expression of maternal genes and repression of Dlk1 or with biallelic expression of Dlk1 and repression of maternal transcripts. In both models, as in WT, monoallelic expression of the genes in this locus is consistent with normal development.
In accordance with that, flipping imprinting on both alleles produced viable offspring, showing that the parental origin of the imprint is irrelevant, provided appropriate balanced gene expression is established and maintained at this locus (Weinberg-Shukron et al., 2022). This has been demonstrated for another imprinted gene as well, where Zdbf2 dosage, regardless of parental origin, regulates postnatal body weight (Glaser et al., 2022). These studies emphasize the importance of exquisite dosage control by genomic imprinting and the adaptability of this epigenetically regulated mechanism in particular developmental contexts (Liao et al., 2021).
The role of the Gtl2-DMR and Gtl2 lncRNA in regional control
Monoallelic expression of Gtl2/Meg3 exclusively from the maternally inherited chromosome is first observed in e3.5 blastocysts (Nowak et al., 2011; Sato et al., 2011). This imprinted expression precedes the acquisition of methylation at the Gtl2 promoter on the paternal chromosome that is not observed until after e5.5. This suggests that transcription from the maternal promoter protects the maternal allele from gaining methylation and that methylation on the paternal chromosome occurs secondary to and in the absence of transcription. Once established, the Gtl2-DMR extends from the promoter into the first intron of the gene. Mouse models deleting the maternal Gtl2-DMR recapitulate the full ICR deletion, with the downregulation of maternally expressed genes, upregulation of paternally expressed genes, and embryos dying in utero (Takahashi et al., 2009; Zhou et al., 2010; Zhu et al., 2019) (Figures 2C–E). Furthermore, a patient with a maternal microdeletion of the Gtl2-DMR presented with features similar to UPD(14)Pat or maternal IG-DMR deletion patients (Kagami et al., 2010). Together, these data indicate that the unmethylated Gtl2-DMR on the maternal chromosome, once established, is able to act as an imprinting control region for the entire domain, but it is unclear whether this is by the Gtl2 lncRNA itself or via direct cis-acting elements within the DMR.
The Gtl2 gene transcribes a long non-coding RNA whose function as a regulatory transcript continues to be explored, and several different roles have been proposed. MEG3, the human ortholog of Gtl2, is downregulated in many forms of cancer and, therefore, is believed to function as a tumor suppressor (Makoukji et al., 2016; Buccarelli et al., 2020). The MEG3 lncRNA has been shown to interact with another tumor suppressor, p53, and influence the expression of p53 target genes (Zhou et al., 2007; Zhu et al., 2015). The lncRNA has also been found to interact with the polycomb repressive complex 2 (PRC2) in both mouse and human cells (Zhao et al., 2010; Mondal et al., 2015). In humans, MEG3 exon3 is thought to contain the region of interaction with PRC2; this exon is conserved with mouse exon3, suggesting a shared function between the two species. MEG3 is then thought to recruit PRC2 to its target genes in trans though interaction with GA-rich repeat regions and formation of RNA–DNA triplexes (Mondal et al., 2015).
In other imprinted regions, lncRNAs have been shown to silence other genes in the domain in cis (Pauler et al., 2012), and in the Dlk1-Dio3 domain, in vivo manipulations that activate Gtl2 on the paternal chromosome result in repression of Dlk1 (Lin et al., 2003; Li et al., 2008; Weinberg-Shukron et al., 2022). Furthermore, knockdown of the Gtl2 lncRNA leads to increased expression of Dlk1 coupled with decreased histone H3K27me3 over the Dlk1 gene in mouse ESCs (Zhao et al., 2010). This suggests that in mice, the Gtl2 lncRNA may facilitate PRC2 recruitment in cis and that one of its major functions is to repress paternally expressed genes on the maternally inherited chromosome (Figure 4A). However, in human iPSCs lacking MEG3 expression, no difference was observed in PRC2 occupancy over the DLK1 promoter, and there were no significant changes in the expression levels of either DLK1 or DIO3 compared with iPSCs that express MEG3 (Kaneko et al., 2014). It should be noted that these experiments were performed in vitro in cell lines where Dlk1 is only weakly expressed.
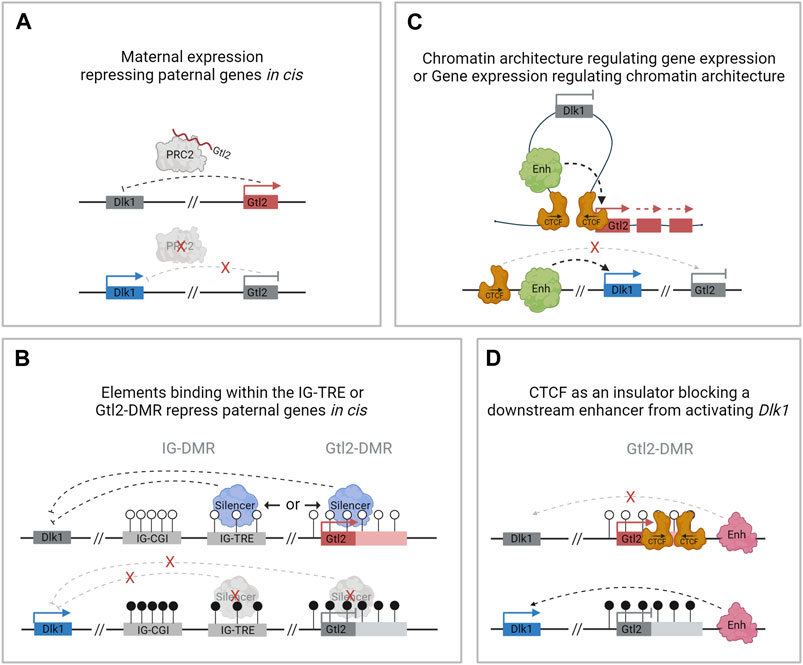
FIGURE 4. Possible mechanisms of regulation at the Dlk1-Dio3 locus by Gtl2. (A). Gtl2 lncRNA facilitates PRC2 recruitment in cis and represses paternally expressed genes on the maternally inherited chromosome. (B). The IG-TRE or the Gtl2-DMR harbors elements that directly repress paternal genes in cis. (C). Differential CTCF binding at the Gtl2-DMR regulates gene expression by restricting access to shared enhancers. (D). Differentially methylated CTCF binding sites at the Gtl2-DMR function as an insulator, preventing Dlk1 from being expressed from the maternal chromosome.
More recently, mouse models have also caused doubt regarding the idea that the Gtl2 lncRNA silences the Dlk1-Dio3 domain in vivo. Whereas a deletion model that removes Gtl2 exons 1–4 leads to the loss of imprinting in the whole domain (Figure 2E), deleting exons 2–4 causes downregulation of Gtl2 but no change in Dlk1 expression in e11.5 embryos (Figure 2F) (Zhu et al., 2019). This suggests that the Gtl2-DMR does not solely function to restrict Gtl2 expression to the maternal chromosome and that the lncRNA does not silence the paternally expressed genes on the maternal chromosome in cis. Instead, these observations suggest that the Gtl2-DMR harbors elements that can directly repress the expression of the paternally expressed genes in cis (Figure 4B). In agreement with this hypothesis are data from ESC deletions. Sanli et al. (2018) made ESC lines lacking either the Gtl2 promoter or intron 1. Intriguingly, the loss of intron 1 alone on the maternal chromosome was sufficient to silence Gtl2 and all the associated maternally expressed non-coding transcripts in ESCs and upon differentiation to NPCs. Furthermore, Dlk1 expression became biallelic in NPCs upon the loss of the maternal intron 1, indicating that Gtl2 intron 1 may be playing a vital role in imprinting control across this domain.
Gtl2 intron 1 is approximately 2.5 kb in length. It contains two CTCF binding sites that are conserved in eutherian mammals and are only able to bind CTCF on the unmethylated maternally inherited chromosome (Lin et al., 2011) (Figure 1D). CTCF plays an important role in genome organization and is frequently found at boundaries of topologically associating domains (TADs) which are self-interacting regions (Merkenschlager and Nora, 2016). Although TADs are thought to be maintained between different tissues, sub-TADs within them are tissue-specific and orchestrate local genomic contacts throughout development (Smith et al., 2016). Data from ESCs indicate that the Gtl2 CTCFs form the boundary of a parent-of-origin-specific sub-TAD on the maternal chromosome (Llères et al., 2019). Thus, another possible mechanism for establishing differential expression profiles between the two parental chromosomes could be that differential CTCF binding contributes to the formation of parental-origin-specific regulatory conformations (Figure 4C). Another possibility is that access to shared enhancers might be insulated through CTCF binding, enabling Gtl2 expression and Dlk1 repression (Figure 4D) consistent with a similar mechanism that is well established for the Igf2/H19 domain, where imprinted gene expression is controlled by differentially methylated CTCF binding sites in the ICR. The H19-CTCF sites are methylated on the paternally inherited chromosome and are thus only able to bind CTCF on the maternal chromosome, where they function as an insulator, preventing Igf2 from being expressed (Bell and Felsenfeld, 2000; Hark et al., 2000). In ESCs, the H19-ICR has been recently shown to form a maternal chromosome-specific sub-TAD boundary that splits the imprinted domain into two, which is similar to what is observed in the Dlk1-Dio3 locus. Deletion of one of the CTCFs in vitro has been shown to cause upregulation of Dlk1 (Llères et al., 2019). However, the extent to which the Gtl2 CTCFs can regulate gene expression in vivo remains to be established.
The Rian-DMR: A paternal chromosome-specific regulatory element?
Rian (RNA imprinted and accumulated in the nucleus) is a lncRNA that has more than 20 predicted alternative transcripts in mice. Its expression from the maternal chromosome has not been dissociated from Gtl2, and an individual promoter for this gene has not been identified or a transcription unit clearly defined; hence, it can be considered as a Gtl2-associated transcript driven by the IG-DMR and Gtl2-DMR. Nonetheless, RNA from this region acts as the host transcript for miRNAs and two clusters of C/D snoRNAs (Cavaillé et al., 2002). In humans, the Rian ortholog, MEG8 (along with MEG3), has been shown to be upregulated in many cancers and is thought to regulate many different pathways by acting as a molecular sponge for various miRNAs (Ghafouri-Fard et al., 2022). A Rian-DMR has been described in the second intron of the gene (Figure 1E; Figure 3A), and as opposed to other DMRs, in the Dlk1-Dio3 domain, this region is methylated in both sperm and oocytes, and then becomes hypomethylated in the blastocyst. The paternally inherited allele remains hypomethylated throughout development, whereas the maternally inherited copy becomes hypermethylated by e6.5 (Zeng et al., 2014). This hypermethylation on the maternal allele may be due to the normal methylation accumulation on actively transcribed gene bodies. Upon maternal transmission of the IG-DMR deletion, the Rian-DMR is lost; however, unlike the Gtl2-DMR, the Rian-DMR becomes biallelically hypomethylated (Figure 3B). This once again illustrates that appropriate methylation of the germline ICR is necessary to establish the epigenotype of the entire domain.
In mice, the DMR consists of a small CpG island that contains 12 copies of a GGCG repeat. This region is conserved and G-rich in eutherian mammals; however, the GGCG repeat is only seen in mice and rats. Upstream of the repeats is a conserved CTCF binding domain. Interestingly, this motif lacks CpG dinucleotides, so binding is not affected by methylation. In agreement with this, CTCF occupancy has been shown to be biallelic at this site (Zeng et al., 2014). Until now, the role of the Rian-DMR has been unclear, but a recent study has thrown some light on its function. Han et al. (2022) have shown that the DMR functions as an insulator in mouse MLTC-1 cells. Intriguingly, the CTCF binding region was only able to act as an insulator in the presence of the repeat element. They further deleted the entire DMR, and the CTCF site repeats individually to assess the role of the region on gene expression. A 661bp deletion of the entire DMR led to reduced Dlk1 and Rtl1 expression and increased expression of Gtl2, Rian, and Mirg. When the CTCF binding site alone was deleted, a similar but less pronounced effect was observed. Interestingly, the tandem repeat deletion only affected the expression of the downstream gene Mirg. These data indicate that the Rian-DMR functions on the unmethylated paternal chromosome to ensure the correct expression of Dlk1 and Rtl1 and the repression of Gtl2 and its associated transcripts (Zeng et al., 2014). However, more recent in vivo data from mice with a 434bp deletion of the CTCF binding site and the GGCG repeats show that the loss of this region has little phenotypic effect as maternal and paternal heterozygotes and homozygotes all survive to adulthood. Furthermore, no effect was observed at e12.5 on Dlk1, Rtl1, Dio3, Gtl2, Rian, or Mirg expression on either maternal or paternal transmission of the deletion. These mice do show increased expression of two miRNAs within the Rian gene, miR-118 and miR-341. Both miRNAs are significantly upregulated in paternal heterozygotes and homozygotes but not in maternal heterozygotes at e12.5. In addition, RNAseq data indicated that many other miRNAs in the region become upregulated on paternal deletion, suggesting a role of the Rian-DMR in preventing miRNA expression on the paternal chromosome (Zhang et al., 2023).
The Dlk1-DMR and Dlk1 isoforms
Both the promoter of Dlk1 and the last exon of Dlk1 contain CpG islands (Takada et al., 2000). Whereas the Dlk1 promoter does not show any parental-origin-specific methylation pattern, the smaller CpG island within the fifth exon is completely unmethylated on the maternal allele and partially methylated on the paternal allele (Takada et al., 2002). This differentially methylated region is termed the Dlk1-DMR (Figure 1B). Similar to the Gtl2-DMR, the Dlk1-DMR acquires paternal allele-specific methylation following fertilization (Gagne et al., 2014). The methylation pattern of this DMR remains dynamic in late embryonic development and into adulthood. Interestingly, the level of Dlk1-DMR methylation does not correlate with the level of Dlk1 expression. Takada et al. (2002) reported a different methylation profile per tissue (lung, muscle, liver, kidney, and brain), suggesting that the methylation is cell-type specific and that allele-specific methylation differences at the Dlk1-DMR may not have a role to play in transcriptional control. The function of the Dlk1-DMR remains to be elucidated.
Alternative splicing at exon 5 generates a membrane-bound and secreted isoform of Dlk1. The secreted isoforms, produced with a longer part of exon 5, include a juxtamembrane motif for cleavage by extracellular proteases, which is absent from constitutively membrane-bound isoforms. In the neurogenic niche, secreted Dlk1 is predominantly expressed by niche astrocytes, whereas neural stem cells (NSCs) express membrane-bound Dlk1 (Ferrón et al., 2011). Interestingly, membrane-bound DLK1 in NSCs is stimulated by astrocyte-secreted DLK1, and communication between these cell types in the neurogenic niche regulates NSC self-renewal.
Selective absence of imprinting of Dlk1 not Gtl2 in the neurogenic niche
Recent evidence suggests that imprinted genes can be selectively “switched on” or “switched off” in particular cell types or at specific developmental time-points to initiate a change in gene dosage that is essential for normal development (Ferrón et al., 2011; Ferrón et al., 2015). Intriguingly, some imprinted genes show a selective absence of imprinting in the neurogenic niche (Lozano-Ureña et al., 2017). The Igf2 gene, which is canonically expressed from the paternally inherited copy, is biallelically expressed in the choroid plexus (DeChiara et al., 1991; Giannoukakis et al., 1993; Lehtinen et al., 2011), and this selective absence of Igf2 imprinting is required for neurogenesis.
The vertebrate-specific atypical Notch ligand gene, Dlk1, is dosage-sensitive with different tissue-specific sensitivities to altered expression levels (Moon et al., 2002; Da Rocha et al., 2009). DLK1 is involved in a range of processes, including non-shivering thermogenesis, metabolism, and behavior (Wallace et al., 2010; Charalambous et al., 2012; García-Gutiérrez et al., 2018; Montalbán-Loro et al., 2021). In humans, DLK1 variants are associated with age at menarche (Day et al., 2017), type I diabetes (Wallace et al., 2010), and a range of cancers, including neural, breast, and liver cancer (Yin et al., 2006; Cai et al., 2016; Makoukji et al., 2016; Buccarelli et al., 2020). DLK1 is, therefore, a biomedically relevant key player in a diverse range of processes. Similar to Igf2, the Dlk1 gene shows selective absence of imprinting in the postnatal neurogenic niche, resulting in the activation of the repressed maternal allele via an unknown mechanism (Figure 5). This absence of imprinting is essential for normal adult neurogenesis (Ferrón et al., 2011). Unlike Dlk1, the neighboring gene Gtl2 keeps its imprinting in the neurogenic niche, suggesting a selective gene-specific regulation (Ferrón et al., 2011).
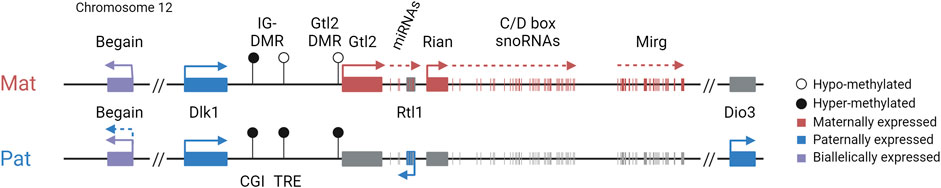
FIGURE 5. Selective absence of imprinting in the neurogenic niche; in the postnatal subventricular zone, niche astrocytes and neuronal stem cells exhibit biallelic expression of Dlk1, whereas Gtl2 retains imprinting and remains exclusively maternally expressed. This selective absence of imprinting is accompanied by increased methylation at the IG-CGI but not at the IG-TRE or Gtl2-DMR.
An important evolutionary question remaining to be elucidated is the time of switch in imprinted gene dosage. Although the dosage change is described as a selective absence of imprinting, it is not known whether biallelic expression of Dlk1 is “switched on” in specific cell types, representing a recently evolved function, or whether imprinting is “switched off” during development, representing an ancestral state prior to the evolution of imprinting, where Dlk1 is biallelically expressed (Edwards et al., 2008). Remarkably, this selective requirement for a double dose for neurogenesis is shared by the imprinted Igf2 gene (Ferrón et al., 2015). This emphasizes the importance of exquisite dosage control of certain genes by genomic imprinting, and the adaptability and flexibility of this epigenetically regulated mechanism in particular developmental contexts.
In conclusion, selective regulation of imprinting is probably a normal mechanism for modulating gene dosage to control stem cell potential in brain development and within the neurogenic niches throughout development and adult life (Perez et al., 2016). The dosage sensitivity of functionally important imprinted genes and the finding of highly selective absence of imprinting at Dlk1 and Igf2 in the brain suggest tight regulation of parental-origin-specific monoallelic expression. Dissecting the molecular players that participate in regulating imprints during postnatal neurogenesis will provide insights into the wider epigenetic control of the neurogenic process and uncover the molecular mechanisms underlying normal NSC function to understand tumoral processes in the adult brain. Therefore, unmasking the mechanism that regulates this time- and tissue-specific change in gene dosage is crucial for expanding our understanding of the physiological pathways regulated by imprinted genes in pathology and health.
Discussion
How to build an imprinted domain
Clearly, appropriate allele- and tissue-specific expression of the Dlk1-Dio3 region is necessary for normal mammalian development. Assessing different models that remove various elements in the region has allowed us to dissect the chain of events that is necessary to establish and maintain imprinted gene expression.
The first stage of the hierarchy is the establishment of the germline-DMR. In sperm, the IG-DMR becomes fully methylated before e19.5 (Hiura et al., 2007), whereas in oocytes, the region remains unmethylated (Figure 6, step 1). After fertilization, the presence of DNA methylation across the paternal IG-CGI allows the recruitment of oocyte-loaded and zygotically expressed ZFP57 to the region, which in turn recruits TRIM28 and DNMTs and ensures that the entire ICR, including the IG-TRE, remains unmethylated on the paternal chromosome (Figure 6, step 2). Meanwhile, on the maternally inherited chromosome, the unmethylated IG-CGI is unable to bind ZFP57 and the IG-TRE remains unmethylated. This allows the IG-TRE to bind transcription factors and act as an enhancer for Gtl2, causing it to be monoallelically expressed from the maternally inherited chromosome from e3.5 (Figure 6, step 3). DNA methylation starts to accumulate over the paternal copy of the Gtl2-DMR at e5.5 and is complete by e6.5 (Figure 6, step 4). At the same time, the other somatic DMRs are also established at Dlk1 and Rian (Figure 6, step 5). Once established, the Gtl2-DMR can control imprinted gene expression on the maternal chromosome either via the lncRNA recruiting PRC2, direct silencing, or insulator activity (Figure 6, step 6). On the paternal chromosome, the unmethylated Rian-DMR regulates paternal miRNA expression, possibly through its insulating properties.
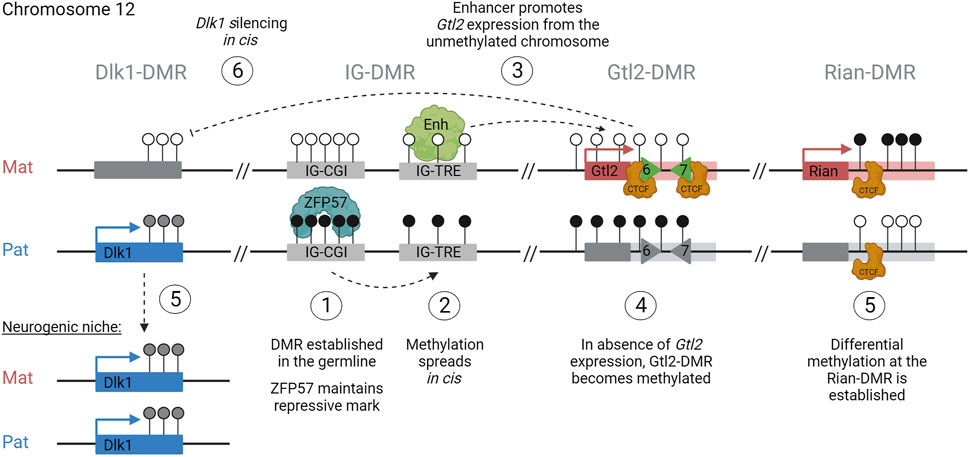
FIGURE 6. How to build an imprint: step 1: establishment of the germline-DMR (IG-CGI), methylated in sperm and unmethylated in oocytes. Step 2: maintenance of methylation across the entire paternal ICR (CGI + TRE). Step 3: monoallelic expression of Gtl2 from the maternal unmethylated allele. Step 4: accumulation of methylation over the paternal Gtl2-DMR. Step 5: establishment of somatic DMRs at Dlk1 and Rian. Step 6: regulation of gene expression profiles in cis, silencing Dlk1 from the maternal copy.
Remaining questions
Although much has been learned about how imprinting is established in this domain, many questions remain. First, it is not clear how the IG-DMR becomes methylated in the male germline yet remains unmethylated in oogenesis and how the maternal copy eludes de novo methylation in later development. Recently, it was shown that a mouse IG-DMR transgene acquired methylation during the post-fertilization period rather than in the sperm (Matsuzaki et al., 2023). This suggests that the transgene is lacking the sequence that initially attracts methylation to the element in the sperm. This “post-fertilization imprinted methylation” was previously reported in mouse and human H19 ICR transgenes as well (Matsuzaki et al., 2009). However, after implantation, the YAC transgene of the IG-DMR became highly methylated from both copies, suggesting that the IG-DMR fragment tested did not protect the maternal IG-DMR from genome-wide de novo DNA methylation. Interestingly, the fragment did not contain most of the IG-TRE, indicating that one function of the IG-TRE may be to protect the maternal sequence from global de novo methylation after implantation. Although maintenance of hypomethylation at the maternal H19 ICR is known to involve CTCF and Sox/Oct factors (Sakaguchi et al., 2013), the mechanism at the maternal IG-DMR is not fully understood; however, this region also contains Sox/Oct binding motifs.
Second, the mechanism by which the maternal IG-TRE directs monoallelic expression in the domain remains to be elucidated. It is known to contain many transcription factor binding motifs and shows low-level expression, suggesting that it most likely functions as an enhancer for Gtl2. However, it is possible that the IG-TRE also contains a silencer element that is capable of directly repressing the paternally expressed genes on the maternally inherited chromosome. Experiments dissecting this region further are necessary to tease apart these options.
Evidence from mice harboring deletions of the Gtl2-DMR and in vitro deletions of the Rian-DMR indicate that somatic DMRs can regulate parent-of-origin-specific expression in later development, but the mechanisms through which this is achieved are not fully understood. Intriguingly, both these DMRs are known to bind CTCF, so they may influence gene expression through mechanisms such as enhancer blocking. The parental-specific sub-TAD identified in vitro with the Gtl2-DMR at the border indicates that it has strong insulator activity. However, whether these parental-specific conformations are a cause or a consequence of differential expression patterns between the two chromosomes is uncertain (Figure 6). The role the Gtl2 lncRNA itself plays in the regulation of gene expression in the region also needs further exploration as it is uncertain whether it recruits PRC2 to the domain in vivo to bring about the epigenetic silencing of genes. Much of the research on conformation and the role of the lncRNA at the Dlk1-Dio3 locus has been performed in vitro, limiting the resolution of information, and thus, findings may not be recapitulated in vivo. For instance, as Dlk1 is lowly expressed in mESCs, experiments designed to look at the effect of perturbation models on paternal gene expression patterns are not informative in culture.
Finally, the Dlk1-Dio3 locus is interesting as the expression varies between tissues and cell types. We recently showed that there are two weakly biased genes at the edge of the Dlk1-Dio3 region: Wdr25 and Wars. Both genes showed a weak skew toward paternal expression, but only in brain tissues. This bias was shown to be under the control of the IG-DMR (Edwards et al., 2023), suggesting that its influence may be more extensive in neuronal tissues. Weakly biased genes were also found at the periphery of other imprinted regions, and further studies are needed to understand the functional and mechanistic implications of this observation. In addition to tissue-specific differences, unique cell types display selective absence of imprinting at the Dlk1-Dio3 domain in a temporal and spatial-specific manner. The mechanism that switches between monoallelic and biallelic expression remains to be elucidated and may provide insights into transcriptional control with wider implications for non-imprinted domains as well (Figure 6). Together, these observations indicate that the mechanisms regulating the imprinting of the Dlk1-Dio3 locus may vary between tissues and time-points in development.
This review highlights the importance of using in vivo models to tease apart the complex chain of epigenetic events that is required to establish and maintain imprinted gene expression throughout development. We also demonstrate that cell-type-specific modulation of this hierarchy is necessary to ensure the correct gene dosage in certain tissues, such as in the neurogenic niche—however, what these mechanistic steps are remains unclear. Together, this work illustrates how studying one imprinted region in detail can add a layer of sophistication to how we think about the epigenetic control of genome function and its consequences for spatial and temporal regulation more generally.
Methods
Southern blot (Figure 3): DNA was isolated by standard techniques (Sambrook et al., 2001). A total of 10 μg of restriction enzyme-digested DNA was separated on a 0.5% TBE gel before transferring to Hybond-N+ (GE Healthcare Life Sciences) nylon membranes. Membranes were pre-hybridized in ULTRAhyb (Ambion) for at least 1 h. The probe was a PCR fragment amplified with 5′-AGTGGCCCAACTTCTATCGG and 5′-GGAACAGAGACCTCCTAAGG, which was labeled with [α-32P]dCTP using the Megaprime DNA labeling system (GE Healthcare Life Sciences) and then purified with ProbeQuant G50 Micro-Columns (GE Healthcare Life Sciences) before being added to the hybridization solution and incubated at 42°C overnight. Filters were washed to a stringency of 0.2X SSC/0.1%SDS at 65°C and then exposed to PhosphorImager Screens (Molecular Dynamics).
Author contributions
AW-S: conceptualization, funding acquisition, visualization, writing–original draft, and writing–review and editing. NY: data curation, investigation, methodology, validation, and writing–review and editing. AF-S: conceptualization, funding acquisition, project administration, resources, supervision, writing–original draft, and writing–review and editing. CE: conceptualization, funding acquisition, project administration, supervision, writing–original draft, and writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AW-S was supported by the Human Frontier Science Program (LT000966/2020-L), the Rothschild Yad Hanadiv Fellowship program, the Humanitarian trust, and the Israel National Postdoctoral Award for Advancing Women in Science. The study was supported by an MRC program grant (MR/X018407/1) awarded to AF-S.
Acknowledgments
Figures were created with BioRender.com. For the purpose of open access, the authors have applied a Creative Commons Attribution (CC BY) license to any author accepted manuscript version arising from this submission.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aman, L. C. S., Manning, K. E., Whittington, J. E., and Holland, A. J. (2018). Mechanistic insights into the genetics of affective psychosis from Prader-Willi syndrome. Lancet Psychiatry 5, 370–378. doi:10.1016/S2215-0366(18)30009-9
Aronson, B. E., Scourzic, L., Shah, V., Swanzey, E., Kloetgen, A., Polyzos, A., et al. (2021). A bipartite element with allele-specific functions safeguards DNA methylation imprints at the Dlk1-Dio3 locus. Dev. Cell 56, 3052–3065.e5. doi:10.1016/j.devcel.2021.10.004
Azzi, S., Brioude, F., Bouc, Y., and Netchine, I. (2014). Human imprinting anomalies in fetal and childhood growth disorders: clinical implications and molecular mechanisms. Curr. Pharm. Des. 20, 1751–1763. doi:10.2174/13816128113199990525
Bartolomei, M. S., and Ferguson-Smith, A. C. (2011). Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 3, a002592. doi:10.1101/cshperspect.a002592
Bell, A. C., and Felsenfeld, G. (2000). Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485. doi:10.1038/35013100
Bondy, C. A. (2006). Genomic imprinting in Turner syndrome. Int. Congr. Ser. 1298, 21–25. doi:10.1016/j.ics.2006.06.008
Buccarelli, M., Lulli, V., Giuliani, A., Signore, M., Martini, M., D'Alessandris, Q. G., et al. (2020). Deregulated expression of the imprinted DLK1-DIO3 region in glioblastoma stemlike cells: tumor suppressor role of lncRNA MEG3. Neuro Oncol. 22, 1771–1784. doi:10.1093/neuonc/noaa127
Cai, C. M., Xiao, X., Wu, B. H., Wei, B. F., and Han, Z. G. (2016). Targeting endogenous DLK1 exerts antitumor effect on hepatocellular carcinoma through initiating cell differentiation. Oncotarget 7, 71466–71476. doi:10.18632/oncotarget.12214
Cavaillé, J., Seitz, H., Paulsen, M., Ferguson-Smith, A. C., and Bachellerie, J. P. (2002). Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 11, 1527–1538. doi:10.1093/hmg/11.13.1527
Charalambous, M., Ferron, S. R., da Rocha, S. T., Murray, A. J., Rowland, T., Ito, M., et al. (2012). Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 15, 209–221. doi:10.1016/j.cmet.2012.01.006
Cleaton, M. A. M., Edwards, C. A., and Ferguson-Smith, A. C. (2014). Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu. Rev. Genomics Hum. Genet. 15, 93–126. doi:10.1146/annurev-genom-091212-153441
Danko, C. G., Hyland, S. L., Core, L. J., Martins, A. L., Waters, C. T., Lee, H. W., et al. (2015). Identification of active transcriptional regulatory elements from GRO-seq data. Nat. Methods 12, 433–438. doi:10.1038/nmeth.3329
Da Rocha, S. T., Charalambous, M., Lin, S. P., Gutteridge, I., Ito, Y., Gray, D., et al. (2009). Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: implications for the evolution of imprinting. PLoS Genet. 5, e1000392. doi:10.1371/journal.pgen.1000392
Day, F. R., Thompson, D. J., Helgason, H., Chasman, D. I., Finucane, H., Sulem, P., et al. (2017). Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet. 49, 834–841. doi:10.1038/ng.3841
DeChiara, T. M., Robertson, E. J., and Efstratiadis, A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859. doi:10.1016/0092-8674(91)90513-x
Demars, J., and Gicquel, C. (2012). Epigenetic and genetic disturbance of the imprinted 11p15 region in Beckwith-Wiedemann and Silver-Russell syndromes. Clin. Genet. 81, 350–361. doi:10.1111/j.1399-0004.2011.01822.x
Edwards, C. A., Mungall, A. J., Matthews, L., Ryder, E., Gray, D. J., Pask, A. J., et al. (2008). The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6, e135–e1305. doi:10.1371/journal.pbio.0060135
Edwards, C. A., Watkinson, W. M. D., Telerman, S. B., Hulsmann, L. C., Hamilton, R. S., and Ferguson-Smith, A. C. (2023). Reassessment of weak parent-of-origin expression bias shows it rarely exists outside of known imprinted regions. Elife 12, e83364. doi:10.7554/eLife.83364
Elena Martinez, M., Charalambous, M., Saferali, A., Fiering, S., Naumova, A. K., St Germain, D., et al. (2014). Genomic imprinting variations in the mouse type 3 deiodinase gene between tissues and brain regions. Mol. Endocrinol. 28, 1875–1886. doi:10.1210/me.2014-1210
Ferguson-Smith, A. C. (2011). Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575. doi:10.1038/nrg3032
Ferguson-Smith, A. C., and Bourc’his, D. (2018). The discovery and importance of genomic imprinting. Elife 7, e42368. doi:10.7554/eLife.42368
Ferrón, S. R., Charalambous, M., Radford, E., McEwen, K., Wildner, H., Hind, E., et al. (2011). Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 475, 381–385. doi:10.1038/nature10229
Ferrón, S. R., Radford, E. J., Domingo-Muelas, A., Kleine, I., Ramme, A., Gray, D., et al. (2015). Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat. Commun. 6, 8265. doi:10.1038/ncomms9265
Flashner, B. M., Russo, M. E., Boileau, J. E., Leong, D. W., and Gallicano, G. I. (2013). Epigenetic factors and autism spectrum disorders. Neuromolecular Med. 15, 339–350. doi:10.1007/s12017-013-8222-5
Furutachi, S., Matsumoto, A., Nakayama, K. I., and Gotoh, Y. (2013). p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 32, 970–981. doi:10.1038/emboj.2013.50
Gagne, A., Hochman, A., Qureshi, M., Tong, C., Arbon, J., McDaniel, K., et al. (2014). Analysis of DNA methylation acquisition at the imprinted Dlk1 locus reveals asymmetry at CpG dyads. Epigenetics Chromatin 7, 9. doi:10.1186/1756-8935-7-9
García-Gutiérrez, M. S., Navarrete, F., Laborda, J., and Manzanares, J. (2018). Deletion of Dlk1 increases the vulnerability to developing anxiety-like behaviors and ethanol consumption in mice. Biochem. Pharmacol. 158, 37–44. doi:10.1016/j.bcp.2018.09.029
Ghafouri-Fard, S., Khoshbakht, T., Hussen, B. M., Taheri, M., and Shojaei, S. (2022). A review on the role of MEG8 lncRNA in human disorders. Cancer Cell Int. 22, 285–289. doi:10.1186/s12935-022-02705-9
Giannoukakis, N., Deal, C., Paquette, J., Goodyer, C. G., and Polychronakos, C. (1993). Parental genomic imprinting of the human IGF2 gene. Nat. Genet. 4, 98–101. doi:10.1038/ng0593-98
Glaser, J., Iranzo, J., Borensztein, M., Marinucci, M., Gualtieri, A., Jouhanneau, C., et al. (2022). The imprinted Zdbf2 gene finely tunes control of feeding and growth in neonates. Elife 11, e65641. doi:10.7554/eLife.65641
Han, X., He, H., Shao, L., Cui, S., Yu, H., Zhang, X., et al. (2022). Deletion of meg8-DMR enhances migration and invasion of MLTC-1 depending on the CTCF binding sites. Int. J. Mol. Sci. 23, 8828. doi:10.3390/ijms23158828
Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M., and Tilghman, S. M. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489. doi:10.1038/35013106
Hiura, H., Komiyama, J., Shirai, M., Obata, Y., Ogawa, H., and Kono, T. (2007). DNA methylation imprints on the IG-DMR of the Dlk1–Gtl2 domain in mouse male germline. FEBS Lett. 581, 1255–1260. doi:10.1016/j.febslet.2007.02.034
Ingason, A., Kirov, G., Giegling, I., Hansen, T., Isles, A. R., Jakobsen, K. D., et al. (2011). Maternally derived microduplications at 15q11-q13: implication of imprinted genes in psychotic illness. Am. J. Psychiatry 168, 408–417. doi:10.1176/appi.ajp.2010.09111660
Ioannides, Y., Lokulo-Sodipe, K., Mackay, D. J. G., Davies, J. H., and Temple, I. K. (2014). Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J. Med. Genet. 51, 495–501. doi:10.1136/jmedgenet-2014-102396
Ishida, M., and Moore, G. E. (2013). The role of imprinted genes in humans. Mol. Asp. Med. 34, 826–840. doi:10.1016/j.mam.2012.06.009
Isles, A. R., Ingason, A., Lowther, C., Walters, J., Gawlick, M., Stöber, G., et al. (2016). Parental origin of interstitial duplications at 15q11.2-q13.3 in schizophrenia and neurodevelopmental disorders. PLoS Genet. 12, e1005993. doi:10.1371/journal.pgen.1005993
Ito, M., Sferruzzi-Perri, A. N., Edwards, C. A., Adalsteinsson, B. T., Allen, S. E., Loo, T. H., et al. (2015). A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development 142, 2425–2430. doi:10.1242/dev.121996
Kagami, M., Kurosawa, K., Miyazaki, O., Ishino, F., Matsuoka, K., and Ogata, T. (2015). Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome). Eur. J. Hum. Genet. 23, 1488–1498. doi:10.1038/ejhg.2015.13
Kagami, M., O'Sullivan, M. J., Green, A. J., Watabe, Y., Arisaka, O., Masawa, N., et al. (2010). The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 6, e1000992. doi:10.1371/journal.pgen.1000992
Kalish, J. M., Jiang, C., and Bartolomei, M. S. (2014). Epigenetics and imprinting in human disease. Int. J. Dev. Biol. 58, 291–298. doi:10.1387/ijdb.140077mb
Kameswaran, V., Bramswig, N. C., McKenna, L. B., Penn, M., Schug, J., Hand, N. J., et al. (2014). Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 19, 135–145. doi:10.1016/j.cmet.2013.11.016
Kaneko, S., Bonasio, R., Saldaña-Meyer, R., Yoshida, T., Son, J., Nishino, K., et al. (2014). Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300. doi:10.1016/j.molcel.2013.11.012
Keverne, E. B. (1997). Genomic imprinting in the brain. Curr. Opin. Neurobiol. 7, 463–468. doi:10.1016/s0959-4388(97)80023-2
Kitazawa, M., Hayashi, S., Imamura, M., Takeda, S., Oishi, Y., Kaneko-Ishino, T., et al. (2020). Deficiency and overexpression of Rtl1 in the mouse cause distinct muscle abnormalities related to Temple and Kagami-Ogata syndromes. Development 147, dev185918. doi:10.1242/dev.185918
Kitazawa, M., Sutani, A., Kaneko-Ishino, T., and Ishino, F. (2021). The role of eutherian-specific RTL1 in the nervous system and its implications for the Kagami-Ogata and Temple syndromes. Genes Cells 26, 165–179. doi:10.1111/gtc.12830
Kojima, S., Shiochi, N., Sato, K., Yamaura, M., Ito, T., Yamamura, N., et al. (2022). Epigenome editing reveals core DNA methylation for imprinting control in the Dlk1-Dio3 imprinted domain. Nucleic Acids Res. 50, 5080–5094. doi:10.1093/nar/gkac344
Labialle, S., Marty, V., Bortolin-Cavaillé, M. L., Hoareau-Osman, M., Pradère, J. P., Valet, P., et al. (2014). The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J. 33, 2216–2230. doi:10.15252/embj.201387038
Lehtinen, M. K., Zappaterra, M. W., Chen, X., Yang, Y. J., Hill, A. D., Lun, M., et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905. doi:10.1016/j.neuron.2011.01.023
Lepage, J. F., Hong, D. S., Mazaika, P. K., Raman, M., Sheau, K., Marzelli, M. J., et al. (2013). Genomic imprinting effects of the X chromosome on brain morphology. J. Neurosci. 33, 8567–8574. doi:10.1523/JNEUROSCI.5810-12.2013
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., et al. (2008). A maternal-zygotic effect gene Zfp57 maintains both maternal and paternal imprints. Dev. Cell 15, 547–557. doi:10.1016/j.devcel.2008.08.014
Liao, J., Zeng, T. B., Pierce, N., Tran, D. A., Singh, P., Mann, J. R., et al. (2021). Prenatal correction of IGF2 to rescue the growth phenotypes in mouse models of Beckwith-Wiedemann and Silver-Russell syndromes. Cell Rep. 34, 108729. doi:10.1016/j.celrep.2021.108729
Lin, S., Ferguson-Smith, A. C., Schultz, R. M., and Bartolomei, M. S. (2011). Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci. Mol. Cell Biol. 31, 3094–3104. doi:10.1128/MCB.01449-10
Lin, S. P., Coan, P., da Rocha, S. T., Seitz, H., Cavaille, J., Teng, P. W., et al. (2007). Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development 134, 417–426. doi:10.1242/dev.02726
Lin, S. P., Youngson, N., Takada, S., Seitz, H., Reik, W., Paulsen, M., et al. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97–102. doi:10.1038/ng1233
Liu, C., Teng, Z. Q., Santistevan, N. J., Szulwach, K. E., Guo, W., Jin, P., et al. (2010). Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444. doi:10.1016/j.stem.2010.02.017
Llères, D., Moindrot, B., Pathak, R., Piras, V., Matelot, M., Pignard, B., et al. (2019). CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains. Genome Biol. 20, 272. doi:10.1186/s13059-019-1896-8
Lozano-Ureña, A., Montalbán-Loro, R., Ferguson-Smith, A. C., and Ferrón, S. R. (2017). Genomic imprinting and the regulation of postnatal neurogenesis. Brain Plast. 3, 89–98. doi:10.3233/BPL-160041
Luo, Z., Lin, C., Woodfin, A. R., Bartom, E. T., Gao, X., Smith, E. R., et al. (2016). Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev. 30, 92–101. doi:10.1101/gad.270413.115
Makoukji, J., Makhoul, N. J., Khalil, M., El-Sitt, S., Aldin, E. S., Jabbour, M., et al. (2016). Gene expression profiling of breast cancer in Lebanese women. Sci. Rep. 6, 36639. doi:10.1038/srep36639
Marty, V., Labialle, S., Bortolin-Cavaillé, M. L., Ferreira De Medeiros, G., Moisan, M. P., Florian, C., et al. (2016). Deletion of the miR-379/miR-410 gene cluster at the imprinted Dlk1-Dio3 locus enhances anxiety-related behaviour. Hum. Mol. Genet. 25, 728–739. doi:10.1093/hmg/ddv510
Matsuzaki, H., Okamura, E., Shimotsuma, M., Fukamizu, A., and Tanimoto, K. (2009). A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independently of its establishment in germ cells. Mol. Cell Biol. 29, 4595–4603. doi:10.1128/MCB.00275-09
Matsuzaki, H., Sugihara, S., and Tanimoto, K. (2023). The transgenic IG-DMR sequence of the mouse Dlk1-Dio3 domain acquired imprinted DNA methylation during the post-fertilization period. Epigenetics Chromatin 16, 7. doi:10.1186/s13072-023-00482-x
Merkenschlager, M., and Nora, E. P. (2016). CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17, 17–43. doi:10.1146/annurev-genom-083115-022339
Messerschmidt, D. M., de Vries, W., Ito, M., Solter, D., Ferguson-Smith, A., and Knowles, B. B. (2012). Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502. doi:10.1126/science.1216154
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Reinius, B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 6 (1), 7743. doi:10.1038/ncomms8743
Montalbán-Loro, R., Lassi, G., Lozano-Ureña, A., Perez-Villalba, A., Jiménez-Villalba, E., Charalambous, M., et al. (2021). Dlk1 dosage regulates hippocampal neurogenesis and cognition. Proc. Natl. Acad. Sci. U. S. A. 118, e2015505118. doi:10.1073/pnas.2015505118
Moon, Y. S., Smas, C. M., Lee, K., Villena, J. A., Kim, K. H., Yun, E. J., et al. (2002). Mice lacking paternally expressed pref-1/dlk1 display growth retardation and accelerated adiposity. Mol. Cell Biol. 22, 5585–5592. doi:10.1128/mcb.22.15.5585-5592.2002
Nicholls, R. D., Saitoh, S., and Horsthemke, B. (1998). Imprinting in prader-willi and angelman syndromes. Trends Genet. 14, 194–200. doi:10.1016/s0168-9525(98)01432-2
Nowak, K., Stein, G., Powell, E., He, L. M., Naik, S., Morris, J., et al. (2011). Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus. Epigenetics 6, 1012–1020. doi:10.4161/epi.6.8.16075
Pauler, F. M., Barlow, D. P., and Hudson, Q. J. (2012). Mechanisms of long range silencing by imprinted macro non-coding RNAs. Curr. Opin. Genet. Dev. 22, 283–289. doi:10.1016/j.gde.2012.02.005
Perez, J. D., Rubinstein, N. D., and Dulac, C. (2016). New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu. Rev. Neurosci. 39, 347–384. doi:10.1146/annurev-neuro-061010-113708
Pinto, C., Souza, R. P., Lioult, D., Semeralul, M., Kennedy, J. L., Warsh, J. J., et al. (2011). Parent of origin effect and allelic expression imbalance of the serotonin transporter in bipolar disorder and suicidal behaviour. Eur. Arch. Psychiatry Clin. Neurosci. 261, 533–538. doi:10.1007/s00406-011-0192-8
Quenneville, S., Verde, G., Corsinotti, A., Kapopoulou, A., Jakobsson, J., Offner, S., et al. (2011). In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372. doi:10.1016/j.molcel.2011.08.032
Rocha, S. T. da, Edwards, C. A., Ito, M., Ogata, T., and Ferguson-Smith, A. C. (2008). Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306–316. doi:10.1016/j.tig.2008.03.011
Saito, T., Hara, S., Kato, T., Tamano, M., Muramatsu, A., Asahara, H., et al. (2018). A tandem repeat array in IG-DMR is essential for imprinting of paternal allele at the Dlk1-Dio3 domain during embryonic development. Hum. Mol. Genet. 27, 3283–3292. doi:10.1093/hmg/ddy235
Sakaguchi, R., Okamura, E., Matsuzaki, H., Fukamizu, A., and Tanimoto, K. (2013). Sox-Oct motifs contribute to maintenance of the unmethylated H19 ICR in YAC transgenic mice. Hum. Mol. Genet. 22, 4627–4637. doi:10.1093/hmg/ddt311
Sanli, I., Lalevée, S., Cammisa, M., Perrin, A., Rage, F., Llères, D., et al. (2018). Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis. Cell Rep. 23, 337–348. doi:10.1016/j.celrep.2018.03.044
Sato, S., Yoshida, W., Soejima, H., Nakabayashi, K., and Hata, K. (2011). Methylation dynamics of IG-DMR and Gtl2-DMR during murine embryonic and placental development. Genomics 98, 120–127. doi:10.1016/j.ygeno.2011.05.003
Seitz, H., Royo, H., Bortolin, M. L., Lin, S. P., Ferguson-Smith, A. C., and Cavaillé, J. (2004). A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 14, 1741–1748. doi:10.1101/gr.2743304
Sekita, Y., Wagatsuma, H., Nakamura, K., Ono, R., Kagami, M., Wakisaka, N., et al. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248. doi:10.1038/ng.2007.51
Smallwood, S. A., Tomizawa, S. I., Krueger, F., Ruf, N., Carli, N., Segonds-Pichon, A., et al. (2011). Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814. doi:10.1038/ng.864
Smith, E. M., Lajoie, B. R., Jain, G., and Dekker, J. (2016). Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am. J. Hum. Genet. 98, 185–201. doi:10.1016/j.ajhg.2015.12.002
Smith, Z. D., and Meissner, A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220. doi:10.1038/nrg3354
Stelzer, Y., Wu, H., Song, Y., Shivalila, C. S., Markoulaki, S., and Jaenisch, R. (2016). Parent-of-Origin DNA methylation dynamics during mouse development. Cell Rep. 16, 3167–3180. doi:10.1016/j.celrep.2016.08.066
Takada, S., Paulsen, M., Tevendale, M., Tsai, C. E., Kelsey, G., Cattanach, B. M., et al. (2002). Epigenetic analysis of the Dlk1–Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2–H19. Hum. Mol. Genet. 11, 77–86. doi:10.1093/hmg/11.1.77
Takada, S., Tevendale, M., Baker, J., Georgiades, P., Campbell, E., Freeman, T., et al. (2000). Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr. Biol. 10, 1135–1138. doi:10.1016/s0960-9822(00)00704-1
Takahashi, N., Coluccio, A., Thorball, C. W., Planet, E., Offner, S., Turelli, P., et al. (2019). ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 33, 49–54. doi:10.1101/gad.320069.118
Takahashi, N., Okamoto, A., Kobayashi, R., Shirai, M., Obata, Y., Ogawa, H., et al. (2009). Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum. Mol. Genet. 18, 1879–1888. doi:10.1093/hmg/ddp108
Tierling, S., Dalbert, S., Schoppenhorst, S., Tsai, C. E., Oliger, S., Ferguson-Smith, A. C., et al. (2006). High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics 87, 225–235. doi:10.1016/j.ygeno.2005.09.018
Tierling, S., Gasparoni, G., Youngson, N., and Paulsen, M. (2009). The Begain gene marks the centromeric boundary of the imprinted region on mouse chromosome 12. Mamm. Genome 20, 699–710. doi:10.1007/s00335-009-9205-6
Tsai, C. E., Lin, S. P., Ito, M., Takagi, N., Takada, S., and Ferguson-Smith, A. C. (2002). Genomic imprinting contributes to thyroid hormone metabolism in the mouse embryo. Curr. Biol. 12, 1221–1226. doi:10.1016/s0960-9822(02)00951-x
Tsan, Y. C., Morell, M. H., and O’Shea, K. S. (2016). miR-410 controls adult SVZ neurogenesis by targeting neurogenic genes. Stem Cell Res. 17, 238–247. doi:10.1016/j.scr.2016.07.003
Tucci, V., Isles, A. R., Kelsey, G., and Ferguson-Smith, A. C.Erice Imprinting Group (2019). Genomic imprinting and physiological processes in mammals. Cell 176, 952–965. doi:10.1016/j.cell.2019.01.043
Tucker, K. L., Beard, C., Dausmann, J., Jackson-Grusby, L., Laird, P. W., Lei, H., et al. (1996). Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10, 1008–1020. doi:10.1101/gad.10.8.1008
Uribe-Lewis, S., Woodfine, K., Stojic, L., and Murrell, A. (2011). Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev. Mol. Med. 13, e2. doi:10.1017/S1462399410001717
Wallace, C., Smyth, D. J., Maisuria-Armer, M., Walker, N. M., Todd, J. A., and Clayton, D. G. (2010). The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 42, 68–71. doi:10.1038/ng.493
Wang, Y., Shen, Y., Dai, Q., Yang, Q., Zhang, Y., Wang, X., et al. (2017). A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron. Nucleic Acids Res. 45, 1177–1185. doi:10.1093/nar/gkw1051
Wang, Z.-X., Teh, C. H. L., Chan, C. M. Y., Chu, C., Rossbach, M., Kunarso, G., et al. (2008). The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells 26, 2791–2799. doi:10.1634/stemcells.2008-0443
Weinberg-Shukron, A., Ben-Yair, R., Takahashi, N., Dunjić, M., Shtrikman, A., Edwards, C. A., et al. (2022). Balanced gene dosage control rather than parental origin underpins genomic imprinting. Nat. Commun. 13, 4391. doi:10.1038/s41467-022-32144-z
Yin, D., Xie, D., Sakajiri, S., Miller, C. W., Zhu, H., Popoviciu, M. L., et al. (2006). DLK1: increased expression in gliomas and associated with oncogenic activities. Oncogene 25, 1852–1861. doi:10.1038/sj.onc.1209219
Youngson, N. A., Kocialkowski, S., Peel, N., and Ferguson-Smith, A. C. (2005). A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481–490. doi:10.1007/s00239-004-0332-0
Zeng, T. B., He, H. J., Han, Z. B., Zhang, F. W., Huang, Z. J., Liu, Q., et al. (2014). DNA methylation dynamics of a maternally methylated DMR in the mouse Dlk1–Dio3 domain. FEBS Lett. 588, 4665–4671. doi:10.1016/j.febslet.2014.10.038
Zhang, L., He, H., Zhang, X., Zhang, M., Li, B., Han, Z., et al. (2023). Meg8-DMR as the secondary regulatory region regulates the expression of MicroRNAs while it does not affect embryonic development in mice. Genes (Basel) 14, 1264. doi:10.3390/genes14061264
Zhao, J., Ohsumi, T. K., Kung, J. T., Ogawa, Y., Grau, D. J., Sarma, K., et al. (2010). Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953. doi:10.1016/j.molcel.2010.12.011
Zhou, Y., Cheunsuchon, P., Nakayama, Y., Lawlor, M. W., Zhong, Y., Rice, K. A., et al. (2010). Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 137, 2643–2652. doi:10.1242/dev.045724
Zhou, Y., Zhong, Y., Wang, Y., Zhang, X., Batista, D. L., Gejman, R., et al. (2007). Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742. doi:10.1074/jbc.M702029200
Zhu, J., Liu, S., Ye, F., Shen, Y., Tie, Y., Zhu, J., et al. (2015). Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One 10, e0139790. doi:10.1371/journal.pone.0139790
Keywords: Dlk1-Dio3 domain, genomic imprinting, CTCF, chromatin architecture, DNA methylation, long non-coding RNA
Citation: Weinberg-Shukron A, Youngson NA, Ferguson-Smith AC and Edwards CA (2023) Epigenetic control and genomic imprinting dynamics of the Dlk1-Dio3 domain. Front. Cell Dev. Biol. 11:1328806. doi: 10.3389/fcell.2023.1328806
Received: 27 October 2023; Accepted: 27 November 2023;
Published: 12 December 2023.
Edited by:
Mellissa Mann, University of Pittsburgh, United StatesReviewed by:
Anthony Isles, Cardiff University, United KingdomYoichi Sekita, Kitasato University, Japan
Copyright © 2023 Weinberg-Shukron, Youngson, Ferguson-Smith and Edwards. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ariella Weinberg-Shukron, YXc5MzdAY2FtLmFjLnVr; Anne C. Ferguson-Smith, YWZzbWl0aEBnZW4uY2FtLmFjLnVr; Carol A. Edwards, Y2FlMjhAY2FtLmFjLnVr
 Ariella Weinberg-Shukron
Ariella Weinberg-Shukron Neil A. Youngson
Neil A. Youngson Anne C. Ferguson-Smith1*
Anne C. Ferguson-Smith1* Carol A. Edwards
Carol A. Edwards