- 1Department of Physiology, Immunology and Pathophysiology, Faculty of Medicine, University of Rijeka, Rijeka, Croatia
- 2University North, University Center Varaždin, Varaždin, Croatia
1 Introduction
Cytomegaloviruses (CMV) cause productive, persistent or latent infections in their hosts. Productive infection requires the establishment of a complete manufacturing chain in the nucleus and cytosol for the successful production and release of infectious virions. This establishment is associated with overproduction of virus-encoded proteins and extensive reorganization of host cell pathways. The cytoplasmic part of the manufacturing chain involves a complex reorganization of the membrane system into a large structure known as the assembly compartment (AC), which adjusts the membrane composition to concentrate all essential components for the final stage of virion production known as the secondary envelopment and provides the exit pathway for the release of assembled virions from the cell (Wofford et al., 2020). These crucial steps in the CMV life cycle (Supplementary Figure S1) are still poorly understood and need to be regularly reconciled with advances in herpesvirus biology and membrane trafficking.
2 Cytoplasmic assembly compartment
The extensive reorganization of the membrane system has been investigated in several electron microscopy (EM) (Maninger et al., 2011; Schauflinger et al., 2013; Read et al., 2019; Taisne et al., 2019; Momtaz et al., 2021; Flomm et al., 2022; Wedemann et al., 2022) and immunofluorescence-based studies (Supplementary Table S1). This reorganization involves displacement the Golgi stacks into a ring-like configuration (outer AC) which surrounds the expanded membranous elements (inner AC), mainly the intermediates of the interface between early endosomes, the endosomal recycling compartment, and the trans-Golgi network (EE-ERC-TGN) (Das et al., 2007; Lučin et al., 2020). The overall structure relocates late endosomes (LE) and the endoplasmic reticulum (ER) to the cell periphery. Membrane expansion in the inner AC is associated with extensive tubulation and sustained recruitment of tubulation machinery. In addition, the AC is rich in vacuolized membrane structures that are often loaded with enveloped virions.
An important goal of membrane reorganization is to create a suitable environment for membrane envelopment of the nucleocapsids and to pave the exit pathway for the enveloped virions. These changes, initiated in the early phase of infection and completed after viral DNA replication, include redistribution and over-recruitment of proteins that regulate membrane trafficking (Beltran et al., 2016; Cook and Cristea, 2019; Lučin et al., 2020), alteration of lipid composition (Liu et al., 2011), retention of membrane trafficking and inhibition of endosomal recycling (Tomaš et al., 2010; Hook et al., 2014; Karleuša et al., 2018; Maschkowitz et al., 2018), and even establishment of heterotypic membranous organelles within the endosomal system in the post-sorting phase of endosomes (Cepeda et al., 2010; Zeltzer et al., 2018). However, little is known about the biogenesis and membrane composition of thousands of membranous units that make up the AC.
At least 14 virus-encoded tegument proteins and 11 glycoproteins are embedded in virions during the secondary envelopment (Varnum et al., 2004), and their concentration at the envelopment organelle is required. Studies focusing on the trafficking of highly conserved glycoproteins (including the gB, gH/gL/gO and gM/gN complexes) (Tugizov et al., 1999; Jarvis et al., 2002; Crump et al., 2003; Krzyzaniak et al., 2009; Maschkowitz et al., 2018) revealed their different maturation pathways. They concentrate in the trans-Golgi and circulate via different routes through the PM, endosomes and TGN. Several studies suggest that the clathrin-dependent endocytic uptake of viral glycoproteins, their delivery into the endosomal system and retrieval at the recycling membranes may be essential for the process of secondary envelopment (Krzyzaniak et al., 2009; Kropff et al., 2010; Wu et al., 2020). The identification of endosomal host cell cargo proteins (i.e., MHC class I proteins and Lamp1), several Rab GTPases (i.e., Rabs 12, 18, 23, and 32), cargo sorting proteins (i.e., sorting nexins 2 and 3), SNARE proteins (i.e., STX12, VAMP2, and VAMP3), and lipid-modifying proteins (i.e., PIK3C2A) as host cell membrane signatures that remain in virions (Mahmutefendić Lučin et al., 2022), support the idea that the distant stage of EE maturation in one of the endosomal recycling pathways may provide the retrieval machinery required for retention of viral glycoproteins. On the other hand, little is known about the concentration of tegument proteins, but they may be retained at the envelopment membrane by direct interaction with the retrieval machinery or by complex formation with retrieved glycoproteins (Seo and Britt, 2006; Wu et al., 2020).
3 Secondary envelopment
The EM studies agree that secondary envelopment occurs within the AC. Enveloped capsids have been observed as single capsids, but the hallmark of CMV infection is their accumulation in large vesicles, often referred to as MVBs or, better, multiviral bodies (MViBs), as recently proposed (Wedemann et al., 2022). Some CMVs, such as murine CMV in fibroblasts, are enveloped as large multicapsid virions (Maninger et al., 2011).
The identity of the envelopment membranes and the mechanism of CMV envelopment are still unknown. Studies of the trafficking of viral envelope proteins point to the trans-Golgi (Jarvis et al., 2002; Homman-Loudiyi et al., 2003; Cepeda et al., 2010), EEs (Tooze et al., 1993; Radsak et al., 1996; Fraile-Ramos et al., 2002), and the ERC (Krzyzaniak et al., 2009; Lučin et al., 2018; Lučin et al., 2020). There are also several lines of evidence that CMV maturation occurs within the membranous system along endocytic flux (Archer et al., 2017; Hasan et al., 2018; Štimac et al., 2021). Knockdown of host cell factors regulating membrane flux suggests that EE maturation is essential for virion envelopment and release (Sharon-Friling and Shenk, 2014; Pavelin et al., 2017; Turner et al., 2020).
Secondary envelopment is initiated by the contact of tegumented capsids with the membranous compartment, causing the membrane to deform and wrap around either as a double membrane or as budding into a larger compartment (Close et al., 2018; Wofford et al., 2020). Advanced EM has shown that CMV envelopment occurs at complex tubular structures that provide sufficient membranes to wrap around the capsids (Maninger et al., 2011; Schauflinger et al., 2013). These events are evenly distributed across the AC, and no preferred site for envelopment has been identified. At a later stage of infection, capsids were observed in large vacuoles at the periphery of the AC, leading to the proposal that envelopment occurs by budding in MVBs (Fraile-Ramos et al., 2002; Flomm et al., 2022; Wedemann et al., 2022). However, suppression of components of the endosomal sorting complex required for transport (ESCRT), which is essential for reverse budding into MVBs and MVB maturation toward the degradation pathway, did not affect CMV maturation (Fraile-Ramos et al., 2007), but further studies showed that the ESCRT-III machinery is involved in the final steps of HCMV replication (Tandon et al., 2009), likely in virion release and virus spread (Streck et al., 2018). A recent study demonstrates that ESCRT-III component CHMPC may be either required for scission of a pool of wrapping membranes derived from recycling endosomes used for envelopment of HSV1 or closure of membranes around virions analogous to the sealing of phagophore in autophagy (Russell et al., 2021). Proteomic and lipidomic analysis of extracellular virions identified several host cell signatures within virions that suggests envelopment at membranes derived from recycling EEs and did not reveal any signatures that might indicate envelopment at MVBs (Mahmutefendić Lučin et al., 2022). Thus, there is little evidence that MVBs can be used for envelopment.
4 Egress route
Alpha-herpesviruses exit cells as single particles via secretory vesicles and exosomes (Bergeman et al., 2023), whereas beta- (Bughio et al., 2015; Momtaz et al., 2021) and gamma-herpesviruses (Peng et al., 2010) appear to collect in large MVB-like membranous organelles before exiting cells. The exit of CMVs has been recently described as intermittent bulk release of many particles that attach to PM prior release into the extracellular fluid (Flomm et al., 2022; Wedemann et al., 2022). Virion-containing MVBs have been observed in several cell types and appear to represent a form of the non-conventional secretion pathway (Turner et al., 2020; Flomm et al., 2022). In fibroblasts, they are likely generated from classical LEs, whereas in endothelial cells they originate from a nonclassical Golgi-mediated exit pathway (Momtaz et al., 2021). The pathway by which enveloped virions are collected and the biogenesis of multivesicular organelles associated with CMV infection remain poorly understood. It is also not known whether CMVs can also envelop into single vesicles and be released as single virions by direct fusion with the PM, as suggested by (Turner and Mathias, 2022).
5 Discussion and further directions
The available information is insufficient to reconstruct the secondary envelopment and exit pathways of CMVs and should be reconciled with the growing knowledge of multiple sorting pathways at the EE-ERC-TGN interface, heterogeneity and redundancy of secretory pathways, and their overlap with secretory autophagy pathways. With this in mind, we propose the following scheme of BHV maturation (Figure 1) and highlight critical steps that should be clarified to fully understand BHV envelopment and egress.
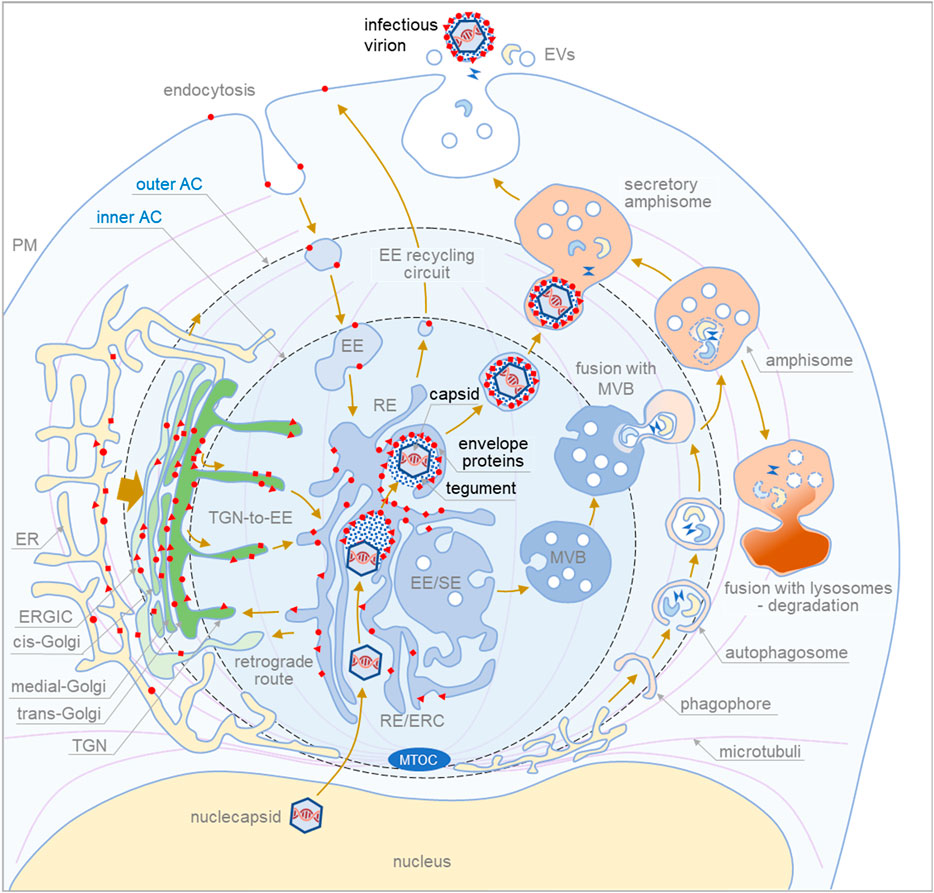
FIGURE 1. Proposed sequence of cytoplasmic events during final envelopment and exit of cytomegaloviruse. The membrane system of the infected cell is reorganized into a large structure called the assembly compartment (AC). The Golgi stacks and trans-Golgi network (TGN) are relocated to the periphery of the AC in the form of a ring called the outer AC. Late endosomes (LE) and the endoplasmic reticulum (ER) remain outside the ring. In contrast, early endosomes (EE), recycling endosomes (RE)/endosomal recycling compartment (ERC) and TGN remain inside the ring (inner AC). Endocytic flux is directed to the inner AC where EE undergoes maturation. CMV infection halts downstream maturation of EEs and expands membrane elements at the EE-RE/ERC-TGN interface, which accumulate in the inner AC. EEs that mature into multivesicular bodies (MVBs) move outside the AC to join LE maturation pathways. Autophagic processes within the AC and fusion with MVBs to form amphisomes occur at the edges of the AC. Degradative amphisomes fuse with lysosomes, and secretory amphisomes dispose of cellular components outside the cell by fusing with the membrane. During CMV infection, virion components are formed in the late phase of infection. Viral envelope proteins are synthesized in the ER, modified, accumulated in the trans-Golgi/TGN (outer AC), and transported to the membrane system of infected cells. Membrane flux transports the envelope proteins to the membranes of the inner AC, and the cargo sorting and cargo retrieval machinery of the EEs retrieves the coat proteins at membranes with a composition suitable for secondary envelopment. This composition, which includes lipid-modifying proteins and curvature-promoting lipids and proteins, is achieved in the final steps of EE maturation in recycling pathways at modified RE-like membranes extending within the AC. Tegument proteins accumulate in the cytosol of the inner AC and form biomolecular condensates on membranes that concentrate envelope proteins. Viral capsids released from the nucleus embed into the tegument matrix and initiate the envelopment of the envelope protein-rich membrane, which wraps around the material containing the capsids, tegument proteins, and host cell proteins (signatures) to generate enveloped virions. The process of envelopment is similar to the growth of phagophores. Enveloped virions within membrane organelles migrate and fuse with secretory amphisomes, which collect multiple virions and form Multi Viral Bodied (MViBs). MViBs migrate to the cell periphery and release many virions simultaneously, which is known as intermittent bulk release.
To form the secondary envelopment site, CMVs should adapt protein and lipid composition that can concentrate envelope and tegument proteins and engulf large tegumented virions (multicapsids in the case of MCMV) into membrane organelle. For concentrating envelope proteins, CMVs can co-opt cargo sorting and retrieval machinery (i.e., sorting nexins, retrieving complexes), a function mostly associated with EEs. Envelopment can occur after the retrieval at membranes that can grow and bend around tegumented capsids, a property that characterize endosomal tubulation. Indeed, high membrane tubulation is a hallmark within the AC (Lučin et al., 2018; Lučin et al., 2020) and may be required for autophagosome-like membrane wrapping around tegumented capsids. However, no single biochemical remnant can be found in released virions as a signature of the conventional autophagy pathway, and on the other hand, many signatures found suggest envelopment at RE-derived membranes (Mahmutefendić Lučin et al., 2022). What triggers membrane wrapping around tegumented capsids remains unknown, and one mechanism could be the formation of tegument-initiated biomolecular condensates at the site of glycoprotein and nucleocapsid concentration, as has been proposed for herpes simplex virus (Metrick et al., 2020). In addition to protein machinery, lipid composition should also facilitate expansion and wrapping of the envelopment membranes. In the inner AC, membranes are rich in phosphatidylserine (PS) (Lučin et al., 2020), a cylindrical phospholipid required for elongation and PS should be replaced by conical phospholipids such as phosphatidylethanolamine (PE), which facilitates negative curvatures (McMahon and Boucrot, 2015). Indeed, proteomic (Turner et al., 2020) and lipidomic (Liu et al., 2011) analyses of virions revealed the absence of phosphoinositides, PS and PS-binding proteins, and enrichment of PE and PE-binding proteins.
Although envelopment on MVB-like structures has been proposed (Flomm et al., 2022), remnants of the machinery that drives the EE to LE maturation, sorts proteins into intralumenal vesicles, and performs the reverse budding process (i.e., ESCRT) on MVBs have been found in virions (Mahmutefendić Lučin et al., 2022). Most LE-derived MVB structures are dislocated outside the AC (Supplementary Table S1; (Lučin et al., 2020), and most envelopment events occur within the AC (Schauflinger et al., 2013). Nevertheless, envelopment on large organelles cannot be ruled out if forces other than the reverse budding machinery are responsible for envelopment. The ESCRT-III may contributes to the final stages Click or tap here to enter text and may be essential for membrane sealing in the wrapping processes, as in autophagosome maturation (Jiang et al., 2021).
To find a way out of the cell, CMV-containing vesicles should fuse with large transport vehicles that travel to the cell surface, as traveling within large MViBs appears to be the main exit route (Flomm et al., 2022). CMV-containing vesicles can fuse either with the continuous stream of large multivesicular organelles known as amphisomes that transport secretory autophagic contents out of the cell (Ganesan and Cai, 2021), or with degradative amphisomes and divert their route to the PM by delivering RE-derived factors, or even with LE- or TGN-derived vessels that form lysosome-related organelles (LRO) (Delevoye et al., 2019). It appears that the large vessels that transport CMV for release are heterogeneous: In fibroblasts, they are more associated with the LE-derived secretory pathway, whereas in endothelial cells, they correspond more to the autophagic pathway (Momtaz et al., 2021). In addition, virion-loaded organelles can undergo a maturation process similar to that described for LRO and be released by a biphasically evoked exocytic mechanism controlled by the GTPases Rab3a/Rab15/Rab35, as described for LRO (Biesemann et al., 2017), or by exocyst and Rab11b as for lysosome-associated exocytosis (Escrevente et al., 2021).
In conclusion, to understand the final steps of CMV production, it would be important to identify the envelopment organelle and the organelle that transports virions out of the cell. The roadmap requires identification of small GTPases (Rab proteins or others), tethering complexes, and SNARE proteins in both organelles. To achieve this, fluorescently labeled capsids would need to be observed throughout the infected cell over a long period of time, using live cell imaging technology with high resolution and very low phototoxicity. This appears to be a difficult task for CMV, and there are only a few reports (Sampaio et al., 2005; Bosse et al., 2012) on capsid-labeled virions. An additional obstacle for more in-depth studies is the heterogeneity of cells, which allows the establishment of the final steps of the assembly chain only in a limited number of cells. Therefore, cytoplasmic MViB transport appears to be a relatively rare event, and single-cell analyzes with labeled capsids are essential for further progress in the field of secondary envelopment and egress of CMVs.
Unlike alpha-herpesviruses and other viruses, CMV extensively remodel the cell’s membrane system to form unique compartments for their envelopment, egress, and possibly signaling (Henaff et al., 2012). Understanding these changes may therefore not only contribute to a better understanding of CMV biology, but also to elucidating the physiology of the membrane system. Accordingly, a detailed understanding of these processes would pave the way for further development of host-directed antiviral therapy, an emerging concept to develop broad-spectrum drugs to counteract the evolution of viral resistance (Kumar et al., 2020).
Author contributions
PL: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. HML: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Writing–review and editing. GBZ: Funding acquisition, Investigation, Methodology, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Croatian Science Foundation (Grants IP-2014-9-9564 and IP-2019-4-3582 to PL, IP-2020-02-2916 to HML, and IP-2020-02-1323 to GBZ), and by the University of Rijeka grants (uniri-biomed-18-88 to PL, uniri-biomed-18-180 to HML, uniri-biomed-18-229 to GBZ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1328751/full#supplementary-material
References
Archer, M. A., Brechtel, T. M., Davis, L. E., Parmar, R. C., Hasan, M. H., and Tandon, R. (2017). Inhibition of endocytic pathways impacts cytomegalovirus maturation. Sci. Rep. 7, 46069. April. doi:10.1038/srep46069
Beltran, P. M. J., Mathias, R. A., Cristea, I. M., Beltran, P. M. J., Mathias, R. A., and Cristea, I. M. (2016). A Portrait of the human organelle proteome in Space and time during cytomegalovirus infection. Cell Syst. 3, 361–373. doi:10.1016/j.cels.2016.08.012
Bergeman, M. H., Hernandez, M. Q., Diefenderfer, J., Drewes, J. A., Velarde, K., Tierney, W. M., et al. (2023). Live-cell fluorescence microscopy of HSV-1 cellular egress by exocytosis. BioRxiv. Available at: https://www.biorxiv.org/content/10.1101/2023.02.27.530373v2.
Biesemann, A., Gorontzi, A., Barr, F., and Gerke, V. (2017). Rab35 protein regulates evoked exocytosis of endothelial Weibel–Palade bodies. J. Biol. Chem. 292 (28), 11631–11640. doi:10.1074/jbc.M116.773333
Bosse, J. B., Bauerfeind, R., Popilka, L., Marcinowski, L., Taeglich, M., Jung, C., et al. (2012). A beta-herpesvirus with fluorescent capsids to study transport in living cells. PLoS ONE 7 (7), e40585. doi:10.1371/journal.pone.0040585
Bughio, F., Umashankar, M., Wilson, J., and Goodrum, F. (2015). Human cytomegalovirus UL135 and UL136 genes are required for postentry tropism in endothelial cells. J. Virology 89 (13), 6536–6550. doi:10.1128/JVI.00284-15
Cepeda, V., Esteban, M., and Fraile-Ramos, A. (2010). Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell. Microbiol. 12 (3), 386–404. doi:10.1111/j.1462-5822.2009.01405.x
Close, W. L., Anderson, A. N., and Pellett, P. E. (2018). “Betaherpesvirus virion assembly and egress,” in Advances in experimental medicine and biology (vol. 1045, pp. 167–207) (New York: Springer New York LLC), 167–207. doi:10.1007/978-981-10-7230-7_9
Cook, K. C., and Cristea, I. M. (2019). Location is everything: protein translocations as a viral infection strategy. Curr. Opin. Chem. Biol. 48, 34–43. doi:10.1016/j.cbpa.2018.09.021
Crump, C. M., Hung, C.-H., Thomas, L., Wan, L., and Thomas, G. (2003). Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virology 77 (20), 11105–11113. doi:10.1128/JVI.77.20.11105-11113.2003
Das, S., Vasanji, A., and Pellett, P. E. (2007). Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virology 81 (21), 11861–11869. doi:10.1128/jvi.01077-07
Delevoye, C., Marks, M. S., and Raposo, G. (2019). Lysosome-related organelles as functional adaptations of the endolysosomal system. Curr. Opin. Cell Biol. 59, 147–158. doi:10.1016/j.ceb.2019.05.003
Escrevente, C., Bento-Lopes, L., Ramalho, J. S., and Barral, D. C. (2021). Rab11 is required for lysosome exocytosis through the interaction with Rab3a, Sec15 and GRAB. J. Cell Sci. 134 (11), jcs246694. doi:10.1242/JCS.246694
Flomm, F. J., Soh, T. K., Schneider, C., Wedemann, L., Britt, H. M., Thalassinos, K., et al. (2022). Intermittent bulk release of human cytomegalovirus. PLoS Pathog. 18 (8), e1010575. doi:10.1371/journal.ppat.1010575
Fraile-Ramos, A., Pelchen-Matthews, A., Kledal, T. N., Browne, H., Schwartz, T. W., and Marsh, M. (2002). Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3 (3), 218–232. doi:10.1034/j.1600-0854.2002.030307.x
Fraile-Ramos, A., Pelchen-Matthews, A., Risco, C., Rejas, M. T., Emery, V. C., Hassan-Walker, A. F., et al. (2007). The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell. Microbiol. 9 (12), 2955–2967. doi:10.1111/j.1462-5822.2007.01024.x
Ganesan, D., and Cai, Q. (2021). Understanding amphisomes. Biochem. J. 478 (10), 1959–1976. doi:10.1042/BCJ20200917
Hasan, M. H., Davis, L. E., Bollavarapu, R. K., Mitra, D., Parmar, R., and Tandon, R. (2018). Dynamin is required for efficient cytomegalovirus maturation and envelopment. J. Virology 92 (24), e01418-18. doi:10.1128/JVI.01418-18
Henaff, D., Radtke, K., and Lippé, R. (2012). Herpesviruses exploit several host compartments for envelopment. Traffic 13 (11), 1443–1449. doi:10.1111/j.1600-0854.2012.01399.x
Homman-Loudiyi, M., Hultenby, K., Britt, W., and Söderberg-Nauclér, C. (2003). Envelopment of human cytomegalovirus occurs by budding into golgi-derived vacuole compartments positive for gB, rab 3, trans-golgi network 46, and mannosidase II. J. Virology 77 (5), 3191–3203. doi:10.1128/jvi.77.5.3191-3203.2003
Hook, L. M., Grey, F., Grabski, R., Tirabassi, R., Doyle, T., Hancock, M., et al. (2014). Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15 (3), 363–373. doi:10.1016/j.chom.2014.02.004
Jarvis, M. A., Fish, K. N., Söderberg-Naucler, C., Streblow, D. N., Meyers, H. L., Thomas, G., et al. (2002). Retrieval of human cytomegalovirus glycoprotein B from cell surface is not required for virus envelopment in astrocytoma cells. J. Virology 76 (10), 5147–5155. doi:10.1128/jvi.76.10.5147-5155.2002
Jiang, W., Chen, X., Ji, C., Zhang, W., Song, J., Li, J., et al. (2021). Key regulators of autophagosome closure. Cells 10 (11), 2814. doi:10.3390/CELLS10112814
Karleuša, L., Mahmutefendić, H., Tomaš, M. I. M. I., Zagorac, G. B. G. B., and Lučin, P. (2018). Landmarks of endosomal remodeling in the early phase of cytomegalovirus infection. Virology 515, 108–122. doi:10.1016/j.virol.2017.12.001
Kropff, B., Koedel, Y., Britt, W., and Mach, M. (2010). Optimal replication of human cytomegalovirus correlates with endocytosis of glycoprotein gpUL132. J. Virology 84 (14), 7039–7052. doi:10.1128/jvi.01644-09
Krzyzaniak, M. A., Mach, M., and Britt, W. J. (2009). HCMV-encoded glycoprotein M (UL100) interacts with rab11 effector protein FIP4. Traffic 10 (10), 1439–1457. doi:10.1111/j.1600-0854.2009.00967.x
Kumar, N., Sharma, S., Kumar, R., Tripathi, B. N., Barua, S., Ly, H., et al. (2020). Host-directed antiviral therapy. Clin. Microbiol. Rev. 33 (3), e00168-19. doi:10.1128/CMR.00168-19
Liu, S. T. H., Sharon-Friling, R., Ivanova, P., Milne, S. B., Myers, D. S., Rabinowitz, J. D., et al. (2011). Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc. Natl. Acad. Sci. U. S. A. 108 (31), 12869–12874. doi:10.1073/pnas.1109796108
Lučin, P., Jug Vučko, N., Karleuša, L., Mahmutefendić Lučin, H., Blagojević Zagorac, G., Lisnić, B., et al. (2020). Cytomegalovirus generates assembly compartment in the early phase of infection by perturbation of host-cell factors recruitment at the early endosome/endosomal recycling compartment/trans-Golgi interface. Front. Cell Dev. Biol. 8, 563607. doi:10.3389/fcell.2020.563607
Lučin, P., Kareluša, L., Blagojević Zagorac, G., Mahmutefendić Lučin, H., Pavišić, V., Jug Vučko, N., et al. (2018). Cytomegaloviruses exploit recycling Rab proteins in the sequential establishment of the assembly compartment. Front. Cell Dev. Biol. 6, 165. DEC. doi:10.3389/fcell.2018.00165
Mahmutefendić Lučin, H., Blagojević Zagorac, G., Marcelić, M., and Lučin, P. (2022). Host cell signatures of the envelopment site within beta-herpes virions. Int. J. Mol. Sci. 23 (17), 9994. doi:10.3390/IJMS23179994
Maninger, S., Bosse, J. B., Lemnitzer, F., Pogoda, M., Mohr, C. A., von Einem, J., et al. (2011). M94 is essential for the secondary envelopment of murine cytomegalovirus. J. Virology 85 (18), 9254–9267. doi:10.1128/jvi.00443-11
Maschkowitz, G., Gärtner, S., Hofmann-Winkler, H., Fickenscher, H., and Winkler, M. (2018). Interaction of human cytomegalovirus tegument proteins ppUL35 and ppUL35A with sorting nexin 5 regulates glycoprotein B (gpUL55) localization. J. Virology 92 (9), e00013-18. doi:10.1128/JVI.00013-18
McMahon, H. T., and Boucrot, E. (2015). Membrane curvature at a glance. J. Cell Sci. 128 (6), 1065–1070. doi:10.1242/JCS.114454
Metrick, C. M., Koenigsberg, A. L., and Heldwein, E. E. (2020). Conserved outer tegument component ul11 from herpes simplex virus 1 is an intrinsically disordered, rna-binding protein. MBio 11 (3), e00810-20. doi:10.1128/mBio.00810-20
Momtaz, S., Molina, B., Mlera, L., Goodrum, F., and Wilson, J. M. (2021). Cell type-specific biogenesis of Novel vesicles containing viral products in human cytomegalovirus infection. J. Virology 95 (11), e02358-20. doi:10.1128/jvi.02358-20
Pavelin, J., McCormick, D., Chiweshe, S., Ramachandran, S., Lin, Y. T., and Grey, F. (2017). Cellular v-ATPase is required for virion assembly compartment formation in human cytomegalovirus infection. Open Biol. 7 (11), 160298. doi:10.1098/rsob.160298
Peng, L., Ryazantsev, S., Sun, R., and Zhou, Z. H. (2010). Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure 18 (1), 47–58. doi:10.1016/j.str.2009.10.017
Radsak, K., Eickmann, M., Mockenhaupt, T., Bogner, E., Kern, H., Eis-Hübinger, A., et al. (1996). Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Archives Virology 141 (3–4), 557–572. doi:10.1007/BF01718317
Read, C., Schauflinger, M., Nikolaenko, D., Walther, P., and von Einem, J. (2019). Regulation of human cytomegalovirus secondary envelopment by a C-terminal tetralysine Motif in pUL71. J. Virology 93 (13), e02244-18. doi:10.1128/jvi.02244-18
Russell, T., Samolej, J., Hollinshead, M., Smith, G. L., Kite, J., and Elliott, G. (2021). Novel role for escrt-iii component chmp4c in the integrity of the endocytic network utilized for herpes simplex virus envelopment. MBio 12 (3), e02183-20. doi:10.1128/mBio.02183-20
Sampaio, K. L., Cavignac, Y., Stierhof, Y.-D., and Sinzger, C. (2005). Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virology 79 (5), 2754–2767. doi:10.1128/jvi.79.5.2754-2767.2005
Schauflinger, M., Villinger, C., Mertens, T., Walther, P., and von Einem, J. (2013). Analysis of human cytomegalovirus secondary envelopment by advanced electron microscopy. Cell. Microbiol. 15 (2), 305–314. doi:10.1111/cmi.12077
Seo, J.-Y., and Britt, W. J. (2006). Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virology 80 (11), 5611–5626. doi:10.1128/jvi.02630-05
Sharon-Friling, R., and Shenk, T. (2014). Human cytomegalovirus pUL37x1-induced calcium flux activates PKCα, inducing altered cell shape and accumulation of cytoplasmic vesicles. Proc. Natl. Acad. Sci. U. S. A. 111 (12), E1140–E1148. doi:10.1073/pnas.1402515111
Štimac, I., Vučko, N. J., Zagorac, G. B., Marcelić, M., Lučin, H. M., and Lučin, P. (2021). Dynamin inhibitors prevent the establishment of the cytomegalovirus assembly compartment in the early phase of infection. Life 11 (9), 876. doi:10.3390/life11090876
Streck, N. T., Carmichael, J., and Buchkovich, N. J. (2018). Nonenvelopment role for the ESCRT-III complex during human cytomegalovirus infection. J. Virology 92 (12), e02096-17. doi:10.1128/jvi.02096-17
Taisne, C., Lussignol, M., Hernandez, E., Moris, A., Mouna, L., and Esclatine, A. (2019). Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci. Rep. 9 (1), 4560. doi:10.1038/s41598-019-41029-z
Tandon, R., AuCoin, D. P., and Mocarski, E. S. (2009). Human cytomegalovirus Exploits ESCRT machinery in the process of virion maturation. J. Virology 83 (20), 10797–10807. doi:10.1128/jvi.01093-09
Tomaš, M. I., Kučić, N., Mahmutefendić, H., Blagojević, G., and Lučin, P. (2010). Murine cytomegalovirus perturbs endosomal trafficking of major histocompatibility complex class I molecules in the early phase of infection. J. Virology 84 (21), 11101–11112. doi:10.1128/JVI.00988-10
Tooze, J., Hollinshead, M., Reis, B., Radsak, K., and Kern, H. (1993). Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60 (1), 163–178.
Tugizov, S., Maidji, E., Xiao, J., and Pereira, L. (1999). An acidic cluster in the cytosolic Domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J. Virology 73 (10), 8677–8688. doi:10.1128/jvi.73.10.8677-8688.1999
Turner, D. L., Korneev, D. V., Purdy, J. G., de Marco, A., and Mathias, R. A. (2020). The host exosome pathway underpins biogenesis of the human cytomegalovirus virion. ELife 9, 582888–e58329. doi:10.7554/ELIFE.58288
Turner, D. L., and Mathias, R. A. (2022). The human cytomegalovirus decathlon: ten critical replication events provide opportunities for restriction. Front. Cell Dev. Biol. 10, 1053139. doi:10.3389/fcell.2022.1053139
Varnum, S. M., Streblow, D. N., Monroe, M. E., Smith, P., Auberry, K. J., Pas, L., et al. (2004). Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virology 78 (20), 10960–10966. doi:10.1128/JVI.78.20.10960-10966.2004
Wedemann, L., Flomm, F. J., and Bosse, J. B. (2022). The unconventional way out-Egress of HCMV through multiviral bodies. Mol. Microbiol. 117 (6), 1317–1323. doi:10.1111/MMI.14946
Wofford, A. S., McCusker, I., Green, J. C., Vensko, T. A., and Pellett, P. E. (2020). “Betaherpesvirus assembly and egress: recent advances illuminate the path,” in Advances in virus research. 1 (Amsterdam, Netherlands: Elsevier Inc). Vol. 108. doi:10.1016/bs.aivir.2020.09.003
Wu, H., Kropff, B., Mach, M., and Britt, W. J. (2020). Human cytomegalovirus envelope protein gpUL132 regulates infectious virus production through Formation of the viral assembly compartment. MBio 11 (5), 020444-20–e2116. doi:10.1128/MBIO.02044-20
Keywords: betaherpesviruses, cytomegalovirus, assembly compartment, secondary envelopment, virion egress, multiviral bodies
Citation: Lučin P, Mahmutefendić Lučin H and Blagojević Zagorac G (2023) Cytomegaloviruses reorganize endomembrane system to intersect endosomal and amphisome-like egress pathway. Front. Cell Dev. Biol. 11:1328751. doi: 10.3389/fcell.2023.1328751
Received: 27 October 2023; Accepted: 08 December 2023;
Published: 19 December 2023.
Edited by:
Roger Schneiter, University of Fribourg, SwitzerlandReviewed by:
Wen Xiong, Baylor College of Medicine, United StatesPhilip E. Pellett, Wayne State University, United States
Copyright © 2023 Lučin, Mahmutefendić Lučin and Blagojević Zagorac. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pero Lučin, cGVyby5sdWNpbkB1bmlyaS5ocg==, cGVyb0B1bmlyaS5ocg==
 Pero Lučin
Pero Lučin Hana Mahmutefendić Lučin
Hana Mahmutefendić Lučin Gordana Blagojević Zagorac
Gordana Blagojević Zagorac